Abstract
This was a household-based prospective cohort study conducted in Rio de Janeiro, in which people with laboratory-confirmed coronavirus disease 2019 (COVID-19) and their household contacts were followed from April 2020 through June 2022. Ninety-eight reinfections were identified, with 71 (72.5%) confirmed by genomic analyses and lineage definition in both infections. During the pre-Omicron period, 1 dose of any COVID-19 vaccine was associated with a reduced risk of reinfection, but during the Omicron period not even booster vaccines had this effect. Most reinfections were asymptomatic or milder in comparison with primary infections, a justification for continuing active surveillance to detect infections in vaccinated individuals. Our findings demonstrated that vaccination may not prevent infection or reinfection with severe acute respiratory syndrome coronavirus 2 (SARS CoV-2). Therefore we highlight the need to continuously update the antigenic target of SARS CoV-2 vaccines and administer booster doses to the population regularly, a strategy well established in the development of vaccines for influenza immunization programs.
Keywords: SARS-CoV-2 infection, COVID-19, Omicron, reinfections, vaccine breakthrough, variants of concern
We followed families in Rio de Janeiro, from April 2020 to June 2022, to characterize the frequency and severity of SARSCoV-2 reinfection. Vaccination prevented severe illness and death throughout the study, but only reduced the risk of reinfection before the Omicron variant.
Billions of vaccine doses against coronavirus disease 2019 (COVID-19) have been administered worldwide, leading to a significant reduction in deaths and changing the course of the pandemic. Nevertheless, coronaviruses are associated with repeated infections [1–3]. Emerging severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants, with different mutations in the spike protein, display increased infectivity and immune escape [4], which is associated with increased risk of reinfection [5, 6].
Prior to the implementation of mass COVID-19 immunization, it was observed that infection-induced immunity resulted in a 10-fold reduction in reinfection risk within 6 months after primary virus exposure [7]. Clinical characterization regarding severity of SARS-CoV-2 reinfection in comparison to primary infection is a controversial subject [8]. Factors influencing SARS-CoV-2 reinfection are not yet fully understood.
Brazil has had the fifth highest absolute number of COVID-19 cases globally and 704 000 deaths reported as of July 2023, ranking second in the number of deaths worldwide [9]. The city of Rio de Janeiro was one of the epicenters of SARS-CoV-2 in Brazil, with 1.2 million cases (the second highest among Brazilian cities) and 37 000 deaths by the end of 2022 [10]. Brazil implemented an immunization program using the following SARS CoV-2 vaccines: BNT162b2 (Pfizer-BioNTech), ChAdOx1 (AstraZeneca/Oxford University), Ad26.COV2.S (Johnson & Johnson/Janssen), and CoronaVac (Sinovac Biotech).
Data on SARS-CoV-2 reinfection confirmed by genomic RNA detection in Brazil is monitored by the Brazilian Ministry of Health. As of January 2023, the most recent month for which data are available, there have been 125 laboratory-confirmed reinfection cases, of which 60 were due to the Omicron variant of concern (VOC) [11].
There is growing interest in identifying the frequency of repeated SARS-CoV-2 infections, including viral, clinical, immune, and social determinants potentially associated with this phenomenon. There is a need for a better understanding of the role of vaccine-induced protective immunity [12, 13]. In this study, we aimed to assess whether age, sex, race, comorbidities, occupation, living conditions, and vaccination status influenced the risk of SARS-CoV-2 reinfections in Rio de Janeiro, before and after the emergence of the Omicron VOC.
METHODS
Study Design
This was a prospective cohort of laboratory-confirmed SARS-CoV-2 infected individuals and their household contacts. Participants with laboratory-confirmed SARS-CoV-2 by a real-time reverse transcription polymerase chain reaction (RT-PCR) test (Kit Molecular SARS-CoV2; Bio-Manguinhos) and their household contacts were recruited at a public, secondary-care hospital in Rio de Janeiro from 7 May 2020 to 30 June 2022. The inclusion criteria were positive RT-PCR or reagent antigen results for SARS-CoV-2 within up to 7 days of symptoms or being a household contact of a confirmed case. Upon recruitment, sociodemographic and clinical variables (age, sex, race, occupation, comorbidities, and vaccination status), and household information (number of rooms and number of people in the household) were collected by phone interview. The first home visit was carried out within 7 days after the onset of symptoms in the index case. Subsequent home visits were conducted 7, 14, 28, and 42 days later, and then quarterly for 2 years. In addition to regularly scheduled visits, sick visits were conducted whenever participants reported new symptoms. During visits, upper respiratory tract (nasopharynx and oropharynx) swabs and saliva samples were collected and tested for SARS-CoV-2 RNA via RT-PCR. Data was recorded in questionnaires through REDCap (Research Electronic Data Capture) at every visit.
SARS-CoV-2 Whole-Genome Sequencing and Lineage Definition
SARS-CoV-2–positive samples with cycle threshold (Ct) up to 27 were selected for whole-genome sequencing (WGS) [5, 14–16] (Supplementary Material). SARS-CoV-2 lineages were classified by the PangoLineages tool [17].
Real-Time RT-PCR Inference for SARS-CoV-2 Variant Assay
The identification of SARS-CoV-2 variants using the real-time RT-PCR inference assay (Kit Molecular SARS-CoV-2; Bio-Manguinhos) was performed through the detection of the target viral nucleocapsid (N) gene and the human ribonuclease P (RP) gene as an internal control. The assays detect the presence or absence of the following deletions: S106del, G107del, and F108del, in the ORF1a gene (nsp6) and the spike gene target failure at positions H69del and V70del. The analysis of the presence or absence of these mutations combined with the epidemiological period allowed us to infer specific SARS-CoV-2 variants, as described in Table 1.
Table 1.
