Abstract
Recently, the focus is on new specific immunotherapies for AML such as cellular therapies employing dendritic cells (DCs) generated from AML blasts. AML-DCs express constitutionally leukemia-associated antigens (LAAs) present in AML blasts they are generated from. Here we investigated whether the generation of AML-DCs would alter the expression level of LAAs. Moreover, we evaluated the presence of HLA and costimulatory molecules on AML blasts versus AML-DCs. Quantitative real-time polymerase chain reaction (PCR) was performed for the following LAAs: preferentially expressed antigen in melanoma (PRAME), the receptor for hyaluronic acid mediated motility (RHAMM/CD168), Wilms’ tumor gene 1 (WT-1) and proteinase 3. The expression of HLA-ABC, HLA-DR, CD40, CD80, CD83 and CD86 was evaluated by flow cytometry. RHAMM protein expression was evaluated by immunocytochemistry, recognition of AML-DCs by PRAME epitope-specific T cells was evaluated in a chromium-release assay. Quantitative real-time PCR for AML-DCs versus AML blasts showed an alteration in mRNA expression of LAAs. An elevated PCR signal for PRAME was detected in 7/12 AML-DC preparations. 6/12 AML-DC preparations showed a significant upregulation of the PCR signal for RHAMM. A stronger WT-1 and proteinase-3 signal was observed in PCR for only 2/12 and 1/12 AML-DCs , respectively. All preparations showed a strong expression of at least one of the LAAs examined. As demonstrated by flow cytometry, AML-DCs strongly upregulated costimulatory molecules like CD40 and CD80 in comparison with AML blasts. AML-DCs tested positive for RHAMM protein. PRAME positive AML-DCs were recognized by specific T cells. AML-DCs might constitute a powerful tool in immunotherapy for AML. Real-time PCR allows a quick and quantitative assessment of immunologically relevant LAA expression with only 105 DCs and might be helpful for the decision whether the AML-DC vaccination strategy is favourable or not.
Keywords: AML, DCs, LAAs/TAAs, PRAME, RHAMM, WT-1, Proteinase 3
Introduction
Acute myeloid leukemia (AML) is a hematological disease, which is characterized by the clonal proliferation of undifferentiated myeloid progenitor cells. Most of the patients with AML achieve a complete hematological remission by chemotherapeutical regimes. However, the long time prognosis for all AML patients is rather poor with a 5 year overall survival of only 20–40% depending on the individual risk profile and the chosen treatment option [36]. Dendritic cells (DCs) are professional antigen-presenting cells with the unique property to prime naive T cells [32]. Hence, DCs are considered to be important elements also in the induction of specific anti-tumor immune responses [2, 3, 32]. Up to now, myeloid leukemias (AML/CML) and ALL are the only malignancies in which DCs can be generated directly from the cancer cells [8–11]. Earlier, we demonstrated [21] that such AML-DCs constitutively express tumor/leukemia-associated antigens (TAAs/LAAs), e.g., the antigen preferentially expressed in melanoma [PRAME; 15, 19, 20, 23, 28, 38], the receptor for hyaluronic acid-mediated motility [RHAMM/CD168, 1, 12, 16, 31, 35, 37, 40, 41] or the Wilms’ tumor gene 1 [WT-1; 5, 7, 18, 24, 29, 30] and proteinase 3 [6, 13, 25–27, 33]. Moreover, we demonstrated that AML-DCs expressed HLA and costimulatory molecules, which are essential for T-cell responses [21].
Specific immunotherapies with LAAs might be an option to enhance an anti-leukemic effect after chemotherapy and or hematopoeitic stem cell transplantation. At present, clinical vaccination trials with LAA peptides are ongoing [22, 25, 27, 33]. The first patients showing both immunological and hematological responses have been reported [25, 27]. At our institution, we are performing a clinical phase-I trial using autologous AML-DC as a vaccine.
To evaluate the immunogenic potential of such AML-DCs for the stimulation of a LAA-specific T-cell response, we investigated in this study both their immunophenotype in terms of antigen-presenting HLA molecules and costimulatory molecules by flow cytometry [21] and their mRNA expression of LAAs by quantitative PCR. Immunocytochemistry for RHAMM was performed, as well as chromium-51 release assays for a PRAME derived epitope peptide.
Material and methods
Cell samples
Peripheral blood samples (median 89% blasts, thereafter called “AML blasts”) were taken from six untreated patients with newly diagnosed AML and from six different patients with relapse at the time when patients were screened for an immunotherapy with autologous AML-DCs. The patients’ characteristics are specified in Table 1. Informed consent to the use of their blood for scientific purposes was obtained from all patients who were treated at our institution, according to clinical study protocols approved by the local ethics committee. Control samples were prepared from 12 healthy volunteers (HV) at our institution after their informed consent was obtained.
Table 1.
Patients’ characteristics
| AML (FAB) | Blasts (%) | Karyotype | |
|---|---|---|---|
| 1 | M1 | 99 | Add(19)(p13) |
| 2 | Secondary AML | 80 | Normal |
| 3 | Secondary AML | 70 | Normal |
| 4 | M5a | 90 | Normal |
| 5 | M3 | 97 | 46, XY del(20)(q11); 46, XY del(20)(q11) add(22)(p11); 47, XY del(20)(q11), +21 |
| 6 | M0 | 99 | 46, XY add8p |
| 7 | M2 | 60 | Normal |
| 8 | M4 | 52 | Normal |
| 9 | M4 | 80 | Normal |
| 10 | M5b | 92 | Normal |
| 11 | M5 | 89 | Normal |
| 12 | M4 | 20 | Normal |
Generation of dendritic cells
Generation of DCs from peripheral blood monocytes or AML blasts was performed as described previously [8–10, 21]. In brief, peripheral blood mononuclear (PBMN) cells were isolated using Ficoll/Paque (Biochrom, Berlin, Germany) density gradient centrifugation of EDTA (Delta-Pharma, Pfullingen, Germany) anticoagulated blood buffy coat preparations from HV or from EDTA anticoagulated blood from AML patients. PBMN were seeded (1×106 cells/cm2) into culture flasks (Nunc, Roslild, Denmark) in RPMI1640 medium (Biochrom AG, Berlin, Germany) supplemented with 2% human AB serum (German Red Cross Blood Center, Ulm, Germany), 2 mM L-glutamine, 100 units/ml penicillin and 100 units/ml streptomycin. After 2 h of incubation at 37°C, non-adherent cells were removed and the adherent blood monocytes (purity>80% of CD14+ cells by FACS analysis) were cultured in RPMI1640 medium supplemented with 100 ng/ml human granulocyte-macrophage colony stimulating factor (GM-CSF, Leukomax, Novartis, Basel Switzerland), 1,000 IU/ml interleukin-4 (Strathmann, Hannover, Germany) and 10% human AB serum. For maturation of the cells, complete medium change was performed on day 6. The new medium contained GM-CSF and IL-4 in the concentrations mentioned above and, in addition, 50 ng/ml TNF-α (Strathmann). The DC cultures were fed with fresh medium and cytokines every three days and cell differentiation was monitored using inverse light microscopy. The expression of cell surface molecules on the DCs was analyzed using flow cytometry after 8 days of culture.
