Abstract
Background:
Early in the pandemic, cruise travel exacerbated the global spread of SARS-CoV-2. We report epidemiologic and molecular findings from an investigation of a cluster of travellers with confirmed COVID-19 returning to the USA from Nile River cruises in Egypt.
Methods:
State health departments reported data on real-time reverse transcription-polymerase chain reaction-confirmed COVID-19 cases with a history of Nile River cruise travel during February–March 2020 to the Centers for Disease Control and Prevention (CDC). Demographic and epidemiologic data were collected through routine surveillance channels. Sequences were obtained either from state health departments or from the Global Initiative on Sharing Avian Flu Data (GISAID). We conducted descriptive analyses of epidemiologic data and explored phylogenetic relationships between sequences.
Results:
We identified 149 Nile River cruise travellers with confirmed COVID-19 who returned to 67 different US counties in 27 states: among those with complete data, 4.7% (6/128) died and 28.1% (38/135) were hospitalized. These individuals travelled on 20 different Nile River cruise voyages (12 unique vessels). Fifteen community transmission events were identified in four states, with 73.3% (11/15) of these occurring in Wisconsin (as the result of a more detailed contact investigation in that state). Phylogenetic analyses supported the hypothesis that travellers were most likely infected in Egypt, with most sequences in Nextstrain clade 20A 93% (87/94). We observed genetic clustering by Nile River cruise voyage and vessel.
Conclusions:
Nile River cruise travellers with COVID-19 introduced SARS-CoV-2 over a very large geographic range, facilitating transmission across the USA early in the pandemic. Travellers who participate in cruises, even on small river vessels as investigated in this study, are at increased risk of SARS-CoV-2 exposure. Therefore, history of river cruise travel should be considered in contact tracing and outbreak investigations.
Keywords: COVID-19, SARS-CoV-2, cruise travel, Egypt
Background
Cruise travel was a source of introductions of COVID-19 in the early phase of the epidemic in the USA. During 3 February–13 March 2020, the US Centers for Disease Control and Prevention (CDC) estimated that 17% of cases were cruise-associated.1 Most of these cases were from large, international cruises,2 but some were from smaller river cruises.3 As the pandemic progressed, phylogenetic analysis was critical to investigate outbreaks and transmission dynamics.4
On 29 February 2020, the County of Santa Clara Public Health Department (PHD) in California confirmed new cases of COVID-19 in a couple who returned home from travel to Egypt on a Nile River Cruise and were hospitalized in a local facility with severe respiratory disease.5 The couple had known exposure to a COVID-19 case, originally identified in Taiwan, on the same Nile River cruise. On March 1, the Santa Clara PHD notified CDC about these cases. On the following day, CDC received separate notifications from the Taiwan Centers for Disease Control (Taiwan CDC) about the case identified in Taiwan, and from the Houston Health Department about multiple suspect COVID-19 cases with a recent history of Nile River cruise travel. On 6 March 2020, the Egyptian Health Ministry and World Health Organization reported that 12 new cases of COVID-19 had been registered on a Nile River cruise ship, with authorities subsequently announcing that the vessel was in quarantine with over 150 tourists and local crew on board.6 Nile River cruises differ from large, ocean-liner cruises in that they are much smaller in size (average of 120 passengers per voyage), and in some cases are shorter in duration (3–7 days, travelling between Luxor and Aswan).7
During 11 February–5 March 2020, 101 Nile River cruise-associated COVID-19 cases in the USA were reported among returning travellers to 18 states, nearly doubling the number of detected COVID-19 cases during that period.8 Phylogenetic evidence confirms that Egyptian sequences belong to the same clade as sequences from Europe, the USA and Australia9; other investigations have linked community transmission within Egypt9 and Japan10 to Nile River cruise travellers.
In this report, we expand upon preliminary characterizations of this cluster8–10 by exploring more detailed phylogenetic data and links between cases. Results from this investigation will contribute to the historical understanding of how Nile River cruise travellers seeded outbreaks across the USA and inform future prevention efforts to improve early detection and surveillance of infections among international travellers.
Methods
Definitions
This cross-sectional investigation involved laboratory-confirmed COVID-19 cases, defined as persons with a positive SARS-CoV-2 test result confirmed by real-time reverse transcription-polymerase chain reaction (rRT-PCR) at state laboratories, academic institutions or CDC.11,12 A ‘primary’ case was defined as a laboratory-confirmed COVID-19 case with reported Nile River cruise travel during the 14 days before symptom onset or, for asymptomatic persons, specimen collection date during February–March 2020. A ‘secondary’ case was defined as a laboratory-confirmed COVID-19 case with reported close contact with a primary case per state or local health department contact investigations. [‘Close contact’ at the time was unspecified by CDC or Council of State and Territorial Epidemiologists (CSTE), and therefore the interpretation of this term was at the discretion of state and local health departments.] A ‘tertiary’ case was a laboratory-confirmed case with reported close contact with a secondary case, and a ‘quaternary’ case was a laboratory-confirmed case with reported close contact with a tertiary case. ‘Community transmission’ collectively refers to instances of secondary, tertiary and quaternary transmission events.
Case finding and epidemiologic data collection
After a confirmed case was identified and linked to a specific cruise ship, CDC contacted the respective USA-based tour company operators to obtain complete lists of their Nile River cruise traveller clients and their addresses [authority 42 Code of Federal Regulations Part 71.32(b) as authorized by 42 U.S. Code § 264]. On 2 March 2020, CDC released the first Epidemic Emergency Information Exchange (Epi-X) notification to state and local health departments regarding known returning US passengers that had travelled on a Nile River cruise with laboratory-confirmed COVID-19 cases. State and local health departments investigated these contacts and in turn notified CDC of any confirmed or suspect cases.
