Abstract
The tetraspanins form a family of about 30 molecules mainly expressed on the cell surface. They have been reported to be involved in many physiological or pathological processes, such as fertilization, immune response, development of the nervous system, and metastasis, as well as in infectious diseases (HCV, malaria, etc.). The tetraspanins may play a role as “organizers” of multimolecular complexes on the cell surface associating numerous proteins, the “tetraspanin web.” To better define the composition of the tetraspanin web, its characterization has been recently performed using mass spectrometry and proteomics. We report the proteomic analysis of tetraspanin complexes on B-lymphoid cells. Immunoprecipitation experiments were performed using mAbs directed against the tetraspanin CD9, and associated molecules were identified by MALDI-TOF (matrix assisted laser desorption ionization time of flight) mass spectrometry. This led to the identification of IgM as a novel component of the complexes. Thus, tetraspanins may connect several types of proteins with Ig domains, including HLA-DR, EWI-2, and IgM, that may play a role in immune responses.
Keywords: Tetraspanin, Immunoglobulin, Proteomics, Mass spectrometry, Immune response
Introduction
The first tetraspanin was reported in 1981 as an antigen expressed on acute lymphoblastic leukemia cells and named CD9 at the first workshop on leukocyte differentiation antigens (Paris, 1982). In the beginning of the 1990s, the cloning of four of such antigens in the same period—CD9, Me491/CD63, TAPA1/CD81, and Sm23 (a schistosome molecule)—established their membership within a family of molecules to which the name of tetraspanins was later given. Indeed, these molecules exhibited similar structure and significant homology suggesting the existence of a novel molecular family. The prediction of structure suggested that these molecules had four transmembrane domains, two extracellular domains of unequal size, and short cytoplasmic ends. The existence of a particular structure in the large extracellular domain reported for CD81 and predictable for the other tetraspanins reinforced this notion of molecular family. In this report, we focus on tetraspanins in the immune response. With respect to the tetraspanin web, a molecular network that these proteins might organize, we describe IgM as a novel component on B-lymphoid cells.
Tetraspanins in the immune response
The tetraspanins are cell surface proteins involved in an astonishing variety of biologic responses. Some of them have been cloned independently, several times for different functional effects and reported to influence adhesion, morphology, activation, proliferation and differentiation of B, T, and other cells [1, 18]. The tetraspanin TAPA1/CD81 was initially discovered on lymphomas in a screening for mAbs able to inhibit cellular proliferation (TAPA1: “target of antiproliferative antibody”) [23]. On B cells, CD81 is part of a complex with CD21, CD19 and Leu13. This complex reduces the threshold (by two orders of magnitude) for B-cell activation via the B-cell receptor by bridging Ag-specific recognition and CD21-mediated complement recognition. Similarly, on T cells, CD81 associates with CD4 and CD8 and provides a costimulatory signal with CD3. The consequences of the absence of CD81 on the immune system were investigated using knockout mice. CD81 knockout leads to a reduction of expression of the B-cell antigen CD19 associated with a decrease of calcium mobilization following CD19 engagement [20, 22, 30]. Two groups observed a reduction of B1 cells in the peritoneum, while Th2-dependent IgG1 production also appeared to be reduced. On the other hand, it has been shown that signaling through CD81 on T cells costimulates both Th1 and Th2 cells, but increases the number of Th2 cells during long-term activation [19]. Finally, a critical role for CD81 in cognate T-B cell interactions leading to Th2 responses has been suggested [9]. Other tetraspanins have been suggested to be involved in the immune system. Indeed, a costimulatory effect on T CD4+ has also been observed with the tetraspanin CD82 and CD3 antibodies [1, 16]. Mice deficient in CD37, a tetraspanin of mature B cells, exhibited a reduced humoral response to T-dependent antigens, suggesting a role for CD37 in mediating B- and T-cell interactions [14].
The tetraspanin web
The biochemical analysis performed on tetraspanins opened a new field of investigation on the molecular organization of the membrane by showing that the tetraspanins formed surface multimolecular complexes. These complexes include various molecules such as CD2, CD3, CD4, CD8, CD19, MHC class I and II, integrins, and growth factor receptors as well as tetraspanins themselves [1]. These molecular complexes are based on the assembly of primary complexes (tetraspanin/partner-specific pairs), which may interact with tetraspanins, thus building larger complexes [4, 24, 25]. The data suggested a model in which the tetraspanins would be “organizers” of multimolecular complexes on the cell surface, each tetraspanin recruiting specifically one or more molecular partners into these complexes; this led to the unifying concept of a network of molecular interactions at the cell surface also called the “tetraspanin web” [24]. In the tetraspanin web, the interaction of tetraspanins with each other seems to be central. It has been recently shown that the stabilization of interaction between tetraspanins involves several levels such as palmitoylation of tetraspanins, divalent cations, and cholesterol [3, 4, 6, 28, 33]. Furthermore, the tetraspanins were also found to partition into a detergent-resistant membrane environment; they may also organize novel types of microdomains in the plasma membrane similar to lipid rafts [3, 7]. Therefore, the characterization of these microdomains in term of composition, organization and functional relevance is a major goal.
