Abstract
The large subunit of herpes simplex virus (HSV) ribonucleotide reductase (RR), RR1, contains a unique amino-terminal domain which has serine/threonine protein kinase (PK) activity. To examine the role of the PK activity in virus replication, we studied an HSV type 2 (HSV-2) mutant with a deletion in the RR1 PK domain (ICP10ΔPK). ICP10ΔPK expressed a 95-kDa RR1 protein (p95) which was PK negative but retained the ability to complex with the small RR subunit, RR2. Its RR activity was similar to that of HSV-2. In dividing cells, onset of virus growth was delayed, with replication initiating at 10 to 15 h postinfection, depending on the multiplicity of infection. In addition to the delayed growth onset, virus replication was significantly impaired (1,000-fold lower titers) in nondividing cells, and plaque-forming ability was severely compromised. The RR1 protein expressed by a revertant virus [HSV-2(R)] was structurally and functionally similar to the wild-type protein, and the virus had wild-type growth and plaque-forming properties. The growth of the ICP10ΔPK virus and its plaque-forming potential were restored to wild-type levels in cells that constitutively express ICP10. Immediate-early (IE) genes for ICP4, ICP27, and ICP22 were not expressed in Vero cells infected with ICP10ΔPK early in infection or in the presence of cycloheximide, and the levels of ICP0 and p95 were significantly (three- to sevenfold) lower than those in HSV-2- or HSV-2(R)-infected cells. IE gene expression was similar to that of the wild-type virus in cells that constitutively express ICP10. The data indicate that ICP10 PK is required for early expression of the viral regulatory IE genes and, consequently, for timely initiation of the protein cascade and HSV-2 growth in cultured cells.
Herpes simplex virus (HSV) expresses a distinct ribonucleotide reductase (RR) that consists of two heterologous protein subunits. The small subunit (RR2) is a 38-kDa protein encoded by UL40; the large subunit (RR1), designated ICP6 and ICP10 for HSV type 1 (HSV-1) and HSV-2, respectively, is a 140-kDa protein encoded by UL39 (3, 6, 24, 45). The two RR subunits have different expression kinetics and can function independently. Thus, RR2 is regulated with characteristic β (also known as delayed-early)-class kinetics. Its expression peaks at 6 to 8 h postinfection (p.i.), and it requires functional ICP4 (73). It imparts β-class kinetics to RR activity (13, 36, 73). By contrast, RR1 expression is regulated with α (also known as immediate-early [IE])-class kinetics, as evidenced by the onset of synthesis at 2 h p.i. and RR1 production in the presence of cycloheximide (3, 29, 69, 75). The RR1 promoter has an octamer/TAATGARAT sequence that responds to the VP16/oct1 complex (18, 70, 77, 78). Basal expression from the RR1 promoter requires AP-1 factors, but not functional ICP4. RR1 is expressed in cells infected with ICP4- or ICP0-defective mutants (17, 43–45, 59). Its expression is enhanced by ICP0, involving the interaction of ICP0 with AP-1 factors (18, 70, 77, 78, 81).
RR1 is a multifunctional protein. It consists of an intrinsic serine/threonine-specific protein kinase (PK) localized at the amino terminus and RR1 localized at the carboxy terminus (10, 11, 14, 16, 41, 42, 46, 50). Sequences homologous to ICP10 PK DNA were cloned from human tissue, suggesting that the PK domain may have evolved from a cellular gene (62). This implies that by participating in the viral life cycle, the cellular gene provided a functional advantage which justified its conservation. Studies of HSV-2 (63) and HSV-1 (25, 26) RR1 mutants led to the conclusion that RR1 is required for virus growth in nondividing cells in culture. Furthermore, HSV-1 RR1 mutants are less neurovirulent (7, 31) and less likely to reactivate from latency (33, 58). Inasmuch as RR activity in infected cells is regulated with β-class kinetics, like the RR2 protein, it seems reasonable to conclude that the IE component of RR1 regulation is required for the role of PK activity early in infection. Indeed, the RR and PK activities of the RR1 proteins can be dissociated by various means, including cellular proteolysis (10, 32, 37, 42). However, PK activity is not required for ribonucleotide reduction (15), and its role in virus growth is still unknown.
Here we describe the results of our studies with an HSV-2 mutant (ICP10ΔPK) with a deletion in the PK domain of ICP10. The data indicate that ICP10 PK activity is required for virus growth in exponential-phase and growth-restricted cells in culture, involving optimal expression of IE genes.
MATERIALS AND METHODS
Cells.
Vero (African green monkey kidney) cells were grown in Eagle’s minimal essential medium (EMEM) supplemented with 10% fetal calf serum (FCS) and antibiotics. JHLa1 cells (which constitutively express ICP10) were previously described (30, 41, 64). They were cultured in EMEM with 10% FCS, 1 mM Na pyruvate (GIBCO-BRL, Gaithersburg, Md.), 1× nonessential amino acids (GIBCO-BRL), and antibiotics. Vero-ICP10 cells were derived by transfection of Vero cells with an ICP10 expression vector that has an SV2-neo cassette (pJW17N) (41). For serum starvation, cells grown to confluency in EMEM containing 10% FCS were washed with phosphate-buffered saline (PBS) at pH 7.0 and grown for 2 days in medium containing 1 or 0.5% FCS.
Construction of ICP10 mutant viruses.
The construction of the ICP10ΔPK virus was described previously (50). Briefly, wild-type sequences in a plasmid (TP101) that contains the HSV-2 BamHI E and T fragments were replaced with the 1.8-kb SalI/BglII fragment from pJHL9 (ICP10 mutant with a deletion in the PK catalytic domain [41]). The resulting plasmid, TP9, contains sequences which code for ICP10 with a deletion in the PK catalytic domain flanked by 4- and 2.8-kb HSV-2 DNA sequences at the 5′ and 3′ ends, respectively. The 10-kb HindIII/EcoRI fragment from TP9 was introduced by marker transfer into ICP10ΔRR, in which the RR domain of ICP10 had been replaced with the lacZ gene. The resulting recombinant virus, designated ICP10ΔPK, was obtained by selecting white plaques on a background of blue plaques after staining with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal). A few white plaques were picked, purified, and grown in Vero cells in EMEM with 10% FCS.
For construction of revertant virus, designated HSV-2(R), Vero-ICP10 cells were cotransfected with 1 μg of infectious viral DNA from ICP10ΔPK, and a 10-fold molar excess of the wild-type BamHI E/T fragment and progeny virus were titrated on serum-starved Vero cells (EMEM–1% FCS). ICP10ΔPK plaque formation is significantly impaired in serum-starved cells (ratio of plaquing efficiencies [expressed as PFU per milliliter] of recombinant virus/wild-type virus, 0.0001), and the plaques are morphologically distinct. Therefore, recombinants with wild-type plaque morphology were easily detected. A few such plaques were picked, purified, and grown in Vero cells. The identity of the revertant virus was confirmed by restored plaquing efficiency (ratio, 0.9) and the ability to express the 140-kDa ICP10 protein. Revertants were not obtained by cotransfection of ICP10ΔPK DNA with a BamHI E/T fragment that had a deletion in the domain which codes for ICP10 PK due to MscI/StuI digestion, nor with a BglII I fragment which contains the VP16 coding sequences.
Plaque assay.
Virus titers were determined by plaque assay as previously described (5). Vero-ICP10 cells were used under an overlay consisting of EMEM supplemented with 10 or 0.5% FCS and 0.3% pooled human serum immunoglobulin G (IgG).
Antibodies.
The production and specificity of the anti-LA-1 antibody against ICP10 amino acids 13 to 26 were previously described (4, 12). Capsid antibody was prepared in rabbits by using HSV-2 nucleocapsids purified as previously described (67). It recognized primarily the 155-kDa major capsid protein (VP5) in an immunoblotting assay (data not shown). The antibody was adsorbed with Vero cells fixed with paraformaldehyde (PFA) and permeabilized with Triton X-100 (30) and used at the highest dilution that gave no signal with uninfected cells. ICP4 and ICP0 monoclonal antibodies were purchased from Advanced Biotechnologies, Columbia, Md.
Southern blot hybridization with an oligonucleotide probe.
Viral DNA was isolated from cytoplasmic virions as previously described (52, 63). Briefly, Vero cells were infected at a multiplicity of infection (MOI) of 5. At 48 h p.i., cells were resuspended (2 × 107/ml) in a buffer consisting of 10 mM Tris-HCl (pH 7.9), 10 mM EDTA, and 0.25% Triton. Following incubation on ice (15 min), NaCl was added at a final concentration of 0.2 M and the nuclei were pelleted by centrifugation at 1,000 × g (10 min, 4°C). The supernatant containing cytoplasmic virions was incubated in 200-μg/ml proteinase K and 0.2% sodium dodecyl sulfate (SDS) (4 h at 37°C), mixed with saturated NaI (final concentration, 1.525 g/ml) and ethidium bromide (final concentration, 3 μg/ml), and centrifuged at 100,000 × g for 16 h.
Viral DNA (5 μg) was digested with BamHI, and the fragments were separated by 1% agarose gel electrophoresis in Tris-acetate-EDTA buffer (40 mM Tris-acetate, 1 mM EDTA) and transferred to GeneScreen membranes (New England Nuclear Corp., Boston, Mass.). The membranes were incubated at 42°C for 2 h in a prehybridization solution containing 5× SSC (750 mM NaCl, 75 mM sodium citrate, pH 7.0), 2% casein, 0.1% N-laurylsarcosine, and 0.02% SDS. The hybridization probes were oligonucleotides AU25 (CAAATGGGATTCATGGACACGTTA) and AU26 (CCCCTTCATCATGTTTAAGGA), which represent sequences in the ICP10 promoter and RR coding regions, respectively. They were 3′ tailed with digoxigenin (DIG)-dUTP by terminal transferase (Boehringer Mannheim, Indianapolis, Ind.) in a 20-μl volume with 1× reaction buffer (5 mM CoCl2, 0.05 mM DIG-dUTP, 5-nmol/ml AU25 or AU26, 0.5 mM dATP, and 2.5-U/μl terminal transferase) at 37°C for 15 min and diluted to a final concentration of 5 pmol/ml in prehybridization solution. Hybridization was done at 42°C for 3 h. Membranes were washed once (room temperature) in a solution containing 2× SSC–0.1% SDS for 5 min and twice in 0.5× SSC–0.1% SDS for 15 min each time. For detection of the hybridized DNA fragments, the membranes were rinsed in buffer 1 (100 mM Tris-HCl [pH 7.5], 150 mM NaCl), incubated in buffer 2 (2% [wt/vol] casein in buffer 1) for 40 min and in buffer 2 containing 3 × 10−4-U/ml alkaline phosphatase-conjugated anti-DIG antibody (Boehringer Mannheim) for 30 min. After washing with buffer 1 (twice) and soaking in buffer 3 (100 mM Tris-HCl [pH 9.5], 100 mM NaCl, 50 mM MgCl2) for 2 min, the membranes were exposed to the chemiluminescent substrate Lumi-Phos 530 (Boehringer Mannheim) and the reaction was developed on X-ray film.
Metabolic labeling and immunoprecipitation.