When additional Nile River cruise-associated cases were identified, states provided the corresponding ID numbers to facilitate querying the surveillance database Data Collation and Integration for Public Health Event Response (DCIPHER). DCIPHER is a platform through which state and local health departments report epidemiologic surveillance data to CDC.13
Information about the voyages, passenger manifests, and travel dates were reported to CDC by the USA-based tour company operators. All epidemiologic information (demographics, travel history, clinical data) was collected by state and local health departments and reported to CDC via DCIPHER. Clinical data included information about whether patients were hospitalized, admitted to an intensive care unit or died, and whether patients were symptomatic or asymptomatic. We compiled existing records from DCIPHER and verified the completeness and accuracy of the information with state and local health departments. Only a subset of Nile River cruise passengers during February–March 2020 were tested; included in this analysis are all cases for which data were available.
Contact tracing
After the Epi-X notices were released, state and local health departments reported to CDC whether contact tracing was completed for each case, and methods used during contact tracing. When available, states provide information about any secondary, tertiary or quaternary cases identified during contact investigations.
RNA sequences and phylogenetic analysis
All laboratory-confirmed cases were positive by rRT-PCR on nasopharyngeal or oropharyngeal swabs. CDC requested that state health departments share sequence data or Global Initiative on Sharing Avian Flu Data (GISAID) accession numbers (www.gisaid.org) for previously sequenced specimens. When available, other existing specimens were shipped to CDC for whole genome sequencing.14
Phylogenetic relationships were inferred through approximate maximum likelihood analyses implemented in TreeTime15 using the NextStrain pipeline.16 High-quality sequences from US Nile River cruise travellers and representative sequences for the USA (collection dates through 30 April 2020) were downloaded from GISAID on 9 April 2021. PANGO lineage for study sequences were assigned on 12 April 2021. For comparison, we obtained 12 background sequences associated with Nile River cruise travel (primary cases), but not associated with importation to the USA. These included 10 background sequences from February to March 2020 from Japanese Nile River cruise travellers,10 one sequence from a Taiwanese traveller,17 and one sequence from an Egyptian who had contact with a relative who had travelled on a Nile River cruise.9 For comparison, we also obtained 51 background sequences from Egypt with no link to Nile River cruise travel. Sequences uploaded by CDC and previously published sequences are listed in Supplementary Table 1, available at JTM online.
Descriptive epidemiology
Descriptive statistics on the demographic and clinical characteristics of primary and community cases were calculated in R.2.21.18 We developed an epidemic curve of symptom onset date for primary cases and calculated the proportion that developed symptoms before, during and after travel. It was not possible to calculate attack rates by vessel or voyage because not all passengers were tested for SARS-CoV-2.We used Quantum GIS19 to develop maps depicting the number of returned primary cases by state, and the number of secondary/community cases by state.
Ethics statement
This activity was reviewed by CDC and determined to be non-research, public health surveillance. The activity was conducted consistent with applicable federal law and CDC policy (See e.g. 45 C.F.R. part 46, 21 C.F.R. part 56; 42 U.S.C. §241(d); 5 U.S.C. §552a; 44U.S.C. §3501 et seq.). Passenger-level surveillance data were deidentified and analysed at the voyage-level.
Results
Descriptive epidemiology of primary cases
We identified 149 primary cases that travelled on 20 different Nile River voyages and 12 unique vessels [resulting in a median of three detected cases/voyage (IQR: 1.0–16.5) or three detected cases/vessel (IQR: 1.0–18.3)] between 12 February–15 March 2020. Among those with complete data, most were female (55.1%, 81/147), over the age of 60 years (81.1%, 116/143), and identified as White (81.5%, 101/124) and non-Hispanic/Latino (93.6%, 103/110) (Table 1). Four individuals were health care workers (3.9%, 4/103). The 149 primary cases were returned to 67 different counties in 27 different states (Figure 1). The largest numbers of primary cases were from California (n = 23), Texas (n = 17), Florida (n = 16), Iowa (n = 15) and Wisconsin (n = 14).
Table 1.
Demographic and clinical characteristics of 149 primary cases with a history of Nile River cruise travel
| Variable | n | (%) |
|---|---|---|
| Sex | ||
| Female | 81 | (55.1) |
| Male | 66 | (44.9) |
| Missing/unknown | 2 | |
| Age | ||
| <30 | 0 | — |
| 31–45 | 2 | (1.4) |
| 46–60 | 25 | (17.5) |
| 61–75 | 106 | (74.2) |
| >75 | 10 | (7.0) |
| Unknown/missing | 6 | |
| Race | ||
| White | 101 | (81.5) |
| Asian | 22 | (17.7) |
| Native Hawaiian/Pacific Islander | 1 | (0.8) |
| Unknown/missing | 25 | |
| Ethnicity | ||
| Non-Hispanic/Latino | 103 | (93.6) |
| Hispanic/Latino | 7 | (6.4) |
| Unknown/missing | 39 | |
| Health care worker | ||
| Yes | 4 | (3.9) |
| No | 99 | (96.1) |
| Unknown/missing | 46 | |
| Symptomatic | ||
| Yes | 139 | (97.2) |
| No | 4 | (2.8) |
| Unknown/missing | 6 | |
| Death as a result of infectiona | ||
| Yes | 6 | (4.7) |
| No | 122 | (95.3) |
| Unknown/missing | 21 | |
| Hospitalized | ||
| Yes | 38 | (28.1) |
| No | 97 | (71.9) |
| Unknown/missing | 14 | |
| Admitted to intensive care unitb | ||
| Yes | 11 | (35.5) |
| No | 20 | (60.5) |
| Unknown/missing | 7 | |
| Required mechanical ventilationb | ||
| Yes | 6 | (25) |
| No | 18 | (75) |
| Unknown/missing | 13 |
Among the 6 patients that died, 6 were hospitalized, 4 were admitted to an intensive care unit and 3 were ventilated.
