Abstract
A detailed map of the transcriptional organization of the CELO virus genome was produced. Recent computer analysis of CELO virus has indicated the presence of 38 putative open reading frames (ORFs). This study, based on analysis of the transcriptional products of CELO in vitro, confirmed the presence of RNAs for 26 of these 38 ORFs. All of the results were obtained by cDNA isolation or specific reverse transcriptase PCR. Observation of ORF transcription kinetics postinfection revealed the existence of early and late expression, with the earliest starting at 2 h postinfection. The 5′ untranslated regions of some RNAs were also studied, and this revealed the existence of a bipartite leader sequence, comparable in structure to the tripartite leader of mastadenovirus. The leader most probably involved in transcriptional activity was observed in most of the structural protein genes of the CELO virus genome. This suggests some homology in transcriptional organization between the avian adenovirus CELO and known mastadenoviruses such as human adenovirus.
Among the various viruses associated with avian species (47), the adenovirus CELO has been the subject of numerous investigations since its initial isolation in 1957 (58). CELO virus is responsible for respiratory pathologies in avian species (18, 38, 39) as well as inclusion body hepatitis in quail and broiler chickens (24, 25, 54, 55). CELO virus is capable of generating tumors when injected into baby hamsters (29, 37, 48) and is also transformant with some cell types in vitro (5).
The structural organization of the avian adenovirus CELO is an icosahedral capsid (19), 60 to 80 nm in diameter (46), made up of hexon and penton structures. Studies of its proteins and physical properties (32, 44, 57) led to identification of similarities with mastadenovirus (mammalian adenovirus) and its classification within the fowl adenoviruses (type 1) in the avian Adenoviridae (6, 38). The CELO virus genome is a double-stranded DNA molecule 43,804 bp in length (13) (GenBank accession no. U46933) and has inverted terminal repeats (ITRs) that are shorter than mastadenovirus ITRs (3, 4, 51, 53). As in human adenovirus, a terminal protein (TP) is covalently attached to the 5′ terminus of the CELO virus genome (33). The CELO virus cycle has been described only by using optical and electron microscopy observations of infected cells (29, 30, 43, 59).
Study of the CELO virus genome has consisted principally of a comparison with known human adenoviruses (1–3). The determination of its complete sequence was very useful in analyzing homology. A gene coding for an endoprotease (EP) was identified. It possesses a nucleic acid sequence with 69% homology to that of the human adenovirus type 2 EP gene (10). Other genes, homologous to human adenovirus genes, that code for structural proteins of the virion have been identified and are at the expected locations on the genome (genes for hexon, penton, protein IIIa [pIIIa], pVI, pVII, pVIII, pX, 52-kDa protein [52K], 100K, and fiber 1) (13, 34–36, 52). The CELO virus sequence also shows homology with genes involved in human adenovirus replication, i.e., those for polymerase (Pol), DNA-binding protein (DBP), and TP.
Nevertheless, there are clear differences between mastadenovirus and CELO virus. (i) The CELO virus genome is on average 7 kb larger than that of human adenovirus. (ii) No equivalent of the E1A- and E1B-type early genes present in human adenovirus have been identified (34). The CELO virus genome also lacks sequences homologous to the mastadenovirus regions E3 and E4 (13). (iii) Moreover, the CELO genome codes for two fibers located on each vertex (called F1 and F2) (26). Both ends of the CELO virus genome carry open reading frames (ORFs) identified, by computer analysis, that have no homology to the mammalian adenovirus genome.
In order to analyze the transcriptional pattern of CELO virus, we screened the CELO virus transcriptional products during infection of chicken embryo kidney cells (28). Sequences of the obtained RNAs were used to make a CELO virus transcriptional organization map. A reverse transcriptase PCR (RT-PCR) protocol was used to elucidate the time schedule of RNA expression. These results give a functional description of the CELO virus genome. These data will facilitate the generation of recombinant vectors for gene delivery or vaccine application.
MATERIALS AND METHODS
Cell cultures and infections.
Nineteen-day-old chicken embryo kidney cells were plated in 75-cm2 flasks at a concentration of 106 cells/ml and maintained in 20 ml of BHK21 medium–5% fetal calf serum at 37°C in 2% CO2. After 24 h, cells were washed and infected with CELO virus at 20 PFU per cell in 4 ml of medium for 1 h at 37°C in 2% CO2. The cells were rinsed with phosphate-buffered saline and then maintained in culture in 20 ml of medium.
Extraction of total RNAs.
The RNAs were extracted by using the RNAnow phenol protocol supplied by Biogentex. RNAnow was used at a concentration of 1 ml/10 cm2 (i.e., 1 × 106 to 5 × 106 cells).
Construction of cDNA banks.
A first bank was constructed by using the SuperScript plasmid system for cDNA synthesis and plasmid cloning kit, supplied by Life Technologies. The cDNA was synthesized by using oligo(dT) primers associated with primer-adapters bearing the SpeI, XbaI, NruI, and NotI restriction sites in the presence of SuperScript II RT. The second strand was synthesized in the presence of Escherichia coli DNA Pol and RNase H. Finally, the SalI adapter was ligated to the 5′ ends after treatment with T4 DNA Pol. The resulting cDNA was then digested with NotI and subcloned in plasmid pSPORT1 by using SalI and NotI.
A second bank was constructed within the pBluescript BS/KS(+) [pBS/KS(+)] plasmid (Stratagene). The first strand was synthesized in the presence of oligo(dT)15 (2.5 M) and hexanucleotide (1 μM) primers. The second strand was prepared as in the previous protocol. An EcoRI-NotI adapter was attached to the ends of the cDNA, which were then subcloned in pBS/KS(+) without orientation.
Bank screening.
The clones were transferred and fixed on special nylon membranes for colony production (Boehringer). The probe was obtained by labeling CELO virus DNA (digested with HindIII or SalI) with digoxigenin-dUTP. Positive clones were detected with the DIG high-prime DNA labeling kit and the detection starter kit II from Boehringer. The hybridization and detection steps were carried out under the conditions recommended by the manufacturer. The hybridization step was performed in a 50% formamide buffer at 42°C overnight. Visualization was by chemiluminescence. Following this, the positive clones were screened a second time and then amplified and sequenced.
Sequence analysis.
The clones were sequenced on an ABI 373 sequencer by using the M13 and 21M13 primers present on the pSPORT and pBS/KS(+) plasmids. The nucleic acid sequences were then analyzed by using MacDNASIS version 3.5 (Hitachi Software) by comparison with the CELO virus sequence.
