Abstract
The enhancer sequences in the Moloney murine leukemia virus (M-MuLV) long terminal repeat (LTR) are of considerable interest since they are crucial for virus replication and the ability of the virus to induce T lymphomas. While extensive studies have identified numerous nuclear factors that can potentially bind to M-MuLV enhancer DNA in vitro, it has not been made clear which of these factors are bound in vivo. To address this problem, we carried out in vivo footprinting of the M-MuLV enhancer in infected cells by in vivo treatment with dimethyl sulfate (DMS) followed by visualization through ligation-mediated PCR (LMPCR) and gel electrophoresis. In vivo DMS-LMPCR footprinting of the upstream LTR revealed evidence for factor binding at several previously characterized motifs. In particular, protection of guanines in the central LVb/Ets and Core sites within the 75-bp repeats was detected in infected NIH 3T3 fibroblasts, Ti-6 lymphoid cells, and thymic tumor cells. In contrast, factor binding at the NF-1 sites was found in infected fibroblasts but not in T-lymphoid cells. These results are consistent with the results of previous experiments indicating the importance of the LVb/Ets and Core sequences for many retroviruses and the biological importance especially of the NF-1 sites in fibroblasts and T-lymphoid cells. No evidence for factor binding to the glucocorticoid responsive element and LVa sites was found. Additional sites of protein binding included a region in the GC-rich sequences downstream of the 75-bp repeats (only in fibroblasts), a hypersensitive guanine on the minus strand in the LVc site (only in T-lymphoid cells), and a region upstream of the 75-bp repeats. These experiments provide concrete evidence for the differential in vivo binding of nuclear factors to the M-MuLV enhancers in different cell types.
Moloney murine leukemia virus (M-MuLV) is a simple retrovirus that has been extensively studied in terms of molecular biology and leukemogenesis. M-MuLV induces T lymphoma when inoculated into neonatal mice. The integrated proviral DNA form of M-MuLV contains long terminal repeats (LTRs) at either end that are generated during the process of reverse transcription. As for all retroviruses, the M-MuLV LTR contains the sequences that govern viral transcription. In particular, the U3 region of the M-MuLV LTR contains a strong transcriptional enhancer that is located approximately 150 bp upstream from the start site of transcription and consists of two directly repeated 75-bp sequences. As in many viral and cellular enhancer elements, each 75-bp repeat contains binding sites for multiple nuclear proteins (see Fig. 1). These binding factors have been defined and characterized by in vitro methods involving incubation of naked DNAs with nuclear extracts or purified proteins (11, 26, 40). Such experiments have identified a complex array of proteins that bind to the M-MuLV enhancer (1, 13, 14, 26, 35, 43, 44, 46–48). The facts that some enhancer sequences have been shown to bind multiple proteins and that binding sites for other proteins overlap previously characterized motifs make it unlikely that all of the known factors simultaneously bind the M-MuLV enhancers in any given infected cell. A better understanding of the biology of M-MuLV will require an understanding of which motifs in the M-MuLV enhancers are occupied by nuclear proteins in different infected cell types. It seems likely that M-MuLV proviral enhancers in different cell types may bind different nuclear factors.
FIG. 1.
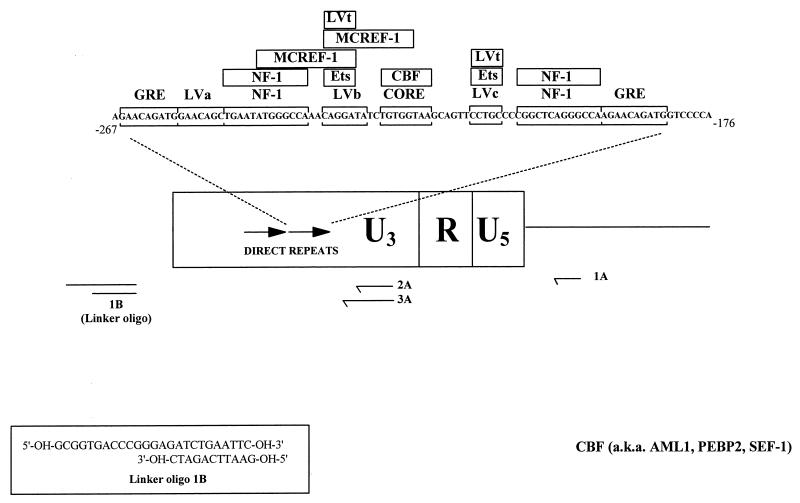
Nuclear factor binding sites in the M-MuLV enhancers. A schematic representation of the M-MuLV LTR with the nuclear protein binding sites (bracketed sequences) and sequence-specific DNA proteins known to bind to them (in boxes) in one copy of the 75-bp direct repeats is shown. (These data are taken from Manley et al. [26]). The nested oligonucleotide primer set (1A, 2A, and 3A) used for LMPCR is shown below. Oligonucleotide 1A enabled preferential analysis of the 5′ LTR without interference from signal due to binding sites at the 3′ LTR. The sequence of the linker oligonucleotide used in the LMPCR (1B) is also shown. Oligo, oligonucleotide.
Previous experiments have identified the LTR enhancers as primary determinants of M-MuLV pathogenesis. Chimeras between M-MuLV and related viruses of different disease specificities (e.g., Friend MuLV [erythroleukemia specific]) or pathogenic potentials (e.g., endogenous ecotropic MuLV [nonleukemogenic]) (2, 3, 5–7, 19, 22, 24) showed that both the disease specificity and leukemogenic potential are largely determined by the enhancers of the different viruses. Further mutagenesis studies implicated individual nuclear protein binding sites within the 75-bp repeats in M-MuLV leukemogenesis. Speck et al. (41) demonstrated that mutations in most binding sites simply increase the time of disease onset without altering disease specificity; however, mutations in the central LVb/Ets and Core elements relaxed the T-cell disease specificity such that a high percentage of erythroleukemias resulted. The importance of the central LVb/Ets-Core region for viral replication was also confirmed by sequence alignment; the LVb/Ets-Core region is highly conserved among 35 members of the type C retrovirus family (12).
Advances have been made in the purification, cloning, and characterization of proteins that bind to the M-MuLV enhancer elements (1, 13, 14, 26, 35, 43, 44, 46–48). For instance, the LVb site has been shown to bind many proteins of the Ets transcription factor family, including Ets-1 and Ets-2 (30), LVt (41), GA binding protein, Fli-1, and a yet to be identified factor the size of Elf-1 (13). In addition, several proteins have been shown to bind the Core site, including activating protein 3 (27), the CAAT-enhancer binding protein (20), and the Core binding factor (CBF), which consists of a heterodimer between AML1 (CBF-α) and CBF-β (25, 48). CBF has been independently characterized for other viral systems and is also known as the SL3 and AKV Core binding factor (1), the polyomavirus enhancer binding protein 2 (38), and the SL3 enhancer factor 1 (45). However, the actual proteins that bind to these individual motifs in infected cells remain to be elucidated for T-lymphoid cells (the target cells for leukemogenesis) and for the cells that are infected by M-MuLV in vivo or in vitro (e.g., fibroblasts).
In this study, we have used in vivo genomic footprinting by dimethyl sulfate (DMS) treatment and ligation-mediated PCR (LMPCR) to investigate which enhancer binding sites are occupied in infected fibroblasts, T lymphocytes, and virus-induced primary thymic tumor cells. We found hallmarks for in vivo protein interactions at the LVb/Ets and Core sites in all cell types analyzed. In addition, striking footprints at the NF-1 sites were observed in M-MuLV-infected fibroblasts but they were absent in infected lymphoid cells. The results were consistent with genetic evidence for the central importance of the LVb/Ets and Core regions in M-MuLV replication, and they indicate that different arrays of proteins bind to the M-MuLV enhancers in different cell types.
MATERIALS AND METHODS
Cell lines.
