Abstract
The development of peptide-based vaccines that are useful in the therapeutic treatment of melanoma and other cancers ultimately requires the identification of a sufficient number of antigenic peptides so that most individuals, regardless of their major histocompatibility complex (MHC)–encoded class I molecule phenotype, can develop a cytotoxic T lymphocyte (CTL) response against one or more peptide components of the vaccine. While it is relatively easy to identify antigenic peptides that are presented by the most prevalent MHC class I molecules in the population, it is problematic to identify antigenic peptides that are presented by MHC class I molecules that have less frequent expression in the population. One manner in which this problem can be overcome is by taking advantage of known MHC class I supertypes, which are groupings of MHC class I molecules that bind peptides sharing a common motif. We have developed a mass spectrometric approach which can be used to determine if an antigenic peptide is naturally processed and presented by any given MHC class I molecule. This approach has been applied to the A3 supertype, and the results demonstrate that some, but not all, A3 supertype family–associated peptides can associate with all A3 supertype family members. The approach also demonstrates the shared nature of several newly identified peptide antigens. The use of this technology negates the need to test peptides for their ability to stimulate CTL responses in those cases where the peptide is not naturally processed and bound to the target MHC class I molecule of interest, thus allowing resources to be focused on the most promising vaccine candidates.
Keywords: Antigen, Major histocompatibility complex class I molecule, Mass spectrometry, Peptide, Supertype
Introduction
Over the past decade, a variety of approaches, including the genetic method [3, 5, 16, 43], motif analysis [20, 21, 26], serological analysis of recombinant cDNA expression libraries (SEREX) [4, 31, 42], and the analytical chemistry approach [6, 13], have been used to identify a large number of tumor antigens that are recognized by cytotoxic T lymphocytes (CTLs) [29, 44]. The identified antigens are most frequently nine amino acids in length and the peptides associated with a particular major histocompatibility complex (MHC)-encoded class I molecule share a common-binding motif that generally distinguishes the peptides capable of binding to one MHC class I molecule from the peptides capable of binding other MHC class I molecules [10]. Many of the peptide antigens are being actively investigated for use in therapeutic cancer vaccines. Because most of the peptide antigens identified to date are presented in association with the most prevalent class I molecules in the population, there are few known peptide antigens with which to vaccinate those individuals who only express relatively rare class I MHC molecules. Likewise, those antigens that are presented in association with relatively rare class I MHC molecules are only useful in the immunization of a small segment of the population.
While the unique peptide-binding grooves of the various MHC class I molecules generally distinguish the peptide antigens that can bind to a given MHC class I molecule with high specificity, it has also been shown that some MHC class I molecules have peptide-binding grooves that bind peptides with a common motif, or supermotif [32–34]. MHC class I molecules that bind peptides of a particular supermotif are in turn classified as members of a common supertype [32–34]. Nine HLA supertypes have been defined including A1, A2, A3, A24, B7, B27, B44, B58, and B62. The A3 supertype, for example, includes HLA-A3, HLA-A11, HLA-A31, HLA-A33, and HLA-A68. Collectively, the A3 supertype covers 37.5% of the Caucasian population, while the individual HLA molecules that constitute the A3 supertype are expressed in as low as 2% of the population [36]. The utility of the supertype concept is that once an antigenic peptide has been identified for one member of a supertype family, that same peptide should then serve as a useful antigen for other members of the same supertype family. Practically, this means that peptide antigens presented in association with prevalent MHC class I molecules such as HLA-A3 (21% of the population) could also function as antigens for less prevalent MHC class I molecules such as HLA-A31 (2% of the population) for which few peptide antigens have been discovered. Conversely, the utility of an HLA-A31–associated peptide antigen would be expanded nearly 20-fold if it can be recognized in association with all A3 supertype family members.
While the sharing of common antigens among supertypes may be generally applicable, at least three observations show that this is not the case for all peptide antigens. First, while peptide-binding studies have clearly established that antigenic peptides can bind to multiple members of the same supertype family, it is also clear that a particular peptide may bind to some, but not all members of a supertype [2, 8, 18, 40]. Second, binding with high to moderate affinity to a particular supertype does not guarantee that a CTL response can be generated to that particular combination of peptide and MHC class I molecule [2, 8, 18, 40]. Third, it has been demonstrated that peptide-specific CTLs that have been stimulated by primary, in vitro stimulation do not always recognize target cells expressing the same MHC class I molecule and the protein from which the peptide is derived [18, 21, 41]. Thus, it cannot be assumed that a given peptide identified in this manner will be a good antigen in the context of all members of a supertype.
While the supertype hypothesis may be generally true, the fact that it cannot be universally applied means that substantial effort must be expended to test each antigenic peptide of interest in combination with each MHC class I molecule of a supertype. To minimize this testing requirement and to restrict the testing to those antigens that are naturally processed and presented on the surface of target cells, we have developed a mass spectrometric approach to directly determine if a specific peptide is bound to a MHC class I molecule on the surface of a target cell.
Materials and methods
Cell lines
The human melanoma cell lines DM14, VMM12, VMM18, VMM19, VMM86, and VMM150 were maintained in RPMI 1640 supplemented with 5–10% fetal bovine serum, 2 mM L-glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin. The hybridomas BB7.2 (γ2b, A2, A69) [24], CR11-351 (γ1, A2, A68, A69) [30], GAP-A3 (γ2a, A3) [1], ME1-1.2 (γ1, B7, B27) [9], SFR8-B6 (γ2b, Bw6) [27], B1.23.2 (γ2a, B-locus, Cw3) [28], and W6/32 (γ2a, monomorphic) [25], were maintained in DMEM with high glucose, 5% FBS, 2 mM L-glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin.
Monoclonal antibody (mAb) purification
Monoclonal antibodies BB7.2, GAP-A3, SFR8-B6, B1.23.2, and W6/32 were purified on a HiTrap rProtein A Column (Pharmacia, Piscataway, NJ, USA), and mAbs CR11-351 and ME1-1.2 were purified on a HiTrap rProtein G Column (Pharmacia). The mAbs were eluted with 100 mM glycine pH 3.0, and immediately neutralized with 1.0 M Tris pH 8.0.
Flow cytometry
The following antibodies were used for the detection of A3 supertype molecules: HLA-A3 was detected with 10 μg/ml of mAb GAP-A3, HLA-A11 was detected with 10 μl of biotinylated mAb HLA-A11 (One Lambda, Canoga Park, CA, USA), HLA-A31 was detected with 10 μl biotinylated mAb HLA-A30,31 (One Lambda), HLA-A33 was detected with 25 μl mAb HLA-A33,B8 (One Lambda), and HLA-A68 was detected with 10 μg/ml of mAb CR11-351. The primary antibodies were added to 2–5×105 cells and incubated for 30 min on ice. The cells were washed twice, 50 μl of a 1:50 dilution of goat-antimouse IgG-FITC (ICN, Irvine, CA, USA) or avidin-FITC (ICN) was then added, and the cells were incubated an additional 30 min on ice. The cells were then washed once and analyzed on a FACSCalibur instrument (BD Biosciences, San Jose, CA, USA).
Total mRNA isolation
Total mRNA was isolated from 5–10×106 tumor cells using the RNeasy Mini Kit (Qiagen, Valencia, CA, USA) as per the kit instructions. RNA was quantified by absorbance at 260 nm.
Polymerase chain reaction
Reverse transcriptase polymerase chain reaction (RT-PCR) was performed using the Access RT-PCR system (Promega, Madison, WI, USA) as per the kit instructions. Gene-specific primers were used to amplify a 598-bp fragment from the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene (forward primer: 5′-CCACCCATGGCAAATTCCATGGCA-3′; reverse primer: 5′-TCTAGACGGCAGGTCAGGTCCACC-3′), a 298-bp fragment from the gp100 gene (forward primer: 5′-TGGTGTCTCAAGGCAACTC-3′; reverse primer: 5′-AGATGCAGGCATCGTCAG-3′), a 458-bp fragment from the NY-ESO-1 gene (forward primer: 5′-GCGGCTTCAGGGCTGAATGGATG-3′; reverse primer: 5′-AAGCCGTCCTCCTCCAGCGACA-3′), and a 355-bp fragment from the TRP-2 gene (forward primer: 5′-GACTCTGATTAGTCGGAACTC-3′; reverse primer: 5′-GAATGTGGCAAAGCGTTTGTC-3′). The cycling conditions for all three primer sets were: 45 min at 48°C; 2 min at 94°C; 40 cycles of (1) 30 s at 94°C, (2) 1 min at 60°C, (3) 1 min at 68°C; 7 min at 68°C. PCR primers and amplification conditions for the TAG were as previously described [15]. The PCR products were visualized on ethidium bromide stained agarose gels.