Kinetics determinations by RT-PCR.
The reactions were performed on total RNA extracted from chicken embryo kidney cells infected with CELO virus. The cDNA was initially synthesized in a 20-μl reaction mixture (27) containing 1× PCR buffer II (Boehringer), 5 mM MgCl2, 1 mM deoxynucleoside triphosphate mix, 1 U of RNase-free DNase I, 20 U of RNase inhibitor, 2.5 μM oligo(dT)15, and 1 μM hexanucleotides per 1 μg of RNA. The DNase I reaction mixture was maintained for 2 h at 37°C, and then the enzyme was inactivated by incubation for 5 min at 75°C. Fifty units of murine leukemia virus (MuLV) RT was added, and the reaction mixture was maintained at 42°C for 30 min and at 90°C for 5 min. In the negative controls, the RT was replaced by 2 μl of RNase-free H2O. The cDNA pools were diluted 1/30 and added to 23 μl of the PCR mixture, to give a final volume of 25 μl containing 1× PCR buffer II, 2 mM MgCl2, 25 pmol of each primer, 1 mM deoxynucleoside triphosphate mix, and 0.5 U of Taq polymerase (Boehringer). The amplification conditions were 2 min at 95°C, followed by 30 cycles of 1 min at 95°C and 1 min at 69°C and one 7-min period at 69°C. All pairs of primers were 25-mers and specific for each ORF.
5′ random amplification of cDNA ends (5′ RACE).
The Marathon cDNA amplification kit (Clontech) was used for these experiments. The single-stranded cDNA was first prepared from 1 μg of RNA extracted from chicken embryo kidney cells infected with CELO virus by using oligo(dT)15 primers bonded to an EcoRI-NotI adapter and MuLV RT. After synthesis of the second strand in the presence of DNA Pol, the cDNA was made blunt with T4 DNA Pol. A NotI-SfrI-SmaI-specific adapter was fixed to the ends. PCR amplification was then carried out with a specific sense primer of this adapter and an antisense primer selected at the 5′ positions of the ORFs (generally in the ATG region). The amplification products were cloned, sequenced, and analyzed by using MacDNasis.
Promoter assays.
LMH cells were transfected with pBS/KS(+)/MLP/βGal or pBS/KS(+)/PORF1/βGal (see Fig. 3) by the calcium phosphate precipitation method with the Calphos maximizer kit from Clontech. DNA was combined with CaCl2 to a final volume of 100 μl. To form the precipitate, the DNA-calcium mixture was added dropwise in 100 μl of 2× HEPES-buffered saline and slowly vortexed. The precipitate was allowed to form for 20 min, and the entire 200 μl was distributed on a 35-mm-diameter tissue culture plate of LMH cells containing 2 ml of William’s medium supplemented with 10% fetal calf serum and l-glutamine. The precipitate was incubated with the cells for 6 h, and then the cells were washed and fresh medium was added. After 48 h, transfected cells were used for β-galactosidase staining (Invitrogen protocol) or harvested with trypsin-EDTA and centrifuged at 250 × g for 5 min for β-galactosidase assay. The pellet was resuspended in lysis buffer (Tris, pH 8). The sample was frozen on dry ice and then thawed in a 37°C water bath. This procedure was repeated twice. After centrifugation, the supernatant was removed for β-galactosidase and protein assays. Protein concentrations were determined by using the DC protein assay from Bio-Rad. The β-Gal assay kit from Invitrogen provides the reagents required to measure the levels of active β-galactosidase expressed on cells transfected with plasmids expressing lacZ. The reaction was based on the hydrolysis of ONPG (ortho-nitrophenyl-β-d-galactopyranoside) to the ONP anion, which produces a bright yellow color with a peak of absorbance at 420 nm that can be quantified with a spectrophotometer. β-Galactosidase activity was calculated by using the formula β-galactosidase units = [(OD420 × 380)/t]/milligram of protein assayed, where t is the time of incubation (in minutes) at 37°C (i.e., 30 min), OD420 is optical density at 420 nm, and 380 is a conversion factor to convert absorbance to millimoles.
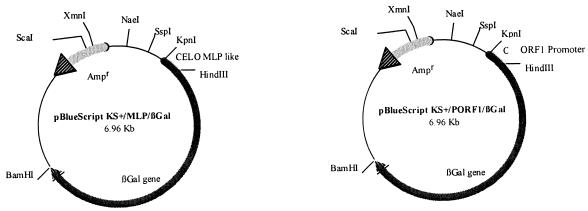
FIG. 3.
Plasmids used for the promoter assays. The CELO virus MLP sequence is between nt 7300 and 7530. The ORF 1 promoter (PORF1) sequence is between nt 204 and 745. These two sequences were amplified by PCR and cloned upstream of the β-galactosidase gene from pCMVβGal.
RESULTS
Analysis of CELO virus transcription products.
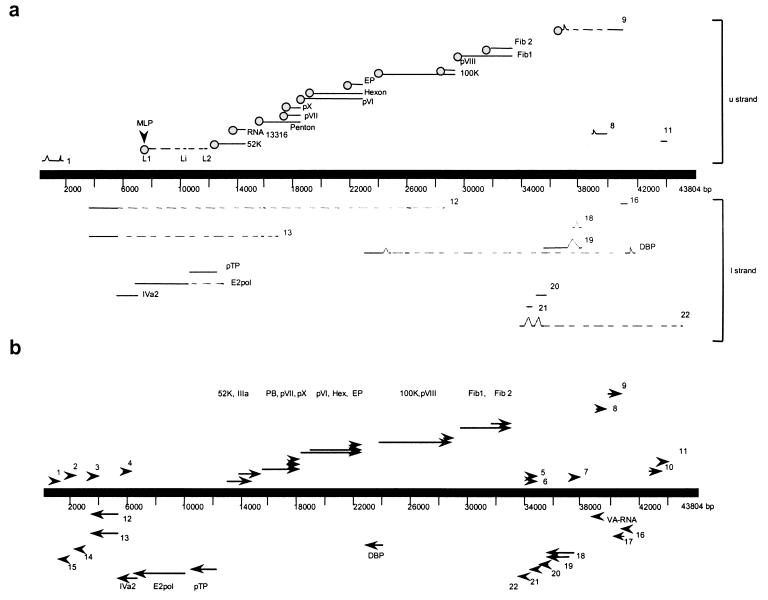
ORFs are described from upper strand (left to right) to lower strand (right to left). Nucleotide positions and ORF names correspond to those for the CELO virus genome sequence (GenBank accession no. U46933) described by Chiocca et al. (13) (Fig. 1).
