Abstract
The emergence of plasmid-mediated tigecycline resistance gene tet(X4) among clinically relevant bacteria has promoted significant concerns, as tigecycline is considered a last-resort drug against serious infections caused by multidrug-resistant bacteria. We herein focused on the isolation and molecular characterization of tet(X4)-positive Klebsiella pneumoniae (K. pneumoniae) and Escherichia coli (E. coli) in wild bird populations with anthropogenic interaction in Faisalabad, Pakistan. A total of 150 birds including black kites (Milvus migrans) and house crows (Corvus splendens) were screened for the presence of tigecycline resistance K. pneumoniae and E. coli. We found two K. pneumoniae and one E. coli isolate carrying tet(X4) originating from black kites. A combination of short- and long-read sequencing strategies showed that tet(X4) was located on a broad host range IncFII plasmid family in K. pneumoniae isolates whereas on an IncFII-IncFIB hybrid plasmid in E. coli. We also found an integrative and conjugative element ICEKp2 in K. pneumoniae isolate KP8336. We demonstrate the first description of tet(X4) gene in the WHO critical-priority pathogen K. pneumoniae among wild birds. The convergence of tet(X4) and virulence associated ICEKp2 in a wild bird with known anthropogenic contact should be further investigated to evaluate the potential epidemiological implications. The potential risk of global transmission of tet(X4)-positive K. pneumoniae and E. coli warrant comprehensive evaluation and emphasizes the need for effective mitigation strategies to reduce anthropogenic-driven dissemination of AMR in the environment.
Keywords: Escherichia coli, Klebsiella pneumoniae, tet(X4), ICEKp2, Birds
Subject terms: Antimicrobial resistance, Ecological epidemiology, Infectious diseases
Introduction
Antimicrobial resistance (AMR) is a growing global threat to human and animal health driven by the selective pressure of extensive antibiotic consumption in multiple sectors including community settings, hospitals, veterinary, agriculture and aquaculture1. The issue has been highlighted in a systematic analysis, where AMR was associated with an estimated 5 million deaths in 2019, with 1.3 million deaths directly attributable to bacterial AMR2. The situation is further aggravated by the widespread distribution of AMR genes in the environment and insufficient investment in the development of new antibiotics3. The emergence of clinically relevant AMR in the environment primarily stems from pollution resulting from human activities. Additionally, this phenomenon is influenced by intricate interactions, including the genetic exchange of resistance genes facilitated by selective mechanisms enhanced by pollutants such as biocides and antibiotics4. The presence of clinically relevant multidrug-resistant (MDR) bacteria in the environment is a growing concern, with wildlife, particularly wild birds, being viewed as important sentinels for AMR surveillance5,6. Although wild birds can acquire antimicrobial-resistant bacteria/genes, likely from foraging in anthropogenically impacted areas including both landfills and WWTPs7–9, the role of wild birds in the dissemination of clinically relevant AMR needs further investigation10. Several studies have suggested that wild birds could be competent vectors of AMR and potentially disperse AMR through their movements8,11,12. Tigecycline is regarded as the last resort antibiotic in the clinical management of infections associated with MDR bacteria, particularly carbapenem and colistin-resistant Enterobacteriaceae. The recently discovered tet(X4) gene on plasmid confers resistance to tigecycline and has been found mainly in E. coli isolated from various sources, including humans, animals, and the environment13. The ability of the tet(X4) gene to be carried on various plasmid types, including hybrid plasmids, can facilitate its spread among bacterial populations and contribute to the emergence and spread of tigecycline-resistant bacteria.
Plasmids, such as IncFII, IncFIB, IncFIA, IncX1, IncQ1, IncA/C, IncHI1, and others, have been discovered to be the key carriers for propagating tet(X4)14,15. Tigecycline resistance in K. pneumoniae is mainly caused by mutations in the genes ramR, soxR, oqxR, rpsJ, and tet(A). However, the recent discovery of tet(X4) in K. pneumoniae is alarming, given K. pneumoniae is listed as a WHO critical-priority pathogen16.
Several studies have reported the presence of tigecycline-resistant E. coli carrying the tet(X4) gene in wild birds17,18, which raises concerns due to its potential spread in the environment. Additionally, this may lead to the transfer of tet(X4) to other bacteria, including those that cause human and animal infections. In this study, we investigated the prevalence and molecular characteristics of tet(X4) in K. pneumoniae and E. coli isolates from wild birds in Pakistan and further described the tet(X4)-harboring plasmids.
