Abstract
The lack of sufficient well-defined tumor-associated antigens is still a drawback on the way to a cytotoxic T-lymphocyte-based immunotherapy of renal cell carcinoma (RCC). We are trying to define a larger number of such targets by a combined approach involving HLA ligand characterization by mass spectrometry and gene expression profiling by oligonucleotide microarrays. Here, we present the results of a large-scale analysis of 13 RCC specimens. We were able to identify more than 700 peptides, mostly from self-proteins without any evident tumor association. However, some HLA ligands derived from previously known tumor antigens in RCC. In addition, gene expression profiling of tumors and a set of healthy tissues revealed novel candidate RCC-associated antigens. For several of them, we were able to characterize HLA ligands after extraction from the tumor tissue. Apart from universal RCC antigens, some proteins seem to be appropriate candidates in individual patients only. This underlines the advantage of a personalized therapeutic approach. Further analyses will contribute additional HLA ligands to this repertoire of universal as well as patient-individual tumor antigens.
Keywords: Renal cell carcinoma, Novel tumor antigens, HLA ligands, Immunotherapy
Introduction
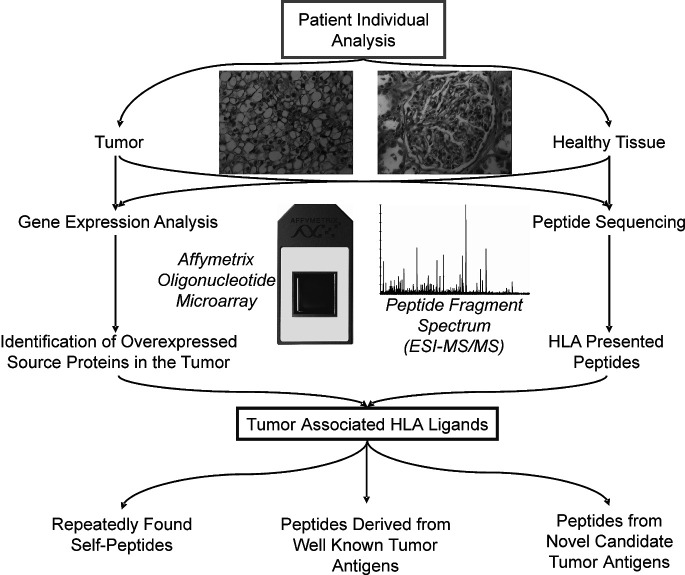
Metastatic renal cell carcinoma (RCC) remains a disease with a fatal prognosis. In 2004, more than 35,000 new cases and more than 12,000 cancer-related deaths were estimated in the US [22]. If metastasis is diagnosed, the 1-year survival rate decreases to approximately 60%. This underlines the dissatisfactory therapeutic situation. Currently, numerous new therapeutical approaches are under investigation. The known, albeit rare, phenomenon of spontaneous regression of metastasis in RCC patients [29] and the existence of tumor-reacting and tumor-infiltrating cytotoxic T-lymphocytes (CTL) suggest that RCC is an immunogenic tumor. Several immunological concepts of therapy have been proposed and several tumor-associated antigens (TAA) defined for RCC in the past. The aim of our investigations was to identify HLA class I-presented peptides characteristic for the tumor in vivo. These peptides, processed from proteins characteristically expressed in the malignancy, may serve as targets for a vaccination-induced CTL response against the tumor. To achieve this goal, we performed mass spectrometry (LC/MS)-based peptide sequencing and patient-individual microarray gene expression profiling (Fig. 1) with surgically resected RCCs. This led to the generation of a data set providing information on the one hand about the sequences of approximately 100 HLA-presented peptides for each tumor specimen of appropriate mass, on the other hand about the level of expression for approximately 14,000 particular genes in every tumor. Overexpressed genes were identified in individual tumors in comparison to a broad set of healthy tissues, covering most human organs. Extensively upregulated genes are expected to give rise to tumor-associated proteins and peptides, which should provide targets for specific CTL recognition of the tumor [40]. We consider such peptides suitable for vaccination. Combining both analytical tools, peptide analysis and gene expression profiling, we are able to identify such potential CTL targets in individual malignancies, which might ultimately find their way into clinical applications.
Fig. 1.
Patient-individual analysis of gene expression patterns and HLA-presented peptides
During our investigations, we were able to sequence peptides from classical TAA such as carbonic anhydrase 9 (CA9) and met proto-oncogene (MET), as well as from constitutively or individually upregulated proteins such as insulin-like growth factor binding protein 3 (IGFBP3), adipophilin (ADFP), and apolipoprotein L1 (APOL1). Here, we describe the results of a systematic analysis of peptide presentation patterns and gene expression profiles in 13 RCC patients.
Materials and methods
Patients and tumor specimens
Surgically removed RCC specimens (Table 1) were provided by the Department of Urology, University of Tübingen, Germany after written informed consent had been obtained from each patient. Specimens were snap frozen in liquid nitrogen immediately after surgery. Pathological staging and grading were performed by the Department of Pathology and HLA typing was done by the Department of Transfusion Medicine, University of Tübingen. This study has been approved by the local ethical review board.
Table 1.
Renal cell carcinoma specimens included in the study
| Specimen | Histology | Grade and stage | HLA typing |
|---|---|---|---|
| RCC01 | Clear cell RCC | T3 Nx Mx (G2) | A*02; A*68; B*18; B*44 |
| RCC13 | Clear cell RCC | T2 Nx Mx (G2) | A*02; A*24; B*07; B*40 |
| RCC44 | Chromophilic RCC | T1 Nx Mx (G2) | A*03; A*11; B*27 |
| RCC68 | Clear cell RCC | T3 N0 Mx (G3) | A*02; A*29; B*15; B*45 |
| RCC70 | Clear cell RCC | T3 N1 M0 (G2) | A*01; A*02; B*07; B*08 |
| RCC73 | Clear cell RCC | T3 N0 (G2) | A*02; A*03; B*07; B*57 |
| RCC75 | Chromophilic RCC | T4 Nx M1 (G2–G3) | A*03; B*07; B*40 |
| RCC98 | Clear cell RCC | T3 Nx M1 (G2–G3) | A*01; A*03; B*07; B*18 |
| RCC103 | Clear cell RCC | T3 N0 Mx (G2) | A*11; A*25; B*15; B*44 |
| RCC112 | Metastasis of clear cell RCC in the adrenal gland | A*01; A*31; B*08; B*27 | |
| RCC115 | Clear cell RCC | T3 N0 Mx (G2) | A*02; A*03; B*15; B*18 |
| RCC116 | Clear cell RCC | T3 N2 Mx (G2) | A*01; A*02; B*27; B*37 |
| RCC130 | Clear cell RCC | T1 N1 Mx (G3) | A*02; A*24; B*07; B*44 |
Peptide data were generated from all samples while gene expression profiling data are available for specimens RCC44–RCC130 only
Peptide isolation and sequencing
The frozen tumor tissue was processed as described previously [44]. Peptides were isolated according to standard protocols [10] using the HLA class I specific antibody W6/32.
For RCC01 and RCC44–75, eluted peptide mixtures were separated offline by reversed-phase high-performance liquid chromatography (SMART system, μRPC C2/C18 SC 2.1/10; Amersham Pharmacia Biotech, Freiburg, Germany) and fractions were analyzed by nano-ESI MS on a hybrid quadrupole orthogonal acceleration time-of-flight mass spectrometer (Q-TOF; Micromass, Manchester, UK), as described previously [44]. For RCC13 and RCC98–130, peptide mixtures were separated and analyzed online by a reversed phase Ultimate HPLC system (Dionex, Amsterdam, Netherlands) coupled directly to the mass spectrometer, as described [28].
