Abstract
Latent membrane protein (LMP)-2 is one of the Epstein–Barr virus (EBV)-encoded proteins consistently expressed by nasopharyngeal carcinoma (NPC). EBV-transformed lymphoblastoid cell lines (LCL) have been used in patients with NPC to induce LMP-2-recognizing T cell lines which have been in turn utilized for protein-wide mapping of T cell epitopes. However, comprehensive mapping of naturally recognized LMP-2 epitopes in non tumor-bearing individuals has not been reported. Here, we applied a low sensitivity epitope-defining technique for the identification of LMP-2 CTL responses detectable ex vivo in EBV-experienced individuals. This screening tool has been previously validated by analyzing memory CTL responses to Flu, cytomegalovirus (CMV), and the melanoma associated antigen gp100/Mel17. Peripheral blood monocytes (PBMC) from ten Caucasian and ten Chinese individuals were stimulated ex vivo with pools of nonamer (9-mer) peptides overlapping in a stepwise fashion each single amino acid of the LMP-2 sequence. No obvious differences were observed between the immune response of the two ethnic groups save for those related to the divergence in the ethnic prevalence of HLA haplotypes. Several novel and known LMP-2 epitopes were identified. Reactivity toward at least one LMP-2 epitope was detected in 18 of the 20 donors but no prevalent human leukocyte antigen (HLA)/epitope combination was observed confirming that LMP-2 reactivity in the context of common HLA alleles is more pleiotropic than that of FLU and CMV. We believe that the usefulness of these epitopes occurring naturally in non-cancer bearing patients as reagents for the immunization of patients with early or advanced stage NPC deserves further evaluation.
Keywords: Tumor immunity, T cells, Cell activation, Antigens/peptides/epitopes
Introduction
Epstein–Barr virus (EBV) is a γ-herpes virus capable, in the carrier state, to induce lymphoproliferative disorders in immune compromised patients or neoplastic disorders in immune competent individuals. Withdrawal or reduction of immune suppression is widely accepted as an effective treatment of post-transplant lymphoproliferative disorders which are characterized by high expression of the full range of viral proteins by the cancer cells [1]. In addition, complete regression of post-transplant lymphoproliferative disorders can be mediated by adoptive transfer of human leukocyte antigen (HLA)-matched EBV-specific cytotoxic T lymphocytes (CTL) [2–4] and prophylactic administration of CTL can prevent their insurgence [5]. These observations suggest that, at least in these settings, CTL play a major role regulating tumor growth. In contrast with the lymphoproliferative disorders, EBV-associated tumors such as Burkitt’s lymphoma, Hodgkin’s disease and nasopharyngeal cancer (NPC) occur in immune competent individuals and display a restricted expression of EBV proteins [6] which may account for the reduced effectiveness of adoptively transferred EBV-specific CTL [6, 7]. Of the three types of NPC (squamous cell, lymphoepithelial and anaplastic) [8–11], the anaplastic form is predominant worldwide and it is the one characterized by consistent expression of LMP-2 [10]. Encouraging results have been recently reported treating patients with NPC with EBV-specific CTL generated by stimulation with autologous EBV cell lines [12]. These CTL demonstrated a predominant recognition of the EBV latent membrane protein (LMP)-2 [12, 13].
Recent reports suggest the LMP-2 epitopes can be used for active specific immunization of patients with NPC [14]. In addition, in vitro exposure of PBMC from NPC patients to autologous EBV-transformed lymphoblastoid cell lines (LCL) has identified a wealth of novel LMP-2 epitopes [13] some of which were associated with tumor regression following adoptive transfer of the LCL-induced T cell lines [12]. Although useful for therapeutic purposes, the in vitro induction of T cell lines through LCL exposure might introduce a bias on our perception of naturally occurring T cell responses to LMP-2 for instance by skewing antigen processing in the presence of a functional immune proteasome [15]. Therefore, characterization of naturally occurring immune responses to LMP-2 by direct ex vivo testing of normal donors may complement previous studies and expand our perception of the immune repertoire against this viral protein. Thus, a goal of the study was a comprehensive mapping of CTL responses against LMP-2 occurring naturally in EBV-experienced non tumor-bearing individuals of Caucasian or Chinese background. Peptide pools spanning the LMP-2 sequence from the B95.8 EBV strain were used to stimulate peripheral blood mononuclear cells (PBMC) obtained from leukapheresis. To cover epitopes of LMP-2 from other strains of EBV, peptides representing variant sequences of the protein known at the time of the study design were also included [16]. CTL reactivity was documented by identification of increments in interferon (IFN)-γ transcript level in response to stimulation using real-time quantitative PCR (qPCR) as previously described [17–19]. This assay utilizes autologous PBMC for antigen presentation and, therefore, relies on the whole autologous HLA repertoire. Putative epitopes were confirmed by intracellular cytokine staining (ICS) and their HLA association using partially matched heterologous LCL.
The prevalence of anaplastic NPC is dramatically dependent upon genetic background. For instance, some regions of Southern Asia suffer about a 100-fold higher prevalence compared with populations not at risk [10]. Although environmental factors could contribute to this predisposition, the high prevalence of NPC in USA Chinese immigrants suggests a genetic influence [9]. Ethnic predisposition to develop NPC is paralleled by a strong association with some HLA class I alleles. Of them, HLA-A2, -B14, -B46, -B58 and -B61 predispose to the disease while HLA-A11, -A31, -B13, -B27, -B39, -B55 exercises a protective effect [20–29]. A meta-analysis noted a consistent association between HLA-A2, A11, B14 and B46 and NPC [30]. With few exceptions [26, 27], most studies employed serologic HLA typing methods that cannot discriminate among alleles within large serologic families [31]. Such discrimination, however, is critical since different ethnic groups display significant differences in frequency of HLA alleles within a serologic family which may have different capacity to bind and present epitopes. For instance, the HLA-A2 serologic family includes close to 100 alleles (http://www.anthonynolan.com/HIG/) whose distribution is different among ethnic groups with HLA-A*0201 (∼90%) and HLA-A*0205 (∼5%) predominant among Caucasian and HLA-A*0203 (∼20%) -A*0206 (∼15%) and -A*0207 (∼40%) in Asians [32–35]. It is possible, therefore, for Asian patients to be more prone to develop NPC because their HLA class I molecules are less efficient in expressing EBV epitopes [26]. This hypothesis has never been tested by comparative epitope mapping of LMP proteins in non tumor-bearing individuals belonging to diverse populations.
Thus, a second goal of this study was to test whether differences in epitope recognition could be identified between Caucasian and Asian/Chinese individuals exposed to EBV infection that could justify further expansion of this line of studies. As a test model, the HLA-A2 serological family was studied in particular (though not uniquely) since it is relatively frequent in both populations (approximately 50%) and it is characterized by different prevalence of HLA-A2 subtypes. The recruited donors of Caucasian or Asian/Chinese background (ten in each group) were HLA typed by sequence-based typing (SBT) [36–38] and the information was used to compare CTL responses in the context of ethnically related allelic diversity.
Materials and methods
Donor accrual
Ten Chinese and ten Caucasian adults who had been exposed to silent primary EBV infection were studied. Individuals who experienced infectious mononucleosis more than ten years prior to enrollment were also eligible. Prior EBV exposure was determined by standard serological identification of IgG antibodies against EBV. Asian/Chinese individuals were defined as those who were born in China (including Taiwan, Hong Kong and Singapore) and immigrated to the USA afterwards as well as the first generation offspring from couples of this ethnicity. Caucasian individuals were defined as any individual born in the USA or Europe of clear Western ancestry. Preferentially, individuals of Northern European ancestry were selected since the frequency of HLA-A*0201 peaks in this ethnic group. Racial selection was not intended to perfectly represent the two ethnic groups since the goal of the study was not to describe genetic differences but rather increase the chances of accruing individuals with HLA-alleles relevant to the immunologic studies to be performed in the two populations. Participants passed the criteria for blood donors established by the American Association of Blood Banks including a WBC ≥ 3,000 per mm3 and a platelet count of ≥90,000 mm3. Individuals who tested positive for hepatitis HBsAG or HIV antibody or HIV or who had any form of active primary or secondary immune deficiency were excluded because of possible immune effects of these conditions. The donor accrual was performed through a National Cancer Institute—Institutional Review Board approved protocol (04-CC-0007) and all individuals signed informed consent before donation.
Lymphocyte separation and peptide stimulation
PBMC were collected by apheresis using the Fenwal CS3000 blood cell separator (Fenwal, Deerfield, IL). Citrate was used as an anticoagulant. During each procedure five liters of whole blood were processed with the goal of obtaining approximately 5 × 109 mononuclear cells. PBMC were separated from granulocytes and red cells using ficoll-hypaque density gradients and frozen in aliquots of 1 × 108 cells. Later, the cells were thawed and a portion of cells was stimulated with pools of peptides from the LMP-2 B95.8 consensus strain plus additional epitopes spanning variant regions of alternate strains of EBV [16]. Each pool contained ten peptides dissolved in DMSO to a final concentration of 1 μM. Stimulation of PBMC was performed following the previous described method [17–19, 39, 40] by plating 2 × 105 thawed PBMC per U-bottom well in a 96 well plate and exposing them to exogenous peptide administration for 3 h before RNA preparation for quantitative real-time PCR (qRT). We have previously discussed extensively the details of the assay, its rationale and possible pitfalls [17–19, 39–42]. A specific CTL response to a peptide pool was based on a confidence interval above a 0.99 divergence over a baseline stimulation with an inert peptide as described in the statistical section [39, 40]. Peptide pools inducing CTL responses were segregated into individual peptides to stimulate in a second assay PBMC of the responsive individual for identification of the minimal determinant. Positive controls included stimulation of PBMC with a pooled peptide library encompassing the complete amino acid sequence of the CMV pp65 protein, the anti-CD3 mAB OKT3, pokeweed mytogen (PWM) and staphylococcal exotoxin B (SEB). The Flu M1:58–66 and the CMV pp65:495–503 epitopes were used as positive controls in HLA-A2 individuals [43] while gp100:209–271 [17, 18] and Flu M1 RLEDVFAGK [44] were used as negative controls to assess respectively epitope and HLA specificity.
Epitope mapping
The epitope library was constructed by synthesis of 9-mer peptides tiled at a one amino acid pace to cover the complete protein sequence of the B95.8 prototype [45]. Nine amino acid lengths were selected to exclude possible binding incompatibility to HLA alleles at the carboxyl terminus as previously observed [13, 46]. Known polymorphic sequences [47] were also included. Epitope mapping was performed by preparing two sets of peptide pools to develop a two-dimensional matrix. The first set consisted of pools including ten peptides each clustered according to a one amino acid tiling selection, therefore, including overlapping portions of LMP-2 (overlapping pools 1–36). The second set consisted of pools that included ten peptides each selected at ten amino acid intervals along the sequence of LMP-2 (spaced pools #A1–A10, #B1-B1- and #C1–C10). This strategy potentially facilitates the identification of putative epitopes by comparing results between the two sets since each pool in one set shares only one peptide present in any pools from the other set.
