Abstract
The bovine papillomavirus E5 protein is a small, homodimeric transmembrane protein that forms a stable complex with the cellular platelet-derived growth factor (PDGF) β receptor through transmembrane and juxtamembrane interactions, resulting in receptor activation and cell transformation. Glutamine 17 in the transmembrane domain of the 44-amino-acid E5 protein is critical for complex formation and receptor activation, and we previously proposed that glutamine 17 forms a hydrogen bond with threonine 513 of the PDGF β receptor. We have constructed and analyzed mutant E5 proteins containing all possible amino acids at position 17 and examined the ability of these proteins to transform C127 fibroblasts, which express endogenous PDGF β receptor. Although several position 17 mutants were able to transform cells, mutants containing amino acids with side groups that were unable to participate in hydrogen bonding interactions did not form a stable complex with the PDGF β receptor or transform cells, in agreement with the proposed interaction between position 17 of the E5 protein and threonine 513 of the receptor. The nature of the residue at position 17 also affected the ability of the E5 proteins to dimerize. Overall, there was an excellent correlation between the ability of the various E5 mutant proteins to bind the PDGF β receptor, lead to receptor tyrosine phosphorylation, and transform cells. Similar results were obtained in Ba/F3 hematopoietic cells expressing exogenous PDGF β receptor. In addition, treatment of E5-transformed cells with a specific inhibitor of the PDGF receptor tyrosine kinase reversed the transformed phenotype. These results confirm the central importance of the PDGF β receptor in mediating E5 transformation and highlight the critical role of the residue at position 17 of the E5 protein in the productive interaction with the PDGF β receptor. On the basis of molecular modeling analysis and the known chemical properties of the amino acids, we suggest a structural basis for the role of the residue at position 17 in E5 dimerization and in complex formation between the E5 protein and the PDGF β receptor.
Bovine papillomavirus type 1 (BPV) is a double-stranded DNA tumor virus that causes benign fibropapillomas in infected cattle and transforms cultured fibroblasts. The transforming activity of BPV resides primarily in the E5 gene, which encodes a small homodimeric protein largely localized to membranes of the endoplasmic reticulum and Golgi apparatus (see reference 7 for a review). The BPV E5 protein forms a stable complex with the endogenous cellular platelet-derived growth factor (PDGF) β receptor in fibroblasts and induces ligand-independent receptor oligomerization, tyrosine phosphorylation, and activation, leading to cell transformation (6, 7, 10, 17, 21, 23–25).
The 44-residue E5 protein consists of a hydrophobic 30-residue segment at the N terminus that spans membranes as an α-helix and a hydrophilic 14-residue segment at the C terminus (7, 30). Extensive mutational analysis has demonstrated that four absolutely conserved residues in the E5 protein are critical for binding and activation of the PDGF β receptor and for cell transformation (13, 20, 22, 27, 28). These residues are glutamine 17, the only hydrophilic residue in the transmembrane region, aspartic acid 33 in the juxtamembrane region, and the two carboxy-terminal cysteines at positions 37 and 39 that are involved in homodimerization of the E5 protein. In addition, the overall hydrophobicity of the central core of the E5 protein, but not the specific amino acid sequence, is required for cell transformation (7).
Complex formation between the E5 protein and the PDGF β receptor is mediated by interactions between the transmembrane and juxtamembrane regions of the two proteins, and removal of the ligand-binding domain of the receptor does not disrupt interaction with the E5 protein (5, 6, 9, 10, 26, 29). A positive charge in the extracellular juxtamembrane region of the receptor and threonine 513 in the transmembrane domain is required for interaction with the E5 protein and for E5-induced receptor activation but not for activation by PDGF. The E5 protein does not interact with the related PDGF α receptor, a difference that maps to the transmembrane/juxtamembrane region of the receptor (10, 25, 29). Strikingly, both a positively charged juxtamembrane residue and the transmembrane threonine are absent from the PDGF α receptor.
The E5 protein is thought to be a type II membrane protein and consequently would be inserted into membranes in an orientation opposite that of the PDGF β receptor (4). This places aspartate 33 of the E5 protein and lysine 499 of the receptor on the extracytoplasmic side of the membrane at the membrane surface, with glutamine 17 and threonine 513 buried in the membrane at roughly the same distance relative to the membrane surface. These considerations have led to the proposal that two pairs of interacting residues, aspartate 33-lysine 499 and glutamine 17-threonine 513, are essential for complex formation between the E5 protein and the PDGF β receptor (20, 22, 26, 30).
There are few structural data about the E5-PDGF β receptor complex. Recently, polarized infrared spectroscopy has shown that the dimeric E5 protein in lipid bilayers is largely α-helical and has a transmembrane orientation (30). Computational searches generated two related structural models for the E5 dimer that account for the role of glutamine 17 and aspartate 33 in complex formation and are consistent with the position of conserved and transformation-sensitive residues (30). In this report, we have constructed and analyzed mutant E5 proteins containing all possible amino acids in place of the required glutamine to establish the role of glutamine 17 in mediating the activities of the E5 protein. Based on the molecular models we previously developed for the E5 dimer and on the chemical properties of the amino acid at position 17, we propose structural explanations for the transforming activity and biochemical properties of the mutants.
MATERIALS AND METHODS
Construction of mutant E5 genes.
Position 17 mutations were constructed by using codon-cassette mutagenesis, a method we previously described in detail (14). Standard subcloning procedures were used to subclone the mutant E5 genes into the retroviral vector pRVY-BPV-E5 (26). The DNA sequence of the entire E5 coding region was confirmed for each mutant. Retroviral DNA containing each mutant was introduced into packaging cell lines, and stable cell lines producing high-titer retrovirus stocks were obtained after selection for hygromycin resistance as described previously (26). Mutant E5 genes encoding glutamic acid, serine, leucine, and glycine at position 17 were subcloned into the vector pPava2, and recombinant BPV/simian virus 40 (SV40) stocks were generated from the resulting plasmids as previously described (22). Details of the mutagenesis and subcloning procedures are available from the authors on request.
Cell lines and tissue culture.
C127 and COS7 cells were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum and antibiotics (DME-10). To assay the mutants for focus-forming activity, 60-mm-diameter dishes of C127 cells were infected with retroviruses (approximately 104 CFU) encoding the mutant or wild-type E5 genes and foci were counted 2 to 3 weeks after infection, as described previously (26). Additional cells were selected in medium containing 300 U of hygromycin B per ml. Cell lines were established from pools of >100 resistant colonies and grown in medium containing 300 U of hygromycin per ml. To calculate focus-forming efficiencies, the number of foci that formed were normalized for the number of hygromycin-resistant colonies that arose in parallel in the same infection. The results shown in Table 1 represent the averages of between two and four independent infections for each mutant. We also transformed C127 cells with retroviruses expressing neu*, an activated version of the ErbB2 protein (1).
TABLE 1.
Summary of position 17 mutants in C127 cells
| Virus or mutant | % C127 cell focus formationa | % PDGF receptor tyrosine phosphorylationb | E5/PDGFR complex formationc | E5 protein dimerizationd |
|---|---|---|---|---|
| Vector alone | 0 | 12 ± 4 | NAe | NA |
| WT | 100 | 100 | ++ | H |
| Q17A | 0 | 19 ± 1 | − | I |
| Q17G | 0 | 17 ± 4 | − | I |
| Q17I | 0 | 13 ± 9 | − | L |
| Q17L | 0 | 5 ± 4 | − | I |
| Q17V | 0 | 28 ± 27 | − | I |
| Q17F | 17 ± 17 | 11 ± 6 | − | I |
| Q17Y | 0 | 28 ± 1 | − | I |
| Q17P | 1 ± 2 | 15 ± 8 | − | NDf |
| Q17R | 3 ± 4 | 2 ± 1 | − | ND |
| Q17C | 4 ± 5 | 21 ± 9 | − | I |
| Q17T | 30 ± 19 | 62 ± 4 | + | I |
| Q17D | 35 ± 11 | 172 ± 153 | +++ | H |
| Q17M | 52 ± 31 | 27 ± 5 | + | I |
| Q17K | 59 ± 14 | 25 ± 3 | + | H |
| Q17W | 103 ± 29 | 176 ± 144 | + | I |
| Q17Q | 127 ± 37 | 122 | ++ | H |
| Q17H | 151 ± 45 | 103 ± 16 | +++ | H |
| Q17N | 158 ± 45 | 181 ± 80 | ++ | H |
| Q17E | 194 ± 54 | 152 ± 61 | +++ | H |
| Q17S | 287 ± 94 | 126 ± 22 | ++ | I |
Average focus formation (corrected for virus titer), expressed as percentage of wild-type +/− standard deviation of the mean. In a typical experiment, the wild-type E5 retrovirus induced 16 transformed foci/1,000 hygromycin-resistant colonies.
