Abstract
Purpose
Recombinant interleukin-21 (rIL-21) is an immune stimulating cytokine recently tested in two Phase 1 trials for immune responsive cancers. A secondary objective of these trials was to characterize pharmacodynamic responses to rIL-21 in patients. Here, we report the effects of systemic rIL-21 on serum markers of immune stimulation.
Experimental design
Recombinant IL-21 was administered by intravenous bolus injection at dose levels from 1 to 100 μg/kg using two distinct treatment regimens: thrice weekly (‘3/w’) for 6 weeks; or once daily for five consecutive days followed by nine dose-free days (‘5 + 9’). In the absence of dose limiting toxicity, additional cycles of dosing were initiated immediately following the nine dose-free days. An array of 70 different proteins was profiled in subject serum samples from several time points during the course of the study. Hierarchical clustering analysis was performed on a normalized subset of these data.
Results
Systemic administration of rIL-21 affected the serum levels of several cytokines, chemokines, acute-phase proteins and cell adhesion proteins. The magnitude and duration of response were dose dependent for a subset of these biomarkers. The 5 + 9 dosing regimen generally produced cyclic changes that were of greater magnitude, as compared to a more chronic stimulation with the 3/w dosing regimen. Despite these differences, rIL-21 effects on many analytes were similar between regimens when averaged over the time of treatment. Based on similar temporal, between-subject and dose response changes, groups of analytes were identified that exhibited distinct components of the rIL-21-mediated immune activation. Biomarkers indicative of lymphocyte activation (increased IL-16, decreased RANTES), acute phase response (increased CRP, ferritin), myeloid activation (increased MDC, MIP-1 alpha), and leukocyte chemotaxis/trafficking (increased sCAMs, MCP-1) were strongly modulated in subjects treated with rIL-21.
Conclusions
Administration of rIL-21 resulted in activation of multiple cell types and immune response pathways. The changes observed in serum proteins were consistent with coincident processes of lymphoid and myeloid cell activation and trafficking, and acute phase response.
Electronic supplementary material
The online version of this article (doi:10.1007/s00262-008-0600-8) contains supplementary material, which is available to authorized users.
Keywords: Immune activation, IL-21, Cytokines, Immunotherapy
Introduction
Interleukin-21 (IL-21) is a class I cytokine produced by activated CD4+ T cells and NKT cells that affects both innate and adaptive immunity. Recombinant IL-21 (rIL-21) enhances proliferation and effector function of multiple immune cell types, activities consistent with the role of this cytokine in immune system activation. Potent effects of rIL-21 have been observed on the growth and functional activity of T, B, NK and NKT cells, particularly when combined with other cytokines or activating stimuli [4, 29]. IL-21 promotes proliferation and differentiation of T cells, particularly CD8 T cells where it stimulates cytokine and antigen-dependent proliferation and development of cytolytic effector function [12, 30, 44]. It also promotes differentiation of NK cells into more effective killer cells [26, 32, 33]. In vitro, IL-21 stimulation leads to enhanced generation of T cells with a memory phenotype [17]. CD4+ T cells stimulated by IL-21 demonstrate greater resistance to regulatory T cell suppression [18, 31]. Furthermore, recent publications suggest that unlike IL-2, IL-21 does not induce proliferation of regulatory T cells [31]. IL-21 has recently been shown to be required for differentiation of Th17 cells, and to act in an autocrine fashion to support function and survival of these cells [15, 45]. Additionally, IL-21 has potent effects on B cell proliferation and antibody production. It enhances B cell proliferation induced by T cell dependent signals and inhibits B cell proliferation induced by T cell independent signals and promotes isotype switching and differentiation of B cells into plasma cells, leading to increased immunoglobulin production [8, 27, 29].
IL-21 has been reported to have anti-tumor effects in various preclinical cancer models through mechanisms that require NK cells and/or CD8+ T cells [6, 23, 34, 38, 39, 42]. Based on the biologic effects demonstrated in preclinical models, rIL-21 is being developed by ZymoGenetics, Inc. and Novo Nordisk A/S as a potential immunotherapeutic agent for cancer indications. Recently, two Phase 1 dose escalation trials have been conducted to explore the safety, pharmacokinetics, and pharmacodynamics of rIL-21 administered intravenously to patients with stage IV metastatic melanoma (MM) or renal cell carcinoma (RCC) [5, 37]. These trials demonstrated a favorable safety profile and signs of anti-tumor activity. In conjunction with these clinical trials, we investigated the pharmacodynamic responses to rIL-21 administration by analysis of changes in serum proteins of study subjects treated with rIL-21. Using multi-analyte profiling, a wide examination of protein expression patterns was evaluated for response to drug administration. The objectives of this study were to broadly classify patterns of change with respect to immune response markers in serum and to identify markers that may be useful in subsequent trials for refining a rIL-21 exposure response model.
Materials and methods
Trial design and patient population
Seventy-two patients were evaluated in two Phase 1 dose escalation studies conducted in Australia and the United States. Both trials were open-label dose escalation studies in which rIL-21 was administered by intravenous bolus injection. In the US Phase 1 study, subjects with MM or RCC received two cycles of the 5 + 9 regimen, where a cycle is defined as 5 days of dosing followed by 9 non-dosing days. This study was conducted in two parts. The maximum tolerated dose (MTD) of rIL-21 was determined in Part A in which subjects were dosed with 3, 10, 30, 50, or 100 μg/kg rIL-21. Upon establishing the MTD, 28 additional subjects were dosed in Part B at the MTD. In the Australian study, subjects with MM were allocated non-randomly in cohorts of two patients into one of two parallel treatment arms: rIL-21 was administered IV thrice weekly (Monday, Wednesday, Friday) for 6 weeks (3/wk); or in three cycles of the 5 + 9 regimen. The primary objective was to assess the safety and tolerability of rIL-21. Among several secondary objectives were assessment of anti-tumor effects and characterization of pharmacodynamic responses to rIL-21. All patients provided written informed consent prior to any study-specific procedures. Additional details on trial design, disease assessment, as well as the primary clinical findings of both studies have been published previously [5, 37]. For both the treatment schedules tested in the Phase 1 trials, the MTD was estimated to be 30 μg/kg.
Blood sample collection and handling
For the US Phase 1 trial during Cycle 1 of treatment, samples were taken at enrollment and prior to dosing on Days 1, 5, and 8. During Cycle 2 of treatment, samples were drawn on Days 15, 19 and 22. These samples were taken consistently, and are included in the statistical analysis. Additionally, banked sera from patients were sent for ad hoc analysis as required from samples drawn on Days 2, 4, 10, and 4 h after dosing on Day 1. Similarly, banked samples collected during Cycle 2 of treatment Days 16, 18, 29, were sent for ad hoc analysis. These ad hoc samples were not included in the clustering analysis. For the Australian Phase 1 trial, samples were taken at enrollment and prior to dosing on Days 1, 2, 5, 15, 19, 26, 29, 36, 38, 43, 61 and 4 and 8 h after dosing on Days 1 and 38. Only samples collected on Days 1, 5, 15 and 19 were in alignment with the US Phase 1, so these samples were included in the clustering analysis. All blood draws were obtained using standard phlebotomy procedures. Peripheral blood with no anti-coagulant was collected into 7-mL red top Vacutainer™ collection tubes. Blood was allowed to clot for 30 to 60 min at ambient temperature (15–25°C) then centrifuged at 2,000g for 15 min at ambient temperature. Resulting serum was aliquoted and immediately frozen at −70 C. Serum samples then were shipped on dry ice for analysis.
Quantification of serum analytes
Serum proteins were analyzed by Rules-Based Medicine, Inc (RBM, Austin, TX) using multi-analyte profiling (MAP). These Luminex-based measurement panels are capable of providing accurate and precise measurement of hundreds of plasma proteins. All samples submitted to RBM were analyzed by the “Human Antigens Only MAP panel”, v1.5 or v1.6. Because v1.6 was used for only a small number of samples, the additional serum analytes quantified with this panel are not reported here.
