Abstract
Infection with the wild-type baculovirus Autographa californica multiple nuclear polyhedrosis virus (AcMNPV) results in complete death of Spodoptera frugiperda (Sf) cells. However, infection of Sf cells with AcMNPV carrying a mutation or deletion of the apoptotic suppressor gene p35 allowed the cloning of surviving Sf cells that harbored persistent viral genomes. Persistent infection established with the virus with p35 mutated or deleted was blocked by stable transfection of p35 in the host genome or by insertion of the inhibitor of apoptosis (iap) gene into the viral genome. These artificially established persistently virus-infected cells became resistant to subsequent viral challenge, and some of the cell lines carried large quantities of viral DNA capable of early gene expression. Continuous release of viral progenies was evident in some of the persistently virus-infected cells, and transfection of p35 further stimulated viral activation of the persistent cells, including the reactivation of viruses in those cell lines without original continuous virus release. These results have demonstrated the successful establishment of persistent baculovirus infections under laboratory conditions and that their establishment may provide a novel continuous, nonlytic baculovirus expression system in the future.
Baculoviruses are a diverse group of common insect pathogens that primarily infect the order Lepidoptera. These viruses contain circular double-stranded DNA genomes of 90 to 160 kb (6, 30, 38, 41). They infect insect cells productively and generate numerous viral progenies.
Persistent baculovirus infection has been studied less than productive infection. Persistent viral infection generates sporadic outbreaks in natural populations of infected insects that appear to be viral reactivation (17, 33) caused by stress factors (26, 30, 33, 42) such as overcrowding, lack of food (43), or thermal shock (24). The ingestion of heterologous viruses has also been shown to activate persistent viral infection (26, 30, 37, 42). Despite these reports of occasional observations of persistently infected insects, the mechanisms that cause persistent baculovirus infection remain unknown.
The p35 gene is known as an apoptotic suppressor gene. The P35 protein can act as an inhibitor of an interleukin-1β-converting enzyme-like protease (1, 3) and prevents cell death in a broad range of hosts, including Drosophila sp. (23), Caenorhabditis elegans nematodes (45), and mammal neural cells (35, 39). p35 is one of the 18 late expression factor (lef) genes involved in the regulation of viral DNA replication and late gene expression in Autographa californica multiple nuclear polyhedrosis virus (AcMNPV)-infected cells (34). The expression of p35 is not only required for optimal late gene expression but is also necessary for blocking premature death of infected Sf21 cells. The apoptotic response of cells to infection with vAcAnh, an AcMNPV mutant defective in p35, results in a significant reduction in viral yield in Spodoptera frugiperda (Sf) cells and larvae, but not in Trichoplusia ni cells or larvae (14). The biological role of p35 is thus suggested to be a host range determinant which can act against host apoptotic defense mechanisms by antagonizing cell death signals.
In the current study, we found that infection of Sf cells with vAcAnh, a p35 null mutant of AcMNPV, results in widespread apoptotic cell death. In contrast to the killing of all infected cells by the wild-type virus, we consistently found that some cell clones survived the infection and became persistently infected, phenotypically resembling cell clones persistently infected with S. frugiperda nuclear polyhedrosis virus (36) and Hz-1 virus (7, 9–11, 25, 31). Hz-1 virus is an unclassified baculovirus-like insect virus (46). This novel, artificially established, persistent viral infection resulting from mutation or deletion of apoptotic suppressor gene p35 was reactivated by transfection of the same gene. Persistent viral genomes propagated in the persistently infected cells through passages, and the cells were resistant to superinfection with both the wild-type and p35 mutant viruses. These findings may provide a novel continuous, nonlytic baculovirus expression system for future biotechnical applications.
MATERIALS AND METHODS
Cells and viruses.
S. frugiperda (fall armyworm) Sf9 and Sf21AE cells and T. ni (cabbage looper) TN368 cells were maintained at 26°C in TNM-FH medium supplemented with 8% fetal bovine serum (Life Technologies). Wild-type AcMNPV, vAcAnh (the annihilator, an AcMNPV mutant which lacks functional P35; 13), vAcZΔp35 (described below; see Fig. 1A), and vAsB6-1 (a recombinant p35 mutant carrying the inhibitor of the apoptosis gene of Cydia pomonella granulosis virus [Cp-iap]; 15) were each propagated in TN368 cells. Titers of the viruses were estimated by plaque assays using TN368 cells.
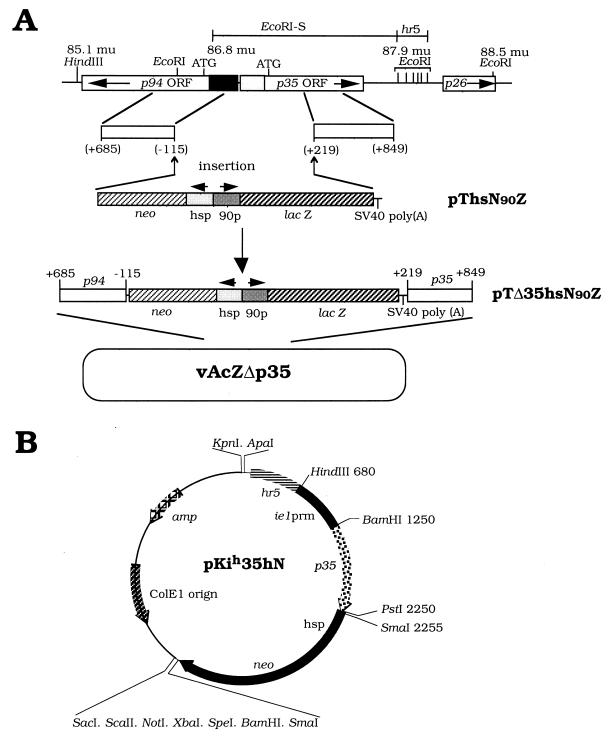
FIG. 1.
Recombinant virus or plasmid construction. (A) Construction of vAcZΔp35, a recombinant AcMNPV which lacks a p35 function. The deleted region between nucleotide −115 relative to the ATG start codon of p94 and nucleotide +219 relative to the ATG start codon of p35 was replaced with both the lacZ and neo genes, and lacZ was driven by an immediate-early-type pag-90 promoter of the Hz-1 virus (10) and the neomycin resistance gene (neo) was driven by a Drosophila hsp70 promoter. These two genes and promoters are shown in newly constructed plasmid pThsN90Z. Further insertion of lateral fragments containing the indicated regions of the p94 and p35 genes into plasmid pThsN90Z resulted plasmid pTΔ35hsN90Z. The final constructed recombinant virus, which contains the exogenous neo and lacZ genes and lacks the capability of P35 expression, was named vAcZΔp35. ORF, open reading frame; SV40, simian virus 40; mu, map units. (B) Organization of a p35-containing plasmid for p35 expression. Plasmid pKih35hN contains the complete open reading frame of the p35 gene driven by the ie1 promoter (prm) of AcMNPV. An hr5 enhancer sequence (677 bp; 22) was inserted upstream of the ie1 promoter. The neo gene is driven by a Drosophila hsp70 promoter.
Construction of a virus with p35 deleted carrying a lacZ gene.
