Abstract
STUDY QUESTION
Is the Tcte1 mutation causative for male infertility?
SUMMARY ANSWER
Our collected data underline the complex and devastating effect of the single-gene mutation on the testicular molecular network, leading to male reproductive failure.
WHAT IS KNOWN ALREADY
Recent data have revealed mutations in genes related to axonemal dynein arms as causative for morphology and motility abnormalities in spermatozoa of infertile males, including dysplasia of fibrous sheath (DFS) and multiple morphological abnormalities in the sperm flagella (MMAF). The nexin–dynein regulatory complex (N-DRC) coordinates the dynein arm activity and is built from the DRC1–DRC7 proteins. DRC5 (TCTE1), one of the N-DRC elements, has already been reported as a candidate for abnormal sperm flagella beating; however, only in a restricted manner with no clear explanation of respective observations.
STUDY DESIGN, SIZE, DURATION
Using the CRISPR/Cas9 genome editing technique, a mouse Tcte1 gene knockout line was created on the basis of the C57Bl/6J strain. The mouse reproductive potential, semen characteristics, testicular gene expression levels, sperm ATP, and testis apoptosis level measurements were then assessed, followed by visualization of N-DRC proteins in sperm, and protein modeling in silico. Also, a pilot genomic sequencing study of samples from human infertile males (n = 248) was applied for screening of TCTE1 variants.
PARTICIPANTS/MATERIALS, SETTING, METHODS
To check the reproductive potential of KO mice, adult animals were crossed for delivery of three litters per caged pair, but for no longer than for 6 months, in various combinations of zygosity. All experiments were performed for wild-type (WT, control group), heterozygous Tcte1+/− and homozygous Tcte1−/− male mice. Gross anatomy was performed on testis and epididymis samples, followed by semen analysis. Sequencing of RNA (RNAseq; Illumina) was done for mice testis tissues. STRING interactions were checked for protein–protein interactions, based on changed expression levels of corresponding genes identified in the mouse testis RNAseq experiments. Immunofluorescence in situ staining was performed to detect the N-DRC complex proteins: Tcte1 (Drc5), Drc7, Fbxl13 (Drc6), and Eps8l1 (Drc3) in mouse spermatozoa. To determine the amount of ATP in spermatozoa, the luminescence level was measured. In addition, immunofluorescence in situ staining was performed to check the level of apoptosis via caspase 3 visualization on mouse testis samples. DNA from whole blood samples of infertile males (n = 137 with non-obstructive azoospermia or cryptozoospermia, n = 111 samples with a spectrum of oligoasthenoteratozoospermia, including n = 47 with asthenozoospermia) was extracted to perform genomic sequencing (WGS, WES, or Sanger). Protein prediction modeling of human-identified variants and the exon 3 structure deleted in the mouse knockout was also performed.
MAIN RESULTS AND THE ROLE OF CHANCE
No progeny at all was found for the homozygous males which were revealed to have oligoasthenoteratozoospermia, while heterozygous animals were fertile but manifested oligozoospermia, suggesting haploinsufficiency. RNA-sequencing of the testicular tissue showed the influence of Tcte1 mutations on the expression pattern of 21 genes responsible for mitochondrial ATP processing or linked with apoptosis or spermatogenesis. In Tcte1−/− males, the protein was revealed in only residual amounts in the sperm head nucleus and was not transported to the sperm flagella, as were other N-DRC components. Decreased ATP levels (2.4-fold lower) were found in the spermatozoa of homozygous mice, together with disturbed tail:midpiece ratios, leading to abnormal sperm tail beating. Casp3-positive signals (indicating apoptosis) were observed in spermatogonia only, at a similar level in all three mouse genotypes. Mutation screening of human infertile males revealed one novel and five ultra-rare heterogeneous variants (predicted as disease-causing) in 6.05% of the patients studied. Protein prediction modeling of identified variants revealed changes in the protein surface charge potential, leading to disruption in helix flexibility or its dynamics, thus suggesting disrupted interactions of TCTE1 with its binding partners located within the axoneme.
LARGE SCALE DATA
All data generated or analyzed during this study are included in this published article and its supplementary information files. RNAseq data are available in the GEO database (https://www.ncbi.nlm.nih.gov/geo/) under the accession number GSE207805. The results described in the publication are based on whole-genome or exome sequencing data which includes sensitive information in the form of patient-specific germline variants. Information regarding such variants must not be shared publicly following European Union legislation, therefore access to raw data that support the findings of this study are available from the corresponding author upon reasonable request.
LIMITATIONS, REASONS FOR CAUTION
In the study, the in vitro fertilization performance of sperm from homozygous male mice was not checked.
WIDER IMPLICATIONS OF THE FINDINGS
This study contains novel and comprehensive data concerning the role of TCTE1 in male infertility. The TCTE1 gene is the next one that should be added to the ‘male infertility list’ because of its crucial role in spermatogenesis and proper sperm functioning.
STUDY FUNDING/COMPETING INTEREST(S)
This work was supported by National Science Centre in Poland, grants no.: 2015/17/B/NZ2/01157 and 2020/37/B/NZ5/00549 (to M.K.), 2017/26/D/NZ5/00789 (to A.M.), and HD096723, GM127569-03, NIH SAP #4100085736 PA DoH (to A.N.Y.). The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
Keywords: TCTE1, N-DRC, male infertility, oligoasthenoteratozoospermia, sperm motility, sperm mitochondria, mt-Co2, Fetub, haploinsufficiency, MMAF
WHAT DOES THIS MEAN FOR PATIENTS?
Abnormal semen parameters (sperm count, motility, and morphology) are the most obvious symptoms that may be related to male fertility problems. The main structural element responsible for sperm motility is the sperm flagellum (tail), which is controlled by a specific sperm structure, the axoneme. This study aimed to determine the role of Tcte1 gene in male infertility using a mouse model of the gene inactivation. The Tcte1 protein is one of the structural elements building N-DRC, a complex within the sperm axoneme that is responsible for the coordination of activities of the sperm tail related to sperm movement. We have found that mutations in the mouse Tcte1 switched-off gene model revealed two phenotypes, dependent on the number of affected alleles (or zygosity): infertile homozygotic males (with two identical switched-off alleles) with oligoasthenoteratozoospermia (reduced sperm number, motility and morphology) and fertile heterozygotic males (with one switched-off and normal allele) with oligozoospermia (reduced sperm number only). A pilot study on human samples with TCTE1 gene variants revealed a wide spectrum in quality of semen including non-obstructive azoospermia (no spermatozoa in ejaculate), cryptozoospermia (few spermatozoa in ejaculate), and severe oligoasthenozoospermia (severely reduced sperm number and motility). Thus, the TCTE1 gene is the next one that should be added to the ‘male infertility list’ because of its crucial role in spermatogenesis, affecting a variety of testicular molecular networks (including energy machinery processing) and proper sperm function.
Introduction
Infertility has been defined by the World Health Organization as a social disease concerning ∼10–18% of couples of reproductive age who are unable to conceive within 1 year of regular unprotected sexual intercourse (WHO, 2020). Approximately 7% of males and ∼12% of females worldwide have reproductive problems, while the male factor is responsible for ∼20–30% of all infertility cases (Barratt et al., 2017; Vander Borght and Wyns, 2018; Agarwal et al., 2021). Male infertility is a disease with multifactorial etiology, including genetic factors, chromosomal aberrations, epigenetic mutations, hormonal abnormalities, infections, or reproductive tract abnormalities (Matzuk and Lamb 2008; Zorrilla and Yatsenko, 2013; Krausz and Riera-Escamilla, 2018; Agarwal et al., 2021). Genetic causes of male infertility can be detected in ∼10–15% of cases, while for 25–50% of infertile males, the etiology remains unexplained (Matzuk and Lamb, 2008; Zorrilla and Yatsenko, 2013; Agarwal et al., 2021). The male infertility phenotype mostly results in abnormal semen parameters, including low sperm concentration, abnormal morphology, or diminished motility. The actual standards for genetic testing of male infertility rely on chromosomal screening for structural and numerical abnormalities, followed by analyses of deletions in the AZF (azoospermia factor) regions of the Y chromosome (Hwang et al., 2018). However, it is still not sufficient due to the fact that ∼2000 genes may be related to spermatogenesis, while only ∼10% of them seem to have a documented fertility role, so far (Krausz and Riera-Escamilla, 2018; Houston et al., 2021). In recent years, the development of NGS technology, including genomic sequencing of DNA (exome or genome for large cohorts of patients) and RNA (RNAseq, scRNAseq on reproductive tissues), followed by animal models of infertility (mostly mouse models with more than 860 genes evaluated, so far (Mouse Genome Informatics, 24 June 2021)) have started the new era of exploration on a massive scale (Wyrwoll et al., 2020; Hardy et al., 2021; Houston et al., 2021; Oud et al., 2021; Xavier et al., 2021).
Besides the genes related to spermatogenesis and low sperm count, also studies of candidates responsible for diminished sperm motility are pending. The main structural element responsible for motility, the sperm flagellum, has to be of high quality to guarantee sperm motility and then fertilization success. It is known that mutations in genes encoding factors involved in the ciliogenesis process, lead to defects in the structure and/or functioning of cilia (Girardet et al., 2019; Sironen et al., 2020; Kumar and Reiter, 2021; Smith et al., 2020). There are plenty of data concerning respiratory cilia; however, still little is known about the axonemal dynein arms functioning in the sperm tail. The latest data have revealed mutations in genes related to axonemal dynein arms as causative for morphology and motility abnormalities in the spermatozoa of infertile males (Girardet et al., 2019; Sironen et al., 2020; Aprea et al., 2021; Lorès et al., 2021; Touré et al., 2021; Martinez et al., 2022). Two definitions were established: (i) for sperm cells with disorganization in axonemal structures, so-called dysplasia of fibrous sheath (DFS) (Chemes et al., 1998), and (ii) for multiple morphological abnormalities in the sperm flagella (MMAF), concerning all possible defects of the sperm tail, mainly short and thick flagella, and a lack of the central pair (CP) of microtubules or dynein arms (Ben Khelifa et al., 2014). The observed spectrum of abnormal sperm tail phenotypes allows the qualification of some of the disorders into both listed groups (DFS and MMAF) (Sironen et al., 2020; Touré et al., 2021).
The basic structural element of motile cilia and flagella is the axoneme which is built from nine microtubules arranged in doublets positioned radially and one central pair (CP) of singlet microtubules. This central pair is connected to radial spokes (RS), which are: protein complexes of the mechanical regulatory network responsible for the regular waveform beating of the cilia (Gui et al., 2019). RS are linked to inner dynein arms (IDA) localized within each outer doublet. Each outer doublet carries outer dynein arms (ODA), which are: multi-subunit motor protein complexes, responsible for the generation and regulation of the sliding between dynein arms leading to ATP-dependent beating (Satir et al., 2014; Wilson et al., 2015). The 9 + 2 basic structure of the axoneme is evolutionary conserved (Ishikawa 2017; Girardet et al., 2019; Touré et al. 2021). Doublets are linked with the nexin–dynein regulatory complex (N-DRC), which coordinates the dynein arm activity and stabilizes the attachment of doublet microtubules. The N-DRC is built from the DRC1–DRC7 proteins, among which DRC1, DRC2, and DRC4 are responsible for physical linkage between the A and B microtubules of doublets (Wirschell et al., 2013; Bower et al., 2018; Gui et al., 2019; Morohoshi et al., 2020). In terms of male infertility, the best studied gene among the N-DRC is DRC7 (MIM: 618769), which is amenable for proper assembly of the N-DRC structure (in Drc7−/− male mice, the length of the sperm tail was reduced, followed by abnormalities of sperm head morphology) (Morohoshi et al., 2020). A mouse model of the Drc6 (Fbxl13) gene (MIM: 609080) revealed no essential role in sperm formation and motility (Morohoshi et al., 2020); however, its disruption together with CEP192 (MIM: 616426) leads to defects in HEK293T cell motility (Fung et al., 2018). Mutations in the Drc3 gene (MIM: 618758) indirectly revealed male infertility as the reason for a lack of pregnancy with wild-type females (Ha et al., 2016).