Isolation of MHC class I–associated peptides
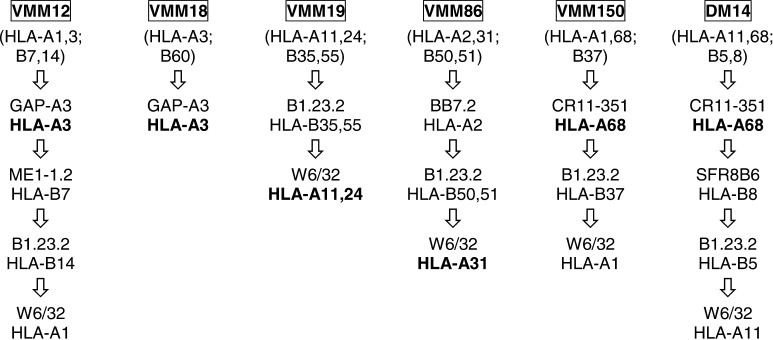
The MHC class I molecules were purified from 6.8×109 VMM12, 5.0×109 VMM18, 2.7×109 VMM19, 16.1×109 VMM86, 3.0×109 VMM150, and 5.8×109 DM14 using immunoaffinity chromatography as previously described [12]. The mAbs BB7.2, GAP-A3, ME1-1.2, B1.23.2, SFR8-B6, and W6/32 were immobilized on protein A sepharose fast-flow, while CR11-351 was covalently linked to hydrazide agarose beads (BioRad Laboratories, Hercules, CA, USA). The columns were sequentially ordered in such a fashion that the MHC class I molecules of interest were either selectively retained with a specific antibody, or they were retained on W6/32 following the selective depletion of the other expressed MHC class I molecules (Fig. 1). The peptides were eluted from the columns with 10% acetic acid, and separated from high molecular weight proteins (e.g., β2-microglobulin, antibody) by spinning through a 5,000-Da cutoff Ultrafree membrane (Millipore, Marlborough, MA, USA). The peptide extract was then concentrated to approximately 200 μl using a Savant SpeedVac Concentrator (Thermo Electron Corporation, Waltham, MA, USA). The peptides were stored at −40°C until further use.
Fig. 1.
Immunoaffinity purification of A3 supertype peptides. Cell lysates obtained from each of the indicated cell lines were sequentially run over immunoaffinity columns in the order indicated. The antibody used in each of the sequential columns is indicated, and is followed by the MHC class I molecule that was retained by that antibody. Peptides obtained from the MHC class I molecules in bold type were analyzed for the presence of specific peptides using SRM mass spectrometry.
Peptide fractionation
Peptide extracts were fractionated using an Applied Biosystems Model 130A Separation System (Foster City, CA, USA) equipped with a C18 Higgins HAISIL 300 column (Higgins, Winter Park, FL, USA). The solvent systems used in the fractionation were solvent A (0.1% trifluoroacetic acid [TFA] in NANOpure water; Barnstead/Thermolyne, Dubuque, IA, USA) and solvent B (0.085% TFA in 60% acetonitrile). Four fifths of each peptide extract was fractionated. Fractions were manually collected into Eppendorf tubes (Sarstedt, Newton, NC, USA) every 40 s during a total gradient time of 75 min at a flow rate of 200 μl/min. The samples were then concentrated to approximately 20 μl using a SpeedVac Concentrator.
Peptide synthesis
Synthetic peptides including ALLAVGATK, LIYRRRLMK, ALNFPGSQK, AAQERRVPR, and ASGPGGGAPR, were synthesized by standard fluorenylmethoxycarbonyl (Fmoc) chemistry using a Gilson model AMS1400 peptide synthesizer and purified by reverse-phase high-performance liquid chromatography (RP-HPLC) using C18 particles. The deuterium-labeled ALLD10AVGD2ATK peptide was synthesized by incorporating the deuterium-labeled amino acids leucine (D10) and glycine (D2) (Cambridge Isotope Laboratories, Andover, MA, USA). In order to include the deuterium-labeled amino acids in the Fmoc synthesis, the free primary amines on the N-terminus were protected with Fmoc. The coupling reaction was performed as described by Lapatsanis et al. [22]. In brief, the deuterium-labeled amino acid was dissolved in Na2CO3 and cooled to 0°C. Succinimidyl 9-fluorenylmethy carbonate was dissolved in dimethylformamide (DMF), added to the solubilized amino acid, and allowed to react at room temperature for 10 min. The solution was extracted with ether and then ethyl acetate to further isolate the product. The product was precipitated with the addition of HCl, and then washed with NaCl and H2O, and dried with sodium sulfate. The Fmoc-protected amino acids were then used to synthesize deuterium-labeled ALLD10AVGD2ATK.
Narrow-bore RP-HPLC fractionation of HLA-A3–supertype synthetic peptides
The RP-HPLC fractions in which the melanoma antigens of interest would elute were determined by establishing an elution profile for the corresponding synthetic peptide. A mixture of the synthetic peptides (~300 pmol of each peptide) was fractionated on the same C-18 column used to fractionate the peptide extracts.
Determination of the selected reaction monitoring (SRM) conditions for the detection of the selected peptides
The synthetic peptides were infused into a TSQ-7000 triple quadrupole mass spectrometer (Finnigan, San Jose, CA, USA) using a 360-μm OD × 75 μm ID-fused silica column. The column contained a ~3 μm tip made using a laser puller (Sutter Instruments, Novato, CA, USA). The column was packed with 1 cm of 5–20 μm irregular ODS-AQ C18 particles (YMC, Wilmington, NC, USA) and rinsed with 0.1% acetic acid. The infusion set-up was similar to the RP-HPLC set-up described by Martin et al. [23], except that the RP-HPLC inlet was replaced with an infusion inlet consisting of 360 μm OD × 100 μm ID-fused silica connected to a 500-μl syringe (Hamilton, Reno, NV, USA). Analytes, at a concentration of 1 pmol/μl, were infused in a solution of 60% acetonitrile (aqueous) with 0.1% acetic acid.
Full scan mass spectra and MS/MS spectra were acquired for each of the peptides of interest. For the MS/MS analysis Q1 scanned the mass/charge range of interest and Q2 was filled with 2.5 m Torr of the collision gas, argon. Typical offset values were −15 to −30 eV depending on the precursor ion charge state. The collision-offset values were optimized for each synthetic peptide to provide the desired fragmentation pattern from SRM experiments.
SRM analysis of peptides obtained from A3 supertype MHC class I molecules
Selected RP-HPLC fractions containing MHC class I–derived peptides were analyzed by nanoflow RP-HPLC/micro-electrospray ionization (μESI) on a TSQ-7000 triple quadrupole mass spectrometer using a C18 microcapillary column constructed from 360 × 50 μm ID-fused silica (5 μm C18 particles, 7 cm length). Flow rates were maintained at 57 nl/min using a spray voltage of 1.6 keV. The precursor, collision offset, and peptide-specific product ions for the peptides were experimentally determined. SRM experiments allowed for the detection of several peptides in one analysis if the peptides eluted in similar RP-HPLC fraction. An aliquot of 5×108 cell equivalents of three consecutive RP-HPLC fractions were loaded onto the one-piece column and washed for 10 min with 0.1% acetic acid. A volume of 16 fmol of ALLD10AVGD2ATK, equivalent to 20 copies per cell, was added to each RP-HPLC fraction and detected in each SRM experiment as a means of monitoring the analysis conditions and as a standard for relative quantitation. The peptides were then eluted into the mass spectrometer using gradient conditions of 0–100% B in 17 min. Duplicate analyses were performed.
Fourier-transform ion cyclotron resonance (FT-ICR) mass spectrometry
Peptides corresponding to approximately 1×107 cell equivalents were loaded onto a C18 microcapillary column constructed from 360 × 50 μm ID-fused silica (5 μm C18 particles, 7 cm length) and washed with 0.1% acetic acid for 10 min. The peptide mixture was gradient-eluted directly into a custom-designed FT-ICR mass spectrometer. The FT-ICR mass spectrometer was fitted with a custom-designed μESI source utilizing nanoflow RP-HPLC columns as described [23]. Nanoflow RP-HPLC gradient conditions were 0–100% solvent B (70% v/v acetonitrile: 0.1 mM acetic acid) in solvent A (0.1 mM acetic acid) over 40 min at a flow rate of 57 nl/min. Full-scan mass spectra (300≤m/z≤1,500) were collected at approximately one scan per second, with typical resolution of 5,000–10,000 atomic mass units (amu).