Cell viability and morphologic studies
The viability of the DCs harvested after 8 days of culture was assessed by trypan blue staining using a Neubauer counting chamber, and the morphology of the cells was analyzed by light microscopy.
Flow cytometry analysis
Harvested cells were washed in FACS medium (PBS containing 1% BSA) and stained at 4°C for 20 min by antibodies directly conjugated with FITC or PE. Thereafter cells were washed three times with PBS and analyzed by FACScan (Becton Dickinson, Heidelberg, Germany) using the CellQuest software (Becton Dickinson). The antibodies used were the following: FITC–labeled anti-mouse IgG, anti-human HLA-DR, CD40 and CD83, as well as PE–labeled anti-mouse IgG, anti-human HLA-ABC, CD 80 and CD86 (Becton Dickinson). Double staining was performed using pairs of PE- and FITC-labeled antibodies. Results were obtained in the dot plot format and transferred from the MacIntosh Apple to the PC format.
Quantitative PCR for LAA
Quantitative PCR was carried out using standard amplification conditions. The LightCycler FastStart DNA SYBR Green I Kit (Roche Diagnostics Mannheim, Germany) was used according to the manufacturer’s protocol. To quantify the mRNA expression of PRAME, RHAMM, WT-1 and proteinase 3, standard curves were established for RHAMM, WT-1 and proteinase 3 by a serial dilution of the products from conventional RT-PCR. A PRAME standard curve was established by a serial dilution of the pcDNA3.1-PRAME vector. The expression of PRAME, RHAMM and WT-1 was evaluated against the standard, and in a second step it was correlated with the mRNA expression of the housekeeping gene TATA-box binding protein (TBP). Primers were designed to produce transcript specific bands for LAA. PRAME: forward: GGA AGG TGC CTG TGA TGA AT; reverse: AAG GTG GGT AGC TTC CAG GT (accession number: NM_006115). RHAMM: forward: 5′ GCT GAA AGG CTG GTC AAG CAA 3′; reverse: 5′ TTC CTG CAA AAG CAG GGT GG 3′ (accession number: NM_012484.1). WT-1: forward: 5′ ATG AGG ATC CCA TGG GCC AGC A 3′; reverse: 5′ CCT GGG ACA CTG AAC GGT CCC CGA 3′ (accession number: NM_000378.2). Proteinase 3: forward: 5′ aac att tgc act ttc gtc cc 3′; reverse: 5′ gcg tga aga agt cag gga aa 3′ (accession number:NM_002777). TBP: forward: 5′ TTA ACT TCG CTT CCG CTG GC 3′; reverse: 5′ GAA ACC CTT GCG CTG GAA CT 3′ (accession number: NM_003194.2). PCR was performed under the following conditions: PRAME: 95°C denaturation for 10 s, 64°C annealing for 10 s, 72°C elongation for 20 s, 40 cycles, 0.5 μM each primer and 2.5 mM MgCl2. RHAMM: 95°C denaturation for 10 s, 62°C annealing for 15 s, 72°C elongation for 20 s, 45 cycles, 0.5 μM each primer and 3 mM MgCl2. WT-1: 95°C denaturation for 10 s, 62°C annealing for 15 s, 72°C elongation for 20 s, 45 cycles, 0.4 μM each primer and 2.25 mM MgCl2. Proteinase 3: 95°C 10 s, 60°C 10 s, 72°C 20 s, 40 cycles. 0.4 μM each primer and 2 mM MgCl2. TBP: 95°C 10 s, 60°C 15 s, 72°C 20 s, 40 cycles. 0.4 μM each primer and 2 mM MgCl2.
The formula for the calculation of copy numbers is as follows:
Copy number (copies/μl)=6×1023 (copies/mol) × concentration (g/μl) /molecular weight (g/mol), while 6×1023 is the Avogadro factor. The expression ratio was calculated as the quotient copy number of LAA/copy number of TBP, TBP being the housekeeping gene for normalization.
Immunocytology for RHAMM (CD168)
The method has been described elsewhere [4]. Briefly, cytospins of PBMN from HV and DCs generated thereof (HV-DCs), as well as AML blasts and DCs generated thereof (AML-DCs) were incubated with a monoclonal anti-CD168 antibody (clone 2D6, Novocastra, Newcastle upon Tyne, UK) at a 1:100 dilution. A polyclonal biotinylated Fab antibody to mouse IgG reactive with all mouse isotypes and a streptavidin-biotinylated peroxidase complex (all obtained from Amersham, High Wycombe, UK) served as a detection system for the primary antibody. 3-Amino-9-ethylcarbazole (Sigma, St. Louis, MO, USA) was used as substrate for the enzyme. The cytospin preparations were faintly counterstained with Harris’ hematoxylin. Negative controls were performed without primary antibody.
Mixed lymphocyte peptide culture
PBMN from a healthy volunteer, negative for RHAMM, were separated by Ficoll and subsequently selected by magnetic beads through a MACS column. CD8-antigen presenting cells (APCs) were irradiated with 30 Gy and pulsed with the PRAME-derived epitope decamer peptide ALYVDSLFFL (14; synthesized at a purity >95% by ThermElectron, Ulm, Germany) at a concentration of 20 μg/ml for 2 h. After co-incubation with CD8+ T lymphocytes overnight, the MLPC was supplemented with IL-2 and IL-7 on day +1. On day +7, the medium was changed and new, irradiated and peptide-pulsed CD8-APCs were added, as it was done one week before. Again, the Mixed lymphocyte peptide culture (MLPC) was supplemented with IL-2 and IL-7. After totally 14 days of culture, T cells were harvested and evaluated for their specific cytotoxicity in a standard Cr-51 release assay against T2 cells pulsed with the PRAME peptide or without peptide, as described earlier [34]. K562, a cell line established from a CML patient in blast crisis, was used as a control for T-cell specificity as K562 expresses PRAME [17], but not HLA class I molecules, thus being a target to natural killer (NK) cells, but not to T lymphocytes. The percentage of specific lysis was calculated as Cr-51 release in the test well minus spontaneous Cr-51 release/maximum Cr-51 release minus spontaneous Cr-51 release.