One of the N-DRC structural genes is TCTE1 (T-Complex-associated-Testis-Expressed 1), also known as DRC5, FAP155, and D6S46 in humans (locus 6p21.1; MIM: 186975) or Tcte1, D17S, Tcte-1, and D17Sil1 in mice (locus 17 B3; 17 22.54 cM; MGI: 98640). TCTE1 is evolutionary conserved in most eukaryotic organisms that contain motility cilia or flagella in their life cycle (Fig. 1) and is required for proper flagella functioning. Expression of TCTE1 is observed to be highest in the testis (with expression starting at the spermatid stage) but is also present in the choroid plexus and Fallopian tube. TCTE1 contains five leucine-rich repeats (LRRs). To date, only two reports have described a role for the Tcte1 gene: as a mice model of infertility, where the Tcte1-null sperm cells exhibited asthenozoospermia, or in a group of patients without a clear explanation of the linkage between the observed variant and sperm phenotype (Castaneda et al., 2017; Zhou et al., 2022). Thus, our understanding of the nature of this specific motility-type defect in Tcte1-null observations has been limited so far.
Figure 1.
Characteristics of the Tcte1 gene. (A) Putative homologs of the Tcte1 gene conserved in Bilateria, according to HomoloGene (8434), including collation of proteins and their conserved domains. (B) Sequence alignment between human, mouse, and rat. (C) NCBI Gene schematic representation of the Tcte1 gene in Mus musculus.
In our study, we aimed to determine the role of Tcte1 in male infertility using a mouse knockout model. We created homo- and heterozygous animals, which were then examined for their reproductive potential, differences in anatomy and histology, and semen parameters. We also performed RNAseq in testis tissue to obtain an answer about the influence of Tcte1 null mutations on the expression pattern of other gonadal genes. In addition, ATP measurements of spermatozoa were performed. Finally, human samples from males with severely decreased sperm counts were screened for TCTE1 variants, followed by protein prediction modeling of the revealed variants. Thus, this study contains novel and comprehensive data concerning the role of TCTE1 in male infertility.
Materials and methods
Animals
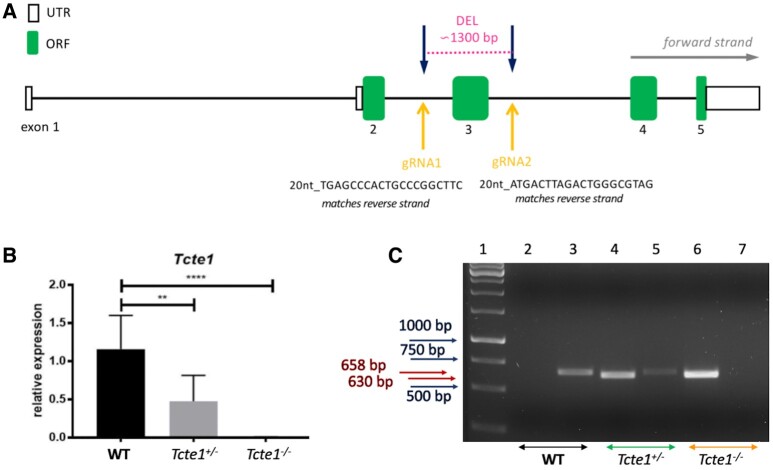
Using the CRISPR/Cas9 genome editing technique, the mouse heterozygous knockout line of the Tcte1 gene (Tcte1+/−) was created on the basis of the C57Bl/6J strain by the Cyagen Biosciences company (Santa Clara, CA, USA). The mouse Tcte1 gene is located on chromosome 17 B3; 17 22.54 cM (GenBank: NM_013688.2; Ensembl: ENSMUSG00000023949). Exon 3 of the Tcte1 gene was selected as the target site (deletion of 1292 bp) (Fig. 2A). gRNAs targeting vectors (constructed, sequenced, and generated by in vitro transcription; Supplementary Data File S1) were injected into fertilized oocytes. The founders were genotyped and those with positive results were bred to the next generation. Then, in the animal facility of the Institute of Human Genetics PAS (Poznan, Poland), adult Tcte1+/− animals were mated to generate homozygous Tcte1−/− mice. Wild-type Tcte1+/+ (WT) animals C57Bl/6J were purchased from Charles River Company (Sulzfeld, Germany). Relative expression of the Tcte1 gene was verified using the real-time PCR method (Fig. 2B; Supplementary Data File S2). All mice were housed in a pathogen-free animal facility with food and water ad libitum, a stable temperature of 22°C, and a light: dark light cycle of 12:12 h. All procedures were in accordance with the guidelines and regulations for animal experimentations and restricted usage of genetically modified organisms (GMO) and were approved by the Local Ethical Committee for Animal Experiments at the Poznan University of Life Sciences (no. 1/2014, 18/2017), and Ministry of Environment (no. 70/2017). Mouse models with infertility phenotypes were reviewed in the Mouse Genome Informatics database (Jackson Laboratory; http://www.informatics.jax.org/) and through literature searches (PubMed; https://pubmed.ncbi.nlm.nih.gov/).
Figure 2.
Characteristics of Tcte1 knockout mice. (A) Schematic representation of gRNA localization in CRISPR/Cas9 KO creation. (B) Relative expression of the Tcte1 gene in wild-type (WT), hetero- (+/−) and homozygous mice (−/−) (Real-time PCR); **** P < 0.0001, ** P < 0.01. (C) Example of genotyping result. Lane 1: 1 kb marker (GeneRuler), lanes 2, 3: wild-type (WT), lanes 4, 5: hetero- (+/−), and lanes 6, 7: homozygous mice (−/−); lanes: 2, 4, 6 identified the lack (0 bp) or presence of mutation (630 bp), lanes: 3, 5, 7 differentiated hetero- (658 bp) and homozygotes (0 bp).
Reproductive potential
To check the reproductive potential of KO mice, adult animals (7–8 weeks old) were crossed for delivery of three litters per caged pair, but no longer than for 6 months. Crossing combinations included: (i) four pairs: homozygous Tcte1−/− male × wild-type (WT) female, (ii) five pairs: heterozygous Tcte1+/− male × heterozygous Tcte1+/− female, and (iii) three pairs: heterozygous Tcte1+/− male × wild-type (WT) female. Vaginal plug presence was checked every morning to confirm mating incidence. The time period from the mating start point to the first pedigree appearance was checked. Numbers of pups were counted on the day after birth. For each crossed pair, the number of litters, pups, and the sex ratio were estimated (also according to pups’ genotypes) (Supplementary Table S1).
Genotyping
Genotyping of the pups delivered was performed via a PCR assay, with two sets of primers: (i) to identify wild-type and positive KO (heterozygous and homozygous) animals (Tcte1-F: CGTTTTAAGAATGTATTGAGGGTTGGG, Tcte1-R: CTCCGTAGGCTCCTGCCAATATG; estimated product size: for WT 1915 bp, for KO ∼630 bp (deleted ∼1300 bp); PCR results analysis: one band of 1915 bp or 0 bp determined for WT, with one band of 630 bp evidenced for hetero- or homozygotes); and (ii) to distinguish between hetero- and homozygotes (Tcte1-WT/He-F: AGCTTGCCACACCCTCAAGGTACTA, Tcte1-R: CTCCGTAGGCTCCTGCCAATATG; estimated product size: 658 bp for heterozygotes and 0 bp for homozygotes; PCR results analysis: one band of 658 bp determined for heterozygotes, with no band (0 bp) indicated for homozygotes (Fig. 2C). The PCR mixtures and reaction conditions are described in Supplementary Data File S3.
Tissue gonadal preparation/collection
Gross anatomy was performed on testis and epididymis of WT (n = 21), Tcte1+/− (n = 22), and Tcte1−/− (n = 18) male animals (10 weeks old). Mice were weighed, and then euthanized with a carbon dioxide (CO2) overdose (euthanasia Minerve Easy-box; Minerve; Esternay, France) with a stable flow rate per minute. Testes and epididymides were collected, weighed, and measured. For histological evaluation, two fixing approaches were applied. Both approaches started with anaesthetization under 4% isoflurane (Baxter, Warsaw, Poland) and transcardial perfusion with 50 ml/animal of ice-cold PBS (Gibco, Paisley, UK), followed by 50 ml/animal of 10% formalin solution, neutrally buffered (Sigma Aldrich, St. Louis, MO, USA, cat. no. HT501850) for three males per zygosity and technical variant. Next, the tissues were immersed in formalin solution for 24 h, 4°C, in darkness. Then, in the first approach, Paraplast-embedded sections were prepared (5 µm thickness) using a Leica 2055 Microtome (Leica, Deer Park, IL, USA). In the second technique, gonads were sequentially immersed in 15%, 20% and 30% of sucrose (Sigma Aldrich, cat. no. S0389), each for 24 h, at room temperature. Thereafter, gonads were coated with Tissue-Tek O.C.T. Compound (Sakura, Alphen aan den Rijn, The Netherlands, cat. no. 4583) and frozen in cold (−80°C) 2-Methylbutane (Sigma Aldrich, cat. no. M32631). Then, cryo-sections were prepared (10 µm thickness), using a Leica CM1950 Cryostat (Leica Biosystems, Nussloch, Germany) and kept at −80°C until further stainings. To obtain the histological picture, slides were stained using the Masson-Goldner protocol (Romeis et al., 2010) for classic paraffin blocks or standard progressive H&E staining method for frozen samples. Briefly, after fixation twice in 100% EtOH (PoCH, Gliwice, Poland) (10 min), slides were first hydrated in an EtOH series (95%, 70% for 5 min), washed in distilled water and stained in Mayer’s hematoxylin (Sigma Aldrich) for 5 min, then washed in distilled water (three times, 5 min) and dehydrated in 50%, 70%, 80%, 96% EtOH (5 min each). Counterstaining with eosin Y (Sigma Aldrich) for 30 seconds was applied, followed by washing in 100% EtOH, clearing in xylene (PoCH) (5 min each, two repeats), and mounting in DPX (Sigma Aldrich). Thereafter, slides were documented with the Leica DM5500 light microscope (×40 dry or ×63 oil immersive objectives, motorized stage), LASX software (with Navigator tool) (Leica Microsystems GmbH, Wetzlar, Germany). All the evaluations were performed blindly by two experienced scientists per technique.
For each zygosity, the geometrical features of the tubule sections were evaluated, including tubule diameter (D) and cell layer thickness (CL) (the mean of two measurements made at opposite sides of the tubule) and lumen diameter (l), using the Fiji (Image J; University of Wisconsin, USA) measurement tool. To check density of the cells, ratios of D: CL, D: l and CL: l were also estimated. In total, for each zygosity, 133–144 tubule sections were evaluated (three random sections per zygosity, each with a minimum of 30 tubules).
Mouse semen samples
Mouse spermatozoa were collected from epididymis post mortem. Collected epididymis were placed immediately in a 2.0 ml test tube with warm 500 µl of a non-capacitating Whitten’s HEPES medium (pH 7.2–7.4, 37°C), then cut into 5–7 fragments, and incubated in 37°C for 15 min to allow the sperm cells to swim out. Whittens-HEPES medium contained: 100.0 mM NaCl, 4.4 mM KCl, 1.2 mM KH2PO4, 1.2 mM MgSO4, 5.4 mM glucose, 0.8 mM pyruvic acid, 4.8 mM lactic acid (hemi-Ca), 20 mM HEPES (all from Sigma Aldrich) (Wertheimer et al., 2013). Then, spermatozoa were collected and seminological analysis performed (concentration, motility, morphology). Next, semen samples were fixed in a fresh fixative solution (methanol: acetic acid, 3:1 v/v, −20°C; 3×; both from PoCH) and stored until further use at −20°C.
