Abstract
In the present study, we established an in vitro system representing the Burkitt’s lymphoma (BL)-type Epstein-Barr virus (EBV) infection which is characterized by expression of EBV-determined nuclear antigen 1 (EBNA-1) and absence of EBNA-2 and latent membrane protein 1 (LMP1) expression. EBV-negative cell clones isolated from the EBV-positive BL line Akata were infected with an EBV recombinant carrying a selectable marker, and the following selection culture easily yielded EBV-infected clones. EBV-reinfected clones showed BL-type EBV expression and restored the capacity for growth on soft agar and tumorigenicity in SCID mice that were originally retained in parental EBV-positive Akata cells and lost in EBV-negative subclones. Moreover, it was found that EBV-positive cells were more resistant to apoptosis than were EBV-negative cells. EBV-infected cells expressed the bcl-2 protein, through which cells might become resistant to apoptosis, at a higher level than did uninfected cells. This is the first report that BL-type EBV infection confers apoptosis resistance even in the absence of expression of LMP1 and BHRF1, both of which are known to have an antiapoptotic function. Surprisingly, transfection of the EBNA-1 gene into EBV-negative Akata clones could not restore malignant phenotypes and apoptosis resistance, thus suggesting that EBNA-1 alone was not sufficient for conferring them. Our results suggest that the persistence of EBV in BL cells is required for the cells to be more malignant and apoptosis resistant, which underlines the oncogenic role of EBV in BL genesis.
Epstein-Barr virus (EBV) is a B-lymphotropic human herpesvirus and, like other herpesviruses, establishes a lifelong presence in the host. The virus infects the vast majority of the world’s adult population and is well known for its association with a broad spectrum of benign and malignant diseases, including infectious mononucleosis (5, 6), Burkitt’s lymphoma (BL) (2, 4), nasopharyngeal carcinoma (11, 26), and B-cell lymphoma in immunocompromised individuals (reviewed in reference 12). In recent years, there has been increasing evidence of the association of EBV with other human malignancies such as gastric carcinoma (7, 16); however, the role of EBV in tumorigenesis remains to be elucidated. It has been difficult to clarify it because a good tool has not been available until the isolation of EBV-negative Akata cells (17).
A BL cell line, Akata, derived from a Japanese patient, has chromosomal translocation t(8;14) and expresses surface immunoglobulin (Ig) of the Gκ class (21). It is now commonly used as a virus source, a targeting host to generate recombinant EBV, and a tool to study viral replication since cross-linking surface IgG with anti-IgG induces viral replication synchronously and efficiently (22). What is unique in Akata is that it retains the type I latency (14) which expresses EBV-associated nuclear antigen 1 (EBNA-1), EBV-encoded nuclear RNAs (EBERs), and a transcript from the BamHI A region (BARF0) even after long-term culture in vitro. By contrast, most BL cell lines convert to type III latency (14), in which all the viral latent genes, including six EBNAs and three latent membrane proteins (LMPs), are expressed.
It was impossible to isolate EBV-negative cells from originally EBV-positive BL cell lines previously. But Akata is also unique in this respect. The parental Akata cells were virtually 100% positive for EBNA; however, some of them became EBV negative after serial passages. We successfully isolated both EBV-positive and -negative clones from the parental Akata cells by the limiting dilution method. The growth characteristics of these clones were subsequently compared. Although both clones proliferated at nearly the same rate in serum-rich conditions, EBV-negative clones could not grow in low-serum conditions. Furthermore, EBV-positive clones could form colonies on soft agar and tumor masses in nude mice, while EBV-negative clones could not. It was suggested that the malignant phenotype of Akata is dependent on the presence of EBV (17).
In this study, we confirmed this hypothesis more directly by reinfecting EBV-negative Akata clones with EBV. First, we demonstrated that reinfection resulted in type I latency. Second, we verified that in reinfected cells two aspects of the parental phenotype, malignancy and apoptosis resistance, were restored. This is the first report that BL cells with type I latency are resistant to apoptosis. Third, we further investigated whether EBNA-1 was responsible for these two characteristics and found that they were not due to EBNA-1 alone.
MATERIALS AND METHODS
Cell culture.
The Akata cell line of BL origin was maintained in RPMI 1640 medium (Nikken or Sigma) supplemented with 10% fetal bovine serum (FBS) (GIBCO BRL), penicillin (40 U/ml), and streptomycin (50 μg/ml) at 37°C in a 5% CO2 humidified atmosphere.
Plasmids, transfection, and cell cloning.
The neomycin resistance gene (neoR) driven by the simian virus 40 promoter was derived from pcdna3 (Invitrogen). pEBO (10) is a mammalian plasmid vector that carries the simian virus 40 promoter-driven neoR gene, the EBNA-1 gene, and the oriP sequence of B95-8 origin. Transfection-grade plasmid DNA was purified by the CsCl-ethidium bromide method or with a FlexiPrep Kit (Pharmacia). Plasmids were introduced into Akata cells by the electroporation method. First, 5 × 106 cells were suspended in serum-free RPMI 1640, washed twice, and resuspended in 400 μl of ice-cold serum-free RPMI 1640 containing 2 to 10 μg of plasmid DNA in a 2-mm-gap electroporation cuvette (BTX). The electroporation was performed with the Electro Cell Manipulator 600 (BTX) set at 180 V, 1 mF, and R1. After transfection, cells were cultured for 2 days and transfectants were selected in the medium containing 700 μg of G418 (GIBCO BRL) per ml. For cloning cells, 10,000 cells were transferred to a well of a flat-bottomed 96-well multiwell plate (Falcon) with 200 μl of selection medium. Half of the medium was changed every 5 days until colonies emerged. Clones were expanded and maintained in selection medium.
