Abstract
The chimeric antibody 806 (Ch806) is a promising antitumor agent that recognizes both the epidermal growth factor receptor variant III (EGFRvIII) and the overexpressed epidermal growth factor receptor (EGFR) in cancer tissues but does not recognize the wild type EGFR in normal tissues. However, passive antibody immunization could not produce effective antitumor titers unless the immunization was administered repeatedly over long periods. To overcome this limitation, we generated epitope mimics that bind to Ch806 and tested whether the peptide mimics could induce the production of similar antibodies when actively immunizing mice with the peptides. We used the PH.D-12 phage display peptide library to identify peptides that bind to the monoclonal antibody (mAb) 12H23, which also recognizes similar epitopes of Ch806. Two mimotopes (WHTEILKSYPHE and LPAFFVTNQTQD) were shown to mimic the mAb 12H23 and Ch806 epitope using immunoassays. The mimotopes were conjugated to immunogenic carrier proteins and used to intraperitoneally immunize BALB/c mice. Interestingly, sera from the mice immunized with the isolated mimotopes not only recognize the recombinant or synthetic 806 eptitope, but can also recognize EGFR that is overexpressed in A431 cells and EGFRvIII expressed in Huh7-EGFRvIII cells, whereas sera from mice immunized with the control peptide-KLH (keyhole limpet hemocyanin) and carrier KLH alone failed to show a similar reactivity. Furthermore, in an antibody-dependent cellular cytotoxicity assay (ADCC), the mimotope-induced antibodies specifically lysed human Huh-7-EGFRvIII cells. Our data indicate that the isolated mimotopes reported here may potentially be used as new alternative agents for treating cancer with EGFRvIII expression or EGFR overexpression.
Electronic supplementary material
The online version of this article (doi:10.1007/s00262-010-0872-7) contains supplementary material, which is available to authorized users.
Keywords: Ch806, EGFR, Cancer immunotherapy, Mimotope, Phage display library
Introduction
The overexpression of EGFR is often caused by amplification or mutation of the egfr gene [1, 2]. The EGFRvIII variant is the most common oncogenic EGFR mutation [3, 4]. The overexpression of EGFR and EGFRvIII has been observed in many epithelial tumors and is often correlated with a worsening of the clinical prognosis [5–10]. As EGFR plays a crucial role in cancer initiation and progression, therapeutic agents that target EGFR have been developed. Two anti-EGFR antibodies (cetuximab and panitumumab) have been approved for treating colorectal cancer. However, both of them recognize EGFR expressed in cancer cells as well as normal cells and thus induce skin rashes and other unwanted side effects [11–13]. Unlike the two antibodies mentioned above, monoclonal antibody 806 is an antibody that preferentially recognizes the untethered form of EGFR (overexpressed EGFR and EGFRvIII) [14]. In phase I clinical trials, specific localization of 111I-Ch806 (chimeric antibody 806) was observed in target lesions of all patients at all dose levels, and notably, no skin rashes or gastrointestinal tract disturbances were observed in any patients [15]. Additionally, Ch806 was found to be better than cetuximab in inhibiting the growth of EGFRvIII positive tumor xenografts [16]. Subsequent epitope analysis indicated that mAb806 recognizes the (287CGADSYEMEEDGVRKC302) epitope (referred to as CC16) of EGFR [17].
Although some therapeutic antibodies have been approved, they still have serious practical limitations, such as obstacles in production and the need for multiple infusions of the antibody to achieve effective titers [18]. Active immunization using epitope peptides to initiate the ongoing production of the desired antibody type would be an attractive alternative, because it can circumvent both the need for multiple administrations and the danger of inducing an immune response against the nonhuman parts of the infused antibodies. However, due to immune tolerance, the use of a natural epitope peptide alone is inefficient at inducing an immune reaction [19]. Mimotopes are small peptides that structurally mimic a given antibody-binding site but are composed of different amino acids [20, 21]. Active immunization using mimotopes would induce antibodies to recognize the mimicked epitope. Notably, mimotopes can be isolated from phage display peptide libraries [22]. For this study, to make a substitution of Ch806, we exploited phage display technology to identify the peptides that can mimic the natural CC16 epitope.
Materials and methods
Cell lines and lysates
The human glioblastoma–astrocytoma, epithelial-like cell line U-87 MG, the human epidermoid carcinoma cell line A-431 (ATCC, USA) capable of overexpressing EGFR and the human hepatocellular carcinoma cell line Huh7-EGFRvIII that stably expresses EGFRvIII [23] were grown in Dulbecco’s modified Eagle’s medium (GIBCO, Grand Island, USA) supplemented with 10% fetal bovine serum and 1% penicillin–streptomycin. The total cell lysates were prepared as described previously [24]. The protein concentration was determined photometrically using bicinchoninic acid (BCA Protein Assay Kit; Pierce, Rockford, IL). The lysates were aliquoted and stored at −80°C.
