Abstract
The human immunodeficiency virus type 1 RNA genome contains a terminal repeat (R) sequence that encodes the TAR hairpin motif, which has been implicated in Tat-mediated activation of transcription. More recently, a variety of other functions have been proposed for this structured RNA element. To determine the replicative roles of the 5′ and 3′ TAR hairpins, we analyzed multiple steps in the life cycle of wild-type and mutant viruses. A structure-destabilizing mutation was introduced in either the 5′, the 3′, or both TAR motifs of the proviral genome. As expected, opening of the 5′ TAR hairpin caused a transcription defect. Because the level of protein expression was not similarly reduced, the translation of this mRNA was improved. No effect of the 3′ hairpin on transcription and translation was measured. Mutations of the 5′ and 3′ hairpin structures reduced the efficiency of RNA packaging to similar extents, and RNA packaging was further reduced in the 5′ and 3′ TAR double mutant. Upon infection of cells with these virions, a reduced amount of reverse transcription products was synthesized by the TAR mutant. However, no net reverse transcription defect was observed after correction for the reduced level of virion RNA. This result was confirmed in in vitro reverse transcription assays. These data indicate that the 5′ and 3′ TAR motifs play important roles in several steps of the replication cycle, but these structures have no significant effect on the mechanism of reverse transcription.
Retroviral RNA genomes contain a sequence repeat (R) that forms the extreme 5′ and 3′ ends of the viral transcripts. This terminal repeat of the genome of human immunodeficiency virus type 1 (HIV-1) is 97 nucleotides in length and contains important cis elements for several steps in viral replication. The TAR RNA hairpin structure within R is important for optimal transcription from the viral promoter in the long terminal repeat (LTR). In particular, the upper part of the TAR structure has been shown to be important for binding of the viral Tat transactivator protein that triggers high-level expression through interaction with the cellular transcription machinery (12, 25, 38, 59, 60). The R region also encodes sequences that are important for polyadenylation of the viral transcripts (4, 18, 28). Whereas the TAR element is functional primarily in the context of the 5′ LTR promoter (41), the polyadenylation signals are used exclusively within the 3′ LTR context. A second structured motif is encoded by the R region, the poly(A) hairpin (10), which is also critical for efficient viral replication, probably at the level of RNA packaging (24, 46). Retroviruses also use the terminal repeat in the process of reverse transcription. This process is initiated near the 5′ end of the genome at the primer-binding site, and a DNA copy of the 5′ R region is synthesized (strong-stop minus-strand cDNA). Upon removal of the 5′ R template strand by RNase H action of reverse transcriptase (RT), this cDNA anneals to the 3′ R region and reverse transcription is resumed. It is currently unknown whether specific sequence or structure motifs within R are required for efficient strand transfer (13).
Although it is generally believed that the TAR element is a critical transcription motif that mediates the Tat response, there have been numerous reports of posttranscriptional effects exerted by this element. A translational component of Tat/TAR-mediated activation of gene expression has been reported initially (22). The 5′ TAR structure was also shown to interfere with mRNA translation in Xenopus oocytes (16, 17) and in cell-free assays (47, 53, 57), and this repression could be overcome by addition of the Tat protein. Two mechanistic explanations have been proposed for TAR-mediated repression of translation. First, the 5′-terminal TAR hairpin may inhibit translation in cis by interfering with the binding of translation initiation factors or ribosomes to the mRNA cap structure (47). Second, TAR may activate the double-stranded RNA-dependent kinase PKR (26, 50, 53). The activated form of this kinase phosphorylates and thereby inactivates the translation initiation factor eIF-2, causing inhibition of translation in trans.
Besides a role of TAR in viral transcription and translation, recent studies have suggested additional functions for the TAR motif, in particular the base-paired stem region, in the viral life cycle (42, 49). Detailed studies indicated that TAR is involved both in packaging of the RNA into viral particles (46) and in reverse transcription (32). Although the exact function of TAR in packaging of the viral genome remains to be determined, it is possible that TAR is part of the packaging signal that is recognized by the viral Gag protein during virion assembly. The packaging function of TAR was shown to be independent of the Tat protein (46). The exact role of TAR in reverse transcription is also presently unknown, but TAR could affect this mechanism at several levels. The TAR structure may affect the process of initiation of reverse transcription. Although binding of the tRNA3Lys primer onto the genomic RNA occurs at the primer-binding site that is located downstream of TAR, additional interactions between the tRNA primer and upstream viral RNA sequences have been proposed (1, 11, 36, 39, 58). In fact, there is some evidence from in vitro studies that deletions within 5′ R reduce the initiation of reverse transcription (3). Alternatively, the TAR hairpin in the RNA template may influence the process of elongation or the first strand transfer reaction. Finally, because it has been reported that the Tat protein can also stimulate reverse transcription (31), the role of TAR may be to tether Tat to the RT enzyme.
Thus, a pleiotropy of functions have been attributed to the TAR motif in a variety of experimental systems. The biologically most relevant assay system is that of the replicating virus, and the importance of TAR is underlined by the observation that mutations within TAR cause severe replication defects (29, 42, 49). However, this experimental system does not allow one to selectively study one of the two TAR motifs that are present at the extreme 5′ and 3′ ends of the viral genome. In particular, mutations introduced in 5′ TAR will be inherited in a dominant manner in both R regions (43). Likewise, 3′ TAR mutations will be lost after a single replication cycle. To gain a better understanding of the role(s) of the 5′ and 3′ TAR motifs, we examined transcription, translation, packaging, and reverse transcription properties of HIV-1 mutants with a mutated 5′ and/or 3′ TAR structure in single-cycle assays. Because replication of TAR-mutated viruses can be restored by the acquisition of additional nucleotide substitutions that restore base pairing of the TAR stem (30, 42), we also included such a revertant genome in this study.
MATERIALS AND METHODS
Cells and viruses.
SupT1 and C8166 T cells were grown in RPMI 1640 medium containing 10% fetal calf serum at 37°C and 5% CO2. Cells were split 1 in 10 twice a week. C33A cervix carcinoma cells (ATCC HTB31) (5) were grown as a monolayer in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum and modified Eagle’s medium nonessential amino acids at 37°C and 5% CO2. C33A cells were transfected by the calcium phosphate method. Cells were grown to 60% confluence in 24-well multidish plates or 75-cm2 tissue culture flasks. For transfection of the 24-well plates, 1 μg of DNA in 22 μl of water was mixed with 25 μl of 2× HBS (50 mM HEPES [pH 7.1], 250 mM NaCl, 1.5 mM Na2HPO4) and 3 μl of 2 M CaCl2. For transfection of 75-cm2 flasks, 40 μg of DNA in 880 μl of water was mixed with 1 ml of 2× HBS and 120 μl of 2 M CaCl2. The mixtures were incubated at room temperature for 20 min and subsequently added to the culture medium. The culture medium was changed after 16 h. The full-length molecular HIV-1 clone pLAI (48) was used to produce wild-type and mutant viruses. The construction of the 5′ and 3′ Xho+10 TAR mutation and selection of the revertant virus were described previously (42).
