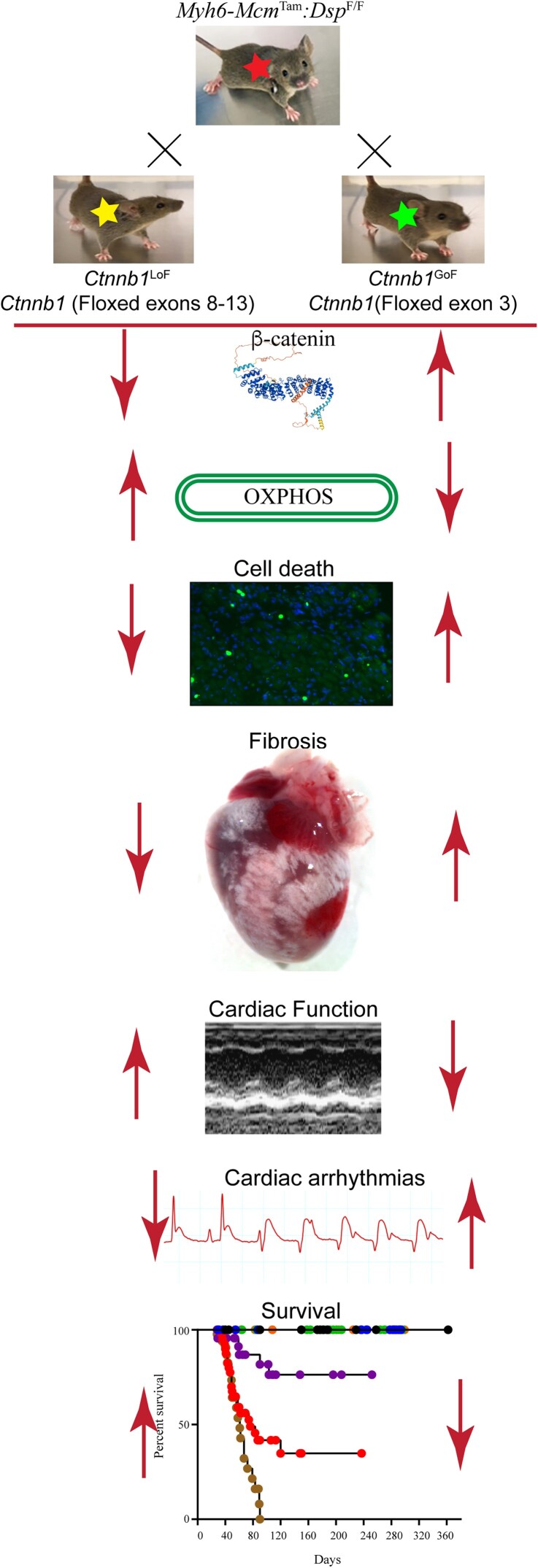
Abstract
Aims
Mutations in the DSP gene encoding desmoplakin, a constituent of the desmosomes at the intercalated discs (IDs), cause a phenotype that spans arrhythmogenic cardiomyopathy (ACM) and dilated cardiomyopathy. It is typically characterized by biventricular enlargement and dysfunction, myocardial fibrosis, cell death, and arrhythmias. The canonical wingless-related integration (cWNT)/β-catenin pathway is implicated in the pathogenesis of ACM. The β-catenin is an indispensable co-transcriptional regulator of the cWNT pathway and a member of the IDs. We genetically inactivated or activated β-catenin to determine its role in the pathogenesis of desmoplakin cardiomyopathy.
Methods and results
The Dsp gene was conditionally deleted in the 2-week-old post-natal cardiac myocytes using tamoxifen-inducible MerCreMer mice (Myh6-McmTam:DspF/F). The cWNT/β-catenin pathway was markedly dysregulated in the Myh6-McmTam:DspF/F cardiac myocytes, as indicated by a concomitant increase in the expression of cWNT/β-catenin target genes, isoforms of its key co-effectors, and the inhibitors of the pathway. The β-catenin was inactivated or activated upon inducible deletion of its transcriptional or degron domain, respectively, in the Myh6-McmTam:DspF/F cardiac myocytes. Genetic inactivation of β-catenin in the Myh6-McmTam:DspF/F mice prolonged survival, improved cardiac function, reduced cardiac arrhythmias, and attenuated myocardial fibrosis, and cell death caused by apoptosis, necroptosis, and pyroptosis, i.e. PANoptosis. In contrast, activation of β-catenin had the opposite effects. The deleterious and the salubrious effects were independent of changes in the expression levels of the cWNT target genes and were associated with changes in several molecular and biological pathways, including cell death programmes.
Conclusion
The cWNT/β-catenin was markedly dysregulated in the cardiac myocytes in a mouse model of desmoplakin cardiomyopathy. Inactivation of β-catenin attenuated, whereas its activation aggravated the phenotype, through multiple molecular pathways, independent of the cWNT transcriptional activity. Thus, suppression but not activation of β-catenin might be beneficial in desmoplakin cardiomyopathy.
Keywords: Desmoplakin, Cardiomyopathy, WNT pathway, β-Catenin, Oxidative phosphorylation, Cell death, Fibrosis
Graphical Abstract
Graphical Abstract.
Time for primary review: 28 days
1. Introduction
Hereditary cardiomyopathies comprise a genetically and phenotypically heterogenous group of myocardial diseases wherein the primary defect is in cardiac myocytes; the ensuing phenotype, however, is multi-cellular involving cellular constituents of the myocardium.1–4 Despite their heterogeneity, hereditary cardiomyopathies exhibit partial genetic and phenotypic overlaps. Among the three most common forms of hereditary cardiomyopathies, namely hypertrophic cardiomyopathy, dilated cardiomyopathy (DCM), and arrhythmogenic cardiomyopathy (ACM), the latter two exhibit considerable overlaps and at times are indistinguishable. Consequently, terms such as arrhythmogenic left dominant cardiomyopathy and arrhythmogenic DCM are used to describe overlapping phenotypes.5,6
DCM is characterized by cardiac dilatation and reduced left ventricular ejection fraction and is caused by mutations in genes coding for the cytoskeletal or sarcomere proteins.1 ACM, in its classic form, predominantly involves the right ventricle and is referred to as arrhythmogenic right ventricular cardiomyopathy and is primarily a disease of the intercalated disc (ID) proteins, mostly desmosome proteins.3 Among the genes known to cause ACM, the DSP gene, which encodes desmoplakin (DSP), causes a somewhat distinct phenotype that straddles between DCM and ACM5,7 The phenotypic overlap likely reflects the topographic location of DSP in the intra-cellular component of the desmosome structures where it interacts with the cytoskeletal proteins, such as cardiac α-actin, which are known to cause DCM.8,9 The phenotype caused by the DSP mutations is somewhat distinct, as it typically involves both ventricles, leading to biventricular dilatation and dysfunction, ventricular tachycardia, severe myocardial fibrosis, and apoptosis.6,7,10,11 Consequently, the phenotype is recognized as DSP cardiomyopathy.6,7,10,11
The cardiac IDs are comprised of multi-functional proteins that attach the adjacent cardiac myocytes and provide mechanical integrity to the myocardium, enable rapid electric conduction throughout the myocardium, and serve as a signalling hub responding to external or internal mechanical stress.12–16 The β-catenin (CTNNB1), which is the indispensable transcriptional co-effectors of the canonical WNT (cWNT) pathway, is an abundant constituent of the adherens junction at the IDs, where it interacts with the mechano-sensing proteins N-cadherin and α-catenin.17,18 Given the intricate interplays between the β-catenin and protein constituents of desmosomes, the cWNT/β-catenin is implicated in the pathogenesis of ACM and has emerged as a target of experimental therapy.16,19–24 However, no consensus has been established because of the inconsistent findings, likely reflective of the differences in the experimental approaches (see Supplementary material online, Figure S1A–D). One set of studies has targeted Glycogen Synthase Kinase 3 (GSK3β), typically using a pharmacological agent that inactivates both GSK3β and GSK3α.19,20,25–27 Inhibition of the GSK3β prevents phosphorylation of the β-catenin and its subsequent degradation following ubiquitinoylation, which leads to the activation of the cWNT pathway (see Supplementary material online, Figure S1C). Inhibition of GSK3β (and GSK3α), which is expected to activate the cWNT pathway, has been shown to improve the phenotype in zebrafish and mouse models of ACM.19,20,25–27 The findings, however, are confounded by the cWNT-independent effects of the GSK3β, which are numerous, as well as the concomitant inhibition of GSK3α, which has functions independent of the cWNT pathway.28,29 The findings are also in contrast with the data showing the deleterious effects of inhibition of the GSK3β on cardiac conduction and arrhythmias, which pertain to ACM.30 In an alternative approach, the cWNT pathway has been targeted by inhibiting porcupine (PORCN), which is the exclusive enzyme involved in the post-translational palmitoylation and the subsequent secretion of the WNT molecule to the extracellular milieu (see Supplementary material online, Figure S1D).22,31 Thus, inhibition of PORCN by preventing secretion of the WNT ligand abrogates receptor–ligand coupling and leads to suppression of the cWNT pathway.31 Suppression of the cWNT pathway upon inhibition of PORCN has been shown to impart salubrious effects and attenuate the phenotype in a mouse model of ACM.22
To address the existing conundrum and gain further insights into the role of the cWNT pathway in the pathogenesis of ACM, we utilized genetic loss-of-function (LoF) and gain-of-function (GoF) approaches to inactivate or activate β-catenin, respectively, in cardiac myocytes in a mouse model of ACM caused by cardiac myocyte-specific DSP deficiency (see Supplementary material online, Figure S1C). We determined the ensuing phenotypic effects and investigated the putative mechanisms.
2. Methods
2.1. Data sharing
RNA-Sequencing (RNA-Seq) data have been submitted to GEO (GSE180972). All data are available from the corresponding authors upon request.
2.2. Regulatory approvals
The use of mice in these studies was approved by the Institutional Animal Care and Use Committee (AWC-21-0015) and as per the NIH Guide for the Care and Use of Laboratory Animals.
2.3. Anaesthesia and euthanasia
Isoflurane was used to induce (3%) and maintain (0.5%) anaesthesia in mice. Euthanasia was accomplished after placing the mice under 100% CO2 inhalation and cervical dislocation.
2.4. The Myh6-Mcm:DspF/F mice
The Dsp gene was specifically deleted in the post-natal cardiac myocytes upon daily intra-peritoneal injection of five consecutive doses of tamoxifen (30 mg/Kg/d) to the Myh6-Mcm:DspF/F mice starting at post-natal day 14, as published.11
2.5. Genetic inducible activation or inactivation of CTNNB1 in cardiac myocytes in the post-natal mice
The mouse models of tamoxifen-inducible Cre-mediated β-catenin activation and inactivation have been published.32 In brief, to activate β-catenin, the floxed exon 3 of the Ctnnb1 gene, which encodes the GSK3β phosphorylation sites (Thr41, Ser37, and Ser33), necessary for its degradation by ubiquitinoylation (degron), was deleted.32–34 The deletion leads to the expression of a stable and a smaller β-catenin protein (lacking 80 amino acids or ∼10 kDa). The mouse represents the GoF of β-catenin (Ctnnb1GoF). To inactivate β-catenin, floxed exons 8–13 of the Ctnnb1 gene, which encodes the trans-activator motif comprised of amino acids 361–692, were deleted.32,35 The deletion leads to the expression of truncated mRNA and protein, which are unstable and degraded.35 The mouse represents the LoF of β-catenin (Ctnnb1LoF).
Inducible activation or inactivation of β-catenin is concomitant with deletion of the Dsp gene in cardiac myocytes in the post-natal mice: To delete Dsp and concomitantly activate or inactivate β-catenin in cardiac myocyte, Myh6-McmTam:DspF/F:Ctnnb1LoF and Myh6-McmTam:DspF/F:Ctnnb1GoF mice were generated and injected with tamoxifen (30 mg/kg/d) starting at P14 for 5 consecutive days (see Supplementary material online, Figure S2). The P14 time point was chosen to avoid embryonic lethality associated with homozygous deletion of the Dsp gene.16,36 It also reduces confounding effects of activation or suppression of the cWNT/β-catenin pathway during the proliferative phase of cardiac myocytes in the early post-natal period.37,38
2.6. Genotypes
The mice were genotyped by polymerase chain reaction (PCR) using genomic DNA isolated from the tail, as described.16,39 The list of oligonucleotide primers used for the genotyping is provided in Supplementary material online, Table S1.
2.7. Survival analysis
Kaplan–Meier survival plots were constructed in the main study groups, namely the wild-type (WT), Myh6-McmTam:DspF/F, Myh6-McmTam:DspF/F:Ctnnb1LoF, and Myh6-McmTam:DspF/F:Ctnnb1GoF using the GraphPad Prism 9 software. The short-term survival data, matching the time frame of the present study, in Myh6-McmTam, Myh6-McmTam:Ctnnb1LoF, and Myh6-McmTam:Ctnnb1GoF have been published and were not included to avoid redundancy.32,40
2.8. Gross morphology
Body and heart weight were measured at 4 weeks of age, which is the time point that most phenotypes were analysed. The heart weight was indexed to body weight and compared among the groups.
