Abstract
We monitored the primary humoral response to human immunodeficiency virus type 1 infection and showed that, in addition to antibodies to p24 and gp41, antigens which form the basis of most diagnostic assays, the response included a significant antibody response directed to the gp120 region of the infecting viral quasispecies. When tested in a recombinant virus neutralization assay, these antibodies were capable of inhibiting viral growth. We found the primary viral quasispecies to solely utilize the CCR-5 chemokine receptor; however, recombinant viruses differed in their cytopathology and in their sensitivity to β-chemokine inhibition of viral growth. Sequence analysis of the gp120 open reading frames showed that amino acid changes in the C1 (D→G at position 62) and C4 (V→A at position 430) regions accounted for the phenotypic differences. These data demonstrate that early in infection, polymorphism exists in envelope glycoprotein coreceptor interactions and imply that therapeutic strategies targeted at this step in the viral life cycle may lead to rapid resistance.
Primary infection with human immunodeficiency virus (HIV) type 1 (HIV-1) is associated with a seroconversion illness characterized by high plasma viral loads and a series of influenza-like symptoms which can vary in severity. In this early phase of infection, in the absence of a detectable immune response, the virus replicates to a high titer, with plasma viral loads in excess of 105 viral RNA (vRNA) copies per ml (15, 17). The severity of the primary infection and its subsequent resolution are prognostic indicators of subsequent disease course (24, 33). This primary viremia is thought to be restricted by the host immune response, in that plasma vRNA levels decrease simultaneously with the first detection of virus-specific antibodies and cytotoxic T cells (CTL) (6, 10, 24, 34). The rate of plasma viral clearance differs between infected individuals; the steady-state or “set-point” vRNA load eventually reached has been reported to be a prognostic marker for subsequent disease progression (24, 27, 77). These observations imply that host factors controlling the early clearance of viremia and the vRNA load at which the set point is established define the subsequent course of disease.
HIV-1 infects CD4+ lymphocytes, monocytes, and dendritic cells in the peripheral blood and lymphoid organs. However, several authors have suggested that during sexual transmission, the primary cell types targeted are Langerhans cells present within the mucosae (20, 54, 58, 70). HIV entry into these cell types is principally defined by the expression of CD4 and chemokine receptors at the cell surface (3, 13, 19, 21–23). Historically, HIV isolates have been classified according to their ability to induce cytopathic effects and have been designated syncytium inducing (SI) or non-syncytium inducing (NSI) (66). SI viruses are generally able to utilize the α-chemokine receptor CXCR-4, which is expressed on naive T cells and the majority of immortalized cell lines, whereas NSI viruses can utilize only members of the β-chemokine receptor family, principally CCR-5 expressed on effector or memory T cells (3, 8, 13, 19, 21–23, 38, 75). However, such NSI viruses have been reported to induce syncytia in cell lines expressing both CD4 and CCR-5 (57, 64); hence, these terms are no longer appropriate, and viruses should be classified according to the coreceptor used. Paxton and colleagues reported that lymphocytes from individuals homozygous for a defective CCR-5 allele (CCR-5 Δ32) were resistant to infection with viruses utilizing CCR-5 but sensitive to infection with viruses utilizing CXCR-4 (37, 55). The relative resistance of individuals homozygous for the CCR-5 Δ32 allele suggests that this receptor is of critical importance for transmission (7, 18, 62). The viral phenotype, defined in terms of chemokine receptor dependency, may help identify the cell types capable of supporting viral replication and hence the tissue distribution of HIV during the primary infection (52). The majority of individuals studied to date harbor viruses of the NSI CCR-5-utilizing phenotype at seroconversion (16, 31, 60). However, the transmission of SI CXCR-4-utilizing viruses has been reported (59, 67, 72); some of these have been associated with a more rapid progression to disease (25, 66).
Several authors have demonstrated that CTL responses are associated with the resolution of the primary viremia (10, 34, 53). However, in one case viruses were shown to “escape” from an early CTL response which was predominantly targeted to single epitopes (11). The data presented here demonstrate that neutralizing antibodies may also be present at seroconversion and may contribute to the early clearance of viremia (36). Antigenic polymorphism was observed between gp120 proteins cloned from a single time point; furthermore, this variation resulted in phenotypic changes with respect to virus-induced cytopathology and sensitivity to neutralization by β-chemokines.
MATERIALS AND METHODS
Patient samples.
A patient (HL60) presenting with influenza-like symptoms was admitted to University College London Medical School and screened for HIV infection by use of four commercially available tests for the detection of HIV-specific antibodies (Wellcozyme Immunometric 1 + 2 enzyme immunoassay [EIA], Wellcozyme competitive EIA, Fujirebio particle agglutination assay, and Launch Diagnostics Anti 1 + 2 EIA). Initial screening was negative; however, 3 days after an admission, EDTA blood sample obtained for genetic screening with the Roche Amplicor PCR test proved positive. The patient had several potentially high-risk HIV exposures 2 to 3 weeks prior to admission. Subsequent quantitative viral load determinations were performed on plasma samples obtained 7, 9, 10, 11, 12, and 13 days after admission (Fig. 1). A very low positive signal was obtained from the Wellcozyme Immunometric test at 9 days postadmission; subsequent testing of later samples demonstrated an increased signal. Results of all other tests performed on later samples were positive.
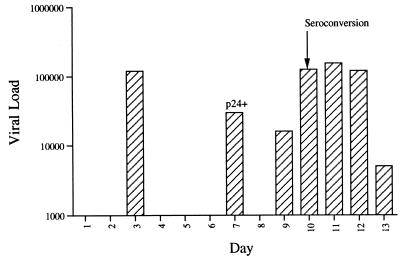
FIG. 1.
vRNA levels in plasma during acute seroconversion. The plasma vRNA copy number was determined (vRNA copies per milliliter) at seven times during the hospital stay by use of the Roche Amplicor assay. In addition, the same samples were quantified for both p24 antigen and the presence of antibodies specific for HIV antigens. Antigenemia was detectable only on day 7. HIV-specific antibodies were detected from day 10 on. The day of seroconversion is noted.
Cloning and expression of gp120 regions from the seroconversion samples.
Only serum and plasma samples were available from the patient. Total RNA was therefore extracted from all available plasma samples (100 μl of plasma), and the gp120 open reading frames (ORFs) were amplified by a nested reverse transcription-PCR (RT-PCR) as described previously (26, 40, 51). PCR products were not obtained from samples in which the vRNA load was below 30,000 copies/ml; however, products were obtained from samples collected 7, 10, 11, and 12 days postadmission. These PCR products were shown to be derived from at least 100 cDNA copies of the gp120 template by limiting-dilution PCR (data not shown). The insensitivity of the gp120 RT-PCR (1.5 kb) compared to the Roche Amplicor PCR (0.4 kb) may be ascribed to the relatively longer cDNA synthesis. PCR products from the day-7 and day-11 samples were cloned into expression vector pcDNA3 and tested for their ability to express gp120 as described previously (51). Fourteen plasmids from each time point were transfected into VTF7.3-infected 293 cells by use of calcium phosphate. Cell supernatants were collected after 72 h, clarified by centrifugation at 1,500 × g, and assayed for gp120 levels in a quantitative gp120 capture enzyme-linked immunosorbent assay (43). Expression-competent clones were identified, and the gp120 ORFs were sequenced with a series of primers by use of a cycle sequencing protocol and an Applied Biosystems 373A automated sequencer (5). Amino acid numbering was based on the HXB2 gp120 sequence, with the first amino acid of the mature protein being designated residue 31.
Antigenic characterization of gp120 proteins.
Saturating concentrations (500 ng/ml) of soluble gp120 were tested for their ability to bind a pool of HIV-positive human sera (QC256); patient HL60 sera; soluble CD4-immunoglobulin (sCD4-Ig) (10 μg/ml) (Genentech, San Francisco, Calif.); and monoclonal antibodies (MAbs) 38.1a, 10.46c, and 39.13g specific for residues overlapping the CD4 binding site in a capture EIA as previously described (39, 43, 68). The strain MN V3 peptide was obtained from the MRC ARP Repository (EVA7019; CTRPNYNKRKRIHIGPGRAFYTTKNIIGTIRQAHC) and bound directly to Immulon II EIA plates at 5 μg/ml in phosphate-buffered saline. Bound ligands were visualized with either anti-human Ig–horseradish peroxidase or anti-rat Ig–horseradish peroxidase (SeraLabs, Crawley, United Kingdom).
