Abstract
A Cucurbita pepo mutant with multiple defects in growth and development has been identified and characterized. The mutant dwfcp displayed a dwarf phenotype with dark green and shrinking leaves, shortened internodes and petioles, shorter but thicker roots and greater root biomass, and reduced fertility. The causal mutation of the phenotype was found to disrupt gene Cp4.1LG17g04540, the squash orthologue of the Arabidopsis brassinosteroid (BR) biosynthesis gene DWF5, encoding for 7-dehydrocholesterol reductase. A single nucleotide transition (G > A) causes a splicing defect in intron 6 that leads to a premature stop codon and a truncated CpDWF5 protein. The mutation co-segregated with the dwarf phenotype in a large BC1S1 segregating population. The reduced expression of CpDWF5 and brassinolide (BL) content in most mutant organs, and partial rescue of the mutant phenotype by exogenous application of BL, showed that the primary cause of the dwarfism in dwfcp is a BR deficiency. The results showed that in C. pepo, CpDWF5 is not only a positive growth regulator of different plant organs but also a negative regulator of salt tolerance. During germination and the early stages of seedling development, the dwarf mutant was less affected by salt stress than the wild type, concomitantly with a greater upregulation of genes associated with salt tolerance, including those involved in abscisic acid (ABA) biosynthesis, ABA and Ca2+ signaling, and those coding for cation exchangers and transporters.
Introduction
Plant architecture is one of the most relevant agronomic traits that affect plant photosynthesis and nutrient distribution, which finally has a great impact on crop yield. The introduction of dwarfing genes into cereal crops was crucial to enhance lodging resistance and improve the harvest index in the so-called Green Revolution. Dwarf varieties, such as the Micro-Tom tomato, are also model cultivars for functional genomics research [1]. Therefore, understanding the genetic mechanisms of plant height is important for plant breeding.
Numerous dwarf mutants have been identified in many species. Most of the identified mutations affect the biosynthesis or signal transmission process of phytohormones, such as gibberellins (GAs), auxins [indole-3-acetic acid (IAA)], ethylene (ETH), strigolactone, and brassinosteroids (BRs). Among the dwarf mutants related to GAs, it is worth mentioning the mutants anther ear1 (an1), dwarf1 (d1), d3, and d5 in maize [2–5]; gid1 in rice [6]; CsCLAVATA1 in cucumber [7]; or rht1 in wheat [8]. Other dwarf mutants, including VT2, Brachytic2, and ZmPIN1a in maize [9–11] and mutants yuc1D, pin, and pgp in Arabidopsis [12–14], were reported to be defective in IAA biosynthesis or polar transport. The dwarf mutant ZmACS7 from maize is defective in ACC synthase 7, a key enzyme in ETH biosynthesis [15]. Strigolactone-related dwarf mutants, such as d27, d14, d53, or d27, have also been identified in rice [16–18].
The role of BRs in plant growth and development is also essential. Currently, it is known that BRs have a key role not only in cell elongation but also in cell division, photomorphogenesis, xylem differentiation, plant reproduction, and responses to abiotic and biotic stress [19]. Numerous mutants related to BR biosynthesis or perception have been isolated in Arabidopsis, tomato, rice, cucumber, barley, and pea. The mutants cpa, scp-1, and scp-2 in cucumber [20–22]; dwf1, cpd/dwf3, dwf4, dwf5, det2/dwf6, stel/dwf7, dwf12, and bri1 in Arabidopsis [23–30]; Ika and Ikb in pea [31]; d, dX, dpy, cu3, and abs in tomato [32–34]; na1, na2, and brd1 in maize [35–37]; or dwarf2, dwarf11, d61, and brd1 in rice [38–41] show a dwarf phenotype associated with a defect in BR biosynthesis or signaling pathways.
Mutants with lesions in intermediates or key enzymes involved in the biosynthesis of sterols, the precursors of BRs, also show a similar dwarf phenotype, including the Arabidopsis mutants dwf1, dwf5, and dwf7 [28, 29, 42]. Mutation dwf1 was identified as a defective flavin adenine dinucleotide (FAD)-dependent oxidoreductase that catalyzes the reduction of C-24 [42], while Arabidopsis dwf7 and dwf5 were reported to be deficient in the Δ7-sterol-C-5 desaturase [28] and sterol Δ7-reductase (S7R) [29], respectively. The phenotype of the Arabidopsis S7R-disrupted mutant dwf5 includes abnormal growth and development with small and dark green leaves, short stems, pedicels, and petioles [29].
No genes related to the biosynthesis of sterol and BRs have been reported in Cucurbita pepo (C. pepo). In this study, the identification and characterization of the squash mutant dwfcp are reported. The mutant displays a dwarf phenotype, affected in plant height, in the length of all vegetative and reproductive organs, and in fertility. However, the mutant shows thicker roots and higher root biomass, as well as enhanced tolerance to salt stress during earlier stages of plant development. The causal mutation of the phenotype was found to disrupt the gene Cp4.1LG17g04540, the squash orthologue of the Arabidopsis BR biosynthesis gene DWF5, encoding for 7-dehydrocholesterol reductase (S7R).
Results
Phenotypic characterization of the dwfcp mutant
A dwarf mutant was isolated from a C. pepo ethyl methanesulfonate (EMS) mutant collection. The so-called dwfcp mutant was backcrossed twice with MUCU16, the background genotype, and then selfed. The wild-type (WT) and mutant phenotypes were found in the resulting BC1S1 and BC2S1 segregating populations in a 3:1 ratio (Supplemental Table S1), indicating that the mutation is recessive. BC2S1 plants were used for phenotyping because the backcrosses are able to remove EMS mutations other than the selected dwfcp.
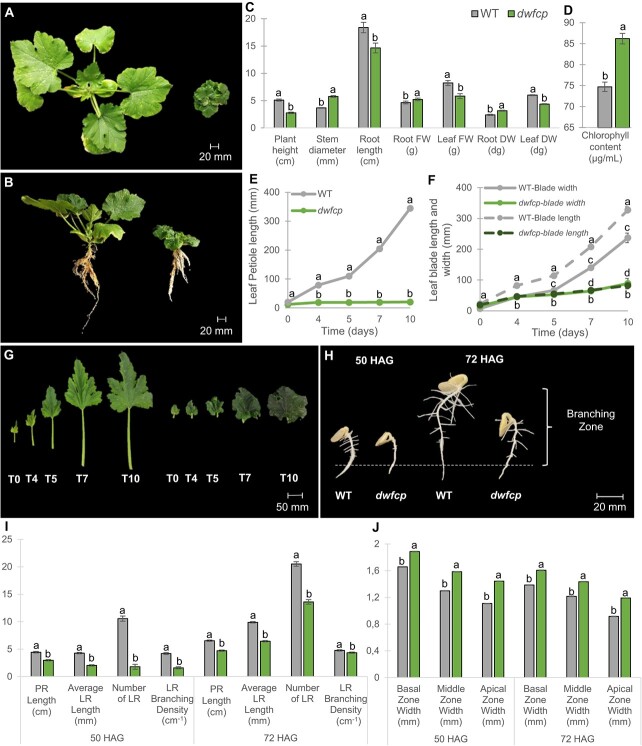
The mutant had a clear dwarf phenotype during vegetative development (Fig. 1A and B). The mutant plants showed shorter but wider stems, smaller, wrinkled, dark green leaves, and shorter petioles (Fig. 1A–F). The roots were also shorter but showed higher fresh weight (FW) and dry weight (DW) than those of the WT (Fig. 1C). However, the FW and DW of the aerial part of the plant were reduced in dwfcp compared to WT (Fig. 1C). The leaf chlorophyll content was significantly higher in the mutant than in the WT (Fig. 1D), which would explain the darker green color of the mutant plants.
Figure 1.
Phenotypic characterization of the dwfcp mutant. Phenotype comparison between WT (left) and dwfcp (right) seedlings at the stage of 7 true leavess. (A) Top view and (B) side view. (C) Comparison of plant height, stem diameter, root length, root biomass, and leaf biomass between WT and dwfcp at 7 true leaves seedling stage. (D) Chlorophyll content in WT and dwfcp leaves. (E) Growth rate of WT and dwfcp leaf petiole over time. (F) Growth rate of WT and dwfcp leaf blade over time. (G) Leaf development in WT (left) and mutant (right) plants. (H) Root growth and development during the establishment of WT and dwfcp seedlings (at 50 and 72 HAG). (I) Statistical comparison of parent root (PR) and LR length, number of LR, and LR branching density between WT and dwfcp at 50 and 72 HAG. (J) Width of the PR in three different zones (basal, middle, and apical) during the establishment of WT and mutant seedlings at 50 and 72 HAG. The LR branching density (cm−1) was calculated as the number of emerged LR per unit length of the root branching zone (in centimeters). The root branching zone includes the zone of the parent root that extends from the most rootward emerged LR to the shoot base. Error bars represent SE. Different letters indicate significant differences (P ≤ 0.05).