In vivo footprinting was performed on the following cell lines: uninfected NIH 3T3 fibroblasts; 43-D, an NIH 3T3 fibroblast cell line productively infected with M-MuLV (23); and Ti-6–M-MuLV (34), a mouse T-cell lymphoma likewise productively infected with M-MuLV. The fibroblast cell lines were grown in Dulbecco modified Eagle medium with the addition of 10% calf serum. The Ti-6–M-MuLV cell line was grown in RPMI 1640 plus 10% fetal bovine serum.
In vivo DMS treatment of M-MuLV-infected cells. (i) DMS treatment of M-MuLV-infected adherent fibroblasts.
To achieve partial methylation of guanines within infected cells in vivo, two 15-cm-diameter plates of subconfluent 43-D cultures were treated with 1% DMS in growth medium at 37°C for 2 min. The treatment was stopped by aspiration of the DMS-containing medium, followed by an immediate rinse with 25 ml of phosphate-buffered saline (PBS) prewarmed to 37°C and two successive 30-s washes with 25 ml of PBS at 37°C. Genomic DNA was harvested by the addition of 3 ml of cell lysis solution (300 mM NaCl, 50 mM Tris-Cl [pH 8.0], 25 mM EDTA [pH 8.0], 0.2% [vol/vol] sodium dodecyl sulfate, 0.2 mg of proteinase K per ml); this mixture was allowed to incubate for 5 min before the cell slurry was removed into a test tube by gentle scraping. As a control, genomic DNA was also harvested from untreated 43-D cultures in parallel. Both samples were incubated at 37°C from 4 h to overnight, after which the genomic DNA was phenol-chloroform extracted and ethanol precipitated.
(ii) DMS treatment of M-MuLV-infected T-lymphoid suspension cultures.
Cells in exponentially growing suspension cultures of M-MuLV-infected Ti-6 cells were counted and harvested by centrifugation at 500 × g. To obtain partial DMS methylation in vivo, three individual samples of 108 cells each were treated with 1 ml of growth medium containing 1% DMS for 1 min at 37°C. Exposure to DMS was stopped by the addition of 49 ml of ice-cold PBS followed by immediate low-speed centrifugation. Residual DMS was removed by an additional PBS wash. The pelleted cells were resuspended in 0.3 ml of PBS, and the genomic DNA was obtained by the addition of 2.7 ml of the cell lysis solution. DNA was also harvested from untreated cells and processed in parallel.
(iii) DMS treatment of M-MuLV-induced thymic tumor cells.
Moribund M-MuLV-infected mice were euthanized, and tumor cell suspensions were obtained from enlarged thymic tissue by extrusion through a stainless steel mesh into ice-cold RPMI 1640 containing 10% fetal bovine serum. The thymic tumor cells were counted, and 108-cell samples were made in triplicate and immediately treated with DMS as described above for the infected Ti-6 cultures.
In vitro DMS treatment of DNA.
Extracted genomic DNA from control cultures was subjected to DMS treatment in vitro by incubation with 1% DMS in H2O for 1 min at 25°C. The reaction was stopped by the addition of ice-cold DMS stop buffer (1.5 M sodium acetate [pH 7.0], 1 M β-mercaptoethanol, 100 μg of Saccharomyces cerevisiae tRNA per ml), which was immediately followed by the addition of 2.5 volumes of ethanol on dry ice. Samples were precipitated by incubation for at least 30 min at −70°C and pelleted by microcentrifugation for 15 min at 4°C. DNA pellets were allowed to air dry for 10 min and resuspended in 200 μl of 1 M piperidine in H2O for 15 min at room temperature prior to cleavage.
Piperidine cleavage.
Following in vivo and in vitro DMS treatment, extracted genomic DNA from each cell type was cleaved at all methylated guanines by incubation in 200 μl of 1 M piperidine for 30 min at 90°C. The piperidine was removed by lyophilization, and the cleaved DNA pellets were resuspended in 360 μl of TE buffer (10 mM Tris-Cl, 1 mM EDTA; pH 7.5). Residual piperidine was removed by two successive ethanol precipitations. The first entailed addition of 40 μl of 3 M sodium acetate followed by 1 ml of 100% ethanol and incubation for 30 min at −70°C. DNA samples were pelleted by microcentrifugation for 15 min at 4°C and resuspended in 500 μl of TE buffer. The DNA pellets were ethanol precipitated a second time by the addition of 170 μl of 8 M ammonium acetate and 670 μl of isopropanol and incubation for at least 30 min at −70°C. The precipitated samples were pelleted by microcentrifugation as described above, washed with 500 μl of 75% ethanol, and microcentrifuged for 5 min at room temperature. The resulting DNA pellets were resuspended in double-distilled water to a final concentration of 0.4 μg/μl.
LMPCR.
Two micrograms of DMS-treated and piperidine-cleaved genomic DNA was used for LMPCR as described previously (10, 29, 32) with minor modifications. Single-stranded DNA fragments with guanine residues at both termini result from the DMS treatment and piperidine cleavage. To provide appropriate substrates for linker ligation, double-stranded, blunt-ended molecules were generated by primer extension from an M-MuLV-specific oligonucleotide (oligonucleotide 1A) (see Fig. 1). This first-strand primer extension was accomplished by incubation of 2 μg of DMS-treated and piperidine-cleaved DNA with 0.3 pmol of oligonucleotide 1A, 1× Vent DNA polymerase buffer (New England Biolabs), 4 mM MgSO4, 0.25 mM each deoxynucleoside triphosphate, and 0.5 U of Vent DNA polymerase (New England Biolabs) in a total volume of 30 μl. The DNA was denatured at 95°C for 5 min, annealed by incubation at 55°C for 20 min, and extended by a subsequent incubation of 10 min at 72°C. Ligation of the unidirectional linker described by Mueller and Wold (linker oligonucleotide 1B) (29) (see Fig. 1) was completed by the addition of 20 μl of 110 mM Tris-Cl (pH 7.5)–17.5 mM MgCl2–50 mM dithiothreitol, 25 μl of 10 mM MgCl2–20 mM DTT–3 mM ATP (pH 7.0)–4 μM unidirectional linker (in 50 mM Tris-Cl [pH 7.7]), and 3 U of T4 DNA ligase (Gibco/BRL). This mixture was incubated at 17°C overnight, after which the DNA was recovered by ethanol precipitation. The precipitated DNA pellet was resuspended in 50 μl of H2O, and PCR amplification was accomplished by the addition of 50 μl of 2× Vent buffer–8 mM MgSO4–5 mM deoxynucleoside triphosphate mix–1 pmol of M-MuLV oligonucleotide 2A (Fig. 1)–1 pmol of oligonucleotide LMPCR.1–1 U of Vent DNA polymerase. These samples were placed in a thermocycler and cycled 17 times with a profile of 95°C for 1 min, 66°C for 2 min, and 72°C for 1 min, with a final extension of 10 min at 72°C. Following amplification, M-MuLV-specific PCR products were labeled by the addition of 5 μl of labeling buffer (2 mM each deoxynucleoside triphosphate, 1× Vent polymerase buffer, 8 mM MgSO4, 1 U of Vent polymerase, 2.3 pmol of an M-MuLV-specific 32P-end-labeled oligonucleotide [oligonucleotide 3A] [Fig. 1]) and subjected to two rounds of 95°C for 1 min, 69°C for 2 min, and 72°C for 1 min. Each reaction mixture was then subjected to phenol-chloroform extraction and ethanol precipitation prior to electrophoresis on a 6% sequencing polyacrylamide gel. The reactions were visualized by autoradiography with Kodak BioMax MR film and also by PhosphorImagery on a Molecular Dynamics 445 SI PhosphorImager.
