Abstract
Previously, we have observed that mutations in proteins 1a and 2a, the two virally encoded components of the brome mosaic virus (BMV) replicase, can affect the frequency of recombination and the locations of RNA recombination sites (P. D. Nagy, A. Dzianott, P. Ahlquist, and J. J. Bujarski, J. Virol. 69:2547–2556, 1995; M. Figlerowicz, P. D. Nagy, and J. J. Bujarski, Proc. Natl. Acad. Sci. USA 94:2073–2078, 1997). Also, it was found before that the N-terminal domain of 2a, the putative RNA polymerase protein, participates in the interactions between 1a and 2a (C. C. Kao, R. Quadt, R. P. Hershberger, and P. Ahlquist, J. Virol. 66:6322–6329, 1992; E. O’Reilly, J. Paul, and C. C. Kao, J. Virol. 71:7526–7532, 1997). In this work, we examine how mutations within the N terminus of 2a influence RNA recombination in BMV. Because of the likely electrostatic character of 1a-2a interactions, five 2a mutants, MF1 to MF5, were generated by replacing clusters of acidic amino acids with their neutral counterparts. MF2 and MF5 retained nearly wild-type levels of 1a-2a interaction and were infectious in Chenopodium quinoa. However, compared to that in wild-type virus, the frequency of nonhomologous recombination in both MF2 and MF5 was markedly decreased. Only in MF2 was the frequency of homologous recombination reduced and the occurrence of imprecise homologous recombination increased. In MF5 there was also a 3′ shift in the positions of homologous crossovers. The observed effects of MF2 and MF5 reveal that the 2a N-terminal domain participates in different ways in homologous and in nonhomologous BMV RNA recombination. This work maps specific locations within the N terminus involved in 1a-2a interaction and in recombination and further suggests that the mechanisms of the two types of crossovers in BMV are different.
RNA recombination contributes significantly to the high level of genetic diversity in animal, plant, and bacterial RNA viruses (5, 8, 20, 35). Studies conducted during the last decade clearly indicate that the exchange of RNA genetic information can occur between viral strains (18, 22), viral species (21), or viral and cellular RNAs (17, 23). In addition to having a role in the evolution of the viral RNA genome and in the generation of new viral strains, RNA recombination can correct errors that arise during RNA replication (5).
In spite of extensive studies, the molecular mechanism of RNA recombination is not well understood. The prevailing model for RNA recombination posits that recombination occurs when the RNA replicase switches strands during RNA synthesis (4, 8, 10, 14, 35). Kirkegaard and Baltimore provided the first experimental evidence in support of this copy-choice model by demonstrating that poliovirus RNA recombination depends on RNA replication (19). For brome mosaic virus (BMV), areas of local complementarity and similarity between viral RNAs have been shown to promote, respectively, nonhomologous and homologous crossovers (24–26). A similar heteroduplex-mediated mechanism may be responsible for homologous recombination in the poliovirus genome (33).
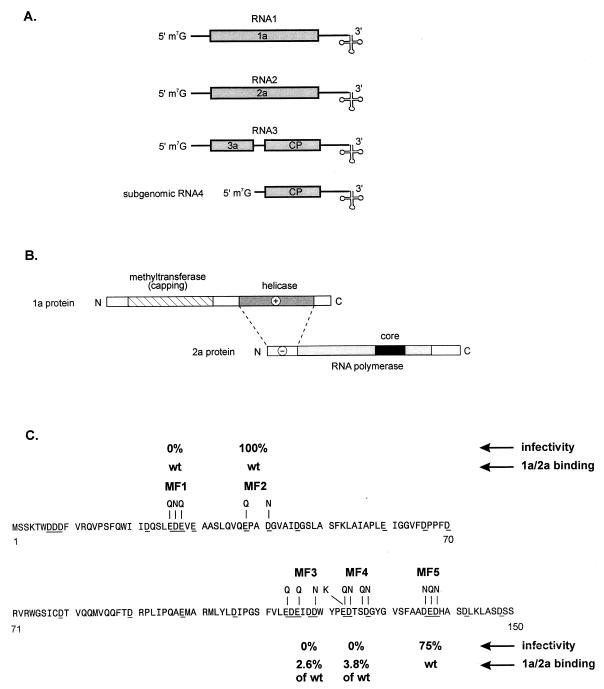
The BMV genome is composed of three RNAs named RNA1 to RNA3 (Fig. 1A). All BMV RNAs share a highly structured 200-nucleotide (nt) tRNA-like sequence at their 3′ ends which directs the synthesis of complementary minus-strand RNAs (2, 7). RNA1 and -2 encode the viral replication proteins and are sufficient for replication activities in the absence of RNA3 in protoplasts (1). RNA1 encodes the 1a protein, whose N-terminal half has homology to known methyltransferases and whose C-terminal half contains all the motifs found in RNA helicases (1). RNA2 encodes the 2a protein, which has a central polymerase-like domain flanked by nonconserved N- and C-terminal domains (Fig. 1B). The C terminus can be deleted without any apparent effect on RNA replication in barley protoplasts (39).
FIG. 1.
Organization of the BMV genome, functional domains of 1a and 2a proteins, and locations of MF mutations. (A) Molecular organization (not to scale) of the BMV genome. RNA1, RNA2, and RNA3 components encode four proteins: 1a, 2a, 3a, and coat protein (CP). The CP is expressed from subgenomic RNA4. Open reading frames are represented by boxes, while terminal and intercistronic noncoding regions are represented by lines. The folding of the tRNA-like structure is shown at the 3′ terminus. (B) Domain organization and interaction between BMV 1a and 2a replicase proteins. Domains for putative methyltransferase (capping), helicase, and RNA polymerase activities are indicated by cross-hatched boxes. Open boxes depict nonconserved regions. The core polymerase domain is marked by a filled box. The negative and positive net charges of 2a N-terminal and helicase domains are indicated by − and +, respectively. (C) Sequence of the first 150 amino acids of the N-terminal domain in 2a protein containing MF mutations. Acidic amino acids bearing negative charges are underlined. The clusters of amino acids that replace the original residues in the MF1 to MF5 mutations are marked by the single-letter code above the wt sequence. The infectivity and 1a-2a interaction data are indicated. For more details, see Table 1.
The BMV replicase is an enzymatic complex that contains the BMV-encoded proteins 1a and 2a, as well as a factor eIF3 associated with the host (32) and possibly other still undefined host factors (16, 32). The interactions in vitro between 1a and 2a have previously been analyzed by coimmunoprecipitation and the yeast two-hybrid assay (15, 16). A portion of the 2a protein N terminus encompassing amino acids 25 to 140 has been demonstrated to bind the 1a helicase-like domain in vitro and by the yeast two-hybrid assay (30a). The domain in 1a that interacts with 2a has been mapped to a protease-resistant structure in the helicase-like region (31). The capacity for protein-protein interaction in vitro is strongly correlated with the ability of the proteins to direct RNA replication in protoplasts (15). The in vitro interaction between 1a and 2a is disrupted by concentrations of KCl greater than 0.5 M (15), suggesting that ionic forces are involved in the interaction of these proteins.
