Abstract
Fabricating three-dimensional (3D) large-scale bone tissue constructs with functional vasculature has been a particular challenge in tissue engineering to repair large bone defects. To address this challenge, a direct-writing bioprinting strategy is utilized to fabricate microstructured bone-like tissue constructs containing a perfusable vascular lumen. The bioprinted constructs are used as biomimetic in vitro matrices to co-culture human umbilical vein endothelial cells (HUVECs) and bone marrow-derived human mesenchymal stem cells (hMSCs) in a naturally derived hydrogel. To form the perfusable blood vessel inside the bioprinted construct, we print the central cylinder with 5% GelMA hydrogel with low methacryloyl substitution (GelMALOW). We also develop cell-laden cylinder elements made of GelMA hydrogel loaded with silicate nanoplatelets to induce osteogenesis, and synthesized hydrogel formulations with chemically conjugated vascular endothelial growth factor (VEGF) to promote vascular spreading. We find that the engineered construct is able to support cell survival and proliferation during the in vitro maturation. Also, the whole construct demonstrates high structural stability during the in vitro culture for 21 days. This method enables the local control of physical and chemical micro-niches and the establishment of gradients in constructs with clinically relevant dimensions.
Keywords: 3D bioprinting, vascularized bone tissue, angiogenic hydrogels, bone-like tissue construct
1. Introduction
Tissue engineers face a daunting challenge when attempting to mimic the complex architecture of biological tissues in terms of physical and chemical characteristics and including their multiscale vasculature [1, 2]. Bone tissue is a particular challenge due to its complex architecture, consisting of organized calcified regions with interpenetrated vasculature that allows the access of nutrients and oxygen to the cells [3]. The incorporation of vasculature is a requirement when engineering bone tissues in order to provide nutrients and factors to avoid tissue necrosis [4]. This is particularly important for large constructs intended for use in the treatment of large bone defects. In this case, the construct should contain vessels with comparable size to host vessels to facilitate their anastomosis and to restore blood supply to the engineered tissue upon its implantation. Therefore, the creation of an organized pattern of constructs containing two separate osteogenic and vasculogenic niches is key to a successful strategy of bone tissue engineering [5]. Creating an intrinsic gradient of biological factors within the engineered constructs can further enhance the formation of microcapillaries [6]. Overall, two strategies have been followed for creating vascularized tissues: 1) engineering of capillaries that can be lined with endothelial cells and 2) formation of vasculatures through biological and self-assembly processes [1]. The first approach is desirable for creating a major vessel, while the latter one is required for the formation of microcapillaries once the larger vessels have been formed [7]. Ideally, these two strategies should be combined to create multi-scale vasculatures within the engineered constructs.
Cell-based approaches for formation of vascular networks from endothelial cells have been investigated [8, 9]. Among these strategies, the use of blood-derived human umbilical vein endothelial cells (HUVECs) and bone marrow-derived human mesenchymal stem cells (hMSCs) is particularly appealing because of the multipotency and extensive in vitro expandability displayed by these cells [10]. Co-culture of HUVECs and hMSCs resulted in the development of a stable and robust vasculature network in a murine model of human cell transplantation. Within the first week after transplantation, the microvascular networks completed its formation, and remained functional and stable; thereafter, the HUVECs formed lumens and the hMSCs surrounded them and differentiated into perivascular cells to stabilize the vessels [10].
The key to controlling this differentiation of HUVECs and hMSCs is the presence of an osteoblastic niche, which exists at the endosteum-stroma interface in the bone. Therefore, creating well-defined patterns of osteogenic and vasculogenic niches is critical for engineering functional bone tissues. Several methods, including stereolithography, bioprinting, textile techniques, and molding, have been used for the microfabrication of three-dimensional (3D) tissue constructs [9, 11]. Among these techniques, 3D bioprinting has emerged as a promising platform capable of engineering finely tuned 3D constructs [12]. In 3D printing, a bioink is extruded or injected through a nozzle and the deposition point is controlled by movement of the substrate or the nozzle [13]. Among the different bioprinting platforms, direct-write printers that generate fibers and assemble them into constructs have been widely used in tissue engineering [14, 15]. The fibers are usually crosslinked at the nozzle outlet or immediately upon extrusion. Among various polymers, hydrogels have proven excellent candidates for printing cell-laden constructs [16].
This 3D bioprinting technology has been widely used for engineering bone tissues. For instance, an extrusion-based system was used to print poly(lactic acid) (PLA) fibers into 3D constructs with embedded vascular-like channels [17]. The resulting scaffolds were then coated with nano-hydroxyapatite (nHA) to enhance osteo-differentiation of the hMSCs. However, the cells were unable to fill the pores of the scaffolds, resulting in a large void volume. Another study printed tricalcium phosphate (TCP) scaffold doped with silicon dioxide (SiO2) and magnesium oxide (MgO) into 3D construct [18]. The incorporation of Mg and Si improved the in vivo bone regeneration; however, the problem of high porosity persisted and the lack of a soft niche to support neo-angiogenesis was a potential limitation to the integration of the engineered tissue. Another example was a 3D printed bioceramic construct containing macropores filled with chitosan and RGD (Arg-Gly-Asp)-expressing phages, which adhered to the construct through electrostatic interactions to form a vasculogenic niche [19]. These constructs were implanted in an animal model and native cells invaded the construct to form vasculature. Unfortunately, the rate of vascularization was slow and this approach was unable to generate sufficient vascular networks to support cell survival within the cell-laden construct. Moreover, a recent study demonstrated that the extrusion bioprinting technique can be adopted to fabricate drug delivery devices that could sequentially release of compounds such as drugs and growth factors in a well-controlled manner, which would enable potential applications in various biomedical applications [20].
In the present study, we demonstrate a 3D bioprinting approach that enables the fabrication of complex bone-like 3D architectures with different physiological niches. We address the key challenge currently limiting the bioprinting of multi-niche constructs that can be interfaced with each other with minimal lamination between different niches by utilizing a naturally derived gelatin-based hydrogel, gelatin methacryloyl (GelMA) [14, 21, 22]. GelMA, a photocrosslinkable form of gelatin, has emerged as a promising biomaterial for engineering bone, blood vessels, muscle, and cardiac tissues [23]. GelMA hydrogels have tunable physical properties that can be tailored to match the targeted application. In addition, the methacryloyl groups form covalent bonds at the interface of two hydrogels to minimize the possibility of lamination and to promote the formation of a unified construct upon fabrication.
The development of vasculature, particularly microvasculature, has been a focus of research in recent years. Many authors have successfully demonstrated the use of different strategies to successfully develop vascular structures [24, 25, 26]. Here, we present an alternative method that could be useful to fabricate larger vascular conduits in ticker cell-laden 3D bone tissue constructs. We aim to develop VEGF functionalized bioactive GelMA bioink to prove the concept of fabricating vascularized bone tissue. We printed this VEGF functionalized GelMA bioink with different methacrylation degrees in one construct and we demonstrated that the use of the GelMA bioink allows the engineering of two different tissues: bone and vascular tissues in one construct due to its tunable mechanical properties through simple direct bioprinting technology. Comparing to the conventional technologies such as thermoresponsive hydrogels [15, 24] or sacrificial templates [26], our bioprinting strategy can fabricate blood vessel through one-step bioprinting without requiring additional treatments such as changing temperature or removing sacrificial materials.
In this study, we first optimize two different types of GelMA hydrogel bioinks to engineer the vasculogenic and osteogenic niches. The central fiber of the construct is printed using a rapidly degradable GelMA hydrogel, which forms a perfusable blood vessel within the bioprinted construct. Around this central vascular fiber, we load silicate nanoplatelets to induce osteogenesis. Moreover, to promote vascular spreading, we introduce chemically conjugated vascular endothelial growth factor (VEGF) with gradient concentrations in the surrounding bone niches. Through optimization of bioprinting conditions, we are able to bioprint well-defined, cell-laden bone tissue constructs with large volumes. Arguably, this strategy is superior to the current technologies available for engineering bone-like construct with vascular channels, suggesting its suitability for the fabrication of large-scale vascularized bone tissue constructs and its clinical application for bone regeneration and bone fracture repair.
2. Results and Discussion
2.1. Engineering a vascularized bone tissue construct
We used a simple bioprinting strategy to mimic the overall architecture of a bone tissue (Figure 1A). To that end, we individually printed GelMA hydrogel cylinders (Figure 1B) using a commercial 3D NovoGen MMX bioprinter. Compared to other bioprinting techniques, this extrusion-based bioprinting system allows rapid deposition of multiple cell-laden materials in a layer-by-layer manner, and also permits the incorporation of growth factors within the bioinks during direct printing. Although the shear stress originated from the extrusion step might lead to decreased cell viability, this side effect could be avoided through careful optimization of bioprinting conditions [27]. However, one of its limitations is the relatively low printing throughput and resolution as compared to other currently available extrusion bioprinting methods. Here, we show that the use of this simple extrusion bioprinter renders great flexibility for tissue engineering of complex constructs. We print individual rods with particular chemical and physical characteristics and then we integrate them to build large and complex architectures with specific micro-niches in a simple fashion. These cylinders were piled up to form pyramidal constructs composed of 28 rods (Figure 1C). The cross-section of the bioprinted construct is presented in Figure 1D. Since the rods are printed individually, the composition of each of them can be customized. This enables a precise control of the spatial composition of the whole construct within the resolution of a single rod and provides a great deal of flexibility for a wide spectrum of tissue engineering applications. For instance, the establishment of any chemical or biological gradients in a large hydrogel construct is possible by printing cylinders with different concentrations and arranging them in a predefined order (i.e., progressively higher or lower concentration from the center to the surface; Figure 1E). As shown in Figure 1E, we printed a chemical gradient using a fluorescent dye-labeled GelMA within a construct. Sulforhodamine 101 (Texas Red) cadaverine, a fluorescent dye with Ex/Em at 480/575 nm, was chemically conjugated onto -COOH modified GelMA bioprinted rods. The dye concentration was evaluated by measuring the fluorescence intensity along an arbitrary straight line in the construct using image analysis techniques (Image J; open source program). Mixing the GelMA-dye at different ratios created a fluorescent gradient, which is indicated by the gray value change along the yellow line in Figure 1E. This demonstration provides direct evidence to support the simplicity, effectiveness, and flexibility of this strategy.
Figure 1.
