Abstract
Because of the high frequency of HLA-DP4 in the Caucasian population, we have selectively delineated HLA-DP4 restricted T cell epitopes in the MAGE-A tumor antigens. We identified 12 good binders to HLA-DP4 and investigated the capacity of the seven best binders to induce in vitro specific CD4+ T cell lines from HLA-DP4 healthy donors. We found that the MAGE-A1 90–104 peptide exhibited a high and constant frequency of CD4+ T cell precursors in all the six tested donors. The MAGE-A1 268–282 peptide was found immunogenic in only two donors but with a high precursor frequency. The MAGE-A12 127–141 peptide was T cell stimulating in six different donors and induced fewer T cell lines. The peptide-specific T cell lines were stimulated by DC loaded with the lysates of cells transfected with MAGE-A1 or MAGE-A12, or loaded with the recombinant protein. We also show that the immunoreactivity of CD4+ T cell epitopes restricted to the same HLA II molecule may vary from one individual to another, as a result of inter-individual variations in the CD4+ T cell repertoire.
Keywords: HLA-DP4, Antigens/peptides/epitopes, CD4+ T lymphocyte, Tumor, MAGE
Introduction
Immune response induced by vaccination against tumor cells remains weak and often insufficient to cure the disease [21]. Tumor regressions have been observed in clinical studies but in most cases it has not been clearly demonstrated that these were due to the immune responses induced by the vaccine [19, 21]. CTL response to tumor antigens has been widely investigated using various reliable tools and methods to enumerate and characterize tumor-specific CD8+ T cells [6, 31, 32]. Multiple CD4+ T cell epitopes from various relevant antigens have been identified [5, 8, 11, 16, 18, 23, 34–36, 38, 39]. Most of these epitopes are restricted to the HLA-DR molecules [5, 8, 11, 16, 18, 34–36, 39], which are highly polymorphic. In contrast to HLA-A molecules, no preponderant HLA-DR molecule exists, each molecule being present at relatively low frequency in the population [4]. Some T cell epitopes derived from tumor antigen sequences are able to bind multiple HLA-DR molecules and therefore could facilitate clinical investigations [11, 16, 18, 36, 39]. Their different affinities and binding registers for the HLA-DR molecules could, however, bias the understanding of inter-individual T cell responses.
In contrast to HLA-DR, HLA-DP4 molecules are advantageously present at a high frequency worldwide [4]. HLA-DPA1*0103/DPB1*0401 (DP401) and HLA-DPA1*0103/DPB1*0402 (DP402) differ by only three amino acids and have a very similar binding motif [3]. In the Caucasian population, HLA-DP401 and HLA-DP402 gene frequency is approximately 40 and 11%, respectively. Together, they are found in approximately 76% of individuals. This high frequency allows easy recruitment of a population of cancer patients and healthy donors, who share a common HLA II molecule. T cell epitopes restricted to HLA-DP4 molecules facilitate the monitoring of the frequency and of the predominance of antigen-specific CD4+ responses in cancer patients in a comprehensive way [1, 22, 40]. They also allow the monitoring of vaccine-induced CD4+ T cell responses raised against defined tumor antigens. Nevertheless, HLA-DP4 restricted T cell epitopes have been found in MAGE-A3 [23] and NY-ESO-1 tumor antigens only [18, 38]. Melanoma patients have been vaccinated with dendritic cells loaded with the HLA-DP4-restricted peptide MAGE-A3 [22]. Recently, functional HLA-DP4 multimers were produced with the MAGE-A3 peptide and have been used to evaluate the frequencies of peptide-specific T cells induced by vaccination [40]. A limited number of HLA-DP4 restricted CD4+ T cell epitopes are therefore available to monitor CD4+ T cell responses specific for tumor antigens.
In this paper, we have tackled the entire MAGE-A family and report three newly discovered epitopes in MAGE-A1 and MAGE-A12. Our motivation of analyzing MAGE-A proteins comes from its broad tumor-specificity. The MAGE-A family belongs to the class of tumor-specific antigens, which are silent in normal tissue, testicular germ cells excepted [13, 31]. These antigens are encoded by 12 closely related genes and include the first identified tumor antigen (MAGE-A1) [32]. Initially found in melanoma cells, MAGE-A1, -A2, -A3, -A4, -A6 and -A10 are also expressed in various tumors, while most of the other MAGE-A proteins have also been detected in different malignant cells [13, 31]. As shown by the multiple CD4+ and CD8+ T cell epitopes identified in MAGE-A proteins [5, 6, 8, 16, 20, 23, 32, 38, 39], they appear to be a favored target of cellular response and are used in multiple vaccination trials [1, 17, 19, 22, 41]. By combining peptide-binding prediction that we have recently set up [10], peptide-binding assays and CD4+ T cell priming experiments, we successfully found new HLA-DP4-restricted T cell epitopes in the MAGE-A antigens.
Materials and methods
Peptides and antigens
Peptides were synthesized by using standard Fmoc chemistry on a multiple peptide synthesizer APEX 396 (Advanced Chemtech, Louisville, KY) as described previously [3]. The MAGE-A1 protein was produced after the infection of insect cells by a recombinant baculovirus. The protein has six histidine residues at the C-terminal for the purification by ion metal affinity chromatography. Bv-PLA2 was produced in E. coli and purified as described previously [2].
HLA-DP4 specific peptide-binding assays
HLA-DP4 molecules were purified by affinity chromatography using B7/21 Mab coupled to Protein A Sepharose CL 4B gel (Pharmacia Biotech) as previously described [3]. Binding assays were performed by competitive ELISA as described previously [3]. Data were expressed as the peptide concentration that prevented binding of 50% of the labeled peptide (IC50). IC50 values of the Oxy 271–287 peptide served as a reference in each experiment.
Blood samples and HLA-DP genotyping
Blood cells were collected at the Etablissement Français du Sang (EFS, Rungis, France) as buffy-coat preparations from anonymous healthy donors after informed consent and following EFS guidelines. Peripheral blood mononuclear cells (PBMC) were isolated by density centrifugation on Ficoll-Hyperpaque gradients (Sigma Aldrich, St Quentin Fallavier, France). Genotyping was performed using the Olerup SSP DPB1 typing kit (Olerup SSP AB, Saltsjobaden, Sweden) according to the manufacturer. HLA-DPB genotyping results were the following: donor 78 (DPB1*0402, DPB1*1301), donor 121 (DPB1*0402, DPB1*401), donor 122 (DPB1*0401), donor 126 (DPB1*0401), donor 129 (DPB1*0402, DPB1*402), donor 156 (DPB1*0401), donor 158 (DPB1*0402), donor 164 (DPB1*0401, DPB1*1101), donor 172 (DPB1*0401, DPB1*301) and donor 177 (DPB1*0401, DPB1*301).
Induction of CD4+ T cells with peptides
Immature and mature DC were generated from plastic-adherent PBMC by a 5-day culture in AIM-V medium supplemented with 1,000 units/ml of rh-GmCSF (R&D System, Lille, France) and 1,000 units/ml of rh-IL-4 (R&D Systems). LPS (Sigma) (1 μg/ml) was used as maturation agent. CD4+ T lymphocytes were isolated from non-adherent PBMC by positive selection using an anti-CD4 monoclonal antibody coupled to magnetic microbeads (Miltenyi Biotech, Paris, France). Mature DC were incubated at 37°C, 5% CO2, for 4 h in IMDM medium (Invitrogen, Cergy Pontoise, France) supplemented with 24 mM glutamine, 55 mM asparagine, 150 mM arginine (all amino acids from Sigma), 50 U/ml penicillin and 50 μg/ml streptomycin (Invitrogen), and 10% human serum (hereafter referred to as complete IMDM medium) with a solution of the MAGE-A peptide mixture (MAGE-A1 90–104, MAGE-A1 268–282, MAGE-A2 111–125, MAGE-A2 219–233, MAGE-A9 68–82, MAGE-A9 153–167, MAGE-A12 127–141)(10 μg/ml of each peptide). Pulsed mature DC cells (104 per round-bottom microwell) were added to 105 autologous CD4+ lymphocytes in 200 μl complete IMDM with 1,000 U/ml of IL-6 (R&D systems, Abingdon, UK) and 10 ng/ml of IL-12 (R&D systems). The CD4+ T lymphocytes were re-stimulated on days 7, 14 and 21 with autologous DC freshly loaded with the MAGE-A peptide mixture, and were grown in complete IMDM medium supplemented with 10 U/ml of IL-2 (R&D systems) and 5 ng/ml of IL-7 (R&D systems). The stimulated CD4+ T cells were analyzed for specificity in enzyme-linked immunospot (ELISpot) assays at least 6 days after the last stimulation.