Proteomic analysis of the tetraspanin web
In order to better define the composition of the tetraspanin web, which contains a lot of proteins still not identified, we characterized the complexes using mass spectrometry. A first strategy was to produce mAbs to proteins present in the complexes. Mice were immunized with tetraspanin complexes, and several monoclonal antibodies were obtained and screened following immunoprecipitation experiments. The antibodies directed against tetraspanin-associated molecules were then selected. Identification of the target was possible following affinity purification using antibodies and mass spectrometry. This methodology has been applied for the identification of the specific partner of CD9 also called CD9P-1 (CD9 Partner 1). An antibody directed against this 135-kDa protein was obtained and the MALDI-TOF mass spectrometry analysis of tryptic peptides from the purified protein led to the identification of the KIAA1436 protein which belongs to a novel subfamily of proteins with Ig domains [2]. Its function is still unknown. A second approach involved isolating tetraspanin complexes after cell lysis with mild detergent and immunoprecipitation experiments, followed by identification of associated molecules using mass spectrometry devices. This approach, using liquid chromatography coupled with tandem mass spectrometry on an ion trap (LC-MS/MS) followed by search in an EST database, led to the identification of human FPRP which is identical to KIAA1436/CD9P-1 [26]. Another protein has also been discovered in tetraspanin complexes by several groups using proteomics. This protein has been identified as EWI-2 or PGRL in different cell lines such as teratocarcinoma cells, T- or B-lymphoid cells [5, 8, 27, 34]. Interestingly, EWI-2 belongs to the same subfamily as CD9P-1 and has been recently reported as a specific partner of CD9 and CD81 [5].
Identification of IgM in CD9-containing complexes
To illustrate and exemplify proteomic analysis of tetraspanin complexes, the identification of IgM as a novel component of the tetraspanin web in B-lymphoid cells is described below. The lymphoid B cell line Daudi (which does not express CD9) and a transfectant Daudi/CD9 were used. Immunoprecipitation experiments were performed using CD9 monoclonal antibodies. CD9 immunoprecipitates collected from Brij 98 extracts of wild-type and CD9-transfected Daudi cells were compared after silver staining (Fig. 1). This permitted the detection of proteins specifically coimmunoprecipitated with CD9 from nonspecific bands that cannot be eliminated by the use of “mild” detergents required to preserve the tetraspanin web. Three major bands specifically detected in the CD9 immunoprecipitate collected from Daudi/CD9 cells were observed. The bands were cut out from the gel and digested with trypsin. The resulting peptides were analyzed by MALDI-TOF mass spectrometry, and the spectrum was used for protein identification in the NCBI database using the MS-FIT search program. We have recently reported the identification of two of these proteins as HLA-DR and EWI-2 [5]. The band exhibiting a high molecular weight was identified as IgM. Indeed, height peptides were consistent with those derived from human IgM covering 18% of the amino acid sequence (Table 1). The presence of IgM in tetraspanin complexes was observed after immunoprecipitation experiments. A band corresponding to the molecular weight of IgM in Daudi cells was observed in tetraspanin complexes following cell surface labeling and immunoprecipitation experiments using mAbs directed against CD53 or CD81 whereas IgM was not detected in CD55 immunoprecipitates. In addition, IgM could be reimmunoprecipitated from tetraspanin immunoprecipitates confirming the association of IgM with tetraspanins in B-lymphoid cells (Fig. 2).
Fig. 1.
