Abstract
We have previously described the development of AIDS in a chimpanzee (C499) infected with human immunodeficiency virus type 1 (HIV-1) and the subsequent pathogenic HIV-1 infection in another chimpanzee (C455) transfused with blood from C499 (F. J. Novembre et al., J. Virol. 71:4086–4091, 1997). In the present study, two virus isolates were derived from these animals: HIV-1JC from peripheral blood mononuclear cells (PBMC) of C499, and HIV-1NC from plasma of C455. These virus isolates were used to generate two infectious molecular clones, termed HIV-1JC16 and HIV-1NC7 (JC16 and NC7, respectively). Comparative analyses of the sequences of the two clones showed that they were highly interrelated but distinct. Based on heteroduplex mobility assays, JC16 and NC7 appear to represent dominant viruses in the uncloned stock population. Compared with amino acid sequences of the parental viruses HIV-1SF2, HIV-1LAV-1b, and HIV-1NDK, JC16 and NC7 showed a number of differences, including insertions, deletions, and point mutations spread throughout the genome. However, insertion/deletion footprints in several genes of both JC16 and NC7 suggested that recombination between SF2 and LAV-1b could have occurred, possibly contributing to the generation of a pathogenic virus. Comparative in vitro analyses of the molecular clones and the uncloned stocks of HIV-1JC and HIV-1NC revealed that these viruses had strikingly similar replicative abilities in mitogen-stimulated PBMC and in macrophages. Compared to the SF2 and LAV-1b isolates of HIV-1, HIV-1JC and HIV-1NC isolates were more similar to LAV-1b with respect to the ability to replicate in mitogen-stimulated PBMC and macrophages. These viruses should prove to be useful in mapping determinants of pathogenesis.
Soon after the isolation of human immunodeficiency virus type 1 (HIV-1) and its association with the development of AIDS in humans (2, 5, 23, 28), attempts to develop relevant animal models for use in pathogenesis, therapy, and vaccine research were made. Among the first species of animals used was the chimpanzee. Successful infection of chimpanzees with various isolates was quickly described by several groups (14, 16, 22). Exposure of chimpanzees to HIV-1 resulted in infection, as determined by repeated virus isolation from peripheral blood mononuclear cells (PBMC) and the development of antiviral antibody (16, 33). However, in contrast to human infection, primary virus infection of chimpanzees seldom resulted in plasma viremia (20, 32, 34), and no pathogenic effects were observed (43). Thus, the use of chimpanzees was abandoned for pathogenesis research. Over the years, a number of groups have used the HIV/chimpanzee system for testing of potential AIDS vaccines (4, 18, 26, 27). These experiments initially focused on recombinant methods of producing vaccines (7, 29) but recently have used more modern methods such as DNA vaccines (4, 40). While these studies have shown variable results including some level of protection, a lack of disease development in these animals has raised questions on appropriateness of this model system.
A number of hypotheses have been put forth to explain the lack of disease development in chimpanzees. These include lack of cytopathic effects of HIV-1 on chimpanzee CD4+ cells (6), inability of chimpanzee macrophages to sustain HIV-1 replication (33), lack of apoptosis of CD4+ cells in chimpanzees infected with HIV-1 (13), and absence of cytotoxic T lymphocytes which lyse uninfected CD4+ cells (an autoimmune phenomenon) (45). The use of T-cell line-adapted strains (and their derivatives) for most chimpanzee inoculations has also led to speculations on the lack of pathogenicity of cell line-adapted strains of HIV-1 for chimpanzees. Most prominent in these reasons is the lack of cytopathic effect observed in vitro with HIV-1 infection of chimpanzee PBMC. Recently two groups have identified primary isolates of HIV-1 which replicate to high titers and induce syncytium formation in chimpanzee PBMC (cPBMC) (25, 42). Both of these isolates also replicate well in chimpanzee macrophages. The DH12 isolate of HIV-1 has been used to inoculate chimpanzees, but development of disease has not been reported.
We previously reported on the first chimpanzee (C499) to develop AIDS, 10 years after infection with HIV-1 (36). The development of AIDS in C499 was associated with opportunistic infection, high viral loads, CD4+ cell decline, and the presence of a cytopathic virus. Transfusion of blood from C499 to an uninfected chimpanzee resulted in high plasma viral loads and persistent decline in CD4+ cell count. Recently, Wei and Fultz described the extensive diversification of env sequences present in C499 in samples obtained 22 months before death (44). These data reveal that significant changes in several regions of Env have occurred over the 9+ years of infection in this animal. The present study extends these observations by reporting on the molecular cloning and complete nucleotide sequence analysis of viruses derived from C499 (at the time of AIDS disease) and C455 (at the time of acute infection). We show that the viruses isolated from these animals have enhanced replication kinetics in cPBMC and that these viruses replicate well in chimpanzee macrophages. In addition, molecular analyses suggest that these viruses may be recombinants between two of the inoculating viruses, HIV-1SF2 and HIV-1LAV-1b.
MATERIALS AND METHODS
Chimpanzees, virus isolates, and genomic DNA preparation.
Chimpanzee C499 and C455 have previously been described (15, 36). Various uninfected chimpanzees were used for blood collection to provide cPBMC and macrophage populations for in vitro sue. For blood collection, animals were anesthetized intramuscularly with Telazol (4 mg/kg of body weight). A virus isolate from C499 (termed HIV-1JC) was generated by coculture of PBMC from this animal with uninfected cPBMC. A virus isolate from C455 (termed HIV-1NC) was generated by incubating plasma, obtained at 1 month postinfection, with uninfected cPBMC. Both stocks of virus were the result of culture expansion, and all virus stocks were prepared as cell-free virus. The LAV-1b strain of HIV-1 (17) was grown in cPBMC. The SF2 strain of HIV-1 (31) was obtained from J. Levy through the AIDS Research and Reference Reagent Program and was grown in human PBMC. The DH12 isolate of HIV-1 (42) was obtained from M. Martin and was grown in cPBMC. cPBMC infected with either HIV-1JC or HIV-1NC were used for isolation of DNA by using a Puregene kit (Gentra Systems, Minneapolis, Minn.) as directed by the manufacturer. The Yerkes Regional Primate Research Center is fully accredited by Association for Assessment and Accreditation of Laboratory Animal Care and all animals were housed in accordance with Animal Welfare Act guidelines.
Long-range PCR and cloning of amplified genome fragments.
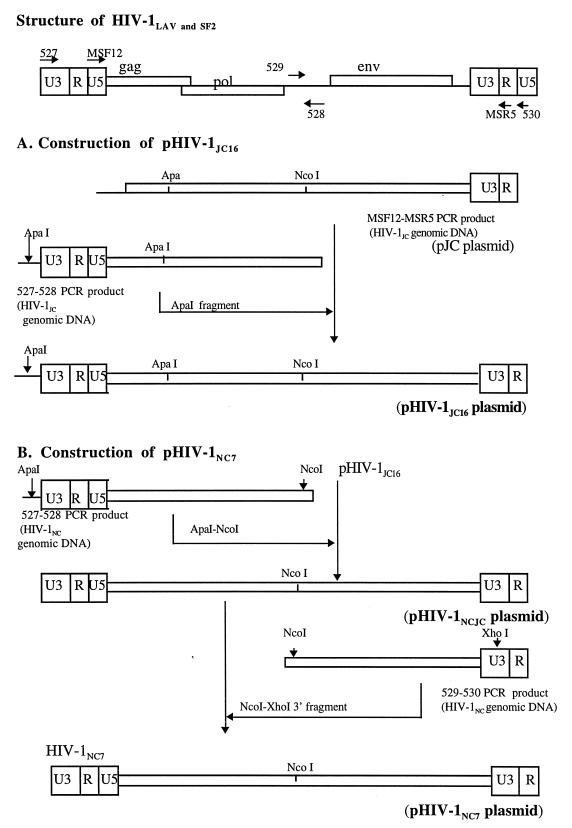
The strategy used for PCR amplification and primer location on the HIV-1 viral genome is shown in Fig. 1. PCR primers MSF12 (5′-AAA TCT CTA GCA GTG GCG CCC GAA CAG-3′, HIV-1LAV nucleotides [nt] 169 to 195) and MSR5 (5′-GCA CTC AAG GCA AGC TTT ATT GAG GCT-3′, HIV-1LAV nt 9225 to 9198) (37) were used to amplify a 9,056-bp product from PBMC genomic DNA prepared from HIV-1JC-infected cells. PCR primers 527 (5′-CAC ACA CAA GGC TAC TTC CCT GAT TGG CAG A-3′, HIV-1LAV nt 8735 to 8765) and 528 (5′-CCA AGT ATC CCC ATA AGT TTC ATA GAT AT-3′, HIV-1LAV nt 5302 to 5274) were used to amplify 5′ long terminal repeat (LTR)-containing fragments (5,699 bp) from the same DNA source. For the HIV-1NC viral genome, PCR primer pairs 527-528 and 529 (5′-ATG GAA CAA GCC CCA GAA GAC CAA GGG CCA CAG-3′, HIV-1LAV nt 5141 to 5173)-530 (5′-GGT CTG AGG GAT CTC TAG TTA CCA GAG TCA C-3′, HIV-1LAV nt 151 to 121) were used to generate the 5′-half (5,699 bp) and 3′-half (4,142 bp) PCR products, respectively, from HIV-1NC genomic PBMC DNA. Primers were synthesized on an Applied Biosystems 392 DNA synthesizer. Briefly, PCR was performed by using reagents from an Expand Long Template kit (Boehringer Mannheim, Indianapolis, Ind.) and 200 ng of DNA template according to the manufacturer’s instructions. After an initial DNA denaturation at 94°C for 2 min, the PCR consisted of 10 cycles of 94°C for 15 s, 61°C for 30 s, and 68°C for 8 min, followed by 20 cycles of 94°C for 15 s, 61°C for 30 s, and 68°C for 8 min, with a 5-s addition to each extension. The samples were incubated at 72°C for 30 min after the last cycle and then cooled to 4°C. Results of PCRs were evaluated on 0.9% agarose gels. PCR products representing the correct-size fragments were isolated from agarose, directly cloned into plasmid, pCR II, and amplified in Escherichia coli (TA cloning kit; Invitrogen Corp., San Diego, Calif.) according to the manufacturer’s protocol. Single bacterial colonies containing plasmids with inserts of the correct size were grown at 30°C overnight, and plasmid DNA was prepared by the alkaline lysis method.