Transfection of COS-7 cells and preparation of cell lysates
cDNA for MAGE-A1 and MAGE-A12 was cloned into the vector pcDNAI/Amp. COS-7 cells were plated in 6-well plates (5 × 105 cells/well) 18 h before transfection. The cells were transiently transfected with 4 μg/well of pcDNA1-MAGE1 or pcDNA1-MAGE12 plasmid using Lipofectamine2000 (Invitrogen). Forty-eight hours later, transfection efficacy was assessed by flow cytometry. Cells were harvested and lysed in AIM V medium at a cellular concentration of 107 cells/ml by five rapid freeze-thaw cycles. Cellular lysates were centrifuged at 14,000 t/min for 10 min. Supernatants were recovered and stocked at −20°C.
IFN-γ ELISpot assays
Multiscreen HA plates (Millipore, St Quentin en Yvelines, France) were coated with 1 μg/ml of mAb anti-human IFN-γ (1-D1K, Mabtech, Stockholm, Sweden) in PBS (Invitrogen) for 1 h at 37°C and saturated with complete IMDM. APC were autologous immature DC or HLA-DP402 transfected L cells (L-DP4 cells). MAGE1 protein (1 μM) and cell lysates (300 μl produced with 3 × 106 cells) were incubated for 4 h at 37°C with immature DC (106/ml) in the presence of 1 μg/ml of LPS. DC were subsequently washed before use. Peptides (10 μg/ml) were directly added to the Multiscreen plates. Immature DC (2 × 104/well) or murine L cells transfected with the HLA-DP402 gene (L-DP4)(3 × 104/well) were distributed in Multiscreen plates together with 5 × 103 T cells. After overnight incubation at 37°C, captured IFN-γ was detected by subsequent addition of biotinylated MAb anti-hIFNγ (7-B6-1; Mabtech) (0.25 μg/ml), Extravidin-phosphatase (Sigma) and NBT/BCIP (Sigma). Spot numbers were automatically determined by the AID EliSpot Reader System (AID, Strassberg, Germany). For statistical evaluation, a t-test was used. Values of P < 0.05 were considered significant.
Results
Twelve good binders to HLA-DP4 molecules were identified in the MAGE-A antigens
We have recently described a new prediction method of binding to HLA-DP4 based on quantitative binding matrices [10]. The prediction matrices were built with the binding data obtained with analogs of a good binder to HLA-DP4, substituted by various amino acids at positions accommodated by the pockets 1, 4, 6 and 9 of the peptide-binding site [3]. To predict the peptide binders to HLA-DP4, we assigned to all 9-mer of a sequence, a predicted IC50 by addition of the values in the P1, P4 P6 and P9 pockets for each corresponding amino acid of the peptide. This method was successfully used to identify HLA-DP4 restricted peptides in the whole genome of HIV [10]. In the present paper, we applied the binding prediction to nine proteins of the MAGE-A tumor antigen family, which are expressed in melanoma cells [25]. Seventeen peptides exhibited a predicted IC50 below 500 nM for HLA-DP401 molecules, but only 12 of them were successfully synthesized (Table 1). Their binding to HLA-DP401 and HLA-DP402 molecules was assessed by ELISA. On the basis of an activity threshold of 1,000 nM [26], they were all found to be good binders for both HLA-DP4 molecules (Table 1). Two and three active peptides derived from MAGE-A1 and MAGE-A2, respectively, displayed very low IC50 values. Only one peptide was active in the MAGE-A3, MAGE-A4 and MAGE-A12 proteins, the MAGE-A3 peptide corresponding to the HLA-DP4 restricted peptide described previously [23]. Finally, two active peptides were found in the MAGE-A9 and MAGE-A10 proteins. As a result, besides the MAGE-A3 peptide, we found 11 new peptide sequences, which are able to bind to HLA-DP4 molecules. The seven best peptides (MAGE-A1 90–104, MAGE-A1 268–282, MAGE-A2 111–125, MAGE-A2 219–233, MAGE-A9 68–82, MAGE-A9 153–167, MAGE-A12 127–141) were retained for T cell priming experiments.
Table 1.
Binding capacity of MAGE-A peptides to HLA DP4 molecules
| Peptide | Sequence | IC50 (DP401) | IC50 (DP402) | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 4 | 6 | 9 | Pred. | Obs. | Pred. | Obs. | ||||||||||||
| MAGE-A1 90–104 | T | S | C | I | L | E | S | L | F | R | A | V | I | T | K | 120 | 155 (±7) | 120 | 100 (±0) |
| MAGE-A1 268–282 | P | R | A | L | A | E | T | S | Y | V | K | V | L | E | Y | 120 | 13 (±1) | 182 | 16 (±1) |
| MAGE-A2 111–125 | R | K | M | V | E | L | V | H | F | L | L | L | K | Y | R | 155 | 6 (±1) | 85 | 4 (±1) |
| MAGE-A2 143–157 | C | Q | D | F | F | P | V | I | F | S | K | A | S | E | Y | 56 | 335 (±7) | 56 | 50 (±5) |
| MAGE-A2 219–233 | E | K | I | W | E | E | L | S | M | L | E | V | F | E | G | 158 | 43 (±14) | 40 | 37 (±4) |
| MAGE-A3 247–261 | T | Q | H | F | V | Q | E | N | Y | L | E | Y | R | Q | V | 79 | 265 (±4) | 1,348 | 400 (±15) |
| MAGE-A4 248–262 | T | Q | D | W | V | Q | E | N | Y | L | E | Y | R | Q | V | 79 | 200 (±0) | 1,348 | 320 (±6) |
| MAGE-A9 68–82 | S | I | S | V | Y | Y | T | L | W | S | Q | F | D | E | G | 219 | 15 (±4) | 602 | 30 (±11) |
| MAGE-A9 153–167 | A | S | E | F | M | Q | V | I | F | G | T | D | V | K | E | 100 | 40 (±11) | 112 | 21 (±7) |
| MAGE-A10 244–258 | E | V | I | W | E | A | L | N | M | M | G | L | Y | D | G | 79 | 245 (±7) | 20 | 64 (±1) |
| MAGE-A10 303–317 | H | A | E | I | R | K | M | S | L | L | K | F | L | A | K | 363 | 730 (±14) | 1,000 | 175 (±14) |
| MAGE-A12 127–141 | R | E | P | F | T | K | A | E | M | L | G | S | V | I | R | 363 | 120 (±11) | 50 | 47 (±4) |
Peptides were investigated for their capacity to bind HLA-DP401 and HLA-DP402 by competitive ELISA. Predicted (Pred.) and observed (Obs.) IC50 are expressed in nM. IC50 means and SD are calculated from two to three independent experiments. The reference peptide used in these assays is Oxy 271–287 (EKKYFAATQFEPLAARL). Its IC50 was 8 and 6 nM for HLA-DP401 and HLA-DP402 molecules, respectively. The P1, P4, P6 and P9 positions are indicated in bold
Six HLA-DP4 restricted MAGE-A peptides are able to prime in vitro human CD4+ T lymphocytes
The CD4+ T cell priming ability of the peptides were investigated based on previously published protocols [8, 35]. Healthy T CD4+ cells were harvested from ten different HLA-DP4+ normal donors and were stimulated by mature DCs loaded with a mixture of the peptides or with the individual peptides. Specificity of the growing T cell lines was assessed by IFN-γ Elispot using L cells transfected by HLA-DP4 molecules as APC. As shown in Table 2, 28 specific T cell lines were derived from four different healthy donors. Their activation by the peptides was HLA-DP4 restricted, as the omission of the HLA-DP4 transfected L cells completely abolished the T cell activation. Assays performed with individual peptides showed that most of the T cell lines were specific for only one peptide, namely MAGE-A1 90–104. Only four T cell lines were specific for other two peptides (MAGE-A2 111–125 and MAGE-A12 127–141). We also derived peptide specific T cell lines from six other healthy donors. Together, we obtained 35 HLA-DP4 restricted T cell lines specific for MAGE-A1 90–104, 12 for MAGE-A1 268–282, 9 for MAGE-A12 127–141, one for MAGE-A2 111–125, MAGE-A9 153–167 and MAGE-A9 68–82 but none for MAGE-A2 219–233. Based on these results, we evaluated the frequency of peptide-specific CD4+ T cell precursors as proposed previously [7, 30] (Table 3). In the conditions of cell distribution we used (105 CD4+ T lymphocytes per well), a minimum of 70% of the wells did not contain T cells specific for MAGE-A peptides, suggesting that no specific T cells precursors were seeded in these wells during the distribution of the CD4+ T cells. As the distribution of cells followed a Poisson distribution, where most of the wells were free of specific T cells, wells that contained specific T cells mostly derived from only one precursor. Accordingly, we observed that the T cell lines were mainly specific for one peptide only. The precursor frequency was estimated on the basis of a Poisson distribution (see legend of Table 3) [7, 30]. The MAGE-A1 90–104 peptide was immunogenic for all the donors and its CD4+ precursor frequency is approximately of 10−6 (Table 3). A lower number of responders or a lower frequency of precursors was observed for the other peptides (Table 3). As a result, we demonstrated that six out of the seven peptides we investigated for T cell stimulating activity were able to prime peptide-specific and HLA-DP4 restricted human T cell lines.