Monoclonal antibodies
The monoclonal antibody 12H23, a mouse immunoglobulin G1 (IgG1) monoclonal antibody, was produced and isolated by our lab. The chimeric antibody 806 was expressed in CHO cells and subsequently purified [25]. Both of these antibodies bind to the CC16 epitope.
Biopanning of PH.D-12 phage display library
The Ph.D-12 library was purchased from New England Bio-Labs Inc. (Beverly, MA, USA). Biopanning procedures were performed according to the manufacturer’s instructions with certain modifications. Briefly, a 96-well plate (JCHS, Shenzhen, China) was coated with 50 μl mAb 12H23 [100 μg/ml in 0.1 M NaHCO3 (pH 8.6)] overnight at 4°C. Wells were then washed 10 times with TBST (50 mM Tris, 150 mM NaCl, pH 7.5 containing 0.1% Tween-20), filled with 300 μl of blocking buffer (0.1 M NaHCO3 pH 8.6, 5 mg/ml BSA) and incubated for 2 h at 37°C. In the first round of biopanning, 10 μl of the phage (1.5 × 1011) isolated from the initial library in 100 μl of TBST was incubated with the coated mAb for 1 h at 37°C. After repeated washes with TBST, the bound phage was eluted with 100 μl of 0.2 M Glycine–HCl (pH 2.2) and neutralized with 15 μl of 1 M Tris–HCl (pH 9.1). The eluate was used for amplification and titration in an Escherichia coli ER2738 culture. The recovered phage was subjected to two additional rounds of biopanning with mAb 12H23 (in the second and third rounds, concentrations of 10 μg/ml and 1 μg/ml of mAb 12H23 were used, respectively). The eluate in the third round of screening was titrated, and blue clones were randomly selected and amplified by infecting ER2738.
DNA sequencing
Single-stranded phage DNA was prepared according to the Ph.D-12 phage display library manufacturer’s instructions and was subsequently sequenced by Invitrogen Inc. (Shanghai, China).
Specificity ELISA
Ninety-six-well ELISA plates were coated with 50 μl mAb 12H23, an isotype-matched control antibody [10 μg/ml in 0.1 M NaHCO3 (pH 8.6)] and 2% BSA overnight at 4°C. The plates were then washed with TBS containing 0.5% Tween-20 and subsequently blocked using TBS containing 5% dry milk. About 1 × 1012 pfu of purified phage in TBS containing 5% dry milk were incubated with the coated antibodies. The bound phage particles were detected with a peroxidase-conjugated mouse anti-phage M13 monoclonal antibody (Pharmacia, Peapack, USA). The reaction was then developed with 2, 2-azino-bis-(3-ethylbenzthiazoline-6-sulphonic acid) (SIGMA, St. Louis, USA) as a substrate. The optical density at 405 nm was measured using an ELISA reader (BioRad model 680). The specificity ELISA was performed in triplicate. The same procedure was also used to examine the binding specificity of the isolated phage to Ch806.
Phage competitive binding assay
Ninety-six-well ELISA plates were coated with 50 μl mAb 12H23 (10 μg/ml in 0.1 M NaHCO3, pH 8.6) overnight at 4°C. The plates were then washed with TBST and blocked by incubation with TBS containing 5% dry milk. About 1 × 1010 pfu of purified phage in TBS containing 5% dry milk were input and incubated for 1 h. After repeated washes, the phage was eluted with six different peptides or proteins (0.2 M Glycine–HCl, pH 2.2, N12-806, S1-GFP, CC16, irrelevant peptide (CKGYEDSRVMEAGDEC) and the isotype-matched control antibody) prior to titration. N12-806 is a fusion protein comprising the CC16 epitope and the N1N2 domain of M13 phage pIII. S1-GFP is a fusion protein comprising the S1 domain of EGFR and the green fluorescent protein (GFP). CC16 was chemically synthesized (Tiansheng, Shanghai, China). Notably, N12-806, S1-GFP and CC16 can be recognized by mAb 12H23 (data not shown). The assay was performed in triplicate.
Synthesis of vaccine constructs
The peptides WHTEILKSYPHEGGGSC (constituting a high-affinity mimotope, GGGSC was used as a linker) and LPAFFVTNQTQDGGGSC (a lower-affinity mimotope) were chemically synthesized (Tiansheng, Shanghai, China). Each peptide was then coupled through its C terminus to an immunogenic carrier, keyhole limpet hemocyanin (KLH) (SIGMA, St. Louis, USA).
Immunization of BALB/c mice
Four groups (n = 6 per group) of BALB/c mice were immunized by repeated intraperitoneal injection with 100 μg of the WHTEILKSYPHE-KLH peptide, the LPAFFVTNQTQD-KLH peptide, the control peptide-KLH, or KLH alone, on days 1, 22 and 43. The complete Freund adjuvant/incomplete Freund adjuvant (CFA/IFA) (SIGMA, St. Louis, USA) was used as an adjuvant in all groups. Blood was obtained from the tail veins on days 8, 29 and 50.
