Summary
Immunotherapy has emerged as a robust approach against cancer, yet its efficacy has varied among individuals, accompanied by the occurrence of immune-related adverse events. As a result, the efficacy of immunotherapy is far from satisfactory, and enormous efforts have been invested to develop strategies to improve patient outcomes. The gut microbiome is now well acknowledged for its critical role in immunotherapy, with better understanding on host-microbes interaction in the context of cancer treatment. Also, an increasing number of trials have been conducted to evaluate the potential and feasibility of microbiome-targeting approaches to enhance efficacy of cancer treatment in patients. Here, the role of the gut microbiome and metabolites (e.g., short-chain fatty acids, tryptophan metabolites) in immunotherapy and the underlying mechanisms are explored. The application of microbiome-targeting approaches that aim to improve immunotherapy efficacy (e.g., fecal microbiota transplantation, probiotics, dietary intervention) is also elaborated, with further discussion on current challenges and suggestions for future research.
Keywords: immunotherapy, microbiome, metabolites, microbiome-targeting approaches
Graphical abstract
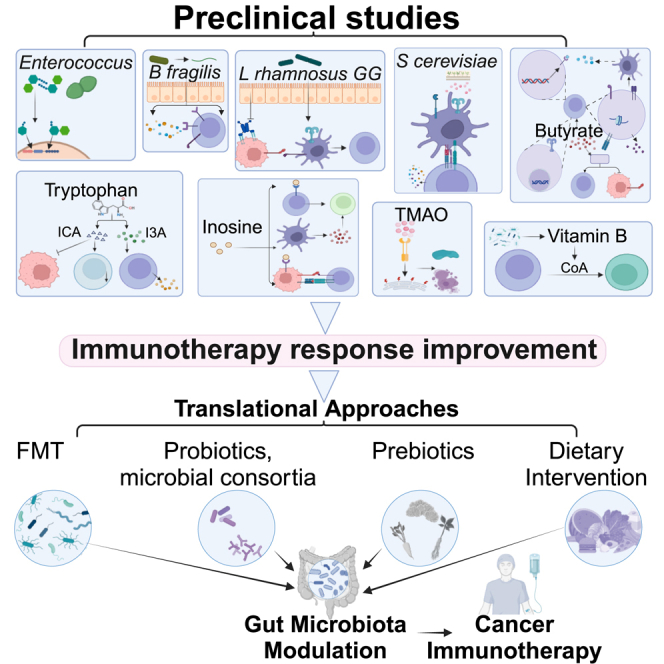
The gut microbiome is closely associated with cancer immunotherapy. Kang et al. explore the roles and underlying mechanisms of gut microbes and their derived metabolites (e.g., tryptophan, short-chain fatty acids) in modulating immunotherapy. Targeting the gut microbiome (e.g., fecal microbiota transplantation, probiotics) yields promising potential to enhance immunotherapy efficacy in clinical settings.
Introduction
The recent advance in immunotherapy has transformed the landscape of cancer treatment. This innovative approach harnesses the host immune system to combat tumor cells, providing a promising alternative to conventional therapies such as chemotherapy and radiotherapy. Cancer immunotherapy generally encompasses two strategies: directly inducing immune response through tumor antigen-targeting antibodies, vaccines, or chimeric antigen receptor (CAR) T cells; and reactivating antitumor immunity by targeting immune checkpoint inhibitors (ICIs), cytokines, or immunosuppressive cells.1,2,3,4,5,6,7,8
Immune checkpoint blockade (ICB) has been the most studied strategy of cancer immunotherapy. ICB refers to the use of antibodies to specifically target ICIs, particularly programmed cell death-1 (PD-1)/PD-ligand 1 (PD-L1) and cytotoxic T lymphocyte-associated protein 4 (CTLA-4), thereby preventing immune evasion by tumor cells.1 While immunotherapy has demonstrated promising results, its objective response rate (ORR) is greatly varied in patients receiving ICB, which can be below 30% in certain cases.9 ICB can also overactivate the host immune system, leading to the occurrence of immune-related adverse events (irAEs).10 Common irAEs include dermatological, gastrointestinal, and endocrine side effects, which can range from mild to severe and even life-threatening conditions.11 In general, the success of immunotherapy largely depends on the characteristics of each patient and their tumors. A personalized approach is thus critical to achieve optimal patient outcomes, which remains challenging in terms of diagnosis, treatment planning, and therapy monitoring. While biomarkers such as PD-L1, microsatellite instability (MSI), and tumor mutational burden have been reported, their predictive performance is far from satisfactory.12
The human gastrointestinal tract harbors a diverse microbial community composed of bacteria, viruses, and fungi, together forming the gut microbiome. Emerging evidence has strongly indicated the significant association of gut microbes with immunotherapy efficacy.13,14 Given its pivotal role, various microbiome-targeting strategies including fecal microbiota transplantation (FMT),15,16,17 prebiotics, and probiotics have been investigated to potentially augment patient responses to immunotherapy. In this article, the role of the gut microbiome in cancer immunotherapy, as well as the underlying mechanisms, is explored. The utilization of microbiome-targeting approaches to improve immunotherapy efficacy is also examined.
Role of the gut microbiome in cancer immunotherapy
Preclinical studies of the impacts of the gut microbiome on immunotherapy
Preclinical studies using mouse models have shown that specific gut microbial populations can affect the response to immunotherapy. For example, mice with depleted gut microbiome (housed in germ-free conditions or treated with antibiotics) demonstrated reduced response to CTLA-4 blockade compared to mice with intact gut microbiome.18 By supplementing specific bacteria, such as Bacteroides fragilis along with Bacteroides thetaiotaomicron or Burkholderia cepacian, these mice restored their response to immunotherapy.18 Other studies also identified several bacterial strains such as Bifidobacterium pseudolongum, Lactobacillus johnsonii, and Olsenella species isolated from ICI-treated tumors that could significantly enhance the efficacy of ICIs in mouse models.19 In particular, Bifidobacterium strains were found to be capable of improving the efficacy of anti-PD-1 immunotherapy through enhancing immune cell function and increasing tumor infiltration.20,21,22 Similarly, newly isolated strains such as Lactobacillus paracasei sh2020 and Lactobacillus kefiranofaciens ZW18 also demonstrated potential in enhancing the efficacy of anti-PD-1 therapy.23,24
Apart from gut bacteria, preclinical studies also reported the significance of microbial metabolites in ICB. For instance, trimethylamine N-oxide (TMAO) was able to stimulate immune activation and enhance the efficacy of ICB in mouse models of pancreatic cancer.25 Butyrate produced by Roseburia intestinalis has been associated with improved anti-PD-1 efficacy against colorectal cancer (CRC) in mouse models.26 Other strains of Clostridiales, including Eubacterium hallii, Faecalibacterium prausnitzii, and Anaerostipes caccae, have been shown to enhance CD8+ T cell activation and their infiltration into tumors, thereby improving the efficacy of anti-PD-1 therapy in solid tumors.27 Moreover, indole-3-carboxylic acid (ICA) derived from Lactobacillus gallinarum also enhanced anti-PD-1 efficacy in mouse models with distinct MSI statuses.28
Human studies of the association between microbiome and immunotherapy
Numerous human studies have investigated the association between microbiome composition and immunotherapy outcomes (Table 1).29,30 Several bacteria were identified as potential biomarkers in multiple cancer types including Akkermansia, which could be applied as an indicator of responders in hepatocellular carcinoma (HCC), non-small cell lung cancer (NSCLC), renal cell carcinoma (RCC), and thoracic cancer.31,32,33,34,35,36,37,38,39,40 The enrichment of Faecalibacterium was also associated with ICB responsiveness in different cancers, involving melanoma, HCC, and NSCLC.36,41,42,43,44,45,46 For a specific cancer type, a meta-analysis of melanoma patients revealed that the presence of Lachnospiraceae species is associated with a more favorable clinical response, while the presence of Streptococcaceae species is linked to an unfavorable response.47 In another study, several bacterial taxa were found to be differentially enriched in melanoma patients who were responders to combined CTLA-4 and PD-1 blockade, including Bacteroides stercoris, Parabacteroides distasonis, and Fournierella massiliensis.48 The enrichment of Bifidobacterium longum, Bifidobacterium adolescentis, Collinsella aerofaciens, and Enterococcus faecium was also observed in patients with metastatic melanoma.49 In gastrointestinal cancer patients, an increased Prevotella/Bacteroides ratio was shown to be correlated with more favorable response to anti-PD-1/PD-L1 treatment.50 Among ICB responders, a specific subgroup had markedly increased abundance of Prevotella, Ruminococcaceae, and Lachnospiraceae, while additional analysis of shotgun metagenomes indicated that gut bacteria capable of producing short-chain fatty acids (SCFAs), such as Eubacterium, Lactobacillus, and Streptococcus, are positively associated with the response to anti-PD-1/PD-L1 therapy across various gastrointestinal cancer types.50
Table 1.
Human gut microbes enriched in responders to immunotherapy
| Cancer type | Microbes enriched in responders | Immunotherapy | Sequencing methods | Sample size | Location | Reference |
|---|---|---|---|---|---|---|
| Breast | Lachnospiraceae, Turicibacteraceae, Bifidobacteriaceae, Prevotellaceae | trastuzumab | 16S rRNA (V3-V4) | 24 | Italy | Di Modica et al.51 |
| GI | Prevotella/Bacteroides ratio, Prevotellaceae, Ruminococcaceae, Lachnospiraceae; Eubacterium, Lactobacillus, Streptococcus | anti-PD-1/PD-L1 | 16S rRNA (V3-V4) and metagenomics | 74 | China | Peng et al.50 |
| HCC | Akkermansia muciniphila, Ruminococcaceae spp. | camrelizumab | metagenomic | 8 | China | Zheng et al.31,32,33,34,35,36,37,38,39,40 |
| HCC | Akkermansia, Citrobacter freundii, Azospirillum sp., Enterococcus durans | nivolumab | 16S rDNA (V3–V4) | 8 | South Korea | Chung et al.31,32,33,34,35,36,37,38,39,40 |
| HCC | Akkermansia | tremelimumab and/or durvalumab | 16S rDNA (V3–V4) | 11 | Italy | Poziani et al.31,32,33,34,35,36,37,38,39,40 |
| HCC | Lachnoclostridium, Lachnospiraceae, Veillonella | nivolumab/pembrolizumab | 16S rRNA (V3-V4) | 74 | Taiwan | Lee et al.52 |
| HCC | Faecalibacterium, Blautia, Lachnospiracea incertae Sedis, Megamonas, Ruminococcus, Coprococcus, Dorea, Haemophilus | anti-PD-1 | 16S rRNA (V3-V4) | 35 | China | Wu et al.36,41,42,43,44,45,46 |
| Hepatobiliary | Lachnospiraceae bacterium GAM79, Alistipes sp. Marseille-P5997, Ruminococcus calidus, Erysipelotichaceae bacterium GAM147 | anti-PD-1 | metagenomic | 65 | China | Mao et al.53 |
| Melanoma | Faecalibacterium, Firmicutes | ipilimumab | 16S rRNA (V3-V4) | 26 | France | Chaput et al.36,41,42,43,44,45,46 |
| Melanoma | Bacteroides caccae | ipilimumab, nivolumab, ipilimumab plus nivolumab, or pembrolizumab | metagenomic | 39 | USA | Frankel et al.36,41,42,43,44,45,46 |
| Melanoma | Faecalibacterium prausnitzii, Bacteroides thetaiotaomicron, Holdemania filiformis | ipilimumab plus nivolumab | metagenomic | 24 | USA | Frankel et al.36,41,42,43,44,45,46 |
| Melanoma | Dorea formicogenerans | pembrolizumab | metagenomic | 13 | USA | Frankel et al.36,41,42,43,44,45,46 |
| Melanoma | Clostridiales, Ruminococcaceae, Faecalibacterium | anti-PD-1 | 16S rRNA | 43 | USA | Gopalakrishnan et al.36,41,42,43,44,45,46 |
| Melanoma | Bifidobacterium longum, Bifidobacterium adolescentis, Collinsella aerofaciens, Enterococcus faecium | anti-PD-1/CTLA-4 | 16S rRNA (V4) and metagenomics | 42 | USA | Matson et al.49 |
| Melanoma | Faecalibacterium prausnitzii, Coprococcus eutactus, Prevotella stercorea, Streptococcus sanguinis, Streptococcus anginosus, Lachnospiraceae bacterium 3 1 46FAA | ICIs | 16S rRNA (V4) and metagenomics | 27 | USA | Peters et al.36,41,42,43,44,45,46 |
| Melanoma | Bacteroides stercoris, Parabacteroides distasonis Fournierella massiliensis | anti-PD-1/CTLA-4 | 16S rRNA (V4) and metagenomics | 77 | USA | Andrews et al.48 |
| NSCLC | Lactobacillus, Clostridium | ICIs | 16S rDNA (V1–V2) | 17 | Japan | Katayama et al.54 |
| NSCLC | Alistipes putredinis, Bifidobacterium longum, Prevotella copri | nivolumab | 16S rDNA (V3–V4) | 37 | China | Jin et al.55 |
| NSCLC | Ruminococcaceae UCG 13, Agathobacter | anti-PD-1/PD-L1 | 16S rRNA (V3-V4) | 70 | Japan | Hakozaki et al.56 |
| NSCLC | Ruminococcus, Akkermansia spp. | nivolumab+ipilimumab | 16S rRNA | 44 | USA | Cascone et al.31,32,33,34,35,36,37,38,39,40 |
| NSCLC | Desulfovibrio, Bifidobacterium, Anaerostipes, Faecalibacterium, Alistipes | anti-PD-1/PD-L1 | 16S rRNA (V3-V4) | 75 | China | Zhang et al.36,41,42,43,44,45,46 |
| NSCLC | Phascolarctobacterium | ICIs | 16S rRNA (V3-V4) | 69 | Spain | Zhang et al.57 |
| NSCLC | Akkermansia muciniphila | pembrolizumab/nivolumab/atezolizumab | metagenomic | 338 | France and Canada | Derosa et al.31,32,33,34,35,36,37,38,39,40 |
| NSCLC | Ruminococcus, Akkermansia, Faecalibacterium | ICIs | 16S rRNA (V1-V3) | 65 | USA | Newsome et al.31,32,33,34,35,36,37,38,39,40 |
| NSCLC | Akkermansiaceae | anti-PD-1/PD-L1 | 16S rRNA (V3-V4) | 47 | Poland | Grenda et al.31,32,33,34,35,36,37,38,39,40 |
| Prostate Cancer | Streptococcus | pembrolizumab | 16S rRNA and qPCR | 23 | USA | Peiffer et al.58 |
| RCC | Akkermansia muciniphila, Bacteroides salyersiae | nivolumab | metagenomic | 58 | France | Derosa et al.31,32,33,34,35,36,37,38,39,40 |
| RCC | Akkermansia muciniphila | nivolumab/pembrolizumab+ipilimumab | metagenomic | 31 | USA | Salgia et al.31,32,33,34,35,36,37,38,39,40 |
| Thoracic carcinoma | Akkermansiaceae, Enterococcaceae, Enterobacteriaceae, Carnobacteriaceae, Clostridiales Family XI | anti-PD-1 | 16S rRNA (V4) | 42 | China | Yin et al.31,32,33,34,35,36,37,38,39,40 |
| Pan-cancer | Trichophyton benhamiae, Cryptococcus amylolentus, Suillus clintonianus, Pseudogymnoascus sp. 05NY08, Schizosaccharomyces octosporus, Podospora anserina, Verticillium longisporum | ICIs | metagenomic | 862 | China | Huang et al.59 |
| Hematologic malignancies | Ruminococcus, Bacteroides, Faecalibacterium | anti-CD19 CAR-T | 16S rRNA | 228 | USA | Smith et al.60 |
| MM, ALL, NHL | Bifidobacterium, Prevotella, Sutterella, Collinsella | CAR-T | 16S rRNA (V4) | 78 | China | Hu et al.61 |
| B cell lymphoma | Bacteroides,, Ruminococcus, Eubacterium, Akkermansia | anti-CD19 CAR-T | metagenomic | 172 | Germany, USA | Stein-Thoeringer et al.62 |
GI, gastrointestinal; MM, multiple myeloma; ALL, acute lymphoblastic leukemia; NHL, non-Hodgkin lymphoma.
