Summary
In eukaryotes, repetitive DNA sequences are transcriptionally silenced through histone H3 lysine 9 tri-methylation (H3K9me3). A loss of silencing of the repeat elements leads to genome instability and human diseases, including cancer and ageing1-3. While the role of H3K9me3 in the establishment and maintenance of heterochromatin silencing has been extensively studied4-6, the pattern and mechanism underlying the partitioning of parental H3K9me3 at replicating DNA strands were unknown. Here, we report that H3K9me3 is preferentially transferred onto the leading strands of replication forks, which occurs predominantly at Long Interspersed Nuclear Element (LINE) retrotransposons that are theoretically transcribed at the head-on (HO) direction with replication fork movement. Mechanistically, the Human Silencing Hub (HUSH) complex interacts with the leading strand DNA polymerase Pol ε, and contributes to the asymmetric segregation of H3K9me3. Cells deficient for Pol ε subunits (POLE3 and POLE4), the HUSH complex (MPP8 and TASOR), or cells expressing a MPP8 mutant defective of H3K9me3 binding, or TASOR mutants with reduced interaction with Pol ε, show compromised H3K9me3 asymmetry and increased LINE expression. These results reveal an unexpected mechanism whereby the HUSH complex functions with Pol ε to promote asymmetric H3K9me3 distribution at HO LINEs to suppress their expression in S phase.
Main
Repetitive DNA, originally coined as ‘junk DNA’, accounts for more than half of the mammalian genome7-9, and plays an important role in gene regulation, chromatin structure organization and maintenance of genomic stability10-12. LINE retrotransposons (also known as LINE-1 or L1) constitute about 17% of the human genome and are distributed throughout the genome, including both eu- and hetero-chromatic regions. Like other transposable elements (TEs), L1 expression is repressed through DNA methylation and histone modifications, including H3K9me310,12. Aberrant expression of L1s and its genomic integration through retrotransposition are likely drivers of multiple human diseases, such as cancer and ageing-associated disorders1-3,13. Importantly, inactivation of genes crucial for LINE silencing such as SETDB1 and the HUSH complex boosts anti-tumor immunity, likely by generating neo-antigens from the transcribed TEs and/or by activating the viral mimicry response14-17. Therefore, a better understanding of LINE repression and regulation is critical.
During mitotic cell divisions, histone modifications, including H3K9me3, could be inherited epigenetically18,19. Parental histones with specific post-translational modifications are proposed to be transferred equally to the leading and lagging strands of DNA replication forks, which then serve as the template to modify nucleosomes containing newly synthesized histones. Consistent with this idea, we and others found that histone H3 lysine 36 tri-methylation (H3K36me3) and histone H4 lysine 20 di-methylation (H4K20me2), two modifications on parental histones, and H4 lysine 5 acetylation (H4K5ac), a marker on newly synthesized H4, are almost equally distributed to the leading and lagging strands in wild-type (WT) cells20,21. This almost symmetric segregation of parental H3-H4 is also observed in budding yeast22,23. Furthermore, two conserved pathways regulating parental histone transfer were discovered. Specifically, MCM2, a subunit of the replicative helicase minichromosome maintenance (MCM) complex, and Pol α, the primase involved in primer synthesis, coordinate with each other and facilitate the transfer of parental H3-H4 to the lagging strands20,23. In contrast, POLE3 and POLE4, two subunits of the leading strand DNA polymerase, Pol ε, promote the transfer of parental histones to the leading strands in both yeast and mammalian cells21,22. Surprisingly, when analyzing the distribution of H3K9me3 at replicating DNA strands, we observed that H3K9me3 is preferentially enriched at the leading strands of DNA replication forks (Figure 1). Therefore, we set out to explore the regulatory mechanisms and biological implications underlying the asymmetric distribution of this critical modification on parental H3 in mammalian cells.
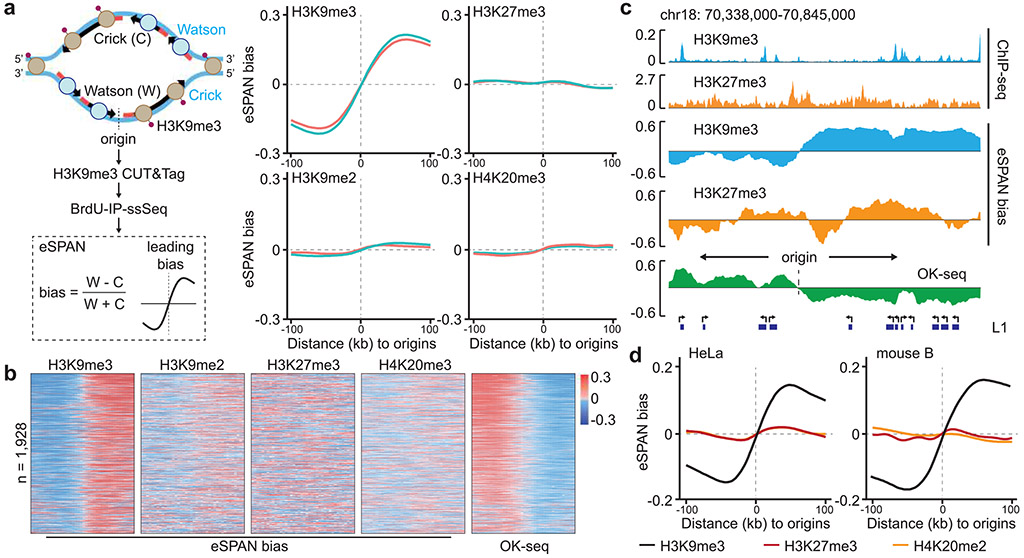
Figure 1. H3K9me3 is transferred preferentially to the leading strands.
a. Left: a schematic diagram of the eSPAN procedure and calculation of eSPAN bias. In this hypothetical model, parental H3K9me3 is transferred to leading strands of DNA replication forks, with two nucleosomes at each leading or lagging strand drawn for simplicity. W and C: sequence reads of Watson and Crick strands, respectively. Right: average bias of H3K9me3, H3K9me2, H3K27me3 and H4K20me3 eSPAN surrounding 1,928 replication origins (−100 kb to 100 kb), with two independent repeats (blue and red) shown.
b. Heatmaps of eSPAN bias for H3K9me3, H3K9me2, H3K27me3 and H4K20me3 centered around each of the 1,928 replication origins sorted based on replication efficiency defined by OK-seq (right).
c. A snapshot of H3K9me3 and H3K27me3 ChIP-seq signals and calculated H3K9me3 and H3K27me3 eSPAN bias, with OK-seq bias used to indicate origin location and DNA replication direction (shown by arrow). L1 elements (≥ 1 kb) at this locus were shown at the bottom with their transcription direction indicated.
d. Average bias of H3K9me3, H3K27me3 and H4K20me2 eSPAN signals in both HeLa cells and primary mouse B cells surrounding 2,809 and 1,073 replication origins (−100 kb to 100 kb), respectively.
H3K9me3 is enriched at leading strands
We employed the enrichment and Sequencing of Protein Associated Nascent DNA (eSPAN) method, which measures the relative amount of a target protein at leading and lagging strands of DNA replication forks21,24, to analyze the distribution of 11 histone modifications at replicating DNA strands in mouse embryonic stem (mES) cells. Briefly, after pulsing cells with 5’-bromo-2’-deoxyuridine (BrdU), a thymidine analog that incorporates into nascent DNA, protein A-fused transposase 5 (pA-Tn5) was targeted to specific chromatin regions via antibodies recognizing a histone modification of interest. When activated, pA-Tn5 locally tagments the genomic DNA. Tagmented DNA was then subjected to immunoprecipitation with antibodies against BrdU to enrich for nascent DNA associated with the modified histones followed by strand-specific sequencing. The eSPAN sequencing reads were aligned to the Watson and Crick strands around replication origins to calculate the eSPAN bias, which reflects the relative amount of a modified histone at the leading and lagging strands of DNA replication forks (Figure 1a). As BrdU is incorporated in place of thymidine into newly synthesized DNA, and BrdU incorporation asymmetry measured by BrdU-IP-ssSeq followed the intrinsic TA strand asymmetry (TA skew) (Extended Data Figure 1a, b), we normalized all eSPAN bias calculations against TA skew or BrdU-IP-ssSeq bias. After removing TA skew, the eSPAN signals for most modifications on parental histone H3-H4 (H3K27me3, H3K9me2, H4K20me3, H4K20me2, H3K36me2, H3K36me3 and H3K4me3) and on newly-synthesized histones (H4K5ac, H4K12ac and H4K5/12ac) displayed little bias towards either leading or lagging strands (Figure 1a and Extended Data Figure 1c), indicating that histones with these modifications are distributed almost evenly on the two sister chromatids arising from leading and lagging strand synthesis.
Strikingly, we found that H3K9me3 eSPAN signals exhibited a strong bias towards the leading strands in mES cells based on the average bias around 1,928 efficient replication origins (Figure 1a) as well as analysis of eSPAN signals at each individual replication origins defined by Okazaki fragment sequencing (OK-seq)25 (Figure 1b, c and Extended Data Figure 1d). The eSPAN bias of H3K9me3 was even stronger than that of MCM2, a subunit of the replicative helicase that travels along the leading strand26 (Extended Data Figure 1c). To rule out potential non-specific effects of H3K9me3 antibodies, we performed the eSPAN experiments using additional H3K9me3 antibodies from two commercial sources (Ab2 and Ab3) and obtained similar results (Extended Data Figure 1e, f). Moreover, the distribution of H3K9me3 on chromatin based on analysis of H3K9me3 CUT&Tag datasets generated using three antibodies correlated well with published H3K9me3 ChIP-seq profiles in mES cells, with Ab1 showing the highest correlation (Extended Data Figure 1g, h), suggesting that the H3K9me3 eSPAN signals detected here reflect genuine H3K9me3 distribution at replicating DNA strands.
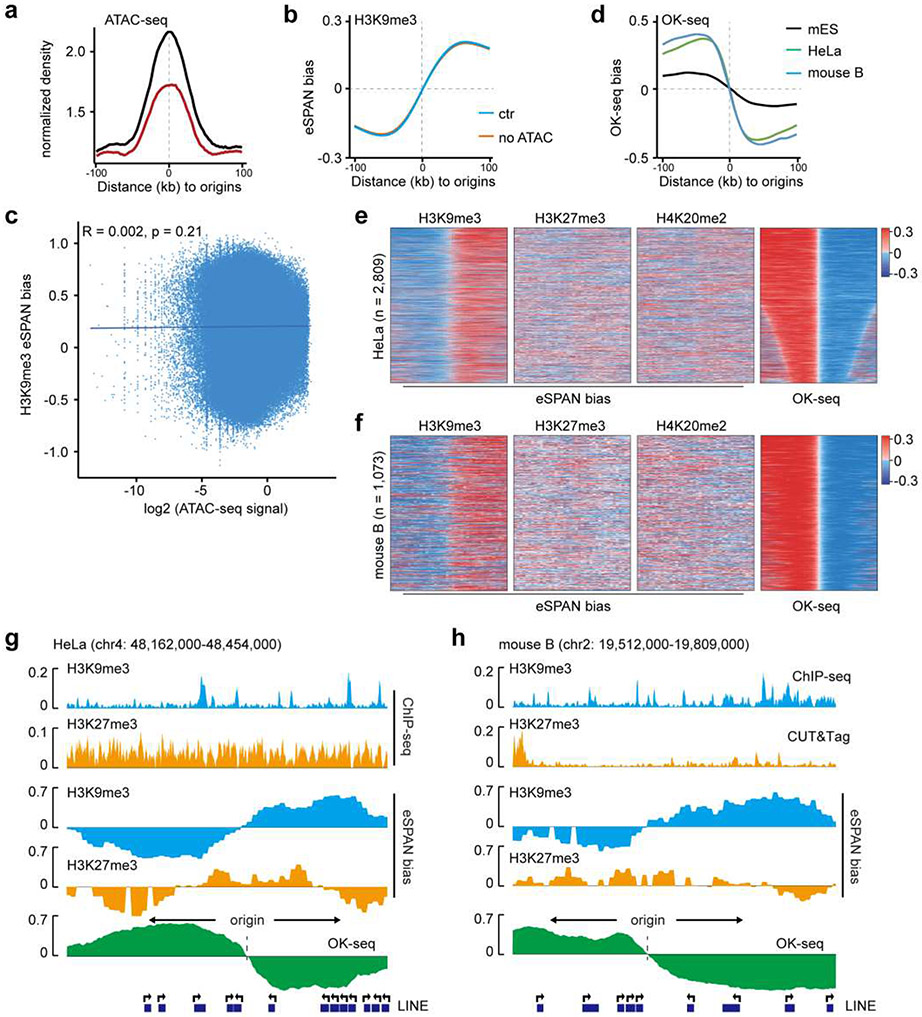
It is possible that the application of pA-Tn5 in the eSPAN procedure gives rise to Assay for Transposase-Accessible Chromatin with sequencing (ATAC-seq) peaks, which may contribute to the H3K9me3 eSPAN bias. To test this idea, we utilized published ATAC-seq datasets and analyzed the signals at replication origins. As expected, ATAC-seq signals were enriched around origins, consistent with the idea that a large fraction of replication origins are located at open chromatin regions (Extended Data Figure 2a). However, H3K9me3 eSPAN signals showed the same bias after the eSPAN sequence reads at regions that overlap with ATAC-seq signals were removed from analysis (Extended Data Figure 2b). In addition, there was no significant correlation between ATAC-seq signals and H3K9me3 eSPAN bias around replication origins (Extended Data Figure 2c), indicating that H3K9me3 eSPAN bias detected in mES cells unlikely arises from ATAC-seq signals.
Next, we tested whether H3K9me3 eSPAN bias could be detected in two other mammalian cell types, in which origins have been defined by OK-seq25,27 (Extended Data Figure 2d). We found that H3K9me3 eSPAN signals in both HeLa cells and activated primary mouse B cells showed a much larger bias towards the leading stand than those of H3K27me3 and H4K20me2 (Figure 1d and Extended Data Figure 2e-h). Together, these results suggest that the asymmetric partition of H3K9me3 is likely conserved in mammalian cells.
H3K9me3 asymmetry occurs mainly at L1s
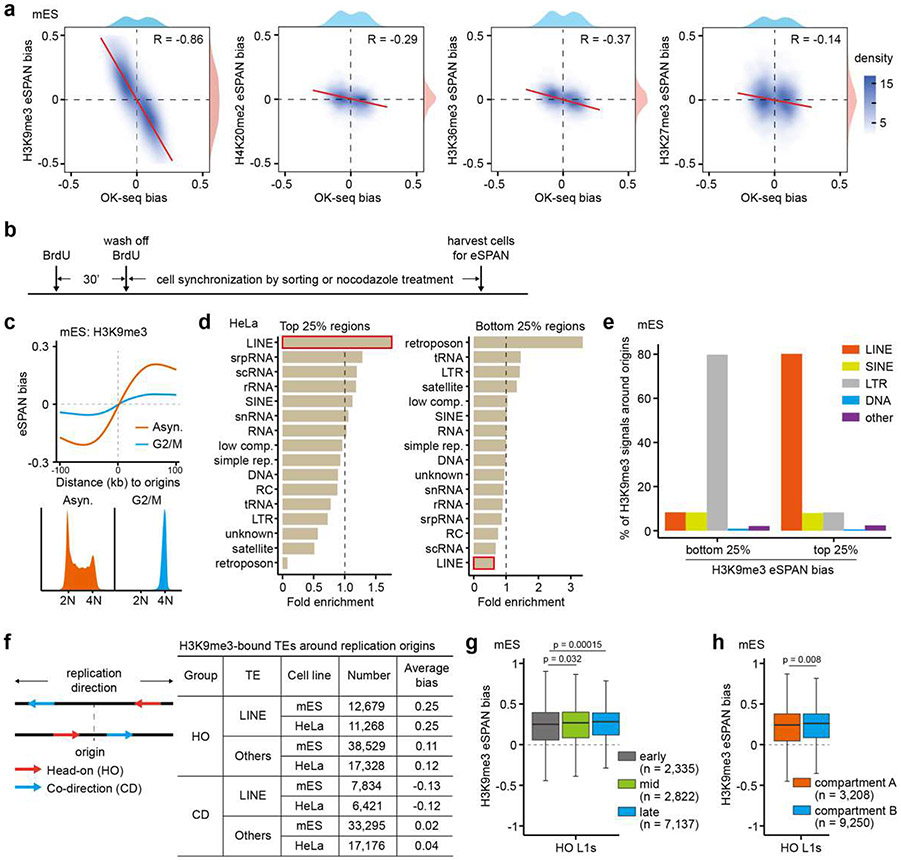
To explore the biological significance of asymmetric H3K9me3 distribution, we first tested whether H3K9me3 asymmetry is linked to DNA replication. Compared to the eSPAN bias of three other histone modifications (H3K27me3, H4K20me2 and H3K36me3), H3K9me3 bias showed the strongest negative correlation with OK-seq bias, an indicator of the efficiency of replication origins (Extended Data Figure 3a), suggesting that H3K9me3 asymmetry is regulated during cell cycle. To test this idea further, we established a mES cell line with ectopic expression of the fluorescence ubiquitination-based cell cycle indicator (FUCCI), in which Cdt1 and Geminin, two DNA replication factors, are fused to fluorescent tags, allowing the isolation of cells at defined cell cycle stages through sorting28. We sorted mES cells pulsed with BrdU at mid S phase and released them into late S or next G1 phase of the cell cycle for H3K9me3 eSPAN analysis (Extended Data Figure 3b). Interestingly, H3K9me3 eSPAN bias peaked at mid S phase and was not detectable at the next G1 phase (Figure 2a). The reduction of H3K9me3 eSPAN bias was also detected in mES cells arrested at the G2/M phase using nocodazole (Extended Data Figure 3c). These results indicate that asymmetric distribution of H3K9me3 occurs in S phase and likely plays certain regulatory roles.
