Abstract
Large language models (LLMs) have shown promise in accelerating radiology reporting by summarizing clinical findings into impressions. However, automatic impression generation for whole-body PET reports presents unique challenges and has received little attention. Our study aimed to evaluate whether LLMs can create clinically useful impressions for PET reporting. To this end, we fine-tuned twelve open-source language models on a corpus of 37,370 retrospective PET reports collected from our institution. All models were trained using the teacher-forcing algorithm, with the report findings and patient information as input and the original clinical impressions as reference. An extra input token encoded the reading physician’s identity, allowing models to learn physician-specific reporting styles. To compare the performances of different models, we computed various automatic evaluation metrics and benchmarked them against physician preferences, ultimately selecting PEGASUS as the top LLM. To evaluate its clinical utility, three nuclear medicine physicians assessed the PEGASUS-generated impressions and original clinical impressions across 6 quality dimensions (3-point scales) and an overall utility score (5-point scale). Each physician reviewed 12 of their own reports and 12 reports from other physicians. When physicians assessed LLM impressions generated in their own style, 89% were considered clinically acceptable, with a mean utility score of 4.08/5. On average, physicians rated these personalized impressions as comparable in overall utility to the impressions dictated by other physicians (4.03, P = 0.41). In summary, our study demonstrated that personalized impressions generated by PEGASUS were clinically useful in most cases, highlighting its potential to expedite PET reporting by automatically drafting impressions.
Keywords: Natural Language Processing, Large Language Models, Informatics, Nuclear Medicine, Positron Emission Tomography, Radiology Report Summarization
Introduction
The radiology report serves as the official interpretation of a radiological examination and is essential for communicating relevant clinical findings amongst reading physicians, the healthcare team, and patients. Compared to other imaging modalities, reports for whole-body PET exams (e.g., skull base to thigh or skull vertex to feet) are notable for their length and complexity [1]. In a typical PET report, the findings section details numerous observations about the study and the impression section summarizes the key findings, offers diagnoses, and provides follow-up recommendations. Given that referring physicians primarily rely on the impression section for clinical decision-making and management [2], it is paramount to ensure its accuracy and completeness. However, creating PET impressions that encapsulate all important findings can be time-consuming and error-prone [3]. Large language models (LLMs) have the potential to accelerate this process by automatically drafting impressions based on the findings.
In recent years, the use of LLMs for summarizing radiology findings has garnered considerable interest. Several research studies [3–8], and even commercial products, have used LLMs to automatically draft impressions based on findings, with the aim of accelerating clinical reporting workflows. Moreover, using LLMs to prepare impressions might help prevent critical findings from being omitted. Previous work has employed LLMs to summarize findings in x-ray, CT, and MRI reports, but no studies have focused on impression generation for PET reports. Compared to CT or MRI findings that often comprise 75–150 words [9], whole-body PET reports are substantially longer, with 250–500 words in the findings section, and contain observations across multiple anatomical regions with cross comparison to available anatomic imaging modalities. This complexity heightens the risk of omissions in the generated impressions. Furthermore, the length of PET impressions can accentuate the unique reporting styles of individual reading physicians, underscoring the need for personalizing impressions to the reading physician’s style. Consequently, adapting LLMs for PET report summarization presents distinct challenges.
Evaluating the performances of LLMs in the task of impression generation is also challenging, given that a single case can have various acceptable impressions. While expert evaluation stands as the gold standard, it is impractical for physicians to exhaustively review outputs of all LLMs to determine the leading model. In recent years, several evaluation metrics designed for general text summarization have been adapted to evaluate summaries of biomedical literature and clinical notes [10–12]. However, it remains unclear how well these metrics could assess the quality of PET impressions. A better understanding of which metrics align most closely with physician judgements is needed.
This study aimed to investigate whether open-source LLMs fine-tuned on a large corpus of PET clinical reports can accurately summarize PET findings and generate clinically useful impressions that are acceptable to physicians. To achieve this, we first determined which model performed best at the PET summarization task. This involved adapting multiple LLMs to the PET domain, benchmarking 30 automatic evaluation metrics against physician preferences, and subsequently using the benchmarked metrics to select the most proficient fine-tuned LLM. Then, we conducted an expert reader study to assess the quality of LLM-generated impressions from the perspective of reading physicians, focusing on the clinical utility, common mistakes, and the importance of tailoring impressions to physician-specific reporting styles. We also performed external testing of the LLM. As an additional evaluation, we assessed the LLM’s reasoning capability within the nuclear medicine (NM) domain by measuring its accuracy in predicting Deauville scores for PET lymphoma reports.
Methods
Dataset Collection
Under a protocol approved by the institutional review board and with a waiver of informed consent, we collected 37,370 retrospective PET reports, dictated by 65 physicians, from our institution between January 2010 and January 2023. Among all internal PET reports, 92.7% (34,655/37,370) pertained to PET/CT whole-body (including skull base to thigh and skull vertex to feet) scans, 1.7% (649/37,370) to PET/MRI whole-body scans, 5.5% (2,066/37,370) to PET limited area (including brain, cardiac and myocardial) scans. Our internal dataset reflects the demographic distribution of patients accepted to our healthcare system in the past 13 years, which consisted of both adult and pediatric cases. We did not exclude any subgroups of patients in model development and evaluation. The findings section in a PET report had 346 [249, 472] (median [25th percentile, 75th percentile]) words, and the impression section had 86 [53, 130] words. Reports were anonymized using NLM-Scrubber [13]. Of 37,370 PET reports, 4000 were randomly selected for internal testing, 2000 were used for validation, and the remaining 31,370 reports were used for training. For external testing, we used data from Children’s Oncology Group (COG) AHOD1331 Hodgkin lymphoma clinical trial (ClinicalTrials.gov number, NCT02166463) [14] as it is the only external dataset with PET reports available to us. We retrieved 100 whole-body PET/CT reports, dictated by 62 physicians. There is no overlap between physicians in the internal and external sets. The AHOD1331 data is archived in NCTN Data Archive, and a data use agreement has been signed between our institution and COG that allows for research use of the radiology reports. All subjects in this COG clinical trial were pediatric patients diagnosed with classical Hodgkin’s lymphoma. Patient information was redacted prior to our access.
Report Preprocessing
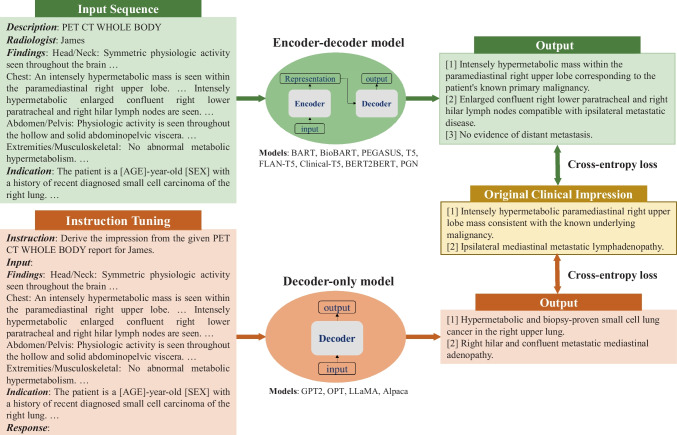
In this work, we investigated both encoder-decoder and decoder-only language models. Considering their different architectures, we customized input templates as illustrated in Fig. 1. For encoder-decoder models, the first lines describe the categories of PET scans, while the second lines encode each reading physician’s identity using an identifier token. The tokens associated with each physician are detailed in Appendix 1. The “Findings” section contains the clinical findings from the PET reports, whereas the “Indications” section encompasses relevant background information, including the patient’s medical history and the reason for the examination. For decoder-only models, we employed the instruction-tuning method [15] and adapted the prompt from [16]. Each case starts with the instruction: “Derive the impression from the given [description] report for [physician].” The PET findings and background information are concatenated to form the “Input” section. The original clinical impressions are used as the reference for model training and evaluation.
Fig. 1.
