Abstract
Among the relatively large number of known tumor-associated antigens (TAA) which are recognized by human CD8 T-cells, Melan-A/MART-1 is one of the most—if not the most—frequently used target for anti-cancer vaccines in HLA-A2 + melanoma patients. In this study, we analyzed the killing of a large panel of melanoma cells by a high avidity, MART-1-specific T-cell clone or a MART-1-specific, polyclonal T-cell culture. Strikingly, we observed that the MART-1-specific T-cells only killed around half of the analyzed melanoma cell lines. In contrast a Bcl-2-specific T-cell clone killed all melanoma cell lines, although the T-cell avidity of this clone was significantly lower. The MART-1-specific T-cell clone expressed NKG-2D and was fully capable of releasing both perforin and Granzyme B. Notably, the resistance to killing by the MART-1-specific T-cells could be overcome by pulsing of the melanoma cells with the MART-1 epitope. Thus, the very frequently used MART-1 epitope was not expressed on the surface of many melanoma cell lines. Our data emphasize that the selected tumor antigens and/or epitopes are critical for the outcome of anti-cancer immunotherapy.
Keywords: Melan-A/MART-1, Cytotox T-cells, Melanoma, Lysis, Bcl-2
Introduction
Harnessing of the immune system for the battle against cancer has been the focus of tremendous research efforts over the past decades. Multiple means to achieve this goal have been investigated, including adoptive transfer of anti-tumor-reactive T-cells, systemic or localized administration of immune modulating cytokines [6]. Likewise, the use of ‘therapeutic’ vaccines has been scrutinized aiming at inducing CTL specific for TAA presented by cancer cells in the context of HLA class I molecules [11]. Surprisingly, until recently only limited attention has been focused on elucidating the most suitable targets for induction of clinically relevant anti-cancer immune responses. Melanoma differentiation antigens such as MART-1 were the first tumor antigens for which T-cell epitopes were identified. MART-1 was identified as a TAA from in vitro expanded TIL cultures [30, 31]. Melan-A/MART-1 is a tumor differentiation antigen which is expressed by melanocytes and malignant melanoma cells from tumors of >95% of patients. Two natural peptide variants derived from Melan-A are presented by HLA-A*0201 and recognized by Melan-A-specific T-cells: the nonapeptide AAGIGILTV, and the decapeptide EAAGIGILTV, differing in glutamic acid (E) at peptide position one [12]. Melan-A-specific CD8 T-cells are readily detectable in the majority of HLA-A*0201 positive normal individuals and in melanoma patients.
Among the relatively large number of known TAA which are recognized by human CD8 T-cells, Melan-A is one of the most—if not the most—frequently used target for anti-cancer vaccines in HLA-A2 + melanoma patients [4]. Both the presence of Melan-A specific CD8 + cells bearing a memory phenotype in cancer patients as well as their cytotoxic activity against various established tumor cell lines have been largely documented [9, 10, 15, 17, 26]. Therefore, it is generally recognized that the Melan-A antigen constitutes one preferential target of the immune system, at least in HLA-A2 + melanoma patients.
To further examine the natural immunity of the Melan-A/MART-1, we generated a high avidity T-cell clone against the immunodominant HLA-A2 restricted MART-1 epitopes and examined its ability to kill a large panel of different melanoma cells. As a control we compared the killing with a completely different target the Bcl-2 protein. Bcl-2 has previously been described as a TAA on the basis of spontaneous immune responses in melanoma as well as other types of cancer patients [3].
Materials and methods
Patients
Peripheral blood lymphocytes (PBL) from HLA-A2 positive cancer patients were obtained from the University Hospital in Herlev, Denmark. The PBL were isolated using Lymphoprep separation and cryopreserved in FCS with 10% DMSO. Tissue typing was conducted at the Department of Clinical Immunology, The State Hospital, Copenhagen, Denmark. Informed consent was obtained from the patients before any of theses measures.
Fluorescence activated cell sorting (FACS)
Peripheral blood lymphocytes and TIL were analyzed by flow cytometry using FACSAria (BD Biosciences, Brøndby, Denmark). The T-cells were stained with PE conjugated MHC-Dextramers (DAKO, Glostrup, Denmark), followed by antibody staining with the fluorochrome-couplet mAbs: CD8-APC-Cy7 or CD8-APC (BD Immunocytometri Systems). Both stainings were performed in RPMI 1640 media (GibcoBRL, Invitrogen, Taastrup, Denmark), for 20 min, 4°C, in the dark. The MHC–Dextramer complexes used were: HLA-A2/Bcl-2208–217 (PLFDFSWLSL), HLA-A2/MART-127–35(AAGIGILTV) and HLA-A2/HIV-1 pol476–484 (ILKEPVHGV). CD8/Bcl-2208–217 positive cells were sorted as single cells for cloning.
The surface expression of NKG2D on antigen-specific CTL clones was analyzed by usage of CD8-APC-Cy7 and NKG2D-PE direct-labeled mAbs. Unstained cells from the clones served as controls in these experiments.
Furthermore, the HLA-A2 expression of cell lines was determined using the FITC conjugated HLA-A2 mAb (BB7.2 clone, BD Immunocytometri Systems). Cells were stained with the antibody for 20 min at 4°C. Before final analysis cells were washed twice and resuspended in FACS buffer (PBS/2% FCS/4% NaN3). Unstained cells served as controls in these experiments. The magnitude of HLA-A2 expression was determined using the staining index (SI) defined as MFIpositive − MFIbackground/2 × SDbackground where MFI is mean fluorescence intensity [21]. This formula accomplishes an algorithm that normalizes the relative brightness of a positive fluorescence signal (stained cells) with the degree of spread in the negative population to which it is to be compared (unstained cells). SI > 3,000 were defined as a high amount of HLA-A2 expression, while 2,000 < SI < 3,000 and 100 < SI < 2,000 defined a medium and low expression, respectively.
Establishment of antigen specific T-cell clones
Specific CTL clones against the MART-1 were developed, as previously described [20]. Briefly, the PBL from a melanoma patient was cloned by limiting dilution in the presence of the irradiated (100 Gy), autologous melanoma cell line ESTDAB-007. Cells were cultured in RPMI-1640 medium containing 10 mM HEPES buffer, penicillin, streptomycin and 10% human serum. CTL clones were repeatedly restimulated with ESTDAB-007 melanoma cells as stimulator cells and JY cells (Epstein-Barr-virus-transformed B lymphoblasts) as feeder cells, as described [20].