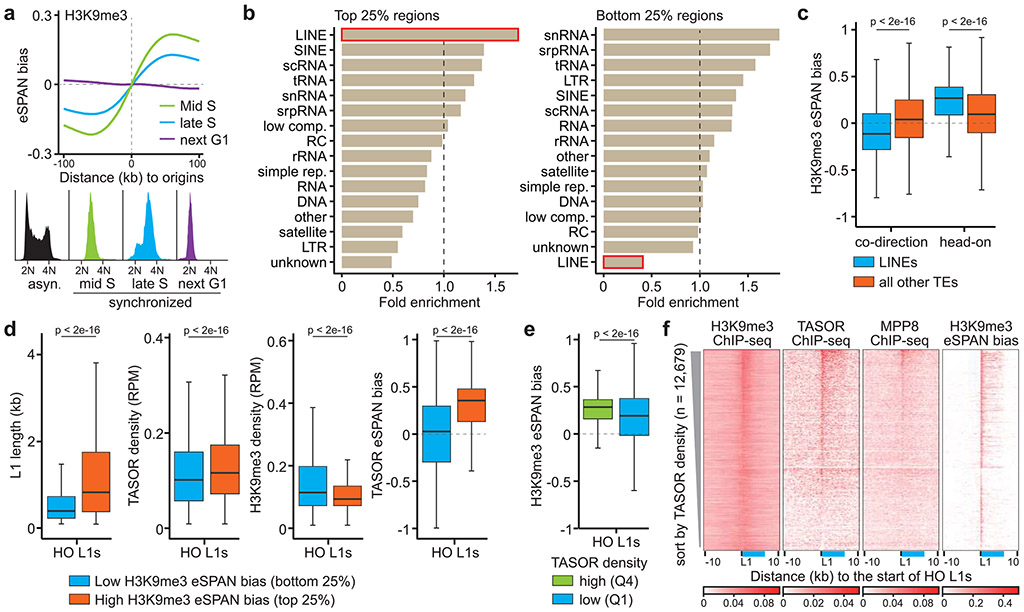
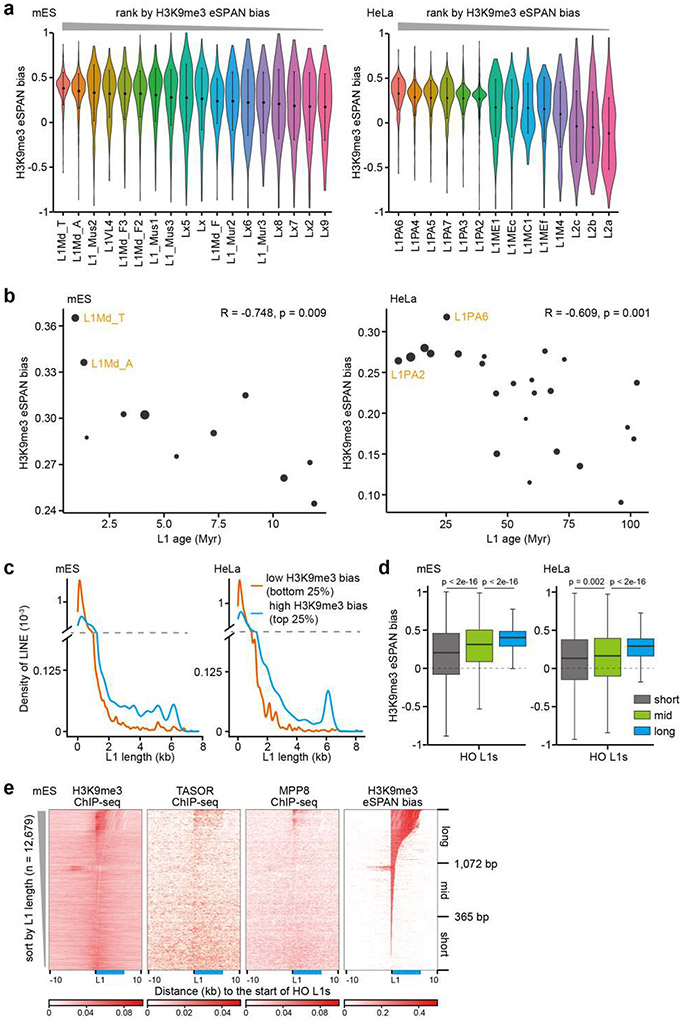
Figure 2. H3K9me3 asymmetry occurs in S phase and at LINE elements.
a. Average H3K9me3 eSPAN bias around origins in mES cells at different cell cycle stages. The respective cell cycle profiles were shown on the bottom.
b. Enrichment of different repetitive elements at 1 kb bins with high (top quartile, 25%) and low (bottom quartile, 25%) H3K9me3 eSPAN bias in mES cells. Fold enrichment is defined as the ratio between calculated and expected enrichment. com., complexity; rep., repeats; LTR, long-terminal repeat; SINE, short interspersed nuclear element; DNA, DNA transposon; scRNA, small cytoplasmic RNA; snRNA, small nuclear RNA; tRNA, transfer RNA; rRNA, ribosomal RNA; srpRNA, signal recognition particle RNA; RC, rolling circle; unknown, repeats without a recognizable TE signature.
c. Box plots of H3K9me3 eSPAN bias at replicated LINEs (n = 20,513) and all other TEs (n = 71,824) that were separated into co-direction (CO) and head-on (HO) groups based on the transcription direction of each TE unit and replication fork direction.
d. Box plots of L1 length, TASOR and H3K9me3 ChIP-seq density, and TASOR eSPAN bias for the top and bottom quartile of HO L1s with the highest and lowest eSPAN bias (total HO L1s = 12,679).
e. Box plots of H3K9me3 eSPAN bias at HO L1s with high TASOR density (Q4, n = 3,170) and low TASOR density (Q1, n = 3,170) based on TASOR ChIP-seq.
f. Heatmaps of H3K9me3, MPP8 and TASOR ChIP-seq density, and H3K9me3 eSPAN bias at HO L1s sorted by TASOR levels. The relative position of a full-length L1 was shown in blue.
Box plots (c-e) show the median, 25% and 75% quartiles and minimal and maximal values with p values by two-sided Mann–Whitney–Wilcoxon tests. c, Bonferroni correction for multiple comparisons. Each panel is a representative of at least two independent experiments. See Materials and Methods for more details.
H3K9me3 is enriched at repetitive DNA elements, which are classified into different categories2,4. To dissect which genomic feature contributed to the H3K9me3 asymmetry, we calculated H3K9me3 eSPAN bias at each 1 kilobase (kb) fragment within the 1,928 initiation zones in mES cells and performed enrichment analysis. Remarkably, L1 elements were the most enriched at the windows with the highest H3K9me3 bias (top 25%), while also being the most under-represented at low bias windows (bottom 25%) (Figure 2b). Similar results were documented in HeLa cells (Extended Data Figure 3d). In addition, the majority of H3K9me3 signals at the 1 kb windows with high H3K9me3 eSPAN bias came from L1 elements, whereas H3K9me3 signals at the bottom 25% regions were mainly from LTRs (Extended Data Figure 3e). Therefore, we identified H3K9me3-bound L1 elements using published datasets and focused our analysis on these L1 elements9.
We observed that most of H3K9me3-bound L1 elements around all the 1,928 origins in mES cells and 2,809 origins in HeLa cells were at a head-on configuration with replication based on their theoretic transcription orientation relative to the direction of replication fork movement (Extended Data Figure 3f). Therefore, we separated all H3K9me3-bound TEs into co-direction (CD) and head-on (HO) groups and directly calculated H3K9me3 eSPAN bias at L1s and all other TEs. The average H3K9me3 eSPAN bias at these HO L1s was much higher than CD L1s or other elements in both mES and HeLa cells (Figure 2c and Extended Data Figure 3f). We therefore focused on H3K9me3-bound HO L1s (12,679 and 11,268 L1s in mES and HeLa cells, respectively) in all following analysis. Together, these results indicate that L1 elements around replication origins are the major contributor to H3K9me3 asymmetry, with H3K9me3 eSPAN at HO L1s showing the strongest bias towards the leading strands.
To understand how the asymmetric distribution of H3K9me3 at HO L1s is regulated, we first analyzed the characteristics of these HO L1s with H3K9me3 bias. HO L1s localized at late replication origins and heterochromatin (compartment B based on Hi-C datasets) displayed a slightly bigger H3K9me3 eSPAN bias than those at early replication origins and euchromatin regions (compartment A), respectively (Extended Data Figure 3g, h). Surprisingly, L1 subfamilies with higher H3K9me3 eSPAN bias tended to be evolutionarily younger (Extended Data Figure 4a, b). Furthermore, when parsing HO L1s by length, we found that HO L1 elements with the highest H3K9me3 eSPAN bias were longer in size than those with the lowest bias (Figure 2d, left panel, and Extended Data Figure 4c-e), suggesting that H3K9me3 asymmetry selectively targets intact young L1s, with a weaker asymmetric distribution of H3K9me3 also detected at old and short L1s.
It is known that the HUSH complex, consisting of TASOR, MPP8 and Periphilin-1 (PPHLN1), works with SETDB1 to silence intact young L1 elements29-31. Using published TASOR ChIP-seq datasets32, we found that HO L1s with the highest H3K9me3 eSPAN bias showed higher TASOR density than those with the lowest bias (Figure 2d). Conversely, by grouping H3K9me3-bound HO L1s based on TASOR density, we detected much higher H3K9me3 eSPAN bias at HO L1s with high TASOR enrichment than those with low TASOR binding (Figure 2e, f). Unexpectedly, the HO L1s with highest H3K9me3 eSPAN bias also contained lower density of H3K9me3 (Figure 2d), which is consistent with previous reports that intact L1s, silenced by the HUSH complex, in general, have lower levels of H3K9me330,32. Together, these results suggest that the HUSH complex contributes to the asymmetric transfer of parental H3K9me3 onto HO L1 elements at the leading strands.
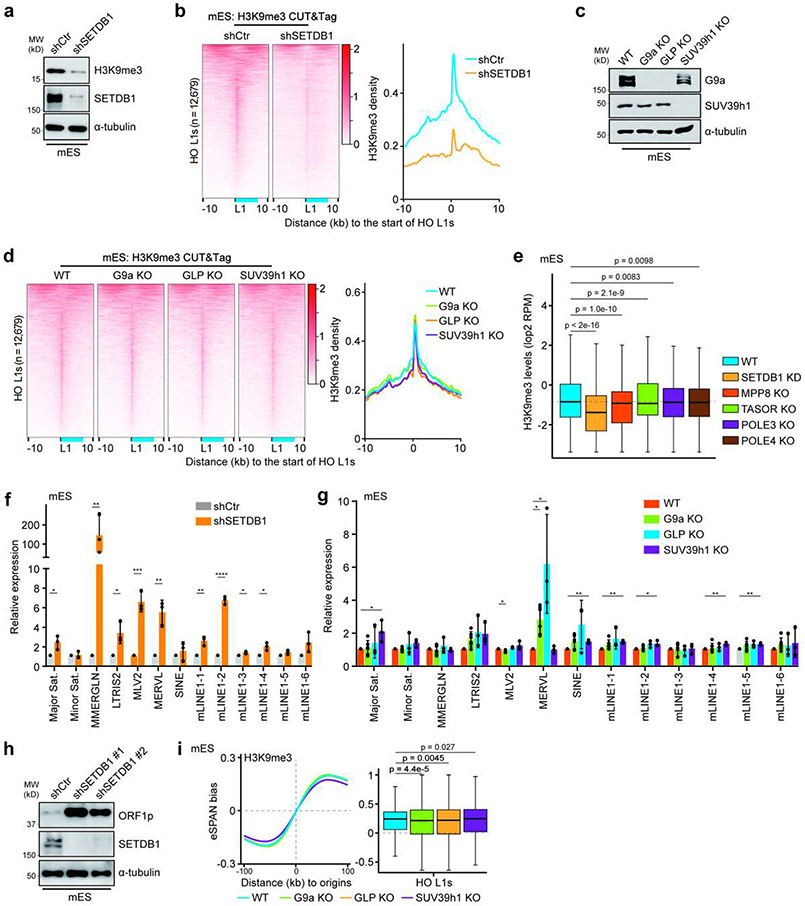
A role of SETDB1 in H3K9me3 asymmetry
SETDB1, SUV39h1, G9a and GLP are the major H3K9 methyltransferases in mammalian cells4. To unravel the mechanisms regulating the H3K9me3 asymmetry, we first aimed to identify the H3K9 methyltransferase responsible for H3K9me3 at HO L1s using unbiased approaches. Consistent with previous reports33,34, SETDB1 knockdown significantly reduced H3K9me3 levels both globally and at L1 elements, yet deletion of SUV39h1, G9a or GLP had no apparent influence (Extended Data Figure 5a-e). Consequently, SETDB1 depletion resulted in increased expression of almost all repetitive elements, including L1 and endogenous retroviruses (ERVs), whereas the effects of other methyltransferases were much weaker or specific to certain TEs (Extended Data Figure 5f-h). Importantly, depletion of SETDB1 dramatically reduced H3K9me3 eSPAN bias around replication origins and at HO L1s (Figure 3a). Intriguingly, deletion of G9a or GLP slightly decreased H3K9me3 eSPAN bias, whereas SUV39h1 deletion resulted in a mild increase, at HO L1s (Extended Data Figure 5i). These results confirm that H3K9me3 at L1 elements is mainly catalyzed by SETDB14. Importantly, the reduction of H3K9me3 eSPAN bias upon SETDB1 depletion is most likely due to the reduced levels of H3K9me3 on parental histone H3.
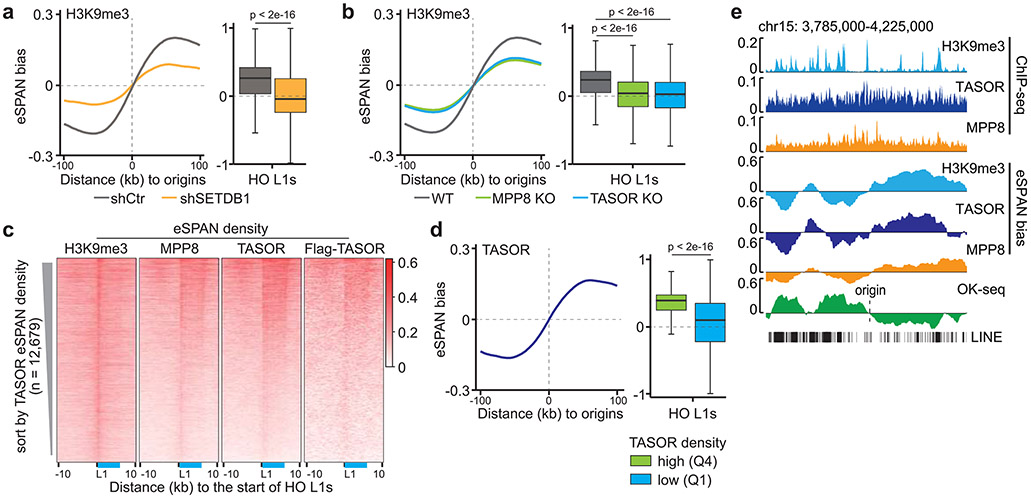
Figure 3. The HUSH complex regulates H3K9me3 asymmetry.
a. H3K9me3 eSPAN bias around replication origins (left) and at HO L1s (right, n = 12,679) in control (shCtr) or SETDB1 knockdown (shSETDB1) mES cells.
b. H3K9me3 eSPAN bias around replication origins (left) and at HO L1s (right, n = 12,679) in WT, TASOR and MPP8 mutant mES cells.
c. Heatmaps of normalized eSPAN density of H3K9me3, MPP8, TASOR and Flag-TASOR at HO L1s, sorted by TASOR eSPAN density in mES cells. The relative position of a full-length L1 was shown in blue.
d. TASOR eSPAN bias around replication origins (left) and at HO L1s (right, n = 12,679) with high and low TASOR ChIP-seq density defined as in Figure 2e, in WT mES cells.
e. A snapshot of H3K9me3, TASOR and MPP8 ChIP-seq signals and calculated eSPAN bias at the indicated locus, with OK-seq bias marking origin location. All L1s at this locus were shown at the bottom.
Box plots (a, b, d) show the median, 25% and 75% quartiles and minimal and maximal values with p values by two-sided Mann–Whitney–Wilcoxon tests. b, Bonferroni correction for multiple comparisons. Each panel is a representative of at least two independent experiments. See Materials and Methods for more details.
HUSH governs H3K9me3 asymmetry
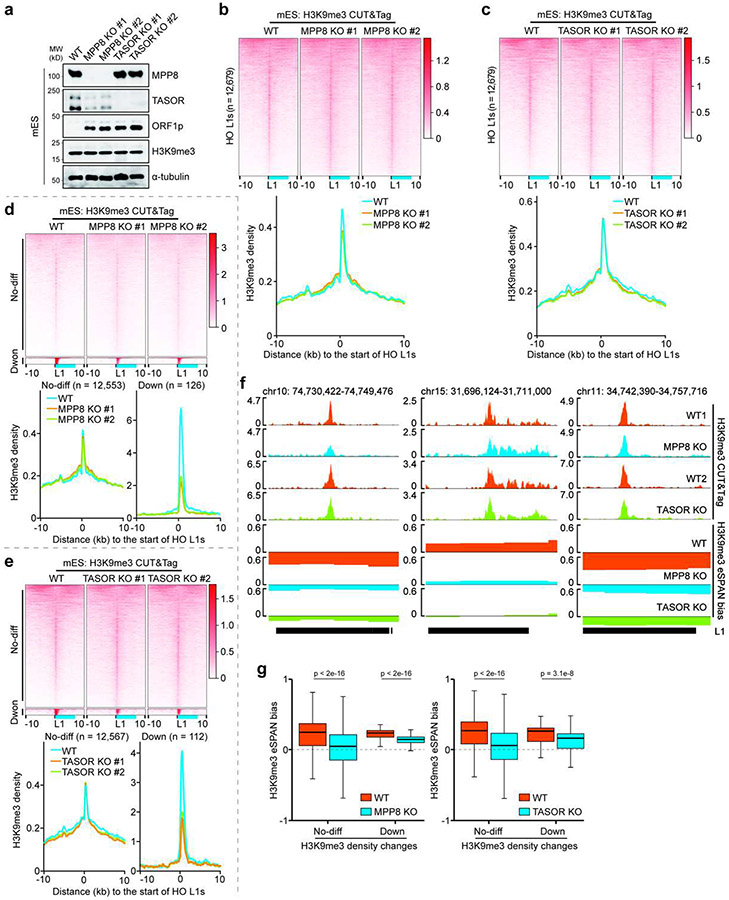
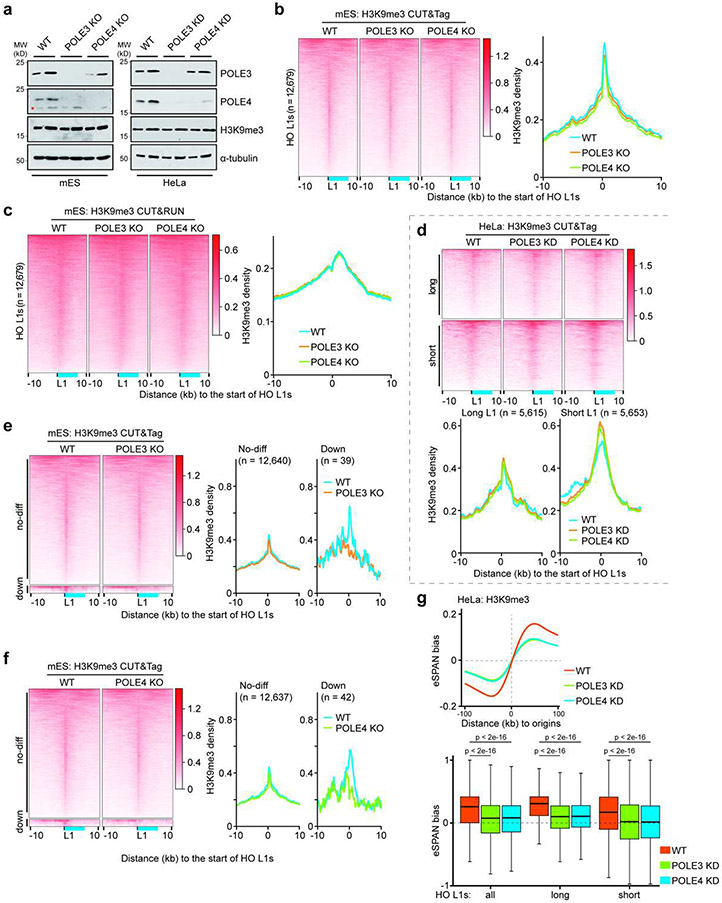
Next, we set out to determine the roles of the HUSH complex in regulating the asymmetric H3K9me3 distribution. Unlike SETDB1 depletion, TASOR or MPP8 knockout (KO) had little impact on H3K9me3 levels globally or at HO L1s at first glance (Extended Data Figure 6a-c). Detailed analysis showed that the average H3K9me3 levels at HO L1 elements were slightly, but significantly reduced in TASOR or MPP8 KO mES cells (Extended Data Figure 5e). However, H3K9me3 levels at the majority of HO L1s were not affected, with reduced H3K9me3 levels detected only at a small number of L1s using a cutoff of 1.5-fold (Extended Data Figure 6d, e). Similar results were obtained in HeLa cells when TASOR or MPP8 was depleted based on analysis of public CUT&RUN and ChIP-seq datasets29,35 (Extended Data Figure 7a, b), and CUT&Tag from this study (Extended Data Figure 7c-e). Together, these results reveal that the HUSH complex is not essential for H3K9me3 levels at the majority of HO L1s.
