Abstract
The Gag proteins of Rous sarcoma virus (RSV) and human immunodeficiency virus (HIV) contain small interaction (I) domains within their nucleocapsid (NC) sequences. These overlap the zinc finger motifs and function to provide the proper density to viral particles. There are two zinc fingers and at least two I domains within these Gag proteins. To more thoroughly characterize the important sequence features and properties of I domains, we analyzed Gag proteins that contain one or no zinc finger motifs. Chimeric proteins containing the amino-terminal half of RSV Gag and various portions of the carboxy terminus of murine leukemia virus (MLV) (containing one zinc finger) Gag had only one I domain, whereas similar chimeras with human foamy virus (HFV) (containing no zinc fingers) Gag had at least two. Mutational analysis of the MLV NC sequence and inspection of I domain sequences within the zinc-fingerless C terminus of HFV Gag suggested that clusters of basic residues, but not the zinc finger motif residues themselves, are required for the formation of particles of proper density. In support of this, a simple string of strongly basic residues was found to be able to substitute for the RSV I domains. We also explored the possibility that differences in I domains (e.g., their number) account for differences in the ability of Gag proteins to be rescued into particles when they are unable to bind to membranes. Previously published experiments have shown that such membrane-binding mutants of RSV and HIV (two I domains) can be rescued but that those of MLV (one I domain) cannot. Complementation rescue experiments with RSV-MLV chimeras now map this difference to the NC sequence of MLV. Importantly, the same RSV-MLV chimeras could be rescued by complementation when the block to budding was after, rather than before, transport to the membrane. These results suggest that MLV Gag molecules begin to interact at a much later time after synthesis than those of RSV and HIV.
Retroviral Gag proteins are capable of directing the production of virus-like particles that are similar in size, shape, and buoyant density to authentic virions. Three small regions of the Gag polyprotein, termed assembly domains, have been found to be important for this budding process (see Fig. 1A) (35). The membrane-binding (M) domain, located within the matrix (MA) sequence, is required for the targeting and binding of the Gag protein to the cytoplasmic face of the plasma membrane (29). The interaction (I) domain, located within the nucleocapsid (NC) sequence, provides a major region of interaction between the 1,500 to 2,000 Gag proteins that make up an individual particle (30) and mediates the production of particles of the proper density (1, 33). The late (L) domain, located in different positions in different Gag proteins, is responsible for a budding event which occurs after the Gag molecules have reached the membrane but before the nascent particle is released from the cell (i.e., during the virus-cell separation step) (20, 34). The primary focus of this report is the mechanism of I domain function.
FIG. 1.
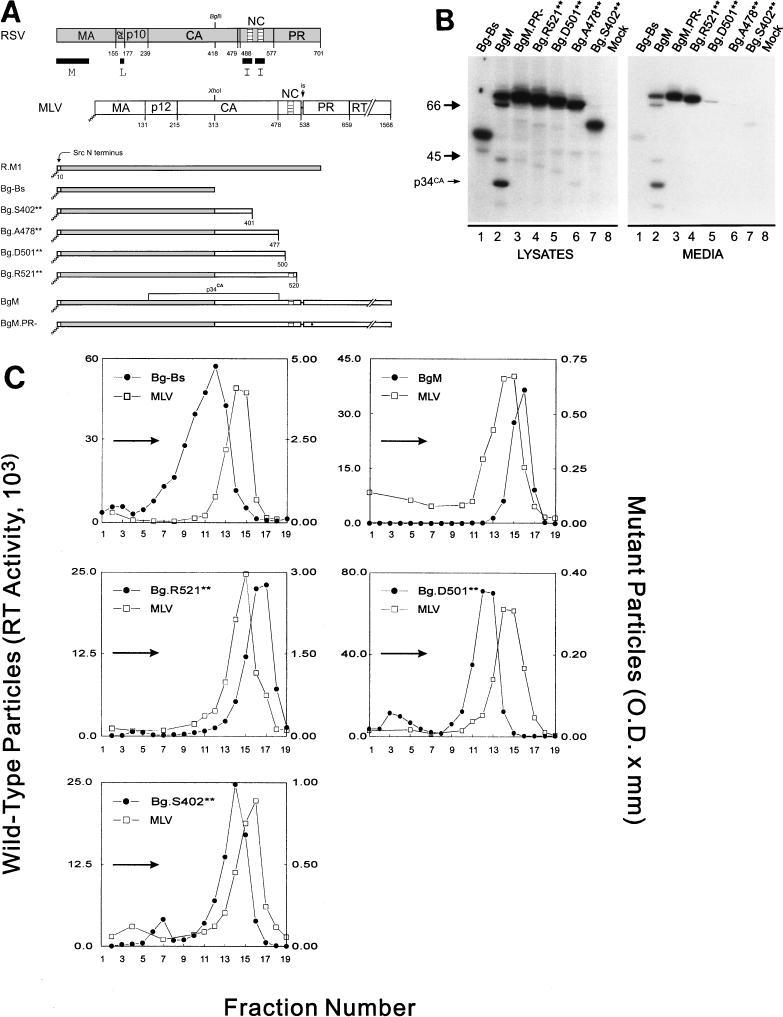
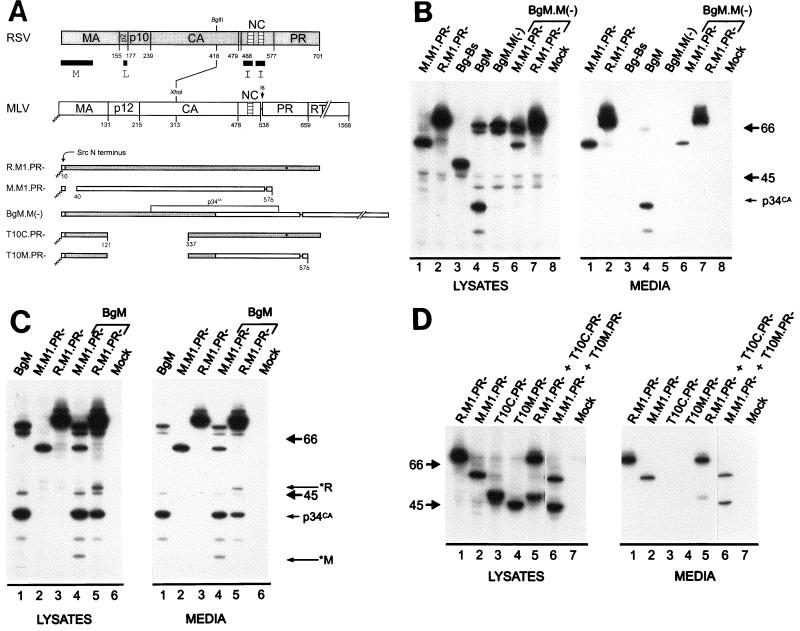
The C terminus of MLV contains at least one I domain. (A) Diagram of RSV-MLV chimeras. The wild-type RSV Gag (shaded) and MLV Gag (unshaded) proteins are aligned at the junction site of the chimeras (made by fusing the XhoI and BglII sites of the corresponding genes). The assembly domains of RSV are indicated by black boxes and the zinc fingers in NC by hatched regions. The site of in-frame suppression of the stop codon separating the MLV gag and pol genes is marked “is.” Sites of proteolytic cleavage are indicated by vertical lines through the proteins, and numbers refer to the last amino acid of the released products. The open box and squiggled line at the N terminus of the chimeras represent replacement of the first 10 RSV amino acids with those of pp60v-src and the consequent addition of myristic acid, respectively. The MLV protease active-site mutation is indicated by a black dot. Chimera R.M1 has been previously referred to as Myr1 (37). (B) Expression of RSV-MLV chimeras. The RSV-MLV constructs were transfected into COS-1 cells, and subsequently labeled for 2.5 h with l-[4,5-3H(N)]leucine. The Gag proteins from the medium and lysates of the transfected cells were immunoprecipitated with a mixture of α-RSV and α-MLV antibodies, separated by SDS-PAGE, and visualized by autoradiography. The numbers to the left are the positions (in kilodaltons) of the molecular mass markers, and the position of the RSV-MLV hybrid CA cleavage product p34CA is indicated. (C) Sucrose density gradient analysis. Cells were transfected and subsequently labeled for 8 h. Medium was collected, cleared of cellular debris, mixed with purified MLV, and centrifuged for 16 h at 83,500 × g through a 10 to 50% sucrose gradient. The gradients were fractionated, and the amount of control or experimental particles in each fraction was determined by reverse transcriptase assays or immunoprecipitation followed by densitometry of autoradiograms, respectively. An arrow in each panel indicates the direction of sedimentation. O.D., optical density.