FIG. 1.
The strategy used in PCR amplification of subgenomic fragments from HIV-1 and the locations and orientations of primers on the viral genome. All primers were designed from the HIV-1LAV nucleotide sequence, and their coordinates are described in Materials and Methods. PCR-amplified fragments were cloned in TA vectors, and the designations for the corresponding recombinant plasmids are shown in parentheses. The 5′ LTR-containing ApaI fragment amplified from HIV-1JC PBMC genomic DNA was subcloned in pJC to generate plasmid pHIV-1JC16, while the ApaI-NcoI fragment containing the 5′ LTR region amplified from HIV-1NC PBMC genomic DNA was subcloned in pHIV-1JC16 to generate chimeric plasmid pHIV-1NCJC. Plasmid pHIV-1NC7 was constructed by subcloning the env-containing NcoI-XhoI fragment amplified from HIV-1NC genomic DNA to chimeric plasmid pHIV-1NCJC. All recombinant plasmids (pHIV-1JC16, pHIV-1NCJC, and pHIV-1NC7) lacked 55 bp at the 5′ end of the genome (U3 region) and all of U5 region in the 3′ LTR region.
Construction of full-length molecular clones of HIV-1.
The strategy for preparing full-length molecular clones is illustrated in Fig. 1. Several restriction enzymes were used to generate restriction maps for the positive clones. The ApaI fragment (1,947 bp) from the 5′-half PCR product was gel purified and subcloned into the large fragment (7,675 bp) of pJC, using standard cloning procedures (39), to generate plasmid pHIV-1JC16 (Fig. 1). A chimeric plasmid (pHIV-1NCJC) was generated by subcloning the PCR-amplified 5′ half of HIV-1NC into pHIV-1JC16. For the HIV-1NC full-length clone, the NcoI-XhoI env-containing fragment from the 3′-half PCR product was gel purified and subcloned into the NcoI-XhoI large fragment of plasmid pHIV-1NCJC containing the 5′ half of HIV-1NC. Multiple restriction enzymes were used for analysis of both viral DNAs to confirm the full-length clones.
Transfection of CEMx174 and 293 cell lines.
CEMx174 cells (5 × 106) in T-25 flasks were transfected with 2 μg of either pHIV-1JC16 or pHIV-1NC7 DNA in transfection buffer (25 mM Tris-HCl [pH 7.5], 140 mM NaCl, 5 mM KCl, 0.7 mM K2HPO43H2O) containing 4 μl of DEAE-dextran (60 mg/ml) for 20 min at room temperature. Five milliliters of complete medium (RPMI 1640 supplemented with 10% fetal bovine serum and 2 mM l-glutamine) was added to stop the reaction, and the cells were centrifuged at 1,000 rpm for 10 min. The cells were washed twice in 10 ml of complete medium and then resuspended in 10 ml of complete medium, transferred to a T-25 flask, and incubated at 37°C (5% CO2). The cells were checked daily for cytopathic effects (syncytium formation), and aliquots of cultures were tested for the presence of reverse transcriptase (RT) activity by using standard assay methods. For 293 cell lines, 2 × 105 cells in six-well plates were transfected with 2 μg of viral DNA by using Lipofectin (Life Technologies, Gaithersburg, Md.) or DOTAP (Boehringer Mannheim) according to the manufacturer’s instructions. After 24 h, the transfected cells were overlaid with uninfected cPBMC (2 × 106/well) previously stimulated with concanavalin A (ConA) for 4 days. After an additional 2-day incubation, the nonadherent cell population (cPBMC) was transferred to a T-25 flask, and additional stimulated cPBMC were added for virus amplification. Culture supernatants were assayed for RT activity, and the cells were observed daily for development of syncytia. Cell-free stocks of molecularly cloned viruses were prepared at peak RT activity, aliquoted, and stored under liquid nitrogen.
Replication kinetics in PBMC and monocyte-derived macrophages (MDM).
A total of 107 freshly isolated or ConA-stimulated PBMC from an HIV-1-negative chimpanzee were infected overnight (at 37°C) with 20 ng of the indicated virus (p24 antigen concentration). The cells were centrifuged at 1,000 rpm for 10 min, resuspended in interleukin-2 (IL-2) medium (10 ml of RPMI 1640 containing 10% fetal bovine serum and IL-2), and incubated at 37°C. Samples of supernatants (1 ml) were harvested on days 3, 7, 10, 14, and 17 postinfection. IL-2 medium was added to the cultures following the sampling to maintain the original volume. Supernatants were used in RT assays to determine the relative amounts of virus produced. Assays were performed three times with PBMC derived from different chimpanzees.
For replication in macrophages, cPBMC were resuspended in macrophage medium (6 × 106 macrophages/well in RPMI 1640 containing 15% human serum [AB+], 1% HEPES, 0.008 ng of granulocyte-macrophage colony-stimulating factor per ml 0.03 ng of macrophage colony-stimulating factor per ml, and 1% antibiotic-antimycotic solution [Sigma, St. Louis, Mo.]), seeded in a 24-well plate, and incubated at 37°C for 4 h. The cells were mixed by pipetting up and down, and then incubation was continued for 4 days. Nonadherent cells were removed by gently washing the wells. Fresh medium (2 ml/well) was added, and the cells were cultured for an additional 3 days to allow full macrophage differentiation. Replicate cultures were tested for the presence of macrophages and T cells by immunohistochemical staining (36) with the Ham56 antibody (macrophages) and an anti-CD3 antibody (T cells). Cells in these cultures were determined to be >99% macrophages. Infections were initiated by adding 10 ng of virus (p24) to the cells in 500 μl of medium and adsorbed overnight. The inoculum was removed, and the cells were washed twice before fresh macrophage medium (2 ml) was added. On days 7 and 14 postinfection, aliquots of 0.5 ml were taken for determination of p24 antigen levels, using an HIV-1 p24 antigen capture enzyme-linked immunosorbent assay (ELISA) kit (Coulter Corp., Miami, Fla.) according to the manufacturer’s instructions. As with the PBMC replication studies, the macrophage studies were performed three times with macrophages derived from different chimpanzees.
Nucleotide sequencing and amino acid analyses.
Primers for sequencing were constructed from conserved regions of aligned sequences of HIV-1LAV and HIV-1SF2 and were synthesized on an Applied Biosystems 392 DNA synthesizer. The DNA sequence of each full-length cloned virus was determined by the dideoxy-chain termination method, using the Sequenase system (Amersham Life Science, Arlington Heights, Ill.) and [35S]dATP. Nucleotide and amino acid sequence alignments were performed with the Intelligenetics (Beaverton, Oreg.) suite of programs and the Lasergene program (DNAStar, Inc., Madison, Wis.).
HMA.