EBV infection.
Preparation of the virus solution was described previously (8, 18, 25). First, 5 × 106 cells were suspended in 2 ml of diluted EBV solution (1:2 to 10) for 60 min at room temperature with continuous mild mixing. Then they were washed three times and cultured for 2 days. Selection and cloning procedures are described above.
Western blot analysis.
Total cell lysate from 5 × 104 cells (corresponding to about 10 μg of protein) was subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Proteins were separated in 8, 10, or 15% polyacrylamide gels and transferred to nitrocellulose membranes (Schleicher & Schuell) by a semidry system (Nihon Eido). Blots were blocked in 5% milk–Tris-buffered saline-Tween. For immunostaining, blots were incubated in a mixture of human sera for EBNAs; S12 monoclonal antibody for LMP1; a mixture of monoclonal antibodies, bcl-2(Ab-1)/100 and bcl-2(Ab-1)/112 for bcl-2; and MAB8188 monoclonal antibody for BHRF1 (Chemicon). The ECL method was used for signal detection according to the manufacturer’s protocol (Amersham).
Reverse transcriptase PCR (RT-PCR).
Total RNA was extracted from Akata cells with Trizol reagent (GIBCO BRL) according to the manufacturer’s instructions. RNA was solubilized in distilled water, treated with DNase I (GIBCO BRL) at 37°C for 15 min, and then incubated at 94°C for 10 min to inactivate the enzyme. For each set of reverse transcription, 2 μg of total RNA was denatured at 94°C for 10 min in the presence of 10 pmol of 3′-specific primer, chilled on ice, and incubated at 37°C for 1 h in the presence of 20 mM Tris HCl (pH 8.3), 75 mM KCl, 3 mM MgCl2, 10 mM dithiothreitol, 0.5 mM deoxynucleoside triphosphate, 10 U of RNasin (Promega), and 200 U of Moloney murine leukemia virus RT (GIBCO BRL) in a total volume of 20 μl. One hundred nanograms of the cDNA was subjected to amplification by PCR. The sequence and the genomic coordinates of oligonucleotides used in the study are listed in Table 1 (1, 3, 19, 23). PCR was carried out in the presence of 10 mM Tris HCl (pH 8.3)–50 mM KCl–1.5 mM MgCl2–0.4 mM deoxynucleoside triphosphate–10 pmol of each primer–5 U of Taq polymerase (Takara) in a total volume of 50 μl. Samples were amplified with a Thermal Cycler 2400 (Perkin-Elmer) for 12 cycles for EBER1 and 35 cycles for the others. Each amplification consisted of denaturation at 94°C for 30 s, annealing at 45 to 50°C for 60 s, and extension at 72°C for 60 s. The amplified products were electrophoresed in 2.0% agarose gels, then vacuum transferred (Pharmacia) to a HyBond N+ nylon membrane (Amersham) under alkaline conditions, and detected by a 3′-end-labeled (Takara) internal probe with the ECL detection system (Amersham).
TABLE 1.
Sequences and coordinates of primers and probes used in this study
| Transcript | Product size (bp) | Type | Sequence (5′ to 3′) | B95-8 genomic coordinates (nucleotides) |
|---|---|---|---|---|
| Qp transcripta | 339 | 5′ primer | AGGCGCGGGATAGCGTGCGCTACCGGA | 62426–62452 |
| 3′ primer | TCCTCGTCCATGGTTATCAC | 108075–108056 | ||
| Probe | AGACCTGGGAGCAGATTCAC | 67608–67627 | ||
| Cp transcripta | 297 | 5′ primer | CACTACAAGACCTACGCCTCTCCATCCATC | 11425–11454 |
| 3′ primer | TCTCCCCTAGGCCCTGAAGGTGAACCGCTT | 14832–14813, 17636–17626 | ||
| Probe | GCGACCGGTGCCTTCTTAGGAGCTGTCCGA | 14708–14737 | ||
| Wp transcripta | 235 | 5′ primer | TCAGAGCGCCAGGAGTCCACACAAAT | 14384–14410 |
| 3′ primer | TCTCCCCTAGGCCCTGAAGGTGAACCGCTT | 14832–14813, 17636–17626 | ||
| Probe | GCGACCGGTGCCTTCTTAGGAGCTGTCCGA | 14708–14737 | ||
| LMP2Ab | 280 | 5′ primer | ATGACTCATCTCAACACATA | 166874–166893 |
| 3′ primer | CATGTTAGGCAAATTGCAAA | 380–361 | ||
| Probe | ATCCAGTATGCCTGCCTGTA | 62–81 | ||
| LMP2Bb | 325 | 5′ primer | CAGTGTAATCTGCACAAAGA | 169819–169838 |
| 3′ primer | CATAGTTAGGCAAATTGCAAA | 380–361 | ||
| Probe | ATCCAGTATGCCTGCCTGTA | 62–81 | ||
| BARF0c | 232 | 5′ primer | AGAGACCAGGCTGCTAAACA | 157154–157174 |
| 3′ primer | AACCAGCTTTCCTTTCCGAG | 159194–159175 | ||
| Probe | AAGACGTTGGAGGCACGCTG | 157359–157378 | ||
| EBER1d | 167 | 5′ primer | AGGACCTACGCTGCCCTAGA | 6648–6629 |
| 3′ primer | AAAACATGCGGACCACCAGC | 6776–6795 |
Soft agar colony assay.