Semen analysis
A 10-μl aliquot of sperm suspension was applied to the Makler counting chamber (Sefi Medical Instrument Ltd, Haifa, Israel) to test the sperm concentration and total sperm count. Also, the ratio of progressive (fast and slow), non-progressive (total and circular), and immotile spermatozoa were calculated (WHO, 2021). For assessment of sperm morphology, Papanicolaou staining was performed. Briefly, the air-dried smears were fixed in ethanol: diethyl ether (1:1 ratio, vol/vol; PoCH) solution for 10 min and left to air-dry. Then, slides were stained by the Papanicolau procedure according to WHO guidelines (WHO, 2021), and mounted in DPX Mountant (Sigma Aldrich, cat. no. 06522). A total of 200 spermatozoa were analyzed using a Leica DM5500 light microscope (×1000 magnification, oil immersion). Various morphological forms of spermatozoa were determined including abnormalities of the head, midpiece, and tail. Also, the length of the sperm tail, midpiece, and the tail:midpiece ratio were measured to check possible differences between males with various zygosities (6 males/per zygosity); for each male, at least 100 spermatozoa were measured with the ‘length measurement’ option by CellSens Dimension software (Olympus BX41, Tokyo, Japan).
Sequencing of RNA
For RNA extraction, testis tissue was collected in RNAlater buffer (Invitrogen, Waltham, MA, USA) and frozen immediately (−80°C). Samples were processed using the AllPrep DNA/RNA/Protein Mini Kit (Qiagen, Hilden, Germany). Quality and quantity of extracted RNA samples were checked using NanoDrop (ND-1000, Thermo Scientific, Waltham, MA, USA) and Quantus (Promega, Madison, WI, USA), and 1 μg of RNA per sample was used for sequencing (Macrogen, Seoul, South Korea). RNA sequencing was performed with the Illumina NovaSeq 6000 platform using TruSeq Stranded mRNA LT Sample Prep Kit (Illumina), with specified conditions of paired-end reads at 100 bp, and throughput 100 M reads per sample. RNASeq raw reads were aligned to the mouse reference genome mm10 using hisat 2.0.5 with Gencode vM13 transcriptome reference. Reads aligned in the coding regions of the genome were counted using Feature Counts. Then, read counts were normalized using DESeq2 and subjected to differential analysis. The differential analysis included computing of median-based fold change, statistical significance using the Students’ t-test, and false discovery rates (correction for multiple testing) in the R/Bioconductor programming environment. Testis tissue expression was retrieved from GTEx (https://gtexportal.org/home/), BioGPS (http://biogps.org/), and AceView (https://www.ncbi.nlm.nih.gov/IEB/Research/Acembly/). All RNAseq expression data have been deposited in Gene Expression Omnibus (GEO), accession number GSE207805.
STRING interactions
For analysis of protein–protein interactions, based on changed expression levels of corresponding genes identified in the mouse testis RNAseq experiments, STRING analysis was performed (version 11.0). The STRING database allows to delineate probability of the interactions between evaluated proteins using different evidence channels (von Mering et al., 2005; Szklarczyk et al., 2019). This tool enables the integration of all data available in public resources concerning protein–protein interactions.
Immunofluorescence on sperm: N-DRC visualization
Immunofluorescence in situ staining was performed to detect the N-DRC complex proteins: Tcte1 (Drc5), Drc7, Fbxl13 (Drc6), and Eps8l1 (Drc3) in mouse spermatozoa. Mouse spermatozoa were collected and fixed as described above. Specific antibodies were applied as primary antibodies: (rabbit anti-human-TCTE1 (cat. no. orb357083), rabbit anti-mouse-Drc7 (orb586958), anti-mouse-Fbxl13 (orb314611), and anti-mouse-Eps8l1 (orb382538; all from Biorbyt, Cambridge, UK)), or the secondary antibody: (goat anti-rabbit-AF594 (Abcam, Cambridge, UK, cat. no. ab150160)). The UniProt IDs and homology of the immunization regions of primary antibodies between human and mouse are described in Supplementary Table S2. Antibodies were diluted in 0.5% PBST (PBS: Gibco; Triton X-100: Sigma Aldrich) (primary: 1:100, secondary: 1:500). First, slides with fixed sperm smears were washed in a series of washes in 1× PBST, and then incubated in 25 mM DTT/1 M Tris-HCl (Sigma Aldrich), pH 9.5, at room temperature for 20 min. Then, washing in 1× PBST, and incubation in 6 N HCl (PoCH) for 30 min was applied, followed by a blocking step with 1% BSA/1× PBST for 30 min (BSA: Sigma Aldrich). Next, overnight incubation with antibodies was performed at 4°C in a humidified container. After washing, the samples with 1× PBST, a secondary AF594-conjugated antibody was applied for 1 h. Next, unconjugated antibodies were washed out (4× in 1× PBST, 5 min each, slight rotation). For the final detection, 20 µl of Fluoroshield with DAPI (Sigma Aldrich, cat. no. F6057) was added to the samples, and microscopic analysis was performed. Images were acquired using a fluorescence microscope with a proper filter set: Leica DM5500, filters: DAPI/TxR/Triple; objectives: 10× and 63× with oil immersion; DFC 7000T camera; software: LASX.
Sperm ATP measurement
For measurement of ATP in spermatozoa, the CellTiter-Glo 2.0 Assay (Promega) was used. This ready-to-use assay allows to determine the quantitation of the ATP amount, indicating the presence of metabolically active cells. The assay is based on the thermostability of luciferase (Ultra-GloTM Recombinant Luciferase) and was performed as described in the manufacturer’s protocol. Briefly, all laboratory work was done in a stable temperature of 22°C. After the collection of mouse spermatozoa (as described above), the amount of ∼1 × 106 of sperm cells was transferred to a black opaque 1.5 ml test tube and the proper amount of CellTiter-Glo kit was added (1:1, v: v). Next, 2 min of orbital mixing of the content was performed to activate cell lysis. Then, after 10 min of incubation (to stabilize the luminescence), the measurement of the luminescent signal was completed using a Glomax 20/20 luminometer (Promega). For each sample, three measurements (repeats) were done, each in two time points of 0 and +60 min. Numbers of animals used were: WT (n = 7), Tcte1+/− (n = 7), and Tcte1−/− (n = 6). Mean luminescence values obtained (represented as RLU: relative luminescence unit) were compared between tested mice groups.
Immunofluorescence on testis
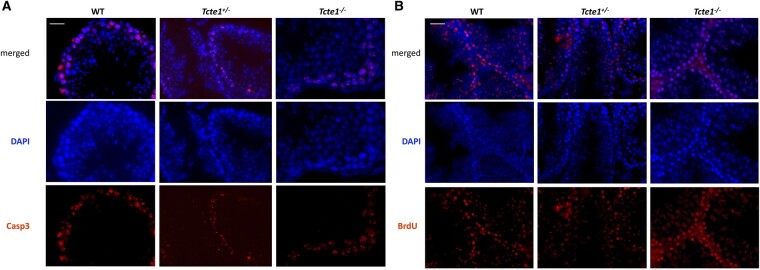
Immunofluorescence in situ staining on testis was performed to check: (i) the mean cell count of spermatogonia and spermatocytes (preleptotene and pachytene) in tubules’ sections (cryo-sections), and (ii) to estimate the level of apoptosis via caspase-3 visualization on mouse testicular samples (paraffin embedded sections) from all three zygosities. Caspases play an important role in the induction, transduction, and amplification of intracellular apoptotic signals, and thus, caspase-3 can be used as one of the apoptotic indicators (Lei et al., 2022). Mouse testes were collected and fixed as described above. Specific antibodies were applied as primary antibodies: (mouse anti-MLH1 (Abcam, cat. no. ab14206), 1:30 in 0.5% PBST, rabbit anti-RAD51 (Abcam, cat. no. ab63801), 1:200 in 0.5% PBST) and: rabbit anti-Casp3, (Biorbyt, cat. no. orb10237), 1:100 in 0.5% PBST) or as secondary antibodies: (goat anti-mouse-FITC (Sigma Aldrich, cat. no. F2012), 1:400 and goat anti-rabbit-AF594 (Abcam, cat. no. 150080), 1:500 in 0.5% PBST). First, slides with paraffin fixed testis cross sections were deparaffinized in three changes of xylene (10 min each), while for cryo-sections, this step was omitted. Then, slides were incubated in a series of ethanol: 100%, 96%, and 70%, twice for 5 mins in each solution. Next, after rinsing in distilled water, tissue sections were incubated sequentially in freshly prepared 2% NaBH4/1× PBS, and in 0.1M glycine (30 min, room temp. each; Sigma Aldrich). Heat-induced epitope retrieval was performed by immersing the slides in 0.01 M sodium citrate/0.05% Tween 20 (Sigma Aldrich), pH 6.0 for 30 min, followed by cooling down at room temperature for the next 20 min. A blocking step was performed in an aliquot of freshly prepared 1% BSA/1× PBST, at 37°C for 1 h. Then, tissue sections were incubated with primary antibody in a humidified container overnight at 4°C. A secondary antibody was applied for 1 h, followed by washing out of the unconjugated antibody (4× in 1× PBST, 5 min each, slight rotation). For the final detection, 20 µl of Fluoroshield with DAPI was added. Images were captured using a fluorescence Leica DM5500 microscope with a proper filter set: DAPI/TxR/SpG/Triple; objectives: 10× and 63× with oil immersion; DFC 7000T camera, LASX software. The analysis was performed for three sections per zygosity, each with a minimum of 30 tubules (for cell count) or two cross-sections per zygosity, 15–20 tubules per tissue section (for Casp-3 evaluation).
TUNEL assay
TUNEL assay was performed to check the level of apoptosis and necrosis on the testicular tissue according to the manufacturer’s protocol (Abcam, ab66110 TUNEL Assay Kit: BrdU-Red). Briefly, paraffin embedded sections (or two cross-sections per zygosity, with 15–20 random tubules analyzed/slide) were deparaffinized in xylene (twice, each 5 min), followed by an EtOH series: 100% (twice, 5 min), 95%, 85%, 70%, and 50% (3 min). Next, slides were incubated in 0.85% NaCl (5 min) and washed in PBS. After incubation with Proteinase K solution (Qiagen) (20 µg/ml in Tris-HCl–EDTA; 5 min) and washing with PBS, slides were immersed in 4% formaldehyde/PBS. Then, Wash Buffer was applied twice for 10 min with a plastic coverslip on the top. Next, DNA Labeling Solution was added followed by incubation in a dark humidified incubator for 1.5 h at 37°C. After the next two washes with PBS, Antibody Solution was applied, and slides were incubated for 30 min at room temperature. The last steps included ddH2O washing (5 min, twice) and the application of 20 µl of Fluoroshield with DAPI for the final detection. Slides were analyzed within 1 h using the fluorescence Leica DM5500 microscope with a proper filter set: DAPI/TxR/Triple; objectives: 10× and 63× with oil immersion; DFC 7000T camera, LASX software.
