Significance
Neuronal communication is mediated by neurotransmitters that are released from synaptic vesicles when they fuse rapidly with the plasma membrane. Proteins called SNAP receptors (SNAREs) mediate vesicle fusion by forming tight helical complexes that are believed to act as semirigid rods, acting mechanically to bring the two membranes together and merging them. This paper describes all-atom molecular dynamics simulations that allow visualization of membrane fusion in atomic detail and suggests that, after the SNAREs bring membranes together, they induce rapid fusion by facilitating local lipid rearrangements rather than through mechanical force exerted by helices. The simulations also suggest that the high speed of synaptic vesicle fusion is favored by polyunsaturated lipids that are known to be important for brain health.
Keywords: SNAREs, membrane fusion, molecular dynamics simulations, neurotransmitter release, juxtamembrane
Abstract
The SNAP receptor (SNARE) proteins syntaxin-1, SNAP-25, and synaptobrevin mediate neurotransmitter release by forming tight SNARE complexes that fuse synaptic vesicles with the plasma membranes in microseconds. Membrane fusion is generally explained by the action of proteins on macroscopic membrane properties such as curvature, elastic modulus, and tension, and a widespread model envisions that the SNARE motifs, juxtamembrane linkers, and C-terminal transmembrane regions of synaptobrevin and syntaxin-1 form continuous helices that act mechanically as semirigid rods, squeezing the membranes together as they assemble (“zipper”) from the N to the C termini. However, the mechanism underlying fast SNARE-induced membrane fusion remains unknown. We have used all-atom molecular dynamics simulations to investigate this mechanism. Our results need to be interpreted with caution because of the limited number and length of the simulations, but they suggest a model of membrane fusion that has a natural physicochemical basis, emphasizes local molecular events over general membrane properties, and explains extensive experimental data. In this model, the central event that initiates fast (microsecond scale) membrane fusion occurs when the SNARE helices zipper into the juxtamembrane linkers which, together with the adjacent transmembrane regions, promote encounters of acyl chains from both bilayers at the polar interface. The resulting hydrophobic nucleus rapidly expands into stalk-like structures that gradually progress to form a fusion pore, aided by the SNARE transmembrane regions and without clearly discernible intermediates. The propensity of polyunsaturated lipids to participate in encounters that initiate fusion suggests that these lipids may be important for the high speed of neurotransmitter release.
Neurotransmitter release by Ca2+-evoked synaptic vesicle exocytosis mediates neuronal communication. This exquisitely regulated process involves tethering of synaptic vesicles to the presynaptic plasma membrane, priming of the vesicles to a release-ready state(s) and fast fusion of the vesicles with the plasma membrane upon Ca2+ influx into the presynaptic terminal (1). These steps are controlled by a complex protein machinery that has been extensively studied (2–4), allowing reconstitution of basic features of synaptic vesicle fusion with purified components (5–7) and definition of their functions. The SNAP receptors (SNAREs) synaptobrevin, syntaxin-1, and SNAP-25 play a central role in membrane fusion by forming tight four-helix bundles that bring the membranes together (8–11). N-ethylmaleimide sensitive factor (NSF) and soluble NSF attachment proteins (SNAPs) disassemble SNARE complexes to recycle the SNAREs (8, 12). Munc18-1 and Munc13-1 organize SNARE complex formation by an NSF-SNAP-resistant mechanism (5, 13) that involves binding of Munc18-1 to a “closed” conformation of syntaxin-1 (14, 15) and to synaptobrevin, thus templating SNARE assembly (16–18) while Munc13-1 opens syntaxin-1 (19) and bridges the vesicle and plasma membranes (6, 20, 21). The resulting partially assembled SNARE complex binds to synaptotagmin-1 (22) and to complexin (23), forming a spring-loaded primed state (24) that is ready to trigger fast membrane fusion when Ca2+ binds to synaptotagmin-1 (25).
While this overall mechanism of neurotransmitter release is well established and it is clear that the neuronal SNAREs alone can induce fusion of reconstituted proteoliposomes (26, 27), a fundamental question remains unanswered: How do the SNAREs induce membrane fusion? Moreover, synaptic vesicle fusion likely occurs in a few microseconds, as the delay between presynaptic Ca2+ influx and postsynaptic currents in rat cerebellar synapses is 60 μs (28) and multiple events occur within this time frame. In contrast, cryoelectron microscopy (cryo-EM) images of SNARE-mediated liposome fusion reactions showed that the SNAREs induce extended bilayer-bilayer interfaces that fuse in seconds or minutes (27). It is thus unclear how the release machinery induces fusion in the microsecond time scale.
The SNARE four-helix bundle is formed by sequences called SNARE motifs (10, 11). SNAP-25 contains two SNARE motifs, whereas synaptobrevin and syntaxin-1 each contain one SNARE motif followed by a short juxtamembrane (jxt) linker and a C-terminal transmembrane (TM) region anchored at the synaptic vesicle or plasma membrane, respectively (SI Appendix, Fig. S1A). X-ray crystallography showed that the SNARE motif, jxt linker, and TM regions of synaptobrevin and syntaxin-1 form continuous α-helices in the cis-SNARE complexes formed after membrane fusion (29) (SI Appendix, Fig. S1B). A widespread textbook model envisions that these helices act as relatively stiff rods that exert mechanical force on the membranes to induce fusion as the complex “zippers” from the N to the C terminus (11, 26, 30). The action of the SNAREs in this model can be rationalized in the framework of elastic continuum models of membrane fusion that are based on the effects of proteins on membrane properties such as curvature, elastic modulus, and tension (31). These models postulate that membrane fusion occurs in several steps that are associated with large energy barriers and involve removal of hydration layers as the two membranes are brought together, bilayer bending to cause protrusions often called nipples, formation of a stalk intermediate in which the proximal leaflets have fused, merger of the distal leaflets to form a fusion pore, and opening of the fusion pore (31, 32). The textbook model was also supported by coarse-grained (CG) molecular dynamics (MD) simulations, which suggested that helix continuity is crucial for the SNAREs to cause fusion and that the mechanical forces arising from SNARE zippering not only bring the membranes together but also facilitate the downstream steps that lead to fusion (33–35).
A considerably different picture emerged from CG MD simulations with two protein-free small (14 to 15 nm) vesicles placed in contact, which suggested that the key step to initiate membrane fusion is the encounter of acyl chains from lipids of the apposed bilayers at the polar interface, leading to formation of a small hydrophobic core that quickly progresses to form a stalk intermediate and later a fusion pore without the help of proteins (36–38). This model was supported by all-atom MD simulations of 15 nm protein-free vesicles (39), but no encounters between lipid acyl chains at the polar interface were observed in MD simulations with 14 nm vesicles using an improved CG force field (40). Interestingly, such encounters were facilitated by lung surfactant protein B, which bridged the two vesicles by binding to them through amphipathic α-helices, and fusion ensued quickly (<500 ns) after the initial acyl chain encounter, with the protein bound only to the vesicle surface (40).
It is important to note that insertion of short helix-breaking sequences between the synaptobrevin jxt and TM sequences still allows robust liposome fusion in vitro and granule exocytosis in chromaffin cells (41–43), and a similar insertion between the syntaxin-1 jxt and TM sequences still allows neurotransmitter release in neurons (44). These results suggested that continuous helices in synaptobrevin and syntaxin-1 are not required for membrane fusion and neurotransmitter release. In contrast, insertion of short helix-breaking sequences between the SNARE motif and jxt linker of synaptobrevin or syntaxin-1 (i.e. before rather than after the jxt linker) strongly impairs liposome fusion (43) or neurotransmitter release (44, 45), respectively. These findings suggested that zippering of the SNARE motif helices into the jxt linkers does play a key role in SNARE action. However, the molecular basis for these observations and the overall mechanism of SNARE-induced membrane fusion remains unclear.
We have addressed these questions using all-atom MD simulations, taking advantage of parallelized computing resources that allow the calculation of trajectories of a few microseconds for systems consisting of millions of atoms, and building on our recent all-atom MD simulations of the neurotransmitter release machinery bridging a flat lipid bilayer and a vesicle with a diameter of 24 nm (24). Importantly, we observed rapid fusion of the vesicle and the flat bilayer in a simulation with the jxt linkers of synaptobrevin or syntaxin-1 fully zippered, allowing us to visualize SNARE-mediated membrane fusion using all-atom MD simulations. No fusion was observed without linker zippering. While further simulations will be required to draw firm conclusions, our results uncover a compelling molecular mechanism that makes a lot of sense from a physicochemical point of view and explains a large body of experimental data. In this mechanism, membrane fusion depends more on local rearrangements than on overall membrane properties, as membrane curvature did not appear to play an important role and no overt nipple formation was observed. Instead, the crucial event that initiated fusion was the facilitation of lipid acyl chain fluctuations out of the bilayers, into the polar membrane-membrane interface, by the zippered linkers and adjacent hydrophobic residues from the TM regions. Encounters of the acyl chains from the two bilayers at the interface formed a hydrophobic core that quickly expanded into stalk-like structures and gradually progressed into a fusion pore, with the help of the TM regions and without going through a clearly defined intermediate. Based on other available data and the importance of the jxt linkers for other types of SNARE-dependent intracellular membrane fusion (e.g. refs. 46 and 47), central aspects of this mechanism are likely to be generally conserved, and the catalysis of lipid acyl chain encounters may be a feature of other types of membrane fusion such as viral:cell or sperm:egg fusion (Discussion).
Results
Four Trans-SNARE Complexes with Fully Assembled Four-Helix Bundles Bridging a Vesicle and a Flat Bilayer.
The extremely fast speed of neurotransmitter release in fast synapses (28) suggests that synaptic vesicle fusion can be triggered in a few microseconds when the release machinery reaches proper configurations. However, the minimum number of trans-SNARE complexes required for fusion and their locations in these configurations are uncertain. In a previous study (24) we performed simulations of a 24 nm vesicle and a flat lipid bilayer bridged by four trans-SNARE complexes that were placed at the periphery of the bilayer-bilayer interface and were generated by a restrained MD simulation started with the crystal structure of the cis-SNARE complex that moved the TM regions to designed target positions and yielded unstructured jxt linkers (29). This was not surprising, as continuous helical conformations throughout the entire syntaxin-1 and synaptobrevin sequences would have required unrealistic bending of the helices and/or dissociation of part of the four-helix bundle, which is very stable (48). During the simulation of the whole system, the vesicle and the flat bilayer quickly formed an extended interface that resembled those observed in cryo-EM images of SNARE-mediated liposome fusion reactions (27), and no fusion was observed after 520 ns at 310 K and 454 ns at 325 K (24).