2.9. Echocardiography
Echocardiography was performed on 4-week-old mice using a Vevo 1100 ultrasound imaging system (FUJIFILM VisualSonics Inc., Toronto, ON, Canada), as published.32,40,41 In brief, cardiac images were obtained in the M mode using a B-mode guide at the level of left ventricular (LV) papillary muscles in mice anaesthetized with 0.5% isoflurane. LV anterior wall thickness, posterior wall thickness, end-systolic diameter, and end-diastolic diameter were measured using the leading-edge method. The LV fractional shortening and mass were calculated from the m-mode images, as published.32,40,41
2.10. Rhythm monitoring
Two surface leads were placed over the mouse’s chest, and the cardiac rhythm was monitored for about an hour in each mouse under 0.5% isoflurane anaesthesia, as published.11,42 A Power Lab 4/30 data acquisition system was used, and the data were analysed using Lab Chart7 software (ADInstruments, Colorado Springs, CO, USA).
2.11. Cardiac myocyte isolation
Cardiac myocytes were isolated from 4-week-old mice as published.11,43 In brief, mice were anaesthetized, and the heart was excised, mounted onto a Langendorff perfusion system, and perfused with a Type 2 collagenase digestion buffer (2.4 mg/mL concentration) at a flow rate of 4 mL/min (Worthington Cat# LS004176). The ventricles were minced in a stop buffer containing 10% calf serum, 12.5 μM CaCl2, and 2 mM adenosine triphosphate, and the cell suspension passed through a 100 µm cell strainer and centrifuged at 20 g to precipitate cardiac myocytes. The isolated cells were treated with CaCl2 in the stop buffer in a stepwise increase from a molar concentration of 100–900 μM. The isolated myocytes were suspended either in a Qiazol reagent (Qiagen Cat# 79306) for RNA extraction or in a protein extraction buffer for immunoblotting.
2.12. Immunoblotting
Ventricular tissues or cardiac myocytes were homogenized and dissolved in a RIPA buffer (Cat# 974821) containing 0.5% SDS.44 The buffers contained protease and phosphatase inhibitors (Roche, Cat#04693159001 and Cat#04906837001, respectively). The lysates were sonicated briefly using a Bioruptor Pico (Diagenode, Denville, NJ) and centrifuged at 13 000 rpm to precipitate the protein. Aliquots of 30–50 μg protein extracts were loaded onto an SDS polyacrylamide gel, electrophoresed, and transferred onto nitrocellulose membranes. The membranes were probed with antibodies against the target proteins followed by incubation with the corresponding secondary antibodies. The signals were detected using the ECL western blotting detection kit (Amersham Cat# RPN2106), and the images were collected using the LiCOR (Odyssey, Denville, NJ) imaging system.
2.13. Reverse transcription–polymerase chain reaction
Total cardiac myocyte RNA was extracted using a miRNeasy Mini Kit (Qiagen, Cat # 217006) and treated with DNase I (Qiagen, Cat# 79254). Reverse transcription was performed on ∼1 μg of total RNA using random primers and a high-capacity cDNA synthesis kit (Applied Biosystems Cat# 4368814). The SYBR green probes or TaqMan assays were used in duplicate to determine the transcript levels of the selected genes, which were normalized to the Gapdh transcript levels. Changes in the transcript levels were calculated using the ΔΔCT method and presented as fold changes relative to the values in the WT mice. The primers used in the study are listed in the Supplementary material online, Table S1.
2.14. RNA-Sequencing
RNA-Seq was performed on ribosome-depleted ventricular cardiac myocyte RNA extracts, as published.11,24,32,40 In brief, total RNA was extracted using the miRNeasy Mini Kit (Qiagen, Cat # 217004), and extracts with an RNA Integrity Number value of >8, determined using an Agilent Bioanalyzer RNA chip, were used to construct sequencing libraries. Strand-specific sequencing libraries were generated using TruSeq stranded total RNA library preparation kit (Illumina Inc. Cat # 20020596) and sequenced as 75 bp paired-end reads on an Illumina instrument.
2.15. Myocardial fibrosis
Myocardial fibrosis was assessed by multiple methods, including calculation of collagen volume fraction (CVF) from thin myocardial sections stained with picrosirius red, quantification of the transcript of genes involved in fibrosis by reverse transcription–PCR (RT–PCR) and selected proteins by western blotting and immunofluorescence staining, as published.11,32,40,45 Likewise, fibrosis pathways were analysed in the cardiac myocyte RNA-Seq dataset.
2.16. Myocyte cross-sectional area
To identify cardiac myocytes, thin myocardial sections were stained with an antibody against pericentriolar material protein 1 (PCM1), which mainly marks the myocyte nuclei in the heart (Sigma Cat# HPA023370), as published.32,39,42,46 The sections were co-stained with wheat germ agglutinin (WGA) conjugated to Texas red [concentration 1 μg/mL, Thermo Fisher Scientific, Cat#W21405)] and 4′,6-diamidino-2-phenylindole (DAPI) to mark the interstitium and nuclei, respectively. The number of myocytes was calculated along with the areas not stained with WGA to determine the myocyte cross-sectional area (CSA).
2.17. Terminal deoxynucleotidyl transferase dUTP nick end labelling assay
The terminal deoxynucleotidyl transferase dUTP nick end labelling (TUNEL) assay was performed to detect apoptosis using the TUNEL assay and in-situ cell death detection Fluorescein kit (Roche Cat#11684795910), as published.41,43,44
2.18. Mitochondrial electron chain transport complexes activities
Enzymatic activity of each mitochondrial electron chain transport (ETC) complex was determined in cardiac myocyte protein extracts by adding specific substrates and inhibitors to homogenates using the commercially available kits (see Supplementary material online, Table S1). In brief, cardiac myocytes were isolated and were gently homogenized on ice using a Dounce homogenizer in a buffer containing 120 mM KCl, 20 mM HEPES, and 1 mM EGTA at pH 7.4. The homogenates were centrifuged at 600 g at 4°C for 10 min, and the supernatants were collected and used in the enzymatic assays. The specific substrate of each ETC complex was added to the supernatant, and the activity was measured by colorimetric assays using a monochromator microplate reader (Tecan M200) at 25°C. Complex I activity was determined upon measuring the oxidation rate of Nicotinamide adenine dinucleotide (NAD) hydrogen in the presence of 1 mM potassium cyanide at 340 nm. Complex II and Complex III activities were determined by the rate of reduction of cytochrome C, measured at 550 nm, in the presence of 1 mM potassium cyanide, 1 mM rotenone, 1 mM antimycin A, and 1 mM thenoyltrifluoroacetone, which were added to inhibit Complex III and Complex II activities, respectively. Complex IV activity was measured by the rate of oxidation of reduced cytochrome C, as determined at an absorbance at 550 nm. Complex V activity was measured by the production of NAD+, measured at 340 nm, based on the utilization of ADP generated by the adenosine triphosphatase upon the reduction of pyruvate to lactate. All activities were calculated as ΔOD/min and expressed as a percentage of WT activity. Experiments were performed on at least five independent samples for each genotype.
2.19. Bioinformatics and statistics
The RNA-Seq reads were aligned to the mouse reference genome build mm10 using STAR.47 The uniquely aligned read pairs were annotated using GENCODE gene model. Read counts were determined using the featureCount tool and normalized using the Remove Unwanted Variation method.48 The differentially expressed genes (DEGs) were identified using the DESeq2 programme.49 Benjamini–Hochberg method was used to calculate the false discovery rate, and a cut-off level of <0.05 was considered significant. Normalized count per million values were used to generate the heatmaps and volcano plots using the Rstudio package. The Circos maps were plotted using the GOCHORD function in the R package.
Gene set enrichment analysis (GSEA, version 2.2.3) was utilized to identify the dysregulated biological pathways. Normalized enrichment score, determined from the molecular signature database (MSigDB) 3.0, was used to curate gene sets to identify the Hallmark canonical pathways. The DEGs were also analysed using the upstream regulator analysis function of the Ingenuity Pathway Analysis software (IPA®, QIAGEN, Redwood City, CA, USA). The targets of the specific transcriptional regulators were identified using the IPA programme, and those with a P-value of <0.05 for overlap with the IPA target genes and a predicted Z score of <−2 or >2 were considered dysregulated.
The conventional statistical analysis was performed as published.11,50 The Gaussian distribution of each set of data was analysed by the Shapiro–Wilk normality test and the normally distributed were compared between two groups by the t-test and among multiple groups by one-way ANOVA followed by Bonferroni pairwise comparisons. Data that deviated from a Gaussian distribution were compared by the Kruskal–Wallis test. The categorical data were compared by the Fisher exact or the χ2 test. Kaplan–Meier survival plots were compared using the Log-rank test. Statistical analyses were performed using Graph pad Prism 9 or STAT IC, 15.1.
3. Results
3.1. Study groups
Seven genotypes were generated, which were comprised of WT, Myh6-McmTam, Myh6-McmTam:DspF/F, Myh6-McmTam:Ctnnb1LoF, Myh6-McmTam:Ctnnb1GoF, Myh6-McmTam:DspF/F:Ctnnb1LoF, and Myh6-McmTam:DspF/F:Ctnnb1GoF. The phenotype in Myh6-McmTam, Myh6-McmTam:DspF/F, Myh6-McmTam:Ctnnb1LoF, and Myh6-McmTam:Ctnnb1GoF have been published.11,32,40 Therefore, to avoid redundancy and for clarity, only the four main groups of WT, Myh6-McmTam:DspF/F, Myh6-McmTam:DspF/F:Ctnnb1LoF, and Myh6-McmTam:DspF/F:Ctnnb1GoF are included.
3.2. Dysregulation of the cWNT pathway in the Myh6-McmTam:DspF/F cardiac myocytes
A total of 12 762 genes were differentially expressed in the Myh6-McmTam:DspF/F as compared to the WT cardiac myocytes, comprised of >10 000 protein-coding and >1000 genes coding for the long coding RNAs.11 Three hundred forty-six genes whose transcript levels were affected by the transient expression and activation of Cre recombinase and tamoxifen injection were removed from the dataset.40 Analysis of the remaining DEGs showed enrichment for the cWNT (TCF7L2/β-catenin) target genes in the Myh6-McmTam:DspF/F myocytes, comprised of 214 up-regulated and 68 down-regulated genes (Figure 1A and B). The dysregulated cWNT target genes predicted the activation of EMT and inflammatory pathways, including TNFα, TP53, and apoptosis, among others (Figure 1C).
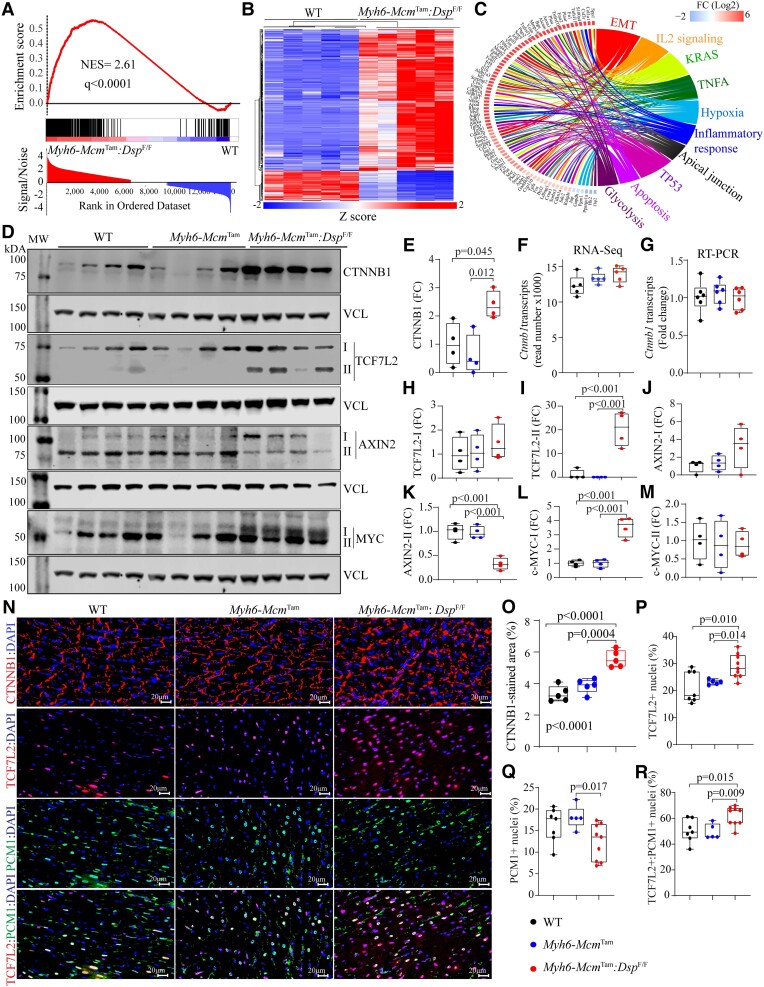
Figure 1.
