Abstract
The surge in reports describing non-coding RNAs (ncRNAs) has focused attention on their possible biological roles and effect on development and disease. ncRNAs have been touted as previously uncharacterized regulators of gene expression and cellular processes, possibly working to fine-tune these functions. The sheer number of ncRNAs identified has outpaced the capacity to characterize each molecule thoroughly and to reliably establish its clinical relevance; it has, nonetheless, created excitement about their potential as molecular targets for novel therapeutic approaches to treat human disease. In this Review, we focus on one category of ncRNAs — long non-coding RNAs — and their expression, functions and molecular mechanisms in cardiac hypertrophy and heart failure. We further discuss the prospects for this specific class of ncRNAs as novel targets for the diagnosis and treatment of these conditions.
ToC
In this Review, Mably and Wang summarize the expression, functions and molecular mechanisms of long non-coding RNAs (lncRNAs) in the cardiac hypertrophy and failure. The authors also discuss lncRNAs as novel targets for the diagnosis and treatment of these conditions.
Introduction
Although only 1% of the human genome is estimated to be composed of protein-coding genes, nearly 80% is transcribed and functional; therefore, only ~2% of transcripts encode proteins, meaning that the vast majority of transcription products are categorized as being non-coding1,2. As a result of this observation, scientists are faced with a conundrum: either the process of transcription is highly non-specific and inefficient, resulting in a huge amount of material that serves no biological purpose, or these non-coding transcripts do indeed have a function that remains to be fully defined. Since the first description of microRNAs (miRNAs) in 19933,4, evidence has continued to accumulate that supports the latter of the two possibilities. Additionally, these studies suggest that miRNAs and other non-coding RNAs (ncRNAs) could have important roles in the regulation of gene expression, including in cardiac disease states5. Multiple studies and biotech start-ups have been initiated to examine the potential use of ncRNAs as therapeutic targets or agents; therefore, a comprehensive understanding of their biology and assessment of their clinical potential are necessary as the field progresses.
Long non-coding RNAs (lncRNAs) are a subgroup of ncRNAs that are broadly defined as being greater than 200 nucleotides in length but they can be as long as 100 kilobases6,7. They are less conserved across species and expressed at lower levels than their coding (mRNA) counterparts. Depending on their genomic environment, synthesis, structure and function, lncRNAs can be further subclassified as intergenic (lincRNA), enhancer (elncRNA), promoter (plncRNA) or antisense (lncRNA-as); some lncRNAs can form a circular structure and form part of the broader circular RNA (circRNA) category. The function and molecular mechanisms of lncRNA action show wide heterogeneity, depending on their subcellular location and interaction partners (Fig. 1). Based on their mechanism of action, they can be categorized into four broad groups: signal lncRNAs, which influence the actions of transcription factors or signalling pathways; decoy lncRNAs, which sequester transcription factors, other proteins or miRNAs to prevent their normal function; guide lncRNAs, which can recruit proteins to target nearby (cis) or distant (trans) target genes; and scaffold lncRNAs, which bring together and/or stabilize multiple proteins to form larger complexes8. They can be further characterized on the basis of their genomic location and context: sense lncRNAs are derived from the sense strand of a protein-coding gene, can include intronic regions and can also overlap with a substantial portion of the protein-coding sequence; antisense lncRNAs are transcribed from the antisense strand of a protein-coding gene and can also overlap with the protein-coding sequence and include intronic sequence; intergenic lncRNAs are transcribed from both strands of DNA between genes; and intronic lncRNAs are transcribed from introns between the exons of protein-coding genes9. These lncRNAs can also function bidirectionally in cis to influence the expression of genes at either their 5ʹ or 3ʹ ends (Fig. 1a)8. In the nucleus, lncRNAs participate in chromatin organization, gene transcription, and the modification and splicing of nascent RNAs (Fig. 1b)10. In the cytoplasm, lncRNAs can modulate mRNA turnover, storage and protein translation (Fig. 1c–e)11. Given the effect of lncRNAs on these essential cellular processes, their influence inevitably extends to broader biological processes and physiological functions.
Fig. 1 |. lncRNA mechanisms of action.
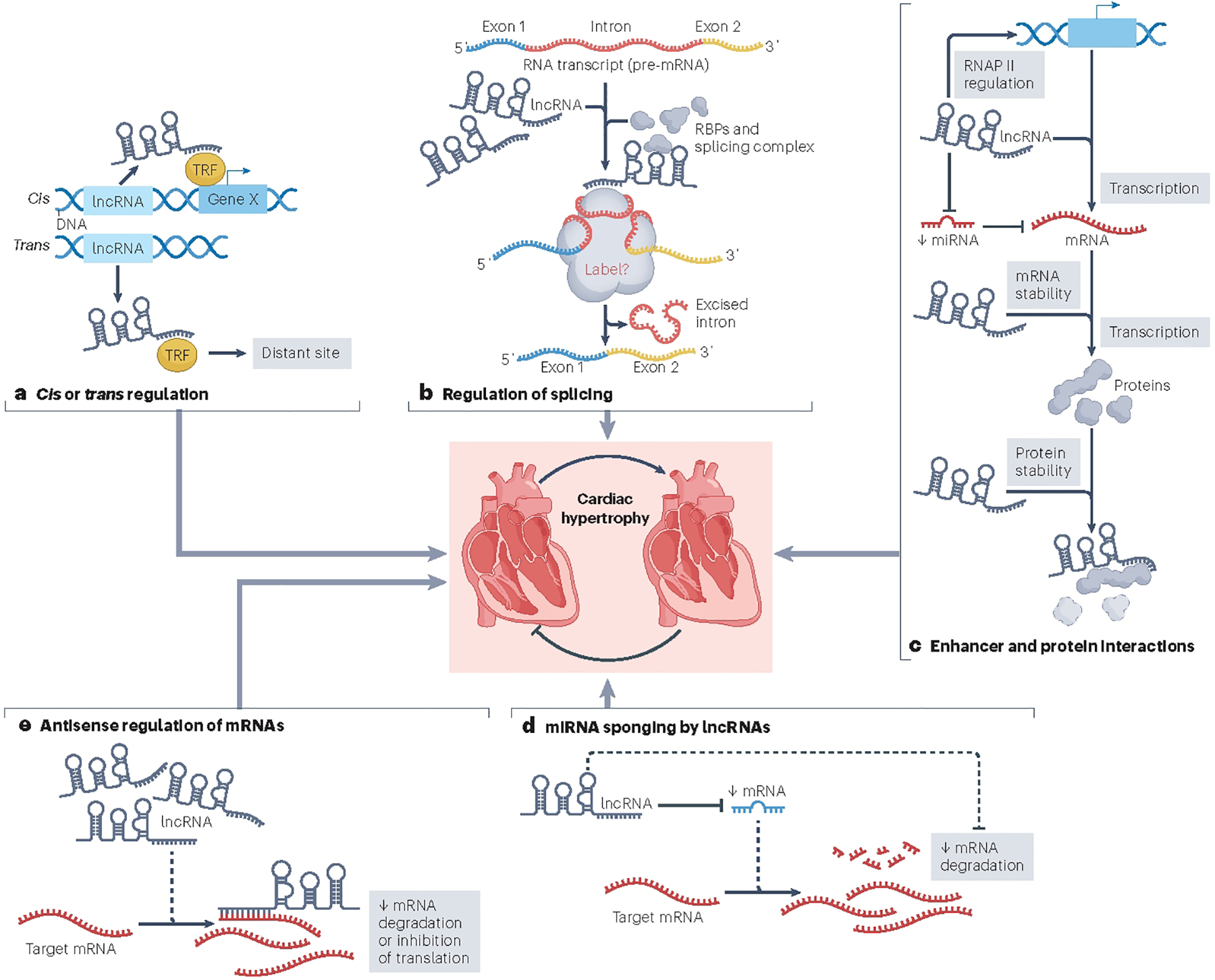
The figure shows mechanism by which long non-coding RNAs (lncRNAs) have been shown to exert their function in the heart and lead to cardiac hypertrophy. a, Cis or trans regulation. lncRNAs can interact with transcription factors (TFs) to facilitate their activity and promote transcription in cis of a nearby gene (gene X). This lncRNA–TF interaction can also facilitate transcription in trans of a gene at a distant site (gene Y). Alternatively, lncRNA–TF interactions can limit the activity of the TF or the lncRNA could participate in the stabilization of a transcriptional repressor complex, thereby reducing transcription. b, Regulation of alternative splicing. lncRNAs can interact with RNA-binding proteins (RBPs) to facilitate the formation of alternative splice variants. c, Protein interactions. lncRNAs can have a direct influence on the stabilization of the RNA polymerase II (Pol II) complex to promote transcription. Some lncRNAs can also stabilize proteins to facilitate or prolong their activity. d, microRNA (miRNA) sponging by lncRNAs. lncRNAs are also able to interact with and limit the activity of miRNAs. In one mechanism, lncRNAs directly sequester and prevent miRNAs from degrading target mRNAs. In a second mechanism, lncRNAs interact with and suppress the ability of miRNAs to inhibit translation. e, Antisense regulation of mRNAs. lncRNAs can influence the stability of an mRNA through direct interaction and by preventing its degradation. Other mechanisms might exist but remain to be validated in the heart.
Two of the aforementioned properties of lncRNAs have created challenges to establishing their functions and mechanisms; their poor sequence conservation makes animal modelling difficult and their characteristically low expression levels is a challenge to assessing their endogenous in vivo functions. Some studies have shown that many lncRNAs have non-essential roles during heart development and in cardiac physiology, and multiple others have demonstrated that the expression of many lncRNAs is altered under pathological cardiac conditions; therefore, lncRNAs have a key role in cardiac remodelling in response to stress. In this Review, we first summarize the expression of lncRNAs and the functional involvement of lncRNAs in cardiac hypertrophy, based on studies using in vitro systems and/or in vivo animal models (see Box 1 for more information about these models). We discuss well-characterized lncRNAs with in vivo cardiac phenotypes and highlight commonalities among these studies. However, we have restricted this Review to a discussion of linear lncRNAs to avoid overlap with other reviews focused on the roles of circular lncRNAs in cardiac hypertrophy12,13. Next, we provide an overview of the mechanisms by which lncRNAs mediate their effects and speculate about their clinical potential. lncRNAs could represent a novel class of molecules that might function as biomarkers and/or as therapeutic targets in the search for new approaches in the treatment of cardiovascular disease.
Box 1 |. Definition and mouse models of cardiac hypertrophy.
Cardiac hypertrophy is an adaptive response by the heart to changes in pressure or volume. This process can be beneficial, such as the hypertrophy associated with exercise observed in athletes. However, multiple disease processes, including genetic mutations in sarcomeric (or other) proteins or loss of cardiac muscle mass as a result of previous damage, can induce pathological hypertrophy. This type of hypertrophy is observed in patients with ischaemic heart disease, hypertension and/or valvular defects. Given that pathological cardiac hypertrophy impairs heart function, an inability to reverse it results in continuing deterioration, that can lead to heart failure and death.
Mice are the most widely used animal models for in vivo studies of roles for long non-coding RNAs (and other molecules) on heart function. One of the most widely used surgical models in mouse is the transverse aortic constriction technique to study pressure overload-induced cardiac hypertrophy and heart failure200. During the early stages, transverse aortic constriction surgery oftens lead to a compensatory hypertrophy associated with a transient increase in cardiac contractility. However, the sustained haemodynamic overload eventually results in a maladaptive response, leading to cardiac dilatation and heart failure. A mouse model of myocardial infarction is also commonly used and involves the surgical ligation of the left anterior descending coronary artery201. The artery ligation can be either permanent or transient (a model of ischaemia–reperfusion injury). As would be expected, permanent ligation results in substantially more damage to the heart tissue, leading to an elevation in apoptosis and scarring. Transient ligation results in less initial damage and is associated with a smaller amount of scarring but is accompanied by necrotic damage associated with reperfusion after removal of the occlusion. Both models are relevant to the pathological processes observed in patients with cardiac disease.
lncRNA expression and association with cardiac hypertrophy and heart disease
Multidimensional comparative transcriptomics analyses across various developmental time points, organs, and species have demonstrated that lncRNAs are preferentially expressed in adult tissues14. Additional integrated transcriptomics and epigenomics studies in developmental models, as well as adult mouse and human hearts, have identified additional novel lncRNAs that are associated with cardiovascular disease15. lncRNAs are expressed at low levels across cell and tissue types; however, lncRNAs that show tissue-restricted and higher expression patterns have been identified. Many of these more tissue-restricted and robustly expressed lncRNAs show greater evolutionary conservation, in particular around their promoter regions16,17, which suggests a paradigm in which tissue specificity and expression level correlate with preserved function across species.
An analysis of the cardiomyocyte transcriptomes of mouse and human hearts revealed that lincRNAs were key nodal regulators of subpopulations of cardiomyocytes18. In this report from 2017, investigators used single-nucleus RNA sequencing to profile the transcriptome from the hearts of adult mice and humans, in both failing and non-failing conditions18. They assessed the function of these lincRNAs by knocking down their levels in mouse ventricular cardiomyocytes and demonstrated changes in the expression patterns of genes involved in dedifferentiation and the cell cycle. These results led the researchers to ascribe possible roles for these lincRNAs in the regulation of regeneration of adult cardiomyocyte subpopulations18. Other annotated lncRNAs have the potential to encode micropeptides, thereby directly contributing to the expressed transcriptome of cardiac cell types19,20. This insight suggests another, possibly complementary, mechanism by which lncRNAs might contribute to the pathophysiology of cardiac hypertrophy and substantially increases the complexity of lncRNA biology19,20.
Mouse models of myocardial infarction (MI) are extensively used to study cardiac damage caused by ischaemia–reperfusion. Investigators studying the heart transcriptome in one of these mouse models used traditional RNA sequencing to determine that the expression of certain lncRNAs was strongly correlated with specific parameters of cardiac function and dimensions21. These data suggest that specific lncRNAs are associated with cardiac-specific genomic enhancers and thereby influence the transcription of genes involved in cardiogenesis and pathological remodelling21.
A systematic analysis of the lncRNA expression profiles between normal embryonic and adult cardiac tissue identified 157 lncRNAs with differential expression22. However, when the expression profiles of adult normal and hypertrophic cardiac tissue were compared, only 17 differentially expressed lncRNAs were observed after transverse aortic constriction (TAC) surgery (analysis performed 1 week and 4 weeks after TAC)22. This finding is intriguing because it shows a lncRNA response that is different from the dogma for the expressed transcriptome in the heart (that many fetal, protein-coding mRNAs (and their products) that are highly expressed in the developing and fetal heart are also induced in the hypertrophic heart23,24). The observation that fetal lncRNAs are not re-expressed in the hypertrophic heart to a high degree suggests an important role during development that is more subtle during the response of the heart to haemodynamic stress22.
In an attempt to characterize the transcriptome changes in diseased hearts, researchers performed next-generation sequencing using tissue collected from human left ventricles; samples collected from non-failing hearts were analysed and compared with samples from paired non-ischaemic and ischaemic failing left ventricular (LV) tissue collected before or after LV assist device implantation25. The investigators found that the expression profiles of lncRNAs, but not those of mRNAs or miRNAs, could discriminate between failing hearts with different pathologies. In particular, lncRNAs were markedly altered in response to LV assist device support, suggesting they could be sensitive markers of heart failure (HF) and might have important roles in the pathogenesis of HF and in the reverse remodelling observed in response to mechanical support25,26. Another interesting finding from this study was the detection of a high abundance of lncRNAs of mitochondrial origin (71%)25. Given that the mitochondrial number and function are dynamically regulated and altered in the heart during various pathophysiological conditions, these mitochondrially encoded lncRNAs might have previously unrecognized roles in the response of the heart to stress. However, due to the small sample size of this analysis (eight in each group), these data need to be validated by further studies to test their reproducibility and whether other tissues and organs respond similarly. The presence of mitochondrial lncRNAs at these elevated levels would introduce another mechanism by which this organelle could actively influence cardiac function and disease. Further investigation to establish the function of these lncRNAs in the heart, as well as their mechanisms of action, will not only contribute to a better understanding of lncRNA biology but will also provide important insights into their roles in cardiovascular disease.
lncRNA mechanisms of action in the heart
lncRNAs are detectable in most cells and tissues, with many showing a tissue-specific or cell-type-specific pattern of expression27. lncRNA transcripts can be found in the nucleus or cytoplasm, and their subcellular localization can provide insights into their function (Fig. 1)28–30. In the nucleus, these functions can involve direct interactions with DNA, RNA and proteins, allowing lncRNAs to modulate chromatin function, regulate the assembly and function of nuclear bodies, and mediate the association between DNA and epigenetic factors, transcription factors and even RNA polymerase (Fig. 1a)31. Alternatively, lncRNAs have been shown to alter the stability and translation of cytoplasmic mRNAs and thereby interfere with signalling pathways (Fig. 1b–e)11,32. lncRNAs might also function as sponges for miRNAs and other RNAs in the cytoplasm (Fig. 1d)33,34. In addition, emerging evidence suggests that many lncRNAs function in a cis-regulatory manner, which can confine their effect on gene expression to specific transcripts, depending on their own genomic context (Fig. 1a)35,36.
Cis-regulatory or trans-regulatory functions
Multiple mechanisms of action have now been described for what has become the broad category of cis-regulatory lncRNAs35,36. However, many of these mechanisms are shared with lncRNAs that do not demonstrate clear cis-regulatory roles. Additionally, many lncRNAs are likely to possess both cis-regulatory and trans-regulatory capacities37 (Fig. 1a). Therefore, in this section, we discuss several lncRNAs with defined mechanisms of action and note for each whether these actions are in cis or trans, if known.
Enhancer interaction.