Ten thousand cells were embedded in 1.5 ml of RPMI 1640 containing 12% FBS and 0.35% Noble agar (Difco) or 0.5% SeaPlaque agarose (FMC) on a base layer made of RPMI 1640 containing 12% FBS and 0.4% Noble agar or 0.5% SeaPlaque agarose in a well of a six-well multiwell plate (Falcon). After 2 to 3 weeks of incubation, all the visible colonies were counted.
Induction of apoptosis.
Cells in the log phase were incubated for 16 h in the presence of 20 μg of cycloheximide (Wako) per ml or for 40 h with 1 mM glucocorticoid (Pharmacia and Upjohn) or exposed to 5 to 10 mJ of UV irradiation in a UV cross-linker (Bio-Rad) and incubated for 16 h. Viability of cells was quantified by a colorimetric assay (Cell Titer 96; Promega) that measured the activity of mitochondrial dehydrogenases. The absorbance at 570 nm (A570) was measured with a plate reader (Bio-Rad). Samples from time zero were used as controls. The survival rate was calculated by the following formula: (% survival rate) = [(A570 of the sample) − (A570 of the blank)]/[(A570 of the control) − (A570 of the blank)] × 100.
DNA extraction for low-molecular-weight DNA.
First, 106 cells were lysed in 100 μl of lysis buffer containing 10 mM EDTA, 10 mM Tris HCl (pH 8.0), and 0.5% Triton X-100 and then kept on ice for 10 min. Supernatants were collected after 20 min by centrifugation of the sample at 16,000 rpm. Then the supernatant was treated with proteinase K (100 μg/ml) (Takara) at 55°C for 30 min, and nucleic acids were precipitated with 2-propanol. Finally, the pellet was resolved in water containing 2 μg of RNase A. DNA corresponding to 4 × 105 cells was applied per lane of agarose gel for electrophoresis.
Statistical analysis.
Statistical significance was assessed by the Mann-Whitney U test and unpaired Student t test on paired values. Values of P < 0.05 were considered significant.
RESULTS
EBV reinfection of EBV-negative Akata cells results in type I latency.
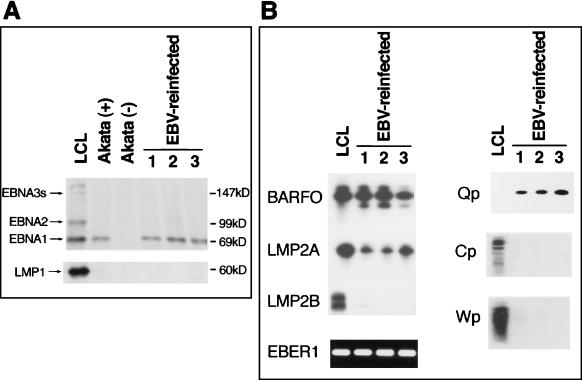
We could not isolate EBV-reinfected Akata clones with either wild-type Akata EBV or B95-8 EBV since the population of infected cells that could stably retain EBV was extremely low. This was a common phenomenon for EBV infection of all EBV-negative B-cell lines tested. Instead of the wild-type virus, we used a recombinant Akata EBV-knockout viral thymidine kinase gene, replaced it with neoR to reinfect EBV-negative clones, and succeeded in isolating reinfected clones in the selection medium (18). All the G418-resistant cells were positive for EBNA as shown by an immunofluorescence assay with human serum (data not shown). Reinfection was ensured by detecting EBV DNA with cellular DNA extracted from G418-resistant cells by Southern blotting (data not shown). From more than a hundred clones, we randomly selected 12 clones which were subjected to immunoblotting and RT-PCR to identify viral latency. EBNA-1 was detected by Western blotting, and BARF0, EBER1, and LMP2A were detected by RT-PCR. The clones were negative for EBNA-2, EBNA-3a, -3b, and -3c (EBNA-3s), and LMP1 by Western blotting and LMP2B by RT-PCR. They used the Q promoter for EBNA-1 transcription, not the C or W promoter. In Fig. 1, the results for three clones are indicated. The pattern of viral gene expression and the promoter usage of reinfected Akata cells were quite similar to those of the parental Akata cells. On the other hand, a lymphoblastoid cell line (LCL) immortalized by the same Akata EBV, a representative for type III latency, expressed EBNA-1, -2, and -3s; LMP1 and -2; EBER1; and BARF0. It utilized the C-W promoter to transcribe EBNAs. These data were consistent with the conclusion that reinfected Akata cells possessed type I latency. In addition, the cells retained the ability to produce virus particles after cross-linking of surface IgG with anti-IgG.