EBV DNA amplification and quantification in donors PBMC
EBV DNA was quantified in patient PBMCs using the Light-Cycler system (Roche Molecular Biochemicals, Indianapolis, IN). In brief, primers EBV FOR.4 (5-AGGAAGCGGGTCTATGGTTGGCTG-3) and EBV REV.5 (5-TAGAACTGACAATTGSCTGCTGTCTG-3, where S–C or G) were used to amplify a segment from the BamHI-W fragment of the EBV genome. Fluorescent resonance energy transfer (FRET) detection probes were commercially synthesized (IT BioChem, Salt Lake City, UT) and labeled with Red 640 as the reporter fluorescent dye: EBVFRETUP.2 (5-GGCCCAAGGGGGTTCGCGTTGCTAG-Fluorescein-3) and EBVFRETDN.2 (5-Red 640-CCACCTTCTCAGTCCAGCGCGTTTAC-3). Real-time PCR was performed in a 20 μL reaction containing a Light-Cycler FastStart DNA Master Hybridization Probes reaction mixture (which contains FastStart Taq polymerase [Roche Molecular Biochemicals], 10 μL of patient sample DNA, deoxynucleoside triphosphate primers [0.5 μM each], FRET probes [0.2 μM each], 4.0 mM MgCl2, and 1 U of uracil-DNA glycosylase). The mix was pre-incubated for 10 min at 30°C, after which PCR was carried out under the following conditions: initial denaturation of 10 min at 95°C, followed by five cycles of 10 s at 95°C and 25 s at 72°C; touch-down procedure, consisting of 6 s at 95°C, annealing for 10 s at temperatures decreasing from 72°C to 55°C during the first nine cycles (with 3°C decremental steps in cycles 2–9), and ending with an extension step at 72°C for 20 s. A total of nine touch-down cycles plus 36 cycles of annealing at 55°C were performed. Amplichek EBV viral DNA control (Advanced Biotechnologies, Columbia, MD) was used as a quantitative reference standard. The EBV DNA control (1 × 104 EBV genome copies per microliter) was diluted to give a standard curve of 5–5,000 EBV genome copies per reaction volume. All fluorescence data were analyzed using LightCycler software (Promega). The LightCycler software generated a best-fit line from the log-linear region of each standard curve that defined the crossing line. The point of intersection between the emitted fluorescence and the crossing line defined the crossing point. The concentration of target DNA was calculated by plotting the crossing point of each sample on the standard curve using the LightCycler software. To detect PCR inhibitors in the patient specimens, a “mimic” sequence amplifiable by the EBV primers described above was constructed by amplifying a segment of the tetracycline resistance gene of pBR322 with primers tailed with the corresponding EBV primers. The resulting sequence, AGGAAGCGGGTCTATGGTTGGCTGTTGCTAACGCAGTCAGGCACCGTGTATGAAATCTAACAATGCGCTCATCGTCATCCTCGGCACCGTCACCCTGGATGCTGTAGGCATAGGCTTGGTTATGCCGGTACTGCCGGGCCTCTTGCGGGATATCGTCCATTCCGACAGCATCGCCAGTCACTATGGCGTGCTGCTAGCGCTATATGCGTTGATGCAATTTCTATGCCAGACAGCAGCCAATTGTCAGTTCTA, was cloned, and the plasmid was propagated and purified. The EBV primers described above were used to amplify the mimic (3000 copies) in a separate reaction that included 10 μL of extracted DNA from each patient sample. The mimic detection FRET probes were commercially synthesized (IT BioChem) and labeled with Red 705: MIMICFRET UP (5-GAT ATC GTC CAT TCC GAC AGC ATC-Fluorescein-3) and MIMICFRET DN (5-Red 705-CCA GTC ACT ATG GCG TGC TGC TAG-Phosphate-3). Real-time PCR was carried out with the Light-Cycler system, as described above. To determine the number of human cellular equivalents tested for the presence of EBV, another aliquot of patient DNA samples was also subjected to real-time PCR analysis for the human β-globin gene using the LightCycler Control DNA kit (Roche Molecular Biochemicals), according to the manufacturer’s recommendations. EBV viral load was calculated by dividing the EBV copy number in a given sample by the number of cellular genome equivalents, based on the human β-globin gene, in the sample. The EBV viral load was expressed per 106 cellular genome equivalents.
Quantitative real-time PCR (qRT)
Quantitative real-time PCR (qRT) was performed as previously described using primers and probes that we have already extensively characterized [43]. Three hours after stimulation of PBMC with peptide pools and relevant controls, RNA was extracted (RNeasy Mini Kit, Qiangen, Valancia, CA) and 1 μl of total RNA was reverse transcribed into cDNA (Invitrogen, Carlsbad, CA). One μl of synthesized cDNA was used as template to measure IFN-γ mRNA abundance by qRT using an ABI Prism 7900 Sequence Detection System (Perkin Elmer, Foster City, CA). Beta-actin (β-actin) was used as internal reference gene.
Intra-cellular cytokine staining (ICS)
Confirmation of epitope specificity was performed by ICS. ICS was performed on independently thawed PBMC or after one round of in vitro sensitization (IVS) to further increase the number of epitope-specific T cells. IVS was performed by plating 1 × 106 per ml PBMC in 48 well plates with 1 ml of complete medium (Iscove, Life Technologies, Grand Island, NY) supplemented with 10% heat inactivated human AB serum, 10 mM HEPES buffer, 100 U/ml penicillin-streptomycin, 0.5 mg/ml amphotericin B and 0.03% glutamine. After resting overnight, the cells were stimulated with 1 μg/ml of pooled or single peptides and cultured for a week in an incubator adding recombinant human IL-2 (rHuIL-2) (Chiron Co, Emeryville, CA) every 2 days at the final concentration of 300 IU/ml. As controls cells were stimulated with gp100:209–217 (melanoma antigen-derived peptide, negative control), OKT3, PWM, SEB and the pooled library of CMV-pp65 epitopes (positive controls). After overnight resting the freshly thawed PBMC (ex vivo assay) or at day 7 (IVS assay) the T cell cultures were stimulated with the relevant pools or peptides. After 1 h from the second stimulation, BrefeldinA was added to the wells at the final concentration of 10 μg/ml and the cells were incubated for extra 5 h at 37°C. The cells were then harvested, washed and fixed in FACS lysing solution (BD Bioscience, San Jose, CA). The day after, cells were permeabilized (Perm2, BD Bioscience), washed and stained with mAb against CD8-PE, CD3-PerCP, CD4-APC and IFNγ-FITC (all from BD Bioscience) for 30 min at 4°C in the dark. Samples were analyzed with FACS sort (BD Bioscience).
Assignment of epitope/HLA associations
The HLA of each donor was determined by sequencing of the HLA-A, -B and -C alleles and ambiguities were resolved as previously discussed [36, 48] (Table 1). Identification of the HLA alleles associated with individual epitopes was deducted by cross-referencing the results in different patients and tested by presenting in separate experiments the test epitopes exogenously pulsed on partially matched heterologous LCL to sensitized T cell cultures. The HLA type of the most frequently used LCL is shown in Table 2.
Table 1.
Sequence-based HLA typing of donors and presence of EBV DNA in their PBMC
| Donor ID | HLA-A | HLA-B | HLA-C | Molecular EBV testing (PBMC) |
|---|---|---|---|---|
| CAU#1 09193-5 | A*0205, 2601 | B*3801, 5801 | C*0701(0706), 12030 | Positive |
| CAU#2 01500-3 | A*0201, 0310 | B*3503, 5701 | C*04, 0602 | Negative |
| CAU#6 23530-1 | A*0201 | B*15 ,44 | C*0102, 05 | Positive |
| CAU#7 19730-0 | A*0101, 3301 | B*1402, 27 | C*0102, 0802 | Negative |
| CAU#8 24311-5 | A*0101, 0201 | B*27, 5701 | C*0202, 0602 | Positive |
| CAU#9 20023-2 | A*0301, 0301 | B*18, 35 | C*04, 07 | Positive |
| CAU#11 20028-1 | A*0101, 2601 | B*0702, 3801 | C*0702, 1203 | Negative |
| CAU#16 25090-9 | A*24, 26 | B*3801, 4002 | C*0202, 1203 | Negative |
| CAU#24 16959-6 | A*0201, 0201 | B*1501, 5701 | C*0102, 0602 | Negative |
| CAU#26 23794-2 | A*0101, 0201 | B*08a, 15a | C*0102, 07 | Positive |
| CHN#3 25088-0 | A*0201, 1101 | B*1301, 5201 | C*0304, 1202 | Negative |
| CHN#4 25097-1 | A*0203, 2402 | B*1502, 4001 | C*0702, 0801 | Positive |
| CHN#5 25100-8 | A*0207, 2402 | B*3802, 4601 | C*0102, 0702 | Negative |
| CHN#10 01772-3 | A*0201, 1101 | B*4001, 4601 | C*0702, 0702 | Positive |
| CHN#12 25299-2 | A*1102, 2402 | B*2704, 4001 | C*1202, 1502 | Negative |
| CHN#13 25249-9 | A*0207, 2402 | B*35, 4601 | C*0102, 0303 | Negative |
| CHN#17 25203-7 | A*1102, 2402 | B*1301, 1302 | C*0304, 0602 | Negative |
| CHN#23 20459-6 | A*1101 | B*1301, 1502 | C*1501, 1602 | Negative |
| CHN#27 25584-1 | A*24, 31 | B*0702, 5801 | C*0302, 0702 | Negative |
| CHN#29 25680-8 | A*1101, 2601 | B*0801, 4001 | C*0304, 0702 | Positive |
Table 2.
HLA of EBV lines used in this study
| ID | A | B | Cw | DRb1 | DRb_ | DQb1 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CL003 | 01 | 6801 | 1402 | 57 | 0602 | 0802 | 01021 | 1305 | 0301 | 0501 | 3*0202 | |
| CL008 | 0301 | 0702 | 0702 | 0401 | 4*01 | 0302 | ||||||
| CL015 | 0205 | 3201 | 1510 | 40 | 0202 | 03 | 0407 | 0806 | 4*01 | 0301 | 602 | |
| CL032 | 2601 | 3801 | 1203 | 0402 | 4*01 | 0302 | ||||||
| CL049 | 0205 | 5801 | 0701 | 0301 | 3*0202 | 0201 | ||||||
| CL057 | 11011 | 3001 | 1302 | 1502 | 0602 | 0801 | 0901 | 1202 | 3*0301 | 04 | 06 | |
| CL068 | 03 | 68 | 42 | 5801 | 0302 | 17 | 03 | 13 | 3*0101 | 04 | 0603 | |
| CL190 | 11 | 13 | 44 | 0403 | 1604 | 11 | 1202 | 3*02 | 3*03 | 03 | ||
| CL216 | 0201 | 570101 | 0602 | 0701 | 01030102 | 030302 | ||||||
| CL224 | 240301 | 3303 | 1512 | 4601 | 0102 | 30302 | 0406 | 1202 | 0301 | 0302 | ||
| CL226 | 110101 | 1102 | 4601 | 5201 | 0102 | 070201 | 090102 | 1410 | 3*0202 | 4*01 | 030302 | 050301 |
| CL228 | 240201 | 520101 | 120202 | 150201 | 5*0102 | 0601 | ||||||
| CL274 | 0201 | 1302 | 0602 | 0701 | 4*0101 | 0202 | ||||||
Data analysis and validation
One goal of the study was to compare ex vivo and in standardized conditions epitope immune dominance between individuals of Caucasian and Chinese ancestry prevalently carrying diverse HLA alleles. In particular, the HLA-A2 system was used as test model. All data were evaluated using the Log2 of individual values and are presented as mean ± SD. Log2 of the absolute IFN-γ copy number and the IFN-γ/β-actin ratios were analyzed obtaining comparable results in all experiments. Therefore, the subsequent analysis was performed only on IFN-γ/β-actin ratios. To account for individual variations in baseline IFN-γ expression, Log2 IFN-γ/β-actin ratios were further adjusted using, as a normalization factor, the average values of all peptide pool stimulations (n = 66) for each donor’s PBMC. This represents a modification of the principle of relative changes in gene expression between paired samples as described by others [49]. This normalization was based on the assumption that peptide pools would be inert in the majority of cases and, therefore, the average of all experiments should approximate a central value in a normal distribution approaching the value for non-stimulated autologous PBMC. Normalized data were subsequently presented as (n) log2 IFN-γ/β-actin. Alternative methods of relative quantification (i.e. the 2–ΔΔCT [49]) yielded concordant results.