To quantify each analyte in test samples, an eight-point standard curve was generated using calibrators of known concentration. The concentration of the analyte in the test sample was extrapolated from the curve using an algorithm that generated a best curve fit. The lowest detectable dose (LDD) is the lower limit at which the system can accurately calculate the concentration of an experimental sample and be certain that the concentration is higher than a blank sample. This value was provided by RBM for each of the measured analytes. Results were reported in Microsoft Excel™ format from RBM.
Analytic methods
Hierarchical clustering was used to investigate grouping in these data by arranging the data into a binary tree, representing the likeness of two or more endpoints from the MAP data set. This tree is not a univariate set of relationships, such as that generated by pairwise correlation methods, but rather a multilevel hierarchy, where groups of similar biomarkers are aggregated. MATLAB Version 7.0.1 (R14) Service Pack 1 with the Statistics Toolbox Version 5.0.1 (R14) Service Pack 1 (Mathworks, Inc., Natick, MA) was used to perform the statistical analysis described below.
The general procedure involved four steps. (1) Data were normalized by log-transformation and application of a z-score algorithm to produce a standard normal deviate dataset. This step also eliminated those analytes with little variation or with a large number of missing values. (2) Similarity or dissimilarity between every pair of analytes was determined to define the distance measure. (3) Groups of similar biomarkers were arranged in a binary, hierarchical cluster tree which defined cluster linkage. (4) A dendrogram was drawn to visually represent correlation between biomarkers and relationships between biomarker clusters. Additional details on these methods are available in the supplementary materials.
Data handling
A number of analytes were removed in the process of data normalization. If a value was reported as “Missing” or was unavailable, the values were set to zero. Analytes having a coefficient of variation (%CV) less than or equal to 1% were excluded from the analysis. The most common cause of lack of variance in the data was that the majority of measurements were below the LDD. Analytes with more than 50% of the measurements at the mode (“Missing” or at the lower limit of quantitation) were excluded from the clustering analysis. The primary cause for this exclusion was that measurements were available on only a subset of the data (e.g., MAP v1.6 analytes). The following v1.5 analytes were excluded following normalization: calcitonin, cancer antigen 125, carcinoembryonic antigen, endothelin-1, erythropoietin, fatty acid binding protein, FGF basic, fibrinogen, GM-CSF, IL-10, IL-12p40, IL-12p70, IL-13, IL-1alpha, IL-1beta, IL-2, IL-3, IL-4, IL-5, IL-6, IL-7, lymphotactin, MMP-2, MMP-9, thrombopoietin, tissue factor, and TNF-beta.
Data normalization, cluster distance, linkage and visualization
Only a subset of the total reported MAP data set was included in the analysis reported herein. Sample values less than the LDD were assigned the LDD for the purpose of further analysis. The data were log-transformed to remove the positive skewness expected and apparent in the data. A z-score algorithm was applied to each analyte, which returns the deviation of each data element from its mean, normalized by its standard deviation. Two vectors of z-score transformed analytes were evaluated for similarities by computing the sample correlation between points when treated as a sequence of values. That is, analytes that correlated well over time, dose, subject and other independent parameters were said to have a small pairwise distance between them. Similar analytes were organized into a binary hierarchical cluster tree using an average linkage algorithm based on their distances. This linkage definition can be interpreted as the unweighted average Pearson correlation coefficient between the members of linked clusters, just as the initial distance definition was the pairwise Pearson correlation coefficient. The hierarchical, binary cluster tree defined by the distance measures were most easily understood when viewed graphically. Distances were visualized using a dendrogram, where the analytes were arranged on the y-axis and the distance between those proteins and clusters of proteins were represented on the x-axis. Large jumps in distances between members can be thought of as natural divisions in the data. Additional details on these methods are available in the supplementary materials.
Results
Dose level comparisons
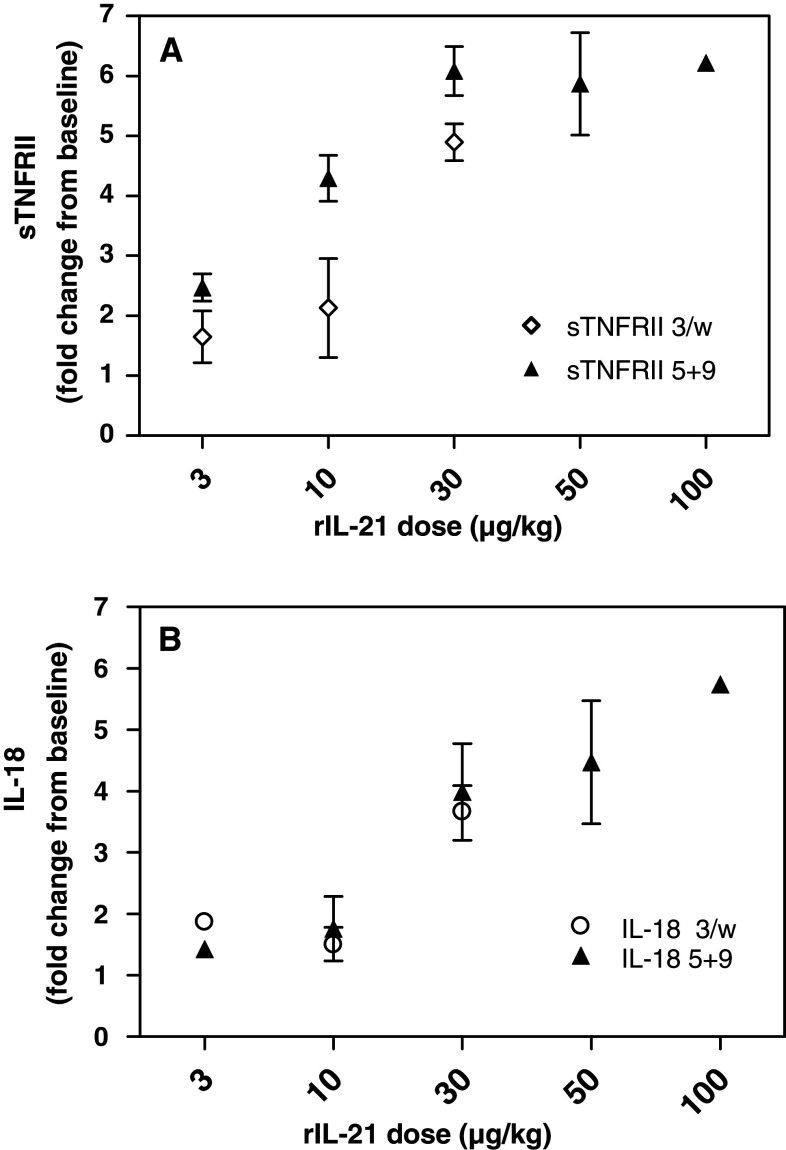
To identify specific changes in serum protein concentrations following treatment with rIL-21, serum samples from subjects treated in two Phase 1 dose escalation trials were analyzed using a multianalyte panel comprising 70 different proteins. Following rIL-21 treatment, changes were detected in over half of these serum proteins, including cytokines, chemokines, acute phase proteins, cell adhesion proteins and other serum proteins involved in metabolism and coagulation. Over the dose range tested in these trials, the majority of analytes that changed significantly with rIL-21 dosing did so in a dose-dependent fashion. The most common indicator of the dose response relationship was a threshold observed for rIL-21 effects on these analytes. The exact dose response relationship for each marker is beyond the scope of this analysis, as limited data were collected at many dose levels. However, for the majority of subjects treated with dose levels ≥10 μg/kg, the magnitude and duration of responses were clearly distinct from baseline or lower dose levels. This threshold effect is exemplified by changes in serum concentration of sTNFRII and IL-18 as a function of rIL-21 dose level shown in Fig. 1. Because robust and consistent responses were observed in subjects treated at the MTD of 30 μg/kg rIL-21 in both the trials, and because limited data were collected at other dose levels, the subsequent analyses are focused on responses at the 30 μg/kg dose level.
