Abstract
“Cancer-germline” genes such as those of the MAGE family are expressed in many tumors and in male germline cells, but are silent in normal tissues. They encode shared tumor-specific antigens that have been used in therapeutic vaccination trials of cancer patients. It was previously demonstrated that MAGE-1 peptide KVLEYVIKV was presented by HLA-A 0201 molecules on the surface of a human breast carcinoma cell line, but no human specific CTL had been isolated so far. Here, we have used HLA-A2/MAGE-1 fluorescent multimers to isolate from blood cells three human CTL clones that recognized the MAGE-1 peptide. These clones killed efficiently HLA-A2 tumor cells expressing MAGE-1, whether or not they were treated with IFN-γ, suggesting that the MAGE-1 antigen is processed efficiently by both the standard proteasome and the immunoproteasome. These results indicate that the MAGE-1.A2 peptide can be used for antitumoral vaccination.
Keywords: CTL, HLA-A2, MAGE-1, Peptide, Tumor
Introduction
Cancer-germline genes potentially code for shared tumor-specific antigens recognized by T cells. An important subset of these genes is the MAGE gene family, which comprises 18 functional genes located on chromosome X [6, 8, 19, 22, 38]. These genes are expressed in many human tumors of different histological types. They are silent in normal cells with the exception of male germline cells, which do not express MHC class I molecules and, therefore, cannot present antigens to cytolytic T lymphocytes (CTL) [12]. It is most likely that the presently identified tumor antigenic peptides encoded by cancer-germline genes constitute only a small subset of those that are encoded by these sequences. The identification of an additional set of antigenic peptides could be useful for the development of new vaccines available for a large set of patients. It will also facilitate the design of vaccines comprising several antigens, which could increase the primary anti-tumor efficacy of the vaccine.
We and others have therefore tried to identify antigenic peptides using the coding sequences as a starting point [27, 31, 40]. One of the approaches to find HLA class I-restricted peptides presented at the surface of tumor cells involves the screening of a protein sequence for the presence of peptides bearing consensus anchor residues, i.e. preferred residues located at the anchoring points of the peptide in the groove of the HLA molecule. The affinity of these peptides is then evaluated and the good HLA binders are used to stimulate blood lymphocytes from donors, with the aim of identifying CTL that lyse target cells pulsed with the peptide. One of the drawbacks is that, sometimes, these peptide-specific CTL prove to be incapable of lysing tumor cells that express the encoding gene because the peptide is processed inefficiently. To reduce this risk, the group of H.–G. Rammensee has introduced an improved approach [29]. Potential antigenic peptides are predicted from the sequences of known proteins and the corresponding synthetic peptides are tested for binding. In parallel, natural peptides are extracted from tumor tissue or tumor cell lines. Candidate antigenic peptides are selected if they co-elute with a natural peptide in high performance liquid chromatography and if the co-eluted natural peptide proves to have the correct sequence as seen in on-line tandem mass spectrometry. Using this approach, they demonstrated that peptide KVLEYVIKV, corresponding to amino acids 278-286 of the MAGE–A1 protein, which will be referred to hereafter as MAGE-1, was presented on the surface of a human breast carcinoma cell line by HLA-A 0201 molecules [26]. The peptide was injected into HLA-A*0201 transgenic mice and a peptide-specific CTL line was isolated. The murine CTL produced IFN-γ upon contact with MAGE-1 transfected cells and lysed HLA-A2 tumor cells pulsed with the peptide. But only a low level of lysis was observed on an HLA-A2 tumor cell line expressing MAGE-1. They failed to obtain human CTL able to lyse relevant tumor targets without addition of peptide and suggested that the human CTL directed against this peptide had a too low affinity or expressed inhibitory receptors [26].
We believed that, before using the MAGE-1.A2 peptide in clinical trials, it remained to be proven that CTL specific for the MAGE-1.A2 peptide exist in the human T cell repertoire, and that these CTL have sufficient avidity to lyse relevant tumor cells. Firstly, it cannot be excluded a priori that the MAGE-1.A2 antigen mimics an ubiquitously expressed antigen, for which high affinity CTL precursors have been purged from the T cell repertoire, or that a certain degree of tolerance for some MAGE proteins exists because the encoding gene is expressed in the thymus [11]. Experiments in mice demonstrated that P1A-transgenic animals were not protected after immunization with P1A-expressing tumors whereas the wild type animals were protected [2]. In p53 transgenic animals also, a considerable degree of tolerance for p53 exists at the CTL level and only low avidity CTL can be found against p53 in the periphery. Several efforts to raise the CTL response, using CTLA-4 blockade or CD40 activation, did result in higher cell numbers, but not in higher avidity [14]. Secondly, a hole in the T cell repertoire could result from the absence of a positive selection in the thymus. The absence of an appropriate self peptide, which is different from the peptide recognized by the mature T cells, could impede the positive selection. It was shown that the inability of bm8 mice to make an H-2K-restricted response to ovalbumin was due to a lack of positive selection of ovalbumin-specific CTL precursors. Stefanski et al. screened a peptide library with ovalbumin-specific CTL and identified candidate peptides recognized by the CTL. The culture of thymic lobes in the presence of one of these candidate peptides restored positive selection of functional anti-ovalbumin CTL [32].