An HMA kit (NIH AIDS Research and Reference Reagent Program) based on the method described by Delwart et al. (9) was used. Briefly, equal amounts (5 μl each) of second-round PCR products (V1-V2 and V3-V5) from infected cPBMC genomic DNA were mixed with the reference PCR products to obtain heteroduplexes. After addition of 1.1 μl of 10× annealing buffer (1 M NaCl, 100 mM Tris [pH 7.8], 20 mM EDTA), the mixed DNAs were denatured at 94°C for 2 min and then reannealed by rapid cooling in ice. Three microliters of loading dye (25% Ficoll, 1% orange G) was added to the cooled DNA mixture, and the samples were loaded onto a 5% polyacrylamide gel in 1× TBE (88 mM Tris-borate, 89 mM boric acid, 2 mM EDTA) buffer and electrophoresed at a constant voltage of 250 V for 2.5 h. The gels were stained with ethidium bromide and visualized under UV light.
Nucleotide sequence accession numbers.
The nucleotide sequences for HIV-1JC16 and HIV-1NC7 reported in this paper have been submitted to GenBank and assigned accession no. AF049494 and AF049495, respectively.
RESULTS
Virus isolates.
As described previously, animal C499 was initially infected with HIV-1SF2 in 1985 (15) and later inoculated with HIV-1LAV-1b and HIV-1NDK in 1986 and 1987, respectively. Superinfection with HIV-1LAV-1b (but not HIV-1NDK) was demonstrated by restriction enzyme analysis of PBMC genomic DNA (17). At the time of disease development in C499, a virus isolate, termed HIV-1JC, was obtained by coculture of C499 PBMC with normal cPBMC. At that time, sequence analysis of the V1-V2 region of env suggested that HIV-1JC was most closely related to HIV-1LAV (36). Also at the time of disease development, blood from C499 was transfused into an uninfected chimpanzee (C455), which resulted in a dramatic decline of CD4+ cells by 2 weeks posttransfusion (36). The depressed CD4+ cell count has been maintained to date in this animal (data not shown). One month posttransfusion, 50 μl of plasma from C455 was used for in vitro infection of normal chimpanzee PBMC, and the resultant virus was designated HIV-1NC.
Generation and analysis of full-length molecular clones of HIV-1JC and HIV-1NC.
To perform a more thorough analysis of the genetic makeup of the HIV-1JC and HIV-1NC, we constructed full-length molecular clones as described in Materials and Methods and shown in Fig. 1. All clones lacked 55 nt at the 5′ end (5′ LTR and U3 region) and all of the U5 region in the 3′ LTR (Fig. 1). Several clones (representing both HIV-1JC and HIV-1NC) which appeared to be the correct size were tested for biological activity by transfection of CEMx174 cells. Supernatants from transfected cells were used in RT assays to monitor virus production. Two clones, one from each group (HIV-1JC [JC16] and HIV-1NC [NC7]), were positive by RT and also showed massive syncytium formation (2 to 3 days posttransfection) similar to that observed with uncloned virus. To prepare stock viruses for use in in vitro assays, 293 cells were transfected with molecularly cloned DNAs followed by amplification with cPBMC as outlined in Materials and Methods.
The complete nucleotide sequences of JC16 and NC7 were determined as described in Materials and Methods. The genomes were determined to be 9,193 nt (JC16) and 9,196 nt (NC7) in length and contained open reading frames for all HIV-1-specific structural, regulatory, and accessory genes. Alignment of JC16 and NC7 DNA sequences revealed that the two genomes were very similar but contained a number of nucleotide changes spread throughout the genome (data not shown). The most divergent region between NC7 and JC16 was the V5 region of the env gene. JC16 contained a 6-bp deletion in the gag gene relative to NC7, while NC7 had a 3-bp deletion in the env gene region relative to JC16. In the LTR region there was 98.7% nucleotide identity between JC16 and NC7, with all changes localized to the U5 region (Table 1).
TABLE 1.
Comparison of chimpanzee HIV-1 isolates with inoculating viruses
| Comparison | HIV-1JC16 vs:
|
Mutations unique to JC and NCa | |||
|---|---|---|---|---|---|
| LAI | SF2 | NDK | NC7 | ||
| Pol | |||||
| % Identityb | 96.6 | 94.8 | 94.0 | 98.8 | |
| Insertions | 12 | 0 | 0 | 0 | 0 |
| Deletions | 0 | 0 | 1 | 0 | 0 |
| Point mutations | 30 | 51 | 56 | 11 | 23 |
| Gag | |||||
| % Identity | 94.2 | 92.2 | 88.8 | 97.4 | |
| Insertions | 12 | 5 | 1 | 2 | 0 |
| Deletions | 0 | 3 | 81 | 0 | 2 |
| Point mutations | 25 | 33 | 42 | 9 | 20 |
| Vif | |||||
| % Identity | 92.2 | 86.0 | 86.5 | 96.9 | |
| Insertions | 0 | 0 | 0 | 0 | 0 |
| Deletions | 0 | 0 | 0 | 0 | 0 |
| Point mutations | 14 | 26 | 25 | 5 | 10 |
| Vpr | |||||
| % Identity | 88.7 | 96.9 | 89.7 | 96.6 | |
| Insertions | 0 | 0 | 0 | 0 | 0 |
| Deletions | 1 | 0 | 1 | 0 | 0 |
| Point mutations | 7 | 2 | 6 | 2 | 0 |
| Tat | |||||
| % Identity | 88.4 | 82.5 | 74.4 | 97.0 | |
| Insertions | 0 | 1 | 0 | 0 | 0 |
| Deletions | 15 | 0 | 15 | 0 | 0 |
| Point mutations | 10 | 18 | 22 | 3 | 4 |
| Nef | |||||
| % Identity | 87.4 | 89.4 | 78.7 | 99.0 | |
| Insertions | 0 | 4 | 1 | 0 | 0 |
| Deletions | 0 | 0 | 0 | 0 | 0 |
| Point mutations | 25 | 18 | 40 | 1 | 8 |
| Rev | |||||
| % Identity | 84.5 | 82.9 | 75.2 | 96.6 | |
| Insertions | 1 | 1 | 1 | 0 | 0 |
| Deletions | 1 | 0 | 0 | 0 | 0 |
| Point mutations | 18 | 19 | 28 | 4 | 11 |
| Env | |||||
| % Identity | 81.9 | 76.1 | 71.4 | 97.6 | |
| Insertions | 9 | 9 | 11 | 0 | 0 |
| Deletions | 7 | 13 | 24 | 1 | 2 |
| Point mutations | 138 | 173 | 203 | 17 | 78 |
| Vpu | |||||
| % Identity | 79.3 | 63.9 | 67.1 | 98.8 | |
| Insertions | 0 | 0 | 0 | 0 | 0 |
| Deletions | 1 | 1 | 1 | 0 | 0 |
| Point mutations | 13 | 24 | 24 | 0 | 10 |
| LTR | |||||
| % Identity | 92.1 | 93.9 | 89.1 | 98.7 | |
| Insertions | 1 | 3 | 1 | 0 | 0 |
| Deletions | 1 | 3 | 1 | 0 | 1 |
| Point mutations | 48 | 29 | 52 | 10 | 14 |
Amino acids or nucleotides not observed in parental viruses.
Amino acid identity for proteins; nucleotide identity for LTR.
Comparative analyses were then performed between JC16, NC7, and the parental inoculating viruses, SF2, LAV, and NDK (the LAV isolate was used for comparison because the complete sequence of LAV-1b has not yet been determined). Compared with the parental inoculating viruses, the LTR sequences of JC16 and NC7 had nucleotide identities of 92.1% (LAV), 93.9% (SF2), and 89.1% (NDK)—lower than that observed upon direct comparison of JC16 and NC7 (Table 1). Most of the host-virus transcription binding factor sequences (sites for AP-1, NF-AT, NF-κB, and Sp-1) and the TAR CORE and the Lys-tRNA sites were conserved (or had single point mutations) between the parental (SF2 and LAV strains) and progeny viruses. However, there were three point mutations unique to JC16 and NC7 at the NRF/NRE binding site.
The deduced amino acid sequences for all proteins of HIV-1JC and HIV-1NC were generated by using the Intelligenetics suite of programs. Using the Lasergene program (DNAStar), multiple alignments of all proteins were constructed to examine similarities between HIV-1NC, HIV-1JC and the parental viruses (Table 1). Based on percent homology calculations, Gag, Pol, Vif, Tat, Rev, Env, and Vpu of JC16 and NC7 were most closely related to LAV, with Vpr and Nef being most closely related to SF2. In no case was it apparent that the NDK isolate was the origin of a protein sequence. While most changes involved amino acid point mutations, several proteins of JC16 and NC7 contained amino acid insertions or deletions relative to the parental inoculating strains. A closer analysis of amino acid alignments revealed that the percent homologies could be misleading with regard to the origin of the protein. For example, in Tat, JC16 and NC7 were more homologous to LAV than to SF2 (based on percent homologies). However, the JC16 and NC7 Tat proteins contained 15 amino acid deletions with respect to LAV, similar to that present in the SF2 isolate. Findings were similar for Gag and Pol.