Formatting of reports for input to encoder-decoder and decoder-only models. For encoder-decoder models, the first two lines describe the examination category and encode the reading physician’s identity. “Findings” contains the clinical findings from the PET report, and “Indication” includes the patient’s background information. For decoder-only models, each case follows a specific format for the instruction: “Derive the impression from the given [description] for [physician]”. “Input” accommodates the concatenation of clinical findings and patient information. The output always starts with the prefix “Response:”. Both model architectures utilize the cross-entropy loss to compute the difference between original clinical impressions and model-generated impressions
Models for PET Report Summarization
We formulated our work as an abstractive summarization task since physicians typically interpret findings in the impression section, rather than merely reusing sentences from the findings section. We fine-tuned 8 encoder-decoder models and 4 decoder-only models, covering a broad range of open-source language models for sequence generation. The encoder-decoder models comprised state-of-the-art (SOTA) transformer-based models, namely BART [17], PEGASUS [18], T5 [19] and FLAN-T5 [20]. These models differ primarily in their pretraining objectives. BART was pretrained as a denoising auto-encoder, aiming to reconstruct original texts from corrupted samples. PEGASUS employed the gap sentence prediction objective, masking key sentences from documents and forcing the model to recover them based on the remaining sentences. T5 used the span-mask denoising objective, involving masking out contiguous spans of text and challenging the model to predict these masked spans. FLAN-T5, in comparison to T5, underwent further instruction fine-tuning in a mixture of tasks. To investigate if the medical-domain adaptation could benefit our task, we fine-tuned 2 biomedical LLMs, BioBART [21] and Clinical-T5 [22]. BioBART was pretrained on the PubMed dataset, while Clinical-T5 was trained using the MIMIC-III dataset [23]. Additionally, we included 2 baseline models, the pointer-generator network (PGN) [3] and BERT2BERT [24], which were considered as the previous SOTA methods in radiology impression generation.
The decoder-only models encompassed GPT2 [25] and OPT [26] as well as LLaMA [27] and Alpaca [16]. All these models are built on the transformer architecture and have been pretrained on vast text corpora for next token prediction.
All twelve language models were trained using the standard teacher-forcing algorithm. The training objective can be written as a maximum likelihood problem:
| 1 |
where denotes the parameters of model , estimates the probability of the next word given the previous sequence in the reference text and the source text . Superscript denotes the word position in the reference text and denotes a single sample. The AdamW optimizer [28] was used to optimize this log-likelihood loss. LLaMA and Alpaca were fine-tuned with low-rank adaptation (LoRA) [29] to allow for training on consumer-level GPUs like NVIDIA A100s, while the other models were subjected to full fine-tuning. We adopted the beam search decoding algorithm to generate impressions and set the number of beams to 4. More comprehensive information regarding the training settings of our models can be found in Appendix 2.
Our models are made available on Hugging Face: https://huggingface.co/xtie/PEGASUS-PET-impression.. The code can be found in the open-source project: https://github.com/xtie97/PET-Report-Summarization.
Benchmarking Evaluation Metrics
To identify the evaluation metrics most correlated with physician preferences, we presented impressions generated by 4 different models (PGN, BERT2BERT, BART, PEGASUS) to two NM physicians. These models represented a wide performance spectrum. One physician (M.S.) reviewed 200 randomly sampled reports in the test set, then scored the quality of model-generated impressions on a 5-point Likert scale (5 best, 1 worst). The definitions of each level are given in Table 1. To assess inter-observer variability, a second physician (S.Y.C.) independently scored 20 of the cases based on the same criterion.
Table 1.
Definition of the 5-point Likert scale for evaluating the quality of model-generated impressions
| Score | Definition |
|---|---|
| 5 | Clinically acceptable impressions. The generated impression is consistent with the key clinical findings and align with the physician’s impression. Well organized and readable |
| 4 | Nearly acceptable impressions. The generated impression is mostly consistent with the key clinical findings and aligns overall with the physician’s impression. Minor additions or subtractions. Organized and readable |
| 3 | Moderately acceptable impressions. The generated impression has some inconsistencies with the key clinical findings and mostly aligns with the physician’s impression. Moderate additions or subtractions |
| 2 | Unacceptable impressions. The generated impression is factually incorrect in parts and/or missing some key clinical findings and may not completely align with the physician’s impression. Major additions or subtractions |
| 1 | Unusable impressions. The generated impression is factually incorrect and/or misses most of the key clinically findings and does not align with the physician’s impression |
Table 2 categorizes the evaluation metrics included in this study. In-depth descriptions of these metrics are provided in Appendix 3. To address the domain gap between general-domain articles and PET reports, we fine-tuned BARTScore on our PET reports using the method described in [30] and named it BARTScore + PET. Following the same approach, we developed PEGASUSScore + PET and T5Score + PET. Unlike the LLMs for impression generation, these three evaluators (available at https://huggingface.co/xtie/BARTScore-PET.) estimated the semantic similarity between generated impressions and their respective references. The Spearman’s ρ correlation quantified how well evaluation metrics correlated with the physicians’ judgments. Metrics with the highest correlations were used to determine the top-performing model.
Table 2.
All evaluation metrics included in this study and their respective categories
| Category | Definition | Corresponding Evaluation Metrics |
|---|---|---|
| Lexical overlap-based metrics | These metrics measure the overlap between the generated text and the reference in terms of textual units, such as n-grams or word sequences | ROUGE-1, ROUGE-2, ROUGE-3, ROUGE-L, ROUGE-LSUM, BLEU, CHRF, METEOR, CIDEr |
| Embedding-based metrics | These metrics measure the semantic similarity between the generated and reference texts using pretrained embeddings | ROUGE-WE-1, ROUGE-WE-2, ROUGE-WE-3, BERTScore, MoverScore |
| Graph-based metrics | These metrics construct graphs using entities and their relations extracted from the sentences, and evaluate the summary based on these graphs | RadGraph |
| Text generation-based metrics | These metrics assess the quality of generated text by framing it as a text generation task using sequence-to-sequence language models | BARTScore, BARTScore + PET PEGASUSScore + PET, T5Score + PET, PRISM |
| Supervised regression-based metrics | These metrics require human annotations to train a parametrized regression model to predict human judgments for the given text | -pyr, -resp |
| Question answering-based metrics | These metrics formulate the evaluation process as a question-answering task by guiding the model with various questions | UniEval |
| Reference-free metrics | These metrics do not require the reference text to assess the quality of the generated text. Instead, they compare the generated text against the source document | SummaQA, BLANC, SUPERT, Stats-compression, Stats-coverage, Stats-density, Stats-novel trigram |
Note that we included 17 different evaluation methods to assess model performance. Given that each method might encompass multiple variants, we have a total of 30 metrics. A detailed overview of these metrics can be found in Appendix 3
Expert Evaluation
To examine the clinical utility of our best LLM, we conducted a reader study involving three physicians: two board-certified in NM (N.I., S.Y.C.) and one board-certified in NM and radiology (A.P.). Blinded to the original interpreting physicians, each reader independently reviewed a total of 24 whole-body PET reports and scored both original clinical impressions and model-generated impressions. Of these, twelve cases were dictated by themselves, and the rest were dictated by other physicians. The LLM impressions were always generated in the style of the interpreting physician by using their specific identifier token. The scoring system included 6 quality dimensions (3-point scale) and an overall utility score (5-point scale). Their definitions are described in Table 3. The application we designed for physician review of test cases can be accessed at https://github.com/xtie97/PET-Report-Expert-Evaluation..
Table 3.
Definitions of six quality dimensions and an overall utility score used in our expert evaluation, along with their corresponding Likert systems
| Evaluation Category | Definition | Likert System |
|---|---|---|
| Additions | The impression is not repetitive and does not include unnecessary findings |
3: No additions 2: Moderate additions 1: Excessive additions |
| Omissions | The impression contains all important findings |
3: No omissions 2: Moderate omissions 1: Significant omissions |
| Factual correctness | The impression accurately represents the findings and is devoid of factual errors |
3: Correct 2: Partially correct 1: Substantially incorrect |
| Clarity and organization | The impression is unambiguous, grammatical, and well-organized |
3: Good 2: Adequate 1: Poor |
| Interpretive and technical jargon | The impression provides appropriate interpretations of the findings and avoids using unnecessary radiologic jargon or details |
3: Appropriate 2: Partially appropriate 1: Inappropriate |
| Recommendations | The recommendations for patient management, if applicable, are clinically valid |
3: Appropriate 2: Partially appropriate 1: Inappropriate |
| Overall utility score | Given the impression as an initial draft, consider how many changes would you make to render it suitable for clinical use |
5: Acceptable with no changes needed 4: Acceptable with minor changes needed 3: Acceptable with moderate changes needed 2: Unacceptable with significant changes needed 1: Unusable |
Additional Analysis
To further evaluate the capability of our fine-tuned LLMs, we conducted three additional experiments.