The oligonucleotide sequences of the nested M-MuLV primer set used for the analysis of the proviral sense strand by LMPCR were oligonucleotide 1A, 5′-TCTCCCGATCCCGGACGA-3′; oligonucleotide 2A, 5′-GGGGCACCCTGGAAACATCTGATGGT-3′; and oligonucleotide 3A, 5′-GGGGCACCCTGGAAACATCTGATGGTTCT-3′. Oligonucleotide 1A is complementary to nucleotides +170 to +153 in the M-MuLV genome, while oligonucleotides 2A and 3A are complementary to nucleotides −118 to −143 and −118 to −146 in the M-MuLV LTR, respectively. Oligonucleotide sequences of the nested primer set used to analyze the minus strand by LMPCR were oligonucleotide 1C, 5′-GACCCCACCTGTAGGTTTGGC-3′ (from −442 to −422); oligonucleotide 2C, 5′-GGCAAGCTAGCTTAAGTAACGCCATTTTGC-3′ (from −424 to −395); and oligonucleotide 3C, 5′-GCAAGCTAGCTTAAGTAACGCCATTTTGCAAGG-3′ (from −423 to −392). The unidirectional linker oligonucleotide sequences have been described by Mueller and Wold (29) and are as follows: LMPCR.1, 5′-GCGGTGACCCGGGAGATCTGAATTC-3′, and LMPCR.2, 5′-GAATTCAGATC-3′.
RESULTS
The nuclear factor binding sites within the M-MuLV direct repeats identified in previous studies are shown in Fig. 1. These sites were identified by incubation in vitro of different DNA fragments with purified proteins or nuclear extracts from different cell types. As shown, at least eight sites for protein binding have been mapped to each copy of the 75-bp repeats. The central LVb/Ets-Core motif is highly conserved in the enhancers of many retroviruses (12), which has suggested that factors bound at these sites are key for enhancer function for these viruses.
To determine those sites within the M-MuLV enhancers that bind nuclear factors in vivo, we employed in vivo footprinting using DMS and LMPCR (see Materials and Methods for details) (10, 28, 29, 31, 36, 37). In this technique, M-MuLV-infected cells were treated in vivo with DMS, which resulted in partial methylation of guanines at the N-7 position. DNA was then extracted from the cells and cleaved at the methylated bases by treatment with piperidine. A primer extension reaction with an M-MuLV-specific oligonucleotide (1A in Fig. 1) resulted in synthesis of a series of double-stranded DNA fragments terminated at formerly methylated guanines within M-MuLV DNA. Blunt-end ligation of a double-stranded primer (oligonucleotide 1B in Fig. 1) yielded a series of fragments that could be extended and further amplified by nested PCR (primer 2A in Fig. 1). The PCR products were labeled by a final primer extension reaction with a 32P-end-labeled M-MuLV-specific oligonucleotide (primer 3A) and visualized by polyacrylamide gel electrophoresis on a DNA sequencing gel followed by autoradiography or by PhosphorImaging. For comparison, DNA was extracted from the same infected cells and the naked DNA was treated in vitro with DMS and processed in parallel.
The results of in vivo footprinting are analogous to the results of in vitro footprinting whereby proteins are bound to specific double-stranded DNAs in vitro and then treated with DMS. If proteins are bound to a specific region of DNA, the guanines within the binding site are protected from DMS methylation. This protection is reflected in specific decreases in the intensities of fragments corresponding to guanines on the sequencing gel relative to the corresponding intensities of guanines of naked DNA treated with DMS. In addition, hypersensitivity to DMS methylation is also observed at specific bases where proteins are bound to DNA. This may have resulted from an increase in the local concentration of DMS created by hydrophobic pockets at the interface between a globular protein domain and DNA (18). In addition, increased DMS access can result from alterations in the local DNA topology induced by protein binding.
In vivo footprinting of M-MuLV proviruses in infected fibroblasts.
The initial in vivo footprinting was carried out on NIH 3T3 fibroblasts productively infected with a molecular clone of M-MuLV (23). The 43-D cells contain multiple copies of provirus and produce high levels of infectious M-MuLV. The 1A, 2A, and 3A oligonucleotide primers were designed for LMPCR analysis of the upper (sense) strand of the M-MuLV LTR. The location of the initial extension oligonucleotide primer (1A) was in viral sequences outside the LTR. Thus, only PCR amplification from the upstream M-MuLV LTR took place and the resulting in vivo footprints reflected proteins bound to the upstream LTR only.
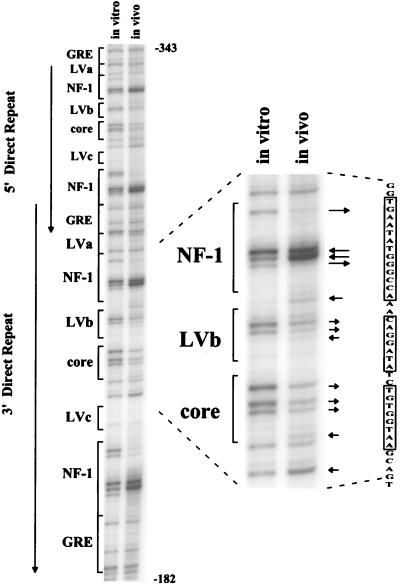
A representative autoradiogram from the in vivo footprint analysis of 43-D cells by LMPCR is shown in Fig. 2. As predicted, the lane containing naked DNA treated with DMS in vitro followed by LMPCR gave a fragment pattern corresponding to an M-MuLV-specific guanine sequencing ladder generated by chemical cleavage (29). In addition, LMPCR amplification from uninfected-cell DNA gave no labeled fragments, which confirmed that M-MuLV proviruses were being analyzed (not shown). As shown, comparison between the in vivo-treated and in vitro-treated DNA samples revealed several footprints in the M-MuLV direct repeats. The most visible footprints were evident at the four NF-1 sites (two in each repeat). The upstream NF-1 sites in both repeats contained two strongly hypersensitive central guanines as well as two strongly protected guanines. The downstream NF-1 sites showed essentially the same pattern of protection and hypersensitivity, with the exception of having three protected guanines instead of two; this was due to the presence of one more guanine in the downstream sites.
FIG. 2.
In vivo DMS footprinting of the M-MuLV 5′ LTR in fibroblasts. An autoradiogram from gel electrophoresis of labeled LMPCR products obtained from M-MuLV-infected 43-D fibroblasts is shown. The autoradiogram is representative of multiple analyses with the same nested oligonucleotide primer set. The portion of the gel shown corresponds to nucleotides −182 to −343 in the upstream M-MuLV LTR. The relative positions of previously characterized nuclear protein binding sites are indicated on the left. Both 75-bp direct repeats of the M-MuLV enhancer are displayed, and the region of interest is expanded at the right. The boxed sequences at the far right correspond to the NF-1, LVb, and Core sites. Comparisons between the in vitro-DMS-treated DNA control in the lane to the left and the in vivo-DMS-treated sample in the lane to the right indicated protection of certain guanine bases and hypersensitivity of other guanine bases in the infected cells. Guanine-specific protection is indicated by arrows pointing away from bands, and guanine or adenine hypersensitivity to DMS is indicated by arrows pointing towards the bands. Other investigators have previously reported hypersensitivity of adenines in in vivo DMS-LMPCR footprinting. The lengths of the arrows indicate the relative magnitudes of the protection or hypersensitivity.
Footprints were also observed over the central LVb/Ets and Core sites as well. The LVb site showed protection of the two central guanines, as well as hypersensitivity of an adjacent adenine and an adenine at the upstream boundary with the NF-1 site. All three guanines in the Core site showed protection as well. The protection of the guanines in the LVb/Ets and Core sites was not absolute, which may have resulted from the sites in some proviruses not being occupied (see Discussion). Similar results were obtained in three independent experiments.
The results indicated that there are proteins bound in vivo at the NF-1, LVb/Ets, and Core sites in the M-MuLV 75-bp repeats in fibroblasts. Moreover, in vivo footprints were not detected at any of the glucocorticoid responsive element (GRE) sites or LVa sites, suggesting that they were not occupied. Occupation of the LVc/Ets site was difficult to ascertain with the sense-strand-specific primer set. The one guanine in this site was methylated poorly even in naked DNA, and in vivo footprinting provided no evidence for further protection at this site in infected cells. Even though the complementary DNA strand contains two centrally located guanines at the LVc/Ets site, no evidence for protein interactions at these bases was obtained when this strand was analyzed (see below). On the other hand, a guanine between the LVc/Ets site and the upstream Core site did show enhanced methylation on the sense strand. This may have resulted from protein binding to the Core site or from a conformational change in the DNA due to binding at a distant site.