We set out to isolate MAGE-1-specific HLA-A2-restricted CTL in blood cells of an individual without cancer using fluorescent multimers of HLA-A 0201 molecules folded with MAGE-1 peptide KVLEYVIKV. We describe here three human CTL clones that show a high degree of lysis on HLA-A2 tumor cells that express MAGE-1.
Material and methods
Cell lines, media, and reagents
The Epstein Barr Virus-transformed B (EBV-B) cell lines and the tumor cell lines were cultured in IMDM (Life Technologies, Gaithersburg, MD, USA) supplemented with 10% fetal calf serum (Life Technologies). COS-7 cells were maintained in DMEM (Life Technologies) supplemented with 5% fetal calf serum. All the media were supplemented with 0.24 mM L-asparagine, 0.55 mM L-arginine, 1.5 mM L-glutamine (AAG), 100 U/ml penicillin and 100 μg/ml streptomycin. Human recombinant IL-2 was purchased from Chiron (Emeryville, CA, USA), IL–6 and IL–7 from Peprotech (Rocky Hill, NJ, USA), and GM-CSF (Leucomax) from Schering-Plough (Brinny, Ireland). Human recombinant IL-12 was received from Wyeth Research (Gosport, United Kingdom). LB1118-EBV.retro-MAGE-1 was obtained by transduction of parental LB1118–EBV-B cells with retroviral vector M1-CSM that encodes the full length MAGE-A1 protein and the truncated form of the human low affinity nerve growth factor receptor (ΔLNGFr). It was produced as previously reported [24]. EBV-B cells were transduced by co-culture with irradiated packaging cell lines producing the M1-CSM vector in the presence of polybrene (8 μg/ml). After 72 h, lymphocytes were harvested and seeded in fresh medium. The percentage of infected cells was evaluated 48 h later by flow cytometry for LNGFr expression with the mAb 20.4 (ATCC, Rockville, MD, USA). The LNGFr positive cells were purified by magnetic cell sorting using rat anti-mouse IgG1-coated beads (Dynabeads M-450, DYNAL A.S. N012, Oslo, Norway).
Dendritic cells
Peripheral blood was obtained from hemochromatosis patient LB2369 as standard buffy coat preparations, which were laid down on a 15-ml Lymphoprep layer (Axis-Shield PoCAS, Oslo, Norway) in 50-ml tubes. The tubes were centrifuged at 2,200 rpm for 20 min at room temperature. The interphase containing the PBMC was harvested and washed three times in cold phosphate buffer solution with 2 mM EDTA in order to eliminate the remaining platelets. To generate autologous dendritic cells, PBMC were left to adhere for 1 h at 37°C in culture flasks (FALCON, BD Biosciences, Erembodegem, Belgium) at a density of 2 × 106 cells per cm2 in RPMI 1640 supplemented with Hepes (2.38 g/liter), AAG, antibiotics, and 1% autologous plasma (hereafter referred to as complete RPMI medium). Non-adherent cells were discarded and adherent cells were cultured in the presence of IL-4 (200 U/ml) and GM-CSF (70 ng/ml) in complete RPMI medium. Cultures were fed on days 2 and 4 by removing 1/3 of the volume and adding fresh medium with cytokines. They were frozen on day 5.
Multimer production and labeling with multimers
Recombinant HLA-A*0201 molecules were folded in vitro with β2-microglobulin and peptide KVLEYVIKV from MAGE-1, or peptide VSDGGPNLY from the EBV lytic cycle antigen BMLF1. They were purified by gel filtration, biotinylated, and mixed as described [1] with Extravidin-PE (Sigma, St Louis, MT, USA) for the HLA-A2/MAGE-1 multimer, or streptavidin-APC (Molecular Probes, Eugene, OR, USA) for the EBV control multimer. For staining and sorting, cells were washed, resuspended at 25 × 106 cells per ml in Hank’s solution modified for flow cytometry [17] with 1% HS and incubated for 15 min at room temperature with HLA-A2 multimers loaded with MAGE-1 peptide (20 nM) or influenza peptide (5 nM). Anti-CD8 antibody coupled to FITC (IQ products, Groningen, The Netherlands) was added and after a further incubation for 15 min, cells were washed. To select the CD4− cells, we have used an anti-CD4 antibody coupled to FITC (SK3, BD Biosciences).
Magnetic sorting and flow cytometry analysis
Multimer-labeled cells (25 × 106 cells/80 μl) were incubated at 4°C with anti-PE microbeads (20 μl) according to the instructions of the manufacturer (Miltenyi Biotec, Bergisch Gladbach, Germany), washed, and sorted through a separation column inserted to a magnet in an AUTOMACSTM at 0.5 ml/min (Miltenyi Biotec). FACSCaliburTM (BD Biosciences) was used for analysis of the T cell clones, using the CellquestTM software (BD Biosciences).
