Summary
Retinitis pigmentosa (RP) is one of the most common forms of hereditary neurodegeneration. It is caused by one or more of at least 3,100 mutations in over 80 genes that are primarily expressed in rod photoreceptors. In RP, the primary rod-death phase is followed by cone death, regardless of the underlying gene mutation that drove the initial rod degeneration. Dampening the oxidation of glycolytic end products in rod mitochondria enhances cone survival in divergent etiological disease models independent of the underlying rod-specific gene mutations. Therapeutic editing of the prolyl hydroxylase domain-containing protein gene (PHD2, also known as Egln1) in rod photoreceptors led to the sustained survival of both diseased rods and cones in both preclinical autosomal-recessive and dominant RP models. Adeno-associated virus-mediated CRISPR-based therapeutic reprogramming of the aerobic glycolysis node may serve as a gene-agnostic treatment for patients with various forms of RP.
Keywords: CRISPR, metabolic reprogramming, AAV, therapeutic editing, rejuvenation, retinal degeneration, prolyl hydroxylase, hypoxia inducible factor, glycolysis, gene therapy
Graphical abstract

Highlights
-
•
PHD antagonists sustain mouse retina in Pde6β preclinical model of RP
-
•
Genetic ablation of PHD confers a stronger rescue effect
-
•
PHD ablation shifts retinal metabolism
-
•
AAV delivery of gene editor rescued two divergent models of retinal degeneration
Recently, the prolyl hydroxylase (PHD) antagonists daprodustat and roxadustat have been approved for anemia treatment. Nolan, Cui et al. demonstrate that genetically ablating this target, PHD, and shifting retinal metabolism rescues retinal degeneration. Reprogramming glycolysis offers a gene-agnostic approach to addressing genetic heterogeneity in retinitis pigmentosa.
Introduction
Retinitis pigmentosa (RP) is a group of inherited retinal dystrophies that affects up to 1 in 4,000 people worldwide.1 RP causes an initial phase of rod photoreceptor loss, followed by secondary loss of cone photoreceptor cells.2,3 Individuals with RP experience symptoms such as night blindness, tunnel vision, and total blindness.4 Together, these vision changes severely impact patients by limiting their ability to conduct activities required for daily living independently. Patients with degenerating photoreceptors, including those with RP, currently have limited therapeutic options. Although defects in over 80 genes are linked to RP, currently only patients with a recessive mutation in RPE65 can opt for precision gene supplementation therapy.5,6,7,8,9,10
Genome surgery that uses CRISPR can precisely repair or replace both dominant and recessive gene mutations, thus offering the best possibility of a lifelong cure. However, CRISPR, in its original, mutation-specific form of homology-directed repair, has a significant limitation: the therapeutic components (the guide RNA [gRNA] and repair template) must be individually custom-designed, engineered, tested, and approved by the US Food and Drug Administration (FDA) for each specific mutation.11 CRISPR repair of each specific mutation that causes a Mendelian disorder would thus be time consuming and prohibitively expensive. To address this, we are developing an alternative therapeutic strategy that targets energy metabolism, a biochemical pathway required for homeostasis that potentially can influence multiple types of photoreceptor degenerative disorders, including RP and age-related macular degeneration (AMD).8,12
Aerobic glycolytic dysregulation: A common therapeutic target pathway
The pathways that link genetic mutations to rod and cone photoreceptor death are only partly understood. Our current knowledge indicates that rods and the retinal pigment epithelium (RPE) are metabolically coupled13 and that most dystrophic retinas exhibit metabolic dysregulation.8,12 In healthy retinas, rods take up glucose from the RPE and convert it to pyruvate and lactate, even in the presence of oxygen, a phenomenon termed aerobic glycolysis or the Warburg effect.14,15,16,17,18,19 This lactate is then supplied to the RPE, where it can suppress consumption of glucose by the RPE.20 In contrast, in rod-primary dystrophies, the lactate concentration decreases as rod cells die, prompting the RPE to use glucose from the choriocapillaris as an energy source. When the RPE consumes too much glucose, cone photoreceptors are starved for fuel.21,22,23 These findings led us to propose that photoreceptor death from RP-induced metabolic imbalance could be addressed by boosting aerobic glycolysis in dystrophic photoreceptors rather than trying to repair each specific type of mutation.24,25 Contrary to other approaches currently being tested in the clinic, our therapeutic design will increase the rod photoreceptor’s metabolic capabilities from a diminished, degenerative state to a more normal, healthy profile. Therapeutic reprogramming approaches such as rod-derived cone viability factor (RdCVF) supplementation have paved the way for our understanding of the importance of glucose import into cones, and our therapeutic strategy similarly aims to modulate the glycolytic capacity of the retina.26
Candidate therapeutic targets: The prolyl hydroxylase-von Hippel-Lindau-hypoxia-inducible factor aerobic axis
The potential impact of boosting aerobic glycolysis to promote photoreceptor survival has been demonstrated by ablating inhibitors of aerobic glycolysis, including TSC1 and SIRT6.4,27,28 The prolyl hydroxylase (PHD)-von Hippel-Lindau (VHL)-hypoxia-inducible factor (HIF) axis has also been suggested as a potentially effective target for boosting aerobic glycolysis in many forms of RP.29,30,31 Under normoxia, PHD and VHL negatively regulate HIF,32 a master aerobic glycolysis regulator, via ubiquitination. Previous studies have shown that high HIF levels are protective in disease states, including in the Pde6βrd10 model of RP33 and after light damage,20 but are damaging in the wild-type retina.34 PHD has three isoenzymes. PHD2 is the most abundantly expressed in most cell types and tissues, including the neuroretina.35 PHD2 limits HIF-1α levels more effectively than PHD1 and PHD3. Additionally, PHD2 is the isoform that is most sensitive to O2 concentration.36
Cell-specific ablation of the PHD-VHL axis may be a potential treatment strategy relevant to several photoreceptor disorders and may be a safer alternative to the pharmacological, orally administered PHD antagonist FG-4592. Because our ablation strategy specifically targets photoreceptors,37 we expect it to have an improved safety profile in RP compared to the systemic oral administration of the PHD inhibitor FG-4592, boosting HIF levels in a cell-autonomous manner.
The rod-specific glycolysis enhancement via PHD-HIF reprogramming proposed in this study is not precision medicine, as it does not represent a mutation-specific treatment.2,38,39 However, this report shows that glycolytic reprogramming may be an efficacious, gene-agnostic therapeutic approach to rescue function and cell survival in RP. An imbalance between glycolysis to lactate versus complete oxidation of glucose to CO2 can contribute to various forms of RP.12,40 As such, our goal is to enhance a metabolic pathway that can generally delay cell death even when caused by a diverse array of underlying mutations. This strategy could offer a therapy for diverse types of photoreceptor degeneration. Previous efforts to reprogram retina metabolism to treat patients regardless of the causative mutation in RP include RdCVF/RdCVFL (full-length RdCVF) enhancement to increase glucose import into cone photoreceptors and mTORC1 (mammalian target of rapamycin complex 1) enhancement to increase glucose uptake, retention, and utilization by photoreceptors, Sirtuin6 inhibition to modulate histone deacetylase and constitutively boosting rod glycolysis, and transforming growth factor-β1 modulation to enhance microglial metabolism in the neuroretina.4,26,27,41,42 Our strategy is fundamentally different. It aims to increase glycolytic flux in rods and cones via key steps and to slow the pace of mitochondrial oxidation. We hypothesize that the inhibition of PHD will increase photoreceptor glycolysis, decrease oxidative activity in mitochondria, slow the rate of deterioration, and prolong residual photoreceptor morphology and function.
Results
Pharmacological inhibition of PHD targets the PHD-VHL-HIF axis, elevates aerobic glycolysis, and prolongs photoreceptor survival
Recent findings suggest that enhancing aerobic glycolysis in rod photoreceptors can improve their resilience to stress.4,27,28 We tested this hypothesis by targeting the PHD-VHL-HIF axis. We administered doses of the PHD inhibitor, FG-4592, by oral gavage to a mouse model for RP that has a PDE6 deficiency (Pde6βH620Q/H620) and degenerates on a timeline similar to that of the Pde6βrd10 model. We delivered doses every other day from postnatal day 5 (P5), boosting aerobic glycolysis in rods and cones. Following our administration of the systemic PHD inhibitor, steady-state levels of some glycolytic intermediates in retinas harvested from FG-4592-treated mice were significantly higher than in retinas from control mice (Figure 1A). Tricarboxylic acid (TCA) cycle intermediates were not significantly different between experimental and control groups (Figure 1A). Histological analysis at age P35 of treated and control eyes showed an increased thickness of the outer nuclear layer (ONL) in the Pde6βH620Q/H620Q RP model treated with 20 mg/kg FG-4592 compared to control untreated mice (Figures 1B and 1C). At 4 weeks of age, phototransduction was evaluated by electroretinography (ERG). Photopic and mixed b-wave responses were significantly enhanced in treated mice compared with untreated controls (Figures 1D and 1E). Rod b-wave responses remained unchanged.
Figure 1.
PHD inhibitor enhances photoreceptor survival and preserves retinal function by increasing retina glycolysis
Pde6βH620Q/H620Q mice were fed the PHD inhibitor, FG-4592, at one dose every 2 days from P5.
(A) Metabolites were extracted from the retinas of mice at P23. 13C-labeled glycolytic intermediates increased in retinas from mice in the treatment group compared to those in the control group. Minimal effects were detected in TCA-cycle intermediates and mitochondrial oxidation metabolites, shown here as treated vs. control. Error bars indicate SEM. ∗p ≤ 0.05, ∗∗p ≤ 0.01; n = 5–6 per group.
(B) H&E staining was performed to analyze retinas from the Pde6βH620Q/H620Q mouse model at P35, with a representative section comparing the treatment group and control group at 500 μm from the optic nerve head. The green bar indicates the outer nuclear layer (ONL). Scale bar, 25 μm.
(C) Spider plot analysis of ONL thickness. Error bars indicate SEM. ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001; n = 6 per group.
(D) Pde6βH620Q/H620Q mice were subjected to three types of serial ERG recordings—scotopic (rod-specific), maximal (rod and cone), and photopic (cone-specific)—at P28. Single tracings of the three-step ERG are shown. Blue represents the ERG tracing from a mouse with FG4592, while the gray tracing is from sham-fed control.
