Abstract
A panel of cytolytic T lymphocyte (CTL) clones was isolated from metastases and blood samples of a melanoma patient vaccinated with MAGE-3.A1-pulsed autologous dendritic cells. We report here the identification of a new antigen encoded by the MAGE-C2 cancer-germline gene. This antigen is recognized by some of these CTL on HLA-B*4403. The sequence of the peptide is SESIKKKVL. It is processed in various melanoma cell lines expressing MAGE-C2 and HLA-B*4403. Because of the expression pattern of gene MAGE-C2, this new antigen is strictly tumor-specific and could therefore be used for peptide-based antitumoral vaccination.
Keywords: Antigenic peptide, Cancer immunotherapy, Cytolytic T lymphocytes, MAGE, Melanoma
Introduction
Among antigens recognized on tumors by autologous T lymphocytes [22], antigens encoded by cancer-germline genes, such as members of the MAGE gene family, represent an interesting category. These genes are expressed in tumors of various histological types but not in normal tissues except male germline cells [23]. Because germline cells do not express MHC molecules [9], they do not display antigenic peptides on their surface. Therefore peptides encoded by cancer-germline genes are strictly tumor-specific and of particular interest for cancer immunotherapy. Based on the use of defined antigens encoded by cancer-germline genes, various clinical trials of therapeutic vaccination have been initiated in melanoma patients. So far, less than 10% of patients have shown an objective clinical response, irrespective of the antigen or the vaccination modality used [2, 18].
Melanoma patient LB2586 received repeated injections of autologous mature dendritic cells pulsed with the MAGE-A3168–176 peptide presented by HLA-A1 for the treatment of multiple in transit skin metastases of the right lower leg. The patient displayed a mixed tumor response involving regression of most and progression of a limited number of cutaneous metastases. A monoclonal cytolytic T lymphocyte (CTL) response to the vaccine antigen was documented in the blood of the patient (J. Carrasco et al., manuscript in preparation). Tumor cell cultures were established from cutaneous metastases that were resected from this patient during the course of the therapeutic vaccination. These cells were used to stimulate autologous tumor infiltrating lymphocytes. Many CTL clones were obtained that lysed specifically the autologous melanoma cells but did not recognize the MAGE-3.A1 vaccine antigen. Some of these tumor-derived CTL clones were found to recognize a new antigen encoded by gene MAGE-C2, which is also recognized by CTL derived from the blood of this patient.
Materials and methods
Patient and cell lines
Melanoma patient LB2586 (HLA-A*0101, -A*2902, -B*27052, -B*4403, -Cw*0202, -Cw*1601) was vaccinated repeatedly with mature, monocyte-derived autologous dendritic cells pulsed with the HLA-A1-restricted MAGE-A3 peptide EVDPIGHLY168–176 [6] and the HLA-DP4-restricted MAGE-A3 peptide KKLLTQHFVQENYLEY243–258 [19]. The detailed vaccination schedule and the analysis of the response will be reported elsewhere (J. Carrasco et al., manuscript in preparation). Cultures of tumor cells were derived from two progressing cutaneous metastases, one resected after two cycles of six vaccinations and the other after two additional cycles. These cultures were established according to the procedure described by Brasseur [3]. Cultured melanoma cells were rapidly obtained in large numbers from the former metastasis, further referred to as LB2586-MEL.B, and could be easily propagated for several passages but a permanent line could not be established. Cultures from the other metastasis gave rise to melanoma line LB2586-MEL.G. Melanoma cells, EBV-transformed B cells and K562 cell line were cultured in Iscove’s modified Dulbecco’s medium (IMDM) (Invitrogen, Carlsbad, CA, USA) supplemented with 10% fœtal calf serum (FCS) (Invitrogen). COS-7 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Invitrogen) supplemented with 10% FCS. WEHI 164 clone 13 cells were cultured in RPMI 1640 medium (Invitrogen) supplemented with 5% FCS. Media were supplemented with l-arginine (116 mg/l), l-asparagine (36 mg/l) and l-glutamine (216 mg/l). Lymphocytes were cultured in IMDM supplemented with 10% human serum (HS) and 5 × 10−5 M 2-mercaptoethanol.
