Abstract
Background
Comprehensive genome profiling (CGP) serves as a guide for suitable genomically matched therapies for patients with cancer. However, little is known about the impact of the timing and types of cancer on the therapeutic benefit of CGP.
Materials and methods
A single hospital-based pan-cancer prospective study (TOP-GEAR; UMIN000011141) was conducted to examine the benefit of CGP with respect to the timing and types of cancer. Patients with advanced solid tumors (>30 types) who either progressed with or without standard treatments were genotyped using a single CGP test. The subjects were followed up for a median duration of 590 days to examine therapeutic response, using progression-free survival (PFS), PFS ratio, and factors associated with therapeutic response.
Results
Among the 507 patients, 62 (12.2%) received matched therapies with an overall response rate (ORR) of 32.3%. The PFS ratios (≥1.3) were observed in 46.3% (19/41) of the evaluated patients. The proportion of subjects receiving such therapies in the rare cancer cohort was lower than that in the non-rare cancer cohort (9.6% and 17.4%, respectively; P = 0.010). However, ORR of the rare cancer patients was higher than that in the non-rare cancer cohort (43.8% and 20.0%, respectively; P = 0.046). Moreover, ORR of matched therapies in the first or second line after receiving the CGP test was higher than that in the third or later lines (62.5% and 21.7%, respectively; P = 0.003). Rare cancer and early-line treatment were significantly and independently associated with ORR of matched therapies in multivariable analysis (P = 0.017 and 0.004, respectively).
Conclusion
Patients with rare cancer preferentially benefited from tumor mutation profiling by increasing the chances of therapeutic response to matched therapies. Early-line treatments after profiling increase the therapeutic benefit, irrespective of tumor types.
Key words: comprehensive genome profiling, precision oncology, cancer, matched therapy
Highlights
-
•
Of 507 patients who underwent CGP, 12.2% received genomically matched therapies with a 32.3% ORR.
-
•
The PFS ratios (>1.3) were observed in 46.3% (19/41) of the evaluated patients.
-
•
Patients with rare cancers benefited most from tumor mutation profiling.
-
•
Early-line treatments following profiling increase the therapeutic benefit, irrespective of tumor type.
Introduction
Next-generation sequencing (NGS)-based comprehensive genomic profiling (CGP) is commonly used to examine somatic and germline mutations of 10 or more cancer-related genes. The technique is carried out to guide suitable genomically matched therapies in patients with advanced cancer through the identification of actionable genomic alterations. The CGP-based precision oncology is often undertaken in a tumor-agnostic manner, based on the prediction that patients would respond to molecular-targeted therapies according to their gene alterations. Several pan-cancer studies, including ours, have demonstrated that ∼18% of patients were enrolled in genomically matched clinical trials based on CGP findings (Supplementary Table S1, available at https://doi.org/10.1016/j.esmoop.2024.102981).1, 2, 3, 4, 5, 6, 7 In a few of the studies, the therapeutic responses of the trials were also reported to be favorable2,4,6,7 (Supplementary Table S1, available at https://doi.org/10.1016/j.esmoop.2024.102981). These findings underpin the authenticity of the strategy for CGP-based precision oncology.
In clinical settings, however, genomically matched therapies are also undertaken mainly through standard treatments (as published by regulatory authorities) and off-label use of the approved drugs other than enrollment in clinical trials of investigational drugs. In fact, CGP is beneficial to patients in such a real-world setting, including enrollments into clinical trials and compassionate and/or off-label use of drugs.8,9 However, it has not yet been fully investigated whether patients with certain types of cancer or on timing (treatment lines) are more likely to benefit from CGP-based precision oncology.
Here, we conducted a hospital-based prospective observational study to determine the impact of the types of cancer and timing (treatment lines) on the derivable therapeutic benefits from CGP.
Materials and methods
Study description
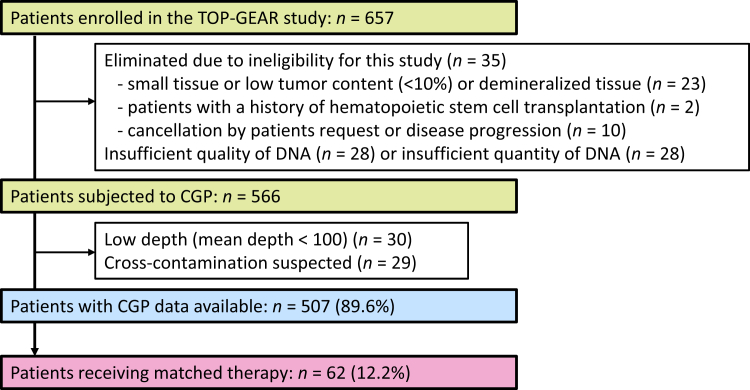
This study was undertaken as a hospital-based prospective observational study (Trial of Onco-panel for Gene-profiling to estimate both adverse events and response during cancer treatment; TOP-GEAR, UMIN 000011141) to address the clinical and genetic factors that can influence clinical benefits from CGP-based precision oncology.10,11 This study was approved by the Institutional Review Board of the National Cancer Center. All patients signed a written informed consent document. Between May 2016 and March 2018, 657 patients were enrolled at the National Cancer Center Hospital (NCCH). A total of 91 patients were eliminated due to ineligibility for this study (n = 35), insufficient quality of DNA (n = 28), or insufficient quantity of DNA (n = 28), and 566 patients were analyzed with NCC Oncopanel.10,11 Details of the enrollment are presented in Figure 1. Patients who met the following criteria were eligible for inclusion: presentation of advanced solid tumors with disease progression during standard therapies or without appropriate standard treatments, rare cancers (with an incidence rate of <6 per 100 000 per year),10 or cancer of unknown primary; and age (≥16 years from May 2016, and ≥1 year from April 2018). There was no limitation on the number of previous treatments taken by the patients.
