Abstract
Objective
Development of clinical phenotypes from electronic health records (EHRs) can be resource intensive. Several phenotype libraries have been created to facilitate reuse of definitions. However, these platforms vary in target audience and utility. We describe the development of the Centralized Interactive Phenomics Resource (CIPHER) knowledgebase, a comprehensive public-facing phenotype library, which aims to facilitate clinical and health services research.
Materials and Methods
The platform was designed to collect and catalog EHR-based computable phenotype algorithms from any healthcare system, scale metadata management, facilitate phenotype discovery, and allow for integration of tools and user workflows. Phenomics experts were engaged in the development and testing of the site.
Results
The knowledgebase stores phenotype metadata using the CIPHER standard, and definitions are accessible through complex searching. Phenotypes are contributed to the knowledgebase via webform, allowing metadata validation. Data visualization tools linking to the knowledgebase enhance user interaction with content and accelerate phenotype development.
Discussion
The CIPHER knowledgebase was developed in the largest healthcare system in the United States and piloted with external partners. The design of the CIPHER website supports a variety of front-end tools and features to facilitate phenotype development and reuse. Health data users are encouraged to contribute their algorithms to the knowledgebase for wider dissemination to the research community, and to use the platform as a springboard for phenotyping.
Conclusion
CIPHER is a public resource for all health data users available at https://phenomics.va.ornl.gov/ which facilitates phenotype reuse, development, and dissemination of phenotyping knowledge.
Keywords: electronic health records, phenomics, algorithms, library
Background and significance
As electronic health record (EHR) systems have become the standard for use in clinical care, secondary use of these data for research and healthcare operations has also grown. The development of “phenotypes” from EHRs and linked data sources involves the creation of algorithms to define health conditions, symptoms, demographics, and other patient characteristics. These algorithms range in complexity, from rules-based logic and code curation by clinicians, to machine learning models that require minimal supervision.1–5 For example, acute ischemic stroke (AIS) could be defined using numerous approaches based on the application. Clinician curated International Classification of Diseases (ICD)-9 and -10 codes can be applied as rules-based algorithms to define an AIS case or control as part of a phenome-wide association study (PheWAS).6 On the other hand, a probabilistic model of AIS (with likelihood ranging from 0 to 1 for each patient) could be generated using supervised or unsupervised machine learning methods, utilizing various cutoffs to define a stroke case.7,8 Regardless of approach, development of a quality phenotype definition can be a resource intensive process requiring computing time, expertise in phenomics science, clinical knowledge, and familiarity with the health system EHR and other data sources.
EHR-based phenotype definitions are used for a variety of purposes including epidemiological research, genomics studies, pragmatic trials, and clinical decision support. However, there are challenges with phenotyping that need to be addressed during the algorithm development process and when reusing existing phenotype definitions. The reuse of administrative data for research purposes can be difficult due to bias in how the data was collected or variability in code usage specific to the healthcare system. A recent study of long coronavirus disease (COVID-19) showed differences in the use of ICD-10 code U09.9 across hospitals that may be impacted by healthcare utilization, including acute hospitalization and visiting a long COVID-19 clinic.9 Portability of phenotype algorithms across systems is also an issue, and certain definitions developed in one healthcare system may not always apply to others. For example, a definition for post-traumatic stress disorder (PTSD) performs well in the Veteran population, but uses unique codes for PTSD compensation data that are only available in the Veterans Affairs (VA) Healthcare System.10 Understanding these nuances is critical for reuse of phenotype definitions.
There is a great unmet need to organize existing phenotype definitions to enable reuse and expedite research. Several phenotype libraries have been established to facilitate reproducibility by capturing and cataloging algorithm metadata, including the Phenotype Knowledgebase (PheKB) by the Electronic MEdical Records and GEnomics (eMERGE) Network and the Health Data Research (HDR) UK Phenotype Library.11,12 However, these libraries vary with regard to content available, contributions accepted, phenotype metadata captured, user interface, and other functionality. Additional features are needed to improve upon existing libraries and facilitate reproducibility of definitions.13–15
With this motivation, we developed the Centralized Interactive Phenomics Resource (CIPHER) knowledgebase, a publicly accessible phenotype library that builds on existing models. Our approach to the development of this library was based on our understanding of the challenges of phenotyping and researcher needs for implementing an algorithm, particularly when porting an algorithm created in a different health system. With this in mind, we developed a knowledgebase resource that takes a broad approach for capturing existing phenotype definitions. Our scope was not to instruct the user how to develop the phenotype, as some have pointed out in the literature, but instead to set standards for phenotype metadata collection that are necessary for users to reconstruct the phenotype according to their data systems and purpose. This broad scope allows us to capture key metadata elements while being flexible, as the field of phenomics science and approaches to phenotyping continue to evolve over time. On top of the knowledgebase, CIPHER integrates applications that assist users in navigating the phenomics knowledgebase and developing phenotypes.
We leveraged clinicians and phenomics leaders in the VA healthcare system, the largest in the United States (US), to develop the phenotype metadata collection standard employed in the CIPHER knowledgebase.16 With the expertise from the Department of Energy (DOE) Oak Ridge National Laboratory in computer science, data management, access, and retrieval, the CIPHER metadata standard and phenotype library has been implemented as a public knowledgebase platform. What was an internal resource for the VA community has now been made available to all health data users via https://phenomics.va.ornl.gov/.
This article describes the resources available on the CIPHER website including the knowledgebase design, phenotype collection process, integrated data visualization tools, and workflows available to users.