Epitope-specific induction of IFN-γ mRNA expression by single or pooled peptides was defined by assessing the confidence interval (CI) of log2 IFN-γ/β-actin values when each donor PBMC was stimulated with an inert peptide (gp100:209–217) never recognized ex vivo by PBMC from non melanoma-bearing, not immunized HLA-A2 expressing or not expressing individuals [17]. Normalized (n) log2 IFN-γ/β-actin in response to stimulation with gp100:209–217 were separately assessed for the Caucasian and the Chinese cohorts. For the ten Caucasian donors the normalized values were 0.881 ± 0.19 resulting in a 0.99 CI of 0.158; therefore, the cutoff to define specific a response to a given pool or peptide was set to a (n) log2 IFN-γ/β-actin ≥ 1.040. For the ten Chinese donors the normalized values were 0.944 ± 0.104 resulting in a 0.99 CI of 0.085 and a specificity cutoff (n) log2 IFN-γ/β-actin ≥ 1.028.
Significant induction of IFN-γ mRNA expression between experimental groups was evaluated by the non-parametric two-tailed Fisher exact test assigning positive or negative values according to the cut off thresholds discussed above. Paired Student t test was used to compare results between unstimulated and stimulated autologous PBMC. In addition, significance was assessed parametrically comparing experimental group results with an unpaired Student t test. Pearson correlation was used to quantify correlations between individual overlapping pools and corresponding spaced pools across the entire donor population.
Positive cut-off values for ICS were considered those whose percent of IFN-γ expressing CD8 or CD4 T cells occurred at a minimum frequency of 0.1% and were at least three fold that of parallel samples stimulated with an irrelevant peptide. This cut-off value also encompassed a previous requirement for values that were above three standard deviation over the distribution of values from samples stimulated with irrelevant peptides [50]. All analyses were confirmed by at least two subsequent independent experiments.
Results
Donors’ characteristics and HLA phenotyping
The ethnicity of individual donors, their HLA class I phenotype and the results of molecular testing for EBV in PBMC are shown in Table 1. Serologically, all donors were VCA IgG positive and IgM negative.
Ex vivo recognition in the context of HLA-A2 of the well-characterized Flu, gp100/PMel17 and CMV epitopes
To validate the methodology, we tested the effect of stimulation with the well characterized HLA-A*0201-associated epitopes Flu M1:58–66 (to which most HLA-A2 expressing patients should be responsive) and gp100:209–217 inert in the same conditions [17].
This analysis was of interest because 4 of the 11 HLA-A2 bearing donors expressed a subtype different from HLA-A*0201 (two were A*0207, one A*0203 and one A*0205, Table 1). Therefore, we stratified the analysis by comparing results from HLA-A*0201 PBMC alone with those obtained from non-HLA-A2 donors or, incrementally adding PBMC from -A*0203 and -A*0207 and -A*0205 donors to the HLA-A2 experimental group. This was done because of the conflicting results previously obtained testing various canonical HLA-A*0201-associated epitopes in the context of different alleles belonging to the HLA-A2 super family [46, 51]. A paired comparison of IFN-γ levels between epitope-stimulated versus non-stimulated PBMC demonstrated significantly higher expression of IFN-γ by HLA-A2 bearing PBMC in response to Flu M1:58–66 stimulation but not gp100:209–217 (Table 3). This observation was corroborated by applying a Fisher’s exact test to (n)log2 IFN-γ/β-actin values observed in HLA-A2 and non HLA-A2-bearing PBMC stimulated with Flu peptide. Epitope-specific responses were considered those above the 0.99 CI of PBMC stimulated with the irrelevant gp100:209–217 epitope as described in the Materials and methods. Ten out of ten HLA-A2 carrying donors with the exclusion of donor #1 (bearing the HLA-A*0205 allele) reacted positively to FLU M1:58–66 while only two out of nine non-HLA-A2 expressing donors’ PBMC resulted positive based on the same criteria (Fisher’s exact test p 2-value < 0.001). The lack of response of the HLA-A*0205 bearing donor was not surprising as we have previously noted a poor response to the Flu M1:58–66 epitope in association with this allele [46, 51]. As a negative control, the gp100 epitope did not elicit epitope-specific increments in IFN-γ mRNA in any HLA-A2 bearing PBMC and one borderline positive response in a non-HLA-2 bearing donor. We also analyzed the reactivity to another immune dominant HLA-A2-associated epitope from the CMV pp65 protein (pp65:495–503). This epitope induced enhancement of IFN-γ expression in HLA-A2 expressing PBMC. The enhancement did not reach statistical significance when only the seven HLA-A*0201 expressing PBMC were tested possibly because two of them were serologically negative for CMV infection reducing the power of the analysis to only five samples. The inclusion of the other HLA-A2 subtypes restored significance to the analysis comparing HLA-A2-associated, CMV specific responses between HLA-A2-bearing versus non HLA-A2-bearing individuals (Table 3). Thus, ex vivo reactivity against classic epitopes known to be recognized with high prevalence in the context of HLA-A2 could be consistently identified by detection of IFN-γ transcript following direct stimulation of PBMC. Since LMP-2 responses are subdominant compared with Flu and CMV, the aim of the study was not to compare such responses but rather to use a validated method for efficient ex vivo screening of T cell epitopes that could consistently identify dominant immune responses in association with specific HLA presentations [52]. Therefore, the LMP-2 responses identified in individual donors by this method could be considered comparable to those of Flu or CMV in such individuals although they might not be as reproducibly identified in other individuals bearing an identical HLA allele due to their sporadic nature.
Table 3.
Validation of ex vivo assay specificity and sensitivity in the context of known immune-dominant T cell epitopes
| (n) log2 IFN-γ/β-act | ||||||||
|---|---|---|---|---|---|---|---|---|
| A*0201 | A*0201;07;03 | A*0201;03;05; 07 | Non A2 | |||||
| N = 7 | N = 10 | N = 11 | N = 9 | |||||
| AVE | p2-value | AVE | p2-value | AVE | p2-value | AVE | p2-value | |
| HLA-A2 controls | ||||||||
| PP65:495-503 (A2) | 1.58 | 0.118 | 1.51 | 0.045 | 1.49 | 0.036 | 0.99 | 0.945 |
| FLU M1:58–66 (A2) | 1.36 | 0.003 | 1.33 | 0.000 | 1.30 | 0.001 | 0.96 | 0.620 |
| GP100:209–217 (A2) | 0.85 | 0.181 | 0.88 | 0.212 | 0.91 | 0.265 | 0.92 | 0.105 |
| FLU (A3) | 1.04 | 0.936 | 1.00 | 0.955 | 1.03 | 0.924 | 1.00 | 0.707 |
| PWM | 2.48 | 0.007 | 2.43 | 0.001 | 2.46 | 0.000 | 1.58 | 0.000 |
| OKT-3 | 1.81 | 0.011 | 1.77 | 0.003 | 1.83 | 0.001 | 1.30 | 0.005 |
Normalized (see statistical section) (n) log2 IFN-γ/β-actin values reflecting the transcriptional response of PBMC to stimulation with immune dominant epitopes from CMV (pp65:495–503) and Flu (Flu M1:58–66) known to be associated with HLA-A*02 alleles are shown. As negative controls for epitope-specificity associated with HLA-A*02 presentation, gp100:209–217 was used as a HLA-A*02-associated epitope known to induce IFN-γ transcripts detectable ex vivo only in HLA-A*0201-bearing individuals with metastatic melanoma who had been previously exposed to the same epitope for anti-cancer immunization purposes [17, 18]. As negative control for HLA-specificity the HLA-A*03 restricted Flu matrix peptide RLEDVFAGK was used [44]. To assess maximum expression of IFN-γ the general T cell stimulators PWM and OKT-3 were used that stimulate T cells (OKT-3) and immune cells (PWM) independently of epitope/HLA restriction; p 2-values refer to paired two-tailed t test between the individual log2 IFN-γ/β-actin values compared to the normalization factor (NF) for each autologous PBMC. NF = average of the IFN-γ/β-actin values for all pools tested in each individual’s PBMC (see statistical section). Significant (≤ 0.05) values are shown underlined with the associated (n) log2 IFN-γ/β-actin values are shown in italics
Ex vivo recognition of previously known EBV epitopes in HLA-A2, A11 and A24 individuals
Contrary to Flu and CMV, analysis of pools in which HLA-A2 associated epitopes of LMP-2 known at the time of the study design were present did not elicit significant differences in IFN-γ transcription between HLA-A2 and non-HLA-A2 expressing PBMC. In addition, no evidence of direct ex vivo activation of PBMC was noted when peptides representative of the variant strains of EBV were tested. It should be clarified that this result does not exclude that in individual donors the reactivity toward a given epitope could be detectable ex vivo. This analysis only allows the conclusion that the prevalence of such responses in a random population of EBV-experienced donors does not reach the consistency observed with other viral model systems such as Flu or CMV to allow the detection of significant difference across the donors’ pool. A similar analysis comparing PBMC from HLA-A11 or HLA-A24-bearing individuals did not identify any prevalent reactivity associated with these alleles. Thus, in general, no prevalent LMP-2 epitope could be identified in association with common HLA class I alleles whose frequency of detection could be compared with that of other viral models.