Culture conditions of cells sorted by flow cytometry
Sorted cells were distributed at 7,900 cells per well in U-bottomed microwells and cultured in 200 μl of IMDM supplemented with AAG, 10% human serum, IL-2 (100 U/ml), and IL-7 (5 ng/ml). They were stimulated on days 0 and 7 with irradiated (100 Gray) peptide pulsed autologous dendritic cells. To prepare the stimulator cells, monocyte-derived immature dendritic cells were incubated for 6 h with 5 μg/ml of peptide KVLEYVIKV, IL-4 (200 U/ml), GM-CSF (70 ng/ml), in the presence of 1 μg/ml of ribomunyl (INAVA, Pierre Fabre Medicament Production, Boulogne, France) and 500 U/ml of IFN-γ (Peprotech) in order to induce their maturation, and washed. Approximately 105 cells from each microculture were stained with multimer and analyzed by flow cytometry. Positive microcultures were either sorted for CD8 positive cells or enriched for multimer-postive cells and selected cells were restimulated every 7-12 days, in the presence of IL-2 (50 U/ml), and IL-7 (5 ng/ml).
Transfection of COS cells and recognition assay based on IFN-γ production
COS-7 cells (1.5 × 104) were distributed in flat-bottomed microwells and cotransfected using 1 μl of Lipofectamine (Invitrogen, Merelbeke, Belgium) with pcDNAI/Amp (50 ng) (Invitrogen) containing either a MAGE-1 cDNA and pcDNA3 (50 ng) containing an HLA-A*0201 cDNA. Transfected cells were incubated for 24 h at 37°C and 8% CO2. The transfectants were then tested for their ability to stimulate the production of IFN-γ by the CTL clone. In total, 5,000 CTL were added in the microwells containing either the COS-7 transfectants or tumor cells, in a total volume of 150 μl of complete IMDM supplemented with 25 U/ml of IL-2. After 24 h, IFN-γ released in the supernatant was measured by ELISA using reagents from Medgenix Diagnostics-Biosource (Fleurus, Belgium).
Cytotoxicity assay
EBV-B cells and tumor cells were labeled with 200 μ Ci of Na(51Cr)O for 1 h, washed and, if indicated, incubated for 15 min with peptide. CTL were then added and chromium release was measured after incubation at 37°C for 4 h.
Lactacystin treatment
To elute the peptides presented at the surface of tumor cells, cells were treated for 10 sec with 500 μl of PBS glycine 300 mM/pH3/1% BSA at room temperature, washed and further treated with 10 μM of lactacystin (Calbiochem, Merck Biosciences, Nottingham, UK) during 1 h at 37°C, and washed. Tumor cells (30,000) were distributed in flat-bottomed microwells and incubated for 1 h with 1 μM lactacystin, with or without 2 μg/ml of peptide KVLEYVIKV, and washed. CTL (5,000) were added to the tumor cells and after 8 h of co-culture, the TNF released in the supernatant was measured by testing its cytotoxic effect on WEHI-164 clone 13 cells in a colorimetric assay [10, 13, 36].
Results
Isolation of CTL clones with HLA-A2 multimers folded with a MAGE-1 peptide
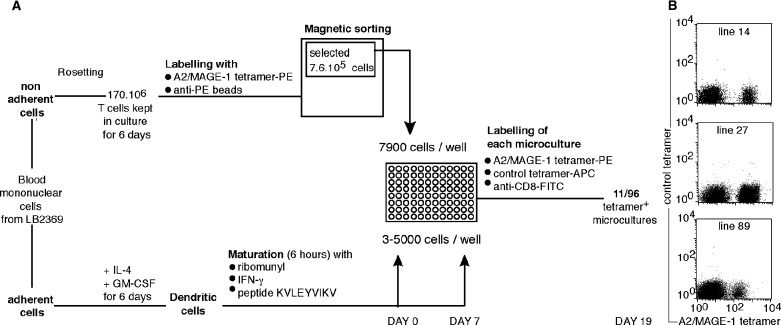
We have developed an approach aimed at obtaining CTL clones from very low frequency precursors: T cells are incubated with HLA/peptide multimers conjugated to phycoerythrin (PE), and with anti-PE antibodies coupled to magnetic beads. The multimer-positive cells are first enriched by magnetic sorting. In our first approach [15, 18], the selected fraction was passed directly through a flow cytometer to further enrich the multimer-positive cells among the CD8 cells. The selected cells were then amplified in clonal conditions. With this approach and an HLA-A1/MAGE-3 multimer, only a fraction of the growing T cell clones proved to be tetramer-positive and a significant fraction of the tetramer-positive clones showed neither lytic activity nor specific cytokine production upon contact with cells pulsed with the peptide or expressing the relevant gene [18]. Presumably, the multimeric nature of the HLA-peptide complexes permitted stable binding on a number of T lymphocytes that have a too low affinity for effective function or proliferation. We have therefore modified our procedure and added a stimulation step in order to amplify only the tetramer-positive cells that proliferate upon stimulation with the peptide. In total, 170 million T cells were labeled with A2/MAGE-1 multimers and the 7.6 × 105 cells selected by magnetic sorting were distributed in microwells at 7,900 cells/well (Fig.1A). Peptide-pulsed autologous mature dendritic cells were used as stimulator cells on days 0 and 7. The microcultures were screened on day 19 for the presence of cells specifically labeled with multimers.
Fig. 1.
