Abstract
Background: In August 2023, the BA.2.86 SARS-CoV-2 variant, with over 30 spike protein mutations, emerged amidst the global dominance of XBB sub-lineages. It evolved into JN.1 by late 2023, spreading across 71 countries. JN.1, distinct for its L455S mutation, significantly dominated global sequences, raising concerns over its transmission and clinical impact. The study investigates JN.1's clinical severity and its effect on hospital admissions in Maharashtra, India.
Methodology: The present study involved 3,150 curated Indian SARS-CoV-2 whole genome sequences with collection dates between 1st August 2023 and 15th January 2024. Lineage and phylogenetic analysis of sequences was performed using Nextclade. Telephonic interviews were conducted to confirm the demographic details and obtain clinical information on the JN.1* (* indicates JN.1 and all its sub-lineages) cases. The obtained data were recorded and analyzed using Microsoft® Excel (Microsoft Corporation, Redmond, WA).
Results: Out of 3,150 sequences analyzed, JN.1* was the most common lineage (2377/3150, 75.46%), followed by XBB.2.3* (281/3150, 8.92%) and XBB.1.16* (187/3150, 5.94%). In India, it was first identified on 6th October 2023, in Kerala. The highest proportion of JN.1* sequences originated from Maharashtra (628/2377, 26.42%), followed by West Bengal (320/2377, 13.46%), Andhra Pradesh (293/2377, 12.33%), Kerala (288/2377, 12.12%), and Karnataka (285/2377, 11.99%). In Maharashtra, the JN.1* variant was first identified on 23rd November 2023. A total of 279 JN.1* cases were included in the clinical study. Of these, 95.34% (266/279) had symptomatic disease with mild symptoms; cold (187/279, 67.03%) being the most common symptom, followed by fever (156/279, 55.91%), cough (114/279, 40.86%), and headache (28/279, 15.64%). Of all the cases, 13.26% (37/279) required institutional quarantine or hospitalization, and the rest were isolated at home. Among the hospitalized patients, 54.05% (20/37) cases were given conservative treatment while 45.95% (17/37) cases required supplemental oxygen therapy. Regarding the vaccination status, 94.26% (263/279) of cases received at least one dose of the COVID-19 vaccine, while 5.02% (14/279) were not vaccinated, of which most were children aged zero to nine years (5/14, 35.71%). The overall recovery rate among JN.1* cases was 98.57% (275/279), with 1.43% (4/279) cases succumbing to the disease.
Conclusion: The JN.1* variant, the dominant variant in India, exhibits clinical features similar to previous circulating variants in Maharashtra without increased severity. Its notable transmissibility underscores the importance of studying the ongoing viral evolution. The pressing necessity for swift identification and the clinical features of new variants is essential for effective public health response.
Keywords: covid-19, severe acute respiratory syndrome coronavirus 2, sars-cov-2, clinical characteristics, covid-19 disease, pirola, ba.2.86, ba.2.86.1.1, jn.1
Introduction
At the time when the sub-lineages of the recombinant SARS-CoV-2 variant, XBB, were predominant globally, a new sub-lineage of BA.2 appeared around August 2023 [1]. This new lineage of SARS-CoV-2 was labeled as BA.2.86 on 17th August 2023 [2]. BA.2.86 had more than 30 spike (S) protein mutations over and above its parent lineage, BA.2 [1]. It was first sequenced in Israel and reported on 13th August 2023 [3]. Alarmed by its potential to evade pre-existing immunity against SARS-CoV-2 developed by the administration of vaccines and from previous infections, the World Health Organization (WHO), within four days, promptly designated it as a variant under monitoring (VUM) on 17th August 2023, the day it was labeled as BA.2.86. Later, due to its growing prevalence, as evidenced by genomic surveillance, the WHO escalated its status to the variant of interest (VoI) on 21st November 2023 [4].
In late 2023, the BA.2.86 SARS-CoV-2 variant evolved, leading to the emergence of the JN.1 (BA.2.86.1.1) variant. It was first identified in the United States in September 2023 [5]. At the same time, it was identified in 12 more countries, with the highest proportions in Canada, France, Singapore, Sweden, the United Kingdom, and the United States [6]. Initially monitored within the BA.2.86 variant, the rapid and extensive spread of JN.1 across various nations prompted WHO to designate it as a VOI on 20th December 2023, thereby acknowledging its distinction from its parent lineage, BA.2.86. According to the WHO reports as of 19th January 2024, JN.1 has emerged as the most reported VOI globally, with detection in 71 countries and accounting for 65.5% of all sequences in the final week of 2023 [7]. By January 2024, the JN.1* variant dominated India, representing 97.25% of Indian sequences on the Global Initiative on Sharing All Influenza Data (GISAID), thus highlighting the significant impact and global concern over its transmission dynamics [8].
While JN.1 is closely related to its precursor, BA.2.86, it features a unique mutation, L455S, in the receptor-binding domain (RBD) of its spike protein, alongside three additional mutations in non-spike proteins. A similar L455F mutation in variants HK.3 and other FLip variants has been shown to enhance transmissibility and immune evasion compared to the original EG.5.1 variant [9]. However, information is scarce regarding the potential impact of JN.1 on clinical outcomes. Therefore, the current study aims to evaluate the clinical severity associated with JN.1 infections and its implications for hospital admissions in Maharashtra.
Materials and methods
The present study is a part of the sequencing activity in Maharashtra under the Indian SARS-CoV-2 Genomics Consortium (INSACOG) to study the evolutionary patterns and epidemiological characteristics of SARS-CoV-2.
SARS-CoV-2 whole genome sequences in India: lineage and phylogenetic analysis
To trace the first appearance and the spread of the JN.1 SARS-CoV-2 variant in India, whole genome sequences of the virus, with sample collection dates from 1st August 2023 to 15th January 2024, submitted by various sequencing laboratories in different states and Union Territories of the country, were downloaded from the Indian Biological Data Centre (IBDC) database [10] with their kind permission. Entries with complete metadata, including geographic locations and sample collection dates, were included in the study and the sequences used are accessible in the supplemental table in the Appendices. The lineage and clade analysis were performed using Nextclade software (version 3.0.1). The phylogenetic tree was constructed using Nextclade Augur and visualized using Auspice (version 2.52.1).
Demographic and clinical data collection of SARS-CoV-2-positive cases in Maharashtra
Demographic details, including age, sex, area of residence, contact number, and date of sample collection and testing, were collected from the metadata provided to the sequencing laboratories by the reverse transcription-polymerase chain reaction (RT-PCR) testing centers. Telephonic interviews were conducted with each patient to confirm their demographic details and obtain clinical information such as symptoms, isolation type, hospitalization, oxygen requirements, treatments, and vaccination status. Those patients who chose not to disclose their clinical history during the interview were documented and excluded from the study.
Also, daily records from 1st August 2023 to 15th January 2024 were collected from the State's District Health Services Department to assess the severity of illness linked to the JN.1 SARS-CoV-2 variant during its surge in Maharashtra and its impact on public health. The data collected included the daily total number of samples tested for SARS-CoV-2, test positivity, hospital admissions, and the demand for oxygen throughout the state.
Statistical analysis
All demographic and clinical data were recorded and analyzed using Microsoft® Excel (Microsoft Corporation, Redmond, WA), and analysis was performed using Microsoft® Excel. Continuous variables were presented as median and interquartile range (IQR). Categorical variables were presented as numbers and percentages.
Results
Distribution of SARS-CoV-2 lineages in India
A total of 3,150 downloaded sequences were included in the study following data curation. A total of 144 different lineages were identified following Nextclade Pangolin nomenclature. JN.1* was the most common lineage (75.46%), followed by XBB.2.3* (8.92%) and XBB.1.16* (5.94%) (Table 1).
Table 1. Distribution of SARS-CoV-2 variants among sequences deposited on IBDC from India.
IBDC: Indian Biological Data Centre.
| Clade | Nextclade Pangolin lineage | Count (%) | ||
| 21K | BA.1* | BA.1 | 2 | 23 (0.73%) |
| BA.1.1 | 21 | |||
| 21L | BA.2* | BA.2 | 31 | 56 (1.78%) |
| BA.2.1 | 4 | |||
| BA.2.10.4 | 1 | |||
| BA.2.12 | 1 | |||
| BA.2.38 | 13 | |||
| BA.2.38.1 | 1 | |||
| BA.2.76 | 1 | |||
| 22C | BA.2.12.1 | 1 | ||
| CH.1.1.24 | 1 | |||
| DV.7.1.3 | 1 | |||
| FK.1.1 | 1 | |||
| 22B | BA.5* | BA.5 | 1 | 5 (0.16%) |
| BA.5.5 | 1 | |||
| BA.5.5.1 | 1 | |||
| BE.1.1 | 1 | |||
| 22E | BQ.1.22 | 1 | ||
| 23A | XBB.1.5* | GF.1 | 1 | 11 (0.35%) |
| JD.1.1 | 2 | |||
| XBB.1.5 | 6 | |||
| XBB.1.5.28 | 1 | |||
| 23G | GK.1.1 | 1 | ||
| 23D | XBB.1.9* | XBB.1.9.1 | 2 | 47 (1.49%) |
| EG.5.2 | 1 | |||
| FL.1.5 | 1 | |||
| FL.1.5.1 | 2 | |||
| FL.13.2 | 1 | |||
| FL.13.4.1 | 1 | |||
| FL.4.8 | 1 | |||
| HN.5 | 1 | |||
| 23F | EG.5.1 | 3 | ||
| EG.5.1.1 | 5 | |||
| EG.5.1.15 | 1 | |||
| EG.5.1.3 | 1 | |||
| EG.5.1.6 | 2 | |||
| EG.5.1.8 | 1 | |||
| HK.19 | 1 | |||
| HK.29 | 1 | |||
| HV.1 | 10 | |||
| HV.1.11 | 2 | |||
| JG.3 | 3 | |||
| JG.3.2 | 1 | |||
| 23H | HK.3 | 4 | ||
| HK.3.1 | 1 | |||
| HK.3.5 | 1 | |||
| 23B | XBB.1.16* | XBB.1.16 | 57 | 187 (5.94%) |
| XBB.1.16.1 | 17 | |||
| XBB.1.16.11 | 37 | |||
| XBB.1.16.12 | 3 | |||
| XBB.1.16.13 | 1 | |||
| XBB.1.16.17 | 32 | |||
| XBB.1.16.18 | 3 | |||
| XBB.1.16.2 | 1 | |||
| XBB.1.16.24 | 28 | |||
| XBB.1.16.8 | 1 | |||
| FU.1 | 3 | |||
| FU.3.1 | 1 | |||
| JF.1.1 | 1 | |||
| JM.2 | 2 | |||
| 23E | XBB.2.3* | GE.1 | 85 | 281 (8.92%) |
| GE.1.1 | 10 | |||
| GJ.1 | 4 | |||
| GJ.1.1 | 8 | |||
| GJ.1.2 | 2 | |||
| GJ.3 | 1 | |||
| GJ.6 | 4 | |||
| GS.1 | 1 | |||
| GZ.1 | 5 | |||
| HH.1 | 1 | |||
| HH.2 | 5 | |||
| HH.6 | 10 | |||
| HH.8 | 1 | |||
| JE.1 | 1 | |||
| JY.1 | 44 | |||
| JY.1.1 | 47 | |||
| XBB.2.3 | 15 | |||
| XBB.2.3.10 | 1 | |||
| XBB.2.3.11 | 2 | |||
| XBB.2.3.12 | 1 | |||
| XBB.2.3.18 | 1 | |||
| XBB.2.3.19 | 2 | |||
| XBB.2.3.2 | 7 | |||
| XBB.2.3.22 | 1 | |||
| XBB.2.3.3 | 20 | |||
| XBB.2.3.5 | 1 | |||
| XBB.2.3.8 | 1 | |||
| 23I | BA.2.86* | BA.2.86 | 15 | 32 (1.02%) |
| BA.2.86.1 | 12 | |||
| JN.2 | 3 | |||
| JN.4 | 1 | |||
| JN.6 | 1 | |||
| JN.1* | JN.1 | 1026 | 2377 (75.46%) | |
| JN.1.1 | 959 | |||
| JN.1.1.1 | 1 | |||
| JN.1.1.3 | 1 | |||
| JN.1.10 | 7 | |||
| JN.1.11 | 304 | |||
| JN.1.2 | 2 | |||
| JN.1.3 | 2 | |||
| JN.1.4 | 36 | |||
| JN.1.5 | 21 | |||
| JN.1.6 | 1 | |||
| JN.1.7 | 1 | |||
| JN.1.8 | 10 | |||
| JN.1.9 | 6 | |||
| 22F | Other XBBs | XBB | 1 | 13 (0.41%) |
| XBB.1 | 1 | |||
| GW.5.1.1 | 3 | |||
| GW.5.3.1 | 2 | |||
| JC.2 | 2 | |||
| KE.2 | 1 | |||
| XBB.1.41.1 | 1 | |||
| XBB.2.4 | 1 | |||
| XBB.2.6 | 1 | |||
| Recombinant | Other recombinant variants | XAC | 1 | 35 (1.11%) |
| XAD | 2 | |||
| XAF | 1 | |||
| XAG | 2 | |||
| XAH | 2 | |||
| XAK | 1 | |||
| XBH | 1 | |||
| XBQ | 1 | |||
| XBW | 1 | |||
| XCG | 1 | |||
| XCH.1 | 1 | |||
| XDA | 12 | |||
| XDA.1 | 1 | |||
| XDB | 3 | |||
| XDD | 4 | |||
| XDK | 1 | |||
| 19A | Other lineages | B | 4 | 44 (1.40%) |
| 20A | B.1 | 7 | ||
| 20B | B.1.1 | 11 | ||
| B.1.1.161 | 4 | |||
| B.1.1.220 | 1 | |||
| B.1.1.519 | 2 | |||
| 20C | B.1.566 | 1 | ||
| 20F | D.2 | 1 | ||
| 20G | B.1.2 | 2 | ||
| 21C | B.1.429 | 3 | ||
| 21J | AY.4 | 2 | ||
| 21M | B.1.1.529 | 5 | ||
| 22A | BA.4.6 | 1 | ||
| Lineage unassigned (due to poor quality sequences) | 39 (1.24%) | |||
| Grand total | 3150 | |||
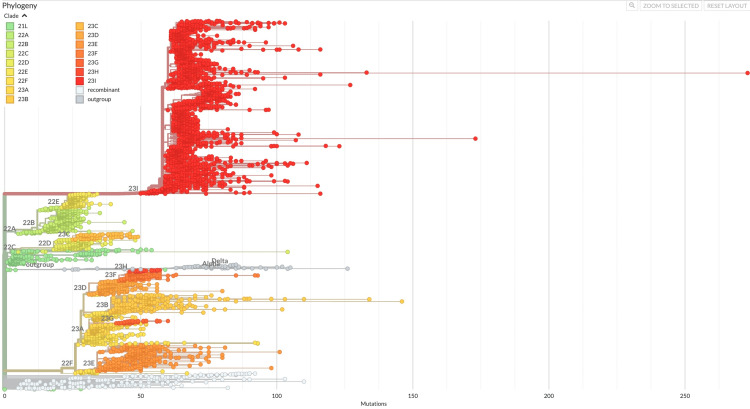
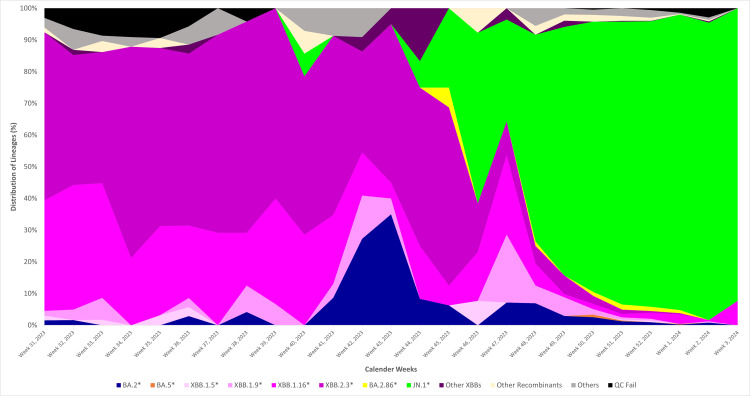
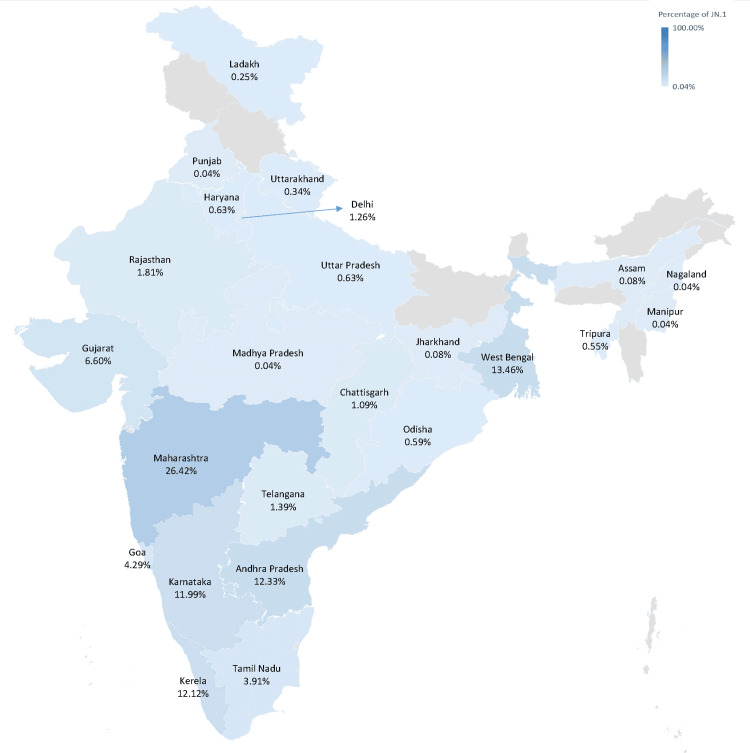
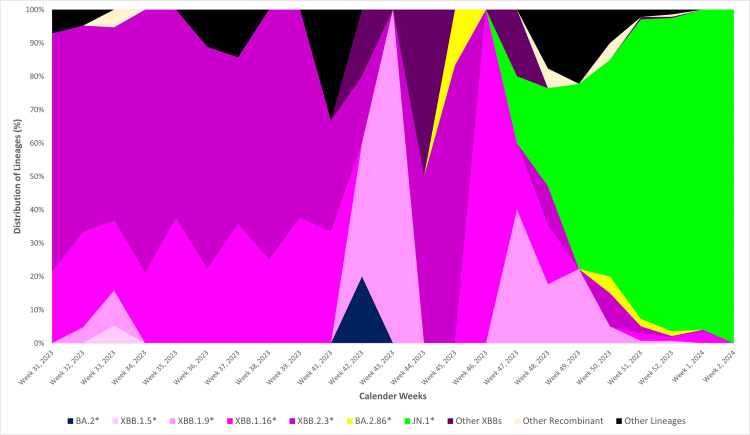
Figure 1 illustrates the evolutionary relationship of the JN.1* variant (clade 23I) in comparison to other variants. In India, the temporal distribution of variants indicates that the XBB variant and its related lineages were predominant from week 31, 2023, until week 45, 2023 (Figure 2). The JN.1* variant was first identified in Indian sequences on 6th October 2023 in Kerala. Subsequently, by December 2023, the JN.1* variant became the most prevalent variant across India. The JN.1* variant has grown from 8.33% since its first detection in week 40, 2023, to 93.83% in week two, 2024. Of all the sequences submitted by various states, 75% are of the JN.1* variant, with the highest proportion of JN.1* sequences originating from Maharashtra (26.42%), followed by West Bengal (13.46%), Andhra Pradesh (12.33%), Kerala (12.12%), and Karnataka (11.99%) (Figure 3).
