Significance
The results of this study establish that removal/salvage of 2-phosphoglycolate occurs in an organism without a CBB cycle, indicating that a system to deal with 2-phosphoglycolate evolved independently of the CBB cycle. As Thermococcales are one of the deep-branching lineages in the tree of life, the results also raise the possibilities that enzymes involved in 2-phosphoglycolate removal/salvage emerged prior to the emergence of cyanobacteria and the Great Oxidation Event.
Keywords: Archaea, metabolism, Rubisco, 2-phosphoglycolate
Abstract
Many organisms that utilize the Calvin–Benson–Bassham (CBB) cycle for autotrophic growth harbor metabolic pathways to remove and/or salvage 2-phosphoglycolate, the product of the oxygenase activity of ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco). It has been presumed that the occurrence of 2-phosphoglycolate salvage is linked to the CBB cycle, and in particular, the C2 pathway to the CBB cycle and oxygenic photosynthesis. Here, we examined 2-phosphoglycolate salvage in the hyperthermophilic archaeon Thermococcus kodakarensis, an obligate anaerobe that harbors a Rubisco that functions in the pentose bisphosphate pathway. T. kodakarensis harbors enzymes that have the potential to convert 2-phosphoglycolate to glycine and serine, and their genes were identified by biochemical and/or genetic analyses. 2-phosphoglycolate phosphatase activity increased 1.6-fold when cells were grown under microaerobic conditions compared to anaerobic conditions. Among two candidates, TK1734 encoded a phosphatase specific for 2-phosphoglycolate, and the enzyme was responsible for 80% of the 2-phosphoglycolate phosphatase activity in T. kodakarensis cells. The TK1734 disruption strain displayed growth impairment under microaerobic conditions, which was relieved upon addition of sodium sulfide. In addition, glycolate was detected in the medium when T. kodakarensis was grown under microaerobic conditions. The results suggest that T. kodakarensis removes 2-phosphoglycolate via a phosphatase reaction followed by secretion of glycolate to the medium. As the Rubisco in T. kodakarensis functions in the pentose bisphosphate pathway and not in the CBB cycle, mechanisms to remove 2-phosphoglycolate in this archaeon emerged independent of the CBB cycle.
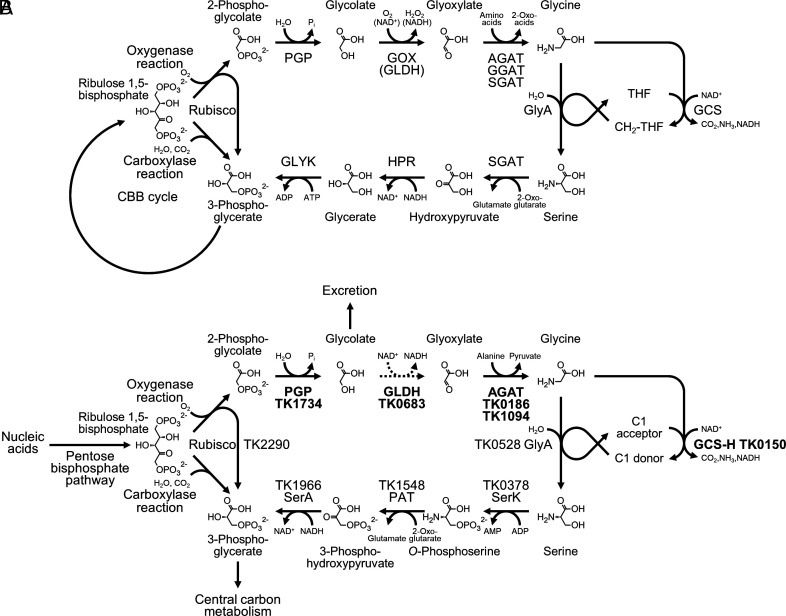
The Calvin–Benson–Bassham (CBB) cycle is responsible for CO2 fixation in plants, algae, cyanobacteria, and many other autotrophic organisms (1–3). In the CBB cycle, the CO2-fixing reaction is catalyzed by ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco). Rubisco is the most abundant enzyme on our planet, and plays an important role in sustaining the global production of organic carbon. The Rubisco carboxylase reaction converts ribulose 1,5-bisphosphate (RuBP), CO2, and H2O to two molecules of 3-phosphoglycerate (3-PGA). Rubisco also displays oxygenase activity, through which RuBP and O2 are converted to one 3-PGA and one 2-phosphoglycolate (2-PG) (2, 4). The oxygenase reaction is considered a wasteful side-reaction, with no apparent use of 2-PG. 2-PG has also been shown to inhibit the CBB pathway enzymes triose phosphate isomerase and sedoheptulose bisphosphatase in addition to phosphofructokinase of glycolysis (5–7). These observations suggest that 2-PG is a toxic compound that must be removed, but also raise the possibilities of 2-PG acting as a regulating metabolite. It has long been known that marine phytoplankton and algae excrete considerable levels of glycolate from their cells, which most likely reflects the disposal of glycolate after dephosphorylation of 2-PG (8, 9). On the other hand, pathways have evolved to remove 2-PG while minimizing the loss of carbon, such as the C2 pathway in photorespiration. The C2 pathway salvages two molecules of 2-PG and converts them into one molecule of 3-PGA while losing one CO2 and consuming one ATP (4, 10–12) (Fig. 1A). The enzymes of the C2 pathway in plants are 2-PG phosphatase, glycolate oxidase (GOX), glyoxylate:amino acid aminotransferase (GAT), the glycine cleavage system, serine hydroxymethyltransferase, serine:2-oxoacid aminotransferase, hydroxypyruvate reductase (HPR), and glycerate kinase (GLYK) (4, 10–12).
Fig. 1.
The classical C2 pathway and 2-PG removal in Thermococcus kodakarensis. (A) The classical photorespiration pathways in plants, algae, and cyanobacteria consist of 2-PG phosphatase (PGP), GOX or glycolate dehydrogenase (GLDH), alanine:glyoxylate aminotransferase (AGAT), glutamate:glyoxylate aminotransferase (GGAT) and/or serine:glyoxylate aminotransferase (SGAT), serine hydroxymethyltransferase (GlyA), glycine cleavage system (GCS), serine:2-oxoglutarate aminotransferase or SGAT, hydroxypyruvate reductase (HPR), and GLYK. (B) 2-PG removal/salvage proposed in T. kodakarensis. Dephosphorylation of 2-PG and excretion of glycolate have been experimentally verified. GLDH that catalyzes the oxidation of glycolate is not a homolog of previously identified GLDH enzymes, and is a member of the lactate dehydrogenase family. Activity levels of GLDH in T. kodakarensis cells are much lower than those of other enzymes, and the conversion is thus indicated by a dotted arrow. The enzymes in a potential route from serine to 3-phosphoglycerate are serine kinase (SerK), phosphoserine aminotransferase (PAT), and 3-phosphoglycerate dehydrogenase (SerA). Distinct to the classical C2 pathway, phosphorylation precedes transamination and reduction. Genes and proteins experimentally validated in this study are indicated in bold.
Photorespiration is tightly linked to the occurrence of oxygenic photosynthesis, which utilizes H2O as an electron donor, generating O2 (4, 10, 11). Although the pathway consumes energy, it is established that photorespiration is necessary in plants, algae, and cyanobacteria (13–18). 2-PG phosphatase (At5g36700) from Arabidopsis and GOX1 from maize are necessary for growth under normal air conditions, and their knockout strains only grow with enriched CO2. Similar observations have been reported in algae studying 2-PG phosphatase from Chlamydomonas reinhardtii and GOX from Cyanidioschyzon merolae. C. reinhardtii utilizes GLDH instead of GOX for the oxidation of glycolate, and this enzyme is necessary for growth under ambient CO2 conditions. In cyanobacteria such as Synechocystis sp. strain PCC6803, 2-PG salvage or removal is accomplished through the cooperation of three metabolic routes diverging from glyoxylate; the C2 pathway, the glycerate pathway and a decarboxylation shunt. In the glycerate pathway, two molecules of 2-PG are converted to one 3-PGA via tartronate semialdehyde. In the decarboxylation shunt, glyoxylate is oxidized to two molecules of CO2 via oxalate. In mutants with defects in one or both of two GLDH genes (glcD1, glcD2), accumulation of glycolate was observed, and the double mutant was unable to grow under low CO2 conditions. The necessity of photorespiration in a wide range of photosynthetic organisms has brought about interest in the evolution of 2-PG salvage pathways and their relationship with that of the CBB cycle (11). Did the pathways coevolve? Did photorespiration evolve after the CBB pathway as a means to cope with increases in Rubisco oxygenase activity in aerobic environments? If so, was this linked to the emergence of terrestrial plants? Interestingly, a previously reported phylogenetic analysis suggested that 2-PG phosphatase may have originated in archaea (11, 12), raising the possibilities of other evolutionary scenarios.
Recent studies have reported important findings on 2-PG and glycolate metabolism. A model chemolithoautotrophic betaproteobacterium Cupriavidus necator H16 (previously Ralstonia eutropha) harbors pathways that utilize and salvage 2-PG generated by its Rubisco (19). C. necator does not utilize the C2 pathway, but mainly utilizes the glycerate pathway, along with a malate cycle to avoid accumulation of 2-PG. Glyoxylate, with acetyl-CoA, is converted to malate by malate synthase. Malate is subsequently converted to pyruvate and then acetyl-CoA, releasing two carbons in the form of CO2. Marine alphaproteobacteria such as Paracoccus denitrificans have been demonstrated to utilize the β-hydroxyaspartate cycle to convert glycolate/glyoxylate to oxaloacetate (20). Elucidating the various systems involved in 2-PG metabolism in different organisms and determining their distribution in nature should provide valuable clues to understand their evolutionary relationship with the CBB cycle.