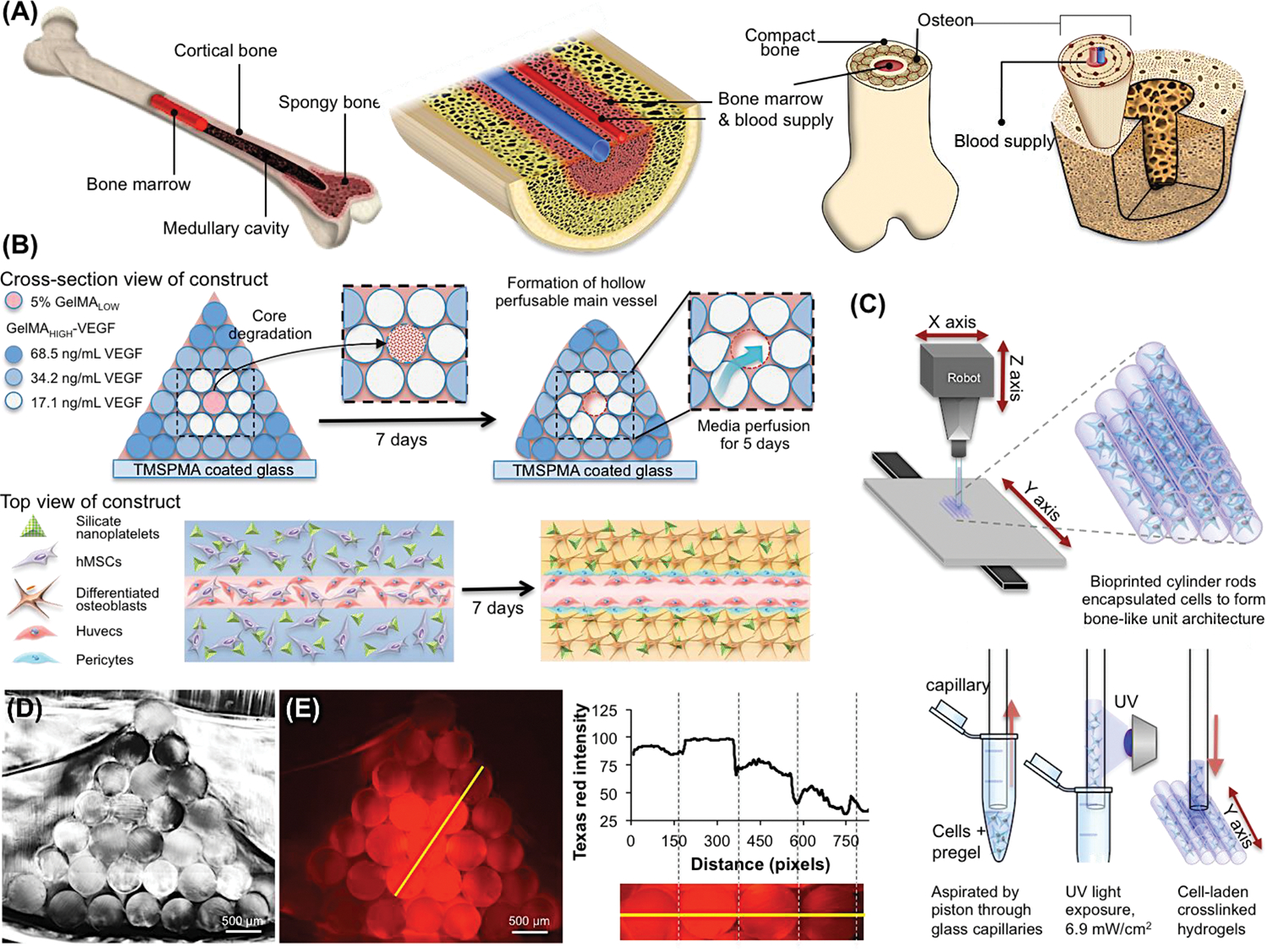
A pyramidal shaped complex large-scaled bone construct architectures fabricated by 3D bioprinted GelMA hydrogel rods with diverse compositions. (A) Schematic illustration of complex bone tissue structure. (B) Illustration of the bioprinting strategy for fabricating complex bone tissue architecture. A perfusable vascular lumen lined with HUVECs can be fabricated within a pyramidal bioprinted construct by arranging individual rods of VEGF-functionalized GelMA bioinks with different mechanical strengths. The hMSCs-laden three outer layers of cylinders were loaded with silicate nanoparticles to induce osteogenic differentiation of hMSCs into bone tissue. The VEGF was covalently conjugated into the three outer layers of the cylindrical hydrogels. The concentrations of conjugated VEGF were determined with ELISA as 17.1, 34.2, and 68.5 ng/ml. (C) Scheme of the 3D printing procedure of independent cell-laden cylinders using an automatized and computer-controlled bioprinter. (D) Cross-section image of the pyramidal bioprinted construct. (E) Chemical conjugation of a gradient sulforhodamine 101 (Texas Red) cadaverine onto –COOH modified GelMA bioprinted fibers. The fluorescence intensity was directly proportional to the conjugated amount of the fluorescent dye.
Next, we explored the fabrication of vascularized bone tissue inspired constructs as a proof-of-concept model. We printed rods with four different compositions (in terms of GelMA mass fraction, cell-laden composition, silicate nanoplatelets concentration, and VEGF-content) to fabricate bone tissue constructs with a specific and predefined architecture (Figure 1A). The central rod of the construct was printed using cell-laden 5% VEGF-conjugated GelMA with low methacryloyl substitution (GelMALOW-VEGF) containing HUVECs and hMSCs. As depicted in Figure 1A, three successive layers of cylinders were printed around this soft core using 10% (w/v) GelMAHIGH loaded with silicate nanoplatelets to induce osteogenic differentiation of hMSCs into osteoblasts, and also containing three different concentrations of covalently conjugated VEGF (68.5, 34.2, 17.1 ng/ml). The softness of the inner core, which was prepared with 5% (w/v) GelMALOW, allowed a degradation of the hydrogel, leaving an open lumen and a perfusable channel of 500 μm after 12 days of in vitro incubation. The HUVECs and hMSCs loaded into the internal cylinder could migrate to and proliferate on the inner surface of this channel. The perfusable channel then functioned as a central blood vessel in a cell-laden construct. The presence of silicate nanoplatelets in the outer layers of the construct and the sustained perfusion of osteogenic medium induced the differentiation of the encapsulated hMSCs into bone tissue. The VEGF gradient also promoted different chemical microenvironments, which can affect the osteogenic differentiation and/or vascularization.
2.2. Formation and physical characterization of VEGF-conjugated GelMA hydrogel
The VEGF growth factor was immobilized and a concentration gradient created within the bioprinted construct by conjugating VEGF to GelMA via EDC/NHS coupling chemistry, following a previously reported procedure [28] with some modifications. Possible side reactions due to the remaining lysine residues in the GelMA were avoided by first reacting the GelMA with an excess amount of succinic anhydride to convert all the amine groups into carboxylic acid groups (Figure 2A). The results from the fluoraldehyde assay [29] indicated that the remaining primary amine groups in GelMAHIGH were almost completely consumed and converted to additional carboxylic acid groups. The 1H NMR spectra also demonstrated additional resonance peaks from the methylene protons from the succinic anhydride modification, as shown in Figure S1 in the Supporting Information. Carboxylic acid groups were then activated by reacting with EDC/NHS at room temperature, and VEGF conjugation was achieved by the formation of amide linkages. Although the conjugated VEGF is not detectable using common molecular characterization techniques such as 1H NMR and FT-IR, the amount of human VEGF conjugated onto GelMAHIGH hydrogel could be quantified using human VEGF ELISA kit [30]. Moreover, the release of VEGF from GelMA-VEGF hydrogels was compared with physically mixed GelMA/VEGF samples (GelMA+VEGF) and pure GelMA (Figure 2D). As expected, the released amount of VEGF from GelMA-VEGF was almost four times lower than that of physically mixed GelMA+VEGF at day 7 (Figure 2D). In addition, the amount of VEGF remaining in the matrix after 7 days was quantified by degrading the protein matrix with collagenase, and quantifying the amount of VEGF released to the solution. Following hydrogel degradation, the amount of released VEGF from GelMA-VEGF was higher as compared to physically mixed GelMA+VEGF, which confirmed the chemical conjugation between VEGF and the hydrogel network (Figure 2E). The ELISA-based bioactivity of conjugated VEGF was 68.5 ng/ml in the 10% hydrogel solution. The resulting GelMA-VEGF material could be mixed with unmodified GelMAHIGH at different ratios for use as the bioink to introduce a VEGF concentration gradient within the printed construct.
Figure 2.
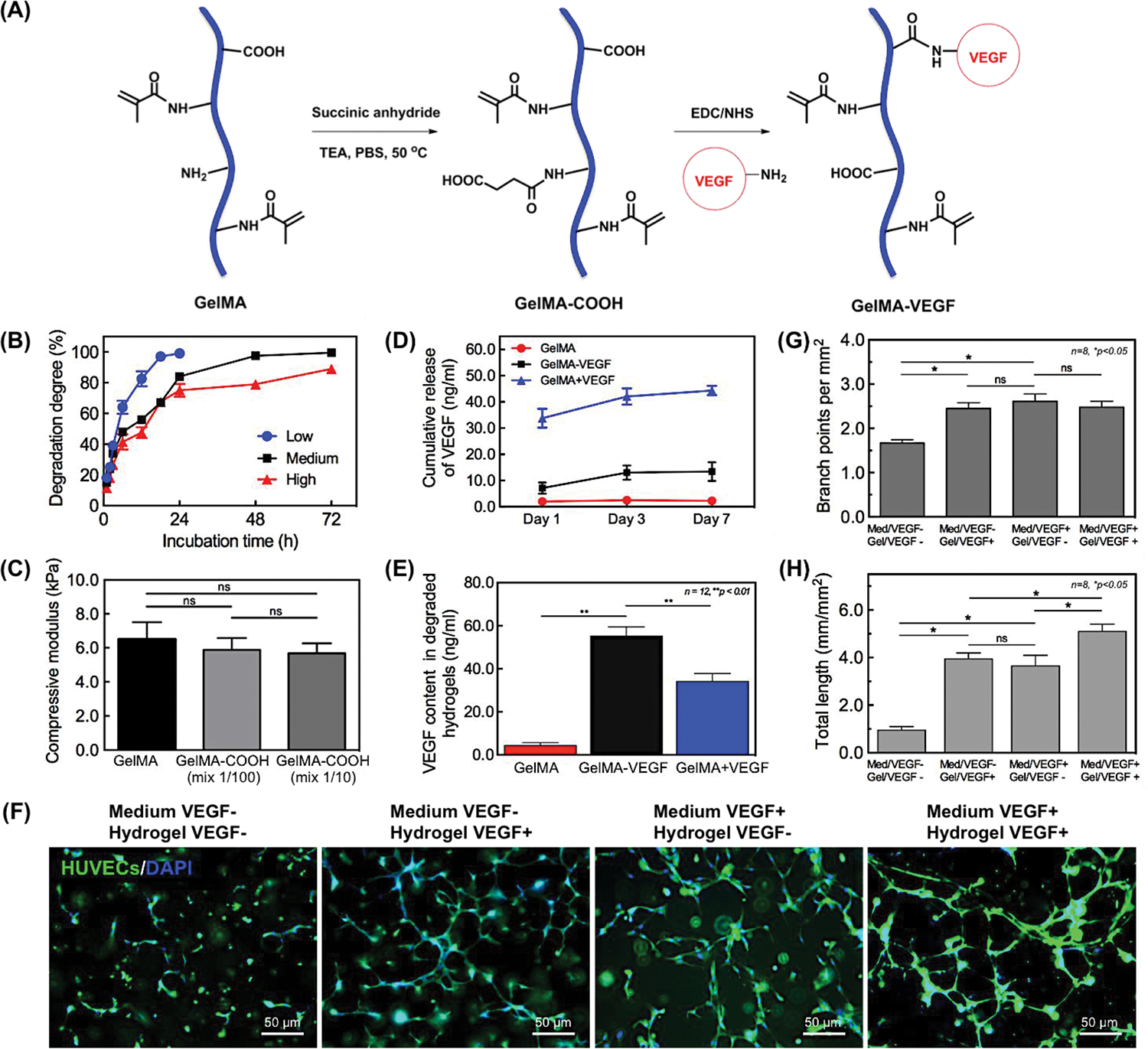
Formation and characterization of VEGF conjugated GelMA hydrogels. (A) Schematic for the preparation of –COOH modified GelMA and chemical conjugation of VEGF onto it. (B) Degradation profiles of GelMA hydrogels upon incubation with 1.0 μg/ml of collagenase at 37°C. (C) Comparison of the compressive modulus of –COOH modified GelMA hydrogel and conventional GelMA hydrogel. (D) Release profiles of VEGF from GelMA-VEGF hydrogels. GelMA hydrogels with/without physically mixed VEGF were used as positive/negative control groups. Release of bioactive VEGF from VEGF conjugated GelMA hydrogel (GelMA-VEGF), physically mixed GelMA/VEGF hydrogel (GelMA+VEGF), and GelMA hydrogel only (GelMA). (E) Quantification of VEGF remaining in different hydrogels after 7 days of release experiments using ELISA. Hydrogel samples were degraded by incubating with 1.0 μg/ml collagenase at 37°C for 72 h. Results are presented as the mean ± standard deviation (n = 12; **p < 0.01). (F) Endothelial cell sprouting inside VEGF-conjugated and non-conjugated bioprinted single GelMA hydrogels on day 5 post culture. (G, H) Quantitative analysis of capillary-like network formation after 5 days of in vitro culture, determined by measuring total capillary-like length and number of branch points per unit of area. Results are presented as the mean ± standard deviation (n = 8; *p < 0.05).