Fig. 1.
Changes in serum concentrations of sTNFRII (a), and IL-18 (b). Mean change from baseline (±SD) on Day 19 of treatment is shown for each rIL-21 treatment cohort
Regimen comparisons
Differences between the two distinct dosing regimens were evaluated: thrice weekly (3/w); or once daily for five consecutive days followed by nine dose-free days (5 + 9). Subjects treated with 30 μg/kg rIL-21 by either regimen showed changes in a similar subset of the serum analytes, and differences were observed primarily in the timing and magnitude of changes. Serum levels of these protein markers are shown in Table 1 for subjects treated with the 5 + 9 dosing regimen. For the majority of markers, maximal changes were observed during the 5 day rIL-21 treatment period, and these returned approximately to baseline levels during the 9 day dose-free period. Similar responses were observed in subsequent treatment cycles. Thus, the cyclic exposure with this dose regimen resulted in discrete pharmacodynamic effects. However, for several of the markers, resolution of effect was not complete within the dose-free period, and persistence or carryover into the subsequent dosing period was observed (i.e., Day 15 baseline was dissimilar to Day 1 baseline). In some cases, this carryover effect led to cumulative changes over time. Serum levels of protein markers for the 3/w dosing regimen with 30 μg/kg rIL-21 are shown in Table 2. In subjects treated with this dose regimen, maximum rIL-21-induced changes were generally lower when compared to the 5 + 9 regimen, but they were maintained at most time points, reflecting the continuous dosing and a more persistent pharmacodynamic effect.
Table 1.
Mean MAP analyte concentrations in the serum of patients treated with 30 μg/kg rIL-21 using the 5 + 9 dosing regimen (n = 12)
| Analyte (units) | Average serum concentration (SD) | |||||
|---|---|---|---|---|---|---|
| Day 1 | Day 5 | Day 8 | Day 15 | Day 19 | Day 22 | |
| Adiponectin (μg/mL) | 3.75 (2.03) | 3.06 (1.67) | 2.63 (1.20) | 3.74 (2.00) | 2.96 (1.46) | 2.49 (1.08) |
| Alpha-1 antitrypsin (mg/mL) | 1.83 (0.375) | 2.97 (0.478) | 3.40 (0.657) | 2.41 (0.579) | 3.27 (0.669) | 3.29 (0.641) |
| Alpha-2 macroglobulin (mg/mL) | 0.728 (0.334) | 0.620 (0.107) | 0.689 (0.372) | 0.600 (0.264) | 0.480 (0.307) | 0.609 (0.341) |
| Alpha-fetoprotein (ng/mL) | 2.50 (1.95) | 2.25 (1.17) | 2.62 (1.67) | 2.48 (2.15) | 1.70 (0.872) | 2.28 (1.47) |
| Apolipoprotein A1 (mg/mL) | 0.342 (0.099) | 0.176 (0.089) | 0.134 (0.062) | 0.320 (0.069) | 0.163 (0.080) | 0.138 (0.051) |
| Apolipoprotein CIII (μg/mL) | 98.4 (47.6) | 76.6 (105) | 74.1 (43.0) | 122 (52.9) | 54.4 (28.8) | 76.3 (21.6) |
| Apolipoprotein H (μg/mL) | 266 (79.3) | 224 (58.0) | 241 (79.1) | 264 (68.1) | 221 (63.0) | 237 (71.8) |
| Beta-2 microglobulin (μg/mL) | 2.48 (0.751) | 4.51 (1.27) | 4.62 (1.37) | 3.14 (0.861) | 5.20 (1.41) | 4.94 (1.17) |
| Brain-derived neurotrophic factor (ng/mL) | 15.6 (3.96) | 7.49 (2.85) | 7.48 (2.70) | 14.5 (5.05) | 7.50 (2.63) | 7.62 (2.42) |
| C Reactive protein (μg/mL) | 4.77 (3.59) | 81.4 (5.85) | 70.4 (21.5) | 14.0 (21.7) | 96.0 (41.6) | 72.0 (15.7) |
| Cancer antigen 19-9 (U/mL) | 1.44 (0.828) | 1.23 (0.774) | 1.09 (0.509) | 1.30 (0.557) | 1.12 (0.449) | 0.979 (0.604) |
| Complement 3 (mg/mL) | 1.20 (0.198) | 1.66 (0.751) | 1.46 (0.250) | 1.30 (0.256) | 1.44 (0.215) | 1.46 (0.214) |
| EGF (pg/mL) | 47.5 (43.4) | 17.3 (12.8) | 42.1 (44.4) | 91.0 (85.4) | 50.4 (49.1) | 50.9 (59.9) |
| ENA-78 (ng/mL) | 1.47 (0.718) | 0.940 (0.636) | 0.667 (0.427) | 2.68 (1.74) | 0.966 (0.736) | 0.776 (0.516) |
| Eotaxin (pg/mL) | 83.7 (32.4) | 35.9 (19.4) | 58.2 (30.6) | 95.1 (32.1) | 44.2 (21.1) | 54.8 (22.2) |
| Factor VII (ng/mL) | 400 (73.5) | 296 (99.9) | 316 (134) | 401 (160) | 298 (157) | 334 (132) |
| Ferritin (ng/mL) | 210 (196) | 688 (330) | 1270 (1070) | 451 (300) | 999 (766) | 1040 (603) |
| ICAM-1 (ng/mL) | 114 (22.8) | 170 (27.7) | 179 (26.7) | 151 (33.0) | 182 (29.3) | 172 (26.2) |
| IgA (mg/mL) | 2.72 (1.26) | 2.99 (1.42) | 3.04 (1.26) | 2.93 (1.32) | 2.79 (1.20) | 3.09 (1.27) |
| IgE (ng/mL) | 130 (212) | 186 (293) | 108 (125) | 215 (397) | 195 (282) | 142 (147) |
| IgM (mg/mL) | 0.972 (0.346) | 1.17 (0.994) | 0.933 (0.290) | 0.862 (0.280) | 0.816 (0.278) | 0.847 (0.325) |
| IL-10 (pg/mL) | 4.17 (3.14) | 123 (71.8) | 14.6 (11.6) | 3.76 (3.22) | 91.1 (35.3) | 6.44 (4.07) |
| IL-15 (ng/mL) | 1.30 (0.808) | 1.31 (1.31) | 4.52 (3.17) | 2.57 (2.09) | 3.48 (3.26) | 3.67 (3.37) |
| IL-16 (pg/mL) | 546 (222) | 902 (373) | 1030 (320) | 781 (188) | 1160 (289) | 1080 (263) |
| IL-18 (pg/mL) | 273 (110) | 551 (179) | 823 (328) | 531 (223) | 913 (335) | 964 (399) |
| IL-8 (pg/mL) | 6.75 (3.83) | 13.3 (9.70) | 8.33 (4.12) | 22.0 (36.3) | 42.7 (91.9) | 28.9 (64.0) |
| Lipoprotein (a) (μg/mL) | 188 (173) | 178 (174) | 109 (96.7) | 106 (81.3) | 107 (92.6) | 83.9 (73.7) |
| MCP-1 (pg/mL) | 170 (98.9) | 418 (289) | 168 (93.4) | 148 (67.2) | 393 (322) | 141 (57.6) |
| MDC (pg/mL) | 478 (117) | 570 (321) | 1340 (1100) | 998 (653) | 1180 (1280) | 1280 (1270) |
| MIP-1alpha (pg/mL) | 15.2 (13.9) | 24.3 (17.5) | 19.9 (6.07) | 30.1 (20.7) | 27.0 (15.6) | 20.0 (8.07) |
| MIP-1beta (pg/mL) | 179 (56.4) | 234 (69.1) | 200 (53.9) | 193 (99.5) | 208 (69.2) | 157 (55.2) |
| MMP-3 (ng/mL) | 8.58 (3.32) | 7.40 (2.75) | 7.89 (3.39) | 7.68 (3.45) | 6.21 (2.22) | 6.79 (2.52) |
| Myoglobin (ng/mL) | 13.3 (7.48) | 15.5 (14.6) | 20.3 (21.9) | 11.7 (4.66) | 17.4 (11.8) | 26.3 (25.3) |
| PAI-1 (ng/mL) | 118 (28.9) | 74.2 (25.4) | 92.6 (42.5) | 150 (47.2) | 76.5 (24.6) | 90.3 (35.0) |
| RANTES (ng/mL) | 22.1 (5.39) | 14.3 (6.18) | 17.4 (8.08) | 40.9 (18.9) | 15.7 (6.90) | 17.5 (5.15) |
| Serum amyloid P (μg/mL) | 36.7 (7.10) | 35.7 (3.19) | 38.0 (4.06) | 33.3 (7.39) | 36.8 (5.45) | 35.4 (7.39) |
| Stem cell factor (pg/mL) | 101 (62.8) | 81.2 (63.5) | 47.0 (22.5) | 170 (150) | 76.8 (51.4) | 49.4 (29.0) |
| TIMP-1 (ng/mL) | 192 (42.4) | 258 (44.5) | 271 (62.6) | 261 (68.3) | 249 (39.9) | 237 (38.8) |
| TNF RII (ng/mL) | 51.9 (51.8) | 255 (249) | 274 (231) | 74.9 (71.8) | 298 (290) | 252 (201) |
| TNF-alpha (pg/mL) | 4.56 (2.08) | 7.79 (2.28) | 7.05 (2.23) | 5.10 (3.04) | 6.82 (2.89) | 6.78 (2.45) |
| VCAM-1 (ng/mL) | 500 (126) | 930 (187) | 1070 (241) | 675 (235) | 1030 (265) | 1040 (238) |
| VEGF (pg/mL) | 355 (147) | 422 (158) | 423 (220) | 497 (241) | 418 (171) | 410 (175) |
| von Willebrand factor (μg/mL) | 26.1 (10.4) | 63.2 (11.4) | 57.4 (13.3) | 34.9 (11.2) | 64.9 (15.4) | 53.3 (11.6) |
Table 2.