Fragment spectra were analyzed manually and database searches (National Center for Biotechnology Information, Expressed Sequence Tag) were carried out using Multiple Alignment System for Protein Sequences Based on Three-way Dynamic Programming (MASCOT, http://www.matrixscience.com).
Peptide synthesis
Synthetic peptides were synthesized in an automated peptide synthesizer EPS221 (Abimed, Langenfeld, Germany) following the 9-fluorenylmethyl-oxycarbonyl/tert-butyl (Fmoc/tBu) strategy, as described [44].
Gene expression analysis by high-density oligonucleotide microarrays
Frozen fragments of tumors RCC44–130 were homogenized with mortar and pestle under liquid nitrogen. Total RNA was prepared from these samples using TRIzol (Invitrogen, Karlsruhe, Germany) according to the manufacturer’s protocol, followed by a cleanup with RNeasy (QIAGEN, Hilden, Germany). Total RNA from healthy human tissues was obtained commercially (Ambion, Huntingdon, UK; Clontech, Heidelberg, Germany; Stratagene, Amsterdam, Netherlands). The RNA from several individuals (between 2 and 62 individuals) was mixed such that RNA from each individual was equally weighted. Quality and quantity were assessed on an Agilent 2100 Bioanalyzer (Agilent, Waldbronn, Germany) using the RNA 6000 Pico LabChip Kit (Agilent).
Gene expression analysis of all RNA samples except RCC130 was performed by Affymetrix Human Genome U133A oligonucleotide microarrays (Affymetrix, Santa Clara, CA, USA). For RCC130, HG-U133 Plus 2.0 was used. The same normal kidney sample was hybridized to both array types to achieve comparability. All steps were carried out according to the Affymetrix manual (http://www.affymetrix.com/support/technical/manual/expression_manual.affx).
Briefly, double-stranded cDNA was synthesized from 5–8 μg of total RNA, using SuperScript RTII (Invitrogen) and the oligo-dT-T7 primer (MWG Biotech, Ebersberg, Germany) as described in the manual. In vitro transcription was performed with the BioArray High Yield RNA Transcript Labeling Kit (ENZO Diagnostics, Inc., Farmingdale, NY, USA) for the U133A arrays or with the GeneChip IVT Labeling Kit (Affymetrix) for the U133 Plus 2.0 arrays, followed by cRNA fragmentation, hybridization, and staining with streptavidin-phycoerythrin and biotinylated anti-streptavidin antibody (Molecular Probes, Leiden, Netherlands). Images were scanned with the Agilent 2500A GeneArray Scanner (U133A) or the Affymetrix GeneChip Scanner 3000 (U133 Plus 2.0), and data were analyzed with the MAS 5.0 (U133A) or GCOS (U133 Plus 2.0) software (Affymetrix), using default settings for all parameters. Pairwise comparisons were calculated using the respective normal kidney array as baseline. For normalization, 100 housekeeping genes provided by Affymetrix were used (http://www.affymetrix.com/support/technical/mask_files.affx). Relative expression values were calculated from the signal log ratios given by the software and the normal kidney sample was arbitrarily set as one.
Results and discussion
Patient-individual analysis of tumor-associated peptides presented on RCC
The LC/MS-based peptide sequencing of HLA ligands, extracted from surgically removed RCC specimens, yielded approximately 100 different peptides per patient. However, many more peptides are expected to be presented by tumor cells, so we estimate that we still detect only the most abundantly presented peptides, which make up just a few percent of the whole HLA “ligandome” [28]. From 13 primary RCC samples, we were able to sequence more than 700 different peptides using fragmentation-induced mass spectrometry (supplementary Table S1, http://www.uni-tuebingen.de/uni/kxi/PaperSupplements/CII_S1.pdf). These peptides derived from more than 500 different source proteins and were presented by various HLA allotypes. The following assignments of peptide sequences to specific allotypes are solely based upon known binding motifs in connection with the HLA typing of the samples and not on direct experimental evidence. All natural HLA ligands will be included in the next update of the HLA ligand database SYFPEITHI (http://www.syfpeithi.de). With regard to their expression profiles, the source proteins of HLA ligands could be divided into three groups. First (1) as expected, only a small percentage of the peptides that were identified were of relevance with regard to tumor immunotherapy. Most peptides derived from structure proteins, constitutively expressed enzymes, and receptors, and represented classical self-peptides. More important (2), we were able to define various peptides derived from well-known TAA such as CA9, MET, and ADFP, which have already been used for vaccination in several patients in an ongoing clinical trial. Additionally (3) we identified several novel antigens such as APOL1, matrix metalloproteinase 7 (MMP7), IGFBP3, regulator of G-protein signalling 5 (RGS5), and acyl-CoA synthetase long-chain family member 4 (ACSL4).
In general, we considered an antigen overexpressed if the mRNA expression of its source protein was increased at least threefold in the respective tumor compared to normal kidney and also markedly increased compared to other healthy tissues. With the knowledge of HLA ligands derived from such overexpressed antigens, vaccination cocktails which aim at individually distinct characteristics of the patient’s malignancy can be designed.
Constitutively expressed structure proteins are a major source of HLA class I-presented peptides
The majority of peptides which were sequenced throughout our analysis derived from housekeeping proteins such as vimentin, actin, or spectrin. For instance, we were able to sequence 14 different peptides from vimentin restricted to several different HLA subtypes (Table 2). So far our peptides cover nearly 26% of the 466 amino acid sequences of vimentin; they were found on 8 of the 13 tumors that were investigated. From no other source protein were more peptides defined, underlining the observation that clear cell RCC express vimentin to a high extent [58]. A median of 3.5-fold overexpression of vimentin in comparison to normal kidney tissue (range 0.2–6.4) was determined, but only a 1.9-fold overexpression when compared to the median of all other healthy tissues (median 1.6; range 0.3–6.1). The ubiquitous expression of vimentin and the resulting widespread presentation of vimentin-derived peptides on healthy tissues exclude vimentin peptides from usage for vaccination. The same is true for other structure proteins: six different peptides were found from β-actin, five from non-erythrocytic beta, and three from alpha spectrin. Adipophilin, a tumor-associated antigen we have recently identified [56, 45], was the only exception which represented a non-structural protein.
Table 2.