Deauville Score (DS) Prediction: To test the reasoning capability of our models within the NM domain, we classified PET lymphoma reports into DS 1–5 based on the exam-level DSs extracted from model-generated impressions. We first identified cases with the term “Deauville” and its common misspellings in model-generated impressions and original clinical impressions. N-gram analysis was then performed to extract the score for each case. If multiple DSs were present in the impression, the highest value was used to represent the exam-level DS [31]. The original clinical impressions served as the reference for the DSs. The evaluation metrics included the 5-class accuracy and the linearly weighted Cohen’s index. For context, our prior study [31] showed that a human expert predicted DSs with 66% accuracy and a Cohen’s of 0.79 when the redacted PET reports and maximum intensity projection images were given.
Encoding Physician-specific Styles: We compared the impressions generated in the styles of two physicians (Physician 1 and Physician 2) who had distinct reporting styles. Physician 1’s impressions tended to be more detailed, whereas Physician 2’s impressions were more concise. To alter the style of the output impression, we directly replaced the original reading physician’s identifier token with the token associated with another physician.
External Testing: We generated the impressions for all cases in the COG AHOD1331 dataset. Since our model was not trained on this cohort, we had to pick physician styles from our internal dataset. We used the styles of three primary physicians (Physician 1, Physician 2, and Physician 3). Their reporting styles range from detailed (Physician 1) to concise (Physician 2), with Physician 3’s style being an intermediate between the two. The model-generated impressions were then compared with clinical impressions originally dictated by external physicians. For the evaluation metrics, the average values across the three physicians’ reporting styles were calculated and used to represent the model’s external performance.
Statistical analysis
Using bootstrap resampling [32], the 95% confidence intervals (CI) for our results were derived from 10,000 repetitive trials. The difference between two data groups was statistically significant at 0.05 only when one group exceeded the other in 95% of trials.
Results
Benchmarking evaluation metrics
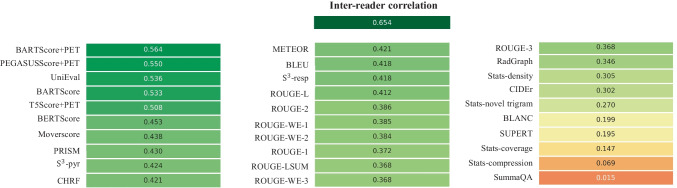
Figure 2 shows the Spearman's ρ correlation between evaluation metrics and quality scores assigned by the first physician (M.S.). BARTScore + PET and PEGASUSScore + PET exhibited the highest correlations with physician judgment (ρ = 0.568 and 0.563, P = 0.30). Therefore, both metrics were employed to determine the top-performing model for expert evaluation. However, their correlation values were still below the degree of inter-reader correlation (ρ = 0.654). Similar results were observed in the correlation between evaluation metrics and the second physician’s scores (available in Appendix 4). Without adaption to PET reports, the original BARTScore showed lower correlation (ρ = 0.474, P < 0.001) compared to BARTScore + PET, but still outperformed traditional evaluation metrics like Recall-Oriented Understudy for Gisting Evaluation-L (ROUGE-L, ρ = 0.398, P < 0.001) [33].
Fig. 2.

Spearman’s ρ correlations between different evaluation metrics and quality scores assigned by the first physician. The top row quantifies the inter-reader correlation. Notably, domain-adapted BARTScore (BARTScore + PET) and PEGASUSScore (PEGASUSScore + PET) demonstrate the highest correlations with physician preferences
The metrics commonly used in radiology report summarization, including ROUGE [33], BERTScore [34] and RadGraph [10], did not demonstrate strong correlation with physician preferences. Additionally, most reference-free metrics, although effective in general text summarization, showed considerably lower correlation compared to reference-dependent metrics.
Model Performance
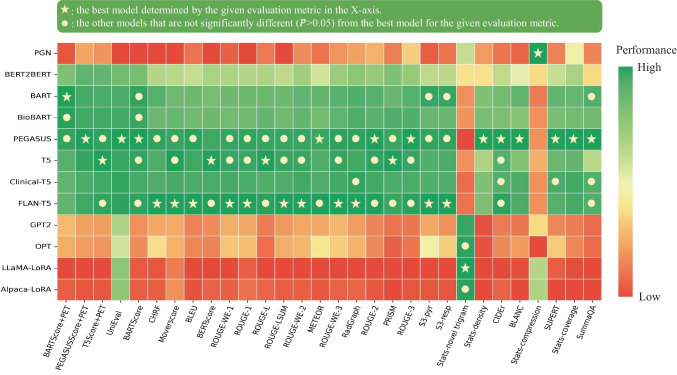
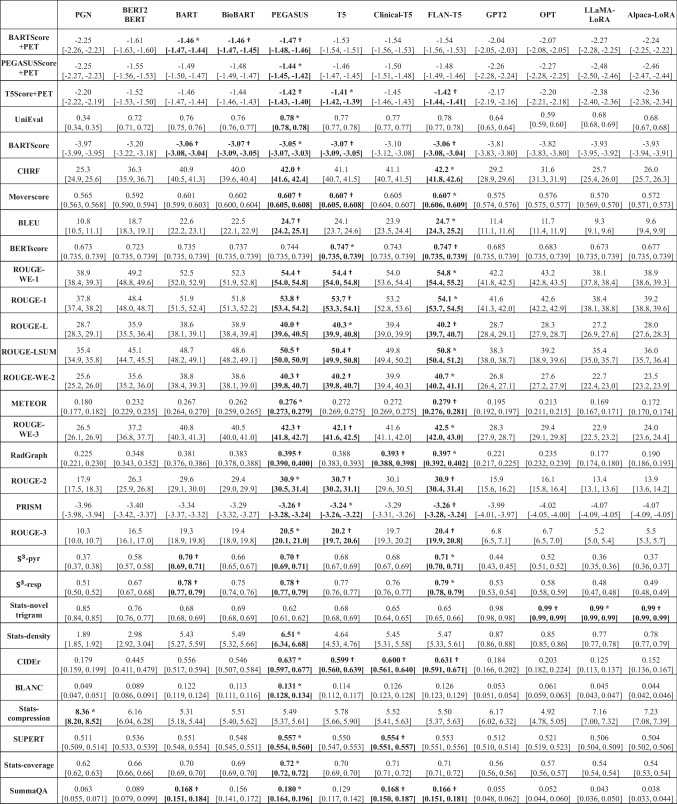
Figure 3 illustrates the relative performance of 12 language models assessed using all evaluation metrics considered in this study. For better visualization, metric values have been normalized to [0, 1], with the original values available in Appendix 5. The SOTA encoder-decoder models, including PEGASUS, BART, and T5, demonstrated similar performance across most evaluation metrics. Since BARTScore + PET and PEGASUSScore + PET identified PEGASUS as the top-performing model, we selected it for further expert evaluation.
Fig. 3.
Performance of 12 language models evaluated by the metrics included in this study. The X-axis displays the metrics arranged in descending order of correlation with physician preferences, with higher correlations on the left and lower correlations on the right. For each evaluation metric, values underwent min–max normalization to allow comparison within a single plot. The actual metric values are referenced in Appendix 5. The star denotes the best model for each metric, and the circle denotes the other models that do not have statistically significant difference (P > 0.05) with the best model
After being fine-tuned on our PET reports, the medical knowledge enriched models, BioBART (BARTScore + PET: -1.46; ROUGE-L: 38.9) and Clinical-T5 (BARTScore + PET: -1.54; ROUGE-L: 39.4), did not show superior performance compared to their base models, BART (BARTScore + PET: -1.46; ROUGE-L: 38.6) and T5 (BARTScore + PET: -1.52; ROUGE-L: 40.3). Additionally, the four decoder-only models included in this study showed significantly lower performance (P < 0.001) compared to the top-tier encoder-decoder LLMs. Interestingly, LLaMA-LoRA (BARTScore + PET: -2.26; ROUGE-L: 27.2) and Alpaca-LoRA (BARTScore + PET: -2.24; ROUGE-L: 28.0), which have been pretrained on one trillion tokens, did not surpass the performance of GPT2 (BARTScore + PET: -2.04, ROUGE-L: 28.7) and OPT (BARTScore + PET: -2.07, ROUGE-L: 28.3).