Among persons hospitalized (n = 38).
Figure 1.
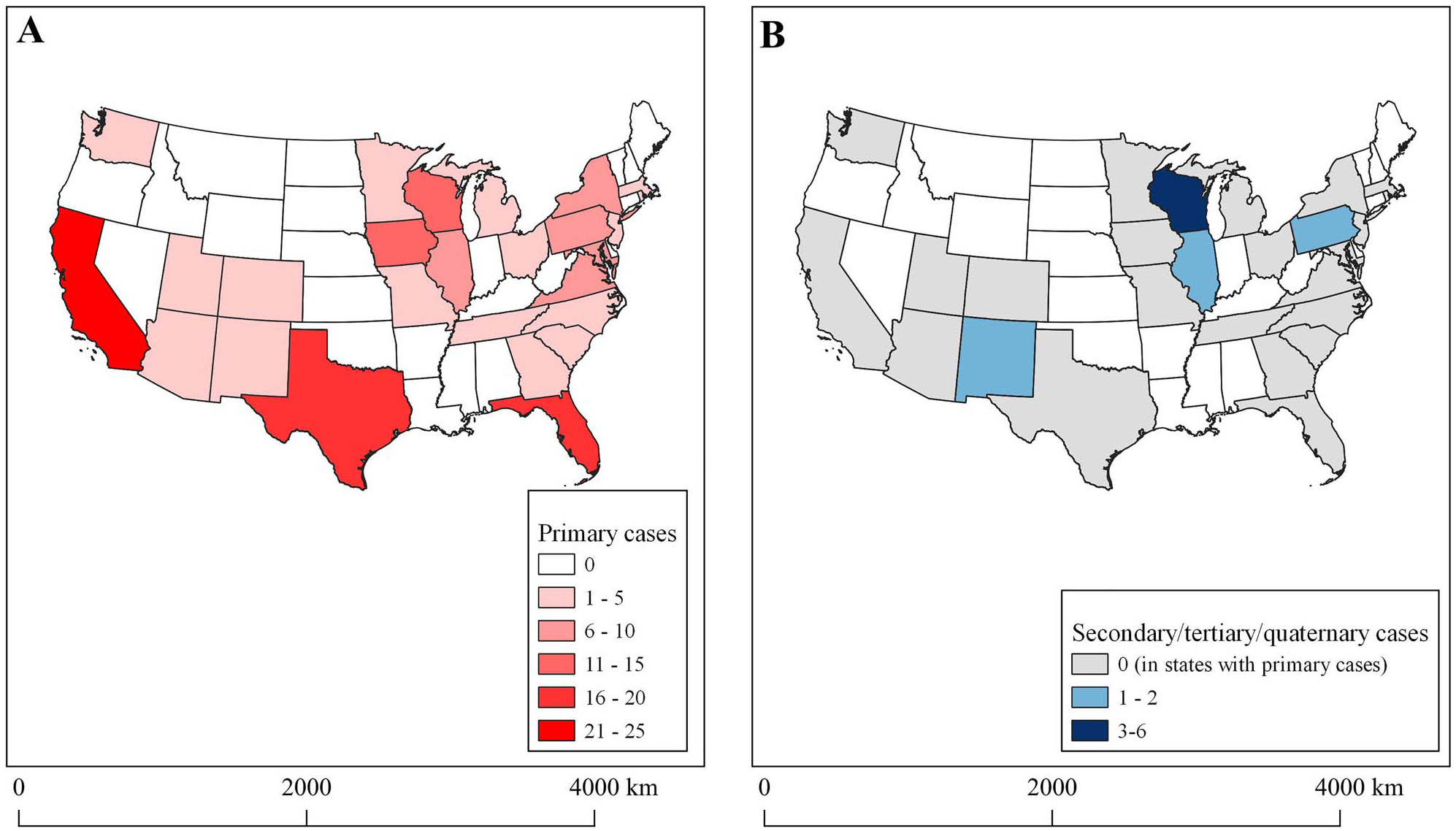
States of residence for 149 primary cases and 15 secondary, tertiary and quaternary cases associated with Nile River cruise travel. A detailed contact investigation was conducted in Wisconsin, resulting in the identification of more community transmission events.
Among those with complete data, 97.2% (139/143) of primary cases were symptomatic and 2.8% (4/143) were asymptomatic (6 unknown). Symptom onset dates ranged from 17 February to 17 March 2020 (Figure 2). About 75% of travellers (94/126) experienced symptom onset after completion of the cruise, 22.2% (28/126) reported symptoms while cruising, and 3.2% (4/126) had symptom onset dates before the cruise (no data for 23 people). Among the 135 primary cases with complete data, 28.1% (38/135) were hospitalized as a result of SARS-CoV-2 infection. Among the primary cases that were hospitalized and had complete data, 35.5% (11/31) were admitted to the ICU and 25% (6/24) required mechanical ventilation. Six patients ultimately died 4.7% (6/128); all six were hospitalized and 50% (3/6) required mechanical ventilation.
Figure 2.