Proteins from which abundant HLA ligands were repeatedly found
| Source protein overexpression > threefold in X/11 RCCs | Entrez Gene ID | Peptides found on X/13 tumors | Sequence | HLA restriction |
|---|---|---|---|---|
| Vimentin (VIM) 6/11 (M 3.5; R 0.2–6.4) | 7431 | 8 | ALRDVRQQY | B*1501 |
| ALRPSTSRSLY | A*03 | |||
| DLERKVESL [11] | A*0201 | |||
| EEIAFLKKL [56] | B*18 | |||
| EENFAVEA | B*45 | |||
| MEENFAVEA | B*45 | |||
| NLRETNLDSLP | NA | |||
| NYIDKVRFL | A*24 | |||
| REKLQEEML | B*40 | |||
| RETNLDSLP | NA | |||
| SLYASSPGGVYATR | A*03 | |||
| SRISLPLPNF | B*27 | |||
| SSVPGVRLLQDSVDF | NA | |||
| SSVPGVRLLQDSVDFSL | NA | |||
| Adipose differentiation-related protein (ADFP) 5/11 (M 2.6; R 0.1–5.5) | 123 | 5 | IARNLTQQL | B*07 |
| MAGDIYSVFR [56] | A*6801 | |||
| MTSALPIIQK [56] | A*6801 | |||
| SLLTSSKGQLQK | A*03 | |||
| SVASTITGV [56] | A*0201 | |||
| TSALPIIQK | A*03 | |||
| VQKPSYYVR | A*31 | |||
| Actin, beta (ACTB) 0/11 (M 0.8; R 0.6–2.5) | 60 | 4 | LRVAPEEHPVL | NA |
| MEKIWHHTF | B*18 | |||
| MQKEITAL | B*1501 | |||
| RVAPEEHPV | A*02 | |||
| RVAPEEHPVL | A*02 | |||
| RVAPEEHPVLLT | A*02 | |||
| Spectrin beta, non-erythrocytic 1 (SPTBN1) 0/11 (M 0.8; R 0.5–2.1) | 6711 | 5 | AVCEVALDY | NA |
| DEKSIITY | B*18 | |||
| DEMKVLVL [56] | B*18 | |||
| EEASLLHQF | B*44 | |||
| KPRDVSSVEL | B*07 | |||
| Myosin light chain alkali non-muscle isoform (MYL6) 0/11 (M 0.7; R 0.4–1.0) | 4637 | 4 | AEIRHVLVTL | B*40 |
| EAFVRHIL | B*08 | |||
| LVRMVLNG | NA | |||
| YEELVRMVL | B*40 | |||
| Catenin (cadherin-associated protein), alpha 1 (CTNNA1) 0/11 (M 0.8; R 0.5–1.1) | 1495 | 3 | FIDASRLVY | A*01 |
| LQHPDVAAY | B*1501 | |||
| NEQDLGIQY [56] | B*44/B*18 | |||
| Spectrin alpha, non-erythrocytic 1 (SPTAN1) 0/11 (M 0.8; R 0.6–1.5) | 6709 | 3 | ADSLRLQQL | B*37 |
| ETFDAGLQAF | A*25 | |||
| RQGFVPAAY | B*1501 |
M median; R range of overexpression
NA not assigned
Investigation of reported TAA in RCC
Only a few TAA have been described to be associated with RCC and suggested to serve as targets in tumor immunotherapy. We specifically searched for reported HLA ligands from these antigens and analyzed their gene expression (Table 3). From RAGE, PRAME, members of the MAGE family, NY-ESO-1, and from shared TAA telomerase, survivin, and MUC-1 we detected no HLA-presented peptides. However, from adipophilin, MET, CA9, and cyclin D1, known and novel HLA ligands were characterized. Upregulation of their genes varied considerably (Table 3). Only three previously described TAA in RCC played a significant role during our analyses; the met proto-oncogene [43, 56], adipophilin [45, 56], and CA9 [15] were upregulated in the majority of the tested specimens and yielded abundant HLA ligands (see below and Tables 2–3). Survivin, cyclin D1, and PRAME were overexpressed in a minority of tumors, but MUC1, hTERT, and RAGE did not fulfill our overexpression criteria in one example.
Table 3.
Expression analysis, known T-cell epitopes, and novel HLA ligands of reported RCC-associated antigens
| RCC-associated tumor antigen overexpression > threefold in X/11 RCCs | Entrez Gene ID | Known T-cell epitopes | HLA restriction | References | Peptides found in this study |
|---|---|---|---|---|---|
| Met proto-oncogene (MET) 11/11 (M 12.3; R 4.3–28.3) | 4233 | YVDPVITSI | A*02 | [43] | YVDPVITSI (A*02) [56] |
| Carbonic anhydrase isoform 9 (CA9) 7/11 (M 4.0; R 0.4–11.3) | 768 | HLSTAFARV | A*02 | [53] | SPRAAEPVQL (B*07) |
| Adipose differentiation-related protein (ADFP) 5/11 (M 2.6; R 0.1–5.5) | 123 | SVASTITGV | A*02 | [45] | See Table 2 |
| Cyclin D1 (CCND1) 4/11 (M 1.8; R 0.7–5.7) | 595 | RLTRFLSRV | A*02 (allo) | [42] | ETIPLTAEKL (A*6801) [56] |
| LLGATCMFV | A*02 (allo) | [42] | |||
| Survivin (BIRC5) 3/11 (M 1.4; R 0.4–0.9) | 332 | ELTLGEFLKL | A*02 | [46] | |
| Preferentially expressed antigen in melanoma (PRAME) 2/11 (M 0.4; R 0.1–4.7) | 23532 | SLLQHLIGL | A*02 | [25] | |
| ALYVDSLFFL | A*02 | [25] | |||
| VLDGLDVLL | A*02 | [25] | |||
| SLYSFPEPEA | A*02 | [25] | |||
| LYVDSLFFL | A*24 | [19] | |||
| Melanoma antigen, family A, 3 (MAGEA3) 1/11 (M 1.3; R 0.4–6.2) | 4102 | FLWGPRALV | A*02 | [12, 52] | |
| KVAELVHFL | A*0201 | [24] | |||
| EVDPIGHLY | A*01, B*35 | [7, 47] | |||
| IMPKAGLLI | A*24 | [51] | |||
| TFPDLESEF | A*2402 | [31] | |||
| MEVDPIGHLY | B*44 | [16] | |||
| Renal tumor antigen (RAGE) 0/11 (M 0.6; R 0.4–1.3) | 5891 | PASKKTDPQK | B*08 | [11] | |
| SPSSNRIRNT | B*07 | [14] | |||
| Cancer/testis antigen 1B (NY-ESO-1) 0/11 (M 0.7; R 0.5–2.6) | 1485 | SLLMWITQC | A*0201 | [21] | |
| Melanoma antigen, family A, 1 (MAGEA1) 0/11 (M 0.1; R 0.0–0.6) | 4100 | KVLEYVIKV and many others | A*0201 | [37] | |
| Mucin 1 (MUC1) 0/11 (M 0.4; R 0.1–0.8) | 4582 | LLLLTVLTV | A*02 | [4] | |
| STAPPVHNV and others | A*0201 | [4, 2] | |||
| Telomerase reverse transcriptase (TERT) 0/11 (M 0.6; R 0.5–1.5) | 7015 | ILAKFLHWL | A*0201 | [55] | |
| KLFGVLRLK | A*03 | [54] | |||
| VYGFVRACL | A*2402 | [3] | |||
| VYAETKHFL | A*2402 | [3] |
M median; R range of overexpression
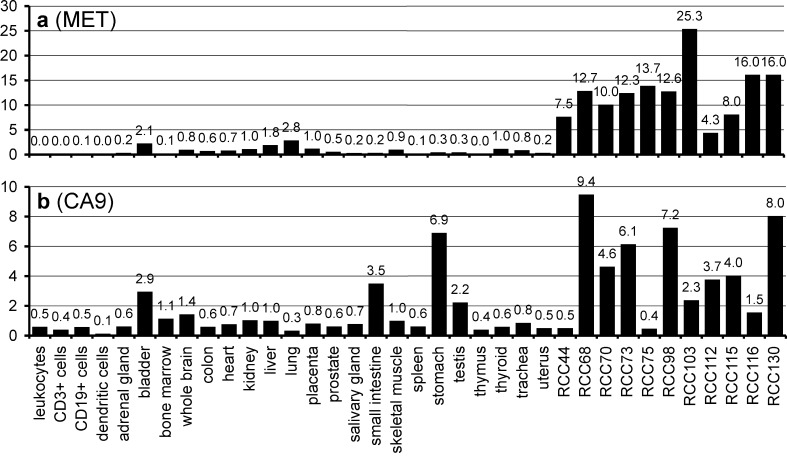
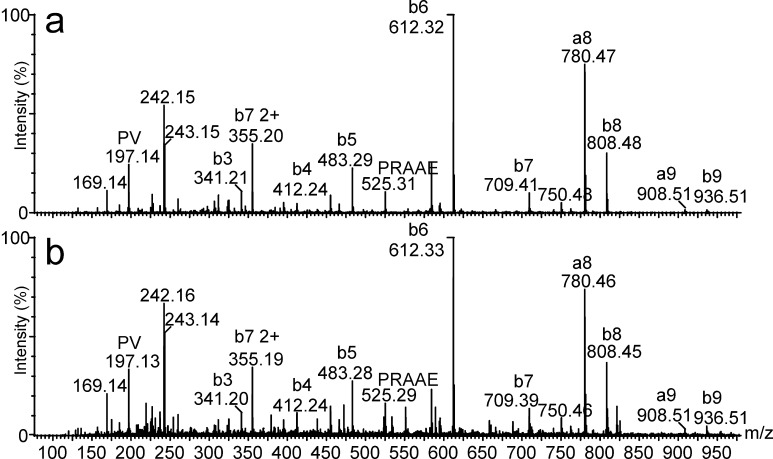