In addition to I domains, the NC region of most retroviral Gag proteins contains one or two zinc finger motifs, characterized by the amino acid sequence CX2CX4HX4C, which are known to be involved in viral RNA packaging (3). Unlike zinc fingers, I domains cannot be identified by a particular sequence motif, but their presence can be confirmed by analyzing the density of particles produced upon expression of a given gag allele. Isopycnic sucrose gradient centrifugation has been used to map the positions of at least two I domains in the NC regions of human immunodeficiency virus (HIV) and Rous sarcoma virus (RSV) (1, 33). Interestingly, both of these Gag proteins have the same number of zinc finger motifs as they do identifiable I domains. Equally interesting is the conspicuous absence of zinc finger motifs in the putative NC region of spumavirus Gag proteins (11). To date, no spumavirus NC region has been analyzed for the presence of specific sequences capable of controlling the density of virus-like particles.
Although there has been a correlation between the number of zinc finger motifs in the NC portion of Gag and the number of identifiable I domains, previous experiments demonstrating that substitutions in the single zinc finger of murine leukemia virus (MLV) do not alter particle density (13) led us to believe that there is no functional significance to this relationship. The experiments described here confirm this hypothesis, identify dual I domains in the human foamy virus (HFV) Gag protein, and implicate basic residues as the primary functional components of I domains. They also suggest that I domains may play a role in the timing and location of the initial Gag protein multimerization.
MATERIALS AND METHODS
Previously constructed gag alleles.
The RSV gag gene was obtained from pATV-8, an infectious molecular clone of the RSV Prague C genome (28). The MLV gag and pol genes were from pRR88, a clone of the integrated Moloney MLV proviral genome in the plasmid pGCcos3neo (13). The HFV gag gene was obtained from pHSRV13, an infectious molecular clone constructed and kindly provided by R. Flugel (18). Several of the plasmids used in these experiments have been previously described, namely, pSV.Myr1 (referred to here as pSV.R.M1) (36), pSV.T10C.PR− (34), pSV.Bg-Bs (32), pSV.p25 (33), and pSV.p25.AD3 (33). Standard protocols were used for all DNA manipulations (25), and all plasmids were propagated in Escherichia coli DH-1 cells in 2× YT medium containing 25 μg of ampicillin/ml.
Newly constructed gag alleles.
Two plasmids were constructed to express the MLV Gag protein. The vector pSV.MLV, which expresses the gag and pol genes of MLV, was constructed by transferring the complete MLV coding sequence (including env) from pRR88 into pSV.R.M1 in place of the RSV gag sequence. The MLV fragment was prepared by digesting pRR88 with AatII (nucleotide [nt] 362), blunt-ending the resulting 3′ overhang with the Klenow fragment of DNA polymerase I (Klenow), and then digesting it with BssHII (nt 8211). This was then ligated with T4 DNA ligase to the large fragment of pSV.R.M1, which had been digested with SstI (nt 255), treated with Klenow, and digested with BssHII (nt 2724). A plasmid expressing MLV Gag and a truncated form of Gag-Pol, pSV.MLV.PR−, was made by first digesting the parent plasmid, pSV.MLV, with DraIII (MLV nt 2330, 5602, and 6656) and treating with Klenow. The 10,078-nt fragment was gel purified and recircularized at a concentration of 20 μg/ml in the presence of XbaI linkers (5′-CTAGTCTAGACTAG-3′), which contain termination codons (underlined) in all three reading frames.
A second set of expression plasmids was generated to replace the membrane-binding domain of MLV with that of the Src oncoprotein (21). The plasmid pSV.MLV.PR− was digested with PstI (MLV nt 738), treated with Klenow, and digested with EagI (pSV.R.M1 nt 3658). The 4,901-nt fragment was then gel-purified and ligated to the 5,774-nt fragment of pSV.R.M1 that had been prepared with MluI, Klenow, and EagI in succession, to create pSV.M.M1.PR−. pSV.M.M1 was constructed to express the MLV Gag and Gag-Pol proteins containing the Src membrane-binding domain in place of the M domain of MLV. For this, the 2,227-nt XhoI-BssHII fragment of pSV.M.M1.PR− was replaced with the 6,553-nt XhoI (nt 1559)-BssHII (nt 8211) fragment from pSV.MLV. An unmyristylated form of the Src-RSV Gag chimera, R.M(−), was constructed by mutagenesis of the R.M1 gag allele by using previously described methods and an oligonucleotide with the sequence 5′-GGATCAAGCATGGAATCCAGCAAAAGC-3′. As a result, the second codon was changed from GGA (glycine) to GAA (glutamic acid) and the expression plasmid was named pSV.Myr1(−). pSV.MLV.Myr(−), which expresses a myristate-minus form of the Src-MLV chimera, was then constructed by joining the 7,473-nt fragment from pSV.MLV that had been prepared with PstI, Klenow, and BssHII with the 6,708-nt fragment derived from pSV.Myr(−) that had been digested with MluI, Klenow treated, and digested with BssHII.
A third set of plasmids was made to express RSV-MLV chimeras. pSV.BgM was generated by digesting plasmids pSV.R.M1 and pRR88 with BglII and XhoI, respectively. The resulting sticky ends were made blunt by using Klenow, and the DNAs were then digested with BssHII. The large fragment of pSV.R.M1 and the 6,652-nt fragment of pRR88 were gel purified and ligated, thereby recreating the XhoI site and producing pSV.BgM. Plasmids which express a myristate-minus derivative of this chimera, pSV.BgM.M(−), or an internally deleted chimera, pSV.T10M.PR−, were created in a similar fashion, except that the parental plasmids used were pSV.Myr(−) and pSV.BgM, or pSV.T10C.PR− and pSV.MLV.PR−, respectively. The MLV protease (PR) active site mutation D32L found in pRR88.2204 (12) was subcloned into pSV.BgM by digesting each plasmid with XhoI and treating it with Klenow and BssHII. The desired fragments were gel purified and ligated to form pSV.BgM.PR−. To create the point mutations (pSV.BgM.C26/29S and pSV.BgM.Y28S), internal deletions (pSV.BgM.ΔK8-R11 and pSV.BgM.ΔR16-R23), and the carboxy-terminal truncations (pSV.BgM.S402**, pSV.BgM.A478**, pSV.BgM.D501**, and pSV.BgM.R521**) of MLV NC, the respective pRR88 mutants were subcloned into pSV.BgM by the same procedure as was used for the protease active-site mutation (see reference 13 for C26/29S and Y28S and reference 22 for ΔK8-R11 and ΔR16-R23; R521** has been previously referred to as R44Ter [22] and S402**, A478**, and D501** are previously unpublished).