Figure 1.
CONSORT diagram for outcome survey in the TOP-GEAR study. CGP, comprehensive genome profiling.
Genomic profiling of tumors
The NCC Oncopanel was designed to examine somatic and germline mutations, amplifications, and homozygous deletions within the entire coding region of 114 genes of clinical or preclinical relevance11,12 (Supplementary Table S2, available at https://doi.org/10.1016/j.esmoop.2024.102981). Moreover, there were fusions of 12 oncogenes in the panel. For analyses, 5 or 10 sections of 10- or 4- to 5-μm sections, respectively, were prepared from archival formalin-fixed and paraffin-embedded tumor specimens. Two additional slides were prepared and hematoxylin eosin-stained to confirm that the tumor cell content was at least 10%. Peripheral blood (2 ml) collected from the same patients was used as a control to allow discrimination of somatic and germline mutations. Sequencing libraries were prepared from 200 ng of DNAs using the SureSelect XT reagent (Agilent Technologies, Santa Clara, CA) and a KAPA Hyper Prep kit (KAPA Biosystems, Woburn, MA), and analyzed on the Illumina NextSeq platform with 150 bp paired-end reads (Illumina, San Diego, CA). NGS reads were mapped to the human reference genome with the Burrows–Wheeler Aligner (BWA) and BWA–Smith–Waterman algorithm (BWA-SW). Variants in the tumor and nontumor DNAs were detected using the cisCall program.13 All the variants studied in the present study were confirmed with manual inspection using the Integrative Genomics Viewer.14 The test system has been approved as the OncoGuideTM NCC Oncopanel System by the Pharmaceuticals and Medical Devices Agency. The cost is reimbursed by the National Health Insurance System of Japan.15
Molecular tumor board and treatment decision
A multidisciplinary molecular tumor board (MTB) was formed to establish the treatment decision. They comprised molecular biologists, medical oncologists, bioinformaticians, pathologists, and genetic counselors, as the ‘expert panel’. The panel met weekly to review NGS reports by prioritizing and selecting the actionable genomic alterations according to the consensus criteria of the Japanese Society of Medical Oncology, Japan Society of Clinical Oncology, and Japanese Cancer Association.16 They also made reference to two publicly available knowledge bases, CIViC17 and OncoKB.18 The MTB evaluated the pathogenicity of the detected variants, recommendations of genomically matched molecular-targeted agents, and the necessity of referral for genetic counseling.
Analysis of clinical outcome
Patients who commenced a genomically matched therapy were followed up according to the clinical trial protocol with which they were referred or according to the routine medical practice. Analyses of clinical data for patients who were enrolled up to 30 March 2018, with molecular analyses, were completed and examined by the MTB before 8 June 2018. The dates were gathered from the medical record and evaluated on 31 March 2022 (data cut-off).
Progression was determined with imaging evaluations or clinician assessments. The best response to the therapy was determined by various medical oncologists via a review of practical medical/radiological records, using the Response Evaluation Criteria in Solid Tumors (RECIST version 1.1). Progression-free survival on matched treatment (PFS2) was referred to as the time from the commencement of treatment to progression (as defined by RECIST 1.1), clinical progression, or death from any cause. Progression-free survival on previous therapy (PFS1) was defined as the time from the commencement of the last treatment to progression (as defined by RECIST 1.1) or clinical progression. The PFS ratio (PFS2/PFS1) is that in which the PFS on matched therapy (PFS2) was compared with the PFS for the most recent therapy on which the patient had just experienced progression (PFS1).19
Statistical analysis
Data analyses involved the use of R software, version 4.0.3 (R Foundation for Statistical Computing, Vienna, Austria). Inferential statistical tests such as the χ2 test, Fisher’s exact test, Wilcoxon rank sum test, and multivariable logistic regression were carried out to analyze the data. Multivariable analysis was carried out on cancer types (rare versus non-rare), the number of regimens received that matched the genetic abnormality (first or second versus third or more), performance status (0 versus non-zero), and type of drug [kinase inhibitor versus immune checkpoint inhibitor (ICI) versus others] as independent variables. Significance level of P value was set at 0.05.