Sequencing of the human samples
A pilot genomic sequencing study of samples from human infertile males was also performed. Bioethical committee approvals were received for the study (Local Bioethics Committee of Poznan University of Medical Sciences, approval no. 1003/18; National Bioethical Committee, Ministry of Health, Warsaw, Poland, approval no. OKB-5-2/15). All participants were notified about the aim of the study and provided written informed consent. All experiments were performed in accordance with relevant guidelines and regulations. DNA from whole blood samples (collected into sterile single test tubes with EDTA) was extracted using standard protocols of the MagCore HF16 Automated Nucleic Acid Extractor (RBC Biosciences, New Taipei City, Taiwan), or Gentra Puregene kit (Qiagen). The total number of sequenced patients was n = 248, including: 137 with non-obstructive azoospermia (lack of spermatozoa in ejaculate) or cryptozoospermia (spermatozoa detectable in ejaculate after centrifugation), and 111 with a spectrum of seminal abnormalities in concentration, and/or motility, and/or morphology. Among those samples, 47 revealed decreased motility (asthenozoospermia). Semen evaluation was performed according to WHO guidelines, followed by AUA and ASRM guidelines for infertile male diagnosis (Schlegel et al., 2021). All men revealed normal hormonal levels, had no chromosomal abnormalities and no AZF microdeletions. For whole exome samples (n = 76 from Pittsburgh), sequencing libraries were constructed using: the SureSelect XT All Exon V4 + UTRs capture kit and the SureSelect XT Target Enrichment System (Agilent Technologies, Santa Clara, CA, USA). Libraries were sequenced with an average read coverage of 62–100× (∼6 Gb per sample), using 100 bp paired-end sequencing with TruSeq PE Cluster Kit v3 cBot HS and 4 × 50 TruSeq SBS V3 HS sequencing kit (Illumina) on the HiSeq2000 (Illumina). Whole-genome sequencing (n = 42, Poznan) was performed using Illumina HiSeqX with the coverage of at least 30× (100–120 Gb per sample). Sanger sequencing for all exomes of TCTE1 gene (n = 130, Poznan) was performed using LightRun96 system of Eurofins Genomics (Eurofins Genomics AT GmbH, Vienna, Austria). Samples were prepared with QIAquick Gel Extraction (cat. no. 28704) or PCR Purification (cat. no. 28104) kits (Qiagen), and analyzed with CLC Main Workbench 8 software (Qiagen).
Filtering of whole-genome sequencing results
The quality of raw NGS data was evaluated using the FastQC and MultiQC packages. Reads were aligned to the reference human genome GRCh37 using bwa-mem aligner included in the Speedseq package (Chiang et al., 2015). For calling single-nucleotide variants (SNVs) and small indels, freeBayes v0.9.21 (speedseq-var) was implemented, following the previously described pipeline (Supernat et al., 2018). Variant disease association and missense mutation prediction effects were determined using: dbSNP (https://www.ncbi.nlm.nih.gov/snp/), OMIM (https://www.ncbi.nlm.nih.gov/omim/), ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/), MutationTaster (http://www.mutationtaster.org/), SIFT (https://sift.bii.a-star.edu.sg/), Polyphen (http://genetics.bwh.harvard.edu/pph2/), CADD (https://cadd.gs.washington.edu/snv), MetaDome (https://stuart.radboudumc.nl/metadome/), REVEL (rare exome variant ensemble learner) (Ioannidis et al., 2016), Ensembl Variant Effect Predictor (McLaren et al., 2016), and the Human Gene Mutation Database (HGMD; Qiagen). SNV pathogenicity was assessed on the basis of American College of Medical Genetics and Genomics (ACMG) guidelines (Richards et al., 2015). PhyloP (Cold Spring Harbor Laboratory) and Clustal Omega were used for the determination of conservation (https://www.ebi.ac.uk/Tools/msa/clustalo/). All NGS variants were confirmed via the Integrative Genomics Viewer (Broad Institute). Variant frequencies have been obtained from the gnomAD database (frequencies from more than 125k exomes and 15k genomes, http://gnomad.broadinstitute.org; v2.1.1), and were filtered in search of rare variants (with minor allele frequency (MAF) <1%).
Protein prediction
The amino acid sequence of TCTE1 was input into Phyre2 (Kelley et al., 2015) for both intensive and fast mode homology modeling, I-TASSER (Roy et al., 2010; Yang et al., 2015, Yang and Zhang, 2015), and AlphaFold (Jumper et al., 2021; Varadi et al., 2022), which utilize sequence alignment-based algorithms for generating predicted secondary and tertiary structures; AlphaFold additionally uses machine learning to integrate biological principles of protein folding with the alignment-based algorithms. Confidence scores were obtained from these programs; AlphaFold additionally generates predicted aligned error plots for assessing the quality of the model’s long-range interactions. Models were inspected manually and aligned using the secondary structure matching (SSM) (Krissinel and Henrick, 2004) tool in COOT (Emsley et al., 2010) to minimize user input bias. Docking of the structures into electron tomography data was performed manually in COOT (Emsley et al., 2010). No refinement was performed to minimize bias. All structure model figures and mutations without post refinement, were generated using Pymol (Schrödinger). I-TASSER was used also for prediction of the exon 3 structure deleted in our mouse KO model (Supplementary Fig. S1).
Statistical analysis
Statistical analyses included the following: normality test (D’Agostino and Pearson or Kolmogorov–Smirnoff), unpaired two-tailed t-test with Welch’s correction or Mann–Whitney test, one-way ANOVA test and Fisher’s exact, and two-tailed Pearson correlation. All tests were performed with a significance level of α = 0.05 using GraphPad Prism (v.7.0e) software.
Results
Reproductive potential of KO mice
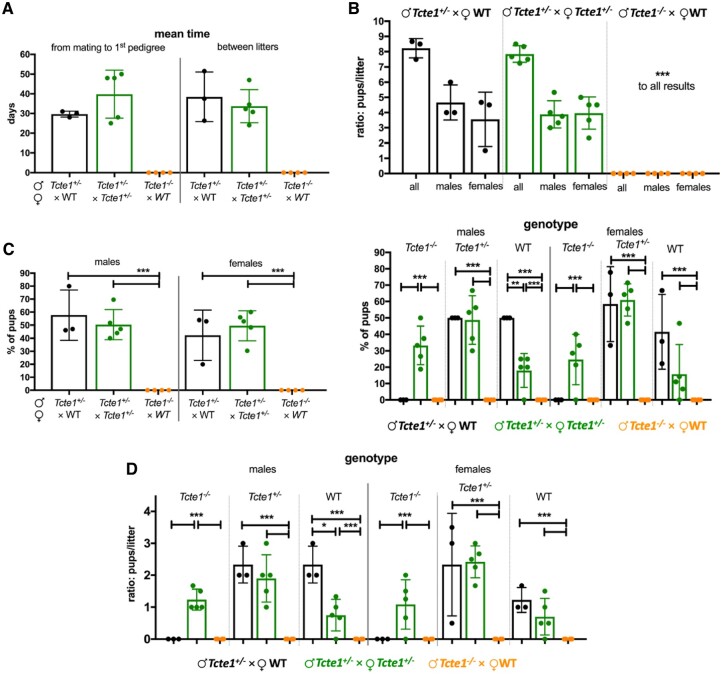
Reproductive potential results are shown in Fig. 3 and Supplementary Table S1. Only pairs with homozygous Tcte1−/− males revealed no progeny (Tcte1−/− male × WT female). Other combinations (Tcte1+/− male × Tcte1+/− female; Tcte1+/− male × WT female) showed similar results when considering numbers of litters, pups, and sex ratio. The mean time of caging to delivery of the first pedigree was longer in the Tcte1+/− male × Tcte1+/− female combination by 25.45% (statistically not significant). When considering genotypes of the pups, statistically significant differences (P < 0.05) were observed in the number of Tcte1−/− male and female pups delivered: no homozygous animals were obtained from the combination of Tcte1+/− male × WT female, while in mating of Tcte1+/− male × Tcte1+/− female, homozygous males (33.25% of all pups) and females (24.67% of all pups) were received.
Figure 3.
Reproductive potential of KO Tcte1 mice. Mating combinations and their results including time of breeding, number of litters and pups, number of male and female pups, and genotypes observed in pups of each combination. (A) Mean time: from mating to first pedigree, and between particular litters. (B) Ratio of males and females per litter in each mating combination. (C) Frequency of males and females and their genotype observed each mating combination; left panel: according to the sex of pups; right panel: according to mating combinations. (D) Ratio of males and females per litter and their genotype observed in each mating combination. ***P < 0.0001, **0.0001 < P < 0.01, *0.01 < P < 0.05.
Histopathology and semen parameters
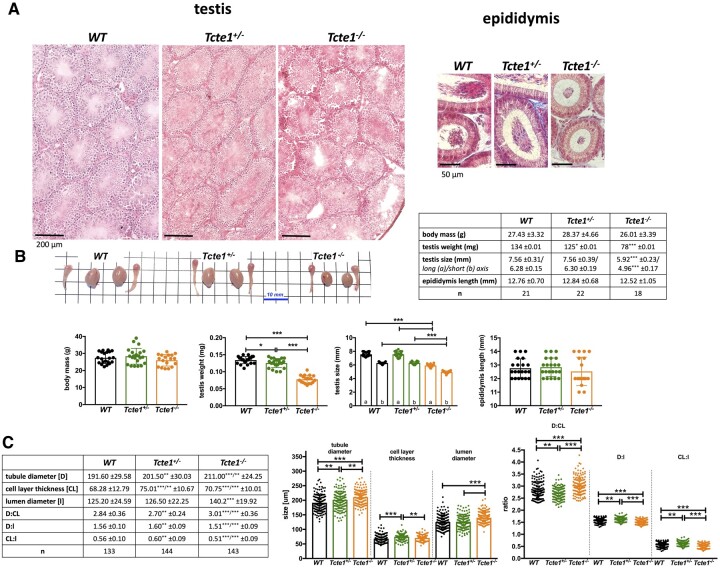
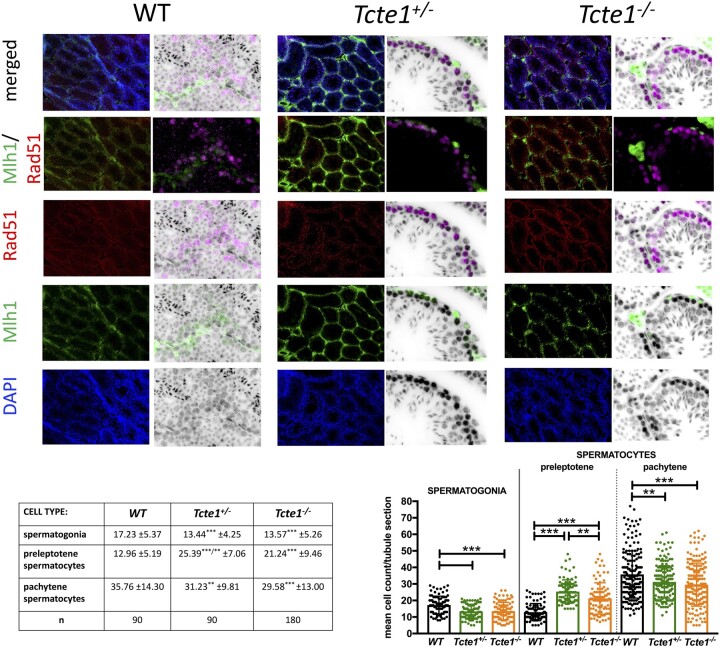
Histopathological evaluation of testicular and epididymal sections revealed no visual structural differences between WT, Tcte1+/−, and Tcte1−/− males, confirming preserved spermatogenesis (Fig. 4A; Supplementary Fig. S2). No differences were found for body mass of the animals and the length of the epididymis (Fig. 4B). In the Tcte1−/− males, testis weight was decreased by ∼41% (P < 0.0001) followed by a ∼22% decline of its size (P < 0.0001), while in Tcte1+/− males, only testis weight was slightly decreased (∼7%; P < 0.05) when compared to control animals. Differences in dimensions of the tubule sections were noted (Fig. 4C), with increasing tubule and lumen diameters and decreasing cell layer thickness in Tcte1−/−. In Tcte+/−, cell layer thickness and tubule diameter were increased. When comparing germ cell counts (Fig. 5), reduced numbers of spermatogonia (∼22%) and pachytene spermatocytes (13–17%) were documented for Tcte1−/−and Tcte+−- (P < 0.001), while the level of preleptotene spermatocytes was elevated (P < 0.001). The mean sum of all evaluated cell types was similar between WT and homozygotes (65.95 and 64.39 of cells/tubule section, respectively). Such data suggest some kind of time shift at the first stages of prophase: germ cells from KO mice seem to be delayed at the preleptotene stage, while WT males enter the pachytene stage at a higher rate.