Dysregulated cWNT pathway in cardiac myocytes in a mouse model of DSP cardiomyopathy. (A) GSEA predicting activation of the cWNT/TCF7L2/β-catenin target genes in the Myh6-McmTam:DspF/F mouse cardiac myocytes. (B) Heat map of the cWNT target genes. (C) Circos map showing pathways predicted to be affected by the cWNT target genes in the Myh6-McmTam:DspF/F mouse cardiac myocytes. (D) Immunoblots showing protein levels of selected key molecules in the cWNT pathway, showing either up-regulation of the main or the alternative isoforms. (E) Quantification of the β-catenin protein levels. (F and G) Transcript levels of the Ctnnb1 gene in the RNA-Seq data and determined by RT–PCR in an independent set of RNA extracts. (H–M) Quantitative data are corresponding to TCF7L2 and MYC proteins and their isoforms, which were shown in (D). (N–R). Immunofluorescence panels showing expression of β-catenin (CTNNB1), TCF7L2, and PCM1 in thin myocardial sections along with the corresponding quantitative data. The mean values among the groups were compared by ANOVA followed by Bonferroni correction for multiple comparisons. Only significant P values (Bonferroni corrected) are depicted in the panels throughout all figures. Sample size: Each dot in the panels represents one independent sample throughout the figures. Whenever a membrane is probed for multiple test proteins, the same blot for the loading control is included in the figures. The same color symbols in all quantitative panels are used for consistency and only one set of symbols is listed in each panel to avoid redundancy.
The level of β-catenin (CTNNB1) protein, a key protein in the cWNT pathway, was increased in the Myh6-McmTam:DspF/F mouse cardiac myocytes, whereas the transcript level of the Ctnnb1 gene was unchanged, indicating a post-transcriptional regulation (Figure 1D–M). In addition, alternative isoforms of TCF7L2 protein, a co-transcriptional regulator of the cWNT pathway, AXIN2 (Axis inhibition), the bona fide target of the pathway, and MYC were increased (Figure 1D–M), whereas the levels of the main isoforms of these proteins were unchanged (Figure 1D–M). Immunofluorescence staining of thin myocardial sections corroborated increased levels of β-catenin at the IDs and increased nuclear localization of TCF7L2 in cardiac myocytes (Figure 1N–R). Moreover, the number of myocardial cells expressing PCM1, which mainly marks cardiac myocytes, was reduced in the Myh6-McmTam:DspF/F mouse hearts (Figure 1N and Q).
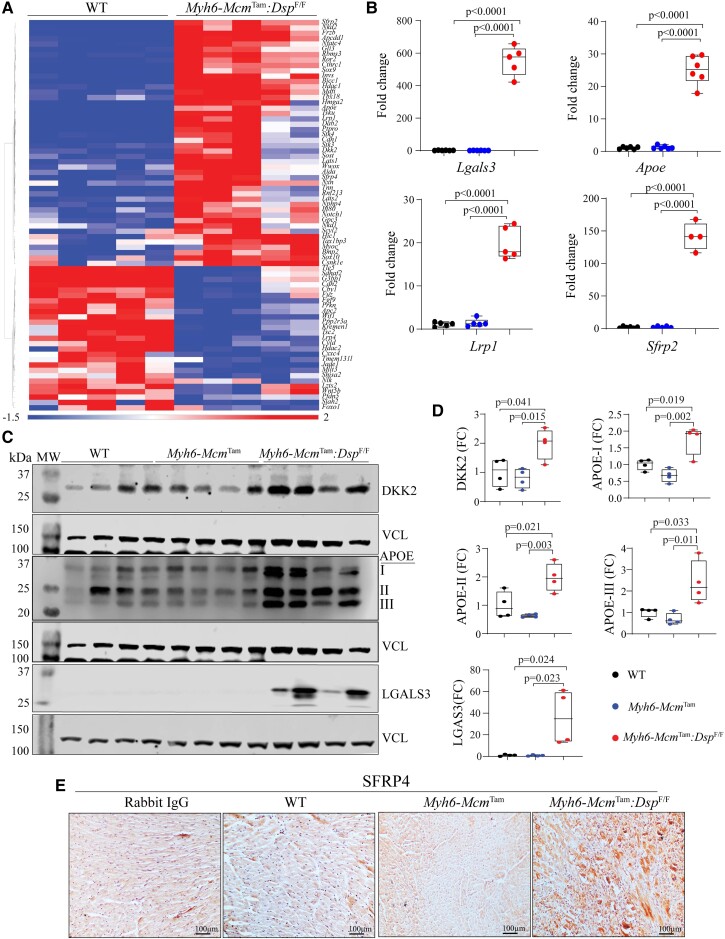
Transcript levels of 73 inhibitors/suppressors of the cWNT/β-catenin pathway were also changed (46 up-regulated and 27 suppressed) in the Myh6-McmTam:DspF/F cardiac myocytes (Figure 2A). Genes encoding secreted cWNT inhibitors Sfrp2 (Secreted frizzled-related protein 2), Dkk2 (Dickkopf-2), Tsku, Apoe, and Sost were up-regulated ,whereas Wnt5b, Wif1, and Fgf9, among others, were down-regulated (Figure 2A). Increased expression of selected inhibitors of the cWNT pathway was corroborated in independent RNA samples by RT–PCR (Figure 2B). In addition, levels of DKK2, APOE (Apolipoprotein E), and LGALS3 (Galectin 3) proteins were also increased in the myocardial protein extracts from the Myh6-McmTam:DspF/F mice (Figure 2C and D). Similarly, immunocytochemical staining of thin myocardial sections showed increased levels of SFRP4, a classic secreted cWNT inhibitor (Figure 2E).
Figure 2.
Up-regulation of the cWNT pathway inhibitors in the Myh6-McmTam:DspF/F cardiac myocytes and myocardium. (A) Heat map of the transcript levels of genes known to encode inhibitors of the cWNT pathways that are differentially expressed in the Myh6-McmTam:DspF/F cardiac myocytes. (B) Transcript levels of selected inhibitors of the cWNT determined by RT–PCR in independent samples. (C and D) Immunoblots showing protein levels of selected cWNT inhibitors and the corresponding quantitative data. (E) Immunohistochemistry panels showing the expression of SFRP4, a classic cWNT inhibitor, in the experimental groups. The mean values among the groups were compared by ANOVA followed by Bonferroni correction for multiple comparisons. Only significant P values (Bonferroni corrected) are depicted in the panels throughout all figures. Sample size: Each dot in the panels represents one independent sample throughout the figures. Whenever a membrane is probed for multiple test proteins, the same blot for the loading control is included in the figures. The same color symbols in all quantitative panels are used for consistency and only one set of symbols is listed in each panel to avoid redundancy.
3.3. Genetic activation or inactivation of β-catenin in cardiac myocytes in the Myh6-McmTam:DspF/F mice
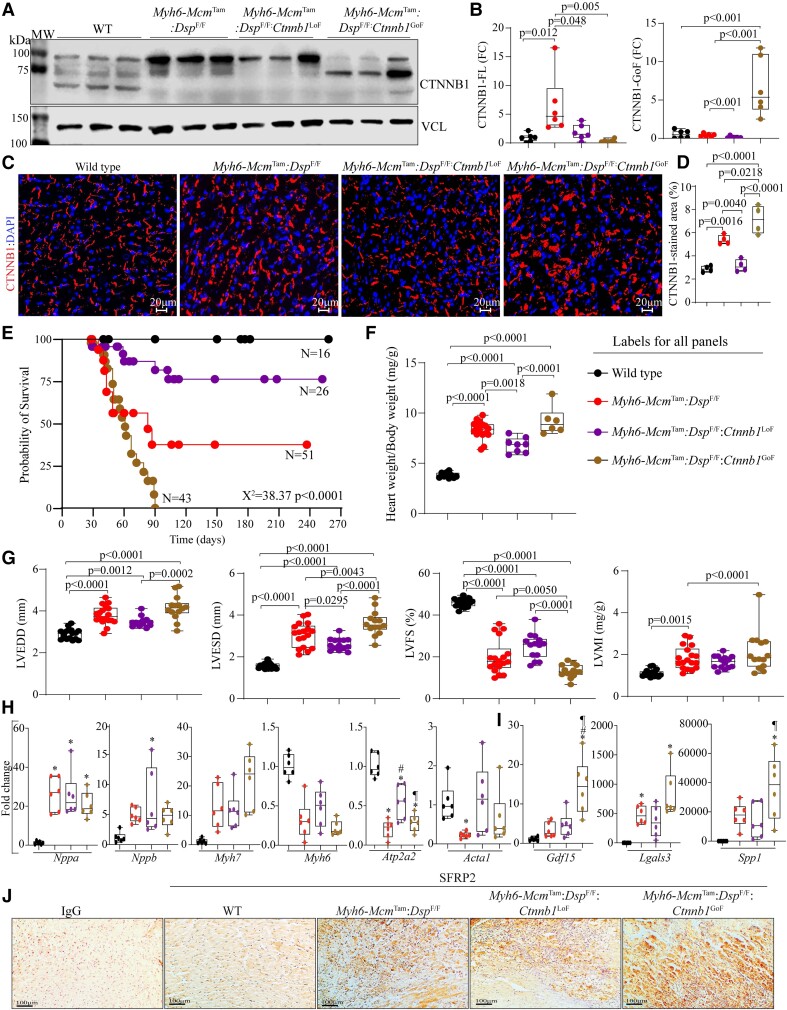
To discern the role of the dysregulated cWNT/β-catenin pathway in the pathogenesis of DSP cardiomyopathy, β-catenin was genetically activated or inactivated in cardiac myocytes in the Myh6-McmTam:DspF/F mice (see Supplementary material online, Figure S2). Consistent with the data in Figure 1, immunoblotting showed increased β-catenin levels in the Myh6-McmTam:DspF/F as compared to the WT mice (Figure 3A and B). Genetic LoF of β-catenin (Myh6-McmTam:DspF/F:Ctnnb1LoF) attenuated the up-regulated β-catenin level as compared to the Myh6-McmTam:DspF/F but did not normalize it as compared to the WT cardiac myocytes (Figure 3A and B). The level of the full-length β-catenin was reduced in the Myh6-McmTam:DspF/F:Ctnnb1GoF cardiac myocytes, but the level of the ∼75 kDa truncated β-catenin was increased, as expected (Figure 3A and B). The residual expression of full-length β-catenin is likely due to incomplete Cre-mediated excision, and the increased levels of the ∼75 kDa β-catenin is the consequence of the deletion of the degron domain (Figure 3A and B). Immunofluorescence staining of thin myocardial sections corroborated the findings by showing increased areas stained for β-catenin expression in Myh6-McmTam:DspF/F mouse hearts as compared to the WT mice, reduced areas in the Myh6-McmTam:DspF/F:Ctnnb1LoF, and augmented β-catenin stained areas in the Myh6-McmTam:DspF/F:Ctnnb1GoF mouse hearts (Figure 3C and D).
Figure 3.
Effects of genetic inactivation and activation of β-catenin on survival and cardiac function. (A) Immunoblot showing β-catenin expression in the WT, Myh6-McmTam:DspF/F, Myh6-McmTam:DspF/F:Ctnnb1LoF, and Myh6-McmTam:DspF/F:Ctnnb1GoF myocytes along with the vinculin (VCL), the latter as a control for loading conditions. (B) Quantitative data showing levels of full-length and stable β-catenin (exon 3 deleted, GoF) protein. (C and D) Immunofluorescence panels show expression and localization of the β-catenin in the myocardial sections along with the corresponding quantitative data. (E) Kaplan–Meier survival plots. (F) Heart weight/body weight ratio. (G) Selected echocardiographic indices, namely LV end-diastolic diameter (LVEDD), LV end-systolic diameter (LVESD), LV fractional shortening (LVFS), and LVMI. (H) Transcript levels of selected markers of hypertrophy and failure, as analysed by RT–PCR. (I) Transcript levels of selected secreted biomarkers of heart failure, as determined by RT–PCR. (J) Immunohistochemistry panels showing expression of SFRP2. Myocardial sections stained with Immunoglobulin G alone served as controls. The mean values among the groups were compared by ANOVA followed by Bonferroni correction for multiple comparisons. Only significant P values (Bonferroni corrected) are depicted in the panels throughout all figures. Sample size: Each dot in the panels represents one independent sample throughout the figures. Whenever a membrane is probed for multiple test proteins, the same blot for the loading control is included in the figures. The same color symbols in all quantitative panels are used for consistency and only one set of symbols is listed in each panel to avoid redundancy. * denotes P < 0.05 compared to WT, # P < 0.05 compared to Myh6-McmTam:DspF/F and ¶ P < 0.05 compared to Myh6-McmTam:DspF/F:Ctnnb1LoF.
