Abstract
Most childhood B cell precursor (BCP) acute lymphoblastic leukaemia (ALL) cases carry the reciprocal translocation t(12;21)(p13;q22) (∼25%), or a high hyperdiploid (HeH) karyotype (30%). The t(12;21) translocation leads to the expression of a novel fusion gene, TEL-AML1 (ETV6-RUNX1), and HeH often involves tri- and tetrasomy for chromosome 21. The presence of TEL-AML1+ and HeH cells in utero prior to the development of leukaemia suggests that these lesions play a critical role in ALL initiation. Based on our previous analysis of HLA-DP in childhood ALL, and evidence from in vitro studies that TEL-AML1 can activate HLA-DP-restricted T cell responses, we hypothesised that the development of TEL-AML1+ ALL might be influenced by the child’s DPB1 genotype. To test this, we analysed the frequency of six HLA-DPB1 supertypes in a population-based series of childhood leukaemias (n = 776) classified by their karyotype (TEL-AML1+, HeH and others), in comparison with newborn controls (n = 864). One DPB1 supertype (GKD) conferred significant protection against TEL-AML1+ ALL (odds ratio (OR), 95% confidence interval (95% CI): 0.42, 0.22–0.81; p < 0.005) and HeH ALL (OR; 95% CI: 0.44, 0.30–0.65; p < 0.0001). These negative associations were almost entirely due to a single allele, DPB1*0101. Our results suggest that DPB1*0101 may afford protection from the development of TEL-AML1+ and HeH BCP ALL, possibly as the result of a DP-restricted immune response to BCP ALL-associated antigen(s), the identification of which could have important implications for the design of prophylactic vaccines.
Keywords: HLA-DPB1, Childhood leukaemia, Immune response, Karyotype, TEL-AML1, Hyperdiploidy, Prevention
Introduction
The majority of childhood leukaemias (85%) in Europe and the USA are acute lymphoblastic leukaemias (ALL), mainly of B cell precursor (BCP) subtype, with a characteristic age-incidence peak between 2 and 5 years [1–3]. Cytogenetic and molecular analyses have revealed that two types of clonally-acquired chromosomal changes predominate in BCP ALL. About 25% of cases carry a reciprocal translocation, t(12;21)(p13;q22), leading to the in-frame fusion of the TEL (ETV6) gene on chromosome 12p13 and the AML1 (RUNX1) gene on 21q22, respectively [4, 5]. Some 30% of cases have a high Hyperdiploid (HeH) karyotype with a modal chromosome number >50, characterised by non-random gain of chromosomes X, 4,6,8,10,14,17,18, and 21 [6, 7]. The TEL-AML1 fusion gene is expressed in BCP ALL as a novel onco-protein [8], and is associated with a favourable treatment outcome [9, 10]. HeH is also associated with a good prognosis [11, 12], and expression profiling suggests that the majority of genes overexpressed by this subtype are localised to chromosome 21 and X [13]. Evidence that TEL-AML1+ and HeH pre-leukaemia cells are present in utero, prior to the onset of overt leukaemia, suggests that these lesions are associated with and perhaps critical to the initiation of BCP ALL leukaemogenesis [14–17].
In vitro studies have shown that a TEL-AML1 nonamer junctional-peptide can activate HLA-DPB1-restricted CD4+ helper and HLA-A2-restricted CD8+ cytotoxic T cell responses [18, 19]. This implies that TEL-AML1 may serve as an onco-antigen in vivo in subjects with HLA alleles that bind this peptide. We previously reported that the frequency of DPB1*0101 was significantly less in childhood BCP ALL cases than in unrelated controls [20]. We also detected a deficit of this allele in cases classified by TEL-AML1 and HeH status, but we did not explore the reason for this putative protective effect. Here we hypothesise that if HLA-DP-restricted T cell responses to TEL-AML1 inhibit the development of BCP ALL, this should be evident from the reduced frequency of the restricting allele(s) in cases, and its predicted ability to bind TEL-AML1. We tested this hypothesis, as part of the UK Childhood Cancer Study [21], by comparing the frequency of HLA-DPB1 peptide-binding supertypes in a population-based series of karyotyped childhood leukaemias with unrelated controls.
Materials and methods
Cases and controls
Blood samples were collected nationally from childhood leukaemia cases (n = 776) between 1992 and 1998, and locally from newborn controls (n = 864), between 1991 and 1997 as part of the national UK Childhood Cancer Study (UKCCS) [21]. Case data (diagnoses, cytogenetics, ages, gender, ethnic background) were validated by the UKCCS data centre at the Epidemiology and Genetics Unit, University of York. Diagnostic immunophenotyping of leukaemia cases was carried out on bone-marrow as previously described [21], according to criteria for UK Medical Research Council (MRC) leukaemia trials. Leukaemia subtypes are defined as Pro-B ALL (CD10−, CD19+); BCP-ALL (CD10+, CD19+); T-ALL (CD2/CD7+, CD19−, DR−), and AML. All procedures were carried out with national and local ethical approval. Controls were drawn from a cross-sectional series of umbilical cord blood samples obtained from normal term newborns born at St Mary’s Hospital, Manchester, UK. Using data obtained at birth (gender, ethnic background), cases and controls were matched for white UK ethnicity. The male:female ratio of the total case series was 1.23:1; for the controls it was 1.01:1.
Conventional cytogenetics
Cytogenetic analysis was carried out on diagnostic bone marrow and/or peripheral blood samples by local UK cytogenetic laboratories according to standard methods [22]. These data were collected and reviewed by the Leukaemia Research Fund Cytogenetics Group [23] and provided to the UKCCS [21].
Molecular cytogenetics
TEL-AML1 transcripts were detected in ALL bone-marrow and peripheral blood using RT-PCR and fluorescence in situ hybridisation (FISH), respectively, as previously described [24]. RT-PCR was carried out on cDNA from reverse transcribed RNA using a single pair of primers [25]. As the t(12;21) is a cryptic translocation, FISH analysis provided confirmation of positive and negative RT-PCR results. HeH was identified either by conventional cytogenetic analysis or, in cases of a failed cytogenetic result, by interphase FISH to indicate extra copies of chromosomes 21 and X, or using a panel of centromeric probes [24]. Based on cytogenetics, RT-PCR, and FISH, cases were classified into three cytogenetic groups: TEL-AML1 positive, high HeH, and combined “other”. The “other” group comprised all cases with a normal or abnormal cytogenetic result that did not include either TEL-AML1 or HeH.
HLA-DPB1 molecular typing
HLA-DPB1 molecular typing was carried out by the method previously described in detail [20]. A 327 bp fragment of DPB1 exon 2 in each DNA sample was amplified using a single pair of DPB1 generic PCR primers, aliquots of each PCR product spotted onto 384 sample nylon filters, and hybridised using a panel of 28 32P-labelled sequence specific oligonucleotide probes (SSOP). Probe hybridisation was detected using real-time autoradiography, and alleles assigned from published DPB1 ideograms.