Results
Patient characteristics
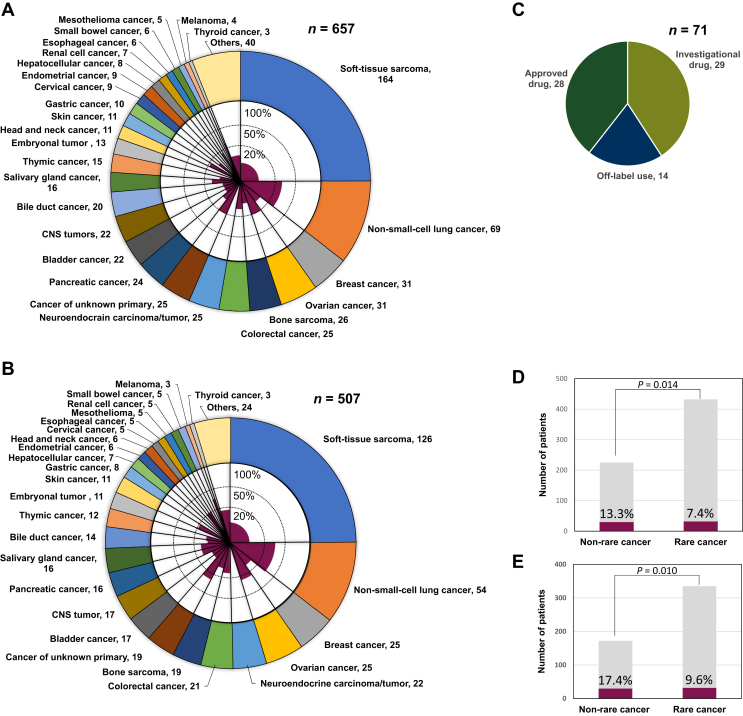
In all 657 enrolled patients, the median age was 53 years (range, 1-90 years). Most patients, 554 (84.3%), had an Eastern Cooperative Oncology Group performance status of 0 or 1. The 657 patients presented with a variety of cancers (>30 tumor types); notably, 432 (65.8%) had rare cancers. The most frequently presented cancer type was soft-tissue sarcoma (25%), followed by non-small-cell lung cancer (NSCLC) (10.5%), breast cancer, and ovarian cancer (both 4.7%) (Figure 2A). The genomic profiling data were obtained from 507 (89.6%) patients. Among the 507 patients, the frequently presented cancer types were also soft-tissue sarcoma (24.9%), NSCLC (10.7%), breast cancer, and ovarian cancer (both 4.9%) (Figure 2B). One or more gene alterations were observed in 390 patients (76.9 %). Among the 507 patients, 332 (65.5%) were followed up in the NCCH after MTB, and the fraction of the patients followed up at NCCH was higher among the patients with rare cancer than patients with non-rare cancer (69.0% versus 58.7%, P = 0.022) (Table 1).
Figure 2.
Cancer type distribution and patients who received genomically matched therapy. (A, B) The outer graph shows the distribution of cancer types in 657 enrolled patients (A) and in 507 genomic profile-obtained patients (B), and the inner graph indicates the percentage of patients who received genomically matched therapy for each cancer type. (C) Treatment methods (n = 71) in 62 patients who received the genomically matched therapy. (D, E) The proportion of patients who received genomically matched therapy in common and rare cancers in 657 enrolled patients (D) and in 507 genomic profile-obtained patients (E). CNS, central nervous system.
Table 1.
Clinical characteristics of patients genotyped in the TOP-GEAR study
| Characteristics | All enrolled cases |
Genomic profile-obtained cases |
Case followed up at NCCH |
Fraction of patients followed up | Genomically matched therapies |
Fraction of patients receiving matched therapy | Response to matched therapies |
Response rate |
|---|---|---|---|---|---|---|---|---|
| n = 657 | n = 507 | n = 332 | n = 62 | n = 20 | ||||
| Age, median (range) | 53 (1-90) | 51 (1-90) | 52 (2-90) | 56 (16-78) | 58 (23-70) | |||
| Sex, n (%) | ||||||||
| Female | 347 (52.8%) | 264 (52.1%) | 176 (53.0%) | 38 (61.3%) | 13 (65.0%) | |||
| Male | 310 (47.2%) | 243 (47.9%) | 156 (47.0%) | 24 (38.7%) | 7 (35.0%) | |||
| ECOG PS, n (%) | ||||||||
| 0-1 | 554 (84.3) | 421 (83.0%) | 285 (85.6%) | 58 (93.5%) | 18 (90.0%) | |||
| 2-4 | 19 (2.9%) | 16 (3.2%) | 9 (2.7%) | 2 (3.2%) | 2 (10.0%) | |||
| N/A | 84 (12.8%) | 70 (13.8%) | 38 (11.4%) | 2 (3.2%) | 0 (0.0%) | |||
| Rare cancer, n (%) | 432 (65.8%) | 335 (66.1%) | 229 (69.0%) | 68.4% | 32 (51.6%) | 9.6% | 14 (70.0%) | 43.8% |
| Non-rare cancer, n (%) | 225 (34.2%) | 172 (33.9%) | 103 (31.0%) | 59.9% | 30 (48.4%) | 17.4% | 6 (30.0%) | 20.0% |
NCCH, National Cancer Center Hospital; ECOG PS, Eastern Cooperative Oncology Group performance status.
Patients receiving genomically matched therapy
Throughout the study period, 12.2% (62/507) of the study sample received 71 genomically matched therapies (Figure 2C, Supplementary Table S3, available at https://doi.org/10.1016/j.esmoop.2024.102981), which consisted of 29 (40.8%) clinical trials of investigational drugs, 28 (39.4%) approved drug treatments, and 14 (19.7%) off-label treatments (Figure 2C). The most common matched regimens were ICIs for the tumor mutation burden (TMB) high (≥10/Mb in all or exon regions) phenotype (n = 17), followed by EGFR tyrosine kinase inhibitors (TKIs) for EGFR mutation (n = 9), ALK-TKIs for ALK fusion (n = 6), and pan-RAF or ERK inhibitors for KRAS mutations (n = 5). Eight patients received more than two regimens of matched therapies, of which three patients each either received ICIs or EGFR-TKIs, while two patients received ALK-TKIs sequentially (Supplementary Table S3, available at https://doi.org/10.1016/j.esmoop.2024.102981).