Increasing evidence also demonstrated the association of treatment outcomes with the gut microbiome. Studies in advanced hepatobiliary cancers receiving anti-PD-1 treatment revealed that patients with higher abundance of Lachnospiraceae bacterium GAM79 and Alistipes sp. Marseille-P5997 had longer progression-free survival (PFS) and overall survival (OS), and patients with higher abundance of Ruminococcus calidus and Erysipelotichaceae bacterium GAM147 also showed improved PFS.53 In HCC patients treated with nivolumab or pembrolizumab, those who achieved an objective response (OR) had a fecal enrichment of Lachnoclostridium, Lachnospiraceae, and Veillonella, along with a higher level of ursodeoxycholic acid and ursocholic acid, which are strongly correlated with Lachnoclostridium. Of note, in an independent validation cohort, patients with a favorable microbial signature had improved PFS and OS.52 In a Japanese study conducted in NSCLC patients, specific bacterial taxa such as Ruminococcaceae UCG 13 and Agathobacter were found to be enriched in patients who had a favorable OR to immunotherapy.56 Another Japanese study demonstrated that NSCLC patients with higher abundance of Lactobacillus and Clostridium tend to take a longer time to reach treatment failure after receiving ICI.54 Additionally, the enrichment of Phascolarctobacterium was observed in ICB responders from a Spanish NSCLC cohort, the presence of which is correlated with prolonged PFS.57 In clinical trials (NCT02613507 and NCT03195491) involving 37 Chinese patients with NSCLC receiving nivolumab treatment, responders showed enrichment of Alistipes putredinis, B. longum, and Prevotella copri.55 A recent study in prostate cancer revealed that the fecal abundance of Streptococcus, particularly the oral bacterium Streptococcus salivarius, was significantly higher in pembrolizumab responders.58 Patients with HER2-positive breast cancer who were unresponsive to trastuzumab had lower β-diversity and abundance of Lachnospiraceae, Turicibacteraceae, Bifidobacteriaceae, and Prevotellaceae.51 The diversity of fecal microbiome in these patients was associated with immune signatures linked to tumor-infiltrating immune cells.51
In addition to bacteria, gut fungi also contribute to ICI responses. In a recent multi-cohort study on 862 fecal metagenomes, differential fungi between ICI responders and non-responders were identified.59 These fungi could be used as biomarkers for predicting ICI response with an average area under the curve of 0.87, and were associated with increased enrichment of exhausted T cells.59 Functional analysis revealed that the central fungus Schizosaccharomyces octosporus in ICI responders may ferment starch into SCFAs, potentially promoting ICI efficacy.59 On the other hand, the role of gut virus in ICI remains uncertain. A recent study revealed a correlation between fecal enterococcal prophage and long-term benefits of PD-1 blockade in renal and lung cancer patients, indicating the therapeutic potential of specific phages to stimulate the host immune system and improve ICI efficacy.63
Although current research on the influence of the gut microbiome on adoptive T cell therapy (ACT) is limited compared to ICIs, emerging evidence suggests that gut microbes do impact ACT outcomes.64 Observational studies reported that prior exposure to antibiotics before CAR-T cell therapy was associated with poorer clinical outcomes. Conversely, a microbiome with higher abundance of Ruminococcus, Bacteroides, and Faecalibacterium has been correlated with better responses to CD19 CAR-T cell therapy.60 In a multi-center cohort of B cell lymphoma patients from Germany and United States, the study reported that prior treatment of wide-spectrum antibiotics before CD19-targeted CAR-T cell therapy could lead to unfavorable outcomes, while several bacteria such as Bacteroides, Ruminococcus, Eubacterium, and Akkermansia play a critical role in CAR-T cell therapy.62 Another clinical trial, ChiCTR1800017404, revealed distinct microbial diversity and composition among patients and treatment stages in relapsed/refractory multiple myeloma. Significant temporal variations in the abundance of Bifidobacterium, Prevotella, Sutterella, and Collinsella were observed between patients in complete remission and those in partial remission.61
Mechanism of the gut microbiome in cancer immunotherapy
Gut microbiome in tumor microenvironment
The tumor microenvironment (TME) refers to the cellular environment in and around a tumor, including immune cells, blood vessels, fibroblasts, and extracellular matrix components. The TME is known to influence tumor growth, invasion, and response to therapy.65,66 Recent studies have demonstrated the presence of intratumoral microbes and their crucial roles in the TME. For instance, immunostimulatory microbes in the gut could activate innate (dendritic cells) and adaptive (cytotoxic CD8+ T cells and interferon-γ [IFN-γ]-producing CD4+ T helper 1 [Th1] cells) immune cells, locally and systemically, to counter inhibitory TME at anatomically distant cancer sites.67
Activation of pattern recognition receptors
The gut microbiome can activate immune responses within the TME by triggering pattern recognition receptors (PRRs), which are proteins expressed by immune cells that recognize specific patterns associated with pathogens or microorganisms. Of note, gut microbes can produce various molecules to activate PRRs, affecting the balance between tumor-promoting and tumor-suppressing immune cells. For example, bacterial peptidoglycan-derived muramyl peptides have been shown to increase the immunosuppressive activity of myeloid-derived suppressor cells and promote tumor progression in CRC through enhancing arginase-1 activity, as detected by nucleotide-binding oligomerization domain 1 (NOD1).68
Molecular mimicry
Molecular mimicry refers to the ability of gut microbes to produce antigens that can be recognized by host immune cells, thereby impacting tumor progression and immune cell activity within the TME. For instance, certain gut bacteria are capable of producing antigens that cross-react with tumor-associated antigens. Such cross-reactivity stimulates immune responses to affect tumor growth. Examples include the cross-reactivity of T cells targeting specific epitopes from commensal bacteria such as Enterococcus hirae and Bifidobacterium breve with melanoma cells, resulting in reduced tumor growth and prolonged survival.63,69 Moreover, several beneficial commensals such as B. fragilis can reverse the defective maturation of PLZF+ innate lymphocytes in germ-free neonatal mice, further highlighting the impact of molecular mimicry.70
Modulation by microbe-derived metabolites
Metabolites produced by the gut microbiome can directly modulate immunity within the TME, especially SCFAs (e.g., butyrate, acetate, and propionate), which play a significant role in regulating immune process. SCFAs are able to regulate the differentiation and activation of inflammatory regulatory T cells (Tregs) and proinflammatory interleukin-17-positive (IL-17+) γδ T cells.71 Other microbial metabolites such as inosine derived from Bifidobacterium promote T cell activation and antitumor immunity by agonizing T cell-specific adenosine 2A receptor (A2AR) signaling.19 The microbe-derived metabolic product of tryptophan metabolism, an essential amino acid, can also influence the TME. For instance, indoleamine 2,3-dioxygenase 1 (IDO1) was able to initiate the transformation of tryptophan into the immunosuppressive metabolite kynurenine (Kyn), while other microbial metabolites such as ICA inhibit IDO2 expression, potentially serving as therapeutic agents in cancer treatment.28,72
Apart from beneficial effects, some microbial metabolites are found to exert opposing effects on cancer. One example is gallic acid, a phenolic acid derived from gut microbes, which is able to switch the tumor-suppressing activity of mutant p53 to an oncogenic state.73 Besides this, succinic acid derived from Fusobacterium nucleatum was shown to inhibit the cyclic GMP-AMP synthase (cGAS)-IFN-β pathway, thereby reducing CD8+ T cell trafficking to the TME and diminishing antitumor immunity.74 It is noteworthy that while the impacts of the gut microbiome on the TME are increasingly acknowledged, further research is needed to fully understand the complex mechanisms involved and to develop targeted approaches for modulating the gut microbiome to enhance cancer immunotherapy.
Mechanisms of how the gut microbiome influences immunotherapy
Microbial interventions with immune checkpoint inhibitors
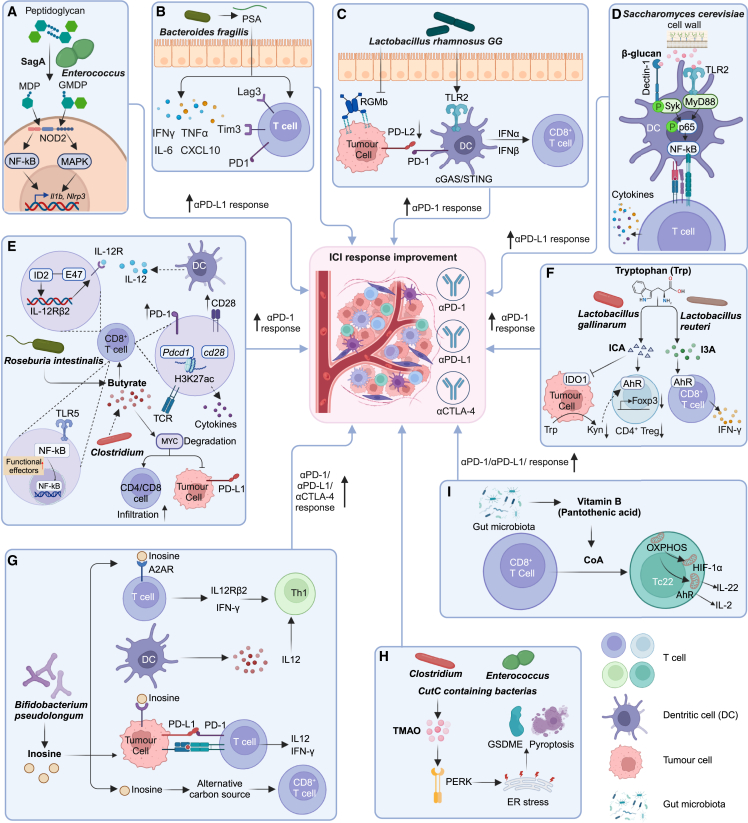
ICIs enhance immune cytotoxicity of T cells by targeting coinhibitory molecules PD-1/PD-L1 to strengthen the natural immune response of the host and prevent tumor cells from evading immune surveillance.1 Notably, gut microbes are capable of modulating host immune regulation, thereby indirectly affecting the efficacy of ICIs in cancer patients.75 A previous study by Griffin et al. demonstrated that enterococci released stimulatory molecule muramyl dipeptide (MDP) fragments through the secretion of NlpC/p60 peptidoglycan hydrolase SagA.76 These fragments activated the innate immune sensor NOD2 and enhanced immunotherapy responses, resulting in the direct activation of macrophages, epigenetic reprogramming of monocytes, generation of conventional type 1 dendritic cells, and priming of dendritic cells for cross-presentation to CD8+ T cells76 (Figure 1A).
Figure 1.
Mechanisms of the gut microbiome and their metabolites in modulating cancer immunotherapy
(A) Enterococci SagA-derived MDP and glucosaminyl-MDP (GMDP) activate NF-κB and mitogen-activated protein kinase (MAPK) pathways.
(B) B. fragilis-derived PSA promotes cytokine secretion and upregulates immune checkpoint markers on T cells.
(C) L. rhamnosus GG triggers IFN production in dendritic cells and downregulates PD-L2 and its binding partner RGMb on tumor cells.
(D) Yeast cell wall β-glucan activates dendritic cells via Dectin-1/Syk and TLR2/MyD88 pathways.
(E) Butyrate inhibits ID2-dependent IL-12 signaling, increases H3K27 acetylation to upregulate PD-1 and CD28, and modulates TCR signaling to stimulate antitumor cytokine secretion. R. intestinalis-derived butyrate binds to TLR5 on CD8+ T cells to activate NF-κB signaling. Butyrate-producing C. butyricum causes MYC degradation to promote immune cell infiltration and downregulate PD-L1 on tumor cells.
(F) L. gallinarum-derived ICA inhibits IDO1 expression to suppress Kyn intratumoral production and compete with Kyn for AhR binding on CD4+ T cells to inhibit Treg differentiation. L. reuteri-derived I3A triggers antitumor IFN-γ+ CD8+ T cell-mediated immunity by AhR activation.
(G) B. pseudolongum-derived inosine activates A2AR on T cells, upregulates IL12Rβ2 and IFN-γ transcription, and promotes Th1 differentiation and accumulation in the TME. Inosine promotes presentation of tumor neoantigens to facilitate the recognition and killing by cytotoxic CD8+ T cells and as an alternative carbon source for CD8+ T cells.
(H) TMAO from CutC-containing bacteria Clostridium and Enterococcus activates PERK to induce ER stress and increase GSDME-mediated pyroptosis of tumor cells.
