Abstract
The movement of bipartite geminiviruses such as squash leaf curl virus (SqLCV) requires the cooperative interaction of two essential virus-encoded movement proteins, BR1 and BL1. While the viral coat protein AR1 is not essential for systemic infection, genetic studies demonstrate that its presence masks the defective phenotype of certain BR1 missense mutants, thus suggesting that coat protein does interact with the viral movement pathway. To further examine the mechanism of this interaction, we have constructed alanine-scanning mutants of AR1 and studied them for the ability to mask the infectivity defects of appropriate BR1 mutants, for the ability to target to the nucleus and to bind viral single-stranded DNA (ssDNA) and multimerize, and for effects on the accumulation of replicated viral ssDNA. We identified a specific region of AR1 required for masking of appropriate BR1 mutants and showed that this same region of AR1 was also important for ssDNA binding and the accumulation of viral replicated ssDNA. This region of AR1 also overlapped that involved in multimerization of the coat protein. We also found that the accumulation in protoplasts of single-stranded forms of a recombinant plasmid that included the SqLCV replication origin but was too large to be encapsidated was dependent on the presence of AR1 but did not appear to require encapsidation. These findings extend our model for SqLCV movement, demonstrating that coat protein affects viral movement through its ability to induce the accumulation of replicated viral ssDNA genomes. They further suggested that encapsidation was not required for the AR1-dependent accumulation of viral ssDNA.
To move cell to cell and systemically infect the host, plant viruses encode movement proteins (MPs) that are essential for infection but not required for viral replication or encapsidation (4, 29). Bipartite geminiviruses, such as squash leaf curl virus (SqLCV), have genomes of covalently closed circular single-stranded DNA (ssDNA) that replicate in the nucleus. This nuclear localization necessitates that these viruses encode two MPs, BR1 and BL1, both of which are essential for systemic infection of all hosts (50). Recent studies of SqLCV (46, 48, 49, 56) and bean dwarf mosaic virus (44) have shown that BR1 and BL1 act in a cooperative manner to move the viral genome intracellularly from the nucleus to the cytoplasm and across the wall cell to cell. BR1 is a nuclear shuttle protein, and it has been proposed to bind newly replicated viral ssDNA genomes and move these between the nucleus and cytoplasm (46, 48). These BR1-genome complexes are then directed to the cell periphery through interactions between BR1 and BL1 (48, 49), where, as the result of BL1 action, the complexes are moved to adjacent uninfected cells (44, 48, 49). The precise mechanism by which BL1 acts to transport these genome complexes across the cell wall, and whether this may differ in different cell types, remains at issue (44, 50, 56). BL1 encoded by the phloem-limited SqLCV has been immunolocalized to unique tubules that extend from and cross the walls of developing phloem cells, and it has been suggested that SqLCV BL1 acts to move BR1-genome complexes along these tubules and into adjacent uninfected phloem cells (56).
Genetic studies of bipartite geminiviruses, including SqLCV, have established that both BR1 and BL1 determine viral infectivity and host range properties and that BL1 determines viral pathogenic properties (25, 30, 31, 55), with BL1 being directly responsible for the production of viral disease symptoms (45). Genetic studies further show that the viral coat protein (CP) AR1 is not required for movement and systemic infection by the bipartite geminiviruses in their natural hosts. AR1 null mutants are infectious by means of agroinoculation or mechanical transmission, and they produce wild-type infection or delayed and attenuated symptoms, depending on the particular virus and host plant (6, 7, 22, 26, 31, 47). The accumulation of viral ssDNA in infected plants or protoplasts is also dramatically reduced when AR1 is absent, although the amount of viral double-stranded DNA (dsDNA) is not affected (47, 54).
These studies on AR1 null mutants were done in the presence of functional BR1 and BL1. However, recent genetic epistasis studies for alanine-scanning mutants of SqLCV MPs demonstrate that the presence of functional AR1 masks the infectivity defects of specific BR1 mutants in cucurbit hosts (31). In the presence of wild-type AR1, alanine-substituted missense mutants BR1K25A/R26A, BR1N201A/K202A/R203A, and BR1N219A are fully infectious and produce wild-type disease symptoms in pumpkin and squash seedlings. These same BR1 mutants are null in infectivity for Nicotiana benthamiana when AR1 is present, and in the absence of functional AR1 these specific BR1 mutants either are noninfectious (BR1K25A/R26A and BR1N201A/K202A/R203A) or have very low infectivity (BR1N219A) for pumpkin and squash (31). AR1 does not appear to substitute for BR1 and provide an alternate path for viral movement since BR1 as well as BL1 null mutants are not infectious when wild-type AR1 is present, and no interaction between AR1 and BL1 has been detected (31, 49). However, AR1, like BR1, is localized to the nucleus and binds ssDNA with high affinity (31, 48, 49). In addition, transient expression studies in tobacco protoplasts show that BR1K25A/R26A, BR1N201A/K202A/R203A, and BR1N219A are defective in nuclear localization, with all three mutants being slow in their targeting to the nucleus and BR1N219A appearing to also be defective in nuclear export (48).
These results have led to the suggestion that AR1 may affect viral movement by signaling a switch to rolling circle replication of viral ssDNA, the substrate that BR1 binds and shuttles between the nucleus and cytoplasm (31, 48). According to this model, AR1 would compensate for the slower nuclear accumulation and concentration of the three mutated BR1 proteins by maintaining high nuclear levels of replicated viral ssDNA. To further investigate this model and elucidate the role of AR1 in viral movement, we generated an extensive set of alanine-scanning AR1 mutants (14) and tested them for the ability to mask the infectivity defects of BR1K25A/R26A and BR1N201A/K202A/R203A in pumpkin seedlings. In this study, we identified a clustered set of AR1 mutations that were no longer infectious with BR1K25A/R26A or BR1N201A/K202A/R203A in pumpkin. Investigations of the AR1-dependent accumulation of ssDNA of viral size and larger, and of the in vitro DNA binding properties, nuclear localization, and multimerization of the CP encoded by these AR1 mutants, support the hypothesis that the ability of AR1 to bind ssDNA, potentially in a cooperative manner, and induce the accumulation of viral ssDNA was essential for AR1 to mask the infectivity of BR1K25A/R26A and BR1N201A/K202A/R203A and appeared not to be coupled to encapsidation.
MATERIALS AND METHODS
Construction of alanine-scanning mutants of AR1 and infectivity assays.
The genomic A and B components (AE and BE) of the extended-host-range strain of SqLCV (SqLCV-E) (35) were used in these studies. The 2.0-kb SstI-BamHI fragment of the SqLCV A component (Fig. 1A, SqLCV-A) containing the entire coding sequence of AR1 was inserted into pAlter-1 (Promega) to create pAlter-AR1. Single-stranded template DNA was prepared and mutagenized by hybridization with a synthetic mutagenic oligonucleotide and an oligonucleotide to restore the ampicillin resistance gene, followed by growth in Escherichia coli BMH-mutS and DH5α, as recommended by the manufacturer. To confirm the presence of the desired mutation and ensure that this mutation was the only one introduced and responsible for any phenotypes observed, a small restriction fragment encompassing each mutation was sequenced in its entirety by the dideoxy-chain termination method (51) and subcloned into the wild-type A-component background. The deduced amino acid sequence of AR1 showing the positions and names of the alanine-scanning mutations used in this study is shown in Fig. 2.
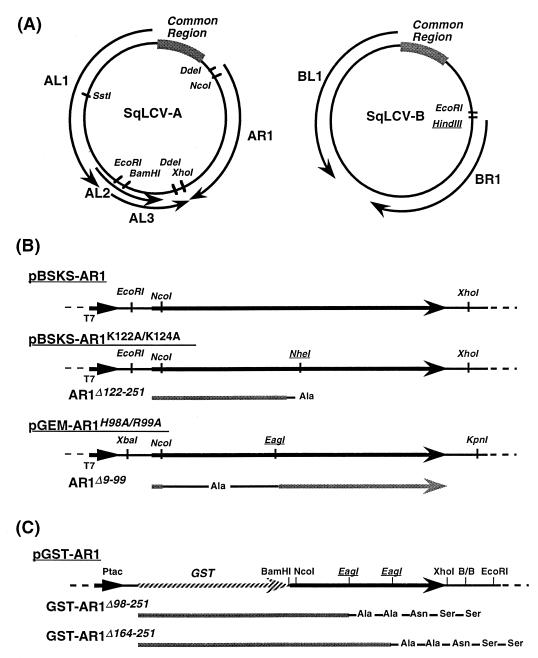
FIG. 1.
Genomic organization of SqLCV-E and construction of expression vectors for wild-type and mutant AR1. (A) Restriction map of SqLCV-E. A HindIII site was introduced in the B component by site-directed mutagenesis. (B) Expression vectors for in vitro transcription and translation. To construct pGEM-AR1, the DdeI-DdeI fragment was excised from SqLCV-AE, blunted, and ligated into SmaI-digested pGEM7Zf+. EagI and NheI sites on pBSKS-AR1 were introduced by site-directed mutagenesis for construction of AR1Δ9-99 and AR1Δ122-251, respectively. T7, T7 promoter. (C) Construction of GST fusions for expression in bacterial cells. The two EagI sites (italicized and underlined) were introduced by site-directed mutagenesis to construct pGST-AR1Δ98-251 and pGST-AR1Δ164-251, respectively. See text for details.
FIG. 2.