The degree of functionalization (DoF) values of the methacryloyl groups in the GelMA samples, defined as the ratio of reacted amino groups to the total amount of original amino groups, was measured by 1H NMR spectroscopy as 34.1, 42.7, and 94.3% for the GelMALOW, GelMAMEDIUM, GelMAHIGH hydrogels, respectively. The degradation kinetics of the GelMA hydrogels with different DoF values were determined in collagenase A, type I solution by measuring the hydrogel residual mass percentage versus time, as previously demonstrated (Figure 2B) [31]. The degradation rate significantly decreased with increased DoF values of GelMA; the GelMALOW hydrogel was completely degraded within 24 h, whereas the GelMAMEDUIM and GelMAHIGH hydrogels were more resistant to enzymatic degradation and took 72 h to degrade (Figure 2B).
Compression tests on the hydrogels of GelMAHIGH and GelMA-COOH revealed that the chemical modifications resulted in no significant alterations in the mechanical strength of the resulting hydrogel (Figure 2C). At the same concentration of 10% (w/v), the measured compressive modulus of GelMAHIGH hydrogels was measured as 6.5 ± 1.0 kPa; this value was not significantly higher than that of a 100:1 mixture with GelMA-COOH (5.9 ± 0.7 kPa) or a 10:1 mixture (5.7 ± 0.6 kPa). This confirmed that the VEGF conjugation had no effect on the printability of GelMA-based bioink or on the mechanical stability of the resulting construct.
2.3. Bioprintability and bioactivity of VEGF-conjugated GelMA-COOH hydrogel
GelMA prepolymers with different DoF can produce hydrogels with various physical properties. Here, we proposed the use of two degrees of functionalization of GelMA prepolymers to print the vascularized osteogenic tissue. GelMA samples with different DoFs were prepared as described previously [21]. GelMALOW was used to print the central vascular fiber, due to its more rapid degradation rate that promoted the core microvessel formation. GelMAHIGH blended with GelMA-VEGF at different ratios was applied to print the surrounding osteogenic fibers. The bioprintability of both GelMA hydrogels as a function of hydrogel concentrations and exposure time is summarized in Table S1 in the Supporting Information. The data shown in Table S1 confirm the feasibility of reproducing fibers using 3D bioprinting with 5% of GelMALOW (for inner core channel) and 10% of GelMAHIGH (outer fibers) prehydrogels with 0.2% PI for 20 s of exposure time. The ELISA results showed that the conjugated amounts of biological active VEGF onto bioprinted GelMA hydrogels were 68.5, 34.2, and 17.1 ng/ml for different locations of the 7-layered construct.
2.4. Characterization of in vitro capillary-like network formation in a VEGF-conjugated bioprinted single fiber
Previously, we have reported that GelMA hydrogels can support formation of capillary networks in vitro [31]. We first examined the biological activity of immobilized VEGF in HUVECs-laden GelMA matrix for supporting vasculogenesis in a single bioprinted fiber. We selected a 10:1 mixture of GelMALOW and GelMA-VEGF at a cell density of 2.0 × 106 cells/ml. Pure GelMA fibers with the same amount of embedded HUVECs cultured in endothelial media without VEGF were used as the negative control. The formation of capillary-like network was revealed by actin/DAPI staining and quantified by measuring the total capillary-like length and the number of branch points per unit of area (Figure 2F). Compared to the control, HUVECs embedded in VEGF-immobilized matrix showed both significantly enhanced capillary formation and increased branch points per unit area (Figure 2G and 2H). The ELISA results and the encapsulated cell culture results shown in Figure 2F–H confirmed the similar biological activity of immobilized VEGF to induce vasculogenesis in HUVEC-laden gels, as observed with stock VEGF dispersed in normal EC media. These results further demonstrated that VEGF is an important growth factor for activating VEGF receptors for regulation of P13K and FAK that play an important role in promoting HUVEC survival and vasculogenesis [32].
Co-culture studies of HUVECs and hMSCs have shown that the synergistic interactions between these endothelial cells and osteoprogenitor cells significantly improved the development of vascular networks [33]. We optimized the stimulatory conditions for enhancing growth and maturation of the vascular network by co-culturing HUVECs and hMSCs in bioprinted single fibers at different cell ratios. Two distinct cell ratios (HUVECs/hMSCs = 1/1 and 2/1) were selected, based on our previous studies [31]. The hydrogel matrix stiffness was also tuned by using GelMA-VEGF (17.1 ng/ml) with different degree of functionalization. Mono-cultured HUVECs in bioprinted single fibers were used as a control. The formation of a capillary-like structure was directly visualized by the green fluorescence of the GFP-labeled HUVECs at days 1, 5, and 7 post culture. The fluorescence microscopy images of co-cultured GFP-labeled HUVECs and hMSCs are shown in Figure 3A. The cells were stained with DAPI to evaluate total cell numbers. Total lengths of the capillary-like structures and the number of capillary branches per unit area were quantified from the images (Figure 3B and 3C). The endothelial cells in co-culture were highly spread and showed higher branch numbers when compared to the mono-culture condition. The hMSCs exhibit a vasculogenic potential and considerable synergism when co-cultured with endothelial cells. Here, the hMSCs, which generally precede the formation of osteoprogenitor cells, could also differentiate towards smooth muscle cells (also pericytes), thereby stabilizing the formed vascular structure in co-culture system with endothelial HUVECs cells in specifically optimized conditions [31, 34]. Additionally, the HUVECs/hMSCs encapsulated in GelMALOW-VEGF showed statistically significant increases in spreading as well as in branch numbers when compared to GelMAHIGH-VEGF (Figure 3B and 3C). Comparison of the different co-culture conditions and mono-culture of HUVECs revealed that the combination of the 2:1 ratio of HUVECs : hMSCs in GelMALOW fibers with lower stiffness resulted in the most profound formation of capillary-like structures by day 7. Therefore, this optimized co-culture condition was applied to fabricate further bioprinted tissue constructs.
Figure 3.
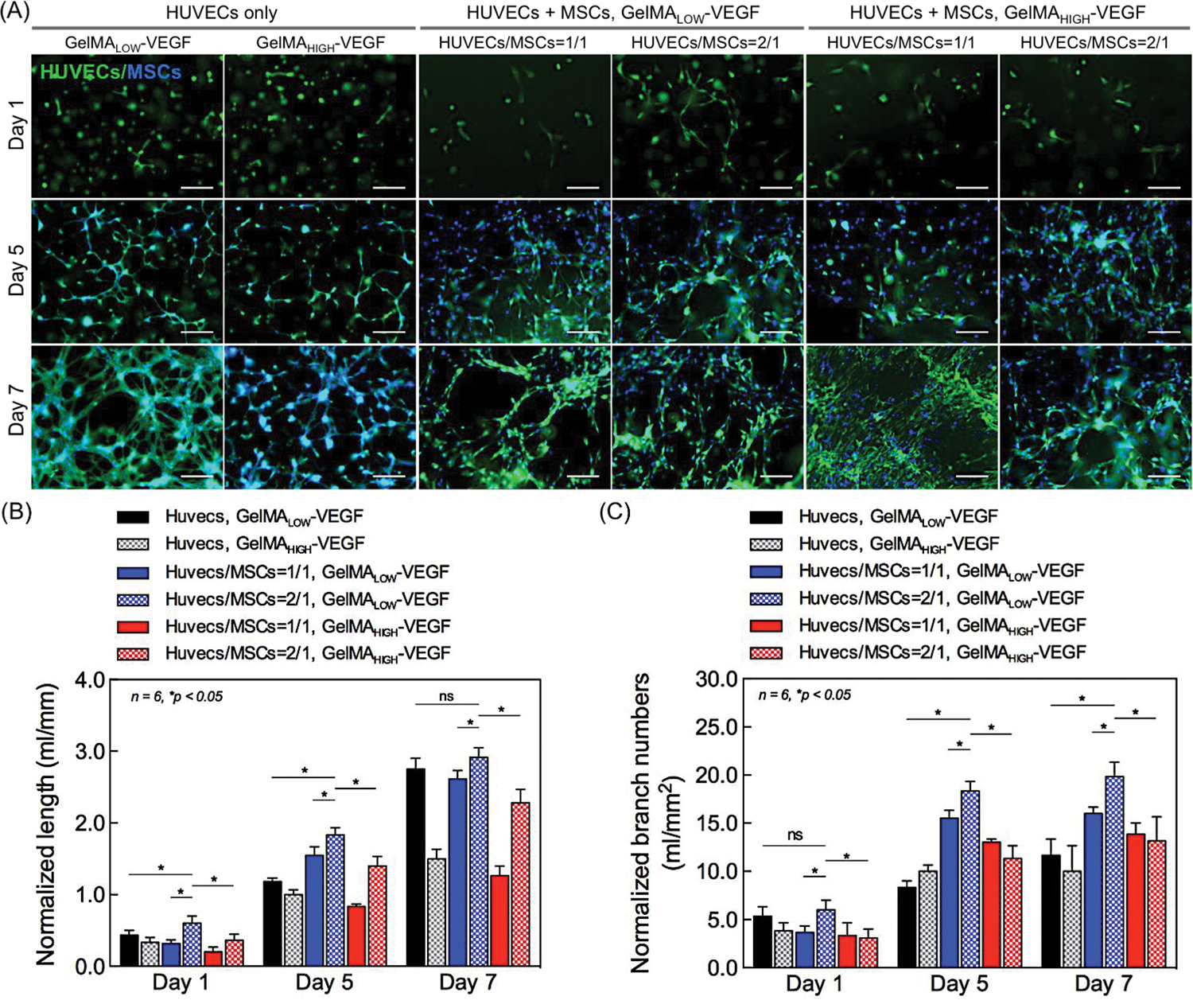
Characterizations of vasculogenic potential from in vitro co-culture in a single bioprinted fiber. (A) The formation of a capillary-like structure was promoted by co-culturing HUVECs and hMSCs in the bioprinted fibers (diameter: 500 μm). Fluorescence images of co-cultured HUVECs (GFP-labeled) and hMSCs, which were stained with DAPI to evaluate cell numbers at 1, 5, and 7 days post culture. Mono-cultured HUVECs (GFP-labeled) in GelMA fibers were used as a control. Scale bar indicates 50 μm. (B, C) Quantitative analysis of capillary-like network formation at 1, 5, and 7 days post culture, determined by measuring (B) total capillary-like length and (C) number of branch points per unit of area. Data normalized by HUVEC cells number. Results are presented as the mean ± standard deviation (n = 6; *p < 0.05).
2.5. Formation of vascular network and osteogenesis in a single GelMA fiber
We evaluated the viability and proliferation of encapsulated cells in bioprinted fibers (GelMALOW-VEGF 17.1 ng/ml). Previous studies demonstrated that the dose range of UV exposure does not compromise the cell viability [21, 31, 35]. The fluorescence microscopy image of Live/Dead staining at day 1 is shown in Figure 4C. The cell viability was calculated using ImageJ software and is plotted in Figure 4A. Although the cell viability in co-culture system was approximately 70% at day 1, the Live/Dead staining results indicated an increase to 93% by day 7, which was statistically similar to the findings observed in bulk control hydrogels (Figure 4A). The viabilities of mono-cultured HUVECs and hMSCs were both relatively low at day 1, at 73% and 50%, respectively, probably due to the external stressed due to the bioprinting process. Nevertheless, after 7 days of culture, the viabilities of HUVECs and hMSCs in mono-culture were increased to 94% and 77% respectively, and remained proliferative in the bioprinted hydrogel fibers.
Figure 4.