Among the 62 patients, NSCLC was the most frequent (31.6%) type of cancer, followed by soft-tissue sarcoma (14.0%), ovarian cancer (10.5%), cancer of unknown primary (8.8%), breast cancer (5.3%), and colorectal cancer (5.3%) (Supplementary Table S4, available at https://doi.org/10.1016/j.esmoop.2024.102981). However, in each type of tumor, the fractions of patients receiving genomically matched therapy were high in NSCLC (33.3%), ovarian cancer (24.0%), cancer of unknown primary (26.3%), breast cancer (12.0%), and colorectal cancer (14.3%), while they were low in soft-tissue sarcoma (6.3%) and other rare cancers (<5%) (Supplementary Table S4, available at https://doi.org/10.1016/j.esmoop.2024.102981). Accordingly, the proportion of matched therapies in rare cancer patients was lower than that in non-rare cancer patients among the 507 patients (17.4% and 9.6%, respectively; P = 0.010) and in all 657 enrolled patients (13.3% and 7.4%, respectively; P = 0.014) (Figure 2D and E).
Clinical and genomic factors affecting response to genomically matched therapy
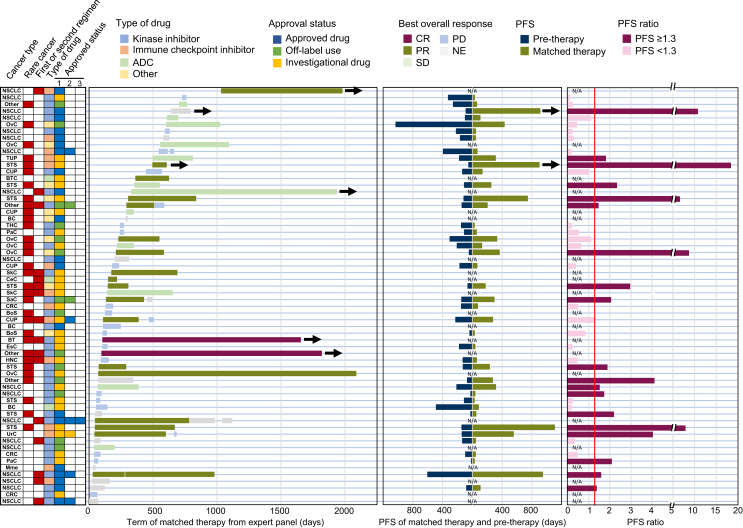
Among 71 genomically matched regimens undertaken by the 62 patients, 2 (BRAF and KIT inhibitors treatments, 2.8%) and 19 (26.8%) patients showed complete and partial responses (PRs), respectively (Figure 3). Therefore, the overall response rate (ORR) estimated by each regimen was 29.6%. Therapeutic response was observed in 20 patients (32.3%, Table 1). PFS ratio of the matched therapy compared with the one just before the matched therapy was evaluated in 41 patients. PFS ratios ranged from 0.09 to 18.32 (Figure 3). Patients with a calculated PFS ratio demonstrated a median of 64.5 days [95% confidence interval (CI) 55.8-92.1 days] and 54.7 days (95% CI 53.2-76.6 days) in these intervals between imaging evaluations for PFS1 and PFS2, respectively. PFS ratio ≥1.3 was observed in 19 (46.3%) patients, and these were also more frequently observed in rare cancer patients than in non-rare cancer patients (52.2% and 38.9%, respectively; P = 0.397). Similarly, the first- or second-line therapies after the CGP test showed more PFS ratios than that in the third and subsequent therapies (50.0% and 45.7%, respectively; P = 0.846), as presented in Figure 3. However, no statistical distinction was observed in the PFS ratio for rare cancer or early treatment.
Figure 3.
Summary of the therapy, duration, and progression-free survival in 62 patients who received genomically matched therapy after the MTB. The left panel shows the cancer type, type of the matched drug, and approval status. The second panel shows the therapy duration after the MTB and its best overall response. The third panel shows the PFS of the matched therapies and pretherapies for patients for whom the PFS ratio could be calculated. The right panel presents the calculated PFS ratio with values of >1.3 (shown as a maroon line). Arrows show the ongoing therapy at the data cut-off in March 2022 (left and center panel). ADC, antibody–drug conjugates; BC, breast cancer; BoS, bone sarcoma; BT, brain tumor; BTC, bile duct cancer; CeC, cervical cancer; CR, complete response; CRC, colorectal cancer; CUP, carcinoma of unknown primary; EsC, esophageal cancer; HNC, head and neck cancer; Mme, malignant melanoma; MTB, molecular tumor board; N/A, not applicable; NE, not evaluable; NSCLC, non-small-cell lung cancer; OvC, ovarian cancer; PaC, pancreatic cancer; PD, progressive disease; PFS, progression-free survival; PR, partial response; SaC, salivary cancer; SD, stable disease; SkC, skin cancer; STS, soft-tissue sarcoma; THC, thymic cancer; TUP, tumor of unknown primary; UrC, urothelial cancer.