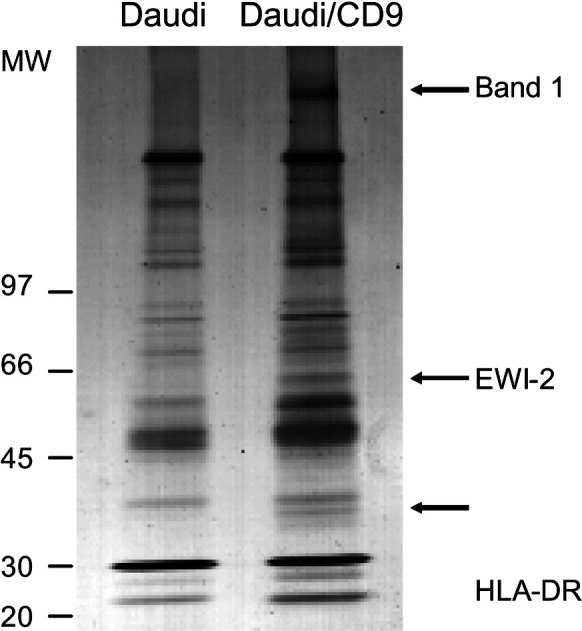
A high molecular weight molecule is a major band detected by silver staining in CD9 immunoprecipitates of Daudi/CD9 cells. CD9-associated proteins collected from Daudi/CD9 cells (8×108 cells) after lysis with the mild detergent Brij98 and immunoprecipitation experiments using sepharose beads coupled to mAb Alb-6 (CD9). The proteins were fractionated by SDS-PAGE under nonreducing conditions and visualized by silver-staining as previously described [5]. To discriminate specifically immunoprecipitated proteins from nonspecific bands, the same procedure was applied to parental CD9-negative Daudi cells
Table 1.
Assignment of peptide masses to the human IgM sequence. Mass spectrometric analyses were performed using a Voyager-DE-STR MALDI-TOF mass spectrometer (PerSeptive Biosystems, Framingham, MA), operated in the delayed extraction mode. Peptide mixtures were analyzed using a saturated solution of 2,5-dihydroxybenzoic acid (DHB) (Aldrich, St Quentin, France) in 10% acetonitrile containing 1% trifluoro-acetic acid (Sigma, St Louis, MO). Peptides were selected in the mass range of 600–4,000 Da. The search program MS-Fit, developed by the University of California at San Francisco (http://prospector.ucsf.edu/), was used for searches in the NCBI database. Typical search parameters were as follows: maximum allowed peptide mass error 150 ppm, consideration of two incomplete cleavages per peptide, MW±50% and full pH range
| M/z submitted | MH+ matched | Delta (ppm) | Residues | Peptide sequence |
|---|---|---|---|---|
| 775.4208 | 775.4466 | −33.3337 | 55–61 | GFPSVLR |
| 900.5354 | 900.5307 | 5.2052 | 113–120 | VSVFVPPR |
| 909.4160 | 909.4219 | −6.4770 | 121–128 | DGFFGNPR |
| 1029.5659 | 1029.5845 | −18.1034 | 143–150 | QIQVSWLR |
| 1249.6267 | 1249.6363 | −7.6955 | 132–142 | LICQATGFSPR |
| 1386.7866 | 1386.8361 | −35.6655 | 100–112 | NVPLPVIAELPPK |
| 1600.7904 | 1600.7794 | 6.9016 | 377–391 | YVTSAPMPEPQAPGR |
| 1773.9787 | 1774.0128 | −19.2207 | 323–338 | GVALHRPDVYLLPPAR |
Fig. 2.
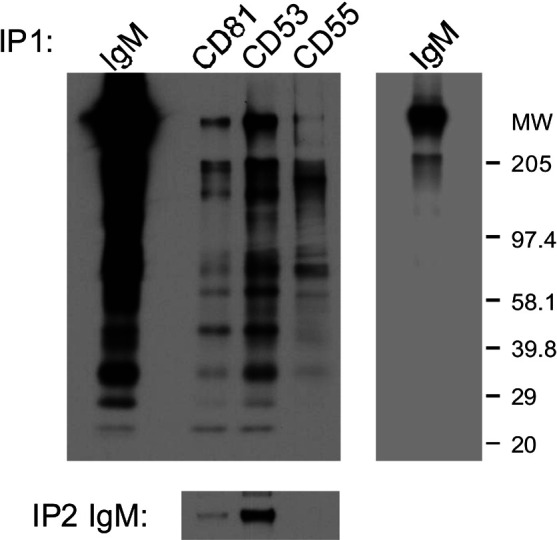
IgM associates with tetraspanins. The proteins on the cell surface of B-lymphoid Daudi cells were labeled with EZ-link-Sulfo-NHS-LC-biotin (Pierce, Rockford, IL). The cells were lysed using the mild detergents Brij98 or Brij97, and immunoprecipitation experiments were performed using mAbs directed against human IgM, the tetraspanins CD9 (Alb-6), CD81 (TS81), CD53 (TS53), or the cell surface molecule CD55 (12A12). The immunoprecipitates were separated by 5–15% SDS-polyacrylamide gel electrophoresis under nonreducing conditions and transferred to a PVDF membrane (Amersham). Blotting on immunoprecipitates was performed using a streptavidin-biotinylated horseradish peroxidase complex (Amersham), which was revealed by enhanced chemiluminescence (NEN, Boston, MA). A band corresponding to the molecular weight of IgM in Daudi cells was observed in tetraspanin complexes (CD53 or CD81) whereas IgM was not detected in the CD55 immunoprecipitate. In addition, IgM could be reimmunoprecipitated from tretraspanin immunoprecipitates confirming the association of IgM with tetraspanins in B-lymphoid cells
Tetraspanins and antigen presentation