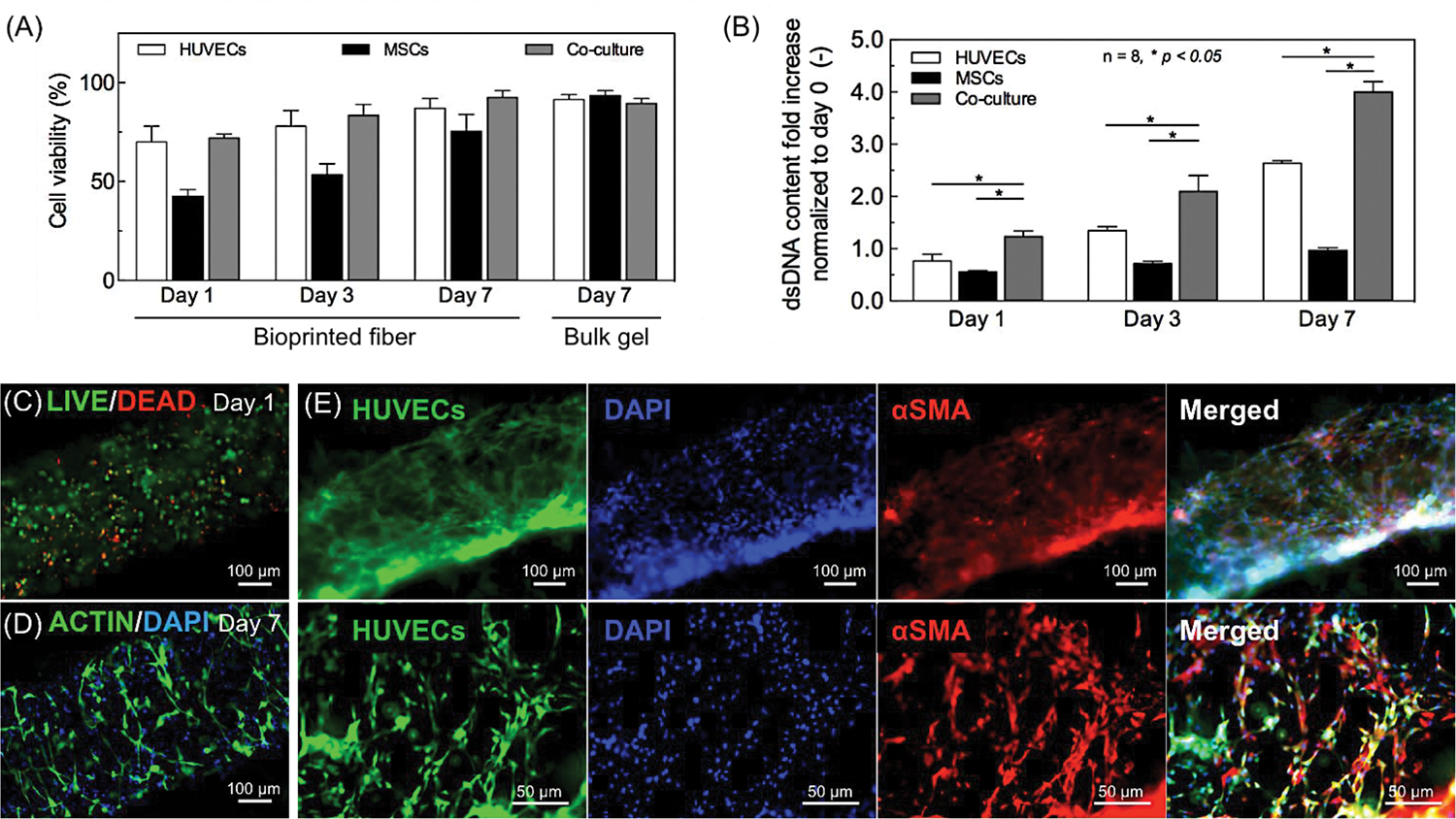
Formation of a stabilized vasculature in a 3D bioprinted construct by hMSC differentiation into perivascular cells in co-culture with HUVECs. (A) Viability of mono- and co-cultured cells in the bioprinted single fiber at days 1, 3, and 7. (B) Cell proliferation in mono- and co-culture system after embedding in a single bioprinted fiber. The dsDNA contents were evaluated by PicoGreen staining at days 1, 3 and 7. Results are presented as the mean ± standard deviation (n = 8; *p < 0.05). (C, D) Representative fluorescence images of (C) LIVE/DEAD staining for cell viability; (D) F-actin/DAPI staining for cellular spreading inside the bioprinted fiber at day 7 in co-culture. (E) Stabilization of the HUVEC capillaries by perivascular cells in the bioprinted fibers with a diameter of 500 μm. Constructs containing both GFP-HUVECs (green fluorescence) and hMSCs cultured for 7 days. The ability of the hMSCs to differentiate into pericytes was analyzed with smooth muscle markers (red fluorescence).
Cell proliferation was quantified by the PicoGreen assay to determine the relative amount of dsDNA collected from a single fiber as compared to that of day 0 (Figure 4B). Importantly, the results in Figure 4B showed that the amount of dsDNA was significantly higher than for mono-cultured HUVECs and hMSCs seeded at the same overall cell density. Therefore, co-culture of HUVECs and hMSCs appears to aid the cell proliferation and vasculogenesis in the bioprinted fibers. F-actin and nuclei (DAPI) staining of the co-cultured cells were performed to monitor the cellular spreading and morphology. By day 7, an elongated spindle morphology was observed due to the cellular spreading within the bioprinted fiber (Figure 4D). Moreover, hMSCs are well known its ability to differentiate into multiple mesodermal lineages under in vitro growth factor stimulation. Among them, the bFGF and TGF-β are commonly used for hMSCs’ differentiation into smooth muscle cells [36], which therefore both were added in EGM™−2 medium in the present study. As a result, the hMSCs were found to differentiate into mature smooth muscle cells in the co-culture with endothelial cells at day 7, as revealed by the extensive expression of α-SMA (Figure 4E), whereas no differentiation was observed in mono-cultures of HUVECs and hMSCs (Supporting Information, Figure S2). As discussed in section 3.4, the hMSCs that differentiated into smooth muscle cells helped in anastomosis to the host vasculature and enhanced the formation of stable vascular networks [37].
The production of a mineralized bone-like extracellular matrix by the hMSCs encapsulated in bioprinted single fiber was studied by ARS staining at days 12 and 21 of culture (Figure 5A). Previously, our group reported that the silicate nanoplatelets significantly induced osteogenic differentiation of encapsulated hMSCs within GelMA hydrogels in growth-factor eluting media [38, 39]. Synthetic silicate nanoplatelets dissociate into nontoxic products (Na+, Mg2+, Si(OH)4, Li+) in aqueous solution, which has potential in triggering specific cellular responses towards osteogenesis induction [40]. For instance, it promotes collagen type I synthesis [41] and activates Wnt-responsive genes by inhibiting the glycogen synthase kinase-3 beta activity that controls osteogenesis via regulating Runt-related transcription factor-2 (RUNX2) activity [42]. Here, we confirmed encapsulated silicate nanoplatelets can induce osteogenic differentiation of hMSCs within bioprinted GelMA hydrogels by day 21 as same as bulk hydrogels that we reported previously (Supporting Information, Figure S3) [38]. We observed a small amount of mineral deposition at day 12, regardless of the hydrogel formulations and culture media. Quantification of the ARS staining at day 21 revealed that production of mineralized extracellular matrix was significantly enhanced (Figure 5B), which showed a clear dependence on the selection of culture media and hydrogel composition. Since the incorporation of silicate nanoplatelets promotes osteogenesis of hMSCs, here we investigated concentration effect of silicate nanoplatelets on calcium deposition at day 21 in absence of osteoinductive growth factors (Figure 5C). Microscopy images of ARS stained samples containing various concentrations of silicate nanoplatelets are shown in Figure S4 in the Supporting Information. As shown in Figure 5C, when compared to pristine hydrogel fibers, the degree of mineralization depended on the content of silicate nanoplatelets. Comparison of the bioprinted constructs encapsulated with 1 and 10 μg/ml of silicate nanoplatelets showed that silicate nanoparticles at 100 μg/ml gave the highest expression of calcium deposition at day 21. Based on these results, GelMAHIGH-VEGF hydrogels containing 100 μg/ml silicate nanoplatelets were selected as the optimized experimental condition for the promotion of osteogenesis of hMSCs in the bioprinted fibers. We developed a functional vascular structure inside these bone tissue like fibers by next evaluating the production of a perfusable lumen formation in the central cylindrical fiber inside the bioprinted construct.
Figure 5.
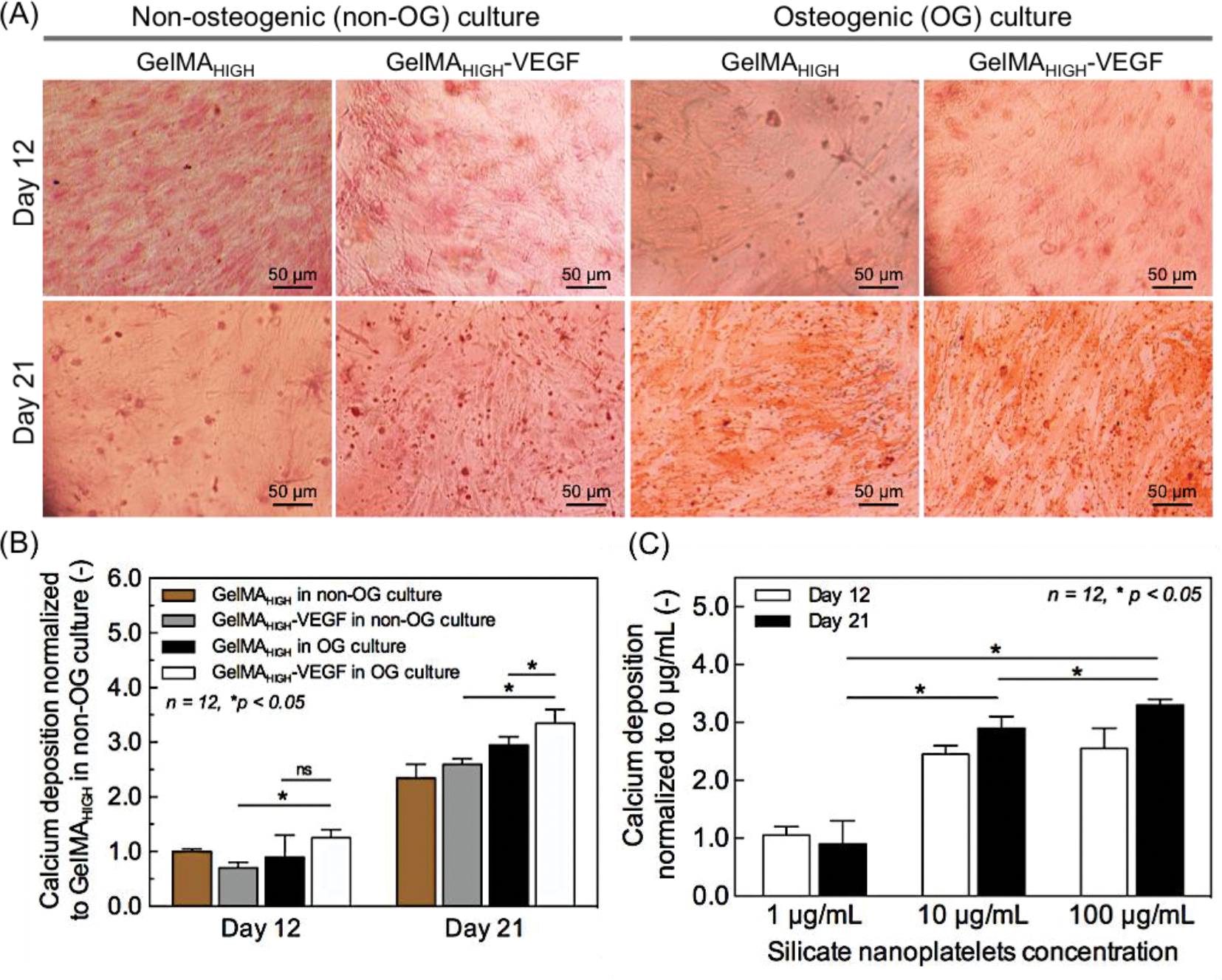
Osteogenic characterization of in vitro mono-culture of hMSCs in a single bioprinted fiber (diameter of 500 μm). (A) Production of mineralized extracellular matrix in GelMAHIGH-VEGF and GelMAHIGH hydrogels loaded with 100 μg/ml of silicate nanoparticles evaluated by Alizarin Red S (ARS) staining on days 12 and 21 post culture. (B) Quantification of the amount of calcium deposition determined by ARS staining, as normalized to that of GelMAHIGH in a normal medium (control) and measured at day 12. When compared to day 12, the production of mineralized matrix was significantly enhanced on day 21 in both osteogenic and normal media. Results are presented as the mean ± standard deviation (n = 12; *p < 0.05). (C) Quantification of the amount of calcium deposition, determined by ARS staining, in osteogenic medium with different silicate nanoplatelet concentrations. The determined amount was normalized to that of silicate nanoplatelets concentration of 0 ng/ml on day 12. The concentration of 100 μg/ml silicate nanoparticles resulted the highest calcium deposition at day 21. Results are presented as the mean ± standard deviation (n = 12; *p < 0.05).