Figure 1. Evolutionary relationship of clade 23I with other clades in India.
The figure represents the phylogenetic relationship of clade 23I with other clades identified in Indian sequences during the study period. The horizontal axis of the phylogenetic tree denotes the number of mutations, with the numbers increasing progressively from left to right. The branches, represented by horizontal lines, signify evolutionary divergence over time. The length of the branch horizontally correlates with the amount of change. The vertical axis on the other hand organizes the clade labels in a spatial manner for clarity. It does not imply any phylogenetic or temporal relationship.
Image credits: Rashmita Das
Figure 2. Temporal distribution of SARS-CoV-2 variants based on sequences submitted to the IBDC in India.
IBDC: Indian Biological Data Centre.
Image credits: Rashmita Das
Figure 3. Geographic distribution of the 2377 JN.1* SARS-CoV-2 sequences based on sequences submitted to the IBDC in India.
IBDC: Indian Biological Data Centre.
Image credits: Rashmita Das
Test positivity and lineage distribution of SARS-CoV-2 in Maharashtra
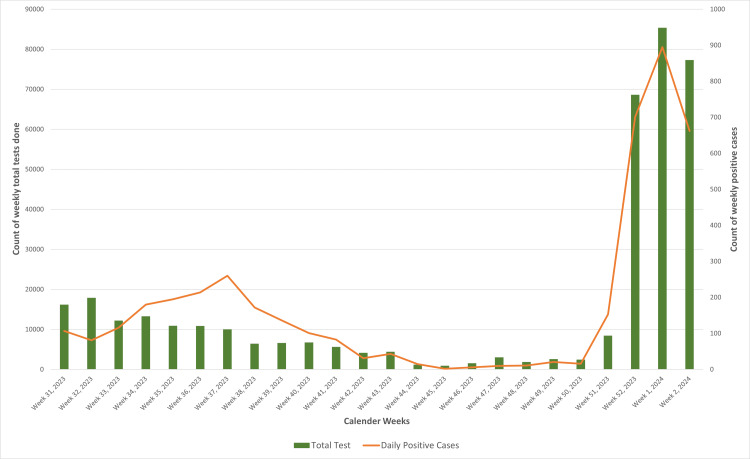
Figure 4 represents the overview of SARS-CoV-2 testing in Maharashtra, tracking the total number of tests performed (including RT-PCR and antigen testing) and the positivity rate from the 31st week of 2023 through the second week of 2024. The total tests performed show significant variability with notable spikes toward the end of the period (in weeks 52 of 2023 and the first and second weeks of 2024), suggesting increased testing efforts during these weeks. Similarly, the weekly positivity rate also experienced fluctuations throughout the period. The highest positivity rate was 2.67% in the 38th week of 2023. Following this peak, the positivity rate gradually declined, reaching its lowest in the 45th week (0.21%) and increasing again to 1.81% in the 51st week. Following the first detection of the JN.1* variant globally, the testing efforts in the state and the country were increased [11], thus explaining the variations in both the number of tests performed and the positivity rate reflecting the changes in the testing strategies and the public health policies in the state.
Figure 4. Weekly tests performed versus weekly count of positive cases in Maharashtra between 1st August 2023 and 15th January 2024.
Image credits: Rashmita Das
Figure 5 presents the weekly distribution of SARS-CoV-2 variants in Maharashtra from week 31, 2023 to week two, 2024, mirroring the trend observed across India. The JN.1* variant was first identified on 23rd November 2023, and its prevalence rose from 20% in week 47, 2023, to 100% by the second week of 2024.
Figure 5. Temporal distribution of SARS-CoV-2 variants based on sequences submitted to the IBDC from Maharashtra.
IBDC: Indian Biological Data Centre.
Image credits: Rashmita Das
Demographic and clinical characteristics of JN.1* SARS-CoV-2 variant in Maharashtra
Out of 628 JN.1* cases in Maharashtra, 550 (87.58%) cases had complete metadata available and were included in the demographic study (Table 2). Of these 550 participants, 55.45% were males and 44.55% were females. The median age of the study population was 42 (IQR: 30-60) years, with individuals aged 60 years and above (25.09%) being predominantly affected.
Table 2. Demographic characteristics of JN.1* SARS-CoV-2 variant in Maharashtra (n = 550).
| Demographic characteristics | Count (%) |
| Gender-wise distribution | |
| Male | 305 (55.45%) |
| Female | 245 (44.55%) |
| Age-wise distribution (in years) | |
| 0-9 | 05 (0.91%) |
| 10-19 | 39 (7.09%) |
| 20-29 | 84 (15.27%) |
| 30-39 | 115 (20.91%) |
| 40-49 | 84 (15.27%) |
| 50-59 | 85 (15.45%) |
| 60 and above | 138 (25.09%) |
| Area-wise distribution | |
| Ahmednagar | 02 (0.36%) |
| Akola | 02 (0.36%) |
| Amravati | 18 (3.27%) |
| Chhatrapati Sambhajinagar | 54 (9.82%) |
| Beed | 03 (0.55%) |
| Bhandara | 01 (0.18%) |
| Hingoli | 03 (0.55%) |
| Jalgaon | 04 (0.73%) |
| Jalna | 02 (0.36%) |
| Kolhapur | 10 (1.82%) |
| Mumbai | 18 (3.27%) |
| Nagpur | 36 (6.55%) |
| Nanded | 02 (0.36%) |
| Nandurbar | 01 (0.18%) |
| Nashik | 02 (0.36%) |
| Osmanabad | 02 (0.36%) |
| Pune | 273 (49.64%) |
| Raigad | 18 (3.27%) |
| Sangli | 01 (0.18%) |
| Satara | 01 (0.18%) |
| Solapur | 13 (2.36%) |
| Thane | 83 (15.09%) |
| Yavatmal | 01 (0.18%) |
From this subset of 550 cases, attempts to contact 253 (46%) participants were unsuccessful due to non-response to calls, and 18 (3.27%) participants declined participation in the study. Therefore, 279 (50.73%) participants were successfully contacted and included in the clinical study. Table 3 summarizes the clinical characteristics, vaccination status, and outcome of JN.1* cases in Maharashtra. Most JN.1* cases had symptomatic disease (95.34%) with mild symptoms. The most common symptoms were cold (67.03%), fever (55.91%), cough (40.86%), headache (15.64%), body ache (9.68%), and fatigue (8.60%). Underlying comorbid conditions, either alone or in combination, were reported in 17.56% (49 out of 279) of cases, with diabetes mellitus (29, alone or in combination; 59.18%), hypertension (22, alone or in combination; 44.90%), and underlying lung pathology (six, alone or in combination; 12.25%) being the most common conditions.
Table 3. Clinical characteristics of JN.1* SARS-CoV-2 variant in Maharashtra (n = 279).
Out of 550 JN.1* cases with complete metadata, 279 participants were successfully contacted and included in the clinical study.
| Clinical characteristics | Count (%) |
| 1. History of previous infection | |
| Present | 41 (14.69%) |
| Absent | 238 (85.31%) |
| 2. Presence of comorbidities | |
| a. Present | 49 (17.56%) |
| i. Diabetes mellitus (DM) | 18 (36.73%) |
| ii. Hypertension (HTN) | 12 (24.29%) |
| iii. Underlying lung disease | 04 (8.16%) |
| iv. Arthritis | 01 (2.04%) |
| v. Tuberculosis | 01 (2.04%) |
| vi. Hypothyroidism | 01 (2.04%) |
| vii. Coronary heart disease (CHD) | 01 (2.04%) |
| viii. HIV/AIDS | 01 (2.04%) |
| ix. DM + HTN | 08 (16.33%) |
| x. DM + underlying lung disease | 01 (2.04%) |
| xi. DM + HTN + CHD | 01 (2.04%) |
| xii. DM + HTN + underlying lung disease | 01 (2.04%) |
| b. Absent | 230 (82.44%) |
| 3. Vaccination status | |
| a. Vaccinated | 263 (94.26%) |
| i. One dose | 12 (4.56%) |
| ii. Two doses | 151 (57.42%) |
| iii. Booster dose (precautionary dose) | 100 (38.02%) |
| b. Not vaccinated | 14 (5.02%) |
| c. Data not available | 02 (0.72%) |
| 4. Symptom status | |
| a. Symptomatic | 266 (95.34%) |
| b. Asymptomatic | 13 (4.66%) |
| 5. Presenting symptoms | |
| a. Fever | 156 (55.91%) |
| b. Cold | 187 (67.03%) |
| c. Cough | 114 (40.86%) |
| d. Breathlessness | 15 (5.38%) |
| e. Headache | 28 (15.64%) |
| f. Fatigue | 24 (8.60%) |
| g. Body ache | 27 (9.68%) |
| h. Diarrhea | 02 (0.72%) |
| i. Sore throat | 16 (5.74%) |
| j. Vomiting | 02 (0.72%) |
| k. Chest pain | 01 (0.36%) |
| l. Loss of taste and smell | 01 (0.36%) |
| m. Joint pain | 01 (0.36%) |
| 6. Type of quarantine | |
| a. Home isolation | 242 (86.74%) |
| b. Hospital isolation/hospitalization | 37 (13.26%) |
| 7. Treatment | |
| a. Received no treatment | 03 (1.075%) |
| b. Received symptomatic treatment | 259 (92.83%) |
| c. Received oxygen therapy | 14 (5.02%) |
| i. Non-invasive ventilation | 09 (64.29%) |
| ii. Invasive ventilation | 05 (35.71%) |
| d. Received antiviral/steroid with oxygen | 03 (1.075%) |
| 8. Outcome | |
| a. Alive | 275 (98.57%) |
| b. Dead | 04 (1.43%) |
Regarding the vaccination status, 94.26% (263 out of 279) of cases received at least one dose of the COVID-19 vaccine, while 5.02% (14 out of 279) were not vaccinated. Most unvaccinated individuals were children aged zero to nine years (five out of 14, 35.71%) and adults aged 30-39 years (three out of 14, 21.43%). Covishield (ChAdOx1nCoV-19 coronavirus vaccine) (242 out of 263, 92.01%) was the predominant vaccine administered, followed by CovaxinTM (BBV152A-a whole inactivated virus-based COVID-19 vaccine) (18 out of 263, 6.84%).
Of all the cases, 13.26% (37 out of 279) required institutional quarantine or hospitalization. Nearly all of the patients who were home-isolated received conservative treatment (240 out of 242, 99.17%), whereas hospitalized patients often received conservative care (20 out of 37, 54.05%) or required supplemental oxygen therapy (17 out of 37, 45.95%). The recovery rate was high; out of 279 cases, 98.57% of cases (275 out of 279) recovered from the disease, while 1.43% (four out of 279) succumbed.
Among the deceased, three were over 60 years of age (75%), and one (25%) was in the age group of 30-39 years. Three out of four cases (75%) suffered from respiratory symptoms and had underlying comorbidities, while one (25%) did not have respiratory symptoms and was admitted to the hospital due to paralysis. Of the three cases with respiratory symptoms, two were aged over 60 years with pre-existing conditions such as chronic obstructive pulmonary disease in one, and diabetes mellitus and hypertension in the other. The third was an immunocompromised individual with HIV/AIDS. Regarding vaccination, two were vaccinated with two doses, one received three doses of vaccine, and the vaccination status was unclear in one.