The lncRNA Uph (upperhand) has been shown to be crucial to heart development, given that blocking its transcription in mice results in right ventricular hypoplasia and embryonic lethality38. However, the process of Uph transcription itself, rather than the mature transcript, was shown to be required for proper Hand2 expression38. This analysis revealed that Uph has a crucial role in maintaining the super-enhancer signature at the Hand2 enhancer locus, thereby promoting RNA polymerase II elongation and ultimately controlling Hand2 gene transcription38.
Antisense lncRNAs.
AIRN (also called AIR) is a lncRNA transcribed in the antisense orientation to the IGF2R gene39,40. Studies in mice and in human cells have shown that AIRN regulates the expression of the nearby imprinted protein-coding genes IGF2R (maternally derived), SLC22A2 and SLC22A3 by a cis-regulatory mechanism41,42. AIRN is highly expressed in the heart43; in a mouse model of MI, its expression is downregulated in non-infarcted regions of the heart, and silencing of Airn induces cell death in cardiomyocytes43. The regulatory activity of Airn is facilitated by its interaction with the product of the nearby gene Igf2bp2, which encodes an RNA-binding protein that regulates the translation of target genes, including that of Ifg2bp2 itself43. These findings are intriguing but were discovered using cell-based assays, without confirmation using in vivo models. Therefore, the role of AIRN in heart development and disease remains to be confirmed. Further studies to determine whether reduced levels of AIRN or its absence is associated with heart defects either in mouse models or in cardiovascular disease in humans would be valuable to assess its contribution to cardiac function.
Alternative splicing.
TRDN-AS is transcribed as an antisense transcript from the TRDN locus encoding the protein triadin (TRDN)44,45, establishing alternative splicing as another process that can be regulated by lncRNAs to influence cardiac function. TRDN is a sarcoplasmic reticulum membrane protein and it is known to have an important role in Ca2+ homeostasis and cardiomyocyte function; mutations in the TRDN gene have been linked to cardiac dysfunction and arrhythmias46,47. TRDN-AS is expressed in cardiomyocytes and co-localizes and interacts with serine/arginine splicing factors in the nucleus, where it efficiently recruits them to the triadin precursor mRNA44,45. Loss of Trdn-as in the heart has been shown to sensitize mice to cardiac arrhythmias in response to catecholamine challenge, at least in part due to a reduction in cardiac triadin transcript and protein levels45. Of note, in contrast to the levels of TRDN, the transcript levels of TRDN-AS are increased in the hearts of patients with HF44, consistent with studies on the influence of Trdn-as deletion on Trdn protein levels in mouse45. These data suggest that this conserved lncRNA has the potential to be a novel therapeutic target for the treatment of HF and arrhythmias. Although analysis of TRDN-AS convincingly shows that it functions as a trans-regulatory factor that modulates the alternative splicing of the TRDN-coding transcripts, further studies are required to determine whether it also functions as a cis-regulatory element to modulate triadin expression.
Protein interactions.
The function of the lncRNA CCRR (cardiac conduction regulatory RNA) has been linked to cardiac arrhythmias48. The expression of Ccrr is downregulated in a mouse model of HF and is associated with slow cardiac conduction and increased arrhythmogenicity48. Furthermore, silencing Ccrr in the heart induces arrhythmias in mice48. Functional analysis of Ccrr determined that its inhibition affects the structure and function of intercalated discs and gap junctions, thereby slowing longitudinal cardiac conduction48. Mechanistically, Ccrr directly binds to the connexin 43 (Cx43)-interacting protein, CIP85, to block endocytic trafficking of Cx43 and prevent its degradation. This lncRNA is conserved in humans, and reduced CCRR expression was found in patients with HF48. Therefore, CCRR would be an interesting target for the design of new approaches to the treatment of pathological arrhythmias.
Caren (cardiomyocyte-enriched non-coding transcript) is a lncRNA that is enriched in cardiomyocytes and located in the cytoplasm49. Given that Caren does not influence the expression levels of neighbouring genes, a cis-regulatory mechanism is unlikely. Instead, Caren transcripts decrease the translation of its downstream target Hint1 (encoding histidine triad nucleotide-binding protein 1)49. In the heart, Caren has an protective role by inactivating the serine-protein kinase ATM-mediated DNA damage response pathway and activating mitochondrial bioenergetics49. This cardioprotective role of Caren suggests that it has the potential to be a novel therapeutic target for the treatment of heart disease.
The lncRNA CARDINAL (myocardin-adjacent lncRNA) is encoded by a genomic locus adjacent to the MYOCD gene, encoding myocardin (a potent transcriptional coactivator of serum response factor (SRF))50. Cardinal and Myocd also share overlapping but distinct cardiac-muscle-specific, smooth-muscle-specific and embryonic expression patterns50. The Cardinal transcript is present in both the nucleus and the cytoplasm of cardiomyocytes50. Nuclear Cardinal forms a complex with SRF to repress the transcription of the pro-mitogenic gene Fos, encoding protein c-Fos50. Cardinal-deficient mice have aberrant expression of TCF–SRF-dependent mitogenic genes and reduced cardiac contractility50; these data suggest that nuclear CARDINAL might function as an RNA cofactor for SRF to regulate the transcription of target genes. However, it remains unclear whether CARDINAL has a cis-regulatory function to regulate the expression of nearby genes, such as myocardin, or how cytosolic CARDINAL functions. Furthermore, it is unknown whether CARDINAL is involved in cardiac remodelling and dysfunction in response to pathophysiological stress.
In a related study, the tissue-specific lncRNA CARMN (or CARMEN, cardiac mesoderm enhancer-associated non-coding RNA) was found to interact directly with MYOCD to modulate its ability to influence cardiac and smooth muscle gene expression51,52. CARMN is encoded by the genomic region that includes the miRNA-143–miRNA-145 cluster; however, the aforementioned function of CARMN seems to be independent of the activity of these miRNAs51,52. Another study reported that CARMN interacts directly with SRF to regulate gene expression and its role in vascular smooth muscle53.
The lncRNA Cpmer (cytoplasmic mesoderm regulator) uses an RNA–RNA pairing mechanism to specifically recognize Eomes mRNA (encoding the protein eomesodermin, a transcription factor involved in early mesoderm development)54. This interaction facilitates binding of the Eomes transcript to the translational elongation factor eEF1A2 and subsequent translation of the eomesodermin protein54. The ability of CPMER to regulate eomesodermin translation is conserved between mice and humans; in addition, Cpmer has been shown to promote the differentiation of both mouse and human embryonic stem cells into cardiomyocytes54.
Molecular sponging.
Overexpression of the lncRNA Plscr4 (phospholipid scramblase 4) in mice can reduce angiotensin II-induced cardiomyocyte hypertrophy, whereas depletion of Plscr4 induces hypertrophy55. Plscr4 overexpression can also attenuate TAC-induced cardiac hypertrophy in mice55. Mechanistically, evidence supports a role for Plscr4 as a natural sequestering agent for miRNA-214, thereby acting as an miRNA-214 sponge. In support of this hypothesis, Plscr4 overexpression in angiotensin II-treated and normal cardiomyocytes decreases the levels of miRNA-21455. This finding was validated in mouse hearts and was proposed as an explanation for the observed reduction in cardiomyocyte hypertrophy55. By contrast, Plscr4 knockdown increases miRNA-214 levels, thereby promoting cardiomyocyte hypertrophy55.
lncRNAs that regulate cardiac hypertrophy
Many lncRNAs expressed in cardiac tissue have been investigated for their influence on gene expression in the heart. As more detailed studies are published, we will gain a better understanding of the roles of these molecules on cardiac function, and certain lncRNAs will inevitably gain prominence because of their effect on processes such as cardiac hypertrophy and remodelling. This trend would be similar to what was observed during research into the roles of miRNAs in the heart, when specific cardiac miRNAs (such as miRNA-1 and miRNA-133) garnered greater attention because of their important roles in cardiac tissue56–60. Studies on some lncRNAs have already provided interesting information into their mechanisms of action, as well as possible links to cardiac disease processes. We highlight some of these in the following section (a more comprehensive list is provided in Box 2, with details provided in Tables 1,2).
Box 2 |. lncRNAs associated with cardiac hypertrophy.
Ahit (antihypertrophic interrelated transcript)163
ANRIL (antisense non-coding RNA in the INK4 locus)150
APF (autophagy promoting factor)174
CAIF (cardiac autophagy inhibitory factor)175
Caren (cardiomyocyte-enriched non-coding transcript)49
Carmen or Carmn (cardiac mesoderm enhancer-associated non-coding RNA)176
CCRR (cardiac conduction regulatory RNA) (also known as AK045950)48
Chaer (cardiac-hypertrophy-associated epigenetic regulator)82
CHAIR (cardiomyocyte hypertrophic associated inhibitory RNA) (also known as 4632428C04Rik)151
CHAR (cardiac hypertrophy-associated regulator)164
Chast (cardiac hypertrophy-associated transcript)81
CPhar (cardiac physiological hypertrophy-associated regulator)165,202
CRRL (cardiomyocyte regeneration-related lncRNA)177
ECRAR (endogenous cardiac regeneration-associated regulator)178
Fendrr (FOXF1 adjacent non-coding developmental regulatory RNA)153
GAS5 (growth arrest-specific 5)166
Gm15834154
HypERlnc (hypoxia-induced endoplasmic reticulum stress regulating long non-coding RNAs)155,156
lncCytB (lncRNA CytB)168
lncDACH1 (long non-coding RNA dachshund homologue 1)157
lncExACT1 (long non-coding exercise associated transcript 1)158,159
lncMYH7b (long non-coding myosin heavy chain 7b)126
Mhrt or Myheart (myosin heavy-chain-associated RNA transcripts)117–119,169,203
NRON (non-coding repressor of nuclear factor of activated T cells)161
OIP5-as1 (Opa-interacting protein 5-antisense 1) (also known as 1700020I14Rik or Cyrano)170
Sirt1-as (silent information regulator 1-antisense)162
Trdn-as (triadin-as)45
Wisper (WISP2 super-enhancer-associated RNA)134
ZNF593-as (zinc finger protein 593-antisense) (also known as RP11-96L14.7 or ENST00000448923.2)171
Table 1 |.
lncRNAs that induce cardiac hypertrophy and/or heart failure
| lncRNA | Models | Functions | Interactions and effects | Ref. |
|---|---|---|---|---|
| ANRIL (antisense non-coding RNA in the INK4 locus) | Rat (diabetic model) | Downregulated ANRIL improved cardiac function index and decreased expression of inflammatory factors, resulting in decreased myocardial collagen deposition area and cardiomyocyte apoptosis and reduced levels of oxidative stress in myocardial tissue | Not determined | 150 |
| Chaer (cardiac-hypertrophy-associated epigenetic regulator) | Mouse | Associated with the development of cardiac hypertrophy | Interacts with the catalytic subunit of PRC2 | 82 |
| CHAIR (cardiomyocyte hypertrophic associated inhibitory RNA; 4632428C04Rik) | Human and mouse | Loss of CHAIR has no effect on normal hearts; however, in response to stress, it accelerates heart functional decline, increases hypertrophy and exacerbates heart failure | Interacts with DNMT3A to inhibit its DNA-binding activity | 151 |
| Chast (cardiac hypertrophy-associated transcript) | Human and mouse | Virus-based overexpression of Chast is sufficient to induce cardiomyocyte hypertrophy in vitro and in vivo; GapmeR-mediated silencing of Chast both prevented and attenuated TAC-induced pathological cardiac remodelling, with no toxicological signs or adverse effects | Negatively regulates PLEKHM1 (located on the opposite strand to Chast), which impedes cardiomyocyte autophagy and drives hypertrophy | 81 |
| CHRF (cardiac hypertrophy related factor)73,152 | Mouse and in vitro model | Small interfering RNA-mediated knockdown of CHRF attenuates ANF and MHCβ levels in the heart | Binds to miRNA-489 to reduce its levels (microRNA sponge) | 73 |
| Fendrr (FOXF1 adjacent non-coding developmental regulatory RNA) | Mouse and in vitro model | Fendrr loss of function reduces cardiac fibrosis induced by TAC | Binds to miRNA-106b | 153 |
| Gm15834 | Mouse (TAC and angiotensin II infusion models) | Forced expression of Gm15834 increases cardiomyocyte autophagy and promotes myocardial hypertrophy; silencing of Gm15834 attenuates autophagy-induced myocardial hypertrophy | Binds to miRNA-30b-3p to function as an endogenous sponge | 154 |
| HypERlnc (hypoxia-induced endoplasmic reticulum stress regulating long non-coding RNAs)155,156 | In vitro | Silencing of HypERlnc decreases cell viability and proliferation and results in pericyte dedifferentiation; associated with increased endothelial permeability in co-cultures consisting of human primary pericyte and human coronary microvascular endothelial cells | Endoplasmic reticulum stress-related transcription factors were prominently activated by HypERlnc knockdown | 155 |
| LncDACH1 (long non-coding RNA-Dachshund homologue 1; DACH1) | Mouse | Transgenic overexpression of LncDACH1 in cardiomyocytes leads to impaired cardiac function, reduced calcium transient and cell shortening, and decreased SERCA2a protein expression; by contrast, conditional knockout of LncDACH1 in TAC-treated mouse cardiomyocytes results in increased calcium transients, cell shortening and SERCA2a protein expression and improved cardiac function | Binds to SERCA2a | 157 |
| lncExACT1 (long non-coding exercise associated transcript 1) | Mouse (exercise model) | lncExACT1 inhibition induced physiological hypertrophy and cardiomyogenesis | Interacts with DCHS2 | 158,159 |
| MIAT (myocardial infarction-associated transcript) | Mouse (myocardial infarction model) | Inhibition of MIAT protects the heart against myocardial infarction | Interacts with miRNA-150 and HOXA4 | 99 |
| Mouse (TAC and angiotensin II infusion models) | Genetic ablation of MIAT attenuates pathological hypertrophy and heart failure | Not available | 97 | |
| Meg3 (maternally expressed gene 3)135,136 | Human (induced pluripotent stem cells) and mouse | Inhibition of Meg3 in vivo after TAC prevented cardiac MMP2 induction, leading to decreased cardiac fibrosis and improved diastolic performance; mostly expressed by cardiac fibroblasts; undergoes transcriptional downregulation during late cardiac remodelling | Interacts with p53 to increase its binding and activity, inducing expression of profibrotic MMP2 gene | 136 |
| In vitro | Silenced Meg3 inhibited cardiomyocyte hypertrophy and reversed other hypertrophic responses | Might regulate miRNA-361–5p and HDAC9 by acting as a competing endogenous RNA; upregulated by the transcription factor STAT3 | 160 | |
| NRON (non-coding repressor of nuclear factor of activated T cells) | Mouse (TAC model) | In a gain-of-function mouse model, hypertrophic cardiomyopathy is worsened; by contrast, loss of function attenuates symptoms | Influences the transcription programme for hypertrophic cardiomyopathy; nuclear localization | 161 |
| Sirt1-as (silent information regulator 1-antisense) | Mouse | Overexpression of Sirt1-as increases cardiomyocyte proliferation, attenuates cardiomyocyte apoptosis, improves cardiac function and decreases mortality after myocardial infarction | Binds to the Sirt1 3′-UTR to increase the stability of Sirtl mRNA and increase abundance at both the mRNA and protein levels | 162 |
Multiple long non-coding RNAs (lncRNAs) have been described with roles in heart development and cardiac cell growth. lncRNAs with defined roles in cardiac hypertrophy and/or heart failure are described here, but this list is rapidly expanding. Some lncRNAs are presented on two rows due to substantial differences in the experimental research design and/or conclusions. miRNA, microRNA; TAC, transverse aortic constriction; UTR, untranslated region.
Table 2 |.
lncRNAs that suppress cardiac hypertrophy and ameliorate cardiac function
| lncRNA | Models | Functions | Interactions and effects | Ref. |
|---|---|---|---|---|
| Ahit (antihypertrophic interrelated transcript) | Mouse and in vitro (rat cardiomyocytes) | Inhibition of Ahit induces cardiac hypertrophy, both in vitro and in vivo | Interacts with SUZ12 | 163 |
| Caren (cardiomyocyte-enriched non-coding transcript) | Mouse | Cardioprotective effects by regulating the translation of a distant gene and maintaining cardiomyocyte homeostasis | Interacts with HINT1 | 49 |
| CHAR (cardiac hypertrophy-associated regulator) | Mouse and in vitro (cardiomyocytes) | CHAR downregulation is sufficient to induce hypertrophic phenotypes in healthy mice; overexpression of CHAR reduced the hypertrophic response | Interacts with miRNA-20b (downregulation) | 164 |
| CPhar (cardiac physiological hypertrophy-associated regulator) | Mouse (exercise model) | Overexpression of CPhar prevents myocardial ischaemia-reperfusion injury and cardiac dysfunction in vivo | CPhar works with DDX17 as a binding partner to sequester C/EBPβ, leading to decreased levels of ATF7 | 165 |
| GAS5 (growth arrest-specific 5) | Mouse (Srsf4 knock-out) | Represses cardiac hypertrophy in Srsf4 knock-out mice | GAS5 is a repressor of the glucocorticoid receptor | 166 |
| H19 | Human and mouse | H19 inhibits E3 ligase-dependent polyubiquitination at Lys3584 of dystrophin (referred to as Ub-DMD), impeding dystrophin protein degradation | Interacts with dystrophin | 71,72 |
| In vitro (cardiomyocytes) | H19–miRNR-675 axis acts as a negative regulator of cardiac hypertrophy by targeting CaMKIIδ | Inhibition of miRNA-675 reversed the increase in cardiomyocyte size associated with H19 overexpression; implicates miRNA-675 derived from H19 in cardiac hypertrophy | 68,167 | |
| lnccytb (lncRNA cytb) | Mouse | Cytosolic lncRNA; function consistent with microRNA sponge | Interacts with miRNA-103–3p | 168 |
| lncMYH7b | In vitro (induced pluripotent stem cell-derived cardiomyocytes) | lncMYH7b regulates the ratio of MHCβ to MHCα, thereby influencing cardiac rhythm | Interacts with TEAD3 | 126 |
| Mhrt or Myheart (myosin heavy-chain-associated RNA transcripts) | Mouse | Highly expressed in adult heart; stress represses Mhrt expression; overexpression of Mhrt protects the heart against cardiac hypertrophy and heart failure in response to stress | Mhrt antagonizes the function of the chromatin remodelling enzyme BRG1 | 117 |
| Mhrt (lncRNA-Mhrt) | Mouse and in vitro (cardiomyocytes) | lncRNA-Mhrt inhibits cardiac hypertrophy by inhibiting myocardin | Interacts with myocardin through miRNA-145a-5p | 169 |
| Mhrt779 | Mouse (exercise and TAC models) | Mhrt779 increases antihypertrophic effects associated with other interventions | Interacts with BRG1-HDAC2 | 120–122 |
| OIP5-AS1 (Opa interacting protein 5-antisense RNA 1; 1700020I14Rik; Cyrano) | Mouse (knock-out model) | Heart failure associated with TAC-induced pressure overload is exacerbated in female (but not male) knock-out mice | Interacts with genes involved with mitochondrial function | 170 |
| Trdn-as (triadin-antisense) | Mouse and in vitro | Role in cardiac conduction through splicing of target genes | Interacts with triadin | 45 |
| ZNF593-as (RP11–96L14.7; ENST00000448923.2) | Mouse | Localized in the cytoplasm of cardiomyocytes; levels are decreased in the failing hearts of patients with dilated cardiomyopathy; improves TAC-induced cardiac dysfunction | ZNF593-as acts as a guide or scaffold for HNRNPC with RYR2 mRNA, thereby stabilizing RYR2 mRNA | 171 |
lncRNA, long non-coding RNA; miRNA, microRNA; TAC, transverse aortic constriction.