3.4. Survival
The Myh6-McmTam:DspF/F mice died prematurely with a median survival of approximately 90 days, as published (Figure 3E).11 Genetic inactivation of β-catenin markedly prolonged survival in the Myh6-McmTam:DspF/F mice, as ∼75% of the Myh6-McmTam:DspF/F:Ctnnb1LoF mice survived up to 9 months (Figure 3E). A small number of mice in the Myh6-McmTam:DspF/F and Myh6-McmTam:DspF/F:Ctnnb1LoF groups survived beyond 90 days, which may reflect technical and experimental conditions, such as inefficient recombination or the intrinsic biological variability among mice in each genotype. In contrast, activation of β-catenin (Myh6-McmTam:DspF/F:Ctnnb1GoF) markedly reduced survival, leading to 100% mortality within 90 days of age (Figure 3E). The Myh6-McmTam, Myh6-McmTam:Ctnnb1LoF, and Myh6-McmTam:Ctnnb1GoF mice survived normally during the first 6 months of life.32,40
3.5. HW/BW
The heart weight (HW)/body weight (BW) ratio, calculated in 4-week-old mice, was increased in the Myh6-McmTam:DspF/F, as compared to the WT mice. It remained unchanged in the Myh6-McmTam:DspF/F:Ctnnb1LoF and Myh6-McmTam:DspF/F:Ctnnb1GoF mice as compared to the Myh6-McmTam:DspF/F but remained increased as compared to the WT mice (Figure 3F).
3.6. Cardiac size and function
The effects of LoF and GoF of β-catenin on cardiac size and function in the Myh6-McmTam:DspF/F mice were determined at 4 weeks of age by echocardiography. As reported before, deletion of Dsp was associated with severe cardiac dilatation and dysfunction consistent with the high mortality of the Myh6-McmTam:DspF/F (Figure 3G and Table 1).11 Genetic inactivation of β-catenin in the Myh6-McmTam:DspF/F mice was associated with the attenuation of cardiac dilatation and dysfunction, whereas its activation had the opposite effects (Figure 3G and Table 1). The LVM index (LVMI) remained increased in all groups with the deletion of the Dsp gene, compared to the WT mice (Figure 3G and Table 1).
Table 1.
Effects of genetic inactivation or activation of β-catenin on echocardiographic indices of cardiac size and function
| WT | Myh6-Mcm Tam:DspF/F | Myh6-Mcm Tam:DspF/F:Ctnnb1LoF | Myh6-Mcm Tam:DspF/F:Ctnnb1GoF | P | |
|---|---|---|---|---|---|
| N | 18 | 18 | 14 | 14 | NA |
| M/F | 9/9 | 9/9 | 7/7 | 7/7 | >0.9999* |
| Age (days) | 28.28 ± 0.46 | 28.06 ± 0.24 | 28.43 ± 0.65 | 28.50 ± 0.85 | 0.1814a |
| Body weight (g) | 17.71 ± 2.12 | 13.72 ± 2.38 | 13.47 ± 1.99 | 12.00 ± 1.83 | <0.0001 |
| HR (b.p.m.) | 657.71 ± 31.20 | 617.54 ± 52.65 | 654.52 ± 45.42 | 603.72 ± 49.84 | 0.022 |
| IVST (mm) | 0.37 ± 0.03 | 0.29 ± 0.03 | 0.30 ± 0.03 | 0.24 ± 0.02 | <0.0001 |
| LVPWT (mm) | 0.36 ± 0.03 | 0.29 ± 0.03 | 0.30 ± 0.03 | 0.25 ± 0.02 | <0.0001 |
| LVEDD (mm) | 2.91 ± 0.24 | 3.80 ± 0.45 | 3.47 ± 0.24 | 4.15 ± 0.58 | <0.0001a |
| LVEDDI (mm/g) | 0.17 ± 0.02 | 0.29 ± 0.07 | 0.26 ± 0.04 | 0.36 ± 0.10 | <0.0001a |
| LVESD (mm) | 1.57 ± 0.14 | 3.06 ± 0.58 | 2.60 ± 0.29 | 3.62 ± 0.59 | <0.0001a |
| LVESDI (mm/mg) | 0.09 ± 0.01 | 0.23 ± 0.07 | 0.20 ± 0.04 | 0.31 ± 0.09 | <0.0001a |
| FS (%) | 45.94 ± 2.01 | 19.94 ± 8.07 | 25.01 ± 6.03 | 13.12 ± 3.07 | <0.0001a |
| LV mass (mg) | 19.74 ± 3.99 | 24.47 ± 5.35 | 21.61 ± 2.85 | 24.21 ± 7.19 | 0.016a |
| LVMI (mg/g) | 1.11 ± 0.16 | 1.84 ± 0.55 | 1.63 ± 0.30 | 2.12 ± 0.96 | <0.0001a |
-Mcm, myosin heavy chain 6; Mer Cre Mer; Ctnnb1, β-catenin gene; LOF, loss of function β-catenin; GOF, gain of function β-catenin; M/F, male/female; HR, heart rate; IVST, diastolic interventricular septum thickness; LVPWT, posterior wall thickness during diastole; LVEDD, LV end-diastolic diameter; LVEDDi, LVEDD divided by the body weight; LVESD, LV end-systolic diameter; LVESDI, LVESD divided by body weight; FS, LV fractional shortening; LVM, LV mass; LVMI, LVM divided by the body weight.
a P value derived by Kruskal–Wallis test.
*χ2 P value.
Cardiac dysfunction is associated with the altered expression of several genes, including those whose protein products are commonly used as heart failure biomarkers. By and large, the LoF and GoF of β-catenin had no significant effects on the transcript levels of the conventionally used molecular markers except Atp2a2 (adenine triphosphate) level, which was reduced in the Myh6-McmTam:DspF/F mice; it was partially rescued in the Myh6-McmTam:DspF/F:Ctnnb1LoF and remained suppressed in the Myh6-McmTam:DspF/F:Ctnnb1GoF mouse hearts (Figure 3H). Similarly, transcript levels of the selected secreted biomarkers of heart failure, namely Gdf15, Lgals3, and Spp1, were elevated in the Myh6-McmTam:DspF/F mouse hearts and remained largely unaffected by the LoF and GoF of β-catenin (Figure 3I). Finally, immunohistochemical staining of thin myocardial sections showed increased levels of the SFRP2 in the Myh6-McmTam:DspF/F mouse myocardium, which remained increased in the myocardium of DSP-deficient mice with the LoF and GoF of β-catenin (Figure 3J).
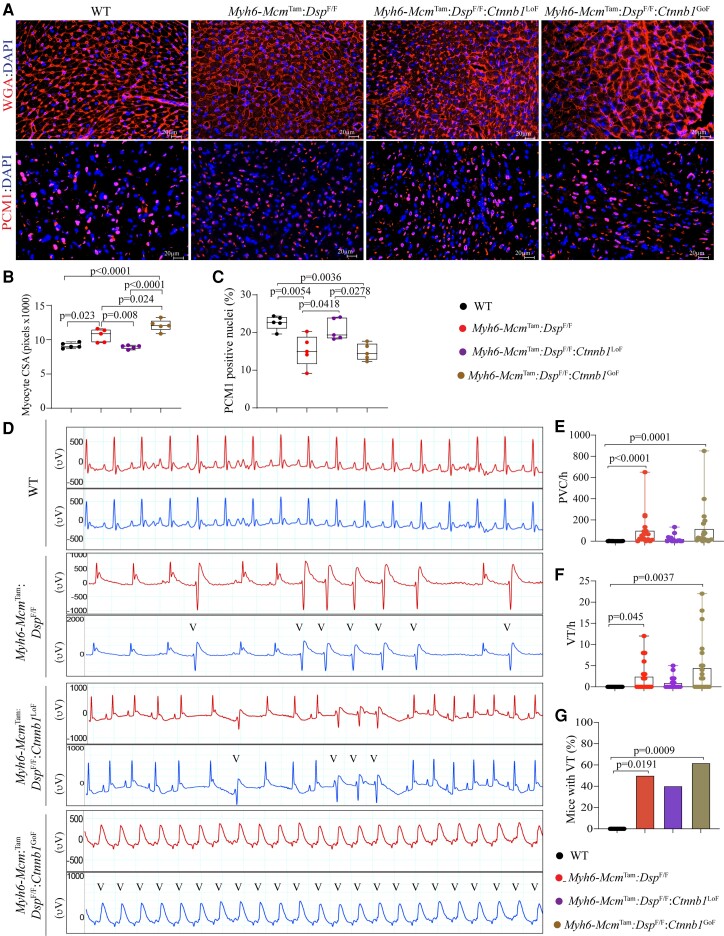
3.7. Myocyte size and number
Cardiac myocyte size and number were assessed upon co-staining of thin myocardial sections with WGA and anti-PCM1 antibody.32,39,42,46 The myocyte CSA was increased in the Myh6-McmTam:DspF/F mouse hearts (Figure 4A and B). It was normalized in the Myh6-McmTam:DspF/F:Ctnnb1LoF and further increased in the Myh6-McmTam:DspF/F:Ctnnb1GoF mouse hearts (Figure 4A and B). The number of cells expressing PCM1, a marker mainly for cardiac myocytes in the myocardium, was reduced in the Myh6-McmTam:DspF/F mouse heart, and it was normalized in Myh6-McmTam:DspF/F:Ctnnb1LoF and remained reduced in the Myh6-McmTam:DspF/F:Ctnnb1GoF mouse hearts (Figure 4A and C).
Figure 4.
Effects of genetic inactivation and activation of β-catenin on cardiac myocyte size and arrhythmias. (A) WGA and PCM1 co-stained thin myocardial sections were used to calculate myocyte CSA, which is depicted in (B). Quantitative data depicting myocyte CSA calculated from the WGA-stained sections and corrected for the number of myocytes. (C) The number of myocardial cells expressing PCM1, a marker mainly of cardiac myocytes in the heart. (D) Illustrates selected two-lead electrocardiograms showing the presence of ventricular ectopic beats in the experimental groups and their absence in the WT mice. (E–G) Quantitative data depict an increased number of PVCs, ventricular tachycardia (VT), and the number of mice exhibiting VT in the experimental but not the control groups. The mean values among the groups were compared by ANOVA followed by Bonferroni correction for multiple comparisons. Only significant P values (Bonferroni corrected) are depicted in the panels throughout all figures. Sample size: Each dot in the panels represents one independent sample throughout the figures. Whenever a membrane is probed for multiple test proteins, the same blot for the loading control is included in the figures.
3.8. Cardiac arrhythmias
Deletion of the Dsp gene in cardiac myocytes was associated with a significant increase in the prevalence of ventricular arrhythmias (Figure 4D and E). Premature ventricular contractions (PVCs) were the most common ventricular arrhythmias, and the number of PVCs per hour was increased by ∼15-fold in the Myh6-McmTam:DspF/F mice, albeit there was a wide range of variability (Figure 4D and E). Likewise, the prevalence of ventricular tachycardia, defined as ≥3 PVCs in the row, was increased markedly in the Myh6-McmTam:DspF/F, and ∼40% of these mice exhibited VT episodes during the rhythm monitoring period (Figure 4D, F, and G). The prevalence of ventricular arrhythmias trended lower in the Myh6-Mcm:DspF/F:Ctnnb1LoF, whereas it was the highest in the Myh6-Mcm:DspF/F:Ctnnb1GoF mice, albeit there was a large variability in the prevalence of arrhythmias (Figure 4D–G).
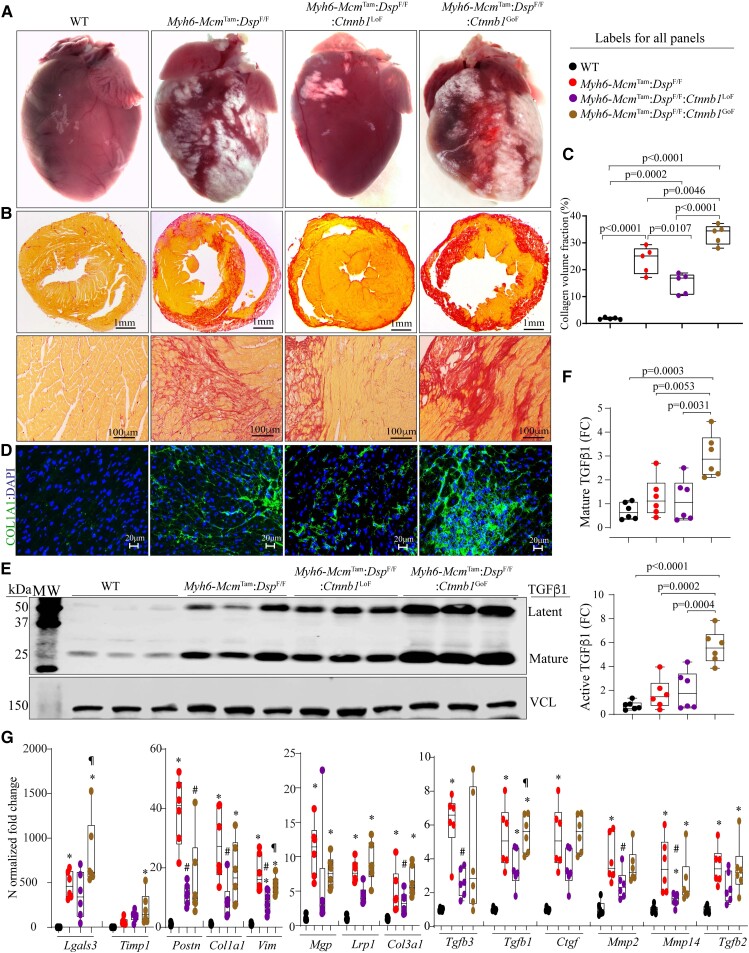
3.9. Myocardial fibrosis
The Myh6-McmTam:DspF/F mice exhibit severe myocardial fibrosis, in agreement with the phenotypic feature of DSP cardiomyopathy, as published.11 Myocardial fibrosis was markedly attenuated in the Myh6-McmTam:DspF/F:Ctnnb1LoF and exacerbated in the Myh6-McmTam:DspF/F:Ctnnb1GoF mice, as determined by multiple complementary methods (Figure 5). For example, CVF comprised ∼25% of the myocardium in the Myh6-McmTam:DspF/F mice; it was reduced to ∼15% in the Myh6-McmTam:DspF/F:Ctnnb1LoF and increased to ∼35% in the Myh6-McmTam:DspF/F:Ctnnb1GoF mice (Figure 5A–C). Immunostaining of thin myocardial sections for the expression of COL1A1, a major myocardial collagen, showed a similar pattern (Figure 5D). Moreover, immunoblotting of myocardial protein extracts probed with an anti-TGFβ1 (transforming growth factor beta 1) antibody showed increased levels of latent and mature TGFβ1 protein in the Myh6-McmTam:DspF/F and Myh6-McmTam:DspF/F:Ctnnb1LoF mice as compared to the WT mice and their further augmentations in the Myh6-McmTam:DspF/F:Ctnnb1GoF mice (Figure 5E and F).