Generation and recovery of chimeric HXB2.gp120 viruses.
A number of pCDNA3 clones expressing gp120 capable of binding sCD4-Ig were chosen for transfer into vector pHXB2-MCSΔenv. Both pHXB2-MCSΔenv and pCDNA3gp120 plasmids were digested with BstEII/MluI, gel purified by the Geneclean procedure, and ligated. Transformants were screened for an insert by PCR, and plasmids were sequenced to verify the presence of the correct insert. All chimeric HXB2.gp120 (HXB2 chimera expressing gp120 ORFs) plasmids (5 μg) were transfected into HEK cells with Lipofectamine (Gibco BRL). At 48 h posttransfection, the HEK cells were cocultured with 107 phytohemagglutinin (PHA)-stimulated peripheral blood lymphocytes (PBL) for 24 h. The PBL were recovered, washed, and cultured in interleukin 2 (IL-2)-containing RPMI medium for 21 days, with the addition of fresh uninfected PBL after 7 and 14 days. The extracellular fluid was tested for the presence of soluble p24 antigen as described previously (39). In addition, the extracellular supernatant from the transfected HEK cells was used to infect U87.CD4 and Hos.CD4 cells expressing a range of chemokine receptors as reported previously (64). Infection was monitored by the production of both soluble and intracellular p24 antigens. Cells were fixed with methanol-acetone (1:1 ratio; stored at −20°C) and incubated with MAbs specific for p24 antigen (38.96K and EF7.1; MRC ARP Repository); bound antibodies were detected with β-galactosidase-conjugated anti-mouse Ig and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) as described previously (14). Cells expressing p24 antigen gave rise to a colored focus; viruses capable of inducing cytopathic effects resulted in multinucleated foci which can be quantified. In addition, virus-infected U87 cells were monitored for gp120-gp41 cell surface expression 48 h postinfection by monitoring the binding of MAb 39.13g by fluorescence-activated cell sorting as described previously (68).
Virus neutralization assays.
Neutralization was assessed by two methods—first by use of PHA-stimulated PBL as the target cells, with the determination of soluble p24 antigen production as the end point (2, 63), and second by infection of U87.CD4.CCR-5 cells, with quantification of intracellular p24 antigen as an indicator of infection (40, 64). Primary viruses W6BC and SF162 (obtained from the MRC ARP Repository) and an HXB2 chimera expressing the SF162 gp120 ORFs were used as positive controls in these assays. Sequential serum samples from patient HL60 were incubated at 37°C for 1 h with a known infectious titer of virus, 100 50% tissue culture infective doses for PBL infection or 100 focus-forming units (FFU) for U87. CD4 cell infection. sCD4 and HIV-positive sera (QC256 and 1785) were used as internal controls in all assays. Following the virus-ligand incubation, the mixture was added to either 100,000 peripheral blood mononuclear cells (mixed from two blood donors) in a final volume of 75 μl of RPMI medium–10% fetal calf serum–IL-2 (5 U/ml) or a 60% confluent monolayer of U87.CD4.CCR-5 cells in a 48-well plate (Nunc). Cells were washed on the following day and incubated at 37°C for 5 or 3 days, respectively. Neutralization was defined as the ability of serum to inhibit virus replication, as assessed by p24 antigen detection, where the sensitivity of the p24 antigen EIA is 10 pg/50 μl. Neutralization therefore was classified as complete inhibition of infection.
RANTES was a gift from L. Czaplewski and was tested for its ability to inhibit HIV infection in PBL assays essentially as described above. With the exception that the chemokine was incubated with the cells for 1 h before viral infection and after washing, the chemokine was added back to the cells for the incubation period of the assay (5 days).
Nucleotide sequence accession numbers.
The nucleotide sequences for the six gp120 clones sequenced here have been deposited with GenBank under accession no. AJ007943 to AJ007948.
RESULTS
Monitoring of the seroconversion process.
Five different serological tests for the presence of both HIV antigen (p24) and HIV-specific antibodies were performed on five separate occasions. All four antibody tests revealed a positive profile at the last two sampling times, with all assays showing an increase in reactivity between days 10 and 13. At the end of the hospital stay, all of the antibody test values were inside the normal range seen for these assays. Viral p24 antigen was detectable only on day 7 of testing. Sequential viral RNA loads were obtained with the Roche Amplicor PCR test and showed peak viremia of 155,000 RNA copies/ml 11 days after admission, declining to 5,000 copies by day 13 (Fig. 1). A significant reduction in loads (>1 log unit) was seen between days 7 and 9; however, no clinical correlate was observed. It is interesting to note that the day on which p24 antigen was detected in plasma did not correlate with the time of peak viremia, supporting the observation that p24 antigenemia may not reflect the presence of virion-associated antigen. Point mutation assays of the RT gene sequence present in plasma RNA showed the virus to be homogeneous for mutations at positions 41 (L) and 215 (Y) of the pol gene, consistent with the presence of an zidovudine-resistant virus (data not shown) (32).
Characterization of plasma-derived gp120 envelope glycoproteins.
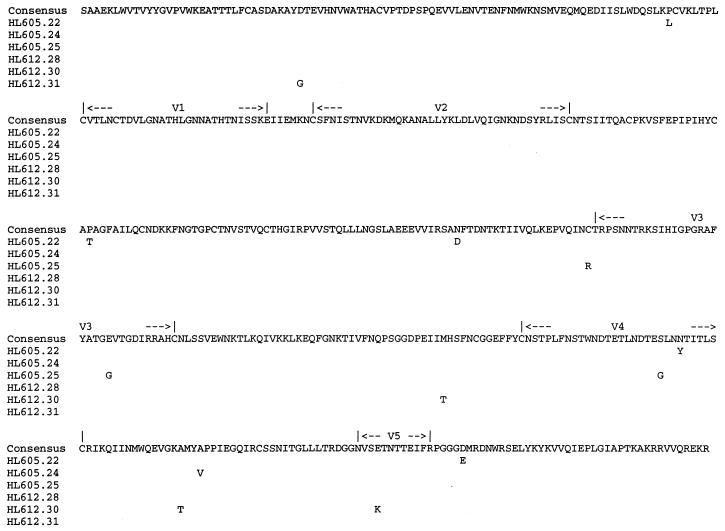
gp120 ORFs derived on days 7 and 11, the time points as close as possible to the date of seroconversion, were chosen for further study and cloned into pCDNA3 as previously described (26, 40, 51). Fourteen plasmids from each time point were chosen for transfection into HEK cells to generate recombinant glycoproteins. In the earlier sample (HL605), 11 of the 14 clones expressed gp120, whereas in the later sample (HL612), 9 of the 14 clones were expression competent (Table 1). Of the 11 glycoproteins expressed from the first sample set, 3, HL605.25, HL605.55, and HL605.58, were unreactive with both sCD4 and 39.13g, suggesting that they may be misfolded. Similarly, of the nine glycoproteins generated from the second sample set, two, HL612.77 and HL612.82, showed minimal recognition of both sCD4 and 39.13g (Table 1). Six gp120 clones were sequenced, three from the first time point (clones 22, 24, and 25) and three from the second (clones 28, 30, and 31), including one of the clones (25) demonstrating poor reactivity with sCD4. The inferred amino acid sequences are shown in Fig. 2. Inspection of the sequences showed a low degree of polymorphism, with no evidence for any systematic change in sequence between the first and second samplings. However, clone 25 was found to lack the N-terminal cysteine residue of the V3 region (C→R), a finding which may result in protein misfolding and altered antigenicity. Sequencing of the other clones (55, 58, 74, 77, and 82) which failed to bind sCD4 showed that they all contained the C→R substitution at residue 305 (data not shown).
TABLE 1.