Weekly trends in isolation bed occupancy in hospitals, oxygen/ventilator support, and COVID-19-positive cases in Maharashtra
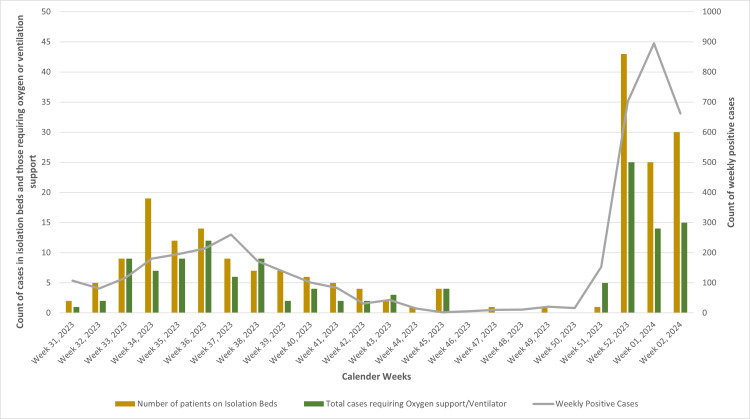
Out of the total positive cases in the state, 8.03% (338 out of 4211) required hospital admission. Among these hospitalized cases, 61.24% (207 out of 338) were managed in isolation beds without the need for oxygen support, while the remaining 38.76% (131 out of 338) needed oxygen or ventilator support. Mirroring the state's positivity rate trend, the patterns of hospital admissions and the need for oxygen support also showed fluctuations during the same timeframe (Figure 6).
Figure 6. Weekly trends in isolation bed occupancy in hospitals, oxygen/ventilator support, and COVID-19-positive cases in Maharashtra.
Image credits: Rashmita Das
Discussion
Although the BA.2.86 variant was first reported in August 2023 globally, it did not exhibit significant growth advantages or abilities to evade the humoral immune response compared to the global dominant variants such as EG.5.1 and HK.3 [12]. Over four weeks, from week 40 to week 44 of 2023, the percentage of BA.2.86 sequences slowly and steadily increased from 1.8% to 8.9% [4]. Conversely, the JN.1 variant, distinguished by a single additional mutation (L455S) in its spike protein compared to BA.2.86, swiftly ascended to global dominance. Within the same duration of four weeks, from week 44 to week 48 of 2023, its global prevalence surged from 3.3% to 27.1% [13], making up 95% of all sequences collected globally in February 2024 [14]. The quick shift from XBB sub-lineages to JN.1* mirrors the transition of Delta to the Omicron variant. In the Omicron wave, Omicron's prevalence increased from under 0.1% to 89.1%, making it the globally dominant variant within nine weeks [15,16]. The rapid emergence and dominance of the JN.1 variant in India and globally can be attributed to its enhanced transmissibility due to spike mutation L455S. In the early stages of its emergence, analysis of JN.1's effective reproductive number (Ro) using sequences from France, the United Kingdom, and Spain indicated that the JN.1 Ro exceeded that of both BA.2.86.1 and HK.3, with the latter being among the XBB lineages with the most significant growth advantage as of late November 2023. This analysis predicted JN.1's trajectory toward global dominance, a status that it had achieved in France and Spain by the close of November 2023 [9]. Presently, the JN.1* variant exhibits a relative growth advantage of 57% (confidence interval: 56-57%) [17].
The in vitro angiotensin-converting enzyme 2 (ACE2)-binding assay [9] and surface plasmon resonance analysis [12] using the RBD (monomeric) of JN.1 and BA.2.86 showed a significant reduction in the affinity of the JN.1 RBD when compared to the BA.2.86 RBD for human ACE2 receptor. Conversely, the pseudovirus assay using trimerized whole spike protein showed that the infectivity rate of JN.1 was substantially greater than that of BA.2.86 [9]. The neutralization assays demonstrated a superior ability of the JN.1 variant to evade the immune response compared to BA.2.86, with a 2.1-fold decrease in neutralization titers against antibodies from individuals reinfected with XBB after BA.5/BF.7 infection, and a 1.1-fold reduction from those with XBB breakthrough infections [9,12]. It was also shown that a small change in the L455, which is predominantly located at the epitope of RBD class 1 antibodies, enhanced JN.1's ability to evade class 1 monoclonal antibodies, making up for the fact that the parent variant, BA.2.86, was susceptible to this antibody group. However, one of the therapeutic antibodies, SA55, was shown to retain its efficacy against JN.1 [12]. As indicated by the pseudovirus assay and neutralization assays mentioned earlier, JN.1 has increased infectivity and immune evasion capabilities compared to BA.2.86.
The JN.1 variant caused mild infections in Maharashtra, mirroring the clinical outcomes observed during the previous waves in the state caused by BA.2.75, XBB, and XBB.1.16, with similar rates of hospital admissions, oxygen requirements, and mortality. In contrast to earlier waves where individuals aged 21-40 years were primarily affected, the current wave has seen a significant shift, with those aged 60 years and above being predominantly affected [18-20]. Despite a notable increase in number of positive cases, health services data showed that the overall hospitalization rate during the study period was 8.03%, and oxygen support was needed in 3.11% of positive cases, reiterating our observations that the uptick in case positivity did not significantly impact the hospitalization or oxygen demand in the state.
This evolutionary pattern of JN.1 arising from BA.2.86 is similar to the previous transition of CH.1.1 (BA.2.75.3.4.1.1.1.1) arising from BA.2.75 [12]. Albeit the addition of a new mutation in JN.1 helped it to overcome the competition faced by the parent BA.2.86 from the existing dominant XBB variants. On the other hand, mutations of CH.1.1 helped to evade the neutralizing antibodies developed in the hosts against the parent lineage, i.e., BA.2.75, which had caused widespread infection in the community. The emergence of XBB due to recombination of BJ.1 (BA.2.10.1) and BM.1.1.1 (BA.2.75.3.1.1.1) also demonstrates the innovative evolutionary strategy of SARS-CoV-2 to evade the existing neutralizing antibodies against the parent lineages. This highlights the importance of closely monitoring variants with high ACE2-binding affinity and distinct antigenic characteristics. Hence, the early detection of these mutations is crucial before they lead to evident health implications, a task that case-based surveillance might not adequately accomplish. Instead, implementing broad, population-level surveillance methods, like wastewater-based surveillance, is essential for uncovering cryptic mutations and variants. For instance, a study in Berlin, Germany, demonstrated the effectiveness of this approach by identifying the JN.1 variant in wastewater three weeks before it was detected in clinical samples, highlighting the value of population-based surveillance in early detection efforts [21]. Therefore, prompt identification of novel SARS-CoV-2 variants is vital for facilitating rapid risk evaluations, timely dissemination of information, and coordinated public health responses.
The study suffers from a few limitations. First, the analysis depends on individuals who tested for COVID-19, allowing their samples to be sequenced, and individuals who responded and consented to the study, suggesting that the reported figures might not represent the actual burden of the disease. Secondly, the study is region-specific, with data from Maharashtra. Therefore, the findings of this study might restrict its generalizability to other geographic areas and populations with different demographic characteristics. Additionally, the clinical data were obtained through telephonic conversation, potentially leading to recall bias among patients. Therefore, more comprehensive studies are needed to enhance representativeness and accuracy, including studies in diverse geographic locations and populations and using a wider range of data collection methods.
Conclusions
The JN.1* variant has emerged as the predominant SARS-CoV-2 variant in India, with clinical presentation similar to previous variants circulating in Maharashtra, India. Notably, while JN.1* has not been associated with more severe disease outcomes, its enhanced transmissibility underscores the importance of studying the ongoing viral evolution. The evolution of the virus by adding novel mutations necessitates prompt detection strategies to prevent potential surges. The pressing necessity for swift identification and the clinical features of new variants will enable public health authorities to predict and mitigate the risks effectively, ensuring preparedness against future outbreaks.
Acknowledgments
We acknowledge the Indian Biological Data Centre (IBDC), Regional Centre for Biotechnology, Department of Biotechnology, Ministry of Science and Technology, Government of India, for providing access to SARS-CoV-2 sequences deposited by INSACOG laboratories. We thank the Maharashtra State Integrated Disease Surveillance Program (IDSP) and District Health Services, Pune, for their valuable contribution to daily COVID-19 statistics for the state. We also acknowledge Poonam Pacharne, Vishal Rajput, and Riyakshi Rajkhowa from Byramjee Jeejeebhoy Government Medical College and Sassoon General Hospitals, Pune, as well as Pratiksha Tulshidas Pawar and Snehal Vasant Jagadale from Indian Council of Medical Research-National Institute of Virology, Pune for their valuable technical assistance throughout the study.
Appendices
Table 4. IDs of the sequences used in the study, downloaded from the Indian Biological Data Centre (IBDC), Regional Centre for Biotechnology, Department of Biotechnology, Ministry of Science and Technology, Government of India, with their kind permission.
| IDs of the sequences used in the study, downloaded from the Indian Biological Data Centre (IBDC), Regional Centre for Biotechnology, Department of Biotechnology, Ministry of Science and Technology, Government of India, with their kind permission |
| INCOV286977 |
| INCOV286992 |
| INCOV287059 |
| INCOV287062 |
| INCOV287064 |
| INCOV287066 |
| INCOV287067 |
| INCOV287068 |
| INCOV287069 |
| INCOV287083 |
| INCOV287086 |
| INCOV287087 |
| INCOV287106 |
| INCOV287107 |
| INCOV287108 |
| INCOV287109 |
| INCOV287110 |
| INCOV287111 |
| INCOV287112 |
| INCOV287113 |
| INCOV287117 |
| INCOV287118 |
| INCOV287119 |
| INCOV287121 |
| INCOV287123 |
| INCOV287124 |
| INCOV287125 |
| INCOV287126 |
| INCOV287127 |
| INCOV287129 |
| INCOV287137 |
| INCOV287138 |
| INCOV287140 |
| INCOV287141 |
| INCOV287146 |
| INCOV287147 |
| INCOV287167 |
| INCOV287168 |
| INCOV287169 |
| INCOV287170 |
| INCOV287171 |
| INCOV287172 |
| INCOV287173 |
| INCOV287174 |
| INCOV287175 |
| INCOV287176 |
| INCOV287177 |
| INCOV287178 |
| INCOV287179 |
| INCOV287180 |
| INCOV287181 |
| INCOV287182 |
| INCOV287183 |
| INCOV287184 |
| INCOV287186 |
| INCOV287187 |
| INCOV287188 |
| INCOV287189 |
| INCOV287190 |
| INCOV287191 |
| INCOV287192 |
| INCOV287193 |
| INCOV287194 |
| INCOV287195 |
| INCOV287196 |
| INCOV287197 |
| INCOV287198 |
| INCOV287199 |
| INCOV287200 |
| INCOV287201 |
| INCOV287202 |
| INCOV287203 |
| INCOV287204 |
| INCOV287205 |
| INCOV287206 |
| INCOV287207 |
| INCOV287208 |
| INCOV287209 |
| INCOV287210 |
| INCOV287211 |
| INCOV287212 |
| INCOV287213 |
| INCOV287214 |
| INCOV287215 |
| INCOV287216 |
| INCOV287217 |
| INCOV287218 |
| INCOV287219 |
| INCOV287220 |
| INCOV287221 |
| INCOV287222 |
| INCOV287223 |
| INCOV287224 |
| INCOV287225 |
| INCOV287226 |
| INCOV287227 |
| INCOV287228 |
| INCOV287229 |
| INCOV287230 |
| INCOV287231 |
| INCOV287232 |
| INCOV287233 |
| INCOV287234 |
| INCOV287235 |
| INCOV287236 |
| INCOV287237 |
| INCOV287238 |
| INCOV287239 |