Alanine-scanning mutations in SqLCV AR1. Shown is the amino acid sequence (one-letter code) of AR1. Mutations are indicated by lines over the amino acid(s) altered to alanine. The name of each mutant is shown above the appropriate mutated peptide sequence, with each mutation named according to the position(s) within AR1 and the amino acid(s) altered to alanine.
To assay infectivity, each mutant A component was dimerized by ligating the 2.57-kb EcoRI-BamHI fragment and 2.0-kb EcoRI-SstI fragment (Fig. 1A) into SstI- and BglII-digested pMON505 and then agroinoculated together with the appropriate wild-type or mutant SqLCV B component (also referred to as SqLCV-B) as previously described (35, 36). Plants were subsequently scored for the appearance of disease symptoms, and the presence of viral A and B components in systemic leaves was detected by the analysis of DNA-containing extracts on Southern blots, using appropriate component-specific probes (35, 40, 52). Probes were labeled by random oligonucleotide-primed synthesis using [α-32P]dCTP (17).
Detection of AR1 protein in infected pumpkin.
Systemic leaves were harvested from symptomatic plants and ground in liquid nitrogen. Ground tissue (0.1 g) was mixed with 300 μl of polyacrylamide gel electrophoresis (PAGE) buffer (0.2 M Tris, 30% glycerol, 3% sodium dodecyl sulfate [SDS]), 3% β-mercaptoethanol, 0.0015% bromophenol blue) by vigorous vortexing, followed by incubation at 100°C for 10 min and centrifugation at 14,000 × g for 5 min. Twenty microliters of each sample was analyzed by SDS-PAGE on 12.5% polyacrylamide gels, using the discontinuous buffer system (33). Resolved proteins were immunoblotted as previously described (45), using a 1:2,000 dilution of polyclonal anti-AR1 antiserum. The generation of rabbit polyclonal antiserum raised against AR1 has been previously described (49).
DNA-cellulose binding assay.
For the synthesis of in vitro-transcribed and -translated AR1, the EcoRI-XhoI fragment containing the coding region of wild-type AR1 was excised from pGEM-AR1 (31) and cloned into pBluescript(KS+) (Stratagene) at the EcoRI and XhoI sites to create a transcriptional fusion of AR1 to the T7 promoter (pBSKS-AR1) (Fig. 1B). For most AR1 mutants, the NcoI-XhoI fragment containing amino acids 8 to 251 of AR1 was substituted for the corresponding fragment of AR1 in pBSKS-AR1. To create AR1Δ122-251, pBSKS-AR1K122A/K124A, which contained a unique introduced NheI site at nucleotides 366 to 371 of the AR1 coding sequence, was digested with NheI and XhoI, blunted by incubation with Klenow fragment in the presence of all four deoxynucleoside triphosphates, and religated (Fig. 1B) (5). To create AR1Δ9-99, an XbaI-KpnI fragment from pBSKS-AR1H98A/R99A, which contained a unique introduced EagI site at nucleotides 293 to 298 in the AR1 coding sequence, was ligated into pGEM7Zf+ (Promega), thus creating pGEM-AR1H98A/R99A (Fig. 1B). pGEM-AR1H98A/R99A was digested with NcoI and EagI, blunted with Klenow fragment, and religated. Plasmid DNA of these AR1 expression constructs was prepared by using the Wizard DNA purification system (Promega) according to the manufacturer’s instructions. Plasmid DNA was further purified by two phenol-chloroform extractions and ethanol precipitation.
[35S]methionine-labeled AR1 was transcribed and translated in vitro by using the TnT coupled reticulocyte lysate system (Promega) according to manufacturer’s instructions. Labeled AR1 (3 μl) was incubated at room temperature for 20 min with 300 μl of ssDNA-coupled cellulose (Pharmacia) equilibrated with buffer Z (50 mM Tris-HCl [pH 8.0], 12.5 mM MgCl2, 1 mM EDTA, 0.1% Nonidet P-40, 20% glycerol, 1 mM phenylmethylsulfonyl fluoride, 1 mM dithiothreitol) containing 50 mM KCl (Z-50 buffer) as described previously (46). Following five washes with 300 μl of Z-50 buffer, bound protein was eluted from the resin by successive washes with 300 μl of Z buffer containing 100 mM, 200 mM, 400 mM, and 1 M KCl. Washes and elutions were performed at the same temperature as the initial binding step. Following the 1 M KCl wash, residual bound proteins were eluted by incubation in 300 μl of PAGE buffer at 100°C for 5 min (46). Fractions of eluted protein (20 μl) were analyzed by SDS-PAGE on 12.5% polyacrylamide gels (33), and gels were impregnated for fluorography prior to exposure to X-ray film (34).
Protein binding assay.
To construct pGST-AR1, the entire coding sequence of SqLCV-E AR1 was synthesized by PCR (43) using the sense-strand primer 5′-CGCGGATCCATGGTTAAGAGAGA-3′ and the complementary-strand primer 5′-GGCAGATCTATGGTAATGGATTTACGC-3′. This PCR fragment was digested with BamHI and BglII and ligated into BamHI-digested pGEX-2T (Pharmacia) to create pGST-AR1(PCR). The NcoI-XhoI fragment of pGST-AR1(PCR) encompassing amino acids 8 to 251 of the 251 amino acids of AR1 was replaced by the corresponding restriction fragment from wild-type AR1, thus creating pGST-AR1 (Fig. 1C). To construct pGST-AR1Δ98-251, the NcoI-XhoI fragment of pGST-AR1 was replaced by the corresponding restriction fragment from pBSKS-AR1H98A/R99A, creating pGST-AR1H98A/R99A, which was then digested with EagI and EcoRI, blunted with Klenow fragment, and religated (Fig. 1C). To construct pGST-AR1Δ164-251, the NcoI-XhoI fragment of pGST-AR1 was replaced by the corresponding restriction fragment from pBSKS-AR1K164A/N165A/D166A to create pGST-AR1K164A/N165A/D166A, containing a unique introduced EagI site at nucleotides 491 to 496 of the AR1 coding sequence. This was then digested with EagI and EcoRI, blunted, and religated (Fig. 1C).
Plasmid DNA of pGST-AR1, pGST-AR1Δ98-251, and pGST-AR1Δ164-251 was transformed into E. coli DH5α. To induce high levels of expression of each glutathione S-transferase (GST) fusion protein, 20 ml of bacterial culture at an optical density at 600 nm of 1.0 to 1.5 was induced in the presence of 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 3 h at 37°C. Cells were pelleted by centrifugation at 8,000 × g for 5 min, resuspended in 5 ml of 1× phosphate-buffered saline (PBS; 140 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4 [pH 7.3]), and sonicated in a Sonifier 200 cell disrupter (Smith-Kline). Triton X-100 was added to a final concentration of 0.5% to aid in solubilization of fusion proteins, and the lysate was gently shaken for 30 min at 4°C. Insoluble protein was pelleted by centrifugation at 30,000 × g for 15 min; the supernatant (0.5-ml aliquot) was mixed with 50 μl of a 50% suspension of glutathione-coupled Sepharose beads (Sigma) in 1× PBS and gently rocked for 30 min at 4°C. Protein bound to the glutathione-Sepharose was pelleted by centrifugation at 16,000 × g for 5 to 10 s, washed three times with 500 μl of 1× PBS, and resuspended in 300 μl of 1× PBS containing 0.1 mg of bovine serum albumin per ml.
For AR1 binding assays, 5 μl of in vitro-translated [35S]methionine-labeled AR1 was incubated with the appropriate GST-AR1-coupled resin (E. coli-expressed GST-AR1, wild type or mutant, bound to glutathione-Sepharose) in 1× PBS containing 0.1 mg of bovine serum albumin per ml and gently agitated for 30 min at 4°C. Following pelleting at 16,000 × g for 5 to 10 s and six washes with 500 μl of 1× PBS containing 0.1% Triton X-100, labeled AR1 bound to the coupled beads was eluted with 300 μl of PAGE buffer and incubated at 100°C for 5 min. Fractions of eluted protein (20 μl) were analyzed by SDS-PAGE on 12.5% acrylamide gels as described above.
Assays for replication or encapsidation.
The EcoRI-SstI fragment (2.0 kb) and EcoRI-BamHI fragment (2.5 kb) of SqLCV-A (Fig. 1A) were cloned into pEMBL19+ which had been digested with SstI and BamHI, creating pBL19-2AE. As constructed, this clone contains a partial tandem direct repeat of the AE genome that includes two copies of the viral common region. The partial tandemly repeated clone of BE, constructed by ligating a full-length SalI linear copy of BE to the SalI-XbaI fragment encompassing the common region (1.44 kb) (Fig. 1A) cloned in pEMBL19+ (pEB8-54; hereafter called pBL19-2BE), has been described elsewhere (36). The EcoRI-HindIII fragment (2.6 kb) of SqLCV-B (Fig. 1A) was cloned into pEMBL19+ to create the 6.8-kb plasmid pBL19-BE, containing the SqLCV common region and replication origin. pBL1-AR1fs8/Δ15-251 (ΔAR1) contains a partial tandem repeat of SqLCV-A with a frameshift mutation in AR1 that results in a nonsense codon at residue 15 and has been described elsewhere (31).
