Abstract
The effectiveness of allogeneic graft-versus-leukemia (GVL) activity in control of acute lymphoblastic leukemia is generally regarded as poor. One possible factor is dynamic adaptation of the leukemia cell to the allogeneic environment. This work tested the hypothesis that the pattern of gene expression in acute lymphoblastic leukemia cells in an allogeneic environment would differ from that in a non-allogeneic environment. Expression microarray studies were performed in murine B lineage acute lymphoblastic leukemia cells recovered from mice that had undergone allogeneic MHC-matched but minor histocompatibility antigen mismatched transplants. A limited number of genes were found to be differentially expressed in ALL cells surviving in the allogeneic environment. Functional analysis demonstrated that genes related to immune processes, antigen presentation, ubiquitination and GTPase function were significantly enriched. Several genes with known immune activities potentially relevant to leukemia survival (Ly6a/Sca-1, TRAIL and H2-T23) were examined in independent validation experiments. Increased expression in vivo in allogeneic hosts was observed, and could be mimicked in vitro with soluble supernatants of mixed lymphocyte reactions or interferon-gamma. The changes in gene expression were reversible when the leukemia cells were removed from the allogeneic environment. These findings suggest that acute lymphoblastic leukemia cells respond to cytokines present after allogeneic transplantation and that these changes may reduce the effectiveness of GVL activity.
Electronic supplementary material
The online version of this article (doi:10.1007/s00262-010-0889-y) contains supplementary material, which is available to authorized users.
Keywords: Leukemia, Bone marrow transplantation, Graft-versus-host disease, Graft-versus-leukemia, Interferon-gamma
Introduction
Allogeneic hematopoietic stem cell transplantation is used to treat patients with acute lymphoblastic leukemia at high risk for relapse. This is based on the observation that alloreactivity will produce a clinically significant graft-versus-leukemia (GVL) effect. Relapse after transplant, however, is the most common cause of treatment failure, with rates approaching 36–60% in some studies [1, 2].
A dynamic inflammatory environment exists following allogeneic hematopoietic stem cell transplantation and is influenced by multiple factors including host disease burden, the toxicity of the conditioning regimen and the presence of high-risk mutations in the leukemia itself [3, 4]. It is this environment that ultimately determines the success of host immune reconstitution, the GVL effect and donor engraftment [5]. While clinical trials have associated these factors with increased rates of relapse, the molecular mechanisms leading to leukemia relapse remain unknown. Understanding the mechanisms of leukemia relapse following allogeneic transplant may have a significant impact on the treatment of patients with high-risk disease.
There is emerging evidence that acute lymphoblastic leukemia cells change their gene expression profile in ways that promote relapse and cell survival. Several recent studies have used whole-genome and single nucleotide polymorphism arrays to show that acute lymphoblastic leukemia cells alter their gene expression in ways that confer resistance to drugs commonly used to treat childhood acute lymphoblastic leukemia [6, 7]. Similar studies have identified changes in gene expression and predicted disease recurrence in leukemia cells carrying the high-risk bcr-abl mutation in ways that potentially confer resistance to targeted imatinib therapy [8, 9]. There is limited data, however, on whether leukemia cells differentially express genes that promote their survival and proliferation following allogeneic transplant.
Because of the genetic diversity of the human population as well as the significant variability of clinical treatment in patients, attempts to answer this question precisely with human clinical samples can be fraught with difficulty. We have chosen an alternate approach with a murine model of human leukemia which allows control of potentially confounding variables. We have developed murine models of precursor B lymphoblastic leukemia in which leukemogenesis is driven by genetic lesions known to be important in human ALL [10]. One set of these ALLs contains the human p210 bcr-abl oncogene and has deletions in the Ink/Arf locus, genetic lesions present in many high risk ALLs; these carry the name “NSTY”. Another ALL line is driven by the human p190 bcr-abl oncogene and carries the name “ASLN”. These ALLs extensively characterized by flow cytometry have a B precursor phenotype and behave like human ALL in vivo with an aggressive growth profile with rapid infiltration of marrow, spleen and the CNS. The murine transplant model is directly relevant to human transplantation, and is one in which the donor and recipient are matched at the major histocompatibility loci but are mismatched at multiple minor histocompatibility antigen loci [11, 12]. Donors are C3.SW (H-2b) and recipients are C57BL/6 (H-2b). This is the situation in human HLA matched related donor and HLA matched unrelated donor transplantation in which allogeneic graft-versus-host disease (GVHD) and GVL reactions are triggered by differences in minor histocompatibility antigen loci. They differ at more than seven defined minor histocompatibility antigen loci, and three of these minor antigens have been genetically and molecularly defined at the peptide epitope level which allows us to exploit the model to study graft-versus-host and GVL immune activity at the minor histocompatibility antigen T cell specific level. This system allowed us to test the hypothesis that leukemia cells change their gene expression profile in response to the alloimmune environment present after transplant. Discovery of such genes and/or pathways may lead to better understanding of the immunobiology of relapse and identify potential therapeutic targets for patients with high-risk leukemias receiving allogeneic transplant.
Materials and methods
Murine leukemia cell lines
We utilized a GFP-labeled murine C57BL/6 pre-B acute lymphoblastic leukemia cell line (NSTY1) carrying the human p210 bcr-abl gene and an Ink/Arf deletion [10]. ASLN, another murine pre-B cell ALL line containing the human p190 bcr-abl gene but lacking an Ink/Arf deletion, was used for comparative analysis in the PCR validation experiments [10].
Murine allogeneic transplant model
C57BL/6 recipients received either syngeneic (B6) or allogeneic C3.SW (SW) donor grafts. Both strains are H-2b (i.e., MHC I and II matched), but differ at multiple minor histocompatibility loci, including the immunodominant minor histocompatibility antigen H7 locus [11]. 106 NSTY1 leukemia cells were incorporated into hematopoietic cell grafts and transplanted into B6 mice following myeloablation with 0.5 mg of 5-fluorouracil (Day-2) and 1,050 cGy irradiation given in two fractions (Days-2, -1). The grafts were comprised of 4 × 106 bone marrow cells and 1 × 107 spleen cells. The syngeneic transplant group (n = 5) received cells from normal C57BL/6 mice. The allogeneic transplant group (n = 5) received cells from C3.SW donors that had been previously primed with 1 × 106 C57BL/6 cells 2 weeks prior to produce a vigorous graft-versus-host reaction in vivo as described previously [12]. Mice were monitored for evidence of progressive leukemia growth and this typically occurred after 3 weeks. At time of leukemia progression the animals were killed and bone marrow harvested. Flow cytometry was performed to measure the number of leukemia cells in each marrow sample. Based on the flow data, 106 leukemia cells were retransplanted into new BMT recipients that underwent myeloablation and infusion of either allogeneic or syngeneic grafts. The leukemia cells recovered from animals were not pooled prior to retransplantation. This cycle was continued for three generations of transplant. 3 weeks after the final serial transplant, bone marrow was harvested and GFP + leukemia cells were purified by flow cytometric sorting for mRNA extraction and in vitro culture assays. Following sorting leukemia cells were 99% of the population. Figure 1 provides a visual schema of the experimental design.
