Abstract
We recently compared the HCV polyprotein to the human proteome in order to test whether amino acid sequences unique to the virus could represent immunodominant epitopic determinants of the human humoral immune response against HCV. We identified a relatively limited number of HCV fragments with no/low similarity to the human host that represented exclusive HCV motifs. In this study, the peptides corresponding to low/zero similarity sequences were synthesized and assayed with HCV-infected sera. With different patterns, the synthetic HCV peptides corresponding to low/zero similarity sequences were found to be immunoreactive. In particular, the HCV E1 (315–323) HRMAWDMMM, HCV E2/NS1 (547–555) NWFGCTWMN, and HCV NS5 (2638–2646) YDTRCFDST sequences were immunodominant in the HCV-infected cohort under study. These three peptides correspond to sequences that are endowed with low-similarity to the human proteome, are highly conserved among various HCV strains, and have, potentially, a scarce susceptibility to proteolytic attacks. These data may be of help in defining the multiple factors which concur in the modulation of the human immune response against HCV, eventually providing information for the design of effective anti-HCV vaccines.
Keywords: Unique peptide sequences, Conserved peptide sequences, Sequence-to-sequence peptide matching, Proteasomal cleavages, HCV-related immunity
Introduction
Recently we undertook a comparative study of HCV and Homo sapiens proteomes to obtain data on peptide sequence (dis)similarity between the HCV polyprotein and human proteins. We found a high degree of amino acid motif sharing between HCV and human proteomes, with only a limited number of viral peptide motifs restricted to HCV with no counterpart in the human proteome [22]. More specifically, 57.6% of the approximately 34,000 human proteins included at the time of the download in the human proteome database at EBI’s Integr8 site contain viral HCV pentamers. High level of similarity persists even when the stringency of the search parameters is increased by extending the HCV peptide length to 6- or 7-mers.
We interpret such ample similarity as a possible factor limiting the immune reactions of the host against HCV, partly explaining the host’s tolerance of the virus responsible for the chronic infection. Indeed, viral peptide sequences identical to those found in self-proteins should be immunotolerated, since autoantigen-specific lymphocytes are deleted during the establishment of self-identity [4, 9, 17]. Consequentially, sharing of common motifs might be one cause of limited immunogenicity of HCV. As a corollary, the data also question a possible direct relationship between HCV molecular mimicry to human proteins and human HCV-associated autoimmune diseases. Molecular mimicry predicts that HCV similarity to human proteins should lead to autoimmune phenomena since immune response against the infectious agent may result in formation of cross-reacting antibodies that bind the shared epitopes on the normal cell and result in the auto-destruction of the cell [30]. The molecular mimicry hypothesis implies that HCV infection should be a practically infinite source of autoimmune diseases since HCV 5-, 6-, and 7-mer matches are widely disseminated throughout more than half of the human proteome while actually HCV-associated autoimmune diseases have a much lower incidence than expected [27].
Based on these observations, we reasoned that a host anti-HCV immune response should be predominantly directed toward amino acid motifs unique to the virus, so collimating with the general acceptance of the preferential immunorecognition of non-self sequences. Therefore, we started this study by testing the potential of HCV peptide sequences not shared with the human proteome to raise the humoral immune response in HCV-infected patients. Synthetic peptides corresponding to HCV polyprotein displaying low sequence similarity to the human proteome were immunoassayed against HCV-infected sera. Synthetic peptides with medium/high similarity to the human proteins were used as controls. We report that, among the HCV peptides we tested, those hosting sequences unique to the HCV proteome were predominant targets of immune recognition by HCV-infected sera. Additionally, we found that the immunoreactivity was further restricted to HCV peptide sequences highly conserved among the different HCV strains and hosting a low number of potential proteolytic sites, so indicating a multiple control pathway in the human anti-HCV response.
Materials and methods
Computational analyses
The HCV polyprotein sequence selected for this study corresponds to the hepatitis C virus, genotype 1a, as reported at http://www.expasy.org, accession P26664. The polyprotein includes the following proteins: capsid C; envelope glycoprotein E1; envelope glycoprotein E2; p7; protease NS2-3; serine protease/NTPase/helicase NS3; non-structural (NS) 4A; NS4B; NS5A; and RNA-directed RNA polymerase, for a total length of 3,010 amino acids. The human proteome was obtained from EBI’s Integr8 site (http://www.ebi.ac.uk/integr8) and consisted of 34,044 nonredundant proteins at the time of download. Hypothetical and/or unidentified protein sequences were included.
Sequence similarity analysis comparing the viral polyprotein sequence to the human proteome was conducted using HCV 5-mers as probes to scan the Homo sapiens proteome looking for exact matches. Overlaps between the human proteome and each viral protein were determined as already described [22, 34].
The comparative peptide similarity analysis was conducted by multiple sequence alignment of the following different HCV isolate genotypes. The HCV types are listed here with the Swiss-Prot accession number in parentheses: 1a, isolate 1 (P26664); 1a, isolate H (P27958); 2a, isolate JFH-1 (Q99IB8); 2a, isolate HC-J6 (P26660); 1b, isolate Taiwan (P29846); 1b, isolate BK (P26663); 1b, isolate Con1 (Q9WMX2); 1b, isolate Japanese (P26662); 1b, isolate HC-JT (Q00269); 2b, isolate HC-J8 (P26661); 2b, isolate JPUT971017 (Q9DHD6). Sequence alignment was conducted using the T-Coffee program (see http://www.expasy.org/tools) [29]. Proteasomal cleavages (made by human proteasome type III) in HCV-1A were predicted using the PAProC program [23].