For the replication assay, cesium chloride gradient-purified pBL19-BE (10 μg) was mixed with 10 μg of pBL19-2AE and 100 μg of carrier salmon sperm DNA, and the mixture was electroporated into Nicotiana tabacum var. Xanthi protoplasts (hereafter referred to as Xanthi protoplasts), using a Bio-Rad GenePulser as previously described (21, 49). Following incubation at 26°C for 72 to 96 h, protoplasts were collected by centrifugation at 100 × g for 5 min, resuspended in extraction buffer containing 1% cetyltrimethylammonium bromide, 50 mM Tris-HCl (pH 8.0), 0.7 M NaCl, 10 mM EDTA, 0.5% polyvinylpyrrolidone, and 0.1% β-mercaptoethanol, and incubated for 1 h at 60°C (40). The lysates were extracted once with chloroform, and DNA was ethanol precipitated and resuspended in 10 mM Tris-HCl–1 mM EDTA (TE; pH 8.0). Viral DNA was detected by analysis of appropriately digested samples on Southern blots (52), using SqLCV component-specific probes as described above.
For the encapsidation assay, 20 μg of cesium chloride gradient-purified pBL19-2AE together with 20 μg of either pBL19-2BE or pBL19-BE was electroporated into Xanthi protoplasts as described above and incubated for 96 h at 26°C. Cells were pelleted at 100 × g, resuspended in 7 ml of reticulocyte standard buffer (10 mM Tris [pH 6.8], 10 mM NaCl, 1.5 mM MgCl2, 1 mM phenylmethylsulfonyl fluoride), and incubated on ice for 20 min. To separate nuclear and cytoplasmic fractions, Nonidet P-40 was added to 0.5%; following disruption by 20 strokes in a tissue homogenizer (Kontes Duall), the lysate was centrifuged at 100 × g. The supernatant (cytoplasmic fraction) was removed, and the pelleted nuclear fraction was ground to a fine powder under liquid nitrogen and resuspended in 0.5 ml of nuclear extraction buffer (10 mM NaPO4 [pH 7.4], 1 mM EDTA). Following clarification at 800 × g, the nuclear extract was loaded onto a linear (10 to 40% [wt/vol]) sucrose gradient and centrifuged at 110,000 × g for 1.5 h at 4°C. Fractions (1 ml) were collected and stored at −20°C prior to analysis. Fifty microliters of each fraction was denatured by adding an equal volume of 1 N NaOH–20 mM EDTA. These preparations were dot blotted onto nylon that had been presoaked in 4× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and neutralized in TE. DNA was crossed-linked to the nylon (UV Stratalinker 1800; Stratagene) and hybridized with strand-specific oligonucleotide probes that had been end labeled with 32P by using T4 polynucleotide kinase (5).
RESULTS
Infectivity phenotypes of AR1 mutants.
To identify regions of AR1 that potentially affected viral movement, we assayed alanine-scanning AR1 mutants (Fig. 2) for the ability to mask the infectivity defects of SqLCV mutants BR1K25A/R26A and BR1N201A/K202A/R203A in pumpkin. As found for AR1 null mutants (31), all of these SqLCV AR1 mutants achieved wild-type levels of 100% infectivity when coinoculated with the wild-type SqLCV B component on pumpkin, producing typical severe disease symptoms with no delay in the onset of their appearance (data not shown). However, these AR1 mutants fell into three broad classes, similar to BR1 and BL1 missense mutants (31), based on their infectivity when coinoculated onto pumpkin with either BR1K25A/R26A or BR1N201A/K202A/R203A (Table 1). Class I mutants had wild-type levels of 100% infectivity and produced severe symptoms similar to those of wild-type SqLCV infection. Constructs with mutations AR1Q74A/R75A/H76A, AR1K122A/K124A, AR1K164A/N165A/D166A, and AR1K213A/Y214A/E215A were in this class. Class II mutants were reduced in infectivity, with 13 to 56% of inoculated plants being infected in the presence of BR1K25A/R26A and 44 to 88% of inoculated plants becoming infected in the presence of BR1N201A/K202A/R203A. All of these class II AR1 mutants were characterized by a delay in the initial appearance of disease symptoms of 3 to 5 days (with BR1K25A/R26A) or 1 to 3 days (with BR1N201A/K202A/R203A) compared to inoculation with wild-type B component, and they produced attenuation of symptoms characterized by a milder mosaic and less epinasty. Mutants AR1R46A/K47A, AR1K102A/R103A, and AR1K240A/R242A were in this second class. Class III mutants were noninfectious in pumpkin in the presence of either BR1K25A/R26A or BR1N201A/K202A/R203A, with no viral DNA detected in systemic leaves from plant inoculated with these mutants (data not shown). This is the same phenotype found for a SqLCV AR1 null mutant and the C-terminal truncation mutant AR1Δ201-251, which produces a protein defective in DNA binding (31). These class III mutants were AR1K68A/Q70A, AR1H98A/R99A, AR1D118A/E119A, AR1R138A/R139A, AR1D155A/N156A/E157A, AR1H175A/R176A, AR1N189A/E190A/Q191A, and AR1R195A/R196A.
TABLE 1.
Infectivity of SqLCV AR1 mutants
| AR1 mutationa | Symptomsb | BR1K25A/R26A
|
BR1N201A/K202A/R203A
|
||
|---|---|---|---|---|---|
| Infectivityc (%) | Dayd | Infectivity (%) | Day | ||
| None (wild type) | wt | 100 | 6 | 100 | 6 |
| Class I | |||||
| Q74A/R75A/H76A | wt | 100 | 12 | 100 | 12 |
| K122A/K124A | wt | 100 | 7 | 100 | 6 |
| K164A/N165A/D166A | wt | 100 | 9 | 100 | 7 |
| K213A/Y214A/E215A | wt | 100 | 10 | 100 | 10 |
| Class II | |||||
| R46A/K47A | att | 31 | 14 | 44 | 14 |
| K102A/R103A | att | 56 | 11 | 88 | 10 |
| K240A/R242A | att | 13 | 9 | 88 | 9 |
| Class III | |||||
| K68A/Q70A | — | 0 | 28 | 0 | 28 |
| H98A/R99A | — | 0 | 28 | 0 | 28 |
| D118A/E119A | — | 0 | 28 | 0 | 28 |
| R138A/R139A | — | 0 | 28 | 0 | 28 |
| D155A/N156A/E157A | — | 0 | 28 | 0 | 28 |
| H175A/R176A | — | 0 | 28 | 0 | 28 |
| N189A/E190A/Q191A | — | 0 | 28 | 0 | 28 |
| R195A/R196A | — | 0 | 28 | 0 | 28 |
SqLCV A component containing the indicated mutation was coinoculated onto pumpkin with the B component containing the BR1 mutation as shown.
Appearance of systemic symptoms in pumpkin: wt, symptoms identical to those for wild-type SqLCV; att, attenuated, characterized by less chlorosis and epinasty; —, no visible symptoms.
Number of symptomatic plants/total number of inoculated plants (average from three independent trials).
Day postinoculation on which infectivity plateaued (average from three independent trials). For noninfectious mutants, this is the final day on which plants were scored.
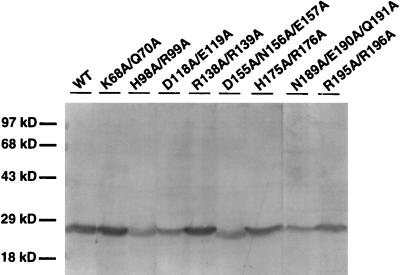
All of these class III AR1 mutants, when coinoculated with wild-type SqLCV B, produced coat protein in infected pumpkin plants that could be detected on immunoblots up to at least 12 days postinoculation. The level of each mutant AR1 detected in infected plants was similar to that of wild-type AR1 (Fig. 3). Thus, it appeared that the phenotypes of these AR1 mutants, particularly those in classes II and III, were not simply due to instability of the mutant proteins produced.
FIG. 3.
Synthesis of AR1 in infected pumpkin plants. Shown are Western blots of AR1 synthesized by each class III AR1 mutant, as indicated above each lane, when coinoculated onto pumpkin with wild-type SqLCV B. Proteins were extracted from the first true leaf at 12 days following inoculation of cotyledons. WT, wild-type SqLCV A component. Sizes of protein molecular mass standards (in kilodaltons) are indicated on the left.
Effects of AR1 mutations on the accumulation of viral ssDNA.
The finding that AR1 null mutants (6, 31, 54) and a C-terminal truncation of AR1 that no longer binds DNA (31) all cause greatly diminished levels of viral ssDNA to accumulate, and that both AR1 and BR1 are located in the nucleus and ssDNA binding proteins (31, 46), has led to the suggestion that the ability of wild-type SqLCV AR1 to mask the infectivity defects of mutants BR1K25A/R26A and BR1N201A/K202A/R203A may involve the ability of AR1 to induce the nuclear accumulation of replicated viral ssDNA, the substrate which BR1 will bind and move (46, 48, 49). Consistent with this hypothesis is the observation that the three BR1 mutants specifically masked by AR1 are defective in their nuclear targeting and, in one case, apparently nuclear export (48, 50). We therefore investigated the effects of AR1 mutants on the accumulation of viral ssDNA in pumpkin plants when coinoculated with wild-type SqLCV B component.