H19
H19 is a lncRNA that is expressed at high levels in both cardiac and skeletal muscle61. The function of H19 has also been extensively characterized, with published studies linking it with the genomic imprinting disorders Silver–Russell syndrome and Beckwith–Wiedemann syndrome62,63 and it has also emerged as a very promising therapeutic target for cancer treatment64. Several studies have examined the function of H19 in cardiac hypertrophy and HF61,65–68. These studies revealed that H19 expression is induced during the early stages of cardiac hypertrophy, then downregulated during decompensation. In response to adeno-associated virus 9 (AAV9)-mediated delivery of H19 to mouse hearts, it was possible to attenuate the development of cardiac hypertrophy, in part by suppression of nuclear factor of activated T cells (NFAT) expression61, a transcription factor associated with the induction of the hypertrophic gene expression programme in the heart. The suppression of NFAT activity was proposed to be mediated by direct interaction between H19 and the polycomb repressive complex 2 (PRC2), which led to a reduction in tescalcin levels and function, resulting in a reduction in NFAT expression61 (Fig. 2).
Fig. 2 |. lncRNAs regulation of cardiac hypertrophy induced by calcium and NFAT.
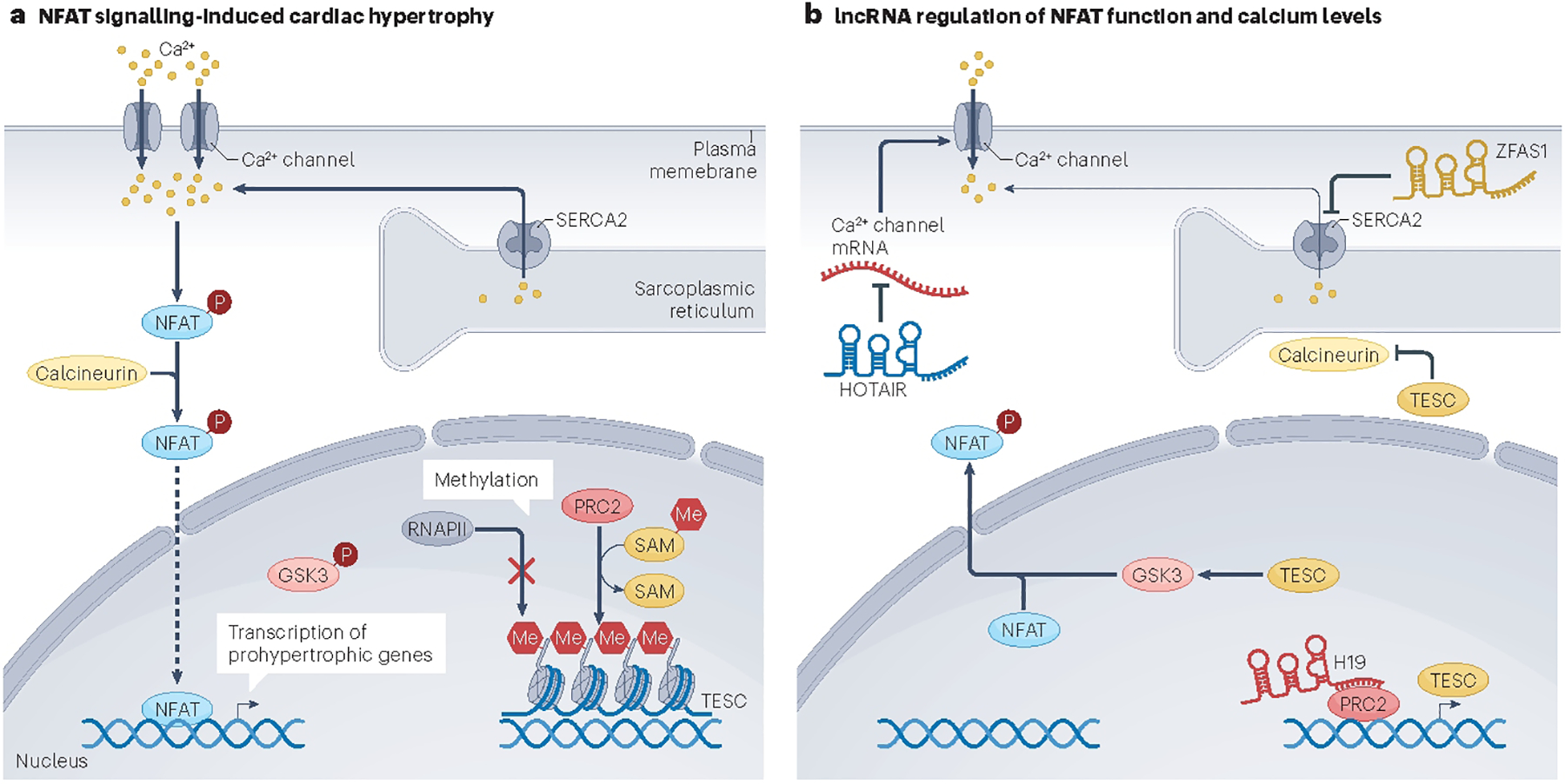
a, Calcium is a key mediator of cardiac hypertrophy. Calcium enters the cell via calcium channels and is sequestered into the sarcoplasmic reticulum via the sarcoplasmic/endoplasmic reticulum calcium ATPase 2 (SERCA2). The long non-coding RNA (lncRNA) ZFAS1 (ZNFX1 antisense RNA 1) binds to and limits the activity of SERCA2 protein, resulting in elevated intracellular calcium levels. In the presence of calcium, calcineurin dephosphorylates nuclear factor of activated T cells (NFAT), which translocates to the nucleus and activates the transcription of prohypertrophic genes, including the lncRNA ZFAS1. b, The lncRNA HOTAIR (HOX antisense intergenic RNA) is proposed to alter calcium homeostasis by decreasing Ca2+ channel activity. Another lncRNA, H19, binds to the protein polycomb repressive complex 2 (PRC2) and thereby promotes the transcription of the TESC gene encoding tescalcin (TESC). The TESC protein inhibits the phosphorylation of glycogen synthase kinase 3 (GSK3), thereby enabling GSK3 to phosphorylate and prevent the DNA-binding function of NFAT. Similarly, TESC suppresses the phosphatase activity of calcineurin A199. This pathway results in suppression of the hypertrophic gene response initiated by NFAT activation. Me, methyl group; P, phosphase group; Pol II, RNA polymerase II; SAM, S-adenosyl methionine.
H19 overexpression has been reported to reduce the size of cardiomyocytes both at baseline and in response to the hypertrophic agonist phenylephrine in vitro; conversely, knockdown of H19 induced cardiomyocyte hypertrophy68. Apart from its role in cardiac hypertrophy, H19 has also been linked to cardiomyocyte proliferation, metabolism and cardiac regeneration through its association with protein lin-28 homologue A (LIN28a)69. The ability of LIN28a (a multifunctional RNA-binding protein) to induce cardiomyocyte metabolism and cell cycle activity were blunted when H19 was inhibited in neonatal rat ventricular cardiomyocytes69. However, the mechanism by which H19 controls these biological processes has not been fully characterized.
The mechanism of action of this lncRNA is further complicated by the fact that another regulatory molecule, miRNA-675, is embedded in the first exon of H1970. Whether H19 regulates cardiomyocyte hypertrophy independently of miRNA-675 or whether the two molecules function together remains unclear. Moreover, the excision of miRNA-675 from H19 is further regulated by the stress-response RNA-binding protein HuR, establishing a multi-tiered regulatory node in the regulation of cardiac hypertrophy70. Evidence suggests that miRNA-675 helps to mediate the function of H19 in regulating hypertrophy, given that an H19 fragment without the pre-miRNA-675 (or with mutant) sequences does not inhibit cardiomyocyte hypertrophy68.
In another study into H19 regulation of the dystrophin protein, a novel mechanism of lncRNA action was discovered. H19 was found to bind to the tail end of the dystrophin cysteine-rich domain, which protected a lysine residue from ubiquitination, thereby stabilizing the dystrophin protein71,72. Moreover, in a mouse model of muscular dystrophy, agrin-conjugated H19 mimics were shown to retain the ability to stabilize dystrophin, resulting in improved muscle function and alleviation of the muscular dystrophy-associated cardiomyopathy71.
Despite these findings, some controversy remains about the role of H19 in the heart, given that some studies suggest it is cardioprotective whereas others suggest it promotes heart disease61. Furthermore, although H19 is more abundant in cardiomyocytes than in cardiac fibroblasts, it is even more highly expressed in endothelial cells than either of those cells types, which suggests possible disparate functions in different cell lineages)61. Although this observation alone does not preclude an important role for H19 in the heart, it does suggest that its mechanism of action could be more complicated; perhaps the influence of H19 on cardiomyocyte function is a result of H19 expression and function in an adjacent cell type. Further study will be required to resolve these questions before consideration of H19 as therapeutic agent would be reasonable.
CHRF
CHRF (cardiac hypertrophy related factor) was among the first-characterized lncRNAs determined to have a role in heart tissue; it was initially identified as a target of miRNA-48973. By binding directly to miRNA-489, CHRF acts as a sponge to prevent miRNA-489 from binding to myeloid differentiation primary response protein 88 (MYD88), its regulatory target; in so doing, CHRF removes the repressive action of miRNA-489 on MYD88 and induces cardiac hypertrophy73 (Fig. 3). This study also demonstrated that exogenous expression of Chrf led to increased apoptosis of cardiomyocytes in mouse hearts73; however, knockdown of Chrf using small interfering RNA (siRNA) also restored atrial natriuretic factor (Anf) and β-myosin heavy chain (β-Mhc) levels in the heart to normal73. Although some aspects of this study indicate that CHRF is a promising lncRNA for regulating cardiac hypertrophy and function by acting as a miRNA sponge, follow-up studies to characterize this function further have not yet been conducted.
Fig. 3 |. lncRNA regulation of miRNA-dependent cardiac hypertrophy.
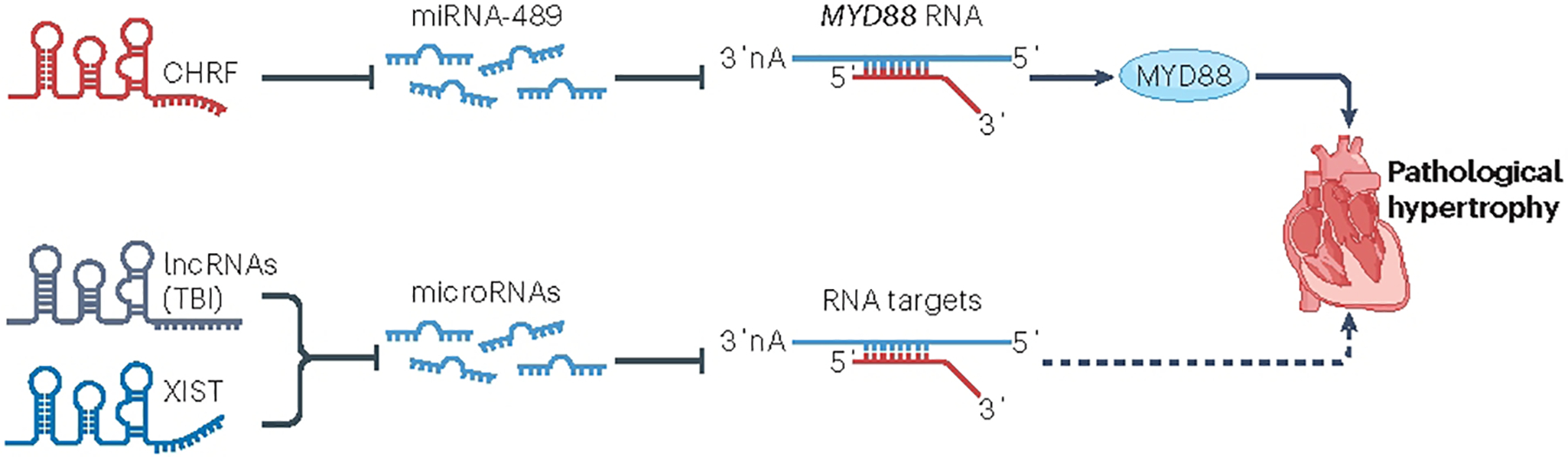
Long non-coding RNAs (lncRNAs), such as CHRF (cardiac hypertrophy related factor) and XIST (X-inactive specific transcript), sequester microRNAs (miRNAs) and prevent their binding to target mRNAs. Myeloid differentiation primary response 88 (MYD88) has been shown to promote inflammatory signalling and lead to cardiac hypertrophy after myocardial infarction. miR-489 suppresses MYD88 activity. Given that CHRF acts as an endogenous sponge of miR-489 and downregulates miR-489 expression levels, CHRF releases MYD88 from the inhibitory influence of this miRNA, enabling the downstream cardiac hypertrophic response. Similarly, the Toll-like receptor 2 (TLR2) is essential for activating the IGF1–PI3K–AKT pathway and promoting cardiac hypertrophy. miR-101 directly targets TLR2 to repress its activity. By suppressing the action of miR-101, the lncRNA XIST activates the TLR2-dependent cardiac hypertrophic response.
ZFAS1
ZFAS1 (ZNFX1 antisense RNA 1) is another lncRNA that seems to participate in the crucial biological process of cardiomyocyte contractility74. ZFAS1 is an antisense lncRNA transcribed from the 5′ end region of the protein-coding gene ZNFX1, and circulating levels of ZFAS1 were measured by quantitative polymerase chain reaction and found to be decreased in whole-blood samples from patients with acute MI75. Knockdown of endogenous Zfas1 in mice partially protects the heart against ischaemia-induced contractile dysfunction, whereas AAV-based overexpression of Zfas1 in the heart impairs contractile function of cardiomyocytes74. Preliminary studies suggest that Zfas1 overexpression results in intracellular Ca2+ overload in cardiomyocytes, providing insights into its mechanism of action74. ZFAS1 is located in the cytoplasm and is associated with the sarcoplasmic reticulum, where it directly binds to the sarcoplasmic/endoplasmic reticulum Ca2+ ATPase 2a (SERCA2a) protein to limit its activity and reduce cardiomyocyte contraction74 (Fig. 2).
This observation raises intriguing possibilities when considered in the light of a previous study that reported the identification of a dwarf open reading frame (DWORF) in an annotated lncRNA gene76. DWORF encodes a micropeptide localized to the sarcoplasmic reticulum membrane and increases SERCA2a activity by displacing inhibitors of the SERCA2a pump, including phospholamban, sarcolipin and myoregulin76. Further investigation to determine whether ZFAS1 and DWORF interact physically and/or functionally to regulate the activity of SERCA2a in cardiomyocytes is ongoing. However, given that the ZFAS1 gene is conserved across species, it might function as a novel biomarker and/or therapeutic target for ischaemic cardiac disease. At this time, studies in mouse and rat have determined that inhibition of endogenous Zfas1 improves cardiac function in response to MI74,77; however, whether ZFAS1 is involved in MI-induced myocardial death and/or cardiac regeneration is uncertain. Furthermore, caution will need to be exercised when considering applications in humans because increased expression of ZFAS1 has been linked with cancer and other diseases78–80.
CHAST
CHAST (cardiac hypertrophy-associated transcript) was originally identified and reported as a lncRNA that was dysregulated during cardiac hypertrophy61,81. Not surprisingly its transcription is regulated by the NFAT signalling pathway, an important component of the cellular processes that govern cardiac hypertrophy and pathological remodelling74. Through a series of gain-of-function and loss-of function studies in mice, Chast overexpression was shown to be sufficient to induce cardiomyocyte hypertrophy both in vitro using isolated cardiomyocytes and in vivo using mouse models81. In an attempt to define a therapeutic approach, GapmeRs (a form of antisense oligonucleotide) were used to inhibit Chast activity and were able to prevent the development of hypertrophy when applied early after TAC81. If treatment was delayed, the use of GapmeRs could still attenuate TAC-induced pathological cardiac remodelling81.