In vivo footprinting of M-MuLV proviruses in infected T-lymphoid cells.
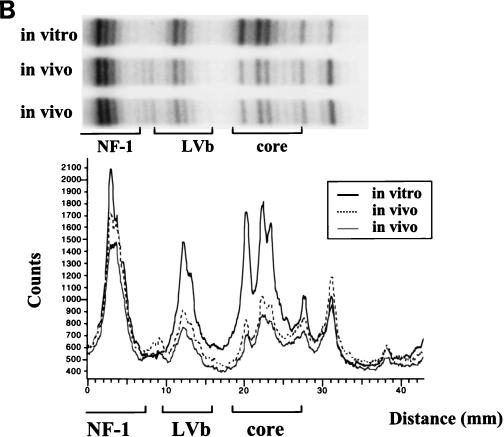
Since M-MuLV induces T-lymphoid tumors in infected mice, and it has also been shown that the M-MuLV enhancers are preferentially active in T-lymphoid cells (39, 42), the nature of proteins bound to the M-MuLV enhancers in T cells was of great interest. Initially, we tested Ti-6 cells infected with M-MuLV. These cells are derived from a chemically induced T lymphoma (34) and are readily infectable by M-MuLV. As shown in Fig. 3A, in vivo DMS footprinting and LMPCR indicated that the LVb/Ets and Core sites in the M-MuLV enhancers were occupied. The patterns of protection observed for these two sites were the same as those observed in the infected 43-D fibroblasts, although the degrees of protection were greater in infected Ti-6 cells than in 43-D fibroblasts. The hypersensitive guanine between the Core and LVc/Ets sites was also evident. However, in contrast to what occurs in 43-D cells, no evidence for in vivo protein binding at the NF-1 sites was found for the infected Ti-6 cells. Neither the hypersensitive nor the protected guanines in these sites were observed. Densitometric scanning of the sequencing gels confirmed the protection in the LVb/Ets and Core sites and the lack of protection or hypersensitivity of the NF-1 sites (Fig. 3B). Thus, M-MuLV proviruses in NIH 3T3 and Ti-6 cells differed in the proteins bound to the upstream enhancers. Proviruses in both cells showed binding at the central LVb/Ets and Core sites, but protein was bound at the NF-1 sites only in the infected fibroblasts.
FIG. 3.
In vivo DMS footprinting of the M-MuLV 5′ LTR in infected lymphoid cells. (A) In vivo DMS-LMPCR footprinting analogous to that described for Fig. 2 was carried out with M-MuLV-infected Ti-6 lymphoid cells. Infected Ti-6 cells were subjected to LMPCR after two independent in vivo DMS treatments. In this figure, the LMPCR fragments were visualized by PhosphorImaging. The same convention as that described for Fig. 2 was used, with arrows indicating protected and hypersensitive sites. (B) Digital densitometric analysis of the PhosphorImaged sequencing gel in panel A was performed. The positions of the NF-1, LVb/Ets, and Core sites are indicated. The portion of the Ti-6 gel analyzed is aligned above the graph.
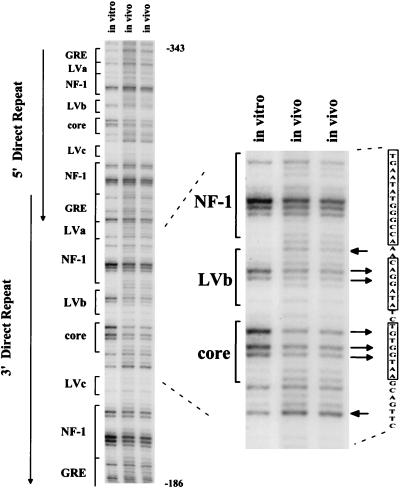
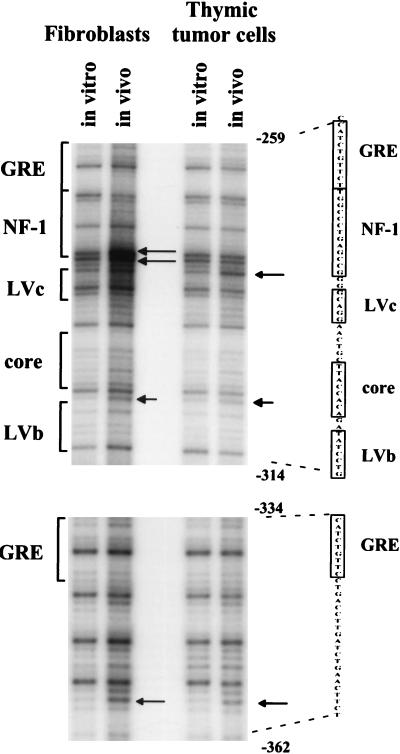
We also directly examined proviruses in primary M-MuLV-induced thymic tumor cells. Moribund mice were sacrificed and single-cell suspensions were prepared from thymic tumors. The suspended cells were then directly treated with DMS and analyzed as described above. As shown in Fig. 4, the in vivo footprint for the primary tumors was the same as for the infected Ti-6 cell line. Thus, the two predominant sites occupied in M-MuLV proviruses in thymic tumors are the central LVb/Ets and Core sites. It should be noted that the in vivo-treated tumor DNA samples had an overall background with low levels of LMPCR fragments corresponding to many nucleotides.
FIG. 4.
In vivo DMS footprinting of the M-MuLV 5′ LTR in primary M-MuLV-induced tumor cells. M-MuLV-induced T-lymphoma cells were treated in vivo with DMS and analyzed in the same manner as described for the 43-D and infected Ti-6 cells shown in Fig. 2 and 3. PhosphorImaging was employed to visualize data. In this experiment, the intensity of the entire in vitro lane was uniformly decreased to equalize the amounts of samples loaded and to aid in footprint visualization.
In vivo footprinting in the GC-rich sequences.
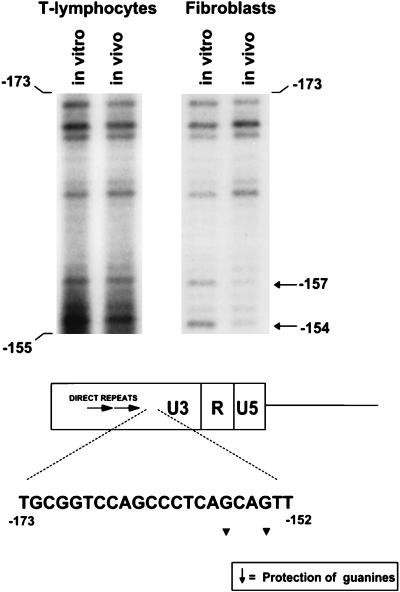
We previously described genetic evidence for the importance of a GC-rich region downstream from the 75-bp repeats for M-MuLV pathogenesis. When this region (−174 to −151) of the M-MuLV LTR was deleted, the disease specificity of M-MuLV was relaxed to include 42% erythroid and myeloid leukemias (15). This result suggested that tissue-specific factors binding to the GC-rich sequences contribute to the T-lymphoid cell specificity of the M-MuLV LTR. Thus, in vivo footprinting of the GC-rich region was of interest. By in vivo footprinting, we found evidence for protein interactions in a novel region located at the downstream border of the GC-rich sequences (Fig. 5). Two guanines at positions −157 and −154 were differentially protected from in vivo methylation within the sequence CAGCAG in fibroblasts but not in the infected T-cell line or primary thymic tumor cells. Although the symmetry of this region is suggestive of a protein recognition sequence, it did not correspond to any known binding sites in the TRANSFAC DNA-binding protein database (16). However, the protected guanine located at position −154 was at the beginning of the sequence AGTTTC, which was found to be highly conserved in 35 members of the type C retrovirus family (12). In addition, the two protected bases are contained within the sequence CAGCAGTTT, which corresponds to a half-site for the MLpal(MCF-13LTR palindrome) binding factor that has been noted in several MuLV LTRs (4, 50).