Anti-tumor CTL clones
Surgically resected progressing cutaneous metastases were used as a source of tumor infiltrating lymphocytes (TIL). A fresh tumor sample was chopped into small pieces that were distributed into wells in 1.5 ml of IMDM containing 1% autologous serum, recombinant human IL-2 (50 U/ml) (Chiron, Amsterdam, The Netherlands), gentamycin (15 μg/ml) and nystatin (200 U/ml). After 14 days, the TIL were pooled in three groups of 4 × 105 lymphocytes and incubated with 2 × 105 irradiated (10,000 rads) LB2586-MEL.B tumor cells pretreated for 48 h with IFN-γ (100 U/ml) (Peprotech, Rocky Hill, NJ, USA), in 2 ml IMDM containing 10% HS, IL-2 (50 U/ml), IL-4 (1.5 ng/ml) (produced in our laboratory) and IL-7 (5 ng/ml) (R&D Systems, Minneapolis, MN, USA). Lymphocytes from the mixed lymphocyte tumor cultures (MLTC) were restimulated at days 7 and 14 in the same conditions. At day 20, they were cloned by limiting dilution in microwells containing irradiated LB2586-MEL.B tumor cells (5,000/well) pretreated with 100 U/ml IFN-γ for 48 h, irradiated allogenic LG2-EBV feeder cells (3 × 104/well) and the cytokine cocktail. Restimulations were done at days 7 and 14 and a 51Cr lysis assay was performed at day 20. CTL clones specific for the tumor were amplified in 2 ml cultures containing irradiated LB2586-MEL.B tumor cells pretreated with IFN-γ (2 × 105/well) and irradiated LG2-EBV feeder cells (106/well) in IMDM containing 10% HS and the cytokine cocktail. IFN-γ treatment of tumor cells upregulated the HLA expression and consistently increased the level of specific lysis in 51Cr assay.
Blood CD8+ lymphocytes purified from thawed PBMC by sorting with anti-CD8 magnetic beads and an autoMACS separator (Miltenyi Biotec, Bergisch Gladbach, Germany) were used to isolate anti-tumor CTL. Lymphocytes were seeded at 200–6,000 cells/well in 48 or 96 V-bottom microwells (Nunc, Roskilde, Denmark) and stimulated by the addition of irradiated (10,000 rads) LB2586-MEL.B cells (5,000/well) pretreated with 100 U/ml IFN-γ in a final volume of 200 μl IMDM supplemented with 10% HS, IDO inhibitor 1-methyl-l-tryptophan (200 μM), IL-2 (50 U/ml), IL-4 (3 ng/ml) and IL-7 (10 ng/ml). The microcultures were restimulated at days 7 and 14 by the addition of irradiated LB2586-MEL.B cells (3,000–5,000/well) and the same cytokine cocktail. Around day 21, aliquots collected from all microcultures were used as effector cells in a cytotoxicity assay, with 51Cr-labeled targets including IFNγ treated LB2586-MEL.B cells, LB2586 EBV-transformed B cells and K562 (1,000 cells/well); a 50-fold excess of unlabeled K562 was added to quench the activity of NK-like effectors. Microcultures displaying a high level of tumor specific cytotoxic activity and prone to be clonal according to the Poisson distribution law were further restimulated in the same conditions and a second lytic assay was performed 2 weeks later in the absence of unlabeled K562 cells. To derive anti-tumor CTL clones from confirmed lytic microcultures, CD8+ cells were sorted and cloned at 1 cell/microwell by use of a FACS Vantage cell sorter (Becton Dickinson, San Jose, CA, USA) and restimulated with irradiated IFN-γ-treated LB2586-MEL.B cells (5 × 103/well), irradiated allogenic LG2-EBV feeder cells (3 × 104/well) in 200 μl medium containing IL-2 (50 U/ml) and IL-7 (10 ng/ml). Cloned CTL were finally grown in 24-well plates (105 cells/well) with irradiated IFN-γ-treated LB2586-MEL.B cells (2 × 105/well), irradiated allogenic LG2-EBV feeder cells (106/well) in 2 ml medium containing IL-2 (50 U/ml) and IL-7 (10 ng/ml).