The genetic sequence of Chast corresponds to the antisense strand of two adjacent genes, Arhgap27 and Plekhm181. Chast has been proposed to act as a cis regulatory factor for Plekhm181, but how these two interact to modulate cardiac hypertrophy has not been determined. Chast is highly conserved across mammalian species, and expression of the CHAST homologue in humans was also substantially upregulated in the hypertrophic hearts of patients with aortic stenosis81. These findings support further research into the possible value of CHAST as a target for therapeutic approaches to treat cardiac hypertrophy, but there is still little independent evidence to support the role of CHAST in cardiac remodelling. Further studies to elucidate the molecular mechanisms underlying the observed function would help to justify follow-up studies to establish the therapeutic potential of this lncRNA for the treatment of cardiac disease.
CHAER
Chaer (cardiac-hypertrophy-associated epigenetic regulator) is another lncRNA that has an important role in cardiac hypertrophy and remodelling82–84. Inhibition of Chaer expression in mouse hearts substantially attenuated pressure overload-induced cardiac hypertrophy and heart dysfunction before the onset of remodelling82; however, inhibition of Chaer was not sufficient to correct the cardiac maladaptive remodelling after the onset of pressure overload82,83. These data support a role for Chaer as an early mediator in the events leading to cardiac hypertrophy and HF. Chaer has been shown to interact directly with PRC2, a key epigenetic regulator that functions to maintain transcriptional repression82. This activity is transient and stress-dependent and modulates the transcription and function of hypertrophy-related genes82 (Fig. 4). This study provides an important piece of evidence as to the mechanism of action of CHAER, but whether it has additional roles in other modes of gene transcription and/or in post-translational modification and protein translation remains to be determined. Studies on possible clinical applications of CHAER have not been conducted.
Fig. 4 |. lncRNA regulation of chromatin-mediated cardiac hypertrophy.
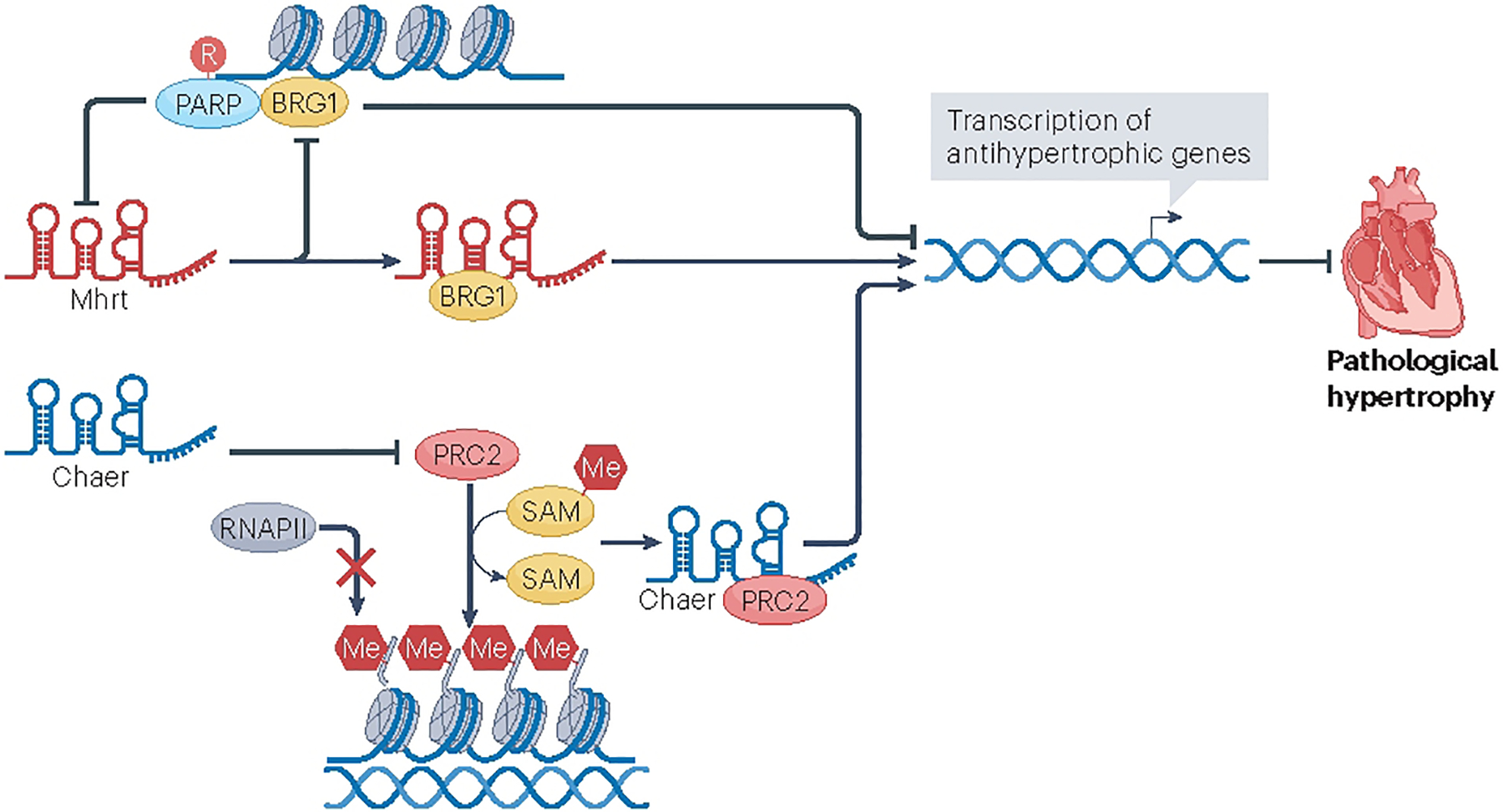
Long non-coding RNAs (lncRNAs) such as Mhrt (myosin heavy-chain-associated RNA transcripts) and Chaer (cardiac-hypertrophy-associated epigenetic regulator) have been shown to regulate the hypertrophic response through their interaction with chromatin-regulatory complexes. Mhrt binds the transcription activator BRG1 and prevents it from recognizing its genomic DNA targets. This releases the chromatin from its inhibitory conformation and facilitates the transcription of an antihypertrophic gene profile. Similarly, Chaer directly binds to polycomb repressive complex 2 (PRC2), releasing chromatin from a repressed state; however, this lncRNA enables the hypertrophic response, suggesting the activation of a hypertrophic gene profile. Me, methyl group; PARP, poly (ADP-ribose) polymerase; Pol II, RNA polymerase II; R, ribosyl group; SAM, S-adenosyl methionine.
MALAT1
MALAT1 (metastasis-associated lung adenocarcinoma transcript 1), also known also NEAT2 (nuclear enriched abundant transcript 2), is a nuclear lncRNA that is widely expressed in multiple tissues, including the heart85. Algorithmic analysis indicates that the MALAT1 transcript has low protein-coding potential86. MALAT1 was also one of the first lncRNAs to be extensively studied in multiple biological systems because its dysregulation has been associated with several disease conditions87. The ~7 kb MALAT1 primary transcript localizes to the nucleus, whereas MALAT1-associated small cytoplasmic RNA (mascRNA) is found exclusively in the cytoplasm88. MALAT1 regulates vascular endothelial cell proliferation and angiogenesis89,90, suggesting that this lncRNA might have a role in cardiac remodelling. However, studies using a loss-of-function genetic mouse model indicated that Malat1 had no significant effect on pathological cardiac hypertrophy, although it remains a topic of active research91.
MIAT
MIAT (myocardial infarction-associated transcript) was originally identified from the mouse nervous system as an mRNA-like non-coding gene called Gomafu92. Further analysis revealed that MIAT is a highly conserved intergenic lncRNA; mutations or single-nucleotide polymorphisms of this lncRNA are associated with increased risks of ischaemic heart disease and/or MI93,94. Furthermore, patients with chronic Chagas disease and dilated cardiomyopathy have increased MIAT expression in heart tissue95, and patients with type 2 diabetes mellitus and maladaptive cardiac remodelling demonstrate similar increases in serum MIAT levels96. Studies in mice show that genetic deletion of Miat attenuates pathological hypertrophy and HF induced by pressure overload or angiotensin II infusion97. Although the molecular mechanism remains unclear, it was observed that the expression levels of SERCA2a and ryanodine receptor 2 in the hearts of these Miat-null mice exposed to pressure overload or angiotensin II were restored to normal levels; restored expression of these, and possibly other, proteins from the reduced levels typically associated with cardiac hypertrophy might contribute to the observed cardiac protection.
A similar finding was reported when small hairpin (shRNA)-mediated inhibition of Miat was used to protect mouse hearts against MI-induced cardiac injury98. The investigators found that Miat directly interacted with translocator protein homologue (TSPO)98, which is primarily found on the outer mitochondrial membrane, suggesting that MIAT might act as a pro-apoptotic lncRNA. The researchers propose that during stress, mitochondria are damaged and the mitochondrial death pathway is triggered by MIAT through TSPO. Another independent study, using both gain-of-function and loss-of-function approaches, found that Miat overexpression worsens cardiac remodelling and that genetic deletion of Miat protects hearts against MI-mediated injury99, consistent with previous findings. The researchers suggest that MIAT functions as a sponge for miRNA-150, consistent with the finding that overexpression of miRNA-150 attenuates the detrimental effects of MIAT in response to MI99.
In addition to its role in cardiomyocytes and cardiomyopathy, the function of MIAT has been linked to smooth muscle cells and atherosclerosis100. Serum levels of MIAT were found to be markedly upregulated in patients with advanced carotid artery atherosclerotic lesions101. In human cultured carotid smooth muscle cells, nuclear MIAT binds to the promoter region of the KLF4 gene to increase its transcription100. GapmeR-based MIAT knockdown led to decreased proliferation and migration of human carotid artery smooth muscle cells in culture but also increased apoptosis100. This finding is in sharp contrast to those in cardiomyocytes, in which MIAT knockdown seems to be beneficial and reduces cardiomyocyte apoptosis in response to stress98. These conflicting tissue-specific functions will require consideration if MIAT is further investigated as a therapeutic target for cardiovascular disease.
XIST
XIST (X-inactive specific transcript) was one of the first lncRNAs to be linked to X chromosome inactivation via an epigenetic mechanism102. Other studies have reported that XIST is expressed in the heart and has a role in the regulation of cardiomyocyte hypertrophy103. Initial studies described high levels of XIST expression in the hearts of female patients with idiopathic dilated cardiomyopathy and the onset of HF103. Although this study did not provide sufficient evidence to establish a firm role for XIST in cardiomyopathy, it does support a role for this lncRNA in the regulation of cardiac function. Multiple reports suggest that XIST might regulate cardiomyocyte hypertrophy by targeting miRNAs104,105 (Fig. 3). These studies used only in vitro models and must, therefore, be interpreted with caution, but the observation that the loss of Xist in the mdx mouse (a model of muscular dystrophy) leads to a delay in the onset of HF supports a role for XIST in the regulation of cardiac function, either directly or indirectly106,107.
HOTAIR
HOTAIR (HOX antisense intergenic RNA) has been well studied in many biological systems and disease states, including cancer108, but has also been linked to cardiomyocyte function and cardiac hypertrophy109–111. HOTAIR can suppress the expression of the calcium channel, CaV1.2 (Fig. 2), whereas knockdown of HOTAIR promotes the expression of CaV1.2 in human cardiomyocytes in vitro109. Other studies have expanded on this work to demonstrate that Hotair is downregulated in heart tissue from TAC-operated mice as well as in cultured cardiomyocytes treated with angiotensin II to induce a cardiac hypertrophy phenotype in vitro110. Overexpression of Hotair in these systems suppresses the cardiac hypertrophy molecular profile110. A role for HOTAIR in cardiac regeneration has also been suggested by a study examining its expression during mouse neonatal development111.
lncRNAs expressed in striated muscle: roles in myogenesis, cardiac function and disease
Cardiac striated muscle and skeletal striated muscle share numerous structural and physiological characteristics, including the sarcomere, the basic contractile unit of the myocyte. In cardiomyocytes, the genes MYH6 and MYH7 encode the myosin heavy chain-α (MHCα) and MHCβ proteins, respectively. In the adult ventricular tissue of large mammals such as humans, MYHβ is the more abundant form, whereas in small mammals such as mice, MYHα is more prevalent112. This difference in the expression patterns of MYH6 and MYH7 in large versus small mammals has been well studied and correlates with the requirement for the higher actin-activated ATPase of MYHα to achieve the higher cardiac contractility rates in small animals. MYH6 is also predominantly expressed in the atria of both large and small mammals113. Subsequent studies of the genomic regions in mice revealed that two microRNAs (miRNA-208a and miRNA-208b) are encoded in the introns of Myh6 and Myh7, respectively114–116. These microRNAs are functional and expressed in a cardiac-specific manner. A cluster of lncRNA transcripts have also been reported as antisense transcripts from the Myh7 locus117. We discuss these below, and others are summarized in Table 3.
Table 3 |.
lncRNAs involved in cardiomyocyte proliferation, cardiac regeneration or cardiac conduction
| lncRNA | Models | Functions | Interactions and effects | Ref. |
|---|---|---|---|---|
| AZIN2-sv (AZIN2-splice variant)172,173 | Rat | AZIN2-sv suppresses endogenous cardiac regeneration by targeting the PTEN-AKT pathway; knock-down of AZIN2-sv attenuates ventricular remodelling and improves cardiac function after myocardial infarction | Stabilizes PTEN through binding; acts as a sponge of miRNA-214 to release PTEN | 172 |
| APF (autophagy promoting factor) | Mouse | Inhibition of APF reduces ischaemia-reperfusion injury after myocardial infarction in vivo | Binds to and regulates miRNA-188–3p | 174 |
| CAIF (cardiac autophagy inhibitory factor) | In vitro (cardiomyocytes) | CAIF suppresses cardiac autophagy and attenuates damage from myocardial infarction | Binds to p53 protein and blocks p53-mediated myocardin transcription | 175 |
| CARMEN (cardiac mesoderm enhancer-associated non-coding RNA) | Human and mouse | Expression of CARMEN is activated during pathological remodelling in mouse and human hearts; necessary for maintaining cardiac identity in differentiated cardiomyocytes; knock-down inhibits specification and differentiation in cardiac precursor cells | Interacts with SUZ12 and EZH2 | 176 |
| CCRR (cardiac conduction regulatory RNA; AK045950) | Human and mouse | CCRR is downregulated in humans with heart failure and a mouse model of heart failure; inhibition of CCRR induces arrhythmias in healthy mice (eliminated by CCRR overexpression); heart failure or CCRR knock-down damages intercalated discs and gap junctions to slow longitudinal cardiac conduction | Binds to CIP85 | 48 |
| CRRL (cardiomyocyte regeneration-related lncRNA) | Mouse | Loss of CRRL attenuates remodelling after myocardial infarction and preserves cardiac function in adult rats; CRRL promotes cardiomyocyte proliferation | CRRL acts as a competing endogenous RNA by binding to miRNA-199a-3p, which results in increased expression of HOPX | 177 |
| ECRAR (endogenous cardiac regeneration-associated regulator) | Human and rat | Promotes DNA synthesis, mitosis and cytokinesis in postnatal day 7 and adult rat cardiomyocytes; overexpression stimulates myocardial regeneration after myocardial infarction; knock-down of ECRAR inhibited postnatal day 1 cardiomyocyte proliferation and prevented recovery after myocardial infarction | ECRAR directly binds and promotes phosphorylation of ERK1/2, resulting in activation of cyclin D1 and cyclin E1, which, in turn, activate E2F1 | 178 |
| Meg3 (maternally expressed gene 3) | Human | Meg3 was increased in samples from patients with heart failure; Meg3 has been shown to have pro-apoptotic properties; mice with Meg3 knock-down show improvement in cardiac function after myocardial infarction | Interacts with the RNA-binding protein FUS | 179 |
| Wisper (WISP2 super-enhancer-associated RNA) | Mouse | Enriched in cardiac fibroblasts; antisense oligonucleotide-mediated silencing of Wisper in vivo attenuates myocardial infarction-induced fibrosis and cardiac dysfunction | Associates with TIAL1 to control expression of a profibrotic form of PLOD2 (which is involved in collagen crosslinking and stabilization of the extracellular matrix) | 134 |
Long non-coding RNAs (lncRNAs) with roles in other cardiac growth processes and functions have also been defined. This list of lncRNAs overlaps with those that affect cardiac hypertrophy. miRNA, microRNA.
MHRT
MHRT (or MYHEART, myosin heavy-chain-associated RNA transcripts) was first identified as a cluster of cardiac-specific lncRNAs that were induced during pathophysiological hypertrophy117. Through a series of experiments conducted both in vitro and in vivo, including the use of mouse models of cardiac hypertrophy, it was determined that repression of Mhrt expression under conditions of stress was essential for the development of cardiomyopathy, whereas restoration of Mhrt to its pre-stress levels could protect the heart against hypertrophy and failure117. The Mhrt–transcription activator BRG1 feedback circuit has been implicated in this process; under pathological stress, the BRG1–HDAC–PARP chromatin-repressing signal is activated, which inhibits Mhrt transcription in the heart117. Conversely, Mhrt binding to BRG1 prevents BRG1 from recognizing its genomic DNA targets, thereby inhibiting chromatin targeting and gene regulation by BRG1. This finding suggests that certain lncRNAs are embedded in protein-coding genomic loci and can regulate cardiac function and disease in response to stress, thereby participating in feedback and/or feedforward regulatory cascades in the cardiac system118,119.