Identification of HLA-associated patterns of LMP-2 reactivity ex vivo
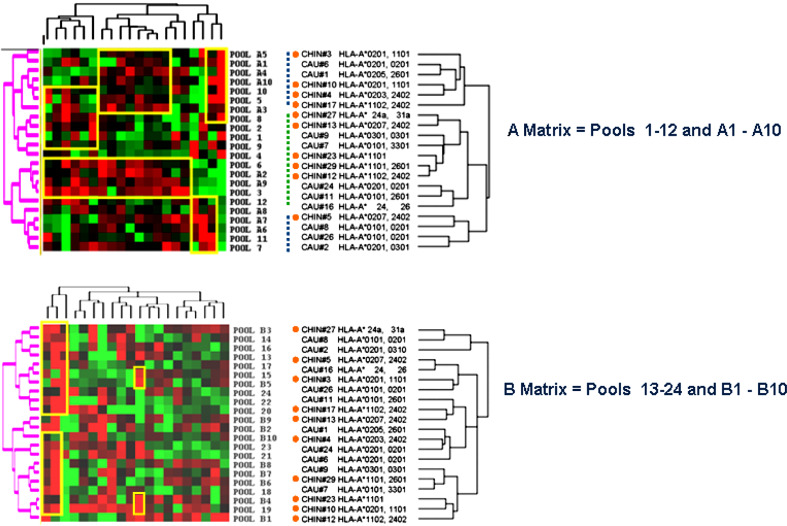
Using the two-dimensional matrix system for each matching pool sub-category we looked for closed correlations between the overlapping (1–12, 13–24 or 25–36) and the correspondent spaced pools (A1–10, B1–10 or C1–10). This was obtained by applying the Eisen’s hierarchical method [53] for unsupervised clustering of PMBC samples and peptide pools (Fig. 1). This allowed an unbiased, global characterization of the immune response to LMP-2 epitopes according to ethnic group and/or HLA-phenotype. The clustering program demonstrated that in each matrix category (only A and B matrixes are shown) there was no significant demarcation of immune response between Chinese and Caucasian donors although some clusters were observed with an enrichment of one population over the other. For instance, in the Matrix A category, clusters were identified with preferential inclusion of Chinese (orange circles) or Caucasian subjects but such grouping was not statistically significant. Groups of donors could be best segregated according to their HLA phenotype. For instance, among the A matrix clusters subgroups could be identified: one predominantly including non HLA-A2 individual (2 out of 10, green dashed vertical line) flanked by two groups in which PBMC bearing HLA-A2 predominated (9 out of 10 individuals, blue dashed vertical lines) (Fisher exact test p 2-value = 0.006, Fig. 1). Interestingly, a sub-class of PBMC in the non-HLA-A2 cluster included a group of 5 PBMC expressing HLA alleles belonging to the HLA-A3-like super family (including HLA-A*0301, HLA-A*1101, HLA-A*3101 and HLA-A*3301) often sharing the presentation of identical peptides [54]. Similarly, in the B matrix, PBMC bearing HLA-A11 or other alleles belonging to the HLA-A3-like super family appeared to segregate separately from the remaining PBMC (Fisher exact test p 2-value = 0.04, Fig. 1b). Interestingly, when the same analysis was applied to evaluate clusters based on the HLA-B or C loci no predominant pattern was observed (data not shown). This suggests that HLA-A alleles exert a predominant role in determining the overall immune response to LMP-2. Obviously, because of the linkage disequilibrium among HLA loci results in their co-expression as ancestral haplotypes, a conclusive segregation of the weight that individual loci play in determining the immune response to LMP-2 could not be done by this approach.
Fig. 1.
Eisen’s clustering analysis of individual peptide pools and PBMC. Normalized (n) log-2 IFN-γ/β-actin ratios are presented according to the central normalization method [83] for the three sub-categories of pool matrixes (A: including pools 1–12 and A1-A10, B: including pools 13–24 and B1–B10). Top panel Matrix A; PBMC from Chinese subjects are underlined by orange circles. Clusters of PBMC predominantly expressing HLA-A2 are outlined by blue dashed vertical bars and the cluster containing PBMC predominantly not expressing HLA-A2 is outlined by a dashed green bar. Bottom panel Matrix B
Ex vivo identification of LMP-2 epitopes in individual PBMC samples using the two-dimensional matrix
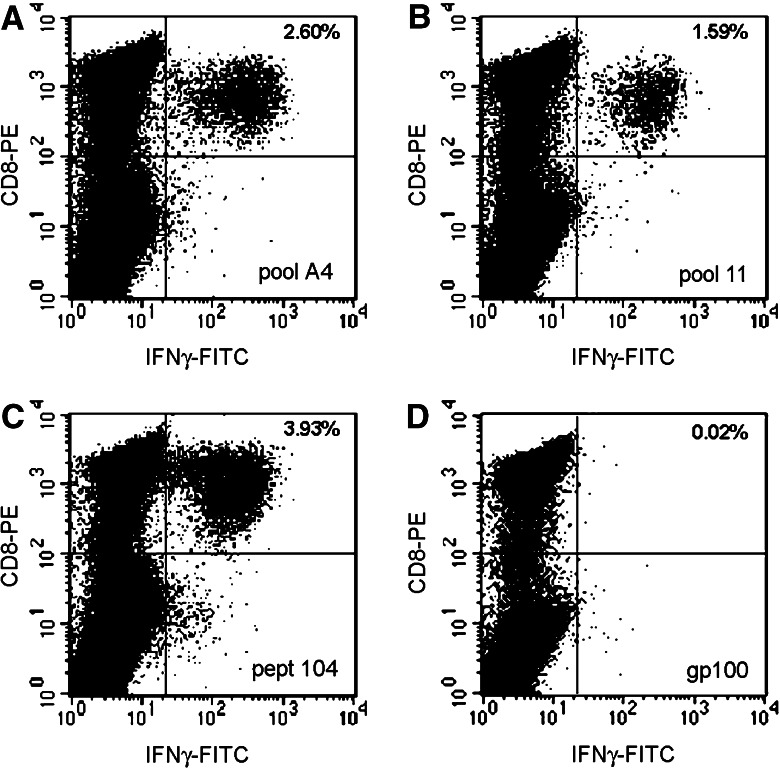
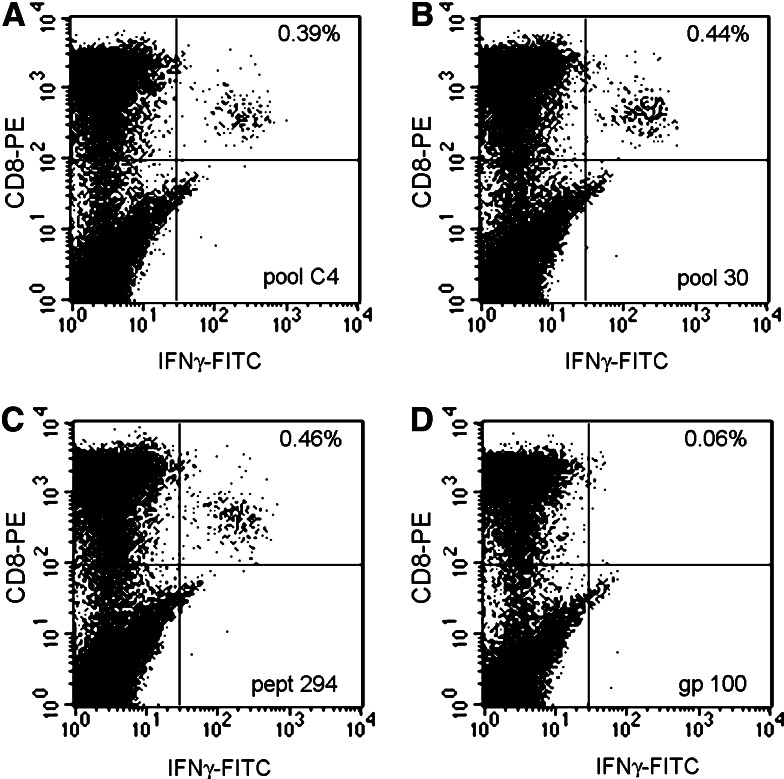
Comparison between overlapping and spaced pools in search of common minimal epitopic determinants was done by ranking the R 2 Pearson’s correlation coefficients between each overlapping peptide pool and the corresponding spaced pool belonging to the same matrix (Matrix A: pools 1–12 and A1–A10; Matrix B: pools 13–24 and B1–B10; Matrix C: pools 25–36 and C1–C10). Correlation was performed by comparing the pattern of induction of IFN-γ mRNA in all 20 samples with the assumption that the strongest correlation should occur most likely between pools sharing the same minimal determinant. Several spaced pools were identified with patterns of IFN-γ mRNA expression strongly correlating with that of overlapping pools indicative of a shared minimal epitopic determinant. Individual PBMC results from these pools were analyzed to identify those that had shown epitope-specific responses according to the previously described parameters: (n) log2 IFNγ/β-actin above 1.04 for Caucasian and 1.028 for Chinese subjects. This approach identified PBMC samples likely to be reactive ex vivo to the epitope suggested by the Matrix analysis. This systematic approach identified reactivity toward at least one LMP-2 epitope in 18 out of 20 donors with the exception of donors 11 and 29 who did not display any reactivity to LMP-2. The epitopes identified by this approach are summarized in Table 4. Several of the epitopes had been previously described by others (marked by asterisks) [13, 15, 47, 55–59] while 18 novel candidate epitopes had not been previously described.
Fig. 3.
Example of epitope validation applying ICS after IVS to increase the frequency of epitopes-specific T cells. PBMC from donor #8 were tested after IVS at day 1 with pool A4 (a). At day 7 the same culture was stimulated with several overlapping pools of which pool 11 tested positive (b). Based on this result, at day 9 the same culture was tested with the corresponding single peptide corresponding according to the two dimensional matrix to LMP2-104 (c). d Negative control (stimulation with gp100:209–217) of the same culture at day 7. In the upper right corner the percent of CD3+CD8+ and IFN-γ+ cells is shown
Fig. 4.
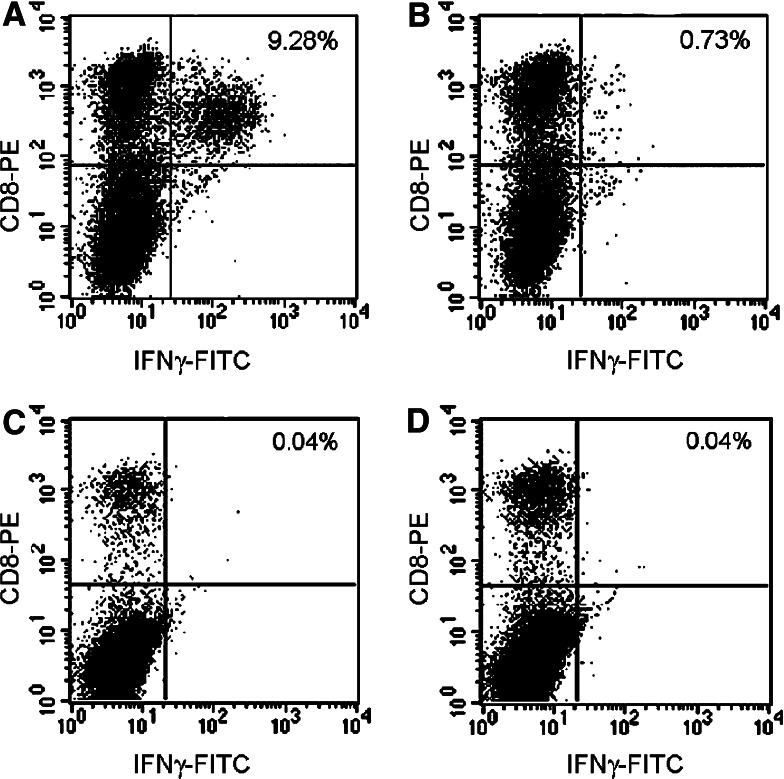
Example of assignment of HLA/epitope association based on parallel stimulation of T cell culture with HLA partially matched and mismatched heterologous LCL exogenously loaded with the target epitope. In this case, PBMC from donor 1 underwent IVS with pool B5. After 7 days the cultures were tested by ICS for reactivity toward a panel of partially matched and unmatched LCL pulsed with LMP:145 peptide. Shown is the response to LCL CL049 pulsed with LMP: 145 (a) or with an irrelevant peptide (b) which shares the HLA-A*0205/B*58 alleles with donor 1. No response could be elicited using other LCL including the unmatched CL274 pulsed with LMP:145 (c) or an irrelevant epitope (d)
Table 4.