Average tyrosine phosphorylation of mature PDGF β receptor as determined by PhosphorImager analysis, expressed as percentage of wild-type +/− standard deviation of the mean. Only one cell line of reconstructed wild-type Q17Q was tested.
Complex formation between the E5 protein and the PDGF β receptor was scored according to the following scale: −, no or trace binding; +, readily detectable binding; ++, levels of binding comparable to that observed with wild-type E5 protein; +++, levels of binding greater than that observed with the wild type.
Symbols: H (high amount of dimer), E5 protein present exclusively or mostly as dimer; I (intermediate), similar levels of monomeric and dimeric E5 protein; L (low), predominantly monomeric E5 protein.
NA, not applicable.
ND, not determined.
The PDGF β receptor kinase inhibitor AG1295 (16) was obtained from Calbiochem, Inc. Subconfluent monolayers of C127 cells were incubated in 50 μM AG1295 in DME-10 for 8 to 10 h, after which the cells were processed for phosphotyrosine blotting as detailed below. Alternatively, cells were maintained in the presence or absence of 50 μM AG1295 for 3 to 5 days and photographed.
Ba/F3 cells were obtained from Alan D’Andrea (Dana Farber Cancer Institute, Boston, Mass.) and maintained as previously described (6) in RPMI 1640 (RPMI) supplemented with 10% heat-inactivated fetal bovine serum, WEHI-conditioned medium as a source of interleukin-3 (IL-3), 0.05 mM β-mercaptoethanol, and antibiotics (RPMI/IL-3). Stable Ba/F3 cell lines expressing the various E5 mutant or wild-type constructs in the absence or presence of the murine PDGF β receptor were established by using recombinant retroviruses to infect Ba/F3-neo or Ba/F3-mPR, respectively, as previously described (6, 26), with minor modifications. Approximately 1.5 × 105 CFU of retrovirus was added to 5 × 106 cells in 10 ml of RPMI/IL-3 containing 4 μg of Polybrene per ml. After 2 days, 1 ml of infected cells was transferred into 10 ml of RPMI/IL-3 containing G418 (1 mg/ml) and hygromycin B (1,000 U/ml). Cells were passaged during drug selection when they reached a density of approximately 106 cells per ml, and after three to five passages, stable cell lines were established.
Assay for IL-3-independent growth.
The ability of Ba/F3 cells to proliferate in the absence of IL-3 was assessed as described previously (6, 26), with some modifications. Drug-resistant cells were grown to a density of approximately 106 cells per ml in complete medium, pelleted, washed with phosphate-buffered saline (PBS; 140 mM NaCl, 27 mM KCl, 1.5 mM KH2PO4, 8.1 mM Na2HPO4), and repelleted. Cells were resuspended in RPMI formulated as described above except that WEHI-conditioned medium was omitted and the serum concentration was reduced to 1% (RPMI/no IL-3). Either 2 × 106 or 5 × 106 cells were seeded into 10 ml of RPMI/no IL-3 and incubated at 37°C, and viable cells were counted in a hemocytometer by trypan blue exclusion each day thereafter.
Metabolic labeling.
C127 cells at 70 to 80% confluence in 10-cm-diameter dishes were rinsed twice in PBS and then incubated for 1 h in leucine-free medium (MEM Select-Amine kit; Gibco). Cells were then incubated for 5 h in 3 ml of leucine-free minimal essential medium containing 16 μCi of [14C]leucine (Amersham) per ml. For harvest, cells were rinsed twice with cold PBS supplemented with 1 mM phenylmethylsulfonyl fluoride (PMSF) and 10 mM iodoacetamide (to prevent postextraction dimer formation). Cells were lysed in 1 ml of cold radioimmunoprecipitation assay (RIPA) buffer (20 mM morpholinepropanesulfonic acid [MOPS; pH 7.0], 150 mM NaCl, 1 mM EDTA, 1% Nonidet P-40, 1% deoxycholate, 0.1% sodium dodecyl sulfate [SDS]) containing 1 mM PMSF, 10 mM iodoacetamide, 10 μg of aprotinin/ml, and 10 μg of leupeptin/ml and then incubated for 20 min on ice. Lysates were cleared by centrifugation at 14,000 × g for 30 min at 4°C and stored at −70°C. To detect 14C-labelled E5 protein, after immunoprecipitation and electrophoresis (see below), the gel was dried and visualized by using a PhosphorImager (Molecular Dynamics).
Immunofluorescence.
COS7 cells grown to 50% confluence on glass coverslips were infected at a multiplicity of ∼1 with BPV/SV40 recombinant viruses containing the wild-type or various mutant E5 genes. Three days after infection, the cells were washed with PBS, fixed with 4% paraformaldehyde in PBS for 20 min and incubated for 20 min in PBS containing 0.01% saponin–0.1% bovine serum albumin (PBS/sB). The cells were then incubated for 1 h with anti-E5 antiserum diluted 1:500 in PBS/sB, washed three times with PBS/sB, and incubated for 45 min with Alexa 488-conjugated goat anti-rabbit antibody (Molecular Probes). The cells were counterstained with a 1:1,000 dilution of 4′,6-diamidino-2-phenylindole hydrochloride (DAPI) in PBS/sB and washed three times with PBS/sB. The coverslips were mounted on slides with Gel/Mount (Biomeda), and fluorescence photomicrographs were obtained.
Protein extracts and immunoprecipitation.
C127 extracts were prepared as described previously (6, 25, 26) in RIPA buffer or CHAPS buffer {15 mM 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate [CHAPS], 30 mM NaCl, 1 mM EDTA, 50 mM Tris-HCl, [pH 7.4]}, both containing protease inhibitors and 2 mM Na3VO4. Extracts of Ba/F3 cells were prepared as described previously (6) in cold RIPA or CHAPS buffer containing vanadate and protease inhibitors.
The PDGF β receptor was immunoprecipitated as previously described (25) by adding 1 μl of anti-PR-C3a antibody (which recognizes the C-terminal 13 amino acids of the receptor) per 100 μg of protein extract. Immunoprecipitation of the E5 protein and associated PDGF β receptor was performed as described previously (25, 26) by adding 1 μl of anti-E5 (which recognizes the 16 C-terminal amino acids of the E5 protein) per 100 μg of RIPA (for E5 immunoblotting) or CHAPS (for coimmunoprecipitation assays) protein extract.
Immunoprecipitates were collected with 60 μl of a 1:1 suspension of protein A-Sepharose beads in Tris-buffered saline (10 mM Tris-HCl [pH 7.4], 165 mM NaCl) containing 10% (wt/vol) bovine serum albumin and washed three to five times with NET-N (20 mM Tris-HCl [pH 8.0], 100 mM NaCl, 1 mM EDTA, 0.5% Nonidet P-40) buffer.
Electrophoresis and immunoblotting.
Samples were boiled in 2× Laemmli sample buffer with β-mercaptoethanol, except for the metabolically labelled samples, which were boiled in sample buffer without any reducing agents. Samples were then electrophoresed in 7.5 or 15% polyacrylamide gels containing SDS, for PDGF β receptor or E5 protein, respectively. Phosphotyrosine, PDGF β receptor, and E5 immunoblotting was performed as described previously (26) by using a 1:500 dilution of antiphosphotyrosine monoclonal antibody 4G10 (Upstate Biotechnology, Inc.), a 1:500 dilution of anti-PR-C3a, or a 1:500 dilution of anti-E5 antiserum, respectively. Proteins were detected with 125I-protein A (ICN) and visualized by using a PhosphorImager. The band corresponding to the mature, tyrosine-phosphorylated PDGF β receptor was quantitated by using ImageQuant software and compared to those of cells transformed by the wild-type E5 protein and analyzed in parallel. The average levels of receptor tyrosine phosphorylation in multiple independent cell lines for each mutant (including Q17Q, a reconstructed wild-type E5 protein) are shown in Table 1.
Molecular modeling.
Conformational searches of the transmembrane region of all 19 mutant E5 dimers were performed to identify likely structures of the mutant E5 proteins. The algorithm and parameters used were the same as those described previously (30), including the sequence length, the helix-helix separation (11.5 Å), and the clustering cutoffs.
The PDGF β receptor transmembrane domain was modeled as a canonical α-helix and manually docked in an antiparallel orientation to the E5 dimer with wild-type glutamine, glutamic acid, or serine at position 17. Docking was carried out using the program MidasPlus. The E5-PDGF β receptor transmembrane helix complex was subjected to 300 rounds of minimization, using the program XPLOR (3). The parameter and topology sets used were modified versions of PARAM19 and TOPH19 (2, 8). The first 100 cycles of Powell minimization were implemented to remove steric clashes. To find the optimal alignment of the two proteins, rigid body minimization was executed for the second 100 cycles of minimization. Another 100 rounds of Powell minimization were added to converge on a low energy structure. Intrahelical restraints were applied during the minimization to maintain helical secondary structure. Restraints were added between residues at position 17 of each E5 monomer to model interhelical hydrogen bonding at position 17, which was predicted by previous computational searches on the E5 dimer (30). Finally, interhelical E5-PDGF β receptor restraints were applied between position 17 and threonine 513 and between aspartate 33 and lysine 499 to establish the proposed E5-receptor contacts.