Synthetic peptides and sera
The HCV synthetic peptides used for immunoassays were: aa315–323 HRMAWDMMM; aa440–448 LFYHHKFNS; aa482–491 RPYCWHYPP; aa547–555 NWFGCTWMN; aa741–750 SQAEAALENL; aa786–794 VYTFYGMWP; aa1603–1611 WDQMWKCLI; aa1628–1637 GAVQNEITLT; aa1760–1768 WAKHMWNFI; aa 2050–2058 TCRNMWSGT; aa 2182–2191 AEAAGRRLAR; aa2638–2646 YDTRCFDST. Peptides were synthesized using standard Fmoc (N-(9-fluorenyl) methoxycarbonyl) solid phase peptide synthesis and were obtained from Primm srl, Milan, Italy. Peptide purity (>90%) was controlled by analytical HPLC, and the molecular mass of purified peptides confirmed by fast atomic bombardment mass spectrometry. Peptides were dissolved in 0.9% NaCl, aliquoted and stored at −20°C.
Serum samples from patients with HCV infection were kindly made available by Dr. Luisa Gennero and Prof. Antonio Ponzetto, Dept. of Internal Medicine, University of Turin, Division of Gastroenterology, San Giovanni Battista Hospital, Turin, Italy. HCV studies had been approved by the Hospital Institutional Review Board. We analyzed only sera from patients who had not yet received therapeutical treatments. Control sera from healthy subjects were available in the lab. Blood donors’ sera were obtained from the Blood Bank, University Hospital of Bari, following donors’ informed and signed consent. Sera were treated with RNase/DNase for 2 h at 37°C, and then partially purified by precipitation with 40% saturated (NH4)2SO4 (x2). The precipitate was dissolved in phosphate-buffered saline (PBS), dialyzed against PBS with several changes for 24 h at 4°C, aliquoted and stored at −20°C until assay. For pre-absorbtion experiments, sera from blood donors were pooled, partially purified by (NH4)2SO4 precipitation and then incubated with peptide 1 (or 4 or 5 or 9) at a threefold excess (w/w) in PBS −1% BSA for 1 h at room temperature.
ELISA immunoassays
Serum reactivity against synthetic HCV peptides was tested by ELISA assays. Multiwell plates were coated with synthetic HCV peptides by adding to each well 100 μl 0.5% glutaraldehyde cross-linked synthetic peptide solution at a concentration of 25 μM. Following incubation for 2 h at RT, wells were blocked with 5% BSA in PBS, and incubated with human partially purified serum. HPR-conjugated goat anti-human antibodies were used as secondary antibody. Peroxidase activity was detected by adding 3,3′,5,5′-tetramethylbenzidine dihydrochloride as substrate. Results are expressed as effective absorbance. The absorbance was measured at wavelengths of 450 and 620 nm. The effective absorbance was calculated subtracting A 620 from A 450.
Results
Identification of HCV-restricted protein sequences and selection of putative epitopes
Similarity analysis identifies amino acid sequences that could represent putative low-similarity epitopes, quantifying the number of perfect peptide matches between the proteomes of the test organisms, in this case, HCV and human. Basic to this analysis is the definition of the length of the minimal immunogenic peptide determinant, i.e., the length of the shortest sequence that can constitute a linear determinant. According to the literature, 5 to 6 amino acids peptides represent sufficient minimal antigenic determinants [13, 14, 25, 28, 37] and, therefore, a putative epitopic peptide is reasonably defined to have a minimal length of five amino acids. Thus, the similarity level of a peptide sequence to a set of proteins can be calculated as the number of times each peptide pentamer occurs in the analysed set of proteins. The similarity level of a peptide is zero when the 5-mers forming the peptide are absent in the proteins under analysis. The similarity level of a peptide is incrementally higher as the 5-mer sequence is repeatedly represented in the proteome investigated.
In this study, the HCV genotype 1a proteome from a previous report [22] was further analysed for similarity to the human proteome searching for low similarity peptide fragments to be evaluated as antigens in immunoassays with sera from HCV-infected patients. A set of nine 9-mer HCV fragments with no more than 2 matches at the 5-mer level were synthesised and used as antigens. The low-similarity peptide sequences thus identified are described in Table 1. The Table also shows three HCV peptides used as controls because of their medium/high similarity to the human proteome.
Table 1.
Description of the HCV peptides used in this study
| Peptide | Aa position | Viral protein | Sequence | Similarity to human proteome (number of 5-mer matches) |
|---|---|---|---|---|
| Controls | ||||
| C1 | 741–750 | E2/p7 | SQAEAALENL | 289 |
| C2 | 1628–1637 | NS3 | GAVQNEITLT | 66 |
| C3 | 2182–2191 | NS5A | AEAAGRRLAR | 263 |
| Low-similarity | ||||
| 1 | 315–323 | E1 | HRMAWDMMM | 1 |
| 2 | 440–448 | E2/NS1 | LFYHHKFNS | 2 |
| 3 | 482–491 | E2/NS1 | RPYCWHYPP | 0 |
| 4 | 547–555 | E2/NS1 | NWFGCTWMN | 1 |
| 5 | 786–794 | NS2 | VYTFYGMWP | 1 |
| 6 | 1603–1611 | NS3 | WDQMWKCLI | 0 |
| 7 | 1760–1768 | NS4A | WAKHMWNFI | 0 |
| 8 | 2050–2058 | NS5 | TCRNMWSGT | 0 |
| 9 | 2638–2646 | NS5 | YDTRCFDST | 2 |
Analysis of peptide sequence variability among distinct HCV Types
This study was focused on the HCV polyprotein sequence corresponding to genotype 1a, since (1) genotype 1a is distributed with a relatively high frequency among the HCV infected italian population [3, 7, 11], and (2) it has been reported that individuals infected with this virus evoke a stronger anti-HCV immune response to several immunodominant epitopes of HCV compared to individuals infected with HCV 1b [5]. On the other hand, it has been well demonstrated that HCV exhibits complex genetic variability that can be classified into four hierarchical strata: genotypes, sub-genotypes, isolates, and quasi-species [39, 40]. Moreover, infected patients may present multiple HCV genotype infections or experience changing patterns of HCV sequence variability over time [33].
Thus, amino acid sequence variability of various HCV types might alter peptide immunoreactivity. To comprehensively define the relationship between HCV sequence variability, its similarity to the human proteome and peptide immunoreactivity, the peptide sequences selected for this study were compared by sequence alignment to those of various HCV types and evaluated for homology level. Table 2 shows that peptides 2, 5 and 8 are highly variable sequences across the HCV type sequences studied while the other peptide sequences tend to be conserved.