Mhrt779 is a member of the Mhrt lncRNA cluster and an antisense RNA of Myh7120. Mechanistically, Mhrt779 seems to participate in epigenetic regulation by directly associating with BRG1. This association might be important to its role in conferring antihypertrophic memory, which refers to a cellular adaptation mechanism in which previous exposure to certain stimuli or stress, such as exercise-induced physiological cardiac hypertrophy, can produce a protective effect on the heart against pathological hypertrophic stressors, such as pressure overload. Given that physiological cardiac hypertrophy produced by exercise hypertrophic preconditioning leads to an increase in the levels of Mhrt779, this lncRNA is thought to be a component in the process to protect the heart from maladaptive remodelling in response to pathological cardiac stress resulting from pressure overload120. These data, which reveal the importance of elevated levels of Mhrt779, partially explains why healthy exercise is good for the heart. However, the finding also raises new questions about whether less-intense exercise can also induce Mhrt779 expression and, therefore, protect the heart. What other factors, in addition to Mhrt779, contribute to cardiac protection remains to be determined121,122.
lncMYH7b (lnc myosin heavy chain 7b) is produced from the MYH7b locus, which encodes a unique member of the myosin heavy chain family that is expressed in striated muscles but does not seem to produce a full-length protein123. Instead, a post-transcriptional exon-skipping mechanism occurs in mammalian cardiac muscle to produce an abridged transcript123. This locus also produces an intronic microRNA, miRNA-499, which has a role in regulating muscle fibre identity by activating slow myofibre and repressing fast myofibre gene programmes115,124. This function is redundant with that of another miRNA, miRNA-208b, which originates from the MYH7 locus115,124. Previous studies suggested that the muscle fibre type-specific function was modulated by miRNA-499 through the regulation of mitochondrial oxidative metabolism125. However, a recent study revealed a new miRNA-499-independent mechanism for the non-coding exon-skipped RNA from the MYH7b locus (lncMYH7b). Mechanistically, lncMYH7b has been suggested to control the activity of a member of the TEA domain transcription factor family (TEAD3), thereby regulating the ratio of MHCβ to MHCα in cardiomyocytes and, in turn, cardiomyocyte function126. Further studies are needed to establish the functional correlation between the protein-coding, miRNA and lncRNA components of this genomic locus. Furthermore, there might be additional, uncharacterized non-coding components in other contractile protein loci that have traditionally been thought to encode only protein-coding transcripts.
CHARME
CHARME (chromatin architect of muscle expression) was originally designated as lnc-31 in mouse because it was derived from the same genomic locus as miRNA-31127,128. CHARME has been characterized as a chromatin-associated muscle-specific RNA that is expressed in striated muscles129. CHARME is conserved between humans and mice, and its expression increases during myogenic differentiation129. CHARME transcripts were detected in the nucleus of myocytes, and its inhibition has been shown to reduce the expression of genes involved in myogenic differentiation and the myogenic programme129. Genetic deletion of Charme results in a reduction in skeletal muscle morphogenesis, as well as a cardiac remodelling phenotype characterized by changes in cardiomyocyte size and structure; these latter changes influence the sculpting of the heart129. Charme is targeted to specific chromosomal sites and is necessary for the disassembly of chromosomal domains and the expression of myogenic genes in skeletal muscle myocytes129. Additionally, Charme has been shown to interact physically with several proteins involved in splicing regulation, including Ptb1 and Matr3, to modulate cardiac gene expression and function130. Interestingly, several genes regulated by CHARME are associated with human cardiomyopathies130, suggesting an important and evolutionarily conserved role in muscle function and disease.
TRDN-AS
TRDN-AS, as discussed in the previous section, is transcribed as an antisense transcript from the locus encoding TRDN44,45. In addition to its function in cardiac tissue discussed above, TRDN-AS was originally discovered to directly regulate the balance between the alternative spliced forms of the triadin gene in cardiac and skeletal muscle45. This lncRNA could, therefore, have important roles and potential therapeutic value for the treatment of diseases affecting both cardiac and skeletal muscle.
lncRNAs expressed in non-cardiomyocyte cells that influence cardiac hypertrophy, remodelling and disease
In addition to roles for cardiomyocyte-expressed lncRNAs in the regulation of cardiac hypertrophy and HF, important insights have been made into the functions of lncRNAs expressed by non-cardiomyocyte cells in the remodelling of the heart in response to stress. In particular, lncRNAs that regulate the expression of genes associated with fibrosis during cardiac remodelling have been found to be crucial to cardiac function during disease progression131,132. Given the severe detrimental effects of cardiac fibrosis on cardiac function, the importance of understanding the mechanisms by which lncRNAs influence this process is paramount.
WISPER
WISPER (WISP2 super-enhancer-associated RNA) is a conserved lncRNA that is enriched in cardiac fibroblasts, and study of WISPER has provided important insights into the important roles of lncRNAs in other cardiac cell types. Examination of Wisper expression under stress conditions using a mouse model of renovascular hypertension133 confirmed that it was elevated in response to volume overload-induced cardiac hypertrophy and fibrosis134. In addition, Wisper silencing reduced the expression of cardiac stress markers134, suggesting a possible approach to reverse the cardiac hypertrophy. Knockdown of Wisper significantly reduces fibroblast proliferation and migration and the expression of cardiac fibroblast gene programmes that are crucial for cell identity, extracellular matrix deposition, proliferation and survival134. Furthermore, Wisper inhibition protected the mouse heart against MI-induced fibrosis and cardiac dysfunction134, suggesting that it might be a therapeutic target for treating heart disease.
MEG3
MEG3 (maternally expressed gene 3) is a genomically imprinted gene that is transcribed in cardiac fibroblasts and has an important role in regulating cardiac remodelling in response to stress135,136. Using gain-of-function and loss-of-function assays both in vitro and in vivo, investigators demonstrated that Meg3 inhibition reduces cardiac hypertrophy and fibrosis in the heart in response to TAC-induced pressure overload136. The mechanism of Meg3 has been studied in the brain as well because of its role in motor neuron development and autophagy after cerebral ischaemia–reperfusion injury137,138. Nuclear MEG3 interacts with the transcription factor p53 through conserved pseudoknot structures (or kissing loops) that facilitate its regulation of p53139. This change in p53 activity influences the expression of a variety of genes involved in cardiac remodelling and fibrosis, including MMP2136.
PFL
Pfl (pro-fibrotic lncRNA) is upregulated in mouse hearts in response to MI140. Further study revealed that Pfl was enriched in cardiac fibroblasts, and its expression increased during cardiac remodelling involving fibrosis140. Adenovirus-mediated shRNA knockdown of Pfl attenuated cardiac interstitial fibrosis and improved cardiac function in response to MI induced in mice by coronary artery ligation140. Overexpression of Pfl in vitro promotes fibroblast proliferation and the fibroblast–myofibroblast transition in mouse cardiac fibroblasts140. Conversely, inhibition of Pfl diminishes transforming growth factor-β1-induced myofibroblast generation and fibrogenesis140. Mechanistically, Pfl was shown to act as a competitive endogenous RNA of let-7d; forced expression of Pfl reduced let-7d expression and activity140,141.
Micropeptides expressed in the heart that regulate cardiac function
lncRNAs are typically defined as RNA transcripts >200 nucleotides in size without protein-coding potential; however, this definition has proven to be inaccurate given that some annotated lncRNAs actually encode micropeptides. As their name suggests, micropeptides typically encode translational products of ≤100 amino acids and are derived from short open reading frames (sORFs) from larger transcripts142. Evidence for the existence of micropeptides in mammalian cells is ever increasing, but their functions remain largely unknown. However, several micropeptides have been identified in striated muscles (cardiac and skeletal muscle) with roles in the regulation of development and function of those tissues19,143. Myomixer, a micropeptide that is enriched in developing and regenerating skeletal muscle, was reported to control the crucial step in myofibre formation during muscle development144. Another muscle-enriched micropeptide, DWORF, was found to be located in the sarcoplasmic reticulum of striated muscles and could activate SERCA to prevent pathological remodelling and Ca2+ dysregulation in a mouse model of HF76,145.
Investigators have attempted to systematically identify micropeptide-coding genes in hypertrophic cardiomyocytes19. Overall, >10,000 open reading frames were detected from the deep sequencing of ribosome-protected fragments19. The investigators identified >100 uncharacterized sORFs in genes that were originally annotated as encoding lncRNAs but were found to encode micropeptides19. Among 15 candidates that were experimentally tested, the investigators verified the coding potential of 11 sORFs. They demonstrated that these micropeptides participate in the regulation of cardiomyocyte hypertrophy by divergent mechanisms, such as modulation of oxidative phosphorylation, the calcium signalling pathway and the mitogen-activated protein kinase pathway19. These microproteins are located in various cell compartments; intriguingly, many are found localized to the mitochondria, possibly related to roles in responding to stress20. Further studies might reveal previously uncharacterized micropeptides that have important roles in cardiomyocyte biology and heart disease.
Clinical implications
The COVID-19 pandemic marked the emergence of RNA-based technologies as a powerful set of tools for the development of new medicines. Although the application was a more traditional use of RNA in its protein-coding capacity, it proved to be a rapid and successful approach for vaccine generation. The interest in RNA-based methodologies that it created will continue to spawn new strategies for the prevention and treatment of disease. The value of lncRNAs as clinical reagents has only just begun to be investigated but their clinical utility is already becoming apparent. One study identified a panel of 2,906 lncRNAs that were either cardiacenriched or differentially expressed between failing and non-failing hearts, and researchers could differentiate between patients with myocarditis and those with acute MI based on the levels of lncRNAs in blood samples146. Other preliminary studies on lncRNA function also report the potential utility of these RNAs as either biomarkers or potential targets for therapeutic study. One study examining the value of lncRNAs as biomarkers for myocardial injury suggested that measuring the circulating levels of these molecules for the detection of cardiac disease or damage could be valuable147; however, the field is too immature to make any clear predictions about their usefulness. Studies characterizing lncRNAs for use as biomarkers of cardiac hypertrophy and disease are summarized in Table 4.
Table 4 |.
lncRNAs that could function as biomarkers for cardiac hypertrophy and cardiac disease
| lncRNAs | Models | Observed change / study findings | Study population parameters | Ref. |
|---|---|---|---|---|
| aHIF, ANRIL, KCNQ1OT1, MALAT1, MIAT | Human | Whole-blood levels of aHIF, KCNQ1OT1 and MALAT1 were higher in patients with MI, whereas the level of ANRIL was lower; the level of MIAT was similar in both groups | Patients with STEMI: n = 274, male/female = 199/75, median age 61 years, range 35–89 years; patients with NSTEMI: n = 140, male/female = 102/38, median age 62 years, range 30–89 years; controls: n = 86, male/female = 70/16, median age 61 years, range 25–82 years | 180 |
| Ahit | Human | Upregulated in serum samples from patients with hypertensive heart disease (compared with individuals with non-hypertrophic hearts) | Patients with cardiac hypertrophy: male/female = 6/5; controls: male/female = 8/7 | 163 |
| BACE1, BACE1-as | Human and mouse | Levels of both transcripts increased in tissue biopsied from left ventricle of patients with HF; transcript levels also increased in a mouse model of ischaemic HF | Patients with HF: n = 18, male/female = 17/1; aged 65.0 ± 0.6 years; controls: n = 17, male/female = 10/6 (noted discrepancy in numbers from report); aged 58.3 ± 3.4 years | 181 |
| Carmen, Fendrr, Mhrt | Human | Left ventricular mass index showed a negative correlation with levels of Mhrt and Fendrr and a positive correlation with levels of Carmen; analysis performed using peripheral blood mononuclear cells from patients with essential hypertension associated with left ventricular hypertrophy | Patients with hypertension: n = 80, male/female = 32/48, aged 67 ± 8 years; controls: n = 25, male/female = 8/17, aged 64 ± 5 years | 182 |
| H19 | Human | Single-nucleotide polymorphisms identified that affect the secondary structure of H19 | Patients with DCM: n = 96, male/female = 70/26, mean age 53 years, range 22–80 years; controls: n = 259, male/female = 129/130, from the 1000 Genomes Project); variants confirmed with cohort of 1,084 patients with DCM and 751 disease-free controls (overall mean age 43 (range 0–83) years, 31% female) | 183 |
| Human, mouse and rat | Dynamically expressed in pathological hearts (changes observed in patient plasma samples, as well as in TAC mouse model of cardiac hypertrophy and rat model of right ventricular failure (monocrotaline treated and pulmonary artery banded)) | Aortic stenosis: n = 24, male/female = 16/8, aged 70 ± 17 years; HCM: n = 12, male/female = 6/6; aged 17 ± 20 years; control hearts: n = 24, male/female = 14/10, aged 39 ± 13 years. Failing hearts: n = 12, male/female = 7/5, aged 72 ± 4 years, fetal hearts: n = 4, aged 12–14 weeks; control (non-failing) hearts: n = 4, male/female = 2/2, aged 48 ± 4 years. LVAD hearts: n = 12, male/female = 9/3, aged 54 ± 6 years; LVAD control hearts: n = 9, male/female = 1/8, aged 40 ± 13 years. 61,67Patients with IPAH: n =52, male/female = 21/31, aged 61 ± 16 years; patients with CTD-IPAH: n = 21, male/female = 5/16, aged 69 ± 8 years; controls: n = 57, male/female = 40/17, aged 46 ± 19 years61,67 | 61,67 | |
| Human | H19 variants associated with elevated risk of developing HCM; genotyped two H19 SNPs in 405 patients with HCM and 550 controls; sequence determined in 100 patients; the incidence of the H19 rs2107425 CC genotype (C homozygous) was higher in patients without sarcomere mutations | Patients with sarcomere-negative HCM: n = 225, male/female = 140/85, aged 56 ± 14 years; patients with sarcomere-positive HCM: n = 180, male/female = 117/63, aged 46 ± 12 years; controls: n = 550, aged 60–85 years | 184 | |
| H19 and LIPCAR | Human | Increased plasma levels associated with increased risk of coronary artery disease in a Chinese population | Patients with coronary artery disease: n = 300, male/female = 188/112, aged 64 ± 11 years; controls: n = 180, male/female = 108/72, aged 63 ± 10 years | 185 |
| H19, HOTAIR, RMRP | Human and mouse | Levels increased in tissue biopsied from left ventricle of patients with ischaemic DCM; also increased in TAC mouse model of cardiac hypertrophy | Patients with ischaemic DCM or HF: n = 18, male/female = 17/1, aged 65 ± 1 years; controls: n = 16, male/female = 10/6, aged 58 ± 3 years | 186 |
| Heat2 | Human | Levels elevated in the blood of patients with HF; enriched in circulating immune cells | Next-generation sequencing cohort (cohort 1): patients with HF: n = 4, male/female = 4/0, aged 62 ± 14.5 years; controls: n = 4, male/female = 4/0, aged 29 ± 2.8 years. First validation cohort (cohort 2): patients with HF: n = 16, male/female = 3/13, aged 65 ± 11.1 years; controls: n = 8, male/female = 2/6, aged 64 ± 5.2 years. Second validation cohort (cohort 3): patients with HF: n = 69, male/female = 58/11, aged 65 ± 10.5 years; controls: n = 38, male/female = 27/11, aged 63.5 ± 12.1 years | 187 |
| LIPCAR188,189 | Human | Plasma levels increased in patients with HF and higher NYHA class | LIPCAR subcohort: patients with HF: n = 967, male/female = 775/192, aged 70 ± 10.8 years | 190 |
| Human | Increased plasma levels in patients with HF after acute MI | Patients with HF after acute MI: n = 59, male/female = 33/26, aged 64 ± 7.2 years; patients without HF after acute MI: n = 68, male/female = 39/29, aged 63 ± 10.5 years | 191 | |
| Human | Plasma levels elevated in patients with HF with reduced ejection fraction and associated with left ventricular remodelling and poor outcomes | Patients with CKD: n = 100, male/female = 41/59, aged 77 ± 8.7 years; patients without CKD: n = 134, male/female = 117/117, aged 72 ± 8.9 years | 192 | |
| Human | Initially downregulated after MI but upregulated during later stages (plasma RNA isolated from three independent cohorts (788 patients) who developed HF with abnormal cardiac remodelling) | Cohort with left ventricular remodelling: patients with acute MI: n = 246, male/female = 200/46, aged 57 ± 14 years. First cohort with HF: patients with ischaemic HF: n = 164, male/female = 136/28; aged 56 ± 11 years; patients with non-ischaemic HF: n = 180, male/female = 139/41, aged 53 ± 11 years. Second cohort with HF: patients with HF and subsequent cardiovascular death: n = 99, male/female = 91/8, aged 59 ± 11 years; patients with HF and no subsequent cardiovascular death: n = 99, male/female = 91/8, aged 59 ± 11 years | 188 | |
| lncCytB | Human and mouse (TAC model) | Plasma levels decreased in patients with HF; cytosolic lncRNA | Patients with HF: n = 12, male/female = 6/6, aged 60 ± 2.8 years; controls: n = 11, male/female = 5/6, aged 54 ± 2.4 years | 168 |
| lncExACT1 | Human | Increased plasma levels in patients with HF | Patients with HF with reduced ejection fraction: n = 18; patients with HF with preserved ejection fraction: n = 16; controls or patients with supraventricular tachycardia but no HF: n = 8 | 158 |
| LncHrt | Human | Levels decreased in the hearts of patients with DCM | Patients with DCM: n = 6, aged 49–68 years; controls: n = 2, aged 15–30 years | 193 |
| lncRNA ENST00000507296 and ENST00000532365 | Human | Plasma levels of lncRNAs ENST00000507296 and ENST00000532365 correlated with cardiac function in patients with DCM | Screening cohort: patients with DCM: n = 10, male/female = 9/1, aged 63–82 years; controls: n = 10, male/female = 10/0, aged 33–56 years. Validation cohort: patients with DCM: n = 64, male/female = 42/22, aged 56 ± 14.6 years; controls: n = 64, male/female = 36/28, aged 56 ± 10.7 years. Replication cohort: patients with DCM: n = 198, male/female = 144/54, aged 55 ± 0.9 years; controls: n = 198, male/female = 133/65, aged 54 ± 0.7 years. Follow-up cohort: patients with DCM (with exclusion of patients with other severe systemic diseases (such as renal failure or hepatic diseases) and malignant tumours or congenital heart diseases or clinically significant valvular diseases): n = 552, male/female = 392/160, aged 55 ± 13 years | 194 |
| Mhrt | Human | Plasma levels of Mhrt were downregulated in patients with chronic HF (compared with healthy controls); potential use of circulating MHRT levels as a diagnostic and prognostic marker for chronic HF | Patients with chronic HF: n = 88, male/female = 50/38, aged 55 ± 6.6 years; controls: n = 65, male/female = 40/25, aged 54 ± 5.9 years | 195 |
| Mhrt, NRON | Human | Plasma levels of both lncRNAs higher in patients with HF than in controls (analysis conducted using real-time RT-PCR) | Patients with HF: n = 72, male/female = 47/25, aged 59 ± 11.2 years; controls: n = 60, male/female = 37/23, aged 60 ± 12 years | 196 |
| NEAT1 | Human | Increased serum levels of NEAT1 associated with decreased microRNA-129–5p expression in patients with chronic HF | Patients with chronic HF: n = 70, male/female = 37/33, aged 67 ± 1.6 years; controls: n = 62, male/female = 34/28, aged 67 ± 1.7 years | 197 |
| NRF | Human | Potential marker to determine the risk of HF after acute MI | Patients with HF after acute MI: n = 76; patients without HF after acute MI: n = 58 | 198 |
CKD, chronic kidney disease; CTD, connective tissue disease-associated; DCM, dilated cardiomyopathy; HF, heart failure; HCM, hypertrophic cardiomyopathy; IPAH, idiopathic pulmonary arterial hypertension; lncRNA, long non-coding RNA; LVAD, left ventricular assist device; MI, myocardial infarction; NSTEMI, non-ST-segment elevation myocardial infarction; TAC, transverse aortic constriction; RT-PCR, reverse transcription polymerase chain reaction; STEMI, ST-segment elevation myocardial infarction;.