Methods
An earlier version of CIPHER was established and piloted as an internal MediaWiki based phenotype library for use within the VA, first by several hundred members of the VA Million Veteran Program (MVP) scientific community.17,18 The platform then expanded to the wider VA community, with thousands of users accessing and contributing phenotype metadata. As our userbase and content expanded, we developed a standardized approach for phenotype metadata collection with input from the user community. The CIPHER phenotype metadata standard has been described in detail previously.16 Briefly, the standard captures information about the algorithm author, rationale for approach, publication and acknowledgements, description of algorithm and components, validation, performance metrics, programming code, and other relevant fields needed to reuse the definition. The metadata captured includes free text, CIPHER defined categories, and existing vocabularies and ontologies such as ICD codes, RxNorm, Logical Observation Identifiers Names and Codes (LOINC), and Medical Subject Headings (MeSH). While developed in the VA, the standard employs a health system agnostic approach.
The aims of launching a publicly accessible website were to create an integrated phenomics knowledgebase that fills information gaps in existing phenotype libraries, collect phenotype metadata using the CIPHER standard, use validation parameters for incoming phenotype metadata content, streamline the phenotype collection and review process, scale content management, enable complex searching of the knowledgebase, and provide tools for developing phenotypes and browsing metadata through visualizing integrated knowledgebase content.
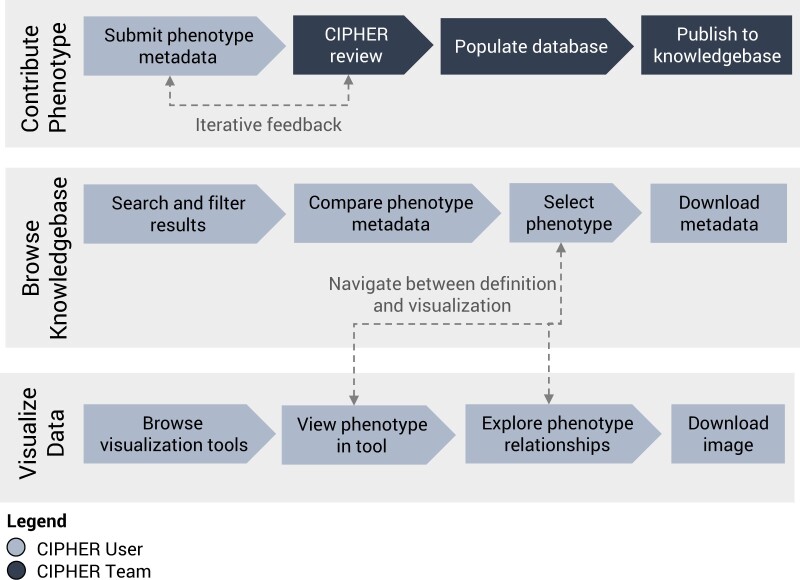
The site was iteratively developed with VA phenomics experts guiding content. Users of the internal VA MediaWiki phenotype library were asked to provide feedback on the front-end user interface and test functionality. All phenotype metadata from the VA MediaWiki library has been structured according to the metadata standard and migrated to the knowledgebase. The new CIPHER platform is comprised of the following features which address the goals of the site: (1) phenotype knowledgebase, (2) algorithm collection workflow, and (3) data visualization tools. Figure 1 depicts the CIPHER website user workflow.
Figure 1.
CIPHER library user workflow. Researchers can contribute phenotype algorithms using our webform, browse definitions in the CIPHER phenotype knowledgebase, and visualize data in connected tools which link back to the knowledgebase.
Results
Phenotype knowledgebase
The knowledgebase stores the metadata about each phenotype according to the CIPHER metadata standard and the information can be searched and linked together through common relationships in the data. By capturing keywords and important contextual information from the user for each phenotype, the website provides users a way to easily search and locate phenotypes with similar characteristics.
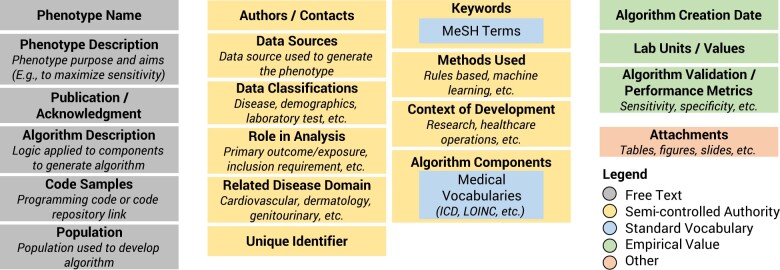
The knowledgebase includes four types of descriptive metadata: standard vocabularies, empirical values, semi-controlled authorities, and free text. Figure 2 illustrates the types of collected metadata components and their metadata category. Standard vocabularies are validated against their definition or format standard to ensure all submissions are semi-validated at entry. Free-text components are arbitrary user provided text. Semi-controlled authorities are short free text labels that are collected into an internal enumeration. Users can add an entry if it does not already exist, and it will be incorporated into the authority once a submission is accepted. Additional unindexed attachments can also be included in an entry for supplemental information. All changes to phenotypes are tracked and recorded to enable internal change tracking.
Figure 2.
Phenotype components in the knowledgebase. ICD, International Classification of Diseases; LOINC, Logical Observation Identifiers Names and Codes; MeSH, Medical Subject Headings. The knowledgebase collects and stores phenotype metadata elements based on the CIPHER standard.
