Abstract
Lipids are essential cellular components forming membranes, serving as energy reserves, and acting as chemical messengers. Dysfunction in lipid metabolism and signaling is associated with a wide range of diseases including cancer and autoimmunity. Heterogeneity in cell behavior including lipid signaling is increasingly recognized as a driver of disease and drug resistance. This diversity in cellular responses as well as the roles of lipids in health and disease drive the need to quantify lipids within single cells. Single-cell lipid assays are challenging due to the small size of cells (~1 pL), and large numbers of lipid species present at concentrations spanning orders of magnitude. A growing number of methodologies enable assay of large numbers of lipid analytes, perform high resolution spatial measurements or permit highly sensitive lipid assays in single cells. Covered in this review are mass spectrometry, Raman imaging, and fluorescence-based assays including microscopy and micro-separations.
Keywords: Single Cell, Lipids, Mass Spectrometry, Raman Imaging, Capillary Electrophoresis, Thin Layer Chromatography
INTRODUCTION
Lipids are a diverse group of organic compounds and can be best defined as molecules that are soluble in non-polar organic solvents but insoluble in water (1). They can be hydrophobic (typically nonpolar) or amphiphilic (a hydrophilic headgroup at one end and a hydrophobic region at another). For example, phospholipids (PL) possess 1 or more phosphate groups in addition to long acyl chains enabling these molecules to align as a bilayer (or two mated monolayers) and form cellular membranes, with the hydrophobic acyl chains facing each other and the hydrophilic phosphate-bearing regions facing the aqueous environment. This bilayer membrane then acts as a barrier surrounding the intracellular constituents and creating a barrier to the extracellular environment. Lipids also provide a multitude of essential biological functions e.g., acting as signaling molecules, providing energy storage, localizing interacting proteins and generating bioactive metabolites. Lipids in eukaryotic cells can be classified in many ways with the most commonly used eight categories developed by the International Lipid Classification and Nomenclature Committee (2). These lipid categories are: fatty acyls, glycerolipids (GL), glycerophospholipids (GPL), sphingolipids (SL), saccharolipids, polyketides and sterol lipids, and prenol lipids. Fatty acyls include fatty acids, fatty alcohols, aldehydes and esters and often act as building blocks for complex lipids such as eicosanoids which act to regulate inflammation. Cellular and organelle membranes are formed from glycerolipids and glycerophospholipids. However some members of these lipid groups such as glycerolipids/free fatty acids act as signaling molecules to regulate energy homeostasis, insulin secretion, gene expression, cell survival and cell proliferation (3). Members of the SL family such as ceramide (Cer), sphingosine (Sph), and their metabolites play a role in signal transduction pathways while other SL members direct protein sorting, mediate cell-to-cell interactions, or form signaling hubs (lipid microdomains and rafts) (4). Members of the remaining four lipid categories (saccharolipids, polyketides, sterol lipids, and prenol lipids) play diverse roles including participation in lipid bilayers, biosynthetic pathways, signaling pathways, and antioxidant functions (1). Considering these varied roles of lipid categories, it is clear that cellular health and function is greatly dependent on lipid metabolism and lipid environment.
Not surprisingly, cellular lipid metabolism is a complex network with an interplay of the various members of the lipid classes, the participation of large numbers of enzymes, and locations throughout a eukaryotic cell (organelles, membranes, lipid droplets). Alterations in this interconnected network can lead to significant consequences for the cell, organ, and/or organism. Dysfunction in lipid metabolism and handling is associated with many diseases, including cancer, cardiovascular disease, diabetes, autoimmunity, and neurodegeneration (5–9). Cancer cells are particularly adept at high jacking lipogenic and lipolytic pathways to support their proliferation and survival (10). For example, both breast and pancreatic cancer cells alter triacylglycerol FA levels by enhancing the activity of acid synthase, which is a key lipogenic enzyme in cancer pathogenesis and a well-known cross-talk node in several cancer-related networks (11). Several inflammatory and autoimmune diseases such as systemic lupus erythematosus (SLE), atherosclerosis, fatty liver disease, and cardio-metabolic disease are associated with impaired lipid metabolism of free fatty acids, triglycerides (TG), lipopolysaccharides, and cholesterol esters (12,13). In all of these diseases, the behaviors of single cells and/or their clonal progeny have profound impacts on disease progression and outcome. Understanding the cellular and molecular mechanisms at single-cell resolution using multi/trans omics approaches reveals information on cellular heterogeneity and progression of disease pathogenesis. This cell-to-cell heterogeneity extends to the information flow through lipid signaling and metabolic pathways. For example, accumulation of fatty acids is correlated with the initiation of pancreatic ductal adenocarcinoma (PDAC) and intracellular heterogeneity in fatty acid distribution results is associated with metastasis which has 5-year survival rate of only 9% (14). Similarly, in metabolomic diseases, heterogeneous thermogenic capacity of a subpopulation of mature adipocytes called beige adipocytes can result in obesity and diabetes (15). Thus, understanding lipid handling at the single cell level is critical to revealing intracellular as well as intercellular heterogeneity and impacts to human health and disease processes.
Lipids have been more challenging to assay than other cellular constituents such as ribonucleotides and proteins primarily due to their lack of repeating residues, high molecular complexity, hydrophobicity, tendency to aggregate, and their propensity to bind to surfaces. Nevertheless, a number of analytical methodologies such as thin-layer chromatography (TLC), high-pressure liquid chromatography (HPLC), gas chromatography (GC) and mass spectrometry (MS) have been successfully applied to build the field of lipidomics providing insights into the diversity of lipid species in humans, their roles as building blocks, signaling agents, structural supports, energy storehouses and temperature regulators. Importantly enzymes within many lipid synthetic or metabolic pathways e.g. phosphoinositide-3 kinase, sphingosine kinase, have become pharmaceutical targets with a goal of modulating disease pathways and further driving the need to understand these important molecules (16). The vast majority of experimental work related to lipid pathways has been performed on bulk tissue specimens or pooled cell lysates due to the ease in extracting intracellular products to provide a large sample size. Only recently have analytical methods achieved the sensitivity and specificity need to assay lipids from a single mammalian cell of picoliter volume. One ongoing analytical challenge is the wide range of lipid analyte concentrations within a cell, for example plasma membranes are enriched in cholesterol, phosphatidylserine (PS), and sphingolipids while the endoplasmic reticulum (ER) is deprived of these lipids. In addition to this heterogeneous lipid distribution between membranes, there are also differences in distribution of lipids across the membrane bilayer. This aspect demands high dynamic range, excellent sensitivity and spatial resolution from an analytical assay which in practice means that most analytical methods target a subset of cellular lipids. Lipids are also found in a range of cellular sub compartments and in varying states e.g., insoluble aggregates, membrane incorporated, and protein-bound, making some lipids readily accessible for analytical assays while other lipids have yet to be assayed from single cells. Innovations in a number of analytical tools (MS, microscopy, Raman spectroscopy, chromatography, electrophoresis and fluorescent probes) now enable a plethora of lipids to be quantified from single cells; however, lipidome characterization in these ultrasmall samples remains far from mature. This review focus on recent developments in single-cell lipid assays including the method’s working principles, advantages, limitations, and applications.
1. MASS SPECTROMETRY OF LIPID ANALYSIS IN SINGLE CELLS
Since mass spectrometry (MS) possesses high-sensitivity and specificity and does not require labels or probes to detect analytes, MS has been widely exploited for single cell analysis (SCA) including the assay of lipids. MS offers a nontargeted assay approach so that unknown lipids can be identified, and assays do not need to focus on a pre-identified lipid species. Single cell mass spectrometry (SCMS) can be performed using different desorption/ionization techniques and sampling environments adding to the versatility of MS. Desorption/ionization strategies include laser desorption/ionization mass spectrometry (MALDI-MS), secondary ion mass spectrometry (SI-MS), and electrospray ionization mass spectroscopy (ESI-MS) (17). Methods which require the sample be placed under vacuum (MALDI-MS and SI-MS) are highly sensitive with a 50 attomole limit of detection and a high spatial resolution of sub-50 nm, but do require sample processing (fixation and dehydration) and so may suffer from unwanted spatial distortions or chemical reactions. In contrast ESI-MS is performed on cells in an ambient environment with minimal sample preparation (18–20). The cellular contents can be introduced directly into the mass spectrometer or utilize a transfer device such as a capillary. More recently, laser microbeams have been used to dissect single cells to provide spatial resolution albeit at a reduced sensitivity (21,22). Imaging mass spectrometry (MALDI-IMS) is increasingly popular displaying a subcellular spatial resolution of 25–50 μm2 pixel size (23). Comprehensive reviews on SCMS are available although focused largely on protein-based assays (24). This section highlights the latest SCMS techniques in lipidomics as well as their advantages, limitations, and opportunities (Figure 1).
Figure 1.
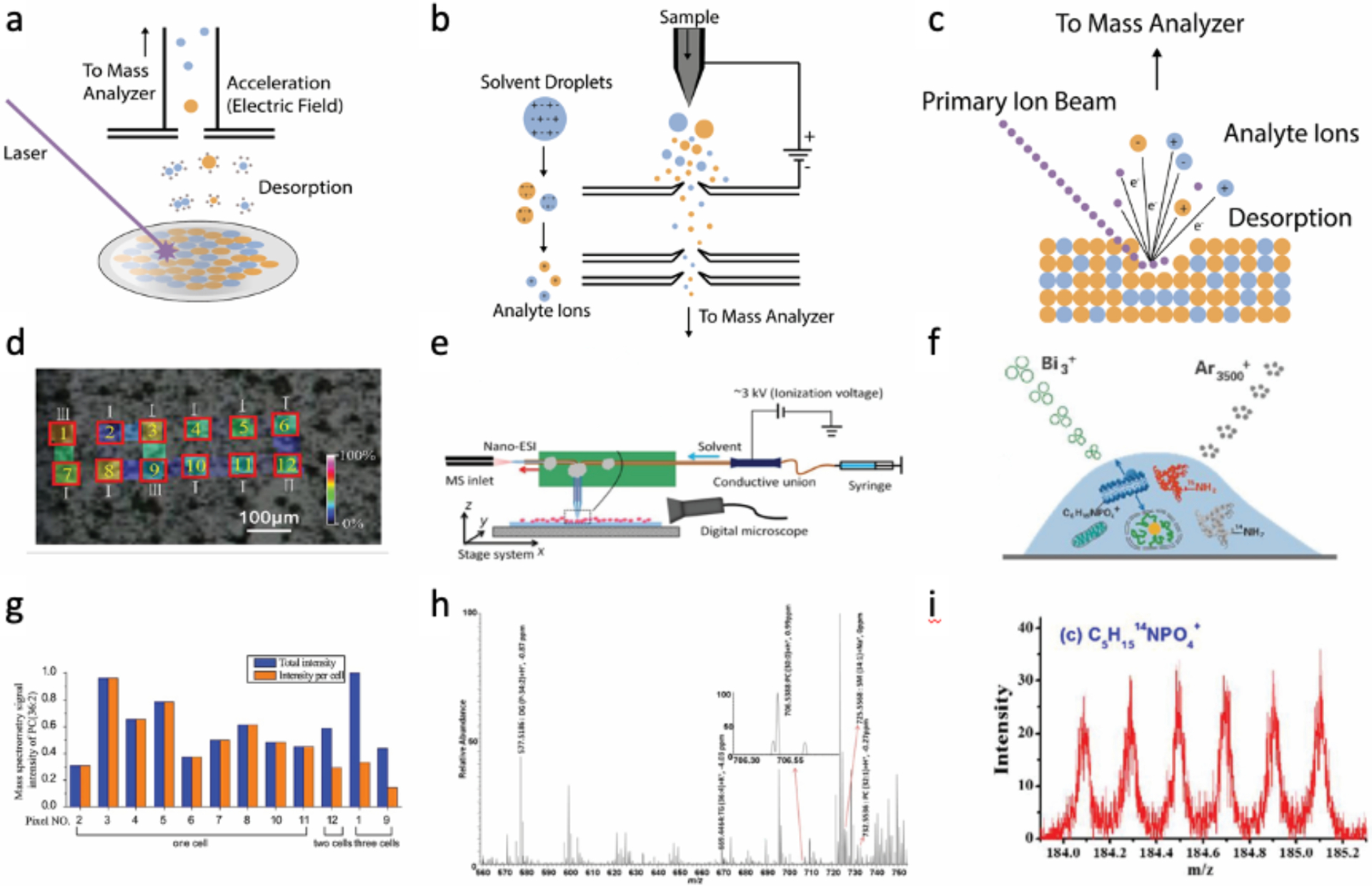
Mass spectrometry assay of lipids in single cells. Schematics describing the working principles of (a) matrix assisted laser desorption ionization mass spectrometry (MALDI-MS), (b) electrospray ionization mass spectrometry (ESI-MS), (c) secondary ion mass spectrometry (SI-MS). (d) A549 cells combined with a 9-aminoacridine (AA) matrix were examined by MALDI-MS Imaging. Pixels positive for PC(36:2) are marked in the yellow numbers and the cell count in every pixel marked in the white Roman numerals in the microscopy image. (e) The schematic demonstrates the use of a single-probe ESI to assay for PC, SM, TG in single HeLa cells. (f) The schematic shows the assay of a single cell by SI-MS. Bi3+ liquid metal ion gun (LMIG) was used to scan across the cell to acquire an XY image while Ar3500+was used to obtain depth or Z profiles. (g) The total and average mass spectrometry signal intensities for PC(36:2) from the extracted pixels in panel D are shown. (h) Displayed is the mass spectrometry data obtained in a mass range from 560 to m/z 750 for the experiment shown in panel E. (i) Data demonstrating the fragment ions obtained from sphingomyelin (C5H15NPO4+) when a cell was imaged at submicrometer resolution by TOF-SI-MS as depicted in panel f. Panels d and g are reprinted with permission from Yang, T., Gao, D., Jin, F., Jiang, Y., & Liu, H. (2016). Rapid Communications in Mass Spectrometry, 30, 73–79. Panels e and h are adapted with permission from Pan, N., Rao, W., Kothapalli, N. R., Liu, R., Burgett, A. W., & Yang, Z. (2014). Analytical chemistry, 86(19), 9376–9380. Copyright (2014) American Chemical Society. Panels f and i are reprinted from the International Journal of Mass Spectrometry, Vol 421, Sheng, L., Cai, L., Wang, J., Li, Z., Mo, Y., Zhang, S., Xu, J.J., Zhang, X. and Chen, H.Y. 2017. pages 238–244. Copyright (2017), with permission from Elsevier.
1.1. Matrix Assisted Laser Desorption Ionization Mass Spectrometry (MALDI-MS)
Laser desorption/ionization mass spectrometry (LDI-MS) techniques for single-cell lipid measurements have evolved greatly in recent years particularly with the development of MALDI-MS for whole cell and subcellular lipid assays (23,25). In MALDI-MS, a sample pre-coated with an energy absorbing matrix is irradiated by a laser beam and energy absorbed by the matrix is then transferred to the sample facilitating sample desorption and ionization (Figure 1a). Ionized sample constituents enter the mass spectrometer followed by measurement of their mass-to-charge (m/z) ratios. The matrix choice typically depends on the analytes to be assayed so as to optimize energy transfer and a variety of different matrices have been employed successfully for lipid measurements by MALDI-MS including α-cyano-4-hydroxy cinnamic acid (CHCA), 2,5-dihydroxybenzoic acid (DHB), 9-aminoacridine (9-AA) (26). The MALDI process is typically coupled to a time-of-flight (TOF) mass spectrometer providing access to a wide ion mass range (m/z) from as low as 100 Da to over 500 kDa but with a lower linear mass resolution of only >5,000 FWHM. An asset of MALDI is the ability to couple to other MS detectors for example tandem MS for structural characterization or FTICR for high precision measurements enhancing MALDI capabilities and applications in single cell lipidomics. However, initial applications of MALDI-MS for lipid-based assays were low in spatial resolution sampling the entirety of a cell and often applied to a large cell type such as an oocyte (27). A wide range of lipids including sphingomyelins (SM), phosphatidylcholines (PC), and triacylglycerols (TAG) were quantifiable revealing important insights into membrane lipid compositions under various environmental conditions and at various developmental stages of an embryo. Although providing significant advancements in knowledge, a challenge in these measurements was that only the most abundant lipids were detectable with many rare species (important in cell signaling) undetectable due to their low abundance, e.g. even with an attomole detection limit, only the most abundant lipid species are detectable (28). These applications, however, pointed the way for improved instrumentation and methods to enhance spatial resolution, throughput, and sensitivity.