Scheme for Interpretation of Real-Time RT-PCR Inference Assay for SARS-CoV-2 Variants Based on the Presence or Absence of Specific Targets and the Epidemiological Scenario
| S106del, G107del, and F108del (nsp6) |
H69del and V70del (Spike) |
Probable VOC/Lineage based on the Molecular Epidemiology of SARS-CoV-2 in Rio de Janeiro |
|---|---|---|
| Presence | Presence | Alpha (samples collected from February 2021 to June 2021) |
| Omicron BA.1 (samples collected from December 2021 to May 2022) Omicron BA.4 or BA.5 (samples collected in or after May 2022) |
||
| Presence | Absence | Gamma (samples collected from January 2021 to June 2021) |
| Omicron BA.2 (samples collected from February 2022 to July 2022) | ||
| Absence | Absence | Pre VOCs lineages (samples collected from March 2020 to February 2021) |
| Delta (samples collected from June 2021 to December 2021) |
Definition of Reinfection
We defined reinfection as 2 SARS-CoV-2 infectious episodes caused by distinct SARS-CoV-2 lineages detected or inferred by laboratory assays (WGS or RT-PCR inference assay). In cases where the suspected reinfection had a viral load too low for performance of WGS, the case was classified as a reinfection when the repeat infection occurred over 90 days after the initial episode, and there was a negative SARS-CoV-2 RT-PCR result between the 2 episodes. We included the time from the subject's last vaccine to reinfection based on the date specimens were collected for RT-PCR. The date of the positive RT-PCR result was chosen instead of the date of symptom onset because the reinfection definition relied on laboratory confirmation while some cases were asymptomatic.
When the viral load was insufficient for WGS (Ct > 27 by real-time RT-PCR), the period when the sample was collected was used to infer the probable SARS-CoV-2 lineage or variant, based on epidemiological data on lineages circulating in the state of Rio de Janeiro. SARS-CoV-2 was first detected in Rio de Janeiro in March 2020. The B.1.1.33 was the predominant lineage until October 2020 [18], followed by the former Zeta (P.2) variant of interest (VOI), which predominated until January 2021 [19], when the Gamma VOC (P.1) was introduced, and vaccination against COVID-19 in health care workers and seniors was initiated. Gamma dominated the epidemiological scenario until June 2021. Subsequently, the Delta VOC replaced Gamma and predominated until November 2021, after which it was replaced by Omicron BA.1/BA.2 VOCs [20]. The cutoff date for Omicron introduction in Rio de Janeiro was defined as 1 December 2021 based on epidemiological curves of cases and hospitalizations [10]. Therefore, for further analysis, the dataset was subdivided into pre-Omicron and Omicron periods with the data censored on 30 June 2022.
Disease Severity
We defined disease severity based on the National Institutes of Health clinical spectrum categories [21]. For the purposes of data analysis, we combined the mild and moderate categories.
Immunization Status
Participants were defined as partially vaccinated when they received only 1 dose of BNT162b2, ChAdOx1, or CoronaVac and as fully vaccinated if they received 2 doses of these vaccines or a single dose of Ad26.COV2.S. Participants were considered boosted if at least 1 booster vaccine dose was administered to a fully vaccinated participant. For statistical analysis, we classified participants as either “fully vaccinated or boosted” or “unvaccinated or partially vaccinated” during the Omicron and pre-Omicron periods.
Statistical Analysis
We tabulated the number of SARS-CoV-2 infections and reinfections by lineages and variants and compared sociodemographic and household characteristics stratified by the pre-Omicron (before December 2021) versus Omicron (December 2021 or later) periods and reinfection status. Categorical variables were reported as frequencies and numeric variables as median, minimum, and maximum. The incidence rate and potential risk factors for reinfection were assessed by random effects Poisson models, in which the time between first infection and reinfection or last negative SARS-CoV-2 real-time RT-PCR result was used as an offset (log scale). These models were adjusted to consider the effect of people per room, which was shown to be the main household-specific risk factor for SARS-CoV-2 infection in a previous study in this population [22]. In addition, the random effects attributed to the household took into account dependence among participants within each household and other potentially relevant unmeasured variables. Common Gaussian random effects were used for participant families evaluating correlations among members of the same family. Gaussian random effects for participants were used because some participants had more than 1 reinfection during the pre-Omicron phase. Age was modelled as a continuous variable using a spline-like approach. To allow a nonlinear interaction between age and the reinfection rate, a second-order Gaussian random walk was used. Vague priors were used for intercept and regression coefficients. Penalized complexity priors were used for the Gaussian random walk precision. The prior probability of a standard deviation for a Gaussian random walk being greater than 2 was 10%. Data were analyzed with R 4.1.3. Risk factor analysis was performed with the R package INLA [23].
Ethics Statement
All participants provided signed informed consent. This study was approved by Brazil's National Research Ethics Committee (number 30639420.0.0000.5262).
RESULTS
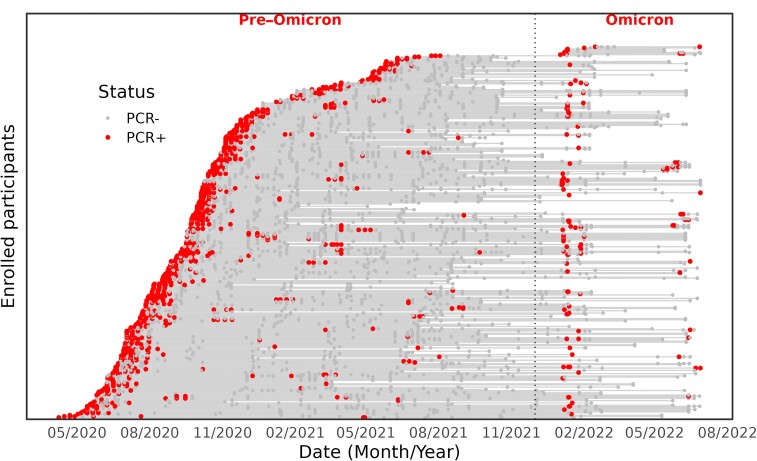
From May 2020 to June 2022, a total of 684 participants were recruited, with 374 confirmed SARS-CoV-2 infections. There were 98 reinfections and 4 cases with a third infectious episode. The median time between infections was 248 days (range, 90–385 days) in the pre-Omicron period, 431 days (range, 132–733 days) during the Omicron period, and 391 days (range, 90–733 days) for both periods. Follow-up of the 94 individuals who were reinfected is depicted in Figure 1.
Figure 1.
Timing of infections for reinfected and nonreinfected individuals; virological results for each participant (n = 331). The follow-up period for each individual in the cohort is represented by a horizontal gray line, starting from the recruitment date. The lines are stacked on each other to form a sloping curve. The vertical dashed line separates the pre-Omicron and Omicron periods. The red dots represent each participant's positive reverse transcription polymerase chain reaction (RT-PCR) result and gray dots represent negative RT-PCR results.