Candidate LMP-2 epitopes identified by ex vivo screening of PBMC or identified in association with HLA alleles relevant to this study [13]
| Candidate epitope | Donor | Candidate HLA-allele | [13] | HLA in [13] |
|---|---|---|---|---|
| Matrix A | ||||
| *LMP-2: - 2 pylfwlaai* | – | – | + | A23/24 |
| LMP-2:4 laaiaascf | 2, 8, 16, (17) | A2, B57, DR07 | – | – |
| LMP-2:6 aiaascfta | 8, 10, 16, 27, (17) | A2, A24, DR07 | - | - |
| LMP-2:10 scftasvst | 2, 27 | B 57/58 | - | - |
| LMP-2:15 svstvvtat | 2, 7, 8 | A1, A11, C6 | – | – |
| LMP-2:25 lalslllla | 8 | A2, B57 | – | – |
| LMP-2:50 vtvltavvt | 2, 9, 26 | A2, B35, C4 | – | – |
| LMP-2:75 sllfallaa | 2, 8 | A 2, B57, C6 | – | – |
| LMP-2:84 agglqgiyvl | (17) | DR07 | – | – |
| LMP-2:94 vmlvllila | 2 | A2 | – | – |
| *LMP-2:104 rrrwrrltv* | 8a, 12, 24 | B27 | + | B27 |
| LMP-2:107 wrrltvcgg | 8a, 12, 24 | B27 | – | – |
| *LMP-2:108 rrltvcggi* | – | – | + | B27 |
| *LMP-2:111 tvcggimfl* | – | – | + | A0101/06 |
| LMP-2:115 gimflacvl | 8 | A2, B27 | – | – |
| Matrix B | ||||
| LMP-2:123 lvlivdavl | 2, 8 | A*0201 | – | – |
| *LMP-2:125 livdavlql* | – | – | + | A0204 or 0217 |
| LMP-2:136 llgavtvvs | 2, 8 | A*0201 | – | – |
| LMP-2: 145 mtllllafv | 1, 2 | A*0205, B57/58 | – | – |
| LMP-2: 155 wlsspgglg | 1, 2, 8, | A*0205/0201 | - | - |
| *LMP-2:161 glgtlgaal* | – | – | + | A2 |
| *LMP-2:197 llwtlvvl* | – | – | + | A*0101 |
| LMP-2:203–206 vllicsscsscp | 2, 3, 8 | A2, B57, C6 | – | – |
| *LMP-2:208 sscsscpls* | – | – | + | A11 |
| *LMP-2:213 cplskilla* | 2, 8, 27 | B 57/58, C 6/7 | - | |
| *LMP-2:224 flyalalll* | - § | A*0201 | + | A0201 |
| Matrix C | ||||
| *LMP-2:287 tygpvfmcl* | 5, 23, 9 | A24 | + | A24 |
| *LMP-2:294–297 clgglltmvaga* | 2, 5, 6 c, 10, 13, 16(?) | A*0201/03/07 | + | A0201/06/07 |
| LMP-2:304–307 gavwltvmtntl | 1, 2, 4, 5 d, 6, 13 | A*0201/03/05/07 | – | – |
| LMP-2:319 wiltagfli | 8, 10, 24, 26 | A*0201 | – | – |
| *LMP-2:321 ltagflifl* | –§ | A2 | + | A2 |
Underlined epitopes that have been confirmed by intra-cellular cytokine staining (ICS)—when overlapping epitopes were identified only the amino acid sequence that yielded the highest frequency of epitope-specific cells by ICS is underlined
The donors in which reactivity was identified are shown and specific reactivity was based on a cut-off (n)log2 IFNγ/β-actin = 1.04 for Caucasian donors and 1.028 for Chinese donors as described in the Materials and methods
*Epitopes already known
§Epitopes identified by ICF only
(17) This donor CD4+ T cells responded to epitope stimulation as detected by ICS
aSee Fig. 3 (pept 104)
bSee Fig. 2 (pept 294)
Validation of previously described and of novel LMP-2 epitopes
To validate the epitopes identified by qRT screening, we re-tested positive PBMC samples by ICS both in ex vivo conditions or after 7 days of IVS by exposing them to the relevant pools. After confirming the best match between spaced and overlapping pools, a candidate epitope could be deduced that was subsequently tested as a single 9-mer peptide by ICS ex vivo and after IVS. In several cases, more than one adjacent spaced pool elicited reactivity in the same PBMC sample. This is not surprising since adjacent spaced pools contain overlapping peptide sequences spanning the same region of LMP-2. In this case, multiple corresponding candidate peptides were tested and the one eliciting the strongest response (higher proportion of CD8+ T cells expressing IFN-γ by ICS over total number of CD8+ T cells) was ranked as the most likely natural epitope (underlined in Table 4). Figure 2 shows an example of an ex vivo analysis of PBMC directly stimulated and analyzed after thawing. Subsequent expansion with IVS confirmed the specificity of the identification. In all cases the epitopes were identified by the two-dimensional matrix. Interestingly, a Chinese donor (#17) displayed strong epitope-specific CD4+ T cell responses to several 9-mer epitopes including LMP:4 through LMP:6 (LAAIAASCFTA) and LMP:84 through LMP:85 (AGGLGGIYVL). Several independent experiments demonstrated and confirmed that the CD4-dependent recognition of these 9-mer peptides was HLA-DR*0701 restricted. The explanation for the unusual identification of CD4 responses elicited by ninemer peptides is presently under investigation. Of the 18 novel LMP-2 epitope were identified by qRT screening, 11 could be confirmed by ICS. The epitope recognition by individual PBMC that tested positive by qRT was strong, clear and easily reproducible by ICS. However, contrary to the commonly studied Flu and CMV responses, these reactivities were restricted to few individuals and not widely detectable in most donors sharing the same HLA haplotypes. In addition, several epitopes identified by this study were different from those identified by stimulation with autologous LCL lines of PBMC from NPC patients as described by Straathof et al. [13] which were concomitantly tested by ICS when their HLA association was relevant to the donors studied here (Table 4). In particular, several epitopes described by the other study were not identified in our study either by qRT or ICS and only in two cases, (LMP:224, FLYALALL and LMP:321, LTAGFLIFL) ICS performed after IVS could identify previously described epitopes that were missed by qRT ex vivo.
Fig. 2.
Example of ex vivo testing of reactivity by ICS. PBMC from donor #6 were stimulated ex vivo and analyzed after 6 h with pool C4 (a), pool 30 (b) or peptide 294 (c). d Negative control (stimulation with gp100:209–217). In the upper right corner is shown the percent of CD3+CD8+ and IFN-γ+ cells
Assignment of HLA/epitope associations
Peptide/HLA binding affinity prediction models were used to deduce the most likely epitope/HLA association. Both Parker et al. [60] (http://www.bimas.dcrt.nih.gov/molbio/hla_bind/) and Rammensee et al. [61] (http://www.syfpeithi.de/) were evaluated as described by Straathof et al. [13] entering 9-mer amino acid sequences in the search engine. This analysis identified predominantly, HLA-A2-associated epitopes. This could be due to the larger proportion of donors expressing HLA-A2. In fact, 39 of the 64 CD8+ T cell responses identified by this study were in PBMC from HLA-A*0201 expressing donors. Importantly, epitopes were identified that elicited strong immune responses in individuals that did not share the expression of any HLA molecule suggesting cross presentation of identical epitopes by different HLA alleles, a phenomenon that we have already observed studying CMV reactivity [42] and has been broadly discussed by others in association with HLA-A locus super families [54, 62–64]. Further characterization of these responses was entertained by presenting candidate epitopes exogenously-pulsed on partially matched LCL to T cell cultures. T cells were expanded with one round of IVS with the relevant epitope and tested against HLA allele matched and not matched LCL. In this fashion most epitope/HLA associations could be defined and verified as exemplified by Fig. 4 and summarized as bolded alleles in Table 4.
Discussion
This investigation was aimed at comprehensively mapping LMP-2 epitopes in normal subjects and testing whether Chinese and Caucasian people carrying a latent EBV infection display different immune reactivity toward the LMP-2 protein. This could in turn provide additional insights about the higher prevalence of NPC among Chinese together with previously described genetic, epidemiologic, socio-economical and environmental explanations [9, 10]. The question was approached by delivering a complete series of minimal epitopic determinants covering the full sequence of LMP-2. The limited number of donors tested in this explorative study (ten in each ethnic group) does not allow a definitive conclusion. However, in our opinion, the lack of distinctive patterns between the two populations (besides those related to the ethnically-related variance of HLA haplotypes) does not justify further expansion of this investigation to larger cohorts. In particular, the sporadic nature of the observed responses (observed in a minor portion of donors sharing the same HLA alleles) and the lack of identification of one or few immune dominant epitope/HLA combinations would require the study of an unjustifiable number of subjects to achieve a sufficient statistical power. On the other hand, more confined investigations focused on particular model systems such as the HLA-A2 or other serologically defined HLA superfamilies [54, 62, 64, 65] might provide useful insights on genetically-determined immune responses to LMP-2.
The results of this study suggest that some level of reactivity toward LMP-2 is identifiable naturally in almost all individuals (18 of 20) independent of ethnic background. This reactivity, however, follows an idiosyncratic pattern characterized by epitope recognition patterns restricted to a limited number of individuals among those sharing a common HLA class I antigen. This confirms others’ finding that LMP reactivity is remarkably different from other viral model systems such as Flu or CMV where recognition of commonly shared immune dominant T cell epitopes can be observed with high prevalence in populations carrying the relevant HLA class I allele [19, 51, 66–69]. Indeed, detailed analysis of HLA-A locus alleles yielded expected results for Flu M1:58–66 and for CMV pp65:495–503 which have been previously extensively characterized in the context of HLA-A2 [19, 51, 66–69]. In both cases, stimulation of HLA-A2-bearing PBMC with the associated epitope induced detectable transcription of IFN-γ in all (Flu) or most (CMV) donors but rare and borderline responses in non-HLA-A2 bearing donors (Table 3). No such consistency could be observed for LMP-2 in the context of any HLA class I allele prevalent in either of the two ethnic groups such as HLA-A1, -A2, -A3, -A11, A24 -B15, -B40 or -B46 [35, 70]. The sporadic nature of the immune responses identified in various donors is intriguing and not easily explainable. It is unlikely that different strains of EBV could have been at the basis of this heterogeneity. Although the genotype of EBV was not tested in this study, it is likely that the B95.8 strain was predominant in the Caucasian population [45] and robustly represented in the Chinese [71]. Recently, Zeng M-S et al. [72] completed the whole genome sequence of an EBV strain derived from the nasopharynx of a NPC patient from Guangdong, China. The EBV strain, named GD1, was characterized by the presence of various deletion and insertions plus 318 missense point mutations scattered throughout the genome. This strain was found to be highly prevalent at least in the Southern Chinese population tested. Although several variants sequences were identified, the study confirmed that LMP-2 is among the most conserved among EBV-encoded proteins.