The growth rate of the WT and dwfcp leaves is shown in Fig. 1E–G. The growth of the leaf petioles, but also the longitudinal and lateral growth of the leaf blade, was significantly lower in the mutant than in the WT. The petioles of the WT leaves grew an average of 35 cm in 10 days, while those of the dwfcp barely grew 1 cm during the same period of time (Fig. 1E). In the WT leaf blade, the longitudinal growth rate was greater than the transverse growth rate, resulting in leaves whose length was greater than their width. However, the longitudinal growth rate and transverse growth rate of the dwfcp leaf blade were similar, resulting in the development of round leaves (Fig. 1F and G). Scanning electron microscopy (SEM) analysis of WT and mutant stems indicated that cell size was significantly shorter but wider in dwfcp than in the WT (Supplemental Fig. S1), indicating that the dwarf phenotype is caused by a reduction in cell elongation.
The development of parental and lateral roots was compared in WT and dwfcp seedlings (Fig. 1H–J). Both the parent root and the lateral roots (LR) were shorter in dwfcp than in WT, and the branching density of LR was also lower in dwfcp seedlings at both 50 and 72 h after germination (Fig. 1H and I). The width of the basal, middle and apical zones of the parent root was found to be greater in the mutant than in the WT, which may explain the higher biomass of mutant roots (Fig. 1H–J). These results indicate that BRs play a positive role in the formation and development of the lateral root in C. pepo.
The mutation dwfcp affected floral development (Fig. 2). The growth rate of the dwfcp corolla was lower than that of WT (Fig. 2A–C). The anthesis time, the period of time taken for a <1 cm floral bud to develop and open, was earlier in female flowers (15 days) than in male flowers (21 days), both in the WT and dwfcp plants, and no significant differences were found between phenotypes (Fig. 2C and D). Significant differences were also detected in the growth rates of the WT and dwfcp ovaries and fruits of the female flowers, and in the pedicels of the male flowers (Fig. 2D). WT and dwfcp ovaries/fruits showed a similar growth rate during the first 11 days. After that time, the ovaries of the WT continued to grow, while mutant ovaries slowed or even stopped their growth. At anthesis, the ovary of the WT reached ~8 cm in length, while the dwfcp ovary reached ~5 cm in length.
Figure 2.
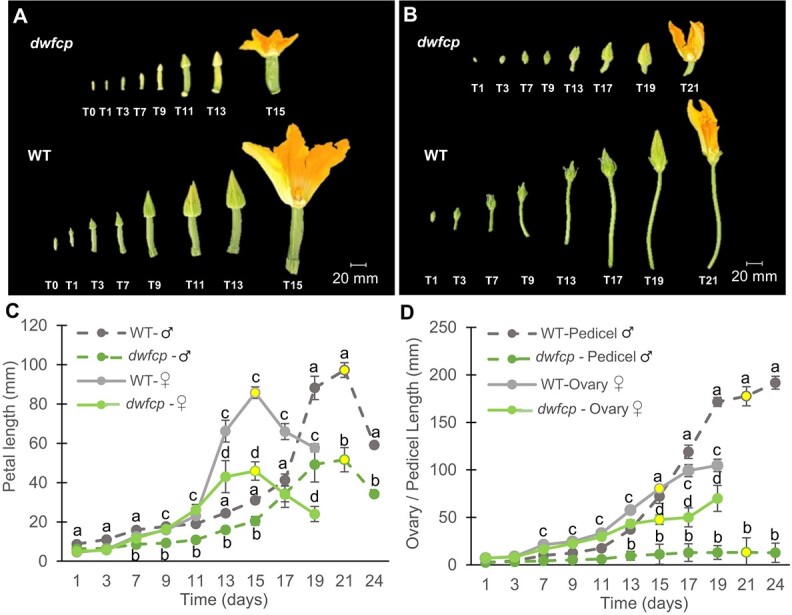
Effect of the dwfcp mutation on floral development. (A) Comparison of the growth of WT and dwfcp female flowers and (B) male flowers. (C) Growth rate of WT and dwfcp corolla over time. (D) Comparison between genotypes of the ovary and pedicel growth rate of female and male flowers, respectively, during a period of 24 days. The growth of flower ovaries was recorded every 2 days for 24 days starting when they were <1 cm length. The time-points at which >80% of the flowers reached anthesis are indicated in yellow. Error bars represent SE. Different letters indicate significant differences (P ≤ 0.05).
The female flowers of dwfcp were pollinated with WT MUCU16 pollen, but most fruits aborted or were unable to set seeds, indicating a lack of female fertility in the mutant. However, male dwfcp flowers produced pollen and were able to plant fruit and seeds when used as pollinators of the background genotype MUCU16 and then selfed to obtain BC1S1 generations, although not all crosses performed were successful, also indicating a reduction in male fertility in the dwfcp male flowers compared to the WT. No differences were found in the size or weight of WT and dwfcp seeds from a BC2S1 segregating population (Supplemental Fig. S2).
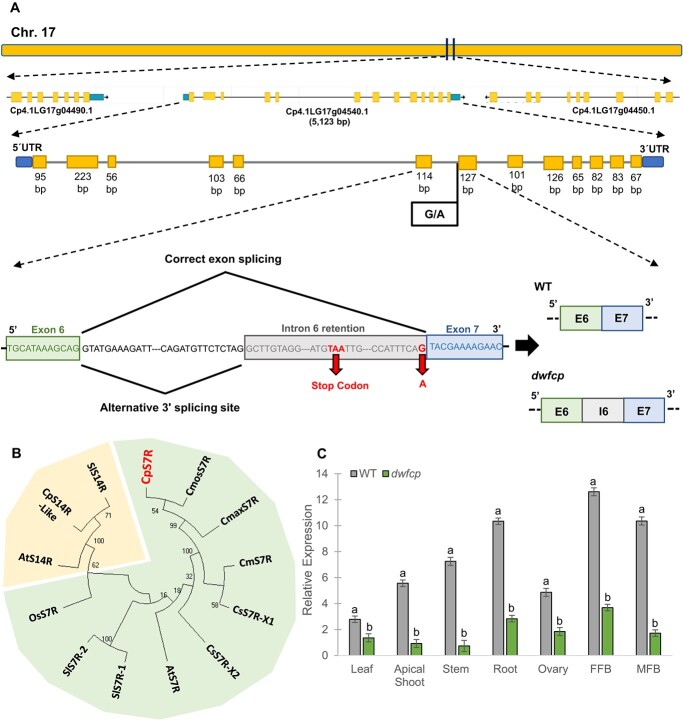
dwfcp is a mutation in the 3′ splicing site of CpDWF5 intron 6
A bulked segregant analysis sequencing (BSA-seq) approach was used to find the causal mutation of dwfcp. Thirty WT plants and thirty dwarf plants from a BC1S1 segregating population were used to construct two DNA bulks. Around 80 million reads in each bulk (>98% of the sequencing reads, representing an average depth of 44.49) were mapped against the C. pepo reference genome (version 4.1), which allowed the identification of more than 400 000 single nucleotide polymorphisms (SNPs) in each bulk (Table 1). The following parameters were established for SNP filtering: (i) transitions G > A and C > T, (ii) alternative allele frequency (AF) = 1 in mutant bulk, and (iii) SNPs with non-synonymous mutations. As the phenotype of WT and heterozygous plants is indistinguishable, we were more flexible with the filter on the AF of the WT bulk. Thus, after filtering for AF ≤ 0.6 in the WT bulk and AF = 1 in the mutant bulk, 1126 SNPs were identified, of which 317 were canonical EMS mutations (C > T and G > A). An SNP found on chromosome 17 was the only one with a significant impact on the protein (Table 1 and Fig. 3). SNP alleles were genotyped in 300 plants from a BC1S1 population for the putative mutation to prove that it was responsible for the dwfcp phenotype, and the results demonstrated 100% co-segregation between the mutant phenotype and the G > A mutation located on chromosome 17.
TABLE 1.
Summary sequencing data for WT and dwfcp
| Sequencing results | WT | dwfcp | ||||
| No. of reads | 95 993 772 | 84 920 814 | ||||
| Mapped reads (%) | 98.09 | 98.28 | ||||
| Average depth | 44.49 | 39.45 | ||||
| Coverage at least 4× (%) | 95.37 | 94.86 | ||||
| SNP filtering | ||||||
| Total no. of SNPs | 425 994 | 417 882 | ||||
| AF (WT) ≤ 0.6; AF (dwfcp) = 1 | 1126 | 1126 | ||||
| EMS SNPs G > A or C > T | 317 | 317 | ||||
| EMS SNPs (GQ > 75; DP > 20) | 2 | 2 | ||||
| High-impact SNPs | 0 | 1 | ||||
| Candidate SNPs | ||||||
| Chr | Position | Ref. | Alt | Gene ID | Effect | Functional annotation |
| 17 | 3.048.864 | G | A | Cp4.1LG17g04540 | Splice site mutation | 7-Dehydrocholesterol reductase |
DP, read depth; GQ, genotype quality.