Isolation of virion and intracellular HIV-1 RNA.
C33A cells were transfected with the proviral clones. At 2 or 3 days after transfection, the medium was centrifuged at 2,750 × g for 5 min. Virion RNA was isolated from 300 μl of the virus-containing supernatant by incubation with 500 μg of proteinase K per ml in the presence of 1% sodium dodecyl sulfate (SDS) and 2.5 mM EDTA at 37°C for 30 min and extracted twice with phenol-chloroform-isoamyl alcohol (25:24:1). After addition of 10 μg of glycogen, the RNA was precipitated with 0.3 M Na-acetate (pH 5.2) and 70% ethanol at −20°C, centrifuged at 16,000 × g for 20 min, washed with 70% ethanol, and dried. The RNA was resuspended in 10 mM Tris-HCl (pH 7.5)–50 mM NaCl–10 mM MgCl2–1 mM dithiothreitol and incubated with 10 U of DNase I (RNase free; Boehringer Mannheim) per 100 μl at 37°C for 30 min to remove any contaminating DNA. After extraction with phenol-chloroform-isoamyl alcohol (25:24:1), the RNA was precipitated with 0.3 M Na-acetate and 70% ethanol. The RNA was pelleted at 16,000 × g for 20 min, washed with 70% ethanol, and dried. Pellets were resuspended in water and stored at −20°C.
Two days after transfection of C33A cells, total cellular RNA was isolated by the acid guanidinium thiocyanate-phenol-chloroform method (19). The RNA was incubated with DNase I (RNase free) and extracted with phenol-chloroform-isoamyl-alcohol (25:24:1) as described above for the virion RNA.
Quantification of viral RNA.
Virion and intracellular RNA was spotted onto nitrocellulose membranes (BA-S 85; Schleicher and Schuell) with a slot blot manifold and hybridized with a 32P-labeled HIV-1 gag-pol probe (PvuII fragment of pLAI, positions +691 to +2881, which is specific for unspliced HIV-1 RNA), as previously described (24). Hybridization signals were quantitated with a PhosphorImager (Molecular Dynamics). To verify the absence of contaminating DNA, RNA was incubated with 0.5 N NaOH at 55°C for 30 min prior to slot blotting. This resulted in a complete loss of the hybridization signals, indicating that the observed hybridization signals correspond exclusively to genomic RNA.
CA p24 levels.
Culture supernatant was heat inactivated (30 min at 56°C) in the presence of 0.05% Empigen-BB (Calbiochem, La Jolla, Calif.). The CA p24 concentration was determined by twin-site enzyme-linked immunosorbent assay (ELISA), as described previously (6).
Reverse transcription analysis upon infection of T cells.
Virus stocks were prepared by transfection of C33A cells. Three days after transfection, the culture medium (20 ml) was centrifuged at 2,750 × g for 30 min to remove cells. The virus-containing supernatant was subsequently filtered through a 0.45-μm-pore-size filter (Schleicher and Schuell) and stored at −70°C. Contaminating plasmid DNA used for transfection was digested by incubation with 100 U of DNase I (RNase free; Boehringer Mannheim) per ml and 10 mM MgCl2 at 37°C for 1 h. SupT1 or C8166 cells (8 × 106 in 5 ml of medium) were infected with the same amount of wild-type and mutant virus (250 ng of CA p24) for 1 h at 37°C. A control sample was placed on ice to block the infection (0-h sample). Viruses were removed from the cells by extensive washing. The cells were either harvested directly (1-h sample) or cultured for 2, 3, 4, and 20 h. Cells were pelleted by centrifugation at 2,750 × g for 4 min and washed with phosphate-buffered saline (10 mM Na-phosphate [pH 7.4], 150 mM NaCl). DNA was solubilized by resuspension of the cells in 10 mM Tris-HCl (pH 8.0)–1 mM EDTA–0.5% Tween 20. The sample was then incubated with 200 μg of proteinase K per ml at 56°C for 30 min and at 95°C for 10 min. Early reverse transcription products formed after the first strand transfer were amplified by PCR with a 5′ primer identical to nef sequences (NEF-B/X; positions 8601 to 8621) and a 3′ primer complementary to U5 sequences (CN1; positions 123 to 151). Late cDNA products were amplified by PCR with a 5′ primer identical to tat sequences (KV1; positions 5367 to 5385) and a 3′ primer complementary to env sequences (WS3; positions 6125 to 6144). PCR products were analyzed by agarose gel electrophoresis and Southern blotted onto a nylon membrane (Zeta probe; Bio-Rad). To quantitate the PCR products, the filters were hybridized with 32P-labeled HIV-1 probes. The filter with early PCR products was treated with an HIV-1 LTR probe (positions −454 to +381). The blot with late PCR signals was probed with an HIV-1 DNA fragment (positions 5821 to 6379), which was obtained by ClaI-Asp718 digestion of pcDNA3-Tat (55). Hybridization was performed in 0.5 M Na-phosphate (pH 7.2)–7% SDS–1 mM EDTA–50 μg of salmon testis DNA per ml at 65°C for 16 h. Membranes were washed in 40 mM Na-phosphate (pH 7.2)–1% SDS at 65°C, three times for 5 min and once for 15 min, and for 5 min in the same buffer without SDS at room temperature. Hybridization signals were quantitated with a PhosphorImager (Molecular Dynamics).
Reverse transcription on virion-extracted RNA genomes.
The virion RNA was isolated by proteinase K treatment and phenol extraction as described previously (23). Reverse transcription was initiated from the associated tRNA primer or from a DNA oligonucleotide primer, which was first annealed to the HIV-1 RNA. This C(N1) primer is complementary to nucleotides +123 to +151 of the viral RNA. Both the oligonucleotide and tRNA primer extension assays were described previously in detail (23).
RESULTS
Mutant and revertant TAR hairpins.