Both met proto-oncogene [1] and CA9 [15] are known to be expressed by the vast majority of RCCs. This was confirmed by our data: MET mRNA was upregulated in all analyzed RCCs by 12.3-fold in average in comparison to healthy kidney tissue (Table 3), with no relevant expression in all other human tissues (Fig. 2a). The HLA-A*02-presented CTL epitope YVDPVITSI [56] was previously shown to mediate tumor cell lysis in vitro [43]. Carbonic anhydrase 9 (CA9; G250), the only known tumor-associated isoform of carbonic anhydrase [15, 35], is also expressed by a set of other malignancies, for example breast cancer [48], non-small-cell lung cancer [50], and squamous-cell head and neck cancer [20, 26]. CA9 expression in general is hypoxia-inducible, and was suggested to be an endogenous marker for tumor hypoxia [50]. The frequent deletion of the von Hippel–Lindau tumor suppressor gene (VHL) in the case of RCC is associated with the upregulation of CA9, a characteristic antigen for RCC [36]. The level of CA9 expression was also shown to be an independent prognostic marker for this disease [5]. In vivo studies show that the monoclonal anti-CA9 antibody G250 exclusively binds to tumor cells, and that CA9 can be used as a therapeutic target [9, 32, 33]. In consequence, it has been targeted in various investigational therapeutic approaches in RCC [17, 23, 27] and is also considered a suitable source of epitopes in CTL-based immunotherapy. Although we could not detect the prominent HLA-A*02-restricted CTL epitope, HLSTAFARV [53], the HLA-B*07-presented CA9 peptide SPRAAEPVQL was sequenced by collision-induced tandem mass spectrometry (Fig. 3) and represents a promising candidate for peptide-based immunotherapy. CA9 was overexpressed in 7/11 RCCs as expected [34]; expression in normal tissue was relevant only in stomach, small intestine, and bladder (Fig. 2b), consistent with previous reports [32, 34].
Fig. 2.
mRNA expression profiles of met proto-oncogene (MET) (a) and carbonic anhydrase 9 (CA9) (b) in 11 analyzed RCCs and various human tissues. Relative expression values are normalized to kidney (expression=1). MET appeared highly overexpressed in all tumors, CA9 was overexpressed over kidney in most tumors, while stomach and small intestine also showed high expression of CA9
Fig. 3.
Fragmentation-induced mass spectra of the HLA-B*07-presented CA9 peptide SPRAAEPVQL. a Synthetic peptide, b peptide extracted after immunoprecipitation of tumor HLA
The most abundant source of HLA ligands in the group of reported tumor antigens was adipose differentiation-related protein adipophilin (ADFP), from which peptides were detected in five of thirteen investigated tumors. This led to the characterization of seven different peptides with different HLA restrictions (Table 2). According to this, adipophilin ranked second among our frequent source protein for peptides after vimentin, suggesting a high abundance of adipophilin peptides on the surface of RCC cells. Adipophilin was also highly overexpressed in most RCCs of the clear cell subtype (data not shown), whereas both chromophilic RCCs, RCC44 and RCC75, showed no upregulation of adipophilin at the mRNA level. The only tissues with relevant adipophilin expression are female mammary glands and placenta. This suggests that adipophilin-derived peptides can be used for vaccination in male patients with RCC, especially in clear-cell-type malignancies. One of the adipophilin-derived HLA ligands, the peptide SVASTITGV presented by HLA-A*02, was recently shown to be a T-cell epitope, which mediates tumor cell lysis in vitro [45]. The novel adipophilin peptides cover a broad range of HLA restrictions (Table 2), and thus represent candidate vaccination peptides in our concept of patient- individual immunotherapy [39].
Complementary analysis of gene expression and peptide presentation leads to the identification of new broadly expressed tumor-associated HLA ligands
A set of proteins was repeatedly found to be upregulated at the mRNA level and a source of HLA ligands in RCC (Table 4). These proteins are, according to their expression profiles, potential sources for vaccination peptides either in all or most patients as for IGFBP3, APOL1, and RGS5, or only in few patients as for MMP7 or ACSL4.
Table 4.
Novel RCC-associated antigens identified by overexpression and the source of HLA ligands presented by several allotypes
| Source protein overexpression > threefold in X/11 RCCs | Entrez Gene ID | Sequence | HLA restriction | References |
|---|---|---|---|---|
| Apolipoprotein L, 1 (APOL1) 9/11 (M 7.1; R 1.1–40.2) | 8542 | FLGENISNFL | A*0201 | [11, 56] |
| ALADGVQKV | A*0201 | [11, 56] | ||
| Insulin-like growth factor binding protein 3 (IGFBP3) 8/11 (M 6.0; R 2.0–10.2) | 3486 | RPTLWAAAL | B*07 | |
| Regulator of G-protein signalling 5 (RGS5) 7/11 (M 7.2; R 0.3–14.9) | 8490 | GLASFKSFLK | A*03 | |
| LAALPHSCL | A*02 | |||
| Matrix metalloproteinase 7, matrilysin (MMP7) 4/11 (M 2.4; R 0.3–12.1) | 4316 | FPNSPKWTSK | A*03 | |
| SLFPNSPKWTSK | A*03 | |||
| Cytochrome P450, family 1, subfamily B, polypeptide 1 (CYP1B1) 3/11 (M 0.6; R 0.3–9.2) | 1545 | FLDPRPLTV | A*02 | |
| Acyl-CoA synthetase long-chain family member 4 (ACSL4) 1/11 (M 1.3; R 0.8–7.1) | 2182 | KLFDHAVSKF | A*03 | |
| VPNQKRLTLL | B*07 |
M median; R range of overexpression
Insulin-like growth factor 1 (IGF1) was shown to be involved in the progression of malignancies derived from proximal tubule epithelial cells, and IGFBP, among them IGFBP3, are known to be upregulated in clear cell RCC [1, 8, 18]. In our investigations, IGFBP3 was upregulated in at least 8 of 11 analyzed specimens (Table 4) with no relevant expression in normal tissues. One in vivo processed peptide from IGFBP3, RPTLWAAAL, was presented by HLA-B*07.