Results
Morphology of AML- and HV-DCs
After 8 days of culture as described in the material and methods section, floating and semi-adherent cells generated from AML blasts (AML-DCs) showed a typical DC morphology with characteristic vails and a kidney-shaped nucleus. Viability of AML- and HV-DC preparations tested > 95% by trypan blue staining (data not shown).
Immunophenotyping of AML blasts and AML-DCs
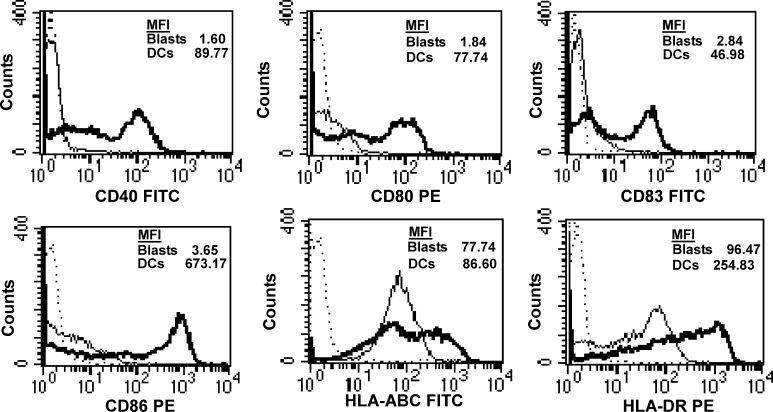
As demonstrated for one exemplary patient in Fig. 1, the expression of CD40, CD80, CD83, CD86 and HLA-DR was upregulated on AML-DCs.
Fig. 1.
The expression of HLA and costimulatory molecules on AML blasts (thin lines) and AML-DCs (bold lines) of patient no. 11 as analysed by flow cytometry. Isocontrols are shown as dotted lines
Quantitative PCR for PRAME, RHAMM, WT-1 and proteinase 3
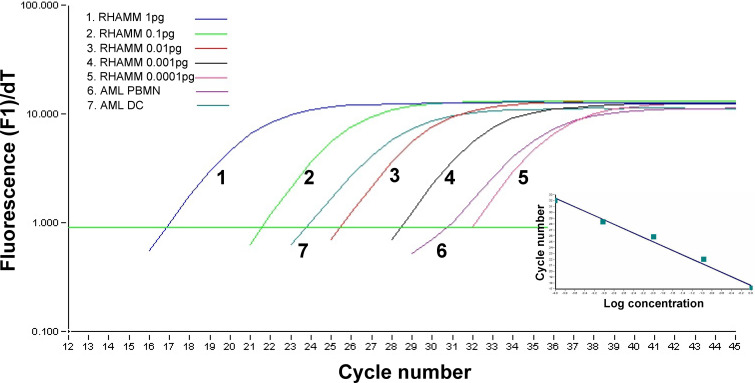
The RT-PCR protocols for the four LAAs (PRAME, RHAMM, WT-1 and proteinase 3) using the Light cycler SYBR green method were optimized as described earlier for PRAME [37] and demonstrated for RHAMM in Fig. 2 and Fig. 3. PCR conditions were chosen to obtain a clear and sharp single peak for the PCR products in the melting curve analysis presented in Fig. 2. For a dilution of a conventional RHAMM PCR product, the light cycler procedure was amplified for one representative analysis in Fig. 3. Subsequently, the regression analysis of these curves was calculated as displayed in the insert of Fig. 3.
Fig. 2.
Normalized amplification curves of different amounts (1 pg down to 0.0001 pg) of the antigen RHAMM cDNA. The inset shows the regression analysis of these curves. Two curves indicate the results obtained for PBMN (blasts>90%) of AML patient no. 3 and for the AML-DCs generated thereof
Fig. 3.
Quantitative real-time PCR analysis of PRAME expression compared between blasts from 12 AML patients and AML-DCs generated thereof. The expression ratio was calculated as PRAME copy number/TBP copy number. For details see the Material and methods section
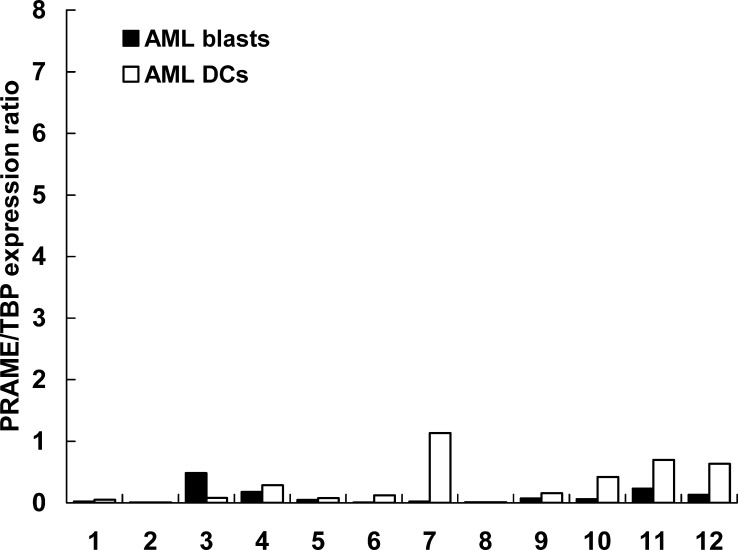
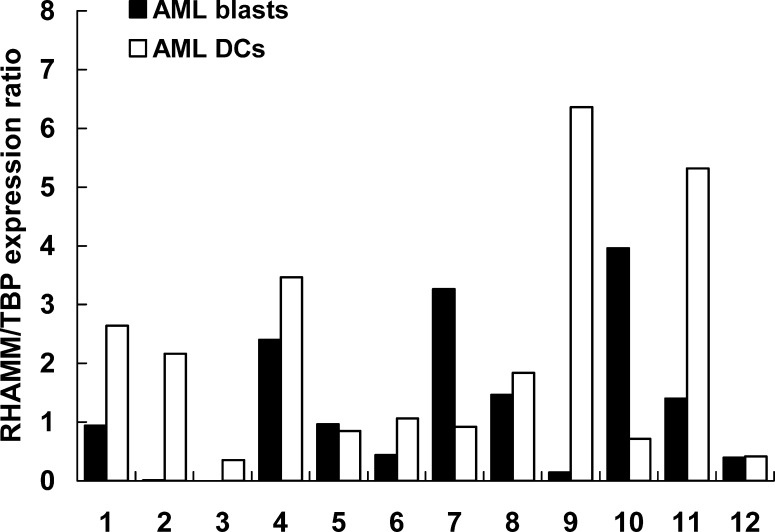
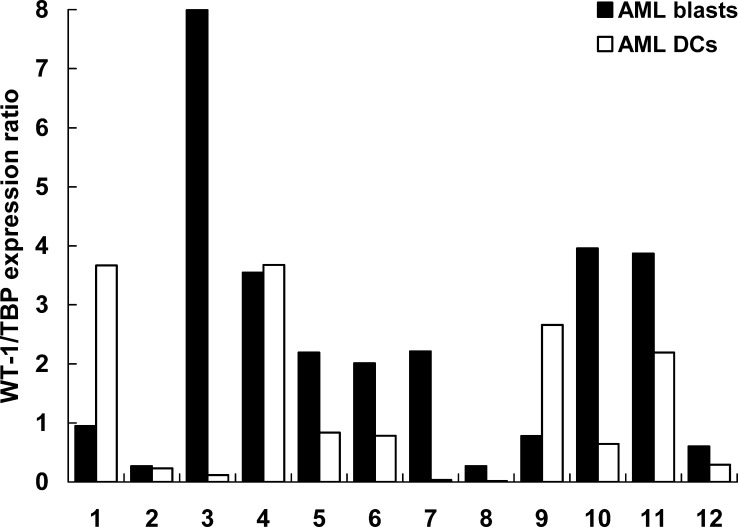
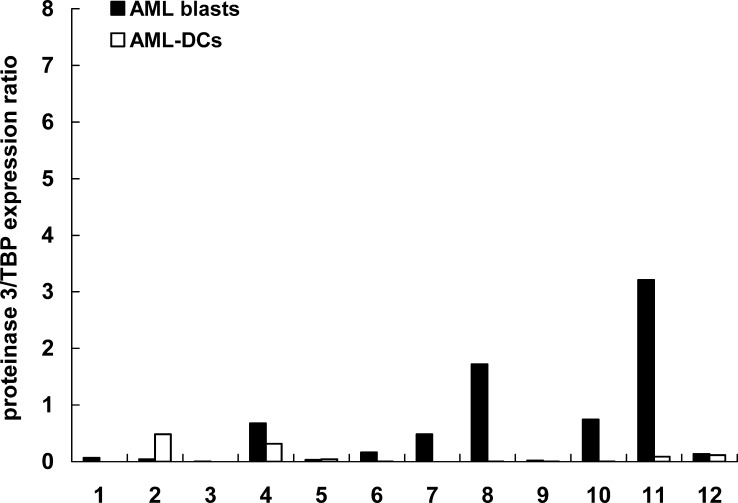