Table 2.
Peptide specificity of T cell lines induced by a mixture of seven HLA-DP4 restricted peptides
| IFN-γ spots/5,000 cells | |||||||
|---|---|---|---|---|---|---|---|
| Donor | T cell lines | Control | Peptide mixture | MAGE-A2 111–125 | MAGE-A1 90–104 | MAGE-A12 127–141 | No APC |
| 78 | 78–13 | 5 (±1) | 98 (±4) | 49 (±3) | 92 (±9) | 10 (±1) | 6 (±1) |
| 78–22 | 27 (±4) | 162 (±13) | 65 (±6) | 177 (±1) | 35 (±2) | 10 (±3) | |
| 78–32 | 1 (±1) | 117 (±11) | 0 (±0) | 185 (±11) | 0 (±0) | 4 (±1) | |
| 121 | 121–6 | 2 (±1) | 92 (±13) | 9 (±4) | 123 (±15) | 3 (±0) | 0 (±0) |
| 121–9 | 3 (±1) | 392 (±19) | 2 (0) | 366 (±8) | 4 (±1) | 0 (±0) | |
| 121–19 | 4 (±3) | 216 (±1) | 5 (±2) | 153 (±6) | 280 (±20) | 0 (±0) | |
| 121–21 | 2 (±1) | 106 (±21) | 4 (±6) | 147 (±17) | 1 (±1) | 0 (±0) | |
| 121–24 | 19 (±0) | 140 (±3) | 23 (±6) | 208 (±3) | 15 (±13) | 1 (±1) | |
| 121–30 | 5 (±0) | 171 (±1) | 4 (±1) | 329 (±1) | 7 (±4) | 2 (±1) | |
| 121–34 | 5 (±5) | 86 (±6) | 23 (±9) | 83 (±1) | 4 (±1) | 1 (±1) | |
| 121–38 | 18 (±10) | 108 (±1) | 15 (±2) | 104 (±1) | 8 (±3) | 0 (±3) | |
| 121–40 | 1 (±1) | 208 (±0) | 4 (±0) | 250 (±9) | 6 (±4) | 0 (±0) | |
| 121–44 | 5 (±0) | 102 (±3) | 15 (±5) | 98 (±8) | 110 (±14) | 1 (±1) | |
| 121–48 | 6 (±0) | 225 (±16) | 6 (±2) | 253 (±1) | 8 (±7) | 0 (±0) | |
| 121–49 | 2 (±1) | 87 (±9) | 5 (±1) | 122 (±8) | 3 (±4) | 0 (±0) | |
| 122 | 122–12 | 9 (±4) | 173 (±18) | 2 (±10) | 178 (±20) | 12 (±6) | 4 (±1) |
| 122–26 | 1 (±1) | 120 (±0) | 11 (±1) | 110 (±11) | 8 (±4) | 3 (±0) | |
| 122–27 | 5 (±7) | 97 (±1) | 4 (±1) | 105 (±7) | 1 (±1) | 0 (±0) | |
| 122–28 | 1 (±1) | 101 (±8) | 4 (±4) | 110 (±7) | 0 (±0) | 0 (±0) | |
| 122–29 | 3 (±2) | 185 (±3) | 7 (±1) | 196 (±15) | 2 (±1) | 1 (±0) | |
| 129 | 129–7 | 5 (±0) | 197 (±20) | 3 (±3) | 1 (±1) | 307 (±17) | 1 (±1) |
| 129–12 | 1 (±1) | 110 (±10) | 2 (±1) | 112 (±13) | 2 (±1) | 1 (±1) | |
| 129–14 | 0 (±0) | 93 (±10) | 1 (±0) | 97 (±19) | 3 (±2) | 1 (±1) | |
| 129–24 | 2 (±3) | 106 (±6) | 21 (±9) | 120 (±4) | 4 (±2) | 0 (±0) | |
| 129–30 | 0 (±0) | 92 (±3) | 0 (±1) | 86 (±17) | 6 (±1) | 1 (±1) | |
| 129–34 | 3 (±3) | 122 (±6) | 115 (±5) | 5 (±3) | 3 (±0) | 2 (±1) | |
| 129–37 | 4 (±1) | 82 (±4) | 1 (±1) | 91 (±0) | 3 (±0) | 0 (±0) | |
| 129–39 | 7 (±2) | 84 (±4) | 4 (±4) | 93 (±18) | 5 (±1) | 1 (±1) | |
CD4+ T cell lines from four tumor-free donors (78, 121, 122 and 129) were obtained after 3 weekly stimulations by autologous mature dendritic cells loaded with a mixture of seven selected peptides (MAGE-A1 90–104, MAGE-A1 268–282, MAGE-A2 111–125, MAGE-A2 219–233, MAGE-A9 68–82, MAGE-A9 153–167, MAGE-A12 127–141). The specificity of the T cell lines was assessed by IFN-γ Elispot. L-DP4 were used as APC. The negative control corresponds to the wells containing no peptides. The positive values are at least three times higher than the negative control with a minimal number of 30 spots (bold)
Table 3.