Cytotoxicity assay
Cytolytic activity was estimated in a chromium release assay as described previously [1]. Briefly, CTL were mixed with 1,000 51Cr-labeled target cells; after 4 h, the radioactivity released in the supernatant was measured. For the peptide assay, 51Cr-labeled autologous EBV-transformed B cells (LB2586-EBV) were incubated with serially diluted peptides for 30 min before addition of the CTL. Peptides were synthesized on solid phase Fmoc for transient N-terminal protection and characterized by mass spectrometry. The 9-mer and 11-mer optimal peptides were further purified by rp-HPLC to give >99% purity. Lyophilized peptides were dissolved at 10 mg/ml in dimethylsulfoxide (DMSO) and stored frozen.
TNF release assay
Target cells (2 × 104/microwell) were incubated with CTL (2 × 103) in 100 μl of culture medium supplemented with IL-2 (25 U/ml). After 24 h of coculture, the supernatant was collected and its TNF content was determined by testing its cytotoxic effect on WEHI 164 clone 13 cells as described [5, 20].
Transient transfection of COS-7 cells
Transfection was performed by the Lipofectamine method according to the manufacturer recommendations (Invitrogen). Briefly, COS-7 cells (1.5 × 104) distributed in flat bottom microwells the day before transfection were cotransfected in triplicate with 50 ng of pcDNAI/Amp or pcDNA3 plasmid containing the cDNA of one of the HLAs corresponding to the patient typing, together with 50 ng of a plasmid containing the cDNA of a gene encoding a tumor antigen, in the presence of 1 μl of Lipofectamine in serum-free OPTIMEM medium (Invitrogen) in a final volume of 50 μl. After a 5-h incubation at 37°C, 200 μl of DMEM medium supplemented with 10% FCS was added and incubation continued for 20 h. The transfection medium was discarded and the CTL to be tested for antigen recognition were added (2 × 103/microwell) together with IL-2 (20 U/ml) in a final volume of 100 μl. After 24 h, the supernatants were collected and their TNF content estimated as described above.
RT-PCR assay for expression of gene MAGE-C2
Expression of gene MAGE-C2 in different cell lines was analyzed by reverse transcription-PCR as described, using primers SL102 and SL103 [14].
TCR analysis of CTL clones
TCR Vα and Vβ chains were assessed by reverse transcription-PCR essentially as described [8], except that fluorescently labeled downstream primers (Cα-HEX and Cβ-NED) (Eurogentec, Liège, Belgium) were used, allowing fast and accurate size determination simultaneously for the Vα and Vβ PCR products that could be distinguished by their different fluorescence using the ABI PRISM 3100 Genetic Analyzer and the GeneScan software (Applied Biosystems, Foster City, CA, USA). PCR products were purified and sequenced using the BigDye Terminator Sequencing Ready Reaction Kit (Applied Biosystems) and the ABI PRISM 3100 Genetic Analyzer to obtain a complete identification of the CDR3 region.
Results
Isolation of anti-tumor CTL from a metastasis
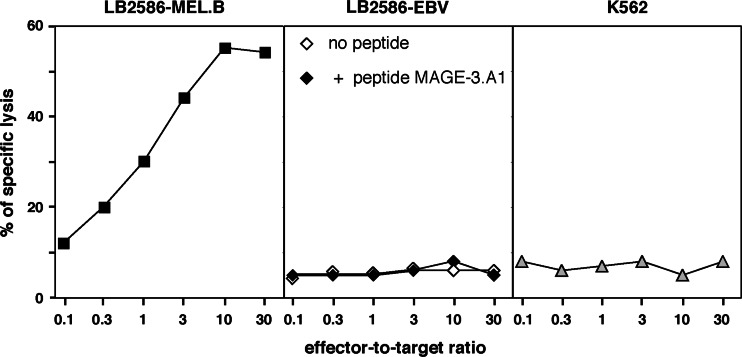