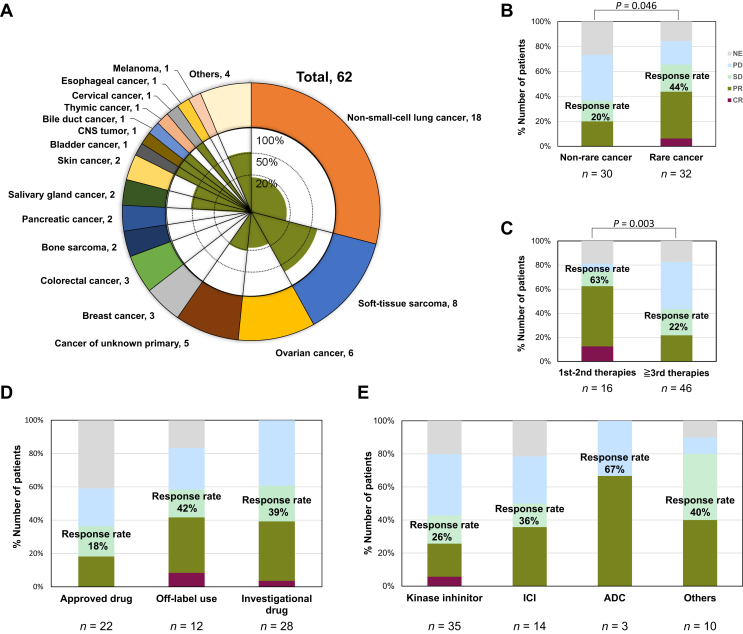
The ORR in patients with rare cancer was higher than those with non-rare cancer (43.8% and 20.0%, respectively; P = 0.046) as represented by results of soft-tissue sarcoma (five of eight; 62.5%) in Figure 4A and B. Regarding the timing of performing the CGP test, the ORR of matched first- or second-line therapies after CGP was higher than that in the third and subsequent therapies (62.5% and 21.7%, respectively; P = 0.003, Figure 4C). Additionally, ORRs with genomically matching therapies using unapproved (off-label use or investigational) drugs were better than those using approved drugs (40.0% and 18.2%, respectively; P = 0.079, Supplementary Figure S1A, available at https://doi.org/10.1016/j.esmoop.2024.102981). Among 14 patients receiving ICI therapy, favorable response occurred in one of six (16.7%) with approved drugs and four of eight (50.0%) with non-approved drugs, and these therapies with non-approved drugs showed a good response compared to ones with approved drugs (Supplementary Figure S1B, available at https://doi.org/10.1016/j.esmoop.2024.102981). In addition, one patient treated with the approved drug (best response: not evaluable) and two patients treated with the non-approved drug (best response: both were PR) were shown to have hyper mutation (≥100 mutations/Mb) or microsatellite instability (MSI)-high (Supplementary Table S3, available at https://doi.org/10.1016/j.esmoop.2024.102981). However, ORRs were not significantly different as regards the treatment (Figure 4D) and type of agents (Figure 4E) (P = 0.208 and P = 0.406, respectively). Multivariable analysis showed that the rare cancer and early treatment line (first or second line) were the parameters significantly impacting the response rate of matched therapies in multivariable analyses [odds ratio (OR) = 6.98, 95% CI 1.55-41.44, P = 0.017 and OR = 10.49, 95% CI 2.41-61.37, P = 0.004, respectively].
Figure 4.
Response of genomically matching therapy in 62 patients. (A) The outer graph exhibits the cancer type distribution of the 62 patients who received genome-matching therapy, and the inner graph shows the percentage of patients whose best overall response was CR or PR by the therapy in each cancer type. (B-E) The proportion of the best overall response in common and rare cancers (B), of matched therapies in the first or second line after CGP (C), in each treatment method (D), and a type of agent (E). ADC, antibody–drug conjugates; CGP, comprehensive genome profiling; CNS, central nervous system; CR, complete response; NE, not evaluable; PD, progressive disease; PR, partial response; SD, stable disease.