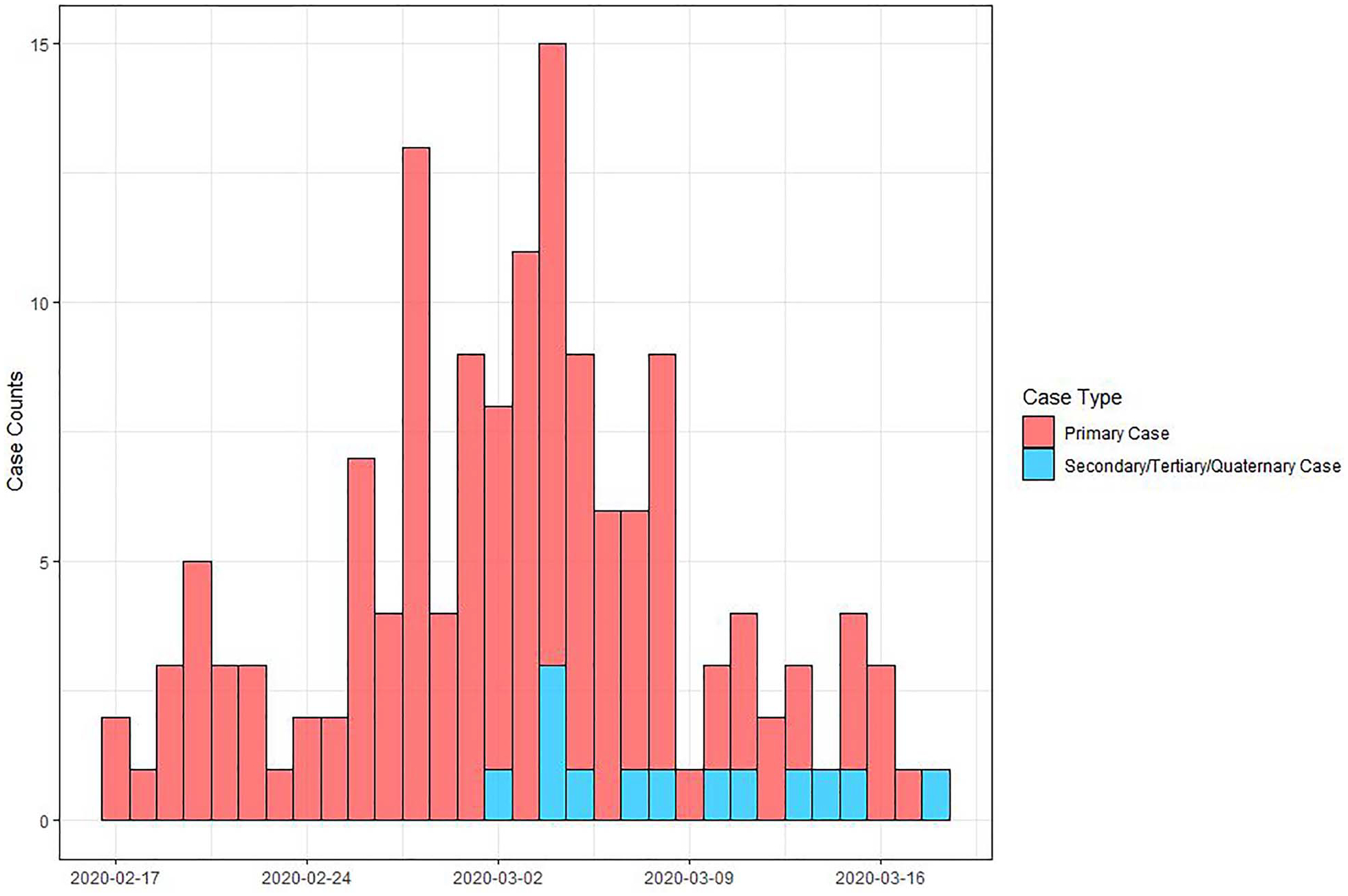
Epidemic curve of symptom onset date for primary and secondary, tertiary, and quaternary cases associated with Nile River cruise travel. Symptom onset date was available for 133 primary cases (missing for 16) and 13 secondary/tertiary/quaternary community cases (missing for 2 cases).
Community transmission
Contact tracing was conducted for 37.6% (56/149) of primary cases. [Answer marked as ‘unknown’ for 15.4% (23/149) of primary cases.] Fifteen states conducted household-level contact investigations, with more extensive investigations conducted in Wisconsin and New Mexico.
We identified 15 community cases (including tertiary and quaternary cases), 11 of which occurred in Wisconsin (Supplementary Table 2 is available at JTM online). Demographic and clinical characteristics among primary and community cases were similar (Supplementary Table 3 is available at JTM online). Phylogenetic analysis supported transmission links for 6/7 household pairs. One out of seven pairs who travelled together had genetically distinct sequences, indicating separate sources of infection. The sequences differed by 6 single nucleotide polymorphisms (SNPs) although one sequence was missing ~7% of its genome. This pair, from California, had travelled together on the same Nile River cruise February 12–19, returned to the USA on February 20, and both developed symptoms on February 21. Nile River cruise-associated transmission chains also occurred in Wisconsin and in New Mexico, described in detail below.
On 11 March 2020, the Wisconsin Department of Health Services (WDHS) was notified of a confirmed case of COVID-19 in a patient who had been hospitalized after returning from a Nile River cruise. In response, detailed case investigations and contact tracing interviews were conducted; identified contacts were also tested through the state public health laboratory if they reported symptoms meeting criteria at the time.11 The investigation revealed that the initial patient had travelled with a tour group of 19 travellers during 20 February–2 March 2020. Among the travellers, 16 were tested and all received positive SARS-CoV-2 RT-PCR results. (Two Wisconsin residents who were primary Nile River cruise cases travelled to Arizona and were reported to CDC via the Arizona Department of Health and Human Services.) A total of seven confirmed secondary cases were identified from Wisconsin (Supplementary Figure 1 is available at JTM online). Events that were linked to secondary infections included two lunches, a dinner party, and social visits with extended family. Contact investigations with the seven secondary cases identified three tertiary cases and one quaternary case. All genome sequences from Wisconsin differed by only 0–1 SNPs, suggesting a possible shared source of infection.