Interestingly, a good proportion of the immune reactivity was observed in association with HLA-A2 and, most specifically, HLA-A*0201. Of the 64 CD8+ T cell responses identified in this study, 39 occurred in PBMC from the HLA-A*0201 expressing donors. This could be partly explained by the relatively high prevalence of HLA-A*0201 expressing donors (7 of 20) in this study. However, it is intriguing and worth exploring in future studies the potential immune relevance of HLA-A*0201-associated immune responses to LMP-2. This may be of particular importance since the strongest association between NPC and HLA phenotype is with this HLA class I super family [30]. In particular, the relatively minor contribution of other HLA-A2 alleles to the immune responses identified in this study suggests that the Chinese population may lack the powerful immune response elicited by LMP-2 in the context of HLA-A*0201. Indeed, the immune responses identified in individuals bearing HLA-A2 alleles different from HLA-A*0201 where few: three in a Caucasian (donor #1) bearing the HLA-A*0205 allele (Table 4) and only six in typical Chinese alleles such as HLA-A*0203 (one response in donor #4) or 0207 (three responses in donor #5 and two in donor #13). A good example was LMP-2:319–327 (identified by the intersection of pool 32 with pool C9) in which all the reactive PBCM were from HLA-A*0201-bearing donors (donor #8, 10, 24 and 26) independently of their ethnic background. The stringency of association with LMP-2:319–327 and HLA-A*0201, however, was specific for this peptide sequence since cross presentation by several HLA-A2 alleles could be observed for peptide LMP-2:304–312 recognized by PBMC from donors 1, 2, 4, 5, 6 and 13 bearing respectively the HLA-A*0205, A*0201, A*0203, A*0207, A*0201, and A*0207 alleles. Thus, we believe that a comprehensive comparison of responses to LMP-2 in the context of the HLA-A2 super family is warranted and we are continuing recruitment of individuals of Chinese and Caucasian background expressing HLA-A2.
The predominant role that HLA-A*0201 may play in the response to LMP-2 (if confirmed) may partly explain in functional terms the epidemiologic observation that NPC is predominant in Chinese patients expressing HLA*A0207, while HLA-A*0201 does not represent a specific risk factor [26]. Possibly, the presence of this allele might be associated with a lower efficiency of presentation of LMP-2 epitopes. Structurally, there could be an explanation for the predominant role that HLA-A*0201 may play in presenting LMP-2 epitopes. LMP-2 is a prevalently transmembrane protein containing hydrophobic sequences. This matches the binding characteristics of HLA-A*0201 characterized by high affinity for hydrophobic peptides [73–77]. Obviously, this hypothesis needs extensive validation since it is against the currently preferred paradigm that HLA class I associations with NPC predisposition depend upon a locus in strong linkage disequilibrium with HLA genes and do not stem from functional characteristics associated with the binding properties of individual HLA molecules [26, 78]. In addition, the hydrophobic nature of LMP-2 versus Flu and CMV peptides might explain partly the preferential identification of epitopes based on their biochemical affinity to HLA alleles in the conditions tested rather their natural presence in vivo based on endogenous antigen processing and presentation. This may provide some bias in the results.
Another interesting finding of this study was the extensive pleiotropism of individual responses to LMP-2. PBMC from all but two donors demonstrated at least one example of strong immune reactivity toward LMP-2 that could be detected ex vivo. Straathof et al. [13] have recently shown that analyses of T cell responses to LMP-2 based on functional parameters may under estimate the frequency of epitope-recognizing T cells in a given specimen as analyzed by non-functional tools such as tetrameric HLA/epitope complex. Thus, it is likely that most individuals infected with EBV carry a significant number of LMP-2-reactive T cells during latent infection. The relevance that these naturally occurring immune responses detectable ex vivo bear on the immune surveillance against the development of EBV-related tumors warrants further investigation.
LMP 1 and, in particular, LMP 2 are consistently expressed by NPC [45, 79, 80] and, therefore, appear to be suitable candidates for immunogenic protocols to treat this cancer. In addition, the protein sequence of LMP-2 appears to be relatively conserved (compared with EBNA and other EBV proteins) across viral strains in various geographical locations [47, 57, 72, 81]. Recently, Straathof et al. [13] have convincingly shown that T cell responses toward conserved sequences of LMP-2 are predominant in T cell lines expanded from NPC patients by exposure to autologous LCL. Such T cell responses are therapeutically relevant as the adoptive transfer of several T cell lines has been associated with regression of cancer [12]. This suggests that the identified epitopes are naturally processed and presented by NPC cells in vivo. Interestingly, only a proportion of the LMP-2 epitopes identified by this ex vivo survey in non-tumor bearing individuals matched the broad repertoire described by Straathof’s study in NPC patients (Table 5). Obviously, this difference could be due to several factors unrelated to the immune biology underlining the transition from a normal non-tumor bearing status to NPC. For instance, the HLA phenotype of the two populations studied could have been significantly different. In addition, the in vitro expansion of T cell lines through LCL stimulation might induce a different pattern of recognition than the one identified by a direct ex vivo analysis of naturally occurring responses and perhaps more representative of the antigen presenting characteristics of NPC cells. Lautscham et al. [15] observed that the immunogenicity of LMP-2 epitopes may be immune proteasome dependent. Thus, LCL cell lines which have an active immune proteasome may display a different library of epitopes compared to other EBV-infected cells occurring in latent conditions in vivo. Finally, the tumor bearing status may alter the type of immune responses in patients through in vivo priming of epitopes more relevant to the antigen processing and presenting machine of tumors cells and/or normal bystander antigen presenting cells. We and others have previously observed a similar phenomenon comparing the recognition of tumor associated antigen in patients with metastatic cutaneous melanoma and HLA matched normal donors [69, 82]. The latter hypothesis is intriguing because it may suggest that the immune response to LMP-2 observable in non tumor-bearing individuals may be partly irrelevant in the context of NPC and, as a consequence, NPC may arise by escaping such immune response to this EBV protein. Obviously, only a direct comparison of the two populations (normal donors and NPC patients) using identical methods will conclusively address this question.
Table 5.
Amino acid sequence of LMP-2 based on the prototype B95.8 strain of EBV and sequences associated with LMP-2 epitopes in NPC patients [13] or in normal EBV-exposed donors (present study)
Underlined sequences correspond to domains of LMP-2 containing epitopes described by Straathof et al. [13]; double underlined sequences correspond to overlapping epitopes; bold sequences correspond to domains of LMP-2 containing epitopes identified by this study
This study was not meant to represent an exhaustive and conclusive analysis of the immune biology of T cell-mediated EBV responses since it is focused only on one, though relevant, EBV product. Because of the usefulness of identifying new LMP-2 associated epitopes in the context of NPC treatment, this study also intended to survey candidate epitopes associated with HLA class I alleles common to different races. The identification of such epitopes could serve two purposes. First, these epitopes could be used to broaden the repertoire of immunogens used to immunize patients with advanced NPC. Second, it would provide reagents to evaluate the prevalence of immune responses in diverse ethnic groups on a larger study comparing the prevalence of the response to the identified epitopes in an adequate NPC patient population. The relatively simplicity in which high throughput testing of peptide libraries could be performed with an automated approach suggests that in the future large cohorts of individuals bearing informative HLA haplotypes could be tested to further our understating of the immunological changes occurring in the transition from a non tumor-bearing status to the development of clinically detectable NPC.
Acknowledgment
This work was partially supported by FIRC (Fondazione Italiana per la Ricerca sul Cancro).
Abbreviations
- CI
Confidence interval
- CMV
Cytomegalovirus
- CTL
Cytotoxic T cells
- EBV
Epstein–Barr virus
- ICS
Intracellular cytokine staining
- HLA
Human leukocyte antigen
- VS
In vitro sensitization
- LMP
Latent membrane protein
- NPC
Nasopharyngeal carcinoma
- PBMC
Peripheral blood mononuclear cells
- PWM
Pokeweed mytogen
- qRT
Quantitative real-time polymerase chain reaction
- SEB
Staphylococcal exotoxin B
Footnotes
Maurizio Provenzano and Silvia Selleri equally contributed to this work.
References
- 1.Green M. Management of Epstein–Barr virus-induced post-tranplant lymphoproliferative disease in recipients of solid organ transplantation. Am J Transplant. 2001;1:103–108. [PubMed] [Google Scholar]
- 2.Heslop HE, Rooney CM. Adoptive cellular immunotherapy for EBV lymphoproliferative disease. Immunol Rev. 1997;157:217–222. doi: 10.1111/j.1600-065X.1997.tb00984.x. [DOI] [PubMed] [Google Scholar]
- 3.Khanna R, Bell S, Sherritt M, Galbraith A, Burrows SR, Rafter L, Clarke B, Slaughter R, Falk MC, Douglass J, Williams T, Elliott SL, Moss DJ. Activation and adoptive transfer of Epstein–Barr virus-specific cytotoxic T cells in solid organ transplant patients with posttransplant lymphoproliferative disease. Proc Natl Acad Sci USA. 1999;96:10391–10396. doi: 10.1073/pnas.96.18.10391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haque T, Taylor C, Wilkie GM, Murad P, Amlot PL, Beath S, McKiernan PJ, Crawford DH. Complete regression of posttransplant lymphoproliferative disease using partially HLA-matched Epstein–Barr virus-specific cytotoxic T cells. Transplantation. 2001;72:1399–1402. doi: 10.1097/00007890-200110270-00012. [DOI] [PubMed] [Google Scholar]