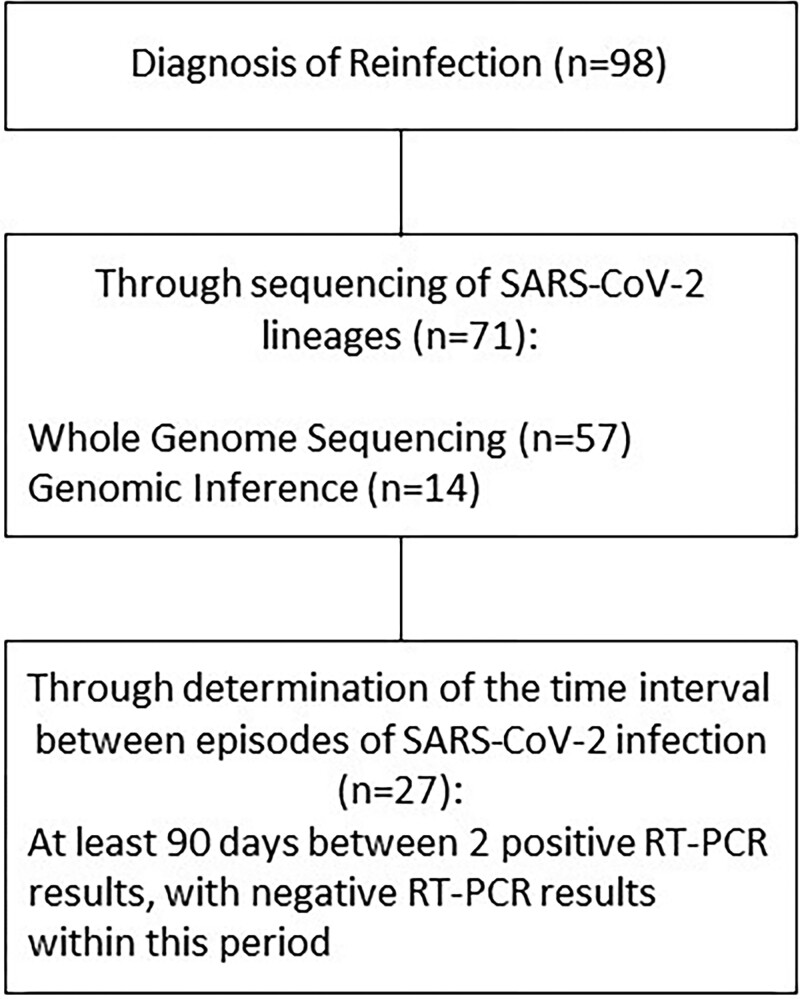
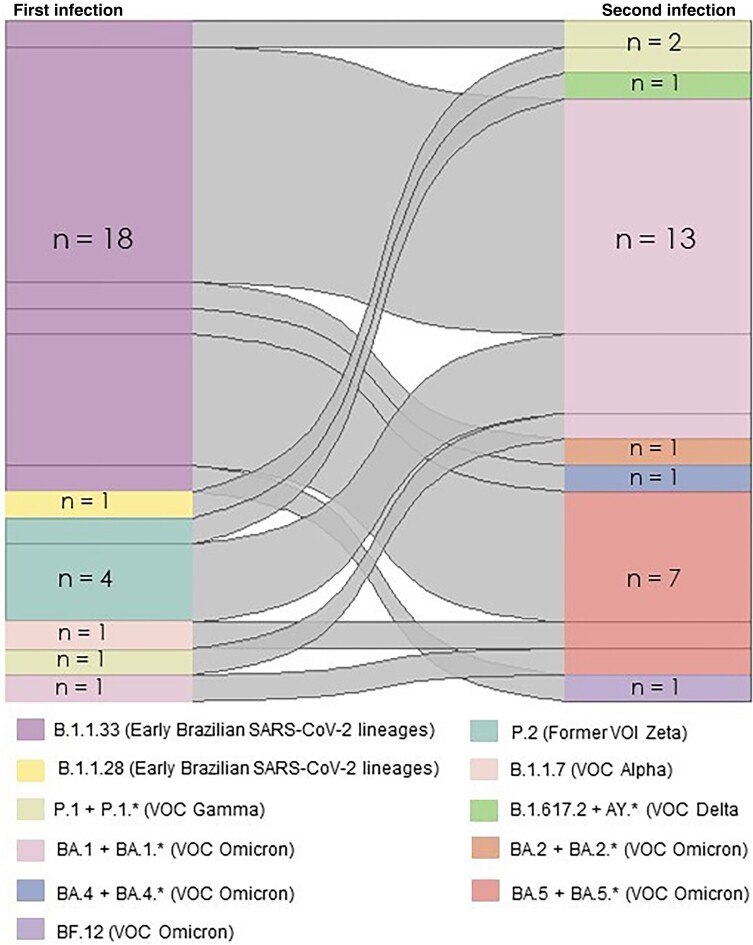
SARS-CoV-2 lineages or variants were genetically characterized in 72.5% (n = 71/98) of samples derived from reinfection cases; 77.46% of these (n = 55/71) had associated variants confirmed by WGS or inference in both primary and reinfection episodes (Figure 2). In 11 reinfection cases, genomic evidence of Zeta, Gamma, Delta, and Omicron variants, which were not circulating at the time of the primary infection, was identified. In 5 cases, only SARS-CoV-2 lineages from the primary infection were identified by WGS, classified as pre-VOC (B.1.1.33, B.1.1.28, and P.2), whereas corresponding reinfections predominantly occurred in the period of VOC emergence, encompassing Gamma (P.1), Delta, and Omicron.
Figure 2.
Characterization of SARS-CoV-2 reinfection episodes. Reinfection was diagnosed by the identification of distinct SARS-CoV-2 lineages in naso/oropharyngeal swabs using whole-genome sequencing or genomic inference. When viral identification was not possible, reinfection was defined as 2 positive RT-PCR results occurring within an interval of at least 90 days, with negative RT-PCR results within this interval. Abbreviations: RT-PCR, reverse transcription polymerase chain reaction; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Cases of reinfection occurred throughout all COVID-19 waves associated with distinct SARS-CoV-2 lineages. Most reinfections occurred during the Omicron BA.1/BA.2 waves (74 of 98, 75.5%). Before that, we observed a reinfection frequency of 7.4% (24 of 327 infections), which increased to 38.7% (74 of 191 infections) after Omicron became dominant. During the Zeta, Gamma, and Delta waves, we found 4, 9, and 10 reinfection episodes, respectively. In contrast, during Omicron BA.1 and BA.2 waves, we observed 57 and 17 episodes of reinfection, respectively (Figure 3). Two primary infection cases occurred during the first Omicron wave, associated with the BA.1 variant, whereas reinfections occurring in the second Omicron wave were associated with BA.2.