Estimate of the number of CD4+ T cell precursors specific for the HLA-DP4 restricted MAGE-A peptides
| Peptides | Positive | CD4+ precursor frequency | ||
|---|---|---|---|---|
| Donors | Min. | Max. | Mean | |
| MAGE-A1 90–104 | 6/6 | 4.1 × 10−7 | 2.7 × 10−6 | 1.4 × 10−6 |
| MAGE-A1 268–282 | 2/6 | 6.9 × 10−7 | 3.1 × 10−6 | 1.9 × 10−6 |
| MAGE-A12 127–141 | 6/10 | 1.7 × 10−7 | 6.9 × 10−7 | 3.1 × 10−7 |
| MAGE-A2 111–125 | 1/5 | – | – | 2.5 × 10−7 |
| MAGE-A9 68–82 | 1/5 | – | – | 1.7 × 10−7 |
| MAGE-A9 153–167 | 1/5 | – | – | 1.7 × 10−7 |
CD4+ T cell precursor frequency was estimated using the Poisson distribution according to the following formula: frequency = −Ln [(number of negative wells/total number of wells tested)]/(number of CD4+ T cells seeded per well). Minima, maxima and means are given for the positive donors only. The number of CD4+ T cells seeded per well were 100,000 cells for all the donors. As an example, the calculations of the precursor frequency for the peptide MAGE-A1 90–104 based on the data presented in Table 1 were the following: donor 78, 3 positive wells for 40 wells seeded (the frequency was −Ln(37/40)/105 = 7.8 × 10−7); donor 121, 12 positive wells for 50 wells seeded (2.7 × 10−6); donor 122, 5 positive wells for 30 wells seeded (1.8 × 10−6) and donor 129, 6 positive wells for 40 wells seeded (1.6 × 10−6). For the peptide MAGE-A12 127–141: donor 121, 2 positive wells for 50 wells seeded (4.1 × 10−7); donor 129, 1 positive well for 40 wells seeded (2.5 × 10−7)
The T cell lines specific for MAGE-A1 90–104, MAGE-A1 268–282 and MAGE-A12 127–142 are specific for the native antigen
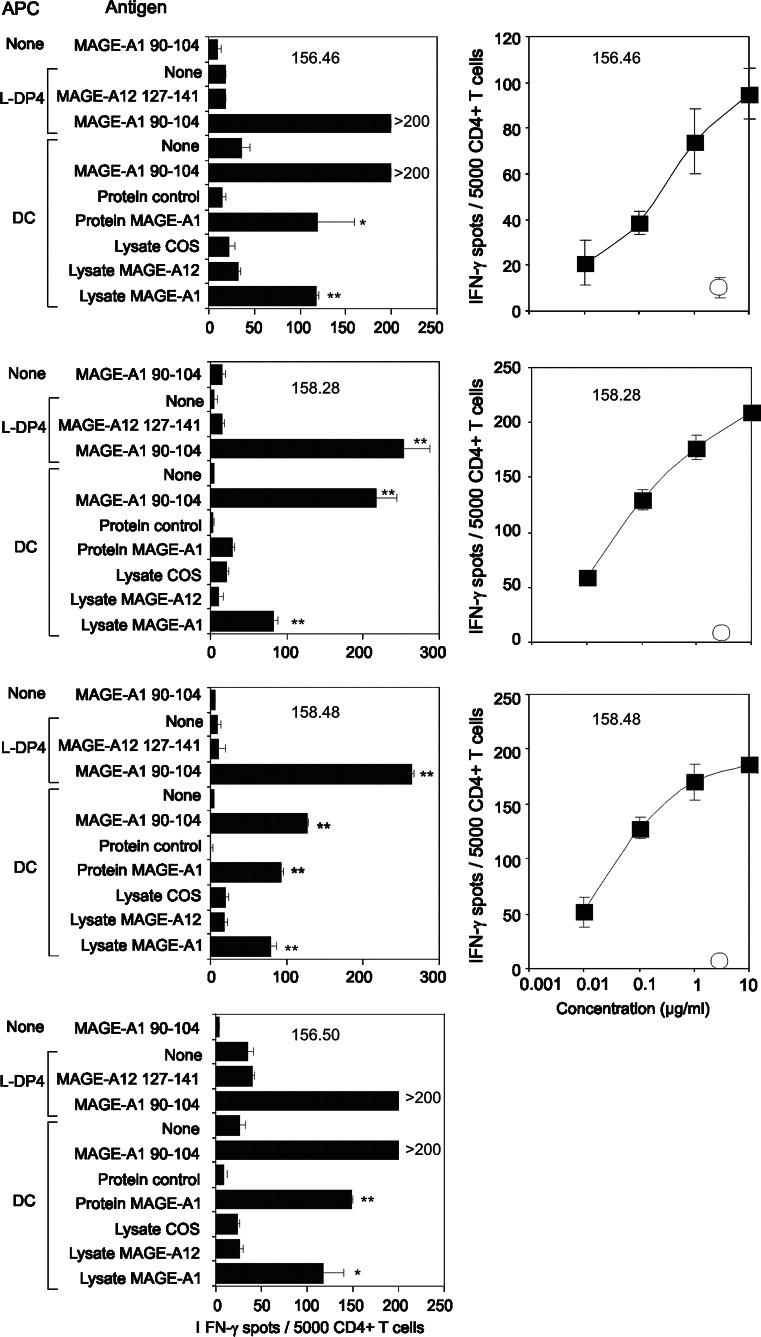
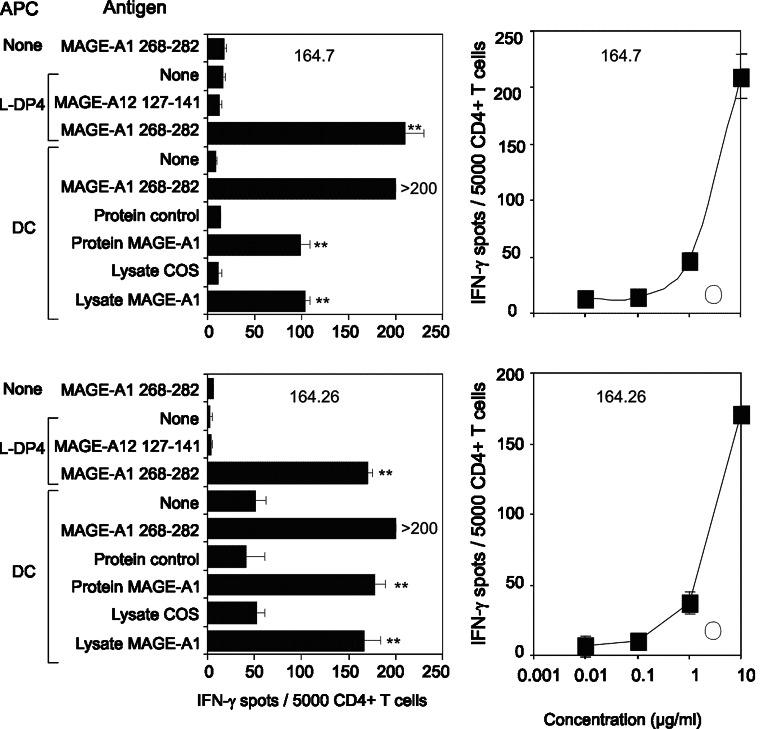
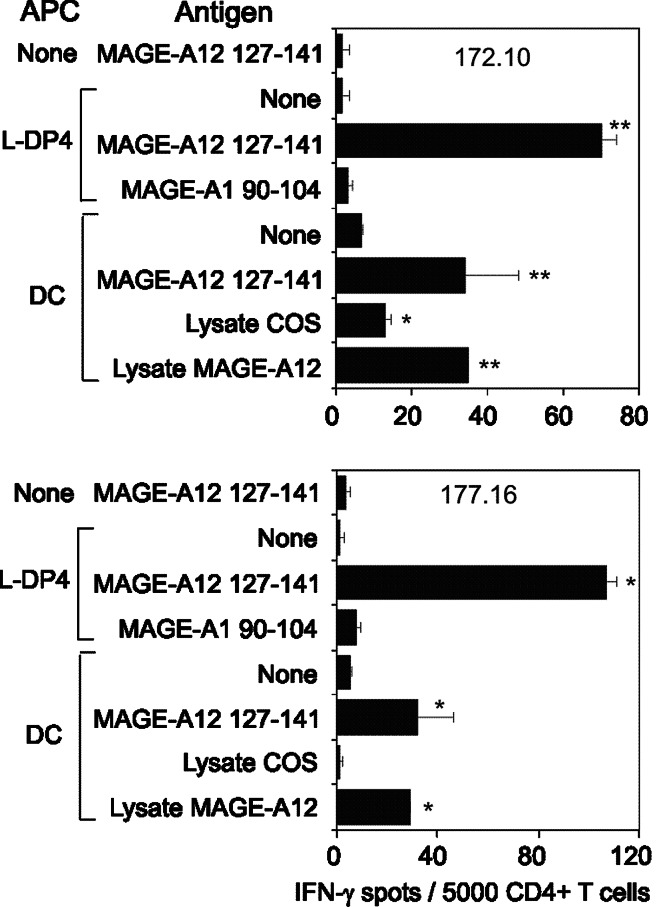
We analyzed the specificity of four T cell lines from two different healthy donors, which were specific for MAGE-A1 90–104 (Fig. 1). These CD4+ T cell lines specifically recognized MAGE-A1 90–104 presented by autologous DC and reacted to autologous DC previously loaded with the recombinant MAGE-A1 protein (Fig. 1 left panels). They were not, however, stimulated by unloaded DC and by DC fed with the recombinant bv-PLA2 (control protein), which was previously used to investigate the T cell response of mice [2] and patients allergic to bee venom [29]. Moreover, T cell lines were activated by DC fed with a lysate of COS-7 cells transfected with the MAGE-A1 gene, but not by DC fed with a lysate of untransfected COS-7 cells or pulsed with a lysate of COS-7 cells transfected with the MAGE-A12 gene. Expression of transfected MAGE-A1 in Cos-7 cells was at a similar level to its natural expression in melanoma cells (Data not shown). Peptide concentration response of three cell lines demonstrated their efficient stimulation by the MAGE-A1 90–104 peptide (Fig. 1, right panels). We also characterized two T cell lines which were specifically stimulated by MAGE-A1 268–282 presented by L-DP4 cells or by autologous DC (Fig. 2). In contrast to the control protein, the recombinant MAGE-A1 protein stimulated the two T cell lines after being captured by autologous DC, while unloaded DC served as baseline control (Fig. 2, left panels). The T cell lines were also activated by a lysate of COS-7 cells transfected with the MAGE-A1 gene but not by a lysate of untransfected COS-7 cells. The peptide concentration response of the T cell lines showed that the activation occurred at a high peptide concentration only, suggesting that the corresponding Tcr had a moderate affinity for the MAGE-A1 268–282 peptide (Fig. 2, right panels). Two T cell lines specific for the MAGE-A12 127–141 peptide exhibited an intense peptide response when L-DP4 was used as APC, although it was of lower intensity on autologous DC (Fig. 3). A significant response was observed for both T cell lines when DC were fed with a lysate of COS-7 cells transfected with the MAGE-A12 gene. It was equivalent to that provoked by the MAGE-A12 127–141 peptide. In contrast, the lysate of untransfected COS-7 cells exhibited a lower, if any, T cell stimulating capacity. The low number of harvested T cells did not allow us to evaluate the peptide concentration response. We concluded from these experiments that the peptides MAGE-A1 90–104, MAGE-A1 268–282 and MAGE-A12 127–141 elicited specific T cell lines, which were also specific for the native protein presented by autologous DC.