FIG. 1.
EBV gene expression in EBV-reinfected Akata cells. (A) Reinfected Akata cell clones are positive for EBNA-1 but negative for EBNA-2, EBNA-3s, and LMP1 by Western blot analysis (arrows at left). The LCL, which represents type III latency, and parental Akata cells (EBV positive), which represent type I latency, are used as positive controls, and EBV-negative cells are used as a negative control. Molecular masses of protein markers are shown at the right. (B) Three EBV-reinfected clones are also positive for BARF0 and EBER1 and weakly positive for LMP2A as demonstrated by RT-PCR. They utilize the Q promoter for transcription of EBNA-1 mRNA but not the C or W promoter. Consequently, reinfected clones were proved to have type I latency. LCL cells are positive for all latent gene expression tested by RT-PCR, and they utilize C and W promoters to transcribe EBNAs.
Growth characteristics of reinfected clones.
As reinfected clones were selected in the presence of G418, we transfected the neoR gene into the same EBV-negative Akata cell clones and isolated more than 100 clones. We selected several G418-resistant clones randomly and compared them with the EBV-reinfected cell clones in terms of growth characteristics. Both proliferated at nearly the same rate in RPMI 1640 medium supplemented with 10% FBS. EBV-positive and -negative clones from the parental Akata cells exhibited distinct differences in growth in low-serum conditions. However, we could hardly observe a significant difference between the reinfected cells and the neoR-transfected cells originating from four independent EBV-negative Akata cell clones with respect to the growth rate in medium with 0.1% FBS, in spite of repeated attempts (data not shown).
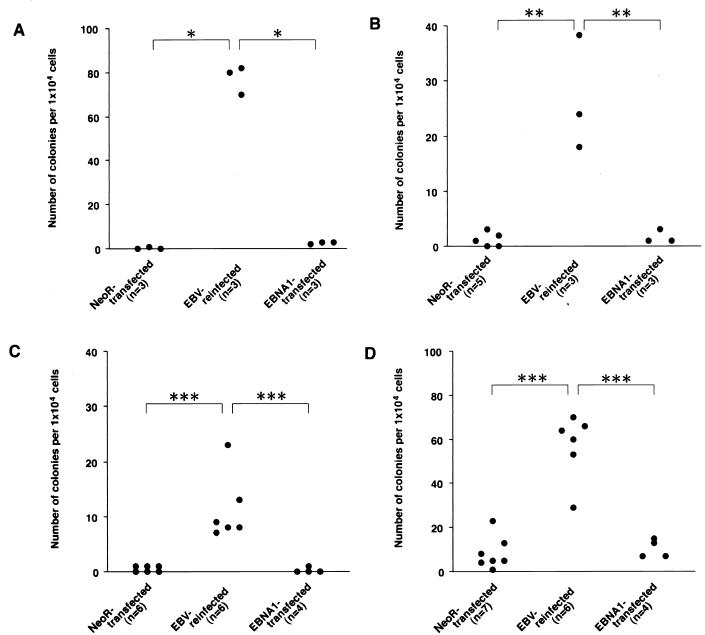
To examine the malignant phenotype, we first performed a soft agar colony assay because our previous study revealed that colony formation in soft agar is an excellent index for the malignant phenotype of Akata. Reinfected cells formed colonies in soft agar, but neoR-transfected clones scarcely did (Fig. 2). As shown in Fig. 2, we tested four pairs of reinfected and neoR-transfected cells, and the results were the same. This phenotypic difference was retained for at least half a year. The absolute numbers of colonies differed, but this was presumed to be due simply to the clonal variation. Furthermore, each of six EBV-reinfected and neoR-transfected cell clones derived from an EBV-negative Akata cell clone was pooled and assayed for tumorigenicity in SCID mice. As shown in Table 2, EBV-reinfected cells produced tumors at the sites of inoculation in four of eight mice. Tumor cells consisted of EBNA-1-positive cells and were negative for EBNA-2 and LMP1. On the other hand, neoR-transfected cells were not tumorigenic in SCID mice.
FIG. 2.
Growth in soft agar of neoR-transfected, EBV-reinfected, and EBNA-1-transfected Akata cell clones. Three sets of clones originating from four independent EBV-negative Akata clones were examined in this study (A, B, C, and D). EBV-reinfected cells could form colonies, but neoR-transfected and EBNA-1-transfected clones could scarcely do so. It is clear that EBNA-1 is not solely responsible for the growth of Akata cells in soft agar. Each dot represents the number of visible colonies of a single clone formed in soft agar. A total of 104 cells were seeded per well. The number of clones tested is noted underneath the graph. Values between the indicated groups (Mann-Whitney U test): ∗, P < 0.005; ∗∗, P < 0.05; ∗∗∗, P < 0.01.
TABLE 2.