The lacZ gene and a neomycin resistance gene were inserted into plasmid pTSV (32), resulting in plasmid pThsN90Z. The neomycin resistance gene was driven by a heat shock promoter (44), and the lacZ gene was driven by the pag-90 promoter. The pag-90 promoter is an immediate-early-type promoter derived from pag1 (the gene which expresses the PAT1 transcript) of the Hz-1 virus (10). Both genes were flanked by p94 and p35 sequences after insertion of homologous regions of baculovirus p94 (801 bp, nucleotides −115 to +685 relative to the ATG start codon) and p35 (631 bp, nucleotides +219 to +849 relative to the ATG start codon; 18) into plasmid pThsN90Z, which produced transfer vector pTΔ35hsN90Z (Fig. 1A). These p94 and p35 specific fragments were generated by PCR and further confirmed by DNA sequencing.
Homologous recombination was carried out by cotransfecting both AcMNPV genomic DNA and plasmid pTΔ35hsN90Z into TN368 cells. The recombinant virus, named vAcZΔp35, was cloned by screening the transfected TN368 cells for blue plaques with occlusion bodies following 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) staining and was further verified in the infected Sf21 cells by the lack of occlusion body formation. The antibiotic G418 (2 mg/ml) was used in all of the screenings to eliminate the wild-type virus. The recombinant viruses were further verified by restriction mapping of their genomic DNAs and functional assay of their induction of apoptosis upon infection of Sf21 cells.
Isolation of a stably transfected cell line expressing P35.
Plasmid pKih35hN contains the full open reading frame of p35 (nucleotides −22 to +1003 relative to the ATG start codon; 18) under the control of the AcMNPV ie1hr5 promoter containing an upstream enhancer hr5 repeat region (40). The plasmid also contains a neomycin resistance gene under the control of the heat shock 70 promoter from pBluescript KSM+ (Stratagene). This plasmid, pKih35hN (Fig. 1B), was transfected into cells by using CellFECTIN in accordance with the manufacturer’s (Life Technologies) instructions. Neomycin-resistant clones were cultured in the presence of 2-mg/ml G418 for 2 weeks, and individual clones of Sf21-p35-1, Sf21-p35-2, and Sf21-p35-3 cells were isolated.
Estimation of the rate of persistently infected clone formation.
Parental cells (Sf9 and Sf21) were seeded at a density of 4 × 104 per well in a 96-well plate and infected individually with Hz-1 virus, AcMNPV, virus vAcAnh or vAcZΔp35 with p35 mutated or deleted, or revertant virus vAsB6-1 at various concentrations. Seven to 10 days after infection, the number of surviving cells was determined by trypan blue exclusion. Colonies containing more than five cells were isolated and used for further propagations into monolayers. Although some of the clones died during propagation, the number of original colonies formed after infection with a certain virus at a certain concentration reflected the colony formation potential of the individual viruses.
Establishment of persistently infected cell lines.
Parental cells (Sf9 and Sf21) at 2 × 105 per well in a 24-well plate were inoculated with a virus with p35 mutated or deleted (vAcAnh or vAcZΔp35) at a multiplicity of infection (MOI) of 50. Two weeks postinoculation, the surviving cell clones became visible. Clones were isolated, transferred into a 96-well plate, and grown for 7 to 10 days with a medium change once every 3 to 4 days. The surviving clones were transferred to a 24-well plate and then, if they grew, to larger plates or flasks. On average, ca. 3 to 6 clones per 10 original clones survived these transfers. Loss of clones during transfers was attributed mainly to apoptotic cell death. Conditioned medium (half fresh and half used) was used to culture these clones. Two groups of persistently infected cell lines were established and used in subsequent experiments. Persistently infected cell lines derived by infection with vAcAnh were Sf9-vAc-1, Sf9-vAc-2, Sf9-vAc-3, and Sf21-vAc-1. Persistently infected cell lines derived by infection with vAcZΔp35 were Sf9-vAcZΔp35-1, Sf9-vAcZΔp35-2, and Sf9-vAcZΔp35-3.
Detection of viral DNA by PCR.
To detect very small amounts of viral DNA in persistently infected cells, DNA was amplified directly from cultured cells by PCR (see Fig. 3A). Cells were washed twice in phosphate-buffered saline and diluted to 106/ml. Ten microliters of this diluted suspension, corresponding to 104 cells, was lysed by adding 90 μl of detergent buffer (50 mM KCl, 10 mM Tris-HCl [pH 8.3], 0.1-mg/ml gelatin, 0.45% Nonidet P-40, 0.45% Tween 20) containing 6 μg of proteinase K. The diluted suspension was then incubated at 60°C for 1 h. After incubation, the proteinase K was inactivated at 95°C for 15 min.
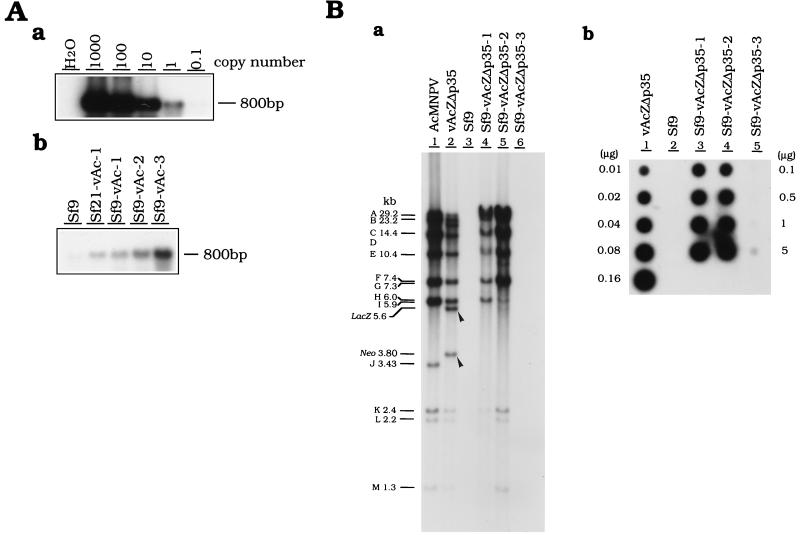
FIG. 3.
Detection of viral DNA in persistently infected cells. (A) Detection of viral DNA in cells persistently infected with vAcAnh. Viral DNA was first PCR amplified and then detected by Southern analysis as described in Materials and Methods. (a) Plasmid pTΔ35hsN90Z was serially diluted and used as a standard for the quantitation of viral DNA (b) in persistently infected cells derived by infection with vAcAnh. (B) Viral DNA detection in cells persistently infected with vAcZΔp35. (a) Southern analysis of the viral genomes in persistently infected cells. Purified genomic DNAs of viruses (wild-type AcMNPV and vAcZΔp35), parental Sf9 cells, and persistently infected cells (Sf9-vAcZΔp35-1, Sf9-vAcZΔp35-2, and Sf9-vAcZΔp35-3) were digested with restriction enzyme XhoI. After fractionation through an agarose gel, genomic DNAs were blotted onto a filter and then hybridized with a probe derived from the total genomic DNA of virus vAcZΔp35. Arrowheads indicate regions which appear in the genome of the original vAcZΔp35 virus and which were later deleted from persistently infected viral genomes during cell culture passages. (b) Quantitation of viral DNA in persistently infected cells by dot hybridization. Various amounts of total genomic DNAs were dotted onto filters. Loading amounts of 0.01 to 0.16 μg were used for DNAs purified from both virus vAcZΔp35 (lane 1) and Sf9 cells (lane 2); the loading amounts for the three persistently infected cells, lanes 3 to 5, were 0.1 to 5 μg. The filter was hybridized with 32P-labeled genomic DNA of vAcZΔp35. Known amounts of vAcZΔp35 DNA (lane 1) were used as standards for calibration of the viral DNA contained in different cell lines. All persistently infected cells were analyzed at 80 to 90 passages.