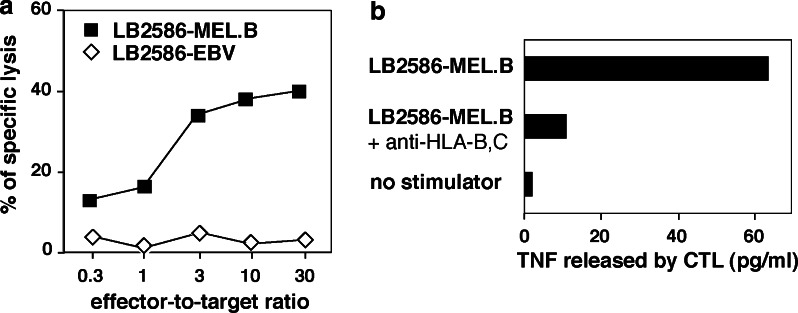
Seven months after the onset of vaccinations with autologous dendritic cells pulsed with the MAGE-3.A1 peptide, a cutaneous metastasis was resected. Part of it was taken for establishment of a tumor cell culture, further referred to as LB2586-MEL.B. Another fragment was chopped into small pieces that were distributed into 20 wells in 1.5 ml culture medium containing autologous serum and IL-2. Two weeks later, lymphocytes which had exited from the tumor pieces and divided, were collected and restimulated in three pools of 4 × 105 cells with the LB2586-MEL.B tumor cells in a MLTC. After 14 days, a 51Cr release assay was performed on each MLTC; Fig. 1 is representative of one of them. It revealed that all three MLTCs contained cells that lysed the autologous tumor cells but did not lyse autologous EBV-transformed B cells and NK target cells K562. Moreover, autologous EBV-B cells pulsed with the vaccine antigen MAGE-3.A1 were not lysed, suggesting that the TIL were not directed against the vaccine antigen. After cloning by limiting dilution, cytotoxic CD8+ clones were obtained. One T cell receptor was observed in several CTL clones derived from two of the three MLTCs, indicating the presence of at least two members of the same clonotype in the tumor. This clonotype is Vβ19-Jβ1.4, with CDR3 sequence tagtTTACGACAGGGCGGAgaaaaa... and Vα9.2-Jα37 with CDR3 sequence tctgGGAGGAtctg... and will be referred to as clonotype A. CTL 2.4 that belonged to this clonotype was able to lyse the autologous LB2586-MEL.B tumor cells but not the autologous EBV-B cells (Fig. 2a). It secreted TNF upon incubation with the autologous tumor cells (Fig. 2b). TNF secretion was inhibited by monoclonal antibody B1.23.2, indicating that the epitope is presented by HLA-B or -C molecules.
Fig. 1.
Lysis of autologous melanoma cells LB2586-MEL.B by TILs maintained in MLTC for 14 days. LB2586-MEL.B melanoma cells were treated with 100 U/ml IFN-γ for 48 h before the test. Autologous EBV-transformed B cell line LB2586-EBV (either pulsed or not with 1 μM MAGE-3.A1 peptide), and K562 cells were used as control targets. The lymphocytes were incubated at various effector-to-target ratios with 1,000 51Cr-labeled targets; unlabeled K562 were added in a 50-fold excess to avoid nonspecific lysis. 51Cr release was measured after 4 h
Fig. 2.
a Specific lysis of autologous melanoma cells LB2586-MEL.B by CTL 2.4. Melanoma cells were treated with 100 U/ml IFN-γ for 48 h before the test. Autologous EBV-transformed B cell line LB2586-EBV was used as control target. 51Cr release was measured after 4 h. b Recognition of autologous melanoma cells LB2586-MEL.B by CTL 2.4 is inhibited by anti-HLA-B,C antibody. CTL 2.4 was cocultured with the IFN-γ-treated autologous melanoma cells in the absence or presence of anti-HLA-B,C antibody clone B1.23.2. After 24 h of coculture, the amount of TNF in the supernatant was measured by its toxicity on WEHI-13 cells
Identification of the antigen recognized by CTL 2.4
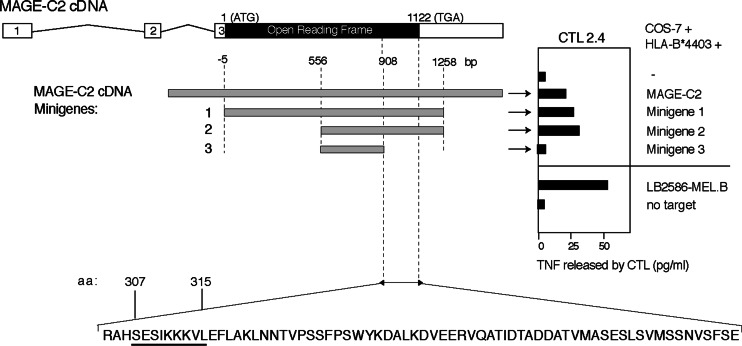