Figure 5.
Effects of genetic inactivation and activation of β-catenin on myocardial fibrosis. (A) Exhibits gross heart morphology, showing extensive epicardial fibrosis (white areas) in the experimental groups. (B) Shows picrosirius red-stained thin myocardial cross-sections (upper panels) and high magnification thin myocardial sections (lower panels), used to calculate CVF. (C) Graph depicts CVF in the study groups. (D) Immunofluorescence panels showing myocardial sections stained with an antibody against COL1A1. (E and F) Immunoblots showing expression of TGFβ1, latent (upper panel), and active (lower panel) in the study groups along with the quantitative data for each isoform. (G) Transcript levels of selected genes involved in fibrosis as quantified by RT–PCR. Symbols as in Figure 4. The mean values among the groups were compared by ANOVA followed by Bonferroni correction for multiple comparisons. Only significant P values (Bonferroni corrected) are depicted in the panels throughout all figures. Sample size: Each dot in the panels represents one independent sample throughout the figures. Whenever a membrane is probed for multiple test proteins, the same blot for the loading control is included in the figures. The same color symbols in all quantitative panels are used for consistency and only one set of symbols is listed in each panel to avoid redundancy. * denotes P < 0.05 compared to WT, # P < 0.05 compared to Myh6-McmTam:DspF/F and ¶ P < 0.05 compared to Myh6-McmTam:DspF/F:Ctnnb1LoF.
Transcript levels of several genes involved in myocardial fibrosis, analysed by RT–PCR, which showed marked increases in the levels of Tgfb1, Tgfb2, Tgfb3, Postn, Pdgfra, Col1a1, Col1a3, Ctgf, Mmp2, Mmp14, Lrp1, Mgp, Vim, Timp1, and Lgals3 in Myh6-McmTam:DspF/F mouse hearts and the pattern of attenuation or augmentation in the Myh6-McmTam:DspF/F:Ctnnb1LoF and Myh6-McmTam:DspF/F:Ctnnb1GoF mice, respectively (Figure 5G).
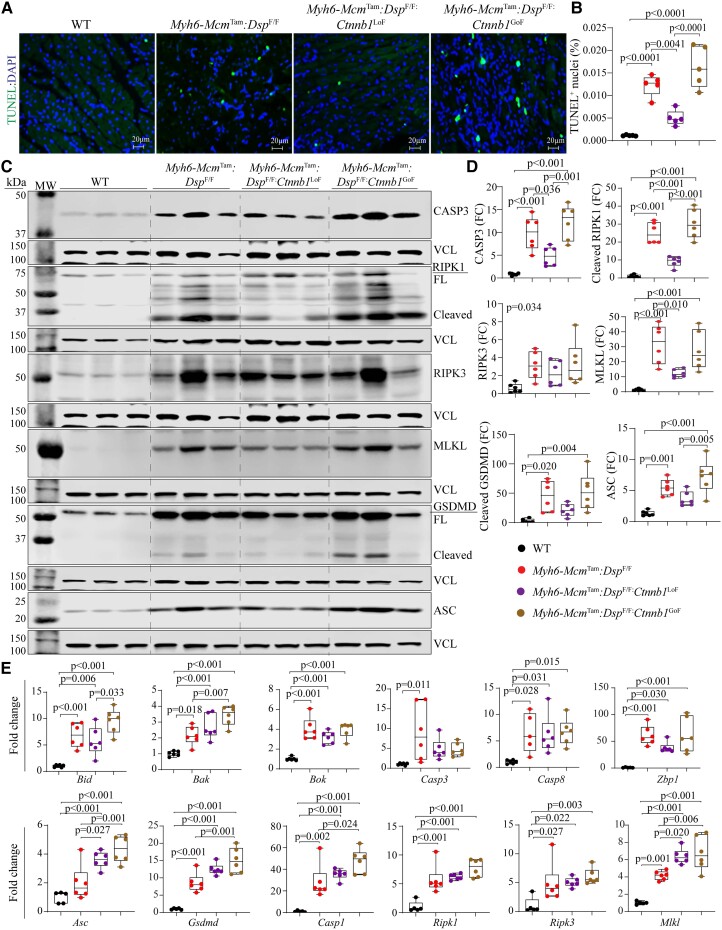
3.10. PANoptosis
PANoptosis, comprised of apoptosis, necroptosis, and pyroptosis, is a prominent phenotypic feature of the Myh6-McmTam:DspF/F mice in agreement with the prominence of cell death in humans with DSP cardiomyopathy.7,11 TUNEL assay showed an increased number of cells stained positive in the Myh6-McmTam:DspF/F mice, an attenuated number in the Myh6-McmTam:DspF/F:Ctnnb1LoF, and an increased number in the Myh6-McmTam:DspF/F:Ctnnb1GoF mouse hearts (Figure 6A and B). Immunoblot analysis of myocardial protein extracts showed increased levels of several proteins involved in apoptosis, necroptosis, and pyroptosis, namely CASP3 (Caspase 3), RIPK1 (Receptor-interacting serine/threonine-protein kinase 3) and 3, MLKL (mixed lineage kinase domain-like), GSDMD (Gasdermin D), and ASC (Apoptosis-associated speck-like protein containing a CARD) in the Myh6-McmTam:DspF/F mouse hearts, consistent with the previous data.11 The LoF of β-catenin attenuated but not normalized up-regulated expression levels of the above proteins, whereas the GoF of β-catenin further augmented the up-regulated proteins involved in selected cell death programmes (Figure 6C and D). The pattern of attenuation upon the LoF of β-catenin and augmented expression upon the GoF of β-catenin were consistent among the selected proteins.
Figure 6.
Effects of genetic inactivation and activation of β-catenin on PANoptosis. (A) Shows thin myocardial sections stained with the TUNEL assay and DAPI, the latter marks the DNA. (B) Quantitative data showing the percentage of myocardial cells stained positive for the TUNEL assay. (C and D) Immunoblot panels showing expression of selected proteins involved in PANoptosis and the corresponding quantitative data. CASP3 is used as a marker of apoptosis; RIPK1, RIPK3, and MLKL are used as markers of necroptosis; and GSDMD and ASC are used as markers of pyroptosis. (E) Shows transcript levels of a dozen genes involved in cell death programmes, as determined by RT–PCR. The mean values among the groups were compared by ANOVA followed by Bonferroni correction for multiple comparisons. Only significant P values (Bonferroni corrected) are depicted in the panels throughout all figures. Sample size: Each dot in the panels represents one independent sample throughout the figures. Whenever a membrane is probed for multiple test proteins, the same blot for the loading control is included in the figures.
Analysis of the transcript levels of a dozen genes involved in PANoptosis in isolated cardiac myocytes showed markedly increased levels in the Myh6-McmTam:DspF/F mice. However, the LoF and GoF of β-catenin did not have consistent effects on the transcript levels, partly reflective of the regulation of the expression levels of the selected markers at the post-transcriptional level (Figure 6E). However, transcript levels of several genes, such as Bak, Casp1, Mlkl, and Asc, were further up-regulated in the GoF group (Figure 6E).
3.11. Expressions and localization of desmosome proteins
The desmosomes undergo extensive remodelling in ACM, including in the Myh6-McmTam:DspF/F mice, which is evidenced by decreased levels of several key protein constituents of desmosomes and altered desmosome structures.11,15,51 To determine whether the LoF and GoF of β-catenin affected expression levels and localization of selected desmosome proteins, myocardial protein extracts were probed, and thin myocardial sections were stained with antibodies against the selected proteins. As reported previously, junction plakoglobin (JUP), Desmoglein 2, Desmocollin 2, and PKP2 (Plakophilin 2) proteins were reduced in the Myh6-McmTam:DspF/F mouse hearts and remained largely suppressed in the Myh6-McmTam:DspF/F:Ctnnb1LoF and Myh6-McmTam:DspF/F:Ctnnb1GoF mouse hearts, except for a modest increase in the JUP and PKP2 expressions in the LoF and further suppression in the GoF of β-catenin groups (see Supplementary material online, Figure S3A and B). The findings of immunofluorescence staining of myocardial sections were largely similar to the immunoblotting data (see Supplementary material online, Figure S3C).
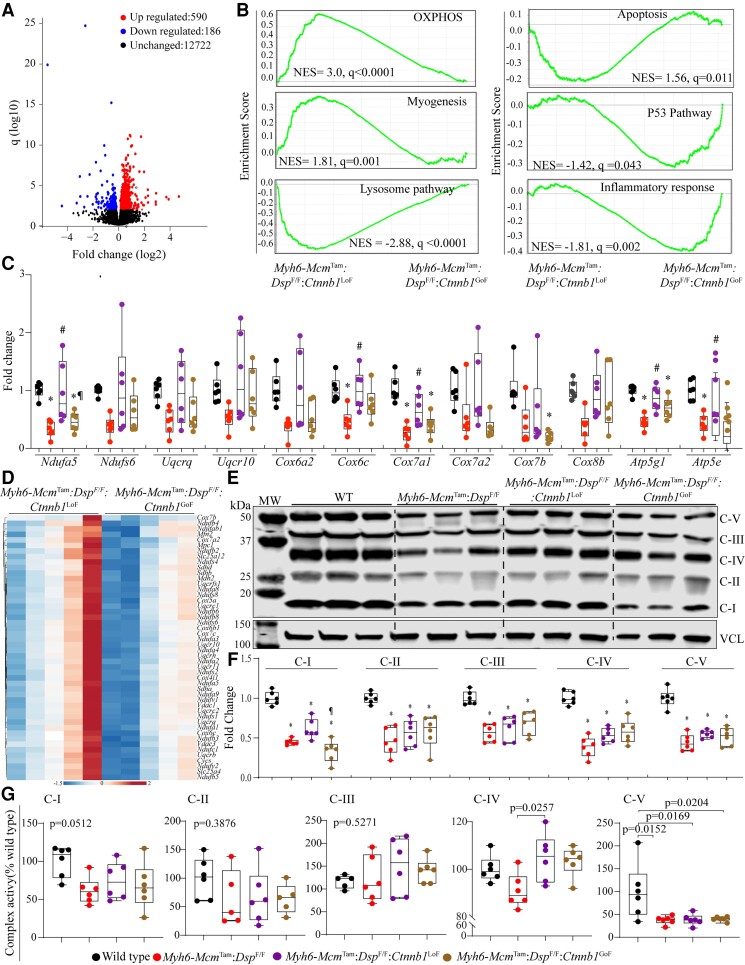
3.12. Transcriptional and biological pathways
To gain further insights into the mechanism(s) by which the LoF of β-catenin attenuates and the GoF exacerbates the phenotype in the Myh6-McmTam:DspF/F mice, transcripts of isolated cardiac myocytes were analysed by RNA-Seq. Genes (n = 346) whose expressions were affected by tamoxifen injection and expression and activation of Cre recombinase, as defined previously, were removed from the analysis.40 The quality control metrics of the RNA-Seq data are shown in Supplementary material online, Table S2, and those in the Myh6-McmTam alone, Myh6-McmTam:DspF/F, Myh6-McmTam:Ctnnb1LoF, and Myh6-McmTam:Ctnnb1GoF mice have been published.32,40
To determine whether phenotypic effects of LoF and GoF of β-catenin were reflective of changes in the transcriptional activity of the cWNT pathway, transcript levels of the recently defined cWNT target genes in the mouse cardiac myocytes were compared among the groups.32 Only a small fraction of the 1075 cWNT target genes in the mouse cardiac myocytes were changed upon LoF (15 genes) or GoF (144 genes) of β-catenin (see Supplementary material online, Figure S4). Comparing the cardiac myocyte transcripts between the Myh6-McmTam:DspF/F:Ctnnb1LoF and Myh6-McmTam:DspF/F:Ctnnb1GoF showed differential expression of only 46 cWNT target genes, comprised of 13 up-regulated and 33 down-regulated genes in the β-catenin LoF genotype (see Supplementary material online, Figure S4).