Titer determination and western blot assays
Titers of sera from the immunized BALB/C mice against KLH, EGFRvIII, the mimotopes and N12-806 were determined using a general ELISA method. For western blot assays, A431 and Huh7-EGFRvIII cell lysates were separated using sodium dodecyl sulfate–polyacrylamide gel electrophoresis and then electrophoretically transferred to nitrocellulose membranes. The membranes were blocked with PBS containing 5% dry milk and then incubated with mAb 12H23 or pooled serum for 2 h at 37°C. Bound antibodies were then detected by a peroxidase-conjugated goat anti-mouse antibody (KeYa, Hangzhou, China). The membranes were then exposed to Kodak (X-OMAT) film at room temperature.
Fluorescent immunostaining
U-87 MG, A431, Huh7-EGFRvIII cells (5 × 103 cells/well) were plated on 24-well tissue culture chamber slides (HaiMen, Haikou, China). The cells were grown overnight at 37°C and 5% CO2. After washing with PBS, the cells were fixed with 4% paraformaldehyde in PBS for 30 min. The chamber slides were then blocked with 10% goat serum in PBS for 1 h, followed by incubation with Ch806, mAb 12H23 (positive control), or the samples from the third immune sera were pooled and incubated at room temperature for 45 min. The bound antibodies were detected using fluorescein isothiocyanate (FITC)-conjugated goat anti-human IgG, FITC-conjugated goat anti-mouse IgG (KangCheng, Shanghai, China) and DAPI (Roche, Shanghai, China). The pooled sera from the peptide-KLH and KLH immunized mice were then used as a control. The cells were observed with a fluorescence microscope (Olympus, Shanghai, China).
Functional assays
Antibody-dependent cellular cytotoxicity (ADCC) and complement-dependent cytotoxicity (CDC) were assessed as described previously [26] with some modifications. Huh7-EGFRvIII cells were used as specific target cells. The third pooled immune sera of the mimotope-immunized mice or the control mice were diluted 1:100 and used in ADCC and CDC assays. For the ADCC experiments, lymphocytes of the naive BALB/c mice (prepared by mashing the spleen and thymus) were prepared as effector cells. The effector to target (E:T) cell ratio was about 50:1. The target cells were incubated in a 96-well plate overnight, and then the pooled immune sera were added to the wells with the effector cells. After co-incubation for 48 h, the supernatants were removed carefully. The reaction was developed with the Cell Counting Kit-8 diluted 1:10. The optical density at 450 nm was measured using an ELISA reader (BioRad model 680). For the CDC assay, the pooled third immune sera served as the source for both antibodies and the complement. A CDC assay was performed in a similar manner as the ADCC assay method. The percentage of cellular cytotoxicity was calculated using the following formula:
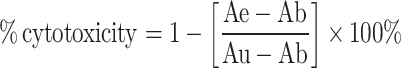 |
where Ab is the background absorbance (only of the pooled immune sera with the effector cells), Ae is the experimental absorbance (target cells + pooled immune sera with the effector cells) and Au is the absorbance of the untreated cultures (target cells + medium with the effector cells). The assay was performed in duplicate.
Statistical analysis
Data from the cytotoxicity experiments were analyzed for statistical significance using the two-tailed Student’s t test by comparing the two-paired groups of data.
Results
Biopanning and mimotope characterization
As mAb 12H23 is a murine monoclonal antibody that can recognize the CC16 epitope (unpublished data), it was used as the target protein for screening peptides that can mimic the epitope. The eluted phage titers increased with each round of panning, indicating that the phage particles might carry epitope mimics. The phage titer increased from 7.9 × 105 pfu/ml (first round) to 106 pfu/ml (second round) and finally to 2.1 × 108 pfu/ml (third round), suggesting the phage bound to mAb 12H23 were well enriched.
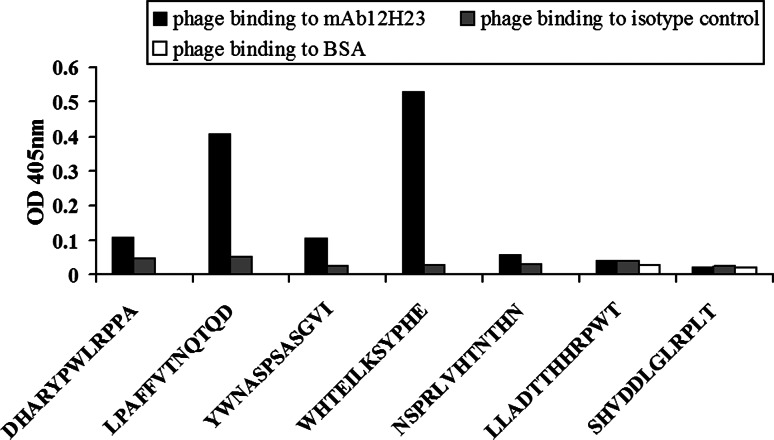
Genomic DNA from 20 phage clones was sequenced, and seven different insert sequences were observed. A specificity ELISA was then performed to independently demonstrate whether the seven different peptides could be recognized by the mAb 12H23. Two (WHTEILKSYPHE and LPAFFVTNQTQD) mimotope candidates were specifically recognized by the selecting antibody 12H23 but not by the isotype-matched control antibody (Fig. 1).