Ten microliters of this lysate, corresponding to 103 cells, was amplified by PCR and analyzed by agarose gel electrophoresis. Serial 10× dilutions of plasmid pTΔ35hsN90Z were used as molecular standards. These plasmid DNAs were amplified simultaneously with the cell lysates to determine the quantity of viral DNA in the cells persistently infected with vAcAnh. Two negative controls, including a reaction mixture minus template DNA and a reaction mixture containing only Sf9 cell lysate, were used. The primers used were complementary to the 5′ and 3′ regions of the 801-bp fragment of the p94 gene (Fig. 1). PCR products were fractionated on a 1.5% agarose gel and transferred by vacuum blotter (Vacu GeneXL; Pharmacia LKB Biotechnology) onto a nylon membrane. The membranes were probed with a p94 gene fragment labeled with [α-32P]dCTP by random priming in accordance with the manufacturer’s (Boehringer Mannheim) instructions.
Dot blot hybridization.
Total genomic DNAs were purified from uninfected and persistently infected cells and blotted onto nylon membranes by using the Hybri-Dot Manifold (Bio-Rad Laboratories) blotting system. The membranes were probed with viral DNA labeled with [α-32P]dCTP by random priming. The blot was visualized by autoradiography and further quantified by using a PhosphorImager (Molecular Dynamics).
RT-PCR and Southern analysis.
Total cellular RNA was prepared from different cell clones by using Ultraspec RNA isolation reagent (Biotecx) in accordance with the manufacturer’s instructions. To detect the expression of various immediate-early genes, 10 μg of total RNA was reverse transcribed by using an oligo(dT) primer and Moloney murine leukemia virus reverse transcriptase. Equal amounts of cDNA were amplified by PCR with specific primers for the viral ie0, ie1, and ie2 genes and the cellular gapdh gene.
The primers used for PCR were as follows: ie0, 5′-GGCAACGCAACATGATAAGAC and 3′-GTTCAAGGGTTGCACAGCTT, complementary to positions −11 to +720 relative to the ATG start codon; ie1, 5′-GATCGTGAACAACCAAGTGA and 3′-GTTCAAGGGTTGCACAGCTT, complementary to positions −22 to +520 relative to the ATG start codon (12); ie2, 5′-AACAGTATCCTACCAGCCA and 3′-CCTCTACTTCTTCTTCGGGT, complementary to positions −23 to +612 relative to the ATG start codon (8); gapdh, 5′-GACGGACCCTCTGGAAAA and 3′-ACCAGCTGATGAGCTTGAC, corresponding to amino acid residues 195 to 310 of the Drosophila melanogaster gapdh gene (34).
Each PCR was carried out for 30 thermal cycles. Samples without Moloney murine leukemia virus reverse transcriptase were also tested to ensure that the fragment was amplified from mRNA. The reverse transcription (RT) products were separated by electrophoreses in 1.2% agarose gels and transferred to a MAGNA nylon transfer membrane (MSI) by a vacuum blotter (Vacu GeneXL). The membrane was hybridized separately with 32P-labeled probes against the ie0, ie1, ie2, and gapdh genes at 60°C overnight by using a hybridization buffer containing 0.25 M Na2HPO4 at pH 7.2, 1 mM EDTA, 7% sodium dodecyl sulfate, 1% bovine serum albumin fraction V, and 10% formamide.
Histochemical staining of β-galactosidase activity.
Cells were fixed for 5 min at room temperature in a solution of 2% formaldehyde and 2% glutaraldehyde in 150 mM NaCl and 15 mM Na2HPO4 buffer. Cells were then washed twice with 150 mM NaCl and 15 mM Na2HPO4 buffer and stained with 1-mg/ml X-Gal in a buffer containing 5 mM K3Fe(CN)6, K4Fe(CN)6, and 5 mM MgCl2. The cells were stained overnight at 37°C before microscopic examination.
Interference assay.
Both the parental cells and persistently infected cells were inoculated with either wild-type AcMNPV or the p35 null mutants of AcMNPV at an MOI of 10, 1, or 0.1. After adsorption, the residual viruses were removed and the cells were incubated with culture medium at 26°C for 3 days. Viability of the cells was estimated by trypan blue exclusion.
Assay of virus release and reactivation in persistently infected cells.
Persistently infected cells (2 × 105/well in 24-well plates) were transfected with 1 μg of plasmid pKih35hN (Fig. 1B) by using CellFECTIN (Life Technologies) for the assay of viral reactivation (see Fig. 7). As a control, parental Sf9 cells were transfected with pKih35hN, and at 24 h posttransfection, these cells were infected with mutant virus vAcZΔp35 at an MOI of 10. The culture media were harvested at 6 days posttransfection, and the titers of released viruses were determined by plaque assay using TN368 cells.
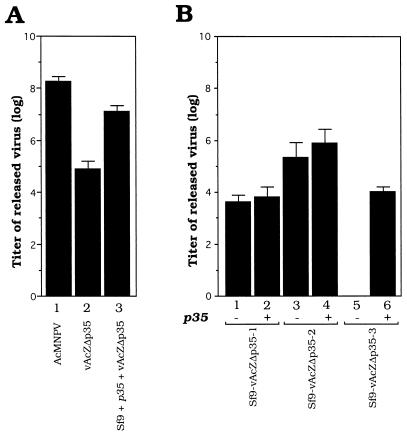
FIG. 7.
Virus release and reactivation in persistently infected cell lines. (A) Productive viral infection of Sf9 cells as a control. Sf9 cells were infected with wild-type AcMNPV (lane 1) and vAcZΔp35 (lane 2) at an MOI of 10. Lane 3 is the virus yield of Sf9 cells first transfected with plasmid pkih35hN and then infected with vAcZΔp35. (B) Release and reactivation of viruses in persistently infected cells. Lanes: 1, 3, and 5, continuous virus-releasing levels of different persistently infected cells; 2, 4, and 6, yields of viruses from these persistently infected cells transfected with pKih35hN. All persistently infected cells were analyzed at 80 to 100 passages.
RESULTS
Mutation or deletion of the p35 gene results in persistent AcMNPV infection.
Two AcMNPV mutants carrying either a mutation or a deletion at the p35 locus were studied, and the correlation of p35 mutation or deletion with apoptosis induction in the infected cells and persistent viral infection was identified. The first p35 mutant virus was vAcAnh, the annihilator. This is an AcMNPV mutant with a 754-bp deletion in the p35 gene resulting in a truncated p35 protein missing 132 amino acids from its carboxyl terminus (13). The other mutant, a vAcZΔp35 virus with p35 deleted, has the promoter and the 5′ end up to +219 bp of the p35 gene replaced with a lacZ gene which is driven by an immediate-early-type promoter, pag-90 (10; Fig. 1A).
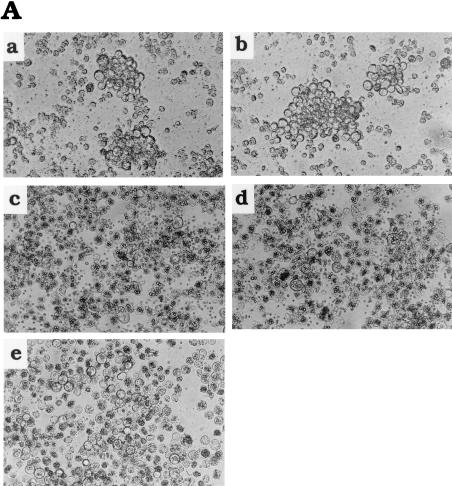
When Sf21 cells were infected with the vAcAnh virus (Fig. 2Aa) or the vAcZΔp35 virus (Fig. 2Ab), most of the cells were lysed by apoptosis, and the remaining cells consistently gave raise to typical persistently infected cell clones 7 days postinfection. Persistent infection did not result from infection with the wild-type virus (Fig. 2Ac) or from infection with a Cp-iap-rescued p35 mutant AcMNPV (named vAsB6-1) (Fig. 2Ad) (5). These results suggest that the repression of persistent viral infection is not due to a specific p35 function but, instead, is more likely due to a general effect related directly or indirectly to the blocking of cellular apoptosis.
