Abstract
Animal models are widely used to study the biological behavior of human tumors in vivo. Murine immunodeficient models are used to test novel human anti-tumor therapies, and humanized mice are employed to study immunotherapeutic protocols. We find that human melanoma cell lines lose HLA class I surface expression after growth in immunodeficient mice and that this phenomenon occurs frequently and is reproducible. This HLA loss is due to a coordinated down-regulation of APM and HLA heavy chain expression at the transcriptional level. It is produced by epigenetic modifications and can be reversed by treatment with histone deacetylase inhibitors or IFN-gamma. These HLA alterations only appear during in vivo growth and not during successive in vitro passages. Interestingly, these new tumor variants with HLA class I loss show higher tumorigenicity per se and may represent a more advanced state of the original tumor. Lack of MHC class I expression on tumor cells represents a frequent escape mechanism from the immune response. Our results indicate that tumor variants with alterations in MHC can also appear in vivo after the immunoescape phase in the absence of anti-tumor immune response. Our findings suggest that any studied parameter, i.e., HLA expression, of malignant cells in xenograft models, has to be evaluated before and after growth in immunodeficient mice, in order to design more appropriate immunotherapy and chemotherapy treatments against tumor cells growing in vivo.
Keywords: HLA alterations, Human tumor, Immunodeficient mice, Oncogenicity
Introduction
One current view of the clonal expansion of tumor cells takes into consideration intrinsic characteristics of these cells (growth signals, ignoring of growth inhibitory signals, avoidance of cell death, and unlimited replication producing angiogenesis and invasion of tissue) and immune selection of tumor variants [4, 5, 12]. Cancer progression occurs despite an active and normal immune response, with the growth of tumor cells that invade and metastasize in a healthy host [17]. The acquisition by cancer cells of genetic and phenotypic alterations allows tumors to escape the anti-tumor immune response [30, 34]. This approach may be useful to explain why acquisition of an immune-escape tumor phenotype is a critical step in the natural progression of human and experimental cancer.
During cancer initiation, tumor cells express tumor-associated antigens that are recognized by the adaptive immune system [22]. Tumor cells have been observed to use a variety of mechanisms to evade a specific T cell immune response [7, 19]. Alterations in MHC expression represent an important escape mechanism that is frequently observed during cancer progression [2, 10, 11]. MHC-altered tumor variants emerge after an immunoselection process in which tumor cells with standard MHC-expression levels are eliminated by the immune system. Several altered HLA class I phenotypes have been detected in human tumors, resulting from the active immune surveillance system in these patients [8]. These findings strongly support the hypothesis that the selection of tumor variants with different MHC class I patterns is dependent on the type and intensity of the immune response of the tumor-bearing host [1]. The immune selection process is followed by an immune-escape step where tumor cells are not recognized by immune system [4, 5].
However, alterations in HLA expression may also occur in absence of autologous immune response. Our earlier studies using Ando-2 human melanoma cells showed that these cells develop a total loss of HLA class I cell surface expression after growth in immunodeficient nude or SCID-Beige mice [23]. In order to learn whether this is just an isolated observation or represents a more general phenomenon and may occur in other cancer cell lines, we compared the HLA phenotype in three different melanoma cell lines (E-033, E-179 and E-195) before and after in vivo growth in immunodeficient mice. Our results indicate that alterations in HLA expression occur frequently in human melanoma cells after growth in nude mice and that these melanoma cells are more tumorigenic in vivo.
Materials and methods
Mice
Athymic 6- to 8-week-old Balb/c nu/nu mice (average weight 20 g) were purchased from Charles River (CRIVER, Barcelona, Spain). Mice were housed under specific pathogen-free conditions, and all work with the animals conformed to guidelines approved by our institution.
Cell lines and reagents
Melanoma cell lines Ando-2, E-033 (FM-93/2), E-179 (URKV-Mel-13) and E-195 (Ma-Mel-48a) were obtained from patients with malignant melanoma. Ando-2 melanoma cell line was kindly provided by Dr. P. Coulie (Universite Louvain (UCL), Brussels, Belgium). The other three melanoma cell lines are publically available at The European Searchable Tumor cell Line Database (ESTDAB) and Cell Bank (http://www.ebi.ac.uk/ipd/estdab) [24]. Cell lines were maintained in ISCOVE tissue culture (Gibco, Paisley, UK) supplemented with 10% fetal bovine serum (Life Technologies, Milan, Italy) and antibiotics. In some experiments, cell lines were treated for 48 h with 800 U/ml IFN-γ (Roche Applied Science, Mannheim, Germany) or Trichostatin A (TSA) (Sigma, St Louis, MO) at concentrations of 50, 250 and 500 nM for 48 h.
HLA typing and microsatellite analysis
Conventional complement-mediated microtoxicity assay was used for serological HLA typing. Sequence specific oligonucleotide analysis was performed using DYNAL RELI® HLA-A, B, C, DR and DQ with DNA obtained from Ando-2 and autologous EBV cell lines. Genomic typing was done using the PMP 5.1 program.
DNA from Ando-2 and autologous EBV cell lines was diluted to 0.50 μg/μl and studied with eight STR markers mapping chromosome 6 (D6s311 located at 6q24; D6s291 at 6p21.2; D6s273, C.1.2.c, C.1.2.5, D6s265, D6s105 at 6p21.3 and D6s276 at 6p22); and two markers located on chromosome 15 (D15S209, D15S126). PCR data were analyzed on an ABI PRISM 377 using ABI PRISM 377 GENESCAN and GENOTYPER programs. LOH was assigned when there was a >25% signal reduction of one allele in tumor versus control sample. Allelic reduction in three or more STR markers in chromosome 6 was defined as haplotype loss.
In vivo tumor growth
5 × 106 cells of each tumor cell line (Ando-2, E-033, E-179 and E-195) were subcutaneously (s.c.) injected into the footpad in groups of five nude mice. When local tumors reached 10 mm in large diameter, they were extirpated and disaggregated and the cells were cultured. These tumor cell lines were designated by adding N to the name of original melanoma cell line, e.g., E-179-N1. All tumor cells were maintained in ISCOVE tissue culture supplemented with 10% fetal bovine serum and antibiotics.
In in vivo oncogenicity assays, different doses of cells, 1- 2.5- and 5 × 106 were s.c. injected into footpad in groups of five nude mice. The large tumor diameter was measured every 2 days.
Surface HLA expression
Surface HLA class I and II expression on cultured cells was determined by indirect immunofluorescence using the appropriate anti-class-I monoclonal antibody (mAb) and fluorescein isothiocyanate-labeled rabbit antimouse Ig (Fab2) fragments (Sigma). Fluorescence was analyzed with a FACScan flow cytometer (Becton-Dickinson, Mountain View, CA, USA) using standard methods. Thus, 5 × 105 cells were washed twice in PBS supplemented with 2% fetal calf serum (FCS) and 0.1% sodium azide. Cells were incubated with the primary mAb at saturating concentration for 30 min at 4°C, using mAbs against: HLA class I (W6/32), HLA-A (Tu-155), HLA-B (42-IB5), HLA-A2 (30.13.38 [Kre-501] and CR11-357), HLA-B40 (HB 115) and HLA-B8 (MRE4). The secondary antibody was used at a 1:80 dilution and incubated with cells for 30 min at 4°C in the dark. In control experiments, a primary antibody was replaced by the isotype-matched non-immune mouse IgG. In addition, cells labeled with only the fluorescein-conjugated antibody were always used as a control. Instrument alignments were checked with caliBrite beads, and the calibration was set by FACSComp software. CellQuest software was used to generate plots. Flow cytometry histograms were generated with the logarithmic amplification of fluorescence emitted by single viable cells. Each sample analyzed consisted of a minimum of 104 cells.
NK cytotoxicity assay
Cytotoxicity assays were performed according to a standard 51Cr-release method as mentioned above. Ando-2 and Ando-nude cell lines were used as target cells. Splenocytes (effector cells) were isolated from all mice by mechanical dissociation and lysing of erythrocytes. Splenocytes were fractionated by density centrifugation at 500×g for 20 min with Ficoll Hypaque and interface lymphocytes were obtained. YAC-1 cells (ATCC), a mouse lymphoma line sensitive to the cytotoxic activity of NK cells, were used as positive control. YAC-1 cells were maintained in RPMI 1640 supplemented with 10% FCS. After 4 h of culture, the supernatant was removed from each well and counted in a gamma counter for the determination of 51Cr release. Lysis was considered significant when >10%.