Table 2.
Peptide sequence and similarity level changes among different HCV isolate genotypes
| HCV type | Peptide 1 | Peptide 2 | Peptide 3 | |||
|---|---|---|---|---|---|---|
| 1a | HRMAWDMMMN | (1) | LFYHHKFNS | (2) | RPYCWHYPP | (0) |
| 1a (H) | HRMAWNMMMN | (1) | LFYQHKFNS | (16) | RPYCWHYPP | (0) |
| 2a (JFH-1) | HRMAWDMMMN | (1) | LFYTNRFNS | (16) | RPYCWHYPP | (0) |
| 2a (HC-J6) | HRMAWDMMMN | (1) | LFYTHSFNS | (14) | RPYCWHYPP | (0) |
| 1b (Taiwan) | HRMAWDMMMN | (1) | LFYAHRFNA | (6) | RPYCWHYAP | (2) |
| 1b (BK) | HRMAWDMMMN | (1) | LFYTHSFNS | (14) | RPYCWHYPP | (0) |
| 1b (Con1) | HRMAWDMMMN | (1) | LFYVHKFNS | (10) | RPYCWHYAP | (2) |
| 1b (Japanese) | HRMAWDMMMN | (1) | LFYAHRFNA | (6) | RPYCWHYAP | (2) |
| 1b (HC-JT) | HRMAWDMMMN | (1) | LFYAHKFNS | (12) | RPYCWHYAP | (2) |
| 2b (HC-J8) | HRMAWDMMLS | (1) | LFYTHKFNS | (14) | RPYCWHYPP | (0) |
| 2b (JPUT971017) | QRMAWDMMLN | (2) | LFYANKFNS | (10) | RPYCWHYPP | (0) |
| HCV type | Peptide 4 | Peptide 5 | Peptide 6 | |||
|---|---|---|---|---|---|---|
| 1a | NWFGCTWMN | (1) | VYTFYGMWP | (1) | WDQMWKCLI | (0) |
| 1a (H) | NWFGCTWMN | (1) | VYALYGMWP | (18) | WDQMRKCLI | (15) |
| 2a (JFH-1) | SWFGCTWMN | (2) | TYCLTGLWP | (25) | WDAMWKCLA | (6) |
| 2a (HC-J6) | SWFGCTWMN | (2) | TYSLTGLWS | (79) | WDVMWKCLT | (0) |
| 1b (Taiwan) | NWFGCTWMN | (1) | AYALYGVWP | (19) | WDQMWKCLT | (0) |
| 1b (BK) | NWFGCTWMN | (1) | TYALYGVWP | (20) | WDQMWKCLI | (0) |
| 1b (Con1) | NWFGCTWMN | (1) | AYALYGVWP | (19) | WDQMWKCLI | (0) |
| 1b (Japanese) | NWFGCTWMN | (1) | AYALYGVWP | (19) | WDQMWKCLI | (0) |
| 1b (HC-JT) | NWFGCTWMN | (1) | AYALYGVWP | (19) | WDQMWKCLI | (0) |
| 2b (HC-J8) | AWFGCTWMN | (1) | TYSVLGLWS | (83) | WDVMWKCLT | (0) |
| 2b (JPUT971017) | AWFGCTWMN | (1) | TYSVLGLWS | (83) | WDVMWKCLT | (0) |
| HCV type | Peptide 7 | Peptide 8 | Peptide 9 | |||
|---|---|---|---|---|---|---|
| 1a | WAKHMWNFI | (0) | TCRNMWSGT | (0) | YDTRCFDST | (2) |
| 1a (H) | WAKHMWNFI | (0) | TCKNMWSGT | (0) | YDTRCFDST | (2) |
| 2a (JFH-1) | WARHMWNFI | (0) | TCMNTWQGT | (6) | YDTRCFDST | (2) |
| 2a (HC-J6) | WAKHMWNFI | (0) | TCMNIWQGT | (1) | YDTRCFDST | (2) |
| 1b (Taiwan) | WANDMWNFI | (0) | TCSNTWHGT | (6) | YDTRCFDST | (2) |
| 1b (BK) | WAKHMWNFI | (0) | TCSNTWHGT | (6) | YDTRCFDST | (2) |
| 1b (Con1) | WAKHMWNFI | (0) | TCSNTWHGT | (6) | YDTRCFDST | (2) |
| 1b (Japanese) | WAKHMWNFI | (0) | TCSNTWHGT | (6) | YDTRCFDST | (2) |
| 1b (HC-JT) | WAKHMWNFI | (0) | TCSNTWHGT | (6) | YDTRCFDST | (2) |
| 2b (HC-J8) | WAKHMWNFI | (0) | TCLNLWQGT | (12) | YDTRCFDST | (2) |
| 2b(JPUT971017) | WARHMWNFI | (0) | TCLNMWQGT | (1) | YDTRCFDST | (2) |
Accession numbers of HCV genotypes are given under “Materials and methods”. The peptide sequences numbered from 1 to 9 are aligned with the corresponding peptide portions present in the different HCV genotypes. Sequences are reported with changed amino acid bold underlined. Peptide similarity level (as total number of pentamer matches between the peptide and human proteome) in parentheses
Proteolytic pattern of low-similarity HCV peptide sequences
The selected low-similarity HCV fragments described in Table 1 were further analyzed for proteolytic sites as a possible additional factor affecting peptide epitopicity. Indeed, it has been reported that mutations within HCV epitopes also cause their destruction by changing the pattern of proteasome digestion [21]. The importance of intracellular antigen processing in the definition of antigenic determinants has been established [38], and cleavage at a single processing site can be crucial for effective antigen presentation [24]. Therefore, to reach a better comprehensive understanding of the molecular basis of HCV antigenicity, the predicted proteolytic pattern of the selected HCV motifs was analyzed. The distribution of potential proteolytic sites in the low-similarity sequences is illustrated in Table 3. It can be seen that sequences 1, 3, 4, 8 and 9 appear to host a low number of potential proteasomal cleavage sites (≤3), suggesting a potential higher stability of the corresponding peptides.