The Rubisco from the hyperthermophilic archaeon T. kodakarensis (Tk-Rubisco) displays both carboxylase and oxygenase activity (21, 22). Tk-Rubisco represents a group of Rubiscos (form III) that can be phylogenetically distinguished from Rubiscos (forms I and II) from organisms that utilize the CBB cycle (21). The functions of many of the form III Rubiscos are not known. Tk-Rubisco does not function in a CBB cycle, but in a pentose bisphosphate pathway for nucleoside/nucleotide conversion (23–25). In nucleoside degradation, the pathway converts the ribose moieties of nucleosides such as adenosine to ribose 1-phosphate, ribose 1,5-bisphosphate (R15P), and ribulose 1,5-bisphosphate (RuBP), the substrate for Rubisco. Adenosine is generated as a by-product when S-adenosylmethionine (SAM) is consumed for biosynthetic purposes. Rubisco activity, using CO2 and H2O, converts RuBP to two molecules of 3-PGA. In terms of pentose degradation, the pathway substitutes for the missing pentose phosphate pathway in Archaea. As T. kodakarensis is an obligate anaerobe, we had previously focused on the carboxylase activity of Tk-Rubisco. However, this archaeon can tolerate microaerobic conditions and displays growth (26), raising the possibility that, although at low levels, the oxygenase reaction of Tk-Rubisco occurs in T. kodakarensis cells, resulting in 2-PG generation. This possibility is also supported by the fact that the τ value or Srel value (kcatC/KmC)/(kcatO/KmO) (parameters for the carboxylase reaction are indicated with a C and those of the oxygenase reaction are indicated with an O) of Tk-Rubisco is not at all high, with a value of 11 ± 1 (21, 22). This poses an interesting question in terms of the evolution of photorespiration and 2-PG salvage, as it provides a situation in which a Rubisco oxygenase reaction occurs in the absence of a CBB pathway and in an obligate heterotroph. In addition, hyperthermophilic archaea are considered one of the deeply branching organisms in evolution, and the presence of a 2-PG salvage pathway in these organisms could provide insight into our understanding of the evolution of 2-PG salvage in nature.
Here, we examined the enzymes involved in the removal/salvage of 2-PG in T. kodakarensis. We identified a phosphatase specific for 2-PG, and demonstrate that its disruption leads to growth defects under microaerobic conditions. An increase in glycolate in the culture supernatant was also observed, suggesting that a pathway involving dephosphorylation of 2-PG and secretion of glycolate occurs in this hyperthermophilic archaeon. Analyses on other enzymes that could be related to this metabolism have also been carried out.
Results
Examination of Enzyme Activities Necessary to Convert 2-PG to Glycine.
T. kodakarensis cells are routinely cultured in media treated with 0.08 mM sodium sulfide (Na2S) prior to cell inoculation to remove O2, designated here as anaerobic conditions. With the addition of 0.08 mM Na2S, the oxygen indicator resazurin in the medium becomes colorless. Adding excess Na2S (e.g., 0.32 mM) results in growth inhibition (see below). Here, medium was prepared, autoclaved, and left still in a microaerobic environment (gas phase ~0.1% O2) for 24 h so that most of the oxygen was removed, and most importantly, that there would be little difference in oxygen concentrations among the individual growth flasks. The medium including resazurin is pink due to the residual oxygen present in the medium. Growth in this medium without addition of Na2S is designated here as microaerobic.
Cell extracts of T. kodakarensis KU216 grown under anaerobic or microaerobic conditions were prepared, and the enzyme activities necessary to convert 2-PG to glycine in the C2 pathway (Fig. 1A) were examined. The activities were 2-PG phosphatase (2-PG to glycolate), GOX/GLDH (glycolate to glyoxylate), and GAT (glyoxylate to glycine). We predicted that the glycolate to glyoxylate conversion would be catalyzed by GLDH, as T. kodakarensis is an anaerobe and concentrations of O2 sufficient to drive an oxidase reaction would not be available. As mentioned above, GLDHs are utilized in cyanobacteria such as Synechocystis sp. PCC6803 (18, 27). The glyoxylate to glycine conversion could, in principle, be carried out by a glycine dehydrogenase instead of an aminotransferase. However, we have previously shown that glycine dehydrogenase activity is not present in T. kodakarensis (28).
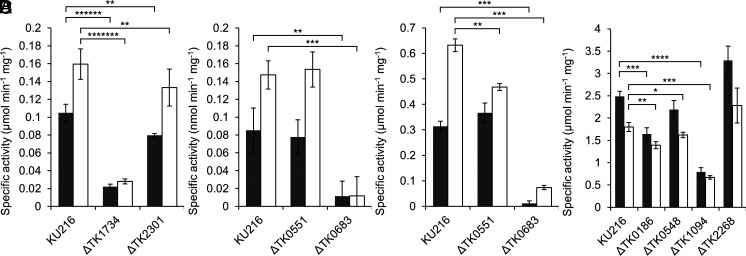
Concerning 2-PG phosphatase, specific activities of 0.10 ± 0.01 and 0.16 ± 0.02 μmol min−1 mg−1 were observed in extracts obtained from cells grown under anaerobic and microaerobic conditions, respectively (Fig. 2A). In the conversion of glycolate to glyoxylate, GLDH activity was 0.09 ± 0.03 and 0.15 ± 0.02 nmol min−1 mg−1, under the respective conditions (Fig. 2B). Higher levels of activity were found in the reverse direction from glyoxylate to glycolate; 0.31 ± 0.02 μmol min−1 mg−1 under anaerobic conditions and 0.63 ± 0.03 μmol min−1 mg−1 under microaerobic conditions (Fig. 2C). For the aminotransferase reaction in the direction of glyoxylate to glycine with alanine as the amino donor, we observed specific activities of 2.5 ± 0.1 and 1.8 ± 0.1 μmol min−1 mg−1 under anaerobic and microaerobic conditions, respectively (Fig. 2D). The 2-PG phosphatase and GLDH activities displayed a 60 and 73% increase under microaerobic conditions, respectively, whereas the aminotransferase activity decreased 28% under microaerobic conditions. The expression of 2-PG phosphatase and GLDH seem to respond to microaerobic conditions. The enzyme activities necessary to convert 2-PG to glycine were detected in the extracts of T. kodakarensis. However, the GLDH activity converting glycolate to glyoxylate was much lower than the phosphatase and aminotransferase activities.
Fig. 2.
Detection of enzyme activity in T. kodakarensis cell extracts. (A) 2-PG phosphatase activity in cell extracts from T. kodakarensis KU216, ΔTK1734, and ΔTK2301 strains. Gray or white bars indicate the specific activities in extracts from cells grown under anaerobic or microaerobic conditions, respectively. (B) GLDH activity from glycolate to glyoxylate in cell extracts from KU216, ΔTK0551, and ΔTK0683 strains. Gray or white bars indicate the specific activities in extracts from cells grown under anaerobic or microaerobic conditions, respectively. (C) GLDH activity from glyoxylate to glycolate in cell extracts from KU216, ΔTK0551, and ΔTK0683 strains. Gray or white bars indicate the specific activities in extracts from cells grown under anaerobic or microaerobic conditions, respectively. (D) AGAT activity in cell extracts from KU216, ΔTK0186, ΔTK0548, ΔTK2268, and ΔTK1094 strains. Gray or white bars indicate the specific activities in extracts from cells grown under anaerobic or microaerobic conditions, respectively. Activity measurements were carried out as described in the Methods section at 80 °C. The data represent the average of three independent experiments and are shown with the SD values. 0.01 < P ≤ 0.05*; 10−3 < P ≤ 10−2**; 10−4 < P ≤ 10−3***; 10−5 < P ≤ 10−4****; 10−7 < P ≤ 10−6******; 10−8 < P ≤ 10−7*******.
Identification of Two Candidate Genes that Encode 2-PG Phosphatase in T. kodakarensis.
We searched for candidate genes that encode 2-PG phosphatase. There is only one report of a protein from archaea exhibiting 2-PG phosphatase activity, the Ta0175 protein from Thermoplasma acidophilum (29). In T. kodakarensis, the TK2301 protein was the most similar to the Ta0175 protein (28% identical). We also searched for protein sequences related to the 2-PG phosphatase from plants. The TK1734 protein is 35% identical to the At5g36700 protein from Arabidopsis thaliana, a 2-PG phosphatase that functions in photorespiration (13). The TK1734 protein is also the most similar to the 2-PG phosphatase (CHLRE_03g168700v5) from C. reinhardtii (36%). Closely related homologs with over 20% identity to the 2-PG phosphatase (slr0458) from Synechocystis sp. PCC6803 were not present on the T. kodakarensis genome. We thus focused on TK2301 and TK1734. The TK2301 and TK1734 proteins are not structurally related to each other (16% identity).
2-PG Phosphatase Activities of the TK1734 and TK2301 Proteins.