To explore potential SNARE configurations that might induce fast membrane fusion, we reasoned that placing the trans-SNARE complexes closer to each other may facilitate the formation of nipples, which is postulated to occur before fusion (31). Moreover, proximity to the center of the bilayer-bilayer interfaces is expected to facilitate the retention of helical conformation in the jxt linkers, which may also facilitate fusion. To generate trans-SNARE complexes for this purpose, we also started with the crystal structure of the cis-SNARE complex (29) and performed a 2 ns restrained MD simulation in which the TM regions were targeted to designed positions using a mild force constant (10 kJ ∗ mol−1 ∗ nm−2) to perturb the helical structure as little as possible. In addition, we carried out a 1 ns restrained simulation with a stronger force constant (50 kJ ∗ mol−1 ∗ nm−2) to force the TM regions closer to the target positions. The resulting trans-SNARE complex was almost completely helical except for kinks in the helices at the jxt linkers (SI Appendix, Fig. S1C). We made four copies of this complex and placed them in designed positions with rotations and translations (SI Appendix, Fig. S1D). The full system was built with these four complexes, a flat lipid bilayer of 30.5 nm × 30.5 nm and the same 24 nm vesicle that we used previously (24) with several lipids moved manually to accommodate the different positions of the synaptobrevin TM regions. The lipid compositions of the vesicle and flat bilayer resembled those of synaptic vesicles and synaptic plasma membranes, respectively (49, 50), and the system (referred to as fusion2g) had 4.4 million atoms after solvation with explicit TIP3P water molecules (Table 1).
Table 1.
Details of the MD simulations*
| CHL1 | POPC | SAGL | SAPE | SAPI2D | SDPE | SDPS | SOPS | Total | |
|---|---|---|---|---|---|---|---|---|---|
| Vesicle | |||||||||
| Outer leaflet | 1,266 | 296 | 0 | 533 | 0 | 283 | 211 | 199 | 2,791 |
| % | 45.4 | 10.6 | 0 | 19.1 | 0 | 10.1 | 7.6 | 7.1 | |
| Inner leaflet | 826 | 669 | 0 | 184 | 0 | 99 | 1 | 1 | 1,780 |
| % | 46.4 | 37.6 | 0 | 10.3 | 0 | 5.6 | 0.1 | 0.1 | |
| Flat bilayer 1 | |||||||||
| Upper leaflet | 748 | 130 | 17 | 115 | 82 | 231 | 165 | 161 | 1,649 |
| % | 45.3 | 7.9 | 1.0 | 7.0 | 5.0 | 14.0 | 10.0 | 9.8 | 100 |
| Lower leaflet | 720 | 688 | 16 | 64 | 0 | 112 | 0 | 0 | 1,600 |
| % | 45.0 | 43.0 | 1.0 | 4.0 | 0 | 7.0 | 0 | 0 | 100 |
| Flat bilayer 2 | |||||||||
| Upper leaflet | 540 | 96 | 12 | 84 | 60 | 156 | 120 | 120 | 1,188 |
| % | 45.4 | 8.1 | 1.0 | 7.1 | 5.1 | 13.1 | 10.1 | 10.1 | |
| Lower leaflet | 540 | 516 | 12 | 36 | 0 | 84 | 0 | 0 | 1,188 |
| % | 45.4 | 43.4 | 1.0 | 3.0 | 0 | 7.1 | 0 | 0 | |
| System name | Flat bilayer | No. of atoms | Initial cell dimensions | Length and temperature | |||||
| 4,390,099 | 35 × 35 × 35 nm3 | 887 ns 310 K / 805 ns 350 K | |||||||
| t350f2e | 5,266,689 | 37.5 × 37.5 × 37.5 nm3 | 780 ns 350 K | ||||||
| t350f4link | 5,276,577 | 37.5 × 37.5 × 37.5 nm3 | 667 ns 350 K | ||||||
| pullf4link | 1 | 5,276,577 | 37.5 × 37.5 × 37.5 nm3 | 1,956 ns 350 K | |||||
| snfreet350 | 2 | 3,960,735 | 32.5 × 37.5 × 32.5 nm3 | 1,000 ns 350 K | |||||
| nscpull4l | 1 | 5,319,378 | 37.7 × 37.7 × 37.7 nm3 | 900 ns 350 K | |||||
*All systems included four trans-SNARE complexes (except snfreet350 and nscpull4l), a 24 nm vesicle, and the indicated flat bilayer.
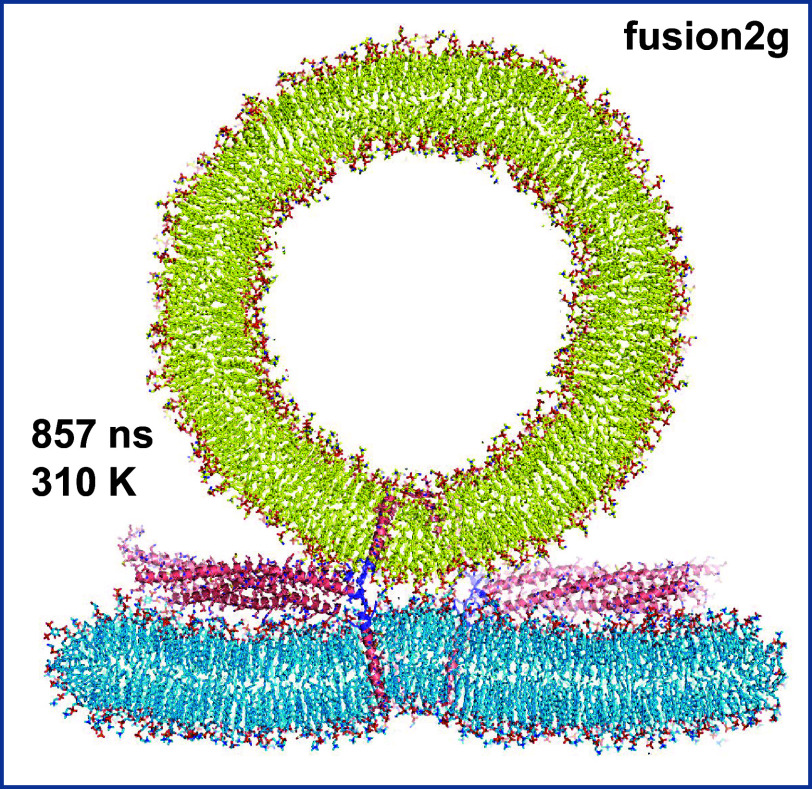
After equilibrating this system, we ran a production simulation at 310 K for 857 ns. During the first 180 ns, the flat bilayer buckled slightly at the center, between the SNARE helices, but the buckling did not progress after this time and there was no initiation of fusion at the end of the simulation (Fig. 1 and SI Appendix, Fig. S2 A and B). Since elevated temperatures can help to overcome energy barriers and are often used in MD simulations of lipid bilayers to increase lipid fluidity (e.g., ref. 33), we used the final configuration of the fusion2g simulation as a starting point to extend the simulation at 350 K (referred to as t350f2). Due to the natural rigid rotation of the system that had already been occurring during fusion2g and continued in t350f2, part of the edge of the flat bilayer came out at the bottom of the cell used for periodic boundary conditions and emerged at the top of the cell, making contact with the vesicle early in the simulation (around 60 ns; SI Appendix, Fig. S2C). As the simulation progressed, the edge of the bilayer merged with the top of the vesicle (SI Appendix, Fig. S2D) and the flat bilayer gradually adopted a semispherical shape to adapt to the vesicle shape as the merger of the two bilayers extended (SI Appendix, Fig. S2E). The merger of the two bilayers was initiated by one 1-stearoyl-2-docosahexaenoyl-sn-glycero-3-phosphoethanolamine (SDPE) molecule from the flat bilayer, which splayed when the polyunsaturated acyl chain moved into the vesicle outer leaflet (SI Appendix, Fig. S3A). This SDPE molecule remained splayed for more than 200 ns and initiated bilayer merger only when its motions facilitated the movement of other acyl chains to the polar interface between the two bilayers, forming a small hydrophobic nucleus that quickly expanded in less than 20 ns (SI Appendix, Fig. S3 B–H).
Fig. 1.
MD simulation of four almost completely helical trans-SNARE complexes bridging a vesicle and a flat bilayer and close to the center of the interface. A slice of the system after 857 ns of simulation at 310 K is shown. Lipids and proteins are shown as stick models with nitrogen atoms in dark blue, oxygens in red, phosphorus in orange, and carbon atoms in yellow for the vesicle, light blue for the flat bilayer and salmon for the SNARE complex except for those of the jxt linkers, which are in slate. The same color-coding was used in all the figures except when noted otherwise. The SNARE complexes are in addition represented by ribbon diagrams.
Although the interaction of the vesicle with the bilayer image at the top can be viewed as an “accident” arising from the rotation and the insufficient thickness of the water layer surrounding the system, it is interesting that the mechanism of merger is reminiscent of that observed previously in CG and all-atom simulations with 14 to 15 nm protein-free vesicles (36–39). In contrast, we did not observe even the initiation of membrane fusion by the SNAREs at the opposite side of the vesicle during the 857 ns at 310 K and the 855 ns at 350 K of the two simulations. To test the possibility that the lack of SNARE-mediated membrane fusion might have arisen because of the changes in shape of the flat bilayer as it was adapting to the vesicle shape, we took the coordinates of the system at 60 ns of the t350f2 simulation, before the membranes merged, reoriented and centered the system in the cell, and built a larger water box to prevent artificial interactions with the periodic image. We ran a 780 ns MD simulation at 350 K of this system (referred to as t350f2e) but again did not observe even the initiation of SNARE-induced membrane fusion (SI Appendix, Fig. S4). Indeed, we did not observe the initiation of membrane fusion between a flat bilayer and a 24 nm vesicle brought into direct contact by fully assembled or close to fully assembled SNARE four-helix bundles in multiple MD simulations that we reported previously (24), that are discussed here and that will be described elsewhere, totaling over 15 μs altogether. These findings are consistent with cryo-EM data showing that neuronal SNARE complexes induce formation of extended contact interfaces that fuse slowly, in the second-minute time scales (27). These observations also suggest that the packing of the lipids in the flat bilayers and the 24 nm vesicles of our simulations is tight enough to strongly hinder the movement of the acyl chains to the polar interface to form nonbilayer structures. Conversely, the observation of such movement and subsequent membrane merger in our t350f2 simulation most likely arose because of poor lipid packing at the edge of the flat bilayer, which facilitated the splaying of the SAPE molecule (SI Appendix, Fig. S3).
jxt Linker Zippering Leads to Fast Vesicle-Flat Bilayer Fusion.
The functional consequences of inserting short helix-disrupting sequences before or after the jxt linker of synaptobrevin or syntaxin-1 suggested that extension of the SNARE motif helices into the jxt linkers (referred to as jxt linker zippering) is crucial for liposome fusion in vitro or neurotransmitter release in neurons, but extension of the helices into the TM regions is not (41–45). Our MD simulations suggest that a substantial energy barrier hinders linker zippering because of the geometry of trans-SNARE complexes, which dictates that the angle between the SNARE four-helix bundle and the TM regions is much smaller than 180° (close to 90° for the geometry of the fusion2g system; SI Appendix, Fig. S1), and leads to unstructured (24) or kinked-helical conformations in the jxt linkers. Zippering of the jxt linker is associated with a substantial folding energy [8 kBT (48)] that can help overcome this energy barrier and may occur spontaneously, albeit slowly, and may be favored by other components of the neurotransmitter release machinery.