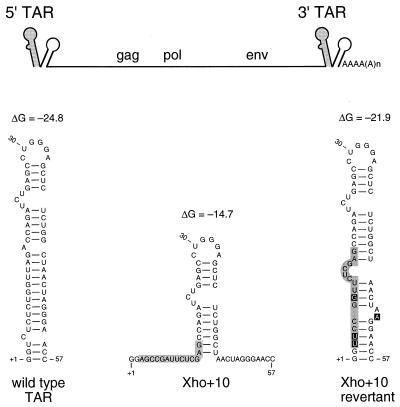
The 57 nucleotides at the 5′ end of the HIV-1 repeat region can fold the relatively stable TAR hairpin (Fig. 1). The lower TAR stem was opened in the Xho+10 mutant by substituting nucleotides +3 to +16 for an unrelated sequence with an XhoI restriction site (Fig. 1). This mutation was previously demonstrated to cause a severe virus replication defect (42). Long-term culturing of this virus resulted in the appearance of phenotypic revertants in which the base pairing of the lower TAR stem was restored by acquisition of additional mutations (Fig. 1). To characterize the replication defect of the Xho+10 virus, proviral plasmids were constructed in which the mutant TAR sequence was introduced into the 5′ LTR (5′ TAR mutant), the 3′ LTR (3′ TAR mutant), or in both LTRs (5′ + 3′ TAR double mutant). Furthermore, the revertant TAR sequence was introduced into the 5′ LTR of this double mutant. Thus, this revertant provirus contains a repaired 5′ TAR hairpin and a destabilized 3′ TAR structure.
FIG. 1.
HIV-1 genomes with 5′ and 3′ TAR mutations. The upper schematic shows HIV-1 genomic RNA with a tandem hairpin motif encoded by the R region. The upstream hairpin is TAR (shaded); the downstream structure is the poly(A) hairpin motif (24). RNA secondary-structure predictions for the wild-type, Xho+10 mutant, and revertant TAR elements are shown below. The free energy for each structure was calculated with the Zuker algorithm (61) and is given in kilocalories per mole. The mutated region of TAR is shaded, and additional mutations in the revertant are boxed. Nucleotide numbers are relative to the RNA start site at +1.
Destabilization of the 5′ TAR structure results in a reduced intracellular HIV-1 RNA level.
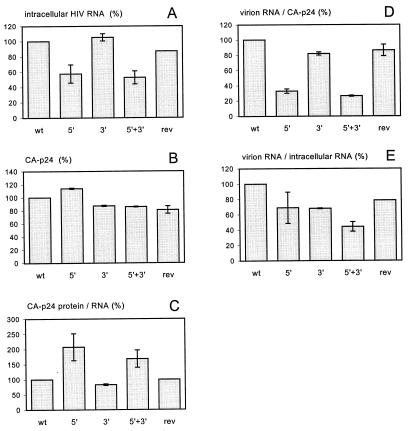
The wild-type and mutant proviruses were transfected into C33A cells (human cervix carcinoma cells not expressing the CD4 receptor), resulting in the synthesis of viral RNAs and proteins and the production of infectious virions. Intracellular RNA was isolated 2 days after transfection, and the level of unspliced HIV-1 RNA was determined by dot blot analysis (Fig. 2A). The intracellular HIV-1 RNA level measured for the wild-type construct was set at 100%. The RNA levels of the 5′ TAR mutant and the 5′ + 3′ TAR double mutant were reduced by approximately 50%, but a wild-type RNA level was measured for the 3′ TAR mutant. Apparently, destabilization of the 5′ TAR hairpin reduces the intracellular HIV-1 RNA content. Repair of the 5′ TAR structure by introduction of the revertant sequence did almost completely restore the RNA level to that of the wild-type construct. These results are in agreement with previous observations and suggest a critical role for the full-length TAR hairpin in optimal Tat-activated LTR transcription (56).
FIG. 2.
Analysis of wild-type, mutant, and revertant TAR constructs. C33A cells were transfected with the proviral constructs, and the intracellular HIV-1 RNA was quantitated by dot blot analysis (A). Virus production was measured in the culture supernatant by CA p24 ELISA (B). These values were used to calculate translational efficiency (C). Virion RNA levels were measured and compared either with the CA p24 values (D) or with the intracellular HIV-1 RNA levels (E). The former value represents the virion RNA content; the latter value represents the RNA packaging efficiency. All parameters were arbitrarily set at 100% for the wild-type HIV-1 construct. Standard errors were calculated for independent transfections (except for the TAR revertant in some panels). For panels C, D, and E, we first calculated the ratio per independent experiment and next calculated the mean value and standard error. wt, wild-type construct; 5′, 5′ TAR mutant, 3′, 3′ TAR mutant; 5′ + 3′, 5′ + 3′ TAR double mutant; rev, revertant construct (see the text).
Destabilization of 5′ TAR enhances the translational efficiency.
The production of viral proteins was assayed by measuring the CA p24 level in the culture supernatant (Fig. 2B). This level is similar for the wild-type, mutant, and revertant viruses. These results were confirmed by Western blot analysis of intracellular HIV-1 proteins (not shown). To determine the translational efficiency of the corresponding mRNAs, the CA p24 values were compared with the intracellular HIV-1 RNA levels, which were reduced for the 5′ and 5′ + 3′ TAR mutants (Fig. 2A). The translational efficiency, expressed as the protein-to-RNA ratio (Fig. 2C), was increased approximately twofold for the two constructs with a destabilized 5′ TAR hairpin. These results are consistent with the idea that the wild-type 5′ TAR hairpin is moderately inhibitory in the process of mRNA translation (47). This partial translational block is relieved by destabilization of the 5′ RNA structure and restored in the 5′ revertant mRNA. No effect of the 3′ TAR element on the translational efficiency of HIV-1 mRNAs was measured.
Both 5′ and 3′ TAR hairpins contribute to RNA packaging.
We next studied the effect of the 5′ and 3′ TAR mutations on packaging of HIV-1 genomic RNA into virions. We therefore measured the virion RNA level by dot blot analysis and calculated the relative RNA content of the virions (virion RNA-to-CA p24 ratio; Fig. 2D). The virion RNA content was reduced significantly for the mutant with an opened 5′ TAR hairpin. A small defect was measured for the 3′ TAR-mutated virus, and the 5′ + 3′ TAR double mutant was affected most severely. This defect was largely restored in the revertant construct that has a repaired 5′ TAR motif.
