Abstract
Adolescent idiopathic scoliosis (AIS) is the most common form of scoliosis, in which spinal curvature develops in adolescence, and 90% of patients are female. Scoliosis is a debilitating disease that often requires bracing or surgery in severe cases. AIS affects 2%–5.2% of the population; however, the biological origin of the disease remains poorly understood. In this study, we aimed to determine the function of a highly conserved genomic region previously linked to AIS using a mouse model generated by CRISPR-CAS9 gene editing to knockout this area of the genome to understand better its contribution to AIS, which we named AIS_CRMΔ. We also investigated the upstream factors that regulate the activity of this enhancer in vivo, whether the spatial expression of the LBX1 protein would change with the loss of AIS-CRM function, and whether any phenotype would arise after deletion of this region. We found a significant increase in mRNA expression in the developing neural tube at E10.5, and E12.5, for not only Lbx1 but also other neighboring genes. Adult knockout mice showed vertebral rotation and proprioceptive deficits, also observed in human AIS patients. In conclusion, our study sheds light on the elusive biological origins of AIS, by targeting and investigating a highly conserved genomic region linked to AIS in humans. These findings provide valuable insights into the function of the investigated region and contribute to our understanding of the underlying causes of this debilitating disease.
Keywords: scoliosis, mouse model, vertebrae, proprioception, gene expression
Background
The most common form of idiopathic scoliosis (IS), which spontaneously arises in individuals without pre-existing comorbidities and primarily affects adolescents, is Adolescent Idiopathic Scoliosis (AIS) [1]. Adolescent-onset scoliosis initially develops between 10–18 years of age and accounts for 85%–90% of IS cases, affecting 2%–3% of adolescents and 2%–5.2% of the total population [2]. Scoliosis is the most common developmental disorder affecting the vertebral column [3].
While AIS is equally common in both sexes early in the disease course, extreme sex bias develops as curve progression increases [4, 5]. As curves progress beyond 40°, females are affected at 10:1 relative to males, with 90% of severely progressive curves requiring surgical intervention. Therefore, AIS appears to be driven by genetic susceptibility to unknown internal and/or extrinsic factors. Intrinsic factors are hypothesized to include hormonal fluctuations during puberty, physical disturbances to stable vertical growth in abnormal muscular and ligament tension, changes in pelvic obliquity and size, particularly in women, and altered dorsal horn/interneuron transmission. Extrinsic factors include environmental factors such as exposure to sunlight, altered vitamin D levels, and nutritional status, which can alter pubertal onset and progression, to name a few [6–8].
Research on the inheritance patterns of AIS, particularly in twin studies, has led to the development of genome-wide association studies (GWAS) [9–11]. Various genes, including LBX1, have been linked to AIS. LBX1 is a highly conserved transcription factor that plays a key role in the development of the spinal cord, heart, and limb muscles [9, 12–14]. The SNP rs11190870 is the most commonly associated variant with an increased risk of AIS. It is located in a highly conserved enhancer element downstream of Lbx1 [13, 15]. A large percentage of disease-associated SNPs, which may be linked to DNA regulation, are found in the non-coding regions of the genome. The onset of diseases in adults is contributed to by tissue- or development-specific roles [16]. Guo et al. [13] found that the DNA fragment had higher activity when the risk SNP (T) was present. Their findings suggest that SNP rs11190870 disrupts the normal function of the enhancer element, leading to an upregulation of LBX1 transcription and, therefore, an increased susceptibility to AIS [13].
Lbx1 was first identified in Drosophila and has since been identified in metazoan genomes [17]. In mice, Lbx1 mRNA expression is seen as early as Embryonic day 9.5 (E9.5) in the developing neural tube, with levels peaking between embryonic days 11.5–12.5 and decreasing steadily by day E16.5 [17]. Lbx1 is crucial for cell fate, migration, and post-mitotic determination during neural tube development. Gross et al. [18] observed that many early-born ventrally migrating cells are Lbx1-derived neurons [18]. These Lbx1+ cells, derived from dI4-6 (Class B) dorsal interneuron progenitors, are born between E10 and 12.5 [18–20], post-mitotically expressing Lbx1 and migrating to populate various regions of the future spinal cord [20]. In mice lacking Lbx1, the substantia gelatinosa within the dorsal horn fails to form, leading to changes in the shape of the gray matter [18, 20]. A second Lbx1-positive lineage, born between E11 and E13, migrates dorsally to the substantia gelatinosa of the dorsal horn and other superficial aspects of the dorsal horn [18, 20]. These late-born cells are called dorsal interneurons late (dIL) A and B (dILA and dILB). Taken together, LBX1 plays a crucial role in the development of interneuron populations in the proprioceptive network.
The proprioceptive system is a subconscious sensory system that allows individuals to perceive and be aware of their body position in space. This system gathers information on fixed body positions through specialized receptors in muscles, tendons, joint capsules, and skin [21]. By contrast, the kinesthetic system provides information on joint and limb movements. The proprioceptive system functions through a series of reflex arcs found in the spinal cord that integrate sensory information and help produce smooth and coordinated movements [22]. Although the role of the proprioceptive system in controlling posture is well understood, recent research has directly implicated proprioceptor dysfunction in maintaining spinal alignment [23, 24]. In peripubertal mice lacking TrkC neurons connecting proprioceptive mechanoreceptors to the spinal cord, spinal curvature development was observed without skeletal dysplasia or muscular asymmetry [24].
Studies have shown that patients with adolescent idiopathic scoliosis (AIS) exhibit proprioceptive deficits, as observed through functional testing of their peripheral joints, such as the knee and elbow [25, 26]. However, the exact location and timing of this dysfunction within neuronal circuitry remain unknown. Maintaining an upright position requires the integration of the somatosensory, visual, and vestibular systems, with most weighting (~70%) on the somatosensory system and the remaining on the vestibular system and some on visual input; however, weighting may change as environmental situations change or there is loss of a system [27]. The reliance on somatosensory input indicates a vital role in proprioception for maintaining spinal alignment and balance, and defects in this system have been linked to AIS [8, 28–30].
Vestibular involvement in scoliosis has been reported in a number of clinical studies, summarized in Hawasli et al. [31], with impaired perception of rotational displacement observed in patients. Haumont et al. (2011) observed worse vestibular function with increased curve progression [32]; however, the findings also reflect overall sensorimotor integration [31, 33]. This has been further confirmed in some animal models, including Lambert et al.‘s (2009) labyrinthectomy of Xenopus laevis [34]. However, not all animal models with vestibular deficits exhibit spinal curvatures. In terrestrial models, the input of limb proprioceptive feedback is compensated, seemingly contributing to curve correction [35]. The contributions of vestibular pathologies have been explored in Scheyerer et al.’s [36] review, with the current literature suggesting a contribution to multifactorial etiology/pathology [36].
The SNP rs11190870 is located in a putative regulatory region, the adolescent idiopathic scoliosis-associated cis-regulatory module (AIS-CRM), which is highly conserved between mice and humans. To better understand the biological cause of AIS, we generated a mouse model using CRISPR-CAS9 gene editing to delete this region of the genome. Our goal was to determine whether the deletion of AIS-CRM affects the expression of nearby genes, especially Lbx1. We also investigated the factors that regulate chromatin structure in vivo and whether there were any changes in the spatial expression of the LBX1 protein or any phenotypic changes after the deletion of AIS-CRM.
Results
Epigenetic analysis of the AIS_CRM genome region
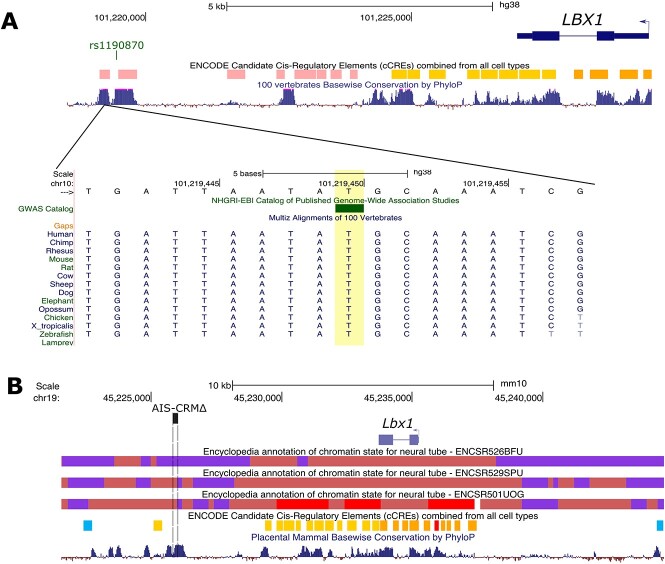
The SNP rs1190870 is located within a highly conserved region near the candidate CRM in human data [14, 37–39]. According to variant analyses in human populations, the T allele (considered high risk) is more prevalent than the C allele, with a ratio of approximately 0.59 (T) to 0.41 (C) in most populations (Table 1). This region is located about 10 kb away from the transcriptional start site (TSS) for LBX1, situated at the edge of a highly conserved genome region (Fig. 1A).
Table 1.
Reference SNP report for rs11190870.
| SNP | rs11190870 |
|---|---|
| Organism | Homo sapiens |
| Position | chr10:101219450 (GRCh38.p14) |
| Alleles | T > A/T > C |
| Variation type | Single nucleotide variation |
| Frequency | C = 0.421317 (137 491/326336, ALFA) C = 0.414077 (109 602/264690, TOPMED) C = 0.411309 (57 408/139574, GnomAD) |
Abbreviations: SNP = single nucleotide polymorph. Data were sourced from (https://www.ncbi.nlm.nih.gov/snp/rs11190870).
Figure 1.
ENCODE regulation data showing candidate CREs and chromatin states for mouse and human LBX1 loci. (A) Human LBX1 gene locus (hg38). The AIS-risk SNP of interest is indicated by the bar (GWAS Catalog track) (rs11190870). The ENCODE registry of candidate cis-regulatory elements (cCREs) based on histone modification, DNase, and CTCF binding data is shown with the corresponding color key. PhyloP score track based on the alignment of 100 vertebrate species to indicate the level of conservation between vertebrates. (B) Mouse Lbx1 locus (mm10 assembly). Chromatin state data (chromHMM 18 state model) for the neural tube (E11.5, E12.5, and E15.5) are displayed, along with the color key. The deleted region in this study is shown (AIS-CRM∆ by the dashed box). PhyloP score track is based on the alignment of placental mammals. Alt. Text. Top panel: A drawing of the human LBX1 gene relative to the location of the SNP of 15 interest, which is ~10 kbp downstream of the gene body. Below is a multiple sequence 16 alignment showing that the SNP rs1190870 is in a region that is 100% identical in other 17 vertebrates including mice. Bottom panel: Overview of the chromatin state of the mouse Lbx1 18 gene locus for neural tube cells. The target region is in an area of repressed by polycomb 19 complex at E11.5, a poised state at E12.5 and an active state at E15.5.