| INCOV287240 |
| INCOV287241 |
| INCOV287242 |
| INCOV287243 |
| INCOV287244 |
| INCOV287245 |
| INCOV287246 |
| INCOV287247 |
| INCOV287248 |
| INCOV287249 |
| INCOV287250 |
| INCOV287251 |
| INCOV287252 |
| INCOV287253 |
| INCOV287257 |
| INCOV287258 |
| INCOV287265 |
| INCOV287266 |
| INCOV287267 |
| INCOV287268 |
| INCOV287269 |
| INCOV287270 |
| INCOV287271 |
| INCOV287272 |
| INCOV287273 |
| INCOV287274 |
| INCOV287275 |
| INCOV287277 |
| INCOV287278 |
| INCOV287279 |
| INCOV287280 |
| INCOV287281 |
| INCOV287282 |
| INCOV287283 |
| INCOV287284 |
| INCOV287286 |
| INCOV287287 |
| INCOV287288 |
| INCOV287289 |
| INCOV287293 |
| INCOV287294 |
| INCOV287295 |
| INCOV287296 |
| INCOV287297 |
| INCOV287298 |
| INCOV287299 |
| INCOV287300 |
| INCOV287302 |
| INCOV287308 |
| INCOV287309 |
| INCOV287310 |
| INCOV287311 |
| INCOV287312 |
| INCOV287313 |
| INCOV287372 |
| INCOV287373 |
| INCOV287374 |
| INCOV287375 |
| INCOV287376 |
| INCOV287377 |
| INCOV287378 |
| INCOV287379 |
| INCOV287380 |
| INCOV287381 |
| INCOV287382 |
| INCOV287383 |
| INCOV287384 |
| INCOV287385 |
| INCOV287386 |
| INCOV287387 |
| INCOV287388 |
| INCOV287389 |
| INCOV287390 |
| INCOV287391 |
| INCOV287392 |
| INCOV287393 |
| INCOV287394 |
| INCOV287395 |
| INCOV287396 |
| INCOV287397 |
| INCOV287398 |
| INCOV287399 |
| INCOV287400 |
| INCOV287401 |
| INCOV287402 |
| INCOV287403 |
| INCOV287404 |
| INCOV287554 |
| INCOV287555 |
| INCOV287556 |
| INCOV287558 |
| INCOV287559 |
| INCOV287560 |
| INCOV287561 |
| INCOV287562 |
| INCOV287567 |
| INCOV287568 |
| INCOV287569 |
| INCOV287570 |
| INCOV287571 |
| INCOV287572 |
| INCOV287573 |
| INCOV287574 |
| INCOV287575 |
| INCOV287576 |
| INCOV287577 |
| INCOV287578 |
| INCOV287579 |
| INCOV287580 |
| INCOV287581 |
| INCOV287585 |
| INCOV287587 |
| INCOV287588 |
| INCOV287589 |
| INCOV287590 |
| INCOV287591 |
| INCOV287592 |
| INCOV287593 |
| INCOV287893 |
| INCOV287894 |
| INCOV287895 |
| INCOV287896 |
| INCOV287897 |
| INCOV287898 |
| INCOV287899 |
| INCOV287900 |
| INCOV287901 |
| INCOV287902 |
| INCOV287903 |
| INCOV287904 |
| INCOV287905 |
| INCOV287906 |
| INCOV287907 |
| INCOV287908 |
| INCOV287909 |
| INCOV287910 |
| INCOV287950 |
| INCOV287951 |
| INCOV287952 |
| INCOV287953 |
| INCOV287954 |
| INCOV287955 |
| INCOV287956 |
| INCOV287957 |
| INCOV287958 |
| INCOV287959 |
| INCOV287961 |
| INCOV287962 |
| INCOV287963 |
| INCOV287964 |
| INCOV287965 |
| INCOV287967 |
| INCOV287968 |
| INCOV287969 |
| INCOV287970 |
| INCOV287971 |
| INCOV287972 |
| INCOV287973 |
| INCOV287974 |
| INCOV287975 |
| INCOV287976 |
| INCOV287977 |
| INCOV287978 |
| INCOV287979 |
| INCOV287980 |
| INCOV287981 |
| INCOV287982 |
| INCOV287983 |
| INCOV287984 |
| INCOV287985 |
| INCOV287986 |
| INCOV287987 |
| INCOV287988 |
| INCOV287989 |
| INCOV287990 |
| INCOV287991 |
| INCOV287992 |
| INCOV287993 |
| INCOV287994 |
| INCOV287995 |
| INCOV287996 |
| INCOV287997 |
| INCOV287998 |
| INCOV287999 |
| INCOV288000 |
| INCOV288001 |
| INCOV288002 |
| INCOV288003 |
| INCOV288004 |
| INCOV288005 |
| INCOV288006 |
| INCOV288007 |
| INCOV288008 |
| INCOV288009 |
| INCOV288012 |
| INCOV288013 |
| INCOV288015 |
| INCOV288016 |
| INCOV288017 |
| INCOV288018 |
| INCOV288019 |
| INCOV288020 |
| INCOV288021 |
| INCOV288022 |
| INCOV288023 |
| INCOV288024 |
| INCOV288025 |
| INCOV288026 |
| INCOV288029 |
| INCOV288030 |
| INCOV288031 |
| INCOV288033 |
| INCOV288034 |
| INCOV288035 |
| INCOV288036 |
| INCOV288037 |
| INCOV288038 |
| INCOV288039 |
| INCOV288040 |
| INCOV288041 |
| INCOV288042 |
| INCOV288043 |
| INCOV288045 |
| INCOV288046 |
| INCOV288047 |
| INCOV288048 |
| INCOV288049 |
| INCOV288050 |
| INCOV288051 |
| INCOV288052 |
| INCOV288053 |
| INCOV288054 |
| INCOV288055 |
| INCOV288056 |
| INCOV288057 |
| INCOV288058 |
| INCOV288059 |
| INCOV288060 |
| INCOV288061 |
| INCOV288062 |
| INCOV288063 |
| INCOV288064 |
| INCOV288065 |
| INCOV288066 |
| INCOV288067 |
| INCOV288068 |
| INCOV288069 |
| INCOV288070 |
| INCOV288071 |
| INCOV288072 |
| INCOV288073 |
| INCOV288074 |
| INCOV288075 |
| INCOV288076 |
| INCOV288077 |
| INCOV288078 |
| INCOV288079 |
| INCOV288080 |
| INCOV288081 |
| INCOV288083 |
| INCOV288084 |
| INCOV288085 |
| INCOV288087 |
| INCOV288088 |
| INCOV288089 |
| INCOV288095 |
| INCOV288096 |
| INCOV288098 |
| INCOV288099 |
| INCOV288114 |
| INCOV288116 |
| INCOV288117 |
| INCOV288120 |
| INCOV288121 |
| INCOV288122 |
| INCOV288123 |
| INCOV288124 |
| INCOV288125 |
| INCOV288126 |
| INCOV288127 |
| INCOV288128 |
| INCOV288129 |
| INCOV288130 |
| INCOV288131 |
| INCOV288132 |
| INCOV288133 |
| INCOV288134 |
| INCOV288135 |
| INCOV288136 |
| INCOV288137 |
| INCOV288138 |
| INCOV288139 |
| INCOV288140 |
| INCOV288141 |
| INCOV288142 |
| INCOV288143 |
| INCOV288144 |
| INCOV288145 |
| INCOV288146 |
| INCOV288147 |
| INCOV288148 |
| INCOV288149 |
| INCOV288150 |
| INCOV288151 |
| INCOV288152 |
| INCOV288153 |
| INCOV288154 |
| INCOV288155 |
| INCOV288156 |
| INCOV288170 |
| INCOV288171 |
| INCOV288172 |
| INCOV288173 |
| INCOV288174 |
| INCOV288175 |
| INCOV288176 |
| INCOV288177 |
| INCOV288178 |
| INCOV288179 |
| INCOV288180 |
| INCOV288181 |
| INCOV288182 |
| INCOV288183 |
| INCOV288184 |
| INCOV288185 |
| INCOV288186 |
| INCOV288207 |
| INCOV288208 |
| INCOV288209 |
| INCOV288210 |
| INCOV288211 |
| INCOV288212 |
| INCOV288213 |
| INCOV288214 |
| INCOV288215 |
| INCOV288216 |
| INCOV288217 |
| INCOV288218 |
| INCOV288219 |
| INCOV288220 |
| INCOV288221 |
| INCOV288222 |
| INCOV288223 |
| INCOV288224 |
| INCOV288225 |
| INCOV288226 |
| INCOV288227 |
| INCOV288228 |
| INCOV288229 |
| INCOV288230 |
| INCOV288231 |
| INCOV288232 |
| INCOV288233 |
| INCOV288234 |
| INCOV288235 |
| INCOV288236 |
| INCOV288237 |
| INCOV288238 |
| INCOV288239 |
| INCOV288240 |
| INCOV288241 |
| INCOV288242 |
| INCOV288243 |
| INCOV288244 |
| INCOV288246 |
| INCOV288247 |
| INCOV288248 |
| INCOV288249 |
| INCOV288250 |
| INCOV288251 |
| INCOV288252 |
| INCOV288253 |
| INCOV288254 |
| INCOV288255 |
| INCOV288256 |
| INCOV288257 |
| INCOV288258 |
| INCOV288259 |
| INCOV288260 |
| INCOV288261 |
| INCOV288262 |
| INCOV288263 |
| INCOV288264 |
| INCOV288265 |
| INCOV288266 |
| INCOV288267 |
| INCOV288268 |
| INCOV288269 |
| INCOV288270 |
| INCOV288271 |
| INCOV288272 |
| INCOV288273 |
| INCOV288274 |
| INCOV288275 |
| INCOV288276 |
| INCOV288277 |
| INCOV288278 |
| INCOV288279 |
| INCOV288280 |
| INCOV288281 |
| INCOV288282 |
| INCOV288283 |
| INCOV288284 |
| INCOV288285 |
| INCOV288286 |
| INCOV288287 |
| INCOV288288 |
| INCOV288289 |
| INCOV288290 |
| INCOV288291 |
| INCOV288292 |
| INCOV288293 |
| INCOV288294 |
| INCOV288295 |
| INCOV288296 |
| INCOV288297 |
| INCOV288298 |
| INCOV288299 |
| INCOV288300 |
| INCOV288301 |
| INCOV288302 |
| INCOV288303 |
| INCOV288304 |
| INCOV288305 |
| INCOV288306 |
| INCOV288307 |
| INCOV288308 |
| INCOV288309 |
| INCOV288310 |
| INCOV288311 |
| INCOV288312 |
| INCOV288313 |
| INCOV288314 |
| INCOV288315 |
| INCOV288316 |
| INCOV288317 |
| INCOV288318 |
| INCOV288319 |
| INCOV288320 |
| INCOV288321 |
| INCOV288322 |
| INCOV288323 |
| INCOV288324 |
| INCOV288325 |
| INCOV288326 |
| INCOV288327 |
| INCOV288328 |
| INCOV288329 |
| INCOV288330 |
| INCOV288331 |
| INCOV288332 |
| INCOV288333 |
| INCOV288334 |
| INCOV288335 |
| INCOV288336 |
| INCOV288337 |
| INCOV288338 |
| INCOV288339 |
| INCOV288340 |
| INCOV288341 |
| INCOV288342 |
| INCOV288343 |
| INCOV288344 |
| INCOV288345 |
| INCOV288346 |
| INCOV288347 |
| INCOV288348 |
| INCOV288349 |
| INCOV288350 |
| INCOV288351 |
| INCOV288352 |
| INCOV288353 |
| INCOV288354 |
| INCOV288355 |
| INCOV288356 |
| INCOV288357 |
| INCOV288358 |
| INCOV288359 |
| INCOV288360 |
| INCOV288361 |
| INCOV288362 |
| INCOV288363 |
| INCOV288364 |
| INCOV288365 |
| INCOV288366 |
| INCOV288367 |
| INCOV288368 |
| INCOV288369 |
| INCOV288370 |
| INCOV288371 |
| INCOV288372 |
| INCOV288373 |
| INCOV288374 |
| INCOV288375 |
| INCOV288376 |
| INCOV288377 |
| INCOV288378 |
| INCOV288379 |
| INCOV288380 |
| INCOV288381 |
| INCOV288382 |
| INCOV288383 |
| INCOV288384 |
| INCOV288385 |
| INCOV288386 |
| INCOV288387 |
| INCOV288388 |
| INCOV288389 |
| INCOV288390 |
| INCOV288391 |
| INCOV288392 |
| INCOV288393 |
| INCOV288394 |
| INCOV288395 |
| INCOV288396 |
| INCOV288397 |
| INCOV288398 |
| INCOV288399 |
| INCOV288400 |
| INCOV288401 |
| INCOV288402 |
| INCOV288403 |
| INCOV288404 |
| INCOV288405 |
| INCOV288406 |
| INCOV288407 |
| INCOV288408 |
| INCOV288409 |
| INCOV288410 |
| INCOV288411 |
| INCOV288412 |
| INCOV288413 |
| INCOV288414 |
| INCOV288415 |
| INCOV288416 |
| INCOV288417 |
| INCOV288418 |
| INCOV288419 |
| INCOV288420 |
| INCOV288421 |
| INCOV288422 |
| INCOV288423 |
| INCOV288424 |
| INCOV288425 |
| INCOV288426 |
| INCOV288427 |
| INCOV288428 |
| INCOV288429 |
| INCOV288430 |
| INCOV288431 |
| INCOV288432 |
| INCOV288433 |
| INCOV288434 |
| INCOV288435 |
| INCOV288436 |
| INCOV288437 |
| INCOV288438 |
| INCOV288439 |
| INCOV288440 |
| INCOV288441 |
| INCOV288442 |
| INCOV288443 |
| INCOV288444 |
| INCOV288445 |
| INCOV288446 |
| INCOV288447 |
| INCOV288448 |
| INCOV288449 |
| INCOV288450 |
| INCOV288451 |
| INCOV288452 |
| INCOV288453 |
| INCOV288454 |
| INCOV288455 |
| INCOV288456 |
| INCOV288457 |
| INCOV288458 |
| INCOV288459 |
| INCOV288460 |
| INCOV288461 |
| INCOV288462 |
| INCOV288463 |
| INCOV288464 |
| INCOV288465 |
| INCOV288466 |
| INCOV288467 |
| INCOV288468 |
| INCOV288469 |
| INCOV288470 |
| INCOV288471 |
| INCOV288472 |
| INCOV288473 |
| INCOV288474 |
| INCOV288475 |
| INCOV288476 |
| INCOV288477 |
| INCOV288478 |
| INCOV288479 |
| INCOV288480 |
| INCOV288481 |
| INCOV288482 |
| INCOV288483 |
| INCOV288484 |
| INCOV288485 |
| INCOV288486 |
| INCOV288487 |
| INCOV288488 |
| INCOV288489 |
| INCOV288490 |
| INCOV288491 |
| INCOV288492 |
| INCOV288493 |
| INCOV288494 |
| INCOV288495 |
| INCOV288496 |
| INCOV288497 |
| INCOV288498 |
| INCOV288499 |
| INCOV288500 |
| INCOV288501 |
| INCOV288502 |
| INCOV288503 |
| INCOV288504 |
| INCOV288505 |
| INCOV288506 |
| INCOV288507 |
| INCOV288508 |
| INCOV288509 |
| INCOV288510 |
| INCOV288511 |
| INCOV288512 |
| INCOV288513 |
| INCOV288514 |
| INCOV288515 |
| INCOV288516 |
| INCOV288517 |
| INCOV288518 |
| INCOV288519 |
| INCOV288520 |
| INCOV288521 |
| INCOV288522 |
| INCOV288523 |
| INCOV288524 |
| INCOV288525 |
| INCOV288526 |
| INCOV288527 |
| INCOV288528 |
| INCOV288529 |
| INCOV288530 |
| INCOV288531 |
| INCOV288532 |
| INCOV288533 |
| INCOV288534 |
| INCOV288535 |
| INCOV288536 |
| INCOV288537 |
| INCOV288538 |
| INCOV288539 |
| INCOV288540 |
| INCOV288541 |
| INCOV288542 |
| INCOV288543 |
| INCOV288544 |
| INCOV288545 |
| INCOV288546 |
| INCOV288547 |
| INCOV288548 |
| INCOV288549 |
| INCOV288550 |
| INCOV288551 |
| INCOV288552 |
| INCOV288553 |
| INCOV288554 |
| INCOV288555 |
| INCOV288556 |
| INCOV288557 |
| INCOV288558 |
| INCOV288559 |
| INCOV288560 |
| INCOV288561 |
| INCOV288562 |
| INCOV288563 |
| INCOV288564 |
| INCOV288567 |
| INCOV288568 |
| INCOV288569 |
| INCOV288570 |
| INCOV288571 |
| INCOV288572 |
| INCOV288573 |
| INCOV288574 |