BMV is the first plant RNA virus for which RNA recombination was observed (6). The RNA3 component bearing a 20-nt deletion at the 3′ end was repaired in infected plants by replacement of the mutated region with wild-type (wt) RNA1 or RNA2 sequences. However, the frequency of recombination (defined as the fraction of local lesions that accumulated recombinant cells) was lower than 5%. Biologically active BMV RNA3 constructs were designed to increase the recombination frequency and to identify the recombinationally active sequences within the 3′ regions of all three BMV RNAs and the intercistronic region of RNA3 (25, 26). Depending on whether or not similar or complementary sequences were required for recombination during BMV RNA replication, as well as whether crossovers occurred within homologous or other regions, the process was described as, respectively, homologous or nonhomologous (30).
For both types of recombination in BMV, experimental systems based on either potential heteroduplex formation or the existence of local homologous regions have been described (30). Results obtained with these systems can be interpreted by assuming the presence of a copy-choice mechanism, which necessitates the participation of viral RNA replication complexes. Consistent with this, mutations introduced into the helicase domain of the 1a protein of BMV can influence the locations of crossover sites and increase the fraction of imprecise heteroduplex-induced recombinants (27) and a single amino acid mutation in the central conserved domain of 2a can inhibit nonhomologous recombination without influencing homologous recombination (12).
We are systematically mapping the domains of BMV replicase proteins involved in recombination. In this work we examine how replacements of clusters of acidic amino acids with their neutral counterparts at five different locations within the 2a N terminus influence 1a-2a interactions and recombinant formation. The lack of 1a-2a interaction is correlated with the loss of virus infectivity, confirming previous observations regarding the role of 1a-2a interaction in BMV replication. Other mutations within the N terminus which do not detectably affect 1a-2a interaction can change the frequencies of homologous and nonhomologous crossovers, as well as the positions at which homologous crossovers occur. One mutant, although noninfectious, exhibited a nearly wt level of 1a-2a interaction. This level of interaction suggests that, in addition to 1a-2a interaction, other features of the 2a N terminus may determine the biological activity of BMV. Overall, our study reveals the multifunctional character of the 2a N terminus and provides new evidence that engineering of modified recombination levels is possible by mutagenizing viral replicase proteins.
MATERIALS AND METHODS
Materials.
Plasmids pB1TP3, pB2TP5, and pB3TP7 (13) were used to synthesize in vitro infectious transcripts of the wt BMV RNA components 1, 2, and 3. Plasmids pB2 MF1 to -5 were used to synthesize RNA2 transcripts bearing the MF1 to -5 mutations. Plasmid pPN8(−) (25) was used to obtain in vitro the transcripts of the PN8(−) derivative of BMV RNA3, which is suitable for study of nonhomologous recombination. Plasmids pPN-H39 and pPN-H66 were used to synthesize transcripts of the PN-H39 and PN-H66 derivatives of BMV RNA3, which are suitable for study of homologous recombination (26). T7 RNA polymerase, Moloney murine leukemia virus reverse transcriptase, and restriction enzymes were from Gibco BRL (Gaithersburg, Md.), and our Sequenase kit was from United States Biochemical Corporation (Cleveland, Ohio).
Preparation of MF RNA2 mutants.
Mutated RNA2 was derived from pB2TP5 containing wt RNA2 cDNA. Appropriate alterations within the cDNA were introduced by using a PCR-based site-directed mutagenesis method with the primers listed below. The primers were used to generate appropriate PCR products that were then used to replace portions of plasmid pB2TP5 by cutting it with specific restriction enzymes. Finally, each new fragment introduced into pB2TP5 was sequenced to verify the introduced changes. Primers and restriction sites used to obtain plasmids pB2MF1 to -5 were as follows (where restriction enzyme sites appear in boldface letters and underlined bases indicate changes introduced into the wt 2a sequence). For construct pB2MF1, primers 1 and 2 were used. Primer 1, 5′-TGAAGGCTAGCAGCCTCCACCTGGTTTTGTAAGGATTG-3′ (38 nt), was complementary to the plus-strand sequence of pB2TP5 between positions 170 and 207 and had an NheI restriction site; primer 2, 5′-CATGCCTGCAGGTCGAC-3′ (17 nt), had a unique PstI restriction site. For construct pB2MF2, primers 3 and 4 were used. Primer 3, 5′-GGCTGCTAGCCTTCAGGTGCAGCAGCCCGCAAACGGAGTTGCC-3′ (43 nt), represented the plus-strand sequence of pB2TP5 between positions 193 and 235 and had an NheI restriction site; primer 4, 5′-GTTGAACCATTTGTTGGACGGTG-3′ (23 nt), represented the plus-strand sequence of pB2TP5 between positions 336 and 362 and had a PflMI restriction site. For construct pB2MF3, primers 5 and 6 were used. Primer 5, 5′-GGGATACCAGTTATCAATTTGATCTTGGAGCACGAAAGAG-3′ (40 nt) (of negative polarity), represented the pB2TP5 sequence between positions 430 and 469; primer 6, 5′-GGGGCTCTATTTGCGACAC-3′ (23 nt) (of positive polarity), represented the pB2TP5 sequence between positions 324 and 342. The PCR product obtained with primers 5 and 6 (about 250 nt long) was purified and used as a primer for the next PCR together with primer 7, 5′-CAAAGTCCATGGAATAATCACC-3′ (22 nt), which was complementary to the pB2TP5 sequence between positions 872 and 898 and had an NcoI restriction site. pB2MF4 and -5 were obtained in the same way except that different mutagenic primers were applied. For construct pBMF4, primer 8, 5′-CCGTAACCATTACTAGTATTCTGGGGATACC-3′ (31 nt) (of negative polarity), representing the pB2TP5 sequence between positions 462 and 492, was used. For construct pBMF5, primer 9, 5′-CGCTCGCATGATTTTGATTGGCGGCAAACG-3′ (30 nt) (of negative polarity) representing the pB2TP5 sequence between positions 498 and 527, was used.
In vivo recombination assays.
Full-length, capped RNA transcripts were made from EcoRI-linearized plasmids, according to previously published procedures (12). Chenopodium quinoa leaves were inoculated with a mixture of the transcribed BMV RNAs, as described before (24). Briefly, a mixture of 1 μg of each transcript in 15 μl of inoculation buffer (10 mM Tris [pH 8.0], 1 mM EDTA, 0.1% Celite, 0.1% bentonite) was inoculated on a fully expanded leaf. In each experiment, six to nine separate leaves (on two to three plants) were inoculated. Each inoculation experiment was repeated two or three times. The inoculated C. quinoa plants were maintained in a standard greenhouse. Local lesions were counted 14 days postinoculation.