Fig. 1.
Experimental design for recovery of leukemia cells from allogeneic or syngeneic transplant environments in vivo. C57BL/6 mice received total body irradiation and 5-fluorouracil chemotherapy as described in “Methods”. In the allogeneic group, they were then infused with allogeneic C3.SW donor bone marrow and spleen cells mixed with NSTY1 leukemia cells. The syngeneic control group received marrow and spleen cells from C57BL/6 mice mixed with leukemia. In each group there were 5 transplant recipients. 3 weeks later bone marrow containing leukemia was harvested from each transplant recipient and following overnight culture was infused into another set of transplant recipients. The re-isolated leukemias from each animal were kept separate. The process was repeated again. Flow cytometry was used to purify leukemia cells for RNA preparation. Leukemia 3 was leukemia that had passed through three transplant recipients and was used for the microarray experiments. In later independent experiments in which quantitative RT-PCR was used to validate upregulation of genes of interest flow sorting was used to purify Leukemia 1 which had passed through one generation of transplant recipients
Oligonucleotide microarray hybridization and analysis
RNA was extracted from leukemia cells using standard Trizol reagents according to the manufacturer’s instructions. RNA submitted to the Functional Genomics Core (University of Rochester, Rochester, NY, USA) was subjected to quality control assays using an Agilent Bioanalyzer, and then processed for hybridization to Affymetrix Mouse 430.2 whole genome arrays (Affymextrix). Invariant set normalization was used to normalize arrays at the probe level, and model-based expression [13, 14] was used for probe selection and computations of expression values. dCHIP was the software used for analysis [13]. Criteria for differential expression were increased or decreased expression by more than 1.5-fold (90% lower confidence bound) with a median false discovery rate less than 5% [15]. Functional annotation clustering of differentially expressed genes was performed using the web-based software at the Database for Annotation, Visualization and Integrated Discovery (DAVID) at the National Institutes of Allergy and Infectious Diseases (http://david.abcc.ncifcrf.gov/home.jsp) [16]. Hierarchical clustering was used to group genes with similar expression patterns as previously described [17]. Microarray CEL files are publicly accessible on the Gene Expression Omnibus (series record GSE17497).
Real-time PCR
Differential gene expression as measured by microarray was validated for genes of interest by real-time polymerase chain reaction, and direct detection of PCR product was detected by measuring the increase in fluorescence caused by the binding of SYBR green fluorochrome to double-stranded DNA (SYBR Green Reagent System, Applied Biosystems, Foster City, CA, USA). Primer sequences for genes were based on published sequences from the NCBI public database. GAPDH, forward: TGCACCACCAACTGCTTAGC, reverse: GGCATGGACTGTGGTCATGAG; Ly6.A (Lymphocyte antigen 6a or Sca-1), forward: AGGAGGCAGCAGTTATTGTGG, reverse: CGTTGACCTTAGTACCCAGGA; H2-T23 (Histocompatibility 2, T region locus 23, or Qa-1), forward: ACAGTCCCGACCCAGAGTAG, reverse: CCACGTAGCCGACAATGATGA; TNFSF10 (Tumor necrosis factor superfamily member 10, or TRAIL), forward: ATGGTGATTTGCATAGTGCTCC, reverse: GCAAGCAGGGTCTGTTCAAGA; IiGP1 (Interferon-inducible GTPase1), forward: GCAATGCCATTCTCCCTAAA, reverse: AATGAAGCAGATGGCAAACC; Mpa2l (Macrophage activation 2 like), forward: GATGGTGCTCATGCTGTTGT, reverse: CTTCTGGACACCGAAGGTCT. Optimum concentrations for each primer were determined by melt dissociation curve to minimize nonspecific amplification. RNA was prepared from NSTY1, ASLN or normal B cells using standard Trizol reagents and chloroform phase separation, with final RNA concentration determined by spectrophotometry. cDNA was synthesized with SYBR Green PCR Master Mix reagents (Applied Biosystems, Foster City, CA, USA) by mixing 1 μg of mRNA, primers and 10 mM dNTP mix with 25 mM MgCl2, 0.1 M DTT, 40 Units/μl RNAse out and 200 U/ml Superscript III reverse transcriptase, according to the manufacturer’s instructions. The primed cDNA synthesis reaction proceeded at 50°C for 50 min and was terminated at 85° for 5 min. PCR amplification was performed for 40 cycles (Light Cycler, Roche Diagnostics). Gene expression was calculated as a ratio between the gene of interest and a GAPDH internal control gene using the Ct method [18]. A paired, two-tailed Student’s t test was used to detect differences in gene expression between control and experimental groups. The number of replicates in experiments is presented in figure legends. P values of 0.05 or less were considered significant.
Interferon-gamma treatment of leukemia cells
To examine the direct effect of interferon-gamma on target gene expression, 106 leukemia cells were cultured for 72 h in murine interferon-gamma (Becton–Dickinson) at concentrations specified in the text and figure legends. Surface expression of Ly6a/Sca-1 was measured with phycoerythrin-labeled anti-Ly6a (BD Pharmingen). Surface expression of H2-T23 was detected by PE-labeled anti-H2-T23. Gene expression was measured by quantitative RT-PCR. Direct ELISA for murine TRAIL was performed using unlabeled goat anti-mouse TRAIL/TNFSF10 antibody (R&D Systems). Detection was performed with biotinylated rabbit anti-goat IgG (R&D Systems) and alkaline phosphatase–streptavidin, with conversion of alkaline phosphatase substrate measured at 405 nm.