Antigenic characterization of glycoproteins
| Antigena | Recognition of gp120 by the following ligand (OD)b:
|
|||
|---|---|---|---|---|
| QC256 | sCD4 | 39.13g | 38.1a | |
| Mock | 0.05 | ND | ND | 0.04 |
| LAI | 1.45 | ND | ND | 1.43 |
| 605.22 | 1.57 | 0.74 | 0.90 | 1.45 |
| 605.24 | 1.50 | 1.34 | 1.41 | 1.47 |
| 605.25 | 1.47 | 0.07 | 0.09 | 1.41 |
| 605.49 | 1.57 | 1.36 | 1.51 | 1.40 |
| 605.51 | 1.65 | 1.14 | 1.32 | 1.60 |
| 605.54 | 1.56 | 1.35 | 1.42 | 1.52 |
| 605.55 | 1.61 | 0.07 | 0.10 | 1.56 |
| 605.56 | 1.61 | 1.31 | 1.50 | 1.62 |
| 605.57 | 1.51 | 0.93 | 1.37 | 1.40 |
| 605.58 | 1.61 | 0.07 | 0.07 | 1.54 |
| 605.59 | 1.66 | 1.35 | 1.45 | 1.58 |
| 612.30 | 1.52 | 1.05 | 1.42 | 1.46 |
| 612.28 | 1.62 | 1.30 | 1.35 | 1.47 |
| 612.31 | 1.55 | 1.24 | 1.39 | 1.55 |
| 612.74 | 1.53 | 0.25 | 1.04 | 1.43 |
| 612.75 | 1.56 | 1.35 | 1.52 | 1.52 |
| 612.77 | 1.40 | 0.08 | 0.14 | 1.37 |
| 612.78 | 1.61 | 1.27 | 1.40 | 1.50 |
| 612.79 | 1.51 | 1.30 | 1.43 | 1.56 |
| 612.82 | 1.36 | 0.07 | 0.09 | 1.33 |
Mock indicates cell-free supernatant from transfection of a pCDNA3 clone with no gp120 insert. LAI indicates cell-free supernatant from transfection of a pCDNA3 clone carrying the HIV strain LAI BH10 sequence. Clone designations are derived from the clinic number. Clones with the 605 prefix were derived from plasma at day 7, and clones with the 612 prefix were derived from day-11 plasma.
Results are shown as the mean optical density (OD) at 450 nm for three replicate samples. ND, not determined. QC256 was tested at a final serum dilution of 1/1,000, and sCD4 and MAbs 39.13g and 38.1a were used at 5 μg/ml.
FIG. 2.
Predicted amino acid alignment of gp120 sequences. Peptide translation of gp120 sequences from clones obtained at days 7 (HL605.22, HL605.24, and HL605.25) and 11 (HL612.28, HL612.30, and HL612.31) is shown. The consensus is shown above the alignment; departures are indicated by the single-letter amino acid code.
Human serum antibody recognition of autologous gp120 envelope glycoproteins.
Serum antibodies from patient HL60 were tested for their recognition of strain LAI and MN gp120, autologous glycoproteins derived from pCDNA3 clones 22, 24, 25, 28, 30, and 31, and the strain MN V3 peptide. In addition, the glycoproteins were tested for their ability to be recognized by an HIV-positive serum pool (QC256) and by two conformation-dependent MAbs, 39.13g and 10.46c, specific for epitopes overlapping the CD4 binding site (Table 2). HL60 serum samples obtained at 3, 9, and 11 days postadmission showed no reactivity with any of the recombinant antigens tested. However, the serum sample obtained from the last time point (day 13) bound to all of the autologous proteins, with the exception of HL605.25 which, as noted above, showed evidence of an altered conformation. It is worth noting that this last serum sample also failed to bind to the MN V3 peptide. In contrast, the HIV-positive serum pool (QC256) bound to all of the proteins equivalently (Table 2 and data not shown).
TABLE 2.
Autologous serum recognition of gp120a
| Antigen | Recognition (OD) by:
|
Serum antigen recognition (OD) on day:
|
|||||
|---|---|---|---|---|---|---|---|
| QC256 | 39.13g | 10.46c | 3 | 7 | 11 | 13 | |
| Mock | 0.04 | 0.05 | 0.06 | 0.12 | 0.10 | 0.08 | 0.02 |
| LAI | 1.56 | 1.48 | 1.38 | 0.11 | 0.10 | 0.11 | 0.01 |
| MN | 1.72 | 1.62 | 1.82 | 0.09 | 0.11 | 0.08 | 0.09 |
| MN V3 | 1.60 | 0.04 | 0.05 | 0.09 | 0.08 | 0.06 | 0.07 |
| 605.22 | 1.64 | 0.85 | 0.76 | 0.10 | 0.09 | 0.08 | 0.48 |
| 605.24 | 1.75 | 1.57 | 1.28 | 0.13 | 0.05 | 0.02 | 0.41 |
| 605.25 | 1.64 | 0.04 | 0.05 | 0.13 | 0.01 | 0.05 | 0.07 |
| 612.28 | 1.71 | 1.48 | 1.31 | 0.02 | 0.08 | 0.05 | 0.29 |
| 612.30 | 1.65 | 1.48 | 1.29 | 0.16 | 0.08 | 0.06 | 0.33 |
| 612.31 | 1.82 | 1.51 | 1.34 | 0.16 | 0.10 | 0.09 | 0.36 |
All gp120 antigens were tested at a concentration of 500 ng/ml, as determined by a quantitative capture EIA (43). Results are shown as the mean optical density (OD) at 450 nm for three replicate samples. The MN V3 peptide was tested at a concentration of 5 μg/ml. All serum samples were tested at a final serum dilution of 1/500.
Phenotypic characterization of chimeric HXB2.gp120 viruses.
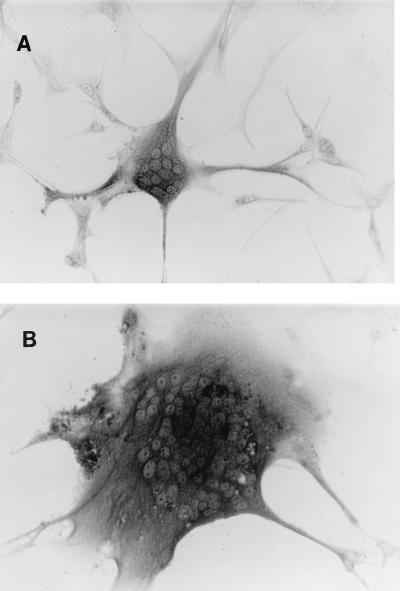
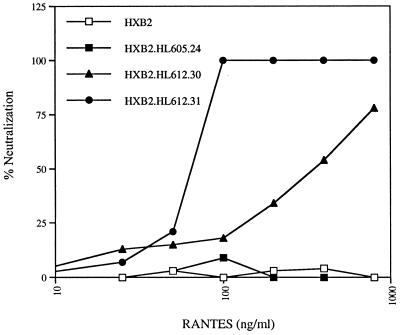
Recombinant viruses were prepared from three of the sequenced clones by transfer of the gp120 sequence into the viable molecular clone pHXB2-MCS as described previously (26, 39, 40). SF162 virus and an HXB2 chimera expressing the SF162 gp120 ORF (derived from a molecular clone kindly provided by C. Cheng-Mayer) were used as additional controls to demonstrate that the transfer of a primary gp120 ORF to HXB2 confers coreceptor usage and neutralization sensitivity of the parental primary virus. Chimeric viruses expressing the gp120 sequences of HL605.24, HL612.30, and HL612.31 were generated and designated HXB2.605.24, HXB2.612.30, and HXB2.612.31, respectively. Virus recovered after transfection of HEK cells was tested for its ability to replicate in PBL and to infect U87.CD4 and Hos.CD4 cells and a variety of chemokine receptors (Table 3). All three viruses (HXB2.605.24, HXB2.612.30, and HXB2.612.31) replicated in PBL with reduced kinetics compared to both the parental HXB2 and chimeric HXB2.SF162 viruses (Table 3). All three viruses were able to replicate only in U87.CD4.CCR-5 cells (Table 4); however, differences in cytopathology between clones was observed (Fig. 3). Virus HXB2.605.24 was less able to induce cell fusion with neighboring cells and to induce multinucleated giant cells than viruses HXB2.612.30 and HXB2.612.31 (Fig. 3). This observation was observed with both HeLa and Hos cells at various multiplicities of infection and hence was not dose dependent (data not shown). To determine if the differences in cytopathology were due to variations in envelope glycoprotein processing and/or expression, U87.CD4.CCR-5 cells were monitored 48 h postinfection for cell surface gp120 expression by fluorescence-activated cell sorting. Mean fluorescence intensity values of 8.4, 68.4, 72.3, and 65.9 were obtained for uninfected cells and cells infected with HXB2.605.24, HXB2.612.30, and HXB2.612.31, respectively. Given the differences in cytopathology, we were interested to know if HXB2.605.24 interacted with either CD4 or CCR-5 less efficiently than HXB2.612.30 and HXB2.612.31. To test this idea, we compared the sensitivity of the viruses to neutralization by the β-chemokine RANTES and sCD4. All of the viruses were resistant to the highest concentration of sCD4 tested (20 μg/ml) (Table 5). HXB2.605.24 was resistant to neutralization by RANTES, whereas HXB2.612.31 was completely neutralized by RANTES at 100 ng/ml. In contrast, HXB2.612.30 demonstrated an intermediate sensitivity to neutralization by the chemokine (Table 5 and Fig. 4). All of these neutralization assays were performed with PBL target cells; we failed to demonstrate chemokine-mediated neutralization of virus infection of U87.CD4.CCR-5 cells (data not shown).