Discussion
The present single hospital-based pan-cancer prospective study (TOP-GEAR) provided evidence on the impact of timing and types of cancer on the benefit of CGP-based precision oncology. Genomically matched therapies were administered with standard treatment of approved drugs, off-label use of the approved drugs, and enrollment in clinical trials of investigational drugs. Therapeutic response was preferentially observed in rare cancers compared to non-rare cancers. It was noted that patients with rare cancers were less frequently treated with genomically matched agents than those diagnosed with non-rare cancers. Matched treatment for rare cancers, such as sarcoma, was also infrequent in recent pan-cancer CGP studies.6,7 However, our finding indicates that the rare cancers responded well to the molecular-targeted agents if treated with matched therapy. In fact, five of eight soft-tissue sarcomas with actionable alterations in our study showed response to agents linked to the alterations, notably MDM2 amplification (n = 2, MDM2 inhibitor), TMB-high (n = 2, anti-PD-L antibody), and BRAF mutation (n = 1, anti-BRAF and MEK inhibitor) (Supplementary Table S5, available at https://doi.org/10.1016/j.esmoop.2024.102981). Our findings are consistent with recent studies, which indicated that CGP-based treatments could lead to clinical benefit in a substantial proportion of patients with advanced rare cancers.8,9 The reason for the difference is still unclear at present. The rare cancers in our cohort have less number of mutations than non-rare cancers, which is consistent with the results of recent genome-wide mutational studies (Supplementary Figure S2, available at https://doi.org/10.1016/j.esmoop.2024.102981). Fewer co-mutations might have reduced the drug resistance by increasing oncogene addiction status. Additionally, recent whole-genome studies revealed that driver mutations for rare cancers, including sarcoma, are more clonal than those of common cancers.20 Therefore, molecular-targeted therapy might be effective for the former due to the low frequency of emergence of drug-resistant clones, enhanced by intratumor heterogeneity.21,22
The present study also indicated that therapies in the first or second line after CGP showed higher responses than those in the third and subsequent therapies. These findings further stress the existing fact that clinical outcomes of patients with early-line enrollments in clinical trials are often better than those with late-line enrollments. Therefore, genomically matched therapy using the appropriate drugs should be done not only for the right patients but also at the right time.23 The present study, on the other hand, showed no significant difference in therapeutic response with reference to the approval status of the treatments and types of agents. This outcome is thought to be influenced by the study period with different available agents and therapeutic approaches used. According to the previous finding, the situation might change as time goes by, since the number of regimens, including ICI agents, could dramatically increase.24
There are several limitations to be acknowledged in this study. Firstly, it was a single-arm nonrandomized study without a control arm of patients without CGP. The clinical benefit of CGP was examined only in a patient cohort with CGP data, and the analysis presented was not based on intent to treat. Furthermore, the study was conducted as a clinical study up to MTB implementation after CGP, after which the selection of treatment options and evaluation of treatment efficacy were conducted in real clinical practice. We hereby suggest a clinical setting where most advanced cancer patients are subjected to CGP to select therapeutic option(s) in future studies. Secondly, molecular analysis was carried out using archival tissue in most patients; thus, some novel molecular profiles, which were acquired by progression and therapies after the CGP, might have influenced the therapeutic response. Thirdly, the outcome of the therapies undertaken outside clinical trials was recorded in the clinical practice setting. Thus, the quality of the clinical data might be different between therapies both within and outside of the clinical trials. Fourthly, this study was carried out with a relatively small cohort of patients with rare cancers, coupled with limited numbers of patients with each type of cancer. Therefore, the present conclusion should be validated in a larger cohort of patients.
Conclusions
Our study demonstrates that certain types of cancer and timing (treatment lines) could influence the clinical benefit derived from CGP-based precision oncology by the patients.
Acknowledgements
The authors would like to thank Shigeki Sekine, Noriko Motoi, Hiroshi Yoshida, Hiroki Kakishima, and Hiromichi Matsushita for operation of CGP test using pathological specimens. We also thank Akihiko Shimomura, Shigehisa Kitano, Noriko Tanabe, Natsuko Okita, and Sachie Kawabata for the implementation of the Molecular Tumor Board and TOP-GEAR project.
Funding