Data analysis
Childhood leukaemia cases typed for HLA-DPB1 alleles were stratified by karyotype into TEL-AML1+, HeH and other karyotypes. Only results from cases with verified cytogenetic data and knowledge of TEL-AML1 status were included in the analysis. DPB1 allele frequencies were determined using the POPGENE (v.1.31) [26] descriptive population genetics programme. Selected DPB1 alleles in cases and controls were then clustered into six peptide binding pocket (PBP) supertypes, based on amino acid di-morphisms at positions 11, 69 and 84 of the DPβ1 domain, corresponding to the P6, P4 and P1 PBP. Using single letter amino-acid codes, these supertypes are designated GEG, LED, GED, GKG, LKD and GKD. Rare supertypes (LRD, GRV, GKV) with a cumulative frequency of ≤5% were excluded. Case-control DPB1 supertype and allele frequencies were compared using univariate statistical analysis to calculate cross-product odds ratios (ORs) and 95% confidence intervals (95% CI) by the Sheehe method with the RERI program from the Linkage Utility Package, LINKUTIL [27]. Statistical significance of case-control supertype and allele frequency differences was determined using Fisher’s Exact test to calculate p values using the 2by2 programme in LINKUTIL. Results are depicted as uncorrected 2-sided p values, with a p value of ≤0.008 being required to correct for six supertype clusters, to give significance (p ≤ 0.05). Other corrections are discussed in the text.
Results
Cytogenetic characteristics of childhood leukaemias
The karyotypes of childhood leukaemia cases recruited during the UKCCS have previously been reported [21]. These included 1,013 (76.5%) of the 1,324 B-lineage ALL cases, comprising TEL-AML1+ (n = 139), HeH (n = 423), and other karyotypes (n = 451). Cytogenetic data were also available for 99 of 137 T ALL (72%), and 160 of 250 AML (72%) cases. Of the 1,272 UKCCS leukaemia cases with cytogenetic data, DNA samples were obtained and typed for DPB1 alleles from 776 (61%) cases (Table 1). The majority of DPB1-typed B-lineage ALL cases were TEL-AML1+ (n = 90) or HeH (n = 276).
Table 1.
Childhood leukaemias classified by cytogenetic status
| Karyotype a | Diagnosisc | |||||
|---|---|---|---|---|---|---|
| Pro-B ALL | BCP ALL | T ALL | AML | OTHER | Total | |
| TEL-AML1 | 1 | 89 | 0 | 0 | 5 | 95 |
| HeH | 3 | 273 | 2 | 0 | 22 | 300 |
| OTHER | 17 | 167 | 72 | 93 | 32 | 381 |
| N b | 21 | 529 | 74 | 93 | 59 | 776 |
aKaryotype based on conventional and molecular cytogenetic analysis on leukaemic bone-marrow or peripheral blood blasts. For details see “Materials and methods”
b N number of diagnosed leukaemia cases with a given karyotype
cLeukaemias diagnosed by immunophenotyping and cytomorphology: Pro-B ALL, pro-B cell acute lymphoblastic leukaemia; BCP ALL, B cell precursor ALL; T ALL, T cell ALL; AML, acute myeloid leukaemia; “other”, leukaemias that could not be classified by immunophenotyping or cytology
DPB1 supertype frequency in relation to leukaemia karyotype
The number and frequency of DPB1 alleles in the karyotyped UKCCS cases and in controls are shown in Table 2. The 3-letter supertype assignment of each allele, based on di-allelic amino acid polymorphisms in the three peptide binding pockets at positions 11, 69 and 84 of the DPβ1 domain, accommodating amino acid residues at P6-P4-P1 of a core peptide, are also shown. A total of 37 DPB1 alleles were identified in the combined case-control series, including 18 in TEL-AML1+, 22 in HeH, 25 in other cases, and 31 in the controls. Six uncommon alleles (2401, 2501, 3301, 3501, 4901, 5101) were confined to the cases, and nine uncommon alleles (1801, 2101, 2701, 3401, 3601, 3901, 4401, 4801, 5001) were found only in controls. Excluding four alleles (1101, 1501, 1801, 3401) with rare supertypes, all other alleles (33) could be clustered into six supertypes, three with β69E (GEG, LED, GED), and three with β69K (GKG, LKD, GKD). The number of alleles with the GKG, LKD, or GKD supertypes in cases (8–12) and controls (12) was similar; the number of alleles with the GEG and GED supertypes in cases and controls (2–3) was also similar, whilst the number with the LED supertype in cases (4–6) differed from the controls (9).
Table 2.