Fig. 1.
Characterization of mAb 12H23 mimotopes. Specific recognition of the selected phage clones by mAb12H23 was examined using ELISA. The M13 phage clones displaying different peptides bound to mAb 12H23 (solid bars), the isotype-matched control antibody (gray bars) and BSA (blank bars) are shown here
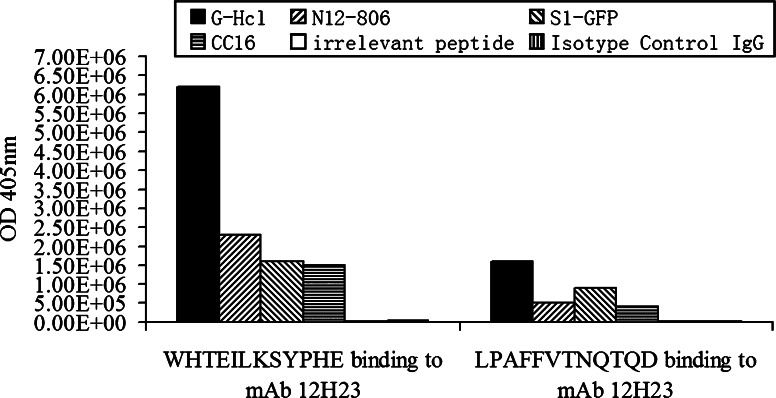
To demonstrate mimicry between the peptides and the original antigen, a phage competitive binding assay was then performed. Different competitors, including 0.2 M Glycine–HCl (pH 2.2), recombinant proteins bearing the CC16 epitope (N12-806 and S1-GFP) and the synthesized CC16 peptide (287CGADSYEMEEDGVRKC302) were used to compete with the binding of phage particles to mAb 12H23. The bound phage particles were detected by titer determination. The isotype-matched control antibody was used as control. The CC16 epitope-bearing proteins or peptide could displace a large amount of bound phage from the mAb 12H23 (Fig. 2), i.e., the phage mimic (WHTEILKSYPHE) bound to mAb 12H23 were displaced, about 6.2 × 106 using 0.2 M Glycine–HCl (pH 2.2), 2.3 × 106 using N12-806, 1.6 × 106 using S1-GFP, 1.5 × 106 using CC16, only 1.0 × 104 and 3.0 × 104 using the irrelevant peptide and isotype-matched control antibody, respectively. The phage mimic (LPAFFVTNQTQD) bound to mAb 12H23 were displaced, about 1.6×106 using 0.2 M Glycine–HCl (pH 2.2), 5.0×105 using N12-806, 9.0×105 using S1-GFP, 4.0×105 using CC16, only 2.0×103 and1.0×103 using the irrelevant peptide and isotype-matched control antibody, respectively.
Fig. 2.
ELISA analysis of the phage competitive binding assay. M13 phage displaying peptides (WHTEILKSYPHE and LPAFFVTNQTQD) bound to mAb 12H23 were competed with different competitors. The competitors included Glycine–HCl (solid bars), N12-806 (right oblique bars), S1-GFP (left oblique bars), the CC16 peptide (horizontally hatched bars), irrelevant peptide (vertically hatched bars) and the isotype-matched control antibody (blank bars), respectively
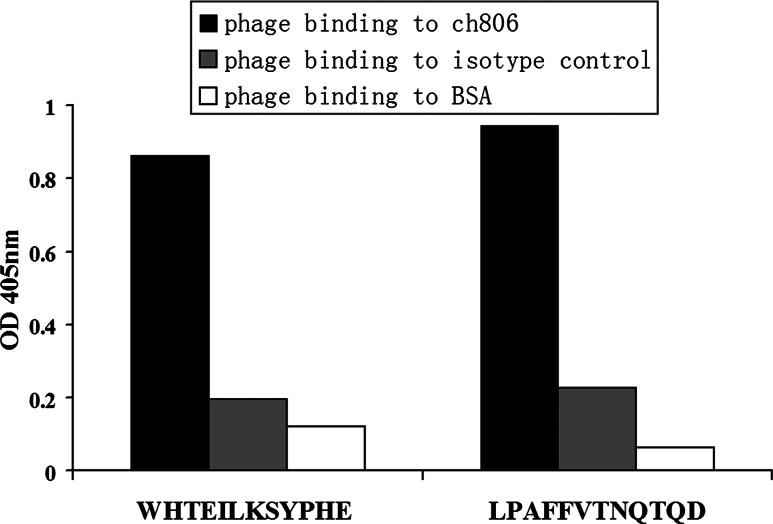
As the mAb 12H23 and Ch806 can bind to the same epitope, ELISA was performed to test whether the peptide mimotopes of mAb 12H23 could also be recognized by Ch806. The result of the ELISA showed that the phage displaying the peptides (WHTEILKSYPHE and LPAFFVTNQTQD) could also bind to Ch806 (Fig. 3). All these data indicate that peptides (WHTEILKSYPHE and LPAFFVTNQTQD) can mimic the structure of the CC16 peptide.