In vitro allogeneic cocultures
An in vitro assay was developed to compare the expression of selected genes in leukemia cells cultured in an allogeneic environment versus a syngeneic control environment. The in vitro assay facilitated recovery of higher replicate numbers for PCR and controlled for in vivo variables that might confound differential expression analysis. 5 × 104 NSTY1 (B6 derived) cells were cultured with either 107 syngeneic BL/6 control splenocytes or 107 C3.SW splenocytes mixed with 107 BL/6 splenocytes in specialized coculture insert plates (Becton–Dickinson Product #35-3180). C3.SW splenocytes were harvested from mice vaccinated with 107 B6 splenocytes 2 weeks earlier. Leukemia cells cultured alone served as negative controls. Cells were cocultured for 72 h, cell inserts containing splenocytes were removed, and remaining NSTY1 leukemia cells processed for RNA extraction and RT-PCR for Ly6a, H2-T23 and TNFSF10 as described above.
Results
Differential gene expression in leukemia cells that survive allogeneic transplant
The central hypothesis of this project was that leukemia cells in vivo in recipients of allogeneic transplants respond to the inflammatory environment and that some aspects of this response may be reflected in the pattern of gene expression in the leukemia cells. To test this hypothesis, the NSTY1 acute lymphoblastic leukemia cell line was inoculated into mice that were undergoing transplantation. Animals that underwent syngeneic transplantation were the control group while the experimental group received allogeneic transplant. Figure 1 presents the experimental schema. As the leukemia can progress rapidly in vivo with death occurring at 3–4 weeks, we designed the experiment to include serial transplantation to allow a greater time of in vivo exposure of leukemia to the allogeneic response. Leukemia cells were recovered after 3 weeks from an animal and then injected into a new animal undergoing transplantation at that time. The leukemias were passed through three recipients and thus exposed to an allogeneic or syngeneic immune environment for 9 weeks in vivo. Following the third transplant bone marrow was harvested and leukemia cells purified by flow cytometry. RNA was immediately extracted and prepared for mRNA gene expression microarray analysis. Gene expression in the leukemia samples recovered from allogeneic transplant recipients was compared to expression in leukemia samples from syngeneic transplant recipients. Lists of genes with expression ≥1.5-fold greater (using expression lower 90% confidence bound of fold change) were generated. Median false discovery rate was estimated to be 3.3%. 455 genes fulfilled these criteria (The complete list is found in supplementary data Table S1).
While properly designed and powered gene expression microarray experiments can generate lists of differentially expressed genes additional work needs to be done to ascertain the biological significance and potential relatedness of the genes. To determine if there were some relationships between these genes or coherent biologic themes in the lists we analyzed the common gene list using DAVID, a publicly accessible bioinformatics tool provided by the NIAID which is designed to identify clustering in gene expression microarray data. This tool integrates a variety of gene annotation databases including GO, KEGG and BioCarta and provides for statistical analysis of the potential significance of clusters of genes identified in expression microarray experiments. The first analysis of this gene list asked the question of whether the pattern of expression was similar to that of any known organs or tissue. Not surprisingly significant clustering of genes normally expressed in activated spleen and bone marrow was observed. In addition, a similarity to gene expression in mammary tumors was found (Table 1). A second analysis asked whether there were functional relationships between the genes found to be differentially expressed. This functional annotation clustering analysis identified clusters related to immune responses and antigen presentation as the most highly statistically significant (Table 1, Functional annotation clusters 1 and 2). Other clusters were also identified including those related to GTPase activities and ubiquitinization (Table 1).
Table 1.
Functional analysis of 455 genes differentially expressed in flow cytometrically purified ALL cells in the allogeneic environment
| Fold enrichment | P value | FDR | |
|---|---|---|---|
| (A) Analysis by tissue | |||
| Tissue | |||
| Activated spleen | 2.5 | 0.00001 | 0.0 |
| Mammary tumor metastatized to lung. Tumor arose spontaneously | 2.8 | 0.00010 | 0.1 |
| Bone marrow | 1.9 | 0.00053 | 0.8 |
| (B) Analysis by Function | |||
| Annotation cluster 1 | Enrichment score 7.04 | ||
| Term | |||
| GO:0002376~immune system process | 3.5 | 0.00000 | 0.0 |
| GO:0006955~immune response | 3.9 | 0.00000 | 0.0 |
| Annotation cluster 2 | Enrichment score 3.47 | ||
| Term | |||
| mmu04940: Type I diabetes mellitus | 10.0 | 0.00000 | 0.0 |
| PIRSF001990: class I histocompatibility antigen | 13.7 | 0.00000 | 0.0 |
| GO:0042612~MHC class I protein complex | 10.5 | 0.00000 | 0.0 |
| IPR001039: MHC class I, alpha chain, alpha1 and alpha2 | 10.5 | 0.00000 | 0.0 |
| IPR011161: MHC class I-like antigen recognition | 9.1 | 0.00001 | 0.0 |
| Immune response | 5.7 | 0.00001 | 0.0 |
| MHC I | 19.2 | 0.00001 | 0.0 |
| GO:0042611~MHC protein complex | 7.4 | 0.00003 | 0.0 |
| GO:0019882~antigen processing and presentation | 6.0 | 0.00004 | 0.1 |
| IPR003006: Immunoglobulin/major histocompatibility complex motif | 5.6 | 0.00008 | 0.2 |
| mmu04612: Antigen processing and presentation | 5.4 | 0.00020 | 0.3 |
| GO:0002474~antigen processing and presentation of peptide antigen via MHC class I | 15.4 | 0.00028 | 0.5 |
| IPR003597: Immunoglobulin C1-set | 5.3 | 0.00029 | 0.6 |
| Region of interest: Alpha-3 | 24.5 | 0.00046 | 1.0 |
| IPR010579: MHC class I, alpha chain, C-terminal | 20.7 | 0.00082 | 1.6 |
| SM00407: IGc1 | 4.4 | 0.00095 | 1.4 |