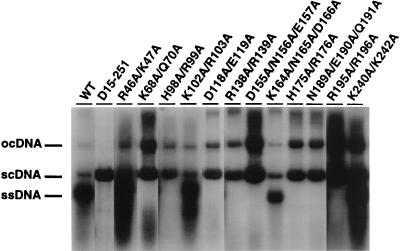
As shown in Fig. 4, high levels of viral ssDNA accumulated in systemic leaves from plants inoculated with class I and class II AR1 mutants. All of these mutants either completely or partially masked the infectivity defects of BR1K25A/R26A and BR1N201A/K202A/R203A (Table 1). In contrast, no viral ssDNA was detected when plants were infected by class III AR1 mutants (Fig. 4), all of which had lost the ability to mask the two SqLCV BR1 mutants (Table 1). Notably, each of these class III mutants still replicated viral dsDNA to high levels (Fig. 4). Similar results were also obtained in Xanthi protoplasts transfected with the different AR1 mutants (data not shown). Thus, there was a correlation between the ability of AR1 mutants to induced high levels of viral ssDNA accumulation and mask the infectivity defects of mutants BR1K25A/R26A and BR1N201A/K202A/R203A.
FIG. 4.
Effects of AR1 mutations on the accumulation of viral ssDNA in infected pumpkin plants. Shown are Southern blots of DNA extracted from symptomatic systemic leaves of pumpkin plants inoculated with wild-type SqLCV-A (WT) or AR1 mutants, as indicated above lanes, in combination with wild-type SqLCV-B. The blot was hybridized with a probe specific for SqLCV-A. scDNA, supercoiled dsDNA; ocDNA, open circular dsDNA.
AR1 mutants with defects in ssDNA binding are defective in masking BR1 mutants.
AR1 is a ssDNA binding protein, and a C-terminal truncation mutant, AR1Δ201-251, that does not mask the infectivity defects of BR1K25A/R26A, BR1N201A/K202A/R203A, or BR1N219A is defective in DNA binding (31). We therefore tested the ability of our AR1 mutant proteins to bind ssDNA.
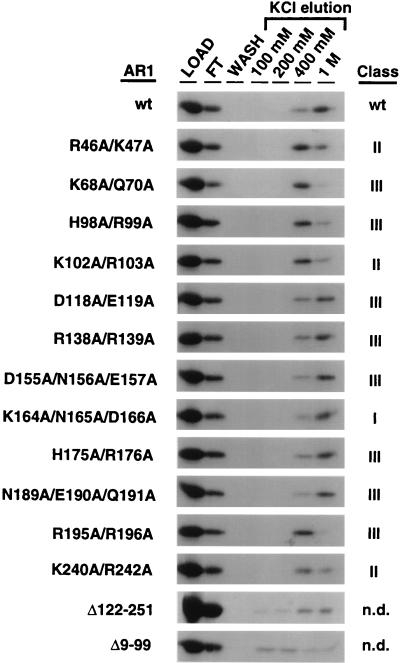
As shown previously (31), wild-type AR1 binds ssDNA with a high affinity, with some AR1 eluting from ssDNA coupled to cellulose at 400 mM KCl but most remaining bound until the 1 M KCl elution (Fig. 5). The four AR1 mutants AR1Q74A/R75A/H76A, AR1K122A/K124A, AR1K164A/N165A/D166A, and AR1K213/Y214/E215, all of which masked BR1K25A/R26A and BR1N201A/K202A/R203A, bound ssDNA with affinities similar to that of wild-type AR1 (Fig. 5 and data not shown). AR1R46A/K47A, AR1K68A/Q70A, AR1H98A/R99A, and AR1K102/R103, each with missense mutations in the N-terminal half of AR1, were all decreased in their affinity for ssDNA, with most of the protein eluting at 400 mM KCl (Fig. 5). AR1K68A/Q70A and AR1H98A/R99A do not mask the infectivity defects of BR1K25A/R26A and BR1N201A/K202A/R203A, and AR1R46A/K47A and AR1K102A/R103A only partially mask these two BR1 mutants (Table 1). C-terminal mutants AR1R195A/R196A and AR1K240A/R242A also had decreased binding affinities for ssDNA (Fig. 5), consistent with the C-terminal truncation mutant AR1Δ201-251 not binding ssDNA or dsDNA (31). Again, AR1R195A/R196A did not mask the infectivity defects of BR1K25A/R26A and BR1N201A/K202A/R203A, and AR1K240A/R242A only partially masked these BR1 mutants. AR1D118A/E119A, AR1R138A/R139A, AR1D155A/N156A/E157A, AR1H175A/R176A, and AR1N189A/E190A/Q191A each bound ssDNA with affinities similar to that of wild-type AR1 (Fig. 5). However, none of these AR1 mutants masked the infectivity defects of BR1K25A/R26A and BR1N201A/K202A/R203A.
FIG. 5.
Binding of wild-type or mutant AR1 to ssDNA-cellulose: SDS-PAGE of AR1 (wild-type [wt] and mutants as indicated) bound to ssDNA-cellulose and eluted at the salt concentrations indicated. [35S]methionine-labeled in vitro-synthesized AR1 was incubated with ssDNA coupled to cellulose resin. Protein was eluted from resins by successive washes with buffer containing increasing concentrations of KCl up to 1 M. Following the 1 M salt elution, no residual bound protein was eluted by boiling in SDS sample buffer (not shown). Equivalent amounts of each eluted fraction, including the LOAD (input protein), FT (flowthrough protein not bound to resin), and WASH (final wash of resin with loading buffer prior to elutions with higher salt) lanes, were analyzed by SDS-PAGE. Not shown were AR1Q74A/R75A/H76A, AR1K122A/K124A, and AR1K213A/Y214A/E215A, all of which retained the ability to mask and had essentially the same profile of ssDNA binding as wild-type AR1 and AR1K164A/N165A/D166A. See text for details.
These DNA binding studies suggested that the ssDNA binding domain of AR1 was located in the N-terminal region of the protein. To further explore this possibility, two truncation mutants of AR1 were constructed. AR1Δ122-251 is a C-terminal truncation mutant that encodes amino acids 1 to 121 of AR1. AR1Δ9-99 is an in-frame deletion mutant that lacks the region encoding residues 9 to 99 of AR1. As shown in Fig. 5, AR1Δ122-251 still bound ssDNA with a fairly high affinity, binding to the ssDNA-cellulose in amounts comparable to that of wild-type AR1, with most of the protein eluting at 400 mM and 1 M KCl. AR1Δ9-99 was defective in DNA binding, binding ssDNA with a very low affinity, as evident from the lower amount binding to the ssDNA-cellulose, and what little was bound eluted at 100 mM and 200 mM KCl (Fig. 5). Thus, it appears that the N-terminal 121 amino acids of AR1 are involved in DNA binding, possibly encompassing the DNA binding domain. The defects in DNA binding of C-terminal mutants AR1R195A/R196A, AR1K240A/R242A, and AR1Δ201-251 may be the consequence of these mutations affecting the conformation of the protein or part of the DNA binding site itself.
Results of our DNA binding studies suggested that the ability of AR1 to bind ssDNA with high affinity was necessary, but not sufficient, to mask BR1K25A/R26A and BR1N201A/K202A/R203A. However, our findings for AR1D118A/E119A, AR1R138A/R139A, AR1D155A/N156A/E157A, AR1H175A/R176A, and AR1N189A/E190A/Q191A, which did not mask these two BR1 mutants even though they all bound ssDNA with a high affinity, would be explained if these mutants were defective in nuclear targeting. To explore this possibility, each of these five mutant AR1 proteins was expressed in transfected Xanthi protoplasts as transcriptional fusions to the cauliflower mosaic virus 35S promoter and then localized by indirect immunofluorescence staining and confocal microscopy.
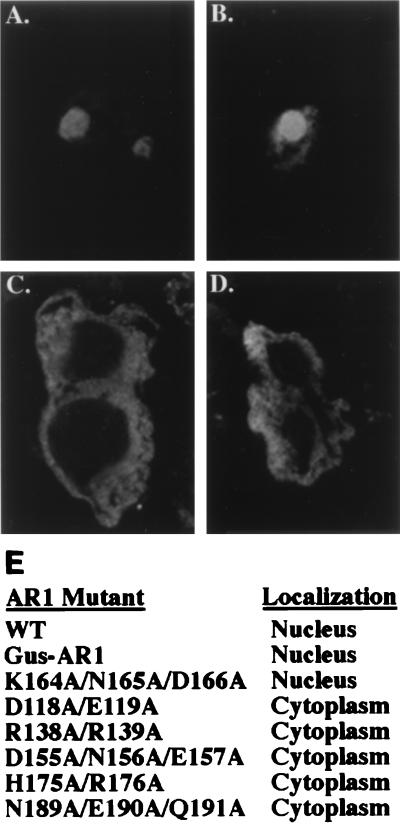
In contrast to wild-type AR1, which localized to the nuclei of expressing protoplasts (Fig. 6A), all five mutant proteins—AR1D118A/E119A, AR1R138A/R139A, AR1D155A/N156A/E157A, AR1H175A/R176A, and AR1N189A/E190A/Q191A—mislocalized to the cytoplasm of the transfected protoplasts (Fig. 6C and D; Table 1). AR1K164A/N165A/D166A, a class I mutant which bound ssDNA with an affinity similar to that of these five nonmasking mutants but did mask the infectivity defects of BR1K25A/R26A and BR1N201A/K202A/R203A, was correctly localized to the nucleus (Fig. 6B). Thus, the AR1D118A/E119A, AR1R138A/R139A, AR1D155A/N156A/E157A, AR1H175A/R176A, and AR1N189A/E190A/Q191A mutations failed to mask BR1K25A/R26A and BR1N201A/K202A/R203A because the mutant AR1 protein encoded by each did not target to the nucleus. These results demonstrate that the nucleus was the site at which AR1 exerted its effect on BR1 function in movement and that the ability of AR1 to bind ssDNA with a high affinity was necessary and sufficient to mask BR1K25A/R26A and BR1N201A/K202A/R203A once AR1 had entered the nucleus. In addition, a fusion protein consisting of AR1 fused to the C terminus of β-glucuronidase (GUS) was also targeted to the nucleus, based on both assay for GUS activity and indirect immunofluorescence staining for GUS protein (Fig. 6). Thus, consistent with out mutant studies, transport of AR1 to the nucleus is an active process mediated by a nuclear targeting signal(s) that appears to be located within the central region of AR1 encompassed by residues 118 to 191.