Improved sample preparation protocols, laser illumination strategies, and image processing and reconstruction methods have enabled significant increases in spatial resolution, throughput and multicomponent analysis. Cell lyophilization applied to large cell types such neuronal ganglion cells opened the door to subcellular sampling of cells with assay of membrane phospholipids such as PCs which could be detected with different profiles in a neurite vs a neuronal cell body (29). While results from this work provided preliminary understanding of neuronal lipid metabolism, the large laser spots utilized for MALDI-MS still yielded poor sensitivity and spatial resolution. The use of overlapping laser spots to oversample as well as complete sample ablation at each laser beam location enabled resolutions greater than that of the microbeam diameter but created tissue distortions such as analyte delocalization ultimately limiting this strategy. Improvements in both sensitivity and spatial resolution were achieved by positioning the laser behind the sample and irradiating from the sample backside (30). This geometry reduced the working distance between the sample and optics enabling a sub-micron laser beam spot on the sample and a submicronmicron spatial resolution during imaging. In this proof-of-concept work, the distribution of intact lipids at a single m/z 782 was imaged in cultured human embryonic kidney and colon cancer cells. Spatial resolution and sensitivity have also been enhanced by optimizing sample preparation, measurement parameters and computational image reconstruction combined with more traditional instrumentation to image a range of lipids [PC, SM, diglycerides, phosphatidylethanolamines (PE), phosphatidylinositol (PI), and/or triglycerides (TGs)] at cell-sized resolution (5–10 mm) (23,31). Three-dimensional (3D) MALDI-IMS has also been developed by stacking and reconstructing traditional two-dimensional (2D) MS images into 3D images (25). A variety of strategies have been developed to enhance the rate of lipid analysis in single cells so that large numbers of cells can be assayed. The use of a laser for sample desorption/ionization provides a speed advantage due to the ability to rapidly scan the beam and very short pulse durations (<1.1 ns). Placing cells into microwell arrays to preposition cells at known locations resulted in 40% single cell capture efficiency, which when followed by MALDI-IMS enabled the assay of 12 lipid species in single cells by extracting relative signal intensity data from every pixel in the images over several minutes (32,33). Integration of cell recognition software with automated instrumentation permitted cells randomly located on a slide to be sequentially examined by MALDI-MS with assay of up to 30,000 cells (34). A feature of MALDI-IMS is the ability to combine MS with other analytical methods, for example, optical microscopy (fluorescence, brightfield) to create information rich data sets as well as co-register the MALDI-MS data with known histologic structures (35). While these methods have improved spatial resolution and analytical throughput, the performance of MALDI-IMS remains focused on high abundance lipid species particularly when combined with TOF or quadrupole-based detection (≤12 lipid species or features).
One reason for the limitation of MALDI-IMS to high abundance lipids in single cells is the low mass resolution of the TOF detectors which ensures that low abundance species are overshadowed by all of the highly abundant lipids. Fourier transform ion cyclotron resonance mass spectrometry (FTICR-MS) offers both high mass resolution and high-mass accuracy enabling the detection of larger numbers of lipid species compared to other MS detectors. When combined with MALDI these assets are retained as well as the MALDI advantage of high single-cell analysis rates, e.g. heterogeneous liposaccharide distributions were measured in >100 RAW 264.7 cells on a timescale of hours using FTICR-MS (36). Up to 670 ions in the lipid mass range could be detected in large numbers of single cells with excellent throughput (100’s of cells/h) (34–37). As another example, up to 500 lipid features were detected within 30,000 rodent brain cells from which 101 significantly distinct cell clusters could be correlated with neuronal or astrocytic lipid markers. Due to the high mass resolution, impressive chemical details were possible with the most common lipids identified as [PC(32:0)+H]+ and [PC(34:1)+H]+ in 98.9 and 89.5% of cells, and [PC(34:1)+K]+ and [PG(40:2(OH))+Na]+ present in <1% of cells (34). To achieve the needed sensitivity, spatial resolution was limited to the size of a cell or larger (25–100 mm) in these studies. A challenge was the need to use reference data banks for lipid identification since tandem MS is not possible on these small-scale samples. However, the ongoing construction of high-quality lipid databases will in the future address this drawback. Finally, the high cost of FTCIR-MS will likely limit this technology to core facilities and centers at large institutions.
1.2. Electrospray Ionization Mass Spectroscopy (ESI-MS)
Electrospray ionization (ESI-MS) is low in energy ionization method which yields minimal fragmentation of analytes. In ESI, charged droplets of analyte solution are produced at the outlet of a capillary tip and are accelerated under an electric field towards the MS detector (Figure 1b). Application of a drying gas or heat progressively evaporates the solvent into charged ions, allowing the analyte to enter the gas phase prior to the MS inlet. ESI possesses high ionization efficiency, is readily coupled to chromatographic devices for sample separation prior to MS analysis but does yield more complex spectra than MALDI with multiply charged species (38). Sufficient volume and quantity of sample is required to accommodate sample loss in the chromatographic step making assay of single cell contents challenging. The complex makeup of biologic samples can lead to matrix effects or ion suppression reducing the accuracy and precision of the MS analysis. Finally, ESI-MS is challenging to integrate with other cellular analysis methods such as microscopy. A plethora of technologies have been developed in recent years to overcome these challenges e.g. nano-ESI-MS, desorption ESI (DESI-MS), laser ablation ESI (LAESI-MS), probe ESI (PESI-MS) and capillary ESI (CESI-MS) (39–43). We will cover a subset of ESI-MS technique in this section: perhaps the most applicable strategy to single cell lipid assays is nano-ESI, a derivative of ESI tailored to accommodate small volume samples with limited analyte concentrations.
Nano-ESI uses an emitter tip of typical diameter ranging from 1 μm internal diameter, employs low solvent flow rates of 20 to 40 nL min−1 (compared to ESI’s typical rate of 100 μL min−1), involves reduced sample consumption and improved detection sensitivity, does not require a drying gas or heating, and applies a high voltage of 1–2 keV for inonization to form an electrospray. A chromatographic step is not required as a first stage for nano-ESI so that samples can be directly injected into the MS detector (eliminating sample loss on a separation column). The dilute sample (~0.5 to 5 pmole/mL) also minimizes matrix effects. Together these features provide a high ionization efficiency and signal to noise relative to that of standard ESI. A convenient aspect is that the nano-tip (most often a pulled capillary) can serve as both a collection device for the cell contents as well as the nano-ESI tip. Precision spatial movement of the nano tip by a micromanipulator is required to precisely collect the cell or its contents. Nano-ESI can be mated with other analytical methods for example microscopy and patch clamp. A significant advantage of nano-ESI is the ability to sample and analyze single cells from within intact tissue slices or organs which provides a more physiologic output then that obtained from disaggregated, isolated cells (44). A variety of capillaries, probes, patch-clamp pipettes, and microfluidic devices under manual or automated operation have been used to collect single cells or their contents as well as function as the nano-ESI tip (39,44–48). When a chromatographic step is not coupled to nano-ESI, a lipid extraction step is often incorporated to enhance lipid introduction into the nano-ESI-MS instrument. Many extraction strategies have been employed successfully including pre-addition of solvent around the cell just prior to cell sampling, inline addition of solvent as the cell or cell lysate flows toward the emitter tip (46,49), use of a dual barrel capillary to introduce solvent through one barrel as a cell is loaded into the other barrel and other strategies (39,50). Cell lysis (and lipid solubilization) during whole-cell collection is accomplished by using the sampling tip and lipid extraction solvent, by incorporating ultrasonic lysis (45), or by addition of inline filters with cellpuncturing spikes (zinc oxide nanothorns) (46). Nano-DESI takes this step to the next level by using a second solvent carrying capillary for lipid extraction forming a liquid bridge connecting to the emitter capillary (51). An alternative version IR-MALDESI is as the name implies a hybrid of MALDI and ESI using a laser to ablate biological samples followed by ESI (52). Nano-ESI and its various forms are also compatible with subcellular sampling depending on the exact sampling strategy with a resolution of ~10–100 mm (47,49). Cytoplasmic sample collection using a patchclamp pipette could in theory leave a living cell after sampling for other assays (although sensitivity will be a challenge) (47). Throughput has been enhanced (520 lipid features from 30,000 cells and 38 cells/min) by automated movement and positioning of the cell-collection tip, extraction steps as well as incorporation of label-free flow cytometry and microfluidics for cell queuing into the emitter tip (20,53,54). Nano-ESI has been applied to detect lipids in a wide range of mammalian cells (HeLa cervical cells, liver, white blood cells, breast cells, neuronal cells, and others) (39,44–48) as well as subcellular components (lipid droplets) (49); however, the majority of lipid analytes detected in cells have been the abundant species such as PC, PL, PE, and PA. Despite the theoretical advantages of direct injection methods, utilizing a pre-ESI separation step has yielded very high performance. Coupling nanoflow liquid chromatography (nLC) to nano-ESI enabled detection of 236 lipids from 4 lipid classes (SL, sterol lipids, glycerolipids, glycerophospholipids) from healthy and diseased mammalian hippocampal slices providing an improved understanding of lipid homeostasis in brain disease (47). Others have used a bulk pre-separation step selectively isolating PL by incubating a single-cell lysate with TiO2-coated Fe3O4 nanoparticles followed by elution from the beads and assay by nano-ESI (55). This strategy permitted detection of eighteen different PLs with limits-of-detection of ~0.01 mg/L in the MS/MS spectra.
1.3. Secondary Ion Mass Spectrometry (SI-MS)
Traditional secondary ion mass spectrometry (SI-MS) employs a high-energy primary ion beam (Ar+, O2+, N2+, and others) to bombard the sample surface releasing charged secondary ions which are then directed into an MS detector (Figure 1c) (56). Like MALDI, SI-MS is performed under vacuum requiring samples to undergo chemical or high pressure/freezing fixation prior to analysis. Since the ion beam can be focused to a tight spot (and is not diffraction limited) the spatial resolution is exceptional at a sub-50 nm range with a depth of about 20 μm (57). While the ion beam efficiently lifts off sample providing excellent sensitivity, the high energy beam also fragments sample molecules into a size range of hundreds of Da making data interpretation more challenging than low energy ionization methods such as MALDI and ESI. However, since the ion beam removes a top layer of sample, 3D imaging is possible by sequential X-Y scanning. To improve SI-MS compatibility with single-cell lipid imaging, recent work has focused on several technological innovations: application of low-energy cluster-ion beams with or without an energy absorbing matrix, optimization of fixation strategies, and enhancement of 3D imaging methods using sample labeling methods. The recent development of cluster ions as the primary beam in SI-MS enables sample surface sputtering at low energy with less sample damage and release of higher molecular weight sample molecules including intact lipid molecules (57,58). For example, C60+ SIMS-MS has been applied to image cells and assay lipid composition of the plasma membrane followed by assay of lipids within the cytosol of the same single cell (59). Using Au32+ as the cluster ion, breast cancer stem cells were demonstrated to have different fatty acid content than non-stem cells (60). Enhanced and validated preservation strategies include plunge freezing in ethane to eliminate sample fracture and characterization of glutaraldehyde-based fixation and the impact on lipids have also advanced single cell lipid assays by SI-MS-MS (61). Ionic liquids (IL) have been applied to samples to perform matrix-enhanced SI-MS (ME-SI-MS) enhancing the sputtering/ionization efficiency and chemical signals attainable from single cells with quantification of PC lipids in neuronal cells (62).
Powerful variants of SI-MS-ES such as multi-isotope imaging MS (MI-MS) use stable isotope labeling of cells and a scanning ion beam to quantitatively image the distribution within cells of a stable-isotope such as 13C. These advances enabled a better understanding of the mechanism of long chain free fatty acid (FFA) transport across adipocyte’s cell membrane (28). Using MI-MS, the localization of intact lipid species such as PC across the surface of single neurons as well as co-localization with vitamin E and cholesterol was assessed (28). Another innovative variant of SIMS-ES is nano-secondary ion mass spectrometry (NanoSI-MS), which uses a high energy focused beam to achieve a spatial resolution down to ~50 nm and is suitable for detecting small (mainly monoatomic and diatomic) ions (63).
2. RAMAN-BASED TECHNOLOGIES
When light impinges upon a molecule, the light can be scattered elastically that is without a change in energy (Rayleigh scattering) or inelastically i.e. with a change in energy (Raman scattering) (64–66). In Raman scattering, the photon’s energy initiates a vibrational change in the molecule and a subsequent scattered photon which can be lower in energy (Stoke’s scattering) or higher in energy (anti-Stoke’s scattering) than the incident radiation. The energy difference is characteristic of the excited chemical bond leading to insights into the molecular species illuminated (64,67,68). Due to the high concentration and/or strong scattering from a number of bonds (e.g. C-H, C-C, C=C) commonly found in organic molecules, Raman scattering has emerged as a powerful tool for cellular imaging (64,65,69). Each chemical bond possesses a characteristic vibrational mode or Raman shift, for example the C-H wavenumber lies in the region of 2700–3100 cm−1, while CH2 in 1400–1500 cm−1, C-C in 600–1300 cm−1, and C=C is 1640–1680 cm−1 (70). Since Raman scattering is based on a vibrational resonance, Raman-based imaging is not impacted by photobleaching or sensitive to the surrounding environment (as in the case of fluorescence). The infrared wavelengths typically used for cellular Raman-imaging confer high sample penetrance with reduced photodamage. In particular Raman scattering has been used to image a wide range of molecules in cells such as lipids, proteins and DNA. Raman spectra can be acquired within seconds especially with the newer Raman methods due to the high total concentration of biological molecules in cells. Lipids in particular with their large number of CH2 groups are well suited for Raman imaging (64,65,69,70) and can be assessed in living or fixed single cells with high contrast and high resolution in a label-free and nondestructive manner (68). However a challenge for all of the Raman-based methods in imaging biological samples is the complex mixture of molecules present with similar vibrational modes making the identification of specific molecular species quite challenging and the detection of low concentration species impossible. Never-the-less Raman microscopy has emerged as a powerful tool for biological imaging to advance our understanding of the mechanisms behind physiological and diseased states.