(E) Analysis of the amplitude of b wave as shown in (D). Each dot or diamond represents the average ERG amplitude of both eyes per mouse. Error bars indicate SEM. ∗p ≤ 0.05; n = 6 per group.
Preclinical model development and tamoxifen-induced ablation of PHD1, PHD2, and PHD3
We then evaluated the effects of PHD inhibition on photoreceptor resilience by eliminating expression in the retina of all three PHD isoenzymes. We probed the ability of this cell-specific ablation to rescue degeneration in Pde6βH620Q/H620Q;Pde6γCreERT2/+ mice, a well-established preclinical model of arRP.4 Pde6βH620Q/H620Q;Pde6γCreERT2/+ mice carry a rod-photoreceptor-specific tamoxifen-inducible Cre driver (Pde6γCreERT2/+).43 Pde6βH620Q/H620Q;Pde6γCreERT2/+ animals were intercrossed with the conditional PHD1f,2f,3f mouse line to generate our experimental mouse line. Tamoxifen treatment blocks expression of PHD2, PHD1, and PHD3 (also known as Egln1, Egln2, and Egln3, respectively), which encode all of the PHD isoenzymes. PHD−/−;Pde6γCreERT2/+;Pde6βH620Q/H620Q mice served as the experimental group and were given an intraperitoneal injection of tamoxifen for three consecutive days (P9, P10, and P11) following the completion of retinal cell differentiation.43,44 Control PHDfl/fl;Pde6γCreERT2/+;Pde6βH620Q/H620Q mice received a sham injection of 10% ethanol (w/w) in sunflower oil at the same age as treated groups.28,43
To demonstrate that PHD1, PHD2, and PHD3 were ablated following tamoxifen treatment, we isolated genomic DNA from mouse retinas and analyzed the PHD genes (Egln1, Egln2, Egln3) with PCR. Genomic PCR revealed a 481-bp truncated band in addition to the 900-bp parental fragments in the treated retinas (Figure S1A). The 900-bp parental product is less intense because, in addition to rods, the whole retina contains cones, bipolar cells, and so forth, in which the Pde6γCreERT2 driver was inactive. Primers for genomic PCR are presented in Table S1. Control retinas in the treated group presented only the non-truncated wild-type fragment (900 bp). Genomic PCR blots were shown to validate PHD2 and three deletions (Figures S1B and S1C).
Upregulation of aerobic glycolysis in PHD1,2,3-deficient retinas
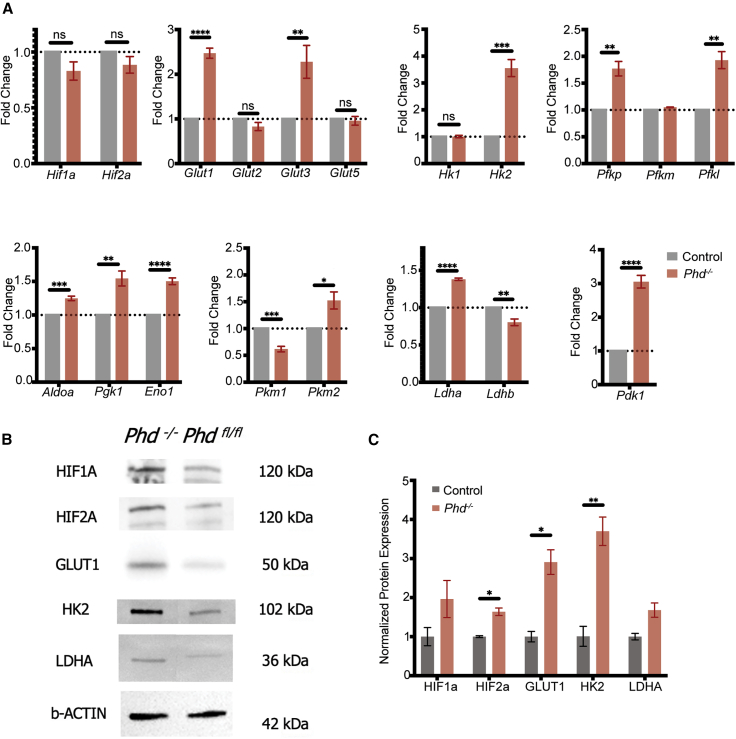
Because of the critical role of PHD1, PHD2, and PHD3 in regulating aerobic glycolysis, extracts from retinas deficient in PHD1, PHD2, and PHD3 were collected, and the transcripts of downstream glycolytic genes regulated by the PHD-VHL-HIF aerobic axis were quantified via qPCR. mRNA expression was quantified to assess changes between PHD1,2,3−/−;Pde6βH620Q/H620Q and PHD1,2,3loxP/loxP;Pde6βH620Q/H620Q at 3 weeks of age, prior to the onset of ONL loss. Quantification of mRNA transcript levels from samples collected from the retinas of experimental and control mice revealed that Glut1, Glut3, Pfk, Hk2, and Pdk1 increased in the PHD1,2,3−/−;Pde6βH620Q/H620Q mice compared with controls (Figure 2A). PHD1,2,3 loxP/loxP;Pde6βH620Q/H620Q degenerate age-matched mice served as a control. β-Actin served as a loading control. Immunoblots were treated with antibodies against downstream targets to PHD1, PHD2, and PHD3, and subsequent analysis of glycolytic regulators in experimental and control retinal extracts revealed higher protein levels of hypoxia-inducible factors (HIF2A), glucose transporters (GLUT1), and hexokinase (HK2) in experimental mice (Figure 2B). Increased aerobic glycolytic shifts were consistent with the expected result of PHD suppression. These data were quantified and magnitudes of protein expression compared (Figure 2C). Efficient, near-complete rod-specific Pde6γCreERT2-mediated downregulation of PHD expression via HIF levels and subsequent aerobic glycolytic shifts were observed (Figure 2C). PHD levels were difficult to assay from tissue, likely due to their rapid degradation in vivo; however, genomic deletion was confirmed previously.45
Figure 2.
PHD deficiency up-regulates key regulators of glycolytic metabolism in photoreceptors
Experimental (PHD−/−;Pde6βH620Q/H620Q;Pde6γCreERT2/+) and control (PHDFL/FL;Pde6βH620Q/H620Q;Pde6γCreERT2/+) mice were treated with tamoxifen or sham solution, respectively, for three consecutive days (P8, P9, and P10). Mice were sacrificed at postnatal 3 weeks, and retinas were collected and snap-frozen in liquid nitrogen until further processing.
(A) mRNA expression of Hif1a and Hif2a and the downstream glycolytic targets (Glut1, Glut2, Glut3, Glut5, Hk1, Hk2, Pfkp, Pfkm, Pfkl, Aldoa, Pgk1, Eno1, Pkm1, 2, Ldha, Ldhb, and Pdk1) were quantified to assess changes before and after PHD was ablated. β-Actin was used as the internal control. Error bars indicate SEM. ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, ∗∗∗∗p ≤ 0.0001; n = 4–5 per group.
(B) Representative immunoblots of glycolytic metabolism enzymes and regulators in the retinas of treated and untreated mice at P21 (before the onset of degeneration) to detect HIF1A, HIF2A, GLUT1, HK2, and LDHA protein levels. β-Actin was used as a loading control. Membrane was stripped and reprobed for all targets.
(C) Quantitative analysis of protein levels shown in (B). Error bars indicate SEM. ∗p ≤ 0.05, ∗∗p ≤ 0.01; n = 3 per group.
Analysis of downstream glycolytic metabolites following PHD1,2,3 ablation
Our qPCR data and immunoblots from tamoxifen-treated versus untreated Pde6βH620Q/H620Q mice suggested that loss of PHD expression could influence glycolytic flux. We measured this metabolic flux through glycolysis and the TCA cycle to clarify the influence of PHD on retinal metabolism. We isolated retinas and incubated them with [U-13C]glucose for 30 s or 90 s. We extracted and derivatized metabolites before using gas chromatography-mass spectrometry (GC-MS) to quantify incorporation of carbons from [13C]glucose into glycolytic and TCA-cycle intermediates (Figure 3). We found more rapid labeling and higher accumulation of glycolytic intermediates all the way through pyruvate in PHD-deficient Pde6βH620Q/H620Q retinas (dashed lines in Figure 3) compared to control Pde6βH620Q/H620Q retinas (solid lines in Figure 3). The most substantial effect is diminished flux into and through the TCA cycle, suggesting that elevated pyruvate dehydrogenase (PDH) kinase (PDK1) expression (Figures 2A and S2) may inhibit oxidation of pyruvate to acetyl-coenzyme A (CoA). Surprisingly, the overall rates of glucose depletion from the medium and lactate release into the medium are unaffected. Pentose phosphate pathway activity was probed and did not indicate an increase in PHD-deficient retinas (Figure S3), while PDH inactivation remains to be further explored for its impact on energy production and availability in the dystrophic photoreceptor population. These data directly support the accumulation of M3-labeled pyruvate and corresponding depletion of M2 citrate seen in Figure 3. Overall levels of recoverin were probed as a proxy for rod survival in our metabolic tracing experiments and indicated that the differences observed were not due to overall rod preservation differences; critical for making statements about the altered cell metabolism as a direct result of our reprogramming seen in Figures 3 and S3 (Figure S4).
Figure 3.
Loss of PHD enhances glycolysis and diminishes mitochondrial oxidation in PDE6-deficient retinas
Eyes from control PHDfl/fl and PHD−/− (homozygous for Pde6βH620Q/H620Q) mice were dissected at P21 before retinal degeneration. Retinas were isolated under ambient illumination, incubated with 5 mM [U-13C]glucose, and harvested at 30-s and 90-s time points. Metabolites were extracted with 80% methanol, derivatized, and quantified by GC-MS. Flux of carbons from [U-13C]glucose through glycolysis is faster in the PHD-deficient retinas, and TCA-cycle activity is slower than controls. The bar graphs in the upper-right section show that the average rates over 40 min at which glucose is consumed and at which lactate is exported are not significantly different between experimental and control retinas. Axis units are indicated in the center field, with the y axis representing picomoles or micrograms of protein and the x axis representing time (seconds). Error bars indicate SEM. ∗p ≤ 0.05, ∗∗p ≤ 0.01; n = 6–9 per group.