Mean MAP analyte concentrations in the serum of patients treated with 30 μg/kg rIL-21 using the 3/w dosing regimen (n = 7)
| Analyte (units) | Average serum concentration (SD) | |||
|---|---|---|---|---|
| Day 1 | Day 5 | Day 15 | Day 19 | |
| Adiponectin (μg/mL) | 3.84 (1.55) | 3.14 (1.29) | 2.87 (0.972) | 2.76 (0.960) |
| Alpha-1 antitrypsin (mg/mL) | 2.18 (0.735) | 2.70 (0.779) | 3.06 (0.765) | 3.30 (0.697) |
| Alpha-2 macroglobulin (mg/mL) | 0.985 (0.235) | 1.03 (0.268) | 0.862 (0.299) | 0.829 (0.335) |
| Alpha-fetoprotein (ng/mL) | 2.68 (0.874) | 2.49 (0.881) | 2.41 (0.953) | 2.05 (0.772) |
| Apolipoprotein A1 (mg/mL) | 0.499 (0.177) | 0.286 (0.132) | 0.208 (0.0730) | 0.211 (0.0921) |
| Apolipoprotein CIII (μg/mL) | 101 (35.6) | 62.2 (23.7) | 81.2 (38.9) | 89.9 (64.8) |
| Apolipoprotein H (μg/mL) | 254 (65.1) | 216 (31.7) | 230 (69.3) | 228 (55.7) |
| Beta-2 microglobulin (μg/mL) | 1.91 (0.281) | 3.16 (0.475) | 3.82 (0.777) | 4.46 (1.34) |
| Brain-derived neurotrophic factor (ng/mL) | 14.7 (7.93) | 5.89 (2.28) | 6.09 (3.70) | 5.38 (2.53) |
| C Reactive protein (μg/mL) | 7.22 (9.94) | 83.7 (38.0) | 45.7 (17.5) | 64.7 (25.5) |
| Cancer antigen 19-9 (U/mL) | 5.12 (4.96) | 4.65 (5.84) | 7.38 (11.9) | 3.87 (4.49) |
| Complement 3 (mg/mL) | 1.45 (0.296) | 1.72 (0.338) | 1.62 (0.253) | 1.74 (0.310) |
| EGF (pg/mL) | 123 (171) | 90.4 (155) | 134 (208) | 107 (191) |
| ENA-78 (ng/mL) | 2.21 (0.747) | 1.62 (0.987) | 1.72 (1.02) | 1.71 (0.969) |
| Eotaxin (pg/mL) | 114 (44.1) | 82.9 (52.1) | 97.1 (54.5) | 91.0 (59.2) |
| Factor VII (ng/mL) | 495 (368) | 313 (157) | 349 (257) | 249 (155) |
| Ferritin (ng/mL) | 226 (239) | 363 (262) | 533 (476) | 669 (583) |
| ICAM-1 (ng/mL) | 135 (22.5) | 164 (20.9) | 179 (36.9) | 155 (16.6) |
| IgA (mg/mL) | 2.76 (1.09) | 2.60 (0.854) | 2.79 (1.01) | 2.98 (1.06) |
| IgE (ng/mL) | 99.5 (164) | 155 (244) | 154 (208) | 154 (259) |
| IgM (mg/mL) | 1.12 (0.452) | 1.08 (0.406) | 0.922 (0.264) | 0.951 (0.377) |
| IL-10 (pg/mL) | 2.41 (1.50) | 12.7 (6.06) | 3.41 (2.68) | 6.53 (5.70) |
| IL-15 (ng/mL) | 0.413 (0.224) | 0.252 (0.232) | 0.485 (0.422) | 0.180 (0.181) |
| IL-16 (pg/mL) | 438 (192) | 600 (292) | 763 (503) | 627 (189) |
| IL-18 (pg/mL) | 229 (129) | 382 (158) | 699 (275) | 822 (457) |
| IL-8 (pg/mL) | 20.0 (16.1) | 28.5 (37.5) | 25.2 (28.5) | 28.6 (27.6) |
| Lipoprotein (a) (μg/mL) | 47.8 (20.7) | 45.9 (17.2) | 38.8 (15.2) | 38.6 (15.5) |
| MCP-1 (pg/mL) | 197 (109) | 231 (97.7) | 184 (165) | 149 (68.2) |
| MDC (pg/mL) | 365 (78.8) | 334 (74.0) | 485 (221) | 382 (128) |
| MIP-1alpha (pg/mL) | 23.7 (9.08) | 30.7 (8.52) | 39.3 (17.5) | 35.3 (17.8) |
| MIP-1beta (pg/mL) | 193 (59.9) | 257 (104) | 171 (67.5) | 173 (63.5) |
| MMP-3 (ng/mL) | 6.86 (2.57) | 6.07 (2.49) | 8.70 (6.68) | 5.49 (2.37) |
| Myoglobin (ng/mL) | 11.1 (4.10) | 11.4 (4.72) | 14.6 (6.74) | 25.4 (14.9) |
| PAI-1 (ng/mL) | 146 (49.8) | 103 (26.6) | 126 (48.2) | 108 (27.9) |
| RANTES (ng/mL) | 26.2 (5.33) | 18.3 (7.06) | 24.0 (17.1) | 21.1 (9.93) |
| Serum amyloid P (μg/mL) | 38.5 (6.10) | 42.5 (5.43) | 42.1 (7.71) | 41.2 (7.21) |
| Stem cell factor (pg/mL) | 188 (98.0) | 136 (97.0) | 140 (101) | 116 (116) |
| TIMP-1 (ng/mL) | 185 (48.3) | 191 (40.8) | 213 (63.0) | 212 (49.0) |
| TNF RII (ng/mL) | 20.9 (31.2) | 73.9 (109) | 77.3 (110) | 102 (144) |
| TNF-alpha (pg/mL) | 3.21 (1.98) | 5.47 (0.748) | 5.23 (3.12) | 4.59 (1.99) |
| VCAM-1 (ng/mL) | 513 (175) | 814 (338) | 986 (291) | 998 (369) |
| VEGF (pg/mL) | 339 (85.8) | 363 (121) | 469 (265) | 356 (131) |
| von Willebrand factor (μg/mL) | 23.5 (7.89) | 55.9 (6.06) | 57.1 (7.54) | 65.2 (14.7) |
Comparison of rIL-21 effects between the different dosing regimens showed similar trends of change in markers, particularly in the initial week of treatment. In general, a cyclic induction pattern was observed with the 5 + 9 regimen while a more continuous induction was seen in subjects treated on the 3/w schedule. Plots of mean change from baseline comparing the two regimens for representative analytes are shown in Fig. 2. For most analytes, changes were similar between the regimens when averaged over the time of treatment, e.g., sTNFRII, sVCAM-1 and IL-18. However, for some analytes such as RANTES, MDC, and MCP-1, notable differences were observed between regimens. In these cases, intermittent dosing with the 5 + 9 regimen led to divergent responses during non-dosing periods or upon re-initiation of treatment.