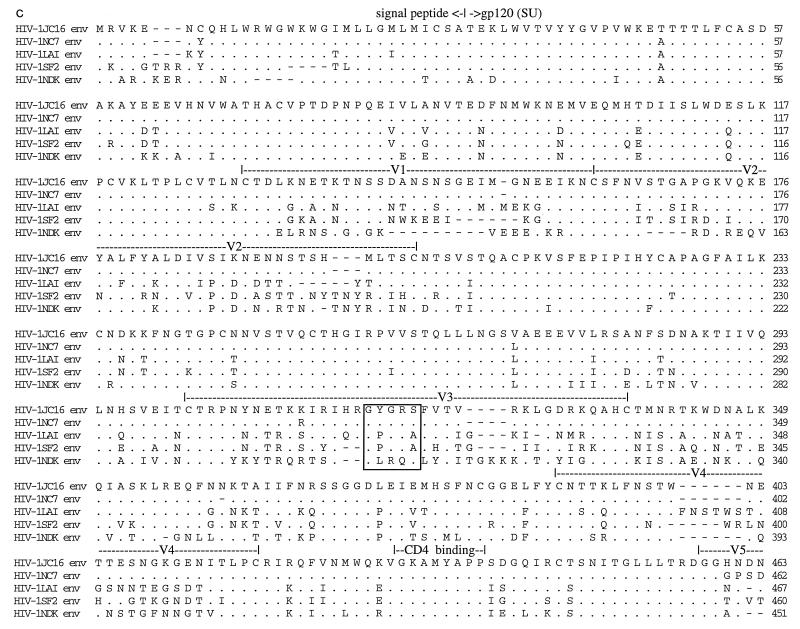
Figure 2 shows amino acid alignments of Gag, Nef, and Env constructed between JC16, NC7, and the inoculating viruses, LAV, SF2, and NDK. There were seven (JC16 and LAV sequences) and five (NC7 sequence) amino acid deletions in the matrix protein (p17) relative to SF2 isolate sequence. At the C-terminal end of Gag polyprotein, the progeny viruses and SF2 virus had 12 amino acid deletions in p6 protein relative to the LAV virus sequence (Fig. 2a). However, the capsid (p24) and nucleocapsid (p7) proteins, including the cysteine residues within the zinc finger domains, were generally well conserved. Point mutations unique to JC16 and NC7 were present in p17, p24, p7, and p6 peptides. The Lck binding domain (proline-rich region) within Nef was well conserved, with only one point mutation in LAV (Fig. 2b). However, there were four amino acid insertions in the SF2 sequence relative to the other viruses at the N-terminal portion of Nef. Sequence analysis of eight other noninfectious clones (four from JC and four from NC) confirmed observations made for the Gag and Nef deletions, suggesting that these characteristics are a general property of the viruses obtained from C499 and C455 (data not shown).
FIG. 2.
Alignment of proteins of HIV-1 isolates. The deduced amino acid sequences for the Gag (a), Nef (b), and Env proteins of HIV-1 were determined by using the Intelligenetics suite of programs, and the CLUSTAL method (DNAStar) was used for the alignment of proteins. Dashes denote amino acid deletion, while dots denote amino acid identity. Numbers at the right show the position of the right-most amino acid in each line. The functional domains on Gag (a) and Nef (b) are indicated. MBD (a) refers to membrane binding domain, while MTD (b) refers to membrane-targeting domain. The hypervariable regions in the Env glycoprotein (c) are shown as V1 to V5, and the CD4 binding domain is shown above the sequence. The GPGRA and GYGRS pentapeptides in the V3 region of Env are boxed. gp120 (SU) is the outer surface membrane Env glycoprotein, while gp41 (TM) is the transmembrane Env glycoprotein.
The vast majority of point mutations, deletions, and insertions in these clones, relative to the parental viruses, were found in the Env glycoprotein region (Fig. 2c; Table 1). Alignment of the Env proteins revealed that all of the cysteine residues resident in the protein were conserved between the viruses (Fig. 2c). There were a total of 30 predicted N-linked glycosylation sequences (Asn-X-Thr or Asn-X-Ser) for the parental strains, compared to 27 (NC7) and 29 (JC16) for the progeny viruses. Most of the glycosylation sites reside in the SU (surface) portion of Env, with HIV-1SF2 containing the highest number and HIV-1 isolate NC the least (25 for SF2, 24 for LAV and JC, and 23 for NC). Glycosylation of glycoproteins has been shown to influence the immune response toward virus infection (1). As expected, the gp120 (SU) glycoprotein contained the highest number of mutations. The V1-V2, V3, V4, and V5 hypervariable regions contained 10, 8, 8, and 3 point mutations, respectively, specific to JC16 and NC7. The V1-V2 region of JC16 and NC7 also contained insertions relative to the other viruses, resulting in amino acid lengths of 72 (progeny viruses), 69 (LAV), 70 (SF2), and 61 (NDK). There were multiple amino acid deletions in the V4 regions and single amino acid insertions in V5 regions of JC16 and NC7 relative to the parental strains. Both the CD4 binding domain and the proteolytic cleavage site (REKR) at the SU/TM (transmembrane) junction were perfectly conserved.
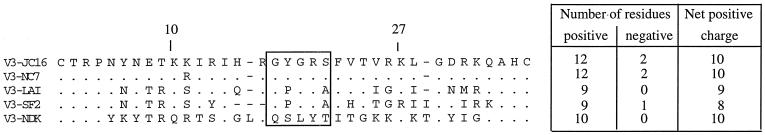
V3 was the most interesting of the hypervariable regions. While the parental strains contained only 9 basic amino acids (Arg, Lys, and His positively charged residues), HIV-1 isolates JC16 and NC7 had 12 basic and 2 negatively charged residues (Asp and Glu) (Fig. 3). This gave the progeny viruses a net positive charge of 10 in the entire V3 region and an overall positive charge of +1 (LAV) or +2 (SF2) compared to the parental strains. Eight of the ten resultant positive charges for isolates JC and NC are located between residues 10 and 27 of V3 (Fig. 3), compared with five of nine (LAV) and four of nine (SF2) in the same region. At least within this region, JC and NC isolates seem to have relatively high net positive charges of 4 (relative to SF2) and 3 (relative to LAV). Other researchers have shown that changes in basic amino acids in the middle portion of V3 loop (residues 10 to 27 in Fig. 3) can alter the syncytium-inducing properties and phenotype of the virus (3, 11, 37).
FIG. 3.
Alignment of the V3 regions of HIV-1 isolates JC16, NC7, LAV, SF2, and NDK. Dots denote identity, while dashes denote deletions. The region most sensitive to changes in basic amino acids is between residues 10 and 27 (36). The GYGRS and GPGRA pentapeptides are boxed.
Replication kinetics in PBMC.
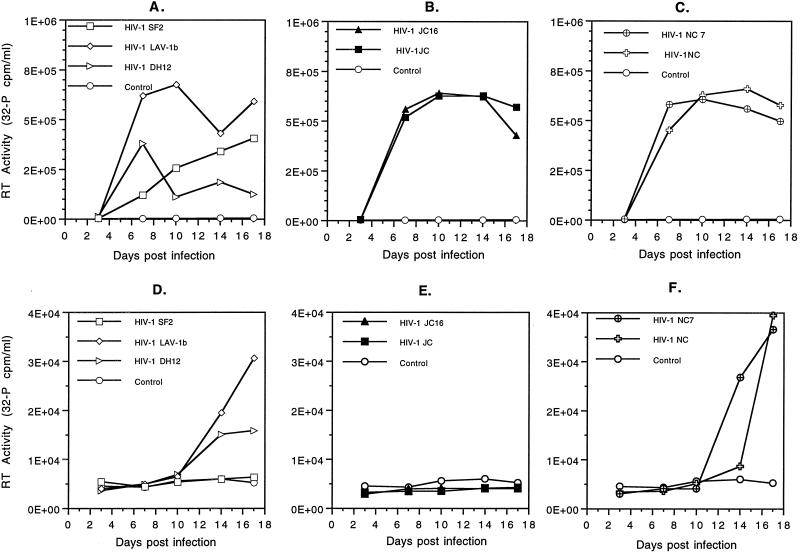
To begin an analysis of the biological activities of the cloned and uncloned viruses derived from C499 and C455, we conducted in vitro replication studies. The results of replication studies in mitogen-stimulated cPBMC are shown in Fig. 4a to c. Two of the three viruses used for inoculation of C499, SF2 and LAV-1b, were able to replicate in stimulated cPBMC, albeit with different kinetics (Fig. 4a). The SF2 isolate grew very slowly and to low titers in cPBMC. In contrast, the LAV-1b isolate grew very well and rapidly, with a high titer already present by day 7 postinfection. The SF2 isolate was unable to induce detectable syncytium formation in cPBMC. Under these conditions, the LAV-1b isolate induced very few syncytia. Included in these analyses was the DH12 isolate of HIV-1 (42). This primary isolate from a human has been shown to be highly cytopathic for cPBMC. While this virus quickly established infection in the stimulated cell population, it did not grow to high titers. Numerous syncytia were formed by infection with DH12 and could account for the lack of growth observed. The uncloned and cloned viruses of JC (Fig. 4b) and NC (Fig. 4c) replicated to levels comparable to those of LAV-1b for the same period of time. The rates of replication for cloned and uncloned viruses were indistinguishable.