Overview of the procedure used to isolate anti-MAGE-1 CTL clones (A) and examples of positive microcultures (B). The control multimer is an HLA-A2 multimer containing an EBV peptide. The cells represented in the figure (B) are CD4− cells.
Eleven of the 96 microcultures were considered as strongly positive because they contained from 4 to 61% of multimer-positive cells in the CD8 fraction. Data obtained with three representative microcultures are shown in Figure 1B. From each of the three microcultures, multimer-positive cells were sorted by flow cytometry or magnetic sorting and stimulated with the antigen. The clonality of each CTL line was verified after a few weeks by sequencing the β chain of the T cell receptor (TCR). The three multimer-positive clones, LB2369-CTL 736/14, 736/27 and 736/89, expressed each a different TCR (Fig.2A). These clones will be referred to hereafter as clones 14, 27 and 89.
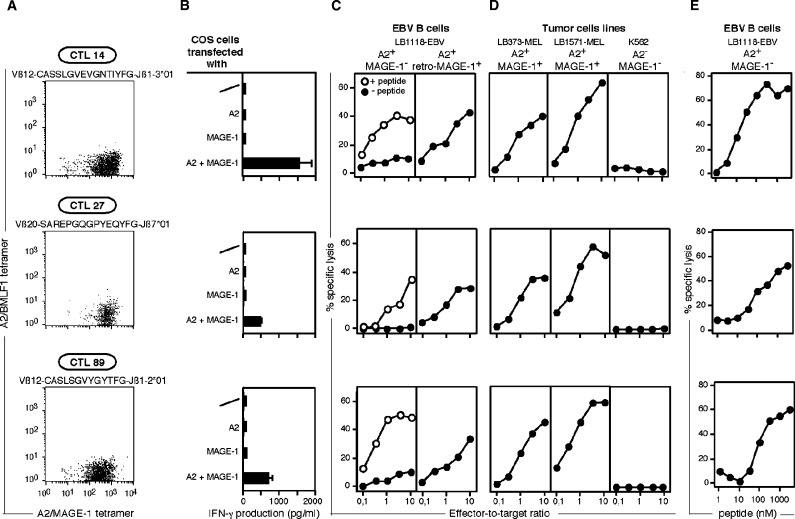
Fig. 2.
Effector functions of the anti-MAGE-1 CTL clones. (A) Labeling of T cell clones with A2/MAGE-1 multimers. The VβJβ usage and the amino-acid sequence of the CDR3 region is indicated for each clone. Cells were labeled for 15 min at room temperature with the A2/MAGE-1 multimer (20 nM) conjugated to PE and the control multimer (5 nM) conjugated to APC. Anti-CD8 antibodies were then added for 15 min. (B) Recognition of cells transiently transfected with MAGE-1. COS-7 cells were transiently transfected using lipofectamine with a MAGE-1 and an HLA-A*0201 coding sequence. One day after transfection, 5,000 CTL were added to the transfected cells. IFN-γ production was measured by ELISA after overnight co-culture. (C) Lysis of HLA-A2 targets loaded with MAGE-1 peptide or transduced with a retrovirus containing the coding sequence of MAGE-1. Target cells were 51 Cr-labeled and, as indicated, pulsed or not for 15 min with 1 μg/ml of peptide KVLEYVIKV. (D) Lysis of HLA-A2 tumor cell lines expressing MAGE-1. Cell line K562 is a target for natural killer cells. Target cells were 51Cr-labeled and incubated for 4 h with CTL at the indicated effector-to-target ratios. Chromium release was measured after 4 h. As indicated, cells were incubated or not with 1 μM of peptide before being put into contact with CTL. (E) Titration of the MAGE-1 peptide. Cells were 51Cr-labeled and incubated for 15 min with threefold dilutions of the synthetic peptide. CTL were subsequently added at an effector-to-target ratio of 10:1. Chromium release was measured after 4 h. The concentrations indicated in the figure correspond to the concentrations during the 4-h incubation.
Effector functions of the anti-MAGE-1 CTL clones
COS-7 cells transfected with both a MAGE-1 and an HLA-A*0201 cDNA construct stimulated the three clones to produce IFN-γ, indicating that the MAGE-1 antigen could be processed in these cells (Fig.2B). The three clones were able to lyse HLA-A2 EBV-B cells loaded with MAGE-1 peptide KVLEYVIKV or transduced with a retrovirus containing the coding sequence of MAGE–1 (Fig.2C). Titration of the peptide revealed that half maximal lysis of A2 target cells was obtained at a peptide concentration of 10-100 nM, depending on the CTL clone (Fig.2E). There was no strict correlation between the intensity of the multimer staining and the avidity of the CTL clone, as estimated by the amount of peptide necessary to induce half-maximal lysis. Two HLA-A2 melanoma cell lines expressing MAGE-1 were lysed by each of the three CTL clones, indicating that the three CTL have enough avidity to lyse tumor targets expressing a physiological level of HLA-A2 molecules and MAGE-1 protein (Fig.2D).