Table 3.
Predicted proteolytic pattern of low-similarity HCV peptides
| Peptide | Aa Position | Viral protein | Sequencea | No. of Proteolytic sites |
|---|---|---|---|---|
| 1 | 315–323 | E1 | ↓HR↓MAWDMMM | 2 |
| 2 | 440–448 | E2/NS1 | ↓LF↓Y↓HHK↓F↓N↓S | 6 |
| 3 | 482–491 | E2/NS1 | ↓RPYCWHYPP | 1 |
| 4 | 547–555 | E2/NS1 | ↓N↓WF↓GCTWMN | 3 |
| 5 | 786–795 | NS2 | VYTFY↓G↓M↓W↓P | 4 |
| 6 | 1603–1611 | NS3 | ↓WD↓Q↓MWK↓C↓LI | 5 |
| 7 | 1760–1768 | NS4A | ↓WA↓K↓H↓M↓WN↓FI | 6 |
| 8 | 2050–2058 | NS5 | T↓CRN↓MWS↓GT | 3 |
| 9 | 2638–2646 | NS5 | ↓Y↓DTRC↓FDST | 3 |
aThe proteolytic sites are indicated by arrows
HCV peptide immunoreactivity assay
Peptides corresponding to the HCV sequences motifs described above were synthesized, and tested as potential antigenic epitopes in ELISA immunoassays using sera from HCV-infected and healthy subjects. The immunoreactivity pattern of the HCV peptides monitored by using sera from five healthy controls, 10 blood donors, and 29 HCV patients, is detailed in Table 4. It can be seen that the majority of the HCV low-similarity peptides showed a clear response, although to different extents. For example, it is evident from Table 4 that peptide 1 (i.e., peptide E1315–323HRMAWDMMM) is immunodominant. This epitope is a conserved sequence among the HCV strains (see Table 2) and hosts a low number of potential proteolytic sites (see Table 3). Analogously, amino acid sequence conservation as well as scarce susceptibility to proteolytic fragmentation, i.e., a low number of proteolytic sites, are shown by the peptides 4 and 9 which are frequently recognized by the HCV-infected sera (Table 4). One exception is peptide no. 3, corresponding to the low-similarity HCV482–491 RPYCWHYPP sequence, which was unreactive with almost all of the sera tested, even if endowed with a high level of sequence conservativeness and low potential to proteolytic degradation. In this context it may be relevant that HCV482–491RPYCWHYPP is located in the viral CD81-binding domain spanning from aa 481 to aa 493 [10], suggesting that the low similarity HCV482–491RPYCWHYPP peptide might potentially represent a cryptic epitope.
Table 4.
HCV peptide antigenic pattern in HCV infected patients
| Serum | HCV peptide | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C1 | C2 | C3 | ||||||||||
| None | – | – | – | 0.27 | – | – | 0.35 | 0.39 | – | – | – | 0.21 |
| Healthy | ||||||||||||
| 1 | – | 0.23 | – | 0.52 | – | – | 0.33 | 0.29 | – | – | – | 0.49 |
| 2 | – | – | – | 0.39 | – | – | 0.24 | 0.30 | – | 0.27 | 0.33 | 0.28 |
| 3 | – | 0.23 | – | 0.48 | – | – | 0.73 | 0.45 | 0.22 | 0.21 | 0.24 | 0.66 |
| 4 | 0.22 | – | – | 0.25 | – | – | 0.44 | 0.28 | – | – | – | 0.94 |
| 5 | – | – | – | 0.30 | – | – | 0.53 | 0.30 | – | – | – | 0.62 |
| Blood donor | ||||||||||||
| 1 | – | 0.22 | – | 0.36 | – | – | 0.34 | – | – | – | – | 0.33 |
| 2 | – | 0.27 | 0.24 | 0.38 | – | – | 0.42 | 0.23 | – | – | – | – |
| 3 | – | – | – | 0.37 | – | – | 0.69 | 0.50 | 0.27 | – | – | 0.48 |
| 4 | – | – | – | 0.35 | – | – | 0.42 | – | – | – | – | 0.64 |
| 5 | – | – | – | 0.21 | – | – | 0.49 | 0.47 | – | – | – | 0.71 |
| 6 | – | – | – | 0.48 | – | – | 0.22 | 0.39 | – | – | – | 0.48 |
| 7 | – | – | – | – | – | – | 0.53 | 0.27 | 0.22 | 0.22 | 0.31 | 0.33 |
| 8 | – | – | – | – | – | – | 0.88 | 0.38 | 0.44 | – | – | 0.73 |
| 9 | 0.21 | – | – | – | – | – | 0.39 | 0.50 | – | – | 0.27 | 0.59 |
| 10 | 0.24 | – | 0.33 | 0.23 | – | – | 0.41 | 0.36 | – | – | – | 0.65 |
| HCV infected | ||||||||||||
| 1 | – | 0.29 | 0.28 | 0.91 | – | – | 0.40 | 0.38 | 0.23 | 0.28 | – | 0.44 |
| 2 | 0.31 | – | – | 0.59 | 0.23 | – | – | – | 0.40 | 0.49 | 0.58 | 0.21 |
| 3 | – | – | – | 1.06 | – | – | 0.33 | 0.27 | – | – | – | 0.60 |
| 4 | – | – | – | 0.93 | 0.30 | – | 0.58 | 0.50 | – | – | – | 0.66 |
| 5 | – | – | – | 0.88 | 0.21 | – | 0.31 | 0.61 | – | – | – | 0.70 |
| 6 | – | – | – | 0.54 | 0.30 | – | 0.24 | 0.29 | 0.30 | 065 | 0.71 | 0.55 |
| 7 | – | 0.21 | – | 1.17 | 0.33 | – | 0.27 | 0.45 | – | – | – | 0.82 |
| 8 | – | – | 0.33 | 1.31 | – | – | 0.60 | 0.23 | – | – | 0.67 | 0.36 |
| 9 | – | – | – | – | – | – | – | 0.21 | 0.22 | – | 0.33 | 0.46 |
| 10 | – | – | – | 0.38 | – | – | 0.33 | 0.48 | 0.39 | – | 0.39 | 0.41 |
| 11 | – | – | – | 0.34 | 0.29 | – | 0.41 | – | – | – | 0.44 | 0.42 |
| 12 | – | – | – | 1.16 | – | – | 0.45 | 0.46 | – | 0.89 | 0.82 | 0.48 |
| 13 | – | – | – | 0.47 | – | – | – | 0.29 | – | – | – | – |
| 14 | – | 0.33 | – | – | – | – | 0.27 | 0.46 | – | – | 0.28 | 0.35 |