The association of MHC class II with tetraspanin complexes and their partitioning in detergent-resistant membrane domains suggest that these proteins may play a role in antigen presentation by compartimentalization and positioning of associated molecules and MHC class II. In dendritic cells (DCs), the most potent antigen-presenting cells, the high capacity of dendritic cells to stimulate T lymphocytes is related to their ability to regulate the distribution of MHC class II molecules. It has been observed that the tetraspanin CD63 associates with peptide-loaded class II molecules intracellularly, whereas the tetraspanins CD9, CD53 and CD81 associate with class II molecules at the plasma membrane. Therefore, selective association of distinct tetraspanins may be involved in the regulation of MHC class II distribution in human dendritic cells [10]. Furthermore, the tetraspanin CD63 was modified posttranslationally during maturation of DCs. This modification was accompanied by a change in morphology of MHC class II compartments suggesting that CD63 may be involved in the functional and morphological changes of MHC class II compartments that occur during DC maturation [11]. It has recently been suggested that microdomains made up of tetraspanin proteins, such as CD9, CD63, CD81, or CD82, may mediate enrichment of MHC class II molecules loaded with a selected set of peptides [15, 31]. In addition, antigen-presenting cells deficient in tetraspaned microdomains have a reduced capacity to activate CD4+ T cells. Thus, the organization of uniformly loaded peptide–MHC class II complexes in tetraspaned domains may be a very early event that determines both the composition of the immunological synapse and the quality of the subsequent T-helper cell response [15]. Furthermore, the dynamic redistribution of tetraspanin CD81 at the central zone of the immune synapse in both T lymphocytes and APCs (antigen-presenting cells) suggests a relevant role for CD81 in the topography of the immune synapse that would explain its functional implication in T cell–B cell collaboration [21].
Interestingly, it has been observed that tetraspanins are enriched in exosomes of different origin. Indeed, tetraspanins CD37, CD53, CD63, CD81 and CD82 have been located in exosomes from B-lymphoid cells as well as from dendritic cells [12, 35]. Exosomes are 50- to 90-nm-diameter vesicles secreted from multivesicular bodies (MVBs) found in a variety of both hematopoietic and tumor cells. Exosomes originating from DCs have been shown to have immunotherapeutic properties through their presentation of biologically relevant antigens and are being developed as an alternative to cellular therapies [17, 35]. These particles contain antigen-presenting molecules (MHC class I, MHC class II, and CD1), tetraspanin molecules (CD9, CD63, CD81), adhesion molecules and costimulatory molecules (CD86) [17, 29]. Exosomes were also shown to be enriched in cholesterol, sphingomyelin and ganglioside GM3, lipids that are typically enriched in detergent-resistant membranes, and the recruitment of membrane proteins from the limiting membranes into the internal vesicles of multivesicular bodies may involve their incorporation into tetraspanin-containing detergent-resistant membrane domains [32]. Finally, exosomes have been proposed to play a role in targeting antigen-presenting cells and it has been suggested that these vesicles provide them the necessary machinery required for generating a potent immune response [13, 17, 29].
Concluding remarks
The characterization of the tetraspanin web is a major goal to better define the function of tetraspanins. Proteomics and mass spectrometry led to the identification of several types of proteins with Ig domains including HLA-DR, EWI-2, CD9P-1 and IgM. The tetraspanin web may connect these molecules with each other and thereby open new insights into antigen uptake and presentation.
Footnotes
This work was presented at the first Cancer Immunology and Immunotherapy Summer School, 8–13 September 2003, Ionian Village, Bartholomeio, Peloponnese, Greece.