Bcl-2-specific cells were isolated and expanded from PBL of cancer patients. CD8/Bcl-2208–217 positive cells were sorted as single cells into 96 well plates (Nunc, Invitrogen, Taastrup, Denmark) containing 105 cloning mix cells/well. The cloning mix was prepared containing 106/ml irradiated (20 Gy) lymphocytes from three healthy donors in X-vivo with 5% heat-inactivated human serum, 25 mM HEPES buffer (GibcoBRL), 1 μg/ml phytohemagglutinin (PHA) (Peprotech, London, UK) and 120 U/ml IL-2 (Chiron, Ratigen, Germany). Every 3–4 days 50 μl fresh media were added containing IL-2 to a final concentration of 120 U/ml. Growing clones were expanded using cloning mix cells (5 × 104 cells/well) and IL-2. The established CTL clones were analyzed for recognition of MART-1 or Bcl-2 in a standard 51Cr-release assay.
Polyclonal, MART-1-specific T-cell culture
Tumor infiltrating lymphocytes (TIL) from a melanoma patient were expanded using high dose IL-2 as described [23], and HLA-A2/MART-127–35
-
specific cells were sorted as a polyclonal cell population. As control cells were stained with the HLA-A2/HIV-1 pol476–484 dextramer complex. The isolated, MART-1-specific cells were used as effector cells in a 51Cr-release assay the following day.
Cytotoxicity assay
Conventional 51Cr-release assays for cytotoxic lymphocyte (CTL)-mediated cytotoxicity were carried out as described elsewhere [1]. Target cells were T2 cells, autologous tumor cells, the HLA-A2 positive breast cancer cell lines MDA-MB-231 and CAMA-1, the HLA-A2 positive colon cancer cell line HCT-116 and the HLA-A2 negative breast cancer cell line ZR-75-1, the HLA-A2-negative melanoma cell line FM9 and the HLA-A2-positive melanoma cell lines FM76, FM92, ESTDAB-001, 006, 014, 017, 019, 021, 026, 027, 028, 029, 033, 100, and 117 [20]. Cancer cell lines were examined for HLA-type and expression of Bcl-2 and MART-1 by RT-PCR. All cancer cells are publically available at the American Type Culture Collection (ATCC), or at The European Searchable Tumour Line Database (ESTDAB) Database and Cell Bank (http://www.ebi.ac.uk/ipd/estdab) [25]. The treatment of melanoma cells with human recombinant IFN-γ (Peprotech EC, London, UK) was done for 2 or 7 days starting the day after cells were seeded into tissue flasks. The melanoma cell lines were stimulated every second day in vitro with 100 U/ml of IFN-γ, prior to analysis in the 51Cr-release assay [28].
In some assays cancer cell lines were loaded with peptide for 2–4 h at RT with 4 μg of the MART-127–35 peptide, before the cell lines were used as targets in the 51Cr-release assay. The assays against unpulsed and peptide-pulsed tumor cells were conducted together. Lysis were blocked using the HLA specific mAb W6/32 (2 μg/100 μl), and in cold targeted inhibition assays using unlabeled T2 cells pulsed with the appropriate peptide. In cold:target assays we often observe a dilution effect [13, 19, 24, 29, 32]. This dilution effect is occurring due to the fact that the addition of 20 times extra target cells to the effectors changes the actual effector:target (E:T) ratio.
IFN-γ ELISPOT assay
The ELISPOT assay was used to quantify peptide epitope-specific IFN-γ releasing effector cells as described previously [2]. Briefly, nitrocellulosebottomed 96-well plates (MultiScreen MAIP N45; Millipore) were coated with anti-IFN-γ Ab (1-D1 K; Mabtech). The wells were washed, blocked by X-vivo medium and the effector cells were added in duplicates at different cell concentrations, together with the target cells. Target cells were peptide loaded/unloaded T2 cells, and the melanoma cell lines ESTDAB-007, 019 and 029. The plates were incubated overnight. The following day, medium was discarded and the wells were washed prior to addition of biotinylated secondary Ab (7-B6-1-Biotin; Mabtech). The plates were incubated at room temperature (RT) for 2 h, washed, and Avidin-enzyme conjugate (AP-Avidin; Calbiochem/Invitrogen Life Technologies) was added to each well. Plates were incubated at RT for 1 h and the enzyme substrate NBT/BCIP (Invitrogen Life Technologies) was added to each well and incubated at RT for 5–10 min. Upon the emergence of dark purple spots, the reaction was terminated by washing with tap water. The spots were counted using the ImmunoSpot Series 2.0 analyzer (CTL Analyzers) and the peptide-specific CTL frequency could be calculated from the numbers of spot-forming cells.
Results
Specificity of clones
The lytic capacity of the MART-127–35-specific CTL clone was analyzed in standard 51Cr-release assays. To this end, T2 cells loaded with either the MART-127–35 peptide or an irrelevant peptide (the high affinity, HLA-A2-restricted HIV-1 pol476–484) served as targets. This assay revealed that only T2 cells pulsed with the MART-127–35 peptide were killed (Fig. 1a). Furthermore, the MART-127-35 CTL clone lysed T2 cells at an E:T ratio at 1:1 pulsed with down to 10−6 mM peptide (Fig. 1a). It has previously been shown that a number of MART-1-specific CTL clones recognize the decapeptide 26–35 (EAAGIGILTV) of MART-1 more efficiently than the corresponding nonapeptide 27–35 [27]. Consequently, we analyzed the recognition of both of these two peptides by the MART-1-specific CTL clone (Fig. 1a). The results showed that the MART-1-specific CTL clone effectively lysed T2 cells loaded with the decapeptide 26-35 (Fig. 1a). Furthermore, the MART-1-specific CTL clone killed T2 cells pulsed with as low as 10−8 mM of the decapeptide MART-126–35.
Fig. 1.
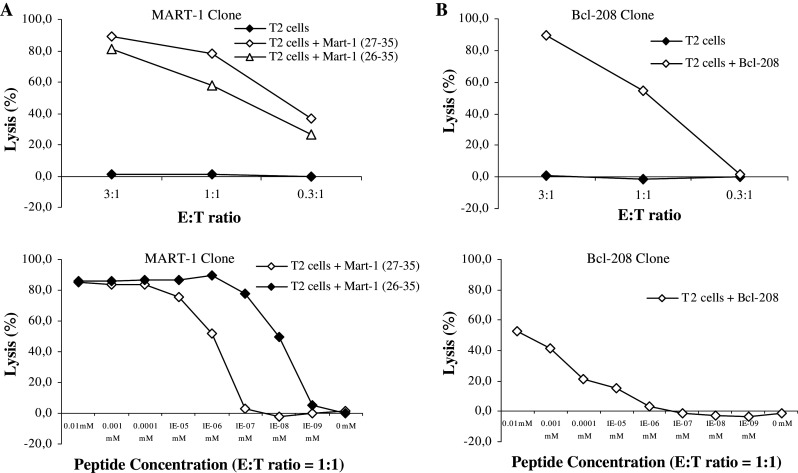
Specificity and cytolytic capacity of antigen specific CTL clones. a The specificity and the cytotoxic potential of the MART-127–35 CTL clones were examined in a 51Cr-release assay measuring cell lysis of T2 cells pulsed with the specific (MART-127–35 and MART-126–35) peptide or an irrelevant peptide (HIV-1 pol476–484) in three E:T ratios. The peptide affinity of the MART-127–35 CTL clone was analyzed further in a 51Cr-release assay measuring cell lysis of T2 cells pulsed with different concentrations of the specific (MART-127–35, and MART-126–35) peptide at the E:T ratio 1:1. b Cytotoxic potential and peptide affinity of the Bcl-2208–217 CTL clone
As a control we compared the killing of the MART-127–35 CTL clone with a Bcl-2208–217 CTL clone (Fig. 1b). The peptide avidity of this clone was lower as compared to the MART-127–35 CTL clone (Fig. 1).