| 15 | – | – | – | 0.28 | – | – | 0.33 | 0.51 | 0.27 | – | – | 0.30 |
| 16 | 0.22 | – | – | 1.28 | 0.40 | – | – | 0.90 | 0.46 | 0.60 | 0.83 | 0.34 |
| 17 | 0.26 | – | – | 0.78 | – | – | 0.30 | 0.48 | 0.47 | – | – | 0.79 |
| 18 | – | – | – | 1.19 | 0.48 | – | 0.48 | – | – | 0.58 | 0.23 | 0.34 |
| 19 | – | 0.27 | – | 0.83 | 0.27 | – | 0.38 | – | 0.33 | 0.73 | 0.85 | 0.53 |
| 20 | – | – | 0.23 | 0.42 | 0.50 | – | 0.77 | 0.43 | – | – | – | 0.48 |
| 21 | – | – | – | 0.46 | – | – | 0.38 | 0.33 | 0.21 | – | 0.49 | 0.58 |
| 22 | – | – | – | 0.31 | – | – | 0.44 | – | – | – | 0.58 | 0.77 |
| 23 | 0.23 | – | – | 1.22 | – | – | 0.45 | 0.60 | – | 0.96 | 0.71 | 0.80 |
| 24 | – | – | – | 0.38 | – | – | 0.24 | 0.23 | – | – | – | 0.31 |
| 25 | 0.22 | – | – | 0.43 | – | – | – | 0.39 | – | – | 0.26 | – |
| 26 | – | – | – | 0.26 | – | – | 0.50 | 0.44 | 0.31 | – | – | 0.36 |
| 27 | – | – | – | 1.38 | 0.38 | 0.23 | – | 0.58 | 0.50 | 0.55 | 0.83 | 0.71 |
| 28 | – | – | – | 0.91 | – | – | 0.48 | 0.56 | 0.46 | – | – | 0.88 |
| 29 | – | – | 0.28 | 1.25 | 0.39 | – | 0.52 | – | – | 0.55 | – | – |
Peptide sequence and similarity level to human proteome are detailed under Table 1. Results are expressed as effective absorbance. The absorbance was measured at wavelengths of 450 and 620 nm. The effective absorbance was calculated subtracting A 620 from A 450. Samples were considered to be negative (-) when effective absorbance was ≤0.20. Data are average values of two independent assays
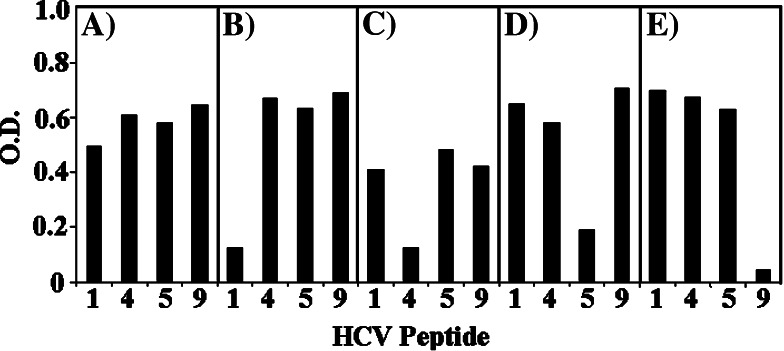
Moreover, positive reactions are observed using sera from healthy subjects and blood donors too. This immunoreactivity has repeatedly been reported in the medical literature and remains unexplained. Among the many possible causes, cross-reactivity between HCV and Influenza A virus [44] as well as the quality of life in volunteer blood donors [42] have been suggested. In this study, the signals produced by peptides 1, 4, 5 and 9 (see Table 4) could be specifically blocked by pAb preabsorption with the relevant peptide (Fig. 1), so indicating specific detection. ELISA data from Table 4 and Fig. 1 are confirmed by immuno-dotblot assays (not shown).
Fig. 1.
Recognition of HCV peptides with low-similarity to the human proteome by pAbs from blood donors. Sera from 10 blood donors were pooled and pAbs were immunoassayed by ELISA (see under “Materials and methods”). HCV peptides were assayed with pAbs a untreated, b pre-treated with peptide n. 1, c pre-treated with peptide n. 4, d pre-treated with peptide n. 5, e pre-treated with peptide n. 9. Peptide numbering is described under Table 1. Results are expressed as optical density (O.D.) and correspond to effective absorbance. The absorbance was measured at wavelengths of 450 and 620 nm. The effective absorbance was calculated subtracting A 620 from A 450. Data are average values of two independent assays
Discussion
The Public Health implications of HCV infection are severe. Chronic hepatitis C, hepatic fibrosis, cirrhosis and hepatocellular carcinoma may sequentially characterize HCV progression [1, 12, 16]. Antiviral treatment of chronic hepatitis C has evolved from interferon monotherapy to a combined regimen (i.e., interferon plus ribavirin) with a concomitant improvement in the clinical management of the viral infection [6, 46]. However, adverse effects remain severe [2, 32] and, most importantly, no therapy has been shown to determine permanent viral eradication [20, 35]. Consequently, great efforts are directed towards therapeutic vaccine approaches by targetting the HCV proteins that play key roles in favoring the continuous presence of the virus in the hepatocytes. One example is the HCV core protein which is characterized by anti-apoptotic activity [19, 26, 41] that can protect the infected cell from immune-mediated destruction. However, the development of an efficient HCV vaccination strategy faces as a main obstacle the high mutational rate of HCV-RNA [39]: the rate of nucleotide variant appearance is high (approximately 10−3 substitutions per site per year), with a large share of HCV sequence variation concentrated within two hypervariable regions, HVR1 and HVR2, located at the N-terminus of the E2 envelope glycoprotein [15]. This explains the many different viral types and subtypes, the resulting heterogeneity of viral antigen proteins and, possibly, the lack of an effective immune response in HCV infection.