(I) Microbe-derived vitamin B5 (pantothenic acid) acts as CoA precursor to reprogram oxidative phosphorylation (OXPHOS), thereby promoting differentiation of CD8+ cytotoxic T cells into IL-22-producing Tc22 cells.
Figure created by BioRender.com.
Another study found that stimulation of T cells with polysaccharide A (PSA) derived from the commensal gut bacteria B. fragilis led to the upregulation and secretion of IFN-γ, tumor necrosis factor α (TNF-α), IL-6, and CXCL10. Importantly, B. fragilis-derived PSA also induced the expression of immune checkpoint markers Lag3, Tim3, and particularly PD-1 on the surface of T cells77 (Figure 1B). Moreover, Lactobacillus rhamnosus GG, a well-known probiotic strain, was able to activate CD8+ T cells by stimulating dendritic cells through the Toll-like receptor 2 (TLR2) pathway.78 This activation was triggered by the production of IFN-α and IFN-β in dendritic cells via the cGAS/stimulator of IFN genes (STING) signaling pathway.79 Furthermore, the gut microbiome was found to be capable of suppressing the expression of PD-L2 and its binding partner repulsive guidance molecule B (RGMb), resulting in enhanced antitumor immunity and increased efficacy of PD-1 inhibitors on dendritic cells80 (Figure 1C).
In addition to bacteria, recent studies reported that the cell wall of yeast, specifically Saccharomyces cerevisiae, contains β-glucan, which could activate dendritic cells through two distinct pathways: the Dectin-1/Syk pathway and the TLR2/MyD88 pathway81 (Figure 1D). The stimulated dendritic cells subsequently trigger the activation of T cells, thereby improving their capacity to fight against tumors.82 Remarkably, when combining PD-L1 blockade with the supplementation of yeast cell wall, significant antitumor efficacy was observed in mice with melanoma, showcasing the potent therapeutic potential of fungal components.81
Boosting immunotherapy efficacy by microbial metabolites
The gut microbiome interacts with host cells mainly through metabolite production. These microbe-derived metabolites can exert local effects on the intestinal epithelium or spread to distant sites and organs via the bloodstream, subsequently acting as signaling molecules or metabolic substrates to impact a wide range of physiological functions.83 To date, a significant amount of research has been conducted to evaluate the role of SCFAs, tryptophan metabolites, inosine, TMAO, and vitamin B5 in cancer immunotherapy (Figures 1E–1I).
SCFAs
SCFAs are a group of microbial metabolites that have been heavily investigated in the context of ICIs. In general, ICI responders tend to have a higher concentration of SCFAs and greater abundance of SCFA-producing bacteria compared to non-responders.84,85,86,87,88
Butyric acid is one of the most studied SCFAs (Figure 1E). A previous study reported that butyric acid produced by Faecalibaculum rodentium PB1 and Hodemanella biformis functions as a histone deacetylase inhibitor to inhibit tumor cell proliferation by increasing acetylation and inhibiting calcineurin-mediated activation of nuclear factor of activated T cells C3 (NFATc3).89 Butyrate can also enhance the antitumor cytotoxicity of CD8+ T cells by inhibiting DNA binding 2 (ID2)-dependent IL-12 signaling to improve the efficacy of anti-PD-1 therapy.90 Butyrate upregulates the expression of PD-1/CD28 by increasing histone 3 lysine 27 acetylation (H3K27ac) at the promoter region of Pdcd1 and Cd28 in human CD8+ T cells, and modulates the T cell receptor (TCR) signaling pathway to promote the expression of antitumor cytokines in cytotoxic CD8+ T cells, all of which could enhance anti-PD-1 efficacy.87 Moreover, butyrate derived from R. intestinalis can directly bind to TLR5 on CD8+ T cells to activate the nuclear factor κB (NF-κB) signaling pathway, leading to the induction of cytotoxic granzyme B+, IFN-γ+, and TNF-α+ CD8+ T cells in CRC with MSI-low CT26 tumors in mice.26 In addition, treating CRC cells with uncharacterized metabolites from Clostridium butyricum, potentially consisting of butyrate, enhances proteasome-mediated ubiquitination, leading to the degradation of the pivotal signal molecule MYC and eventually increases the efficacy of anti-PD-1 therapy by promoting CD8/CD4 cell infiltration and downregulating the expression of PD-L1 on tumor cells.91,92 Of note, high concentrations of SCFAs such as butyrate and propionate have been associated with reduced efficacy of CTLA-4 blockade, which could inhibit dendritic cell maturation and T cell accumulation in the TME.93
Tryptophan metabolites
Various metabolites are produced by microbe-mediated tryptophan metabolism, and some have shown their impacts on immunotherapy (Figure 1F). For instance, ICA derived from L. gallinarum improved the efficacy of anti-PD-1 therapy by inhibiting IDO1 expression and Kyn production within tumors, competing with Kyn for binding to the aryl hydrocarbon receptor (AhR) and antagonizing Kyn binding on CD4+ T cells.28 These events inhibit the differentiation of Tregs to enhance the efficacy of anti-PD-1 therapy.28 Daily administration of Lactobacillus reuteri also led to the translocation of gut microbes to B16-F0 melanoma tumors in mice, triggering antitumor IFN-γ+ CD8+ T cell-mediated immunity in the TME through the metabolism of dietary tryptophan to indole-3-aldehyde (I3A) by intratumoral L. reuteri and subsequent activation of AhR, enhancing ICI efficacy in murine models.94
Inosine
Mager et al. discovered that intestinal B. pseudolongum-produced inosine significantly boosts the efficacy of anti-PD-L1 and anti-CTLA-4 in mouse models with various cancer types, including CRC, bladder cancer, and melanoma.19 Mechanistically, inosine promotes immune cell activation by activating adenosine A2AR on T cells, leading to the upregulation of IL12Rβ2 and IFN-γ transcription, promoting Th1 cell differentiation and accumulation in the TME.19 Inosine was reported to also enhance the immunogenicity of tumor cells by stimulating dendritic cells and adenosine receptors on T cells, resulting in increased tumor antigen display and improved function of tumor-specific T cells with enhanced production of IFN-γ and IL-12, thereby improving responses to anti-PD-1 therapy.95 Moreover, inosine acts as an energy source for CD8+ T cells, providing them with energy to enhance their effectiveness in combating tumor cells96 (Figure 1G).
TMAO
Gut microbiome-derived metabolite TMAO enhances the efficacy of ICIs by promoting the infiltration of immunostimulatory macrophages and CD8+ T cells into the TME.25,97 Previous studies have reported the correlation between the presence of bacteria containing CutC (e.g., Clostridium and Enterococcus), the enzyme responsible for generating the TMAO precursor trimethylamine, and enhanced survival in cancer patients.25 Mechanistically, TMAO activates the protein kinase R-like ER kinase (PERK) pathway, inducing endoplasmic reticulum (ER) stress and subsequently triggering gasdermin-E (GSDME)-mediated pyroptosis, thereby enhancing CD8+ T cell-mediated antitumor immunity97 (Figure 1H).
Vitamin B5
Intestinal bacteria-produced vitamin B5, also known as pantothenic acid, acts as a precursor of coenzyme A (CoA) found in food. Vitamin B5 was found to promote the differentiation of CD8+ cytotoxic T cells into IL-22-producing Tc22 cells, which exhibit strong antitumor effects and are associated with enhanced responses to immunotherapy.98,99 Tc22 cells upregulated the pantothenate/CoA pathway and relied on oxidative phosphorylation for differentiation, which can be reprogrammed by exogenous CoA administration through HIF-1α and AhR, promoting CD8+ Tc22 phenotype regardless of polarizing conditions99 (Figure 1I). In murine tumor models, pantothenate treatment improved the efficacy of anti-PDL1 antibody therapy, while in melanoma patients, higher pretreatment plasma pantothenic acid levels correlated with a positive response to anti-PD-1 therapy.99
Other microbe-derived molecules
Hippuric acid, in combination with butyrylcarnitine, cysteine, and glutathione disulfide, was shown to be associated with a greater response in NSCLC patients receiving PD-1 blockade, potentially due to their association with T cell metabolism.100 Muropeptides generated by Enterococcus faecium enhance anti-PD-L1 therapy by activating innate immune sensing protein NOD2, increasing CD8+ T cells expressing granzyme B, improving the TME, and enhancing the efficacy of immunotherapeutic monoclonal antibodies.76 Apart from metabolites, microbial exopolysaccharides produced by Lactobacillus delbrueckii subsp. Bulgaricus OLL1073R-1 (EPS-R1) also enhanced the efficacy of anti-CTLA-4 or anti-PD-1 therapy by inducing the expression of CCR6 in CD8+ T cells, thereby promoting T cell function.101
Modulation of adoptive T cell therapy
The administration of the SCFAs pentanoate and butyrate reprograms the metabolism and epigenetics of cytotoxic T cells and CAR-T cells, leading to heightened production of effector molecules (CD25, IFN-γ, and TNF-α), improved mammalian target of rapamycin efficacy, and inhibition of class I histone deacetylase activity, ultimately enhancing the antitumor activity of these cells in murine melanoma and pancreatic cancer models.102 Notably, further investigation is needed to understand the mechanisms underlying these effects on ACT efficacy.
Translational approaches for modulating the gut microbiome to enhance immunotherapy response
Given the crucial role of gut microbes in cancer immunotherapy, there has been growing interest in developing various targeted approaches to modulate the gut microbiome to enhance treatment efficacy. These interventions include FMT, live biotherapeutic products, and probiotic and prebiotic supplements.103,104
Fecal microbiota transplantation in immunotherapy
FMT is a direct approach to manipulate the gut microbiome. It involves the transfer of stools from a donor to the recipient through oral administration of lyophilized or frozen capsules or direct delivery by colonoscopy or gastroscopy. Numerous studies have investigated the potential of FMT to improve patients’ response to immunotherapy.15,16 For example, clinical studies (NCT03353402 and NCT03341143) conducted by Baruch et al. and Davar et al. have shown promising results of using FMT to increase the efficacy of anti-PD-1 immunotherapy in metastatic melanoma patients, with enhanced activity of CD8+ T cells and reduced frequency of immunosuppressive IL-8-expressing myeloid cells.15,16 A phase 1 clinical trial (NCT03772899) evaluated the efficacy of combined treatment with FMT and anti-PD-1 immunotherapy against advanced melanoma and observed a promising ORR of 65% (13 out of 20 patients), including four complete responses, thus emphasizing the need for further investigations of FMT.105 To date, only one completed clinical trial combining FMT with cancer immunotherapy has been registered on ClinicalTrials.gov (NCT04056026), involving a mesothelioma patient receiving pembrolizumab. Meanwhile, multiple clinical trials are also ongoing (e.g., NCT04924374, NCT05286294, NCT05251389), which aim to provide further evidence regarding the impact of FMT on ICI response as well as the associated immune and transcriptomic changes in the gut and tumor tissues.106 These ongoing trials primarily investigate the combination of FMT with immunotherapy in patients with melanoma, while some also explore other cancer types (e.g., RCC, NSCLC, CRC) (Table 2).
Table 2.
Clinical trials of FMT manipulating the gut microbiome in immunotherapy
| Cancer type | Enrollment | Immunotherapy | Microbial intervention | Phase | Location | Status | |
|---|---|---|---|---|---|---|---|
| NCT03341143 | melanoma | 18 | pembrolizumab | FMT via colonoscopy from ICI responders | 2 | USA | active, not recruiting |
| NCT03353402 | melanoma | 40 | anti-PD-1 | FMT capsules from ICI responders | 1 | Israel | unknown |
| NCT03772899 | melanoma | 20 | pembrolizumab/nivolumab | FMT capsules from healthy donors | 1 | Canada | active, not recruiting |
| NCT03819296 | melanoma, genitourinary, malignant solid neoplasm, lung | 800 | ICIs | FMT from healthy donors | 1/2 | USA | recruiting |
| NCT04038619 | genitourinary, melanoma, lung, ovarian, uterine, breast, cervical | 40 | loperamide | FMT via colonoscopy | 1 | USA | recruiting |
| NCT04056026 | mesothelioma | 1 | pembrolizumab (Keytruda) | single-dose FMT infusion | 1 | USA | completed |
| NCT04116775 | prostate | 32 | pembrolizumab | FMT via endoscopy | 2 | USA | recruiting |
| NCT04130763 | gastrointestinal system | 10 | anti-PD-1 | FMT capsules | 1 | China | unknown |
| NCT04163289 | RCC | 20 | ipilimumab+nivolumab | FMT capsules | 1 | Canada | recruiting |
| NCT04264975 | solid carcinoma | 60 | immunotherapy | FMT | N/A | Korea | unknown |
| NCT04521075 | melanoma, NSCLC | 42 | nivolumab | FMT capsules | 1/2 | Israel | unknown |
| NCT04577729 | melanoma | 5 | ICIs | allogenic FMT; autologous FMT | N/A | Austria | terminated |
| NCT04729322 | CRA, small intestinal adenocarcinoma, CRC | 14 | pembrolizumab/nivolumab | FMT capsules | 2 | USA | recruiting |
| NCT04758507 | RCC | 50 | ICIs | FMT capsules | 1/2 | Italy | recruiting |
| NCT04883762 | solid tumors | 4 | ICIs | FMT via colonoscopy | 1 | USA | active, not recruiting |
| NCT04924374 | lung cancer | 20 | pembrolizumab, nivolumab, atezolizumab | FMT capsules | N/A | Spain | recruiting |
| NCT04951583 | NSCLC, melanoma | 70 | pembrolizumab, ipilimumab+nivolumab | investigational FMT capsules | 2 | Canada | recruiting |
| NCT04988841 | melanoma | 60 | ipilimumab+nivolumab | fecal microbiotherapy (MaaT013): pooled-donor, full-ecosystem intestinal microbiome | 2 | France | recruiting |
| NCT05008861 | NSCLC | 20 | anti-PD-1/PD-L1 | FMT capsules | 1 | China | unknown |
| NCT05251389 | melanoma | 24 | ICIs | FMT from ICI non-responders/responders | 1/2 | Netherlands | recruiting |
| NCT05273255 | malignancies | 30 | ICIs | FMT via endoscopy | N/A | Switzerland | recruiting |
| NCT05279677 | CRC | 30 | sintilimab+fruquintinib | FMT | 2 | China | recruiting |
| NCT05286294 | melanoma, HNSCC, CSCC, MSI-high, clear cell RCC, NSCLC | 20 | ICIs | FMT | 2 | Norway | recruiting |
| NCT05502913 | lung | 80 | ICIs | FMT capsules | 2 | Israel | recruiting |
| NCT05690048 | liver | 48 | atezolizumab+bevacizumab | FMT capsules | 2 | Germany | not recruiting |
| NCT05750030 | HCC | 12 | atezolizumab+bevacizumab | FMT | 2 | Austria | recruiting |
CRA, colorectal adenocarcinoma; HNSCC, head and neck squamous cell carcinoma; CSCC, cutaneous squamous cell carcinoma.