FIG. 1.
Transcription map and genome organization of CELO virus. (a) Transcriptional organization of CELO virus. Lines above and below the central line represent mRNAs from the upper (u) and lower (l) strands, respectively. RNA sequences are represented by solid lines, and introns are represented by dashed lines. RNAs with the bipartite leader (L1-L2) and MLP are identified. (b) Genomic organization of the CELO virus. Arrows and arrowheads show the positions of the ORFs as described previously (13).
ORF 1.
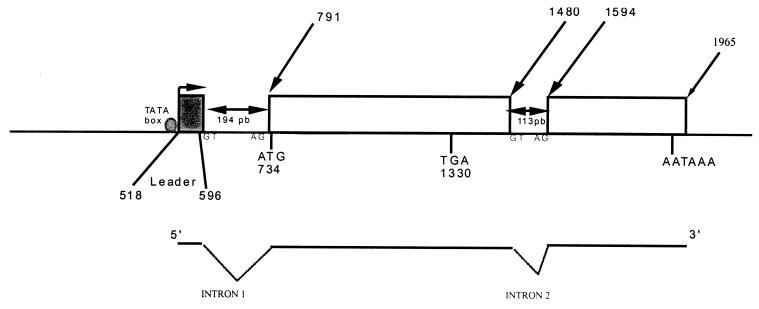
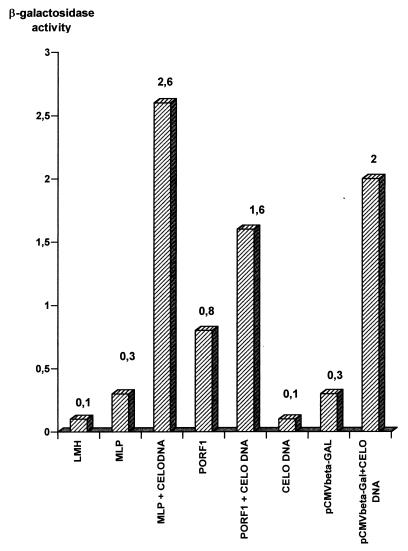
The ATG is at nucleotide (nt) 794, and the stop codon is at nt 1330. The 178 amino acids encoded showed strong homology with the dUTPase amino acid sequence. The RNA was spliced before the ATG and after the stop codon (Fig. 2). The first splice was located between a leader (nt 518 to 596) and the ATG position. The leader (nt 518 to 596) was downstream from a promoter-like sequence (nt 470 to 490) containing a TATA box. This sequence was tested in transfection experiments with β-galactosidase assays. Cells expressing β-galactosidase were observed after transfection with the pBS/KS(+)/PORF1/βGal plasmid (Fig. 3, 4, and 5). It was noteworthy that β-galactosidase activity was twofold greater after cotransfection with CELO virus DNA.
FIG. 2.
Transcriptional organization of ORF 1. The upper diagram shows the positions of noteworthy sites within the CELO virus genome, including the ATG initiation codon, TGA stop codon, and AATAAA polyadenylation site. The TATA box is represented by a circle upstream from the leader sequence. The open squares are the segments present on the RNA. The lower diagram represents the splicing of the mature RNA.
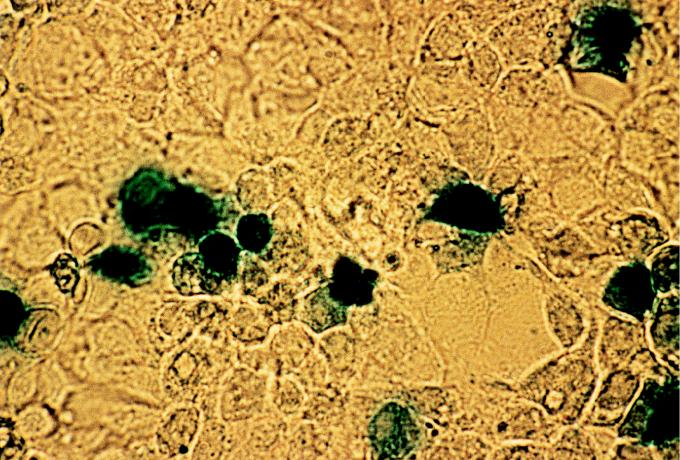
FIG. 4.
β-Galactosidase assay on LMH cells. Cells were transfected with plasmid pBlueScript KS(+)/PORF1/βGal. The results were observed 48 h later.
FIG. 5.
β-Galactosidase activity in transfected LMH cells. Cells were transfected with pBlueScript KS(+)/PORF1/βGal or pBlueScript KS(+)/MLP/βGal with or without purified CELO virus DNA. After 48 h, the cells were assayed for β-galactosidase activity (see Materials and Methods).
MLP sequence.
A sequence with strong homology to the mastadenovirus major late promoter (MLP) was found between positions 7310 and 7530, and a putative TATA box was at nt 7488 (13). LMH cells were transfected with the pBS/KS(+)/MLP/βGal plasmid (Fig. 3) and tested 48 h later in β-galactosidase assays. The experiment showed blue (positive) cells expressing the β-galactosidase. The experiment was done a second time with cotransfection with CELO virus DNA. The results demonstrated that the MLP had an activity in β-galactosidase transcription (Fig. 5). The observed activity was 3.7-fold greater in cotransfection experiments with CELO virus DNA.
A bipartite leader.
A bipartite sequence (L1-L2) was found in the 5′ untranslated regions of 13 ORFs (Table 1 and Fig. 1). This structure was homologous with the tripartite leader sequence of mastadenovirus (7, 17). The first part (L1) consisted of a 22-bp fragment between nt 7533 and 7554, and the second part (L2) was 129 bp long, starting from nt 11285. These two regions were 3,730 nt apart and framed by eukaryote splice acceptor and donor sites (GT…AG) (40). Splice donor and acceptor sites (GT…AG) were present at the end of the L2 sequence and upstream from the transcribed ORF.
TABLE 1.