Fig. 1.
Characterization of MAGE-A1 90–104 specific T cell lines. Left panels MAGE-A1 90–104 specific T cell lines (156.46, 158.28, 158.48 and 156.50) were incubated (5 × 103 cells/well) in an Elispot assay using L-DP4 or immature DC as APC (2 × 104/well). DCs were previously loaded with a lysate of transfected or untransfected COS-7 cells or proteins (1 μM). Right panels T cell lines were incubated in the presence of L-DP4 cells (3 × 104/well) with a concentration range of the peptide (closed squares) or without any peptide (open circle). Each value represents the average spot number of the duplicates. Double asterisk and asterisk indicate P < 0.01 and P < 0.05, respectively
Fig. 2.
Characterization of MAGE-A1 268–282 specific T cell lines. Two MAGE-A1 268–282 specific T cell lines (164.7 and 164.28) were submitted to an IFN-γ EliSpot assay as described for Fig. 1
Fig. 3.
Characterization of MAGE-A12 128–142 specific T cell lines. Two MAGE-A12 127–141 specific T cell lines (172.10 and 177.16) were submitted to an IFN-γ EliSpot assay as described for Fig. 1
MAGE-A12 127–141 specific T cell lines recognize other MAGE-A sequences besides the native one
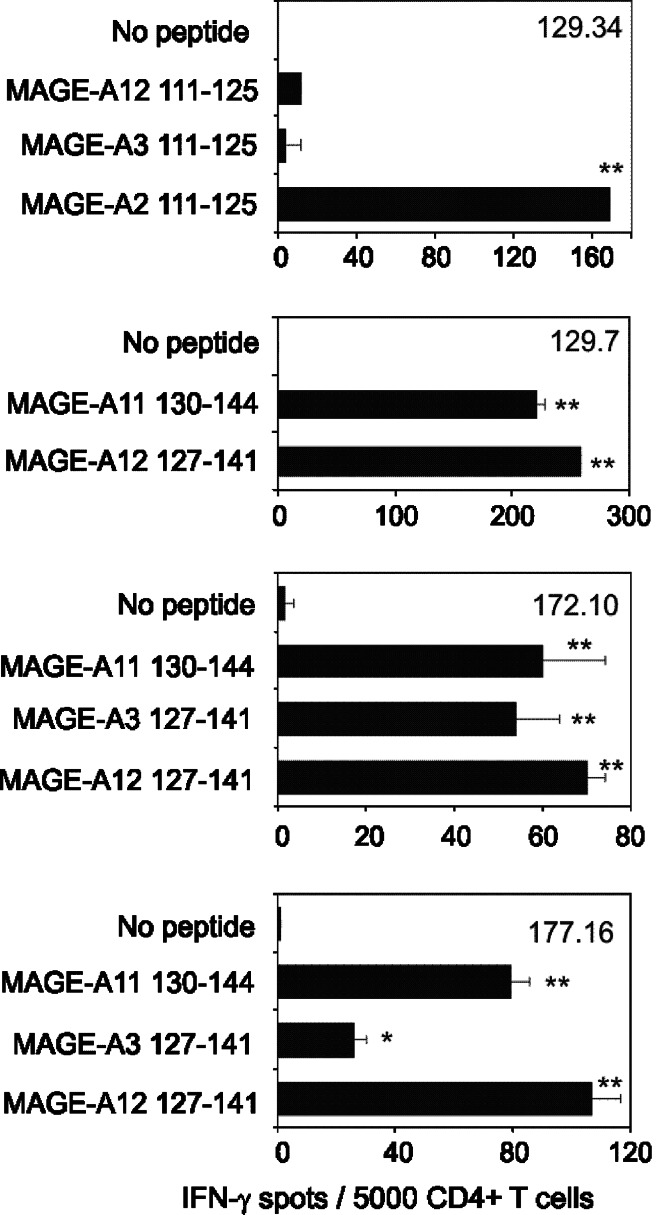
Among immunogenic peptides we identified in this study, MAGE-A2 111–125 and MAGE-A12 127–141 exhibited a good level of conservation with homologous peptides encoded by the other MAGE-A genes. We therefore investigated the peptide-binding activity (Table 4) and T cell reactivity of corresponding analogs (Fig. 4). Two natural analogs of MAGE-A2 111–125, namely MAGE 3 111–125 and MAGE 12 111–125, bound well to both HLA-DP4 molecules (Table 4). However, they were not antigenic for the MAGE-A2 111–125 specific T cell line 129.34 (Fig. 4). Among the natural analogs of MAGE-A12 127–141, MAGE-A11 130–144 bound to both HLA-DP4 molecules, while the six other bound to HLA-DP402 only (Table 4). MAGE-A11 130–144 stimulated three T cell lines specific for MAGE-A12 127–141, while MAGE-A3 127–141 stimulated the 172.10 and 177.16 T cell lines only (Fig. 4).
Table 4.
Capacity of MAGE-A analogs of MAGE-A2 111–125 and MAGE-12 127–141 to bind to HLA DP4 molecules
| Peptide | Sequence | IC50 (DP401) | IC50 (DP402) | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 4 | 6 | 9 | Pred. | Obs. | Pred. | Obs. | ||||||||||||
| MAGE-A3 111-125 a | R | K | V | A | E | L | V | H | F | L | L | L | K | Y | R | 562 | 100 (±0) | 100 | 35 (±2) |
| MAGE-A12 111-125 a | R | K | M | A | E | L | V | H | F | L | L | L | K | Y | R | 562 | 32 (±14) | 100 | 18 (±2) |
| MAGE-A11 130-144 b | K | G | L | I | T | K | A | E | M | L | G | S | V | I | K | 1,071 | 260 (±0) | 100 | 184 (±21) |
| MAGE-A1 120-134 b | R | E | P | V | T | K | A | E | M | L | E | S | V | I | K | 3,981 | 3,800 (±424) | 302 | 424 (±35) |
| MAGE-A2 130-144 b | R | E | P | V | T | K | A | E | M | L | E | S | V | I | R | 3,981 | 2,850 (±778) | 302 | 310 (±14) |
| MAGE-A3 127-141 b | R | E | P | V | T | K | A | E | M | L | G | S | V | I | G | 3,981 | 26,500 (±2121) | 302 | 4,250 (±354) |
| MAGE-A4 128-142 b | K | E | L | V | T | K | A | E | M | L | E | R | V | I | K | 21,877 | 2,400 (±566) | 2,398 | 414 (±49) |
| MAGE-A9 126-140 b | K | E | P | V | T | K | A | E | M | L | E | S | V | I | K | 3,981 | 2,900 (±707) | 302 | 639 (±35) |
| MAGE-A10 152-166 b | K | E | P | I | T | K | A | E | I | L | E | S | V | I | K | 1,905 | 1,500 (±0) | 602 | 570 (±35) |
aThe MAGE-A analogs of MAGE-A2 111–125 and bMAGE-12 127–141 were submitted to HLA-DP4 competitive ELISA under the same conditions as described in Table 1. Predicted (Pred.) and observed (Obs.) IC50 are expressed in nM. Experimental values are the means of two to three experiments. The P1, P4, P6 and P9 positions are indicated in bold
Fig. 4.