The wt, MF2, or MF5 mutant RNA2 was coinoculated with wt RNA1 and one of the three recombinationally active RNA3 derivatives [PN8(−), PN-H39, or PN-H66]. For each RNA2-RNA3 mutant combination, total RNA was isolated from separate local lesions, as described previously (24), and the 3′ regions in RNA3 recombinants were identified by amplification of their 3′ regions by a reverse transcription-PCR (RT-PCR) procedure, as described previously (24). Primer 10 for first-strand cDNA synthesis (5′-CAGTGAATTCTGGTCTCTTTTAGAGATTTACAG-3′ [EcoRI site in bold]) was complementary to the 3′-terminal 23 nt of all of the BMV virion RNAs (the rest of the primer represents a 5′ overhang [see reference 24]), while primer 11 for second-strand cDNA synthesis (5′-CTGAAGCAGTGCCTGCTAAGGCGGTC-3′) corresponded to nt 392 to 367 upstream of the 3′ end of wt BMV RNA3. By comparing the sizes of the obtained RT-PCR products with those synthesized with the RNA inoculation mixture, it was determined whether accumulating RNA3 represented a recombinant or parental molecule (input RNA3 mutants are normally capable of replication at lower levels [9, 25, 26]). The resulting cDNA products were digested with EcoRI and XbaI restriction enzymes and ligated into the pGEM3zf(−) cloning vector (Promega). Sequencing of the cloned cDNAs localized the sites of crossovers. A similar methodology was used to check if the MF2 and -5 mutants maintained their modified amino acids during infection. Primer 12 (5′-GTTGAACCATTTGTTGGACGGTGTCGC-3′) was complementary to pB2TP5 between positions 336 and 362. Primers 2 and 12 were used with MF2, while primers 6 and 7 were used with MF5.
To rule out the possibility of recombinant artifacts being generated during RT-PCR amplifications, control experiments were performed as follows. Total RNA extracts isolated from separate local lesions of C. quinoa, after the plants were infected with selected mutants, were amplified separately five times by RT-PCR. Four to six separate local lesions were analyzed per infection. Such reactions reproducibly led to the isolation of the same parental cDNA sequence (data not shown), as demonstrated by sequencing of cloned cDNA fragments.
Northern blot hybridization.
The accumulation of recombinant RNAs was analyzed by Northern blot hybridization. Total RNA extracts from separate local lesions were separated by electrophoresis in a 1% agarose gel and blotted to a Nylon membrane (Hybond N+; Amersham). The membranes were probed with a 32P-labeled RNA complementary to the 3′ end of the plus strand of BMV RNA3. Based on the differences in the sizes of the parental and recombinant RNA species, we were able to confirm that the recombinants were not generated by RT-PCR but were formed during infection in planta.
1a-2a interaction in the yeast two-hybrid system.
DNAs encoding the MF1 to -5 mutations were generated by PCR with a 5′ primer (5′-AGAATTCGTCTTTCCAATGGATC-3′) containing an EcoRI site and BMV cDNA2 (nt 148 to 163) and a 3′ primer (5′-AGGATCCATCAGATCGCTCGCATGA-3′) containing a BamHI site and sequence complementary to BMV cDNA2 (nt 531 to 517). After PCR with templates containing the MF1 to -5 mutations, the products were first cloned into the pCRII vector (Stratagene Inc.) and then digested with EcoRI and BamHI and cloned behind the GAL4 activation domain in plasmid pGAD424 (kind gift of Stan Fields [11]). The resultant plasmids, pGAD2aN-MF1 to -MF5, were transformed into Saccharomyces cerevisiae Y835 (lys2::lexAop-HIS3 ura3::lexAop-lacZ trp1-901 his3 leu2-3 ade2 Δgal14 Δgal80) by electroporation (3) along with 812 H (31). Qualitative and quantitative assays for β-galactosidase activity were performed as previously described (31).
RESULTS
Infectivity and stability of 2a mutants.
Several clusters of acidic amino acids can be distinguished within the 2a N-terminal domain (Fig. 1C). To examine their role in 1a-2a interaction and in RNA recombination, we decided to mutagenize the 2a N terminus. The mutations within the N terminus were expected to affect not only the interaction of 1a and 2a but also, indirectly, the activities of 1a and 2a. The MF series of mutants (MF1 to MF5) were made in the N terminus of 2a by site-directed mutagenesis, as described in Materials and Methods. The amino acids changed are between residues 26 and 28 (MF1), 38 and 41 (MF2), 114 and 123 (MF3), 123 and 127 (MF4), and 136 and 138 (MF5). Each mutant contains changes of two to four amino acids, with aspartate usually being changed to asparagine and glutamate usually being changed to glutamine (Table 1; Fig. 1C). These changes were predicted to reduce the overall negative charge within the N-terminal domain.
TABLE 1.
Mutations introduced into BMV RNA2 within the region encoding the 2a protein N terminus and their influence on virus infectivity
| RNA2 | Mutations in RNA2 | Amino acid substitution in 2a protein | RNA2 infectivity (%)a |
|---|---|---|---|
| wt | 100 | ||
| MF1 | G179 to C | Glu26 to Gln | 0 |
| G182 to A | Asp27 to Asn | ||
| G185 to C | Glu28 to Gln | ||
| MF2 | G215 to C | Glu38 to Gln | 100 |
| G224 to A | Asp41 to Asn | ||
| MF3 | G443 to C | Glu114 to Gln | 0 |
| G449 to C | Glu116 to Gln | ||
| G458 to A | Asp119 to Asn | ||
| G470 to A | Glu123 to Lys | ||
| MF4 | G470 to C | Glu123 to Gln | 0 |
| G473 to A | Asp124 to Asn | ||
| G480 to A | Ser126 to Asn | ||
| G482 to A | Asp127 to Asn | ||
| MF5 | G509 to A | Asp136 to Asn | 75 |
| G512 to C | Glu137 to Gln | ||
| G515 to A | Asp138 to Asn |
Infectivity is defined as the percentage of local lesions relative to that in wt infection. Four C. quinoa plants were inoculated, and the total numbers of lesions were compared.
The effects of MF mutations on RNA replication and virus infectivity were tested by inoculating BMV transcripts into a local-lesion host of BMV, C. quinoa. For these inoculations, wt RNA1 and recombinationally active RNA3 derivatives were mixed with either wt RNA2 or mutant MF RNA2. In this system the mutated RNA3 itself, without the need for recombination, can support the formation of local lesions and the number of local lesions does not depend on the frequency of recombination (24, 25). Fourteen days after inoculation, the number of the BMV-characteristic local lesions was counted and compared relative to that of the positive control, as presented in Table 1. We found that MF1, MF3, and MF4 did not produce any lesions but that MF2 and MF5 generated, respectively, 100 and 75% of the number of lesions obtained with the wt RNA2.
To determine whether mutations in MF2 and MF5 RNA2s were maintained during infection, total RNAs were extracted from 10 separate local lesions per infection and the corresponding RNA2 region, which contained the MF mutations, was amplified by RT-PCR with RNA2-specific primers. The cDNAs were cloned individually and sequenced. The results showed that the mutations in MF2 and MF5 were stably maintained at least 2 weeks after inoculation (data not shown).
The effect of MF mutations on 1a-2a interactions.
Whether mutations in the acidic residues of 2a will affect the 1a-2a protein-protein interaction was tested by the yeast two-hybrid assay (31). For these assays, DNA encoding the N-terminal 140 residues of wt or mutant 2a were fused to DNA encoding the GAL4 activation domain in a LEU2+ plasmid (see Materials and Methods). The 1a helicase-like sequence was fused with the LexA protein in a TRP1+ plasmid, p1aH (31). Plasmids were then transformed into yeast, and the interactions between both proteins were detected and quantified as previously described (31). The levels of specific activity obtained for MF1, MF2, and MF5 were similar to that of the wt sequence (Table 2), but there was no detectable interaction obtained for MF3 and MF4 with the 1a helicase-like domain. This demonstrates that the 2a amino acids within residues 114 and 127 are important for 1a-2a interaction. Interestingly, MF1 was not infectious despite the wt level of 1a-2a interaction. Apparently, factors other than the level of 1a-2a interaction are also important for virus infectivity.