Raman imaging of lipids within single cells typically employs either spontaneous Raman scattering, SRS (stimulated Raman scattering) or CARS (coherent anti-Stokes Raman scattering) although other strategies are possible (64,65). With spontaneous Raman-based microscopy, a single laser beam illuminates the biological sample, exciting molecules to a higher vibrational state, followed by light emission at a longer wavelength (Figure 2a). Due to the very small scattering cross section (~10−30 cm2 steradian−1) for spontaneous Raman scattering, light emission is weak (~1/1016 that of fluorescence) resulting in long imaging times (many minutes/image) with use of high laser powers even for high concentration analytes such as cellular lipids. The high required illumination intensities also yield significant background in the form of sample autofluorescence and Raleigh scattering. SRS and CARS address many of these challenges providing stronger signals with lower background. For SRS two laser beams (pump and Stokes beams) are incident on the sample. When the frequency difference between the two beams matches a molecular vibration, stimulated excitation of the vibrational transition occurs with an intensity loss at the scattered pump wavelength and an intensity gain at the scattered Stokes wavelength (Figure 2b). Measurement of this energy gain/loss can then be accomplished albeit with sophisticated equipment. An advantage of SRS is the proportionality of the signal to molecular concentration, and the spectral match to spontaneous Raman scattering. CARS which can be accomplished using a multitude of strategies also employs two beams (pump and Stokes) but in this instance molecules are stimulated while in their vibrational state yielding a higher-energy emission (anti-Stokes shifted) (Figure 2c) (67,71). CARS provides fast, high sensitivity measurements due to emission at a shorter wavelength (with facile spectral separation from longer-wavelength sample autofluorescence). CARS however requires high intensity beams which can lead to multi-photon absorption and material damage. The required complex instrumentation also requires careful beam synchronization. A number of compound microscopes that combine Raman with traditional optical microscopes have demonstrated added value to that of Raman imaging alone (72–78). These compound microscopes include Raman imaging with bright field, fluorescence or confocal microscopy to provide simultaneous information on both lipids as well as other cellular constituents such as proteins including their relative spatial locations (75,79–84). As an example, SRS combined with confocal fluorescent microscopy enabled high-speed multicolor imaging providing insights into lipid droplet (LD) biology and other markers associated with these LDs at the single cell and even subcellular level (79,81–83). All of the Raman imaging methods can now be configured to readout continuous spectra e.g. hyperspectral Raman, enabling a greater characterization of the complex lipid mixtures within cells (85–89). Comprehensive reviews with a sole focus on Raman microscopy are available and this section highlights spontaneous, SRS, and CARS with a focus on their advantages, limitations, and opportunities in the measurement of lipids in single cells (Figure 2) (84,90,91).
Figure 2.
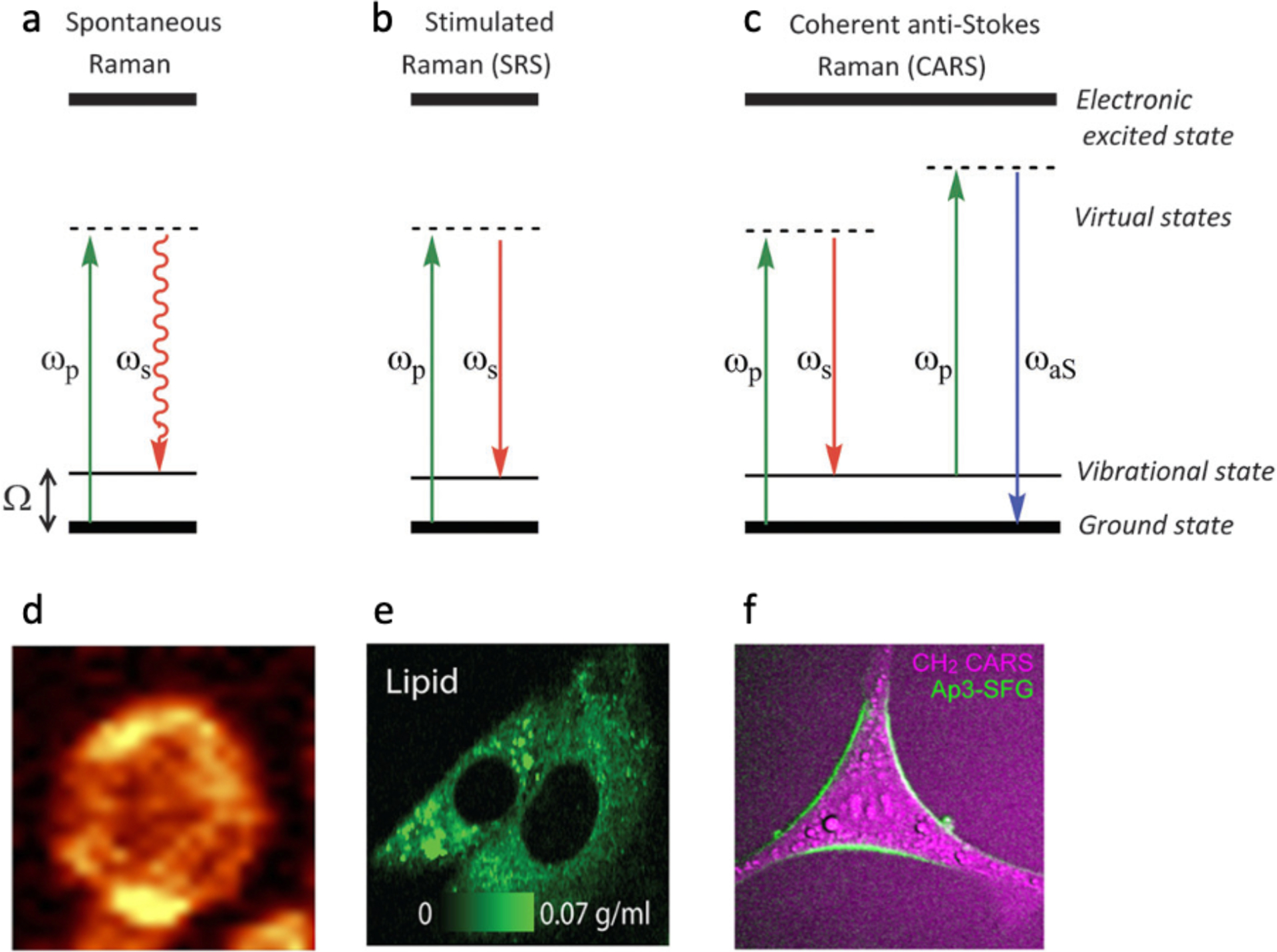
Raman imaging of single cells. (a-c) Schematics demonstrating the energy level diagrams in Raman imaging. (a) Spontaneous Raman scattering. The straight line (green) indicates the pump beam at ωP, and the curved line (red) denotes the scattered light at a longer wavelength ωS. (b) Stimulated Raman scattering (SRS). Two lasers, a pump (ωP) and a Stokes beam (ωS) impinge upon a sample with stimulated emission occurring when △ω (Dω = ωP - ωS) equals to a molecular vibration frequency. (c) Coherent anti-stokes Raman scattering (CARS). In CARS, a pump (ωP) and a Stokes beam (ωS) illuminate the sample and when ωP - ωS matches the molecular vibration frequency an anti-Stokes signal at 2ωP - ωS = ωaS is generated. (d) Distribution of lipids (1420–1460 cm−1) in a pulmonary cell obtained by spontaneous Raman imaging. (e) An SRS image showing the concentration of cytoplasmic membrane lipids in live melanoma cells. (f) CARS image of a neuronal cell. The purple color shows CH2 bonds while the green color is the plasma membrane dye, Ap3-SFG. Panel a-c was reprinted with permission from Tipping WJ, Lee M, Serrels A, Brunton VG, Hulme AN. Chem Soc Rev. 2016;45(8):2075–89. Panel d was reprinted with permission from Tott S, Grosicki M, Glowacz J, Mohaissen T, Wojnar-Lason K, Chlopicki S, et al. Biochimica et Biophysica Acta 2021;1867(9):166180. Panel e was reprinted with permission (CC BY-NC-ND) from Oh S, Lee C, Yang W, Li A, Mukherjee A, Basan M, et al. Proc Natl Acad Sci USA. 2022;119(17):e2117938119. Panel f was reprinted with permission from Mizuguchi T, Momotake A, Hishida M, Yasui M, Yamamoto Y, Saiki T, et al. Anal Chem. 2020;92(8):5656–60. Copyright 2020 American Chemical Society.
2.1. Spontaneous Raman Imaging
Spontaneous Raman spectroscopy has enabled lipid assays in single cells and has many advantages relative to the newer Raman imaging methods including simpler instrumentation with a single laser that is readily focused to submicron spots sizes easily scanned across a sample. This enables a typical imaging spatial resolution in the range of ~0.3–0.6 mm (92,93). A strength of Raman in lipid analysis is the ability to assess the degree of lipid unsaturation, as well as the ratio of cis/trans isomers in lipid samples (94). The emission spectral resolution is typically of ~1.5–3 cm−1 (92,95–97). The collection of spectra at each image pixel provides high value chemical information but can require up to 1 s/pixel yielding long times to image entire samples (92,98–100). The availability of commercial microscopes (WITec and Renishaw, for example) that combine spontaneous Raman and confocal microscopy also make the technology accessible to nonexperts. Due to the inherently weak signal of spontaneous Raman and the complex nature of cellular structures, the method generally focuses on organelles with high lipid concentration such as lipid droplets (LDs) or incorporates Raman labels such as deuterated or alkyne labelled compounds to enhance analyte detectability.
The combination of spontaneous Raman with other microscopy methods such as confocal, atomic force and even other Raman techniques has emerged as critical enablers to measure lipids within LDs and other subcellular organelles (72–78,80,101–103). A number of studies have combined Raman and fluorescence microscopy with fluorescent lipid probes such as Nile Red, Oil-O-Red, ReZolve-L1™ or BODIPY 493/503 with Raman imaging to label lipids especially LDs directly enhancing sensitivity, lipid characterization and spatial precision (98,104,105). Another example of the advantages of compound microscopes is the spatial targeting of Raman measurements to fluorescently labeled organelles (mitochondria or endoplasmic reticulum) for fast, efficient subcellular measurements as opposed to the time-consuming imaging of an entire cell (80). This approach termed “micro-Raman assay” has enabled assay of the lipid unsaturation, cis/trans isomer ratio, sphingolipids and cholesterol levels in live cells. Another compound system has been demonstrated to acquire Raman hyperspectral images with a four-minute temporal resolution followed by comparison of the Raman data to that of quantitative phase microscopy to distinguish living cell types (72).
Cellular constituents have little Raman scattering between 1800–2800 cm−1 aka the “silent region” and Raman tags with spectra in this region have been developed to track intracellular molecules without interference from the Raman signal of the endogenous cellular components (104,106–108). Commonly used tags such as alkynes, nitrile and deuterium provide a strong Raman scattering peak in the silent region and have facilitated identification of organelles or specific molecules within the complex cellular environment (104). A series of alkyne-tagged coenzyme Q (AltQ4) analogues label the mitochondria permitted visualization of the spatial location of this Raman tag (109). An alkyne-tagged cholesterol labelled LDs in a liver cancer cell line yielding the molecular identification of as many as seven different constituents of LDs, as well as the spectral isolation of three structurally different lipid species (110). Falcarinol, an anti-inflammatory polyacetylene which naturally scatters in the silent region has been used to investigate cellular changes resulting from induced endothelial dysfunction (111). Deuterium is also commonly used as a Raman tag to visualize lipids (112,113). Advances in the understanding of lipid physiology created through the use of deuterium tags include the visualization of lipid translocation between endocytic vesicles and lipid droplets in macrophages (114), intracellular lipid metabolism in macrophages (113), time-dependent investigation of fatty acid distribution in macrophages (115), and the quantification of stearic acid uptake and accumulation in lipid droplets of cat oocytes (116). One recent innovation incorporated H-alkyne and D-alkyne labels into long chain fatty acid probes to distinguish between two structurally similar small molecules in living cells (117).
2.2. Stimulated Raman Scattering (SRS) Imaging
Stimulated Raman scattering (SRS) is a nonlinear optical process for which the scattered signal is generated at the focal plane of the sample enabling intrinsic three-dimensional sectioning by scanning in the x, y, and z axes with submicron to micron spatial resolution (65,70). SRS is characterized by a low background (orders of magnitude lower than spontaneous Raman) since non-resonant signal is not present i.e. when the frequency difference between the pump and Stokes beams does not match a vibrational transition then no energy exchange occurs between the scattered signals. This translates into detection limits of greater than ~0.1 mM (often 10’s of mM) depending on the molecule. High speed imaging of entire samples (ms/pixel) is possible especially when measuring at a single wavenumber. Since an intensity difference is the measured signal riding on the pump and probe Raman-scattered photons, optical modulation (>2 MHz) and phasesensitive detection are needed to differentiate the scattered signal from the incoming excitation beams. This requires sophisticated instrumentation (in addition to the lasers) such as a lock-in amplifier to detect the stimulated Raman loss or gain but does enable the measured signal to be of high intensity relative to that measured for spontaneous Raman or CARS (65,70). Although low in magnitude relative to other Raman methods, SRS does have a background which is of complex origin and thus challenging to mitigate; however frequency modulation of the incident light can be employed to minimize this background (118). Other innovations such as hyperspectral SRS enable imaging over a range of wave numbers (300 cm−1) at each pixel to obtain detailed spectral information. The collection of Raman spectra permits deconvolution of overlapping Raman signals due to the presence of multiple molecules. This is particularly useful in reconstructing lipid types in an imaged sample (85,119,120). Moreover, hyperspectral in combination with broadband imaging offers an imaging range as great as 600 cm−1 with a fast speed of ~8 μs/pixel (85). A major application of SRS lipid imaging in single cells has focused on LDs and other high-concentration lipids due to the poor sensitivity relative to other methods e.g. mass spectrometry or fluorescence (121–124).
Exciting advances are underway in SRS imaging of single cells. For example, customized equipment paired with new computational tools enables the measurement of protein mass, lipid mass and water content in the same single cell with 3D resolution permitting internal comparison of concentrations (123). This system provided a sensitivity of ~0.015 g/mL, a spatial resolution of 1.18 and 1.90 μm in lateral and axial dimensions respectively. Hyperspectral SRS imaging has been utilized to identify lipid content differences in cancerous vs normal cells to understand fundamental differences in lipid biology of these cells (81,119,120). Even lipid droplets within the two types of cells are observed to have distinct lipid signatures (120). A growing trend is the integration of SRS with other types of imaging methods either instrumentation or sample preparation strategies (79,122,125–128). For example, expansion microscopy, a type of sample preparation that expands a sample along all axes so that the sample can be imaged in finer detailed has been used to prepare samples for hyperspectral SRS. This combination provided nanoscale spatial resolution of endogenous lipids (and other molecules) within a sample (126). SRS has been combined with fluorescence microscopy to yield a high speed multiplexed imaging system with a temporal resolution of seconds so that the dynamics of LDs in living cells could be tracked (79). Notably, this system offered 20-color vibrational contrast with the ability to be increased to 26 colors creating a powerful tool to investigate intercellular molecules. Since this compound instrument is also applicable to molecules other than lipids, single-cell multiomics becomes possible on a single platform. The use of Raman-active probes including photoactivatable probes further expands the utility of SRS imaging (82,83,106,129,130). Multicolor photoactivatable alkyne Raman reporters utilizing cyclopropenone caging have been applied to image and track live cells (130). These light activatable probes are relatively small compared to fluorescent probes and minimally perturb cellular physiology. The probes can be multiplexed with other methods providing multicomponent imaging while measuring cellular dynamics, for example, simultaneous imaging of mitochondria, lysosomes, and LDs.
2.3. Coherent anti-Stokes Raman Scattering (CARS) Imaging
As with SRS, CARS is a nonlinear optical process with submicron spatial resolution when applied to imaging single cells. The signal of CARS is up to 5 orders of magnitude stronger than that of spontaneous Raman scattering, and it is highly effective for detection of lipids (131). Compared to SRS, CARS has a simpler signal detection system since the emitted signal is at a different energy than that of the pump/probe scattered photons and so signal detection requires only spectral filters (64,69,91,119). But CARS typically has a poorer signal-to-noise ratio compared to SRS, and the CARS signal is not proportional to analyte concentration (unlike the SRS signal), making CARS more challenging in terms of analyte quantification (84,91,119). CARS possesses an acquisition time two orders of magnitude lower than SR (90). Although CARS and SRS both permit nondestructive imaging in a label-free manner, both methods can induce significant photodamage due to the high peak powers of the ps or fs lasers. Like SRS, spatial resolutions in the 100’s of nm are achievable with CARS (65). These attributes have enabled CARS to have a tremendous impact on the field of single cell lipid measurements particularly in the dynamics of LDs (132–134). For example, recent technological innovations have enabled high speed, submicron visualization of LD movements within single cells (132).