We analyzed the ChromHMM data for the mouse genome (ENCODE phase 3 data) in the developing neural tube, focusing on the Lbx1 locus. We found that this region of the genome, which is rich in histone marks, exhibits a bivalent transcriptional start site (TSSBiv) state interspersed between regions repressed by polycomb complex chromatin (PRC2) (Fig. 1B; Indian red). TSSBiv is frequently associated with tissue-specific transcription factor genes and is often assigned to areas with PRC2-bound silencers [40]. We observed similar chromatin landscapes at E11.5 and E12.5. However, by E15.5, this region shifted to the TSS state (indicating the removal of the repressive mark H3K27me3), suggesting a loss of repressive marks over time (Fig. 1B). This change could potentially affect the expression of nearby genes, such as Lbx1. The region has multiple candidate proximal and distal cCREs, most located near the Lbx1 gene. CTCF-only cCREs were located at each end, with high CTCF signals and low H3K4me3 and H3K27ac candidate sites for chromatin looping or insulators. Although the mouse genomic region corresponding to the AIS-linked SNP in humans is not predicted to lie in cCREs (based on currently available datasets), it lies in a region that shifts from a repressed to bivalent state, indicating the loss of repressive histone marks (Fig. 1B).
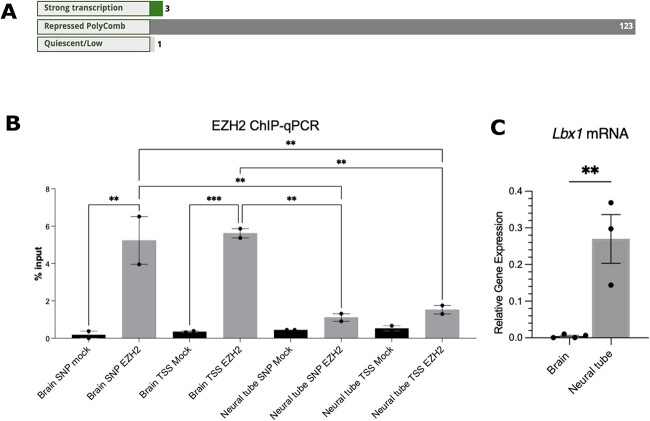
We used RegulomeDB [41] to annotate SNP-associated regions. This database employs various public databases and literature sources to annotate SNP regions. In 123 different tissues, the region containing rs1190870 was marked as repressed polycomb. The tissues include “spinal cord”, “embryo”, “brain”, and “gonad” (for the complete list, see File S1). Chromatin immunoprecipitation data from RegulomeDB revealed that enhancer of zeste homolog 2 (EZH2), SUZ12, and RNF2 significantly bind to 10 cell lines derived from a variety of tissues, including embryos (H1 cell line), the spinal cord (neuronal cells), and blood (B cell) (File S1). These proteins are part of the PRC2.
EZH2 is responsible for H3K27 trimethylation and functions with multiple TFs to repress gene expression [42]. To determine whether EZH2 binds to E12.5 brain and neural tube tissues in vivo, ChIP-qPCR was utilized because the RegulomeDB data above were generated with human tissues (Fig. 2B). These tissues were selected because Lbx1 mRNA is expressed in the neural tube but is barely detectable in the developing brain (Fig. 2C, P = 0.005), suggesting repression in the CNS. The AIS-CRM and Lbx1 TSS areas were targeted. In the brain, EZH2 was significantly enriched in both the AIS-CRM region (Fig. 2B, P = 0.0011), and TSS (Fig. 2B, P = 0.008). However, no significant enrichment was observed in the neural tube compared to the mock control (Fig. 2B, P = 0.096; P = 0.081). In addition, there was significantly more pulldown at these sites within the E12.5 brain than in the neural tube (Fig. 2B, P = 0.0042 and P = 0.0045, respectively).
Figure 2.
EZH2 targets the Lbx1 locus for repression. (A) RegulomeDB (2.0) analysis for chr10:102979206-102 979 207 found that this region is in a repressed chromatin state for most cell types and tissues. (B) ChIP-qPCR for EZH2 (n = 2). (C) The expression of Lbx1 mRNA was higher in the neural tube at E12.5, compared to that in the developing brain (n = 3-4), two-tailed unpaired t-test. One-way ANOVA with Tukey’s multiple comparison test. ***P < 0.001, **P < 0.01. Alt. Text. A. Three horizontal bar graphs showing the chromatin state for 127 tissues and cell 29 types. For 123 of these, the region of interest is in a region repressed by polycomb. Panel B: 30 vertical bar graph showing the percentage pull-down of EZH2 bound to the AIS-CRM genomic 31 region or the transcriptional start site. The data shows ~5-fold increase in bound EZH2 for 32 chromatin extracted from brain tissue compared to the neural tube. Panel C: Vertical bar 33 graph showing that Lbx1 mRNA expression is near 0 for the brain and ~0.25 (relative gene 34 expression) in the neural tube.
Together, these epigenetic data suggest that the target region of the mouse genome, which corresponds to the area containing rs1190870 in the human genome, is within the region of the PRC2-bound repressed chromatin.
Gene expression changes in the AIS-CRM∆ mouse line
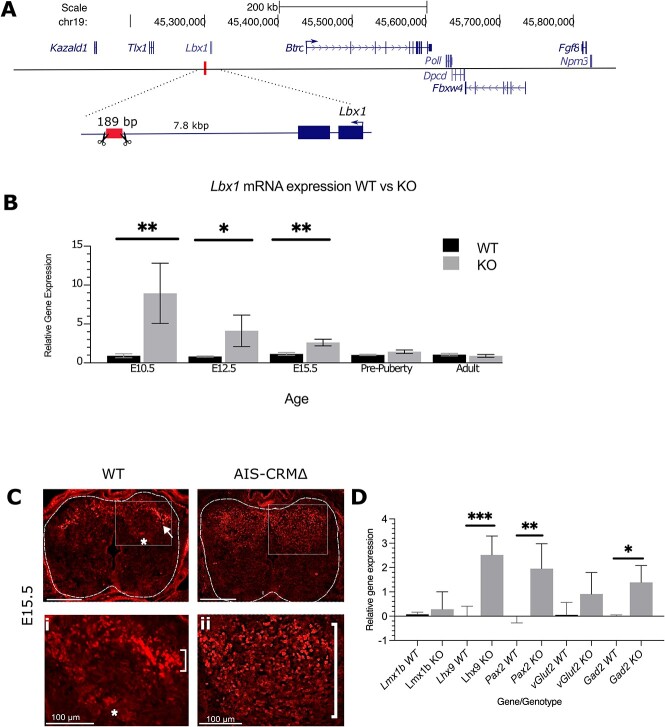
AIS-CRM∆ mice were created using CRISPR-Cas9 genome-editing technology. RNA-guided nucleases cleave DNA phosphodiester bonds at specific sites in the genome, resulting in the deletion or insertion of these regions [43]. The AIS-CRM∆ mouse line removed 189 bp of the mouse genomic region corresponding to the AIS risk SNP rs11190870 (Figs 1B and 3A). The AIS-CRM∆ adult mice were found to breed normally, suggesting unaffected fertility, and the offspring were viable and displayed typical sex ratios and litter sizes (data not shown).
Figure 3.
Lbx1 mRNA and protein expression in WT and AIS-CRM∆ mice. (A) Schema showing the location of the deleted region in the mouse genome. The box (with scissors) indicates the deleted region in the mouse model. (B) RT-qPCR analysis for Lbx1 transcripts reveals a significant increase in mRNA expression at E10.5, E12.5 and E15.5 in the AIS-CRM∆ embryos relative to WT counterparts. The data are presented as 2-∆∆CT relative to WT (mean ± SEM), and statistical significance was determined using an unpaired t-test (*P < 0.05, **P < 0.001). The sample sizes for each age group were as follows: E10.5 n = 10 (WT), n = 3 (AIS-CRM∆); E12.5 n = 11 (WT), n = 8 (AIS-CRM∆), E15.5 n = 17 (WT), n = 7 (AIS-CRM∆); pre-puberty n = 9 (WT), n = 8 (AIS-CRM∆); and adult n = 6 (WT), n = 7 (AIS-CRM∆). (C) Spatial expression of LBX1 in the neural tube and spinal cord of WT and KO mice. (C) Representative image of LBX1 protein in the developing neural tubes of embryos at E15.5. (Ci and Cii) Higher magnification images of the boxed areas in C for WT and KO embryos. Scale bar = 200 μm unless otherwise indicated. (D) RT-qPCR analysis for markers of dorsal interneuron populations and neurotransmitters. Lmx1b (dl5), Lhx9 (dI1i), Pax2 (dI4 and dI6), vGlut (glutamatergic), and Gad2 (GABAergic) the data are presented as 2-∆∆CT relative to WT (mean ± SD), and statistical significance was determined using an unpaired t-test (*P < 0.05, **P < 0.001, ***P < 0.0001, n = 4 WT, n = 3-5 KO). Alt. Text. (A) The deleted region is ~7.8 kbp downstream from the mouse Lbx1 gene. Upstream 9 there are six genes shown: In order—Btrc, Poll, Dpcd, Fbxw4, Fgf8 and Npm3. Panel B: 10 vertical bar graph comparing expression values for Lbx1 mRNA for WT and KO tissues. E10.5 11 mean 0.997 WT, 8.93 KO; E12.5 mean of 0.775 WT, 2.468 KO, means of 0.943 WT, 2.62 KO, 12 pre-puberty means of 1 and 1.4 KO. Panel C. LBX1 protein in a section of the neural tube for 13 WT and KO. In the WT tissue, LBX1 is detected in the nucleus of cells that form an arch shape 14 in the dorsal region of the neural tube. In the KO, LBX1 is detected in a much broader region 15 of the dorsal neural tube. Panel D. Bar graph showing the relative expression of selected 16 markers for WT and KO. Mean values are WT, KO: Lmx1b 0.076, 0.28; Lhx9 0.01, 2.5; Pax2 17 0.001, 1.95; vGlut2 0.052, 0.91; Gad2 0.0007, 1.39.