Expert Evaluation
The distributions of overall utility scores and 6 specific quality scores are illustrated in Fig. 4. Each plot compares four types of impressions: original clinical impressions dictated by the physicians themselves (Orig., own), PEGASUS-generated impressions in the physician’s own style (LLM, own), original clinical impressions dictated by other physicians (Orig, other), and PEGASUS-generated impressions in other physicians’ styles (LLM, other). In total, 83% (60/72) of the PEGASUS-generated impressions were scored as clinically acceptable (scores 3–5), with 60% (43/72) scoring 4 or higher, and 28% (20/72) receiving a score of 5.
Fig. 4.
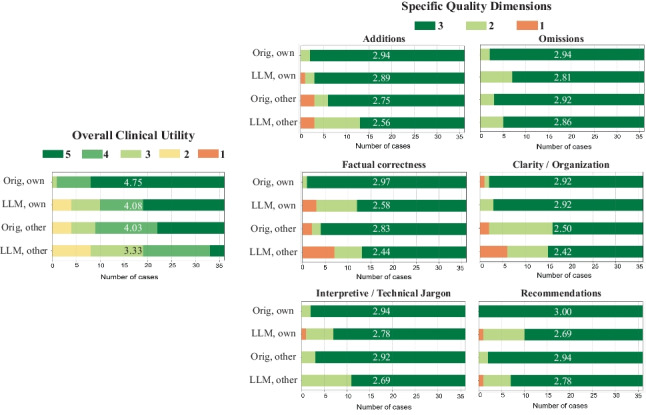
Expert evaluation consisting of an overall utility score and 6 specific quality dimensions. “Orig, own”: original clinical impressions from the physician’s own reports; “LLM, own”: PEGASUS-generated impressions for the physician’s own reports; “Orig, other”: original clinical impressions from other physicians’ reports; “LLM, other”: PEGASUS-generated impressions for other physicians’ reports. The average score for each group is provided to highlight the differences
When the physicians reviewed their own reports, 89% (32/36) of the PEGASUS-generated impressions (LLM, own) were clinically acceptable, with a mean utility score of 4.08 (95% CI, 3.72, 4.42). This score was significantly (P < 0.001) lower than the mean utility score (4.75, 95% CI, 4.58, 4.89) of the clinical impressions originally dictated by themselves (Orig., own). The discrepancy was primarily attributable to 3 quality dimensions: “factual correctness” (Clinical vs. PEGASUS: 2.97 vs. 2.58, P = 0.001), “interpretive and technical jargon” (2.94 vs. 2.78, P = 0.034) and “recommendations” (3.00 vs. 2.69, P = 0.001).
When the physicians evaluated clinical impressions dictated by other physicians (Orig., other), the mean utility score (4.03, 95% CI, 3.69, 4.33) was significantly lower than the score they assigned to their own impressions (Orig., own) (4.75, P < 0.001), indicating a strong preference for their individual reporting style. The primary quality dimensions contributing to such difference included “additions” (Physician’s own impressions vs. Other physicians’ impressions: 2.94 vs. 2.75, P = 0.039) and “clarity and organization” (2.92 vs. 2.50, P < 0.001).
On average, the physicians considered the overall utility of PEGASUS-generated impressions in their own style (LLM, own) to be comparable to the clinical impressions dictated by other physicians (Orig., other) (mean utility score: 4.08 vs. 4.03, P = 0.41).
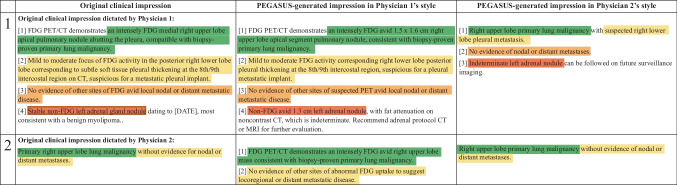
Figure 5 presents four PEGASUS-generated impressions with overall utility scores ranging from 2 to 5. For each case, PEGASUS successfully identified the salient findings, offered interpretations, and provided recommendations. Notably, a majority of cases (60%, 43/72) were problem-free or had minor issues (i.e., scores 4 and 5), similar to cases 1 and 2. Among the cases with low utility scores, the main problem was factual incorrectness, which manifested as misinterpretation of findings and inconsistent statements in the impressions, as evidenced in case 4. Additionally, there were cases where the model could give overly definite diagnoses, as observed in case 3.
Fig. 5.
A side-by-side comparison of clinical impressions and PEGASUS-generated impressions (overall utility scores range from 2 to 5). The last column presents comments from the physicians in our expert reader study. Sentences with similar semantic meanings in the original clinical impressions and the PEGASUS-generated impressions are highlighted using identical colors. Protected health information (PHI) has been anonymized and denoted with [X], where X may represent age or examination date
Additional Analysis
Deauville Score Prediction: Of the 4,000 test cases, 405 PET lymphoma reports contained DSs in the impression sections. Table 4 presents the DS classification results for all evaluated models. PEGASUS achieved the highest 5-class accuracy (76.7%, 95% CI, 72.0%, 81.0%). Among decoder-only models, GPT2 demonstrated the best performance, with an accuracy of 71.3% (95% CI, 65.8%, 76.4%).
Table 4.
Performance of 12 language models on Deauville score prediction
| Models | 5-Class Accuracy (%) | Weighted Cohen’s κ |
|---|---|---|
| PGN | 53.5 [46.9, 60.2] | 0.528 [0.445, 0.605] |
| BERT2BERT | 69.7 [64.9, 74.3] | 0.762 [0.716, 0.805] |
| BART | 75.3 [70.6, 79.7] † | 0.806 [0.760, 0.846] † |
| BioBART | 73.9 [69.7, 78.1] † | 0.802 [0.761, 0.840] † |
| PEGASUS | 76.7 [72.0, 81.0] * | 0.811 [0.767, 0.852] † |
| T5 | 76.3 [72.0, 80.6] † | 0.814 [0.772, 0.853] * |
| Clinical-T5 | 72.5 [67.7, 77.0] † | 0.788 [0.745, 0.829] † |
| FLAN-T5 | 72.6 [68.0, 77.2] † | 0.798 [0.757, 0.837] † |
| GPT2 | 71.3 [65.8, 76.4] | 0.768 [0.715, 0.817] † |
| OPT | 63.1 [57.7, 68.6] | 0.718 [0.665, 0.767] |
| LLaMA-LoRA | 62.9 [56.8, 68.7] | 0.708 [0.647, 0.763] |
| Alpaca-LoRA | 70.6 [64.9, 75.8] | 0.754 [0.696, 0.805] |
Note that data are shown as mean [2.5th percentile, 97.5th percentile]. “*” denotes the best model for each metric, and “†” denotes the other models that have no statistically significant difference (P > 0.05) with the best model
Encoding Physician-specific Styles:Fig. 6 shows the PEGASUS-generated impressions in two physicians’ styles: Physician 1 and Physician 2. Altering a single token in the input could lead to a drastic change in the output impressions. For each case, both impressions managed to capture the salient findings and delivered similar diagnoses, however, their length, level of detail, and phrasing generally reflected the respective physician’s style. This reveals the model’s ability to tailor the impressions to individual physicians.
Fig. 6.
Examples of PEGASUS-generated impressions customized for the physician’s reporting style. The first column shows the original clinical impressions: the first example from Physician 1 and the second from Physician 2. Subsequent columns present impressions generated in the style of Physician 1 and Physician 2, respectively
External Testing: When PEGASUS was applied to the external test set, a significant drop (P < 0.001) was observed in the evaluation metrics, as shown in Table 5. Averaged across the reporting styles of Physicians 1, 2, and 3, BARTScore + PET in the external set was 15% worse than in the internal test set (internal vs. external: -1.47 vs. -1.69, P < 0.001). Similarly, ROUGE-L decreased by 29% in the external set (40.0 vs. 28.5, P < 0.001). Four sample cases from the external dataset are provided in Appendix 6.