Results of quantitative real-time PCR for AML-DCs versus AML blasts showed an up to one log alteration in mRNA expression of LAAs in AML-DCs. As displayed in Fig. 4, a stronger PCR signal for PRAME was detected in 7/12 AML-DC preparations, in 4/12 AML-DC preparations the expression level was maintained and reduced only in 1/12 AML-DC preparations. In 6/12 AML-DC preparations, a significant upregulation of the PCR signal for RHAMM could be observed. In 4/12 AML-DC preparations, the RHAMM mRNA expression was maintained and in 2/12 AML-DC preparations it was reduced (Fig. 5). A stronger PCR signal for WT-1 was observed in 2/12 AML-DC preparation, 3/12 AML-DC preparations maintained the level of WT-1 expression. In 7/12 cases, the WT-1 mRNA expression was downregulated in the AML-DCs (Fig. 6). A stronger PCR signal for proteinase 3 was observed for only 1/12 AML-DC preparation, 4/12 AML-DC preparations maintained the level of proteinase 3 expression. In 7/12 cases, the proteinase 3 mRNA expression was downregulated (Fig. 7). In PBMN from HV and in HV-DCs, none of the four LAAs revealed to be expressed (all LAA/TBP copy number ratios<0.01; data not shown).
Fig. 4.
Quantitative real-time PCR analysis of RHAMM expression compared between blasts from 12 AML patients and AML-DCs generated thereof. The expression ratio was calculated as RHAMM copy number/TBP copy number. For details see the Material and methods section
Fig. 5.
Quantitative real-time PCR analysis of WT-1 expression compared between blasts from 12 AML patients and AML-DCs generated thereof. The expression ratio was calculated as WT-1 copy number/TBP copy number. For details see the Material and methods section
Fig. 6.
Quantitative real-time PCR analysis of proteinase 3 expression compared between blasts from 12 AML patients and AML-DCs generated thereof. The expression ratio was calculated as proteinase 3 copy number/TBP copy number. For details see the Material and methods section
Fig. 7.
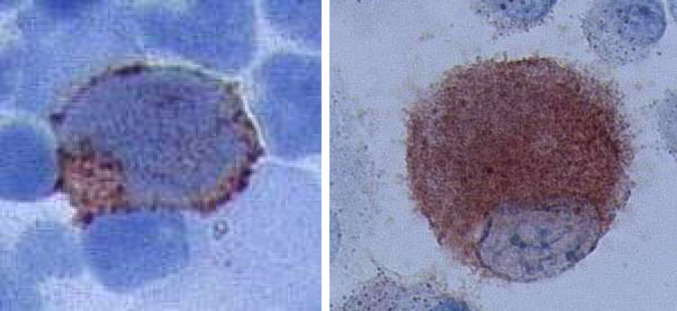
Immunocytological staining for RHAMM/CD168 was performed for AML blasts (upper left) and AML-DCs (upper right). Positive cells were found in both cell population. These findings prove the origin of AML-DCs from RHAMM-positive AML blasts (patient no. 4)
Simultaneous expression of LAAs
Table 2 gives a summarized comparison of the LAAs expression data obtained in both AML blasts and AML-DCs. The majority of LAAs is upregulated in most of the patients during AML-DC generation. LAAs are upregulated (for example PRAME, RHAMM and WT-1 in patient no. 1), while another LAA is downregulated in the same patient (proteinase 3). In all patients at least one LAA was maintained or even upregulated (Table 3).
Table 2.