FIG. 4.
Replication of cloned and uncloned HIV-1 isolates in ConA-stimulated and unstimulated cPBMC. Stimulated cPBMC infected with LAV-1b and SF2 parental HIV-1 strains and the highly cytopathic DH12 isolate (A), JC (uncloned) and JC16 (cloned) isolates of HIV-1 (B), and NC (uncloned) and NC7 (cloned) isolates of HIV (C). Unstimulated cPBMC were infected with LAV-1b, SF2, and DH12 (D), JC and JC16 (E), and NC and NC7 (F). Chimpanzee PBMC (107) in T-25 flasks were infected with 20 ng of either HIV-1JC16 (molecular clone), HIV-1JC (uncloned), HIV-1NC7 (molecular clone), HIV-1NC (uncloned), HIV-1SF2, HIV-1LAV-1b, or HIV-1DH12 and incubated for a total of 17 days at 37°C. Supernatant aliquots were made on 3, 7, 10, 14, and 17 days postinfection. RT assays were performed as outlined in Materials and Methods. Data shown are representative of three separate experiments yielding similar results.
The ability of HIV-1 virus isolates to replicate in unstimulated cPBMC was evaluated in a similar manner. Results of this assay (Fig. 4d to f) showed that only the NC (cloned and uncloned) and the LAV-1b isolates of HIV-1 were capable of significant replication in unstimulated cPBMC. Peak titers of these viruses in unstimulated cells were less than 10% of those observed in stimulated cPBMC. Additionally, virus production in unstimulated cells was much slower than that observed in stimulated cPBMC. Interestingly, JC (cloned and uncloned) isolates of HIV-1 failed to replicate in cPBMC, which suggests an inherent biological difference between the JC and NC viruses. The DH12 isolate was able to replicate in unstimulated cells, but to levels much lower than those of the other viruses. The SF2 isolate was unable to replicate in unstimulated PBMC. In general, virus recovered from the molecular clones displayed the intrinsic replicative properties exhibited by the viruses from which they were derived.
Macrophage tropism of viruses.
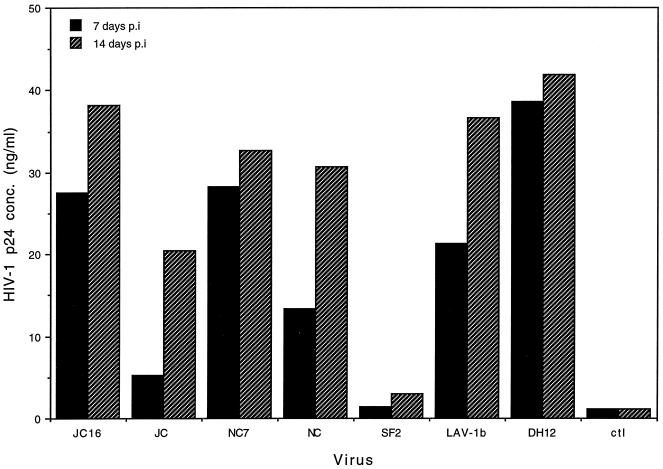
The ability of HIV-1 isolates to replicate well in chimpanzee macrophages has been controversial and the source of some hypotheses concerning the lack of HIV-1 pathogenicity in chimpanzees. To investigate the abilities of these viruses and their respective clones to replicate in macrophages, we conducted in vitro assays using purified chimpanzee MDM. Figure 5 shows virus production in macrophages at 7 and 14 days postinfection. Both JC and NC virus isolates (cloned and uncloned) replicated in MDM, as determined by ELISA analysis of the levels of p24 antigen produced. However, the amount of virus produced by the HIV-1JC16 molecular clone at 14 days postinfection was twice that produced by the HIV-1JC uncloned stock. The titers for cloned and uncloned HIV-1NC isolates were comparable for the same period of time. Among the viruses that were tropic for MDM, HIV-1JC (uncloned) produced the least amount of virus. HIV-1LAV-1b and HIV-1DH12 also infected MDM and produced virus, which is consistent with previous observations of Gendelman et al. (24) and Shibata et al. (42), respectively. Unlike HIV-1LAV-1b, the other parental virus, HIV-1SF2, did not replicate in MDM.
FIG. 5.
Replication of HIV-1 isolates in chimpanzee MDM. Purified PBMC (6 × 106/well) were used to obtain MDM. Ten nanograms of virus (HIV-1JC16 [molecular clone], HIV-1JC [uncloned], HIV-1NC7 [molecular clone], HIV-1NC [uncloned], HIV-1SF2, HIV-1LAV-1b, and HIV-1DH12) was used for infection; on days 7 and 14 postinfection (p.i.), supernatants were harvested. The amount of virus in the supernatants was determined by HIV-1 p24 antigen capture ELISA (Coulter). ctl, supernatants from control uninfected cultures. Data shown are representative of three separate experiments yielding similar results.
Heterogeneity of HIV-1 in PBMC of chimpanzees with AIDS.
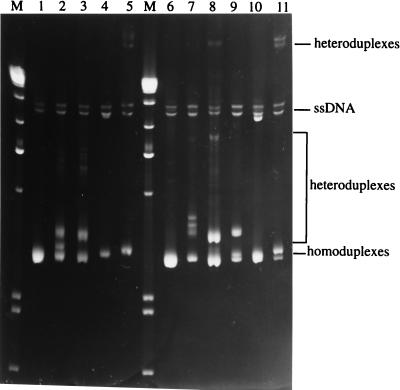
Finally, because the in vitro biologic activities of the cloned viruses were so similar to those of the uncloned stocks, we examined whether these viruses represented major species in the viral mix. Genomic DNA was isolated, and nested PCR was used to amplify the V1-V2 and V3-V5 regions, the hypervariable regions of the HIV-1 env gene with which most viral heterogeneity has been associated. Equal amounts of PCR products were mixed, heat denatured, and then reannealed with analogous fragments derived from standards (SF162 subtype B3 and ZM18 subtype C2) or from JC16 and NC7 molecular clones. Electrophoresis of these fragments on nondenaturing polyacrylamide gels revealed differences in migration rates, with homoduplexes moving faster in the gel than heteroduplexes. Homoduplexes based on genomic DNA (HIV-1JC or HIV-1NC) resulted in homoduplex (lower) and heteroduplex (upper) bands that had comparable intensities and were close together (Fig. 6, lanes 3 [HIV-1JC] and 8 [HIV-1NC]). The HIV-1JC/HIV-1JC16 (but not HIV-1NC/HIV-1NC7) heteroduplex band was between the homoduplex and heteroduplex bands obtained with genomic DNA homoduplexes (Fig. 6, lanes 2 and 7). The distance between homoduplex and heteroduplex bands formed between JC16 and NC7 molecular clones (lane 9) was larger than that between bands formed from homoduplexes derived from genomic DNA (lanes 3 and 8). The SF162 (subtype B3) heteroduplex bands migrated at almost the same rate as the single-stranded DNA (lanes 4 and 10), whereas the ZM18 (subtype C2) heteroduplex bands migrated most slowly (lanes 5 and 11).
FIG. 6.
HMA for the V3-V5 region of Env. The V3-V5 regions of Env for HIV-1 isolates were PCR amplified as outlined in Materials and Methods. Ten microliters of PCR product were used for homoduplex formation, while equal amounts (5 μl) of PCR products derived from different HIV-1 virus DNA were mixed to form heteroduplexes. The PCR products were denatured for 2 min at 94°C followed by rapid cooling in ice water. The samples were resolved on a nondenaturing polyacrylamide gel (6%) at a constant voltage of 250 V for 2.5 h. The gel was stained in ethidium bromide solution and visualized under UV light. Lanes: M, 1-kb molecular weight markers; 1, HIV-1JC16 homoduplex; 2, HIV-1JC16-HIV-1JC (genomic DNA) heteroduplex; 3, HIV-1JC (genomic DNA) homoduplex; 4, HIV-1JC16-HIV-1SF162 heteroduplex; 5, HIV-1JC16-HIV-1ZM18 heteroduplex; 6, HIV-1NC7 homoduplex; 7, HIV-1NC7-HIV-1NC (genomic DNA) heteroduplex; 8, HIV-1NC (genomic DNA) homoduplex; 9, HIV-1JC16-HIV-1NC7 heteroduplex; 10, HIV-1NC7-HIV-1SF162 heteroduplex; 11, HIV-1NC7-HIV-1ZM18 heteroduplex. HIV-1SF162 (HIV-1 subtype B3) and HIV-1ZM18 (subtype C2) plasmid DNAs are provided in the HMA kit as control DNAs. ssDNA, single-stranded DNA.