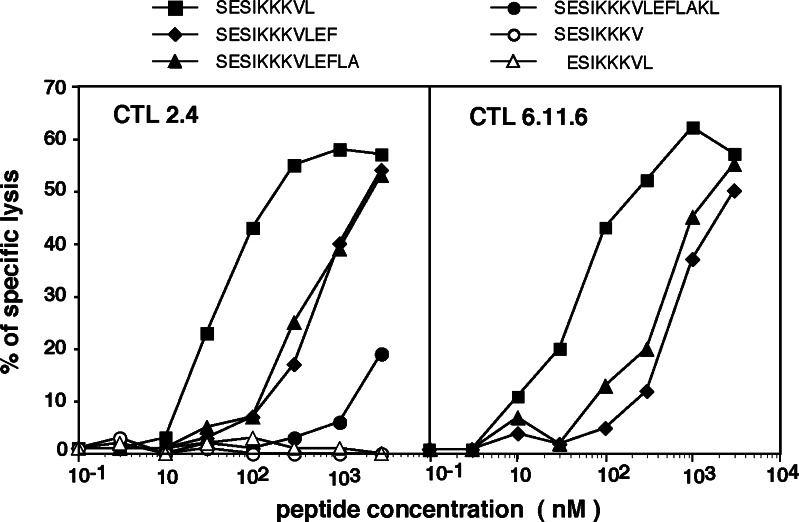
CTL 2.4 was incubated with COS-7 cells transiently cotransfected with plasmids containing cDNA of HLA-B or C corresponding to the patient alleles, together with the cDNA obtained from a set of genes known to encode tumor antigens. Secretion of TNF by the CTL was observed with COS cells cotransfected with cDNAs of HLA-B*4403 and MAGE-C2 (Fig. 3). To delimit the peptide-coding sequence, COS-7 cells were cotransfected with HLA-B*4403 cDNA and three different expression plasmids containing parts of the MAGE-C2 cDNA sequence [16]. The results indicated that the peptide recognized by CTL 2.4 was encoded within the last 211 nucleotides of the MAGE-C2 open reading frame (Fig. 3). Several peptides encoded within this region and bearing the HLA-B*4403 binding motif with E in position 2 and/or Y or F at the carboxyterminus, were synthesized. They were tested for recognition by CTL 2.4 onto autologous EBV-B cells. Peptide307–317 and analogs elongated at the C-terminus were recognized, albeit at a relatively high concentration (Fig. 4). The undecapeptide contained two overlapping candidates, a nonapeptide starting at position 307 and a decapeptide starting at position 308. Only nonapeptide307–315 SESIKKKVL was recognized, with half-maximum lysis obtained at 40 nM (Fig. 4). This nonapeptide appeared to be the optimal peptide, as peptides truncated by one residue either at the C-terminus or the N-terminus were not efficiently recognized.
Fig. 3.
Representation of the MAGE-C2 cDNA and minigenes used to identify the region coding for the antigenic peptide. Exons appear as open boxes, spliced introns as kinky lines, and the open reading frame as a black box. The minigene constructs corresponding to truncated fragments of the MAGE-C2 cDNA obtained by PCR are presented as gray boxes, with nucleotide positions numbered starting from the first ATG of the ORF. These constructs were cotransfected into COS-7 cells with the HLA-B*4403 cDNA. Twenty-four hours later, recognition of the transfected cells by CTL 2.4 was tested in a TNF production assay. The antigenic peptide sequence (see Fig. 4) is underlined
Fig. 4.
Lysis by CTL 2.4 and CTL 6.11.6 of autologous EBV-transformed B cell line LB2586-EBV pulsed with the indicated peptides. The 51Cr-labeled LB2586-EBV B cells were pulsed for 30′ with various concentrations of peptides. CTL 2.4 and CTL 6.11.6 were then added at an effector-to-target ratio of 10 and 3, respectively. 51Cr release was measured after 4 h
Many additional independent CTL clones recognizing peptide SESIKKKVL on autologous EBV-B cells and expressing the same TCR (clonotype A) were further isolated, either from other skin metastases resected at later time points, or from blood lymphocytes collected in the course of vaccination. One of these clones is CTL 6.11.6 isolated from blood taken after one cycle of six vaccinations. This CTL recognized the antigen with affinity similar to CTL 2.4 (Fig. 4). It proliferated better in vitro and was used in further experiments.