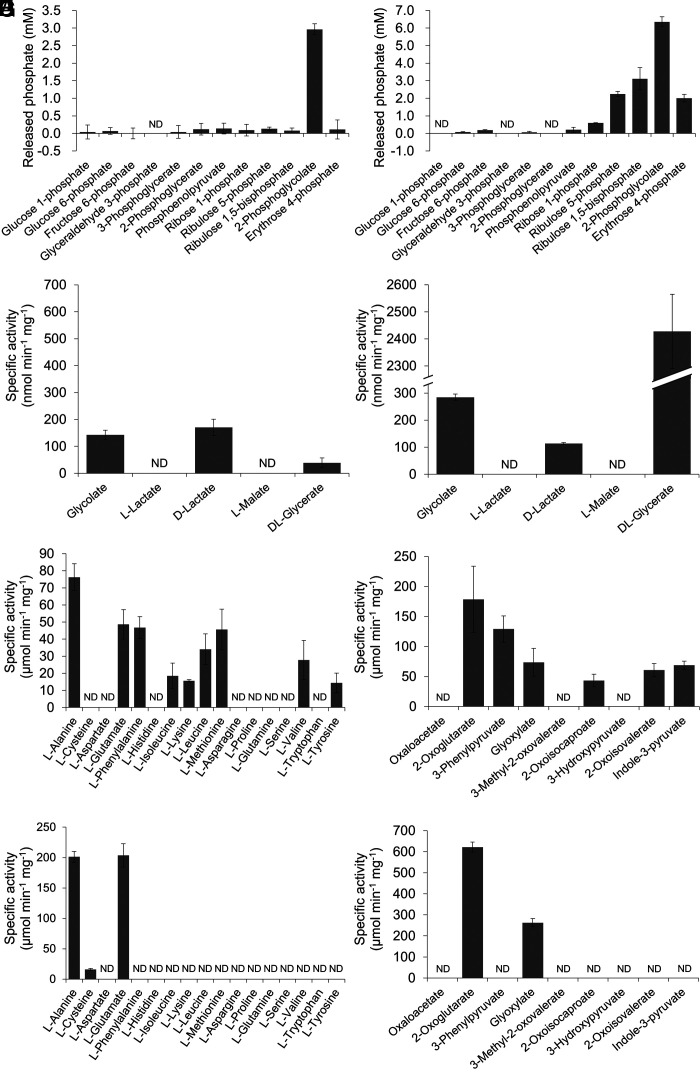
Recombinant TK1734 and TK2301 proteins were produced in Escherichia coli BL21-CodonPlus(DE3)-RIL and purified to apparent homogeneity (SI Appendix, Fig. S1 A and B). Proteins were individually incubated with 10 mM 2-PG, and the concentration of released Pi after 10 min incubation at 80 °C was measured. In the case of the TK1734 protein, generation of 2.96 ± 0.16 mM Pi was observed, while reactions with the TK2301 protein led to 6.36 ± 0.29 mM Pi (Fig. 3 A and B), suggesting that both proteins displayed 2-PG phosphatase activity. We also examined the activity of each protein toward various phosphorylated metabolites (10 mM) from glycolysis/gluconeogenesis (G1P, glucose 1-phosphate; G6P, glucose 6-phosphate; F6P, fructose 6-phosphate; GAP, glyceraldehyde 3-phosphate; 3-PGA, 3-phosphoglycerate; 2-PGA, 2-phosphoglycerate; PEP, phosphoenolpyruvate) and pentose metabolism (R5P, ribose 5-phosphate; Ru5P, ribulose 5-phosphate; RuBP, ribulose 1,5-bisphosphate; E4P, erythrose 4-phosphate). Among the substrates examined, the TK1734 protein only displayed significant levels of activity with 2-PG (Fig. 3A), whereas the TK2301 protein displayed activity with a number of substrates including 2-PG, RuBP, Ru5P, and E4P (Fig. 3B). The results suggest that the TK1734 protein is a phosphatase specific to 2-PG, whereas the TK2301 protein recognizes a relatively broad range of phosphorylated compounds.
Fig. 3.
Phosphatase, dehydrogenase, or aminotransferase activity of recombinant proteins with various substrates. Phosphatase activities of the purified TK1734 (A) and TK2301 (B) proteins were measured toward various phosphorylated metabolites. Dehydrogenase activities of the purified TK0551 (C) and TK0683 (D) proteins were examined toward various 2-hydroxyacids. Aminotransferase activities of the purified TK0186/TK1094 proteins with various amino donor compounds in the presence of glyoxylate as the amino acceptor (E/G) or various amino acceptor compounds in the presence of alanine as the amino donor (F/H) were evaluated. In all cases (A–H), activity measurements were carried out as described in the Methods section at 80 °C. ND: not detected. The data represent the average of three independent experiments and are shown with the SD values.
We performed kinetic analyses with the substrates that resulted in a relatively high level of activity; 2-PG for the TK1734 protein and 2-PG, RuBP, Ru5P, and E4P for the TK2301 protein (Table 1). Reactions were carried out at 80 °C and at optimal pH values (SI Appendix, Fig. S2). The kcat/Km value of the TK1734 protein toward 2-PG was 37.5 s−1 mM−1, higher than that of the TK2301 protein (17.4 s−1 mM−1). As for the TK2301 protein, the enzyme displays a significantly higher specificity toward RuBP (126 s−1 mM−1) among the substrates examined.
Table 1.
Kinetic parameters of the PGP candidates
| Enzyme | Substrate |
K m (mM) |
k cat (s−1) |
kcat/Km (s−1 mM−1) |
|---|---|---|---|---|
| TK1734 | 2-PG | 1.75 ± 0.22 | 65.6 ± 2.5 | 37.5 |
| TK2301 | 2-PG | 1.22 ± 0.09 | 21.2 ± 0.4 | 17.4 |
| E4P | 1.25 ± 0.28 | 17.1 ± 1.0 | 13.7 | |
| Ru5P | 0.979 ± 0.157 | 16.3 ± 0.5 | 16.6 | |
| RuBP | 0.238 ± 0.044 | 30.1 ± 1.3 | 126 |
All analyses were carried out at 80 °C.
Gene Disruption of TK1734 and TK2301 and Their Effects on 2-PG Phosphatase Activity.
To examine the contribution of TK1734 and TK2301 to the 2-PG phosphatase activity observed in T. kodakarensis cell extracts, the genes were individually disrupted. T. kodakarensis KU216 was used as the host strain. Gene disruption was examined by PCR analysis (SI Appendix, Fig. S3 A and B), followed by confirmation with DNA sequencing.
2-PG phosphatase activities were measured in cell extracts of the individual gene disruption strains grown under anaerobic or microaerobic conditions (Fig. 2A). Compared to the specific activities observed in cell extracts of KU216, we observed a dramatic decrease (79 to 82%) in activity in extracts of ΔTK1734 under both conditions. In contrast, activity in cell extracts of ΔTK2301 was only 16 to 24% lower than that of KU216. The results indicate that, at least under our conditions for growth and activity measurements, the TK1734 protein is responsible for the majority of 2-PG phosphatase activity in T. kodakarensis cells. It is worthy to note that the increase in 2-PG phosphatase activity in cells grown under microaerobic conditions can be attributed to the increase in activity of the TK1734 protein. Together with the high specificity of the TK1734 protein toward 2-PG, our results suggest that TK1734 encodes the physiologically relevant 2-PG phosphatase in this organism.
Identification of Two GLDH Candidate Genes.
We next searched for a protein that converts glycolate to glyoxylate. On the T. kodakarensis genome, there was no homolog of the GOX (GOX, At3g14415) from A. thaliana. Cyanobacteria and some algae such as C. reinhardtii use a GLDH. As we observed NAD+-dependent GLDH activity in cell extracts, we searched for homologs of characterized GLDH proteins. We could not find homologs of the GLDHs from cyanobacteria Synechocystis sp. PCC 6803 (sll0404) (18) nor algae C. reinhardtii (CHLRE_10g434900v5) (16). Taking the structures of glycolate and glyoxylate into account, we then searched for putative 2-hydroxyacid dehydrogenases. Three candidate genes (TK0551, TK0683, and TK1966), all encoding proteins of the d-lactate dehydrogenase (LDH) family, were found on the T. kodakarensis genome. TK0551 and TK0683 displayed 42% identity, TK0683 and TK1966 displayed 45% identity, and TK1966 and TK0551 displayed 37% identity to one another. Among the three, the TK1966 protein was previously identified as a 3-PGA dehydrogenase (SerA), involved in the conversion of 3-PGA to serine (30). Among the two remaining genes, TK0551 and TK0683, proteins encoded by TK0683 homologs from Thermococcales have been biochemically characterized. The OCC_02245 protein from Thermococcus litoralis and the PH0597 protein from Pyrococcus horikoshii were described as reductases preferring glyoxylate (31, 32), while another report concluded that the proteins encoded by PH0597, PF0319 from Pyrococcus furiosus, and PYCH_09300 from Pyrococcus yayanosii are actually HPRs (33). TK0551 homologs in Thermococcales have not yet been characterized and the physiological roles of TK0551 and TK0683 are still unclear.
GLDH Activities of the TK0551 and TK0683 Proteins.
The TK0551 and TK0683 proteins were produced in E. coli BL21-CodonPlus(DE3)-RIL and purified to apparent homogeneity (SI Appendix, Fig. S1 C and D). The proteins were individually incubated with 10 mM glycolate and 10 mM NAD+ at 80 °C. Both proteins showed detectable levels of NADH generation dependent on glycolate, 142 ± 18 nmol min−1 mg−1 for the TK0551 protein and 285 ± 12 nmol min−1 mg−1 for the TK0683 protein (Fig. 3 C and D). We further examined the dehydrogenase activity of each protein toward other 2-hydroxyacids (d- or l-lactate, l-malate, dl-glycerate). Among the substrates examined, the TK0551 protein exhibited relatively high activity with glycolate and d-lactate (Fig. 3C) while the TK0683 protein showed activity with glycolate, d-lactate and dl-glycerate (Fig. 3D).