Figure 3.
Identification and characterization of the DWFCP locus. (A) Identification of dwfcp causal mutation by BSA-sequencing. The DWFCP locus corresponds to gene Cp4.1LG17g04540.1 (CpDWF5), located in the chromosome 17. The mutation produces a 3′-splice site mutation of intron 6, a premature stop codon and a truncated CpDWF5 protein. (B) Phylogenetic tree of sterol Δ7 reductases (S7R) and sterol Δ14 reductases (S14R) from different cucurbit species, C. maxima, C. melo, Cucumis sativus, C. moschata, and C. pepo, together with those of the most studied model species, A. thaliana, O. sativa, and S. lycopersicum. Bootstrap values for the main branches are depicted on the tree. (C) Relative gene expression of CpDWF5 in different tissues of WT and dwfcp plants measured by qRT-PCR. FFB, female floral bud excluding the ovary; MFB, male floral bud. Error bars represent SE. Different letters indicate significant differences (P ≤ 0.05).
The identified mutation affected a single nucleotide at the 3′ splicing site of the sixth intron of the gene Cp4.1LG17g04540, encoding for a 7-dehydrocholesterol reductase, an enzyme in the cholesterol and BR biosynthesis pathway. This gene was homologous to the Arabidopsis DWF5 gene, so it was named CpDWF5. To see whether the EMS mutation disrupts intron 6 splicing, the WT and mutant gene transcripts were amplified by reverse transcription polymerase chain reaction (RT-PCR) and then sequenced. The mutant transcript had an additional fragment of 76 nucleotides that corresponded to part of the sequence of the sixth intron of the gene, which generates a premature stop codon and a truncated protein (Fig. 3A). Although the genome of C. pepo is duplicated [43], the gene CpDWF5 on chromosome 17 does not have any other paralog in the genome. The gene encoded a putative protein of 435 amino acids. The protein sequence was used in BLAST searches against the NCBI protein database, which demonstrated a high identity with homologous proteins in cucurbits, including Cucurbita moschata (99.08%), Cucurbita argyrosperma (98.62%), Cucurbita maxima (98.39%), Momordica charantia (91.71%), or Cucumis melo (91.24%), but also with other crops, including legumes (85.51% homology with Glycine max), Solanaceae (82.56% homology with Solanum lycopersicum), or Cruciferae (85.28% homology with Brassica oleracea). By using the 7-dehydrocholesterol reductase (S7R) protein sequences from different cucurbit species, together with Arabidopsis thaliana, Oryza sativa, and S. lycopersicum, a multiple sequence alignment was conducted using the software Omega (Supplemental Fig. S3). A phylogenetic tree was then inferred using not only these S7R protein sequences but also some S14R (Fig. 3B). As expected, squash CpDWF5 clustered together with other S7R enzymes in other plants but separated from S14R.
The expression of CpDWF5 in different WT and dwfcp tissues is shown in Fig. 3C. CpDWF5 was found to be expressed in all tissues, although the transcript accumulated the most in the roots and flower buds. These results suggest that CpDWF5 plays a crucial role in the development and growth of most tissues in zucchini squash. The transcription level of CpDWF5 was significantly lower in the mutant than in the WT, suggesting that the mutation was able to downregulate the accumulation of the transcript in the different organs analyzed.
Exogenous BR treatment can partially rescue the dwfcp phenotype
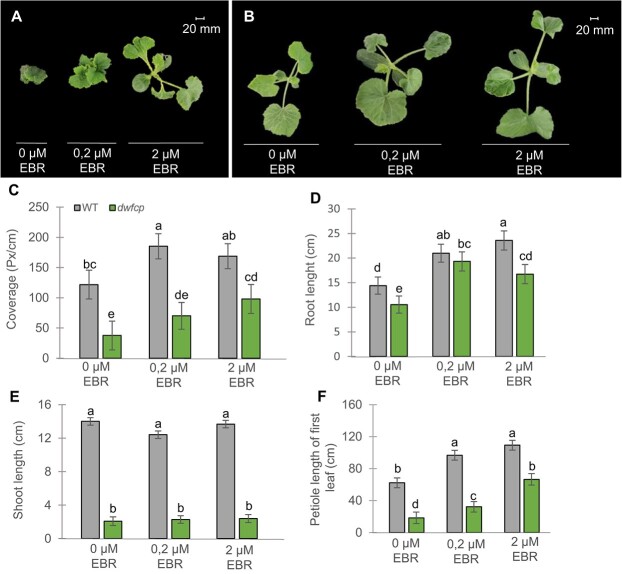
Since the dwfcp mutation affects the BR biosynthesis pathway, we wondered how an exogenous treatment with BRs would affect the mutant phenotype. WT and mutant seedlings were sprayed with two concentrations of epibrassinolide (EBR). Both WT and mutant seedlings responded to treatment, increasing leaf petiole, leaf blade growth (represented by leaf coverage parameter), and root length (Fig. 4C, D, and F). However, the length of shoots of WT or mutant plants was not affected by the treatments (Fig. 4E). The dosage effect on leaf growth was observed in the mutant but not in the WT plants (Fig. 4C), suggesting a deficiency of BRs in the mutant and hormone saturation in the WT under the most concentrated EBR treatment. Taken together, these results indicated that BRs were able to partially rescue the dwfcp phenotype, increasing the size of the leaf petiole and the leaf blade and the length of the root.
Figure 4.
Partial rescue of dwfcp phenotype by exogenous application of EBR. (A–B) Top view of dwfcp (A) and WT (B) seedlings after 1 week of treatment with 0, 0.2, and 2 μM of EBR. (C–F) Effect of exogenous treatment with EBR on leaf coverage, and the length of shoots, roots, and leaf petioles of WT and dwfcp plants. Error bars represent SE from the treatments of nine plants per genotype and treatment. Different letters indicate significant differences between samples from different treatments and genotypes (P ≤ 0.05).
BR deficiency in dwfcp was also confirmed by measuring the content of brassinolide (BL) in different organs of WT and dwfcp plants. As expected, most of the organs analyzed, including the apical shoot, the leaf, and reproductive organs such as female and male flowers and the ovary, showed lower BL content in the mutant than in WT. However, the stems and roots of the WT and dwfcp plants did not show significant differences in BL content (Supplemental Fig. S4).
Response of dwfcp to salt stress during germination and radicle growth
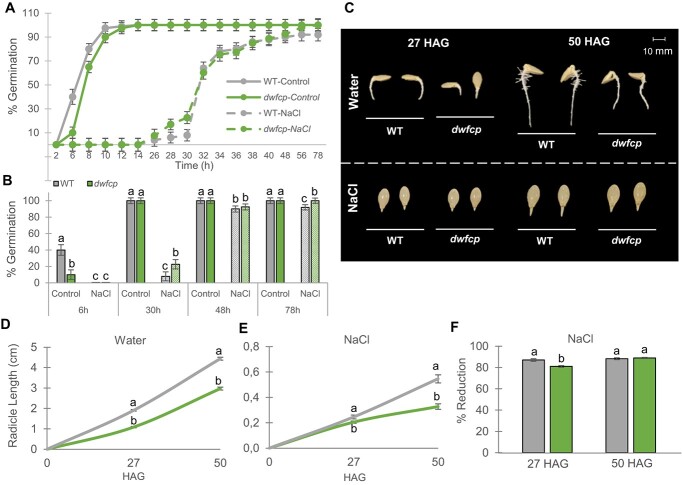
To study the role of BRs in the germination of C. pepo, WT and dwfcp seeds were germinated under control (water) and salt conditions for 78 h, recording the germination as the time of radicle protrusion every 2 h (Fig. 5). Based on a dose–response study between NaCl concentration and germination stress on the seed germination of the genetic background of the mutant collection MUCU16 (Supplemental Fig. S5), a final concentration of 200 mM NaCl was selected for this assay since it led to a delay in germination of 1 day difference compared to control conditions, which would allow a clear distinction between sensitive and tolerant to salt stress phenotypes during seed germination.
Figure 5.
Effect of NaCl on the germination and radicle growth of WT and dwfcp seeds. (A) Germination rate of WT and dwfcp mutant seeds under control and saline conditions. (B) Effect of 200 mM NaCl on the germination percentage at 6, 30, 48, and 78 h. (C–E) Radicle length of WT and dwfcp seeds at different HAG under control and saline conditions. (F) Effect of salt on the percentage of reduction of radicle length. Error bars represent SE. Different letters indicate significant differences between samples (P ≤ 0.05).