TABLE 2.
Effects of MF mutations on the interaction between BMV 1a and 2a proteins in the yeast two-hybrid system
| Tested sample(s) | Sp acta | Relative activityb |
|---|---|---|
| 1a | 3.28 | 1.00 |
| 2a | 3.00 | 0.91 |
| MF1 | 4.10 | 1.25 |
| MF2 | 5.30 | 1.62 |
| MF3 | 3.00 | 0.91 |
| MF4 | 3.40 | 1.05 |
| MF5 | 3.90 | 1.19 |
| 1a + 2a | 166.0 | 50.60 |
| 1a + MF1 | 168.5 | 51.40 |
| 1a + MF2 | 164.5 | 50.20 |
| 1a + MF3 | 4.4 | 1.34 |
| 1a + MF4 | 6.3 | 1.90 |
| 1a + MF5 | 170.0 | 51.80 |
Micromoles of o-nitrophenol galactoside produced per milligram of protein lysate per minute.
Relative activity is relative to the specific activity of the yeast strain harboring only the plasmid bearing the gene expressing 1a protein.
The effect of MF mutations on the frequency of recombination.
Since MF2 and MF5 were infectious, they were used for further studies of RNA recombination. For this reason, two previously elaborated in vivo recombination systems were employed (Fig. 2). We have found previously that, with both systems, practically all local lesions accumulated only one type (though different types among lesions) of recombinant.
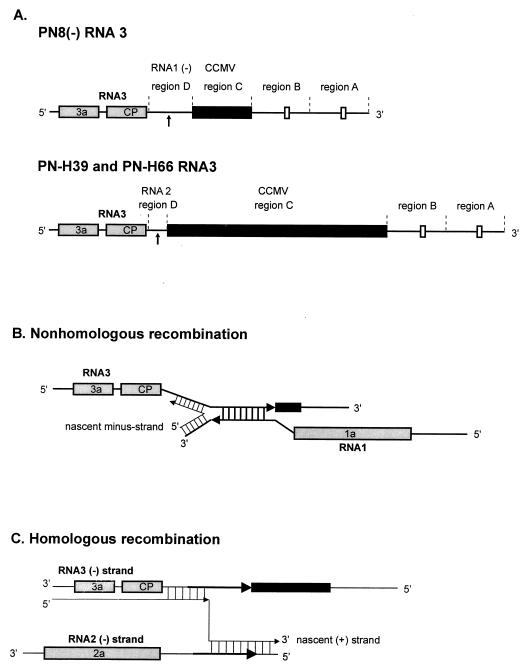
FIG. 2.
Schematic diagrams (not to scale) of BMV RNA3 recombination vectors used in this study. (A) General organizations of PN8(−) RNA3 (used for the study of nonhomologous recombination) and PN-H39 and PN-H66 RNA3 (used for the study of homologous recombination). Each RNA3 consists of four regions, A, B, C, and D. In PN8(−) RNA3, region A is 216 nt long and consists of the conserved BMV 3′ end (positions 1 to 236 from the 3′ end, counting an internal 20-nt deletion [see below]), and region B constitutes a partial duplication of region A between positions 7 and 200. Additionally, both regions A and B have 20-nt deletions between positions 81 and 100 (marked with small boxes). The 197-nt region C (black box) is derived from all but the last 23 nt of the 3′-terminal sequence of cowpea chlorotic mottle virus (CCMV) RNA3, and region D is the 141-nt sequence complementary to the RNA1 3′ noncoding fragment between positions 249 and 389 (counted from the 5′ end). PN-H39 and PN-H66 RNA3s have the same regions A and B as PN8(−) RNA3. However, their region C has a 765-nt-long CCMV RNA3 sequence (positions 24 to 788, counted from the 3′ end). Also, region D includes wt RNA2 sequence between positions 160 and 219 (for PN-H39) or between positions 197 and 219 (for PN-H66). (B) During nonhomologous (heteroduplex-mediated) recombination, PN8(−) RNA3 and wt RNA1 can form a heteroduplex structure. The proposed mechanism of nonhomologous recombination assumes that crossovers take place during minus-strand synthesis initially of RNA1 and that when viral replicase reaches the stable double-stranded region, the enzyme complex switches from one RNA template to another. Repaired recombinant RNA3 is generated if replicase starts RNA synthesis from the nonmodified RNA1 3′ end and after crossover resumes nascent-strand elongation with RNA3 as a template. (C) The PN-H39 and PN-H66 RNA3-based vectors, which have 23 to 60 nt of sequence homology with wt RNA2, can efficiently direct homologous RNA2-RNA3 crossovers. In such instances, the generated recombinant has a nonmodified 3′ end derived from RNA2 and the coding region and noncoding 5′ end derived from RNA3. According to a proposed mechanism (29, 30), the enzyme complex initiates on the minus-strand RNA3 template and continues the synthesis of the nascent (plus) strand on the RNA2 template. CP, coat protein.
Besides wt RNA1 and either wt or MF RNA2, the recombinationally active RNA3 derivatives were used in both recombination systems. For nonhomologous recombination, a PN8(−) RNA3 derivative that included a 141-nt-long sequence complementary to wt RNA1 was used (Fig. 2A). These sequences could form an extended imperfect heteroduplex structure (ensured by mismatches at three sites of the heteroduplex) between PN8(−) RNA3 and wt RNA1. The mismatches were introduced to destabilize the heteroduplex structure so that the crossovers could occur at the inner parts of this double-stranded region (25). PN8(−) RNA3 supported recombinant RNA formation in 78% of local lesions (Table 3), when it was coinoculated with wt RNA1 and wt RNA2. (The results of Table 3 show the averaged data of three inoculation experiments. The observed frequencies in each experiment were consistent within a 5% range.) The crossovers are concentrated within the stable, left-side-proximal part of the heteroduplex (as displayed in Fig. 3A). For homologous recombination, PN-H39 and PN-H66 RNA3 derivatives were employed. These constructs have, respectively, 60- and 23-nt-long sequences homologous with wt RNA2 (Fig. 2A) and in previous experiments supported homologous recombination in 83 and 53% of local lesions, respectively (Table 3), with crossovers occurring between the homologous or similar sequences of RNA2 and RNA3 during infection with wt RNA1 and -2 (reference 26 and Fig. 4). (Based on our data and considerations described previously [27]), a 20% difference in such defined recombination frequencies can be considered statistically significant).
TABLE 3.