| INCOV288575 |
| INCOV288576 |
| INCOV288577 |
| INCOV288578 |
| INCOV288579 |
| INCOV288580 |
| INCOV288581 |
| INCOV288582 |
| INCOV288583 |
| INCOV288584 |
| INCOV288585 |
| INCOV288586 |
| INCOV288587 |
| INCOV288588 |
| INCOV288589 |
| INCOV288590 |
| INCOV288591 |
| INCOV288592 |
| INCOV288593 |
| INCOV288594 |
| INCOV288595 |
| INCOV288596 |
| INCOV288597 |
| INCOV288598 |
| INCOV288599 |
| INCOV288600 |
| INCOV288601 |
| INCOV288602 |
| INCOV288603 |
| INCOV288604 |
| INCOV288605 |
| INCOV288606 |
| INCOV288607 |
| INCOV288608 |
| INCOV288609 |
| INCOV288610 |
| INCOV288611 |
| INCOV288612 |
| INCOV288613 |
| INCOV288614 |
| INCOV288615 |
| INCOV288616 |
| INCOV288617 |
| INCOV288618 |
| INCOV288619 |
| INCOV288620 |
| INCOV288621 |
| INCOV288622 |
| INCOV288623 |
| INCOV288624 |
| INCOV288625 |
| INCOV288626 |
| INCOV288627 |
| INCOV288628 |
| INCOV288629 |
| INCOV288630 |
| INCOV288631 |
| INCOV288632 |
| INCOV288633 |
| INCOV288634 |
| INCOV288635 |
| INCOV288636 |
| INCOV288637 |
| INCOV288638 |
| INCOV288639 |
| INCOV288640 |
| INCOV288641 |
| INCOV288642 |
| INCOV288643 |
| INCOV288644 |
| INCOV288645 |
| INCOV288646 |
| INCOV288647 |
| INCOV288648 |
| INCOV288649 |
| INCOV288650 |
| INCOV288651 |
| INCOV288652 |
| INCOV288653 |
| INCOV288654 |
| INCOV288655 |
| INCOV288656 |
| INCOV288657 |
| INCOV288658 |
| INCOV288659 |
| INCOV288660 |
| INCOV288661 |
| INCOV288662 |
| INCOV288663 |
| INCOV288664 |
| INCOV288665 |
| INCOV288666 |
| INCOV288667 |
| INCOV288668 |
| INCOV288669 |
| INCOV288670 |
| INCOV288671 |
| INCOV288672 |
| INCOV288673 |
| INCOV288674 |
| INCOV288675 |
| INCOV288676 |
| INCOV288677 |
| INCOV288678 |
| INCOV288679 |
| INCOV288680 |
| INCOV288681 |
| INCOV288682 |
| INCOV288683 |
| INCOV288684 |
| INCOV288685 |
| INCOV288686 |
| INCOV288687 |
| INCOV288688 |
| INCOV288689 |
| INCOV288690 |
| INCOV288691 |
| INCOV288692 |
| INCOV288693 |
| INCOV288694 |
| INCOV288695 |
| INCOV288696 |
| INCOV288697 |
| INCOV288698 |
| INCOV288699 |
| INCOV288700 |
| INCOV288701 |
| INCOV288702 |
| INCOV288703 |
| INCOV288704 |
| INCOV288705 |
| INCOV288706 |
| INCOV288707 |
| INCOV288708 |
| INCOV288709 |
| INCOV288710 |
| INCOV288711 |
| INCOV288712 |
| INCOV288713 |
| INCOV288714 |
| INCOV288715 |
| INCOV288716 |
| INCOV288717 |
| INCOV288718 |
| INCOV288719 |
| INCOV288720 |
| INCOV288721 |
| INCOV288722 |
| INCOV288723 |
| INCOV288724 |
| INCOV288725 |
| INCOV288726 |
| INCOV288727 |
| INCOV288728 |
| INCOV288729 |
| INCOV288730 |
| INCOV288731 |
| INCOV288732 |
| INCOV288733 |
| INCOV288734 |
| INCOV288735 |
| INCOV288736 |
| INCOV288737 |
| INCOV288738 |
| INCOV288739 |
| INCOV288740 |
| INCOV288741 |
| INCOV288742 |
| INCOV288743 |
| INCOV288744 |
| INCOV288745 |
| INCOV288746 |
| INCOV288747 |
| INCOV288748 |
| INCOV288749 |
| INCOV288750 |
| INCOV288751 |
| INCOV288752 |
| INCOV288753 |
| INCOV288754 |
| INCOV288755 |
| INCOV288756 |
| INCOV288757 |
| INCOV288758 |
| INCOV288759 |
| INCOV288760 |
| INCOV288761 |
| INCOV288762 |
| INCOV288763 |
| INCOV288764 |
| INCOV288765 |
| INCOV288766 |
| INCOV288767 |
| INCOV288768 |
| INCOV288769 |
| INCOV288770 |
| INCOV288771 |
| INCOV288773 |
| INCOV288774 |
| INCOV288775 |
| INCOV288776 |
| INCOV288777 |
| INCOV288778 |
| INCOV288779 |
| INCOV288780 |
| INCOV288781 |
| INCOV288782 |
| INCOV288783 |
| INCOV288784 |
| INCOV288785 |
| INCOV288786 |
| INCOV288787 |
| INCOV288788 |
| INCOV288789 |
| INCOV288790 |
| INCOV288791 |
| INCOV288792 |
| INCOV288793 |
| INCOV288794 |
| INCOV288795 |
| INCOV288796 |
| INCOV288797 |
| INCOV288798 |
| INCOV288799 |
| INCOV288800 |
| INCOV288801 |
| INCOV288802 |
| INCOV288803 |
| INCOV288804 |
| INCOV288805 |
| INCOV288806 |
| INCOV288807 |
| INCOV288808 |
| INCOV288809 |
| INCOV288810 |
| INCOV288811 |
| INCOV288812 |
| INCOV288813 |
| INCOV288814 |
| INCOV288815 |
| INCOV288816 |
| INCOV288817 |
| INCOV288818 |
| INCOV288819 |
| INCOV288820 |
| INCOV288821 |
| INCOV288822 |
| INCOV288823 |
| INCOV288824 |
| INCOV288825 |
| INCOV288826 |
| INCOV288827 |
| INCOV288828 |
| INCOV288829 |
| INCOV288830 |
| INCOV288831 |
| INCOV288832 |
| INCOV288833 |
| INCOV288834 |
| INCOV288835 |
| INCOV288836 |
| INCOV288837 |
| INCOV288838 |
| INCOV288839 |
| INCOV288840 |
| INCOV288841 |
| INCOV288842 |
| INCOV288843 |
| INCOV288844 |
| INCOV288845 |
| INCOV288846 |
| INCOV288847 |
| INCOV288848 |
| INCOV288849 |
| INCOV288850 |
| INCOV288851 |
| INCOV288852 |
| INCOV288853 |
| INCOV288854 |
| INCOV288855 |
| INCOV288856 |
| INCOV288857 |
| INCOV288860 |
| INCOV288861 |
| INCOV288862 |
| INCOV288863 |
| INCOV288864 |
| INCOV288865 |
| INCOV288866 |
| INCOV288867 |
| INCOV288868 |
| INCOV288869 |
| INCOV288870 |
| INCOV288871 |
| INCOV288872 |
| INCOV288873 |
| INCOV288874 |
| INCOV288875 |
| INCOV288876 |
| INCOV288877 |
| INCOV288878 |
| INCOV288879 |
| INCOV288880 |
| INCOV288881 |
| INCOV288882 |
| INCOV288883 |
| INCOV288884 |
| INCOV288885 |
| INCOV288886 |
| INCOV288887 |
| INCOV288888 |
| INCOV288889 |
| INCOV288890 |
| INCOV288891 |
| INCOV288892 |
| INCOV288893 |
| INCOV288894 |
| INCOV288895 |
| INCOV288896 |
| INCOV288897 |
| INCOV288898 |
| INCOV288899 |
| INCOV288900 |
| INCOV288901 |
| INCOV288902 |
| INCOV288903 |
| INCOV289049 |
| INCOV289050 |
| INCOV289051 |
| INCOV289053 |
| INCOV289054 |
| INCOV289055 |
| INCOV289056 |
| INCOV289057 |
| INCOV289058 |
| INCOV289059 |
| INCOV289060 |
| INCOV289061 |
| INCOV289062 |
| INCOV289063 |
| INCOV289064 |
| INCOV289065 |
| INCOV289066 |
| INCOV289067 |
| INCOV289068 |
| INCOV289069 |
| INCOV289070 |
| INCOV289071 |
| INCOV289072 |
| INCOV289073 |
| INCOV289074 |
| INCOV289075 |
| INCOV289076 |
| INCOV289077 |
| INCOV289078 |
| INCOV289079 |
| INCOV289080 |
| INCOV289081 |
| INCOV289082 |
| INCOV289083 |
| INCOV289084 |
| INCOV289085 |
| INCOV289086 |
| INCOV289087 |
| INCOV289088 |
| INCOV289089 |
| INCOV289090 |
| INCOV289091 |
| INCOV289092 |
| INCOV289093 |
| INCOV289094 |
| INCOV289095 |
| INCOV289096 |
| INCOV289097 |
| INCOV289098 |
| INCOV289099 |
| INCOV289100 |
| INCOV289101 |
| INCOV289102 |
| INCOV289103 |
| INCOV289104 |
| INCOV289105 |
| INCOV289106 |
| INCOV289107 |
| INCOV289108 |
| INCOV289109 |
| INCOV289110 |
| INCOV289111 |
| INCOV289112 |
| INCOV289113 |
| INCOV289114 |
| INCOV289115 |
| INCOV289116 |
| INCOV289117 |
| INCOV289118 |
| INCOV289119 |
| INCOV289120 |
| INCOV289121 |
| INCOV289122 |
| INCOV289123 |
| INCOV289124 |
| INCOV289125 |
| INCOV289126 |
| INCOV289127 |
| INCOV289128 |
| INCOV289129 |
| INCOV289130 |
| INCOV289131 |
| INCOV289132 |
| INCOV289133 |
| INCOV289134 |
| INCOV289135 |
| INCOV289136 |
| INCOV289137 |
| INCOV289138 |
| INCOV289139 |
| INCOV289140 |
| INCOV289141 |
| INCOV289142 |
| INCOV289143 |
| INCOV289144 |
| INCOV289145 |
| INCOV289146 |
| INCOV289147 |
| INCOV289148 |
| INCOV289149 |
| INCOV289150 |
| INCOV289151 |
| INCOV289152 |
| INCOV289153 |
| INCOV289154 |
| INCOV289155 |
| INCOV289156 |
| INCOV289157 |
| INCOV289158 |
| INCOV289159 |
| INCOV289160 |
| INCOV289161 |
| INCOV289162 |
| INCOV289163 |
| INCOV289164 |
| INCOV289165 |
| INCOV289166 |
| INCOV289167 |
| INCOV289168 |
| INCOV289169 |
| INCOV289170 |
| INCOV289171 |
| INCOV289172 |
| INCOV289173 |
| INCOV289174 |
| INCOV289175 |
| INCOV289176 |
| INCOV289177 |
| INCOV289178 |
| INCOV289179 |
| INCOV289180 |
| INCOV289181 |
| INCOV289182 |
| INCOV289183 |
| INCOV289184 |
| INCOV289185 |
| INCOV289186 |
| INCOV289187 |
| INCOV289188 |
| INCOV289189 |
| INCOV289190 |
| INCOV289191 |
| INCOV289192 |
| INCOV289193 |
| INCOV289194 |
| INCOV289195 |
| INCOV289196 |
| INCOV289197 |
| INCOV289198 |
| INCOV289199 |
| INCOV289200 |
| INCOV289201 |
| INCOV289202 |
| INCOV289203 |
| INCOV289204 |
| INCOV289205 |
| INCOV289206 |
| INCOV289207 |
| INCOV289208 |
| INCOV289209 |
| INCOV289210 |
| INCOV289211 |
| INCOV289212 |
| INCOV289213 |
| INCOV289214 |
| INCOV289215 |
| INCOV289216 |
| INCOV289217 |
| INCOV289218 |
| INCOV289219 |
| INCOV289220 |
| INCOV289221 |
| INCOV289222 |
| INCOV289223 |
| INCOV289224 |
| INCOV289225 |
| INCOV289226 |
| INCOV289227 |
| INCOV289228 |
| INCOV289229 |
| INCOV289230 |
| INCOV289231 |
| INCOV289232 |
| INCOV289233 |
| INCOV289234 |
| INCOV289235 |
| INCOV289236 |
| INCOV289237 |
| INCOV289238 |
| INCOV289239 |
| INCOV289240 |
| INCOV289241 |
| INCOV289242 |
| INCOV289243 |
| INCOV289244 |
| INCOV289245 |
| INCOV289246 |
| INCOV289247 |
| INCOV289248 |
| INCOV289249 |
| INCOV289250 |
| INCOV289251 |
| INCOV289252 |
| INCOV289253 |
| INCOV289254 |
| INCOV289255 |
| INCOV289256 |
| INCOV289257 |
| INCOV289258 |
| INCOV289259 |
| INCOV289263 |
| INCOV289264 |
| INCOV289265 |
| INCOV289266 |
| INCOV289286 |
| INCOV289287 |
| INCOV289288 |
| INCOV289289 |
| INCOV289291 |
| INCOV289292 |
| INCOV289293 |
| INCOV289294 |
| INCOV289295 |
| INCOV289296 |
| INCOV289297 |
| INCOV289298 |
| INCOV289299 |
| INCOV289300 |
| INCOV289301 |
| INCOV289302 |
| INCOV289303 |
| INCOV289304 |
| INCOV289305 |
| INCOV289306 |
| INCOV289307 |
| INCOV289308 |
| INCOV289309 |
| INCOV289310 |
| INCOV289394 |
| INCOV289395 |
| INCOV289396 |
| INCOV289397 |
| INCOV289398 |
| INCOV289399 |
| INCOV289400 |
| INCOV289401 |
| INCOV289402 |
| INCOV289403 |
| INCOV289404 |
| INCOV289405 |
| INCOV289406 |
| INCOV289407 |
| INCOV289408 |
| INCOV289409 |
| INCOV289410 |
| INCOV289411 |
| INCOV289412 |
| INCOV289413 |
| INCOV289414 |
| INCOV289415 |
| INCOV289416 |
| INCOV289417 |
| INCOV289418 |
| INCOV289419 |
| INCOV289420 |
| INCOV289421 |
| INCOV289422 |
| INCOV289423 |
| INCOV289424 |
| INCOV289425 |
| INCOV289426 |
| INCOV289427 |
| INCOV289428 |
| INCOV289429 |
| INCOV289430 |
| INCOV289431 |
| INCOV289432 |
| INCOV289433 |
| INCOV289434 |
| INCOV289435 |
| INCOV289436 |
| INCOV289437 |
| INCOV289438 |
| INCOV289439 |
| INCOV289440 |
| INCOV289441 |
| INCOV289442 |
| INCOV289443 |
| INCOV289444 |
| INCOV289445 |
| INCOV289446 |
| INCOV289447 |
| INCOV289448 |
| INCOV289449 |
| INCOV289450 |
| INCOV289451 |
| INCOV289452 |
| INCOV289453 |
| INCOV289454 |
| INCOV289455 |
| INCOV289456 |
| INCOV289457 |
| INCOV289458 |
| INCOV289459 |
| INCOV289460 |
| INCOV289461 |
| INCOV289462 |
| INCOV289463 |
| INCOV289464 |
| INCOV289465 |
| INCOV289466 |
| INCOV289467 |
| INCOV289468 |
| INCOV289469 |
| INCOV289470 |
| INCOV289471 |
| INCOV289472 |
| INCOV289473 |
| INCOV289474 |
| INCOV289475 |
| INCOV289476 |
| INCOV289499 |
| INCOV289500 |
| INCOV289501 |
| INCOV289502 |
| INCOV289503 |
| INCOV289504 |
| INCOV289505 |
| INCOV289506 |
| INCOV289507 |
| INCOV289508 |
| INCOV289509 |
| INCOV289510 |
| INCOV289511 |
| INCOV289512 |
| INCOV289513 |
| INCOV289514 |
| INCOV289515 |
| INCOV289516 |
| INCOV289517 |
| INCOV289518 |
| INCOV289519 |
| INCOV289520 |
| INCOV289542 |
| INCOV289543 |
| INCOV289554 |
| INCOV289555 |
| INCOV289556 |
| INCOV289558 |
| INCOV289564 |
| INCOV289565 |
| INCOV289566 |
| INCOV289567 |
| INCOV289568 |
| INCOV289569 |
| INCOV289570 |
| INCOV289571 |
| INCOV289572 |
| INCOV289573 |
| INCOV289574 |
| INCOV289575 |
| INCOV289576 |
| INCOV289577 |
| INCOV289578 |
| INCOV289579 |