Remarkably, H3K9me3 eSPAN bias was dramatically reduced in MPP8 or TASOR KO mES cells (Figure 3b and Extended Data Figure 6f, g). The reduction of H3K9me3 eSPAN bias in MPP8 KO and TASOR KO cells was observed at L1s regardless of the degree of H3K9me3 density changes (Extended Data Figure 6g). Similar observations were made in HeLa cells (Extended Data Figure 7f, g), suggesting that the bias change is unlikely caused solely by the reduction of H3K9me3 levels at these L1 elements. Interestingly, depletion of TASOR or MPP8 reduced H3K9me3 eSPAN bias at L1s independent of their length (Extended Data Figure 7h). These results indicate that the HUSH complex is important for the asymmetric distribution of H3K9me3 at most, if not all, HO L1s.
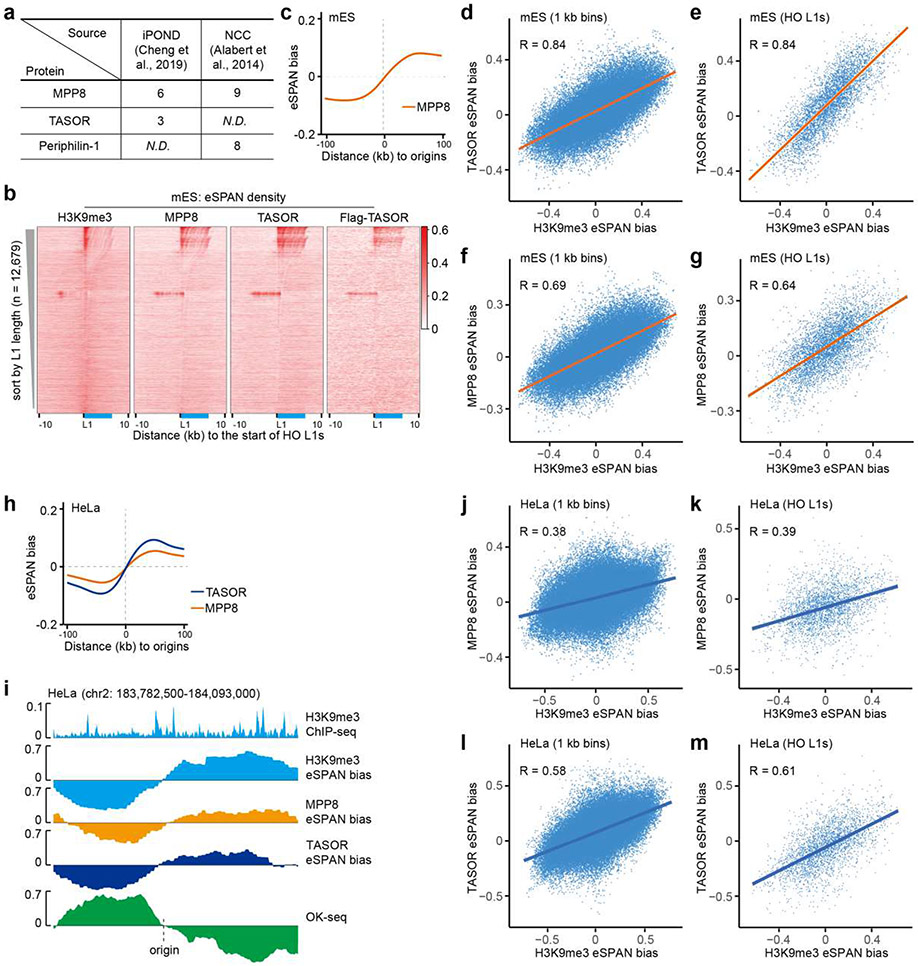
The HUSH complex was detected at replication forks based on published proteomic datasets36,37 (Extended Data Figure 8a). To understand how the HUSH complex facilitates the transfer of H3K9me3 onto HO L1s at the leading strands, we analyzed the distribution of MPP8 and TASOR at DNA replication forks by eSPAN using antibodies against endogenous TASOR/MPP8 or the Flag tag fused with endogenous TASOR (Flag-TASOR) in mES cells. We detected strong TASOR and MPP8 eSPAN signals at HO L1s, which exhibited a pattern similar to H3K9me3 eSPAN density when sorted by TASOR eSPAN density (Figure 3c) or L1 length (Extended Data Figure 8b). Importantly, both TASOR and MPP8 displayed a strong bias towards the leading strand (Figure 3d, e and Extended Data Figure 8c), with the biggest TASOR eSPAN bias detected at HO L1s with the highest levels of TASOR (Figure 3d, right panel). In addition, the eSPAN bias of TASOR and MPP8 was highly correlated with that of H3K9me3 around origins as well as at HO L1s (Figure 2d, right panel, and Extended Data Figure 8d-g). We obtained similar results when analyzing the distribution of MPP8 and TASOR at replicating DNA strands in HeLa cells (Extended Figure 8h-m). These results indicate that the HUSH complex travels along the leading strands of DNA replication forks, which in turn promotes the transfer of existing H3K9me3 onto HO L1s at the leading strands.
Pol ε coordinates with HUSH
We have shown that POLE3 and POLE4, two subunits of the leading strand DNA polymerase Pol ε, facilitate the transfer of parental histones to the leading strands21,22. Similar as in the HUSH mutants, POLE3 or POLE4 depletion did not affect global H3K9me3 levels in either mES or HeLa cells (Extended Data Figure 9a). At HO L1s of mES cells, POLE3 or POLE4 KO reduced the average H3K9me3 levels significantly, but only slightly (Extended Data Figure 5e, 9b and 9c). In HeLa cells, the effects of POLE3 or POLE4 deletion on H3K9me3 levels at HO L1s were comparable regardless of the length (Extended Data Figure 9d). Furthermore, with the cutoff of 1.5-fold, only ~40 of the 12,679 HO L1s showed reduced H3K9me3 density in POLE3 or POLE4 KO mES cells (Extended Data Figure 9e, f). These results indicated that, similar to TASOR and MPP8 KO, mutating POLE3 or POLE4 had minor effects on overall H3K9me3 levels at HO L1 elements in both mES and HeLa cells.
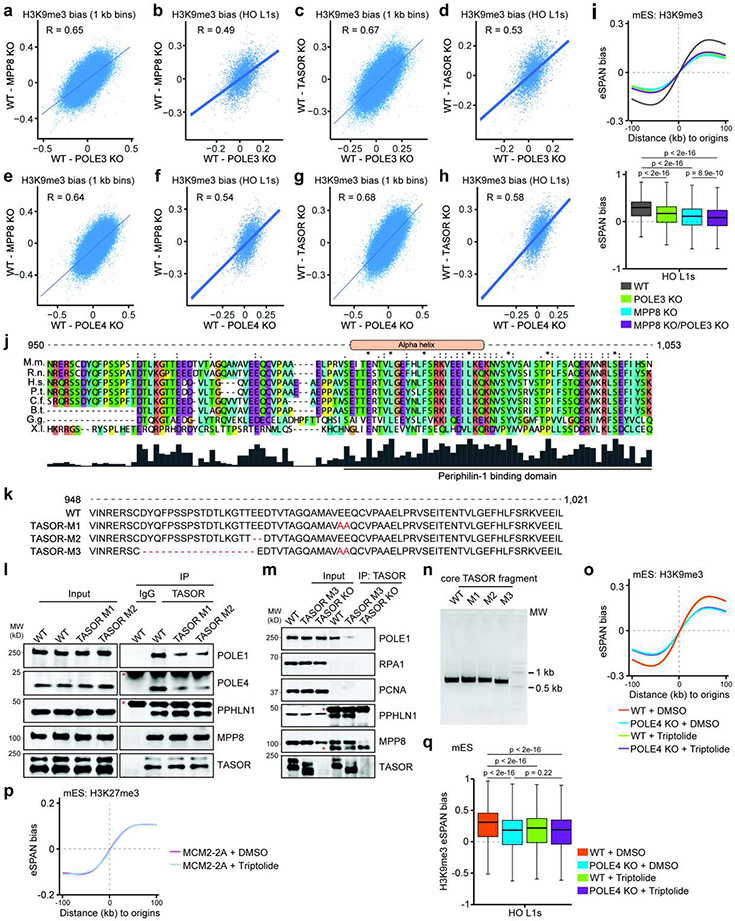
Remarkably, deletion of POLE3 or POLE4 markedly decreased H3K9me3 eSPAN bias based on analysis of average bias at replication origins and at HO L1s in mES and HeLa cells, irrespective of L1 length (Figure 4a and Extended Data Figure 9g), suggesting that Pol ε promotes asymmetric H3K9me3 transfer to HO L1s at leading strands, in a manner akin to the HUSH complex. Supporting this idea, the effects of POLE3 or POLE4 KO on H3K9me3 eSPAN bias changes were highly correlated with those of TASOR/MPP8 deletion in mES cells (Extended Data Figure 10a-h). Importantly, double mutant cells that are depleted of both POLE3 and MPP8 showed no further decrease of H3K9me3 bias compared to single mutants based on the average bias at replication origins, but only a mild additive effect at HO L1s (Extended Data Figure 10i). These results suggest that Pol ε and the HUSH complex function in a largely overlapped manner, but also to some extent independently, to promote asymmetric H3K9me3 distribution.
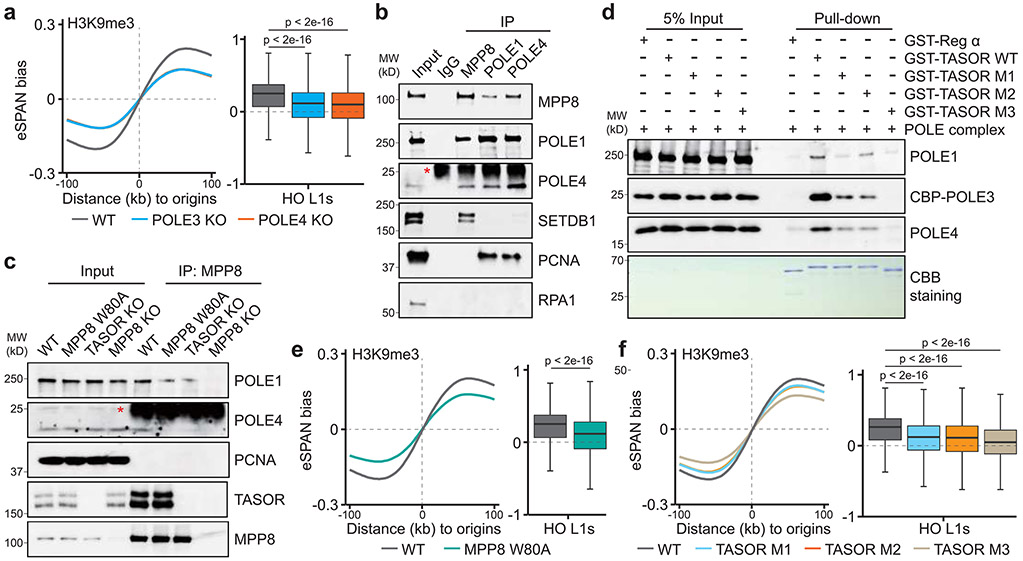
Figure 4. Pol ε coordinates with the HUSH complex for asymmetric H3K9me3 transfer.
a. H3K9me3 eSPAN bias around replication origins (left) and at HO L1s (right, n = 12,679) in WT, POLE3 KO and POLE4 KO mES cells.
b. Interactions between MPP8 and Pol ε determined by co-immunoprecipitations. Anti-Rabbit IgG was used as a negative control. * indicates the IgG light chain. n = 3.
c. TASOR KO or MPP8 chromodomain mutation (W80A) compromised the MPP8-Pol ε interaction. MPP8 KO was used as a negative control. * indicates the IgG light chain. n = 3.
d. TASOR site-specific mutations (M1, M2, and M3) compromised the TASOR-Pol ε interaction as determined by in vitro GST pull-down assays. GST-Reg α was used as a negative control. n = 5.
e, f. H3K9me3 eSPAN bias around replication origins (left) and at HO L1s (right, n = 12,679) in WT and MPP8 W80A (e) or three TASOR mutants (f) mES cells.
Box plots (a, e, f) show the median, 25% and 75% quartiles and minimal and maximal values with p values by two-sided Mann–Whitney–Wilcoxon tests. a, f, Bonferroni correction for multiple comparisons. Each panel is a representative of at least two independent experiments. See Materials and Methods for more details.
For gel source data, see Supplementary Figure 1.
These results prompted us to ask whether Pol ε and the HUSH complex directly associate with each other. Indeed, we found that MPP8 interacted with several subunits of Pol ε, but not PCNA or RPA, which are enriched at the lagging strand (Figure 4b). The interaction between HUSH and Pol ε was also detected via immunoprecipitation of POLE1 or POLE4, two subunits of Pol ε (Figure 4b), but not by immunoprecipitation of MPP8 in MPP8 KO cells (Figure 4c). Interestingly, point mutation of MPP8 chromodomain (W80A), which abolishes MPP8 binding to H3K9me3, as well as TASOR KO, compromised this interaction (Figure 4c). These results suggest that Pol ε binding to HUSH is dynamically regulated in part through the recognition of H3K9me3 by the HUSH complex. To further dissect the interactions between these two complexes, we serendipitously made two mutations (M1, E986A/E987A; and M2, ΔE973/E974) at the acidic residues localized within a highly conserved region of TASOR, which is adjacent to the reported PPHLN1-binding domain38, and a third one combining M1 and a deletion of 18 amino acids at this conserved region (M3) (Extended Data Figure 10j, k). These mutations did not affect TASOR binding to other HUSH subunits (MPP8 and PPHLN1), but reduced its interaction with Pol ε in mES cells (Extended Data Figure 10l, m). To test whether these TASOR mutants affect binding Pol ε in vitro, we cloned a short TASOR fragment surrounding the mutated regions (amino acids 857-1,080) from both WT and TASOR mutant cells (Extended Data Figure 10n). In vitro binding assays using the recombinant human Pol ε complex showed that these mutations reduced the binding of TASOR fragment to Pol ε, with mutant M3 showing the most defects (Figure 4d). Importantly, we found that MPP8 W80A and all of the three TASOR mutations significantly reduced H3K9me3 eSPAN bias around origins and at HO L1s (Figure 4e, f). Thus, both abilities of HUSH to bind H3K9me3 and Pol ε are important for the transfer of parental H3K9me3 onto HO L1s at the leading strands.
Transcription and H3K9me3 asymmetry
Transcription of L1 elements and intronless genes plays an important role in the initiation of H3K9me3-mediated silencing by the HUSH complex30,35,39. We, therefore, asked whether transcription also has a role in the asymmetric distribution of H3K9me3 at replicating DNA strands. To this end, we performed eSPAN analysis in cells treated with triptolide, a widely-used inhibitor of global transcription. Interestingly, average H3K9me3 eSPAN bias around replication origins was largely unaffected by triptolide in both WT and POLE4 KO cells (Extended Data Figure 10o). In addition, triptolide did not affect the leading strand bias of H3K27me3 eSPAN in mES cells carrying the MCM2-2A mutation, which impairs the transfer of parental H3K27me3 to the lagging strands20,21 (Extended Data Figure 10p). These results suggest that triptolide treatment unlikely compromised DNA replication or caused global defects of parental histone transfer. Surprisingly, H3K9me3 eSPAN bias at HO L1s was significantly reduced, with no additive effects when combined with POLE4 KO under the treatment conditions (Extended Data Figure 10q). Together, these results indicate that transcription likely also regulates asymmetric H3K9me3 segregation at HO L1 elements.