Upon exclusion of the cWNT transcriptional activity, cardiac myocyte transcripts were compared between the Myh6-McmTam:DspF/F and Myh6-McmTam:DspF/F:Ctnnb1LoF, which showed differential expression of 205 genes (see Supplementary material online, Figure S5A and B). The DEGs predicted increased activities of transcriptional regulators TP53 and NCOA1 and reduced activities of MYC, TP63, RB1, MYCN, and TP73 in the Myh6-McmTam:DspF/F:Ctnnb1LoF as compared to Myh6-McmTam:DspF/F myocytes (see Supplementary material online, Figure S5C). Given the relatively small number of DEGs, no further analysis was performed. A similar comparison between Myh6-McmTam:DspF/F and Myh6-McmTam:DspF/F:Ctnnb1GoF myocytes identified differential expression of 2610 genes (see Supplementary material online, Figures S6A and B). KDM5A, TP53, BACH1, and several other transcriptional regulators were predicted to be activated in the Myh6-McmTam:DspF/F:Ctnnb1GoF myocytes, whereas RB1, NFE2L2, and several others were predicted to be suppressed (see Supplementary material online, Figure S6C and D). The GSEA predicted activation of Interferon gamma response, EMT, and apoptosis and suppression of oxidative phosphorylation (OXPHOS), adipogenesis, and myogenesis in the Myh6-McmTam:DspF/F:Ctnnb1GoF myocytes (see Supplementary material online, Figure S6E).
To gain further insights into the pathways regulated by the β-catenin, which might contribute to the partial phenotypic rescue in the Myh6-McmTam:DspF/F mice, gene expression between the Myh6-McmTam:DspF/F:Ctnnb1LoF and Myh6-McmTam:DspF/F:Ctnnb1GoF were compared. There were 880 DEGs, which were comprised of 652 up-regulated and 228 down-regulated genes in the Myh6-McmTam:DspF/F:Ctnnb1LoF compared to the Myh6-McmTam:DspF/F:Ctnnb1GoF myocytes (Figure 7A and Supplementary material online, Figure S7A). The GSEA predicted suppression of the cWNT/β-catenin signalling pathway, indicating the fidelity of the LoF and GoF models and suppression of transcriptional regulators TP53 and KDM5A and activation of RB1 and PPARGC1A, among others in the Myh6-McmTam:DspF/F:Ctnnb1LoF myocytes (see Supplementary material online, Figure S7B and C). The DEGs also predicted suppression of the inflammatory pathways and activation of OXPHOS and metabolic pathways in the Myh6-McmTam:DspF/F:Ctnnb1LoF myocytes (see Supplementary material online, Figure S7D). Analysis of the transcripts by GSEA in the Myh6-McmTam:DspF/F:Ctnnb1LoF predicted enrichment of genes involved in OXPHOS, myogenesis, and suppression of the apoptosis, TP53, inflammatory responses, and lysosomal pathways, as compared to the Myh6-McmTam:DspF/F:Ctnnb1GoF myocytes (Figure 7B). Transcript levels of several genes involved in inflammation and lysosomal pathways were analysed by RT–PCR in independent samples, which showed increased levels of the transcripts in the Myh6-McmTam:DspF/F, attenuation of several but not all transcript levels in the Myh6-McmTam:DspF/F:Ctnnb1LoF, and further up-regulation in the Myh6-McmTam:DspF/F:Ctnnb1GoF (see Supplementary material online, Figure S8).
Figure 7.
Effects of genetic inactivation and activation of β-catenin on biological pathways. (A) Volcano plot showing DEGs between the Myh6-McmTam:DspF/F:Ctnnb1LoF and Myh6-McmTam:DspF/F:Ctnnb1GoF myocytes. (B) GSEA plots showing biological pathways (Hallmark signature) that were predicted to be differential between the Myh6-McmTam:DspF/F:Ctnnb1LoF and Myh6-McmTam:DspF/F:Ctnnb1GoF myocytes. (C) Data illustrate transcript levels of selected genes involved in OXPHOS in independent RNA extracts, as determined by RT–PCR. (D) Heat map of genes involved in OXPHOS, which were shown to be enriched in the Hallmark OXPHOS gene signature and were differentially expressed between the two groups. (E) Shows an OXPHOS blot of one selected protein in each of the five mitochondrial OXPHOS complexes detected using an antibody cocktail. (F) Quantitative data of mitochondrial proteins shown in (E) blots. Complex I is represented by NDUFB8, Complex II by SDHB, Complex III by UQCRC2, Complex IV by MTCO1, and Complex V by ATP5A. (G) Quantitative data of each OXPHOS protein in (F), normalized to vinculin (VCL). Symbols as in Figure 4. (G) Dot plot showing activities of five mitochondrial ECT in the experimental groups. Data are normalized to the corresponding activity of each complex in the WT cardiac myocytes. The mean values among the groups were compared by ANOVA followed by Bonferroni correction for multiple comparisons. Only significant P values (Bonferroni corrected) are depicted in the panels throughout all figures. Sample size: Each dot in the panels represents one independent sample throughout the figures. Whenever a membrane is probed for multiple test proteins, the same blot for the loading control is included in the figures. * denotes P < 0.05 compared to WT, # P < 0.05 compared to Myh6-McmTam:DspF/F and ¶ P < 0.05 compared to Myh6-McmTam:DspF/F:Ctnnb1LoF.
3.13. Mitochondrial OXPHOS
Given that the GSEA and pathway analysis predicted a partial rescue of OXPHOS as one of the several pathways involved in the phenotypic attenuation upon LoF of β-catenin in the Myh6-McmTam:DspF/F mice, transcript levels of a dozen genes involved in OXPHOS in cardiac myocytes were determined in independent samples, which showed suppressed levels in the Myh6-McmTam:DspF/F mouse myocytes as compared to the levels in the WT myocytes (Figure 7C). Likewise, a heat map of transcript levels of selected genes involved in OXPHOS in the Myh6-McmTam:DspF/F:Ctnnb1LoF and Myh6-McmTam:DspF/F:Ctnnb1GoF myocytes largely corroborated the partial rescues of genes involved in OXPHOS in the Myh6-McmTam:DspF/F:Ctnnb1LoF myocytes (Figure 7D). Overall, the LoF of β-catenin partially restored suppressed transcript levels of the OXPHOS genes; however, the transcript levels remained suppressed in the Myh6-McmTam:DspF/F:Ctnnb1GoF mice (Figure 7C and D). Moreover, immunoblotting of selected proteins in five OXPHOS complexes largely corroborated the findings of RNA-Seq and RT–PCR, showing reduced levels of the selected proteins in the Myh6-McmTam:DspF/F mouse hearts and a modest rescue of OXPHOS Complex I protein in the Myh6-McmTam:DspF/F:Ctnnb1LoF, whereas levels of the selected protein in other OXPHOS complexes remained suppressed (Figure 7E and F).
Finally, the activities of mitochondrial ECT Complexes I–V against the specific substrate of each complex were determined using commercial assays. Activities of Complexes 1, IV, and V were markedly reduced in the Myh6-McmTam:DspF/F mouse cardiac myocytes (Figure 7G). The LoF of β-catenin was associated with a modest restoration of Complex IV activity, whereas functions of the remaining OXPHOS complexes were unchanged (Figure 7G). The GoF of β-catenin did not significantly alter the activities of the mitochondrial OXPHOS complexes as compared to the Myh6-McmTam:DspF/F mouse cardiac myocytes and remained suppressed (Figure 7G).
4. Discussion
The findings are notable for the beneficial effects of reduced expression of β-catenin and the deleterious effects of increased expression of a stable β-catenin in a mouse model of ACM generated by tamoxifen-inducible deletion of the Dsp gene in the post-natal cardiac myocytes. The contrasting effects of LoF and GoF of β-catenin were remarkable for several phenotypes, including survival, cardiac function, arrhythmias, myocardial fibrosis, and the cell death programmes (PANaptosis). The beneficial effects of the suppression of β-catenin, however, were partial, as would be expected, given the multi-farity of the pathways involved in the pathogenesis of ACM and incomplete tamoxifen-induced Cre-mediated recombination. Likewise, the effects were judged to be independent of the role of the β-catenin in transcriptional activity of the cWNT pathway, as too few cWNT target genes were changed upon the LoF of β-catenin in cardiac myocytes, which contrasts with marked phenotypic benefits. The salubrious effects likely reflect the cumulative effects of incomplete restoration of multiple pathways, including cell death programmes and mitochondrial function.
The cWNT pathway, which is one of the most conserved pathways across the animal kingdom, is comprised of a large number of protein constituents that converge on the β-catenin/TCF7L2 transcriptional machinery to induce gene expression.31 The majority of the previous studies, including ours, have reported suppression of the cWNT pathway in ACM.16,19–21 Data in the present portrait a complex picture of marked dysregulation of the cWNT pathway, as indicated by concomitant up-regulation of the cWNT target genes as well as the well-established inhibitors of the cWNT pathway.52 In addition, isoforms of TCF7L2, the co-transcriptional effector of the cWNT pathway, and AXIN2, the bona fide target of the cWNT pathway, whose functions are poorly defined, were up-regulated. The data collectively indicate that the cWNT pathway is markedly dysregulated in the Myh6-McmTam:DspF/F cardiac myocytes.
Fidelity of the genetic LoF and GoF of β-catenin in cardiac myocytes in mice has been established.32–35 Likewise, the phenotypic consequences of short-term genetic activation or suppression of the β-catenin in mouse cardiac myocytes have been published.32 Overall, the LoF and GoF of β-catenin had no discernible effect on cardiac function, myocardial fibrosis, and cell death at 4 weeks old mice, the time point that corresponds to the mouse age in the present study.32 The findings were corroborated using complementary methods, which showed mainly concordant results. In addition, the contrasting phenotypic directions between the GoF and LoF of the β-catenin mice provide further credence to the findings. Finally, several quality control measures were incorporated into the study design, including the removal of the genes whose expressions were affected by the expression of Cre recombinase and the administration of tamoxifen from the RNA-Seq data before additional analysis of the RNA-Seq data.40
β-Catenin is an indispensable and non-redundant member of the cWNT pathway.31,53 Consequently, it has emerged as the most effective target for genetic manipulation of the cWNT pathway.31,53 Pharmacological inhibition of GSK3β, which phosphorylates β-catenin at its degron sites, have been reported to improve the ACM phenotype in mouse and zebrafish models.19–21 Likewise, pharmacological inhibition of the cWNT pathway by WNT974, which inactivates PORCN, responsible for palmitoylation and subsequent secretion of WNT ligand, exerts salutary effects.22 The findings of the present study, which is distinct from the previous studies and utilizes complementary genetic LoF and GoF approaches, provide multiple layers of evidence that LoF of β-catenin is beneficial and its GoF is deleterious in a mouse model of DSP cardiomyopathy.
β-Catenin is not only a co-transcriptional regulator of the cWNT signalling pathway but also has several non-canonical functions, including regulation of the Hippo, SRY-related HMG box, Forkhead box, T-box transcription factor 5, GATA4, Krüppel-like factor 15, Neurogenic locus notch homolog protein 1, and Hypoxia-inducible factor alpha pathways.15,24,54,55 It is also a major constituent of the IDs, where its C-terminal domain interacts with the cadherins and plays a crucial role in cell–cell adhesion and mechanotransduction.17,18 The increase in the β-catenin levels in the Myh6-McmTam:DspF/F mice likely reflects its junctional localization secondary to molecular remodelling of the IDs.11 β-Catenin and JUP are homologous proteins with a high level of amino acid identity and are partially interchangeable at the IDs and the nucleus.23,56,57 Moreover, β-catenin functions are regulated by extensive post-translational modifications, including phosphorylation in over 2-dozen serine, tyrosine, and threonine amino acids throughout the length of the protein by over a dozen kinases, acetylation, and ubiquitylation, among others.33,58 The regulatory role of the post-translational modification of β-catenin is well recognized for its flux through the AXIN-Adenomatous polyposis coli destruction scaffold, wherein co-ordinated phosphorylation of β-catenin degron by casein kinase 1, and GSK3β facilitate its ubiquitylation and degradation, whenever the WNT signalling is inactive.58 Pertinent to the present study, β-catenin is implicated in the regulation of OXPHOS, and inducible deletion of β-catenin has been shown to up-regulate expression of genes encoding protein constituents of the OXPHOS, particularly the Complex I proteins.32,59,60 The findings of the present study implicate partial recovery of suppressed OXPHOS, as a putative mechanism in the partial phenotypic rescue of the Myh6-McmTam:DspF/F:Ctnnb1LoF mice. Given the scant number of altered cWNT/β-catenin target gene expressions in the Myh6-McmTam:DspF/F:Ctnnb1LoF as compared to the Myh6-McmTam:DspF/F cardiac myocytes, the partial rescue upon inactivation of the β-catenin is likely independent of the transcriptional activity of the cWNT/β-catenin pathway.
