Abstract
Purpose
Uveitis is a heterogenous group of inflammatory eye disease for which current cytokine-targeted immune therapies are effective for only a subset of patients. We hypothesized that despite pathophysiologic nuances that differentiate individual disease states, all forms of eye inflammation might share common mechanisms for immune cell recruitment. Identifying these mechanisms is critical for developing novel, broadly acting therapeutic strategies.
Design
Experimental study.
Subjects
Biospecimens from patients with active or inactive uveitis and healthy controls.
Methods
Protein concentration and single cell gene expression were assessed in aqueous fluid biopsies and plasma samples from deidentified patients with uveitis or healthy controls.
Main Outcome Measures
The concentration of 31 inflammatory proteins was measured in all aqueous samples, as well as plasma samples from patients with active uveitis. Chemokine and cytokine ligand and receptor expression were assessed in individual cell types from aqueous biopsies obtained from patients with active uveitis.
Results
We identified 6 chemokines that were both elevated in active uveitis compared with controls and enriched in aqueous compared with plasma during active uveitis (C-C motif chemokine ligand [CCL]2, C-X-C motif chemokine ligand [CXCL]10, CXCL9, CXCL8, CCL3, and CCL14), forming potential gradients for migration of immune cells from the blood to the eye. Of these, CCL2 and CXCL10 were consistently enriched in the aqueous of all patients in our cohort, as well as in a larger cohort of patients from a previously published study. These data suggest that CCL2 and CXCL10 are key mediators in immune cell migration to the eye during uveitis. Next, single cell RNA sequencing suggested that macrophages contribute to aqueous enrichment of CCL2 and CXCL10 during human uveitis. Finally, using chemokine ligand and receptor expression mapping, we identified a broad signaling network for macrophage-derived CCL2 and CXCL10 in human uveitis.
Conclusions
These data suggest that ocular macrophages may play a central role, via CCL2 and CXCL10 production, in recruiting inflammatory cells to the eye in patients with uveitis.
Financial Disclosure(s)
Proprietary or commercial disclosure may be found in the Footnotes and Disclosures at the end of this article.
Keywords: Chemokine, Cytokine, Macrophage, Single cell RNA sequencing, Uveitis
Uveitis is a heterogeneous group of inflammatory eye diseases for which targeted immune therapies fail in 30% to 50% of patients.1, 2, 3 Multiple studies have been undertaken to quantify specific inflammatory molecules present in the eye during uveitis, many aimed at characterizing specific disease subtypes. Several molecules have been thus identified and targeted therapeutically with varying success; however, an inflammatory mediator that is common to all forms of eye inflammation has not been identified. Despite clinical differences, a common pathologic feature between all types of uveitis is infiltration of inflammatory cells into the normally immune-privileged eye. Therefore, characterizing the mechanisms by which immune cells enter the eye may reveal broadly applicable therapeutic targets for uveitis.
Cytokines are the signaling molecules that mediate inflammation. Cytokines may be produced locally or systemically and engage receptors on inflammatory cells to induce activation, proliferation, and differentiation. These proteins have cell type-dependent effects and function in specific combinations for either pro- or antiinflammatory responses in specific contexts (e.g., infection, cancer, and others) and during specific stages of an inflammatory response (i.e., induction and resolution). A related group of proteins, called chemokines, or chemotactic cytokines, are produced locally and diffuse from their source to generate molecular gradients. These gradients function during both inflammation and homeostasis to recruit immune cells from the circulation into a tissue.
Previous analyses of the cytokines present in ocular fluid during uveitis revealed a number of proteins at elevated levels compared with nonuveitic eyes. These include interferon-gamma (IFNγ), tumor necrosis factor-alpha (TNFα), and interleukin-6 (IL-6), as well as IL-17, IL-15, IL-10, and IL-2. Despite their abundance in eyes with uveitis, targeted therapy using inhibitors to cytokines such as TNFα, IL-6, and IL-17 have been successful for only a portion of patients.1,3, 4, 5, 6, 7, 8, 9 One possible interpretation of these outcomes are that the contribution of the targeted cytokine to disease pathogenesis can vary with disease subtype,10 chronicity, or severity5,11, an interpretation that is consistent with the context-dependent nature of cytokine signaling networks.12
In contrast to the highly context-dependent nature of cytokines, chemokine signals operate by more conserved mechanisms to direct the trafficking of immune cells13,14 and therefore may represent more broadly applicable therapeutic targets. Prior studies have compared the concentration of chemokines in the ocular compartment and blood; however, 2 chemokines, C-C motif chemokine ligand (CCL)2 (monocyte chemoattractant protein-1) and C-X-C motif chemokine ligand (CXCL)10 (interferon gamma-induced protein 10), have been measured higher in the eye compared with the peripheral blood during human uveitis.6,15 In line with these findings, mechanistic studies in murine uveitis suggest a fundamental role for CCL2 in driving ocular inflammation.16
Drawing on these studies, we hypothesized that local enrichment of chemokines in the eye compared with peripheral blood may be a shared feature of uveitis with a key role in recruiting immune cells during ocular inflammation. To test this, we compared ocular and peripheral blood protein levels in patients with uveitis to identify chemokines with ocular concentrations exceeding those in the blood. We then performed single cell transcriptomic analysis to identify both the producing and responding cell types for these chemokine gradients.
Methods
Patient Classification and Sample Isolation
Patients were enrolled after providing signed informed consent in accordance with the tenets of the Declaration of Helsinki and the institutional review board of Washington University in St. Louis. Aqueous fluid and blood samples were collected in clinic from patients with active uveitis defined according to the Standardization of Uveitis Nomenclature criteria as ≥ 1+ anterior chamber (AC) cell (> 6 cells/high-powered field), and aqueous fluid was collected intraoperatively during cataract surgery for patients with low aqueous cellularity (i.e., inactive uveitis) as well as for healthy controls, and stored as previously described.17 In the active uveitis group, 1 patient with human leukocyte antigen-B27 disease was sampled twice during subsequent disease flares. One patient with idiopathic uveitis was sampled during both disease activity and inactivity and coded once in each group. In the inactive uveitis group, 1 patient with human leukocyte antigen-B27 disease had both eyes sampled during disease quiescence.
Luminex of Aqueous Fluid and Plasma
The aqueous samples were centrifuged at 400 × g for 5 minutes to remove cells, and the fluid supernatant was frozen at –80°C. Plasma was isolated from blood samples collected by venipuncture into ethylenediaminetetraacetic acid-coated tubes and stored at –80°C. A Luminex FLEXMap 3D analyzer (APX1342 Luminex Corp), Milliplex Map Kit Human Cytokine/Chemokine Magnetic Bead Panel (Sigma-Aldrich), and 96-well Plate Assay (HCYTOMAG-60K-25, HCYP3MAG-63K-06) were used in this study. The approximate minimum detection level is indicated with a dashed line.
Animals and Experimental Uveitis Induction
The animal study protocol was approved by the Animal Care and Use Committee of the University of Washington (animal study protocol # 4481-02) and was compliant with the Association for Research in Vision and Ophthalmology Statement for the Use of Animals in Ophthalmic and Vision Research. Male and female C57BL/6J mice weighing ≥ 18 g and between the ages of 6 and 10 weeks at the time of uveitis initiation were used in all experiments. Unprimed mycobacterial uveitis was induced and uveitis severity scored as previously described. On day 0, the right eye of each animal receives an intravitreal injection of 5 μg of a suspension of killed Mycobacterium tuberculosis H37Ra antigen in 1 to 1.5 μl of phosphate buffered saline.