Methods
Sample collection and bacterial isolates
In this cross-sectional study, faecal droppings of 150 wild birds with known anthropogenic interaction (n = 100 from black kites and n = 50 from house crows) were collected from various public parks in Faisalabad, Pakistan during June 2022. A single fresh isolated faecal dropping sample was taken from an individual bird using sterile charcoal swabs and transported to the lab for microbiological examination. To prevent contamination, only the top surface of each dropping was swabbed, avoiding contact with the ground beneath. In addition, we confirmed the origin of the avian species by the direct visual observation of crows and kites in a public park. Collected samples were cultivated on Simmons citrate agar supplemented with amoxicillin and myo-inositol at concentrations of 10 µg/mL and 10%, respectively for the isolation of Klebsiella spp.19.
For the isolation of E. coli, UTI ChromoSelect agar (Merck, Darmstadt, Germany) was used. For the isolation of tigecycline-resistant colonies, sub-culturing of both E. coli and Klebsiella spp. was performed on UTI ChromoSelect agar supplemented with tigecycline (4 µg/mL). All the phenotypically resistant isolates were confirmed for species using API 20E biochemical strips (bioMérieux, Marcy l'Etoile, France). For the detection of tet(X4)-positive E. coli and K. pneumoniae isolates, PCR was performed using primers described earlier20. Briefly tet(X4)-gene was amplified using primer pairs tetX4-F (5ʹ-CCGATATTCATCATCCAGAGG-3ʹ) and tetX4-R (5ʹ CGCTTACTTTTCCAAGACTTACC-3ʹ) as forward and reverse primers with 32 cycles of denaturation at 95 °C for 30 s; annealing at 55 °C for 30 s; extension at 72° for 30 s20.
Conjugation experiments
Conjugation assays were conducted to investigate the transferability of the tet(X4) positive isolates with sodium azide resistant E. coli J53 as the recipient strain. Transconjugants were selected on MacConkey agar containing (4 µg/mL) tigecycline combined with 100 µg/mL sodium azide. Subsequent carriage of tet(X4) bearing plasmids in the original parental strains and corresponding transconjugants was confirmed by PCR.
Whole-genome sequencing and bioinformatics analysis
The total genomic DNA of isolates from overnight cultures was prepared using MagnaPure compact total NA kit according to the manufacturer’s instructions (Roche, Sweden). Library preparation was performed with the Illumina Nextera XT kit (Illumina, USA). Libraries were verified with the bioanalyzer high sensitivity DNA method (Agilent, USA). Paired-end sequencing (2 × 250 bp) of genomic DNA using a V3 run kit (Illumina) was performed on a MiSeq instrument (Illumina, San Diego, CA, United States). Short-read Illumina raw sequence reads were de novo assembled into contigs using SPAdes21 and contigs less than 500 bp were discarded. Analysis of multilocus sequence typing (MLST), acquired resistance genes, and plasmid replicons were conducted by the online tools MLST, ResFinder, and PlasmidFinder at the web service of Center for Genomic Epidemiology (http://www.genomicepidemiology.org/. Kleborate, which was designed specifically for K. pneumoniae, was used to identify virulence factors and ICEKp structures, and further determine the sequence types (STs) of K. pneumoniae22.
To explore the evolutionary relationship of tet(X4)-positive K. pneumoniae between this study and other isolates in the NCBI database, all the tet(X4)-positive K. pneumoniae isolates were retrieved and downloaded from the NCBI Pathogen detection database. The isolates were retrieved with the search criteria ‘AMR_genotypes: tet(X4)’ and ‘Organism_group: Klebsiella pneumoniae’ as of March 2023. The draft assembled contigs were annotated using Prokka23, and then applied for phylogenetic analysis using Roary24 and FastTree25 based on single nucleotide polymorphisms (SNPs) of core genomes. The phylogenetic tree was visualized by iTOL (https://itol.embl.de/itol.cgi)26.
Genomic DNA was extracted using MagAttract HMW DNA kit according to the manufacturer’s instructions (Qiagen, Sweden) and was then subjected to long-read sequencing to obtain the complete sequences27,28. Library preparation and sample barcoding were performed using Rapid sequencing gDNA-barcoding chemistry and protocol (Oxford Nanopore Technologies, UK, SQK-RBK110.96, version RPK_9126_v110_revK_24Mar2021). Sequencing was performed using MinION™ MK-1B with FLO-MIN106 R9.4.1 flow cell and high-accuracy basecalling with read filtering out at Q score < 9 and trimming of barcodes, MinKnow 22.03.6 and Guppy 6.0.7. Short-read Illumina data and long-read Nanopore data were subjected to perform de novo assembly by Unicycler with the hybrid strategy. The rapid annotation website server (https://rast.nmpdr.org/rast.cgi) was then used to annotate the complete genome sequences29. Circular comparisons between tet(X4)-bearing plasmids and homologous plasmids available in the NCBI database were performed using the BRIG tool30. To visualize the genetic comparison features of ICEKp2, Easyfig was used to generate linear comparison figures31.