Tamoxifen-induced ablation of PHD2 enhances photoreceptor survival
After validating tamoxifen-induced excision of PHD1, PHD2, and PHD3 and confirming that parts of glycolytic flux are enhanced, we analyzed neurological function by ERG. At 3 weeks post treatment, experimental mice responded with significantly larger b-wave amplitudes than control mice (Figures 4A–4C). Representative traces are shown in Figure S5A. Histological analysis revealed that the ONL in experimental mice is significantly thicker than in control mice at 500 μm from the optic nerve head (Figure 4D). Comprehensive spider plots indicated that the rescue effect was strongest near the optic nerve head but was preserved throughout almost the entirety of the retina at both 4 and 6 weeks of age (Figures 4E and 4F). Fluorescent flat-mount staining of cone photoreceptors in the retinas of experimental and control mice revealed more significant numbers and a greater density of cone cells labeled with anti-peanut agglutinin antibodies in experimental mice retina than in controls (Figures 4G and 4H). No safety concerns such as neovascularization were detected in any of our Pde6βH620Q/H620Q retinas lacking PHD1, PHD2, or PHD3 up to 1 year of age. Fluorescein angiography images from 1-year-old, subretinally transduced (at an age of 1 month) RHOC110R/+ mice demonstrate no patterns of neovascularization and support the lack of safety concerns held by our group (Figure S6).
Figure 4.
Upregulation of glycolysis through ablation of PHD enhances cone survival in a preclinical Pde6βH620Q/H620Q RP model
PHD deficiency improves rod and cone survival in the Pde6βH620Q/H620Q mutant background. Experimental (PHD−/−;Pde6βH620Q/H620Q;Pde6γCreERT2/+) and control (PHDFL/FL;Pde6βH620Q/H620Q;Pde6γCreERT2/+) mice were treated with tamoxifen or sham solution for three consecutive days (P9, P10, and P11).
(A–C) Analysis of amplitudes of electroretinogram tracings comparing experimental and control mice. Mice were subjected to three types of serial ERG recordings: scotopic rod (A), maximal rod and cone (B), and photopic cone (C) at 6, 8, and 10 weeks postnatally. Experimental mice with precise PHD1,2,3 ablation in rod photoreceptors are shown in red, while the signal from control mice is in gray. The average ERG amplitude of both eyes per mouse was used for analysis. Error bars indicate SEM. ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, ∗∗∗∗p ≤ 0.0001; n = 5–6 in each group per time point.
(D) Representative H&E-stained central retinal sections from experimental and control mice at 4 and 6 weeks at a distance of 500 μm from the optic nerve head. Yellow bars indicate ONL thickness. Scale bars, 25 μm.
(E) Spider plot analysis of ONL thickness at 4 weeks. Error bars indicate SEM. ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001; n = 6 per group.
(F) Spider plot analysis of ONL thickness at 6 weeks. Error bars indicate SEM. ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001; n = 6 per group.
(G) Peanut agglutinin (PNA) staining of cones in 12-week-old PHDfl/fl control and PHD−/− (deficient) mice. The inset shows representative central retinal cones.
(H) Bar chart of cone cell counts, quantified by percent area of the total area measured in both groups in (G). Error bars indicate SEM. ∗∗p ≤ 0.01; n = 3 in each group.
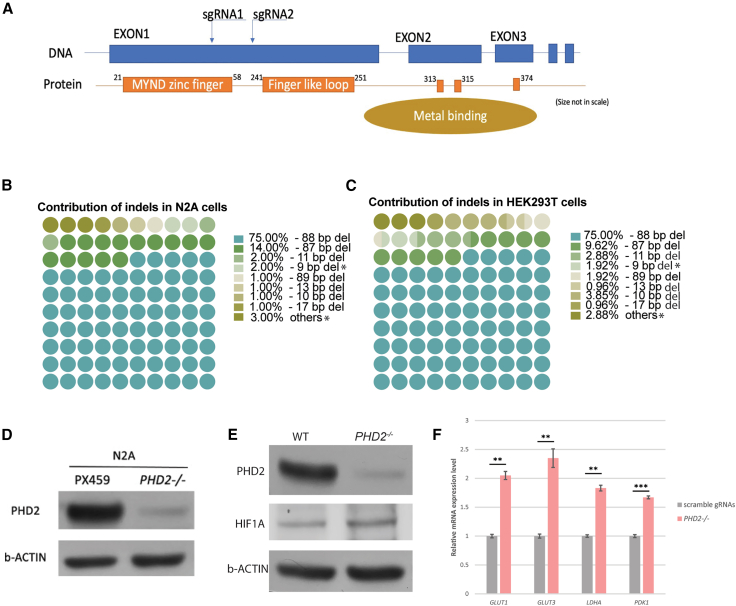
Therapeutic genome editing of PHD2 in mouse and human cells
Using a dual-gRNA-guided CRISPR-Cas9 system, exon 1 of PHD2 was ablated in mouse Neuro-2a cells (N2A) and human HEK293 cells (Figure 5A). The overwhelming majority of resultant quantified edits were indels that were not divisible by 3, thereby causing a frameshift-mediated premature stop codon, ultimately truncating the gene expression (Figures 5B and 5C), where unsuccessful edits are indicated with an asterisk. Cells transfected with experimental gRNAs directed by CRISPR-Cas9 excision had markedly lower PHD2 protein expression than cells transfected with control PX459:scramble gRNAs (Figures 5D and 5E). The results indicated successful PHD2 ablation and associated increased HIF levels as anticipated, compared to a non-transfected wild-type control (Figure 5E). Bands were quantified in ImageJ and on comparison showed a ∼1.7-fold increase in the treated cells compared to non-edited control cells. Subsequent quantification of mRNA levels revealed that the cells transfected with experimental gRNAs also had higher levels of LDHA, PDK1, GLUT1, and GLUT 3, indicating that PHD2 ablation may contribute to an increase in glycolysis (Figure 5F).
Figure 5.
Ablation of PHD2 with gRNAs is sufficient for Hif1a augmentation and target engagement in vitro
(A) Schematic summary of the outcomes produced by gRNA therapeutic deletion. The gRNAs used target exon 1 in human PHD2 sequence.
(B and C) Ninety-five percent of non-homologous end-joining insertions and deletions (indels) resulted in a frameshift mutation in both mouse N2A (B) and human HEK293 (C) cells. Indels that are indivisible by 3 represent a successful insertion of a frameshift-mediated stop codon, truncating gene function. Unsuccessful edits are indicated with an asterisk and account for approximately 5% of the cases in both N2A (B) and HEK293 (C) cells.
(D) Immunoblotting was performed, and the level of PHD2 from mouse N2A cells with control (PX459) or plasmids containing gRNAs targeting PHD2 was determined. β-Actin was used as a loading control.
(E) Immunoblot was performed, and the level of PHD2 from human HEK293 cells with control (PX459) or plasmids containing gRNAs targeting PHD2 was quantified. β-Actin was used as a loading control. After transfection with scramble gRNAs or gRNAs targeting PHD2, HEK293 cells were cultured in standard media. Immunoblots revealed lower levels of PHD2 after gRNA therapeutic gene deletion of PHD2 compared to scramble gRNA controls.
(F) Increased expression of LDHA, PDK1, GLUT1, and GLUT3 (glycolytic markers of the Warburg effect) after ablation of PHD2 in normoxic conditions. Control transduced with plasmids containing scramble gRNAs. Error bars indicate SEM. ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001; n = 3 in each group.
Therapeutic genome editing of PHD2 in arRP and adRP preclinical models
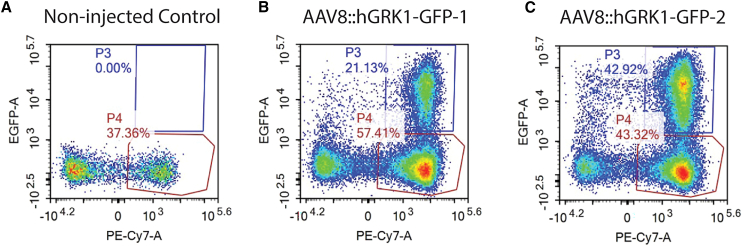
Following the confirmation of the therapeutic PHD2 editing in human cells, wild-type C57BL/6J mice were transduced subretinally with dual gRNA adeno-associated virus (AAV) vectors targeting rod photoreceptors to validate the transduction efficiency of AAV8 delivery. We dissociated whole retinas and used fluorescence-activated cell sorting to identify AAV-transduced rod photoreceptors, utilizing CD73+ markers as a proxy for this cell type. Approximately 35% of cells positive for CD73+, a rod-specific cell-surface marker, also were positive for GFP, indicating that they had been transduced by AAV8:hGRK1-GFP (Figures 6A–6C). Few other CD73-negative GFP-positive cells were present, confirming that this AAV8 had driven expression specifically in rod photoreceptors. GFP+/CD73+ cells were absent in control-injected mice. The specificity of this AAV8 vector has been confirmed.46 Similar pathway modulation has been achieved previously, demonstrating how metabolic shifts could be tolerable and safe in treated individuals.46
Figure 6.
Approximately 35% of CD73+ rod photoreceptors were transduced by AAV8::hGRK1-GFP
(A–C) Mouse retinas receiving a subretinal injection (A–C) of AAV8:hGRK1-GFP for 1 month were dissociated, labeled with the rod photoreceptor marker anti-CD73, and analyzed by flow cytometry. BL6 wild-type mice were used here as a control. (A) Representative results in control mice with no injection and (B and C) in two injected mice. The GFP+/CD73+ cells are gated with the visible blue square and labeled as “P3”; the GFP−/CD73+ cells were gated with the red square and labeled as “P4.” x axis, CD73; y axis, GFP.
After confirming the specificity and efficiency of our AAV8 in transducing rod photoreceptors, the next step in developing this therapeutic approach was to evaluate efficacy in the Pde6βH620Q//Pde6βCas9 arRP model. This model harbors Cas9 expressed in photoreceptors and degenerates on a timeline akin to that of homozygous Pde6βH620Q/H620Q mice. Importantly, Pde6βCas9/+ photoreceptors have no detectable deleterious effects on their own (Figure S7). We used ERG at 8 weeks of age to quantify photoresponses from Pde6βH620Q/H620Q mice transduced with our therapeutic PHD2 editing vector, AAV8:U6-gRNAs_PHD2 at 2 weeks of age. Co-injection of AAV8:hGRK1-GFP revealed that the subretinal transduction area covered ∼25% of the retina. Contralateral eyes served as non-injected controls (Figure 7A). ERG tracings showed that the therapeutic PHD2 editing vector enhanced photoreceptor function (Figure 7B). These traces were analyzed for maximal rod and cone responses, a metric routinely used as a hallmark of retinal function, which revealed that magnitudes of a and b waves from the injected eyes generated significantly larger ERG responses than the control non-injected contralateral eyes (Figure 7C). Eight-week-old wild-type responses can be perused in Figure S5B, which helps better appreciate the discrepancy between the treated and control groups. The histology preservation effect remains to be further characterized in this mouse model.