This work was supported in part by grants-in-aid from the Japan Agency for Medical Research and Development (AMED) [grant numbers JP18kk0205004, JP19lk1403003], and the National Cancer Center Research and Development Fund [grant numbers 27-A-1, 30-A-6], and NCC Biobank.
Disclosure
TK declares payment for expert testimony from Sysmex. KS declares grants from Sysmex; honoraria from Sysmex, Illumina, Guardant Health Japan, Chugai, Pfizer, and Eisai. TK declares grants from PACT Pharma, Chugai, Daiichi-Sankyo, AstraZeneca, Novartis, Eli Lilly, Takeda, and Zymworks; honoraria from Sysmex, Chugai, and AstraZeneca. YF declares grants from AbbVie, AstraZeneca, Amgen, AnHeart, BMS, Chugai, Eli Lilly, Incyte, and MSD; honoraria from Amgen, AstraZeneca, BMS, Chugai, Daiichi-Sankyo, Eli Lilly, Merck Biopharma, MSD, Novartis, Pfizer, Takeda, Taiho, and Yakult; payment for expert testimony from AstraZeneca, Daiichi-Sankyo, Micron, and Ono Pharmaceutical. SK declares grants from Abbi, Astellas Pharma, AstraZeneca, Eisai, ASLAN Pharmaceuticals, MSD, and Lilly Japan; consulting fees from Takeda; honoraria from Chugai, Incyte, and Eisai. KY declares grants Merck Sharp & Dohme LLC, Daiichi-Sankyo, AstraZeneca, Taiho, Boehringer Ingelheim, Kyowa Hakko Kirin, Nihon Kayaku, Sanofi, and Haihe; consulting fees from Novartis, Eisai, AstraZeneca, Chugai, Takeda, Genmab, Sanofi, and OncXerna; honoraria from Pfizer, Eisai, AstraZeneca, Eli Lilly, Takeda, Chugai, MSD, FujiFilm Pharma, Bayer, Astellas, Boehringer Ingelheim, Daiichi-Sankyo, Ono, Bristol-Myers, PDR Pharma, and Sanofi. EN declares honoraria from AstraZeneca, Chugai Pharma Eisai, Eli Lilly, Novartis, and Pfizer. CM declares grants for Institution from Eisai, Yakult Honsha, Ono Pharmaceutical, Taiho Pharmaceutical, J-Pharma, AstraZeneca, Merck biopharma, Daiichi Sankyo, HITACHI, and Boehringer Ingelheim; consulting fees from Yakult Honsha, MSD K.K., SERVIER, Boehringer Ingelheim, and Taiho; honoraria from Novartis, Yakult Honsha, Teijin Pharma, Taiho Pharmaceutical, Eisai, MSD K.K., and AstraZeneca. YG declares grants from AZK and Pfizer to the clinical trial group, from AbbVie, Eli Lilly, Pfizer, Bristol-Myers Squibb, Ono, Novartis, Kyorin, Daiichi-Sankyo, and Preferred Networks to Institution; honoraria from Eli Lilly, Chugai, Taiho, Boehringer Ingelheim, Ono, Bristol-Myers Squibb, Pfizer, MSD, Novartis, Merck, and Thermo Fisher; participation on advisory board for AstraZeneca, Chugai, Boehringer Ingelheim, Eli Lilly, Taiho, Pfizer, Novartis, Guardant Health Inc., Illumina, Daiichi-Sankyo, Ono Pharmaceutical, Bristol-Myers Squibb, and MSD; leadership/fiduciary roles in Cancer Net Japan and JAMT. SI declares grants from Daiichi-Sankyo, Bristol-Myers Squibb, Eisai, Merck Serono, Bayer, Ono Pharmaceutical, Astellas Pharma, Pfizer, Seagen, Zymeworks, Taiho Pharmaceutical, and AstraZeneca to Institution; honoraria from Taiho Pharmaceutical, Lilly Japan, Chugai Pharma, Daiichi-Sankyo, and Bristol-Myers Squibb Japan; has been an employee of Chugai Pharmaceutical Co. Ltd. since October 2022. TH declares grants from Daiichi-Sankyo, Taiho Pharmaceutical, Ono Pharmaceutical, Chugai Pharma, BeiGene, Astellas Pharma, AstraZeneca, and Pfizer to Institution; honoraria from Chugai Pharma, Merck Serono, Takeda, Taiho Pharmaceutical, Yakult, Eli Lilly, Bristol-Myers Squibb Japan, Bayer, and Daiichi-Sankyo/UCB Japan. AK declares honoraria from Novartis, Taiho Pharmaceutical, Eisai, Daiichi-Sankyo, and Bayer. KN declares grants from Ono Pharmaceutical, Bristol-Myers Squibb, Novartis, and Amgen to Institution; honoraria from Ono Pharmaceutical, Novartis, Bristol-Myers Squibb, and MSD; participation on advisory board in Novartis and MSD. AA declares grants from Takeda Pharmaceutical and Chugai Pharmaceutical. MO declares grants from Sumitomo Pharma, Eisai, and DANBA to Institution. MY declares consulting fees from Lilly Japan and Roche Japan; honoraria from Agilent Technologies, Chugai Pharma, Ono Yakuhin, MSD, and Daiichi-Sankyo. DN declares has a concurrent job at Chugai Pharmaceutical Co. Ltd. MKa declares patents planned, issued, or pending for sequence analysis method, sequence analysis apparatus, reference sequence generation method, reference sequence generation apparatus, program, and storage medium. HI declares grants from Chugai Pharmaceutical, Eisai, Healios, and Ono Pharmaceutical to Institution. TK declares grants from National Cancer Center Research and Development Fund and Sysmex; honoraria from Sysmex. NY declares funding from AMED; grants from Chiome Bioscience, Otsuka, and to Institution from Chugai Pharma, Taiho Pharmaceutical, Eisai, Astellas Pharma, Novartis, Daiichi-Sankyo, Lilly Japan, Boehringer Ingelheim, Takeda, Kyowa Hakko, Kirin, Bayer, Pfizer, Ono Pharmaceutical, Janssen, MSD, AbbVie Bristol-Myers Squibb, Merck Serono, GlaxoSmithKline, Sumitomo Dainippon, Carna Biosciences, Genmab/Seattle Genetics, Shionogi, Toray industries, Kaken Pharmaceutical, AstraZeneca, and CMIC; consulting fees from Eisai, Takeda, Boehringer Ingelheim, CMIC, Chugai Pharma, Healios, and Merck; honoraria from Chugai Pharma, Ono Pharmaceutical, Daiichi-Sankyo/UCB Japan, and Eisai. All other authors have declared no conflicts of interest.