Enhancements in CARS instrumentation have yielded additional advances multi-plexed and hyperspectral imaging at high spatial resolution to enhance analyte identification in single cells, as well as the integration of CARS with other microscopy methods for multimodal imaging (86–89,131,135–141). As an example, hyperspectral CARS has been applied to assay lipids, proteins and DNA in single osteosarcoma cells, and track their concentration changes over time during mitosis. Similar methodology was used to quantify lipid concentration and composition in individual lipid droplets in living cells over time when supplied with medium possessing different fatty acids (142). Multiplexed CARS permitting imaging over a range of wavelengths (or vibrational energies) and combined with fluorescence microscopy has enabled tracking of lipid storage in LDs vs the endoplasmic reticulum before and after receptors (TrkB) stimulation in living single colorectal cells (135). Other CARS advances include the use of greater than 2 lasers to access a broader range of Raman bands i.e. broadband CARS (143) probing a wavelength range of >3000 cm−1 with a resolution <10 cm−1 for assay of lipids in single murine pancreatic duct cells. Another compound microscope example is the integration of sum-frequency generation (SFG) imaging with CARS (136). In this instance SFG imaging of an exogenously added dye enabled selective visualization of the plasma membrane of cells coupled to CARS based-visualization of cellular lipids in single neurons.
3. FLUORESCENT-BASED TECHNOLOGIES
Fluorescence involves the absorption of a photon with excitation of a molecule from a ground electronic state into a higher energy electronic state (first or second excited singlet state). Energy is then dissipated by internal conversion with return to the lowest excited singlet state. Return to the ground state can occur by emission of a photon (fluorescence) now at a lower energy than that originally absorbed (144). Fluorescence-based measurements offer many advantages including high sensitivity, and proportionality of fluorescence intensity to fluorophore concentration. The unique spectral properties of a fluorophore confer specificity in fluorophore identification as well as enabling multiplexed assays utilizing many fluorophores. Additionally, a large number of fluorophores with a wide range of emission and excitation spectra are available. Fluorescence-based detection is compatible with a wide range of assay types often using simple, robust instrumentation. Fluorophores are readily conjugated to other molecules using a variety of chemistries. Major limitations of fluorescent-based measurements include fluorophore photodegradation or loss over time, environment (pH, solvent, etc) sensitivity of the photon emission energy, and potential for quenching (return to ground state without fluorophore emission). Since cellular lipids are not typically fluorescent, a strategy to incorporate a fluorophore into the assay i.e. a labelling strategy is also required. This section focuses on microscopy methods and micro-separation techniques that utilize fluorescence as a component of the lipid detection strategy with a focus on the quantification of the lipid species as well as lipid metabolic pathways in single cells.
3.1. Fluorescence Microscopy
Fluorescent microscopy offers significant advantages over other methods including the ease of accessibility and wide range of available microscopy methods in addition to standard fluorescence microscopy, for example, confocal, 2-photon, light-sheet and supper resolution microscopes. Fluorescence microscopy typically achieves high sensitivity to measure very low concentrations (nanomolar) of cellular lipids with a high signal to noise ratio and excellent spatial resolution (often sub-micron). Fluorescence labelling can be accomplished by using pre-labelled lipids added to the cells, staining by immunofluorescence methods, employing lipid-binding or reactive fluorophores, or applying other strategies. These labeling methods while highlighting the desired lipids may also increase background noise through nonspecific staining. Additionally the reagents are critically dependent on their design for specificity as to the molecules labelled (with either high or low specificity possible). The methods frequently require addition of exogenous reagents to cells so that the probes must be membrane permeant or the cells fixed to allow probe access to intracellular lipids. Another strategy is gene-engineering of cells to express protein constructs tagged with a fluorescent protein. In this section, we present three different fluorescent strategies to assay lipids in single cells: lipid partitioning reagents, lipid binding proteins, and functionalized lipids (Figure 3).
Figure 3.
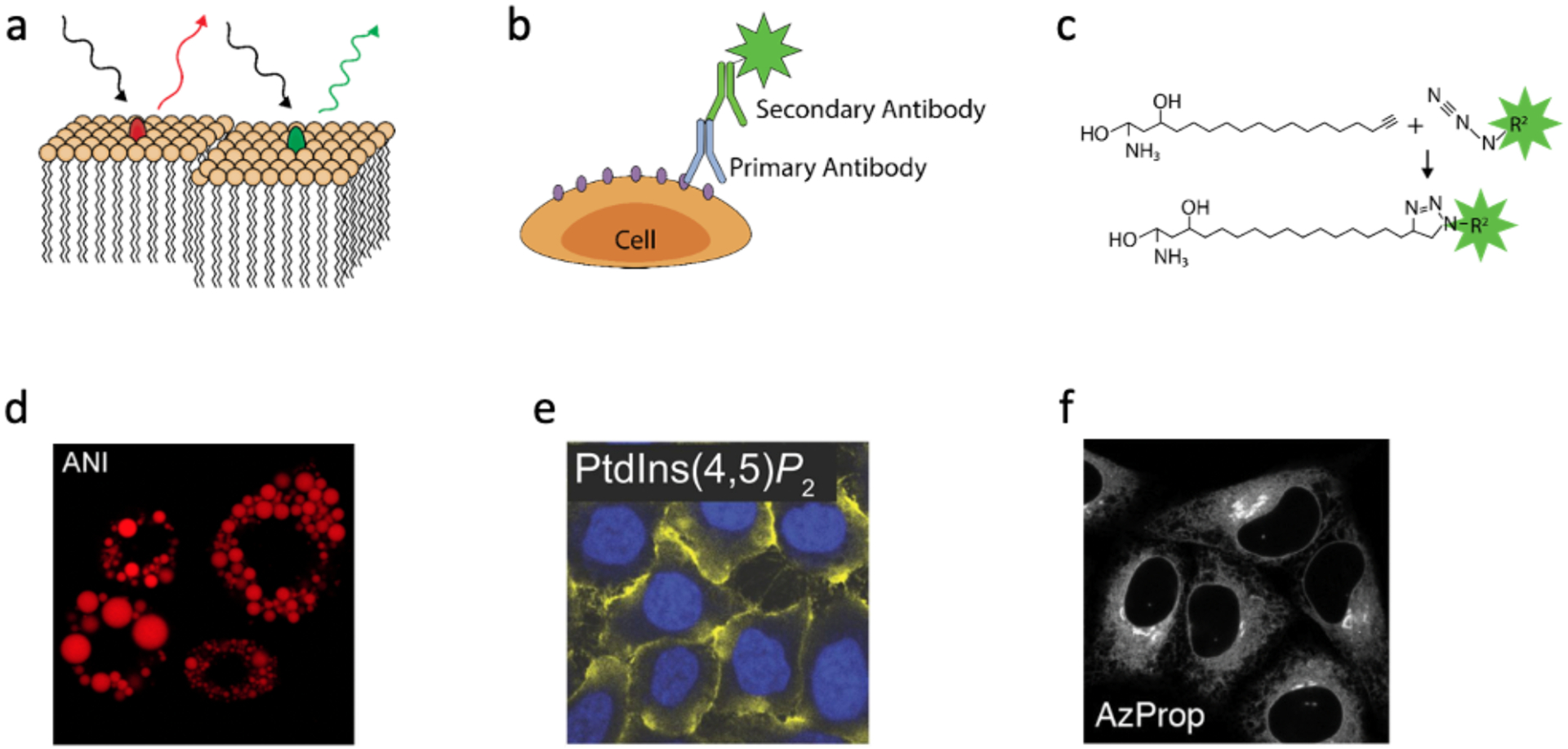
Fluorescence microscopy-based measurement of lipids. Schematics show the principles of (a) dyes that partition into lipids; (b) antibody-based lipid probes; (c) clickable lipid probes. In panel a, the red and green ovals represent fluorophores of different wavelengths present in different membrane regions. In panels b and c, the green star depicts a fluorescent molecule placed onto an antibody or added via a click reaction. (d) Confocal laser scanning image of lipid droplets in preadipocyte cells stained by a solvatochromic probe. (e) Immunofluorescent staining of PtdIns(4,5)P2 in HeLa cells. (f) The location of phospholipase D activity was highlighted by adding a clickable substrate to cells. The product formed was made visible using a “clickable” fluorophore. Panel c was adapted with permission from Gallion LA, Wang Y, Massaro A, Yao M, Petersen BV, Zhang Q, et al. Anal Chem. 2022;94(3):1594–600. Copyright 2022 American Chemical Society. Panel d was reprinted with permission from Wu CJ, Li XY, Zhu T, Zhao M, Song Z, Li S, et al. Anal Chem. 2022;94(9):3881–7. Copyright 2022 American Chemical Society. Panel e was reprinted with permission from Hammond GRV, Schiavo G, Irvine RF. Biochemical Journal. 2009 Aug 15;422(1):23–35. Panel f was reprinted with permission from Bumpus TW, Baskin JM. ACS Cent Sci. 2017 Oct 25;3(10):1070–7. Copyright 2017 American Chemical Society.
Cells possess a number of organelles such as LD’s, lipid bodies and other structures greatly enriched in lipid content relative the majority of the intracellular environment. This attribute has been capitalized on through the use of hydrophobic dyes such as BODIPY493/503, FD13 and Nile Red that when added to cells will partition into and mark these hydrophobic or other unique microenvironments (145–158). Due to the very high concentration of dye loaded into the compartments, the signal sensitivity is exceptional and spatial resolution of 100’s of nm is attainable for detailed imaging and tracking of these subcellular structures in living cells (152,159). The use of molecules with differing partitioning behaviors and fluorescence properties also enables multiple structures to be tracked over time with minimal photodamage. Properly designed probes can report multiple attributes of the microenvironment including membrane fluidity, local polarity, lipid composition, domain sizes, and temporal properties (145–158,160). Newer probes such as PIE1 possess high fluorescent quantum yield and photostability further improving upon measurement attributes including imaging speed for live cell imaging (147). Organelle-specific probes permit imaging of the polarity and lipid order in LDs, lysosomes, golgi, and mitochondria in the same cell (149). LD-specific probes with enhanced attributes have also been developed for standard or two-photon imaging with high spatial resolution but low cellular toxicity (152,158). Energy transfer between pairs of these dyes and/or other molecules can be used to investigate the degree of order or disorder in lipid domains as well as lipid packing in living cells (161,162). For example, these types of studies have revealed that living cell membranes are comprised of a mixture of both ordered and disordered lipid domains (163). While these probes have enabled breakthrough advances in the understanding of lipid-rich subcellular organelles and their structure, the lack of specificity of the probes in terms of labeling specific lipid species has limited their application.
Protein-based staining reagents such as antibodies and binding domains have long been a mainstay for protein identification or sensors in single cells and have been used with success for lipids (164–172). Antibodies have the advantage of being easy to apply reagents using standard protocols and are often commercially available. It can be quite challenging, however, to raise antibodies against lipids due to their hydrophobic character and ubiquitous presence throughout species (lessening their immunogenicity). Antibodies can be quite specific for the targeted molecule but a wellrecognized challenge with antibodies is that they often bind to molecules other than the intended target. Immunofluorescence-based probes have been used most often to assay phosphoinositide and sphingolipid location within single cells (171). An advantage of these reagents is the ability to perform multiplexed assays by also performing simultaneously immunofluorescence labeling of proteins, organelle markers and other fluorescent probes with complementary spectral properties. Gene engineering of cells to express lipid binding domains (subunits of a larger protein) linked to a fluorescence protein (enhanced green fluorescent protein or other) has also found success. For example, a peptide-based sensor was developed to detect anionic phospholipids in live HeLa cells which was compatible with two-photon excitation (167).
One strategy to improve the specificity of lipid probes is to label or functionalize the lipids themselves and then incorporate these labelled molecules into the cells to assay lipid localization, metabolism, transport, compartmentalization and other properties within single cells. A wide array of fluorescently labelled or otherwise functionalized (biotinylated, photoactivatable, clickable, etc) lipids are now commercially available (173–175). These probes have the advantage of a having a known molecular structure and often undergoing some but not all of the endogenous metabolic reactions of the native molecule. A challenge, however, can be the efficient loading of these molecules into cells given the poor water solubility of lipids although carriers such as polyamines and dispersants have been utilized to facilitate lipid delivery to cells (173–175). Exogenous loading can lead to mis-localization of these lipids; never-the-less these functionalized lipids have led to many novel insights into cellular lipid biology. An example is the use of BODIPY FL Ganglioside GM1 to label, characterize and track lipid rafts in the plasma membrane of cells (176). Fluorescent sphingomyelin analogs were also developed to investigate the formation and function of raft domains (175). A mitochondrial-specific photoactivatable sphingosine has been used to show the rapid conversion of sphingosine into sphingosine-1 phosphate in HeLa cells (174). An obstacle present for all of the fluorescent lipids is the presence of the bulky fluorophore that can block binding to intracellular proteins including metabolic enzymes. A significant innovation addressing these impediments is the development of click lipids (177–182). Instead of a bulky fluorescent moiety, these reagents possess either a small azido group or a terminal alkyne making the click lipids structurally very close to the endogenous counterpart (180,183). The fluorophore used for detection is then added at experimental completion enabling the click lipid to participate in normal cellular physiology. For example, an alkyne-oleate probe was used to track fatty acid metabolism in cells and thern clicked with an azide fluorophore (184). Azidoalcohols can be used in the phospholipase D-catalyzed (PLD) transphosphatidylation reaction to produce azidolipids that can then be fluorescently labelled to track the subcellular location of phospholipase D activity and phosphatidic acid (PA) production (182). A related activity-based imaging strategy employs cyclooctene-containing primary alcohols to act as a nucleophile in the phospholipase D reaction with phosphatidylcholine providing near real-time imaging of PA production in living cells (181,182).
3.2. Fluorescent-Based Micro-Separation Methods
Many separation techniques have been applied to assay single cells, including thin layer chromatography (TLC), capillary electrophoresis (CE), and even high performance liquid chromatography (HPLC) (4,185–188). To date, CE is the most prevalent method used for lipid assays in single cells with TLC as an emerging technique. With CE, analytes are separated in a solvent (typically aqueous) under an electric field. The dominant parameter for separation is the charged to mass of the analytes although other separation strategies are possible such as micellar electrokinetic chromatography which separates analytes based on partitioning into micelles while under an electric field (185). In contrast TLC separation relies on the capillary forces of an organic solvent moving through a matrix (typically silica based) and separates analytes based on their differential interactions with the matrix and organic solvent (189). Separation techniques have common advantages in lipid analysis. The methods typically have low background noise since the analyte is separated from other cellular material at the time of detection. The separation step also provides some degree of specificity since different analytes will possess characteristic migration times or arrival times at the detection zone. CE provides exceptionally high resolution (peak capacities of 50–100), separation efficiency (>1 million theoretical plates) and sensitivity (≤10−21 moles or ≤1 nM in a 1 pL cell) (185). Some CE systems have even achieved limits of detection of 10−23 moles (190). Use of multiple fluorophores with different spectral properties further increases the number of analytes that can be simultaneously measured in CE. As long as the analytes remain fluorescent, CE can be used to track downstream metabolites as they form from a reporter molecule (4,187). Single cell lipid assays by CE can also be fully automated and integrated with microscopy methods (185). Microfluidic-based CE assays enable further integration and miniaturization of complete workflows. TLC, on the other hand, offers a major advantage of low-cost and ease of accessibility but possesses low resolution and separation (189). However, both CE and TLC applied to single cell lipid assays have drawbacks. It is quite difficult to measure endogenous lipids in cell samples using CE and fluorescence since lipids are not typically fluorescent and so would need to be derivatized prior to detection. For this reason, both methods utilize a pre-labelled substrate that is loaded into the cells and this loading step can be facile or quite challenging depending on the properties of the molecule. Both techniques are cell destructive and consume the cellular contents making it difficult to multiplex CE or TLC based single-cell lipid assays with additional downstream assays. Additionally this means that for the most part the methods do not possess a subcellular spatial resolution. Finally most of the demonstrated technologies for singlecell lipid assay are moderate in throughput (≤5 cells/min) at best (4,185).