Processing of the antigenic peptide by the two types of proteasome
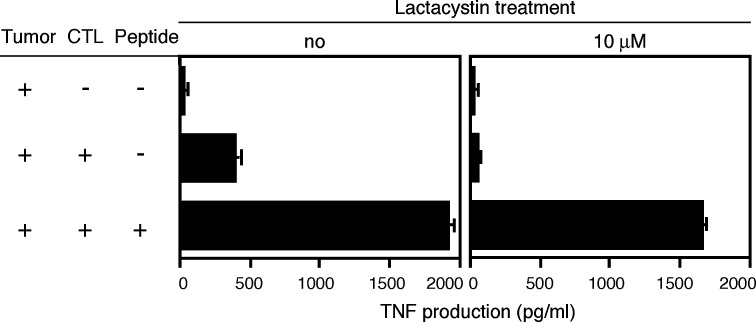
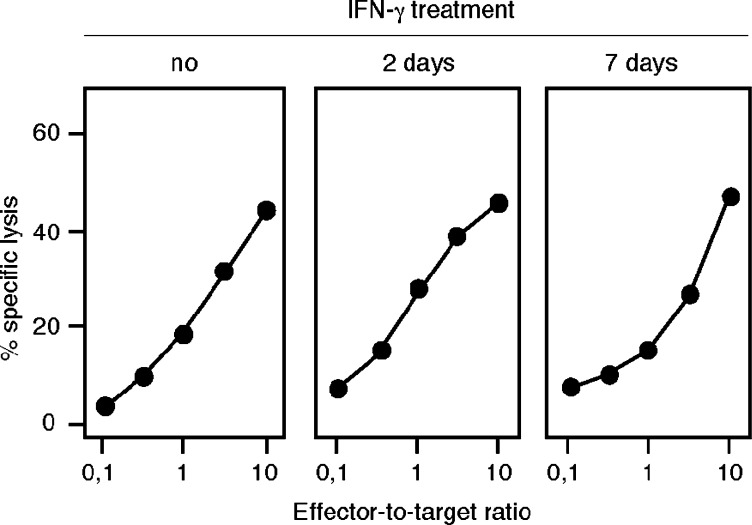
Peptides presented by MHC class I molecules usually derive from intracellular proteins that are degraded by the proteasome, a proteolytic complex [28]. Most non-lymphoid cells, be they normal or tumoral, constitutively express standard proteasomes, and switch to immunoproteasomes when exposed to IFN-γ. The immunoproteasome, compared to the standard proteasome, contains three different catalytic subunits, and therefore has different cleavage specificities [9, 16, 34]. Therefore, a number of antigenic peptides are not processed with the same efficiency by the immunoproteasome and by the standard proteasome [37]. The recognition by CTL 89 of an A2 melanoma line expressing MAGE-1 was abolished after treatment with lactacystin, indicating that the processing of the antigen is dependent on the proteasome (Fig.3). Lysis by CTL 89 of LB373-MEL tumor cells remained unchanged when targets were incubated for 2 or 7 days with IFN–γ, suggesting that the processing of the MAGE-1 antigen can be done efficiently by both the standard and the immunoproteasome (Fig.4).
Fig. 3.
Loss of recognition by CTL clone 89 of a tumor cell line treated by lactacystin. HLA-A2 tumor cell line CP50-MEL expresses MAGE-1. Cells were first treated at pH3 for 10 sec to remove peptides bound to HLA molecules on the surface. Cells were then incubated for 1 h with 10 μM of lactacystin, followed by overnight incubation with 1 μM of lactacystin. In total, 30,000 tumor cells were co-cultured for 8 h with 5,000 CTL of the clone 89. To establish the viability of the cells, they were also pulsed with 2 μg/ml of peptide and tested for recognition by the CTL.
Fig. 4.
Lysis by CTL clone 89 of a tumor cell line treated with IFN-γ. The tumor cell line LB1017-SCCHN is a squamous cell carcinoma of the head and neck. The cell line was treated with 50 U/ml of IFN-γ for either 2 or 7 days. Targets were 51Cr-labeled and chromium release was measured after 4 h.
Discussion
We demonstrate here that several CTL against the MAGE-1.A2 complex exist in the human T cell repertoire and that these CTL have enough avidity to kill efficiently human A2-positive tumor cells expressing MAGE-1. If we consider that 1/3 of the 170 million T cells that were labeled with the multimers are CD8 T cells (57 million) and that the eleven positive microcultures all contained anti-MAGE-1.A2 CTL that lyse relevant tumor targets, the frequency of naive precursors can be estimated at 2 × 10−7 of the CD8 T cells (1 precursor for 5 million CD8 T cells). This frequency is close to the frequency of 4 × 10−7 estimation for the anti-MAGE-3.A1 CTL [18].
MAGE-1 is expressed in 53% of esophageal carcinomas, 49% of non-small lung carcinomas, 46% of metastatic melanomas, 32% of infiltrating bladder carcinomas, and 31% of head and neck tumors [40]. Several MAGE-1 peptides that are recognized by CD8 or CD4 T cells have already been identified [3–5, 7, 20, 21, 33, 35, 39]. They are presented by HLA class I molecules A1, A3, A68, B7, B35, B37, B53, B57, Cw2, Cw3 and Cw16, and class II molecules DR13 and DR15. The MAGE-1 antigenic peptide described here is an interesting target for cancer immunotherapy because it is presented by HLA-A2 molecules, which are widely expressed in the different major ethnic groups: Oriental (47%), Caucasoid (44%), Amerindian (44%), Black (34%) [23].