Figure 3.
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) lineages responsible for the first SARS-CoV-2 infectious episode and corresponding SARS-CoV-2 lineages responsible for the second infection. Different lineages, identified by whole-genome sequencing, are represented by different tones; the width of the linkages is proportional to the number of people infected with each SARS-CoV-2 lineage.
When comparing reinfected and nonreinfected individuals, no significant association was found with sex, age, race, occupation as a health care worker, number of rooms in the house, and the family size in the pre-Omicron or Omicron periods (Table 2). No differences were noted when assessing the presence of most comorbidities (obesity, hypertension, diabetes, cardiovascular disease, asthma and other pulmonary diseases, and chronic renal diseases), except for chronic rhinitis, which differed between reinfected and nonreinfected individuals in the pre-Omicron period.
Table 2.
Characteristics of Participants Reinfected From April 2020 to June 2022, Stratified by the Pre-Omicron and Omicron Periods
| Pre-Omicrona (n = 327 Infections) |
Omicronb (n = 191 Infections) |
|||
|---|---|---|---|---|
| Characteristic | No Reinfection (n = 303) |
Reinfection (n = 24) |
No Reinfection (n = 117) |
Reinfection (n = 74) |
| Time since infection, d, median (min–max)c | 377 (92–766) | 262 (90–385) | 525 (122–766) | 454 (132–733) |
| Sex | ||||
| Female | 171 (56) | 14 (58) | 61 (52) | 43 (58) |
| Male | 132 (44) | 10 (42) | 56 (48) | 31 (42) |
| Age, y | ||||
| Median (min–max) | 40 (0–94) | 44 (5–80) | 37 (0–91) | 42 (5–87) |
| 0–4 | 12 (4.0) | 0 (0) | 9 (7.7) | 0 (0) |
| 5–11 | 16 (5.3) | 2 (8.3) | 7 (6.0) | 2 (2.7) |
| 12–17 | 13 (4.3) | 2 (8.3) | 6 (5.1) | 3 (4.1) |
| 18–29 | 45 (15) | 5 (21) | 18 (15) | 14 (19) |
| 30–59 | 164 (54) | 13 (54) | 53 (45) | 45 (61) |
| >60 | 53 (17) | 2 (8.3) | 24 (21) | 10 (14) |
| Race | ||||
| Black or more than one race | 122 (41) | 8 (33) | 38 (33) | 39 (53) |
| White | 179 (59) | 16 (67) | 78 (67) | 35 (47) |
| Unknown | 2 | 0 | 1 | 0 |
| Comorbidities | ||||
| Chronic rhinitis | 31 (10) | 5 (21) | 15 (13) | 11 (15) |
| Obesity | 40 (13) | 4 (17) | 12 (10) | 10 (14) |
| Diabetes | 28 (9.2) | 2 (8.3) | 12 (10) | 7 (9.5) |
| Hypertension | 80 (26) | 6 (25) | 31 (26) | 19 (26) |
| Cardiovascular disease | 10 (3.3) | 1 (4.2) | 5 (4.3) | 4 (5.4) |
| Chronic lung disease | 16 (5.3) | 4 (17) | 6 (5.1) | 3 (4.1) |
| Chronic kidney disease | 2 (0.7) | 0 (0) | 1 (0.9) | 1 (1.4) |
| Health care worker | 62 (20) | 7 (29) | 23 (20) | 14 (19) |
| Time since vaccination, d, median (min–max; IQR)c | 71 (15–528; 41–113) | 46 (18–92; 26–70) | 133 (15–274; 76–178) | 104 (27–281; 75–149) |
| No. of vaccine dosesc | ||||
| Unvaccinated or partially vaccinated | 110 (36) | 20 (83) | 18 (15) | 4 (5) |
| Fully vaccinated | 144 (48) | 4 (17) | 35 (30) | 33 (45) |
| Booster | 49 (16) | 0 | 64 (55) | 37 (50) |
| Vaccines receivedc | ||||
| AstraZeneca | 81 (27) | 4 (17) | 25 (21) | 9 (12) |
| Coronavac | 40 (13) | 3 (12) | 5 (4.3) | 7 (9.5) |
| Janssen | 8 (2.6) | 0 | 4 (3.4) | 2 (2.7) |
| Pfizer | 99 (33) | 1 (4.2) | 68 (58) | 53 (72) |
| None | 75 (25) | 16 (67) | 15 (13) | 3 (4.1) |
| People per room in the household, median (min–max) | 0.6 (0.2–2.0) | 0.6 (0.3–2.0) | 0.5 (0.2–2.0) | 0.6 (0.25–2.0) |
Data are No. (%) except where indicated.
Abbreviations: IQR, interquartile range; PCR, polymerase chain reaction.
aParticipants = 323; families = 152.
bParticipants = 191; families = 101.
cFor the nonreinfected group, based on the date of the last PCR assay performed for the study and for the reinfected participants, based on the date of the last positive PCR.
In the pre-Omicron period, 83% of reinfected participants were unvaccinated or partially vaccinated, 17% were fully vaccinated (with 1 dose of the Ad26.COV2.S or 2 doses of any other vaccine authorized in Brazil), and none had received a booster dose (Table 2). In the Omicron period, 5% of reinfected patients were unvaccinated (or partially vaccinated), and 45% were fully vaccinated, with 50% of fully vaccinated participants having received at least 1 booster dose. The median time between the last vaccination and reinfection pre-Omicron was 46 days (interquartile range [IQR], 26–70 days) and during Omicron was 104 days (IQR, 75–149 days). The median time between the first and second dose was 84 days (range, 20–148 days), and the median time between the second dose and the booster dose was 165 days (range, 70–534 days).
In the pre-Omicron period, at least 1 vaccine dose was protective against reinfection (relative risk: 12.70; 95% credible interval [CI], 5.06–36.21), and chronic rhinitis was a risk factor for reinfection. However, in the Omicron period, booster doses did not show a protective effect against reinfection (0.73; CI, .40–1.29). In both periods, persons per room and being a health care worker were not associated with reinfection (Table 3).
