Abstract
Nucleotide sequences of the reverse transcriptase (RT) coding region have been compared in four new human immunodeficiency virus type 1 (HIV-1) group O isolates. Phylogenetic analysis of this pol region highlights a cluster of these four HIV-1 group O sequences with seven other group O isolates (5% intracluster nucleotide sequence diversity) similar to clusters classified as subtypes in HIV-1 group M (an average of 4.9% intrasubtype sequence diversity). Based on these analyses, this group O cluster has been designated subtype A-O. A longitudinal study of a heterosexual couple infected with group O (ESP1 and ESP2) allowed a detailed analysis of RT sequences (amino acids 28 to 219). Directed evolution and a slightly higher mutation frequency was observed in the RT sequences of patient ESP2, treated with antiretroviral drugs, than that from the untreated patient ESP1. Antiretroviral treatment also selected for specific substitutions, M184V and T215Y in the RT coding region, conferring resistance to 3′-dideoxy-3′-thiacytidine and zidovudine, respectively. A Gly98 to Glu RT substitution identified in the treated patient suggests a possible reversion of a nonnucleoside RT inhibitor-resistant phenotype. Using RT clones from this longitudinal study, both heteroduplex tracking assay and cloning-sequencing techniques were employed for an extensive genetic analysis of pol gene quasispecies. Amino acid substitutions (i.e., Phe-77 to Leu, Lys-101 to Glu, and Val-106 to Iso) associated with antiretroviral resistance were identified in RT clones from HIV-1 group O-infected patients not subjected to drug therapy or treated with unrelated drugs. Finally, phylogenetic relationships between RT clones of the treated ESP2 patient and those of the untreated ESP1 patient show how drug pressure can direct evolution of viral pol gene quasispecies independently of direct drug-resistant substitutions.
Human immunodeficiency virus (HIV) is subdivided into two types, HIV type 1 (HIV-1) being the pathogen most omnipresent in the epidemic and exhibiting the highest degree of genetic diversity. Based on genetic variability in the envelope (env) gene, HIV-1 can be subdivided into at least 10 distinct subtypes (designated A to J) responsible for separate geographic pandemics (39). Phylogenetic analyses have shown that each subtype in this major group (group M) is approximately equidistant from the others, as if arising from a common ancestor (33, 38, 39). In contrast, a few divergent HIV-1 strains form a cluster distinct from group M and have been categorized as members of the outlier group (group O) (7, 10, 19, 58). Lack of a clearly defined phylogenetic tree topology for group O viruses has prevented further division into subtypes as described for group M (27, 30). Group O virus was first isolated from HIV-1-infected individuals in West Central Africa (10, 19, 58) and has been subsequently identified in Europe (7, 10, 21, 25, 30, 55), throughout Africa (4, 19, 20, 22, 23, 35, 41, 44, 58), and, to a lesser extent, in the United States (5). Paradoxically, group O virus and not the predominant group M HIV-1 appears responsible for the earliest case of HIV-1 infection reported in Europe (25).
Although similar in overall genomic arrangement, a large genetic distance at the level of nucleotide sequence separates groups M and O (24 to 32%, 33 to 37%, and 39 to 49% divergence in the gag, pol, and env genes, respectively) (13, 30, 50). Because of this diversity, commercial diagnostic assays for HIV-1 detection, commonly derived from group M subtype B strains or from HIV-2, often failed to identify HIV-1 group O samples (32, 52). Even with the detection of group O infections, it is difficult to monitor the effectiveness of treatments since viral loads cannot be determined with conventional assays (31). Finally, HIV-1 group O isolates appear to be sensitive to most nucleoside analogs and protease inhibitors but may be intrinsically resistant to some nonnucleoside reverse transcriptase (RT) inhibitors (NNRTIs) (13, 14, 50).
Essentially, the extremely high genetic variation of HIV-1 is a consequence of a relatively low fidelity of the RT (47) and the rapid turnover of virus (46). A remarkable intrapatient variability suggests that HIV-1 disease progression is associated with a wide distribution of viral quasispecies rather than active replication of a single isolate. Consequently, the host immune response or treatment with antiretroviral drugs may simply select for those HIV-1-resistant variants preexisting in the intrapatient population of quasispecies (40, 59). Like group M strains, a high interpatient variation has been described among group O viruses: 1.9 to 14.3% for the gag gene (20, 30), 3.3 to 12.2% for the pol gene (13), and 4 to 30% and 7 to 24.5% for the C2V3 (23, 30, 34) and the gp41 immunodominant (4) regions of the env gene, respectively. However, only one study has described intrapatient variation in the C2V3 env region of epidemiologically linked HIV-1 group O infections (mother-to-infant transmission) (6). Recently, the pathogenic course (virologic, immunologic, and clinical changes) of HIV-1 group O infection was described in a married couple (17), but no data on group O HIV-1 heterogeneity was reported in that study.
To date, the emergence of amino acid substitutions associated with resistance to RT inhibitors has been well characterized in patients infected with HIV-1 group M (2, 36, 43). In this report, we have performed evolutionary and drug resistance studies of the pol gene quasispecies in samples obtained over a year and half from an epidemiologically linked HIV-1 group O-infected couple, the female being treated with antiretroviral agents, while the infected male remained untreated. Sequence analysis revealed the presence of two drug-resistant substitutions (M184V and T215Y) in the RT-coding region during the course of antiretroviral therapy of the female. We adapted the heteroduplex tracking analysis (HTA) to then analyze the intrapatient heterogeneity in the complete RT-coding region (a 1.7-kb genomic fragment) of four HIV-1 group O-infected individuals living in Spain, the infected couple and two other unrelated individuals. Quasispecies diversity as measured by HTA matched the actual nucleotide sequence diversity of the individual pol gene clones of each patient sample. Finally, we compared the phylogenetic relationship of the entire RT-coding region (1,680 bp) in samples collected from these four patients with that sequence of other group O viruses. Based on the analysis of pol sequences, it appears that group O virus can be further separated into at least two different phylogenetic clusters.
MATERIALS AND METHODS
Patients and samples.
Four HIV-1 group O-infected patients living in Spain were included in this study. They were selected for their HIV serological profiles, i.e., atypical Western blots, showing weak or absent reactivity to the group M env glycoproteins and stronger reactivity to a synthetic peptide assay which contains three synthetic V3 loop peptides of the HIV-1 ANT70 isolate (Inno-Lia HIV type O, Ghent, Belgium). As summarized in Table 1, the first two patients (ESP1 and ESP2), a Spanish-born couple, were identified in Madrid. The 35-year-old male (ESP1) was believed to be infected while traveling to Equatorial Guinea and Cameroon. Sample ESP1/0 from this patient was previously described as the first case of HIV-1 group O in Spain (55). Patient ESP2 initiated antiretroviral treatment 14 months after HIV diagnosis (June 1996), whereas her partner (ESP1), infected with the founder virus, has refused any antiretroviral therapy (Table 1). The other two patients (ESP3 and ESP4) were identified in Madrid and Barcelona (651 km from Madrid), respectively, and are of African origin. No epidemiological relationship could be established between the couple from Madrid and these two individuals. Patient ESP4 was receiving antiretroviral drugs during the study (Table 1) but would not participate in the longitudinal study.
TABLE 1.
Epidemiological and virological data
| Patienta | Age/sex/citya | Sample/dateb | CD4 count (cells/μl) | p24 Ag (pg/ml)c | Treatment |
|---|---|---|---|---|---|
| ESP1 | 35/M/Madrid | 0/Sept. ’95 | 290 | 15 | Noned |
| +4/Jan. ’96 | 182 | 21 | None | ||
| +11/Aug. ’96 | 152 | 34 | None | ||
| +19/Apr. ’97 | 144 | 25 | None | ||
| ESP2 | 34/F/Madrid | +4/Jan. ’96 | 48 | 390 | None |
| +11/Aug. ’96 | 24 | 131 | AZT + ddIe | ||
| +17/Feb. ’97 | 165 | 50 | d4T + 3TC + IDVf | ||
| ESP3 | 45/M/Madrid | ESP3/Jan. ’97 | 96 | 105 | None |
| ESP4 | 15/M/Barcelona | ESP4/Feb. ’97 | 16 | 0 | AZT + 3TC |
Age is given in years. Sex: M, male; F, female. City, location where the sample was collected.
The number following the patient’s code in the two related isolates (ESP1 and ESP2) indicates months after the first HIV-1 group O isolate described (ESP1/0) (50) at the time of sampling of lymphocytes. Samples ESP3 and ESP4 are unique.
p24 antigen (Ag) was measured using a commercial kit (HIV-1 p24 Antigen Assay; Coulter, Miami, Fla.).
No treatment with antiretroviral inhibitors.
Patient received AZT + ddI therapy from June to September 1996.
Patient received d4T + 3TC + indinavir (IDV) from September 1996 to the present.
Proviral DNA purification, PCR, and molecular cloning.
Proviral DNA was extracted directly from lysed peripheral blood mononuclear cells (PBMCs), obtained uncultured from the patient as described previously (48). Genomic regions encoding the RT (pol gene) were PCR amplified using a set of nested oligonucleotide primers (50). Briefly, 1 μg of template DNA and primers RTO1 and RTO2 (100 pmol each) were used in the first external amplification. The PCRs were carried out in a 100-μl reaction mixture containing 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 0.01% gelatin, a 0.2 mM concentration of each of the four deoxynucleoside triphosphates, 200 ng of each primer, and 2.5 U of Taq polymerase (Boehringer). Cycling temperatures and times have been previously described (50).
Using the same PCR conditions, the products (5 μl) of the external amplification were reamplified using nested inner primers RTO3 and RTO4 (50). Negative controls (distilled H2O used in the other reaction mixtures) were PCR amplified with each set of sample amplifications and never resulted in detectable product. PCR-amplified products were separated in agarose gels and then purified using a commercially available kit (PCR Clean Up kit; Boehringer). The purified DNA obtained from the nested PCR (approximately 1.7 kb) was cleaved with NcoI and EcoRI and cloned into the vector pRT6 as previously described (50). At least two independent products of nested PCR amplifications per sample were mixed to increase the number of amplified quasispecies and to avoid possible analysis of mutations introduced by Taq polymerase. Previously, it has been shown that mutations introduced by Taq polymerase in HIV-1 account for less than 1.2 × 10−4 substitutions per nucleotide (s/nt) observed in each PCR product (40). Clones with inserts of the appropriate size, as judged by restriction enzyme digestion and gel electrophoresis, were used for DNA sequencing.