Recognition of allogenic melanoma cell lines expressing MAGE-C2
CTL 6.11.6 was tested on the LB2586-MEL.G autologous melanoma cell line and on four allogenic melanoma lines expressing MAGE-C2 and HLA-B44. Two of them expressed B*4403 like the autologous line, one expressed B*4404 and one expressed B*4402. Recognition by CTL 6.11.6 was restricted to the cell lines expressing MAGE-C2 and HLA-B*4403 (Fig. 5). This restricted recognition was confirmed in lysis assay showing that CTL 6.11.6 was able to lyse MAGE-C2+ HLA-B*4403+ tumor cells exclusively, with 60% specific lysis at an effector-to-target ratio of 10 (data not shown).
Fig. 5.
Recognition of melanoma cell lines expressing MAGE-C2 by CTL 6.11.6 is restricted by HLA-B*4403. The melanoma cell lines, expressing various HLA-B44 molecules, were treated with IFN-γ and further incubated in the presence of CTL 6.11.6. After 24 h, the production of TNF was measured
Discussion
We report the molecular definition of a new antigen that is encoded by gene MAGE-C2 and presented by HLA-B44. This identification relies on the recognition by CTL clones obtained either from metastases or from the blood of a melanoma patient enrolled in a vaccination protocol in which autologous mature dendritic cells pulsed with the MAGE-3.A1 peptide were injected. Their emergence in the course of patient LB2586 vaccination will be discussed elsewhere (J. Carrasco et al., manuscript in preparation). The clinical outcome of our patient who remains in complete remission more than 3 years after the initiation of therapeutic vaccination is encouraging and supports further investigation of the SESIKKKVL peptide for the therapeutic vaccination of HLA-B*4403 positive patients with MAGE-C2 expressing cancers.
Gene MAGE-C2 is a member of the MAGE gene family, which comprises several genes located on chromosome X and which displays a cancer-germline expression pattern [4]. MAGE-C2 is expressed in 40% of melanomas, 30% of bladder carcinomas, 20% of head and neck carcinomas and 10% of nonsmall cells lung carcinomas while silent in normal tissues, except in male germline cells, which are devoid of MHC class I molecules [14]. Genes of the MAGE-A cluster were found to encode a large number of tumor-specific antigens recognized by T lymphocytes [22]. Recently, three antigenic peptides were found to be encoded by gene MAGE-C2, one of the four genes comprised in the MAGE-C cluster: two presented by HLA-A2 and one by HLA-B57 [7, 16].
HLA-B44 is expressed by 24% of Caucasians, thus being the most frequent HLA-B allele in this population [11, 13]. This allelic group comprises at least eight subtypes, among which predominate B*4402 (61%) and B*4403 (36%). The new MAGE-C2 antigenic peptide described here needs to be presented to clonotype A CTL in the context of HLA-B*4403. This may be due to the fact that the peptide binds specifically to HLA-B*4403 and not to the other B44 subtypes tested, as shown for other antigenic peptides [10]. Alternatively, assuming the peptide binds to various subtypes, the specificity of the CTL clone could be due to a particular conformation adopted by the peptide when bound to B*4403, for previous reports have shown that CTL are able to distinguish between various conformations adopted by the same peptide in two B44 or B35 subtypes [10, 21]. A third potential explanation is that clonotype A CTL, which is B*4403-restricted, shows strict specificity for this presenting HLA molecule although it differs from B*4402 or B*4404 only at residue 156 or residues 156 and 163, respectively; such strict specificity for one B44 allele has already been reported for an anti-MAGE-3 CTL derived from a B*4403 patient [10].
MAGE-C2 antigenic peptide SESIKKKVL presented by HLA-B*4403 is the fourth MAGE-C2 antigenic peptide which has been identified. The first three MAGE-C2 peptides have been identified in another melanoma patient who displayed impressive CTL responses against these antigens, reaching frequencies in the tumor of several percent of CD8 T cells. Some of these responses occurred spontaneously before vaccination [7, 15]. High spontaneous responses have also been observed for antigens encoded by LAGE-1 and NY-ESO-1 [12, 17]. Whether this makes these antigens particularly useful for anti-tumoral vaccines will have to be determined by clinical experimentation.