DPB1 allele frequency in childhood leukaemia in relation to karyotype
| DPB1* allele | Frequency (f ×100)b | ||||
|---|---|---|---|---|---|
| Supertypea | TEL-AML1 | HeH | OTHER | CONTROL | |
| 0101 | GKD | 2.55 | 3.92 | 6.04 | 7.99 |
| 0201 | GEG | 9.69 | 9.97 | 9.58 | 5.96 |
| 0202 | GEG | 1.53 | 0.82 | 1.05 | 0.64 |
| 0301 | LKD | 10.71 | 9.48 | 10.50 | 11.46 |
| 0401 | GKG | 44.39 | 47.06 | 46.46 | 45.25 |
| 0402 | GKG | 12.76 | 13.07 | 12.07 | 11.52 |
| 0501 | GKD | 2.04 | 1.47 | 1.97 | 3.01 |
| 0601 | LED | 3.06 | 1.63 | 2.49 | 0.35 |
| 0801 | GED | 0.33 | 0.26 | 0.06 | |
| 0901 | LED | 1.02 | 0.98 | 0.52 | 1.10 |
| 1001 | LED | 1.53 | 2.78 | 1.44 | 1.45 |
| 1101 | [LRD] | 3.57 | 2.29 | 1.31 | 3.76 |
| 1301 | LED | 0.51 | 1.63 | 2.23 | 1.74 |
| 1401 | LKD | 2.55 | 0.49 | 0.39 | 0.81 |
| 1501 | [GRV] | 0.51 | 0.98 | 0.79 | 0.81 |
| 1601 | GED | 1.02 | 0.06 | ||
| 1701 | LED | 1.14 | 0.92 | 0.23 | |
| 1801 | [GKV] | 0.41 | |||
| 1901 | GED | 1.53 | 1.14 | 0.66 | 0.35 |
| 2001 | LKD | 0.16 | 0.13 | 0.06 | |
| 2101 | LED | 0.17 | |||
| 2301 | GKG | 0.26 | 0.46 | ||
| 2401 | GKG | 0.13 | |||
| 2501 | LKD | 0.51 | 0.13 | ||
| 2601 | LKD | 0.16 | 0.13 | 0.58 | |
| 2701 | LKD | 0.51 | 0.23 | ||
| 3001 | LED | 0.13 | 0.06 | ||
| 3301 | GEG | 0.13 | |||
| 3401 | [GKV] | 0.17 | |||
| 3501 | LKD | 0.16 | 0.13 | ||
| 3601 | LED | 0.06 | |||
| 3901 | GKG | 0.64 | |||
| 4401 | LED | 0.06 | |||
| 4801 | GEG | 0.17 | |||
| 4901 | GKG | 0.16 | |||
| 5001 | GKD | 0.06 | |||
| 5101 | GKG | 0.16 | |||
| N | 95 | 300 | 381 | 864 | |
aSupertypes designated by polymorphic amino acid residue (aa) in the P6, P4 and P1 peptide binding pockets (PBP). Single letter amino acid (aa) codes are used throughout: G, glycine; E, glutamic acid; K, lysine; D, aspartic acid; V, valine; R, arginine; L, leucine. Supertypes in parentheses are not analysed further
bDPB1 allele frequencies (f x 100) calculated using POPGENE (see “Materials and methods”)
The frequencies of all 6 DPB1 supertype clusters in cases classified by karyotype compared with controls are shown in Table 3. Only one (GKD) of the six supertypes had OR values significantly <1.0 in all three karyotyped case series (Odds Ratio, 95% confidence interval: TEL-AML1: 0.42, 0.22–0.81; HeH: 0.44, 0.30–0.65; other: 0.72, 0.53–0.97). Correction for six supertypes gives significant p values for TEL-AML1 (0.03) and HeH (0.00006), but not other leukaemias (0.18). Cases with two other supertypes having lysine at position 69 (GKG, LKG) were not significantly different from controls. Odds ratios were significantly >1.0 for the three β69E supertypes, GEG (all case series), for LED (HeH and other), and GED (TEL-AML1 and HeH). P values were significant after correction for six supertypes for GEG in HeH (0.048), and other cases (0.006).
Table 3.
HLA-DPB1 supertype frequency in relation to leukaemia karyotype
| DPB1 supertypea | Control % |
TEL-AML1 | HeH | OTHER | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| % | OR | 95% CIb | 2 sided p | % | OR | 95% CI | 2 sided p | % | OR | 95% CI | 2 sided p | ||
| GKD | 11.1 | 4.7 | 0.42 | 0.22–0.81 | 0.005 | 5.2 | 0.44 | 0.30–0.65 | 0.00001 | 8.1 | 0.72 | 0.53–0.97 | 0.03 |
| GKG | 58.0 | 57.4 | 0.97 | 0.72–1.31 | 0.91 | 61.2 | 1.14 | 0.94–1.38 | 0.19 | 58.9 | 1.04 | 0.87–1.23 | 0.71 |
| LKD | 13.2 | 13.2 | 1.01 | 0.65–1.56 | 0.99 | 10.8 | 0.80 | 0.60–1.07 | 0.15 | 11.4 | 0.85 | 0.65–1.11 | 0.24 |
| GED | 0.5 | 2.6 | 6.00 | 2.11–17.05 | 0.01 | 1.5 | 3.25 | 1.31–8.04 | 0.03 | 0.9 | 2.01 | 0.77–5.23 | 0.28 |
| GEG | 6.8 | 11.6 | 1.83 | 1.14–2.94 | 0.03 | 10.3 | 1.59 | 1.15- 2.19 | 0.008 | 10.8 | 1.66 | 1.24–2.23 | 0.001 |
| LED | 5.2 | 6.3 | 1.28 | 0.70–2.34 | 0.58 | 7.7 | 1.54 | 1.07–2.21 | 0.03 | 7.7 | 1.55 | 1.10–2.17 | 0.01 |
| N c | 864 | 95 | 300 | 381 | |||||||||
aDPB1 supertype designations as shown in Table 2
bOR, 95% CI: Odds ratios and 95% confidence intervals
c N number of subjects
Only 3 DPB1 alleles (0101, 0501, 5001) with the “protective” GKD supertype were present in the case-control series (Table 2). Table 4 compares the frequency of two of these alleles (0101, 0501) with other alleles having lysine at β69 (0401, 0402, 0301). DPB1*0101 was significantly reduced in TEL-AML1 (OR, 95% CI: 0.34, 0.15–0.78; uncorrected p = 0.005) and HeH cases (OR, 95% CI: 0.45, 0.28–0.70; uncorrected p = 0.0002), but not in the other karyotyped cases. Correction for 37 DP alleles removes significance from TEL-AML1 (p = 0.18), but not HeH (p = 0.007). The odds ratios for DPB1*0501 in TEL-AML1 and HeH were <1.0, but were not significant.
Table 4.
DPB1 allele-associated relative risk of leukaemia in relation to leukaemia karyotype
| DPB1* allele/ Supertype | Control % |
TEL-AML1 | HeH | OTHER | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| % | OR | 95% CI | 2-sided p | % | OR | 95% CI | 2- sided p | % | OR | 95% CI | 2 sided p | ||
| 0101/GKD | 8.0 | 2.6 | 0.34 | 0.15–0.78 | 0.005 | 3.7 | 0.45 | 0.28–0.70 | 0.0002 | 6.0 | 0.75 | 0.53–1.05 | 0.09 |
| 0501/GKD | 3.0 | 2.1 | 0.77 | 0.30–1.95 | 0.67 | 1.5 | 0.51 | 0.26–1.01 | 0.05 | 2.0 | 0.66 | 0.38–1.17 | 0.17 |
| 0401/GKG | 45.3 | 45.3 | 1.00 | 0.74–1.35 | 0.99 | 48.3 | 1.13 | 0.94–1.36 | 0.21 | 46.5 | 1.05 | 0.88–1.25 | 0.60 |
| 0402/GKG | 11.5 | 12.1 | 1.08 | 0.68–1.69 | 0.88 | 12.5 | 1.10 | 0.83–1.46 | 0.56 | 12.1 | 1.06 | 0.81–1.37 | 0.73 |
| 0301/LKD | 11.5 | 10.0 | 0.88 | 0.54–1.43 | 0.64 | 9.8 | 0.85 | 0.62–1.15 | 0.31 | 10.5 | 0.91 | 0.69–1.20 | 0.53 |
Other details as in Table 3
Discussion
We previously reported that the frequency of DPB1*0101 in common (B cell precursor) ALL, and in TEL-AML1+ and HeH cases was reduced compared with newborn controls [20], but we did not enlarge upon the possible reason. Yun et al. [19] reported that a TEL-AML1 junctional nonamer onco-peptide, RIAECILGM, can activate an HLA-DPB1*0501 restricted CD4+ T cell response in vitro. Since DPB1*0101 has the same supertype as 0501, we hypothesised that the deficit of 0101 might be related to similar peptide-binding motifs in these two alleles. One testable consequence of our result is that children typing for DPB1*0101 may be more likely than children with other DPB1 genotypes to generate T cell responses to the TEL-AML1 peptide. In support of this hypothesis, we show that the TEL-AML1 junctional peptide contains amino acid residues that we predict bind to appropriate peptide binding pockets of DPB1*0101. A similar hypothesis was proposed by Postuma et al. [28] to explain the deficit of HLA-A3, A11, and B8 in chronic myeloid leukaemias expressing the BCR-ABL p210 fusion protein.