FIG. 5.
In vivo DMS footprinting in the GC-rich sequences. The regions from DMS footprinting gels analogous to those shown in Fig. 2 and 4 and corresponding to the GC-rich sequences downstream of the 75-bp repeats are shown for thymic tumor DNA and 43-D infected fibroblasts. Evidence for protein interactions at positions −153 and −155 in the sequence CAGCAG was obtained for fibroblasts but not for M-MuLV-infected Ti-6 cells or the primary thymic tumor cells. Infected Ti-6 cells showed the same pattern as the thymic tumor cells (no protection [data not shown]). The relative positions of the protected bases in the GC-rich sequences are shown.
A summary of the in vivo footprints of the 75-bp repeats for the sense strand of M-MuLV proviruses in fibroblasts and T-lymphoid cells is shown in Fig. 6. The footprints at the LVb/Ets and Core sites were the same in infected NIH 3T3 cells, Ti-6 cells, and M-MuLV-induced thymic tumor cells. In addition, both NF-1 sites in each repeat showed strong footprints in infected NIH 3T3 cells (43-D) but not in the lymphoid cells.
FIG. 6.
Summary of DNA-protein interactions on the sense strand obtained by in vivo footprinting by LMPCR. The same convention described for Fig. 2 was used to summarize all interactions observed in Fig. 2 to 5. Arrows in the LVb-Core region represent interactions observed in both lymphoid cells and fibroblasts infected with M-MuLV; those pointing upward indicate sites hyperactive to DMS, and those pointing downward indicate protection of specific guanines. Other interactions were observed only in M-MuLV-infected fibroblasts.
Comparison of in vivo footprinting and in vitro methylation interference.
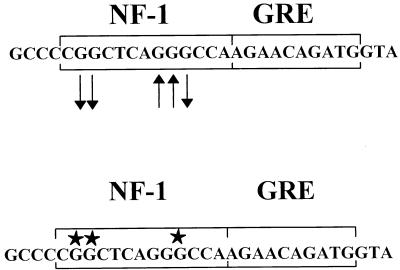
In light of the detection of in vivo footprints in the M-MuLV proviruses studied, it was of interest to determine if the in vivo DMS footprinting detected protection at the same bases shown to be protein-DNA contact points by in vitro methylation interference reactions. Figure 7 shows a comparison between the in vivo DMS footprints of the downstream NF-1 sites detected in Fig. 2 and the results obtained by Speck and Baltimore (40) for in vitro methylation interference of the same DNA sequence with extracts from WEHI 231 cells (a mouse B-lymphoid cell line). The same three guanines that were shown to be protein contact points by in vitro methylation interference showed protection from methylation in the in vivo footprinting experiments. Moreover, the two adjacent guanines that were not protein-DNA contact points were hypersensitive to methylation in vivo. Thus, the patterns of in vivo methylation protection and in vitro methylation interference were very consistent and suggested that the same factor (putatively NF-1) detected in the previous in vitro experiments was bound to the NF-1 sites in vivo in 43-D cells.
FIG. 7.
Comparison of in vivo footprints and in vitro methylation interference results. Nuclear protein interactions at the NF-1 site reported by Speck and Baltimore (40) detected by methylation interference with WEHI 231 nuclear extracts are shown in the lower sequence (stars indicate G residues that interacted with protein). The results of in vivo DMS footprinting are shown in the upper sequence. Guanine bases protected in vivo (arrows pointing away from bases) correspond to guanines shown to be protein contact points by methylation interference.
With regard to the LVb/Ets and Core sites, all guanines in these two sites showed protection from methylation in vivo. In in vitro experiments with naked DNA containing the LVb/Ets and Core sites, the same guanines were found to be protein-DNA contact points by methylation interference when nuclear extracts from multiple cell types (including T-lymphoid cells) were used (40). In addition, the pattern of in vivo protection at the LVb and Core sites obtained here agreed with the in vitro methylation interference data generated when recombinant Ets-1 and CBF were used (49).
In vivo footprinting on the minus strand of the M-MuLV LTR.
The in vivo footprinting analyses described in the preceding paragraphs all characterized protein-DNA contacts on the sense (plus) strands of the M-MuLV enhancers. It was also of interest to examine the minus strands for protein-DNA contacts. However, there were two technical issues. First, the known nuclear factor binding sites in the M-MuLV 75-bp repeats have relatively few guanines on the minus strand, so that in vivo DMS-LMPCR footprinting is less likely to detect contacts here than on the plus strand. In addition, it was not possible to design an initial oligonucleotide primer (equivalent to oligonucleotide 1A) (Fig. 1) that would allow analysis of the minus strand in the upstream LTR only.
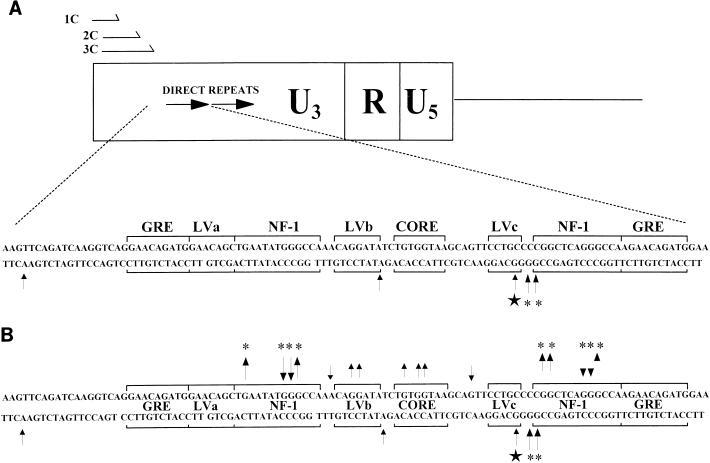
In any event, a set of primers that allowed analysis of the lower strand of the M-MuLV LTR was designed as shown in Fig. 8. These primers were able to produce LMPCR amplification products from both the upstream and downstream LTRs. In vivo DMS-LMPCR footprinting with this primer set was carried out on M-MuLV-infected 43-D fibroblasts and M-MuLV-infected primary thymic tumor cells as shown in Fig. 9. No in vivo DMS protection was detected on the lower strand. However, several minus-strand guanines and adenines showed in vivo hypersensitivity to DMS methylation. Two hypersensitive A bases (one present in each 75-bp repeat) were located between the LVb/Ets and Core sites, and they were detected in both infected fibroblasts and thymic tumor cells. In addition, a hypersensitive adenine was present in sequences upstream of the 75-bp repeats and was detected in both infected lymphocytes and fibroblasts. Two other hypersensitive guanines (one present in each 75-bp repeat) were located on the downstream side of the LVc sites and were present in infected thymic tumor cells but not in infected 43-D fibroblasts. Furthermore, four hypersensitive guanines (two in each 75-bp repeat) were located at the start of the downstream NF-1 sites in fibroblasts and not in T-lymphoid cells. The upstream NF-1 sites in each 75-bp repeat did not show hypersensitive sites at the equivalent positions, most likely due to the absence of guanines at those positions. These results indicated that protein contacts were detected on the lower strand of the M-MuLV LTR, although only hypersensitive sites were observed. Moreover, there was evidence for the presence of an LTR binding factor in M-MuLV-induced primary thymic tumor cells that was not detected in infected fibroblasts. Figure 8B shows a composite of the in vivo DMS footprints for both strands in the enhancer region of the M-MuLV LTR.
FIG. 8.
DNA-protein interactions on the minus strand obtained by in vivo footprinting. (A) The nested oligonucleotide primer set used for the minus-strand in vivo footprinting (1C, 2C, and 3C) is shown. A summary of the minus (lower)-strand bases hypersensitive to in vivo DMS methylation in fibroblasts and primary thymic tumor cells was compiled analogously to that shown in Fig. 6. These results were taken from the in vivo footprinting analysis shown in Fig. 9. Asterisks indicate bases hypersensitive to DMS methylation in infected fibroblasts, the star indicates a hypersensitive base specific to primary thymic tumor cells, and arrows indicate sites hyperactive to DMS. (B) A summary of in vivo protein-DNA interactions observed for both DNA strands is shown. Arrows pointing away from bases on either strand indicate DMS protection, while arrows pointing towards bases indicate DMS-hypersensitive sites. Note that protein interactions on the minus strand were evident only as hypersensitive sites. The data for the sense (upper) strand are specific for the upstream LTR, while the data for the minus strand are composites of the upstream and downstream LTRs. Stars indicate T-lymphoid-cell-specific bases, and asterisks indicate fibroblast-specific bases.