RESULTS
Construction of the mutants.
Saturation mutagenesis of position 17 in the E5 gene was performed by using codon cassette mutagenesis (14). This method utilized a set of 11 universal mutagenic oligodeoxyribonucleotide cassettes which were temporarily inserted at position 17 of the E5 gene. Subsequently, most of the cassette was removed, leaving three-base cohesive overhangs that were ligated to yield the desired codon substitution. The mutants were subcloned into a retroviral vector and into a BPV/SV40 recombinant virus vector, and virus stocks were produced. Mutant genes encoding all possible substitutions at position 17 were obtained and analyzed.
Studies with C127 cells.
To gain a comprehensive picture of the role of the amino acid at position 17 in productive interaction between the E5 protein and the PDGF β receptor and in cell transformation, we examined the biological activities and biochemical properties of all possible position 17 mutants in C127 mouse fibroblasts. To assess the transforming activity of the mutants, focus-formation assays were performed by infecting cells with retroviruses expressing the mutant or wild-type E5 protein. The number of foci obtained with each viral stock was normalized to the number of drug-resistant colonies that developed from a portion of the infected cells, in order to correct for differences in titers of the viral stocks. In addition, stable cell lines for biochemical analysis were established by pooling drug-resistant colonies.
(i) Transforming activity of the position 17 mutants in C127 cells.
A wide range of transformation activities was seen in focus-formation assays with the position 17 mutants (Table 1). Most E5 mutants with nonpolar hydrophobic substitutions, namely, alanine, glycine, leucine, valine, isoleucine, and phenylalanine, lacked significant transforming ability. In the three infections averaged for the phenylalanine mutant, it induced foci at a moderate level in one experiment, but averaged less than 5% wild-type activity in two additional experiments. In addition, in two transfection experiments with the mutant cloned in a different vector (not included in the average shown in the table), it failed to induce foci. Moreover, a stable cell line expressing this mutant was morphologically flat. Thus, we conclude that the phenylalanine mutant has very little transforming activity and that the 17% level shown in the table is misleadingly high. The transforming activity of the rest of the mutants ranged from completely defective to hypertransforming compared to that of the wild type. The cysteine, tyrosine, proline, and arginine mutants displayed little, if any, focus-forming activity, whereas the lysine, aspartic acid, methionine, and threonine mutants possessed significant activity that was clearly reduced compared to that of the wild type. The tryptophan, asparagine, and histidine mutants transformed cells efficiently, and the serine and glutamic acid mutants reproducibly displayed approximately two to three times the focus-forming activity of the wild-type E5 protein. There was an excellent correlation between the transforming activities of the various mutants as measured by focus formation and by the morphology of stable cell lines expressing these mutants (data not shown; see also Fig. 8). These results confirmed that the identity of the residue at position 17 of the E5 protein is important in C127 cell transformation and highlighted the ability of several amino acids to substitute for the glutamine at position 17 and allow efficient C127 cell transformation. As discussed later, one feature common to all the amino acids that allowed transformation is their potential to form hydrogen bonds.
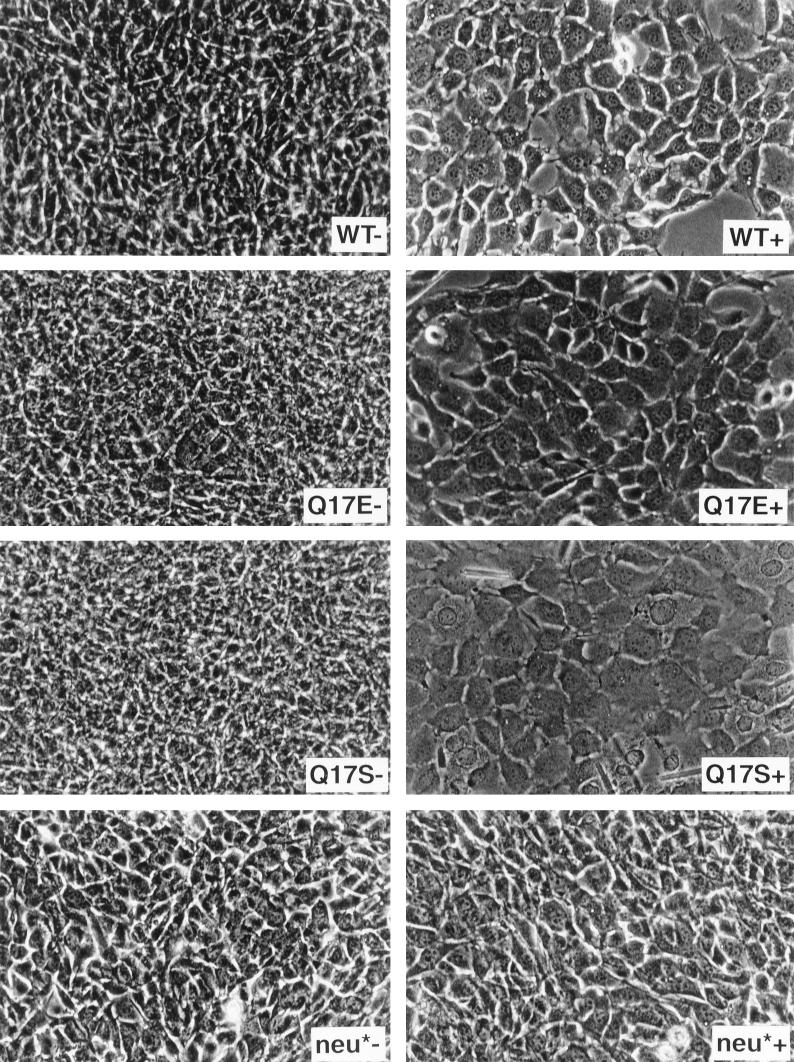
FIG. 8.
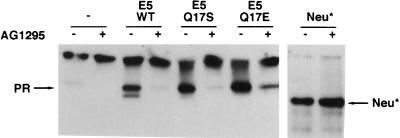
Photomicrographs of C127 cells in the presence or absence of a PDGF receptor-specific kinase inhibitor. Transformed C127 cells stably expressing wild-type E5 protein, the glutamic acid or serine E5 mutant, or the neu* oncoprotein were seeded in 24-well plates and grown in the presence (+) or absence (−) of AG1295 for several days and then photographed.
(ii) Expression, localization, and dimerization of the position 17 mutants.
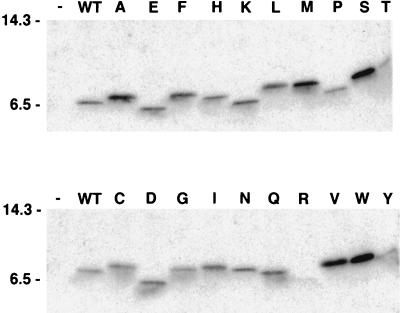
Stable C127 cell lines generated with each of the mutant viruses were analyzed biochemically. The E5 protein was immunoprecipitated from each of the cell lines by using an anti-E5 antiserum which recognizes the hydrophilic carboxy terminus of the protein distal to position 17. After electrophoresis under reducing conditions to disrupt disulfide bonds, the E5 protein was detected by immunoblotting with the same antiserum. As shown in Fig. 1, the great majority of the mutant E5 proteins accumulated to levels similar to those of the wild-type protein or to higher levels. The one exception was the mutant containing arginine, which was present at markedly reduced levels. Thus, with the exception of the arginine mutant, the differences noted above in transforming activities were not due to differences in the level of E5 protein expressed in the cells. It is striking that the mobility of the various E5 mutants differed markedly upon SDS-polyacrylamide gel electrophoresis, even though they all contained the same number of amino acids as the wild-type protein and differed from one another only in the identity of the amino acid at position 17.
FIG. 1.
Expression of the E5 protein in C127-derived cell lines. RIPA extracts of stable C127 cell lines were immunoprecipitated with anti-E5 antibodies. After electrophoresis under reducing conditions and transfer to membranes, the samples were probed with anti-E5 antibodies. The letter at the top of each lane indicates the amino acid at position 17 of each mutant E5 protein. Extracts from cell lines established by infection with the empty retrovirus vector (−) or with retrovirus expressing the wild-type (WT) E5 gene are at the left of each panel. Migration of molecular size markers (in kilodaltons) is shown on the left side. The following abbreviations are used in this and subsequent figures to identify the amino acid at position 17: A, alanine; C, cysteine; D, aspartic acid; E, glutamic acid; F, phenylalanine; G, glycine; H, histidine; I, isoleucine; K, lysine; L, leucine; M, methionine; N, asparagine; P, proline; Q, glutamine; R, arginine; S, serine; T, threonine; V, valine; W, tryptophan; Y, tyrosine.