FIG. 6.
Nuclear localization of AR1. Wild-type (WT) AR1, a fusion of GUS to AR1, and AR1 alanine-scanning mutants were expressed in Xanthi protoplasts and localized by indirect immunofluorescence staining with anti-CP antiserum and a Texas red-conjugated secondary antibody and confocal microscopy. (A) Wild-type AR1 localized to nuclei in two protoplasts. (B) Nuclear localization of AR1K164A/N164A/D166A, a mutant that masks the defective phenotypes of BR1K25A/R26A and BR1N201A/K202A/R203A. (C) Cytoplasmic mislocalization of AR1H175A/R176A in a recently divided protoplasts. (D) Cytoplasmic mislocalization of AR1D118A/E119A. (E) Results of localization studies for these proteins as well as AR1R138A/R139A, AR1D155A/N156A/E157A, and AR1N189A/E190A/Q191A.
Encapsidation is not essential for the accumulation of viral ssDNA.
There is precedence in animal virus and phage systems for DNA replication and encapsidation in virion assembly being coupled events (3, 16, 32, 38, 42). Thus, the accumulation of viral ssDNA in SqLCV-infected plants could be the consequence of encapsidation into virions protecting the genome from nuclease degradation. Alternatively, AR1 could signal a switch from semiconservative viral dsDNA replication to the replication of viral ssDNA by a rolling circle mechanism without requiring packaging of the genomic ssDNA into mature capsids or sequester viral ssDNA from the replication pool in the absence of encapsidation. We therefore investigated whether encapsidation was required for the AR1-dependent accumulation of viral ssDNA.
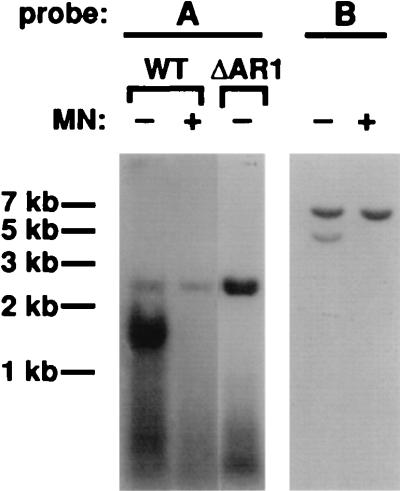
Xanthi protoplasts were cotransfected with SqLCV-A, cloned as a tandem direct repeat in pEMBL19+ (pBL19-2AE; see Materials and Methods), and SqLCV-B, cloned as a single-copy 2.6-kb EcoRI-HindIII fragment (Fig. 1) into pEMBL19+ (pBL19-BE; see Materials and Methods). As expected, SqLCV-A was efficiently excised from pBL19-2AE by rolling circle replication in the transfected protoplasts (53, 54), and both dsDNA and ssDNA replicated forms of 2.6 kb were found to accumulate to high levels (Fig. 7). Identification of the ssDNA was confirmed by mung bean nuclease digestion (Fig. 7). In contrast, the single cloned insert of SqLCV in pBL19-BE cannot excise from the plasmid, either by rolling circle replication or by homologous recombination (37, 53), and was replicated as the 6.8-kb recombinant plasmid (Fig. 7). Both dsDNA and ssDNA replicated forms of pBL19-BE 6.8 kb in size were detected in the transfected protoplasts (Fig. 7). Identification of the ssDNA was again confirmed by mung bean nuclease digestion (Fig. 7). Earlier studies show that large recombinant plasmids that contain the SqLCV replication origin are replicated in an AL1-dependent manner, although less efficiently than the smaller 2.6-kb SqLCV genomic components, and only the recombinant dsDNA form is detected in the absence of AR1 (37). As the replicated 6.8-kb pBL19-BE ssDNA would be too large to be encapsidated within the geminivirus icosahedral capsid, which by analogy with the packaging limits established for other icosahedral phage and animal viruses would maximally package a nucleic acid of 2.6 kb ± 15 to 20% (2, 8, 9, 11, 41), these results suggested that packaging of the viral ssDNA into a fully assembled capsid is not required for the AR1-dependent accumulation of viral ssDNA.
FIG. 7.
Replication of pBL19-BE ssDNA and dsDNA in the presence of wild-type AR1. Shown are Southern blots of DNA extracted from Xanthi protoplasts that were cotransfected with pBL19-2AE (wild type [WT]) and pBL19-BE or with pBL1-AR1fs8/Δ15-251 (ΔAR1) containing SqLCV-A with a frameshift mutation that results in a nonsense codon at residue 15 (31). DNA was digested with EcoRI. Blots were probed with an SqLCV-A- or SqLCV-B-specific probe, as indicated. −, not treated with mung bean nuclease (MN); +, treated with mung bean nuclease. No pBL19-BE ssDNA was replicated in the presence of the frameshift mutant pBL1-AR1fs8/Δ15-251 (data not shown).
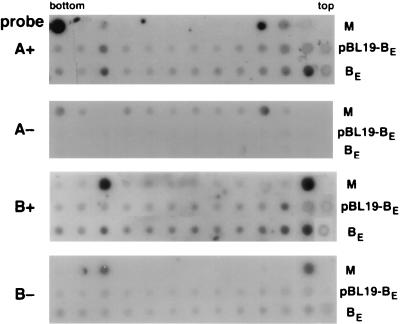
To demonstrate this directly, nuclear extracts from Xanthi protoplasts transfected with the tandem repeat of the SqLCV A component (pBL19-2AE) and either the cloned tandem repeat of the wild-type B component (pBL19-2BE) or the 6.8-kb pBL19-BE clone were prepared, and encapsidated forms of the viral ssDNA were isolated by differential centrifugation on sucrose gradients (see Materials and Methods). Using strand-specific probes for the SqLCV A and B components, dot blot analysis of gradient fractions from protoplasts transfected with pBL19-2AE and pBL19-2BE, which would replicate the genomic 2.6-kb AE and BE, detected the virion positive-sense strands of the A and B components in a peak sedimenting at the expected density for encapsidated virions (Fig. 8) (1). In contrast, for protoplasts transfected with pBL19-2AE and the 6.8-kb pBL19-BE, only the virion positive-sense strand of AE was detected sedimenting at the density of encapsidated virions; neither the virion nor the complementary strand of the 6.8-kb pBL19-BE was detected in this peak (Fig. 8). Thus, although pBL19-BE ssDNA is replicated in the presence of the wild-type SqLCV A component, it appears that it is not encapsidated into virions.
FIG. 8.
Velocity gradient centrifugation analysis of replicated 6.8-kb pBL19-BE ssDNA and wild-type 2.6-kb AE and BE. Nuclear extracts from transfected protoplasts were analyzed on 10 to 40% (wt/vol) sucrose gradients, and fractions were dotted onto nylon and hybridized with ssDNA probes specific for the virion positive-sense (+) or complementary negative-sense (−) strands of SqLCV-A and -B. M, SqLCV AE and BE dsDNA dotted as controls for specificity of ssDNA probes. AE is dotted above the position of the fourth fraction from the tops of the gradients and, in the top panel, above the bottom gradient fraction as well. BE is dotted above the second fraction from the top of the gradients and the third fraction from the bottom of the gradients. pBL19-BE, fractions from protoplasts replicating 2.6-kb AE and 6.8-kb pBL19-BE. AE virion positive-strand ssDNA is detected in a peak three fractions from the bottom of the gradient, but neither positive- nor negative-strand B-component probes detect the replicated pBL19-BE in this fraction; pBL19-BE is detected only at the top of the gradient. BE, fractions from protoplasts replicating wild-type 2.6-kb AE and BE. Note that virion positive-strand ssDNA for both the AE and BE components is detected in a peak three fractions from the bottom of the gradient.
The ssDNA binding region of AR1 overlaps a multimerization domain.
Proteins that bind single-stranded nucleic acids tend to bind cooperatively as homomultimeric complexes (10, 12, 13, 39). In addition, the finding that AR1 is the only subunit of the virion capsid (24) suggests that it multimerizes to form higher order complexes. We therefore tested whether potential AR1 multimerization correlated with the ability of AR1 to bind ssDNA and mask the infectivity defects of BR1 mutants.