Figure 4.
Characteristics of Tcte1 wild-type (WT), hetero- (+/ − ) and homozygous mice ( − / − ) . (A) Histopathology of testes and epididymis (caput); staining: classic hematoxylin–eosin staining (testis) or Masson–Goldner protocol (epididymis); microscope: Leica DM5500, objective 40, LASX software with Navigator function. (B) Comparison of body mass, testis weight and size, and epididymis length, followed by gross morphology of gonads (n: no. of mice analyzed). (C) Comparison of the dimensions of testicular tubule sections (n: no. of sections analyzed); ***P < 0.0001, **0.0001 < P < 0.01, *0.01 < P < 0.05.
Figure 5.
Evaluation of germ cell types in testes of Tcte1 wild-type (WT), heterozygous (+/ − ) and homozygous ( − / − ) mice. Three germ cell types were evaluated: spermatogonia (stained with intensive green color), preleptotene spermatocytes (intensive red color), and pachytene spermatocytes (medium green and red). Immunofluorescence staining with antibodies: primary: mouse anti-MLH1 (Abcam, Cambridge, UK, cat. no. ab14206), 1:30, rabbit anti-RAD51 (Abcam, Cambridge, UK, cat. no. ab63801), 1:200; secondary: goat anti-mouse-FITC (Sigma Aldrich, St. Louis, MO, USA, cat. no. F2012), 1:400 and goat anti-rabbit-AF594 (Abcam, Cambridge, UK, cat. no. 150080), 1:500.
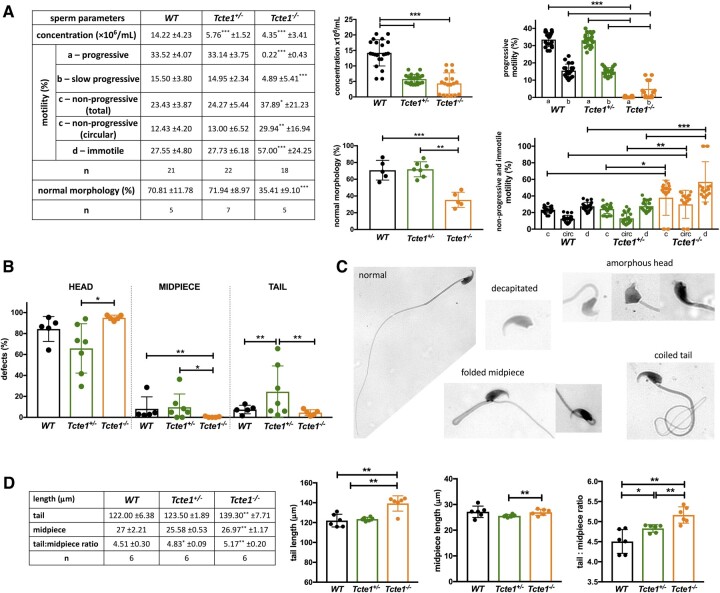
When considering seminal parameters, both in Tcte1−/− as well as in Tcte1+/− males, the sperm concentration was importantly decreased (3.27, and 2.47-fold, respectively), when compared to control results (P < 0.0001; Fig. 6A). Additionally, in Tcte1−/− males, the frequency of progressively motile spermatozoa was residual, with the progressive type of motility in only 0.22% of spermatozoa (decreased ∼150-fold versus control 33.52%, (P < 0.0001)), followed by 4.89% being slow progressive spermatozoa (decreased ∼3-fold versus control 15.50%) (P < 0.0001). Interestingly, an ∼2.4-fold increase in the circular type of motility (around own axis and non-progressive) was observed for Tcte1−/− males when compared to WT and Tcte1+/− animals (P < 0.01) (Supplementary Video S1, S2, and S3). The frequency of immotile spermatozoa was also increased in Tcte1−/− males (2-fold to control; P < 0.0001). The frequency of morphologically normal spermatozoa in Tcte1−/− animals was significantly decreased (2-fold to control; P < 0.0001; Fig. 6A–C; Supplementary Data File S4). The majority of morphological defects was observed for the sperm head, mostly of decapitated spermatozoa in Tcte1−/− animals: 52.55% versus 19.16% for Tcte1+/− and 23.33% for WT, and amorphous head shape with decreased frequency in Tcte1−/− (26.11%) and Tcte1+/− (33.72%) males versus WT animals (48.88%) (Supplementary Data File S4). Midpiece defects were found in Tcte1+/− (9.71%) and WT (8.20%) animals, while Tcte1−/− mice revealed only a low frequency (0.38%) of such defects. On the other hand, an increased frequency (∼5-fold) of coiled tail was observed for Tcte1+/−/− animals when compared to other mice zygosity (P < 0.01) (Supplementary Data File S4). To summarize, seminological changes in mice with Tcte1 mutations resulted in oligoasthenoteratozoospermia in homozygous Tcte1−/− or oligozoospermia in heterozygous Tcte1+/− animals.
Figure 6.
Characteristics of spermatozoa of Tcte1 wild-type (WT), heterozygous (+/ − ) and homozygous ( − / − ) mice. (A) Sperm parameters: concentration, motility, and morphology comparisons; statistical marks indicate differences according to WT animals; n: number of mice. (B) Frequency of morphological defects according to three parts of the spermatozoa. (C) Examples of the main sperm morphological abnormalities found (all abnormalities observed available in Supplementary Data File S4); Leica DM5500 microscope, objective 63×, CytoVision software. (D) Comparison of the mean length of sperm tail, midpiece and ratio tail:midpiece; n: no. of animals, each with at least 100 spermatozoa analyzed; ***P < 0.0001, **0.0001 < P < 0.01, *0.01 < P < 0.05.
Increased mean lengths of tail and tail:midpiece ratios in Tcte1−/− animals (139.30 ± 7.71 µm; 5.16, respectively) were measured when comparing to WT (122.00 ± 6.375 µm; 4.51; P < 0.002) and Tcte1+/− (123.50 ± 1.89 µm; 4.83; P < 0.008) (Fig. 6D). Also, in the Tcte1+/−, the midpiece length differed from Tcte1−/− animals (P = 0.0334), with tail:midpiece ratio differences when compared to WT (P = 0.0435).
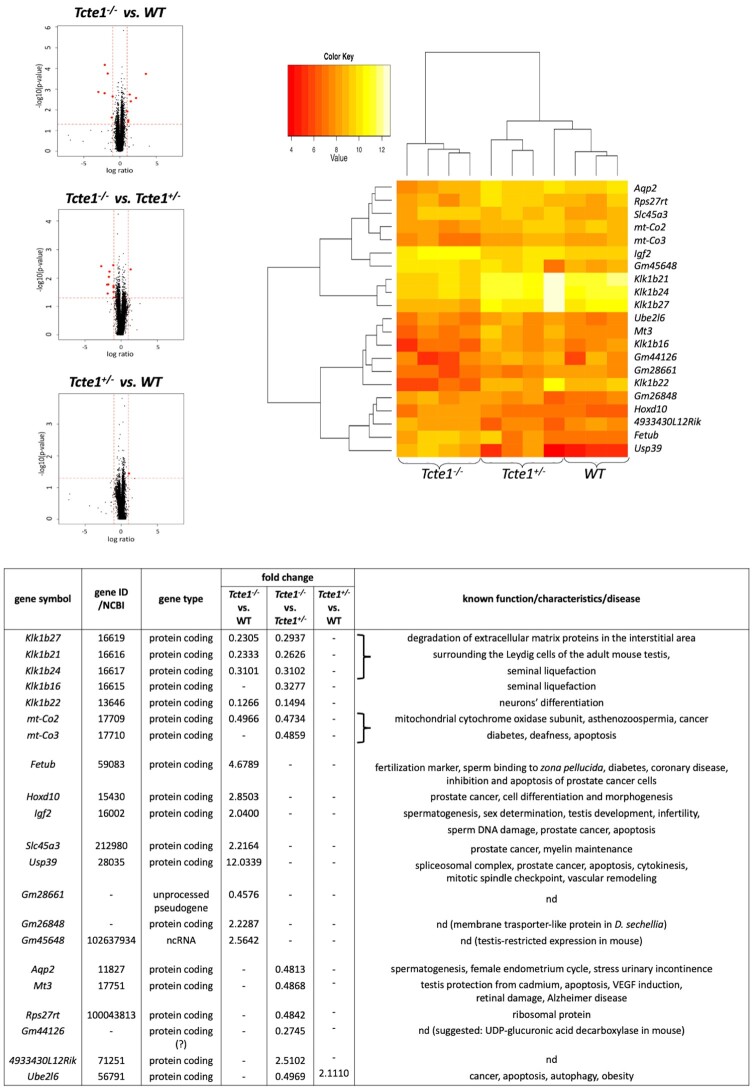
RNAseq and STRING pathway analysis
RNA sequencing analysis of gene expression in testes of KO mice revealed statistically significant changes (P < 0.05) in 21 genes, including five genes both for Tcte1−/− versus Tcte1+/− and WT, eight genes only for Tcte1−/− versus WT, seven genes only for Tcte1−/− versus Tcte1+/−, and 1 gene both for Tcte1+/− versus Tcte1−/− and WT (Fig. 7). In Tcte1−/− animals versus both Tcte1+/− and WT, the expression level of four genes from the kallikrein subfamily was decreased 2–8-fold, including genes involved in semen liquefaction and degradation of extracellular matrix proteins in the interstitial area surrounding Leydig cells of the adult mouse testes (Klk1b27, Klk1b21, Klk1b24) or neuronal activity (Klk1b22) (Fig. 7; Supplementary Data File S5) (Fink et al., 2007; Du Plessis et al., 2013; Marques et al., 2016; Michaelis et al., 2017). A 2-fold decrease of expression of genes related to mitochondrial cytochrome oxidase subunits and asthenozoospermia was documented (mt-Co2, mt-Co3) (Fig. 7) (Baklouti-Gargouri et al., 2013; Heidari et al., 2016; Mostafa et al., 2016). In Tcte1−/− versus WT, eight genes revealed changes in expression level, including a 4.7-fold increase for the Fetub gene, which is known to be required for oocyte fertilization (via specific inhibitor activity of the zona pellucida (ZP) ovastacin, the cysteine protease responsible for ZP2 cleavage) (Karmilin et al., 2019; Binsila et al., 2021), and plays also a role of a secreted factor during reprogramming (Bansho et al., 2017).
Figure 7.
RNAseq results of gene expression levels found in testes of KO Tcte1+/ − and Tcte1 − / − mice versus control (WT) mice. Volcano plots: black dots indicate all analyzed genes within the genome, red dots represent genes with significantly changed expression levels. A heatmap and a table describing the genes are also shown. Filtering criteria: P <0.05, log ratio >1 or <−1 (adequate to fold change value of 2 (expression level increased) or 0.5 (expression level decreased)), expression level >8. nd: no data concerning function or disease.