Fig. 2.
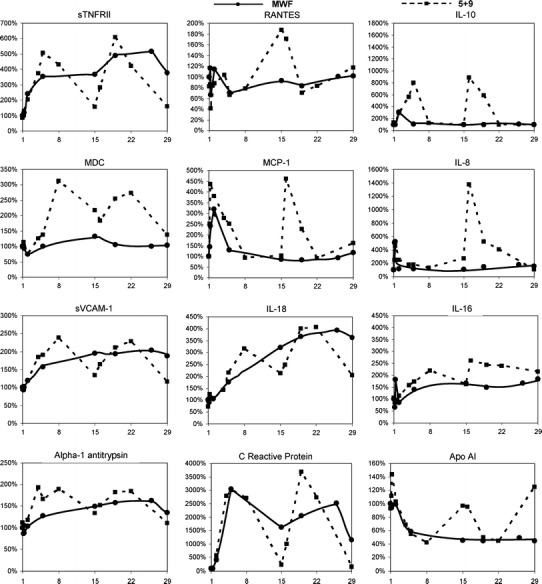
Changes in serum concentration relative to baseline for selected analytes in subjects treated with 30 μg/kg rIL-21. Effects of the different dose regimens, 5 + 9 (dashed line) and 3/w (solid line), are compared
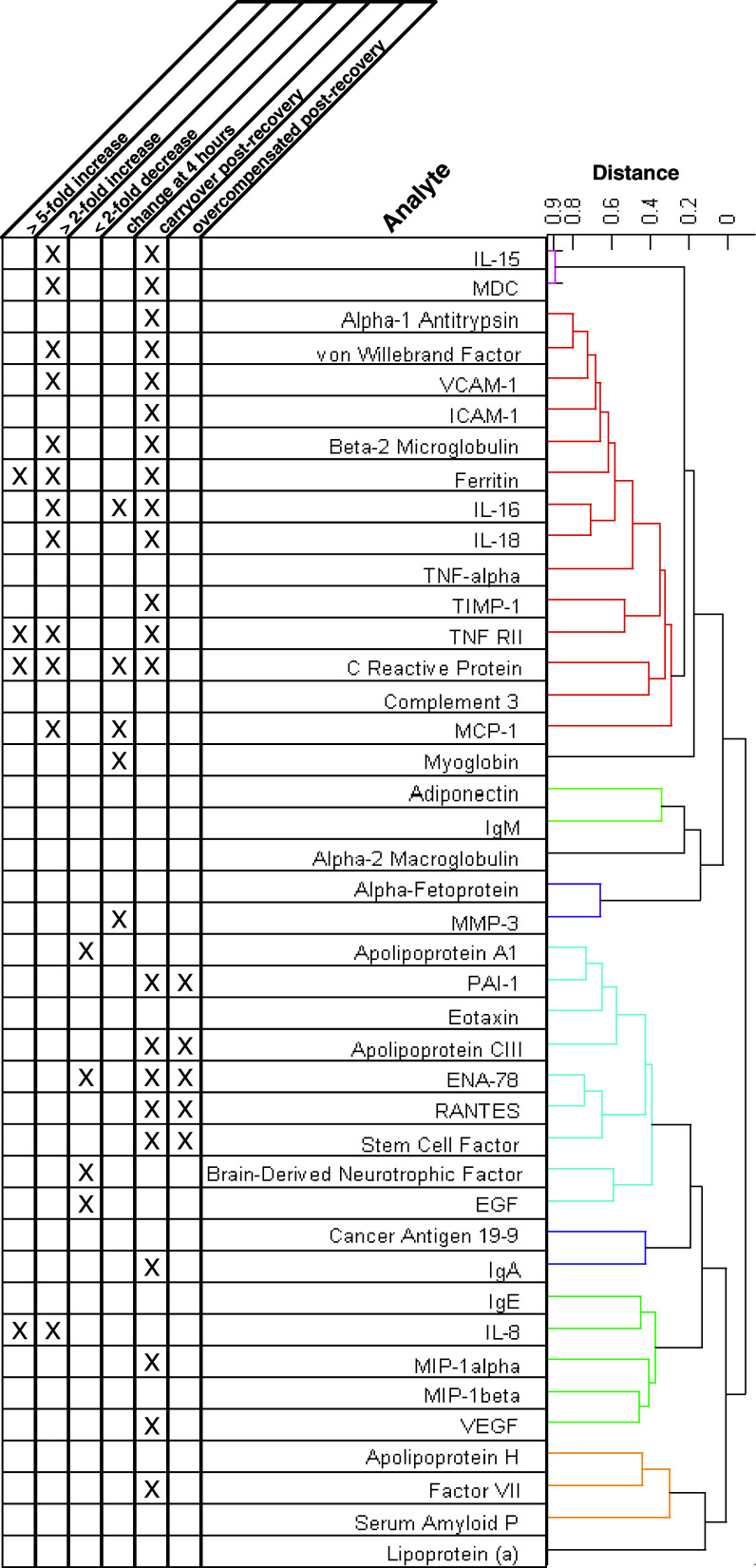
Biomarker clusters
To help discern distinct and similar patterns of change in the serum protein biomarkers, hierarchical clustering analysis was performed on a subset of the normalized data. This analysis focused on subjects treated with 30 μg/kg rIL-21 using the 5 + 9 dosing regimen because this represented the largest (n = 12) and most dynamic subset of the response data. The results of the clustering analysis are shown in Fig. 3. Several clusters of serum markers were identified based on changes relative to baseline levels. The degree of correlation between markers or groups of markers is indicated by the linkage distances shown in the dendrogram. Primary associations were driven by general upward or downward trends during the dosing periods. Categories of response type are indicated in the table accompanying the dendrogram in Fig. 3. Analytes in each of six main clusters are plotted together in Fig. 4, demonstrating similar patterns of change in response to treatment with rIL-21 (30 μg/kg dose level with 5 + 9 regimen). A heat map showing the relative changes in each of the markers for all subjects and sampling times is provided in the supplementary materials (Fig. A1). This diagram also shows a dendrogram representing the degree of correlation between subjects and sampling times.
Fig. 3.
Dendrogram of the distance measures (unweighted average Pearson correlation coefficient between the members of linked cluster) of the serum markers for each Subject treated with 30 μg/kg rIL-21 on the 5 + 9 regimen
Fig. 4.