2.6. In vitro formation of a perfusable vascular lumen within the 3D bioprinted bone construct
Once the viability and proliferation of different cell types in individual GelMA rods were established, we printed a larger and more complex architecture by simply assembling individual cylinders using the 3D bioprinter. A few drops of GelMA prepolymer solution (approximately 100 μl volume) was dropped into bioprinted construct to attach one bioprinted cylinder to another and exposed for further 5 s. The integrity of whole construct was confirmed by simple visual inspection of bioprinted construct in different culture time courses, which was concluded that the bioprinted construct was maintaining its structure and stability at least for 21 days in culture and 5 days of media perfusion. In particular, we here demonstrated the fabrication of a perfusable central lumen structure within a relatively large GelMA structure with a size of 16.5 cm3. This vessel-like structure had a closer resemblance to bone tissue architecture and could overcome the eventual nutrient diffusional limitations inherent in tick constructs (Figure 1A). The confocal micrographs in Figure 6 reveal the different aspects of the process of induced formation of HUVEC-lined lumen structures in pyramidal GelMA constructs consisting of an assembly of individual cylinders with different cellular and chemical compositions. The bioprinting process conditions were fine-tuned to render thick pyramidal constructs (3 cm in thickness) with cell viabilities higher than 70%, as validated by staining using Live/Dead technique (Figure 6A). The central cylinder in the pyramidal construct, which was printed using a hydrogel with lower stiffness (5% w/v GelMALOW), encapsulated the co-culture of GFP-HUVECs and hMSCs and served as a rapidly biodegradable core to fabricate the internal lumen. The rest of the cylinder fibers were printed with a hydrogel with high stiffness (10% w/v GelMAHIGH) and preloaded with hMSCs and osteoinductive silicate nanoplatelets. The construct was incubated at 37°C for seven days in static conditions. Due to its lower mechanical properties compared to the rest of the construct, the central cylinder underwent a degradation during this incubation period, which is consistent with the results of our previous studies [14, 22, 23, 31]. This resulted in the formation of a lumen-like hollow structure with a well-defined cylindrical shape and a cell-lined wall presumably conformed by a HUVECs/hMSCs layer (Figure 6B).
Figure 6.
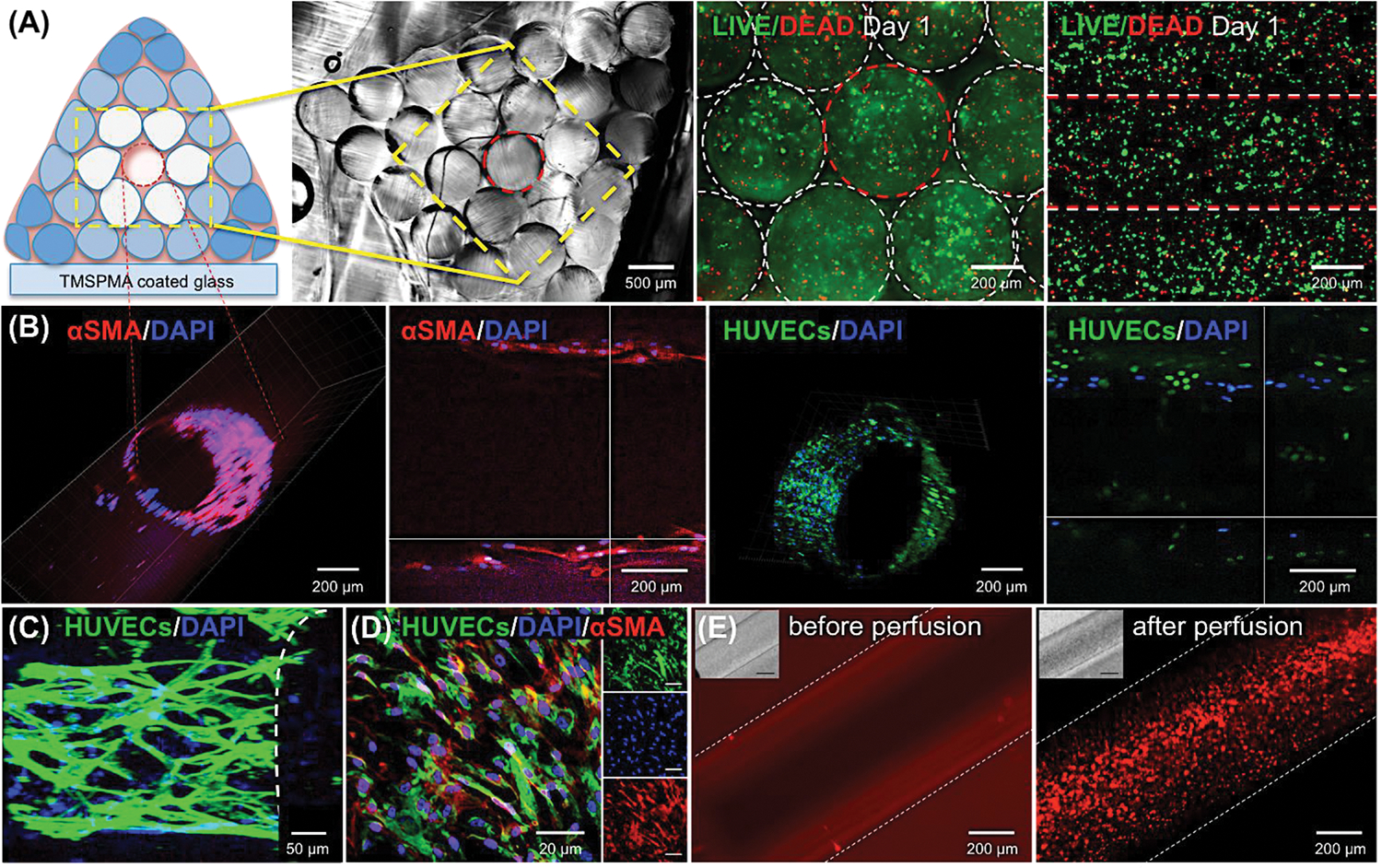
Formation of HUVECs/hMSCs-lined perfusable hollow lumen structure in the bioprinted bone construct. The construct, containing both GFP-HUVECs and hMSCs, was co-cultured for 7 days, followed by media perfusion for 5 days. (A) The cross-sectional view of the whole bioprinted construct. Cross- and top-view of the encapsulated cells stained with LIVE/DEAD inside the whole construct. (B) Demonstration of a HUVEC-lined vessel-like lumen structure within the bioprinted construct. Cross- and top-sectioned confocal micrographs of central vessel within the construct. The central vessel was stained with DAPI and α-SMA at day 12 post culture. Encapsulated endothelial cells were lined the vascular walls (green fluorescence) and hMSC cells were differentiated into pericytes (red fluorescence). (C) Formation and lining of endothelial cells inside the central channel. (D) Immunostaining of endothelial cells and α-SMA-expressing hMSCs in the inner part of the lumen. (E) Vascular lumen network perfused with a fluorescent microbeads suspension at day 7 post culture. Images of the 3D hydrogel construct before and after microbeads perfusion through the core hollow lumen.
The cell layer was stained with F-actin/DAPI and anti-α-SMA, a crucial marker for a stable and mature perivascular cells differentiated from hMSCs [22, 37]. The vessel lumen was continuously perfused for 5 days. Both the construct and the central vascular structure retained their integrity during the whole perfusion period. The cells forming the wall monolayer spread and developed a densely packed network along the channel. Moreover, immunostaining of the cell monolayer in the interior wall of the lumen (Figure 6C and 6D) revealed the sustained active expression of α-SMA, which indicates an early-stage of vessel maturation resembling in vasculature. The presence of a perivascular cell phenotype at the vascular layer is relevant for closely mimicking the structure and important aspects of native blood vessel (Supporting Information, Movie S1). Multiple studies have discussed that the contact between endothelial cells and smooth muscle cells is required for vascular maturation [43]. Perivascular cells might provide structural support to the HUVEC network, enabling longer stability and adequate tissue flexibility under shear flow conditions. Here, the differentiation of hMSCs into prevascular cells accelerated the formation and maturation of vascular network within the bioprinted construct. Although HUVEC cell lines are commonly used in studies of in vitro expression of endothelial phenotypes due to its low cost, simple-handling advantages such as well-characterized, unlimited life span, and faster proliferation rate comparing to endothelial progenitor cells (EPCs), it also has some challenges such as limited clinical applications due to its maturely differentiated properties [44]. The technology presented in the present study could be furtherly improved by incorporating less differentiated EPC cell lines for the potential clinical applications. We further verified the functionality of the vessel-like structure by continuously flowing a suspension of fluorescent particles throughout the construct (Figure 6E, and Supporting Information, Movie S2). Our results demonstrated the feasibility of this simple strategy for fabricating functional cellularized vessel-like lumen structure within thick bioprinted constructs. In turn, this will enable the continuous supply of culture media (and possibly gases) to multi-layer tissue-like structures. We believe this result is highly relevant in tissue engineering field, where the fabrication of a facile, a perfusable, and a long-term mechanically stable construct continues to be a challenge.
2.7. Formation of vascularized bone niche in the bioprinted construct
As described in section 3.1, the outer cylinders surrounding central lumen fiber were bioprinted with a high stiffness hydrogel, 10% (w/v) GelMAHIGH-VEGF, which was loaded with hMSCs and osteoinductive silicate nanoplatelets. The whole bioprinted construct was cultured under static conditions for one week. Subsequent maturation of the construct was achieved by perfusing the medium through the core vessel for 5 days, and further 9 days of static culture (Figure 7A). In accordance with previous findings, the addition of VEGF to the osteogenic medium enhanced the osteogenic differentiation of hMSCs due to growth factor interactions [45]. Additionally, the efficiency of osteogenic differentiation of the hMSCs was further improved by pre-treating the hMSCs in osteogenic media for a week before use in bioprinting.
Figure 7.