To investigate the sequence requirements for dense particle formation, two constructs which expressed proteins encoding foreign amino acids at the end of p25 were made. pSV.p25.AD3 was digested with BssHII, treated with Klenow, and digested with BglII. Oligonucleotides with the sequences 5′-GATCGTAAGAAAGGTCGCAAAAAGGTCTAGA-3′ and 5′-TCTAGACCTTTTTGCGACCTTTCTTAC-3′ or 5′-GATCACCATCACCATCACCACGTCTAGA-3′ and 5′-TCTAGACGTGGTGATGGTGATGGT-3′ were annealed and ligated to the prepared pSV.p25.AD3 vector to create pSV.p25.RKK and pSV.p25.H6, respectively.
To determine whether the HFV Gag protein contains I domains, several plasmids were constructed to express RSV-HFV chimeras. These constructs were made by using PCR to amplify the desired sequences of HFV from the plasmid pHSRV13. The oligonucleotides used in the PCR were designed to incorporate BglII and BssHII restriction endonuclease sites (underlined) flanking the HFV sequences. The amplified product was digested with the respective enzymes and then cloned into the plasmid pSV.G1P (7) in place of the small fragment generated by double digestion with BglII and BssHII. Amplifications of fragments for pSV.Bg.1,2,3, pSV.Bg.1,2, and pSV.Bg.1 all used 5′-TTGAAGATCTGATGCTTTCTGGACAAAATTA-3′ as the forward primer and 5′-TACTAGGCGCGCGTTAACACCCCTTGTTT-3′, 5′-TACTAGGCGCGCTGATTTGGCCTAGGAGTTT-3′, and 5′-TACTAGGCGCGCGGAAGATTATATCCTCCTTGA-3′ as the respective reverse primers. Amplifications of fragments for pSV.Bg.2 and pSV.Bg.2,3 both used 5′-TTGAAGATCTATCTAGTACTCAGAATCAAAAT-3′ as the forward primer and 5′-TACTAGGCGCGCTGATTTGGCCTAGGAGTTT-3′ and 5′-GTCGCGCGCTTAGGATGATCGTTGGTTT CGGTT-3′ as the respective reverse primers. Finally, amplification of the fragment for pSV.Bg.3 used 5′-TTGAAGATCTTCAAACTCCTAGGCCAAAT-3′ as the forward primer and 5′-GTCGCGCGCTTAGGATGATCGTTGGTTTCGGTT-3′ as the reverse primer.
Transfection and labeling of cells and immunoprecipitation of Gag proteins.
COS-1 cells were transfected by the DEAE-dextran/chloroquine method, as previously described (36). Approximately 48 h after transfection, the cells were labeled for 2.5 h with either l-[4,5-3H(N)]leucine (150 μCi, 60 Ci/mmol) or 50 μCi (>1,000 Ci/mmol) of l-[35S]methionine (RSV-HFV chimeras). The cells and growth medium from each labeled culture were separated and mixed with lysis buffer containing protease inhibitors (36). The Gag proteins were immunoprecipitated with polyclonal rabbit serum against whole RSV (32) (reacts with MA, capsid [CA], NC, and PR) or a combination of α-RSV serum and polyclonal goat serum against MLV CA. The immunoprecipitated proteins were resolved by electrophoresis in sodium dodecyl sulfate (SDS)–12% polyacrylamide gels and detected by fluorography (36).
Isopycnic sucrose gradient analysis.
To determine the density of the particles produced by our various constructs, transfected COS-1 cells were labeled for 8 h with either 500 μCi of l-[4,5-3H(N)]leucine or 100 μCi of l-[35S]methionine (RSV-HFV chimeras). The growth medium was collected and centrifuged for 1 min at 15,000 × g to remove cellular debris before being layered onto a 10 to 50% sucrose gradient. An internal control was included in each gradient and consisted of either purified, unlabeled Moloney MLV or labeled COS-cell-produced RSV particles of wild-type density. The gradients were spun for 16 h at 83,500 × g in a Beckman SW41Ti rotor, and 0.6-ml fractions were collected from the bottom of each tube. The relative amount of MLV in each fraction was determined by the incorporation of [α-32P]TTP during synthesis of DNA on a poly(A) template as described previously (1). The amount of labeled Gag protein in each fraction was determined by immunoprecipitating with the appropriate antibody followed by SDS-polyacrylamide gel electrophoresis (PAGE), fluorography, and densitometry of the Gag-specific bands.
RESULTS
When expressed in mammalian cells, the RSV Gag protein directs the production of virus-like particles. It is still able to produce particles when its M domain is replaced by that of the Src oncoprotein (37). However, the RSV Gag derivative Bg-Bs (Fig. 1A), which is truncated after residue 418 in CA, produces low amounts of particles (Fig. 1B) that are light in density (1.14 g/ml) relative to the wild type (Fig. 1C; see also reference 32). The low density of this mutant is due to the lack of I domains within the NC sequence, which are responsible for proper Gag-Gag interactions (33). Therefore, by fusing foreign sequences to Bg-Bs and assaying for restoration of normal density to the particles produced, I domains from other retroviral Gag proteins can be identified. This gain-of-function approach has been used to successfully locate two I domains in the NC region of the HIV Gag protein (1). To gain a better understanding of how I domains work, we initially set out to map the I domains of MLV and HFV Gag proteins and to characterize their important sequence features.
Identification of an MLV I domain.
Several chimeric proteins were made to identify I domains in the MLV NC region (Fig. 1A). When amino acid residues 314 to 538 from the MLV Gag protein were fused to Bg-Bs to form BgM (Fig. 1A), efficient release of protein into the medium was restored (Fig. 1B). Since this construct contains most of the adjacent pol gene, including the sequences encoding the MLV protease, Gag cleavage products were also evident, including the chimeric capsid species p34CA (Fig. 1B, lanes 2). Protease activity was not required for budding, however, since BgM.PR− was released just as efficiently (Fig. 1B, lanes 3). When particles made from BgM were analyzed in sucrose gradients, they were found to be of higher density (1.19 g/ml; Fig. 1C) than those from Bg-Bs (Fig. 1C), indicating that amino acids either in the C terminus of MLV Gag or in MLV Pol were able to provide I domain activity. The particles from BgM were of slightly higher density than the control MLV particles (1.16 to 1.18 g/ml), which is in agreement with previous results for Env-less particles of RSV (37). To more specifically map the region of MLV that contains this I domain activity, we constructed truncations of BgM that ended after MLV amino acid 401, 477, 500, or 520. Of these constructs, only the largest (Bg.R521**) produced particles efficiently (Fig. 1B, lanes 4), and these were of high density (1.19 g/ml; Fig. 1C). Low amounts of particles (less than 10% of wild type) were obtained from Bg.D501** and Bg.S402** (Fig. 1B, lanes 5 and 7) but were light in density (Fig. 1C). Bg.A478** did not produce enough particles to test in density gradients. These data reveal the presence of at least one I domain in the NC region of MLV Gag whereas similar analyses of RSV and HIV had revealed two (1, 8, 33).
Importance of basic residues for I domain function.