H3K9me3 asymmetry silences L1
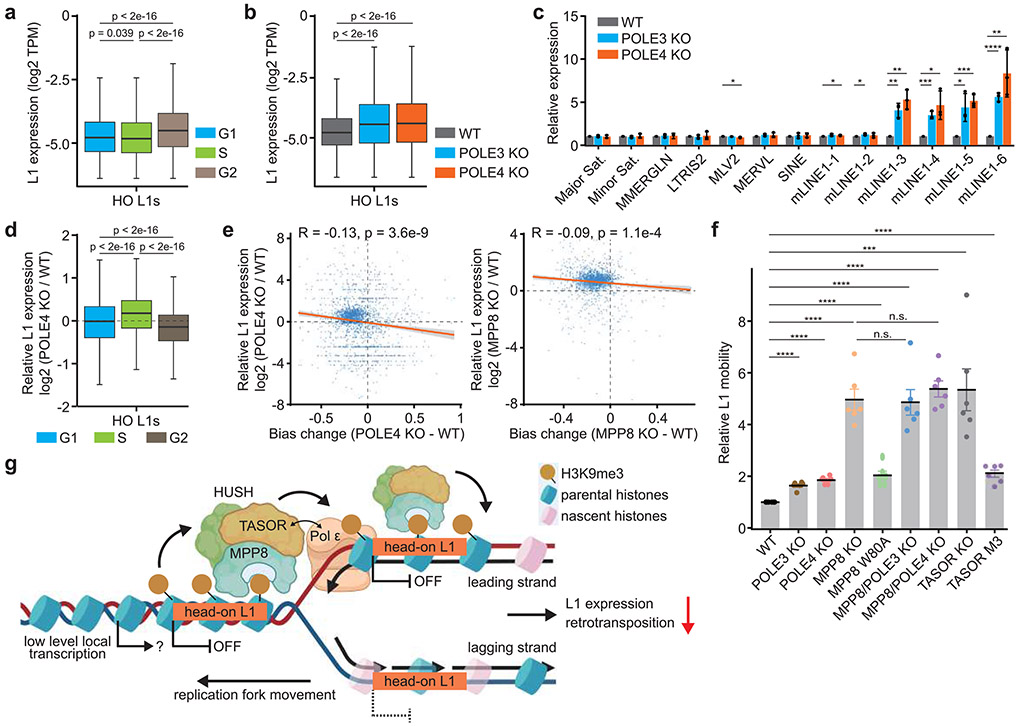
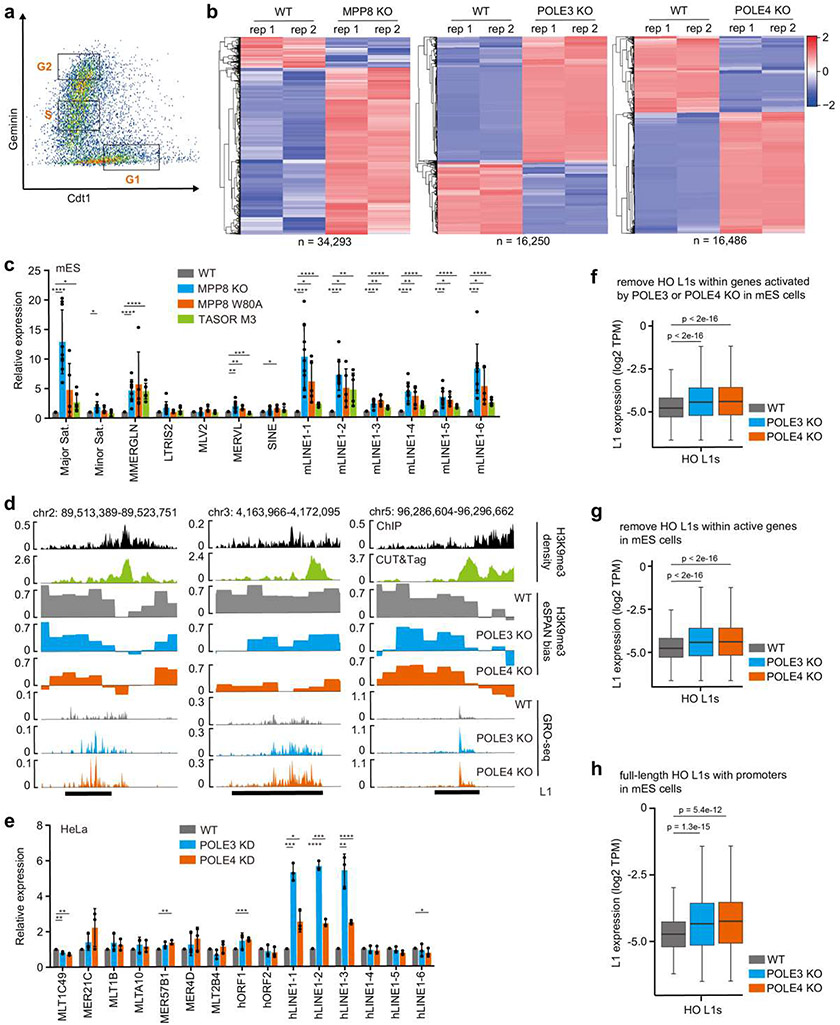
Because the H3K9me3 asymmetry occurs predominantly in S phase, we hypothesized that the asymmetric distribution of H3K9me3 silences L1 expression during DNA replication. To test this idea, we first asked whether the L1 expression is regulated during the cell cycle. Using the FUCCI reporter system, we sorted mES cells into G1, S and G2 phases and analyzed nascent transcription using GRO-seq (global run-on sequencing)40 (Extended Data Figure 11a). The expression of HO L1s in S phase was lower than G1 or G2/M phase cells (Figure 5a), suggesting an S phase specific silencing mechanism. Next, we examined nascent transcription of L1s in selected mutant cells that showed compromised asymmetric H3K9me3 segregation. As expected, MPP8 KO resulted in a dramatic L1 activation (Extended Data Figure 11b, c), echoing its role as a bona fide L1 suppressor. MPP8 W80A and TASOR M3 mutants also showed varying degrees of increased L1 expression (Extended Data Figure 11c), suggesting that the ability of the HUSH complex to bind H3K9me3, as well as Pol ε is important for L1 silencing.
Figure 5. Effects of asymmetric H3K9me3 distribution on L1 silencing and retrotransposition.
a. Expression of HO L1s (n = 2,681) in WT mES cells at G1, S or G2 phase of the cell cycle detected by GRO-seq. Note that HO L1s with expression lower than the cutoff (CPM = 0.1) were excluded from analysis. The same number of L1s was used for analysis in panels b, d.
b. Expression of HO L1s (n = 2,681) in WT and mutant mES cells detected by GRO-seq.
c. Relative expression of representative repetitive elements of different classes in WT and mutant mES cells by RT-qPCR. The expression was normalized against WT (mean ± SEM. n = 3). Sat., satellite. mLINE1, mouse L1 elements.
d. Relative expression of HO L1s (n = 2,681) in POLE4 KO versus WT mES cells at G1, S or G2 phase of the cell cycle. The dashed line indicates no changes compared to WT (0).
e. H3K9me3 eSPAN bias changes were negatively correlated with L1 activation in the mutant cells. Each dot represents an expressed HO L1 within the 1,928 initiation zones.
f. Relative L1 mobility in WT and mutant mES cells (mean ± SEM. n = 6).
g. Schematic working model. In WT cells, asymmetric H3K9me3 distribution to head-on L1s at the leading strand is regulated by low level transcription, the HUSH complex and Pol ε, which is important to inhibit L1 transcription and retrotransposition during S phase.
Box plots (a, b, d) show the median, 25% and 75% quartiles and minimal and maximal values with p values by two-sided Mann–Whitney–Wilcoxon tests, and Bonferroni correction for multiple comparisons. Each panel is a representative of at least two independent experiments. c, f, Two-sided Student’s t test. ****, p < 0.0001. ***, p < 0.001. **, p < 0.01. *, p < 0.05. n.s., not significant, p > 0.05. See Materials and Methods for more details.
Interestingly, L1 expression in POLE3 or POLE4 KO cells was also significantly elevated compared to WT cells (Figure 5b and Extended Data Figure 11b, d). The de-repression of selected L1s in POLE3- and POLE4-depleted cells was confirmed using RT-qPCR in mES and HeLa cells (Figure 5c and Extended Data Figure 11e). Because it is known that a large fraction of L1s is expressed through the “read-through transcription”41, we performed additional analyses by removing HO L1s within the transcribed regions (TSS − 5 kb to TTS + 5 kb) of up-regulated genes (defined by RNA-seq) in POLE3 or POLE4 KO cells (Extended Data Figure 11f), or any genes that are actively transcribed in WT mES cells (Extended Data Figure 11g) and still observed an increased expression of HO L1s in the mutant cells. Moreover, POLE3 or POLE4 KO resulted in increased expression of full-length head-on L1s with a promoter (Extended Data Figure 11h), which could be transcribed via their own promoters and are rarely affected by read-through transcription41. These results suggest that L1 activation in POLE3 or POLE4 KO is unlikely due to the increased expression of genes, which could in turn drive the expression of neighboring L1s through read-through transcription. Intriguingly, double mutant cells depleted of both MPP8 and POLE3 showed a weak, yet significant increase in L1 expression, compared to MPP8 KO single mutant alone (Extended Data Figure 12a). Nevertheless, we observed a significant overlap between HO L1s activated by MPP8 KO and POLE3 KO, or POLE4 KO (Extended Data Figure 12b), supporting the idea that HUSH and Pol ε play a largely overlapped, but partially independent, role in silencing HO L1 elements.
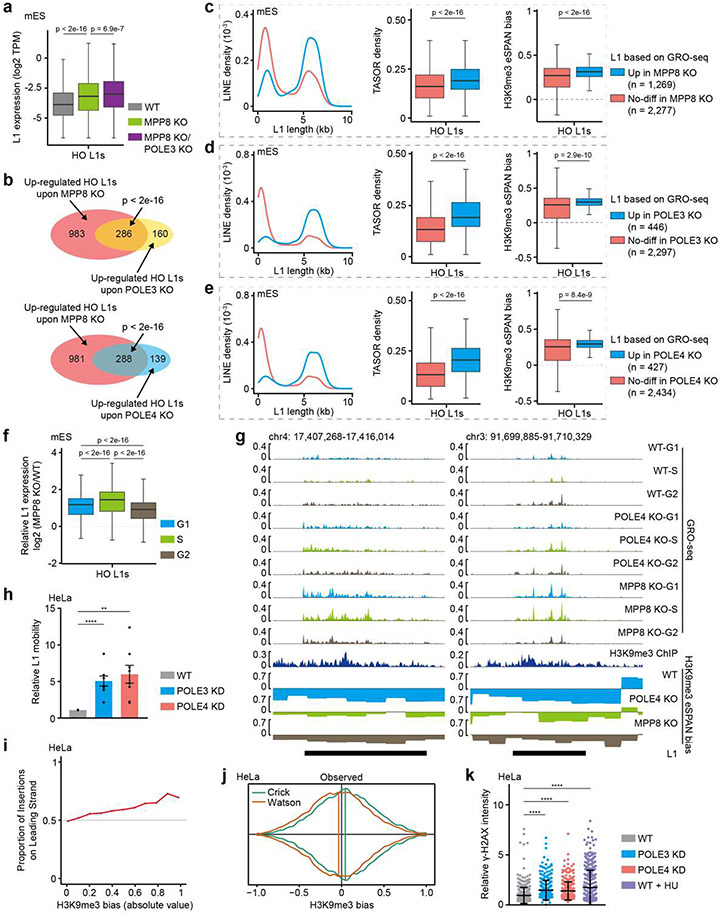
To identify the characteristics of L1s whose expression was affected by MPP8 depletion, we grouped HO L1s based on the effects of MPP8 KO measured by GRO-seq. Consistent with previous reports30, reactivated L1s in MPP8 KO mES cells were longer in size and enriched with higher levels of TASOR, compared to L1 elements whose expression was not affected to a detectable degree (Extended Data Figure 12c, left two panels). Interestingly, these HO L1s also exhibited higher H3K9me3 eSPAN bias (Extended Data Figure 12c, right panel), suggesting that HUSH-repressed L1s are subjected to regulation by H3K9me3 asymmetry. Importantly, HO L1s with increased expression in POLE3 or POLE4 KO cells also showed similar properties (Extended Data Figure 12d, e). Notably, MPP8 KO affected more L1s than POLE3 or POLE4 KO, suggesting that besides regulating asymmetric H3K9me3 distribution, HUSH plays additional roles in L1 silencing.
To further probe the link between H3K9me3 asymmetry and L1 silencing, we analyzed the effects of POLE4 KO and MPP8 KO on L1 expression at different cell cycle stages using the same approaches outlined in Figure 5a. Compared to WT cells, both POLE4 KO and MPP8 KO led to a more pronounced increase of L1 expression in S phase than other cell cycle stages, although the overall increase induced by MPP8 KO was much higher than POLE4 KO (Figure 5d and Extended Data Figure 12f, g). Importantly, H3K9me3 eSPAN bias changes induced by POLE4 or MPP8 KO were moderately, but significantly, correlated with changes in L1 expression (Figure 5e). The relatively low coefficient likely reflected the redundant L1 silencing pathways as well as the transient nature of H3K9me3 asymmetry in S phase. Taken together, these results support the idea that the asymmetric distribution of H3K9me3 at HO L1s suppresses their transcription in the S phase of the cell cycle.
The expression of intact L1 elements can result in retrotransposition and genome instability. Using a dual-luciferase reporter42, we detected increased L1 retrotransposition in mutant cells that showed defective asymmetric H3K9me3 distribution, although to varying degrees in mES cells (Figure 5f) and HeLa cells (Extended Data Figure 12h), suggesting that H3K9me3 asymmetry at HO L1s prevents L1 retrotransposition. Furthermore, no additive effects were detected in double mutant cells depleted of both MPP8 and POLE3 or POLE4 (Figure 5f), consistent with the idea that Pol ε and the HUSH complex work largely overlappingly to silence L1s. Recent reports suggest that L1 integration prefers the leading template strand43,44. Therefore, we analyzed the public datasets of L1 integration in HeLa cells, and found that the fraction of L1 integration into the leading strand steadily increased across H3K9me3 eSPAN bias intervals from 0 to 1 (Extended Data Figure 12i, j), suggesting that both L1 integration and H3K9me3 recycling at head-on L1s prefer the leading strand. Finally, POLE3 or POLE4 deleted cells expressed higher levels of γ-H2AX (Extended Data Figure 12k), an indicator of genome instability. Together, these results indicate that Pol ε- and HUSH-regulated asymmetric H3K9me3 distribution also suppresses L1 retrotransposition.
Discussion
We found that parental histone mark H3K9me3 is asymmetrically distributed on LINE retrotransposons, with head-on L1 elements at the leading strands showing the highest H3K9me3 enrichment during S phase of the cell cycle. Multiple mechanisms likely regulate this unique pattern of H3K9me3 partitioning at the head-on L1s, which constitute of the majority of H3K9me3-bound L1 elements at replication forks. First, the HUSH complex, which binds H3K9me3 released from disassembled nucleosomes ahead of DNA replication forks via its MPP8 subunit, interacts with the leading strand DNA polymerase, Pol ε. This interaction facilitates the transfer of parental H3K9me3 onto L1 elements on nascent chromatin regions synthesized through the leading strands (Figure 5g). Supporting this idea, we found that the interaction between HUSH and Pol ε is regulated by the ability of MPP8 to bind H3K9me3. In addition, both TASOR and MPP8 travel along DNA replication forks on the leading strands. Second, transcription likely also contributes to the asymmetric transfer of H3K9me3 (Extended data Figure 10q). Third, other factors may also participate in this process, as H3K9me3 eSPAN bias was reduced, but not abolished at HO L1s in cells lacking the HUSH complex, POLE3 or POLE4. Previous studies demonstrated that transcription plays a key role in the establishment of H3K9me3-mediated silencing of L1 elements by the HUSH complex45. Furthermore, transcription is also required for the establishment and maintenance of H3K9me3 at heterochromatin in fission yeast5,46. Therefore, more studies are warranted to dissect the roles of transcription and other factors in the transfer of H3K9me3 onto HO L1s during chromatin replication.
The uneven distribution of parental H3K9me3 at HO L1 elements between the two sister chromatids raises the question about how this mark at HO L1 elements is restored following DNA replication. The current paradigm for the restoration of parental histone marks assumes that they are evenly distributed onto the leading and lagging strands of DNA replication forks, a presumption held true in most cases. The classic model, best exemplified by PRC2 and H3K27me3, features an enzymatic complex that binds its cognate histone mark (read) and then modifies neighboring nucleosomes formed with newly synthesized H3-H4 (write) on the same sister chromatid18 (Supplementary Figure 2). Because H3K9me3 is preferentially transferred onto the head-on L1s at the leading strands, this raises the possibility that SETDB1 and/or the HUSH complex recognize H3K9me3-containing nucleosomes on one sister chromatid and methylates neighboring nucleosomes on the other sister chromatid.
Following DNA replication, H3K9me3 levels on replicating chromatin are reduced due to the incorporation of newly synthesized and unmodified histone H3, which undergoes methylation until they reach parental levels in the next G1 phase or subsequent cell cycles47. Surprisingly, we found that L1 expression during S phase is low when compared to the G2/M phase, suggesting an S-phase specific silencing event. Indeed, our results indicate that the asymmetric distribution of H3K9me3 at head-on L1s contributes to their suppression during DNA replication. Supporting this idea, we observed a significant increase of L1 expression in POLE4 or MPP8 KO cells, particularly during S phase of the cell cycle. Furthermore, the activation of L1 transcription correlated with a reduction in H3K9me3 eSPAN bias. Given the intrinsic complexity in lagging-strand DNA synthesis, we postulate that nascent chromatin consisting of the leading strands likely “matures earlier” than the corresponding regions of the lagging strands, creating a time window for these “leading strand regions” to become transcriptionally competent faster, even though both sister chromatids carry the same L1 elements (Figure 5g). Therefore, the transfer of parental H3K9me3 to the head-on L1 elements at leading strands offers a temporary solution to safeguard cells from the harmful activities such as L1 reactivation and retrotransposition during S phase of the cell cycle. Indeed, we found unexpectedly that H3K9me3 levels are lower at HO L1s with the largest eSPAN bias, corroborating previous reports that H3K9me3 levels at full-length L1s, on which we observed high H3K9me3 eSPAN bias, are low30,32. Therefore, we suggest that the transfer of H3K9me3 onto these HO L1 at the leading strand during S phase allow cells to prioritize the limited H3K9me3 to silence L1s on the leading strand first. Alternatively, but not mutually exclusively, activation of L1s at the leading strands is likely more detrimental to cells compared to L1s at the lagging strands during S phase. Indeed, it is known that L1 activation could collide with replication forks, causing so-called ‘LINE-1 toxicity’ and genome instability48-50. Recently, the HUSH complex was reported to silence the transcription of intronless mobile elements, with a strong preference for those with the enrichment of the A nucleotide at the sense strand39. In the future, it would be interesting to determine whether the asymmetric distribution of H3K9me3 mediated by the HUSH complex during S phase, as we observed in this study, is associated with the strand preference of HUSH-mediated silencing.
Materials and Methods
Cell culture
Mouse E14 ES cells (a gift from Dr. Thomas Fazzio, University of Massachusetts Medical School, Worcester, MA) were cultured in DMEM (Corning) medium supplemented with 15% (vol/vol) fetal bovine serum (FBS), 1% penicillin/streptomycin (Invitrogen), 1 mM sodium pyruvate (Cellgro), 2 mM L-glutamine (Cellgro), 1% MEM non-essential amino acids (Invitrogen), 55 μM β-Mercaptoethanol (Sigma) and 10 ng/mL mouse leukemia inhibitory factor (mLIF) on gelatin-coated dishes in a 37°C incubator with a humidified, 5% CO2 atmosphere. HeLa and HEK 293T cells (purchased from American Tissue Culture Collection) were cultured in DMEM medium supplemented with 10% (vol/vol) FBS and 1% penicillin/streptomycin. Primary mouse B cells were isolated from the spleen of wild-type C57BL/6 mice (6-8 weeks old, The Jackson Laboratory). B cells were activated and cultured as described27. Sf9 insect cells were purchased from Thermo Fisher and cultured in suspension in Sf-900 III serum-free medium (Gibco) in a 26°C incubator with gentle shaking, per the manufacturer’s standard protocol. No commonly misidentified cell lines were used in this study. The cell lines were not authenticated. All cells were routinely tested for mycoplasma contamination and the results were always negative.
Mice
The study used 6-8 weeks old wild-type C57BL/6 mice as the donor for splenic B cells. The experimental animals were housed in Specific Pathogen-Free facilities at the Columbia University Irving Medical Center, with a 12/12-h dark/light cycle in environmentally controlled rooms (25 °C, 50% to 55% humidity). The relevant animal care and procedures were conducted according to the protocol AC-AAAY5452 (to Dr. Shan Zha) approved by the Institutional Animal Care and Use Committee (IACUC) of Columbia University. All animal experiments were complied with the ethical regulations according to the NIH Guide for the Care and Use of Laboratory Animals.