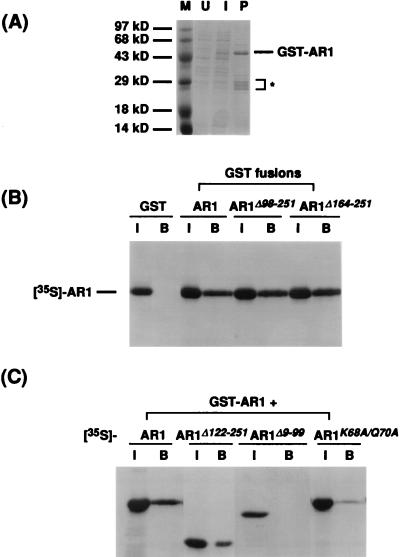
The fusion protein GST-AR1 was expressed in E. coli and purified by binding to glutathione-Sepharose (Fig. 9A). When [35S]methionine-labeled in vitro-synthesized AR1 was incubated with the GST-AR1 bound to glutathione-Sepharose, the AR1 was bound to the bead complexes (Fig. 9B). This interaction was specific for the AR1 sequence of the GST-AR1 fusion protein since AR1 did not bind to GST alone coupled to the beads (Fig. 9B).
FIG. 9.
AR1-AR1 in vitro binding assay. (A) Induction and purification of GST-AR1 from E. coli. GST-AR1 was purified from E. coli extracts by binding to and elution from glutathione-Sepharose beads. U, lysate from uninduced bacteria; I, lysate from bacteria induced with IPTG; P, GST-AR1 fusion protein bound to and eluted from glutathione-Sepharose beads. The position of the GST-AR1 fusion protein was verified by immunoblotting with anti-AR1 antiserum (data not shown). The asterisk indicates GST-AR1 degradation products. Sizes of protein molecular mass markers (M) are indicated. See text for details. (B) Binding of wild-type AR1 to GST, GST-AR1, and C-terminal truncations of AR1 fused to GST (GST-AR1Δ98-251 and GST-AR1Δ164-251). [35S]methionine-labeled in vitro-synthesized wild-type AR1 protein was incubated with the GST fusion proteins, as indicated, bound to glutathione-Sepharose beads. Shown are SDS-polyacrylamide gels of labeled AR1 bound. I, input [35S]methionine-labeled AR1; B, bound [35S]methionine-labeled AR1. (C) Binding of [35S]methionine-labeled AR1, AR1Δ122-251, AR1Δ9-99, and AR1K68A/Q70A to GST-AR1. In vitro-transcribed and -translated wild-type or mutant AR1 (as indicated) was incubated with GST-AR1 bound to glutathione-Sepharose beads. Shown are SDS-polyacrylamide gels of each AR1 protein bound. I, input [35S]methionine-labeled AR1 wild-type or mutant protein; B, bound [35S]methionine-labeled AR1 wild-type or mutant protein.
To identify the region of AR1 responsible for multimerization, two C-terminally truncated forms of AR1 were fused to GST, and the resulting fusion proteins, GST-AR1Δ98-251 and GST-AR1Δ164-251, were bound to glutathione-Sepharose and tested for the ability to bind AR1. Both GST-AR1Δ98-251 and GST-AR1Δ164-251 bound 35S-labeled AR1 (Fig. 9B). Thus, the N-terminal 97 amino acids of AR1 appeared to include the multimerization domain of AR1. To confirm this, GST-AR1 bound to glutathione-Sepharose was tested for the ability to bind in vitro-synthesized [35S]methionine-labeled AR1Δ122-251 and AR1Δ9-99. As shown in Fig. 9C, AR1Δ122-251 bound to GST-AR1, but AR1Δ9-99 did not. AR1K68A/Q70A, mutated within this N-terminal region of AR1, was also found to bind GST-AR1 very poorly (Fig. 9C). AR1K68A/Q70A did not mask BR1K25A/R26A and BR1N201A/K202A/R203A and also had a decreased binding affinity for ssDNA. Thus, the multimerization domain of AR1 is within the N-terminal 97 residues of the protein, overlapping the region of AR1 that contains the ssDNA binding domain.
DISCUSSION
The movement of bipartite geminiviruses such as SqLCV to systemically infect a host is a complex process that requires the presence of the two virus-encoded MPs BR1 and BL1 and the viral genomic DNA. BR1 and BL1 act cooperatively to move the viral genome (44, 49, 50), with BR1 functioning as a nuclear shuttle protein to bind the viral ssDNA genome and transport it into and out of the nucleus (46, 48). Genetic studies demonstrate that the SqLCV coat protein AR1, although not essential for movement, interacts with the viral movement pathway (31)—a conclusion based on the finding that the presence of wild-type CP masks the infectivity defects in cucurbits of certain BR1 missense mutants, namely, BR1K25A/R26A, BR1N201A/K202A/R203A, and BR1N219A. We have investigated here the mechanism of this masking by AR1.
Our mutational studies of AR1 identified a set of CP mutants that no longer masked the infectivity defects of BR1K25A/R26A and BR1N201A/K202A/R203A in pumpkin. As all of these mutants accumulated levels of CP in infected pumpkin and in protoplasts comparable to that of wild-type SqLCV, it did not appear that simple instability of the mutant CPs was responsible for this lack of masking. None of these class III masking-defective AR1 mutants induced the accumulation of viral ssDNA in infected plants or in protoplasts, demonstrating that accumulation of viral ssDNA was essential to fully mask the mutant phenotypes of BR1K25A/R26A and BR1N201A/K202A/R203A. Of these class III mutants, the N-terminal mutants AR1K68A/Q70A and AR1H98A/R99A had a decreased binding affinity for ssDNA. This was also true for the N-terminal class II mutants AR1R46A/R47A and AR1K102/R103, both of which were partially defective in their ability to mask the defects of BR1K25A/R26A and BR1N201A/K202A/R203A. Thus, the ability to efficiently bind ssDNA was necessary for AR1 to mask the defects of BR1K25A/R26A and BR1N201A/K202A/R203A; however, it did not appear to be sufficient.
This latter conclusion was based on the finding that class III mutants in the central region of AR1 between residues 118 and 191 all retained wild-type affinities for binding to ssDNA, despite their inability to mask BR1K25A/R26A and BR1N201A/K202A/R203A. Interestingly, these mutants—A R 1D 1 1 8 A / E 1 1 9 A, A R 1R138A / R139A, A R 1D155A / N156A / E157A, AR1H175A/R176A, and AR1N189A/E190A/Q191A — all failed to accumulate SqLCV ssDNA in infected plants or protoplasts. This finding suggested that these AR1 mutants may be defective in nuclear targeting, since if the mutant CPs could not enter the nucleus, the site of viral DNA replication, then they could not induce accumulation of viral ssDNA despite their ability to bind ssDNA. That this was the case was shown by transient expression of each of these AR1 mutants in tobacco protoplasts. All five mutants mislocalized to the cytoplasm, which explained their inability to induce viral ssDNA replication. Thus, the nucleus is the site at which AR1 exerts its positive effect in aiding BR1 function in movement, and the ability of AR1 to bind viral ssDNA is necessary and sufficient for AR1 to mask the defects of BR1K25A/R26A and BR1N201A/K202A/R203A. Since all of these AR1 mutants still bound ssDNA as efficiently as did wild-type CP, it is unlikely that the specific mutations have caused global misfolding of the protein. Thus, these findings further suggest that this central region of AR1 encompassing residues 118 to 191 either contains the nuclear localization signals of AR1 or affects their function in nuclear targeting. Further studies in which fragments of AR1 are tested directly in fusion constructs for the ability to target proteins to the nucleus and/or localize to nuclear pores are required to define the nuclear targeting signals in AR1.
The DNA binding defect of the N-terminal mutants AR1K68A/Q70A, AR1H98A/R99A, AR1R46A/R47A, and AR1K102/R103, but wild-type binding affinities for mutants between residues 118 and 191, suggested that the DNA binding domain of AR1 might be located within the N-terminal 118 amino acids of the protein. This conclusion was supported by our finding that a peptide containing the N-terminal 121 residues of AR1 retained the ability to bind ssDNA with a high affinity, while an in-frame deletion mutant containing the first eight amino acids of AR1 and the C-terminal residues from positions 100 to 251 did not. However, the N-terminal 121-amino-acid peptide of AR1 did not bind ssDNA as efficiently as intact AR1. The simplest explanation for these findings is that the DNA binding domain of AR1 is located within the N-terminal 121 amino acids of the protein and that as a peptide this fragment has a somewhat lower affinity for ssDNA than intact AR1, since it is not the native protein and thus is likely to contain some conformational alterations. This would suggest that the decreased ssDNA binding affinities of the C-terminal mutants AR1R195A/R196A and AR1R240A/R242A (this study) and AR1Δ201-251 (31) were due to these particular mutations affecting the overall conformation of CP. Alternatively, the DNA binding domain of AR1 may be formed by the association of sequences within the first 121 amino acids and C-terminal ∼50 amino acids of the protein, with the N-terminal 121-amino-acid peptide retaining only partial binding activity. In either case, the N-terminal 121 amino acids of AR1 encompass at least part of the DNA binding domain, and this domain is required to induce the accumulation of viral ssDNA.