Acknowledgments
We thank V. Ha Thi, M. Panagiotakopoulos and M. Swinarska for technical assistance, W. Ma for providing the MAGE-C2 minigenes, and V. Stroobant for peptide synthesis. We thank P. van der Bruggen for critical reading, and N. Krack for editorial assistance. This work was supported by the Belgian Programme on Interuniversity Poles of Attraction initiated by the Belgian State, Prime Minister’s Office, Science Policy Programming, and by a grant of the Fédération Belge contre le Cancer. Javier Carrasco was supported by a grant FNRS-TELEVIE.
References
- 1.Boon T, Van Snick J, Van Pel A, Uyttenhove C, Marchand M. Immunogenic variants obtained by mutagenesis of mouse mastocytoma P815 II T lymphocyte-mediated cytolysis. J Exp Med. 1980;152:1184–1193. doi: 10.1084/jem.152.5.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boon T, Coulie PG, Van den Eynde B, van der Bruggen P. Human T cell responses against melanoma. Annu Rev Immunol. 2006;24:6–1–634. doi: 10.1146/annurev.immunol.24.021605.090733. [DOI] [PubMed] [Google Scholar]
- 3.Brasseur F (1999) Melanoma: Brussels Melanoma cell lines. In: Masters JRW, Palsson B (eds) Human cell culture, vol 1. Kluwer, Dordrecht, pp 275–282
- 4.Chomez P, De Backer O, Bertrand M, De Plaen E, Boon T, Lucas S. An overview of the MAGE gene family with the identification of all human members of the family. Cancer Res. 2001;61:5544–5551. [PubMed] [Google Scholar]
- 5.Espevik T, Nissen-Meyer J. A highly sensitive cell line, WEHI 164 clone 13, for measuring cytotoxic factor/tumor necrosis factor from human monocytes. J Immunol Methods. 1986;95:99–105. doi: 10.1016/0022-1759(86)90322-4. [DOI] [PubMed] [Google Scholar]
- 6.Gaugler B, Van den Eynde B, van der Bruggen P, Romero P, Gaforio JJ, De Plaen E, Lethé B, Brasseur F, Boon T. Human gene MAGE-3 codes for an antigen recognized on a melanoma by autologous cytolytic T lymphocytes. J Exp Med. 1994;179:921–930. doi: 10.1084/jem.179.3.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Germeau C, Ma W, Schiavetti F, Lurquin C, Henry E, Vigneron N, Brasseur F, Lethé B, De Plaen E, Velu T, Coulie PG. High frequency of anti-tumor T cells in the blood of melanoma patients before and after vaccination with tumor antigens. J Exp Med. 2005;201:241–248. doi: 10.1084/jem.20041379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Godelaine D, Carrasco J, Lucas S, Karanikas V, Schuler-Thurner B, Coulie PG, Schuler G, Boon T, Van Pel A. Polyclonal CTL responses observed in melanoma patients vaccinated with dendritic cells pulsed with a MAGE-3.A1 peptide. J Immunol. 2003;171:4893–4897. doi: 10.4049/jimmunol.171.9.4893. [DOI] [PubMed] [Google Scholar]
- 9.Haas GG, Jr, D’Cruz OJ, De Bault LE. Distribution of human leukocyte antigen-ABC and -D/DR antigens in the unfixed human testis. Am J Reprod Immunol Microbiol. 1988;18:47–51. doi: 10.1111/j.1600-0897.1988.tb00234.x. [DOI] [PubMed] [Google Scholar]
- 10.Herman J, Jongeneel V, Kuznetsov D, Coulie PG. Differences in the recognition by CTL of peptides presented by the HLA-B*4402 and the HLA-B*4403 molecules which differ by a single amino acid. Tissue Antigens. 1999;53:111–121. doi: 10.1034/j.1399-0039.1999.530201.x. [DOI] [PubMed] [Google Scholar]
- 11.Imanishi T, Akaza T, Kimura A, Tokunaga K, Gojobori T (1991) Allele and haplotype frequencies for HLA and complement loci in various ethnic groups. In: Tsuji K, Aizawa M, Sasazuki T (eds) XIth international histocompatibility workshop and conference, vol 1. Oxford University Press, New York, p 1065