Generation of FUCCI cell lines
The pBOB-EF1-FastFUCCI-Puro vector was a gift from Kevin Brindle & Duncan Jodrell (Addgene #86849)51. To generate the cell cycle reporter mES cell lines, the pBOB-EF1-FastFUCCI-Puro plasmid was packaged into HEK 293T cells to produce lentiviruses, which were used to infect WT or mutant mES cells. After puromycin treatment (1 μg/ml) for 2 days, the pooled cells were sorted into single cells in 96-well plates based on their fluorescent signals using Influx Cell Sorter (BD Biosciences). After growing for ~5 days, cell colonies expressing cell cycle indicators were selected under the microscope and confirmed by flow cytometry. All flow cytometry profiles were analyzed using FlowJo (version 10.0, see gating details in Supplementary Figure 3).
Cell cycle synchronization in mES cells
To analyze H3K9me3 eSPAN in different cell cycle phases, WT mES cells expressing a FUCCI reporter were sorted on a flow cytometer (BD Biosciences) based on Cdt1/Geminin expression levels. Briefly, asynchronous cells pulsed with 50 μM BrdU for 30 min were harvested and sorted into mid S phase. Cells were either immediately harvested as mid S phase samples, or released into culture medium for 2 h and collected as the late S sample. Alternatively, cells released into late S phases were treated with 0.1 mM mimosine for 14 h to allow cells to reach next G1 phase, while preventing them from entering next S phase of the cell cycle. To synchronize cells at G2/M phase, BrdU pulsed mES cells were treated with 50 ng/mL nocodazole for 14 h. Cell synchronization was confirmed by flow cytometry using DAPI or PI staining.
CUT&Tag and eSPAN
eSPAN was performed as previously described52. Briefly, mES cells were pulsed with 50 μM BrdU for 20-40 min. HeLa cells were synchronized with double thymidine block, released into S phase and pulsed with 50 μM BrdU for 1 h. Primary mouse B cells were activated for 48 h and pulsed with 50 μM BrdU for 30-40 min. For each sample, 1-2 million cells were harvested and bound to Concanavalin A-coated magnetic beads, incubated with antibodies (diluted at 1: 200 for primary antibodies) and the pre-assembled pA-Tn5 complex. After extensive washing, the tagmentation reaction was initiated by adding magnesium and incubating at 37°C for 1 h with gentle shaking. After tagmentation, 95% DNA was purified and subjected to oligo-replacement reaction and BrdU IP, and ~5% DNA was saved before BrdU IP as the CUT&Tag samples53. Indexing PCR was performed using standard Illumina Nextera primers with a unique set of barcodes for each sample, and the resulting CUT&Tag and eSPAN libraries were pooled and sequenced using paired-end sequencing on Illumina NextSeq 500/550 or NovaSeq 6000 platforms at Columbia University Genome Center.
CUT&RUN
CUT&RUN was performed as described54. Briefly, 0.1 million mES cells were bound to Concanavalin A-coated magnetic beads, incubated with antibodies (diluted at 1: 200 for primary antibodies) and protein A-micrococcal nuclease (pA-MNase). After extensive washing, the reaction was initiated by adding 2 mM calcium and incubated at 0°C for 30 min. After quenching the reaction with 2× stop buffer (340 mM NaCl, 20 mM EDTA, 4 mM EGTA, 0.05% Digitonin, 50 μg/mL RNase A, 50 μg/mL glycogen) and incubation at 37°C for 30 min, the released DNA fragments were purified and processed for library preparation using Swift Accel-NGS 1S Plus DNA Library Kit (Swift Biosciences). The libraries were pooled and sequenced using paired-end sequencing on Illumina NextSeq 500/550 or NovaSeq 6000 platforms at Columbia University Genome Center.
CRISPR-Cas9-mediated gene editing
CRISPR-Cas9-guided mutation or knockout was performed following published procedures55. Briefly, oligos were synthesized from Integrated DNA Technologies (IDT) and cloned into the p×459 or pLenti-CRISPR vector. Gene editing oligo details are in Supplementary Table 2. To generate TASOR/MPP8/POLE3/POLE4 KO mES cells, plasmids expressing the corresponding sgRNAs were transfected using Lipofectamine 3000 (Invitrogen). To generate knock-in mutations of MPP8/TASOR (MPP8 W80A and TASOR M1/M2/M3) or Flag-TASOR mES cells, plasmids expressing the corresponding sgRNAs were co-transfected with synthesized single-stranded oligos as repair templates for homologous recombination. After puromycin treatment at 1 μg/mL for 2-3 days, cells were re-seeded at single cell density. After growing for 5-6 more days, colonies from single cells were then picked under a microscope. Please note that when we attempted to introduce M2 mutation in mES cells carrying M1, we isolated a clone instead containing deletion of 18 amino acids as well as the M1 mutation (M3). To generate POLE3/POLE4/TASOR/MPP8 knockdown HeLa cells, the pLenti-CRISPR vectors expressing the corresponding sgRNAs were packaged into HEK293T cells to produce lentiviruses. HeLa cells were then infected at high MOI (multiplicity of infection) using spin-infection at 1,200 rpm for 1 h. After puromycin selection at 2 μg/mL for 4-5 days, cells were harvested to confirm the knockdown. All genetic mutations were confirmed by Sanger sequencing of the genomic DNA and/or immunoblotting.
Depletion of SETDB1 in mES cells using shRNA
shRNA-guided depletion of SETDB1 in mES cells was performed using commercially available lentiviral shRNA plasmids (Sigma, see details in Supplementary Table 1). Briefly, the pLKO.5 vectors expressing SETDB1 shRNAs or a non-targeting control were packaged into HEK293T cells to produce lentiviruses. mES cells were then infected at a high MOI using spin infection at 1,200 rpm for 1 h. After puromycin selection at 1 μg/mL for 5 days, cells were harvested to confirm the knockdown effects using immunoblotting or perform eSPAN analysis.
GRO-seq
GRO-seq was performed according to published procedures40. Briefly, about 20 million cells were harvested for each sample. After nuclei isolation, nuclear run-on reactions were assembled and incubated at 30°C for 7 min to label nascent RNA with 4-thiouridine (4sU). Total RNAs were purified and fragmented with mild sonication using the Diagenode Bioruptor system, which was then processed for the biotinylation reaction at 24°C for 2 h in the dark. The resulting RNAs were collected and bound by streptavidin beads to capture biotinylated nascent RNA. After extensive washes, the nascent RNA was purified and processed for library preparation using Swift RNA library preparation kit (Swift Biosciences). Strand-specific libraries were pooled and sequenced on Illumina NextSeq 500/550 or NovaSeq 6000 platforms at Columbia University Genome Center.
Expression and purification of the Pol ε complex
Human Pol ε complex was purified using a baculoviral construct, gifted by Dr. Joseph T. P. Yeeles of MRC Laboratory of Molecular Biology (Cambridge, UK). As previously reported56, 2 liters of Sf9 insect cells infected with a single bacmid expressing all the four Pol ε subunits were harvested and lysed in lysis buffer (45 mM HEPES-KOH pH 7.6, 100 mM NaCl, 10% glycerol, 0.5 mM TCEP, 0.02% NP-40-S, supplemented with protease inhibitors) by Dounce homogenization. After centrifugation, the supernatant was incubated with 1 mL Calmodulin Affinity Resin (+ 2 mM CaCl2) for 1 h at 4°C. The unbound proteins were applied to 1 mL HiTrap Heparin column (GE Healthcare) equilibrated in the lysis buffer. The protein was then eluted with a 30CV gradient from 100 to 1000 mM NaCl. Peak fractions were pooled and incubated with 1 mL Calmodulin Affinity Resin (+ 2 mM CaCl2) for 2 h at 4°C. After incubation, the resin was collected and washed twice with lysis buffer (+ 2 mM CaCl2) and the bound proteins were eluted with lysis buffer (+ 2 mM EDTA and 2 mM EGTA). The eluents were applied to MonoQ PC 1.6/5 (GE Healthcare) column equilibrated in lysis buffer and eluted with a 30CV gradient from 100 to 600 mM NaCl. Peak fractions were pooled, confirmed by CBB staining on an SDS-PAGE gel and dialyzed overnight against 1 liter of dialysis buffer (25 mM HEPES-KOH pH 7.6, 10% glycerol, 1 mM DTT, 0.005% Tween, 10% glycerol, 300 mM KOAc). Proteins were aliquoted, snap-frozen in liquid nitrogen and kept at −80 °C until used for in vitro binding assays.
In vitro binding assay using recombinant core TASOR fragment
A core TASOR fragment encoding amino acids 857-1080 was amplified from the cDNA of WT or mutant (M1, M2 and M3) mES cells and cloned into the pGST-parallel vector, using primers listed in Supplementary Table 2. Proteins were expressed in Rosetta 2 (DE3) competent cells under the induction of 0.25 mM IPTG at 16 °C for 16 h. GST-tagged TASOR fragments were then purified with glutathione agarose and eluted with 20 mM reduced glutathione following standard procedures. The eluted proteins were dialyzed overnight against 1 liter of dialysis buffer (20 mM Tris-HCl pH 8.0, 150 mM NaCl, 10% glycerol, 1 mM DTT, 0.01% Triton X-100, supplemented with protease inhibitors) and further purified using a size exclusion column (Superdex 200, GE Healthcare). Peak fractions were pooled and confirmed by CBB staining on an SDS-PAGE gel. For GST pull-down assays, equal molars of GST-TASOR fragments (WT, M1, M2 and M3) or GST-Reg α (as a control) were mix with glutathione agarose in binding buffer (20 mM Tris-HCl pH 8.0, 150 mM NaCl, 5% glycerol, 1 mM DTT, 0.01% Triton X-100, supplemented with protease inhibitors) and incubated at 4 °C for 1 h. Then equal amount of Pol ε complex were added to each reaction and incubated at 4 °C overnight with gentle rotation. After extensively washing the glutathione agarose with binding buffer, the bound proteins were eluted with 1× SDS sample buffer by boiling and analyzed by immunoblotting.
Immunoprecipitation
To immunoprecipitate proteins associated with TASOR/MPP8/POLE1/POLE4, 10-20 million mES cells were harvested for each sample and nuclear extracts were prepared in nuclear extraction buffer (50 mM HEPES pH=7.4, 200 mM NaCl, 10% glycerol, 1 mM EDTA, 0.5% NP-40, supplemented with protease inhibitors). Nuclear lysates were sonicated at low settings to increase protein yield from the chromatin fraction. After centrifugation at max speed for 15 min, the supernatants were pre-cleared with protein G sepharose beads at 4°C for 1 h. After removing the beads, 2-4 μg of the indicated antibodies or IgG (as a control) were added to the supernatants and incubated at 4°C overnight. Protein G sepharose beads were then added and incubated at 4°C for 2 h. The beads were extensively washed with wash buffer (50 mM HEPES pH=7.4, 100 mM NaCl, 10% glycerol, 1 mM EDTA, 0.01% NP-40, supplemented with protease inhibitors) and boiled in 1× SDS protein sample buffer. Immunoprecipitated proteins were then analyzed by immunoblotting.
Immunoblotting
Whole cell lysates were prepared by resuspending cell pellets in 1× SDS protein sample buffer. Proteins were resolved on SDS-PAGE gels and detected by standard Western blot procedures using antibodies listed in Supplementary Table 1. Primary antibodies were diluted as following: all three anti-H3K9me3 antibodies, 1: 2000; anti-POLE1, GTX132100, 1: 1000; anti-POLE3, A6469, 1: 1000; anti-POLE4, A9882, 1: 1000; anti-MPP8, 16796-1-AP, 1: 1000; anti-SETDB1, 11231-1-AP, 1: 1000; anti-PCNA, 307902, 1: 2000; anti-RPA1, NA13, 1: 1000; anti-TASOR, GTX66177, 1: 1000; anti-CBP, 07-482, 1: 2000; anti-α-tubulin, 12G10, 1: 2000; anti-G9a, 3306, 1: 1000; anti-SUV39h1, 8729, 1: 1000; anti-ORF1p, NBP2-66934, 1: 1000; anti-ORF1p, 88701, 1: 1000; anti-PPHLN1, A17706, 1: 1000. All secondary antibodies were diluted at 1: 5000. Relative intensity of immunoblot signals were quantified by ImageJ when necessary. Representative images of at least three independent repeats for each experiment were shown. Raw and uncropped immunoblot results were provided in Supplementary Figure 1.
Retrotransposition assay
To measure L1 retrotransposition in mES and HeLa cells, a dual-luciferase reporter system, gifted by Dr. Wenfeng An of South Dakota State University (Brookings, SD), was employed. Briefly, as reported42, mES and HeLa cells were transfected with a single-vector reporter containing a hyperactive synthetic mouse L1 (ORFeus) and a human L1 element (L1RP), respectively, using Lipofectamine 3000 (Invitrogen), per the manufacturer’s protocol. As a control, another single-vector reporter expressing a retrotransposition deficient L1 (JM111) was separately transfected as the reference vector for normalization. After selection with puromycin at 1.5 μg/mL for 2 days to remove the un-transfected cells, cells were re-seeded on 96-well plates, with at least three replica wells for each sample. The luciferase activities were detected using the Dual-Glo Luciferase Assay System (Promega) in a GloMax plate reader (Promega) per the manufacturer’s procedures. Relative L1 mobility was first calculated as the ratio between firefly and Renilla luciferase values (Fluc/Rluc), and then normalize against Fluc/Rluc values from the JM111 control, which was set as 1.
RT-qPCR
Total RNA was purified with RNeasy Plus Mini kit (Qiagen). After reverse transcription with random hexamers (Invitrogen), real-time quantitative polymerase chain reaction (RT-qPCR) was performed in triplicates for each sample with SYBR Green PCR Master Mix on a CFX96 platform (Bio-Rad Laboratories). Primers for detection of endogenous retroviral elements were described in21,57 and others were listed in Supplementary Table 2.
Immunofluorescence (IF)
To detect γ-H2AX expression by IF staining, cells were seeded onto glass coverslips, fixed with 4% paraformaldehyde (PFA) and permeabilized with 0.25% Triton X-100. After blocking with 5% BSA for 1 h, cells were incubated with γ-H2AX antibody at 1: 500 dilutions at 4°C overnight. Samples were washed with PBS and incubated with secondary antibody conjugated to Alexa Fluro 594 (diluted at 1: 500) at room temperature for 1 h in the dark. Samples were then counterstained with DAPI and the coverslips were mounted onto microscope slides. Images were collected with Nikon Eclipse TS100 microscope and data was processed and analyzed using Image J/Fiji58.
Analysis of CUT&Tag, CUT&RUN, ChIP-seq and eSPAN datasets
DNA libraries for CUT&Tag, CUT&RUN and eSPAN were sequenced using the paired-end method on Illumina NextSeq 500/550 or NovaSeq 6000 platforms. Public ChIP-seq datasets were downloaded from the GEO (Gene Expression Omnibus) and ENCODE (see Supplementary Table 1). Raw reads were filtered by trimming Illumina adapters and removing low-quality reads using Trim Galore (version 0.6.7) with default parameters. Trimmed reads (more than 20 nt) were aligned to human (hg19) or mouse (mm10) reference genomes using Bowtie2 (version 2.2.4)59 with the following flags: with --no-mixed --no-discordant --no-dovetail --no-contain --local --maxins 1000. Sequence alignment map files including uniquely mapped and multi-mapped reads were transformed into BAM files using SAMtools (version 1.11)60. Duplicate reads were removed using the MarkDuplicates function of Picard (version 2.23.8) (https://github.com/broadinstitute/picard) with REMOVE_DUPLICATES = true. Alignment files in the BAM format were converted to read coverage files in Bigwig format using deepTools bamCoverage (version 3.2.1)61 with RPM normalization and 10 bp window size after removing blacklisted regions annotated from ENCODE. The consistent paired-end reads mapped to the Watson (W) and Crick (C) strands of the reference genome were separated by BEDTools (version 2.29.2)62 bamtobed function and in-house Perl programs.
To calculate the eSPAN bias, each bin was computed from the separated Watson and Crick reads within a 5 kb window size using the formula: across the whole genome. The bins with less than 6 sequencing reads were discarded. To reduce potential contributions of TA skew, as defined by the formula , to eSPAN bias, we normalized all eSPAN biases against TA skew or BrdU-IP-ssSeq bias. The eSPAN bias was smoothed by flanking five bins for further visualization around the origins expanding 100 kb on both left and right sides (-100 kb to 100 kb). TE annotations of mouse (mm10) and human (hg19) were obtained from the UCSC Genome Browser annotation track database. To identify the enrichment of repetitive elements associated with highest and lowest H3K9me3 bias, replicated regions (origins ± 100 kb) were separated into 1 kb sliding window bins, the absolute H3K9me3 eSPAN bias was calculated at each window, and then all bins grouped at replicated regions into four quartiles, with Q4 representing top 25% bins with high H3K9me3 eSPAN bias and Q1 representing the lowest 25% H3K9me3 eSPAN bias, respectively, based on the H3K9me3 bias of each bin. Bins with highest or lowest H3K9me3 bias were then used independently to calculate the significance and enrichment score for each class of TE elements using the Genomic Association Tester (GAT) tool (version 1.3.4) (https://github.com/AndreasHeger/gat).
To correlate the H3K9me3 eSPAN bias with L1 integration in HeLa cells, the human reference genome (hg19) was divided into 2 kb bins and H3K9me3 eSPAN bias was calculated for each bin using the formula: . Correlation was then performed as described with minor modifications43. Briefly, we performed wavelet smoothing with option -j 3 on a subset of genome bins. The smoothing was applied to bins with total read counts ranging from 20 to 500. Adjacent smoothed bins were merged if they exhibited the same trend for delta H3K9me3 eSPAN bias. The merged bins were used to calculate the slope and identify ascending and descending segments using SEGMENT_THRESHOLD=0.35, FUSE_MAX_FRAC=0.33, and FUSE_MAX_BINS=10 parameters. Linear regression analysis of H3K9me3 eSPAN bias was conducted over each called segment, weighted by the total read count in each bin. We optimized the linear regression analysis and estimated the final linear model of H3K9me3 eSPAN bias using the method provided by Dr. Diane A. Flasch43. To explore the association between L1 integration and local H3K9me3 eSPAN bias, we incorporated L1 integration data from Dr. Diane A. Flasch and counted the insertions within each weighted segment.