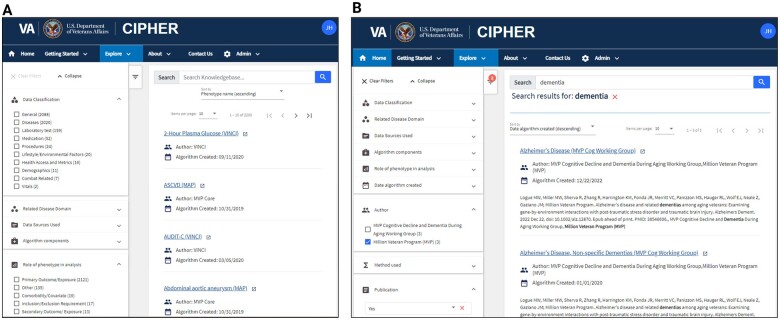
Users can take advantage of search suggestions and complex searching to locate entries based on validation status, publications, and many other criteria. Figure 3 demonstrates some of the filtering components (Figure 3A) and the application of multiple filters and text search related to dementia (Figure 3B). Each category of descriptive metadata can be used as a filter independently. Free text is utilized for full text search. Standard vocabularies and semi-controlled authorities can be used for full text search, but also filtering in the search interface. Empirical values are used to enable comparison operators, such as selecting phenotypes created in a certain date range. Additional comparison filters will be implemented in the future.
Figure 3.
CIPHER phenotype knowledgebase search interface. Researchers can use text search (A) and filters (B) to browse phenotypes.
Individual phenotype results appear as a knowledgebase article that displays the metadata for a particular phenotype. The phenotype information is organized by sections providing the user an overview of the algorithm, details about its components, the logic and any validation or associated publications. Users may freely view the results; however, some licensed content may require authentication to view particular metadata fields. In these instances, the website allows users to login and automatically authenticate to access fields such as licensed Unified Medical Language System (UMLS) content. Author emails can be found on the phenotype article, facilitating acknowledgement in studies, feedback from users, and collaboration.
Phenotype collection workflow
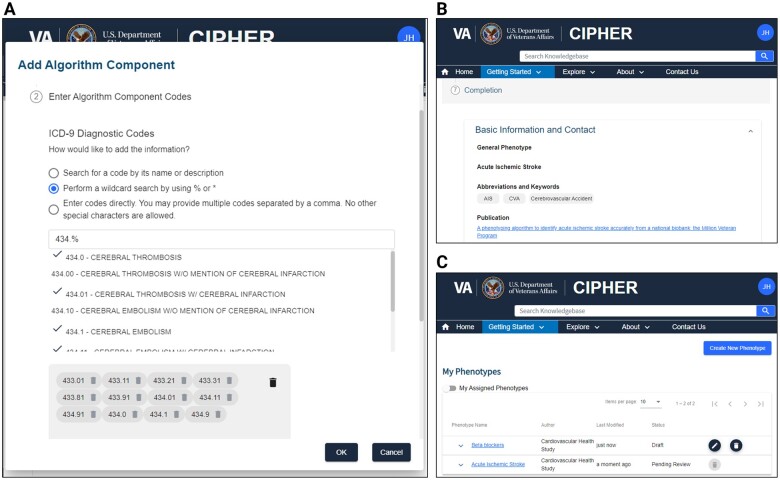
The phenotype collection workflow standardizes the collection of phenotype definition metadata and enables review of submitted phenotypes by the CIPHER team (Figure 1). All health data users are welcome to contribute their phenotypes regardless of the healthcare system used to build the phenotype algorithm, validation status, or presence of the condition in the knowledgebase. A user who would like to contribute phenotypes to CIPHER, or “contributor,” begins the process by registering for an account on the website. The phenotype webform is then enabled and the contributor proceeds with entering metadata (Figure 4A and B). At any time, the contributor may save the phenotype metadata and return later to complete it. A contributor registered with the site may have multiple phenotype entries in progress and can view the status of all submissions in their personal dashboard (Figure 4C).
Figure 4.
Phenotype entry user interface. The phenotype webform allows validation of standard vocabularies (A), collection of phenotype metadata using the CIPHER standard (B), and viewing of phenotype submission status in the user’s personal phenotype dashboard (C).
The web-based phenotype entry form is a multi-step interface for collection of phenotype metadata using the CIPHER standard. The collection form is divided into 5 sections: basic phenotype and contributor information, algorithm overview, algorithm components, validation, and additional information. The webform interface minimizes entry errors via automated validation of metadata fields, use of required fields, and suggestion of standard vocabularies or previously submitted values where appropriate. The latter feature is most useful for the collection of algorithm components using standard vocabularies. Figure 4A is an example of this feature for retrieving ICD-9 codes using a wildcard. Custom algorithm components can also be entered by the user for EHR specific fields used in the definition.
Other highlights of the phenotype entry form include the ability to store multiple sets of performance metrics for one phenotype definition. Users of the final phenotype definition may contact CIPHER to share their performance metrics for the phenotype if it was validated in a secondary cohort or health system. Programming code is shared on the site by entering short code or sharing a code repository link. We encourage the sharing of code repositories given that the user can manage their own code.
After the form is completed, the contributor submits the phenotype to CIPHER for review. The contributor can view the status as “pending review” in the interface for managing submissions. (Figure 4C) The phenotype submission is then reviewed by the CIPHER team for the completeness and clarity of the information provided, and we do not judge the quality of the phenotype. Instead, CIPHER aims to collect phenotypes developed using various methods and metrics that may be suitable for different settings to allow flexibility for users to identify algorithms most appropriate for their projects. If CIPHER has clarifying questions about the definition for the contributor, comments can be sent between the parties directly in the platform. Contributors receive an email notification when questions are asked by the CIPHER team and when the phenotype is published. The CIPHER team reviews the form submission and upon approval the data is incorporated into the knowledgebase.