Organization of detected RNAsa
| ORF | Position (nt)
|
Transcription start | ||||||
|---|---|---|---|---|---|---|---|---|
| Leader sequence:
|
Transcription signals
|
|||||||
| 1 | 2 | 3 | 4 | ATG | Stop (TAA, TGA, TAG) | Poly(A)+ (AATAAA, GATAAA) | ||
| ORF 1 | 518 | 789 | 794 | 1330 | 1943 | 791 | ||
| 596 | 793 | |||||||
| 52K | L1 | L2 | 12193 | 13329 | 13858 | 12183 | ||
| RNA13316 | L1 | L2 | 13316 | 15043 | 13858? | 13246 | ||
| Penton | L1 or Li | L2 | 15110 | 16657 | 17526 | 15099 | ||
| pVII | L1 | L2 | 16679 | 16897 | 17526? | 16556 | ||
| pX | L1 | L2 | 16929 | 17495 | 17526? | 16702 or 16952 | ||
| pVI | L1 | L2 | 17559 | 18230 | 21767? | 17545 | ||
| 21826? | ||||||||
| 21836? | ||||||||
| Hexon | L1 | L2 | 18289 | 21717 | 21767? | 18263 or 18284 | ||
| 21836? | ||||||||
| EP | L1 | L2 | 21134 | 21754 | 21767? | 21104 | ||
| 21826? | ||||||||
| 21836? | ||||||||
| 100K | L1 | L2 | 23680 | 26634 | 27920 | 23178 | ||
| pVIII | L1 | L2 | 27149 | 27866 | 27920 | 27137 | ||
| Fiber 1 | 28363 | 30495 | 31775 | 37389 | ||||
| Fiber 2 | L1 | L2 | 30536 | 31768 | 31775 | 30514 | ||
| ORF 8 | L1 | L2 | ? | 37391 | 38239 | 38250 | 37389 | |
| ORF 9 | L1 | L2 | ? | ? | ? | 41002 | 41017? | 40133 |
| 41544? | ||||||||
| ORF 11 | 41958 | 42365 | 43660 | |||||
| 43670? | ||||||||
| ORF 16 | 39705 | 39286 | ? | ? | ||||
| ORF 18 | 38432 | 36144 | 35536 | 33924 | 36201 | |||
| 36349 | ||||||||
| ORF 19 | 38432 | 35599 | 34238 | 33924 | 36201 | |||
| 36349 | ||||||||
| ORF 20 | 33707 | 32892 | ? | |||||
| ORF 21 | 33058 | 32735 | ? | |||||
| ORF 22 | 43330 | 33936 | 33204 | 32429 | 31811 | 31811 | 32473 | |
| 43059 | 33887 | 33070 | ||||||
| DBP | 39815 | 39291 | 24716 | 23455 | 23224 | 21899 | 21880 or 21820 | 23340 |
| 39853 | 39716 | 24780 | 23667 | |||||
| TP | 11996 | 10269 | ? | ? | ||||
| Pol | 9866 | 6501 | ? | ? | ||||
| IVa2 | 6685 | 5366 | ? | ? | ||||
| ORF 12 | 27068 | 24716 | 15081 | 5094 | 4462 | 3596 or 3550 | 5385 | |
| 27095 | 24780 | 15272 | ||||||
| ORF 13 | 15977 | 15081 | 4568 | 3549 | 3550 | 5385 | ||
| 16037 | 15272 | |||||||
| RNA6278 | 6278 | 17348 | 19151 | ? | ? | ? | ? | |
| 6288 | 17883 | 19671 | ||||||
| RNA12191 | 12191 | 15380 | ? | ? | ? | ? | ||
| 12752 | 15997 | |||||||
| RNA7810 | 7810 | 17022 | ? | ? | ? | ? | ||
| 8160 | 17541 | |||||||
| RNA18133 | 18133 | 22147 | ? | ? | ? | ? | ||
| 18392 | 22517 | |||||||
Nucleotide positions correspond to the CELO virus genome sequence under GenBank accession no. U46933. Nucleotide numbers represent a site location (transcription signals and transcription start positions) or a sequence between two nucleotides (leader sequence positions). L1, Li, and L2 are the sequences involved in the bipartite leader described in the text. Only detected ORFs are listed.
ORFs for 52K and RNA13316.
cDNA for 52K was isolated at 24 h postinfection (hpi) (Table 2). RT-PCR enabled the identification of expression at 12 hpi.
TABLE 2.
Time of first detection of the ORF products by the cDNA bank method and by RT-PCR
| ORF | Method(s)a by which detected at:
|
|||
|---|---|---|---|---|
| 2 hpi | 4 hpi | 12 hpi | 24 hpi | |
| ORF 1 | R | C | ||
| 52K | R | C | ||
| RNA13316b | C, R | |||
| Penton | C, R | |||
| pVII | R | C | ||
| pX | R | C | ||
| pVI | R | C | ||
| Hexon | C, R | |||
| EP | R | C | ||
| 100K | R | C | ||
| pVIII | R | C | ||
| Fiber 1 | C, R | |||
| Fiber 2 | R | C | ||
| ORF 8 | R | |||
| ORF 9 | R | C | ||
| ORF 11 | C, R | |||
| ORF 16 | R | |||
| ORF 18 | R | C | ||
| ORF 19 | R | C | ||
| ORF 20 | R | |||
| ORF 21 | R | |||
| ORF 22 | R | C | ||
| DBP | R | C | ||
| TP | R | C | ||
| Pol | R | C | ||
| IVa2 | R | |||
| ORF 12 | R | C | ||
| ORF 13 | R | C | ||
| RNA6278b | C | |||
| RNA12191b | C | |||
| RNA7810b | C | |||
| RNA18133b | C | |||
C, cDNA bank method; R, RT-PCR.
RNA only isolated in bank and not tested by RT-PCR.
RNA13316 was detected in the cDNA bank at 24 hpi. RNA13316 was polyadenylated, with a GATAAA sequence at nt 13858. RT-PCR with a primer corresponding to the bipartite leader and an antisense primer corresponding to the RNA13316 sequence showed the first signal at 24 hpi. The RNA13316 ATG was the same as the ATG of the expected ORF for pIIIa described by Chiocca et al. (13), but the RNA13316 sequence was shorter. No cDNA which corresponded to the complete expected sequence of the ORF for pIIIa was isolated.