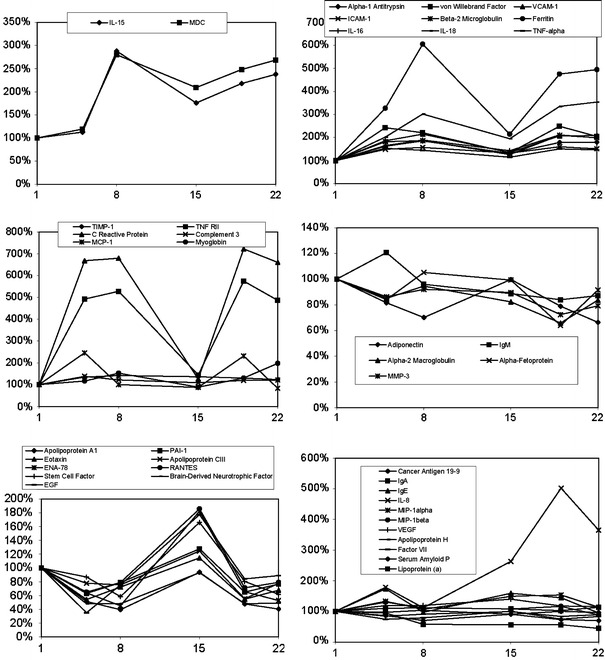
Group average percent change from group average baseline (y-axis, %) over time (x-axis, days) for serum markers grouped in clusters identified in Fig. 3 for Subjects treated with 30 μg/kg rIL-21 on the 5 + 9 regimen (n = 12)
Serum markers that trended strongly upward (fivefold induction or more) during dosing were IL-8, TNFRII, Ferritin and C Reactive Protein (CRP). Serum markers that were more moderately increased (two to fivefold induction) were MDC, IL-15, von Willebrand Factor, VCAM-1, Beta-2 Microglobulin, IL-16, IL-18 and MCP-1. Another major cluster of serum markers that trended downward was identified, including some that were reduced by a factor of 2 or more (Apo A1, ENA-78, Brain-Derived Neurotrophic Factor and EGF). Secondary associations were driven by temporal similarities (i.e., timing or duration of response) and occasionally by excessive or atypical responses in a few subjects. Serum proteins that increased at the earliest timepoint, 4 h after rIL-21 dosing, include IL-16, CRP, MCP-1, Myoglobin and MMP-3. Although IL-10 was excluded from clustering analysis due to many subjects having baseline levels below the LDD, the predominant pattern observed with rIL-21 dosing was similar to these rapidly-induced analytes. The concentration of IL-10 increased rapidly following rIL-21 dosing and continued to rise during the dosing period in subjects treated with the 5 + 9 dose regimen (Fig. 2). In contrast, IL-10 was more transiently induced by the 3/w dose regimen and was below LDD in most subjects following Day 2 of treatment.
Carryover effects were observed with numerous markers. This was most common in the strongly induced cluster that included IL-15, MDC, alpha-1 antitrypsin, von Willebrand factor, sVCAM-1, sICAM-1, beta-2 microglobulin, ferritin, IL-16, IL-18, TNF alpha, TIMP-1, TNF-RII, CRP, complement 3 and MCP-1, but was also observed for markers that were reduced. In some cases, carryover effects in markers reduced during the dosing period were characterized by over-compensation between dosing periods. This is exemplified by several markers in the cluster including Apo AI, PAI-1, eotaxin, APO CIII, ENA-78, RANTES, stem cell factor, BDNF and EGF.
Discussion
Analysis of serum samples collected from subjects dosed with rIL-21 demonstrated a complex array of changes indicative of strong immune modulation. The overall pattern of change for these serum biomarkers indicates that rIL-21 induced multiple concurrent pharmacodynamic effects. Hierarchical clustering analysis identified strong signals for factors associated with leukocyte activation and migration, and acute phase response. While these processes comprise interdependent and overlapping effects that are common to many other circumstances of immune activation, the pattern of changes induced by systemic administration of rIL-21 likely represents a unique serum protein signature.
In a larger treated population, hierarchical clustering methods could be useful in analysis of patient subsets. This would be particularly valuable for identifying pharmacodynamic responses common among the responding population. In this study, however, the frequency of disease response (i.e., partial response or better by RECIST criteria) was not sufficient to determine such associations. Considering subjects treated at the 30 μg/kg dose level, 1 of the 7 treated with the 3/w regimen and 2 of the 12 treated with the 5 + 9 regimen were evaluable by this clinical response criteria. Coupled with the observation that the pharmacodynamic effects described occurred consistently among subjects treated with the same rIL-21 dosing regimen, no strong correlations could be made between tumor response and pharmacodynamic markers in this limited analysis.
Dose dependent changes were observed for a subset of the analytes measured. Given the limitation that few subjects were treated at dose levels other than 30 μg/kg, the principal indicator of dose response was a threshold for detecting rIL-21 response between dose levels of 10 and 30 μg/kg. Clustering analysis focused on subjects treated at 30 μg/kg with the 5 + 9 dosing regimen because this represented the largest and most dynamic subset of the response data. However, it should be noted that it is not known which of the two dosing regimens tested in the Phase 1 is more likely to be associated with clinical efficacy. The 5 + 9 dosing regimen produced a more discrete pharmacodynamic effect, with changes generally occurring during or directly after dosing periods and subsequent reversal to baseline levels during resting periods. In contrast, response patterns observed with the 3/w dose regimen reflect a more moderate, continuous pharmacodynamic effect. A direct comparison between the two dosing regimens was limited by different sampling schedules in the two regimens. Despite differences in timing of rIL-21 pharmacologic effects, time-averaged effects for many markers, particularly those with longer serum residence time (sTNFRII, sVCAM-1 and IL-18), were similar between dosing regimens.
Cytokines and chemokines that were strongly increased include IL-18, TNF-alpha, IL-16, and MCP-1. The increase in IL-18 is especially interesting since this cytokine is known to co-stimulate IL-12-induced production of IFN-γ, a cytokine that is required for generation of a Th1 responses and T cell- and NK cell-mediated antitumor activity [21, 25]. Thus, it is likely that the induction of IL-18 may play a role in rIL-21 mediated anti-tumor response. In this context, it is noteworthy that recombinant human IL-18 has been tested in Phase 2 trials in patients with malignant melanoma and is currently being tested in other malignancies (http://www.clinicaltrials.gov). The evidence of elevated TNF-alpha, including sTNFRII in the serum, suggests another mechanism of anti-tumor activity for rIL-21, as TNF-alpha is known to activate T cells, to have direct anti-tumor effects, and has shown some anti-tumor activity in clinical trials [22]. In contrast to TNF-alpha, IL-10 functions to prevent the development of pathological lesions that result from exacerbated immune responses during infections [20]. Increased production of IL-10 may result from a compensatory mechanism to counter-balance strong pro-inflammatory stimuli. Consistent with this, the highest levels of IL-10 were observed in subjects that experienced dose limiting toxicity in these trials (data not shown). Also, IL-10 levels have been shown to progressively increase during aggressive high-dose IL-2 treatment of patients with malignant melanoma [7]. Conversely, compelling evidence supports an anti-tumor effect of IL-10 in malignant melanoma patients [1, 13].
The patterns of sICAM-1 and sVCAM-1 response to rIL-21 dosing correspond to that previously described for the lymphocyte activation marker soluble IL-2Rα (sCD25) [9, 37]. As these factors reflect augmented interactions between activated lymphocytes and endothelium that may result in adhesion and transmigration of these cells into the extravascular space, the patterns suggest a plausible mechanism for the previously described redistribution of lymphocyte subsets upon rIL-21 dosing [9, 37]. Increased concentrations of sICAM-1 and sVCAM-1 have been reported in other investigations of cytokine treatment of cancer patients, including high dose IL-2, TNF alpha, and IL-12 [19, 24, 28]. Large increases in sICAM-1 and sVCAM-1 are thought to be associated with development of vascular leak syndrome with high-dose IL-2 [3, 28, 36]. Vascular leak has not been reported in rIL-21 clinical trials.