Fig. 3.
ELISA of selected phage clones bound to Ch806. M13 phage displaying peptides (WHTEILKSYPHE and LPAFFVTNQTQD) bound to Ch806 (solid bars), the isotype-matched control antibody (gray bars) and BSA (blank bars) are shown here
Vaccine constructs and immune responses induced by mimotope vaccination
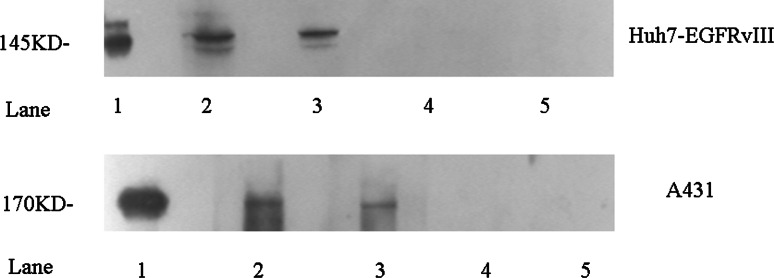
From the specificity ELISA and the competitive binding assay results, the higher reactive peptide WHTEILKSYPHE and the less reactive peptide LPAFFVTNQTQD were chosen for chemical synthesis. The immunogenicity of the mimotope conjugates was then evaluated by immunizing BALB/c mice. The serum titers were successfully induced, as indicated by the high anti-antigen titers. Mice immunized with the mimotope conjugates not only showed a humoral immune response toward the KLH and the respective peptide but also, more importantly, developed antibodies that recognize the CC16 epitope-bearing recombinant proteins, N12-806 and EGFRvIII (Table 1). Moreover, the results of the western blots revealed that the induced mouse antibodies could specifically recognize the overexpressing EGFR protein in the A431 cell line and the EGFRvIII protein in the Huh7-EGFRvIII cell line (Fig. 4).
Table 1.
Titers of the four mouse groups determined by ELISA (the third immune serum, day 50), against KLH, mimotope and original antigen
| Antigen | Titer | |||
|---|---|---|---|---|
| Anti-KLH | Anti-mimotype | Anti-N12-806 | Anti-EGFRvIII | |
| WHTEILKSPHE-KLH | 1:364,500 | 1:364,500 | 1:1,500 | 1:4,500 |
| LPAFFVTNQTQD-KLH | 1:364,500 | 1:364,500 | 1:1,500 | 1:4,500 |
| Control peptide-KLH | 1:364,500 | |||
| KLH | 1:364,500 | |||
Fig. 4.
Western blot assays with mAb 12H23 or mimotope-induced antibodies for EGFRvIII and EGFR recognition in Huh7-EGFRvIII and A431 cell lysates. EGFRvIII and EGFR detection using mAb 12H23 (lane 1, positive control) and the sera from mice immunized with the WHTEILKSYPHE-KLH conjugate (lane 2), the LPAFFVTNQTQD-KLH conjugate (lane 3), the control peptide-KLH conjugate (lane 4), or the carrier KLH alone (lane 5)
Immunofluorescence
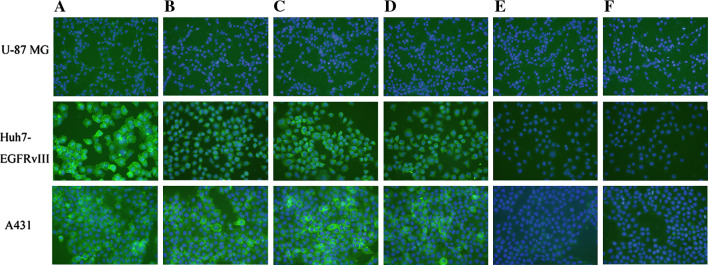
To further demonstrate whether the induced antibodies can bind to normal EGFR expressed in U-87 MG, EGFR overexpressed in A431 and EGFRvIII expressed in Huh7-EGFRvIII cells, an immunofluorescence staining assay was performed. Here, Ch806 and mAb 12H23 was employed as a positive control. The sera from mice immunized with WHTEILKSYPHE-KLH and LPAFFVTNQTQD-KLH showed intense staining in A431 and Huh7-EGFRvIII cells, whereas the sera from mice immunized with KLH and the control peptide-KLH displayed only background staining levels (Fig. 5). For comparability, membranes and nuclei were stained in all panels the same way. These results further supported that the induced antibodies can recognize EGFR overexpressed in A431 cells and EGFRvIII expressed in Huh7-EGFRvIII cells, but can not recongnize normal EGFR expressed in U-87 MG.