DISCUSSION
Earlier studies showed that chimpanzees could easily be infected with a number of strains of HIV-1. Despite this fact, the use of chimpanzees in pathogenesis research was all but abandoned because of the lack of evidence of disease development. Indeed, studies then focused on determining why these animals did not develop AIDS. Recently, we reported on the incidence of AIDS in one chimpanzee (C499) and on another (C455) that suffered a dramatic and sustained decline in CD4+ cell count following transfusion of blood from C499 (36). It was our hypothesis that the virus present in C499 was a mutant of the original infecting viruses and that the changes led to a virus more pathogenic than either of the parental strains. To begin to investigate this hypothesis, virus stocks were generated from both C499 and C455-(HIV-1JC and HIV-1NC, respectively), and molecular clones were prepared from both viruses. These viruses and clones were used for various biological and molecular studies.
Nucleotide sequence analysis of these molecular clones revealed that HIV-1JC and HIV-1NC were very similar. For the entire genome, all mutations between isolates JC16 and NC7 were due to point mutations except for two amino acid insertions in the matrix protein (p17) and a single amino acid deletion in Env of NC7 relative to JC16. The Env glycoprotein had the most, i.e., 17, point mutations, followed by Pol, while the Vpu protein was the most conserved (Table 1). The virus functional domains (zinc finger motifs in Gag, Lck binding domain in Nef, CD4 binding domain in Env, and transcription and regulatory sequences in LTR) were well conserved.
Several regions of JC and NC isolates had specific sequences inserted or deleted (in addition to point mutations) relative to the parental strains, as outlined in Results. From the alignment analysis data, it appears that specific regions of Gag had amino acid sequences derived from isolates LAV-1b (matrix protein) and SF2 (nucleocapsid protein), suggested by the presence of similar deletion patterns with these parental viruses. The N-terminal portions of Nef of JC16 and NC7 also had deletions of amino acid sequences, relative to the SF2 isolate, that indicated a greater similarity with LAV in this region. Because of the role of Env in a number of functional aspects of HIV pathogenesis, we focused on a detailed analysis of its origins. In the Env protein, there were no obvious sequences that could be directly associated with either of the parental strains. However, comparison of the individual hypervariable regions of JC16 and NC7 with those of parental strains indicated that there was more homology with the LAV strain than with the SF2 strain. The signal peptide of progeny viruses was clearly derived from the LAV-1b strain, as the deletion and insertion patterns were similar. Since the Env proteins of JC16 and NC7 had higher homologies with the LAV (81.9%) than with the SF2 (76.1%) isolate, it is logical to conclude that the majority of Env was derived from the LAV-1b isolate.
Although the V3 regions of JC16 and NC7 were identical, there were profound differences in this region compared with the parental strains. Elevated basic charge in the V3 region has been associated with conversion of non-syncytium-inducing to syncytium-inducing phenotype in HIV-1 (3, 11, 36), a possible explanation for syncytium-inducing properties observed for JC and NC isolates. Additionally, it is reasonable to hypothesize that the shift from the GPGRA pentapeptide in the V3 region of parental strains to GYGRS in JC16 and NC7 could alter the architecture of the principal neutralizing antibody epitope (12) with the possible outcome of the generation of neutralization variant viruses. Finally, changes in the V3 area may also have a potential effect on coreceptor usage of viruses. While all of these hypotheses will require additional investigations, it is possible to speculate that the evolution of mutant viruses escaping neutralization and with syncytium-inducing capabilities in vivo may have resulted in more aggressive disease progression that led to the development of clinical AIDS in C499.
Recently, Wei and Fultz (44) described changes in several genomic regions of the virus derived from C499. These data are based on a sampling obtained 22 months before the death of C499 and 17 months prior to our sampling for the studies presented here. The data shown here and in our previous publication (36) are derived from viruses obtained during the disease state. Comparative analyses of the amino acid sequence contained in the C2-V5 regions of Env of JC16 and NC7 with those samples of Wei and Fultz show that these sequences are closely related (Fig. 7) The alignment presented in Fig. 7 shows sequences of JC16 and NC7 along with those of the closest and most distant clones of Wei and Fultz. JC16 and NC7 contain point mutations not observed in any of the clones obtained by Wei and Fultz, which suggests that the isolates diverged further between the time of samplings. Indeed, results previously published by us found no evidence of SF2-related V1-V2 sequences in the general population, despite the use of conserved PCR primers.
FIG. 7.
Alignment of C2-V5 regions of Env proteins derived from chimpanzee C499. By using the Lasergene program (DNAStar), the deduced C2-V5 regions of Env from JC16, NC7, and the most closely related (#19; GenBank accession no. U56872 and most distant (#25; GenBank accession no. U56887) clones derived by Wei and Fultz (44) were aligned.
In agreement with Wei and Fultz, our data on alignment of env clones derived from C499 genomic DNA suggest extensive diversity of HIV-1. However, sequence alignments of Nef and the V4-V5 regions of JC16 and NC7 did not indicate the parental sequences from which they were derived, contrary to data presented by Wei and Fultz (44). The V3 region of Env for JC16 has 63.9% homology with that of LAV and 40.0% homology with that of SF2, suggesting that the progeny viruses may have derived this sequence from LAV-1b and not from SF2. The discrepancy in the results of Wei and Fultz (44) and the data presented here could be due to the quasi-species nature of HIV-1, where the major species isolated at the time was different. Another factor could be the differences in time at which samples were taken. It is possible that our data are based on a much more aggressive and pathogenic virus, since the animal showed clinical signs of AIDS and deteriorated soon thereafter.
Because lentiviruses are replicated by RT that lacks DNA editing functions, a swarm of related modified viruses that are referred to as quasi-species result in vivo. Indeed, results of nucleotide sequence analysis of proviral clones from the env region (data not shown and reference 36) indicated that several similar but different genomes were present in the sample. To evaluate whether the molecularly cloned viruses (JC16 and NC7) represented the major virus species in cPBMC, the V1-V2 and V3-V5 regions of env were amplified from genomic DNA and subjected to HMA. The vast majority of heteroduplexes formed between JC16 and NC7 molecular clones and PBMC genomic DNA were close to homoduplexes of JC16 and NC7, indicating that the variation between viruses in the regions analyzed were minimal. Since there is 97.4% homology between JC16 and NC7 in the env gene region (Table 1) and heteroduplexes between JC16 and NC7 had migration distances similar to those between them and the genomic DNA from which they were derived (Fig. 6), we conclude that the molecular clones of HIV-1JC and HIV-1NC represented virus of the major species in PBMC at the time of sampling.
In vitro analysis of biological characteristics of the uncloned and cloned virus stocks illustrated the highly related nature of these viruses. The replication rates for the JC- and NC-derived viruses were very impressive in cPBMC and were much higher than for the SF2 isolate (as shown here). However, while the replicative abilities of NC- and JC-derived viruses were similar to that of the LAV-1b isolate, the former induced higher levels of cytopathic effects (syncytium formation), suggestive of a more pathogenic virus. Although LAV-1b has been previously found to induce syncytia, (43), it induced very little syncytium formation in cPBMC in our hands. The comparable replicative abilities of the cloned viruses, JC16 and NC7, and the uncloned viruses are consistent with results obtained in HMA analyses.
Replication experiments using unstimulated PBMC showed that HIV-1NC and its clone were able to grow. This is similar to the well-known ability of the acutely lethal simian immunodeficiency virus variant SIVsmmPBj14 to grow in unstimulated macaque PBMC (19). The exact consequences of this ability are unknown in this instance but could possibly contribute to pathogenesis by providing a more rapid expansion of productively infected cells. This hypothesis will need to be tested in vivo.