Recognition of homologous MAGE peptides by MAGE2 111–125 and MAGE-A12 127–141 T cell lines. T cell lines specific for MAGE-A2 111–125 (129.34) or MAGE-A12 127–141 (129.7, 172.10 and 177.16) were incubated in an Elispot assay using L-DP4 as APC (2 × 104/well). Peptides were added to the culture at a concentration of 10 μg/ml. Each value represents the average spot number of the duplicates. Double asterisk and asterisk indicate P < 0.01 and P < 0.05, respectively
Discussion
The high frequency of HLA-DP4 in the population facilitates the recruitment of patients for clinical trials and the development of new approaches of cancer immunotherapy. As an example, HLA-DP4 restricted T cell epitopes discovered in MAGE-A3 [23] and NY-ESO-1 tumor antigens [18, 38] have been proposed as vaccine candidates [22, 24, 37] and immunomonitoring reagents [1, 40]. However, these epitopes were discovered by chance, since no reliable methods existed to selectively identify HLA-DP4 restricted T cell epitopes. We have therefore recently set up a binding prediction method to selectively identify HLA-DP4-restricted T cell epitopes in any kind of antigens [10]. Considering the interest of targeting the MAGE-A family for cancer immunotherapy, this approach has been applied to this tumor-specific gene family. We identified 12 peptides, which were good binders to HLA-DP401, ranked six different peptides for their T cell stimulating properties, and identified two peptides from MAGE-A1 and one peptide from MAGE-A12, which were demonstrated to be naturally processed epitopes of the native antigens.
Based on the frequency of responders and on the frequency of peptide-specific precursors, the six T cell stimulating peptides we identified may be classified into three different categories. The peptide MAGE-A1 90–104 was stimulating for all six healthy donors tested. The frequency of precursors ranged from 0.4 to 2.7 per million CD4+ T cells (Table 3). Tcr capable of reacting with this peptide were present in all the individuals and at a high frequency in comparison to previous studies [12, 17, 30, 40]. In particular, its precursor frequency was similar to that of the MAGE-A3/DP4 epitope found in melanoma patients before vaccination [40]. The second category is described by the peptide MAGE-A1 268–282. T cell reactivity against this peptide was found in two of the six donors at a frequency of precursors of 0.7 to 3.1 per million CD4+ T cells (Table 3). Therefore, it was present probably in a limited number of the donors but at a comparable precursor frequency to MAGE-A1 90–104. In congenic mice, it has been shown that a T cell response raised against an antigen involves a “public” Vβ repertoire found in all animals and a “private” one, which is specific to each individual [9]. According to these experiments MAGE-A1 268–282 seemed to be recognized by Tcr belonging to the private repertoire, while MAGE-A1 90–104 recruited Tcr from the public repertoire. The third category is composed by the peptides that elicited a T cell response in some donors only because of their low precursor frequencies. They are close to the detection threshold of the method (0.1 per million CD4+ T cells) (Table 3). This category comprises the MAGE-A2 111–125, MAGE-A9 68–82, MAGE-A9 153–167 and MAGE-A12 127–141 peptides. The latter was active in six out of ten donors tested and its precursor frequency ranged from 0.2 to 0.7 per million CD4+ T cells. Interestingly, we observed that the most T cell stimulating peptides are not the best binders to HLA-DP4. MAGE-A1 90–104 was less efficient to bind HLA-DP4 molecules than MAGE-A2 111–125, MAGE-A1 268–282 and MAGE-A9 153–167. In many studies [14, 27], including ours [28], affinity for MHC molecules appeared to be a limiting factor for foreign antigens to elicit CD4+ T cell response. In this paper, we compared T cell stimulating efficiency of peptides that displayed a good affinity and did not submit low binders to T cell stimulation assays. Our data strongly suggest that good binders recruit CD4+ T cells at different levels of efficacy and that in these conditions affinity does not constitute a limiting factor. We also showed that the capacity of peptides to stimulate T cells could vary greatly from one individual to another, even in HLA class II controlled conditions. Thus, this sustains the interest in investigating the T cell response in multiple donors.
Two of the peptides we identified as HLA-DP4 restricted T cell epitopes derive from the MAGE-A1 gene. MAGE-A1 was initially identified in a human melanoma cell line [32]. It is expressed in approximately half of metastatic melanomas, esophageal carcinomas and non-small cell lung carcinomas and in 80% of hepatocellular carcinomas [13, 31]. To our knowledge, only two MAGE-1 specific CD4+ T cell epitopes have been previously described [5, 6], namely MAGE-A1 281–292 and MAGE-A1 121–134. These two peptides are restricted to HLA-DR15 [5] and HLA-DR13 [6], respectively. Moreover, MAGE-A3 267–282 is restricted to HLA-DR1 and induces T cells, which recognize MAGE-A1 260–275 [39]. Two of these epitopes are closed to the MAGE-A1 268–282 peptide we identified, suggesting that this sequence region comprises multiple CD4+ T cell epitopes. Like MAGE-A3 267–282, the MAGE-A1 268–282 peptide is only slightly different from the homologous sequences of MAGE-A2, -A3, -A4, -A6, -A12, suggesting that these sequences may also contain HLA-DP4 restricted peptides. However, we did not assess their cross-reactivity. In contrast, MAGE-A1 90–104 we identified as an HLA-DP4 restricted T cell epitope is not a conserved sequence. In particular, the I94, which is accommodated in the P1 pocket of HLA-DP4, is substituted by a D94 in most of the MAGE-A gene family. This substitution is not expected to favor the binding of the peptides to HLA-DP4 according to the HLA-DP4 binding motif [3]. It is noteworthy that the 90–104 encompasses the 96–104 sequence which is an HLA-A3 restricted T cell epitope [6]. This is reminiscent of the observations made on the NY-ESO-1 157–170 peptide which contains HLA-DP4 and HLA-A2 restricted T cell epitopes [37]. It therefore suggests that MAGE-A1 90–104 could be able to induce both CD4+ and CD8+ responses, as shown for the NY-ESO-1 157–170 peptide [37].
We demonstrated that MAGE-A12 127–141 is an HLA-DP4 restricted T cell epitope of the MAGE-A12 antigen. Moreover, T cells primed by this peptide recognized homologous sequences as MAGE-A11 130–144 and MAGE-A3 127–141. MAGE-A12 is frequently expressed in melanomas [13, 25] and could be the target of tumor-specific CTL [20]. Only the HLA-DR1 restricted MAGE-A3/12 267–282 peptide has been previously described as a CD4 T cell epitope of MAGE-A12 [39].
Finally, we describe in this paper potential HLA-DP4 restricted CD4+ T cell epitopes which require further investigations. We show that MAGE-A2 111–125, MAGE-A9 68–82 and MAGE-A9 153–167 were able to elicit peptide-specific T cell lines. Particularly, MAGE-A2 111–125 is worthy of investigation as it encompasses the sequence 112–120, which is an HLA-A*0201 restricted epitope in HLA-A2 transgenic mouse [33]. This peptide is homologous to the HLA-DR restricted T cell epitope MAGE-A3 111–125 [11]. We also show that the MAGE-A2 143–157, MAGE-A4 248–262, MAGE-A10 244–258 and MAGE-A10 303–317 peptides had a good affinity for HLA-DP4 molecules. This might be especially interesting for the MAGE-A10 244–258 peptide as it overlaps an HLA-A2 restricted CD8+ epitope [15]. The T cell priming ability of these peptides has not been investigated but their capacity to bind HLA-DP4 does not preclude the possibility that they are immunogenic.
In conclusion, we present in this paper the first selective identification of HLA-DP4 restricted T cell epitopes performed on the MAGE-A gene family. We mainly identified new epitopes in the MAGE-A1 antigen, one of them being stimulating for all the tested donors. The other sequences were also identified in multiple MAGE-A antigens, including MAGE-A12. Considering the high frequency of HLA-DP4 in the population, these peptide sequences are of major interest for vaccine trials and immunomonitoring of the CD4+ T cell response raised against this major family of tumor-specific antigens.