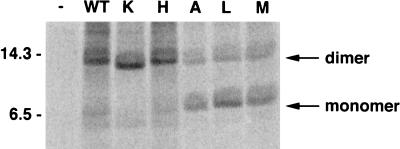
The wild-type E5 protein exists in C127 cells largely as a disulfide-linked dimer, with low amounts of monomer present as well. Here, we examined the relative amounts of monomeric and dimeric E5 protein in the stable C127 cell lines metabolically labelled with [14C]leucine. The E5 protein was immunoprecipitated, subjected to gel electrophoresis under nonreducing conditions, and detected by PhosphorImager analysis. The results for a representative set of mutant proteins are shown in Fig. 2, and the results for all tested mutants are summarized in Table 1. The 14-kDa band disappeared under reducing conditions, leaving only a 7-kDa band (data not shown), indicating that the 14-kDa band is dimeric E5 protein. The ratio of dimeric to monomeric E5 protein varied considerably among the position 17 mutants, but all of the mutant E5 proteins displayed some dimer formation. In general, the mutants with charged residues at position 17, as well as the uncharged asparagine mutant, were similar to the wild-type E5 protein in that greater than 90% of the protein was present as dimer. Other mutants, such as those containing the serine and threonine substitutions, had much lower levels of dimer, and the isoleucine mutant was predominantly monomeric.
FIG. 2.
Effect of the residue at position 17 on E5 dimerization. RIPA extracts of [14C]leucine-labelled C127 cells stably expressing representative E5 mutants were prepared as described in Materials and Methods. Extracts (normalized for incorporation of label) were immunoprecipitated with anti-E5 antibody, and the immunoprecipitates were resolved on a 15% SDS-polyacrylamide gel under nonreducing conditions. The gel was dried, and labeled proteins were detected by using a PhosphorImager. The letter at the top of each lane indicates the amino acid at position 17 of the mutant E5 protein. Extracts from cells infected with empty retrovirus vector (−) or the retrovirus expressing wild-type (WT) E5 protein are also shown, and the positions of monomeric and dimeric wild-type E5 protein are indicated. Sizes of molecular size markers (in kilodaltons) are shown on the left.
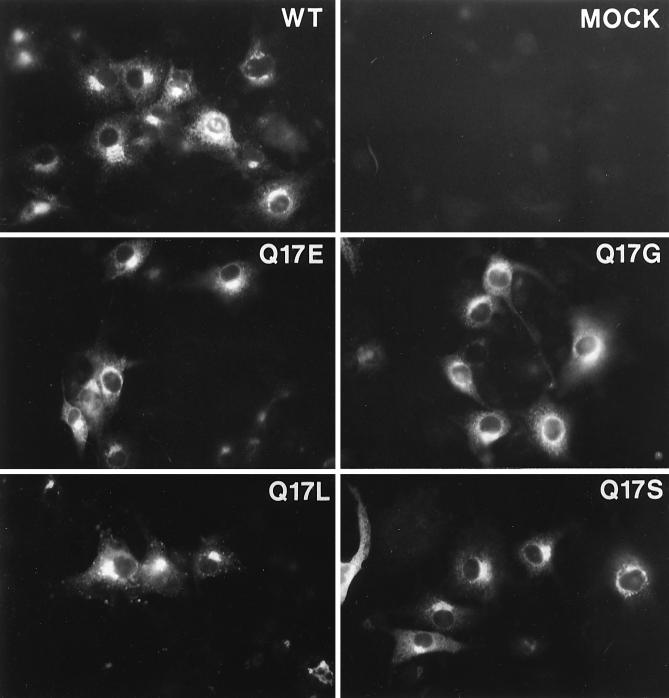
To determine whether aberrant localization of the mutant E5 proteins might account for the observed phenotypes, we used immunofluorescence to determine the intracellular localization of the wild-type protein and representative transformation-competent (glutamic acid and serine) and transformation-defective (glycine and leucine) proteins. We used COS monkey cells for these experiments, because we were not able to detect E5 expression in C127 cells by immunofluorescence. Recombinant BPV/SV40 stocks expressing the various E5 genes were generated and used to infect COS cells. Three days after infection, cells were fixed and permeabilized and E5 protein was detected by indirect immunofluorescence with the anti-E5 antiserum (Fig. 3). There was low background staining in mock-infected cells and bright staining at a predominantly perinuclear location for cells expressing the wild-type E5 protein (panel labelled WT). This staining pattern has been observed previously and is thought to signify Golgi localization (4). All four mutants showed staining indistinguishable from that of the wild type. Thus, the position 17 mutations did not appear to affect localization of the E5 protein.
FIG. 3.
Localization of mutant E5 proteins in COS cells. COS7 cells were mock infected or infected with BPV/SV40 recombinant viruses expressing the wild-type or indicated mutant E5 proteins. After 3 days, the cells were fixed, permeabilized, stained with anti-E5 antibody, and visualized by immunofluorescence.
(iii) PDGF β receptor activation and binding.
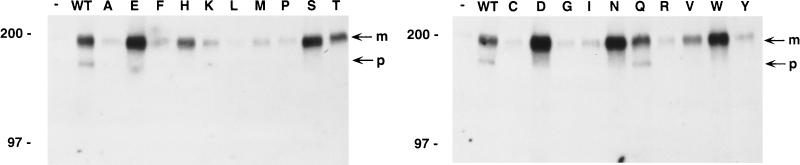
To examine the ability of the various mutants to activate the PDGF β receptor, the receptor was immunoprecipitated from extracts of the stable cell lines by using a PDGF β receptor-specific antiserum and tyrosine phosphorylation of the receptor was determined by immunoblotting with a monoclonal antibody that recognizes phosphotyrosine. Results of a typical experiment are shown in Fig. 4. As described previously, the wild-type E5 protein induced tyrosine phosphorylation of both the slowly migrating form of the PDGF β receptor with mature carbohydrates and a more rapidly migrating intracellular precursor form of the receptor containing incompletely processed carbohydrates (23). In contrast, the transformation-competent mutants preferentially induced tyrosine phosphorylation of the mature form of the receptor and had little effect on the tyrosine phosphorylation of the precursor. The average results from the examination and quantitation of multiple independently derived cell lines expressing each mutant are tabulated in Table 1. Although the results of this assay were somewhat variable and the PDGF β receptor in untransformed cells showed background levels of tyrosine phosphorylation, several trends emerged. All of the E5 mutant proteins which efficiently transformed C127 cells, including the serine mutant, induced abundant receptor tyrosine phosphorylation. In contrast, the transformation-defective mutants induced little or no receptor tyrosine phosphorylation. (Additional, independently derived C127 cell lines expressing the valine mutant, as well as Ba/F3 cells expressing this mutant [see Fig. 10, top], displayed less tyrosine phosphorylation than in the example shown in Fig. 4.) All four of the mutants with intermediate transforming activity induced more receptor phosphorylation than did the most defective hydrophobic mutants. In the analysis of the aspartic acid mutant, one of three cell lines tested (shown in Fig. 4) displayed high receptor tyrosine phosphorylation, but tyrosine phosphorylation in the other cell lines expressing this mutant was approximately one-half of the wild-type levels. In summary, for the mutants with clearcut transformation phenotypes, i.e., displaying either marked transformation defects or robust transforming activity, there was an excellent correlation between transforming activity and the extent of receptor tyrosine phosphorylation.
FIG. 4.
Tyrosine phosphorylation of the PDGF β receptor in C127-derived cell lines. RIPA extracts (500 μg of protein) of C127 cells expressing no E5 protein (−), the wild-type (WT) E5 protein or the position 17 E5 mutants (indicated by the letter at the top of each lane) were precipitated with anti-PDGF receptor antibody. Proteins were resolved by electrophoresis, transferred to membranes, and probed with antiphosphotyrosine antibodies for detection of tyrosine-phosphorylated receptors. Bands corresponding to the mature (m) and precursor (p) forms of the PDGF β receptor are indicated by arrows at the right. Sizes of markers (in kilodaltons) are shown on the left.
FIG. 10.
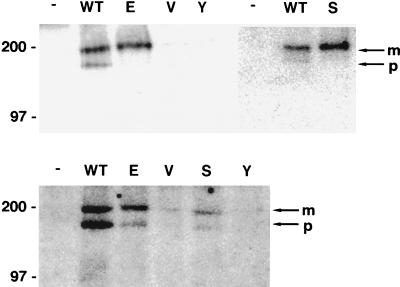
PDGF β receptor tyrosine phosphorylation and complex formation in Ba/F3 cells. (Top) CHAPS extracts (900 μg of protein) from Ba/F3-derived cell lines expressing the murine PDGF β receptor either alone (−) or with the wild-type E5 protein (WT) or the indicated position 17 mutants were immunoprecipitated with anti-PDGF receptor antibody. Proteins were resolved by electrophoresis, transferred to membranes, and probed with antiphosphotyrosine antibody for detection of tyrosine-phosphorylated receptors. The figure is a composite of two independent immunoprecipitations, but the positive (WT) and negative (−) controls processed in parallel were included with both sets of immunoprecipitations. (Bottom) CHAPS extracts (1,500 μg of protein) from Ba/F3-derived cell lines were immunoprecipitated with anti-E5 antibodies. Precipitated proteins were resolved by electrophoresis and transferred, and membranes were probed with anti-PDGF receptor antibody for the detection of receptors associated with the E5 protein. Bands corresponding to the mature (m) and precursor (p) forms of the PDGF β receptor are indicated by arrows at the right, and the sizes of markers (in kilodaltons) are shown on the left.