Serum concentrations of both ENA-78 and RANTES were reduced during the dosing period and increased over baseline levels between dosing periods. The decrease during dosing periods may result from rIL-21-mediated increase in expression of receptors for these chemokines and/or the numbers of cells bearing these receptors. ENA-78 is a potent chemoattractant for neutrophils [41]. RANTES is chemotactic for T cells, monocytes, eosinophils and basophils and causes activation of these cell types [16].
Macrophage-derived chemokine (MDC) and IL-15 displayed a very high degree of correlation, suggesting production by a common source. Both of these factors are produced by macrophages, in addition to other cell types, and their production is increased by cell activation through a variety of stimuli [11, 35, 43]. IL-15 and MDC may contribute to rIL-21-induced NK-cell mediated anti-tumor effects through NK cell survival, activation and chemotaxis, in concert with other factors induced by rIL-21 treatment [2, 11]. Considering the potential sources of MDC and IL-15 and observations from preclinical studies that myeloid activation is an important and consistent component of rIL-21 pharmacodynamic response in vivo [40], their common source is likely activated macrophages.
The majority of the cytokines included in the MAP profiles were below the lower limit of detection. This was true for most of the interleukins, notably IL-2, IL-5, IL-6, IL-12 (p40 and p70) and many of the baseline measurements for IL-10. However, since cytokines generally have short half-lives and are often sequestered in the local area of their production, the lack of detectable serum levels does not exclude the presence of local and biologically significant levels of these cytokines in peripheral tissues.
Several of the factors that were highly induced by rIL-21 increased in association with acute phase response: CRP, alpha-1-antitrypsin, beta-2 Microglobulin, ferritin and von Willebrand factor are all positive acute phase proteins [10]. In contrast, serum concentrations of apolipoproteins AI, CIII and H trend lower over the course of a dosing cycle, consistent with their behavior as negative acute phase proteins [10, 14]. Modulation of these factors corresponds with observed clinical sequelae of acute phase response in these trials (i.e., flu-like symptoms) [9, 37]. While these data reinforce standard clinical observations and clearly establish an association of rIL-21 administration with acute phase response, it is not clear whether this is a direct effect of rIL-21 or an indirect effect mediated by a secondary cascade of cytokine release that produces critical levels of TNF-alpha, IL-1, and IL-6. In either case, the acute phase response is an early and prominent effect of rIL-21 administration. The strong temporal association of innate immune cell activation (macrophage, NK cells), cellular responses to early cytokine induction, and leukocyte-vascular interaction is a very significant driver of associations in the overall cluster.
Comparison of these results with a similar analysis conducted for high dose IL-2 therapy in cancer patients shows similarities as well as clear differences between the treatments [28]. Therapy with rIL-21 resulted in a similar induction of TNF-alpha, MIP-1 alpha, sTNFRII and IL-18, as was observed with high-dose IL-2. In contrast, a smaller increase in serum proteins IL-6, IL-8, IL-16, MCP-1, MMP-3, sVCAM-1 and sICAM-1 was observed with rIL-21. Differences in effect on sCAM levels, and possibly MMP-3, may reflect the greater safety and tolerability of rIL-21 regimens tested in these trials, which were designed to be administered on an outpatient basis. Increased levels of these factors have been associated with development of vascular leak syndrome, a dose-limiting hemodynamic side effect associated with high-dose IL-2 administration [28]. The relatively moderate increases in these markers of vascular reactivity that occurred with the regimens tested in rIL-21 Phase 1 trials are consistent with the general lack of hemodynamic effects observed.
In summary, this pilot study identified major patterns of change in serum proteins during systemic rIL-21 treatment of advanced cancer patients. These changes were consistent with coincident processes of lymphoid and myeloid cell activation and trafficking, and acute phase response. This work offers opportunities to understand sources of rIL-21 mechanism of action as well as variation among subjects to identify early biomarkers predictive of beneficial disease response or adverse drug effects. The data demonstrate that administration of rIL-21 to patients with metastatic disease induces a variety of pharmacodynamic responses related to immune effector functions that warrant further investigation in larger patient populations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
References
- 1.Alonso R, et al. Influence of interleukin-10 genetic polymorphism on survival rates in melanoma patients with advanced disease. Melanoma Res. 2005;15(1):53–60. doi: 10.1097/00008390-200502000-00009. [DOI] [PubMed] [Google Scholar]
- 2.Carson WE, et al. A potential role for interleukin-15 in the regulation of human natural killer cell survival. J Clin Invest. 1997;99(5):937–943. doi: 10.1172/JCI119258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Damle NK, Doyle LV. IL-2-activated human killer lymphocytes but not their secreted products mediate increase in albumin flux across cultured endothelial monolayers. Implications for vascular leak syndrome. J Immunol. 1989;142(8):2660–2669. [PubMed] [Google Scholar]
- 4.Davis ID, et al. Interleukin-21 signaling: functions in cancer and autoimmunity. Clin Cancer Res. 2007;13(23):6926–6932. doi: 10.1158/1078-0432.CCR-07-1238. [DOI] [PubMed] [Google Scholar]
- 5.Davis ID, et al. An open-label, two-arm, phase I trial of recombinant human interleukin-21 in patients with metastatic melanoma. Clin Cancer Res. 2007;13(12):3630–3636. doi: 10.1158/1078-0432.CCR-07-0410. [DOI] [PubMed] [Google Scholar]
- 6.Di Carlo E, et al. IL-21 induces tumor rejection by specific CTL and IFN-gamma-dependent CXC chemokines in syngeneic mice. J Immunol. 2004;172(3):1540–1547. doi: 10.4049/jimmunol.172.3.1540. [DOI] [PubMed] [Google Scholar]
- 7.Engelhardt M, et al. Clinical and immunomodulatory effects of repetitive 2-day cycles of high-dose continuous infusion IL-2. Eur J Cancer. 1997;33(7):1050–1054. doi: 10.1016/S0959-8049(96)00530-8. [DOI] [PubMed] [Google Scholar]
- 8.Ettinger R. IL-21 induces differentiation of human naive and memory B cells into antibody-secreting plasma cells. J Immunol. 2005;175(12):7867–7879. doi: 10.4049/jimmunol.175.12.7867. [DOI] [PubMed] [Google Scholar]
- 9.Frederiksen KS, et al. IL-21 induces in vivo immune activation of NK cells and CD8(+) T cells in patients with metastatic melanoma and renal cell carcinoma. Cancer Immunol Immunother. 2008;57(10):1439–1449. doi: 10.1007/s00262-008-0479-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gabay C, Kushner I. Acute-phase proteins and other systemic responses to inflammation. N Engl J Med. 1999;340(6):448–454. doi: 10.1056/NEJM199902113400607. [DOI] [PubMed] [Google Scholar]