The CIPHER interface also supports administrators with workflows and dashboards for curation and management of knowledgebase content, user requests, and performance metrics. Website usage and traffic can be evaluated via software that tracks unique views, approximate location where users are logging in, and monitors page views to measure content engagement. These features are vital to managing the website’s continued growth.
Data visualization tools
The integration of data visualizations into the CIPHER platform enhances the user experience and facilitates further exploration of phenotype metadata. Two applications contributed by the MVP Phenomics Core in collaboration with Harvard School of Public Health and Mass General Brigham are currently available on CIPHER.
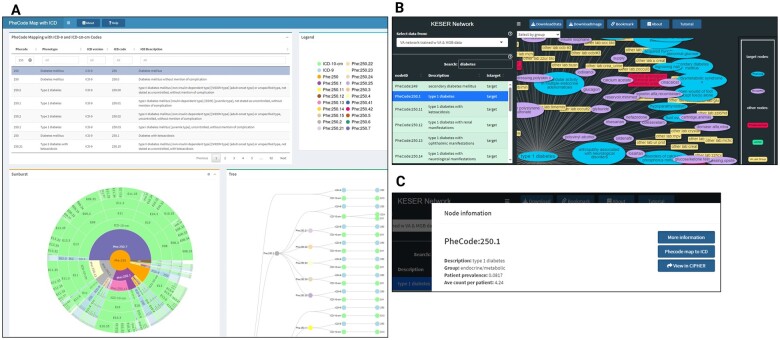
The Phecode to ICD Map was created to facilitate the use of phecodes in EHR-based studies. Phecodes are mappings of ICD-9 and -10 billing codes into clinically relevant concepts developed by Denny et al.6 (Figure 5A) The Phecode to ICD Map visually displays these mappings in an interactive sunburst plot and tree diagram. Users can search for a phecode or condition of interest to view the hierarchical structure and compare relationships of ICD-9 and -10 code mappings within a phecode as well as compare relationships of ICD codes mapped to similar phecodes. Phecodes were initially created for use in phenome wide association studies or PheWAS, but this mapping has been widely used in other phenotype applications and to use as a starting point for further phenotype development.8
Figure 5.
Data visualization tools in CIPHER. The Phecode to ICD Map facilitates the understanding of ICD-9 and -10 codes to phecode mappings (A). The KESER network visualizes the network structure of EHR codes, learned using co-occurrence summary level data from VA and Mass General Brigham (B). Users can navigate from a phenotype present in the KESER network to the phenotype article describing the algorithm in the knowledgebase (C).
The Knowledge Extraction via Sparse Embedding Regression (KESER) Network was created to identify and visualize network structure of EHR codes, learned using co-occurrence summary level data from VA and Mass General Brigham.19 (Figure 5B) The KESER algorithm was used to construct knowledge networks for code concepts including diagnoses (phecodes), medications (RxNorm), procedures and laboratory tests (Logical Observation Identifiers Names and Codes and VA local lab codes). This interactive knowledge map enables users to automatically select important concepts related to a condition and bypass manual feature identification. Users may search for a node in the network, such as a phecode, and view related neighboring nodes.
A key, novel feature of CIPHER is its capability of linking these visualization tools to the knowledgebase. When viewing the phenotype articles throughout the knowledgebase, users can click on relevant links that navigate to nodes or concepts embedded within visualization tools whenever the phenotype is being used utilized in the tools. The linkage is bidirectional so users exploring visualization tools can also click on nodes to automatically navigate users back to the knowledgebase. This facilitates retrieval of phenotype metadata related to concepts in the tools. (Figure 5C) This is accomplished by generating a set of system identifiers that uniquely identifies each phenotype. The unique identifier is used as a crosswalk between the knowledgebase and the visualization tools.
The implementation of visualization tools aims to enhance the user experience, phenotype browsability, and expedite knowledge retrieval. Our goal is to expand the tools available to users and integrate the growing CIPHER phenotype knowledgebase into them. While the first two tools available serve to facilitate the understanding of data mappings (Phecode to ICD Map) and development of phenotypes (KESER), we plan to expand tool offerings to include display of phenotype prevalence and visual knowledgebase browsing.
Use of CIPHER
The CIPHER website was initially released in June 2023. As of January 2023, there have been over 5900 definitions stored in the CIPHER knowledgebase (Table 1). As the CIPHER user community grows, we anticipate further expansion of CIPHER knowledgebase. For example, in the VA, the CIPHER website is now used to capture all phenotypes from VA funded projects and definitions used in research and healthcare operations. Since CIPHER’s launch, there have been 1036 site visitors and 155 accounts created.
Table 1.