| INCOV289580 |
| INCOV289581 |
| INCOV289582 |
| INCOV289583 |
| INCOV289584 |
| INCOV289585 |
| INCOV289586 |
| INCOV289587 |
| INCOV289588 |
| INCOV289589 |
| INCOV289590 |
| INCOV289591 |
| INCOV289592 |
| INCOV289593 |
| INCOV289594 |
| INCOV289595 |
| INCOV289596 |
| INCOV289597 |
| INCOV289598 |
| INCOV289599 |
| INCOV289600 |
| INCOV289601 |
| INCOV289602 |
| INCOV289603 |
| INCOV289604 |
| INCOV289605 |
| INCOV289606 |
| INCOV289607 |
| INCOV289608 |
| INCOV289609 |
| INCOV289610 |
| INCOV289611 |
| INCOV289612 |
| INCOV289613 |
| INCOV289614 |
| INCOV289615 |
| INCOV289616 |
| INCOV289617 |
| INCOV289618 |
| INCOV289619 |
| INCOV289620 |
| INCOV289621 |
| INCOV289622 |
| INCOV289623 |
| INCOV289624 |
| INCOV289625 |
| INCOV289626 |
| INCOV289627 |
| INCOV289628 |
| INCOV289629 |
| INCOV289630 |
| INCOV289631 |
| INCOV289632 |
| INCOV289633 |
| INCOV289634 |
| INCOV289635 |
| INCOV289636 |
| INCOV289637 |
| INCOV289638 |
| INCOV289639 |
| INCOV289640 |
| INCOV289641 |
| INCOV289642 |
| INCOV289643 |
| INCOV289644 |
| INCOV289645 |
| INCOV289646 |
| INCOV289647 |
| INCOV289648 |
| INCOV289649 |
| INCOV289650 |
| INCOV289651 |
| INCOV289652 |
| INCOV289653 |
| INCOV289654 |
| INCOV289655 |
| INCOV289656 |
| INCOV289657 |
| INCOV289658 |
| INCOV289659 |
| INCOV289660 |
| INCOV289661 |
| INCOV289662 |
| INCOV289663 |
| INCOV289664 |
| INCOV289665 |
| INCOV289666 |
| INCOV289667 |
| INCOV289668 |
| INCOV289669 |
| INCOV289670 |
| INCOV289671 |
| INCOV289672 |
| INCOV289673 |
| INCOV289674 |
| INCOV289675 |
| INCOV289676 |
| INCOV289677 |
| INCOV289678 |
| INCOV289679 |
| INCOV289680 |
| INCOV289681 |
| INCOV289683 |
| INCOV289684 |
| INCOV289685 |
| INCOV289686 |
| INCOV289687 |
| INCOV289688 |
| INCOV289689 |
| INCOV289690 |
| INCOV289691 |
| INCOV289692 |
| INCOV289693 |
| INCOV289694 |
| INCOV289695 |
| INCOV289696 |
| INCOV289697 |
| INCOV289698 |
| INCOV289699 |
| INCOV289700 |
| INCOV289701 |
| INCOV289702 |
| INCOV289703 |
| INCOV289704 |
| INCOV289705 |
| INCOV289706 |
| INCOV289707 |
| INCOV289708 |
| INCOV289709 |
| INCOV289710 |
| INCOV289711 |
| INCOV289712 |
| INCOV289713 |
| INCOV289714 |
| INCOV289715 |
| INCOV289716 |
| INCOV289717 |
| INCOV289718 |
| INCOV289719 |
| INCOV289720 |
| INCOV289721 |
| INCOV289722 |
| INCOV289723 |
| INCOV289724 |
| INCOV289725 |
| INCOV289726 |
| INCOV289727 |
| INCOV289728 |
| INCOV289729 |
| INCOV289730 |
| INCOV289731 |
| INCOV289732 |
| INCOV289733 |
| INCOV289734 |
| INCOV289735 |
| INCOV289736 |
| INCOV289737 |
| INCOV289738 |
| INCOV289739 |
| INCOV289740 |
| INCOV289741 |
| INCOV289742 |
| INCOV289743 |
| INCOV289744 |
| INCOV289745 |
| INCOV289746 |
| INCOV289747 |
| INCOV289748 |
| INCOV289749 |
| INCOV289750 |
| INCOV289751 |
| INCOV289752 |
| INCOV289753 |
| INCOV289754 |
| INCOV289755 |
| INCOV289756 |
| INCOV289757 |
| INCOV289758 |
| INCOV289759 |
| INCOV289760 |
| INCOV289761 |
| INCOV289762 |
| INCOV289763 |
| INCOV289764 |
| INCOV289765 |
| INCOV289766 |
| INCOV289767 |
| INCOV289768 |
| INCOV289769 |
| INCOV289770 |
| INCOV289771 |
| INCOV289772 |
| INCOV289773 |
| INCOV289774 |
| INCOV289775 |
| INCOV289776 |
| INCOV289777 |
| INCOV289778 |
| INCOV289779 |
| INCOV289780 |
| INCOV289781 |
| INCOV289782 |
| INCOV289783 |
| INCOV289784 |
| INCOV289785 |
| INCOV289788 |
| INCOV289791 |
| INCOV289793 |
| INCOV289794 |
| INCOV289795 |
| INCOV289796 |
| INCOV289797 |
| INCOV289798 |
| INCOV289799 |
| INCOV289800 |
| INCOV289801 |
| INCOV289802 |
| INCOV289803 |
| INCOV289804 |
| INCOV289805 |
| INCOV289806 |
| INCOV289807 |
| INCOV289808 |
| INCOV289809 |
| INCOV289810 |
| INCOV289811 |
| INCOV289822 |
| INCOV289823 |
| INCOV289824 |
| INCOV289825 |
| INCOV289826 |
| INCOV289827 |
| INCOV289828 |
| INCOV289829 |
| INCOV289830 |
| INCOV289846 |
| INCOV289847 |
| INCOV289848 |
| INCOV289849 |
| INCOV289850 |
| INCOV289851 |
| INCOV289852 |
| INCOV289853 |
| INCOV289897 |
| INCOV289898 |
| INCOV289899 |
| INCOV289900 |
| INCOV289901 |
| INCOV289902 |
| INCOV289904 |
| INCOV289905 |
| INCOV289978 |
| INCOV289983 |
| INCOV289984 |
| INCOV289985 |
| INCOV289986 |
| INCOV289987 |
| INCOV289988 |
| INCOV289989 |
| INCOV289990 |
| INCOV289991 |
| INCOV289993 |
| INCOV289994 |
| INCOV289995 |
| INCOV289996 |
| INCOV289997 |
| INCOV289998 |
| INCOV289999 |
| INCOV290000 |
| INCOV290002 |
| INCOV290004 |
| INCOV290005 |
| INCOV290006 |
| INCOV290007 |
| INCOV290008 |
| INCOV290014 |
| INCOV290015 |
| INCOV290016 |
| INCOV290017 |
| INCOV290018 |
| INCOV290019 |
| INCOV290020 |
| INCOV290021 |
| INCOV290022 |
| INCOV290023 |
| INCOV290024 |
| INCOV290025 |
| INCOV290026 |
| INCOV290027 |
| INCOV290028 |
| INCOV290029 |
| INCOV290030 |
| INCOV290031 |
| INCOV290032 |
| INCOV290033 |
| INCOV290034 |
| INCOV290035 |
| INCOV290036 |
| INCOV290037 |
| INCOV290038 |
| INCOV290039 |
| INCOV290040 |
| INCOV290041 |
| INCOV290042 |
| INCOV290043 |
| INCOV290044 |
| INCOV290045 |
| INCOV290046 |
| INCOV290047 |
| INCOV290048 |
| INCOV290049 |
| INCOV290050 |
| INCOV290051 |
| INCOV290052 |
| INCOV290053 |
| INCOV290054 |
| INCOV290055 |
| INCOV290056 |
| INCOV290057 |
| INCOV290058 |
| INCOV290059 |
| INCOV290060 |
| INCOV290061 |
| INCOV290062 |
| INCOV290063 |
| INCOV290064 |
| INCOV290065 |
| INCOV290066 |
| INCOV290067 |
| INCOV290068 |
| INCOV290069 |
| INCOV290070 |
| INCOV290071 |
| INCOV290072 |
| INCOV290073 |
| INCOV290074 |
| INCOV290075 |
| INCOV290076 |
| INCOV290077 |
| INCOV290078 |
| INCOV290079 |
| INCOV290080 |
| INCOV290081 |
| INCOV290082 |
| INCOV290084 |
| INCOV290086 |
| INCOV290087 |
| INCOV290088 |
| INCOV290089 |
| INCOV290090 |
| INCOV290091 |
| INCOV290092 |
| INCOV290093 |
| INCOV290094 |
| INCOV290095 |
| INCOV290096 |
| INCOV290097 |
| INCOV290098 |
| INCOV290099 |
| INCOV290100 |
| INCOV290101 |
| INCOV290102 |
| INCOV290103 |
| INCOV290104 |
| INCOV290105 |
| INCOV290106 |
| INCOV290107 |
| INCOV290108 |
| INCOV290109 |
| INCOV290110 |
| INCOV290111 |
| INCOV290113 |
| INCOV290114 |
| INCOV290115 |
| INCOV290116 |
| INCOV290117 |
| INCOV290118 |
| INCOV290119 |
| INCOV290120 |
| INCOV290121 |
| INCOV290122 |
| INCOV290123 |
| INCOV290124 |
| INCOV290125 |
| INCOV290126 |
| INCOV290127 |
| INCOV290128 |
| INCOV290129 |
| INCOV290130 |
| INCOV290131 |
| INCOV290132 |
| INCOV290133 |
| INCOV290134 |
| INCOV290135 |
| INCOV290136 |
| INCOV290137 |
| INCOV290138 |
| INCOV290139 |
| INCOV290140 |
| INCOV290141 |
| INCOV290142 |
| INCOV290143 |
| INCOV290144 |
| INCOV290146 |
| INCOV290147 |
| INCOV290148 |
| INCOV290149 |
| INCOV290150 |
| INCOV290151 |
| INCOV290153 |
| INCOV290154 |
| INCOV290155 |
| INCOV290156 |
| INCOV290158 |
| INCOV290159 |
| INCOV290160 |
| INCOV290161 |
| INCOV290162 |
| INCOV290163 |
| INCOV290164 |
| INCOV290165 |
| INCOV290166 |
| INCOV290167 |
| INCOV290168 |
| INCOV290169 |
| INCOV290170 |
| INCOV290171 |
| INCOV290172 |
| INCOV290173 |
| INCOV290174 |
| INCOV290175 |
| INCOV290176 |
| INCOV290177 |
| INCOV290178 |
| INCOV290179 |
| INCOV290180 |
| INCOV290181 |
| INCOV290182 |
| INCOV290183 |
| INCOV290184 |
| INCOV290185 |
| INCOV290186 |
| INCOV290187 |
| INCOV290188 |
| INCOV290189 |
| INCOV290190 |
| INCOV290194 |
| INCOV290195 |
| INCOV290196 |
| INCOV290197 |
| INCOV290198 |
| INCOV290199 |
| INCOV290200 |
| INCOV290201 |
| INCOV290203 |
| INCOV290204 |
| INCOV290206 |
| INCOV290207 |
| INCOV290208 |
| INCOV290209 |
| INCOV290210 |
| INCOV290211 |
| INCOV290213 |
| INCOV290214 |
| INCOV290215 |
| INCOV290216 |
| INCOV290217 |
| INCOV290218 |
| INCOV290219 |
| INCOV290220 |
| INCOV290221 |
| INCOV290222 |
| INCOV290223 |
| INCOV290224 |
| INCOV290225 |
| INCOV290226 |
| INCOV290227 |
| INCOV290228 |
| INCOV290229 |
| INCOV290230 |
| INCOV290231 |
| INCOV290232 |
| INCOV290234 |
| INCOV290235 |
| INCOV290236 |
| INCOV290237 |
| INCOV290238 |
| INCOV290939 |
| INCOV290940 |
| INCOV290941 |
| INCOV290942 |
| INCOV290943 |
| INCOV290944 |
| INCOV290945 |
| INCOV290946 |
| INCOV290947 |
| INCOV290948 |
| INCOV290949 |
| INCOV290950 |
| INCOV290951 |
| INCOV290952 |
| INCOV290953 |
| INCOV290954 |
| INCOV290955 |
| INCOV290956 |
| INCOV290957 |
| INCOV290958 |
| INCOV290959 |
| INCOV290960 |
| INCOV290961 |
| INCOV290962 |
| INCOV290963 |
| INCOV290964 |
| INCOV290965 |
| INCOV290966 |
| INCOV290967 |
| INCOV290968 |
| INCOV290969 |
| INCOV290970 |
| INCOV290971 |
| INCOV290974 |
| INCOV290975 |
| INCOV290976 |
| INCOV290977 |
| INCOV290978 |
| INCOV290979 |
| INCOV290981 |
| INCOV290982 |
| INCOV290983 |
| INCOV290984 |
| INCOV290985 |
| INCOV290986 |
| INCOV290987 |
| INCOV290988 |
| INCOV290989 |
| INCOV290990 |
| INCOV290991 |
| INCOV290992 |
| INCOV290993 |
| INCOV290995 |
| INCOV290996 |
| INCOV290997 |
| INCOV290998 |
| INCOV290999 |
| INCOV291000 |
| INCOV291001 |
| INCOV291002 |
| INCOV291003 |
| INCOV291004 |
| INCOV291005 |
| INCOV291006 |
| INCOV291007 |
| INCOV291008 |
| INCOV291009 |
| INCOV291010 |
| INCOV291011 |
| INCOV291012 |
| INCOV291013 |
| INCOV291014 |
| INCOV291015 |
| INCOV291016 |
| INCOV291017 |
| INCOV291025 |
| INCOV291026 |
| INCOV291032 |
| INCOV291033 |
| INCOV291034 |
| INCOV291035 |
| INCOV291036 |
| INCOV291037 |
| INCOV291038 |
| INCOV291039 |
| INCOV291040 |
| INCOV291041 |
| INCOV291042 |
| INCOV291043 |
| INCOV291044 |
| INCOV291045 |
| INCOV291046 |
| INCOV291047 |
| INCOV291048 |
| INCOV291049 |
| INCOV291050 |
| INCOV291051 |
| INCOV291060 |
| INCOV291061 |
| INCOV291062 |
| INCOV291063 |
| INCOV291064 |
| INCOV291065 |
| INCOV291066 |
| INCOV291067 |
| INCOV291068 |
| INCOV291069 |
| INCOV291070 |
| INCOV291071 |
| INCOV291072 |
| INCOV291073 |
| INCOV291074 |
| INCOV291075 |
| INCOV291076 |
| INCOV291077 |
| INCOV291078 |
| INCOV291079 |
| INCOV291080 |
| INCOV291081 |
| INCOV291082 |
| INCOV291083 |
| INCOV291084 |
| INCOV291085 |
| INCOV291086 |
| INCOV291087 |
| INCOV291088 |
| INCOV291089 |
| INCOV291090 |
| INCOV291091 |
| INCOV291092 |
| INCOV291093 |
| INCOV291094 |
| INCOV291095 |
| INCOV291096 |
| INCOV291097 |
| INCOV291098 |
| INCOV291099 |
| INCOV291100 |
| INCOV291101 |
| INCOV291102 |
| INCOV291103 |
| INCOV291104 |
| INCOV291105 |
| INCOV291106 |
| INCOV291107 |
| INCOV291108 |
| INCOV291109 |
| INCOV291110 |
| INCOV291111 |
| INCOV291112 |
| INCOV291113 |
| INCOV291114 |
| INCOV291115 |
| INCOV291116 |
| INCOV291117 |
| INCOV291118 |
| INCOV291119 |
| INCOV291120 |
| INCOV291121 |
| INCOV291122 |
| INCOV291123 |
| INCOV291124 |
| INCOV291125 |
| INCOV291126 |
| INCOV291127 |
| INCOV291128 |
| INCOV291129 |
| INCOV291130 |
| INCOV291131 |
| INCOV291132 |
| INCOV291133 |
| INCOV291134 |
| INCOV291135 |
| INCOV291136 |
| INCOV291137 |
| INCOV291138 |
| INCOV291139 |
| INCOV291140 |
| INCOV291141 |
| INCOV291142 |
| INCOV291143 |
| INCOV291144 |
| INCOV291145 |
| INCOV291146 |
| INCOV291147 |
| INCOV291148 |
| INCOV291149 |
| INCOV291150 |
| INCOV291151 |
| INCOV291152 |
| INCOV291153 |
| INCOV291154 |
| INCOV291155 |
| INCOV291156 |
| INCOV291157 |
| INCOV291158 |
| INCOV291159 |
| INCOV291160 |
| INCOV291161 |
| INCOV291162 |
| INCOV291163 |
| INCOV291164 |
| INCOV291165 |
| INCOV291166 |