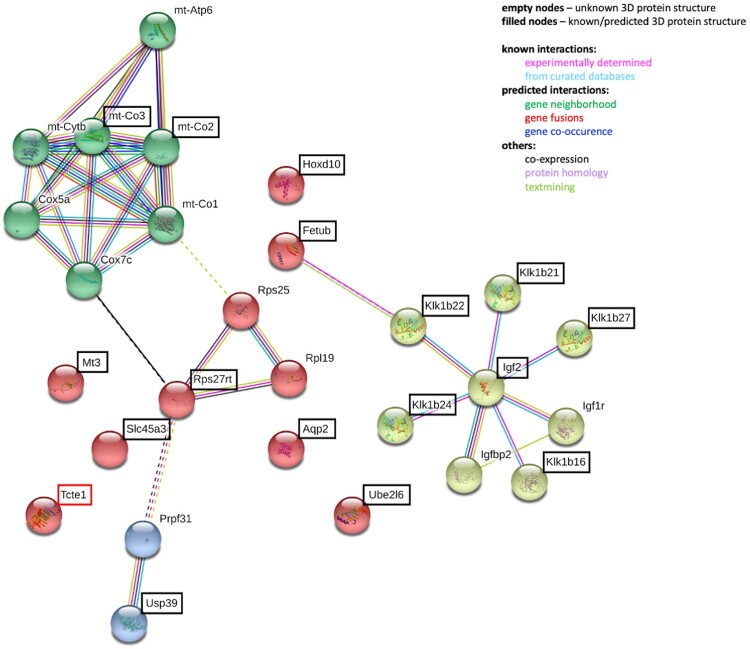
A two-fold increase for the Igf2 gene (strongly linked to spermatogenesis, sex determination, testis development, infertility and prostate cancer; Damaschke et al., 2017; Neirijnck et al., 2019; Cannarella et al., 2020; Matsuzaki et al., 2020) has been also observed. Igf2 and Klk proteins are enclosed in a common network consisting of proteins directly linked to fertility (Fig. 8; Supplementary Data File S5 and Fig. S3). One of them is Ambp, a fusion protein, from which proteolysis generates bikunin, a protein whose deficiency results in infertility, due to diminished formation of the stable cumulus–oocyte complex required for maturation and ovulation (Sato and Kan, 2001; Naz and Rajesh, 2005). Bikunin is one of the elements of ITIH domains (Inter-Trypsin Inhibitor Heavy-chain), responsible for hyaluronian metabolism (Salustri et al., 2019; Hull et al., 2021). ITIH is also linked to Plg, which has protease activity and activates metalloproteins, such as Mmp8 and Mmp9, also included in the network. Mmp8 is engaged in degradation and stabilization of occluding and blood–testis barrier (Chen et al., 2018), while Mmp9 (with Igf1 and Igf2) is important for normal placentation (Ghosh et al., 2011). Igf1 and Igf2, are related to Igfbp3, which interacts with humanin expressed in Leydig cells. Humanin together with IGFBP3 and Bax prevents the activation of germ cell apoptosis (Jia et al., 2015; Lue et al., 2021). Importantly, expression of IGFBP3 is regulated by the HOXD10 gene (Xue et al., 2013; Pan et al., 2021), whose expression was found to increase by 2.9-fold in our study. Hoxd10 is involved in prostate cancer processing, cell differentiation and morphogenesis (Mo et al., 2017; Zhang et al., 2019; Jonkers et al., 2020).
Figure 8.
STRING analyses of potential interactions between proteins with observed differences in gene expression level in RNAseq from mouse testis of the KO model for Tcte1 gene. Genes revealed by the RNAseq are bracketed. (STRING database, 24 February 2021).
Another gene with a 2.2-fold expression rise is Slc45a3, a well-known marker of prostate cancer progression, whose loss determines poorer prognosis for cancer patients (Perner et al., 2013; Hernández-Llodrà et al. 2017; Pin et al., 2017). Overexpression of another gene identified in our study (a 12-fold rise for the Usp39 gene) is linked with promotion of prostate or ovarian tumourigenesis. Usp39 is also involved in pre-mRNA splicing, cytokinesis and spindle checkpoint function, and is crucial for apoptosis (Huang et al., 2016; Zhao et al., 2016; Yuan et al., 2021).
Furthermore, a 2.2–2.5-fold increase for ncRNA genes (Gm26848, Gm45648; no data concerning their role) and a 2-fold reduction for one pseudogene (Gm28661; also no data) were found (Fig. 7). Following the latest data (November 2021) from the Gene NCBI database, Gm45648 has been annotated as having testis-restricted expression in mice, but no function has been suggested. The second ncRNA Gm26848 found is suggested as an organic cation transporter-like protein across membranes in Drosophila sechellia. There were also two other genes with expression changes: a 3.6-fold decrease in pseudogene Gm44126, and a 2.5-fold increase in 4933430L12Rik, which is probably involved in antisense transcription). ‘Gene’ database data showed that Gm44126 in mice is a gene coding a protein with the function of UDP-glucuronic acid decarboxylase and contains two domains involved in NAD(P)H processing. Thus, it can be suggested that this gene is a novel candidate involved in the mitochondrial respiratory chain. Our data with 3.6-fold expression decrease of this gene seem to fit into the observed diminished functioning of mitochondrial machinery in Tcte1−/− mice.
Another three genes manifested 2-fold decreases of their testis expression when comparing Tcte1−/− versus Tcte1+− (but not to WT): Aqp2 (aquaporin-2, which is involved in osmotic transportation of water), with well-known roles in spermatogenesis, the female endometrium cycle, and urinary incontinence (Klein et al., 2013; Ramli et al., 2019); Mt3 (metallothionein-3, committed in homeostasis and detoxification of toxic metals, also in testis) which is important in apoptosis regulation of Bax and Caspase pathways (Honda et al., 2010; Gao et al., 2021)); and Rps27rt, a ribosomal protein (Fig. 7; Supplementary Data File S5). It was also found that Rps27rt may interact indirectly with Usp39 and mitochondrial respiratory complex linking mitochondrial energy processing with apoptosis (Supplementary Data File S5). The last gene, Ube2l6, was found to have a 2-fold increased expression level in the testis in Tcte1+/− animals versus Tcte1−/− and WT. Its role has been established as a ubiquitin enzyme engaged in: stress damage response, cell cycle progression, embryo development, immune response, and factors linked to ATP machinery (Fig. 7) (Huang et al., 2016; Zhao et al., 2016; Sandy et al., 2020; Cai et al., 2021; Yuan et al., 2021). Increased expression of Ube2l6 is linked to degradation/suppression of cancer cells and affected downstream apoptotic factors (i.e. Caspase 3, Caspase 9, Bcl-2, Bax) (Li et al., 2018).
STRING analyses of potential interactions between proteins with observed differences in gene expression levels in RNAseq in mouse testis of the KO model for the Tcte1 gene are shown in Fig. 8.
Immunofluorescence in sperm
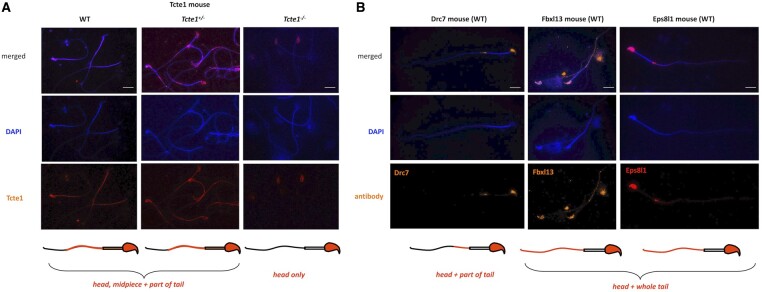
An example of immunofluorescence in situ staining on mouse spermatozoa used to detect the N-DRC proteins has been presented in Fig. 9. WT mouse spermatozoa showed Tcte1 protein presence in the sperm head nucleus, midpiece, and a part of the tail (Fig. 9A). Immunofluorescence signals of Tcte1 in Tcte1+/− mice were similar to that in WT animals, while in Tcte1−/− were localized only in the nucleus (Fig. 9A). It seems to confirm that in Tcte1−/− males, the protein has not been produced properly, revealing only residual amounts in the sperm head nucleus, with no transport to the sperm flagella. The other N-DRC components (Drc7, Fbxl13, and Eps8l1) were localized in the sperm nucleus and among the whole sperm tail (Fbxl13, Eps8l1) or only within its fragment (Drc7), with exception of the midpiece (Fig. 9B). The results obtained seem to suggest co-localization of evaluated N-DRC proteins in various combinations, not common among the midpiece and the terminal part of the sperm tail.
Figure 9.
Immunolocalization of the nexin–dynein regulatory complex (N-DRC) proteins: Tcte1 (Drc5), Drc7, Fbxl13 (Drc6), and Eps8l1 (Drc3) within mouse sperm cells. (A) Localization of Tcte1 in spermatozoa from wild-type (WT), hetero- (+/−) and homozygous (−/−) animals. (B) Localization of other proteins (Drc7, Fbxl13, Eps8l1) building the nexin–dynein regulatory complex (N-DRC), responsible for the coordination of the dynein arm activity and stabilization of the doublet microtubules attachment, in wild-type (WT) spermatozoa. Antibodies: primary (all anti-rabbit 1:100; Biorbyt, Cambridge, UK): Tcte1 (orb357083); Drc7 (orb58695); Fbxl13 (orb678278); Eps8l1 (orb382538); secondary: goat anti-rabbit-AF594 conjugated, 1:500, ab150160, Abcam, Cambridge, UK. Fluorescence microscope: Leica DM5500, filters: DAPI, TxR, FITC, BGR, magnification 630 (with immersion); software: LASX or CytoVision. Bar represents 10 µm.
Sperm ATP levels
Measurements performed at two time points (0 and +60 min) revealed that in Tcte1−/− samples, the luminescence level was ∼2.4-fold lower when compared to WT and Tcte1+/− mice (P < 0.0001) (Fig. 10; Supplementary Table S3). There were no differences between WT and Tcte1+/− males. Additionally, the decrease in the luminescent signal after 60 min was similar in all three groups studied: ∼35% (WT: 35.27%, Tcte1+/−: 33.88%, Tcte1−/− 37.76%).
Figure 10.
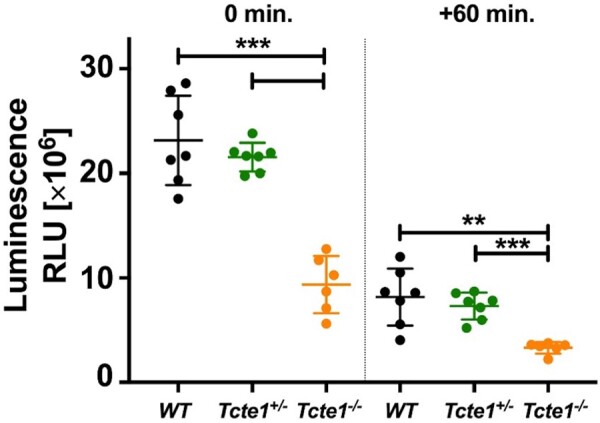
ATP levels in wild-type (WT), heterozygous (+/ − ) and homozygous ( − / − ) Tcte1 mice. Measurements made at two time points (0 and +60 min) showed a decrease of ∼35% in luminescence in all three groups over time. Mean measured values in the homozygous group were ∼2.4-fold lower. Dots: particular animals; long dash: mean value for the group; short dashes, standard deviation; RLU, relative luminescence unit. ***P < 0.0001, **0.0001 < P < 0.01.
Apoptosis in testicular tissue
Evaluation of the immunofluorescence staining results revealed Casp3-positive signals (indicating apoptosis) in spermatogonia only, in all three mouse genotypes (Fig. 11A). In each zygosity, the numbers of tubules and positively stained spermatogonia (intensity of the signal) were similar, suggesting similar levels of apoptosis. Also, the lack of differences between zygosities was found in TUNEL assays, for all testicular cell types (Fig. 11B). Apoptosis-positive signals were observed in all tubules with both techniques.
Figure 11.