Table 5.
Performance of PEGASUS in the external test set
| BARTScore + PET (↑) |
PEGASUSScore + PET (↑) |
ROUGE-1 (↑) |
ROUGE-2 (↑) |
ROUGE-L (↑) |
BLEU (↑) |
BERTScore (↑) |
|
|---|---|---|---|---|---|---|---|
| Internal test |
-1.47 [-1.48, -1.46] |
-1.44 [-1.45, -1.42] |
53.8 [53.4, 54.2] |
30.9 [30.5, 31.4] |
40.0 [39.6, 40.5] |
24.7 [24.2, 25.1] |
0.747 [0.735, 0.739] |
| External test using Physician 1’s style |
-1.66 [-1.70, -1.62] |
-1.72 [-1.77, -1.67] |
38.6 [36.9, 40.2] |
14.8 [13.5, 16.1] |
26.2 [24.9, 27.6] |
11.1 [9.9, 12.3] |
0.671 [0.662, 0.679] |
| External test using Physician 2’s style |
-1.68 [-1.73, -1.63] |
-1.67 [-1.72, -1.61] |
38.5 [36.5, 40.5] |
15.9 [14.1, 17.8] |
29.2 [27.2, 31.3] |
11.5 [9.8, 13.4] |
0.679 [0.668, 0.691] |
| External test using Physician 3’s style |
-1.73 [-1.78, -1.68] |
-1.75 [-1.81, -1.69] |
42.2 [40.6, 43.8] |
18.1 [16.5, 19.7] |
30.0 [28.4, 31.8] |
13.3 [11.8, 14.9] |
0.688 [0.679, 0.697] |
Note that BARTScore and PEGASUSScore compute the log-probability of generating one text given another text, with a range of negative infinity to 0. Other metrics, including ROUGE, BLEU and BERTScore, compute the F1 score of n-gram overlap or semantic similarity, ranging from 0 to 1 (or 0 to 100% when converted to a percentage). A higher value (less negative or more positive) indicates better performance for all these metrics. Data are shown as mean [2.5th percentile, 97.5th percentile]
Discussion
In this work, we trained 12 open-source language models on the task of PET impression generation. After benchmarking various automatic evaluation metrics against physician preferences, we found that the fine-tuned PEGASUS model performed best. Our reader study revealed that the large majority of PEGASUS-generated impressions were rated as clinically acceptable. Moreover, we showed that LLMs were able to learn different reporting styles, and that personalizing impressions to the style of the reading physician had a considerable impact on how they scored the clinical utility of the impression. When physicians assessed impressions generated in their own style, they considered these impressions to be of comparable overall utility to the impressions dictated by other physicians, but of lower quality to impressions that they had dictated themselves.
Past research on text summarization has introduced numerous evaluation metrics for assessing the quality of AI-generated summaries. However, when these metrics were employed to evaluate PET impressions, the majority did not align closely with physician judgments. This observation is consistent with findings from other works that evaluated medical literature [35] or clinical note summarization [12]. In general, we found that model-based metrics slightly outperformed lexical-based metrics, although better evaluation metrics are needed.
Based on our comparison of 12 language models, we observed that the biomedical-domain pretrained LLMs did not outperform their base models. This could be attributed to two reasons. First, our large training set diminished the benefits of medical-domain adaptation. Second, the corpora, such as MIMIC-III and PubMed, likely had limited PET related content, making pretraining less effective for our task. Additionally, we found that the large decoder-only models showed inferior performance in summarizing PET findings compared to the SOTA encoder-decoder models. It likely stems from their lack of an encoder mechanism that can distill the essence of input sequences.
In this study, we focused on open-source models instead of proprietary models like GPT4. This was due to data ownership concerns and the inability to fine-tune proprietary models for personalized impressions. Moreover, with open-source LLMs, institutions can deploy their own solutions for impression generation in clinical workflows, and even share them (which would not be practical with current proprietary models). Recent works [7, 8] explored the capability of proprietary LLMs in radiology report summarization using the in-context learning technique on the publicly available dataset. The question of whether this approach could surpass the full fine-tuning method for open-sourced LLMs and its suitability for clinical use remains to be answered. In the future, we will explore opportunities to work with large proprietary models in a closed and controllable environment to further investigate this question. Additionally, we did not compare our fine-tuned LLMs with commercial products for automatic impression generation. This was primarily due to the unavailability of such products in our institution. However, our findings highlight the importance of considering physicians’ reporting styles in impression generation. Therefore, commercial products should ideally be tailored to individual institutions, otherwise, in-house solutions may offer some advantages.
While most PEGASUS-generated impressions were deemed clinically acceptable in expert evaluation, it is crucial to understand what mistakes are commonly committed by the LLM. First, the main problem in model-generated impressions is factual inaccuracies, which manifest as misinterpretation of findings or contradictory statements. Second, the diagnoses given by the LLM could sometimes be overly definite without considering differential diagnoses. Third, some recommendations for clinical follow-up were non-specific, failing to offer detailed guidance for patient management. It is worth mentioning that final diagnoses and recommendations are usually not included in the report findings and must be inferred by the model. These observations underscore the need for review and appropriate editing by physicians before report finalization. Of note, LLM-based impression generation can be akin to preliminary impression drafts by radiology resident trainees provided for review by the radiology faculty in an academic training setting.
In this work, we personalized impressions to the styles of individual reading physicians. This allows the LLM to create impressions that are more likely to be adopted by the reading physician. It could be seen as antithetical to efforts that aim to standardize reporting. However, we find that physicians interpret reporting guidelines differently, and place different levels of importance on different aspects of reporting. Therefore, in our goal of expediting clinical PET reporting, we focused on accommodating the preferences of reading physicians. We acknowledge that the idea of standardizing impressions is compelling as it might be beneficial to report recipients, including oncologists and patients. Our current method could allow for more consistent impressions by specifying a single reporting style for all physicians; however, whether impressions with a similar style could facilitate better patient care remains to be answered.
To evaluate the performance drop that might occur when models are shared with outside institutions, we performed an external testing and observed a moderate decrease in the evaluation metrics. A critical challenge in this external evaluation is the inability of our personalized LLM to automatically adapt to the external physicians’ styles. Instead, a specific style from the internal dataset must be selected, which may be suboptimal. The differences in reporting styles between our internal and external physicians likely contributed to the observed performance decrease. However, we cannot exclude the possibility that other factors may also play a role, such as differences between the populations (pediatric vs. all ages, lymphoma vs. multiple diseases, etc.). The current results set a lower bound for external performance, and we expect that fine-tuning, or style-matching, would further improve the effectiveness of our LLM. Future research will explore methods for efficiently adapting LLMs to new readers with limited data.
This study had several limitations. First, when fine-tuning LLaMA and Alpaca, we only investigated a lightweight domain adaptation method, LoRA, constrained by computational resources. Second, our current model generates impressions based solely on reporting findings and patient information. However, the PET images themselves could be used to fact-check the generated impressions using vision-language modeling. Third, the number of reports assessed in our reader study was not large enough. Each physician reviewed only 24 reports due to the difficulty of the task. For most cases, scoring impressions took 15–20 min per report. Considering practical limitations in physician time, we decided to have multiple readers so that more reports (N = 72) can be evaluated. Lastly, our training dataset was restricted to a single institution. Future work should be expanding our research to a multi-center study.
In conclusion, we systematically investigated the potential of LLMs to draft impressions for whole-body PET reports. Our reader study showed that the top-performing LLM, PEGASUS, produced clinically useful and personalized impressions for the majority of cases.
Appendix 1: Input Template to the Model
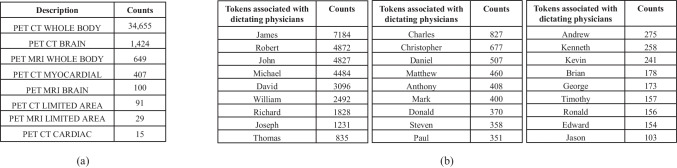
In the input template, “Description” denotes the categories of PET scans, with their frequencies provided in Fig. 7(a). “Radiologist” consists of a single token that encodes the reading physician’s identity. The list of these tokens as well as their frequencies are given in Fig. 7(b).