| INCOV291167 |
| INCOV291168 |
| INCOV291169 |
| INCOV291170 |
| INCOV291171 |
| INCOV291172 |
| INCOV291173 |
| INCOV291174 |
| INCOV291175 |
| INCOV291176 |
| INCOV291177 |
| INCOV291178 |
| INCOV291179 |
| INCOV291180 |
| INCOV291181 |
| INCOV291182 |
| INCOV291183 |
| INCOV291184 |
| INCOV291185 |
| INCOV291186 |
| INCOV291187 |
| INCOV291188 |
| INCOV291189 |
| INCOV291190 |
| INCOV291191 |
| INCOV291192 |
| INCOV291193 |
| INCOV291194 |
| INCOV291195 |
| INCOV291196 |
| INCOV291197 |
| INCOV291202 |
| INCOV291203 |
| INCOV291204 |
| INCOV291205 |
| INCOV291206 |
| INCOV291216 |
| INCOV291217 |
| INCOV291218 |
| INCOV291220 |
| INCOV291221 |
| INCOV291222 |
| INCOV291224 |
| INCOV291225 |
| INCOV291314 |
| INCOV291315 |
| INCOV291316 |
| INCOV291317 |
| INCOV291318 |
| INCOV291319 |
| INCOV291320 |
| INCOV291321 |
| INCOV291322 |
| INCOV291323 |
| INCOV291324 |
| INCOV291325 |
| INCOV291326 |
| INCOV291327 |
| INCOV291328 |
| INCOV291329 |
| INCOV291330 |
| INCOV291331 |
| INCOV291332 |
| INCOV291333 |
| INCOV291336 |
| INCOV291341 |
| INCOV291342 |
| INCOV291345 |
| INCOV291346 |
| INCOV291347 |
| INCOV291348 |
| INCOV291349 |
| INCOV291350 |
| INCOV291351 |
| INCOV291352 |
| INCOV291353 |
| INCOV291354 |
| INCOV291355 |
| INCOV291356 |
| INCOV291357 |
| INCOV291358 |
| INCOV291359 |
| INCOV291360 |
| INCOV291361 |
| INCOV291362 |
| INCOV291363 |
| INCOV291364 |
| INCOV291365 |
| INCOV291366 |
| INCOV291367 |
| INCOV291368 |
| INCOV291369 |
| INCOV291370 |
| INCOV291371 |
| INCOV291372 |
| INCOV291373 |
| INCOV291374 |
| INCOV291375 |
| INCOV291376 |
| INCOV291377 |
| INCOV291378 |
| INCOV291379 |
| INCOV291380 |
| INCOV291381 |
| INCOV291382 |
| INCOV291383 |
| INCOV291384 |
| INCOV291385 |
| INCOV291386 |
| INCOV291387 |
| INCOV291388 |
| INCOV291389 |
| INCOV291390 |
| INCOV291391 |
| INCOV291392 |
| INCOV291393 |
| INCOV291394 |
| INCOV291395 |
| INCOV291396 |
| INCOV291397 |
| INCOV291398 |
| INCOV291399 |
| INCOV291405 |
| INCOV291411 |
| INCOV291420 |
| INCOV291422 |
| INCOV291427 |
| INCOV291605 |
| INCOV291606 |
| INCOV291607 |
| INCOV291630 |
| INCOV291631 |
| INCOV291632 |
| INCOV291633 |
| INCOV291634 |
| INCOV291635 |
| INCOV291636 |
| INCOV291639 |
| INCOV291648 |
| INCOV291649 |
| INCOV291650 |
| INCOV291651 |
| INCOV291652 |
| INCOV291653 |
| INCOV291654 |
| INCOV291655 |
| INCOV291656 |
| INCOV291657 |
| INCOV291658 |
| INCOV291659 |
| INCOV291660 |
| INCOV291661 |
| INCOV291662 |
| INCOV291663 |
| INCOV291664 |
| INCOV291665 |
| INCOV291666 |
| INCOV291667 |
| INCOV291668 |
| INCOV291754 |
| INCOV291755 |
| INCOV291756 |
| INCOV291757 |
| INCOV291758 |
| INCOV291759 |
| INCOV291761 |
| INCOV291762 |
| INCOV291781 |
| INCOV291782 |
| INCOV291783 |
| INCOV291784 |
| INCOV291785 |
| INCOV291786 |
| INCOV291787 |
| INCOV291788 |
| INCOV291789 |
| INCOV291794 |
| INCOV291795 |
| INCOV291807 |
| INCOV291808 |
| INCOV291809 |
| INCOV291810 |
| INCOV291811 |
| INCOV291812 |
| INCOV291817 |
| INCOV291823 |
| INCOV291824 |
| INCOV291825 |
| INCOV291826 |
| INCOV291827 |
| INCOV291829 |
| INCOV291831 |
| INCOV291832 |
| INCOV291833 |
| INCOV291837 |
| INCOV291838 |
| INCOV291839 |
| INCOV291855 |
| INCOV291856 |
| INCOV291880 |
| INCOV291889 |
| INCOV291890 |
| INCOV291974 |
| INCOV292069 |
| INCOV292070 |
| INCOV292071 |
| INCOV292078 |
| INCOV292079 |
| INCOV292150 |
| INCOV292157 |
| INCOV292158 |
| INCOV292159 |
| INCOV289887 |
| INCOV289260 |
| INCOV289261 |
| INCOV289262 |
| INCOV289267 |
| INCOV289268 |
| INCOV289269 |
| INCOV289270 |
| INCOV289271 |
| INCOV289272 |
| INCOV289273 |
| INCOV289274 |
| INCOV289275 |
| INCOV289276 |
| INCOV289277 |
| INCOV289278 |
| INCOV289521 |
| INCOV289522 |
| INCOV289540 |
| INCOV289541 |
| INCOV289557 |
| INCOV289682 |
| INCOV289789 |
| INCOV289812 |
| INCOV289813 |
| INCOV289814 |
| INCOV290003 |
| INCOV290145 |
| INCOV290152 |
| INCOV290157 |
| INCOV290191 |
| INCOV290192 |
| INCOV290193 |
| INCOV290212 |
| INCOV291018 |
| INCOV291019 |
| INCOV291020 |
| INCOV291021 |
| INCOV291022 |
| INCOV291023 |
| INCOV291024 |
| INCOV291052 |
| INCOV291053 |
| INCOV291054 |
| INCOV291055 |
| INCOV291198 |
| INCOV291223 |
| INCOV291226 |
| INCOV291313 |
| INCOV291343 |
| INCOV291401 |
| INCOV291402 |
| INCOV291403 |
| INCOV291406 |
| INCOV291642 |
| INCOV291677 |
| INCOV291678 |
| INCOV291679 |
| INCOV291680 |
| INCOV291681 |
| INCOV291682 |
| INCOV291708 |
| INCOV291710 |
| INCOV291711 |
| INCOV291712 |
| INCOV291713 |
| INCOV291714 |
| INCOV291715 |
| INCOV291716 |
| INCOV291760 |
| INCOV291763 |
| INCOV291764 |
| INCOV291765 |
| INCOV291770 |
| INCOV291796 |
| INCOV291797 |
| INCOV291798 |
| INCOV291840 |
| INCOV291857 |
| INCOV291868 |
| INCOV291891 |
| INCOV291920 |
| INCOV291952 |
| INCOV292008 |
| INCOV292009 |
| INCOV292010 |
| INCOV292117 |
| INCOV289280 |
| INCOV289290 |
| INCOV289523 |
| INCOV289786 |
| INCOV289787 |
| INCOV289790 |
| INCOV289815 |
| INCOV289816 |
| INCOV289817 |
| INCOV289818 |
| INCOV289819 |
| INCOV289820 |
| INCOV289821 |
| INCOV289831 |
| INCOV289832 |
| INCOV289833 |
| INCOV289834 |
| INCOV289835 |
| INCOV289836 |
| INCOV289837 |
| INCOV290001 |
| INCOV290205 |
| INCOV290233 |
| INCOV290239 |
| INCOV290240 |
| INCOV290972 |
| INCOV290980 |
| INCOV291056 |
| INCOV291057 |
| INCOV291058 |
| INCOV291059 |
| INCOV291199 |
| INCOV291200 |
| INCOV291207 |
| INCOV291344 |
| INCOV291400 |
| INCOV291404 |
| INCOV291407 |
| INCOV291408 |
| INCOV291410 |
| INCOV291428 |
| INCOV291438 |
| INCOV291637 |
| INCOV291641 |
| INCOV291717 |
| INCOV291718 |
| INCOV291719 |
| INCOV291766 |
| INCOV291767 |
| INCOV291768 |
| INCOV291769 |
| INCOV291776 |
| INCOV291778 |
| INCOV291779 |
| INCOV291790 |
| INCOV291791 |
| INCOV291792 |
| INCOV291820 |
| INCOV291821 |
| INCOV291843 |
| INCOV291858 |
| INCOV291869 |
| INCOV291870 |
| INCOV291882 |
| INCOV291942 |
| INCOV291943 |
| INCOV291956 |
| INCOV292118 |
| INCOV292154 |
| INCOV292155 |
| INCOV289279 |
| INCOV289281 |
| INCOV289282 |
| INCOV289283 |
| INCOV289284 |
| INCOV289285 |
| INCOV289838 |
| INCOV289839 |
| INCOV289840 |
| INCOV289841 |
| INCOV289854 |
| INCOV289855 |
| INCOV289856 |
| INCOV289857 |
| INCOV289858 |
| INCOV289859 |
| INCOV289860 |
| INCOV289893 |
| INCOV289894 |
| INCOV289903 |
| INCOV290009 |
| INCOV290010 |
| INCOV290011 |
| INCOV290973 |
| INCOV291027 |
| INCOV291028 |
| INCOV291029 |
| INCOV291201 |
| INCOV291212 |
| INCOV291213 |
| INCOV291214 |
| INCOV291215 |
| INCOV291219 |
| INCOV291334 |
| INCOV291409 |
| INCOV291413 |
| INCOV291424 |
| INCOV291439 |
| INCOV291638 |
| INCOV291669 |
| INCOV291670 |
| INCOV291720 |
| INCOV291721 |
| INCOV291722 |
| INCOV291723 |
| INCOV291724 |
| INCOV291740 |
| INCOV291771 |
| INCOV291772 |
| INCOV291773 |
| INCOV291774 |
| INCOV291775 |
| INCOV291777 |
| INCOV291780 |
| INCOV291816 |
| INCOV291867 |
| INCOV291871 |
| INCOV291892 |
| INCOV291893 |
| INCOV291921 |
| INCOV291944 |
| INCOV291945 |
| INCOV291979 |
| INCOV291980 |
| INCOV291981 |
| INCOV291982 |
| INCOV291983 |
| INCOV292067 |
| INCOV292072 |
| INCOV292080 |
| INCOV292119 |
| INCOV289524 |
| INCOV289525 |
| INCOV289526 |
| INCOV289527 |
| INCOV289528 |
| INCOV289529 |
| INCOV289530 |
| INCOV289531 |
| INCOV289532 |
| INCOV289533 |
| INCOV289536 |
| INCOV289538 |
| INCOV289539 |
| INCOV289842 |
| INCOV289843 |
| INCOV289844 |
| INCOV289882 |
| INCOV289883 |
| INCOV289884 |
| INCOV289885 |
| INCOV289886 |
| INCOV289888 |
| INCOV289889 |
| INCOV289890 |
| INCOV289891 |
| INCOV290012 |
| INCOV290013 |
| INCOV291030 |
| INCOV291031 |
| INCOV291208 |
| INCOV291209 |
| INCOV291210 |
| INCOV291211 |
| INCOV291412 |
| INCOV291414 |
| INCOV291415 |
| INCOV291416 |
| INCOV291417 |
| INCOV291430 |
| INCOV291538 |
| INCOV291539 |
| INCOV291540 |
| INCOV291541 |
| INCOV291542 |
| INCOV291543 |
| INCOV291544 |
| INCOV291545 |
| INCOV291546 |
| INCOV291547 |
| INCOV291556 |
| INCOV291557 |
| INCOV291558 |
| INCOV291559 |
| INCOV291593 |
| INCOV291594 |
| INCOV291595 |
| INCOV291596 |
| INCOV291643 |
| INCOV291644 |
| INCOV291645 |
| INCOV291646 |
| INCOV291671 |
| INCOV291672 |
| INCOV291673 |
| INCOV291725 |
| INCOV291726 |
| INCOV291727 |
| INCOV291793 |
| INCOV291813 |
| INCOV291828 |
| INCOV291841 |
| INCOV291844 |
| INCOV291864 |
| INCOV291873 |
| INCOV291876 |
| INCOV291883 |
| INCOV291894 |
| INCOV291922 |
| INCOV291935 |
| INCOV291946 |
| INCOV291953 |
| INCOV291957 |
| INCOV291958 |
| INCOV291984 |
| INCOV291985 |
| INCOV292011 |
| INCOV292012 |
| INCOV292036 |
| INCOV292056 |
| INCOV292057 |
| INCOV289845 |
| INCOV289992 |
| INCOV291335 |
| INCOV291418 |
| INCOV291419 |
| INCOV291421 |
| INCOV291423 |
| INCOV291535 |
| INCOV291536 |
| INCOV291537 |
| INCOV291548 |
| INCOV291549 |
| INCOV291561 |
| INCOV291562 |
| INCOV291563 |
| INCOV291564 |
| INCOV291565 |
| INCOV291566 |
| INCOV291567 |
| INCOV291568 |
| INCOV291569 |
| INCOV291577 |
| INCOV291578 |
| INCOV291579 |
| INCOV291580 |
| INCOV291581 |
| INCOV291597 |
| INCOV291598 |
| INCOV291647 |
| INCOV291674 |
| INCOV291675 |
| INCOV291676 |
| INCOV291701 |
| INCOV291702 |
| INCOV291703 |
| INCOV291704 |
| INCOV291705 |
| INCOV291706 |
| INCOV291728 |
| INCOV291729 |
| INCOV291730 |
| INCOV291731 |
| INCOV291732 |
| INCOV291733 |
| INCOV291734 |
| INCOV291735 |
| INCOV291736 |
| INCOV291737 |
| INCOV291738 |
| INCOV291739 |
| INCOV291741 |
| INCOV291742 |
| INCOV291743 |
| INCOV291818 |
| INCOV291830 |
| INCOV291834 |
| INCOV291842 |
| INCOV291851 |
| INCOV291854 |
| INCOV291859 |
| INCOV291860 |
| INCOV291865 |
| INCOV291874 |
| INCOV291878 |
| INCOV291879 |
| INCOV291884 |
| INCOV291923 |
| INCOV291924 |
| INCOV291936 |
| INCOV291937 |
| INCOV291947 |
| INCOV291959 |
| INCOV291960 |
| INCOV291986 |
| INCOV291987 |
| INCOV291988 |
| INCOV292013 |
| INCOV292014 |
| INCOV292015 |
| INCOV292016 |
| INCOV292017 |
| INCOV292018 |
| INCOV292019 |
| INCOV292020 |
| INCOV292065 |
| INCOV292073 |
| INCOV292075 |
| INCOV292081 |
| INCOV292082 |
| INCOV289861 |
| INCOV289862 |
| INCOV289863 |
| INCOV289864 |
| INCOV289865 |
| INCOV289866 |
| INCOV289867 |
| INCOV289868 |
| INCOV289869 |
| INCOV289870 |
| INCOV289871 |
| INCOV289872 |
| INCOV289873 |
| INCOV289874 |
| INCOV289875 |
| INCOV289876 |
| INCOV289877 |
| INCOV289878 |
| INCOV289879 |
| INCOV289880 |
| INCOV289881 |
| INCOV289892 |
| INCOV290994 |
| INCOV291425 |
| INCOV291426 |
| INCOV291433 |
| INCOV291434 |
| INCOV291550 |
| INCOV291551 |
| INCOV291553 |
| INCOV291554 |
| INCOV291555 |
| INCOV291560 |
| INCOV291570 |
| INCOV291571 |
| INCOV291582 |
| INCOV291583 |
| INCOV291584 |
| INCOV291585 |
| INCOV291586 |
| INCOV291587 |
| INCOV291588 |
| INCOV291608 |
| INCOV291640 |
| INCOV291695 |
| INCOV291696 |
| INCOV291697 |
| INCOV291698 |
| INCOV291819 |
| INCOV291835 |
| INCOV291845 |
| INCOV291861 |
| INCOV291881 |
| INCOV291885 |
| INCOV291925 |
| INCOV291926 |
| INCOV291948 |
| INCOV291961 |
| INCOV291989 |
| INCOV291990 |
| INCOV291991 |
| INCOV291992 |
| INCOV292021 |
| INCOV292022 |
| INCOV292023 |
| INCOV292024 |
| INCOV292066 |
| INCOV292074 |
| INCOV292083 |
| INCOV292099 |
| INCOV292100 |
| INCOV292120 |