However, because destabilization of the 5′ TAR structure did also affect the intracellular HIV-1 RNA level (Fig. 2A), the virion RNA-to-CA p24 ratio (Fig. 2D) may not accurately reflect the RNA packaging efficiency. As a more appropriate measure of the packaging efficiency, we therefore calculated the ratio of virion RNA to intracellular HIV-1 RNA (Fig. 2E). This packaging efficiency was reduced in both the 5′ and the 3′ TAR mutants to 70% of the wild-type packaging efficiency, and only 40% packaging was calculated for the 5′ + 3′ TAR double mutant. Upon introduction of a revertant hairpin at the 5′ position in this double mutant, the packaging ratio was found to increase to the level of the two constructs with a single mutant TAR element. These results show that both the 5′ and the 3′ TAR hairpins contribute to RNA packaging and that destabilization of either one of these hairpins reduces the packaging efficiency by approximately 30%.
TAR mutations do not affect the stability of virion RNA.
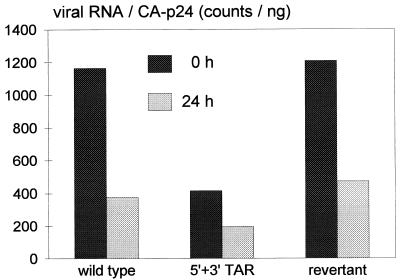
The TAR hairpin is present at both the 5′ and the 3′ ends of the RNA genome. Because this structure may protect the RNA molecule from degradation by exonucleases, one could argue that the reduced packaging efficiency measured for mutants with an opened 5′ or 3′ TAR structure does in fact reflect reduced stability of the virion RNA (51). We performed an additional experiment to test the stability of virion RNA. The wild-type, double-mutant, and revertant proviral constructs were transfected into C33A cells, and virions produced between 48 and 64 h posttransfection were harvested. The RNA content of these virions was determined either directly (0-h sample) or after an additional 24-h incubation of the cell-free virions at 37°C (Fig. 3). As observed previously (Fig. 2E), the RNA level of the freshly made virions was reduced for the 5′ + 3′ TAR mutant and restored in the revertant. The three viruses showed similar, approximately 2.5-fold drops in RNA content following the 24-h incubation, indicating that the presence of a truncated TAR stem at either end of HIV-1 RNA does not affect the intravirion genome stability.
FIG. 3.
Stability of TAR-mutated RNA within virions. Wild-type, mutant (5′ + 3′ TAR), and revertant constructs were used to produce virions in transfected C33A cells. Virus particles produced between 48 and 64 h after transfection were harvested, and RNA was quantitated either directly (0-h sample) or after incubation of the virions for 24 h at 37°C in RPMI medium. The data are absolute PhosphorImager counts divided by the CA p24 levels.
TAR hairpin mutations do not affect reverse transcription.
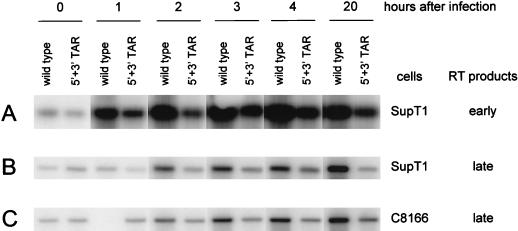
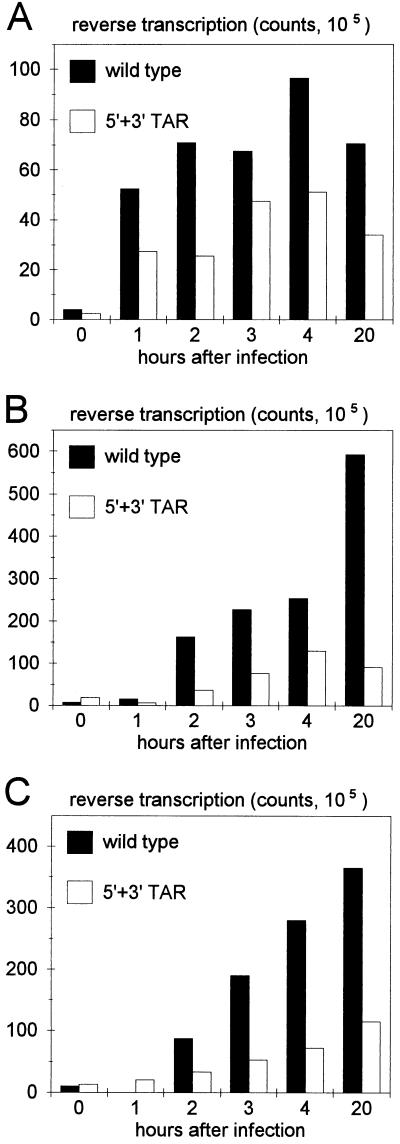
We analyzed the effect of the TAR hairpin mutations on the process of reverse transcription in infected cells. This assay was performed only with the 5′ + 3′ TAR double mutant because all other mutants have nonidentical 5′ and 3′ R regions, which will cause problems in the first strand transfer step (13). Equal amounts of wild-type and 5′ + 3′ TAR mutant viruses were used to infect the SupT1 and C8166 T-cell lines. Viruses were incubated with the cells for 1 h at 37°C and then removed by extensive washing. Cells were cultured for a prolonged period. Control cells were harvested before the 37°C incubation (0-h sample); the other samples were taken at 1, 2, 3, 4, or 20 h after infection. Total cellular DNA was extracted, and reverse-transcribed HIV-1 DNA was PCR amplified with different primer sets. Early reverse transcription products formed after the first strand transfer were amplified with the 5′ nef and 3′ U5 primers, and cDNA products formed after continued reverse transcription were detected with the 5′ tat and 3′ env primers. These early and late cDNA products were analyzed by Southern blotting (Fig. 4) and quantitated by PhosphorImager analysis (Fig. 5). Early cDNAs were detected as early as 1 h after infection of SupT1 cells with either the wild-type or mutant virus. This early signal increased in abundance over the next 3 h (Fig. 4A and 5A). At all times, a reduced cDNA level was measured for the 5′ + 3′ TAR double mutant. Similar results were obtained in the analysis of late cDNA products, which were first detected 2 h after infection of SupT1 cells (Fig. 4B and 5B), and in the infection of C8166 cells (Fig. 4C and 5C). This kinetic analysis of the reverse transcription reaction revealed a significant reduction of cDNA products for the TAR-mutated virus compared with the wild-type virus. However, this reduced cDNA level correlated with the reduced RNA content of the mutant virus particles (Fig. 2D). These results suggest that the efficiency of reverse transcription is not significantly affected by the 5′ + 3′ TAR mutations.
FIG. 4.