For CE, the ability to measure multiple lipid metabolites from a single cell has enabled fundamental investigations into cell-to-cell heterogeneity in signaling behavior as well as responses to drugs (Figure 4a) (4,185,186). Sphingolipids, glycosphingolipids, phosphoinositides as well as ganglioside metabolic pathways have been assessed in single cells by CE (185,186,191,192) For example when cells were loaded with sphingosine fluorescein and then assayed by CE, greater than 5 metabolic products can be identified in cells with sphingosine-1-phospate fluorescein and hexadecinal fluorescein comprising the majority of products formed. Most interesting is that the single cells readily clustered into two groups, one group cells with high and one with low sphingosine kinase (SK) activity (186). After loading with a fluorescent lipid, single cells can be assayed while living as in the prior example or fixed and stored prior to assay since the lipids remain partitioned in the fixed cells (187,192,193). The bulky fluorophore placed on the lipids can alter lipid location and metabolism in cells, and recent studies have used click lipids for assay of single-cell lipid metabolism. In this strategy termed “fix and click”, cells are loaded with a clickable lipid and after incubation for varying times with or without agonists or inhibitors, the cells are fixed and the click reaction performed to create fluorescent analytes from the intracellular molecules possessing the clickable moiety (4). This strategy enables the tracking of signal pathway activity in single cells by CE using a near native lipid substrate loaded into cells with greater measurement fidelity.
Figure 4.
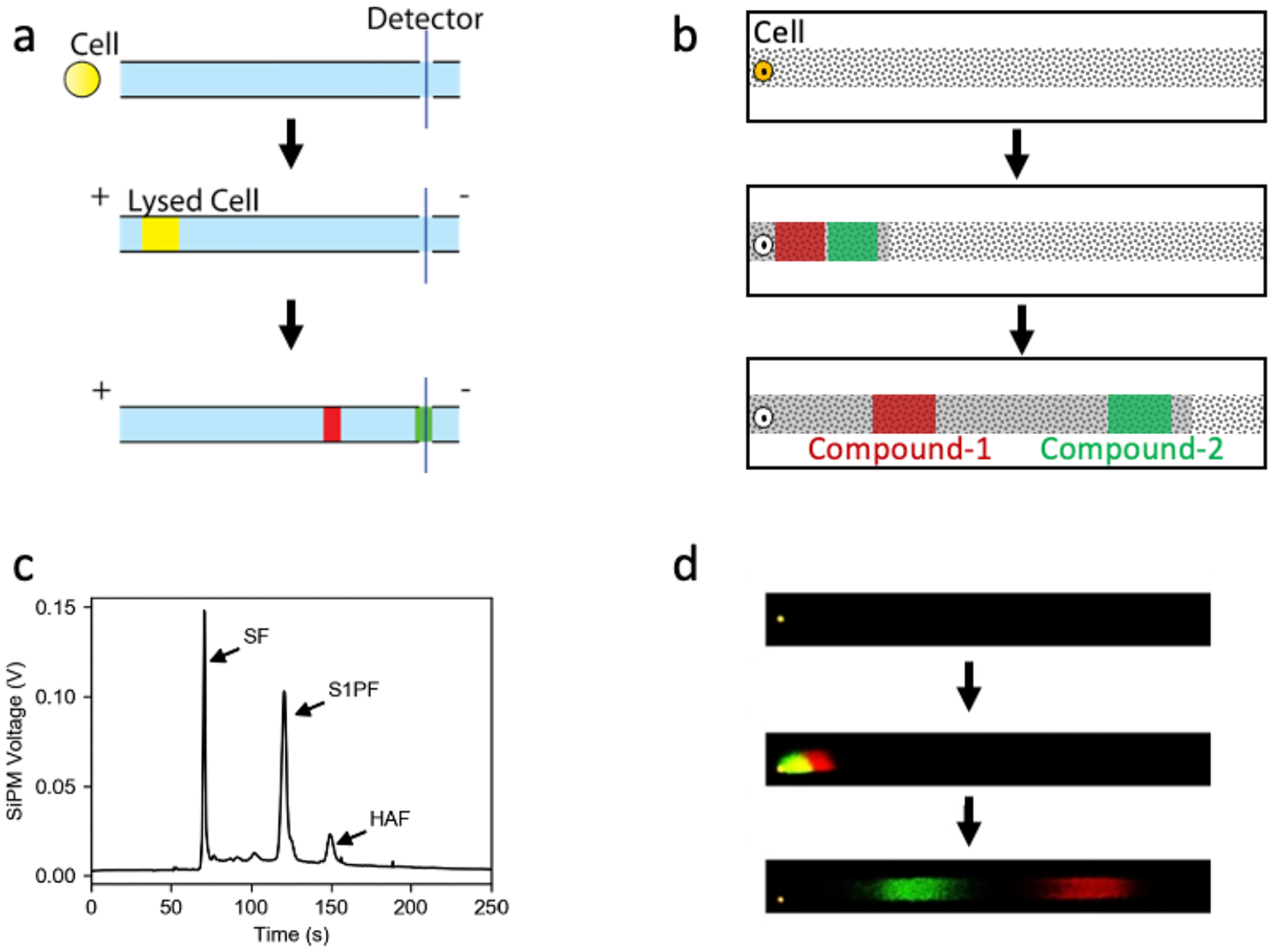
Fluorescent-based separation techniques for single-cell lipid assays. Schematics showing the separation of cellular analytes using CE (a) or pTLC (b). (c) Electropherogram of the CE separation from a single leukemia cell loaded with sphingosine fluorescein (SF). Sphingosine kinase activity was measured by quantifying the conversion of reporter SF to downstream metabolites, sphingosine-1-phosphate fluorescein (S1PF) or hexadecenoic acid fluorescein (HAF). (d) A single leukemic cell was loaded with 3,3’-dioctadecyloxacarbocyanine perchlorate (DiO) and 1,1’-dioctadecyl-3,3,3’,3’-tetramethylindodicarbocyanine, 4-chlorobenzenesulfonate salt (DiD) and the cell assayed by pTLC. Panel b was reprinted with permission from [Please see letter to the editor - permission will be obtained]. Panel c was reprinted with permission from Petersen BV, Gallion L, Allbritton NL. Anal Chem. 2020;92(20):13683–7. Copyright 2020 American Chemical Society. Panel d was preprinted with permission from [Please see letter to the editor - permission will be obtained].
To overcome the throughput and cost limitations of CE, several groups have worked to improve the rates of both serial and parallel throughput to increase the number of cells that can be assayed, as well as to automate and diminish instrumental costs. For example, a system with a hybrid CE-microfluidic device (Figure 4b) enabled 100 cells placed on an array to be sequentially and rapidly assayed with a sensitivity of 10−21 moles and throughput of 3.5 cells/min (186,191). A parallel CE system with an array of five capillaries coupled to an array of microwells demonstrated the simultaneous separation of ceramides and gangliosides from single cells (194). This system was also adapted to incorporate a multi-colored fluorescence detection system to further expand the numbers of detectable analytes and assay simultaneously glycolipid catabolism and anabolism from a single cell with detection limits of 10−22 to 10−23 moles (188,192). Other innovations developed low-cost ($130), yet highly sensitive detection systems using silicon photomultipliers (SiPMs) (185).
TLC has in the past been viewed as a low sensitivity method requiring large sample volumes (10−7 L) and so not suitable for assay of a 1 pL volume cell. This is due in part to the plate-based format (cms in length and width with a depth of ~100–250 mm) for TLC which enables diffusional spreading (or dilution) of analytes in three dimensions. Recent innovations in the microfabrication of arrays of miniature TLC separation lanes or pTLC has created opportunities for assay of lipids in single cells (Reference to be added later). Microchannels (width ~50 μm and depth ~13 μm) were fabricated on a surface and then filled with a monolithic microporous silica gel to confine the movement of analytes along the microchannels limiting diffusion to one dimension. These devices enable the TLC-based separation of pL to nL volume samples with excellent fluorescence detection limits. When single cells loaded with fluorescent lipids e.g. a fluorescent sphingosine were spotted at the entry way of the microchannels and TLC initiated, the lipid and its downstream metabolites (ceramide and sphingosine-1 phosphate) were detectable. While the separation resolution is low (~2–4) compared to CE, the advantages of the pTLC platform are its operational simplicity, minimal equipment needs and robustness, making the technology suitable for many applications including single cell assays of lipases and lipid kinases.
Conclusion
Single cell omics is progressing rapidly with impressive technological developments in the past decade in proteomics, genomics, and transcriptomics. These high throughput technologies have revealed fundamentally new insights into cell physiology. Given the importance of lipids in both health and disease, it is increasingly critical to understand the full range of lipids within a single cell as has now become possible with other cellular constituents. These attributes will include not only lipid chemical identity and spatial and temporal distribution, but also the flow of signals through the various lipid pathways. This review covers the impressive advances that have been made in the measurement of lipids from a single cell including innovations in mass spectrometry, Raman microscopy, fluorescence microscopy and micro-separation methods. These methods are in general complementary in their properties with some providing high spatial or temporal resolution, excellent specificity, precise quantification, impressive sensitivity, or multiplexed lipid measurements yet none of the methods possess all or even several of these characteristics. This is largely due to the challenges in lipid measurement (hydrophobicity as well as chemical diversity) combined with the extremely small size of a typical mammalian cell and wide concentration range of lipid species spanning within a cell. However these attributes create enormous opportunities for engineers, biologists and chemists to develop improved technologies propelling the field of single cell lipidomics to new heights and rivaling the assays performed in the other “omics” fields.
Acknowledgements
This work was supported by the National Institutes of Health under Award CA233811 and CA224763. The authors thank Gregery Woss for assistance with illustrations.
Abbreviations
- MS
mass spectrometry
- MALDI-MS
matrix assisted laser desorption ionization-imaging mass spectrometry
- ESI-MS
electrospray ionization mass spectroscopy
- SI-MS
secondary ion mass spectrometry
- SRS
stimulated Raman scattering
- CARS
coherent anti-Stokes Raman scattering
- CE
capillary electrophoresis
- TLC
thin layer chromatography
- LD
lipid droplet
Footnotes
Disclosure Statement
N.L.A is an inventor of pTLC (PCT/US2022/019103). N.L.A and M.Y disclose a financial interest in Piccolo Biosystems, Inc. M.V. discloses no conflicts of interest.
References:
- 1.Cockcroft S Mammalian lipids: structure, synthesis and function. Essays in Biochemistry. 2021; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fahy E, Subramaniam S, Brown HA, Glass CK, Merrill AH, Murphy RC, et al. A Comprehensive Classification System for Lipids 1. In: Handbook of Biochemistry and Molecular Biology. 5th ed. CRC Press; 2018. [Google Scholar]
- 3.Olzmann JA, Carvalho P. Dynamics and functions of lipid droplets. Nature reviews Molecular cell biology. 2019;20(3):137–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gallion LA, Wang Y, Massaro A, Yao M, Petersen BV, Zhang Q, et al. “Fix and Click” for Assay of Sphingolipid Signaling in Single Primary Human Intestinal Epithelial Cells. Anal Chem. 2022. Jan 25;94(3):1594–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hussain G, Anwar H, Rasul A, Imran A, Qasim M, Zafar S, et al. Lipids as biomarkers of brain disorders. Critical reviews in food science and nutrition. 2020;60(3):351–74. [DOI] [PubMed] [Google Scholar]
- 6.Dumas F, Haanappel E. Lipids in infectious diseases–The case of AIDS and tuberculosis. Biochimica Et Biophysica Acta (BBA)-Biomembranes. 2017;1859(9):1636–47. [DOI] [PubMed] [Google Scholar]
- 7.Liu Q, Luo Q, Halim A, Song G. Targeting lipid metabolism of cancer cells: a promising therapeutic strategy for cancer. Cancer letters. 2017;401:39–45. [DOI] [PubMed] [Google Scholar]
- 8.Kohno S, Keenan AL, Ntambi JM, Miyazaki M. Lipidomic insight into cardiovascular diseases. Biochemical and biophysical research communications. 2018;504(3):590–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Perrotti F, Rosa C, Cicalini I, Sacchetta P, Del Boccio P, Genovesi D, et al. Advances in lipidomics for cancer biomarkers discovery. International journal of molecular sciences. 2016;17(12):1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Snaebjornsson MT, Janaki-Raman S, Schulze A. Greasing the wheels of the cancer machine: the role of lipid metabolism in cancer. Cell metabolism. 2020;31(1):62–76. [DOI] [PubMed] [Google Scholar]
- 11.Fhu CW, Ali A. Fatty acid synthase: an emerging target in cancer. Molecules. 2020;25(17):3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun W, Li P, Cai J, Ma J, Zhang X, Song Y, et al. Lipid Metabolism: Immune Regulation and Therapeutic Prospectives in Systemic Lupus Erythematosus. Front Immunol. 2022. Mar 18;13:860586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bernardi S, Marcuzzi A, Piscianz E, Tommasini A, Fabris B. The Complex Interplay between Lipids, Immune System and Interleukins in Cardio-Metabolic Diseases. IJMS. 2018. Dec 14;19(12):4058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu Q, Ge W, Wang T, Lan J, Martínez-Jarquín S, Wolfrum C, et al. High-Throughput Single-Cell Mass Spectrometry Reveals Abnormal Lipid Metabolism in Pancreatic Ductal Adenocarcinoma. Angewandte Chemie. 2021;133(46):24739–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rajbhandari P, Arneson D, Hart SK, Ahn IS, Diamante G, Santos LC, et al. Single cell analysis reveals immune cell–adipocyte crosstalk regulating the transcription of thermogenic adipocytes. eLife. 2019. Oct 23;8:e49501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sethi S, Brietzke E. Recent advances in lipidomics: Analytical and clinical perspectives. Prostaglandins & other lipid mediators. 2017;128:8–16. [DOI] [PubMed] [Google Scholar]
- 17.Feider CL, Krieger A, DeHoog RJ, Eberlin LS. Ambient ionization mass spectrometry: recent developments and applications. Analytical chemistry. 2019;91(7):4266–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu R, Pan N, Zhu Y, Yang Z. T-probe: an integrated microscale device for online in situ single cell analysis and metabolic profiling using mass spectrometry. Analytical chemistry. 2018;90(18):11078–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duncan KD, Fyrestam J, Lanekoff I. Advances in mass spectrometry based single-cell metabolomics. Analyst. 2019;144(3):782–93. [DOI] [PubMed] [Google Scholar]
- 20.Huang L, Fang M, Cupp-Sutton KA, Wang Z, Smith K, Wu S. Spray-capillary-based capillary electrophoresis mass spectrometry for metabolite analysis in single cells. Analytical Chemistry. 2021;93(10):4479–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Van Acker T, Buckle T, Van Malderen SJ, van Willigen DM, van Unen V, van Leeuwen FW, et al. High-resolution imaging and single-cell analysis via laser ablation-inductively coupled plasma-mass spectrometry for the determination of membranous receptor expression levels in breast cancer cell lines using receptor-specific hybrid tracers. Analytica chimica acta. 2019;1074:43–53. [DOI] [PubMed] [Google Scholar]
- 22.Vaysse PM, Heeren RM, Porta T, Balluff B. Mass spectrometry imaging for clinical research–latest developments, applications, and current limitations. Analyst. 2017;142(15):2690–712. [DOI] [PubMed] [Google Scholar]
- 23.Capolupo L, Khven I, Mazzeo L, Glousker G, Russo F, Montoya JP, et al. Sphingolipid control of fibroblast heterogeneity revealed by single-cell lipidomics. bioRxiv. 2021; [Google Scholar]
- 24.Yin L, Zhang Z, Liu Y, Gao Y, Gu J. Recent advances in single-cell analysis by mass spectrometry. Analyst. 2019;144(3):824–45. [DOI] [PubMed] [Google Scholar]
- 25.Dueñas ME, Essner JJ, Lee YJ. 3D MALDI mass spectrometry imaging of a single cell: spatial mapping of lipids in the embryonic development of zebrafish. Scientific reports. 2017;7(1):1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leopold J, Popkova Y, Engel KM, Schiller J. Recent developments of useful MALDI matrices for the mass spectrometric characterization of lipids. Biomolecules. 2018;8(4):173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marsico TV, de Sousa Sales JN, Ferreira CR, Sudano MJ, Viana JHM, de Almeida Camargo LS, et al. Characteristic MALDI-MS lipid profiles of Gir, Holstein and crossbred (Gir x Holstein) oocytes recovered by ovum pick-up. Livestock Science. 2021;243:104380. [Google Scholar]
- 28.Agüi-Gonzalez P, Jähne S, Phan NT. SIMS imaging in neurobiology and cell biology. Journal of Analytical Atomic Spectrometry. 2019;34(7):1355–68. [Google Scholar]
- 29.Schnackenberg LK, Thorn DA, Barnette D, Jones EE. MALDI imaging mass spectrometry: an emerging tool in neurology. Metab Brain Dis. 2022. Jan 1;37(1):105–21. [DOI] [PubMed] [Google Scholar]