Nucleotide sequence analysis.
Direct sequencing of the PCR-amplified DNA (average sequences) and of 10 clones per sample (individual quasispecies) was performed using the fmol method (Promega), followed by treatment of the reaction mixture with terminal deoxynucleotidyl transferase (48). Primers used in the sequencing reactions have been previously described (50). The full RT-coding region (1,680 bp) from samples ESP2/+4 (obtained 4 months after the description of sample ESP1/0), ESP3, and ESP4 (Table 1) was sequenced. The RT sequence for ESP1/0 has been previously reported (50). Using ESP1 and ESP2 samples from all time points, we also sequenced a 576-bp fragment encoding codons 28 to 219 of RT. Finally, a 267-bp fragment, RT codons 28 to 116, was sequenced from each individual genomic clone. Nucleotide sequences were edited and translated by EDITSEQ software (DNASTAR, Inc.) and then aligned using the CLUSTAL X version 1.63b program (57). Pairwise DNA matrices, generated using the Kimura two-parameter model (26) and phylogenetic analyses, were determined with the use of the MEGA version 1.02 program (28). Tree topologies were inferred by the neighbor-joining method (51) with the Kimura two-parameter distance matrices. Bootstrap resampling (1,000 data sets) of the multiple alignment tested the statistical robustness of the trees.
HTA.
Nested PCR products of the complete RT-coding region (1,680 bp of pol gene) were analyzed using HTA (11, 12). The same region of RT from the first HIV-1 group O Spanish sample (ESP1/0, Table 1 [50]) was PCR amplified and used as radioactive probe DNA. Briefly, primers RTO3 (end labeled using T4 polynucleotide kinase and 2 μCi of [γ-32P]ATP) and RTO4 were used to amplify the corresponding fragment from the homogeneous p66RTO clone, derived from an ESP1/0 RT fragment (50). This PCR-amplified probe was separated on agarose gel and then purified using the Agarose Gel DNA Extraction kit (Boehringer). Conditions for heteroduplex formation as previously described (11) have been slightly modified for these analyses. The reaction mixture contained DNA annealing buffer (100 mM NaCl, 10 mM Tris-HCl [pH 7.8], and 2 mM EDTA), 10 μl of PCR-amplified sample DNA, and 1 μl of radioactive probe DNA (prepared by diluting the radiolabeled PCR-amplified probe 20-fold in annealing buffer). The reaction mixtures were denatured at 95°C for 5 min and rapidly annealed in wet ice. After 30 min on ice, the 1,680-bp DNA heteroduplexes were resolved on 5% nondenaturing polyacrylamide gels (30:0.8, acrylamide-bis) in a model V16 vertical gel apparatus (Gibco BRL, Gaithersburg, Md.) with 1× Tris-borate-EDTA buffer (TBE) at 200 V for 7 h. Gels were dried onto Whatman 3MM paper under vacuum and exposed to X-ray film (Eastman Kodak Co., Rochester, N.Y.). Films were scanned for presentation and analysis. Actual migration distances of different heteroduplexes are directly related to the genetic distances separating different samples. For a controlled analysis of migration distances, we calculated heteroduplex mobility ratios for each sample by measuring the migration distance traveled by the heteroduplexes (distance from the well in millimeters) and dividing by the distance traveled by the radiolabeled single-stranded DNA probe.
GenBank accession numbers.
Nucleotide sequences reported in this study have been submitted to GenBank under the following accession numbers: complete RT of ESP2 (AF068947), complete RT of ESP3 (AF068948), complete RT of ESP4 (AF068949), and ESP1 and ESP2 sequential samples encoding amino acids 28 to 219 of RT (AF068950 to AF068952 and AF068953 to AF068954, respectively). Accession numbers of the cloning sequences encoding amino acids 28 to 116 of the RT are as follows: ESP1 (AF068955 to AF068984), ESP2 (AF068985 to AF069014), ESP3 (AF069015 to AF069024), and ESP4 (AF069025 to AF069034).
RESULTS
Clinical data and antiviral therapy.
Following the first report of HIV-1 group O infection in Spain (55) and characterization of its env gene (34), the RT of this isolate was cloned, sequenced, and biochemically characterized (50). Recombinant RT expressed from this HIV-1 group O RT clone possessed biochemical characteristics similar to those of the recombinant HIV-1 RT from subtype B virus. In this longitudinal study, the RT-coding region was sequenced and analyzed to determine the effects of antiretroviral therapy on the development of drug-resistant mutations and heterogeneity in HIV-1 group O isolates. For these analyses, we obtained three PBMC samples from the untreated patient (ESP1) at 4, 11, and 19 months and three from his infected female partner at 4, 11, and 17 months (Table 1). Additionally, we analyzed one sample from two new HIV-1 group O patients in Spain (ESP3 and ESP4) (Table 1). Patient ESP1, in spite of a moderate decrease in CD4 cell counts, remained asymptomatic and with a constant viral load as estimated by p24 antigen capture assays (Table 1). ESP2, the female partner infected by ESP1, had developed AIDS and was subsequently treated with 2′,3′-dideoxynosine (ddI or didanosine; Bristol-Myers-Squibb, Wallingford, Conn.) and 3′-azido-3′-deoxythimidine (AZT or zidovudine; Glaxo-Wellcome, Dartford, United Kingdom) after 8 months of the study (Table 1). After 3 months of this treatment regimen, she was prescribed a more aggressive therapy: a triple combination regimen containing 2′3′-didehydro-2,3′-dideoxythimidine (d4T or stavudine; Bristol-Myers-Squibb), 2′,3′-dideoxy-3′-thiacytidine (3TC or lamivudine; Glaxo-Wellcome), plus a protease inhibitor, indinavir (Merck & Co., Inc.). As a likely consequence of this triple combination therapy, her CD4 cell counts increased over 6 months, while levels of HIV-1 p24 antigen in her plasma dropped precipitously (Table 1). Difficulties in PCR amplification of the RT region from patient ESP2 after 7 months (sample +19) of triple combination therapy suggest a significant reduction in viremia. Finally, patient ESP3 had not been treated, but patient ESP4, having a low CD4 cell count, had started antiretroviral therapy (AZT plus 3TC) prior to the time of sample collection. HIV-1 p24 levels were used as a crude measure of viral load determination, since conventional HIV-1 RNA load assays, e.g., Amplicor HIV monitor test (Roche Diagnostics, Basel, Switzerland), NASBA (Organon Teknika, Boxtel, The Netherlands), or branched DNA (Chiron, Emeryville, CA), do not efficiently detect HIV-1 group O RNA.
Phylogenetic relationship and nucleotide sequence analysis. A potential subtype definition within group O viruses?
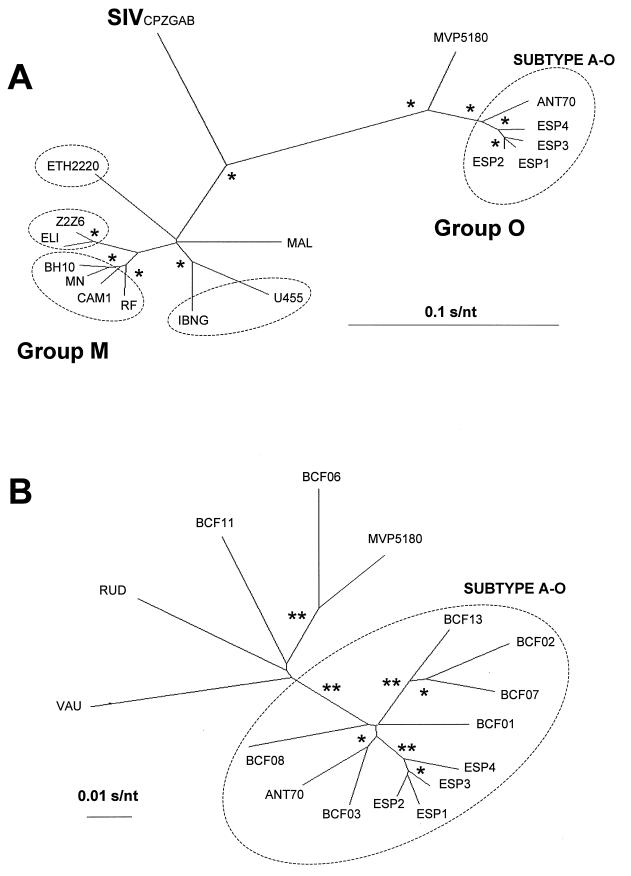
Currently, there are only three complete nucleotide sequences for the RT-coding region of HIV-1 group O reported in the HIV database (39): two RT group O sequences from Cameroon (ANT70 [10] and MVP5180 [19]) and one from Spain (ESP1 [50]). In this study, we have sequenced the complete RT-coding region of three new HIV-1 group O isolates from patients ESP2, ESP3, and ESP4. A phylogenetic tree was constructed from pairwise comparisons of the RT sequences (1,680 nucleotides) from four different HIV-1 group O Spanish isolates (ESP1, ESP2, ESP3, and ESP4), two group O viruses (ANT70 and MVP5180) from Cameroon, and 10 HIV-1 strains clustering into group M (U455, IBNG, CAM1, RF, MN, BH10, ETH2220, ELI, Z2Z6, and MAL, arbitrarily selected to represent subtypes A, B, C, and D as defined by the env gene) (39) (Fig. 1A). It is important to note that no complete RT sequences have been reported for subtypes E to J (as defined by the env gene) to the Los Alamos database (39). Three major branches, 100% supported after 1,000 bootstrap trees, could be identified in this phylogenetic tree (Fig. 1A), representing the two previously defined HIV-1 genotypic groups (M and O) and the outgroup CPZGAB. The three new Spanish sequences (ESP2, ESP3, and ESP4) cluster together with the previously described ESP1 isolate (50) and the ANT70 isolate, with 100% of bootstrap resampling values.
FIG. 1.