- 12.Jäger E, Chen Y-T, Drijfhout JW, Karbach J, Ringhoffer M, Jäger D, Arand M, Wada H, Noguchi Y, Stockert E, Old LJ, Knuth A. Simultaneous humoral and cellular immune response against cancer-testis antigen NY-ESO-1: definition of human histocompatibility leukocyte antigen (HLA)-A2-binding peptide epitopes. J Exp Med. 1998;187:265–270. doi: 10.1084/jem.187.2.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee TD. Distributions of HLA antigens. In: Lee J, editor. The HLA system A new approach. Berlin Heidelberg New York: Springer; 1990. pp. 141–178. [Google Scholar]
- 14.Lucas S, De Plaen E, Boon T. MAGE-B5, MAGE-B6, MAGE-C2 and MAGE-C3: four new members of the MAGE family with tumor-specific expression. Int J Cancer. 2000;87:55–60. doi: 10.1002/1097-0215(20000701)87:1<55::AID-IJC8>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 15.Lurquin C, Lethé B, Corbière V, Théate I, van Baren N, Coulie PG, Boon T. Contrasting frequencies of anti-tumor and anti-vaccine T cells in metastases of a melanoma patient vaccinated with a MAGE tumor antigen. J Exp Med. 2005;201:249–257. doi: 10.1084/jem.20041378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ma W, Germeau C, Vigneron N, Maernoudt A-S, Morel S, Boon T, Coulie PG, Van den Eynde B. Two new tumor-specific antigenic peptides encoded by gene MAGE-C2 and presented to cytolytic T lymphocytes by HLA-A2. Int J Cancer. 2004;109:698–702. doi: 10.1002/ijc.20038. [DOI] [PubMed] [Google Scholar]
- 17.Rimoldi D, Rubio-Godoy V, Dutoit V, Liénard D, Salvi S, Guillaume P, Speiser D, Stockert E, Spagnoli G, Servis C, Cerottini J-C, Lejeune F, Romero P, Valmori D. Efficient simultaneous presentation of NY-ESO-1/LAGE-1 primary and nonprimary open reading frame-derived CTL epitopes in melanoma. J Immunol. 2000;165:7253–7261. doi: 10.4049/jimmunol.165.12.7253. [DOI] [PubMed] [Google Scholar]
- 18.Rosenberg SA, Yang JC, Restifo NP. Cancer immunotherapy: moving beyond current vaccines. Nat Med. 2004;10:909–915. doi: 10.1038/nm1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schultz ES, Lethé B, Cambiaso CL, Van Snick J, Chaux P, Corthals J, Heirman C, Thielemans K, Boon T, van der Bruggen P. A MAGE-A3 peptide presented by HLA-DP4 is recognized on tumor cells by CD4+ cytolytic T lymphocytes. Cancer Res. 2000;60:6272–6275. [PubMed] [Google Scholar]
- 20.Traversari C, van der Bruggen P, Van den Eynde B, Hainaut P, Lemoine C, Ohta N, Old L, Boon T. Transfection and expression of a gene coding for a human melanoma antigen recognized by autologous cytolytic T lymphocytes. Immunogenetics. 1992;35:145–152. doi: 10.1007/BF00185107. [DOI] [PubMed] [Google Scholar]
- 21.Tynan FE, Elhassen D, Purcell AW, Burrows JM, Borg NA, Miles JJ, Williamson NA, Green KJ, Tellam J, Kjer-Nielsen L, McCluskey J, Rossjohn J, Burrows SR. The immunogenicity of a viral cytotoxic T cell epitope is controlled by its MHC-bound conformation. J Exp Med. 2005;202:1249–1260. doi: 10.1084/jem.20050864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van den Eynde B, van der Bruggen P. Peptide database of T-cell defined tumor antigens. URL: http://www.cancerimmunity.org/peptidedatabase/Tcellepitopes.htm [DOI] [PubMed]
- 23.van der Bruggen P, Zhang Y, Chaux P, Stroobant V, Panichelli C, Schultz ES, Chapiro J, Van den Eynde BJ, Brasseur F, Boon T. Tumor-specific shared antigenic peptides recognized by human T cells. Immunol Rev. 2002;188:51–64. doi: 10.1034/j.1600-065X.2002.18806.x. [DOI] [PubMed] [Google Scholar]