Cells were mock infected with PBS (pH 7.4) or infected with 200-PFU/cell HSV-2, ICP10ΔPK, or HSV-2(R). They were labeled with [35S]methionine (100 μCi/ml) (specific activity, 1,120 Ci/mmol; Dupont, NEN) in methionine-free EMEM with 10% dialyzed FCS (64). In some experiments, infection was done in the presence of cycloheximide (50 μg/ml) for 6 h. At that time, the cycloheximide was removed and the cells were washed extensively with PBS and incubated (3 h) in the presence of 10-μg/ml actinomycin D and 100-μCi/ml [35S]methionine (69). For immunoprecipitation, cell lysates were incubated in cold radioimmunoprecipitation assay buffer (0.01 M Tris-HCl [pH 8.0], 0.1% SDS, 1% Nonidet P-40, 1% deoxycholate, 0.15 M NaCl) with 1 mM phenylmethylsulfonyl fluoride and aprotinin at 100 kallikrein U/ml (Sigma, St. Louis, Mo.) for 15 min on ice and cleared of cell debris by centrifugation for 30 min at 20,000 × g. They were incubated (1 h, 4°C) with 15 to 20 μl of antibody (30 min, 4°C) and with 100 μl of protein A-Sepharose CL4B beads (10 mg; Sigma) in a buffer consisting of 0.1 M Tris-HCl (pH 8.0), 0.15 M NaCl, and 0.5% Nonidet P-40. Beads were washed extensively with ice-cold radioimmunoprecipitation assay buffer, and bound proteins were eluted by boiling (5 min) in 100 μl of denaturing solution (150 mM Tris-HCl [pH 7.0], 5.7% SDS, 14% 2-mercaptoethanol, 17% sucrose, 0.04% bromothymol blue). Proteins were resolved by SDS-polyacrylamide gel electrophoresis (PAGE) on 7 or 8.5% polyacrylamide gels and visualized by autoradiography as previously described (10, 42, 64). In some experiments, cells were resuspended directly in denaturing solution, boiled for 5 min, and analyzed by SDS-PAGE.
Immunocomplex PK assay.
Immunoprecipitates of cell extracts normalized for protein concentration by the bicinchoninic acid protein assay kit (Pierce, Rockford, Ill.) were washed with TS buffer containing 20 mM Tris-HCl (pH 7.4) and 0.15 M NaCl, suspended in 50 μl of kinase reaction buffer consisting of 20 mM Tris-HCl (pH 7.4), 5 mM MgCl2, 2 mM MnCl2 and 10 μCi of [γ-32P]ATP (3,000 Ci/mmol; Dupont, NEN), and incubated at 30°C for 15 min. The beads were washed once with 1 ml of TS buffer, resuspended in 100 μl of denaturing solution, and boiled for 5 min. The proteins were resolved by SDS-PAGE on 7% polyacrylamide gels as previously described (10, 11, 41, 50, 64).
Western blot assay.
Cell extracts or immunoprecipitates were subjected to SDS-PAGE on 7% polyacrylamide gels. Proteins were electrotransferred onto nitrocellulose membranes, and immunoblotting was performed by incubation for 1 h each at room temperature with the respective antibodies, followed by protein A-peroxidase (Sigma). Detection was done with ECL reagents (Amersham, Chicago, Ill.) as previously described (64).
Immunofluorescence staining.
Vero cells were grown on coverslips for 1 to 2 days, until they reached 70% confluency, and infected with HSV-2 or ICP10ΔPK at an MOI of 100 PFU/cell. They were fixed with 3% PFA for 20 min, and then the remaining fixative was quenched with 50 mM NH4Cl for 10 min and the cells were permeabilized with 0.1% Triton X-100 for 4 min (30, 66). Cells were washed with PBS (pH 7.4), exposed to 10% normal goat serum for 30 min, and stained with the primary antibody in 10% goat serum for 20 min. The coverslips were rinsed three times (5 min each time) and incubated with a fluorescein-conjugated secondary antibody in 10% goat serum for 30 min. After extensive washing in PBS and one short wash in water, the coverslips were mounted in Mowiol containing 2.5% (wt/vol) 1,4-diazabicyclo-[2.2.2]octane on glass slides and examined with a Zeiss fluorescence microscope (30, 66).
RR assay.
RR activity was assayed as previously described (12, 63). Extracts from 12-h-infected cells or mock-infected cells were resuspended in HD buffer (100 mM HEPES buffer [pH 7.6], 2 mM dithiothreitol) at 2 × 107 cell equivalents/ml, incubated on ice for 15 min, disrupted by sonication (30 to 60 s at the maximum setting of an Ultrasonics 220F Sonifier), and clarified of cell debris by centrifugation (100,000 × g; 1 h, 4°C). The HSV RR activity was precipitated with crystalline ammonium sulfate at 45% saturation (0.258 g/ml). Following dialysis and centrifugation (16,000 × g, 30 min), the partially purified enzyme preparations were incubated (37°C, 10 min) with equal volumes of a 2× standard reaction mixture containing 400 mM HEPES buffer (pH 8.0), 20 mM dithiothreitol, and 0.2 mM [3H]CDP (17.8 Ci/mmol; Amersham). The reaction was terminated by addition of 100 mM hydroxyurea with 10 mM EDTA (pH 8.0) and boiling for 3 min. Crotalus atrox venom (Sigma) was added (0.5 mg/ml in 12 mM Tris-HCl [pH 9.0]–4 mM MgCl2–1 mM deoxycytidine), and the mixture was incubated for 30 min at 37°C, boiled for 3 min, and applied to a 0.5-ml Dowex-1 borate column (Sigma). The column was washed with 2.5 ml of H2O, and 0.5-ml eluate fractions were mixed with Biofluor (NEN) for scintillation counting. RR activity is expressed as units per milligram, where 1 U represents the conversion of 1 nmol of [3H]CDP to dCDP/h/mg of protein.
Northern blot hybridization.
The guanidinium isothiocyanate-cesium chloride gradient method was used to isolate and purify RNA from Vero cells infected with HSV-2, ICP10ΔPK, or HSV-2(R) (MOI, 200 PFU/cell). Northern blot hybridization was done as previously described (22). Hybridization was for 16 h at 42°C with a 32P-labeled ICP4 or ICP0 DNA probe in a solution containing 50% formamide, 5× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7]), 2× Denhardt’s solution, 0.1% SDS, and 250-μg/ml salmon sperm DNA. The ICP4 probe was a 1.9-kb BamHI DNA fragment derived from pXhoI-C (22). The ICP0 probe was a 1.7-kb NruI fragment derived from pGH15 (47). The human GAPDH probe was a 40-mer oligonucleotide purchased from Oncogene Science (catalog no. ON407). Probes were [α-32P]dCTP labeled by the random priming method using an oligonucleotide kit (Pharmacia, Uppsala, Sweden) in accordance with the manufacturer’s instructions. Blots were washed twice in 2× SSC–0.1% SDS and twice in 0.1× SSC–0.01% SDS for 10 min each time at ambient temperature and then washed once in 0.1× SSC–0.1% at 50°C and visualized by autoradiography.
RESULTS
Characterization of the ICP10ΔPK and HSV-2(R) viruses.
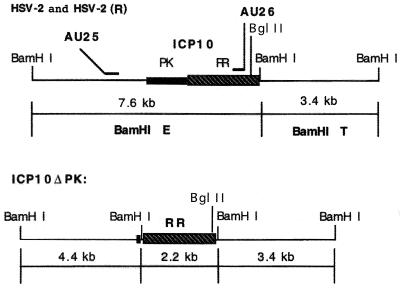
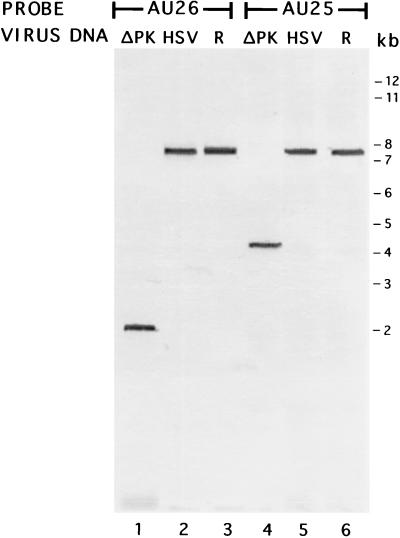
The AU25 and AU26 probes, which recognize sequences within the ICP10 promoter and its RR coding region, respectively (Fig. 1), were used to confirm the construction of the ICP10ΔPK mutant and its revertant [HSV-2(R)]. DNA (5 μg) from HSV-2, ICP10ΔPK, or HSV-2(R) was digested with BamHI, separated on a 1% agarose gel, and used for hybridization. Bands of 7.6 kb (representing the BamHI E fragment) were observed for DNAs from HSV-2 (Fig. 2, lanes 2 and 5) and HSV-2(R) (Fig. 2, lanes 3 and 6) hybridized with AU26 and AU25, respectively. The AU26-hybridizing band seen for ICP10ΔPK DNA was 2.2 kb (Fig. 2, lane 1), and the AU25-hybridizing band was 4.4 kb (Fig. 2, lane 4), as predicted from deletion of the PK coding region.
FIG. 1.
Schematic representation of ICP10ΔPK DNA. Oligonucleotide probe AU26 recognizes the 7.6-kb BamHI E fragment from HSV-2 or HSV-2(R) DNA and a 2.2-kb BamHI fragment from ICP10ΔPK DNA. Oligonucleotide probe AU25 recognizes the 7.6-kb BamHI E fragment from HSV-2 or HSV-2(R) DNA and a 4.4-kb BamHI fragment from ICP10ΔPK DNA.
FIG. 2.
Southern blot hybridization of BamHI-digested DNA from ICP10ΔPK (lanes 1 and 4), HSV-2 (lanes 2 and 5), or HSV-2(R) (lanes 3 and 6) with a DIG-labeled AU26 (lanes 1 to 3) or AU25 (lanes 4 to 6) oligonucleotide probe. Size markers are shown in the right margin.
Expression of the ICP10 protein with a deletion in the PK domain (p95).
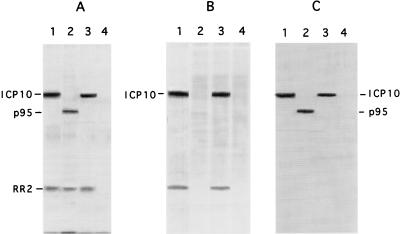
We have previously shown (41) that the size of the ICP10 protein with a deletion in its PK domain is 95 kDa (p95). To determine whether the ICP10ΔPK virus expresses p95, Vero cells were infected with 100 PFU/cell and labeled with [35S]methionine (100 μCi/ml) from 6 to 16 h p.i. Cells similarly infected with HSV-2 or HSV-2(R) served as controls. Cell extracts were precipitated with ICP10 antibody, and the proteins were resolved by SDS-PAGE on 7% polyacrylamide gels. A 140-kDa protein was precipitated from cells infected with HSV-2 (Fig. 3A, lane 1) or HSV-2(R) (Fig. 3A, lane 3), while a 95-kDa protein (p95) was precipitated from cells infected with ICP10ΔPK (Fig. 3A, lane 2). A 38-kDa protein, consistent with RR2, was equally coprecipitated from cells infected with all three viruses, indicating that p95 can complex with RR2. Preimmune serum was negative (Fig. 3A, lane 4).