Phylogenetic tree analysis of the RT-coding region from four HIV-1 group O isolates from Spain compared to this region in group M and other group O HIV-1 strains (39). (A) Full-length RT-coding region (1,680-bp) sequences were utilized to construct a neighbor-joining consensus tree as described in Materials and Methods. Group M (using sequences from A, B, C, and D subtypes) and group O isolates as well as the simian immunodeficiency virus (SIV) CPZGAB strain, used as an outgroup, are indicated in this panel. GenBank accession numbers for reference sequences are as follows: HIV-1 group M subtype A, U455 (M62320), IBNG (L39106); subtype B, MN (M17449), CAM1 (D10112), RF (M17451), BH10 (M15654); subtype C, ETH2220 (U46016); subtype D, ELI (K03454), Z2Z6 (M22639); undefined, MAL (K03456). HIV-1 group O, ANT70 (L20587), MVP5180 (L20571), ESP1 (U97171), ESP2 (AF068947), ESP3 (AF068948), ESP4 (AF068949), and SIV CPZGAB (X52154). Encircled are the sequences corresponding to different HIV-1 group M subtypes, as well as the HIV-1 group O subtype A. An asterisk indicates bootstrap resampling values (1,000 sets) of 100%. (B) A phylogenetic consensus tree was constructed using the 12 available pol sequences (a 792-bp fragment encoding amino acids 1 to 264 of RT) and this segment in the complete RT sequence from the 4 Spanish isolates. GenBank accession numbers for these sequences are as follows: BCF01 (Y14496), BCF02 (Y14497), BCF03 (Y14498), BCF06 (Y14499), BCF07 (Y14500), BCF08 (Y14501), BCF011 (Y14502), BCF013 (Y14503), RUD (Y14504), VAU (Y14505), ANT70 (L20587), MVP5180 (L20571), ESP1 (U97171), ESP2 (AF068947), ESP3 (AF068948), and ESP4 (AF068949). The 11 sequences forming subtype A-O are encircled. Bootstrap resampling values (1,000 sets) >80% and >99% are indicated by one and two asterisks, respectively. All phylogenetic relationships were determined and edited using the MEGA version 1.02 (28) and TreeView version 1.5.0 (42) programs, respectively. The distance between two sequences in each tree is obtained by summing the length of the connecting branches, using the corresponding scale (in nucleotide substitutions per nucleotide [s/nt]).
Interisolate nucleotide sequence diversities were calculated from a distance matrix based on the Kimura two-parameter model (26), while point mutation frequencies were calculated for each set of sequences relative to the corresponding consensus. An expected average of genetic distances was obtained between group O and M sequences (33.2%; range 31.7 to 35.1%), while the intragroup diversities ranged from 1.6 to 10.7% for group O and from 2.4 to 13.5% for group M. Using this large pol gene fragment (approximately 17% of the HIV-1 genome), it may be possible to establish intragroup O subtypes. The average point mutation frequency for the Spanish isolates is 1.1 × 10−2 s/nt, while divergence between isolates in the entire RT nucleotide sequence ranged from 1.6 to 2.8% (Table 2). RT sequences from these group O isolates form a cluster with the ANT70 isolate (average intracluster divergence, 3.3%) which is separate from the MVP5180 isolate (9.9% divergence from the cluster in group O) (Fig. 1A). The average genetic distance for intrasubtype classification in group M (based on the same isolates used to construct the phylogenetic tree; Fig. 1A) was 4.9%, greater than the distance used to establish a group O subtype. In addition, the average intersubtype diversity in group M was 11.4%, resembling the distance between MVP5180 and the other group O viruses found in this cluster.
TABLE 2.
Comparison of the genetic variability between the pol sequences of HIV-1 groups O and M
| HIV-1 group, isolate, source, and no. | Subtype classification | Regiona | Mutation frequencyb | % Nucleotide diversityc
|
|
|---|---|---|---|---|---|
| Average ± SD | Range | ||||
| O | |||||
| 4 Spanish isolates (this study) | Total | 1–560 | 1.1 × 10−2 | 2.2 ± 0.5 | 1.6–2.8 |
| 4.4 × 10−2 | 7.8 ± 2.8 | 1.4–12.9 | |||
| 16 isolates (13) (this study) | Subtype A-O | 1–264 | 2.9 × 10−2 | 5.0 ± 1.4 | 1.4–7.1 |
| Non-subtype A-O | 7.8 × 10−2 | 9.0 ± 1.5 | 5.7–12.9 | ||
| Quasispecies (this study) | 28–116 | 6.5 × 10−3 | 1.1 ± 0.6 | 0–2.4 | |
| M | |||||
| Subtype B (49) | Total | 1–560 | 1.9 × 10−2 | 3.5 ± 0.7 | 0.8–5.4 |
| 5.0 × 10−2 | 8.9 ± 3.3 | 1.8–14.4 | |||
| 10 isolates (40) | Subtype B | 1–264 | 2.7 × 10−2 | 4.9 ± 1.1 | 1.8–4.1 |
| Non-subtype B | 6.5 × 10−2 | 10.6 ± 2.7 | 2.8–14.4 | ||
| Quasispecies (41, 49) | 41–108/181–219 | 7.9 × 10−3 | 1.4 ± 1.0 | 0–5.4 | |
Numbers correspond to codons in the RT. The full RT is 560 amino acids in length, while the polymerase domain spans codons 1 to 266.
Mutation frequency is defined as the proportion of mutant positions relative to the consensus nucleotide sequence for each sample group; the frequencies have been calculated by dividing the number of mutations (relative to the consensus) by the total number of nucleotides sequenced in the corresponding group.
Corresponds to the average proportion of substituted nucleotides when pairs of sequences were compared by applying the Kimura two-parameter model (26).
To corroborate a possible subtype definition in group O, a phylogenetic tree was constructed using a 792-bp fragment in the RT-coding region from the 4 Spanish isolates and the same fragment from 12 HIV-1 group O pol gene sequences available in the database (39). Figure 1B shows a neighbor-joining tree derived from RT sequences of the polymerase domain (nucleotides 1 to 792, coding for RT amino acids 1 to 264). It is evident that 11 of the pol sequences analyzed (including the 4 isolates sequenced in this study) form a cluster in group O similar to that observed with the complete RT-coding region. This cluster, supported by 100% of the bootstrap trees, is designated clade A of HIV-1 group O (subtype A-O). The other five group O RT sequences (BCF06, BCF11, MVP5180, RUD, and VAU) shown in this tree (Fig. 1B) are more divergent and represent an internal tree topology statistically less significant. The average point mutation frequency for all the group O RT sequences is 4.4 × 10−2 s/nt, with an overall divergence of 7.8% (Table 2), whereas a frequency of only 2.9 × 10−2 s/nt (nucleotide diversity 5%) was calculated for the proposed subtype A-O (Table 2). In contrast, the average point mutation frequency is 7.8 × 10−2 s/nt (nucleotide diversity, 9%) for pol sequences of those group O isolates not clustering with subtype A-O. Intrasubtype A diversity in group O can be compared with that of subtype B in group M (average point mutation frequency of 2.7 × 10−2 s/nt; divergence of 4.9%) (Table 2). When comparing subtype B-M isolates with non-subtype B isolates in group M, a mutation frequency of 6.5 × 10−2 s/nt and a sequence divergence of 10.6% were observed. Furthermore, the pairwise genetic distances between any member of subtype A-O and the other group O isolates ranged from 8.6 to 12.9% (average, 10.3%), similar to those genetic distances for group M intersubtype comparisons (data not shown). Together, these data suggest that the RT sequences of 11 group O isolates form a clade designated subtype A within HIV-1 group O viruses, similar to those subtypes described for group M viruses.
Finally, a 432-bp pol gene sequence fragment from the earliest known HIV-1 group O isolate (HIV1T29) (25) was used to construct a neighbor-joining phylogenetic tree with the 16 pol gene sequences available in the database (including the three new isolates from this study). The HIV-1 group O Norwegian isolate did not cluster with the subtype A-O viruses; being placed with the more divergent sequences (data not shown). However, the topology and cluster found in the former tree construction (Fig. 1B) were retained and supported by 978 of 1,000 bootstrap trees (data not shown).
Deduced amino acid sequences of the RT from all group O isolates and identification of drug-resistant genotypes over the course of infection and treatment.
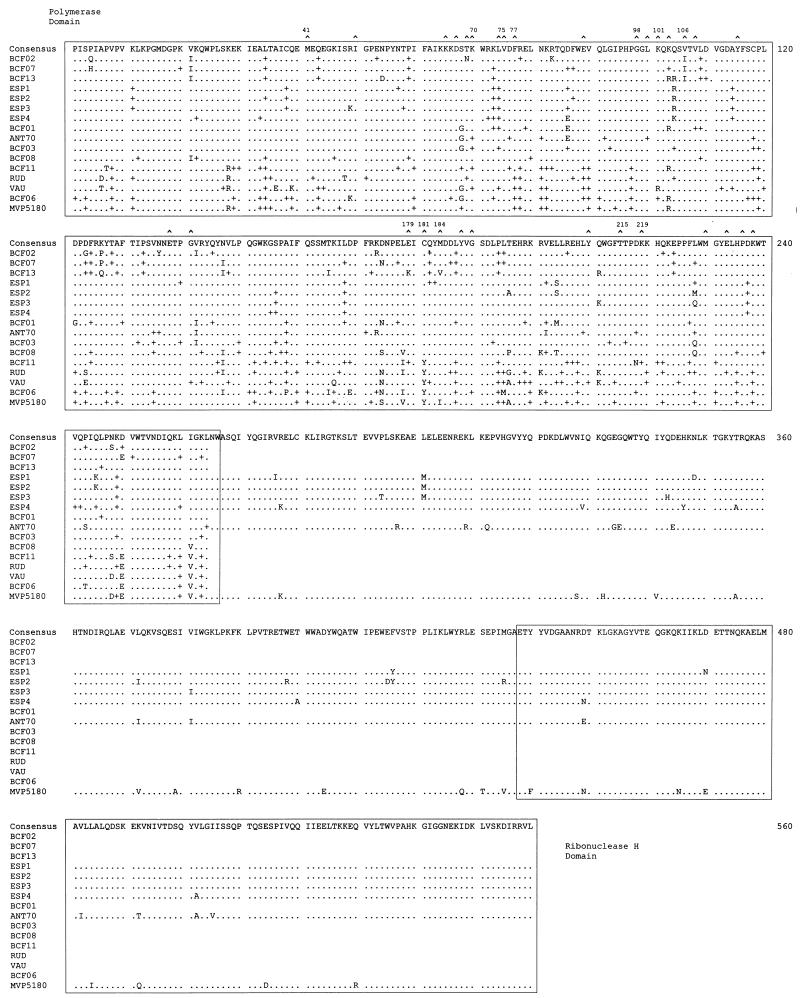
The consensus RT amino acid sequence of our four group O isolates was aligned with 12 HIV-1 group O sequences in the Los Alamos National Laboratory Database (39) (Fig. 2). Several amino acid substitutions are found scattered along the entire analyzed region. When comparing the only six full RT sequences available (ESP1-4, ANT70, and MVP5180), a higher degree of heterogeneity was observed in the “connection” subdomain and RNase H domain (residues 267 to 560) than in the DNA polymerase domain of RT (3.1 × 10−2 and 1.9 × 10−2 substitutions per amino acid, respectively). This observation is consistent with comparisons of RT sequences from group M viruses (39).