Influence of MF2 and MF5 mutations on nonhomologous [parental PN8(−) RNA3] and homologous (parental PN-H39 and PN-H66 RNA3s) recombination frequencies
| RNA3 derivative | RNA2 | No. of samples analyzed | No. of recombinants identified | Recombination frequency (%) |
|---|---|---|---|---|
| PN8(−) | wt | 36 | 28 | 78 |
| MF2 | 50 | 3 | 6 | |
| MF5 | 40 | 6 | 15 | |
| PN-H39 (60-nt homology) | wt | 36 | 30 | 83 |
| MF2 | 36 | 23 | 64 | |
| MF5 | 36 | 31 | 86 | |
| PN-H66 (23-nt homology) | wt | 36 | 19 | 53 |
| MF2 | 36 | 6 | 17 | |
| MF5 | 36 | 22 | 61 |
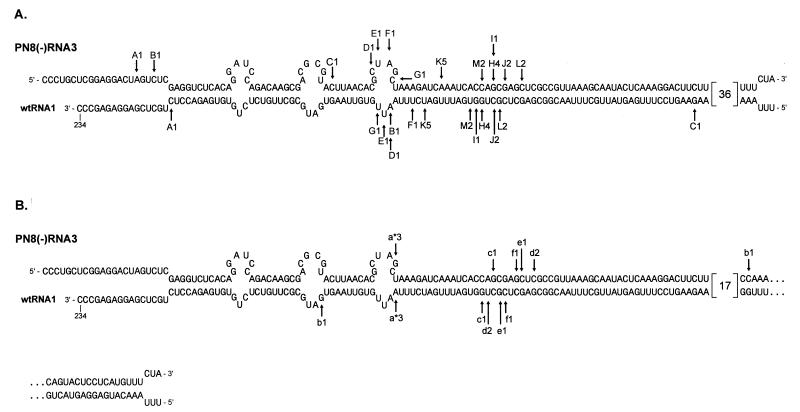
FIG. 3.
Distribution of crossover sites in nonhomologous recombinants generated between PN8(−) RNA3 and nonmutated RNA2 (A) or with MF2 and MF5 RNA2 mutants (B). Each recombinant was isolated from a separate local lesion. The upper lines of sequences represent the positive strand of PN8(−) RNA3 that contains the RNA1 sequences of negative polarity (segment D), while the lower lines represent the corresponding (complementary) sequences of the positive strand of wt RNA1. Junctions are marked by arrows with letters (uppercase letters for wt RNA2 and lowercase letters for MF mutants). Asterisks indicate recombinants found with the MF2 mutant. The numbers show how many recombinants of a given type have been identified. The numbers 36 and 17 in large brackets indicate the numbers of bases in unknown wt BMV sequences.
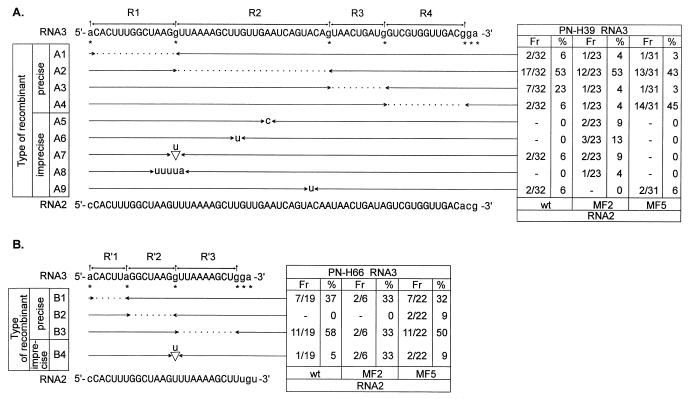
FIG. 4.
Diagram summarizing the recombination frequencies (Fr) and distribution of crossover sites in homologous recombinants. (A) Locations of crossover sites and of marker nucleotide substitutions within the common sequence in homologous RNA2-RNA3 recombinants isolated from infections with wt RNA1; either wt, MF2, or MF5 RNA2; and PN-H39 RNA3 (A) and with PN-H66 RNA3 (B). Uppercase letters indicate homologous nucleotides. Marker mutations are indicated by asterisks, whereas regions between marker mutations are named R1 to R4 or R′1 to R′3. Each recombinant contains 3′-terminal sequences derived from RNA2 on the right side and 5′-terminal sequences from RNA3 on the left side. The incidence of each RNA3 recombinant is shown in the boxes immediately to the right of the arrows (the Fr columns give the numbers of particular recombinants over the total numbers of identified recombinants, and the % columns give those values expressed as percentages). Each entry represents an RNA2-RNA3 recombinant isolated from a separate local lesion. The names of individual recombinants are shown on the left. Each leftward-pointing arrowhead marks the last nucleotide derived from RNA2, and each rightward-pointing arrowhead marks the first nucleotide derived from RNA3. Dotted lines show ambiguous regions that may be derived from either RNA2 or RNA3 in the precise homologous recombinants. Nontemplated nucleotides and nucleotide substitutions generated during the crossover events are shown by lowercase letters between the arrows. The nucleotide sequences in the imprecise recombinants with ambiguous crossovers were arbitrarily placed with the upstream junction.
The above-described system was used in this work to analyze the effect of MF2 and MF5 mutants on homologous and nonhomologous recombination in C. quinoa. Since the RNA3 recombinants contained a 3′ noncoding region shorter than those of the parental RNA3 molecules, their presence could be determined by the sizes of the obtained PCR cDNA products (24). Identical RT-PCRs were performed with the in vitro-transcribed inoculation mixture and used as a control to guard against recombinants generated during RT-PCR amplifications. To ensure that the multiple RT-PCRs (see Materials and Methods) reflected real BMV RNA species existing in the local lesions, the extracted total RNAs were analyzed by Northern blot hybridization. These assays demonstrated the presence of recombinant BMV RNAs in local lesions (as reflected by the size of the RNA3 component [data not shown]). Also, these Northern blot analyses revealed that the overall levels of the accumulated BMV RNAs in local lesions were similar for MF2, MF5, and the wt virus (data not shown). We conclude that the identified recombinants were not RT-PCR artifacts but that they were formed by recombination during BMV infection in planta.
Compared to the 78% (Table 3) frequency of nonhomologous recombination observed with wt RNA1, wt RNA2, and PN8(−) RNA3 (24), MF2 and MF5 RNA2s recombined with markedly decreased frequencies of, respectively, 6 and 15% (Table 3). For homologous recombination, PN-H39 RNA3 underwent recombination at frequencies of 83 and 86% with wt and MF5 RNA2s, respectively, but at only 64% with MF2. Reduced frequencies of homologous recombination for MF2 were more pronounced with PN-H66 RNA3, where the corresponding percentages were 53% with wt RNA2, 17% with MF2, and 61% with MF5 (Table 3).
The effect of MF mutations on the distribution of crossover sites.
Locations of the crossover sites were determined by sequencing of the corresponding RT-PCR-generated cDNA clones. With the nonhomologous recombinants it was possible to determine precisely the locations of the junction sites. Sequencing revealed that the nonhomologous junction sites obtained with MF2 and MF5 occurred within the same portion of the heteroduplex as those obtained with wt RNA2 (Fig. 3). For MF2 RNA2 we found only three recombinants. Interestingly, all of them were identical even though they were from independent infections. At present we do not know whether this is fortuitous, reflects changes in overall selection pressure, or is an effect of the particular 2a mutation on selection of crossover sites. For MF5 RNA2, six recombinants were identified; five of them were located within the recombination hotspot region, while only one recombinant (b1) was highly asymmetrical, i.e., the crossovers occurred at distant locations on both recombining RNA components (Fig. 3).