Figure 7.
AAV8::U6-gRNAs_PHD2 CRISPR-Cas9 therapy improves photoreceptor function and structure in Pde6βH620Q/Cas9 arRP and RhoC110R/+ adRP mouse models
Experiments were conducted using Pde6βH620Q/Cas9 arRP (A–C) and RhoC110R/+ adRP (D–H) mouse models.
(A) Injection with a single dual-vector, AAV8:U6-gRNAs_PHD2;hGRK1-GFP, allowed marking of ventral subretinal transduction sites. Cas9 expression was transgenic in this line.
(B) Representative global electroretinography (ERG) traces of AAV-treated right eyes (red) and untreated left fellow eyes (black) from the experimental groups. There were improvements in the scotopic ERG b-wave, mixed rod-cone ERG a- and b-wave, and photopic ERG b-wave recordings (μV) from the AAV2/8(Y733F)-gRNA transduced eyes compared with uninjected fellow eyes at 8 weeks of age. The subretinal injection was performed on one eye of each mouse at P14. ERG was performed 8 weeks post injection.
(C) Gray bars represent untreated fellow eyes, red bars represent vector-transduced eyes, and black dots represent each mouse after virus treatment. AAV8-transduced and uninjected fellow eyes were compared at each time point. Error bars indicate SEM. ∗p < 0.05, ∗∗p < 0.01; n = 4 for all groups.
(D) Co-injection of two vectors AAV8:U6-gRNAs_PHD2;hGRK1-GFP and AAV8:hGRK1-Cas9 allowed marking of subretinal transduction sites in dominant RhoC110R/+ at 1 month post injection.
(E) Representative global ERG traces of AAV-treated right eyes (red) and untreated left fellow eyes (black) from the experimental groups. Subretinal injection was performed on one eye of each mouse at P28. There were statistically significant improvements in the scotopic ERG b-wave, mixed rod-cone ERG a- and b-wave, and photopic ERG b-wave recordings (μV) from the AAV2/8(Y733F)-gRNA transduced eyes compared with the uninjected fellow eyes at 5 months of age. Gray bars represent untreated fellow eyes, and red bars represent vector-injected eyes (black dots represent each mouse after virus treatment).
(F) AAV8-transduced and uninjected fellow eyes were compared at each time point. Error bars indicate SEM. ∗∗p ≤ 0.01, ∗∗∗p ≤0.001; n = 4 for all groups.
(G) H&E-stained retinal sections taken from peripheral RhoC110R/+ retina transduced ventrally with AAV8:U6-gRNAs_PHD2 in the right eye. Yellow bar denotes ONL thickness. Scale bar (black, top left), 25 μm.
(H) Quantification of ONL thickness in RhoC110R/+ mice injected with the dual AAV8 compared to the non-injected fellow eye as seen in (G). Error bars indicate SEM. ∗p ≤ 0.05; n = 4 in each group.
Following the increased functional preservation from therapeutic PHD2 editing in an arRP preclinical model, we applied the AAV therapeutic PHD2 editing vector to an alternative degeneration model. We then used an approach identical to the one first used for the arRP model (Pde6β deficiency), this time as a treatment for an adRP model caused by a rhodopsin mutation, RhoC110R/+.37,46 We performed subretinal injections of AAV8:U6-gRNAs_PHD2 into 4-week-old RhoC110R/+ mice and observed a similar ventral area of AAV transduction (Figure 7D), readily identifiable by co-injection with a GFP cassette containing AAV, similarly to our approach in the previous model, ERG analyses showed that photoresponse amplitudes from rods in treated eyes were significantly improved compared to non-injected control eyes at 6 months (representative recordings are shown in Figure 7E). ERG responses were quantified, averaged, and compared, revealing a statistically significant rescue of function (Figure 7F). Lastly, histology showed increased structural preservation in our AAV8-transduced peripheral retina compared to the non-injected contralateral retina, suggesting that the rescue effect extended to retinal function and survival (Figures 7G and 7H). Interestingly, the preservation effect seemed to spread to the central retina (500 μm from optic nerve head) as seen in Figure 7G, even though the area of GFP seen in Figure 7D is further away from the central retinal region. This is likely accountable to a fluctuation in the area of viral transduction and some degree of variability in this platform. These findings demonstrate that PHD2 ablation can rescue degeneration caused by two distinct types of genetic deficiency, an essential factor for a truly gene-agnostic therapy.
Discussion
Barring a single gene-specific therapy for RPE65-related RP, there are no curative treatments for hereditary retinal degenerations.47 This report explores CRISPR therapeutic editing to reprogram aerobic glycolysis intrinsic to photoreceptors and make them more robust to stress. Such therapeutic intervention has the potential to be effective against retinal disorders caused by a diverse array of strain or genetic deficiencies.
Metabolomic reprogramming to favor glycolysis over mitochondrial oxidation can confer two critical advantages over current mutation-specific CRISPR-based homology-directed repair. First, because it may be used to treat diseases and disorders caused by more than one mutation or gene, a strategy focused on glycolytic reprogramming could dramatically lower treatment costs. Second, cell-specific therapeutic editing may treat both dividing and non-dividing cells, as the metabolic pathway of interest is shared among all cell types.31,48
Both pharmacological inhibition with FG-4592 (Figure 1) and genetic ablation (Figures 2, 3, and 4) of PHD1, PHD2, and PHD3 (Egln2, Egln1, and Egln3) increase the ratio of glycolytic to mitochondrial activity and preserve rod and cone survival and function in the Pde6βH620Q/H620Q arRP model. Interestingly, the disconnect seen between functional and morphological data in the pharmacological inhibition experiment in Figure 1 is almost erased in the loxP-Cre mouse model seen in Figure 4. When using a cell-specific therapy, we are more able to preserve the metabolism of a young healthy retina in all cell types. This allows more cells that verge on the boundary of death to survive and convey a thicker ONL and stronger ERG response. Systemic PHD antagonist administration is commonly associated with blurred vision, swelling of the eyelids, face, lips, and hands, joint pain, stiffness of feet, chest tightness, headache, nausea, hypertension, hyperkalemia, slow or fast heartbeat, stomach pain, coffee-ground emesis, black or tarry stools, hoarseness, and difficulty swallowing.
The genetic ablation strategy offered in this article more specifically targets photoreceptors and likely will provide a safer and more effective treatment effect when further refined. Cell-specific therapeutic editing of PHD could be relevant for treating several metabolic disorders and provide a safe, efficacious therapy to address a dire unmet clinical need. The therapeutic editing gRNAs we tested in this study can be used in clinical trials for the RP subgroup of secondary cone-degenerative diseases caused by primary rod dysfunction. Therapeutic editing of PHD also may provide a safer alternative to the pharmacological orally administered PHD antagonist, FG-4592 (Evrenzo), which is currently approved to treat anemia in the European Union.49 The PHD inhibitor FG-4592 was approved in 2020 by the European Medicines Agency50 and the Pharmaceuticals and Medical Devices Agency in Japan for the treatment of anemia,51 and daprodustat (Jesduvroq) was approved by the FDA in 2023 for the once-daily treatment of anemia,52 substantiating the notion that inhibition of PHD is not only a practical but also a safe therapeutic approach.
In contrast to the pharmacological approach, our gene-editing strategy enhances the ratio of glycolysis to mitochondrial oxidation, specifically in rods. This reprogrammed rod metabolism conveys a survival advantage against the background of Pde6β degeneration, as indicated by our ERG functional and morphological outcomes. Rods rescued by enhancing aerobic glycolysis still have their original phototransduction deficiency, so they cannot be entirely functional. However, their preservation contributes to non-cell-autologous preservation of cone photoreceptors. Aerobic glycolysis can retard progression in the arRP Pde6βH620Q/H620Q preclinical model and preserve cone function for the mouse equivalent of ten human years.25 Therapeutic PHD2 editing increases the concentrations of glycolytic intermediates and diminishes flux through the TCA cycle.
We chose to focus on PHD2 because we found that ablating PHD2 alone is as effective as a combined knockout of all three PHD isoforms. PHD2 also is the most abundantly expressed isoform in rods. Other isoforms are expressed more strongly in other organs, such as the testes. Accumulated glycolytic intermediates enhance rod and cone survival in our preclinical arRP model (Figures 4 and 7). Remarkably, therapeutic PHD2 editing also can rescue rod degeneration and maintain cone viability in an adRP model with a completely different underlying mutation (Figure 7), further substantiating the notion of our gene-agnostic medicine approach to treating various forms of RP. Interestingly, most of the effect of PHD modulation on HIF levels is at post-transcriptional proteolysis. PHD hydroxylates HIF and directs the physical protein toward proteasomal degradation. When we reprogram PHD, this has a limited effect on HIF transcription, as expected (Figure 2A), yet results in accumulation of glycolytic intermediates and less mitochondrial activity, likely accounting for the preservation of visual function and histology seen in this study.
Enhanced HIF is protective in diseased states but damaging in wild-type retinas. Elevated HIF causes relatively slow degeneration of photoreceptors on a wild-type background,53,54 but it retards the degeneration that occurs in a preclinical RP model (Pde6βH620Q/H620Q)20 or in a model for light-induced retinopathy.55 It is essential to note that while the rescue effects in preclinical models may be on the time span of weeks to months, this translates to clinically significant preservation of equivalent human vision.25
Overall, our glycolytic reprogramming can significantly enhance neural function and phototransduction function in divergent models of retinal degeneration via CRISPR therapeutic editing. We showed that rod-specific aerobic glycolysis enhancement via PHD-HIF reprogramming is an effective therapeutic editing approach that does not induce toxicity in both recessive and dominant preclinical mouse models of RP. Our study suggests that the balance between glycolytic and mitochondrial oxidation of glucose may influence photoreceptor death in patients with either recessive or dominant forms of RP.8,12 Our findings also suggest that metabolomic reprogramming could be a gene-agnostic therapy that can be applied to preserve cone function in heterogeneous forms of RP (Figures 4 and 7) as well as in AMD-affected photoreceptors and other neurodegenerative disorders, including Alzheimer’s disease.31,48 We explored constitutive Cas9 expression (Figures 7A–7C) as well as AAV delivery (Figures 7D–7H) in this article, and interestingly found a similar level of response between the two approaches in two divergent models of retinal degeneration. Importantly, in CRISPR-Cas9-based therapies in humans, the Cas9 enzyme will likely need to be co-delivered via a second AAV, as seen in the RHOC110R/+ treatment in Figures 7D–7H.