Tumorigenicity of EBV-reinfected Akata cells in SCID micea
| Expt no. and cell type | No. of mice with tumor/total no. | Ht/width/depth of tumor (mm) |
|---|---|---|
| Expt 1 | ||
| EBV positive | 3/4 | 25/20/11 |
| 24/18/7 | ||
| 21/21/11 | ||
| EBV negative | 0/4 | |
| EBV reinfected | 2/4 | 20/16/5 |
| 25/20/7 | ||
| neoR transfected | 0/4 | |
| EBNA-1 transfected | 0/4 | |
| Expt 2 | ||
| EBV reinfected | 2/4 | 22/17/6 |
| 12/4/2 | ||
| neoR transfected | 0/4 | |
| EBNA-1 transfected | 0/4 |
Since there was some clonal variation in the assay of soft agar, we pooled six cell clones for the generation of tumors so that we could test many clones and minimize the effect of clonal variation. A total of 107 cells each of EBV-positive, EBV-negative, EBV-reinfected, neoR-transfected, and EBNA-1-transfected Akata cell clones were inoculated into the thigh subcutis of the 6-week-old male SCID mice. The SCID mouse strains was FOX CHASE C.B-17/Icr-scid Jcl (CLEA, Tokyo, Japan). Six weeks after inoculation, tumors developed at the site of inoculation. Mice were killed 8 weeks after inoculation, and the developed tumors were measured.
Resistance to apoptosis is dependent on the presence of EBV.
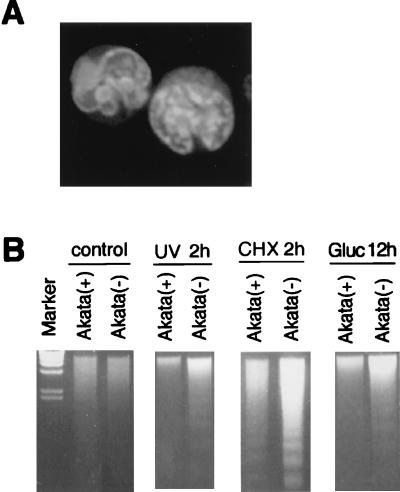
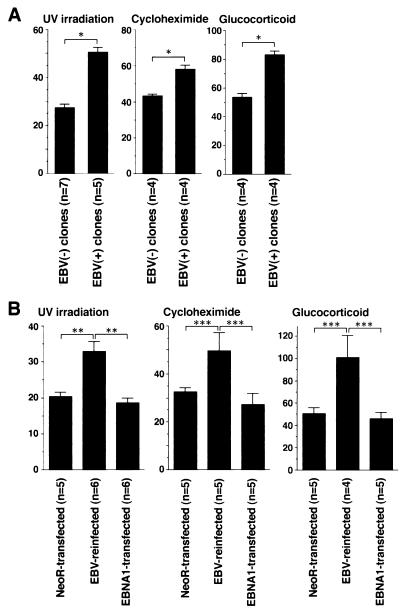
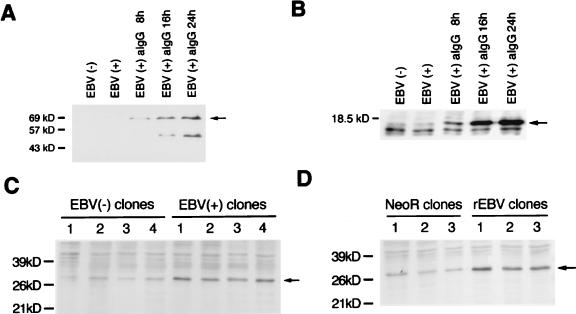
The typical morphology and degradation of genomic DNA into nucleosomal units define apoptosis. Akata cells underwent apoptosis after treatment of cells with various inducers such as cycloheximide, UV light, and glucocorticoid. We examined whether there was any difference in the susceptibility to apoptotic cell death between EBV-infected and uninfected Akata clones. The percentages of cells that had apoptotic morphology were 20 to 40% of EBV-positive cells and 50 to 80% of EBV-negative cells, when cells were treated with cycloheximide for 18 h (Fig. 3A). After treatment with apoptotic stimuli, EBV-positive Akata cells showed a more prominent DNA laddering than did EBV-negative Akata cells (Fig. 3B). The survival rate was examined by a colorimetric assay that measured the activity of mitochondrial dehydrogenases. As a result, EBV-positive clones from parental Akata cells were found to be more resistant to cell death than were EBV-negative clones (Fig. 4A). In addition, restoration of the resistance to apoptosis was observed in the EBV-reinfected Akata clones, as shown in Fig. 4B. This phenotypic difference was stable for more than 1 year. These findings made it clear that resistance to apoptosis of Akata cells was dependent on the presence of EBV. It should be noted that EBV-infected Akata cells were negative for LMP1 and BHRF1, which are known to be antiapoptotic genes. They were expressed only when virus replication was induced by incubation of cells with anti-IgG (Fig. 5A and B).
FIG. 3.
Apoptosis in Akata cells. (A) Typical morphology of apoptotic cells induced by cycloheximide in EBV-negative Akata cells. Cells were stained with acridine orange (2 μg/ml) and photographed by laser scanning fluoromicroscopy (Molecular Dynamics). (B) DNA laddering occurs in response to the treatment with UV radiation (UV), cycloheximide (CHX), and glucocorticoid (Gluc). DNA from 4 × 105 cells was subjected to 2% agarose gel electrophoresis and stained with ethidium bromide. Incubation times are given above the gels. The intensity of the DNA ladder of EBV-negative cells [Akata(−)] is stronger than that from EBV-positive cells [Akata(+)], suggesting that EBV-negative cells are more liable to die of apoptosis than are EBV-positive cells.