FIG. 3.
Expression and PK activity of the p95 protein from ICP10ΔPK-infected cells. (A) Vero cells were infected with HSV-2 (lane 1), ICP10ΔPK (lanes 2 and 4), or HSV-2(R) (lane 3) and labeled with [35S]methionine from 6 to 16 h p.i. Cell extracts obtained at this time were immunoprecipitated with ICP10 antibody (recognizes amino acids 13 to 26) (lanes 1 to 3) or preimmune serum (lane 4). (B) Immunocomplex PK assays with ICP10 antibody (lanes 1 to 3) or preimmune serum (lane 4) of extracts from Vero cells infected for 16 h with HSV-2 (lane 1), ICP10ΔPK (lanes 2 and 4), or HSV-2(R) (lane 3). (C) Immunoprecipitates from panel B immunoblotted with ICP10 antibody.
p95 lacks kinase activity.
Studies of RR and PK expression vectors have shown that the PK activity is associated with the 57- to 60-kDa amino-terminal domain of RR1 and is independent of its 90- to 95-kDa carboxy-terminal domain (10, 41). To determine whether this is also true within the context of the virus, extracts of cells infected for 16 h with HSV-2, ICP10ΔPK, or HSV-2(R) (MOI, 200 PFU/cell) were immunoprecipitated with ICP10 antibody and subjected to immunocomplex PK assays. The resolved proteins were transferred to a nitrocellulose membrane and immunoblotted with ICP10 antibody to determine the protein levels in the immunoprecipitates. A phosphorylated 140-kDa protein consistent with ICP10 was observed in HSV-2 (Fig. 3B, lane 1)- or HSV-2(R) (Fig. 3B, lane 3)-infected cells, but p95 was no phosphorylated (Fig. 3B, lane 2). This is not due to low levels of protein in the precipitates, since the levels of p95 detected by immunoblotting of the precipitates from ICP10ΔPK-infected cells with ICP10 antibody (Fig. 3C, lane 2) were similar to those of ICP10 in HSV-2 (Fig. 3C, lane 1) and HSV-2(R) (Fig. 3C, lane 3) precipitates. A phosphorylated 38-kDa protein, consistent with RR2, was also seen in the ICP10 precipitates from HSV-2 (Fig. 3B, lane 1) and HSV-2(R) (Fig. 3B, lane 3)-infected cells, but it was not seen in those from ICP10ΔPK-infected cells (Fig. 3B, lane 2). Preimmune serum was negative (Fig. 3B, C, and lanes 4). These data are consistent with previous reports that ICP10 PK phosphorylates both viral and cellular substrates (2, 6, 10, 47, 50) and indicate that the PK coding region is required for kinase activity, also within the context of virus infection.
The ICP10ΔPK virus has RR activity.
Although p95 coprecipitates with RR2 (Fig. 3A, lane 2), the question arises of whether the loss of the ICP10 PK domain affects RR activity. To address this question, RR assays were performed on extracts from cells infected with ICP10ΔPK, HSV-2, or HSV-2(R) (MOI, 20 PFU/cell) for 12 h as previously described (12, 63). As shown in Table 1, the RR activity of the ICP10ΔPK virus (8.4 U) was similar to those of HSV-2 and HSV-2(R) (10.2 and 8.8 U, respectively), supporting the conclusion that the PK and RR activities can be functionally dissociated (10, 32, 37, 42).
TABLE 1.
RR activity of ICP10ΔPK virus
| Virus | Radioactivity (cpm)a | RR sp act (U)b |
|---|---|---|
| HSV-2 | 11,534 | 10.2 |
| HSV-2(R) | 10,037 | 8.8 |
| ICP10ΔPK | 9,540 | 8.4 |
| Mock infected | 3,060 | 2.7 |
In cpm/270 mg of protein.
One RR unit equals conversion of 1 nmol of CDP to dCDP/h/mg of protein.
The ICP10ΔPK virus is defective for growth in culture.
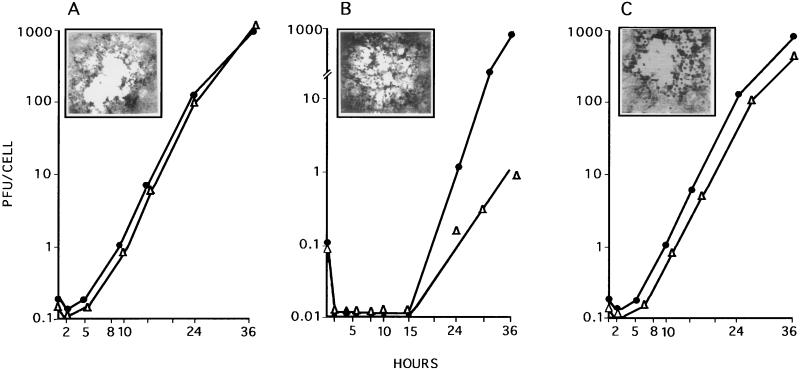
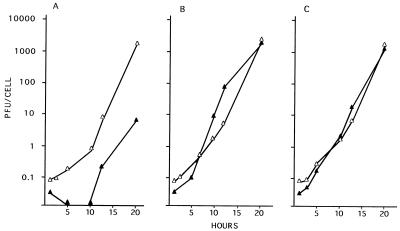
In a first series of experiments, we examined the growth of the ICP10ΔPK virus in dividing (10% serum) and nondividing (0.5% serum) Vero cells infected with 2 PFU/cell. Adsorption was for 2 h (0 h on the growth curve), and virus titers were determined at 2 to 36 h after adsorption. Cells similarly infected with HSV-2 or HSV-2(R) served as controls. As shown in Fig. 4A, HSV-2 grew equally well in dividing and nondividing cells. Virus replication began at 2 h after adsorption. The burst size was 1,000 PFU/cell at 36 h after adsorption. A similar growth pattern was evidenced by HSV-2(R) (Fig. 4C). By contrast, onset of ICP10ΔPK replication was not seen until 15 h after adsorption in both exponentially growing and serum-starved cells (Fig. 4B). In dividing cells, the rate of virus growth between 15 and 36 h and the virus titers at 36 h after adsorption were similar to those seen for HSV-2 (burst size, 1,000 PFU/cell). However, virus titers were significantly (1,000-fold) lower in serum-starved cells (burst size, 1 PFU/cell at 36 h).
FIG. 4.
Virus growth in dividing and nondividing cells. Vero cells were grown and infected (MOI, 2 PFU/cell) with HSV-2 (A), ICP10ΔPK (B), or HSV-2(R) (C) in 10% (•) or 0.5% (▵) FCS. Adsorption was for 2 h at 37°C (0 h of the growth curve). Virus titers were determined at 2 to 36 h after adsorption, and results are expressed as PFU per cell (burst size). The insets in each panel show the morphologies of the respective virus plaques in Vero cells overlaid with EMEM–10% FCS and 0.3% IgG and stained with Giemsa at 48 h. ICP10ΔPK plaques in Vero-ICP10 cells were identical to those of HSV-2 in Vero cells.
In a second series of experiments, we considered the possibility that growth defects may be MOI dependent and repeated the growth curve measurement with cells infected at a significantly higher MOI (200 PFU/cell). The growth of the ICP10ΔPK virus in dividing cells was somewhat improved by infection under these conditions in that virus replication began at 10 to 12 h after adsorption, compared to 15 h at a low MOI. However, virus titers in serum-starved cells were still approximately 1,000-fold lower than in dividing cells (burst sizes, 1 and 980 PFU/cell, respectively). The growth defect does not appear to be cell type determined, as similar results were obtained with HeLa, L, BHK (data not shown), and 293 (Fig. 5A) cells.
FIG. 5.
Virus growth in cells that constitutively express ICP10. JHLa1 cells, which constitutively express ICP10 (▵), and 293 cells, which were used to establish JHLa1 (▴), were infected with ICP10ΔPK (A), HSV-2 (B), or HSV-2(R) (C) at an MOI of 200 PFU/cell and overlaid with medium containing 1% FCS. Adsorption was for 2 h (0 h of the growth curve), and virus titers were assayed at 2 to 20 h after adsorption. Results are expressed as PFU per cell (burst size). Onset of ICP10ΔPK replication in 293 cells infected in 10% FCS is similarly delayed.
Single step growth kinetics indicate that ICP10ΔPK grows as well as HSV-2 and HSV-2(R) in cells which supply ICP10 PK activity in trans, such as JHLa1 (41, 64). 293 and JHLa1 cells were infected with the ICP10ΔPK virus at 200 PFU/cell in EMEM containing 1% FCS, a condition which does not support efficient virus growth. In JHLa1 cells, replication was first seen at 2 h after adsorption, and the burst size at 20 h was 2,500 PFU/cell. This compares to growth onset at 10 h after adsorption in 293 cells and a burst size of 8 PFU/cell at 20 h after adsorption (Fig. 5A). By contrast, HSV-2 grew equally well in JHLa1 and 293 cells. Replication began at 2 h, and the burst sizes at 20 h after adsorption were 2,800 and 2,750 PFU/cell in JHLa1 and 293 cells, respectively (Fig. 5B). Similar results were obtained for HSV-2(R), with replication beginning at 2 h after adsorption and burst sizes of 2,570 and 2,610 PFU/cell in JHLa1 and 293 cells, respectively (Fig. 5C). We interpret these findings to indicate that ICP10ΔPK evidences two growth defects: (i) delayed growth onset, which is seen in both dividing and nondividing cells, and (ii) impaired replication, which is seen only in nondividing cells. An induced or activated cellular function(s) compensates for the missing viral protein in dividing cells but not in serum-starved cells.
ICP10ΔPK has altered plaque morphology and compromised plaquing ability.
To analyze the plaque-forming ability of the ICP10ΔPK virus, we used Vero and Vero-ICP10 cells grown in 10 or 0.5% serum. ICP10ΔPK plaque-forming ability was severely compromised in serum-starved Vero cells but not in dividing cells, in which it was similar to that of HSV-2. Plaque-forming ability was also normal in Vero-ICP10 cells, which supply ICP10 PK activity (Table 2). The size of the ICP10ΔPK plaques was similar to that of HSV-2 or HSV-2(R) plaques. However, in both dividing and nondividing Vero cells, the ICP10ΔPK plaques differed from those of HSV-2 or HSV-2(R) in that they were hazy, apparently reflecting incomplete cell lysis (Fig. 4B, inset). The extent of cell lysis differed somewhat from one experiment to the next, but it was never as complete as that seen for HSV-2 (Fig. 4A, inset) or HSV-2(R) (Fig. 4C, inset). The morphology of the ICP10ΔPK plaques in Vero-ICP10 cells was similar to that of HSV-2 and HSV-2(R) plaques (data not shown).
TABLE 2.