Phenotype of DC generated from blasts of AML patients on day 8 of culture
| Patient no. | CD40 | CD80 | CD83 | CD86 | HLA-ABC | HLA-DR |
|---|---|---|---|---|---|---|
| 1 | 81.9 | 80.0 | 59.2 | 95.7 | 97.6 | 98.1 |
| 2 | 23.9 | 17.5 | 10.8 | 77.2 | 95.4 | 92.2 |
| 3 | 70.1 | 68.1 | 1.3 | 38.8 | 91.6 | 84.2 |
| 4 | 60.5 | 21.6 | 6.8 | 67.0 | 77.3 | 85.0 |
| 5 | 40.6 | 36.4 | 13.5 | 80.9 | 90.7 | 95.2 |
| 6 | 92.8 | 89.0 | 44.2 | 96.5 | 98.0 | 98.6 |
| 7 | 95.1 | 97.5 | 49.1 | 98.8 | 98.9 | 99.7 |
| 8 | 90.3 | 85.5 | 64.3 | 97.3 | 98.7 | 99.8 |
| 9 | 75.6 | 55.9 | 61.1 | 77.2 | 91.1 | 98.8 |
| 10 | 79.6 | 27.0 | 42.0 | 91.1 | 88.5 | 93.8 |
| 11 | 87.2 | 79.9 | 74.5 | 90.9 | 98.1 | 98.9 |
| 12 | 97.2 | 92.0 | 81.1 | 100.0 | 97.1 | 99.0 |
Table 3.
The distribution of LAAs expressed on AML-DCs compared with those on AML blasts
| Patient no. | PRAME | RHAMM | WT-1 | Proteinase 3 |
|---|---|---|---|---|
| 1 | ↑ | ↑ | ↑ | ↓ |
| (0.020) | (0.938) | (0.950) | (0.069) | |
| 2 | ~ | ↑ | ~ | ↑ |
| (0.006) | (0.012) | (0.267) | (0.043) | |
| 3 | ↓ | ↑ | ↓ | ~ |
| (0.483) | (0.000) | (7.990) | (0.006) | |
| 4 | ~ | ~ | ~ | ↓ |
| (0.176) | (2.400) | (3.550) | (0.675) | |
| 5 | ~ | ~ | ↓ | ~ |
| (0.044) | (0.961) | (2.190) | (0.030) | |
| 6 | ↑ | ↑ | ↓ | ↓ |
| (0.006) | (0.438) | (2.010) | (0.163) | |
| 7 | ↑ | ↓ | ↓ | ↓ |
| (0.020) | (3.260) | (2.210) | (0.483) | |
| 8 | ~ | ~ | ↑ | ↑ |
| (0.009) | (1.460) | (0.267) | (1.720) | |
| 9 | ↑ | ↑ | ↑ | ~ |
| (0.070) | (0.143) | (0.776) | (0.021) | |
| 10 | ↑ | ↓ | ↓ | ↓ |
| (0.059) | (3.960) | (3.960) | (0.745) | |
| 11 | ↑ | ↑ | ~ | ↓ |
| (0.231) | (1.400) | (3.870) | (3.210) | |
| 12 | ↑ | ~ | ↓ | ~ |
| (0.128) | (0.395) | (0.600) | (0.138) |
↑ upregulation, ↓ downregulation, ~ maintenance of the LAA expression; the expression ration for the respective LAA in AML blasts is indicated in brackets below the respective symbol
Immunocytology for RHAMM/CD168
The AML blasts, staining positive for RHAMM, were detected in the Ficoll preparation of the peripheral blood from AML patients (Fig. 7; left). AML blasts identified by morphology showed staining positive for RHAMM at different levels of intensity maybe reflecting a different stage of maturation or a different stage of cell cycle. Furthermore, we detected CD168-positive DCs generated from AML blasts (Fig. 7; right). False positive reactions due to intrinsic peroxidase activity were excluded by negative controls without primary antibody. PBMN from HV and DCs generated thereof tested negative for RHAMM (data not shown).
Mixed lymphocyte peptide culture
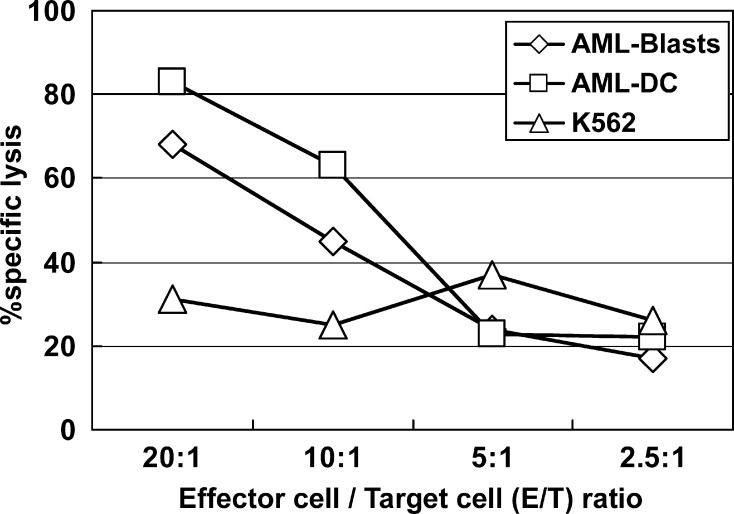
The CD8+ T cells, presensitized with the PRAME-derived peptide P3, were able to recognize AML blasts testing positive for this LAA in quantitative RT-PCR assays. AML-DCs generated thereof were recognized at a higher lytic level by the P3-primed T lymphocytes (Fig. 8).
Fig. 8.
The PRAME expressing AML blasts from patient no. 11 were maturated into AML-DCs with sustained PRAME expression. CD8+ T lymphocytes were presensitized for one week with the PRAME-derived peptide P3. Such primed T cells were able to recognize PRAME-positive AML blasts to a certain level. The lysis of AML-DCs through P3-primed T cells was much stronger, thus indicating the preservation of the T-cell epitope through the DC maturation and the highly immunogenic potential of LAA expressing/presenting AML-DCs. K562, a cell line established from a CML patient in blast crisis, was used as a control for T cell specificity as K562 expresses PRAME (16), but not HLA class I molecules, thus being a target to natural killer (NK) cells, but not to T lymphocytes
Discussion
Autologous AML-DCs generated from AML blasts might be used as a vaccine potentially eliciting LAA-specific T-cell responses. To design such experiments and clinical trials, the origin of DCs from leukemic blasts and the maintenance of LAA expression have to be demonstrated. The origin of AML-DCs has been proved by other groups using fluorescence in situ hybridization (FISH) [9, 10]. However, AML-DC samples for laboratory tests are rather restricted in clinical trials. Moreover, only about 50% of AML patients (not only of those included in the present study) show chromosomal aberrations detectable by the FISH probes available [14]. To ensure a rapid method for the evaluation of LAAs expression in AML-DCs with only 105 cells, we established RT-PCR for testing LAAs in AML [21].