Murine Cytokine Analysis
As previously described,18 aqueous fluid was collected in ethylenediaminetetraacetic acid-containing capillary tubes (Sarstedt) after corneal paracentesis with a 30-gauge needle (Becton, Dickinson and Company). A volume of 1 to 5 μl of aqueous was collected from each eye, and stored at –80°C in combination with 1X protease inhibitor (Sigma-Aldrich Corp) until assayed. Aqueous protein was quantified using Pierce 660 nm Protein Assay Reagent (Thermo Scientific) for colorimetric detection on the Nanodrop ND-1000 spectrophotometer (Thermo Scientific).
After aqueous collection, the eye was enucleated and frozen on dry ice. The frozen eye was bisected at the limbus. Serum from all animals was collected by cardiac puncture immediately after death. Serum samples were not pooled. The concentration of 32 cytokines was determined using the MilliplexMAP mouse cytokine/chemokine premixed 32 plex immunology multiplex assay (EMD MilliporeCorp). The cytokines measured were as follows: Eotaxin, CSF1/2/3, IFNγ, IL-1A, IL-1B, IL-2, IL-3, IL-4, IL-5, IL-6, IL-7, IL-9, IL-10, IL-12B, IL-12, IL-13, IL-15, IL-17, CXCL10, CXCL1, LIF, CXCL5, CCL2, CXCL9, CCL3, CCL4, CXCL2, VEGF, TNFα, and CCL5. Samples were analyzed using the MAGPIX system (Luminex) with xPonent software version 4.2 (EMD Millipore). Data analysis was performed using Milliplex Analyst Standard version 5.1 software (EMD Millipore).
Single Cell RNA Sequencing
We performed single cell RNA sequencing (scRNA-Seq) of aqueous fluid immune cells using the 10x Genomics platform following the manufacturer’s protocol. Each patient sample was run individually without pooling. We used the Chromium Single Cell 5′ v2 Reagent Kit. We generated an emulsion of individual droplets with each droplet containing a barcoded gel bead and a single cell. We then lysed cells within each droplet and reverse-transcribed RNA to cDNA. We then used recovery agent to break the emulsion, amplified and fragmented the cDNA, and added Illumina adapters. We then sequenced the samples on the Illumina NovaSeq 6000 platform at the McDonnell Genome Institute at Washington University School of Medicine in St. Louis.
Single Cell RNA Sequencing Analysis
Import, Quality Control, and Cell Type Annotation of Data Set
We processed the raw FASTQ sequencing files using CellRanger with alignment to the 10x Genomics human reference genome (refdata-gex-GRCh38-2020-A). We imported the filtered count matrices into Seurat v4,19 and assigned each cell a unique identifier to prevent overlap of barcodes between different samples. Using default parameters, we normalized, log-transformed, and scaled the count matrices to remove unwanted sources of variation such as discrepancies in sequencing depth. We identified the top 2000 highly variable genes for principal component analysis using default parameters. We integrated across samples to account for any sample- or experiment-specific batch effects using the Harmony package. We utilized these harmony embeddings to run Uniform Manifold Approximation and Projection dimensional reduction to 2 dimensions. We clustered cells according to the Louvain algorithm and identified marker genes for each cluster using Wilcoxon rank sum tests comparing each cluster to all other clusters. Marker genes for each cell type are provided as Table S1 (available at www.ophthalmologyscience.org).
RNA-Protein Correlation Analysis
We pseudobulked gene expression by patient for each cell type. Then, we exported the normalized gene expression from the “data” slot. We logarithmized the protein concentrations of the aqueous fluid before running simple linear regression between gene expression and protein abundances.
Ligand-Receptor Analysis
We used the package SingleCellSignalR on our pseudobulked scRNA-Seq data set with the function cell_signaling() and the following parameters: int.type = “paracrine,” s.score = 0.60, and tol = 0.05. We excluded any interactions involving sparsely detected cell types (plasmablasts and plasmacytoid dendritic cells [pDCs]) and proliferating cells. Graphs of intercellular networks were generated using Cytoscape.
Statistics
We describe the statistical analyses used for scRNA-Seq in prior sections. For all other data, we performed statistical analyses using GraphPad Prism 9. We first assessed the normality of our data graphically and by using a Kolmogorov–Smirnov test, using nonparametric alternatives when appropriate. When comparing a single variable between 2 different groups, we used 2-tailed t tests or 2-tailed Mann–Whitney tests. For other analyses, we assessed statistical significance using the appropriate parametric or nonparametric test, as indicated in figure legends. A P value < 0.05 was considered statistically significant.
Data Availability
Genomic data have been deposited in the Gene Expression Omnibus under the following accession number: GSE229166 (scRNA-Seq).
Study Approval
This study was approved by the Human Research Protection Office of Washington University School of Medicine in St. Louis. We obtained written informed consent from all subjects before enrollment in the study.
Results
Profiling of Aqueous Fluid Proteins in Uveitis
We measured 31 proteins in aqueous fluid samples taken from patients with active uveitis disease (n = 26), as well as in samples from patients in the quiescent phase of uveitis without active inflammation (n = 17), and healthy controls without history of uveitis (n = 3) (Table 2). We identified 21 proteins that were elevated in samples from patients with active uveitis compared with inactive disease and healthy controls (Fig S1, available at www.ophthalmologyscience.org). These included the proinflammatory cytokines IL-6 and TNFα (Fig S1). Importantly, none of the cytokines and chemokines we measured in the aqueous fluid were significantly different between healthy patients and uveitis patients with inactive disease (Fig S1), though sample size may be limiting the power to detect any minor differences.
Table 2.
Patient Demographics and Clinical Characteristics
| Active Uveitis (n = 26 Patients) | Quiescent Uveitis (n = 17 Patients) | Healthy Controls (n = 3 Patients) | |
|---|---|---|---|
| Mean age ± standard deviation | 47.5 ± 19.1 | 58.3 ± 13.8 | 59 ± 23 |
| Females/males (ratio) | 19/6 (3.3) | 7/8 (0.9) | 3:0 |
| Disease associations (n) | Idiopathic (15) HLA-B27 (4) Birdshot chorioretinitis (2) Vogt-Koyanagi-Harada (2) Juvenile idiopathic arthritis (1) Ulcerative colitis (1) |
Idiopathic (8) Birdshot chorioretinitis (2) HLA-B27 (1) Sarcoidosis (1) Herpetic (2) Fungal (1) Traumatic iritis (1) |
Cataract (3) |
HLA = human leukocyte antigen.