CTL cytotoxicity assay
Cytotoxicity of CTL was assessed using the standard 51Chromium-release assay. Briefly, one million target cells were labeled at 37°C for 1 h with 100 μCi Na2 51CrO4 (New England Nuclear, Boston, MA, USA). Target cells were washed and resuspended in CM at 5 × 104 cells/ml. Five thousand target cells per well (100 μl) were added to a 96 well plate (Costar, Cambridge, MA, USA) following the appropriate number of effector cells (100 μl/well). Cells at the defined effector:target (E:T) ratios were plated in triplicate. Cytotoxicity assays were performed at 37°C for 4 or 12 h. After incubation, cell-free supernatants were collected using a Skatron harvester and analyzed in a gamma counter (LKB Wallac CliniGamma 1272, Wallac, Finland). The percentage of the specific lysis was calculated using the following equation: (ER−SR)/(MR−SR) × 100, where ER = experimental release, SR = spontaneous release and MR = maximum release. For maximal release, target cells were treated with 0.3% Triton X-100 (Sigma). Spontaneous release of radioactivity by target cells was determined in the absence of effector cells. Results are shown as an average percentage of the specific lysis ±SE of triplicate determinations.
For the first screening assay, the labeled-target cells were mixed with cold (unlabeled) K562 cells at a 1:20 labeled-target:cold-target ratio to decrease the impact of the nonspecific killing by the natural killer cells.
Ando-2 and Ando-nude cell lines were used as target cells. CTL clones used as effector cells were obtained from co-cultures of CD8+ lymphocytes (purified by negative selection with CD8+ T Cell Isolation Kit, human, Myltenyi-Biotech, Germany) of melanoma patient Ando-2 with irradiated (10,000 rads) autologous melanoma cells Ando-2. Cultures were weekly stimulated with irradiated autologous tumor cells in RPMI 1640 medium-containing IL-2 (50 U/ml) and IL-7 (10 ng/ml). After 4 weeks of stimulation, 4 h cytotoxic assays were performed against Ando-2 and Ando-2 EBV cells and those cultures that exhibited specific cytotoxic activity were selected and expanded in cell culture. The CTL clones were restimulated weekly by addition of feeder cells and irradiated autologous melanoma cells Ando-2.
RT and quantitative real-time PCR
The mRNA isolation kit (Myltenyi-Biotech) was used to extract mRNA from tumor cell lines under basal conditions and after 48-h IFN-γ treatment. First-strand cDNA was synthesized with 100 ng of mRNA using a High Capacity Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA) in a total volume of 20 μl. These cDNAs were diluted to a final volume of 100 μl. Real-time quantitative PCR analyses for β2-microglobulin, HLA-A, HLA-B, HLA-C, TAP1, TAP2, LMP2, LMP7 and Tapasin genes were performed by means of the 7500 Fast System (Applied Biosystems), using GAPDH and TBP genes (Kit from Applied Biosystems) as housekeeping genes. PCR reactions were performed in quadruplicate, and values obtained were expressed as mean ± SD (standard deviation). Quantitative PCR was performed with the Power SYBR Green Master mix (Applied Biosystems). Primers, amplicon size and annealing temperature for each gene are shown in Table 1. PCR conditions were 40 cycles of 15 s of denaturation at 95°C and 60 s at 60°C.
Table 1.
Oligonucleotides used in Q-PCR
| Gene | Oligo | Sequence | Position | Fragment size |
|---|---|---|---|---|
| Beta-2m | Forward | TGACTTTGTCACAGCCCAAGAT | 374–459 | 86 |
| Reverse | CAATCCAAATGCGGCATCTTCA | |||
| TAP1 | Forward | GCCTCACTGACTGGATTCTAC | 964–1123 | 160 |
| Reverse | TCTCCCTGCAAGTGGCTGTG | |||
| TAP2 | Forward | GGTCGTGTGATTGACATCCTG | 642–870 | 229 |
| Reverse | TCAGCTCCCCTGTCTTAGTCT | |||
| Tapasin | Forward | TCCAGCCTCTTGCGACCACA | 577–713 | 137 |
| Reverse | CTCAAGTCCAGCAGAGCATCT | |||
| LMP2 | Forward | ATGGGTTCTGATTCCCGAGTG | 139–260 | 122 |
| Reverse | GCTTGGGCATCAGCAGCTGA | |||
| LMP7 | Forward | CCTTCAAGTTCCAGCATGGAG | 740–875 | 136 |
| Reverse | GCTGCACAGCCAGACATGGT | |||
| HLA-A | Forward A2 | CTCTTTGGAGCTGTGATCACT | 949–1147 | 198 |
| Forward A32 | TCTCTTTGGAGCTATGTTCGCT | 948–1147 | 199 | |
| Reverse | GAAGGGCAGGAACAAMTCTTG | |||
| HLA-B | Forward | GTCCTAGCAGTTGTGGTCATC | 949–1089 | 140 |
| Reverse | TCAAGCTGTGAGAGACACATCA | |||
| HLA-C | Forward | TCCTGGCTGTCCTAGCTGTC | 950–1100 | 150 |
| Reverse | CAGGCTTTACAAGTGATGAGAG |
Results
HLA genotype and phenotype of human melanoma cell lines
HLA genomic typing of human melanoma cell lines Ando-2, E-179, E-195 and E-033, are presented in Table 2. Analysis of microsatellites of chromosome 6 showed loss of one chromosome 6 in Ando-2, E-179 and E-195 cell lines (data not shown).These melanoma cell lines present the loss of one HLA haplotype. The E-033 melanoma cell line does not show losses and maintains the two HLA haplotypes.
Table 2.
HLA class I genomic typing of melanoma cell lines
| Melanoma cell line | HLA-A | HLA-B | HLA-C |
|---|---|---|---|
| E-033 | 0201, 2601 | 4001, 4402 | 0304, 0501 |
| Ando-2 | 3201 | 4001 | 0602 |
| E-179 | 0201 | 4001 | 0304 |
| E-195 | 0101 | 0801 | 0701 |
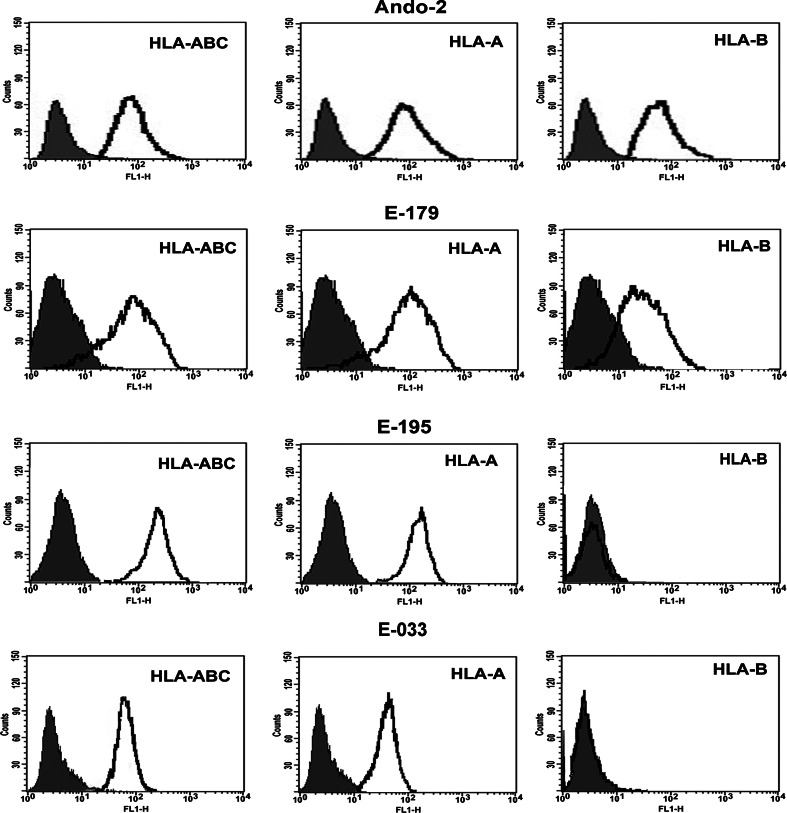
HLA class I surface expression on these melanoma cell lines was measured by indirect immunofluorescence and flow cytometry. Since the isotype-matched antibody control and FITC-conjugated secondary Ab showed identical results, we showed only one control in the histograms and plots. The Ando-2 melanoma cell line showed surface expression of one HLA class I haplotype, expressing HLA-A32 and HLA-B40 alleles (Fig. 1). E-179 melanoma cell line showed surface expression of one HLA class I haplotype, corresponding to HLA-A2 and HLA-B40 alleles (Fig. 1). E-195 melanoma cell line showed surface expression of one HLA-A allele, HLA-A1, but no surface expression of locus B (Fig. 1). In all melanoma cell lines, treatment with IFN-γ induced surface expression of HLA class I alleles expressed in baseline conditions (data not shown). E-033 melanoma cell line showed surface expression of both HLA-A alleles but no surface expression of locus B (Fig. 1).The HLA class I phenotype of these melanoma cell lines was assessed at different time points during in vitro cell culture. HLA phenotypes were identical and did not change during in vitro passages.