Calculation of peptide abundance
Peptide abundance, or the average number of copies of a particular peptide per cell, was calculated from the measured ion current, which is linear over a dynamic range of greater than five orders of magnitude on the TSQ-7000. Abundance was calculated by comparing the averaged ion current from the measured product ions of 16 fmol of each of the synthetic test peptides with the same amount of the internal standard ALLD10AVGD2ATK. The ratio of these average ion currents was then used to calculate the relative ionization efficiency of each of the test peptides in comparison to the ALLD10AVGD2ATK peptide. Because the average ion current is proportional to the peptide concentration, the average copy number of peptide per cell could be calculated by including a known amount of the ALLD10AVGD2ATK peptide in each test sample and correcting for the ionization efficiency of the measured peptides. If a peptide was not detected it was reported as being below the detection limit for that peptide.
Results
MHC class I protein expression, and gp100 and NY-ESO gene expression, in A3 supertype–expressing melanoma cell lines
The MHC class I typing of peripheral blood mononuclear cells obtained from the melanoma patients from which the melanoma cell lines VMM12, VMM18, VMM19, VMM86, VMM150, and DM14 were derived, previously demonstrated that these individuals express MHC class I molecules belonging to the A3 supertype. To ensure that the melanoma cell lines expressed the relevant MHC class I molecules, the expression levels of HLA-A3, HLA-A11, HLA-A31, and HLA-A68 on the respective melanoma cell lines were assessed by flow cytometry. VMM12 and VMM18 express HLA-A3, VMM19 expresses HLA-A11, VMM86 expresses HLA-A31, and both DM14 and VMM150 express HLA-A68 (data not shown).
The melanoma cell lines were further assessed by PCR for the expression of the gp100 and NY-ESO-1 genes (Table 1). Five of the six melanoma cell lines expressed gp100, while VMM12, VMM18, and VMM86, but not VMM19, DM14, or VMM150, expressed NY-ESO-1. A previous study has shown that DM14 was PCR-negative for the expression of gp100, although the same study showed that the cells were weakly positive for the expression of the protein when examined by immunohistochemistry [38]. Our present results showed that PCR amplification at 35 or 40 cycles was required to obtain a visible band, while 30 cycles was insufficient, thus further demonstrating that gp100 is expressed at very low levels in the melanoma tumor cell line DM14 (data not shown).
Table 1.
Expression of gp100 and NY-ESO-1 genes in A3 supertype–expressing cell lines
| Gene | Cell line | |||||
|---|---|---|---|---|---|---|
| VMM12 (HLA-A3) | VMM18 (HLA-A3) | VMM19 (HLA-A11) | VMM86 (HLA-A31) | VMM150 (HLA-A68) | DM14 (HLA-A68) | |
| GAPDH | +a | + | + | + | + | + |
| gp100 | + | + | + | + | − | + |
| NY-ESO-1 | + | + | − | + | − | − |
aPlus sign Indicates that a band of an appropriate molecular weight was observed on a agarose gel following RT-PCR. A negative sign indicates that no such band was observed
Isolation and fractionation of A3 supertype peptides from DM14, VMM12, VMM18, VMM19, VMM86, and VMM150
To isolate peptides from the A3 supertype molecules, the tumor cells were detergent lysed and the MHC class I molecules purified by immunoaffinity chromatography (Fig. 1). HLA-A3 was purified from the VMM12 and VMM18 melanoma cell lines with the HLA-A3–specific mAb, GAP-A3, and HLA-A68 was purified from the DM14 and VMM150 melanoma cell lines with the HLA-A2,-28 (-A68, -A69)–specific mAb, CR11-351. As there are no mAbs specific for HLA-A11 or HLA-A31, these molecules were isolated with W6/32, a mAb with specificity for a monomorphic epitope present on all MHC class I molecules. Sequential immunoaffinity columns were used to eliminate some or all of the MHC class I, which we did not desire to retain with W6/32. Thus, HLA-A31 was purified from the VMM86 melanoma tumor cell line by first passing the lysate over mAb BB7.2 to remove HLA-A2, and then mAb B1.23.2 to remove HLA-B50 and HLA-B51. mAb W6/32 was then used to retain HLA-A31. Similarly HLA-A11 was purified from VMM19 by first passing the lysate over mAb B1.23.2 to remove HLA-B35 and HLA-B55. mAb W6/32 was then used to purify HLA-A11 along with HLA-A24. Four fifths of the isolated A3 supertype peptides from each cell line were RP-HPLC fractioned into approximately 96 fractions and stored at −40°C until used.
Determination of the RP-HPLC elution profiles and product ion profiles for the selected A3 supertype melanoma peptide antigens
The HLA-A3–restricted peptides ALLAVGATK [37], ALNFPGSQK [19], LIYRRRLMK [17], RLSNRLLLR [15], and SQNFPGSQK [14], and the HLA-A31–restricted peptides AAQERRVPR [46], ASGPGGGAPR [46], and LLGPGRPYR [45] were selected for study. The deuterated peptide, ALLD10AVGD2ATK, was used as an internal standard. Each of these peptides was synthesized and its RP-HPLC elution profile determined on a narrow-bore C18 Higgins HAISIL 300 column. This information was then used to select the appropriate RP-HPLC fractions (Table 2) for SRM scanning.
Table 2.
Experimentally determined SRM conditions used to scan for the ALLD10AVGD2ATK, ALLAVGATK, LIYRRRLMK, ALNFPGSQK, ASGPGGGAPR, and AAQERRVPR peptides in complex mixtures
| Source protein | Peptides | RP-HPLC fractions | Precursor ion (Da) | Product ions (Da) | Collision energy (kV) | |||
|---|---|---|---|---|---|---|---|---|
| Internal standard | ALLD10AVGD2ATK | – | 428.27 (+2)a | 185.4 | 477.3 | 671.5 | −19 | |
| gp100 | ALLAVGATK | 30 | 422.27 (+2) | 185.4 | 474.9 | 659.1 | −19 | |
| gp100 | ALNFPGSQK | 30 | 481.26 (+2) | 298.9 | 664.0 | 777.8 | −22 | |
| gp100 | LIYRRRLMK | 24 | 416.93 (+3) | 495.0 | 560.1 | −20 | ||
| 313.36 (+4) | 86.4 | −17 | ||||||
| NY-ESO-1 | ASGPGGGAPR | 15 | 413.71 (+2) | 306.4 | 334.5 | 514.0 | −22 | |
| NY-ESO-1 | AAQERRVPR | 18 | 541.62 (+2) | 115.4 | 228.5 | 462.5 | 497.5 | −21 |
| TAG | RLSNRLLLR | 43 | 380.92 (+3) | 175.3 | 288.6 | 492.8 | −22 | |
| TRP-2 | LLGPGRPYR | 28 | 514.80 (+2) | 199.0 | 401.8 | 458.3 | −15 | |
| Unknown | SQNFPGSQK | 18 | 496.74 (+2) | 515.9 | 663.3 | 777.7 | −22 | |
aThe number in parentheses refers to the charge state of the precursor ion
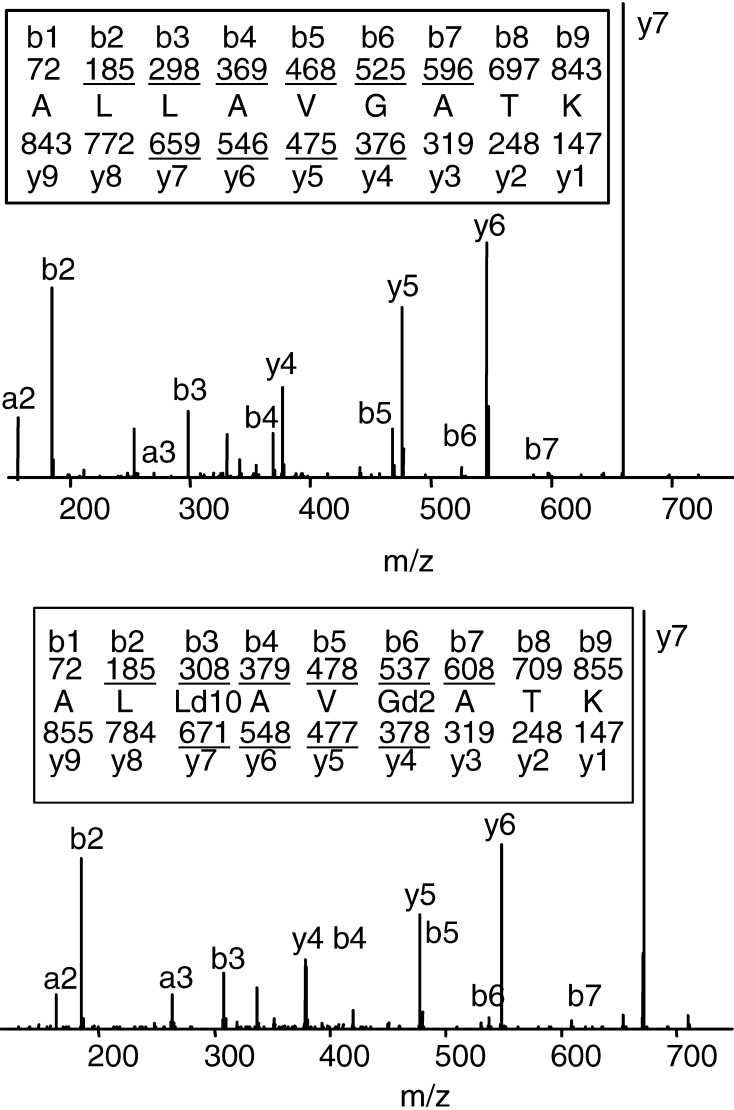
To establish an SRM scanning profile for each peptide of interest, the CAD spectra of the corresponding synthetic peptide were determined on the TSQ-7000. As illustrated for the ALLAVGATK peptide, distinct product ions can be detected that correspond to predicted bn and yn masses (Fig. 2). Moreover, several product ions of the deuterated ALLD10AVGD2ATK peptide differ from those of the corresponding ALLAVGATK peptide, making it possible to simultaneously detect and quantify both peptides in the same sample. For example, when comparing the deuterated and nondeuterated ALLAVGATK peptides, the y5 and y7 product ions differ in mass, while the b2 product ions have the same mass. A similar analysis was performed for each of the peptides of interest and was used to determine the appropriate collision energy to use for the analysis, as well as the profile of product ions that would be generated from each precursor ion (Table 2).