| mmu04514: Cell adhesion molecules (CAMs) | 3.7 | 0.00120 | 1.5 |
| Transplantation antigen | 17.6 | 0.00137 | 2.1 |
| Region of interest: Alpha-2 | 15.3 | 0.00201 | 4.2 |
| Region of interest: Alpha-1 | 15.3 | 0.00201 | 4.2 |
| Annotation cluster 3 | Enrichment Score 2.3 | ||
| Term | |||
| IPR000626: Ubiquitin | 7.9 | 0.00091 | 1.8 |
| SM00213: UBQ | 6.3 | 0.00254 | 3.7 |
| Annotation cluster 4 | Enrichment Score 1.92 | ||
| Term | |||
| GO:0003924~GTPase activity | 5.3 | 0.00004 | 0.1 |
| IPR003191: Guanylate-binding protein, C-terminal | 30.0 | 0.00025 | 0.5 |
| IPR015894:Guanylate-binding protein, N-terminal | 22.5 | 0.00063 | 1.2 |
| GO:0017111~nucleoside-triphosphatase activity | 2.3 | 0.00328 | 5.7 |
| Annotation cluster 5 | Enrichment Score 1.87 | ||
| Term | |||
| GO:0046914~transition metal ion binding | 1.5 | 0.00259 | 4.5 |
| Annotation cluster 6 | Enrichment Score 1.83 | ||
| Term | |||
| IPR000215: Protease inhibitor I4, serpin | 7.2 | 0.00042 | 0.8 |
| PIRSF001630: serpin | 7.8 | 0.00090 | 1.6 |
| SM00093: SERPIN | 5.9 | 0.00113 | 1.7 |
Probe set identifiers of genes found to be differentially expressed flow cytometrically purified ALL cells (supplementary data) were submitted to the DAVID program. (A) Analysis by tissue: “Fold enrichment” refers to fraction of genes associated with a tissue in the experimental list divided by the fraction of genes associated with a tissue in the murine population background. “P value” is a modified Fisher’s exact test evaluating the significance of the tissue or gene term enrichment. “FDR” is the estimated percent false discovery rate. (B) Analysis by gene function: “Enrichment score” is calculated as the geometric mean of the P values of each term in an annotation group, and is a measure of the overall enrichment of an annotation group in the experimental sample gene list. Enrichment scores of greater than 1.3 are similar to non-log P values of 0.05 or less. For both analysis by tissue (A) and function (B), only those tissues or annotation functions with P values < 0.01 and a FDR < 5% are listed in the table
Rationale for selection for further study of differentially expressed genes
The microarray studies showed that in the leukemia cells in the allogeneic environment many of the differentially expressed genes clustered into known annotation groups related to immune function. This allowed us to focus further consideration on genes related to these groups. We then reviewed the functional annotations of individual genes from these clusters in the NCBI Entrez Gene database. Based on review of the known functions of the differentially expressed genes, we chose to focus further experimentation on three genes with potential physiologic relevance to the survival of leukemia cells in the allogeneic environment.
The first gene chosen was lymphocyte antigen 6 complex locus A (Ly6a, or Sca-1), a hematopoietic stem cell marker and regulator of lymphocyte activation [19, 20]. In the microarray experiment, Ly6a/Sca-1 exhibited 5.96-fold increased expression (supplementary data Table S1). We reasoned that this gene might be related to capacity of a cell to maintain self-renewal properties in an inflammatory environment.
A second gene chosen for further study was histocompatibility 2, T region locus 23 (H2-T23, or Qa-1) (supplementary data Table S1). In the microarray experiment, H2-T23 exhibited 2.3-fold increased expression. This gene is associated with regulatory T cell induction and cell protection from NK cell mediated lysis [21]. Early in the course of hematopoietic and immunologic reconstitution following bone marrow transplantation, NK cells emerge and are believed to exert some antileukemia activity. We reasoned that increased expression of H2-T23 by leukemia cells might provide some protection against activated NK cells.
The third gene chosen for further study was TRAIL (TNFSF10, tumor necrosis factor ligand superfamily member 10), a TNF superfamily member that preferentially induces apoptosis in cells that display the DR5 death receptor such as activated T cells and many malignant cells [22, 23]. In the microarray experiment, TRAIL exhibited 5.87-fold increased expression (supplementary data Table S1). We reasoned that TRAIL production by leukemia cells could conceivably provide some measure of local protection against activated T cells that play a role in GVL activity.
Focusing on these three candidate genes, we performed a series of experiments designed to (1) test the reproducibility of the differential expression observed in the microarray studies, (2) determine if these changes were reversible, (3) determine if the changes could be induced by soluble cytokines, and (4) determine that changes in mRNA levels led to increased protein expression.
Changes induced by the alloreactive environment are reversible
We hypothesized that the changes in gene expression in the genes of interest might be quite dynamic, with changes occurring relatively early in the allogeneic environment and reversible once the leukemia cells were removed from the allogeneic environment. Thus, we performed an independent experiment in which the leukemia cells were in the alloreactive environment for only 2 weeks without serial transplantation. At this point they were purified by flow cytometry. RNA was immediately prepared from 1 aliquot, while the rest of the leukemia cells were placed in culture for 96 h before RNA was extracted. Quantitative RT-PCR was performed for Ly6a, H2-T23 and TNFSF10/TRAIL. mRNA levels in leukemia cells were normalized to mRNA levels of GAPDH which is a widely expressed housekeeping gene. Confirming what had been seen in the microarray experiments, all three genes were expressed in higher levels in the leukemia cells obtained from the allogeneic transplant recipients compared to syngeneic transplant recipients (Fig. 2). The increased gene expression was rapidly reversible since following short term in vitro culture away from the inflammatory in vivo environment the differences in gene expression were no longer observed (Fig. 2).
Fig. 2.