Ikeda et al. [19] reported PRAME as a melanoma associated tumor antigen recognized by cytotoxic T lymphocytes (CTLs) in an HLA-A24- restricted manner. Expression of the gene PRAME was described in melanoma, sarcoma, lung squamous-cell carcinoma, renal-cell carcinoma [19, 20, 28], and in 35% of AML patients [23], whereas no expression was found in PBMN and bone marrow cells from HV [15, 38]. In this study, PRAME was expressed at a rather low level in AML blasts when compared with the housekeeping gene TBP. However, a remarkable upregulation of the gene was detected in 7/12 AML-DCs, suggesting that these cells might be a useful tool to elicit T cell responses to PRAME, which were otherwise difficult to obtain (J.H. Kessler/C.J. Melief, Leiden University Medical Center, NL, Ref. 14 and personal communication). This finding was confirmed by our data demonstrating recognition of PRAME-expressing AML blasts and AML-DCs by P3-primed CD8+ lymphocytes.
The RHAMM, also designated CD168, is a receptor for hyaluronan, a glycoaminoglycan that plays a fundamental role in cell growth, differentiation, and motility [40]. CD168 is a cell-surface receptor and belongs to the group of extracellular matrix molecules [35, 41]. The molecule is highly expressed in different tumor cell lines [1, 37] in multiple myeloma [12] and AML/CML [16], but not in PBMN of health volunteers [31]. RHAMM was upregulated in 6/12 AML-DCs. We could confirm the RHAMM protein expression both on the surface and in the cytoplasm of these AML-DCs by immunocytology.
The Wilms’ tumor antigen WT-1, a differentiation gene first described in pediatric kidney tumors [7], is expressed in the majority (about 80%) of AML patients, but also in CD34 positive cells of the early hematopoiesis and its expression is correlated with a poor clinical prognosis [5, 24, 29]. Because of the expression in CD34 positive cells, which has been a matter of debate for several years [7, 18, 24, 29, 30], vaccination against WT-1 might stimulate autoimmune responses.
Our quantitative mRNA expression data suggest that RHAMM might have a similar potential as WT-1 to elicit T cell responses, at least as far as AML-DCs are used for vaccination of patients. The down-regulation of WT-1 in AML-DCs when compared with AML blasts might be related to the maturation process of DCs as WT-1 is also downregulated during the maturation of CD34+ stem cells [24].
Proteinase 3 is a myeloid tissue-restricted 26 kDa serine protease that is abundantly expressed in azurophilic granules in normal myeloid cells, is overexpressed by two- to fivefold in some leukemia cells, and may be important for maintenance of the leukemic phenotype [6, 13]. Evidence that proteinase 3 is immunogenic was recently obtained in patients with CML and AML [25–27, 33]. The quantitative RT-PCR for proteinase 3 revealed an expression of the gene in 8/12 of the AML blasts when compared with the housekeeping gene TBP. This characterizes proteinase 3 as a gene with an inhomogenous mRNA expression level in our cohort of AML patients. Only in 2/12 samples, an expression ratio bigger than 1 (compared with TBP) was revealed. When AML-DCs where generated from the AML blasts, only 1/12 samples upregulated the mRNA expression of proteinase 3. In most of the cases (7/12), proteinase 3 mRNA was dramatically downregulated. This indicates that generating AML-DCs might not constitute a method to induce effectively T-cell answers against proteinase 3. It remains to be elucidated whether the loss of proteinase 3 expression is associated with the maturation process as it is the case for WT-1.
In this study, quantitative RT-PCR showed an upregulation of the LAAs PRAME and RHAMM in AML-DCs compared with AML blasts in the majority of the patients. This might be due to the maturation process of the DCs or due to the higher percentage of AML-DCs in the AML-DC preparation when compared to the percentage of blasts in the peripheral blood of the patients. The stronger expression of LAAs by AML-DCs when compared to AML blasts make AML-DCs better APCs for a stronger stimulation of a specific T-cell response. In addition, vaccination with AML-DCs can circumvent the problem of low expression of a certain LAA, as AML-DCs might still express another LAA efficiently (Table 1).
In summary, we have proven the origin of DCs from AML blasts and the maintained or even upregulated expression of LAAs in AML-DCs using a technique, which requires only a small number of DCs. The results were confirmed for RHAMM on the protein level by immunocytology. As HV-DCs did not show any expression of the LAAs tested, we conclude that their expression is not related to differentiation into DCs per se. Taken together with the expression of HLA and costimulatory molecules [21, 39], the expression of multiple LAAs in AML-DCs is a prerequisite for the stimulation of a LAA-specific T-cell reaction as we demonstrated for the PRAME derived peptide P3 in cytotoxicity assays. LAA expressing autologous AML-DCs might be a promising vaccine for AML patients for maintenance therapy in complete remission or as a specific immunotherapy for minimal residual disease (MRD).
For the selection of patients for clinical trials with AML-DCs, the production of antibodies against LAAs of interest for immunocytology, as well as the definition of T-cell epitopes of more LAAs (e.g., RHAMM/CD168) for immuno-monitoring would be highly desirable. Such studies are ongoing in our laboratory.