In addition, the feasibility of using FMT as a preventive measure for treatment-related complications and to reduce treatment toxicity is also being evaluated. For instance, several clinical trials (NCT03819296, NCT04038619, NCT04163289, and NCT04883762) have been conducted with the aim of enhancing patient outcomes by combining FMT with immunotherapy (Table 2).
Probiotics, prebiotics, and dietary interventions
Probiotics and microbial consortia
Probiotics are live microorganisms or blends of microorganisms that can bring positive impacts on individuals’ well-being when consumed in sufficient quantity. Certain probiotic strains have been studied for their potential to modulate the gut microbiome and enhance immune responses in the context of cancer immunotherapy. In a retrospective multi-center study in Japan, the use of probiotics such as Bifidobacterium and C. butyricum was associated with better outcomes in patients with advanced or recurrent NSCLC undergoing anti-PD-1 monotherapy.107 A randomized phase 1 trial (NCT03829111) investigated the impact of CBM588 (C. butyricum strain MIYAIRI 588) in combination with nivolumab-ipilimumab immunotherapy for patients with metastatic RCC.108 This trial demonstrated that CBM588 supplementation combined with ICB significantly extends PFS compared to immunotherapy alone (12.7 months vs. 2.5 months). CBM588 also showed significant extension of PFS and OS in another study of patients with NSCLC, including those who received antibiotic therapy.109 Mechanistically, CBM588 was able to increase the abundance of other probiotics and stimulate the expansion of IL-17A-producing cells such as γδT cells and CD4 cells in preclinical mouse models.110 These findings collectively suggest the potential of CBM588 in modulating the gut microbiome to enhance immunotherapy efficacy in cancer patients.
Akkermansia muciniphila is another candidate probiotic that may boost immunotherapy efficacy in patients. In a retrospective analysis by Derosa et al., the result revealed a correlation between the presence of fecal A. muciniphila and improved ICI outcomes in patients with NSCLC.35 Subsequent investigation identified that supplementation of lyophilized encapsulated A. muciniphila (Akkp2611) can benefit patients who were exposed to antibiotics and lacked endogenous A. muciniphila.35 In general, further research is needed to confirm the observational findings and gain more understanding of the mechanistic functions of probiotics on the gut microbiome and immune system. Currently, there are multiple ongoing clinical trials investigating the manipulation of the gut microbiome in immunotherapy against various cancer types using different probiotic strains, including Bifidobacterium trifidum live powder BiFico, Probio-M9 (L. rhamnosus), and Bifidobacterium bifidum (Table 3).
Table 3.
Clinical trials of probiotics, bacteria consortia, prebiotics. and dietary interventions manipulating the gut microbiome in immunotherapy
| Cancer type | Enrollment | Immunotherapy | Microbial intervention | Phase | Location | Status | |
|---|---|---|---|---|---|---|---|
| Probiotics and bacteria consortia | |||||||
| NCT03595683 | melanoma | 8 | pembrolizumab | Bifidobacterium longum EDP1503 | 2 | USA | suspended |
| NCT03637803 | NSCLC, RCC, melanoma, bladder cancer | 63 | pembrolizumab | MRx0518 (a lyophilized proprietary bacterium strain) | 1/2 | USA | terminated |
| NCT03686202 | all solid tumors | 65 | anti-PD-1/PD-L1 | MET-4 | 2/3 | Canada | active, not recruiting |
| NCT03775850 | CRC, triple-negative breast cancer, NSCLC, bladder, gastroesophageal, RCC | 69 | pembrolizumab | Bifidobacterium longum EDP1503 | 1 | USA, Canada | completed |
| NCT03817125 | melanoma | 14 | nivolumab | SER-401 | 1 | USA | completed |
| NCT03829111 | RCC | 30 | nivolumab+ipilimumab | CBM588 | 1 | USA | active, not recruiting |
| NCT04208958 | melanoma, gastric, gastroesophageal junction adenocarcinoma, CRC | 111 | nivolumab | VE800 | 1/2 | USA | completed |
| NCT04601402 | solid tumor, NSCLC, HNSCC, urothelial carcinoma | 11 | avelumab | live biotherapeutic product GEN-001 | 1 | USA | completed |
| NCT04699721 | NSCLC | 40 | nivolumab+paclitaxel+carboplatin | BiFico | 1 | China | active, not recruiting |
| NCT04909034 | NSCLC | 30 | pembrolizumab | fermented soybean extract MicrSoy-20 (MS-20) | 2 | Taiwan | recruiting |
| NCT05032014 | liver | 46 | anti-PD-1 | Probio-M9 | N/A | China | recruiting |
| NCT05094167 | NSCLC | 46 | carrilizumab+platinum | Kex02 (Lactobacillus Bifidobacterium V9) | N/A | China | recruiting |
| NCT05122546 | RCC | 31 | nivolumab+cabozantinib S-malate | CBM588 | 1 | USA | active, not recruiting |
| NCT05220124 | bladder, urothelial | 190 | immunotherapy | live combined Bifidobacterium, Lactobacillus and Enterococcus capsules | 4 | China | recruiting |
| NCT05354102 | NSCLC, melanoma, RCC | 12 | nivolumab | BMC128 | 1 | Israel | recruiting |
| NCT05620004 | advanced HCC | 30 | carrilizumab+apatinib mesylate | Bifidobacterium bifidum | 1/2 | China | recruiting |
| Prebiotics | |||||||
| NCT01829373 | lung | 5 | lung cancer vaccine | yeast-derived β-glucan | 1 | USA | completed |
| NCT04552418 | solid tumor | 12 | ipilimumab+nivolumab | potato starch (Bob’s Red Mill) | 1 | USA | completed |
| NCT06049576 | RCC | 30 | nivolumab+ipilimumab | camu camu | 1 | USA | recruiting |
| Dietary intervention | |||||||
| NCT03340935 | Cancer | 101 | standard-of-care treatment | FMD: a 5-day plant-based, low-calorie, low-protein, low-carbohydrate diet | N/A | Italy | completed |
| NCT03595540 | breast, CRC | 90 | nivolumab (Opdivo), pembrolizumab (Keytruda) | FMD Prolon | N/A | Italy | completed |
| NCT03700437 | NSCLC | 12 | pembrolizumab | FMD Chemolieve | N/A | USA | completed |
| NCT03709147 | advanced LKB1-inactive Lung adenocarcinoma | 64 | pembrolizumab | metformin hydrochloride/metformin hydrochloride + FMD | 2 | Italy | unknown |
| NCT04316520 | metastatic RCC | 20 | nivolumab+ipilimumab, pembrolizumab+axitinib, sunitinib or pazopanib | ketogenic diet 2:1 | N/A | France | recruiting |
| NCT04645680 | melanoma | 42 | pembrolizumab/nivolumab | isocaloric high-fiber diet | 2 | USA | recruiting |
| NCT04866810 | melanoma | 80 | relatlimab+nivolumab | high-fiber, plant-based diet + exercise | N/A | USA | recruiting |
| NCT05119010 | metastatic RCC | 60 | nivolumab+ipilimumab | ketogenic diet, BHB (β-hydroxybutyrate) | N/A | France | recruiting |
| NCT05083416 | HNSCC | 29 | nivolumab, pembrolizumab, atezolizumab, avelumab, or durvalumab | prolonged nightly fasting | N/A | USA | active, not recruiting |
| NCT05356182 | integrative oncology | 30 | anti-PD-1/PD-L1/CTLA-4 | low-protein diet (10%) | N/A | USA | recruiting |
| NCT05384873 | metastatic NSCLC | 180 | immunotherapy | immunonutrients (Oral Impact): high-calorie, high-protein nutritional liquid supplement | N/A | No data | not recruiting |
| NCT05703997 | SCLC | 20 | atezolizumab | cyclic, 5-day, calorie-restricted, plant-based, low-protein, low-carbohydrate diet | 2 | Italy | not recruiting |
| NCT05763992 | triple-negative breast cancer | 145 | pembrolizumab | fasting-like approach (FLA): a plant-based, low-calorie, low-protein, low-carbohydrate diet | 2 | Italy | recruiting |
HNSCC, head and neck squamous cell carcinoma; SCLC, small cell lung cancer.
Apart from probiotics, the use of bacteria consortia as biotherapeutic products has shown promising results in cancer treatment. For example, an ongoing phase 2/3 trial (NCT03686202) is studying the efficacy of providing a defined bacterial mixture, Microbial Ecosystem Therapeutic 4 (MET-4), which consists of a defined mixture of pure live gut bacterial culture isolated from stools of a healthy individual for administration to patients with various solid tumors. In a clinical assessment (NCT03817125), SER-401, an experimental microbiome-based treatment containing a high concentration of Ruminococcaceae and other spore-forming microorganisms, was investigated in combination with nivolumab in a group of 14 patients with first-line multiple myeloma. Additionally, a completed clinical trial (NCT04208958) involving 111 patients with different gastrointestinal cancers assessed the combination of nivolumab with VE800, composed of 11 non-pathogenic commensal bacterial strains. Altogether these studies highlight the feasibility of developing bacterial formulations and consortia as potential adjuncts to cancer immunotherapy. Nevertheless, the optimal strain, dosage, and duration of microbial intervention, still require extensive investigations.
Prebiotics
Recent research has highlighted the benefits of certain prebiotics in enhancing the efficacy of cancer immunotherapy by modulating the metabolism of gut microbes.111 One major example is inulin, which can improve response of T cells and enhance anti-PD-1 efficacy by regulating the gut microbiome.112,113 Similarly, ginseng polysaccharides were found to impact IDO activity and enhance anti-PD-1 antitumor response by modulating microbial metabolites and promoting effector T cells while suppressing Tregs.114 Ganoderma lucidum polysaccharide also showed tumor-suppressive benefits by alleviating gut dysbiosis, increasing SCFA production, and mitigating endotoxemia by suppressing the TLR4/MyD88/NF-κB pathway.115
Clinical trials have been conducted to investigate the use of prebiotics as adjuvants for cancer treatment. In a completed phase 1 trial (NCT01829373), the efficacy and safety of an oral β-glucan prebiotic in combination with a lung cancer vaccine was assessed. Another ongoing clinical trial (NCT04552418) is investigating the effect of resistant starch supplement in patients with advanced or metastatic solid tumors and aims to evaluate its impact on patient outcomes. Although no results have been reported, these trials signify the growing interest in exploring the potential of prebiotics combined with cancer treatment.
Dietary intervention
Given that gut microbes are readily affected by diet, dietary intervention has gained interest as a potential strategy to enhance immunotherapy efficacy by modulating the gut microbiome.116 Several dietary interventions, such as fasting-mimicking diets (FMDs),117 ketogenic diets,118 high-fiber diets,119 low-protein diets, and prolonged nightly fasting, are being investigated for their ability to shape the microbiome and create a more favorable environment for cancer treatment.
Multiple clinical trials (NCT03340935, NCT03595540, and NCT03700437) have evaluated the synergistic antitumor effect of FMD in combination with immunotherapy. In a clinical trial (NCT03340935), 5 out of 101 patients with advanced solid neoplasms and poor prognosis achieved complete and durable tumor response after treatment of cyclic FMD combined with standard systemic treatments.117 Integrated transcriptomic and deep-phenotyping analyses revealed that FMD markedly enhances anticancer immunity by reducing immunosuppressive myeloid cells and Tregs in peripheral blood, enhancing Th1/cytotoxic responses in the TME and upregulating immune signatures (e.g., IFN-γ), all of which are associated with improved patient outcomes.120 These findings thus prompt further clinical investigations to assess the therapeutic potential of cyclic FMD combined with standard cancer treatment in clinical settings.
A ketogenic diet aims to promote fat metabolism to override glucose utilization. In particular, a ketogenic diet induces the production of ketone bodies, which have the ability to regulate the microbiome and decrease the proportion of proinflammatory Th17 cells in the gut lamina propria.121 Preclinical studies have demonstrated the potential of ketogenic dietary intervention and the ketone body 3-hydroxybutyrate in inhibiting tumor growth and enhancing ICB efficacy.118 In humans, an ongoing clinical trial (NCT04316520) is currently evaluating the use of a ketogenic diet in patients receiving first-line treatment for metastatic RCC. Also under way is another pilot study (NCT05119010) aiming to assess the efficacy of a ketogenic diet or ketone supplements in combination with nivolumab and ipilimumab in patients with metastatic RCC. These investigations may provide critical insights into the discovery of dietary strategies that could be utilized in standard cancer treatment regimens to improve patient outcomes.
A high-fiber diet has been associated with positive effects in cancer patients receiving immunotherapy. This diet can promote the enrichment of beneficial commensal bacteria and improve antitumor immune response while reducing the risk of irAEs. In melanoma patients receiving combined neoadjuvant therapy and ICIs, a high-fiber diet was demonstrated to increase the abundance of Ruminococcaceae, leading to improved antitumor immune response and decreased risk of irAEs during immunotherapy.119,122 In another study by Spencer et al., increased dietary fiber intake was found to be significantly associated with improved PFS in patients receiving ICIs, especially for those who consumed sufficient dietary fiber and did not use probiotics.119 Currently, two clinical trials (NCT04645680 and NCT04866810) are investigating the effect of a high-fiber diet in patients with melanoma and RCC, respectively.
A low-protein diet is another dietary intervention and has been studied in a clinical trial (NCT05356182) focusing on head and neck squamous cell carcinoma. Prolonged nightly fasting is the focus of another clinical trial (NCT05083416), which aims to evaluate the effect of fasting on cancer progression, treatment response, and survival outcomes in patients with triple-negative breast cancer. Collectively, all these clinical trials promise to uncover the potential benefits of various dietary interventions as adjuncts to cancer immunotherapy, thereby providing more options to improve patient outcomes.