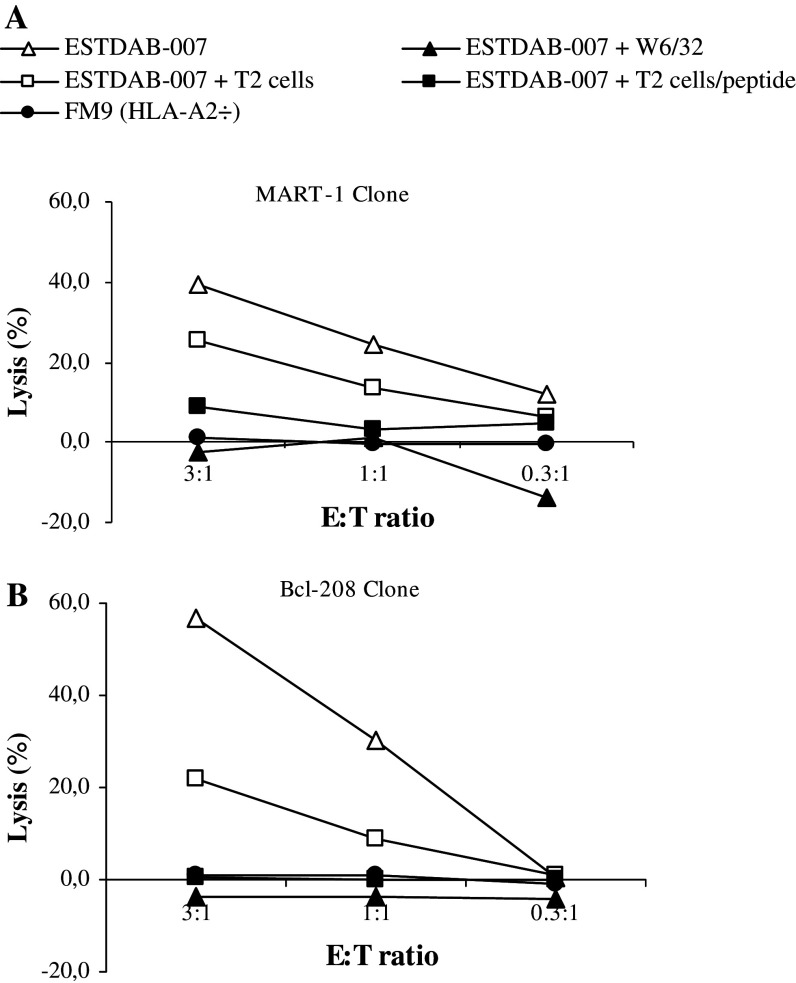
To further examine the antigen specificity and HLA-restriction of the MART-127–35 CTL clone we examined if lysis could be inhibited by the HLA specific mAb W6/32. First, we examined the killing of the ESTDAB-007 melanoma cell line. The CTL clone effectively lysed ESTDAB-007 melanoma cells, and lysis could be completely blocked by incubation of target cells with antibody (Fig. 2a). Likewise, the addition of cold (unlabeled) T2 cells pulsed with the specific MART-127–35 peptide completely abrogated the killing of ESTDAB-007 melanoma cells, whereas the addition of T2 cells without peptide only showed a limited dilution effect (Fig. 2a). As a supplementary control, we used the HLA-A2 negative melanoma cell line FM9 as target cells. No cytotoxicity was observed against this cell line (Fig. 2a). The peptide specificity and HLA-restriction of the control Bcl-2208–217 CTL clone was confirmed likewise (Fig. 2b). Thus, the addition of cold (unlabeled) T2 cells pulsed with the specific Bcl-2208–217 peptide completely abrogated the killing of ESTDAB-007 melanoma cells, whereas the addition of T2 cells without peptide showed a reduced lysis of ESTDAB-007 (Fig. 2b). This dilution effect is occurring because the addition of 20 times as many target cells to the effectors completely changes the actual E:T ratio. This dilution effect is a well-described effect and is almost always seen when performing cold:target inhibition assay (except for very high avidity T-cell clones) [13, 19, 24, 29, 32]. The dilution effect can also be seen for the MART-1-specific clone, although it is a higher avidity clone than the Bcl-2 clone. Hence, what is important in a cold:target assay is the complete inhibition of the lysis using peptide-pulsed T2 cells in contrast to the dilution effect when using cold T2 cells without peptide. In this regard it should be noted that it would have been preferable to perform the cold target assay with irrelevant T2 cells instead of T2 cells without peptide; however, we could not perform this due to the limited amount of T-cell clones.
Fig. 2.
Antigen specific lysis of human melanoma cell lines by CTL clones. Cell lysis of the HLA-A2 positive melanoma cell line ESTDAB-007 without and with the HLA-specific antibody W6/32. In addition, unlabeled T2 cells either with or without the relevant peptide a MART-127–35, b Bcl-2208–217 were added to ESTDAB-007 cells at a ratio of inhibitor to target cells of 20:1. Moreover, cell lysis of the HLA-A2 negative melanoma cell line FM9. All 51Cr-release assays were performed in three E:T ratios
Killing of melanoma cell lines
The MART-127–35 CTL clone was used to test the capacity of MART-127–35-specific CTL to kill melanoma cell lines. Subsequently, the HLA-A2 positive melanoma cell lines FM76, FM92, ESTDAB-001, 006, 014, 017, 019, 021, 026, 027, 028, 029, 033, 100, and 117 were used as target cells (Fig. 3a). Surprisingly, this clone was only capable of killing a limited number (7/15) of these melanoma cell lines. The Bcl-2208–217-specific CTL clone effectively lysed all the melanoma cell lines except FM76 (Fig. 3b). Furthermore, the percentages of lysis observed for the MART-1 CTL clone were lower compared to the lysis for the Bcl-2208–217-specific CTL clone. As expected the MART-127–35-specific CTL clone only killed melanoma cells (Fig. 3c), whereas the Bcl-2208–217-specific CTL clone effectively lysed the HLA-A2 positive breast cancer cell lines MDA-MB-231 and CAMA-1, as well as the HLA-A2 positive colon cancer cell line HCT-116 (Fig. 3c). The HLA-A2 negative breast cancer cell line ZR-75-1 was used as a control of the HLA-A2 restricted killing of the Bcl-2208–217-specific CTL clone.
Fig. 3.