References
- 1.Abetamann V, Kern HF, Elsasser HP. Differential expression of the hyaluronan receptors CD44 and RHAMM in human pancreatic cancer cells. Clin Cancer Res. 1996;2:1607–1618. [PubMed] [Google Scholar]
- 2.Banchereau J, Brière F, Caux C, Davoust J, Lebecque S, Liu YJ, Pulendran B, Palucka K. Immunobiology of dendritic cells. Annu Rev Immunol. 2000;18:767–811. doi: 10.1146/annurev.immunol.18.1.767. [DOI] [PubMed] [Google Scholar]
- 3.Banchereau J, Schuler-Thurner B, Palucka AK, Schuler G. Dendritic cells as vectors for therapy. Cell. 2001;106:271–274. doi: 10.1016/S0092-8674(01)00448-2. [DOI] [PubMed] [Google Scholar]
- 4.Barth TF, Rinaldi N, Bruderlein S, Mechtersheimer G, Strater J, Altevogt P, Moller P. Mesothelial cells in suspension expose an enriched integrin repertoire capable of capturing soluble fibronectin and laminin. Cell Commun Adhes. 2002;9:1–14. doi: 10.1080/15419060212184. [DOI] [PubMed] [Google Scholar]
- 5.Bergmann L, Miething C, Maurer U, Brieger J, Karakas T, Weidmann E, Hoelzer D. High levels of Wilms’ tumor gene (wt1) mRNA in acute myeloid leukemias are associated with a worse long-term outcome. Blood. 1997;90:1217–1225. [PubMed] [Google Scholar]
- 6.Bories D, Raynal MC, Solomon DH, Darzynkiewicz Z, Cayre YE. Down-regulation of a serine protease, myeloblastin, causes gwoth arrest and differentiation of promyelocytic leukemia cells. Cell. 1989;59:959–968. doi: 10.1016/0092-8674(89)90752-6. [DOI] [PubMed] [Google Scholar]
- 7.Call KM, Glaser T, Ito CY, Buckler AJ, Pelletier J, Haber DA, Rose EA, Kral A, Yeger H, Lewis WH. Isolation and characterization of a zinc finger polypeptide gene at the human chromosome 11 Wilms’ tumor locus. Cell. 1990;60:509–520. doi: 10.1016/0092-8674(90)90601-A. [DOI] [PubMed] [Google Scholar]
- 8.Choudhury A, Gajewski JL, Liang JC, Popat U, Claxton DF, Kliche KO, Andreeff M, Champlin RE. Use of leukemic dendritic cells for the generation of anti-leukemic cellular cytotoxicity against Philadelphia chromosome-positive chronic myelogenous leukemia. Blood. 1997;89:1133–1142. [PubMed] [Google Scholar]
- 9.Choudhury A, Toubert A, Sutaria S, Charron D, Champlin RE, Claxton DF. Human leukemia-derived dendritic cells: ex-vivo development of specific antileukemic cytotoxicity. Crit Rev Immunol. 1998;18:121–131. doi: 10.1615/critrevimmunol.v18.i1-2.130. [DOI] [PubMed] [Google Scholar]
- 10.Choudhury BA, Liang JC, Thomas EK, Flores-Romo L, Xie QS, Agusala K, Sutaria S, Sinha I, Champlin RE, Claxton DF. Dendritic cells derived in vitro from acute myelogenous leukemia cells stimulate autologous antileukemic T cell responses. Blood. 1999;93:780–786. [PubMed] [Google Scholar]
- 11.Cignetti A, Bryant E, Allione B, Vitale A, Foa R, Cheever MA. CD34(+) acute myeloid and lymphoid leukemic blasts can be induced to differentiate into dendritic cells. Blood. 1999;94:2048–2055. [PubMed] [Google Scholar]
- 12.Crainie M, Belch AR, Mant MJ, Pilarski LM. Overexpression of the receptor for hyaluronan- mediated motility (RHAMM) characterizes the malignant clone in multiple myeloma: identification of three distinct RHAMM variants. Blood. 1999;93:1684–1696. [PubMed] [Google Scholar]
- 13.Dengler R, Munstermann U, al-Batran S, Hausner I, Faderl S, Nerl C, Emmerich B. Immunocytochemical and flow cytometric detection of proteinase 3 (myeloblastin) in normal and leukaemic myeloid cells. Br J Haematol. 1995;89:250–257. doi: 10.1111/j.1365-2141.1995.tb03297.x. [DOI] [PubMed] [Google Scholar]
- 14.Frohling S, Skelin S, Liebisch C, Scholl C, Schlenk RF, Dohner H, Dohner K. Comparison of cytogenetic and molecular cytogenetic detection of chromosome abnormalities in 240 consecutive adult patients with acute myeloid leukemia. J Clin Oncol. 2002;20:2480–2485. doi: 10.1200/JCO.2002.08.155. [DOI] [PubMed] [Google Scholar]
- 15.Greiner J, Ringhoffer M, Taniguchi M, Hauser T, Schmitt A, Dohner H, Schmitt M. Characterization of several leukemia-associated antigens inducing humoral immune responses in acute and chronic myeloid leukemia. Int J Cancer. 2003;106:224–231. doi: 10.1002/ijc.11200. [DOI] [PubMed] [Google Scholar]
- 16.Greiner J, Ringhoffer M, Taniguchi M, Schmitt A, Kirchner D, Krahn G, Heilmann V, Gschwend J, Bergmann L, Dohner H, Schmitt M. Receptor for hyaluronan acid-mediated motility (RHAMM) is a new immunogenic leukemia- associated antigen in acute and chronic myeloid leukemia. Exp Hematol. 2002;30:1029–1035. doi: 10.1016/S0301-472X(02)00874-3. [DOI] [PubMed] [Google Scholar]
- 17.Greiner J, Ringhoffer M, Szmaragowska A (eds) (2002) Quantitative measurement of the mRNA expression of the tumor-associated antigen PRAME by real-time RT-PCR using the lightcycler and SYBR green I technology. Rapid cycle real-time PCR methods and applications. Genetics and oncology. Springer, Berlin Heidelberg New York, pp 177–186
- 18.Hosen N, Sonoda Y, Oji Y, Kimura T, Minamiguchi H, Tamaki H, Kawakami M, Asada M, Kanato K, Motomura M, Murakami M, Fujioka T, Masuda T, Kim EH, Tsuboi A, Oka Y, Soma T, Ogawa H, Sugiyama H. Very low frequencies of human normal CD34+ haematopoetic progenitor cells express the Wilms’ tumour gene WT1 at levels similar to those in leukaemia cells. Br J Haematol. 2002;116:409–420. doi: 10.1046/j.1365-2141.2002.03261.x. [DOI] [PubMed] [Google Scholar]
- 19.Ikeda H, Lethe B, Lehmann F, van Baren N, Baurain JF, de Smet C, Chambost H, Vitale M, Moretta A, Boon T, Coulie PG. Characterization of an antigen that is recognized on a melanoma showing partial HLA loss by CTL expressing an NK inhibitory receptor. Immunity. 1997;6:199–208. doi: 10.1016/S1074-7613(00)80426-4. [DOI] [PubMed] [Google Scholar]