Reverse transcription of TAR-mutated viral transcripts in infected cells. Wild-type and 5′ + 3′ TAR mutant virus stocks (normalized by CA p24 levels) were used to infect SupT1 cells (A and B) or C8166 cells (C). Cell samples were harvested at the times indicated. Total cellular DNA was extracted, and HIV-1 cDNA was PCR amplified with primers that detect early or late DNA products of reverse transcription. PCR products were visualized on a Southern blot probed with a 32P-labeled HIV-1 fragment. Hybridization signals were quantitated with a PhosphorImager and are presented in Fig. 5. The same PCR protocol was performed on various amounts of the pLAI HIV-1 plasmid to check that the amplification was performed within the linear range.
FIG. 5.
Normal reverse transcription efficiency of TAR-mutated viral transcripts. See the legend to Fig. 4 for details.
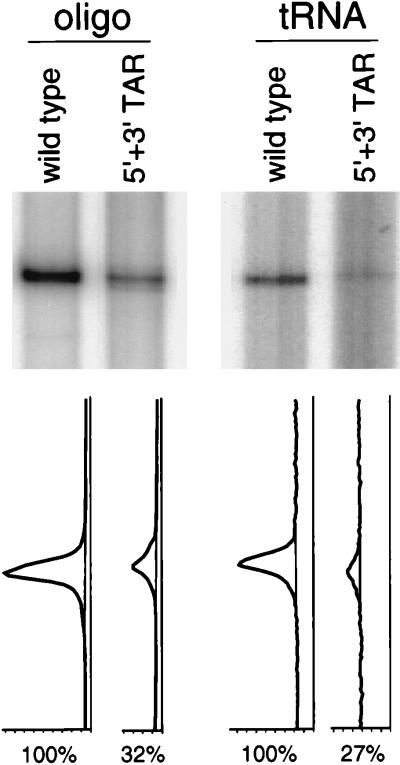
To further confirm these results, we performed reverse transcription assays with the wild-type and mutant HIV-1 RNA genomes that were extracted from virion particles. The same amount of virions (based on CA p24) was used for this RNA isolation. In a primer extension reaction with a DNA oligonucleotide primer complementary to the HIV-1 leader RNA (Fig. 6), a reduced level of the cDNA product was obtained for the 5′ + 3′ TAR mutant (32% of the wild-type level). This reduced cDNA production correlates precisely with, and thus confirms, the reduced RNA content of the mutant virus as plotted in Fig. 2D. We also performed reverse transcription in the absence of an exogenous DNA primer. In this case, a relatively weak extension product, primed by the natural tRNA3Lys primer that is coextracted with the virion RNA genome, can be detected (Fig. 6). The tRNA-primed cDNA signal obtained for the mutant genome was 27% compared with the wild-type signal. Given the similar reduction in RNA content, we conclude that there is no significant effect of the TAR mutations on the tRNA-primed reverse transcription reaction.
FIG. 6.
In vitro reverse transcription assays with the TAR-mutated transcript. The RNA genomes of the wild-type and 5′ + 3′ TAR mutant virus stocks (normalized by CA p24 levels) were phenol extracted and analyzed in vitro. Reverse transcription from the associated tRNA3Lys primer was initiated by addition of RT and deoxynucleoside triphosphates. In the oligonucleotide-primed reaction, an exogenous DNA-oligonucleotide primer was annealed to the virion RNA and extended by reverse transcription. The oligonucleotide-primed and tRNA extension signals were analyzed with a PhosphorImager, and their profiles are shown below. The relative level of cDNA production was set at 100% for the wild-type template in both primer extension assays.
DISCUSSION
Our data indicate that the structured TAR RNA motif plays several roles in the HIV-1 replication cycle. Besides the well-known transcriptional function of the 5′ TAR hairpin, this motif was found to partially repress translation of the viral RNA and to stimulate packaging of the RNA genome. The 3′ TAR motif also activated the encapsidation of RNA into virions, and this contribution to packaging was quantitatively similar to that of the 5′ TAR element. Reduced reverse transcription levels were demonstrated to correlate with the reduced level of RNA template within the mutant virions. Although all reported TAR effects seem to depend on the base-paired structure of this RNA signal, it cannot be excluded that important nucleotide sequences within TAR are also critical for some of these functions. However, the finding that the TAR functions can be restored by the additional nucleotide changes observed in a revertant TAR element with a repaired stem suggests that the actual sequence of the lower TAR stem is not important for these functions.
We observed that mutant HIV-1 mRNAs with a destabilized 5′ TAR structure can be translated more efficiently than the wild-type transcript. This indicates that there is a repressive effect of the wild-type TAR hairpin, which is consistent with previous reports (26, 47, 50, 53, 57). The TAR effect may operate either in cis by restricting the accessibility of the mRNA cap structure to the translational machinery or in trans through induction of the PKR protein kinase system. Whatever the mechanism, it is possible that translational repression observed for wild-type HIV-1 RNA is a viral strategy to balance the processes of translation and packaging. This may provide the optimal amount of genomic RNA and viral proteins, and therefore ultimately control the production of infectious virus. Previous findings with murine leukemia virus suggested that full-length viral RNA was routed to either a pool for translation or a pool for packaging and that the RNA bound by ribosomes could not be packaged (44). For Rous sarcoma virus, it has been reported recently that this sorting mechanism is mediated by the viral Gag proteins (54).
In this study, we measured reduced virion RNA levels with both 5′ and 3′ TAR mutants. Part of this reduction is directly due to the lower levels of intracellular HIV-1 RNA, suggesting that the RNA packaging efficiency is exquisitely sensitive to changes in the concentration of intracellular RNA. Therefore, the ratio of virion RNA to intracellular HIV-1 RNA (Fig. 2E) is a better measure of the packaging efficiency than the ratio of virion RNA to virion protein (Fig. 2D). After correction for the reduced amount of intracellular RNA, a packaging defect remained for the mutants. The contribution of the 5′ TAR motif to packaging is consistent with a previous study (46), and we now report a quantitatively similar contribution of the 3′ TAR element. With the results of other studies in mind (2, 20, 21, 33, 45), it is appropriate to consider the entire untranslated 5′ leader region as the packaging signal (8). It is likely that the complete leader region is required to fold a specific tertiary RNA structure that forms the actual packaging signal. It is also possible that the TAR motif affects RNA dimerization (14, 34), which in turn may affect the process of RNA packaging (27). Therefore, the TAR motif will reflect the superimposed demands of multiple essential processes, which could be difficult to separate experimentally.