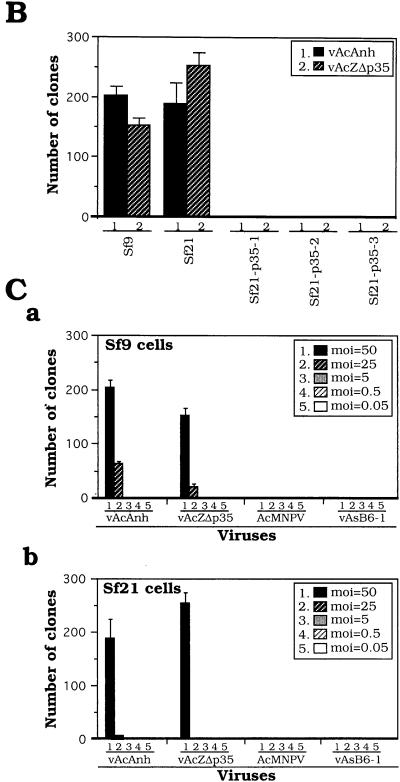
To confirm the effect of p35 blocking of persistent viral infection, Sf21 cell lines stably transfected with p35 (driven by the ie1 promoter [Fig. 1B]) were isolated. Viral occlusion bodies formed upon infection of these cells with vAcAnh (Fig. 2Ae), suggesting that functional P35 was produced in these cells and the phenotype of the mutant virus was restored. Further results showed that the ability of these cell lines to form persistently infected clones after infection with vAcAnh was inhibited in all stably p35-transfected cell lines (Fig. 2B). Collectively, the results suggest that the inhibition of persistent viral infection by p35 is due to the function of p35 or iap through its protein product rather than to any cis effect of the p35 gene in the viral genome.
FIG. 2.
Persistently infected cell clones established by infection with AcMNPV lacking functional p35. (A) Colony formation by Sf9 cells infected with AcMNPV and mutant AcMNPVs. Colonies formed by Sf9 cells infected with vAcAnh (a) and vAcZΔp35 (b). (c and d) show that colony formation was suppressed by infection with either wild-type AcMNPV (c) or revertant virus vAsB6-1 (d). (e) Formation of colonies by Sf9 cells stably transfected with p35 prior to infection with vAcAnh was suppressed. Cells (4 × 104/well) in 96-well plates were infected with viruses at an MOI of 50 and photographed at 7 days postinfection. Colony formation can be observed in parts a and b, and occlusion body formation is evident in parts c, d, and e. (B) Colony formation tests of Sf21 cells stably transfected with p35 prior to infection with viruses vAcAnh and vAcZΔp35 with p35 mutated or deleted. The formation of clones in parental Sf9 and Sf21 cells was used as a control. Cells (4 × 104/well) in 96-well plates were infected (MOI = 50) with either vAcAnh or vAcZΔp35. Sf21-p35-1, Sf21-p35-2, and Sf21-p35-3 are three cell lines stably transfected with p35. (C) Colony formation by Sf9 (a) and Sf21 (b) cells infected with different viruses at various MOIs. Cells (4 × 104/well) in 96-well plates were used for viral infection. Numbers of growing cell clones were determined by trypan blue exclusion tests at 7 days postinfection. Data (means ± standard deviations) were collected from triplicate assays of three independent experiments.
The ability of various p35 mutants to establish persistently infected cell clones was tested. We found that only vAcAnh and vAcZΔp35 could generate persistently infected clones. These clones were not obtained by infection with the wild type or the vAsB6-1 revertant at a wide range of titers (Fig. 2C). In these experiments, higher MOIs generated more colonies, suggesting that a specific virus gene product(s) enhanced the formation of persistently infected clones. When Sf21 cells are infected with AcMNPV with p35 mutated or deleted, there is a delay in the transcription and translation of early and late viral genes, followed by a lack of very late gene expression (14). Therefore, the specific virus gene product that enhances the increasing number of persistently infected cell clones must be either an early or a late viral gene product or a virion protein which migrates into the cell upon viral infection. The specific virus gene product for persistent clone formation seems not to be propagative and could only be enriched by infection at a higher MOI.
Detection of various amounts of viral DNAs with different gene expressions in persistently infected cells.
In all of the cells persistently infected with vAcAnh, viral DNA was not detectable by Southern analysis (80 passages; data not shown) unless the viral DNA was first amplified by PCR and then subjected to Southern hybridization (Fig. 3A). The persistently infected cells established by infection with vAcZΔp35 behaved differently from those derived by infection with vAcAnh in terms of viral DNA content and gene expression in some of the established cell lines. After prolonged serial passages, higher viral DNA content was still detectable by Southern analysis in the Sf9-vAcZΔp35-1, Sf9-vAcZΔp35-2, and Sf9-vAcZΔp35-3 cell lines than in cell lines established by infection with vAcAnh. Although the last tested cell line, Sf9-vAcZΔp35-3, contained much less viral DNA than other lines established by vAcZΔp35, its DNA content was still higher than that found in cells established by infection with vAcAnh. During 80 to 90 passages, the pattern of restriction fragments showed that both the lacZ and neomycin resistance genes in the persistent viral genomes of Sf9-vAcZΔp35-1 and Sf9-vAcZΔp35-2 cells (Fig. 3Ba, arrowheads) were deleted. In addition to these deletions, the viral genome in Sf9-vAcZΔp35-2 may have a partially deleted XhoI H or I fragment, or there may be a mixture of virus populations with different genomic deletions. Dot blot hybridizations showed that the genomes of Sf9-vAcZΔp35-1, Sf9-vAcZΔp35-2, and Sf9-vAcZΔp35-3 cells still contained relatively large amounts of viral DNA (8, 14, and 0.3%, respectively; Fig. 3Bb) compared with those of persistently infected lines established by infection with vAcAnh.
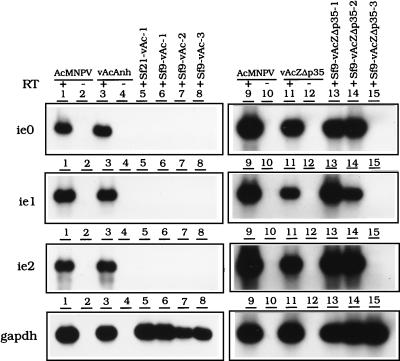
The expression of viral early genes was monitored by using RT-PCR. No early gene expression was detectable, even by RT-PCR followed by Southern hybridization, in cells persistently infected with vAcAnh (Fig. 4, lanes 5 to 8; the Sf21-vAc-1, Sf9-vAc-1, Sf9-vAc-2, and Sf9-vAc-3 cell lines). Nevertheless, the expression of these early genes was detectable in Sf9 cells initially infected with wild-type AcMNPV (Fig. 4, lanes 1 and 9) or the annihilator vAcAnh that had not yet turned persistent (Fig. 4, lanes 3 and 11). In contrast, most of the viral early genes were expressed in the persistently infected cells established by infection with vAcZΔp35, e.g., the Sf9-vAcZΔp35-1 and Sf9-vAcZΔp35-2 cell lines (Fig. 4, lanes 13 and 14). The expression of all of the early genes tested from these two cell lines was relatively strong and not distinguishable from the early gene expression of cells productively infected with the viruses, suggesting that the persistent viral genome was still “alive” and that with the transfection of the p35 gene, mature virus particles may be generated. The expression of early genes was not detectable in Sf9-vAcZΔp35-3 cells, probably due to low viral DNA content or weak viral gene expression (Fig. 4, lanes 15).