Phenotypes available in CIPHER.
| Metadata field | Number of phenotypes |
|---|---|
| Data classification | |
| Demographics | 16 |
| Diseases | 5315 |
| Health access and metrics | 20 |
| Labs | 198 |
| Lifestyle/environmental factors | 264 |
| Medications | 59 |
| Procedures | 31 |
| Vitals | 7 |
| Related disease domain | |
| Cardiovascular | 646 |
| Dermatology | 268 |
| Endocrine/metabolic | 525 |
| ENT/ophthalmology | 258 |
| Gastrointestinal | 551 |
| Genitourinary | 372 |
| Hematology | 235 |
| Infectious disease | 156 |
| Injuries/poisonings | 158 |
| Mental/behavioral health | 152 |
| Musculoskeletal | 368 |
| Neurology | 225 |
| Obstetrics/gynecology | 12 |
| Oncology/neoplasms | 330 |
| Respiratory | 301 |
| Symptoms | 148 |
| Women’s health | 9 |
| Algorithm components | |
| ICD-9 code | 5214 |
| ICD-10 codes | 4796 |
| ICD-9 procedure codes | 16 |
| ICD-10 procedure codes | 13 |
| Lab tests | 206 |
| Medications | 76 |
| Text snippets | 1767 |
| VA clinic stop code | 12 |
| Method used | |
| Rules-based | 4022 |
| Machine learning | 1865 |
| Published | 5597 |
| Algorithm programming code provided | 262 |
| Validated | 62 |
| Total phenotypes | 5904 |
Abbreviations: ENT = ear, nose, and throat, ICD = International Classification of Diseases.
Use case: VA Million Veteran Program
The Million Veteran Program (MVP) is an observational cohort study and biobank embedded in the VA Healthcare System, and its design, goals, and infrastructure have been described elsewhere.17,20 Consented participants complete surveys, contribute blood samples for genotyping and storage, and allow access to EHR data for use in research. MVP has a large research portfolio covering a wide range of research domains, and to date, there have been hundreds of publications. MVP has made major contributions to CIPHER’s development and content for the knowledgebase. MVP uses CIPHER as their primary phenomics metadata library and has custom built content for their research community. Researchers approved to conduct MVP projects are required to contribute their phenotype definitions to CIPHER upon publication of a phenotype definition. CIPHER partners with MVP to implement methods and tools including two data visualization tools currently available. Resources related to the phenome-wide genome-wide association study of 635 969 MVP participants will also be hosted on CIPHER, including phenotype definitions and tools for visual results browsing.21 As MVP’s work products and resources continue to grow, CIPHER will be part of MVP’s scalable solution for information dissemination and platform for sharing knowledge to the wider health data community.
Use case: Boston Musculoskeletal Clinical Research Collaboratory
The Boston Musculoskeletal Clinical Research Collaboratory (MCRC) is affiliated with the Boston University School of Medicine comprised of researchers with backgrounds in clinical rheumatology, physical therapy and biomechanics, epidemiology and biostatistics, as well as visiting scholars. The Boston MCRC has historically used internal processes to track phenotype definition metadata but was looking for a more efficient and scalable solution to manage their growing body of definitions. Stakeholder meetings were conducted with MCRC members, indicating that key features of interest in a phenotype library included the ability to store phenotypes based on diagnostic code, procedure, medication, laboratory, imaging or combination definitions. Phenotype metadata of interest included phenotype creation date, lexicon date, creator, validation information, and performance characteristics. MCRC users anticipated searching the library at the start of any new study using health data, and identified need for access by both current group members and collaborators at outside institutions to access their own phenotypes for future work. The Boston MCRC is contributing phenotype definitions to CIPHER, with the expectation to revise definitions are they are updated with subsequent versions of the relevant health code lexicons.
Discussion
CIPHER is a phenotyping resource that standardizes phenotype metadata collection, provides a searchable knowledgebase, and connects to data visualization tools. The CIPHER knowledgebase brings the culmination of phenomics expertise and contribution of the VA community to the public, and it provides a platform that serves all health care data users. CIPHER has been adopted as the central phenotype library of the largest healthcare system in the US and our successful internal pilot with the MVP and external pilot with the Boston MCRC demonstrates its utility beyond the VA. As journals and research funders expand requirements for sharing data and metadata, including the National Institutes of Health Data Management and Sharing Policy, CIPHER provides a solution for public dissemination of phenotype metadata documentation.22 Sharing phenotypes via CIPHER enhances the impact of funded research and the reproducibility of studies by ensuring that phenotype definitions are publicly available. CIPHER also improves the rigor of phenotypes through searchable code lists, and by ensuring that relevant metadata is documented. As the knowledgebase grows and matures, the platform will improve efficiency of health data research by making phenotype definitions ready for use and obviating the need for investigators to search published literature for definitions, or to request such definitions from other investigators.
There are several public-facing phenotype libraries currently in use which vary in terms of audience, phenotype metadata collected, user workflows, and site features. PheKB was created to develop, store, and implement EHR-based algorithms at eMERGE network sites across the US.11 The site hosts 85 phenotype algorithms with browsable metadata and additional functionality is available to network members with an account. The HDR UK Phenotype Library is affiliated with HDR UK and stores 1053 definitions for phenotypes developed in UK EHRs.12 The Observational Health Data Sciences and Informatics (OHDSI) Gold Standard Phenotype Library differs from the aforementioned libraries in that it hosts definitions using the Observational Medical Outcomes Partnership (OMOP) Common Data Model (CDM) and its 569 algorithms are stored in an R package.23,24 The functionality of the CIPHER platform differs substantially from these libraries. The integration of data visualization tools allows users to explore relationships between phenotypes and data elements, while seamlessly linking back to phenotype articles in the knowledgebase. The pairing of the knowledgebase with visualization tools is a unique feature not found in existing libraries. The CIPHER metadata standard builds upon the fields used in PheKB and HDR UK libraries and a comparison has been previously described.16 CIPHER uses these fields in complex searching, a feature unavailable in PheKB or OHDSI libraries and minimally available in HDR UK. One key differentiator is that CIPHER captures validation metrics (such as positive predictive value), which is critical for reuse of a phenotype. CIPHER also allows researchers reusing a definition to contribute validation metrics to the phenotype article, enabling evaluation of phenotype portability across health systems.