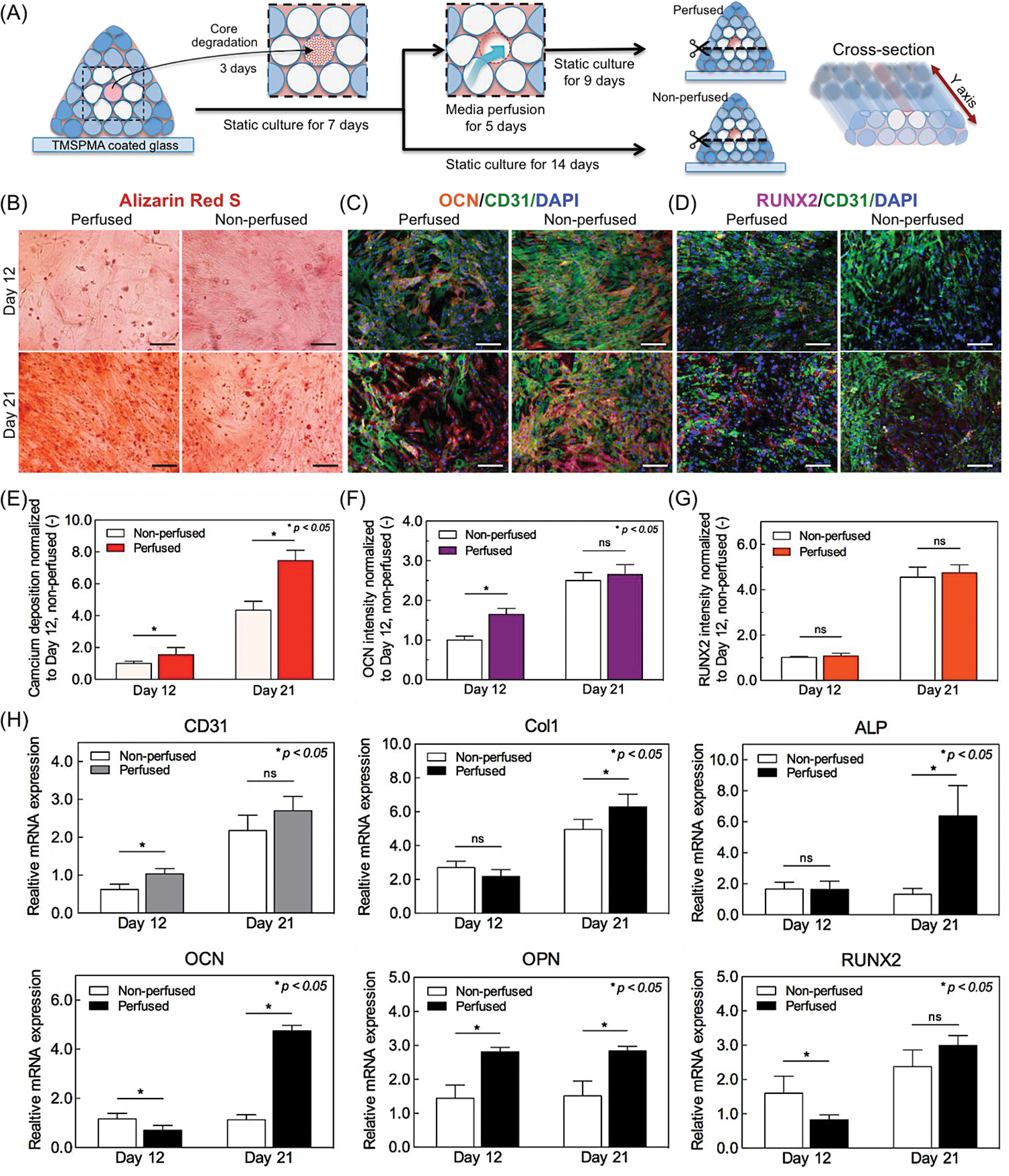
Osteogenic and vasculogenic characterizations of vascularized bone-like tissue bioprinted construct. (A) Schematic illustration of cross-sectioned samples for ARS and immunostaining to evaluate the osteogenesis and vasculogenesis in bioprinted construct. (B) Representative microscopic images of ARS staining of mineralized extracellular matrix. Scale bar indicates 100 μm. (C, D) Representative fluorescence microscopic images of bioprinted construct with double-immunostaining of OCN/CD31/DAPI and RUNX2/CD31/DAPI on days 12 and 21. Scale bar indicates 50 μm. (E-G) Image analysis quantification of the amount of calcium deposition (ARS) and expression levels on osteogenic-related OCN and RUNX2 markers as normalized to that of non-perfused construct at day 12. (H) RT-qPCR assay analysis for mRNA expression of typical angiogenic (CD31) and osteogenic genes (Col1, ALP, OCN, OPN, RUNX2) in the bioprinted construct at days 12 and 21 post culture (n=16).
ARS staining was used to investigate the osteogenic activity of hMSCs of calcified bioprinted matrix (Figure 7B). For this purpose, the whole construct was sliced through as illustrated in Figure 7A, and stained with ARS on days 12 and 21 post culture. The effect of hollow lumen perfusability on osteogenesis (perfused construct) was compared with that of control (non-perfused) construct. The quantification of ARS mineralization assay evidenced the production of mineralized bone-like extracellular matrix was significantly enhanced in perfused construct comparing to that of non-perfused construct on both days 12 and 21 (Figure 7E), which clearly indicates the positive effect of medium perfusion on bone tissue formation.
The osteogenic differentiation of hMSCs in silicate nanoplatelet-laden bioprinted construct was evaluated by double-immunostaining of key osteogenic and vasculogenic markers: OCN/CD31 and RUNX2/CD31 on days 12 and 21 (Figure 7C and 7D). According to the quantitative analysis of immunostaining, we observed a strong expression of OCN, a late osteoblast marker, in perfused construct on day 12 (Figure 7F), which demonstrates medium perfusion promoted osteoblastic maturation in bioprinted construct by day 12. By contrast, only negligible differences were observed on the expression of RUNX2, a central transcription factor of bone formation, between perfused and non-perfused constructs on days 12 and 21 (Figure 7G). These findings were validated by real-time PCR gene expression results. The result of RT-qPCR analysis revealed an increased expression level of angiogenesis-related gene of CD31 on day 12, which indicates the HUVECs/hMSCs-laden core GelMA fiber held a great potential for generation of an organized and mature vascular niche within the construct (Figure 7H). Moreover, medium perfusion affected significant increases in gene expression levels of early osteogenic differentiation markers: Col1, ALP, and late osteogenic differentiation markers: OCN, OPN of hMSCs as compared with non-perfused construct on day 21 (Figure 7H). These results clearly confirm that the medium perfusion also promoted the maturation of hMSCs differentiation into osteoblasts in bioprinted construct. Nevertheless, there was only insignificant difference in gene expression of RUNX2 between perfused and non-perfused constructs. Though RUNX2 is a crucial for osteoblast differentiation, multiple genes regulate its activity and effectiveness on stimulating osteoblast differentiation [46]. Thus, it is probably not surprising that earlier detection of RUNX2 expression in immature osteoprogenitor cells generated from hMSCs in non-perfused construct. Taken together, both immunostaining and quantitative gene expression results demonstrate that the formation and maturation of bone tissue are further promoted by the presence of a central angiogenic vessel, furthermore, key osteogenic genes such as Col1, ALP, OCN, and OPN were upregulated by medium perfusion throughout bioprinted construct over 21 days of culture. These results demonstrated that our 3D bioprinting strategy could engineer vascularized bone-like tissue units with a stable controlled architecture, which can be expected to generate a new paradigm in bone-related tissue engineering approaches. However, we also observed some difficulties maintaining mechanical (structural) stability and perfusability after 21 days of culture, mainly due to the degradation of GelMA hydrogel. Further modification of bioink and designing of printed construct might address this issue more effectively in a future study.
3. Conclusion
Mimicking the complex architecture of tissues that also include multiscale vasculature networks continues to be a challenge for tissue engineers. Bone has a complex architecture consisting of calcified regions and interpenetrating vasculature; therefore, engineering functional bone requires the presence of distinct niches that can support vasculogenesis as well as osteogenesis. Moreover, an intrinsic gradient of biological factors within the engineered constructs can enhance the formation of microcapillaries. In this study, we synthesized and optimized two different GelMA-based hydrogels - one that supported vasculogenesis and the other that supported osteogenesis. GelMA hydrogels containing different levels of VEGF were bioprinted into well-defined and controlled 3D architectures to create a gradient of vasculogenic factors. The bioprinting and incorporation of a rapidly degradable GelMA hydrogel resulted in the formation of a perfusable lumen with an endothelial lining at the center of the construct. The co-cultured hMSCs in the inner fiber differentiated into smooth muscle cells that promoted the formation, stabilization, and maturation of vascular vessels, as well as aiding endothelial cell proliferation. Three outer layers were printed with 10% (w/v) of hMSCs and bioactive silicate nanoplatelets encapsulated in GelMAHIGH-VEGF, which induced osteogenic differentiation in vitro. After 5 days in static culture, perfusion the construct with culture medium through the inner core vessel supported the survival of the cells in the bone niche. The results of immunostaining and RT-qPCR confirmed that the encapsulated hMSCs formed a mature bone niche after 21 days of culture under the medium perfused condition. This approach can pave the way for engineering bone constructs that can be used for treatment of large bone defects.
4. Experimental Section
Materials:
Methacrylic anhydride (MA), gelatin (Type A, 300 bloom from porcine skin), 2-hydroxy-4’-(2-hydroxyethoxy)-2-methylpropiophenone (Irgacure 2959), N-hydroxysuccinimide (NHS), triethylamine, and N-(3-dimethylaminopropyl)-N’-ethylcarbodiimide hydrochloride (EDC) were purchased from Sigma Aldrich (St. Louis, MO, USA). Human umbilical vein endothelial cells (HUVECs) and bone marrow-derived human mesenchymal stem cells (hMSCs) were purchased from ATCC (Manassas, VA, USA). Minimum essential medium alpha (α-MEM), Dulbecco’s modified Eagle’s medium (DMEM), fetal bovine serum (FBS), and the cell dissociation reagent TrypLE™ Express were provided by GIBCO®/Life Technologies (Carlsbad, CA, USA).
Chemical conjugation of VEGF with GelMA:
High degree of methacryloyl substitution GelMA (GelMAHIGH, 1.0 g) was fully dissolved in 20 mL PBS at 50°C with magnetic stirring, followed by the addition of an excess amount of succinic anhydride (0.5 g) dissolved in 10 mL dimethyl sulfoxide (DMSO) and triethylamine (0.5 mL). The mixture was stirred overnight, diluted with 50 mL PBS, and then dialyzed against DI water using dialysis tubing (3.5 kDa cutoff) for ten days at room temperature to eliminate impurities. The resulting solution was lyophilized to generate GelMAHIGH-COOH as a white porous foam. For VEGF conjugation, GelMAHIGH-COOH (300 mg) was dissolved in 5 mL PBS, EDC (0.5 mg) and NHS (0.5 mg) were added and kept under agitation for 30 min, and VEGF was added thereafter. This mixture was kept under agitation at room temperature for 6 h before dialysis against DI water using 3.5 kDa cut-off dialysis tubing and lyophilization to generate GelMA-VEGF. For 1H NMR analysis, the hydrogels were dissolved in 30 mg/ml of deuterium oxide (D2O). 1H NMR spectrum was determined using a Bruker 300M NMR instrument and the data were processed using the ACD LABS 12.0 software.
Hydrogel formulation and preparation:
GelMA (5 to 10% (w/v)) was fully dissolved in PBS at 80°C and 0.1% (w/v) photoinitiator was added. Photopolymerization was induced by UV light exposure of the GelMA prepolymer at 6.9 mW/cm2 for 20 s (Omnicure S2000, 360–480 nm).
Fabrication of GelMA cylinders and mechanical testing:
Cylindrical samples were fabricated by mixing 150 μL without cells, 5 to 10% (w/v) GelMA prepolymer solutions and photoinitiator and pipetting the mixture carefully into a poly(dimethylsiloxane) (PDMS) mold (3 mm thickness; 8 mm diameter). The prepolymer solutions were exposed to UV light (wavelengths between 360 and 480 nm; 6.9 mW/cm2) for 5 to 90 s and incubated in PBS for 24 h at room temperature. Compression test were conducted on the GelMA cylinders through the procedures previously described [21] and the compressive modulus was calculated from the slope of the initial linear region corresponding to 0 to 10% strain. The cylinders were dried superficially before mechanical testing an Instron 5542 at a crosshead speed of 0.1 mm/min.
Enzymatic degradation test:
The degradation properties of GelMAHIGH, GelMAMEDIUM, and GelMALOW were evaluated in vitro by incubation in a PBS solution containing collagenase A, type I (1.0 μg/ml, Roche Diagnostics) for 72 at 37°C. For each condition, the samples were frozen, and freeze dried. The extent of mass loss was determined at different time points by the ratio of the weight to the original weight.