We analysed the frequency of 6 HLA-DPB1 supertype clusters defined by amino acid dimorphisms in three critical peptide binding pockets (P6-P4-P1) at positions β11- β69- β84 (single amino acid code: GEG, GED, LED, GKG, GKD, LKD) in a population-based series of karyotyped childhood leukaemia cases in comparison with newborn controls. By clustering DPB1 alleles into three pairs of supertypes, we were able to distinguish between allele groups with glutamic acid or lysine at β69. Castelli et al. [29] clustered DPB1 alleles into three peptide-binding supertypes, based on amino acid dimorphisms at β84 (pocket 1) and β11 (pocket 6), but others have emphasised the importance of β69 (pocket 4) in antibody binding [30], allorecognition [31], peptide binding [32] and disease susceptibility [33]. Using in silico analysis Doytchinova and Flower [34] proposed a DP-supertype classification that takes account of β69. Our supertype classification is based on a synthesis of these findings, and distinguishes 3 β69E (GEG, LED, GED) from 3 β69K supertypes (GKG, LKD, GKD) (Taylor et al. submitted for publication). Supertype clusters defined by DP peptide binding predictions [35, 36] provide a better insight into the functional role of DPB1 in BCP ALL than classical alleles [37].
Because of the large number of DPB1 alleles, we previously used Monte-Carlo (CLUMP) analysis [38] to show that several alleles with a glutamic acid residue at position 69 (β69E) were associated with susceptibility to childhood BCP ALL [20]. In this study, prior clustering of alleles allowed the number of comparisons to be reduced to 6. Univariate analysis showed that the frequency of the GKD supertype was significantly less in TEL-AML1+ and HeH ALL than controls. This represents a 57% reduction of this supertype in TEL-AML1+ cases. Because we clustered alleles into six supertypes, we corrected only for six comparisons rather than the total number of DPB1 alleles. However, the reduction of DPB1*0101 in HeH cases was significant (p = 0.007) even after correction for 37 alleles, whilst lack of significance for DPB1*0101 in TEL-AML1 cases (n = 95), can be ascribed to an insufficient number of cases. Breslow and Day [39] pointed to the conservatism of multiple allele (Bonferroni) corrections in HLA studies, a critical issue in studies of DP and disease where there is a large number of uncommon alleles that have a disproportionate effect on significance tests. Such limitations could lead to type II error, as discussed in more general terms by Perneger [40]. The key message of our study is, therefore, that our analysis requires confirmation in other karyotyped leukaemia series, particularly in populations with a high frequency of DPB1*0101.
The case-control differences in DPB1 supertype and allele frequency that we report here are unlikely to be explained by ethnic stratification. We previously discussed this in detail in relation to the use of local controls for the UKCCS case series [20]. These controls were matched for white UK ethnic background with UKCCS cases. Had we chosen to ignore ethnicity, the frequency of DPB1*0101 in our total controls (8.4%), consisting of 864 white UK and 442 newborns with other ethnic backgrounds (n = 1,306), would have differed little from the white UK series (8.0%). Furthermore the frequency of DPB1*0101 in the previously reported UKCCS series of solid tumour cases (n = 409) [20] was not significantly different from the white UK newborns (OR, 95% CI: 0.8, 0.6–1.1).
The blood samples for this study were obtained in remission from UKCCS cases. This might have influenced allele frequencies if there had been selection of cases that favoured survival. With very few exceptions, however, UKCCS cases were entered into the United Kingdom UKALL XI trial (1990–1997), and this had a remission rate of 98%, only 2% deaths in remission, and an overall survival rate of 85% at 5 years [41]. The median time from diagnosis to blood sampling in our series was 7 months, >25% of samples having been collected within 2 months, and 70% within 12 months. It is thus unlikely that there was an allele bias due to delayed blood sampling. We are proposing to examine the relationship between DPB1 supertypes and long-term survival in a further study.
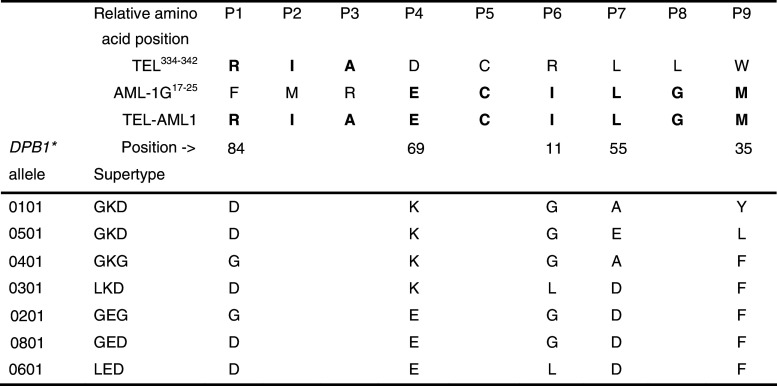
The lysine residue at position 69 (β69K) of the GKD supertype creates a positively charged P4 pocket with an affinity for ligands with a negative charge, including glutamic acid (E). Yun et al. [19] generated two DPB1 allele-restricted T cell clones to TEL-AML1 fusion peptides, one of which carried the 0501 allele. They found that this recognised the TEL-AML1 nonapeptide RIAECILGM, which has a negatively charged E in the P4 position. The positively charged K residue in the P4 PBP of DPB1*0501 and 0101 (Fig. 1) should bind the E residue of the TEL-AML1 nonapeptide [35, 36]. Whether differences between DPB1*0101 and 0501 in the P9 pocket (β9: 0101 = Y; 0501 = L; β55: 0101 = A; 0501 = E) significantly affect binding remains to be determined. Two other DPB1 supertypes with β69K (GKG and LKD) account for 71% of the supertype frequency in the controls, but were not deficient in TEL-AML1+ ALL. This suggests that a glycine (G) at position 11 and aspartic acid (D) at position 84, as well as lysine at position 69 of GKD could be important in protection from TEL-AML1+ ALL.
Fig. 1.