FIG. 4.
Detection of viral transcripts in persistently infected cells by RT-PCR. Total RNAs from productively infected and persistently infected cells were reverse transcribed and amplified by PCR with the specific primers ie0, ie1, ie2, and gapdh as indicated in Materials and Methods. The RT-PCR products were separated on a 1.5% agarose gel and transferred to a nylon membrane. The 32P-labeled AcMNPV ie0, ie1, ie2, and S. frugiperda gapdh DNAs were used separately as probes. Lanes 1 to 4 and 9 to 12 are RT-PCR products from cells infected directly with the wild type or a virus with p35 mutated or deleted (MOI = 10 at 3 h postinfection). The reverse transcriptase was omitted in lanes 2, 4, 10, and 12, which served as negative controls. Lanes 5 to 8 are RT-PCR products from cells persistently infected with vAcAnh, and lanes 13 to 15 are RT-PCR products from cells persistently infected with vAcZΔp35. All persistently infected cells were analyzed at 80 to 90 passages. The RT-PCR product of the gapdh transcript was used as a control for mRNA expression.
Expression of the engineered lacZ gene by persistently infected cells.
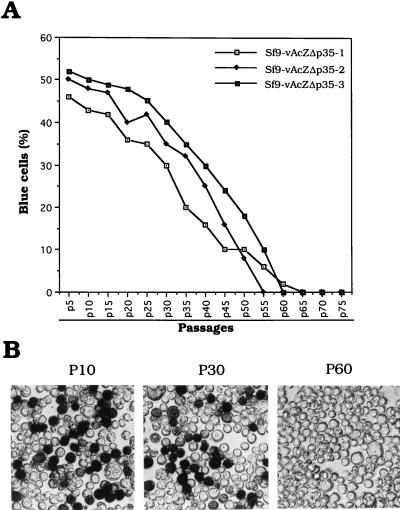
LacZ activity of persistent viruses originally derived by infection with lacZ gene-carrying virus vAcZΔp35 was studied. Essentially all of the infected cells exhibited LacZ activity upon initial viral infection. Once the persistently infected cell clones were isolated, the cells went through an unstable early passage stage. During these early passages, some 10 to 20% of the cells went through apoptosis and died during each passage. At passage 5, the percentage of apoptotic cells began to decrease and the percentage of cells with LacZ activity was recorded. A high percentage (ca. 50%) of the cell lines expressed LacZ activity during the early stage of culturing. These persistently infected cells grew slower than did the parental cells, with a doubling time of roughly 27 to 30 h. The LacZ activities were observed for 50 to 60 passages (Fig. 5) over a 5- to 6-month period. After 65 passages, there was no longer detectable LacZ activity; however, large amounts of viral genomic DNAs were still detectable and early gene expression was evident in all of the cell lines (Fig. 3 and 4).
FIG. 5.
lacZ expression in cells persistently infected with virus vAcZΔp35. (A) Sf9 cells were infected with virus vAcZΔp35 at an MOI of 50. Two weeks postinfection, clones of persistently infected cells were isolated. Three individual clones, Sf9-vAcZΔp35-1, Sf9-vAcZΔp35-2, and Sf9-vAcZΔp35-3, were established, and percent lacZ expression was calculated through long passages. (B) Histochemical staining of Sf9-vAcZΔp35-2 cells by X-Gal at 10, 30, and 60 passages. P10, 10 passages; P30, 30 passages; P60, 60 passages.
Persistently infected cells are resistant to superinfection with wild-type or mutant AcMNPV.
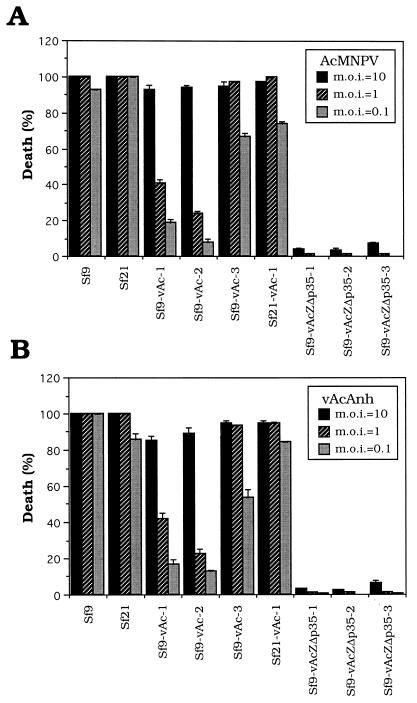
Parental and persistently infected cells were challenged with either vAcAnh or wild-type AcMNPV at passage 90. Resistance to infection with these two viruses was observed in two persistently infected cell lines, Sf9-vAc-1 and Sf9-vAc-2, whereas virus resistance was less evident in the Sf9-vAc-3 and Sf21-vAc-1 cell lines. Significant virus resistance was observed in cells persistently infected with vAcZΔp35. Among these cells, only a few were killed by infection with vAcAnh or wild-type AcMNPV at different viral dosages (Fig. 6, Sf9-vAcZΔp35-1, Sf9-vAcZΔp35-2, and Sf9-vAcZΔp35-3).
FIG. 6.
Analysis of the virus resistance of different cell lines. The death percentages of both parental Sf9 and Sf21 cells and persistently infected Sf9-vAc-1, Sf9-vAc-2, Sf9-vAc-3, Sf21-vAc-1, Sf9-vAcZΔp35-1, Sf9-vAcZΔp35-2, and Sf9-vAcZΔp35-3 cells after a challenge with either wild-type AcMNPV (A) or p35 mutant virus vAcAnh (B) were recorded. Cellular monolayers (4 × 104 cells/well) at passages 90 to 100 were infected with viruses at an MOI of 10, 1, or 0.1 in 96-well plates. Percentage of cell death was calculated based on the total number of cells in each field determined by trypan blue exclusion at 72 h postinfection. Each field contained 300 to 400 cells, and the death percentages in three such fields were analyzed for the infection of individual viruses.
In these experiments, challenges with viruses at higher MOIs were found to kill higher percentages of persistently infected cells than those with lower MOIs. Although a similar phenomenon was observed in parental cells productively infected with these viruses, the differences between high and low MOIs in productively infected cells were small. Such differences in cellular survivability between parental and persistently infected cells challenged with viruses suggest that persistently infected, but not parental, cells restrict the amplification of a factor(s) derived from the invading viruses which can successfully kill the host cells. In addition, cells established by infection with vAcAnh and vAcZΔp35 are significantly different in terms of the ability to resist infection upon further viral challenges. This is likely due to the differences in viral DNA content and gene expression between these two types of persistently infected cells, which may interfere with the attachment, entrance, uncoating, gene expression, or maturation of the invading viruses.
Release and reactivation of persistent viruses.