Current challenges and future directions
Limitations and challenges
Uncertain clinical relevance
The characteristics of the human gut microbiome can vary significantly due to inter-individual differences, including genetics, dietary habits, age, sex, accompanying diseases, and ethnicity.123 Such variability can yield inconsistent findings among different studies, therefore challenging the clinical evaluation of microbiome-targeting strategies in immunotherapy. Moreover, as the gut microbiome varies greatly, it remains difficult to identify universal microbial biomarkers that are applicable for patients from the global population.
Lack of mechanistic understanding
Although the correlation between the gut microbiome and cancer immunotherapy has been extensively studied, uncovering the causality and underlying mechanisms remains challenging. In addition to the gut microbiome, it is crucial to explore the mechanisms involved in the translocation of intratumoral microbes and their impacts on immunotherapy. Further research is needed to elucidate how particular microbes interact with the host’s antitumor immunity, ultimately influencing the efficacy of immunotherapy.
Lack of standardization
It is inevitable that distinct approaches of sample collection, storage, and processing methods were used by different studies. Such inter-study methodological variation can introduce biases and affect the reproducibility of microbiome studies. Standardizing protocols is therefore crucial for meaningful comparisons across studies and institutions. Moreover, in regard of live biotherapy such as FMT, there is also a lack of standardized dosage and frequency protocols as well as insufficient preclinical and clinical evidence.
Safety of microbial intervention
FMT has been proposed as a potential approach to address ICI resistance. However, adverse events related to FMT have been reported, with an incidence of 19% and serious adverse events accounting for approximately 1.4% of all cases.124 It is important to improve and establish stricter criteria during donor screening and testing protocols, thereby ensuring the safety of FMT in the recipient cancer patients. Meanwhile, more high-quality clinical data are needed to determine the safety and effectiveness of FMT as an adjuvant for cancer immunotherapy.
Strategies to overcome challenges and improve clinical translation
Large-scale and longitudinal studies
The variability of the gut microbiome has been a challenge to identifying universal microbial signatures of immunotherapy response. To address this issue, conducting large-scale clinical trials with diverse patient populations is crucial to determine the clinical significance of microbiome signatures and interventions. Indeed, while most previous trials involved relatively small cohorts of patients, several clinical trials with large cohort size are currently under way (Tables 1, 2, and 3). For example, the MITRE trial (NCT04107168) aims to recruit 1,800 participants across three cancer types, while another multi-center observational study (UMIN000046428) involving 400 lung cancer patients leverages artificial intelligence to identify microbial predictive biomarkers of immunotherapy response.125,126 In addition, longitudinal studies that track the changes in the gut microbiome before, during, and after immunotherapy can also offer valuable insights into its dynamic nature and interaction with the treatment.
Multidisciplinary collaboration
The role of the gut microbiome in cancer immunotherapy involves intricate correlations among multiple factors including microbes, host immunity, and tumor cells. Therefore, effective collaboration among professionals from multiple aspects, including microbiologists, immunologists, oncologists, and bioinformaticians, is essential for gaining a comprehensive understanding and translation of microbiome research into clinical practice. In particular, by bringing experts together it becomes possible for integrative analysis on microbiome data with clinical parameters and immune profiling, hence facilitating a more holistic approach for studying the role of the gut microbiome as well as identifying microbial biomarkers that could accurately predict immunotherapy response.
Mechanistic investigation
Prior to the translation of preclinical findings into clinical practice, it is necessary to fully understand the mechanistic role of the gut microbiome in cancer immunotherapy. Animal models such as gnotobiotic mice remain the most robust tool for mechanistic investigation, as they allow researchers to study the effects of specific microbial populations on immunotherapy. Techniques such as FMT and selective colonization can also introduce specific microbes into these preclinical models to examine their effects. In vitro cell culture is another common methodology that involves coculture of immune cells with microbes to investigate the direct interaction and underlying signaling between microbes and the immune system. In terms of bioinformatics, integrative analysis of multi-omics data such as metagenome, metabolome, transcriptome, and proteome data can provide a more comprehensive understanding of the gut microbiome and its functional interaction with host cells in the context of immunotherapy.
Standardization of protocols
Standardizing protocols for sample collection, storage, and processing is crucial to ensure the comparability of gut microbiome studies. Collaborative efforts among researchers and institutions are necessary to establish consensus guidelines for these protocols, thereby maintaining the reproducibility of microbiome studies on cancer immunotherapy.
Regarding the safety of FMT, it is important to establish a universally recognized and consistent protocol for handling the donor stools. To enhance the clinical accessibility of FMT, an international consensus was convened and formulated a guideline to standardize operating manuals of stool banks by donor material handling, storage, and donor screening.127 Rigorous donor screening and testing protocols should be implemented to enhance the safety of FMT, with careful assessment for infectious diseases and potential pathogens. Apart from safety, it is also essential to optimize the efficacy of FMT. This can be achieved by emphasizing donor-recipient selection through rational strategies, for instance by analyzing microbiome profiles to identify suitable matches, functional assessment, longitudinal monitoring, and enhancing donor diversity. By implementing more precise strategies, the challenges in studying the gut microbiome can be mitigated, leading to more reliable and accurate clinical findings.
Future directions to optimize microbial interventions in immunotherapy
Advances in sequencing
Currently, the most used sequencing methods for studying the gut microbiome are 16S rRNA sequencing and shotgun metagenomic sequencing. Although the latter is much more costly, it is capable of profiling microbes at deeper taxonomic resolution. Meanwhile, integrative analysis of metagenomic data with different sequencing such as transcriptomic, proteomic, and metabolomic sequencing can provide additional functional insights. Recent advances in sequencing technology also enable analysis of newer aspects, including single-cell imaging and spatial transcriptomics, which allow characterization of spatial distribution and interaction among gut microbes.128 Using more diverse sequencing methods, future research can reach a more detailed and deeper understanding of the gut microbiome and its role in the TME and immunotherapy.
Engineered and surface-modified bacteria
Advances in synthetic biology and microbial engineering offer the potential to design and develop engineered microbes with specific functions to enhance immunotherapy response. For example, genetically engineered Escherichia coli Nissle 1917 has been used to produce and release nanobodies that target immune checkpoint molecules in the TME, thereby enhancing systemic antitumor immunity.129 Another study reported that modifying E. coli SYNB1891 to express immunostimulatory molecules (e.g., STING agonists) can stimulate IFN expression to exhibit antitumor effects.130 Loading genes expressing antibodies into Salmonella also showed promising results, with improved drug delivery and enhanced treatment efficacy.131 Another strategy is surface modification of bacteria, which works by altering the structure of the bacterial envelope to confer a new biological property.132 Although this technique is relatively new, a study demonstrated that surface decoration of bacteria with checkpoint-blocking antibodies and tumor-specific antigens improved antitumor efficacy in preclinical models.133
Bioengineered bacterial extracellular vesicles
Current research is actively investigating bacterial extracellular vesicles (BEVs) as a viable alternative to using entire bacteria to mediate immune response at systemic humoral and cellular levels. Recent studies have successfully developed genetically modified BEVs with the insertion of the ectodomain of PD-1 antibody, which offers a significant advantage by binding to PD-L1 on tumor cells to facilitate the reduction of PD-L1 level and protecting T cells from the immunosuppressive PD-1/PD-L1 axis.134 This genetic modification is able to enhance antitumor efficacy through the intratumoral accumulation of effector T cells and comprehensive TME modulation, ultimately surpassing the efficacy of native BEVs and PD-L1 monotherapy. However, large-scale production of safe and efficient BEVs, which remains challenging, will be necessary prior to the implementation of these novel cancer immunotherapeutic agents in clinical practice.135,136
Conclusions
The importance of the gut microbiome in cancer immunotherapy, as well as irAEs, is now well acknowledged. Since various gut microbes can influence systemic immune responses, the TME, and immunotherapy efficacy, increasing evidence has demonstrated that harnessing the gut microbiome can lead to improved treatment outcomes. However, there are still challenges and limitations that need to be addressed, especially regarding the lack of standardized protocols for sample collection, storage, and analysis, which are crucial for ensuring reproducibility and comparability across different studies. Further research is also needed to establish causality and develop microbial interventions that can be applied in actual clinical settings. Multidisciplinary collaboration, mechanistic investigations, and large-scale clinical trials are therefore necessary to advance the current understanding of the gut microbiome in cancer immunotherapy.
It is important that personalized approaches considering individual microbiome profiles, immune status, and treatment regimens should be a focus in future research. Identifying robust microbial biomarkers, refining microbial interventions, and exploring microbial engineering techniques may provide further insights into the enhancement of cancer immunotherapy. In particular, comprehensively unraveling the role of the gut microbiome may yield potential to optimize treatment strategy and improve patient outcomes. To date, the gut microbiome represents a promising therapeutic target and an exciting area of research. Further investigations of gut microbes and clinical applications that aim to modulate the gut microbiome can contribute to the development of personalized and more effective strategies for cancer patients receiving immunotherapy.
Acknowledgments
This study was supported by the Research Talent Hub-Innovation and Technology Fund Hong Kong (ITS/177/21FP), RGC Research Impact Fund Hong Kong (R4032-21F), and RGC Collaborative Research Fund (C4039-19GF).
Author contributions
X.K. collected the data, drafted the manuscript, and prepared the figures and tables. H.C.-H.L. revised the manuscript. J.Y. supervised the study and revised the manuscript.
Declaration of interests
The authors declare no competing interests.
References
- 1.Robert C. A decade of immune-checkpoint inhibitors in cancer therapy. Nat. Commun. 2020;11:3801. doi: 10.1038/s41467-020-17670-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aparicio C., Acebal C., González-Vallinas M. Current approaches to develop "off-the-shelf" chimeric antigen receptor (CAR)-T cells for cancer treatment: a systematic review. Exp. Hematol. Oncol. 2023;12:73. doi: 10.1186/s40164-023-00435-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buonaguro L., Tagliamonte M. Peptide-based vaccine for cancer therapies. Front. Immunol. 2023;14 doi: 10.3389/fimmu.2023.1210044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee K.W., Yam J.W.P., Mao X. Dendritic Cell Vaccines: A Shift from Conventional Approach to New Generations. Cells. 2023;12 doi: 10.3390/cells12172147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ramirez F., Zambrano A., Hennis R., Holland N., Lakshmanaswamy R., Chacon J. Sending a Message: Use of mRNA Vaccines to Target the Tumor Immune Microenvironment. Vaccines (Basel) 2023;11 doi: 10.3390/vaccines11091465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Welty N.E., Gill S.I. Cancer Immunotherapy Beyond Checkpoint Blockade: JACC: CardioOncology State-of-the-Art Review. JACC. CardioOncol. 2022;4:563–578. doi: 10.1016/j.jaccao.2022.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shi Y., Tomczak K., Li J., Ochieng J.K., Lee Y., Haymaker C. Next-Generation Immunotherapies to Improve Anticancer Immunity. Front. Pharmacol. 2020;11 doi: 10.3389/fphar.2020.566401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Waldman A.D., Fritz J.M., Lenardo M.J. A guide to cancer immunotherapy: from T cell basic science to clinical practice. Nat. Rev. Immunol. 2020;20:651–668. doi: 10.1038/s41577-020-0306-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qin S., Ren Z., Meng Z., Chen Z., Chai X., Xiong J., Bai Y., Yang L., Zhu H., Fang W., et al. Camrelizumab in patients with previously treated advanced hepatocellular carcinoma: a multicentre, open-label, parallel-group, randomised, phase 2 trial. Lancet Oncol. 2020;21:571–580. doi: 10.1016/S1470-2045(20)30011-5. [DOI] [PubMed] [Google Scholar]