Acknowledgments
This work was supported by the CEA (BM), a grant from Association pour la Recherche contre le Cancer (ARC) (BM), National Institutes of Health (NIH)/ National Cancer Institute (NCI) Grants CA90360 (HZ and BM) and CA112198 (HZ).
References
- 1.Atanackovic D, Altorki NK, Stockert E, Williamson B, Jungbluth AA, Ritter E, Santiago D, Ferrara CA, Matsuo M, Selvakumar A, Dupont B, Chen YT, Hoffman EW, Ritter G, Old LJ, Gnjatic S. Vaccine-induced CD4+ T cell responses to MAGE-3 protein in lung cancer patients. J Immunol. 2004;172:3289–3296. doi: 10.4049/jimmunol.172.5.3289. [DOI] [PubMed] [Google Scholar]
- 2.Buhot C, Chenal A, Sanson A, Pouvelle-Moratille S, Gelb MH, Menez A, Gillet D, Maillere B. Alteration of the tertiary structure of the major bee venom allergen Api m 1 by multiple mutations is concomitant with low IgE reactivity. Protein Sci. 2004;13:2970–2978. doi: 10.1110/ps.04885404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Castelli FA, Buhot C, Sanson A, Zarour H, Pouvelle-Moratille S, Nonn C, Gahery-Segard H, Guillet JG, Menez A, Georges B, Maillere B. HLA-DP4, the most frequent HLA II molecule, defines a new supertype of peptide-binding specificity. J Immunol. 2002;169:6928–6934. doi: 10.4049/jimmunol.169.12.6928. [DOI] [PubMed] [Google Scholar]
- 4.Charron D, Fauchet R, Albert E, Bodmer J, Cambon-Thomsen A, Degos L, Hors J, Piazza A, Schreuder I (1997) (eds) Genetic diversity of HLA, functional and medical implication. In: Twelfth international histocompatibility workshop and conference, Charron D Paris, France
- 5.Chaux P, Lethe B, Van Snick J, Corthals J, Schultz ES, Cambiaso CL, Boon T, van der Bruggen P. A MAGE-1 peptide recognized on HLA-DR15 by CD4(+) T cells. Eur J Immunol. 2001;31:1910–1916. doi: 10.1002/1521-4141(200106)31:6<1910::AID-IMMU1910>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 6.Chaux P, Luiten R, Demotte N, Vantomme V, Stroobant V, Traversari C, Russo V, Schultz E, Cornelis GR, Boon T, van der Bruggen P. Identification of five MAGE-A1 epitopes recognized by cytolytic T lymphocytes obtained by in vitro stimulation with dendritic cells transduced with MAGE-A1. J Immunol. 1999;163:2928–2936. [PubMed] [Google Scholar]
- 7.Chaux P, Vantomme V, Coulie P, Boon T, van der Bruggen P. Estimation of the frequencies of anti-MAGE-3 cytolytic T-lymphocyte precursors in blood from individuals without cancer. Int J Cancer. 1998;77:538–542. doi: 10.1002/(SICI)1097-0215(19980812)77:4<538::AID-IJC11>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 8.Chaux P, Vantomme V, Stroobant V, Thielemans K, Corthals J, Luiten R, Eggermont AM, Boon T, van der Bruggen P. Identification of MAGE-3 epitopes presented by HLA-DR molecules to CD4(+) T lymphocytes. J Exp Med. 1999;189:767–778. doi: 10.1084/jem.189.5.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cibotti R, Cabaniols JP, Pannetier C, Delarbre C, Vergnon I, Kanellopoulos JM, Kourilsky P. Public and private V beta T cell receptor repertoires against hen egg white lysozyme (HEL) in nontransgenic versus HEL transgenic mice. J Exp Med. 1994;180:861–872. doi: 10.1084/jem.180.3.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cohen WM, Pouvelle-Moratille S, Wang XF, Farci S, Munier G, Charron D, Menez A, Busson M, Maillere B. Scanning the HIV genome for CD4+ T cell epitopes restricted to HLA-DP4, the most prevalent HLA class II molecule. J Immunol. 2006;176:5401–5408. doi: 10.4049/jimmunol.176.9.5401. [DOI] [PubMed] [Google Scholar]
- 11.Consogno G, Manici S, Facchinetti V, Bachi A, Hammer J, Conti-Fine BM, Rugarli C, Traversari C, Protti MP. Identification of immunodominant regions among promiscuous HLA-DR-restricted CD4+ T-cell epitopes on the tumor antigen MAGE-3. Blood. 2003;101:1038–1044. doi: 10.1182/blood-2002-03-0933. [DOI] [PubMed] [Google Scholar]
- 12.Coulie PG, Karanikas V, Colau D, Lurquin C, Landry C, Marchand M, Dorval T, Brichard V, Boon T. A monoclonal cytolytic T-lymphocyte response observed in a melanoma patient vaccinated with a tumor-specific antigenic peptide encoded by gene MAGE-3. Proc Natl Acad Sci USA. 2001;98:10290–10295. doi: 10.1073/pnas.161260098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Plaen E, Arden K, Traversari C, Gaforio JJ, Szikora JP, De Smet C, Brasseur F, van der Bruggen P, Lethe B, Lurquin C, et al. Structure, chromosomal localization, and expression of 12 genes of the MAGE family. Immunogenetics. 1994;40:360–369. doi: 10.1007/BF01246677. [DOI] [PubMed] [Google Scholar]
- 14.Diepolder HM, Gerlach JT, Zachoval R, Hoffmann RM, Jung MC, Wierenga EA, Scholz S, Santantonio T, Houghton M, Southwood S, Sette A, Pape GR. Immunodominant CD4+ T-cell epitope within nonstructural protein 3 in acute hepatitis C virus infection. J Virol. 1997;71:6011–6019. doi: 10.1128/jvi.71.8.6011-6019.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang LQ, Brasseur F, Serrano A, De Plaen E, van der Bruggen P, Boon T, Van Pel A. Cytolytic T lymphocytes recognize an antigen encoded by MAGE-A10 on a human melanoma. J Immunol. 1999;162:6849–6854. [PubMed] [Google Scholar]
- 16.Kobayashi H, Song Y, Hoon DS, Appella E, Celis E. Tumor-reactive T helper lymphocytes recognize a promiscuous MAGE-A3 epitope presented by various major histocompatibility complex class II alleles. Cancer Res. 2001;61:4773–4778. [PubMed] [Google Scholar]
- 17.Lonchay C, van der Bruggen P, Connerotte T, Hanagiri T, Coulie P, Colau D, Lucas S, Van Pel A, Thielemans K, van Baren N, Boon T. Correlation between tumor regression and T cell responses in melanoma patients vaccinated with a MAGE antigen. Proc Natl Acad Sci USA. 2004;101(Suppl 2):14631–14638. doi: 10.1073/pnas.0405743101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mandic M, Castelli F, Janjic B, Almunia C, Andrade P, Gillet D, Brusic V, Kirkwood JM, Maillere B, Zarour HM. One NY-ESO-1-derived epitope that promiscuously binds to multiple HLA-DR and HLA-DP4 molecules and stimulates autologous CD4+ T cells from patients with NY-ESO-1-expressing melanoma. J Immunol. 2005;174:1633–1640. doi: 10.4049/jimmunol.174.3.1751. [DOI] [PubMed] [Google Scholar]
- 19.Marchand M, van Baren N, Weynants P, Brichard V, Dreno B, Tessier MH, Rankin E, Parmiani G, Arienti F, Humblet Y, Bourlond A, Vanwijck R, Lienard D, Beauduin M, Dietrich PY, Russo V, Kerger J, Masucci G, Jager E, De Greve J, Atzpodien J, Brasseur F, Coulie PG, van der Bruggen P, Boon T. Tumor regressions observed in patients with metastatic melanoma treated with an antigenic peptide encoded by gene MAGE-3 and presented by HLA-A1. Int J Cancer. 1999;80:219–230. doi: 10.1002/(SICI)1097-0215(19990118)80:2<219::AID-IJC10>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 20.Panelli MC, Bettinotti MP, Lally K, Ohnmacht GA, Li Y, Robbins P, Riker A, Rosenberg SA, Marincola FM. A tumor-infiltrating lymphocyte from a melanoma metastasis with decreased expression of melanoma differentiation antigens recognizes MAGE-12. J Immunol. 2000;164:4382–4392. doi: 10.4049/jimmunol.164.8.4382. [DOI] [PubMed] [Google Scholar]