While the glycolytic end products pyruvate and lactate do not increase to the degree that we expected, the levels of several key glycolytic intermediaries increase, and overall metabolic flux through the TCA cycle decreases. PDH kinase is a metabolic regulatory enzyme whose expression is enhanced significantly by HIF stabilization in this study. PDH kinase inhibits mitochondrial flux by phosphorylating and inhibiting PDH, preventing pyruvate’s entry into the mitochondrial TCA cycle as acetyl-CoA. Diminished flux of metabolites through mitochondria, caused by enhanced PDK (Figures 2A, 5F,and S2), could be as influential as enhanced glycolysis because it can diminish formation of reactive oxygen species produced by electron transport activity. Pentose phosphate pathway activity appears to be unaffected by our reprogramming (Figure S3), while overall levels of rods appear to be on par between treatment and control groups at the time of experiment (Figure S4). This is an important observation, as it indicates any differences between the treatment and control group are due to inherent changes in intracellular metabolism rather than the number of photoreceptors present in the analyzed sample. Investigating alternative pathways such as lipid metabolism and reactive oxygen species generation and management will be essential to understanding the fundamental cause of retinal degeneration and the mechanism of action of our therapeutic intervention in upcoming studies.
Limitations of the study
While this study is immensely informative on how metabolic reprogramming approaches may help mitigate some aspects of retinal degeneration associated with conditions such as RP, some limitations remain. First, the various mouse lines and therapeutic interventions administered and evaluated at multiple time points could be extrapolated to better understand the timing of the studied metabolic processes and how they contribute, or not, to the progression of retinal degeneration. Additionally, more targets associated with various metabolic pathways (both direct and indirect) could be evaluated via spatial genomics to understand better their respective contribution to the degenerative process and its rescue. This would allow a more comprehensive, targeted metabolomics analysis. The number of rod cells could have been attempted to be counted, both from a flat-mount or cross-sectional view, to gain a better understanding of how the directly targeted cell population responds to our therapeutic intervention. A more thorough evaluation of the effect of our gRNA system is in order. We are planning to complete this as we continue to study this therapeutic intervention and its impact on the prevention of the progression of retinal degeneration. Comprehensive histology from the Pde6βH620Q/H620Q model evaluated in Figure 7 should be reviewed to see the direct impact of our AAV-delivered therapeutic intervention on histology preservation. While this study deals with two of the more prevalent causative mutations in RP, reviewing the rescue effect on a third animal model, harboring a mutation in an independent gene, would be interesting.
In addition, this study deals mainly with mouse biology, and it remains to be explored how well this therapy could be translated into humans. Some examples of studies that could be pursued include functional and histological preservation assays in larger animal models that more closely mimic the human retina, such as pig or non-human primate. Lastly, the effect of sex on the results seen in our animal models should be further probed to evaluate the response in relation to this variable.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| HIF1a | Novus Biologicals | Cat no. NB100-105 Lot no. BS; RRID:AB_10001154 |
| HIF2a | Novus Biologicals | Cat no. NB100-122 Lot no. CO-2; RRID:AB_350059 |
| GLUT1 | Abcam | Cat no. ab40084 Lot no. GR3213392-4; RRID:AB_2190927 |
| HK2 | Cell Signaling | Cat no. C2867S Lot no. 5; RRID:AB_2232946 |
| LDHA | Cell Signaling | Cat no. C2012S Lot no. 2; RRID:AB_2137173 |
| β-Actin | Cell Signaling | Cat no. C3700S Lot no. 18; RRID:AB_2242334 |
| PHD2/EGLN1 | Novus Biologicals | Cat no. NB100-137 Lot no. A4; RRID:AB_10003054 |
| Bacterial and virus strains | ||
| AAV8-Y733F.GRK1.eGFP.U6.SG168.U6.SG166 | Penn Vector Core | Lot no. V7471S |
| AAV8-Y733F.GRK1.NLS-Cas9-NLS.synPA | Penn Vector Core | Lot no. V7471S |
| Chemicals, peptides, and recombinant proteins | ||
| Roxadustat | MedChemExpress | Cat no. HY-13426 |
| Tamoxifen | Sigma-Aldrich | Cat no. T5648 |
| D-Glucose (U-13 C₆, 99%) | Cambridge Isotope Laboratories, Inc. | Cat no. CLM-1396-PK |
| D-Glucose (U-12 C₆, 99.9%) | Cambridge Isotope Laboratories, Inc. | Cat no. CLM-4819-PK |
| Experimental models: Cell lines | ||
| HEK293 Cells | ATCC | Cat no. CRL-1573; RRID:CVCL_0045 |
| N2A Cells | ATCC | Cat no. CCL-131; RRID:CVCL_0470 |
| Experimental models: Organisms/strains | ||
| Mouse: Egln2tm2Fong Egln1tm2Fong Egln3tm2Fong/J | Jackson Laboratory | Cat no. 028097; RRID:IMSR_JAX:028097 |
| Mouse: Pde6γCreERT2 | Tsang Laboratory | N/A |
| Mouse: RhoC110R/+ | Tsang Laboratory | N/A |
| Mouse: Pde6βH620Q/H620Q | EMMA | Cat no. EM:01292; RRID:IMSR_EM:01292 |
| Oligonucleotides | ||
| PCR Primers | See Table S1 for all PCR Primers used in this manuscript. | N/A |
| qPCR Primers | See Table S1 for qPCR Primers used in this manuscript. | N/A |
| Software and algorithms | ||
| Excel 2016 | Microsoft | N/A |
Resource availability
Lead contact
Further information and requests for resources and or reagents should be directed to and will be fulfilled by the lead contact, Stephen H. Tsang (sht2@columbia.edu).
Materials availability
Plasmids generated in this study have been included in Table S2, provided as a separate excel file as requested. All unique/stable reagents generated in this study are available from the lead contact with a completed Materials Transfer Agreement.
Data and code availability
All data reported in this paper will be shared by the lead contact upon request. Any non-publicly available datasets can be obtained through the lead contact upon request. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental model and study participant details
Preclinical models
Pde6βH620Q/H620Q; Pde6γCreERT2/+ is a well-established preclinical model of arRP;4,43,44,56 RhoC110R/+ is a well-established preclinical model of adRP.37,46 Pde6γCreERT2/+ mice were generated at Jonas Children’s Vision Care laboratory28,43 PHD1,2,3 mouse model PHD1,2,3 fL/fL mice [Egln2tm2Fong Egln1tm2Fong Egln3tm2Fong/J] were obtained from Jackson Laboratory (stock no. 028097). The resulting progeny were bred with PHD1f,2f,3f mice. We used Pde6βH620Q/H620Q and Pde6γCreERT2 mice rederived using previously published methods.28 Pde6βH620Q/H620Q and Pde6γCreERT2 lines were crossed to yield experimental lines in this research study.
All mice were housed in the Edward S. Harkness Eye Institute at Columbia University Irving Medical Center Facility under a 12-h light and 12-h dark cycle.4,57,58 When applicable, mice were euthanized following the Columbia University IACUC guidelines and using methods reported in previous studies.4,57,58 Mice were used in accordance with the Statement for the Use of Animals in Ophthalmic and Vision Research of the Association for Research in Vision and Ophthalmology and the Policy on the Use of Animals in Neuroscience Research of the Society for Neuroscience.
Method details
PHD inhibitor
5 mg FG-4592 (M.C.E.; HY-13426) was dissolved in 100 μL DMSO (Sigma-Aldrich; 67-68-5) at 42°C to generate a stock solution and subsequently mixed at a ratio of 1:9 in Sunflower oil (Sigma-Aldrich; 8001-21-6). 20 mg/kg BW of FG-4592 was administered via oral gavage to the Pde6βH620Q/H620Q mouse model as treatment group. The same volume of DMSO was mixed 1:9 with sunflower oil and fed to the retinal degenerating mice for the control group.
Histology
Experimental and control retinae were harvested from euthanized mice to observe retinal morphology, and retinae were stained with H&E as previously described.4,43,44,56 The H&E-stained samples were used to quantify the thickness of the retinal ONL. ONL layers are labeled with a yellow or green bar in figures within this manuscript.
Electroretinography
ERG tests were administered to both eyes of all mice using previously described methods.4,38,59 ERG testing was performed 6-, 8-, and 10-week post-injection. Mice were dark-adapted for 12 h prior to anesthetization with 0.1 mL/10 g B.W. of 1 mL of 100 mg/mL ketamine and 0.1 mL of 20 mg/mL xylazine in 8.9 mL PBS, which was injected i.p., as previously described.4 During anesthetization, mice were placed on heating pads to maintain their body temperature at 37°C. Mouse eyes were dilated with one drop per eye of Tropicamide Ophthalmic Solution (1%; Akorn). Electrodes were placed on the corneas, and Gonak Hypromellose Ophthalmic Demulcent Solution (2.5%; Akorn) was applied to the eyes to prevent corneal scarring.
ERG recordings were simultaneously for both eyes. As previously described, pulses of 0.00130 cd/m2 and 3 cd/m2 (White-6500K) were employed.4,43,44,56,60 This allowed the specific testing of rod and cone functionality so that the individual and combined function could be analyzed.
Fluorescein angiography (FA)
FA was conducted at 1-year post subretinal AAV transduction of dual gRNA and Cas9 plasmids. Mice were anesthetized with 0.1 mL/10 g B.W. of 1 mL of 100 mg/mL ketamine and 0.1 mL of 20 mg/mL xylazine in 8.9 mL PBS, injected i.p. Following anesthesia, Tropicamide Opthlamic Solution (1%; Akorn) was used to dilate pupils. After proper dilation, 100 μL of Fluorescein Sodium (25%; Akorn) was injected i.p. and a Heidelberg Spectralis machine was used to capture angiographic images in the infrared and FA channels.