Fig. 5.
Immunofluorescence staining of EGFRvIII, EGFR overexpressed and normal EGFR expressed in Huh7-EGFRvIII, A431 and U-87 MG cells. Cells were incubated with anti-EGFR antibodies followed by FITC-labeled secondary antibodies where the nuclei were stained with DAPI. Cells were visualized with a fluorescence microscope. a Ch806 (positive control), b mAb 12H23 (positive control), c sera from mice immunized with WHTEILKSYPHE-KLH, d sera from mice immunized with LPAFFVTNQTQD-KLH, e sera from mice immunized with the control peptide-KLH, f sera from mice immunized with KLH alone
Functional assays
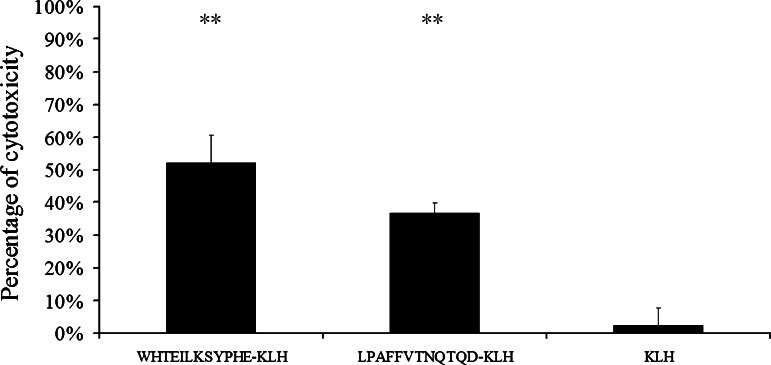
To assess whether the antibodies induced by the mimotopes can exhibit specific lysis of the target cells, we performed the ADCC assay. In the ADCC assay with Huh7-EGFRvIII cells, the anti-WHTEILKSYPHE-KLH antibodies and the anti-LPAFFVTNQTQD-KLH antibodies elicited specific lysis of 53 and 37% of these cells, respectively, while no specific cell lysis was observed with antibodies from the KLH immunized group (Fig. 6). However, for the CDC assay tested on the Huh7-EGFRvIII cells, almost no specific cytotoxicity was observed (data not shown).
Fig. 6.
Antibody-mediated cytotoxicity against Huh7-EGFRvIII cells elicited by antibodies induced after vaccination with mimotopes (WHTEILKSYPHE-KLH and LPAFFVTNQTQD-KLH) and keyhole limpet hemocyanin. Statistical analysis was performed using the Student’s t test (**P ≤ 0.005; exact P values: comparison of ADCC mediated by anti-WHTEILKSYPHE-KLH antibodies with anti-KLH antibodies, P = 0.0011; comparison of ADCC mediated by anti-LPAFFVTNQTQD-KLH antibodies to anti-KLH antibodies, P = 0.0032)
Discussion
One way to overcome the serious practical limitations of the monoclonal antibody approach could be to perform immunization with peptide mimics (mimotopes) of a naturally occurring epitope that is recognized by the desired antibody. To isolate mimotopes of the Ch806 antibody, which has been regarded as a promising agent for treating cancer that overexpresses EGFR or EGFRvIII, the phage display was exploited in this study. After three rounds of biopanning, we isolated two peptide mimics (WHTEILKSYPHE and LPAFFVTNQTQD) of the epitope that were specifically recognized by mAb 12H23 and Ch806. We aligned the mimotope sequences and have not found the homology between the two clones (and previously published mimotopes).
Titer determination was performed to evaluate the immunogenicity of the isolated mimotopes. As small peptide molecules are not sufficient enough to stimulate the immune system and may even induce tolerance [27], we coupled the mimotopes to an immunogenic carrier, KLH. Mice immunized with WHTEILKSYPHE-KLH and LPAFFVTNQTQD-KLH successfully developed high antibody titers against KLH as well as the mimotopes. The induced sera also recognized N12-806 and the EGFRvIII protein. Immunofluorescence staining further supported that the antibodies induced by the two isolated mimotopes can bind to overexpressed EGFR and EGFRvIII. The antibodies induced by the phage particles (WHTEILKSYPHE and LPAFFVTNQTQD) show a similar capacity for binding to the overexpressed EGFR and EGFRvIII (data not shown). Unlike the mimotopes of C225 [28], the mimotopes in this study can not induced anti-sera with detectable CDC effect. In general, the effect of antibodies in the sense of complement activation is dependent on the Fc portion of antibody isotypes. So the isotypes of antibodies determined whether the antibodies can activate CDC. For instance, IgG1 in mice is non-complement fixing. Therefore, whether the CDC could be activated may be ascribed to the different isotypes of the induced antibodies.