Results
Identification of tet(X)-positive isolates and transferability
We identified two K. pneumoniae isolates (KP8333 and KP8336) and one E. coli isolate (EC8331) originating from black kites (Milvus migrans) with phenotypic resistance to tigecycline. All three isolates were also found to be positive for the carriage of tet(X4) gene and successfully transferred the tet(X4) gene into host E. coli J53 by conjugation.
The phylogenetic analysis
To understand the evolutionary relationship of tet(X4)-carrying K. pneumoniae isolates, phylogenetic analysis of 34 tet(X4)-positive K. pneumoniae isolates (including two isolates in this study and 32 K. pneumoniae isolates in the NCBI Pathogen detection database) was performed. The phylogenetic analysis revealed that all 34 K. pneumoniae isolates were divided into two major clusters and none of the isolates displayed any clonal relationship with KP8333 or KP8336 (Fig. 1A). Among the tet(X4)-carrying K. pneumoniae, the majority was distributed in China (23/34), followed by Singapore (7/34). Only K. pneumoniae KP8333 and KP8336 in this study were from Pakistan. In addition, tet(X4)-carrying K. pneumoniae strains were prevalent in pigs (15/34), humans (9/34), and pork (5/34). The distribution of STs was diverse and only ST414-1LV K. pneumoniae strains from the pig in China demonstrate obvious clonal relationship (Fig. 1A). Virulence genes encoding yersiniabactin (ybt), colibactin (clb), aerobactin (iuc), salmochelin (iro), hypermucoidy (rmp and rmpA2) were detected whereas clb and rmpA2 was not found in any tet(X4)-positive K. pneumoniae strains in this phylogenetic tree. Notably, five ICEKp variants (ICEKp1, ICEKp2, ICEKp4, ICEKp5, and ICEKp12) carrying ybt locus were found in six K. pneumoniae strains, and ICEKp2 appeared in KP8336.
Figure 1.
Phylogenetic tree of tet(X4)-positive K. pneumoniae strains and the ICEKp2 structure in KP8336. (A) Phylogenetic tree of 34 tet(X4)-positive K. pneumoniae strains, including two isolates in this study and 32 K. pneumoniae strains in the NCBI Pathogen detection database. The blue squares indicate the presence of virulence genes. (B) Alignment of the virulence-encoding region carried by KP8336, ICEKp2 structure in K. pneumonia 2485STDY5477980 and other chromosome regions of K. pneumonia 2485STDY5477980 (ERR314530).
The ICEKp structure in KP8336
The boundaries of ICEKp in KP8336 was identified by the 17 bp direct repeats (5ʹ-CCAGTCAGAGGAGCCAA-3ʹ) formed upon integration (Fig. 1B). Comparative analysis indicated that the ICEKp structure in KP8336 was one ICEKp2 variant, a 63 kb chromosomal island. The ICEKp2 structure mainly included the P4-like integrase gene int, the ybt locus (29 kb), and the sequence (14 kb) encoding the virB-type IV secretion system (T4SS) that is responsible for DNA transfer (Fig. 1B). In addition, the chromosome flanking ICEKp2 in KP8336 exhibited extremely high homology with the chromosome of K. pneumoniae 2485STDY5477980 isolated from human in UK (Fig. 1B).
Genetic environment of tet(X4) in E. coli
The isolate EC8331 was ST746 E. coli containing three plasmids: pEC8331-tetX, pEC8331-155 kb, and pEC8331-119 kb (Table 1). The tet(X4) gene was located on the IncFII-IncFIB (AP001918) plasmid pEC8331-tetX (178,255 bp), which is an MDR plasmid harboring tet(X4), floR, fosA4, mph(A), dfrA12 and blaTEM-215 (Table 1 and Fig. 2). Sequence analysis revealed that pEC8331-tetX shared 99.85% identity at 86% coverage with IncFII-IncFIB (AP001918) plasmid pCTXM-2271 (MF589339) in E. coli 2271 from China (Fig. 2). The plasmid pEC8331-tetX also exhibited 99.99% identity at 43% coverage with IncFIB(AP001918) plasmid pTEM (CP047003) in E. coli J-8 from China, and 98.05% identity at 63% coverage with tet(X4)-bearing IncFII plasmid pPK8261-tetX (CP080156) in E. coli PK8261 isolated in chicken from Pakistan (Fig. 2). Evidently, it can be assumed that pEC8331-tetX was an evolved tet(X4)-positive IncFII-IncFIB (AP001918) hybrid plasmid and all antibiotic resistance genes on this plasmid were concentrated in the IncFII plasmid backbone structure region. The large repeat structure IS26-hp-hp-fosA4-hp-IS26-virD2-floR-lysR-ISCR2-hp-abh-tet(X4)-ISCR2-IS26 with 13 kb in length was found in pEC8331-tetX (Fig. 2). The repeat structure was mediated by IS26, which has previously been reported to mediate tandem multiplication of genes within plasmids. In addition, tet(X4) was flanked by two copies of ISCR2 with the structure ISCR2-hp-abh-tet(X4)-ISCR2.