- 5.Rooney CM, Roskrow MA, Smith CA, Brenner MK, Heslop HE. Immunotherapy of Epstein–Barr virus-associated cancer. J Natl Cancer Inst Monogr. 1998;23:89–93. doi: 10.1093/oxfordjournals.jncimonographs.a024180. [DOI] [PubMed] [Google Scholar]
- 6.Gottschalk S, Heslop HE, Rooney CM. Treatment of Epstein–Barr virus-associated malignancies with specific T cells. Adv Cancer Res. 2002;84:175–201. doi: 10.1016/S0065-230X(02)84006-4. [DOI] [PubMed] [Google Scholar]
- 7.Chua D, Huang J, Zheng B, Lau SY, Luk W, Kwong DL, Sham JS, Moss D, Yuen KY, Im SW, Ng MH. Adoptive transfer of autologous Epstein–Barr virus-specific cytotoxic T cells for nasopharyngeal carcinoma. Int J Cancer. 2001;94:73–80. doi: 10.1002/ijc.1430. [DOI] [PubMed] [Google Scholar]
- 8.Vokes EE, Liebowitz DN, Weichselbaum RR. Nasopharyngeal carcinoma. Lancet. 1997;350:1087–1091. doi: 10.1016/S0140-6736(97)07269-3. [DOI] [PubMed] [Google Scholar]
- 9.McDermott AL, Dutt SN, Watkinson JC. The aetiology of nasopharyngeal carcinoma. Clin Otolaryngol. 2001;26:82–92. doi: 10.1046/j.1365-2273.2001.00449.x. [DOI] [PubMed] [Google Scholar]
- 10.Chan ATC, Teo PML, Johnson PJ. Nasopharyngeal carcinoma. Ann Oncol. 2002;13:1007–1015. doi: 10.1093/annonc/mdf179. [DOI] [PubMed] [Google Scholar]
- 11.Mould RF, Tai THP. Nasopharyngeal carcinoma: treatments and outcomes in the 20th century. Br J Radiol. 2002;75:307–339. doi: 10.1259/bjr.75.892.750307. [DOI] [PubMed] [Google Scholar]
- 12.Straathof KC, Bollard CM, Popat U, Huls MH, Lopez T, Morriss MC, Gresik MV, Gee AP, Russell HV, Brenner MK, Rooney CM, Heslop HE. Treatment of nasopharyngeal carcinoma with Epstein–Barr virus-specific T lymphocytes. Blood. 2005a;105:1898–1904. doi: 10.1182/blood-2004-07-2975. [DOI] [PubMed] [Google Scholar]
- 13.Straathof KC, Leen AM, Buza EL, Taylor G, Huls MH, Heslop HE, Rooney CM, Bollard CM. Characterization of latent membrane protein 2 specificity in CTL lines from patients with EBV-positive nasopharyngeal carcinoma and lymphoma. J Immunol. 2005b;175:4137–4147. doi: 10.4049/jimmunol.175.6.4137. [DOI] [PubMed] [Google Scholar]
- 14.Lin C-L, Lo W-F, Lee T-H, Yi R, Hwang S-L, Cheng Y-F, Chen C-L, Chang Y-S, Lee SP, Rickinson AB, Tam PKH. Immunization with Epstein–Barr virus (EBV) peptide-pulsed dendritic cells induces functional CD8+ T-cell immunity and may lead to tumor regression in patients with EBV-positive nasopharyngeal carcinoma. Cancer Res. 2002;62:6952–6958. [PubMed] [Google Scholar]
- 15.Lautscham G, Haigh T, Mayrhofer S, Taylor G, Croom-Carter D, Leese A, Gadola S, Cerundolo V, Rickinson A, Blake N. Identification of a TAP-independent, immunoproteasome-dependent CD8+ T-cell epitope in Esptein–Barr virus latent membrane protein 2. J Virol. 2003;77:2757–2761. doi: 10.1128/JVI.77.4.2757-2761.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sung NS, Edwards RH, Seillier-Moiseiwitsch F, Perkins AG, Zeng Y, Raab-Traub N. Epstein–Barr virus strain variation in nasopharyngeal carcinoma from the endemic and non-endemic regions of China. Int J Cancer. 1998;76:207–215. doi: 10.1002/(SICI)1097-0215(19980413)76:2<207::AID-IJC7>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 17.Kammula US, Lee K-H, Riker A, Wang E, Ohnmacht GA, Rosenberg SA, Marincola FM. Functional analysis of antigen-specific T lymphocytes by serial measurement of gene expression in peripheral blood mononuclear cells and tumor specimens. J Immunol. 1999;163:6867–6879. [PubMed] [Google Scholar]
- 18.Kammula US, Marincola FM, Rosenberg SA. Real-time quantitative polymerase chain reaction assessment of immune reactivity in melanoma patients after tumor peptide vaccination. J Natl Cancer Inst. 2000;92:1336–1344. doi: 10.1093/jnci/92.16.1336. [DOI] [PubMed] [Google Scholar]
- 19.Provenzano M, Mocellin S, Bettinotti M, Preuss J, Monsurro’ V, Marincola FM, Stroncek D. Identification of immune dominant cytomegalovyrus epitopes using quantitative real-time PCR to measure interferon-γ production by peptide stimulated peripheral blood mononuclear cells. J Immunother. 2002;25:342–351. doi: 10.1097/00002371-200207000-00006. [DOI] [PubMed] [Google Scholar]
- 20.Simons MJ, Day NE, Wee GB, Shanmugaratnam K, Ho HC, Wong SH, Ti TK, Yong NK, Darmalingam S, De-The G. Nasopharyngeal carcinoma V: immunogenetic studies of Southeast Asian ethnic groups with high and low risk for the tumor. Cancer Res. 1974;34:1192–1195. [PubMed] [Google Scholar]
- 21.Jing J, Louie E, Henderson BE, Terasaki IP. Histocompatibility leukocyte antigen patterns in nasopharyngeal carcinoma cases from California. Natl.Cancer Inst.Monogr. 1977;47:156. [PubMed] [Google Scholar]
- 22.Cui SH, Lin Y. Apparent correlation between nasopharyngeal carcinoma and HLA phenotype. Zhonghua Zhong Liu Za Zhi. 1982;4:249–253. [PubMed] [Google Scholar]
- 23.Chan SH, Day NE, Kunaratnam N, Chia KB, Simons MJ. HLA and nasopharyngeal carcinoma in Chinese—a further study. Int J Cancer. 1983;32:171–176. doi: 10.1002/ijc.2910320206. [DOI] [PubMed] [Google Scholar]
- 24.Ou B-X, Ruan H-Y, Fan Y. Study on association between HLA-A, B, C DR, nasopharyngeal carcinoma in Guangzhou area. Ai Zheng. 1985;4:5–8. [Google Scholar]
- 25.Zhang JZ. Correlation between nasopharyngeal carcinoma (NPC) and HLA in Hunan Province. Zhonghua Zhong Liu Za Zhi. 1986;8:170–172. [PubMed] [Google Scholar]
- 26.Hildesheim A, Apple RJ, Chen C-J, Wang SS, Cheng Y-J, Klitz W, Mack SJ, Chen I-H, Hsu M-M, Yang C-S, Brinton LA, Levine PH, Erlich HA. Association of HLA class I and II alleles and extended haplotypes with nasopharyngeal carcinoma in Taiwan. J Natl Cancer Inst. 2002;94:1780–1789. doi: 10.1093/jnci/94.23.1780. [DOI] [PubMed] [Google Scholar]
- 27.Pimtanothai N, Charoenwongse P, Mutirangura A, Hurley CK. Distribution of HLA-B alleles in nasopharyngeal carcinoma patients and normal controls in Thailand. Tissue Antigens. 2002;59:223–225. doi: 10.1034/j.1399-0039.2002.590308.x. [DOI] [PubMed] [Google Scholar]
- 28.Lu C-C, Chen J-C, Jin Y-T, Yang H-B, Chan S-H, Tsai S-T. Genetic susceptibility to nasopharyngeal carcinoma within the HLA-A locus in Taiwanese. Int J Cancer. 2003;103:745–751. doi: 10.1002/ijc.10861. [DOI] [PubMed] [Google Scholar]
- 29.Hu SP, Day NE, Li DR, Luben RN, Cai KL, Ou-Yang T, Li B, Lu XZ, Ponder BA. Further evidence for an HLA-related recessive mutation in nasopharyngeal carcinoma among the Chinese. Br J Cancer. 2005;92:967–970. doi: 10.1038/sj.bjc.6602347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goldsmith DB, West TM, Morton R. HLA associations with nasopharyngeal carconoma in Southern Chinese: a meta-analysis. Clin Otolaryngol. 2002;27:61–67. doi: 10.1046/j.0307-7772.2001.00529.x. [DOI] [PubMed] [Google Scholar]
- 31.Krausa P, Browning MJ. A comprehensive PCR-SSP typing system for identification of HLA-A locus alleles. Tissue Antigens. 1996;47:237–244. doi: 10.1111/j.1399-0039.1996.tb02547.x. [DOI] [PubMed] [Google Scholar]
- 32.Browning M, Krausa P. Genetic diversity of HLA-A2: evolutionary and functional significance. Immunol.Today. 1996;17:165–170. doi: 10.1016/0167-5699(96)80614-1. [DOI] [PubMed] [Google Scholar]
- 33.Simonis TB, Barracchini KC, Hackett JA, Maurer D, Siauw P, Tonai R, Marincola FM. HLA-A*02 allele frequencies of different ethnic groups. Hum Immunol. 1997;55:99. [Google Scholar]
- 34.Krausa P, Brywka M, Savage D, Hui KM, Bunce M, Ngai JL, Teo DL, Ong YW, Barouch D, Allsop CE, et al. Genetic polymorphism within HLA-A*02: significant allelic variation revealed in different populations. Tissue Antigens. 1995;45:223–231. doi: 10.1111/j.1399-0039.1995.tb02444.x. [DOI] [PubMed] [Google Scholar]
- 35.Chang YW, Hawkins BR. HLA class I and class II frequencies of a Hong Kong Chinese population based on bone marrow donor registry data. Human Immunol. 1997;56:125–135. doi: 10.1016/S0198-8859(97)00108-0. [DOI] [PubMed] [Google Scholar]
- 36.Adams SD, Barracchini KC, Chen D, Robbins F, Wang L, Larsen P, Luhm R, Stroncek DF. Ambiguous allele combinations in HLA Class I and Class II sequence-based typing: when precise nucleotide sequencing leads to imprecise allele identification. J Transl Med. 2004;2:30. doi: 10.1186/1479-5876-2-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Adams SD, Barracchini KC, Simonis TB, Stroncek D, Marincola FM. High throughput HLA sequence-based typing utilizing the ABI prism 3700 analyzer. Tumori. 2001;87:s41–s44. [PubMed] [Google Scholar]
- 38.McGinnis M, Stein JB, Adams SD, Marincola FM, Krausa P. Implementing high throughput HLA SBT on a 96-capillary DNA sequencer. Hum Immunol. 2001;s146:74. [Google Scholar]
- 39.Keilholz U, Weber J, Finke J, Gabrilovich D, Kast WM, Disis N, Kirkwood J, Scheibenbogen C, Schlom J, Maino V, Lyerly K, Lee PJ, Storkus WJ, Marincola FM, Worobec A, Atkins M. Immunologic monitoring of cancer vaccine therapy: results of a workshop sponsored by the Society of Biological Therapy. J Immunother. 2002;25:97–138. doi: 10.1097/00002371-200203000-00001. [DOI] [PubMed] [Google Scholar]
- 40.Panelli MC, Wang E, Monsurro’ V, Marincola FM. The role of quantitative PCR for the immune monitoring of cancer patients. Exp Opin Biol Ther. 2002;2:557–564. doi: 10.1517/14712598.2.5.557. [DOI] [PubMed] [Google Scholar]
- 41.Mocellin S, Rossi CR, Pilati P, Nitti D, Marincola FM. Quantitative real-time PCR: a powerful ally in cancer research. Trends Mol Med. 2003;9:189–195. doi: 10.1016/S1471-4914(03)00047-9. [DOI] [PubMed] [Google Scholar]
- 42.Ghei M, Stroncek DF, Provenzano M. Analysis of memory T lymphocyte activity following stimulation with overlapping HLA-A*2402, A*0101 and Cw*0402 restricted CMV pp65 peptides. J Transl Med. 2005;3:23. doi: 10.1186/1479-5876-3-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Provenzano M, Mocellin S, Bonginelli P, Nagorsen D, Kwon SW, Stroncek D Ex vivo screening for immunodominant viral epitopes by quantitative real time polymerase chain reaction (qRT-PCR). J Transl Med 1:11, 3 A.D [DOI] [PMC free article] [PubMed]
- 44.Trojan A, Urosevic M, Hummerjohann J, Giger R, Schanz U, Stahel RA. Immune reactivity against a novel HLA-A3-restricted influenza virus peptide identified by predictive algorithms and interferon-gamma quantitative PCR. J Immunother. 2003;26:41–46. doi: 10.1097/00002371-200301000-00005. [DOI] [PubMed] [Google Scholar]