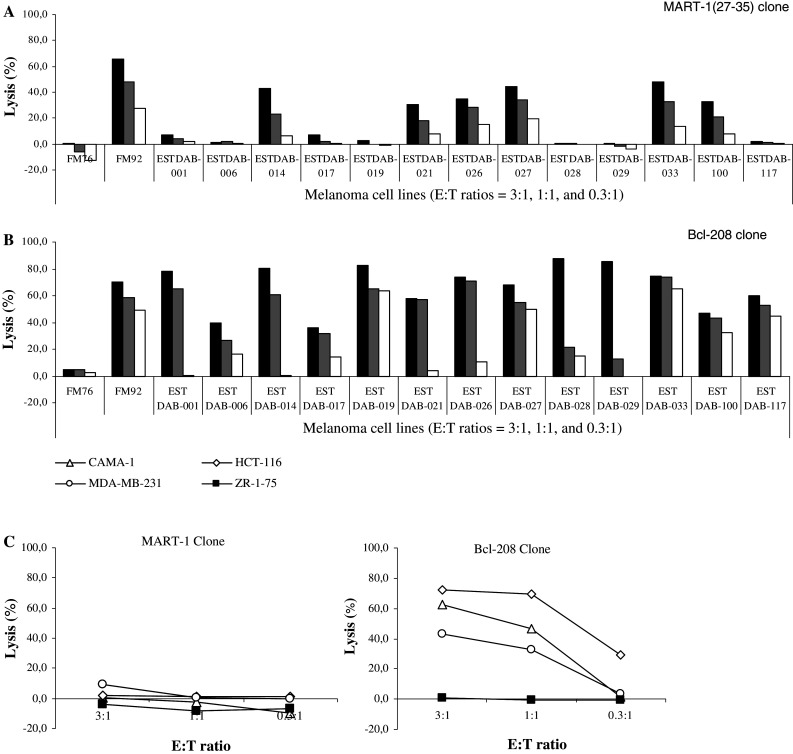
Lysis of human cancer cell lines by CTL clones. Cell lysis of the HLA-A2 positive melanoma cell lines FM76, FM92, ESTDAB-001, 006, 014, 017, 019, 021, 026, 027, 028, 029, 033, 100, and 117 by the a MART-127–35 and the b Bcl-2208–217-specific CTL clones. c Killing of non-melanoma cell lines (i.e., the HLA-A2 positive breast cancer cell lines MDA-MB-132 and CAMA-1, the HLA-A2 positive colon cancer cell line HCT-116, and the HLA-A2 negative breast cancer cell line ZR-75-1) by the MART-127–35 and Bcl-2208–217-specific CTL clones. All 51Cr-release assays were performed in three E:T ratios
NKG2D expression and granzymeB/perforin
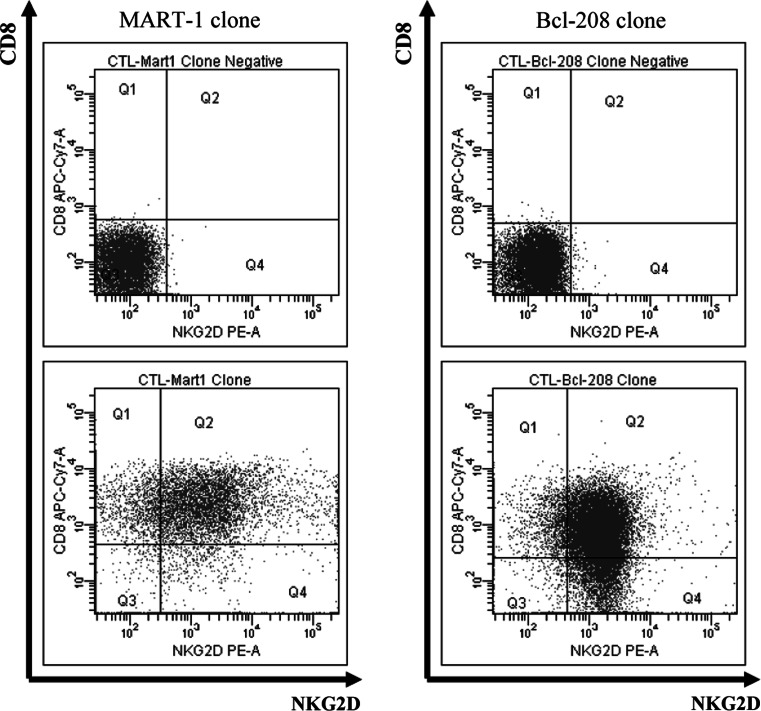
We examined if the difference in killing of melanoma cells (MART-127–35 vs. Bcl-2208–217) were due to the lack of NKG2D expression on the cell surface. Thus, the CTL clones were analyzed by FACS to determine the surface expression of the NKG2D NK-activating cell receptor. However, both CTL clones expressed the NKG2D receptor (Fig. 4). In addition, the ability of the MART-127–35-specific CTL clone to release both granzyme B and perforin was analyzed by ELISPOT using peptide-loaded T2 cells. These CTL clone was capable of releasing both granzymes and perforin (data not shown).
Fig. 4.
NKG2D receptor expression on CTL clones. The FACS diagrams show the CD8 (CD8-APC-Cy7) and NKG2D (NKG2D-PE) surface expression of the MART-127–35 and Bcl-2208–217-specific CTL clones. Unstained cells from the CTL clones served as control
Presentation of T-cell epitopes
Most cancer cell lines used were examined for Bcl-2 (data not shown) and MART-1 (Table 1) expression by RT-PCR. All examined melanoma cell lines expressed Bcl-2 and MART-1 RNA (Table 1). In addition, the cell lines were examined for HLA-A and HLA-A2 expression (Table 1). The expression of HLA-A were obtained from the ESTDAB-database (http://www.ebi.ac.uk/ipd/estdab) whereas the HLA-A2 expression was analyzed by FACS.
Table 1.