Plaquing efficiency of ICP10ΔPK virus in dividing and serum-starved cells
| Virus | Cells (serum concn [%])a | Virus titerb (wild-type/mutant ratio) |
|---|---|---|
| HSV-2 | Vero (10) | 5.0 × 107 |
| ICP10ΔPK | Vero (10) | 2.8 × 107 (1.8) |
| HSV-2 | Vero (0.5) | 4.7 × 107 |
| ICP10ΔPK | Vero (0.5) | 3.5 × 104 (1.3 × 103) |
| HSV-2 | Vero-ICP10 (10) | 4.9 × 107 |
| ICP10ΔPK | Vero-ICP10 (10) | 2.2 × 107 (2.2) |
| HSV-2 | Vero-ICP10 (0.5) | 4.6 × 107 |
| ICP10ΔPK | Vero-ICP10 (0.5) | 2.0 × 107 (2.3) |
Plaque assays were done in medium containing 10 or 0.5% serum.
In PFU per milliliter.
ICP10ΔPK has wild-type adsorption-and-penetration kinetics and is not defective in capsid transport to the nucleus.
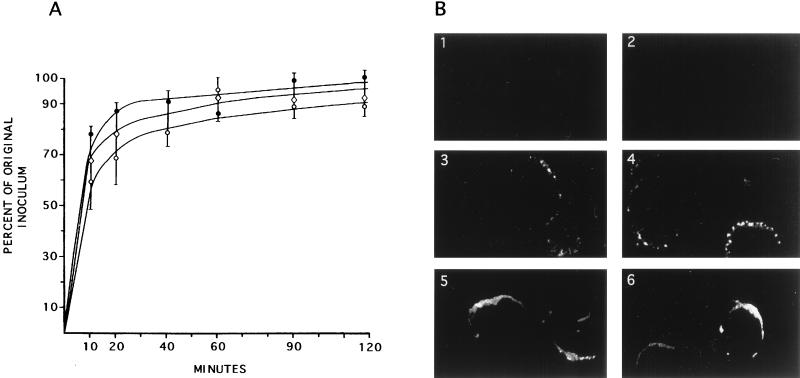
One possible interpretation for the growth and plaquing patterns evidenced by the ICP10ΔPK virus is that it is defective in the ability to adsorb to and penetrate target cells. To address this question, we exposed Vero cells in six-well plates to 5 or 200 PFU of HSV-2, ICP10ΔPK, or HSV-2(R) for 0, 10, 30, 60, 90, and 120 min at 4°C. At this time, the plates were extensively washed with PBS and overlaid with EMEM–10% FCS and 0.3% IgG. The plates were reincubated at 37°C for 48 h and then scored for the number of plaques. Under these conditions, each plaque represents the progeny of 1 adsorbed PFU. As shown in Fig. 6A, the number of HSV-2 plaques increased as a function of exposure time, reaching maximal levels at 20 to 30 min and plateauing thereafter. Similar patterns were seen for ICP10ΔPK and HSV-2(R) and at both MOIs, suggesting that ICP10ΔPK is not defective for adsorption and penetration.
FIG. 6.
Virus adsorption and penetration of cells and capsid transport to the nucleus. (A) Six-well plates of Vero cells were exposed to 200 PFU of HSV-2 (•), ICP10ΔPK (○), or HSV-2(R) (◊) at 4°C for 0, 10, 30, 60, 90, and 120 min. At this time, the cells were overlaid with EMEM–10% FCS and 0.3% IgG and reincubated at 37°C. They were scored for plaque numbers at 48 h. (B) Vero cells were infected at an MOI of 100 PFU/cell with HSV-2 (panels 1, 3, and 5) or ICP10ΔPK (panels 2, 4, and 6), and adsorption was allowed to occur for 2 h at 4°C. The cells were fixed in 3% PFA, permeabilized with 0.1% Triton X-100, and stained by immunofluorescence with capsid antibody at 0 (panels 1 and 2), 2 (panels 3 and 4), or 4 (panels 5 and 6) h after adsorption.
Another interpretation for the growth and plaquing defect evidenced by ICP10ΔPK is that PK activity is required for the transport of capsids from the cell periphery to the nucleus. To address this possibility, Vero cells were exposed to HSV-2 or ICP10ΔPK at 100 PFU/cell and virus adsorption was allowed to occur at 4°C for 2 h. At this time (0 h), the cultures were transferred to 37°C. They were stained with capsid antibody at 0, 2, 3, and 4 h thereafter. For both HSV-2 and ICP10ΔPK, staining was not seen at 0 h, indicating that capsids present in surface-bound intact viruses are not recognized by the antibody (Fig. 6B, panels 1 and 2). Presumably, this reflects antigen inaccessibility due to epitope masking in the intact virus particles by envelope and/or tegument components. By 2 h after adsorption, isolated labeled spots were seen at the nuclear membrane, with a similar distribution in HSV-2 (Fig. 6B, panel 3)- and ICP10ΔPK (Fig. 6B, panel 4)-infected cells. Their number appeared to increase with time, such that by 4 h they had coalesced into rings localizing around the nuclear membrane. The distribution and intensity of the labeled spots, and the number of staining cells, were similar in HSV-2 (Fig. 6B, panel 5)- and ICP10ΔPK (Fig. 6B, panel 6)-infected cells. Similar results were obtained for HSV-2(R) (data not shown). While we do not exclude the possibility that the number of capsids represented by a fluorescent spot differs for ICP10ΔPK and HSV-2, the data suggest that capsid transport to the nucleus is not significantly different for the two viruses. The transport of tegument proteins was not studied.
p95 expression in ICP10ΔPK-infected cells and its intracellular localization.
These studies sought to examine whether the kinetics of p95 expression in ICP10ΔPK-infected cells and its intracellular localization are similar to those of ICP10. Vero cells were infected with HSV-2, HSV-2(R), or ICP10ΔPK for 6, 8, 12, or 18 h (in 10% serum) and stained by an indirect immunofluorescence assay with ICP10 antibody. The results are shown in Fig. 7. Approximately 70% of the HSV-2- and HSV-2(R)-infected cells stained with the ICP10 antibody at 6 h p.i., and the proportion reached 100% at 8 h p.i., as previously reported for HSV-2-infected cells (10). Staining was localized in the cytoplasm and the perinuclear region and had a diffuse distribution pattern. By contrast, cells infected with ICP10ΔPK for 6 h did not stain with the ICP10 antibody. Staining was first seen at 8 h p.i. and in only 5 to 10% of the infected cells. It was in the perinuclear space and in restricted cytoplasmic granules. Diffuse cytoplasmic staining was not observed at this time (Fig. 7). The proportion of staining cells increased with time p.i., reaching levels of 15 to 20% and 85 to 100% at 12 and 18 h p.i., respectively. These findings indicate that the expression of p95 is delayed relative to that of ICP10 and, at least during the first 12 h p.i., appears to be partially sequestered within granular structures in the cytoplasm. The delay in p95 expression and its sequestration in granular structures were also seen in cells infected with ICP10ΔPK in the presence of 1% FCS (data not shown).
FIG. 7.
p95 synthesis and intracellular localization. Vero cells were infected with HSV-2 for 6 h or with ICP10ΔPK for 6 or 8 h and stained with ICP10 antibody.
Onset of protein synthesis is delayed in ICP10ΔPK-infected cells.
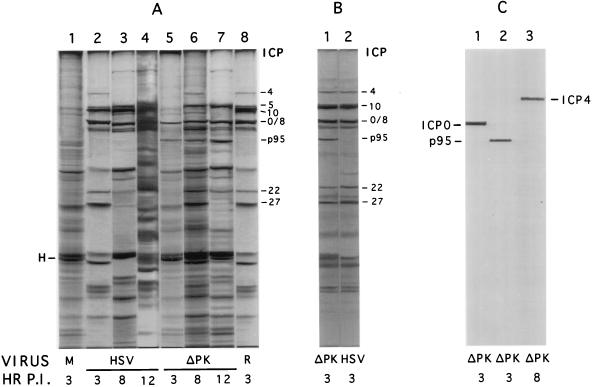
Delayed onset of p95 expression and ICP10ΔPK replication may reflect the failure to initiate the protein synthesis cascade. To examine the validity of this interpretation, Vero cells were mock infected (with PBS) or infected with HSV-2, ICP10ΔPK, or HSV-2(R) (MOI, 200 PFU/cell) in medium containing 10% FCS. At 2, 7, or 11 h p.i., the cultures were pulse labeled with [35S]methionine for 60 min. Proteins in the cell extracts obtained at that time were resolved by SDS-PAGE. As previously described (51, 68, 76), protein profiles in cells infected with HSV-2 for 3 h included IE species ICP4, ICP0, ICP22, and ICP27, as well as ICP10 (Fig. 8A, lane 2). Host protein synthesis was significantly decreased relative to that of mock-infected cells (Fig. 8A, lane 1), as exemplified by host cell protein H (Fig. 8A, lane 2). Additional viral proteins were seen in cells infected with HSV-2 for 8 h (Fig. 8A, lane 3) or 12 h (Fig. 8B, lane 4). Similar protein profiles were seen in cells infected with HSV-2(R), as shown in Fig. 8A (lane 8) for 3-h-infected cells.
FIG. 8.
Protein profiles of HSV-2- and ICP10ΔPK-infected cells. (A) Vero cells were mock infected (lane 1) or infected with HSV-2 (lanes 2 to 4), ICP10ΔPK (lanes 5 to 7), or HSV-2(R) (lane 8) in EMEM containing 10% FCS. They were labeled with [35S]methionine from 2 to 3 h p.i. (lanes 1, 2, 5, 8), 7 to 8 h p.i. (lanes 3 and 6), or 11 to 12 h p.i. (lanes 4 and 7), and proteins were resolved by SDS–8.5% PAGE. H, host protein. (B) JHLa1 cells, which express ICP10 constitutively, were infected with ICP10ΔPK (lane 1) or HSV-2 (lane 2) for 2 h and labeled with [35S]methionine from 2 to 3 h p.i. in EMEM containing 1% FCS. Proteins were resolved by SDS–8.5% PAGE. (C) Duplicate samples of the extracts from cells infected with ICP10ΔPK for 3 h (lanes 1 and 2) or 8 h (lane 3) were immunoblotted with antibody to ICP0 (lane 1), ICP10 (lane 2), or ICP4 (lane 3). Protein profiles in Vero-ICP10 cells were similar to those in JHLa1 cells; profiles in 293 cells were similar to those in Vero cells.