Table 1.
Characteristics of three tet(X4)-carrying isolates recovered from birds in Pakistan.
| Strain IDs | Species | MLST | Components | Replicon types | Size (bp) | Resistance genes |
|---|---|---|---|---|---|---|
| EC8331 | E. coli | ST746 | EC8331-chromosome | 4,685,870 | None | |
| pEC8331-tetX | IncFII, IncFIB(AP001918) | 178,255 | tet(X4), floR, fosA4, mph(A), dfrA12, blaTEM-215 | |||
| pEC8331-155 kb | p0111 | 155,907 | aph(3'')-Ib, aph(6)-Id, tet(A), qnrS13, dfrA15, blaTEM-220 | |||
| pEC8331-119 kb | IncI1-I(Alpha) | 119,809 | aph(3')-Ia, aph(3'')-Ib, aph(6)-Id, blaTEM-126, dfrA15, qnrS13, tet(A) | |||
| KP8333 | K. pneumoniae | ST281 | KP8333-chromosome | 5,147,119 | fosA5, blaSHV-108, oqxA, oqxB | |
| pKP8333-tetX | IncFII | 100,799 | tet(X4), floR, fosA4, mph(A), dfrA12, blaTEM-215 | |||
| pKP8333-201 kb | IncFIB(K)(pCAV1099-114) | 201,997 | None | |||
| pKP8333-48 kb | IncFIA(HI1), repB(R1701) | 48,790 | aph(6)-Id, aph(3'')-Ib, sul1, qacE, blaDHA-1, qnrB4 | |||
| pKP8333-2 kb | ColRNAI | 2563 | none | |||
| KP8336 | K. pneumoniae | ST41-4LV | KP8336-chromosome | 5,456,436 | fosA5, blaSHV-108, oqxA, oqxB | |
| pKP8336-tetX | IncFII(29) | 66,571 | tet(X4) | |||
| pKP8336-167 kb | IncFIB(K), IncFII(K) | 167,021 | None |
Figure 2.
Circular comparison of tet(X4)-bearing plasmid in EC8331 and other similar plasmids in NCBI database. The pEC8331-tetX in this study were compared with plasmids pCTXM-2271, pTEM, and pPK8261-tetX in the NCBI database. The outermost circle indicates the plasmid pEC8331-tetX with genes annotated.
Genetic environment of tet(X4) in K. pneumoniae
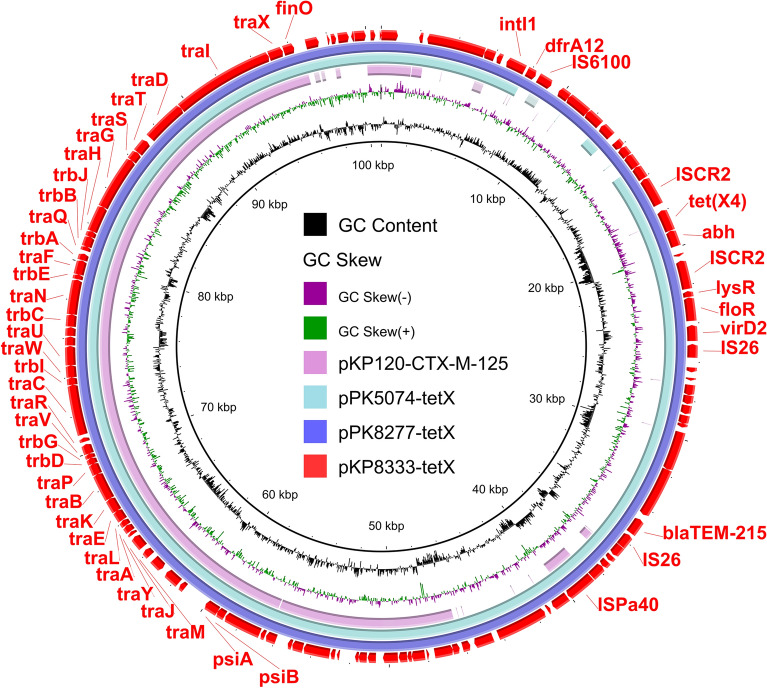
The K. pneumoniae isolate, KP8333, belonged to ST281 and harbored four plasmids: pKP8333-tetX, pKP8333-201 kb, pKP8333-48 kb, and pKP8333-2 kb (Table 1). The plasmid pKP8333-tetX was a tet(X4)-positive IncFII plasmid containing similar antibiotic resistance genes as pEC8331-tetX (Table 1 and Fig. 3). BLASTn search revealed that the plasmid pKP8333-tetX shared a high degree of genetic identity with the reported plasmids pPK8277-tetX (CP080134) (99.98% identity at 100% coverage) in chicken derived E. coli PK8277 and pPK5074-tetX (CP072807) (100.00% identity at 93% coverage) in human derived E. coli PK5074, which were from Pakistan (Fig. 3). Among K. pneumoniae, the plasmid pKP8333-tetX exhibited the highest similarity (97.77% identity at 55% coverage) to pKP120-CTX-M-125 (CP060746) (Fig. 3), suggesting that pKP8333-tetX is a newly emerging plasmid in K. pneumoniae.