Since jxt linker zippering may be a slow step that may be difficult to reach in the time scales that we can simulate, we used restrained MD simulations to induce zippering and test its effects on membrane fusion. Since the TM regions of the trans-SNARE complexes of the fusion2g simulation were close to each other and jxt linker zippering would bring them even closer, we moved the trans-SNARE complexes manually from the fusion2g simulation further from each other and used restrained MD simulations to move the TM regions to new designed positions. This procedure stretched the conformations of the jxt linkers because the TM regions of synaptobrevin and syntaxin-1 are located further from each other in the new locations (SI Appendix, Fig. S5A). We used additional restrained MD simulations to induce helical conformations in the jxt linkers, up to residue 94 of synaptobrevin and 265 of syntaxin-1, using the structure of the cis-SNARE complex (29) as a model (SI Appendix, Fig. S6A). The trans-SNARE complexes were merged with the same flat bilayer and vesicle used for the fusion2g simulation, and lipids of both membranes were moved manually to accommodate the new TM positions. After equilibrating the system first at 310 K and then at 350 K, we ran a production simulation at 350 K for 667 ns (referred to as t350f4link; SI Appendix, Fig. S6 B–F). The two membranes were brought into close contact during the simulation and some of the TM regions came close to each other, buckling the flat bilayer slightly (SI Appendix, Fig. S6 C–F), but without initiating fusion. The jxt linkers remained partially helical throughout the simulation, with some oscillations in the degree of helicity and separation between the helix C termini, which varied among the complexes at the end of the simulation (SI Appendix, Fig. S6B).
Zippering of the jxt linkers is energetically favorable [8 kBT(48)] but pulls the TM regions toward the membrane interface, which is energetically unfavorable. Hence, there is likely a tug-of-war that results in zippering and unzippering of the jxt linkers, and fast membrane fusion may be triggered in a stochastic manner during one of the transient events of full zippering. To facilitate permanent full zippering of the jxt linkers and investigate whether fast membrane fusion might ensue, we generated a similar system in which the trans-SNARE complexes were induced to adopt helical conformation a little beyond the linkers (up to residue 99 of synaptobrevin and 268 of syntaxin-1) using also the cis-SNARE complex structure (29) as a model (SI Appendix, Fig. S5B) and the initial equilibrated configuration of the t350f4link system as the starting point (i.e. including the flat bilayer and the vesicle). Compared to the trans-SNARE complexes before forcing any helical conformations in the linkers (SI Appendix, Fig. S5A), this procedure brought the TM regions of synaptobrevin molecules closer to the syntaxin-1 TM regions, as expected, and stretched them such that their N termini became partially unstructured (SI Appendix, Fig. S5B). Correspondingly, the C termini of synaptobrevin and syntaxin-1 were brought inside their corresponding bilayers (SI Appendix, Fig. S5 C and D). We then ran a production simulation of 1,956 ns at 350 K while using the pull code from Gromacs (51, 52) to restrain the distance between the geometric centers of the atoms of residues 94 to 97 of synaptobrevin and of the atoms of residues 263 to 266 of syntaxin-1 to within 1 nm, which is their distance in the cis-SNARE complex structure (referred to as pullf4link simulation). Importantly, as illustrated by full views of the system in Fig. 2 and SI Appendix, Fig. S5 E and F, the system slices of Fig. 3 and Movie S1, this simulation did lead to full fusion of the vesicle and the flat bilayer. The two membranes were in close contact early in the simulation and retained their bilayer structures until about 400 ns (Fig. 3B). A stalk-like structure began to form shortly afterward (SI Appendix, Fig. S5E and Fig. 3C) and lipids at this region kept rearranging (e.g., Fig. 3D) during over 1,000 ns, leading to a full merger of the two bilayers to initiate formation of a fusion pore (SI Appendix, Fig. S5F and Fig. 3E) that opened very quickly (Figs. 2B and 3F).
Fig. 2.
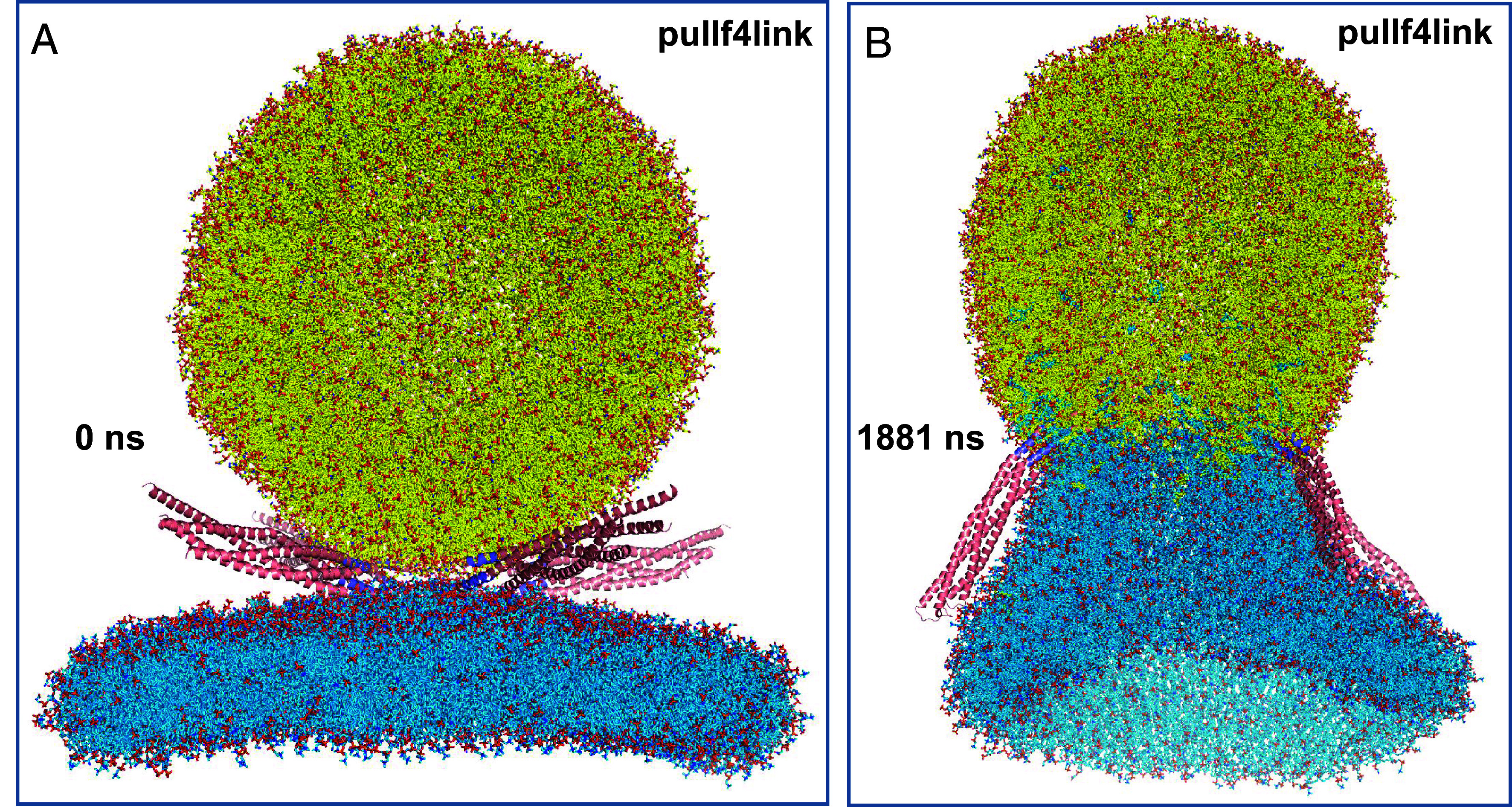
MD simulation of four trans-SNARE complexes bridging a vesicle and a flat bilayer, with the linkers zippered and a pulling force to keep them zippered. (A and B) Full views of the system in the beginning (A) and end (B) of the pullf4link simulation. Lipids and proteins are shown as stick models, and SNARE complexes are in addition represented by ribbon diagrams. The color code is the same as in Fig. 1.
Fig. 3.
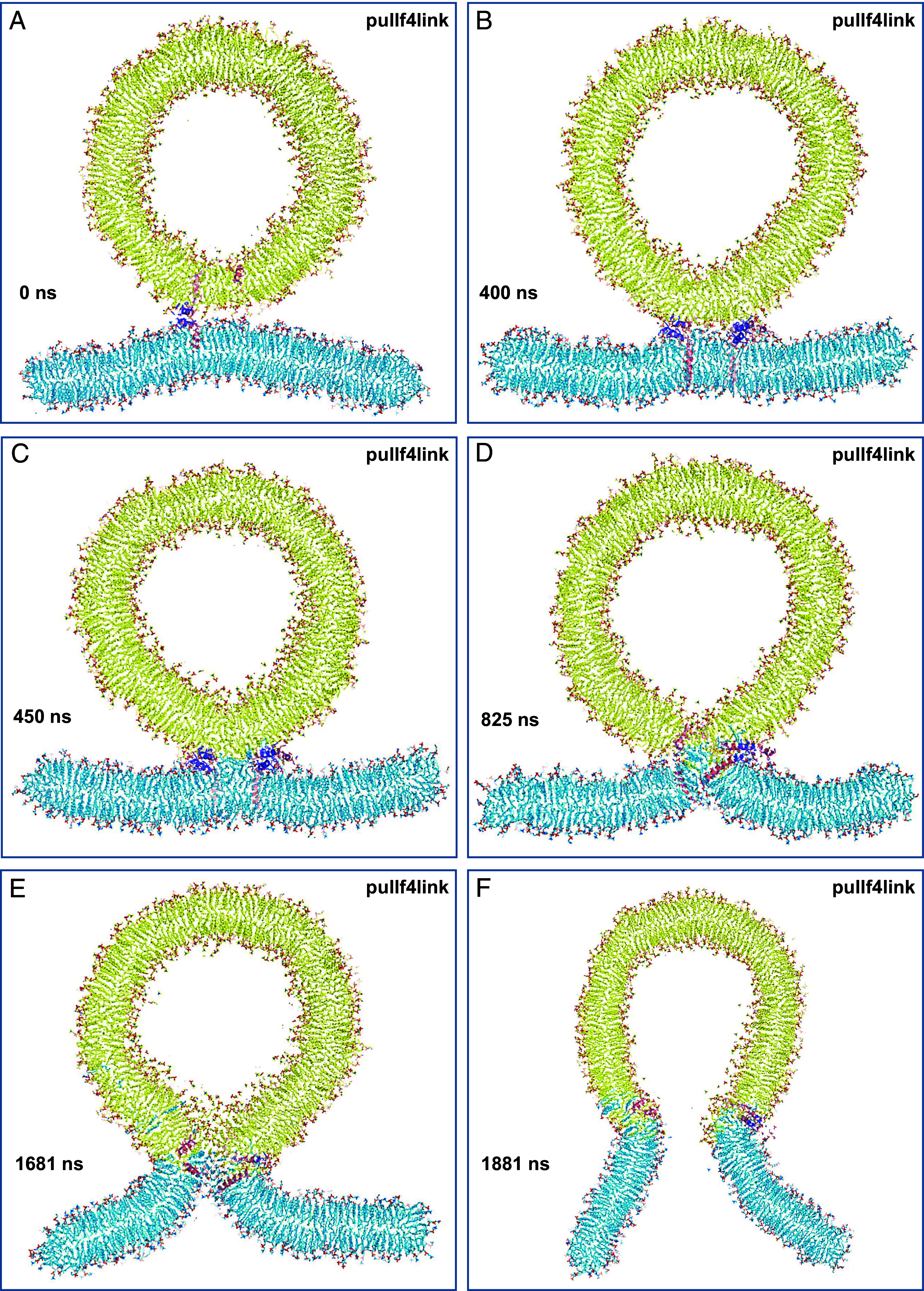
Slices taken at the indicated time points along the pullf4link simulation. The slices illustrate how the membranes retained their bilayer structure from 0 to 400 ns (A and B), a small stalk-like structure had formed by 450 ns (C), and the interface kept evolving (D) until a fusion pore was formed (E) and expanded (F). Lipids and proteins are shown as stick models, and SNARE complexes are in addition represented by ribbon diagrams. Note that the slices shown are thin and only small portions of the SNARE complexes, if any, are present in the slices. The color code is the same as in Fig. 1.