TABLE 3.
Chimeric virus replication
| Virus | Replication in PHA-stimulated PBL (p24, ng/ml) at day postinfectiona:
|
||
|---|---|---|---|
| 3 | 5 | 8 | |
| SF162 | 3.4 | 12.3 | 18.5 |
| HXB2 | 0.4 | 4.6 | 8.9 |
| HXB2.SF162 | 1.2 | 9.3 | 12.3 |
| HXB2.605.24 | 0.3 | 1.2 | 3.0 |
| HXB2.612.30 | 0.4 | 2.0 | 5.2 |
| HXB2.612.31 | 0.4 | 2.8 | 6.1 |
Transfected HEK cell supernatants were normalized for p24 antigen levels (1.5 ng) and used to infect 3-day-old PHA- and IL-2-activated PBL. Extracellular fluid was harvested at 3, 5, and 8 days postinfection and assayed for p24 antigen.
TABLE 4.
Chemokine receptor utilization
| Virusa | Infection of the following CD4+ cells expressing the indicated receptorb:
|
||||||
|---|---|---|---|---|---|---|---|
| U87-CD4
|
Hos-CD4
|
||||||
| CCR-1 | CCR-2b | CCR-3 | CCR-5 | CXCR-4 | Bonzo | BOB | |
| W6BC | − | + | ++ | ++++ | ++++ | − | − |
| SF162 | − | − | − | +++ | − | − | + |
| HXB2 | − | − | − | − | ++++ | − | − |
| HXB2.SF162 | − | − | − | +++ | − | − | + |
| HXB2.605.24 | − | − | − | + | − | − | − |
| HXB2.612.30 | − | − | − | +++ | − | − | − |
| HXB2.612.31 | − | − | − | +++ | − | − | − |
W6BC was used as a control for a virus that utilizes multiple receptors. In contrast, SF162 and the HXB2 chimera expressing the SF162 gp120 ORFs were able to utilize only CCR-5 and BOB.
Infected cells were stained for intracellular p24 antigen. −, no detectable infection of cells; +, >10 to <50 FFU/ml; ++, >50 to <200 FFU/ml; +++, >200 to <1,000 FFU/ml; ++++, >1,000 FFU/ml.
FIG. 3.
Chimeric virus-induced cytopathology. U87.CD4.CCR-5 cells were infected with 100 FFU of HXB2.605.24 (A) or HXB2.612.31 (B) per ml and fixed with methanol-acetone at 72 h postinfection. Infected cells were visualized by the detection of intracellular p24 antigen. Multinucleated foci are apparent in both fields of view and are representative of the cytopathology observed throughout the infected-cell population.
TABLE 5.
Neutralization sensitivity of chimeric virusesa
| Virus | Neutralization titer with:
|
ID50 (ng/ml) of RANTESc | ||||||
|---|---|---|---|---|---|---|---|---|
| Patient serum samples on day postadmission:
|
Control ligands
|
|||||||
| 3 | 9 | 11 | 13 | QC256 | 1785 | sCD4b | ||
| W6BC | <8 | <8 | <8 | <8 | 40 | 160 | 10.0 | |
| SF162 | <8 | <8 | <8 | <8 | 80 | 320 | >20.0 | 65 |
| HXB2 | <8 | <8 | <8 | <8 | 320 | 640 | 0.5 | |
| HXB2.SF162 | <8 | <8 | <8 | <8 | 80 | 160 | >20.0 | 50 |
| HXB2.HL605.24 | <8 | <8 | <8 | 160 | 40 | 320 | >20.0 | >800 |
| HXB2.HL612.30 | <8 | <8 | <8 | 80 | 20 | 320 | >20.0 | 400 |
| HXB2.HL612.31 | <8 | <8 | <8 | 80 | 20 | 320 | >20.0 | 70 |
Viruses were tested for their sensitivity to neutralization by patient sera, control sera QC256 and 1785 (74), sCD4, and RANTES in a PBL assay. Serum neutralization was classified as the reciprocal dilution of serum capable of inhibiting p24 antigen production.
sCD4 neutralization was classified as the concentration (micrograms per milliliter) of ligand required to inhibit p24 antigen production.
Concentration of RANTES capable of reducing extracellular p24 antigen production by 50% (Fig. 4).
FIG. 4.
Sensitivity of chimeric viruses to RANTES neutralization. PHA-activated PBL were infected with HXB2 and chimeric viruses in the presence of increasing concentrations of RANTES. At 5 days postinfection, extracellular p24 antigen levels were measured, and the percentage of neutralization relative to that for viral infection in the absence of the chemokine was determined. In the absence of RANTES, p24 antigen levels were in the range of 0.8 to 4.5 ng/ml.
We were interested to know if the autologous antibody recognition observed in Table 2 was capable of neutralizing infection by these chimeric viruses. We therefore tested sera from days 7, 11, and 13 postadmission for their ability to inhibit virus infection of both PHA- and IL-2-activated PBL and U87.CD4.CCR-5 cells. The serum sample obtained after 13 days was able to neutralize all of the chimeric viruses; however, it failed to neutralize W6BC, SF162, HXB2, and the chimeric virus HXB2.SF162 (29) (Table 5). All viruses were sensitive to neutralization by serum from an individual who had been shown to have a high-titer broadly cross-reactive neutralizing-antibody response (74).
DISCUSSION
The results obtained from this patient clearly demonstrate a seroconversion illness. Viral load estimates during seroconversion were in excess of 105 copies per ml and subsequently declined to 5,000 copies per ml over a 48-h period. Such a reduction implies a strong immune response to the virus and potent neutralization of infection (36). However, previous studies on neutralization titers for sera derived from recently infected individuals showed negligible activity against the strains of HIV-1 tested (6, 44). Binley and colleagues (6) reported the detection of HIV-specific serum antibodies shortly after viral clearance, concluding that the humoral response was not a significant component of this clearance. In contrast, several studies have reported CTL activity in the CD8+ population concurrent with a decrease in plasma vRNA levels (11, 34, 53). However, the data presented here suggest that type-specific neutralizing antibodies can also be detected at seroconversion and may play a role in reducing the primary viremic phase.
In agreement with previous reports (1, 35, 76, 77), the viral quasispecies present at seroconversion were found to be very limited in polymorphisms, with mean pairwise intersequence distances of <1% (0.05 to 1.4%). The differences between sequences included a number of polymorphisms which were previously reported, a number of which occurred in more than one sequence. Since the 14 clones analyzed were derived from an initial PCR template of >100 cDNA molecules, these sequences are unlikely to have been derived from the same molecule. Thirteen of the 19 polymorphisms were associated with amino acid sequence changes; the average ratio of synonymous to nonsynonymous substitutions (Ds/Dn) seen between samples was 0.3, similar to that reported previously for other sample sets of this type (4, 35, 76, 77). However, the possibility cannot be ruled out that some of the changes seen may have been due to PCR error. Eight plasmid constructs failed to produce gp120 upon transfection into HEK cells; this failure may have been the result of defective gp120 sequences in vivo or an RT-PCR-derived mutation(s) in vitro. However, PCR amplification, cloning, and expression of gp120 from 100 copies of the plasmid template failed to show any detectable antigenic polymorphisms (47). Direct sequencing of PCR products derived by limiting-dilution amplification of single molecules of HIV-1 RNA showed that 10 to 30% of the sequences obtained from plasma demonstrated defects (frameshifts and/or stop codons) (76). Similar frequencies of defective clones were reported when methods based on the cloning of PCR products were used (12, 28, 61). Taken together, these experiments suggest that the majority of changes observed reflect those present in vivo.