Fig. 7.
a shows the descriptions of examination categories in our internal dataset. b lists the reading physicians’ unique identifier tokens
Appendix 2: Models for PET Report Summarization
Table 6 summarizes the training settings for each model. The last column includes links to the original implementations or the pretrained weights of the large language models (LLMs). For LLaMA and Alpaca, we chose the model with 7B parameters and used LoRA [29] to accelerate training and reduce memory usage. The hyperparameters of the LoRA module are listed as follows: the rank of the low-rank factorization is 8, the scaling factor for the rank is 16, the dropout rate is 0.05, the target modules for LoRA are projection layers in query (q_proj) and value (v_proj). The learning environment requires at least 2 NVIDIA A100 GPUs and the following Python (3.8.8) libraries: PyTorch (1.13.1), transformer (4.30.0), fastAI (2.7.11), deepspeed (0.9.2).
Table 6.
Training settings of language models investigated in this study
| Language models | Number of trainable parameters | Learning rate | Total batch size | Number of training epochs | Implementations and pretrained weighted |
|---|---|---|---|---|---|
| PGN | 8.3 M | 1e-3 * | 25 * | 30 * | https://github.com/yuhaozhang/summarize-radiology-findings |
| BERT2BERT | 301.7 M | 1e-4 | 32 | 15 | https://huggingface.co/yikuan8/Clinical-Longformer |
| BART | 406.3 M | 5e-5 | 32 | 15 | https://huggingface.co/facebook/bart-large |
| BioBART | 406.3 M | 5e-5 | 32 | 15 | https://huggingface.co/GanjinZero/biobart-large |
| PEGASUS | 568.7 M | 2e-4 | 32 | 15 | https://huggingface.co/google/pegasus-large |
| T5 | 783.2 M | 4e-4 | 32 | 15 | https://huggingface.co/google/t5-v1_1-large |
| Clinical-T5 | 737.7 M | 4e-4 | 32 | 15 | https://huggingface.co/luqh/ClinicalT5-large |
| FLAN-T5 | 783.2 M | 4e-4 | 32 | 15 | https://huggingface.co/google/flan-t5-large |
| GPT2 | 1.5 B | 5e-5 | 32 | 15 | https://huggingface.co/gpt2-xl |
| OPT | 1.3 B | 1e-4 | 32 | 15 | https://huggingface.co/facebook/opt-1.3b |
| LLaMA-LoRA | 4.2 M | 2e-4 | 128 | 20 | available upon request |
| Alpaca-LoRA | 4.2 M | 2e-4 | 128 | 20 | https://huggingface.co/tatsu-lab/alpaca-7b-wdiff |
Note that “*” denotes the hyperparameters directly taken from the original paper. Total batch size = training batch size per device × number of GPU devices × gradient accumulation steps
Appendix 3: Benchmarking Evaluation Metrics
We investigated a broad spectrum of evaluation metrics, comprising 17 different methods.
ROUGE [33]: It measures the number of overlapping textual units between generated and reference texts. ROUGE-N (N = 1,2,3) measures the overlap of N-grams, and ROUGE-L measures the overlap of longest common subsequence. ROUGE-LSUM extends ROUGE-L by computing the ROUGE-L for each sentence, and then summing them up.
BLEU [36]: It computes the precision of n-gram overlap (n ranges from 1 to 4) between generated and reference texts with a brevity penalty.
CHRF [37]: It computes the character-based n-gram overlap between the output sequence and the reference sequence. In this study, we set the n-gram length to 10.
METEOR [38]: It computes an alignment of the generated text and the reference text based on synonymy, stemming, and exact word matching.
CIDEr [39]: It computes the term frequency-inverse document frequency (TF-IDF) vectors for both human and machine-generated texts based on the n-gram (n ranges from 1 to 4) co-occurrence, and then measures the cosine similarity of the two vectors.
ROUGE-WE [40]: It is an extension of the ROUGE metric, designed to assess the semantic similarity between generated and reference texts using pretrained word embeddings.
BERTScore [34]: It evaluates the cosine similarity of contextual embeddings from BERT for each token in the output and reference sequences.
MoverScore [41]: Similar to BERTScore, it leverages the power of BERT’s contextual embeddings to measure the semantic similarity between generated and reference texts. Instead of token-level cosine similarity, MoverScore calculates the Earth Mover’s Distance between the embeddings of the two texts.
RadGraph [10]: It is a specialized evaluation metric tailored for radiology report summarization. RadGraph works by initially extracting clinical entities and their relations from the model-generated impression and the original clinical impression. Leveraging this data, it constructs knowledge graphs to compare the content coverage and structural coherence between the two impressions.
BARTScore [30]: It leverages a pretrained BART model to compute the log probability of generating one text conditioned on another text. In this study, BARTScore is the BART model finetuned on the CNN Daily Mail dataset. BARTScore + PET is the BART model finetuned on our internal PET report dataset. PEGASUSScore + PET is the PEGASUS model finetuned on our internal dataset. T5Score + PET is the FLAN-T5 model finetuned on our internal dataset.
PRISM [42]: It is an evaluation metric used in multilingual machine translation. PRISM employs a sequence-to-sequence model to score the machine-generated output conditioned on the human reference.
S3 [43]: It uses previously proposed evaluation metrics, including ROUGE and ROUGE-WE, as input features for a regression model to estimate the quality score of the generate text. S3-resp is based on a model trained with human annotations following the responsiveness scheme, while S3-pyr follows the pyramid scheme.
UniEval [44]: It first constructs pseudo summaries by perturbing reference summaries, then defines evaluation dimensions using different prompt templates. The model is trained to differentiate pseudo data from reference data in a Boolean question-answering framework. While UniEval evaluates coherence, consistency, fluency, and relevance, we only present the overall score which is the average of these 4 dimensions.
SummaQA [45]: It creates questions from the source document by masking entities. The generated text is then evaluated by a question-answering BERT model, with results reported in terms of the F1 overlap score.
BLANC [46]: It measures how well a generated summary can help improve the performance of a pretrained BERT model in understanding each sentence from the source document with masked tokens.
SUPERT [47]: It creates pseudo-reference summaries by extracting important sentences from the source document and then measures the semantic similarity between the generated text and this pseudo reference.
Stats (Data Statistics) [48]: (1) Stats-compression refers to the word ratio of the source document to its summary. A higher compression indicates a shorter summary. (2) Stats-coverage measures the proportion of words in the generated text that also appear in the source document. Higher coverage implies that more words in the generated text are directly from the source document. (3) Stats-density is the average length of the fragment (e.g., sentence in the source document) from which each summary word is extracted. A higher density suggests that more content from the source is being reused in the generated text. (4) Stats-novel trigram is the percentage of trigrams present in the summary but absent in the source document. A higher novel trigram score indicates the inclusion of more new words or phrases in the generated text.
For the metrics that have precision, recall and F1, we only presented the F1 score, which is the harmonic mean of precision and recall. Metrics such as ROUGE, BLEU, CHRF, METEOR, CIDEr, ROUGE-WE, BERTScore, MoverScore, RadGraph, S3, BLANC, SUPERT and SummaQA typically range from 0 to 1 (or 0 to 100% when converted to a percentage). A higher score, closer to 1 (or 100%), indicates better similarity (n-gram overlap or semantic similarity) between the evaluated text and its reference.
BARTScore and PRISM compute the log-probability of generating one text given another text, with a range from negative infinity to 0. A higher (less negative) BARTScore (or PRISM) indicates a greater similarity, hence a better quality of the generated text.
UniEval computes the scores along four quality dimensions (coherence, consistency, fluency, and relevance). These scores range from 0 to 1, and a higher score means better quality. The overall score is the average of four quality scores.
The evaluation codes are partially adapted from [49] and made available on GitHub: https://github.com/xtie97/PET-Report-Summarization/tree/main/evaluation_metrics.
Appendix 4: Correlation of Evaluation Metrics with the Second Physician’s Scores
Figure 8 presents the Spearman’s ρ correlation between evaluation metrics and quality scores assigned by the second physician (S.Y.C.)
Fig. 8.
Spearman’s ρ correlations between different evaluation metrics and quality scores assigned by the second physician.