Mechanism of Vesicle-Flat Bilayer Fusion.
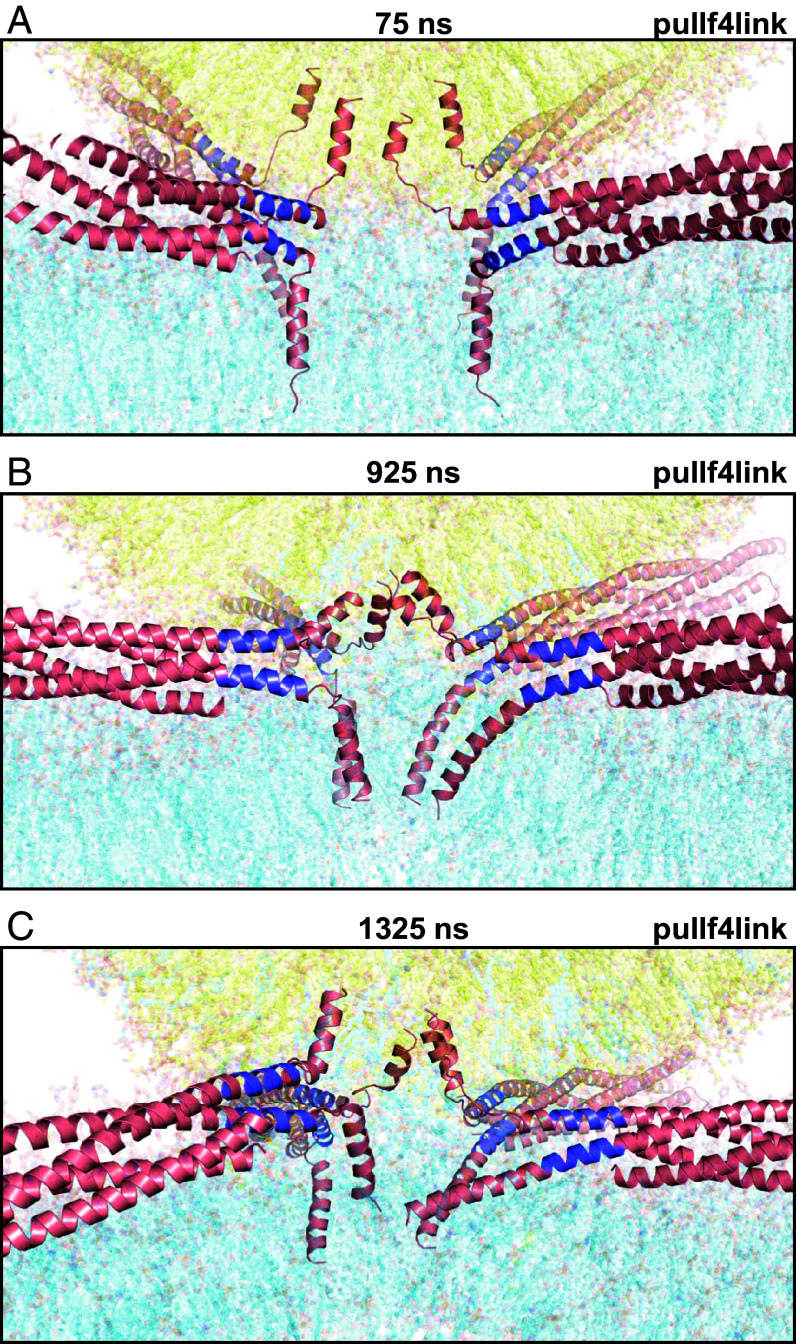
Zippering of the linkers was crucial to initiate the merger of the vesicle and the flat bilayer because it brought the hydrophobic residues of the TM regions to the polar interface, where they facilitate the transition of hydrophobic acyl chains out of the two bilayers. In the trajectory, it became apparent that these transitions are particularly likely for lipids containing polyunsaturated acyl chains that are abundant in the vesicle and plasma membranes (49, 50). The vesicle and flat bilayer contained three types of such lipids (number of carbons and unsaturated bonds indicated in parenthesis): SDPE (18:0, 22:6), 1-stearoyl-2-arachidonoyl-sn-glycero-3-phosphoethanolamine (SAPE; 18:0, 20:4) and 1-stearoyl-2-docosahexaenoyl-sn-glycero-3-phospho-L-serine (SDPS; 18:0, 22:6). SI Appendix, Fig. S7 illustrates how the 22:6 acyl chain of an SDPS molecule began to emerge from the flat bilayer already at 75 ns and was fully splayed by 80 ns. These events were facilitated by contacts of this acyl chain with the hydrophobic side chains of the TM regions and the methylene groups of lysine side chains from the jxt linkers of one SNARE complex. However, there was no initiation of lipid mixing in this area of the vesicle-flat bilayer interface, suggesting that lipid splaying itself is not sufficient to induce membrane merger.
Membrane merger was initiated by another SNARE complex (Fig. 4) when an acyl chain from an SDPS molecule from the flat bilayer (2,501) emerged at the polar interface and made contact with acyl chains of two SAPE molecules from the vesicle (8,037 and 8,045) that also emerged at the interface at 402 ns (Fig. 4B). These acyl chains also made contacts with hydrophobic side chains of the TM regions and methylene groups of lysine side chains from the jxt linkers of this SNARE complex. The acyl chain of an SDPE molecule from the flat bilayer (2,012) soon emerged to join these hydrophobic contacts with the vesicle (Fig. 4C) and additional hydrophobic contacts were formed quickly at the interface (Fig. 4D) to initiate the formation of a small stalk-like structure in which the proximal leaflets of the flat bilayer and the vesicle merged. This mini stalk occurred on one side of the flat bilayer-vesicle interface (on the right in the orientation of SI Appendix, Fig. S8A) and gradually expanded toward the other side (SI Appendix, Fig. S8B), but polar groups from both bilayers remained at the interface and a full stalk spanning the entire interface never formed. Lipid rearrangements continued and led to structures in which the vesicle formed a V-shape and the flat bilayer formed an inverted V-shape as expected for the stalk intermediate (31) (SI Appendix, Fig. S8 C and D). These shapes helped to minimize the volume of the hydrophobic area at the center of the interface and facilitated the packing of the lipids in these nonbilayer structures. The lipids at the center of these structures were somewhat disordered and some polar groups remained at the hydrophobic interface, likely because they can interact with the backbone amide groups of residues from the synaptobrevin TM regions that are not helical (SI Appendix, Fig. S8 C and D). The nonsymmetrical nature of these structures is illustrated by the views from different angles of the pose obtained at 1,125 ns (SI Appendix, Fig. S8 D–F).
Fig. 4.
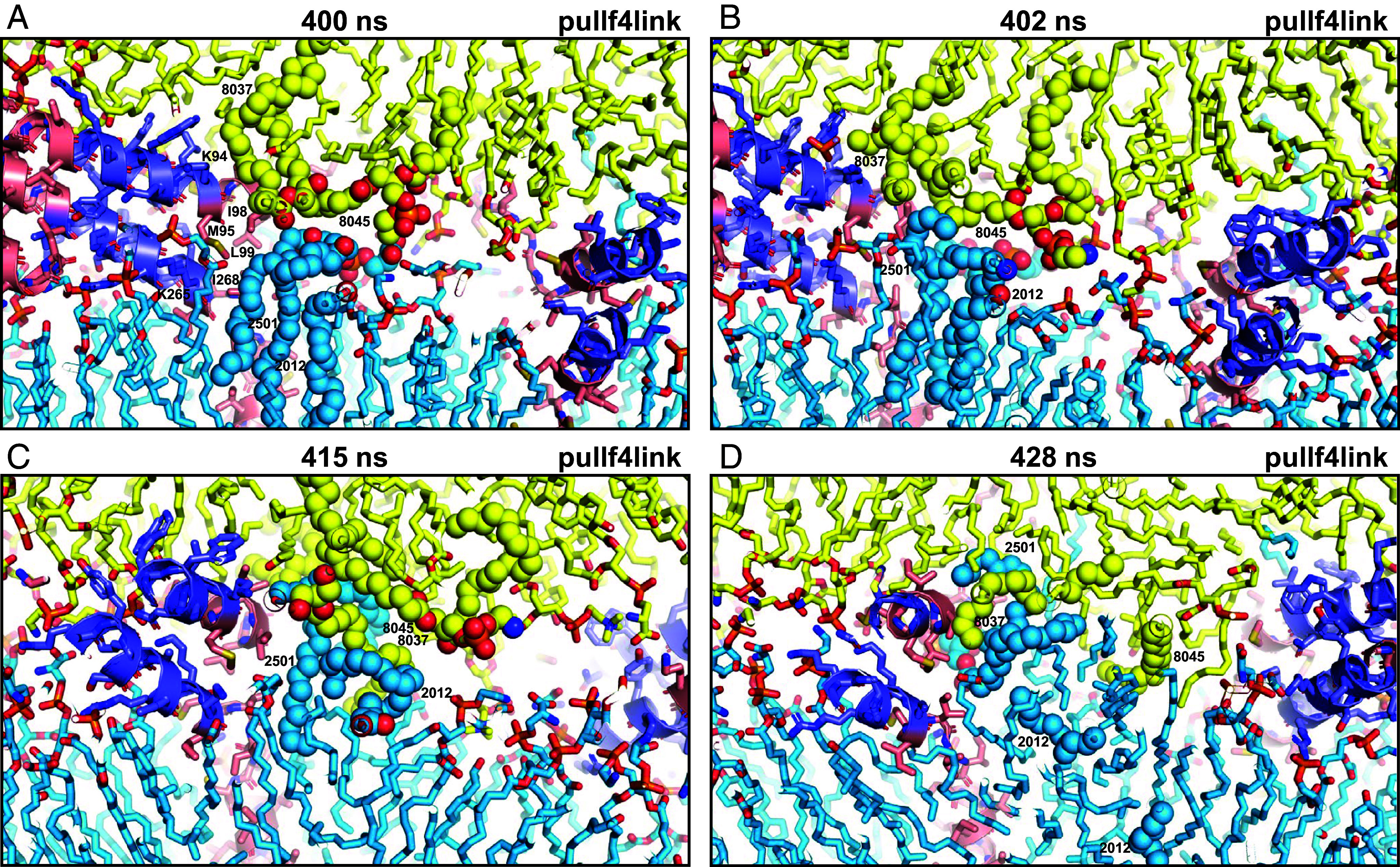
Snapshots taken at the indicated time points of the pullf4link simulation showing the formation of a small hydrophobic nucleus at the polar interface. The diagrams illustrate how there were no hydrophobic contacts in this region of the polar interface between the two bilayers at 400 ns (A). Acyl chains of polyunsaturated lipids from the two bilayers came into contact at the polar interface at 402 ns (B), when they came out of the bilayers due to fluctuations next to the methylene groups of lysine side chains from the linkers and hydrophobic side chains from the TM regions [labeled with the single letter amino acid residue abbreviation and residue number in (A)]. Additional contacts kept forming next to these residues (C) and facilitated the formation of hydrophobic acyl chain contacts, forming a small hydrophobic nucleus (D). Lipids and proteins are shown as stick models, and SNARE complexes are in addition represented by ribbon diagrams. The color code is the same as in Fig. 1. The positions of SDPE2012, SDPS2501, SAPE8037, and SAPE8045 (shown as spheres), which were involved in the initial contacts, are indicated. Note that parts of these lipids cannot be seen in the slice of the system shown. This is why the two acyl chains of SDPE2012 look disconnected in panel (D).