The observed amino acid differences between the clones were not localized to any one region within gp120 (Fig. 2). Several unique changes were seen and could be used to map the residues important for defining differences in virus phenotype. We were surprised to find that pCDNA3 clones 22 and 31, both with sequence changes in the C4 region within the epitope recognized by MAb 38.1a (39), bound the MAb with affinities similar to those of the other glycoproteins tested, suggesting that these substitutions have little effect on epitope recognition by 38.1a or sCD4. We identified a common disruption in glycoprotein conformation, exemplified by negligible sCD4 binding and due to mutation of the N-terminal V3 cysteine (C→R), in 5 of the 20 clones expressing gp120 (Table 1). These glycoproteins were likely to have been misfolded. This conclusion was further supported by their inability to bind a number of conformation-dependent MAbs independent of the CD4 binding site (data not shown). This polymorphism was reported previously; however, the authors did not analyze the possible effects of the mutation on glycoprotein folding (35). Patient HL60 serum (day 13) recognized only autologous gp120 proteins, with the exception of HL605.25, suggesting that this early antibody response was both type specific and sensitive to antigen conformation. The ability of this serum to inhibit the growth of recombinant viruses carrying homologous primary gp120 sequences also suggests that this early immune response is neutralizing. Several reports suggest the V3 region to be immunodominant and to be the primary target for the first detectable antibody response (71) (reviewed in reference 45). We failed to detect HL60 serum (day 13) reactivity with the MN V3 peptide. However, this finding may have been due to differences in amino acid sequences between viruses (the MN V3 sequence differs from the V3 sequence of the viruses studied here at 11 positions); alternatively, the V3 peptide may be unable to present conformation-dependent epitopes. Similarly, Bolognesi reported a type-specific V3-independent neutralizing response elicited in chimpanzees after infection by a primary HIV-1 strain, DH012 (9). These observations and others (41, 42, 56, 73) suggest that the primary neutralizing response to HIV-1 is type specific and may be independent of the V3 region.
All of the chimeric viruses were able to infect only U87.CD4.CCR-5 cells, suggesting that they were able to utilize only the CCR-5 coreceptor. These data are consistent with reports suggesting that the CCR-5 molecule is of critical importance for viral transmission (7, 18, 62). However, differences were noted in virus-induced cytopathology in U87.CD4.CCR-5 cells, whereby HXB2.605.24 appeared to be less cytopathic than either HXB2.612.30 or HXB2.612.31 (Fig. 3). No cytopathic effects were observed upon infection of PBL (data not shown). The inability of CCR-5-utilizing viruses to induce cytopathic effects in PBL has been proposed to be due to the low level of CCR-5 expression on PBL, suggesting that coreceptor density may partially define cytopathology (57). Since CXCR-4-utilizing primary viruses are cytopathic in PBL, despite similarly low levels of CXCR-4 expression (data not shown), there may be differences in the affinity of the viral glycoprotein interaction between the CCR-5 and CXCR-4 coreceptors. The differences in cytopathology noted between the chimeric clones in this study are likely to reflect differences in the interaction of gp120 with CCR-5. To further investigate this conclusion, we tested the sensitivity of the chimeric viruses to neutralization by RANTES. Surprisingly, HXB2.605.24 was resistant to RANTES at concentrations of up to 800 ng/ml (Table 5 and Fig. 4). Previous authors also reported differences in the sensitivity of primary viruses to neutralization by β-chemokines (64). Schols and colleagues (65) recently reported the in vitro selection of a stroma-derived factor 1-α-resistant virus, suggesting that chemokine-resistant viruses can be selected for; however, such resistance was not associated with a change in coreceptor usage. Preliminary comparison of the ability of gp120 from HXB2.605.24 and HXB2.612.31 to compete with biotinylated MIP-1β for binding to CCR-5-expressing cells suggests that HXB2.605.24 gp120 is less able to compete for ligand binding, implying either a reduced affinity or an interaction with a different site on CCR-5 (data not shown). We are presently comparing the abilities of HXB2.605.24 and HXB2.612.31 to infect CD4+ cells expressing a series of mouse or human CCR-5 chimeric receptors to identify the interactive regions of the CCR-5 molecule.
Such polymorphisms in the early viral population imply that selection for resistance to ligands which neutralize viral growth via CCR-5 may be very rapid. Given the low level of genetic polymorphisms between the clones, it is possible to locate the genetic changes responsible for the phenotypic differences between the chimeric viruses. Despite the differences in cytopathology and RANTES neutralization observed between the HXB2.605.24 and HXB2.612.31 clones (Fig. 4), the gp120 sequences of these clones differed only at two positions, 62 (D→G) in the C1 region and 430 (A→V) in the C4 region. The C1 region has been reported to interact with the C5 region, to define gp120-gp41 interactions, and to influence the sensitivity of virus to neutralization by sCD4 (30, 50). Interestingly, Orloff and colleagues (50) reported mutations at residue 62 (D→A) together with changes at residues 63, 68, and 95 in the C1 region of a primary isolate selected for the ability to replicate in C8166 cells and for subsequent increased sensitivity to neutralization by sCD4. These authors inferred that changes in the C1 region affect gp120-gp41 associations and thereby indirectly influence sensitivity to sCD4 neutralization. It should be noted that all of the chimeric viruses were resistant to the highest concentration of sCD4 tested (20 μg/ml) (Table 5); hence, we were unable to demonstrate any effect of this mutation on sCD4 neutralization. In addition, the C1 region has been reported also to interact with both the C2 and the V3 regions, suggesting that changes in these regions may affect the global conformation of the glycoprotein (46). The C4 region has been reported to be a component of the CD4 binding site (49). Chimeric virus HXB2.612.30 showed reduced sensitivity to RANTES neutralization compared to HXB2.612.31 (Fig. 4); this virus had the same amino acid sequence as the resistant clone (HXB2.605.24) in the C1 region and differed from the sensitive clone by two mutations, A→T in the C4 region and K→E in the V5 region (Fig. 2). These data imply that both the C1 and the C4 regions are important components of the chemokine receptor binding site, in agreement with the recent observations of Smyth and colleagues (69).
It should be noted that the distribution of coding changes within the gp120 ORFs differs from the distribution reported later in disease, when changes accumulate within the V2 and V3 regions in response to immune selection (40, 48). Furthermore, the Ds/Dn ratio observed between the clones is unusually low, indicating that these changes are unlikely to be the result of random genetic drift. It is therefore interesting to speculate that the amino acid changes observed in the early gp120 sequences may be defined by forces different from those operating later in disease, within the context of a virus-specific immune response. The amino acid changes observed in the sequences may be associated with selection for alterations in growth rate and/or tissue tropism.
ACKNOWLEDGMENTS
J.A.M. was supported the Medical Research Council, The Wellcome Trust, and the Lister Institute for Preventive Medicine. P.B., S.K., and R.S.T. were supported by the Medical Research Council and The UCL Trustees.
U87.CD4 and Hos.CD4 cells expressing various chemokine receptors were kindly provided by Dan Littman and Hong Kui Deng (New York University Medical Center). HIV-positive human serum 1785 was provided by E. M. Fenyö (Karolinska Institute, Stockholm, Sweden). RANTES was a gift from L. Czaplewski (British Biotech Ltd., Oxford, United Kingdom). We also thank the MRC AIDS Repository for considerable help with reagent requests.
REFERENCES
- 1.Ait-Khaled M, Emery V C. Sequence variation within the human immunodeficiency virus V3 loop at seroconversion. J Med Virol. 1993;41:270–274. doi: 10.1002/jmv.1890410403. [DOI] [PubMed] [Google Scholar]
- 2.Albert J, Abrahamsson B, Nagy K, Aurelius E, Gaines H, Nystrom G, Fenyö E M. Rapid development of isolate-specific neutralizing antibodies after primary HIV-1 infection and consequent emergence of virus variants which resist neutralization by autologous sera. AIDS. 1990;4:107–112. doi: 10.1097/00002030-199002000-00002. [DOI] [PubMed] [Google Scholar]
- 3.Alkhatib G, Combadiere C, Broder C C, Feng Y, Kennedy P E, Murphy P M, Berger E A. CC-CKR-5—a RANTES, MIP-1-alpha, MIP-1-beta receptor as a fusion cofactor for macrophage-tropic HIV-1. Science. 1996;272:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 4.Arnold C. Ph.D. thesis. London, United Kingdom: University College London; 1996. [Google Scholar]
- 5.Arnold C, Balfe P, Clewley J P. Sequence distances between Env genes of HIV-1 from individuals infected from the same source—implications for the investigation of possible transmission events. Virology. 1995;211:198–203. doi: 10.1006/viro.1995.1391. [DOI] [PubMed] [Google Scholar]
- 6.Binley J M, Klasse P J, Cao Y Z, Jones I, Markowitz M, Ho D D, Moore J P. Differential regulation of the antibody responses to Gag and Env proteins of human immunodeficiency virus type 1. J Virol. 1997;71:2799–2809. doi: 10.1128/jvi.71.4.2799-2809.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Biti R, French R, Young J, Bennetts B, Stewart G. HIV-1 infection in an individual homozygous for the CCR5 deletion allele. Nat Med. 1997;3:252–253. doi: 10.1038/nm0397-252. [DOI] [PubMed] [Google Scholar]
- 8.Bleul C C, Wu L J, Hoxie J A, Springer T A, Mackay C R. The HIV coreceptors CXCR-4 and CCR-5 are differentially expressed and regulated on human T lymphocytes. Proc Natl Acad Sci USA. 1997;94:1925–1930. doi: 10.1073/pnas.94.5.1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bolognesi, D. 1994. Humoral immune responses to primary HIV isolates: implications for vaccine development. Neuvieme Colloque des Cent Gardes Paris 1994:285–291.