Expression of MART-1 and class I HLA in tumor cell lines
| Cell line | MART-1 expressiona | HLA-A expressiona | HLA-A2 expressionb |
|---|---|---|---|
| FM9 | ND | ND | − |
| FM76 | ND | ND | ND |
| FM92 | ND | ND | + |
| ESTDAB-001 | +++ | ++ | ND |
| ESTDAB-006 | +++ | + | + |
| ESTDAB-007 | + | ++ | ++ |
| ESTDAB-014 | +++ | + | ++ |
| ESTDAB-017 | +++ | ND | ND |
| ESTDAB-019 | ++ | ++ | +++ |
| ESTDAB-021 | ++ | + | + |
| ESTDAB-026 | +++ | +++ | + |
| ESTDAB-027 | +++ | ++ | ND |
| ESTDAB-028 | +++ | ND | ++ |
| ESTDAB-029 | ++ | ND | + |
| ESTDAB-033 | +++ | ++ | + |
| ESTDAB-100 | +++ | ++ | ND |
| ESTDAB-117 | +++ | ND | ND |
| CAMA-1 | ND | ND | ND |
| HCT-166 | ND | ND | ND |
| MDA-MB-231 | ND | ND | ND |
| ZR-1-75 | ND | ND | – |
aData obtained from the ESTDAB-database (http://www.ebi.ac.uk/ipd/estdab)
bThe magnitude of HLA-A2 expression was determined using the staining index (SI) defined as MFIpositive − MFIbackground/2 × SDbackground where MFI is mean fluorescence intensity. SDbackground is the degree of spread in the negative population (unstained cells) to which the cells stained with HLA-A2 mAb is to be compared
+ 100 < SI < 2,000
++ 2,000 < SI < 3,000
+++ SI > 3,000
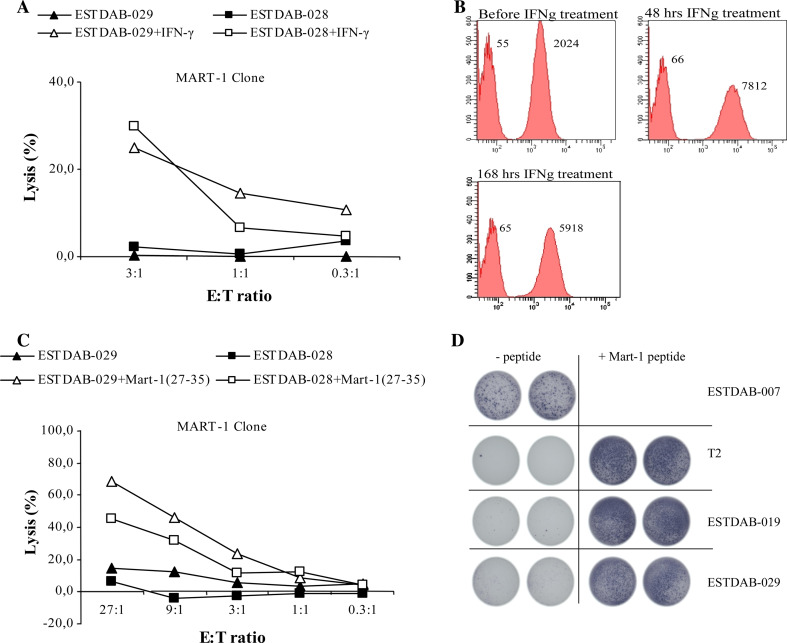
To examine if the resistance to killing by the MART-127–35-specific CTL clone was due to a low or absent presentation of the MART-127–35 epitope on the cell surface by HLA-A2, we treated the resistant melanoma cell lines, ESTDAB-028 and 029, with IFN-γ for 48 h to increase susceptibility to CTL lysis, since IFN-γ treatment upregulates HLA-expression on the cell surface. After IFN-γ treatment the MART-127–35 CTL clones were capable of killing these cells (Fig. 5a). To find the optimal expression of HLA on the melanoma cells we analyzed HLA-A2 expression of ESTDAB-029 cells before IFN-γ treatment, as well as after 48 and 168 h of IFN-γ treatment (Fig. 5b). We observed an increased expression of HLA-A2 after 48 h of IFN-γ treatment compared to before and after 168 h IFN-γ treatment (Fig. 5b).
Fig. 5.
Lysis of human melanoma cell lines treated with IFN-γ, or loaded with peptide on the cell surface by CTL clones. a The melanoma cell lines ESTDAB-028 and 029 were treated with IFN-γ for 48 hours before being used as target cells in the 51Cr-release assay. Effector cells were the MART-127-35-specific CTL clone. The 51Cr-release assays were performed in three E:T ratios. b Histograms show HLA-A2 expression on the surface of the melanoma cell line ESTDAB-029 before, 48 and 168 h of IFN-γ treatment analyzed by FACS. Negative controls are unstained cells (left histograms). Median values are given above the histograms. The magnitude of HLA-A2 expression before or after IFN-γ treatment was determined using the staining index (SI) defined as MFIpositive − MFIbackground/2 × SDbackground where MFI is mean fluorescence intensity. SDbackground is the degree of spread in the negative population (unstained cells) to which the cells stained with HLA-A2 mAb is to be compared. c The melanoma cell lines ESTDAB-028 and 029 were loaded with the MART-127–35 peptide before being used as target cells in c a 51Cr-release assay or d in an ELISPOT assay. The 51Cr-release assays were performed in five E:T ratios, and the ELISPOT were performed in an E:T ratio at 3:1. As shown in the figure was T2 cells and the melanoma cell line ESTDAB-007 included as target cells in the ELISPOT assay
In addition, we loaded the killing resistant melanoma cell lines, ESTDAB-028 and 029, with the MART-127–35 peptide (Fig. 5c), and subsequently used these as target cells in a 51Cr-release assay. Indeed, after peptide loading specific lysis of ESTDAB-028 and 029 by the MART-127–35 CTL clone was observed (Fig. 5c). In addition, lysis of ESTDAB-028 and 029 was carried out with higher E:T ratios, i.e., 9:1 and 27:1. However, at these E:T ratios the melanoma cell line ESTDAB-028 were still not killed by the MART–127–35-specific CTL clone without peptide added, whereas the ESTDAB-029 were only killed weakly (Fig. 5c). Furthermore, we analyzed the killing of the resistant melanoma cell lines, ESTDAB-028 and 029 with and without peptide loading, in the more sensitive IFN-γ ELISPOT assay (Fig. 5d). Indeed, we only observed IFN-γ secretion if the target cells, ESTDAB-028 and 029, were loaded with the MART-127–35 peptide (Fig. 5d). As a control we used MART-127–35 peptide loaded/unloaded T2 cells as well as the melanoma cell line ESTDAB-007 as target cells in the ELISPOT assay. IFN-γ secretion was observed against both unloaded ESTDAB-007 and MART-127–35 loaded T2 cells (Fig. 5d).
Polyclonal MART-127–25-specific CTL
Since observations based only on one single MART-127–35-specific CTL clone is not sufficient to conclude that many melanoma cell lines are not efficiently presenting the MART-127–35 HLA-A2 restricted epitope, we included a polyclonal MART-127–35 culture from another patient. Hence, tumor infiltrating lymphocytes (TIL) from melanoma patients were expanded using high dose IL-2 as described [23] and the MART-1-specific cells were isolated using a HLA-A2/MART-127–35 dextramer (Fig. 6a). The specificity of the dextramer was confirmed by staining of the MART-1 specific T-cell clone (Fig. 6b). The resulting MART-127–35-specific polyclonal cell culture was examined in a 51Cr-release assay. As depicted in Fig. 6c the results using this polyclonal culture are similar to the results using the MART-127–35-specific CTL clone; the melanoma cell lines FM92, ESTDAB-33 as well as autologous tumor cells were lysed by the polyclonal MART–127–35-specific cells, whereas ESTDAB–019, 028, and 029, were not lysed (Fig. 6c). In addition, loading of MART-127–35 peptide on the resistant melanoma cell lines ESTDAB-019 and 029 induced lysis, whereas the percentage of killing of the autologous tumor cells was unchanged upon peptide loading (Fig. 6c).