Fig. 1.
Surface expression of HLA class I antigens on Ando-2, E-033, E-179 and E-195 human melanoma cell lines. Melanoma cells were stained with specific HLA antibodies and isotype controls and analyzed by flow cytometry. Isotype controls are shown as shaded peaks, and heavy lines represent expression determined by specific antibody staining. Data represent more than three independent experiments
Changes in HLA class I phenotype after growth in nude mice
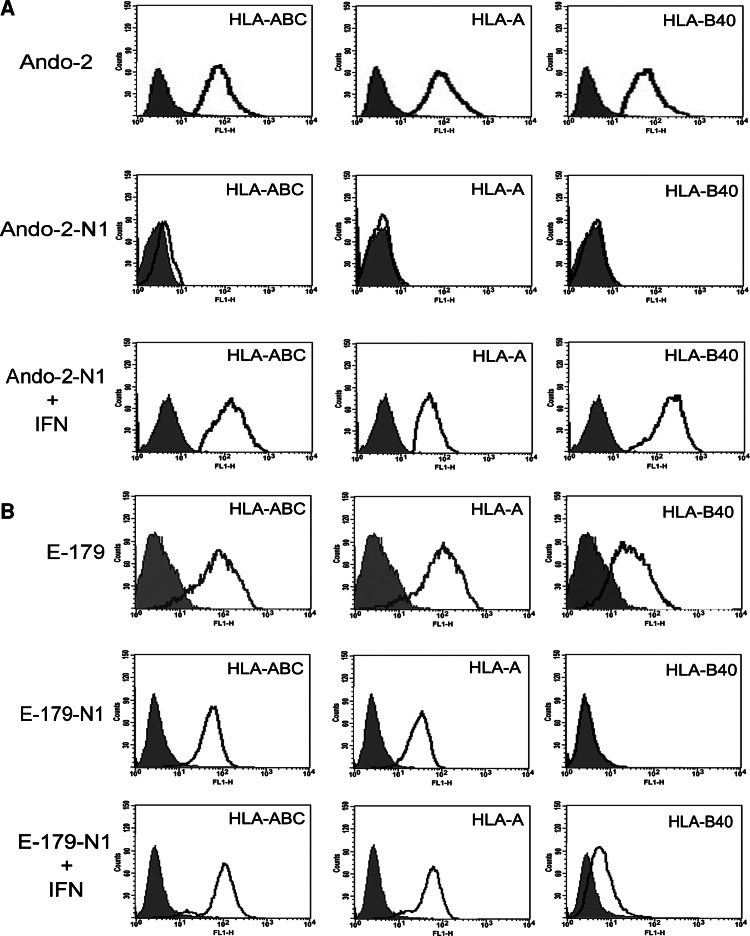
5 × 106 cells of each melanoma cell line were injected in groups of five nude mice and after local growth of the tumors, they were extirpated and the cells adapted to tissue culture, thus each tumor generated a new melanoma cell line. Each human melanoma cell line obtained after growth in nude mice was labeled with suffix -N. The HLA phenotype of tumor cells was analyzed immediately after extirpation and again after adaptation to tissue culture. The same HLA phenotype was found in all of the melanoma cell lines. Ando-2 melanoma cells showed total loss of HLA class I expression after growth in nude mice (Ando-2-N1 human melanoma cells) (Fig. 2). Both HLA-A and HLA-B alleles were induced after IFN-γ treatment (Fig. 2). The same HLA class I phenotype was found in other studied human melanoma cells, from Ando-2-N2 to -N5 (Table 3).
Fig. 2.
Growth in nude mice decreases surface expression of MHC class I antigens. Ando-2-N1 and E-179-N1 melanoma cells derived from nude mice (corresponding to mouse 1 of each group) were stained with specific antibodies and isotype controls and analyzed by flow cytometry. Isotype controls are shown as shaded peaks, and heavy lines represent expression determined by specific antibody staining. Treatment with IFN-γ produces an increase in HLA-A and HLA-B expression. Data for other melanoma cell lines, -N2 to -N5 (obtained from mouse 2–5) presented very similar results (Table 3). Data represent more than three independent experiments
Table 3.
Mean fluorescence of HLA class I surface expression
| Fluorescence intensity (mean) | |||||
|---|---|---|---|---|---|
| Cell linesa | HLA-ABC | HLA-A | HLA-A2 | HLA-B40 | HLA-B8 |
| Ando-2 | 87 | 60 | – | 66 | − |
| Ando-2-N1 | 6 | 0 | − | 0 | − |
| Ando-2-N2 | 8 | 0 | − | 1 | − |
| Ando-2-N3 | 5 | 1 | − | 0 | − |
| Ando-2-N4 | 5 | 1 | − | 1 | − |
| Ando-2-N5 | 3 | 0 | − | 0 | − |
| E-179 | 84 | 84 | 151 | 102 | − |
| E-179-N1 | 46 | 28 | 41 | 1 | − |
| E-179-N2 | 46 | 56 | 29 | 0 | − |
| E-179-N3 | 69 | 56 | 61 | 2 | − |
| E-179-N4 | 48 | 53 | 35 | 1 | − |
| E-179-N5 | 41 | 35 | 25 | 0 | − |
| E-195 | 185 | 169 | − | − | 1 |
| E-195-N1 | 3 | 1 | − | − | 2 |
| E-195-N2 | 79 | 68 | – | − | 1 |
| E-195-N3 | 4 | 0 | – | − | 0 |
| E-195-N4 | 3 | 3 | – | − | 1 |
| E-195-N5 | 46 | 28 | – | − | 2 |
| E-033 | 78 | 56 | 47 | 2 | − |
| E-033-N1 | 80 | 60 | 50 | 3 | − |
| E-033-N2 | 72 | 55 | 40 | 1 | − |
| E-033-N3 | 70 | 50 | 41 | 1 | − |
| E-033-N4 | 65 | 48 | 39 | 0 | − |
| E-033-N5 | 70 | 55 | 45 | 2 | − |
aN1–N5 melanoma cell lines obtained after in vivo growth in nude mice
Tumor cells grown in nude mice after injection of E-179 melanoma cells showed total loss of surface expression of locus B and strong down-regulation of locus A (Fig. 2) and only weak surface expression of one HLA-A allele, HLA-A2. HLA-A and HLA-B alleles were induced after IFN-γ treatment (Fig. 2). The same HLA phenotype was found in the five melanoma cell lines E-179-N (Table 3).