FIG. 2.
Multiple alignment of amino acid sequences of the RT of 16 HIV-1 group O isolates. The top line corresponds to the group O consensus amino acid sequence. Six full-length HIV-1 group O RT sequences, 3 from the database and 3 from this study, as well as 12 annotated RT sequences containing just the polymerase domain, were used in this alignment. Only those amino acids that differ from the consensus are given. Dots indicate the same residues compared with the group O consensus sequence, and plus signs denote synonymous substitutions in the RT polymerase domain. Residues involved in resistance to RT inhibitors (16, 36) are marked at the top (∧), and some of them are numbered. The polymerase and RNase H domains (24) are boxed.
As previously described (50), three amino acids (Gly-98, Glu-179, and Cys-181) in the group O Spanish sequences have been characterized in group M isolates as amino acid substitutions conferring resistance to NNRTIs (36, 43) (Fig. 2). Gly-98 and Glu-179 have been found in the RT regions of all group O isolates (except the BCF13 isolate containing Lys-179) (13, 50). In all average RT sequences analyzed, including that of patient ESP4 treated with nucleoside analogs, there are no substitutions commonly associated with nucleoside analog resistance (Fig. 2). It is important to note that patient ESP2 had received antiretroviral therapy only after this sample (ESP2/+4) was obtained and used for these RT sequence analyses.
In addition to the single ESP3 and ESP4 samples, we have sequenced and analyzed the RT-coding regions of proviral DNA from the untreated ESP1 and treated ESP2 patients in a longitudinal study. The consensus nucleotide sequences encoding amino acids 28 to 219 of RT were sequenced from PCR-amplified pol gene products from the first sample (50) and from 4-, 11-, and 19-month samples of patient ESP1 and from the 4-, 11-, and 17-month samples of patient ESP2. As expected, a longitudinal analysis of the consensus RT-coding region of all four samples of untreated ESP1 showed no substitutions conferring drug resistance (Fig. 3). However, an alanine was found at position 75 in the sample ESP1/+11. This substitution has not been associated with loss of sensitivity to any drug. However, only a single transition mutation (G-223→A) is necessary for an A75T substitution, a reduced genetic distance from the V75T substitution associated with phenotypic resistance to ddI, 2′-3′dideoxycytidine (ddC), and d4T in group M HIV-1 isolates (36). Patient ESP2, infected by patient ESP1, initiated antiretroviral therapy 14 months after HIV diagnosis (Table 1). No amino acid substitutions were detected in the sample ESP2/+4, i.e., prior to antiretroviral therapy (Fig. 3). However, 7 months later (ESP2/+11 sample, 2 months after the start of AZT + ddI therapy), five substitutions were identified in this 192-amino-acid fragment (2.6% sequence variation from that of ESP2/+4), suggesting an accelerated evolution to permit or compensate for the emergence of subsequent drug-resistant substitutions in the RT.
FIG. 3.
Amino acid sequence alignment of an RT fragment from sequential samples of two HIV-1 group O-infected individuals. A fragment of 192 amino acids (spanning codons 28 to 219) from two sets of longitudinal samples (ESP1 and ESP2) is shown. The top line is the consensus amino acid sequence from the first group O pol sequence described in Spain (ESP1/0) (50) and corresponds with time zero for the longitudinal samples. Symbols are described in the legend to Fig. 2. Those residues involved in resistance to RT inhibitors (16) for which amino acid substitutions were found are numbered. Boxes enclose the sequences from longitudinal samples.
In ESP2/+11, the T215S substitution, resulting from an A562→T nucleotide transition, appears to be an intermediate of the T215Y substitution found in ESP2/+17 (Fig. 3). In patient ESP2, this AZT resistance substitution (T215Y) appears after a switch at month 12 from an AZT plus ddI regimen to a d4T plus 3TC plus indinavir treatment regimen. However, the appearance of an intermediate substitution, T215S in ESP2/+11, suggests that the T215Y change emerged prior to the switch in treatment strategies and remained in the virus population for another 5 to 6 months in the absence of AZT pressure. Interestingly, the M184V substitution, associated with resistance to 3TC and cross-resistance to ddC and ddI, did appear after 5 months of d4T + 3TC + indinavir therapy (ESP2/+17) (Fig. 3). Many factors, including poor adherence to the treatment regimen, poor drug tolerance, and/or high viral loads at the start of therapy, may have contributed to the rapid emergence of the M184V substitution. In contrast to the appearance of drug resistance mutations, the Gly at position 98, highly conserved among all group O isolates previously described (13, 50) and associated with NNRTI resistance (36), had reverted to Glu in sample ESP2/+17 (Fig. 3). Finally, a stop codon at position 88 was found in this sample, altering the open reading frame, the significance of which will be discussed later.
pol gene quasispecies evolution as determined by HTA and cloning-sequence analysis.
More or less sequence heterogeneity can be distinguished by HTA via the number of mismatches between the probe and the test sequences, resulting in slower or faster electrophoretic migration, respectively (11, 12). Previous applications of HTA employed HIV-1 PCR products from the long terminal repeat (37) and the env gene (9, 11, 12). Although never applied to pol gene studies, HTA is an appropriate qualitative assay to study heterogeneity in the RT-coding region of HIV-infected patients treated with nucleoside analogs or NNRTI, i.e., drugs that affect RT evolution. Therefore, we decided to use the full HIV-1 RT-coding region for HTA and qualitative variability studies. A 1,680-bp fragment of the pol gene was PCR amplified directly from DNA extracts of patient PBMCs. Products of three different PCR amplifications of the same sample were mixed to prevent overrepresentation of single isolates. Clones were then generated by ligation of the PCR-amplified RT region into the pRT6 vector (50). The ESP1/0 RT clone (50) of the earliest group O Spanish isolate was used as a template to generate a 32P-labeled probe for HTA.
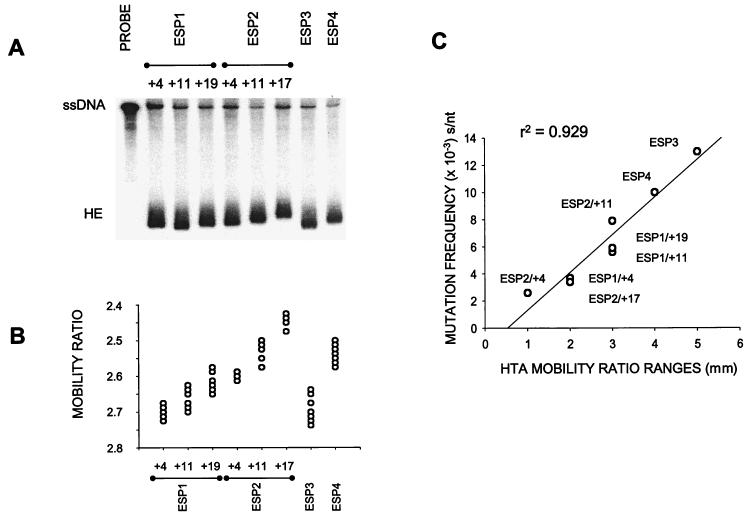
HTA was then applied to compare the genetic diversity in the average PCR-amplified products and that between 10 individual clones from each patient sample (Table 1). Figure 4A shows the heteroduplexes of probe DNA with average RT fragments of each sample. Even though we did not observe a clear range of heteroduplexes using the average PCR product of one sample, the genetic distances between a particular sample and ESP1/0 as determined by sequence analysis correlated with the actual migration distance of that sample DNA hybridized to the ESP1/0 probe DNA (data not shown). A heteroduplex mobility ratio (see Materials and Methods) correcting for any inaccuracies due to electrophoresis was used for comparison of the sequence diversity between RT clones from each patient sample. Figure 4B shows a plot of mobility ratios for all clones of each analyzed sample. The distribution of dots (clones) represents the pol sequence heterogeneity within the corresponding sample and is proportional to the complexity of quasispecies. The RT clones of patient ESP1 showed a more consistent distribution of heteroduplexes at 4, 11, and 19 months. In patient ESP2, the diversity of pol sequence quasispecies appears to increase after the initiation of triple-drug combination therapy (sample ESP2/+11), compared to a more restricted heterogeneity in clones of sample ESP2/+17 (Fig. 4B). In sample ESP2/+17, pol sequence heterogeneity may be biased by those variants harboring drug resistance substitutions (M184V and T215Y), resulting in a narrower distribution of quasispecies. Interestingly, HTA revealed that samples ESP3 and ESP4 contained a wider distribution of pol gene quasispecies than in those of patients ESP1 and ESP2 at any sample time.
FIG. 4.