Peptide binding motifs defined by HLA-DPB1 alleles/supertypes in relation to the TEL-AML1 junctional 9-mer core peptide (RIAECILGM) with a negatively charged E in the P4 peptide binding pocket. This peptide is known to bind to DPB1*0501. The homologous wild-type TEL and AML1G peptide sequences are shown for comparison. The peptide binding frame for RIAECILGM is numbered P1–P9. The TEL and AML1G peptides have been positioned so as to show the same amino acid sequences as present in TEL-AML1, but it is not known whether they have the same binding frame. Note that AML-1G shares six of the nine amino acids with TEL-AML1, including an E at P4. DPB1*0101 and 0501 are identical for PBP P6, P4 and P1, but differ for the P7 and P9 PBP. All other supertypes differ from GKD at P6, P4 and/or P1
The GKD supertype frequency in HeH ALL was significantly less (5.2%) than in the controls (11.1%; OR, 95% CI: 0.44, 0.30–0.65; p < 0.0001), equivalent to a 50% reduction in DPB1*0101. It is unlikely that this can be explained by undetected TEL-AML1 transcripts in HeH cases since we used interphase FISH [24] as well as RT-PCR to detect this rearrangement. This suggests that HeH ALL might also express a DPB1*0101-restricted onco-antigen, of which the AML1 protein whose expression is amplified in HeH ALL [42, 43] is a candidate. Six of the 9 TEL-AML1 junctional amino acid residues (RIAECILGM) are coded by the alternately spliced native AML1 variant, AML1G, including the negatively charged E with an affinity for the β69K in the P4 PBP of DPB1*0101 (Fig. 1). In the absence of a suitable peptide-binding prediction algorithm for DPB1*0101, we scanned AML1G in silico for DRB1-binding peptides using TEPITOPE [44] and identified several putative DR-restricted T cell epitopes. Thus, it is possible that AML1 overexpression in HeH could lead to loss of immunological tolerance, resulting in the development of a protective DPB1-restricted T cell response.
We observed a marginally significant reduction in DPB1*0101 in leukaemias with other (i.e. TEL-AML1-, HeH-) karyotypes, but since this series was cytogenetically heterogeneous, it could have included small numbers of undetected TEL-AML1+ and HeH cases. DPB1-restricted T cell responses to other onco-proteins might also explain this mildly protective effect.
Our result does not prove a causal relationship between DPB1*0101 and a T cell response to TEL-AML+ BCP ALL. This will require evidence for the expression of this allele by TEL-AML1+ leukaemias, the 0101-restricted binding of this onco-peptide, and the generation of an 0101-restricted T cell response in vivo, all of which require further study. Expression of HLA-DR is used as a diagnostic aid in BCP ALL [45, 46], and recent evidence suggests that DR expression is significantly greater in TEL-AML1+ than TEL-AML1- BCP ALL or normal pre-B cells, implying that TEL-AML1 leukaemias may have a distinctive antigen-presenting phenotype [47], which may be true for DP. Thus, although expression of HLA-DP by B-ALL is about 25% that of HLA-DR, DP molecules are effectively recognised by DP allo-specific cytotoxic T cells [48].
Human leukaemias express an impressive array of leukaemia-associated fusion genes and onco-fetal proteins [49], and we cannot rule out an effect due to immune responses to other leukaemia antigens. The Wilm’s tumour onco-fetal protein, WT1 is expressed by a majority of childhood leukaemias [50, 51] and is known to elicit HLA class I and II-restricted T cell responses [52, 53]. Guo et al. [54] showed that the WT1337-347 peptide elicited a DP5-restricted cytotoxic response to AML cells, but the highly variable expression of WT1 by BCP ALL [55] may limit its value as a target for cytotoxic T cells.
Our results showed that K → E substitution at β69 significantly increased the risk of TEL-AML1+ ALL in cases with the GED supertype (OR, 95% CI: 6, 2.1–17.0). This represents a 14-fold difference in relative risk compared to GKD, and raises the possibility that alleles with the GED supertype may interact with ligands, perhaps derived from infections [56], in which the negatively charged E at P4 of the TEL-AML1 nona-peptide is replaced by a positively charged K.
There is increasing evidence to suggest that the emergence of pre-malignant cells may be prevented by the immune system [57]. Molecular evidence that TEL-AML1+ positive B cell precursors are present in cord blood of newborns at 100 times the rate of overt leukaemia [58, 59] raises the possibility of host control over the development of ALL. However, the frequency of DPB1*0101 in our controls is too low to explain this difference and it may be that other HLA alleles restricting the immune responses to TEL-AML1 contribute to protection. This is supported by evidence that the TEL-AML1 nonamer elicits an HLA*A2-restricted cytotoxic T cell response [18]. Further studies are needed to determine whether there are other T cell epitopes in TEL-AML1 and AML1 peptides. However our data together with the results of in vitro studies [18, 19] suggest that modelling and testing of TEL-AML1 and AML1 peptides could be a useful adjunct to the design of a prophylactic vaccine for BCP ALL.
Acknowledgments
This study was funded by grants from the Kay Kendall Leukaemia Fund (to GMT and MFG), by support from Cancer Research UK (JMB, TE), and by the Leukaemia Research Fund (MFG, CJH). We are indebted to the children and families who took part in the UK Childhood Cancer Study for enabling us to carry out this work. We thank J. Simpson and Professor E. Roman at the Epidemiology and Genetics Unit, University of York for providing diagnostic and other patient information, Mrs R. Carter for blood sample documentation and the midwives at St Mary’s Hospital, Manchester for cord blood samples. We are grateful to M. D. Robinson, Dr C. Watson, Dr D. A. Gokhale, S. P. Dearden for sample processing, and DPB1 typing. The United Kingdom Childhood Cancer Study (UKCCS) was sponsored and administered by the United Kingdom Co-ordinating Committee on Cancer Research, and has been conducted by 12 teams of investigators (ten clinical and epidemiological and two biological) based in university departments, hospitals, research institutes and the Scottish health service. The work is co-ordinated by a Management Committee and in Scotland by a Steering Group. It is supported by the United Kingdom Children’s Cancer Study Group, consisting of paediatric oncologists, and by the National Radiological Protection Board. Funding has been provided by a consortium of statutory bodies, cancer charities and industrial sponsors. A complete list of UKCCS investigators is given in: UK Childhood Cancer Study Investigators. The United Kingdom Childhood Cancer Study: objectives, materials and methods. Br. J. Cancer, 82, 1073-1102 (2000).