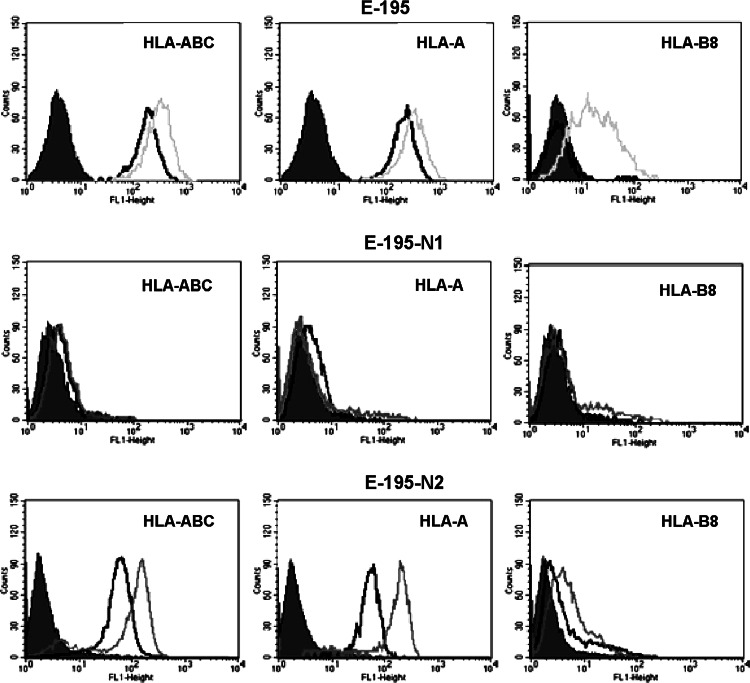
E-195 melanoma cells showed surface expression of one HLA-A allele, HLA-A1 and treatment with IFN-γ induced HLA-A and HLA-B surface expression (Fig. 3). After growth in nude mice, the cells had two different HLA class I phenotypes: (1) total loss of HLA class I expression and no induction of HLA expression after IFN-γ treatment (E-195-N1 cells; Fig. 3); and (2) down-regulation of surface expression of HLA-A1 allele, no expression of HLA-B8 allele and induction of HLA-A and HLA-B alleles with IFN-γ treatment (E-195-N2 cells; Fig. 3). This is the only melanoma cell line showing two different HLA phenotypes after growth in nude mice. In five melanoma cell lines E-195-N, three showed total loss of HLA class I expression and two had HLA class I down-regulation (Table 3). All assays were repeated at least three times with similar results.
Fig. 3.
E-195 human melanoma cells show two different altered HLA class I phenotypes after growth in nude mice. Isotype controls are shown as shaded peaks, expression levels without treatment (baseline conditions) are shown as heavy lines, and dotted lines represent expression after IFN-γ treatment. E-195-N1 (derived from mouse 1) do not present HLA class I surface expression under baseline conditions or after IFN-γ treatment. E-195-N2 (derived from mouse 2) show down-regulation of HLA-A class I allele and IFN-γ induced enhancement of HLA-A and HLA-B alleles. Data for other melanoma cell lines, -N3 to -N5 (obtained from mouse 3–5) presented very similar results (Table 3). Data represent more than three independent experiments
Table 3 depicts the mean fluorescence of the melanoma cell lines before and after growth in nude mice. Three melanoma cell lines (Ando-2, E-179 and E-195) lost HLA class I surface expression after growth in nude mice. E-033 melanoma cells showed no changes in HLA class I phenotype after growth in nude mice, expressing only locus A (Fig. 1, Table 3).
In vitro 4 h cytotoxic assays were performed to determine whether Ando-2 or Ando-2-N cells might be recognized in vitro by mononuclear cells from spleen of nude mice. The results showed that none of the studied melanoma cell lines were recognized by the spleen cells (lysis <10%; data not shown). These cytotoxic assays were also performed using effector cells derived from nude mice previously injected with Ando-2 or Ando-2-N melanoma cells for different periods of time (1–4 weeks), and the results were negative once again. According to these findings, neither Ando-2 nor Ando-2N melanoma cells are recognized in vitro by mononuclear cells derived from spleen of nude mice. Similar results were found in case of E-179, E-195 and E-033 melanoma cells.
Molecular mechanisms implicated in the loss of HLA class I expression
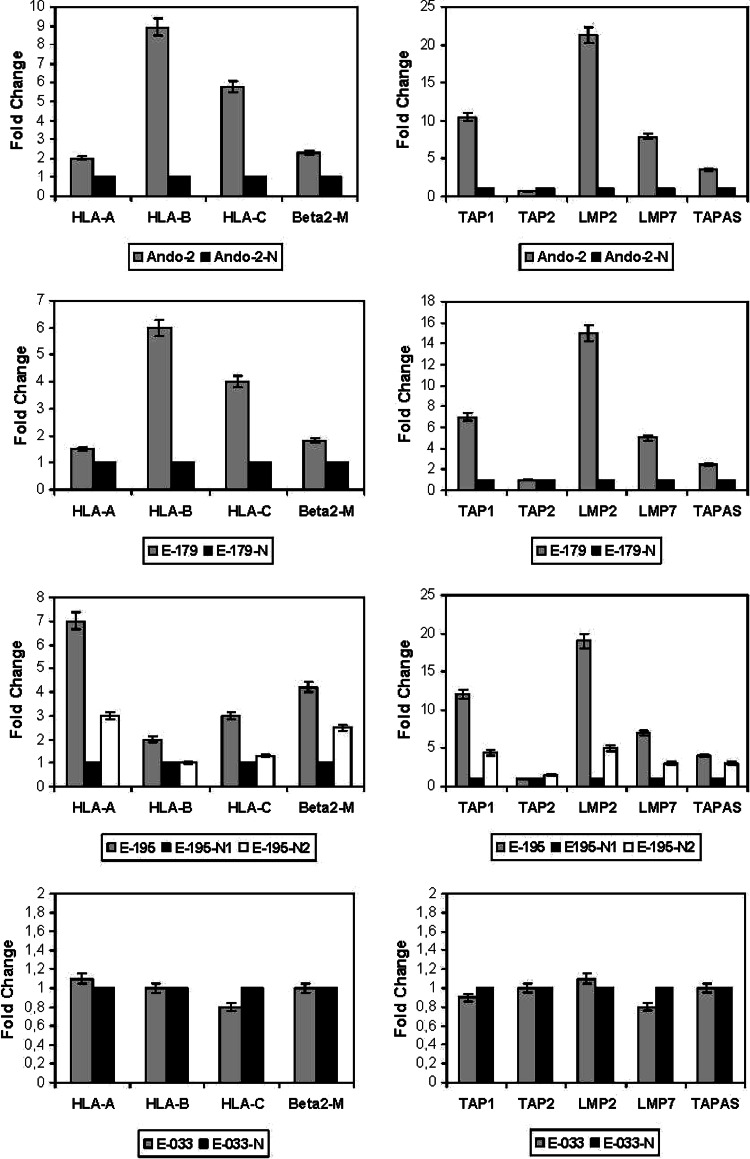
The mRNA expression of HLA class I heavy chains (HLA-A, -B and -C), was determined by quantitative RT-PCR to explore whether transcriptional mechanisms underlie the loss of HLA class I surface expression after growth in nude mice. A strong transcriptional down-regulation of all three HLA loci was observed in Ando-2-N, E-179-N, E-195-N1 and -N2 cells as compared to Ando-2, E-179 and E-195 melanoma cells, respectively. Figure 4 presents the mean values ±SD measured in these three melanoma systems. The down-regulation was around 10-fold for locus B, 6-fold for locus C, and 2-fold for locus A. In E-033 system no changes were detected (Fig. 4).
Fig. 4.
Detection of HLA class I, β2-m and APM component mRNAs by quantitative RT-PCR. This figure depicts the results for melanoma cell lines -N1 to -N5 as mean ± SD in each case (values corresponding to data with -N), except for E-195 where two different means are determinate according to different HLA phenotypes found (N1 and N2). After growth in nude mice of Ando-2, E- 179 and E-195, the human melanoma cells show approximately two- to ninefold decrease in mRNA of HLA-class I heavy chains and β2-microglobulin. The decrease is approximately 5- to 20-fold in APM genes. Only TAP2 gene showed no change in expression. In E-033 system, we did not detect any change at mRNA levels. These RT-PCR experiments were repeated at least three times with similar results
The mRNA expression of APM components (TAPs, LMPs and Tapasin) was also determined by RT-PCR, finding a strong down-regulation of TAP1, LMP2, LMP7, and Tapasin after growth in nude mice for Ando-2-N, E-179-N and E-195-N cells (Fig. 4). The down-regulation was greatest for LMP2, at around 20-fold, followed by TAP1, LMP7 and Tapasin. No change in Tap2 mRNA expression was observed (Fig. 4). The transcriptional expression of β2-microglobulin was also modified but at a lesser degree, with the tumor cells showing half of their original mRNA expression after growth in nude mice (Fig. 4). In E-033 tumor system, we did not find any changes in the transcriptional level of neither APM components nor β2-microglobulin gene (Fig. 4). According to these findings, the loss of HLA surface expression is mainly due to a strong coordinated transcriptional down-regulation of HLA class I heavy chains and APM components.