In water, dwfcp germination was slightly delayed with respect to WT (Fig. 5A and B), indicating a positive role for BRs in the germination of C. pepo. However, the final percentage of germination was 100% for both genotypes. NaCl treatment delayed the initiation of germination in both WT and dwfcp compared to control conditions, although the mutant started to germinate earlier than WT and reached a higher percentage of germination under salinity conditions (Fig. 5A and B).
The effect of NaCl on the growth rate of WT and dwfcp radicle length was evaluated 27 and 50 h after germination (HAG) (Fig. 5C). The radicle length in water was always larger in the WT than in the mutant, both at 27 and 50 HAG (Fig. 5D), and the same was true under NaCl treatment (Fig. 5E), which was expected given the dwarf phenotype of the mutant. However, the percentage of reduction of the radicle length was significantly lower in dwfcp than in the WT at 27 HAG under saline conditions, which means that the dwfcp tolerated salinity better than the WT during this first stage of radicle development. At a later stage, 50 HAG, no significant differences were observed in the percentage of reduction of radicle length between WT and mutant (Fig. 5F).
Response of dwfcp to salt stress during etiolation
To study the effect of the BR-deficient mutation dwfcp of cell elongation, we measured seedling growth in darkness for 72 h under control and salinity conditions (Fig. 6). Similar to seed germination, a study of dose–response relationship between NaCl stress and seedling growth in line MUCU16 was carried out to select the optimal concentration for this assay (Supplemental Fig. S5). All saline concentrations (30, 60, 100, and 150 mM) reduced the analyzed parameters, except 60 mM NaCl, which did not reduce the hypocotyl length. This is why a concentration of 100 mM NaCl was finally selected, since it would allow a clear distinction between sensitive and tolerant-to-salinity phenotypes during seedling establishment.
Figure 6.
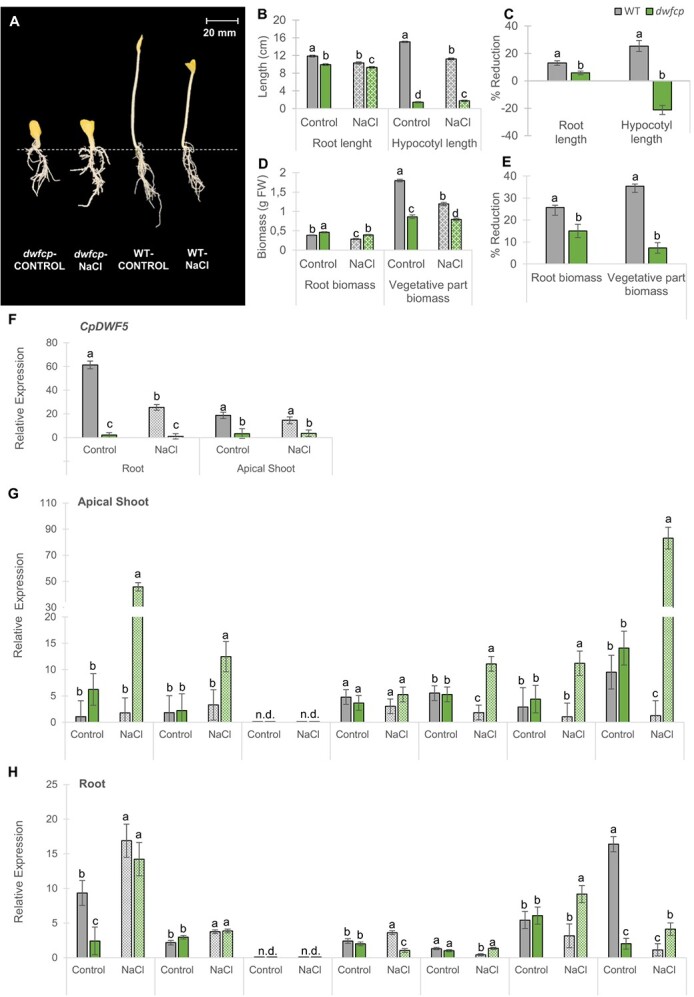
Effect of NaCl on growth parameters and gene expression of WT and dwfcp seedlings grown in darkness for 72 h. (A) WT and dwfcp seedlings under normal and saline conditions. (B) Effect of NaCl on root and hypocotyl length. (D) Effect of NaCl on root and vegetative part biomass. (C, E) Percentages of reduction of each parameter in response to salt. (F) Relative expression of CpDWF5. (G, H) Relative expression of genes involved in salt stress tolerance. N.d., not detected. Error bars represent SE. Different letters indicate statistically significant differences (P < 0.05) of the same gene expression between samples.
The dwfcp mutant showed a de-etiolation phenotype, lacking the hypocotyl elongation observed in the WT-etiolated seedlings (Fig. 6A). NaCl treatment affected the length of the roots, as well as the biomass of the roots and aerial parts both in the WT and dwfcp, but the sensitivity of mutant seedlings to salt was lower than in the WT (Fig. 6B–D). Furthermore, though saline treatment reduced the length of hypocotyl in the WT, in the mutant it was greater than in control plants (Fig. 6B and C), meaning that the salt even stimulated the growth of mutant hypocotyl. In fact, under salt stress, the root length of the WT seedlings was reduced by ~13%, while the dwfcp mutant exhibited a reduction of only 5.8%. Hypocotyl length was reduced 25% in WT, while that of the mutant increased by 21% (Fig. 6C). In the same way, the biomass of the aerial part was reduced by 35% in WT, but only by 7% in the mutant (Fig. 6E). Lastly, the root biomass was higher in dwfcp than in the WT under control conditions (Fig. 6D), unlike the rest of the parameters, which is probably a consequence of the increased thickness of the mutant roots. As in the rest of the parameters analyzed, the reduction in root biomass was more pronounced in the WT (26%) than in the dwfcp (15%) (Fig. 6E). These data indicate that dwfcp was more tolerant to salinity than WT, not only during germination and radicle growth, but also during etiolation at seedling stage.
Comparison of gene expression in WT and dwfcp in response to salt stress
To better understand the mechanisms that coordinate the enhanced salt tolerance of dwfcp, the expression of CpDWF5, but also genes related to salt tolerance in other species were compared in the apical shoots and roots of WT and dwfcp seedlings grown for 72 h in darkness under control and salinity conditions (Fig. 6F–H). The following genes were selected because they had already been reported to have a role in salinity tolerance in different species, including C. pepo [44]. CpNCED2/5A and CpPP2C-A are ABA biosynthesis and signaling genes, respectively, while CpCYP707A2 is involved in ABA catabolism. CpKEA4-2A encodes for a K+/H+ efflux antiporter, CpKUP6-1B for a potassium transporter, and CpNHK1-3B for a Na+/H+ exchanger. The calcium signaling pathway is represented by the calmodulin-binding receptor-like cytoplasmic kinase gene CpCRCK2B.
In WT plants, the CpDWF5 gene was expressed higher in the root than in the apical shoot, and it was significantly downregulated in response to salt treatment in the roots but not in the apical shoots (Fig. 6F). These data suggest that the response of C. pepo to salinity is associated with a reduction in the biosynthesis of BRs. As expected, the dwfcp mutant showed a very reduced expression of CpDWF5 under both control and salinity stress (Fig. 6F).
Under control conditions, genes that in other species mediate the salt tolerance response, including genes that encode ion transporters, ABA biosynthesis, and ABA and Ca2+ signaling, showed a similar expression level in the apical shoots and roots of WT and dwfcp (Fig. 6G and H). However, under salt stress, some of these genes were expressed much higher in the mutant than in WT seedlings, especially in the apical shoot (Fig. 6G and H). This induction may explain the enhanced response of dwfcp to salt stress. In the apical shoot, K+ uptake permease CpKUP6-1B gene, Na+/H+ exchanger CpNHK1-3B, and the Ca2+ signaling gene for calmodulin-binding receptor-like cytoplasmic kinase CpCRCK2B were only upregulated by salt stress in dwfcp (Fig. 6G). The same was true for roots (Fig. 6H). Similarly, genes involved in the ABA biosynthesis and signaling pathway, including CpNCED2/5A and CpPP2C-A, were not regulated under salt stress in the WT apical shoot, while they were upregulated in the mutant (Fig. 6G). In roots, ABA genes were induced in both WT and dwfcp in response to salt stress, but no significant differences were found between genotypes (Fig. 6H). Of particular interest were the CpNCED2/5A and CpCRCK2B genes, whose expression was increased >45 and 80 times, respectively, in response to salinity in the apical shoot of dwfcp (Fig. 6G).