Identification of H3K9me3-bound L1 in mES and HeLa cells
To identify H3K9me3-bound L1 elements in mES cells, we obtained H3K9me3 ChIP-seq data from ENCODE (ENCSR857MYS) and GEO database (GSE100168, GSE199040)9,63-65. The ChIP-seq data was aligned against the mm10 reference genome to generate BAM file including both uniquely mapped and multi-mapped reads. Peak calling was performed using the SICER2 software (version 1.0.3) for ChIP-seq data66. The analysis aimed to identify broad peaks, with a false discovery rate (FDR) cutoff of 0.05, window size of 200 bp, and gap size of 600 bp. The corresponding control samples were used as the background. Similar analysis was conducted in HeLa cells using publicly available H3K9me3 ChIP-seq data from ENCODE (ENCSR000AQO) and GEO database (GSE198978)67. For CUT&Tag data, we employed GoPeaks (version 1.0.0)68 with default parameters to identify peaks. To overcome limitations associated with regions with low signal intensity in individual datasets, we adopted a strategy of extracting union peaks from multiple datasets. In our analysis, we utilized a total of 103,013 H3K9me3 peaks in mES cells and 74,098 peaks in HeLa cells, obtained from the union of H3K9me3 peaks of the datasets mentioned above across the whole genome. This approach allowed us to maximize the number of H3K9me3-bound TEs for analysis. Importantly, all TEs, without any length cutoff were included in the analysis, unless length is the factor to separate L1s into different groups. We specifically focused on replicated H3K9me3-bound TEs located within ±100 kb of replication regions around the efficient origins identified by OK-seq. These replicated H3K9me3-bound TEs were further categorized into co-directional and head-on TEs based on their orientation of transcription with respect to the direction of replication fork movement. After confirming a good consistency among all repeats of the eSPAN data, we proceeded to merge them to improve the sequencing coverage on all TEs. Notably, H3K9me3 eSPAN bias exhibited the highest magnitude on LINEs, compared to other TEs, such as LTR retrotransposons, SINE, DNA transposons, and others. Furthermore, most of the replicated H3K9me3-bound LINE elements were identified as head-on LINEs, displaying a prominent leading strand bias. In contrast, co-directional LINEs exhibited a much weaker bias towards the lagging strands. Based on the reasons listed above, we have decided to focus our in-depth analysis on replicated H3K9me3-bound head-on LINEs, with 12,679 and 11,268 L1s identified in mES and in HeLa cells, respectively. To identify bias, head-on LINEs with fewer than 6 mapped read counts were excluded. Furthermore, the H3K9me3 eSPAN bias at head-on LINEs and at replication origins was normalized against the TA skew before proceeding with further investigations to eliminate the potential contribution of TA skew to the eSPAN bias.
Analysis of enrichment for TASOR peaks or H3K9me3 eSPAN bias
Due to the limited number of TASOR ChIP-seq peaks that could be identified using published datasets, we identified TASOR-High and TASOR-Low HO L1 elements in mouse ES cells using TASOR ChIP-seq data from the GEO database (GSE208748)32. We calculated the average TASOR ChIP-seq signals across the HO L1 elements by normalizing the reads per million and ranked them in descending order using WiggleTools (version 1.2.11) (https://github.com/Ensembl/WiggleTools) and java-genomics-toolkit (https://github.com/timpalpant/java-genomics-toolkit). The top 25% of ranked elements were classified as TASOR-High L1, while the bottom 25% were classified as TASOR-Low L1. Heatmaps illustrating the absolute H3K9me3 eSPAN bias across the head-on L1s were generated following established protocols30 with minor modifications. Only the detectable bias at the TEs, while excluding the noise from non-TE regions using BEDTools (version 2.29.2), was considered for constructing the corresponding Bigwig file used in the enrichment analysis. Heatmaps and profiles were generated around the HO L1 using the plotHeatmap and plotProfile functions from the deepTools package (version 3.2.1).
Differential H3K9me3 binding analysis
Sequencing read counts within each H3K9me3-bound LINE were calculated using BEDTools (version 2.29.2) and subsequently normalized to reads per million by total mapped reads to the corresponding reference genome. To identify H3K9me3-bound LINEs with significantly reduced H3K9me3 signals in different mutant samples compared to control, we employed the edgeR package (version 3.34.0)69. The threshold for significance was set at log2 (Fold change) < −0.585 and FDR < 0.05.
Correlation analysis between L1 age and H3K9me3 eSPAN bias
The age data of mouse and human L1s were acquired from previously published studies70,71. The average age of each group classified by Repeatmasker was used to indicate the age of each family for mouse L1. Pearson correlation analysis was performed to examine the relationship between the average H3K9me3 eSPAN bias and age of each family for head-on L1 in both mES and HeLa cells, taking into consideration of the assumption of normal distributions.
OK-seq and Repli-seq analysis
While many methods are available to identify and analyze replication origins in mammalian cells, we decided to choose OK-seq, as it offers high resolution and strand-specific information of initiation zones, which further helps us to clearly define efficient origins and distinguish leading and lagging strands. OK-seq datasets of mES cells, activated primary mouse B cells, and HeLa cells were obtained from the GEO database (see Supplementary Table 1)21,25,27. Identification of replication initiation zones followed the method described previously25. Downloaded or processed OK-seq reads were mapped to the reference genomes corresponding with the species using Burrows–Wheeler Aligner (version 0.7.17)72 software with default parameters. As previously described73, a 4-state HMM model (Up, regions of predominant initiation (IZ); Down, regions of predominant termination (TZ); Flat1 and Flat2, two intermediate transition states) was used in the segmentation process to call initiation and termination zones based on 1 kb bins across all chromosomes setting read coverage threshold = 10 and the smoothing window size = 30 kb. Further, origin expanding efficiency (OEM) of fixed window sizes is defined as: , in which can be custom defined by users as a list of windows (e.g. [1, 10, 20, 50, 100]), and ranges from 1 to the total length of the data. and correspond to the number of reads mapped to the Watson and Crick strands within the corresponding windows, respectively. It can be used to directly determine the transition states of replication and to validate the pre-identified IZs under the 100 kb scale. Pre-identified IZs were filtered if the maximum OEM across the IZs was less than 0.1. The distance from the boundary of initiation zone to origin position was at least 50 kb. The final initiation zones were the overlapped IZ regions between two repeats. The average lines and heatmaps of bias from eSPAN or OK-seq were generated around the defined origins in different cell lines. Public Repli-seq datasets of E14 mES cells were downloaded and analyzed as described74. The pre-defined origins were divided into early, mid, late origins based on the average BrdU density during different stages of S phase.
GRO-seq and RNA-seq analysis
GRO-seq libraries were sequenced using the paired-end methods on Illumina NextSeq 500/550 platforms. Published RNA-seq datasets in mES cells were downloaded and re-analyzed75. Raw reads were trimmed using Trim Galore (version 0.4.4, https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/) to remove Illumina adaptors and low-quality bases with Phred score lower than 30, and reads shorter than 20 nt. The genome sequence and coding gene annotation of mm10 was derived from GENCODE76. The repetitive elements annotations were obtained from RepeatMasker (version UCSC mm10) and L1Base 277. The clean fastq files of GRO-seq experiments were processed using the previously described PEPPRO pipeline (version 0.10.1) to perform quality control78. The resulting fastq files were then “pre-aligned” against the mouse rDNA genome library constructed by refgenie (version 0.12.1) (https://github.com/refgenie/refgenie) to siphon off these unwanted reads using Bowtie2 (version 2.2.4). The downstream analysis of GRO-seq was performed as previously described40 with slight modifications as deemed necessary. Briefly, after quality control assessment, the rDNA-removed fastq files were re-aligned to mm10 using STAR (version 2.7.6a)79 with the following parameters: “--outFilterMultimapNmax 100 --winAnchorMultimapNmax 100” in which reads with maximum multi-mapped alignment of no more than 100 were kept. Using SAMtools (version 1.11)60, we extracted unique mapped reads with ‘-q 40’ parameter. Unique mapped reads were counted in the gene body using featureCounts (version 2.0.1)80. Read counts of TEs at the locus level were calculated by overlapping with SAF format of TEs annotation transformed from BED format using featureCounts with ‘-p -M -O -s 2’ parameters which can quantify TEs by using both uniquely mapped and multi-mapped reads identified as reversely stranded library using the ‘infer_experiment.py’ function from the RSeQC package (version 4.0.0)81. The matrix of read counts at coding genes and TEs were combined for analysis of differential gene expression using edgeR (version 3.34.0)69,82 with default parameters. Batch effect was removed using ComBat-seq83 with group=NULL parameter. Expression of coding genes and TEs was quantified by transcripts per kilobase million (TPM), which was calculated from counts of all samples using TPM calculator84. TEs with zero or extremely low counts in both WT and the mutant cells were filtered using counts per millions (CPM) values instead of raw counts, as CPM considers the variations in sequencing depth across different samples. Specifically, only TEs with a CPM value greater than 0.1 in at least two samples were included. TEs with a BH-adjusted P value less than 0.05 and a fold change greater than 1.5 (up-regulated) or less than 0.67 (down-regulated) were considered as differentially expressed. For visualization, the strand specific genomic coverage was calculated and normalized to library size using deepTools bamCoverage (version 3.2.1)61.
Evaluate the effects of read-through transcription on L1 expression in POLE3 KO and POLE4 KO cells
To eliminate potential read-through effects in the L1 expression analysis, HO LINEs identified in this study were filtered if they were located within the genic regions of any active genes. Specifically, HO L1s localized within a range of 5 kb upstream of transcription start site and 5 kb downstream of transcription termination site (TSS − 5 kb to TTS + 5 kb) of actively transcribed genes were removed from the analysis. The threshold to define a gene as active is TPM > 0.5 in any GRO-seq samples. Alternatively, we also removed HO L1s located within this same range of up-regulated genes in POLE3 or POLE4 KO mES cells. Briefly, total RNA-seq data in WT, POLE3 KO and POLE4 KO mES cells were obtained from GSE142996 and GSE183065 datasets. After trimming adaptors and low-quality reads using Trim Galore (version 0.6.7), the clean reads were aligned to the mouse genome (GENCODE mm10 primary assembly) using STAR (version 2.7.6a). Reads within exonic regions, as defined by GENCODE’s vM17 version, were counted using featureCounts (version 2.0.1). After generating a count matrix using a custom Perl script, differentially expressed genes were identified using the edgeR (version 3.34.0) with a BH-adjusted P value threshold of <0.05 and a fold change threshold of >1.5. The total number of up-regulated genes was 433 and 687 upon POLE3 KO and POLE4 KO, respectively. HO L1s located within of these up-regulated genes (TSS − 5 kb to TTS + 5 kb) were removed, and only those HO L1s outside this range were selected to assess the effects of POLE3 KO and POLE4 KO on L1 expression using GRO-seq datasets. Full-length (≥ 6 kb) head-on L1s were extracted from L1Base 277, a specialized database that focuses on putatively active LINE-1 elements. Subsequently, a BEDTools INTERSECT comparison was performed between the RepeatMasker LINE annotation and the L1Base annotation to identify full-length L1s with additional regions that contain the promoter, 5’-UTR and 3’-UTR. A total of 703 full-length (≥ 6 kb) head-on L1 elements within the 1,928 initiation zones in the mm10 reference genome were identified. GRO-seq signals from these 703 L1s in WT, POLE3 KO and POLE4 KO mES cells were examined.
Statistics and reproducibility
All experiments involved in deep sequencing were independently repeated at least twice. The remaining experiments were repeated at least three times, and data were presented as the mean ± SD, or mean ± SEM, as indicated. We utilized the nortest package (version 1.0.4) (https://CRAN.R-project.org/package=nortest) to perform a normality test on the H3K9me3 eSPAN bias, expression, and H3K9me3 signals of head-on LINEs. Our analysis revealed that these data deviated from a normal distribution significantly (p < 0.05). Consequently, the p values were calculated by two-sided Mann–Whitney–Wilcoxon test or two-sided Student’s t test, unless otherwise specified. To account for multiple comparisons, we applied the ‘bonferroni’ method to adjust the p values. For sequencing related datasets, significance was shown with adjusted p values, calculated by two-sided Mann–Whitney–Wilcoxon tests. Significance for two-sided Student’s t tests was indicated as: ****, p < 0.0001. ***, p < 0.001. **, p < 0.01. *, p < 0.05. n.s., not significant, p > 0.05. Spearman’s rank correlation was utilized for datasets that exhibited non-normal distributions. In the figures of the genomics data, the box plots were displayed in R 3.6.3 to denote the medians and the interquartile ranges, with upper and lower whiskers. The boundary of the box shows values from Q1 to Q3 (25% to 75%) of the values. The central line in the box is the median value. The Interquartile range (IQR) is defined as the distance between Q3 and Q1 (Q3 − Q1). The upper whisker extends to the maximal value defined as 1.5 times IQR above Q3 (Q3 + 1.5 × IQR). The lower whisker extends to the minimal value defined as 1.5 times IQR below Q1 (Q1 − 1.5 × IQR). Outliers were omitted from box plots to improve clarity, but they were still considered when calculating the p values. When applicable, statistical parameters, statistical tests used, error bar definitions and sample sizes are reported in the figures and corresponding figure legends.
Extended Data
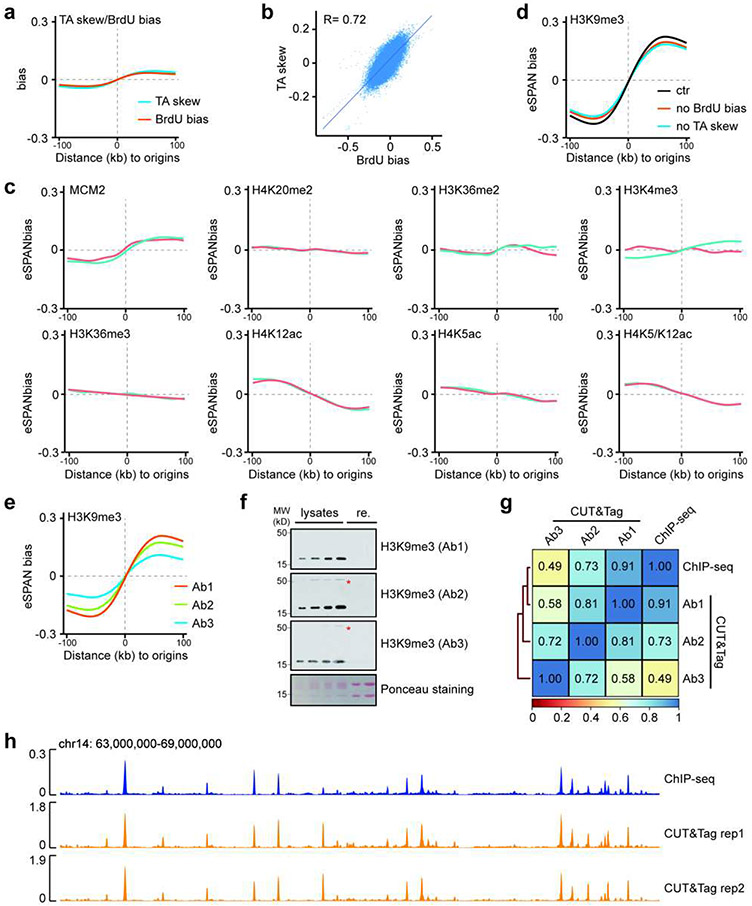
Extended Data Figure 1. Leading strand bias of H3K9me3 eSPAN is detected by H3K9me3 antibodies from three different sources.
a. The TA skew and average BrdU-IP-ssSeq bias (BrdU bias) around replication origins in mES cells.
b. The TA skew was correlated with BrdU bias. The BrdU bias, reflecting the relative amount of DNA synthesis at leading and lagging strands, was calculated using formula . and represent sequencing reads of Watson and Crick strands, respectively. Spearman’s rank correlation coefficient was shown. Each dot represents a 1 kb bin within the 1,928 initiation zones in mES cells. p < 2.2e-16.
c. Average eSPAN bias of MCM2, a subunit of the CMG replicative helicase, and 7 histone modifications (H4K20me2, H3K36me3, H3K36me2, H3K4me3, H4K12ac, H4K5ac and H4K5/K12ac) in mES cells. Two independent repeats, indicating by blue and red lines, for each eSPAN experiment shown.
d. Raw average H3K9me3 eSPAN bias (ctr) and after normalizing against BrdU bias (no BrdU bias) or TA skew (no TA skew) in mES cells.
e. Average bias of H3K9me3 eSPAN bias generated using three H3K9me3 antibodies from different sources.
f. Immunoblots of H3K9me3 in different amounts of mES cell lysates by three different H3K9me3 antibodies used in e. Recombinant histone H3/H4 (re.) were used as negative controls. * indicates non-specific signals detected by antibody 2 (Ab2, Diagenode) and antibody 3 (Ab3, Active motif) after heavy exposure. Ab1: self-made in the laboratory and used in this study. n = 3.
g. Genome-wide correlations of ENCODE H3K9me3 ChIP-seq with H3K9me3 CUT&Tag signals generated with three different antibodies Ab1, Ab2 and Ab3, with a window size of 20 kb. Note that Ab1 showed the strongest correlation with published ChIP-seq datasets, consistent with the immunoblotting results. These differences in performances of the three H3K9me3 antibodies likely contribute to the different H3K9me3 eSPAN biases observed in e.
h. A snapshot of ENCODE H3K9me3 ChIP-seq signals and two repeats of H3K9me3 CUT&Tag signals generated by Ab1 H3K9me3 antibodies at the indicated mouse Chr14 region.
For gel source data, see Supplementary Figure 1.
Extended Data Figure 2. The enrichment of H3K9me3 at the leading strands is also detected in HeLa and primary mouse B cells.
a. Normalized density of ATAC-seq signals around replication origins in mES cells. Black and red lines indicate two independent datasets.
b. Raw average H3K9me3 eSPAN bias (ctr) and after removing eSPAN sequencing reads at regions that also contain ATAC-seq peaks (no ATAC) from analysis.
c. Correlations between H3K9me3 eSPAN bias and normalized published ATAC-seq signals. Each dot represents a 1 kb bin within the 1,928 initiation zones in mES cells. Spearman’s rank correlation coefficient and p value were shown.
d. OK-seq biases at origins in mES (n = 1,928), HeLa (n = 2,809) and primary mouse B cells (n = 1,073) used in this study.
e, f. Heatmaps of eSPAN biases of H3K9me3, H3K27me3 and H4K20me2 and OK-seq bias in HeLa (e) and activated mouse B cells (f) at each individual replication origin, with the number of origins used for analysis shown. The heatmap was sorted based on replication efficiency defined by OK-seq.
g, h. A snapshot of ChIP-seq and CUT&Tag signals and calculated eSPAN bias for H3K9me3 and H3K27me3 in HeLa (g) and activated mouse B cells (h). OK-seq bias indicates origin location and DNA replication direction (shown by arrow), and L1 elements (≥ 1 kb) with their transcription direction at each locus were shown.
Extended Data Figure 3. LINE retrotransposons contribute to the asymmetric H3K9me3 distribution.
a. Correlations between OK-seq bias and eSPAN bias of H3K9me3, H4K20me2, H3K36me3, or H3K27me3. Spearman’s rank correlation coefficient and the density distribution were shown.
b. Experimental schemes for eSPAN analysis in synchronized mES cells shown in Figure 2a and Extended Data Figure 3c. After pulsing cells with BrdU for 30 min, cells were either sorted by flow cytometry based on the FUCCI reporters (Figure 2a) or treated with nocodazole (Extended Data Figure 3c). See Materials and Methods section for more details.
c. Average H3K9me3 eSPAN in asynchronized mES cells or cells synchronized at G2/M phase. Bottom: flow cytometry analysis of cell cycle of asynchronized mES cells and cells arrested at G2/M by nocodazole.
d. The relative enrichment of different repetitive elements at 1kb bins with high or low H3K9me3 eSPAN bias surrounding DNA replication origins in HeLa cells. DNA sequences around replication origins were fragmented into 1 kb bin, and ranked based on H3K9me3 eSPAN bias. The top 25% of regions with the highest H3K9me3 eSPAN bias and the bottom 25% with the lowest H3K9me3 eSPAN bias were then used for calculating the enrichment of each indicated DNA element. Fold enrichment is defined as the ratio between the calculated and expected enrichment. com., complexity; rep., repeats.
e. Percentage of the accumulative H3K9me3 ChIP-seq signals at different TEs around replication origins with highest (top quartile) and lowest (bottom quartile) H3K9me3 eSPAN bias defined in Figure 2b.
f. A schematic representation showing L1 elements whose transcription direction is head-on (HO) and co-direction (CD) with the direction of replication fork movement (left). The numbers and average H3K9me3 eSPAN bias of different TEs within the (−100 kb to 100 kb) regions of 1,928 origins in mES cells and 2,809 origins in Hela cells were counted and shown. Others: all other TEs excluding LINEs.
g. Box plots of H3K9me3 eSPAN bias at HO L1s separated by their locations in the early, mid or late replicating origins, which are defined based on the replication timing data in mES cells.
h. Box plots of H3K9me3 eSPAN bias at HO L1s separated by their locations in genome compartment A or B based on Hi-C datasets in mES cells.