Figure 3.
Circular comparison of tet(X4)-bearing plasmid in KP8333 and other similar plasmids in NCBI database. The pEC8331-tetX in this study were compared with plasmids pPK8277-tetX, pPK5074-tetX, and pKP120-CTX-M-125 in the NCBI database. The outermost circle indicates the plasmid pKP8333-tetX with genes annotated.
The isolate KP8336 belonged to the ST41-4LV K. pneumoniae. Two plasmids pKP8336-tetX and pKP8336-167 kb were identified in KP8336 (Table 1). The pKP8336-tetX-66 kb was a tet(X4)-bearing IncFII(29) plasmid and no other resistance gene was found in this plasmid (Table 1 and Fig. 4). BLASTn analysis showed that pKP8336-tetX exhibited high homology (99.80% identity at 100% coverage) to the plasmid pPK8241-tetX (CP080140) in E. coli PK8241 from chicken in Pakistan (Fig. 4). The pKP8336-tetX also showed 94.96% identity at 83% coverage with plasmid pRHB34-C05_2 (CP057177) in E. coli RHB34-C05 and 94.96% identity at 83% coverage with plasmid pJUNP054 (LC506717) in K. pneumoniae JUNP054 (Fig. 4). This indicated that E. coli and K. pneumoniae are important host bacteria of tet(X4)-bearing IncFII(29) plasmids.
Figure 4.
Circular comparison of tet(X4)-bearing plasmid in KP8336 and other similar plasmids in NCBI database. The pEC8331-tetX in this study were compared with plasmids pPK8241-tetX, pRHB34-C05_2, and pJUNP054 in the NCBI database. The outermost circle indicates the plasmid pKP8336-tetX with genes annotated.
Discussion
In this study, we isolated two tet(X4)-positive K. pneumoniae and one tet(X4)-positive E. coli isolate from black kites (Milvus migrans) in Faisalabad, Pakistan and performed a comparative genomic analysis on these isolates. Until now, only a few studies have reported the existence of tet(X4)-positive K. pneumoniae in human16, animal, and food specimens32 predominantly from China. This is the first report of the presence of tet(X4)-positive K. pneumoniae in Pakistan indicating the spread of tet(X4) beyond E. coli. Black kite is an abundant bird and an opportunistic and scavenging feeder providing ecosystem functions such as nutrient cycling and pest control etc. In Pakistan, black kites frequently visit anthropogenic affected areas such as landfills, food-producing animals and agricultural land33. One more indigenous reason for anthropogenic interaction is that a fraction of the population from a dominant religious group believes in giving pieces of flesh (cow/goat/chicken) as a means of warding off calamities and hardships34. Recent findings suggest a link between certain wild bird species and the acquisition of clinically relevant AMR. Notably, black kites have been reported to harbor carbapenem-resistant NDM-5-producing E. coli, in Pakistan7. Our finding of tet(X4)-positive K. pneumoniae in this wild bird species along with findings from previous studies is an indication that plasmid-mediated tigecycline resistance has the potential to disseminate within the one health framework like previously described with both ESBLs and carbapenem resistance genes7,13,17,18,32,35. Wild birds can acquire clinically relevant MDR bacteria, likely from foraging in anthropogenically impacted areas as reported earlier36,37. Tigecycline is rarely used in human medicine and is not used in food animals in Pakistan. However, plasmid-mediated tet(X4)-positive E. coli have been reported both from clinical and non-clinical settings16,18, which could be linked to excessive use of early generations of tetracycline antibiotics in food animals as proposed by several researchers15,38. However, the link between the long-term use of tetracyclines in food animals and the emergence of tigecycline resistance in bacteria needs to be ascertained.
Genome data found that both the K. pneumoniae isolates have different STs and showed no clonal relationship with the global tet(X4)-positive K. pneumoniae strains in the NCBI database indicative of genetic diversity. Additionally, a single tet(X4)-positive E. coli isolate belonged to ST746 which has been previously associated with carbapenem resistance in Korea39 and China40. It has been suggested that mobile genetic elements, not the clones, play an important role in tet(X4) transmission32. We found an integrative and conjugative elements ICEKp2 in K. pneumoniae KP8336 which carried ybt locus encoding yersiniabactin. It has been reported that ICEKp variants could form an extrachromosomal circular intermediate and integrate into the chromosomes of recipient cells41,42. These results suggest that the formation of tet(X4)-positive hypervirulent K. pneumoniae KP8336 may be due to the horizontal transfer of ICEKp2 and IncFII(29) plasmid carrying tet(X4). The finding of ICEKp2 in K. pneumoniae from wild birds with known anthropogenic interaction is concerning. K. pneumoniae with ICEKp2 has been associated with clinical outbreaks16 and therefore, further investigative studies are important to be able to identify dissemination routes of AMR within the one health context. Additionally, mitigation efforts should be encouraged for already identified anthropogenic-driven AMR dissemination through waste, sewage and industrial pollution.