The study has several limitations. Notable among them is the restriction of the findings to a single mouse model of cardiomyopathy whereby the Dsp gene was almost completely deleted. This contrasts with most human cardiomyopathy, which is typically caused by heterozygous variants in the DSP gene, albeit homozygous mutations in the DSP and other genes have been reported in humans with ACM.61–63 Likewise, the Dsp gene was deleted conditionally at post-natal day 14, in contrast to the human genotype whereby the mutation is present since the single-cell embryo formation. This approach was taken to avoid the confounding effects of activation or suppression of the cWNT/β-catenin pathway during embryogenesis, as the cWNT/β-catenin is known to affect cardiac development.16,36 Furthermore, DSP mutations cause a partially distinct phenotype notable for marked fibrosis and LV dysfunction, which was also observed in the present mouse model, but the findings might not be fully applicable to ACM caused by mutations in other genes. Moreover, the phenotypic rescue upon the LoF of β-catenin in the Myh6-McmTam:DspF/F mice was incomplete, in part because of the multi-farity of the involved mechanisms in the pathogenesis of DC and partly of partial deletion of the Ctnnb1 gene because of incomplete recombination event and residual expression of the full size β-catenin. Furthermore, no single dominant mechanism accounted for the salubrious effects of the LoF and the deleterious effects of GoF of β-catenin in these studies. The findings invoke multiple likely mechanisms, each imparting a partial effect, and the involvements of multiple biological pathways, including reduced cell death, increased myocyte number, improved mitochondrial function, and restoration of transcriptional regulators of gene expression. Thus, the findings do not lend themselves to targeting a specific mechanism to identify the main and the ancillary effects of the genetic LoF and GoF of β-catenin in the mouse model of ACM. Finally, the β-catenin is implicated in regulating energy metabolism and mitochondrial function, which were only partially investigated in the present study.64
In conclusion, the findings suggest that the genetic suppression of the β-catenin imparts beneficial effects on survival, cardiac function, arrhythmias, myocardial fibrosis, and cell death, whereas its activation is deleterious in a mouse model of DSP cardiomyopathy. The findings set the stage for pilot interventional studies to explore the effects of the suppression of the β-catenin on the phenotypic expression of ACM in larger animal models and subsequently in humans.
Supplementary material
Supplementary material is available at Cardiovascular Research online.
Supplementary Material
Acknowledgement
The authors thank the technical support from the Cancer Genomics Core funded by the Cancer Prevention and Research Institute of Texas (CPRIT RP180734).
Contributor Information
Melis Olcum, The Brown Foundation Institute of Molecular Medicine, McGovern Medical School, The University of Texas Health Science Center, Houston, TX 77030, USA; The Department of Internal Medicine, McGovern Medical School, The University of Texas Health Science Center, Houston, TX 77030, USA.
Siyang Fan, The Brown Foundation Institute of Molecular Medicine, McGovern Medical School, The University of Texas Health Science Center, Houston, TX 77030, USA; The Department of Internal Medicine, McGovern Medical School, The University of Texas Health Science Center, Houston, TX 77030, USA.
Leila Rouhi, The Brown Foundation Institute of Molecular Medicine, McGovern Medical School, The University of Texas Health Science Center, Houston, TX 77030, USA; The Department of Internal Medicine, McGovern Medical School, The University of Texas Health Science Center, Houston, TX 77030, USA.
Sirisha Cheedipudi, The Brown Foundation Institute of Molecular Medicine, McGovern Medical School, The University of Texas Health Science Center, Houston, TX 77030, USA; The Department of Internal Medicine, McGovern Medical School, The University of Texas Health Science Center, Houston, TX 77030, USA.
Benjamin Cathcart, The Brown Foundation Institute of Molecular Medicine, McGovern Medical School, The University of Texas Health Science Center, Houston, TX 77030, USA; The Department of Internal Medicine, McGovern Medical School, The University of Texas Health Science Center, Houston, TX 77030, USA.
Hyun-Hwan Jeong, The Brown Foundation Institute of Molecular Medicine, McGovern Medical School, The University of Texas Health Science Center, Houston, TX 77030, USA; The Department of Internal Medicine, McGovern Medical School, The University of Texas Health Science Center, Houston, TX 77030, USA.
Zhongming Zhao, The Brown Foundation Institute of Molecular Medicine, McGovern Medical School, The University of Texas Health Science Center, Houston, TX 77030, USA; The Department of Internal Medicine, McGovern Medical School, The University of Texas Health Science Center, Houston, TX 77030, USA.
Priyatansh Gurha, The Brown Foundation Institute of Molecular Medicine, McGovern Medical School, The University of Texas Health Science Center, Houston, TX 77030, USA; The Department of Internal Medicine, McGovern Medical School, The University of Texas Health Science Center, Houston, TX 77030, USA.
Ali J Marian, The Brown Foundation Institute of Molecular Medicine, McGovern Medical School, The University of Texas Health Science Center, Houston, TX 77030, USA; The Department of Internal Medicine, McGovern Medical School, The University of Texas Health Science Center, Houston, TX 77030, USA.
Authors’ contributions
M.O. performed most of the molecular biology and histological experiments, echocardiography, and electrocardiographic rhythm monitoring, analysed the data, and edited the manuscript. S.F. generated and established the mouse colony, performed molecular biology experiments, echocardiography, and cardiac rhythm monitoring, and prepared the samples for RNA-Seq. L.R. performed part of the RT–PCR, immunoblotting, and immunofluorescence studies and edited the manuscript. S.C. performed immunofluorescent staining. B.C. performed part of the molecular biology experiments. H.-H.J. mapped the RNA-Seq data to the mouse reference genome and performed the initial bioinformatics analysis of the RNA-Seq data. Z.Z. supervised the bioinformatics analysis of the RNA-Seq data. P.G. performed the secondary analysis of the RNA-Seq data, interpreted the findings, and edited the manuscript. A.J.M. developed the concept, supervised the experiments, interpreted the findings, and wrote the manuscript.
Funding
A.J.M. is supported by the National Heart, Lung, and Blood Institute (HL151737 and HL132401). P.G. is supported by R56HL165334-01.
Translational perspective.
The canonical WNT pathway is implicated in the pathogenesis of arrhythmogenic cardiomyopathy. Pharmacological activation of the canonical WNT pathway has shown beneficial effects in model organisms raising the prospect for the activation of this pathway as a therapeutic option. We provide compelling data that genetic activation of β-catenin is deleterious, whereas its genetic inactivation is salubrious by improving survival, cardiac function, arrhythmia, fibrosis, and cell death in desmoplakin cardiomyopathy. The mechanisms involved alterations in several pathways, including cell death programmes.
References
- 1. McNally EM, Mestroni L. Dilated cardiomyopathy: genetic determinants and mechanisms. Circ Res 2017;121:731–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Marian AJ. Molecular genetic basis of hypertrophic cardiomyopathy. Circ Res 2021;128:1533–1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Elliott PM, Anastasakis A, Asimaki A, Basso C, Bauce B, Brooke MA, Calkins H, Corrado D, Duru F, Green KJ, Judge DP, Kelsell D, Lambiase PD, McKenna WJ, Pilichou K, Protonotarios A, Saffitz JE, Syrris P, Tandri H, Te Riele A, Thiene G, Tsatsopoulou A, van Tintelen JP. Definition and treatment of arrhythmogenic cardiomyopathy: an updated expert panel report. Eur J Heart Fail 2019;21:955–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Marian AJ. Pathogenesis of diverse clinical and pathological phenotypes in hypertrophic cardiomyopathy. Lancet 2000;355:58–60. [DOI] [PubMed] [Google Scholar]
- 5. Bariani R, Cason M, Rigato I, Cipriani A, Celeghin R, De Gaspari M, Bueno Marinas M, Mattesi G, Pergola V, Rizzo S, Zorzi A, Giorgi B, Rampazzo A, Thiene G, Iliceto S, Perazzolo Marra M, Corrado D, Basso C, Pilichou K, Bauce B. Clinical profile and long-term follow-up of a cohort of patients with desmoplakin cardiomyopathy. Heart Rhythm 2022;19:1315–1324. [DOI] [PubMed] [Google Scholar]
- 6. Graziosi M, Ditaranto R, Rapezzi C, Pasquale F, Lovato L, Leone O, Parisi V, Potena L, Ferrara V, Minnucci M, Caponetti AG, Chiti C, Ferlini A, Gualandi F, Rossi C, Berardini A, Tini G, Bertini M, Ziacchi M, Biffi M, Galie N, Olivotto I, Biagini E. Clinical presentations leading to arrhythmogenic left ventricular cardiomyopathy. Open Heart 2022;9:e001914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Smith ED, Lakdawala NK, Papoutsidakis N, Aubert G, Mazzanti A, McCanta AC, Agarwal PP, Arscott P, Dellefave-Castillo LM, Vorovich EE, Nutakki K, Wilsbacher LD, Priori SG, Jacoby DL, McNally EM, Helms AS. Desmoplakin cardiomyopathy, a fibrotic and inflammatory form of cardiomyopathy distinct from typical dilated or arrhythmogenic right ventricular cardiomyopathy. Circulation 2020;141:1872–1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mohammed F, Chidgey M. Desmosomal protein structure and function and the impact of disease-causing mutations. J Struct Biol 2021;213:107749. [DOI] [PubMed] [Google Scholar]
- 9. Olson TM, Michels VV, Thibodeau SN, Tai YS, Keating MT. Actin mutations in dilated cardiomyopathy, a heritable form of heart failure. Science 1998;280:750–752. [DOI] [PubMed] [Google Scholar]
- 10. Di Lorenzo F, Marchionni E, Ferradini V, Latini A, Pezzoli L, Martino A, Romeo F, Iorio A, Bianchi S, Iascone M, Calo L, Novelli G, Mango R, Sangiuolo F. DSP-related cardiomyopathy as a distinct clinical entity? Emerging evidence from an Italian cohort. Int J Mol Sci 2023;24:2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Olcum M, Rouhi L, Fan S, Gonzales MM, Jeong HH, Zhao Z, Gurha P, Marian AJ. PANoptosis is a prominent feature of desmoplakin cardiomyopathy. J Cardiovasc Aging 2023;3:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yeruva S, Waschke J. Structure and regulation of desmosomes in intercalated discs: lessons from epithelia. J Anat 2023;242:81–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Vermij SH, Abriel H, van Veen TA. Refining the molecular organization of the cardiac intercalated disc. Cardiovasc Res 2017;113:259–275. [DOI] [PubMed] [Google Scholar]