When cells persistently infected with virus vAcZΔp35 were newly established, infectious viruses could be detected in the media (data not shown). To analyze whether these cells could still release virus after long passages, the titers of infectious viruses present in the culture media of different cell lines at 80 to 100 passages were estimated. No infectious viruses could be detected in persistently infected cell lines Sf21-vAc-1, Sf9-vAc-1, Sf9-vAc-2, and Sf9-vAc-3 (data not shown), which were established by infection with vAcAnh. The production of low viral titers could be detected in two persistently infected cell lines, Sf9-vAcZΔp35-1 and Sf9-vAcZΔp35-2 (Fig. 7B, lanes 1 and 3), but not in Sf9-vAcZΔp35-3 cells (Fig. 7B, lane 5), which were established by infection with vAcZΔp35.
To further study whether infectious virus could be reactivated from these persistently infected cells, we transfected plasmid pKih35hN (Fig. 1B), in which the p35 gene was driven by an immediate-early ie1 promoter, into persistently infected cells. As a control, plasmid pKih35hN was first transfected into parental Sf9 cells, and then the cells were infected with virus vAcZΔp35. This experiment showed that the p35 product significantly restored higher yields of vAcZΔp35 viral progeny (Fig. 7A, lane 3) compared with vAcZΔp35 infection of Sf9 cells without p35 transfection (Fig. 7A, lane 2). Six days after transfection of plasmid pKih35hN into persistently infected cell lines, the culture media were harvested and the yields of released viruses were determined by plaque assays (Fig. 7B, lanes 2, 4, and 6). Compared with those of untransfected persistently infected cell lines (Fig. 7B, lanes 1 and 3), yields of viral progeny from Sf9-vAcZΔp35-1 and Sf9-vAcZΔp35-2 cells were increased, although the difference was not significant when a log scale was used. Interestingly, after p35 gene transfection, production of viruses by non-virus-producing line Sf9-vAcZΔp35-3 was observed (Fig. 7B, lane 6). Another non-virus-producing persistently infected cell line, Sf9-vAcZΔp35-4, was established later by infection with the vAcZΔp35 virus, and further experiments showed that persistently infected viruses were also significantly activated by transfection with p35 (data not shown). This result further confirms that these four cell lines were persistently infected cells which were subject to viral reactivation upon stimulation by p35 transfection.
DISCUSSION
In this study, we demonstrated that deletion of the p35 gene from AcMNPV results in long-term association of the virus (or the viral genome) with host cells. Although it appears that deletion of p35 damages the virus and renders the viral genome inactive in the infected cells, several lines of evidence suggest that the infected cell and the viral genome actually exist in a state resembling persistent viral infection. The viral genomes were found to persist in persistently infected cells after a number of passages, and the results of Southern analysis showed that the viral genome, although it had one or two deletions, persisted in an intact form (Fig. 3Ba). Therefore, long-term persistence of viral DNAs in persistently infected cells was not due to random insertions of fragmented viral DNAs into host genomes. The persistent viral genome is still actively expressed and propagated following replication of the host genome during cellular passages. Infectious viral particles were continuously produced in many of the persistently infected cells, and infectious viruses were further stimulated or reactivated by p35 transfection in all of the cells tested (Fig. 7). The persistently infected cells were partially or highly resistant to superinfection with the wild-type or annihilator virus (Fig. 6). Resistance to infection with homologous viruses is a characteristic frequently observed in cells persistently infected with viruses (4, 16, 19, 20, 31, 47). This further strengthens our conclusion that cells were persistently infected. Collectively, many of the phenomena previously described are characteristics indistinguishable from those of Sf cells persistently infected with naturally derived S. frugiperda nuclear polyhedrosis virus (36).
Because infection with viruses with p35 mutated or deleted results in apoptotic cell death, it seems unlikely that persistent viral infection could be established. p35 has been referred to as one of the lef genes which are involved in expression of late baculovirus promoters in transient expression assays (34). We do not know the mechanism by which some cells are not susceptible to apoptotic death upon initial infection with mutant viruses. Our results showed that replication of viral genomes was still evident in persistently infected cells during passages; however, since the function of p35 is required for late gene expression, as a result, the maturation of the viruses in these survival cells was largely inhibited. Therefore, the generation of surviving cell clones may be the direct result of selection of Sf cells that express higher levels of host apoptotic suppressors. If those host apoptotic suppressors were sufficient to block or tolerate a weak apoptotic signal resulting from a low level of virus replication, persistent viral infection would be established. Those cells harboring viral genomes which replicate in a manner relatively similar to that of host genomes would then behave in the same way as persistently infected cells.
In our experiments, although both viruses vAcAnh and vAcZΔp35 could induce the establishment of persistent viral infection, early gene expression was only detectable in the cells infected with the latter virus. This is probably due to the following two reasons. First, there is a difference between vAcAnh and vAcZΔp35. vAcAnh can still produce the P35 protein, although it is missing 132 amino acids from its carboxyl terminus. vAcZΔp35 has no ability whatsoever to produce P35. Thus, the truncated P35 produced by vAcAnh may still have some effect on the establishment of a persistent viral infection, viral DNA replication, or gene expression in the persistently infected viral genome. Second, the viral genome content in persistently infected cells as a result of vAcAnh infection is extremely low compared with that from vAcZΔp35 infection. Therefore, even if early gene products are expressed by infection with vAcAnh, they would likely not be detectable. Certainly, we cannot rule out the possibility that other differences between these two viruses exist; however, these two viruses have important features in common with respect to the ability to induce persistent viral infection, and this ability can be similarly blocked by p35.
Persistently infected cells were found to resist superinfection with either the wild type or a virus with p35 mutated or deleted. During persistent infection, the existence of viral genomes or the expression of viral genes in the cell may make superinfected viral expression difficult due to the competition for cellular elements required for viral gene expression or DNA replication. It is also possible that the resistance of persistently infected cells to superinfection is due to factors other than the existence of persistent viral DNA or viral gene expression. The persistently infected cells may become resistant to superinfection due to the elimination of virus-binding sites (47) or mutations in the existing viral genome (2). Deletions other than the p35 gene in the viral genome were evident in Sf cells persistently infected with the virus with p35 deleted (Fig. 3Ba). It is possible that further mutations take place somewhere else in the viral genome. These extra deletions and mutations may cause resistance to the challenge of these cells with AcMNPV. All possible mechanisms for viral resistance will be examined in future studies by using these newly established persistent baculovirus infection cell lines.
In one of our previous studies, the promoter and the 5′-end coding region of p35 were replaced with the insertion of a LacZ coding region which is driven by an immediate-early-type pag1 promoter. pag1 is the only detectable gene expressed during persistent Hz-1 virus infection (10). Thus, the pag1 promoter may have a better chance of expressing the gene product during persistent baculovirus infection as well. In the current study, LacZ was expressed for long periods of time in cells persistently infected with vAcZΔp35 (Fig. 5). Although LacZ activity was not detectable after 65 passages, we have demonstrated the successful establishment of a continuous baculovirus expression system by using persistent viral infection. In subsequent experiments, we found that the lack of lacZ gene expression after 65 passages was not due to diminished total viral genomes in persistently infected cells, as very large amounts of viral DNAs were still detectable. We determined that the real cause for elimination of lacZ gene expression was deletion of the lacZ gene in at least two of the persistently infected cell lines after repeated passages (Fig. 3Ba).