In trials of cancer vaccination with defined antigens, we have to consider three stages where antigen needs to be expressed. At the vaccine site or in the draining lymph node the antigenic peptide should be expressed by mature dendritic cells in order to stimulate efficiently the CTL precursors. Once activated, those CTL should migrate to the tumor site and attack malignant cells, which should also present the peptide. The tumor cells are normally equipped with standard proteasomes, as it is suggested by tumor cell lines analyzed in vitro for their ability to present antigenic peptides that are destroyed by either the immunoproteasome or the standard proteasome [25, 30]. Finally, once a response has been initiated at the tumor site, IFN-γ may be released and induce expression of immunoproteasomes in the tumor cells. Antigenic peptides that are produced by the two proteasome types, such as the MAGE-1.A2 peptide described here, will be presented at all three stages and may therefore be preferred as candidate antigens for vaccination.
Acknowledgements
We thank Dr Benoît Van den Eynde for critical reading and Mrs Nathalie Krack for editorial assistance. This work was supported by the Belgian Programme on Interuniversity Poles of Attraction initiated by the Belgian State, Prime Minister’s Office, Science Policy Programming, by a grant from the Fédération Belge contre le Cancer (Belgium), by grant n° QLK3-CT-1999-00064 from the European Community, Fifth Framework programme, and by the TELEVIE fund (Belgium).
References
- 1.Altman JD, Moss PAH, Goulder PJR, Barouch DH, McHeyzer-Williams MG, Bell JI, McMichael AJ, Davis MM. Phenotypic analysis of antigen-specific T lymphocytes. Science. 1996;274:94–96. [PubMed] [Google Scholar]
- 2.Brändle D, Bilsborough J, Rülicke T, Uyttenhove C, Boon T, Vanden Eynde BJ. The shared tumor-specific antigen encoded by mouse gene P1A is a target not only for cytolytic T lymphocytes but also for tumor rejection. Eur J Immunol. 1998;28:4010–4019. doi: 10.1002/(SICI)1521-4141(199812)28:12<4010::AID-IMMU4010>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 3.Chaux P, Luiten R, Demotte N, Vantomme V, Stroobant V, Traversari C, Russo V, Schultz E, Cornelis GR, Boon T, van der Bruggen P. Identification of five MAGE-A1 epitopes recognized by cytolytic T lymphocytes obtained by in vitro stimulation with dendritic cells transduced with MAGE-A1. J Immunol. 1999;163:2928–2936. [PubMed] [Google Scholar]
- 4.Chaux P, Vantomme V, Stroobant V, Thielemans K, Corthals J, Luiten R, Eggermont AM, Boon T, van der Bruggen P. Identification of MAGE-3 epitopes presented by HLA-DR molecules to CD4(+) T lymphocytes. J Exp Med. 1999;189:767–777. doi: 10.1084/jem.189.5.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chaux P, Lethé B, Van Snick J, Corthals J, Schultz ES, Cambiaso CL, Boon T, van der Bruggen P. A MAGE-1 peptide recognized on HLA-DR15 by CD4+ T cells. Eur J Immunol. 2001;31:1910–1916. doi: 10.1002/1521-4141(200106)31:6<1910::aid-immu1910>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 6.Chomez P, De Backer O, Bertrand M, De Plaen E, Boon T, Lucas S. An overview of the MAGE gene family with the identification of all human members of the family. Cancer Res. 2001;61:5544–5551. [PubMed] [Google Scholar]
- 7.Corbière V, Nicolay H, Russo V, Stroobant V, Brichard V, Boon T, van der Bruggen P. Identification of a MAGE-1 peptide recognized by cytolytic T lymphocytes on HLA-B*5701 tumors. Tissue Antigens. 2004;63:453–457. doi: 10.1111/j.0001-2815.2004.00203.x. [DOI] [PubMed] [Google Scholar]
- 8.De Plaen E, Arden K, Traversari C, Gaforio JJ, Szikora J-P, De Smet C, Brasseur F, van der Bruggen P, Lethé B, Lurquin C, Brasseur R, Chomez P, De Backer O, Cavenee W, Boon T. Structure, chromosomal localization and expression of twelve genes of the MAGE family. Immunogenetics. 1994;40:360–369. doi: 10.1007/BF01246677. [DOI] [PubMed] [Google Scholar]
- 9.Driscoll J, Brown M, Finley D, Monaco J. MHC-linked LMP gene products specifically alter peptidase activities of the proteasome. Nature. 1993;365:262–264. doi: 10.1038/365262a0. [DOI] [PubMed] [Google Scholar]