- 10.Borrow P, Lewicki H, Hahn B H, Shaw G M, Oldstone M B A. Virus-specific CD8+ cytotoxic T-lymphocyte activity associated with control of viremia in primary human immunodeficiency virus type 1 infection. J Virol. 1994;68:6103–6110. doi: 10.1128/jvi.68.9.6103-6110.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Borrow P, Lewicki H, Wei X P, Horwitz M S, Peffer N, Meyers H, Nelson J A, Gairin J E, Hahn B H, Oldstone M B A, Shaw G M. Antiviral pressure exerted by HIV-1-specific cytotoxic T lymphocytes (CTLs) during primary infection demonstrated by rapid selection of CTL escape virus. Nat Med. 1997;3:205–211. doi: 10.1038/nm0297-205. [DOI] [PubMed] [Google Scholar]
- 12.Cheynier R, Henrichwark S, Hadida F, Pelletier E, Oksenhendler E, Autran B, Wain Hobson S. HIV and T-cell expansion in splenic white pulps is accompanied by infiltration of HIV-specific cytotoxic T-lymphocytes. Cell. 1994;78:373–387. doi: 10.1016/0092-8674(94)90417-0. [DOI] [PubMed] [Google Scholar]
- 13.Choe H, Farzun M, Sun Y, Sullivan N, Rollins B, Ponath P D, Wu L, Mackay C R, LaRosa G, Newman W, Gerard N. The β-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell. 1996;85:1136–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 14.Clapham P R, McKnight A, Weiss R A. Human immunodeficiency virus type 2 infection and fusion of CD4-negative human cell lines: induction and enhancement by soluble CD4. J Virol. 1992;66:3531–3537. doi: 10.1128/jvi.66.6.3531-3537.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clark S J, Saag M S, Decker W D, Campbellhill S, Robertson J L, Veldkamp P J, Kappes J C, Hahn B H, Shaw G M. High titers of cytopathic virus in plasma of patients with symptomatic primary HIV-1 infection. N Engl J Med. 1991;324:954–960. doi: 10.1056/NEJM199104043241404. [DOI] [PubMed] [Google Scholar]
- 16.Connor R I, Ho D D. Transmission and pathogenesis of human immunodeficiency virus type-1. AIDS Res Hum Retroviruses. 1994;10:321–323. doi: 10.1089/aid.1994.10.321. [DOI] [PubMed] [Google Scholar]
- 17.Daar E S, Moudgil T, Meyer R D, Ho D D. Transient high levels of viremia in patients with primary human immunodeficiency virus type-1 infection. N Engl J Med. 1991;324:961–964. doi: 10.1056/NEJM199104043241405. [DOI] [PubMed] [Google Scholar]
- 18.Dean M, Carrington M, Winkler C, Huttley G A, Smith M W, Allikmets R, Goedert J J, Buchbinder S P, Vittinghof E, Gomperts E, Donfield S, Vlahov D, Kaslow R, Saah A, Rinaldo C, Detels R, O’Brien S J Hemophilia Growth and Development Study; Multicenter AIDS Cohort Study; Multicenter Hemophilia Cohort Study; San Francisco City Cohort; ALIVE Study. Genetic restriction of HIV-1 infection and progression to AIDS by a deletion allele of the CKR-5 structural gene. Science. 1996;273:1856–1861. doi: 10.1126/science.273.5283.1856. [DOI] [PubMed] [Google Scholar]
- 19.Deng H K, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, Dimarzio P, Marmon S, Sutton R E, Hill C M, Davis C B, Peiper S C, Schall T J, Littman D R, Landau N R. Identification of a major coreceptor for primary isolates of HIV-1. Nature. 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 20.Dittmar M T, Simmons G, Hibbitts S, O’Hare M, Louisirirotchanakul S, Beddows S, Weber J, Clapham P R, Weiss R A. Langerhans cell tropism of human immunodeficiency virus type 1 subtype A through F isolates derived from different transmission groups. J Virol. 1997;71:8008–8013. doi: 10.1128/jvi.71.10.8008-8013.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Doranz B J, Rucker J, Yi Y, Smyth R J, Samson M, Peiper S C, Parmentier M, Collman R G, Doms R W. A dual-tropic primary HIV-1 isolate that uses fusin and the β-chemokine receptors CKR-5, CKR-3, and CKR-2b as fusion cofactors. Cell. 1996;85:1149–1158. doi: 10.1016/s0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- 22.Dragic T, Litwin V, Allaway G P, Martin S R, Huang Y, Nagashima K A, Cayanan C, Maddon P J, Koup R A, Moore J P, Paxton W A. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature. 1996;381:667–671. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 23.Feng Y, Broder C C, Kennedy P E, Berger E A. HIV-1 entry cofactor—functional cDNA cloning of a 7-transmembrane, G-protein-coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 24.Ferbas J, Daar E S, Grovitferbas K, Lech W J, Detels R, Giorgi J V, Kaplan A H. Rapid evolution of human immunodeficiency virus strains with increased replicative capacity during the seronegative window of primary infection. J Virol. 1996;70:7285–7289. doi: 10.1128/jvi.70.10.7285-7289.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fiore J R, Bjorndal A, Peipke K A, Distefano M, Angarano G, Pastore G, Gaines H, Fenyö E M, Albert J. The biological phenotype of HIV-1 is usually retained during and after sexual transmission. Virology. 1994;204:297–303. doi: 10.1006/viro.1994.1534. [DOI] [PubMed] [Google Scholar]
- 26.Fox D G, Balfe P, Palmer C, May J, Arnold C, McKeating J. Length polymorphism within the second variable region of the human immunodeficiency virus type 1 envelope glycoprotein affects accessibility of the receptor binding site. J Virol. 1997;71:759–765. doi: 10.1128/jvi.71.1.759-765.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goudsmit J, De Ronde A, De Rooij E, De Boer R. Broad spectrum of in vivo fitness of human immunodeficiency virus type 1 subpopulations differing at reverse transcriptase codons 41 and 215. J Virol. 1997;71:4479–4484. doi: 10.1128/jvi.71.6.4479-4484.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goudsmit J, Zwart G, Wolfs T. Assessment of HIV-1 antigenic diversification using serum as sample source. AIDS Res Hum Retroviruses. 1992;8:1473. doi: 10.1089/aid.1992.8.1473. [DOI] [PubMed] [Google Scholar]
- 29.Groenink M, Andeweg A C, Fouchier R A M, Broersen S, Vanderjagt R C M, Schuitemaker H, De Goede R E Y, Bosch M L, Huisman H G, Tersmette M. Phenotype-associated env gene variation among eight related human immunodeficiency virus type 1 clones: evidence for in vivo recombination and determinants of cytotropism outside the V3 domain. J Virol. 1992;66:6175–6180. doi: 10.1128/jvi.66.10.6175-6180.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Helseth E, Olshevsky U, Furman C, Sodroski J. Human immunodeficiency virus type 1 gp120 envelope glycoprotein regions important for association with the gp41 transmembrane glycoprotein. J Virol. 1991;65:2119–2123. doi: 10.1128/jvi.65.4.2119-2123.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jurriaans S, Vangemen B, Weverling G J, Vanstrijp D, Nara P, Coutinho R, Koot M, Schuitemaker H, Goudsmit J. The natural history of HIV-1 infection—virus load and virus phenotype independent determinants of clinical course. Virology. 1994;204:223–233. doi: 10.1006/viro.1994.1526. [DOI] [PubMed] [Google Scholar]
- 32.Kaye S, Loveday C, Tedder R S. A microtitre format point mutation assay—application to the detection of drug resistance in human immunodeficiency virus type-1 infected patients treated with zidovudine. J Med Virol. 1992;37:241–246. doi: 10.1002/jmv.1890370402. [DOI] [PubMed] [Google Scholar]
- 33.Keet I P M, Krijnen P, Koot M, Lange J M A, Miedema F, Goudsmit J, Coutinho R A. Predictors of rapid progression to AIDS in HIV-1 seroconverters. AIDS. 1993;7:51–57. doi: 10.1097/00002030-199301000-00008. [DOI] [PubMed] [Google Scholar]