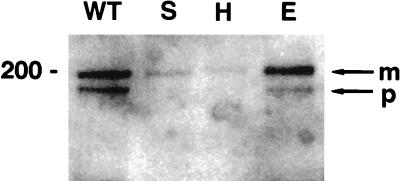
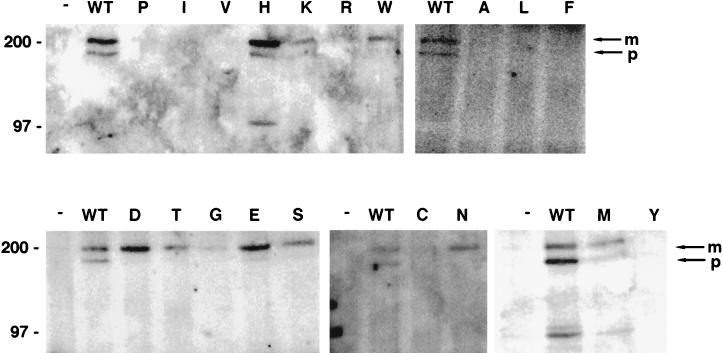
To assess the ability of the mutant E5 proteins to form a stable complex with the PDGF β receptor, coimmunoprecipitation analysis was carried out. Extracts prepared in RIPA buffer were immunoprecipitated with the E5 antiserum, and PDGF β receptor in the immunoprecipitate was detected by immunoblotting with antiserum that recognized the receptor (Fig. 5). No PDGF β receptor was immunoprecipitated with the E5 antiserum from cells infected with the empty vector (data not shown), and both mature and precursor forms of the receptor were coimmunoprecipitated from cells expressing the wild-type E5 protein. The mature form and a relatively small amount of the precursor form of the PDGF β receptor were coimmunoprecipitated from cells expressing the glutamic acid mutant, the mutant that consistently displayed very high transforming activity. However, little PDGF β receptor was coimmunoprecipitated from cells expressing the other mutants, including those, such as the histidine and serine mutants, that efficiently transformed cells and induced receptor tyrosine phosphorylation (Fig. 5 and data not shown). To test the possibility that the stability of receptor-E5 complexes containing these mutants was reduced compared to the stability of complexes containing the wild-type E5 protein or the glutamic acid mutant, extracts were also prepared using the gentler detergent, CHAPS. In contrast to the results obtained with RIPA buffer, the E5 antiserum coimmunoprecipitated the PDGF β receptor from CHAPS extracts prepared from cells expressing several mutants (Fig. 6). Under these conditions, all of the transformation-competent mutants were present in a stable complex with the mature form of the PDGF β receptor, whereas none of the nontransforming mutants was able to bind significant amounts of the PDGF β receptor. The aspartic acid mutant bound more receptor than the other mutants with intermediate transforming activity. Relatively low amounts of the precursor form of the receptor were present in complexes containing the transformation-competent mutant E5 proteins compared to complexes containing the wild-type E5 protein.
FIG. 5.
RIPA extraction buffer preserves complex formation between glutamic acid mutant E5 protein and the PDGF β receptor. RIPA extracts from C127 cells stably expressing various E5 proteins were immunoprecipitated with anti-E5 antibody, and precipitated proteins were resolved by electrophoresis and transferred to membranes. Membranes were probed with anti-PDGF receptor antibody to detect receptor associated with the wild-type E5 protein or the indicated position 17 mutants. Bands corresponding to the mature (m) and precursor (p) forms of the PDGF β receptor are indicated by arrows at the right, and the size of the marker (in kilodaltons) is shown on the left.
FIG. 6.
Complex formation between position 17 mutant E5 proteins and the PDGF β receptor in C127 cells. CHAPS extracts of C127 cells stably expressing various E5 proteins were immunoprecipitated with anti-E5 antibody, and precipitated proteins were resolved by electrophoresis and transferred to membranes. Membranes were probed with anti-PDGF receptor antibody to detect receptors associated with the E5 protein. The letter at the top of each lane indicates the amino acid at position 17 of each mutant E5 protein. Extracts from cells expressing the wild-type (WT) E5 protein or no E5 protein (−) are also shown. Bands corresponding to the mature (m) and precursor (p) forms of the PDGF β receptor are indicated by arrows at the right, and the sizes of markers (in kilodaltons) are shown on the left. The figure is a composite of several independent immunoprecipitations, but positive and negative controls processed in parallel with each set of immunoprecipitations were included.
The results of the phosphotyrosine blots and receptor coimmunoprecipitation analysis are summarized in Table 1 and compared to transforming activity. In general, the correlation between C127 cell transformation, receptor activation, and complex formation was excellent and in no case did transformation occur in the absence of receptor phosphorylation and binding.
(iv) Kinase inhibitor studies.
Our results with the serine mutant are particularly interesting since Sparkowski et al. reported that this mutant transformed mouse fibroblasts in the absence of PDGF β receptor activation (28). To explore explicitly the requirement for PDGF β receptor signaling in C127 cell transformation, we treated transformed cells with AG1295, a specific inhibitor of PDGF receptor tyrosine kinase activity (16). As a control, we also examined C127 cells transformed by neu*, an unrelated activated receptor tyrosine kinase. AG1295 treatment of C127 cells transformed by the wild-type E5 protein caused rapid dephosphorylation of the endogenous PDGF β receptor but did not decrease tyrosine phosphorylation of neu* (Fig. 7). Moreover, AG1295 treatment caused morphologic reversion of E5-transformed cells so that they resembled parental C127 cells, but had no effect on neu*-transformed cells (Fig. 8). Treatment of C127 cells transformed with the glutamic acid or serine mutant also caused receptor dephosphorylation and reversion of the transformed phenotype. Therefore, not only do these mutants bind and activate the endogenous PDGF β receptor in C127 fibroblasts, but sustained receptor kinase activity is required to maintain the transformed phenotype.
FIG. 7.
Tyrosine phosphorylation of the PDGF β receptor in the presence or absence of a PDGF receptor-specific kinase inhibitor. C127 cells stably expressing wild-type E5 protein, the glutamic acid or serine E5 mutant, or empty vector were exposed to AG1295 in DME-10 (+) or to medium alone (−) for 8 h and lysed in RIPA buffer. C127 cells transformed by activated neu* were included as a control. Extracts were immunoprecipitated with anti-PDGF receptor or anti-p185Neu antibody, and precipitated proteins were resolved by electrophoresis and transferred to membranes. Membranes were probed with antiphosphotyrosine antibodies. Bands corresponding to the mature form of the PDGF β receptor and p185Neu* are indicated by arrows.
Studies with Ba/F3 cells.
Ba/F3 cells are murine hematopoietic cells that do not express the endogenous PDGF β receptor or other receptor tyrosine kinases proposed to interact with the E5 protein. These cells are normally dependent on IL-3 for survival and proliferation, but coexpression of the wild-type BPV E5 protein and the PDGF β receptor resulted in complex formation between the two proteins, constitutive tyrosine phosphorylation of the receptor, and IL-3-independent proliferation (6). To examine the role of the amino acid at position 17 in Ba/F3 cells, cell lines expressing no PDGF β receptor or exogenous murine PDGF β receptor were infected with retroviruses carrying the wild-type E5 gene or a mutant gene encoding glutamic acid, serine, valine, or tyrosine at position 17. Following selection for a cotransduced hygromycin resistance gene, pooled drug-resistant cell lines were established. Expression of the E5 proteins in these stable cell lines was confirmed by immunoblotting, as was expression of similar levels of PDGF β receptor in the appropriate cell lines (data not shown).
(i) Transformation of Ba/F3 cells by the position 17 mutants.