Fig. 2.
The CAD mass spectra of ALLAVGATK and ALLD10AVGD2ATK. The predicted masses for the product ions of types bn and yn are written above and below the peptide sequence, respectively. Ions of type bn and yn that are observed in the spectrum are underlined.
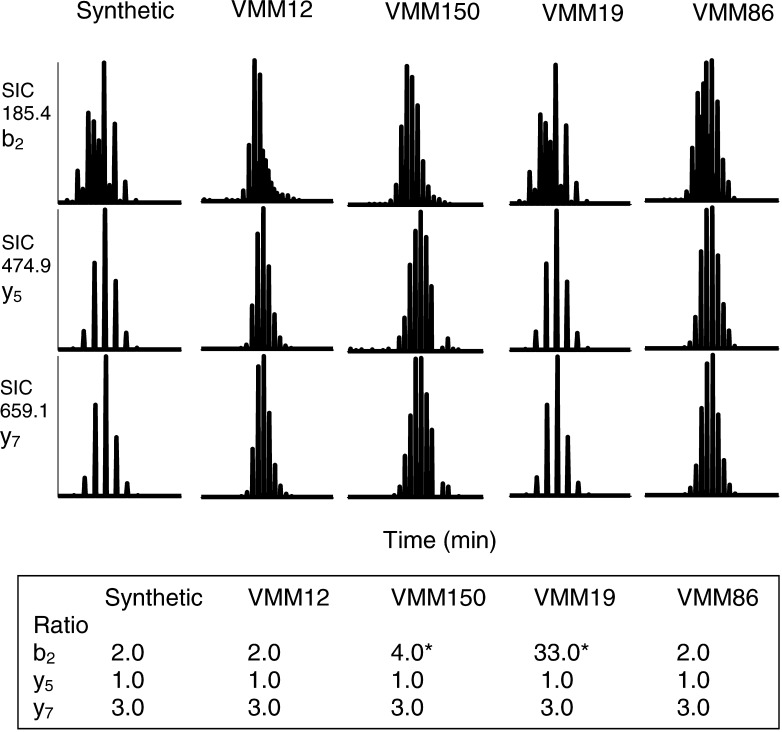
In combination with the RP-HPLC fractionation profile and the precursor ion mass, the ratios of the relative abundance of the specific product ion masses, as well as their retention times, represent a fingerprint for each of the peptides as is demonstrated in an SRM scanning experiment in which ALLAVGATK is the targeted peptide and ALLD10AVGD2ATK is used as an internal standard (Fig. 3). The ALLAVGATK and ALLD10AVGD2ATK peptides have nearly identical retention times and when in a given sample they elute at slightly different times, their measured b2, y5, and y7 ions are present in a ratio of 2:1:3 (see the synthetic ALLAVGATK, as well as the VMM12 and VMM86 samples). If in a given sample the deuterated and nondeuterated samples coelute, the degree to which coelution occurs means that the b2 product ions of both species will simultaneously elute, thus altering the ratio of the b2 product ion to the y5 and y7 product ions (see VMM150 and VMM19 samples). The ratios of the product ions that differ in mass will, however, be unchanged.
Fig. 3.
SRM mass spectrometric detection of ALLAVGATK. Single ion chromatograms (SIC) for three product ions of ALLAVGATK are shown. The product ions observed for the synthetic peptide and each of the purified peptide samples have equal retention times and b2/y5/y7 ratios, indicating that ALLAVGATK is present in each of the samples. *Indicates that the ratio is high due to ion contribution from the internal standard deuterium-labeled ALLAVGATK (the contribution of the internal standard is observed in the b2 product ion because the mass of b2 for both the deuterium-labeled and non-deuterium-labeled peptides is 185.4).
Use of ALLD10AVGD2ATK as an internal standard to estimate the number of copies per cell of each detected peptide
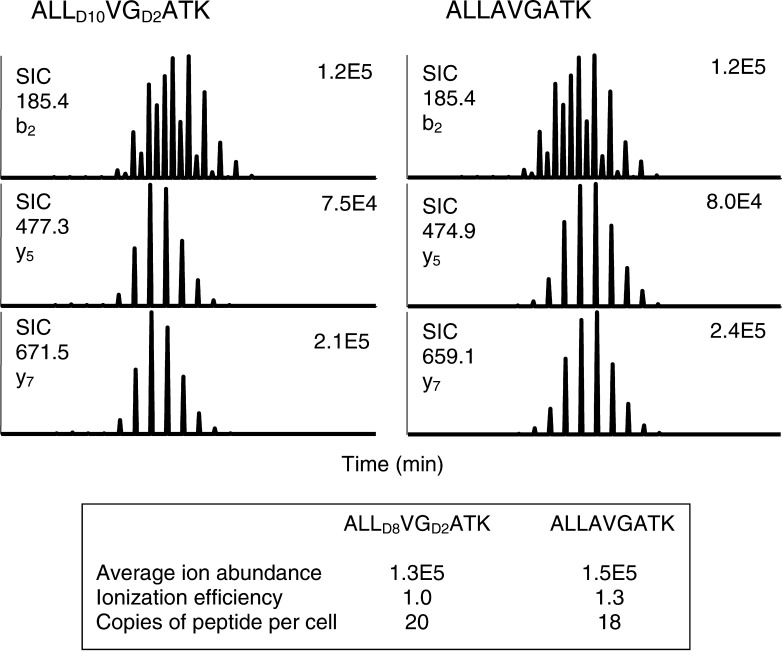
Peptide detection was made semiquantitative by the inclusion of the deuterated peptide, ALLD10AVGD2ATK, as an internal standard in each RP-HPLC fraction. A volume of 16 fmol of the standard, an amount equivalent to that found on 5×108 cells at 20 copies per cell, was included in each analyzed fraction. The average ion abundance of the product ions and the ionization efficiency of the precursor ion was determined for each peptide including the deuterium-labeled ALLD10AVGD2ATK peptide. Once these parameters were established for each of the test peptides, it was then possible to select a given mass in a test sample, determine the abundance of the selected product ions, and normalize the abundance to a known quantity of the ALLD10AVGD2ATK peptide used as a standard within each test sample. The detection and quantification of ALLAVGATK is demonstrated in Fig. 4.
Fig. 4.
Peptide quantification by comparison to the deuterium-labeled ALLAVGATK peptide. The average ion abundance was calculated from the three measured product ions. The ionization efficiency was determined from each peptide compared to the deuterium-labeled ALLAVGATK. The average ion abundance and ionization efficiencies were used to calculate the abundance of the peptides.