On 11 March 2020, the New Mexico Department of Health reported cases with a history of domestic air travel to the CDC Quarantine Station in San Diego, CA. An air contact tracing investigation was launched (authority 42 Code of Federal Regulations Part 71.32(b) as authorized by 42 U.S. Code § 264) and the airline was required to provide CDC with passenger contact information.20,21 All passengers and crew were contacted for questioning about symptoms, travel history, and whether they had been tested for SARS-CoV-2.
The investigation identified two primary cases that travelled on a 5-h flight from New York City to Albuquerque on 27 February 2020, the last leg of their return home from Egypt. The two primary cases were symptomatic at the time of the flight. The investigation uncovered two additional cases, who were seated in the row behind the secondary cases. Sequences from these four travellers differed by 0–4 SNPS, suggesting that they were linked.
Phylogenetic analysis
A total of 94 sequences were available for analysis, including 82 Nile River cruise-associated sequences from the USA and 12 non-USA Nile River cruise-associated sequences that were previously published. Most sequences (93%, 87/94) were in Nextstrain clade 20A, but two sequences (from California and Virginia) were in clade 19A, and one (from Utah) was in 20C (Figure 3).
Figure 3.
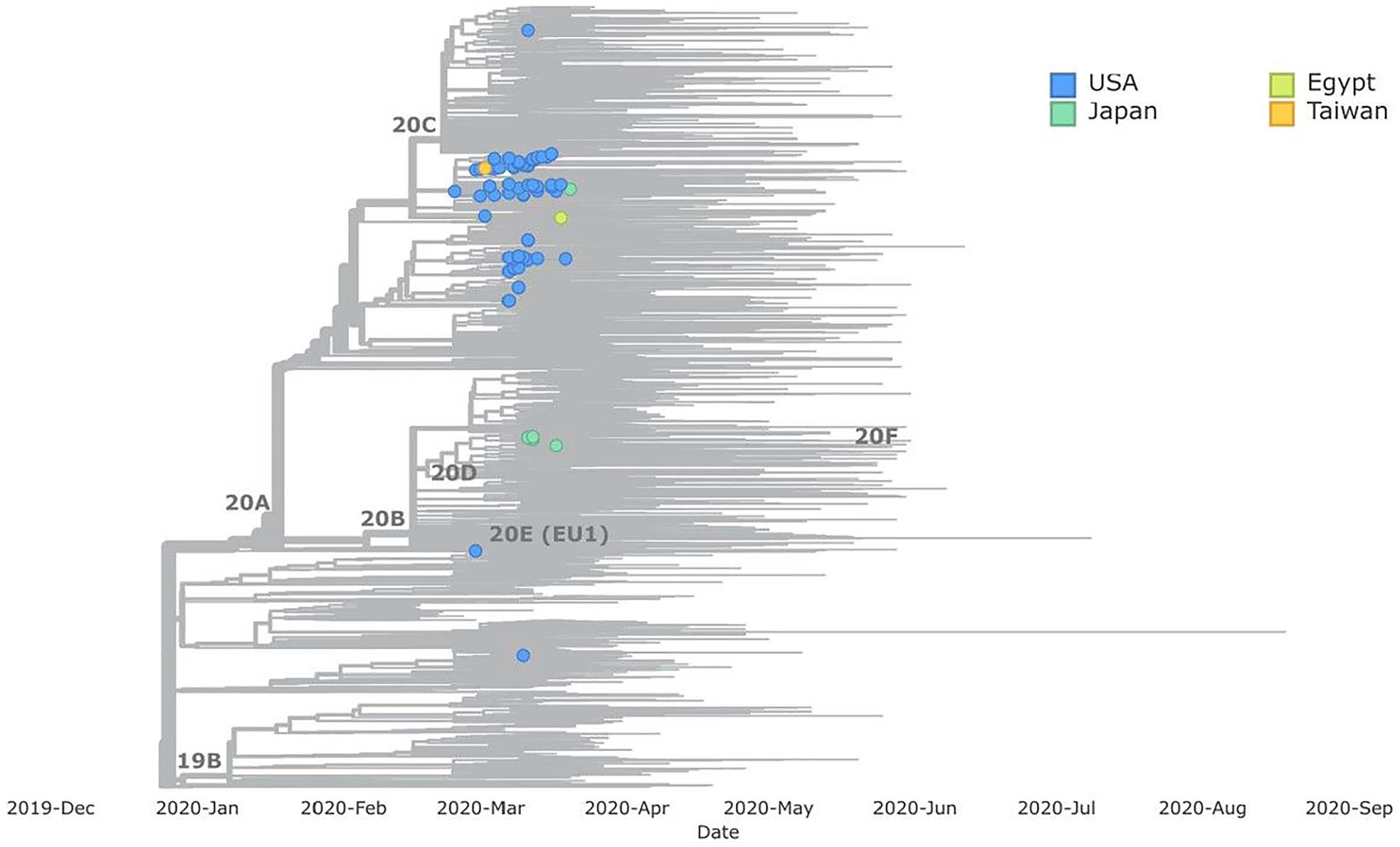
Phylogenetic tree including sequences from Nile River cruise travellers returning to the USA (n = 82), sequences from Egypt (n = 1), Japan (n = 10) and Taiwan (n = 1). Global background sequences are shown in grey. Sequences were only available for 82/164 primary and community cases associated with Nile cruise travel.