Next, we sought to determine how deletion of this putative regulatory region influences gene expression in the neural tube and spinal cord, as this genomic region is predicted to alter the expression of nearby genes. Tissue was collected at three embryonic time points (E10.5, 12.5, and 15.5), representing critical time points during embryonic development, where LBX1 is known to affect neuronal migration and identity [18, 20, 44]. Additionally, spinal cord mRNA from two postnatal time points (PN28 pre-puberty and PN120 mature) was used to assess changes in the spinal cord before puberty and at maturity.
First, we focused on Lbx1 mRNA expression, as this gene is predicted to have altered expression in vivo because it is located near the target genomic region (Fig. 3A). Total RNA was isolated from either pre- (neural tube) or post-natal (spinal cord) time points for WT and AIS-CRMΔ littermates for RT-qPCR. We observed a significant increase in Lbx1 mRNA levels in the AIS-CRMΔ mice at E10.5 (~9-fold; P = 0.0024) and at E12.5 (5-fold; P = 0.033) and E15.5 (2.3-fold; P = 0.0024) (Fig. 3B. There was no difference in Lbx1 mRNA expression between WT and AIS-CRMΔ for the pre-puberty and adult spinal cord (Fig. 3B; P = 0.13, P = 0.45).
Following the observation of increased Lbx1 mRNA expression in the embryonic AIS-CRM∆ neural tube, we assessed the spatial distribution of the LBX1 protein. Immunofluorescence experiments were performed on transverse sections of E15.5 thoracic samples (Fig. 3C–E). In E15.5 mice, LBX1 protein was found in the substantia gelationsa dorsal horn (Fig. 3C, arrow) and nucleus proprius (Fig. 3C, asterisk). In KO mice, the expression was detected throughout the dorsal horn substantia gelatinosa.
Genes essential for specifying different dorsal interneuron populations in the NT were analyzed at E12.5 to determine if the overexpression of Lbx1 resulted in changes to their expression levels (Fig. 4.1). This was because Lbx1 overexpression peaks at E12.5 (112). As specific genes are found in specific dorsal interneuron populations, changes in their expression profile will directly result in changes to the composition of that population and thus, functional consequences. No significant differences were observed in the expression levels of Lmx1b between the Lbx1EH∆−/− and WT mice (P = 0.8843, WT n = 3, KO n = 5). A statistically significant upregulation of Lhx9 mRNA expression was observed in the spinal cord of Lbx1EH∆−/− mice compared to the WT mice (P = 0.0007, WT n = 4, KO n = 5). Finally, a statistically significant upregulation of Pax2 mRNA relative to the WT mice was found (P = 0.0082, WT n = 4, KO n = 5) (Fig. 3C). These changes in gene expression suggest the AIS-CRMΔ deletion likely results in a dysregulation of specific dorsal interneuron populations.
Figure 4.
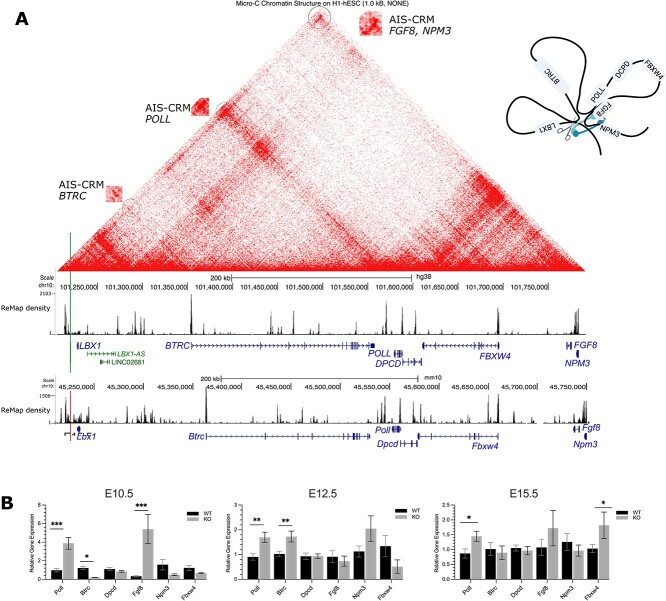
Transcript expression of genes predicted to be in physical contact with the AIS-associated regulatory region in vivo. (A) Human genome (hg38) browser, with micro-C data from human embryonic stem cells (ESC) shown at 1.0 kb resolution and ReMap data. The green vertical line indicates the position of rs1119870 SNP. The red line indicates the deleted region of AIS-CRM. The contact points are circled and enlarged. The density track (ReMap) shows the density of overlapping ChIP-seq peaks. These regions were enriched in CTCF peak regions. In both mice and humans. TAD and internal sub-TAD. The red range indicates the interaction frequency (0 = none, red = high). (B) Poll and Fgf8 mRNA expression significantly increased in the AIS-CRM∆ group compared to the WT group during neural tube development at E10.5 (P < 0.001). At E12.5, the AIS-CRM∆ cohort showed approximately 1.8-fold higher levels of Poll and Btrc transcripts than those in the WT group (P < 0.05). Similarly, at E15.5, there were significant increases in the mRNA expression levels of Poll and Fbx4 (P < 0.05). The results are presented as mean ± SEM. Statistical significance was determined using two-way ANOVA, and the results are presented as *P < 0.05, ***P < 0.001. The sample size for each group was as follows: E10.5 n = 10 (WT), n = 3 (AIS-CRM∆); E12.5 n = 11 (WT), n = 8 (AIS-CRM∆), E15.5 n = 17 (WT), n = 7 (AIS-CRM∆); pre-puberty n = 9 (WT), n = 8 AIS-CRM∆); and adult n = 7 (WT), n = 6 (AIS-CRM∆). Alt. Text: (A) Physical contact map for the LBX1 locus, includes 6 genes upstream and 9 genes 1 downstream. Contacts are shown as a geometric pattern, and the TADs are highlighted—These 2 are shown as triangular shapes, with one corner corresponding to the position of rs11190870; 3 the top of the triangle displays a dense signal suggesting frequent long-range contacts with the 4 regions located on the right bottom corner, ~550 kbp downstream from the SNP. Panel b: 5 vertical bar graph showing the relative expression in WT and KO samples. The mean values 6 for those comparisons that were significant: E10.5—Poll 0.97, 3.88; Btrc 1.22, 0.21; Fgf8 7 0.334, 5.41. E12.5 Poll 0.91, 1.69, Btrc 1.03, 1.73. E15.5 Poll 0.87, 1.45; Fbxw4 1.04, 1.81.
An additional role of LBX1 in spinal cord development is determining GABAergic inhibitory neuron cell fates in the spinal cord [20]. Analyzing the expression of the associated markers VGLUT2, which specifies excitatory glutamatergic neurons, and GAD2, which specifies inhibitory GABAergic neurons, may highlight any dysregulation. While there was a large variation in the expression of vGlut2 mRNA between biological replicates, there was no significant difference between WT and KO (P = 0.16). A statistically significant increase in Gad2 was observed in the AIS-CRMΔ mice compared to their WT counterparts (P = 0.026) (Fig. 3D). This suggests that the AIS-CRMΔ deletion results in a disequilibrium of excitatory and inhibitory neuronal cell fates in the spinal cord with an increase in inhibitory GABAergic neurons.
AIS-CRM is located at the boundary of the TAD
Additionally, we investigated whether AIS-CRM knockout affects the expression of nearby genes. The 3D structure of chromatin and distal regulatory elements allows them to interact with genes across long distances. The genome is organized into topologically associated domains (TADs) within the nucleus. These domains are enriched in intradomain chromatin interactions, isolated from the surrounding chromatin, and bounded by narrow segments known as boundary regions, appearing as triangles (as shown in Fig. 4). TADs are highly conserved and play crucial roles crucial for determining cell fate during development [45, 46].
After analyzing the available HiC and Micro-C chromatin structure data, we discovered potential contacts and a local TAD structure [47] (Fig. 4A and Fig. S1). LBX1 and six other genes were identified within a single TAD, including Fibroblast growth factor 8 (FGF8), F-box and WD-repeat domain containing 4 (FBXW4), β-transducin repeat containing e3 ubiquitin-protein ligase (BTRC), deleted in primary ciliary dyskinesia (DPCD), and DNA polymerase lambda (POLL) (Table S2). Additionally, there are sub/internal TAD interactions, such as contact between the AIS-CRM/TAD boundary and POLL and BTRC.
Thus, the rs11190870 variant is located at the TAD boundary, which was also confirmed by ReMap data. This extensive ChIP-seq database can be accessed using the UCSC browser [48]. The data showed several dense peak areas, including a similar pattern between mouse and human genomes (Fig. 4), regions near the SNP enriched with CTCF, cohesion proteins (SMC3, RAD21), and Polycomb complex (CBX7, EZH2, JARID2) proteins (zoomed-in view, see Fig. S2). Cohesin subunit proteins and CTCF overlap at boundary regions [49]. PRC proteins (EZH2, CBX7, and JARID7) are also associated with genome organization and chromatin looping [50]. They are enriched here, consistent with the previous results (Fig. 2 and Fig. S2).
Similar to Lbx1, the RT-qPCR data showed a significant increase in Poll and Fgf8 mRNA expression at E10.5 in AIS-CRM∆ mice (Fig. 4B). This increase was four-fold for Poll and 16-fold for Fgf8 (Fig. 4B). Btrc mRNA expression decreased by approximately 5.9-fold in AIS-CRM∆ embryos. At E12.5, the Poll and Btrc mRNA levels were approximately 1.8 times higher in the AIS-CRM∆ group than in the WT group (Fig. 4B). Poll mRNA levels remained higher than those in WT at E15.5, ~1.7-fold (Fig. 4B). Additionally, the expression of Fbxw4 mRNA was significantly increased at this stage, ~1.8-fold (Fig. 4B).
There were no significant differences in the expression of any of the tested genes between the WT and KO mice in the pre/post-puberty spinal cord (Fig. S3; P > 0.05). In addition, we also tested a gene just outside the TAD region, Nucleophosmin/nucleoplasmin 3 (Npm3); this gene showed no change in mRNA expression (Fig. 4B). Taken together, these results suggest that the AIS-CRM region of the mouse genome can influence the expression of other genes located within the same TAD during development.
Phenotypic changes to the vertebral column
To assess gross spine morphology, microCT scans were examined using the 3D Viewer plugin for FIJI. No apparent differences were observed in the shape or profile of the vertebral bodies, pedicles/lamina, vertebral foramen, or spinous processes (Fig. S4). We could not find obvious evidence of ultrastructural changes to the vertebrae at 34 μm resolution (data not shown). Vertebral rotation is a feature of scoliosis, and in the absence of obvious lateral curvature, we sought to determine whether any alternative phenotypic characteristics were present [51–54]. Therefore, we determined vertebral rotation in our mouse line. To ensure accurate and comparable results, the 5th Lumbar vertebrae (L5) was selected as the “anchor” with its rotation set as 0°, allowing us to measure vertebral rotation relative to L5 in all samples.