Polar lipid head groups gradually moved toward the center of the interface (SI Appendix, Fig. S9 A–D) until two bilayers that resulted from merging the flat bilayer and the vesicle were clearly formed, resulting in a nascent fusion pore (SI Appendix, Fig. S9E) that quickly expanded (SI Appendix, Fig. S9F and Fig. 3F). This expansion released the high curvature existing at the neck of the pore and may have been facilitated by the geometry of our system, as the small flat bilayer readily adapted to the shape of the vesicle (Fig. 2B), much as the flat bilayer accommodated to the shape of the vesicle when they merged in the t350f2 simulation (SI Appendix, Fig. S2E). The movement of polar head groups to the center of the interface again appeared to be facilitated by interactions of the head groups with backbone amide bonds from unstructured residues of the synaptobrevin TM regions, which were interspersed with the lipids at the interface throughout the entire process of merging the membranes, as were the syntaxin-1 TM regions (Fig. 4 and SI Appendix, Figs. S8 and S9). Interestingly, conformational flexibility in the synaptobrevin TM region has been shown to be crucial for efficient Ca2+-triggered exocytosis (53, 54). These observations correlate with the fact that, while the C-terminal half of the synaptobrevin TM regions (residues 106 to 116) were helical throughout the simulation, residues 100 to 105 remained largely unstructured (Fig. 5 and SI Appendix, Fig. S10).
Fig. 5.
Snapshots taken at 75 ns (A), 925 ns (B) and 1325 ns (C) of the pullf4link simulation illustrating the conformational changes in the SNAREs as the system evolved from bilayer-bilayer contact to the fusion pore. SNARE complexes are represented by ribbon diagrams and lipids are represented by 90% transparent sticks to allow visualization of the ribbons. Note that the thickness of the slices shown in these panels is much larger than those of the slices shown in Figs. 3 and 4 and SI Appendix, Figs. S8 and S9 so that all four trans-SNARE complexes can be observed, but the lipids then need to be transparent to allow visualization of the SNAREs. Additional snapshots are shown in SI Appendix, Fig. S10. The color code is the same as in Fig. 1.
A substantially different behavior was observed for the syntaxin-1 TMs. Although the N-terminal regions of the TMs, close to the jxt linkers, became unstructured after the restrained MD simulation used to set up the initial configuration of this system (SI Appendix, Fig. S5B), they quickly regained most of the helicity (Fig. 5A). Two of the syntaxin-1 TM regions became fully helical by 675 ns, with a slight kink so that the helices were mostly parallel to the flat bilayer lipids (both on the right side of SI Appendix, Fig. S10B). Both helices gradually straightened such that the entire syntaxin-1 sequences spanning the SNARE motif, jxt linker and TM region formed continuous helices (Fig. 5 B and C and SI Appendix, Fig. S10 C–F). Such straightening of the helices likely contributes to the progress from the stalk-like structures to the fusion pore, as the neighboring lipids generally have parallel orientations to the TM regions and therefore reorient from their initial perpendicular orientation with respect to the flat bilayer to their parallel orientation in the fusion pore (e.g., compare SI Appendix, Fig. S8A with Fig. S9 B and F). At the same time, the reorientation of lipids from the distal leaflets of the flat bilayer also brings their polar head groups to the middle to form the fusion pore. The notion that helix continuity from the jxt linker into the TM region of syntaxin-1 plays a role in completing rather than initiating synaptic vesicle fusion is supported by the observation that insertion of a short helix-breaking sequence between the jxt linker and TM region decreases the charge of Ca2+-evoked excitatory postsynaptic currents (EPSCs) much less severely than their amplitude because it slowed down the kinetics of release (44).
It is also worth noting that the four-helix bundles of the four SNARE complexes remained bound to the flat bilayer throughout this simulation (Fig. 2 and SI Appendix, Fig. S5 E and F), as they did in the fusion2g/t350f2 simulations (SI Appendix, Fig. S2). This feature can be attributed to the multiple positively charged residues of the four-helix bundle that can simultaneously mediate ionic interactions with negatively charged lipids of the flat bilayer, including PIP2 (24), and likely dictates the proper orientation of the four-helix bundle with respect to the two membranes (from the point of view of rotation around its long axis) such that the jxt linkers and TM regions of synaptobrevin and syntaxin-1 are suitably projected toward the membrane-membrane interface to efficiently mediate membrane fusion.
The SNAREs Facilitate Stalk Expansion and Fusion Pore Formation.
The mechanism of SNARE-mediate membrane fusion observed in the pullf4link simulation suggests that the SNAREs not only act to bring the membranes together and initiate bilayer merger but also play active roles in promoting the formation of stalk-like intermediates and generating the fusion pore. This proposal contrasts with results from CG and all-atom simulations with 14 to 15 nm vesicles suggesting that, once membrane merger is initiated by encounters between acyl chains of the two bilayers at the polar interface, the lipids by themselves can rapidly progress toward formation of the stalk and the fusion pore (36–40). To distinguish between these two models, we first performed all-atom MD simulations of a 24 nm vesicle and a flat bilayer (SI Appendix, Fig. S11). Although the vesicle is still smaller than typical synaptic vesicles [ca. 40 nm (49)], this geometry resembles better that of synaptic vesicle fusion than fusion between two highly curved, 15 nm vesicles.
The simulation was initiated with the vesicle close to the flat bilayer (SI Appendix, Fig. S11A). The distance between them increased by about 1 nm during 1 ns temperature equilibration to 350 K and 1 ns pressure equilibration to 1 atm (SI Appendix, Fig. S11B), which can be attributed to repulsion between the two membranes. We then ran a 100 ns production simulation at 350 K while applying a pulling force between the acyl chain of one flat bilayer lipid and the acyl chain of a vesicle lipid to promote their encounter at the interface and potentially initiate bilayer fusion. Instead, this pulling force caused the splaying of the vesicle lipid such that one of its acyl chains remained in the vesicle and the other was in the flat bilayer, and the two bilayers came into close contact (SI Appendix, Fig. S11 C and E). To investigate whether such lipid splaying together with the bilayer-bilayer contact might lead to fusion, we extended this simulation for 900 ns but without the pulling force between the acyl chains (referred to as snfreet350). The splayed lipid remained in this configuration for most of the simulation but was fully in the vesicle at the end (SI Appendix, Fig. S11 E–G). There was no initiation of fusion but, intriguingly, the flat bilayer curved to adapt to the vesicle shape such that one entire side of the flat bilayer made contact with the vesicle (SI Appendix, Fig. S11 D, F, and G). This extended interface resembles those observed for SNARE-mediated liposome fusion reactions by cryo-EM (27) and the extended interface that we observed in our first MD simulation with four SNAREs bridging a flat bilayer and a vesicle (figure 2J of ref. 24), but is larger because there are no SNAREs in the periphery. This phenomenon may be entropically driven, as water molecules become immobilized at membrane interfaces (55) (Discussion).
The snfreet350 simulation suggests that lipid splaying is not sufficient to initiate bilayer merger, which is consistent with the observation that the SNARE-mediated splaying of one lipid in the pullf4link simulation also failed to start fusion (SI Appendix, Fig. S7). Instead, fusion was initiated by encounters between acyl chains of the flat bilayer and the vesicle promoted by the SNAREs (Fig. 4). Hence, we asked whether the hydrophobic core induced by the SNAREs that initiated fusion in that simulation might be able to progress unabated toward formation of stalk-like structures, generation of a fusion pore and full fusion without the help of SNARE proteins. For this purpose, we took the 475 ns frame of the pullf4link simulation, in which a substantial hydrophobic core had already formed at the interface, removed the SNARE complexes, replaced their TM regions with lipids, and used the resulting system as the starting point (SI Appendix, Fig. S12 A and B) for equilibration and production simulation for 900 ns at 350 K (referred to as nscpull4l simulation). Early in the simulation, the flat bilayer curved downward in the orientation of SI Appendix, Fig. S12C, away from the vesicle, and the contact between the two bilayers was expanded (SI Appendix, Fig. S12D). There was some transfer of lipids from the flat bilayer to the vesicle, but no clear expansion of the hydrophobic core was observed by 300 ns of simulation (SI Appendix, Fig. S12D). Indeed, there was no expansion of the core even at 900 ns, when the size of the core appeared to be smaller and the flat bilayer had wrapped around the vesicle (SI Appendix, Fig. S12 E and F) as we observed for the snfreet350 simulation (SI Appendix, Fig. S11 D and G). Although we cannot rule out the possibility that the hydrophobic core might have expanded at longer time scales, particularly if the curving of the flat bilayer to wrap around the vesicle were avoided, these results reinforce the notion that SNARE proteins facilitate all the steps that lead to fast membrane fusion at a synapse, not only bringing the two membranes together and initiating bilayer merger, but also accelerating formation of stalk-like structures and generating a fusion pore.