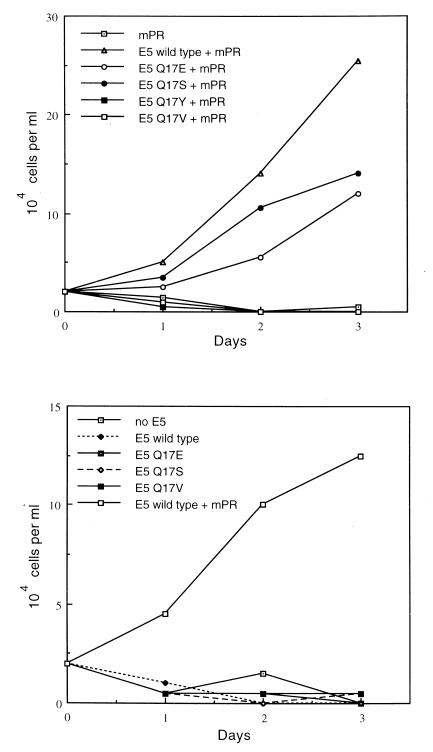
Neither the wild-type E5 protein nor any of the mutants supported IL-3-independent proliferation in Ba/F3 cells not expressing the PDGF β receptor, whereas IL-3-independent proliferation occurred in cells coexpressing the PDGF β receptor and the wild-type E5 protein (Fig. 9, bottom panel). When exogenous PDGF β receptor was expressed in these cells, the glutamic acid and the serine substitution mutants, which efficiently transformed C127 cells, supported IL-3 independence, whereas two transformation-defective mutants, the valine and the tyrosine substitutions, did not (Fig. 9, top panel). Thus, expression of the PDGF β receptor was required for transformation of Ba/F3 cells by the serine mutant, as well as by the wild-type E5 protein and the glutamic acid mutant. Treatment with AG1295, the PDGF receptor kinase inhibitor, prevented IL-3-independent proliferation of cells coexpressing the PDGF β receptor and either the wild-type E5 protein or the serine mutant but had no effect on the growth of these cells in the presence of IL-3 (data not shown).
FIG. 9.
Proliferative effects of wild-type and mutant E5 proteins in Ba/F3 cells. Ba/F3 cells expressing either the murine PDGF β receptor (mPR) (top panel) or vector alone (bottom panel) were infected with retroviruses encoding the indicated mutant or wild-type E5 genes or with the empty vector. The resulting cell lines were seeded in medium lacking IL-3, and viable cells were counted each day by trypan blue exclusion. For comparison, the bottom panel also shows cells coexpressing the wild-type E5 protein and the PDGF β receptor.
(ii) Biochemical analysis of Ba/F3 cells.
To further characterize the interaction of the E5 protein and the PDGF β receptor in Ba/F3 cells, extracts were prepared from cells grown in the presence of IL-3. As shown in Fig. 10, the glutamic acid and serine mutants, which supported IL-3-independent growth when coexpressed with the PDGF β receptor, led to tyrosine phosphorylation of the mature form of the PDGF β receptor (top panel) and formed a stable complex with the receptor (bottom panel). In contrast, the two defective mutants, valine and tyrosine, did not bind the PDGF β receptor or lead to its tyrosine phosphorylation. Thus, there was an excellent correlation between the ability of mutants to transform Ba/F3 cells and to bind to and activate the mature form of the PDGF β receptor in these cells.
Molecular modeling.
To explore the structural basis for the activities of the various position 17 mutants, computational searches were performed of the E5 dimer with different substitutions at position 17. For the wild-type E5 protein, two low-energy dimer structures are found through computational searches (30). In both conformations, the E5 dimer is formed by two long transmembrane α-helices that pack together in a left-handed coiled-coil geometry. The dimer is stabilized largely by van der Waals interactions along the interface. In one conformation, the side chain of glutamine 17 is directed away from the helix interface, while in the second conformation glutamine 17 is packed in the interface, forming interhelical hydrogen bonds. When other residues were substituted at position 17, these two conformations were frequently found in the computational searches (data not shown), indicating that the residue at position 17 did not make the dominant contribution to dimer stability. These results agree with the observation that E5 dimerization was not completely disrupted in the glutamine 17 mutants.
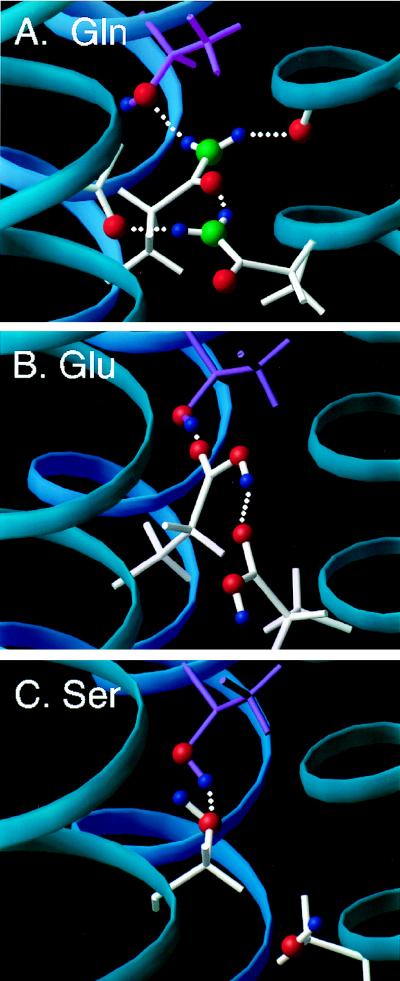
To evaluate the ability of the residue at position 17 and aspartate 33 to interact directly with threonine 513 and lysine 499 of the PDGF β receptor, we docked the PDGF β receptor transmembrane domain to both of the structural models of the E5 dimer found previously. The transmembrane portion of the PDGF β receptor was modeled as a canonical α-helix, oriented to form favorable interactions with residues on the E5 dimer, and energy was minimized. It was possible to obtain simultaneous aspartate 33-lysine 499 and glutamine 17-threonine 513 interactions with the glutamine oriented either away from or in the dimer interface (Fig. 11A and data not shown). Figure 11A presents a model of the E5 dimer in which glutamine 17 is packed in the interface and interacts with the PDGF β receptor transmembrane domain. The side chain of each glutamine can form three distinct hydrogen bonds and thus has the potential to hydrogen bond across the E5 dimer interface and to threonine 513 of the PDGF β receptor. In the model shown in Fig. 11A, each glutamine forms two hydrogen bonds across the E5 dimer interface, one to the other glutamine and the other to a backbone carbonyl oxygen on the opposite E5 monomer, for a total of three hydrogen bonds stabilizing the E5 dimer. The remaining functional group on each glutamine forms a hydrogen bond with the threonine side chain hydroxyl group of a different PDGF β receptor molecule. The side chain of glutamic acid also has a hydrogen bond donor (C—OH) and an acceptor (C⩵O) and can also interact with the other E5 monomer across the E5 interface as well as with threonine 513. In Fig. 11B, interhelical hydrogen bonding restraints were placed between the glutamic acid side chain and the threonine β-hydroxyl proton. The two glutamic acid side chains are hydrogen bonded to one another, leaving a free functional group on each glutamic acid to hydrogen bond to threonine 513 on a different PDGF β receptor molecule. In the case of serine, which has a smaller side chain than glutamine and glutamic acid, it is also possible to dock the transmembrane domain of the PDGF β receptor and form interhelical hydrogen bonds with threonine 513 (Fig. 11C). In the calculations for the mutant E5 dimer alone, the serine side chain is hydrogen bonded back to the i-4 carbonyl of the same helix (data not shown), whereas it rotates outward in the PDGF β receptor-E5 complex to hydrogen bond with the receptor threonine. Thus, in the docked models, each of the three residues that support PDGF receptor complex formation and transformation can form hydrogen bonds with threonine 513 of the PDGF β receptor.
FIG. 11.
Models of the E5 dimer with residue 17 interacting with threonine 513 in the transmembrane domain of the PDGF β receptor. To simplify the figure, only one PDGF β receptor transmembrane helix (dark blue) docked to the two helices of E5 dimer (light blue) is shown. Threonine 513, depicted in purple, is the uppermost residue in each panel, shown hydrogen bonded to glutamine (A), glutamic acid (B), and serine (C). Hydrogen bonds are denoted by dotted lines, and key oxygens (red), nitrogens (green), and protons (blue) are color coded.
DISCUSSION
Our experiments determined the effect of all possible amino acids at position 17 on the ability of the E5 protein to bind to and activate the PDGF β receptor and to transform cells. In both C127 fibroblasts and Ba/F3 cells, all of the E5 mutant proteins that transformed cells formed complexes with the receptor and induced receptor autophosphorylation, whereas the transformation-defective mutant proteins did not efficiently bind or activate the receptor. The wild-type and transformation-competent mutant E5 proteins induced growth factor independence in Ba/F3 cells only when coexpressed with exogenous PDGF β receptor. Furthermore, a PDGF receptor-specific kinase inhibitor reduced receptor tyrosine phosphorylation, led to reversion of the transformed phenotype in C127 cells, and prevented IL-3-independent growth in Ba/F3 cells. Thus, the wild-type and transformation-competent mutant E5 proteins require the presence and sustained activation of the PDGF β receptor to transform cells. These results confirm the importance of position 17 of the E5 protein in PDGF β receptor binding and activation, imply that complex formation with the E5 protein is required for receptor activation, and provide compelling evidence that the PDGF β receptor is the primary target of the E5 protein.