Kinetic analyses were performed on both proteins toward glycolate and d-lactate, and dl-glycerate for the TK0683 protein (Table 2). The kcat/Km value of the TK0683 protein toward glycolate was 0.486 s−1 mM−1 which is 31.2 times higher than that of the TK0551 protein (0.0156 s−1 mM−1). However, the highest kcat/Km value of TK0683 was obtained with d- or l-glycerate (6.56 s−1 mM−1), which was calculated with the assumption that the protein recognizes only one chiral species. When the reaction in the reverse direction (reduction) was examined, we observed activity toward various substrates other than glyoxylate with the TK0551 protein. The highest kcat/Km value was observed with glyoxylate. In the case of the TK0683 protein, relevant levels of activity were only observed with glyoxylate and 3-hydroxypyruvate, with reactions with the latter substrate displaying higher kcat/Km values. As in the forward reaction, the kcat/Km value of the TK0683 protein (1.36 × 103) toward glyoxylate was higher than that of the TK0551 protein (64.8).
Table 2.
Kinetic parameters of the GLDH candidates
| Enzyme | Substrate |
K m (mM) |
k cat (s−1) |
kcat/Km (s−1 mM−1) |
|---|---|---|---|---|
| TK0551 | Glycolate* | 24.1 ± 1.26 | 0.376 ± 0.009 | 0.0156 |
| d-Lactate* | 9.90 ± 1.15 | 0.324 ± 0.013 | 0.0327 | |
| Glyoxylate** | 2.87 ± 0.39 | 186 ± 2 | 64.8 | |
| 3-Hydroxypyruvate** | 3.73 ± 0.25 | 104 ± 3 | 27.9 | |
| 3-Methyl-2-oxovalerate** | 1.93 ± 0.25 | 18.3 ± 2.2 | 9.48 | |
| 2-Oxoisovalerate** | 1.90 ± 0.44 | 15.9 ± 2.2 | 8.37 | |
| 2-Oxovalerate** | 1.44 ± 0.09 | 14.0 ± 0.3 | 9.72 | |
| Pyruvate** | 6.89 ± 0.81 | 12.4 ± 0.6 | 1.78 | |
| TK0683 | Glycolate* | 0.44 ± 0.06 | 0.213 ± 0.005 | 0.49 |
| d-Lactate* | 0.44 ± 0.06 | 0.105 ± 0.003 | 0.24 | |
| dl-Glycerate* | 0.66 ± 0.06*** | 4.31 ± 0.10 | 6.6*** | |
| Glyoxylate** | 1.49 ± 0.80 | 2030 ± 92 | 1.36 × 103 | |
| 3-Hydroxypyruvate** | 0.31 ± 0.07 | 3130 ± 462 | 1.0 × 104 |
*Analyses were carried out at 80 °C under the presence of 10 mM NAD+.
**Analyses were carried out at 85 °C under the presence of 0.3 mM NADH.
***The values were calculated under an assumption that the protein recognizes only one of chiral species.
Gene Disruption of TK0551 and TK0683 and Their Effects on GLDH Activity.
We individually disrupted TK0551 and TK0683 from the host strain T. kodakarensis KU216. Gene disruption was confirmed by PCR analysis (SI Appendix, Fig. S3 C and D) and DNA sequencing. We measured GLDH activity converting glycolate to glyoxylate in the cell extracts of the KU216, ΔTK0551, and ΔTK0683 strains grown under anaerobic and microaerobic conditions. In extracts from cells grown in either condition, a large decrease in GLDH activity was observed only in ΔTK0683 cells (Fig. 2B). This was further supported by measuring activities of the reverse reaction, which were much higher. A much larger decrease in activity was observed in ΔTK0683 compared to ΔTK0551 (Fig. 2C). The results indicate that the TK0683 protein is responsible for a major portion (~90%) of GLDH activity detected in T. kodakarensis.
Identification of Glyoxylate Aminotransferase Candidate Genes.
We next searched for genes predicted to encode aminotransferases that would convert glyoxylate to glycine. In plants such as A. thaliana, multiple enzymes are involved in this conversion; AGAT, GGAT, and SGAT (34). An aminotransferase specific toward glyoxylate and alanine from T. litoralis, encoded by OCC_02240, has been reported (35). A BLAST search using the amino acid sequence of OCC_02240 as the query against the T. kodakarensis proteome resulted in the identification of multiple proteins (encoded by TK0186, TK2268, TK1094, TK0548, TK0260, and TK0250) with varying degrees of homology, suggesting that multiple proteins might exhibit aminotransferase activity with glyoxylate as the amino acceptor. Among the six candidate genes, TK0250 and TK0260 are included in the biosynthesis gene clusters of His and Tyr/Phe, respectively, suggesting that these genes are specifically involved in the biosynthesis of these amino acids (36). Among the remaining four candidates, the biochemical properties of two have recently been reported (36). The remaining two, encoded by TK0186 and TK1094, were examined here in detail. The TK0186 protein was most related (41% identical) to the OCC_02240 protein specific toward glyoxylate and alanine.
Glyoxylate Aminotransferase Activity of the TK0186 and TK1094 Proteins.
The TK0186 and TK1094 genes were expressed in E. coli BL21-CodonPlus(DE3)-RIL and the recombinant proteins were purified. Concerning the TK0186 protein, we consistently observed low-molecular-weight contaminants in the SDS-PAGE gels. We examined whether these contaminants were degradation products of the TK0186 protein by varying the periods of time the protein was heated after mixing with sample buffer (SI Appendix, Fig. S1E). The extent of degradation of the TK0186 protein increased with time and the degradation patterns matched those observed in our original gels, indicating that the TK0186 protein was purified to apparent homogeneity. The TK1094 protein was also purified, and its homogeneity is shown in SI Appendix, Fig. S1F.
AGAT activity was measured by monitoring glycine generation upon incubating the TK0186 or TK1094 protein, along with the previously characterized TK0548 and TK2268 proteins, with 10 mM alanine and 10 mM glyoxylate. AGAT activity was observed only in the TK0186 and TK1094 proteins. The amino donor selectivities of the TK0186 and TK1094 proteins using glyoxylate as the amino acceptor were evaluated by monitoring glycine formation in the presence of 17 individual amino acids. For the TK0186 protein, l-alanine, l-glutamate, l-phenylalanine, l-isoleucine, l-lysine, l-leucine, l-methionine, l-valine, and l-tyrosine were recognized as substrates, and the most preferred amino donor was l-alanine (Fig. 3E). In the case of the TK1094 protein, activity was observed with l-alanine and l-glutamate, along with low activity with l-cysteine (Fig. 3G). The amino acceptor selectivity was then examined with various 2-hydroxyacid compounds using l-alanine as the amino donor, and the generation of the corresponding amino acids was monitored (Fig. 3 F and H). With the TK0186 protein, we observed product generation with 2-oxoglutarate, 3-phenylpyruvate, glyoxylate, 2-oxoisocaproate, 2-oxoisovalerate, and indole-3-pyruvate, and the most preferred amino acceptor was 2-oxoglutarate. The TK1094 protein accepted only 2-oxoglutarate and glyoxylate.
Gene Disruption and Its Effect on AGAT Activity.
To evaluate the contribution of each aminotransferase gene on AGAT activity in the cell, individual gene disruption strains were constructed. The disruption of TK1094 (37), TK0548, and TK2268 (36) have previously been reported. The intergenic regions between TK0186 and TK0185, and TK0186 and TK0187, are 240 bp and 67 bp, respectively, with only short distances separating the genes. Disruption of TK0186 was designed so that the transcription of adjacent genes was not affected (SI Appendix, Fig. S3E). Gene disruption was confirmed by PCR analysis and DNA sequencing.
AGAT activities in the cell extracts of the gene disruption strains grown under anaerobic and microaerobic conditions were examined. Compared to the AGAT activity in extracts from KU216 cells grown microaerobically, those in ΔTK0186 (23%) and ΔTK1094 (63%) displayed the largest decreases, followed by ΔTK0548 (10%) (Fig. 2D). The same trend was observed when cells were grown anaerobically. The results suggest that multiple proteins contribute in the AGAT activity in T. kodakarensis, particularly TK1094, TK0186, and to some extent TK0548.
Identification of Candidate Genes Encoding a Glycine Cleavage System.
In the classical photorespiration system, the glycine generated from 2-PG is converted by two enzymes. One glycine molecule is converted by serine hydroxymethyltransferase (GlyA). In T. kodakarensis, TK0528 encodes GlyA, and is responsible for the transfer of a hydroxymethyl group to glycine to form serine (30). The other enzyme is the glycine cleavage system (GCS), which catalyzes the conversion of glycine and NAD+ to CO2, NH4+, NADH, and a methylene group bound to tetrahydrofolate (THF) (SI Appendix, Fig. S4A). GCS is composed of four loosely interacting components: T-protein (aminomethyltransferase), P-protein (glycine decarboxylase), L-protein (dihydrolipoyl dehydrogenase), and H-protein, a protein modified with lipoic acid that interacts with all other components (SI Appendix, Fig. S4A) (38). Homologs in T. kodakarensis similar to GCS genes from Synechocystis sp. PCC6803 were T-protein (TK2035, 33% identical to sll0171), P-protein subunit 1 and 2 (TK1380 and TK1379, 37% and 41% identical to the corresponding domains of slr0293, respectively) and H-protein (TK0150, 46% identical to slr0879). There were multiple genes whose proteins displayed limited homology with the L-protein (slr1096). An enzyme responsible for lipoic acid biosynthesis has been identified, suggesting the function of GCS in T. kodakarensis (39, 40).
Gene Disruption of TK0150 and Its Effect on Growth.
As the components of GCS only loosely interact with one another and might be difficult to reconstitute in vitro, we carried out a genetic examination. The TK0150 gene predicted to encode the H-protein was disrupted. Gene disruption was confirmed by PCR analysis (SI Appendix, Fig. S3F) and DNA sequencing.