By contrast, the protein profile in cells infected with ICP10ΔPK for 3 h (Fig. 8A, lane 5) was not significantly different from that in mock-infected cells (Fig. 8A, lane 1). ICP4 (identity confirmed by immunoblotting [Fig. 8C, lane 3]), ICP22, and ICP27 were not seen in ICP10ΔPK-infected cells, and the levels of ICP0 (identity confirmed by immunoblotting [Fig. 8C, lane 1]) were fourfold lower than in cells infected with HSV-2 or HSV-2(R) [3,130, 3,099 and 782 densitometric integration U for HSV-2, HSV-2(R), and ICP10ΔPK, respectively]. The levels of p95 (identity confirmed by immunoblotting [Fig. 8C, lane 2]) were also lower (sevenfold) than the ICP10 levels in HSV-2- or HSV-2(R)-infected cells [3,567, 3,630, and 480 densitometric integration U for HSV-2, HSV-2(R), and ICP10ΔPK, respectively]. At 8 h p.i., with ICP10ΔPK, the levels of ICP0 and p95 were higher, and bands consistent with ICP4, ICP22, and ICP27 were also seen (Fig. 8A, lane 6). Protein profiles in cells infected with ICP10ΔPK for 12 h (Fig. 8A, lane 7) were similar to those seen in HSV-2-infected cells at 8 h p.i. (Fig. 8A, lane 3). This is consistent with the growth kinetics in cells infected at a high MOI in that virus replication under these conditions begins at 10 to 12 h after adsorption both in 10 and 1% FCS (Fig. 4 and 5). In JHLa1 cells, expression of the major IE genes (those for ICP4, ICP22, ICP27, and ICP0) was seen as early as 3 h p.i. with ICP10ΔPK (Fig. 8B, lane 1), and their levels were comparable to those seen in HSV-2-infected cells (Fig. 8B, lane 2).
ICP10 PK is required for expression of ICP4, ICP27, and ICP22.
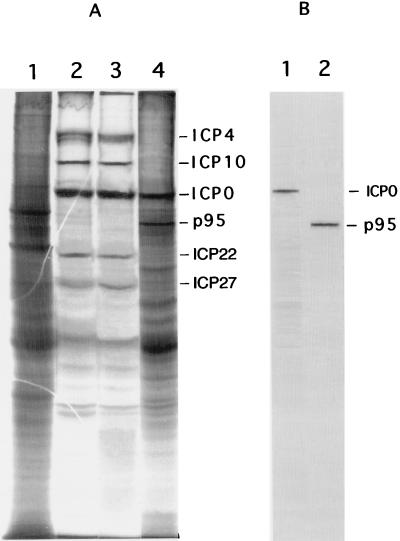
To further examine the synthesis of IE proteins in ICP10ΔPK-infected cells, we used a cycloheximide block, a condition which allows expression of IE but not other viral genes (29, 69). Cells were infected with ICP10ΔPK, HSV-2, or HSV-2(R) at 200 PFU/cell in the presence of 50-μg/ml cycloheximide (6 h) and labeled with [35S]methionine for 3 h in medium containing 10-μg/ml actinomycin D. Proteins consistent with ICP4, ICP10, ICP0, ICP22, and ICP27 were seen in cells infected with HSV-2 (Fig. 9A, lane 2) or HSV-2(R) (Fig. 9A, lane 3) under these conditions. ICP4, ICP22, and ICP27 were not seen in cells similarly infected with ICP10ΔPK (Fig. 9A, lane 4). A 110-kDa protein which is recognized by anti-ICP0 antibody (Fig. 9B, lane 1) was seen in the ICP10ΔPK-infected cells (Fig. 9A, lane 4), but its levels were twofold lower than in HSV-2 (Fig. 9A, lane 2)- or HSV-2(R) (Fig. 9A, lane 3)-infected cells (1,760 and 3,520 densitometric integration U for ICP10ΔPK- and HSV-2-infected cells, respectively). p95 was also seen in ICP10ΔPK-infected cells (Fig. 9A, lane 4, and B, lane 2), but its levels were fivefold lower than those of ICP10 in HSV-2-infected cells (413 and 2,200 densitometric integration U for p95 and ICP10, respectively). The data support the conclusion that ICP10 PK is required for optimal IE gene expression. Significantly, host protein synthesis was not shut off in ICP10ΔPK-infected cells (Fig. 9A, lane 4), although the infection was at the same MOI as for HSV-2 or HSV-2(R). This may reflect the role of ICP10 PK in the phosphorylation of vhs (55), which is responsible for host shutoff (54).
FIG. 9.
IE protein synthesis in HSV-2- and ICP10ΔPK-infected cells. (A) Vero cells were mock infected (lane 1) or infected with HSV-2 (lane 2), HSV-2(R) (lane 3), or ICP10ΔPK (lane 4) in the presence of 50-μg/ml cycloheximide (6 h) and labeled with [35S]methionine for 3 h in medium containing 10-μg/ml actinomycin D. Proteins were resolved by SDS–8.5% PAGE. (B) Duplicate samples of extracts from cells infected with ICP10ΔPK were immunoblotted with ICP0 antibody (lane 1) or ICP10 antibody (lane 2).
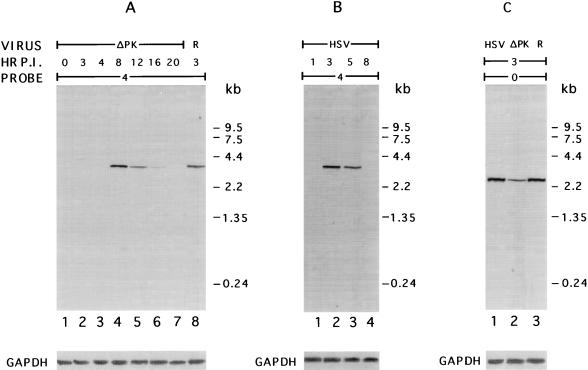
IE gene transcription is delayed in ICP10ΔPK-infected cells.
Northern hybridization was used to examine whether the defect in IE gene expression in ICP10ΔPK-infected cells is at the level of transcription. RNA was obtained from Vero cells infected with HSV-2, ICP10ΔPK, or HSV-2(R) (in 10% serum), and ICP4 and ICP0 DNAs were used as probes. GAPDH served as a control transcript. The relative abundance of ICP4 and ICP0 mRNAs was estimated by first normalizing to the value of GAPDH mRNA in each sample. The kinetics of ICP4 expression in HSV-2-infected cells were similar to those previously described for HSV-1-infected cells (27). Optimal levels were seen at 3 h p.i. (Fig. 10B, lane 2), and the transcript was no longer detectable at 8 h p.i. (Fig. 10B, lane 4). By contrast, in cells infected with ICP10ΔPK, ICP4 mRNA was not seen at 3 and 4 h p.i. (Fig. 10A, lanes 2 and 3). It was first seen at 8 h p.i. (Fig. 10A, lane 4), at which time its levels were similar to those seen in HSV-2-infected cells at 3 h p.i. (Fig. 10B, lane 2). The ICP4 transcript was still seen at 12 h p.i. (Fig. 10A, lane 5), indicating that the eventual expression of ICP4 correlates with the rise of virus growth late in infection. The transcript was no longer detected at 20 h p.i. with ICP10ΔPK (Fig. 10A, lane 7). Similar results were obtained for ICP27 (data not shown). ICP0 mRNA was seen at 3 h p.i. with ICP10ΔPK (Fig. 10C, lane 2), but its relative abundance (expressed as ICP0/GAPDH mRNA) was threefold lower than in cells similarly infected with HSV-2 (Fig. 10C, lane 1) or HSV-2(R) (Fig. 10C, lane 3) (ICP0/GAPDH ratios of 0.32, 1.0, and 0.95, respectively). We interpret these data to indicate that ICP10 PK is required for early transcription of the IE genes.
FIG. 10.
ICP4 and ICP0 RNA synthesis in ICP10ΔPK-infected cells. (A) RNA was isolated from cells infected (in 10% FCS) with ICP10ΔPK at 0 to 20 h p.i. (lanes 1 to 7) and from cells infected with HSV-2(R) at 3 h p.i. (lane 8). It was hybridized with a 32P-labeled ICP4 DNA probe or GAPDH oligonucleotide (bottom). Molecular size marker positions are indicated in the margin. (B) RNA was isolated from cells infected with HSV-2 at 1 to 8 h p.i. (lanes 1 to 4) and hybridized with a 32P-labeled ICP4 DNA probe or GAPDH oligonucleotide (bottom). Molecular size marker positions are indicated in the margin. (C) RNA was isolated from cells infected with HSV-2 (lane 1), ICP10ΔPK (lane 2), or HSV-2(R) (lane 3) at 3 h p.i. and hybridized with a 32P-labeled ICP0 DNA probe or GAPDH oligonucleotide (bottom). Molecular size marker positions are indicated in the margin.
DISCUSSION
Studies of deletion and temperature-sensitive mutants showed that RR1 is required for virus growth in nondividing cells in culture (25, 26, 63), as well as for optimal neurovirulence (7, 31) and latency reactivation (33, 58) in infected animals. However, these studies did not differentiate between the respective contributions of the PK and RR domains of the multifunctional RR1 proteins. In nondividing cells, the RR domain presumably functions to supply the RR activity which is necessary for virus growth (25, 26, 63). The PK activity is not required for ribonucleotide reduction (15), and it can be dissociated from the RR activity (10, 15, 32, 37, 42). Because the PK domain likely evolved from a cellular gene and was conserved through many evolutionary cycles (62), it seems reasonable to conclude that the PK activity is critical for virus growth. The studies described in this report were designed to test this hypothesis. The following comments seem pertinent with respect to our findings.
The virus used in these studies (ICP10ΔPK) is an HSV-2 mutant with a deletion in the RR1 PK domain. It expresses a 95-kDa protein (p95) that lacks PK activity but retains the ability to complex with RR2, giving rise to an RR activity similar to that of the wild-type virus. This is consistent with our previous finding that ICP10 residues which complex with RR2 are at the C terminus (amino acids 1096 to 1144) (12). ICP10ΔPK is unlikely to have defects other than a PK-deficient ICP10, since a revertant virus [HSV-2(R)] was generated by recombination of ICP10ΔPK DNA with the wild-type BamHI E/T fragment that encompasses the ICP10 coding sequences, but not with the BamHI E/T fragment with the sequences which code for ICP10 PK deleted. Because VP16 activates the expression of IE genes (8, 53) as well as that of RR1 (18, 70, 77, 78), we considered the possibility that growth defects evidenced by ICP10ΔPK may be due to a mutated VP16 gene. However, this is unlikely, since the wild-type phenotype was not restored in a rescue experiment with VP16-encoding DNA, and the levels of VP16 were similar in ICP10ΔPK- and HSV-2-infected cells (data not shown). Nonetheless, we do not exclude the possibility that VP16 is involved in the timely expression of IE genes in cells infected with ICP10ΔPK, because VP16 is phosphorylated on serine residues (48) and it may be a substrate for ICP10 PK.
Single-step growth curve analyses indicated that ICP10ΔPK has two, apparently distinct, growth defects. First, onset of virus replication was significantly delayed (10 to 15 h) in both dividing and nondividing cells. Second, in nondividing cells, virus titers were approximately 1,000-fold lower than in dividing cells, and plaquing ability was severely compromised. Similar defects were observed in various cell lines, indicating that they are not determined by the cell type. They were not evidenced by the restored virus [HSV-2(R)] or in cells that constitutively express ICP10 (JHLa1 or Vero-ICP10), indicating that they are due to the lack of a functional ICP10 PK.