HTA of eight HIV-1 group O samples from Spain. A 1,680-bp fragment corresponding to full-length encoding RT sequences was analyzed by HTA. (A) A comparison of quasispecies diversity in eight average sequences (Table 1) using HTA. PCR-amplified pol gene products from samples were hybridized to the probe, a PCR-amplified 32P-labeled clone, separated on a nondenaturing polyacrylamide gel, and then autoradiographed as described in Materials and Methods. Samples corresponding to those described in Table 1 are indicated above each lane. The lane labeled PROBE contains only PCR-amplified, radiolabeled probe DNA in the heteroduplex reaction. ssDNA and HE designate the single-stranded probe DNA and heteroduplexes, respectively. (B) Mobility ratios calculated by dividing the migration distance of each heteroduplex band by the migration distance of the single-stranded probe were plotted for each HTA performed on the 10 clones of each sample. The distribution of dots represents the relative heterogeneity of quasispecies in each sample. (C) Comparison between the heterogeneity of quasispecies inferred by HTA for each sample and those calculated by sequencing the corresponding set of clones. Mobility ratio ranges (in millimeters) of the heteroduplexes from each group of 10 clones were plotted against the point mutation frequency (in substitutions per nucleotide). Frequencies were obtained by sequencing a 267-bp fragment, RT codons 28 to 116, from each individual genomic clone of the corresponding group (see Fig. 5) and were calculated as described in footnote b of Table 2. A coefficient of regression was obtained (r2 = 0.929), with a 99% degree of confidence.
Although HTA provides a qualitative assessment of heterogeneity and a relative measure of sequence diversity, it cannot define the direction of quasispecies evolution or indicate specific substitutions responsible for this heterogeneity. Thus, to corroborate HTA analyses and to investigate specific amino acid changes, we have sequenced a 267-bp fragment of RT (amino acids 28 to 116) from each clone analyzed by HTA. The nucleotide sequences of the 10 clones from each viral sample are shown in Fig. 5. Mutation frequencies of the quasispecies in each sample were directly related to the distribution of heteroduplexes with HTA (r2 = 0.929; Fig. 4C). Quasispecies in samples ESP1/+4, +11, and +/19 had a point mutation frequency of 3.7 × 10−3, 5.6 × 10−3, and 5.9 × 10−3 s/nt, respectively. These mutation frequencies are comparable to those previously described for the pol gene of HIV-1 group M samples isolated from untreated patients (Table 2) (41, 49). In support of earlier qualitative analyses by HTA, we did observe a modest twofold decrease in mutation frequency from sample ESP2/+11 (7.9 × 10−3 s/nt) to sample ESP2/+17 (3.4 × 10−3 s/nt) or after the initiation of triple-drug therapy. This difference can also be seen in Fig. 4C in which the distribution of quasispecies by HTA or sequence analysis for sample ESP2/+17 falls below sample ESP2/+11 on the curve. Higher point mutation frequencies, as inferred by HTA, were obtained for ESP3 (1.3 × 10−2 s/nt) and ESP4 (1.0 × 10−2 s/nt).
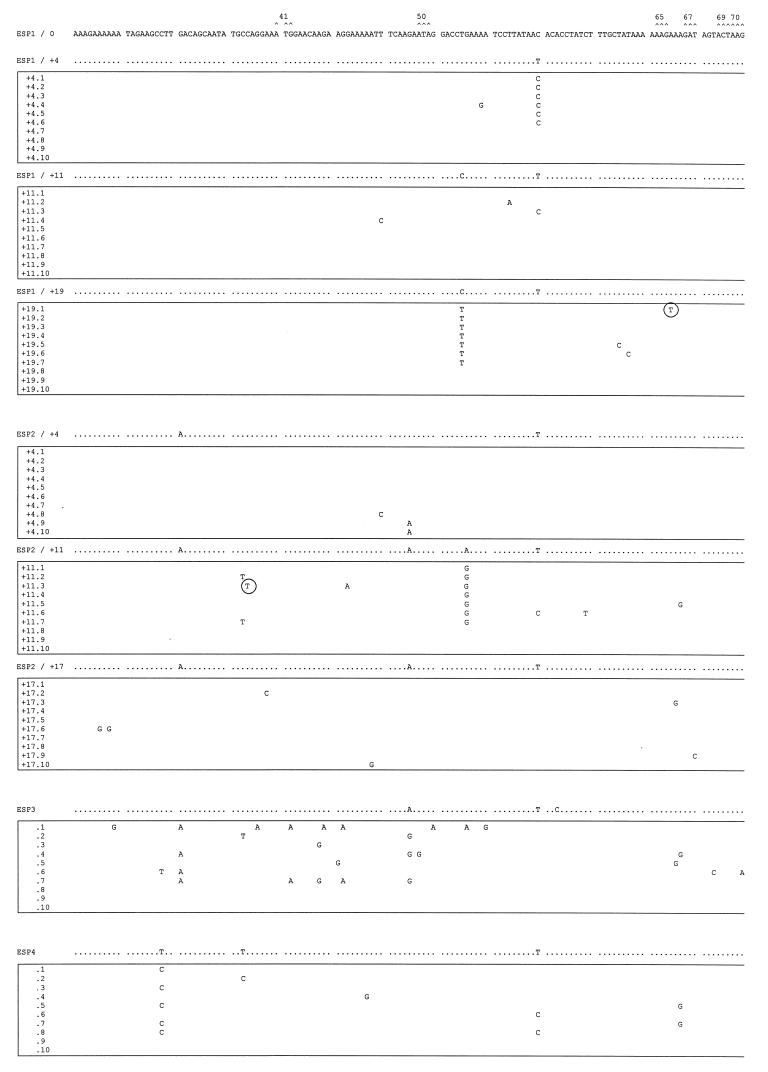
FIG. 5.
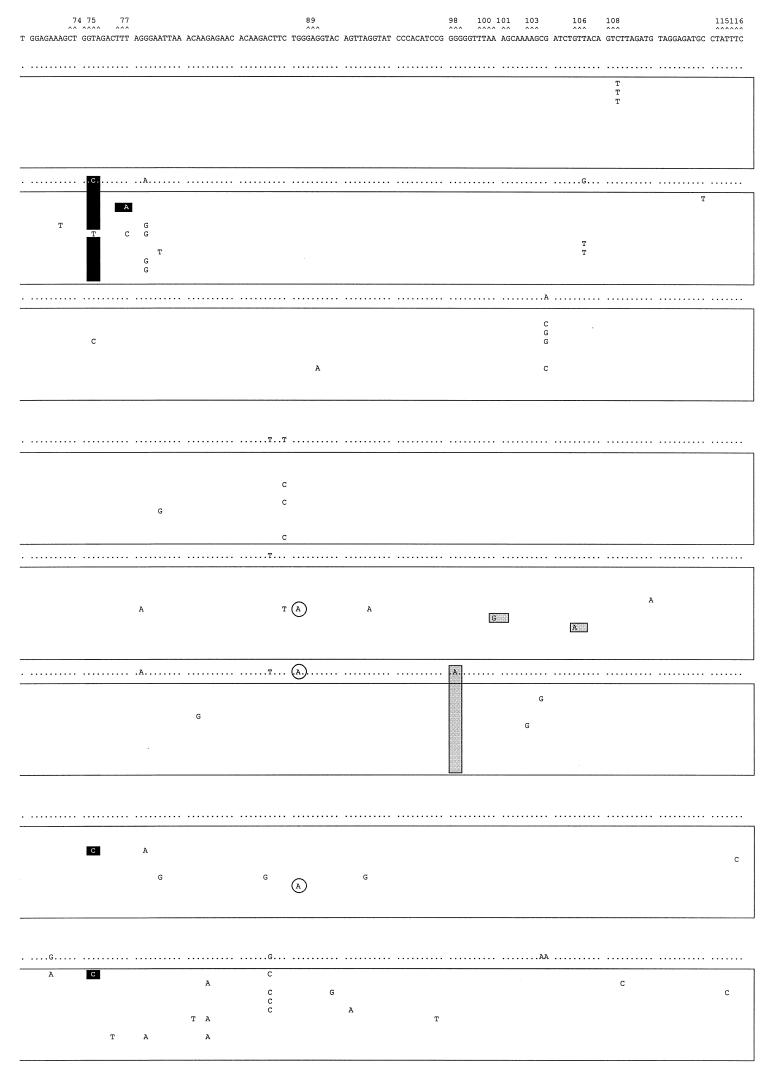
Mutant spectra of pol gene quasispecies in samples from the longitudinal study and the other two HIV-1 group O isolates. Nucleotide sequences (positions 82 to 348, encoding amino acids 28 to 116 of the RT) of clones from each sample are boxed. The uppermost sequence corresponds with the sample used as reference (ESP1/0). The nucleotide sequence at the top of each box represents the consensus nucleotides (determined experimentally by the direct sequencing of the average uncloned PCR product) for each sample. Only those nucleotides that differ from the ESP1/0 sequence are indicated. The mutant spectrum of each quasispecies is represented by the number of mutations from the corresponding consensus sequence. Triplets encoding residues involved in resistance to RT inhibitors (16) are numbered and marked (∧) above the ESP1/0 sequence. Shaded and solid boxes designate nucleotide mutations encoding amino acid substitutions associated with resistance to NNRTIs or nucleoside analogs, respectively. Encircled nucleotides are those mutations leading to stop codons in the pol gene.
In the more homogeneous quasispecies of the untreated patient ESP1, one clone (ESP1/+11.2, Fig. 5) contained a T231→A mutation resulting in an F77L substitution (black boxed nucleotide in Fig. 5), one of five substitutions associated with multiple nucleoside analog resistance (54). Nine of 10 ESP1/+11 clones (Fig. 5) and the consensus sequence (Fig. 3) had a T224→C mutation generating the V75A substitution. This mutation may affect deoxynucleoside triphosphate binding or inhibition by ddI, ddC, and d4T (36), considering that the V75T substitution may confer resistance to these nucleoside analogs. As described previously for group M viruses (40), substitutions conferring resistance to antiretroviral agents can also be found in group O isolates from patients exposed to unrelated RT inhibitors. Two of the RT clones of patient sample ESP2/+11 have amino acid substitutions associated with NNRTI resistance (solid boxed nucleotide in Fig. 5), e.g., clone ESP2/+11.6 contains a A301→G mutation corresponding to the K101E substitution, while clone ESP2/+11.7 had a G316→A mutation resulting in a V106I substitution (Fig. 5). As predicted by the average RT sequence of sample ESP2/+17, the G98E substitution was identified in all clones of this sample, suggesting a possible reversion in an NNRTI-resistant mutation, present on the RT-coding region of all reported group O isolates. In spite of a broad distribution of quasispecies in samples ESP3 and ESP4, only clones ESP3.3 and ESP4.1 had the substitution V75A, which was even remotely associated with a nucleoside analog-resistant phenotype (see above). Amber stop codons were identified in one clone of several samples (i.e., clones ESP1/+19.1, ESP2/+11.3, ESP2/+11.5, and ESP3.7) (circled nucleotides in Fig. 5). However, all clones of sample ESP2/+17 as well as the consensus RT sequence contained a G263→A mutation, resulting a premature amber stop codon at amino acid position 88. Although this result suggests that sample ESP2/+17 contained RT-deficient uninfectious virus, it is important to note that a minor population of infected cells producing infectious virus may not be represented in this set of clones.