- 10.Espevik T, Nissen-Meyer J. A highly sensitive cell line, WEHI 164 clone 13, for measuring cytotoxic factor/tumor necrosis factor from human monocytes. J Immunol Methods. 1986;95:99–105. doi: 10.1016/0022-1759(86)90322-4. [DOI] [PubMed] [Google Scholar]
- 11.Gotter J, Brors B, Hergenhahn M, Kyewski B. Medullary epithelial cells of the human thymus express a highly diverse selection of tissue-specific genes colocalized in chromosomal clusters. J Exp Med. 2004;199:155–166. doi: 10.1084/jem.20031677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haas GG, Jr, D’Cruz OJ, De Bault LE. Distribution of human leukocyte antigen-ABC and -D/DR antigens in the unfixed human testis. Am J Reprod Immunol Microbiol. 1988;18:47–51. doi: 10.1111/j.1600-0897.1988.tb00234.x. [DOI] [PubMed] [Google Scholar]
- 13.Hansen MB, Nielsen SE, Berg K. Re-examination and further development of a precise and rapid dye method for measuring cell growth/cell kill. J Immunol Methods. 1989;119:203–210. doi: 10.1016/0022-1759(89)90397-9. [DOI] [PubMed] [Google Scholar]
- 14.Hernandez J, Ko A, Sherman LA. CTLA-4 blockade enhances the CTL responses to the p53 self-tumor antigen. J Immunol. 2001;166:3908–3914. doi: 10.4049/jimmunol.166.6.3908. [DOI] [PubMed] [Google Scholar]
- 15.Kobayashi T, Lonchay C, Colau D, Demotte N, Boon T, van der Bruggen P. New MAGE-4 antigenic peptide recognized by cytolytic T lymphocytes on HLA-A1 tumor cells. Tissue Antigens. 2003;62:426–432. doi: 10.1034/j.1399-0039.2003.00123.x. [DOI] [PubMed] [Google Scholar]
- 16.Kuckelkorn U, Frentzel S, Kraft R, Kostka S, Groettrup M, Kloetzel PM. Incorporation of major histocompatibility complex-encoded subunits LMP2 and LMP7 changes the quality of the 20S proteasome polypeptide processing products independent of interferon-gamma. Eur J Immunol. 1995;25:2605–2611. doi: 10.1002/eji.1830250930. [DOI] [PubMed] [Google Scholar]
- 17.Lehmann F, Marchand M, Hainaut P, Pouillart P, Sastre X, Ikeda H, Boon T, Coulie PG. Differences in the antigens recognized by cytolytic T cells on two successive metastases of a melanoma patient are consistent with immune selection. Eur J Immunol. 1995;25:340–347. doi: 10.1002/eji.1830250206. [DOI] [PubMed] [Google Scholar]
- 18.Lonchay C, van der Bruggen P, Connerotte T, Hanagiri T, Coulie P, Colau D, Lucas S, Van Pel A, Thielemans K, van Baren N, Boon T. Correlation between tumor regression and T cell responses in melanoma patients vaccinated with a MAGE antigen. Proc Natl Acad Sci USA. 2004;101:14631–14638. doi: 10.1073/pnas.0405743101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lucas S, De Smet C, Arden KC, Viars CS, Lethé B, Lurquin C, Boon T. Identification of a new MAGE gene with tumor-specific expression by representational difference analysis. Cancer Res. 1998;58:743–752. [PubMed] [Google Scholar]
- 20.Luiten R, van der Bruggen P. A MAGE-A1 peptide is recognized on HLA-B7 human tumors by cytolytic T lymphocytes. Tissue Antigens. 2000;55:149–152. doi: 10.1034/j.1399-0039.2000.550206.x. [DOI] [PubMed] [Google Scholar]
- 21.Luiten RM, Demotte N, Tine J, van der Bruggen P. A MAGE-A1 peptide presented to cytolytic T lymphocytes by both HLA-B35 and HLA-A1 molecules. Tissue Antigens. 2000;56:77–81. doi: 10.1034/j.1399-0039.2000.560110.x. [DOI] [PubMed] [Google Scholar]
- 22.Lurquin C, De Smet C, Brasseur F, Muscatelli F, Martelange V, De Plaen E, Brasseur R, Monaco AP, Boon T. Two members of the human MAGEB gene family located in Xp.21.3 are expressed in tumors of various histological origins. Genomics. 1997;46:397–408. doi: 10.1006/geno.1997.5052. [DOI] [PubMed] [Google Scholar]
- 23.Marsh SGE, Parham P, Barber LD, editors. The HLA FactsBook. London: Academic Press; 2000. [Google Scholar]
- 24.Mavilio F, Ferrari G, Rossini S, Nobili N, Bonini C, Casorati G, Traversari C, Bordignon C. Peripheral blood lymphocytes as target cells of retroviral vector-mediated gene transfer. Blood. 1994;83:1988–1997. [PubMed] [Google Scholar]
- 25.Morel S, Lévy F, Burlet-Schiltz O, Brasseur F, Probst-Kepper M, Peitrequin A-L, Monsarrat B, Van Velthoven R, Cerottini J-C, Boon T, Gairin JE, Vanden Eynde BJ. Processing of some antigens by the standard proteasome but not by the immunoproteasome results in poor presentation by dendritic cells. Immunity. 2000;12:107–117. doi: 10.1016/s1074-7613(00)80163-6. [DOI] [PubMed] [Google Scholar]