It is possible that deletion of the viral genome was caused by a strong and continuous expression of the lacZ gene that may subsequently conflict with regional DNA replication in the persistent viral genome. This possibility seems unlikely, however, considering that continuous early gene expression did not result in deletion of these genes. Another possibility for specific deletion of the lacZ gene may be attributed to the p35 locus, where the lacZ gene is located. The p35 locus could be an unstable and easily deleted genomic region during viral persistence. Experiments intended to address this possibility are in progress in our laboratory.
The baculovirus expression vector system (BEVS) is a widely accepted tool for high-level protein expression. It is a productive viral infection system by which the protein of interest is produced by very late promoters accompanied by cell death. A continuous protein expression system created by stable transfection of a foreign gene into living insect cells has been reported to produce intact recombinant glycoproteins without obvious degradation, and the glycoproteins were secreted more completely than with a conventional lytic BEVS (28). It was later shown that biologically active eucaryotic secretory pathway proteins could be produced by stable transfection of a foreign gene with yields equivalent to that of a conventional BEVS (29). Our newly established persistent baculovirus infection could provide an alternative continuous protein expression system that may compare favorably, in some respects, with the stable transfection of insect cells.
Although improvements are still required before persistent baculovirus infection can be used as an efficient protein expression system, continuous protein expression using persistent viral infection may have several advantages over stable transfection of genes of interest into cells. Persistent baculovirus infection could use all of the versatile techniques and tools developed for a BEVS with only minor modifications. Its establishment requires no antibiotics or selection markers, and with future improvement of the persistent infection, it is possible that a much higher percentage of cells will become persistently infected upon initial viral infection. The persistently infected cell lines may be able to harbor a high level of genomic copies of the virus (more than 10%), thus providing a basis for high-level foreign protein expression. Immediate-early-type genes like ie1 are expressed during persistent infection; thus, the use of stronger intermediate or even late promoters of the persistent viral genomes for foreign protein expression will be possible. Since the capacity of the viral genome is enormous, it is possible to use persistent viral infection for introduction of multiple genes into cells, which is difficult to achieve by conventional stable DNA transfection.
Systems for the study of persistent baculoviral infections in insects are limited in number and rely on occasional observations of persistently virus-infected insects from field collections (26, 27, 37). Thus, our findings should provide a workable system for a more specific and detailed molecular analysis of persistent baculovirus infection. If persistent viral infection can be established in insects, the persistent viruses could be used to introduce foreign genes into insects for engineering of beneficial insects or analysis of insect physiology.
ACKNOWLEDGMENTS
We thank L. K. Miller for kindly providing the p35 gene and the vAcAnh and vAsB6-1 viruses and C. C. Wang and Douglas Platt for critical reading of the manuscript.
This research was supported by Academia Sinica and by grant NSC 87-2311-B-001-124 from the National Science Council, Taiwan, Republic of China.
REFERENCES
- 1.Ahmad M, Srinivasula S M, Wang L, Litwack G, Fernandes T, Alnemri E S. Spodoptera frugiperda caspase-1, a novel insect death protease that cleaves the nuclear immunophilin FKBP46, is the target of the baculovirus antiapoptotic protein p35. J Biol Chem. 1997;272:1421–1424. doi: 10.1074/jbc.272.3.1421. [DOI] [PubMed] [Google Scholar]
- 2.Berkowitz R D, Goff S P. Point mutations in Moloney murine leukemia virus envelope protein: effects on infectivity, virion association, and superinfection resistance. Virology. 1993;196:748–757. doi: 10.1006/viro.1993.1532. [DOI] [PubMed] [Google Scholar]
- 3.Bertin J, Mendrysa S M, LaCount D J, Gaur S, Krebs J F, Armstrong R C, Tomaselli K J, Friesen P D. Apoptotic suppression by baculovirus P35 involves cleavage by and inhibition of a virus-induced CED-3/ICE-like protease. J Virol. 1996;70:6251–6259. doi: 10.1128/jvi.70.9.6251-6259.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Billecocq A, Vialat P, Bouloy M. Persistent infection of mammalian cells by Rift Valley fever virus. J Gen Virol. 1996;77:3053–3062. doi: 10.1099/0022-1317-77-12-3053. [DOI] [PubMed] [Google Scholar]
- 5.Birnbaum M J, Clem R J, Miller L K. An apoptosis-inhibiting gene from a nuclear polyhedrosis virus encoding a polypeptide with Cys/His sequence motifs. J Virol. 1994;68:2521–2528. doi: 10.1128/jvi.68.4.2521-2528.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blissard G W, Rohrmann G F. Baculovirus diversity and molecular biology. Annu Rev Entomol. 1990;35:127–155. doi: 10.1146/annurev.en.35.010190.001015. [DOI] [PubMed] [Google Scholar]
- 7.Burand J P, Kawanishi C Y, Huang Y S. Persistent baculovirus infections. In: Granados R R, Federici B A, editors. The biology of baculovirus. Boca Raton, Fla: CRC Press, Inc.; 1986. pp. 159–175. [Google Scholar]
- 8.Carson D D, Summer M D, Guarino L A. Molecular analysis of a baculovirus regulatory gene. Virology. 1991;182:279–286. doi: 10.1016/0042-6822(91)90671-w. [DOI] [PubMed] [Google Scholar]
- 9.Chao Y-C, Hamblin M, Wood H A. The physical map of Hz-1 viral genome from standard and defective interference viral particles. J Gen Virol. 1990;71:1265–1270. [Google Scholar]
- 10.Chao Y-C, Lee S-T, Chang M-C, Chen H-H, Chen S-S, Wu T-Y, Liu F-H, Hsu E-L, Hou R F. A 2.9-kilobase noncoding nuclear RNA functions in the establishment of persistent Hz-1 viral infection. J Virol. 1998;72:2233–2245. doi: 10.1128/jvi.72.3.2233-2245.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chao Y-C, Wood H A, Chang C-Y, Lee H-J, Shen W-C, Lee H-T. Differential expression of Hz-1 baculovirus genes during productive and persistent viral infections. J Virol. 1992;66:1442–1448. doi: 10.1128/jvi.66.3.1442-1448.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chisholm G E, Henner D J. Multiple early transcripts and splicing of the Autographa californica nuclear polyhedrosis virus IE-1 gene. J Virol. 1988;62:3193–3200. doi: 10.1128/jvi.62.9.3193-3200.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clem R J, Fechheimer M, Miller L K. Prevention of apoptosis by a baculovirus gene during infection of insect cells. Science. 1991;254:1388–1390. doi: 10.1126/science.1962198. [DOI] [PubMed] [Google Scholar]
- 14.Clem R J, Miller L K. Apoptosis reduces both the in vitro replication and the in vivo infectivity of a baculovirus. J Virol. 1993;67:3730–3738. doi: 10.1128/jvi.67.7.3730-3738.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crook M E, Clem R J, Miller L K. An apoptosis-inhibiting baculovirus gene with a zinc finger-like motif. J Virol. 1993;67:2168–2174. doi: 10.1128/jvi.67.4.2168-2174.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doller E, Aucker J, Weissbach A. Persistence of herpes simplex virus type 1 in rat neurotumor cells. J Virol. 1979;29:43–50. doi: 10.1128/jvi.29.1.43-50.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Evans H F, Harrap K A. Persistence of insect viruses. In: Mahy B W J, Minson A C, Darby G K, editors. Virus persistence SGM Symposium. Cambridge, England: Cambridge University Press; 1982. pp. 57–96. [Google Scholar]