Given the essential function of CP in virion assembly, it seemed logical that AR1 could interact with itself in a cooperative manner to form an oligomeric structure (the capsid). Using specific [35S]Met-labeled in vitro-synthesized segments of AR1 in a GST-AR1 binding assay, we showed that AR1 could bind to itself and that this AR1 multimerization region was located within the N-terminal 97 amino acids of the protein. Although our assay does not formally distinguish the formation of simple dimers from the formation of higher-order multimeric structures, our results demonstrate the involvement of this region in self-association of CP and, by inference, capsid assembly. Given the overlap of this region with the ssDNA binding domain and the ability to induce the accumulation of viral ssDNA, these results raised the question of whether the accumulation of ssDNA required encapsidation into virions as opposed to AR1 acting as a switch to signal the start of rolling circle replication or to sequester ssDNA from the replication pool in the absence of encapsidation. Detailed studies of the encapsidation of DNA by the icosahedral phages φX174, T4, T7, λ, Mu-1, P1, and P22 have established that in contrast to helical viruses, which can encapsidate nucleic acids of potentially indeterminate length, the size of the packaged genome is limited by the icosahedral head to between ∼80 and ∼115% of that of the wild-type viral genome normally packaged (2, 3, 8). Similar results have been obtained in studies on simian virus 40, adenoviruses, and herpesviruses (9, 15, 16, 23, 27, 32, 38). We found that 6.8-kb ssDNA copies of recombinant pBL-19BE were replicated in the presence, but not the absence, of wild-type SqLCV AR1. This ssDNA is 2.6 times the size of the encapsidated SqLCV 2.6-kb genomic components and thus is unlikely to be encapsidated. Consistent with this conclusion, the replicated 6.8-kb pBL-19BE virion (positive-strand) ssDNA did not sediment at the position of encapsidated virions, although the 2.6-kb AE virion (positive-strand) ssDNA did. Thus, these results suggest that the AR1-dependent accumulation of viral ssDNA is not coupled to encapsidation into virions.
Our results suggest that AR1 acts to signal the switch from viral dsDNA replication to the replication of viral ssDNA by a rolling circle mechanism or to sequester virion ssDNA from the replication pool without fully encapsidating it, and it is this action of AR1 that affects the movement pathway, as revealed by the masking of the infectivity defects of mutants BR1K25A/R26A, BR1N201A/K202A/R203A, and BR1N219A. Given the defects in nuclear localization of these particular BR1 mutants (48), and the AR1 mutants between residues 118 and 191 that still bind viral ssDNA but fail to target to the nucleus (this study), our findings support our hypothesis that this ability of AR1 to induce the accumulation of high levels of viral ssDNA in the nucleus, the substrate to which BR1 binds, acts to compensate for the lower accumulated levels of these BR1 mutant proteins in the nucleus and thereby masks the defective phenotypes of these specific BR1 mutants. It appears that cooperative binding of AR1 to viral ssDNA, a common property of ssDNA binding proteins (10, 13, 39), may be involved in this switch or sequestration, even though assembly of virion capsids is apparently not required. This may be analogous mechanistically to the coating of φX174 or f1 ssDNA by the phage-encoded ssDNA binding gene A or gene V protein, respectively (28), or to the coating of negative-sense RNA genomes by capsid protein in viruses such as the paramyxo-, rhabdo-, and myxoviruses, which signals a switch from the transcription of monocistronic mRNAs to the replication of full-length genomes (18).
We suggest that AR1 acts as this signal for the switch to viral ssDNA replication or sequestration of viral ssDNA from the replication pool early during infection of a cell, binding to single-strand regions of the replicating DNA—possibly involving the conserved hairpin within the positive-strand origin of replication (19, 20, 37) or viral ssDNA as its synthesis is initiated—when the concentration of AR1 is low and insufficient to assemble capsids. According to this model, BR1 would have a higher affinity for the replicating viral ssDNA and would displace AR1, much in the same way as the capsid protein of f1 phage displaces the gene V ssDNA binding protein during f1 assembly and extrusion from the cell (57–59). The ability of BR1 to be exported from the nucleus would remove these ssDNA genomes from the nuclear pool targeting them for movement. As infection of the cell progressed and increased amounts of AR1 accumulated, these would encapsidate the viral ssDNA into virions, characteristically seen as arrays in the nuclei of infected cells (24). However, at this later stage, intracellular and intercellular movement of the viral ssDNA genome mediated by BR1 would have already occurred to propagate the infection within the plant. Whether BR1 has a higher binding affinity than AR1 for viral ssDNA and can displace AR1 from the viral genomic DNA and the precise sequences within the N-terminal 121 residues of AR1 that are required for DNA binding and multimerization are all important issues that can now be addressed.
ACKNOWLEDGMENTS
The first two authors contributed equally to this work.
We thank current and past members of our laboratory—Tony Sanderfoot, David Ingham, Erica Pascal, Brian Ward, and Janet Hill—for insightful comments and helpful suggestions during the course of these studies.
These studies were supported by USDA NRI CRG grant 95-37303-1710 to S.G.L. and funds from the University of Illinois Research Board.
REFERENCES
- 1.Abouzid A M, Hiebert E, Strandberg J O. Cloning, identification and partial sequencing of the genomic components of a geminivirus infecting the Brassicaceae. Phytopathology. 1992;82:1070. [Google Scholar]
- 2.Aoyama A, Hayashi M. Effects of genome size on bacteriophage φX174 DNA packaging in vitro. J Biol Chem. 1985;260:11033–11038. [PubMed] [Google Scholar]
- 3.Aoyama A, Hayashi M. Synthesis of bacteriophage φX174: mechanism of switch from DNA replication to DNA packaging. Cell. 1986;47:99–106. doi: 10.1016/0092-8674(86)90370-3. [DOI] [PubMed] [Google Scholar]
- 4.Atabekov J G, Taliansky M E. Expression of a plant virus-coded transport function by different virus genomes. Adv Virus Res. 1990;38:201–248. doi: 10.1016/s0065-3527(08)60863-5. [DOI] [PubMed] [Google Scholar]
- 5.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1989. [Google Scholar]
- 6.Azzam O, Frazer J, De La Rosa D, Beaver J S, Ahlquist P, Maxwell D P. Whitefly transmission and efficient ssDNA accumulation of bean golden mosaic geminivirus require functional coat protein. Virology. 1994;204:289–296. doi: 10.1006/viro.1994.1533. [DOI] [PubMed] [Google Scholar]
- 7.Brough C L, Hayes R J, Morgan A J, Coutts R H, Buck K W. Effects of mutagenesis in vitro on the ability of cloned tomato golden mosaic virus DNA to infect Nicotiana benthamiana plants. J Gen Virol. 1988;69:503–514. [Google Scholar]
- 8.Casjens S, Hendrix R. Control mechanisms in dsDNA bacteriophage assembly. In: Calendar R, editor. The bacteriophages. New York, N.Y: Plenum; 1988. pp. 15–91. [Google Scholar]
- 9.Chang X-B, Wilson J H. Formation of deletions after initiation of simian virus 40 replication: influence of packaging limit of the capsid. J Virol. 1986;58:393–401. doi: 10.1128/jvi.58.2.393-401.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chase J W, Williams K R. Single-stranded DNA binding proteins required for DNA replication. Annu Rev Biochem. 1986;55:103–136. doi: 10.1146/annurev.bi.55.070186.000535. [DOI] [PubMed] [Google Scholar]
- 11.Chauthaiwale V M, Therwath A, Deshpande V V. Bacteriophage lambda as a cloning vector. Microbiol Rev. 1992;56:577–591. doi: 10.1128/mr.56.4.577-591.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Citovsky V, Knorr D, Schuster G, Zambryski P. The P30 movement protein of tobacco mosaic virus is a single-stranded nucleic acid binding protein. Cell. 1990;60:637–647. doi: 10.1016/0092-8674(90)90667-4. [DOI] [PubMed] [Google Scholar]
- 13.Citovsky V, Wong M L, Zambryski P. Cooperative interaction of Agrobacterium VirE2 protein with single stranded DNA: implication for the T-DNA transfer process. Proc Natl Acad Sci USA. 1989;86:1193–1197. doi: 10.1073/pnas.86.4.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cunningham B C, Wells J A. High-resolution epitope mapping of hGH-receptor interactions by alanine-scanning mutagenesis. Science. 1989;244:1081–1085. doi: 10.1126/science.2471267. [DOI] [PubMed] [Google Scholar]
- 15.Daniell E, Mullenbach T. Synthesis of defective viral DNA in HeLa cells infected with adenovirus type 3. J Virol. 1978;26:61–70. doi: 10.1128/jvi.26.1.61-70.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deiss L P, Frenkel N. Herpes simplex virus amplicon: cleavage of concatemeric DNA is linked to packaging and involves amplification of the terminally reiterated a sequence. J Virol. 1986;57:933–941. doi: 10.1128/jvi.57.3.933-941.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feinberg A, Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1984;137:266–267. doi: 10.1016/0003-2697(84)90381-6. [DOI] [PubMed] [Google Scholar]
- 18.Fields B N, Knipe D M, Howley P M, editors. Virology. 3rd ed. New York, N.Y: Lippincott-Raven; 1996. [Google Scholar]