We noted considerable variation in vertebral rotation between the genotypes (Fig. 5). However, the results showed that the variation between the two groups (over the entire vertebral column) was not significantly different, and that pre-puberty WT and AIS-CRM∆ mice exhibited similar variation in vertebral rotation (Fig. 5; P = 0.43). When we examined variation at the individual vertebral level, that is, comparing the variation in standard errors of the mean between WT and AIS-CRM∆ mice, we observed a greater propensity for rotation in AIS-CRM∆ mice. Thus, KO mice showed greater variation than WT mice did.
Figure 5.
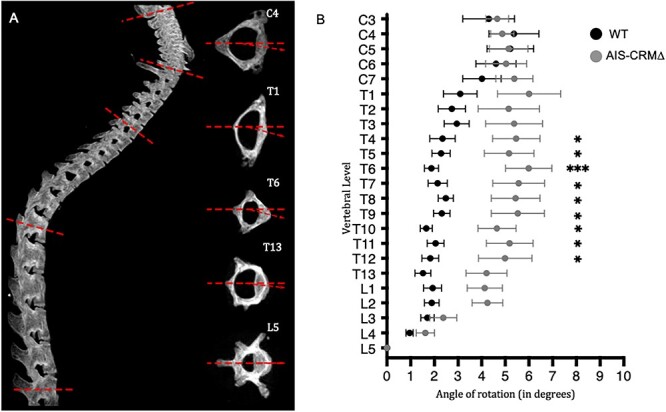
Vertebral rotation in adult WT and AIS-CRM∆ knockout mice using microCT. (A) Lateral view of an example spine (left) and the acquisition of rotational measurements using individually reconstructed vertebrae (right). The dashed line shows baseline, 0° rotation, the arrow shows deviation of the vertebra. (B) Degree of rotation measured across C3-L5 in the mouse samples, combining sexes. Wild-type (WT) mouse measurements are represented in black (n = 20) and AIS-CRM∆ in gray (n = 10). Measurements are presented as mean rotational deviation; error bars represent ± SEM; statistical significance is represented as P < 0.05, *P < 0.01, **P < 0.001, *** using multiple unpaired t-tests with multiple comparison correction (two-stage step-up (Benjamini, Krieger, and Yekutieli)).
In human AIS, curved apices are found in the central vertebrae of the affected region (i.e. the cervical apex is typically C3-4, thoracic T6-T7, thoracolumbar T12-L1) [6, 7, 55]. Thoracic and thoracolumbar curves have been observed in 88% of AIS cases [1, 56, 57]. This led us to focus on the thoracic region in the mouse cohort. We found a significant rotational deviation in the thoracic region, T4-T12, in the AIS-CRM cohort (Fig. 5). Considering rotational variation across all samples, the most considerable degree of rotational deviation is at T6, with a mean of 6° (±2.94; P < 0.001) (Fig. 5). Vertebral rotations for T4–T12 were all significantly more rotated than their WT counterparts (P < 0.05), with a larger variation in AIS-CRM samples than in their WT counterparts, as evidenced by the larger SEM (Fig. S5). Subsequently, two-way ANOVA (pooled) was performed to determine whether the genotype had a similar effect at all vertebral levels, and 4.3% of the variation (P = 0.02) suggested an interaction.
Owing to the sex-biased nature of AIS, we sought to determine whether this difference between WT and AIS-CRM was influenced by biological sex (Fig. S6). The largest rotation angle was nearly identical between our male (6.416° ± 3.491; T6) and female cohorts (6.415° ± 2.824; T8) (Fig. S5). We conducted a comprehensive 3-way ANOVA analysis (File S2) to investigate potential interactions among genotype (WT/KO), sex, and vertebral level. Notably, our findings revealed a significant effect of genotype (P = 0.0001), indicating that the presence or absence of the conserved genomic region associated with AIS in humans plays a pivotal role in vertebral rotation. However, we did not observe any significant interactions between sex, genotype, and vertebral level. This implies that the impact of the AIS-CRM deletion on vertebral rotation remains consistent regardless of sex.
Vertebral rotation was also analyzed in prepubertal mice (< PN28). There were no statistically significant differences between the two cohorts (Fig. S6; P = 0.44). These results (Fig. 5 and Fig. S6) suggested that the onset of vertebral rotation in AIS-CRM mice occurred following pubertal onset.
Simple proprioceptive behavior was affected in AIS-CRM∆ mice relative to their WT littermates
Having demonstrated rotational instability of the vertebrae in AIS-CRM∆ mice, we sought to determine whether these mice exhibited proprioceptive deficits following the findings of Blecher et al [24], who linked proprioceptive system disruption to scoliotic/vertebral instability [24]. This was particularly interesting given the influence of LBX1 on neural tube interneuron development and the potential for the proprioceptive system to be influenced by the altered expression observed above.
Initially, we performed a modified SNAP test (Simple Neuroassessment for asymmetric impairment), as previously described [58], at four time points: 4 weeks (pre-puberty) and 16, 20, and 24 weeks of age. We found that across all time points, AIS-CRM∆ mice scored higher than their WT littermates; this difference was statistically significant at 4 (P = 0.0043) and 16 weeks (P = 0.0093) (Fig. 6). The statistically significant increase in the SNAP score from 4 to 16-week (P = 0.012) for knockout mice coincided with pubertal transition; however, this was not observed in the WT cohort (P = 0.478) (Fig. 6). At 20 weeks of age, WT and AIS-CRM mice performed equally well in the SNAP test. This trend continued until the end of testing at 24 weeks of age (P = 0.12). Most notably, the AIS-CRM∆ cohorts exhibited a wide, splayed stance and splayed hindlimbs when attempting the hind limb baton grip test. Together, this suggests the possibility of a premature decline in proprioceptive function for AIS-CRM∆ and confirms the previously reported changes in age-related proprioceptive functions.
Figure 6.
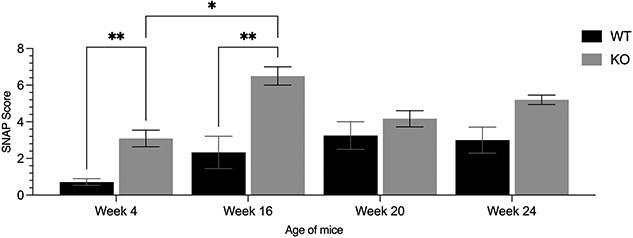
SNAP testing of AIS-CRM∆ revealed premature proprioceptive deficits. The results of the SNAP score suggest that at 4 and 16 weeks of age, AIS-CRM∆ mice performed worse in simple proprioceptive tasks than their WT counterparts (P = 0.0043; P = 0.0124). However, at 20 weeks, the mice performed equally in the proprioceptive tests (P > 0.05), which continued until 24 weeks of age. Data are presented as the mean ± SEM. Statistical significance was determined using two-way ANOVA where **P < 0.01. Abbreviations: SNAP, simple assessment of asymmetric imPairment; WT, wild-type; AIS-CRM. Alt. Text. Vertical bar graph comparing WT and KO SNAP scores for WT and KO. Mean values 16 are—Week 4: 0.71, 3.09, week 16 2.33, 6.5, week 20 3.25, 4.16, week 24 3, 5.2.
We used the cylinder test to evaluate mouse CNS circuits that control simple reflexes and motor function [59]. We wanted to determine if AIS-CRM mice had problems with basic motor skills, including raising, choosing their paws, controlled paw removal, and other noticeable behaviors such as stumbling or falling (Fig. S7). However, between 8 and 24 weeks, we only noticed a modest reduction in the time spent rearing in the WT and AIS-CRM groups, which was not statistically significant (Fig. S7; P = 0.28, P = 0.09, respectively).
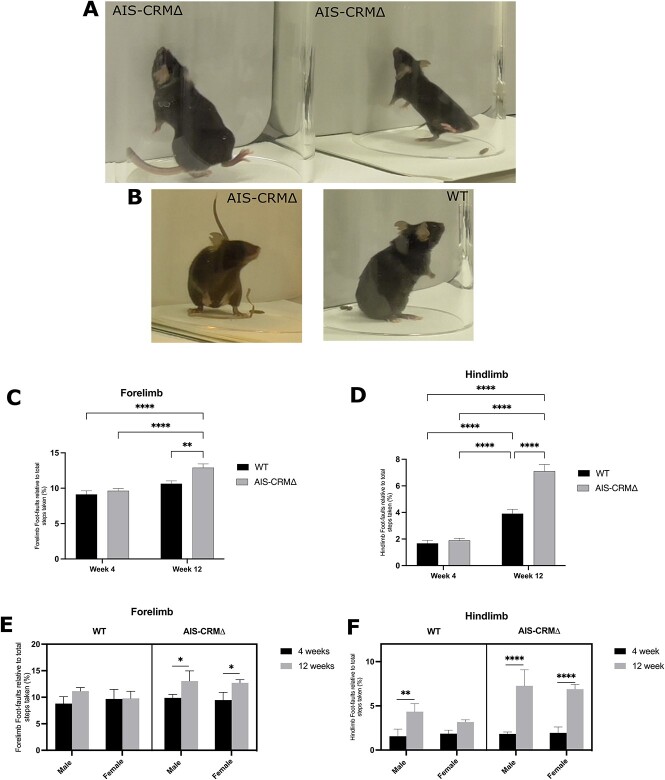
Although AIS-CRM∆ mice spent a similar amount of time rearing as mice without the AIS-CRM∆ mutation, they exhibited some functional impairments in their forelimbs. This was further confirmed through the grid walk and grip strength tests (Figs 7 and S8). Additionally, when standing and reared, AIS-CRM mice exhibited a propensity to fall forwards to the ground because of hindlimb instability (Fig. 7A). Furthermore, their hindlimbs had a wider stance and were more splayed than their WT counterparts (Fig. 7B).
Figure 7.