Appendix 5: Model Performance
Figure 9 presents the performance evaluation of 12 language models across all 30 metrics (17 different methods) considered in this study. All numbers in this figure are actual metric values. In the first column, we sort the metrics in descending order of correlation with the first physician’s (M.S.) preference.
Fig. 9.
Assessment of 12 language models using all evaluation metrics included in this study. Displayed numbers are actual metric values, and the 95% confidence intervals were determined via bootstrap resampling.
Appendix 6: External Testing
Figure 10
Fig. 10.
Examples of PEGASUS-generated impressions for the external whole-body PET/CT reports. The first column shows the reference clinical impressions. Subsequent columns present impressions generated in the styles of Physician 1, 2, and 3 from our internal dataset
Funding
We acknowledge funding support from Imaging and Radiology Oncology Core Rhode Island (U24CA180803), Biomarker, Imaging and Quality of Life Studies Funding Program (BIQSFP), NCTN Operations Center Grant U10CA180886, NCTN Statistics & Data Center Grant U10CA180899 and St. Baldrick's Foundation. Research reported in this publication was supported by the National Institute Of Biomedical Imaging And Bioengineering of the National Institutes of Health under Award Number R01EB033782.
Data Availability
The radiology reports used in this study are not publicly available due to privacy concerns related to HIPAA. However, upon reasonable request and approval of a data use agreement, they can be made available for research purposes. COG AHOD1331 clinical trial data is archived in NCTN Data Archive
Declarations
Ethics Approval
Institutional Review Board approval was obtained. Informed consent was waived by IRB due to minimal risk to subjects.
Disclaimer
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Competing Interests
Dr. Sharon M. Castellino is on SeaGen Inc. advisory board (no honorarium); Bristol Meyers Squibb advisory board; MJH Health Sciences – CME sponsored speaker. Dr. Ali Pirasteh receives Departmental Research Support from GE Healthcare and Bracco Diagnostics. No products from any of the aforementioned companies were used in this study. Other authors have no relevant conflicts of interest to declare.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Niederkohr RD, et al. Reporting Guidance for Oncologic 18 F-FDG PET/CT Imaging. J Nucl Med. 2013;54(5):756–761. doi: 10.2967/jnumed.112.112177. [DOI] [PubMed] [Google Scholar]
- 2.Hartung MP, Bickle IC, Gaillard F, Kanne JP. How to Create a Great Radiology Report. RadioGraphics. 2020;40(6):1658–1670. doi: 10.1148/rg.2020200020. [DOI] [PubMed] [Google Scholar]
- 3.Y. Zhang, D. Y. Ding, T. Qian, C. D. Manning, and C. P. Langlotz, “Learning to Summarize Radiology Findings,” in Proceedings of the Ninth International Workshop on Health Text Mining and Information Analysis, Brussels, Belgium: Association for Computational Linguistics, 2018, pp. 204–213. 10.18653/v1/W18-5623.
- 4.J. Hu, Z. Li, Z. Chen, Z. Li, X. Wan, and T.-H. Chang, “Graph Enhanced Contrastive Learning for Radiology Findings Summarization.” arXiv, Jun. 08, 2022. Accessed: Mar. 02, 2023. [Online]. Available: http://arxiv.org/abs/2204.00203
- 5.J.-B. Delbrouck, M. Varma, and C. P. Langlotz, “Toward expanding the scope of radiology report summarization to multiple anatomies and modalities.” arXiv, Nov. 18, 2022. Accessed: Mar. 02, 2023. [Online]. Available: http://arxiv.org/abs/2211.08584
- 6.Z. Liu et al., “Radiology-GPT: A Large Language Model for Radiology.” arXiv, Jun. 14, 2023. Accessed: Jul. 17, 2023. [Online]. Available: http://arxiv.org/abs/2306.08666
- 7.Sun Z, et al. Evaluating GPT4 on Impressions Generation in Radiology Reports. Radiology. 2023;307(5):e231259. doi: 10.1148/radiol.231259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.C. Ma et al., “ImpressionGPT: An Iterative Optimizing Framework for Radiology Report Summarization with ChatGPT.” arXiv, May 03, 2023. Accessed: Aug. 14, 2023. [Online]. Available: http://arxiv.org/abs/2304.08448
- 9.Johnson AEW, et al. MIMIC-III, a freely accessible critical care database. Sci Data. 2016;3(1):160035. doi: 10.1038/sdata.2016.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.J. Hu et al., “Word Graph Guided Summarization for Radiology Findings.” arXiv, Dec. 18, 2021. Accessed: Mar. 02, 2023. [Online]. Available: http://arxiv.org/abs/2112.09925
- 11.A. Smit, S. Jain, P. Rajpurkar, A. Pareek, A. Y. Ng, and M. P. Lungren, “CheXbert: Combining Automatic Labelers and Expert Annotations for Accurate Radiology Report Labeling Using BERT.” arXiv, Oct. 18, 2020. Accessed: Aug. 27, 2023. [Online]. Available: http://arxiv.org/abs/2004.09167
- 12.A. B. Abacha, W. Yim, G. Michalopoulos, and T. Lin, “An Investigation of Evaluation Metrics for Automated Medical Note Generation.” arXiv, May 27, 2023. Accessed: Aug. 27, 2023. [Online]. Available: http://arxiv.org/abs/2305.17364
- 13.M. Kayaalp, A. C. Browne, Z. A. Dodd, P. Sagan, and C. J. McDonald, “De-identification of Address, Date, and Alphanumeric Identifiers in Narrative Clinical Reports”. AMIA Annu Symp Proc; 2014; 2014: 767–776. PMID: 25954383; PMCID: PMC4419982. [PMC free article] [PubMed]
- 14.Castellino SM, et al. Brentuximab Vedotin with Chemotherapy in Pediatric High-Risk Hodgkin’s Lymphoma. N Engl J Med. 2022;387(18):1649–1660. doi: 10.1056/NEJMoa2206660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Y. Wang et al., “Self-Instruct: Aligning Language Models with Self-Generated Instructions.” arXiv, May 25, 2023. Accessed: Aug. 14, 2023. [Online]. Available: http://arxiv.org/abs/2212.10560
- 16.T. Rohan, G. Ishaan, Z. Tianyi, et al. Stanford Alpaca: An Instruction-following LLaMA model. Available at https://github.com/tatsu-lab/stanford_alpaca. Accessed June 20, 2023
- 17.M. Lewis et al., “BART: Denoising Sequence-to-Sequence Pre-training for Natural Language Generation, Translation, and Comprehension.” arXiv, Oct. 29, 2019. Accessed: Mar. 07, 2023. [Online]. Available: http://arxiv.org/abs/1910.13461
- 18.J. Zhang, Y. Zhao, M. Saleh, and P. J. Liu, “PEGASUS: Pre-training with Extracted Gap-sentences for Abstractive Summarization.” arXiv, Jul. 10, 2020. Accessed: Mar. 07, 2023. [Online]. Available: http://arxiv.org/abs/1912.08777
- 19.C. Raffel et al., “Exploring the Limits of Transfer Learning with a Unified Text-to-Text Transformer.” arXiv, Jul. 28, 2020. Accessed: Aug. 14, 2023. [Online]. Available: http://arxiv.org/abs/1910.10683
- 20.J. Wei et al., “Finetuned Language Models Are Zero-Shot Learners.” arXiv, Feb. 08, 2022. Accessed: Aug. 15, 2023. [Online]. Available: http://arxiv.org/abs/2109.01652
- 21.H. Yuan, Z. Yuan, R. Gan, J. Zhang, Y. Xie, and S. Yu, “BioBART: Pretraining and Evaluation of A Biomedical Generative Language Model.” arXiv, Apr. 22, 2022. Accessed: Aug. 15, 2023. [Online]. Available: http://arxiv.org/abs/2204.03905
- 22.Q. Lu, D. Dou, TH. Nguyen. ClinicalT5: A Generative Language Model for Clinical Text. Findings of the Association for Computational Linguistics: EMNLP 2022, pages 5436–5443, Abu Dhabi, United Arab Emirates. Association for Computational Linguistics. 10.18653/v1/2022.findings-emnlp.398.