Aqueous versus Plasma Protein Enrichment in Uveitis
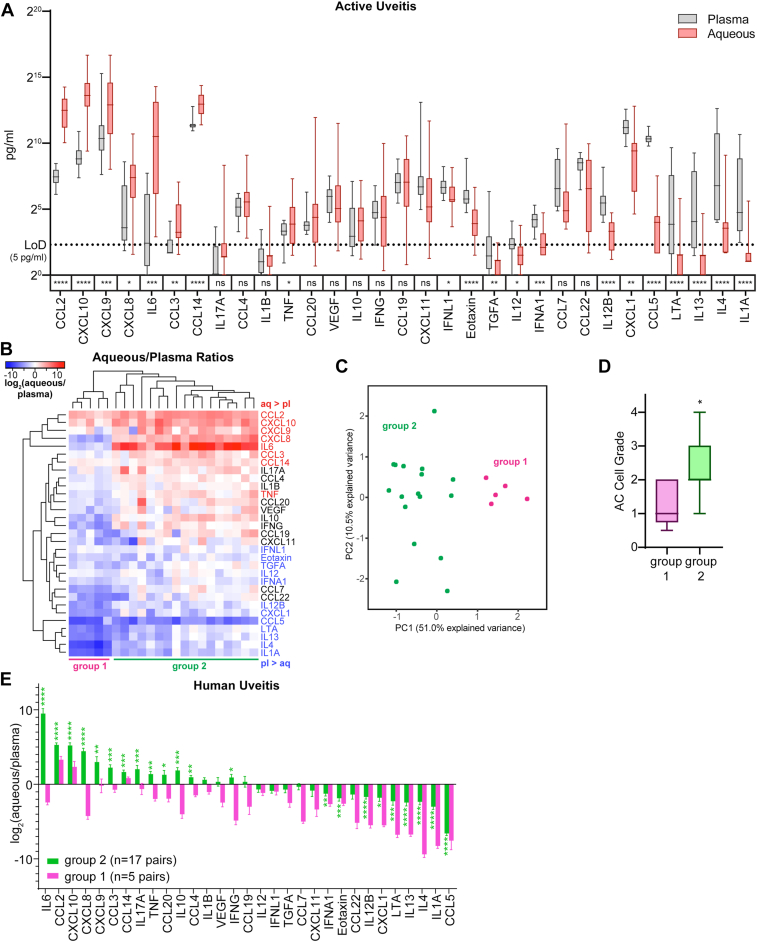
Using paired-sample analysis of aqueous and plasma proteins (n = 22 patients with active uveitis), we identified 8 proteins that demonstrated significant ocular enrichment in actively inflamed eyes including 6 chemokines (CCL2, CXCL10, CXCL9, CXCL8, CCL3, and CCL14) and 2 cytokines (IL-6 and TNFα) (Fig 2A). On a per patient basis, the chemokines CCL2 and CXCL10 were aqueous-enriched in all 22 patients with active uveitis, suggesting these chemokines may be a conserved signal that recruits inflammatory cells to the eye during active phase of disease (Fig 2B). Unsupervised clustering of the aqueous/plasma ratio for each cytokine (Fig 2B) as well as principal component analysis (Fig 2B, C) identified 2 groups of patients based on aqueous/plasma ratios. Group 1 patients were more likely to have panuveitis and to be on systemic immunosuppression, whereas group 2 were more likely to have either anterior or anterior-intermediate uveitis and to be untreated along with high IL-6 enrichment in the aqueous (Table S3, available at www.ophthalmologyscience.org and Fig 2B). Not surprisingly, group 2 patients exhibited significantly higher AC cell grades than group 1 patients (Fig 2D) and had more cytokines and chemokines with increased aqueous/plasma ratios (Fig 2E). Although most molecules only demonstrated aqueous enrichment in the more inflamed group 2 patients, CCL2 and CXCL10 were elevated in the group 1 patients as well (Fig 2E), suggesting they play conserved roles in recruiting immune cells to the eye during uveitis.
Figure 2.
CCL2 and CXCL10 are among the most aqueous-enriched proteins during active human uveitis. A, Paired measurements of chemokines and cytokines in aqueous fluid and plasma from n = 22 patients with active uveitis. We assessed for enrichment in either tissue using Wilcoxon matched-pairs signed rank test. The limit of detection (LoD) was protein- and assay-dependent but was approximately 5 pg/ml. Protein concentrations are plotted on a log scale. B, Heatmap showing the logarithmized ratios for each protein measured in aqueous to that measured in the plasma. Protein names are colored based on whether they were more abundant in aqueous than plasma (red) or more abundant in plasma than aqueous (blue). C, Principal component analysis plot of aqueous/plasma protein ratios. D, Comparison of anterior chamber cell grades between groups 1 and 2. Statistical significance was assessed using Mann–Whitney test. E, Aqueous-to-plasma protein ratios in our cohort of human uveitis patient separated by group. We assessed for enrichment in either tissue using Wilcoxon matched-pairs signed rank test. Statistical significance is indicated as follows: ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001; and ∗∗∗∗P < 0.0001. AC = anterior chamber; CCL = C-C motif chemokine ligand; CXCL = C-X-C motif chemokine ligand; IFNA = interferon-alpha; IFNG = interferon-gamma; IFNL = interferon-lambda; IL = interleukin; LTA = lymphotoxin-alpha; TGFA = transforming growth factor-alpha; TNF = tumor necrosis factor.
We next asked whether CCL2 and CXCL10 were enriched in other cases of uveitis. To do this, we analyzed the aqueous/plasma concentration ratios in a separate cohort of patients with uveitis from a previously published manuscript by Errera et al7 (Fig S3A, available at www.ophthalmologyscience.org). Consistent with our cohort, CCL2 and CXCL10 were also enriched in the aqueous compared with serum in the Errera cohort (Fig S3A). Interleukin-6 and CXCL8 were highly enriched in patients with high cellularity in our cohort and also aqueous-enriched in the Errera cohort (Fig S3A). Because the Errera cohort included aqueous and serum data for healthy patients, we further assessed whether these aqueous- or blood-enrichment patterns were specific to disease. Notably, CCL2 was also enriched, albeit much lower, in the aqueous of healthy patients, suggesting there may be a role for CCL2 during immune surveillance of the eye (Fig S3A).
To evaluate whether this phenomenon is relevant to murine uveitis, we analyzed the aqueous/serum chemokine ratios in murine unprimed mycobacterial uveitis, a model in which mycobacterial antigen is injected intraocularly in the absence of systemic inflammatory stimuli. We found that CCL2 and CXCL10 trended toward being enriched in the aqueous versus serum (Fig S3B, C, available at www.ophthalmologyscience.org), suggesting that these chemokines may form a conserved gradient from the blood to the eye during ocular inflammation.