- 10.Cheng Y., Ling F., Li J., Chen Y., Xu M., Li S., Zhu L. An updated review of gastrointestinal toxicity induced by PD-1 inhibitors: from mechanisms to management. Front. Immunol. 2023;14 doi: 10.3389/fimmu.2023.1190850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dora D., Bokhari S.M.Z., Aloss K., Takacs P., Desnoix J.Z., Szklenárik G., Hurley P.D., Lohinai Z. Implication of the Gut Microbiome and Microbial-Derived Metabolites in Immune-Related Adverse Events: Emergence of Novel Biomarkers for Cancer Immunotherapy. Int. J. Mol. Sci. 2023;24 doi: 10.3390/ijms24032769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin X., Zong C., Zhang Z., Fang W., Xu P. Progresses in biomarkers for cancer immunotherapy. MedComm. 2023;4:e387. doi: 10.1002/mco2.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lione L., Salvatori E., Petrazzuolo A., Massacci A., Maggio R., Confroti A., Compagnone M., Aurisicchio L., Ciliberto G., Palombo F. Antitumor efficacy of a neoantigen cancer vaccine delivered by electroporation is influenced by microbiota composition. OncoImmunology. 2021;10 doi: 10.1080/2162402X.2021.1898832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reens A.L., Cabral D.J., Liang X., Norton J.E., Jr., Therien A.G., Hazuda D.J., Swaminathan G. Immunomodulation by the Commensal Microbiome During Immune-Targeted Interventions: Focus on Cancer Immune Checkpoint Inhibitor Therapy and Vaccination. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.643255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davar D., Dzutsev A.K., McCulloch J.A., Rodrigues R.R., Chauvin J.M., Morrison R.M., Deblasio R.N., Menna C., Ding Q., Pagliano O., et al. Fecal microbiota transplant overcomes resistance to anti-PD-1 therapy in melanoma patients. Science. 2021;371:595–602. doi: 10.1126/science.abf3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baruch E.N., Youngster I., Ben-Betzalel G., Ortenberg R., Lahat A., Katz L., Adler K., Dick-Necula D., Raskin S., Bloch N., et al. Fecal microbiota transplant promotes response in immunotherapy-refractory melanoma patients. Science. 2021;371:602–609. doi: 10.1126/science.abb5920. [DOI] [PubMed] [Google Scholar]
- 17.Huang J., Zheng X., Kang W., Hao H., Mao Y., Zhang H., Chen Y., Tan Y., He Y., Zhao W., Yin Y. Metagenomic and metabolomic analyses reveal synergistic effects of fecal microbiota transplantation and anti-PD-1 therapy on treating colorectal cancer. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.874922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vétizou M., Pitt J.M., Daillère R., Lepage P., Waldschmitt N., Flament C., Rusakiewicz S., Routy B., Roberti M.P., Duong C.P.M., et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science. 2015;350:1079–1084. doi: 10.1126/science.aad1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mager L.F., Burkhard R., Pett N., Cooke N.C.A., Brown K., Ramay H., Paik S., Stagg J., Groves R.A., Gallo M., et al. Microbiome-derived inosine modulates response to checkpoint inhibitor immunotherapy. Science. 2020;369:1481–1489. doi: 10.1126/science.abc3421. [DOI] [PubMed] [Google Scholar]
- 20.Shi Y., Zheng W., Yang K., Harris K.G., Ni K., Xue L., Lin W., Chang E.B., Weichselbaum R.R., Fu Y.X. Intratumoral accumulation of gut microbiota facilitates CD47-based immunotherapy via STING signaling. J. Exp. Med. 2020;217 doi: 10.1084/jem.20192282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee S.H., Cho S.Y., Yoon Y., Park C., Sohn J., Jeong J.J., Jeon B.N., Jang M., An C., Lee S., et al. Bifidobacterium bifidum strains synergize with immune checkpoint inhibitors to reduce tumor burden in mice. Nat. Microbiol. 2021;6:277–288. doi: 10.1038/s41564-020-00831-6. [DOI] [PubMed] [Google Scholar]
- 22.Yoon Y., Kim G., Jeon B.N., Fang S., Park H. Bifidobacterium Strain-Specific Enhances the Efficacy of Cancer Therapeutics in Tumor-Bearing Mice. Cancers. 2021;13 doi: 10.3390/cancers13050957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang S.L., Han B., Mao Y.Q., Zhang Z.Y., Li Z.M., Kong C.Y., Wu Y., Chen G.Q., Wang L.S. Lacticaseibacillus paracasei sh2020 induced antitumor immunity and synergized with anti-programmed cell death 1 to reduce tumor burden in mice. Gut Microb. 2022;14 doi: 10.1080/19490976.2022.2046246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao J., Wang Y., Wang J., Lv M., Zhou C., Jia L., Geng W. Lactobacillus kefiranofaciens ZW18 from Kefir enhances the anti-tumor effect of anti-programmed cell death 1 (PD-1) immunotherapy by modulating the gut microbiota. Food Funct. 2022;13:10023–10033. doi: 10.1039/d2fo01747d. [DOI] [PubMed] [Google Scholar]
- 25.Mirji G., Worth A., Bhat S.A., El Sayed M., Kannan T., Goldman A.R., Tang H.Y., Liu Q., Auslander N., Dang C.V., et al. The microbiome-derived metabolite TMAO drives immune activation and boosts responses to immune checkpoint blockade in pancreatic cancer. Sci. Immunol. 2022;7 doi: 10.1126/sciimmunol.abn0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kang X., Liu C., Ding Y., Ni Y., Ji F., Lau H.C.H., Jiang L., Sung J.J., Wong S.H., Yu J. Roseburia intestinalis generated butyrate boosts anti-PD-1 efficacy in colorectal cancer by activating cytotoxic CD8(+) T cells. Gut. 2023;72:2112–2122. doi: 10.1136/gutjnl-2023-330291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Montalban-Arques A., Katkeviciute E., Busenhart P., Bircher A., Wirbel J., Zeller G., Morsy Y., Borsig L., Glaus Garzon J.F., Müller A., et al. Commensal Clostridiales strains mediate effective anti-cancer immune response against solid tumors. Cell Host Microbe. 2021;29:1573–1588.e7. doi: 10.1016/j.chom.2021.08.001. [DOI] [PubMed] [Google Scholar]
- 28.Fong W., Li Q., Ji F., Liang W., Lau H.C.H., Kang X., Liu W., To K.K.W., Zuo Z., Li X., et al. Lactobacillus gallinarum-derived metabolites boost anti-PD1 efficacy in colorectal cancer by inhibiting regulatory T cells through modulating IDO1/Kyn/AHR axis. Gut. 2023;72:2272–2285. doi: 10.1136/gutjnl-2023-329543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tao J., Li S., Gan R.Y., Zhao C.N., Meng X., Li H.B. Targeting gut microbiota with dietary components on cancer: Effects and potential mechanisms of action. Crit. Rev. Food Sci. Nutr. 2020;60:1025–1037. doi: 10.1080/10408398.2018.1555789. [DOI] [PubMed] [Google Scholar]
- 30.Aron-Wisnewsky J., Vigliotti C., Witjes J., Le P., Holleboom A.G., Verheij J., Nieuwdorp M., Clément K. Gut microbiota and human NAFLD: disentangling microbial signatures from metabolic disorders. Nat. Rev. Gastroenterol. Hepatol. 2020;17:279–297. doi: 10.1038/s41575-020-0269-9. [DOI] [PubMed] [Google Scholar]
- 31.Zheng Y., Wang T., Tu X., Huang Y., Zhang H., Tan D., Jiang W., Cai S., Zhao P., Song R., et al. Gut microbiome affects the response to anti-PD-1 immunotherapy in patients with hepatocellular carcinoma. J. Immunother. Cancer. 2019;7:193. doi: 10.1186/s40425-019-0650-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chung M.W., Kim M.J., Won E.J., Lee Y.J., Yun Y.W., Cho S.B., Joo Y.E., Hwang J.E., Bae W.K., Chung I.J., et al. Gut microbiome composition can predict the response to nivolumab in advanced hepatocellular carcinoma patients. World J. Gastroenterol. 2021;27:7340–7349. doi: 10.3748/wjg.v27.i42.7340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ponziani F.R., De Luca A., Picca A., Marzetti E., Petito V., Del Chierico F., Reddel S., Paroni Sterbini F., Sanguinetti M., Putignani L., et al. Gut Dysbiosis and Fecal Calprotectin Predict Response to Immune Checkpoint Inhibitors in Patients With Hepatocellular Carcinoma. Hepatol. Commun. 2022;6:1492–1501. doi: 10.1002/hep4.1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cascone T., William W.N., Jr., Weissferdt A., Leung C.H., Lin H.Y., Pataer A., Godoy M.C.B., Carter B.W., Federico L., Reuben A., et al. Neoadjuvant nivolumab or nivolumab plus ipilimumab in operable non-small cell lung cancer: the phase 2 randomized NEOSTAR trial. Nat. Med. 2021;27:504–514. doi: 10.1038/s41591-020-01224-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Derosa L., Routy B., Thomas A.M., Iebba V., Zalcman G., Friard S., Mazieres J., Audigier-Valette C., Moro-Sibilot D., Goldwasser F., et al. Intestinal Akkermansia muciniphila predicts clinical response to PD-1 blockade in patients with advanced non-small-cell lung cancer. Nat. Med. 2022;28:315–324. doi: 10.1038/s41591-021-01655-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Newsome R.C., Gharaibeh R.Z., Pierce C.M., da Silva W.V., Paul S., Hogue S.R., Yu Q., Antonia S., Conejo-Garcia J.R., Robinson L.A., Jobin C. Interaction of bacterial genera associated with therapeutic response to immune checkpoint PD-1 blockade in a United States cohort. Genome Med. 2022;14:35. doi: 10.1186/s13073-022-01037-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grenda A., Iwan E., Chmielewska I., Krawczyk P., Giza A., Bomba A., Frąk M., Rolska A., Szczyrek M., Kieszko R., et al. Presence of Akkermansiaceae in gut microbiome and immunotherapy effectiveness in patients with advanced non-small cell lung cancer. Amb. Express. 2022;12:86. doi: 10.1186/s13568-022-01428-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Derosa L., Routy B., Fidelle M., Iebba V., Alla L., Pasolli E., Segata N., Desnoyer A., Pietrantonio F., Ferrere G., et al. Gut Bacteria Composition Drives Primary Resistance to Cancer Immunotherapy in Renal Cell Carcinoma Patients. Eur. Urol. 2020;78:195–206. doi: 10.1016/j.eururo.2020.04.044. [DOI] [PubMed] [Google Scholar]
- 39.Salgia N.J., Bergerot P.G., Maia M.C., Dizman N., Hsu J., Gillece J.D., Folkerts M., Reining L., Trent J., Highlander S.K., Pal S.K. Stool Microbiome Profiling of Patients with Metastatic Renal Cell Carcinoma Receiving anti-PD-1 Immune Checkpoint Inhibitors. Eur. Urol. 2020;78:498–502. doi: 10.1016/j.eururo.2020.07.011. [DOI] [PubMed] [Google Scholar]
- 40.Yin H., Yang L., Peng G., Yang K., Mi Y., Hu X., Hao X., Jiao Y., Wang X., Wang Y. The commensal consortium of the gut microbiome is associated with favorable responses to anti-programmed death protein 1 (PD-1) therapy in thoracic neoplasms. Cancer Biol. Med. 2021;18:1040–1052. doi: 10.20892/j.issn.2095-3941.2020.0450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chaput N., Lepage P., Coutzac C., Soularue E., Le Roux K., Monot C., Boselli L., Routier E., Cassard L., Collins M., et al. Baseline gut microbiota predicts clinical response and colitis in metastatic melanoma patients treated with ipilimumab. Ann. Oncol. 2017;28:1368–1379. doi: 10.1093/annonc/mdx108. [DOI] [PubMed] [Google Scholar]
- 42.Frankel A.E., Coughlin L.A., Kim J., Froehlich T.W., Xie Y., Frenkel E.P., Koh A.Y. Metagenomic Shotgun Sequencing and Unbiased Metabolomic Profiling Identify Specific Human Gut Microbiota and Metabolites Associated with Immune Checkpoint Therapy Efficacy in Melanoma Patients. Neoplasia. 2017;19:848–855. doi: 10.1016/j.neo.2017.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gopalakrishnan V., Spencer C.N., Nezi L., Reuben A., Andrews M.C., Karpinets T.V., Prieto P.A., Vicente D., Hoffman K., Wei S.C., et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science. 2018;359:97–103. doi: 10.1126/science.aan4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Peters B.A., Wilson M., Moran U., Pavlick A., Izsak A., Wechter T., Weber J.S., Osman I., Ahn J. Relating the gut metagenome and metatranscriptome to immunotherapy responses in melanoma patients. Genome Med. 2019;11:61. doi: 10.1186/s13073-019-0672-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu H., Zheng X., Pan T., Yang X., Chen X., Zhang B., Peng L., Xie C. Dynamic microbiome and metabolome analyses reveal the interaction between gut microbiota and anti-PD-1 based immunotherapy in hepatocellular carcinoma. Int. J. Cancer. 2022;151:1321–1334. doi: 10.1002/ijc.34118. [DOI] [PubMed] [Google Scholar]
- 46.Zhang C., Wang J., Sun Z., Cao Y., Mu Z., Ji X. Commensal microbiota contributes to predicting the response to immune checkpoint inhibitors in non-small-cell lung cancer patients. Cancer Sci. 2021;112:3005–3017. doi: 10.1111/cas.14979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McCulloch J.A., Davar D., Rodrigues R.R., Badger J.H., Fang J.R., Cole A.M., Balaji A.K., Vetizou M., Prescott S.M., Fernandes M.R., et al. Intestinal microbiota signatures of clinical response and immune-related adverse events in melanoma patients treated with anti-PD-1. Nat. Med. 2022;28:545–556. doi: 10.1038/s41591-022-01698-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Andrews M.C., Duong C.P.M., Gopalakrishnan V., Iebba V., Chen W.S., Derosa L., Khan M.A.W., Cogdill A.P., White M.G., Wong M.C., et al. Gut microbiota signatures are associated with toxicity to combined CTLA-4 and PD-1 blockade. Nat. Med. 2021;27:1432–1441. doi: 10.1038/s41591-021-01406-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Matson V., Fessler J., Bao R., Chongsuwat T., Zha Y., Alegre M.L., Luke J.J., Gajewski T.F. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science. 2018;359:104–108. doi: 10.1126/science.aao3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Peng Z., Cheng S., Kou Y., Wang Z., Jin R., Hu H., Zhang X., Gong J.F., Li J., Lu M., et al. The Gut Microbiome Is Associated with Clinical Response to Anti-PD-1/PD-L1 Immunotherapy in Gastrointestinal Cancer. Cancer Immunol. Res. 2020;8:1251–1261. doi: 10.1158/2326-6066.CIR-19-1014. [DOI] [PubMed] [Google Scholar]