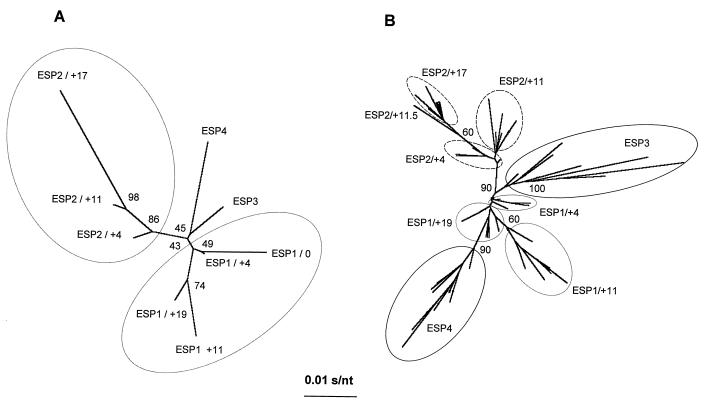
Finally, a phylogenetic tree was constructed using average sequences of the 576-bp pol fragment of the ESP1 and ESP2 longitudinal samples as well as those of ESP3 and ESP4. Average sequences from the ESP2 isolate at times 4, 11, and 17 months show a directional evolution gradually diverging from that of the ESP1/0 virus and supported by bootstrap analyses (Fig. 6A). In contrast, the average pol sequences of ESP1 isolates over this same time period displayed an unpredictable pattern of evolution (Fig. 6A). These data on average sequence analyses were compared to the phylogenetic relationships among the 80 clones from these Spanish isolates (Fig. 6B). ESP2 clones showed the same directional evolution as the average sequences of ESP2 (Fig. 6A), while ESP1 clones were grouped in the center of the tree, with a poorer statistical robustness. The eight different clusters of clones corresponding to the sample of origin (except clone ESP2/+11.5) (Fig. 6B) had an average branch length proportional to the heterogeneity determined by both sequence analysis and HTA (Fig. 4C).
FIG. 6.
Phylogenetic relationships among eight HIV-1 group O Spanish samples. (A) Consensus 576-bp fragments encoding amino acids 28 to 219 of the RT (Fig. 3) were utilized to construct a neighbor-joining tree as described in Materials and Methods. Enclosed are the longitudinal samples corresponding to the related viruses (ESP1 and ESP2). (B) A phylogenetic tree was constructed from a 267-bp fragment (codons 28 to 116) in the RT-coding region of 80 individual clones from these eight samples. Each set of quasispecies is encircled. Bootstrap resampling percentage values (1,000 sets) are indicated.
DISCUSSION
The discovery of two distinct HIV-1 lineages, groups M and O, suggests an origin from two different zoonotic infections by the same primate retrovirus (27, 38, 58). Even though most group O strains originate from Central Africa (10, 19, 58), HIV-1 group O infections have been reported as early as 1960 (25) and from three different continents (5, 7, 21, 30, 55), suggesting a wider distribution of group O than of many subtypes of HIV-1 group M. Similar to results of many studies of group M isolates, most genotypic and phylogenetic descriptions of group O virus are based on sequences from the gag and env genes (4, 7, 20, 23, 27, 30, 34, 35), with less emphasis on the pol gene (13, 50). The relatively conserved pol gene (39) displays only two- to threefold lower variability than the env gene (49) and can be used to reconstruct the same phylogenetic relationships for different HIV-1 group M isolates as determined by analysis of the gag or env genes (1, 48, 53, 56).
The lack of subtype definition for group O strains is due to the limited number and/or length of group O sequences described for any gene (27). Our study has provided the nucleotide sequences of the complete RT-coding region (1,680 bp) of the pol gene from three new HIV-1 group O isolates. A phylogenetic study of six RT-coding sequences of group O viruses, four from Spain (50, this study) and two from Cameroon (10, 19), showed a cluster of five of six isolates. Using a smaller RT fragment (792 bp) for this phylogenetic analysis, we were able to both confirm this subtyping and further define a statistically significant cluster (11 of 16 isolates) in the same phylogenetic position and with a genetic diversity similar to that described for the subdivision of the pol gene into group M subtypes (39, 49). Previous phylogenetic studies of gag, pol, and env sequences from a subset of these group O isolates (4, 13, 30, 35) and recent env genotypic analyses of the new HIV-1 group O isolates (34a) further support the subdivision of group O in at least two collections of sequences. Based on these analyses, this cluster is now designated subtype A in group O (or clade A-O).
As a consequence of significant sequence diversity (24 to 49%) (19, 30, 58), groups M and O differ at genotypic sites, influencing phenotypic characteristics of HIV-1 replication and structure. For example, most if not all HIV-1 group O isolates contain the genetic information encoding resistance to several NNRTIs (13, 14, 50), suggesting that these drugs may not be effective in HIV-1 group O-infected individuals. Our first objective was the identification of any drug-resistant genotypes in pol gene quasispecies and in the average pol sequences, in the absence of treatment with the related antiretroviral drug. These Spanish group O isolates contained RT amino acids Gly-98, Glu-179, and Cys-181, thought to confer NNRTI resistance in group M viruses. Interestingly, the ESP2 patient receiving antiretroviral treatment lost the glycine at position 98 (highly conserved residue in all characterized group O isolates (references 13 and 50 and this study) to a glutamate in RT at 17 months, perhaps to compensate for the drug-resistant substitutions M184V and T215Y. An A98G substitution in group M isolates confers resistance to nevirapine, pyridinones, and thiocarboxanilides (16, 36). Finally, some individual clones from ESP1 and ESP2 patients contained substitutions (K101E and V106I) associated with NNRTI resistance or nucleoside analog resistance (F77V) (16, 36) previously unidentified in group O viruses. As described in earlier studies with group M viruses (40), the findings presented herein prove that drug-resistant substitutions arise in a portion of evolving group O quasispecies in the absence of drug pressure.
Using the same samples from this infected couple, we also screened for the appearance of nucleoside analog-resistant substitutions related to therapy. Over 20 months, virus from the untreated patient ESP1 developed few amino acid substitutions in the consensus pol sequence, none associated with antiretroviral drug resistance. However, two drug-resistant substitutions (M184V and T215Y) were identified at month 17 in viral sequences from patient ESP2. These results suggest that treatment with AZT and 3TC (in different regimens) could select for the same RT substitutions, T215Y and M184V, respectively, in group O viruses as those found in AZT- or 3TC-resistant group M viruses. However, the emergence of the M184V substitution in group M virus may reverse the AZT resistance conferred by the T215Y substitution (29). Considering that (i) the T215Y substitution may not confer resistance in the presence of the M184V substitution and (ii) most drug-resistant substitutions should revert to the wild type in the absence of drug pressure (8), it is surprising that a T215Y substitution was found in the consensus sequence and in all RT clones (data not shown) 5 months after patient ESP2 had switched from an AZT + ddI to a d4T + 3TC + indinavir treatment regimen. This situation is not unprecedented in group M infections (3, 18, 29) but does occur with a frequency low enough to suggest that the T215Y substitution in the presence of valine at position 184 may be more stable in the group O virus. Any verification of these hypotheses and observations would require the study of several HIV-1 group O-infected individuals treated with antiretroviral drugs. Unfortunately, the difficulties of identifying HIV-1 group O infections and the current lack of therapy for even HIV-1 group M infections in the areas where these viruses are most prevalent (e.g., Cameroon) imply that extensive studies of the treatment of HIV-1 group O-infected patients is nearly impossible. Finally, a substitution leading to an amber stop codon at position 88 was found in the consensus pol sequence and in all clones of patient ESP2 at 17 months. We were unable to obtain plasma samples from these patients to screen for this premature stop codon or other substitutions in the RT-coding region of HIV-1 RNA in plasma. However, this stop codon was not found in the consensus RT sequence of a subsequent sample (22 months) (34a).
Genetic variability in a defined genomic region (i.e., RT-coding region) may be influenced by the viability of specific nucleotide mutations and by specific host-environmental factors such as the immune response (15). In the absence of treatment, pol gene quasispecies of patient ESP1 may have evolved to increase viral fitness and/or to avoid the host immune response (e.g., a cell-mediated cytotoxic CD8+ T-cell response). However, the evolution of the RT-coding region appears to be erratic, without a clear pattern of development, perhaps owing to an adaptation in other genomic regions (i.e., in the env gene) or a weak immune pressure on pol gene products. In approximately the same period of time, a disparate evolution in pol gene quasispecies was observed in patient ESP2, infected by patient ESP1. Differences in virulence of the infecting strain and/or a weaker immune response may explain the rapid development of clinical symptoms in patient ESP2. Despite a decrease in viral load after the initiation of antiretroviral therapy, the pol gene quasispecies continued to expand in patient ESP2, suggesting that this directed evolution was not due to nonspecific convergent evolution by the drug-mediated reduction in viral loads as previously described by others (45). This expansion likely contributed to the subsequent emergence of drug-resistant substitutions. The switch in therapy would have imposed new restrictions on the selection of quasispecies with decreased viral fitness (15). However, alternating therapies may also maintain a pressure for the development of drug-resistant and compensatory mutations and prevent the appearance of nonspecific substitutions found in the absence of therapy.