Tamoxifen injection and DNA recombination assay
Tamoxifen (Sigma-Aldrich; T5648) was injected intraperitoneally (i.p.) 3 times at a concentration of 100 μg/g B.W. as the treatment group, as established in our previous study.4,28 For the control group, a sham injection was administered with 10% Ethanol (w/w) in sunflower oil (Sigma-Aldrich; S5007) using the methods in Koch et al. and Zhang et al.4,43,44,56 Mice were injected at P7 and tested 4-, 6-, 8-, 10-, and 12-week post-injection.
To assay for recombination, DNA was extracted from the whole retina by using a DNeasy Blood & Tissue Kit (Qiagen, #69506). PCR was performed as previously described.45 Primers that target Egln1, Egln2, and Egln3 segments of DNA are listed in Table S1. The protein was extracted from the whole retina to assay for successful recombination. Immunoblots were performed as previously described45 to compare the protein levels of PHD1, PHD2, and PHD3 between treatment and control groups.
Mouse retinae collection
Retinae were harvested from mice at P21-P23 and snap-frozen in liquid nitrogen before being stored at −80°C for further processing; DNA, RNA, protein, or mass spectrometry.
RNA extraction and qRT-PCR
Total RNA was extracted from retinae or cell lines using an RNeasy mini kit (QIAGEN, #74104) and was reverse transcribed using SuperScript III First-Strand Synthesis SuperMix (ThermoFisher, 18080-400). The reactions were run as previously described.45 Transcript levels of each target gene were determined by SYBR Green-based qPCR (BIO-RAD, 1725271) and were standardized to β-Actin.
Immunoblotting of retinal lysates
At postnatal week 3 (P21) before the onset of ONL loss, retinae were harvested and prepared for immunoblot using previously described experimental procedures.4 The protein lysate was resuspended, and the supernatant was collected and subject to SDS-polyacrylamide gel electrophoresis using 4–15% BIO-RAD TGX pre-cast gels (#4561083). The proteins were transferred to nitrocellulose membranes for western blotting analysis. Whole-cell proteins were normalized to β-Actin (Cell Signaling, #3700). Membrane was stripped and re-probed for all targets (singular membrane). Immunoblotting signals were visualized by an iBright FL 1500 Imaging System (ThermoFisher Scientific). Whole blot images have been included where available as a separate supplemental file.
Metabolomics
To validate the specificity and, thus, the safety of our PHD-targeting strategy, flux and steady-state metabolite levels were quantified using stable isotope-resolved metabolomics coupled with mass spectrometry.
Steady-state levels of metabolites
Eyes were collected from 2-month-old experimental and control mice (i.e., before the onset of degeneration), and retinae were harvested and processed as previously described.4,60
Metabolic flux
To confirm the metabolic efficacy of glycolysis following the loss of PHD, we have quantified metabolic flux levels in PHD-deficient (PHD−/−; Pde6γCreERT2/+; Pde6βH620Q/H620Q) and control (PHDFL/F.L.; Pde6γCreERT2/+; Pde6βH620Q/H620Q) retinae using stable isotope-resolved metabolomics coupled with gas-chromatography/mass spectrometry (GC-MS), as in our previous Sirt6 studies.4,28,60 Both in vivo and ex vivo samples were analyzed to reveal how the loss of PHD affected glycolysis, mitochondrial activity, and other potentially relevant metabolic pathways. Raw metabolomics analysis data can be seen in Table S3.
Protocol A)
Quantify rates at which PHD-deficient and control retinae consume glucose medium and release lactate
Retinae were isolated and placed in a Krebs-Ringer Bicarbonate buffer (K.R.B.) supplemented with 5 mM glucose. Every 10 min, an aliquot of media was collected and subsequently analyzed for glucose and other metabolite levels with a simple glucose oxidase assay. Figure 3 shows an example, comparing rates of glucose consumption by retinae versus eyecups wherein the neuroretina has been removed (primarily RPE tissue being analyzed).
Protocol B)
Quantify flux of carbons from universally labeled (U-)13C-glucose into glycolytic and tricarboxylic acid (TCA) cycle intermediates in PHD-deficient vs. control retinae
Retinae were isolated, incubated with 5 mM U-13C-glucose, and harvested at 30 s, 90 s, 20 min, and 40 min to analyze glucose consumption and lactate production. Metabolites were extracted with 80% methanol, derivatized for gas chromatography-MS, and quantified as previously described.4,28 Figure 3 shows an example of the quantification of flux into pyruvate, citrate, and lactate. Our computational analysis quantified various parameters of isotope enrichment in downstream metabolites including flux of 13C from [U-13C] glucose into intermediates of glycolysis and the citric acid cycle and [U-13C] glucose into the pentose phosphate pathway.
Gene-ablation analysis
Human HEK293 and mouse N2A cells were transfected with the plasmid harboring the gRNAs targeting Egln1 or scrambled gRNAs unspecific to any segment of DNA. Genomic DNA from human HEK293 cells and mouse tails was extracted using the DNeasy Blood & Tissue Kit (Qiagen, 69506). DNA was extracted from Human HEK293 and mouse N2A cells after three days of selection, and PCR was performed to validate the gRNA-targeting specificity. gRNA sequences are included in Table S1. Plasmid sequences are included in Table S2.
Genomic DNA from retinae for subsequent gene-editing analyses was extracted using TRIzol reagent (Thermo Fisher Scientific, 15596018). Genomic PCR was performed using Phusion DNA polymerase (Thermo Fisher Scientific, F549S). Primers for detecting CRISPR-mediated editing in cultured human cells and degenerative mice are listed in Table S1. For deep sequencing, 436- to 267-bp PCR amplicons of sgA + sgB-treated HEK293 cells and AAV-treated mouse retinae were generated using primers with partial Illumina adapter sequences (hEGLN1-F-EZ + hEGLN1-R-EZ for HEK293, and mEGLN1-F-EZ + mEGLN1-R-EZ for mouse retinae (Table S1) and purified using the QIAquick PCR Purification Kit (Qiagen, 28106). Samples were sequenced using an Illumina-based Amplicon-EZ service offered by Genewiz. Between 70,000 and 100,000 next-generation sequencing reads for each sample were generated on 2 × 250 bp paired-end reads. Cas-analyzer software was used to analyze the next-generation sequencing data.61 The SigmaPlot 14 software package was used to represent our data graphically. All cell lines were validated for chromosome stability and the lack of mycoplasma contamination.
Therapeutic genome editing of PHD2 in mouse and human cells
Using a dual gRNA, exon 1 of Egln1 was ablated in mouse Neuro-2a cells (N2A) and human HEK293 cells. Immunoblots confirmed downregulation of PHD2 (Figures 5D and 5E). Cells transfected with experimental gRNAs directed by CRISPR-Cas9 excision had higher levels of HIF1A expression than cells transfected with control PX459: scramble gRNAs in normoxic conditions. Quantification of mRNA levels was then performed.
Cone densities
Whole eyes were processed for cryosection and subsequent staining, mounting, and imaging. Whole retinas were dissected and flat-mounted, as previously described.37,62,63 Cone numbers were averaged, assessed, and compared.44 Regions of the central retina were sampled as this is the region most susceptible to degeneration.
Adeno-associated viral vectors
Two sgRNAs, termed here as sgA and sgB (Table S1), together with a GRK1 promoter-driven GFP cassette, were cloned into the pZac2.1 vector (Penn Vector Core PL-C-PV0100, University of Pennsylvania) to form a dual AAV8:U6-gRNAs_PHD2; hGRK1 GFP construct. For the control vector, the two sgRNA-expressing cassettes of the U6-gRNAs_PHD2 were replaced with the scrambled sequences, sgC and sgD, which are not homologous to the human or mouse genome. For our AAV-Cas9 construct, a human codon-optimized pyogenes Cas9(SpCas9) was cloned into pZac2.1 between the hGrk1 (sCMV) promoter and the synthetic poly(A) sequence. Injection of AAV8:hGRK1-GFP (first virus) allowed the clear marking of ventral subretinal transduction sites. These vectors were then packaged into an AAV8 capsid with a Y733F modification by the Penn Vector Core, University of Pennsylvania. All plasmid sequences are included in Table S2.
Subretinal injection
2- to 4-week-old mice were anesthetized via i.p. injection with 10 mg/mL ketamine and 1 mg/mL xylazine in PBS at a dosage of 0.1 mL/10 g B.W. Subretinal injections were performed using a surgical microscope (Zeiss). An incision was made through the cornea from the posterior portion of the right eye using a 31-gauge needle. A pre-pulled glass micropipette (FIVEphoton Biochemicals, M.G.M. 1D) connected to a 1 mL syringe (B.D., 309623) by the tubing of a butterfly infusion set was inserted through the corneal incision and pushed through the choroid and retinal pigment epithelial cell layer. Each eye received 1.5 μL of dual AAV mixture (5 × 1012 vg/ml of each virus) or PBS. In each group, 1/10 volume of AAV8-CMV::H2B-EGFP (5 × 1012 vg/ml) was co-injected to ensure successful delivery.
Study approval
All animal studies were conducted according to the Institutional Animal Care and Use Committee Protocol Number AABU2668 at Columbia University Irving Medical Center.
Quantification and statistical analysis
Statistics
All statistical tests utilized a standardized two-sample t-test with equal variance assumptions, where appropriate. An exception is when a contralateral eye is a control, wherein a paired t-test was implemented instead. All error bars are S.E.M. unless explicitly denoted otherwise. All ERG results are averaged, with each mouse representing n = 1. The exception is in the case of the contralateral eye serving as a control, wherein the paired t-test is implemented, and each eye is taken as n = 1. All RNA and Protein analysis considers a single retinal extract as n = 1. Metabolomic analysis was performed in Microsoft Excel 2016. All statistical data were analyzed in PRISM GraphPad comprehensive statistical analysis software. No methods were used to determine whether the data met extraneous assumptions of the particular statistical approach, outside of normal requirements of a particular test.
Acknowledgments
We thank Prof. Guo-Hua Fong, Siyuan Liu, Yong-Shi Li, Salvatore M. Caruso, Marilyn Rodriguez, John Peregrin, Georgy Komissarov, Jimmy Duong, Li-Juan Duan, Anders Knudsen, and members of Jonas Children’s Vision Care (JCVC) for sharing mice and ideas; the JCVC is supported by the National Institutes of Health U01 EY030580, U54OD020351, R24EY028758, R24EY027285, 5P30EY019007, R01EY018213, R01EY024698, R01EY033770, R21AG050437, F31EY033660, NYEE Foundation, the Foundation Fighting Blindness TA-GT-0321-0802-COLU-TRAP, Lynette & Richard Jaffe Foundation, Nancy & Kobi Karp, the Crowley Family Funds, the Rosenbaum Family Foundation, Alcon Research Institute, the Gebroe Family Foundation, the Piyada Phanahat fund, and Research to Prevent Blindness (RPB) Physician-Scientist Award, unrestricted funds from RPB, New York, NY, USA. The funding organizations had no role in the design or conduct of this research. BioRender.com was used to create the graphical abstract.