The finding that the 3′ end of the viral genome also contributes to packaging may suggest that both ends of the RNA interact, a phenomenon that has been proposed for cellular transcripts to explain the effects that the poly(A) tail and 3′ untranslated region can have on translation initiation (7, 37). There is no direct biochemical evidence for such a 5′-to-3′ interaction in HIV-1 RNA, but the close proximity of the two R regions may be particularly advantageous in the strand transfer step of reverse transcription. In fact, there is some recent evidence that the native dimer conformation of HIV-1 virion RNA is required for this strand transfer step (9). Finally, the presence of multiple accessory packaging signals in parts of the genome that are present in spliced HIV-1 RNAs can explain the relative abundance of such subgenomic RNAs in viral particles (45, 46).
Our results are not consistent with a previous report on the effect of TAR on reverse transcription (32). It remains possible that these conflicting results are caused by differences in the TAR mutants used in the two studies. For instance, we analyzed TAR mutants with an opened lower stem, whereas changes in the upper TAR domain that forms the Tat binding site may be important for reverse transcription. However, the previous study (32) reported that the Tat-binding domain within TAR is dispensable for TAR-mediated activation of reverse transcription. In fact, most dramatic effects were observed with stem mutants, which are very similar to the Xho+10 mutant used in this study. It is possible that the level of HIV-1 RNA in the virion particles was not accurately measured in the former study (32). Specifically, an RT-PCR protocol was used that does not discriminate between spliced and unspliced HIV-1 mRNAs. Spliced HIV-1 RNAs are encapsidated with high efficiency compared with nonviral RNA (15), and mutants that package less full-length RNA genome seem to compensate this defect by increased packaging of spliced HIV-1 RNAs (52). Thus, the RT-PCR measure may have underestimated the packaging defect of TAR-mutated transcripts. Indeed, whereas we measured a virion RNA content of only 30% for the mutant with an opened 5′ and 3′ TAR motif, no such packaging defect has been reported for similar TAR mutants in the former study. We measured the reverse transcription efficiency both in assays with infected cells and in in vitro assays with the virion-extracted RNA genome. When the reduced level of cDNA production was corrected for this diminished concentration of RNA template, no net effect of TAR on reverse transcription was measured. It has been reported that the viral Tat protein may exert additional functions in the virus life cycle (35), and a putative role in reverse transcription was proposed (31). We did not address this issue in the current study, but some evidence was recently presented against a Tat function other than its role in activated transcription from the viral LTR promoter (40, 55).
ACKNOWLEDGMENTS
We thank Koen Verhoef for critical reading of the manuscript and Wim van Est for photography.
This work was supported by grants from the Dutch Cancer Society (KWF), the European Community (EU 950675), and the Dutch AIDS Fund.
REFERENCES
- 1.Aiyar A, Cobrinik D, Ge Z, Kung H J, Leis J. Interaction between retroviral U5 RNA and the TYC loop of the tRNATrp primer is required for efficient initiation of reverse transcription. J Virol. 1992;66:2464–2472. doi: 10.1128/jvi.66.4.2464-2472.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aldovini A, Young R A. Mutations of RNA and protein sequences involved in human immunodeficiency virus type 1 packaging results in production of noninfectious virus. J Virol. 1990;64:1920–1926. doi: 10.1128/jvi.64.5.1920-1926.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arts E J, Li X, Gu Z, Kleiman L, Parniak M A, Wainberg M A. Comparison of deoxyoligonucleotide and tRNA(Lys-3) as primers in an endogenous human immunodeficiency virus-1 in vitro reverse transcription/template-switching reaction. J Biol Chem. 1994;269:14672–14680. [PubMed] [Google Scholar]
- 4.Ashe M P, Pearson L H, Proudfoot N J. The HIV-1 5′ LTR poly(A) site is inactivated by U1 snRNP interaction with the downstream major splice donor site. EMBO J. 1997;16:5752–5763. doi: 10.1093/emboj/16.18.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Auersperg N. Long-term cultivation of hypodiploid human tumor cells. J Natl Cancer Inst. 1964;32:135–163. [PubMed] [Google Scholar]
- 6.Back N K T, Nijhuis M, Keulen W, Boucher C A B, Oude Essink B B, van Kuilenburg A B P, Van Gennip A H, Berkhout B. Reduced replication of 3TC-resistant HIV-1 variants in primary cells due to a processivity defect of the reverse transcriptase enzyme. EMBO J. 1996;15:4040–4049. [PMC free article] [PubMed] [Google Scholar]
- 7.Beelman C A, Parker R. Degradation of mRNA in eukaryotes. Cell. 1995;81:179–183. doi: 10.1016/0092-8674(95)90326-7. [DOI] [PubMed] [Google Scholar]
- 8.Berkhout B. Structure and function of the human immunodeficiency virus leader RNA. Prog Nucleic Acid Res Mol Biol. 1996;54:1–34. doi: 10.1016/s0079-6603(08)60359-1. [DOI] [PubMed] [Google Scholar]
- 9.Berkhout B, Das A T, van Wamel J L B. The native structure of the HIV-1 RNA genome is required for the first strand-transfer of reverse transcription. Virology. 1998;249:211–218. doi: 10.1006/viro.1998.9321. [DOI] [PubMed] [Google Scholar]
- 10.Berkhout B, Klaver B, Das A T. A conserved hairpin structure predicted for the poly(A) signal of human and simian immunodeficiency viruses. Virology. 1995;207:276–281. doi: 10.1006/viro.1995.1077. [DOI] [PubMed] [Google Scholar]
- 11.Berkhout B, Schoneveld I. Secondary structure of the HIV-2 leader RNA comprising the tRNA-primer binding site. Nucleic Acids Res. 1993;21:1171–1178. doi: 10.1093/nar/21.5.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berkhout B, Silverman R H, Jeang K T. Tat trans-activates the human immunodeficiency virus through a nascent RNA target. Cell. 1989;59:273–282. doi: 10.1016/0092-8674(89)90289-4. [DOI] [PubMed] [Google Scholar]
- 13.Berkhout B, van Wamel J, Klaver B. Requirements for DNA strand transfer during reverse transcription in mutant HIV-1 virions. J Mol Biol. 1995;252:59–69. doi: 10.1006/jmbi.1994.0475. [DOI] [PubMed] [Google Scholar]