Validation of differential expression of genes of interest by quantitative RT-PCR in freshly flow sorted leukemia cells and in cells placed in short term culture. 2 weeks after allogeneic or syngeneic transplant NSTY1 leukemia cells were purified by flow sorting. RNA was prepared immediately from 1 aliquot of the sorted cells, while the remaining aliquot was placed in culture for 96 h. After this short term culture RNA was prepared. Quantitative RT-PCR was performed for the genes Ly6a, H2-T23 and TNFSF10 using the fluorescence threshold detection method. mRNA copy numbers are expressed after normalization to the copy number of a GAPDH internal housekeeping gene. Because of limited cell numbers a single PCR reaction was performed for each gene and condition. The white bars represent gene expression in leukemia cells processed for RNA immediately at the time of cell sorting while the black bars represent gene expression following short term culture in normal tissue culture medium
Increased expression of genes of interest can be induced by soluble factors produced by allogeneic reaction supernatants in vitro
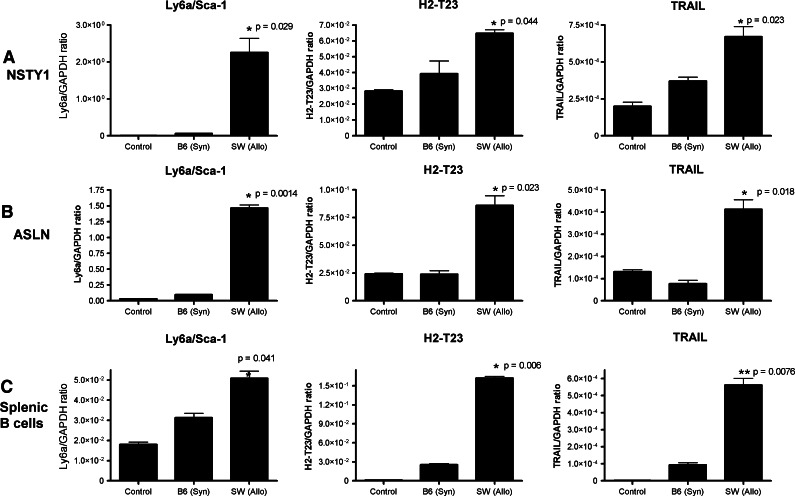
Studies in humans and mice have documented that after allogeneic transplantation a “cytokine storm” occurs. Based on this, we hypothesized that the changes observed in the leukemia cells in the allogeneic environment might be mediated by soluble cytokines released by T cells. To explore this possibility we exposed NSTY1 leukemia cells in vitro to supernatants of donor/recipient mixed lymphocyte reactions. In the experimental group, NSTY1 leukemia cells were cocultured in one partition of a transwell plate that allowed diffusion of medium but not the donor SW and recipient B6 cells present in the other partition. Leukemia cells cultured alone and in control transwell cocultures in which there were only naïve B6 splenocytes served as negative controls. After 72 h, gene expression levels were determined by quantitative RT-PCR. Replicating the observations made in the microarrays and quantitative RT-PCR from leukemia cells directly isolated from allogeneic transplant recipients, NSTY1 leukemia cells significantly increased expression of Ly6a/Sca-1 (P = 0.029), H2-T23 (P = 0.044) and TRAIL (P = 0.023) when cultured with alloreactive splenocytes compared to coculture with syngeneic (B6) controls (Fig. 3a). We extended the studies to determine if this was a response idiosyncratic to the NSTY1 ALL line. Identical coculture experiments were performed with another independently derived C57BL/6 pre-B ALL line, ASLN (Fig. 3b), and the same changes in gene expression were observed. The same changes were observed in two similar experiments in which leukemia cells were incubated in conditioned medium from allogeneic mixed lymphocyte reaction cultures (C3.SW splenocytes vs. C57BL/6 splenocytes) compared to medium from syngeneic (C57BL/6) splenocyte cultures (data not shown).
Fig. 3.
Soluble factors from allogeneic mixed lymphocyte reactions induce similar changes in gene expression. Membrane inserts were placed in tissue culture wells that allowed free diffusion of soluble molecules but prevented passage of cells. NSTY1 leukemia cells (a), ASLN leukemia cells (b) or normal recipient B cells (c) were placed in the one chamber. In the other chamber were placed either a mixture of allogeneic donor C3.SW splenocytes and recipient C57BL/6 splenocytes (SW Allo), only syngeneic C57BL/6 splenocytes (B6 Syn), or only tissue culture medium (Control). After 72 h, leukemia cells and splenic B cells were removed from their chamber, RNA prepared and RT-PCR performed for the selected genes Ly6a, H2-T23 and TNFSF10 using the fluorescence threshold detection method. mRNA copy numbers are expressed after normalization to the copy number of a GAPDH internal housekeeping gene. PCR reactions were performed in duplicate and differences in mRNA expression between cells cultured in an allogeneic environment were compared to syngeneic controls using a paired Student’s t test. Significant P values are presented for the comparison of allogeneic to syngeneic wells. There was no significant difference (P > 0.05, values not shown) between Ly6a, H2-T23 or TNFSF10 expression in NSTY1, ASLN or splenic B cells cultured with control versus B6 syngeneic splenocytes. Data are presented from representative experiments. The number of independent experiments for each cell type is: (NSTY1, 3 experiments), (ASLN 1 experiment) and (nonmalignant B cells, 2 experiments)
We hypothesized that the observed changes may be related to normal B cell immunophysiology rather than phenomena restricted to leukemia cells. To test this hypothesis, normal C57BL/6 B cells were placed in transwells adjacent to allogeneic mixed lymphocyte reaction cells or syngeneic splenocytes. Again in quantitative RT-PCR assays we observed a similar increase in expression of Ly6a/Sca-1 (P = 0.041), H2-T23 (P = 0.006) and TRAIL (P = 0.0076) when compared to syngeneic controls (Fig. 3c).
Interferon-gamma induces increases in expression of the genes of interest
Among the cytokines present in the post-transplant “cytokine storm” interferon-gamma is prominent. In the list of genes overexpressed in the leukemia cells, among the most highly differentially expressed genes were interferon-inducible GTPase (Iigp1, 146.87-fold increased expression, supplemental data Table S1) and macrophage activation 2 like (Mpa2l, 50.87-fold increased expression, supplemental data Table S1). Both Iigp1 [24, 25] and Mpa2l [26] are known to play roles in innate immunity and are regulated by interferon-gamma. Given this observation we hypothesized that interferon-gamma may be the cytokine that induced our genes of interest. To test this hypothesis, the expression of the Ly6a/Sca-1, H2-T23, and TRAIL were measured in NSTY1 and ASLN leukemia cells cultured in 1 ng/ml of interferon-gamma. This concentration is similar to concentrations that have been reported in clinically significant GVHD [27]. In vitro interferon-gamma does not inhibit the proliferation of or induce apoptosis in either of our ALL models (data not shown). Figure 4 demonstrates that interferon-gamma exposure increased both mRNA for Ly6a/Sca-1 as measured by quantitative RT-PCR (Fig. 4a) and cell surface Ly6a/Sca-1 as measured by flow cytometry (Fig. 4b). In similar experiments, NYST1 and ASLN leukemia cells were incubated for 48 h in 1 ng/ml interferon-gamma, and expression of H2-T23 and TRAIL/TNFSF10 evaluated. In the presence of interferon-gamma, both leukemia lines increase H2-T23 message (Fig. 4c) as well as cell surface H2-T23 (Fig. 4d). Similarly interferon-gamma increased TRAIL/TNFSF10 message (Fig. 4e) as well as TRAIL/TNFSF10 secretion as measured by ELISA (Fig. 4f). Additional quantitative RT-PCR experiments examined in vitro with interferon-gamma treatment on the expression of interferon-inducible GTPase 1 (Iigp1) and macrophage activation 2 like (Mpa2l) revealed two additional genes identified as highly expressed in leukemia cells in allogeneic environments. Again upregulation was seen, results consistent with the microarray data (Fig. 4g, h).