The role that macrophages play in the pathogenesis of AIDS is still not fully understood. However, macrophages are an important reservoir of HIV-1 in infected humans. In addition, macrophages have important regulatory immune functions which could be compromised by HIV-1 infection (reviewed in references 30 and 35). The inability of HIV-1 isolates to replicate well in chimpanzee macrophages has been postulated as one reason why infected chimpanzees do not develop AIDS (33). While two recently described primary HIV-1 isolates (DH12 and SG3) have been shown to replicate to high titers in human macrophages as well as in chimpanzee PBMC (25, 42), the ability of these isolates to cause disease in chimpanzees has yet to be demonstrated. The development of AIDS in C499 and the subsequent rapid decline in CD4+ cells in an animal transfused with blood from C499 strongly suggests that the virus derived from C499 is pathogenic for chimpanzees. Thus, we tested the abilities of the JC- and NC-derived viruses to replicate in chimpanzee macrophages. Both the uncloned stocks and the viruses derived from molecular clones replicated well in macrophage populations. These replicative abilities were similar to those of the DH12 isolate mentioned above. The SF2 isolate failed to replicate in chimpanzee macrophages.
Taken together, the results presented in these studies suggest that a long-term accumulation of mutations has resulted in the generation of a virus which is pathogenic for chimpanzees. While the results presented here are suggestive of a recombination event, the length of time of virus replication (and mutation) in C499 does not permit the conclusion that recombination occurred. Additional genetic analyses of earlier isolates will be more informative in this area. The in vitro replicative abilities of viruses isolated from C499 and C455, and their respective molecular clones, appear to differ from those of the parental viruses used for inoculation. The fact that the viruses derived from the molecular clones behave similarly to the uncloned parental viruses in in vitro assays and are indistinguishable from the major species in an uncloned population in HMA suggests that these viruses may be pathogenic in vivo. The molecular clones described here will be valuable in helping define the genetic components involved in the phenotypic changes observed with isolates JC and NC. Additionally, genetic analysis of earlier viral isolates derived from C499 may help in elucidating the genetic changes associated with the pathogenic events leading to the development of AIDS.
ACKNOWLEDGMENTS
The HMA kit (contributed by Eric Delwart, Belinda Herangi, and James Mullins) and HIV-1SF2 isolate (contributed by Jay Levy) were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIH. We thank Malcolm Martin for the generous gift of the HIV-1DH12 isolate. We thank Harriet Robinson for critical review of the manuscript. We also thank Shawn O’Neil, Michelle Saucier, Terri Gibson, and Juliette deRosayro for technical help, and we are grateful for the expert care provided to the chimpanzees by the primate care technicians at the Yerkes Center.
This work was supported by NIH grant RO1 AI-40879 to F.J.N. and by grant RR-00165.
REFERENCES
- 1.Alexander S, Elder J H. Carbohydrate dramatically influences immune reactivity of antisera to viral glycoprotein antigens. Science. 1984;226:1328–1332. doi: 10.1126/science.6505693. [DOI] [PubMed] [Google Scholar]
- 2.Barre-Sinoussi F, Chermann J C. The etiologic agent of AIDS. Mt Sinai J Med. 1986;53:598–608. [PubMed] [Google Scholar]
- 3.Bhattacharyya D, Brooks B R, Callahan L. Positioning of positively charged residues in the V3 loop correlates with HIV-1 type 1 syncytium-inducing phenotype. AIDS Res Hum Retroviruses. 1996;12:83–90. doi: 10.1089/aid.1996.12.83. [DOI] [PubMed] [Google Scholar]
- 4.Boyer J D, Wang B, Ugen K E, Agadjanyan M, Javadian A, Frost P, Dang K, Carrano R A, Ciccarelli R, Coney L, Williams W V, Weiner D B. In vivo protective anti-HIV immune responses in non-human primates through DNA immunization. J Med Primatol. 1996;25:242–250. doi: 10.1111/j.1600-0684.1996.tb00022.x. [DOI] [PubMed] [Google Scholar]
- 5.Broder S, Gallo R C. A pathogenic retrovirus (HTLV-III) N Engl J Med. 1984;311:1292–1297. doi: 10.1056/NEJM198411153112006. [DOI] [PubMed] [Google Scholar]
- 6.Camerin D, Seed B. A CD4 domain important for HIV-mediated syncytium formation lies outside the virus binding site. Cell. 1990;60:747–754. doi: 10.1016/0092-8674(90)90089-w. [DOI] [PubMed] [Google Scholar]
- 7.Corey L, McElrath M J, Weinhold K, Matthews T, Stablein D, Graham B, Keefer M, Schwartz D, Gorse G. Cytotoxic T cell and neutralizing antibody responses to human immunodeficiency virus type 1 envelope with a combination vaccine regimen. AIDS Vaccine Evaluation Group. J Infect Dis. 1998;177:301–309. doi: 10.1086/514202. [DOI] [PubMed] [Google Scholar]
- 8.Dawn P W, Smith R A, Czajak S, Desrosiers R C. Direct demonstration of retroviral recombination in a rhesus monkey. J Virol. 1997;71:9650–9653. doi: 10.1128/jvi.71.12.9650-9653.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Delwart E L, Shpaer E G, Louwagie J, McCutchan F E, Grez M, Rubsamen-Waigmann H, Mullins J I. Genetic relationships determined by a DNA heteroduplex mobility assay: analysis of HIV-1 env genes. Science. 1993;262:1257–1261. doi: 10.1126/science.8235655. [DOI] [PubMed] [Google Scholar]
- 10.De Mareuil J, Salaun D, Chermann J C, Hirsch I. Fusogenic determinants of highly cytopathic subtype D Zairean isolate HIV-1 NDK. Virology. 1995;209:649–653. doi: 10.1006/viro.1995.1298. [DOI] [PubMed] [Google Scholar]
- 11.De Wolf F, Hogervorst E, Goudsmit J, Fenyo E M, Rubsamen-Waigmann H, Holmes H, Galvao-Castro B, Karita E, Wasi C, Sempala S D, et al. Syncytium-inducing and non-syncytium-inducing capacity of human immunodeficiency virus type 1 subtypes other than B: phenotypic and genotypic characteristics. WHO Network for HIV Isolation and Characterization. AIDS Res Hum Retroviruses. 1994;10:1387–1400. doi: 10.1089/aid.1994.10.1387. [DOI] [PubMed] [Google Scholar]
- 12.di Marzo Veronese F, Reitz M S, Jr, Gupta G, Robert-Guroff M, Boyer-Thompson C, Louie A, Gallo R C, Lusso P. Loss of a neutralizing epitope by a spontaneous point mutation in the V3 loop of HIV-1 isolated from an infected laboratory worker. J Biol Chem. 1993;268:25894–25901. [PubMed] [Google Scholar]
- 13.Estaquier J, Idziorek T, DeBels F, Barre-Sinoussi F, Hurtel B, Aubertin A M, Venet A, Mehtali M, Muchmore E, Michel P, Mouton Y, Girard M, Ameisen J C. Programmed cell death and AIDS: significance of T-cell apoptosis in pathogenic and nonpathogenic primate lentiviral infections. Proc Natl Acad Sci USA. 1994;91:9431–9435. doi: 10.1073/pnas.91.20.9431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Francis D P, Feorino P M, Broderson J R, McClure H M, Getchell J P, McGrath C R, Swenson B, McDougal J S, Palmer E L, Harrison A K, et al. Infection of chimpanzees with lymphoadenopathy-associated virus. Lancet. 1984;ii:1276–1277. doi: 10.1016/s0140-6736(84)92824-1. [DOI] [PubMed] [Google Scholar]
- 15.Fultz P N, McClure H M, Swenson R B, McGrath C R, Brodie A, Getchell J P, Jensen F C, Anderson D C, Broderson J R, Francis D P. Persistent infection of chimpanzees with human T-lymphotropic virus type III/lymphadenopathy-associated virus: a potential model for acquired immunodeficiency syndrome. J Virol. 1986;58:116–124. doi: 10.1128/jvi.58.1.116-124.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fultz P N, McClure H M, Daugharty H, Brodie A, McGrath C R, Swenson B, Francis D P. Vaginal transmission of human immunodeficiency virus (HIV) to a chimpanzee. J Infect Dis. 1986;154:896–900. doi: 10.1093/infdis/154.5.896. [DOI] [PubMed] [Google Scholar]