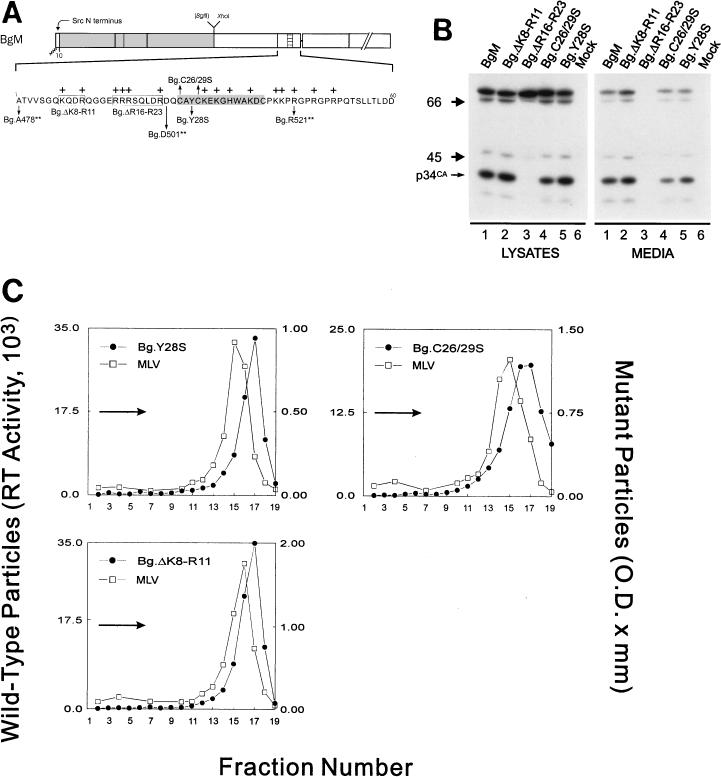
The zinc fingers of Gag proteins have long been known to be dispensable for particle assembly and, in the case of MLV, particle density (13). To ensure that this was true for the RSV-MLV chimeras, we constructed zinc finger point mutations in the context of full-length BgM (Fig. 2A). In mutant Bg.C26/29S, the cysteine residues at positions 26 and 29 of MLV NC were replaced with serine residues, while in mutant Bg.Y28S, the tyrosine residue at position 28 was replaced with serine. Neither of these mutations reduced the number of particles released (Fig. 2B, lanes 4 and 5) nor the densities (1.18 g/ml) relative to wild type (Fig. 2C). This confirms that an intact zinc finger motif is not required for dense particle formation.
FIG. 2.
An intact zinc finger motif is not required for MLV I domain activity. (A) Diagram of the chimeric BgM protein and various mutants derived from it. The precise locations of the changes made in the 60 amino acids of the MLV NC sequence are shown along with the positions of basic residues (marked with +). Substitutions are designated with arrows and deletions are boxed. The residues comprising the zinc finger motif are highlighted in gray. (B) Gag proteins were labeled and visualized and (C) sucrose gradient analysis was performed as described in the legend for Fig. 1.
The only feature of retroviral NC sequences that is more conserved than the zinc fingers is the presence of numerous basic amino acids. To investigate the potential role of basic residues in the production of dense particles, two internal regions in NC were deleted (Fig. 2A). The first deletion tested, Bg.ΔK8-R11, lacked two basic residues and one acidic residue but had no effect on particle release (Fig. 2B, lanes 2) or density (1.18 g/ml; Fig. 2C). The second deletion, Bg.ΔR16-R23, lacked eight amino acids, including four basic residues, and completely (limit of detection, <2% of wild type) blocked particle release (Fig. 2B, lanes 3). The nature of the assembly defect of this construct is not clear, but the lack of proteolytic processing in the cells (Fig. 2B, lanes 3) suggests that the mutant proteins were either not arriving at the plasma membrane or not interacting with the mutant Gag-Pol molecules once they arrived to allow protease activation. Because the removal of the second set of basic residues had a detrimental effect on particle assembly, we were unable to make a density determination for this mutant and therefore cannot draw a conclusion from this result about the contribution of these residues to the MLV I domain. However, unpublished results (21a) indicate that MLV Gag proteins with this deletion form particles of normal density and this, taken together with the results from the C-terminal truncation mutants, suggests that the MLV I domain maps somewhere between Gag residues 501 and 520 (residues 24 to 43 of NC).
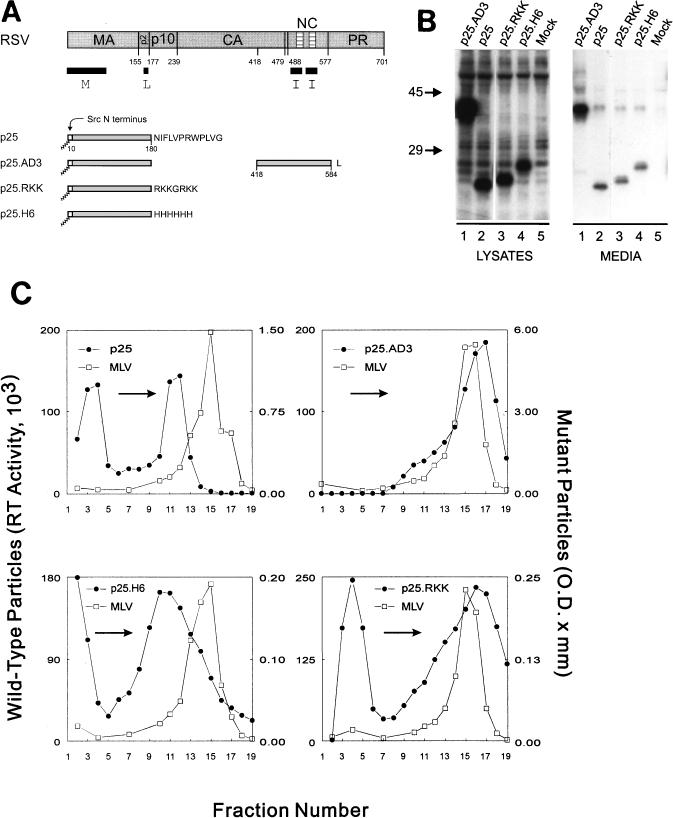
To test the hypothesis that basic residues alone are sufficient for I domain function, we investigated artificial sequences of basic residues in our gain-of-function assay (Fig. 3). Since CA is not required for budding and might actually interfere with the function of a small basic extension, we used the shorter RSV Gag derivative named p25 (33). p25 contains only 180 Gag residues and, like Bg-Bs, contains the M and L assembly domains but lacks both I domains. Therefore, unless an I domain is added to its C terminus, p25 releases few particles (down 10-fold from wild type; Fig. 3B, lanes 2), all of which exhibit lighter than normal density (1.14 g/ml; Fig. 3C). To decide which sequence of basic residues to use, we examined the NC regions of RSV, HIV, and MLV. Within the regions identified as I domains of these proteins, the following sequences are found: PKKRK (RSV), PRKK (HIV), and PKKP (MLV). We wanted to use a sequence similar but not identical to any of these, so we chose the sequence RKKGRKK (p25.RKK). We also constructed a p25 chimera with a stretch of six histidine residues, which is slightly basic but does not resemble basic regions found within NC, at its C terminus (p25.H6). Both chimeric proteins expressed well and released particles (down 10-fold from wild type; Fig. 3B, lanes 3 and 4). As previously described (33), the addition of residues 418 to 584 from RSV Gag to the p25 truncation (p25.AD3) restored normal density to the produced particles (1.18 g/ml; Fig. 3C). Similarly, p25.RKK (1.18 g/ml), but not p25.H6 (1.13 g/ml), was found to be capable of producing dense particles (Fig. 3C). A portion of the extracellular Gag protein from p25, p25.H6, and p25.RKK is found in the top fractions of the gradient. We have previously shown (33) that the p25 Gag proteins that sediment into the gradient (fractions 11 and 12, Fig. 3C) are membrane enclosed and that the proteins at the top of a gradient containing particles from a ΔNC construct (which also lacks I domains) are not particulate (data not shown). As expected, the upper material fails to shift in the gradients relative to the internal control while the material that moves well into the gradient changes density in a construct-dependent manner. It should be noted that the particles formed by p25.RKK are more heterogeneous in density than those formed by the BgM mutations (Fig. 1C and 2C). However, the heterogeneity of the p25.RKK particles is only slightly more pronounced than that of the p25.AD3 particles and the bulk of the peak is clearly shifted relative to the peak of p25 particles. Thus, from this experiment, it seems that a simple string of strongly basic residues is sufficient to provide I domain function.