Table 3.
Risk Factors for SARS-CoV-2 Reinfection
| Variable | Pre-Omicron | Omicron |
|---|---|---|
| People per room | 0.59 (0.13–2.60) | 0.99 (0.49–1.97) |
| Chronic rhinitis | 2.93 (1.01–8.64) | 1.06 (0.55–2.08) |
| Health care worker | 2.58 (0.95–7.13) | 0.70 (0.37–1.33) |
| Vaccination status: unvaccinated or partially vaccinateda | 17.81 (5.72–56.91) | 0.49 (0.17–1.44) |
The model incorporated participant age and family and individual random effects to account for dependence among observations. The data indicate the relative risk of reinfection associated with each variable (+/- 95% credible interval). Variables in bold were associated with a significant increase in the risk of reinfection.
aFully vaccinated or booster were aggregated and used as baseline in both pre-Omicron and Omicron models.
Most infections in both periods were mild/moderate (67.4%), followed by asymptomatic infections (29.3%), and severe/critical disease (3.3%). Symptomatic infections occurred in 71% of participants in the pre-Omicron period and in 68% of participants in the Omicron period. Most cases in both periods were mild/moderate: 66.6% and 68.6%, respectively. In the pre-Omicron period, 5.8% of infections were severe/critical, while no severe cases occurred in the Omicron period.
Regarding primary infections, the proportion of asymptomatic infections was 32.7%, mild/moderate disease 64.7%, and severe disease 2.6%, whereas among reinfections, 46.5% were asymptomatic infections, 53.5% were mild/moderate, and none were severe.
When comparing unvaccinated and vaccinated participants, a higher proportion of asymptomatic infections among fully vaccinated participants was noted (41.5%) in contrast to unvaccinated or partially vaccinated participants (27.5%). Among unvaccinated/partially vaccinated participants, 5.7% of infections were severe, whereas only 1.2% of infections were severe in fully vaccinated participants.
DISCUSSION
The introduction of the SARS-CoV-2 Omicron VOC in December 2021 increased the risk of reinfection with SARS-CoV-2 in Rio de Janeiro. During the Omicron period, when half of participants had taken the first booster vaccination, the occurrence of reinfection (38.7%) was approximately 5 times higher than in the pre-Omicron period (7.5%), when one-third of participants were not yet vaccinated. The higher risk of reinfection in the Omicron period was also observed in Italy [24] and in South Africa, but in a scenario of lower vaccine coverage and a higher infection rate in the latter [25].
Among nonreinfected participants in the pre-Omicron period, two-thirds (64%) were fully vaccinated or had received a booster dose. In this group, protection against reinfection was likely due to hybrid immunity (vaccination and infection), which stimulates greater antibody and T-cell responses than infection or vaccination alone [26]. Among reinfected participants, in the same period, secondary episodes occurred on average 8 months after the initial infection, while 83% of reinfected individuals were unvaccinated or partially vaccinated, and only 17% were fully vaccinated. During Omicron, nonreinfected participants had a median duration of follow-up since the last vaccine dose of approximately 4 months. Among participants who were reinfected, the duration of follow-up after vaccination was also 4 months. In this group, reinfections occurred on average 14 months after the first SARS CoV-2 infection and less than 4 months after the last vaccine dose. During the Omicron period, reinfected and nonreinfected participants did not differ with respect to vaccines administered or the number of vaccine doses. Monovalent vaccines did not confer long-lasting protection against infection with Omicron lineages, likely due to Omicron's antigenic divergence and higher immune evasion [27, 28]. Our previous findings demonstrated that, in this population, neutralizing antibodies elicited by pre-Omicron VoCs had limited ability to neutralize Omicron BA.1 [29]. Most reinfections occurred in vaccinated individuals and were asymptomatic or milder in comparison to primary infections. This provides further support for the finding that vaccination prevents severe illness and death [27, 30] but may not prevent infection or reinfection.
It is unclear whether the selective pressure imposed by immunization in combination with high rates of virus cocirculation after removal of mitigation policies favored the emergence of new SARS-CoV-2 lineages. Omicron is known to have 15 mutations in the receptor-binding domain, while Delta has only 4 mutations in the same region [4, 31]. Although the origin of Omicron is unclear, it has been hypothesized that novel SARS-CoV-2 lineages may evolve during prolonged infection in immunocompromised individuals [32]. Its antigenic divergence has made Omicron more successful in evading host immune defenses than previous VOCs. Our results support the hypothesis that Omicron's selective advantage and increased ability to infect previously exposed individuals is related to antigenic divergence and waning levels of neutralizing antibodies after infection and/or immunization. The demonstration of higher susceptibility of vaccinated individuals to reinfection by Omicron VOCs even after vaccine boosting was corroborated by Andeweg et al [33], who showed that protection against COVID-19 conferred by previous infection or vaccination was lower for VOCs Omicron BA.1 and BA.2 than for Delta. In addition, similar to our results, these authors showed that vaccine boosting increased protection against Omicron infection, although it rapidly decreased thereafter, thus creating the opportunity for reinfection with emerging SARS-CoV-2 variants.
On average, 391 days elapsed between primary infection and the reinfection episode. This is in accordance with data shown by Almadhi et al [34], who reported that most reinfections occurred at least 9 months after prior infection. It is known that the risk of infection increases with the time elapsed since vaccination and/or previous infection, which is directly influenced by antibody decay [35]. The effectiveness of previous SARS-CoV-2 infection against reinfection with Omicron was shown in one study to be reduced to 25% at 12 months, while protection conferred by hybrid immunity waned to 42%, with booster vaccination unable to restore protection [30]. We observed 2 reinfection cases in our study population during the Omicron wave, within an interval of 90 days, coinciding with the transition in the circulation of the BA.1 to the BA.2 Omicron variant, confirming the immune escape of BA.2 from specific responses targeting the BA.1 variant, as demonstrated by others [36, 37]. The variant-specific risk of SARS-CoV-2 infection was recently evaluated by Nilles et al [38], who showed that increasing anti-spike levels are necessary to protect against symptomatic infection by B.1.621 (Mu), Delta, BA.1, BA.2, and BA.4/5 variants, in order of increasing level of anti-spike required.