ORFs for penton base, pVII, and pX.
cDNA of the penton base was isolated in the 24-hpi banks. Its late expression was confirmed by the RT-PCR method (Table 2). Two types of RNA were observed. A major form included the L1-L2 bipartite leader and the coding sequence. A rarer form used an intermediate leader sequence, Li, plus L2 upstream of the coding sequence. The Li sequence was 34 bp long, starting at nt 9759. A splice donor was observed at the end of the leader sequence. In the two cases the splice acceptor for the coding sequence was the same, at position 15099 (Table 1).
pVII ORF cDNA was isolated in the 24-hpi bank. The RT-PCR results showed the first expression earlier, at 2 hpi (Table 2).
pX cDNA was observed in the 24-hpi bank, but it was expressed earlier, as it was amplified by RT-PCR from 2 hpi. The 5′ RACE experiment with one primer 70 bp downstream from the ATG codon of pX and one sense primer corresponding to L2 gave two fragments. The longer one had the predicted ATG codon conserved, whereas in the shorter one this codon was spliced. It was clear that the longer fragment corresponded to the pX RNA structure, whereas the other represents an unknown sequence, as no ORF was identified.
ORFs for pVI, hexon, and EP.
pVI RNA was isolated in the 24-hpi bank but was detected by RT-PCR at 12 hpi.
RNA corresponding to the hexon was the most frequently isolated in the 12- and 24-hpi banks (Table 2). The polyadenylation signal was not defined, as there were three possibilities. Nevertheless, the majority of RNA possessed the last signal at position 21836, and the poly(A) tail started at position 21862.
The EP RNA was not clearly identified in the cDNA bank. RT-PCR detected the transcripts at 12 and 24 hpi. In the absence of complete cDNA, the polyadenylation signal remained uncertain (Table 1).
ORFs for 100K and pVIII.
The 100K and pVIII ORFs appeared very early in RT-PCR experiments. They used the same polyadenylation signal, at nt 27920.
ORFs for fiber 1 and fiber 2.
Fiber 1 cDNA was observed in the 12- and 24-hpi banks. RT-PCR detected RNA at 12 hpi (Table 2). The 5′ RACE analysis of the untranslated region of fiber 1 was unsuccessful. The presence of leaders was not demonstrated and remains unknown. In contrast, the fiber 2 ORF possessed the bipartite leader. Its expression was first detected earlier, at 4 hpi, by RT-PCR.
ORFs 8 and 9.
ORF 8 cDNA was observed in the RT-PCR experiment at 24 hpi. The bipartite leader was present on transcripts. The 5′ RACE experiments revealed the presence of various splicings between ATG and the bipartite leader.
ORF 9 cDNA was detected in the 24-hpi bank and was first observed at 12 hpi by RT-PCR (Table 2). The ORF 9 splicing started with the bipartite leader and used two other intermediate leaders (Table 1). It was noteworthy that after the 5′ untranslated region, the RNA was spliced up to nucleotide 40133, whereas the ORF 9 ATG was at position 40037 (13). The sequence analysis did not show any other reading frames than ATG40037 to TAA41002.
ORF 11.
ORF 11 cDNA was observed in the 24-hpi bank, at the same time as the first detection by RT-PCR (Table 2). The 5′ RACE experiments did not give any more information.
ORF 16.
RT-PCR enabled the detection of ORF 16 product at 4 hpi.
ORFs 18 and 19.
All of the results obtained were the same for ORFs 18 and 19. cDNAs isolated from the 12- and 24-hpi banks had the same sequence. They enabled the identification of a leader of 84 bp (Table 1). In 5′ RACE experiments, the same result was obtained with primers for ORF 18 or ORF 19. The RNA sequence contained the ATG of the two ORFs, so it was impossible to know which one was identified.
ORF 22.
ORF 22 was complex. cDNA was found in the 24-hpi bank, whereas RT-PCR detected the first expression at 4 hpi. Three leaders were observed in the RNA (Table 1). The first leader was situated on the extreme right of the genome near the ITR.
ORFs for DBP, TP, Pol, and IVa2.
DBP cDNA was found in the 12- and 24-hpi banks. RT-PCR demonstrated that the first RNA expression was around 4 hpi. The RNA was composed of four leaders. The first one was more than 14,000 bp upstream from the ORF.
The TP ORF had an early transcription, detected at 4 hpi by RT-PCR. The RNA structure remained unknown, as no results were obtained in 5′ RACE experiments.
The Pol ORF was also transcribed early after infection, as RNA was present at 4 hpi.
IVa2 ORF transcription appeared at 4 hpi. No cDNA was isolated in the banks, and the RNA structure remained unknown.
ORFs 12 and 13.
ORFs 12 and 13 were isolated in 24-hpi banks. The RNAs were composed of three leaders for ORF 12 and two leaders for ORF 13 (Table 1). Their expression started at 4 hpi.
ORFs 2, 3, 4, 5, 6, 7, 10, 14, 15, 17, 20, and 21.
ORFs 2, 3, 4, 5, 6, 7, 10, 14, 15, 17, 20, and 21 need to be further analyzed by other methods, such as Northern blotting or RNase protection. No conclusions can be made about their expression and structures until further experiments have been carried out.
DISCUSSION
Most of the available molecular knowledge on adenoviruses concerns the mastadenoviruses (31, 45), whereas little is known about the molecular biology of avian adenoviruses. Numerous studies have been published on their epidemiological aspects (55), their tumorigenic properties (29, 48), and cellular behavior during infection (21, 29, 30). The complete CELO virus sequence was published only 2 years ago (13). Information on its homology with human adenoviruses has been based solely on data bank analysis and computer comparisons (3, 4, 13, 33). The present study provides essential information on transcription of the predicted ORFs. In addition, this is the first time that a model of the transcriptional architecture of an adenovirus other than the mastadenovirus has been reported. The transcription of the previously described ORFs has been mostly confirmed. No transcriptional products could be detected in the case of 12 ORFs. To date, it has not been possible to determine if this absence of transcripts is due to our system of analysis. This work was carried out by infection of chicken embryo kidney cells with CELO virus, a widely used model in CELO virus studies (28, 30). As CELO virus develops preferentially in the hepatic tissues of adult chickens, leading to the formation of inclusion body hepatitis (24, 25, 55), the cellular context might not be favorable for the transcription of ORFs 2, 3, 4, 5, 6, 7, 10, 14, 15, 17, 20, and 21. In addition it should be noted that the replication of CELO virus was performed in vitro in the absence of regulatory mechanisms which are present during the infection of an entire organism. Other experiments with different cell cultures, such as LMH avian hepatocytes, could give information about tissue-specific expression or in vivo specific responses. Also, our experiments were done at the specific postinfection times 2, 4, 6, 12, and 24 hpi but not earlier or between these times. It is possible that some ORFs were expressed during short periods that were not explored.