Discussion
BRs control vegetative and reproductive development in C. pepo
We have shown that the locus DWFCP corresponds to the gene CpDWF5, which encodes 7-dehydrocholesterol reductase (S7R), a key enzyme in the biosynthesis of sterols, the precursors of BRs [45]. Given the reduced level of BL (the most bioactive BR) in most of the dwfcp organs and the partial rescue of the mutant phenotype by exogenous application of BL, we concluded that the primary cause of the dwarfism in dwfcp is a deficiency in BRs. Furthermore, plant sterols are important structural components of cellular membranes and regulate membrane fluidity [46], and they also participate in cellulose biosynthesis [47]. This BR-independent role of sterols may also explain some of the phenotypic effects of dwfcp and why the dwfcp phenotype was only partially rescued by the application of BRs. Therefore, this mutant offers a novel genetic tool to study the effect of sterols and BRs in squash, a horticultural species of great importance.
The phenotypic effect of dwfcp on vegetative development, including shorter leaves, stems, pedicels, petioles, roots, and fruits, coincides with the defects reported in other BR-deficient mutants [27–29]. An increase in chlorophyll content and a darker green color was also found in cpa, a cucumber mutant similarly altered in an S7R gene [22]. BRs have been reported to maintain the stability of the thylakoid membrane and regulate chlorophyll molecules by controlling the activity of the chlorophyllase enzyme [48]. As in other BR mutants [49, 50], the shorter and wider roots of dwfcp indicate that BRs have an opposite effect on longitudinal and radial growth, and that the increased fresh and dry biomass of dwfcp roots is likely due to the increased thickness of the parental and lateral roots. The reduced branching of the mutant roots also suggests that BRs play a major role in lateral root initiation and development, possibly via its interaction with the auxin gradient in the root [51]. The BR-insensitive mutant bri1 also displays a decreased number of lateral roots [52]. Understanding how BRs interact with other signals to regulate root growth and development in C. pepo represents a logical follow-up research target.
The reduced length of the dwfcp organs is due to a failure in the elongation of the cells. In fact, the number of cells in the mutant stem is greater than that in the WT, and the cells are shorter but wider than those of the WT, indicating that the dwfcp phenotype is caused by an alteration in cell size and shape. As in other BR mutants [22, 39, 53], this cell shape makes the width of leaf blades and roots, for example, not reduced nearly as much as their length, giving rise to rounder leaves and wider roots.
BRs also have a role in plant reproductive development [54]. Arabidopsis BR mutants cpd, bin2, and bri1-201 have little or no male fertility due to reduced filament elongation or a failure in pollen tube elongation [24, 55, 56]. The male flowers of the dwfcp plants were less fertile than those of the WT, but not completely sterile. As in other BR mutants [57], we found that the seeds of dwfcp developed normally and germinated well. However, the slight delay in dwfcp germination compared to WT suggests that BRs may have a minor stimulating effect on the germination of C. pepo. Steber and McCourt [58] reported that Arabidopsis BR mutants det2-1 and bri1-1 germinate normally, but show increased sensitivity to ABA during germination and that BRs partially rescue the germination of GA-deficient and GA-insensitive mutants.
dwfcp enhances tolerance to salt stress during earlier stages of plant development
Our physiological and molecular data have shown that sterols and BRs are involved in the squash salt stress response. The reduction in the germination rate, radicle growth, and hypocotyl and root elongation of seedlings in response to salt stress was found to be lower in dwfcp than in WT, suggesting that the mutant exhibits a higher tolerance to salinity than the WT, especially at early stages of seedling development growing in darkness. These are novel results indicating that sterols and BRs would play a negative role in C. pepo salt tolerance, at least under the tested conditions. Furthermore, ABA and Ca2+ biosynthesis and signaling pathways can mediate the tolerance response of dwfcp to salt, since the dwfcp mutation upregulated some of the genes involved in these two pathways, which are already known to control salt tolerance in C. pepo and other species [44]. The expression of ion transport genes, as well as the influx of Ca2+ in guard cells that leads to closure of the stomata, is known to be controlled by ABA [59, 60], but also ion homoeostasis and osmotic adjustment under salt stress [61]. We found that the KUP6-1B gene, which has a high affinity with the Arabidopsis KUP6 gene, an ABA-responsive K+ transporter [60], was upregulated in the apical shoot and root of dwfcp exposed to salt. This large KUP/HAK/KT family of high-affinity K+ transporters was reported to function in the acquisition and translocation of K+ from roots to shoots and facilitates K+ efflux from the vacuole to regulate osmotic adjustment [60, 62].
The negative role of CpDWF5 and BRs in salt tolerance was not previously reported. Exogenous BRs have been shown to mitigate the negative effects of salinity in many plants [63–65], and the Arabidopsis BR mutants det2–1 and bin2–1 were more sensitive to salt stress than the WT at germination and early seedling growth, suggesting that endogenous BRs play a positive role in salt tolerance [66]. However, recent studies have revealed a hormonal stress level-dependent biphasic effect (SLDB), whereby both the level of BRs and the level of stress can have an adverse effect on the tolerance of the plant to salt [67]. Furthermore, environmental factors such as temperature and light can also affect hormone signaling and transcriptional responses [68]. Given that our germination and seedling etiolation assays were performed under dark conditions and at a single NaCl concentration, we cannot exclude the possibility that the dwfcp mutant may respond differentially to other environmental and NaCl conditions. Alternatively, it is also possible that the higher tolerance of dwfcp to NaCl is not dependent on BRs, but on changes in the sterol profile, which is known to also regulate the plant response to abiotic stresses [69, 70]. Future studies determining the response of dwfcp to different doses of stress and environmental conditions will help to better understand the role of CpDWF5 gene and BRs in C. pepo salt tolerance.
Materials and methods
Plant materials
In this study, we present the identification and characterization of a novel dwarf mutant line called dwfcp. It was identified from a direct phenotypic screening of a C. pepo EMS collection [71]. The M2 plants of the collection lines were planted and evaluated under standard greenhouse conditions. BC1S1 and BC2S1 segregating populations from the crosses of dwfcp (male) with the background genotype MUCU16 (female) and their subsequent self-pollination were developed for phenotyping and gene mapping.
Phenotypic characterization of dwfcp
For the characterization and morphological identification of the dwfcp line, the weight and height of the plant, the diameter of the stem, as well as the growth of the vegetative and reproductive organs, the color of the leaf, and the cell size were evaluated in seedlings at the 7- leaf stage. The measurement of chlorophyll content was carried out on second true leaf samples according to the Warren procedure [72], and the calculation formula was based on the method described by Ritchie [73]. The analysis was conducted in three biological replicates per genotype and three plants per replicate. In addition, three technical replicates of each biological replicate were realized. Samples (5-mm sections from leaves of 7- leaf stage seedlings) for SEM were fixed in FAE (formaldehyde 3.7%: acetic acid 5%: absolute ethanol 50%), dehydrated through a graded ethanol series, critical point dried with liquid carbon dioxide in a pressure chamber BAL-TEC CPD 030 (Central Research Services, UAL, Spain), sputter coated with gold in a LEICA EM ACE 200 (Central Research Services, UAL, Spain), and photographed with a Zeiss Sigma 300 VP High-Resolution Field Emission Scanning Electron Microscope (Central Research Services, UAL, Spain).
The growth rates of vegetative and reproductive organs of WT and dwfcp plants were assessed by measuring the length of leaves, ovaries, and petals in both male and female flowers every 2 days for a total of 10 days in leaves or 24 days in flowers. A total of 10 leaves—15 male and 15 female flowers of each genotype—were evaluated, starting with leaves <3 cm and flower buds <1 cm in length. The anthesis time was estimated as the number of days taken for a <1-cm female or male floral bud to reach anthesis. The size and weight of the WT and dwfcp seeds from a segregating BC2S1 population were also evaluated. After the evaluation of the parameters, seeds were germinated under standard conditions, and the identification of WT and mutant seeds was carried out after genotyping for the dwfcp mutation using individual DNA from radicles.
Finally, the growth and development of the roots during the establishment of the WT and dwfcp seedlings were evaluated, including the width of the parent root in three different zones (basal, middle, and apical) and the formation and growth of the LR. ImageJ® was used to measure the elongation of both the parent and the LR of WT and dwfcp from digital images taken 50 and 72 h after the protrusion of the radicle (radicle length of ~1 mm). The LR branching density (cm−1) was calculated as the number of emerged LR per unit length in the root branching zone (in centimeters). The root branching zone includes the zone of the parent root that extends from the most rootward emerged LR to the shoot base. Since the seeds were selected from a BC2S1 population, the identification of WT and mutant seedlings was performed after genotyping for the dwfcp mutation using individual DNA from radicles.