Previous studies have reported a wide range of SARS-CoV-2 reinfection rates, which likely resulted from differences in study design, the definition used for reinfection, and characteristics of the study populations. Flacco et al [39] reported an overall reinfection rate of 0.97% but a much higher reinfection rate of 3.31% during the Omicron period. In Iceland, Eythorsson et al [40] reported a reinfection rate of 11.7% with Omicron in individuals who were unvaccinated or who received 1 vaccine dose, and a 10.9% reinfection rate for those who had received 2 or more doses. The frequency of reinfection in our study (7.5% pre-Omicron and 38.7% during the Omicron period) was higher than what has been previously reported. Our reinfection rate may be higher because of our systematic search for SARS-CoV-2 infection by PCR testing throughout the pandemic period.
While previous studies have reported that allergic conditions such as rhinitis reduce the risk of SARS-CoV-2 infection [41], we found it was associated with a greater risk of reinfection in the pre-Omicron period. However, comorbidities were not risk factors for reinfection in the Omicron period. According to a recent meta-analysis, the risk of reinfection with SARS-COV-2 is no higher among health care workers than other occupations [7, 39]. This agrees with our finding that health care workers did not have a significantly higher rate of reinfection. The infection risk among health care workers seemed to be related to virus circulation in the community and not occupational exposure.
The limitations of our study are related to sample size. Due to limited numbers of participants, it may not have been possible to detect the impact of comorbidities on the risk of reinfection, nor to run the model stratified by time among participants vaccinated more than 4 months prior to infection. Because most of our cases were mild, we could not estimate the risk of reinfection by disease severity. Nor was it possible to determine whether recurrent SARS-CoV-2 infections are more transmissible than primary infections. Another limitation is that we did not perform serologic assays measuring vaccine titers. Such data could have helped determine if reinfection was associated with waning antibody levels. The strengths of our study include the long-term follow-up of participants infected with SARS-CoV-2 and the stored genetic material from the first infectious episode, which allowed confirmation of reinfections through viral genomic sequencing, the gold standard to define reinfection. We performed genetic characterization of specimens from reinfection episodes and we had negative RT-PCR results in the period between infections. This approach enabled greater certainty that reinfection episodes were in fact new infections and not prolonged viral shedding from the first infection.
A further strength is that we have followed this cohort since the beginning of the pandemic in Brazil, during consecutive waves of COVID-19 associated with emerging SARS-CoV-2 variants (including Gamma, which became predominant in Brazil in early 2021 but was less abundant in the United States and Europe). The follow-up period also included the rollout of vaccination.
A contribution of this study is the demonstration that there were more cases of asymptomatic infection during the Omicron variant. This finding provides a justification for continuing active surveillance to detect infection in vaccinated individuals without clinical signs and symptoms. In addition, few studies have reported results of inactivated virus vaccines (Coronavac). We demonstrated that reinfections occurred after mRNA vaccines (Pfizer-BioNTech), after the adenovirus vector vaccines ChAdOx1 (AstraZeneca/Oxford University) and Ad26.COV2.S (Johnson & Johnson/Janssen), as well as after an inactivated vaccine CoronaVac (Sinovac Biotech). The latter was not associated with more breakthrough infections than the others. In the present study, individuals who were infected during the circulation of the BA.1 lineage were later reinfected when BA.2 was dominant. This immune escape has implications for the production of future vaccines. In light of our findings, we believe it is necessary to continuously update the antigenic target of SARS CoV-2 vaccines and administer booster doses to the population regularly, a strategy well established for influenza vaccines.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Supplementary Material
Contributor Information
Stephanie L S Penetra, Evandro Chagas National Institute of Infectious Diseases, Oswaldo Cruz Foundation, Rio de Janeiro, Rio de Janeiro, Brazil.
Heloisa F P Santos, Evandro Chagas National Institute of Infectious Diseases, Oswaldo Cruz Foundation, Rio de Janeiro, Rio de Janeiro, Brazil.
Paola Cristina Resende, Laboratory of Respiratory Viruses and Measles National Influenza Centre, Americas Regional Reference Lab for Measles and Rubella, Reference Laboratory for COVID-19 World Health Organization, Oswaldo Cruz Institute, Oswaldo Cruz Foundation, Rio de Janeiro, Rio de Janeiro, Brazil.
Leonardo Soares Bastos, Scientific Computing Program, Oswaldo Cruz Foundation, Rio de Janeiro, Rio de Janeiro, Brazil.
Michele F B da Silva, Evandro Chagas National Institute of Infectious Diseases, Oswaldo Cruz Foundation, Rio de Janeiro, Rio de Janeiro, Brazil.
Anielle Pina-Costa, Evandro Chagas National Institute of Infectious Diseases, Oswaldo Cruz Foundation, Rio de Janeiro, Rio de Janeiro, Brazil.
Renata Serrano Lopes, Laboratory of Respiratory Viruses and Measles National Influenza Centre, Americas Regional Reference Lab for Measles and Rubella, Reference Laboratory for COVID-19 World Health Organization, Oswaldo Cruz Institute, Oswaldo Cruz Foundation, Rio de Janeiro, Rio de Janeiro, Brazil.
Leonardo Saboia-Vahia, Laboratory of Respiratory Viruses and Measles National Influenza Centre, Americas Regional Reference Lab for Measles and Rubella, Reference Laboratory for COVID-19 World Health Organization, Oswaldo Cruz Institute, Oswaldo Cruz Foundation, Rio de Janeiro, Rio de Janeiro, Brazil.
Any Caroline Alves de Oliveira, Laboratory of Respiratory Viruses and Measles National Influenza Centre, Americas Regional Reference Lab for Measles and Rubella, Reference Laboratory for COVID-19 World Health Organization, Oswaldo Cruz Institute, Oswaldo Cruz Foundation, Rio de Janeiro, Rio de Janeiro, Brazil.
Elisa Cavalcante Pereira, Laboratory of Respiratory Viruses and Measles National Influenza Centre, Americas Regional Reference Lab for Measles and Rubella, Reference Laboratory for COVID-19 World Health Organization, Oswaldo Cruz Institute, Oswaldo Cruz Foundation, Rio de Janeiro, Rio de Janeiro, Brazil.
Fernando Medeiros Filho, Evandro Chagas National Institute of Infectious Diseases, Oswaldo Cruz Foundation, Rio de Janeiro, Rio de Janeiro, Brazil.
Mayumi D Wakimoto, Evandro Chagas National Institute of Infectious Diseases, Oswaldo Cruz Foundation, Rio de Janeiro, Rio de Janeiro, Brazil.