The delayed onset of virus growth is consistent with previous conclusions that the IE component of RR1 regulation is required for early expression of PK activity (18, 70, 77, 78, 81). Presumably, growth onset at 10 to 15 h p.i. reflects compensation for the missing ICP10 PK by a cellular function which may be induced or activated by virion structural proteins. Such an interpretation is consistent with the finding that replication begins 3 to 5 h earlier, when the cells are infected at a high (200 PFU/cell), rather than a low (2 PFU/cell), MOI. Virion proteins that could induce or activate such a cellular function include VP16, which was previously shown to activate cellular promoters such as beta interferon (38) and Gal4 (79), and the promoter-independent promiscuous transactivator ICP0 (20, 39), which was recently shown to be a virion protein (80). Inasmuch as ICP10ΔPK growth began at the same time in dividing and nondividing cells, the putative compensatory function presumably does not require de novo protein synthesis and may involve the activation of a PK cascade. It is noteworthy that a mutant defective in both the PK and RR activities did not replicate, even in dividing cells, suggesting that the RR domain may be involved in induction or activation of the PK compensatory function (data not shown). Implicit in such an interpretation is the conclusion that, in dividing cells, sufficient levels of RR activity are produced early in infection with ICP10ΔPK to induce or activate the PK compensatory function. Ongoing studies were designed to examine the validity of this interpretation.
What is the function of ICP10 PK in the early onset of virus growth? Our data suggest that ICP10ΔPK is not defective in adsorption and penetration or in the transport of incoming capsids to the nucleus. However, IE gene expression is selectively delayed. Thus, ICP4, which is required for synthesis of early and late viral proteins (17, 19, 47), was first seen in cells infected with ICP10ΔPK at 8 h p.i., compared to 3 h p.i. for HSV-2 and HSV-2(R). ICP4 mRNA was not seen until 8 h p.i., and neither ICP4 mRNA nor protein was seen with a cycloheximide block, a condition that allowed IE gene expression in cells infected with HSV-2 or HSV-2(R). Cells infected with ICP10ΔPK for less than 8 h or in the presence of cycloheximide were also negative for ICP27, which often functions together with ICP4 to initiate early gene expression (60), and ICP22, which is required for late gene expression (40, 57, 61). ICP0 was expressed early after infection with ICP10ΔPK (3 h) and in the presence of cycloheximide, but its levels were significantly lower than those in HSV-2- or HSV-2(R)-infected cells. p95 was also expressed early after infection with ICP10ΔPK and in the presence of cycloheximide, but is levels were lower than those of ICP10 in cells similarly infected with HSV-2 or HSV-2(R), suggesting that the PK domain is involved in ICP10 self-regulation. p95 expression in the absence of ICP4 is consistent with previous findings that basal expression from the RR1 promoter requires AP-1 transcription factors and is independent of ICP4 (18, 77, 78, 81).
Shutoff of host protein synthesis was delayed in ICP10ΔPK-infected cells, also in the presence of cycloheximide, and this is consistent with the incomplete lysis of the infected cells and the hazy appearance of the ICP10ΔPK plaques. Because vhs phosphorylation affects its ability to induce mRNA degradation (55), impaired host shutoff may reflect the role of ICP10 PK in vhs phosphorylation. Indeed, vhs is not phosphorylated by another HSV PK species (UL13) (49), and a phosphorylated 57- to 59-kDa species consistent with vhs was not seen in ICP10ΔPK-infected cells (data not shown). If vhs is phosphorylated by ICP10 PK, ICP10ΔPK grown in JHLa1 cells may contain a vhs-encoded protein which is relatively more activated (has higher mRNA degradation activity) than the vhs-encoded protein of ICP10ΔPK propagated in Vero cells. Ongoing studies were designed to test this interpretation.
The exact role of ICP10 PK in IE gene expression is unknown. It is unlikely that it functions as a transactivator of IE gene expression, because transactivating activity was not observed in transient transfection assays with pICP4-cat constructs (unpublished data). Because ICP10 is located in the virion tegument (65), its PK domain could be involved in the transport of incoming VP16 to the nucleus, for example, by maintaining tegument integrity. In addition, ICP10 PK could phosphorylate and consequently activate VP16, thereby determining early onset of IE gene expression. Implicit in the interpretation that ICP10 PK functions at the level of VP16 is the conclusion that VP16 is not required for early expression of ICP0, which is seen as early as 3 h p.i. with ICP10ΔPK. However, previous studies showed that VP16 is required for expression of ICP0 but not ICP4 (1). Also, inasmuch as the kinetics of synthesis of the IE proteins and their levels were restored to wild-type patterns in ICP10ΔPK-infected cells, which provide ICP10 PK activity in trans, it is unlikely that ICP10 PK is required for transport of tegument proteins to the nucleus. Nonetheless, ongoing studies were designed to test this hypothesis.
Delayed onset of ICP10ΔPK growth could be due to the inhibition of IE gene expression by PK-defective ICP10. This implies that in the wild-type virus, ICP10 PK downregulates a protein which inhibits early onset of IE gene expression. A function that represses accumulation of ICP4 transcripts was recently described in mouse neurons latently infected with HSV-1, but it was attributed to the latency-associated transcript (LAT) locus (9). It may also be that the PK domain is not involved in IE gene expression but, rather, that in its absence, the RR domain causes transdominant inhibition of IE gene expression and virus growth. If this were the case, a virus with deletions in both the PK and RR domains should grow as well as HSV-2 in 10% FCS. However, a mutant with a deletion in ICP10 failed to replicate under these conditions (data not shown), suggesting that the RR domain does not have transdominant downregulatory activity. Finally, ICP10 PK could phosphorylate, and thereby activate, one or more factors involved in IE gene transcription. For example, carboxy-terminal domain (CTD) kinase(s), a component(s) of transcription factor IIH, is activated by phosphorylation (23, 28) and, in turn, phosphorylates the CTD of the large subunit of polymerase II. If their phosphorylation is altered by the direct or indirect contribution of virion-associated PKs, polymerase II might be redirected from cellular to viral IE promoters (56). This possibility has been excluded for HSV-1 virions. They contain only one trans-phosphorylating kinase activity (UL13), and it does not phosphorylate CTD kinases (57). However, ICP10 PK is structurally and functionally different from ICP6 PK (14, 16, 46). ICP10 is located within the tegument fractions of HSV-2 virions and has trans-phosphorylating activity (65). It is therefore in a position to be involved in alterations of the phosphorylation of CTD kinases. Implicit in this interpretation is the conclusion that ICP10 PK functions in the nucleus. While the available data indicate that ICP10 is localized in the cytoplasm and on the surfaces of cells infected with HSV-2 for at least 6 h (10), we recently found nuclear staining with monoclonal antibodies to epitopes within the ICP10 PK (but not RR) domain in cells infected with HSV-2 for 1 to 3 h, i.e., before the onset of viral DNA synthesis (4a).
The finding that p95 is localized primarily within restricted cytoplasmic compartments in cells infected with ICP10ΔPK for 8 to 12 h suggests that, in addition to its role in the early onset of virus growth, the PK domain is involved in RR1 intracellular localization. RR1 sequestration may render it unavailable for complexation with RR2 and the generation of appropriate levels of RR activity, thereby explaining the reduced virus titers in nondividing cells.
What is the role of ICP10 PK in virus pathogenesis? In vivo, ICP10 PK might be required for virus replication at the site of infection and, thereby, efficient latency establishment, and/or for reactivation from latency. It has been proposed that in addition to IE genes, early genes involved in viral DNA synthesis must be turned on by the reactivating stimuli resulting in limited DNA replication. This, in turn, stimulates a viral function that upregulates IE gene expression, leading to the lytic cascade and the production of infectious virus (35). However, reactivating stimuli upregulate AP-1 transcription factors (21, 34, 72), and RR1 is the only viral promoter that contains AP-1 cis response elements (77, 78, 81). Because the ICP10 promoter responds to AP-1 with basal expression which is independent of VP16, ICP0, or ICP4 (18, 77, 78, 81), we propose that ICP10 is an early response to latency-reactivating stimuli. An AP-1 amplification loop is further provided by the ability of ICP10 PK to activate the ras signaling pathway (30, 64). RR1 is uniquely compatible with a virus-reactivating function because it provides both the PK activity which is necessary for IE gene expression and the RR activity which further upregulates IE gene expression by inducing viral DNA synthesis in nondividing neuronal cells (25, 26). ICP0, the expression of which is thus increased, cooperates with AP-1 to further activate expression from the ICP10 promoter (18, 77, 78, 81). It also upregulates other HSV IE genes. The outcome is initiation of the lytic cascade and the production of infectious virus. Indeed, recent studies indicate that the HSV-2 LATs, generally assumed to be the only viral transcripts involved in latency reactivation, are inefficient and weak determinants of HSV-2 reactivation, at least as reflected by the quantity of the major LATs in the ganglia (74). Furthermore, studies of the mouse trigeminal model indicate that during reactivation, early viral transcripts, notably, RR1 and TK, are detected before IE transcripts (71). Consistent with these interpretations, HSV-2 was not reactivated from latently infected ganglia explanted in the presence of an antisense oligonucleotide that inhibits ICP10 expression (65a). Ongoing studies were designed to examine the role of ICP10 PK in latency reactivation.
REFERENCES
- 1.Ace C I, McKee T A, Ryan M, Cameron J M, Preston C M. Construction and characterization of a herpes simplex virus type 1 mutant unable to transinduce immediate-early gene expression. J Virol. 1989;63:2260–2269. doi: 10.1128/jvi.63.5.2260-2269.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ali M A, Prakash S S, Jariwalla R J. Localization of the antigenic sites and intrinsic protein kinase domain within a 300 amino acid segment of the ribonucleotide reductase large subunit from herpes simplex virus type 2. Virology. 1992;187:360–367. doi: 10.1016/0042-6822(92)90328-m. [DOI] [PubMed] [Google Scholar]
- 3.Anderson K P, Frink R J, Devi G B, Gaylord B H, Wagner E K. Detailed characterization of the mRNA mapping in the HindIII fragment K region of the herpes simplex virus type 1 genome. J Virol. 1981;37:1011–1027. doi: 10.1128/jvi.37.3.1011-1027.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aurelian L, Terzano P, Smith C C, Chung T D, Shamsuddin A, Costa S, Orlandi C. Amino-terminal epitope of herpes simplex virus type 2 ICP10 protein as a molecular diagnostic marker for cervical intraepithelial neoplasia. Cancer Cells. 1989;7:187–191. [Google Scholar]
- 4a.Aurelian, L., et al. Unpublished data.