Supplementary data
References
- 1.Zehir A., Benayed R., Shah R.H., et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med. 2017;23:703–713. doi: 10.1038/nm.4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tanabe Y., Ichikawa H., Kohno T., et al. Comprehensive screening of target molecules by next-generation sequencing in patients with malignant solid tumors: guiding entry into phase I clinical trials. Mol Cancer. 2016;15:73. doi: 10.1186/s12943-016-0553-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Flaherty K.T., Gray R.J., Chen A.P., et al. Molecular landscape and actionable alterations in a genomically guided cancer clinical trial: National Cancer Institute molecular analysis for therapy choice (NCI-MATCH) J Clin Oncol. 2020;38:3883–3894. doi: 10.1200/JCO.19.03010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schwaederle M., Parker B.A., Schwab R.B., et al. Precision oncology: the UC San Diego Moores Cancer Center PREDICT experience. Mol Cancer Ther. 2016;15:743–752. doi: 10.1158/1535-7163.MCT-15-0795. [DOI] [PubMed] [Google Scholar]
- 5.Belin L., Kamal M., Mauborgne C., et al. Randomized phase II trial comparing molecularly targeted therapy based on tumor molecular profiling versus conventional therapy in patients with refractory cancer: cross-over analysis from the SHIVA trial. Ann Oncol. 2017;28:590–596. doi: 10.1093/annonc/mdw666. [DOI] [PubMed] [Google Scholar]
- 6.Trédan O., Wang Q., Pissaloux D., et al. Molecular screening program to select molecular-based recommended therapies for metastatic cancer patients: analysis from the ProfiLER trial. Ann Oncol. 2019;30:757–765. doi: 10.1093/annonc/mdz080. [DOI] [PubMed] [Google Scholar]
- 7.Tuxen I.V., Rohrberg K.S., Oestrup O., et al. Copenhagen prospective personalized oncology (CoPPO)-clinical utility of using molecular profiling to select patients to phase I trials. Clin Cancer Res. 2019;25:1239–1247. doi: 10.1158/1078-0432.CCR-18-1780. [DOI] [PubMed] [Google Scholar]
- 8.Horak P., Heining C., Kreutzfeldt S., et al. Comprehensive genomic and transcriptomic analysis for guiding therapeutic decisions in patients with rare cancers. Cancer Discov. 2021;11:2780–2795. doi: 10.1158/2159-8290.CD-21-0126. [DOI] [PubMed] [Google Scholar]
- 9.Cobain E.F., Wu Y.M., Vats P., et al. Assessment of clinical benefit of integrative genomic profiling in advanced solid tumors. JAMA Oncol. 2021;7:525–533. doi: 10.1001/jamaoncol.2020.7987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tamaki T., Dong Y., Ohno Y., Sobue T., Nishimoto H., Shibata A. The burden of rare cancer in Japan: application of the RARECARE definition. Cancer Epidemiol. 2014;38:490–495. doi: 10.1016/j.canep.2014.07.014. [DOI] [PubMed] [Google Scholar]
- 11.Arakawa A., Ichikawa H., Kubo T., et al. Vaginal transmission of cancer from mothers with cervical cancer to infants. N Engl J Med. 2021;384:42–50. doi: 10.1056/NEJMoa2030391. [DOI] [PubMed] [Google Scholar]
- 12.Sunami K., Ichikawa H., Kubo T., et al. Feasibility and utility of a panel testing for 114 cancer-associated genes in a clinical setting: a hospital-based study. Cancer Sci. 2019;110:1480–1490. doi: 10.1111/cas.13969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kato M., Nakamura H., Nagai M., et al. A computational tool to detect DNA alterations tailored to formalin-fixed paraffin-embedded samples in cancer clinical sequencing. Genome Med. 2018;10:44. doi: 10.1186/s13073-018-0547-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Robinson J.T., Thorvaldsdóttir H., Winckler W., et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ebi H., Bando H. Precision oncology and the universal health coverage system in Japan. JCO Precis Oncol. 2019;3 doi: 10.1200/PO.19.00291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Naito Y., Aburatani H., Amano T., et al. Clinical practice guidance for next-generation sequencing in cancer diagnosis and treatment (edition 2.1) Int J Clin Oncol. 2021;26:233–283. doi: 10.1007/s10147-020-01831-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Griffith M., Spies N.C., Krysiak K., et al. CIViC is a community KnowledgeBase for expert crowdsourcing the clinical interpretation of variants in cancer. Nat Genet. 2017;49:170–174. doi: 10.1038/ng.3774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chakravarty D., Gao J., Phillips S.M., et al. OncoKB: a precision oncology knowledge base. JCO Precis Oncol. 2017;2017 doi: 10.1200/PO.17.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Von Hoff D.D., Stephenson J.J., Jr., Rosen P., et al. Pilot study using molecular profiling of patients’ tumors to find potential targets and select treatments for their refractory cancers. J Clin Oncol. 2010;28:4877–4883. doi: 10.1200/JCO.2009.26.5983. [DOI] [PubMed] [Google Scholar]
- 20.Dentro S.C., Leshchiner I., Haase K., et al. Characterizing genetic intra-tumor heterogeneity across 2,658 human cancer genomes. Cell. 2021;184:2239–2254.e39. doi: 10.1016/j.cell.2021.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McGranahan N., Swanton C. Clonal heterogeneity and tumor evolution: past, present, and the future. Cell. 2017;168:613–628. doi: 10.1016/j.cell.2017.01.018. [DOI] [PubMed] [Google Scholar]
- 22.Sasaki N., Clevers H. Studying cellular heterogeneity and drug sensitivity in colorectal cancer using organoid technology. Curr Opin Genet Dev. 2018;52:117–122. doi: 10.1016/j.gde.2018.09.001. [DOI] [PubMed] [Google Scholar]
- 23.Johnson A., Zeng J., Bailey A.M., et al. The right drugs at the right time for the right patient: the MD Anderson precision oncology decision support platform. Drug Discov Today. 2015;20:1433–1438. doi: 10.1016/j.drudis.2015.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haslam A., Kim M.S., Prasad V. Updated estimates of eligibility for and response to genome-targeted oncology drugs among US cancer patients, 2006-2020. Ann Oncol. 2021;32:926–932. doi: 10.1016/j.annonc.2021.04.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.