Our study demonstrated that active immunization induced by a mimotope in vivo can produce the desired antibody. With active immunization, the main obstacles of passive immunotherapy, specifically the comparatively short antibody half-life, may be overcome. Immunological memory will be induced to provide ongoing protection for vaccinated individuals. Meanwhile, the immunogenicity of artificial antibodies may be solved because the antibodies are produced by the patients themselves. Thus, the mimotopes identified for the overexpressed EGFR and EGFRvIII may represent a novel option for treating cancer.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Western blot assays with Ch806 for EGFRvIII and EGFR recognition in Huh7-EGFRvIII and A431 cell lysates (Tif 76.3 kb)
Acknowledgments
This study was supported by the Supporting Program of the “Eleventh Five-year Plan” for Sci & Tech Research of China (Grant No. 2008ZX10002-024).
References
- 1.Frederick L, Wang XY, Eley G, James CD. Diversity and frequency of epidermal growth factor receptor mutations in human glioblastomas. Cancer Res. 2000;60:1383–1387. [PubMed] [Google Scholar]
- 2.Malden LT, Novak U, Kaye AH, Burgess AW. Selective amplification of the cytoplasmic domain of the epidermal growth factor receptor gene in glioblastoma multiforme. Cancer Res. 1988;48:2711–2714. [PubMed] [Google Scholar]
- 3.Wong AJ, Ruppert JM, Bigner SH, Grzeschik CH, Humphrey PA, Bigner DS, Vogelstein B. Structural alterations of the epidermal growth-factor receptor gene in human gliomas. Proc Natl Acad Sci USA. 1992;89:2965–2969. doi: 10.1073/pnas.89.7.2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sugawa N, Ekstrand AJ, James CD, Collins VP. Identical splicing of aberrant epidermal growth-factor receptor transcripts from amplified rearranged genes in human glioblas. Proc Natl Acad Sci USA. 1990;87:8602–8606. doi: 10.1073/pnas.87.21.8602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mendelsohn J. Targeting the epidermal growth factor receptor for cancer therapy. J Clin Oncol. 2002;20:1S–13S. [PubMed] [Google Scholar]
- 6.Arteaga CL. Overview of epidermal growth factor receptor biology and its role as a therapeutic target in human neoplasia. Semin Oncol. 2002;29:3–9. doi: 10.1053/sonc.2002.35642. [DOI] [PubMed] [Google Scholar]
- 7.Nicholson RI, Gee JM, Harper ME. EGFR and cancer prognosis. Eur J Cancer. 2001;37:S9–S15. doi: 10.1016/S0959-8049(01)00231-3. [DOI] [PubMed] [Google Scholar]
- 8.Nishikawa R, Ji XD, Harmon RC, et al. A mutant epidermal growth factor receptor common in human glioma confers enhanced tumorigenicity. Proc Natl Acad Sci USA. 1994;91:7727–7731. doi: 10.1073/pnas.91.16.7727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Olapade-Olaopa EO, Moscatello DK, MacKay EH, Horsburgh T, Sandhu DP, Terry TR, Wong AJ, Habib FK. Evidence for the differential expression of a variant EGF receptor protein in human prostate cancer. Br J Cancer. 2000;82:186–194. doi: 10.1054/bjoc.1999.0898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moscatello DK, Holgado-Madruga M, Godwin AK, Ramirez G, Gunn G, Zoltick PW, Biegel JA, Hayes RL, Wong AJ. Frequent expression of a mutant epidermal growth factor receptor in multiple human tumors. Cancer Res. 1995;55:5536–5539. [PubMed] [Google Scholar]
- 11.Cartenì G, Fiorentino R, Vecchione L, Chiurazzi B, Battista C. Panitumumab a novel drug in cancer treatment. Ann Oncol. 2007;18:vi16–vi21. doi: 10.1093/annonc/mdm218. [DOI] [PubMed] [Google Scholar]
- 12.Cunningham D, Humblet Y, Siena S, Khayat D, Bleiberg H, Santoro A, Bets D, Mueser M, Harstrick A, Verslype C, Chau I, Van Cutsem E. Cetuximab monotherapy and cetuximab plus irinotecan in irinotecan-refractory metastatic colorectal cancer. N Engl J Med. 2004;351:337–345. doi: 10.1056/NEJMoa033025. [DOI] [PubMed] [Google Scholar]
- 13.Zhang W, Gordon M, Lenz HJ. Novel approaches to treatment of advanced colorectal cancer with anti-EGFR monoclonal antibodies. Ann Med. 2006;38:545–551. doi: 10.1080/09546630601070812. [DOI] [PubMed] [Google Scholar]