FIG. 3.
Basic residues can substitute for the RSV I domain. (A) p25 fusion constructs are shown with foreign amino acids denoted by their single letter code. (B) Gag proteins were labeled and visualized and (C) sucrose gradient analysis was performed as described in the legend for Fig. 1.
Absence of zinc fingers in HFV I domains.
Although the experimental evidence shows that zinc finger motifs can be disrupted without inhibiting I domain function (13), there remained an apparent correlation between the number of I domains present in a given Gag protein and the number of zinc finger motifs. That is, our low-resolution mapping of I domains revealed two in RSV Gag and two in HIV Gag, but only one in MLV Gag. To eliminate the possibility that some aspect of the zinc finger other than zinc binding was contributing to I domain function, we decided to examine the HFV Gag protein, which lacks zinc fingers, for the presence of I domains.
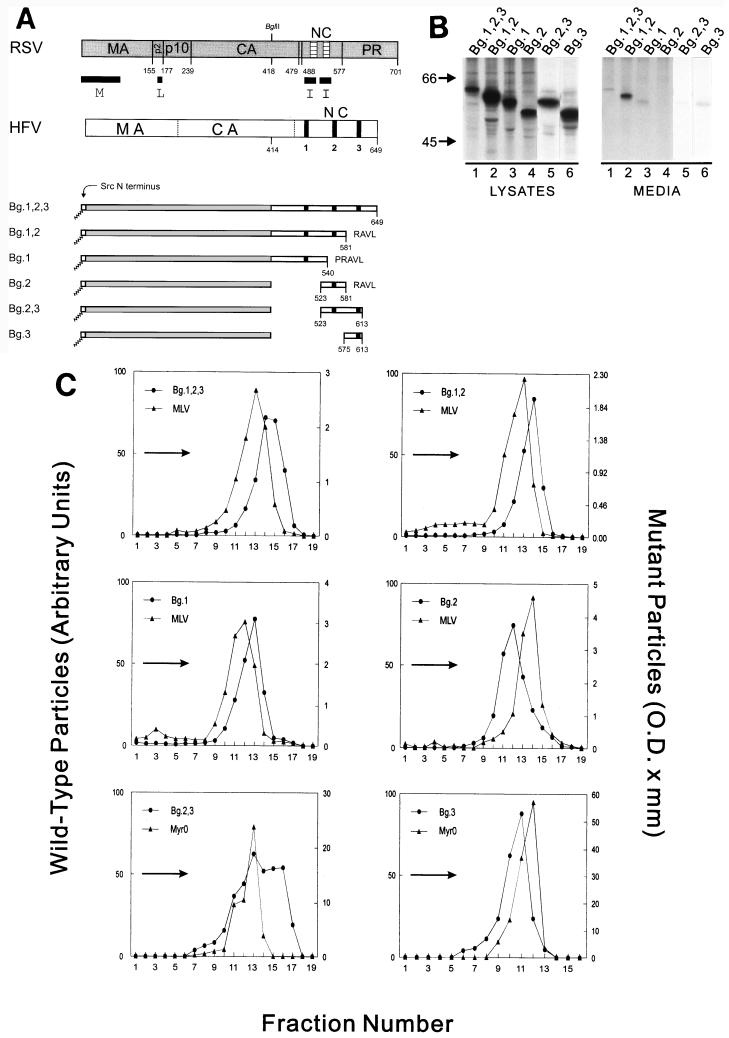
Within the HFV NC sequence are three glycine-rich (GR) regions containing large numbers of basic residues (previously designated GR boxes I, II, and III; see reference 25). Since our previous experiments indicate that basic residues are likely to be important, we made Bg-Bs/HFV chimeric proteins that contained various combinations of these three regions (Fig. 4A). Although each RSV-HFV chimera expressed well, the amount of protein released into the media varied considerably (from 2 to 35% of wild type; Fig. 4B). In each case, however, enough extracellular material was available to perform sucrose density gradient analysis. When the entire NC region of HFV (amino acids 414 to 780) was fused after residue 418 of RSV Gag to form Bg.1,2,3, dense particles were produced (1.19 g/ml; Fig. 4C). This was also true when residues 414 to 581 (Bg.1,2, 1.19 g/ml) or only residues 414 to 540 (Bg.1) were added to Bg-Bs (Fig. 4C). These results show that an I domain is present within amino acids 414 to 540 of HFV Gag. To determine whether additional I domains were present in the HFV Gag sequence downstream of residue 540, we examined particles from Bg.2 (containing residues 523 to 581), Bg.3 (residues 574 to 780), and Bg.2,3 (residues 523 to 780) (Fig. 4C). Although particles from Bg.2 and Bg.3 were lighter than normal (1.16 g/ml), when the two regions were combined, the resultant protein (Bg.2,3) was able to form dense particles (1.19 to 1.22 g/ml). This result indicates the presence of an additional I domain downstream of the one contained in Bg.1. From these experiments, we can conclude that HFV NC contains at least two I domains in the absence of zinc finger motifs. Although the regions of HFV Gag that contain the interaction domains are also rich in basic residues, the importance of these residues for HFV I domain function requires further investigation.
FIG. 4.
The HFV Gag protein contains at least two I domains. (A) RSV and HFV Gag proteins are illustrated. The vertical dotted lines separate HFV Gag into regions which are analogous to the cleavage products of other retroviral Gag proteins. The black vertical bars in HFV NC indicate the position of the three GR boxes. The numbers below the chimeric proteins indicate the HFV Gag residues contained in the constructs and the letters at the end of the molecules are the single letter codes of any nonviral residues present. (B) Expression of RSV-HFV chimeras was performed as described in the legend for Fig. 1 except that l-[35S]methionine was used to label the cells, and the relevant proteins were immunoprecipitated with α-RSV antibody. (C) Sucrose gradient analysis was performed as described in the legend for Fig. 1 except that the RSV-HFV chimeras were labeled with l-[35S]methionine and immunoprecipitated with α-RSV antibody, and two constructs (Bg.2,3 and Bg.3) were mixed with labeled RSV Gag-only particles (rather than authentic MLV) before layering on the gradient.
Complementation analysis.