Reproducibility of SRM scanning measurements
To determine the reproducibility of SRM scanning in determining the number of copies of peptide per cell, triplicate SRM scans were made to detect the ALLAVGATK and ALNFPGSQK peptides among the peptides isolated from HLA-A3 purified from the melanoma tumor cell line VMM12. SRM scanning experiments with RP-HPLC fractions 29–31 (ALLAVGATK and ALNFPGSQK) were performed to detect the three product ions for each peptide, as well as the product ion from the internal standard peptide, ALLD10AVGD2ATK. The presence of a given peptide was indicated by the detection of the three product ions at the appropriate retention time. The average abundance of the three product ions for each peptide was then normalized against the internal standard. In the first analysis, the average ion abundance for ALLAVGATK was 1.5×105, and for 20 copies/cell of ALLD10AVGD2ATK, the average ion abundance was 1.3×105 (Fig. 4). By comparing the ion abundance of ALLAVGATK to ALLD10AVGD2ATK and correcting for the ionization efficiency it was then possible to determine that the sample contained approximately 18 copies of ALLAVGATK per cell [1.5×105/(1.3×105/1.3×20)=18]. Two subsequent analyses detected ALLAVAGTK at 17 and 12 copies per cell, for an average 16 copies per cell (Table 3). A similar analysis was performed for the ALNFPGSQK peptide (Table 3).
Table 3.
Reproducibility of SRM scanning for the determination of the copy number of peptides per cell
| Peptidea | Copies of peptide per cell | Average copies of peptide per cell ± SDc | ||
|---|---|---|---|---|
| Experiment 1b | Experiment 2 | Experiment 3 | ||
| ALLAVGATK | 18 | 17 | 12 | 15.7±3.2 |
| ALNFPGSQK | 3 | 2 | 3 | 2.7±0.6 |
aThe peptides were eluted from HLA-A3 obtained from the melanoma tumor cell line VMM12
bThe spectra for ALLAVGATK in experiment 1 are shown in Fig. 4
cSD Standard deviation
Detection limit determination
To determine the detection limit of the SRM scanning methodology, approximately 100 amol of ALLD10AVGD2ATK was spiked into an irrelevant RP-HPLC fraction containing 4×108 cell equivalents of VMM12. The peptide was detected at this level in the presence of a complex peptide mixture. One copy of peptide per cell in a mixture of peptides derived from 4×108 cells is equivalent to 660 amol. The detection of 100 amol of peptide thus corresponds to a detection limit of one peptide per 6.6 cells, or approximately 0.15 copies per cell. Thus, it is possible to detect peptide at physiologically significant levels that are as low as one copy per cell, even allowing for losses in peptide extraction and purification.
Detection of A3 supertype–associated peptides with different members of the A3 supertype family
Having established appropriate scanning parameters for each of the peptides of interest, we next sought to detect the peptides on four different A3 supertype molecules including HLA-A3, HLA-A11, HLA-A31, and HLA-A68. Six different melanoma cell lines, each expressing one of the MHC class I molecules of interest, were used for the analysis. PCR was used to detect the expression of the gp100 and NY-ESO-1 in the cell lines, with expression being observed in some, but not all of the lines (Table 4).
Table 4.
Detection of A3 supertype–associated peptides on different A3 supertype molecules
| Source protein | Peptide sequence | Known restriction element | Copy level of peptide per cell | |||||
|---|---|---|---|---|---|---|---|---|
| VMM12 (HLA-A3) | VMM18 (HLA-A3) | VMM19 (HLA-A11) | VMM86 (HLA-A31) | VMM150 (HLA-A68) | DM14 (HLA-A68) | |||
| gp100 | ALLAVGATK | HLA-A3 | ++a [+]b | +++ [+] | + [+] | ++ [+] | + [−]c | + [+] |
| ALNFPGSQK | HLA-A3 | + [+] | NTd | + [+] | − [+] | − [−] | + [+] | |
| LIYRRRLMK | HLA-A3 | +++ [+] | ++ [+] | − [+] | − [+] | − [−] | − [+] | |
| NY-ESO-1 | AAQERRVPR | HLA-A31 | −[+] | NT | − [−] | − [+] | − [−] | − [−] |
| ASGPGGGAPR | HLA-A31 | + [+] | + [+] | − [−] | − [+] | − [−] | − [−] | |
a−, <1 copy per cell; +, one to ten copies per cell; ++, greater than ten copies per cell and equal to or less than 20 copies per cell; +++, greater than 20 copies per cell
b[+] Indicates that the cell line was positive by PCR for the expression of the corresponding tumor antigen gene
c[−] Indicates that the cell line was negative by PCR for the expression of the corresponding tumor antigen gene
dNT Not tested
Three different gp100-derived peptides known to be presented by HLA-A3, were targeted for analysis by SRM on A3 supertype family members. The ALLAVGATK peptide was detected on all four A3 supertype molecules tested, although it was expressed at significantly higher levels on HLA-A3 (VMM12) and HLA-A68 (VMM31) than it was on HLA-A11 (VMM19) and HLA-A68 (VMM150, DM14) (Table 4). Interestingly, the ALLAVGATK peptide could be detected among the HLA-A68–associated peptides obtained from the VMM150 melanoma tumor cell line, which is negative for the expression of gp100 by PCR. The ALNFPGSQK peptide was found at low levels on HLA-A3 (VMM12), HLA-A11 (VMM19), and HLA-A68 (DM14), but was not associated with HLA-A31 (VMM86) (Table 4). Finally, the LIYRRRLMK peptide was detected among HLA-A3–associated peptides, but not among the peptides obtained from the other A3 supertype family members (Table 4).
Two different NY-ESO-1–derived peptides known to be presented by HLA-A31 were also targeted for analysis by SRM analysis among HLA-A3 and HLA-A31–associated peptides obtained from the NY-ESO-1+ melanoma cell lines VMM12, VMM18, and VMM86 (Table 4). Although originally identified as HLA-A31–restricted antigens, neither of the peptides was found among the peptides obtained from VMM86. The inability to detect the peptides may be due to the low level of expression of HLA-A31 on VMM86 (data not shown) and/or the possibility that the melanoma cell line is heterogeneous for the expression of NY-ESO-1. The former issue will reduce the total number of HLA-A31/peptide complexes available on the cell surface, while the latter issue may result in only a low percentage of the cells expressing the protein. In either case, a consequence of the low level of expression is that the peptide copy number may drop below the threshold detection limit (~1 copy/cell). Importantly, however, the ASGPGGGAPR peptide was found among the peptides associated with HLA-A3 on both the VMM12 and VMM18 melanoma cell lines.
The utility of the SRM methodology for detecting the presence of selected peptides was further demonstrated by scanning for three additional melanoma peptide antigens among the HLA-A3–associated peptides obtained from the VMM12 and VMM18 melanoma cell lines (Table 5). The TRP-2–derived, HLA-A31–restricted peptide LLGPGRPYR, was found to associate with HLA-A3 obtained from the TRP-2+ (by PCR) melanoma tumor cell line VMM12. The TAG-derived, HLA-A3–restricted peptide RLSNRLLLR, and the HLA-A3–restricted peptide SQNFPGSQK derived from an as-yet-unidentified gene, were originally identified as antigens on VMM18 recognized by melanoma reactive CTLs [14, 15]. Both of these antigens were also detected among the HLA-A3–associated peptides obtained from the TAG+ VMM12 melanoma tumor cell line, thus demonstrating the shared nature of these peptide antigens.
Table 5.
Detection of selected A3 supertype–associated peptides among HLA-A3 peptide extracts obtained from two different melanoma cell lines
| Source protein | Peptide sequence | Known restriction element | Copy level of peptide per cell | |
|---|---|---|---|---|
| VMM12 (HLA-A3) | VMM18 (HLA-A3) | |||
| TAG | RLSNRLLLR | HLA-A3 | + [+]b | ++ [+] |
| Unknown | SQNFPGSQK | HLA-A3 | ++ [?]c | ++ [?] |
| TRP-2 | LLGPGRPYR | HLA-A31 | ++ [+] | NTd |
a+, One to ten copies per cell; ++, greater than ten copies per cell and equal to or less than 20 copies per cell
b[+] Indicates that the cell line was positive by PCR for the expression of the corresponding tumor antigen gene
c[?] Indicates that the tumor antigen is derived from an unknown source protein, and thus it is not possible to determine by PCR those cell lines expressing the corresponding tumor antigen gene
dNT not tested
Discussion
The phenomenon of MHC restriction poses an impediment to the design and testing of peptide-based vaccines that may be useful in the treatment of cancer and infectious diseases in humans. Most of the peptide antigens that have been discovered are associated with the most prevalent MHC class I molecules in the population, an attribute that makes it relatively easy to test these antigens in clinical trials. Peptide antigens associated with less prevalent MHC class I molecules in the population are difficult both to identify and to test because of the small numbers of individuals that express the MHC molecule of interest within a population. This in turn restricts access to clinical trials for a subset of patients, and specifically for certain ethnic and racial subpopulations. The potential utility of the supertype concept is that peptide antigens known to associate with prevalent MHC class I molecules in the population will also be useful in the treatment of individuals expressing rare MHC class I molecules of the corresponding supertype. Likewise, the usefulness of those peptide antigens known to associate with rare MHC class I molecules may be greatly expanded if the peptide is recognized in association with the more prevalent members of the supertype.