References
- 1.Greaves MF, Colman SM, Beard MEJ, Bradstock K, Cabrera ME, Chen P-M, Jacobs P, Lam-Po-Tang PRL, MacDougal LG, Williams CKO, Alexander FE. Geographical distribution of acute lymphoblastic leukaemia subtypes: second report of the collaborative study group. Leukemia. 1993;7:27–34. [PubMed] [Google Scholar]
- 2.Buckley JD, Buckley CM, Ruccione K, Sather HN, Waskerwitz MJ, Woods WG, Robison LL. Epidemiological characteristics of childhood acute leukemia. Analysis by immunophenotype. Leukemia. 1994;8:856–864. [PubMed] [Google Scholar]
- 3.Steliarova-Foucher E, Stiller C, Kaatsch P, Berrino F, Coeburgh J-M, Lacour B, Parkin M. Geographical patterns and time trends of cancer incidence and survival among children and adolescents in Europe since the 1970s (the ACCIS project): an epidemiological study. Lancet. 2004;364:2097–2105. doi: 10.1016/S0140-6736(04)17550-8. [DOI] [PubMed] [Google Scholar]
- 4.Golub TR, Barker GF, Bohlander SK, Hiebert SW, Ward DC, Bray-Ward P, Morgan E, Raimondi SC, Rowley JD, Gilliland DG. Fusion of the TEL gene on 12p13 to the AML1 gene on 21q22 in acute lymphoblastic leukaemia. Proc Nat Acad Sci. 1995;92:4917–4921. doi: 10.1073/pnas.92.11.4917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Romana SP, Mauchauffé M, Le Coniat M, Chumakov I, Le Paslier D, Berger R, Bernard OA. The t (12; 21) of acute lymphoblastic leukaemia results in a tel-AML1 gene fusion. Blood. 1995;85:3662–3670. [PubMed] [Google Scholar]
- 6.Raimondi SC, Pui C-H, Hancock ML, Behm FG, Filatov L, Rivera GK. Heterogeneity of hyperdiploid (51–67) childhood acute lymphoblastic leukemia. Leukemia. 1996;10:213–224. [PubMed] [Google Scholar]
- 7.Paulsson K, Mörse H, Fioretos T, Behrendtz M, Strömbeck B, Johanssen B. Evidence for a single-step mechanism in the origin of hyperdiploid childhood acute lymphoblastic leukaemia. Genes Chromosomes Cancer. 2005;44:113–122. doi: 10.1002/gcc.20222. [DOI] [PubMed] [Google Scholar]
- 8.Poirel H, Lacronique V, Mauchauffé M, Le Coniat M, Raffoux E, Daniel M-T, Erickson P, Drabkin H, MacLeod RAF, Drexler HG, Ghysdael J, Berger R, Bernard OA. Analysis of TEL proteins in human leukemias. Oncogene. 1998;16:2895–2903. doi: 10.1038/sj.onc.1201817. [DOI] [PubMed] [Google Scholar]
- 9.McLean TW, Ringold S, Neuberg D, Stegmeier K, Tantravahi R, Ritz J, Koeffler HP, Takeuchi S, Janssen JWG, Seriu T, Bartram CR, Sallan SE, Gilliland DG, Golub TR. TEL-AML1 dimerizes and is associated with a favourable outcome in childhood acute lymphoblastic leukaemia. Blood. 1996;88:4252–4258. [PubMed] [Google Scholar]
- 10.Rubnitz JE, Shuster JJ, Land VJ, Link MP, Pullen DJ, Camitta BM, Pui C-H, Downing JR, Behm FG. Case-control study suggests a favorable impact of TEL rearrangement in patients with B-lineage acute lymphoblastic leukaemia treated with antimetabolite-based therapy: a Pediatric Oncology Group Study. Blood. 1997;89:1143–1146. [PubMed] [Google Scholar]
- 11.Onadera N, McCabe NR, Rubin CM. Formation of a hyperdiploid karytoype in childhood acute lymphoblastic leukemia. Blood. 1992;80:203–208. [PubMed] [Google Scholar]
- 12.Moorman AV, Richards SM, Martineau M, Cheung KL, Robinson HM, Jalali GR, Broadfield ZJ, Harris RL, Taylor KE, Gibson BES, Hann IM, Hill FGH, Kinsey SE, Eden TO, Mitchell CD, Harrison CJ. Outcome heterogeneity in childhood high-hyperdiploid acute lymphoblastic leukemia. Blood. 2003;102:2756–2762. doi: 10.1182/blood-2003-04-1128. [DOI] [PubMed] [Google Scholar]
- 13.Yeoh E-J, Ross ME, Shurtleff SA, Williams WK, Patel D, Mahfouz R, Behm FG, Raimondi SC, Relling MV, Patel A, Cheng C, Campana D, Wilkins D, Zhou X, Li J, Liu H, Pui C-H, Evans WE, Naeve C, Wong L, Downing JR. Classification, subtype discovery, and prediction of outcome in pediatric acute lymphoblastic leukemia by gene expression profiling. Cancer Cell. 2002;1:133–143. doi: 10.1016/S1535-6108(02)00032-6. [DOI] [PubMed] [Google Scholar]
- 14.Ford AM, Bennett CA, Price CM, Bruin MCA, Van Wering ER, Greaves M. Fetal origins of the TEL-AML1 fusion gene in identical twins with leukaemia. Proc Nat Acad Sci. 1998;95:4584–4588. doi: 10.1073/pnas.95.8.4584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wiemels JL, Cazzaniga G, Daniotti M, Eden OB, Addison GM, Masera G, Saha V, Biondi A, Greaves MF. Prenatal origin of acute lymphoblastic leukaemia in children. Lancet. 1999;354:1499–1503. doi: 10.1016/S0140-6736(99)09403-9. [DOI] [PubMed] [Google Scholar]
- 16.Wiemels JL, Ford AM, Van Wering AM, Postma ER, Greaves M. Protracted and variable latency of acute lymphoblastic leukaemia after TEL-AML1 gene fusion in utero. Blood. 1999;94:1057–1062. [PubMed] [Google Scholar]
- 17.Maia AT, Tussiwand R, Cazzaniga G, Rebulla P, Colman S, Biondi A, Greaves M. Identification of preleukemic precursors of hyperdiploid acute lymphoblastic leukaemia in cord blood. Genes Chromosomes Cancer. 2004;40:38–43. doi: 10.1002/gcc.20010. [DOI] [PubMed] [Google Scholar]
- 18.Yotdna P, Garcia F, Peuchmaur M, Grandchamp B, Duval M, Lemonnier F, Vilmer E, Langlade-Demoyen P. Cytotoxic T cell response against the chimeric ETV6-AML1 protein in childhood acute lymphoblastic leukemia. J Clin Invest. 1998;102:455–462. doi: 10.1172/JCI3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yun C, Senju S, Fujita H, Tsuji Y, Irie A, Matsushita S, Nishimura Y. Augmentation of immune responses by altered peptide ligands of the antigenic peptide in a human CD4+ T-cell clone reacting to TEL/AML1 fusion protein. Tissue Antigens. 1999;54:153–161. doi: 10.1034/j.1399-0039.1999.540206.x. [DOI] [PubMed] [Google Scholar]