Expression of Chemokines by Aqueous Immune Cells
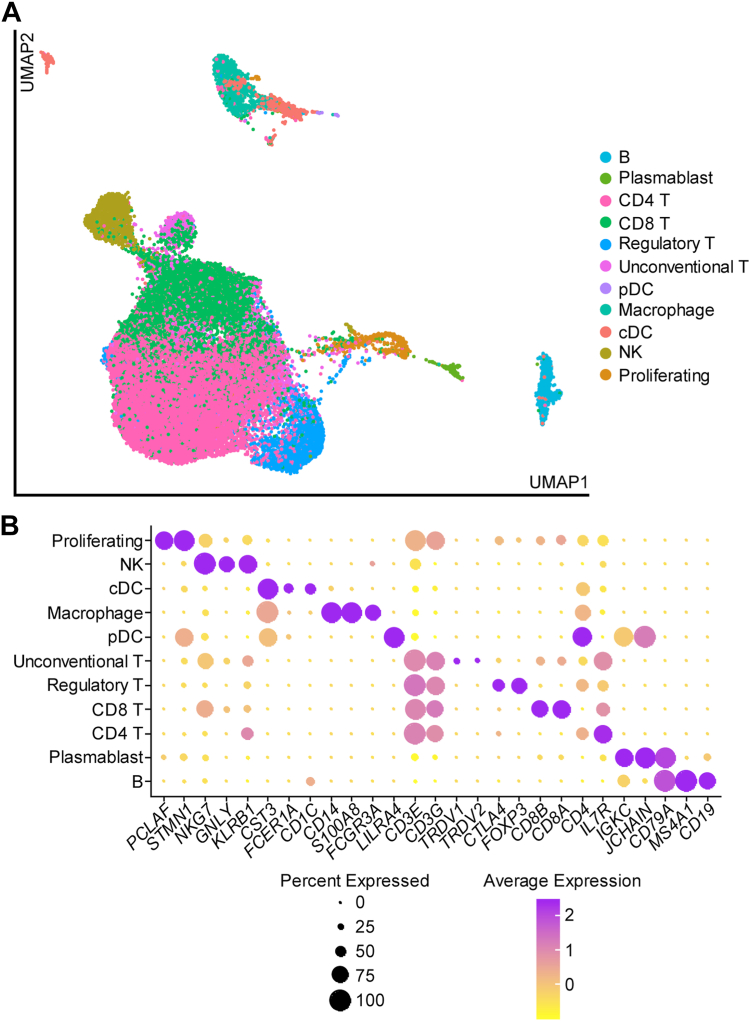
Given the significant association between enrichment of specific chemokines and cytokines in the aqueous and aqueous cellularity (Fig 2D), we hypothesized that these proteins were produced by immune cells localized to the AC. To test this, we profiled cells in the aqueous fluid of patients with uveitis (n = 23) using scRNA-Seq. In total, we analyzed transcriptomes for 37 672 cells (Fig 4A). These included: B cells, plasmablasts, CD4 T cells, CD8 T cells, regulatory T cells, unconventional T cells, pDCs, macrophages, conventional dendritic cells, natural killer cells, and proliferating cells (Fig 4A, B). Because of the heterogeneous nature of human uveitis, the distribution of cell types was highly variable between patients (Table 4). Cell populations that were rare with a median of < 5 cells per patient were plasmablasts and pDCs (Table 4).
Figure 4.
Transcript-based classification of aqueous immune cells in active human uveitis. A, UMAP plot showing the cell type heterogeneity of aqueous immune cells in uveitis. B, Dot plot showing marker gene expression for each named cell type. cDC = conventional dendritic cell; NK = natural killer; pDC = plasmacytoid dendritic cell; UMAP = uniform manifold aproximation and projection.
Table 4.
Frequencies of Aqueous Singlet Cells for Gene Expression Analysis
| Frequency |
Relative Frequency |
|||||
|---|---|---|---|---|---|---|
| Median | Min | Max | Median (%) | Min (%) | Max (%) | |
| B | 12 | 0 | 277 | 0.7 | 0.0 | 24.0 |
| Plasmablast | 2 | 0 | 115 | 0.1 | 0.0 | 12.5 |
| CD4 T | 561 | 17 | 3208 | 37.8 | 12.5 | 68.2 |
| CD8 T | 359 | 13 | 1067 | 22.0 | 7.8 | 56.0 |
| Regulatory T | 44 | 2 | 1190 | 4.1 | 0.6 | 20.5 |
| Unconventional T | 66 | 0 | 394 | 5.0 | 0.0 | 15.9 |
| pDC | 0 | 0 | 12 | 0.0 | 0.0 | 0.5 |
| Macrophage | 40 | 0 | 189 | 2.7 | 0.0 | 50.7 |
| cDC | 26 | 3 | 213 | 2.3 | 0.2 | 13.6 |
| NK | 34 | 1 | 397 | 3.7 | 0.3 | 22.0 |
| Proliferating | 25 | 0 | 91 | 1.9 | 0.0 | 6.5 |
cDC = conventional dendritic cell; NK = natural killer; pDC = plasmacytoid dendritic cell.
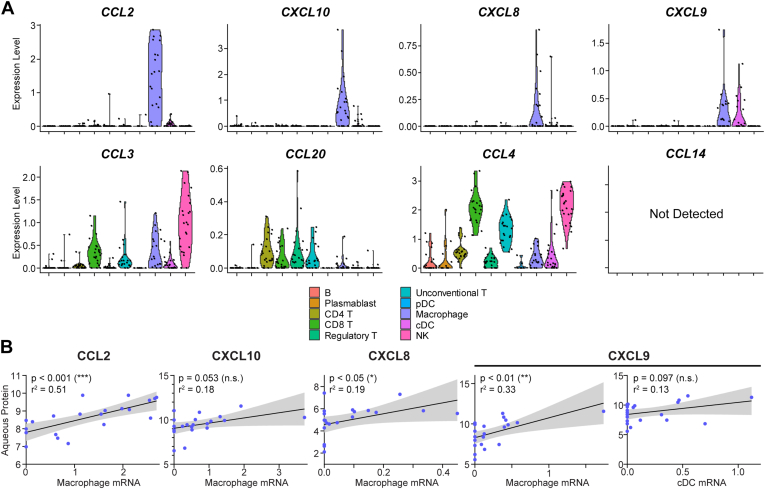
We first assessed the gene expression for the 8 chemokines that were aqueous-enriched in group 2 of our cohort: CCL2, CXCL10, CXCL8, CXCL9, CCL3, CCL14, CCL20, and CCL4 (Fig 2E). CCL14 was not detected in the scRNA-Seq data (Fig 5A), suggesting that CCL14 may be produced by ocular cells present in the tissue stroma rather than the aqueous we sampled. The remaining 7 chemokines were expressed either broadly by several aqueous immune cell populations or had cell type-specific expression restricted to 1 to 2 populations (Fig 5A). Chemokines with cell type-specific expression were CCL2 (macrophages), CXCL10 (macrophages), CXCL8 (macrophages), and CXCL9 (macrophages and conventional dendritic cells) (Fig 5A). Broadly expressed aqueous-enriched chemokines were CCL3, CCL20, and CCL4 (Fig 5A).
Figure 5.
Macrophages express aqueous-enriched chemokines, including CCL2 and CXCL10. A, Violin plots mRNA expression of chemokines by aqueous immune cells. Gene expression measured by single cell RNA sequencing was pseudobulked by patient, and each dot represents a single patient. B, Scatter plots comparing mRNA expression by cell type indicated in parentheses versus protein abundance in aqueous fluid. Transcript expression was pseudobulked by patient, and protein concentrations were logarithmized. Each point represents a single patient. We assessed for correlation using linear regression and plotted the 95% confidence interval. Statistical significance is indicated as follows: ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001. CCl = C-C motif chemokine ligand; cDC = conventional dendritic cell; CXCL = C-X-C motif chemokine ligand; NK = natural killer; pDC = plasmacytoid dendritic cell.