- 23.Johnson AEW, et al. MIMIC-CXR, a de-identified publicly available database of chest radiographs with free-text reports. Sci Data. 2019;6(1):317. doi: 10.1038/s41597-019-0322-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.C. Chen et al., “bert2BERT: Towards Reusable Pretrained Language Models,” in Proceedings of the 60th Annual Meeting of the Association for Computational Linguistics (Volume 1: Long Papers), Dublin, Ireland: Association for Computational Linguistics, 2022, pp. 2134–2148. 10.18653/v1/2022.acl-long.151.
- 25.D. M. Ziegler et al., “Fine-Tuning Language Models from Human Preferences.” arXiv, Jan. 08, 2020. Accessed: Aug. 14, 2023. [Online]. Available: http://arxiv.org/abs/1909.08593
- 26.S. Zhang et al., “OPT: Open Pre-trained Transformer Language Models.” arXiv, Jun. 21, 2022. Accessed: Feb. 22, 2023. [Online]. Available: http://arxiv.org/abs/2205.01068
- 27.H. Touvron et al., “LLaMA: Open and Efficient Foundation Language Models.” arXiv, Feb. 27, 2023. Accessed: Aug. 14, 2023. [Online]. Available: http://arxiv.org/abs/2302.13971
- 28.I. Loshchilov and F. Hutter, “Decoupled Weight Decay Regularization.” arXiv, Jan. 04, 2019. Accessed: Aug. 31, 2023. [Online]. Available: http://arxiv.org/abs/1711.05101
- 29.E. J. Hu et al., “LoRA: Low-Rank Adaptation of Large Language Models.” arXiv, Oct. 16, 2021. Accessed: Aug. 15, 2023. [Online]. Available: http://arxiv.org/abs/2106.09685
- 30.W. Yuan, G. Neubig, and P. Liu, “BARTScore: Evaluating Generated Text as Text Generation.” arXiv, Oct. 27, 2021. Accessed: Aug. 15, 2023. [Online]. Available: http://arxiv.org/abs/2106.11520
- 31.Z. Huemann, C. Lee, J. Hu, S. Y. Cho, and T. Bradshaw, “Domain-adapted large language models for classifying nuclear medicine reports.” arXiv, Mar. 01, 2023. Accessed: Mar. 17, 2023. [Online]. Available: http://arxiv.org/abs/2303.01258 [DOI] [PMC free article] [PubMed]
- 32.Smith L, et al. Overview of BioCreative II gene mention recognition. Genome Biol. 2008;9(S2):S2. doi: 10.1186/gb-2008-9-s2-s2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.C.-Y. Lin, ROUGE: A package for automatic evaluation of summaries. In Text Summarization Branches Out, Barcelona, Spain, July 2004. Association for Computational Linguistics, 2004; 74–81. https://aclanthology.org/W04-1013/.
- 34.T. Zhang, V. Kishore, F. Wu, K. Q. Weinberger, and Y. Artzi, “BERTScore: Evaluating Text Generation with BERT.” arXiv, Feb. 24, 2020. Accessed: Aug. 22, 2023. [Online]. Available: http://arxiv.org/abs/1904.09675
- 35.L. L. Wang et al., “Automated Metrics for Medical Multi-Document Summarization Disagree with Human Evaluations.” arXiv, May 23, 2023. Accessed: Aug. 22, 2023. [Online]. Available: http://arxiv.org/abs/2305.13693
- 36.K. Papineni, S. Roukos, T. Ward, and W.-J. Zhu, “BLEU: a method for automatic evaluation of machine translation,” in Proceedings of the 40th Annual Meeting on Association for Computational Linguistics - ACL ’02, Philadelphia, Pennsylvania: Association for Computational Linguistics, 2001, p. 311. 10.3115/1073083.1073135.
- 37.M. Popović, “chrF: character n-gram F-score for automatic MT evaluation,” in Proceedings of the Tenth Workshop on Statistical Machine Translation, Lisbon, Portugal: Association for Computational Linguistics, 2015, pp. 392–395. 10.18653/v1/W15-3049.
- 38.Banerjee, S. and Lavie, A. METEOR: An Automatic Metric for MT Evaluation with Improved Correlation with Human Judgments. Proceedings of Workshop on Intrinsic and Extrinsic Evaluation Measures for MT and/or Summarization. Ann Arbor, Michigan: Association of Computational Linguistics, 2005. p. 65–72.
- 39.R. Vedantam, C. L. Zitnick, and D. Parikh, “CIDEr: Consensus-based Image Description Evaluation.” arXiv, Jun. 02, 2015. Accessed: Aug. 31, 2023. [Online]. Available: http://arxiv.org/abs/1411.5726
- 40.J.-P. Ng and V. Abrecht, “Better Summarization Evaluation with Word Embeddings for ROUGE.” arXiv, Aug. 25, 2015. Accessed: Aug. 31, 2023. [Online]. Available: http://arxiv.org/abs/1508.06034
- 41.W. Zhao, M. Peyrard, F. Liu, Y. Gao, C. M. Meyer, and S. Eger, “MoverScore: Text Generation Evaluating with Contextualized Embeddings and Earth Mover Distance,” in Proceedings of the 2019 Conference on Empirical Methods in Natural Language Processing and the 9th International Joint Conference on Natural Language Processing (EMNLP-IJCNLP), Hong Kong, China: Association for Computational Linguistics, 2019, pp. 563–578. 10.18653/v1/D19-1053.
- 42.B. Thompson and M. Post, “Automatic Machine Translation Evaluation in Many Languages via Zero-Shot Paraphrasing,” in Proceedings of the 2020 Conference on Empirical Methods in Natural Language Processing (EMNLP), Online: Association for Computational Linguistics, 2020, pp. 90–121. 10.18653/v1/2020.emnlp-main.8.
- 43.M. Peyrard, T. Botschen, and I. Gurevych, “Learning to Score System Summaries for Better Content Selection Evaluation.,” in Proceedings of the Workshop on New Frontiers in Summarization, Copenhagen, Denmark: Association for Computational Linguistics, 2017, pp. 74–84. 10.18653/v1/W17-4510.
- 44.M. Zhong et al., “Towards a Unified Multi-Dimensional Evaluator for Text Generation,” in Proceedings of the 2022 Conference on Empirical Methods in Natural Language Processing, Abu Dhabi, United Arab Emirates: Association for Computational Linguistics, 2022, pp. 2023–2038. 10.18653/v1/2022.emnlp-main.131.
- 45.T. Scialom, S. Lamprier, B. Piwowarski, and J. Staiano, “Answers Unite! Unsupervised Metrics for Reinforced Summarization Models.” arXiv, Sep. 04, 2019. Accessed: Aug. 31, 2023. [Online]. Available: http://arxiv.org/abs/1909.01610
- 46.L. V. Lita, M. Rogati, and A. Lavie, “BLANC: learning evaluation metrics for MT,” in Proceedings of the conference on Human Language Technology and Empirical Methods in Natural Language Processing - HLT ’05, Vancouver, British Columbia, Canada: Association for Computational Linguistics, 2005, pp. 740–747. 10.3115/1220575.1220668.
- 47.Y. Gao, W. Zhao, and S. Eger, “SUPERT: Towards New Frontiers in Unsupervised Evaluation Metrics for Multi-Document Summarization,” in Proceedings of the 58th Annual Meeting of the Association for Computational Linguistics, Online: Association for Computational Linguistics, 2020, pp. 1347–1354. 10.18653/v1/2020.acl-main.124.
- 48.M. Grusky, M. Naaman, and Y. Artzi, “Newsroom: A Dataset of 1.3 Million Summaries with Diverse Extractive Strategies,” in Proceedings of the 2018 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, Volume 1 (Long Papers), New Orleans, Louisiana: Association for Computational Linguistics, 2018, pp. 708–719. 10.18653/v1/N18-1065.
- 49.Fabbri AR, Kryściński W, McCann B, Xiong C, Socher R, Radev D. “SummEval: Re-evaluating Summarization Evaluation”, Transactions of the Association for. Computational Linguistics. 2021;9:391–409. doi: 10.1162/tacl_a_00373. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The radiology reports used in this study are not publicly available due to privacy concerns related to HIPAA. However, upon reasonable request and approval of a data use agreement, they can be made available for research purposes. COG AHOD1331 clinical trial data is archived in NCTN Data Archive