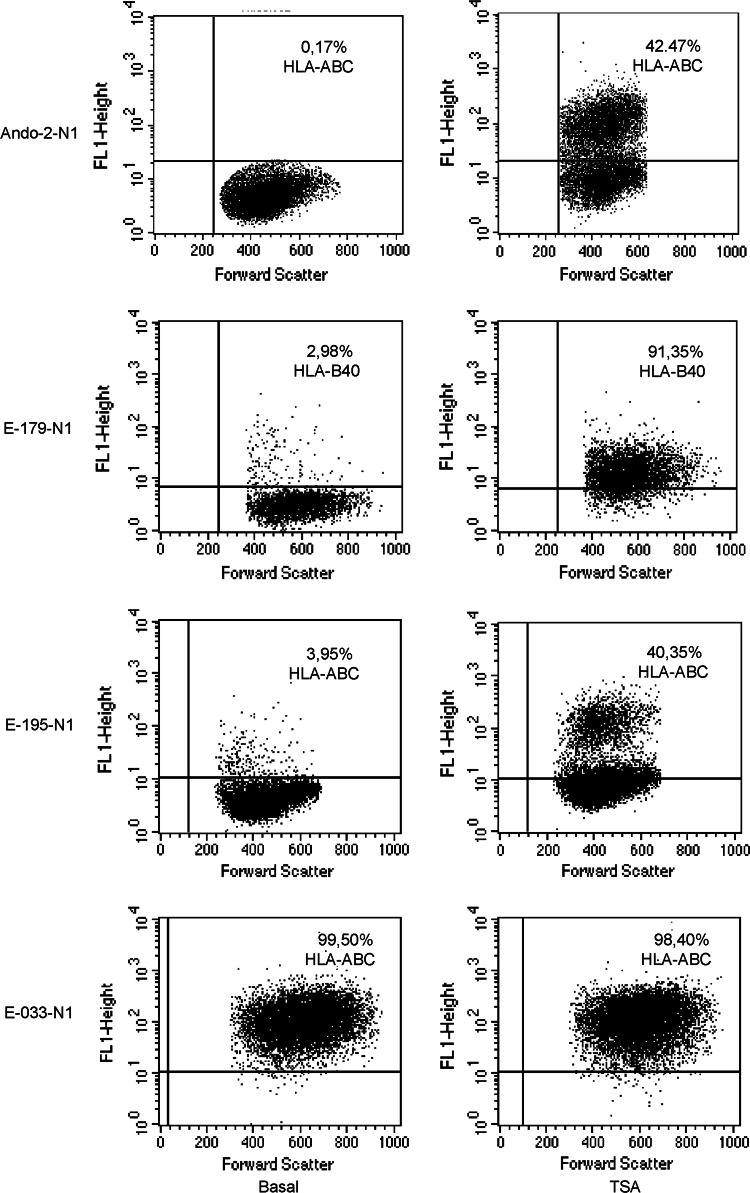
Previous studies showed that surface MHC class I expression on tumor cell lines can be enhanced by treatment with histone deacetylase inhibitors (HDCAi) [14] and that HDCAi induces coordinated mRNA expression of APM and MHC class I heavy chains [13, 16]. The role of histone deacetylation in the loss of MHC class I expression after growth in nude mice was examined by treating the Ando-2-N1 tumor cells with different doses of TSA (50, 250 and 500 nM) for 48 h. All treatments induced HLA class I surface expression in the Ando-2-N1 cells at various levels. Figure 5 depicts, as an example, the results of 48-h treatment with 500 nM TSA, after which positive expression of HLA class I molecules was detected in 42% of tumor cells. HLA-A and HLA-B allele expression was enhanced after TSA treatment. Similar results were obtained with other Ando-2-N human melanoma cells. E-179-N1 melanoma cells do not have cell surface expression of locus B, but after treatment with TSA practically all cells recovered surface expression of allele HLA-B40 (Fig. 5). Similar results were found for other E-179-N melanoma cells. A 40% of E-195-N1 melanoma cells showed positive surface expression of HLA class I molecules after treatment with TSA (Fig. 5). These findings suggest that histone deacetylase-mediated epigenetic mechanisms may be involved in the down-regulation of the MHC class I expression in these tumor cells. When E-033-N1 to -N5 melanoma cells were treated with TSA, no changes in HLA surface expression were found (Fig. 5).
Fig. 5.
TSA treatment enhances expression of down-regulated MHC class I molecules. Ando-2-N1, E179-N1, E195-N1 and E-033-N1 melanoma cells were stained with specific antibodies (W6/32 or HB 115) and isotype controls before and after treatment with TSA (500 nM for 48 h) and analyzed by flow cytometry. Values in the dot-plot are the percentage of cells positive for the respective antibody relative to untreated cells. E-033-N1 cells did not show changes after TSA treatment. The results found for the other melanoma cell lines, named -N2 to -N5, were identical. Data represent more than three independent experiments
Absence of recognition by CTLs and increased in vivo oncogenicity of tumor cell lines after growth in immunodeficient mice
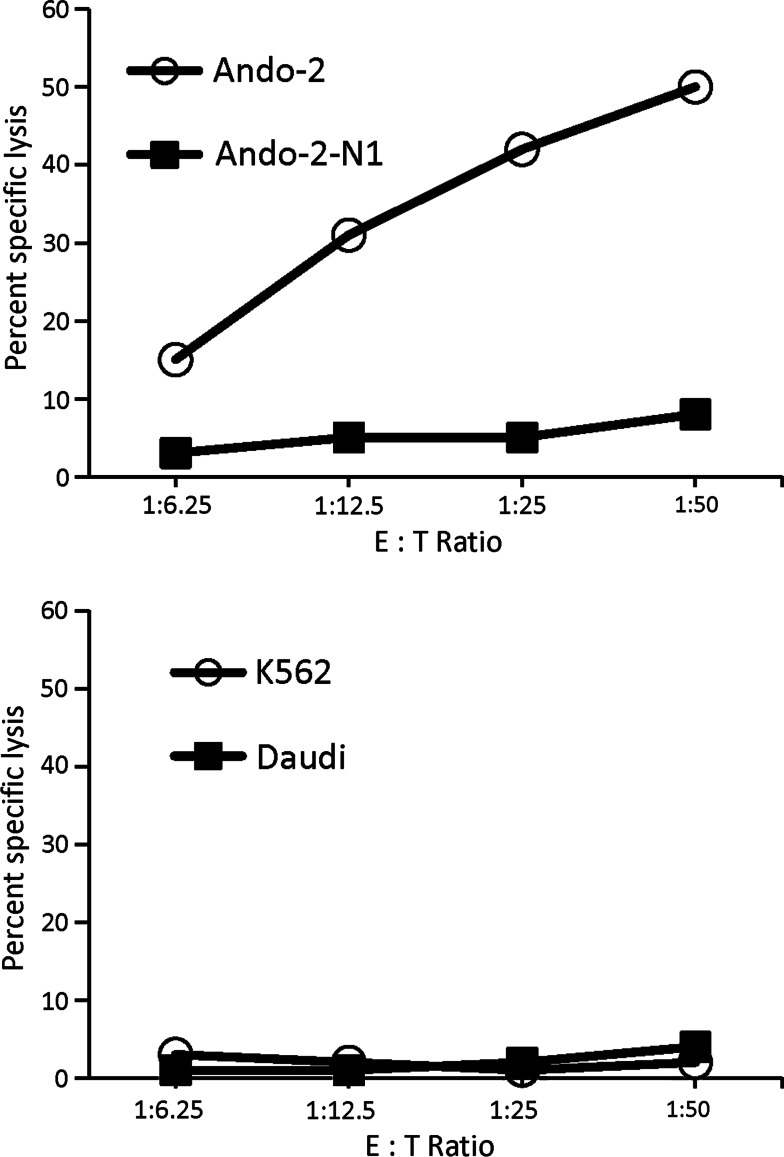
The Ando-2-N cells are not expected to be recognized by the autologous CTLs generated against Ando-2 melanoma cells due to the total cell surface loss of HLA class I molecules. We tested this possibility in cytotoxic assays using different CTL clones as effector cells against Ando-2 and different Ando-2-N melanoma cells as target cells. These CTLs specifically recognized Ando-2 cells but did not recognize Ando-2-N1 cells (Fig. 6). As controls, NK-susceptible target (K562) and LAK-sensitive target (Daudi) were used (Fig. 6). None of the other melanoma cell lines (Ando-2-N2 to-N5) were recognized by these CTLs. The Ando-2 human melanoma cells are not recognized by autologous CTLs after growth in nude mice.