In homologous recombination, the locations of junction sites could be assigned only to (short) regions enclosed within marker mutations. The results, presented in Fig. 4, reveal that the inhibition of recombination frequency, which was observed for MF2 RNA2 (Table 3), did not correlate with any significant effects on the locations of crossover sites in either the PN-H39 or the PN-H66 RNA3 derivative. However, homologous crossovers were affected with MF5 RNA2, especially during infection with wt RNA1 and PN-H39 RNA3. Here, the most frequently identified recombinants belonged to the A4 type (14 recombinants, i.e., 45%), whereas infections with wt and MF2 RNA2 resulted in 6 and 4%, respectively, of recombinants with junction sites located within region R4. A shift in crossover location with MF5 was less obvious with PN-H66 RNA3 (which has a sequence homologous to RNA2 shorter than that in MF5 RNA3). However, we could still observe that during infection with MF5 RNA2, two recombinants with junction sites within region R′2 (type B2 in Fig. 4B) were formed. These novel recombinant types were not observed for wt or MF2 RNA2.
Precision of homologous recombination.
Analysis of recombinant sequences allowed us to determine whether mutations introduced into the N terminus of 2a protein can influence the precision of homologous crossovers. Four imprecise homologous recombinants (types A5 to A9) were identified during infection with wt RNA1, wt RNA2, and PN-H39 RNA3, while one imprecise recombinant (type B4) was observed with PN-H66 RNA3 (Fig. 4). Imprecise recombinants constituted 12 and 5% of the total number of recombinants generated with PN-H39 and PN-H66, respectively. Similar results were obtained with MF5 RNA2. Here, two imprecise recombinants were obtained with PN-H39 RNA3 and two others were obtained with PN-H66 RNA3 (respectively, 6 and 9% of identified recombinants). The situation markedly changed if, instead of wt RNA2, MF2 RNA2 was used. MF2 RNA2 gave eight (35%) imprecise recombinants when it was used with wt RNA1 and PN-H39 RNA3 (Fig. 4A) or 2 (33%) imprecise recombinants with wt RNA1 and PN-H66 RNA3 (Fig. 4B). As with precise recombinants, MF2 mutation (Fig. 4A) did not influence the locations of imprecise crossover sites for PN-H39 RNA3 (situated within region R2).
DISCUSSION
Several lines of evidence demonstrate that RNA recombination in BMV is a process intimately associated with viral RNA synthesis. Mutations in the BMV-encoded 1a protein, and especially a temperature-sensitive mutation in its helicase-like domain, were shown to influence the frequency and positions of nonhomologous crossovers (27). Also, we demonstrated previously that a single amino acid substitution (designated DR7) within the central core polymerase domain of 2a can inhibit nonhomologous RNA-RNA recombination (12). DR7 also influenced the location but not the frequency of homologous crossovers, suggesting that homologous and nonhomologous recombination are the result of different mechanisms.
To assess the role of various functional domains on BMV replicase proteins in recombination, in this work we study mutations within the N-terminal domain of 2a. Our findings have several implications. First, we show that replacements of selected clusters of negatively charged amino acids into their neutral counterparts reduce the levels of 1a-2a interactions. The N-terminal domain has been shown previously to participate in 1a-2a interactions (15, 16). Our data not only map the participating regions of the N terminus but also suggest that these interactions have an electrostatic character. Also, the role of 1a-2a interactions in virus infectivity is confirmed by observing a correlation between the presence of negatively charged amino acids and the electrostatic character of the regions in which they are found.
Second, one purpose of our study was to test whether 1a-2a interactions were involved in guiding recombination events. We expected to observe a quantitative correlation between levels of 1a-2a interaction and recombination. However, of the two infectious MF mutations, neither displayed conditional properties. Thus, it was not possible to test the influence of the 1a-2a interaction on recombination, even though it was possible to test that on replication. Further mutagenesis efforts are necessary.
Third, we now confirm a previously reported property of DR7 (see above), namely, that it is possible to decouple replication and recombination events. While the replication levels of MF2 and MF5 reach those of the wt virus, the ability of both mutants to support crossovers is deeply reduced. Apparently, there are some regions within the viral RNA polymerase which are responsible for recombinant generation and at the same time are not involved in RNA replication. By studying the progeny BMV RNAs obtained with mutants of different recombination activities, one should be able to determine the role of recombination in maintaining viral genomic sequences during infection.
Fourth, it is certain that decoupling of replication and recombination functions can be achieved not only by mutagenizing directly the putative catalytic domain of RNA polymerase (mutant DR7) but also by changing more distant regions of the 2a protein. This finding suggests that the N-terminal domain is directly involved in the process of RNA recombination. Alternatively, indirect steric effects of N-terminal mutations on the active centers of the enzyme cannot be excluded.
Fifth, MF2 and MF5 display markedly reduced frequencies of nonhomologous recombination, and, in addition, MF2 shows a reduced frequency of homologous recombination. Mutant DR7 can affect only the frequency of nonhomologous recombination (12). Thus, we show that different sites on 2a participate in the two kinds of recombination events during the virus life cycle. Our data further confirm that the mechanisms responsible for both types of recombination are different.
Considering these findings together, we have shown that the N-terminal domain has a multifunctional character, as it can influence the levels of RNA replication, the levels of nonhomologous RNA recombination, and the levels of homologous RNA recombination. Whether this domain has other functions and whether or how all these functions are interrelated remain to be determined. Below we discuss more details regarding the role of the N-terminal domain in 1a-2a interaction and in RNA recombination.
Role of the 2a N terminus in 1a-2a interaction.
It was shown previously by using both the in vitro and the yeast two-hybrid system (11, 15, 16, 30a, 31) that the nonconserved 2a N terminus is at least partially responsible for interaction with 1a through the helicase domain. In this work we confirm the involvement of this domain in 1a-2a interactions. Specifically, we observe that the acidic residues within positions 114 and 127 are important for 1a-2a interaction. However, since MF1, MF2, and MF5 did not affect 1a-2a interactions while MF3 and MF4 did, we conclude that acidic amino acids located in regions both downstream and upstream from MF3 and MF4 sites are not needed for interaction with 1a. Other data show that single acidic amino acid changes within residues 70 and 124 can abolish 1a-2a interaction in the yeast two-hybrid system (38). Residues 26 to 28, which are modified in MF1, are known to be dispensable for 1a-2a interaction (31), and deletions in this region can reduce, but do not abolish, RNA replication in barley protoplasts (39). It is then interesting that MF1 is not infectious. Perhaps this mutation affects directly the structure of 2a (or indirectly the structure of 1a), which can then affect the RNA replication process. Thus, MF1 mutation may cause reduction of putative interactions of negatively charged amino acids with the phosphate backbones of replicating RNAs, which are mediated by metal cations (37).