- 11.Godiska R, et al. Human macrophage-derived chemokine (MDC), a novel chemoattractant for monocytes, monocyte-derived dendritic cells, and natural killer cells. J Exp Med. 1997;185(9):1595–1604. doi: 10.1084/jem.185.9.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He H, et al. Combined IL-21 and low-dose IL-2 therapy induces anti-tumor immunity and long-term curative effects in a murine melanoma tumor model. J Transl Med. 2006;4:24. doi: 10.1186/1479-5876-4-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Howell WM, et al. IL-10 promoter polymorphisms influence tumour development in cutaneous malignant melanoma. Genes Immun. 2001;2(1):25–31. doi: 10.1038/sj.gene.6363726. [DOI] [PubMed] [Google Scholar]
- 14.Jahoor F, et al. The acute-phase protein response to human immunodeficiency virus infection in human subjects. Am J Physiol. 1999;276(6 Pt 1):E1092–E1098. doi: 10.1152/ajpendo.1999.276.6.E1092. [DOI] [PubMed] [Google Scholar]
- 15.Korn T, et al. IL-21 initiates an alternative pathway to induce proinflammatory T(H)17 cells. Nature. 2007;448(7152):484–487. doi: 10.1038/nature05970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuna P, et al. RANTES, a monocyte and T lymphocyte chemotactic cytokine releases histamine from human basophils. J Immunol. 1992;149(2):636–642. [PubMed] [Google Scholar]
- 17.Li Y, Bleakley M, Yee C. IL-21 influences the frequency, phenotype, and affinity of the antigen-specific CD8 T cell response. J Immunol. 2005;175(4):2261–2269. doi: 10.4049/jimmunol.175.4.2261. [DOI] [PubMed] [Google Scholar]
- 18.Li Y, Yee C. IL-21 mediated Foxp3 suppression leads to enhanced generation of antigen-specific CD8+ cytotoxic T lymphocytes. Blood. 2008;111(1):229–235. doi: 10.1182/blood-2007-05-089375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lieuw-a-Fa M, Schalkwijk C, van Hinsbergh VW. Distinct accumulation patterns of soluble forms of E-selectin, VCAM-1 and ICAM-1 upon infusion of TNFalpha in tumor patients. Thromb Haemost. 2003;89(6):1052–1057. [PubMed] [Google Scholar]
- 20.Mege JL, et al. The two faces of interleukin 10 in human infectious diseases. Lancet Infect Dis. 2006;6(9):557–569. doi: 10.1016/S1473-3099(06)70577-1. [DOI] [PubMed] [Google Scholar]
- 21.Micallef MJ, et al. Interferon-gamma-inducing factor enhances T helper 1 cytokine production by stimulated human T cells: synergism with interleukin-12 for interferon-gamma production. Eur J Immunol. 1996;26(7):1647–1651. doi: 10.1002/eji.1830260736. [DOI] [PubMed] [Google Scholar]
- 22.Mocellin S, et al. Tumor necrosis factor, cancer and anticancer therapy. Cytokine Growth Factor Rev. 2005;16(1):35–53. doi: 10.1016/j.cytogfr.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 23.Moroz A, et al. IL-21 enhances and sustains CD8+ T cell responses to achieve durable tumor immunity: comparative evaluation of IL-2, IL-15, and IL-21. J Immunol. 2004;173(2):900–909. doi: 10.4049/jimmunol.173.2.900. [DOI] [PubMed] [Google Scholar]
- 24.Mortarini R, et al. Peripheral burst of tumor-specific cytotoxic T lymphocytes and infiltration of metastatic lesions by memory CD8+ T cells in melanoma patients receiving interleukin 12. Cancer Res. 2000;60(13):3559–3568. [PubMed] [Google Scholar]
- 25.Nakanishi K, et al. Interleukin-18 regulates both Th1 and Th2 responses. Annu Rev Immunol. 2001;19:423–474. doi: 10.1146/annurev.immunol.19.1.423. [DOI] [PubMed] [Google Scholar]
- 26.Nutt SL, et al. Interleukin 21: a key player in lymphocyte maturation. Crit Rev Immunol. 2004;24(4):239–250. doi: 10.1615/CritRevImmunol.v24.i4.20. [DOI] [PubMed] [Google Scholar]
- 27.Ozaki K, et al. A critical role for IL-21 in regulating immunoglobulin production. Science. 2002;298(5598):1630–1634. doi: 10.1126/science.1077002. [DOI] [PubMed] [Google Scholar]
- 28.Panelli MC, et al. Forecasting the cytokine storm following systemic interleukin (IL)-2 administration. J Transl Med. 2004;2(1):17. doi: 10.1186/1479-5876-2-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parrish-Novak J, et al. Interleukin 21 and its receptor are involved in NK cell expansion and regulation of lymphocyte function. Nature. 2000;408(6808):57–63. doi: 10.1038/35040504. [DOI] [PubMed] [Google Scholar]
- 30.Parrish-Novak J, et al. Interleukin-21 and the IL-21 receptor: novel effectors of NK and T cell responses. J Leukoc Biol. 2002;72(5):856–863. [PubMed] [Google Scholar]
- 31.Peluso I, et al. IL-21 counteracts the regulatory T cell-mediated suppression of human CD4 + T lymphocytes. J Immunol. 2007;178(2):732–739. doi: 10.4049/jimmunol.178.2.732. [DOI] [PubMed] [Google Scholar]
- 32.Perez SA, et al. Effect of IL-21 on NK cells derived from different umbilical cord blood populations. Int Immunol. 2006;18(1):49–58. doi: 10.1093/intimm/dxh348. [DOI] [PubMed] [Google Scholar]
- 33.Skak K, Frederiksen KS, Lundsgaard D. Interleukin-21 activates human natural killer cells and modulates their surface receptor expression. Immunology. 2008;123(4):575–583. doi: 10.1111/j.1365-2567.2007.02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sondergaard H, et al. Interleukin 21 therapy increases the density of tumor infiltrating CD8+ T cells and inhibits the growth of syngeneic tumors. Cancer Immunol Immunother. 2007;56(9):1417–1428. doi: 10.1007/s00262-007-0285-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tagaya Y, et al. IL-15: a pleiotropic cytokine with diverse receptor/signaling pathways whose expression is controlled at multiple levels. Immunity. 1996;4(4):329–336. doi: 10.1016/S1074-7613(00)80246-0. [DOI] [PubMed] [Google Scholar]
- 36.Thom AK, et al. Cytokine levels and systemic toxicity in patients undergoing isolated limb perfusion with high-dose tumor necrosis factor, interferon gamma, and melphalan. J Clin Oncol. 1995;13(1):264–273. doi: 10.1200/JCO.1995.13.1.264. [DOI] [PubMed] [Google Scholar]
- 37.Thompson JA, et al. Phase I study of recombinant interleukin-21 in patients with metastatic melanoma and renal cell carcinoma. J Clin Oncol. 2008;26(12):2034–2039. doi: 10.1200/JCO.2007.14.5193. [DOI] [PubMed] [Google Scholar]
- 38.Ugai S, et al. Expression of the interleukin-21 gene in murine colon carcinoma cells generates systemic immunity in the inoculated hosts. Cancer Gene Ther. 2003;10(3):187–192. doi: 10.1038/sj.cgt.7700552. [DOI] [PubMed] [Google Scholar]
- 39.Ugai S, et al. Transduction of the IL-21 and IL-23 genes in human pancreatic carcinoma cells produces natural killer cell-dependent and -independent antitumor effects. Cancer Gene Ther. 2003;10(10):771–778. doi: 10.1038/sj.cgt.7700630. [DOI] [PubMed] [Google Scholar]
- 40.Van Ness K et al. (2008) Preclinical evaluation of immunomodulatory activity and safety of recombinant human interleukin-21. In: The toxicologist. Society of Toxicology, Seattle, WA. p. Abstract No. 1928
- 41.Walz A, et al. Regulation and function of the CXC chemokine ENA-78 in monocytes and its role in disease. J Leukoc Biol. 1997;62(5):604–611. doi: 10.1002/jlb.62.5.604. [DOI] [PubMed] [Google Scholar]
- 42.Wang G, et al. In vivo antitumor activity of interleukin 21 mediated by natural killer cells. Cancer Res. 2003;63(24):9016–9022. [PubMed] [Google Scholar]
- 43.Xiao T, et al. Both IL-4 and IL-13 inhibit the TNF-alpha and IFN-gamma enhanced MDC production in a human keratinocyte cell line, HaCaT cells. J Dermatol Sci. 2003;31(2):111–117. doi: 10.1016/S0923-1811(02)00149-4. [DOI] [PubMed] [Google Scholar]
- 44.Zeng R, et al. Synergy of IL-21 and IL-15 in regulating CD8+ T cell expansion and function. J Exp Med. 2005;201(1):139–148. doi: 10.1084/jem.20041057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou L, et al. IL-6 programs T(H)-17 cell differentiation by promoting sequential engagement of the IL-21 and IL-23 pathways. Nat Immunol. 2007;8(9):967–974. doi: 10.1038/ni1488. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.