Box plots (g, h) show the median, 25% and 75% quartiles and minimal and maximal values with p values by two-sided Mann–Whitney–Wilcoxon tests. Each panel is a representative of at least two independent experiments. See Materials and Methods for more details.
Extended Data Figure 4. Strong asymmetric H3K9me3 distribution is detected at “young” and long L1s.
a. All HO L1 families with more than 195 copies in mES (left) and HeLa cells (right) were ranked from left to right based on average H3K9me3 eSPAN bias. See Source Data for numbers of L1s in each family. Data were plotted as mean ± SD. Note that L1Md_T and L1Md_A in mES and L1PA in HeLa cells, which have been reported as young and full-length L1s in mouse and human, respectively, show bigger bias.
b. Pearson correlations between H3K9me3 eSPAN bias at HO L1s surrounding origins and their corresponding ages in mES (left) and HeLa (right) cells. Each dot represents a L1 subfamily and the size of the dot is proportional to its copy numbers around replication origins. Note that some of the youngest L1s with high H3K9me3 eSPAN bias were highlighted in orange.
c. Distribution of the length of H3K9me3-bound HO L1s with the lowest H3K9me3 eSPAN bias (red, bottom 25%) and highest H3K9me3 eSPAN bias (blue, top 25%) in mES (left) and HeLa (right) cells. The Y axis was fragmented to better show the details of LINE distribution. Note that L1s with the lowest H3K9me3 bias were shorter than L1s with the highest bias.
d. Box plots of H3K9me3 eSPAN bias at HO L1s that were separated into three groups based on their size in mES and HeLa cells. HO L1s were ranked from short to long according to their lengths. The shortest 1/3 was grouped as short (HeLa, n = 3,747; mES, n = 4,225), the middle 1/3 as mid (HeLa, n = 3,767; mES, n = 4,230) and the longest 1/3 as long (HeLa, n = 3,754; mES, n = 4,224).
e. Heatmaps of H3K9me3, MPP8 and TASOR ChIP-seq density and H3K9me3 eSPAN bias at HO L1s sorted by L1 length in mES cells, with the size range for long, mid and short L1 groups indicated. The relative position of a full-length L1 was shown in blue.
Box plots (d) show the median, 25% and 75% quartiles and minimal and maximal values with p values by two-sided Mann–Whitney–Wilcoxon tests, and Bonferroni correction for multiple comparisons. Each panel is a representative of at least two independent experiments. See Materials and Methods for more details.
Extended Data Figure 5. Effects of mutating H3K9 methyltransferases on H3K9me3 density and eSPAN bias at L1s in mES cells.
a. SETDB1 depletion reduced H3K9me3 levels dramatically, as detected by immunoblotting. n = 3.
b. Heatmaps (left) and average density (right) of H3K9me3 CUT&Tag signals at all the HO L1s in control (shCtr) and SETDB1 knockdown (shSETDB1) mES cells. Heatmaps were sorted by the average H3K9me3 signals of each row in the control sample. The relative position of a full-length L1 was shown in blue.
c. Immunoblots of G9a and SUV39h1 to confirm the knockout of G9a, GLP and SUV39h1. Note that antibodies against GLP were not working, but GLP knockout also dramatically reduced the levels of its binding partner, G9a. n = 3.
d. Heatmaps (left, sorted as in b) and average density (right) of H3K9me3 CUT&Tag signals at all HO L1s in WT, G9a KO, GLP KO and SUV39h1 KO mES cells.
e. Box plots of H3K9me3 density at all HO L1s (n = 12,679) in WT and mutant mES cells. The dashed line indicates the median of H3K9me3 levels in WT cells.
f, g. Relative expression of representative repetitive elements in SETDB1 KD (f) and G9a, GLP or SUV39h1 KO (g) mES cells compared to control (shCtr) or WT cells by RT-qPCR analysis. Expression was normalized against shCtr or WT. Data were plotted as mean ± SEM. n = 3-6.
h. Immunoblots of ORF1p, the translational products of full-length L1s, in mES cells treated with control or two SETDB1 shRNAs. n = 3.
i. H3K9me3 eSPAN bias around replication origins and at HO L1s (n = 12,679) in WT, G9a KO, GLP KO and SUV39h1 KO mES cells.
Box plots (e, i) show the median, 25% and 75% quartiles and minimal and maximal values with p values by two-sided Mann–Whitney–Wilcoxon tests, and Bonferroni correction for multiple comparisons. f, g, Two-sided Student’s t test. ****, p < 0.0001. ***, p < 0.001. **, p < 0.01. *, p < 0.05. Each panel is a representative of at least two independent experiments. See Materials and Methods for more details.
For gel source data, see Supplementary Figure 1.
Extended Data Figure 6. TASOR or MPP8 deletion reduced H3K9me3 eSPAN bias in mES cells, while having little effects on H3K9me3 levels.
a. Immunoblots to confirm the knockout of MPP8 and TASOR in mES cells. Note that ORF1p were markedly up-regulated, while total H3K9me3 levels didn’t change to a detectable degree. n = 3.
b, c. H3K9me3 CUT&Tag signals at HO L1s in MPP8 KO (b) and TASOR KO (c) mES cells, compared to WT cells. Two repeats for each mutant were shown in the heatmaps (top), with the average H3K9me3 density shown at the bottom. Heatmaps were sorted by the average H3K9me3 signals of each row in WT cells. The relative position of a full-length L1 was shown in blue.
d, e. H3K9me3 CUT&Tag signals at HO L1s in MPP8 KO (b) and TASOR KO (c) mES cells, compared to WT cells. Two repeats for each mutant were shown in the heatmaps (top, sorted as in b), with average density shown at the bottom. L1s with reduced H3K9me3 levels for more than 1.5-fold were grouped as Down and those without significant changes were grouped as no-difference (No-diff). Note that less than 130 (~1%) L1s showed significant reduction of H3K9me3 density.
f. Snapshots of H3K9me3 CUT&Tag signals and eSPAN bias in WT, MPP8 KO and TASOR KO mES cells at three loci. Note that H3K9me3 CUT&Tag in MPP8 KO and TASOR KO were performed in separate batches with their corresponding H3K9me3 CUT&Tag in WT cells (WT1 and WT2 shown for more accurate comparisons.
g. Box plots of H3K9me3 eSPAN bias at two groups of HO L1s (n = 12,679) in WT, MPP8 KO (left) and TASOR KO (right) mES cells. An average of two independent repeats were shown and L1s were grouped as in d, e.
Box plots (g) show the median, 25% and 75% quartiles and minimal and maximal values with p values by two-sided Mann–Whitney–Wilcoxon tests, and Bonferroni correction for multiple comparisons. Each panel is a representative of at least two independent experiments. See Materials and Methods for more details.
For gel source data, see Supplementary Figure 1.
Extended Data Figure 7. TASOR or MPP8 depletion reduced H3K9me3 eSPAN bias at HO L1s in HeLa cells.
a. H3K9me3 density at HO L1s in WT and TASOR KO HeLa cells based on published CUT&RUN datasets35. Heatmaps (left) were sorted by the average H3K9me3 signals of each row in WT cells, with average density shown at the right. L1s were separated based on the effects TASOR KO on H3K9me3 levels. Of the 393 TASOR regulated H3K9me3 loci identified by Douse et al.35 using a cutoff of log2 fold-change < −1, we found that 119 HO L1s identified in this study were located at these loci and defined them as the Down group. All other HO L1s were grouped as No-diff.
b. H3K9me3 density at HO L1s in WT, MPP8 KO and TASOR KO HeLa cells based on published ChIP-seq datasets29, with heatmaps (top, sorted as in a) and average density (bottom) shown. HO L1s were grouped as in a.
c. Immunoblots to confirm the knockdown (KD) of MPP8 and TASOR in HeLa cells. Note that while sgRNAs targeting MPP8 or TASOR were used to generate these cells, cells were pooled after selection, instead of cloned. Therefore, MPP8 and TASOR were only depleted and labeled as KD in HeLa cells. Note that ORF1p were markedly up-regulated, while total H3K9me3 levels didn’t change to a detectable degree. n = 3.
d. H3K9me3 CUT&Tag signals at HO L1s in WT, MPP8 KD and TASOR KD HeLa cells. The datasets were generated in this study and HO L1s were grouped as in a.
e. H3K9me3 CUT&Tag signals at HO L1s separated based on L1 length in WT, MPP8 KD and TASOR KD HeLa cells. HO L1s longer and shorter than the medium length were grouped as long (n = 5,615) and short (n = 5,653), respectively.
f. Average H3K9me3 eSPAN bias around all 2,809 replication origins in WT, MPP8 KD and TASOR KD HeLa cells.
g, h. Box plots of H3K9me3 eSPAN bias at HO L1s in WT, MPP8 KD and TASOR KD HeLa cells. HO L1s (n = 11,268) were grouped based on the effects of MPP8/TASOR KD on H3K9me3 density defined in a (g), or based on L1 length, as defined in e (h).
Box plots (g, h) show the median, 25% and 75% quartiles and minimal and maximal values with p values by two-sided Mann–Whitney–Wilcoxon tests, with Bonferroni correction for multiple comparisons. Each panel is a representative of at least two independent experiments. See Materials and Methods for more details.
For gel source data, see Supplementary Figure 1.
Extended Data Figure 8. The HUSH complex is enriched at the leading strands of DNA replication forks.
a. Detection of the HUSH complex subunits at replication forks based on published iPOND (isolation of proteins on nascent DNA)37 and NCC (nascent chromatin capture)36 datasets. Numbers of peptides identified were shown. N.D., not detected.
b. Heatmaps of normalized eSPAN density of H3K9me3, MPP8, TASOR and Flag-TASOR at HO L1s, sorted by L1 length. The relative position of a full-length L1 was shown in blue.
c. Average MPP8 eSPAN bias around all 1,928 replication origins in mES cells.
d-g. Correlations between the biases of TASOR eSPAN and H3K9me3 eSPAN (d, e) or the biases between MPP8 eSPAN and H3K9me3 eSPAN (f, g) in mES cells. Each dot represents a 1 kb bin (d, f) or a HO L1 (e, g) within the 1,928 initiation zones (−100 kb, 100 kb). Spearman’s rank correlation coefficient was shown. p < 2.2e-16.
h. Average MPP8 and TASOR eSPAN bias around all 2,809 replication origins in HeLa cells.
i. A snapshot of H3K9me3 ChIP-seq and calculated eSPAN biases of H3K9me3, MPP8 and TASOR in HeLa cells. OK-seq bias was shown to mark origin location.
j-m. Correlations between the biases of MPP8 eSPAN and H3K9me3 eSPAN (j, k) or between TASOR and H3K9me3 (l, m) in HeLa cells. Each dot represents a 1 kb bin (j, l) or a HO L1(k, m) within the 2,809 initiation zones in HeLa cells (−100 kb, 100 kb). Spearman’s rank correlation coefficient was shown. p < 2.2e-16.
Extended Data Figure 9. Effects of POLE3 or POLE4 deletion/depletion on H3K9me3 density and H3K9me3 eSPAN bias in mES and HeLa cells.
a. Immunoblots of POLE3 and POLE4 to confirm their deletion in mES and depletion in HeLa cells. Note that cloned ES cells (KO) and pooled HeLa cells (KD) were used for analysis and that H3K9me3 levels remained largely unaffected in the mutant cells. n = 3.
b, c. H3K9me3 CUT&Tag (b) or CUT&RUN (c) signals at HO L1s in WT, POLE3 KO and POLE4 KO mES cells. Heatmaps (left) were sorted by the average H3K9me3 signals of each row in WT cells, with average density shown at the bottom. Note that very little changes of H3K9me3 levels were observed in the mutants.
d. H3K9me3 CUT&Tag signals at HO L1s in WT, POLE3 KD and POLE4 KD HeLa cells, with the heatmaps (top, sorted as in b) and average density at HO L1s (bottom) shown. HO L1s were grouped as long and short, as defined in Extended Data Figure 7e.
e, f. H3K9me3 CUT&Tag signals at HO L1s in WT, POLE3 KO (e) and POLE4 KO (f) mES cells. Heatmaps (left, sorted as in b) and average density (right) were shown. HO L1s were separated into two groups based on the effects POLE3 or POLE4 KO on H3K9me3 levels at HO L1s, with a reduction of more than 1.5-fold defined as the Down group and the rest of L1s within this cutoff being grouped as No-diff group. Note that less than 50 (~0.4%) HO L1s showed a marked reduction of H3K9me3 density and therefore the eSPAN bias was not calculated at this group separately.
g. H3K9me3 eSPAN bias around replication origins (top) and at HO L1s (bottom, n = 11,268) in WT, POLE3 KD and POLE4 KD HeLa cells. Long and short HO L1 elements were defined as in Extended Data Figure 7.
Box plots show (g) the median, 25% and 75% quartiles and minimal and maximal values with p values by two-sided Mann–Whitney–Wilcoxon tests, and Bonferroni correction for multiple comparisons. Each panel is a representative of at least two independent experiments. See Materials and Methods for more details.
For gel source data, see Supplementary Figure 1.
Extended Data Figure 10. Pol ε coordinates with the HUSH complex for asymmetric H3K9me3 distribution.
a-d. A correlation of the reduction of H3K9me3 eSPAN bias between MPP8 KO (a, b) or TASOR KO (c, d) and POLE3 KO, with each mutant compared to WT mES cells. Each dot represents a 1 kb bin (a, c) or a HO L1(b, d) within the 1,928 initiation zones. Spearman’s rank correlation coefficient was shown. p < 2.2e-16.
e-h. Correlation of the reduction of H3K9me3 eSPAN bias between MPP8 KO (e, f) or TASOR KO (g, h) and POLE4 KO compared to WT mES cells. Each dot represents a 1 kb bin (e, g) or a HO L1(f, h) within the 1,928 initiation zones. Spearman’s rank correlation coefficient was shown. p < 2.2e-16.
i. H3K9me3 eSPAN bias around replication origins (top) and at HO L1s (bottom, n = 12,679) in WT, POLE3 KO, MPP8 KO and MPP8 KO/POLE3 KO double mutant mES cells.
j. Alignment of the protein sequences surrounding an unstructured region of TASOR in 8 different species. M.m., Mus musculus, R.n., Rattus norvegicus, H.s., Homo sapiens, P.t., Pan troglodytes, C.f., Canis familiaris, B.t., Bos taurus, G.g., Gallus gallus, X.l., Xenopus laevis. A predicted alpha helix was indicated and conservancy scores were shown at the bottom. Note that a reported domain that is responsible for the binding of Periphilin-, another HUSH subunit, is in the region.
k. Amino acid sequences of the TASOR mutations generated to analyze their effects of mutations on the TASOR-Pol ε binding. The mutated or deleted amino acids were highlighted in red.
l, m. TASOR M1/M2 (l) and M3 (m) mutations compromised TASOR interaction with Pol ε subunits, but not MPP8 or PPHLN1. TASOR KO or IgG was used as a negative control. * indicates bands from IgG light or heavy chains. Note that M3 mutation, which contains 18-amino acid deletion in TASOR, caused a major shift of the TASOR band on the gel. n = 3.
n. cDNA products of a TASOR fragment amplified from WT or TASOR mutant mES cells for expression TASOR fragments used in the GST pull down assays in Figure 4d.
o. Average H3K9me3 eSPAN bias around all 1,928 replication origins in WT or POLE4 KO mES cells treated with triptolide (0.5 μM for 45 min). DMSO was added as a control.
p. Average H3K27m3 eSPAN bias around all 1,928 replication origins in MCM2–2A mutant mES cells treated with triptolide (0.5 μM for 45 min). DMSO was added as a control. Note that H3K27me3 eSPAN bias towards the leading strand in MCM2-2A cells is much bigger than that in WT cells, due to the defective transfer of parental histones to the lagging strand, as previously reported20,23.
q. H3K9me3 eSPAN bias at HO L1s (n = 12,679) in WT or POLE4 KO mES cells treated with triptolide. Note that while triptolide didn’t affect overall H3K9me3 bias around origins, H3K9me3 bias at HO L1s at replicating origins was reduced, suggesting that triptolide affects asymmetric H3K9me3 distribution at selective genomic loci.
Box plots (i, q) show the median, 25% and 75% quartiles and minimal and maximal values with p values by two-sided Mann–Whitney–Wilcoxon tests, and Bonferroni correction for multiple comparisons. Each panel is a representative of at least two independent experiments. See Materials and Methods for more details.
For gel source data, see Supplementary Figure 1.
Extended Data Figure 11. Linking asymmetric H3K9me3 segregation at HO Ls to their silencing during S phase.
a. Isolation of mES cells at G1, S and G2 phases based on the expression of Cdt1-mKO2 and Geminin-mAG1 by flow cytometry. Cells were sorted based on the expression of these two cell cycle indicators. G1 phase cells only express Cdt1, but not Geminin. S phase cells express medium levels of Geminin, but not Cdt1, and G2 phase cells express the highest levels of Geminin. To increase the purity of S and G2 phase cells, we used a stringent gating strategy as shown with isolated G1, S and G2 phase of cells accounting for ~10%, ~15% and ~10% of total cells, respectively.
b. Heatmaps of differentially expressed L1 elements in MPP8 KO, POLE3 KO and POLE4 KO versus WT mES cells based on GRO-seq analysis. Numbers of total differentially expressed L1s in each mutant were shown. Please note that all L1 elements were used in the analysis.
c. Relative expression of representative repetitive elements in MPP8 KO, MPP8 W80A or TASOR M3 mutant cells compared to WT mES cells by RT-qPCR. The expression was normalized against WT. Data were plotted as mean ± SEM. n = 5-9.
d. Snapshots of H3K9me3 signals (both ChIP-seq and CUT&Tag), H3K9me3 eSPAN bias and GRO-seq signals at three L1 elements in WT or mutant mES cells. The three up-regulated L1s were highlighted.
e. Relative expression of representative repetitive elements in POLE3 KD and POLE4 KD cells compared to WT HeLa cells detected by RT-qPCR. The expression was normalized against WT. Data were plotted as mean ± SEM. n = 3.
f. The expression of HO L1s (n = 2,662) in WT, POLE3 KO and POLE4 KO mES cells detected by GRO-seq after excluding the ones located within the transcribed regions of up-regulated genes in POLE3 KO or POLE4 KO mES cells defined by RNA-seq.
g. The expression of HO L1s (n = 2,309) in WT, POLE3 KO and POLE4 KO mES cells detected by GRO-seq after excluding the ones located within any actively transcribed genes from analysis (cutoff: TPM > 0.5).
h. The expression of full-length (≥ 6 kb) HO L1s (n = 703) with their own promoters in WT, POLE3 KO and POLE4 KO mES cells. See Materials and Methods section for more details for panels f-h.
c, e, Two-sided Student’s t test. ****, p < 0.0001. ***, p < 0.001. **, p < 0.01. *, p < 0.05. Box plots (f-h) show the median, 25% and 75% quartiles and minimal and maximal values with p values by two-sided Mann–Whitney–Wilcoxon tests, and Bonferroni correction for multiple comparisons. Each panel is a representative of at least two independent experiments. See Materials and Methods for more details.