- 20.Taylor GM, Dearden S, Ravetto P, Ayres M, Watson P, Hussain A, Greaves M, Alexander F, Eden OB, Investigators UKCCS. Genetic susceptibility to childhood common acute paediatric lymphoblastic leukaemia is associated with polymorphic peptide-binding pocket profiles in HLA-DPB1*0201. Hum Mol Genet. 2002;11:1585–1597. doi: 10.1093/hmg/11.14.1585. [DOI] [PubMed] [Google Scholar]
- 21.UK Childhood Cancer Study Investigators The United Kingdom Childhood Cancer Study: objectives, materials and methods. Br J Cancer. 2000;82:1073–1102. doi: 10.1054/bjoc.1999.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rooney DE, Czepulkowski BH. Human cytogenetics. A practical approach. Vol II. Malignancy and acquired abnormalities. 2nd edn. New York: Oxford University; 1992. [Google Scholar]
- 23.Harrison CJ, Martineau M, Secker-Walker LM. The Leukaemia Research Fund/United Kingdom Cancer Cytogenetics Group Karyotype Database in acute lymphoblastic leukaemia: a valuable resource for patient management. Br J Haematol. 2001;113:3–10. doi: 10.1046/j.1365-2141.2001.02643.x. [DOI] [PubMed] [Google Scholar]
- 24.Harrison CJ, Moorman AV, Barber KE, Broadfield ZJ, Cheung KL, Harris RL, et al. Interphase molecular cytogenetic screening for chromosomal abnormalities of prognostic significance in childhood acute lymphoblastic leukaemia: a UK Cancer Cytogenetics Group Study. Br J Haematol. 2005;129:520–530. doi: 10.1111/j.1365-2141.2005.05497.x. [DOI] [PubMed] [Google Scholar]
- 25.Romana SP, Poirel H, Leconiat M, Flexor MA, Mauchauffé M, Jonveaux P, Macintyre EA, Berger R, Bernard OA. High frequency of t (12; 21) in childhood B-lineage acute lymphoblastic leukaemia. Blood. 1995;86:4263–4269. [PubMed] [Google Scholar]
- 26.POPGENE version 1.31: Microsoft Window-based freeware for population genetic analysis: http://www.ualberta.ca/∼fyeh
- 27.LINKUTIL: http://linkage.rockefeller.edu/soft/linkutil/
- 28.Postuma EFM, Falkenburg JHF, Apperley JF, Gratwohl A, Roosnek E, Hertenstein B, Schipper RF, Schreuder GMT, D’Amaro J, Oudshoorn M, v. Biezen JH, Hermans J, Willemze R, Niederwieser D, on behalf of the Chronic Leukaemia Working Party of the EBMT. Blood 93:3863–3865 [PubMed]
- 29.Castelli FA, Buhot C, Sanson A, Zarour H, Pouvelle-Moratille S, Nonn C, Gahery-Segard H, Guillet J-G, Menez A, Georges B, Maillere B. HLA-DP4, the most frequent HLA II molecule, defines a new supertype of peptide-binding specificity. J Immunol. 2002;169:6928–6934. doi: 10.4049/jimmunol.169.12.6928. [DOI] [PubMed] [Google Scholar]
- 30.Arroyo J, Alvarez AM, Nombela C, Sanchez-Perez M. The role of HLA- DPβ residue 69 in the definition of antibody-binding epitopes. Hum Immunol. 1995;43:219–226. doi: 10.1016/0198-8859(95)00022-V. [DOI] [PubMed] [Google Scholar]
- 31.Diaz G, Catalfamo M, Coiras MT, Alvarez AM, Jaraquemamda D, Nombela C, Sanchez-Perez M, Arroyo J. HLA- DPβresidue 69 plays a crucial role in allorecognition. Tissue Antigens. 1998;52:27–36. doi: 10.1111/j.1399-0039.1998.tb03020.x. [DOI] [PubMed] [Google Scholar]
- 32.Berretta F, Butler RH, Diaz G, Sanarico N, Arroyo J, Fraziano M, Aichinger G, Wucherpfennig KW, Colizzi V, Saltini C, Amicosante M. Detailed analysis of the effects of Glu/Lys b69 human leukocyte antigen-DP polymorphism on peptide-binding specificity. Tissue Antigens. 2003;62:459–471. doi: 10.1046/j.1399-0039.2003.00131.x. [DOI] [PubMed] [Google Scholar]
- 33.Wang Z, White PS, Petrovic M, Tatum OL, Newman LS, Maier LA, Marrone BL. Differential susceptibilities to chronic beryllium disease contributed by different Glu69 HLA-DPB1 and –DPA1 alleles. J Immunol. 1999;163:1647–1653. [PubMed] [Google Scholar]
- 34.Doytchinova IA, Flower DR. f identification of supertypes LA ligand HLA-Ligand for class II MHCs. J Imunol. 2005;174:7085–7095. doi: 10.4049/jimmunol.174.11.7085. [DOI] [PubMed] [Google Scholar]
- 35.Diaz G, Amicosante M, Jaraquemada D, Butler RH, Guillen MV, Sancez M, Nombela C, Arroyo J. Functional analysis of HLA-DP polymorphism: a crucial role for DPβ residues 9, 11, 35, 55, 69, and 84–87 in T cell allorecognition and peptide binding. Int Immunol. 2003;15:565–576. doi: 10.1093/intimm/dxg057. [DOI] [PubMed] [Google Scholar]
- 36.Diaz G, Canas B, Vazquez J, Nombela C, Arroyo J. Characterization of natural peptide ligands from HLA-DP2: new insights into HLA-DP peptide-binding motifs. Immunogenetics. 2005;56:754–759. doi: 10.1007/s00251-004-0735-5. [DOI] [PubMed] [Google Scholar]
- 37.Sette A, Sidney J. HLA supertypes and supermotfis: a functional perspective on HLA polymorphism. Curr Opin Immunol. 1998;10:478–482. doi: 10.1016/S0952-7915(98)80124-6. [DOI] [PubMed] [Google Scholar]
- 38.Sham PC, Curtis D. Monte Carlo tests for associations between disease and alleles at highly polymorphic loci. Ann Hum Genet. 1995;59:97–105. doi: 10.1111/j.1469-1809.1995.tb01608.x. [DOI] [PubMed] [Google Scholar]
- 39.Breslow NE, Day NE. Statistical Methods in Cancer Research. Lyon: IARC Scientific Publications; 1980. [PubMed] [Google Scholar]