Author contributions
Conceptualization, N.D.N., X.C., and S.H.T.; methodology, N.D.N., X.C., B.M.R., A.D., K.P., W.-H.W., H.F.H., L.A.J., C.-S.L., D.T.H., J.D., J.B.H., and S.H.T.; investigation, N.D.N. and X.C.; visualization, N.D.N. and X.C.; funding acquisition, S.H.T. and J.B.H.; project administration, S.H.T.; supervision, N.D.N., X.C., J.B.H., and S.H.T.; writing – original draft, N.D.N., X.C., A.D., L.A.J., and S.H.T.; writing – review & editing, N.D.N., X.C., B.M.R., A.D., L.A.J., D.T.H., J.B.H., and S.H.T.
Declaration of interests
S.H.T. receives research support from Emendo. He is on the scientific and clinical advisory board for Emendo, Medical Excellence Capital, and Nanoscope Therapeutics. S.H.T. and X.C. hold a patent related to this work: WO2018232227A1.
Published: March 21, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xcrm.2024.101459.
Contributor Information
James B. Hurley, Email: jbhhh@uw.edu.
Stephen H. Tsang, Email: sht2@columbia.edu.
Supplemental information
References
- 1.O'Neal T.B., Luther E.E. Retinitis Pigmentosa. 2022. https://www.statpearls.com/ [PubMed]
- 2.Wert K.J., Lin J.H., Tsang S.H. General pathophysiology in retinal degeneration. Dev. Ophthalmol. 2014;53:33–43. doi: 10.1159/000357294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nuzbrokh Y., Ragi S.D., Tsang S.H. Gene therapy for inherited retinal diseases. Ann. Transl. Med. 2021;9:1278. doi: 10.21037/atm-20-4726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang L., Du J., Justus S., Hsu C.W., Bonet-Ponce L., Wu W.H., Tsai Y.T., Wu W.P., Jia Y., Duong J.K., et al. Reprogramming metabolism by targeting sirtuin 6 attenuates retinal degeneration. J. Clin. Invest. 2016;126:4659–4673. doi: 10.1172/JCI86905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Russell S., Bennett J., Wellman J.A., Chung D.C., Yu Z.F., Tillman A., Wittes J., Pappas J., Elci O., McCague S., et al. Efficacy and safety of voretigene neparvovec (AAV2-hRPE65v2) in patients with RPE65-mediated inherited retinal dystrophy: a randomised, controlled, open-label, phase 3 trial. Lancet. 2017;390:849–860. doi: 10.1016/S0140-6736(17)31868-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maguire A.M., Simonelli F., Pierce E.A., Pugh E.N., Jr., Mingozzi F., Bennicelli J., Banfi S., Marshall K.A., Testa F., Surace E.M., et al. Safety and efficacy of gene transfer for Leber's congenital amaurosis. N. Engl. J. Med. 2008;358:2240–2248. doi: 10.1056/NEJMoa0802315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bainbridge J.W.B., Smith A.J., Barker S.S., Robbie S., Henderson R., Balaggan K., Viswanathan A., Holder G.E., Stockman A., Tyler N., et al. Effect of gene therapy on visual function in Leber's congenital amaurosis. N. Engl. J. Med. 2008;358:2231–2239. doi: 10.1056/NEJMoa0802268. [DOI] [PubMed] [Google Scholar]
- 8.Duncan J.L., Pierce E.A., Laster A.M., Daiger S.P., Birch D.G., Ash J.D., Iannaccone A., Flannery J.G., Sahel J.A., Zack D.J., et al. Inherited Retinal Degenerations: Current Landscape and Knowledge Gaps. Transl. Vis. Sci. Technol. 2018;7:6. doi: 10.1167/tvst.7.4.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cideciyan A.V., Jacobson S.G., Beltran W.A., Sumaroka A., Swider M., Iwabe S., Roman A.J., Olivares M.B., Schwartz S.B., Komáromy A.M., et al. Human retinal gene therapy for Leber congenital amaurosis shows advancing retinal degeneration despite enduring visual improvement. Proc. Natl. Acad. Sci. USA. 2013;110:E517–E525. doi: 10.1073/pnas.1218933110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bainbridge J.W.B., Mehat M.S., Sundaram V., Robbie S.J., Barker S.E., Ripamonti C., Georgiadis A., Mowat F.M., Beattie S.G., Gardner P.J., et al. Long-term effect of gene therapy on Leber's congenital amaurosis. N. Engl. J. Med. 2015;372:1887–1897. doi: 10.1056/NEJMoa1414221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maeder M.L., Stefanidakis M., Wilson C.J., Baral R., Barrera L.A., Bounoutas G.S., Bumcrot D., Chao H., Ciulla D.M., DaSilva J.A., et al. Development of a gene-editing approach to restore vision loss in Leber congenital amaurosis type 10. Nat. Med. 2019;25:229–233. doi: 10.1038/s41591-018-0327-9. [DOI] [PubMed] [Google Scholar]
- 12.Bramall A.N., Wright A.F., Jacobson S.G., McInnes R.R. The genomic, biochemical, and cellular responses of the retina in inherited photoreceptor degenerations and prospects for the treatment of these disorders. Annu. Rev. Neurosci. 2010;33:441–472. doi: 10.1146/annurev-neuro-060909-153227. [DOI] [PubMed] [Google Scholar]
- 13.Kanow M.A., Giarmarco M.M., Jankowski C.S., Tsantilas K., Engel A.L., Du J., Linton J.D., Farnsworth C.C., Sloat S.R., Rountree A., et al. Biochemical adaptations of the retina and retinal pigment epithelium support a metabolic ecosystem in the vertebrate eye. Elife. 2017;6 doi: 10.7554/eLife.28899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.MacIver N.J., Michalek R.D., Rathmell J.C. Metabolic regulation of T lymphocytes. Annu. Rev. Immunol. 2013;31:259–283. doi: 10.1146/annurev-immunol-032712-095956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vander Heiden M.G., Cantley L.C., Thompson C.B. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vander Heiden M.G., DeBerardinis R.J. Understanding the Intersections between Metabolism and Cancer Biology. Cell. 2017;168:657–669. doi: 10.1016/j.cell.2016.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu J., Thompson C.B. Metabolic regulation of cell growth and proliferation. Nat. Rev. Mol. Cell Biol. 2019;20:436–450. doi: 10.1038/s41580-019-0123-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Warburg O., Schroder W., Gewitz H.S., Volker W. Manometric x-ray actinometer and the effect of x-rays on the fermentation of cancer cells. Z. Naturforsch. B. 1958;13B:591–596. [PubMed] [Google Scholar]
- 19.Warburg O. On respiratory impairment in cancer cells. Science. 1956;124:269–270. [PubMed] [Google Scholar]
- 20.Nolan N.D., Caruso S.M., Cui X., Tsang S.H. Renormalization of metabolic coupling treats age-related degenerative disorders: an oxidative RPE niche fuels the more glycolytic photoreceptors. Eye (Lond) 2022;36:278–283. doi: 10.1038/s41433-021-01726-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hurley J.B., Lindsay K.J., Du J. Glucose, lactate, and shuttling of metabolites in vertebrate retinas. J. Neurosci. Res. 2015;93:1079–1092. doi: 10.1002/jnr.23583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang N.K., Tosi J., Kasanuki J.M., Chou C.L., Kong J., Parmalee N., Wert K.J., Allikmets R., Lai C.C., Chien C.L., et al. Transplantation of reprogrammed embryonic stem cells improves visual function in a mouse model for retinitis pigmentosa. Transplantation. 2010;89:911–919. doi: 10.1097/TP.0b013e3181d45a61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cepko C., Punzo C. Cell metabolism: Sugar for sight. Nature. 2015;522:428–429. doi: 10.1038/522428a. [DOI] [PubMed] [Google Scholar]
- 24.Rohowetz L.J., Kraus J.G., Koulen P. Reactive Oxygen Species-Mediated Damage of Retinal Neurons: Drug Development Targets for Therapies of Chronic Neurodegeneration of the Retina. Int. J. Mol. Sci. 2018;19 doi: 10.3390/ijms19113362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Caruso S.M., Quinn P.M., da Costa B.L., Tsang S.H. CRISPR/Cas therapeutic strategies for autosomal dominant disorders. J. Clin. Invest. 2022;132 doi: 10.1172/JCI158287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Byrne L.C., Dalkara D., Luna G., Fisher S.K., Clérin E., Sahel J.A., Léveillard T., Flannery J.G. Viral-mediated RdCVF and RdCVFL expression protects cone and rod photoreceptors in retinal degeneration. J. Clin. Invest. 2015;125:105–116. doi: 10.1172/JCI65654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Venkatesh A., Ma S., Le Y.Z., Hall M.N., Rüegg M.A., Punzo C. Activated mTORC1 promotes long-term cone survival in retinitis pigmentosa mice. J. Clin. Invest. 2015;125:1446–1458. doi: 10.1172/JCI79766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang L., Justus S., Xu Y., Pluchenik T., Hsu C.W., Yang J., Duong J.K., Lin C.S., Jia Y., Bassuk A.G., et al. Reprogramming towards anabolism impedes degeneration in a preclinical model of retinitis pigmentosa. Hum. Mol. Genet. 2016;25:4244–4255. doi: 10.1093/hmg/ddw256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moslehi J., Rathmell W.K. The 2019 Nobel Prize honors fundamental discoveries in hypoxia response. J. Clin. Invest. 2020;130:4–6. doi: 10.1172/JCI134813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Griffioen A.W., Bischoff J. Oxygen sensing decoded: a Nobel concept in biology. Angiogenesis. 2019;22:471–472. doi: 10.1007/s10456-019-09692-y. [DOI] [PubMed] [Google Scholar]