- 21.Rosenberg SA, Yang JC, Restifo NP. Cancer immunotherapy: moving beyond current vaccines. Nat Med. 2004;10:909–915. doi: 10.1038/nm1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schuler-Thurner B, Schultz ES, Berger TG, Weinlich G, Ebner S, Woerl P, Bender A, Feuerstein B, Fritsch PO, Romani N, Schuler G. Rapid induction of tumor-specific type 1 T helper cells in metastatic melanoma patients by vaccination with mature, cryopreserved, peptide-loaded monocyte-derived dendritic cells. J Exp Med. 2002;195:1279–1288. doi: 10.1084/jem.20012100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schultz ES, Lethe B, Cambiaso CL, Van Snick J, Chaux P, Corthals J, Heirman C, Thielemans K, Boon T, van der Bruggen P. A MAGE-A3 peptide presented by HLA-DP4 is recognized on tumor cells by CD4+ cytolytic T lymphocytes. Cancer Res. 2000;60:6272–6275. [PubMed] [Google Scholar]
- 24.Schultz ES, Schuler-Thurner B, Stroobant V, Jenne L, Berger TG, Thielemanns K, van der Bruggen P, Schuler G. Functional analysis of tumor-specific Th cell responses detected in melanoma patients after dendritic cell-based immunotherapy. J Immunol. 2004;172:1304–1310. doi: 10.4049/jimmunol.172.2.1304. [DOI] [PubMed] [Google Scholar]
- 25.Serrano A, Lethe B, Delroisse JM, Lurquin C, De Plaen E, Brasseur F, Rimoldi D, Boon T. Quantitative evaluation of the expression of MAGE genes in tumors by limiting dilution of cDNA libraries. Int J Cancer. 1999;83:664–669. doi: 10.1002/(SICI)1097-0215(19991126)83:5<664::AID-IJC16>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 26.Southwood S, Sidney J, Kondo A, del Guercio MF, Appella E, Hoffman S, Kubo RT, Chesnut RW, Grey HM, Sette A. Several common HLA-DR types share largely overlapping peptide binding repertoires. J Immunol. 1998;160:3363–3373. [PubMed] [Google Scholar]
- 27.Tangri S, Mothe BR, Eisenbraun J, Sidney J, Southwood S, Briggs K, Zinckgraf J, Bilsel P, Newman M, Chesnut R, Licalsi C, Sette A. Rationally engineered therapeutic proteins with reduced immunogenicity. J Immunol. 2005;174:3187–3196. doi: 10.4049/jimmunol.174.6.3187. [DOI] [PubMed] [Google Scholar]
- 28.Texier C, Hervé M, Pouvelle S, Ménez A, Maillere B. On the diversity and heterogeneity of H-2d restricted determinants and T cell epitopes from the major bee venom allergen. Int Immunol. 1999;11:1313–1325. doi: 10.1093/intimm/11.8.1313. [DOI] [PubMed] [Google Scholar]
- 29.Texier C, Pouvelle-Moratille S, Buhot C, Castelli FA, Pecquet C, Menez A, Leynadier F, Maillere B. Emerging principles for the design of promiscuous HLA-DR-restricted peptides: an example from the major bee venom allergen. Eur J Immunol. 2002;32:3699–3707. doi: 10.1002/1521-4141(200212)32:12<3699::AID-IMMU3699>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 30.Valmori D, Souleimanian NE, Hesdorffer CS, Old LJ, Ayyoub M. Quantitative and qualitative assessment of circulating NY-ESO-1 specific CD4(+) T cells in cancer-free individuals. Clin Immunol. 2005;117:161–167. doi: 10.1016/j.clim.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 31.Van den Eynde BJ, van der Bruggen P. T cell defined tumor antigens. Curr Opin Immunol. 1997;9:684–693. doi: 10.1016/S0952-7915(97)80050-7. [DOI] [PubMed] [Google Scholar]
- 32.van der Bruggen P, Traversari C, Chomez P, Lurquin C, De Plaen E, Van den Eynde B, Knuth A, Boon T. A gene encoding an antigen recognized by cytolytic T lymphocytes on a human melanoma. Science. 1991;254:1643–1647. doi: 10.1126/science.1840703. [DOI] [PubMed] [Google Scholar]
- 33.Visseren MJ, van der Burg SH, van der Voort EI, Brandt RM, Schrier PI, van der Bruggen P, Boon T, Melief CJ, Kast WM. Identification of HLA-A*0201-restricted CTL epitopes encoded by the tumor-specific MAGE-2 gene product. Int J Cancer. 1997;73:125–130. doi: 10.1002/(SICI)1097-0215(19970926)73:1<125::AID-IJC19>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 34.Wang HY, Lee DA, Peng G, Guo Z, Li Y, Kiniwa Y, Shevach EM, Wang RF. Tumor-specific human CD4+ regulatory T cells and their ligands: implications for immunotherapy. Immunity. 2004;20:107–118. doi: 10.1016/S1074-7613(03)00359-5. [DOI] [PubMed] [Google Scholar]
- 35.Zarour HM, Kirkwood JM, Kierstead LS, Herr W, Brusic V, Slingluff CL, Jr, Sidney J, Sette A, Storkus WJ. Melan-A/MART-1(51–73) represents an immunogenic HLA-DR4-restricted epitope recognized by melanoma-reactive CD4(+) T cells. Proc Natl Acad Sci USA. 2000;97:400–405. doi: 10.1073/pnas.97.1.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zarour HM, Maillere B, Brusic V, Coval K, Williams E, Pouvelle-Moratille S, Castelli F, Land S, Bennouna J, Logan T, Kirkwood JM. NY-ESO-1 119–143 is a promiscuous major histocompatibility complex class II T-helper epitope recognized by Th1- and Th2-type tumor-reactive CD4+ T cells. Cancer Res. 2002;62:213–218. [PubMed] [Google Scholar]
- 37.Zeng G, Li Y, El-Gamil M, Sidney J, Sette A, Wang RF, Rosenberg SA, Robbins PF. Generation of NY-ESO-1-specific CD4+ and CD8+ T cells by a single peptide with dual MHC class I and class II specificities: a new strategy for vaccine design. Cancer Res. 2002;62:3630–3635. [PMC free article] [PubMed] [Google Scholar]
- 38.Zeng G, Wang X, Robbins PF, Rosenberg SA, Wang RF. CD4(+) T cell recognition of MHC class II-restricted epitopes from NY-ESO-1 presented by a prevalent HLA DP4 allele: association with NY-ESO-1 antibody production. Proc Natl Acad Sci USA. 2001;98:3964–3969. doi: 10.1073/pnas.061507398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Y, Chaux P, Stroobant V, Eggermont AM, Corthals J, Maillere B, Thielemans K, Marchand M, Boon T, Van Der Bruggen P. A MAGE-3 peptide presented by HLA-DR1 to CD4(+) T cells that were isolated from a melanoma patient vaccinated with a MAGE-3 protein. J Immunol. 2003;171:219–225. doi: 10.4049/jimmunol.171.1.219. [DOI] [PubMed] [Google Scholar]
- 40.Zhang Y, Renkvist N, Sun Z, Schuler-Thurner B, Glaichenhaus N, Schuler G, Boon T, van der Bruggen P, Colau D. A polyclonal anti-vaccine CD4 T cell response detected with HLA-DP4 multimers in a melanoma patient vaccinated with MAGE-3.DP4-peptide-pulsed dendritic cells. Eur J Immunol. 2005;35:1066–1075. doi: 10.1002/eji.200425847. [DOI] [PubMed] [Google Scholar]
- 41.Zhang Y, Sun Z, Nicolay H, Meyer RG, Renkvist N, Stroobant V, Corthals J, Carrasco J, Eggermont AM, Marchand M, Thielemans K, Wolfel T, Boon T, van der Bruggen P. Monitoring of anti-vaccine CD4 T cell frequencies in melanoma patients vaccinated with a MAGE-3 protein. J Immunol. 2005;174:2404–2411. doi: 10.4049/jimmunol.174.4.2404. [DOI] [PubMed] [Google Scholar]