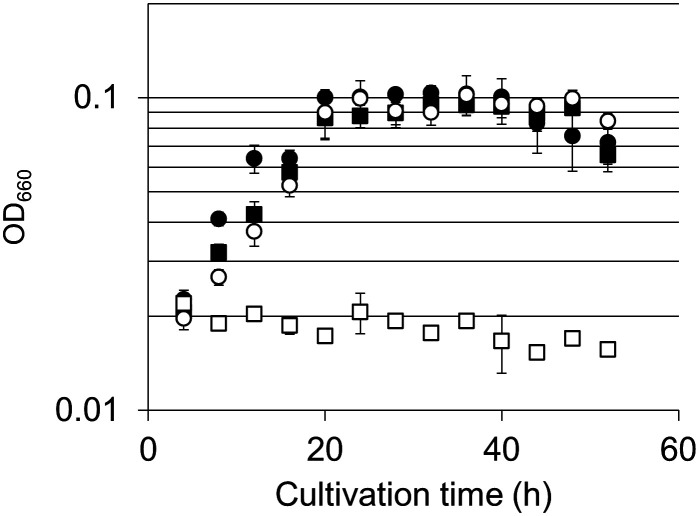
Growth characteristics of the ΔTK0150 strain and host strain KU216 were evaluated. It has previously been demonstrated that disruption of the serine hydroxymethyltransferase gene (glyA, TK0528) results in a strain that displays serine auxotrophy in ASW-AA-S0 medium. We thus presumed that if the TK0150 protein acts as an H-protein of GCS, disruption of TK0150 would also result in serine auxotrophy due to a shortage in supply of C1 units (5,10-methylenetetrahydrofolate) for the serine hydroxymethyltransferase reaction. Growth of ΔTK0150 and KU216 in ASW-AA-S0 with or without serine was examined. In the case of KU216, growth in ASW-AA-S0 medium was not affected by the presence or absence of serine. With the ΔTK0150 strain, growth similar to KU216 was observed in medium with serine. However, in medium without serine, we could not observe growth of the ΔTK0150 strain, indicating that disruption of TK0150 resulted in serine auxotrophy (Fig. 4 and SI Appendix, Fig. S5A). The serine auxotrophy observed in the ΔTK0150 strain was due to a shortage of 5,10-methylenetetrahydrofolate and not an impairment in the serine biosynthesis pathway per se, as described in SI Appendix. The results strongly suggest that TK0150 functions as the H-protein of GCS in T. kodakarensis, contributing to 5,10-methylenetetrahydrofolate generation in this organism. The presence of activities of 2-PG phosphatase, GLDH, AGAT, serine hydroxymethyltransferase, and GCS suggests that T. kodakarensis harbors the activities to salvage 2-PG and convert the compound to serine.
Fig. 4.
Growth properties of T. kodakarensis KU216 and ΔTK0150 strains. Serine auxotrophy was examined by cultivating cells in ASW-AA-S0-Ura+ medium with or without serine. Symbols: KU216 with (closed circles) or without serine (closed squares), ΔTK0150 with (open circles) or without (open squares) serine. Error bars indicate the SD values of three independent culture experiments.
Growth Effects of Disrupting 2-PG Salvage.
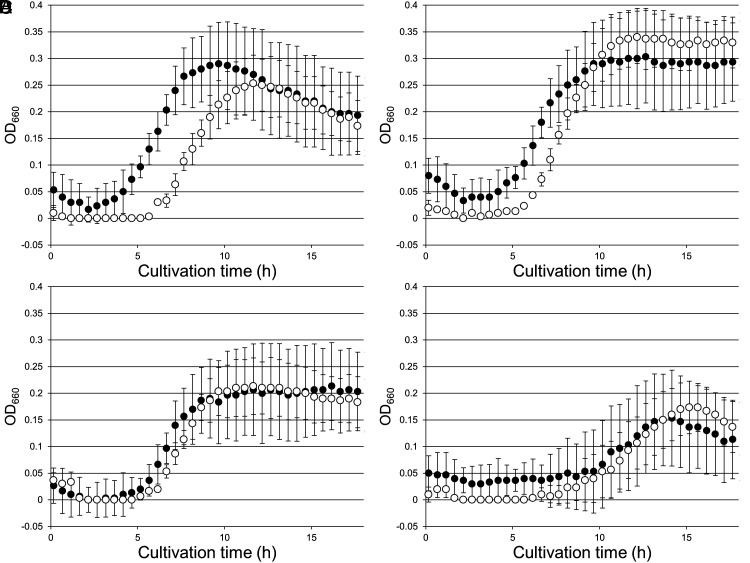
To observe the effects of interrupting 2-PG salvage, we first examined the gene disruption strain of TK1734, encoding 2-PG phosphatase. We examined growth with the addition of different concentrations of Na2S, which we routinely use to remove O2 until the resazurin in the medium becomes colorless. The medium without addition of Na2S used for growth experiments in Fig. 5A was pink, indicating that residual oxygen was present in the medium. The host strain KU216 did not display growth impairment in this medium, as addition of 0.08 mM Na2S did not significantly enhance growth (Fig. 5 A and B). We did however observe that cell lysis occurs more rapidly in cells grown without Na2S. On the other hand, addition of 0.16 mM Na2S resulted in a slight decrease in cell yield (Fig. 5C). Higher concentrations (0.32 mM) of Na2S had stronger, negative effects on growth (Fig. 5D). With the ΔTK1734 strain, we observed impaired growth compared to KU216 cells in the absence of Na2S (Fig. 5A). When Na2S was added at a concentration of 0.08 mM, growth of the ΔTK1734 strain was significantly enhanced and was comparable to that of the host strain (Fig. 5B). Addition of higher concentrations of Na2S had a negative effect on the growth of both strains (Fig. 5 C and D), but there were no longer large differences in growth between KU216 and ΔTK1734. This suggests that the impairment in growth of the ΔTK1734 strain was due to the residual oxygen in the medium in the absence of Na2S, most likely due to an increase in the oxygenase activity of Rubisco.
Fig. 5.
Growth properties of T. kodakarensis KU216 and ΔTK1734 strains under microaerobic conditions. KU216 (closed circles) and ΔTK1734 (open circles) strains were cultivated in ASW-YT-m1-Pyr medium. All media were left in an anaerobic box for 24 h prior to inoculation to allow dissolved gases to equilibrate with the atmosphere in the anaerobic box (oxygen concentration: ~0.1%). Media for (A) were prepared without Na2S (microaerobic conditions), and the concentration of Na2S in (B–D) were 0.08 mM, 0.16 mM, and 0.32 mM, respectively. Error bars indicate the SD values of three independent culture experiments.
To further examine whether this was the case, we designed an artificial situation aimed to increase the intracellular concentration of RuBP, the Rubisco substrate. Nucleosides are readily taken up by T. kodakarensis (41), and metabolized by the pentose bisphosphate pathway, generating RuBP (25). We grew cells in a medium with or without Na2S, but with high concentrations of uridine, 25 g L−1. As shown in SI Appendix, Fig. S6A, the difference in growth observed in Fig. 5A was dramatically amplified, supporting the notion that the growth impairment observed with residual oxygen is indeed due to an increase in the oxygenase reaction of Rubisco and the absence of a mechanism to remove 2-PG. Although not complete, adding Na2S enhanced the growth of ΔTK1734 cells with uridine to an extent that it became comparable with that of KU216, again suggesting that the growth impairment was dependent on oxygen (SI Appendix, Fig. S6 B–D).
We next examined the growth of ΔTK0683 cells (lacking GLDH) under anaerobic and microaerobic conditions in comparison with KU216 (SI Appendix, Fig. S8). ΔTK0683 cells displayed a slight decrease in growth rate and yield under microaerobic conditions compared to KU216 cells. However, in contrast to the ΔTK1734 strain (lacking 2-PG phosphatase), we did not observe recovery of growth with the addition of Na2S, suggesting that the change in phenotype may not be related to glycolate metabolism. We also examined the growth of ΔTK0150 (lacking GCS:H-protein). With this strain, a much more severe growth defect was observed. As GCS is responsible for providing C1 units to tetrahydrofolate, and can be considered to be involved in many biosynthesis pathways, the degree of impairment in growth was not surprising. As the addition of Na2S had little effect on the growth defect, the role of this enzyme is clearly not limited to glycolate metabolism.
Taking into account the low levels of activity of GLDH converting glycolate to glyoxylate in the cell extracts of T. kodakarensis, and the lack of growth impairment responding to oxygen in the ΔTK0683 strain, we considered the possibility that T. kodakarensis does not actively assimilate glycolate, but excretes the compound after the 2-PG phosphatase reaction. In order to detect 2-PG assimilation, we grew the cells in the presence of 13C-labeled glycolate. If T. kodakarensis were able to take up exogenous 13C-labeled glycolate, we would be able to monitor the assimilation pathway by tracer-based metabolomics using the methods described in SI Appendix. However, in protein-derived amino acids from the T. kodakarensis KU216 cells grown in ASW-YT-m1-Pyr medium supplemented with 1,2-13C-labeled glycolate, 13C-labeled amino acids were not detected. In order to detect whether glycolate is excreted from cells, we measured the concentration of glycolate present in the culture medium of T. kodakarensis grown under microaerobic conditions (SI Appendix, Methods and Fig. S9). Using LC–MS/MS, we observed increases in concentrations of glycolate (8.2 ± 2.5 μM) in the medium compared to those observed in medium without cell inoculation. Using LC–MS/MS and LC–high resolution MS, we further confirmed that the analyzed compound exhibited the same exact mass as that of commercially available glycolate. Although we do not rule out the possibility of glycolate assimilation to glycine and/or serine, the results suggest the major direction of 2-PG removal proceeds toward excretion after dephosphorylation.