- 40.Perneger TV. What’s wrong with Bonferroni adjustments. Br Med J. 1998;316:1236–1238. doi: 10.1136/bmj.316.7139.1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eden OB, Harrison G, Richards S, Lilleyman JS, Bailey CC, Chessells JM, Hann IM, Hill FGH. Long-terrm follow-up of the United Kingdom Medical Research Council protocols for childhood acute lymphoblastic leukaemia, 1980–1997. Leukemia. 2000;14:2307–2320. doi: 10.1038/sj.leu.2401962. [DOI] [PubMed] [Google Scholar]
- 42.Niini T, Kanerva J, Vettenranta K, Saarinen-Pihkala UM, Knuutila S. AML1 gene amplification: a novel finding in childhood acute lymphoblastic leukemia. Haematoligica. 2000;85:362–366. [PubMed] [Google Scholar]
- 43.Mikhail FM, Serry KA, Hatem N, Mourad ZI, Farawela HM, El Kaffash DM, Coignet L, Nucifora G. AML1 gene over-expression in childhood acute lymphoblastic leukemia. Leukemia. 2002;16:658–668. doi: 10.1038/sj.leu.2402399. [DOI] [PubMed] [Google Scholar]
- 44.Sturniolo T, Bono E, Ding J, Raddrizzani L, Tuereci O, Sahin U, Braxenthaler M, Gallazzi F, Protti MP, Sinigaglia F, Hammer J. Generation of tissue-specific and promiscuous HLA ligand databases using DNA microarrays and virtual HLA class II matrices. Nat Biotechnol. 1999;17:555–561. doi: 10.1038/9858. [DOI] [PubMed] [Google Scholar]
- 45.Chan LC, Pegram SM, Greaves MF. Contribution of immunophenotype to the classification and differential diagnosis of acute leukaemia. Lancet. 1985;325:475–479. doi: 10.1016/S0140-6736(85)92085-9. [DOI] [PubMed] [Google Scholar]
- 46.Jennings CD, Foon KA. Recent advances in flow cytometry: application to the diagnosis of hematologic malignancy. Blood. 1997;90:2863–2892. [PubMed] [Google Scholar]
- 47.Alessandri AJ, Reid GSD, Bader SA, Massing BG, Sorensen PHB, Schultz KR. ETV6 (TEL)-AML1 pre-B acute lymphoblastic leukaemia cells are associated with a distinct antigen-presenting phenotype. Br J Haematol. 2002;116:266–272. [PubMed] [Google Scholar]
- 48.Ibisch C, Gallot G, Vivien R, Diez E, Jotereau F, Garand R, Vie H. Recognition of leukemic blasts by HLA-DPB1-specific cytotoxic T cell clones: a perspective for adjuvant immunotherapy post-bone marrow transplantation. Bone Marrow Transplant. 1999;23:1153–1159. doi: 10.1038/sj.bmt.1701768. [DOI] [PubMed] [Google Scholar]
- 49.Renkvist N, Castelli C, Robbins PF, Parmiani G. A listing of human tumor antigens recognised by T cells. Cancer Immunol Immunother. 2001;50:3–15. doi: 10.1007/s002620000169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Niegemann E, Wehner S, Kornhuber B, Schwabe D, Ebener U. Wt1 expression in childhood leukemias. Acta Haematolgica. 1999;102:72–76. doi: 10.1159/000040973. [DOI] [PubMed] [Google Scholar]
- 51.Spanaki A, Linardakis E, Perdikogianni C, Stiakaki E, Morotti A, Cilloni D, Kalmanti M. Quantitative assessment of WT1 expression in diagnosis of childhood acute leukaemia. Leuk Res. 2007;31:569–575. doi: 10.1016/j.leukres.2006.10.015. [DOI] [PubMed] [Google Scholar]
- 52.Oka Y, Elisseeva OA, Tsuboi A, Ogawa H, Tamaki H, Li H, Oji Y, Kim EH, Soma T, Asada M, Ueda K, Maruya E, Saji H, Kishimoto T, Udaka K, Sugiyama H. Human cytotoxic T-lymphocyte responses specific for peptides of the wild-type Wilm’s tumor gene (WT1) product. Immunogenetics. 2000;51:99–107. doi: 10.1007/s002510050018. [DOI] [PubMed] [Google Scholar]
- 53.Dubrovina ES, Doubrovin MM, Lee S, Shieh J-H, Heller G, Pamer E, O’Rielly RJ. In vitro stimulation with WT1 peptide-loaded Epstein-Barr virus-positive B cells elicits high frequencies of WT1 peptide-specific T cells with in vitro and in vivo tumoricidal activity. Clin Cancer Res. 2004;10:7207–7219. doi: 10.1158/1078-0432.CCR-04-1040. [DOI] [PubMed] [Google Scholar]
- 54.Guo Y, Niiya H, Azuma T, Uchida N, Yakushijn Y, Sakai I, Hato T, Takahashi M, Senju S, Nishimura Y, Yasukawa M. Direct recognition and lysis of leukaemia cell by WT1-specific CD4+ T lymphocytes in an HLA-restricted manner. Blood. 2005;106:1415–1418. doi: 10.1182/blood-2005-01-0413. [DOI] [PubMed] [Google Scholar]
- 55.Boublikova L, Kalinova M, Ryan J, Quinn F, O’Marcaigh A, Smith O, Browne P, Stary J, McCann SR, Trka J, Lawler M. Wilm’s tumor gene (WT1) expression in childhood acute lymphoblastic leukaemia: a wide range of WT1 expression levels, its impact on prognosis and minimal residual disease monitoring. Leukemia. 2006;20:254–263. doi: 10.1038/sj.leu.2404047. [DOI] [PubMed] [Google Scholar]
- 56.Greaves M. Infection, immune responses, and the aetiology of childhood leukaemia. Nat Rev Genet. 2006;6:193–203. doi: 10.1038/nrc1816. [DOI] [PubMed] [Google Scholar]
- 57.Spadaro M, Lanzardo S, Curcio C, Forni G, Cavallo F. Immunological inhibition of carcinogenesis. Cancer Immunol Immunother. 2004;53:204–216. doi: 10.1007/s00262-003-0483-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mori H, Colman SM, Xiao Z, Ford AM, Healy LE, Donaldson C, Hows JM, Navarette C, Greaves M. Chromosome translocations and covert leukemic clones are generated during normal fetal development. Proc Nat Acad Sci. 2002;99:8242–8247. doi: 10.1073/pnas.112218799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ravetto PF, Agarwal R, Chiswick ML, D’Souza SW, Eden OB, Taylor GM. Absence of leukaemic fusion gene transcripts in preterm infants exposed to diagnostic x rays. Arch Dis Child. 2003;88:F237–F244. doi: 10.1136/fn.88.3.F237. [DOI] [PMC free article] [PubMed] [Google Scholar]