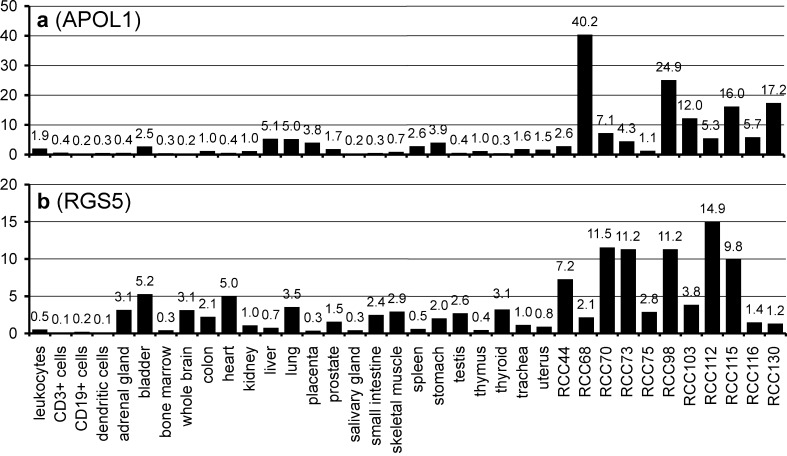
The expression profile of APOL1 also suggests tumor association: 9 of 11 clear cell carcinomas that were analyzed showed an upregulation of APOL1 in comparison to normal kidney (Table 4), although the factors of overexpression were rather heterogeneous (Fig. 4a). The repeated detection of HLA ligands derived from APOL1 and their extensive overexpression in tumors RCC68, RCC98, RCC115, and RCC130 justifies the usage of APOL1 peptides for vaccination in these patients according to our criteria.
Fig. 4.
mRNA expression profiles of a apolipoprotein L1 (APOL1) and b regulator of G-protein signalling 5 (RGS5). APOL1 is extensively upregulated in RCC68, RCC98, RCC115, and RCC130. RGS5 appears overexpressed in RCC70, RCC73, RCC98, RCC112, and RCC115
Regulator of G-protein signalling 5, from which two HLA ligands were detected, was upregulated in 7 of the 11 tumors that were tested (Table 4) and reported to be overexpressed in RCCs previously [1, 13]. However, in comparison to the other healthy tissues, RGS5 shows a very heterogeneous pattern of expression (Fig. 4b), which necessitates an individual expression analysis of each tumor before its peptides are used for vaccination.
Some HLA ligands are tumor-associated candidates in individual cases only
Met proto-oncogene, CA9, ADFP, IGFBP3, and APOL1 represent antigens overexpressed in all or most RCCs. Such antigens provide a source of vaccination peptides per se, even if only few naturally processed peptides are known. However, individual patterns of gene expression may be found in individual cancer specimens. Therefore, we place our emphasis on a patient-individual concept of immunotherapy and perform individualized gene expression and peptide analysis [39]. In this individual approach we also use peptides derived from genes that are exclusively upregulated in one or only a few patients.
One example of such an antigen is MMP7 (matrilysin), which was shown to be expressed in cancer cells of various origins and to play a role in the process of metastasis [38, 41, 57]. Apart from high expression levels in RCC68, RCC98, and RCC116 (Table 4), we detected relevant MMP7 expression only in the bladder. The HLA-A*03 ligand SLFPNSPKWTSK and its shorter variant FPNSPKWTSK were found on RCC75 and RCC98. It has to be mentioned that for these peptides and for all other HLA ligands described in this section and the preceding section, no data on T-cell reactions exist so far.
ACSL4 was overexpressed in one patient of the chromophilic subtype, RCC75 (Table 4). ACSL4 overexpression was recently reported to be associated with colon adenocarcinoma [6] and hepatocellular carcinoma [49]. From RCC75, the HLA-A*03-presented peptide KLFDHAVSKF was characterized and later also found in RCC98. ACSL4 stands for an antigen which might be used for vaccination only in particular cases.
Gene expression profiles from 11 RCCs allow for the identification of novel candidate RCC antigens
While gene expression analysis yields comprehensive data, HLA ligand characterization does not: from the estimated over 10,000 peptides making up the HLA class I ligandome of a given cell, only a very low percentage can be identified with current tools and strategies. In contrast, almost every gene of the human genome can be assessed by gene expression profiling.
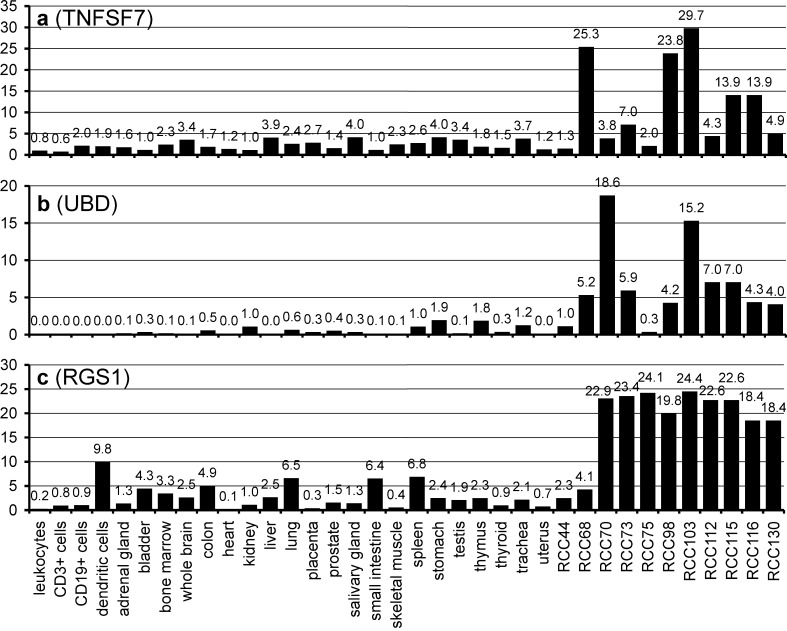
Therefore, we searched our gene expression data for genes upregulated in most tumors in relation to healthy tissues, even without identified HLA ligands. Here, we present three promising antigens that emerged from these analyses. The tumor necrosis factor (ligand) superfamily, member 7 (TNFSF7), was overexpressed in 5 of 11 tumors (Fig. 5a). Ubiquitin D (UBD) is characterized by undetectable expression in most healthy tissues but strong expression in clear cell RCCs (Fig. 5b). One peptide from UBD, DANPYDSVKKI (HLA-B*51), has recently been identified [30]. Our third example of a consistently overexpressed protein in RCC (9 of the 11 tumors tested with a more than 18-fold overexpression) is RGS1 (Fig. 5c). In contrast to RGS5, which has already been described as upregulated in RCC [1, 13], RGS1 has not yet been mentioned in the context of RCC. Interestingly, no other members of the RGS family were overexpressed in our RCC samples. Unfortunately, no HLA-presented peptides from RGS1 are known so far.
Fig. 5.