As a matter of fact, we still do not understand the molecular mechanisms of antibody-mediated neutralization and its impact for HCV pathogenesis. Indeed, the acute phase of HCV infection is often subclinical, and 70% of infected individuals develop a chronic infection. This suggests that an anti-HCV immune response is weak or absent. In particular, the role of humoral immune response is unclear. Previous experiments showed that serum from a chronically infected patient could neutralize HCV infectivity in a chimpanzee model, so suggesting the existence of specific anti-HCV antibodies [8]. However this report has remained an isolated datum. In general, whereas neutralizing antibodies directed against HCV are present in Igs made from anti-HCV-positive plasma, nonetheless these HCV-specific Igs are largely ineffective in vivo. The mechanism for the poor effectiveness is currently unknown. Of interest is the report of a close association between sequence diversity in the hypervariable region 1 and the appearance of HCV specific antibodies in the sera of subjects with acute infection [18]. This association together with the analysis of the rate of hypervariable region evolution suggest that variation is a function of the immune pressure exerted by the Ab response [43, 45]. Finally, recently it has been proposed that enrichment of neutralization epitope-specific antibodies might permit a better definition of neutralizing antibodies in immune prophylaxis of HCV infection [47].
In this context, the present study offers support to possible anti-HCV immunotherapeutical approaches. Indeed, the similarity profile of HCV proteins to the human proteome may allow the definition of a HCV peptidome platform theoretically able to evoke humoral immune response without concomitant harmful cross-reactions. Moreover, utilizing highly conserved sequences among HCV-specific motifs would offer the potential therapeutic advantage of a potentially broader vaccine effectiveness. In particular, the highly conserved HCV E1315−323HRMAWDMMM sequence interestingly displays an immunodominant role among HCV-infected sera confirming previous reports about its central role in modulating anti-HCV humoral responses [31, 34, 36]. As an additional notation, screening for proteasomal cleavages would provide peptidome sets endowed with the necessary stability so further increasing the potential vaccine efficacy.
References
- 1.Alter HJ. HCV natural history: the retrospective and prospective in perspective. J Hepatol. 2005;43:550–552. doi: 10.1016/j.jhep.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 2.Bagheri H, Fouladi A, Barange K, Lapeyre-Mestre M, Payen JL, Montastruc JL, Vinel JP. Follow-up of adverse drug reactions from peginterferon alfa-2b-ribavirin therapy. Pharmacotherapy. 2004;24:1546–1553. doi: 10.1592/phco.24.16.1546.50947. [DOI] [PubMed] [Google Scholar]
- 3.Bortolotti F, Resti M, Marcellini M, Giacchino R, Verucchi G, Nebbia G, Zancan L, Marazzi MG, Barbera C, Maccabruni A, Zuin G, Maggiore G, Balli F, Vajro P, Lepore L, Molesini M, Guido M, Bartolacci S, Noventa F. Hepatitis C virus (HCV) genotypes in 373 Italian children with HCV infection: changing distribution and correlation with clinical features and outcome. Gut. 2005;54:852–857. doi: 10.1136/gut.2004.053744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burnet FM. The clonal selection theory of acquired immunity. Nashville: Vanderbilt University Press; 1959. [Google Scholar]
- 5.Carlos MP, Yamamura Y, Vu Q, Conzen K, Anderson DE, Torres JV. Humoral immunity to immunodominant epitopes of hepatitis C virus in individuals infected with genotypes 1a or 1b. Clin Immunol. 2004;111:22–27. doi: 10.1016/j.clim.2003.11.012. [DOI] [PubMed] [Google Scholar]
- 6.Causse X, Si Ahmed SN, Gros J, Loustaud-Ratti V, Bacq Y, Abergel A, Silvain C, Veillon P, Labarriere D, Giraudeau B, Fontevraud Group Treatment of chronic hepatitis C in patients unresponsive to interferon. Interest of re-treatment combining interferon induction therapy and ribavirin (a multicenter pilot study) Gastroenterol Clin Biol. 2005;29:117–121. doi: 10.1016/S0399-8320(05)80713-4. [DOI] [PubMed] [Google Scholar]
- 7.Coppola RC, Masia G, Pradat P, Trepo C, Carboni G, Argiolas F, Rizzetto M. Impact of hepatitis C virus infection on healthy subjects on an Italian island. J Viral Hepat. 2000;7:130–137. doi: 10.1046/j.1365-2893.2000.00199.x. [DOI] [PubMed] [Google Scholar]
- 8.Farci P, Alter HJ, Wong DC, Miller RH, Govindarajan S, Engle R, Shapiro M, Purcell RH. Prevention of hepatitis C virus infection in chimpanzees after antibody-mediated in vitro neutralization. Proc Natl Acad Sci USA. 1994;91:7792–7796. doi: 10.1073/pnas.91.16.7792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fenner F, Ada G. Frank MacFarlane Burnet: two personal views. Nat Immunol. 2007;8:111–113. doi: 10.1038/ni0207-111. [DOI] [PubMed] [Google Scholar]