The protein encoded by ORF 8 has recently been identified as a novel antiapoptotic protein called GAM-1 (12). This 30-kDa nuclear protein functions similarly to Bcl-2 and adenovirus E1B 19K in blocking apoptosis. The results presented in this paper demonstrated that the first expression of ORF 8 was late, at around 24 hpi. The presence of the bipartite leader on the RNA suggested the gene was under the control of the putative CELO virus MLP. The antiapoptotic activity indicated a rapid expression of the protein, like for E1B 19K, a product of early genes. It is possible that the in vitro system with LMH cells was not optimal for the antiapoptotic response and that another regulatory system for ORF 8 expression exists.
The existence of RNAs transcribed from undescribed ORFs was demonstrated. The ORF13316 translation product exhibited homology with pIIIa but was truncated in its 3′ segment in relation to the initially predicted size. RNA13316 bears the predicted initiation codon at position 13316 and the polyadenylation signal at nt 13858. It was 953 nt in size, compared with the predicted 3,130 nt. This shows better correlation with the human model, in which all genes of the L1 group use the same polyadenylation signal. It is possible that two forms of pIIIa exist. Nevertheless, it should be emphasized that no clones corresponding to the complete pIIIa RNA have been isolated to determine the accuracy of the pIIIa ORF position.
Homology between the transcriptional organizations of the genomes of avian adenovirus and the mastadenoviruses was demonstrated. In particular, the splice system operating in the central region of the upper strand was identical to that observed in human adenovirus (8, 14, 20, 41, 61). This regulation is no doubt the result of an equivalent to the human MLP occurring between positions 7300 and 7500 (49–51). A bipartite leader sequence, L1-L2 or Li-L2, occurs after the MLP, which is reminiscent of the tripartite equivalent in mammalian adenovirus. These observations suggest the presence of a pre-RNA in the central region cleaved into transcriptional subunits by alternative splicing mechanisms to give the final RNA, i.e., leaders plus coding region. The existence of splicing was confirmed by the presence of eukaryote-type splice acceptor (…AG) and donor (…GT) sites and by PCR analysis of cDNA molecules. Moreover, the polyadenylation signals seem to be common to groups of genes, as in mastadenovirus (7, 15, 60).
Nevertheless, there is approximately 5 kb of sequence on the left end and 15 kb of sequence on the right end of the CELO virus that have no specific transcriptional organization. It is now important to localize the promoters which control the transcription in these regions. We are at present testing some putative promoters for their in vitro activity.
The first β-galactosidase assays demonstrated the activity of the putative CELO virus promoter sequences in LMH cells. The ORF 1 promoter and the putative MLP have an activity dependent on viral factors, as β-galactosidase levels are higher in cotransfections with CELO virus DNA. These transcriptional factors remain unknown at present. In the case of the human adenovirus MLP, there is basic activity during early infection and an important late activity on late genes in association with viral factors.
The RT-PCR analysis distinguished groups of transcription, ranging between very early (at 2 hpi) to late (at 24 hpi). Comparison with human adenovirus shows that the two viruses possess early, intermediate, and late groups of gene expression (11, 16). It is noteworthy that only 8 RNAs appear late, at 24 hpi, in the CELO virus, and 17 RNAs are early. These observations on transcription kinetics suggest that transcription regulation in human adenovirus, and CELO virus could be different (23). It would be interesting to analyze the role of the CELO virus MLP sequence in the viral cycle in comparison to that of the human adenovirus MLP. Quantitative information about transcription during infection will determine if all of the ORFs are transcribed at the same level immediately after infection and, if not, which ones are essential at the beginning of the viral cycle.
This study has provided information about the organization of the avian CELO adenovirus genome. In subsequent experiments, we intend to examine the roles of specific ORFs of CELO virus in the viral cycle. Information on the CELO virus genome organization and essential regions facilitates our approach to generate recombinant vectors (22, 56). It also enables the mapping on the genome of the complete structures of some ORFs with their promoter leaders and coding sequences. This is a great advantage for insertion or deletion strategies (9, 42), as in the construction of complementing avian cell lines.
REFERENCES
- 1.Akopian T A, Kaverina E N, Naroditsky B S, Tikhonenko T I. Nucleotide sequence analysis of the avian adenovirus CELO (FAV1) DNA fragment (92–100%) Mol Microbiol Virol. 1992;11:19–23. [Google Scholar]
- 2.Akopian T A, Kruglyak V A, Rivkina M B, Naroditsky B S, Tikhonenko T I. Sequence of an avian adenovirus (CELO) DNA fragment (0–11.2%) Nucleic Acids Res. 1990;18:2825. doi: 10.1093/nar/18.9.2825. . (Erratum, 19:424, 1991.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aleström P, Stenlund A, Li P, Pettersson U. A common sequence in the inverted terminal repetitions of human and avian adenovirus. Gene. 1982;18:193–197. doi: 10.1016/0378-1119(82)90117-2. [DOI] [PubMed] [Google Scholar]
- 4.Aleström P, Stenlund A, Li P, Bellett A, Pettersson U. Sequence homology between avian and human adenoviruses. J Virol. 1982;42:306–310. doi: 10.1128/jvi.42.1.306-310.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anderson J V, Yates V J, Jasty V, Mancini O. The in vitro transformation by an avian adenovirus (CELO). III. Human amnion cell cultures. J Natl Cancer Inst. 1969;43:575–580. [PubMed] [Google Scholar]