Discussion
The notion that SNARE proteins execute intracellular membrane fusion is well established, but the underlying molecular mechanism has remained enigmatic. Elastic continuum models explained the roles of SNAREs and other proteins in membrane fusion in terms of their effects on macroscopic properties such as membrane curvature, elastic moduli, and tension (31), and the widespread textbook model envisions that the SNAREs act as semirigid rods that exert mechanical force on the membranes to fuse them as they zipper into SNARE complexes from the N to the C terminus (11, 26, 30), which was supported by CG MD simulations (33, 34). Conversely, MD simulations with SNARE-free membranes suggested that fusion is initiated by local encounters of lipid acyl chains at the interface, which may be facilitated by proteins, and membrane fusion can then ensue rapidly without proteins (36–40). Our current study describes an all-atom MD simulation that incorporates SNARE proteins and leads to fusion of two lipid bilayers. Our results need to be interpreted with caution because of the caveats mentioned below but, together with existing experimental data, they suggest a mechanism of how the neuronal SNAREs induce membrane fusion that has a clear, sound physicochemical basis and emphasizes local events at atomic/molecular level over macroscopic membrane features. In this model, assembly of the SNARE four-helix bundle brings the membranes into contact, which may be aided at the last stages by entropically driven exclusion of water from the interface. The key event that initiates membrane fusion occurs when the jxt linkers zipper and, together with adjacent hydrophobic residues from the TM regions, they catalyze acyl chain fluctuations out of the bilayers to favor hydrophobic encounters at the polar interface. The resulting small hydrophobic core gradually expands into stalk-like structures and develops into a fusion pore, aided by the TM regions and without a clear intermediate. Our results also suggest that polyunsaturated lipids facilitate the high speed of membrane fusion, which is critical for fast neurotransmission.
Elastic continuum theory has been very helpful in understanding how membranes fuse (31), and the stalk mechanism has been a guiding light since it was first formulated (56), even though the nature of the stalk may need to be revised (see below). CG MD simulations have also been very insightful to understand membrane fusion (36–38, 40) but are limited by insufficient representation of atomic interactions and cannot simulate substantial conformational changes in proteins (reviewed in refs. 35 and 57). All-atom MD simulations allow the study of structural changes in proteins but still involve approximations and are computationally very intensive, which limited the number and length of our simulations, and led us to use high temperature to help overcome energy barriers as well as an artificial constraint to maintain the linkers zippered (SI Appendix, Supplementary Discussion). Despite these limitations, it is remarkable that the key event that initiated membrane fusion in our pullf4link simulation is the encounter of hydrophobic acyl chains from the two bilayers at the polar interface, in analogy with results from previous CG and all-atom MD simulations using SNARE-free vesicles (36–40). This convergence of data from diverse simulations performed with different force fields and under distinct conditions strongly suggests that acyl chain encounters at the interface generally constitute the seed that initiates membrane fusion, although the encounters may be facilitated by diverse mechanisms. The relevance of our results is also supported by the fact that they correlate with extensive experimental data available on neurotransmitter release and liposome fusion, providing a natural physicochemical basis for the catalysis of acyl chain encounters by the SNAREs and for the functional importance of linker zippering.
Initiation of SNARE-Mediated Membrane Fusion.
Overwhelming evidence shows that SNAREs cannot be viewed as preformed fully helical rods that span the SNARE motifs, jxt linkers and TM regions, and that exert mechanical force as they zipper from the N termini to the TM C termini (SI Appendix, Supplementary Discussion). Zippering of the four-helix bundle formed by the SNARE motifs could be viewed as a mechanical process that overcomes the repulsion between the two membranes and removes the hydration layers between them, events believed to impose high energy barriers (e.g., refs. 31 and 32). However, the energetic cost of bringing two membranes into contact does not appear to be high, as electrostatic repulsion is weak at distances beyond 3 nm, cations can shield the charges of lipid head groups and exclusion of water molecules may become favorable once two membranes are close. The latter notion is suggested by the extended membrane interfaces induced by SNARE complexes in our MD simulations (24), which have been observed by cryo-EM (27), and the way flat bilayers wrap around vesicles when held together by lipid bridges (SI Appendix, Figs. S11 and S12). Moreover, our previous MD simulations suggested that two bilayers could be readily brought into contact even by SNARE four-helix bundles that were not fully assembled (24), also consistent with cryo-EM data (27). It seems likely that water exclusion from the space between two membranes is driven by gains in entropy, as all-atom MD simulations showed that water molecules become ordered at membrane interfaces (55). Mechanical force exerted by four-helix bundle assembly may still be important to push away membrane proteins of both membranes from the interface, but our results suggest that the most important role for the very high stability of the SNARE four-helix bundle is to provide a stable framework to nucleate jxt linker zippering.
Indeed, the most compelling insight from our MD simulations is that linker zippering constitutes a key event to initiate membrane fusion because it draws the TM regions toward the polar bilayer-bilayer interface, where the hydrophobic TM side chains and methylene groups of jxt linker lysines catalyze movement of the hydrophobic acyl chains from the lipids to the polar interface (SI Appendix, Fig. S7 and Fig. 4). When acyl chains from the two bilayers come into contact, they form a small hydrophobic nucleus that initiates bilayer merger. Without linker zippering, these encounters are very unlikely, as shown by the fact that we did not observe initiation of membrane merger in MD simulations where SNARE complexes brought two bilayers in contact but the linkers did not zipper (totaling over 15 μs between those described here, those reported in ref. 24, and others that will be described elsewhere). Note also that the extended contact interfaces between liposomes induced by the SNAREs and observed by cryo-EM fuse in seconds/minutes (27), showing that high energy barriers hinder linker zippering or spontaneous acyl chain encounters independent of linker zippering. We did observe acyl chain encounters and merger of the flat bilayer with the vesicle far from the SNARE complex-bridged interface in the t350f2 simulation (SI Appendix, Fig. S2) because lipids are not well packed at the edge of the flat bilayer. Similarly, the observation of acyl chain encounters and fusion between closely apposed 14 to 15 nm vesicles in CG and all-atom MD simulations (36–40) may have arisen from poor lipid packing characteristic of highly curved membranes. The fact that we did not observe such events in our simulations without linker zippering indicates that the lipids are sufficiently well-packed in the 24 nm vesicles and flat bilayers to make acyl chain encounters at the polar interface very unlikely. Thus, the relatively strong curvature of 24 nm vesicles does not appear to be a determining factor for the fusion with the flat bilayer observed in the pullf4link simulation.
Formation of Stalk-Like Structures and Progression to a Fusion Pore.
Ample evidence supports the notion that a central step in membrane fusion is the formation of a stalk intermediate in which the proximal leaflets of two bilayers have merged but not the distal leaflets (31). In our pullf4link simulation, the small hydrophobic nucleus formed by lipid acyl chain encounters at the polar interface between the vesicle and the flat bilayer (Fig. 4D) gradually expanded, first at one side of the interface and later toward the middle, forming structures that resemble the postulated stalk intermediate (SI Appendix, Fig. S8). However, the interior of these structures was never fully hydrophobic (SI Appendix, Fig. S8 D–F), and kept evolving gradually as lipid head groups moved to the center of the interface and eventually a fusion pore was formed (SI Appendix, Fig. S9). Substantial lipid disorder was observed at the interface such that some lipids could not be assigned to a leaflet. Such disorder likely was facilitated by the interspersed TM regions of the SNAREs, particularly those of synaptobrevin that were flexibly linked to the jxt linkers throughout the simulation. The exposed polar amide groups of the flexible TM residues likely facilitated the movement of polar head groups to the center of the interface to form the fusion pore. Conversely, the TM regions of two syntaxin-1 molecules formed continuous helices with the jxt linkers and the SNARE motifs relatively early in the simulation, when these helices were bent due to the geometry of the system (Fig. 5B and SI Appendix, Fig. S10 B–D). Straightening of these helices is expected to be energetically favorable and hence is likely to drag the nearby lipids from their initial orientations perpendicular to the flat bilayer to their parallel orientations in the fusion pore. These observations suggest that, while the TM regions of synaptobrevin and syntaxin-1 may both influence all the events that lead to membrane fusion, the former plays a more preponderant role in facilitating dynamic motions that help the lipids to reorient during these events, whereas the syntaxin-1 TM region may be more important for ordering the lipids toward formation of the fusion pore structure.
This mechanism does not contradict the stalk hypothesis, as the structures that lead to fusion pore formation do resemble stalks, but they refine the common view of the stalk in three ways. First, there was more lipid disorder than commonly depicted in cartoons of the stalk, which is not surprising because lipid packing is not optimal and is natural that lipids keep moving in search for stable orientations. Second, the interior of the stalk-like structures was not purely hydrophobic because there are exposed backbone amide groups that interact with lipid head groups. Third, there was no clear intermediate as the system progressed gradually to fusion pore formation. These observations suggest that SNARE-mediated membrane fusion does not involve a unique stalk intermediate corresponding to a well-defined local free energy minimum but rather proceeds through a “continuum” of structures that lead to fusion pore formation and might be viewed collectively as a dynamic stalk. We note that fusion pore expansion was very fast in our simulations (Fig. 3 E and F), but expansion was likely facilitated by the small size of the flat bilayer, which easily adapted to the vesicle shape after fusion. Expansion is expected to be more energetically costly for vesicle fusion to an extended flat membrane (31, 32).
CG and all-atom MD simulations suggested that the defining moment for membrane fusion is the splaying of lipids between the two bilayers and/or the encounter of acyl chains from the two bilayers at the interface such as those that we observe, and fusion ensues unabated and quickly (less than 1 μs in most of the simulations) without the help of proteins (36–40). However, these results may also have arisen from poor packing of the highly curved 14-15 nm vesicles used in these studies, as we did not observe fusion of the flat bilayer to the 24 nm vesicle when they were bridged by a splayed lipid or by a hydrophobic nucleus involving multiple contacts between acyl chains from the two bilayers (SI Appendix, Figs. S11 and S12). While it is plausible that fusion might have been observed at much longer time scales, these results support the notion emerging from our pullf4link simulation that the SNARE TMs play an active role in accelerating all the steps of membrane fusion from initial bilayer-bilayer contact to fusion pore formation, as was suggested previously by CG MD simulations (58).
Experimental Support and Implications for the Mechanisms Underlying Neurotransmitter Release and Membrane Fusion.
It is satisfying that the mechanism of SNARE-mediated membrane fusion suggested by our MD simulations correlates with and explains multiple lines of experimental evidence. Particularly important are the findings that insertion of a helix-breaking proline–proline sequence before the jxt linker of synaptobrevin (85PP) strongly impaired liposome fusion (43) and insertion of another helix-breaking sequence (glycine–serine–glycine) before the jxt linker of syntaxin-1 (259GSG) abolished neurotransmitter release (44, 45). These results suggested that jxt linker zippering plays a crucial role for liposome fusion in vitro and neurotransmitter release in neurons, in correlation with the key importance of linker zippering for the fusion mechanism that we observed. In contrast, a PP insertion after the jxt linker of synaptobrevin (93PP) had no or small effects on liposome fusion in vitro (43) and Ca2+-triggered exocytosis in chromaffin cells (42), indicating that zippering into the synaptobrevin TM region is not functionally important and consistent with the observation that TM residues close to the jxt linker remained flexible throughout our pullf4link simulation. In fact, the functional defects caused by mutations in the helix-breaking glycine of the synaptobrevin TM region (G100) or by helix stabilizing mutations suggested that flexibility in the synaptobrevin TM region is crucial for efficient Ca2+-triggered exocytosis (53, 54). Conversely, a GSG insertion after the jxt linker of syntaxin-1 (265GSG) dramatically decreased the amplitude of EPSCs in neurons while having little effect on the EPSC charge because it slowed down the kinetics of release (44). These results can be naturally explained by the observation that extension of the jxt linker helix into the TM of syntaxin-1 was not important to initiate membrane fusion but such helical extension likely facilitated formation of the fusion pore in our pullf4link simulation. Furthermore, the strong impairment of Ca2+-triggered exocytosis in chromaffin cells caused by addition of two charged residues (lysines or glutamates) at the C terminus of synaptobrevin led to the conclusion that the C terminus needs to be pulled inside the vesicle membrane during exocytosis (59), and such pulling is a direct consequence of linker zippering as observed in the pullf4link simulation.