Hybrid genome assembly of E. coli EC8331 revealed that tet(X4) was located on a large MDR hybrid plasmid (~ 178 kb) IncFII-IncFIB (AP001918) plasmid harbouring additional resistance genes for quinolones, fosfomycin, macrolides, aminoglycosides and β-lactams. The hybrid plasmid pEC8331-tetX has large repeat structures surrounded by IS26, which has previously been reported to mediate tandem multiplication of genes within plasmids43. In addition, tet(X4) was flanked by two copies of ISCR2, forming a structure ISCR2-hp-abh-tet(X4)-ISCR2, which was found in several tet(X4)-positive isolates20,44. In previous studies from Pakistan, IncFII was the most common tet(X4) bearing plasmid ranging from 66 to 119 kbp in size15,18. tet(X4) bearing hybrid plasmids are being increasingly reported from China and are considered a new threat41. Therefore, the emergence of novel MDR hybrid plasmids in Pakistan is a serious concern because of their ability to contribute to the resistance and virulence genes co-translocation and demands continuous surveillance of AMR. Genomic comparison of tet(X4)-positive IncFII plasmid from K. pneumoniae KP8333 showed it has a similar genetic environment as that of E. coli isolated in this study and of chicken and human origins reported earlier in Pakistan15,45. These results indicate that E. coli and K. pneumoniae are important host bacteria of tet(X4)-bearing IncFII plasmids in Pakistan.
Conclusion
The prevalence and molecular features of the tet(X4) positive bacteria in wild birds demonstrate that this gene has disseminated within the One Health framework and is yet an example of wild birds as potential carriers of novel plasmid-mediated resistance genes together with hypervirulent traits. This emphasizes the need for mitigation strategies for anthropogenic-driven dissemination of AMR in the environment.
As part of a comprehensive One Health strategy, we advocate for increased environmental AMR surveillance. To effectively monitor these efforts, we propose more detailed studies on wildlife and their interactions with known anthropogenic sources of AMR. By leveraging animal movement data from GPS telemetry, we can gain valuable insights into dynamics of AMR at the human–wildlife interface. Obvious mitigation strategies would be to expanding already existing interventions aimed at reducing AMR dissemination targeting landfills and wastewater treatment plants. Future strategies might also include reducing wildlife access to identified point sources of AMR.
A better understanding of the human–animal–wildlife interface will guide the development of evidence-based and effective One Health interventions that ultimately can reduce the AMR crisis.
Author contributions
M.H.M., A.S., F.H., J.A. collected the data, X.L., H.V., R.L. performed the whole data integration and sequence analysis, M.M. conceived and designed the article, R.Li., J.B., M.M. revised the manuscript. All authors approved the submitted version.
Funding
Open access funding provided by Linköping University. This work was funded by Region Kalmar County and Linköpings Universitet.
Data availability
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at DDBJ/ENA/GenBank under BioProject ID PRJNA972190.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jonas Bonnedahl, Email: jonas.bonnedahl@liu.se.
Mashkoor Mohsin, Email: mashkoormohsin@uaf.edu.pk.