CIPHER integrates desired components for phenotype libraries outlined in recent articles. Almowil et al. interviewed and led focus groups with health data researchers to understand the needs of phenotype library users.15 Their discussions showed that researchers are interested in using publicly available libraries for phenotype development, but that significant barriers to their usage and utility exist including searchability, limited user interface, and quality of code lists. CIPHER sought to address user needs by integrating phenomics experts into the development and testing of the site. Spotnitz et al. developed a desired list of metadata fields within a phenomics working group.14 Of the 38 metadata fields identified by Spotnitz, 29 can be found in the CIPHER metadata standard. Chapman et al. outline 14 desired components for phenotype libraries.13 The CIPHER library aligns with the expert recommendation’s focus on sharing validation metrics, providing accessible metadata, and logging the evolution of a definition. These desiderata also emphasize use of modelling languages to capture algorithms and facilitate implementation. CIPHER does not require the use of modelling languages in order to allow flexibility in metadata capture. Given the rapid pace of methods development in the phenomics field and the increasing breadth of data integrated into algorithms such as imaging and wearables, our goal is to be future oriented and ensure we can adapt to yet unknown advances in technology.
There are several limitations of the CIPHER platform. Licensed data elements and proprietary elements may have restrictions on what we can share. For example, UMLS requires authorization with an existing account to view certain data elements on our site. The addition of further data elements to CIPHER requires evaluation to ensure licensing requirements are met. Integration of data elements from the OMOP CDM is planned but validation of OMOP concepts is not currently available. The CIPHER platform is not configured to link to patient level data or enable development of computable phenotypes within the platform.
Enhancement of the CIPHER platform continues in response to the needs of the health data community. We encourage users to submit feedback via https://phenomics.va.ornl.gov/web/cipher/about#contact-us. Future directions include tools for visualizing phenotype prevalence, side by side comparison of metadata from multiple phenotype algorithms, and building an expanded knowledge network. Further expansion of the resources pertaining to other data types such as omics and imaging data is also in planning. The workflows available to users will also expand as we show the user changes in phenotype metadata between versions, enable self-service updating of phenotype definitions, and iteratively improve the entry workflow and metadata elements captured based on feedback.
The goals of CIPHER are to provide an easy-to-use platform to develop, collect, store, and share computable phenotypes, metadata, and resources; maintain a standardized and scalable metadata collection process to optimize phenotype reproducibility and interoperability across health systems, and to encourage collaboration across the data science community. CIPHER’s growing knowledgebase and platform can play a role in many applications where computable phenotypes are used. We invite health data users to contribute to CIPHER and provide feedback to continue our expansion and enhancement of the site.
Conclusion
The CIPHER knowledgebase provides a publicly accessible phenotype library with a design supporting front end tools and scalable expansion. It fills gaps in available phenotyping resources and provides a solution for phenotype definition sharing. The CIPHER platform is designed to evolve with advances in phenomics, facilitate collaboration, and enable interoperability with other healthcare systems. What once was an internal resource to the VA is now a publicly accessible information resource that welcomes all health data users.
Acknowledgments
The contents do not represent the views of the US Department of Veterans Affairs or the US Government. This manuscript has been authored by UT-Battelle, LLC, under contract DE-AC05-00OR22725 with the US DOE. The US government retains and the publisher, by accepting the article for publication, acknowledges that the US government retains a nonexclusive, paid-up, irrevocable, worldwide license to publish or reproduce the published form of this manuscript, or allow others to do so, for US government purposes. DOE will provide public access to these results of federally sponsored research in accordance with the DOE Public Access Plan (https://www.energy.gov/doe-public-access-plan).
Contributor Information
Jacqueline Honerlaw, Centralized Interactive Phenomics Resource (CIPHER), Office of Research and Development, Veterans Health Administration, Washington, DC 20002, United States; VA Boston Healthcare System, Boston, MA 02111, United States.
Yuk-Lam Ho, Centralized Interactive Phenomics Resource (CIPHER), Office of Research and Development, Veterans Health Administration, Washington, DC 20002, United States; VA Boston Healthcare System, Boston, MA 02111, United States.
Francesca Fontin, Centralized Interactive Phenomics Resource (CIPHER), Office of Research and Development, Veterans Health Administration, Washington, DC 20002, United States; VA Boston Healthcare System, Boston, MA 02111, United States.
Michael Murray, Centralized Interactive Phenomics Resource (CIPHER), Office of Research and Development, Veterans Health Administration, Washington, DC 20002, United States; VA Boston Healthcare System, Boston, MA 02111, United States.
Ashley Galloway, Centralized Interactive Phenomics Resource (CIPHER), Office of Research and Development, Veterans Health Administration, Washington, DC 20002, United States; VA Boston Healthcare System, Boston, MA 02111, United States.
David Heise, Oak Ridge National Laboratory (ORNL), Oak Ridge, TN 37830, United States.
Keith Connatser, Oak Ridge National Laboratory (ORNL), Oak Ridge, TN 37830, United States.
Laura Davies, Oak Ridge National Laboratory (ORNL), Oak Ridge, TN 37830, United States.
Jeffrey Gosian, Centralized Interactive Phenomics Resource (CIPHER), Office of Research and Development, Veterans Health Administration, Washington, DC 20002, United States; VA Boston Healthcare System, Boston, MA 02111, United States.