The 259GSG and 265GSG insertions in syntaxin-1 had milder effects on slower forms of neurotransmitter release than on Ca2+-evoked release (44) (SI Appendix, Supplementary Discussion), suggesting that zippering of the syntaxin-1 linker is particularly crucial for evoked release because it helps to trigger membrane fusion in microseconds, as observed in our MD simulations. Linker zippering is expected to be hindered by energy barriers, as it pulls the TM regions toward the membrane interface and involves conformational changes in the linkers, which should be unstructured or form kinked helices in the primed state [(24) and SI Appendix, Fig. S1D]. Moreover, linker zippering likely requires remodeling of interactions between the lipids and the abundant basic and aromatic residues present in the jxt linkers (24, 60), which is supported by the effects of linker mutations on release (61, 62). The substantial folding energy associated with linker zippering [8 kBT (48)] likely helps to overcome these energy barriers, but it is tempting to speculate that linker zippering is also facilitated during Ca2+-triggering of release by other components of the neurotransmitter release machinery (e.g. synaptotagmin-1) through an as yet unknown mechanism.
Linker zippering also facilitates fusion at longer time scales such as those involved in sucrose-induced release (44, 45) and bulk liposome fusion assays (43). Moreover, reconstitution experiments with the vacuolar fusion machinery have shown that the jxt linkers of the vacuolar SNAREs also play a key role in liposome fusion (46) and the jxt linker of the exocytotic syntaxin homologue in yeast is important for secretion (47). Hence, the notion that linker zippering promotes hydrophobic encounters of the lipid acyl chains of two bilayers at the interface to initiate bilayer merger likely constitutes a general principle underlying intracellular membrane fusion. The earlier finding that surfactant-associated protein B also promoted such encounters to initiate fusion of 14 nm vesicles in CG MD simulations (40) suggests that these principles likely apply also to other forms of membrane fusion, including virus-induced fusion.
Our MD simulations do not support models predicting a proteinaceous fusion pore (e.g., refs. 63 and 64) but indicate that the SNARE TM regions play active roles during membrane fusion and hence provide a framework to reinterpret the mutagenesis data that suggested that these regions line the fusion pore in these studies. Note also that the importance of the TM regions for fusion has been controversial, as replacing them with lipid anchors allowed neurotransmitter release or SNARE-mediated liposome fusion in some studies but not in others (45, 65–67). The mechanism of SNARE-mediated membrane fusion uncovered by our MD simulations suggests that the ability of lipid anchors to support fusion may depend on the way they are attached to the SNAREs and whether they attach hydrophobic moieties close enough to the jxt linkers such that they can facilitate lipid acyl chain encounters at the interface.
An additional insight provided by our pullf4link simulation is that the acyl chain encounters at the interface primarily involved lipids with polyunsaturated acyl chains. Such lipids are highly abundant in synaptic vesicles (49) and brain membranes in general (50), are known to be important for brain health (68) and they were shown to enhance liposome fusion in vitro (69), which likely arises because of mismatch with the length and conformational preferences of the saturated acyl chain commonly present in the same lipid together with the polyunsaturated chain. Our results suggest that polyunsaturated lipids may be important for the fast speed of Ca2+-evoked neurotransmitter release.
Clearly, further research will be necessary to test the overall model of SNARE-mediated membrane fusion and other ideas presented here. Our results provide a foundation to pursue this research and a vivid illustration of the power of all-atom MD simulations to make further progress in this field and to elucidate complex biological problems.
Methods
High-Performance Computing.
All-atom MD simulations were performed using Gromacs (51, 52) with the CHARMM36 force field (70). Most high-performance computing, including all production MD simulations were carried out on Frontera at TACC. System setup, including solvation, ion addition, minimizations, and equilibration steps were performed at the BioHPC supercomputing facility of UT Southwestern. System visualization and manual manipulation were performed with Pymol (Schrödinger, LLC).
System Setup and MD Simulations.
The methodology used in system setups, equilibration, and production MD simulations was analogous to that described previously (24). The vesicle generated previously (24) was adapted for each system by moving lipids manually to accommodate different positions of the SNARE TM regions or their absence. A flat square bilayer of 30.5 nm × 30.5 nm was built at the CHARMM-GUI website (71) (https://charmm-gui.org/) for the fusion2g system and was adapted for all other systems (also moving lipids manually to accommodate SNARE TM regions) except for the snfreet350 system, which used a smaller bilayer adapted from the qscv simulation of ref. 24. The lipid compositions of the original vesicle and flat bilayers are described in Table 1, and those of each system were only slightly altered by adding or removing a few lipids as needed. The number of atoms, box dimensions, and simulation temperatures of each system are also listed in Table 1. All systems were solvated with explicit water molecules (TIP3P model), adding potassium and chloride ions as needed to reach a concentration of 145 mM and make the system neutral.
All systems were energy minimized using double precision, heated to the desired temperature over the course of a 1 ns MD simulation in the NVT ensemble with 1 fs steps, and equilibrated to 1 atm for 1 ns in the NPT ensemble using isotropic Parrinello–Rahman pressure coupling and 2 fs steps (72). NPT production level MD simulations were performed for the times indicated in Table 1 using 2 fs steps, isotropic Parrinello–Rahman pressure coupling, and a 1.1 nm cutoff for nonbonding interactions. Nose–Hoover temperature coupling (73) was used separately for three groups: i) protein atoms, ii) lipid atoms, and iii) water and KCL. Periodic boundary conditions were imposed with Particle Mesh Ewald (PME) (74) summation for long-range electrostatics. The default mixed precision was used in all MD simulations. The speeds of the production simulations ran on Frontera at TACC were typically about 28 ns/day for a typical system of about 5.2 million atoms using 40 nodes.
Supplementary Material
Appendix 01 (PDF)
Movie showing slices of the pullf4link simulation from 0 to 1956 ns, illustrating the events that lead to membrane fusion.
Acknowledgments
We thank Carsten Kutzner and Helmut Grubmüller for advice on how to optimize the parameters of our MD simulations. Most of the work presented in this paper was performed through Pathways and Leadership Computing Resource allocations on Frontera at the Texas Advanced Computing Center of The University of Texas at Austin (http://www.tacc.utexas.edu) (projects MCB20033 and IBN23002). This research also used computational resources provided by the BioHPC supercomputing facility located in the Lyda Hill Department of Bioinformatics, UT Southwestern Medical Center, TX (https://portal.biohpc.swmed.edu). This work was supported by grant I-1304 from the Welch Foundation (to J.R.) and by NIH Research Project Award R35 NS097333 (to J.R.).
Author contributions
J.R., L.S., K.J., C.R., and M.M.L. designed research; J.R. performed research; J.R. and L.S. contributed new reagents/analytic tools; J.R., L.S., and M.M.L. analyzed data; and J.R. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
Most files corresponding to the MD simulations are available in the dryad database (10.5061/dryad.9kd51c5rc) (75). Because of the very large size of trajectory files, it was not practical to deposit them in this database, but these files are available from the corresponding author upon reasonable request.
Supporting Information
References
- 1.Sudhof T. C., Neurotransmitter release: The last millisecond in the life of a synaptic vesicle. Neuron 80, 675–690 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rizo J., Molecular mechanisms underlying neurotransmitter release. Annu. Rev. Biophys. 51, 377–408 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brunger A. T., Leitz J., The core complex of the Ca(2+)-triggered presynaptic fusion machinery J. Mol. Biol. 435, 167853 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jahn R., Cafiso D. C., Tamm L. K., Mechanisms of SNARE proteins in membrane fusion. Nat. Rev. Mol. Cell Biol. 25, 101–118 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ma C., Su L., Seven A. B., Xu Y., Rizo J., Reconstitution of the vital functions of Munc18 and Munc13 in neurotransmitter release. Science 339, 421–425 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu X., et al. , Functional synergy between the Munc13 C-terminal C1 and C2 domains. eLife 5, e13696 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lai Y., et al. , Molecular mechanisms of synaptic vesicle priming by Munc13 and Munc18. Neuron 95, 591–607.e510 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sollner T., Bennett M. K., Whiteheart S. W., Scheller R. H., Rothman J. E., A protein assembly-disassembly pathway in vitro that may correspond to sequential steps of synaptic vesicle docking, activation, and fusion. Cell 75, 409–418 (1993). [DOI] [PubMed] [Google Scholar]
- 9.Hanson P. I., Roth R., Morisaki H., Jahn R., Heuser J. E., Structure and conformational changes in NSF and its membrane receptor complexes visualized by quick-freeze/deep-etch electron microscopy. Cell 90, 523–535 (1997). [DOI] [PubMed] [Google Scholar]
- 10.Poirier M. A., et al. , The synaptic SNARE complex is a parallel four-stranded helical bundle. Nat. Struct. Biol. 5, 765–769 (1998). [DOI] [PubMed] [Google Scholar]
- 11.Sutton R. B., Fasshauer D., Jahn R., Brunger A. T., Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 A resolution. Nature 395, 347–353 (1998). [DOI] [PubMed] [Google Scholar]
- 12.Mayer A., Wickner W., Haas A., Sec18p (NSF)-driven release of Sec17p (alpha-SNAP) can precede docking and fusion of yeast vacuoles. Cell 85, 83–94 (1996). [DOI] [PubMed] [Google Scholar]
- 13.Prinslow E. A., Stepien K. P., Pan Y. Z., Xu J., Rizo J., Multiple factors maintain assembled trans-SNARE complexes in the presence of NSF and alphaSNAP. eLife 8, e38880 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dulubova I., et al. , A conformational switch in syntaxin during exocytosis: Role of munc18. EMBO J. 18, 4372–4382 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Misura K. M., Scheller R. H., Weis W. I., Three-dimensional structure of the neuronal-Sec1-syntaxin 1a complex. Nature 404, 355–362 (2000). [DOI] [PubMed] [Google Scholar]