To test whether gene expression of chemokines by aqueous immune cells was correlated with protein abundance, we performed linear regression comparing aqueous protein concentration with mean RNA expression in specific cell types for the 4 chemokines that demonstrated cell type-specific expression. Aqueous protein concentrations of CCL2, CXCL8, and CXCL9 exhibited statistically significant correlations with macrophage mRNA expression (Fig 5B), suggesting a central role for macrophages in the ocular enrichment of chemokines in uveitis.
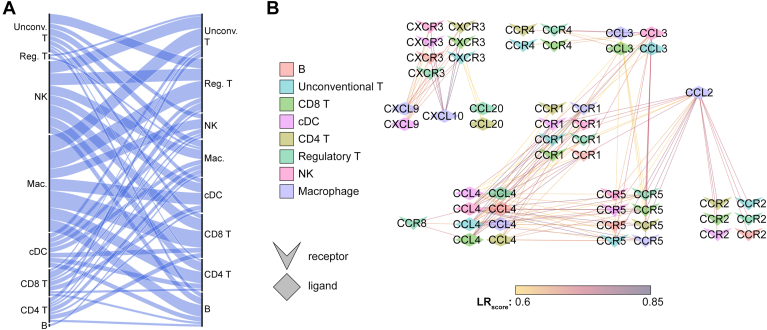
To assess for the potential for intercellular chemokine signaling, we performed ligand-receptor scoring of the scRNA-Seq data using the tool SingleCellSignalR.20 This computes LRscore (ranging from 0 to 1) for known ligand-receptor pairs from the expression of ligand gene in one cell population and the expression of its cognate receptor gene in another cell population. Ligand-receptor pairs that have high expression in 2 distinct cell populations will have high LRscore, suggesting that there may be paracrine signaling between these 2 cell types. We excluded any interactions involving sparsely detected cell types (plasmablasts and pDCs) and also excluded proliferating cells. We assessed potential cell–cell interactions involving any of the 8 aqueous-enriched chemokines: CCL2, CXCL10, CXCL8, CXCL9, CCL3, CCL14, CCL20, and CCL4. Consistent with macrophage-specific expression for many of these proteins (Fig 5A), macrophages appeared to play a central role in this intraocular chemokine signaling network (Fig 6A). We identified potential signaling between macrophage-expressed CCL2 and the receptors CCR1, CCR2, and CCR5 expressed by many immune cells in the aqueous fluid (Fig 6B) and similarly between macrophage CXCL10 and CXCL9 and multiple aqueous immune cells via the receptor CXCR3 (Fig 6B). Taken together, these analyses suggests that macrophages play a key role in recruiting inflammatory cells to the eye during human uveitis by expressing ocular-enriched chemokines.
Figure 6.
Macrophage-expressed chemokines can signal to all aqueous immune cell types. Potential ligand-receptor pairs were identified based on cell type-specific gene expression using SingleCellSignalR. A, Alluvial diagram showing high number of predicted interactions (wide bands) between macrophage-expressed ligands and receptors expressed by other cells. Cell types expressing ligand genes are on the left and cell types expressing receptor genes are on the right. The thickness of the connecting bands indicates the number of ligand–receptor interactions between the 2 connected cell types. B, Network plot showing strong predicted ligand–receptor interaction (LR score, line color) between macrophage ligands (purple diamonds) and receptors (chevrons) expressed by other aqueous cell types. Cell types are indicated by the color of the icon. Chevron icons indicate receptor genes and diamond icons indicate ligand genes. CCL = C-C motif chemokine ligand; CCR = C-C motif chemokine receptor; cDC = conventional dendritic cell; CXCL = C-X-C motif chemokine ligand; NK = natural killer; pDC = plasmacytoid dendritic cell.
Expression of Cytokines by Aqueous Immune Cells
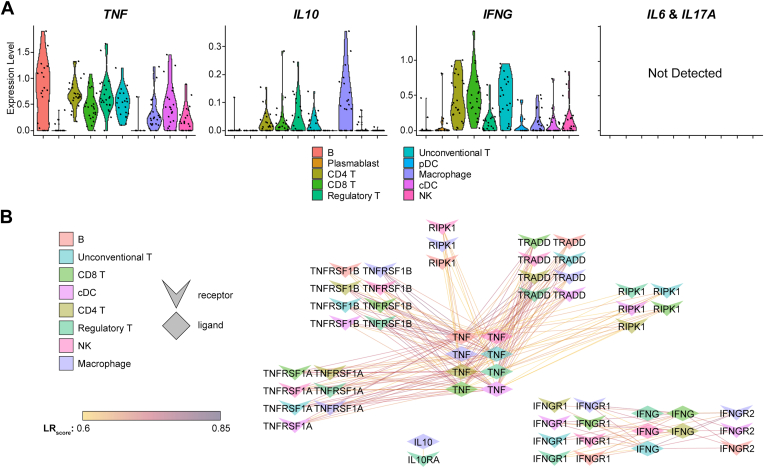
Next, we assessed which of the 5 aqueous-enriched cytokines were expressed at the mRNA level by aqueous immune cells (Fig 7A). IL6 and IL17A were not detected in the scRNA-Seq data set (Fig 7A), suggesting that these transcripts for these cytokines may be low in aqueous immune cells, or they may be expressed by other cell types in the ocular environment. The remaining 3 cytokines—TNF, IL10, and IFNG—were expressed broadly by aqueous immune cell types (Fig 7A).
Figure 7.
Aqueous-enriched cytokines and their receptors are broadly expressed. A, Violin plots mRNA expression of cytokines by aqueous immune cells. Gene expression was pseudobulked by patient, and each dot represents a single patient. Potential ligand-receptor pairs were identified based on cell type-specific gene expression using SingleCellSignalR. B, Network plot showing cytokine signaling among immune cells of the aqueous fluid. Cell types are indicated by the color of the icon. Chevron icons indicate receptor genes and diamond icons indicate ligand genes. cDC = conventional dendritic cell; IL = interleukin; NK = natural killer; pDC = plasmacytoid dendritic cell; TNF = tumor necrosis factor.
We next assessed for potential cytokine signaling interactions between aqueous immune cells involving these aqueous-enriched cytokines (Fig 7B). TNF was expressed by nearly all immune cell populations (Fig 7A) and was predicted to signal to a multitude of immune cell populations via TRADD, TNFRSF1A, TNFRSF1B, and RIPK1 (Fig 7B). The abundance of TNF interactions is consistent with the central role for TNFα in ocular inflammation21,22 and the efficacy of TNFα inhibitors to treat uveitis in some patients.1 IFNG was expressed by several cell populations and predicted to interact with IFNGR1 and IFNGR2 on multiple cell types (Fig 7B), suggesting a role for Th1-type inflammation in human uveitis.
Discussion
Uveitis is clinically heterogeneous, and patients with similar presentations have variable severity, chronicity, and sequelae from ocular inflammation. Paralleling this clinical heterogeneity, the cytokine composition of ocular fluid during uveitis also varies between etiologies and by disease severity. Not surprising, therapies that target individual cytokines in uveitis are also variably effective and a therapeutic gap remains for 30% to 50% of patients with uveitis.