- 51.Di Modica M., Gargari G., Regondi V., Bonizzi A., Arioli S., Belmonte B., De Cecco L., Fasano E., Bianchi F., Bertolotti A., et al. Gut Microbiota Condition the Therapeutic Efficacy of Trastuzumab in HER2-Positive Breast Cancer. Cancer Res. 2021;81:2195–2206. doi: 10.1158/0008-5472.CAN-20-1659. [DOI] [PubMed] [Google Scholar]
- 52.Lee P.C., Wu C.J., Hung Y.W., Lee C.J., Chi C.T., Lee I.C., Yu-Lun K., Chou S.H., Luo J.C., Hou M.C., Huang Y.H. Gut microbiota and metabolites associate with outcomes of immune checkpoint inhibitor-treated unresectable hepatocellular carcinoma. J. Immunother. Cancer. 2022;10 doi: 10.1136/jitc-2022-004779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mao J., Wang D., Long J., Yang X., Lin J., Song Y., Xie F., Xun Z., Wang Y., Wang Y., et al. Gut microbiome is associated with the clinical response to anti-PD-1 based immunotherapy in hepatobiliary cancers. J. Immunother. Cancer. 2021;9 doi: 10.1136/jitc-2021-003334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Katayama Y., Yamada T., Shimamoto T., Iwasaku M., Kaneko Y., Uchino J., Takayama K. The role of the gut microbiome on the efficacy of immune checkpoint inhibitors in Japanese responder patients with advanced non-small cell lung cancer. Transl. Lung Cancer Res. 2019;8:847–853. doi: 10.21037/tlcr.2019.10.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jin Y., Dong H., Xia L., Yang Y., Zhu Y., Shen Y., Zheng H., Yao C., Wang Y., Lu S. The Diversity of Gut Microbiome is Associated With Favorable Responses to Anti-Programmed Death 1 Immunotherapy in Chinese Patients With NSCLC. J. Thorac. Oncol. 2019;14:1378–1389. doi: 10.1016/j.jtho.2019.04.007. [DOI] [PubMed] [Google Scholar]
- 56.Hakozaki T., Richard C., Elkrief A., Hosomi Y., Benlaïfaoui M., Mimpen I., Terrisse S., Derosa L., Zitvogel L., Routy B., Okuma Y. The Gut Microbiome Associates with Immune Checkpoint Inhibition Outcomes in Patients with Advanced Non-Small Cell Lung Cancer. Cancer Immunol. Res. 2020;8:1243–1250. doi: 10.1158/2326-6066.CIR-20-0196. [DOI] [PubMed] [Google Scholar]
- 57.Zhang F., Ferrero M., Dong N., D'Auria G., Reyes-Prieto M., Herreros-Pomares A., Calabuig-Fariñas S., Duréndez E., Aparisi F., Blasco A., et al. Analysis of the Gut Microbiota: An Emerging Source of Biomarkers for Immune Checkpoint Blockade Therapy in Non-Small Cell Lung Cancer. Cancers. 2021;13 doi: 10.3390/cancers13112514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Peiffer L.B., White J.R., Jones C.B., Slottke R.E., Ernst S.E., Moran A.E., Graff J.N., Sfanos K.S. Composition of gastrointestinal microbiota in association with treatment response in individuals with metastatic castrate resistant prostate cancer progressing on enzalutamide and initiating treatment with anti-PD-1 (pembrolizumab) Neoplasia. 2022;32 doi: 10.1016/j.neo.2022.100822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huang X., Hu M., Sun T., Li J., Zhou Y., Yan Y., Xuan B., Wang J., Xiong H., Ji L., et al. Multi-kingdom gut microbiota analyses define bacterial-fungal interplay and microbial markers of pan-cancer immunotherapy across cohorts. Cell Host Microbe. 2023;31:1930–1943.e4. doi: 10.1016/j.chom.2023.10.005. [DOI] [PubMed] [Google Scholar]
- 60.Smith M., Dai A., Ghilardi G., Amelsberg K.V., Devlin S.M., Pajarillo R., Slingerland J.B., Beghi S., Herrera P.S., Giardina P., et al. Gut microbiome correlates of response and toxicity following anti-CD19 CAR T cell therapy. Nat. Med. 2022;28:713–723. doi: 10.1038/s41591-022-01702-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hu Y., Li J., Ni F., Yang Z., Gui X., Bao Z., Zhao H., Wei G., Wang Y., Zhang M., et al. CAR-T cell therapy-related cytokine release syndrome and therapeutic response is modulated by the gut microbiome in hematologic malignancies. Nat. Commun. 2022;13:5313. doi: 10.1038/s41467-022-32960-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stein-Thoeringer C.K., Saini N.Y., Zamir E., Blumenberg V., Schubert M.L., Mor U., Fante M.A., Schmidt S., Hayase E., Hayase T., et al. A non-antibiotic-disrupted gut microbiome is associated with clinical responses to CD19-CAR-T cell cancer immunotherapy. Nat. Med. 2023;29:906–916. doi: 10.1038/s41591-023-02234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fluckiger A., Daillère R., Sassi M., Sixt B.S., Liu P., Loos F., Richard C., Rabu C., Alou M.T., Goubet A.G., et al. Cross-reactivity between tumor MHC class I-restricted antigens and an enterococcal bacteriophage. Science. 2020;369:936–942. doi: 10.1126/science.aax0701. [DOI] [PubMed] [Google Scholar]
- 64.Schubert M.L., Rohrbach R., Schmitt M., Stein-Thoeringer C.K. The Potential Role of the Intestinal Micromilieu and Individual Microbes in the Immunobiology of Chimeric Antigen Receptor T-Cell Therapy. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.670286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Helmink B.A., Reddy S.M., Gao J., Zhang S., Basar R., Thakur R., Yizhak K., Sade-Feldman M., Blando J., Han G., et al. B cells and tertiary lymphoid structures promote immunotherapy response. Nature. 2020;577:549–555. doi: 10.1038/s41586-019-1922-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Petitprez F., de Reyniès A., Keung E.Z., Chen T.W.W., Sun C.M., Calderaro J., Jeng Y.M., Hsiao L.P., Lacroix L., Bougoüin A., et al. B cells are associated with survival and immunotherapy response in sarcoma. Nature. 2020;577:556–560. doi: 10.1038/s41586-019-1906-8. [DOI] [PubMed] [Google Scholar]
- 67.Villemin C., Six A., Neville B.A., Lawley T.D., Robinson M.J., Bakdash G. The heightened importance of the microbiome in cancer immunotherapy. Trends Immunol. 2023;44:44–59. doi: 10.1016/j.it.2022.11.002. [DOI] [PubMed] [Google Scholar]
- 68.Maisonneuve C., Tsang D.K.L., Foerster E.G., Robert L.M., Mukherjee T., Prescott D., Tattoli I., Lemire P., Winer D.A., Winer S., et al. Nod1 promotes colorectal carcinogenesis by regulating the immunosuppressive functions of tumor-infiltrating myeloid cells. Cell Rep. 2021;34 doi: 10.1016/j.celrep.2020.108677. [DOI] [PubMed] [Google Scholar]
- 69.Bessell C.A., Isser A., Havel J.J., Lee S., Bell D.R., Hickey J.W., Chaisawangwong W., Glick Bieler J., Srivastava R., Kuo F., et al. Commensal bacteria stimulate antitumor responses via T cell cross-reactivity. JCI Insight. 2020;5 doi: 10.1172/jci.insight.135597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ennamorati M., Vasudevan C., Clerkin K., Halvorsen S., Verma S., Ibrahim S., Prosper S., Porter C., Yeliseyev V., Kim M., et al. Intestinal microbes influence development of thymic lymphocytes in early life. Proc. Natl. Acad. Sci. USA. 2020;117:2570–2578. doi: 10.1073/pnas.1915047117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lee J., d'Aigle J., Atadja L., Quaicoe V., Honarpisheh P., Ganesh B.P., Hassan A., Graf J., Petrosino J., Putluri N., et al. Gut Microbiota-Derived Short-Chain Fatty Acids Promote Poststroke Recovery in Aged Mice. Circ. Res. 2020;127:453–465. doi: 10.1161/CIRCRESAHA.119.316448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Le Naour J., Galluzzi L., Zitvogel L., Kroemer G., Vacchelli E. Trial watch: IDO inhibitors in cancer therapy. OncoImmunology. 2020;9 doi: 10.1080/2162402X.2020.1777625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kadosh E., Snir-Alkalay I., Venkatachalam A., May S., Lasry A., Elyada E., Zinger A., Shaham M., Vaalani G., Mernberger M., et al. The gut microbiome switches mutant p53 from tumour-suppressive to oncogenic. Nature. 2020;586:133–138. doi: 10.1038/s41586-020-2541-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jiang S.S., Xie Y.L., Xiao X.Y., Kang Z.R., Lin X.L., Zhang L., Li C.S., Qian Y., Xu P.P., Leng X.X., et al. Fusobacterium nucleatum-derived succinic acid induces tumor resistance to immunotherapy in colorectal cancer. Cell Host Microbe. 2023;31:781–797.e9. doi: 10.1016/j.chom.2023.04.010. [DOI] [PubMed] [Google Scholar]
- 75.Morad G., Helmink B.A., Sharma P., Wargo J.A. Hallmarks of response, resistance, and toxicity to immune checkpoint blockade. Cell. 2022;185:576. doi: 10.1016/j.cell.2022.01.008. [DOI] [PubMed] [Google Scholar]
- 76.Griffin M.E., Espinosa J., Becker J.L., Luo J.D., Carroll T.S., Jha J.K., Fanger G.R., Hang H.C. Enterococcus peptidoglycan remodeling promotes checkpoint inhibitor cancer immunotherapy. Science. 2021;373:1040–1046. doi: 10.1126/science.abc9113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Alvarez C.A., Jones M.B., Hambor J., Cobb B.A. Characterization of Polysaccharide A Response Reveals Interferon Responsive Gene Signature and Immunomodulatory Marker Expression. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.556813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Owens J.A., Saeedi B.J., Naudin C.R., Hunter-Chang S., Barbian M.E., Eboka R.U., Askew L., Darby T.M., Robinson B.S., Jones R.M. Lactobacillus rhamnosus GG Orchestrates an Antitumor Immune Response. Cell. Mol. Gastroenterol. Hepatol. 2021;12:1311–1327. doi: 10.1016/j.jcmgh.2021.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Si W., Liang H., Bugno J., Xu Q., Ding X., Yang K., Fu Y., Weichselbaum R.R., Zhao X., Wang L. Lactobacillus rhamnosus GG induces cGAS/STING- dependent type I interferon and improves response to immune checkpoint blockade. Gut. 2022;71:521–533. doi: 10.1136/gutjnl-2020-323426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Park J.S., Gazzaniga F.S., Wu M., Luthens A.K., Gillis J., Zheng W., LaFleur M.W., Johnson S.B., Morad G., Park E.M., et al. Targeting PD-L2-RGMb overcomes microbiome-related immunotherapy resistance. Nature. 2023;617:377–385. doi: 10.1038/s41586-023-06026-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Xu J., Ma Q., Zhang Y., Fei Z., Sun Y., Fan Q., Liu B., Bai J., Yu Y., Chu J., et al. Yeast-derived nanoparticles remodel the immunosuppressive microenvironment in tumor and tumor-draining lymph nodes to suppress tumor growth. Nat. Commun. 2022;13:110. doi: 10.1038/s41467-021-27750-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Geller A., Shrestha R., Yan J. Yeast-Derived beta-Glucan in Cancer: Novel Uses of a Traditional Therapeutic. Int. J. Mol. Sci. 2019;20 doi: 10.3390/ijms20153618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Krautkramer K.A., Fan J., Bäckhed F. Gut microbial metabolites as multi-kingdom intermediates. Nat. Rev. Microbiol. 2021;19:77–94. doi: 10.1038/s41579-020-0438-4. [DOI] [PubMed] [Google Scholar]
- 84.Danne C., Sokol H. Butyrate, a new microbiota-dependent player in CD8+ T cells immunity and cancer therapy? Cell Rep. Med. 2021;2 doi: 10.1016/j.xcrm.2021.100328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ghini V., Laera L., Fantechi B., Monte F.D., Benelli M., McCartney A., Leonardo T., Luchinat C., Pozzessere D. Metabolomics to Assess Response to Immune Checkpoint Inhibitors in Patients with Non-Small-Cell Lung Cancer. Cancers. 2020;12 doi: 10.3390/cancers12123574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Botticelli A., Vernocchi P., Marini F., Quagliariello A., Cerbelli B., Reddel S., Del Chierico F., Di Pietro F., Giusti R., Tomassini A., et al. Gut metabolomics profiling of non-small cell lung cancer (NSCLC) patients under immunotherapy treatment. J. Transl. Med. 2020;18:49. doi: 10.1186/s12967-020-02231-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhu X., Li K., Liu G., Wu R., Zhang Y., Wang S., Xu M., Lu L., Li P. Microbial metabolite butyrate promotes anti-PD-1 antitumor efficacy by modulating T cell receptor signaling of cytotoxic CD8 T cell. Gut Microb. 2023;15 doi: 10.1080/19490976.2023.2249143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nomura M., Nagatomo R., Doi K., Shimizu J., Baba K., Saito T., Matsumoto S., Inoue K., Muto M. Association of Short-Chain Fatty Acids in the Gut Microbiome With Clinical Response to Treatment With Nivolumab or Pembrolizumab in Patients With Solid Cancer Tumors. JAMA Netw. Open. 2020;3 doi: 10.1001/jamanetworkopen.2020.2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zagato E., Pozzi C., Bertocchi A., Schioppa T., Saccheri F., Guglietta S., Fosso B., Melocchi L., Nizzoli G., Troisi J., et al. Endogenous murine microbiota member Faecalibaculum rodentium and its human homologue protect from intestinal tumour growth. Nat. Microbiol. 2020;5:511–524. doi: 10.1038/s41564-019-0649-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.He Y., Fu L., Li Y., Wang W., Gong M., Zhang J., Dong X., Huang J., Wang Q., Mackay C.R., et al. Gut microbial metabolites facilitate anticancer therapy efficacy by modulating cytotoxic CD8(+) T cell immunity. Cell Metabol. 2021;33:988–1000.e7. doi: 10.1016/j.cmet.2021.03.002. [DOI] [PubMed] [Google Scholar]
- 91.Xu H., Luo H., Zhang J., Li K., Lee M.H. Therapeutic potential of Clostridium butyricum anticancer effects in colorectal cancer. Gut Microb. 2023;15 doi: 10.1080/19490976.2023.2186114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chung Y.M., Khan P.P., Wang H., Tsai W.B., Qiao Y., Yu B., Larrick J.W., Hu M.C.T. Sensitizing tumors to anti-PD-1 therapy by promoting NK and CD8+ T cells via pharmacological activation of FOXO3. J. Immunother. Cancer. 2021;9 doi: 10.1136/jitc-2021-002772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Coutzac C., Jouniaux J.M., Paci A., Schmidt J., Mallardo D., Seck A., Asvatourian V., Cassard L., Saulnier P., Lacroix L., et al. Systemic short chain fatty acids limit antitumor effect of CTLA-4 blockade in hosts with cancer. Nat. Commun. 2020;11:2168. doi: 10.1038/s41467-020-16079-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bender M.J., McPherson A.C., Phelps C.M., Pandey S.P., Laughlin C.R., Shapira J.H., Medina Sanchez L., Rana M., Richie T.G., Mims T.S., et al. Dietary tryptophan metabolite released by intratumoral Lactobacillus reuteri facilitates immune checkpoint inhibitor treatment. Cell. 2023;186:1846–1862.e26. doi: 10.1016/j.cell.2023.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Allard B., Allard D., Buisseret L., Stagg J. The adenosine pathway in immuno-oncology. Nat. Rev. Clin. Oncol. 2020;17:611–629. doi: 10.1038/s41571-020-0382-2. [DOI] [PubMed] [Google Scholar]