Observational functional deficits were observed in AIS-CRM∆ mice. (A) AIS-CRM∆ mice showed a loss of hindlimb balance when reared and falling forward towards the ground after failing to touch the cylinder wall. (B) The AIS-CRM∆ mouse (left) was observed with a wider hindlimb stance when reared and on the ground than their WT counterparts (right). Effect of the AIS-CRM∆ deletion on the number of limb foot-faults over 8 weeks. (C) Forelimb foot faults relative to the total steps taken for the WT and AIS-CRM∆. (D) Hindlimb foot faults relative to the total steps taken by WT and AIS-CRM∆. All data are expressed as the mean ± SEM (n = 8 WT and n = 7-13 AIS-CRM∆), and statistical analysis using two-way ANOVA is represented as *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. (E) Forelimb data separated by sex (n = 5 WT and n = 4–5 AIS-CRM∆). (F) Hindlimb foot faults separated by sex (n = 3 WT and n = 3-8 AIS-CRM∆). The data are shown as a three-way plot separated by AIS-CRM genotype, age, and sex. Statistical analysis was performed using three-way ANOVA (mixed effects) and Tukey”s multi-comparison test. Abbreviations: WT, wild-type; AIS-CRM, ∆ knockout mice. Alt. Text. Panel A and B have three examples of KO mice losing balance while performing a 35 cylinder task, whereas the WT mouse has a stable-reared stance. (C–F) Vertical bar graphs 36 comparing WT and KO foot faults at weeks 4 and 12, separated by sex. Mean hindfoot faults 37 are week 4 (WT, KO): 1.67, 1.89; week 12 3.9, 7.1. Mean forelimb foot faults: Week 4 9.14, 9.6; 38 week 12 10.65, 12.91.
The grid walk test evaluates the locomotor function and motor deficits in rodents with CNS disorders [60–62]. It is also commonly used to assess the sensorimotor abilities of forelimbs and hindlimbs [63]. Our goal was to determine if the deletion of certain genes affected the functional abilities of rodents, specifically by measuring the number of foot faults relative to the total number of steps taken per limb (Fig. 7C and D). We found a modest increase in forelimb foot faults in AIS-CRM∆ mice from 4 to 12 weeks of age, which was statistically significant. This increase was not observed in WT littermates over the same period. Additionally, at 12 weeks, there was a significant difference in the number of forelimb foot faults between WT and AIS-CRM∆ mice (1.3-fold; P = 0.0087; Fig. 7C). This suggests that deletion of AIS-CRM∆ results in sensorimotor dysfunction of the forelimb during puberty, which leads to an increase in the number of foot faults.
The WT and AIS-CRM∆ cohorts showed a significant increase in hindlimb foot faults from 4 to 12 weeks of age. The WT cohort showed a 2.3-fold increase (P < 0.0001), whereas the AIS-CRM∆ cohort showed a 3.7-fold increase (P < 0.0001). Although both cohorts experienced an increase in hindlimb foot faults, AIS-CRM∆ mice had a 1.8-fold increase (P < 0.0001, Fig. 7D) in the total number of hindlimb foot faults compared to WT mice. This indicates that, while there is a natural decline in function during this period, the deletion of AIS-CRM leads to a more significant decrease in sensorimotor function.
We conducted a three-way ANOVA using the mixed-effects model REML to examine how sex, AIS-CRM genotype, and age (in weeks) interacted. The results are summarized in File S2. Genotype and age significantly affected forelimb foot faults (P = 0.048; Fig. 7C), whereas sex did not (File S2). When we separated the results by sex, male and female AIS-CRM∆ mice showed a significant increase in forelimb foot faults from four to 12 weeks of age (File S2). For the hind limb, there was a significant interaction between genotype and age (P = 0.0003), but not with sex. Both sexes showed significant increases in hindlimb foot faults (File S2). Female WT mice showed no significant changes in the number of hindlimb foot faults from week 4 to week 12 (P = 0.57, File S2). However, male WT mice experienced a significant increase in hindlimb foot faults (2.8-fold, P = 0.0047; File S2) over 8 weeks. This suggests that male WT mice may experience a decline in hindlimb locomotor function over time. Male and female AIS-CRM∆ mice also showed a significant increase in hindlimb foot faults over eight weeks (P < 0.0001) (Fig. 7F; File S2). This trend continued at 12 weeks of age, with 1.7- and 2.2-fold increases in male and female AIS-CRM∆ mice, respectively (P < 0.0001; Fig. 7F; File S2).
The grip strength test is used to evaluates motor function and deficits in mice with CNS disorders (Maurissen et al [72]). WT and AIS-CRM mice showed a decline in forelimb grip strength over time, with no significant difference in strength between the two groups (Fig. S8, P = 0.08, P = 0.65, respectively). However, there was a significant decrease in forelimb grip strength in WT mice from 4 to 14 weeks (P = 0.004) and from 4 to 24 weeks (P = 0.002), with 38% and 47.8% declines in strength, respectively (Fig. S8). These decreases likely reflect the natural decline in motor function with age, with no apparent differences in grip strength between the WT and AIS-CRM mice. During grip strength testing, AIS-CRM∆ mice behaved differently from WT mice, struggling to hold onto the bar and twisting and turning. This behavior was consistent with observations during SNAP testing, where AIS-CRM∆ mice repeatedly failed to perform the baton grip task appropriately (Fig. 6).
Discussion
In this study, we investigated the genomic region linked to AIS to understand its function. By analyzing data from a mouse model and using bioinformatics and ChIP-qPCR, we found that EZH2 binds to this region near the TAD boundary. Deleting this area in mice causes changes in gene expression in the developing neural tube. This affected the nearby Lbx1 gene and other genes within the same TAD. Our analysis of AIS-CRM∆ mice showed significant vertebral rotation and variation compared with WT mice. Before any morphological changes occurred, these mice displayed proprioceptive deficits and sensorimotor decline. We believe that deleting this genomic region disrupts connectivity in the developing spinal cord. This disruption leads to proprioceptive deficits and spinal misalignments.
Gene regulation and AIS-CRM
According to previous studies, AIS-CRM regulates Lbx1 expression during critical stages of embryonic development, contributing to neural tube patterning and migration. An in vitro luciferase reporter assay conducted by Guo et al. [13] showed that the risk SNP increased luciferase activity and that spinal curvature was caused by lbx1 overexpression in zebrafish [13]. We also observed an increase in mouse Lbx1 expression when the region containing the SNP was knocked out. This increase could be attributed to either an increase in the number of cells expressing Lbx1 and their protein expression, or the detection of cells expressing increased levels of Lbx1. Gross et al. used both antibody and GFP lines and found a broader expression in the GFP line, which they suggested was more sensitive to Lbx1 expression [18]. The observed increase in expression could be due to an increase in the number of cells that normally express lower levels of Lbx1, now detectable with the antibody, or an increase in the number of cells that express Lbx1 and their relative increase in protein expression. We also found that deletion of this region increased the expression of nearby genes.
We initially hypothesized that this region plays a regulatory role in the presence of a PRC2 repressor. PRC2-bound silencers are distal regulatory elements that mediate long-range chromatin interactions with target genes, and disruptions such as CRISPR-Cas9 mediated deletion can lead to the activation of target gene expression [64]. In addition, PRC2-bound silencers can transition to enhancers in specific cell lineages [64]. Changes in TAD boundaries are associated with changes in gene expression in cell culture. TAD boundaries restrict the interactions of cis-regulatory elements with genes within the TAD, and the loss of TAD boundaries results in ectopic gene expression in in vitro cell culture [65]. Furthermore, TAD boundaries often contain housekeeping genes and transcriptional start sites [45].
Possible consequences of changes to Lbx1 gene expression
We observed significant differences in Lbx1 mRNA expression between WT and AIS-CRM∆ mice during spinal cord development; these differences were no longer evident in our adult cohorts (Fig. 3A). We hypothesize that changes in Lbx1 expression during development likely led to changes in downstream effectors, consequently triggering changes in neuronal wiring, neuron identity, and overall functionality.
It has been established that Lbx1 is essential for the specification and migration of the Class B neurons dI4-6 in the E12.5 neural tube [20]. Additionally, Lbx1 antagonizes the differentiation of Class A neurons dI1-3, and the misexpression of Lbx1 causes ectopically expressed Class B neurons and reduced Class A neuronal numbers [20]. Our study supported these findings, whereby the broad Class B domain extensively migrated ventrally and laterally into the mantle zone, implying a large excess of Class B neurons occupying the adult spinal cord (Fig. 3C). Due to the diversity of dI4-6, the marker Lmx1b was further analyzed in the Lbx1EH∆−/− spinal cord to determine the patterning of a single Class B dorsal interneuron population. Surprisingly, Lmx1b had no significant change in gene expression levels in the E12.5 compared to the WT (Fig. 3D). This suggests that the overexpression and migratory increase of Class B neurons may only profoundly affect dI4 and dI6, potentially observed as a lack of uniformity in staining intensity (Fig. 3C). This is supported by the statistically significant upregulation of both Lbx1 and Pax2 gene expression, markers of both dI4 and dI6, respectively (Fig. 3D).
Additionally, dI4/6 are inhibitory GABAergic neuronal types, utilizing GABA as a neurotransmitter, whilst Lmx1b-expressing dI5 is glutamatergic, utilizing VGLUT2 as a neurotransmitter [66]. Results further support this, with a statistically significant upregulation of Gad2, a marker of GABAergic neurons, but no significant difference in vGlut2 or Lmx1b expression (Fig. 3D). As GABAergic neurons are inhibitory, its excessive overexpression relative to vGlut2 means an excessive inhibitory gating of incoming proprioceptive afferents, resulting in reduced proprioceptive feedback to the brain. Furthermore, the functionality of dI5 does not contribute to proprioception, but rather pain, itch, and thermoception. Functional studies of dI6 indicate that loss of its postmitotic marker Wt1, and therefore dI6, results in direct motor deficits involved in stride frequency and length, and uncoordinated forelimb and hindlimb coordination [67, 68]. Thus, the overexpression of Pax2 may induce a similar dysregulation in gait characteristics for Lbx1EH∆−/− mice, supporting the hypothesis that a proprioceptive and motor decline, with notable hindlimb deficits, is due to changes in spinal cord neuronal composition or numbers.
Impaired proprioception as a mechanism for AIS
The changes in vertebral stability, as demonstrated by increased rotation in AIS-CRM∆ mice, with AIS-CRM∆ mice performing poorly on simple proprioceptive tasks, provide an initial possible mechanism for further investigation and further understanding of the role of proprioception in AIS. Although these findings are not as striking as those in the Runx3 mutant described by Blecher et al. [24], they provide a plausible link between proprioceptive dysfunction and vertebral rotation.