The E5 protein also binds to a 16-kDa transmembrane subunit of the H+-ATPase in some cell types (e.g., see reference 10). We and others have not been able to detect this interaction in C127 cells (28). Furthermore, in mouse NIH 3T3 cells, the ability of mutant E5 proteins to bind this protein shows no apparent correlation with transforming efficiency and appears to require only a positive charge at position 17 and a negative charge on the ATPase subunit (28).
These experiments extend the work of two other groups. Meyer et al. (20) examined the focus-forming efficiencies of several position 17 mutants in NIH 3T3 cells. Since transformation appeared to require a residue at position 17 capable of forming hydrogen bonds, they speculated that the main function of glutamine 17 was to form interhelical hydrogen bonds to stabilize the E5 dimer. However, E5 expression and dimerization by these mutants were not assessed, nor were PDGF β receptor binding and activation examined. Sparkowski et al. analyzed a panel of position 17 mutants in NIH 3T3 and C127 cells and reported that most of the transformation-competent mutants induced PDGF β receptor phosphorylation without binding the receptor and that the serine mutant transformed cells without inducing PDGF receptor activation (27, 28). In addition, they saw no effect of the position 17 mutations on E5 dimerization. Several factors are likely to account for the differences from our results. First, the basal level of phosphorylation of the mature form of the receptor was high in the experiments of Sparkowski and colleagues, precluding further analysis of its phosphorylation and focusing their attention on the immature form of the receptor (28). However, in our experiments, the active position 17 mutant E5 proteins were impaired in their ability to bind and induce autophosphorylation of the immature form of the receptor, so conclusions about the activities of various mutant E5 proteins cannot be based on study of this receptor form alone. The basis for this impaired interaction of the mutants with the precursor form of the receptor is not known. Second, we found that the stringent buffer RIPA disrupts complexes containing most transformation-competent mutants, whereas the gentle lysis buffer CHAPS preserves these interactions. Thus, a possible explanation for the findings of Sparkowski et al. (28) was the use of extraction buffers that disrupted complexes containing these mutants. Finally, they constructed their position 17 substitutions in a mutant, epitope-tagged version of the E5 protein (27), whereas we analyzed the authentic E5 protein, differing from the wild-type protein only by the identity of the amino acid at position 17. It is possible that the presence of the epitope affected the properties of the E5 mutants.
In the experiments reported here, we constructed and analyzed all possible position 17 E5 mutants to gain a comprehensive picture of the role of the residue at this position. The residue at position 17 affected the ability of the E5 protein to dimerize and form stable complexes with the PDGF β receptor. Representative mutants localized normally in cells, and almost all of the mutant E5 proteins accumulated in cells, suggesting that the mutations did not affect appropriate partitioning of the E5 protein into the hydrophobic membrane environment. However, the transformation-defective arginine mutant was present in low levels. It is possible that the arginine side chain was unable to deprotonate to form the neutral species and partition into the membrane, resulting in decreased stability of this mutant.
Previous analyses of mutants with substitutions at the cysteines, which mediate E5 dimerization, imply that dimerization is required for complex formation and transformation (13, 20, 22), and we have proposed that dimer formation is required to generate the binding site on the E5 protein for the PDGF β receptor (30). The results reported here provide new insight into the role of dimerization in transformation and the role of position 17 in dimer formation. In general, the mutants with strongly polar residues at position 17 showed high levels of dimer, similar to those of the wild-type protein, whereas the residues with less-polar side chains, including serine and threonine, resulted in higher levels of monomer (Table 1). However, all the position 17 mutants formed some dimers and there was no strict correlation between the extent of dimer formation and transforming activity. For example, the serine mutant, which transformed cells very efficiently, showed intermediate levels of dimer, similar to those of most nontransforming mutants. This shows that relatively low levels of dimer are sufficient for receptor binding and activation and suggests that the transformation defects of various mutants are not due to the inability of these mutants to dimerize efficiently. However, it is possible that the position 17 mutations caused local perturbations in the structure of the E5 dimer that affected receptor binding and activation.
The residue at position 17 may influence complex formation, and hence transformation, by directly affecting interactions that stabilize the E5-PDGF β receptor complex, as well as by affecting dimerization of the E5 protein. All of the mutants containing hydrophobic residues with side chains unable to form hydrogen bonds were defective for transformation, a result consistent with the proposed hydrogen bond between the residue at position 17 and threonine 513 in the transmembrane domain of the PDGF β receptor. The tyrosine, proline, and cysteine mutants, although capable of hydrogen bonding, were also defective for complex formation and transformation. Proline and tyrosine are largely excluded from the hydrophobic transmembrane region of proteins having single-membrane-spanning helices, and tyrosine occurs preferentially in the region of the polar headgroups in membrane proteins (18). Proline disrupts the regular hydrogen bonding pattern along the helix backbone, causing kinks in transmembrane helices (31), and tyrosine substitutions in the transmembrane domain of glycophorin A disrupted dimerization even when located at positions not in the helix interface (19). Thus, tyrosine and proline may interfere with proper E5 dimerization or interaction with the PDGF β receptor.
Several polar amino acids at position 17 supported complex formation and transformation. Of these, glutamine and glutamic acid appeared to impart the most stability to the E5-receptor complex, based on its ability to withstand RIPA buffer extraction. This may be due to the length and strongly polar nature of these side chains which have great conformational flexibility and facilitate the formation of strong hydrogen bonds. Tryptophan and methionine, residues with substantial hydrophobic character, also allowed transformation. Tryptophan differs from the nontransforming hydrophobic residues in that it has an NH group on the side chain indole ring which is capable of hydrogen bonding. Similarly, methionine is able to hydrogen bond through its side chain sulfur atom (12). For example, there is a hydrogen bond between a hydroxyl group of a threonine residue and a methionine sulfur in the crystal structure of myohemerythrin (11). The aspartic acid mutant displayed intermediate transforming activity even though it bound the receptor well and, at least in some cell lines, induced a high level of receptor tyrosine phosphorylation. It is possible that this mutant induced the formation of aberrant complexes such as, for example, ones in which the sites of receptor phosphorylation differed from the sites in complexes containing the wild-type E5 protein.
Pairwise comparisons of mutants containing similar amino acid substitutions also highlight the importance of hydrogen bonds involving the side chain at position 17. For example, valine and threonine are isosteric β-branched amino acids. The threonine hydroxyl group and each valine side chain methyl group have roughly the same molecular volume, but the hydroxyl-group, unlike the methyl group, is capable of hydrogen bonding interactions. The valine mutant did not form complexes with the PDGF β receptor or lead to transformation, whereas the threonine mutant transformed cells far better than the valine mutant, formed complexes with the receptor, and induced intermediate levels of receptor tyrosine phosphorylation. The different activities of these mutants did not reflect differences in dimerization. Rather, these results strongly argue that hydrogen bonding by the amino acid at position 17 is an essential element in stabilizing the E5-receptor complex. Similarly, the serine mutant transformed at high efficiency, while the cysteine mutant was transformation defective. These different phenotypes do not appear related to dimerization efficiency and are likely to result from differences in the chemical properties of the cysteine sulfhydryl group and the serine hydroxyl group, such as the ability of the oxygen atom in the serine to form stronger hydrogen bonds than the cysteine sulfur atom (15).
The effects of the mutations on dimerization suggest that the position 17 side chain forms contacts between E5 monomers, while the data on complex formation suggest that it forms contacts with the receptor. In both proposed conformations of the wild-type and representative transformation-competent E5 dimers, it was possible to dock aspartate 33 of the E5 protein with lysine 499 of the receptor and the residue at position 17 of the E5 protein with threonine 513 of the receptor. Therefore, these models can account for the ability of the residue at position 17 to influence both E5 dimer formation and complex formation with the PDGF receptor.
In summary, these studies provide a coherent view of the role of glutamine 17 in cell transformation by the E5 protein. This analysis of all possible substitutions at position 17 has revealed an excellent correlation between the ability of these mutants to form a stable complex with the PDGF β receptor, induce receptor activation, and cause transformation. The activities of the individual mutants and molecular modeling studies provide further evidence that the residue at position 17 contributes to transformation by stabilizing the E5 dimer and by interacting with threonine 513 in the PDGF β receptor transmembrane domain.
ACKNOWLEDGMENTS
We thank S. Courtneidge for an initial sample of AG1295; New England Biolabs, Inc., for oligonucleotides used for mutagenesis; P. Irusta, L. Petti, and E. Goodwin for helpful discussions; J. McMenamin-Balano and D. Stern for the neu* retrovirus and anti-neu antibodies; J. Su, E. Caler, and V. Reddy for technical assistance; and J. Zulkeski for assistance in preparing the manuscript.
O.K. was supported in part by an MSTP grant from the NIH. This work was supported by grant CA37157 from the NIH.