- 17.Fultz P N, Srinivasan A, Greene C R, Butler D, Swenson R B, McClure H M. Superinfection of chimpanzee with a second strain of human immunodeficiency virus. J Virol. 1987;61:4026–4029. doi: 10.1128/jvi.61.12.4026-4029.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fultz P N, McClure H M, Swenson R B, Anderson D C. HIV-1-infection of chimpanzees as a model for testing chemotherapeutics. Intervirology Suppl. 1989;1:51–58. doi: 10.1159/000150124. [DOI] [PubMed] [Google Scholar]
- 19.Fultz P N. Replication of an acutely lethal simian immunodeficiency virus activates and induces proliferation of lymphocytes. J Virol. 1991;65:4902–4909. doi: 10.1128/jvi.65.9.4902-4909.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fultz P N. Nonhuman primate models for AIDS. Clin Infect Dis. 1993;17:S230–S235. doi: 10.1093/clinids/17.supplement_1.s230. [DOI] [PubMed] [Google Scholar]
- 21.Fultz P N, Yue L, Wei Q, Girard M. Human immunodeficiency virus type 1 intersubtype (B/E) recombination in a superinfected chimpanzee. J Virol. 1997;71:7990–7995. doi: 10.1128/jvi.71.10.7990-7995.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gajdusek D C, Amyx H L, Gibbs C J, Jr, Asher D M, Rodgers-Johnson P, Epstein L G, Sarin P S, Gallo R C, Maluish A, Arthur L O, et al. Infection of chimpanzee by human T-lymphotropic retroviruses in brain and other tissues from AIDS patients. Lancet. 1985;i:55–56. doi: 10.1016/s0140-6736(85)91011-6. [DOI] [PubMed] [Google Scholar]
- 23.Gallo R C, Wong-Staal F. A human T-lymphotropic retrovirus (HTLV-III) as the cause of the acquired immunodeficiency syndrome. Ann Intern Med. 1985;103:679–689. doi: 10.7326/0003-4819-103-5-679. [DOI] [PubMed] [Google Scholar]
- 24.Gendelman H E, Ehrlich G D, Baca L M, Conley S, Ribas J, Kalter D C, Meltzer M S, Poiesz B J, Nara P. The inability of human immunodeficiency virus to infect chimpanzee monocytes can be overcome by serial viral passage in vivo. J Virol. 1991;65:3853–3863. doi: 10.1128/jvi.65.7.3853-3863.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ghosh S K, Fultz P N, Keddie E, Saag M S, Sharp P M, Hahn B H, Shaw G M. A molecular clone of HIV-1 tropic and cytopathic for human and chimpanzee lymphocytes. Virology. 1993;194:858–864. doi: 10.1006/viro.1993.1331. [DOI] [PubMed] [Google Scholar]
- 26.Girard M, Barre-Sinoussi F, van der Ryst E. Vaccination of chimpanzees against HIV-1. Antibiot Chemother. 1996;48:121–124. doi: 10.1159/000425166. [DOI] [PubMed] [Google Scholar]
- 27.Girard M, Meignier B, Barre-Sinoussi F, Kieny M P, Matthews T, Muchmore E, Nara P L, Wei Q, Rimsky L, Weinhold K, et al. Vaccine-induced protection of chimpanzees against infection by a heterologous human immunodeficiency virus type 1. J Virol. 1995;69:6239–6248. doi: 10.1128/jvi.69.10.6239-6248.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hahn B H, Shaw G M, Arya S K, Popovic M, Gallo R C, Wong-Staal F. Molecular cloning and characterization of the HTLV-III virus associated with AIDS. Nature. 1984;312:166–169. doi: 10.1038/312166a0. [DOI] [PubMed] [Google Scholar]
- 29.Hanke T, Blanchard T J, Schneider J, Ogg G S, Tan R, Becker M, Gilbert S C, Hill A V, Smith G L, McMichael A. Immunogenicities of intravenous and intramuscular administrations of modified vaccinia virus Ankara-based multi-CTL epitope vaccine for human immunodeficiency virus type 1 in mice. J Gen Virol. 1998;79:83–90. doi: 10.1099/0022-1317-79-1-83. [DOI] [PubMed] [Google Scholar]
- 30.Horrowitz M C, Friedlaender G E, Qian G Y. The immune system: the efferent arm. Clin Orthop. 1996;326:25–34. doi: 10.1097/00003086-199605000-00004. [DOI] [PubMed] [Google Scholar]
- 31.Levy J A, Hoffman A D, Kramer S M, Landis J A, Shimabukuro J M, Oshiro L S. Isolation of lymphocytopathic retroviruses from San Francisco patients with AIDS. Science. 1984;225:840–842. doi: 10.1126/science.6206563. [DOI] [PubMed] [Google Scholar]
- 32.Morrow W J, Homsy J, Eichberg J W, Krowka J, Pan L Z, Gaston I, Legg H, Lerche N, Thomas J, Levy J A. Long-term observation of baboons, rhesus monkeys, and chimpanzees inoculated with HIV and given periodic immunosuppressive treatment. AIDS Res Hum Retroviruses. 1989;5:233–245. doi: 10.1089/aid.1989.5.233. [DOI] [PubMed] [Google Scholar]
- 33.Nara P L, Hutch W, Kessler J, Kelliher J, Carter S. The biology of human immunodeficiency virus-1 IIIB infection in the chimpanzee: in vivo and in vitro correlations. J Med Primatol. 1989;18:343–355. [PubMed] [Google Scholar]
- 34.Nara P L, Robey W G, Arthur L O, Asher D M, Wolff A V, Gibbs C J, Gajdusek D C, Fischinger P J. Persistent infection of chimpanzees with human immunodeficiency virus: serological responses and properties of reisolated viruses. J Virol. 1987;61:3173–3180. doi: 10.1128/jvi.61.10.3173-3180.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nathan C F. Secretory products of macrophages. J Clin Invest. 1987;79:319–326. doi: 10.1172/JCI112815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Novembre F J, Saucier M, Anderson D C, Klumpp S A, O’Neil S P, Brown II C R, Hart C E, Guenthner P C, Swenson R B, McClure H M. Development of AIDS in a chimpanzee infected with human immunodeficiency virus type 1. J Virol. 1997;71:4086–4091. doi: 10.1128/jvi.71.5.4086-4091.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Okada T, Patterson B K, Otto P A, Gurney M E. HIV-1 type 1 infection of CD4+ T cells depends critically on basic amino acid residues in the V3 domain of envelope glycoprotein 120. AIDS Res Hum Retroviruses. 1994;10:803–811. doi: 10.1089/aid.1994.10.803. [DOI] [PubMed] [Google Scholar]
- 38.Salminen M O, Koch C, Sanders-Buell E, Ehrenberg P E, Michael N L, Carr J K, Burke D S, McCutchan F E. Recovery of virtually full length HIV-1 provirus of diverse subtypes from primary virus cultures using the polymerase chain reaction. Virology. 1995;213:80–86. doi: 10.1006/viro.1995.1548. [DOI] [PubMed] [Google Scholar]
- 39.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 40.Sasaki S, Fukushima J, Arai H, Kusakabe K I, Hamajima K, Ishii N, Hirahara F, Okuda K, Kawamoto S, Ruysschaert J M, Vandenbranden M, Wahren B, Okuda K. Human immunodeficiency virus type 1-specific immune responses induced by DNA vaccination are greatly enhanced by mannan-coated diC14-amidine. Eur J Immunol. 1997;27:3121–3129. doi: 10.1002/eji.1830271207. [DOI] [PubMed] [Google Scholar]
- 41.Schwartz D, Sharma U, Bucsch M, Weinhold K, Matthews T, Lieberman J, Birx D. Lack of recoverable infectious virus and unique immune responses in an asymptomatic HIV+ long-term survivor. AIDS Res Hum Retroviruses. 1994;10:1703–1711. doi: 10.1089/aid.1994.10.1703. [DOI] [PubMed] [Google Scholar]
- 42.Shibata R, Hoggan M D, Broscius C, Englund G, Theodore T S, Buckler-White A, Arthur L O, Israel Z, Schultz A, Lane H C, Martin M A. Isolation and characterization of a syncytium-inducing, macrophage/T-cell line-tropic human immunodeficiency virus type 1 isolate that readily infects chimpanzee cells in vitro and in vivo. J Virol. 1995;69:4453–4462. doi: 10.1128/jvi.69.7.4453-4462.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Watanabe M, Ringler D J, Fultz P N, Mackey J J, Boyson J E, Levine C G, Letvin N L. A chimpanzee-passaged human immunodeficiency virus isolate is cytopathic for chimpanzee cells but does not induce disease. J Virol. 1991;65:3344–3348. doi: 10.1128/jvi.65.6.3344-3348.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wei Q, Fultz P N. Extensive diversification of human immunodeficiency virus type 1 subtype B strains during dual infection of a chimpanzee that progressed to AIDS. J Virol. 1998;72:3005–3017. doi: 10.1128/jvi.72.4.3005-3017.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zarling J M, Ledbetter J A, Sias J, Fultz P, Eichberg J, Gjerset G, Moran P A. HIV-infected humans, but not chimpanzees have circulating cytotoxic T lymphocytes that lyse uninfected CD4+ cells. J Immunol. 1990;144:2992–2998. [PubMed] [Google Scholar]
- 46.Zhang J, Novembre F, Rabson A B. Simian immunodeficiency viruses containing mutations in the long terminal repeat NF-kappa B or Sp1 binding sites replicate efficiently in T cells and PHA-stimulated PBMCs. Virus Res. 1997;49:205–213. doi: 10.1016/s0168-1702(97)01462-7. [DOI] [PubMed] [Google Scholar]