- 10.Flint M, Maidens C, Loomis-Price LD, Shotton C, Dubuisson J, Monk P, Higginbottom A, Levy S, McKeating JA. Characterization of hepatitis C virus E2 glycoprotein interaction with a putative cellular receptor, CD81. J Virol. 1999;73:6235–6244. doi: 10.1128/jvi.73.8.6235-6244.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Franchini M, Rossetti G, Tagliaferri A, Capra F, de Maria E, Pattacini C, Lippi G, Lo Cascio G, de Gironcoli M, Gandini G. The natural history of chronic hepatitis C in a cohort of HIV-negative Italian patients with hereditary bleeding disorders. Blood. 2001;98:1836–1841. doi: 10.1182/blood.V98.6.1836. [DOI] [PubMed] [Google Scholar]
- 12.Fung SK, Lok AS. Update on viral hepatitis in 2004. Curr Opin Gastroenterol. 2005;21:300–307. doi: 10.1097/01.mog.0000158109.13722.36. [DOI] [PubMed] [Google Scholar]
- 13.Gautam AM, Pearson CI, Smilek DE, Steinman L, McDevitt HO. A polyalanine peptide with only five native myelin basic protein residues induces autoimmune encephalomyelitis. J Exp Med. 1992;176:605–609. doi: 10.1084/jem.176.2.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hemmer B, Kondo T, Gran B, Pinilla C, Cortese I, Pascal J, Tzou A, McFarland HF, Houghten R, Martin R. Minimal peptide length requirements for CD4(+) T cell clones-implications for molecular mimicry and T cell survival. Int Immunol. 2000;12:375–383. doi: 10.1093/intimm/12.3.375. [DOI] [PubMed] [Google Scholar]
- 15.Hijikata M, Kato N, Ootsuyama Y, Nakagawa M, Ohkoshi S, Shimotohno K. Hypervariable regions in the putative glycoprotein of hepatitis C virus. Biochem Biophys Res Commun. 1991;175:220–228. doi: 10.1016/S0006-291X(05)81223-9. [DOI] [PubMed] [Google Scholar]
- 16.Idilman R, De Maria N, Colantoni A, Van Thiel DH. Pathogenesis of hepatitis B and C-induced hepatocellular carcinoma. J Viral Hepat. 1998;5:285–299. doi: 10.1046/j.1365-2893.1998.00116.x. [DOI] [PubMed] [Google Scholar]
- 17.Jerne NK. The somatic generation of immune recognition. 1971. Eur J Immunol. 2004;34:1234–1242. doi: 10.1002/eji.200425132. [DOI] [PubMed] [Google Scholar]
- 18.Kato N, Ootsuyama Y, Sekiya H, Ohkoshi S, Nakazawa T, Hijikata M, Shimotohno K. Genetic drift in hypervariable region 1 of the viral genome in persistent hepatitis C virus infection. J Virol. 1994;68:4776–4784. doi: 10.1128/jvi.68.8.4776-4784.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kato N, Yoshida H, Ono-Nita S, Kato J, Goto T, Otsuka M, Lan K, Matsushima K, Shiratori Y, Omata M. Activation of intracellular signaling by hepatitis B and C viruses: C-viral core is the most potent signal inducer. Hepatology. 2000;32:405–412. doi: 10.1053/jhep.2000.9198. [DOI] [PubMed] [Google Scholar]
- 20.Kelleher TB, Afdhal N. Maintenance therapy for chronic hepatitis C. Curr Gastroenterol Rep. 2005;7:50–53. doi: 10.1007/s11894-005-0066-1. [DOI] [PubMed] [Google Scholar]
- 21.Kimura Y, Gushima T, Rawale S, Kaumaya P, Walker CM. Escape mutations alter proteasome processing of major histocompatibility complex class I-restricted epitopes in persistent hepatitis C virus infection. J Virol. 2005;79:4870–4876. doi: 10.1128/JVI.79.8.4870-4876.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kusalik A, Bickis M, Lewis C, Li Y, Lucchese G, Marincola FM, Kanduc D. Widespread and ample peptide overlapping between HCV and Homo sapiens proteomes. Peptides. 2007;28:1260–1267. doi: 10.1016/j.peptides.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 23.Kuttler C, Nussbaum AK, Dick TP, Rammensee HG, Schild H, Hadeler KP. An algorithm for the prediction of proteasomal cleavages. J Mol Biol. 2000;298:417–429. doi: 10.1006/jmbi.2000.3683. [DOI] [PubMed] [Google Scholar]
- 24.Lemberg MK, Bland FA, Weihofen A, Braud VM, Martoglio B. Intramembrane proteolysis of signal peptides: an essential step in the generation of HLAE epitopes. J Immunol. 2001;167:6441–6446. doi: 10.4049/jimmunol.167.11.6441. [DOI] [PubMed] [Google Scholar]
- 25.Lucchese G, Stufano A, Trost B, Kusalik A, Kanduc D. Peptidology: short amino acid modules in cell biology and immunology. Amino Acids. 2007;33:703–707. doi: 10.1007/s00726-006-0458-z. [DOI] [PubMed] [Google Scholar]
- 26.Marusawa H, Hijikata M, Chiba T, Shimotohno K. Hepatitis C virus core protein inhibits Fas- and tumor necrosis factor alpha-mediated apoptosis via NF-kappaB activation. J Virol. 1999;73:4713–4720. doi: 10.1128/jvi.73.6.4713-4720.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meyer zum Buschenfelde KH, Lohse AW, Gerken G, Treichel U, Lohr HF, Mohr H, Grosse A, Dienes HP. The role of autoimmunity in hepatitis C infection. J Hepatol. 1995;22:S93–S96. doi: 10.1016/0270-9139(95)94144-4. [DOI] [PubMed] [Google Scholar]
- 28.Niman HL, Houghten RA, Walker LE, Reisfeld RA, Wilson IA, Hogle JM, Lerner RA. Generation of protein-reactive antibodies by short peptides is an event of high frequency: implications for the structural basis of immune recognition. Proc Natl Acad Sci USA. 1983;80:4949–4953. doi: 10.1073/pnas.80.16.4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Notredame C, Higgins DG, Heringa J. T-Coffee: a novel method for multiple sequence alignments. J Mol Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 30.Oldstone MAB. Molecular mimicry and autoimmune disease. Cell. 1987;50:819. doi: 10.1016/0092-8674(87)90507-1. [DOI] [PubMed] [Google Scholar]