Future studies will continue to characterize novel lncRNAs and the molecular mechanisms they use to influence gene and protein expression and function. The definition of these mechanisms alone might lead to the development of new therapeutic strategies, as well as identifying new potential molecular targets. Approaches to exploit lncRNAs for clinical use will also need to be developed, although some tools are already available. The use of AAVs has been labelled as a disruptive innovation in the pharmaceutical industry for gene delivery. In addition to their traditional use for the expression of mRNAs to restore or augment protein function, they could be adopted for use in expression of ncRNAs, such as the lncRNAs described in this Review, if the efficacy of lncRNAs to improve cardiac function is validated in clinical studies. There are already several pharmaceutical companies heavily invested in the use of antisense oligonucleotides for therapeutic strategies, and this approach could similarly be used to reduce the level of a lncRNA (a GapmeR approach) if its expression is detrimental to heart function.
One approach that warrants a much more substantial discussion than is possible here is use of the CRISPR/Cas9 genome editing system to target lncRNAs in patients with cardiovascular disease148. In addition to the use of CRISPR/Cas9 genome editing to delete part or all of a locus, more sophisticated CRISPR/Cas9 techniques are available and are being refined that would allow more precise targeting and intervention. Using such an approach to target a promoter region could allow the expression of a lncRNA to be fine-tuned to either increase or decrease its activity or possibly even alter its function. Whereas the use of a reagent such as a GapmeR would be transient, a CRISPR/Cas9 intervention would be longer lasting (although this durability might not be desirable under all circumstances). In addition, the potential off-target DNA changes associated with the use of CRISPR/Cas9 also have to be considered149. Gene editing is also not trivial when targeting non-coding genes because they lack an open reading frame148.
The results of the studies we have described examining the effects of increasing or decreasing the levels of lncRNAs in animal models are very encouraging. However, one caveat of this body of work is that the poorly conserved nature of ncRNAs across species means that the interpretation of animal studies in the context of human disease must be carefully considered. The gap between these preclinical studies and use in humans is large and will require questions about the specificity and sensitivity of lncRNAs and standard criteria required for any small-molecule therapy to be thoroughly addressed. As with most drugs, the ability to target a specific organ or tissue is often the greatest obstacle; therefore, lncRNAs that are enriched in cardiac tissue might prove the most amenable to clinical applications.
Conclusions
Many early studies suggested that lncRNAs might be biologically unimportant because they did not have profound effects on heart development. However, subsequent studies suggested that lncRNAs have important effects on heart function during cardiac stress. This realization is a reason for optimism for the clinical potential of lncRNAs, as they could emerge as major regulators during cell stress responses and in pathophysiological heart conditions.
Key points.
Long non-coding RNAs (lncRNAs) are a class of non-coding RNAs (ncRNAs) that can regulate gene expression at multiple levels, including transcription, RNA splicing, and protein translation.
There are a variety of lncRNA classes, based on their structures and/or mechanisms of action but their study has been limited by their low levels of expression and relatively poor conservation between species.
LncRNAs are critical regulators of cell differentiation, development, and disease; in the heart, their expression is often associated with stress conditions and they participate in the pathophysiological remodelling associated with cardiac hypertrophy and heart failure.
Studies have shown that dysregulation of lncRNAs is associated with a variety of cardiac diseases, including coronary artery disease (CAD), myocardial infarction (MI), heart failure (HF), and arrhythmias.
Indications from studies in animal models and humans reveal that lncRNAs have great potential for disease diagnosis as biomarkers and as novel targets for the treatment of cardiac disease.
Footnotes
Competing interests
The authors declare no competing interests.
References
- 1.Consortium EP An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74, doi: 10.1038/nature11247 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pennisi E Genomics. ENCODE project writes eulogy for junk DNA. Science 337, 1159, 1161, doi: 10.1126/science.337.6099.1159 (2012). [DOI] [PubMed] [Google Scholar]
- 3.Lee RC, Feinbaum RL & Ambros V The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75, 843–854, doi: 10.1016/0092-8674(93)90529y (1993). [DOI] [PubMed] [Google Scholar]
- 4.Wightman B, Ha I & Ruvkun G Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 75, 855–862, doi: 10.1016/00928674(93)90530-4 (1993). [DOI] [PubMed] [Google Scholar]
- 5.Neppl RL & Wang DZ The myriad essential roles of microRNAs in cardiovascular homeostasis and disease. Genes Dis 1, 18–39, doi: 10.1016/j.gendis.2014.06.003 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Amaral PP, Clark MB, Gascoigne DK, Dinger ME & Mattick JS lncRNAdb: a reference database for long noncoding RNAs. Nucleic Acids Res 39, D146–151, doi: 10.1093/nar/gkq1138 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Quek XC et al. lncRNAdb v2.0: expanding the reference database for functional long noncoding RNAs. Nucleic Acids Res 43, D168–173, doi: 10.1093/nar/gku988 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang KC & Chang HY Molecular mechanisms of long noncoding RNAs. Mol Cell 43, 904–914, doi: 10.1016/j.molcel.2011.08.018 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ma L, Bajic VB & Zhang Z On the classification of long non-coding RNAs. RNA Biol 10, 925–933, doi: 10.4161/rna.24604 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Statello L, Guo CJ, Chen LL & Huarte M Gene regulation by long non-coding RNAs and its biological functions. Nat Rev Mol Cell Biol 22, 96–118, doi: 10.1038/s41580-020-00315-9 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Noh JH, Kim KM, McClusky WG, Abdelmohsen K & Gorospe M Cytoplasmic functions of long noncoding RNAs. Wiley Interdiscip Rev RNA 9, e1471, doi: 10.1002/wrna.1471 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qin T, Li J & Zhang KQ Structure, Regulation, and Function of Linear and Circular Long Non-Coding RNAs. Front Genet 11, 150, doi: 10.3389/fgene.2020.00150 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu S, Chen L & Zhou X Circular RNAs in the regulation of cardiac hypertrophy. Mol Ther Nucleic Acids 27, 484–490, doi: 10.1016/j.omtn.2021.12.025 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Darbellay F & Necsulea A Comparative Transcriptomics Analyses across Species, Organs, and Developmental Stages Reveal Functionally Constrained lncRNAs. Mol Biol Evol 37, 240–259, doi: 10.1093/molbev/msz212 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao J et al. Integrated transcriptomics and epigenomics reveal chamber-specific and species-specific characteristics of human and mouse hearts. PLoS Biol 19, e3001229, doi: 10.1371/journal.pbio.3001229 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Derrien T et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res 22, 1775–1789, doi: 10.1101/gr.132159.111 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carninci P et al. The transcriptional landscape of the mammalian genome. Science 309, 1559–1563, doi: 10.1126/science.1112014 (2005). [DOI] [PubMed] [Google Scholar]
- 18.See K et al. Single cardiomyocyte nuclear transcriptomes reveal a lincRNA-regulated de-differentiation and cell cycle stress-response in vivo. Nature communications 8, 225, doi: 10.1038/s41467-017-00319-8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yan Y et al. The cardiac translational landscape reveals that micropeptides are new players involved in cardiomyocyte hypertrophy. Mol Ther 29, 2253–2267, doi: 10.1016/j.ymthe.2021.03.004 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Heesch S et al. The Translational Landscape of the Human Heart. Cell 178, 242–260 e229, doi: 10.1016/j.cell.2019.05.010 (2019). [DOI] [PubMed] [Google Scholar]
- 21.Ounzain S et al. Genome-wide profiling of the cardiac transcriptome after myocardial infarction identifies novel heart-specific long non-coding RNAs. European heart journal 36, 353–368a, doi: 10.1093/eurheartj/ehu180 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matkovich SJ, Edwards JR, Grossenheider TC, de Guzman Strong C & Dorn GW 2nd. Epigenetic coordination of embryonic heart transcription by dynamically regulated long noncoding RNAs. Proc Natl Acad Sci U S A 111, 12264–12269, doi: 10.1073/pnas.1410622111 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Izumo S et al. Myosin heavy chain messenger RNA and protein isoform transitions during cardiac hypertrophy. Interaction between hemodynamic and thyroid hormone-induced signals. J Clin Invest 79, 970–977, doi: 10.1172/JCI112908 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Izumo S, Nadal-Ginard B & Mahdavi V Protooncogene induction and reprogramming of cardiac gene expression produced by pressure overload. Proc Natl Acad Sci U S A 85, 339–343 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang KC et al. Deep RNA sequencing reveals dynamic regulation of myocardial noncoding RNAs in failing human heart and remodeling with mechanical circulatory support. Circulation 129, 1009–1021, doi: 10.1161/CIRCULATIONAHA.113.003863 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xiao XG, Touma M & Wang Y Decoding the noncoding transcripts in human heart failure. Circulation 129, 958–960, doi: 10.1161/CIRCULATIONAHA.114.007548 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Z et al. LncExpDB: an expression database of human long non-coding RNAs. Nucleic Acids Res 49, D962–D968, doi: 10.1093/nar/gkaa850 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mas-Ponte D et al. LncATLAS database for subcellular localization of long noncoding RNAs. RNA 23, 1080–1087, doi: 10.1261/rna.060814.117 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bridges MC, Daulagala AC & Kourtidis A LNCcation: lncRNA localization and function. J Cell Biol 220, doi: 10.1083/jcb.202009045 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carlevaro-Fita J & Johnson R Global Positioning System: Understanding Long Noncoding RNAs through Subcellular Localization. Mol Cell 73, 869–883, doi: 10.1016/j.molcel.2019.02.008 (2019). [DOI] [PubMed] [Google Scholar]
- 31.Yu B & Shan G Functions of long noncoding RNAs in the nucleus. Nucleus 7, 155–166, doi: 10.1080/19491034.2016.1179408 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rashid F, Shah A & Shan G Long Non-coding RNAs in the Cytoplasm. Genomics Proteomics Bioinformatics 14, 73–80, doi: 10.1016/j.gpb.2016.03.005 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Olgun G, Sahin O & Tastan O Discovering lncRNA mediated sponge interactions in breast cancer molecular subtypes. BMC genomics 19, 650, doi: 10.1186/s12864-018-5006-1 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Y et al. Long non-coding RNA DSCR8 acts as a molecular sponge for miR-485–5p to activate Wnt/beta-catenin signal pathway in hepatocellular carcinoma. Cell Death Dis 9, 851, doi: 10.1038/s41419-018-0937-7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gil N & Ulitsky I Regulation of gene expression by cis-acting long non-coding RNAs. Nat Rev Genet 21, 102–117, doi: 10.1038/s41576-019-0184-5 (2020). [DOI] [PubMed] [Google Scholar]
- 36.Ma W, Liu Y, Ma H, Ren Z & Yan J Cis-acting: A pattern of lncRNAs for gene regulation in induced pluripotent stem cells from patients with Down syndrome determined by integrative analysis of lncRNA and mRNA profiling data. Exp Ther Med 22, 701, doi: 10.3892/etm.2021.10133 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yan P, Luo S, Lu JY & Shen X Cis- and trans-acting lncRNAs in pluripotency and reprogramming. Curr Opin Genet Dev 46, 170–178, doi: 10.1016/j.gde.2017.07.009 (2017). [DOI] [PubMed] [Google Scholar]
- 38.Anderson KM et al. Transcription of the non-coding RNA upperhand controls Hand2 expression and heart development. Nature 539, 433–436, doi: 10.1038/nature20128 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yotova IY et al. Identification of the human homolog of the imprinted mouse Air non-coding RNA. Genomics 92, 464–473, doi: 10.1016/j.ygeno.2008.08.004 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wutz A et al. Imprinted expression of the Igf2r gene depends on an intronic CpG island. Nature 389, 745–749, doi: 10.1038/39631 (1997). [DOI] [PubMed] [Google Scholar]
- 41.Zwart R, Sleutels F, Wutz A, Schinkel AH & Barlow DP Bidirectional action of the Igf2r imprint control element on upstream and downstream imprinted genes. Genes Dev 15, 2361–2366, doi: 10.1101/gad.206201 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Santoro F et al. Imprinted Igf2r silencing depends on continuous Airn lncRNA expression and is not restricted to a developmental window. Development 140, 1184–1195, doi: 10.1242/dev.088849 (2013). [DOI] [PubMed] [Google Scholar]
- 43.Hosen MR et al. Airn Regulates Igf2bp2 Translation in Cardiomyocytes. Circ Res 122, 1347–1353, doi: 10.1161/CIRCRESAHA.117.312215 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang L et al. A heart-enriched antisense long non-coding RNA regulates the balance between cardiac and skeletal muscle triadin. Biochim Biophys Acta Mol Cell Res 1865, 247–258, doi: 10.1016/j.bbamcr.2017.11.002 (2018). [DOI] [PubMed] [Google Scholar]
- 45.Zhao Y et al. Cardiomyocyte-Specific Long Noncoding RNA Regulates Alternative Splicing of the Triadin Gene in the Heart. Circulation 146, 699–714, doi: 10.1161/CIRCULATIONAHA.121.058017 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roux-Buisson N et al. Absence of triadin, a protein of the calcium release complex, is responsible for cardiac arrhythmia with sudden death in human. Hum Mol Genet 21, 2759–2767, doi: 10.1093/hmg/dds104 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Altmann HM et al. Homozygous/Compound Heterozygous Triadin Mutations Associated With Autosomal-Recessive Long-QT Syndrome and Pediatric Sudden Cardiac Arrest: Elucidation of the Triadin Knockout Syndrome. Circulation 131, 2051–2060, doi: 10.1161/CIRCULATIONAHA.115.015397 (2015). [DOI] [PubMed] [Google Scholar]
- 48.Zhang Y et al. Long non-coding RNA CCRR controls cardiac conduction via regulating intercellular coupling. Nature communications 9, 4176, doi: 10.1038/s41467-018-06637-9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sato M et al. The lncRNA Caren antagonizes heart failure by inactivating DNA damage response and activating mitochondrial biogenesis. Nature communications 12, 2529, doi: 10.1038/s41467-02122735-7 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Anderson DM et al. A myocardin-adjacent lncRNA balances SRF-dependent gene transcription in the heart. Genes Dev 35, 835–840, doi: 10.1101/gad.348304.121 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vacante F et al. CARMN Loss Regulates Smooth Muscle Cells and Accelerates Atherosclerosis in Mice. Circ Res 128, 1258–1275, doi: 10.1161/CIRCRESAHA.120.318688 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dong K et al. CARMN Is an Evolutionarily Conserved Smooth Muscle Cell-Specific LncRNA That Maintains Contractile Phenotype by Binding Myocardin. Circulation 144, 1856–1875, doi: 10.1161/CIRCULATIONAHA.121.055949 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ni H et al. A Smooth Muscle Cell-Enriched Long Noncoding RNA Regulates Cell Plasticity and Atherosclerosis by Interacting With Serum Response Factor. Arterioscler Thromb Vasc Biol 41, 2399–2416, doi: 10.1161/ATVBAHA.120.315911 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lyu Y et al. Cpmer: A new conserved eEF1A2-binding partner that regulates Eomes translation and cardiomyocyte differentiation. Stem Cell Reports 17, 1154–1169, doi: 10.1016/j.stemcr.2022.03.006 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lv L et al. The lncRNA Plscr4 Controls Cardiac Hypertrophy by Regulating miR-214. Mol Ther Nucleic Acids 10, 387–397, doi: 10.1016/j.omtn.2017.12.018 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen J et al. Induction of microRNA-1 by myocardin in smooth muscle cells inhibits cell proliferation. Arterioscler Thromb Vasc Biol 31, 368–375, doi: 10.1161/atvbaha.110.218149 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huang ZP et al. Loss of microRNAs in neural crest leads to cardiovascular syndromes resembling human congenital heart defects. Arterioscler Thromb Vasc Biol 30, 2575–2586, doi: 10.1161/ATVBAHA.110.213306 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kurdi M & Booz GW Carvedilol protects the infarcted heart by upregulating miR-133: first evidence that disease state affects beta-adrenergic arrestin-biased signaling? J Mol Cell Cardiol 76, 12–14, doi: 10.1016/j.yjmcc.2014.08.004 (2014). [DOI] [PubMed] [Google Scholar]