References
- 1.Allel K, et al. Global antimicrobial-resistance drivers: an ecological country-level study at the human–animal interface. Lancet Planet. Heal. 2023;7:e291–e303. doi: 10.1016/S2542-5196(23)00026-8. [DOI] [PubMed] [Google Scholar]
- 2.Murray CJL, et al. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet. 2022;399:629–655. doi: 10.1016/S0140-6736(21)02724-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Årdal C, et al. Antibiotic development—Economic, regulatory and societal challenges. Nat. Rev. Microbiol. 2020;18:267–274. doi: 10.1038/s41579-019-0293-3. [DOI] [PubMed] [Google Scholar]
- 4.Wellington EMH, et al. The role of the natural environment in the emergence of antibiotic resistance in gram-negative bacteria. Lancet Infect. Dis. 2013;13:155–165. doi: 10.1016/S1473-3099(12)70317-1. [DOI] [PubMed] [Google Scholar]
- 5.Ramey AM. Antimicrobial resistance: Wildlife as indicators of anthropogenic environmental contamination across space and through time. Curr. Biol. 2021;31:R1385–R1387. doi: 10.1016/j.cub.2021.08.037. [DOI] [PubMed] [Google Scholar]
- 6.Dolejska M, Literak I. Wildlife is overlooked in the epidemiology of medically important antibiotic-resistant bacteria. Antimicrob. Agents Chemother. 2019;63:1–5. doi: 10.1128/AAC.01167-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ahlstrom CA, et al. Genomically diverse carbapenem resistant Enterobacteriaceae from wild birds provide insight into global patterns of spatiotemporal dissemination. Sci. Total Environ. 2022;824:153632. doi: 10.1016/j.scitotenv.2022.153632. [DOI] [PubMed] [Google Scholar]
- 8.Ahlstrom CA, et al. Acquisition and dissemination of cephalosporin-resistant E. coli in migratory birds sampled at an Alaska landfill as inferred through genomic analysis. Sci. Rep. 2018;8:1–11. doi: 10.1038/s41598-018-25474-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Woksepp H, et al. Dissemination of carbapenemase-producing Enterobacterales through wastewater and gulls at a wastewater treatment plant in Sweden. Sci. Total Environ. 2023;886:163997. doi: 10.1016/j.scitotenv.2023.163997. [DOI] [PubMed] [Google Scholar]
- 10.Ramey AM, Ahlstrom CA. Antibiotic resistant bacteria in wildlife: Perspectives on trends, acquisition and dissemination, data gaps and future directions. J. Wildl. Dis. 2020;56:1–15. doi: 10.7589/2019-04-099. [DOI] [PubMed] [Google Scholar]
- 11.Ahlstrom CA, et al. Evidence for continental-scale dispersal of antimicrobial resistant bacteria by landfill-foraging gulls. Sci. Total Environ. 2021;764:144551. doi: 10.1016/j.scitotenv.2020.144551. [DOI] [PubMed] [Google Scholar]
- 12.Sandegren L, et al. Long-term carriage and rapid transmission of extended spectrum beta-lactamase-producing E. coli within a flock of Mallards in the absence of antibiotic selection. Environ. Microbiol. Rep. 2018;10:576–582. doi: 10.1111/1758-2229.12681. [DOI] [PubMed] [Google Scholar]
- 13.Li Y, Sun X, Xiao X, Wang Z, Li R. Global distribution and genomic characteristics of tet(X)-positive Escherichia coli among humans, animals, and the environment. Sci. Total Environ. 2023;887:164148. doi: 10.1016/j.scitotenv.2023.164148. [DOI] [PubMed] [Google Scholar]
- 14.Xiao X, et al. Persistence of plasmid and tet(X4) in an Escherichia coli isolate coharboring blaNDM-5 and mcr-1 after acquiring an IncFII tet(X4)-positive plasmid. Front. Microbiol. 2022;13:1–10. doi: 10.3389/fmicb.2022.1010387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li R, et al. Widespread prevalence and molecular epidemiology of tet(X4) and mcr-1 harboring Escherichia coli isolated from chickens in Pakistan. Sci. Total Environ. 2022;806:150689. doi: 10.1016/j.scitotenv.2021.150689. [DOI] [PubMed] [Google Scholar]
- 16.Zhai W, et al. Presence of mobile tigecycline resistance gene tet(X4) in clinical Klebsiella pneumoniae. Microbiol. Spectr. 2022;10:1–5. doi: 10.1128/spectrum.01081-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang W, et al. Molecular epidemiology and population genomics of tet(X4), blaNDM or mcr-1 positive Escherichia coli from migratory birds in southeast coast of China. Ecotoxicol. Environ. Saf. 2022;244:114032. doi: 10.1016/j.ecoenv.2022.114032. [DOI] [PubMed] [Google Scholar]
- 18.Mohsin M, et al. Emergence of plasmid-mediated tigecycline resistance tet(X4) gene in Escherichia coli isolated from poultry, food and the environment in South Asia. Sci. Total Environ. 2021;787:147613. doi: 10.1016/j.scitotenv.2021.147613. [DOI] [PubMed] [Google Scholar]
- 19.Kregten EV, Westerdaal NA, Willers JM. New, simple medium for selective recovery of Klebsiella pneumoniae and Klebsiella oxytoca from human feces. J. Clin. Microbiol. 1984;20:936–941. doi: 10.1128/jcm.20.5.936-941.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.He T, et al. Emergence of plasmid-mediated high-level tigecycline resistance genes in animals and humans. Nat. Microbiol. 2019;4:1450–1456. doi: 10.1038/s41564-019-0445-2. [DOI] [PubMed] [Google Scholar]