Discussion
The results presented here indicate that T. kodakarensis harbors a 2-PG phosphatase that functions to remove 2-PG which is generated from the oxygenase activity of Rubisco. We observed that the expression of 2-PG phosphatase (60%) and GLDH (70%) respond to the presence of oxygen. However, GLDH activity in the direction of glyoxylate formation was much lower than that of 2-PG phosphatase. Gene disruption of 2-PG phosphatase displayed a phenotype responding to oxygen, whereas that of GLDH did not. Furthermore, glycolate was detected in the culture supernatant of T. kodakarensis grown under microaerobic conditions. As we also could not detect incorporation of exogenous glycolate into protein-derived amino acids, the current data suggest that 2-PG removal in T. kodakarensis is mainly achieved via dephosphorylation and glycolate excretion.
Further analyses will be necessary to establish whether a portion of glycolate is converted to glycine and other compounds in T. kodakarensis. Downstream of the GLDH reaction, the glyoxylate:amino acid aminotransferase activities in T. kodakarensis cell extracts were relatively high. The functions of GCS and serine hydroxymethyltransferase have been experimentally verified here and in a previous report (30), respectively. In addition, the T. kodakarensis genome harbors homologs of 3-phosphoglycerate dehydrogenase (TK1966), phosphoserine aminotransferase, and phosphoserine phosphatase. Using a TK1966 gene disruption strain, it was genetically demonstrated that 3-PGA is a precursor for serine biosynthesis (30). As T. kodakarensis harbors a serine kinase (30), and 3-phosphoglycerate dehydrogenase and phosphoserine aminotransferase are known to be reversible, the results suggested that serine and 3-PGA are metabolically linked. This implies that glycine can be converted to 3-PGA, which, although through different chemistry, corresponds to the downstream portion of the C2 pathway of photorespiration (Fig. 1B).
TK1734 encodes the physiologically relevant 2-PG phosphatase in T. kodakarensis. Interestingly, the gene encoding rubrerythrin, known to be involved in oxidative stress tolerance, is immediately downstream of the TK1734 gene and may constitute an operon that responds to oxidative stress. Among the Thermococcales genomes, this same gene orientation is seen in 31 genomes among 40 examined. Although the ΔTK1734 strain displayed growth impairment responding to residual oxygen and nucleosides (Fig. 5), we do not regard 2-PG removal as a major antioxidant system, as it contributes only to the removal and conversion of the Rubisco biproduct 2-PG generated in the presence of O2. Proteins such as superoxide reductase and a putative flavodiiron protein have been shown to be the major players against oxygen stress in another member of the Thermococcales, Pyrococcus furiosus (42), whose homologs are also present in T. kodakarensis.
We examined the presence of TK1734 homologs in archaea and compared their distribution with that of Rubisco. The presence of both Rubisco and TK1734 homologs (E-value < 10−5) is observed throughout members of Thermococcales with the exception of a single species Thermococcus piezophilus, which lacks a Rubisco. We also find the two genes in various members of Archaeoglobales, halophilic archaea, Thermoplasmatales, Lokiarchaeota, and some members of Thermoproteales. There are also a large number of methanogens that harbor a Rubisco but not a 2-PG phosphatase. This may reflect the fact that the methanogens occupy the most anaerobic environments, and that the oxygenase activity of Rubisco in these cells is negligible. An interesting observation is that the Rubisco from the methanogen Methanocaldococcus jannaschii is itself oxygen-sensitive (43). In addition to methanogens, all genera that harbor Rubisco but do not possess PGP (Halodesulfurarchaeum, Halanaeroarchaeum, Desulfurococcus, Thermosphaera, Ignisphaera, Thermogladius, Hyperthermus, Thermofilum, and Infirmifilum) are strictly anaerobic, while at least one copy of a PGP homolog is found on the genomes of all genera that harbor a Rubisco but are aerobic. This tendency suggests an evolutionary linkage between the occurrence of Rubisco oxygenase activity and 2-PG phosphatases.
As described above, TK1734 homologs are conserved throughout Thermococcales, suggesting that a system for removal or salvage of 2-PG was present in the ancestor of this order, emerging independently of the CBB cycle. Concerning when 2-PG salvage occurred in archaea, there is a possibility that the enzymes/genes involved in 2-PG salvage were obtained after the occurrence of photorespiration in plants, algae, and/or cyanobacteria. However, phylogenetic studies on 2-PG phosphatase indicate that the enzymes utilized by photosynthetic organisms have an archaeal origin (11). This raises the possibility that removal of 2-PG by 2-PG phosphatase and excretion evolved in archaea prior to their counterparts in photosynthetic organisms such as those reported for algae and marine phytoplankton (8, 9).
It is presumed that oxygen levels were less than 10−5 times that of the present atmospheric levels before the Great Oxidation Event (44). However, there is a growing amount of geological evidence that, coinciding with the early emergence of cyanobacteria, there were “whiffs” of oxygen late in the Archaean (>2.3 Gyr ago) prior to the Great Oxidation Event (45, 46). If the ancestors of the Thermococcales emerged prior to the emergence of cyanobacteria on Earth, our results support the notion that molecular oxygen was present at levels that allowed the selection and emergence of a mechanism to remove 2-PG prior to the Great Oxidation Event.
Methods
Chemicals, Strains, and Medium.
Unless mentioned otherwise, chemical reagents were purchased from Nacalai Tesque (Kyoto, Japan), Fujifilm Wako Pure Chemicals (Osaka, Japan), or MilliporeSigma (Burlington, MA). Strains and plasmids used in this study are listed in SI Appendix, Table S2. T. kodakarensis KOD1 (47, 48) (wild type), KU216 (ΔpyrF mutant), and its derivative strains were cultivated under anaerobic conditions at 85 °C in a nutrient-rich medium (ASW-YT and ASW-YT-m1) or a synthetic medium (ASW-AA and ASW-AA-m1). Compositions of liquid and solid media are indicated in SI Appendix. In routine cultures, Na2S was added prior to inoculation to remove O2. E. coli DH5α (TaKaRa, Ohtsu, Japan) and BL21-CodonPlus(DE3)-RIL (Agilent Technologies, Santa Clara, CA) strains were used for plasmid construction and heterologous gene expression, respectively. Cells were cultivated at 37 °C in Lysogeny Broth (LB) medium containing 100 mg L−1 ampicillin.
Gene Expression in E. coli and Purification of the Recombinant Proteins.
The TK0186, TK0551, TK0683, TK1094, TK1734, and TK2301 genes were individually inserted into pET21a(+) expression plasmid (Merck KGaA, Darmstadt, Germany), and expressed in E. coli. Recombinant proteins were purified from cell extracts through combinations of heat treatment, anion exchange, hydrophobic interaction, and/or gel-filtration chromatography. Details for each protein are indicated in SI Appendix.
Phosphatase Activity of Purified TK1734 and TK2301 Proteins.
Phosphatase activity of the TK1734 and TK2301 proteins was measured with substrates shown in Fig. 3 A and B. Reaction mixtures contained 10 mM substrate, 10 mM MgCl2, and 10 μg mL−1 protein in 50 mM Tris (pH 8.0). Reactions were carried out for 10 min at 80 °C and initiated by addition of phosphorylated metabolite after 3 min preincubation of the other components. As for all other enzyme assays, reactions were terminated by cooling on ice, and proteins were removed with an Amicon Ultra centrifugal filter unit (MWCO 3000). The concentration of released inorganic phosphate was determined using a Malachite Green Phosphate Detection Kit (R&D Systems, Minneapolis, MN).
To examine the effects of pH, acetate-NaOH (pH 4.0 to 5.5), MES-NaOH (pH 5.5 to 7.0), HEPES-NaOH (pH 7.0 to 8.0), and Bicine-NaOH (pH 8.0 to 9.0) were used. Reaction mixtures contained 10 mM 2-PG, 10 mM MgCl2, and 1.0 μg mL−1 protein in 50 mM buffer. Reactions were carried out as described above at 80 °C for 1, 2, and 3 min to calculate reaction rates. In kinetic analyses, reaction mixtures contained various concentrations of phosphorylated substrate, 10 mM MgCl2, and 1.0 μg mL−1 protein in 50 mM MES-NaOH (pH 7.0 for the TK1734 protein, pH 6.5 for the TK2301 protein). Reactions were carried out at 80 °C for 0, 1, 2, and 3 min. Reactions were initiated as described above. Substrates examined are shown in Table 1. Kinetic parameters were calculated with IGOR Pro v. 5.03 (Wave-Metrics, Lake Oswego, OR)
Dehydrogenase Activity of Purified TK0551 and TK0683 Proteins.
Activity of the TK0551 and TK0683 proteins in the direction of oxidation was measured with the sodium salts of the 2-hydroxyacids glycolate, d- or l-lactate, l-malate, and dl-glycerate. The reaction mixture contained 10 mM substrate, 10 mM NAD+, and 10 μg mL−1 protein in 50 mM Tris (pH 7.5 at 80 °C). Reactions were carried out at 80 °C and initiated by addition of 2-hydroxyacid after 3 min preincubation of the other components. The rate of NADH generation was measured by monitoring absorbance at 340 nm. Kinetic analyses were carried out using various concentrations of 2-hydroxyacid. For kinetic analyses in the direction of reduction, reaction mixtures contained various concentrations of 2-oxoacid (Table 2), 0.3 mM NADH, and 0.5 μg mL−1 TK0551 protein or 0.05 μg mL−1 TK0683 protein in 50 mM Tris (pH 7.5 at 85 °C). Reactions were carried out at 85 °C and initiated by addition of NADH after 2 min preincubation of the other components. The rate of NADH consumption was measured by monitoring absorbance at 340 nm. Kinetic parameters were calculated with IGOR Pro v. 5.03.