Identification of the causal mutation of dwfcp phenotype by whole genome resequencing
To identify the causal mutation of the dwfcp phenotype, 120 BC1S1 seeds were grown in trays until the seedling stage, where the phenotype was verified. Young leaves of 30 WT and 30 dwfcp plants were collected and pooled in two separate bulks. Genomic DNA was isolated using the CTAB method described previously [74] and then subjected to whole genome resequencing (WGS). The NEBNext® DNA Library Prep Kit (https://international.neb.com) was used to create a library of short fragments of ~350 bp randomly sheared from WT and mutant DNA bulks. Fragments were then briefly PCR-enriched with indexed oligos, and pair-end sequencing, with a read length of PE150 bp at each end, was performed into the Illumina® sequencing platform. The BWA software was used to align effective sequencing data with the reference genome of C. pepo v.4.1, and GATK HAPLOTYPECALLER and ANNOVAR were used to detect and annotate SNPs. Sequencing data from other lines of the EMS mutant squash collection were used to discard common variants between lines, as they represent variants already present in the background genotype MUCU16 before sequencing of the reference genome. The following parameters were taken into account to filter the sequencing data: [1] selection of EMS-induced canonical transitions C > T and G > A; [2] selection of variants with an alternative AF = 1 in the mutant bulk (dwfcp/dwfcp); [3] selection of SNPs with nonsynonymous mutations. Finally, Integrative Genomics Viewer (IGV) software and the Cucurbit Genomics Database (CuGenDB) were used to determine the impact of the final set of EMS SNPs on gene function (http://cucurbitgenomics.org/).
Validation of the identified mutations by high-throughput genotyping of individual segregating plants
Kompetitive allele-specific PCR (KASP) technology was used to genotype 322 plants from a BC1S1 segregating population and to validate the causal mutation of the dwfcp phenotype. The LGC protocol and the FX96 Touch real-time PCR detection system (Bio-Rad®) were used to perform the KASP assay. LGC Genomics® synthesized the primers. A final reaction volume of 10 μl was used for multiplex PCRs, containing 5 μl KASP V4.0 2x Master Mix Standard ROX (LCG Genomics®), 0.138 μl KASP-by-Design Primer Mix (LCG Genomics®), and 4.862 μl of 20–50 ng/μl genomic DNA. SNP genotypes were finally identified using CFX Maestro™ software (Bio-Rad®).
Sanger sequencing of CpDWF5 cDNA in WT and dwfcp
Given that the dwfcp mutation was identified at the 3′ splicing site of the gene CpDWF5, the cDNA sequence of the gene in WT and dwfcp plants was compared. Total RNA from WT and dwfcp leaves was isolated according to the CTAB protocol. The cDNA RevertAid™ Kit (Thermo Fisher®) was used to convert the total RNA to cDNA. Templates for sequencing were amplified by PCR from cDNA (sequences of the forward and reverse primers in Supplemental Table S2) and NZYTaq II DNA polymerase (Nzytech®), according to the manufacturer’s protocol. A GeneJET PCR Purification Kit (Thermo Fisher®) was used to purify the PCR products and visualized on a gel. BigDye XTerminator™ Purification Kit (Thermo Fisher®) was used to purify the products of the sequencing reactions and the ABI 3500 Genetic Analyzer (Applied Biosystems®) to perform sequencing. Chromas software was used to analyze the sequencing results.
Assessment of relative gene expression by quantitative RT-PCR
Transcript levels of the candidate gene CpDWF5 were analyzed in different WT and dwfcp tissues, including male and female floral buds, ovaries, leaves, stems, apical shoots, and roots. Quantitative reverse transcription (qRT)-PCR assays were also performed to assess the relative expression of different genes involved in salt stress tolerance, both in the apical shoot and roots of WT and dwfcp seedlings grown under control and saline conditions for 72 h in darkness. The analyses were conducted in three biological replicates per genotype and growing condition, each derived from a pool of samples from five to six plants. Plant samples were collected and pulverized in liquid nitrogen, and total RNA was isolated according to the GeneJET Plant RNA Purification Kit (Thermo Fisher®) and converted into cDNA with the cDNA RevertAid™ Kit (Thermo Fisher Scientific®). qRT-PCR was performed in a 96-well plate using the CFX96 Touch Real-Time PCR Detection System Thermocycler (Bio-Rad®) in a 10 μl total volume with SYBR Green PCR Master Mix (BioRad®). The 2-ΔΔCT method [75] was used to calculate gene expression values. Constitutive CpEF1α and Cp18S genes were used as reference genes to normalize the results of gene expression. For expression assessment in genes related to salt tolerance, the calibrator samples were those coming from plants under control conditions. For CpDWF5 gene expression assessment, the calibrator sample was the one with the lowest transcription level. Supplemental Table S2 shows the sequences of the primers used for each qRT-PCR reaction.
BR treatments
Given that the dwfcp mutation is in an enzyme of the BR biosynthesis pathway, the mutant phenotype was tried to be rescued by external BR application. Seeds were germinated in 96-well trays, and seedlings were transferred to pots and grown in a phytotron with 60% relative humidity (RH), 24°C, and a photoperiod of 16 h day/8 h night. The apical shoots of both WT and mutant plants were sprayed with 0, 0.2, and 2 μM EBR once a day for a week, since the plants had four true leaves. For hormone treatment, a stock solution of EBR (Sigma-Aldrich, CAS number: 78821-43-9) was prepared by dissolving 10 mg of EBR in N,N-dimethylformamide. Dilutions of the desired concentrations were prepared from the stock solution with distilled water. For control conditions, distilled water was used with an equivalent concentration of N,N-dimethylformamide. All solutions were supplemented with 0.1% Tween-20.
Protein sequence alignment and phylogenetic analysis
The BLAST alignment tools at NCBI (http://www.blast.ncbi.nlm.nih.gov/) and Clustal Omega at EMBL-EBI (https://www.ebi.ac.uk/Tools/msa/clustalo/) were used to perform the alignments. The phylogenetic relations between different cucurbit species, A. thaliana, O. sativa, and S. lycopersicum S7R and sterol Δ14 reductase (S14R) proteins were assessed. Protein sequences were aligned using MEGA X software. The construction of phylogenetic trees was performed using MUSCLE and the maximum likelihood method based on the Poisson correction model, with 2000 bootstrap replicates. Protein sequences (Supplemental Table S3) used for multiplex alignment and phylogenetic study were obtained from the Arabidopsis Information Resource (https://www.arabidopsis.org/) and the Cucurbit Genomics Database (CuGenDB, http://cucurbitgenomics.org/). Gene Structure Display Server (GSDS) was used to visualize the CpDWF5 gene structure, including the number and position of introns and exons (http://gsds.gao-lab.org/Gsds_about.php).
Seed germination under water and salinity stress
Seed germination of WT and dwfcp was tested under salinity stress. Seeds were incubated in 50-ml Falcon tubes containing 25 ml of distilled water (control) or 200 mM NaCl for 16 h at 24°C in darkness under continuous shaking. After 16 h of imbibition, seeds were seeded in Petri plates between two filter papers moistened with the corresponding solution. The Petri plates with seeds were then incubated in a dark growth chamber at 24°C and 80% RH for 78 h. Two independent experiments were performed to study seed germination. Every 2 h for 80 h, germination seed was recorded. ImageJ® was used to compare root elongation from digital images taken 27 and 50 HAG. Before treatment, WT and mutant seeds were subjected to the same conditions since they were selected from BC2S1 seeds developed in the same fruit. The identification of WT and mutant seeds was done after genotyping for the causal mutation in the gene CpDWF5 using individual DNA from radicles. The concentration of NaCl used for this assay was selected after analyzing the effect of different concentrations of NaCl (85, 150, 200, 250, and 300 mM) on the seed germination of the genetic background of the collection MUCU16 following the same protocol described (Supplemental Fig. S5).
Seedling etiolation under control and salinity stress
Etiolation in darkness of WT and dwfcp seedlings was evaluated under 100 mM NaCl in two independent experiments. A total of 247 seeds from the segregating population BC2S1 were incubated in 50-ml Falcon tubes containing 25 ml of distilled water for 16 h at 24°C in darkness under continuous shaking. The seeds were then seeded in Petri dishes between two filter papers moistened with distilled water and incubated in a dark growth chamber at 24°C and 80% RH for 72 h. After this time, the germinated seeds were seeded in pots containing vermiculite and then randomly distributed in the growth chamber. The control and saline solutions were prepared in distilled water supplemented with nutrient solution (1 g/l), and the pH was adjusted to 5.8. 100 mM NaCl was added to the saline solution. The conductivities of the control and salt solutions were 1.725 and 11.573 dS/m, respectively. The pots with the germinated seeds were incubated in a growth chamber under the same conditions described for germination. After 72 h, the length and weight of hypocotyl and roots were measured in etiolated seedlings after extracting them from the pot and washing them to remove vermiculite. The identification of WT and mutant seeds was done after genotyping for the causal mutation in the gene CpDWF5 using individual DNA from cotyledons. Apical shoot and root samples were collected to measure the relative expression of a set of salt stress-associated genes. The concentration of NaCl used for this assay was selected after analyzing the effect of different concentrations of NaCl (30, 60, 100, and 150 mM) on seedling growth of the genetic background of the mutant collection MUCU16 following the same protocol described (Supplemental Fig. S5).