Monika Maripuri, Centralized Interactive Phenomics Resource (CIPHER), Office of Research and Development, Veterans Health Administration, Washington, DC 20002, United States; VA Boston Healthcare System, Boston, MA 02111, United States.
John Russo, Centralized Interactive Phenomics Resource (CIPHER), Office of Research and Development, Veterans Health Administration, Washington, DC 20002, United States; VA Boston Healthcare System, Boston, MA 02111, United States; Department of Computer Science, Landmark College, Putney, VT 05346, United States.
Rahul Sangar, Centralized Interactive Phenomics Resource (CIPHER), Office of Research and Development, Veterans Health Administration, Washington, DC 20002, United States; VA Boston Healthcare System, Boston, MA 02111, United States.
Vidisha Tanukonda, Centralized Interactive Phenomics Resource (CIPHER), Office of Research and Development, Veterans Health Administration, Washington, DC 20002, United States; VA Atlanta Healthcare System, Decatur, GA 30033, United States.
Edward Zielinski, Centralized Interactive Phenomics Resource (CIPHER), Office of Research and Development, Veterans Health Administration, Washington, DC 20002, United States; VA Boston Healthcare System, Boston, MA 02111, United States.
Maureen Dubreuil, VA Boston Healthcare System, Boston, MA 02111, United States; Section of Rheumatology, Boston University Chobanian and Avedisian School of Medicine, Boston, MA 02118, United States.
Andrew J Zimolzak, Center for Innovations in Quality, Effectiveness and Safety, Michael E. DeBakey VA Medical Center, Houston, TX 77030, United States; Department of Medicine, Baylor College of Medicine, Houston, TX 77030, United States.
Vidul A Panickan, VA Boston Healthcare System, Boston, MA 02111, United States; Department of Biomedical Informatics, Harvard Medical School, Boston, MA 02115, United States.
Su-Chun Cheng, VA Boston Healthcare System, Boston, MA 02111, United States; Department of Biomedical Informatics, Harvard Medical School, Boston, MA 02115, United States.
Stacey B Whitbourne, VA Boston Healthcare System, Boston, MA 02111, United States; Million Veteran Program (MVP) Coordinating Center, VA Boston, Boston, MA 02111, United States; Division of Aging, Department of Medicine, Brigham and Women’s Hospital, Boston, MA 02115, United States; Department of Medicine, Harvard Medical School, Boston, MA 02115, United States.
David R Gagnon, VA Boston Healthcare System, Boston, MA 02111, United States; Department of Biostatistics, Boston University School of Public Health, Boston, MA 02118, United States.
Tianxi Cai, VA Boston Healthcare System, Boston, MA 02111, United States; Department of Biomedical Informatics, Harvard Medical School, Boston, MA 02115, United States; Department of Biostatistics, Harvard T. H. Chan School of Public Health, Boston, MA 02115, United States.
Katherine P Liao, VA Boston Healthcare System, Boston, MA 02111, United States; Department of Medicine, Harvard Medical School, Boston, MA 02115, United States; Division of Rheumatology, Inflammation, and Immunity, Brigham and Women’s Hospital, Boston, MA 02115, United States; Department of Biomedical Informatics, Harvard Medical School, Boston, MA 02115, United States.
Rachel B Ramoni, Office of Research and Development, Veterans Health Administration, Washington, DC 20002, United States.
J Michael Gaziano, VA Boston Healthcare System, Boston, MA 02111, United States; Million Veteran Program (MVP) Coordinating Center, VA Boston, Boston, MA 02111, United States; Division of Aging, Department of Medicine, Brigham and Women’s Hospital, Boston, MA 02115, United States; Department of Medicine, Harvard Medical School, Boston, MA 02115, United States.
Sumitra Muralidhar, Office of Research and Development, Veterans Health Administration, Washington, DC 20002, United States.
Kelly Cho, Centralized Interactive Phenomics Resource (CIPHER), Office of Research and Development, Veterans Health Administration, Washington, DC 20002, United States; VA Boston Healthcare System, Boston, MA 02111, United States; Million Veteran Program (MVP) Coordinating Center, VA Boston, Boston, MA 02111, United States; Division of Aging, Department of Medicine, Brigham and Women’s Hospital, Boston, MA 02115, United States; Department of Medicine, Harvard Medical School, Boston, MA 02115, United States.
Author contributions
Jacqueline Honerlaw and Yuk-Lam Ho equally contributed to the conceptualization, data curation, methodology, visualization, working draft, and final approval. Francesca Fontin, Michael Murray, Ashley Galloway, Jeffrey Gosian, Monika Maripuri, John Russo, Rahul Sangar, Vidisha Tanukonda, and Edward Zielinski contributed to data curation, methodology, and manuscript editing. David Heise, Keith Connatser, and Laura Davies contributed to methodology and manuscript editing. Vidul A. Panickan and Su-Chun Cheng contributed to methodology. Andrew J. Zimolzak, Tianxi Cai, and Katherine P. Liao contributed to methodology and editing. Stacey B. Whitbourne and Maureen Dubreuil contributed to manuscript editing. David R. Gagnon and J. Michael Gaziano contributed to conceptualization and methodology. Sumitra Muralidhar and Rachel B. Ramoni provided program resources, project administration, edits, and final approval. Kelly Cho led manuscript supervision, contributed to conceptualization, methodology, project administration, visualization, manuscript drafting, editing, and final approval.