Of the 10 publicly available Nile River cruise-associated background sequences from Japan, 5 fell into Nextstrain clade 20A and 5 fell into 20B. Most (86%, 44/51) background sequences from Egypt with no known affiliation with Nile River cruise travel were in clade 20A. Among the US cruise passengers, more than half of the sequences belonged to the B.1.255 PANGO lineage, followed by the B.1.5 lineage (Supplementary Figure 2 is available at JTM online).
Some clustering was observed by voyage and vessel (Figure 4). Sequences from the vessels M/S Esplanade and Oberoi Zahra grouped together, while sequences from the M/S A’Sara, M/S Sonesta, M/S Queen of Hansa and Steigenerger Oman grouped together. Sequences from sequential voyages on the M/S A’Sara and M/S Esplanade were similar, supporting that there were continued outbreaks on the same vessel.
Figure 4.
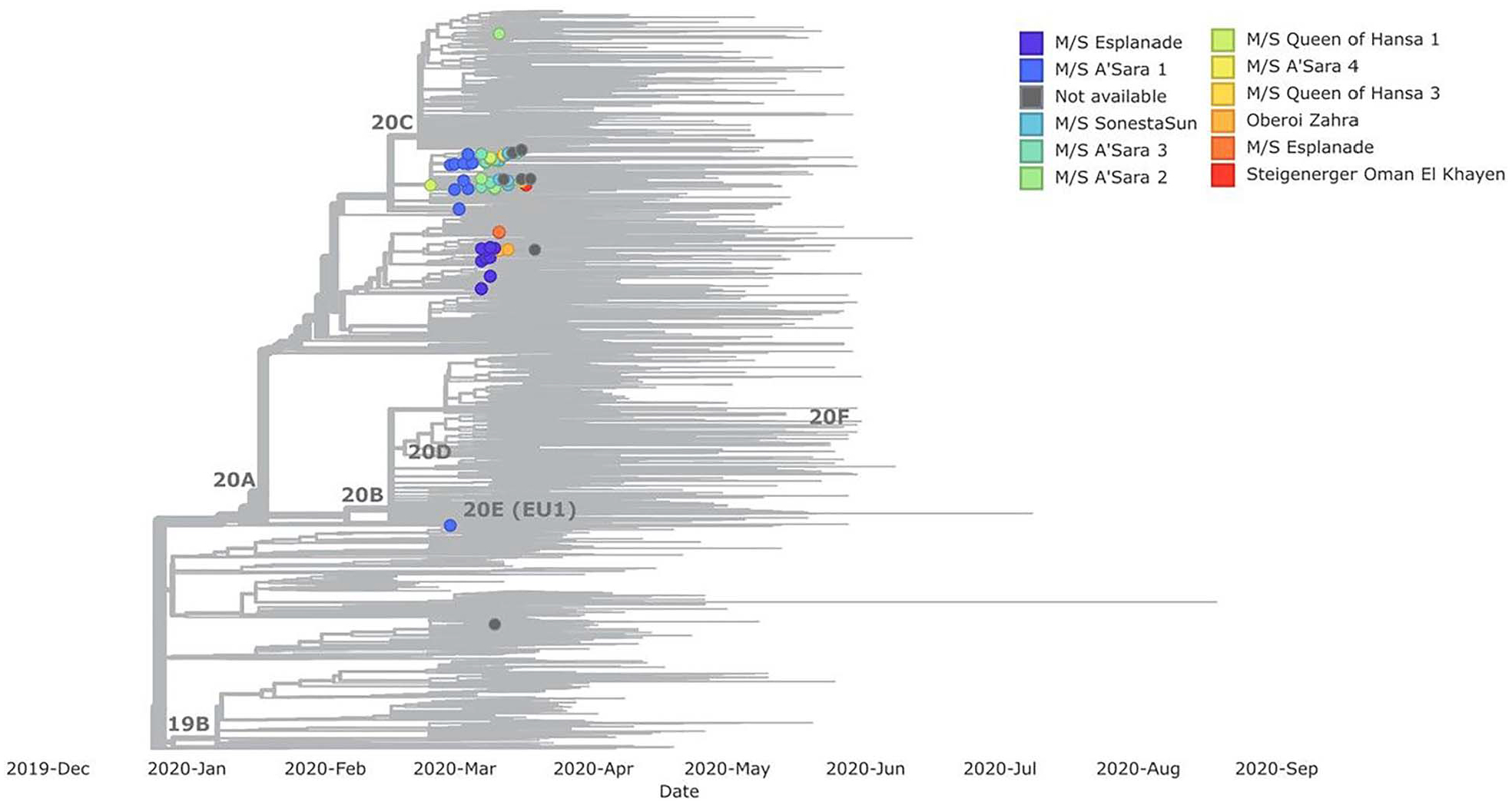
Phylogenetic tree of sequences from Nile River cruise travellers (n = 69), organized by voyage. Among the 82 Nile River cruise travellers, vessel information was not available for 13 people
Discussion
We detected 149 individuals who had a history of Nile River cruise travel during February–March 2020 and also tested positive for SARS-CoV-2. These individuals returned to 67 counties in 27 states, potentially spreading SARS-CoV-2 throughout the US early in the pandemic. However, it is likely that many more cases that engaged in Nile River cruise travel went undetected upon return to the USA.