- 31.Olenina LV, Nikolaeva LI, Sobolev BN, Blokhina NP, Archakov AI, Kolesanova EF. Mapping and characterization of B cell linear epitopes in the conservative regions of hepatitis C virus envelope glycoproteins. J Viral Hepat. 2002;9:174–182. doi: 10.1046/j.1365-2893.2002.00358.x. [DOI] [PubMed] [Google Scholar]
- 32.Pawlotsky JM. Current and future concepts in hepatitis C therapy. Semin Liver Dis. 2005;25:72–83. doi: 10.1055/s-2005-864783. [DOI] [PubMed] [Google Scholar]
- 33.Pol S, Thiers V, Nousbaum JB, Legendre C, Berthelot P, Kreis H, Brechot C. The changing relative prevalence of hepatitis C virus genotypes: evidence in emodialyzed patients and kidney recipients. Gastroenterology. 1995;108:581–583. doi: 10.1016/0016-5085(95)90088-8. [DOI] [PubMed] [Google Scholar]
- 34.Polimeno L, Mittelman A, Gennero L, Ponzetto A, Lucchese G, Stufano A, Kusalik A, Kanduc D (2007) Sub-epitopic dissection of HCV E1 (315–328) HRMAWDMMMNWSPT sequence by similarity analysis. Amino Acids. doi:10.1007/s00726-007-0539-7 [DOI] [PubMed]
- 35.Radkowski M, Laskus T. Persistence of hepatitis C virus after successful treatment of chronic hepatitis C: is hepatitis C infection for life? Liver Transpl. 2005;11:114–116. doi: 10.1002/lt.20335. [DOI] [Google Scholar]
- 36.Ray R, Khanna A, Lagging LM, Meyer K, Choo QL, Ralston R, Houghton M, Becherer PR. Peptide immunogen mimicry of putative E1 glycoprotein-specific epitopes in hepatitis C virus. J Virol. 1994;68:4420–4426. doi: 10.1128/jvi.68.7.4420-4426.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reddehase MJ, Rothbard JB, Koszinowski UH. A pentapeptide as minimal antigenic determinant for MHC class I-restricted T lymphocytes. Nature. 1989;337:651–653. doi: 10.1038/337651a0. [DOI] [PubMed] [Google Scholar]
- 38.Sercarz EE, Lehmann PV, Ametani A, Benichou G, Miller A, Moudgil K. Dominance and crypticity of T cell antigenic determinants. Annu Rev Immunol. 1993;11:729–766. doi: 10.1146/annurev.iy.11.040193.003501. [DOI] [PubMed] [Google Scholar]
- 39.Simmonds P. Variability of hepatitis C virus. Hepatology. 1995;21:570–583. doi: 10.1002/hep.1840210243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Simmonds P, Bukh J, Combet C, Deleage Enomoto Feinstone Halfon Inchauspe G N S P, Kuiken C, Maertens G, Mizokami M, Murphy DG, Okamoto H, Pawlotsky JM, Penin F, Sablon E, Shin-I T, Stuyver LJ, Thiel HJ, Viazov S, Weiner AJ, Widell A. Consensus proposals for a unified system of nomenclature of hepatitis C virus genotypes. Hepatology. 2005;42:962–973. doi: 10.1002/hep.20819. [DOI] [PubMed] [Google Scholar]
- 41.Tai D, Tsai S, Chen Y, Chuang Y, Peng C, Sheen I, Yeh C, Chang K, Huang S, Kuo G, Liaw Y. Activation of nuclear factor kappaB in hepatitis C virus infection: implications for pathogenesis and hepatocarcinogenesis. Hepatology. 2000;31:656–664. doi: 10.1002/hep.510310316. [DOI] [PubMed] [Google Scholar]
- 42.Teixeira MC, Ribeiro Mde F, Gayotto LC, Chamone Dde A, Strauss E. Worse quality of life in volunteer blood donors with hepatitis C. Transfusion. 2006;46:278–283. doi: 10.1111/j.1537-2995.2006.00712.x. [DOI] [PubMed] [Google Scholar]
- 43.Timm J, Roggendorf M. Sequence diversity of hepatitis C virus: Implications for immune control and therapy. World J Gastroenterol. 2007;13:4808–4817. doi: 10.3748/wjg.v13.i36.4808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wedemeyer H, Mizukoshi E, Davis AR, Bennink JR, Rehermann B. Cross-reactivity between hepatitis C virus and Influenza A virus determinant-specific cytotoxic T cells. J Virol. 2001;75:11392–11400. doi: 10.1128/JVI.75.23.11392-11400.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weiner AJ, Geysen HM, Christopherson C, Hall JE, Mason TJ, Saracco G, Bonino F, Crawford K, Marion CD, Crawford KA. Evidence for immune selection of hepatitis C virus (HCV) putative envelope glycoprotein variants: potential role in chronic HCV infections. Proc Natl Acad Sci USA. 1992;89:3468–3472. doi: 10.1073/pnas.89.8.3468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zeuzem S, Hultcrantz R, Bourliere M, Goeser T, Marcellin P, Sanchez-Tapias J, Sarrazin C, Harvey J, Brass C, Albrecht J. Peginterferon alfa-2b plus ribavirin for treatment of chronic hepatitis C in previously untreated patients infected with HCV genotypes 2 or 3. J Hepatol. 2004;40:993–999. doi: 10.1016/S0168-8278(04)00060-1. [DOI] [PubMed] [Google Scholar]
- 47.Zhang P, Wu CG, Mihalik K, Virata-Theimer ML, Yu MY, Alter HJ, Feinstone SM. Hepatitis C virus epitope-specific neutralizing antibodies in Igs prepared from human plasma. Proc Natl Acad Sci USA. 2007;104:8449–8454. doi: 10.1073/pnas.0703039104. [DOI] [PMC free article] [PubMed] [Google Scholar]