Fig. 6.
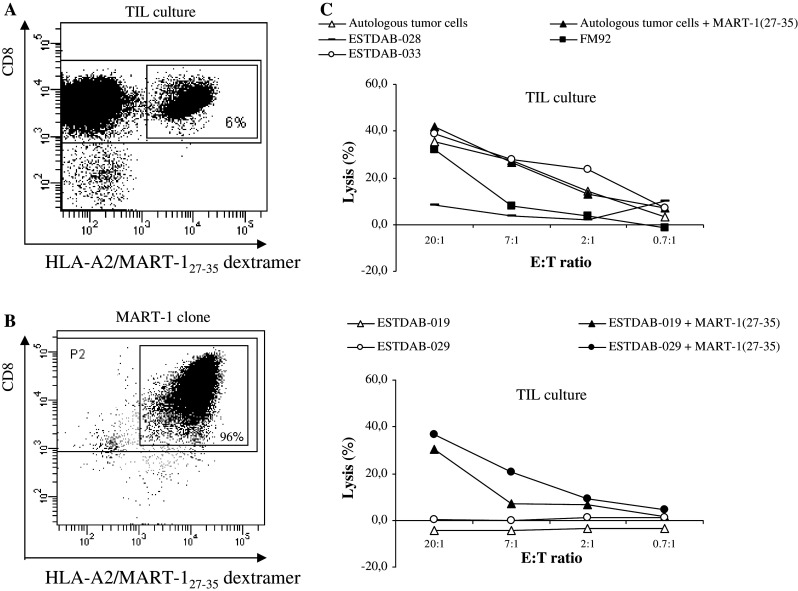
Functionality and cytotoxic potential of polyclonal MART-127–25 specific CTL. a The FACS diagrams show the percentage of CD8/MART-127–35 specific cells in a TIL culture. b The MART-127–35-specific CTL clone stained with the same dextramer as used in a as a control of the HLA-A2/MART-127–35 dextramer. c Lysis of the HLA-A2 positive melanoma cell lines FM92, ESTDAB-028, ESTDAB-033, and of the autologous melanoma cells, ESTDAB-019 and 029 with and without the MART-127–35 peptide
Discussion
A critical parameter for the success or failure of anti-cancer vaccines is the definition of the best-suited antigens. In addition, it is important to make use of the most effective epitopes within these antigens, especially when using small defined epitopes for therapy. Immunotherapy of melanoma patients very often applies the Melan-A decapeptide EAAGIGILTV and/or the nonapeptide AAGIGILTV [7]. This may be due to the fact the MART-1 was one of the first TAA described, and because Melan-A/MART-1-specific CD8 T-cells are readily detectable in the majority of HLA-A2 positive normal individuals and in melanoma patients. The high frequency of specific T-cells allows a closer monitoring of spontaneous and therapy induced T-cell responses. In this regard, it should be mentioned that the interpretation of the results obtained from immunomonitoring of clinical trials is a difficult task due to the variety of methods and protocols available to detect vaccine-specific T-cell responses [5]. In the present study, we generated a high avidity T-cell clone against the MART-127–35 epitope. Like most previously described MART-1-specific T-cell clones this clone recognized both the nonapeptide MART-127–35 as well as the decapeptide MART-126–35. Strikingly, when analyzing the killing of a large panel of melanoma cell lines, we observed that the MART-1-specific T-cell clone only killed around half of the melanoma cells lines. Hence, 7/15 melanoma cell lines were killed by the MART-1-specific T-cell clone compared to the killing of 14/15 of the same melanoma cell lines by a Bcl-2208–217-specific T-cell clone. The killing of the melanoma cells in a HLA-restricted manner by the Bcl-2 clone showed that the analyzed melanoma cells were not resistance to killing by T-cells, and that that the melanoma cells expressed HLA-A2 on the surface. Only one melanoma cell line (FM76) was not recognized by the Bcl-2-specific T-cell clone. This was indeed due to the lack of HLA-A2 on the surface. IFN-γ treatment increased HLA-A2 expression on the surface of this cell line, which made the cell line vulnerable for T-cell killing. In addition, the Bcl-2-specific CTL clone efficiently killed cancer cells of different origin, whereas the MART-1-specific clone naturally killed only melanoma cells.
We examined if the better recognition of melanoma cell lines by the Bcl-2-specific T-cell clone were due to a higher T-cell avidity of this clone compared to the MART-1-specific T-cell clone. T-cell avidity can be defined as the amount of peptide needed for the activation of the CTL. At an effector:target ratio at 1:1, it was possible to detect target lysis as low as at peptide concentrations 10−8 mM for the MART-1-specific T-cell clone. In contrast, the Bcl-2208–217-specific T-cell clone only killed target cells at this E:T ratio at a peptide concentration of 10−5 mM. Thus, the avidity of this clone was considerably lower compared to the MART-1 clone.
It has been described how NKG2D expression is up-regulated upon activation and expansion of human CD8 T-cells. Multiple studies support the concept that NKG2D ligation results in different functional outcomes on T-cells. NKG2D signaling does augment cytotoxic and proliferative responses of T-cells on antigen encounter, thus qualifying NKG2D as a T-cell co-stimulatory molecule. Using CMV-specific CTL, Groh et al. [14] demonstrated that on TCR triggering, NKG2D stimulation enhances cytokine production, T-cell proliferation, and cytolysis. Notably, NKG2D signaling augmented cytotoxicity only when TCR triggering occurred. However, the MART-1-specific T-cell clone as well as the Bcl-2-specific T-cell clone expressed NGK2D. Hence, the difference in the killing of melanoma cells was not due to differences in the expression of this co-stimulatory molecule. In addition, we by ELISPOT concluded that the MART-1 specific T-cell clone was fully capable of releasing both perforin as well as granzyme B in response to the MART-127–35 epitope.
Next, we examined if the deficient killing were due to lack of the MART-1 peptide on the cell surface of the melanoma cells. After adding MART-1 peptide to the resistant melanoma cells, these were killed by the MART-1-specific T-cell clone suggesting that the lack of killing were due to absence or limitation of the HLA-peptide complex on the cell surface. We, in addition, examined the expression of HLA-A2/MART-127–35 on the surface of melanoma cells using the more sensitive IFN-γ ELISPOT. The ELISPOT verified that the melanoma cell lines ESTDAB-019 and ESTDAB-029 did not activate MART-1-specific T-cells unless pulsed with the peptide epitope.