- 14. Nielsen MS, van Opbergen CJM, van Veen TAB, Delmar M. The intercalated disc: a unique organelle for electromechanical synchrony in cardiomyocytes. Physiol Rev 2023;103:2271–2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chen SN, Gurha P, Lombardi R, Ruggiero A, Willerson JT, Marian AJ. The Hippo pathway is activated and is a causal mechanism for adipogenesis in arrhythmogenic cardiomyopathy. Circ Res 2014;114:454–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Garcia-Gras E, Lombardi R, Giocondo MJ, Willerson JT, Schneider MD, Khoury DS, Marian AJ. Suppression of canonical Wnt/beta-catenin signaling by nuclear plakoglobin recapitulates phenotype of arrhythmogenic right ventricular cardiomyopathy. J Clin Invest 2006;116:2012–2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Buckley CD, Tan J, Anderson KL, Hanein D, Volkmann N, Weis WI, Nelson WJ, Dunn AR. Cell adhesion. The minimal cadherin-catenin complex binds to actin filaments under force. Science 2014;346:1254211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chopra A, Tabdanov E, Patel H, Janmey PA, Kresh JY. Cardiac myocyte remodeling mediated by N-cadherin-dependent mechanosensing. Am J Physiol Heart Circ Physiol 2011;300:H1252–H1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Asimaki A, Kapoor S, Plovie E, Karin Arndt A, Adams E, Liu Z, James CA, Judge DP, Calkins H, Churko J, Wu JC, MacRae CA, Kleber AG, Saffitz JE. Identification of a new modulator of the intercalated disc in a zebrafish model of arrhythmogenic cardiomyopathy. Sci Transl Med 2014;6:240ra274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chelko SP, Asimaki A, Andersen P, Bedja D, Amat-Alarcon N, DeMazumder D, Jasti R, MacRae CA, Leber R, Kleber AG, Saffitz JE, Judge DP. Central role for GSK3beta in the pathogenesis of arrhythmogenic cardiomyopathy. JCI Insight 2016;1:e85923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Padron-Barthe L, Villalba-Orero M, Gomez-Salinero JM, Dominguez F, Roman M, Larrasa-Alonso J, Ortiz-Sanchez P, Martinez F, Lopez-Olaneta M, Bonzon-Kulichenko E, Vazquez J, Marti-Gomez C, Santiago DJ, Prados B, Giovinazzo G, Gomez-Gaviro MV, Priori S, Garcia-Pavia P, Lara-Pezzi E. Severe cardiac dysfunction and death caused by arrhythmogenic right ventricular cardiomyopathy type 5 are improved by inhibition of glycogen synthase kinase-3beta. Circulation 2019;140:1188–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cheedipudi SM, Fan S, Rouhi L, Marian AJ. Pharmacological suppression of the WNT signaling pathway attenuates age-dependent expression of the phenotype in a mouse model of arrhythmogenic cardiomyopathy. J Cardiovasc Aging 2021;1:10.20517/jca.2021.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lombardi R, da Graca Cabreira-Hansen M, Bell A, Fromm RR, Willerson JT, Marian AJ. Nuclear plakoglobin is essential for differentiation of cardiac progenitor cells to adipocytes in arrhythmogenic right ventricular cardiomyopathy. Circ Res 2011;109:1342–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rouhi L, Fan S, Cheedipudi SM, Braza-Boils A, Molina MS, Yao Y, Robertson MJ, Coarfa C, Gimeno JR, Molina P, Gurha P, Zorio E, Marian AJ. The EP300/TP53 pathway, a suppressor of the Hippo and canonical WNT pathways, is activated in human hearts with arrhythmogenic cardiomyopathy in the absence of overt heart failure. Cardiovasc Res 2022;118:1466–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Calore M, Lorenzon A, Vitiello L, Poloni G, Khan MAF, Beffagna G, Dazzo E, Sacchetto C, Polishchuk R, Sabatelli P, Doliana R, Carnevale D, Lembo G, Bonaldo P, De Windt L, Braghetta P, Rampazzo A. A novel murine model for arrhythmogenic cardiomyopathy points to a pathogenic role of Wnt signalling and miRNA dysregulation. Cardiovasc Res 2019;115:739–751. [DOI] [PubMed] [Google Scholar]
- 26. Giuliodori A, Beffagna G, Marchetto G, Fornetto C, Vanzi F, Toppo S, Facchinello N, Santimaria M, Vettori A, Rizzo S, Della Barbera M, Pilichou K, Argenton F, Thiene G, Tiso N, Basso C. Loss of cardiac Wnt/beta-catenin signalling in desmoplakin-deficient AC8 zebrafish models is rescuable by genetic and pharmacological intervention. Cardiovasc Res 2018;114:1082–1097. [DOI] [PubMed] [Google Scholar]
- 27. Roberts JD, Murphy NP, Hamilton RM, Lubbers ER, James CA, Kline CF, Gollob MH, Krahn AD, Sturm AC, Musa H, El-Refaey M, Koenig S, Aneq MA, Hoorntje ET, Graw SL, Davies RW, Rafiq MA, Koopmann TT, Aafaqi S, Fatah M, Chiasson DA, Taylor MR, Simmons SL, Han M, van Opbergen CJ, Wold LE, Sinagra G, Mittal K, Tichnell C, Murray B, Codima A, Nazer B, Nguyen DT, Marcus FI, Sobriera N, Lodder EM, van den Berg MP, Spears DA, Robinson JF, Ursell PC, Green AK, Skanes AC, Tang AS, Gardner MJ, Hegele RA, van Veen TA, Wilde AA, Healey JS, Janssen PM, Mestroni L, van Tintelen JP, Calkins H, Judge DP, Hund TJ, Scheinman MM, Mohler PJ. Ankyrin-B dysfunction predisposes to arrhythmogenic cardiomyopathy and is amenable to therapy. J Clin Invest 2019;129:3171–3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Beurel E, Grieco SF, Jope RS. Glycogen synthase kinase-3 (GSK3): regulation, actions, and diseases. Pharmacol Ther 2015;148:114–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wang L, Li J, Di LJ. Glycogen synthesis and beyond, a comprehensive review of GSK3 as a key regulator of metabolic pathways and a therapeutic target for treating metabolic diseases. Med Res Rev 2022;42:946–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Li G, Brumback BD, Huang L, Zhang DM, Yin T, Lipovsky CE, Hicks SC, Jimenez J, Boyle PM, Rentschler SL. Acute glycogen synthase kinase-3 inhibition modulates human cardiac conduction. JACC Basic Transl Sci 2022;7:1001–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nusse R, Clevers H. Wnt/beta-catenin signaling, disease, and emerging therapeutic modalities. Cell 2017;169:985–999. [DOI] [PubMed] [Google Scholar]
- 32. Olcum M, Cheedipudi SM, Rouhi L, Fan S, Jeong HH, Zhao Z, Gurha P, Marian AJ. The WNT/beta-catenin pathway regulates expression of the genes involved in cell cycle progression and mitochondrial oxidative phosphorylation in the postmitotic cardiac myocytes. J Cardiovasc Aging 2022;2:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Liu C, Li Y, Semenov M, Han C, Baeg GH, Tan Y, Zhang Z, Lin X, He X. Control of beta-catenin phosphorylation/degradation by a dual-kinase mechanism. Cell 2002;108:837–847. [DOI] [PubMed] [Google Scholar]
- 34. Harada N, Tamai Y, Ishikawa T, Sauer B, Takaku K, Oshima M, Taketo MM. Intestinal polyposis in mice with a dominant stable mutation of the beta-catenin gene. EMBO J 1999;18:5931–5942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Messerschmidt D, de Vries WN, Lorthongpanich C, Balu S, Solter D, Knowles BB. β-catenin-mediated adhesion is required for successful preimplantation mouse embryo development. Development 2016;143:1993–1999. [DOI] [PubMed] [Google Scholar]
- 36. Gallicano GI, Kouklis P, Bauer C, Yin M, Vasioukhin V, Degenstein L, Fuchs E. Desmoplakin is required early in development for assembly of desmosomes and cytoskeletal linkage. J Cell Biol 1998;143:2009–2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Soonpaa MH, Kim KK, Pajak L, Franklin M, Field LJ. Cardiomyocyte DNA synthesis and binucleation during murine development. Am J Physiol 1996;271:H2183–H2189. [DOI] [PubMed] [Google Scholar]
- 38. Senyo SE, Lee RT, Kuhn B. Cardiac regeneration based on mechanisms of cardiomyocyte proliferation and differentiation. Stem Cell Res 2014;13:532–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cheedipudi SM, Hu J, Fan S, Yuan P, Karmouch J, Czernuszewicz G, Robertson MJ, Coarfa C, Hong K, Yao Y, Campbell H, Wehrens X, Gurha P, Marian AJ. Exercise restores dysregulated gene expression in a mouse model of arrhythmogenic cardiomyopathy. Cardiovasc Res 2020;116:1199–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rouhi L, Fan S, Cheedipudi SM, Olcum M, Jeong HH, Zhao Z, Gurha P, Marian AJ. Effects of tamoxifen inducible MerCreMer on gene expression in cardiac myocytes in mice. J Cardiovasc Aging 2022;2:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Auguste G, Rouhi L, Matkovich SJ, Coarfa C, Robertson MJ, Czernuszewicz G, Gurha P, Marian AJ. BET bromodomain inhibition attenuates cardiac phenotype in myocyte-specific lamin A/C-deficient mice. J Clin Invest 2020;130:4740–4758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rouhi L, Cheedipudi SM, Chen SN, Fan S, Lombardi R, Chen X, Coarfa C, Robertson MJ, Gurha P, Marian AJ. Haploinsufficiency of Tmem43 in cardiac myocytes activates the DNA damage response pathway leading to a late-onset senescence-associated pro-fibrotic cardiomyopathy. Cardiovasc Res 2021;117:2377–2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Auguste G, Gurha P, Lombardi R, Coarfa C, Willerson JT, Marian AJ. Suppression of activated FOXO transcription factors in the heart prolongs survival in a mouse model of laminopathies. Circ Res 2018;122:678–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chen SN, Lombardi R, Karmouch J, Tsai JY, Czernuszewicz G, Taylor MRG, Mestroni L, Coarfa C, Gurha P, Marian AJ. DNA damage response/TP53 pathway is activated and contributes to the pathogenesis of dilated cardiomyopathy associated with LMNA (lamin A/C) mutations. Circ Res 2019;124:856–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rouhi L, Auguste G, Zhou Q, Lombardi R, Olcum M, Pourebrahim K, Cheedipudi SM, Asghar S, Hong K, Robertson MJ, Coarfa C, Gurha P, Marian AJ. Deletion of the lmna gene in fibroblasts causes senescence-associated dilated cardiomyopathy by activating the double-stranded DNA damage response and induction of senescence-associated secretory phenotype. J Cardiovasc Aging 2022;2:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bergmann O, Zdunek S, Alkass K, Druid H, Bernard S, Frisen J. Identification of cardiomyocyte nuclei and assessment of ploidy for the analysis of cell turnover. Exp Cell Res 2011;317:188–194. [DOI] [PubMed] [Google Scholar]
- 47. Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Risso D, Ngai J, Speed TP, Dudoit S. Normalization of RNA-seq data using factor analysis of control genes or samples. Nat Biotechnol 2014;32:896–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-Seq data with DESeq2. Genome Biol 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Cheedipudi SM, Asghar S, Marian AJ. Genetic ablation of the DNA damage response pathway attenuates lamin-associated dilated cardiomyopathy in mice. JACC Basic Transl Sci 2022;7:1232–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Liang Y, Lyon RC, Pellman J, Bradford WH, Lange S, Bogomolovas J, Dalton ND, Gu Y, Bobar M, Lee MH, Iwakuma T, Nigam V, Asimaki A, Scheinman M, Peterson KL, Sheikh F. Desmosomal COP9 regulates proteome degradation in arrhythmogenic right ventricular dysplasia/cardiomyopathy. J Clin Invest 2021;131:e137689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Cruciat CM, Niehrs C. Secreted and transmembrane Wnt inhibitors and activators. Cold Spring Harb Perspect Biol 2013;5:a015081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Rim EY, Clevers H, Nusse R. The Wnt pathway: from signaling mechanisms to synthetic modulators. Annu Rev Biochem 2022;91:571–598. [DOI] [PubMed] [Google Scholar]
- 54. Jin T, George Fantus I, Sun J. Wnt and beyond Wnt: multiple mechanisms control the transcriptional property of beta-catenin. Cell Signal 2008;20:1697–1704. [DOI] [PubMed] [Google Scholar]
- 55. Acar A, Hidalgo-Sastre A, Leverentz MK, Mills CG, Woodcock S, Baron M, Collu GM, Brennan K. Inhibition of Wnt signalling by Notch via two distinct mechanisms. Sci Rep 2021;11:9096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Butz S, Stappert J, Weissig H, Kemler R. Plakoglobin and beta-catenin: distinct but closely related. Science 1992;257:1142–1144. [DOI] [PubMed] [Google Scholar]
- 57. Bierkamp C, Schwarz H, Huber O, Kemler R. Desmosomal localization of beta-catenin in the skin of plakoglobin null-mutant mice. Development 1999;126:371–381. [DOI] [PubMed] [Google Scholar]
- 58. Verheyen EM, Gottardi CJ. Regulation of Wnt/beta-catenin signaling by protein kinases. Dev Dyn 2010;239:34–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Quaife-Ryan GA, Mills RJ, Lavers G, Voges HK, Vivien CJ, Elliott DA, Ramialison M, Hudson JE, Porrello ER. β-Catenin drives distinct transcriptional networks in proliferative and nonproliferative cardiomyocytes. Development 2020;147:dev193417. [DOI] [PubMed] [Google Scholar]
- 60. Balatskyi VV, Sowka A, Dobrzyn P, Piven OO. WNT/beta-catenin pathway is a key regulator of cardiac function and energetic metabolism. Acta Physiol (Oxf) 2023;237:e13912. [DOI] [PubMed] [Google Scholar]
- 61. Norgett EE, Hatsell SJ, Carvajal-Huerta L, Cabezas JC, Common J, Purkis PE, Whittock N, Leigh IM, Stevens HP, Kelsell DP. Recessive mutation in desmoplakin disrupts desmoplakin-intermediate filament interactions and causes dilated cardiomyopathy, woolly hair and keratoderma. Hum Mol Genet 2000;9:2761–2766. [DOI] [PubMed] [Google Scholar]
- 62. Alcalai R, Metzger S, Rosenheck S, Meiner V, Chajek-Shaul T. A recessive mutation in desmoplakin causes arrhythmogenic right ventricular dysplasia, skin disorder, and woolly hair. J Am Coll Cardiol 2003;42:319–327. [DOI] [PubMed] [Google Scholar]
- 63. McKoy G, Protonotarios N, Crosby A, Tsatsopoulou A, Anastasakis A, Coonar A, Norman M, Baboonian C, Jeffery S, McKenna WJ. Identification of a deletion in plakoglobin in arrhythmogenic right ventricular cardiomyopathy with palmoplantar keratoderma and woolly hair (Naxos disease). Lancet 2000;355:2119–2124. [DOI] [PubMed] [Google Scholar]
- 64. Balatskyi VV, Vaskivskyi VO, Myronova A, Avramets D, Abu Nahia K, Macewicz LL, Ruban TP, Kucherenko DY, Soldatkin OO, Lushnikova IV, Skibo GG, Winata CL, Dobrzyn P, Piven OO. Cardiac-specific beta-catenin deletion dysregulates energetic metabolism and mitochondrial function in perinatal cardiomyocytes. Mitochondrion 2021;60:59–69. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.