Statistical analyses
An analysis of variance (ANOVA) was performed in Statgraphic Centurion XVIII statistical software. The least significant difference (LSD) at a significance level of P ≤ 0.05 was used for multiple comparisons between genotypes and treatments.
Acknowledgements
This work was supported by grants PID2020-118080RB-C21 and P20_00327 funded by the Spanish Ministry of Science and Innovation and the Junta de Andalucía. J.I.-M. and G.C. gratefully acknowledge the FPI and FPU Scholarship Program from the Spanish Ministry of Science and Innovation.
Data Availability
The data underlying this article are available in the article and in its online supplementary material.
Conflict of Interests
The authors have no conflicts of interest to declare.
Supplementary Data
Supplementary data is available at Horticulture Research online.
Supplementary Material
Contributor Information
Sonsoles Alonso, Department of Biology and Geology, Agrifood Campus of International Excellence (CeiA3), and Research Center CIAMBITAL, University of Almería, Ctra. Sacramento s/n, 04120 Almería, Spain.
Gustavo Cebrián, Department of Biology and Geology, Agrifood Campus of International Excellence (CeiA3), and Research Center CIAMBITAL, University of Almería, Ctra. Sacramento s/n, 04120 Almería, Spain.
Keshav Gautam, Department of Biology and Geology, Agrifood Campus of International Excellence (CeiA3), and Research Center CIAMBITAL, University of Almería, Ctra. Sacramento s/n, 04120 Almería, Spain.
Jessica Iglesias-Moya, Department of Biology and Geology, Agrifood Campus of International Excellence (CeiA3), and Research Center CIAMBITAL, University of Almería, Ctra. Sacramento s/n, 04120 Almería, Spain.
Cecilia Martínez, Department of Biology and Geology, Agrifood Campus of International Excellence (CeiA3), and Research Center CIAMBITAL, University of Almería, Ctra. Sacramento s/n, 04120 Almería, Spain.
Manuel Jamilena, Department of Biology and Geology, Agrifood Campus of International Excellence (CeiA3), and Research Center CIAMBITAL, University of Almería, Ctra. Sacramento s/n, 04120 Almería, Spain.
REFERENCES
- 1. Shikata M, Ezura H. Micro-tom tomato as an alternative plant model system: mutant collection and efficient transformation. Methods Mol Biol. 2016;1363:47–55 [DOI] [PubMed] [Google Scholar]
- 2. Bensen RJ, Johal GS, Crane VC. et al. Cloning and characterization of the maize An1 gene. Plant Cell. 1995;7:75–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Winkler RG, Helentjaris T. The maize Dwarf3 gene encodes a cytochrome P450-mediated early step in gibberellin biosynthesis. Plant Cell. 1995;7:1307–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chen Y, Hou M, Liu L. et al. The maize DWARF1 encodes a gibberellin 3-oxidase and is dual localized to the nucleus and cytosol. Plant Physiol. 2014;166:2028–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fu J, Ren F, Lu X. et al. A tandem array of ent-kaurene synthases in maize with roles in gibberellin and more specialized metabolism. Plant Physiol. 2016;170:742–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tanaka N, Matsuoka M, Kitano H. et al. gid1, a gibberellin-insensitive dwarf mutant, shows altered regulation of probenazole-inducible protein (PBZ1) in response to cold stress and pathogen attack. Plant Cell Environ. 2010;29:619–31 [DOI] [PubMed] [Google Scholar]
- 7. Xu L, Wang C, Cao W. et al. CLAVATA1-type receptor-like kinase CsCLAVATA1 is a putative candidate gene for dwarf mutation in cucumber. Mol Gen Genomics. 2018;293:1393–405 [DOI] [PubMed] [Google Scholar]
- 8. Gale MD, Marshall GA. The chromosomal location of Gai 1 and Rht 1, genes for gibberellin insensitivity and semi-dwarfism, in a derivative of Norin 10 wheat. Heredity. 1976;37:283–9 [Google Scholar]
- 9. Multani DS, Briggs SP, Chamberlin MA. et al. Loss of an MDR transporter in compact stalks of maize br2 and sorghum dw3 mutants. Science. 2003;302:81–4 [DOI] [PubMed] [Google Scholar]
- 10. Carraro N, Forestan C, Canova S. et al. ZmPIN1a and ZmPIN1b encode two novel putative candidates for polar auxin transport and plant architecture determination of maize. Plant Physiol. 2006;142:254–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Phillips KA, Skirpan AL, Liu X. et al. Vanishing tassel2 encodes a grass-specific tryptophan aminotransferase required for vegetative and reproductive development in maize. Plant Cell. 2011;23:550–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Friml J. Subcellular trafficking of PIN auxin efflux carriers in auxin transport. Eur J Cell Biol. 2010;89:231–5 [DOI] [PubMed] [Google Scholar]
- 13. Peer WA, Blakeslee JJ, Yang H. et al. Seven things we think we know about auxin transport. Mol Plant. 2011;4:487–504 [DOI] [PubMed] [Google Scholar]
- 14. Mano Y, Nemoto K. The pathway of auxin biosynthesis in plants. J Exp Bot. 2012;63:2853–72 [DOI] [PubMed] [Google Scholar]
- 15. Li H, Wang L, Liu M. et al. Maize plant architecture is regulated by the ethylene biosynthetic gene ZmACS7. Plant Physiol. 2020;183:1184–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hao L, Renxiao W, Qian Q. et al. DWARF27, an iron-containing protein required for the biosynthesis of strigolactones, regulates rice tiller bud outgrowth. Plant Cell. 2009;21:1512–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Song X, Lu Z, Yu H. et al. IPA1 functions as a downstream transcription factor repressed by D53 in strigolactone signaling in rice. Cell Res. 2017;27:1128–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yao R, Wang L, Li Y. et al. Rice DWARF14 acts as an unconventional hormone receptor for strigolactone. J Exp Bot. 2018;69:2355–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nolan TM, Vukasinović N, Liu D. et al. Brassinosteroids: multidimensional regulators of plant growth, development, and stress responses. Plant Cell. 2020;32:295–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hou S, Niu H, Tao Q. et al. A mutant in the CsDET2 gene leads to a systemic brassinosteriod deficiency and super compact phenotype in cucumber (Cucumis sativus L.). Theor Appl Genet. 2017;130:1693–703 [DOI] [PubMed] [Google Scholar]
- 21. Wang H, Li W, Qin Y. et al. The cytochrome P450 gene CsCYP85A1 is a putative candidate for super compact-1 (scp-1) plant architecture mutation in cucumber (Cucumis sativus L.). Front Plant Sci. 2017;8:266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhang M, Song M, Cheng F. et al. Identification of a putative candidate gene encoding 7-dehydrocholesterol reductase involved in brassinosteroids biosynthesis for compact plant architecture in cucumber (Cucumis sativus L.). Theor Appl Genet. 2021;134:2023–34 [DOI] [PubMed] [Google Scholar]
- 23. Clouse SD, Langford M, Mcmorris TC. A brassinosteroid-insensitive mutant in Arabidopsis thaliana exhibits multiple defects in growth and development. Plant Physiol. 1996;111:671–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Szekeres MS, Né K. Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell. 1996;85:171–82 [DOI] [PubMed] [Google Scholar]
- 25. Fujioka S, Li J, Choi YH. et al. The Arabidopsis deetiolated2 mutant is blocked early in brassinosteroid biosynthesis. Plant Cell. 1997;9:1951–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Choe S, Dilkes BP, Fujioka S. et al. The DWF4 gene of Arabidopsis encodes a cytochrome P450 that mediates multiple 22α-hydroxylation steps in brassinosteroid biosynthesis. Plant Cell. 1998;10:231–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Choe S, Dilkes BP, Gregory BD. et al. The Arabidopsis dwarf1 mutant defective in the conversion of 24-methylenecholesterol to campesterol in brassinosteroid biosynthesis. Plant Physiol. 1999;119:897–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Choe S, Noguchi T, Fujioka S. et al. The Arabidopsis dwf7/ste1 mutant is defective in the Δ7 sterol C-5 desaturation step leading to brassinosteroid biosynthesis. Plant Cell. 1999;11:207–21 [PMC free article] [PubMed] [Google Scholar]
- 29. Choe S, Tanaka A, Noguchi T. et al. Lesions in the sterol Δ7 reductase gene of Arabidopsis cause dwarfism due to a block in brassinosteroid biosynthesis. Plant J. 2000;21:431–43 [DOI] [PubMed] [Google Scholar]