- 34.Koup R A, Safrit J T, Cao Y Z, Andrews C A, McLeod G, Borkowsky W, Farthing C, Ho D D. Temporal association of cellular immune responses with the initial control of viremia in primary human immunodeficiency virus type 1 syndrome. J Virol. 1994;68:4650–4655. doi: 10.1128/jvi.68.7.4650-4655.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kuiken C L, Lukashov V V, Baan E, Dekker J, Leunissen J A M, Goudsmit J. Evidence for limited within-person evolution of the V3 domain of the HIV-1 envelope in the Amsterdam population. AIDS. 1996;10:31–37. doi: 10.1097/00002030-199601000-00005. [DOI] [PubMed] [Google Scholar]
- 36.Lathey J L, Pratt R D, Spector S A. Appearance of autologous neutralizing antibody correlates with reduction in virus load and phenotype switch during primary infection with human immunodeficiency virus type 1. J Infect Dis. 1997;175:231–232. doi: 10.1093/infdis/175.1.231. [DOI] [PubMed] [Google Scholar]
- 37.Liu R, Paxton W, Choe S, Ceradini D, Martin S R, Horuk R, MacDonald M E, Stuhlman H, Koup R A, Landau N R. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiple-exposed individuals to HIV-1 infection. Cell. 1996;86:367–377. doi: 10.1016/s0092-8674(00)80110-5. [DOI] [PubMed] [Google Scholar]
- 38.Loestcher P, Uguccioni M, Bordoli L, Baggiolini M, Moser B, Chizzolini C, Dayer J-M. CCR-5 is characteristic of Th1 lymphocytes. Nature. 1998;391:344–345. doi: 10.1038/34814. [DOI] [PubMed] [Google Scholar]
- 39.McKeating J A, Bennett J, Zolla Pazner S, Schutten M, Ashelford S, Leigh Brown A, Balfe P. Resistance of a human serum-selected human immunodeficiency virus type 1 escape mutant to neutralization by CD4 binding site monoclonal antibodies is conferred by a single amino-acid change in gp120. J Virol. 1993;67:5216–5225. doi: 10.1128/jvi.67.9.5216-5225.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McKeating J A, Zhang Y J, Arnold C, Frederiksson R, Fenyö E M, Balfe P. Biological characterization of T-cell adapted clones of human immunodeficiency virus type 1 expressing primary envelope glycoproteins. Virology. 1996;220:450–460. doi: 10.1006/viro.1996.0332. [DOI] [PubMed] [Google Scholar]
- 41.McKnight A, Clapham P R, Goudsmit J, Cheingsongpopov R, Weber J N, Weiss R A. Development of HIV-1 group-specific neutralizing antibodies after seroconversion. AIDS. 1992;6:799–802. doi: 10.1097/00002030-199208000-00005. [DOI] [PubMed] [Google Scholar]
- 42.Moog C, Fleury H J A, Pellegrin I, Kirn A, Aubertin A M. Autologous and heterologous neutralizing antibody responses following initial seroconversion in human immunodeficiency virus type 1-infected individuals. J Virol. 1997;71:3734–3741. doi: 10.1128/jvi.71.5.3734-3741.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moore J P. Simple methods for monitoring HIV-1 and HIV-2 gp120 binding to soluble CD4 by enzyme-linked immunosorbent assay: HIV-2 has a 25-fold lower affinity than HIV-1 for soluble CD4. AIDS. 1990;4:297–305. doi: 10.1097/00002030-199004000-00003. [DOI] [PubMed] [Google Scholar]
- 44.Moore J P, Cao Y Z, Ho D D, Koup R A. Development of the anti-gp120 antibody response during seroconversion to human immunodeficiency virus type 1. J Virol. 1994;68:5142–5155. doi: 10.1128/jvi.68.8.5142-5155.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moore, J. P., and P. L. Nara. 1991. The role of the V3 loop of gp120 in HIV infection. AIDS 5(Suppl. 2):S21–S33. [DOI] [PubMed]
- 46.Moore J P, Willey R L, Lewis G K, Robinson J, Sodroski J. Immunological evidence for interactions between the first, second, and fifth conserved domains of the gp120 surface glycoprotein of human immunodeficiency virus type 1. J Virol. 1994;68:6836–6847. doi: 10.1128/jvi.68.11.6836-6847.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Murphy A. Ph.D. thesis. Reading, United Kingdom: University of Reading; 1998. [Google Scholar]
- 48.Myers G, Korber B, Wain-Hobson S, Jeang K T, Henderson L E, Pavlakis G N. Human retroviruses and AIDS: a compilation and analysis of nucleic acid and amino acid sequences. Los Alamos, N.Mex: Los Alamos National Laboratory; 1994. [Google Scholar]
- 49.Olshevsky U, Helseth E, Furman C, Li J, Haseltine W, Sodroski J. Identification of individual human immunodeficiency virus type 1 gp120 amino acids important for CD4 receptor binding. J Virol. 1990;64:5701–5707. doi: 10.1128/jvi.64.12.5701-5707.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Orloff S L, Bandea C I, Kennedy M S, Allaway G P, Maddon P J, McDougal J S. Increase in sensitivity to soluble CD4 by primary HIV type-1 isolates after passage through C8166 cells—association with sequence differences in the first constant (C1) region of glycoprotein-120. AIDS Res Hum Retroviruses. 1995;11:335–342. doi: 10.1089/aid.1995.11.335. [DOI] [PubMed] [Google Scholar]
- 51.Palmer C, Balfe P, Fox D G, May J C, Frederiksson R, Fenyö E M, McKeating J A. Functional characteristics of the V1V2 region of human immunodeficiency virus type 1. Virology. 1996;220:436–449. doi: 10.1006/viro.1996.0331. [DOI] [PubMed] [Google Scholar]
- 52.Pantaleo G, Cohen O J, Schacker T, Vaccarezza M, Graziosi C, Paolo Rizzardi G, Kahn J, Fox C H, Schnittman S M, Schwartz D H, Corey L, Fauci A S. Evolutionary pattern of human immunodeficiency virus (HIV) replication and distribution in lymph nodes following primary infection: implications for antiviral therapy. Nat Med. 1998;4:341–345. doi: 10.1038/nm0398-341. [DOI] [PubMed] [Google Scholar]
- 53.Pantaleo G, Graziosi C, Demarest J F, Cohen O J, Vaccarezza M, Gantt K, Murocacho C, Fauci A S. Role of lymphoid organs in the pathogenesis of human immunodeficiency virus (HIV) infection. Immunol Rev. 1994;140:105–130. doi: 10.1111/j.1600-065x.1994.tb00867.x. [DOI] [PubMed] [Google Scholar]
- 54.Patterson S, English N R, Longhurst H, Balfe P, Helbert M, Pinching A J, Knight S C. Analysis of human immunodeficiency virus type 1 (HIV-1) variants and levels of infection in dendritic and T cells from symptomatic HIV-1-infected patients. J Gen Virol. 1998;79:247–257. doi: 10.1099/0022-1317-79-2-247. [DOI] [PubMed] [Google Scholar]
- 55.Paxton W A, Martin S R, Tse D, O’Brien T R, Skurnick J, Van-Devanter N L, Padian N, Braun J F, Kotler D P, Wolinsky S M, Koup R A. Relative resistance to HIV-1 infection of CD4 lymphocytes from persons who remain uninfected despite multiple high-risk sexual exposure. Nat Med. 1996;2:412–417. doi: 10.1038/nm0496-412. [DOI] [PubMed] [Google Scholar]
- 56.Pilgrim A K, Pantaleo G, Cohen O J, Fink L M, Zhou J Y, Zhou J T, Bolognesi D P, Fauci A S, Montefiori D C. Neutralizing antibody responses to human immunodeficiency virus type 1 in primary infection and long-term-nonprogressive infection. J Infect Dis. 1997;176:924–932. doi: 10.1086/516508. [DOI] [PubMed] [Google Scholar]