Previously, it was shown that CTL epitopes from Melan/MART-1 are not processed efficiently by the immunoproteasome, but only by the standard proteasome [22]. Yet, we examined the effect of treating the melanoma cells with IFN-γ, which is known among others to induce the immunoproteasome and to upregulate HLA class I on the cell surface. The IFN-γ treatment resulted in increased recognition of the melanoma cell lines by the MART-1 specific CTL clone, which is in conflict with previous data that the immunoproteasome in contrast to the standard proteasome are able to process and present the MART-127–35 epitope [22]. It should be noted, however, that IFN-γ has several positive effects on recognition of the target cells including increased HLA-expression [18]. However, the melanoma cells could be killed by the Bcl-2-specific T-cell clone without IFN-γ treatment, and, in addition, the melanoma cells had measurable HLA-A2 surface expression without IFN-γ treatment.
As a final control, using a polyclonal, MART-1-specific culture isolated from TIL from a melanoma patient we obtained similar results as with the MART-1 specific T-cell clone.
The anti-apoptotic protein Bcl-2 is expressed at high levels in melanoma [16]. However, a rather paradoxical role of the anti-apoptosis protein Bcl-2 have been described, showing an association between high Bcl-2 expression and improved survival in melanoma [8]. The Bcl-2 epitope was identified on the basis of spontaneous immune responses in cancer patients, but not in healthy individuals [3]. The loss of the Bcl-2 expression during progression from primary to metastatic melanoma in patients suggests an active immune selection of the respective melanoma clones by the tumor-bearing host (e.g., via a specific immune response). The frequent expression of the Bcl-2 epitope in melanoma cell presented in this study support this notion. The MART-1 epitope was originally identified as a target from in vitro expanded TIL cultures [30, 31]. The first method might give a more accurate picture of which peptide epitopes actual are presented on the cell surface of cancer cells. Most melanoma patients also harbor spontaneous immune responses to MART-1, however, so do most healthy individuals.
In conclusion, the Bcl-2 clone killed melanoma cells must more efficiently than MART-1-specific T-cell clones despite the lower avidity of the former. The main explanation for this was related to the target cells. Thus, the very frequently used HLA-A2-restricted MART-1 nona- and decapeptide epitopes were not expressed on the surface of some of the melanoma cells. Our data emphasize that the selected tumor antigens and/or epitopes are critical for the outcome of anti-cancer immunotherapy.
Acknowledgments
We would like to thank Merete Jonassen for excellent technical assistance. We further extend our thanks to all the patients who donated blood to perform these studies. This study was supported by grants from the Danish Medical Research Council, The Novo Nordisk Foundation, The Danish Cancer Society, The John and Birthe Meyer Foundation and Herlev Hospital. This article was supported by grants from the Danish Medical Research Council, The Novo Nordisk Foundation, The Danish Cancer Society, The John and Birthe Meyer Foundation, Danish Cancer Research Foundation, ESTAB.
References
- 1.Andersen MH, Bonfill JE, Neisig A, Arsequell G, Sondergaard I, Valencia G, Neefjes J, Zeuthen J, Elliott T, Haurum JS. Phosphorylated peptides can be transported by TAP molecules, presented by Class I MHC molecules, and recognized by phosphopeptide-specific CTL. J Immunol. 1999;163:3812–3818. [PubMed] [Google Scholar]
- 2.Andersen MH, Pedersen LO, Becker JC, Thor Straten P. Identification of a cytotoxic T lymphocyte response to the apoptose inhibitor protein survivin in cancer patients. Cancer Res. 2001;61:869–872. [PubMed] [Google Scholar]
- 3.Andersen MH, Svane IM, Kvistborg P, Nielsen OJ, Balslev E, Reker S, Becker JC, Thor SP. Immunogenicity of Bcl-2 in cancer patients. Blood. 2005;15:728–734. doi: 10.1182/blood-2004-07-2548. [DOI] [PubMed] [Google Scholar]
- 4.Boon T, Coulie PG, Van den Eynde BJ, van der BP. Human T cell responses against melanoma. Annu Rev Immunol. 2006;24:175–208. doi: 10.1146/annurev.immunol.24.021605.090733. [DOI] [PubMed] [Google Scholar]
- 5.Britten CM, Gouttefangeas C, Welters MJ, Pawelec G, Koch S, Ottensmeier C, Mander A, Walter S, Paschen A, Muller-Berghaus J, Haas I, Mackensen A, Kollgaard T, Thor SP, Schmitt M, Giannopoulos K, Maier R, Veelken H, Bertinetti C, Konur A, Huber C, Stevanovic S, Wolfel T, van der Burg SH. The CIMT-monitoring panel: a two-step approach to harmonize the enumeration of antigen-specific CD8(+) T lymphocytes by structural and functional assays. Cancer Immunol Immunother. 2008;57:289–302. doi: 10.1007/s00262-007-0378-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chattopadhyay U. Tumour immunotherapy: developments and strategies. Immunol Today. 1999;20:480–482. doi: 10.1016/S0167-5699(99)01526-1. [DOI] [PubMed] [Google Scholar]
- 7.Dalgleish AG, Whelan MA. Cancer vaccines as a therapeutic modality: the long trek. Cancer Immunol Immunother. 2006;55:1025–1032. doi: 10.1007/s00262-006-0128-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Divito KA, Berger AJ, Camp RL, Dolled-Filhart M, Rimm DL, Kluger HM. Automated quantitative analysis of tissue microarrays reveals an association between high Bcl-2 expression and improved outcome in melanoma. Cancer Res. 2004;64:8773–8777. doi: 10.1158/0008-5472.CAN-04-1387. [DOI] [PubMed] [Google Scholar]
- 9.Dunbar PR, Chen JL, Chao D, Rust N, Teisserenc H, Ogg GS, Romero P, Weynants P, Cerundolo V. Cutting edge: rapid cloning of tumor-specific CTL suitable for adoptive immunotherapy of melanoma. J Immunol. 1999;162:6959–6962. [PubMed] [Google Scholar]
- 10.Dunbar PR, Smith CL, Chao D, Salio M, Shepherd D, Mirza F, Lipp M, Lanzavecchia A, Sallusto F, Evans A, Russell-Jones R, Harris AL, Cerundolo V. A shift in the phenotype of melan-A-specific CTL identifies melanoma patients with an active tumor-specific immune response. J Immunol. 2000;165:6644–6652. doi: 10.4049/jimmunol.165.11.6644. [DOI] [PubMed] [Google Scholar]