- 14.Berkhout B, van Wamel J L B. Role of the DIS hairpin in replication of human immunodeficiency virus type 1. J Virol. 1996;70:6723–6732. doi: 10.1128/jvi.70.10.6723-6732.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berkowitz R D, Ohagen A, Hoglund S, Goff S P. Retroviral nucleocapsid domains mediate the specific recognition of genomic viral RNAs by chimeric Gag polyproteins during RNA packaging in vivo. J Virol. 1995;69:6445–6456. doi: 10.1128/jvi.69.10.6445-6456.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Braddock M, Powell R, Blanchard A D, Kingsman A J, Kingsman S M. HIV-1 TAR RNA-binding proteins control TAT activation of translation in Xenopus oocytes. FASEB J. 1993;7:214–222. doi: 10.1096/fasebj.7.1.8422967. [DOI] [PubMed] [Google Scholar]
- 17.Braddock M, Thorburn A M, Chambers A, Elliot G D, Anderson G J, Kingsman A J, Kingsman S M. A nuclear translational block imposed by the HIV-1 U3 region is relieved by the Tat-TAR interaction. Cell. 1990;62:1123–1133. doi: 10.1016/0092-8674(90)90389-v. [DOI] [PubMed] [Google Scholar]
- 18.Brown P H, Tiley L S, Cullen B R. Efficient polyadenylation within the human immunodeficiency virus type 1 long terminal repeat requires flanking U3-specific sequences. J Virol. 1991;65:3340–3343. doi: 10.1128/jvi.65.6.3340-3343.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 20.Clavel F, Orenstein J M. A mutant of human immunodeficiency virus with reduced RNA packaging and abnormal particle morphology. J Virol. 1990;64:5230–5234. doi: 10.1128/jvi.64.10.5230-5234.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clever J, Sassetti C, Parslow T G. RNA secondary structure and binding sites for gag gene products in the 5′ packaging signal of human immunodeficiency virus type 1. J Virol. 1995;69:2101–2109. doi: 10.1128/jvi.69.4.2101-2109.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cullen B R. Trans-activation of human immunodeficiency virus occurs via a bimodal mechanism. Cell. 1986;46:973–982. doi: 10.1016/0092-8674(86)90696-3. [DOI] [PubMed] [Google Scholar]
- 23.Das A T, Klaver B, Berkhout B. Reduced replication of human immunodeficiency virus type 1 mutants that use reverse transcription primers other than the natural tRNA3Lys. J Virol. 1995;69:3090–3097. doi: 10.1128/jvi.69.5.3090-3097.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Das A T, Klaver B, Klasens B I F, van Wamel J L B, Berkhout B. A conserved hairpin motif in the R-U5 region of the human immunodeficiency virus type 1 RNA genome is essential for replication. J Virol. 1997;71:2346–2356. doi: 10.1128/jvi.71.3.2346-2356.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dingwall C, Ernberg I, Gait M J, Green S M, Heaphy S, Karn J, Lowe A D, Singh M, Skinner M A, Valerio R. Human immunodeficiency virus 1 tat protein binds trans-activating-responsive region (TAR) RNA in vitro. Proc Natl Acad Sci USA. 1989;86:6925–6929. doi: 10.1073/pnas.86.18.6925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Edery I R, Petryshyn R, Sonenberg N. Activation of double-stranded RNA dependent kinase (dsI) by the TAR region of HIV-1 mRNA: a novel translational control mechanism. Cell. 1989;56:303–312. doi: 10.1016/0092-8674(89)90904-5. [DOI] [PubMed] [Google Scholar]
- 27.Fu W, Gorelick R J, Rein A. Characterization of human immunodeficiency virus type 1 dimeric RNA from wild-type and protease-defective virions. J Virol. 1994;68:5013–5018. doi: 10.1128/jvi.68.8.5013-5018.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gilmartin G M, Fleming E S, Oetjen J, Graveley B R. CPSF recognition of an HIV-1 mRNA 3′-processing enhancer: multiple sequence contacts involved in poly(A) site definition. Genes Dev. 1995;9:72–83. doi: 10.1101/gad.9.1.72. [DOI] [PubMed] [Google Scholar]
- 29.Harrich D, Hsu C, Race E, Gaynor R B. Differential growth kinetics are exhibited by human immunodeficiency virus type 1 TAR mutants. J Virol. 1994;68:5899–5910. doi: 10.1128/jvi.68.9.5899-5910.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harrich D, Mavankal G, Mette-Snider A, Gaynor R B. Human immunodeficiency virus type 1 TAR element revertant viruses define RNA structures required for efficient viral gene expression and replication. J Virol. 1995;69:4906–4913. doi: 10.1128/jvi.69.8.4906-4913.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harrich D, Ulich C, Garcia-Martinez L F, Gaynor R B. Tat is required for efficient HIV-1 reverse transcription. EMBO J. 1997;16:1224–1235. doi: 10.1093/emboj/16.6.1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harrich D, Ulich C, Gaynor R B. A critical role for the TAR element in promoting efficient human immunodeficiency virus type 1 reverse transcription. J Virol. 1996;70:4017–4027. doi: 10.1128/jvi.70.6.4017-4027.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harrison G P, Lever A M L. The human immunodeficiency virus type 1 packaging signal and major splice donor region have a conserved stable secondary structure. J Virol. 1992;66:4144–4153. doi: 10.1128/jvi.66.7.4144-4153.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hoglund S, Ohagen A, Goncalves J, Panganiban A T, Gabuzda D. Ultrastructure of HIV-1 genomic RNA. Virology. 1997;233:271–279. doi: 10.1006/viro.1997.8585. [DOI] [PubMed] [Google Scholar]
- 35.Huang L M, Joshi A, Willey R, Orenstein J, Jeang K T. Human immunodeficiency viruses regulated by alternative trans-activators: genetic evidence for a novel non-transcriptional function of Tat in virion infectivity. EMBO J. 1994;13:2886–2896. doi: 10.1002/j.1460-2075.1994.tb06583.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Isel C, Ehresmann C, Keith G, Ehresmann B, Marquet R. Initiation of reverse transcription of HIV-1: secondary structure of the HIV-1 RNA/tRNA(3Lys) (template/primer) J Mol Biol. 1995;247:236–250. doi: 10.1006/jmbi.1994.0136. [DOI] [PubMed] [Google Scholar]
- 37.Jackson R J, Standart N. Do the poly(A) tail and 3′ untranslated region control mRNA translation? Cell. 1990;62:15–24. doi: 10.1016/0092-8674(90)90235-7. [DOI] [PubMed] [Google Scholar]