Previous experiments with the Gag protein of RSV have shown that all types of mutants defective for membrane binding can be rescued into particles by complementation, but this requires the presence of functional I domains (2, 34, 37). In contrast, the unmyristylated form of the MLV Gag protein cannot be rescued by functional MLV Gag, even when the single I domain is intact (27). These observations suggest the possibility that differences between the I domains of RSV and MLV are responsible for the different rescue phenotypes. To test this hypothesis, we examined the ability of a myristate-minus form of the BgM chimera to be rescued. Because its I domain is derived from MLV, we predicted that this chimeric protein would not be rescued into particles when its membrane-binding domain (here derived from Src) was inactivated. We used the protease-proficient form of BgM.M(−) and a protease-deficient form of the rescuing molecule. Thus, the appearance of Gag cleavage products in the medium would be an indicator of rescue.
When BgM.M(−) was expressed, it was unable to release particles on its own (Fig. 5B, lanes 5) and, as expected for a non-membrane-targeted Gag protein (36, 37), exhibited reduced processing by the MLV protease (compare with Fig. 5B, lanes 4). Upon coexpression of BgM.M(−) with a protease-deficient, budding-competent MLV protein, M.M1.PR−, both full-length products could be observed in the cell lysates but only the MLV Gag precursor, in the absence of any cleavage products, was evident in the media (Fig. 5B, lane 6). Similarly, a budding-competent RSV protein, R.M1.PR−, was unable to rescue BgM.M(−), and again, only uncleaved RSV Gag is seen in the medium (Fig. 5B, lane 7). Thus, it appears that BgM.M(−) cannot interact with other Gag proteins to be rescued into particles and therefore behaves like the unmyristylated MLV Gag protein (27), consistent with the hypothesis that the determinant of rescuability resides in the C terminus of Gag. The trivial explanation that the myristate-minus form of BgM (and MLV Gag) is globally misfolded and therefore not rescued for reasons unrelated to MLV I domain function seems unlikely because mutants having the same lack of myristate in the context of the RSV-Src chimera can be rescued with ease (37).
FIG. 5.
Membrane localization is required for the rescue of molecules containing the MLV I domain. (A) To assess the involvement of I domains in complementation rescue, budding-defective RSV-MLV chimeras were examined. All constructs shown here contain either two RSV I domains or the single MLV I domain. T10C.PR− and T10M.PR−, which lack an L domain, are blocked at a late step during budding, and BgM.M(−), which lacks myristate, is not targeted to the plasma membrane. Cells were transfected and labeled as described in the legend for Fig. 1. Cotransfection is denoted by brackets above the lanes and the cotransfected DNA above the bracket. (B) Unmyristylated BgM.M(−) cannot be rescued either by molecules containing RSV I domains or by the MLV I domain. However, BgM, which is targeted to the membrane, is copackaged with molecules containing either RSV or MLV I domains. (C) RSV-specific (*R) and MLV-specific (*M) cleavage products are indicated. (D) A chimera containing the MLV I domain, but no L domain (T10M), is targeted to the membrane and rescued by either RSV or MLV Gag.
We also considered the possibility that BgM Gag molecules can interact only with themselves to form particles (Fig. 1B and C) because their chimeric CA region is misfolded in a way that perturbs interactions with other Gag proteins (i.e., those with normal CA sequences). If this were true, then the assembly- competent chimera BgM would not be able to interact and be copackaged with protease-minus forms of RSV or MLV Gag. However, if the folding of the chimeric CA sequence was irrelevant and these proteins are able to interact with BgM and be copackaged into the same particle, then the MLV protease provided by BgM should be able to recognize and cleave the MLV and RSV Gag proteins in trans. This was in fact observed as coexpression of the Gag proteins resulted in particles that contained BgM and its cleavage products as well as cleavage products specific for MLV Gag (*M; Fig. 5C, lanes 4) or RSV (*R; Fig. 5C, lanes 5). These successful copackaging results suggest that there is great plasticity in Gag-Gag interactions, even among heterologous Gag sequences. Taken together, the experiments described above support the idea that the difference in ability of M-domain mutants of RSV and MLV to be rescued maps to their I domains. Further, we propose that the difference in rescue phenotype is because RSV Gag proteins initiate interactions in a cytosolic compartment while MLV Gag interactions occur first at the plasma membrane.
If MLV Gag molecules truly interact only at the plasma membrane, then any MLV Gag protein with a mutant L domain, which would be targeted to the membrane but otherwise be budding defective, should be capable of being rescued into particles. Since the L domain of MLV has not been mapped, we decided to examine a derivative of BgM that contains a deletion of the RSV L domain. For this, we made use of the RSV Gag mutant T10C.PR− (Fig. 5A), which lacks amino acids 122 to 336, including the entire L domain, but retains the M and I domains (34). As shown in Fig. 5D (lanes 3), this molecule is budding incompetent but is rescuable by full-length RSV Gag (Fig. 5D, lanes 5). This well-characterized RSV L-domain mutation was introduced into BgM to create chimera T10M.PR− (lanes 4). When expressed by itself, this recombinant, like T10C.PR−, was unable to bud from the cell (Fig. 5D, lanes 4). The presence of the strong membrane-binding sequence of Src and the high proteolytic activity of the PR+ form (data not shown) suggest that T10M is targeted to the plasma membrane. When coexpressed with an assembly-competent molecule (M.M1.PR−), T10M.PR− was readily rescued into particles (Fig. 5D, lanes 6), which further indicates that it is not severely disrupted by the T10 deletion. This evidence supports the idea that interactions among MLV Gag proteins occur after the molecules are targeted to, and concentrated on, the plasma membrane (see Discussion).
DISCUSSION
I domains are essential for the production of retroviral particles of the proper density. The sequence characteristics and mechanism of action of I domains are, however, largely unknown. The data presented here show that a simple artificial string of basic residues is sufficient for I domain function. In addition, we provide evidence that I domains may play a role in the timing of initial Gag-Gag interactions and that RSV Gag proteins may interact earlier during assembly than MLV Gag proteins.
Mechanism of I domain function.
I domains are currently thought to influence particle density by interacting with RNA and using it as a scaffold to facilitate the subsequent Gag-Gag protein interactions mediated by CA (1, 14, 16, 17). This model is supported by in vitro assembly data from RSV (6) and HIV Gag (6, 14) that shows formation of multimeric structures dependent on, and proportional in length to, added RNA. Although the genomic viral RNA is used during infection, it is not required for in vitro Gag multimerization (6, 14) or for the production of dense, Gag-only particles (36). Collectively, these results suggest that the nonspecific RNA binding activity of NC is involved in I-domain function. Further evidence that the Cys-His boxes, which are required for the specific incorporation of viral RNA into the particle (3), are not part of the I domain comes from studies of point mutations within the single zinc finger of MLV (13). The experiments reported here have extended this finding by showing that particle density is not affected by additional NC mutations in the context of RSV-MLV chimeras. They have also strengthened the above model by demonstrating that a simple string of basic residues, which should interact with RNA but should not exhibit binding specificity for the viral nucleic acid, can substitute for NC’s density-determining properties. However, while it is clear that basic residues can replace the RSV I domains, it may be that not just any basic array will suffice.
Conservation of RNA-mediated assembly.
In addition to identifying the functional component of I domains, we also sought to determine the extent to which I domains are maintained between distantly related retroviruses. Although spumavirus NC sequences do not contain zinc fingers, they do contain three GR boxes which have been implicated in nucleic acid binding (38). Since sequence inspection is not informative for identifying I domains, we used a gain-of-function approach to identify the presence of at least two I domain equivalents within HFV Gag. The location of the HFV I domains is consistent with previous reports indicating that GR box 1 is the primary determinant of HFV NC nucleic acid binding (38). RNA binding by GR box 2 or 3 alone is drastically reduced (38) but, when present together, they cooperate sufficiently to provide I domain function.