- 96.Wang T., Gnanaprakasam J.N.R., Chen X., Kang S., Xu X., Sun H., Liu L., Rodgers H., Miller E., Cassel T.A., et al. Inosine is an alternative carbon source for CD8(+)-T-cell function under glucose restriction. Nat. Metab. 2020;2:635–647. doi: 10.1038/s42255-020-0219-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wang H., Rong X., Zhao G., Zhou Y., Xiao Y., Ma D., Jin X., Wu Y., Yan Y., Yang H., et al. The microbial metabolite trimethylamine N-oxide promotes antitumor immunity in triple-negative breast cancer. Cell Metabol. 2022;34:581–594.e8. doi: 10.1016/j.cmet.2022.02.010. [DOI] [PubMed] [Google Scholar]
- 98.Bourgin M., Kepp O., Kroemer G. Immunostimulatory effects of vitamin B5 improve anticancer immunotherapy. OncoImmunology. 2022;11 doi: 10.1080/2162402X.2022.2031500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.St Paul M., Saibil S.D., Han S., Israni-Winger K., Lien S.C., Laister R.C., Sayad A., Penny S., Amaria R.N., Haydu L.E., et al. Coenzyme A fuels T cell anti-tumor immunity. Cell Metabol. 2021;33:2415–2427.e6. doi: 10.1016/j.cmet.2021.11.010. [DOI] [PubMed] [Google Scholar]
- 100.Hatae R., Chamoto K., Kim Y.H., Sonomura K., Taneishi K., Kawaguchi S., Yoshida H., Ozasa H., Sakamori Y., Akrami M., et al. Combination of host immune metabolic biomarkers for the PD-1 blockade cancer immunotherapy. JCI Insight. 2020;5 doi: 10.1172/jci.insight.133501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kawanabe-Matsuda H., Takeda K., Nakamura M., Makino S., Karasaki T., Kakimi K., Nishimukai M., Ohno T., Omi J., Kano K., et al. Dietary Lactobacillus-Derived Exopolysaccharide Enhances Immune-Checkpoint Blockade Therapy. Cancer Discov. 2022;12:1336–1355. doi: 10.1158/2159-8290.CD-21-0929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Luu M., Riester Z., Baldrich A., Reichardt N., Yuille S., Busetti A., Klein M., Wempe A., Leister H., Raifer H., et al. Microbial short-chain fatty acids modulate CD8(+) T cell responses and improve adoptive immunotherapy for cancer. Nat. Commun. 2021;12:4077. doi: 10.1038/s41467-021-24331-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Fong W., Li Q., Yu J. Gut microbiota modulation: a novel strategy for prevention and treatment of colorectal cancer. Oncogene. 2020;39:4925–4943. doi: 10.1038/s41388-020-1341-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Liu L., Shah K. The Potential of the Gut Microbiome to Reshape the Cancer Therapy Paradigm: A Review. JAMA Oncol. 2022;8:1059–1067. doi: 10.1001/jamaoncol.2022.0494. [DOI] [PubMed] [Google Scholar]
- 105.Routy B., Lenehan J.G., Miller W.H., Jr., Jamal R., Messaoudene M., Daisley B.A., Hes C., Al K.F., Martinez-Gili L., Punčochář M., et al. Fecal microbiota transplantation plus anti-PD-1 immunotherapy in advanced melanoma: a phase I trial. Nat. Med. 2023;29:2121–2132. doi: 10.1038/s41591-023-02453-x. [DOI] [PubMed] [Google Scholar]
- 106.Li L., McAllister F. Too much water drowned the miller: Akkermansia determines immunotherapy responses. Cell Rep. Med. 2022;3 doi: 10.1016/j.xcrm.2022.100642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Takada K., Shimokawa M., Takamori S., Shimamatsu S., Hirai F., Tagawa T., Okamoto T., Hamatake M., Tsuchiya-Kawano Y., Otsubo K., et al. Clinical impact of probiotics on the efficacy of anti-PD-1 monotherapy in patients with nonsmall cell lung cancer: A multicenter retrospective survival analysis study with inverse probability of treatment weighting. Int. J. Cancer. 2021;149:473–482. doi: 10.1002/ijc.33557. [DOI] [PubMed] [Google Scholar]
- 108.Dizman N., Meza L., Bergerot P., Alcantara M., Dorff T., Lyou Y., Frankel P., Cui Y., Mira V., Llamas M., et al. Nivolumab plus ipilimumab with or without live bacterial supplementation in metastatic renal cell carcinoma: a randomized phase 1 trial. Nat. Med. 2022;28:704–712. doi: 10.1038/s41591-022-01694-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Tomita Y., Ikeda T., Sakata S., Saruwatari K., Sato R., Iyama S., Jodai T., Akaike K., Ishizuka S., Saeki S., Sakagami T. Association of Probiotic Clostridium butyricum Therapy with Survival and Response to Immune Checkpoint Blockade in Patients with Lung Cancer. Cancer Immunol. Res. 2020;8:1236–1242. doi: 10.1158/2326-6066.CIR-20-0051. [DOI] [PubMed] [Google Scholar]
- 110.Hagihara M., Kuroki Y., Ariyoshi T., Higashi S., Fukuda K., Yamashita R., Matsumoto A., Mori T., Mimura K., Yamaguchi N., et al. Clostridium butyricum Modulates the Microbiome to Protect Intestinal Barrier Function in Mice with Antibiotic-Induced Dysbiosis. iScience. 2020;23 doi: 10.1016/j.isci.2019.100772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Marco M.L., Sanders M.E., Gänzle M., Arrieta M.C., Cotter P.D., De Vuyst L., Hill C., Holzapfel W., Lebeer S., Merenstein D., et al. The International Scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on fermented foods. Nat. Rev. Gastroenterol. Hepatol. 2021;18:196–208. doi: 10.1038/s41575-020-00390-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Li Y., Elmén L., Segota I., Xian Y., Tinoco R., Feng Y., Fujita Y., Segura Muñoz R.R., Schmaltz R., Bradley L.M., et al. Prebiotic-Induced Anti-tumor Immunity Attenuates Tumor Growth. Cell Rep. 2020;30:1753–1766.e6. doi: 10.1016/j.celrep.2020.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Han K., Nam J., Xu J., Sun X., Huang X., Animasahun O., Achreja A., Jeon J.H., Pursley B., Kamada N., et al. Generation of systemic antitumour immunity via the in situ modulation of the gut microbiome by an orally administered inulin gel. Nat. Biomed. Eng. 2021;5:1377–1388. doi: 10.1038/s41551-021-00749-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Huang J., Liu D., Wang Y., Liu L., Li J., Yuan J., Jiang Z., Jiang Z., Hsiao W.W., Liu H., et al. Ginseng polysaccharides alter the gut microbiota and kynurenine/tryptophan ratio, potentiating the antitumour effect of antiprogrammed cell death 1/programmed cell death ligand 1 (anti-PD-1/PD-L1) immunotherapy. Gut. 2022;71:734–745. doi: 10.1136/gutjnl-2020-321031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Guo C., Guo D., Fang L., Sang T., Wu J., Guo C., Wang Y., Wang Y., Chen C., Chen J., et al. Ganoderma lucidum polysaccharide modulates gut microbiota and immune cell function to inhibit inflammation and tumorigenesis in colon. Carbohydr. Polym. 2021;267 doi: 10.1016/j.carbpol.2021.118231. [DOI] [PubMed] [Google Scholar]
- 116.Taylor S.R., Falcone J.N., Cantley L.C., Goncalves M.D. Developing dietary interventions as therapy for cancer. Nat. Rev. Cancer. 2022;22:452–466. doi: 10.1038/s41568-022-00485-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ligorio F., Fucà G., Provenzano L., Lobefaro R., Zanenga L., Vingiani A., Belfiore A., Lorenzoni A., Alessi A., Pruneri G., et al. Exceptional tumour responses to fasting-mimicking diet combined with standard anticancer therapies: A sub-analysis of the NCT03340935 trial. Eur. J. Cancer. 2022;172:300–310. doi: 10.1016/j.ejca.2022.05.046. [DOI] [PubMed] [Google Scholar]
- 118.Ferrere G., Tidjani Alou M., Liu P., Goubet A.G., Fidelle M., Kepp O., Durand S., Iebba V., Fluckiger A., Daillère R., et al. Ketogenic diet and ketone bodies enhance the anticancer effects of PD-1 blockade. JCI Insight. 2021;6 doi: 10.1172/jci.insight.145207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Spencer C.N., McQuade J.L., Gopalakrishnan V., McCulloch J.A., Vetizou M., Cogdill A.P., Khan M.A.W., Zhang X., White M.G., Peterson C.B., et al. Dietary fiber and probiotics influence the gut microbiome and melanoma immunotherapy response. Science. 2021;374:1632–1640. doi: 10.1126/science.aaz7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Vernieri C., Fucà G., Ligorio F., Huber V., Vingiani A., Iannelli F., Raimondi A., Rinchai D., Frigè G., Belfiore A., et al. Fasting-Mimicking Diet Is Safe and Reshapes Metabolism and Antitumor Immunity in Patients with Cancer. Cancer Discov. 2022;12:90–107. doi: 10.1158/2159-8290.CD-21-0030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ang Q.Y., Alexander M., Newman J.C., Tian Y., Cai J., Upadhyay V., Turnbaugh J.A., Verdin E., Hall K.D., Leibel R.L., et al. Ketogenic Diets Alter the Gut Microbiome Resulting in Decreased Intestinal Th17 Cells. Cell. 2020;181:1263–1275.e16. doi: 10.1016/j.cell.2020.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Simpson R.C., Shanahan E.R., Batten M., Reijers I.L.M., Read M., Silva I.P., Versluis J.M., Ribeiro R., Angelatos A.S., Tan J., et al. Diet-driven microbial ecology underpins associations between cancer immunotherapy outcomes and the gut microbiome. Nat. Med. 2022;28:2344–2352. doi: 10.1038/s41591-022-01965-2. [DOI] [PubMed] [Google Scholar]
- 123.Kurilshikov A., Medina-Gomez C., Bacigalupe R., Radjabzadeh D., Wang J., Demirkan A., Le Roy C.I., Raygoza Garay J.A., Finnicum C.T., Liu X., et al. Large-scale association analyses identify host factors influencing human gut microbiome composition. Nat. Genet. 2021;53:156–165. doi: 10.1038/s41588-020-00763-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Marcella C., Cui B., Kelly C.R., Ianiro G., Cammarota G., Zhang F. Systematic review: the global incidence of faecal microbiota transplantation-related adverse events from 2000 to 2020. Aliment. Pharmacol. Ther. 2021;53:33–42. doi: 10.1111/apt.16148. [DOI] [PubMed] [Google Scholar]
- 125.Thompson N.A., Stewart G.D., Welsh S.J., Doherty G.J., Robinson M.J., Neville B.A., Vervier K., Harris S.R., Adams D.J., Dalchau K., et al. The MITRE trial protocol: a study to evaluate the microbiome as a biomarker of efficacy and toxicity in cancer patients receiving immune checkpoint inhibitor therapy. BMC Cancer. 2022;22:99. doi: 10.1186/s12885-021-09156-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Shoji F., Yamashita T., Kinoshita F., Takamori S., Fujishita T., Toyozawa R., Ito K., Yamazaki K., Nakashima N., Okamoto T. Artificial intelligence-derived gut microbiome as a predictive biomarker for therapeutic response to immunotherapy in lung cancer: protocol for a multicentre, prospective, observational study. BMJ Open. 2022;12 doi: 10.1136/bmjopen-2022-061674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Keller J.J., Ooijevaar R.E., Hvas C.L., Terveer E.M., Lieberknecht S.C., Högenauer C., Arkkila P., Sokol H., Gridnyev O., Mégraud F., et al. A standardised model for stool banking for faecal microbiota transplantation: a consensus report from a multidisciplinary UEG working group. United European Gastroenterol. J. 2021;9:229–247. doi: 10.1177/2050640620967898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.McCallum G., Tropini C. The gut microbiota and its biogeography. Nat. Rev. Microbiol. 2024;22:105–118. doi: 10.1038/s41579-023-00969-0. [DOI] [PubMed] [Google Scholar]
- 129.Gurbatri C.R., Lia I., Vincent R., Coker C., Castro S., Treuting P.M., Hinchliffe T.E., Arpaia N., Danino T. Engineered probiotics for local tumor delivery of checkpoint blockade nanobodies. Sci. Transl. Med. 2020;12 doi: 10.1126/scitranslmed.aax0876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Leventhal D.S., Sokolovska A., Li N., Plescia C., Kolodziej S.A., Gallant C.W., Christmas R., Gao J.R., James M.J., Abin-Fuentes A., et al. Immunotherapy with engineered bacteria by targeting the STING pathway for anti-tumor immunity. Nat. Commun. 2020;11:2739. doi: 10.1038/s41467-020-16602-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Guo Y., Chen Y., Liu X., Min J.J., Tan W., Zheng J.H. Targeted cancer immunotherapy with genetically engineered oncolytic Salmonella typhimurium. Cancer Lett. 2020;469:102–110. doi: 10.1016/j.canlet.2019.10.033. [DOI] [PubMed] [Google Scholar]
- 132.Wu L., Bao F., Li L., Yin X., Hua Z. Bacterially mediated drug delivery and therapeutics: Strategies and advancements. Adv. Drug Deliv. Rev. 2022;187 doi: 10.1016/j.addr.2022.114363. [DOI] [PubMed] [Google Scholar]
- 133.Li J., Xia Q., Guo H., Fu Z., Liu Y., Lin S., Liu J. Decorating Bacteria with Triple Immune Nanoactivators Generates Tumor-Resident Living Immunotherapeutics. Angew. Chem., Int. Ed. Engl. 2022;61 doi: 10.1002/anie.202202409. [DOI] [PubMed] [Google Scholar]
- 134.Li Y., Zhao R., Cheng K., Zhang K., Wang Y., Zhang Y., Li Y., Liu G., Xu J., Xu J., et al. Bacterial Outer Membrane Vesicles Presenting Programmed Death 1 for Improved Cancer Immunotherapy via Immune Activation and Checkpoint Inhibition. ACS Nano. 2020;14:16698–16711. doi: 10.1021/acsnano.0c03776. [DOI] [PubMed] [Google Scholar]
- 135.Won S., Lee C., Bae S., Lee J., Choi D., Kim M.G., Song S., Lee J., Kim E., Shin H., et al. Mass-produced gram-negative bacterial outer membrane vesicles activate cancer antigen-specific stem-like CD8(+) T cells which enables an effective combination immunotherapy with anti-PD-1. J. Extracell. Vesicles. 2023;12 doi: 10.1002/jev2.12357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Qing S., Lyu C., Zhu L., Pan C., Wang S., Li F., Wang J., Yue H., Gao X., Jia R., et al. Biomineralized Bacterial Outer Membrane Vesicles Potentiate Safe and Efficient Tumor Microenvironment Reprogramming for Anticancer Therapy. Adv. Mater. 2020;32 doi: 10.1002/adma.202002085. [DOI] [PubMed] [Google Scholar]