Funding
This work was supported by the Department of Veterans Affairs Office of Research and Development, the Office of Science of the US Department of Energy under Contract No. DE-AC05-00OR22725 and the Department of Veterans Affairs Office of Information Technology Inter-Agency Agreement with the Department of Energy under IAA No. VA118-16-M-1062.
Conflict of interests
The authors have no competing interests to declare.
Data availability
The CIPHER phenotype library and tools described in this article are available at https://phenomics.va.ornl.gov.
References
- 1. Slaby I, Hain HS, Abrams D, et al. An electronic health record (EHR) phenotype algorithm to identify patients with attention deficit hyperactivity disorders (ADHD) and psychiatric comorbidities. J Neurodev Disord. 2022;14(1):37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Psaty BM, Delaney JA, Arnold AM, et al. Study of cardiovascular health outcomes in the era of claims data: the cardiovascular health study. Circulation. 2016;133(2):156-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Huang H, Turner M, Raju S, et al. Identification of acute decompensated heart failure hospitalizations using administrative data. Am J Cardiol. 2017;119(11):1791-1796. [DOI] [PubMed] [Google Scholar]
- 4. Alexander N, Alexander DC, Barkhof F, Denaxas S.. Identifying and evaluating clinical subtypes of Alzheimer's disease in care electronic health records using unsupervised machine learning. BMC Med Inform Decis Mak. 2021;21(1):343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hellwege JN, Dorn C, Irvin MR, et al. Predictive models for abdominal aortic aneurysms using polygenic scores and PheWAS-derived risk factors. Pac Symp Biocomput. 2023;28:425-436. [PMC free article] [PubMed] [Google Scholar]
- 6. Denny JC, Bastarache L, Ritchie MD, et al. Systematic comparison of phenome-wide association study of electronic medical record data and genome-wide association study data. Nat Biotechnol. 2013;31(12):1102-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Imran TF, Posner D, Honerlaw J, et al. A phenotyping algorithm to identify acute ischemic stroke accurately from a national biobank: the Million Veteran Program. Clin Epidemiol. 2018;10:1509-1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Liao KP, Sun J, Cai TA, et al. High-throughput multimodal automated phenotyping (MAP) with application to PheWAS. J Am Med Inform Assoc. 2019;26(11):1255-1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhang HG, Honerlaw JP, Maripuri M, et al. Potential pitfalls in the use of real-world data for studying long COVID. Nat Med. 2023;29(5):1040-1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Harrington KM, Quaden R, Stein MB, et al. Validation of an electronic medical record-based algorithm for identifying posttraumatic stress disorder in U.S. veterans. J Trauma Stress. 2019;32(2):226-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kirby JC, Speltz P, Rasmussen LV, et al. PheKB: a catalog and workflow for creating electronic phenotype algorithms for transportability. J Am Med Inform Assoc. 2016;23(6):1046-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Denaxas S, Gonzalez-Izquierdo A, Direk K, et al. UK phenomics platform for developing and validating electronic health record phenotypes: CALIBER. J Am Med Inform Assoc. 2019;26(12):1545-1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chapman M, Mumtaz S, Rasmussen LV, et al. Desiderata for the development of next-generation electronic health record phenotype libraries. Gigascience. 2021;10(9):giab059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Spotnitz M, Acharya N, Cimino JJ, et al. A metadata framework for computational phenotypes. JAMIA Open. 2023;6(2):ooad032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Almowil Z, Zhou SM, Brophy S, Croxall J.. Concept libraries for repeatable and reusable research: qualitative study exploring the needs of users. JMIR Hum Factors. 2022;9(1):e31021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Honerlaw J, Ho YL, Fontin F, et al. Framework of the Centralized Interactive Phenomics Resource (CIPHER) standard for electronic health data-based phenomics knowledgebase. J Am Med Inform Assoc. 2023;30(5):958-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gaziano JM, Concato J, Brophy M, et al. Million Veteran Program: a mega-biobank to study genetic influences on health and disease. J Clin Epidemiol. 2016;70:214-223. [DOI] [PubMed] [Google Scholar]
- 18. Knight KE, Honerlaw J, Danciu I, et al. Standardized architecture for a mega-biobank phenomic library: the Million Veteran Program (MVP). AMIA Jt Summits Transl Sci Proc. 2020;2020:326-334. [PMC free article] [PubMed] [Google Scholar]
- 19. Hong C, Rush E, Liu M, et al. Clinical knowledge extraction via sparse embedding regression (KESER) with multi-center large scale electronic health record data. NPJ Digit Med. 2021;4(1):151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Department of Veterans Affairs. 2023. Accessed September 19, 2023. https://www.mvp.va.gov/pwa/.
- 21. Verma A, Huffman JE, Rodriguez A, et al. 2023. Diversity and scale: genetic architecture of 2,068 traits in the VA Million Veteran Program. medRxiv;2023.06.28.23291975, June 29, 2023, preprint: not peer reviewed. [DOI] [PubMed]
- 22. National Institutes of Health. 2023. Accessed September 19, 2023. https://grants.nih.gov/grants/guide/notice-files/NOT-OD-21-013.html
- 23. Stang PE, Ryan PB, Racoosin JA, et al. Advancing the science for active surveillance: rationale and design for the observational medical outcomes partnership. Ann Intern Med. 2010;153(9):600-606. [DOI] [PubMed] [Google Scholar]
- 24. Observational Health Data Sciences and Informatics (OHDSI). 2023. Accessed September 19, 2023. https://github.com/OHDSI/PhenotypeLibrary
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The CIPHER phenotype library and tools described in this article are available at https://phenomics.va.ornl.gov.