The most common way of determining if a peptide is presented by multiple members of a supertype is to first perform a binding assay to determine if the peptide is capable of binding to a given member of the supertype, to then determine if CTLs can be generated to the peptide when bound to the corresponding MHC class I molecule, and ultimately to determine if the CTLs recognize cells expressing both the relevant MHC class I molecule and the parent protein from which the peptide is derived [32]. It has been shown, however, that binding does not always correlate with the ability to induce a peptide-specific CTL response [2, 8, 18, 40], and such CTLs, even when generated, do not always recognize targets expressing the appropriate MHC class I molecule and precursor protein [18, 21, 41]. Thus, a substantial amount of work can be invested in peptides that ultimately are not naturally occurring epitopes. To overcome the limitations of this approach, we developed an alternative mass spectrometric approach that has the major advantage of being able to determine if a defined peptide is naturally processed and presented with a particular MHC class I molecule.
Determining if a particular peptide is associated with a given MHC class I molecule on the surface of a cell is a difficult analytical problem, as it has been estimated that greater than 10,000 different peptides are presented by a particular MHC class I molecule on the cell surface [11]. We have developed a new mass spectrometric approach involving SRM scanning that can be used in combination with RP-HPLC fractionation to search for predefined peptides associated with any MHC class I molecule. The sensitivity of the analytical approach described here requires a modest amount of cellular material and can readily be performed with peptides obtained from 2×109 cells. The approach is applicable to peptides derived from any MHC class I molecule, although the availability of monoclonal antibodies with specificity for HLA-A2/A69 (BB7.2), HLA-A2/A68/A69 (CR11-351), HLA-A3 (GAP-A3), and HLA-B7/B27 (ME1-1.2) simplifies the isolation of these particular MHC class I molecules. Sequential immunoaffinity purification with combinations of antibodies with broader specificities such as W6/32 (all MHC class I molecules), B1.23.2 (B-locus specific), and SFR8-B6 (Bw6) can be used either to selectively eliminate or to retain particular MHC class I molecules selectively as was done in this study (Fig. 1).
The SRM scanning makes use of a triple quadrupole mass spectrometer coupled with nRP-HPLC/μESI. When operated in SRM scanning mode, the triple quadrupole mass spectrometer is set so that the first quadrupole (Q1) passes only a predetermined precursor ion of a particular mass to charge (m/z) ratio into the second quadrupole (Q2) where it collides with argon molecules. The collisions result in dissociation of the precursor ion into product ions. Thus, the presence of a particular precursor ion is confirmed by only allowing product ions of a defined m/z ratio to pass from Q2 to the third quadrupole (Q3) where they are subsequently detected. Prior to subjecting the purified peptides to SRM scanning, they are first fractionated by RP-HPLC, which serves two purposes. First, it simplifies the mixture that will be scanned, and second, it serves as an additional identifying characteristic of the peptide being sought. The peptide can be synthesized and run on the same column as the test sample, thus allowing one to determine the fraction(s) in which the peptide of interest will elute. Additionally, each precursor ion will have a characteristic elution time from the nRP-HPLC/μESI source as it is being eluted into the mass spectrometer. Thus, four criteria must be fulfilled for a sample to test positive for the presence of a particular peptide: (1) the peptide must be detected in the same RP-HPLC fraction(s) as the synthetic equivalent; (2) the precursor ion in the sample must elute from nRP-HPLC/μESI source at the same time point as the synthetic peptide; (3) the characteristic product ions from the prerequisite precursor ion must be detected in Q3; and (4) the relative abundance of each of the product ions must be the same as found for the obtained from the synthetic peptide.
Through the use of an internal standard, SRM can also be used to provide quantitative information on a particular peptide, with the sensitivity of the method typically in the range of 0.1–1.0 copies of peptide per cell. The actual number of copies of peptide per cell is likely to be higher by a factor of at least two, as previous work has shown that the efficiency of peptide recovery following extraction and RP-HPLC separation is about 50% [7]. As CTLs have been shown to recognize target cells with as few as one copy of peptide on their surface [39], this detection is more than adequate to search for potential CTL epitopes. SRM scanning is also highly reproducible, with multiple runs of the same sample demonstrating similar results (Table 3). As the m/z ratios of both the precursor and product ions of the deuterated peptide ALLD10AVGD2ATK differ from those of the nondeuterated peptide, ALLAVGATK, the deuterated peptide can be used as a standard, even when searching for the ALLAVGATK peptide as both species can be discriminated on the mass spectrometer.
The gp100-derived peptide, ALLAVGATK, has previously been shown to be naturally processed and presented in association with HLA-A3, and melanoma tumor cells expressing this epitope are recognized and lysed by CTLs that are reactive with this epitope [37]. Binding assays demonstrated that ALLAVGATK binds to HLA-A3 and HLA-A11 with reasonably high affinity, but binds poorly or not at all to HLA-31 and HLA-A68 [19]. The results presented here demonstrate that ALLAVGATK is naturally bound to HLA-A3, HLA-A11, HLA-A31, and HLA-A68, with the highest copy per cell on HLA-A3 and HLA-A31. Interestingly, the binding affinity of the peptides for a given MHC class I molecule does not correlate with the copy number of peptides associated with that peptide. This could reflect differences in the copies of peptide within a cell that are available for binding, or it might reflect the fact that class I MHC molecules may play a role in directing the trimming of the N-terminal end of the peptide [35].
Although the melanoma tumor cell line VMM150 tested negative by PCR for the expression of the gp100 gene, the ALLAVGATK peptide (but not the ALNFPGSQK and LIYRRRLMK peptides) was nonetheless detected among the peptides associated with HLA-A68 isolated from the cell line. The gp100 mRNA results from the splicing of 11 exons, with the sequence in the first exon coding for the ALLAVGATK peptide, the third exon coding for the ALNFPGSQK peptide, and the tenth and 11th exons coding for the LIYRRRLMK peptide. Because the PCR primer set used for the detection of the gp100 gene targeted the second and fourth exons, a likely explanation for this result is that the gene has been aberrantly rearranged in the VMM150 tumor cells, such that the ALLAVGATK peptide, but not the ALNFPGSQK and LIYRRRLMK peptides, can still be expressed from the gene.
A second gp100-derived peptide, ALNFPGSQK, was found to be associated with HLA-A3, HLA-A11, and HLA-A68, but not with HLA-A31. This peptide was previously identified by first establishing that it bound with high affinity to HLA-A3, and then by establishing that it could be used for the primary, in vitro stimulation of CTLs that could in turn recognize HLA-A3+/gp100+ tumor cells [19]. In that same study, it was also shown that the peptide bound to HLA-A11 with high affinity, and bound poorly or not at all to HLA-A31 and HLA-A68. The present work confirms that the ALNFPGSQK peptide is naturally processed and presented in association with both HLA-A3 and HLA-A11.
The gp100-derived peptide LIYRRRLMK was originally identified by the genetic approach as an HLA-A3–restricted antigen recognized by melanoma tumor-infiltrating lymphocytes [17]. The identification by SRM scanning of this peptide among HLA-A3-associated peptides obtained from the melanoma cell lines VMM12 and VMM18 confirms that this peptide is processed and presented in association with HLA-A3. The peptide is not, however, processed and presented in association with HLA-A11, HLA-A31, or HLA-A68.
The NY-ESO-1–derived peptides, AAQERRVPR and ASGPGGGAPR, have previously been shown to be recognized by CTLs in association with HLA-A31 [46]. Among the A3 supertype melanoma cells that we had available for testing, only VMM12 (HLA-A3+), VMM18 (HLA-A3+), and VMM86 (HLA-A31+) expressed the NY-ESO-1 gene. SRM scanning detected both peptides in the peptide extracts obtained from HLA-A3, indicating that these peptides can be presented in association with at least one additional A3 supertype family member. Interestingly, neither peptide was detected in the HLA-A31 peptide extracts obtained from the NY-ESO-1+ melanoma tumor cell line VMM86. One possible explanation for this discrepancy is that while VMM86 tested positive by PCR for the expression of the NY-ESO-1 gene, it is possible that the line is heterogeneous in its expression of the gene, with a large proportion being negative for expression. If, for example, 1% of the cells in the line were positive for NY-ESO-1 expression and the positive cells had an average of ten copies per cell of peptide, the overall average would be 0.1 copies per cell and thus undetectable following purification. This problem can most easily be overcome by assessing protein expression in individual cells rather than gene expression in a population.