Extended Data Figure 12. Effects of POLE3 KO, POLE4 KO and TASOR/MPP8 mutants on L1 expression and retrotransposition.
a. The expression of HO L1s (n = 2,681) in WT, MPP8 KO and MPP8 KO/POLE3 KO double mutant mES cells detected by GRO-seq.
b. Overlaps between the up-regulated HO L1s in MPP8 KO and POLE3 KO (top) or between MPP8 KO and POLE4 KO (bottom) mES cells detected by GRO-seq. P values by hypergeometric test.
c-e. Comparison of properties (L1 length, TASOR density, and H3K9me3 eSPAN bias) of HO L1s whose expression is up-regulated in MPP8 KO (c), POLE3 KO (d) or POLE4 KO (e) mES cells to those HO L1s without changes in expression in the corresponding mutants. L1s with more than 1.5-fold increase in expression were grouped as Up and those within the 1.5-fold threshold were grouped as No-diff.
f. Relative expression of HO L1s (n = 2,681) in MPP8 KO versus WT mES cells at G1, S or G2 phase of the cell cycle detected by GRO-seq. The dashed line indicates no changes compared to WT cells (0).
g. Snapshots of GRO-seq signals at the indicated L1 elements in G1, S and G2 phases of WT, POLE4 KO and MPP8 KO mES cells. H3K9me3 ChIP-seq signals and eSPAN biases at these two loci in both WT and mutant cells were also shown.
h. Relative L1 mobility in WT, POLE3 KD and POLE4 KD HeLa cells as measured by dual-luciferase reporter assays. Data were plotted as mean ± SEM. n = 8.
i. The H3K9me3 eSPAN bias correlates with L1 integration at the leading strands. Absolute values of H3K9me3 eSPAN bias were separated into eleven equal intervals from 0 to 1 (X axis). The fraction of insertions where (+) strand of L1 cDNA integrated into the predominant leading strand template (Y axis) was plotted at each of the matching H3K9me3 bias interval.
j. Overlaid violin plots of H3K9me3 eSPAN bias frequency distributions for L1 integrations into the reference genome. Observed L1 insertions in HeLa cells were stratified by the integration strand. The colored lines identify L1 integration into the Watson (orange) and Crick (green) strands of human genome, which means that L1 endonuclease cleaved the opposite strands, i.e., the Crick and Watson strands, respectively. All violin plots were adjusted to have the same total area and vertical lines denote the distribution medians.
k. Relative γ-H2AX signal intensity measured by immunofluorescence in WT (n = 551), POLE3 KD (n = 286) and POLE4 KD (n = 362) HeLa cells. WT cells were treated with 1 mM hydroxyurea (HU, n = 361) for 1 h as a positive control. Data were plotted as mean ± SD.
Box plots show the median, 25% and 75% quartiles and minimal and maximal values. a, c-f, k, p values by two-sided Mann–Whitney–Wilcoxon tests, and Bonferroni correction for multiple comparisons. h, Two-sided Student’s t test. ****, p < 0.0001. **, p < 0.01. Each panel is a representative of at least two independent experiments. See Materials and Methods for more details.
Supplementary Material
Acknowledgements
We greatly appreciate Dr. Songtao Jia of Columbia University (New York, NY) for general discussions and suggestions, Drs. Alessandro Gardini and Connor Hill of the Wistar Institute (Philadelphia, PA) for GRO-seq, Drs. Diane A. Flasch of St. Jude Children’s Research Hospital (Memphis, TN), Thomas E. Wilson and John V. Moran of University of Michigan (Ann Arbor, MI) for assistance in the L1 integration analysis, Dr. Sandra Richardson of University of Queensland (Queensland, Australia) for analysis of L1 age, and Drs. Xiang Feng of Van Andel Institute (Grand Rapids, MI) and Dheva Setiaputra of Lunenfeld-Tanenbaum Research Institute (Ontario, Canada) for assistance with protein structural analysis. The authors also thank Dr. Rebecca Burgess of UT Southwestern Medical Center (Dallas, TX) for proofreading and editing the manuscript. All graphic illustrations were created with BioRender.com.
Footnotes
Competing interests
The authors declare no conflicts of interests.
Additional information
Supplementary material is available in the online version.
Data availability
All raw and processed sequencing data generated in this study have been deposited in GEO under the accession number GSE211192. All other data needed to evaluate the conclusions in this study are available in the Article and its Supplementary Information. The following public databases were used in this study (see Materials and Methods and Supplementary Table 1 for more details): the GENCODE database (https://www.gencodegenes.org/, mm10, GENCODE release M27, and hg19, GENCODE release v19), the ENCODE database (https://www.encodeproject.org/, datasets ENCSR857MYS, ENCSR059MBO, ENCSR000AQO, and ENCSR000APB), the GEO database (https://www.ncbi.nlm.nih.gov-/geo/, datasets GSE211192, GSE100168, GSE199040, GSE95374, GSE208748, GSE113592, GSE198978, GSE63116, GSE155693, GSE202066, GSE82144, GSE137764, GSE142996, GSE99741, GSE126477, GSE116319, and SRP065949), the UCSC Genome Browser database (https://genome.ucsc.edu/cgi-bin/hgTables), and the L1base 2 database (https://l1base.charite.de/l1base.php). Source data are provided with the paper. All other data and materials are available from the corresponding author (Z. Z.) upon reasonable request.
References
- 1.Burns KH Repetitive DNA in disease. Science 376, 353–354 (2022). [DOI] [PubMed] [Google Scholar]
- 2.Kazazian HH Jr. & Moran JV Mobile DNA in Health and Disease. N Engl J Med 377, 361–370 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gorbunova V et al. The role of retrotransposable elements in ageing and age-associated diseases. Nature 596, 43–53 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Padeken J, Methot SP & Gasser SM Establishment of H3K9-methylated heterochromatin and its functions in tissue differentiation and maintenance. Nat Rev Mol Cell Biol (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grewal SI & Jia S Heterochromatin revisited. Nat Rev Genet 8, 35–46 (2007). [DOI] [PubMed] [Google Scholar]
- 6.Grewal SI & Moazed D Heterochromatin and epigenetic control of gene expression. Science 301, 798–802 (2003). [DOI] [PubMed] [Google Scholar]
- 7.Charlesworth B, Sniegowski P & Stephan W The evolutionary dynamics of repetitive DNA in eukaryotes. Nature 371, 215–220 (1994). [DOI] [PubMed] [Google Scholar]
- 8.Lander ES et al. Initial sequencing and analysis of the human genome. Nature 409, 860–921 (2001). [DOI] [PubMed] [Google Scholar]
- 9.Consortium EP An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fueyo R, Judd J, Feschotte C & Wysocka J Roles of transposable elements in the regulation of mammalian transcription. Nat Rev Mol Cell Biol 23, 481–497 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chuong EB, Elde NC & Feschotte C Regulatory activities of transposable elements: from conflicts to benefits. Nat Rev Genet 18, 71–86 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Slotkin RK & Martienssen R Transposable elements and the epigenetic regulation of the genome. Nat Rev Genet 8, 272–285 (2007). [DOI] [PubMed] [Google Scholar]
- 13.Burns KH Transposable elements in cancer. Nat Rev Cancer 17, 415–424 (2017). [DOI] [PubMed] [Google Scholar]
- 14.Gu Z et al. Silencing of LINE-1 retrotransposons is a selective dependency of myeloid leukemia. Nat Genet 53, 672–682 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Griffin GK et al. Epigenetic silencing by SETDB1 suppresses tumour intrinsic immunogenicity. Nature 595, 309–314 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang SM et al. KDM5B promotes immune evasion by recruiting SETDB1 to silence retroelements. Nature 598, 682–687 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen R, Ishak CA & De Carvalho DD Endogenous Retroelements and the Viral Mimicry Response in Cancer Therapy and Cellular Homeostasis. Cancer Discov 11, 2707–2725 (2021). [DOI] [PubMed] [Google Scholar]
- 18.Escobar TM, Loyola A & Reinberg D Parental nucleosome segregation and the inheritance of cellular identity. Nat Rev Genet 22, 379–392 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ragunathan K, Jih G & Moazed D Epigenetic inheritance uncoupled from sequence-specific recruitment. Science 348, 1258699 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Petryk N et al. MCM2 promotes symmetric inheritance of modified histones during DNA replication. Science 361, 1389–1392 (2018). [DOI] [PubMed] [Google Scholar]
- 21.Li Z et al. DNA polymerase alpha interacts with H3-H4 and facilitates the transfer of parental histones to lagging strands. Sci Adv 6, eabb5820 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu C et al. A mechanism for preventing asymmetric histone segregation onto replicating DNA strands. Science 361, 1386–1389 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gan H et al. The Mcm2-Ctf4-Polalpha Axis Facilitates Parental Histone H3-H4 Transfer to Lagging Strands. Mol Cell 72, 140–151 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu CH et al. Strand-Specific Analysis Shows Protein Binding at Replication Forks and PCNA Unloading from Lagging Strands when Forks Stall. Mol Cell 56, 551–563 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Petryk N et al. Replication landscape of the human genome. Nat Commun 7, 10208 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fu YV et al. Selective bypass of a lagging strand roadblock by the eukaryotic replicative DNA helicase. Cell 146, 931–941 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tubbs A et al. Dual Roles of Poly(dA:dT) Tracts in Replication Initiation and Fork Collapse. Cell 174, 1127–1142 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sakaue-Sawano A et al. Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell 132, 487–498 (2008). [DOI] [PubMed] [Google Scholar]
- 29.Tchasovnikarova IA et al. GENE SILENCING. Epigenetic silencing by the HUSH complex mediates position-effect variegation in human cells. Science 348, 1481–1485 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu N et al. Selective silencing of euchromatic L1s revealed by genome-wide screens for L1 regulators. Nature 553, 228–232 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Robbez-Masson L et al. The HUSH complex cooperates with TRIM28 to repress young retrotransposons and new genes. Genome Res 28, 836–845 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Spencley AL et al. Co-transcriptional genome surveillance by HUSH is coupled to termination machinery. Mol Cell 83, 1623–1639 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karimi MM et al. DNA methylation and SETDB1/H3K9me3 regulate predominantly distinct sets of genes, retroelements, and chimeric transcripts in mESCs. Cell Stem Cell 8, 676–687 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matsui T et al. Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature 464, 927–931 (2010). [DOI] [PubMed] [Google Scholar]
- 35.Douse CH et al. TASOR is a pseudo-PARP that directs HUSH complex assembly and epigenetic transposon control. Nat Commun 11, 4940 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alabert C et al. Nascent chromatin capture proteomics determines chromatin dynamics during DNA replication and identifies unknown fork components. Nat Cell Biol 16, 281–293 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cheng L et al. Chromatin Assembly Factor 1 (CAF-1) facilitates the establishment of facultative heterochromatin during pluripotency exit. Nucleic Acids Res 47, 11114–11131 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prigozhin DM et al. Periphilin self-association underpins epigenetic silencing by the HUSH complex. Nucleic Acids Res 48, 10313–10328 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seczynska M, Bloor S, Cuesta SM & Lehner PJ Genome surveillance by HUSH-mediated silencing of intronless mobile elements. Nature 601, 440–445 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barbieri E et al. Rapid and Scalable Profiling of Nascent RNA with fastGRO. Cell Rep 33, 108373 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Deininger P et al. A comprehensive approach to expression of L1 loci. Nucleic Acids Res 45, e31 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xie Y, Rosser JM, Thompson TL, Boeke JD & An W Characterization of L1 retrotransposition with high-throughput dual-luciferase assays. Nucleic Acids Res 39, e16 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Flasch DA et al. Genome-wide de novo L1 Retrotransposition Connects Endonuclease Activity with Replication. Cell 177, 837–851 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sultana T et al. The Landscape of L1 Retrotransposons in the Human Genome Is Shaped by Pre-insertion Sequence Biases and Post-insertion Selection. Mol Cell 74, 555–570 (2019). [DOI] [PubMed] [Google Scholar]
- 45.Seczynska M & Lehner PJ The sound of silence: mechanisms and implications of HUSH complex function. Trends Genet 39, 251–267 (2023). [DOI] [PubMed] [Google Scholar]
- 46.Buhler M & Moazed D Transcription and RNAi in heterochromatic gene silencing. Nat Struct Mol Biol 14, 1041–1048 (2007). [DOI] [PubMed] [Google Scholar]
- 47.Alabert C et al. Two distinct modes for propagation of histone PTMs across the cell cycle. Genes Dev 29, 585–590 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gasior SL, Wakeman TP, Xu B & Deininger PL The human LINE-1 retrotransposon creates DNA double-strand breaks. J Mol Biol 357, 1383–1393 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ardeljan D et al. Cell fitness screens reveal a conflict between LINE-1 retrotransposition and DNA replication. Nat Struct Mol Biol 27, 168–178 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mita P et al. BRCA1 and S phase DNA repair pathways restrict LINE-1 retrotransposition in human cells. Nat Struct Mol Biol 27, 179–191 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Koh SB et al. A quantitative FastFUCCI assay defines cell cycle dynamics at a single-cell level. J Cell Sci 130, 512–520 (2017). [DOI] [PubMed] [Google Scholar]
- 52.Li Z, Hua X, Serra-Cardona A, Xu X & Zhang Z Efficient and strand-specific profiling of replicating chromatin with enrichment and sequencing of protein-associated nascent DNA in mammalian cells. Nat Protoc 16, 2698–2721 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kaya-Okur HS et al. CUT&Tag for efficient epigenomic profiling of small samples and single cells. Nat Commun 10, 1930 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Skene PJ & Henikoff S An efficient targeted nuclease strategy for high-resolution mapping of DNA binding sites. Elife 6, e21856 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ran FA et al. Genome engineering using the CRISPR-Cas9 system. Nat Protoc 8, 2281–2308 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Baris Y, Taylor MRG, Aria V & Yeeles JTP Fast and efficient DNA replication with purified human proteins. Nature 606, 204–210 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Roulois D et al. DNA-Demethylating Agents Target Colorectal Cancer Cells by Inducing Viral Mimicry by Endogenous Transcripts. Cell 162, 961–973 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schindelin J et al. Fiji: an open-source platform for biological-image analysis. Nat Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Langmead B & Salzberg SL Fast gapped-read alignment with Bowtie 2. Nat Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Danecek P et al. Twelve years of SAMtools and BCFtools. Gigascience 10, giab008 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ramirez F et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res 44, W160–W165 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Quinlan AR & Hall IM BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sloan CA et al. ENCODE data at the ENCODE portal. Nucleic Acids Res 44, D726–D732 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li H et al. Remodeling of H3K9me3 during the pluripotent to totipotent-like state transition. Stem Cell Rep 18, 449–462 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhao N et al. Critically short telomeres derepress retrotransposons to promote genome instability in embryonic stem cells. Cell Discov 9, 45 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xu S, Grullon S, Ge K & Peng W Spatial clustering for identification of ChIP-enriched regions (SICER) to map regions of histone methylation patterns in embryonic stem cells. Methods Mol Biol 1150, 97–111 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lukasak BJ et al. TGM2-mediated histone transglutamination is dictated by steric accessibility. Proc Natl Acad Sci U S A 119, e2208672119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yashar WM et al. GoPeaks: histone modification peak calling for CUT&Tag. Genome Biol 23, 144 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Robinson MD, McCarthy DJ & Smyth GK edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Khan H, Smit A & Boissinot S Molecular evolution and tempo of amplification of human LINE-1 retrotransposons since the origin of primates. Genome Res 16, 78–87 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sookdeo A, Hepp CM, McClure MA & Boissinot S Revisiting the evolution of mouse LINE-1 in the genomic era. Mob DNA 4, 3 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li H & Durbin R Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26, 589–595 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liu Y, Wu X, d'Aubenton-Carafa Y, Thermes C & Chen CL OKseqHMM: a genome-wide replication fork directionality analysis toolkit. Nucleic Acids Res 51, e22 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhao PA, Sasaki T & Gilbert DM High-resolution Repli-Seq defines the temporal choreography of initiation, elongation and termination of replication in mammalian cells. Genome Biol 21, 76 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Xu X et al. Stable inheritance of H3.3-containing nucleosomes during mitotic cell divisions. Nat Commun 13, 2514 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Frankish A et al. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res 47, D766–D773 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Penzkofer T et al. L1Base 2: more retrotransposition-active LINE-1s, more mammalian genomes. Nucleic Acids Res 45, D68–D73 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Smith JP, Dutta AB, Sathyan KM, Guertin MJ & Sheffield NC PEPPRO: quality control and processing of nascent RNA profiling data. Genome Biol 22, 155 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dobin A et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Liao Y, Smyth GK & Shi W featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014). [DOI] [PubMed] [Google Scholar]
- 81.Wang LG, Wang SQ & Li W RSeQC: quality control of RNA-seq experiments. Bioinformatics 28, 2184–2185 (2012). [DOI] [PubMed] [Google Scholar]
- 82.McCarthy DJ, Chen Y & Smyth GK Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res 40, 4288–4297 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang Y, Parmigiani G & Johnson WE ComBat-seq: batch effect adjustment for RNA-seq count data. NAR Genom Bioinform 2, lqaa078 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Vera Alvarez R, Pongor LS, Marino-Ramirez L & Landsman D TPMCalculator: one-step software to quantify mRNA abundance of genomic features. Bioinformatics 35, 1960–1962 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw and processed sequencing data generated in this study have been deposited in GEO under the accession number GSE211192. All other data needed to evaluate the conclusions in this study are available in the Article and its Supplementary Information. The following public databases were used in this study (see Materials and Methods and Supplementary Table 1 for more details): the GENCODE database (https://www.gencodegenes.org/, mm10, GENCODE release M27, and hg19, GENCODE release v19), the ENCODE database (https://www.encodeproject.org/, datasets ENCSR857MYS, ENCSR059MBO, ENCSR000AQO, and ENCSR000APB), the GEO database (https://www.ncbi.nlm.nih.gov-/geo/, datasets GSE211192, GSE100168, GSE199040, GSE95374, GSE208748, GSE113592, GSE198978, GSE63116, GSE155693, GSE202066, GSE82144, GSE137764, GSE142996, GSE99741, GSE126477, GSE116319, and SRP065949), the UCSC Genome Browser database (https://genome.ucsc.edu/cgi-bin/hgTables), and the L1base 2 database (https://l1base.charite.de/l1base.php). Source data are provided with the paper. All other data and materials are available from the corresponding author (Z. Z.) upon reasonable request.