- 38.Jeang K T, Chun R, Lin N H, Gatignol A, Glabe C G, Fan H. In vitro and in vivo binding of human immunodeficiency virus type 1 Tat protein and Sp1 transcription factor. J Virol. 1993;67:6224–6233. doi: 10.1128/jvi.67.10.6224-6233.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kang S-M, Wakefield J K, Morrow C D. Mutations in both the U5 region and the primer-binding site influence the selection of the tRNA used for the initiation of HIV-1 reverse transcription. Virology. 1996;222:401–414. doi: 10.1006/viro.1996.0437. [DOI] [PubMed] [Google Scholar]
- 40.Kim V N, Mitrophanous K, Kingsman S M, Kingsman A J. Minimal requirements for a lentivirus vector based on human immunodeficiency virus type 1. J Virol. 1998;72:811–816. doi: 10.1128/jvi.72.1.811-816.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Klaver B, Berkhout B. Comparison of 5′ and 3′ long terminal repeat promoter function in human immunodeficiency virus. J Virol. 1994;68:3830–3840. doi: 10.1128/jvi.68.6.3830-3840.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Klaver B, Berkhout B. Evolution of a disrupted TAR RNA hairpin structure in the HIV-1 virus. EMBO J. 1994;13:2650–2659. doi: 10.1002/j.1460-2075.1994.tb06555.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Klaver B, Berkhout B. Premature strand transfer by the HIV-1 reverse transcriptase during strong-stop DNA synthesis. Nucleic Acids Res. 1994;22:137–144. doi: 10.1093/nar/22.2.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Levin J G, Rosenak M J. Synthesis of murine leukemia virus proteins associated with virions assembled in actinomycin-D-treated cells: evidence for persistence of viral messenger RNA. Proc Natl Acad Sci USA. 1976;73:1154–1158. doi: 10.1073/pnas.73.4.1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McBride M S, Panganiban A T. The human immunodeficiency virus type 1 encapsidation site is a multipartite RNA element composed of functional hairpin structures. J Virol. 1996;70:2963–2973. doi: 10.1128/jvi.70.5.2963-2973.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McBride M S, Schwartz M D, Panganiban A T. Efficient encapsidation of human immunodeficiency virus type 1 vectors and further characterization of cis elements required for encapsidation. J Virol. 1997;71:4544–4554. doi: 10.1128/jvi.71.6.4544-4554.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Parkin N T, Cohen E A, Darveau A, Rosen C, Haseltine W, Sonenberg N. Mutational analysis of the 5′ noncoding region of human immunodeficiency virus type 1: effects of secondary structure. EMBO J. 1988;7:2831–2837. doi: 10.1002/j.1460-2075.1988.tb03139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peden K, Emerman M, Montagnier L. Changes in growth properties on passage in tissue culture of viruses derived from infectious molecular clones of HIV-1LAI, HIV-1MAL, and HIV-1ELI. Virology. 1991;185:661–672. doi: 10.1016/0042-6822(91)90537-l. [DOI] [PubMed] [Google Scholar]
- 49.Rounseville M P, Lin H C, Agbottah E, Shukla R R, Rabson A B, Kumar A. Inhibition of HIV-1 replication in viral mutants with altered TAR RNA stem structures. Virology. 1996;216:411–417. doi: 10.1006/viro.1996.0077. [DOI] [PubMed] [Google Scholar]
- 50.Roy S, Agy M, Hovanessian A G, Sonenberg N, Katze M G. The integrity of the stem structure of human immunodeficiency virus type 1 Tat-responsive sequence of RNA is required for interaction with the interferon-induced 68,000-Mr protein kinase. J Virol. 1991;65:632–640. doi: 10.1128/jvi.65.2.632-640.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sakuragi J-I, Panganiban A T. Human immunodeficiency virus type 1 RNA outside the primary encapsidation and dimer linkage region affects RNA dimer stability in vivo. J Virol. 1997;71:3250–3254. doi: 10.1128/jvi.71.4.3250-3254.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schwartz M D, Fiore D, Panganiban A T. Distinct functions and requirements for the Cys-His boxes of the human immunodeficiency virus type 1 nucleocapsid protein during RNA encapsidation and replication. J Virol. 1997;71:9295–9305. doi: 10.1128/jvi.71.12.9295-9305.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.SenGupta D N, Berkhout B, Gatignol A, Zhou A M, Silverman R H. Direct evidence for translational regulation by leader RNA and Tat protein of human immunodeficiency virus type 1. Proc Natl Acad Sci USA. 1990;87:7492–7496. doi: 10.1073/pnas.87.19.7492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sonstegard T S, Hackett P B. Autogenous regulation of RNA translation and packaging by Rous sarcoma virus Pr76gag. J Virol. 1996;70:6642–6652. [PMC free article] [PubMed] [Google Scholar]
- 55.Verhoef K, Koper M, Berkhout B. Determination of the minimal amount of Tat activity required for human immunodeficiency virus type 1 replication. Virology. 1997;237:228–236. doi: 10.1006/viro.1997.8786. [DOI] [PubMed] [Google Scholar]
- 56.Verhoef K, Tijms M, Berkhout B. Optimal Tat-mediated activation of the HIV-1 LTR promoter requires a full-length TAR RNA hairpin. Nucleic Acids Res. 1997;25:496–502. doi: 10.1093/nar/25.3.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Viglianti G A, Rubinstein E C, Graves K L. Role of the TAR RNA splicing in translational regulation of simian immunodeficiency virus from rhesus macaques. J Virol. 1992;66:4824–4833. doi: 10.1128/jvi.66.8.4824-4833.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wakefield J K, Kang S-M, Morrow C D. Construction of a type 1 human immunodeficiency virus that maintains a primer binding site complementary to tRNAHis. J Virol. 1996;70:966–975. doi: 10.1128/jvi.70.2.966-975.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wu-Baer F, Lane W S, Gaynor R B. Identification of a group of cellular cofactors that stimulate the binding of RNA polymerase II and TRP-185 to human immunodeficiency virus 1 TAR RNA. J Biol Chem. 1996;271:4201–4208. doi: 10.1074/jbc.271.8.4201. [DOI] [PubMed] [Google Scholar]
- 60.Wu-Baer F, Sigman D, Gaynor R B. Specific binding of RNA polymerase II to the human immunodeficiency virus trans-activating region RNA is regulated by cellular cofactors and Tat. Proc Natl Acad Sci USA. 1995;92:7153–7157. doi: 10.1073/pnas.92.16.7153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zuker M. On finding all suboptimal foldings of an RNA molecule. Science. 1989;244:48–52. doi: 10.1126/science.2468181. [DOI] [PubMed] [Google Scholar]