Fig. 6.
Cytotoxicity of autologous CTL in the 51Cr-release assay. CTL cell line obtained against Ando-2 was utilized as effector cells. Various numbers of CTL cells were tested against 5 × 103 Ando-2 or Ando-2-N1 cells (target cells) for 4 h at 37°C. Results are expressed as the mean percentage specific lysis of triplicate samples, in which the SEs of the means were consistently below 10% of the value of the means. Data is representative of three experiments. Similar results were obtained for other Ando-2-N melanoma cell lines and other different autologous CTLs
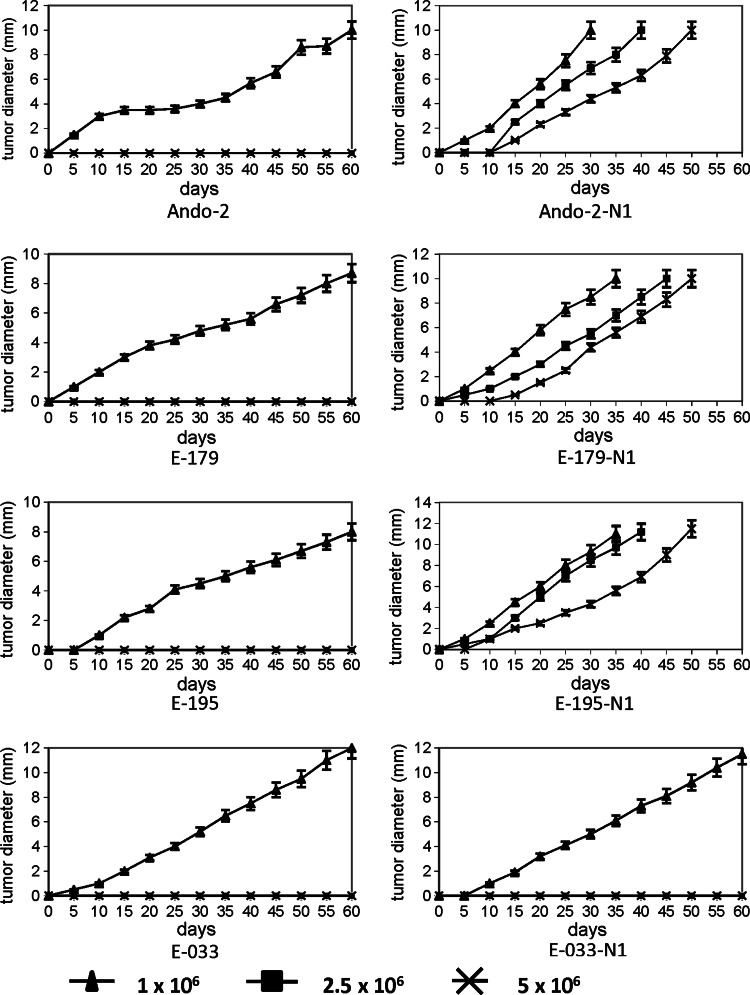
We evaluated the in vivo oncogenicity of the human melanoma cells obtained before and after growth in nude mice. 106, 2.5 × 106 and 5 × 106 cells were injected into different groups of immunodeficient mice, five mice per group and the local tumor growth rate was measured every 2 days. Figure 7 shows the results of local tumor growth as a mean value ±SD from each group of five mice. Ando-2 melanoma cells generated a solid tumor only at a dose of 5 × 106 cells, with no tumor growth when fewer cells were injected (Fig. 7). The Ando-2 cells initially grew very slowly, with an increased growth rate after 20 days, reaching a diameter of 10 mm at day 60 after the injection (Fig. 7). In contrast, Ando-2-N1 cells grew into a solid tumor at all cell doses, even when only 106 cells were injected (Fig. 7). Ando-2-N1 melanoma cells grew rapidly, reaching a diameter of 10 mm by day 30 (Fig. 7). In E-179 and E-195 tumor system, we observed similar results, E-179-N1 and E-195-N1 cells presented a higher in vivo oncogenicity and local growth than E-179 and E-195 cells, respectively (Fig. 7). In addition, we obtained very similar results when we used SCID-Beige mice (data not shown). Therefore, the studied human melanoma cells obtained after growth in immunodeficient mice showed a more rapid local growth and higher oncogenic potential in vivo. On the contrary, E-033 and E-033-N1 human melanoma cells showed the same in vivo oncogenicity and local growth (Fig. 7).
Fig. 7.
In vivo growth of human melanoma cell lines derived from nude mice. Three different doses of human melanoma cells (1 × 106, 2.5 × 106 and 5 × 106) were implanted in nude mice and the tumor growth rate was assessed. Data depict mean ± SD from five mice from each group. a Ando-2 tumor cells grew only after injection of 5 × 106 cells, reaching a tumor diameter of 10 mm in 60 days. b Ando-2-N1 tumor cells grew at all cell doses, reaching a tumor diameter of 10 mm at 30–50 days. Very similar results were obtained for E-179, E-179-N1, E-195 and E-195-N1 human melanoma cells. E-033 and E-033-N1 melanoma cells did not present differences in in vivo growth in nude mice. The assays were performed in groups of five mice and were repeated at least three times
Discussion
Mouse models of cancer have consistently been used to qualify new antitumor therapies for study in human clinical trials. The most frequently used models include transplantable murine tumors grown in syngeneic hosts and xenografts of human tumors grown in immunodeficient mice or humanized mice [29, 32]. However, the phenotype of human tumor cells growing in immunodeficient mice is not usually analyzed or compared with their previous phenotype. Our results describe differences in the HLA phenotype and tumorigenicity of human melanoma cells before and after their transplantation into immunodeficient mice. Three melanoma cell lines showed various types of HLA class I loss after growth in nude mice. One of them, Ando-2, showed a total loss of HLA class I surface expression recoverable after IFN-γ treatment [23]. Another cell line, E-179, presented a total loss of HLA locus B expression and a strong down-regulation of HLA-A locus. The third line, E-195, showed two distinct patterns of changes in the HLA class I expression: down-regulation of locus A or total loss of HLA class I surface expression. Only one melanoma cell line, E-033, showed no changes in HLA class I surface expression after growth in nude mice, indicating that these HLA alterations in human melanoma cell lines are frequent and reproducible but do not always occur. Interestingly, unlike the other studied cell lines, E-033 did not have HLA haplotype loss before being injected into nude mice, while the other investigated melanoma cell lines had developed this escape mechanism to avoid anti-tumor immune attack. These results might indicate that E-033 melanoma cells might have developed a different escape mechanism. In all melanoma cell lines, except for E-195-N1, -N3 and N-4 the HLA class I expression was inducible after IFN-γ treatment. According to our results, the loss of HLA class I surface expression in the studied cell lines was due to a transcriptional mechanism, i.e., the coordinated downregulation of LMP, TAP, Tapasin and HLA class I heavy chain.