REFERENCES
- 1.Bargmann C I, Weinberg R A. Increased tyrosine kinase activity associated with the protein encoded by the activated neu oncogene. Proc Natl Acad Sci USA. 1988;85:5394–5398. doi: 10.1073/pnas.85.15.5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brooks B R, Bruccoleri R E, Olafson B D, States D J, Swaminathan S, Karplus M. CHARMM: a program for macromolecular energy, minimization, and dynamics calculations. J Comp Chem. 1983;4:187–217. [Google Scholar]
- 3.Brünger A T. X-PLOR, version 3.1. New Haven, Conn: Yale University; 1992. [Google Scholar]
- 4.Burkhardt A, Willingham M, Gay C, Jeang K-T, Schlegel R. The E5 oncoprotein of bovine papillomavirus is oriented asymmetrically in Golgi and plasma membranes. Virology. 1989;170:334–339. doi: 10.1016/0042-6822(89)90391-7. [DOI] [PubMed] [Google Scholar]
- 5.Cohen B D, Goldstein D J, Rutledge L, Vass W C, Lowy D R, Schlegel R, Schiller J. Transformation-specific interaction of the bovine papillomavirus E5 oncoprotein with the platelet-derived growth factor receptor transmembrane domain and the epidermal growth factor receptor cytoplasmic domain. J Virol. 1993;67:5303–5311. doi: 10.1128/jvi.67.9.5303-5311.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Drummond-Barbosa D, Vaillancourt R R, Kazlauskas A, DiMaio D. Ligand-independent activation of the platelet-derived growth factor β receptor: requirements for bovine papillomavirus E5-induced mitogenic signaling. Mol Cell Biol. 1995;5:2570–2581. doi: 10.1128/mcb.15.5.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drummond-Barbosa D, DiMaio D. Virocrine transformation. Biochim Biophys Acta. 1997;1332:M1–M17. doi: 10.1016/s0304-419x(96)00034-0. [DOI] [PubMed] [Google Scholar]
- 8.Engh R A, Huber R. Accurate bond and angle parameters for X-ray protein-structure refinement. Acta Cryst. 1991;A47:392–400. [Google Scholar]
- 9.Goldstein D J, Andresson T, Sparkowski J J, Schlegel R. The BPV-1 E5 protein, the 16 kDa membrane pore-forming protein and the PDGF receptor exist in a complex that is dependent on hydrophobic transmembrane interactions. EMBO J. 1992;11:4851–4859. doi: 10.1002/j.1460-2075.1992.tb05591.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goldstein D J, Li W, Wang L-M, Heidaran M A, Aaronson S A, Shinn R, Schlegel R, Pierce J H. The bovine papillomavirus type 1 E5 transforming protein specifically binds and activates the β-type receptor for platelet-derived growth factor but not other tyrosine kinase-containing receptors to induce cellular transformation. J Virol. 1994;68:4432–4441. doi: 10.1128/jvi.68.7.4432-4441.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gregoret L M, Rader S D, Fletterick R J, Cohen F E. Hydrogen bonds involving sulfur atoms in proteins. Proteins Struct Funct Genet. 1991;9:99–107. doi: 10.1002/prot.340090204. [DOI] [PubMed] [Google Scholar]
- 12.Honig B H, Hubbell W L. Stability of “salt bridges” in membrane proteins. Proc Natl Acad Sci USA. 1984;81:5412–5416. doi: 10.1073/pnas.81.17.5412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Horwitz B H, Burkhardt A L, Schlegel R, DiMaio D. 44-amino-acid E5 transforming protein of bovine papillomavirus requires a hydrophobic core and specific carboxyl-terminal amino acids. Mol Cell Biol. 1988;8:4071–4078. doi: 10.1128/mcb.8.10.4071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kegler-Ebo D M, Docktor C M, DiMaio D. Codon cassette mutagenesis: a general method to insert or replace individual codons by using universal mutagenic cassettes. Nucleic Acids Res. 1994;22:1593–1599. doi: 10.1093/nar/22.9.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kollman P, McKelvey J, Johansson A, Rothenberg S. Theoretical studies of hydrogen-bonded dimers. Complexes involving HF, H2O, NH3, HCl, H2S, PH3, HCN, HNC, HCP, CH2NH, H2CS, H2CO, CH4, CF3H, C2H2, C2H4, C6H6, F−, and H3O+ J Am Chem Soc. 1975;97:955–965. [Google Scholar]
- 16.Kovalenko M, Gazit A, Böhmer A, Rorsman C, Rönnstrand L, Heldin C-H, Waltenberger J, Böhmer F-D, Levitzki A. Selective platelet-derived growth factor receptor kinase blockers reverse sis-transformation. Cancer Res. 1994;54:6106–6114. [PubMed] [Google Scholar]
- 17.Lai, C. C., C. Henningson, and D. DiMaio. Bovine papillomavirus E5 protein induces oligomerization and trans-phosphorylation of the platelet-derived growth factor receptor. Submitted for publication. [DOI] [PMC free article] [PubMed]
- 18.Landolt-Marticorena C, Williams K A, Deber C M, Reithmeier R A F. Non-random distribution of amino acids in the transmembrane segments of human type I single span membrane proteins. J Mol Biol. 1993;229:602–608. doi: 10.1006/jmbi.1993.1066. [DOI] [PubMed] [Google Scholar]
- 19.Lemmon M A, Flanagan J M, Treutlein H R, Zhang J, Engelman D M. Sequence specificity in the dimerization of transmembrane α-helices. Biochemistry. 1992;31:12719–12725. doi: 10.1021/bi00166a002. [DOI] [PubMed] [Google Scholar]
- 20.Meyer A N, Xu Y-F, Webster M K, Smith A S, Donoghue D J. Cellular transformation by a transmembrane peptide: structural requirements for the bovine papillomavirus E5 oncoprotein. Proc Natl Acad Sci USA. 1994;91:4634–4638. doi: 10.1073/pnas.91.11.4634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nilson L, DiMaio D. Platelet-derived growth factor receptor can mediate tumorigenic transformation by the bovine papillomavirus E5 protein. Mol Cell Biol. 1993;13:4137–4145. doi: 10.1128/mcb.13.7.4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nilson L A, Gottlieb R, Polack G W, DiMaio D. Mutational analysis of the interaction between the bovine papillomavirus E5 transforming protein and the endogenous β receptor for platelet-derived growth factor in mouse C127 cells. J Virol. 1995;69:5869–5874. doi: 10.1128/jvi.69.9.5869-5874.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Petti L, Nilson L A, DiMaio D. Activation of the platelet-derived growth factor receptor by the bovine papillomavirus E5 transforming protein. EMBO J. 1991;10:845–855. doi: 10.1002/j.1460-2075.1991.tb08017.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Petti L, DiMaio D. Stable association between the bovine papillomavirus E5 transforming protein and activated platelet-derived growth factor receptor in transformed mouse cells. Proc Natl Acad Sci USA. 1992;89:6736–6740. doi: 10.1073/pnas.89.15.6736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Petti L, DiMaio D. Specific interaction between the bovine papillomavirus E5 transforming protein and the β receptor for platelet-derived growth factor in stably transformed and acutely transfected cells. J Virol. 1994;68:3582–3592. doi: 10.1128/jvi.68.6.3582-3592.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Petti L M, Reddy V, Smith S O, DiMaio D. Identification of amino acids in the transmembrane and juxtamembrane domains of the platelet-derived growth factor receptor required for productive interaction with the bovine papillomavirus E5 protein. J Virol. 1997;71:7318–7327. doi: 10.1128/jvi.71.10.7318-7327.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sparkowski J, Anders J, Schlegel R. Mutation of the bovine papillomavirus E5 oncoprotein at amino acid 17 generates both high- and low-transforming variants. J Virol. 1994;68:6120–6123. doi: 10.1128/jvi.68.9.6120-6123.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sparkowski J, Mense M, Anders J, Schlegel R. E5 oncoprotein transmembrane mutants dissociate fibroblast transforming activity from 16-kilodalton protein binding and platelet-derived growth factor receptor binding and phosphorylation. J Virol. 1996;70:2420–2430. doi: 10.1128/jvi.70.4.2420-2430.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Staebler A, Pierce J H, Brazinski S, Heidaran M A, Li W, Schlegel R, Goldstein D J. Mutational analysis of the β-type platelet-derived growth factor receptor defines the site of interaction with the bovine papillomavirus type 1 E5 transforming protein. J Virol. 1995;69:6507–6517. doi: 10.1128/jvi.69.10.6507-6517.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Surti T, Klein O, Ascheim K, DiMaio D, Smith S O. Structural models of the bovine papillomavirus E5 protein. 1998. Proteins Struct. Funct. Genet., in press. [PubMed] [Google Scholar]
- 31.Williams K A, Deber C M. Proline residues in transmembrane helices: structural or dynamic role? Biochemistry. 1991;30:8919–8923. doi: 10.1021/bi00101a001. [DOI] [PubMed] [Google Scholar]