- 20.Kessler JH, Beekman NJ, Bres-Vloemans SA, Verdijk P, van Veelen PA, Kloosterman-Joosten AM, Vissers DC, ten Bosch GJ, Kester MG, Sijts A, Wouter Drijfhout J, Ossendorp F, Offringa R, Melief CJ. Efficient identification of novel HLA-A*0201-presented cytotoxic T lymphocyte epitopes in the widely expressed tumor antigen PRAME by proteasome-mediated digestion analysis. J Exp Med. 2001;193:73–88. doi: 10.1084/jem.193.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li L, Schmitt A, Reinhardt P, Greiner J, Ringhoffer M, Vaida B, Bommer M, Vollmer M, Wiesneth M, Dohner H, Schmitt M. Reconstitution of CD40 and CD80 in dendritic cells generated from blasts of patients with acute myeloid leukemia. Cancer Immun. 2003;3:8. doi: 10.1016/S0008-8749(03)00155-2. [DOI] [PubMed] [Google Scholar]
- 22.Mailander V, Scheibenbogen C, Thiel E, Letsch A, Blau IW, Keilholz U. Complete remission in a patient with recurrent acute myeloid leukemia induced by vaccination with WT1 peptide in the absence of hematological or renal toxicity. Leukemia. 2004;18:165–166. doi: 10.1038/sj.leu.2403186. [DOI] [PubMed] [Google Scholar]
- 23.Matsushita M, Yamazaki R, Ikeda H, Kawakami Y. Preferentially expressed antigen of melanoma (PRAME) in the development of diagnostic and therapeutic methods for hematological malignancies. Leuk Lymphoma. 2003;44:439–444. doi: 10.1080/1042819021000035725. [DOI] [PubMed] [Google Scholar]
- 24.Maurer U, Weidmann E, Karakas T, Hoelzer D, Bergmann L. Wilms tumor gene (wt1) mRNA is equally expressed in blast cells from acute myeloid leukemia and normal CD34+ progenitors. Blood. 1997;90:4230–4232. [PubMed] [Google Scholar]
- 25.Molldrem Abstract. 2002;8:92. [Google Scholar]
- 26.Molldrem JJ, Lee PP, Kant S, Wieder E, Jiang W, Lu S, Wang C, Davis MM. Chronic myelogenous leukemia shapes host immunity by selective deletion of high-avidity leukemia-specific T cells. J Clin Invest. 2003;111:639–647. doi: 10.1172/JCI200316398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Molldrem JJ, Lee PP, Wang C, Felio K, Kantarjian HM, Champlin RE, Davis MM. Evidence that specific T lymphocytes may participate in the elimination of chronic myelogenous leukemia. Nat Med. 2000;6:1018–1023. doi: 10.1038/79526. [DOI] [PubMed] [Google Scholar]
- 28.Neumann E, Engelsberg A, Decker J, Storkel S, Jaeger E, Huber C, Seliger B. Heterogeneous expression of the tumor-associated antigens RAGE-1, PRAME, and glycoprotein 75 in human renal cell carcinoma: candidates for T-cell-based immunotherapies. Cancer Res. 1998;58:4090–4095. [PubMed] [Google Scholar]
- 29.Ohminami H, Yasukawa M, Fujita S. HLA class I-restricted lysis of leukemia cells by a CD8+ cytotoxic T-lymphocyte clone specific for WT1 peptide. Blood. 2000;95:286–293. [PubMed] [Google Scholar]
- 30.Oka Y, Udaka K, Tsuboi A, Elisseeva OA, Ogawa H, Aozasa K, Kishimoto T, Sugiyama H. Cancer immunotherapy targeting Wilms’ tumor gene WT1 product. J Immunol. 2000;164:1873–1880. doi: 10.4049/jimmunol.164.4.1873. [DOI] [PubMed] [Google Scholar]
- 31.Pilarski LM, Masellis-Smith A, Belch AR, Yang B, Savani RC, Turley EA. RHAMM, a receptor for hyaluronan-mediated motility, on normal human lymphocytes, thymocytes and malignant B cells: a mediator in B cell malignancy? Leuk Lymphoma. 1994;14:363–374. doi: 10.3109/10428199409049691. [DOI] [PubMed] [Google Scholar]
- 32.Sallusto F, Lanzavecchia A. Efficient presentation of soluble antigen by cultured human dendritic cells is maintained by granulocyte/macrophage colony-stimulating factor plus interleukin 4 and downregulated by tumor necrosis factor. J Exp Med. 1994;179:1109–1118. doi: 10.1084/jem.179.4.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scheibenbogen C, Letsch A, Thiel E, Schmittel A, Mailaender V, Baerwolf S, Nagorsen D, Keilholz U. CD8 T-cell responses to Wilms tumor gene product WT1 and proteinase 3 in patients with acute myeloid leukemia. Blood. 2002;100:2132–2137. doi: 10.1182/blood-2002-01-0163. [DOI] [PubMed] [Google Scholar]
- 34.Schmitt M, Ikeda H, Nagata Y, Gu X, Wang L, Kuribayashi K, Shiku H. Involvement of T-cell subsets and natural killer (NK) cells in the growth suppression of murine fibrosarcoma cells transfected with interleukin-12 (IL-12) genes. Int J Cancer. 1997;72:505–511. doi: 10.1002/(SICI)1097-0215(19970729)72:3<505::AID-IJC20>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 35.Sherman L, Sleeman J, Herrlich P, Ponta H. Hyaluronate receptors: key players in growth, differentiation, migration and tumor progression. Curr Opin Cell Biol. 1994;6:726–733. doi: 10.1016/0955-0674(94)90100-7. [DOI] [PubMed] [Google Scholar]
- 36.Stockerl-Goldstein KG, Blume KG (eds) (1999) Allogeneic hematopoietic cell transplantation for adult patients with acute myeloid leukemia. In: Thomas ED, Blume KG, Forman SJ (eds) Hematopoietic cell transplantation, 2nd edn, pp 823–834
- 37.Teder P, Bergh J, Heldin P. Functional hyaluronan receptors are expressed on a squamous cell lung carcinoma cell line but not on other lung carcinoma cell lines. Cancer Res. 1995;55:3908–3914. [PubMed] [Google Scholar]
- 38.van Baren N, Chambost H, Ferrant A, Michaux L, Ikeda H, Millard I, Olive D, Boon T, Coulie PG. PRAME, a gene encoding an antigen recognized on a human melanoma by cytolytic T cells, is expressed in acute leukaemia cells. Br J Haematol. 1998;102:1376–1379. doi: 10.1046/j.1365-2141.1998.00982.x. [DOI] [PubMed] [Google Scholar]
- 39.Vollmer M, Li L, Schmitt A, Greiner J, Reinhardt P, Ringhoffer M, Wiesneth M, Dohner H, Schmitt M. Expression of human leucocyte antigens and and costimulatory antigens on blasts of patients with acute myeloid leukaemia. Br J Haematol. 2003;120:1000–1008. doi: 10.1046/j.1365-2141.2003.04212.x. [DOI] [PubMed] [Google Scholar]
- 40.Wang C, Entwistle J, Hou G, Li Q, Turley EA. The characterization of a human RHAMM cDNA: conservation of the hyaluronan-binding domains. Gene. 1996;174:299–306. doi: 10.1016/0378-1119(96)00080-7. [DOI] [PubMed] [Google Scholar]
- 41.Yang B, Zhang L, Turley EA. Identification of two hyaluronan-binding domains in the hyaluronan receptor RHAMM. J Biol Chem. 1993;268:8617–8623. [PubMed] [Google Scholar]