Fig. 4.
Interferon gamma induces increased expression of Ly6a/Sca-1, H2-T23 and TRAIL in ALL cells. NSTY1 and ASLN acute lymphoblastic leukemia cells were incubated in interferon gamma 1 ng/ml for 48 h. At that time, cells were analyzed for expression of the genes by reverse transcriptase quantitative PCR (a, c and e) and for protein expression by flow cytometry or ELISA (b, d and f). RT-quantitative PCR was also performed for interferon-inducible GTPase 1 (Iigp1, g) and for macrophage activation 2 like, (Mpa2l, h). Data are presented from representative experiments. The number of independent experiments for each panel is: a, 6 experiments; b, 6 experiments; c, 2 experiments; d, 2 experiments; e, 3 experiments; f, 3 experiments; g, 3 experiments and h, 1 experiment
Discussion
These studies tested the hypothesis that acute lymphoblastic leukemia cells surviving in an allogeneic transplant environment have a different pattern of gene expression compared to leukemia cells surviving in a syngeneic environment. Microarray studies demonstrated significant differences in patterns of gene expression, with changes in clusters of genes related to immune function, interferon signaling, and GTPase functions. In vitro studies that focused on three genes of immunological interest validated the microarray findings at both an RNA and protein level, and the changes could be reproduced by interferon-gamma alone. In these latter studies, we observed similar changes in the expression of Ly6a/Sca-1, H2-T23, and TRAIL in nonmalignant recipient strain B cells as well. The changes in gene expression are reversible when cells are removed from the inflammatory cytokines.
These observations raise the important question of whether the altered gene expression in the leukemia cells in the allogeneic environment is important to leukemia cell survival. It is unlikely that these responses are unique adaptations of malignant lymphoid cells for survival. Our observation that normal B cells in vivo experienced similar changes in the expression of the genes of interest is compatible with the hypothesis that the malignant lymphoid cells are using normal adaptive patterns of B lymphocytes for homeostasis in inflammatory environments. The studies reported here do not establish the mechanistic significance of the changes but provide a credible basis for further studies to test the hypothesis that increased expression of some of these genes may enhance survival of leukemia cells in vivo. To address this question, we have initiated studies in which we are using integrating gene vectors to either force high level expression or knockdown expression of genes in the leukemia cells with the goal of determining whether this alters leukemia cell survival in vivo.
In these studies, we chose to focus confirmatory studies on several genes that conceivably could be mechanistically related to survival of leukemia cells. Ly6a/Sca-1 was selected for study because of its prominence in the parallel literatures of murine stem cell biology (where Sca-1 is the common gene name) and lymphocyte activation immunology (where Ly6a is the gene’s name). The functions of Sca-1, as a marker of hematopoietic and other murine tissue stem cells and Ly6a as a marker of activated lymphocytes that promote Th2 cytokine production and the generation of regulatory T cells have generally been described separately [28, 29]. Recent evidence suggests that Ly6a/Sca-1, anchored in membrane lipid rafts, acts as a coordinator of multiple downstream signaling pathways rather than by a single receptor-ligand interaction [30]. Our data suggest the hypothesis that with increased expression of Ly6a/Sca-1 in leukemia cells may alter signaling through a number of downstream pathways whose combined activities may affect leukemia cell survival.
Expression of surface H2-T23 (Qa-1) protects dendritic cells and CD4+ T cells from natural killer-mediated direct lysis [21]. In models of allergic encephalomyelitis, the disruption of H2-T23 expression allows increased CD8+ regulatory T cell activity that completely protects against immune-mediated autoreactivity [31]. H2-T23 is homologous to human HLA-E, a nonclassical MHC I molecule thought to play a role in protecting the fetus from maternal allogeneic immune responses. Our findings suggest the hypothesis that increased expression of H2-T23 may represent a protective mechanism that helps leukemia cells evade direct cell killing by natural killer cells and cytolytic T cells present in the early post-transplant allogeneic environment.
Tumor-necrosis factor-related apoptosis-inducing ligand (TRAIL) is a well-known inducer of apoptosis in cells that express cognate death receptors (DR5 in mice). TRAIL has been examined as an antitumor agent because many malignant cells express death receptors [32] while most normal cells do not. However, normal activated T cells do express the DR5 death receptor and a growing body of evidence suggests that TRAIL plays a role in immune regulation [33]. Our findings suggest the hypothesis that leukemia-secreted TRAIL may induce apoptosis in alloactivated donor T cells in the microenvironment of the leukemia cell. We are currently exploring this possibility.
There are important limitations to these studies. First, while interferon-gamma does play a significant role in the changes in gene expression it is almost certain that multiple other cytokines and chemokines present in the inflammatory environment are also exerting effects on the leukemia cells. Dissection of the multiple potential effects and identification of other dominant cytokines will be technically complex. We are currently attempting to knockdown various cytokine receptors in our leukemia models with the goal of identifying the ones responsible for modulating the expression of genes in the leukemia cells in the allogeneic environment. A second important limitation of these studies is that while they provide insight into the syndrome of acute GVHD, they do not shed light on how leukemia cells adapt in the allogeneic environment in hosts with chronic GVHD. Since chronic GVHD differs from acute GVHD in terms of cytokine profiles and is characterized by less inflammation, it is likely that different genes would be differentially expressed in leukemia cells and different mechanisms of leukemia survival in this environment would be important.
The role of interferon-gamma in GVHD and overall survival following allogeneic transplant is an area of active investigation. Donor-derived interferon-gamma acts as an immune modulator, and is required for the inhibition of GVHD by IL-12 dependent mechanisms [34]. In addition, CD8+ T cells from interferon-gamma knockout donor mice induce more severe GVHD by attenuating activation and cell division [35]. Similarly, high endogenous levels of interferon-gamma have been associated with early mortality in allogeneic stem cell recipients, likely due to lymphohematopoetic graft-versus-host reactions [36]. Further, recent studies of tumor vaccines in IFN gamma knockout mice show enhanced immunogenicity and tumor protection while avoiding GVHD [37]. There is limited data, however, on the direct effect of high exogenous interferon-gamma on leukemia cells themselves. In our models, interferon-gamma does not increase apoptosis in the leukemia cells nor does it reduce leukemia cell proliferation. The uniform upregulation of interferon-gamma-associated GTPases and other interferon-regulated proteins observed in leukemia cells surviving allogeneic transplants in a high GVHD environment suggest that a variety of complex signaling pathways in leukemia cells are significantly affected by this inflammatory cytokine. Further studies are required to determine how these changes may affect acute leukemia progression after transplant and whether these changes identify genes that may be targeted to increase the GVL effect in allogeneic transplantation.