- 6.Andrews C H, Pereira H G. Virus of vertebrates. 2nd ed. London, United Kingdom: Bailliere, Tindall, and Cassell; 1967. [Google Scholar]
- 7.Berget S M, Moore C, Sharp P A. Spliced segments at the 5′ terminus of adenovirus 2 late mRNA. Proc Natl Acad Sci USA. 1977;74:3171–3175. doi: 10.1073/pnas.74.8.3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berk A J, Sharp P A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease digested hybrids. Cell. 1977;12:721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- 9.Bett A J, Krougliak V, Graham F L. DNA sequence of the deletion/insertion in early region 3 of the Ad5 dl309. Virus Res. 1995;39:75–82. [PubMed] [Google Scholar]
- 10.Cai F, Weber J M. Organization of the avian adenovirus genome and the structure of its endopeptidase. Virology. 1993;196:358–362. doi: 10.1006/viro.1993.1489. [DOI] [PubMed] [Google Scholar]
- 11.Challberg M D, Kelly T J., Jr Animal virus DNA replication. Annu Rev Biochem. 1989;58:671–717. doi: 10.1146/annurev.bi.58.070189.003323. [DOI] [PubMed] [Google Scholar]
- 12.Chiocca S, Cotten M. Identification of a novel antiapoptotic protein, GAM1, encoded by the CELO adenovirus. J Virol. 1997;71:3168–3177. doi: 10.1128/jvi.71.4.3168-3177.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chiocca S, Kurzbauer R, Schaffner G, Baker A, Mautner V, Cotten M. The complete DNA sequence and genomic organization of the avian adenovirus CELO. J Virol. 1996;70:2939–2949. doi: 10.1128/jvi.70.5.2939-2949.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chow L T, Broker T R, Lewis J B. Complex splicing patterns of RNAs from the early regions of adenovirus 2. J Mol Biol. 1979;134:265–303. doi: 10.1016/0022-2836(79)90036-6. [DOI] [PubMed] [Google Scholar]
- 15.Chow L T, Gellinas R E, Broker T R, Roberets R J. An amazing sequence arrangement at the 5′ ends of adenovirus 2 messenger RNA. Cell. 1977;12:1–8. doi: 10.1016/0092-8674(77)90180-5. [DOI] [PubMed] [Google Scholar]
- 16.Chow L T, Roberts J M, Lewis J B, Broker T R. A map of cytoplasmic RNA transcripts from lytic adenovirus type 2, determined by electron microscopy of RNA:DNA hybrids. Cell. 1977;11:819–836. doi: 10.1016/0092-8674(77)90294-x. [DOI] [PubMed] [Google Scholar]
- 17.Dolph P J, Huang J, Schneider R J. Translation by the adenovirus tripartite leader element, which determines independence from cap-binding protein complex. J Virol. 1990;64:2669–2677. doi: 10.1128/jvi.64.6.2669-2677.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dubose R J, Grumbles L C. The relationship between quail bronchitis virus and chicken embryo lethal orphan virus. Avian Dis. 1959;3:321–344. [Google Scholar]
- 19.Dutta S K, Pomeroy B S. Electron microscopic structure of chicken embryo lethal orphan virus. Proc Soc Exp Biol Med. 1963;114:539–541. doi: 10.3181/00379727-114-28726. [DOI] [PubMed] [Google Scholar]
- 20.Evans R M, Fraser N, Ziff E, Weber J, Wilson M, Darnell J E. The initiation sites for RNA transcription in Ad2 DNA. Cell. 1977;12:733–739. doi: 10.1016/0092-8674(77)90273-2. [DOI] [PubMed] [Google Scholar]
- 21.Grabko V I, Lunin V G, Naroditsky B S, Khilko S N, Tickhonenko T I, Makhov A M, Karpova O V. Study of avian adenovirus DNA infectivity in chick embryos. Acta Virol. 1987;31:97–102. [PubMed] [Google Scholar]
- 22.Graham F L. Adenoviruses as expression vectors and recombinant vaccines. Trends Biotechnol. 1990;8:85–87. doi: 10.1016/0167-7799(90)90144-m. [DOI] [PubMed] [Google Scholar]
- 23.Green M, Daesh G E. Biochemical studies on adenovirus multiplication. I. Kinetics of nucleic acid and protein in suspension cultures. Virology. 1961;13:169–176. doi: 10.1016/0042-6822(61)90051-4. [DOI] [PubMed] [Google Scholar]
- 24.Grewal G S, Singh A, Singh B, Oberoi M S. Inclusion body hepatitis in Japanese quail (Coturnix coturnix japonica) Indian J Anim Sci. 1994;64:665–667. [Google Scholar]
- 25.Grewal G S, Sharma S N, Deka B C. Inclusion body hepatitis in broiler chickens. Indian J Poultry Sci. 1981;16:51–56. [Google Scholar]
- 26.Hess M, Cuzange A, Ruigrok R W H, Chroboczek J, Jacrot B. The avian adenovirus penton: two fibres and one base. J Mol Biol. 1995;252:379–385. doi: 10.1006/jmbi.1995.0504. [DOI] [PubMed] [Google Scholar]
- 27.Huang Z, Fasco M J, Kaminsky L S. Optimization of DNase I removal of contaminating DNA from RNA for use in quantitative RNA-PCR. BioTechniques. 1996;20:1012–1020. doi: 10.2144/96206st02. [DOI] [PubMed] [Google Scholar]
- 28.Ishibashi M. Temperature sensitive conditional-lethal mutants of an avian adenovirus (CELO) Virology. 1971;45:42–52. doi: 10.1016/0042-6822(71)90111-5. [DOI] [PubMed] [Google Scholar]
- 29.Jasty V, Pendola R, Yates V J. Cell-to-cell attachments and associations in tumors induced by celo virus or virus transformed cells in hamsters. Am J Vet Res. 1975;36:1643–1647. [PubMed] [Google Scholar]
- 30.Jasty V, Yates V J, Anderson J, Fry D, Chang P W, Pendola R. Replication of an avian adenovirus (CELO) large-plaque mutant in chick kidney cells. Avian Dis. 1973;17:49–65. [PubMed] [Google Scholar]
- 31.Larsson S, Bellett A, Akusjarvi G. VA RNAs from avian and human adenoviruses: dramatic differences in length, sequence, and gene location. J Virol. 1986;58:600–609. doi: 10.1128/jvi.58.2.600-609.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Laver W G, Younghusband H B, Wrigley N G. Purification and properties of chick embryo lethal orphan virus (an avian adenovirus) Virology. 1971;45:598–614. doi: 10.1016/0042-6822(71)90175-9. [DOI] [PubMed] [Google Scholar]
- 33.Li P, Bellett A J D, Parish C R. A comparison of the terminal protein and hexon polypeptides of avian and human adenoviruses. J Genet Virol. 1983;64:1375–1379. doi: 10.1099/0022-1317-64-6-1375. [DOI] [PubMed] [Google Scholar]