We did observe a directed evolution in the consensus pol gene sequence from patient ESP2 over the course of therapy but a more random evolution in the untreated ESP1 patient. Statistically significant clusters of all RT clones from each ESP2 sample followed this same directed evolution. In contrast, a lack of statistical robustness was found for the clusters of sample clones from the untreated ESP1 patient. These results were derived from both heteroduplex tracking and sequence analysis of the pol gene contained in specific clones or the average PCR-amplified products. Interestingly, the small differences in heteroduplex migration with all clones correlated directly with the calculated nucleotide sequence diversity among the same clones. A direct relationship between heteroduplex migration in an HTA and sequence analysis supports the use of HTA over sequencing for a rapid assessment of quasispecies diversity.
In conclusion, we have identified in pol gene quasispecies of group O viruses several substitutions associated with drug resistance in the absence of the corresponding inhibitor. Even with similar point mutation frequencies and inter- and intrapatient divergence, HIV-1 group O viruses have an advantage over the group M strains in that they are naturally resistant to NNRTIs. In addition to this intrinsic resistance shared with HIV-2 and all other retroviruses, treatment of an HIV-1 group O infection resulted in directed evolution in the pol gene as well as the emergence of specific drug-resistant substitutions. A slow but continuous worldwide spread of HIV-1 group O, difficulties in its detection, and its potential resistance to some antiretroviral drugs provides a strong justification for these analyses and subsequent studies of the evolution of HIV-1 group O.
ACKNOWLEDGMENTS
This research was supported by developmental and supplemental funds from the Center for AIDS Research (NIH A1-36219) at Case Western Reserve University (E.J.A.). Research performed in the laboratory of V. Soriano was supported by funds from AIES and CAM-project 08.2/0014/1997.
REFERENCES
- 1.Apetrei C, Descamps D, Collin G, Loussert-Ajaka I, Damond F, Duca M, Simon F, Brun-Vezinet F. Human immunodeficiency virus type 1 subtype F reverse transcriptase sequence and drug susceptibility. J Virol. 1998;72:3534–3538. doi: 10.1128/jvi.72.5.3534-3538.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arts E J, Wainberg M A. Mechanisms of nucleoside analog antiviral activity and resistance during human immunodeficiency virus reverse transcription. Antimicrob Agents Chemother. 1996;40:527–540. doi: 10.1128/aac.40.3.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Batisse D, Karmochkine M, Si Mohamed A, Piketty C, Kazatchkine M D, Belec L. Persistence of HIV-1 variants harboring the zidovudine resistance mutation at pol codon 215 in patients who respond to triple combination therapy. AIDS. 1998;12:824–825. [PubMed] [Google Scholar]
- 4.Brennan C A, Hackett J, Jr, Zekeng L, Lund J K, Vallari A S, Hickman R K, Gürtler L, Kaptue L, Von Overbeck J, Hampl H, Devare S G. Sequence of gp41env immunodominant region of HIV type 1 group O from West Central Africa. AIDS Res Hum Retroviruses. 1997;13:901–904. doi: 10.1089/aid.1997.13.901. [DOI] [PubMed] [Google Scholar]
- 5.Britvan L, Gould K, Dryjanski J, Kerndt P, Mascola L, Sun R. Identification of HIV-1 group O infection—Los Angeles County, California, 1996. Morbid Mortal Weekly Rep. 1996;45:561–565. [PubMed] [Google Scholar]
- 6.Chaix-Baudier M-L, Chappey C, Burgard M, Letourneur F, Igual J, Saragosti S, Rouzioux C the French HIV Pediatric Cohort Study Group. First case of mother-to-infant HIV type 1 group O transmission and evolution of C2V3 sequences in the infected child. AIDS Res Hum Retroviruses. 1998;14:15–23. doi: 10.1089/aid.1998.14.15. [DOI] [PubMed] [Google Scholar]
- 7.Charneau P, Borman A M, Quillent C, Guetard D, Chamaret S, Cohen J, Remy G, Montagnier L, Clavel F. Isolation and envelope sequence of a highly divergent HIV-1 isolate: definition of a new HIV-1 group. Virology. 1994;205:247–253. doi: 10.1006/viro.1994.1640. [DOI] [PubMed] [Google Scholar]
- 8.Coffin J M. HIV population dynamics in vivo: implications for genetic variation, pathogenesis, and therapy. Science. 1995;267:483–489. doi: 10.1126/science.7824947. [DOI] [PubMed] [Google Scholar]
- 9.Contag C H, Ehrnst A, Duda J, Bohlin A-B, Lindgren S, Learn G H, Mullins J I. Mother-to-infant transmission of human immunodeficiency virus type 1 involving five envelope sequence subtypes. J Virol. 1997;71:1292–1300. doi: 10.1128/jvi.71.2.1292-1300.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Leys R, Vanderborght B, Vanden Haesevelde M, Heyndrickx L, van Geel A, Wauters C, Bernaerts R, Saman E, Nijs P, Willems H, Taelman H, van der Groen G, Piot P, Tersmette T, Huisman J G, van Heuverswyn H. Isolation and partial characterization of an unusual human immunodeficiency retrovirus from two persons of West-Central African origin. J Virol. 1990;64:1207–1216. doi: 10.1128/jvi.64.3.1207-1216.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delwart E L, Shpaer E G, Louwagie J, McCutchan F E, Grez M, Rübsamen-Waigmann H, Mullins J I. Genetic relationships determined by a DNA heteroduplex mobility assay: analysis of HIV-1 env genes. Science. 1993;262:1257–1261. doi: 10.1126/science.8235655. [DOI] [PubMed] [Google Scholar]
- 12.Delwart E L, Sheppard H W, Walker B D, Goudsmit J, Mullins J I. Human immunodeficiency virus type 1 evolution in vivo tracked by DNA heteroduplex mobility assays. J Virol. 1994;68:6672–6683. doi: 10.1128/jvi.68.10.6672-6683.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Descamps D, Collin G, Letourneur F, Apetrei C, Damond F, Loussert-Ajaka I, Simon F, Saragosti S, Brun-Vezinet F. Susceptibility of human immunodeficiency virus type 1 group O isolates to antiretroviral agents: in vitro phenotypic and genotypic analyses. J Virol. 1998;71:8893–8898. doi: 10.1128/jvi.71.11.8893-8898.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Descamps D, Collin G, Loussert-Ajaka I, Saragosti S, Simon F, Brun-Vézinet F. HIV-1 group O sensitivity to antiretroviral drugs. AIDS. 1995;9:977–978. [PubMed] [Google Scholar]
- 15.Domingo E, Holland J J. RNA virus mutations and fitness for survival. Annu Rev Microbiol. 1997;51:151–178. doi: 10.1146/annurev.micro.51.1.151. [DOI] [PubMed] [Google Scholar]
- 16.Domingo E, Menendez-Arias L, Quiñones-Mateu M E, Holguín A, Gutierrez-Rivas M, Martinez M A, Quer J, Novella I S, Holland J. Viral quasispecies and the problem of vaccine-escape and drug-resistant mutants. Prog Drug Res. 1997;48:99–128. doi: 10.1007/978-3-0348-8861-5_4. [DOI] [PubMed] [Google Scholar]
- 17.Gervaix A, Li X, Kraus G, Wong-Staal F. Multigene antiviral vectors inhibit diverse human immunodeficiency virus type 1 clades. J Virol. 1997;71:3048–3053. doi: 10.1128/jvi.71.4.3048-3053.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Günthard H F, Wong J K, Ignacio C C, Guatelli J C, Riggs N L, Havlir D V, Richman D D. Human immunodeficiency virus replication and genotypic resistance in blood and lymph nodes after a year of potent antiretroviral therapy. J Virol. 1998;72:2422–2428. doi: 10.1128/jvi.72.3.2422-2428.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gürtler L G, Hauser P H, Eberle J, von Brunn A, Knapp S, Zekeng L, Tsague J M, Kaptue L. A new subtype of human immunodeficiency virus type 1 (MVP-5180) from Cameroon. J Virol. 1994;68:1581–1585. doi: 10.1128/jvi.68.3.1581-1585.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hackett J, Jr, Zekeng L, Brennan C A, Lund J K, Vallari A S, Hickman R K, Gurtler L, Kaptue L, Devare S G. Genetic analysis of HIV type 1 group O p24gag sequences from Cameroon and Equatorial Guinea. AIDS Res Hum Retroviruses. 1997;13:1155–1158. doi: 10.1089/aid.1997.13.1155. [DOI] [PubMed] [Google Scholar]
- 21.Hampl H, Sawitzky D, Stoffler-Meilicke M, Groh A, Schmitt M, Eberle J, Gurtler L. First case of HIV subtype O infection in Germany. Infection. 1995;23:369–370. doi: 10.1007/BF01713567. [DOI] [PubMed] [Google Scholar]
- 22.Heyndrickx L, Alary M, Janssens W, Davo N, van der Groen G. HIV-1 group O and group M dual infection in Bénin. Lancet. 1996;347:902–903. doi: 10.1016/s0140-6736(96)91383-5. [DOI] [PubMed] [Google Scholar]
- 23.Hunt J C, Golden A M, Lund J K, Gurtler L G, Zekeng L, Obiang J, Kaptue L, Hampl H, Vallari A, Devare S G. Envelope sequence variability and serologic characterization of HIV type 1 group O isolates from Equatorial Guinea. AIDS Res Hum Retroviruses. 1997;13:995–1005. doi: 10.1089/aid.1997.13.995. [DOI] [PubMed] [Google Scholar]
- 24.Jacobo-Molina A, Arnold E. HIV reverse transcriptase structure-function relationships. Biochemistry. 1991;30:6351–6361. doi: 10.1021/bi00240a001. [DOI] [PubMed] [Google Scholar]
- 25.Jonassen T O, Stene-Johansen K, Berg E S, Hungnes O, Lindboe C F, Froland S S, Grinde B. Sequence analysis of HIV-1 group O from Norwegian patients infected in the 1960s. Virology. 1997;231:43–47. doi: 10.1006/viro.1997.8510. [DOI] [PubMed] [Google Scholar]
- 26.Kimura M. A simple model for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 27.Korber B, Loussert-Ajaka I, Blouin J, Saragosti S. A comparison of HIV-1 group M and group O functional and immunogenic domains in the gag p24 protein and the C2V3 region of the envelope protein, part 4. Los Alamos, N. Mex: Theoretical Biology and Biophysics Group, Los Alamos National Laboratory; 1997. pp. 63–79. [Google Scholar]
- 28.Kumar S, Tamura K, Nei M. MEGA: molecular evolutionary genetic analysis, version 1.0. University Park, Pa: The Pennsylvania State University; 1993. [Google Scholar]
- 29.Larder B A, Kemp S D, Harrigan P R. Potential mechanism for sustained antiretroviral efficacy of AZT-3TC combination therapy. Science. 1995;269:696–699. doi: 10.1126/science.7542804. [DOI] [PubMed] [Google Scholar]
- 30.Loussert-Ajaka I, Chaix M-L, Korber B, Letourneur F, Gomas E, Allen E, Ly T-D, Brun-Vézinet F, Simon F, Saragosti S. Variability of human immunodeficiency virus type 1 group O strains isolated from Cameroonian patients living in France. J Virol. 1995;69:5640–5649. doi: 10.1128/jvi.69.9.5640-5649.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Loussert-Ajaka I, Descamps D, Simon F, Brun-Vézinet F, Ekwalanga M, Saragosti S. Genetic diversity and HIV detection by polymerase chain reaction. Lancet. 1995;346:912–913. doi: 10.1016/s0140-6736(95)92762-x. [DOI] [PubMed] [Google Scholar]
- 32.Loussert-Ajaka I, Ly T D, Chaix M L, Ingrand D, Saragosti S, Couroucé A M, Brun-Vézinet F. HIV-1/HIV-2 seronegativity in HIV-1 subtype O infected patients. Lancet. 1994;343:1393–1394. doi: 10.1016/s0140-6736(94)92524-0. [DOI] [PubMed] [Google Scholar]
- 33.Louwagie J, McCutchan F E, Peeters M, Brennan T P, Sanders-Buell E, Eddy G A, van der Groen G, Fransen K, Gershy-Damet G-M, Deleys R, Burke D S. Phylogenetic analysis of gag genes from 70 international HIV-1 isolates provides evidence for multiple genotypes. AIDS. 1993;7:769–780. doi: 10.1097/00002030-199306000-00003. [DOI] [PubMed] [Google Scholar]
- 34.Mas A, Quiñones-Mateu M E, Soriano V, Domingo E. env gene characterization of the first HIV-1 group O isolate from Spain. AIDS Res Hum Retroviruses. 1996;12:1647–1649. doi: 10.1089/aid.1996.12.1647. [DOI] [PubMed] [Google Scholar]
- 34a.Mas, A., and V. Soriano. Unpublished data.