Multiple epigenetic mechanisms have been described to underlie changes in HLA antigens on tumor cells and they have been shown to impair the recognition of tumor cells by the components of the adaptive immunity [3]. It was recently reported that TSA treatment of murine cells induces MHC class I surface expression by enhancing the coordinated expression of TAP and LMP molecules [13, 16]. In our experiments, TSA treatment recovered HLA class I expression on human melanoma cell lines derived from nude mice, suggesting that HDAC-mediated chromatin regulation is involved in the suppression of class I antigen processing genes in these cells.
These alterations in HLA class I expression did not occur during in vitro culture, they are produced solely when human tumor cells were grown in vivo in the absence of an autologous immune response. B lymphocytes and NK cells of the mice do not appear to be implicated, since the HLA expression changes were also observed in SCID-Beige mice [23]. Recently, it has been reported that stromal cells may have a major influence on the growth and progression of tumors [33], and that their genetic characteristics determine the prognosis and malignancy of tumors [6, 25]. The HLA alterations observed in the present study might have been caused by interaction with stromal cells. In vitro cell culture maintains the HLA phenotype of tumor cells, whereas in vivo growth in immunodeficient mice can produce phenotypic changes in tumor cells, possibly due to interactions with fibroblasts, epithelial cells and macrophages in the stroma.
Importantly, in all cases where we found alterations in MHC after growth in immunodeficient mice, the melanoma cells acquired a phenotype more tumorigenic per se with higher oncogenic capacity. Based on the obtained results, we can propose a hypothetical path of the step-by-step changes that might take place in the studied melanoma cells during the course of cancer progression in the patients. First, the tumor cells with HLA haplotype loss were selected during the autologous immunoselection stage in the patient. Indeed, the haplotype loss of HLA class I antigen has been described widely as escape mechanism in different tumors [15, 21, 26, 31]. During the next stage, called immunoescape, these cells evade the immune reactivity due to the growth advantage that gives them altered HLA phenotype. After the extirpation from patient and an in vitro culture, these tumor cells maintained their HLA phenotype. However, when these tumor cells were then injected into immunodeficient mice, new HLA alterations had appeared during their in vivo growth indicating that loss of HLA expression can also occur after the immunoescape step. This hypothesis is supported by previous reports describing that two successive mechanisms are implicated in total loss of MHC class I expression in tumor cells: loss of one MHC haplotype and downregulation of APM and MHC heavy chains [9, 15, 18, 27]. This new phase that comes after the immunoescape phase, we have called “Immunoblindness” phase, since such tumor cells are invisible to immune system. We believe that this is an important stage of the tumor progression because the human melanoma cells obtained after growth in nude mice were more oncogenic in vivo than their respective original melanoma cells, indicating a higher grade of malignancy and a more evolved state of the tumor.
The observed HLA alterations have great importance in immunotherapeutic procedures used in humanized-immunodeficient mice against human tumor cells, where immune response component must be evaluated. Moreover, it also must be considered in chemotherapy protocols given the importance of the immune system in this type of therapy [35, 36]. Recently, it has been reported that chemotherapy enhances vaccine-induced antitumor immunity in melanoma patients [20]. The dacarbazine administration before peptides vaccination was able to induce a long-lasting enhancement of memory CD8+ T cell responses to cancer vaccines. We propose that it is crucial to apply a combined anti-tumor treatment including chemo- and radiotherapy along with immunotherapy treatment at that particular stage. This approach might prevent an outgrowth of the most aggressive tumor cell variants and allow prediction of the progression of a specific tumor.
In addition, recently it has been reported a direct relation between defective MHC class I expression and cellular survival promoting resistance to apoptosis [28]. Our results show that human melanoma cells with lower MHC class I expression are more tumorigenic in vivo in immunodeficient mice, suggesting in this xenogenic model an inverse relation between HLA class I expression and tumor oncogenicity per se.
Our findings suggest that any studied parameter, i.e., HLA expression, of malignant cells in xenograft models, has to be evaluated before and after transplantation into immunodeficient mice. These changes in tumor cell phenotype must be taken into account in order to design more appropriate immunotherapy and chemotherapy treatments against tumor cells growing in vivo. Moreover, our results could indicate new implications of HLA losses in oncogenicity and survival of tumor cells. These experimental tumor models will also be useful to study additional functions of HLA class I molecules in tumor progression and the underlying molecular mechanisms.
Acknowledgments
The authors thank I. Linares and E. Arias for technical advice. They also thank Dr. Natalia Aptsiauri for helpful discussion. This work was partially supported by grants from the Fondo de Investigaciones Sanitarias (FIS, CP03/0111), Red Genómica del Cancer RTICC (RETIC RD 06/020), Consejeria de Salud and by Plan Andaluz de Investigación (PAI, Group CTS-143 and project CTS-695) from the Junta de Andalucia, in Spain; by the ENACT project (LSHC-CT-2004-503306), and by the Cancer Immunotherapy project (OJ 2004/c158,18234) from the European Community. A. G. L. was supported by FIS-Research Contract CP03/0111 and C. G. was supported by FPU grant from MICINN (AP2007-03102).
Abbreviations
- HLA
Human leukocyte antigens
- APM
Antigen processing machinery
- TSA
Trichostatin A
- HDCAi
Histone deacetylase inhibitors
References
- 1.Algarra I, García-Lora A, Cabrera T, Ruiz-Cabello F, Garrido F. The selection of tumor variants with altered expression of classical and nonclassical MHC class I molecules: implications for tumor immune escape. Cancer Immunol Immunother. 2004;53:904–910. doi: 10.1007/s00262-004-0517-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aptsiauri N, Cabrera T, Garcia-Lora A, Lopez-Nevot MA, Ruiz-Cabello F, Garrido F. MHC class I antigens and immune surveillance in transformed cells. Int Rev Cytol. 2007;256:139–189. doi: 10.1016/S0074-7696(07)56005-5. [DOI] [PubMed] [Google Scholar]
- 3.Campoli M, Ferrone S. HLA antigen changes in malignant cells: epigenetic mechanisms and biologic significance. Oncogene. 2008;27:5869–5885. doi: 10.1038/onc.2008.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dunn GP, Old LJ, Schreiber RD. The immunobiology of cancer immunosurveillance and immunoediting. Immunity. 2004;21:137–148. doi: 10.1016/j.immuni.2004.07.017. [DOI] [PubMed] [Google Scholar]
- 5.Dunn GP, Old LJ, Schreiber RD. The three Es of cancer immunoediting. Annu Rev Immunol. 2004;22:329–360. doi: 10.1146/annurev.immunol.22.012703.104803. [DOI] [PubMed] [Google Scholar]
- 6.Finak G, Bertos N, Pepin F, Sadekova S, Souleimanova M, Zhao H, Chen H, Omeroglu G, Meterissian S, Omeroglu A, Hallett M, Park M. Stromal gene expression predicts clinical outcome in breast cancer. Nat Med. 2008;14:518–527. doi: 10.1038/nm1764. [DOI] [PubMed] [Google Scholar]
- 7.Foss FM. Immunologic mechanisms of antitumor activity. Semin Oncol. 2002;29:5–11. doi: 10.1053/sonc.2002.33076. [DOI] [PubMed] [Google Scholar]