Immunolocalization of caspase 3 and TUNEL assay results within mouse testicular tissue of the KO Tcte1 model. Caspase 3 (A) is one of the apoptotic indicators because of its engagement in the induction, transduction, and amplification of intracellular apoptotic signals. Caspase 3 signals were found only in spermatogonia of all three genotypes (wild-type (WT), hetero- (+/−), and homozygous (−/−)). No differences between the level of fluorescent signals were found between evaluated mouse genotypes, indicating similar levels of apoptosis. Antibodies: primary (1:100) rabbit anti-Casp3 (Biorbyt, Cambridge, UK); secondary (1:500): goat anti-rabbit-AF594 conjugated (ab150160, Abcam, Cambridge, UK). (B) TUNEL assay revealed no differences between zygosities (positive BrdU signal). Kit used: ab66110 TUNEL Assay Kit: BrdU-Red; Abcam, Cambridge, UK. Fluorescent microscope: Leica DM5500, filters: DAPI, TxR, magnification 630 (with immersion); software: LASX. Bar represents 5 µm. Analyzed: three independent males per zygosity, on 15–20 tubules each.
Sequencing: human male samples
There were six potentially associated or important heterozygous SNVs for the TCTE1 gene found in 15 out of 248 (6.05%) infertile males with drastically decreased semen parameters, ranging from azoospermia to severe oligoasthenozoospermia (Table 1). Three variants were identified with ultrarare allele frequency (MAF ≤0.0005 in the general population; gnomAD) in three males with NOA: c.862C>T (p.Arg288Ter), predicted as a stop-gained mutation leading to non-mediated decay, and thus to protein loss of function with high probability; c.185G>A (p.Arg62His) and c.1048G>A, (p.Gly350Ser), defined as missense ones and probably damaging. The next two SNVs were found as rare missense ones with MAF <0.005: c.397C>T (p.Arg133Cys) (one NOA case) and c.470A>C (p.Glu157Ala) (four NOA cases), and were defined as probably damaging. The last SNV (found in one male with NOA, in one with cryptozoospermia, and in five with severe oligoasthenozoospermia) was not found in the databases, and thus was specified as a novel SNV (c.374T>G; p. Ile125Arg) with missense prediction (Table 1; Supplementary Fig. S4).
Table 1.
Potentially disease-causing novel and ultrarare heterozygous TCTE1 variants found in patients with disturbed spermatogenesis.
| Sample ID | Phenotype [if available—sperm count: concentration/volume/total//motility (%): a + b/total | Origin | NT change | cDNA position | Prediction | Mutation T@ster | GnomAD | rs | Length of protein | CADD | Position(s) of altered AA [HGVSp] | Polyphen/Sift | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Novel variants | |||||||||||||
| P52 | SOAs | 4.3 × 106/ml/3.0 ml/12.9 × 106//15.0/20.0 | Poland | c.374T>G | cDNA.497T>G | Disease causing | Amino acid sequence changed; protein features (might be) affected; splice site changes | Not found | — | Normal | Missense | p.Ile125Arg | Unknown |
| P88 | SOAs | 3.4 × 106/ml/1.5 ml/5.1 × 106//2.0/5.0 | |||||||||||
| P105 | SOAs | 0.2 × 106/ml/1.5 ml/0.3 × 106//17.0/23.0 | |||||||||||
| P111 | SOAs | 3.7 × 106/ml/3.0 ml/11.1 × 106//15.0/20.0 | |||||||||||
| P161 | SOAs | 2.3 × 106/ml/3.0 ml/6.90 × 106//10.0/13.0 | |||||||||||
| P94 | Crypto | 0.05 × 106/ml/2.0 ml/0.1 × 106//0.0/0.0 | |||||||||||
| P149 | NOA | 0.0 × 106/ml/2.0 ml/0.0 × 106//0/0 | |||||||||||
| Ultrarare variants (MAF <0.005) | |||||||||||||
|
NOA | Low volume | USA | c.185G>A | cDNA.308G>A | Disease causing | Amino acid sequence changed | 0.00007778 | rs201144307 | Normal | Missense | p.Arg62His | Probably damaging/deletorious |
| U17 | NOA | Late meiotic arrest | USA | c.397C>T | cDNA.520C>T | Disease causing | Amino acid sequence changed; heterozygous in TGP or ExAC; protein features (might be) affected; splice site changes | 0.001722 | rs145304614 | Normal | Missense | p.Arg133Cys | Probably damaging/deletorious |
| U50, U57, U117, U235 | NOA | Low volume | USA | c.470A>C | cDNA.593A>C | Disease causing | Amino acid sequence changed; heterozygous in TGP or ExAC; protein features (might be) affected; splice site changes | 0.002681 | rs45591736 | Normal | Missense | p.Glu157Ala | Probably damaging/deletorious |
| U11 | NOA | Early meiotic arrest | USA | c.862C>T | cDNA.985C>T | Disease causing | NMD amino acid sequence changed; protein features (might be) affected; splice site changes | 0.0002840 | rs138414421 | NMD | Stop_gained | p.Arg288Ter | Stopgain SNV; PLoF: high confidence |
| U76 | NOA | USA | c.1048G>A | cDNA.1171G>A | Disease causing | Amino acid sequence changed; protein features (might be) affected; splice site changes | 0.00006382 | rs570334918 | Normal | Missense | p.Gly350Ser | Probably damaging/deletorious | |
Results obtained from the screening of n = 248 participants. Minor allele frequency data obtained from GnomAD v2.1.1.
SOAs, severe oligoasthenozoospermia; NOA, non-obstructive azoospermia; Crypto, cryptozoospermia; NMD, non-mediated decay; NT, nucleotide; AA, amino acid; SNV, single-nucleotide variant; PLoF, loss of function probability; rs, reference SNP accession number; CADD: Combined Annotation Dependent Depletion tool.
Protein prediction
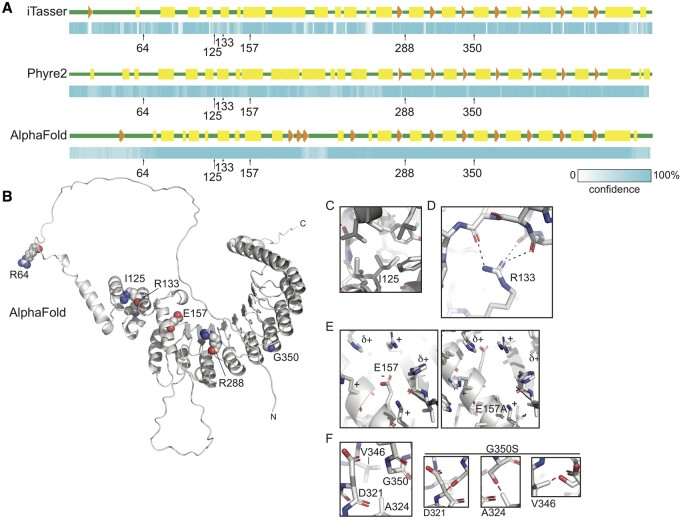
To understand the molecular implications of the identified mutations on the structure and function of TCTE1, we performed homology modeling experiments using the Phyre2, I-TASSER, and AlphaFold tools (Roy et al., 2010; Kelley et al., 2015; Yang et al., 2015; Yang and Zhang, 2015; Tunyasuvunakool et al., 2021). TCTE1 is predicted to contain at least five tandem LRRs in its C-terminus (UniProt Consortium, 2019), while no known three-dimensional protein folds have been assigned to its N-terminus. All homology searches yielded a polypeptide with tandem LRRs with nearly 100% confidence (Fig. 12A; Supplementary Fig. S5). Modeling the N-terminal region of TCTE1 was more difficult; although both Phyre2 and I-TASSER confidently predicted a largely alpha-helical secondary structure for the N-terminal half of TCTE1 (Fig. 12A; Supplementary Fig. S5). Neither tertiary structure was predicted with high confidence; indeed, a comparison of the two demonstrates vastly different folds for the N-terminus from the programs. However, the confidence of the AlphaFold-generated model was high for the region just N-terminal to the LRR. Further, based on the predicted aligned error plot calculated for its models, the confidence of the predicted structure of residues 100–200 was high relative to the position of residues 220–480, providing support for the modeled through-space three-dimensional interactions. That said, we interpreted these models very conservatively.
Figure 12.
Homology modeling of TCTE1. (A) Secondary structure diagram and associated confidence scheme generated by I-TASSER (Roy et al., 2010; Yang et al., 2015; Yang and Zhang, 2015], Phyre2 (Kelley et al., 2015], and AlphaFold intensive mode. Positions of amino acids of interest are indicated. Green, predicted unstructured regions; yellow rectangles, predicted helices; orange arrows and arrowheads, predicted strands. The confidence scale ranges from white to cyan, with the probability of the secondary structure increasing with increasing intensity of cyan. (B) Cartoon diagram of the homology model of human TCTE1 generated by AlphaFold with residues of interest shown as spheres and labeled. N, amino terminus. C, carboxy terminus. (C–F) Detailed views of indicated residues and mutations: (C) Ile-125 resides deep within a hydrophobic pocket, which would be disturbed upon substitution with arginine. (D) Mutation R133C would impact the hydrogen bond interactions that organize the architecture of the globular domain N-terminal to the tandem leucine-rich repeats. (E) Mutation of Glu-157 to alanine would destabilize the charge of the surface of this globular region, likely leading to unfolding. (F) Gly-350 allows for the compact interactions of tandem helices of the LRR. Depending on the rotamer it adopted, a serine side chain in this position would sterically clash with residues D321, A324, or V346 in the neighboring helix.
We attempted to hypothesize how the mutations of interest could affect the tandem LRRs and putative secondary structure of the N-terminus of the protein. Residue R64 resides in a predicted unstructured region and, as such, most likely is solvent exposed in the tertiary structure (Fig. 12A and B). Mutation of this residue from arginine to histidine could therefore affect the surface potential of this disordered region, making it less positively charged. Indeed, this mutation would be expected to impact the interactions that the N-terminus of TCTE1 can have with other molecules, including other members of the N-DRC. The interaction of microtubule-associated proteins, including the N-DRC, with binding partners, is known to be largely charge-based (Kubo and Oda, 2017; Drechsler et al., 2019).
Three mutations of interest occur in the globular region just N-terminal to the LRR. The AlphaFold model clearly shows how mutation I125R would affect the structure of this region (Fig. 12A and C). Residue isoleucine 125 is predicted to be nestled within the hydrophobic core including L97 L109, L112, F91, W131, L197, Y130, and the methyl group of the T121 sidechain. A substitution of positively charged arginine for isoleucine in this position would not be accommodated in this hydrophobic region, likely resulting in local unfolding that could potentially propagate beyond the pocket. Likewise, residue R133 supports the structure of this putative globular domain by forming hydrogen bonds with the backbone of residues 109, 110, and/or 112, thereby possibly contributing to the overall organization of this domain and its positioning relative to the LRR (Fig. 12D). Mutation of this residue to a cysteine would remove the hydrogen bonding groups supporting these interactions, likely leading to a local unfolding that could possibly propagate further. Interestingly, residue E157 is predicted to interact with residues in the LRR, likely stabilizing positively charged residues around it, including several from the linker leading up to the first alpha-strand of the LRR (Fig. 12B and E). Mutation of this residue to alanine would lead to a tertiary destabilization, most likely affecting the overall structure of the N-terminal region of the LRR and the globular region just N-terminal to it.
Two mutations of interest affect the LRR helices. Mutation R288* results in premature truncation, eliminating most of the LRR region from the final protein product, although it is predicted that this premature termination codon would lead to nonsense-mediated decay. Mutation G350S would be expected to result in the destabilization of inter-repeat interactions (Fig. 12F), possibly even unfolding the protein from that position on; the lack of a side chain in glycine allows for the close approach of these tandem structural elements, whereas the addition of the serine side chain would abrogate the tight packing of residues, regardless of which rotamer it adopts. This could result in a restructuring of its C-terminus and/or deforming the surface of the module, therefore affecting its interactions with its binding partners.