Research has found that proprioception develops and functions from an early age, reaches maturity in adulthood, and subsequently declines with age [69, 70]. This study showed that AIS-CRM∆ mice exhibit an early proprioceptive deficit, which precedes the onset of rotational deviation in the vertebral column. This deficit explains the vertebral deviation exhibited and follows previous studies showing the development of vertebral rotation and scoliosis upon loss of proprioceptive neurons [24]. Proprioceptive deficits have also been observed in individuals with and without AIS (Lau et al. [29]). Although not evident in all populations, a similar mechanism involving other genes leading to proprioceptive deficits may contribute to AIS. Notably, the current GWAS associating rs11190870 with AIS did not include proprioceptive testing in their studies because of the large cohort sizes [8, 29].
The study did not show any notable differences between sexes. The relationship between genetics and biological sex is intricate, with approximately 37% of genes showing some level of sex-specific expression in at least one tissue [71]. Additionally, environmental factors contribute to the variability in the effects of sex hormones on individuals and populations. Although we investigated the influence of sex in AIS-CRM∆ mice, we did not examine the individual effects of hormones in this context. To gain insight into the sex-specific effects during puberty in the AIS-CRM∆ line, gonadectomy (orchiectomy or ovariectomy) was performed on male and female mice before puberty.
Future directions
Our study used whole tissue samples, which limited our ability to identify changes at the cellular level. To address this limitation, future investigations should use single-cell RNA-sequencing to examine changes in specific cell populations within different spinal cord regions during critical developmental periods. Analyzing these cell population differences may provide valuable insights into the cellular profiles and subsequent spinal cord functionality when AIS-CRM is deleted.
Our findings indicate that AIS-CRM∆ mice show increased vertebral rotation and poor proprioceptive task performance, suggesting a possible mechanism that requires further investigation into the role of proprioception in AIS. Altered spinal circuitry, favoring increased GABAergic/inhibitory signaling, may have functional consequences, such as chronic stimulation of associated paraspinal muscles and subsequent muscle atrophy, contributing to the phenotypic differences observed between WT and AIS-CRM∆ mice. Investigating the possible consequences of changes in Lbx1 gene expression is also crucial, as it regulates neuronal population numbers, migration, and cell fate, and its loss leads to changes in neurotransmitter identity, such as the switch from GABAergic to glutamatergic. Our study focused primarily on Lbx1; however, other genes within the same TAD, such as Fgf8, Poll, Btrc, and Fbxw4, should be further investigated for changes in neuronal expression.
Summary
Our findings suggest that AIS-CRM influences gene expression during a crucial embryonic period in the developing spinal cord (E10.5-E15.5). These effects were not limited to Lbx1, but also extended to other nearby genes such as Poll, Fgf8, Fbxw4, and Btrc. Together, these results suggest that the spinal cord contributes to AIS through changes in Lbx1, and that the embryonic stages of neural tube development are the time points likely to influence spinal cord function postnatally. Further research into changes in neural tube patterning and specific neuron subtypes and how these persist postnatally remains to be investigated. Nevertheless, this is the first evidence linking the genetic variant of Lbx1, rs11190870, with the role of Lbx1 in spinal cord development and subsequent functional deficits, resulting in a phenotype in mice that aligns with one aspect of the human phenotype of AIS.
Materials and methods
Generation of the deletion line and genotyping
To examine the effects of the human gene variant rs11190870 on spinal cord development and its possible contribution to AIS, a homologous, highly conserved region in the mouse was deleted using CRISPR-Cas9. A custom mouse line was obtained from Australian BioResources at the Garvan Institute (Australia). The deleted sequences are shown in Fig. S9. Two founder breeding pairs were sent, and the male and female founders were heterozygous. For genotyping, tissue samples were incubated in cell/tissue lysis buffer (10 mM NaCl, 10 mM Tris-HCl) with 20 mg/ml proteinase K at 55°C for a minimum of 2 h until the tissue was dissolved, and proteinase K was inactivated by incubating the samples at 85°C for 1 h. The samples were then spun at 14 000 g for 3 min to pellet any cellular/tissue debris, and the supernatant was transferred to a tube containing 500 μl of isopropanol and spun for 3 min to pellet the DNA. Finally, DNA pellets were resuspended in 300 μl of nuclease-free water. Polymerase chain reaction was performed using DreamTaq Hot Start Green DNA Polymerase (ThermoFisher), with an annealing temperature of 60°C. Oligonucleotide primer sequences can be found in Table S1. The PCR products were analyzed on a 3% agarose gel; the WT amplicon was 639 bp, the AIS-CRM∆ amplicon was 450 bp, and HET bands were present.
In silico analysis of rs11190870
HiC and 4C data around SNP rs11190870 was accessed from http://3dgenome.fsm.northwestern.edu/. ENCODE data from www.encodeproject.org/. Data tracks were added to the UCSC genome browser for mice (mm10) and humans (hg19) to view the target genome region. RegulomeDB https://regulomedb.org/ was additionally used for chromatin state analysis.
ReMap (2022) was loaded into the UCSC browser, which is a public database [48]. For biotypes, ChIP-seq data from mouse and human tissues, including neuronal tissues, cell lines (NPC, NSC, and ESC), and fetal tissues, were chosen. The following transcriptional regulators were selected: CTCF, EZH2, RAD21, SMC1, SCM3, SMC4, STAG1, and STAG2 (Cohesin proteins). The selection was limited to available data, and not all the selected TFs were analyzed for each tissue type.
RNA collection from embryos
After euthanizing the dams, the uterus was placed in ice-cold 1x PBS. The neural tube was carefully dissected from the base of the developing brain vesicle to the caudal-most region, and alternative tissues were collected for genotyping. The samples were homogenized in 1 mL of TRIzol (Thermo Fisher, Cat. No. 15596026). Chloroform (200 μl) was added to the sample and incubated at R.T. 2–3 min before centrifugation. The aqueous layer was then transferred to a new RNase-free tube and combined with 500 μl of isopropanol to precipitate the RNA before spinning for 15 min at 4°C to pellet the RNA. The pellet was washed in ice-cold 70% ethanol twice, and the air-dried RNA pellet was resuspended in 50 μl of nuclease-free water. RNA samples were stored at −20°C for short-term storage or at −80°C for long-term storage. RNA concentration and purity were measured using Nanodrop2000 (Thermo Fisher Scientific). Samples with a 260:280 ratio between 1.90–2.10 were deemed acceptable for downstream applications.
RT-qPCR
Following RNA extraction, complementary DNA (cDNA) was synthesized from 1 μg RNA in a 20 μl reaction mix using the qScript XLT supermix (QuantaBio) as per the manufacturer’s instructions. RT-qPCR was performed using a QuantStudio3 Real-Time PCR system (ThermoFisher). Reactions consisted of 5 μl PowerUp™ SYBR™ Green Master Mix (Applied Biosystems), 1 μL primer working stock at 20 pmol/μl (10 pmol forward, 10 pmol reverse) and 3 μl nuclease-free H2O, and 1 μl of cDNA. All reactions were run in triplicate per biological replicate. Relative expression was calculated using two reference genes, Pgk1 and Sdha. Fold change in expression was calculated using the Livak method (2-ΔΔCt), relative to WT.
MicroCT imaging of the vertebral column
We used a post-mortem micro-CT scan to provide a detailed 3D view of the vertebral column. Previous studies have used plastic straws and similar objects to secure their spines in place; however, there was a concern that a plastic straw or similar object would interfere with any observable phenotype. Therefore, a “stand” was developed (Fig. S11). This ensured that there was little spine movement during the setup and imaging; it provided a humid environment that stopped desiccation.
The SkyScan 1172 source voltage was set to 50 kV and the source current to 200 uA with a 0.5 mm aluminum filter. Image pixel size (resolution) was set to 34.78 μm with a 0.5-degree rotation step. Following imaging, 3D scans were reconstructed using NRecon (1.7.4.6), allowing for FIJI (ImageJ) analysis. The stand and other materials were filtered and excluded from the 3-dimensional reconstructions during reconstruction. Vertebral bodies were individually reconstructed from the source image, and the rotation angle was measured relative to the L5 vertebrae. Angular measurements were compared using the GraphPad Prism software.
Behavioral testing
Upon entry into the study, the animals were housed in the behavioral phenotyping unit at The University of Otago (BPU) for 2–3 days before the first testing day for acclimation. The animals were weighed and moved from their housing space at the BPU (University of Otago, N.Z.) to a testing room before testing. Baseline recordings of the behavioral tests were conducted to obtain the initial behavioral readings for each litter. The baseline grid walk, open field, and SNAP tests were carried out on postnatal day 28 (P28), while the baseline cylinder and grip strength tests were carried out two days later at P30. After obtaining baseline readings, behavioral testing was repeated every two weeks for two testing days for each litter. Mice were weighed before every testing session to monitor their health and for later use in behavioral analysis. Observers performing behavioral analysis were blinded to the genotype of the mice.
Adapting simple Neuroassessment for asymmetric impairment (SNAP) testing to assess proprioceptive dysfunction
The Simple Neuroassessment for Asymmetric Impairment (SNAP) is an observational assessment used to quantify neurological deficits in a mouse model of traumatic brain injury (TBI) mouse model [58]. It consists of seven tests to evaluate emotional and physical behaviors in mice. Shelton et al. [58] observed proprioceptive deficits in their model of TBI; as such, this was adapted to score the observed proprioceptive defects in our cohort. A detailed description of the scoring metrics is presented in Table S3. Briefly, we assessed six behavioral tasks: general activity, interactions (with handlers), cage grasp, visual placement, gait/posture, and baton/grip test. Animals were scored from to 0–5, with 0 being “typical/no defect” and 5 being severe impairment/deficit. The scores were summed, averaged across the cohorts, and plotted to provide an overall SNAP score for the mice comparing WT and AIS-CRM∆ littermates. All tests were performed in a blinded manner. In addition, the data were analyzed using two-way ANOVA to account for age and genotype.
Grid walk test
The grid walk test assesses sensorimotor function by assessing motor coordination and foot-placing deficits during locomotion (Russel et al. [63]). It is also a simple and effective way to assess proprioceptive dysfunction by comparing the number of foot faults with the total number of steps taken. Mice were placed in a plexiglass box on a gridded area (32 × 20 × 50 cm) with 11 × 11 mm diameter openings. Behavior was recorded using a camera placed on a tripod facing down at a mirror beneath the grid to assess the animal’s stepping errors (i.e. foot faults). The animals were allowed 5 min to walk around the gridded area. The foot faults for each limb were counted and compared with the number of steps performed by the limbs. A step was considered a foot fault if it did not provide support and the foot slipped through the grid hole. The percentage of foot faults out of the total number of steps was calculated for each limb and used to compare sensorimotor function between limbs and animals.