- 57.Platt E J, Wehrly K, Kuhmann S E, Chesebro B, Kabat D. Effects of CCR5 and CD4 cell surface concentrations on infections by macrophage-tropic isolates of human immunodeficiency virus type 1. J Virol. 1998;72:2855–2864. doi: 10.1128/jvi.72.4.2855-2864.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pope M, Frankel S S, Mascola J R, Trkola A, Isdell F, Birx D L, Burke D S, Ho D D, Moore J P. Human immunodeficiency virus type 1 strains of subtypes B and E replicate in cutaneous dendritic cell–T-cell mixtures without displaying subtype-specific tropism. J Virol. 1997;71:8001–8007. doi: 10.1128/jvi.71.10.8001-8007.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Roos M T L, Lange J M A, De Goede R E Y, Coutinho R A, Schellekens P T A, Miedema F, Tersmette M. Viral phenotype and immune response in primary human immunodeficiency virus type-1 infection. J Infect Dis. 1992;165:427–432. doi: 10.1093/infdis/165.3.427. [DOI] [PubMed] [Google Scholar]
- 60.Safrit J T, Andrews C A, Zhu T F, Ho D D, Koup R A. Characterization of human immunodeficiency virus type 1 specific cytotoxic T-lymphocyte clones isolated during acute seroconversion—recognition of autologous virus sequences within a conserved immunodominant epitope. J Exp Med. 1994;179:463–472. doi: 10.1084/jem.179.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Salminen M, Nykanen A, Leinikki P. Solid-phase direct genomic sequencing and phylogenetic analysis reveal 4 major HIV-1 lineages in Finland. J Cell Biochem. 1993;S17E SIE:85. [Google Scholar]
- 62.Samson M, Libert F, Doranz B J, Rucker J, Liesnard C, Farber C M, Saragosti S, Lapoumeroulie C, Cognaux J, Forceille C, Muyldermans G, Verhofsteede C, Burtonboy G, Georges M, Imai T, Rana S, Yi Y, Smyth R J, Collman R G, Doms R W, Vassart G, Parmentier M. Resistance to HIV-1 infection in caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature. 1996;382:722–725. doi: 10.1038/382722a0. [DOI] [PubMed] [Google Scholar]
- 63.Scarlatti G, Albert J, Rossi P, Hodara V, Biraghi P, Muggiasca L, Fenyö E M. Mother-to-child transmission of human immunodeficiency virus type-1—correlation with neutralizing antibodies against primary isolates. J Infect Dis. 1993;168:207–210. doi: 10.1093/infdis/168.1.207. [DOI] [PubMed] [Google Scholar]
- 64.Scarlatti G, Tresoldi E, Björndal A, Frederiksson R, Colognesi C, Deng H K, Malnati M S, Plebani A, Siccardi A G, Littman D R, Fenyö E M, Lusso P. In vivo evolution of HIV-1 co-receptor usage and sensitivity to chemokine-mediated suppression. Nat Med. 1997;3:1259–1265. doi: 10.1038/nm1197-1259. [DOI] [PubMed] [Google Scholar]
- 65.Schols D, Esté J A, Cabrera C, De Clercq E. T-cell-line-tropic human immunodeficiency virus type 1 that is made resistant to stromal cell-derived factor 1α contains mutations in the envelope gp120 but does not show a switch in coreceptor use. J Virol. 1998;72:4032–4037. doi: 10.1128/jvi.72.5.4032-4037.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schuitemaker H, Koot M, Kootstra N A, Dercksen M W, De Goede R E Y, Vansteenwijk R P, Lange J M A, Schattenkerk J, Miedema F, Tersmette M. Biological phenotype of human immunodeficiency virus type 1 clones at different stages of infection: progression of disease is associated with a shift from monocytotropic to T-cell-tropic virus populations. J Virol. 1992;66:1354–1360. doi: 10.1128/jvi.66.3.1354-1360.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schuitemaker H, Kootstra N A, De Goede R E Y, De Wolf F, Miedema F, Tersmette M. Monocytotropic human immunodeficiency virus type 1 (HIV-1) variants detectable in all stages of HIV-1 infection lack T-cell-line tropism and syncytium-inducing ability in primary T-cell culture. J Virol. 1991;65:356–363. doi: 10.1128/jvi.65.1.356-363.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shotton C, Arnold C, Sattentau Q J, Sodroski J, McKeating J A. Identification and characterization of monoclonal antibodies specific for polymorphic antigenic determinants within the V2 region of the human immunodeficiency virus type 1 envelope glycoprotein. J Virol. 1995;69:222–230. doi: 10.1128/jvi.69.1.222-230.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Smyth R J, Yi Y, Singh A, Collman R G. Determinants of entry cofactor utilization and tropism in a dualtropic human immunodeficiency virus type 1 primary isolate. J Virol. 1998;72:4478–4484. doi: 10.1128/jvi.72.5.4478-4484.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Soto Ramirez L E, Renjifo B, McLane M F, Marlink R, O’Hara C, Sutthent R, Wasi C, Vithayasai P, Vithayasai V, Apichartpiyakul C, Auewarakul P, Cruz V P, Chui D S, Osathanondh R, Mayer K, Lee T H, Essex M. HIV-1 Langerhans cell tropism associated with heterosexual transmission of HIV. Science. 1996;271:1291–1293. doi: 10.1126/science.271.5253.1291. [DOI] [PubMed] [Google Scholar]
- 71.Spear G T, Takefman D M, Sharpe S, Ghassemi M, Zolla Pazner S. Antibodies to the HIV-1 V3 loop in serum from infected persons contribute a major proportion of immune effector functions including complement activation, antibody-binding, and neutralization. Virology. 1994;204:609–615. doi: 10.1006/viro.1994.1575. [DOI] [PubMed] [Google Scholar]
- 72.Tersmette M, Gruters R A, De Wolf F, De Goede R E, Lange J M, Schellekens P T, Goudsmit J, Huisman H G, Miedema F. Evidence for a role of virulent human immunodeficiency virus (HIV) variants in the pathogenesis of acquired immunodeficiency syndrome: studies on sequential HIV isolates. J Virol. 1989;63:2118–2125. doi: 10.1128/jvi.63.5.2118-2125.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tsang M L, Evans L A, McQueen P, Hurren L, Byrne C, Penny C D. Neutralizing antibodies against sequential autologous human immunodeficiency virus type-1 isolates after seroconversion. J Infect Dis. 1994;170:1141–1147. doi: 10.1093/infdis/170.5.1141. [DOI] [PubMed] [Google Scholar]
- 74.Weber J, Fenyö E M, Beddows S, Kaleebu P, Björndal A, Osmanov S, Heyward W, Esparza J, Galvaocastro B, Vandeperre P, Karita E, Wasi C, Sempala S, Tugume B, Biryahwaho B, Rübsamen Waigmann H, Von Briesen H, Esser R, Grez M, Holmes H, Newberry A, Ranjbar S, Tomlinson P, Bradac J, McCutchan F, Louwagie J, Hegerich P, Lopez Galindez C, Olivares I, Dopazo J, Mullins J I, Delwart E L, Bachmann H M, Goudsmit J, De Wolf F, Hahn B H, Gao F, Yue L, Saragosti S, Schochetman G, Kalish M, Luo C C, George R, Pau C P, Cheingsongpopov R, Nara P, Albert J, Myers G, Korber B. Neutralization serotypes of human immunodeficiency virus type 1 field isolates are not predicted by genetic subtype. J Virol. 1996;70:7827–7832. doi: 10.1128/jvi.70.11.7827-7832.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wu L J, Paxton W A, Kassam N, Ruffing N, Rottman J B, Sullivan N, Choe H, Sodroski J, Newman W, Koup R A, Mackay C R. CCR5 levels and expression pattern correlate with infectability by macrophage-tropic HIV-1, in vitro. J Exp Med. 1997;185:1681–1691. doi: 10.1084/jem.185.9.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang L Q, Mackenzie P, Cleland A, Holmes E C, Leigh Brown A J, Simmonds P. Selection for specific sequences in the external envelope protein of human immunodeficiency virus type 1 upon primary infection. J Virol. 1993;67:3345–3356. doi: 10.1128/jvi.67.6.3345-3356.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhu T F, Wang N, Carr A, Nam D S, Moorjankowski R, Cooper D A, Ho D D. Genetic characterization of human immunodeficiency virus type 1 in blood and genital secretions: evidence for viral compartmentalization and selection during sexual transmission. J Virol. 1996;70:3098–3107. doi: 10.1128/jvi.70.5.3098-3107.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]