Overall, these results illustrate both commonality and disparity in the ability of A3 supertype–derived peptides to be presented in association with multiple members of the A3 supertype. The ALLAVGATK peptide was found to be naturally associated with all four members tested, the ALNFPGSQK peptide was found to be associated with only HLA-A3 and HLA-A11, and the LIYRRRLMK peptide was found to be associated with only HLA-A3. The AAQERRVPR, ASGPGGGAPR, LLGPGRPYR, and LLGPOGRPYR peptides, originally identified as HLA-A31–restricted antigens, were found to associate with HLA-A3 (the lack of HLA-A11+ or HLA-A68+ cell lines that also expressed NY-ESO-1 precludes the ability to make a statement about the ability of the first two peptides to associate with HLA-A11 or HLA-A68; SRM scanning for the third and fourth peptides was only performed with HLA-A3 peptide extracts). These results indicate that the members of the A3 supertype present similar, yet distinct subsets of peptides. In the absence of a substantially larger dataset, it is not yet possible to predict the members of the A3 supertype to which any given A3 supertype–associated peptide will bind.
In summary, we have developed a highly sensitive technique to search for defined MHC class I–associated peptides. This methodology can be used to determine if a known peptide antigen is naturally processed and presented with multiple MHC class I molecules within a supertype, and it can be used to determine if newly predicted epitopes are naturally processed and presented on any particular MHC class I molecule. Once it is established that a defined peptide is naturally processed and presented in association with a given MHC class I molecule, efforts can be made to determine if the particular peptide/MHC class I complex is recognized by CTLs. Because these latter efforts need only be directed against those antigens that are known to be expressed on a tumor, the need to perform peptide-binding experiments is eliminated and in vitro lymphocyte stimulation efforts can be focused only on those peptides which are biologically relevant.
Acknowledgements
The authors would like to thank Mitsu Fink, Julie Fitzgerald, and Chantel Tracy for their assistance with the cell culture and isolation of the peptides bound to the A3 supertype molecules. This work was supported by RO1CA90815 (KTH), K08CA91995 (KUC), RO1AI33993 (DFH), and RO1CA57653 (CLS).
References
- 1.Berger AE, Davis JE, Cresswell P. Monoclonal antibody to HLA-A3. Hybridoma. 1982;1:87. doi: 10.1089/hyb.1.1982.1.87. [DOI] [PubMed] [Google Scholar]
- 2.Bertoni R, Sidney J, Fowler P, Chesnut RW, Chisari FV, Sette A. Human histocompatibility leukocyte antigen-binding supermotifs predict broadly cross-reactive cytotoxic T lymphocyte responses in patients with acute hepatitis. J Clin Invest. 1997;100:503. doi: 10.1172/JCI119559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brichard V, Van Pel A, Wolfel T, Wolfel C, De Plaen E, Lethe B, Coulie P, Boon T. The tyrosinase gene codes for an antigen recognized by autologous cytolytic T lymphocytes on HLA-A2 melanomas. J Exp Med. 1993;178:489. doi: 10.1084/jem.178.2.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen YT, Scanlan MJ, Sahin U, Tureci O, Gure AO, Tsang S, Williamson B, Stockert E, Pfreundschuh M, Old LJ. A testicular antigen aberrantly expressed in human cancers detected by autologous antibody screening. Proc Natl Acad Sci U S A. 1997;94:1914. doi: 10.1073/pnas.94.5.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coulie PG, Brichard V, Van Pel A, Wolfel T, Schneider J, Traversari C, Mattei S, De Plaen E, Lurquin C, Szikora JP, et al. A new gene coding for a differentiation antigen recognized by autologous cytolytic T lymphocytes on HLA-A2 melanomas. J Exp Med. 1994;180:35. doi: 10.1084/jem.180.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cox AL, Skipper J, Chen Y, Henderson RA, Darrow TL, Shabanowitz J, Engelhard VH, Hunt DF, Slingluff CL., Jr Identification of a peptide recognized by five melanoma-specific human cytotoxic T cell lines. Science. 1994;264:716. doi: 10.1126/science.7513441. [DOI] [PubMed] [Google Scholar]
- 7.Crotzer VL, Christian RE, Brooks JM, Shabanowitz J, Settlage RE, Marto JA, White FM, Rickinson AB, Hunt DF, Engelhard VH. Immunodominance among EBV-derived epitopes restricted by HLA-B27 does not correlate with epitope abundance in EBV-transformed B-lymphoblastoid cell lines. J Immunol. 2000;164:6120. doi: 10.4049/jimmunol.164.12.6120. [DOI] [PubMed] [Google Scholar]
- 8.Doolan DL, Hoffman SL, Southwood S, Wentworth PA, Sidney J, Chesnut RW, Keogh E, Appella E, Nutman TB, Lal AA, Gordon DM, Oloo A, Sette A. Degenerate cytotoxic T cell epitopes from P. falciparum restricted by multiple HLA-A and HLA-B supertype alleles. Immunity. 1997;7:97. doi: 10.1016/S1074-7613(00)80513-0. [DOI] [PubMed] [Google Scholar]
- 9.Ellis SA, Taylor C, McMichael A. Recognition of HLA-B27 and related antigen by a monoclonal antibody. Hum Immunol. 1982;5:49. doi: 10.1016/0198-8859(82)90030-1. [DOI] [PubMed] [Google Scholar]
- 10.Engelhard VH. Structure of peptides associated with class I and class II MHC molecules. Annu Rev Immunol. 1994;12:181. doi: 10.1146/annurev.iy.12.040194.001145. [DOI] [PubMed] [Google Scholar]
- 11.Engelhard VH. Structure of peptides associated with MHC class I molecules. Curr Opin Immunol. 1994;6:13. doi: 10.1016/0952-7915(94)90028-0. [DOI] [PubMed] [Google Scholar]
- 12.Hendrickson RC, Skipper JC, Shabanowitz J, Slingluff CL Jr (1996) Use of tandem mass spectrometry for MHC ligand analysis. In: Lefkovits I (ed) Immunology methods manual, vol 2. Academic, New York, p 605
- 13.Hogan KT, Eisinger DP, Cupp SB, III, Lekstrom KJ, Deacon DD, Shabanowitz J, Hunt DF, Engelhard VH, Slingluff CL, Jr, Ross MM. The peptide recognized by HLA-A68.2-restricted, squamous cell carcinoma of the lung-specific cytotoxic T lymphocytes is derived from a mutated elongation factor 2 gene. Cancer Res. 1998;58:5144. [PubMed] [Google Scholar]
- 14.Hogan KT, Coppola MA, Gatlin CL, Thompson LW, Shabanowitz J, Hunt DF, Engelhard VH, Slingluff CL, Ross MM. Identification of a shared epitope recognized by melanoma-specific, HLA-A3-restricted cytotoxic T lymphocytes. Immunol Lett. 2003;90:131. doi: 10.1016/j.imlet.2003.08.003. [DOI] [PubMed] [Google Scholar]
- 15.Hogan KT, Coppola MA, Gatlin CL, Thompson LW, Shabanowitz J, Hunt DF, Engelhard VH, Ross MM, Slingluff CL. Identification of novel and widely expressed cancer/testis gene isoforms that elicit spontaneous cytotoxic T lymphocyte reactivity to melanoma. Cancer Res. 2004;64:1157. doi: 10.1158/0008-5472.can-03-2209. [DOI] [PubMed] [Google Scholar]
- 16.Kawakami Y, Eliyahu S, Delgado CH, Robbins PF, Rivoltini L, Topalian SL, Miki T, Rosenberg SA. Cloning of the gene coding for a shared human melanoma antigen recognized by autologous T cells infiltrating into tumor. Proc Natl Acad Sci U S A. 1994;91:3515. doi: 10.1073/pnas.91.9.3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kawakami Y, Robbins PF, Wang X, Tupesis JP, Parkhurst MR, Kang X, Sakaguchi K, Appella E, Rosenberg SA. Identification of new melanoma epitopes on melanosomal proteins recognized by tumor infiltrating T lymphocytes restricted by HLA-A1, -A2, and -A3 alleles. J Immunol. 1998;161:6985. [PubMed] [Google Scholar]
- 18.Kawashima I, Hudson SJ, Tsai V, Southwood S, Takesako K, Appella E, Sette A, Celis E. The multi-epitope approach for immunotherapy for cancer: identification of several CTL epitopes from various tumor-associated antigens expressed on solid epithelial tumors. Hum Immunol. 1998;59:1. doi: 10.1016/S0198-8859(97)00255-3. [DOI] [PubMed] [Google Scholar]