- 30.Niehaus M, Soltwisch J, Belov ME, Dreisewerd K. Transmission-mode MALDI-2 mass spectrometry imaging of cells and tissues at subcellular resolution. Nature methods. 2019;16(9):925–31. [DOI] [PubMed] [Google Scholar]
- 31.Martín-Saiz L, Mosteiro L, Solano-Iturri JD, Rueda Y, Martín-Allende J, Imaz I, et al. High-Resolution Human Kidney Molecular Histology by Imaging Mass Spectrometry of Lipids. Analytical Chemistry. 2021; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xie W, Gao D, Jin F, Jiang Y, Liu H. Study of phospholipids in single cells using an integrated microfluidic device combined with matrix-assisted laser desorption/ionization mass spectrometry. Analytical chemistry. 2015;87(14):7052–9. [DOI] [PubMed] [Google Scholar]
- 33.Yang T, Gao D, Jin F, Jiang Y, Liu H. Surface-printed microdot array chips coupled with matrix-assisted laser desorption/ionization mass spectrometry for high-throughput singlecell patterning and phospholipid analysis: Surface-printed microdot array chips coupled with MALDI-TOF MS. Rapid Commun Mass Spectrom. 2016. Aug;30:73–9. [DOI] [PubMed] [Google Scholar]
- 34.Neumann EK, Ellis JF, Triplett AE, Rubakhin SS, Sweedler JV. Lipid Analysis of 30 000 Individual Rodent Cerebellar Cells Using High-Resolution Mass Spectrometry. Anal Chem. 2019. Jun 18;91(12):7871–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Neumann EK, Comi TJ, Rubakhin SS, Sweedler JV. Lipid Heterogeneity between Astrocytes and Neurons Revealed by Single-Cell MALDI-MS Combined with Immunocytochemical Classification. Angewandte Chemie. 2019;131(18):5971–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang B, Patterson NH, Tsui T, Caprioli RM, Norris JL. Single-cell mass spectrometry reveals changes in lipid and metabolite expression in RAW 264.7 cells upon lipopolysaccharide stimulation. Journal of The American Society for Mass Spectrometry. 2018;29(5):1012–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Do TD, Ellis JF, Neumann EK, Comi TJ, Tillmaand EG, Lenhart AE, et al. Optically guided single cell mass spectrometry of rat dorsal root ganglia to profile lipids, peptides and proteins. Chemphyschem: a European journal of chemical physics and physical chemistry. 2018;19(10):1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zenobi R Single-cell metabolomics: analytical and biological perspectives. Science. 2013;342(6163). [DOI] [PubMed] [Google Scholar]
- 39.Pan N, Rao W, Kothapalli NR, Liu R, Burgett AW, Yang Z. The single-probe: a miniaturized multifunctional device for single cell mass spectrometry analysis. Analytical chemistry. 2014;86(19):9376–80. [DOI] [PubMed] [Google Scholar]
- 40.Li Z, Cheng S, Lin Q, Cao W, Yang J, Zhang M, et al. Single-cell lipidomics with high structural specificity by mass spectrometry. Nature communications. 2021;12(1):1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen F, Lin L, Zhang J, He Z, Uchiyama K, Lin JM. Single-cell analysis using drop-on-demand inkjet printing and probe electrospray ionization mass spectrometry. Analytical chemistry. 2016;88(8):4354–60. [DOI] [PubMed] [Google Scholar]
- 42.Zhu Y, Wang W, Yang Z. Combining Mass Spectrometry with Paternò–Büchi Reaction to Determine Double-Bond Positions in Lipids at the Single-Cell Level. Analytical chemistry. 2020;92(16):11380–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cahill JF, Riba J, Kertesz V. Rapid, untargeted chemical profiling of single cells in their native environment. Analytical chemistry. 2019;91(9):6118–26. [DOI] [PubMed] [Google Scholar]
- 44.Pedro L, Rudewicz PJ. Analysis of Live Single Cells by Confocal Microscopy and High-Resolution Mass Spectrometry to Study Drug Uptake, Metabolism, and Drug-Induced Phospholipidosis. Analytical Chemistry. 2020;92(24):16005–15. [DOI] [PubMed] [Google Scholar]
- 45.Hiyama E, Ali A, Amer S, Harada T, Shimamoto K, Furushima R, et al. Direct lipidometabolomics of single floating cells for analysis of circulating tumor cells by live singlecell mass spectrometry. Analytical Sciences. 2015;31(12):1215–7. [DOI] [PubMed] [Google Scholar]
- 46.Xu ST, Yang C, Yan XP. Nanothorn Filter-Facilitated Online Cell Lysis for Rapid and Deep Intracellular Profiling by Single-Cell Mass Spectrometry. Anal Chem. 2021. Nov 30;93(47):15677–86. [DOI] [PubMed] [Google Scholar]
- 47.Merrill CB, Basit A, Armirotti A, Jia Y, Gall CM, Lynch G, et al. Patch clamp-assisted single neuron lipidomics. Scientific reports. 2017;7(1):1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang R, Zhao H, Zhang X, Zhao X, Song Z, Ouyang J. Metabolic discrimination of breast cancer subtypes at the single-cell level by multiple microextraction coupled with mass spectrometry. Analytical chemistry. 2019;91(5):3667–74. [DOI] [PubMed] [Google Scholar]
- 49.Zhao Y, Chen Z, Wu Y, Tsukui T, Ma X, Zhang X, et al. Separating and profiling phosphatidylcholines and triglycerides from single cellular lipid droplet by in-tip solvent microextraction mass spectrometry. Analytical chemistry. 2019;91(7):4466–71. [DOI] [PubMed] [Google Scholar]
- 50.Sun M, Yang Z. Metabolomic studies of live single cancer stem cells using mass spectrometry. Analytical chemistry. 2018;91(3):2384–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bergman HM, Lanekoff I. Profiling and quantifying endogenous molecules in single cells using nano-DESI MS. Analyst. 2017;142(19):3639–47. [DOI] [PubMed] [Google Scholar]
- 52.Xi Y, Tu A, Muddiman DC. Lipidomic profiling of single mammalian cells by infrared matrix-assisted laser desorption electrospray ionization (IR-MALDESI). Analytical and Bioanalytical Chemistry. 2020;412(29):8211–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang L, Xu T, Zhang J, Wong SCC, Ritchie M, Hou HW, et al. Single Cell Metabolite Detection Using Inertial Microfluidics-Assisted Ion Mobility Mass Spectrometry. Analytical chemistry. 2021;93(30):10462–8. [DOI] [PubMed] [Google Scholar]
- 54.Yao H, Zhao H, Zhao X, Pan X, Feng J, Xu F, et al. Label-free Mass Cytometry for Unveiling Cellular Metabolic Heterogeneity. Anal Chem. 2019. Aug 6;91(15):9777–83. [DOI] [PubMed] [Google Scholar]
- 55.Liu A, Zhang H, Ding J, Kou W, Yan F, Huang K, et al. Enrichment of phospholipids using magnetic Fe3O4/TiO2 nanoparticles for quantitative detection at single cell levels by electrospray ionization mass spectrometry. Talanta. 2020;212:120769. [DOI] [PubMed] [Google Scholar]
- 56.Popczun NJ, Breuer L, Wucher A, Winograd N. On the SIMS ionization probability of organic molecules. Journal of The American Society for Mass Spectrometry. 2017;28(6):1182–91. [DOI] [PubMed] [Google Scholar]
- 57.Sheng L, Cai L, Wang J, Li Z, Mo Y, Zhang S, et al. Simultaneous imaging of newly synthesized proteins and lipids in single cell by TOF-SIMS. International Journal of Mass Spectrometry. 2017;421:238–44. [Google Scholar]
- 58.Hua X, Li HW, Long YT. Investigation of silver nanoparticle induced lipids changes on a single cell surface by time-of-flight secondary ion mass spectrometry. Analytical chemistry. 2018;90(2):1072–6. [DOI] [PubMed] [Google Scholar]
- 59.Chang-Fang S, Yao Z, Kui WU, Fei-Fei JIA, Qun LUO, Zhe LIU, et al. Correlated Secondary Ion Mass Spectrometry-Laser Scanning Confocal Microscopy Imaging for Single Cell-Principles and Applications. Chinese Journal of Analytical Chemistry. 2018;46(7):1005–16. [Google Scholar]
- 60.Waki M, Ide Y, Ishizaki I, Nagata Y, Masaki N, Sugiyama E, et al. Single-cell time-offlight secondary ion mass spectrometry reveals that human breast cancer stem cells have significantly lower content of palmitoleic acid compared to their counterpart non-stem cancer cells. Biochimie. 2014;107:73–7. [DOI] [PubMed] [Google Scholar]
- 61.Milijaš Jotić M, Panevska A, Iacovache I, Kostanjšek R, Mravinec M, Skočaj M, et al. Dissecting Out the Molecular Mechanism of Insecticidal Activity of Ostreolysin A6/Pleurotolysin B Complexes on Western Corn Rootworm. Toxins 2021, 13, 455. 2021; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Do TD, Comi TJ, Dunham SJ, Rubakhin SS, Sweedler JV. Single cell profiling using ionic liquid matrix-enhanced secondary ion mass spectrometry for neuronal cell type differentiation. Analytical chemistry. 2017;89(5):3078–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.He C, Fong LG, Young SG, Jiang H. NanoSIMS imaging: an approach for visualizing and quantifying lipids in cells and tissues. Journal of Investigative Medicine. 2017;65(3):669–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang C, Zhang D, Cheng JX. Coherent Raman Scattering Microscopy in Biology and Medicine. Annu Rev Biomed Eng. 2015. Dec 7;17(1):415–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tipping WJ, Lee M, Serrels A, Brunton VG, Hulme AN. Stimulated Raman scattering microscopy: an emerging tool for drug discovery. Chem Soc Rev. 2016;45(8):2075–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gomes da Costa S, Richter A, Schmidt U, Breuninger S, Hollricher O. Confocal Raman microscopy in life sciences. Morphologie. 2019. Mar;103(341):11–6. [DOI] [PubMed] [Google Scholar]
- 67.Müller M, Zumbusch A. Coherent anti-Stokes Raman Scattering Microscopy. ChemPhysChem. 2007. Oct 22;8(15):2156–70. [DOI] [PubMed] [Google Scholar]
- 68.Eberhardt K, Stiebing C, Matthäus C, Schmitt M, Popp J. Advantages and limitations of Raman spectroscopy for molecular diagnostics: an update. Expert Review of Molecular Diagnostics. 2015. Jun 3;15(6):773–87. [DOI] [PubMed] [Google Scholar]
- 69.Yue S, Cheng JX. Deciphering single cell metabolism by coherent Raman scattering microscopy. Current Opinion in Chemical Biology. 2016. Aug;33:46–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yu Y, Ramachandran PV, Wang MC. Shedding new light on lipid functions with CARS and SRS microscopy. Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids. 2014. Aug;1841(8):1120–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li S, Li Y, Yi R, Liu L, Qu J. Coherent Anti-Stokes Raman Scattering Microscopy and Its Applications. Front Phys. 2020. Dec 17;8:598420. [Google Scholar]
- 72.Kang JW, Nguyen FT, Lue N. Temporal Imaging of Live Cells by High-Speed Confocal Raman Microscopy. Materials. 2021. Jul 3;14(13):3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Xu Y, Hou X, Zhu Q, Mao S, Ren J, Lin J, et al. Phenotype Identification of HeLa Cells Knockout CDK6 Gene Based on Label-Free Raman Imaging. Anal Chem. 2022. Jun 28;94(25):8890–8. [DOI] [PubMed] [Google Scholar]
- 74.Surmacki JM, Quiros-Gonzalez I, Bohndiek SE. Evaluation of Label-Free Confocal Raman Microspectroscopy for Monitoring Oxidative Stress In Vitro in Live Human Cancer Cells. Antioxidants. 2022. Mar 17;11(3):573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Roman M, Wrobel TP, Panek A, Paluszkiewicz C, Kwiatek WM. Lipid droplets in prostate cancer cells and effect of irradiation studied by Raman microspectroscopy. Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids. 2020. Sep;1865(9):158753. [DOI] [PubMed] [Google Scholar]
- 76.Uematsu M, Shimizu T. Raman microscopy-based quantification of the physical properties of intracellular lipids. Commun Biol. 2021. Dec;4(1):1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Beton K, Wysocki P, Brozek-Pluska B. Mevastatin in colon cancer by spectroscopic and microscopic methods – Raman imaging and AFM studies. Spectrochimica Acta Part A: Molecular and Biomolecular Spectroscopy. 2022. Apr;270:120726. [DOI] [PubMed] [Google Scholar]
- 78.Feuerer N, Marzi J, Brauchle EM, Carvajal Berrio DA, Billing F, Weiss M, et al. Lipidome profiling with Raman microspectroscopy identifies macrophage response to surface topographies of implant materials. Proc Natl Acad Sci USA. 2021. Dec 28;118(52):e2113694118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shou J, Oda R, Hu F, Karasawa K, Nuriya M, Yasui M, et al. Super-multiplex imaging of cellular dynamics and heterogeneity by integrated stimulated Raman and fluorescence microscopy. iScience. 2021. Aug;24(8):102832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lita A, Kuzmin AN, Pliss A, Baev A, Rzhevskii A, Gilbert MR, et al. Toward Single-Organelle Lipidomics in Live Cells. Anal Chem. 2019. Sep 3;91(17):11380–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wei L, Chen Z, Shi L, Long R, Anzalone AV, Zhang L, et al. Super-multiplex vibrational imaging. Nature. 2017. Apr;544(7651):465–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hu F, Zeng C, Long R, Miao Y, Wei L, Xu Q, et al. Supermultiplexed optical imaging and barcoding with engineered polyynes. Nat Methods. 2018. Mar;15(3):194–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wei L, Hu F, Shen Y, Chen Z, Yu Y, Lin CC, et al. Live-cell imaging of alkyne-tagged small biomolecules by stimulated Raman scattering. Nat Methods. 2014. Apr;11(4):410–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lima C, Muhamadali H, Goodacre R. The Role of Raman Spectroscopy Within Quantitative Metabolomics. Annual Rev Anal Chem. 2021. Jul 27;14(1):323–45. [DOI] [PubMed] [Google Scholar]
- 85.Figueroa B, Fu W, Nguyen T, Shin K, Manifold B, Wise F, et al. Broadband hyperspectral stimulated Raman scattering microscopy with a parabolic fiber amplifier source. Biomed Opt Express. 2018. Dec 1;9(12):6116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Boorman D, Pope I, Masia F, Langbein W, Hood S, Borri P, et al. Hyperspectral CARS microscopy and quantitative unsupervised analysis of deuterated and non-deuterated fatty acid storage in human cells. J Chem Phys. 2021. Dec 14;155(22):224202. [DOI] [PubMed] [Google Scholar]
- 87.Nahmad-Rohen A, Regan D, Masia F, McPhee C, Pope I, Langbein W, et al. Quantitative Label-Free Imaging of Lipid Domains in Single Bilayers by Hyperspectral Coherent Raman Scattering. Anal Chem. 2020. Nov 3;92(21):14657–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Masia F, Pope I, Watson P, Langbein W, Borri P. Bessel-Beam Hyperspectral CARS Microscopy with Sparse Sampling: Enabling High-Content High-Throughput Label-Free Quantitative Chemical Imaging. Anal Chem. 2018. Mar 20;90(6):3775–85. [DOI] [PubMed] [Google Scholar]