Most previously reported examples of COVID-19 outbreaks among returning travellers were detected when large groups of people returned to a single location within the USA—for example, outbreaks were detected among college students returning from Mexico to Austin, Texas,22 and among travellers from Wuhan, China who returned to California and Seattle.8 In contrast, the Nile River cruise-associated cases reported here consisted of smaller travel groups that returned to many different locations. CDC was only able to identify these cases via a detailed investigation involving collaboration across 67 public health jurisdictions. The US public health system necessitates coordination across many state and local health departments and federal bodies, posing major challenges to case tracking and timely detection of novel variants. Future efforts aimed at SARS-CoV-2 genomic surveillance among air travellers to the US could help fill this gap by providing an early warning system.23
Travellers on cruises are exposed to transmission risk during the cruise, but also during travel to and from the cruise embarkation and disembarkation ports, often via airplane. In this cohort of Nile River cruise travellers, we identified two individuals that were likely infected with SARS-CoV-2 during a 5-h cross-country flight from New York City to New Mexico (in addition to the two primary cases). Points of disembarkation during which travellers mingle with local communities might also serve as opportunities for infection—this could possibly explain why one pair of travellers from California were infected with genetically distinct viral sequences.
Our results showed that the mutations we identified from Nile River cruise sequences were similar to sequences from throughout the USA and therefore are not unique to just Nile River cruise travellers. Consequently, we are unable to determine the extent to which Nile River cruise travellers seeded outbreaks. Still, the travellers returned to 67 counties, with high probability of introducing the virus over a large geographic area. Other works have concluded that outbreaks were caused by Nile River cruise travellers returning to Japan,10 although this analysis is subject to the same limitations we describe here.
Some limitations of this analysis should be noted. Not all individuals aboard the 12 affected boats were tested, and therefore we were unable to estimate vessel-specific attack rates. Access to testing at the time was extremely limited and usually reserved for symptomatic cases. It is estimated that up to 45% of infections are asymptomatic,24 and it is therefore probable that many asymptomatic cases with a history of Nile River cruise travel were never identified. There are an estimated 200 riverboats operating on the Nile River,7 and the current report only captured 12 of the affected vessels. Further, CDC was also only able to request Nile River cruise manifests from USA-based tour company operators, and consequently cases associated with other tour operators were not captured. Because resources available for contact-tracing across the USA were highly variable at the time, our capacity to identify long transmission chains in many states was limited. In Wisconsin, a detailed contact investigation was conducted, and more instances of community transmission were identified in that state.
Between 1 February 2020 and 6 April 2020, COVID-19 outbreaks on cruise ships triggered 27 CDC notifications to state, local and international health departments for over 11 000 cruise ship passengers.24 Although shorter duration cruises (<7 days) like those touring the Nile River are associated with smaller SARS-CoV-2 outbreaks,3 our data suggest that short cruises are still vulnerable to onboard transmission, given that travellers from all over the world congregate in these confined spaces. Other available data suggest that outbreaks on smaller cruise vessels (e.g. Greg Mortimer) might result in higher attack rates25 because of the increased density of people on board. The median number of detected cases per vessel on Nile River cruises (median = 3, IQR: 1.0–18.3) is similar to that reported previously on large cruise ships (median = 3, IQR: 1.0–17.8).26 Some cruise ships experienced very large outbreaks affecting hundreds of people (e.g. the Diamond Princess had 712 reported cases),3,26 whereas the maximum number of cases observed on any single Nile River voyage was 20.
Because cruises involve large groups of people sharing confined spaces for long periods of time, layered mitigation efforts (i.e. pre-embarkation testing, reduction of duration and port visits) can help reduce SARS-CoV-2 transmission. CDC’s No Sail Order to restrict passenger operations on cruise ships under US jurisdiction (March 2020–October 2020) applied to large passenger vessels (250 or more individuals).25 Similarly any conditional sail orders or subsequent programmes did not apply to international river cruises as described in this report. River cruises in the USA fall under the jurisdiction of the Food and Drug Administration (FDA), under Section 361 of the Public Health Service Act, which states that FDA may inspect US flagged vessels engaged in interstate traffic to prevent the introduction and transmission of communicable diseases. However, cruises that operate overseas on the Nile River are not subject to US regulations.
River cruises are smaller in size, shorter in duration, and are often overlooked during outbreak investigations or medical histories. Yet they could be responsible for widespread introductions of virus in the early phases of disease emergence. Surveillance and contact tracing during respiratory virus outbreaks should therefore include travellers on river cruises or should ask patients about history of river cruise travel.
Supplementary Material
Acknowledgements
We thank Seema Jain (California Department of Public Health), Amanda Kamali (California Department of Public Health), Kimberly Mueller (Fond du Lac County, Wisconsin), Tye Harlow (Colorado Department of Public Health and Environment), and Brittany Liebhard (CDC).
Funding
This work was supported by routine funding from the Centers for Disease Control and Prevention.
Footnotes
Conflict of interest: The authors have declared no conflicts of interest.
Disclaimer
The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of CDC.
Data availability
The data underlying this article cannot be shared publicly in order to protect to the privacy of individuals associated with this study. Aggregate data will be shared on reasonable request to the corresponding author.
References
- 1.Coronavirus Disease 2019 (COVID-19): Cases in U.S 2020. https://covid.cdc.gov/covid-data-tracker/?CDC_AA_refVal=https%3A%2F%2Fwww.cdc.gov%2Fcoronavirus%2F2019-ncov%2Fcases-in-us.html#datatracker-home (10 August 2022, date last accessed).