We hypothesized that, despite the heterogeneity inherent in uveitis, there may exist conserved mechanisms governing the recruitment of inflammatory cells to this organ. To identify chemokines with the potential to recruit immune cells to the eye during uveitis, we tested whether there was local ocular enrichment of chemokines compared with the peripheral blood. Our results confirmed previous reports that CCL2 and CXCL10 are enriched in aqueous6 or vitreous15 during uveitis; in fact, these molecules were enriched in the eyes of every patient in our small cohort regardless of the etiology, disease severity, or treatment, and were also trended toward aqueous enrichment in a murine model of uveitis induced by intraocular mycobacterial extract. If validated in a larger cohort of patients, our data suggest that CCL2 and CXCL10 form a conserved gradient during uveitis.
Previous studies identified significant heterogeneity in the concentration of various aqueous inflammatory proteins. Although our study was not powered to discover disease-specific cytokine patterns, we did note a correlation between the aqueous cellularity and the ocular enrichment of several chemokines, including CXCL8, CXCL9, CCL3, CCL4, CCL14, and CCL20. Similarly, Bonacini et al6 found that multiple proteins, including IL-6, CXCL8, CXCL10, TNFα, and IFNγ, positively correlated with total leukocyte count, and El-Asrar et al11 found a correlation between disease activity and multiple chemokines including CXCL1, CXCL9, and CXCL10. In agreement with these human observations, Pepple et al23 found that multiple aqueous proteins, including CXCL10, reflected disease severity in 2 rat models of uveitis. These correlations suggest that either immune cells infiltrate the eye in proportion to the concentration of aqueous cytokine, or that the infiltrating immune cells are producing the cytokines and raise the possibility that controlling for severity/aqueous cellularity may improve the disease-specificity of certain cytokines, as demonstrated by studies lead by Ahn et al10 and El-Asrar et al.5
To test whether ocular immune cells express locally enriched cytokines/chemokines, we performed scRNA-Seq. We found that several chemokines exhibited macrophage-specific expression including CCL2, CXCL10, CXCL8, and CXCL9. Furthermore, we found that for CCL2, CXCL8, and CXCL9, the average mRNA expression in macrophages correlated with the total aqueous protein concentration across the patients in our cohort. This scRNA-Seq analysis suggests that macrophages play a central role in the production of ocular-enriched chemokines in uveitis.
Finally, we looked for potential intercellular chemokine networks using ligand-receptor analysis of scRNA-Seq data. In line with the finding that macrophages preferentially expressed the chemokines in our study with the greatest eye-blood gradients, we identified many putative interactions between chemokine-expressing macrophages and receptor-expressing immune cells. This analysis implicates a central role for aqueous macrophages in recruiting nearly every other ocular immune cell type to the inflamed eye during uveitis. Our results are consistent with previous findings that macrophages are among the first cells to recruited early during the ocular inflammatory response.24,25
These findings on ocular chemokine enrichment in uveitis are complementary to existing literature. C-C motif chemokine ligand 2 (also known as monocyte chemoattractant protein-1) is a central chemokine in many immune responses and can be secreted by multiple cell types including macrophages as well as endothelial cells and fibroblasts. C-C motif chemokine ligand 2 is best known for its role in recruiting monocytes and macrophages early in an immune response and has been implicated in inflammatory diseases of multiple organs including rheumatoid arthritis26 and multiple sclerosis.27 Elevated ocular levels of CCL2 relative to blood have been noted in human uveitis,15,28 and blockade of CCL2 signaling prevents immune cell infiltration in several murine models of uveitis.29,30 Previous studies have also demonstrated a role for CCL2 in endophthalmitis.31 C-X-C motif chemokine ligand 10 (also known interferon gamma-induced protein 10) is another key chemokine in inflammatory cell recruitment in diverse inflammatory diseases including viral infections and cancer. Ocular enrichment of CXCL10 has been noted in human uveitis subtypes including birdshot chorioretinitis28 and sarcoiditis,15 and was a shared feature of 2 rodent uveitis models, experimental autoimmune uveitis and primed mycobacterial uveitis.23 C-X-C motif chemokine ligand 10 has also been shown to play an important role in the ocular response to herpes simplex virus infection.32,33 Though CCL2 and CXCL10 have been previously implicated in uveitis before, our study highlights that these 2 chemokines may be conserved ocular inflammatory signals and implicates macrophages as a key cellular source of these chemotactic gradients in uveitis.
C-C motif chemokine ligand 2 was also enriched, albeit at lower levels, in the nonuveitic aqueous, suggesting that a CCL2 chemokine gradient precedes ocular inflammation in uveitis. Recently, a single nucleus RNA-Seq atlas of the anterior segment was generated which reveals that the healthy eye contains lymphocytes in addition to the macrophages revealed by previous histologic analysis.34 Exploration of this data set (data not shown) revealed that CCL2 was expressed by multiple epithelial cell types in the healthy eye, including ciliary body pigmented epithelial cells. The receptors for CCL2, primarily CCR2 (but also CCR5 and CCR1), were expressed by lymphocytes and macrophages in this study of normal noninflamed eyes. This suggests that, in addition to driving the influx of immune cells during uveitis, CCL2 may drive immune surveillance by macrophages and lymphocytes in the anterior segment.
Limitations
One limitation of the current study is that we had few (n = 3) aqueous samples from healthy patients. We endeavored to expand the generalizability of the findings in our small cohort by comparing with aqueous/blood ratios derived from a published data set which included a larger cohort of healthy patient samples.7 Although CCL2, CXCL10, CXCL8, and IL-6 were similarly enriched in the aqueous in both cohorts, several aqueous-enriched molecules in our cohort were not enriched in the aqueous in the Errera cohort. Most notably, IL-17, TNFα, IL-1B, and IFNγ were enriched in the aqueous in our cohort, but present at higher levels in the blood in the Errera cohort. This discrepancy could be related to the predominance of AC-involving disease in our cohort (our inclusion criteria included > 1+ AC cell) versus a predominance of posterior uveitis in the Errera cohort; inclusion of a large number infectious uveitis (toxoplasmosis) samples in the Errera cohort; regional differences in genetic or environmental contributions (France vs. St. Louis, MO); as well as technical differences in sample collection, storage, or protein measurement.
Several chemokines in our analysis did not show a strong correlation between protein levels and cell type-specific mRNA expression. Gene expression of CXCL10 by macrophages along with CCL3 by natural killer cells and CD8 T cells did not strongly correlate with aqueous protein levels (not shown). C-C motif chemokine ligand 3 was present at lower levels compared with CCL2, CXCL8, CXCL9, and CXCL10, suggesting that this analysis may be most robust for highly abundant proteins, or those proteins may be produced by cells in the tissue stroma, which we did not sample. Notably, we found very high levels of IL-6 protein in most aqueous samples yet were unable to detect IL6 mRNA in any cell type. One possibility is that IL6 transcript expression is very low or transient, but that the protein accumulates and is later released by aqueous immune cells. In this case, our mRNA analysis of single cells would not detect mRNA expression of IL6. Another explanation may be that IL-6 is produced primarily by cells within the inflamed ocular stroma which were not captured in our scRNA-Seq analysis. This scenario would be consistent with IL-6 production by other inflamed tissues, as well as the robust detection of ocular and systemic IL-6 described in other ocular diseases which do not feature a significant number of aqueous immune cells.35,36
Our cohort was limited to a relatively small number of patients with heterogeneous clinical disease subtypes and we were not able to identify disease-specific cytokine patterns. Errera et al7 similarly found a lack of correlation with specific uveitis etiologies in their larger analysis. The results of other cytokine analyses have been somewhat heterogenous; however, a few trends emerge, particularly when cytokine levels were controlled for severity of inflammation. For example, Ahn et al10 found higher levels of TNFα and IFNγ in Behcet’s-associated uveitis, as did El-Asrar et al.5 Given the correlation that we and others have found between aqueous cellularity and inflammatory protein levels, future analysis aimed at identifying cytokine patterns associated with specific disease states may be enhanced by considering or controlling for aqueous cellularity or severity of inflammation.