- 34.Li P, Bellett A J D, Parish C R. DNA-binding protein of chick embryo lethal orphan virus: lack of complementation between early proteins of avian and human adenoviruses. J Virol. 1984;65:1817–1825. doi: 10.1099/0022-1317-65-10-1817. [DOI] [PubMed] [Google Scholar]
- 35.Li P, Bellett A J D, Parish C R. Structural organization and polypeptide composition of the avian adenovirus core. J Virol. 1984;52:638–649. doi: 10.1128/jvi.52.2.638-649.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li P, Bellett A J D, Parish C R. The structural proteins of chick embryo lethal orphan virus (fowl adenovirus type 1) J Genet Virol. 1984;65:1803–1815. doi: 10.1099/0022-1317-65-10-1803. [DOI] [PubMed] [Google Scholar]
- 37.Mancini L O, Yates V J, Jasty V, Anderson J. Ependymomas induced in hamsters inoculated with an avian adenovirus (CELO) Nature. 1968;222:190–191. doi: 10.1038/222190a0. [DOI] [PubMed] [Google Scholar]
- 38.McCracken W, Adair B M. Avian adenovirus, 123–144. In: McFerran M, editor. Viral infections of vertebrates. 3. Viral infections of birds. Amsterdam, The Netherlands: Elsevier Scientific; 1993. [Google Scholar]
- 39.McFerran J B, Adair B M. Avian adenovirus. Avian Pathol. 1977;6:189–217. doi: 10.1080/03079457708418228. [DOI] [PubMed] [Google Scholar]
- 40.Mount S M. A catalogue of splice junction sequences. Nucleic Acids Res. 1982;10:459–472. doi: 10.1093/nar/10.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nevin J R, Darnell J E. Groups of adenovirus type 2 mRNAs derived from a large primary transcript: probable nuclear origin and possible common 3′ ends. J Virol. 1978;25:811–823. doi: 10.1128/jvi.25.3.811-823.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Parkas G. A helper-dependent adenovirus vector system: removal of helper virus by Cre mediated excision of the viral packaging system. Proc Natl Acad Sci USA. 1996;93:13565–13570. doi: 10.1073/pnas.93.24.13565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Petek M, Felluga B, Zoletto R, Bersani G. Further studies on CELO virus: its relationship to the adenovirus group. Arch Gesamte Virusforsch. 1964;14:637–649. doi: 10.1007/BF01555121. [DOI] [PubMed] [Google Scholar]
- 44.Petek M, Felluga B, Zoletto R. Biological properties of CELO virus: stability to various agents, and electron microscopic study. Avian Dis. 1963;7:38–49. [PubMed] [Google Scholar]
- 45.Petterson U, Roberts R J. Adenovirus gene expression and replication: a historical review. Cancer Cells. 1986;4:37–57. [Google Scholar]
- 46.Potter C W, Oxford J S, Downie J C, Atwood M M, Hardy R D. Chick embryo lethal orphan (CELO) virus: some physical and immunological properties. Virology. 1971;44:418–424. doi: 10.1016/0042-6822(71)90272-8. [DOI] [PubMed] [Google Scholar]
- 47.Prier J E. Prier basic medical virology. Baltimore, Md: Williams and Wilkins; 1966. pp. 625–651. [Google Scholar]
- 48.Sarma P S, Huebner R J, Lane W T. Induction of tumors in hamster with avian adenovirus (CELO) Science. 1965;149:1108. doi: 10.1126/science.149.3688.1108. [DOI] [PubMed] [Google Scholar]
- 49.Sawadago M, Roeder R G. Interaction of a gene specific transcription factor with the adenovirus major late promoter upstream of the TATA box region. Cell. 1985;43:165–175. doi: 10.1016/0092-8674(85)90021-2. [DOI] [PubMed] [Google Scholar]
- 50.Shaw A R, Ziff E B. Transcripts from the adenovirus 2 late promoter yield a single early family of 3′ coterminal mRNAs and five late families. Cell. 1980;22:905–916. doi: 10.1016/0092-8674(80)90568-1. [DOI] [PubMed] [Google Scholar]
- 51.Sheppard M, Erny K M. DNA sequence analysis of the inverted terminal repeats of a non-oncogenic avian adenovirus. Nucleic Acids Res. 1989;17:3995. doi: 10.1093/nar/17.10.3995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sheppard M, McCoy R, Werner W. Genomic mapping and sequence analysis of the fowl adenovirus serotype 10 hexon gene. J Gen Virol. 1995;76:2595–2600. doi: 10.1099/0022-1317-76-10-2595. [DOI] [PubMed] [Google Scholar]
- 53.Shinagawa M, Ishiyama T, Padmanabhan R V, Fujinaga K, Kamada M, Sato G. Comparative sequence analysis of the inverted terminal repetition in the genome of animal and avian adenoviruses. Virology. 1983;125:491–495. doi: 10.1016/0042-6822(83)90221-0. [DOI] [PubMed] [Google Scholar]
- 54.Singh A, Oberoi M S, Singh B. Pathogenicity of quail’s inclusion body hepatitis virus (avian adenovirus-1) for Japanese quails and broiler chicks. Vet Res Commun. 1995;19:545–551. doi: 10.1007/BF01839342. [DOI] [PubMed] [Google Scholar]
- 55.Singh A, Oberroi M S, Singh B. Epidemiology of inclusion body hepatitis in poultry in northern India from 1990 to 1994. Rev Sci Tech O I E. 1996;15:1053–1060. doi: 10.20506/rst.15.3.976. [DOI] [PubMed] [Google Scholar]
- 56.Trapnell B C, Gorziglia M. Gene therapy using adenoviral vectors. Curr Opin Biotechnol. 1994;5:617–625. doi: 10.1016/0958-1669(94)90084-1. [DOI] [PubMed] [Google Scholar]
- 57.Yasue H, Ishibashi M. Chick embryo lethal orphan (CELO) virus-induced early and late polypeptides. Virology. 1977;78:216–233. doi: 10.1016/0042-6822(77)90093-9. [DOI] [PubMed] [Google Scholar]
- 58.Yates V J, Fry D E. Observation of a chicken embryo lethal orphan (CELO) virus. Am J Vet Res. 1957;18:657–660. [PubMed] [Google Scholar]
- 59.Yates V J, Rhee Y O, Fry D E. Comments on adenoviral antigens (Celo, QBV, Gal) Am J Vet Res. 1975;36:530–531. [PubMed] [Google Scholar]
- 60.Ziff E, Fraser N W. Adenovirus type 2 mRNA: structural evidence for 3′ coterminal species. J Virol. 1978;25:897–906. doi: 10.1128/jvi.25.3.897-906.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ziff E B, Evans R M. Coincidence of the promoter and capped 5′ terminus of RNA from the adenovirus 2 major late promoter unit. Cell. 1978;15:1463–1476. doi: 10.1016/0092-8674(78)90070-3. [DOI] [PubMed] [Google Scholar]