FIG. 4.
EBV-infected cells are resistant to apoptosis. EBV-negative [EBV(−)] and EBV-positive [EBV(+)] clones derived from the parental Akata cells (A) and neoR-transfected, EBV-reinfected, and EBNA-1-transfected clones derived from an EBV-negative Akata clone (B) were exposed to apoptotic stimuli. The average relative survival rates (percent) and error bars (standard errors of the means) are shown. (A) The average relative survival rate of the EBV-positive clones is significantly higher than that of the EBV-negative clones (∗, < 0.001), suggesting that EBV-positive cells are resistant to apoptosis because of the presence of EBV in cells. The data presented here are typical results from three independent experiments. (B) The average relative survival rates of reinfected clones are significantly higher than those of NeoR- and EBNA-1-transfected clones (∗∗, < 0.005; ∗∗∗, < 0.05), suggesting that EBV-infected cells are resistant to apoptosis because of the presence of EBV in cells. The data presented here are typical results from three independent experiments. The numbers of clones tested are noted beneath the graph. Significance values between the indicated groups were determined by unpaired Student’s t test.
FIG. 5.
Western blot analysis of LMP1, BHRF1, and bcl-2 protein. (A and B) LMP1 (A) and BHRF1 (B) (arrows) are detected only after cells are treated with anti-IgG, which induces the virus lytic cycle, indicating that these proteins are lytic gene products rather than latent gene products. Minor bands appear to be degraded proteins. (C) bcl-2 protein expression (arrow) of EBV-positive and -negative cell clones derived from parental Akata cells (four clones each). (D) bcl-2 protein expression (arrow) of EBV-reinfected and neoR-transfected clones derived from an EBV-negative Akata clone (three clones each). bcl-2 protein is expressed at a higher level in EBV-infected clones than in uninfected clones. Molecular masses of protein markers are shown at the left of each panel.
We then examined expression of bcl-2 protein, an oncogene having an antiapoptotic function, in EBV-infected and uninfected cells. In Western blotting, EBV-positive clones from the parental Akata cells were shown to express a higher level of bcl-2 protein than did EBV-negative clones (Fig. 5C). Furthermore, expression of bcl-2 protein was higher in reinfected clones than in neoR-transfected clones (Fig. 5D).
EBNA-1 alone cannot induce malignant phenotype and apoptosis resistance in Akata cells.
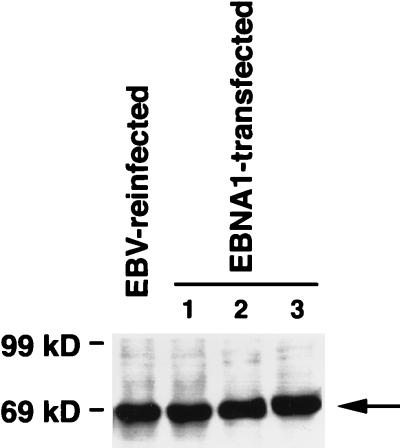
To investigate whether EBNA-1 was responsible for the three phenotypes observed in the EBV-infected Akata cells, anchorage-independent growth in soft agar, tumorigenicity in SCID mice, and apoptosis resistance, we isolated EBNA-1-expressing clones by transfecting EBNA-1/oriP vector, which contains neoR also, into EBV-negative Akata clones and selected stable transformants in the selection medium. Because all the G418-resistant cells should carry this mammalian plasmid vector, 100% of the G418-resistant cells were positive for EBNA-1 as shown by immunofluorescence (data not shown). We isolated more than 100 G418-resistant clones and selected several clones randomly. Those EBNA-1-transfected clones expressed amounts of EBNA-1 protein almost equivalent to those of reinfected clones, as shown in Fig. 6; however, they did not acquire the capacity to grow in soft agar, tumorigenicity in SCID mice, and resistance to apoptosis (Table 2) (Fig. 2, 4, and 6). EBNA-1-transfected cells expressed as little bcl-2 protein as did neoR-transfected cells as a matter of course (data not shown). These data suggested that EBNA-1 alone was responsible for neither the malignant phenotype nor the resistance to apoptosis of Akata cells.
FIG. 6.
Detection of EBNA-1 protein in an EBV-reinfected Akata clone and three EBNA-1-transfected Akata clones by Western blot analysis. EBNA-1 (arrow) is expressed in the EBNA-1-transfected clones in an amount nearly equal to that of the EBV-reinfected clone. Molecular masses of protein markers are shown at the left.
DISCUSSION
In the present study, we compared two pairs of cell clones: EBV-positive and -negative clones from parental Akata cells and neoR-transfected and EBV-reinfected clones from EBV-negative Akata cell clones. This system made it possible to confirm whether any phenotypic difference between EBV-infected and uninfected Akata cell clones was due to EBV. In other words, this system provided an excellent and powerful tool to investigate the pathogenic role of EBV in BL. The unique characteristics of Akata cells, that is, spontaneous loss of EBV DNA from cells during long-term culture and restoration of type I latency by reinfection with EBV, made it possible to establish this system. Other EBV-negative B-cell lines such as BJAB are not ideal hosts for infection study since EBV infection leads to type III latency (unpublished observations), which does not represent in vivo BL latency.