Grip strength test
The grip strength test is often used to evaluate motor function and deficits in mice with CNS disorders such as proprioceptive dysfunction [72]. This test was used to quantify the skeletal and muscular strength of forelimbs and hindlimbs in mice. The grip strength apparatus (Model BIO-GS3, Bioseb) comprised a bar connected to an isometric force transducer (dynamometer) for forelimb testing and a wire grid (8 × 8 cm). The force transducer meters were set to zero and reset before each measurement to allow the detection of proper values. The force was measured in grams (g). After visually verifying that the grip was symmetric and tight, the mice were gently pulled horizontally by the tail until the grasp of the bar was broken [73]. To measure the combined limb grip strength, the mice were allowed to rest on an angled wire grid and were pulled by the tail until their grip was broken. A dynamometer reduces the value of maximal force/grip [73]. Each test was repeated thrice to obtain the maximal forelimb and combined limb grip strength for each mouse. Hind limb grip strength was calculated by subtracting the maximal forelimb grip strength from the maximal combined grip strength. Body weight was measured before each testing session and used to normalize grip strength results [73].
Cylinder test
The cylinder test assessed CNS and proprioceptive function by measuring rearing frequency and paw preference. The animals were placed in an upright clear plexiglass cylinder (100 mm diameter × 150 mm height) with a mirror positioned behind the cylinder, allowing all movements to be captured. Videos (5 min) of animal activity were recorded and assessed for forelimb placement, slips, and other rearing behaviors. Mice often rear up to a standing position to explore and press the cylinder wall with their left, right, or both forelimbs for support. The ratio of the time spent on the left forelimb relative to the right forelimb was also calculated to measure forelimb asymmetry.
Immunofluorescence
Transverse cryosections of neural tubes and adult spinal cords were collected and mounted directly onto slides. The slides were defrosted and allowed to dry to room temperature (R. T.). Immunofluorescence for LBX1 was carried out as described in [74].
ChIP-qPCR
Tissue for chromatin extraction was prepared from the embryos (E12.5). Brain and neural tubes were dissected from the embryos and cross-linked for 10 min with methanol-free formaldehyde (ThermoFisher #28906). The cross-linking reactions were stopped with 0.125 M glycine before spinning down, and the tissue pellet was frozen at −80°C for storage.
To prepare the beads, 50 μl of protein G Dynabeads (ThermoFisher) was placed into a microfuge tube with 1 ml of block solution (0.5% bovine serum albumin in PBS). Beads were washed with block solution three times, re-suspended in 1.5 ml of block solution, and split between two microfuge tubes. The EZH2 antibody (10 mg) was added to one tube, and the other tube received IgG (pre-immune serum) as a mock control. The tubes were topped up to 1.5 ml with block solution and then rocked overnight at 4°C.
Following overnight incubation, 1 ml of membrane extraction buffer with 1% proteinase inhibitor (ThermoFisher) was added, and tissue was broken up with an 18 g syringe. The tissue was spun at 9000 × g for 3 min. The supernatant was removed, and tissue was resuspended in 500 μl of digestion buffer with 1 μl of 500 nM Dithiothreitol (DTT). The tissue was digested with 1ul MNase (ThermoFisher; 100 units) in 500 μl volume at 37°C for 15 min. 50 ml of stop solution was added, and the sample was left on ice for 5 min. The mixture was centrifuged at 9000 × g for 3 min, and the supernatant was removed. Nuclear extraction buffer (250 ml) was added to 2.5 ml of proteinase inhibitor, and the sample was left on ice for 15 min and vortexed for 15 s every 5 min. The sample was spun at 9000 × g for 5 min, and the supernatant was collected. Fifty μl of supernatant was reserved for input.
The unbound antibody was removed by washing the beads with blocking solution. The beads were then re-suspended in 1.25 ml of block solution. The chromatin solution was split equally between the beads, and the chromatin-antibody samples were rocked at 4°C overnight. Beads were washed four times with wash buffer 1 (50 mM HEPES-KOH pH 7.5, 500 mM LiCl, 1 mM EDTA, 1% Nonident P-40, 0.7% Sodium deoxycholate) for 15 min each, rotated between washes, and then rinsed in wash buffer 2 (TE pH 8.0, 50 mM NaCl) for 5 min. Elution buffer (100 μl) was added, and the samples were incubated at 65°C for 10 min. Chromatin (50 μl) was reserved for a input control during chromatin preparation. Samples were incubated with 5 M NaCl and Proteinase K at 65°C for 2 h and followed by phenol: chloroform precipitation and ethanol precipitation. The resulting pellets were dissolved in 50 μl of H2O.
ChIP-qPCR reactions were repeated in triplicate, and amplification of the region of interest was deemed successful if the melt curve produced a single peak and Ct values in triplicate were within < 0.5. The resulting Ct values were then used to calculate the percentage pulldown compared to the no antibody/mock control using the following formula: % pulldown = 100 × 2(Ct adjusted input—Ct antibody)/2(Ct adjusted input—Ct mock).
Statistical analysis
All statistical tests were performed using Prism 9. The effect of three variables (AIS-CRM genotype, age, and sex) was determined using three-way analysis of variance (ANOVA) with multiple comparisons (Tukey’s multiple comparisons test). A two-way ANOVA with multiple comparison testing was conducted to determine the effect of two variables, AIS-CRM genotype and age. Pairwise comparisons were performed using unpaired t-tests.
Supplementary Material
Acknowledgements
We thank Andrew McNaughton from the Otago Micro and Nano Imaging Unit, who helped establish the microCT protocol. The authors wish to thank Pr. Carmen Birchmeier and Dr Thomas Müller for the LBX1 antibody.
Contributor Information
Jeremy McCallum-Loudeac, Department of Anatomy, School of Biomedical Sciences, University of Otago, P.O. Box 56, Dunedin 9054, New Zealand.
Edward Moody, Department of Anatomy, School of Biomedical Sciences, University of Otago, P.O. Box 56, Dunedin 9054, New Zealand.
Jack Williams, Department of Anatomy, School of Biomedical Sciences, University of Otago, P.O. Box 56, Dunedin 9054, New Zealand.
Georgia Johnstone, Department of Anatomy, School of Biomedical Sciences, University of Otago, P.O. Box 56, Dunedin 9054, New Zealand.
Kathleen J Sircombe, Department of Anatomy, School of Biomedical Sciences, University of Otago, P.O. Box 56, Dunedin 9054, New Zealand.
Andrew N Clarkson, Department of Anatomy, School of Biomedical Sciences, University of Otago, P.O. Box 56, Dunedin 9054, New Zealand.
Megan J Wilson, Department of Anatomy, School of Biomedical Sciences, University of Otago, P.O. Box 56, Dunedin 9054, New Zealand.
Conflict of interest statement: The authors have no conflict of interest to declare.
Funding
This research project was funded by a University of Otago Research Grant (UORG) to MJW. JML was supported by a postgraduate scholarship from the University of Otago.
References
- 1. Weinstein SL, Dolan LA, Cheng JC. et al. Adolescent idiopathic scoliosis. Lancet 2008;371:1527–37. [DOI] [PubMed] [Google Scholar]
- 2. de Souza FI, Di Ferreira RB, Labres D. et al. Epidemiologia da escoliose idiopática do adolescente em alunos da rede pública de Goiânia-GO. Acta Ortop Bras 2013;21:223–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wise CA, Sepich D, Ushiki A. et al. The cartilage matrisome in adolescent idiopathic scoliosis. Bone Res 2020;8:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Konieczny MR, Senyurt H, Krauspe R. Epidemiology of adolescent idiopathic scoliosis. J Child Orthop 2013;7:3–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Grauers A, Einarsdottir E, Gerdhem P. Genetics and pathogenesis of idiopathic scoliosis. Scoliosis Spinal Disord 2016;11:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lenssinck ML, Frijlink AC, Berger MY. et al. Effect of bracing and other conservative interventions in the treatment of idiopathic scoliosis in adolescents: a systematic review of clinical trials. Phys Ther 2005;85:1329–39. [PubMed] [Google Scholar]
- 7. Wong C. Mechanism of right thoracic adolescent idiopathic scoliosis at risk for progression; a unifying pathway of development by normal growth and imbalance. Scoliosis 2015;10:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Assaiante C, Mallau S, Jouve JL. et al. Do adolescent idiopathic scoliosis (AIS) neglect proprioceptive information in sensory integration of postural control? PLoS One 2012;7:e40646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fadzan M, Bettany-Saltikov J. Etiological theories of adolescent idiopathic scoliosis: past and present. Open Orthop J 2017;11:1466–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Simony A, Carreon LY, Jmark KH. et al. Concordance rates of adolescent idiopathic scoliosis in a Danish twin population. Spine (Phila Pa 1976) 2016;41:1503–7. [DOI] [PubMed] [Google Scholar]
- 11. Grauers A, Rahman I, Gerdhem P. Heritability of scoliosis. Eur Spine J 2012;21:1069–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liu S, Wu N, Zuo Y. et al. Genetic polymorphism of LBX1 is associated with adolescent idiopathic scoliosis in northern Chinese Han population. Spine 2017;42:1125–9. [DOI] [PubMed] [Google Scholar]
- 13. Guo L, Yamashita H, Kou I. et al. Functional investigation of a non-coding variant associated with adolescent idiopathic scoliosis in zebrafish: elevated expression of the ladybird Homeobox gene causes body Axis deformation. PLoS Genet 2016;12:e1005802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Takahashi Y, Kou I, Takahashi A. et al. A genome-wide association study identifies common variants near LBX1 associated with adolescent idiopathic scoliosis. Nat Genet 2011;43:1237–40. [DOI] [PubMed] [Google Scholar]
- 15. Chen S, Zhao L, Roffey DM. et al. Association of rs11190870 near LBX1 with adolescent idiopathic scoliosis in east Asians: a systematic review and meta-analysis. Spine J 2014;14:2968–75. [DOI] [PubMed] [Google Scholar]
- 16. Maurano MT, Humbert R, Rynes E. et al. Systematic localization of common disease-associated variation in regulatory DNA. Science 2012;337:1190–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jagla K, Dolle P, Mattei MG. et al. Mouse Lbx1 and human LBX1 define a novel mammalian homeobox gene family related to the drosophila lady bird genes. Mech Dev 1995;53:345–56. [DOI] [PubMed] [Google Scholar]
- 18. Gross MK, Dottori M, Goulding M. Lbx1 specifies somatosensory association interneurons in the dorsal spinal cord. Neuron 2002;34:535–49. [DOI] [PubMed] [Google Scholar]
- 19. Kruger M, Schafer K, Braun T. The homeobox containing gene Lbx1 is required for correct dorsal-ventral patterning of the neural tube. J Neurochem 2002;82:774–82. [DOI] [PubMed] [Google Scholar]