Recent work of O’Reilly et al. (30a, 31) demonstrates with the yeast two-hybrid system that 2a, in order to interact with the helicase domain of 1a, has to compete with intramolecular interactions between the helicase and capping domains of 1a. Also, 1a can apparently interact with another 1a molecule through their capping domains (31a). Data obtained with the two-hybrid system have been shown to correlate well with viral replication in barley protoplasts (31). The nature of all the above-described interactions is not known. A counting of charged amino acids shows that the N-terminal domain of 2a contains more acidic amino acids than that of 1a and that the helicase domain of 1a has more basic residues, suggesting the electrostatic nature of 1a-2a interactions. Almost total elimination of the 1a-2a interaction in MF3 and MF4 mutants seems to confirm that both MF variants contain drastic substitutions of three to four amino acid clusters, which changes them into neutral analogues. Perhaps, then, MF3 and MF4 2a proteins cannot compete effectively for binding with the intramolecular interactions within 1a.
A recent publication by Smirnyagina et al. (36) suggests that the N terminus is dispensable for RNA replication, in contrast to the previously published in vivo replication results of Traynor et al. (39). O’Reilly et al. (31) demonstrated that an additional domain of 1a-2a interaction exists in the 2a central domain. Perhaps the overexpression of viral proteins in a DNA-mediated system with a strong 35S promoter, used by Smirnyagina et al. (36), abrogated the need for the 2a N terminus.
The role of the N-terminal domain in recombination.
Mutants MF2 and MF5 altered the frequency of nonhomologous recombination, but no effect on the locations of junction sites was observed. We speculate that such properties of the MF2 and MF5 mutants in nonhomologous recombination can be explained by a mechanism presented in Fig. 2, according to which the existence of a strong heteroduplex (140 nt long) that forms between wt RNA1 and PH8(−) RNA3 may ensure that the crossovers occur at specific locations (12, 25). This might explain the observations that the locations of crossovers were affected only by mutations within the helicase domain of 1a (27) and not within the domains of 2a (this paper and reference 12). Mutated helicase may have lower efficiency in unwinding the double-stranded RNA, which, in turn, may cause the changes in the positions of crossovers (27). On the other hand, the stability of the overall recombination complex (designated the recombinosome complex [see references 12 and 29]), which is formed by viral replicase, the RNA template (donor), the nascent strand, and the donor-associated acceptor RNA, might be important for the ability of both viral replicase and the nascent RNA strand to approach the acceptor RNA molecule during the switching event. This, in turn, may affect the frequency of crossover events at heteroduplex structures. Accordingly, mutants MF2 and MF5 are expected to influence the stability of the recombinosome complex. At present, we cannot unambigously determine whether changes introduced to the 2a N-terminal domain affect protein-protein or protein-RNA interactions. The data obtained with the yeast two-hybrid system may indicate that protein-RNA binding is disrupted, as both MF2 and MF5 mutants display the wt level of 1a-2a interaction.
As far as homologous recombination is concerned, MF2 altered the precision of homologous crossovers with both PN-H39 and PN-H66 RNA3. This might reflect an increased level of addition of nontemplated bases by MF2 replicase and/or reduction of precise alignment between the nascent and the acceptor strands during the crossover events (28, 34). Unlike MF2, MF5 did not influence either the precision or the frequency of homologous crossovers. However, the locations of homologous crossover sites were affected: the replacement of wt RNA2 with MF5 RNA2 (in combination with wt RNA1 and PN-H39 RNA3) caused an even distribution of crossovers within regions R2 (42%) and R4 (45%). The observation that MF2 decreased the precision and the frequency of RNA recombination and MF5 caused a significant shift in the locations of crossover sites during homologous recombination may be the consequence of changes in protein-RNA interactions. For instance, according to a proposed model of homologous recombination (30), the frequency and the precision of recombination and the locations of the crossovers can be altered as a result of the (i) instability of the replicase-nascent strand complex, (ii) weaker replicase-RNA template interaction, (iii) reduced ability of the replicase–nascent-strand complex to “land” on the acceptor RNA molecule and to reinitiate nascent-strand elongation, and (iv) reduced ability of the replicase–nascent-strand complex to leave the donor RNA molecule. Modifications in these functions might affect recombination events directly or might cause subtle alterations in levels of RNA synthesis, which then might influence recombination profiles. The determination of the crystal structure of 2a should help us significantly in understanding how particular regions of 2a participate in recombination.
ACKNOWLEDGMENTS
We thank Stuart MacFarlane and the Cereal Killers of Indiana University for helpful comments and discussions.
M.F., P.D.N., and J.J.B. were supported by grants from the National Institute of Allergy and Infectious Diseases (3RO1 AI26769), the National Science Foundation (MCB-9630794), the U.S. Department of Agriculture (96-39210-3842), and the Plant Molecular Biology Center at Northern Illinois University. In addition, M.F. was supported by the Polish government through a grant (6P04A 02712) from the State Committee for Scientific Studies. N.T. and C.C.K. were supported by the National Science Foundation (MCB-9507344).
REFERENCES
- 1.Ahlquist P. Bromovirus RNA replication and transcription. Curr Opin Genet Dev. 1992;2:71–76. doi: 10.1016/s0959-437x(05)80325-9. [DOI] [PubMed] [Google Scholar]
- 2.Ahlquist P, Bujarski J J, Kaesberg P, Hall T C. Localization of replicase recognition site within brome mosaic virus RNA by hybrid arrested RNA synthesis. Plant Mol Biol. 1985;3:37–44. doi: 10.1007/BF00023414. [DOI] [PubMed] [Google Scholar]
- 3.Becker D M, Guarante L. High efficiency transformation of yeast by electroporation. Methods Enzymol. 1991;194:186–210. doi: 10.1016/0076-6879(91)94015-5. [DOI] [PubMed] [Google Scholar]
- 4.Buck K W. Comparison of the replication of positive-stranded RNA viruses of plants and animals. Adv Virus Res. 1996;47:159–251. doi: 10.1016/S0065-3527(08)60736-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bujarski J J. Genetic RNA recombination and defective RNA formation in RNA viruses. Semin Virol. 1996;7:361–362. [Google Scholar]
- 6.Bujarski J J, Kaesberg P. Genetic recombination in a multipartite plant virus. Nature. 1986;321:528–531. doi: 10.1038/321528a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bujarski J J, Dreher T W, Hall T C. Deletion in the 3′-terminal tRNA-like structure of brome mosaic virus RNA differentially affects aminoacylation and replication in vitro. Proc Natl Acad Sci USA. 1985;82:5636–5640. doi: 10.1073/pnas.82.17.5636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bujarski J J, Nagy P D. Genetic RNA-RNA recombination in positive-stranded RNA viruses of plants. In: Paszkowski J, editor. Homologous recombination in plants. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1994. pp. 1–24. [Google Scholar]