- 35.Mauclere P, Loussert-Ajaka I, Damond F, Fagot P, Souquieres S, Lobe M M, Keou F-X M, Barré-Sinoussi F, Saragosti S, Brun-Vézinet F, Simon F. Serological and virological characterization of HIV-1 group O infection in Cameroon. AIDS. 1997;11:445–453. doi: 10.1097/00002030-199704000-00007. [DOI] [PubMed] [Google Scholar]
- 36.Mellors J W, Schinazi R F, Larder B A. Mutations in retroviral genes associated with drug resistance, part 3. Los Alamos, N. Mex: Theoretical Biology and Biophysics Group, Los Alamos National Laboratory; 1996. pp. 206–241. [Google Scholar]
- 37.Montano M A, Novitsky V A, Blackard J T, Cho N L, Katzenstein D A, Essex M. Divergent transcriptional regulation among expanding human immunodeficiency virus type 1 subtypes. J Virol. 1997;71:8657–8665. doi: 10.1128/jvi.71.11.8657-8665.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Myers G. Tenth anniversary perspectives on AIDS. HIV: between past and future. AIDS Res Hum Retroviruses. 1994;10:1317–1324. doi: 10.1089/aid.1994.10.1317. [DOI] [PubMed] [Google Scholar]
- 39.Myers G, Korber B, Hahn B H, Jeang K-T, Mellors J W, McCutchan F E, Henderson L E, Pavlakis G N. Human retroviruses and AIDS 1995. A compilation and analysis of nucleic acid and amino acid sequences. Los Alamos, N. Mex: Theoretical Biology and Biophysics Group, Los Alamos National Laboratory; 1996. [Google Scholar]
- 40.Nájera I, Holguín A, Quiñones-Mateu M E, Muñoz-Fernández M A, Nájera R, López-Galíndez C, Domingo E. The pol gene quasispecies of human immunodeficiency virus: mutations associated with drug resistance in virus from patients undergoing no drug therapy. J Virol. 1995;69:23–31. doi: 10.1128/jvi.69.1.23-31.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nkengasong J N, Janssens W, Heyndrickx L, Fransen K, Ndumbe P M, Motte J, Leonaers A, Ngolle M, Ayuk J, Piot P, van der Groen G. Genotypic subtypes in HIV-1 in Cameroon. AIDS. 1994;8:1405–1412. doi: 10.1097/00002030-199410000-00006. [DOI] [PubMed] [Google Scholar]
- 42.Page R D M. TREEVIEW: an application to display phylogenetic trees on personal computers. Comp Appl Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- 43.Palmer S, Alaeus A, Albert J, Cox S. Drug susceptibility of subtypes A, B, C, D, and E human immunodeficiency virus type 1 primary isolates. AIDS Res Hum Retroviruses. 1998;14:157–162. doi: 10.1089/aid.1998.14.157. [DOI] [PubMed] [Google Scholar]
- 44.Peeters M, Gueye A, Mboup S, Bibollet-Ruche F, Ekaza E, Mulanga C, Ouedrago R, Gandji R, Mpele P, Dibanga G, Koumare B, Saidou M, Esu-Williams E, Lombart J-P, Badombena W, Luo N, Vanden Haesevelde M, Delaporte E. Geographical distribution of HIV-1 group O viruses in Africa. AIDS. 1997;11:493–498. doi: 10.1097/00002030-199704000-00013. [DOI] [PubMed] [Google Scholar]
- 45.Pelletier E, Saurin W, Cheynier R, Letvin N L, Wain-Hobson S. The tempo and mode of SIV quasispecies development in vivo calls for massive viral replication and clearance. Virology. 1995;208:644–652. doi: 10.1006/viro.1995.1195. [DOI] [PubMed] [Google Scholar]
- 46.Perelson A S, Neumann A U, Markowitz M, Leonard J M, Ho D D. HIV-1 dynamics in vivo: virion clearance rate, infected cell life-span, and viral generation time. Science. 1996;271:1582–1586. doi: 10.1126/science.271.5255.1582. [DOI] [PubMed] [Google Scholar]
- 47.Preston B D, Poiesz B J, Loeb L A. Fidelity of HIV-1 reverse transcriptase. Science. 1988;242:1168–1171. doi: 10.1126/science.2460924. [DOI] [PubMed] [Google Scholar]
- 48.Quiñones-Mateu M E, Dopazo J, Esté J A, Rota T R, Domingo E. Molecular characterization of human immunodeficiency virus type 1 isolates from Venezuela. AIDS Res Hum Retroviruses. 1995;11:605–616. doi: 10.1089/aid.1995.11.605. [DOI] [PubMed] [Google Scholar]
- 49.Quiñones-Mateu M E, Holguín A, Dopazo J, Nájera I, Domingo E. Point mutant frequencies in the pol gene of human immunodeficiency virus type 1 are about as high as those in the env gene. AIDS Res Hum Retroviruses. 1996;12:1117–1128. doi: 10.1089/aid.1996.12.1117. [DOI] [PubMed] [Google Scholar]
- 50.Quiñones-Mateu M E, Soriano V, Domingo E, Menendez-Arias L. Characterization of the reverse transcriptase of a human immunodeficiency virus type 1 group O isolate. Virology. 1997;236:364–373. doi: 10.1006/viro.1997.8748. [DOI] [PubMed] [Google Scholar]
- 51.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 52.Schable C, Zekeng L, Pau C-P, Hu D, Kaptue L, Gurtler L, Dondero T, Tsague J-M, Schochetman G, Jaffe H, George J R. Sensitivity of United States HIV antibody tests for detection of HIV-1 group O infections. Lancet. 1994;344:1333–1334. doi: 10.1016/s0140-6736(94)90695-5. [DOI] [PubMed] [Google Scholar]
- 53.Shafer R W, Eisen J A, Merigan T C, Katzenstein D A. Sequence and drug susceptibility of subtype C reverse transcriptase from human immunodeficiency virus type 1 seroconverters in Zimbabwe. J Virol. 1997;71:5441–5448. doi: 10.1128/jvi.71.7.5441-5448.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shirasaka T, Kavlick M F, Ueno T, Gao W-Y, Kojima E, Alcaide M L, Chokekijchai S, Roy B M, Arnold E, Yarchoan R, Mitsuya H. Emergence of human immunodeficiency virus type 1 variants with resistance to multiple dideoxynucleosides in patients receiving therapy with dideoxynucleosides. Prog Nucleic Acid Res Mol Biol. 1995;92:2398–2402. doi: 10.1073/pnas.92.6.2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Soriano V, Gutierrez M, Garcia-Lerma G, Aguilera O, Mas A, Bravo R, Pérez-Labad M L, Baquero M, González-Lahoz J. First case of HIV-1 group O infection in Spain. Vox Sang. 1996;71:66. doi: 10.1046/j.1423-0410.1996.7110066.x. [DOI] [PubMed] [Google Scholar]
- 56.Soto-Ramirez L E, Tripathy S, Renjifo B, Essex M. HIV pol sequences from India fit distinct subtype pattern. J AIDS Hum Retrovirol. 1996;13:299–307. doi: 10.1097/00042560-199612010-00001. [DOI] [PubMed] [Google Scholar]
- 57.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vanden Haesevelde M, Decourt J-L, de Leys R, Vanderborght B, van der Groen G, van Heuverswijn H, Saman E. Genomic cloning and complete sequence analysis of a highly divergent African human immunodeficiency virus isolate. J Virol. 1994;68:1586–1596. doi: 10.1128/jvi.68.3.1586-1596.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang I, Diaz R S, Ho D D, Mosley J W, Busch M P, Mayer A. Host-specific driving force in human immunodeficiency virus type 1 evolution in vivo. J Virol. 1997;71:2555–2561. doi: 10.1128/jvi.71.3.2555-2561.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]