- 19.Kawashima I, Tsai V, Southwood S, Takesako K, Celis E, Sette A. Identification of gp100-derived, melanoma-specific cytotoxic T-lymphocyte epitopes restricted by HLA-A3 supertype molecules by primary in vitro immunization with peptide-pulsed dendritic cells. Int J Cancer. 1998;78:518. doi: 10.1002/(SICI)1097-0215(19981109)78:4<518::AID-IJC20>3.3.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 20.Kawashima I, Tsai V, Southwood S, Takesako K, Sette A, Celis E. Identification of HLA-A3-restricted cytotoxic T lymphocyte epitopes from carcinoembryonic antigen and HER-2/neu by primary in vitro immunization with peptide-pulsed dendritic cells. Cancer Res. 1999;59:431. [PubMed] [Google Scholar]
- 21.Keogh E, Fikes J, Southwood S, Celis E, Chesnut R, Sette A. Identification of new epitopes from four different tumor-associated antigens: recognition of naturally processed epitopes correlates with HLA-A*0201-binding affinity. J Immunol. 2001;167:787. doi: 10.4049/jimmunol.167.2.787. [DOI] [PubMed] [Google Scholar]
- 22.Lapatsanis L, Milias G, Froussios K, Kolovos M. Synthesis of N-2,2,2-(trichloroethoxycarbonyl)-L-amino acids and N-(9-Fluorenylmethoxycarbonyl)-L-amino acids involving succinimidoxy anion as a leading group in amino acid protection. Synthesis. 1983;8:671. doi: 10.1055/s-1983-30468. [DOI] [Google Scholar]
- 23.Martin SE, Shabanowitz J, Hunt DF, Marto JA. Subfemtomole MS and MS/MS peptide sequence analysis using nano-HPLC micro-ESI fourier transform ion cyclotron resonance mass spectrometry. Anal Chem. 2000;72:4266. doi: 10.1021/ac000497v. [DOI] [PubMed] [Google Scholar]
- 24.Parham P, Brodsky FM. Partial purification and some properties of BB7.2. A cytotoxic monoclonal antibody with specificity for HLA-A2 and a variant of HLA-A28. Hum Immunol. 1981;3:277. doi: 10.1016/0198-8859(81)90065-3. [DOI] [PubMed] [Google Scholar]
- 25.Parham P, Barnstable CJ, Bodmer WF. Use of a monoclonal antibody (W6/32) in structural studies of HLA-A,B,C, antigens. J Immunol. 1979;123:342. [PubMed] [Google Scholar]
- 26.Parkhurst MR, Salgaller ML, Southwood S, Robbins PF, Sette A, Rosenberg SA, Kawakami Y. Improved induction of melanoma-reactive CTL with peptides from the melanoma antigen gp100 modified at HLA-A*0201-binding residues. J Immunol. 1996;157:2539. [PubMed] [Google Scholar]
- 27.Radka SF, Kostyu DD, Amos DB. A monoclonal antibody directed against the HLA-Bw6 epitope. J Immunol. 1982;128:2804. [PubMed] [Google Scholar]
- 28.Rebai N, Malissen B. Structural and genetic analyses of HLA class I molecules using monoclonal xenoantibodies. Tissue Antigens. 1983;22:107. doi: 10.1111/j.1399-0039.1983.tb01176.x. [DOI] [PubMed] [Google Scholar]
- 29.Renkvist N, Castelli C, Robbins PF, Parmiani G. A listing of human tumor antigens recognized by T cells. Cancer Immunol Immunother. 2001;50:3. doi: 10.1007/s002620000169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Russo C, Ng AK, Pellegrino MA, Ferrone S. The monoclonal antibody CR11-351 discriminates HLA-A2 variants identified by T cells. Immunogenetics. 1983;18:23. doi: 10.1007/BF00401353. [DOI] [PubMed] [Google Scholar]
- 31.Sahin U, Tureci O, Schmitt H, Cochlovius B, Johannes T, Schmits R, Stenner F, Luo G, Schobert I, Pfreundschuh M. Human neoplasms elicit multiple specific immune responses in the autologous host. Proc Natl Acad Sci U S A. 1995;92:11810. doi: 10.1073/pnas.92.25.11810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sette A, Sidney J. HLA supertypes and supermotifs: a functional perspective on HLA polymorphism. Curr Opin Immunol. 1998;10:478. doi: 10.1016/S0952-7915(98)80124-6. [DOI] [PubMed] [Google Scholar]
- 33.Sette A, Sidney J. Nine major HLA class I supertypes account for the vast preponderance of HLA-A and -B polymorphism. Immunogenetics. 1999;50:201. doi: 10.1007/s002510050594. [DOI] [PubMed] [Google Scholar]
- 34.Sette A, Newman M, Livingston B, McKinney D, Sidney J, Ishioka G, Tangri S, Alexander J, Fikes J, Chesnut R. Optimizing vaccine design for cellular processing, MHC binding and TCR recognition. Tissue Antigens. 2002;59:443. doi: 10.1034/j.1399-0039.2002.590601.x. [DOI] [PubMed] [Google Scholar]
- 35.Shastri N, Schwab S, Serwold T. Producing nature’s gene-chips: the generation of peptides for display by MHC class I molecules. Annu Rev Immunol. 2002;20:463. doi: 10.1146/annurev.immunol.20.100301.064819. [DOI] [PubMed] [Google Scholar]
- 36.Sidney J, Grey HM, Kubo RT, Sette A. Practical, biochemical and evolutionary implications of the discovery of HLA class I supermotifs. Immunol Today. 1996;17:261. doi: 10.1016/0167-5699(96)80542-1. [DOI] [PubMed] [Google Scholar]
- 37.Skipper JC, Kittlesen DJ, Hendrickson RC, Deacon DD, Harthun NL, Wagner SN, Hunt DF, Engelhard VH, Slingluff CL., Jr. Shared epitopes for HLA-A3-restricted melanoma-reactive human CTL include a naturally processed epitope from Pmel-17/gp100. J Immunol. 1996;157:5027. [PubMed] [Google Scholar]
- 38.Slingluff CL, Jr, Colella TA, Thompson L, Graham DD, Skipper JC, Caldwell J, Brinckerhoff L, Kittlesen DJ, Deacon DH, Oei C, Harthun NL, Huczko EL, Hunt DF, Darrow TL, Engelhard VH. Melanomas with concordant loss of multiple melanocytic differentiation proteins: immune escape that may be overcome by targeting unique or undefined antigens. Cancer Immunol Immunother. 2000;48:661. doi: 10.1007/s002620050015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sykulev Y, Joo M, Vturina I, Tsomides TJ, Eisen HN. Evidence that a single peptide-MHC complex on a target cell can elicit a cytolytic T cell response. Immunity. 1996;4:565. doi: 10.1016/S1074-7613(00)80483-5. [DOI] [PubMed] [Google Scholar]
- 40.Threlkeld SC, Wentworth PA, Kalams SA, Wilkes BM, Ruhl DJ, Keogh E, Sidney J, Southwood S, Walker BD, Sette A. Degenerate and promiscuous recognition by CTL of peptides presented by the MHC class I A3-like superfamily: implications for vaccine development. J Immunol. 1997;159:1648. [PubMed] [Google Scholar]
- 41.Tsai V, Southwood S, Sidney J, Sakaguchi K, Kawakami Y, Appella E, Sette A, Celis E. Identification of subdominant CTL epitopes of the GP100 melanoma-associated tumor antigen by primary in vitro immunization with peptide-pulsed dendritic cells. J Immunol. 1997;158:1796. [PubMed] [Google Scholar]
- 42.Tureci O, Sahin U, Schobert I, Koslowski M, Scmitt H, Schild HJ, Stenner F, Seitz G, Rammensee HG, Pfreundschuh M. The SSX-2 gene, which is involved in the t(X;18) translocation of synovial sarcomas, codes for the human tumor antigen HOM-MEL-40. Cancer Res. 1996;56:4766. [PubMed] [Google Scholar]
- 43.van der Bruggen P, Traversari C, Chomez P, Lurquin C, De Plaen E, Van den Eynde B, Knuth A, Boon T. A gene encoding an antigen recognized by cytolytic T lymphocytes on a human melanoma. Science. 1991;254:1643. doi: 10.1126/science.1840703. [DOI] [PubMed] [Google Scholar]
- 44.Wang RF, Rosenberg SA. Human tumor antigens for cancer vaccine development. Immunol Rev. 1999;170:85. doi: 10.1111/j.1600-065x.1999.tb01331.x. [DOI] [PubMed] [Google Scholar]
- 45.Wang RF, Appella E, Kawakami Y, Kang X, Rosenberg SA. Identification of TRP-2 as a human tumor antigen recognized by cytotoxic T lymphocytes. J Exp Med. 1996;184:2207. doi: 10.1084/jem.184.6.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang RF, Johnston SL, Zeng G, Topalian SL, Schwartzentruber DJ, Rosenberg SA. A breast and melanoma-shared tumor antigen: T cell responses to antigenic peptides translated from different open reading frames. J Immunol. 1998;161:3598. [PubMed] [Google Scholar]