- 11.Gouttefangeas C, Stenzl A, Stevanovic S, Rammensee HG. Immunotherapy of renal cell carcinoma. Cancer Immunol Immunother. 2007;56:117–128. doi: 10.1007/s00262-006-0172-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Griffioen M, Borghi M, Schrier PI, Osanto S, Schadendorf D. Analysis of T-cell responses in metastatic melanoma patients vaccinated with dendritic cells pulsed with tumor lysates. Cancer Immunol Immunother. 2004;53:715–722. doi: 10.1007/s00262-004-0514-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Griffiths SD, Cawley JC. The beneficial effects of alpha-interferon in hairy cell leukemia are not attributable to NK cell-mediated cytotoxicity. Leukemia. 1987;1:372–376. [PubMed] [Google Scholar]
- 14.Groh V, Rhinehart R, Randolph-Habecker J, Topp MS, Riddell SR, Spies T. Costimulation of CD8alphabeta T cells by NKG2D via engagement by MIC induced on virus-infected cells. Nat Immunol. 2001;2:255–260. doi: 10.1038/85321. [DOI] [PubMed] [Google Scholar]
- 15.Guichard G, Zerbib A, Le Gal FA, Hoebeke J, Connan F, Choppin J, Briand JP, Guillet JG. Melanoma peptide MART-1(27–35) J Med Chem. 2000;43:3803–3808. doi: 10.1021/jm000909s. [DOI] [PubMed] [Google Scholar]
- 16.Hussein MR, Haemel AK, Wood GS. Apoptosis and melanoma: molecular mechanisms. J Pathol. 2003;199:275–288. doi: 10.1002/path.1300. [DOI] [PubMed] [Google Scholar]
- 17.Jager E, Hohn H, Necker A, Forster R, Karbach J, Freitag K, Neukirch C, Castelli C, Salter RD, Knuth A, Maeurer MJ. Peptide-specific CD8+ T-cell evolution in vivo: response to peptide vaccination with Melan-A/MART–1. Int J Cancer. 2002;20:376–388. doi: 10.1002/ijc.10165. [DOI] [PubMed] [Google Scholar]
- 18.Kaplan BL, Norell H, Callender GG, Ohlum T, Kiessling R, Nishimura MI. Interferon-gamma renders tumors that express low levels of Her-2/neu sensitive to cytotoxic T cells. Cancer Immunol Immunother. 2006;55:653–662. doi: 10.1007/s00262-005-0050-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kiessling R, Klein E, Wigzell H. “Natural” killer cells in the mouse. I. Cytotoxic cells with specificity for mouse Moloney leukemia cells. Specificity and distribution according to genotype. Eur J Immunol. 1975;5:112–117. doi: 10.1002/eji.1830050208. [DOI] [PubMed] [Google Scholar]
- 20.Kirkin AF, Reichert Petersen T, Olsen AC, Li L, Thor Straten P, Zeuthen J. Generation of human-melanoma specific T lymphocyte clones defining novel cytolytic targets with panels of newly established melanoma cell lines. Cancer Immunol Immunother. 1995;41:71–81. doi: 10.1007/BF01527402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maecker HT, Frey T, Nomura LE, Trotter J. Selecting fluorochrome conjugates for maximum sensitivity. Cytometry A. 2004;62:169–173. doi: 10.1002/cyto.a.20092. [DOI] [PubMed] [Google Scholar]
- 22.Morel S, Levy F, Burlet-Schiltz O, Brasseur F, Probst-Kepper M, Peitrequin AL, Monsarrat B, Van Velthoven R, Cerottini JC, Boon T, Gairin JE, Van den Eynde BJ. Processing of some antigens by the standard proteasome but not by the immunoproteasome results in poor presentation by dendritic cells. Immunity. 2000;12:107–117. doi: 10.1016/S1074-7613(00)80163-6. [DOI] [PubMed] [Google Scholar]
- 23.Morgan RA, Dudley ME, Wunderlich JR, Hughes MS, Yang JC, Sherry RM, Royal RE, Topalian SL, Kammula US, Restifo NP, Zheng Z, Nahvi A, de Vries CR, Rogers-Freezer LJ, Mavroukakis SA, Rosenberg SA. Cancer regression in patients after transfer of genetically engineered lymphocytes. Science. 2006;314:126–129. doi: 10.1126/science.1129003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parkinson DR, Brightman RP, Waksal SD. Altered natural killer cell biology in C57BL/6 mice after leukemogenic split-dose irradiation. J Immunol. 1981;126:1460–1464. [PubMed] [Google Scholar]
- 25.Pawelec G, Marsh SG. ESTDAB: a collection of immunologically characterised melanoma cell lines and searchable databank. Cancer Immunol Immunother. 2006;55:623–627. doi: 10.1007/s00262-005-0117-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pittet MJ, Valmori D, Dunbar PR, Speiser DE, Lienard D, Lejeune F, Fleischhauer K, Cerundolo V, Cerottini JC, Romero P. High frequencies of naive Melan-A/MART–1-specific CD8(+) T cells in a large proportion of human histocompatibility leukocyte antigen (HLA)-A2 individuals. J Exp Med. 1999;190:705–715. doi: 10.1084/jem.190.5.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Romero P, Gervois N, Schneider J, Escobar P, Valmori D, Pannetier C, Steinle A, Wolfel T, Lienard D, Brichard V, Van Pel A, Jotereau F, Cerottini JC. Cytolytic T lymphocyte recognition of the immunodominant HLA-A*0201-restricted Melan-A/MART-1 antigenic peptide in melanoma. J Immunol. 1997;159:2366–2374. [PubMed] [Google Scholar]
- 28.Schultz ES, Chapiro J, Lurquin C, Claverol S, Burlet-Schiltz O, Warnier G, Russo V, Morel S, Levy F, Boon T, Van den Eynde BJ, BP van der. The production of a new MAGE-3 peptide presented to cytolytic T lymphocytes by HLA-B40 requires the immunoproteasome. J Exp Med. 2002;195:391–399. doi: 10.1084/jem.20011974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sorensen RB, Hadrup SR, Kollgaard T, Svane IM, Thor SP, Andersen MH. Efficient tumor cell lysis mediated by a Bcl-X(L) specific T cell clone isolated from a breast cancer patient. Cancer Immunol Immunother. 2007;56:527–533. doi: 10.1007/s00262-006-0200-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spagnoli GC, Schaefer C, Willimann TE, Kocher T, Amoroso A, Juretic A, Zuber M, Luscher U, Harder F, Heberer M. Peptide-specific CTL in tumor infiltrating lymphocytes from metastatic melanomas expressing MART-1/Melan-A, gp100 and tyrosinase genes: a study in an unselected group of HLA-A2.1-positive patients. Int J Cancer. 1995;20:309–315. doi: 10.1002/ijc.2910640505. [DOI] [PubMed] [Google Scholar]
- 31.Stevens EJ, Jacknin L, Robbins PF, Kawakami Y, el Gamil M, Rosenberg SA, Yannelli JR. Generation of tumor-specific CTLs from melanoma patients by using peripheral blood stimulated with allogeneic melanoma tumor cell lines. Fine specificity and MART-1 melanoma antigen recognition. J Immunol. 1995;154:762–771. [PubMed] [Google Scholar]
- 32.Teng JM, Hogan KT. Both major and minor peptide-binding pockets in HLA-A2 influence the presentation of influenza virus matrix peptide to cytotoxic T lymphocytes. Mol Immunol. 1994;31:459–470. doi: 10.1016/0161-5890(94)90065-5. [DOI] [PubMed] [Google Scholar]