Novel potential RCC-associated antigens, identified by their overexpression in RCC. a Tumor necrosis factor (ligand) superfamily, member 7 (TNFSF7), b Ubiquitin D (UBD), c Regulator of G-protein signalling 1 (RGS1). HLA-presented peptides have been identified only for UBD so far
Conclusions
In this report, we present data resulting from a systematic large-scale analysis of HLA-peptide presentation patterns and mRNA-expression profiles in RCC. We identified a number of novel HLA ligands from reported RCC antigens such as adipophilin and CA9 and confirmed the constitutively high expression of the classical antigens CA9, ADFP, and MET, whereas no evidence was revealed for a concurrent elevated expression level of most other previously suggested TAA. Various proteins constitutively or sporadically overexpressed in RCC were suggested TAA, for example RGS5, RGS1, IGFBP3, and APOL1. From some of these proteins, novel HLA class I peptides were characterized that might turn out to represent target epitopes for CTL responses. Future T-cell work will have to reveal the immunogenicity of these peptides. The therapeutic impact of a vaccination treatment with the peptides mentioned is currently under intensive investigation.
Acknowledgements
This work was supported by the Deutsche Forschungsgemeinschaft (SFB 510 and Graduiertenkolleg 794), the European Union (LSHB-CT-2003-503231 GenomesToVaccines), and by the German Federal Ministry of Education and Research (Fö. 01KS9602) in connection with the Interdisciplinary Center of Clinical Research, Tübingen (IZKF, Project S.04.00088). We thank Lynne Yakes for critically reading the manuscript and Patricia Hrstić for perfect technical assistance.
Footnotes
Tobias Krüger and Oliver Schoor contributed equally to this work.
References
- 1.Amatschek S, Koenig U, Auer H, Steinlein P, Pacher M, Gruenfelder A, Dekan G, Vogl S, Kubista E, Heider KH, Stratowa C, Schreiber M, Sommergruber W. Tissue-wide expression profiling using cDNA subtraction and microarrays to identify tumor-specific genes. Cancer Res. 2004;64:844. doi: 10.1158/0008-5472.can-03-2361. [DOI] [PubMed] [Google Scholar]
- 2.Apostolopoulos V, Karanikas V, Haurum JS, McKenzie IF. Induction of HLA-A2-restricted CTLs to the mucin 1 human breast cancer antigen. J Immunol. 1997;159:5211. [PubMed] [Google Scholar]
- 3.Arai J, Yasukawa M, Ohminami H, Kakimoto M, Hasegawa A, Fujita S. Identification of human telomerase reverse transcriptase-derived peptides that induce HLA-A24-restricted antileukemia cytotoxic T lymphocytes. Blood. 2001;97:2903. doi: 10.1182/blood.V97.9.2903. [DOI] [PubMed] [Google Scholar]
- 4.Brossart P, Heinrich KS, Stuhler G, Behnke L, Reichardt VL, Stevanovic S, Muhm A, Rammensee HG, Kanz L, Brugger W. Identification of HLA-A2-restricted T-cell epitopes derived from the MUC1 tumor antigen for broadly applicable vaccine therapies. Blood. 1999;93:4309. [PubMed] [Google Scholar]
- 5.Bui MH, Seligson D, Han KR, Pantuck AJ, Dorey FJ, Huang Y, Horvath S, Leibovich BC, Chopra S, Liao SY, Stanbridge E, Lerman MI, Palotie A, Figlin RA, Belldegrun AS. Carbonic anhydrase IX is an independent predictor of survival in advanced renal clear cell carcinoma: implications for prognosis and therapy. Clin Cancer Res. 2003;9:802. [PubMed] [Google Scholar]
- 6.Cao Y, Dave KB, Doan TP, Prescott SM. Fatty acid CoA ligase 4 is up-regulated in colon adenocarcinoma. Cancer Res. 2001;61:8429. [PubMed] [Google Scholar]
- 7.Celis E, Tsai V, Crimi C, DeMars R, Wentworth PA, Chesnut RW, Grey HM, Sette A, Serra HM. Induction of anti-tumor cytotoxic T lymphocytes in normal humans using primary cultures and synthetic peptide epitopes. Proc Natl Acad Sci USA. 1994;91:2105. doi: 10.1073/pnas.91.6.2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheung CW, Vesey DA, Nicol DL, Johnson DW. The roles of IGF-I and IGFBP-3 in the regulation of proximal tubule, and renal cell carcinoma cell proliferation. Kidney Int. 2004;65:1272. doi: 10.1111/j.1523-1755.2004.00535.x. [DOI] [PubMed] [Google Scholar]
- 9.Divgi CR, Bander NH, Scott AM, O’Donoghue JA, Sgouros G, Welt S, Finn RD, Morrissey F, Capitelli P, Williams JM, Deland D, Nakhre A, Oosterwijk E, Gulec S, Graham MC, Larson SM, Old LJ. Phase I/II radioimmunotherapy trial with iodine-131-labeled monoclonal antibody G250 in metastatic renal cell carcinoma. Clin Cancer Res. 1998;4:2729. [PubMed] [Google Scholar]
- 10.Falk K, Rotzschke O, Stevanovic S, Jung G, Rammensee HG. Allele-specific motifs revealed by sequencing of self-peptides eluted from MHC molecules. Nature. 1991;351:290. doi: 10.1038/351290a0. [DOI] [PubMed] [Google Scholar]
- 11.Flad T, Spengler B, Kalbacher H, Brossart P, Baier D, Kaufmann R, Bold P, Metzger S, Bluggel M, Meyer HE, Kurz B, Muller CA. Direct identification of major histocompatibility complex class I-bound tumor-associated peptide antigens of a renal carcinoma cell line by a novel mass spectrometric method. Cancer Res. 1998;58:5803. [PubMed] [Google Scholar]
- 12.Fleischhauer K, Tanzarella S, Russo V, Sensi ML, van der BP, Bordignon C, Traversari C. Functional heterogeneity of HLA-A*02 subtypes revealed by presentation of a MAGE-3-encoded peptide to cytotoxic T cell clones. J Immunol. 1997;159:2513. [PubMed] [Google Scholar]
- 13.Furuya M, Nishiyama M, Kimura S, Suyama T, Naya Y, Ito H, Nikaido T, Ishikura H. Expression of regulator of G protein signalling protein 5 (RGS5) in the tumour vasculature of human renal cell carcinoma. J Pathol. 2004;203:551. doi: 10.1002/path.1543. [DOI] [PubMed] [Google Scholar]
- 14.Gaugler B, Brouwenstijn N, Vantomme V, Szikora JP, Van der Spek CW, Patard JJ, Boon T, Schrier P, Van den Eynde BJ. A new gene coding for an antigen recognized by autologous cytolytic T lymphocytes on a human renal carcinoma. Immunogenetics. 1996;44:323. doi: 10.1007/s002510050133. [DOI] [PubMed] [Google Scholar]
- 15.Grabmaier K, Vissers JL, De Weijert MC, Oosterwijk-Wakka JC, Van Bokhoven A, Brakenhoff RH, Noessner E, Mulders PA, Merkx G, Figdor CG, Adema GJ, Oosterwijk E. Molecular cloning and immunogenicity of renal cell carcinoma-associated antigen G250. Int J Cancer. 2000;85:865. doi: 10.1002/(SICI)1097-0215(20000315)85:6<865::AID-IJC21>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 16.Herman J, van der Bruggen P, Luescher IF, Mandruzzato S, Romero P, Thonnard J, Fleischhauer K, Boon T, Coulie PG. A peptide encoded by the human MAGE3 gene and presented by HLA-B44 induces cytolytic T lymphocytes that recognize tumor cells expressing MAGE3. Immunogenetics. 1996;43:377. doi: 10.1007/s002510050078. [DOI] [PubMed] [Google Scholar]