Bioprinting of GelMA-based 3D constructs:
A NovoGen MMX Bioprinter™ (Organovo) was used for bioprinting to print the constructs. The cell-laden prehydrogel (2.0×106 cells/mL) was aspirated by piston into an immersed glass capillary (500 μm diameter, 65 mm long). A UV light guide (Omnicure S2000, 6.9 mW/cm2) was connected to the bioprinter to perform photo-induced polymerization of the aspirated prehydrogels in the glass capillaries. Thereafter, the inner piston was used to push down the photocrosslinked hydrogel with a controlled dispensing speed and the coordinated changes on positions of the X–Z-Y stage resulted in the dispensation of 10 mm fibers, as described previously [14]. The printability of the construct was determined by the reproducibility of hydrogels with different hydrogel concentrations (5 to 10% (w/v)) and UV exposure times (5 to 60 s). Printing was defined as successful and reproducible in case all the dispensed fibers (n=10) were printed while keeping its cylindrical shape.
Cell culture:
The HUVECs were cultured in EGM™−2 (Lonza, Inc.) medium. The hMSCs were cultured in α-MEM added with 1 ng/ml basic fibroblast growth factor (bFGF) and 10% fetal bovine serum (FBS). Osteogenic differentiation of hMSCs was induced in osteoinductive medium containing 2 mM L-ascorbic acid, 50 mM β-glycerophosphate, and 100 nM dexamethasone. The conditioned medium was changed with fresh medium every two days.
Cell encapsulation in bioprinted fibers and cell viability/proliferation assay:
The cell mixtures were molded and crosslinked inside the glass capillary. Cell viability was determined using a Live/Dead assay. Briefly, cells were stained with calcein AM (0.5 μL/mL) and ethidium homodimer-1 (EthD-1, 2 μL/mL) in PBS. The cells were incubated at 37°C for 20 min, thoroughly washed three times with PBS, and immediately observed under the fluorescence microscope (Zeiss Axiovert 135). Image J was used to estimate the number of living and dead cells; cell viability was determined as the ratio of the number of living cells and the total cell number. For testing cell proliferation with the PicoGreen assay, 20 μl of working solution was added to 20 μl of cell sample, and the sample was measured at Ex/Em=485±10 nm/528±10 nm on a spectrophotometer (BioTek). Calibration curves of a known control dsDNA sample were used to calculate the amount of dsDNA. Cell viability was quantified in a co-culture experiment by labeling the hMSCs with red fluorescent CellTracker CMFDA (Invitrogen, Inc.) for easier identification. The GFP-HUVECs were identified by green fluorescence.
Characterization of in vitro capillary-like structure visualization and perfusion through inner vessels:
The capillary-like structure was visualized by the green fluorescence of the GFP-HUVECs. The total capillary-like length and the number of capillary-like branches were quantified on fluorescent images using Image J software (NIH). For image analysis, 4 representative images of 6 bioprinted samples, which formed separately (total of 24 images,) were randomly taken for each condition. The formation of the inner perfusable channel was characterized by imaging a bioprinted construct with 7 layer architectures in brightfield using a Nikon TE optical microscope. The perfusability of the inner vessel was examined by loading the bioprinted construct with a suspension of rhodamine B conjugated fluorescent microbeads (0.1% (v/v)) and imaging using a fluorescence microscope (AxioCam MRc5, Carl Zeiss).
Immunostaining of the inner perfusable hollow channel:
For immunostaining of alpha-smooth muscle actin (α-SMA), the cells inside bioprinted construct were fixed with 4% (w/v) paraformaldehyde for 20 min, and treated with 0.5% (v/v) Triton X-100 for a further 20 min. The samples were treated with blocking solution of 1% (w/v) BSA for 60 min and incubated in mouse anti-α-SMA primary antibodies (1/100 dilution) for 3 h at 37°C. After three times of PBS washing, the bioprinted construct was incubated overnight in 1/100 diluted Alexa Fluor-488 conjugated rabbit anti-mouse secondary antibody at 4°C. Thereafter, the construct was stained with 4’,6-diamidino-2-phenylindole (DAPI) for 5 min and a confocal microscopic images were obtained (LSM 880, Zeiss Carl).
Characterization of osteogenic differentiation of embedded hMSC cells in the bioprinted construct:
For Alizarin Red S staining (ARS), the bioprinted construct sample was fixed with 4% paraformaldehyde for 10 min and washed thrice with PBS. Any remaining salt residues were removed by washing the sample with distilled water and then adding a 2% ARS solution (pH = 4.2) for 20 min at room temperature. After three washes with distilled water and 15 min incubations in a shaker, bright field images were obtained. For quantification of orange-red coloration of ARS, 10% acetic acid was added to the sample and incubated overnight. The samples with the acetic acid were centrifuged for 30 min at 20,000×g, then the supernatant was neutralized with 100 μl of 10% ammonium hydroxide and the OD405 was measured.
RT-qPCR:
TRIzol (Invitrogen, Inc.) was utilized to extract the total RNA from samples and 1 μg of extracted RNA was utilized for cDNA synthesis with SuperScript™ III First-Strand Synthesis SuperMix (Invitrogen, Inc.). Quantitative real-time PCR analysis was conducted using primers and PCR conditions for glyceraldehyde 3-phosphate dehydrogenase (GAPDH), Col1, CD31, osteocalcin (OCN), osteopontin (OPN), alkaline phosphatase (ALP), and runt-related transcription factor 2 (RUNX2) as shown in Table 1. Amplifications were conducted in reaction mixtures (20 μl) containing approximately 1 μl of template, 500 nmol/l of gene-specific primers, 1 nmol/l of Bio-Rad SYBR Green Master Mix (Bio-Rad Laboratories, Inc.). We used a sequence of PCR steps consisting on: an initial denaturation at 95°C for 1 min; 45 cycles of 95°C (15 sec), 56°C (15 sec), and 72°C (15 sec); and a final extension at 72°C for 5 min. The baseline and threshold levels were selected using CFX Manager Software (Bio-Rad Laboratories, Inc.). The 2−ΔΔCt method [47] was used for the relative quantification of gene expression.
Table 1.
Oligonucleotide primers used in RT-qPCR
| Target cDNA | Orientation | Primer sequence | |
|---|---|---|---|
|
| |||
| Col1 | Forward | GTC ACC CAC CGA CCAAGAAAC C | |
| Angiogenic genes | Col1 | Reverse | AAG TCC AGG CTG TCC AGG GAT G |
| CD31 | Forward | TCT ATG ACC TCG CCC TCC AC AAA | |
| CD31 | Reverse | GAA CGG TGT CTT CAG GTT GGT ATT TCA | |
|
| |||
| RUNX2 | Forward | GGT TAATCT CCG CAG GTC ACT | |
| RUNX2 | Reverse | CAC TGT GCT GAA GAG GCT GTT | |
| OCN | Forward | TCA CAC TCC TCG CCC TAT TG | |
| Osteogenic genes | OCN | Reverse | TCG CTG CCC TCC TGC TTG |
| ALP | Forward | GGA CAT GCA GTA CGA GCT GA | |
| ALP | Reverse | GCA GTG AAG GGC TTC TTG TC | |
| OPN | Forward | CTC AGG CCA GTT GCA GCC | |
| OPN | Reverse | CAA AAG CAAATC ACT GCA ATT CTC | |
|
| |||
| Housekeeping gene | GAPDH | Forward | GCT CTC TGC TCC TCC TGT TC |
| GAPDH | Reverse | ACG ACC AAA TCC GTT GAC TC | |
Statistical analysis:
All assays were performed with a minimum of n=3 per group. The results for each measurement were defined in terms of mean values and standard deviation. One-way analysis of variance (ANOVA) was used for the statistical analysis of quantitative values. The least significant difference (LSD) post hoc test was utilized for all pairwise comparisons among groups. Student t-test was used to compare means between groups. A level of statistical significance (p<0.05) was used across the study.
Supplementary Material
Acknowledgements
The authors acknowledge funding from the National Science Foundation (EFRI-1240443), the Office of Naval Research Young Investigator award, ONR PECASE Award, and the National Institutes of Health (HL092836, DE019024, EB012597, AR057837, DE021468, HL099073, EB008392). B.B. was supported by postdoctoral funding from the Khuree Rotary Club, International Rotary District 3450. M.M.A. gratefully acknowledge the funding provided by Consejo Nacional de Ciencia y Tecnología (CONACyT) in the form of a sabbatical scholarship (262130). GTdS would like to acknowledge the funding received by CONACyT (scholarship 234713) and Fundación México in Harvard. This research has been partially funded by the MIT-Tecnológico de Monterrey Nanotechnology Program, and the MIT International Science and Technology Initiatives (MISTI). The authors declare no competing financial interest.
Footnotes
Supporting Information
Supporting Information is available from the Wiley Online Library or from the author.
Contributor Information
Batzaya Byambaa, Biomaterials Innovation Research Center, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, Boston, MA 02139, USA.; Harvard-MIT Division of Health Sciences and Technology, Massachusetts Institute of Technology, Cambridge, MA 02139, USA.
Nasim Annabi, Biomaterials Innovation Research Center, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, Boston, MA 02139, USA.; Harvard-MIT Division of Health Sciences and Technology, Massachusetts Institute of Technology, Cambridge, MA 02139, USA. Wyss Institute for Biologically Inspired Engineering, Harvard University, Boston, MA 02115, USA. Department of Chemical Engineering, Northeastern University, Boston, MA, 02115-5000, USA.
Kan Yue, Biomaterials Innovation Research Center, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, Boston, MA 02139, USA.; Harvard-MIT Division of Health Sciences and Technology, Massachusetts Institute of Technology, Cambridge, MA 02139, USA.
Grissel Trujillo de Santiago, Biomaterials Innovation Research Center, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, Boston, MA 02139, USA.; Harvard-MIT Division of Health Sciences and Technology, Massachusetts Institute of Technology, Cambridge, MA 02139, USA.
Mario Moisés Alvarez, Biomaterials Innovation Research Center, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, Boston, MA 02139, USA.; Harvard-MIT Division of Health Sciences and Technology, Massachusetts Institute of Technology, Cambridge, MA 02139, USA. Centro de Biotecnología-FEMSA, Tecnológico de Monterrey at Monterrey, CP 64849, Monterrey, Nuevo León, México.
Weitao Jia, Biomaterials Innovation Research Center, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, Boston, MA 02139, USA.; Harvard-MIT Division of Health Sciences and Technology, Massachusetts Institute of Technology, Cambridge, MA 02139, USA. Department of Orthopedic Surgery, Shanghai Jiaotong University Affiliated Sixth People’s Hospital, Shanghai Jiaotong University, Shanghai 200233, P.R. China
Mehdi Kazemzadeh-Narbat, Biomaterials Innovation Research Center, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, Boston, MA 02139, USA.; Harvard-MIT Division of Health Sciences and Technology, Massachusetts Institute of Technology, Cambridge, MA 02139, USA.
Su Ryon Shin, Biomaterials Innovation Research Center, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, Boston, MA 02139, USA.; Harvard-MIT Division of Health Sciences and Technology, Massachusetts Institute of Technology, Cambridge, MA 02139, USA. Wyss Institute for Biologically Inspired Engineering, Harvard University, Boston, MA 02115, USA.
Ali Tamayol, Biomaterials Innovation Research Center, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, Boston, MA 02139, USA.; Harvard-MIT Division of Health Sciences and Technology, Massachusetts Institute of Technology, Cambridge, MA 02139, USA. Wyss Institute for Biologically Inspired Engineering, Harvard University, Boston, MA 02115, USA.
Ali Khademhosseini, Biomaterials Innovation Research Center, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, Boston, MA 02139, USA.; Harvard-MIT Division of Health Sciences and Technology, Massachusetts Institute of Technology, Cambridge, MA 02139, USA. Wyss Institute for Biologically Inspired Engineering, Harvard University, Boston, MA 02115, USA. Department of Physics, King Abdulaziz University, Jeddah 21569, Saudi Arabia WPI-Advanced Institute for Materials Research, Tohoku University, Sendai, Japan; Department of Bioindustrial Technologies, College of Animal Bioscience and Technology, Konkuk University, Seoul, Republic of Korea.