- 5.Aurelian L. Herpes simplex viruses. In: Specter S, Lancz G, editors. Clinical virology manual. 2nd ed. New York, N.Y: Elsevier Science Publishers; 1992. pp. 473–494. [Google Scholar]
- 6.Bacchetti S, Evelegh M J, Muirhead B, Sartari C S, Huszar D. Immunological characterization of herpes simplex virus type 1 and 2 polypeptide(s) involved in viral ribonucleotide reductase activity. J Gen Virol. 1984;49:591–593. doi: 10.1128/jvi.49.2.591-593.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cameron J M, McDougall I, Marsden H W, Preston V G, Ryan D M, Subak-Sharpe S H. Ribonucleotide reductase encoded by herpes simplex virus is a determinant of the pathogenicity of the virus in mice and a valid antiviral target. J Virol. 1988;69:2607–2612. doi: 10.1099/0022-1317-69-10-2607. [DOI] [PubMed] [Google Scholar]
- 8.Campbell M E M, Palfreyman J W, Preston C M. Identification of herpes simplex virus DNA sequences which encode a trans-acting polypeptide responsible for stimulation of immediate early transcription. J Mol Biol. 1984;180:1–19. doi: 10.1016/0022-2836(84)90427-3. [DOI] [PubMed] [Google Scholar]
- 9.Chen S H, Kramer M F, Schaffer P A, Coen D M. A viral function represses accumulation of transcripts from productive-cycle genes in mouse ganglia latently infected with herpes simplex virus. J Virol. 1997;71:5878–5884. doi: 10.1128/jvi.71.8.5878-5884.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chung T D, Wymer J P, Smith C C, Kulka M, Aurelian L. Protein kinase activity associated with the large subunit of the herpes simplex virus type 2 ribonucleotide reductase (ICP10) J Virol. 1989;63:3389–3398. doi: 10.1128/jvi.63.8.3389-3398.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chung T D, Wymer J P, Kulka M, Smith C C, Aurelian L. Myristylation and polylysine-mediated activation of the protein kinase domain of the large subunit of herpes simplex virus type 2 ribonucleotide reductase (ICP10) Virology. 1990;179:168–178. doi: 10.1016/0042-6822(90)90286-z. [DOI] [PubMed] [Google Scholar]
- 12.Chung T D, Luo J H, Wymer J P, Smith C C, Aurelian L. Leucine repeats in the large subunit of herpes simplex virus type 2 ribonucleotide reductase (RR; ICP10) are involved in RR activity and subunit complex formation. J Gen Virol. 1991;72:1139–1144. doi: 10.1099/0022-1317-72-5-1139. [DOI] [PubMed] [Google Scholar]
- 13.Cohen G H. Ribonucleotide reductase activity of synchronized KB cells infected with herpes simplex virus. J Virol. 1972;9:408–418. doi: 10.1128/jvi.9.3.408-418.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Conner J, Cooper J, Furlong J, Clements J B. An autophosphorylating but not transphosphorylating activity is associated with the unique N terminus of the herpes simplex virus type 1 ribonucleotide reductase large subunit. J Virol. 1992;66:7511–7516. doi: 10.1128/jvi.66.12.7511-7516.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Conner J, Macfarlene J, Lankinen H, Marsden H. The unique N-terminus of the herpes simplex virus type 1 large subunit is not required for ribonucleotide reductase activity. J Gen Virol. 1992;73:103–112. doi: 10.1099/0022-1317-73-1-103. [DOI] [PubMed] [Google Scholar]
- 16.Cooper J, Conner J, Clements J B. Characterization of the novel protein kinase activity present in the R1 subunit of herpes simplex virus ribonucleotide reductase. J Virol. 1995;69:4979–4985. doi: 10.1128/jvi.69.8.4979-4985.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DeLuca N A, Schaffer P A. Activation of immediate-early, early, and late promoters by temperature-sensitive and wild-type forms of herpes simplex virus type 1 protein ICP4. Mol Cell Biol. 1985;5:1997–2008. doi: 10.1128/mcb.5.8.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Desai P, Ramakrishnan R, Lin Z W, Osak B, Glorioso J C, Levine M. The RR1 gene of herpes simplex virus type 1 is uniquely trans activated by ICP0 during infection. J Virol. 1993;67:6125–6135. doi: 10.1128/jvi.67.10.6125-6135.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dixon R A F, Schaffer P A. Fine-structure mapping and functional analysis of temperature-sensitive mutants in the gene encoding the herpes simplex virus type 1 immediate early protein VP175. J Virol. 1980;36:189–203. doi: 10.1128/jvi.36.1.189-203.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Everett R E, Preston C M, Stow N W. Functional and genetic analysis of the role of Vmw110 in herpes simplex virus replication. In: Wagner E K, editor. Herpesvirus transcription. Boca Raton, Fla: CRC Press, Inc.; 1991. pp. 49–76. [Google Scholar]
- 21.Fawl R L, Roizman B. Induction of reactivation of herpes simplex virus in murine sensory ganglia in vivo by cadmium. J Virol. 1993;67:7025–7031. doi: 10.1128/jvi.67.12.7025-7031.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feng C P, Kulka M, Smith C C, Aurelian L. Herpes simplex virus-mediated activation of human immunodeficiency virus is inhibited by oligonucleotide methylphosphonates that target immediate-early mRNAs 1 and 3. Antisense Nucleic Acid Drug Dev. 1996;6:25–35. doi: 10.1089/oli.1.1996.6.25. [DOI] [PubMed] [Google Scholar]
- 23.Fisher R P, Jin P, Chamberlin H M, Morgan D O. Alternative mechanisms of CAK assembly require an assembly factor or an activating kinase. Cell. 1995;83:47–57. doi: 10.1016/0092-8674(95)90233-3. [DOI] [PubMed] [Google Scholar]
- 24.Frame M C, Marsden H S, Dutia B M. The ribonucleotide reductase induced by herpes simplex virus type 1 involves minimally a complex of two polypeptides (136K and 38K) J Gen Virol. 1985;66:1581–1587. doi: 10.1099/0022-1317-66-7-1581. [DOI] [PubMed] [Google Scholar]
- 25.Goldstein D J, Weller S K. Herpes simplex virus type 1-induced ribonucleotide reductase activity is dispensable for virus growth and DNA synthesis: isolation and characterization of an ICP6 lacZ insertion mutant. J Virol. 1988;62:196–205. doi: 10.1128/jvi.62.1.196-205.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goldstein D J, Weller S K. Factor(s) present in the herpes simplex virus type-1 infected cells can compensate for the loss of the large subunit of the viral ribonucleotide reductase: characterization of an ICP6 deletion mutant. Virology. 1988;166:41–51. doi: 10.1016/0042-6822(88)90144-4. [DOI] [PubMed] [Google Scholar]
- 27.Harris-Hamilton E, Bachenheimer S L. Accumulation of herpes simplex virus type 1 RNAs of different kinetic classes in the cytoplasm of infected cells. J Virol. 1985;53:144–151. doi: 10.1128/jvi.53.1.144-151.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hermann C H, Gold M O, Rice A P. Viral transactivators specifically target distinct cellular protein kinases that phosphorylate the RNA polymerase II C-terminal domain. Nucleic Acids Res. 1995;24:501–504. doi: 10.1093/nar/24.3.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Honess R W, Roizman B. Regulation of herpesvirus macromolecular synthesis. I. Cascade regulation of the synthesis of three groups of viral proteins. J Virol. 1974;14:8–19. doi: 10.1128/jvi.14.1.8-19.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hunter J C R, Smith C C, Bose D, Kulka M, Broderick R, Aurelian L. Intracellular internalization and signaling pathways triggered by the large subunit of HSV-2 ribonucleotide reductase (ICP10) Virology. 1995;210:345–360. doi: 10.1006/viro.1995.1351. [DOI] [PubMed] [Google Scholar]
- 31.Idowu A D, Fraser-Smith E B, Paffenberger K L, Herman R C. Deletion of herpes simplex virus type 1 ribonucleotide reductase gene alters virulence and latency in vitro. Antiviral Res. 1992;17:145–156. doi: 10.1016/0166-3542(92)90048-a. [DOI] [PubMed] [Google Scholar]
- 32.Ingemarson R, Lankinen H. The herpes simplex virus type 1 ribonucleotide reductase is a tight complex of the type alpha 2 and beta 2 composed of 40K and 140K proteins, of which the latter shows multiple forms due to proteolysis. Virology. 1987;156:417–422. doi: 10.1016/0042-6822(87)90422-3. [DOI] [PubMed] [Google Scholar]
- 33.Jacobson J G, Leib D A, Goldstein D J, Bogard C L, Schaffer P A, Weller S K, Coen D M. A herpes simplex virus ribonucleotide reductase deletion mutant is defective for productive acute and reactivatable latent infections of mice and for replication in mouse cells. Virology. 1989;173:276–283. doi: 10.1016/0042-6822(89)90244-4. [DOI] [PubMed] [Google Scholar]
- 34.Jin P, Ringertz N R. Cadmium induces transcription of proto-oncogenes c-jun and c-myc in rat L6 myoblasts. J Biol Chem. 1990;265:14061–14064. [PubMed] [Google Scholar]
- 35.Kosz-Vnenchak M, Jacobson J, Coen D M, Knipe D M. Evidence for a novel regulatory pathway for herpes simplex virus gene expression in trigeminal ganglion neurons. J Virol. 1993;67:5383–5393. doi: 10.1128/jvi.67.9.5383-5393.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Langelier Y, Buttin G. Characterization of ribonucleotide reductase induction in BHK-21/C13 Syrian hamster cell line upon infection by herpes simplex virus. J Gen Virol. 1981;57:21–31. doi: 10.1099/0022-1317-57-1-21. [DOI] [PubMed] [Google Scholar]
- 37.Lankinen H, Telford E, MacDonald D, Marsden H. The unique N-terminal domain of the large subunit of herpes simplex virus ribonucleotide reductase is preferentially sensitive to proteolysis. J Gen Virol. 1989;70:3159–3169. doi: 10.1099/0022-1317-70-12-3159. [DOI] [PubMed] [Google Scholar]
- 38.LeBlanc J F, Hiscott J. Differential response of human interferon-beta promoter elements to trans-activation by HSV VP16 and IRF-1. Virology. 1992;186:760–763. doi: 10.1016/0042-6822(92)90043-o. [DOI] [PubMed] [Google Scholar]
- 39.Lees-Miller S P, Long M C, Kilvert M A, Lam V, Rice S A, Spencer C A. Attenuation of DNA-dependent protein kinase activity and its catalytic subunit by the herpes simplex virus type 1 transactivator ICP0. J Virol. 1996;70:7471–7477. doi: 10.1128/jvi.70.11.7471-7477.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leopardi R, Riozman B. Functional interaction and colocalization of the herpes simplex virus 1 major regulatory protein with EAP, a nucleolar-ribosomal protein. Proc Natl Acad Sci USA. 1996;93:4562–4576. doi: 10.1073/pnas.93.10.4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luo J, Aurelian L. The transmembrane helical segment but not the invariant lysine is required for the kinase activity of the large subunit of herpes simplex virus type 2 ribonucleotide reductase (ICP10) J Biol Chem. 1992;267:9645–9653. [PubMed] [Google Scholar]
- 42.Luo J H, Smith C C, Kulka M, Aurelian L. A truncated protein kinase domain of the large subunit of herpes simplex virus type 2 ribonucleotide reductase (ICP10) expressed in Escherichia coli. J Biol Chem. 1991;266:20976–20983. [PubMed] [Google Scholar]
- 43.Marsden H S, Crombie I K, Subak-Sharpe J H. Control of protein synthesis in herpesvirus-infected cells: analysis of the polypeptides induced by wild type and sixteen temperature sensitive mutants of strain 17. J Gen Virol. 1976;31:347–372. doi: 10.1099/0022-1317-31-3-347. [DOI] [PubMed] [Google Scholar]