- 17.Hernandez JM, Bui MH, Han KR, Mukouyama H, Freitas DG, Nguyen D, Caliliw R, Shintaku PI, Paik SH, Tso CL, Figlin RA, Belldegrun AS. Novel kidney cancer immunotherapy based on the granulocyte-macrophage colony-stimulating factor and carbonic anhydrase IX fusion gene. Clin Cancer Res. 2003;9:1906. [PubMed] [Google Scholar]
- 18.Hintz RL, Bock S, Thorsson AV, Bovens J, Powell DR, Jakse G, Petrides PE. Expression of the insulin like growth factor-binding protein 3 (IGFBP-3) gene is increased in human renal carcinomas. J Urol. 1991;146:1160. doi: 10.1016/s0022-5347(17)38031-x. [DOI] [PubMed] [Google Scholar]
- 19.Ikeda H, Lethe B, Lehmann F, van Baren N, Baurain JF, De Smet C, Chambost H, Vitale M, Moretta A, Boon T, Coulie PG. Characterization of an antigen that is recognized on a melanoma showing partial HLA loss by CTL expressing an NK inhibitory receptor. Immunity. 1997;6:199. doi: 10.1016/S1074-7613(00)80426-4. [DOI] [PubMed] [Google Scholar]
- 20.Ivanov S, Liao SY, Ivanova A, Danilkovitch-Miagkova A, Tarasova N, Weirich G, Merrill MJ, Proescholdt MA, Oldfield EH, Lee J, Zavada J, Waheed A, Sly W, Lerman MI, Stanbridge EJ. Expression of hypoxia-inducible cell-surface transmembrane carbonic anhydrases in human cancer. Am J Pathol. 2001;158:905. doi: 10.1016/S0002-9440(10)64038-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jager E, Chen YT, Drijfhout JW, Karbach J, Ringhoffer M, Jager D, Arand M, Wada H, Noguchi Y, Stockert E, Old LJ, Knuth A. Simultaneous humoral and cellular immune response against cancer-testis antigen NY-ESO-1: definition of human histocompatibility leukocyte antigen (HLA)-A2-binding peptide epitopes. J Exp Med. 1998;187:265. doi: 10.1084/jem.187.2.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jemal A, Tiwari RC, Murray T, Ghafoor A, Samuels A, Ward E, Feuer EJ, Thun MJ. Cancer statistics, 2004. CA Cancer J Clin. 2004;54:8. doi: 10.3322/canjclin.54.1.8. [DOI] [PubMed] [Google Scholar]
- 23.Jongmans W, van den Oudenalder K, Tiemessen DM, Molkenboer J, Willemsen R, Mulders PF, Oosterwijk E. Targeting of adenovirus to human renal cell carcinoma cells. Urology. 2003;62:559. doi: 10.1016/S0090-4295(03)00378-9. [DOI] [PubMed] [Google Scholar]
- 24.Kawashima I, Hudson SJ, Tsai V, Southwood S, Takesako K, Appella E, Sette A, Celis E. The multi-epitope approach for immunotherapy for cancer: identification of several CTL epitopes from various tumor-associated antigens expressed on solid epithelial tumors. Hum Immunol. 1998;59:1. doi: 10.1016/s0198-8859(97)00255-3. [DOI] [PubMed] [Google Scholar]
- 25.Kessler JH, Beekman NJ, Bres-Vloemans SA, Verdijk P, van Veelen PA, Kloosterman-Joosten AM, Vissers DC, ten Bosch GJ, Kester MG, Sijts A, Wouter DJ, Ossendorp F, Offringa R, Melief CJ. Efficient identification of novel HLA-A(*)0201-presented cytotoxic T lymphocyte epitopes in the widely expressed tumor antigen PRAME by proteasome-mediated digestion analysis. J Exp Med. 2001;193:73. doi: 10.1084/jem.193.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koukourakis MI, Giatromanolaki A, Sivridis E, Simopoulos K, Pastorek J, Wykoff CC, Gatter KC, Harris AL. Hypoxia-regulated carbonic anhydrase-9 (CA9) relates to poor vascularization and resistance of squamous cell head and neck cancer to chemoradiotherapy. Clin Cancer Res. 2001;7:3399. [PubMed] [Google Scholar]
- 27.Kranenborg MH, Boerman OC, Oosterwijk-Wakka JC, De Weijert MC, Corstens FH, Oosterwijk E. Development and characterization of anti-renal cell carcinoma x antichelate bispecific monoclonal antibodies for two-phase targeting of renal cell carcinoma. Cancer Res. 1995;55:5864s. [PubMed] [Google Scholar]
- 28.Lemmel C, Weik S, Eberle U, Dengjel J, Kratt T, Becker HD, Rammensee HG, Stevanovic S. Differential quantitative analysis of MHC ligands by mass spectrometry using stable isotope labeling. Nat Biotechnol. 2004;22:450. doi: 10.1038/nbt947. [DOI] [PubMed] [Google Scholar]
- 29.Lokich J. Spontaneous regression of metastatic renal cancer. Case report and literature review. Am J Clin Oncol. 1997;20:416. doi: 10.1097/00000421-199708000-00020. [DOI] [PubMed] [Google Scholar]
- 30.Luckey CJ, Marto JA, Partridge M, Hall E, White FM, Lippolis JD, Shabanowitz J, Hunt DF, Engelhard VH. Differences in the expression of human class I MHC alleles and their associated peptides in the presence of proteasome inhibitors. J Immunol. 2001;167:1212. doi: 10.4049/jimmunol.167.3.1212. [DOI] [PubMed] [Google Scholar]
- 31.Oiso M, Eura M, Katsura F, Takiguchi M, Sobao Y, Masuyama K, Nakashima M, Itoh K, Ishikawa T. A newly identified MAGE-3-derived epitope recognized by HLA-A24-restricted cytotoxic T lymphocytes. Int J Cancer. 1999;81:387. doi: 10.1002/(SICI)1097-0215(19990505)81:3<387::AID-IJC12>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 32.Oosterwijk E, Bander NH, Divgi CR, Welt S, Wakka JC, Finn RD, Carswell EA, Larson SM, Warnaar SO, Fleuren GJ. Antibody localization in human renal cell carcinoma: a phase I study of monoclonal antibody G250. J Clin Oncol. 1993;11:738. doi: 10.1200/JCO.1993.11.4.738. [DOI] [PubMed] [Google Scholar]
- 33.Oosterwijk E, Debruyne FM. Radiolabeled monoclonal antibody G250 in renal-cell carcinoma. World J Urol. 1995;13:186. doi: 10.1007/BF00184877. [DOI] [PubMed] [Google Scholar]
- 34.Oosterwijk E, Debruyne FM, Schalken JA. The use of monoclonal antibody G250 in the therapy of renal-cell carcinoma. Semin Oncol. 1995;22:34. [PubMed] [Google Scholar]
- 35.Oosterwijk E, Ruiter DJ, Hoedemaeker PJ, Pauwels EK, Jonas U, Zwartendijk J, Warnaar SO. Monoclonal antibody G 250 recognizes a determinant present in renal-cell carcinoma and absent from normal kidney. Int J Cancer. 1986;38:489. doi: 10.1002/ijc.2910380406. [DOI] [PubMed] [Google Scholar]
- 36.Pantuck AJ, Zeng G, Belldegrun AS, Figlin RA. Pathobiology, prognosis, and targeted therapy for renal cell carcinoma: exploiting the hypoxia-induced pathway. Clin Cancer Res. 2003;9:4641. [PubMed] [Google Scholar]