Aminotransferase Activity of Purified TK0186 and TK1094 Proteins.
Amino donor selectivity was evaluated by monitoring glycine formation from glyoxylate with the amino donors shown in Fig. 3 E and G. Reaction mixtures contained 10 mM amino acid, 10 mM sodium glyoxylate, 20 μM pyridoxal 5′-phosphate (PLP) and 1.0 μg mL−1 protein in 50 mM HEPES (pH 7.5). Reactions were initiated by addition of amino acid after 3 min preincubation of the other components at 80 °C and continued for 0, 1, 2, and 3 min. After terminating the reaction and removing proteins, the concentration of glycine was measured with HPLC (Nexera X2 system, Shimadzu, Kyoto, Japan). Samples were diluted eightfold in 400 mM Na2BO4 buffer (pH 10.4) and 5 min prior to applying to HPLC, mixed with 1/4 equivalent volume of labeling solution (8 mg mL−1 o-phthalaldehyde, 10 mg mL−1 N-acetylcysteine in methanol) to derivatize glycine for fluorescent detection. Derivatized samples were applied to a C18 column, COSMOSIL 5C18-PAQ (Nacalai Tesque), and separated with an equal volume mixture of 20 mM sodium acetate (pH 5.6) and methanol as the mobile phase. Column temperature was set at 40 °C. Labeled glycine was excited by UV (350 nm) and fluorescence (450 nm) was detected. Concentrations of glycine were determined with a standard curve generated with defined concentrations of glycine.
Amino acceptor selectivity was examined by monitoring the generation of aspartate, glutamate, phenylalanine, glycine, isoleucine, leucine, serine, valine, and tryptophan from their respective 2-oxoacids (Fig. 3 F and H) using l-alanine as the amino donor. The reaction mixture contained 10 mM alanine, 10 mM amino acceptor (sodium salt), 20 μM PLP and 1.0 μg mL−1 protein in 50 mM HEPES (pH 7.5) and incubated at 80 °C for 0, 1, 2, and 3 min. The reaction was initiated by addition of amino acceptor after 3 min preincubation of other components. After terminating the reaction and removing proteins, the concentration of generated amino acids was measured with HPLC. Samples were derivatized and analyzed by HPLC as described above. Concentrations of amino acids were determined with a standard curve generated with defined concentrations of each amino acid.
Construction of Gene Disruption Strains.
Disruption plasmids for TK0150, TK0186, TK0551, TK0683, TK1734, and TK2301 were constructed and introduced into T. kodakarensis KU216. Details for plasmid construction and transformation of T. kodakarensis are described in SI Appendix.
Investigations of Growth Properties of Gene Disruption Strains.
For investigation of C1 donor auxotrophy, the ΔTK0150 and host strains were precultivated in ASW-YT-S0 medium. Equivalent amounts of cells were inoculated into 20 mL ASW-AA-S0 with or without serine, valine, or 2 mM ketopantoate, and cultivated at 85 °C.
To evaluate growth in the presence of oxygen, the ΔTK1734, ΔTK0683, ΔTK0150, and host strains were precultivated in ASW-YT-m1-Pyr medium without Na2S. The medium was left in an anaerobic chamber for at least 24 h prior to inoculation to allow dissolved gases to equilibrate with the atmosphere in the anaerobic chamber (oxygen concentration ~0.1%). Equivalent amounts of cells were inoculated into anaerobic (with 0.08, 0.16, or 0.32 mM Na2S) or microaerobic (without Na2S) 10 mL ASW-YT-m1-Pyr in vials (Fig. 5 and SI Appendix, Fig. S6) or test tubes (SI Appendix, Fig. S8) with or without 25 g L−1 uridine, and cultivated at 85 °C. Growth was examined by monitoring OD660 using OD-BR-43FH-VH (TAITEC, Saitama, Japan) for Fig. 5 and SI Appendix, Fig. S6, and mini-photo518 (TAITEC) for SI Appendix, Fig. S8.
Preparation of Cell-Free Extracts.
Cell-free extracts were prepared from T. kodakarensis cells cultivated in 400 mL ASW-YT-m1-Pyr medium for 15 h at 85 °C under anaerobic (with 0.08 mM Na2S) or microaerobic (without Na2S). Cells were harvested (4 °C, 5,000×g, 15 min) and washed with 12 mL 0.8 × ASW-m1 [modified version of previously defined ASW solution (49) supplemented with an additional 3.32 mg L−1 KI, 1.236 mg L−1 H3BO3, and 2.38 mg L−1 NiCl2 6H2O]. After centrifugation (4 °C, 5,000×g, 15 min) and removal of supernatant, cells were resuspended in 12 mL of an appropriate buffer; 50 mM MES-NaOH (pH 7.0) for ΔTK0551, ΔTK0683, ΔTK1734, and ΔTK2301, and 50 mM HEPES-NaOH (pH 7.5) for ΔTK0186, ΔTK0548, ΔTK1094, and ΔTK2268. After sonication with TOMY UD-201 (OUTPUT:4, DUTY:50, 30 min) and centrifugation (4 °C, 20,400×g, 15 min), supernatants were used as cell-free extracts.
Measurement of Enzyme Activities in Cell-Free Extracts.
Reaction mixtures to measure 2-PG phosphatase activities in cell-free extracts contained 10 mM 2-PG, 10 mM MgCl2, and 1.0 mg mL−1 cell-free extract in 50 mM MES (pH 7.0). Reactions were carried out at 80 °C for 1, 2, and 3 min and initiated by addition of 2-PG after 3 min preincubation of the other components. Reaction termination and subsequent procedures are described above.
The reaction mixture to measure GLDH activities in cell-free extracts contained 50 mM sodium glycolate, 10 mM NAD+, and 1.0 mg mL−1 cell-free extract in 100 mM MES (pH 7.0). Reactions were carried out at 80 °C. The reaction was initiated by addition of sodium glycolate after 3 min preincubation of the other components. NADH generation was measured by monitoring absorbance at 340 nm from 3 to 10 min.
AGAT activities in cell-free extracts were examined by measuring the concentration of generated glycine. The reaction mixture contained 10 mM alanine, 10 mM sodium glyoxylate, 20 μM PLP, and 100 μg mL−1 cell-free extract in 50 mM HEPES (pH 7.5). Reactions were carried out at 80 °C for 0, 1, 2, 3 min. The reaction was initiated by addition of sodium glyoxylate after 3 min preincubation of the other components. After terminating the reaction and removing proteins, the concentration of generated glycine was determined as described above.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
H.A. expresses sincere thanks to Drs. Sung Gyun Kang and Hyun Sook Lee of the Korea Institute of Ocean Science and Technology (KIOST) for fruitful discussions throughout the project. This study was partially supported by JSPS KAKENHI grant numbers JP19H05679 (Post-Koch Ecology) and JP19H05684, and the Green Innovation Fund Project, JPNP22010, commissioned by the New Energy and Industrial Technology Development Organization (NEDO). This work was partially supported by JST, the establishment of university fellowships toward the creation of science technology innovation, Grant Nos. JPMJFS2123 and JST SPRING, and JPMJSP2110.
Author contributions
H.A. designed research; Y. Michimori, R.I., Y.S., Y.F., S.S., K.N., Y. Miwa, S.H., T. Shimosaka, Y. Makino, R.T., and I.C. performed research; H.B. and T.K. contributed new reagents/analytic tools; Y. Michimori, Y.F., S.S., T. Sato, T.N., and H.A. analyzed data; H.B. and T.K. constructed and provided strains; and Y. Michimori, T.N., and H.A. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
Protein sequences are designated by entries used in the KEGG database (https://www.genome.jp/kegg/kegg_ja.html) (50). All other data are included in this manuscript and/or SI Appendix.