- 19.Fontes E P B, Gladfelter H J, Schaffer R L, Petty I T D, Hanley-Bowdoin L. Geminivirus replication origins have a modular organization. Plant Cell. 1994;6:405–416. doi: 10.1105/tpc.6.3.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fontes E P B, Luckow V A, Hanley-Bowdoin L. A geminivirus replication protein is a sequence-specific DNA binding protein. Plant Cell. 1992;4:597–608. doi: 10.1105/tpc.4.5.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fromm M E, Taylor L P, Walbot V. Stable transformation of maize after gene transfer by electroporation. Nature. 1986;319:791–793. doi: 10.1038/319791a0. [DOI] [PubMed] [Google Scholar]
- 22.Gardiner W E, Senter G, Brand L, Elmer J S, Rogers S G, Bisaro D M. Genetic analysis of tomato golden mosaic virus: the coat protein is not required for systemic spread or symptom development. EMBO J. 1988;7:899–904. doi: 10.1002/j.1460-2075.1988.tb02894.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ghosh-Choudhury G, Haj-Ahmad Y, Graham F L. Protein IX, a minor component of the human adenovirus capsid, is essential for packaging of full length genomes. EMBO J. 1987;6:1733–1739. doi: 10.1002/j.1460-2075.1987.tb02425.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goodman R M. Geminiviruses. In: Kurstak E, editor. Handbook of plant virus infection and comparative diagnosis. New York, N.Y: Elsevier/North Holland Biomedical Press; 1981. pp. 879–910. [Google Scholar]
- 25.Haley A, Richardson K, Zhan X, Morris B. Mutagenesis of the BC1 and BV1 genes of African cassava mosaic virus identifies conserved amino acids that are essential for spread. J Gen Virol. 1995;76:1291–1298. doi: 10.1099/0022-1317-76-5-1291. [DOI] [PubMed] [Google Scholar]
- 26.Hayes R J, Buck K W. Replication of tomato golden mosaic virus DNA B in transgenic plants expressing open reading frames (ORFs) of DNA A: requirement of ORF AL2 for production of single-stranded DNA. Nucleic Acids Res. 1989;17:10213–10222. doi: 10.1093/nar/17.24.10213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hearing P, Samulski R J, Wishart W L, Shenk T. Identification of a repeated sequence element required for efficient encapsidation of the adenovirus type 5 chromosome. J Virol. 1987;61:2555–2558. doi: 10.1128/jvi.61.8.2555-2558.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heidekamp F, Bass P D, Jansz H S. Nucleotide sequence at the φX gene A protein cleavage site in replicative form I DNAs of bacteriophages U3, G14, and α3. J Virol. 1982;42:91–99. doi: 10.1128/jvi.42.1.91-99.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hull R. The movement of viruses within plants. Semin Virol. 1991;2:89–95. [Google Scholar]
- 30.Ingham D J, Lazarowitz S G. A single missense mutation in the BR1 movement protein alters the host range of the squash leaf curl geminivirus. Virology. 1993;196:694–702. doi: 10.1006/viro.1993.1526. [DOI] [PubMed] [Google Scholar]
- 31.Ingham D J, Pascal E, Lazarowitz S G. Both geminivirus movement proteins define viral host range, but only BL1 determines viral pathogenicity. Virology. 1995;207:191–204. doi: 10.1006/viro.1995.1066. [DOI] [PubMed] [Google Scholar]
- 32.Ladin B F, Blankenship M L, Ben-Porat T. Replication of herpesvirus DNA. V. The maturation of concatemeric DNA of pseudorabies virus of genome length is related to capsid formation. J Virol. 1980;33:1151–1164. doi: 10.1128/jvi.33.3.1151-1164.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 34.Laskey R A. The use of intensifying screens or organic scintillators for visualizing radioactive molecules resolved by gel electrophoresis. Methods Enzymol. 1980;65:363–371. doi: 10.1016/s0076-6879(80)65047-2. [DOI] [PubMed] [Google Scholar]
- 35.Lazarowitz S G. Molecular characterization of two bipartite geminiviruses causing squash leaf curl disease: role of viral replication and movement functions in determining host range. Virology. 1991;180:70–80. doi: 10.1016/0042-6822(91)90010-9. [DOI] [PubMed] [Google Scholar]
- 36.Lazarowitz S G, Lazdins I B. Infectivity and complete nucleotide sequence of the cloned genomic components of a bipartite squash leaf curl geminivirus with a broad host range phenotype. Virology. 1991;180:58–69. doi: 10.1016/0042-6822(91)90009-z. [DOI] [PubMed] [Google Scholar]
- 37.Lazarowitz S G, Wu C. L, Rogers S G, Elmer J S. Sequence specific interaction with the viral AL1 protein identifies a geminivirus DNA replication origin. Plant Cell. 1992;4:799–809. doi: 10.1105/tpc.4.7.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee J Y, Irmiere A, Gibson W. Primate cytomegalovirus assembly: evidence that DNA packaging occurs subsequent to B capsid assembly. Virology. 1988;167:87–96. doi: 10.1016/0042-6822(88)90057-8. [DOI] [PubMed] [Google Scholar]
- 39.Lohman T M, Ferrari M E. Escherichia coli single-stranded DNA-binding protein: multiple DNA-binding modes and cooperatives. Annu Rev Biochem. 1994;63:527–570. doi: 10.1146/annurev.bi.63.070194.002523. [DOI] [PubMed] [Google Scholar]
- 40.McGarvey P, Kaper J M. A simple and rapid method for screening transgenic plants using the PCR. BioTechniques. 1991;11:428–432. [PubMed] [Google Scholar]
- 41.McGeogh D J. The genome of herpes simplex virus: structure, replication and evolution. J Cell Sci Suppl. 1987;7:67–94. doi: 10.1242/jcs.1987.supplement_7.6. [DOI] [PubMed] [Google Scholar]
- 42.Mosig G, Colowick N, Gruidl M E, Chang A, Harvey A J. Multiple initiation mechanisms adapt phage T4 DNA replication to physiological changes during T4’s development. FEMS Microbiol Rev. 1995;17:83–98. doi: 10.1111/j.1574-6976.1995.tb00190.x. [DOI] [PubMed] [Google Scholar]
- 43.Mullis K B, Faloona F A. Specific synthesis of DNA in vitro via a polymerase-catalyzed chain reaction. Methods Enzymol. 1987;155:335–350. doi: 10.1016/0076-6879(87)55023-6. [DOI] [PubMed] [Google Scholar]
- 44.Noueiry A O, Lucas W J, Gilbertson R L. Two proteins of a plant DNA virus coordinate nuclear and plasmodesmatal transport. Cell. 1994;76:925–932. doi: 10.1016/0092-8674(94)90366-2. [DOI] [PubMed] [Google Scholar]
- 45.Pascal E, Goodlove P E, Wu L C, Lazarowitz S G. Transgenic tobacco plants expressing the geminivirus BL1 protein exhibit symptoms of viral disease. Plant Cell. 1993;5:795–807. doi: 10.1105/tpc.5.7.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pascal E, Sanderfoot A A, Ward B M, Medville R, Turgeon R, Lazarowitz S G. The geminivirus BR1 movement protein binds single-stranded DNA and localizes to the cell nucleus. Plant Cell. 1994;6:995–1006. doi: 10.1105/tpc.6.7.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pooma W, Gillette W K, Jeffrey J L, Petty I T D. Host and viral factors determine the dispensability of coat protein for bipartite geminivirus systemic movement. Virology. 1996;218:264–268. doi: 10.1006/viro.1996.0189. [DOI] [PubMed] [Google Scholar]
- 48.Sanderfoot A A, Ingham D J, Lazarowitz S G. A viral movement protein as a nuclear shuttle: the geminivirus BR1 movement protein contains domains essential for interaction with BL1 and nuclear localization. Plant Physiol. 1996;110:23–33. doi: 10.1104/pp.110.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sanderfoot A A, Lazarowitz S G. Cooperation in viral movement: the geminivirus BL1 movement protein interacts with BR1 and redirects it from the nucleus to the cell periphery. Plant Cell. 1995;7:1185–1194. doi: 10.1105/tpc.7.8.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sanderfoot A A, Lazarowitz S G. Getting it together in plant virus movement: cooperative interactions between bipartite geminivirus movement proteins. Trends Cell Biol. 1996;6:353–358. doi: 10.1016/0962-8924(96)10031-3. [DOI] [PubMed] [Google Scholar]
- 51.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Southern E. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 53.Stenger D C, Revington G, Stevenson M C, Bisaro D M. Replicational release of geminivirus genomes from tandemly repeated copies: evidence for rolling circle replication of a plant viral DNA. Proc Natl Acad Sci USA. 1991;88:8029–8033. doi: 10.1073/pnas.88.18.8029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sunter G, Hartitz M D, Hormuzdi S G, Brough C L, Bisaro D M. Genetic anaysis of tomato golden mosaic virus: ORF AL2 is required for coat protein accumulation while ORF AL3 is necessary for efficient DNA replication. Virology. 1990;179:69–77. doi: 10.1016/0042-6822(90)90275-v. [DOI] [PubMed] [Google Scholar]
- 55.von Arnim A, Stanley J. Determinants of tomato golden mosaic virus symptom development located on DNA B. Virology. 1992;186:286–293. doi: 10.1016/0042-6822(92)90083-2. [DOI] [PubMed] [Google Scholar]
- 56.Ward B M, Medville R, Lazarowitz S G, Turgeon R. The geminivirus BL1 movement protein is associated with endoplasmic reticulum-derived tubules in developing phloem cells. J Virol. 1997;71:3726–3733. doi: 10.1128/jvi.71.5.3726-3733.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Webster R E, Cashman J S. Morphogenesis of the filamentous single-stranded DNA phages. In: Denhardt D T, Dressler D, Ray D S, editors. The single-stranded DNA phages. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1978. pp. 557–569. [Google Scholar]
- 58.Webster R E, Grant R A, Hamilton L W. Orientation of the DNA in the filamentous bacteriophage f1. J Mol Biol. 1983;152:357–374. doi: 10.1016/0022-2836(81)90247-3. [DOI] [PubMed] [Google Scholar]
- 59.Zinder N, Horiuchi K. Multiregulatory element of filamentous bacteriophages. Microbiol Rev. 1985;49:101–106. doi: 10.1128/mr.49.2.101-106.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]