- 37.Pascolo S, Schirle M, Guckel B, Dumrese T, Stumm S, Kayser S, Moris A, Wallwiener D, Rammensee HG, Stevanovic S. A MAGE-A1 HLA-A A*0201 epitope identified by mass spectrometry. Cancer Res. 2001;61:4072. [PubMed] [Google Scholar]
- 38.Powell WC, Knox JD, Navre M, Grogan TM, Kittelson J, Nagle RB, Bowden GT. Expression of the metalloproteinase matrilysin in DU-145 cells increases their invasive potential in severe combined immunodeficient mice. Cancer Res. 1993;53:417. [PubMed] [Google Scholar]
- 39.Rammensee HG, Weinschenk T, Gouttefangeas C, Stevanovic S. Towards patient-specific tumor antigen selection for vaccination. Immunol Rev. 2002;188:164. doi: 10.1034/j.1600-065X.2002.18815.x. [DOI] [PubMed] [Google Scholar]
- 40.Renkvist N, Castelli C, Robbins PF, Parmiani G. A listing of human tumor antigens recognized by T cells. Cancer Immunol Immunother. 2001;50:3. doi: 10.1007/s002620000169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rudolph-Owen LA, Chan R, Muller WJ, Matrisian LM. The matrix metalloproteinase matrilysin influences early-stage mammary tumorigenesis. Cancer Res. 1998;58:5500. [PubMed] [Google Scholar]
- 42.Sadovnikova E, Jopling LA, Soo KS, Stauss HJ. Generation of human tumor-reactive cytotoxic T cells against peptides presented by non-self HLA class I molecules. Eur J Immunol. 1998;28:193. doi: 10.1002/(SICI)1521-4141(199801)28:01<193::AID-IMMU193>3.3.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 43.Schag K, Schmidt SM, Muller MR, Weinschenk T, Appel S, Weck MM, Grunebach F, Stevanovic S, Rammensee HG, Brossart P. Identification of C-met oncogene as a broadly expressed tumor-associated antigen recognized by cytotoxic T-lymphocytes. Clin Cancer Res. 2004;10:3658. doi: 10.1158/1078-0432.CCR-03-0640. [DOI] [PubMed] [Google Scholar]
- 44.Schirle M, Keilholz W, Weber B, Gouttefangeas C, Dumrese T, Becker HD, Stevanovic S, Rammensee HG. Identification of tumor-associated MHC class I ligands by a novel T cell-independent approach. Eur J Immunol. 2000;30:2216. doi: 10.1002/1521-4141(2000)30:8<2216::AID-IMMU2216>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 45.Schmidt SM, Schag K, Muller MR, Weinschenk T, Appel S, Schoor O, Weck MM, Grunebach F, Kanz L, Stevanovic S, Rammensee HG, Brossart P. Induction of adipophilin-specific cytotoxic T lymphocytes using a novel HLA-A2-binding peptide that mediates tumor cell lysis. Cancer Res. 2004;64:1164. doi: 10.1158/0008-5472.can-03-2538. [DOI] [PubMed] [Google Scholar]
- 46.Schmitz M, Diestelkoetter P, Weigle B, Schmachtenberg F, Stevanovic S, Ockert D, Rammensee HG, Rieber EP. Generation of survivin-specific CD8+ T effector cells by dendritic cells pulsed with protein or selected peptides. Cancer Res. 2000;60:4845. [PubMed] [Google Scholar]
- 47.Schultz ES, Zhang Y, Knowles R, Tine J, Traversari C, Boon T, van der Bruggen P. A MAGE-3 peptide recognized on HLA-B35 and HLA-A1 by cytolytic T lymphocytes. Tissue Antigens. 2001;57:103. doi: 10.1034/j.1399-0039.2001.057002103.x. [DOI] [PubMed] [Google Scholar]
- 48.Span PN, Bussink J, Manders P, Beex LV, Sweep CG. Carbonic anhydrase-9 expression levels and prognosis in human breast cancer: association with treatment outcome. Br J Cancer. 2003;89:271. doi: 10.1038/sj.bjc.6601122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sung YK, Hwang SY, Park MK, Bae HI, Kim WH, Kim JC, Kim M. Fatty acid-CoA ligase 4 is overexpressed in human hepatocellular carcinoma. Cancer Sci. 2003;94:421. doi: 10.1111/j.1349-7006.2003.tb01458.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Swinson DE, Jones JL, Richardson D, Wykoff C, Turley H, Pastorek J, Taub N, Harris AL, O’Byrne KJ. Carbonic anhydrase IX expression, a novel surrogate marker of tumor hypoxia, is associated with a poor prognosis in non-small-cell lung cancer. J Clin Oncol. 2003;21:473. doi: 10.1200/JCO.2003.11.132. [DOI] [PubMed] [Google Scholar]
- 51.Tanaka F, Fujie T, Tahara K, Mori M, Takesako K, Sette A, Celis E, Akiyoshi T. Induction of antitumor cytotoxic T lymphocytes with a MAGE-3-encoded synthetic peptide presented by human leukocytes antigen-A24. Cancer Res. 1997;57:4465. [PubMed] [Google Scholar]
- 52.van der BP, Bastin J, Gajewski T, Coulie PG, Boel P, De Smet C, Traversari C, Townsend A, Boon T. A peptide encoded by human gene MAGE-3 and presented by HLA-A2 induces cytolytic T lymphocytes that recognize tumor cells expressing MAGE-3. Eur J Immunol. 1994;24:3038. doi: 10.1002/eji.1830241218. [DOI] [PubMed] [Google Scholar]
- 53.Vissers JL, De Vries IJ, Schreurs MW, Engelen LP, Oosterwijk E, Figdor CG, Adema GJ. The renal cell carcinoma-associated antigen G250 encodes a human leukocyte antigen (HLA)-A2.1-restricted epitope recognized by cytotoxic T lymphocytes. Cancer Res. 1999;59:5554. [PubMed] [Google Scholar]
- 54.Vonderheide RH, Anderson KS, Hahn WC, Butler MO, Schultze JL, Nadler LM. Characterization of HLA-A3-restricted cytotoxic T lymphocytes reactive against the widely expressed tumor antigen telomerase. Clin Cancer Res. 2001;7:3343. [PubMed] [Google Scholar]
- 55.Vonderheide RH, Hahn WC, Schultze JL, Nadler LM. The telomerase catalytic subunit is a widely expressed tumor-associated antigen recognized by cytotoxic T lymphocytes. Immunity. 1999;10:673. doi: 10.1016/S1074-7613(00)80066-7. [DOI] [PubMed] [Google Scholar]
- 56.Weinschenk T, Gouttefangeas C, Schirle M, Obermayr F, Walter S, Schoor O, Kurek R, Loeser W, Bichler KH, Wernet D, Stevanovic S, Rammensee HG. Integrated functional genomics approach for the design of patient-individual antitumor vaccines. Cancer Res. 2002;62:5818. [PubMed] [Google Scholar]
- 57.Wilson CL, Heppner KJ, Labosky PA, Hogan BL, Matrisian LM. Intestinal tumorigenesis is suppressed in mice lacking the metalloproteinase matrilysin. Proc Natl Acad Sci USA. 1997;94:1402. doi: 10.1073/pnas.94.4.1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Young AN, Amin MB, Moreno CS, Lim SD, Cohen C, Petros JA, Marshall FF, Neish AS. Expression profiling of renal epithelial neoplasms: a method for tumor classification and discovery of diagnostic molecular markers. Am J Pathol. 2001;158:1639. doi: 10.1016/S0002-9440(10)64120-X. [DOI] [PMC free article] [PubMed] [Google Scholar]