- 30. Choe S, Schmitz RJ, Fujioka S. et al. Arabidopsis brassinosteroid-insensitive dwarf12 mutants are semidominant and defective in a glycogen synthase kinase 3β-like kinase. Plant Physio. 2002;130:1506–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nomura T, Kitasaka Y, Takatsuto S. et al. Brassinosteroid/sterol synthesis and plant growth as affected by lka and lkb mutations of pea. Plant Physiol. 1999;119:1517–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bishop GJ, Nomura T, Yokota T. et al. The tomato DWARF enzyme catalyses C-6 oxidation in brassinosteroid biosynthesis. Plant Biol. 1999;96:1761–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Koka CV, Cerny RE, Gardner RG. et al. A putative role for the tomato genes DUMPY and CURL-3 in brassinosteroid biosynthesis and response. Plant Physiol. 2000;122:85–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Montoya T, Nomura T, Farrar K. et al. Cloning the tomato curl3 gene highlights the putative dual role of the leucine-rich repeat receptor kinase tBRI1/ SR160 in plant steroid hormone and peptide hormone signaling. Plant Cell. 2002;14:3163–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hartwig T, Chuck GS, Fujioka S. et al. Brassinosteroid control of sex determination in maize. Proc Natl Acad Sci USA. 2011;108:19814–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Makarevitch I, Thompson A, Muehlbauer GJ. et al. Brd1 gene in maize encodes a brassinosteroid C-6 oxidase. PLoS One. 2012;7:e30798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Best NB, Hartwig T, Budka J. et al. Nana plant2 encodes a maize ortholog of the Arabidopsis brassinosteroid biosynthesis gene DWARF1, identifying developmental interactions between brassinosteroids and gibberellins. Plant Physiol. 2016;171:2633–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yamamuro C, Ihara Y, Wu X. et al. Loss of function of a rice brassinosteroid insensitive 1 homolog prevents internode elongation and bending of the lamina joint. Plant Cell. 2000;12:1591–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mori M, Nomura T, Ooka H. et al. Isolation and characterization of a rice dwarf mutant with a defect in brassinosteroid biosynthesis. Plant Physiol. 2002;130:1152–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hong Z, Ueguchi-Tanaka M, Fujioka S. et al. The rice brassinosteroid-deficient dwarf2 mutant, defective in the rice homolog of Arabidopsis DIMINUTO/DWARF1 is rescued by the endogenously accumulated alternative bioactive brassinosteroid, dolichosterone. Plant Cell. 2005;17:2243–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tanabe S, Ashikari M, Fujioka S. et al. A novel cytochrome P450 is implicated in brassinosteroid biosynthesis via the characterization of a rice dwarf mutant, dwarf11, with reduced seed length. Plant Cell. 2005;17:776–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Klahre U, Noguchi T, Fujioka S. et al. The Arabidopsis DIMINUTO/DWARF1 gene encodes a protein involved in steroid synthesis. Plant Cell. 1998;10:1677–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Montero-Pau J, Blanca J, Bombarely A. et al. De novo assembly of the zucchini genome reveals a whole-genome duplication associated with the origin of the Cucurbita genus. Plant Biotechnol J. 2018;16:1161–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cebrián G, Iglesias-Moya J, García A. et al. Involvement of ethylene receptors in the salt tolerance response of Cucurbita pepo. Horticulture Research. 2021;8:73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Fujioka S, Yokota T. Biosynthesis and metabolism of brassinosteroids. Annu Rev Plant Biol. 2003;54:137–64 [DOI] [PubMed] [Google Scholar]
- 46. Bach TJ, Benveniste P. Cloning of cDNAs or genes encoding enzymes of sterol biosynthesis from plants and other eukaryotes: heterologous expression and complementation analysis of mutations for functional characterization. ProgLipid Res. 1997;36:197–226 [DOI] [PubMed] [Google Scholar]
- 47. Peng LC, Kawagoe Y, Hogan P. et al. Sitosterol-β-glucoside as primer for cellulose synthesis in plants. Science. 2002;295:147–50 [DOI] [PubMed] [Google Scholar]
- 48. Hafeez MB, Zahra N, Zahra K. et al. Brassinosteroids: molecular and physiological responses in plant growth and abiotic stresses. Plant Stress. 2021;2:100029 [Google Scholar]
- 49. Müssig C, Shin GH, Altmann T. Brassinosteroids promote root growth in Arabidopsis. Plant Physiol. 2003;133:1261–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Fridman Y, Strauss S, Horev G. et al. The root meristem is shaped by brassinosteroid control of cell geometry. Nat Plants. 2021;7:1475–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wei Z, Li J. Brassinosteroids regulate root growth, development, and symbiosis. Mol Plant. 2016;9:86–100 [DOI] [PubMed] [Google Scholar]
- 52. Bao F, Shen J, Brady SR. et al. Brassinosteroids interact with auxin to promote lateral root development in A. thaliana. Plant Physiol. 2004;134:1624–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kwon M, Choe S. Brassinosteroid biosynthesis and dwarf mutants. J Plant Biol. 2005;48:1–15 [Google Scholar]
- 54. Altmann T. A tale of dwarfs and drugs: brassinosteroids to the rescue. Trends Genet. 1998;14:490–5 [DOI] [PubMed] [Google Scholar]
- 55. Bouquin T, Meier C, Foster R. et al. Control of specific gene expression by gibberellin and brassinosteroid. Plant Physiol. 2001;127:450–8 [PMC free article] [PubMed] [Google Scholar]
- 56. Li J, Nam KH, Vafeados D. et al. BIN2, a new brassinosteroid-insensitive locus in Arabidopsis. Plant Physiol. 2001;127:14–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Clouse SD, Sasse JM. BRASSINOSTEROIDS: essential regulators of plant growth and development. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:427–51 [DOI] [PubMed] [Google Scholar]
- 58. Steber CM, McCourt P. A role for brassinosteroids in germination in Arabidopsis. Plant Physiol. 2001;125:763–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. DeFalco TA, Bender KW, Snedden WA. Breaking the code: Ca2+ sensors in plant signalling. Biochem J. 2010;425:27–40 [DOI] [PubMed] [Google Scholar]
- 60. Osakabe Y, Arinaga N, Umezawa T. et al. Osmotic stress responses and plant growth controlled by potassium transporters in Arabidopsis. Plant Cell. 2014;25:609–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Verslues PE, Bray EA. Role of abscisic acid (ABA) and Arabidopsis thaliana ABA-insensitive loci in low water potential-induced ABA and proline accumulation. J Exp Bot. 2006;57:201–12 [DOI] [PubMed] [Google Scholar]
- 62. Grabov A. Plant KT/KUP/HAK potassium transporters: single family-multiple functions. Ann Bot. 2007;99:1035–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Hayat S, Hasan SA, Yusuf M. et al. Effect of 28-homobrassinolide on photosynthesis, fluorescence and antioxidant system in the presence or absence of salinity and temperatura in Vigna radiata. Environ Exp Bot. 2010;69:105–12 [Google Scholar]
- 64. Yuan L, Yuan Y, Du J. et al. Effects of 24-epibrassinolide on nitrogen metabolism in cucumber seedlings under Ca(NO3)2 stress. Plant Physiol Biochem. 2012;61:29–35 [DOI] [PubMed] [Google Scholar]
- 65. Yue J, You Y, Zhang L. et al. Exogenous 24-epibrassinolide alleviates effects of salt stress on chloroplasts and photosynthesis in Robinia pseudoacacia L. seedlings. J Plant Growth Regul. 2018;38:669–82 [Google Scholar]
- 66. Zeng H, Tang Q, Hua X. Arabidopsis brassinosteroid mutants det2-1 and bin2-1 display altered salt tolerance. J Plant Growth Regul. 2010;29:44–52 [Google Scholar]
- 67. Liu J, Yang R, Jian N. et al. Putrescine metabolism modulates the biphasic effects of brassinosteroids on canola and Arabidopsis salt tolerance. Plant Cell Environ. 2020;43:1348–59 [DOI] [PubMed] [Google Scholar]
- 68. Oh E, Zhu JY, Wang ZY. Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat Cell Biol. 2012;14:802–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Clouse SD. Arabidopsis mutants reveal multiple roles for sterols in plant development. Plant Cell. 2002;14:1995–2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Rogowska A, Szakiel A. The role of sterols in plant response to abiotic stress. Phytochem Rev. 2020;19:1525–38 [Google Scholar]
- 71. García A, Aguado E, Parra G. et al. Phenomic and genomic characterization of a mutant platform in Cucurbita pepo. Front Plant Sci. 2018;9:1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Warren CR. Rapid measurement of chlorophylls with a microplate reader. J Plant Nutr. 2008;31:1321–32 [Google Scholar]
- 73. Ritchie RJ. Consistent sets of spectrophotometric chlorophyll equations for acetone, methanol and ethanol solvents. Photosynth Res. 2006;89:27–41 [DOI] [PubMed] [Google Scholar]
- 74. Doyle JJ, Doyle JL. Isolation of plant DNA from fresh tissue. Focus. 1990;12:13–5 [Google Scholar]
- 75. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–8 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article are available in the article and in its online supplementary material.