- 21.Bankevich A, et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lam MMC, et al. A genomic surveillance framework and genotyping tool for Klebsiella pneumoniae and its related species complex. Nat. Commun. 2021;12:1–16. doi: 10.1038/s41467-021-24448-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seemann T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 24.Page AJ, et al. Roary: Rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015;31:3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Price MN, Dehal PS, Arkin AP. FastTree: Computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 2009;26:1641–1650. doi: 10.1093/molbev/msp077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Letunic I, Bork P. Interactive tree of life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021;49:W293–W296. doi: 10.1093/nar/gkab301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wick RR, Judd LM, Gorrie CL, Holt KE. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017;13:1–22. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li R, et al. Efficient generation of complete sequences of MDR-encoding plasmids by rapid assembly of MinION barcoding sequencing data. Gigascience. 2018;7:1–9. doi: 10.1093/gigascience/gix132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Overbeek R, et al. The SEED and the rapid annotation of microbial genomes using subsystems technology (RAST) Nucleic Acids Res. 2014;42:D206–D214. doi: 10.1093/nar/gkt1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alikhan NF, Petty NK, Ben Zakour NL, Beatson SA. BLAST ring image generator (BRIG): Simple prokaryote genome comparisons. BMC Genom. 2011;12:1–10. doi: 10.1186/1471-2164-12-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sullivan MJ, Petty NK, Beatson SA. Easyfig: A genome comparison visualizer. Bioinformatics. 2011;27:1009–1010. doi: 10.1093/bioinformatics/btr039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Y, et al. Emergence of tet(X4)-positive hypervirulent Klebsiella pneumoniae of food origin in China. Lwt. 2023;173:114280. doi: 10.1016/j.lwt.2022.114280. [DOI] [Google Scholar]
- 33.Noreen Z, Sultan K. Hegemony of two avian species: Black kites and cattle egrets on two distant landfills during COVID-19 pandemic in Gujranwala. Pak. Pure Appl. Biol. 2022;11:11–25. [Google Scholar]
- 34.Shafiq, A. A meaty issue. In The News. 1–5. https://www.thenews.com.pk/tns/detail/558784-a-meaty-issue-roadsidevendors-of-sadka-gosht (2015).
- 35.Bonnedahl J, et al. Dissemination of Escherichia coli with CTX-M type ESBL between humans and yellow-legged gulls in the south of France. PLoS One. 2009;4:1–6. doi: 10.1371/journal.pone.0005958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martín-Maldonado B, et al. Urban birds: An important source of antimicrobial resistant Salmonella strains in Central Spain. Comp. Immunol. Microbiol. Infect. Dis. 2020;72:101519. doi: 10.1016/j.cimid.2020.101519. [DOI] [PubMed] [Google Scholar]
- 37.Umair M, et al. International manufacturing and trade in colistin, its implications in colistin resistance and One Health global policies: A microbiological, economic, and anthropological study. Lancet Microbe. 2023;4:e264–e276. doi: 10.1016/S2666-5247(22)00387-1. [DOI] [PubMed] [Google Scholar]
- 38.Zhang S, et al. Dissemination and prevalence of plasmid-mediated high-level tigecycline resistance gene tet(X4) Front. Microbiol. 2022;13:1–20. doi: 10.3389/fmicb.2022.969769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shin H, Kim Y, Han D, Hur HG. Emergence of high level carbapenem and extensively drug resistant Escherichia coli ST746 producing NDM-5 in influent of wastewater treatment plant, Seoul South Korea. Front. Microbiol. 2021;12:1–7. doi: 10.3389/fmicb.2021.645411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Teo JQ-M, et al. Genomic surveillance of carbapenem-resistant Klebsiella pneumoniae from a major public health hospital in Singapore. Microbiol. Spectr. 2022;10:1–16. doi: 10.1128/spectrum.00957-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bortolaia V, et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 2020;75:3491–3500. doi: 10.1093/jac/dkaa345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin TL, Lee CZ, Hsieh PF, Tsai SF, Wang JT. Characterization of integrative and conjugative element ICEKp1-associated genomic heterogeneity in a Klebsiella pneumoniae strain isolated from a primary liver abscess. J. Bacteriol. 2008;190:515–526. doi: 10.1128/JB.01219-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Raj CTD, Suryavanshi MV, Kandaswamy S, Ramasamy KP, James RA. Whole genome sequence analysis and in-vitro probiotic characterization of Bacillus velezensis FCW2 MCC4686 from spontaneously fermented coconut water. Genomics. 2023;115:110637. doi: 10.1016/j.ygeno.2023.110637. [DOI] [PubMed] [Google Scholar]
- 44.Li R, et al. Deciphering the structural diversity and classification of the mobile tigecycline resistance gene tet(X)-bearing plasmidome among bacteria. mSystems. 2020;5:1–17. doi: 10.1128/mSystems.00134-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li R, et al. Emergence of plasmid-mediated resistance genes tet(X) and mcr-1 in Escherichia coli clinical isolates from Pakistan. mSphere. 2021;6:1–8. doi: 10.1128/mSphere.00695-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at DDBJ/ENA/GenBank under BioProject ID PRJNA972190.