- 44.McCarthy A M, McMahan L, Schaffer P A. Herpes simplex virus type 1 ICP27 deletion mutants exhibit altered patterns of transcription and are DNA deficient. J Virol. 1989;63:18–27. doi: 10.1128/jvi.63.1.18-27.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McLauchlan J, Clements J B. DNA sequence homology between two co-linear loci on the HSV genome which have different transforming abilities. EMBO J. 1983;2:1953–1961. doi: 10.1002/j.1460-2075.1983.tb01684.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nelson J W, Zhu J, Smith C C, Kulka M, Aurelian L. ATP and SH3 binding sites in the protein kinase of the large subunit of herpes simplex virus type 2 of ribonucleotide reductase (ICP10) J Biol Chem. 1996;271:17021–17027. doi: 10.1074/jbc.271.29.17021. [DOI] [PubMed] [Google Scholar]
- 47.O’Hare P, Hayward G S. Evidence for a direct role for both the 175,000- and 110,000-molecular-weight immediate-early proteins of herpes simplex virus in the transactivation of delayed-early promoters. J Virol. 1985;53:751–760. doi: 10.1128/jvi.53.3.751-760.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.O’Reilly D, Hanscombe O, O’Hare P. A single serine residue at position 375 of VP16 is critical for complex assembly with Oct-1 and HCF and is a target of phosphorylation by casein kinase II. EMBO J. 1997;16:2420–2430. doi: 10.1093/emboj/16.9.2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Overton H, McMillan D, Hope L, Wong-Kai-In P. Production of host shutoff-defective mutants of herpes simplex virus type 1 by inactivation of the UL13 gene. Virology. 1994;202:97–106. doi: 10.1006/viro.1994.1326. [DOI] [PubMed] [Google Scholar]
- 50.Peng T, Hunter J R C, Nelson J W. The novel protein kinase of the RR1 subunit of herpes simplex virus has autophosphorylation and transphosphorylation activity that differs in its ATP requirements for HSV-1 and HSV-2. Virology. 1996;216:184–196. doi: 10.1006/viro.1996.0045. [DOI] [PubMed] [Google Scholar]
- 51.Pereira L, Wolff M H, Fenwick M, Roizman B. Regulation of herpesvirus macromolecular synthesis. V. Properties of α polypeptides made in HSV-1 and HSV-2 infected cells. Virology. 1977;77:733–749. doi: 10.1016/0042-6822(77)90495-0. [DOI] [PubMed] [Google Scholar]
- 52.Pignatti P F, Cassai E, Meneguzzi G, Chencinger N, Milanesi G. Herpes simplex virus DNA isolation from infected cells with a novel procedure. Virology. 1979;93:260–264. doi: 10.1016/0042-6822(79)90295-2. [DOI] [PubMed] [Google Scholar]
- 53.Post L E, Mackem S, Roizman B. Regulation of α genes of herpes simplex virus: expression of chimeric genes produced by fusion of thymidine kinase with α gene promoters. Cell. 1981;24:555–565. doi: 10.1016/0092-8674(81)90346-9. [DOI] [PubMed] [Google Scholar]
- 54.Read G S, Frenkel N. Herpes simplex virus mutants defective in the virion-associated shutoff of host polypeptide synthesis and exhibiting abnormal synthesis of α (immediate early) viral polypeptides. J Virol. 1983;46:498–512. doi: 10.1128/jvi.46.2.498-512.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Read G S, Karr B M, Knight K. Isolation of a herpes simplex virus type 1 mutant with a deletion in the virion host shutoff gene and identification of multiple forms of the vhs (UL41) polypeptide. J Virol. 1993;67:7149–7160. doi: 10.1128/jvi.67.12.7149-7160.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rice S A, Long M C, Lam V, Spencer C A. RNA polymerase II is aberrantly phosphorylated and localized to viral replication compartments following herpes simplex virus infection. J Virol. 1994;68:988–1001. doi: 10.1128/jvi.68.2.988-1001.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rice S A, Long M C, Lam V, Schaffer P A, Spencer C. Herpes simplex virus immediate-early protein ICP22 is required for viral modification of host RNA polymerase II and establishment of the normal viral transcription program. J Virol. 1995;69:5550–5559. doi: 10.1128/jvi.69.9.5550-5559.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Russell J, Stow N D, Stow E C, Preston C M. Herpes simplex virus genes involved in latency in vitro. J Gen Virol. 1987;68:3009–3018. doi: 10.1099/0022-1317-68-12-3009. [DOI] [PubMed] [Google Scholar]
- 59.Sacks W R, Schaffer P A. Deletion mutants in the gene encoding the herpes simplex virus type 1 immediate-early protein ICP0 exhibit impaired growth in cell culture. J Virol. 1987;61:829–839. doi: 10.1128/jvi.61.3.829-839.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Samaniego L A, Webb A L, DeLuca N A. Functional interactions between herpes simplex virus immediate-early proteins during infection: gene expression as a consequence of ICP27 and different domains of ICP4. J Virol. 1995;69:5705–5715. doi: 10.1128/jvi.69.9.5705-5715.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Smith C C, Aurelian L, Reddy M P, Miller P S, Ts’o P O P. Antiviral effect of an oligo(nucleoside methylphosphonate) complementary to the splice junction of herpes simplex virus type 1 immediate early pre-mRNAs 4 and 5. Proc Natl Acad Sci USA. 1986;83:2787–2791. doi: 10.1073/pnas.83.9.2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Smith C C, Wymer J P, Luo J H, Aurelian L. Genomic sequences homologous to the protein kinase region of the bifunctional herpes simplex virus type 2 protein ICP10. Virus Genes. 1991;5(3):215–226. doi: 10.1007/BF00568971. [DOI] [PubMed] [Google Scholar]
- 63.Smith C C, Kulka M, Wymer J P, Chung T D, Aurelian L. Expression of the large subunit of herpes simplex virus type 2 ribonucleotide reductase (ICP10) is required for virus growth and neoplastic transformation. J Gen Virol. 1992;73:1417–1428. doi: 10.1099/0022-1317-73-6-1417. [DOI] [PubMed] [Google Scholar]
- 64.Smith C C, Luo J H, Hunter J C R, Ordonez J V, Aurelian L. The transmembrane domain of the large subunit of HSV-2 ribonucleotide reductase (ICP10) is required for the transformation-related signaling pathways that involve ras activation. Virology. 1994;200:598–612. doi: 10.1006/viro.1994.1223. [DOI] [PubMed] [Google Scholar]
- 65.Smith C C, Aurelian L. The large subunit of herpes simplex virus type 2 ribonucleotide reductase (ICP10) is associated with the virion tegument and has PK activity. Virology. 1997;234:235–242. doi: 10.1006/viro.1997.8645. [DOI] [PubMed] [Google Scholar]
- 65a.Smith, C. C., et al. Unpublished data.
- 66.Sodeik B, Ebersold M W, Helenius A. Microtubule-mediated transport of incoming herpes simplex virus 1 capsids to the nucleus. J Cell Biol. 1997;136:1007–1021. doi: 10.1083/jcb.136.5.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Strnad B, Aurelian L. Proteins of herpesvirus type 2. III. Isolation and immunologic characterization of a large molecular weight viral protein. Virology. 1978;87:401–415. doi: 10.1016/0042-6822(78)90144-7. [DOI] [PubMed] [Google Scholar]
- 68.Strnad B C, Aurelian L. Proteins of herpesvirus type 2. I. Virion, non-virion and antigenic polypeptides in infected cells. Virology. 1976;69:438–452. doi: 10.1016/0042-6822(76)90475-x. [DOI] [PubMed] [Google Scholar]
- 69.Strnad B C, Aurelian L. Proteins of herpesvirus type 2. II. Studies demonstrating a correlation between a tumor-associated antigen (AG-4) and a virion protein. Virology. 1976;73:244–258. doi: 10.1016/0042-6822(76)90078-7. [DOI] [PubMed] [Google Scholar]
- 70.Sze P, Herman R C. The herpes simplex virus type 1 ICP6 gene is regulated by a “leaky” early promoter. Virus Res. 1992;2:141–152. doi: 10.1016/0168-1702(92)90153-z. [DOI] [PubMed] [Google Scholar]
- 71.Tal-Singer R, Lasner T M, Podrzucki W, Skokotas A, Laeary J J, Berger S L, Fraser N W. Gene expression during reactivation of herpes simplex virus type 1 from latency in the peripheral nervous system is different from that during lytic infection of tissue cultures. J Virol. 1997;71:5268–5276. doi: 10.1128/jvi.71.7.5268-5276.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Valyi-Nagy T, Deshmane S, Dillner A, Fraser N W. Induction of cellular transcription factors in trigeminal ganglia of mice by corneal scarification, herpes simplex virus type 1 infection, and explantation of trigeminal ganglia. J Virol. 1991;65:4142–4152. doi: 10.1128/jvi.65.8.4142-4152.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wagner E. Transcription patterns in HSV infections. In: Klein G, editor. Advances in viral oncology. New York, N.Y: Raven Press; 1983. pp. 239–276. [Google Scholar]
- 74.Wang K, Pesnicak L, Straus S E. Mutations in the 5′ end of the herpes simplex virus type 2 latency-associated transcript (LAT) promoter affect LAT expression in vivo but not the rate of spontaneous reactivation of genital herpes. J Virol. 1997;71:7903–7910. doi: 10.1128/jvi.71.10.7903-7910.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Watson R J, Preston C M, Clements J B. Separation and characterization of herpes simplex virus type 1 immediate-early mRNA’s. J Virol. 1979;31:42–52. doi: 10.1128/jvi.31.1.42-52.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wilcox K W, Kohn A, Sklyanskaya E, Roizman B. Herpes simplex virus phosphoproteins. I. Phosphate cycles on and off some viral polypeptides and can alter their affinity for DNA. J Virol. 1980;33:167–182. doi: 10.1128/jvi.33.1.167-182.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wymer J P, Chung T D, Chang Y-N, Hayward G S, Aurelian L. Identification of immediate-early-type cis-response elements in the promoter for the ribonucleotide reductase large subunit from herpes simplex virus type 2. J Virol. 1989;63:2773–2784. doi: 10.1128/jvi.63.6.2773-2784.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wymer J P, Aprhys C M J, Chung T C, Feng T P, Kulka M, Aurelian L. Immediate early and functional AP-1 cis-response elements are involved in the transcriptional regulation of the large subunit of herpes simplex virus type 2 ribonucleotide reductase (ICP10) Virus Res. 1992;23:253–270. doi: 10.1016/0168-1702(92)90112-m. [DOI] [PubMed] [Google Scholar]
- 79.Yankulov K, Blau J, Purton T, Roberts S, Bentley D L. Transcriptional elongation by RNA polymerase II is stimulated by transactivators. Cell. 1994;77:749–759. doi: 10.1016/0092-8674(94)90058-2. [DOI] [PubMed] [Google Scholar]
- 80.Yao F, Courtney R J. Association of ICP0 but not ICP27 with purified virions of herpes simplex virus type 1. J Virol. 1992;66:2709–2716. doi: 10.1128/jvi.66.5.2709-2716.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhu J, Aurelian L. AP-1 cis-response elements are involved in basal expression and Vmw110 transactivation of the large subunit of herpes simplex virus type 2 ribonucleotide reductase (ICP0) Virology. 1997;231:301–312. doi: 10.1006/viro.1997.8522. [DOI] [PubMed] [Google Scholar]