Electronic supplementary material
Acknowledgments
This work was supported in part by grant support from the National Institutes of Health (1R01CA10628) (C.A.M.) and the Brockport High School Leukemia Dance Marathon (C.A.M.).
Footnotes
J. C. Shand and J. Jansson contributed equally to this work.
References
- 1.Dahlke J, Kroger N, Zabelina T, Ayuk F, Fehse N, Wolschke C, Waschke O, Schieder H, Renges H, Kruger W, Kruell A, Hinke A, Erttmann R, Kabisch H, Zander AR. Comparable results in patients with acute lymphoblastic leukemia after related and unrelated stem cell transplantation. Bone Marrow Transplant. 2006;37:155–163. doi: 10.1038/sj.bmt.1705221. [DOI] [PubMed] [Google Scholar]
- 2.Woolfrey AE, Anasetti C, Storer B, Doney K, Milner LA, Sievers EL, Carpenter P, Martin P, Petersdorf E, Appelbaum FR, Hansen JA, Sanders JE. Factors associated with outcome after unrelated marrow transplantation for treatment of acute lymphoblastic leukemia in children. Blood. 2002;99:2002–2008. doi: 10.1182/blood.V99.6.2002. [DOI] [PubMed] [Google Scholar]
- 3.Munoz A, Diaz-Heredia C, Diaz MA, Badell I, Verdeguer A, Martinez A, Gomez P, Perez-Hurtado JM, Bureo E, Fernandez-Delgado R, Gonzalez-Valentin ME, Maldonado MS. Allogeneic hemopoietic stem cell transplantation for childhood acute lymphoblastic leukemia in second complete remission-similar outcomes after matched related and unrelated donor transplant: a study of the Spanish Working Party for Blood and Marrow Transplantation in Children (Getmon) Pediatr Hematol Oncol. 2008;25:245–259. doi: 10.1080/08880010802016557. [DOI] [PubMed] [Google Scholar]
- 4.Bhojwani D, Kang H, Moskowitz NP, Min DJ, Lee H, Potter JW, Davidson G, Willman CL, Borowitz MJ, Belitskaya-Levy I, Hunger SP, Raetz EA, Carroll WL. Biologic pathways associated with relapse in childhood acute lymphoblastic leukemia: a Children’s Oncology Group study. Blood. 2006;108:711–717. doi: 10.1182/blood-2006-02-002824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anderson LDMS, Mann S, Savary CA, Mullen CA. Pretransplant tumor antigen-specific immunization of allogeneic bone marrow transplant donors enhances graft-versus-host disease. Cancer Res. 2000;60:5797–5802. [PubMed] [Google Scholar]
- 6.Bacher U, Kohlmann A, Haferlach T. Current status of gene expression profiling in the diagnosis and management of acute leukaemia. Br J Haematol. 2009;145:555–568. doi: 10.1111/j.1365-2141.2009.07656.x. [DOI] [PubMed] [Google Scholar]
- 7.Yang JJ, Bhojwani D, Yang W, Cai X, Stocco G, Crews K, Wang J, Morrison D, Devidas M, Hunger SP, Willman CL, Raetz EA, Pui CH, Evans WE, Relling MV, Carroll WL. Genome-wide copy number profiling reveals molecular evolution from diagnosis to relapse in childhood acute lymphoblastic leukemia. Blood. 2008;112:4178–4183. doi: 10.1182/blood-2008-06-165027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zembutsu H, Yanada M, Hishida A, Katagiri T, Tsuruo T, Sugiura I, Takeuchi J, Usui N, Naoe T, Nakamura Y, Ohno R. Prediction of risk of disease recurrence by genome-wide cDNA microarray analysis in patients with Philadelphia chromosome-positive acute lymphoblastic leukemia treated with imatinib-combined chemotherapy. Int J Oncol. 2007;31:313–322. [PubMed] [Google Scholar]
- 9.Iacobucci I, Lonetti A, Messa F, Cilloni D, Arruga F, Ottaviani E, Paolini S, Papayannidis C, Piccaluga PP, Giannoulia P, Soverini S, Amabile M, Poerio A, Saglio G, Pane F, Berton G, Baruzzi A, Vitale A, Chiaretti S, Perini G, Foa R, Baccarani M, Martinelli G. Expression of spliced oncogenic Ikaros isoforms in Philadelphia-positive acute lymphoblastic leukemia patients treated with tyrosine kinase inhibitors: implications for a new mechanism of resistance. Blood. 2008;112:3847–3855. doi: 10.1182/blood-2007-09-112631. [DOI] [PubMed] [Google Scholar]
- 10.Young FM, Campbell A, Emo KL, Jansson J, Wang P-Y, Jordan CT, Mullen CA. High-risk acute lymphoblastic leukemia cells with bcr-abl and INK4A/ARF mutations retain susceptibility to alloreactive T cells. Biol Blood Marrow Transplant. 2008;14:622–630. doi: 10.1016/j.bbmt.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mori S, El-Baki H, Mullen CA. An analysis of immunodominance among minor histocompatibility antigens in allogeneic hematopoietic stem cell transplantation. Bone Marrow Transplant. 2003;31:865–875. doi: 10.1038/sj.bmt.1704021. [DOI] [PubMed] [Google Scholar]
- 12.Anderson LJ, Petropoulos D, Everse LA, Mullen CA. Enhancement of graft-versus-tumor activity and graft-versus-host disease by pretransplant immunization of allogeneic bone marrow donors with a recipient-derived tumor cell vaccine. Cancer Res. 1999;59:1525–1530. [PubMed] [Google Scholar]
- 13.Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci USA. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gentleman R, Carey VJ, Huber W, Irizarry RA, Dudroit S. Bioinformatics and computational biology solutions using R and bioconductor. New York: Springer; 2005. [Google Scholar]