Guilherme A Calvet, Evandro Chagas National Institute of Infectious Diseases, Oswaldo Cruz Foundation, Rio de Janeiro, Rio de Janeiro, Brazil.
Trevon L Fuller, Evandro Chagas National Institute of Infectious Diseases, Oswaldo Cruz Foundation, Rio de Janeiro, Rio de Janeiro, Brazil; University of California, Los Angeles, Los Angeles, California, USA.
Jimmy Whitworth, International Public Health, London School of Hygiene and Tropical Medicine, London, United Kingdom.
Christopher Smith, International Public Health, London School of Hygiene and Tropical Medicine, London, United Kingdom; School of Tropical Medicine and Global Health, Nagasaki University, Nagasaki, Japan.
Karin Nielsen-Saines, University of California, Los Angeles, Los Angeles, California, USA.
Marilia Sá Carvalho, Scientific Computing Program, Oswaldo Cruz Foundation, Rio de Janeiro, Rio de Janeiro, Brazil.
Otávio M Espíndola, Evandro Chagas National Institute of Infectious Diseases, Oswaldo Cruz Foundation, Rio de Janeiro, Rio de Janeiro, Brazil.
Lusiele Guaraldo, Evandro Chagas National Institute of Infectious Diseases, Oswaldo Cruz Foundation, Rio de Janeiro, Rio de Janeiro, Brazil.
Marilda M Siqueira, Laboratory of Respiratory Viruses and Measles National Influenza Centre, Americas Regional Reference Lab for Measles and Rubella, Reference Laboratory for COVID-19 World Health Organization, Oswaldo Cruz Institute, Oswaldo Cruz Foundation, Rio de Janeiro, Rio de Janeiro, Brazil.
Patricia Brasil, Evandro Chagas National Institute of Infectious Diseases, Oswaldo Cruz Foundation, Rio de Janeiro, Rio de Janeiro, Brazil.
Notes
Acknowledgements. This paper is dedicated to the memory of Dr. Fernando Medeiros Filho.
Author contributions. P. B., L. G., L. S. B., S. L. S. P., H. F. P. S., F. M. F., and M. S. C. conceived and designed the analyses. M. F. B. S., A. P. C., and H. F. P. S. collected the data. L. S. B. and H. F. P. S. performed the statistical analyses. A. C. A. O., P. C. R., M. M. S., R. S. L., and L. S. V. conducted the laboratory analyses. S. L. S. P., H. F. P. S., P. B., L. G., T. L. F., P. C. R., and M. M. S. wrote the paper. All the authors have reviewed the manuscript.
Financial support. This work was supported by Ministério da Saúde/Fundo Nacional de Desenvolvimento Científico e Tecnológico/Secretaria de Ciência, Tecnologia e Insumos Estratégicos/Departamento de Ciência e Tecnologia (grant numbers 402457/2020-9 and 403276/2020-9); Inova Fiocruz/Fundação Oswaldo Cruz (grant numbers VPPCB-007-FIO-18-2-30 and VPPCB-005-FIO-20-2-87); the Carlos Chagas Foundation for the Advancement of Science of the State of Rio de Janeiro (grant numbers E-26/200.935/2022, E-26/210.149/2020, E-26/211.565/2019, E-26/211.125/2021, and E-26/201.277/2021); National Council for Scientific and Technological Development (grant numbers 311759/2022-0, 423857/2021-5, 311562/2021-3, 409108/2022-7, and 310530/2021-0); the National Institutes of Allergy and Infectious Diseases, National Institutes of Health (grant numbers AI129534 and AI140718); the Simons Foundation Autism Research Initiative (grant number 866410); and the UK Medical Research Council (grant number MR/V033530/1).
References
- 1. Bont L, Versteegh J, Swelsen WT, et al. Natural reinfection with respiratory syncytial virus does not boost virus-specific T-cell immunity. Pediatr Res 2002; 52:363–7. [DOI] [PubMed] [Google Scholar]
- 2. Patel MM, York IA, Monto AS, Thompson MG, Fry AM. Immune-mediated attenuation of influenza illness after infection: opportunities and challenges. Lancet Microbe 2021; 2:E715–25. [DOI] [PubMed] [Google Scholar]
- 3. Ringlander J, Nilsson S, Westin J, Lindh M, Martner A, Hellstrand K. Low incidence of reinfection with endemic coronaviruses diagnosed by real-time PCR. J Infect Dis 2021; 223:2013–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Guo YF, Han JJ, Zhang Y, et al. SARS-CoV-2 Omicron variant: epidemiological features, biological characteristics, and clinical significance. Front Immunol 2022; 13:877101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Naveca FG, Nascimento V, de Souza VC, et al. COVID-19 in Amazonas, Brazil, was driven by the persistence of endemic lineages and P.1 emergence. Nat Med 2021; 27:1230. [DOI] [PubMed] [Google Scholar]
- 6. Resende PC, Bezerra JF, Vasconcelos RHT, et al. Severe acute respiratory syndrome coronavirus 2 P.2 lineage associated with reinfection case, Brazil, June October 2020. Emerg Infect Dis 2021; 27:1789–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Leidi A, Berner A, Dumont R, et al. Occupational risk of SARS-CoV-2 infection and reinfection during the second pandemic surge: a cohort study. Occup Environ Med 2022; 79:116–9. [DOI] [PubMed] [Google Scholar]
- 8. Abrokwa SK, Muller SA, Mendez-Brito A, Hanefeld J, El Bcheraoui C. Recurrent SARS-CoV-2 infections and their potential risk to public health—a systematic review. PLoS One 2021; 16:e0261221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. World Health Organization . WHO COVID-19 dashboard. https://covid19.who.int/. Accessed 16 July 2023.
- 10. Municipal Health Secretariat of Rio de Janeiro . Covid-19 epidemiological bulletin, 2023 [in Portuguese]. https://coronavirus.rio/boletim-epidemiologico/. Accessed 21 August 2023.
- 11. Brazilian Ministry of Health . COVID-19 epidemiological bulletin, 2023. [in Portuguese]. https://www.gov.br/saude/pt-br/centrais-de-conteudo/publicacoes/boletins/epidemiologicos/covid-19/2022/boletim-epidemiologico-no-146-boletim-coe-coronavirus/view. Accessed 21 August 2023.