Spumaviruses have been observed to exhibit strong nuclear localization in infected cells (10, 15). Although our RSV-HFV chimeras were capable of producing dense particles, they did not exhibit the strong nuclear localization (data not shown) previously seen for HFV NC sequences (26). We considered the possibility that the myristylated amino terminus of our chimeras was a dominant plasma membrane-targeting signal and made myristate-minus forms of our chimeric spumavirus constructs. Again, however, strong nuclear localization was not observed by either immunofluorescence or cell fractionation (data not shown). The difference in nuclear localization may be the result of using different expression systems (vaccinia-virus-expressed [26] versus COS-cell-expressed chimeras) or may be related to the fact that, during infection, nuclear localization is transient (26). However, the relevance of nuclear localization is questionable since recent results indicate that nuclear targeting is not essential for HFV infectivity (38). Regardless of the issue of nuclear localization, our finding of sequences in the NC region of HFV that can provide the interactions necessary to restore dense particle production to an RSV mutant Gag protein suggests that spumaviruses, even in the absence of zinc finger domains, may utilize RNA during assembly to initiate protein-protein contacts.
Timing of initial Gag-Gag interactions.
Although I domains play a major role in the formation of proper Gag-Gag interactions, the location and timing of the initial contacts remain unknown. Because type B and D retroviruses, such as Mason-Pfizer monkey virus (M-PMV), assemble electron-dense cores in the cytoplasm of an infected cell (9), we know that Gag proteins from these viruses are able to interact prior to membrane transport. However, since a single point mutation in the matrix region of M-PMV Gag causes core assembly to take place at the plasma membrane rather than in the cytoplasm (23), it appears that the membrane-binding and targeting domain, rather than the I domain, determines the cellular site of assembly. Although the Gag proteins of C-type retroviruses, such as RSV, do not become visible until after their localization to the plasma membrane (4, 5), it is quite possible that they begin to interact prior to membrane transport but in a manner not visible by electron microscopy. In fact, complementation analyses showing rescue of membrane-binding mutants suggest that RSV Gag proteins may interact prior to membrane localization (34, 37). In contrast to RSV, unmyristylated MLV Gag proteins, which also exhibit C-type plasma membrane assembly, are unable to be rescued in vivo (27). This indicates either that MLV Gag proteins do not interact with each other in the cytoplasm or that any pre-membrane-localization interactions are not of sufficient strength for rescue to occur.
Although the observations that cytoplasmic MLV and RSV Gag molecules have different rescue phenotypes were made some time ago, the molecular basis of those phenotypes remains unexplained. Our results map this difference to the C terminus of Gag. Membrane-binding mutants having the RSV C terminus can be rescued (37) while those with MLV C-terminal sequences cannot. Since the inability to be rescued is a negative result, a number of trivial explanations could account for our observations. As has been previously suggested for unmyristylated MLV (27), the lack of rescue of our RSV-MLV chimera could be due to low expression levels, rapid protein degradation and/or misfolding, or an inability of the protein to interact with the rescuing Gag molecule. Low expression or rapid degradation of BgM.M(−) does not appear to be a problem since large amounts (i.e., similar to M.M1 levels) of this protein are present in the cell lysates. Although BgM.M(−) could be globally misfolded, this is unlikely because loss of myristate is not known to severely disrupt the structure of other Gag proteins. For example, myristate-minus Gag proteins of M-PMV are still able to associate into cytoplasmic cores (24) and studies with myristylation inhibitors have shown that HIV particles can form even when only a few of the Gag proteins are myristylated (19). Likewise, even RSV Gag proteins that have become dependent on the Src membrane-binding sequence (e.g., R.M1) can be rescued when myristate is absent (37). Furthermore, chimeras containing the large T10C deletion (i.e., T10M) are readily rescued, indicating that the BgM chimera is not overly sensitive to mutations. This plasticity of Gag is emphasized further by the fact that BgM can be packaged with either MLV or RSV Gag, even though BgM contains a highly distorted capsid sequence, and this represents the first evidence for copackaging of Gag proteins from different retroviruses. Experiments demonstrating that copackaging of MLV and RSV Gag proteins requires that each protein contain the same membrane-binding domain will be presented elsewhere (2). Thus, the simplest interpretation of our results is that the MLV Gag protein (but not that of RSV) requires concentration at the membrane in order for Gag-Gag interactions to occur.
One of the most striking differences between the C termini of RSV and MLV is the presence of two readily identified I domains in the former and one in the latter. This observation leads us to hypothesize that there may be a dosage effect in which two I domains provide interactions strong enough to allow Gag proteins to associate in the cytoplasm. The presence of a single I domain would allow interactions only after concentration at the plasma membrane. Consistent with this idea is data showing that the myristate-minus form of the Gag protein of spleen necrosis virus (SNV) cannot be rescued into particles by wild-type SNV Gag (31). SNV is closely related to MLV (∼40% identity) and also has a single Cys-His box. The ultimate test of our hypothesis, however, will be to find mutants of MLV, perhaps containing a duplicated NC region with two I domains, that can be rescued when myristate is removed.
ACKNOWLEDGMENTS
This research was sponsored by the National Cancer Institute, DHHS, under contract with ABL (A.R.), by NIH grants to J.B.B. (training grant CA60395) and J.W.W. (CA47482), and by a grant from the American Cancer Society to J.W.W. (FRA-427).