- 26.Pascolo S, Schirle M, Guckel B, Dumrese T, Stumm S, Kayser S, Moris A, Wallwiener D, Rammensee HG, Stevanovic S. A MAGE-A1 HLA-A A*0201 epitope identified by mass spectrometry. Cancer Res. 2001;61:4072–4077. [PubMed] [Google Scholar]
- 27.Rammensee H-G, Weinschenk T, Gouttefangeas C, Stevanovic S. Towards patient-specific tumor antigen selection for vaccination. Immunol Rev. 2002;188:164–176. doi: 10.1034/j.1600-065x.2002.18815.x. [DOI] [PubMed] [Google Scholar]
- 28.Rock KL, Goldberg AL. Degradation of cell proteins and the generation of MHC class I-presented peptides. Annu Rev Immunol. 1999;17:739–779. doi: 10.1146/annurev.immunol.17.1.739. [DOI] [PubMed] [Google Scholar]
- 29.Schirle M, Keilholz W, Weber B, Gouttefangeas C, Dumrese T, Becker HD, Stevanovic S, Rammensee H-G. Identification of tumor-associated MHC class I ligands by a novel T cell-independent approach. Eur J Immunol. 2000;30:2216–2225. doi: 10.1002/1521-4141(2000)30:8<2216::AID-IMMU2216>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 30.Schultz ES, Chapiro J, Lurquin C, Claverol S, Burlet-Schiltz O, Warnier G, Russo V, Morel S, Levy F, Boon T, Vanden Eynde BJ, van der Bruggen P. The production of a new MAGE-3 peptide presented to cytolytic T lymphocytes by HLA-B40 requires the immunoproteasome. J Exp Med. 2002;195:391–399. doi: 10.1084/jem.20011974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sette A, Fikes J. Epitope-based vaccines: an update on epitope identification, vaccine design and delivery. Curr Opin Immunol. 2003;15:461–470. doi: 10.1016/s0952-7915(03)00083-9. [DOI] [PubMed] [Google Scholar]
- 32.Stefanski HE, Jameson SC, Hogquist KA. Positive selection is limited by available peptide-dependent MHC conformations. J Immunol. 2000;164:3519–3526. doi: 10.4049/jimmunol.164.7.3519. [DOI] [PubMed] [Google Scholar]
- 33.Tanzarella S, Russo V, Lionello I, Dalerba P, Rigatti D, Bordignon C, Traversari C. Identification of a promiscuous T cell epitope encoded by multiple members of the MAGE family. Cancer Res. 1999;59:2668–2674. [PubMed] [Google Scholar]
- 34.Toes REM, Nussbaum AK, Degermann S, Schirle M, Emmerich NPN, Kraft M, Laplace C, Zwinderman A, Dick TP, Müller J, Schönfisch B, Schmid C, Fehling H-J, Stevanovic S, Rammensee HG, Schild H. Discrete cleavage motifs of constitutive and immunoproteasomes revealed by quantitative analysis of cleavage products. J Exp Med. 2001;194:1–12. doi: 10.1084/jem.194.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Traversari C, van der Bruggen P, Luescher IF, Lurquin C, Chomez P, Van Pel A, De Plaen E, Amar-Costesec A, Boon T. A nonapeptide encoded by human gene MAGE-1 is recognized on HLA-A1 by cytolytic T lymphocytes directed against tumor antigen MZ2-E. J Exp Med. 1992;176:1453–1457. doi: 10.1084/jem.176.5.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Traversari C, van der Bruggen P, Vanden Eynde B, Hainaut P, Lemoine C, Ohta N, Old L, Boon T. Transfection and expression of a gene coding for a human melanoma antigen recognized by autologous cytolytic T lymphocytes. Immunogenetics. 1992;35:145–152. doi: 10.1007/BF00185107. [DOI] [PubMed] [Google Scholar]
- 37.Vanden Eynde BJ, Morel S. Differential processing of class-1-restricted epitopes by the standard proteasome and the immunoproteasome. Curr Opin Immunol. 2001;13:147–153. doi: 10.1016/s0952-7915(00)00197-7. [DOI] [PubMed] [Google Scholar]
- 38.van der Bruggen P, Traversari C, Chomez P, Lurquin C, De Plaen E, Vanden Eynde B, Knuth A, Boon T. A gene encoding an antigen recognized by cytolytic T lymphocytes on a human melanoma. Science. 1991;254:1643–1647. doi: 10.1126/science.1840703. [DOI] [PubMed] [Google Scholar]
- 39.van der Bruggen P, Szikora J-P, Boël P, Wildmann C, Somville M, Sensi M, Boon T. Autologous cytolytic T lymphocytes recognize a MAGE-1 nonapeptide on melanomas expressing HLA-Cw*1601. Eur J Immunol. 1994;24:2134–2140. doi: 10.1002/eji.1830240930. [DOI] [PubMed] [Google Scholar]
- 40.van der Bruggen P, Zhang Y, Chaux P, Stroobant V, Panichelli C, Schultz ES, Chapiro J, Vanden Eynde BJ, Brasseur F, Boon T. Tumor-specific shared antigenic peptides recognized by human T cells. Immunol Rev. 2002;188:51–64. doi: 10.1034/j.1600-065x.2002.18806.x. [DOI] [PubMed] [Google Scholar]