- 12. Tillett RL, Sevinsky JR, Hartley PD, et al. Genomic evidence for reinfection with SARS-CoV-2: a case study. Lancet Infect Dis 2021; 21:52–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang JZ, Kaperak C, Sato T, Sakuraba A. COVID-19 reinfection: a rapid systematic review of case reports and case series. J Investig Med 2021; 69:1253–5. [DOI] [PubMed] [Google Scholar]
- 14. Dezordi FZ, Neto AMD, Campos TD, et al. Viralflow: a versatile automated workflow for SARS-CoV-2 genome assembly, lineage assignment, mutations and intrahost variant detection. Viruses-Basel 2022; 14:217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Singaporean Agency for Science . Coronavirus surveillance server. https://mendel.bii.a-star.edu.sg/METHODS/corona/beta/. Accessed 16 July 2023.
- 16. Aksamentov I, Roemer C, Hodcroft EB, Neher RA. Nextclade: clade assignment, mutation calling and quality control for viral genomes. J Open Source Softw 2021; 6:3773. [Google Scholar]
- 17. Rambaut A, Holmes EC, O'Toole A, et al. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat Microbiol 2020; 5:1403–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Resende PC, Delatorre E, Graf T, et al. Evolutionary dynamics and dissemination pattern of the SARS-CoV-2 lineage B.1.1.33 during the early pandemic phase in Brazil. Front Microbiol 2021; 11:615280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Voloch CM, Francisco RD, de Almeida LGP, et al. Genomic characterization of a novel SARS-CoV-2 lineage from Rio de Janeiro, Brazil. J Virol 2021; 95:e00119-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Menezes D, Fonseca PLC, de Araujo JLF, de Souza RP. SARS-CoV-2 genomic surveillance in Brazil: a systematic review with scientometric analysis. Viruses (Basel) 2022; 14:2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. National Institutes of Health . Clinical spectrum of SARS-CoV-2 infection. Last updated: 6 March 2023. https://www.covid19treatmentguidelines.nih.gov/overview/clinical-spectrum/. Accessed 21 August 2023.
- 22. Carvalho MS, Bastos LS, Fuller T, et al. Incidence of SARS-CoV-2 over four epidemic waves in a low-resource community in Rio de Janeiro, Brazil: a prospective cohort study. Lancet Reg Health Am 2022; 12:100283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rue H, Riebler A, Sørbye SH, Illian JB, Simpson DP, Lindgren FK. Bayesian computing with INLA: a review. Annu Rev Stat Appl 2017; 4:395–421. [Google Scholar]
- 24. Sacco C, Petrone D, Del Manso M, et al. Risk and protective factors for SARS-CoV-2 reinfections, surveillance data, Italy, August 2021 to March 2022. Euro Surveill 2022; 27:2200372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pulliam JRC, van Schalkwyk C, Govender N, et al. Increased risk of SARS-CoV-2 reinfection associated with emergence of Omicron in South Africa. Science 2022; 376:596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Keeton R, Tincho MB, Suzuki A, et al. Impact of SARS-CoV-2 exposure history on the T-cell and IgG response. Cell Rep Med 2023; 4:100898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shao W, Chen X, Zheng C, et al. Effectiveness of COVID-19 vaccines against SARS-CoV-2 variants of concern in real-world: a literature review and meta-analysis. Emerg Microbes Infect 2022; 11:2383–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wu N, Joyal-Desmarais K, Ribeiro PAB, et al. Long-term effectiveness of COVID-19 vaccines against infections, hospitalisations, and mortality in adults: findings from a rapid living systematic evidence synthesis and meta-analysis up to December, 2022. Lancet Respir Med 2023; 11:439–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Espíndola OM, Fuller TL, de Araújo MF, et al. Reduced ability to neutralize the Omicron variant among adults after infection and complete vaccination with BNT162b2, ChAdOx1, or CoronaVac and heterologous boosting. Sci Rep 2023; 13:7437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bobrovitz N, Ware H, Ma X, et al. Protective effectiveness of previous SARS-CoV-2 infection and hybrid immunity against the Omicron variant and severe disease: a systematic review and meta-regression. Lancet Infect Dis 2023; 23:556–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. He XM, Hong WQ, Pan XY, Lu GW, Wei XW. SARS-CoV-2 Omicron variant: characteristics and prevention. Medcomm 2021; 2:838–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Avanzato VA, Matson MJ, Seifert SN, et al. Case study: prolonged infectious SARS-CoV-2 shedding from an asymptomatic immunocompromised individual with cancer. Cell 2020; 183:1901–12.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Andeweg SP, De Gier B, Eggink D, et al. Protection of COVID-19 vaccination and previous infection against Omicron BA.1, BA.2 and Delta SARS-CoV-2 infections. Nat Commun 2022; 13:4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Almadhi M, Alsayyad AS, Conroy R, et al. Epidemiological assessment of SARS-CoV-2 reinfection. Int J Infect Dis 2022; 123:9–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Goldberg Y, Mandel M, Bar-On YM, et al. Protection and waning of natural and hybrid immunity to SARS-CoV-2. N Engl J Med 2022; 386:2201–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nguyen NN, Houhamdi L, Delorme L, Colson P, Gautret P. Reinfections with different SARS-CoV-2 Omicron subvariants, France. Emerg Infect Dis 2022; 28:2341–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Winchester NE, Shrestha NK, Kim P, Tereshchenko LG, Rothberg MB. Protection conferred by Delta and BA.1/BA.2 infection against BA.4/BA.5 infection and hospitalization: a retrospective cohort study. J Infect Dis 2023; 227:800–5. [DOI] [PubMed] [Google Scholar]
- 38. Nilles EJ, Paulino CT, de St Aubin M, et al. Tracking immune correlates of protection for emerging SARS-CoV-2 variants. Lancet Infect Dis 2023; 23:153–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Flacco ME, Martellucci CA, Baccolini V, et al. Risk of reinfection and disease after SARS-CoV-2 primary infection: meta-analysis. Eur J Clin Invest 2022; 52:e13845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Eythorsson E, Runolfsdottir HL, Ingvarsson RF, Sigurdsson MI, Palsson R. Rate of SARS-CoV-2 reinfection during an Omicron wave in Iceland. JAMA Netw Open 2022; 5:e2225320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Holt H, Talaei M, Greenig M, et al. Risk factors for developing COVID-19: a population-based longitudinal study (COVIDENCE UK). Thorax 2022; 77:900–12. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.