References
- 1.Boucheix Cell Mol Life Sci. 2001;58:1189. doi: 10.1007/PL00000933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Charrin J Biol Chem. 2001;276:14329. doi: 10.1074/jbc.M011297200. [DOI] [PubMed] [Google Scholar]
- 3.Charrin FEBS Lett. 2002;516:139. doi: 10.1016/S0014-5793(02)02522-X. [DOI] [PubMed] [Google Scholar]
- 4.Charrin Biochem Biophys Res Commun. 2003;304:107. doi: 10.1016/S0006-291X(03)00545-X. [DOI] [PubMed] [Google Scholar]
- 5.Charrin S, Le Naour F, Billard M, Labas V, Le Caer JP, Emile JF, Petit MA, Boucheix C, Rubinstein E. EWI-2 is a new component of the tetraspanin web in hepatocytes and lymphoid cells. Biochem. J. 2003;373:409–421. doi: 10.1042/BJ20030343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Charrin Eur J Immunol. 2003;33:2479. doi: 10.1002/eji.200323884. [DOI] [PubMed] [Google Scholar]
- 7.Claas J Biol Chem. 2001;276:7974. doi: 10.1074/jbc.M008650200. [DOI] [PubMed] [Google Scholar]
- 8.Clark J Immunol. 2001;167:5115. doi: 10.4049/jimmunol.167.9.5115. [DOI] [PubMed] [Google Scholar]
- 9.Deng Int Immunol. 2002;14:513. doi: 10.1093/intimm/14.5.513. [DOI] [PubMed] [Google Scholar]
- 10.Engering Int Immunol. 2001;13:127. doi: 10.1093/intimm/13.2.127. [DOI] [PubMed] [Google Scholar]
- 11.Engering Eur J Biochem. 2003;270:2412. doi: 10.1046/j.1432-1033.2003.03609.x. [DOI] [PubMed] [Google Scholar]
- 12.Escola J Biol Chem. 1998;273:20121. doi: 10.1074/jbc.273.32.20121. [DOI] [PubMed] [Google Scholar]
- 13.Fritzsching J Immunol. 2002;169:5531. doi: 10.4049/jimmunol.169.10.5531. [DOI] [PubMed] [Google Scholar]
- 14.Knobeloch Mol Cell Biol. 2000;20:5363. doi: 10.1128/MCB.20.15.5363-5369.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kropshofer Nat Immunol. 2002;3:61. doi: 10.1038/ni750. [DOI] [PubMed] [Google Scholar]
- 16.Lagaudrière-Gesbert Cellular Immunol. 1997;182:105. doi: 10.1006/cimm.1997.1223. [DOI] [PubMed] [Google Scholar]
- 17.Lamparski J Immunol Methods. 2002;270:211. doi: 10.1016/s0022-1759(02)00330-7. [DOI] [PubMed] [Google Scholar]
- 18.Levy Annu Rev Immunol. 1998;16:89. doi: 10.1146/annurev.immunol.16.1.89. [DOI] [PubMed] [Google Scholar]
- 19.Maecker BMC Immunol. 2003;4:1. doi: 10.1186/1471-2172-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maecker J Exp Med. 1997;185:1505. doi: 10.1084/jem.185.8.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mittelbrunn J Immunol. 2002;169:6691. doi: 10.4049/jimmunol.169.12.6691. [DOI] [PubMed] [Google Scholar]
- 22.Miyazaki EMBO J. 1997;16:4217. doi: 10.1093/emboj/16.14.4217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oren Mol Cell Biol. 1990;10:4007. doi: 10.1128/mcb.10.8.4007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rubinstein Eur J Immunol. 1996;26:2657. doi: 10.1002/eji.1830261117. [DOI] [PubMed] [Google Scholar]
- 25.Serru Biochem J. 1999;340:103. doi: 10.1042/0264-6021:3400103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stipp J Biol Chem. 2001;276:4853. doi: 10.1074/jbc.M009859200. [DOI] [PubMed] [Google Scholar]
- 27.Stipp J Biol Chem. 2001;276:40545. doi: 10.1074/jbc.M107338200. [DOI] [PubMed] [Google Scholar]
- 28.Stipp Trends Biochem Sci. 2003;28:106. doi: 10.1016/S0968-0004(02)00014-2. [DOI] [PubMed] [Google Scholar]
- 29.Thery J Cell Biol. 1999;147:599. doi: 10.1083/jcb.147.3.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsitsikov Proc Natl Acad Sci U S A. 1997;94:10844. doi: 10.1073/pnas.94.20.10844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vogt Immunol Rev. 2002;189:136. doi: 10.1034/j.1600-065X.2002.18912.x. [DOI] [PubMed] [Google Scholar]
- 32.Wubbolts J Biol Chem. 2003;278:10963. doi: 10.1074/jbc.M207550200. [DOI] [PubMed] [Google Scholar]
- 33.Yang Mol Biol Cell. 2002;13:767. doi: 10.1091/mbc.01-05-0275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Cancer Res. 2003;200363:2665. [PubMed] [Google Scholar]
- 35.Zitvogel Nat Med. 1998;4:594. doi: 10.1038/nm0598-594. [DOI] [PubMed] [Google Scholar]