Conclusion
In summary, our results suggest that the chemokines CCL2 and CXCL10 form a conserved gradient in uveitis and implicate macrophages as key in immune cell recruitment during uveitis. These findings suggest that macrophages, and the chemokines they produce, may represent more globally applicable therapeutic targets for patients with uveitis. An important question is whether the CCL2 and CXCL10 signaling networks are important, and targetable, in other diseases that feature ocular inflammation, such as diabetic retinopathy, macular degeneration, and during gene therapy for inherited retinal degeneration.
Acknowledgments
The authors thank patients and their families for participating in their study.
Manuscript no. XOPS-D-23-00136R2.
Footnotes
Supplemental material available atwww.ophthalmologyscience.org.
Disclosure(s):
All authors have completed and submitted the ICMJE disclosures form.
The author(s) have made the following disclosures:
K.L.P.: Meeting and travel support – Genetech; Data safety monitoring board – Janssen.
M.A.P.: Consultant – AbbVie, Priovant Therapeutics, and USB; Patent – T-020486, Quantification of YeiH-specific CD8 T cells in HLA-B∗27 Positive Persons.
G.L.P.: Equipment – Alcon Vision LLC, Genmab US Inc., Allergan, Inc., Johnson & Johnson Surgical Vision, Inc., AbbVie Inc., Oyster Point Pharma, Inc., Sun Pharmaceutical Industries Inc., Sight Sciences Inc.
L.M.H.: Consultant – Eyepoint Pharmaceuticals and Priovant Therapeutics; Honoraria – University of Wisconsin and Flaum Eye Institute; Meeting and travel support – Federation of Clinical Immunology Societies 2023; Other – AbbVie.
The other authors have no proprietary or commercial interest in any materials discussed in this article.
This work was supported by the National Institutes of Health grants K08 EY033045 (L.M.H.), R01 EY019287 (R.S.A.), R01 EY030431 (K.L.P.), and P30 EY02687 (Vision Core Grant), Jeffery T. Fort Innovation Fund (R.S.A.), Starr Foundation AMD Research Fund (R.S.A.), Siteman Retina Research Fund (R.S.A.), Retina Associates Research Fund (R.S.A.), and an unrestricted grant from Research to Prevent Blindness to the John F. Hardesty, MD, Department of Ophthalmology and Visual Sciences at Washington University School of Medicine in St. Louis. J.B.L. was supported by National Institutes of Health grant F30 DK130282 and the Washington University in St. Louis Medical Scientist Training Program (National Institutes of Health grant T32 GM07200), and M.A.P. was supported by the Rheumatology Research Foundation (Scientist Development Award). Research reported in this publication was supported by the Washington University Institute of Clinical and Translational Sciences (National Institutes of Health grant UL1 TR002345). Experimental support was also partially provided by the Bursky Center for Human Immunology and Immunotherapy Programs at Washington University Immunomonitoring Laboratory, in support of the Rheumatic Diseases Core Center (NIH WLC6313040077). The sponsors or funding organizations had no role in the design or conduct of this research.
HUMAN SUBJECTS: Human subjects were included in this study. Patients were enrolled after obtaining signed informed consent in accordance with the tenets of the Declaration of Helsinki and the institutional review board of Washington University in St. Louis.
ANIMAL SUBJECTS: Animal subjects were included in this study. The animal study protocol was approved by the Animal Care and Use Committee of the University of Washington (animal study protocol # 4481-02) and was compliant with the Association for Research in Vision and Ophthalmology Statement for the Use of Animals in Ophthalmic and Vision Research.
Author Contributions:
Conception and design: Lin, Pepple, Concepcion, Hassman
Data collection: Lin, Pepple, Korshunova, M.A. Paley, G.L. Paley, Laurent, Hassman
Analysis and interpretation: Lin, Concepcion, Korshunova, Apte, Hassman
Obtained funding: Hassman
Overall responsibility: Lin, Pepple, Concepcion, Korshunova, Hassman
Supplementary Data
References
- 1.Jaffe G.J., Dick A.D., Brézin A.P., et al. Adalimumab in patients with active noninfectious uveitis. N Engl J Med. 2016;375:932–943. doi: 10.1056/NEJMoa1509852. [DOI] [PubMed] [Google Scholar]
- 2.Ramanan A.V., Dick A.D., Guly C., et al. Tocilizumab in patients with anti-TNF refractory juvenile idiopathic arthritis-associated uveitis (APTITUDE): a multicentre, single-arm, phase 2 trial. Lancet Rheumatol. 2020;2:e135–e141. doi: 10.1016/S2665-9913(20)30008-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dick A.D., Tugal-Tutkun I., Foster S., et al. Secukinumab in the treatment of noninfectious uveitis: results of three randomized, controlled clinical trials. Ophthalmology. 2013;120:777–787. doi: 10.1016/j.ophtha.2012.09.040. [DOI] [PubMed] [Google Scholar]
- 4.Curnow S.J., Falciani F., Durrani O.M., et al. Multiplex bead immunoassay analysis of aqueous humor reveals distinct cytokine profiles in uveitis. Invest Ophthalmol Vis Sci. 2005;46:4251–4259. doi: 10.1167/iovs.05-0444. [DOI] [PubMed] [Google Scholar]
- 5.El-Asrar A.M.A., Struyf S., Kangave D., et al. Cytokine profiles in aqueous humor of patients with different clinical entities of endogenous uveitis. Clin Immunol. 2011;139:177–184. doi: 10.1016/j.clim.2011.01.014. [DOI] [PubMed] [Google Scholar]
- 6.Bonacini M., Soriano A., Cimino L., et al. Cytokine profiling in aqueous humor samples from patients with non-infectious uveitis associated with systemic inflammatory diseases. Front Immunol. 2020;11:358. doi: 10.3389/fimmu.2020.00358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Errera M.H., Pratas A., Fisson S., et al. Cytokines, chemokines and growth factors profile in human aqueous humor in idiopathic uveitis. PLOS ONE. 2022;17 doi: 10.1371/journal.pone.0254972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vegas-Revenga N., Calvo-Río V., Mesquida M., et al. Anti-IL6-receptor tocilizumab in refractory and noninfectious uveitic cystoid macular edema: multicenter study of 25 patients. Am J Ophthalmol. 2019;200:85–94. doi: 10.1016/j.ajo.2018.12.019. [DOI] [PubMed] [Google Scholar]
- 9.Ramanan A.V., Dick A.D., Jones A.P., et al. A phase II trial protocol of tocilizumab in anti-TNF refractory patients with JIA-associated uveitis (the APTITUDE trial) BMC Rheumatol. 2018;2:4. doi: 10.1186/s41927-018-0010-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ahn J.K., Yu H.G., Chung H., Park Y.G. Intraocular cytokine environment in active Behçet uveitis. Am J Ophthalmol. 2006;142:429–434. doi: 10.1016/j.ajo.2006.04.016. [DOI] [PubMed] [Google Scholar]