- 20. Müller T, Brohmann H, Pierani A. et al. The homeodomain factor lbx1 distinguishes two major programs of neuronal differentiation in the dorsal spinal cord. Neuron 2002;34:551–62. [DOI] [PubMed] [Google Scholar]
- 21. Proske U, Gandevia SC. The proprioceptive senses: their roles in signaling body shape, body position and movement, and muscle force. Physiol Rev 2012;92:1651–97. [DOI] [PubMed] [Google Scholar]
- 22. Dietz V. Proprioception and locomotor disorders. Nat Rev Neurosci 2002;3:781–90. [DOI] [PubMed] [Google Scholar]
- 23. Allum JH, Bloem BR, Carpenter MG. et al. Proprioceptive control of posture: a review of new concepts. Gait Posture 1998;8:214–42. [DOI] [PubMed] [Google Scholar]
- 24. Blecher R, Krief S, Galili T. et al. The proprioceptive system masterminds spinal alignment: insight into the mechanism of scoliosis. Dev Cell 2017;42:388–399.e3. [DOI] [PubMed] [Google Scholar]
- 25. Pialasse JP, Descarreaux M, Mercier P. et al. The vestibular-evoked postural response of adolescents with idiopathic scoliosis is altered. PLoS One 2015;10:e0143124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kouwenhoven JW, Castelein RM. The pathogenesis of adolescent idiopathic scoliosis: review of the literature. Spine (Phila Pa 1976) 2008;33:2898–908. [DOI] [PubMed] [Google Scholar]
- 27. Peterka RJ. Sensorimotor integration in human postural control. J Neurophysiol 2002;88:1097–118. [DOI] [PubMed] [Google Scholar]
- 28. Blecher R, Heinemann-Yerushalmi L, Assaraf E. et al. New functions for the proprioceptive system in skeletal biology. Philos Trans R Soc Lond Ser B Biol Sci 2018;373:20170327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lau KKL, Law KKP, Kwan KYH. et al. Timely revisit of proprioceptive deficits in adolescent idiopathic scoliosis: a systematic review and meta-analysis. Global Spine J 2022;12:1852–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Le Berre M, Guyot MA, Agnani O. et al. Clinical balance tests, proprioceptive system and adolescent idiopathic scoliosis. Eur Spine J 2017;26:1638–44. [DOI] [PubMed] [Google Scholar]
- 31. Hawasli AH, Hullar TE, Dorward IG. Idiopathic scoliosis and the vestibular system. Eur Spine J 2015;24:227–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Haumont T, Gauchard GC, Lascombes P, Perrin PP. Postural instability in early-stage idiopathic scoliosis in adolescent girls. Spine (Phila Pa 1976) 2011:36:E847–54. [DOI] [PubMed] [Google Scholar]
- 33. Simoneau M, Lamothe V, Hutin E. et al. Evidence for cognitive vestibular integration impairment in idiopathic scoliosis patients. BMC Neurosci 2009;10:102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lambert FM, Malinvaud J. Glaunès C et al. Vestibular asymmetry as the cause of idiopathic scoliosis: A possible answer from Xenopus. J Neurosci. 2009;29:12477–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Curthoys IS. Vestibular compensation and substitution. Curr Opin Neurol 2000;13:27–30. [DOI] [PubMed] [Google Scholar]
- 36. Scheyerer MJ, Rohde A, Stuermer KJ. et al. Impact of the vestibular system on the formation and progression to idiopathic scoliosis: a review of literature. Asian Spine J 2021;15:701–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jiang H, Yang Q, Liu Y. et al. Association between ladybird homeobox 1 gene polymorphisms and adolescent idiopathic scoliosis: a MOOSE-compliant meta-analysis. Medicine (Baltimore) 2019;98:e16314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gao W, Peng Y, Liang G. et al. Association between common variants near LBX1 and adolescent idiopathic scoliosis replicated in the Chinese Han population. PLoS One 2013;8:e53234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Londono D, Kou I, Johnson TA. et al. A meta-analysis identifies adolescent idiopathic scoliosis association with LBX1 locus in multiple ethnic groups. J Med Genet 2014;51:401–6. [DOI] [PubMed] [Google Scholar]
- 40. van der Velde A, Fan K, Tsuji J, Moore JE, Purcaro MJ, Pratt HE, Weng Z. Annotation of chromatin states in 66 complete mouse epigenomes during development. Commun Biol 2021:4;239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Boyle AP, Hong EL, Hariharan M. et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res 2012;22:1790–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gan L, Yang Y, Li Q. et al. Epigenetic regulation of cancer progression by EZH2: from biological insights to therapeutic potential. Biomark Res 2018;6:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lau CH, Suh Y. CRISPR-based strategies for studying regulatory elements and chromatin structure in mammalian gene control. Mamm Genome 2018;29:205–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cheng L, Samad OA, Xu Y. et al. Lbx1 and Tlx3 are opposing switches in determining GABAergic versus glutamatergic transmitter phenotypes. Nat Neurosci 2005;8:1510–5. [DOI] [PubMed] [Google Scholar]
- 45. Dixon JR, Selvaraj S, Yue F. et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 2012;485:376–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Li D, He M, Tang Q. et al. Comparative 3D genome architecture in vertebrates. BMC Biol 2022;20:99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wang Y, Song F, Zhang B. et al. The 3D genome browser: a web-based browser for visualizing 3D genome organization and long-range chromatin interactions. Genome Biol 2018;19:151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hammal F, de Langen P, Bergon A. et al. ReMap 2022: a database of human, mouse, drosophila and Arabidopsis regulatory regions from an integrative analysis of DNA-binding sequencing experiments. Nucleic Acids Res 2022;50:D316–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zuin J, Dixon JR, van der Reijden MI. et al. Cohesin and CTCF differentially affect chromatin architecture and gene expression in human cells. Proc Natl Acad Sci U S A 2014;111:996–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kheradmand Kia S, Solaimani Kartalaei P, Farahbakhshian E. et al. EZH2-dependent chromatin looping controls INK4a and INK4b, but not ARF, during human progenitor cell differentiation and cellular senescence. Epigenetics Chromatin 2009;2:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Janssen MM, Vincken KL, Kemp B. et al. Pre-existent vertebral rotation in the human spine is influenced by body position. Eur Spine J 2010;19:1728–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Xiong B, Sevastik J, Hedlund R. et al. Segmental vertebral rotation in early scoliosis. Eur Spine J 1993;2:37–41. [DOI] [PubMed] [Google Scholar]
- 53. Lee CS, Hwang CJ, Jung HS. et al. Association between vertebral rotation pattern and curve morphology in adolescent idiopathic scoliosis. World Neurosurg 2020;143:e243–52. [DOI] [PubMed] [Google Scholar]
- 54. Lam GC, Hill DL, Le LH. et al. Vertebral rotation measurement: a summary and comparison of common radiographic and CT methods. Scoliosis 2008;3:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Busscher I, Wapstra FH, Veldhuizen AG. Predicting growth and curve progression in the individual patient with adolescent idiopathic scoliosis: design of a prospective longitudinal cohort study. BMC Musculoskelet Disord 2010;11:93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Greiner KA. Adolescent idiopathic scoliosis: radiologic decision-making. Am Fam Physician 2002;65:1817–22. [PubMed] [Google Scholar]
- 57. Schlosser TP, van der Heijden GJ, Versteeg AL. et al. How 'idiopathic' is adolescent idiopathic scoliosis? A systematic review on associated abnormalities. PLoS One 2014;9:e97461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Shelton SB, Pettigrew DB, Hermann AD. et al. A simple, efficient tool for assessment of mice after unilateral cortex injury. J Neurosci Methods 2008;168:431–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Roome RB, Vanderluit JL. Paw-dragging: a novel, sensitive analysis of the mouse cylinder test. J Vis Exp 2015;98:e52701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Schaar KL, Brenneman MM, Savitz SI. Functional assessments in the rodent stroke model. Exp Transl Stroke Med 2010;2:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lin SH, Cheng YR, Banks RW. et al. Evidence for the involvement of ASIC3 in sensory mechanotransduction in proprioceptors. Nat Commun 2016;7:11460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Baskin YK, Dietrich WD, Green EJ. Two effective behavioral tasks for evaluating sensorimotor dysfunction following traumatic brain injury in mice. J Neurosci Methods 2003;129:87–93. [DOI] [PubMed] [Google Scholar]
- 63. Russell KL, Kutchko KM, Fowler SC. et al. Sensorimotor behavioral tests for use in a juvenile rat model of traumatic brain injury: assessment of sex differences. J Neurosci Methods 2011;199:214–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ngan CY, Wong CH, Tjong H. et al. Chromatin interaction analyses elucidate the roles of PRC2-bound silencers in mouse development. Nat Genet 2020;52:264–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. McArthur E, Capra JA. Topologically associating domain boundaries that are stable across diverse cell types are evolutionarily constrained and enriched for heritability. Am J Hum Genet 2021;108:269–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Lai HC, Seal RP, Johnson JE. Making sense out of spinal cord somatosensory development. Development 2016;143:3434–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Schnerwitzki D, Perry S, Ivanova A. et al. Neuron-specific inactivation of Wt1 alters locomotion in mice and changes interneuron composition in the spinal cord. Life Sci Alliance 2018;1:e201800106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Gosgnach S. Spinal inhibitory interneurons: regulators of coordination during locomotor activity. Front Neural Circuits 2023;17:1167836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Martel M, Finos L, Koun E. et al. The long developmental trajectory of body representation plasticity following tool use. Sci Rep 2021;11:559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Viel S, Vaugoyeau M, Assaiante C. Adolescence: a transient period of proprioceptive neglect in sensory integration of postural control. Mot Control 2009;13:25–42. [DOI] [PubMed] [Google Scholar]
- 71. Oliva M, Munoz-Aguirre M, Kim-Hellmuth S. et al. The impact of sex on gene expression across human tissues. Science 2020;369:eaba3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Maurissen JP, Marable BR, Andrus AK. et al. Factors affecting grip strength testing. Neurotoxicol Teratol 2003;25:543–53. [DOI] [PubMed] [Google Scholar]
- 73. Takeshita H, Yamamoto K, Nozato S. et al. Modified forelimb grip strength test detects aging-associated physiological decline in skeletal muscle function in male mice. Sci Rep 2017;7:42323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Decourtye L, McCallum-Loudeac JA, Zellhuber-McMillan S. et al. Characterization of a novel Lbx1 mouse loss of function strain. Differentiation 2022;123:30–41. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.