- 59.Malizia AP & Wang DZ MicroRNAs in cardiomyocyte development. Wiley Interdiscip Rev Syst Biol Med 3, 183–190, doi: 10.1002/wsbm.111 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Townley-Tilson WH, Callis TE & Wang D MicroRNAs 1, 133, and 206: critical factors of skeletal and cardiac muscle development, function, and disease. Int J Biochem Cell Biol 42, 1252–1255, doi: 10.1016/j.biocel.2009.03.002 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Viereck J et al. Targeting muscle-enriched long non-coding RNA H19 reverses pathological cardiac hypertrophy. European heart journal 41, 3462–3474, doi: 10.1093/eurheartj/ehaa519 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Reik W et al. Allelic methylation of H19 and IGF2 in the Beckwith-Wiedemann syndrome. Hum Mol Genet 3, 1297–1301, doi: 10.1093/hmg/3.8.1297 (1994). [DOI] [PubMed] [Google Scholar]
- 63.Miyoshi N et al. Identification of the Meg1/Grb10 imprinted gene on mouse proximal chromosome 11, a candidate for the Silver-Russell syndrome gene. Proc Natl Acad Sci U S A 95, 1102–1107, doi: 10.1073/pnas.95.3.1102 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wu B et al. Long Noncoding RNA H19: A Novel Therapeutic Target Emerging in Oncology Via Regulating Oncogenic Signaling Pathways. Front Cell Dev Biol 9, 796740, doi: 10.3389/fcell.2021.796740 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pagiatakis C, Hall IF & Condorelli G Long non-coding RNA H19: a new avenue for RNA therapeutics in cardiac hypertrophy? European heart journal 41, 3475–3476, doi: 10.1093/eurheartj/ehaa663 (2020). [DOI] [PubMed] [Google Scholar]
- 66.Fernandez-Ruiz I H19 in cardiac hypertrophy. Nat Rev Cardiol 17, 612, doi: 10.1038/s41569-0200434-4 (2020). [DOI] [PubMed] [Google Scholar]
- 67.Omura J et al. Identification of Long Noncoding RNA H19 as a New Biomarker and Therapeutic Target in Right Ventricular Failure in Pulmonary Arterial Hypertension. Circulation 142, 1464–1484, doi: 10.1161/CIRCULATIONAHA.120.047626 (2020). [DOI] [PubMed] [Google Scholar]
- 68.Liu L et al. The H19 long noncoding RNA is a novel negative regulator of cardiomyocyte hypertrophy. Cardiovasc Res 111, 56–65, doi: 10.1093/cvr/cvw078 (2016). [DOI] [PubMed] [Google Scholar]
- 69.Rigaud VOC et al. RNA-Binding Protein LIN28a Regulates New Myocyte Formation in the Heart Through Long Noncoding RNA-H19. Circulation 147, 324–337, doi: 10.1161/CIRCULATIONAHA.122.059346 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Keniry A et al. The H19 lincRNA is a developmental reservoir of miR-675 that suppresses growth and Igf1r. Nat Cell Biol 14, 659–665, doi: 10.1038/ncb2521 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang Y et al. The lncRNA H19 alleviates muscular dystrophy by stabilizing dystrophin. Nat Cell Biol 22, 1332–1345, doi: 10.1038/s41556-020-00595-5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ritso M & Rudnicki MA H19 lncRNA to dystrophin’s rescue. Nat Cell Biol 22, 1289–1290, doi: 10.1038/s41556-020-00598-2 (2020). [DOI] [PubMed] [Google Scholar]
- 73.Wang K et al. The long noncoding RNA CHRF regulates cardiac hypertrophy by targeting miR-489. Circ Res 114, 1377–1388, doi: 10.1161/CIRCRESAHA.114.302476 (2014). [DOI] [PubMed] [Google Scholar]
- 74.Zhang Y et al. LncRNA ZFAS1 as a SERCA2a Inhibitor to Cause Intracellular Ca(2+) Overload and Contractile Dysfunction in a Mouse Model of Myocardial Infarction. Circ Res 122, 1354–1368, doi: 10.1161/CIRCRESAHA.117.312117 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang Y et al. Reciprocal Changes of Circulating Long Non-Coding RNAs ZFAS1 and CDR1AS Predict Acute Myocardial Infarction. Scientific reports 6, 22384, doi: 10.1038/srep22384 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nelson BR et al. A peptide encoded by a transcript annotated as long noncoding RNA enhances SERCA activity in muscle. Science 351, 271–275, doi: 10.1126/science.aad4076 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zou B et al. Knockdown of ZFAS1 improved the cardiac function of myocardial infarction rats via regulating Wnt/beta-catenin signaling pathway. Aging (Albany NY) 13, 12919–12928, doi: 10.18632/aging.202961 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Thorenoor N et al. Long non-coding RNA ZFAS1 interacts with CDK1 and is involved in p53dependent cell cycle control and apoptosis in colorectal cancer. Oncotarget 7, 622–637, doi: 10.18632/oncotarget.5807 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Askarian-Amiri ME et al. SNORD-host RNA Zfas1 is a regulator of mammary development and a potential marker for breast cancer. RNA 17, 878–891, doi: 10.1261/rna.2528811 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ghafouri-Fard S, Kamali MJ, Abak A, Shoorei H & Taheri M LncRNA ZFAS1: Role in tumorigenesis and other diseases. Biomed Pharmacother 142, 111999, doi: 10.1016/j.biopha.2021.111999 (2021). [DOI] [PubMed] [Google Scholar]
- 81.Viereck J et al. Long noncoding RNA Chast promotes cardiac remodeling. Science translational medicine 8, 326ra322, doi: 10.1126/scitranslmed.aaf1475 (2016). [DOI] [PubMed] [Google Scholar]
- 82.Wang Z et al. The long noncoding RNA Chaer defines an epigenetic checkpoint in cardiac hypertrophy. Nat Med 22, 1131–1139, doi: 10.1038/nm.4179 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Viereck J & Thum T Long Noncoding RNAs in Pathological Cardiac Remodeling. Circ Res 120, 262–264, doi: 10.1161/CIRCRESAHA.116.310174 (2017). [DOI] [PubMed] [Google Scholar]
- 84.Wang S EZH2 Dynamically Associates With Non-coding RNAs in Mouse Hearts After Acute Angiotensin II Treatment. Front Cardiovasc Med 8, 585691, doi: 10.3389/fcvm.2021.585691 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ji P et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene 22, 8031–8041, doi: 10.1038/sj.onc.1206928 (2003). [DOI] [PubMed] [Google Scholar]
- 86.Arun G, Aggarwal D & Spector DL MALAT1 Long Non-Coding RNA: Functional Implications. Noncoding RNA 6, doi: 10.3390/ncrna6020022 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lorenzen JM & Thum T Long noncoding RNAs in kidney and cardiovascular diseases. Nat Rev Nephrol 12, 360–373, doi: 10.1038/nrneph.2016.51 (2016). [DOI] [PubMed] [Google Scholar]
- 88.Gast M et al. Long noncoding RNA MALAT1-derived mascRNA is involved in cardiovascular innate immunity. J Mol Cell Biol 8, 178–181, doi: 10.1093/jmcb/mjw003 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Michalik KM et al. Long noncoding RNA MALAT1 regulates endothelial cell function and vessel growth. Circ Res 114, 1389–1397, doi: 10.1161/CIRCRESAHA.114.303265 (2014). [DOI] [PubMed] [Google Scholar]
- 90.Thum T & Fiedler J LINCing MALAT1 and angiogenesis. Circ Res 114, 1366–1368, doi: 10.1161/CIRCRESAHA.114.303896 (2014). [DOI] [PubMed] [Google Scholar]
- 91.Peters T et al. Long Non-Coding RNA Malat-1 Is Dispensable during Pressure Overload-Induced Cardiac Remodeling and Failure in Mice. PloS one 11, e0150236, doi: 10.1371/journal.pone.0150236 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sone M et al. The mRNA-like noncoding RNA Gomafu constitutes a novel nuclear domain in a subset of neurons. J Cell Sci 120, 2498–2506, doi: 10.1242/jcs.009357 (2007). [DOI] [PubMed] [Google Scholar]
- 93.Ohnishi Y et al. Identification of 187 single nucleotide polymorphisms (SNPs) among 41 candidate genes for ischemic heart disease in the Japanese population. Hum Genet 106, 288–292, doi: 10.1007/s004390051039 (2000). [DOI] [PubMed] [Google Scholar]
- 94.Ishii N et al. Identification of a novel non-coding RNA, MIAT, that confers risk of myocardial infarction. J Hum Genet 51, 1087–1099, doi: 10.1007/s10038-006-0070-9 (2006). [DOI] [PubMed] [Google Scholar]
- 95.Frade AF et al. Myocardial Infarction-Associated Transcript, a Long Noncoding RNA, Is Overexpressed During Dilated Cardiomyopathy Due to Chronic Chagas Disease. J Infect Dis 214, 161–165, doi: 10.1093/infdis/jiw095 (2016). [DOI] [PubMed] [Google Scholar]
- 96.de Gonzalo-Calvo D et al. Circulating long-non coding RNAs as biomarkers of left ventricular diastolic function and remodelling in patients with well-controlled type 2 diabetes. Scientific reports 6, 37354, doi: 10.1038/srep37354 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yang L et al. Ablation of lncRNA Miat attenuates pathological hypertrophy and heart failure. Theranostics 11, 7995–8007, doi: 10.7150/thno.50990 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bai X et al. LncRNA MIAT impairs cardiac contractile function by acting on mitochondrial translocator protein TSPO in a mouse model of myocardial infarction. Signal Transduct Target Ther 6, 172, doi: 10.1038/s41392-021-00538-y (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Aonuma T et al. MiR-150 Attenuates Maladaptive Cardiac Remodeling Mediated by Long Noncoding RNA MIAT and Directly Represses Profibrotic Hoxa4. Circ Heart Fail 15, e008686, doi: 10.1161/CIRCHEARTFAILURE.121.008686 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Fasolo F et al. Long Noncoding RNA MIAT Controls Advanced Atherosclerotic Lesion Formation and Plaque Destabilization. Circulation 144, 1567–1583, doi: 10.1161/CIRCULATIONAHA.120.052023 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Fasolo F, Di Gregoli K, Maegdefessel L & Johnson JL Non-coding RNAs in cardiovascular cell biology and atherosclerosis. Cardiovasc Res 115, 1732–1756, doi: 10.1093/cvr/cvz203 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lee JT Epigenetic regulation by long noncoding RNAs. Science 338, 1435–1439, doi: 10.1126/science.1231776 (2012). [DOI] [PubMed] [Google Scholar]
- 103.Heidecker B et al. The gene expression profile of patients with new-onset heart failure reveals important gender-specific differences. European heart journal 31, 1188–1196, doi: 10.1093/eurheartj/ehp549 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Chen Y et al. The long noncoding RNA XIST protects cardiomyocyte hypertrophy by targeting miR330–3p. Biochem Biophys Res Commun 505, 807–815, doi: 10.1016/j.bbrc.2018.09.135 (2018). [DOI] [PubMed] [Google Scholar]
- 105.Xiao L The long noncoding RNA XIST regulates cardiac hypertrophy by targeting miR-101. J Cell Physiol 234, 13680–13692, doi: 10.1002/jcp.28047 (2019). [DOI] [PubMed] [Google Scholar]
- 106.van Putten M et al. Low dystrophin levels increase survival and improve muscle pathology and function in dystrophin/utrophin double-knockout mice. FASEB J 27, 2484–2495, doi: 10.1096/fj.12224170 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.van Putten M et al. Low dystrophin levels in heart can delay heart failure in mdx mice. J Mol Cell Cardiol 69, 17–23, doi: 10.1016/j.yjmcc.2014.01.009 (2014). [DOI] [PubMed] [Google Scholar]
- 108.Liu SJ, Dang HX, Lim DA, Feng FY & Maher CA Long noncoding RNAs in cancer metastasis. Nat Rev Cancer 21, 446–460, doi: 10.1038/s41568-021-00353-1 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Tan WL et al. HOTAIR inhibited intracellular Ca2+ via regulation of Cav1.2 channel in human cardiomyocytes. Cell Mol Biol (Noisy-le-grand) 61, 79–83 (2015). [PubMed] [Google Scholar]
- 110.Lai Y et al. HOTAIR functions as a competing endogenous RNA to regulate PTEN expression by inhibiting miR-19 in cardiac hypertrophy. Mol Cell Biochem 432, 179–187, doi: 10.1007/s11010-0173008-y (2017). [DOI] [PubMed] [Google Scholar]
- 111.Fei Q et al. Downregulation of Hotair or LSD1 Impaired Heart Regeneration in the Neonatal Mouse. DNA Cell Biol 40, 1177–1184, doi: 10.1089/dna.2021.0095 (2021). [DOI] [PubMed] [Google Scholar]
- 112.Weiss A & Leinwand LA The mammalian myosin heavy chain gene family. Annu Rev Cell Dev Biol 12, 417–439, doi: 10.1146/annurev.cellbio.12.1.417 (1996). [DOI] [PubMed] [Google Scholar]
- 113.Gorza L et al. Myosin types in the human heart. An immunofluorescence study of normal and hypertrophied atrial and ventricular myocardium. Circ Res 54, 694–702, doi: 10.1161/01.res.54.6.694 (1984). [DOI] [PubMed] [Google Scholar]
- 114.van Rooij E et al. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science 316, 575–579, doi: 10.1126/science.1139089 (2007). [DOI] [PubMed] [Google Scholar]
- 115.van Rooij E et al. A family of microRNAs encoded by myosin genes governs myosin expression and muscle performance. Dev Cell 17, 662–673, doi: 10.1016/j.devcel.2009.10.013 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Callis TE et al. MicroRNA-208a is a regulator of cardiac hypertrophy and conduction in mice. The Journal of clinical investigation 119, 2772–2786, doi: 10.1172/JCI36154 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Han P et al. A long noncoding RNA protects the heart from pathological hypertrophy. Nature 514, 102–106, doi: 10.1038/nature13596 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Liu J & Wang DZ An epigenetic “LINK(RNA)” to pathological cardiac hypertrophy. Cell Metab 20, 555–557, doi: 10.1016/j.cmet.2014.09.011 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wang Z & Wang Y Dawn of the Epi-LncRNAs: new path from Myheart. Circ Res 116, 235–236, doi: 10.1161/CIRCRESAHA.114.305490 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lin H et al. Antihypertrophic Memory After Regression of Exercise-Induced Physiological Myocardial Hypertrophy Is Mediated by the Long Noncoding RNA Mhrt779. Circulation 143, 2277–2292, doi: 10.1161/CIRCULATIONAHA.120.047000 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.He M, Lin H & Liao Y Response by He et al to Letter Regarding Article, “Antihypertrophic Memory After Regression of Exercise-Induced Physiological Myocardial Hypertrophy Is Mediated by the Long Noncoding RNA Mhrt779”. Circulation 144, e271–e272, doi: 10.1161/CIRCULATIONAHA.121.056310 (2021). [DOI] [PubMed] [Google Scholar]
- 122.Wu G, Gao F & Zhang X Letter by Wu et al Regarding Article, “Antihypertrophic Memory After Regression of Exercise-Induced Physiological Myocardial Hypertrophy Is Mediated by the Long Noncoding RNA Mhrt779c”. Circulation 144, e270, doi: 10.1161/CIRCULATIONAHA.121.055528 (2021). [DOI] [PubMed] [Google Scholar]
- 123.Lee LA, Broadwell LJ, Buvoli M & Leinwand LA Nonproductive Splicing Prevents Expression of MYH7b Protein in the Mammalian Heart. J Am Heart Assoc 10, e020965, doi: 10.1161/JAHA.121.020965 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Bell ML, Buvoli M & Leinwand LA Uncoupling of expression of an intronic microRNA and its myosin host gene by exon skipping. Mol Cell Biol 30, 1937–1945, doi: 10.1128/MCB.01370-09 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Liu J Coupling of mitochondrial function and skeletal muscle fiber type by a miR499/Fnip1/AMPK circuit. EMBO Mol Med 8, 1212–1228, doi: 10.15252/emmm.201606372 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Broadwell LJ et al. Myosin 7b is a regulatory long noncoding RNA (lncMYH7b) in the human heart. J Biol Chem 296, 100694, doi: 10.1016/j.jbc.2021.100694 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ballarino M et al. Novel long noncoding RNAs (lncRNAs) in myogenesis: a miR-31 overlapping lncRNA transcript controls myoblast differentiation. Mol Cell Biol 35, 728–736, doi: 10.1128/MCB.01394-14 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Ballarino M et al. Correction for Ballarino et al., “Novel Long Noncoding RNAs (lncRNAs) in Myogenesis: a miR-31 Overlapping lncRNA Transcript Controls Myoblast Differentiation”. Mol Cell Biol 38, doi: 10.1128/MCB.00167-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Ballarino M et al. Deficiency in the nuclear long noncoding RNA Charme causes myogenic defects and heart remodeling in mice. EMBO J 37, doi: 10.15252/embj.201899697 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Desideri F et al. Intronic Determinants Coordinate Charme lncRNA Nuclear Activity through the Interaction with MATR3 and PTBP1. Cell Rep 33, 108548, doi: 10.1016/j.celrep.2020.108548 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Zhang F et al. Long noncoding RNA Cfast regulates cardiac fibrosis. Mol Ther Nucleic Acids 23, 377–392, doi: 10.1016/j.omtn.2020.11.013 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Huang ZP et al. Long non-coding RNAs link extracellular matrix gene expression to ischemic cardiomyopathy. Cardiovasc Res 112, 543–554, doi: 10.1093/cvr/cvw201 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Wiesel P, Mazzolai L, Nussberger J & Pedrazzini T Two-kidney, one clip and one-kidney, one clip hypertension in mice. Hypertension 29, 1025–1030, doi: 10.1161/01.hyp.29.4.1025 (1997). [DOI] [PubMed] [Google Scholar]