- 11.El-Asrar A.M.A., Al-Obeidan S.S., Kangave D., et al. CXC chemokine expression profiles in aqueous humor of patients with different clinical entities of endogenous uveitis. Immunobiology. 2011;216:1004–1009. doi: 10.1016/j.imbio.2011.03.007. [DOI] [PubMed] [Google Scholar]
- 12.Moraga I., Spangler J., Mendoza J.L., Garcia K.C. Multifarious determinants of cytokine receptor signaling specificity. Adv Immunol. 2014;121:1–39. doi: 10.1016/B978-0-12-800100-4.00001-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zlotnik A., Yoshie O. The chemokine superfamily revisited. Immunity. 2012;36:705–716. doi: 10.1016/j.immuni.2012.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kunkel E.J., Butcher E.C. Chemokines and the tissue-specific migration of lymphocytes. Immunity. 2002;16:1–4. doi: 10.1016/s1074-7613(01)00261-8. [DOI] [PubMed] [Google Scholar]
- 15.Nagata K., Maruyama K., Uno K., et al. Simultaneous analysis of multiple cytokines in the vitreous of patients with sarcoid uveitis. Invest Ophthalmol Vis Sci. 2012;53:3827–3833. doi: 10.1167/iovs.11-9244. [DOI] [PubMed] [Google Scholar]
- 16.Tuaillon N., Shen D.F., Berger R.B., et al. MCP-1 expression in endotoxin-induced uveitis. Invest Ophthalmol Vis Sci. 2002;43:1493–1498. [PubMed] [Google Scholar]
- 17.Hassman L.M., Paley M.A., Esaulova E., et al. Clinicomolecular identification of conserved and individualized features of granulomatous uveitis. Ophthalmol Sci. 2021;1 doi: 10.1016/j.xops.2021.100010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pepple K.L., John S., Wilson L., et al. Systemic prime exacerbates the ocular immune response to heat-killed Mycobacterium tuberculosis. Exp Eye Res. 2022;223 doi: 10.1016/j.exer.2022.109198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hao Y., Hao S., Andersen-Nissen E., et al. Integrated analysis of multimodal single-cell data. Cell. 2021;184:3573–3587.e29. doi: 10.1016/j.cell.2021.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cabello-Aguilar S., Alame M., Kon-Sun-Tack F., et al. SingleCellSignalR: inference of intercellular networks from single-cell transcriptomics. Nucleic Acids Res. 2020;48:e55. doi: 10.1093/nar/gkaa183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dick A.D., McMenamin P.G., Körner H., et al. Inhibition of tumor necrosis factor activity minimizes target organ damage in experimental autoimmune uveoretinitis despite quantitatively normal activated T cell traffic to the retina. Eur J Immunol. 1996;26:1018–1025. doi: 10.1002/eji.1830260510. [DOI] [PubMed] [Google Scholar]
- 22.Dick A.D., Forrester J.V., Liversidge J., Cope A.P. The role of tumour necrosis factor (TNF-alpha) in experimental autoimmune uveoretinitis (EAU) Prog Retin Eye Res. 2004;23:617–637. doi: 10.1016/j.preteyeres.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 23.Pepple K.L., Rotkis L., Van Grol J., et al. Primed mycobacterial uveitis (PMU): histologic and cytokine characterization of a model of uveitis in rats. Invest Ophthalmol Vis Sci. 2015;56:8438–8448. doi: 10.1167/iovs.15-17523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Forrester J.V., Huitinga I., Lumsden L., Dijkstra C.D. Marrow-derived activated macrophages are required during the effector phase of experimental autoimmune uveoretinitis in rats. Curr Eye Res. 1998;17:426–437. doi: 10.1080/02713689808951224. [DOI] [PubMed] [Google Scholar]
- 25.Epps S.J., Boldison J., Stimpson M.L., et al. Re-programming immunosurveillance in persistent non-infectious ocular inflammation. Prog Retin Eye Res. 2018;65:93–106. doi: 10.1016/j.preteyeres.2018.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moadab F., Khorramdelazad H., Abbasifard M. Role of CCL2/CCR2 axis in the immunopathogenesis of rheumatoid arthritis: latest evidence and therapeutic approaches. Life Sci. 2021;269 doi: 10.1016/j.lfs.2021.119034. [DOI] [PubMed] [Google Scholar]
- 27.Cui L.Y., Chu S.F., Chen N.H. The role of chemokines and chemokine receptors in multiple sclerosis. Int Immunopharmacol. 2020;83 doi: 10.1016/j.intimp.2020.106314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuiper J.J.W., Mutis T., de Jager W., et al. Intraocular interleukin-17 and proinflammatory cytokines in HLA-A29–associated birdshot chorioretinopathy. Am J Ophthalmol. 2011;152:177–182.e1. doi: 10.1016/j.ajo.2011.01.031. [DOI] [PubMed] [Google Scholar]
- 29.London A., Benhar I., Mattapallil M.J., et al. Functional macrophage heterogeneity in a mouse model of autoimmune central nervous system pathology. J Immunol. 2013;190:3570–3578. doi: 10.4049/jimmunol.1202076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen Y.F., Zhou D., Metzger T., et al. Spontaneous development of autoimmune uveitis is CCR2 dependent. Am J Pathol. 2014;184:1695–1705. doi: 10.1016/j.ajpath.2014.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mursalin M.H., Astley R., Coburn P.S., et al. Roles of CCL2 and CCL3 in intraocular inflammation during Bacillus endophthalmitis. Exp Eye Res. 2022;224 doi: 10.1016/j.exer.2022.109213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wickham S., Ash J., Lane T.E., Carr D.J.J. Consequences of CXCL10 and IL-6 induction by the murine IFN-alpha1 transgene in ocular herpes simplex virus type 1 infection. Immunol Res. 2004;30:191–200. doi: 10.1385/IR:30:2:191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mursalin M.H., Coburn P.S., Miller F.C., et al. C-X-C chemokines influence intraocular inflammation during Bacillus endophthalmitis. Invest Ophthalmol Vis Sci. 2021;62:14. doi: 10.1167/iovs.62.14.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Zyl T., Yan W., McAdams A.M., et al. Cell atlas of the human ocular anterior segment: tissue-specific and shared cell types. Proc Natl Acad Sci U S A. 2022;119 doi: 10.1073/pnas.2200914119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kauffmann D.J., van Meurs J.C., Mertens D.A., et al. Cytokines in vitreous humor: interleukin-6 is elevated in proliferative vitreoretinopathy. Invest Ophthalmol Vis Sci. 1994;35:900–906. [PubMed] [Google Scholar]
- 36.Funatsu H., Yamashita H., Ikeda T., et al. Vitreous levels of interleukin-6 and vascular endothelial growth factor are related to diabetic macular edema. Ophthalmology. 2003;110:1690–1696. doi: 10.1016/S0161-6420(03)00568-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Genomic data have been deposited in the Gene Expression Omnibus under the following accession number: GSE229166 (scRNA-Seq).