Supporting Information
References
- 1.Bassham J. A., Benson A. A., Calvin M., The path of carbon in photosynthesis. J. Biol. Chem. 185, 781–787 (1950). [PubMed] [Google Scholar]
- 2.Hartman F. C., Harpel M. R., Structure, function, regulation, and assembly of D-ribulose-1,5-bisphosphate carboxylase/oxygenase. Annu. Rev. Biochem. 63, 197–234 (1994). [DOI] [PubMed] [Google Scholar]
- 3.Shively J. M., van Keulen G., Meijer W. G., Something from almost nothing: Carbon dioxide fixation in chemoautotrophs. Annu. Rev. Microbiol. 52, 191–230 (1998). [DOI] [PubMed] [Google Scholar]
- 4.Tolbert N. E., The C2 oxidative photosynthetic carbon cycle. Annu. Rev. Plant. Physiol. Plant Mol. Biol. 48, 1–25 (1997). [DOI] [PubMed] [Google Scholar]
- 5.Anderson L. E., Chloroplast and cytoplasmic enzymes. II. Pea leaf triose phosphate isomerases. Biochim. Biophys. Acta 235, 237–244 (1971). [DOI] [PubMed] [Google Scholar]
- 6.Flügel F., et al. , The photorespiratory metabolite 2-phosphoglycolate regulates photosynthesis and starch accumulation in Arabidopsis. Plant Cell 29, 2537–2551 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kelly G. J., Latzko E., Inhibition of spinach-leaf phosphofructokinase by 2-phosphoglycollate. FEBS Lett. 68, 55–58 (1976). [DOI] [PubMed] [Google Scholar]
- 8.Tolbert N. E., Zill L. P., Excretion of glycolic acid by algae during photosynthesis. J. Biol. Chem. 222, 895–906 (1956). [PubMed] [Google Scholar]
- 9.Hellebust J. A., Excretion of some organic compounds by marine phytoplankton1. Limnol. Oceanogr. 10, 192–206 (1965). [Google Scholar]
- 10.Fernie A. R., Bauwe H., Wasteful, essential, evolutionary stepping stone? The multiple personalities of the photorespiratory pathway. Plant J. 102, 666–677 (2020). [DOI] [PubMed] [Google Scholar]
- 11.Hagemann M., et al. , Evolution of photorespiration from cyanobacteria to land plants, considering protein phylogenies and acquisition of carbon concentrating mechanisms. J. Exp. Bot. 67, 2963–2976 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Bauwe H., Photorespiration—Rubisco’s repair crew. J. Plant Physiol. 280, 153899 (2023). [DOI] [PubMed] [Google Scholar]
- 13.Schwarte S., Bauwe H., Identification of the photorespiratory 2-phosphoglycolate phosphatase, PGLP1, Arabidopsis. Plant Physiol. 144, 1580–1586 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zelitch I., Schultes N. P., Peterson R. B., Brown P., Brutnell T. P., High glycolate oxidase activity is required for survival of maize in normal air. Plant Physiol. 149, 195–204 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Suzuki K., Marek L. F., Spalding M. H., A photorespiratory mutant of Chlamydomonas reinhardtii. Plant Physiol. 93, 231–237 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakamura Y., Kanakagiri S., Van K., He W., Spalding M. H., Disruption of the glycolate dehydrogenase gene in the high-CO2-requiring mutant HCR89 of Chlamydomonas reinhardtii. Can. J. Bot. 83, 820–833 (2005). [Google Scholar]
- 17.Rademacher N., et al. , Photorespiratory glycolate oxidase is essential for the survival of the red alga Cyanidioschyzon merolae under ambient CO2 conditions. J. Exp. Bot. 67, 3165–3175 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eisenhut M., et al. , The photorespiratory glycolate metabolism is essential for cyanobacteria and might have been conveyed endosymbiontically to plants. Proc. Natl. Acad. Sci. U.S.A. 105, 17199–17204 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Claassens N. J., et al. , Phosphoglycolate salvage in a chemolithoautotroph using the Calvin cycle. Proc. Natl. Acad. Sci. U.S.A. 117, 22452–22461 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schada von Borzyskowski L., et al. , Marine Proteobacteria metabolize glycolate via the beta-hydroxyaspartate cycle. Nature 575, 500–504 (2019). [DOI] [PubMed] [Google Scholar]
- 21.Ezaki S., Maeda N., Kishimoto T., Atomi H., Imanaka T., Presence of a structurally novel type ribulose-bisphosphate carboxylase/oxygenase in the hyperthermophilic archaeon, Pyrococcus kodakaraensis KOD1. J. Biol. Chem. 274, 5078–5082 (1999). [DOI] [PubMed] [Google Scholar]
- 22.Yoshida S., Atomi H., Imanaka T., Engineering of a type III rubisco from a hyperthermophilic archaeon in order to enhance catalytic performance in mesophilic host cells. Appl. Environ. Microbiol. 73, 6254–6261 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sato T., Atomi H., Imanaka T., Archaeal type III RuBisCOs function in a pathway for AMP metabolism. Science 315, 1003–1006 (2007). [DOI] [PubMed] [Google Scholar]
- 24.Aono R., et al. , Enzymatic characterization of AMP phosphorylase and ribose-1,5-bisphosphate isomerase functioning in an archaeal AMP metabolic pathway. J. Bacteriol. 194, 6847–6855 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aono R., Sato T., Imanaka T., Atomi H., A pentose bisphosphate pathway for nucleoside degradation in Archaea. Nat. Chem. Biol. 11, 355–360 (2015). [DOI] [PubMed] [Google Scholar]
- 26.Fukushima E., Shinka Y., Fukui T., Atomi H., Imanaka T., Methionine sulfoxide reductase from the hyperthermophilic archaeon Thermococcus kodakaraensis, an enzyme designed to function at suboptimal growth temperatures. J. Bacteriol. 189, 7134–7144 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eisenhut M., et al. , The plant-like C2 glycolate cycle and the bacterial-like glycerate pathway cooperate in phosphoglycolate metabolism in cyanobacteria. Plant Physiol. 142, 333–342 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yokooji Y., Sato T., Fujiwara S., Imanaka T., Atomi H., Genetic examination of initial amino acid oxidation and glutamate catabolism in the hyperthermophilic archaeon Thermococcus kodakarensis. J. Bacteriol. 195, 1940–1948 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim Y., et al. , Structure- and function-based characterization of a new phosphoglycolate phosphatase from Thermoplasma acidophilum. J. Biol. Chem. 279, 517–526 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Makino Y., et al. , An archaeal ADP-dependent serine kinase involved in cysteine biosynthesis and serine metabolism. Nat. Commun. 7, 13446 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ohshima T., Nunoura-Kominato N., Kudome T., Sakuraba H., A novel hyperthermophilic archaeal glyoxylate reductase from Thermococcus litoralis. Characterization, gene cloning, nucleotide sequence and expression in Escherichia coli. Eur. J. Biochem. 268, 4740–4747 (2001). [DOI] [PubMed] [Google Scholar]
- 32.Yoshikawa S., et al. , Structure of archaeal glyoxylate reductase from Pyrococcus horikoshii OT3 complexed with nicotinamide adenine dinucleotide phosphate. Acta Crystallogr. D, Biol. Crystallogr. 63, 357–365 (2007). [DOI] [PubMed] [Google Scholar]
- 33.Lassalle L., et al. , New insights into the mechanism of substrates trafficking in Glyoxylate/Hydroxypyruvate reductases. Sci. Rep. 6, 20629 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liepman A. H., Olsen L. J., Alanine aminotransferase homologs catalyze the glutamate:glyoxylate aminotransferase reaction in peroxisomes of Arabidopsis. Plant Physiol. 131, 215–227 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sakuraba H., Kawakami R., Takahashi H., Ohshima T., Novel archaeal alanine:glyoxylate aminotransferase from Thermococcus litoralis. J. Bacteriol. 186, 5513–5518 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Su Y., Michimori Y., Atomi H., Biochemical and genetic examination of two aminotransferases from the hyperthermophilic archaeon Thermococcus kodakarensis. Front. Microbiol. 14, 1126218 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kanai T., et al. , Distinct physiological roles of the three [NiFe]-hydrogenase orthologs in the hyperthermophilic archaeon Thermococcus kodakarensis. J. Bacteriol. 193, 3109–3116 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kikuchi G., Motokawa Y., Yoshida T., Hiraga K., Glycine cleavage system: reaction mechanism, physiological significance, and hyperglycinemia. Proc. Jpn. Acad. B: Phys. Biol. Sci. 84, 246-263 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jin J. Q., Hachisuka S. I., Sato T., Fujiwara T., Atomi H., A structurally novel lipoyl synthase in the hyperthermophilic archaeon Thermococcus kodakarensis. Appl. Environ. Microbiol. 86, e01359-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jin J. Q., Sato T., Hachisuka S. I., Atomi H., A lipoate-protein ligase is required for de novo lipoyl-protein biosynthesis in the hyperthermophilic archaeon Thermococcus kodakarensis. Appl. Environ. Microbiol. 88, e0064422 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Orita I., et al. , The ribulose monophosphate pathway substitutes for the missing pentose phosphate pathway in the archaeon Thermococcus kodakaraensis. J. Bacteriol. 188, 4698–4704 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thorgersen M. P., Stirrett K., Scott R. A., Adams M. W., Mechanism of oxygen detoxification by the surprisingly oxygen-tolerant hyperthermophilic archaeon, Pyrococcus furiosus. Proc. Natl. Acad. Sci. U.S.A. 109, 18547–18552 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Watson G. M., Yu J. P., Tabita F. R., Unusual ribulose 1,5-bisphosphate carboxylase/oxygenase of anoxic Archaea. J. Bacteriol. 181, 1569–1575 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bekker A., et al. , Dating the rise of atmospheric oxygen. Nature 427, 117–120 (2004). [DOI] [PubMed] [Google Scholar]
- 45.Anbar A. D., et al. , A whiff of oxygen before the great oxidation event? Science 317, 1903–1906 (2007). [DOI] [PubMed] [Google Scholar]
- 46.Lyons T. W., Reinhard C. T., Planavsky N. J., The rise of oxygen in Earth’s early ocean and atmosphere. Nature 506, 307–315 (2014). [DOI] [PubMed] [Google Scholar]
- 47.Morikawa M., Izawa Y., Rashid N., Hoaki T., Imanaka T., Purification and characterization of a thermostable thiol protease from a newly isolated hyperthermophilic Pyrococcus sp. Appl. Environ. Microbiol. 60, 4559–4566 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Atomi H., Fukui T., Kanai T., Morikawa M., Imanaka T., Description of Thermococcus kodakaraensis sp. nov., a well studied hyperthermophilic archaeon previously reported as Pyrococcus sp. KOD1. Archaea 1, 263–267 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sato T., Fukui T., Atomi H., Imanaka T., Targeted gene disruption by homologous recombination in the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1. J. Bacteriol. 185, 210–220 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kanehisa M., Goto S., KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 28, 27–30 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
Protein sequences are designated by entries used in the KEGG database (https://www.genome.jp/kegg/kegg_ja.html) (50). All other data are included in this manuscript and/or SI Appendix.