FIG. 9.
In vivo footprinting of the M-MuLV LTR minus strand. Infected 43-D fibroblasts and thymic tumor cells treated in vivo or in vitro with DMS were analyzed by cleavage and LMPCR by using the minus-strand-specific primers shown in Fig. 8A. Arrows indicate bases hypersensitive to in vivo DMS methylation in each respective cell type. DNA templates were the same as those used for the sense-strand analysis.
DISCUSSION
In this study, we analyzed by in vivo DMS footprinting the M-MuLV LTR enhancers in fibroblasts, thymic tumor cells, and a T-lymphoid cell line to identify nuclear factor binding sites that were occupied in vivo. We obtained evidence for in vivo binding to three of the multitude of binding sites identified by in vitro binding of nuclear extracts to naked DNA. The conserved central LVb/Ets and Core sites showed in vivo DMS footprints in all cells tested. In addition, strong footprints at all four NF-1 sites were observed in NIH 3T3 cells but not in the lymphoid Ti-6 cell line or in primary M-MuLV-induced T-lymphomas. No evidence for in vivo factor binding at the LVa or GRE site was obtained, but it was possible that binding at LVc occurred, particularly in T-lymphoid cells.
The binding of factors at the central LVb/Ets and Core sites in both lymphoid cells and fibroblasts was consistent with results of previous in vitro studies. As mentioned above, the LVb/Ets and Core sites are highly conserved both in sequence and in their proximity to each other among different retroviral LTRs (12). Moreover, cooperative in vitro binding of Ets-1 and CBF-α and -β to the LVb-Core sequences has been demonstrated (49). These previous results have suggested that binding at the central LVb/Ets and Core sites is essential for LTR activity of many retroviruses. The finding of in vivo footprinting in this region of the M-MuLV LTR in both fibroblasts and lymphoid cells supports this notion.
It was particularly noteworthy that the in vivo footprinting indicated occupation of the NF-1 binding sites in fibroblasts but not in lymphoid cells. This finding was consistent with results obtained by mutational analysis of the M-MuLV enhancers. In a study performed by Speck et al. (42), when two or all four NF-1 sites were mutated, transcriptional activity was decreased to a greater extent in NIH 3T3 fibroblasts than in T-cell lines. In addition, in SL3-3 MuLV, which also induces T-lymphoid leukemias, the NF-1 sites have been shown to stimulate transcription in fibroblasts and inhibit transcription in T cells (8). Furthermore, feline T-lymphoid lines from feline leukemia virus-induced tumors were found to express NF-1 transcripts but no NF-1 DNA binding activity was detectable. This disparity was apparently due to lymphoid-cell-specific posttranslational modification of NF-1 (33). Taken together, these results all suggest that the NF-1 binding sites are not important for M-MuLV infection of T-lymphoid cells. When M-MuLV with NF-1 mutations in the LTRs were tested for leukemogenicity in mice, T lymphoma was still induced (41). On the other hand, these mutants showed extended latency of disease (41). One explanation for this may be that in animals, cells other than T-lymphoid cells that are important for propagation of M-MuLV may use the NF-1 binding sites. Thus, during preleukemic times, the M-MuLV mutant in NF-1 might not establish viral loads as high as those acheived by wild-type M-MuLV, which in turn may extend the latency of the disease.
It should also be noted that for several of the binding sites in the M-MuLV 75-bp repeats, more than one protein may have bound the same site. For instance, the central LVb site has been reported to bind several Ets family proteins in vitro, including Ets-1, GABP, FLI-1, LVt (expressed predominantly in T lymphocytes), and MCREF-1 (13, 30, 43). Since some of these factors (LVt and Ets-1) show the same pattern of in vitro methylation interference as observed in the in vivo footprinting, it was not possible to deduce which of these factors bound in vivo. However, in the case of MCREF-1, one of the possible binding modes observed in vitro overlaps the upstream NF-1 and LVb sites (Fig. 1). In T lymphocytes, it is clear that MCREF-1 binding to the upstream site did not occur in vivo, since no footprint over the NF-1 sites was observed. The in vivo footprinting experiments also did not distinguish between binding of potential NF-1 isoforms (21).
The results also provided evidence for binding of a factor to the GC-rich sequences downstream of the 75-bp repeats in the M-MuLV LTR. The positions of the protected guanines did not correspond to any known binding patterns for proteins binding to potential motifs in these sequences (e.g., E-box or Sp-1). Thus, a novel binding factor might have been bound. It was also somewhat surprising that the footprint was found in fibroblasts but apparently not in primary thymic tumor cells. In previous mutational analyses, we and others have suggested that the GC-rich sequences may increase the T-lymphoid-cell specificities of the M-MuLV enhancers, since their deletion or mutation yields viruses that show relaxed disease specificities (11, 15). No protected or hypersensitive sites on the minus strands of the GC-rich sequences were observed, subject to the limitations discussed below.
In vivo DMS-LMPCR footprinting of the minus strands of the M-MuLV LTRs also provided interesting information. As mentioned in Results, the limitations of the analysis were that it was not possible to design lower-strand oligonucleotide primers that would amplify fragments only from the upstream LTR without amplifying sequences from the downstream LTR as well. Thus, if the upstream and downstream LTRs had different arrays of proteins bound (or not bound), the resulting LMPCR patterns combined the data for both of them. For instance, if the upstream LTR had nuclear factors bound at some of the sites but the downstream LTR did not, then any protected sites in the upstream LTR needed to be visualized above a background of unprotected fragments emanating from the downstream LTR. This might explain the lack of observed guanine protection in the lower-strand analysis. On the other hand, a site that gave hypersensitive guanine methylation would probably be readily visible even in the presence of fragments that do not show hypersensitivity. This possibility would be compatible with the detection of hypersensitive guanines shown in the in vivo footprinting of Fig. 9.
The location of hypersensitive guanines on the minus strands complemented and extended the upper-strand analysis. The presence of a hypersensitive adenine between the LVb/Ets and Core sites would be consistent with the sense-strand protection indicating occupancy of both of those sites. Moreover, the presence of the hypersensitive adenines in the LTRs from both infected fibroblasts and thymic tumor cells was consistent with occupation of those sites in both cell types. On the other hand, the presence of a hypersensitive guanine on the minus strand of the LVc site in thymic tumor cells but not in fibroblasts suggested that this site is occupied by a factor in T lymphocytes but not in fibroblasts. As discussed above, the presence of a hypersensitive sense-strand guanine between the Core site and the LVc site might have reflected binding of some factor to the LVc sequences. However, the upper-strand hypersensitive guanine was detected in both infected primary thymic tumor cells and fibroblasts but the minus-strand hypersensitive guanine was detected only in infected thymic tumor cells. Thus, either the LVc site is occupied by different factors in fibroblasts and T lymphocytes or the LVc site is occupied only in T lymphocytes and the hypersensitive sense-strand guanine does not reflect factor binding at LVc. In fibroblasts, but not in thymic tumor cells, there were two hypersensitive minus-strand guanines immediately upstream of the downstream NF-1 site in each 75-bp repeat. This finding strongly supported the conclusion from the plus-strand analysis that the NF-1 sites are occupied in vivo in fibroblasts but not in T-lymphoid cells.
The in vivo lower-strand analysis also identified a hypersensitive A base in sequences upstream of the 75-bp repeats. The hypersensitive A base was present in both fibroblasts and lymphoid cells. Sequences surrounding this base were scanned for known or potential factor binding sites, but none was identified in the immediate vicinity. A site for binding of a negative regulatory factory approximately 30 bp upstream from the hypersensitive A base has been reported (9), but it seems unlikely that binding at such a distant site would induce hypersensitive methylation. It is possible that an unidentified factor binds in the vicinity of the upstream hypersensitive guanine.