- 9.Bujarski J J, Nagy P D, Flasinski S. Molecular studies of genetic RNA-RNA recombination in brome mosaic virus. Adv Virus Res. 1994;43:275–302. doi: 10.1016/S0065-3527(08)60051-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cascone P J, Carpenter C D, Li X H, Simon A E. Recombination between satellite RNAs of turnip crinkle virus. EMBO J. 1990;9:1709–1715. doi: 10.1002/j.1460-2075.1990.tb08294.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fields S, Sternglanz R. The two-hybrid system: an assay for protein-protein interactions. Trends Genet. 1994;10:286–292. doi: 10.1016/0168-9525(90)90012-u. [DOI] [PubMed] [Google Scholar]
- 12.Figlerowicz M, Nagy P D, Bujarski J J. A mutation in the putative RNA polymerase gene inhibits nonhomologous, but not homologous, genetic recombination in an RNA virus. Proc Natl Acad Sci USA. 1997;94:2073–2078. doi: 10.1073/pnas.94.5.2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Janda M, French R, Ahlquist P. High efficiency T7 polymerase synthesis of infectious RNA from cloned brome mosaic virus cDNA and effects of 5′ extensions of transcript infectivity. Virology. 1987;158:259–262. doi: 10.1016/0042-6822(87)90265-0. [DOI] [PubMed] [Google Scholar]
- 14.Jarvis T C, Kirkegaard K. The polymerase in its labyrinth: mechanisms and implications of RNA recombination. Trends Genet. 1991;7:186–191. doi: 10.1016/0168-9525(91)90434-R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kao C C, Ahlquist P. Identification of the domains required for direct interaction of the helicase-like and polymerase-like RNA replication proteins of brome mosaic virus. J Virol. 1992;66:7293–7302. doi: 10.1128/jvi.66.12.7293-7302.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kao C C, Quadt R, Hershberger R P, Ahlquist P. Brome mosaic virus RNA replication proteins 1a and 2a form a complex in vitro. J Virol. 1992;66:6322–6329. doi: 10.1128/jvi.66.11.6322-6329.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khatchikian D, Orlich M, Rott R. Increased viral pathogenicity after insertion of a 28S ribosomal RNA sequence into the haemagglutinin gene of an influenza virus. Nature. 1989;340:156–157. doi: 10.1038/340156a0. [DOI] [PubMed] [Google Scholar]
- 18.King A M Q, McCahon D, Saunders K, Newtoman J W I, Slade W R. Multiple sites of recombination within the RNA genome of foot-and-mouth disease virus. Virus Res. 1985;3:373–384. doi: 10.1016/0168-1702(85)90437-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kirkegaard K, Baltimore D. The mechanism of RNA recombination in poliovirus. Cell. 1986;47:433–443. doi: 10.1016/0092-8674(86)90600-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lai M C M. RNA recombination in animal and plant viruses. Microbiol Rev. 1992;56:61–79. doi: 10.1128/mr.56.1.61-79.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luytjes W, Bredenbeek P J, Noten A F H, Horzinek M C, Spaan W J. Sequence of mouse hepatitis virus A59 mRNA: indications for RNA recombination between coronavirus and influenza C virus. Virology. 1988;166:415–422. doi: 10.1016/0042-6822(88)90512-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.MacCahon D. The genetics of aphthoviruses. Arch Virol. 1981;69:1–23. doi: 10.1007/BF01315261. [DOI] [PubMed] [Google Scholar]
- 23.Mayers G, Tautz N, Dubovi E J, Thiel H-J. Viral cytopathogenicity correlated with integration of ubiquitin-coding sequence. Virology. 1991;180:602–616. doi: 10.1016/0042-6822(91)90074-L. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nagy P D, Bujarski J J. Genetic recombination in brome mosaic virus: effect of sequence and replication of RNA on accumulation of recombinants. J Virol. 1992;66:6824–6828. doi: 10.1128/jvi.66.11.6824-6828.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nagy P D, Bujarski J J. Targeting the site of RNA-RNA recombination in brome mosaic virus with antisense sequences. Proc Natl Acad Sci USA. 1993;90:6390–6394. doi: 10.1073/pnas.90.14.6390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nagy P D, Bujarski J J. Efficient system of homologous RNA recombination in brome mosaic virus: sequence and structure requirements and accuracy of crossovers. J Virol. 1995;69:131–140. doi: 10.1128/jvi.69.1.131-140.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nagy P D, Dzianott A, Ahlquist P, Bujarski J J. Mutations in the helicase-like domain of protein 1a alter the sites of RNA-RNA recombination in brome mosaic virus. J Virol. 1995;69:2547–2556. doi: 10.1128/jvi.69.4.2547-2556.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nagy P D, Bujarski J J. Homologous RNA recombination in brome mosaic virus: AU-rich sequences decrease the accuracy of crossovers. J Virol. 1996;70:415–426. doi: 10.1128/jvi.70.1.415-426.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nagy P D, Bujarski J J. Engineering of homologous recombination hotspots with AU-rich sequences in brome mosaic virus. J Virol. 1997;71:3799–3810. doi: 10.1128/jvi.71.5.3799-3810.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nagy P D, Bujarski J J. Different mechanisms of homologous and nonhomologous recombination in brome mosaic virus. Semin Virol. 1996;7:363–372. [Google Scholar]
- 30a.O’Reilly E, Tang N, Ahlquist P, Kao C. Biochemical and genetic analysis of the interaction between the helicase-like and polymerase-like proteins of the brome mosaic virus. Virology. 1995;214:59–71. doi: 10.1006/viro.1995.9954. [DOI] [PubMed] [Google Scholar]
- 31.O’Reilly E K, Paul J D, Kao C C. Analysis of the interaction of viral RNA replication proteins by using the yeast two-hybrid assay. J Virol. 1997;71:7526–7532. doi: 10.1128/jvi.71.10.7526-7532.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31a.O’Reilly E K, Wang Z, French R, Kao C C. Interactions between the structural domains of the RNA replication proteins of plant-infecting RNA viruses. J Virol. 1998;72:7160–7169. doi: 10.1128/jvi.72.9.7160-7169.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Quadt R, Kao C C, Browning K S, Hershberger R P, Ahlquist P. Characterization of a host protein associated with brome mosaic virus RNA-dependent RNA polymerase. Proc Natl Acad Sci USA. 1993;90:1498–1502. doi: 10.1073/pnas.90.4.1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Romanova L I, Blinov V M, Tolskaya E A, Victorova E G, Kolesnikova M S, Guseva L A, Agol V I. The primary structure of crossover regions of intertypic poliovirus recombination: a model of recombination between RNA genomes. Virology. 1986;155:202–213. doi: 10.1016/0042-6822(86)90180-7. [DOI] [PubMed] [Google Scholar]
- 34.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 35.Simon A E, Bujarski J J. RNA-RNA recombination and evolution in virus infected plants. Annu Rev Phythopathol. 1994;32:337–362. [Google Scholar]
- 36.Smirnyagina S, Lin N-S, Ahlquist P. The polymerase-like core of brome mosaic virus 2a protein lacking a region interacting with viral 1a protein in vitro maintains activity and selectivity in RNA replication. J Virol. 1996;70:4729–4736. doi: 10.1128/jvi.70.7.4729-4736.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Steitz T. Structural studies of protein-nucleic acid interaction: the source of sequence-specific interaction. Q Rev Biophys. 1990;23:205–280. doi: 10.1017/s0033583500005552. [DOI] [PubMed] [Google Scholar]
- 38.Tang, N., and C. C. Kao. Unpublished results.
- 39.Traynor P, Young B M, Ahlquist P. Deletion analysis of brome mosaic virus 2a protein: effects on RNA replication and systemic spread. J Virol. 1991;65:2807–2815. doi: 10.1128/jvi.65.6.2807-2815.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]