- 15.Norris AW, Kahn CR. Analysis of gene expression in pathophysiological states: balancing false discovery and false negative rates. Proc Natl Acad Sci USA. 2006;103:649–653. doi: 10.1073/pnas.0510115103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 17.Bhattacharjee A, Richards WG, Staunton J, Li C, Monti S, Vasa P, Ladd C, Beheshti J, Bueno R, Gillette M, Loda M, Weber G, Mark EJ, Lander ES, Wong W, Johnson BE, Golub TR, Sugarbaker DJ, Meyerson M. Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proc Natl Acad Sci USA. 2001;98:13790–13795. doi: 10.1073/pnas.191502998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cikos S, Bukovska A, Koppel J. Relative quantification of mRNA: comparison of methods currently used for real-time PCR data analysis. BMC Mol Biol. 2007;8:113. doi: 10.1186/1471-2199-8-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henderson SC, Kamdar MM, Bamezai A. Ly-6A.2 expression regulates antigen-specific CD4+ T cell proliferation and cytokine production. J Immunol. 2002;168:118–126. doi: 10.4049/jimmunol.168.1.118. [DOI] [PubMed] [Google Scholar]
- 20.van de Rijn M, Heimfeld S, Spangrude GJ, Weissman IL. Mouse hematopoietic stem-cell antigen Sca-1 is a member of the Ly-6 antigen family. Proc Natl Acad Sci USA. 1989;86:4634–4638. doi: 10.1073/pnas.86.12.4634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu L, Ikizawa K, Hu D, Werneck MB, Wucherpfennig KW, Cantor H. Regulation of activated CD4+ T cells by NK cells via the Qa-1-NKG2A inhibitory pathway. Immunity. 2007;26:593–604. doi: 10.1016/j.immuni.2007.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ashkenazi A, Holland P, Eckhardt SG. Ligand-based targeting of apoptosis in cancer: the potential of recombinant human apoptosis ligand 2/Tumor necrosis factor-related apoptosis-inducing ligand (rhApo2L/TRAIL) J Clin Oncol. 2008;26:3621–3630. doi: 10.1200/JCO.2007.15.7198. [DOI] [PubMed] [Google Scholar]
- 23.Fakler M, Loeder S, Vogler M, Schneider K, Jeremias I, Debatin KM, Fulda S. Small molecule XIAP inhibitors cooperate with TRAIL to induce apoptosis in childhood acute leukemia cells and overcome Bcl-2-mediated resistance. Blood. 2009;113:1710–1722. doi: 10.1182/blood-2007-09-114314. [DOI] [PubMed] [Google Scholar]
- 24.Uthaiah RC, Praefcke GJ, Howard JC, Herrmann C. IIGP1, an interferon-gamma-inducible 47-kDa GTPase of the mouse, showing cooperative enzymatic activity and GTP-dependent multimerization. J Biol Chem. 2003;278:29336–29343. doi: 10.1074/jbc.M211973200. [DOI] [PubMed] [Google Scholar]
- 25.Boehm U, Guethlein L, Klamp T, Ozbek K, Schaub A, Futterer A, Pfeffer K, Howard JC. Two families of GTPases dominate the complex cellular response to IFN-gamma. J Immunol. 1998;161:6715–6723. [PubMed] [Google Scholar]
- 26.Degrandi D, Konermann C, Beuter-Gunia C, Kresse A, Wurthner J, Kurig S, Beer S, Pfeffer K. Extensive characterization of IFN-induced GTPases mGBP1 to mGBP10 involved in host defense. J Immunol. 2007;179:7729–7740. doi: 10.4049/jimmunol.179.11.7729. [DOI] [PubMed] [Google Scholar]
- 27.Yang YG, Wang H, Asavaroengchai W, Dey BR. Role of interferon-gamma in GVHD and GVL. Cell Mol Immunol. 2005;2:323–329. [PubMed] [Google Scholar]
- 28.Bradfute SB, Graubert TA, Goodell MA. Roles of Sca-1 in hematopoietic stem/progenitor cell function. Exp Hematol. 2005;33:836–843. doi: 10.1016/j.exphem.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 29.Zhang Z-X, Stanford WL, Zhang L. Ly-6A is critical for the function of double negative regulatory T cells. Eur J Immunol. 2002;32:1584–1592. doi: 10.1002/1521-4141(200206)32:6<1584::AID-IMMU1584>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 30.Holmes C, Stanford WL. Concise review: stem cell antigen-1: expression, function, and enigma. Stem Cells. 2007;25:1339–1347. doi: 10.1634/stemcells.2006-0644. [DOI] [PubMed] [Google Scholar]
- 31.Lu L, Kim HJ, Werneck MB, Cantor H. Regulation of CD8+ regulatory T cells: interruption of the NKG2A-Qa-1 interaction allows robust suppressive activity and resolution of autoimmune disease. Proc Natl Acad Sci USA. 2008;105:19420–19425. doi: 10.1073/pnas.0810383105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Henson ES, Johnston JB, Gibson SB. The role of TRAIL death receptors in the treatment of hematological malignancies. Leuk Lymphoma. 2008;49:27–35. doi: 10.1080/10428190701713655. [DOI] [PubMed] [Google Scholar]
- 33.Chen JJ, Chou CW, Chang YF, Chen CC. Proteasome inhibitors enhance TRAIL-induced apoptosis through the intronic regulation of DR5: involvement of NF-kappa B and reactive oxygen species-mediated p53 activation. J Immunol. 2008;180:8030–8039. doi: 10.4049/jimmunol.180.12.8030. [DOI] [PubMed] [Google Scholar]
- 34.Yang YG, Dey BR, Sergio JJ, Pearson DA, Sykes M. Donor-derived interferon gamma is required for inhibition of acute graft-versus-host disease by interleukin 12. J Clin Invest. 1998;102:2126–2135. doi: 10.1172/JCI4992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Asavaroengchai W, Wang H, Wang S, Wang L, Bronson R, Sykes M, Yang YG. An essential role for IFN-gamma in regulation of alloreactive CD8 T cells following allogeneic hematopoietic cell transplantation. Biol Blood Marrow Transplant. 2007;13:46–55. doi: 10.1016/j.bbmt.2006.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang H, Asavaroengchai W, Yeap BY, Wang MG, Wang S, Sykes M, Yang YG. Paradoxical effects of IFN-gamma in graft-versus-host disease reflect promotion of lymphohematopoietic graft-versus-host reactions and inhibition of epithelial tissue injury. Blood. 2009;113:3612–3619. doi: 10.1182/blood-2008-07-168419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Capitini CM, Herby S, Milliron M, Anver MR, Mackall CL, Fry TJ. Bone marrow deficient in IFN-{gamma} signaling selectively reverses GVHD-associated immunosuppression and enhances a tumor-specific GVT effect. Blood. 2009;113:5002–5009. doi: 10.1182/blood-2008-11-187385. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.