- 16.Parisotto D., et al. , An extended helical conformation in domain 3a of Munc18-1 provides a template for SNARE (soluble N-ethylmaleimide-sensitive factor attachment protein receptor) complex assembly. J. Biol. Chem. 289, 9639–9650 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baker R. W., et al. , A direct role for the Sec1/Munc18-family protein Vps33 as a template for SNARE assembly. Science 349, 1111–1114 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stepien K. P., Xu J., Zhang X., Bai X. C., Rizo J., SNARE assembly enlightened by cryo-EM structures of a synaptobrevin-Munc18-1-syntaxin-1 complex. Sci. Adv. 8, eabo5272 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ma C., Li W., Xu Y., Rizo J., Munc13 mediates the transition from the closed syntaxin-Munc18 complex to the SNARE complex. Nat. Struct. Mol. Biol. 18, 542–549 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu J., et al. , Mechanistic insights into neurotransmitter release and presynaptic plasticity from the crystal structure of Munc13-1 C1C2BMUN. eLife 6, e22567 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Quade B., et al. , Membrane bridging by Munc13-1 is crucial for neurotransmitter release. eLife 8, e42806 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou Q., et al. , Architecture of the synaptotagmin-SNARE machinery for neuronal exocytosis. Nature 525, 62–67 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen X., et al. , Three-dimensional structure of the complexin/SNARE complex. Neuron 33, 397–409 (2002). [DOI] [PubMed] [Google Scholar]
- 24.Rizo J., Sari L., Qi Y., Im W., Lin M. M., All-atom molecular dynamics simulations of Synaptotagmin-SNARE-complexin complexes bridging a vesicle and a flat lipid bilayer. eLife 11, e76356 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fernandez-Chacon R., et al. , Synaptotagmin I functions as a calcium regulator of release probability. Nature 410, 41–49 (2001). [DOI] [PubMed] [Google Scholar]
- 26.Weber T., et al. , SNAREpins: Minimal machinery for membrane fusion. Cell 92, 759–772 (1998). [DOI] [PubMed] [Google Scholar]
- 27.Hernandez J. M., et al. , Membrane fusion intermediates via directional and full assembly of the SNARE complex. Science 336, 1581–1584 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sabatini B. L., Regehr W. G., Timing of neurotransmission at fast synapses in the mammalian brain. Nature 384, 170–172 (1996). [DOI] [PubMed] [Google Scholar]
- 29.Stein A., Weber G., Wahl M. C., Jahn R., Helical extension of the neuronal SNARE complex into the membrane. Nature 460, 525–528 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hanson P. I., Heuser J. E., Jahn R., Neurotransmitter release—Four years of SNARE complexes. Curr. Opin. Neurobiol. 7, 310–315 (1997). [DOI] [PubMed] [Google Scholar]
- 31.Chernomordik L. V., Kozlov M. M., Mechanics of membrane fusion. Nat. Struct. Mol. Biol 15, 675–683 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cohen F. S., Melikyan G. B., The energetics of membrane fusion from binding, through hemifusion, pore formation, and pore enlargement. J. Membr. Biol. 199, 1–14 (2004). [DOI] [PubMed] [Google Scholar]
- 33.Risselada H. J., Kutzner C., Grubmuller H., Caught in the act: Visualization of SNARE-mediated fusion events in molecular detail. Chembiochem 12, 1049–1055 (2011). [DOI] [PubMed] [Google Scholar]
- 34.Sharma S., Lindau M., Molecular mechanism of fusion pore formation driven by the neuronal SNARE complex. Proc. Natl. Acad. Sci. U.S.A. 115, 12751–12756 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Risselada H. J., Grubmuller H., How proteins open fusion pores: Insights from molecular simulations. Eur. Biophys. J. 50, 279–293 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stevens M. J., Hoh J. H., Woolf T. B., Insights into the molecular mechanism of membrane fusion from simulation: Evidence for the association of splayed tails. Phys. Rev. Lett. 91, 188102 (2003). [DOI] [PubMed] [Google Scholar]
- 37.Marrink S. J., Mark A. E., The mechanism of vesicle fusion as revealed by molecular dynamics simulations. J. Am. Chem. Soc. 125, 11144–11145 (2003). [DOI] [PubMed] [Google Scholar]
- 38.Kasson P. M., et al. , Ensemble molecular dynamics yields submillisecond kinetics and intermediates of membrane fusion. Proc. Natl. Acad. Sci. U.S.A. 103, 11916–11921 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kasson P. M., Lindahl E., Pande V. S., Atomic-resolution simulations predict a transition state for vesicle fusion defined by contact of a few lipid tails. PLoS Comput. Biol. 6, e1000829 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baoukina S., Tieleman D. P., Direct simulation of protein-mediated vesicle fusion: Lung surfactant protein B. Biophys. J. 99, 2134–2142 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McNew J. A., Weber T., Engelman D. M., Sollner T. H., Rothman J. E., The length of the flexible SNAREpin juxtamembrane region is a critical determinant of SNARE-dependent fusion. Mol. Cell 4, 415–421 (1999). [DOI] [PubMed] [Google Scholar]
- 42.Kesavan J., Borisovska M., Bruns D., v-SNARE actions during Ca(2+)-triggered exocytosis Cell 131, 351–363 (2007). [DOI] [PubMed] [Google Scholar]
- 43.Hu Y., Zhu L., Ma C., Structural roles for the juxtamembrane linker region and transmembrane region of synaptobrevin 2 in membrane fusion. Front. Cell Dev. Biol. 8, 609708 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vardar G., Salazar-Lazaro A., Zobel S., Trimbuch T., Rosenmund C., Syntaxin-1A modulates vesicle fusion in mammalian neurons via juxtamembrane domain dependent palmitoylation of its transmembrane domain. eLife 11, e78182 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou P., Bacaj T., Yang X., Pang Z. P., Sudhof T. C., Lipid-anchored SNAREs lacking transmembrane regions fully support membrane fusion during neurotransmitter release. Neuron 80, 470–483 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Orr A., Song H., Wickner W., Fusion with wild-type SNARE domains is controlled by juxtamembrane domains, transmembrane anchors, and Sec17. Mol. Biol. Cell 33, ar38 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Van Komen J. S., Bai X., Rodkey T. L., Schaub J., McNew J. A., The polybasic juxtamembrane region of Sso1p is required for SNARE function in vivo. Eukaryot. Cell 4, 2017–2028 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gao Y., et al. , Single reconstituted neuronal SNARE complexes zipper in three distinct stages. Science 337, 1340–1343 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Takamori S., et al. , Molecular anatomy of a trafficking organelle. Cell 127, 831–846 (2006). [DOI] [PubMed] [Google Scholar]
- 50.Chan R. B., et al. , Comparative lipidomic analysis of mouse and human brain with Alzheimer disease. J. Biol. Chem. 287, 2678–2688 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pronk S., et al. , GROMACS 4.5: A high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics 29, 845–854 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Van Der Spoel D., et al. , GROMACS: Fast, flexible, and free. J. Comput. Chem. 26, 1701–1718 (2005). [DOI] [PubMed] [Google Scholar]
- 53.Dhara M., et al. , v-SNARE transmembrane domains function as catalysts for vesicle fusion. eLife 5, e17571 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hastoy B., et al. , A central small amino acid in the VAMP2 transmembrane domain regulates the fusion pore in exocytosis. Sci. Rep. 7, 2835 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kasson P. M., Lindahl E., Pande V. S., Water ordering at membrane interfaces controls fusion dynamics. J. Am. Chem. Soc. 133, 3812–3815 (2011). [DOI] [PubMed] [Google Scholar]
- 56.Kozlov M. M., Markin V. S., Possible mechanism of membrane fusion. Biofizika 28, 242–247 (1983). [PubMed] [Google Scholar]
- 57.Risselada H. J., Mayer A., SNAREs, tethers and SM proteins: How to overcome the final barriers to membrane fusion? Biochem. J. 477, 243–258 (2020). [DOI] [PubMed] [Google Scholar]
- 58.Risselada H. J., Grubmuller H., How SNARE molecules mediate membrane fusion: Recent insights from molecular simulations. Curr. Opin. Struct. Biol. 22, 187–196 (2012). [DOI] [PubMed] [Google Scholar]
- 59.Ngatchou A. N., et al. , Role of the synaptobrevin C terminus in fusion pore formation. Proc. Natl. Acad. Sci. U.S.A. 107, 18463–18468 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Brewer K. D., Li W., Horne B. E., Rizo J., Reluctance to membrane binding enables accessibility of the synaptobrevin SNARE motif for SNARE complex formation. Proc. Natl. Acad. Sci. U.S.A. 108, 12723–12728 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Maximov A., Tang J., Yang X., Pang Z. P., Sudhof T. C., Complexin controls the force transfer from SNARE complexes to membranes in fusion. Science 323, 516–521 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fang Q., Zhao Y., Lindau M., Juxtamembrane tryptophans of synaptobrevin 2 control the process of membrane fusion. FEBS Lett. 587, 67–72 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Han X., Wang C. T., Bai J., Chapman E. R., Jackson M. B., Transmembrane segments of syntaxin line the fusion pore of Ca2+-triggered exocytosis. Science 304, 289–292 (2004). [DOI] [PubMed] [Google Scholar]
- 64.Bao H., et al. , Exocytotic fusion pores are composed of both lipids and proteins. Nat. Struct. Mol. Biol. 23, 67–73 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.McNew J. A., et al. , Close is not enough: SNARE-dependent membrane fusion requires an active mechanism that transduces force to membrane anchors. J. Cell Biol. 150, 105–117 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xu H., Zick M., Wickner W. T., Jun Y., A lipid-anchored SNARE supports membrane fusion. Proc. Natl. Acad. Sci. U.S.A. 108, 17325–17330 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chang C. W., Chiang C. W., Gaffaney J. D., Chapman E. R., Jackson M. B., Lipid-anchored synaptobrevin provides little or no support for exocytosis or liposome fusion. J. Biol. Chem. 291, 2848–2857 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Manni M. M., et al. , The fatty acids of sphingomyelins and ceramides in mammalian tissues and cultured cells: Biophysical and physiological implications. Chem. Phys. Lipids 217, 29–34 (2018). [DOI] [PubMed] [Google Scholar]
- 69.Francois-Martin C., Bacle A., Rothman J. E., Fuchs P. F. J., Pincet F., Cooperation of conical and polyunsaturated lipids to regulate initiation and processing of membrane fusion. Front. Mol. Biosci. 8, 763115 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Best R. B., et al. , Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone phi, psi and side-chain chi(1) and chi(2) dihedral angles. J. Chem. Theory Comput. 8, 3257–3273 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jo S., Kim T., Iyer V. G., Im W., CHARMM-GUI: A web-based graphical user interface for CHARMM. J. Comput. Chem. 29, 1859–1865 (2008). [DOI] [PubMed] [Google Scholar]
- 72.Parrinello M., Rahman A., Polymorphic transitions in single crystals: A new molecular dynamics method. J. Appl. Phys. 52, 7182–7190 (1981). [Google Scholar]
- 73.Hoover T., Canonical dynamics: Equilibrium phase-space distributions. Phys. Rev. A: Gen. Phys. 31, 1695–1697 (1985). [DOI] [PubMed] [Google Scholar]
- 74.Darden T., York D., Pedersen L., Particle mesh Ewald: An N·log(N) method for Ewald sums in large systems. J. Chem. Phys. 98, 10089–10092 (1993). [Google Scholar]
- 75.Rizo J., Molecular mechanism underlying SNARE-mediated membrane fusion enlightened by all-atom molecular dynamics simulations. Dryad. 10.5061/dryad.9kd51c5rc. Deposited 20 March 2024. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Movie showing slices of the pullf4link simulation from 0 to 1956 ns, illustrating the events that lead to membrane fusion.
Data Availability Statement
Most files corresponding to the MD simulations are available in the dryad database (10.5061/dryad.9kd51c5rc) (75). Because of the very large size of trajectory files, it was not practical to deposit them in this database, but these files are available from the corresponding author upon reasonable request.