- 18.Friesen P D, Miller L K. Divergent transcription of early 35- and 94-kilodalton protein genes encoded by the HindIII K genome fragment of the baculovirus Autographa calofornica nuclear polyhedrosis virus. J Virol. 1987;61:2264–2272. doi: 10.1128/jvi.61.7.2264-2272.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.GilFernandez C, Garcia-Villalon D. A model virus-cell system to study the persistence of African swine fever virus. Arch Virol. 1988;100:161–169. doi: 10.1007/BF01487680. [DOI] [PubMed] [Google Scholar]
- 20.Gorska-Flipot I, Huang M, Cantin M, Rassart E, Massé G, Jolicoeur P. U3 long terminal repeat-mediated induction of intracellular immunity by a murine retrovirus: a novel model of latency for retroviruses. J Virol. 1992;66:7201–7210. doi: 10.1128/jvi.66.12.7201-7210.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Granados R R, Nguyen T, Cato B. An insect cell line persistently infected with a baculovirus-like particle. Intervirology. 1978;10:309–317. doi: 10.1159/000148993. [DOI] [PubMed] [Google Scholar]
- 22.Guarino L A, Summer M D. Interspersed homologous DNA of Autographa californica nuclear polyhedrosis virus enhances delayed-early gene expression. J Virol. 1986;60:215–223. doi: 10.1128/jvi.60.1.215-223.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hay B A, Wolff T, Rubin G M. Expression of baculovirus p35 prevents cell death in Drosophila. Development. 1994;120:2121–2129. doi: 10.1242/dev.120.8.2121. [DOI] [PubMed] [Google Scholar]
- 24.Himeno M, Matsubara F, Hayashiya K. The occult virus of nuclear polyhedrosis of the silkworm larvae. J Invertebr Pathol. 1973;22:292–295. [Google Scholar]
- 25.Huang Y-S, Hedberg M, Kawanishi C Y. Characterization of the DNA of a nonoccluded baculovirus, Hz-1V. J Virol. 1982;43:174–181. doi: 10.1128/jvi.43.1.174-181.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hughes D S, Possee R D, King L A. Activation and detection of a latent baculovirus resembling Mamestra brassicae nuclear polyhedrosis virus in M. brassicae insects. Virology. 1993;194:608–615. doi: 10.1006/viro.1993.1300. [DOI] [PubMed] [Google Scholar]
- 27.Hughes D S, Possee R D, King L A. Evidence for the presence of a low-level, persistent baculovirus infection of Mamestra brassicae insects. J Gen Virol. 1997;78:1801–1805. doi: 10.1099/0022-1317-78-7-1801. [DOI] [PubMed] [Google Scholar]
- 28.Jarvis D L, Fleming J G W, Kovacs G R, Summers M D, Guarino L A. Use of early baculovirus promoters for continuous expression and efficient processing of foreign gene products in stably transformed lepidopteran cells. Bio/Technology. 1990;8:956–958. doi: 10.1038/nbt1090-950. [DOI] [PubMed] [Google Scholar]
- 29.Jarvis D L, Weinkauf C, Guarino L A. Immediate-early baculovirus vectors for foreign gene expression in transformed or infected insect cells. Protein Expr Purif. 1996;8:191–203. doi: 10.1006/prep.1996.0092. [DOI] [PubMed] [Google Scholar]
- 30.King L A, Possee R D. Advances in insect virology. Adv Insect Physiol. 1994;25:2–73. [Google Scholar]
- 31.Lee J-C, Chen H-H, Wei H-L, Chao Y-C. Superinfection-induced apoptosis and its correlation with the reduction of viral progeny in cells persistently infected with Hz-1 baculovirus. J Virol. 1993;67:6989–6994. doi: 10.1128/jvi.67.12.6989-6994.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee S-T, Yu S-M, Hsu E-L, Chao Y-C. Identification of a very early promoter of Hz-1 virus using a novel dual-expression shuttle vector. Nucleic Acids Res. 1995;22:4683–4689. doi: 10.1093/nar/23.22.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Longworth J F, Cunningham J C. The activation of occult nuclear-polyhedrosis viruses by foreign nuclear polyhedra. J Invertebr Pathol. 1968;10:361–367. [Google Scholar]
- 34.Lu A, Miller L K. The role of eighteen late expression factor genes in transcription and DNA replication. J Virol. 1995;69:975–982. doi: 10.1128/jvi.69.2.975-982.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martinou I, Fernandez P A, Missotten M, White E, Allet B, Sadoul R, Martinou J C. Viral proteins E1B19K and p35 protect sympathetic neurons from cell death induced by NGF deprivation. J Cell Biol. 1995;128:201–208. doi: 10.1083/jcb.128.1.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McIntosh A H, Ignoffo C M. Establishment of a persistent baculovirus infection in a lepidopteran cell line. J Invertebr Pathol. 1981;38:395–403. [Google Scholar]
- 37.McKinley D J, Brown D A, Payne C C, Harrap K A. Cross infectivity and activation studies with four baculoviruses. Entomophaga. 1981;26:79–90. [Google Scholar]
- 38.Miller L K, Dawes K P. Physical map of the DNA genome of Autographa californica nuclear polyhedrosis virus. J Virol. 1979;29:1044–1045. doi: 10.1128/jvi.29.3.1044-1055.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rabizadeh S, LaCount D J, Friesen P D, Bredesen D E. Expression of the baculovirus p35 gene inhibits mammalian cell death. J Neurochem. 1993;61:2318–2321. doi: 10.1111/j.1471-4159.1993.tb07477.x. [DOI] [PubMed] [Google Scholar]
- 40.Rodems S M, Friesen P D. The hr5 transcriptional enhancer stimulates early expression from the Autographa californica nuclear polyhedrosis virus genome but is not required for virus replication. J Virol. 1993;67:5776–5785. doi: 10.1128/jvi.67.10.5776-5785.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith G E, Summers M D. Restriction maps of five Autographa californica MNPV variants, Trihoplusia ni MNPV, and Galleria mellonella MNPV DNAs with endonucleases SmaI, KpnI, BamHI, SalcI, XhoI, and EcoRI. J Virol. 1979;30:828–838. doi: 10.1128/jvi.30.3.828-838.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smith K M. The cytoplasmic virus diseases. In: Steinhaus E A, editor. Insect pathology: an advanced treatise. Vol. 1. New York, N.Y: Academic Press, Inc.; 1963. pp. 457–497. [Google Scholar]
- 43.Steinhaus E A, Dineen J P. Observations on the role of stress in a granulosis of the variegated cutworm. J Insect Pathol. 1960;2:55–65. [Google Scholar]
- 44.Steller H, Pirrotta V. P transposons controlled by the heat shock promoter. Mol Cell Biol. 1986;6:1640–1649. doi: 10.1128/mcb.6.5.1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sugimoto A, Friesen P D, Rothman J H. Baculovirus p35 prevents developmentally programmed cell death and rescues a ced-9 mutant in the nematode Caenorhabditis elegans. EMBO J. 1994;13:2023–2028. doi: 10.1002/j.1460-2075.1994.tb06475.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Volkman, L. E., G. W. Blissard, P. Friesen, B. A. Keddie, R. Possee, and D. A. Theilmann. 1995. Virus taxonomy: the classification and nomenclature of viruses. Arch. Virol. 6(Suppl. 3):104–113.
- 47.Wang H, Klamo E, Kuhmann S E, Kozak S L, Kavanaugh M P, Kabat D. Modulation of ecotropic murine retroviruses by N-linked glycosylation of the cell surface receptor/amino acid transporter. J Virol. 1996;70:6884–6891. doi: 10.1128/jvi.70.10.6884-6891.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]