References
- [1].Annabi N, Tamayol A, Uquillas JA, Akbari M, Bertassoni LE, Cha C, Camci-Unal G, Dokmeci MR, Peppas NA, Khademhosseini A, Adv Mater 2014, 26, 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Akbari M, Tamayol A, Bagherifard S, Serex L, Mostafalu P, Faramarzi N, Mohammadi MH, Khademhosseini A, Advanced healthcare materials 2016, 5, 751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Salgado AJ, Coutinho OP, Reis RL, Macromolecular Bioscience 2004, 4, 743. [DOI] [PubMed] [Google Scholar]
- [4].Santos MI, Reis RL, Macromolecular bioscience 2010, 10, 12. [DOI] [PubMed] [Google Scholar]
- [5].Nguyen LH, Annabi N, Nikkhah M, Bae H, Binan L, Park S, Kang Y, Yang Y, Khademhosseini A, Tissue Engineering Part B: Reviews 2012, 18, 363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Discher DE, Mooney DJ, Zandstra PW, Science 2009, 324, 1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Jain RK, Au P, Tam J, Duda DG, Fukumura D, Nature biotechnology 2005, 23, 821; [DOI] [PubMed] [Google Scholar]; Rafii S, Lyden D, Nature medicine 2003, 9, 702. [DOI] [PubMed] [Google Scholar]
- [8].Koike N, Fukumura D, Gralla O, Au P, Schechner JS, Jain RK, Nature 2004, 428, 138; [DOI] [PubMed] [Google Scholar]; Samuel R, Daheron L, Liao S, Vardam T, Kamoun WS, Batista A, Buecker C, Schafer R, Han X, Au P, Scadden DT, Duda DG, Fukumura D, Jain RK, Proceedings of the National Academy of Sciences of the United States of America 2013, 110, 12774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Bae H, Puranik AS, Gauvin R, Edalat F, Carrillo-Conde B, Peppas NA, Khademhosseini A, Sci Transl Med 2012, 4, 160ps23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Chen Y-C, Lin R-Z, Qi H, Yang Y, Bae H, Melero-Martin JM, Khademhosseini A, Advanced Functional Materials 2012, 22, 2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Tamayol A, Najafabadi AH, Aliakbarian B, Arab-Tehrany E, Akbari M, Annabi N, Juncker D, Khademhosseini A, Advanced healthcare materials 2015; [DOI] [PubMed] [Google Scholar]; Murphy SV, Atala A, Nature biotechnology 2014, 32, 773. [DOI] [PubMed] [Google Scholar]
- [12].Mironov V, Boland T, Trusk T, Forgacs G, Markwald RR, Trends in Biotechnology 2003, 21, 157. [DOI] [PubMed] [Google Scholar]
- [13].Pfister A, Landers R, Laib A, Hübner U, Schmelzeisen R, Mülhaupt R, Journal of Polymer Science Part A: Polymer Chemistry 2004, 42, 624. [Google Scholar]
- [14].Luiz EW Bertassoni JCC, Manoharan Vijayan, Cristino Ana L, Bhise Nupura S, Araujo Wesleyan A, Zorlutuna Pinar, Vrana Nihal E, Ghaemmaghami Amir M, Dokmeci Mehmet R, Khademhosseini Ali, Biofabrication 2014, 6, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Kolesky DB, Truby RL, Gladman AS, Busbee TA, Homan KA, Lewis JA, Adv Mater 2014, 26, 3124. [DOI] [PubMed] [Google Scholar]
- [16].Ghorbanian S, Qasaimeh MA, Akbari M, Tamayol A, Juncker D, Biomedical microdevices 2014, 16, 387. [DOI] [PubMed] [Google Scholar]
- [17].Holmes B, Bulusu K, Plesniak M, Zhang LG, Nanotechnology 2016, 27, 064001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Bose S, Tarafder S, Bandyopadhyay A, Annals of Biomedical Engineering 2016, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Wang J, Yang M, Zhu Y, Wang L, Tomsia AP, Mao C, Advanced Materials 2014, 26, 4961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Do AV, Akkouch A, Green B, Ozbolat I, Debabneh A, Geary S, Salem AK, Annals of biomedical engineering 2017, 45, 297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Nichol JW, Koshy ST, Bae H, Hwang CM, Yamanlar S, Khademhosseini A, Biomaterials 2010, 31, 5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Jia W, Gungor-Ozkerim PS, Zhang YS, Yue K, Zhu K, Liu W, Pi Q, Byambaa B, Dokmeci MR, Shin SR, Khademhosseini A, Biomaterials 2016, 106, 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Yue K, Trujillo-de Santiago G, Alvarez MM, Tamayol A, Annabi N, Khademhosseini A, Biomaterials 2015, 73, 254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Lee VK, Kim DY, Ngo H, Lee Y, Seo L, Yoo SS, Vincent PA, Dai G, Biomaterials 2014, 35, 8092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Kolesky DB, Homan KA, Skylar-Scott MA, Lewis JA, Proceedings of the National Academy of Sciences of the United States of America 2016, 113, 3179; [DOI] [PMC free article] [PubMed] [Google Scholar]; Blache U, Metzger S, Vallmajo-Martin Q, Martin I, Djonov V, Ehrbar M, Advanced healthcare materials 2016, 5, 489. [DOI] [PubMed] [Google Scholar]
- [26].Miller JS, Stevens KR, Yang MT, Baker BM, Nguyen DH, Cohen DM, Toro E, Chen AA, Galie PA, Yu X, Chaturvedi R, Bhatia SN, Chen CS, Nature materials 2012, 11, 768; [DOI] [PMC free article] [PubMed] [Google Scholar]; Bertassoni LE, Cecconi M, Manoharan V, Nikkhah M, Hjortnaes J, Cristino AL, Barabaschi G, Demarchi D, Dokmeci MR, Yang Y, Khademhosseini A, Lab on a chip 2014, 14, 2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Do AV, Khorsand B, Geary SM, Salem AK, Advanced healthcare materials 2015, 4, 1742; [DOI] [PMC free article] [PubMed] [Google Scholar]; Ozbolat IT, Hospodiuk M, Biomaterials 2016, 76, 321. [DOI] [PubMed] [Google Scholar]
- [28].Shen YH, Shoichet MS, Radisic M, Acta biomaterialia 2008, 4, 477. [DOI] [PubMed] [Google Scholar]
- [29].Hume PS, Anseth KS, Biomaterials 2010, 31, 3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Askarinam A, James AW, Zara JN, Goyal R, Corselli M, Pan A, Liang P, Chang L, Rackohn T, Stoker D, Zhang X, Ting K, Peault B, Soo C, Tissue engineering. Part A 2013, 19, 1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Chen YC, Lin RZ, Qi H, Yang Y, Bae H, Melero-Martin JM, Khademhosseini A, Advanced functional materials 2012, 22, 2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Gupta VK, Jaskowiak NT, Beckett MA, Mauceri HJ, Grunstein J, Johnson RS, Calvin DA, Nodzenski E, Pejovic M, Kufe DW, Posner MC, Weichselbaum RR, Cancer journal 2002, 8, 47. [DOI] [PubMed] [Google Scholar]
- [33].Choong CS, Hutmacher DW, Triffitt JT, Tissue engineering 2006, 12, 2521; [DOI] [PubMed] [Google Scholar]; Ma J, Both SK, Yang F, Cui FZ, Pan J, Meijer GJ, Jansen JA, van den Beucken JJ, Stem cells translational medicine 2014, 3, 98; [DOI] [PMC free article] [PubMed] [Google Scholar]; Correia C, Grayson WL, Park M, Hutton D, Zhou B, Guo XE, Niklason L, Sousa RA, Reis RL, Vunjak-Novakovic G, PloS one 2011, 6, e28352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Vattikuti R, Towler DA, American Journal of Physiology - Endocrinology and Metabolism 2004, 286, E686; [DOI] [PubMed] [Google Scholar]; García J, García A, Drug Deliv. and Transl. Res. 2015, 1;25787334 [Google Scholar]; Kang Y, Kim S, Fahrenholtz M, Khademhosseini A, Yang Y, Acta biomaterialia 2013, 9, 4906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Moon JJ, Saik JE, Poche RA, Leslie-Barbick JE, Lee SH, Smith AA, Dickinson ME, West JL, Biomaterials 2010, 31, 3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Ng F, Boucher S, Koh S, Sastry KS, Chase L, Lakshmipathy U, Choong C, Yang Z, Vemuri MC, Rao MS, Tanavde V, Blood 2008, 112, 295; [DOI] [PubMed] [Google Scholar]; Watabe T, Miyazono K, Cell research 2009, 19, 103. [DOI] [PubMed] [Google Scholar]
- [37].Rouwkema J, Rivron NC, van Blitterswijk CA, Trends in biotechnology 2008, 26, 434. [DOI] [PubMed] [Google Scholar]
- [38].Gaharwar AK, Mihaila SM, Swami A, Patel A, Sant S, Reis RL, Marques AP, Gomes ME, Khademhosseini A, Adv Mater 2013, 25, 3329; [DOI] [PubMed] [Google Scholar]; Paul A, Manoharan V, Krafft D, Assmann A, Uquillas JA, Shin SR, Hasan A, Hussain MA, Memic A, Gaharwar AK, Khademhosseini A, Journal of materials chemistry. B, Materials for biology and medicine 2016, 4, 3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Xavier JR, Thakur T, Desai P, Jaiswal MK, Sears N, Cosgriff-Hernandez E, Kaunas R, Gaharwar AK, ACS nano 2015, 9, 3109. [DOI] [PubMed] [Google Scholar]
- [40].Dudley W Thompson JTB, J. Colloid Interface Sci. 1992, 151, 8. [Google Scholar]
- [41].Reffitt DM, Ogston N, Jugdaohsingh R, Cheung HF, Evans BA, Thompson RP, Powell JJ, Hampson GN, Bone 2003, 32, 127. [DOI] [PubMed] [Google Scholar]
- [42].Kugimiya F, Kawaguchi H, Ohba S, Kawamura N, Hirata M, Chikuda H, Azuma Y, Woodgett JR, Nakamura K, Chung UI, PloS one 2007, 2, e837; [DOI] [PMC free article] [PubMed] [Google Scholar]; Kubota T, Michigami T, Ozono K, Journal of bone and mineral metabolism 2009, 27, 265. [DOI] [PubMed] [Google Scholar]
- [43].Baranski JD, Chaturvedi RR, Stevens KR, Eyckmans J, Carvalho B, Solorzano RD, Yang MT, Miller JS, Bhatia SN, Chen CS, Proceedings of the National Academy of Sciences of the United States of America 2013, 110, 7586; [DOI] [PMC free article] [PubMed] [Google Scholar]; Isayama N, Matsumura G, Sato H, Matsuda S, Yamazaki K, Biomaterials 2014, 35, 3589; [DOI] [PubMed] [Google Scholar]; Mehdi Kazemzadeh-Narbat JR, Annabi Nasim, Cheng Hao, Ghaderi Masoumeh, Cha Byung-Hyun, Aparnathi Mansi, Khalilpour Akbar, Byambaa Batzaya, Jabbari Esmaiel, Tamayol Ali, Khademhosseini Ali, Advanced healthcare materials 2017. [DOI] [PubMed] [Google Scholar]
- [44].Trounson A, DeWitt ND, Nature reviews. Molecular cell biology 2016, 17, 194; [DOI] [PubMed] [Google Scholar]; Asahara T, Murohara T, Sullivan A, Silver M, van der Zee R, Li T, Witzenbichler B, Schatteman G, Isner JM, Science 1997, 275, 964. [DOI] [PubMed] [Google Scholar]
- [45].Kanczler JM, Ginty PJ, White L, Clarke NM, Howdle SM, Shakesheff KM, Oreffo RO, Biomaterials 2010, 31, 1242; [DOI] [PubMed] [Google Scholar]; Vater C, Kasten P, Stiehler M, Acta biomaterialia 2011, 7, 463; [DOI] [PubMed] [Google Scholar]; Bose S, Roy M, Bandyopadhyay A, Trends in biotechnology 2012, 30, 546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Karsenty G, Wagner EF, Developmental cell 2002, 2, 389; [DOI] [PubMed] [Google Scholar]; Huang W, Yang S, Shao J, Li YP, Frontiers in bioscience : a journal and virtual library 2007, 12, 3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Livak KJ, Schmittgen TD, Methods 2001, 25, 402. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