The status of in vivo binding site occupation in the downstream LTR is also of great interest. The downstream LTRs of retroviruses are much less transcriptionally active than the upstream LTRs in vivo (17), so it seems possible that fewer or different proteins are bound to them. Experiments to in vivo footprint the plus and minus strands of the downstream M-MuLV LTR are being designed.
It should be noted that the cell lines and tumors used in these experiments had more than one integrated M-MuLV provirus. Some of these proviruses are transcriptionally active (since the cells are productively infected), but others might not be transcribed. The analyses carried out in this study represent the average state of occupation for the binding sites for all of the proviruses. Where no footprints or extremely strong footprints were detected, the results probably reflect the status for most of the proviruses. Thus, the NF-1 sites in most of the proviruses are occupied in fibroblasts but not in lymphocytes. However, the in vivo footprints over the LVb/Ets and Core sites were not complete, particularly for fibroblasts. This might reflect the fact that some but not all of the proviruses contain factors bound at these sites in the infected cells; a possible distinction may be the transcription state of the proviruses. Alternatively, all of the proviruses might have factors bound at the LVb/Ets-Core sites, but in vivo protection from DMS methylation by these factors might not be absolute. It will be interesting to design experiments where the in vivo footprints over the LTRs of individual proviruses can be examined.
ACKNOWLEDGMENTS
This work was supported by grant CA32455 from the National Cancer Institute. S.W.G. was supported by grant 5 T32 CA09054 from the National Cancer Institute. The support of the UCI Cancer Research Institute and the Chao Family Comprehensive Cancer Center is gratefully acknowledged.
We thank Jeanne M. LeBon and Barbara Graves for advice and suggestions.
REFERENCES
- 1.Boral A L, Okenquist S A, Lenz J. Identification of the SL3-3 virus enhancer core as a T-lymphoma cell-specific element. J Virol. 1989;63:76–84. doi: 10.1128/jvi.63.1.76-84.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chatis P A, Holland C A, Hartley J W, Rowe W P, Hopkins N. Role for the 3′ end of the genome in determining disease specificity of Friend and Moloney murine leukemia viruses. Proc Natl Acad Sci USA. 1983;80:4408–4411. doi: 10.1073/pnas.80.14.4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chatis P A, Holland C A, Silver J E, Frederickson T N, Hopkins N, Hartley J W. A 3′ end fragment encompassing the transcriptional enhancers of nondefective Friend virus confers erythroleukemogenicity on Moloney leukemia virus. J Virol. 1984;52:248–254. doi: 10.1128/jvi.52.1.248-254.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen H, Yoshimura F K. Identification of a region of a murine leukemia virus long terminal repeat with novel transcriptional regulatory activities. J Virol. 1994;68:3308–3316. doi: 10.1128/jvi.68.5.3308-3316.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DesGroseillers L, Jolicoeur P. Mapping the viral sequences conferring leukemogenicity and disease specificity in Moloney and amphotropic murine leukemia viruses. J Virol. 1984;52:448–456. doi: 10.1128/jvi.52.2.448-456.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DesGroseillers L, Jolicoeur P. The tandem direct repeats within the long terminal repeat of murine leukemia viruses are the primary determinant of their leukemogenic potential. J Virol. 1984;52:945–952. doi: 10.1128/jvi.52.3.945-952.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DesGroseillers L, Rassart E, Jolicoeur P. Thymotropism of murine leukemia virus is conferred by its long terminal repeat. Proc Natl Acad Sci USA. 1983;80:4203–4207. doi: 10.1073/pnas.80.14.4203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ethelberg S, Hallberg B, Lovmand J, Schmidt J, Luz A, Grundstrom T, Pedersen F S. Second-site proviral enhancer alterations in lymphomas induced by enhancer mutants of SL3-3 murine leukemia virus: negative effect of nuclear factor 1 binding site. J Virol. 1997;71:1196–1206. doi: 10.1128/jvi.71.2.1196-1206.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flanagan J R, Becker K G, Ennist D L, Gleason S L, Driggers P H, Levi B Z, Appella E, Ozato K. Cloning of a negative transcription factor that binds to the upstream conserved region of Moloney murine leukemia virus. Mol Cell Biol. 1992;12:38–44. doi: 10.1128/mcb.12.1.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garrity P A, Wold B J. Effects of different DNA polymerases in ligation-mediated PCR: enhanced genomic sequencing and in vivo footprinting. Proc Natl Acad Sci USA. 1992;89:1021–1025. doi: 10.1073/pnas.89.3.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Golemis E, Li Y, Fredrickson T N, Hartley J W, Hopkins N. Distinct segments within the enhancer region collaborate to specify the type of leukemia induced by nondefective Friend and Moloney viruses. J Virol. 1989;63:328–337. doi: 10.1128/jvi.63.1.328-337.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Golemis E A, Speck N A, Hopkins N. Alignment of U3 region sequences of mammalian type C viruses: identification of highly conserved motifs and implications for enhancer design. J Virol. 1990;64:534–542. doi: 10.1128/jvi.64.2.534-542.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gunther C V, Graves B J. Identification of ETS domain proteins in murine T lymphocytes that interact with the Moloney murine leukemia virus enhancer. Mol Cell Biol. 1994;14:7569–7580. doi: 10.1128/mcb.14.11.7569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gunther C V, Nye J A, Bryner R S, Graves B J. Sequence-specific DNA binding of the proto-oncoprotein ets-1 defines a transcriptional activator sequence within the long terminal repeat of the Moloney murine sarcoma virus. Genes Dev. 1990;4:667–679. doi: 10.1101/gad.4.4.667. [DOI] [PubMed] [Google Scholar]
- 15.Hanecak R, Pattengale P K, Fan H. Deletion of a GC-rich region flanking the enhancer element within the long terminal repeat sequences alters the disease specificity of Moloney murine leukemia virus. J Virol. 1991;65:5357–5363. doi: 10.1128/jvi.65.10.5357-5363.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heinemeyer T, Wingender E, Reuter I, Hermjakob H, Kel A E, Kel O V, Ignatieva E V, Ananko E A, Podkolodnaya O A, Kolpakov F A, Podkolodny N L, Kolchanov N A. Databases on transcriptional regulation: TRANSFAC, TRRD and COMPEL. Nucleic Acids Res. 1998;26:362–367. doi: 10.1093/nar/26.1.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Herman S A, Coffin J M. Differential transcription from the long terminal repeats of integrated avian leukosis virus DNA. J Virol. 1986;60:497–505. doi: 10.1128/jvi.60.2.497-505.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hornstra I K, Yang T P. In vivo footprinting and genomic sequencing by ligation-mediated PCR. Anal Biochem. 1993;213:179–193. doi: 10.1006/abio.1993.1407. [DOI] [PubMed] [Google Scholar]
- 19.Ishimoto A, Adachi A, Sakai K, Matsuyama M. Long terminal repeat of Friend-MCF virus contains the sequence responsible for erythroid leukemia. Virology. 1985;141:30–42. doi: 10.1016/0042-6822(85)90180-1. [DOI] [PubMed] [Google Scholar]
- 20.Johnson P F, Landschulz W H, Graves B J, McKnight S L. Identification of a rat liver nuclear protein that binds to the enhancer core element of three animal viruses. Genes Dev. 1987;1:133–146. doi: 10.1101/gad.1.2.133. [DOI] [PubMed] [Google Scholar]
- 21.Johnson P F, McKnight S L. Eukaryotic transcriptional regulatory proteins. Annu Rev Biochem. 1989;58:799–839. doi: 10.1146/annurev.bi.58.070189.004055. [DOI] [PubMed] [Google Scholar]