- 89.Karuna A, Masia F, Wiltshire M, Errington R, Borri P, Langbein W. Label-Free Volumetric Quantitative Imaging of the Human Somatic Cell Division by Hyperspectral Coherent Anti-Stokes Raman Scattering. Anal Chem. 2019. Feb 19;91(4):2813–21. [DOI] [PubMed] [Google Scholar]
- 90.Syed A, Smith EA. Raman Imaging in Cell Membranes, Lipid-Rich Organelles, and Lipid Bilayers. Annual Rev Anal Chem. 2017. Jun 12;10(1):271–91. [DOI] [PubMed] [Google Scholar]
- 91.Manifold B, Fu D. Quantitative Stimulated Raman Scattering Microscopy: Promises and Pitfalls. Annual Rev Anal Chem. 2022. Jun 13;15(1):269–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Szafraniec E, Kus E, Wislocka A, Kukla B, Sierka E, Untereiner V, et al. Raman spectroscopy–based insight into lipid droplets presence and contents in liver sinusoidal endothelial cells and hepatocytes. J Biophotonics. 2019. Apr;12(4):e201800290. [DOI] [PubMed] [Google Scholar]
- 93.Denbigh JL, Perez-Guaita D, Vernooij RR, Tobin MJ, Bambery KR, Xu Y, et al. Probing the action of a novel anti-leukaemic drug therapy at the single cell level using modern vibrational spectroscopy techniques. Sci Rep. 2017. Dec;7(1):2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kochan K, Maslak E, Krafft C, Kostogrys R, Chlopicki S, Baranska M. Raman spectroscopy analysis of lipid droplets content, distribution and saturation level in Non-Alcoholic Fatty Liver Disease in mice. J Biophoton. 2015. Jul;8(7):597–609. [DOI] [PubMed] [Google Scholar]
- 95.Tott S, Grosicki M, Glowacz J, Mohaissen T, Wojnar-Lason K, Chlopicki S, et al. Raman imaging-based phenotyping of murine primary endothelial cells to identify diseaseassociated biochemical alterations. Biochimica et Biophysica Acta (BBA) - Molecular Basis of Disease. 2021. Sep;1867(9):166180. [DOI] [PubMed] [Google Scholar]
- 96.Janik-Olchawa N, Drozdz A, Wajda A, Sitarz M, Planeta K, Setkowicz Z, et al. Biochemical changes of macrophages and U87MG cells occurring as a result of the exposure to iron oxide nanoparticles detected with the Raman microspectroscopy. Spectrochimica Acta Part A: Molecular and Biomolecular Spectroscopy. 2022. Oct;278:121337. [DOI] [PubMed] [Google Scholar]
- 97.Sitarz K, Czamara K, Bialecka J, Klimek M, Szostek S, Kaczor A. Dual Switch in Lipid Metabolism in Cervical Epithelial Cells during Dysplasia Development Observed Using Raman Microscopy and Molecular Methods. Cancers. 2021. Apr 21;13(9):1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Pliss A, Kuzmin AN, Prasad PN, Mahajan SD. Mitochondrial Dysfunction: A Prelude to Neuropathogenesis of SARS-CoV-2. ACS Chem Neurosci. 2022. Feb 2;13(3):308–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Notarstefano V, Pisani M, Bramucci M, Quassinti L, Maggi F, Vaccari L, et al. A vibrational in vitro approach to evaluate the potential of monoolein nanoparticles as isofuranodiene carrier in MDA-MB 231 breast cancer cell line: New insights from Infrared and Raman microspectroscopies. Spectrochimica Acta Part A: Molecular and Biomolecular Spectroscopy. 2022. Mar;269:120735. [DOI] [PubMed] [Google Scholar]
- 100.Bakar J, Michael-Jubeli R, Tfaili S, Assi A, Baillet-Guffroy A, Tfayli A. Biomolecular modifications during keratinocyte differentiation: Raman spectroscopy and chromatographic techniques. Analyst. 2021;146(9):2965–73. [DOI] [PubMed] [Google Scholar]
- 101.Wenzel Carvajal Berrio, Reisenauer Layland, Koch Wallwiener, et al. Trans-Mucosal Efficacy of Non-Thermal Plasma Treatment on Cervical Cancer Tissue and Human Cervix Uteri by a Next Generation Electrosurgical Argon Plasma Device. Cancers. 2020. Jan 22;12(2):267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bik E, Orleanska J, Mateuszuk L, Baranska M, Majzner K, Chlopicki S. Raman and fluorescence imaging of phospholipidosis induced by cationic amphiphilic drugs in endothelial cells. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research. 2022. Mar;1869(3):119186. [DOI] [PubMed] [Google Scholar]
- 103.Pacia MZ, Chorazy N, Sternak M, Fels B, Pacia M, Kepczynski M, et al. Rac1 regulates lipid droplets formation, nanomechanical, and nanostructural changes induced by TNF in vascular endothelium in the isolated murine aorta. Cell Mol Life Sci. 2022. Jun;79(6):317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lin J, Graziotto ME, Lay PA, New EJ. A Bimodal Fluorescence-Raman Probe for Cellular Imaging. Cells. 2021. Jul 5;10(7):1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Bader CA, Shandala T, Carter EA, Ivask A, Guinan T, Hickey SM, et al. A Molecular Probe for the Detection of Polar Lipids in Live Cells. Witt SN, editor. PLoS ONE. 2016. Aug 23;11(8):e0161557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chen Z, Paley DW, Wei L, Weisman AL, Friesner RA, Nuckolls C, et al. Multicolor Live-Cell Chemical Imaging by Isotopically Edited Alkyne Vibrational Palette. J Am Chem Soc. 2014. Jun 4;136(22):8027–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Gala de Pablo J, Chisholm DR, Ambler CA, Peyman SA, Whiting A, Evans SD. Detection and time-tracking activation of a photosensitiser on live single colorectal cancer cells using Raman spectroscopy. Analyst. 2020;145(17):5878–88. [DOI] [PubMed] [Google Scholar]
- 108.Watson MD, Lee JC. Genetically Encoded Aryl Alkyne for Raman Spectral Imaging of Intracellular α-Synuclein Fibrils. Journal of Molecular Biology. 2022. Jul 2;167716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Yamakoshi H, Dodo K, Palonpon A, Ando J, Fujita K, Kawata S, et al. Alkyne-Tag Raman Imaging for Visualization of Mobile Small Molecules in Live Cells. J Am Chem Soc. 2012. Dec 26;134(51):20681–9. [DOI] [PubMed] [Google Scholar]
- 110.Samuel AZ, Miyaoka R, Ando M, Gaebler A, Thiele C, Takeyama H. Molecular profiling of lipid droplets inside HuH7 cells with Raman micro-spectroscopy. Commun Biol. 2020. Dec;3(1):372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Matuszyk E, Adamczyk A, Radwan B, Pieczara A, Szcześniak P, Mlynarski J, et al. Multiplex Raman imaging of organelles in endothelial cells. Spectrochimica Acta Part A: Molecular and Biomolecular Spectroscopy. 2021. Jul;255:119658. [DOI] [PubMed] [Google Scholar]
- 112.Hekmatara M, Heidari Baladehi M, Ji Y, Xu J. D 2 O-Probed Raman Microspectroscopy Distinguishes the Metabolic Dynamics of Macromolecules in Organellar Anticancer Drug Response. Anal Chem. 2021. Feb 2;93(4):2125–34. [DOI] [PubMed] [Google Scholar]
- 113.Matthäus C, Krafft C, Dietzek B, Brehm BR, Lorkowski S, Popp J. Noninvasive Imaging of Intracellular Lipid Metabolism in Macrophages by Raman Microscopy in Combination with Stable Isotopic Labeling. Anal Chem. 2012. Oct 16;84(20):8549–56. [DOI] [PubMed] [Google Scholar]
- 114.Stiebing C, Schmölz L, Wallert M, Matthäus C, Lorkowski S, Popp J. Raman imaging of macrophages incubated with triglyceride-enriched oxLDL visualizes translocation of lipids between endocytic vesicles and lipid droplets. Journal of Lipid Research. 2017. May;58(5):876–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Stiebing C, Matthäus C, Krafft C, Keller AA, Weber K, Lorkowski S, et al. Complexity of fatty acid distribution inside human macrophages on single cell level using Raman micro-spectroscopy. Anal Bioanal Chem. 2014. Nov;406(27):7037–46. [DOI] [PubMed] [Google Scholar]
- 116.Ranneva SV, Okotrub KA, Amstislavsky SY, Surovtsev NV. Deuterated stearic acid uptake and accumulation in lipid droplets of cat oocytes. Archives of Biochemistry and Biophysics. 2020. Oct;692:108532. [DOI] [PubMed] [Google Scholar]
- 117.Egoshi S, Dodo K, Ohgane K, Sodeoka M. Deuteration of terminal alkynes realizes simultaneous live cell Raman imaging of similar alkyne-tagged biomolecules. Org Biomol Chem. 2021;19(38):8232–6. [DOI] [PubMed] [Google Scholar]
- 118.Hill AH, Munger E, Francis AT, Manifold B, Fu D. Frequency Modulation Stimulated Raman Scattering Microscopy through Polarization Encoding. J Phys Chem B. 2019. Oct 10;123(40):8397–404. [DOI] [PubMed] [Google Scholar]
- 119.Tipping WJ, Wilson LT, An C, Leventi AA, Wark AW, Wetherill C, et al. Stimulated Raman scattering microscopy with spectral phasor analysis: applications in assessing drug–cell interactions. Chem Sci. 2022;13(12):3468–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hislop EW, Tipping WJ, Faulds K, Graham D. Label-Free Imaging of Lipid Droplets in Prostate Cells Using Stimulated Raman Scattering Microscopy and Multivariate Analysis. Anal Chem. 2022. Jun 28;94(25):8899–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Stiebing C, Meyer T, Rimke I, Matthäus C, Schmitt M, Lorkowski S, et al. Real-time Raman and SRS imaging of living human macrophages reveals cell-to-cell heterogeneity and dynamics of lipid uptake. J Biophoton. 2017. Sep;10(9):1217–26. [DOI] [PubMed] [Google Scholar]
- 122.Du J, Su Y, Qian C, Yuan D, Miao K, Lee D, et al. Raman-guided subcellular pharmacometabolomics for metastatic melanoma cells. Nat Commun. 2020. Dec;11(1):4830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Oh S, Lee C, Yang W, Li A, Mukherjee A, Basan M, et al. Protein and lipid mass concentration measurement in tissues by stimulated Raman scattering microscopy. Proc Natl Acad Sci USA. 2022. Apr 26;119(17):e2117938119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ranjan R, Ferrara MA, Filograna A, Valente C, Sirleto L. Femtosecond Stimulated Raman microscopy: home-built realization and a case study of biological imaging. J Inst. 2019. Sep 12;14(09):P09008–P09008. [Google Scholar]
- 125.Li J, Condello S, Thomes-Pepin J, Ma X, Xia Y, Hurley TD, et al. Lipid Desaturation Is a Metabolic Marker and Therapeutic Target of Ovarian Cancer Stem Cells. Cell Stem Cell. 2017. Mar;20(3):303–314.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Shi L, Klimas A, Gallagher B, Cheng Z, Fu F, Wijesekara P, et al. Super-Resolution Vibrational Imaging Using Expansion Stimulated Raman Scattering Microscopy. Advanced Science. 2022. Jul;9(20):2200315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Fung AA, Hoang K, Zha H, Chen D, Zhang W, Shi L. Imaging Sub-Cellular Methionine and Insulin Interplay in Triple Negative Breast Cancer Lipid Droplet Metabolism. Front Oncol. 2022. Mar 10;12:858017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Adams WR, Gautam R, Locke A, Masson LE, Borrachero-Conejo AI, Dollinger BR, et al. Visualizing the lipid dynamics role in infrared neural stimulation using stimulated Raman scattering. Biophysical Journal. 2022. Apr;121(8):1525–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Hong S, Chen T, Zhu Y, Li A, Huang Y, Chen X. Live-Cell Stimulated Raman Scattering Imaging of Alkyne-Tagged Biomolecules. Angew Chem Int Ed. 2014. Jun 2;53(23):5827–31. [DOI] [PubMed] [Google Scholar]
- 130.Du J, Wei L. Multicolor Photoactivatable Raman Probes for Subcellular Imaging and Tracking by Cyclopropenone Caging. J Am Chem Soc. 2022. Jan 19;144(2):777–86. [DOI] [PubMed] [Google Scholar]
- 131.Vukosavljevic B, Hittinger M, Hachmeister H, Pilger C, Murgia X, Gepp MM, et al. Vibrational spectroscopic imaging and live cell video microscopy for studying differentiation of primary human alveolar epithelial cells. J Biophotonics [Internet]. 2019. Jun [cited 2022 Jul 24];12(6). Available from: https://onlinelibrary.wiley.com/doi/10.1002/jbio.201800052 [DOI] [PubMed] [Google Scholar]
- 132.Zhang C, Boppart SA. Dynamic Signatures of Lipid Droplets as New Markers to Quantify Cellular Metabolic Changes. Anal Chem. 2020. Dec 15;92(24):15943–52. [DOI] [PubMed] [Google Scholar]
- 133.Levchenko SM, Kuzmin AN, Pliss A, Ohulchanskyy TY, Prasad PN, Qu J. Cellular transformations in near-infrared light-induced apoptosis in cancer cells revealed by labelfree CARS imaging. J Biophotonics [Internet]. 2019. Dec [cited 2022 Jul 24];12(12). Available from: https://onlinelibrary.wiley.com/doi/10.1002/jbio.201900179 [DOI] [PubMed] [Google Scholar]
- 134.Levchenko SM, Peng X, Liu L, Qu J. The impact of cell fixation on coherent anti-stokes Raman scattering signal intensity in neuronal and glial cell lines. J Biophotonics. 2019. Jan;12(1):e201800203. [DOI] [PubMed] [Google Scholar]
- 135.Guerenne-Del Ben T, Couderc V, Duponchel L, Sol V, Leproux P, Petit JM. Multiplex coherent anti-Stokes Raman scattering microspectroscopy detection of lipid droplets in cancer cells expressing TrkB. Sci Rep. 2020. Dec;10(1):16749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Mizuguchi T, Momotake A, Hishida M, Yasui M, Yamamoto Y, Saiki T, et al. Multimodal Multiphoton Imaging of the Lipid Bilayer by Dye-Based Sum-Frequency Generation and Coherent Anti-Stokes Raman Scattering. Anal Chem. 2020. Apr 21;92(8):5656–60. [DOI] [PubMed] [Google Scholar]
- 137.Takei Y, Hirai R, Fukuda A, Miyazaki S, Shimada R, Okamatsu-Ogura Y, et al. Visualization of intracellular lipid metabolism in brown adipocytes by time-lapse ultra-multiplex CARS microspectroscopy with an onstage incubator. J Chem Phys. 2021. Sep 28;155(12):125102. [DOI] [PubMed] [Google Scholar]
- 138.Mukherjee P, Aksamitiene E, Alex A, Shi J, Bera K, Zhang C, et al. Differential Uptake of Antisense Oligonucleotides in Mouse Hepatocytes and Macrophages Revealed by Simultaneous Two-Photon Excited Fluorescence and Coherent Raman Imaging. Nucleic Acid Therapeutics. 2022. Jun 1;32(3):163–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Matuszyk E, Sierka E, Rodewald M, Bae H, Meyer T, Kus E, et al. Differential response of liver sinusoidal endothelial cells and hepatocytes to oleic and palmitic acid revealed by Raman and CARS imaging. Biochimica et Biophysica Acta (BBA) - Molecular Basis of Disease. 2020. Jun;1866(6):165763. [DOI] [PubMed] [Google Scholar]