Using this system, we demonstrated that reinfection of EBV-negative Akata clones with EBV restores the ability to grow on soft agar, tumorigenicity in SCID mice, and resistance to apoptosis. Accordingly, it was concluded that these three phenotypes are due to EBV infection. Under the ordinary culture conditions in the medium with 10% FBS, EBV-reinfected Akata cells lose EBV plasmids very easily, because cells possibly do not require EBV for growth under such conditions (unpublished observation). Therefore, it is noteworthy that almost all tumor cells in SCID mice retained EBV as shown by EBNA-1 expression, suggesting that the presence of EBV is critical for tumor formation.
This is the first report providing evidence that EBV-infected BL cells with type I latency are resistant to apoptosis. This may be due to increased expression of bcl-2 protein. The bcl-2 family of proteins appears to be located downstream of any signaling cascades of apoptosis determining the life-death balance of a cell. In Akata cells, a viral gene or genes other than LMP1 must upregulate expression of bcl-2. Our findings imply the presence of an unknown pathway by which EBV modulates the life of host cells.
Required for the maintenance of EBV DNA in a cell, EBNA-1 is expressed in all EBV-infected cells with any type of latency. Whether EBNA-1 plays an oncogenic role in the human cancer cell was an open question. EBNA-1 has been shown to be associated with the development of lymphoma in a transgenic mouse study (24). EBNA-1 has also been reported to increase tumorigenicity and metastatic capability of nasopharyngeal carcinoma cells in a mouse model (15). Those reports suggest that EBNA-1 may have an oncogenic function. If so, it is likely that EBNA-1 is responsible for the malignant phenotype and the resistance to apoptosis of the human BL cell line Akata. However, our results indicated that EBNA-1 alone was not responsible for those two phenotypes.
Then which virus gene is responsible for those two phenotypes of Akata cells? Three other virus gene products—EBERs, BARF0, and LMP2A—are expressed in the cells. It is not yet known whether they have any functions in BL genesis. It has been reported that they are not necessary for immortalization of primary B cells by EBV infection (9, 13, 20). They or a novel virus gene(s) might be required for those two phenotypes of Akata cells. We are currently attempting to identify the responsible virus gene(s).
ACKNOWLEDGMENTS
We thank all the members of our laboratory, especially K. Adachi for her excellent technical assistance. We thank R. F. Margolskee for pEBO.
This work was supported in part by grants-in-aid from the Ministry of Education, Science, Sports, and Culture, Japan, and from the Vehicle Racing Commemorative Foundation and the Suhara Foundation.
REFERENCES
- 1.Brooks L, Yao Q Y, Rickinson A B, Young L S. Epstein-Barr virus latent gene transcription in nasopharyngeal carcinoma cells: coexpression of EBNA1, LMP1, and LMP2 transcripts. J Virol. 1992;66:2689–2697. doi: 10.1128/jvi.66.5.2689-2697.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burkitt D. A sarcoma involving the jaws in African children. Br J Surg. 1958;46:218–219. doi: 10.1002/bjs.18004619704. [DOI] [PubMed] [Google Scholar]
- 3.Deacon E M, Pallesen G, Niedobitek G, Crocker J, Brooks L, Rickinson A B, Young L S. Epstein-Barr virus and Hodgkin’s disease: transcriptional analysis of virus latency in the malignant cells. J Exp Med. 1993;177:339–349. doi: 10.1084/jem.177.2.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Epstein M A, Achong B G, Barr Y M. Virus particles in cultured lymphoblasts from Burkitt’s lymphoma. Lancet. 1964;i:702–703. doi: 10.1016/s0140-6736(64)91524-7. [DOI] [PubMed] [Google Scholar]
- 5.Henle G, Henle W, Diehl V. Relation of Burkitt’s tumor-associated herpes-type virus to infectious mononucleosis. Proc Natl Acad Sci USA. 1968;59:94–101. doi: 10.1073/pnas.59.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Henle W, Henle G. The virus as the etiologic agent of infectious mononucleosis. In: Epstein M A, Achong B G, editors. The Epstein-Barr virus. Berlin, Germany: Springer-Verlag; 1979. pp. 297–320. [Google Scholar]