REFERENCES
- 1.Bennett R P, Nelle T D, Wills J W. Functional chimeras of the Rous sarcoma virus and human immunodeficiency virus Gag proteins. J Virol. 1993;67:6487–6498. doi: 10.1128/jvi.67.11.6487-6498.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bennett, R. P., and J. W. Wills. Conditions for co-packaging Rous sarcoma virus and murine leukemia virus Gag proteins during retroviral budding. Submitted for publication. [DOI] [PMC free article] [PubMed]
- 3.Berkowitz R, Fisher J, Goff S P. RNA packaging. Curr Top Microbiol Immunol. 1996;214:177–218. doi: 10.1007/978-3-642-80145-7_6. [DOI] [PubMed] [Google Scholar]
- 4.Bernhard W. Electron microscopy of tumor cells and tumor viruses: a review. Cancer Res. 1958;18:491–509. [PubMed] [Google Scholar]
- 5.Bernhard W. The detection and study of tumor viruses with the electron microscope. Cancer Res. 1960;20:712–727. [PubMed] [Google Scholar]
- 6.Campbell S, Vogt V M. Self assembly in vitro of purified CA-NC proteins from Rous sarcoma virus and human immunodeficiency virus type 1. J Virol. 1995;69:6487–6497. doi: 10.1128/jvi.69.10.6487-6497.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Craven R C, Bennett R P, Wills J W. Role of the avian retroviral protease in the activation of reverse transcriptase during virion assembly. J Virol. 1991;65:6205–6217. doi: 10.1128/jvi.65.11.6205-6217.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Craven R C, Leure-duPree A E, Weldon R A, Jr, Wills J W. Genetic analysis of the major homology region of the Rous sarcoma virus Gag protein. J Virol. 1995;69:4213–4227. doi: 10.1128/jvi.69.7.4213-4227.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fine D, Schochetman G. Type D primate retroviruses: a review. Cancer Res. 1978;38:3123–3139. [PubMed] [Google Scholar]
- 10.Fleming W A, Clarke J K. Fluorescence assay of foamy virus. J Gen Virol. 1970;6:277–284. doi: 10.1099/0022-1317-6-2-277. [DOI] [PubMed] [Google Scholar]
- 11.Flugel R M. Spumaviruses: a group of complex retroviruses. J Acquired Immune Defic Syndr. 1991;4:739–750. [PubMed] [Google Scholar]
- 12.Fu W, Rein A. Maturation of dimeric viral RNA of Moloney murine leukemia virus. J Virol. 1993;67:5443–5449. doi: 10.1128/jvi.67.9.5443-5449.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gorelick R J, Henderson L E, Hanser J P, Rein A. Point mutations of Moloney murine leukemia virus that fail to package viral RNA: evidence for specific RNA recognition by a “zinc finger-like” protein sequence. Proc Natl Acad Sci USA. 1988;85:8420–8424. doi: 10.1073/pnas.85.22.8420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gross I, Hohenberg H, Krausslich H-G. In vitro assembly properties of purified bacterially expressed capsid proteins of human immunodeficiency virus. Eur J Biochem. 1997;249:592–600. doi: 10.1111/j.1432-1033.1997.t01-1-00592.x. [DOI] [PubMed] [Google Scholar]
- 15.Hooks J J, Gibbs C J. The foamy viruses. Bacteriol Rev. 1975;39:169–185. doi: 10.1128/br.39.3.169-185.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jowett J B, Hockley D J, Nermut M V, Jones I M. Distinct signals in human immunodeficiency virus type 1 Pr55 necessary for RNA binding and particle formation. J Gen Virol. 1992;73:3079–3086. doi: 10.1099/0022-1317-73-12-3079. . (Erratum, 74:943, 1993.) [DOI] [PubMed] [Google Scholar]
- 17.Krishna N K, Campbell S, Vogt V M, Wills J W. Genetic determinants of Rous sarcoma virus particle size. J Virol. 1998;72:564–577. doi: 10.1128/jvi.72.1.564-577.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lochelt M, Zentgraf H, Flugel R M. Construction of an infectious DNA clone of the full-length human spumaretrovirus genome and mutagenesis of the bel 1 gene. Virology. 1991;184:43–54. doi: 10.1016/0042-6822(91)90820-2. [DOI] [PubMed] [Google Scholar]
- 19.Morikawa Y, Hinata S, Tomoda H, Goto T, Nakai M, Aizawa C, Tanaka H, Omura S. Complete inhibition of human immunodeficiency virus Gag myristoylation is necessary for inhibition of particle budding. J Biol Chem. 1995;271:2868–2873. doi: 10.1074/jbc.271.5.2868. [DOI] [PubMed] [Google Scholar]
- 20.Parent L J, Bennett R P, Craven R C, Nelle T D, Krishna N K, Bowzard J B, Wilson C B, Puffer B A, Montelaro R C, Wills J W. Positionally independent and exchangeable late budding functions of the Rous sarcoma virus and human immunodeficiency virus Gag proteins. J Virol. 1995;69:5455–5460. doi: 10.1128/jvi.69.9.5455-5460.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pellman D, Garber E A, Cross F R, Hanafusa H. An N-terminal peptide from p60src can direct myristylation and plasma membrane localization when fused to heterologous proteins. Nature (London) 1985;314:374–377. doi: 10.1038/314374a0. [DOI] [PubMed] [Google Scholar]
- 21a.Rein, A. Unpublished results.
- 22.Rein A, Harvin D P, Mirro J, Ernst S M, Gorelick R J. Evidence that a central domain of nucleocapsid protein is required for RNA packaging in murine leukemia virus. J Virol. 1994;68:6125–6129. doi: 10.1128/jvi.68.9.6124-6129.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rhee S S, Hunter E. A single amino acid substitution within the matrix protein of a type D retrovirus converts its morphogenesis to that of a type C retrovirus. Cell. 1990;63:77–86. doi: 10.1016/0092-8674(90)90289-q. [DOI] [PubMed] [Google Scholar]
- 24.Rhee S S, Hunter E. Myristylation is required for intracellular transport but not for assembly of D-type retrovirus capsids. J Virol. 1987;61:1045–1053. doi: 10.1128/jvi.61.4.1045-1053.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 26.Schliephake A W, Rethwilm A. Nuclear localization of foamy virus Gag precursor protein. J Virol. 1994;68:4946–4954. doi: 10.1128/jvi.68.8.4946-4954.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schultz A M, Rein A. Unmyristylated Moloney murine leukemia virus Pr65gag is excluded from virus assembly and maturation events. J Virol. 1989;63:2370–2373. doi: 10.1128/jvi.63.5.2370-2373.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schwartz D E, Tizard R, Gilbert W. Nucleotide sequence of Rous sarcoma virus. Cell. 1983;32:853–869. doi: 10.1016/0092-8674(83)90071-5. [DOI] [PubMed] [Google Scholar]
- 29.Verderame M F, Nelle T D, Wills J W. The membrane-binding domain of the Rous sarcoma virus Gag protein. J Virol. 1996;70:2664–2668. doi: 10.1128/jvi.70.4.2664-2668.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vogt V M, Eisenman R, Diggleman H. Generation of avian myeloblastosis virus structural proteins by proteolytic cleavage of a precursor polypeptide. J Mol Biol. 1975;96:471–493. doi: 10.1016/0022-2836(75)90174-6. [DOI] [PubMed] [Google Scholar]
- 31.Weaver T A, Panganiban A T. N myristoylation of the spleen necrosis virus matrix protein is required for correct association of the Gag polyprotein with intracellular membranes and for particle formation. J Virol. 1990;64:3995–4001. doi: 10.1128/jvi.64.8.3995-4001.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weldon R A, Jr, Erdie C R, Oliver M G, Wills J W. Incorporation of chimeric Gag protein into retroviral particles. J Virol. 1990;64:4169–4179. doi: 10.1128/jvi.64.9.4169-4179.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weldon R A, Jr, Wills J W. Characterization of a small (25-kilodalton) derivative of the Rous sarcoma virus Gag protein competent for particle release. J Virol. 1993;67:5550–5561. doi: 10.1128/jvi.67.9.5550-5561.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wills J W, Cameron C E, Wilson C B, Xiang Y, Bennett R P, Leis J. An assembly domain of the Rous sarcoma virus Gag protein required late in budding. J Virol. 1994;68:6605–6618. doi: 10.1128/jvi.68.10.6605-6618.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wills J W, Craven R C. Form, function, and use of retroviral Gag proteins. AIDS. 1991;5:639–654. doi: 10.1097/00002030-199106000-00002. [DOI] [PubMed] [Google Scholar]
- 36.Wills J W, Craven R C, Achacoso J A. Creation and expression of myristylated forms of Rous sarcoma virus Gag protein in mammalian cells. J Virol. 1989;63:4331–4343. doi: 10.1128/jvi.63.10.4331-4343.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wills J W, Craven R C, Weldon R A, Jr, Nelle T D, Erdie C R. Suppression of retroviral MA deletions by the amino-terminal membrane-binding domain of p60src. J Virol. 1991;65:3804–3812. doi: 10.1128/jvi.65.7.3804-3812.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu S F, Edelmann K, Strong R K, Moebes A, Rethwilm A, Linial M L. The carboxy terminus of the human foamy virus Gag protein contains separable nucleic acid binding and nuclear transport domains. J Virol. 1996;70:8255–8262. doi: 10.1128/jvi.70.12.8255-8262.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]