- 14.Luwor RB, Johns TG, Murone C, Su Huang H-J, Cavenee WK, Ritter G, Old LJ, Burgess AW, Scott AM. Monoclonal antibody 806 inhibits the growth of tumor xenografts expressing either the de2–7 or amplified epidermal growth factor receptor EGFR but not wild-type EGFR. Cancer Res. 2001;61:5355–5361. [PubMed] [Google Scholar]
- 15.Scott AM, Lee F-T, Tebbutt N, Herbertson R, Gill SS, Liu Z, et al. A phase I clinical trial with monoclonal antibody Ch806 targeting transitional state and mutant epidermal growth factor receptors. Proc Natl Acad Sci USA. 2007;104:4071–4076. doi: 10.1073/pnas.0611693104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rocha-Lima CM, Soares HP, Raez LE, Singal R. EGFR targeting of solid tumors. Cancer Control. 2007;14:295–304. doi: 10.1177/107327480701400313. [DOI] [PubMed] [Google Scholar]
- 17.Johns TG, Adams TE, Cochran JR, Hall NE, Hoyne PA, Olsen MJ, et al. Identification of the epitope for the epidermal growth factor receptor-specific monoclonal antibody 806 reveals that it preferentially recognizes an untethered form of the receptor. J Biol Chem. 2004;29:30375–30384. doi: 10.1074/jbc.M401218200. [DOI] [PubMed] [Google Scholar]
- 18.Riemer AB, Jensen-Jarolim E. Mimotope vaccines: epitope mimics induce anti-cancer antibodies. Immunol Lett. 2007;113:1–5. doi: 10.1016/j.imlet.2007.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adams TE. Tolerance to self-antigens in transgenic mice. Mol Biol Med. 1990;7:341–357. [PubMed] [Google Scholar]
- 20.Felici F, Luzzago A, Folgori A, Cortese R. Mimicking of discontinuous epitopes by phage-displayed peptides, II. Selection of clones recognized by a protective monoclonal antibody against the Bordetella pertussis toxin from phage peptide libraries. Gene. 1993;128:21–27. doi: 10.1016/0378-1119(93)90148-V. [DOI] [PubMed] [Google Scholar]
- 21.Luzzago A, Felici F, Tramontano A, Pessi A, Cortese R. Mimicking of discontinuous epitopes by phage-displayed peptides, I. Epitope mapping of human H ferritin using a phage library of constrained peptides. Gene. 1993;128:51–57. doi: 10.1016/0378-1119(93)90152-S. [DOI] [PubMed] [Google Scholar]
- 22.Cwirla SE, Peters EA, Barrett RW, Dower WJ. Peptides on phage: a vast library of peptides for identifying ligands. Proc Natl Acad Sci USA. 1990;87:6378–6382. doi: 10.1073/pnas.87.16.6378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang H, Jiang H, Zhou M, Xu Z, Liu S, Shi B, Yao X, Yao M, Gu J, Li Z. Epidermal growth factor receptor vIII enhances tumorigenicity and resistance to 5-fluorouracil in human hepatocellular carcinoma. Cancer Lett. 2009;279:30–38. doi: 10.1016/j.canlet.2009.01.019. [DOI] [PubMed] [Google Scholar]
- 24.Parmley SF, Smith GP. Antibody-selectable filamentous fd phage vectors: affinity purification of target genes. Gene. 1988;73:305–318. doi: 10.1016/0378-1119(88)90495-7. [DOI] [PubMed] [Google Scholar]
- 25.Panousis C, Rayzman VM, Johns TG, Renner C, Liu Z, Cartwright G, Lee F-T, Wang D, Gan H, Cao D, Kypridis A, Smyth FE, Brechbiel MW, Burgess AW, Old LJ, Scott AM. Engineering and characterisation of chimeric monoclonal antibody 806 (Ch806) for targeted immunotherapy of tumours expressing de2–7 EGFR or amplified EGFR. Br J Cancer. 2005;92:1069–1077. doi: 10.1038/sj.bjc.6602470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sato N, Yabuki Y, Toh K, Ishii Y, Kikuchi K. Separation of cell-dependent antibody (CDA) and inhibitory antibody by protein-A affinity chromatography and the effect of fractions on antibody-dependent cellular cytotoxicity (ADCC) Immunology. 1979;36:421–426. [PMC free article] [PubMed] [Google Scholar]
- 27.Wauben MH. Immunological mechanisms involved in experimental peptide immunotherapy of T-cell-mediated diseases. Crit Rev Immunol. 2000;20:451–469. [PubMed] [Google Scholar]
- 28.Riemer AB, Kurz H, Klinger M, Scheiner O, Zielinski CC, Jensen-Jarolim E. Vaccination with cetuximab mimotopes and biological properties of induced anti-epidermal growth factor receptor antibodies. J Natl Cancer Inst. 2005;97:1663–1670. doi: 10.1093/jnci/dji373. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Western blot assays with Ch806 for EGFRvIII and EGFR recognition in Huh7-EGFRvIII and A431 cell lysates (Tif 76.3 kb)