- 8.Garcia-Lora A, Algarra I, Garrido F. MHC class I antigens, immune surveillance, and tumor immune escape. J Cell Physiol. 2003;195:346–355. doi: 10.1002/jcp.10290. [DOI] [PubMed] [Google Scholar]
- 9.Garcia-Lora A, Martinez M, Algarra I, Gaforio JJ, Garrido F. MHC class I-deficient metastatic tumor variants immunoselected by T lymphocytes originate from the coordinated down-regulation of APM components. Int J Cancer. 2003;106:521–527. doi: 10.1002/ijc.11241. [DOI] [PubMed] [Google Scholar]
- 10.Garrido F, Cabrera T, Concha A, Glew S, Ruiz-Cabello F, Stern PL. Natural history of HLA expression during tumor development. Immunol Today. 1993;14:491–499. doi: 10.1016/0167-5699(93)90264-L. [DOI] [PubMed] [Google Scholar]
- 11.Garrido F, Ruiz-Cabello F, Cabrera T, Pérez-Villar JJ, López-Botet M, Duggan-Keen M, Stern PL. Implications for immunosurveillance of altered HLA class I phenotypes in human tumors. Immunol Today. 1997;18:89–95. doi: 10.1016/S0167-5699(96)10075-X. [DOI] [PubMed] [Google Scholar]
- 12.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/S0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 13.Khan AN, Gregorie CJ, Tomasi TB. Histone deacetylase inhibitors induce TAP, LMP, Tapasin genes and MHC class I antigen presentation by melanoma cells. Cancer Immunol Immunother. 2008;57:647–654. doi: 10.1007/s00262-007-0402-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Magner WJ, Kazim AL, Stewart C, Romano MA, Catalano G, Grande C, Keiser N, Santaniello F, Tomasi TB. Activation of MHC class I, II, and CD40 gene expression by histone deacetylase inhibitors. J Immunol. 2000;165:7017–7024. doi: 10.4049/jimmunol.165.12.7017. [DOI] [PubMed] [Google Scholar]
- 15.Maleno I, Lopez-Nevot MA, Cabrera T, Salinero J, Garrido F. Multiple mechanisms generate HLA class I altered phenotypes in laryngeal carcinomas: high frequency of HLA haplotype loss associated with loss of heterozygosity in chromosome region 6p21. Cancer Immunol Immunother. 2002;51:389–396. doi: 10.1007/s00262-002-0296-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Manning J, Indrova M, Lubyova B, Pribylova H, Bieblova J, Hejnar J, Simova J, Jandlova T, Bubenik J, Reinis M. Induction of MHC class I molecule cell surface expression and epigenetic activation of antigen-processing machinery components in a murine model for human papilloma virus 16-associated tumors. Immunology. 2008;123:218–227. doi: 10.1111/j.1365-2567.2007.02689.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mareel M, Leroy A. Clinical, cellular, and molecular aspects of cancer invasion. Physiol Rev. 2003;83:337–376. doi: 10.1152/physrev.00024.2002. [DOI] [PubMed] [Google Scholar]
- 18.Méndez R, Rodríguez T, Del Campo A, Monge E, Maleno I, Aptsiauri N, Jiménez P, Pedrinaci S, Pawelec G, Ruiz-Cabello F, Garrido F. Characterization of HLA class I altered phenotypes in a panel of human melanoma cell lines. Cancer Immunol Immunother. 2008;57:719–729. doi: 10.1007/s00262-007-0411-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nagorsen D, Scheibenbogen C, Marincola FM, Letsch A, Keilholz U. Natural T cell immunity against cancer. Clin Cancer Res. 2003;9:4296–4303. [PubMed] [Google Scholar]
- 20.Nisticò P, Capone I, Palermo B, Del Bello D, Ferraresi V, Moschella F, Aricò E, Valentini M, Bracci L, Cognetti F, Ciccarese M, Vercillo G, Roselli M, Fossile E, Tosti ME, Wang E, Marincola F, Imberti L, Catricalà C, Natali PG, Belardelli F, Proietti E. Chemotherapy enhances vaccine-induced antitumor immunity in melanoma patients. Int J Cancer. 2009;124:130–139. doi: 10.1002/ijc.23886. [DOI] [PubMed] [Google Scholar]
- 21.Norell H, Carlsten M, Ohlum T, Malmberg KJ, Masucci G, Schedvins K, Altermann W, Handke D, Atkins D, Seliger B, Kiessling R. Frequent loss of HLA-A2 expression in metastasizing ovarian carcinomas associated with genomic haplotype loss and HLA-A2-restricted HER-2/neu-specific immunity. Cancer Res. 2006;66:6387–6394. doi: 10.1158/0008-5472.CAN-06-0029. [DOI] [PubMed] [Google Scholar]
- 22.Novellino L, Castelli C, Parmiani G. A listing of human tumor antigens recognized by T cells: March 2004 update. Cancer Immunol Immunother. 2005;54:187–207. doi: 10.1007/s00262-004-0560-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paco L, Garcia-Lora AM, Casares C, Cabrera C, Algarra I, Collado A, Maleno I, Garrido F, Lopez-Nevot MA. Total loss of HLA class I expression on a melanoma cell line after growth in nude mice in absence of autologous antitumor immune response. Int J Cancer. 2007;121:2023–2030. doi: 10.1002/ijc.22925. [DOI] [PubMed] [Google Scholar]
- 24.Pawelec G, Marsh SG. ESTDAB: a collection of immunologically characterised melanoma cell lines and searchable databank. Cancer Immunol Immunother. 2006;55:623–627. doi: 10.1007/s00262-005-0117-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Postovit LM, Margaryan NV, Seftor EA, Kirschmann DA, Lipavsky A, Wheaton WW, Abbott DE, Seftor RE, Hendrix MJ. Human embryonic stem cell microenvironment suppresses the tumorigenic phenotype of aggressive cancer cells. Proc Natl Acad Sci USA. 2008;105:4329–4334. doi: 10.1073/pnas.0800467105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rodriguez T, Méndez R, Roberts CH, Ruiz-Cabello F, Dodi IA, López Nevot MA, Paco L, Maleno I, Marsh SG, Pawelec G, Garrido F. High frequency of homozygosity of the HLA region in melanoma cell lines reveals a pattern compatible with extensive loss of heterozygosity. Cancer Immunol Immunother. 2005;54:141–148. doi: 10.1007/s00262-004-0561-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Romero JM, Jiménez P, Cabrera T, Cózar JM, Pedrinaci S, Tallada M, Garrido F, Ruiz-Cabello F. Coordinated downregulation of the antigen presentation machinery and HLA class I/beta2-microglobulin complex is responsible for HLA-ABC loss in bladder cancer. Int J Cancer. 2005;113:605–610. doi: 10.1002/ijc.20499. [DOI] [PubMed] [Google Scholar]
- 28.Sabapathy K, Nam SY. Defective MHC class I antigen surface expression promotes cellular survival through elevated ER stress and modulation of p53 function. Cell Death Differ. 2008;15:1364–1374. doi: 10.1038/cdd.2008.55. [DOI] [PubMed] [Google Scholar]
- 29.Sausville EA, Burger AM. Contributions of human tumor xenografts to anticancer drug development. Cancer Res. 2006;66:3351–3354. doi: 10.1158/0008-5472.CAN-05-3627. [DOI] [PubMed] [Google Scholar]
- 30.Scheel C, Onder T, Karnoub A, Weinberg RA. Adaptation versus selection: the origins of metastatic behavior. Cancer Res. 2007;67:11476–11479. doi: 10.1158/0008-5472.CAN-07-1653. [DOI] [PubMed] [Google Scholar]
- 31.So T, Takenoyama M, Mizukami M, Ichiki Y, Sugaya M, Hanagiri T, Sugio K, Yasumoto K. Haplotype loss of HLA class I antigen as an escape mechanism from immune attack in lung cancer. Cancer Res. 2005;65:5945–5952. doi: 10.1158/0008-5472.CAN-04-3787. [DOI] [PubMed] [Google Scholar]
- 32.Talmadge JE, Singh RK, Fidler IJ, Raz A. Murine models to evaluate novel and conventional therapeutic strategies for cancer. Am J Pathol. 2007;170:793–804. doi: 10.2353/ajpath.2007.060929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tisty TD, Coussens LM. Tumor stroma and regulation of cancer development. Annu Rev Pathol. 2006;1:119–150. doi: 10.1146/annurev.pathol.1.110304.100224. [DOI] [PubMed] [Google Scholar]
- 34.Villunger A, Strasser A. The great escape: is immune evasion required for tumor progression? Nat Med. 1999;5:874–875. doi: 10.1038/11311. [DOI] [PubMed] [Google Scholar]
- 35.Zitvogel L, Apetoh L, Ghiringhelli F, Kroemer G. Immunological aspects of cancer chemotherapy. Nat Rev Immunol. 2008;8:59–73. doi: 10.1038/nri2216. [DOI] [PubMed] [Google Scholar]
- 36.Zitvogel L, Apetoh L, Ghiringhelli F, André F, Tesniere A, Kroemer G. The anticancer immune response: indispensable for therapeutic success? J Clin Invest. 2008;118:1991–2001. doi: 10.1172/JCI35180. [DOI] [PMC free article] [PubMed] [Google Scholar]