- 45.Busson P, McCoy R, Sadler R, Gilligan K, Tursz T, Raab-Traub N. Consistent transcription of the Epstein–Barr virus LMP2 gene in nasopharyngeal carcinoma. J Virol. 1992;66:3257–3262. doi: 10.1128/jvi.66.5.3257-3262.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bettinotti M, Kim CJ, Lee K-H, Roden M, Cormier JN, Panelli MC, Parker KC, Marincola FM. Stringent allele/epitope requirements for MART-1/Melan A immunodominance: implications for peptide-based immunotherapy. J Immunol. 1998;161:877–889. [PubMed] [Google Scholar]
- 47.Lee SP, Thomas WA, Murray RJ, Khanim F, Kaur S, Young LS, Rowe M, Kurilla M, Rickinson AB. HLA A2.1-restricted cytotoxic T cells recognizing a range of Epstein–Barr virus isolates through a defined epitope in latent membrane protein LMP 2. J Virol. 1993;67:7428–7435. doi: 10.1128/jvi.67.12.7428-7435.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Adams S, Robbins FM, Chen D, Wagage D, Holbeck SL, Morse HC, III, Stroncek D, Marincola FM. HLA class I and II genotype of the NCI-60 cell lines. J Transl Med. 2005;3:11. doi: 10.1186/1479-5876-3-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 50.Nielsen M-B, Monsurro’ V, Miguelse S, Wang E, Perez-Diez A, Lee K-H, Kammula Rosenberg US SA, Marincola FM. Status of activation of circulating vaccine-elicited CD8+ T cells. J Immunol. 2000;165:2287–2296. doi: 10.4049/jimmunol.165.4.2287. [DOI] [PubMed] [Google Scholar]
- 51.Rivoltini L, Loftus DJ, Barracchini K, Arienti F, Mazzocchi A, Biddison WE, Salgaller ML, Appella E, Parmiani G, Marincola FM. Binding and presentation of peptides derived from melanoma antigens MART-1 and gp100 by HLA-A2 subtypes: implications for peptide-based immunotherapy. J Immunol. 1996;156:3882–3891. [PubMed] [Google Scholar]
- 52.Provenzano M, Panelli MC, Mocellin S, Bracci L, Sais G, Stroncek DF, Spagnoli GC, Marincola FM (2006) MHC-peptide specificity and T-cell epitope mapping: where immunotherapy starts. Trends Mol Med [DOI] [PubMed]
- 53.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Threlkeld SC, Wentworth PA, Kalams SA, Wilkes BM, Ruhl DJ, Keogh E, Sidney J, Southwood S, Walker BD, Sette A. Degenerate and promiscuous recognition by CTL of peptides presented by the MHC class I A3-like superfamily: implications for vaccine development. J Immunol. 1997;159:1648–1657. [PubMed] [Google Scholar]
- 55.Gordon S. Alternative activation of macrophages. Nature Rev. 2003;3:23–35. doi: 10.1038/nri978. [DOI] [PubMed] [Google Scholar]
- 56.Khanna R, Burrows SR, Thomson SA, Moss DJ, Cresswell P, Poulsen LM, Cooper L. Class I processing-defective Burkitt’s lymphoma cells are recognized efficiently by CD4+ EBV-specific CTLs. J Immunol. 1997;158:3619–3625. [PubMed] [Google Scholar]
- 57.Lee SP, Tierney RJ, Thomas WA, Brooks JM, Rickinson AB. Conserved CTL epitopes within EBV latent membrane protein 2. A potential target for CTL-based tumor therapy. J Immunol. 1997;158:3325–3334. [PubMed] [Google Scholar]
- 58.Meij P, Leen A, Rickinson AB, Varkoeijen S, Vervoort MBHJ, Bloemena E, Middeldorp JM. Identification and prevalence of CD8+ T-cell responses directed against Epstein–Barr virus-encoded latent membrane protein 1 and latent membrane protein 2. Int J Cancer. 2002;99:93–99. doi: 10.1002/ijc.10309. [DOI] [PubMed] [Google Scholar]
- 59.Whitney BM, Chan A. T. C, Rickinson AB, Lee SP, Lin CK, Johnson PJ. Frequency of Epstein–Barr virus-specific cytotoxic T lymphocytes in the blood of Southern Chinese blood donors and nasopharyngeal carcinoma patients. J Med Virol. 2002;67:359–363. doi: 10.1002/jmv.10073. [DOI] [PubMed] [Google Scholar]
- 60.Parker KC, Bednarek MA, Coligan JE. Scheme for ranking potential HLA-A2 binding peptides based on independent binding of individual peptide side-chains. J Immunol. 1994;152:163–175. [PubMed] [Google Scholar]
- 61.Rammensee H, Bachmann J, Emmerich NP, Bachor OA, Stevanovic S. SYFPEITHI: database for MHC ligands and peptide motifs. Immunogenetics. 1999;50:213–219. doi: 10.1007/s002510050595. [DOI] [PubMed] [Google Scholar]
- 62.del Guercio MF, Sidney J, Hermanson G, Perez C, Grey HM, Kubo RT, Sette A. Binding of a peptide antigen to multiple HLA alleles allows definition of an A2-like supertype. J Immunol. 1995;154:685–693. [PubMed] [Google Scholar]
- 63.Kan-Mitchell J, Bajcz M, Schaubert KL, Price DA, Brenchley JM, Asher TE, Douek DC, Ng HL, Yang OO, Rinaldo CR, Jr, Benito JM, Bisikirska B, Hegde R, Marincola FM, Boggiano C, Wilson D, Abrams J, Blondelle SE, Wilson DB. Degeneracy and repertoire of the human HIV-1 Gag p1777–85 CTL response. J Immunol. 2006;176:6690–6701. doi: 10.4049/jimmunol.176.11.6690. [DOI] [PubMed] [Google Scholar]
- 64.Sidney J, Grey HM, Southwood S, Celis E, Wentworth PA, del Guercio MF, Kubo RT, Chesnut RW, Sette A. Definition of an HLA-A3-like supermotif demonstrates the overlapping peptide-binding repertoires of common HLA molecules. Hum.Immunol. 1996;45:79–93. doi: 10.1016/0198-8859(95)00173-5. [DOI] [PubMed] [Google Scholar]
- 65.DiBrino M, Parker KC, Shiloach J, Turner RV, Tsuchida T, Garfield M, Biddison WE, Coligan JE. Endogenous peptides with distinct amino acid anchor residue motifs bind to HLA-A1 and HLA-B8. J.Immunol. 1994;152:620–631. [PubMed] [Google Scholar]
- 66.Parker KC, Bednarek MA, Hull LK, Utz U, Cunningham B, Zweerink HJ, Biddison WE, Coligan JE. Sequence motifs important for peptide binding to the human MHC class I molecule, HLA-A2. J Immunol. 1992;149:3580–3587. [PubMed] [Google Scholar]
- 67.Sauma SY, Gammon MC, Bednarek MA, Cunningham B, Biddison WE, Hermes JD, Porter G, Tamhankar S, Hawkins JC, Bush BL, et al. Recognition by HLA-A2-restricted cytotoxic T lymphocytes of endogenously generated and exogenously provided synthetic peptide analogues of the influenza A virus matrix protein. Hum Immunol. 1993;37:252–258. doi: 10.1016/0198-8859(93)90508-X. [DOI] [PubMed] [Google Scholar]
- 68.Utz U, Koenig S, Coligan JE, Biddison WE. Presentation of three different viral peptides, HTLV-1 Tax, HCMV gB, and influenza virus M1, is determined by common structural features of the HLA-A2.1 molecule. J Immunol. 1992;149:214–221. [PubMed] [Google Scholar]
- 69.Marincola FM, Rivoltini L, Salgaller ML, Player M, Rosenberg SA. Differential anti-MART-1/MelanA CTL activity in peripheral blood of HLA-A2 melanoma patients in comparison to healthy donors: evidence for in vivo priming by tumor cells. J Immunother. 1996;19:266–277. doi: 10.1097/00002371-199607000-00003. [DOI] [PubMed] [Google Scholar]
- 70.Marincola FM, Stroncek D, Simonis T. Population studies. Caucasian US melanoma cancer. In: Terasaki IP, Gjertson DW, editors. HLA 1998. 2. Kansas: Lanexa; 1998. pp. 276–277. [Google Scholar]
- 71.Zhang XS, Wang HH, Hu LF, Li A, Zhang RH, Mai HQ, Xia JC, Chen LZ, Zeng YX. V-val subtype of Epstein–Barr virus nuclear antigen 1 preferentially exists in biopsies of nasopharyngeal carcinoma. Cancer Lett. 2004;211:11–18. doi: 10.1016/j.canlet.2004.01.035. [DOI] [PubMed] [Google Scholar]
- 72.Zeng M-S, Li D-J, Liu Q-L, Song L-B, Li M-Z, Zhang R-H, Yu X-J, Wang H-M, Ernberg I, Zeng YX (2005) Genomic sequence analysis of an Epstein–Barr virus strain DG1 from a nasopharyngeal carcinoma patient. J Virol (in press) [DOI] [PMC free article] [PubMed]
- 73.Bjorkman PJ, Saper MA, Samraoui B, Bennett WS, Strominger JL, Wiley DC. Structure of the human class I histocompatibility antigen, HLA-A2. Nature. 1987;329:506–512. doi: 10.1038/329506a0. [DOI] [PubMed] [Google Scholar]
- 74.Bjorkman PJ, Parham P. Structure, function, and diversity of class I major histocompatibility complex molecules. Annu Rev Biochem. 1990;59:253–288. doi: 10.1146/annurev.bi.59.070190.001345. [DOI] [PubMed] [Google Scholar]
- 75.Altuvia Y, Sette A, Sidney J, Southwood S, Margalit HA. Structure-based algorithm to predict potential binding peptides to MHC molecules with hydrophobic binding pockets. Hum Immunol. 1997;58:1–11. doi: 10.1016/S0198-8859(97)00210-3. [DOI] [PubMed] [Google Scholar]
- 76.Meng WS, von Grafenstein H, Haworth IS. Water dynamics at the binding interface of four different HLA-A2-peptide complexes. Int Immunol. 2000;12:949–957. doi: 10.1093/intimm/12.7.949. [DOI] [PubMed] [Google Scholar]
- 77.Desmet J, Meersseman G, Boutonnet N, Pletinckx J, De Clercq K, Debulpaep M, Braeckman T, Lasters I. Anchor profiles of HLA-specific peptides: analysis by a novel affinity scoring method and experimental validation. Proteins. 2005;58:53–69. doi: 10.1002/prot.20302. [DOI] [PubMed] [Google Scholar]
- 78.Simons MJ. HLA and nasopharyngeal carcinoma:30 years on. ASHI Q. 2003;27:52–55. [Google Scholar]
- 79.Pallesen G, Hamilton-Dutoit SJ, Rowe M, Young SL. Expression of Epstein–Barr virus latent gene products in tumour cells of Hodgkin’s disease. Lancet. 1991;337:320–322. doi: 10.1016/0140-6736(91)90943-J. [DOI] [PubMed] [Google Scholar]
- 80.Niedobitek G, Kremmer E, Herbst H, Whitehead L, Dawson CW, Niedobitek E, von Ostau C, Rooney N, Grasser FA, Young LS. Immunohistochemical detection of the Epstein–Barr virus-encoded latent membrane protein 2A in Hodgkin’s disease and infectious mononuclosis. Blood. 1997;90:1664–1672. [PubMed] [Google Scholar]
- 81.Berger C, Rothenberger S, Bachmann E, McQuain C, Nadal D, Kneth H. Sequence polymorphisms between latent membrane proteins LMP1 and LMP2A do not correlate in EBV-associated reactive and malignant lympho-proliferations. Int J Cancer. 1999;81:371–375. doi: 10.1002/(SICI)1097-0215(19990505)81:3<371::AID-IJC10>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 82.D’Souza S, Rimoldi D, Lienard D, Lejeune F, Cerottini JC, Romero P. Circulating Melan-A/Mart-1 specific cytolytic T lymphocyte precursors in HLA-A2+ melanoma patients have a memory phenotype. Int J Cancer. 1998;78:699–706. doi: 10.1002/(SICI)1097-0215(19981209)78:6<699::AID-IJC6>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 83.Ross DT, Scherf U, Eisen MB, Perou CM, Rees CA, Spellman PT, Iyer V, Jeffrey SS, van de Rijn M, Waltham M, Pergamenschikov A, Lee JC, Lashkari D, Shalon D, Myers TG, Weinstein JN, Botstein D, Brown PO. Systematic variation in gene expression patterns in human cancer cell lines. Nat Genet. 2000;24:227–235. doi: 10.1038/73432. [DOI] [PubMed] [Google Scholar]