- 140.Borek-Dorosz A, Grosicki M, Dybas J, Matuszyk E, Rodewald M, Meyer-Zedler T, et al. Identification of inflammatory markers in eosinophilic cells of the immune system: fluorescence, Raman and CARS imaging can recognize markers but differently. Cell Mol Life Sci. 2022. Jan;79(1):52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Xu D, Liang S, Xu L, Bourdakos KN, Johnson P, Read J, et al. Widely-tunable synchronisation-free picosecond laser source for multimodal CARS, SHG, and two-photon microscopy. Biomed Opt Express. 2021. Feb 1;12(2):1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Di Napoli C, Pope I, Masia F, Langbein W, Watson P, Borri P. Quantitative Spatiotemporal Chemical Profiling of Individual Lipid Droplets by Hyperspectral CARS Microscopy in Living Human Adipose-Derived Stem Cells. Anal Chem. 2016. Apr 5;88(7):3677–85. [DOI] [PubMed] [Google Scholar]
- 143.Camp CH Jr, Lee YJ, Heddleston JM, Hartshorn CM, Walker ARH, Rich JN, et al. High-speed coherent Raman fingerprint imaging of biological tissues. Nature Photon. 2014. Aug;8(8):627–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Lakowicz JR. Principles of Fluorescence Spectroscopy [Internet]. Springer; US; 2007. Available from: https://books.google.com/books?id=-PSybuLNxcAC [Google Scholar]
- 145.de la Rosa Rodriguez MA, Deng L, Gemmink A, van Weeghel M, Aoun ML, Warnecke C, et al. Hypoxia-inducible lipid droplet-associated induces DGAT1 and promotes lipid storage in hepatocytes. Molecular Metabolism. 2021. May;47:101168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Liu C, Zhang D, Ye S, Chen T, Liu R. D-π-A structure fluorophore: NIR emission, response to viscosity, detection cyanide and bioimaging of lipid droplets. Spectrochimica Acta Part A: Molecular and Biomolecular Spectroscopy. 2022. Feb;267:120593. [DOI] [PubMed] [Google Scholar]
- 147.Niu J, Liu Y, Wang W, Lin W. Novel two-photon fluorescent probe with high fluorescence quantum yields for tracking lipid droplets in biological systems. Spectrochimica Acta Part A: Molecular and Biomolecular Spectroscopy. 2019. Jun;216:35–44. [DOI] [PubMed] [Google Scholar]
- 148.Ramosaj M, Madsen S, Maillard V, Scandella V, Sudria-Lopez D, Yuizumi N, et al. Lipid droplet availability affects neural stem/progenitor cell metabolism and proliferation. Nat Commun. 2021. Dec;12(1):7362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Danylchuk DI, Jouard PH, Klymchenko AS. Targeted Solvatochromic Fluorescent Probes for Imaging Lipid Order in Organelles under Oxidative and Mechanical Stress. J Am Chem Soc. 2021. Jan 20;143(2):912–24. [DOI] [PubMed] [Google Scholar]
- 150.Wang J, Fang N, Xiong J, Du Y, Cao Y, Ji WK. An ESCRT-dependent step in fatty acid transfer from lipid droplets to mitochondria through VPS13D−TSG101 interactions. Nat Commun. 2021. Dec;12(1):1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Lorent JH, Levental KR, Ganesan L, Rivera-Longsworth G, Sezgin E, Doktorova M, et al. Plasma membranes are asymmetric in lipid unsaturation, packing and protein shape. Nat Chem Biol. 2020. Jun 1;16(6):644–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Lee HW, Lee IJ, Lee SJ, Kim YR, Kim HM. Highly Sensitive Two-Photon Lipid Droplet Tracker for In Vivo Screening of Drug Induced Liver Injury. ACS Sens. 2022. Apr 22;7(4):1027–35. [DOI] [PubMed] [Google Scholar]
- 153.Xue H, Ge E, Ge W, Li J, Tian M. Single Fluorescent Probe for Zero-Crosstalk Discrimination of Lipid Droplets and the Endoplasmic Reticulum Based on Reversible Cyclization Reaction. Anal Chem. 2022. Jun 28;94(25):9158–65. [DOI] [PubMed] [Google Scholar]
- 154.Wu CJ, Li XY, Zhu T, Zhao M, Song Z, Li S, et al. Exploiting the Twisted Intramolecular Charge Transfer Effect to Construct a Wash-Free Solvatochromic Fluorescent Lipid Droplet Probe for Fatty Liver Disease Diagnosis. Anal Chem. 2022. Mar 8;94(9):3881–7. [DOI] [PubMed] [Google Scholar]
- 155.Lai C, Zhao Y, Liang Y, Zou X, Lin W. BF2 group chelated AIE fluorescent probe for polarity mapping of lipid droplets in cells and in vivo. Spectrochimica Acta Part A: Molecular and Biomolecular Spectroscopy. 2022. Mar;268:120637. [DOI] [PubMed] [Google Scholar]
- 156.Jung Y, Jin JH, Kim Y, Oh JH, Moon H, Jeong H, et al. Development of a fluorescent nanoprobe based on an amphiphilic single-benzene-based fluorophore for lipid droplet detection and its practical applications. Org Biomol Chem. 2022;20(27):5423–33. [DOI] [PubMed] [Google Scholar]
- 157.Li X, Yang Z, Bian J, Fu M, Zhang Y, Jiang N, et al. Fluorescent probes based on multifunctional encapsulated perylene diimide dyes for imaging of lipid droplets in live cells. Analyst. 2022;147(7):1410–6. [DOI] [PubMed] [Google Scholar]
- 158.Öberg E, Appelqvist H, Nilsson KPR. Non-fused Phospholes as Fluorescent Probes for Imaging of Lipid Droplets in Living Cells. Front Chem. 2017. Apr 25;5:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Huang B, Bates M, Zhuang X. Super-Resolution Fluorescence Microscopy. Annu Rev Biochem. 2009. Jun 1;78(1):993–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Yin J, Peng M, Lin W. Two-photon fluorescence imaging of lipid drops polarity toward cancer diagnosis in living cells and tissue. Sensors and Actuators B: Chemical. 2019. Jun;288:251–8. [Google Scholar]
- 161.Kreder R, Pyrshev KA, Darwich Z, Kucherak OA, Mély Y, Klymchenko AS. Solvatochromic Nile Red Probes with FRET Quencher Reveal Lipid Order Heterogeneity in Living and Apoptotic Cells. ACS Chem Biol. 2015. Jun 19;10(6):1435–42. [DOI] [PubMed] [Google Scholar]
- 162.Yamaguchi E, Wang C, Fukazawa A, Taki M, Sato Y, Sasaki T, et al. Environment-Sensitive Fluorescent Probe: A Benzophosphole Oxide with an Electron-Donating Substituent. Angew Chem Int Ed. 2015. Apr 7;54(15):4539–43. [DOI] [PubMed] [Google Scholar]
- 163.Owen DM, Williamson DJ, Magenau A, Gaus K. Sub-resolution lipid domains exist in the plasma membrane and regulate protein diffusion and distribution. Nat Commun. 2012. Jan;3(1):1256. [DOI] [PubMed] [Google Scholar]
- 164.Sohn M, Toth DJ, Balla T. Monitoring Non-vesicular Transport of Phosphatidylserine and Phosphatidylinositol 4-Phosphate in Intact Cells by BRET Analysis. In: Drin G, editor. Intracellular Lipid Transport: Methods and Protocols [Internet]. New York, NY: Springer; 2019. [cited 2022 Aug 1]. p. 13–22. (Methods in Molecular Biology). Available from: 10.1007/978-1-4939-9136-5_2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Tóth JT, Gulyás G, Hunyady L, Várnai P. Development of Nonspecific BRET-Based Biosensors to Monitor Plasma Membrane Inositol Lipids in Living Cells. In: Drin G, editor. Intracellular Lipid Transport: Methods and Protocols [Internet]. New York, NY: Springer; 2019. [cited 2022 Aug 1]. p. 23–34. (Methods in Molecular Biology). Available from: 10.1007/978-1-4939-9136-5_3 [DOI] [PubMed] [Google Scholar]
- 166.Wilhelm LP, Voilquin L, Kobayashi T, Tomasetto C, Alpy F. Intracellular and Plasma Membrane Cholesterol Labeling and Quantification Using Filipin and GFP-D4. In: Drin G, editor. Intracellular Lipid Transport: Methods and Protocols [Internet]. New York, NY: Springer; 2019. [cited 2022 Aug 1]. p. 137–52. (Methods in Molecular Biology). Available from: 10.1007/978-1-4939-9136-5_11 [DOI] [PubMed] [Google Scholar]
- 167.Chandra A, Datta A. A Peptide-Based Fluorescent Sensor for Anionic Phospholipids. ACS Omega. 2022. Mar 29;7(12):10347–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Moser von Filseck J, Čopič A, Delfosse V, Vanni S, Jackson CL, Bourguet W, et al. Phosphatidylserine transport by ORP/Osh proteins is driven by phosphatidylinositol 4-phosphate. Science. 2015. Jul 24;349(6246):432–6. [DOI] [PubMed] [Google Scholar]
- 169.Chung J, Torta F, Masai K, Lucast L, Czapla H, Tanner LB, et al. PI4P/phosphatidylserine countertransport at ORP5- and ORP8-mediated ER–plasma membrane contacts. Science. 2015. Jul 24;349(6246):428–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Sohn M, Korzeniowski M, Zewe JP, Wills RC, Hammond GRV, Humpolickova J, et al. PI(4,5)P2 controls plasma membrane PI4P and PS levels via ORP5/8 recruitment to ER–PM contact sites. Journal of Cell Biology. 2018. May 7;217(5):1797–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Capasso S, D’Angelo G. Imaging Lipid Metabolism at the Golgi Complex. In: Drin G, editor. Intracellular Lipid Transport: Methods and Protocols [Internet]. New York, NY: Springer New York; 2019. p. 47–56. Available from: 10.1007/978-1-4939-9136-5_5 [DOI] [PubMed] [Google Scholar]
- 172.Hammond GRV, Schiavo G, Irvine RF. Immunocytochemical techniques reveal multiple, distinct cellular pools of PtdIns4 P and PtdIns(4,5) P 2. Biochemical Journal. 2009. Aug 15;422(1):23–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Zhang J, Nie J, Sun H, Li J, Andersen JP, Shi Y. De novo labeling and trafficking of individual lipid species in live cells. Molecular Metabolism. 2022. Jul;61:101511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Feng S, Harayama T, Montessuit S, David FP, Winssinger N, Martinou JC, et al. Mitochondria-specific photoactivation to monitor local sphingosine metabolism and function. eLife. 2018. Jan 29;7:e34555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Kinoshita M, Suzuki KGN, Matsumori N, Takada M, Ano H, Morigaki K, et al. Raft-based sphingomyelin interactions revealed by new fluorescent sphingomyelin analogs. Journal of Cell Biology. 2017. Apr 3;216(4):1183–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Brameshuber M, Weghuber J, Ruprecht V, Gombos I, Horváth I, Vigh L, et al. Imaging of Mobile Long-lived Nanoplatforms in the Live Cell Plasma Membrane. Journal of Biological Chemistry. 2010. Dec;285(53):41765–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Haberkant P, Stein F, Höglinger D, Gerl MJ, Brügger B, Van Veldhoven PP, et al. Bifunctional Sphingosine for Cell-Based Analysis of Protein-Sphingolipid Interactions. ACS Chem Biol. 2016. Jan 15;11(1):222–30. [DOI] [PubMed] [Google Scholar]
- 178.Höglinger D, Nadler A, Haberkant P, Kirkpatrick J, Schifferer M, Stein F, et al. Trifunctional lipid probes for comprehensive studies of single lipid species in living cells. Proc Natl Acad Sci USA. 2017. Feb 14;114(7):1566–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Gaebler A, Penno A, Kuerschner L, Thiele C. A highly sensitive protocol for microscopy of alkyne lipids and fluorescently tagged or immunostained proteins. Journal of Lipid Research. 2016. Oct;57(10):1934–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Bumpus TW, Liang D, Baskin JM. IMPACT: Imaging phospholipase d activity with clickable alcohols via transphosphatidylation. In: Methods in Enzymology [Internet]. Elsevier; 2020. [cited 2021 Dec 18]. p. 75–94. Available from: https://linkinghub.elsevier.com/retrieve/pii/S0076687920301658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Liang D, Wu K, Tei R, Bumpus TW, Ye J, Baskin JM. A real-time, click chemistry imaging approach reveals stimulus-specific subcellular locations of phospholipase D activity. Proc Natl Acad Sci USA. 2019. Jul 30;116(31):15453–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Bumpus TW, Baskin JM. Clickable Substrate Mimics Enable Imaging of Phospholipase D Activity. ACS Cent Sci. 2017. Oct 25;3(10):1070–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Gaebler A, Milan R, Straub L, Hoelper D, Kuerschner L, Thiele C. Alkyne lipids as substrates for click chemistry-based in vitro enzymatic assays. Journal of Lipid Research. 2013. Aug;54(8):2282–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Thiele C, Papan C, Hoelper D, Kusserow K, Gaebler A, Schoene M, et al. Tracing Fatty Acid Metabolism by Click Chemistry. ACS Chem Biol. 2012. Dec 21;7(12):2004–11. [DOI] [PubMed] [Google Scholar]
- 185.Petersen BV, Gallion L, Allbritton NL. Silicon Photomultipliers as a Low-Cost Fluorescence Detector for Capillary Electrophoresis. Anal Chem. 2020. Oct 20;92(20):13683–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Dickinson AJ, Hunsucker SA, Armistead PM, Allbritton NL. Single-cell sphingosine kinase activity measurements in primary leukemia. Anal Bioanal Chem. 2014. Nov;406(27):7027–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Proctor A, Sims CE, Allbritton NL. Chemical fixation to arrest phospholipid signaling for chemical cytometry. Journal of Chromatography A. 2017. Nov;1523:97–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Essaka DC, Prendergast J, Keithley RB, Palcic MM, Hindsgaul O, Schnaar RL, et al. Metabolic Cytometry: Capillary Electrophoresis with Two-Color Fluorescence Detection for the Simultaneous Study of Two Glycosphingolipid Metabolic Pathways in Single Primary Neurons. Anal Chem. 2012. Mar 20;84(6):2799–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Ciura K, Dziomba S, Nowakowska J, Markuszewski MJ. Thin layer chromatography in drug discovery process. Journal of Chromatography A. 2017. Oct;1520:9–22. [DOI] [PubMed] [Google Scholar]
- 190.Whitmore CD, Olsson U, Larsson EA, Hindsgaul O, Palcic MM, Dovichi NJ. Yoctomole analysis of ganglioside metabolism in PC12 cellular homogenates. Electrophoresis. 2007. Sep;28(17):3100–4. [DOI] [PubMed] [Google Scholar]
- 191.Dickinson AJ, Meyer M, Pawlak EA, Gomez S, Jaspers I, Allbritton NL. Analysis of sphingosine kinase activity in single natural killer cells from peripheral blood. Integr Biol. 2015;7(4):392–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Keithley RB, Rosenthal AS, Essaka DC, Tanaka H, Yoshimura Y, Palcic MM, et al. Capillary electrophoresis with three-color fluorescence detection for the analysis of glycosphingolipid metabolism. Analyst. 2013;138(1):164–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Proctor A, Allbritton NL. “Fix and assay”: separating in-cellulo sphingolipid reactions from analytical assay in time and space using an aldehyde-based fixative. Analyst. 2019;144(3):961–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Boardman A, Chang T, Folch A, Dovichi NJ. Indium−Tin Oxide Coated Microfabricated Device for the Injection of a Single Cell into a Fused Silica Capillary for Chemical Cytometry. Anal Chem. 2010. Dec 1;82(23):9959–61. [DOI] [PMC free article] [PubMed] [Google Scholar]