| INCOV292122 |
| INCOV292123 |
| INCOV292124 |
| INCOV292148 |
| INCOV291436 |
| INCOV291552 |
| INCOV291599 |
| INCOV291836 |
| INCOV291852 |
| INCOV291872 |
| INCOV291875 |
| INCOV291877 |
| INCOV291886 |
| INCOV291899 |
| INCOV291917 |
| INCOV291927 |
| INCOV291928 |
| INCOV291949 |
| INCOV291962 |
| INCOV291993 |
| INCOV292149 |
| INCOV292151 |
| INCOV289544 |
| INCOV289545 |
| INCOV291429 |
| INCOV291431 |
| INCOV291432 |
| INCOV291435 |
| INCOV291437 |
| INCOV291572 |
| INCOV291573 |
| INCOV291574 |
| INCOV291575 |
| INCOV291576 |
| INCOV291589 |
| INCOV291590 |
| INCOV291591 |
| INCOV291592 |
| INCOV291600 |
| INCOV291601 |
| INCOV291602 |
| INCOV291603 |
| INCOV291604 |
| INCOV291610 |
| INCOV291622 |
| INCOV291801 |
| INCOV291814 |
| INCOV291846 |
| INCOV291847 |
| INCOV291848 |
| INCOV291862 |
| INCOV291863 |
| INCOV291866 |
| INCOV291888 |
| INCOV291895 |
| INCOV291896 |
| INCOV291900 |
| INCOV291901 |
| INCOV291902 |
| INCOV291903 |
| INCOV291904 |
| INCOV291908 |
| INCOV291938 |
| INCOV291963 |
| INCOV291975 |
| INCOV291994 |
| INCOV291995 |
| INCOV291996 |
| INCOV292037 |
| INCOV292038 |
| INCOV292039 |
| INCOV292058 |
| INCOV292059 |
| INCOV292060 |
| INCOV292061 |
| INCOV292062 |
| INCOV292063 |
| INCOV292064 |
| INCOV292076 |
| INCOV292216 |
| INCOV292217 |
| INCOV292218 |
| INCOV292219 |
| INCOV292220 |
| INCOV291609 |
| INCOV291611 |
| INCOV291612 |
| INCOV291614 |
| INCOV291623 |
| INCOV291624 |
| INCOV291628 |
| INCOV291629 |
| INCOV291683 |
| INCOV291684 |
| INCOV291685 |
| INCOV291686 |
| INCOV291687 |
| INCOV291688 |
| INCOV291689 |
| INCOV291691 |
| INCOV291692 |
| INCOV291694 |
| INCOV291699 |
| INCOV291707 |
| INCOV291709 |
| INCOV291744 |
| INCOV291752 |
| INCOV291753 |
| INCOV291802 |
| INCOV291849 |
| INCOV291850 |
| INCOV291853 |
| INCOV291887 |
| INCOV291897 |
| INCOV291905 |
| INCOV291906 |
| INCOV291907 |
| INCOV291910 |
| INCOV291929 |
| INCOV291930 |
| INCOV291931 |
| INCOV291939 |
| INCOV291964 |
| INCOV291976 |
| INCOV292025 |
| INCOV292026 |
| INCOV292034 |
| INCOV292040 |
| INCOV292068 |
| INCOV292077 |
| INCOV292098 |
| INCOV292125 |
| INCOV292126 |
| INCOV292135 |
| INCOV292136 |
| INCOV292221 |
| INCOV292222 |
| INCOV292223 |
| INCOV292228 |
| INCOV291613 |
| INCOV291615 |
| INCOV291616 |
| INCOV291617 |
| INCOV291618 |
| INCOV291619 |
| INCOV291620 |
| INCOV291625 |
| INCOV291803 |
| INCOV291898 |
| INCOV291911 |
| INCOV291912 |
| INCOV291918 |
| INCOV291932 |
| INCOV291933 |
| INCOV291940 |
| INCOV291965 |
| INCOV291966 |
| INCOV291997 |
| INCOV291998 |
| INCOV292027 |
| INCOV292028 |
| INCOV292029 |
| INCOV292041 |
| INCOV292042 |
| INCOV292043 |
| INCOV292121 |
| INCOV292127 |
| INCOV292143 |
| INCOV292144 |
| INCOV292145 |
| INCOV292146 |
| INCOV292147 |
| INCOV292152 |
| INCOV292153 |
| INCOV292156 |
| INCOV291626 |
| INCOV291690 |
| INCOV291693 |
| INCOV291700 |
| INCOV291745 |
| INCOV291746 |
| INCOV291804 |
| INCOV291815 |
| INCOV291913 |
| INCOV291914 |
| INCOV291915 |
| INCOV291934 |
| INCOV291950 |
| INCOV291954 |
| INCOV291955 |
| INCOV291967 |
| INCOV291968 |
| INCOV291972 |
| INCOV291973 |
| INCOV291977 |
| INCOV291978 |
| INCOV291999 |
| INCOV292000 |
| INCOV292001 |
| INCOV292002 |
| INCOV292030 |
| INCOV292031 |
| INCOV292044 |
| INCOV292045 |
| INCOV292046 |
| INCOV292047 |
| INCOV292048 |
| INCOV292049 |
| INCOV292101 |
| INCOV292102 |
| INCOV292103 |
| INCOV292104 |
| INCOV292105 |
| INCOV292106 |
| INCOV292107 |
| INCOV291747 |
| INCOV291748 |
| INCOV291805 |
| INCOV291909 |
| INCOV291916 |
| INCOV291941 |
| INCOV291951 |
| INCOV291969 |
| INCOV291970 |
| INCOV291971 |
| INCOV292003 |
| INCOV292004 |
| INCOV292005 |
| INCOV292032 |
| INCOV292033 |
| INCOV292050 |
| INCOV292051 |
| INCOV292108 |
| INCOV292109 |
| INCOV292110 |
| INCOV292111 |
| INCOV292112 |
| INCOV292113 |
| INCOV291621 |
| INCOV291749 |
| INCOV291750 |
| INCOV291751 |
| INCOV291806 |
| INCOV292006 |
| INCOV292007 |
| INCOV292052 |
| INCOV292053 |
| INCOV292054 |
| INCOV291627 |
| INCOV291919 |
| INCOV292114 |
| INCOV292115 |
| INCOV292116 |
| INCOV292128 |
| INCOV292129 |
| INCOV292130 |
| INCOV292131 |
| INCOV292132 |
| INCOV292133 |
| INCOV292134 |
| INCOV292137 |
| INCOV292138 |
| INCOV292139 |
| INCOV292140 |
| INCOV292141 |
| INCOV292035 |
| INCOV292055 |
| INCOV292226 |
| INCOV292227 |
| INCOV292231 |
| INCOV291822 |
| INCOV292142 |
| INCOV292232 |
| INCOV292224 |
| INCOV292229 |
| INCOV292230 |
| INCOV292233 |
| INCOV292225 |
The authors have declared that no competing interests exist.
Author Contributions
Concept and design: Rajesh P. Karyakarte, Rashmita Das
Acquisition, analysis, or interpretation of data: Rajesh P. Karyakarte, Rashmita Das, Mansi V. Rajmane, Sonali Dudhate, Jeanne Agarasen, Praveena Pillai, Priyanka M. Chandankhede, Rutika S. Labhshetwar, Yogita Gadiyal, Preeti P. Kulkarni, Safanah Nizarudeen, Sushma Yanamandra, Nyabom Taji, Suvarna Joshi, Varsha Potdar
Drafting of the manuscript: Rajesh P. Karyakarte, Rashmita Das
Critical review of the manuscript for important intellectual content: Rajesh P. Karyakarte, Rashmita Das, Mansi V. Rajmane, Sonali Dudhate, Jeanne Agarasen, Praveena Pillai, Priyanka M. Chandankhede, Rutika S. Labhshetwar, Yogita Gadiyal, Preeti P. Kulkarni, Safanah Nizarudeen, Sushma Yanamandra, Nyabom Taji, Suvarna Joshi, Varsha Potdar
Supervision: Rajesh P. Karyakarte, Rashmita Das
Human Ethics
Consent was obtained or waived by all participants in this study. Institutional Ethics Committee, Byramjee Jeejeebhoy Government Medical College (BJGMC), Pune issued approval BJGMC/IEC/Pharmac/0721233-233
Animal Ethics
Animal subjects: All authors have confirmed that this study did not involve animal subjects or tissue.
References
- 1.Virological characteristics of the SARS-CoV-2 BA.2.86 variant. Tamura T, Mizuma K, Nasser H, et al. Cell Host Microbe. 2024;32:170–180. doi: 10.1016/j.chom.2024.01.001. [DOI] [PubMed] [Google Scholar]
- 2.2nd-generation BA.2 saltation lineage, >30 spike mutations (3 seq, 2 countries, Aug 14) #2183. [ Mar; 2024 ]. 2024. https://github.com/cov-lineages/pango-designation/issues/2183 https://github.com/cov-lineages/pango-designation/issues/2183
- 3.Early detection and surveillance of the SARS-CoV-2 variant BA.2.86 — worldwide, July-October 2023. [ Feb; 2024 ];Lambrou AS, South E, Ballou ES, et al. https://www.cdc.gov/mmwr/volumes/72/wr/mm7243a2.htm. MMWR Morb Mortal Wkly Rep. 2023 72:1162–1167. doi: 10.15585/mmwr.mm7243a2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Initial risk evaluation of BA.2.86 and its sublineages, 21 November 2023. [ Feb; 2024 ]. 2023. https://www.who.int/docs/default-source/coronaviruse/21112023_ba.2.86_ire.pdf?sfvrsn=8876def1_3 https://www.who.int/docs/default-source/coronaviruse/21112023_ba.2.86_ire.pdf?sfvrsn=8876def1_3
- 5.Update on SARS-CoV-2 variant JN.1 being tracked by CDC. [ Feb; 2024 ]. 2023. https://www.cdc.gov/ncird/whats-new/SARS-CoV-2-variant-JN.1.html?CDC_AA_refVal=https%3A%2F%2Fwww.cdc.gov%2Frespiratory-viruses%2Fwhats-new%2FSARS-CoV-2-variant-JN.1.html https://www.cdc.gov/ncird/whats-new/SARS-CoV-2-variant-JN.1.html?CDC_AA_refVal=https%3A%2F%2Fwww.cdc.gov%2Frespiratory-viruses%2Fwhats-new%2FSARS-CoV-2-variant-JN.1.html
- 6.COVID-19: WHO adds JN.1 as new variant of interest. Looi MK. BMJ. 2023;383:0. doi: 10.1136/bmj.p2975. [DOI] [PubMed] [Google Scholar]
- 7.COVID-19 epidemiological update - 19 January 2024: edition 163. [ Feb; 2024 ]. 2024. https://www.who.int/publications/m/item/covid-19-epidemiological-update---19-january-2024 https://www.who.int/publications/m/item/covid-19-epidemiological-update---19-january-2024
- 8.GISAID. hCoV-19 variants dashboard. [ Feb; 2024 ]. 2024. https://gisaid.org/hcov-19-variants-dashboard/ https://gisaid.org/hcov-19-variants-dashboard/
- 9.Virological characteristics of the SARS-COV-2 JN.1 variant. Kaku Y, Okumura K, Padilla-Blanco M, et al. Lancet Infect Dis. 2024;24:0. doi: 10.1016/S1473-3099(23)00813-7. [DOI] [PubMed] [Google Scholar]
- 10.Indian Biological Data Centre (IBDC) [ Jan; 2024 ]. https://ibdc.rcb.res.in/ https://ibdc.rcb.res.in/
- 11.Ministry of Health and Family Welfare. Centre issues advisory to states in view of a recent upsurge in COVID-19 cases and detection of first case of JN.1 variant in India. [ Feb; 2024 ]. 2023. https://pib.gov.in/PressReleasePage.aspx?PRID=1987840 https://pib.gov.in/PressReleasePage.aspx?PRID=1987840
- 12.Fast evolution of SARS-COV-2 BA.2.86 to JN.1 under heavy immune pressure. Yang S, Yu Y, Xu Y, et al. Lancet Infect Dis. 2024;24:0–2. doi: 10.1016/S1473-3099(23)00744-2. [DOI] [PubMed] [Google Scholar]
- 13.Executive summary. Initial risk evaluation of JN.1, 19 December 2023. [ Feb; 2024 ]. 2024. https://www.who.int/docs/default-source/coronaviruse/18122023_jn.1_ire_clean.pdf?sfvrsn=6103754a_3 https://www.who.int/docs/default-source/coronaviruse/18122023_jn.1_ire_clean.pdf?sfvrsn=6103754a_3
- 14.SARS-CoV-2 variant reports. [ Mar; 2024 ]. 2024. https://github.com/neherlab/SARS-CoV-2_variant-reports/blob/main/reports/variant_report_latest_draft.md https://github.com/neherlab/SARS-CoV-2_variant-reports/blob/main/reports/variant_report_latest_draft.md
- 15.WHO. Weekly epidemiological update on COVID-19 - 30 November 2021. [ Feb; 2024 ]. 2021. https://www.who.int/publications/m/item/weekly-epidemiological-update-on-covid-19---30-november-2021 https://www.who.int/publications/m/item/weekly-epidemiological-update-on-covid-19---30-november-2021
- 16.WHO. Weekly epidemiological update on COVID-19 - 25 January 2022. [ Feb; 2024 ]. 2022. https://www.who.int/publications/m/item/weekly-epidemiological-update-on-covid-19---25-january-2022 https://www.who.int/publications/m/item/weekly-epidemiological-update-on-covid-19---25-january-2022
- 17.covSPECTRUM. JN.1* (Nextclade) [ Feb; 2024 ]. 2024. https://cov-spectrum.org/explore/World/AllSamples/Past6M/variants?nextcladePangoLineage=JN.1%2A& https://cov-spectrum.org/explore/World/AllSamples/Past6M/variants?nextcladePangoLineage=JN.1%2A&
- 18.An early and preliminary assessment of the clinical severity of the emerging SARS-COV-2 Omicron variants in Maharashtra, India. Karyakarte RP, Das R, Taji N, et al. Cureus. 2022;14:0. doi: 10.7759/cureus.31352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clinical characteristics and outcomes of laboratory-confirmed SARS-CoV-2 cases infected with Omicron subvariants and the XBB recombinant variant. Karyakarte RP, Das R, Dudhate S, et al. Cureus. 2023;15:0. doi: 10.7759/cureus.35261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chasing SARS-COV-2 XBB.1.16 recombinant lineage in India and the clinical profile of XBB.1.16 cases in Maharashtra, India. Karyakarte RP, Das R, Rajmane MV, et al. Cureus. 2023;15:0. doi: 10.7759/cureus.39816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Timely monitoring of SARS-CoV-2 RNA fragments in wastewater shows the emergence of JN.1 (BA.2.86.1.1, clade 23I) in Berlin, Germany. Bartel A, Grau JH, Bitzegeio J, et al. Viruses. 2024;16:102. doi: 10.3390/v16010102. [DOI] [PMC free article] [PubMed] [Google Scholar]