- 7.Imai S, Koizumi S, Sugiura M, Tokunaga M, Uemura Y, Yamamoto N, Tanaka S, Sato E, Osato T. Gastric carcinoma: monoclonal epithelial malignant cells expressing Epstein-Barr virus latent infection protein. Proc Natl Acad Sci USA. 1994;91:9131–9135. doi: 10.1073/pnas.91.19.9131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Imai S, Nishikawa J, Takada K. Cell-to-cell contact as an efficient mode of Epstein-Barr virus infection of diverse human epithelial cells. J Virol. 1998;72:4371–4378. doi: 10.1128/jvi.72.5.4371-4378.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Longnecker R, Miller C L, Tomkinson B, Miao X Q, Kieff E. Deletion of DNA encoding the first five transmembrane domains of Epstein-Barr virus latent membrane proteins 2A and 2B. J Virol. 1993;67:5068–5074. doi: 10.1128/jvi.67.8.5068-5074.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Margolskee R F, Kavathas P, Berg P. Epstein-Barr virus shuttle vector for stable episomal replication of cDNA expression libraries in human cells. Mol Cell Biol. 1988;8:2837–2847. doi: 10.1128/mcb.8.7.2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nonoyama M, Huang C H, Pagano J S, Kline G, Singh S. DNA of Epstein-Barr virus detected in tissue of Burkitt’s lymphoma and nasopharyngeal carcinoma. Proc Natl Acad Sci USA. 1973;70:3265–3268. doi: 10.1073/pnas.70.11.3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rickinson A B, Kieff E. Epstein-Barr virus. In: Fields B N, Knipe D M, Howley P M, editors. Virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 2397–2446. [Google Scholar]
- 13.Robertson E S, Tomkinson B, Kieff E. An Epstein-Barr virus with a 58-kilobase-pair deletion that includes BARF0 transforms B lymphocytes in vitro. J Virol. 1994;68:1449–1458. doi: 10.1128/jvi.68.3.1449-1458.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rowe M, Rowe D T, Gregory C D, Young L S, Farrell P J, Rupani H, Rickinson A B. Differences in B cell growth phenotype reflect novel patterns of Epstein-Barr virus latent gene expression in Burkitt’s lymphoma cells. EMBO J. 1987;6:2743–2751. doi: 10.1002/j.1460-2075.1987.tb02568.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sheu L F, Chen A, Meng C L, Ho K C, Lee W H, Leu F J, Chao C F. Enhanced malignant progression of nasopharyngeal carcinoma cells mediated by the expression of Epstein-Barr nuclear antigen 1 in vivo. J Pathol. 1996;180:243–248. doi: 10.1002/(SICI)1096-9896(199611)180:3<243::AID-PATH655>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 16.Shibata D, Weiss L M. Epstein-Barr virus-associated gastric adenocarcinoma. Am J Pathol. 1992;140:769–774. [PMC free article] [PubMed] [Google Scholar]
- 17.Shimizu N, Tanabe-Tochikura A, Kuroiwa Y, Takada K. Isolation of Epstein-Barr virus (EBV)-negative cell clones from the EBV-positive Burkitt’s lymphoma (BL) line Akata: malignant phenotypes of BL cells are dependent on EBV. J Virol. 1994;68:6069–6073. doi: 10.1128/jvi.68.9.6069-6073.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shimizu N, Yoshiyama H, Takada K. Clonal propagation of Epstein-Barr virus (EBV) recombinants in EBV-negative Akata cells. J Virol. 1996;70:7260–7263. doi: 10.1128/jvi.70.10.7260-7263.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sugiura M, Imai S, Tokunaga M, Koizumi S, Uchizawa M, Okamoto K, Osato T. Transcriptional analysis of Epstein-Barr virus gene expression in EBV-positive gastric carcinoma: unique viral latency in the tumor cells. Br J Cancer. 1996;74:625–631. doi: 10.1038/bjc.1996.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Swaminathan S, Tomkinson B, Kieff E. Recombinant Epstein-Barr virus with small RNA (EBER) genes deleted transforms lymphocytes and replicates in vitro. Proc Natl Acad Sci USA. 1991;88:1546–1550. doi: 10.1073/pnas.88.4.1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takada K, Horinouchi K, Ono Y, Aya T, Osato T, Takahashi M, Hayasaka S. An Epstein-Barr virus-producer line Akata: establishment of the cell line and analysis of viral DNA. Virus Genes. 1991;5:147–156. doi: 10.1007/BF00571929. [DOI] [PubMed] [Google Scholar]
- 22.Takada K, Ono Y. Synchronous and sequential activation of latently infected Epstein-Barr virus genomes. J Virol. 1989;63:445–449. doi: 10.1128/jvi.63.1.445-449.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tierney R J, Steven N, Young L S, Rickinson A B. Epstein-Barr virus latency in blood mononuclear cells: analysis of viral gene transcription during primary infection and in the carrier state. J Virol. 1994;68:7374–7385. doi: 10.1128/jvi.68.11.7374-7385.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wilson J B, Bell J L, Levine A J. Expression of Epstein-Barr virus nuclear antigen-1 induces B cell neoplasia in transgenic mice. EMBO J. 1996;15:3117–3126. [PMC free article] [PubMed] [Google Scholar]
- 25.Yoshiyama H, Shimizu N, Takada K. Persistent Epstein-Barr virus infection in a human T-cell line: unique program of latent virus expression. EMBO J. 1995;14:3706–3711. doi: 10.1002/j.1460-2075.1995.tb00040.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.zur Hausen H, Schulte-Holthausen H, Kline G, Henle W, Henle G, Clifford P, Santesson L. EBV DNA in biopsies of Burkitt tumours and anaplastic carcinomas of the nasopharynx. Nature. 1970;228:1056–1058. doi: 10.1038/2281056a0. [DOI] [PubMed] [Google Scholar]