- 2.Moriarty LF, Plucinski MM, Marston BJ et al. Public health responses to COVID-19 outbreaks on cruise ships - worldwide, February–March 2020. MMWR Morb Mortal Wkly Rep 2020; 69:347–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guagliardo SAJ, Prasad PV, Rodriguez A et al. Cruise ship travel in the era of COVID-19: a summary of outbreaks and a model of public health interventions. Clin Infect Dis 2022; 74:490–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lemieux JE, Siddle KJ, Shaw BM et al. Phylogenetic analysis of SARS-CoV-2 in Boston highlights the impact of superspreading events. Science 2021; 371:eabe3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Villarino E, Deng X, Kemper CA et al. Introduction, transmission dynamics, and fate of early severe acute respiratory syndrome coronavirus 2 lineages in Santa Clara County. California J Infect Dis 2021; 224:207–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Raghavan S, Kornfield M. The Tale of How a Nile Cruise Boosted the International Coronavirus Outbreak. Washington, DC.: The Washington Post, 2021. https://www.washingtonpost.com/world/middle_east/the-tale-of-a-nile-cruise-that-spawned-an-international-coronavirus-outbreak/2020/03/13/6ab633fc-6314-11ea-8a8e-5c5336b32760_story.html (30 May 2022, date last accessed). [Google Scholar]
- 7.Buff AM, Reaves EJ. “Egypt & Nile River Cruises.” Chapter 10: popular itineraries Africa & the Middle East, In: CDC Yellow Book. New York: Health Information for International Travel. Oxford University Press. New York, 2020. [Google Scholar]
- 8.Schuchat A,CDC COVID-19 Response Team. Public health response to the initiation and spread of pandemic COVID-19 in the United States, February 24-April 21, 2020. MMWR Morb Mortal Wkly Rep 2020; 69:551–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kandeil A, Mostafa A, Rabeh E-S et al. Coding-complete genome sequences of two SARS-CoV-2 isolates from Egypt. Microbiol Resour Announc 2020; 9:e00489–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sekizuka T, Kuramoto S, Nariai E et al. SARS-CoV-2 genome analysis of Japanese Travelers in Nile River cruise. Front Microbiol 2020; 11:1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Council of State and Territorial Epidemiologists. Interim Position Statement: Update to COVID-19 Case Definition. 2020. https://www.cste.org/news/520707/CSTE-Interim-Position-Statement-Update-to-COVID-19-Case-Definition.htm (18 May 2021, date last accessed).
- 12.Council of State and Territorial Epidemiologists. Coronavirus Disease 2019 (COVID-19) 2020 Interim Case Definition, 2020 CSTE Position Statement; 2020. https://ndc.services.cdc.gov/case-definitions/coronavirus-disease-2019-2020/ (18 May 2021, date last accessed). [Google Scholar]
- 13.Gold JAW, DeCuir J, Coyle JP et al. COVID-19 case surveillance: trends in person-level case data completeness, United States, April 5–September 30, 2020. Public Health Rep 2021; 136: 466–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Centers for Disease Control and Prevention. CDC Comprehensive SARS-CoV-2 Sequencing Protocols. 2020. https://github.com/CDCgov/SARS-CoV-2_Sequencing/tree/master/protocols/CDC-Comprehensive (1 June 2022, date last accessed).
- 15.Sagulenko P, Puller V, Neher RA. TreeTime: maximum-likelihood phylodynamic analysis. Virus Evol 2018; 4:vex042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hadfield J, Megill C, Bell SM et al. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics 2018; 34:4121–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang JT, Lin YY, Chang SY et al. The role of phylogenetic analysis in clarifying the infection source of a COVID-19 patient. J Infect 2020; 81:147–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.R Core Development Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing, 2019. [Google Scholar]
- 19.QGIS Development Team. Quantum Geographic Information System. Open Source Geospatial Foundation Project, 2019. [Google Scholar]
- 20.Regan JJ, Jungerman MR, Lippold SA et al. Tracing airline Travelers for a public health investigation: Middle East respiratory syndrome coronavirus (MERS-CoV) infection in the United States, 2014. Public Health Rep 2016; 131: 552–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vogt TM, Guerra MA, Flagg EW, Ksiazek TG, Lowther SA, Arguin PM. Risk of severe acute respiratory syndrome-associated coronavirus transmission aboard commercial aircraft. J Travel Med 2006; 13:268–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Centers for Disease Control and Prevention. COVID-19 outbreak among college students after a spring break trip to Mexico—Austin, Texas, March 26–April 5, 2020. MMWR Morb Mortal Wkly Rep 2020; 69:830–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wegrzyn RD, Appiah GD, Morfino R et al. Early detection of SARS-CoV-2 variants using traveler-based genomic surveillance at four US airports, September 2021–January 2022. Clin Infect Dis, 2022, p. ciac461. https://pubmed.ncbi.nlm.nih.gov/35686436/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Centers for Disease Control and Prevention; No Sail Order and Suspension of Further Embarkation; Notice of Modification and Extension and Other Measures Related to Operations. 2020. https://www.federalregister.gov/documents/2020/04/15/2020-07930/no-sail-order-and-suspension-of-further-embarkation-notice-of-modification-and-extension-and-other. (11 June 2022, date last accessed).
- 25.Ing AJ, Cocks C, Green JP. COVID-19: in the footsteps of Ernest Shackleton. Thorax 2020; 75:693–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Willebrand KS, Pischel L, Malik AA, Jenness SM, Omer SB. A review of COVID-19 transmission dynamics and clinical outcomes on cruise ships worldwide, January to October 2020. Euro Surveill 2022; 27:2002113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article cannot be shared publicly in order to protect to the privacy of individuals associated with this study. Aggregate data will be shared on reasonable request to the corresponding author.