- 31.Park K.S., Xu C.L., Cui X., Tsang S.H. Reprogramming the metabolome rescues retinal degeneration. Cell. Mol. Life Sci. 2018;75:1559–1566. doi: 10.1007/s00018-018-2744-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Epstein A.C., Gleadle J.M., McNeill L.A., Hewitson K.S., O'Rourke J., Mole D.R., Mukherji M., Metzen E., Wilson M.I., Dhanda A., et al. C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell. 2001;107:43–54. doi: 10.1016/s0092-8674(01)00507-4. [DOI] [PubMed] [Google Scholar]
- 33.Olivares-Gonzalez L., Martinez-Fernandez de la Camara C., Hervas D., Millan J.M., Rodrigo R. HIF-1alpha stabilization reduces retinal degeneration in a mouse model of retinitis pigmentosa. FASEB J. 2018;32:2438–2451. doi: 10.1096/fj.201700985R. [DOI] [PubMed] [Google Scholar]
- 34.Lange C., Heynen S.R., Tanimoto N., Thiersch M., Le Y.Z., Meneau I., Seeliger M.W., Samardzija M., Caprara C., Grimm C. Normoxic activation of hypoxia-inducible factors in photoreceptors provides transient protection against light-induced retinal degeneration. Invest. Ophthalmol. Vis. Sci. 2011;52:5872–5880. doi: 10.1167/iovs.11-7204. [DOI] [PubMed] [Google Scholar]
- 35.Yang M., Su H., Soga T., Kranc K.R., Pollard P.J. Prolyl hydroxylase domain enzymes: important regulators of cancer metabolism. Hypoxia. 2014;2:127–142. doi: 10.2147/HP.S47968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nguyen T.L., Durán R.V. Prolyl hydroxylase domain enzymes and their role in cell signaling and cancer metabolism. Int. J. Biochem. Cell Biol. 2016;80:71–80. doi: 10.1016/j.biocel.2016.09.026. [DOI] [PubMed] [Google Scholar]
- 37.Tsai Y.T., Wu W.H., Lee T.T., Wu W.P., Xu C.L., Park K.S., Cui X., Justus S., Lin C.S., Jauregui R., et al. Clustered Regularly Interspaced Short Palindromic Repeats-Based Genome Surgery for the Treatment of Autosomal Dominant Retinitis Pigmentosa. Ophthalmology. 2018;125:1421–1430. doi: 10.1016/j.ophtha.2018.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Davis R.J., Tosi J., Janisch K.M., Kasanuki J.M., Wang N.K., Kong J., Tsui I., Cilluffo M., Woodruff M.L., Fain G.L., et al. Functional rescue of degenerating photoreceptors in mice homozygous for a hypomorphic cGMP phosphodiesterase 6 b allele (Pde6bH620Q) Invest. Ophthalmol. Vis. Sci. 2008;49:5067–5076. doi: 10.1167/iovs.07-1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wert K.J., Davis R.J., Sancho-Pelluz J., Nishina P.M., Tsang S.H. Gene therapy provides long-term visual function in a pre-clinical model of retinitis pigmentosa. Hum. Mol. Genet. 2013;22:558–567. doi: 10.1093/hmg/dds466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wright A.F., Jacobson S.G., Cideciyan A.V., Roman A.J., Shu X., Vlachantoni D., McInnes R.R., Riemersma R.A. Lifespan and mitochondrial control of neurodegeneration. Nat. Genet. 2004;36:1153–1158. doi: 10.1038/ng1448. [DOI] [PubMed] [Google Scholar]
- 41.Noel J., Jalligampala A., Marussig M., Vinot P.A., Marie M., Butler M., Lorget F., Boissel S., Leveillard T.D., Sahel J.A., McCall M.A. SPVN06, a Novel Mutation-Independent AAV-based Gene Therapy, Protects Cone Degeneration in a Pig Model of Retinitis Pigmentosa. Invest. Ophthalmol. Vis. Sci. 2021;62 [Google Scholar]
- 42.Wang S.K., Xue Y., Cepko C.L. Microglia modulation by TGF-beta1 protects cones in mouse models of retinal degeneration. J. Clin. Invest. 2020;130:4360–4369. doi: 10.1172/JCI136160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Koch S.F., Tsai Y.T., Duong J.K., Wu W.H., Hsu C.W., Wu W.P., Bonet-Ponce L., Lin C.S., Tsang S.H. Halting progressive neurodegeneration in advanced retinitis pigmentosa. J. Clin. Invest. 2015;125:3704–3713. doi: 10.1172/JCI82462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Davis R.J., Hsu C.W., Tsai Y.T., Wert K.J., Sancho-Pelluz J., Lin C.S., Tsang S.H. Therapeutic margins in a novel preclinical model of retinitis pigmentosa. J. Neurosci. 2013;33:13475–13483. doi: 10.1523/JNEUROSCI.0419-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Takeda K., Ho V.C., Takeda H., Duan L.J., Nagy A., Fong G.H. Placental but not heart defects are associated with elevated hypoxia-inducible factor alpha levels in mice lacking prolyl hydroxylase domain protein 2. Mol. Cell Biol. 2006;26:8336–8346. doi: 10.1128/MCB.00425-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu W.H., Tsai Y.T., Huang I.W., Cheng C.H., Hsu C.W., Cui X., Ryu J., Quinn P.M.J., Caruso S.M., Lin C.S., Tsang S.H. CRISPR genome surgery in a novel humanized model for autosomal dominant retinitis pigmentosa. Mol. Ther. 2022;30:1407–1420. doi: 10.1016/j.ymthe.2022.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Daiger S.P. The University of Texas Health Science Center; 2020. RetNet Retinal Information Network.https://web.sph.uth.edu/RetNet/ [Google Scholar]
- 48.Lin M.K., Kim S.H., Zhang L., Tsai Y.T., Tsang S.H. Rod metabolic demand drives progression in retinopathies. Taiwan J. Ophthalmol. 2015;5:105–108. doi: 10.1016/j.tjo.2015.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dhillon S. Roxadustat: First Global Approval. Drugs. 2019;79:563–572. doi: 10.1007/s40265-019-01077-1. [DOI] [PubMed] [Google Scholar]
- 50.EMA . European Medicines Agency; 2021. Evrenzo.https://www.ema.europa.eu/en/medicines/human/EPAR/evrenzo [Google Scholar]
- 51.Dhillon S. Daprodustat: First Approval. Drugs. 2020;80:1491–1497. doi: 10.1007/s40265-020-01384-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Singh A.K., Carroll K., Perkovic V., Solomon S., Jha V., Johansen K.L., Lopes R.D., Macdougall I.C., Obrador G.T., Waikar S.S., et al. Daprodustat for the Treatment of Anemia in Patients Undergoing Dialysis. N. Engl. J. Med. 2021;385:2325–2335. doi: 10.1056/NEJMoa2113379. [DOI] [PubMed] [Google Scholar]
- 53.Barben M., Schori C., Samardzija M., Grimm C. Targeting Hif1a rescues cone degeneration and prevents subretinal neovascularization in a model of chronic hypoxia. Mol. Neurodegener. 2018;13:12. doi: 10.1186/s13024-018-0243-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Barben M., Ail D., Storti F., Klee K., Schori C., Samardzija M., Michalakis S., Biel M., Meneau I., Blaser F., et al. Hif1a inactivation rescues photoreceptor degeneration induced by a chronic hypoxia-like stress. Cell Death Differ. 2018;25:2071–2085. doi: 10.1038/s41418-018-0094-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Grimm C., Wenzel A., Groszer M., Mayser H., Seeliger M., Samardzija M., Bauer C., Gassmann M., Remé C.E. HIF-1-induced erythropoietin in the hypoxic retina protects against light-induced retinal degeneration. Nat. Med. 2002;8:718–724. doi: 10.1038/nm723. [DOI] [PubMed] [Google Scholar]
- 56.Koch S.F., Duong J.K., Hsu C.W., Tsai Y.T., Lin C.S., Wahl-Schott C.A., Tsang S.H. Genetic rescue models refute nonautonomous rod cell death in retinitis pigmentosa. Proc. Natl. Acad. Sci. USA. 2017;114:5259–5264. doi: 10.1073/pnas.1615394114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tanabe T., Tsang S.H., Kjeldbye H., Berns K., Goff S., Gouras P. Adeno-associated virus mediated gene transfer into pde g knockout mouse. Invest Ophth Vis Sci. 1998:S5153. [Google Scholar]
- 58.Tsang S.H., Burns M.E., Calvert P.D., Gouras P., Baylor D.A., Goff S.P., Arshavsky V.Y. Role for the target enzyme in deactivation of photoreceptor G protein in vivo. Science. 1998;282:117–121. doi: 10.1126/science.282.5386.117. [DOI] [PubMed] [Google Scholar]
- 59.Salchow D.J., Gouras P., Doi K., Goff S.P., Schwinger E., Tsang S.H. A point mutation (W70A) in the rod PDE-gamma gene desensitizing and delaying murine rod photoreceptors. Invest. Ophthalmol. Vis. Sci. 1999;40:3262–3267. [PMC free article] [PubMed] [Google Scholar]
- 60.Wert K.J., Velez G., Kanchustambham V.L., Shankar V., Evans L.P., Sengillo J.D., Zare R.N., Bassuk A.G., Tsang S.H., Mahajan V.B. Metabolite therapy guided by liquid biopsy proteomics delays retinal neurodegeneration. EBioMedicine. 2020;52 doi: 10.1016/j.ebiom.2020.102636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Park J., Lim K., Kim J.S., Bae S. Cas-analyzer: an online tool for assessing genome editing results using NGS data. Bioinformatics. 2017;33:286–288. doi: 10.1093/bioinformatics/btw561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li Y., Wu W.H., Hsu C.W., Nguyen H.V., Tsai Y.T., Chan L., Nagasaki T., Maumenee I.H., Yannuzzi L.A., Hoang Q.V., et al. Gene therapy in patient-specific stem cell lines and a preclinical model of retinitis pigmentosa with membrane frizzled-related protein defects. Mol. Ther. 2014;22:1688–1697. doi: 10.1038/mt.2014.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wert K.J., Sancho-Pelluz J., Tsang S.H. Mid-stage intervention achieves similar efficacy as conventional early-stage treatment using gene therapy in a pre-clinical model of retinitis pigmentosa. Hum. Mol. Genet. 2014;23:514–523. doi: 10.1093/hmg/ddt452. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data reported in this paper will be shared by the lead contact upon request. Any non-publicly available datasets can be obtained through the lead contact upon request. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.