- 22.Lenz J, Celander D, Crowther R L, Patarca R, Perkins D W, Haseltine W A. Determination of the leukaemogenicity of a murine retrovirus by sequences within the long terminal repeat. Nature. 1984;308:467–470. doi: 10.1038/308467a0. [DOI] [PubMed] [Google Scholar]
- 23.Li Q X, Fan H. Combined infection by Moloney murine leukemia virus and a mink cell focus-forming virus recombinant induces cytopathic effects in fibroblasts or in long-term bone marrow cultures from preleukemic mice. J Virol. 1990;64:3701–3711. doi: 10.1128/jvi.64.8.3701-3711.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Y, Golemis E, Hartley J W, Hopkins N. Disease specificity of nondefective Friend and Moloney murine leukemia viruses is controlled by a small number of nucleotides. J Virol. 1987;61:693–700. doi: 10.1128/jvi.61.3.693-700.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu P, Tarle S A, Hajra A, Claxton D F, Marlton P, Freedman M, Siciliano M J, Collins F S. Fusion between transcription factor CBF beta/PEBP2 beta and a myosin heavy chain in acute myeloid leukemia. Science. 1993;261:1041–1044. doi: 10.1126/science.8351518. [DOI] [PubMed] [Google Scholar]
- 26.Manley N R, O’Connell M, Sun W, Speck N A, Hopkins N. Two factors that bind to highly conserved sequences in mammalian type C retroviral enhancers. J Virol. 1993;67:1967–1975. doi: 10.1128/jvi.67.4.1967-1975.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mercurio F, Karin M. Transcription factors AP-3 and AP-2 interact with the SV40 enhancer in a mutually exclusive manner. EMBO J. 1989;8:1455–1460. doi: 10.1002/j.1460-2075.1989.tb03528.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mueller P R, Salser S J, Wold B. Constitutive and metal-inducible protein:DNA interactions at the mouse metallothionein I promoter examined by in vivo and in vitro footprinting. Genes Dev. 1988;2:412–427. doi: 10.1101/gad.2.4.412. [DOI] [PubMed] [Google Scholar]
- 29.Mueller P R, Wold B. In vivo footprinting of a muscle specific enhancer by ligation mediated PCR. Science. 1989;246:780–786. doi: 10.1126/science.2814500. . (Erratum, 248:802, 1990.) [DOI] [PubMed] [Google Scholar]
- 30.Nye J A, Petersen J M, Gunther C V, Jonsen M D, Graves B J. Interaction of murine ets-1 with GGA-binding sites establishes the ETS domain as a new DNA-binding motif. Genes Dev. 1992;6:975–990. doi: 10.1101/gad.6.6.975. [DOI] [PubMed] [Google Scholar]
- 31.Pfeifer G P, Steigerwald S D, Hansen R S, Gartler S M, Riggs A D. Polymerase chain reaction-aided genomic sequencing of an X chromosome-linked CpG island: methylation patterns suggest clonal inheritance, CpG site autonomy, and an explanation of activity state stability. Proc Natl Acad Sci USA. 1990;87:8252–8256. doi: 10.1073/pnas.87.21.8252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pfeifer G P, Steigerwald S D, Mueller P R, Wold B, Riggs A D. Genomic sequencing and methylation analysis by ligation mediated PCR. Science. 1989;246:810–813. doi: 10.1126/science.2814502. [DOI] [PubMed] [Google Scholar]
- 33.Plumb M, Fulton R, Breimer L, Stewart M, Willison K, Neil J C. Nuclear factor 1 activates the feline leukemia virus long terminal repeat but is posttranscriptionally down-regulated in leukemia cell lines. J Virol. 1991;65:1991–1999. doi: 10.1128/jvi.65.4.1991-1999.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rassart E, Sankar-Mistry P, Lemay G, DesGroseillers L, Jolicoeur P. New class of leukemogenic ecotropic recombinant murine leukemia virus isolated from radiation-induced thymomas of C57BL/6 mice. J Virol. 1983;45:565–575. doi: 10.1128/jvi.45.2.565-575.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reisman D. Nuclear factor-1 (NF-1) binds to multiple sites within the transcriptional enhancer of Moloney murine leukemia virus. FEBS Lett. 1990;277:209–211. doi: 10.1016/0014-5793(90)80846-b. [DOI] [PubMed] [Google Scholar]
- 36.Saluz H P, Jost J P. Approaches to characterize protein-DNA interactions in vivo. Crit Rev Eukaryot Gene Expr. 1993;3:1–29. [PubMed] [Google Scholar]
- 37.Saluz H P, Jost J P. In vivo DNA footprinting by linear amplification. Methods Mol Biol. 1994;31:317–329. doi: 10.1385/0-89603-258-2:317. [DOI] [PubMed] [Google Scholar]
- 38.Satake M, Inuzuka M, Shigesada K, Oikawa T, Ito Y. Differential expression of subspecies of polyomavirus and murine leukemia virus enhancer core binding protein, PEBP2, in various hematopoietic cells. Jpn J Cancer Res. 1992;83:714–722. doi: 10.1111/j.1349-7006.1992.tb01971.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Short M K, Okenquist S A, Lenz J. Correlation of leukemogenic potential of murine retroviruses with transcriptional tissue preference of the viral long terminal repeats. J Virol. 1987;61:1067–1072. doi: 10.1128/jvi.61.4.1067-1072.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Speck N A, Baltimore D. Six distinct nuclear factors interact with the 75-base-pair repeat of the Moloney murine leukemia virus enhancer. Mol Cell Biol. 1987;7:1101–1110. doi: 10.1128/mcb.7.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Speck N A, Renjifo B, Golemis E, Fredrickson T N, Hartley J W, Hopkins N. Mutation of the core or adjacent LVb elements of the Moloney murine leukemia virus enhancer alters disease specificity. Genes Dev. 1990;4:233–242. doi: 10.1101/gad.4.2.233. [DOI] [PubMed] [Google Scholar]
- 42.Speck N A, Renjifo B, Hopkins N. Point mutations in the Moloney murine leukemia virus enhancer identify a lymphoid-specific viral core motif and 1,3-phorbol myristate acetate-inducible element. J Virol. 1990;64:543–550. doi: 10.1128/jvi.64.2.543-550.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sun W, O’Connell M, Speck N A. Characterization of a protein that binds multiple sequences in mammalian type C retrovirus enhancers. J Virol. 1993;67:1976–1986. doi: 10.1128/jvi.67.4.1976-1986.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thornell A, Hallberg B, Grundstrom T. Binding of SL3-3 enhancer factor 1 transcriptional activators to viral and chromosomal enhancer sequences. J Virol. 1991;65:42–50. doi: 10.1128/jvi.65.1.42-50.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thornell A, Hallberg B, Grundstrom T. Differential protein binding in lymphocytes to a sequence in the enhancer of the mouse retrovirus SL3-3. Mol Cell Biol. 1988;8:1625–1637. doi: 10.1128/mcb.8.4.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thornell A, Holm M, Grundstrom T. Purification of SEF1 proteins binding to transcriptional enhancer elements active in T lymphocytes. J Biol Chem. 1993;268:21946–21954. [PubMed] [Google Scholar]
- 47.Wang S, Wang Q, Crute B E, Melnikova I N, Keller S R, Speck N A. Cloning and characterization of subunits of the T-cell receptor and murine leukemia virus enhancer core-binding factor. Mol Cell Biol. 1993;13:3324–3339. doi: 10.1128/mcb.13.6.3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang S W, Speck N A. Purification of core-binding factor, a protein that binds the conserved core site in murine leukemia virus enhancers. Mol Cell Biol. 1992;12:89–102. doi: 10.1128/mcb.12.1.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wotton D, Ghysdael J, Wang S, Speck N A, Owen M J. Cooperative binding of Ets-1 and core binding factor to DNA. Mol Cell Biol. 1994;14:840–850. doi: 10.1128/mcb.14.1.840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yoshimura F K, Diem K, Chen H, Tupper J. A protein-binding site with dyad symmetry in the long terminal repeat of the MCF13 murine leukemia virus that contributes to transcriptional activity in T lymphocytes. J Virol. 1993;67:2298–2304. doi: 10.1128/jvi.67.4.2298-2304.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]