- 134.Micheletti R et al. The long noncoding RNA Wisper controls cardiac fibrosis and remodeling. Science translational medicine 9, doi: 10.1126/scitranslmed.aai9118 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Uchida S Besides Imprinting: Meg3 Regulates Cardiac Remodeling in Cardiac Hypertrophy. Circ Res 121, 486–487, doi: 10.1161/CIRCRESAHA.117.311542 (2017). [DOI] [PubMed] [Google Scholar]
- 136.Piccoli MT et al. Inhibition of the Cardiac Fibroblast-Enriched lncRNA Meg3 Prevents Cardiac Fibrosis and Diastolic Dysfunction. Circ Res 121, 575–583, doi: 10.1161/CIRCRESAHA.117.310624 (2017). [DOI] [PubMed] [Google Scholar]
- 137.Li TH et al. Long non-coding RNA MEG3 regulates autophagy after cerebral ischemia/reperfusion injury. Neural Regen Res 17, 824–831, doi: 10.4103/1673-5374.322466 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Vangoor VR, Gomes-Duarte A & Pasterkamp RJ Long non-coding RNAs in motor neuron development and disease. J Neurochem 156, 777–801, doi: 10.1111/jnc.15198 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Uroda T et al. Conserved Pseudoknots in lncRNA MEG3 Are Essential for Stimulation of the p53 Pathway. Mol Cell 75, 982–995 e989, doi: 10.1016/j.molcel.2019.07.025 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Liang H et al. LncRNA PFL contributes to cardiac fibrosis by acting as a competing endogenous RNA of let-7d. Theranostics 8, 1180–1194, doi: 10.7150/thno.20846 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Leisegang MS LET’s sponge: How the lncRNA PFL promotes cardiac fibrosis. Theranostics 8, 874–877, doi: 10.7150/thno.23364 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Mackowiak SD et al. Extensive identification and analysis of conserved small ORFs in animals. Genome Biol 16, 179, doi: 10.1186/s13059-015-0742-x (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Anderson DM et al. A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell 160, 595–606, doi: 10.1016/j.cell.2015.01.009 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Bi P et al. Control of muscle formation by the fusogenic micropeptide myomixer. Science 356, 323–327, doi: 10.1126/science.aam9361 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Makarewich CA et al. The DWORF micropeptide enhances contractility and prevents heart failure in a mouse model of dilated cardiomyopathy. eLife 7, doi: 10.7554/eLife.38319 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Firat H et al. FIMICS: A panel of long noncoding RNAs for cardiovascular conditions. Heliyon 9, e13087, doi: 10.1016/j.heliyon.2023.e13087 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Schulte C et al. Comparative Analysis of Circulating Noncoding RNAs Versus Protein Biomarkers in the Detection of Myocardial Injury. Circ Res 125, 328–340, doi: 10.1161/CIRCRESAHA.119.314937 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Haemmig S & Feinberg MW Targeting LncRNAs in Cardiovascular Disease: Options and Expeditions. Circ Res 120, 620–623, doi: 10.1161/CIRCRESAHA.116.310152 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Guo C, Ma X, Gao F & Guo Y Off-target effects in CRISPR/Cas9 gene editing. Front Bioeng Biotechnol 11, 1143157, doi: 10.3389/fbioe.2023.1143157 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Dai W & Lee D Interfering with long chain noncoding RNA ANRIL expression reduces heart failure in rats with diabetes by inhibiting myocardial oxidative stress. J Cell Biochem 120, 18446–18456, doi: 10.1002/jcb.29162 (2019). [DOI] [PubMed] [Google Scholar]
- 151.Qian Y et al. A long noncoding RNA CHAIR protects the heart from pathological stress. Clin Sci (Lond) 134, 1843–1857, doi: 10.1042/CS20200149 (2020). [DOI] [PubMed] [Google Scholar]
- 152.Dorn GW 2nd & Matkovich SJ Menage a Trois: intimate relationship among a microRNA, long noncoding RNA, and mRNA. Circ Res 114, 1362–1365, doi: 10.1161/CIRCRESAHA.114.303786 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Gong L, Zhu L & Yang T Fendrr involves in the pathogenesis of cardiac fibrosis via regulating miR-106b/SMAD3 axis. Biochem Biophys Res Commun 524, 169–177, doi: 10.1016/j.bbrc.2020.01.062 (2020). [DOI] [PubMed] [Google Scholar]
- 154.Song C et al. Inhibition of lncRNA Gm15834 Attenuates Autophagy-Mediated Myocardial Hypertrophy via the miR-30b-3p/ULK1 Axis in Mice. Mol Ther 29, 1120–1137, doi: 10.1016/j.ymthe.2020.10.024 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Bischoff FC et al. Identification and Functional Characterization of Hypoxia-Induced Endoplasmic Reticulum Stress Regulating lncRNA (HypERlnc) in Pericytes. Circ Res 121, 368–375, doi: 10.1161/CIRCRESAHA.116.310531 (2017). [DOI] [PubMed] [Google Scholar]
- 156.Cooke JP & Leeper NJ A Missing LNC in Vascular Diseases. Circ Res 121, 320–322, doi: 10.1161/CIRCRESAHA.117.311493 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Cai B et al. Long Noncoding RNA-DACH1 (Dachshund Homolog 1) Regulates Cardiac Function by Inhibiting SERCA2a (Sarcoplasmic Reticulum Calcium ATPase 2a). Hypertension 74, 833–842, doi: 10.1161/HYPERTENSIONAHA.119.12998 (2019). [DOI] [PubMed] [Google Scholar]
- 158.Li H et al. lncExACT1 and DCHS2 Regulate Physiological and Pathological Cardiac Growth. Circulation 145, 1218–1233, doi: 10.1161/CIRCULATIONAHA.121.056850 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Makarewich CA & Thum T Exercise-Induced Long Noncoding RNAs As New Players in Cardiac Hypertrophy. Circulation 145, 1234–1237, doi: 10.1161/CIRCULATIONAHA.122.059278 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Zhang J et al. STAT3-induced upregulation of lncRNA MEG3 regulates the growth of cardiac hypertrophy through miR-361–5p/HDAC9 axis. Scientific reports 9, 460, doi: 10.1038/s41598-01836369-1 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Hoepfner J et al. The long non-coding RNA NRON promotes the development of cardiac hypertrophy in the murine heart. Mol Ther 30, 1265–1274, doi: 10.1016/j.ymthe.2021.11.018 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Li B et al. Sirt1 Antisense Long Noncoding RNA Promotes Cardiomyocyte Proliferation by Enhancing the Stability of Sirt1. J Am Heart Assoc 7, e009700, doi: 10.1161/JAHA.118.009700 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Yu J et al. Long Noncoding RNA Ahit Protects Against Cardiac Hypertrophy Through SUZ12 (Suppressor of Zeste 12 Protein Homolog)-Mediated Downregulation of MEF2A (Myocyte Enhancer Factor 2A). Circ Heart Fail 13, e006525, doi: 10.1161/CIRCHEARTFAILURE.119.006525 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Zhang M et al. Long non-coding RNA cardiac hypertrophy-associated regulator governs cardiac hypertrophy via regulating miR-20b and the downstream PTEN/AKT pathway. Journal of cellular and molecular medicine 23, 7685–7698, doi: 10.1111/jcmm.14641 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Gao R et al. Long Noncoding RNA Cardiac Physiological Hypertrophy-Associated Regulator Induces Cardiac Physiological Hypertrophy and Promotes Functional Recovery After Myocardial Ischemia-Reperfusion Injury. Circulation 144, 303–317, doi: 10.1161/CIRCULATIONAHA.120.050446 (2021). [DOI] [PubMed] [Google Scholar]
- 166.Larrasa-Alonso J et al. The SRSF4-GAS5-Glucocorticoid Receptor Axis Regulates Ventricular Hypertrophy. Circ Res 129, 669–683, doi: 10.1161/CIRCRESAHA.120.318577 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Matkovich SJ A balancing act in cardiac hypertrophy. Cardiovasc Res 111, 8–9, doi: 10.1093/cvr/cvw109 (2016). [DOI] [PubMed] [Google Scholar]
- 168.Zhang X et al. Overexpression of cytosolic long noncoding RNA cytb protects against pressure-overload-induced heart failure via sponging microRNA-103–3p. Mol Ther Nucleic Acids 27, 1127–1145, doi: 10.1016/j.omtn.2022.02.002 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Xu Y, Luo Y, Liang C & Zhang T LncRNA-Mhrt regulates cardiac hypertrophy by modulating the miR-145a-5p/KLF4/myocardin axis. J Mol Cell Cardiol 139, 47–61, doi: 10.1016/j.yjmcc.2019.12.013 (2020). [DOI] [PubMed] [Google Scholar]
- 170.Zhuang A et al. Loss of the long non-coding RNA OIP5-AS1 exacerbates heart failure in a sex-specific manner. iScience 24, 102537, doi: 10.1016/j.isci.2021.102537 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Fan J et al. LncRNA ZNF593-AS Alleviates Contractile Dysfunction in Dilated Cardiomyopathy. Circ Res 128, 1708–1723, doi: 10.1161/CIRCRESAHA.120.318437 (2021). [DOI] [PubMed] [Google Scholar]
- 172.Li X et al. Loss of AZIN2 splice variant facilitates endogenous cardiac regeneration. Cardiovasc Res 114, 1642–1655, doi: 10.1093/cvr/cvy075 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Meloni M, Riley PR & Baker AH A new “lnc” between non-coding RNA and cardiac regeneration. Cardiovasc Res 114, 1569–1570, doi: 10.1093/cvr/cvy153 (2018). [DOI] [PubMed] [Google Scholar]
- 174.Wang K et al. APF lncRNA regulates autophagy and myocardial infarction by targeting miR-188-3p. Nature communications 6, 6779, doi: 10.1038/ncomms7779 (2015). [DOI] [PubMed] [Google Scholar]
- 175.Liu CY et al. LncRNA CAIF inhibits autophagy and attenuates myocardial infarction by blocking p53-mediated myocardin transcription. Nature communications 9, 29, doi: 10.1038/s41467-01702280-y (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Ounzain S et al. CARMEN, a human super enhancer-associated long noncoding RNA controlling cardiac specification, differentiation and homeostasis. J Mol Cell Cardiol 89, 98–112, doi: 10.1016/j.yjmcc.2015.09.016 (2015). [DOI] [PubMed] [Google Scholar]
- 177.Chen G et al. Loss of long non-coding RNA CRRL promotes cardiomyocyte regeneration and improves cardiac repair by functioning as a competing endogenous RNA. J Mol Cell Cardiol 122, 152–164, doi: 10.1016/j.yjmcc.2018.08.013 (2018). [DOI] [PubMed] [Google Scholar]
- 178.Chen Y et al. Long Non-coding RNA ECRAR Triggers Post-natal Myocardial Regeneration by Activating ERK1/2 Signaling. Mol Ther 27, 29–45, doi: 10.1016/j.ymthe.2018.10.021 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Wu H et al. Long noncoding RNA Meg3 regulates cardiomyocyte apoptosis in myocardial infarction. Gene Ther 25, 511–523, doi: 10.1038/s41434-018-0045-4 (2018). [DOI] [PubMed] [Google Scholar]
- 180.Vausort M, Wagner DR & Devaux Y Long noncoding RNAs in patients with acute myocardial infarction. Circ Res 115, 668–677, doi: 10.1161/CIRCRESAHA.115.303836 (2014). [DOI] [PubMed] [Google Scholar]
- 181.Greco S et al. Increased BACE1-AS long noncoding RNA and beta-amyloid levels in heart failure. Cardiovasc Res 113, 453–463, doi: 10.1093/cvr/cvx013 (2017). [DOI] [PubMed] [Google Scholar]
- 182.Kontaraki JE et al. The long non-coding RNAs MHRT, FENDRR and CARMEN, their expression levels in peripheral blood mononuclear cells in patients with essential hypertension and their relation to heart hypertrophy. Clin Exp Pharmacol Physiol 45, 1213–1217, doi: 10.1111/1440-1681.12997 (2018). [DOI] [PubMed] [Google Scholar]
- 183.Martens L et al. A genetic variant alters the secondary structure of the lncRNA H19 and is associated with dilated cardiomyopathy. RNA Biol 18, 409–415, doi: 10.1080/15476286.2021.1952756 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Gomez J et al. Genetic variation at the long noncoding RNA H19 gene is associated with the risk of hypertrophic cardiomyopathy. Epigenomics 10, 865–873, doi: 10.2217/epi-2017-0175 (2018). [DOI] [PubMed] [Google Scholar]
- 185.Zhang Z et al. Increased plasma levels of lncRNA H19 and LIPCAR are associated with increased risk of coronary artery disease in a Chinese population. Scientific reports 7, 7491, doi: 10.1038/s41598-017-07611-z (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Greco S et al. Long noncoding RNA dysregulation in ischemic heart failure. J Transl Med 14, 183, doi: 10.1186/s12967-016-0926-5 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Boeckel JN et al. Identification and regulation of the long non-coding RNA Heat2 in heart failure. J Mol Cell Cardiol 126, 13–22, doi: 10.1016/j.yjmcc.2018.11.004 (2019). [DOI] [PubMed] [Google Scholar]
- 188.Kumarswamy R et al. Circulating long noncoding RNA, LIPCAR, predicts survival in patients with heart failure. Circ Res 114, 1569–1575, doi: 10.1161/CIRCRESAHA.114.303915 (2014). [DOI] [PubMed] [Google Scholar]
- 189.Dorn GW 2nd. LIPCAR: a mitochondrial lnc in the noncoding RNA chain? Circ Res 114, 1548–1550, doi: 10.1161/CIRCRESAHA.114.304028 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Meessen J et al. LIPCAR Is Increased in Chronic Symptomatic HF Patients. A Sub-Study of the GISSI-HF Trial. Clin Chem 67, 1721–1731, doi: 10.1093/clinchem/hvab197 (2021). [DOI] [PubMed] [Google Scholar]
- 191.Yan L et al. Circulating LIPCAR is a potential biomarker of heart failure in patients post-acute myocardial infarction. Exp Biol Med (Maywood) 246, 2589–2594, doi: 10.1177/15353702211036055 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Santer L et al. Circulating Long Noncoding RNA LIPCAR Predicts Heart Failure Outcomes in Patients Without Chronic Kidney Disease. Hypertension 73, 820–828, doi: 10.1161/HYPERTENSIONAHA.118.12261 (2019). [DOI] [PubMed] [Google Scholar]
- 193.Liu N et al. LncRNA LncHrt preserves cardiac metabolic homeostasis and heart function by modulating the LKB1-AMPK signaling pathway. Basic Res Cardiol 116, 48, doi: 10.1007/s00395021-00887-3 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Zhang X et al. Circulating Long Non-coding RNA ENST00000507296 Is a Prognostic Indicator in Patients with Dilated Cardiomyopathy. Mol Ther Nucleic Acids 16, 82–90, doi: 10.1016/j.omtn.2019.02.004 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Zhang L, Wu YJ & Zhang SL Circulating lncRNA MHRT predicts survival of patients with chronic heart failure. J Geriatr Cardiol 16, 818–821, doi: 10.11909/j.issn.1671-5411.2019.11.006 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Xuan L et al. Circulating long non-coding RNAs NRON and MHRT as novel predictive biomarkers of heart failure. Journal of cellular and molecular medicine 21, 1803–1814, doi: 10.1111/jcmm.13101 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Zhang H, Zhang N, Jiang W & Lun X Clinical significance of the long non-coding RNA NEAT1/miR-129–5p axis in the diagnosis and prognosis for patients with chronic heart failure. Exp Ther Med 21, 512, doi: 10.3892/etm.2021.9943 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Yan L, Zhang Y, Zhang W, Deng SQ & Ge ZR lncRNA-NRF is a Potential Biomarker of Heart Failure After Acute Myocardial Infarction. Journal of cardiovascular translational research 13, 1008–1015, doi: 10.1007/s12265-020-10029-0 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Gutierrez-Ford C et al. Characterization of tescalcin, a novel EF-hand protein with a single Ca2+binding site: metal-binding properties, localization in tissues and cells, and effect on calcineurin. Biochemistry 42, 14553–14565, doi: 10.1021/bi034870f (2003). [DOI] [PubMed] [Google Scholar]
- 200.Rockman HA et al. Segregation of atrial-specific and inducible expression of an atrial natriuretic factor transgene in an in vivo murine model of cardiac hypertrophy. Proc Natl Acad Sci U S A 88, 8277–8281, doi: 10.1073/pnas.88.18.8277 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.De Villiers C & Riley PR Mouse models of myocardial infarction: comparing permanent ligation and ischaemia-reperfusion. Dis Model Mech 13, doi: 10.1242/dmm.046565 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Lara-Pezzi E Understanding Cardiac Physiological Hypertrophy in a LncRNA Way. Journal of cardiovascular translational research 15, 3–4, doi: 10.1007/s12265-022-10212-5 (2022). [DOI] [PubMed] [Google Scholar]
- 203.Han P & Chang CP Myheart hits the core of chromatin. Cell Cycle 14, 787–788, doi: 10.1080/15384101.2015.1010963 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]