Since the N-DRC is conserved from single-celled eukaryotes to mammals, we took advantage of published cryo-electron tomography studies examining the N-DRC from model organism Chlamydomonas (Gui et al., 2019). This study was of great interest since it explicitly located the position of the C-terminus of DRC5 with a nanogold particle; importantly, it was consistent with earlier cryo-ET experiments positioning the N-DRC (Heuser et al., 2009). While the resolution of this structure is only 40 Å, it was still useful for placing the modeled TCTE1 structure in the context of the entire N-DRC (Supplementary Fig. S5B). Manual docking of the AlphaFold predicted TCTE1 structure is consistent with the cryo-EM tomography data (Heuser et al., 2009; Gui et al., 2019).
Discussion
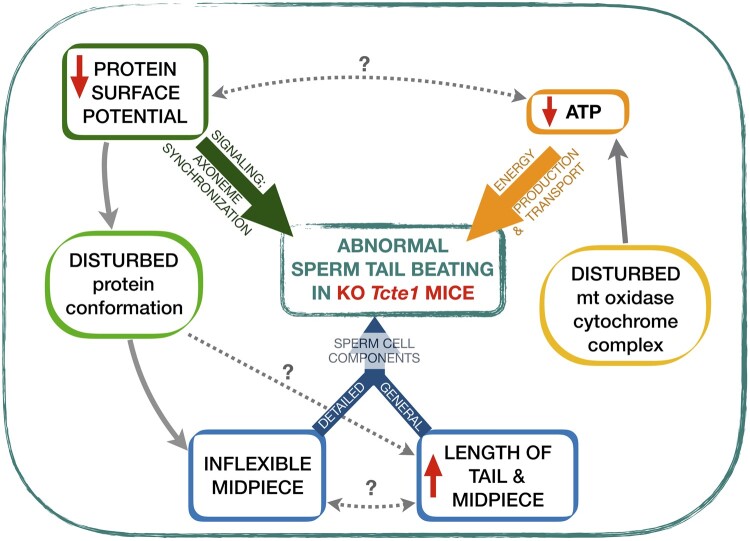
In this study, we have investigated the role of Tcte1 in male infertility using a mouse knockout model. Using homo- and heterozygous animals, their reproductive potential has been examined, and no progeny were produced by homozygous males. The size of the gonads varied between the zygosity of animals, with two semen phenotypes revealed by sperm quality evaluation. RNAseq analysis of the mutant testicular tissue demonstrated the influence of Tcte1 mutations on the expression pattern of genes related to mitochondrial cytochrome oxidase activity, apoptosis and spermatogenesis. The ATP level estimates of spermatozoa showed a decrease in homozygous males. Additionally, protein prediction modeling of identified variants in human samples revealed changes in the protein surface charge potential, leading to disruption in helix flexibility or dynamics, and thus, suggesting a disrupted TCTE1 interaction with its binding partners within the axoneme. The major findings concerning observed abnormal sperm tail beating in KO Tcte1 mice are summarized in Fig. 13. We have also identified and discussed novel functions of genes with testicular expression significantly disturbed in the Tcte1 knockout. Therefore, this study contains novel and comprehensive data concerning the role of TCTE1 in male infertility.
Figure 13.
Summary of findings revealed for abnormal sperm tail beating in the mouse knockout model of the Tcte1 gene, a component of the N-DRC complex in the sperm tail. Three major groups of factors required for proper sperm tail beating are marked with bold color arrows (green, yellow, and blue). Documented connections (solid grey arrows) were revealed on the basis of RNAseq data for testis tissues, protein in silico predictions for mutated Tcte1 protein, and measurements of ATP level and sperm cell components in mouse spermatozoa. Unresolved possible linkages (cause/effect) are marked with a question (?) mark.
Phenotypes of homozygotes and heterozygotes
There was only one previously published study concerning a knockout mouse model of Tcte1 (Castaneda et al., 2017). It was shown that the expression of Tcte1 starts at the stage of early haploid round spermatids and continues into the elongated spermatids. The authors showed the infertility of homozygous males due to asthenozoospermia derived from limited and circular flagellar beating, followed by fertile heterozygotes. The study did not find any differences between homozygous and WT males in the testicular histology, followed by similar testis weight, sperm concentration, and sperm morphology (Castaneda et al., 2017). We also did not find any structural differences in the testicular and epididymal tissue histology. However, the weight of the testes decreased significantly (∼41% in Tcte1−/−, 7% in Tcte1+/−), followed also by testis size reduction (by 22%) and lowered germ cell layer thickness in Tcte1−/− animals (Fig. 4B and C). When comparing semen parameters (Fig. 6A–C), we found an enormous decline in sperm concentration in both Tcte1− (3.3-fold) or Tcte1+/− (2.5-fold) males, and a double increase in abnormal sperm morphology in Tcte1−/− animals. Similar to Castaneda et al. (2017), we also documented the impaired sperm motility representing a circular mode of the sperm tail beating. In addition, the increased ratio of immotile spermatozoa in homozygous males was documented in our study. We can suggest that these discrepancies may result from the various number of animals used for evaluation: three males in the study of Castaneda et al., (2017) versus at least 18 per each zygosity in our study when comparing semen parameters and tissue characteristics (except for sperm morphology evaluation with at least 5 animals used). Furthermore, the differences in knockout construction used could also be causative for the observed variances: reporter-tagged insertion with conditional potential versus the exon 3 deletion. Thus, we have observed two various phenotypes dependent on the zygosity of the animals: homozygous males manifested oligoasthenoteratozoospermia (OAT) and were infertile, while heterozygous males demonstrated oligozoospermia (O) and remained fertile. Data collected in the International Mouse Phenotypic Consortium (IMPC; a repository of all available mouse phenotypes created, followed by the possible linkage to human disease defects; https://www.mousephenotype.org), concern only one Tcte1 mouse study (Castaneda et al., 2017), while we reveal an unmatched phenotype of decreased sperm count.
We suggest that a similar effect, decreased sperm count linked to TCTE1 mutations, could be observed in humans. Our pilot mutation screening of 248 human infertile males with severely decreased sperm count (from oligo-, via crypto-, to azoospermia) revealed six heterogeneous SNVs (one novel, three ultrarare, and two rare; all predicted as disease-causing) with a total frequency of 6.05% (Table 1). We propose that molecular epidemiology studies of infertile patients should reveal the existence of homozygous or compound heterozygous TCTE1 variants with clear direct downgrading effects on spermatogenesis. However, we claim that also heterozygous variants can be causative, similar to SPINK2 whose deficiency has been documented as responsible for oligoasthenozoospermia in heterozygous mice versus azoospermia in homozygous animals, as well as azoospermia in human homozygous brothers versus their heterozygous father with oligozoospermia (Kherraf et al., 2017). It can be explained by the probable occurrence of haploinsufficiency; when the single copy of the WT allele is in a heterozygous combination, the variant allele is insufficient to produce the WT phenotype. Our observations, in addition to the SPINK2 data and data related to infertility (i.e. P1/P2 protamines, Pde8b, Trim28, Tcfl5), and to the other diseases (i.e. PIKFYVE in cataract, NFIA, SCN1A in neurological or epilepsy disturbances, KLF1 in hemoglobin persistence, NR5A2 in pancreatic cancer), seem to confirm that the severity of the phenotype depends on the gene expression level (Jodar and Oliva, 2014; Tan et al., 2020; Leal et al., 2021; Sandhu et al., 2021; Heshusius et al., 2022; Mei et al., 2022; Ogura et al., 2022; Valassina et al., 2022; Xu et al., 2022). We have documented variable phenotypes for a novel TCTE1 variant c.374T>G (p.Ile125Arg) that has been documented in seven males, with azoospermia, cryptozoospermia, or severe oligoasthenozoospermia. Thus, it seems to suggest the reasonable candidacy of haploinsufficiency as the mechanism responsible for the observed phenotypic spectra also in the case of the TCTE1 gene. There is only one study available documenting TCTE1−/− in a human case, so far (Zhou et al., 2022). Among 130 asthenozoospermic patients, the authors found a frameshift mutation c.396-397insTC (p.Arg133Serfs*33) in one of them that resulted in the premature translational arrest of the forming peptide. Successful delivery was possible only after in vitro fertilization (IVF). Importantly, the authors also checked the IVF rate in homozygous mice but showed no pregnancy success. The unresolved question was whether homozygous TCTE1 mutation in humans causes only asthenozoospermia or rather the semen quality phenotype is mixed, including also abnormal morphology, but this was not evaluated by the authors.
Kallikreins, aquaporin 2
Our pathway analysis revealed a network including two genes with changed expression level in testis: Igf2 (2-fold increased) and Klk1b22 (8-fold decreased), supported by an interesting set of proteins linked to apoptosis and cellular processing (Figs 7 and 8; Supplementary Data File S5 and Fig. S3). We can suggest that the observed Igf2 overexpression in our study can be one of the associated factors leading to decreased sperm count and testis parameters in the evaluated KO mice model because of its known relation to spermatogenesis and testis development (Damaschke et al., 2017; Neirijnck et al., 2019; Cannarella et al., 2020; Matsuzaki et al., 2020). Documented reductions can be supported by the observed constrained expression of Klk1b22; kallikrein is mostly known for its neuronal activity; however, it cannot be excluded that we have revealed an additional role of this protein, similar to the other members of the kallikrein family with roles in semen liquefaction and degradation of extracellular matrix proteins in the interstitial area surrounding Leydig cells of the adult mouse testes (Klk1b27, Klk1b21, Klk1b24) (Fink et al., 2007; Marques et al., 2016, Du Plessis et al., 2013; Michaelis et al. 2017). Notably, Klk1b22 expression is significantly enriched in testis. Expression of all four kallikreins was decreased by 2.5 to 8-fold in our Tcte1 KO model, indicating their participation in the reduction of the sperm parameters measured (Fig. 5).
We have also documented a 2-fold decrease in the expression of Aqp2 (Aquaporin 2), which is involved in fluid resorption and water movement within the duct system, with a known role in spermatogenesis (Klein et al., 2013; Ramli et al., 2019). It was documented that in stallions, Aqp2 is expressed in Leydig cells, round and elongated spermatids (Klein et al., 2013). In addition, this protein works within a network of molecules that are linked to cell membrane trafficking (i.e. Rab11a, Myo5b), followed by activity of protein kinases (Prkacb, Prkaca) that are known as regulators of cell proliferation, chromatin condensation/decondensation, and nuclear envelope disassembly/reassembly, and all is supported by ATP metabolism (Hspa8) (Supplementary Data File S5) (Markou et al., 2022; Wagner et al., 2022). Considering the literature data, we suggest Aqp2 plays an important role in the reduction of the spermatid volume during spermatogenesis and in proper sperm morphology assessment and the observed morphological differences between Tcte1 mice zygosities in our study. However, such measurements require high-resolution microscopy, thus, we can only speculate the Aqp2 role in our model.
Disruptions in sperm cell structure
The TCTE1 protein contains five LRRs that are known to be directly involved in receptor-mediated signaling on a structural basis for interaction between proteins (Kobe and Kajava 2001; Ng et al., 2011). It was documented that variants in genes encoding LRRs are commonly implicated in the pathogenesis of more than 60 human diseases (Ng et al., 2011; Matsushima et al., 2019). LRR mutations mostly occur within regions that are protective for the hydrophobic core of the LRR domains. Thus, mutations in LRRs may affect the charge of the surface potential, leading to conformational changes of the protein or induction of protein cumulation. Our prediction model of potential changes in the TCTE1 protein resulting from mutations found in human samples seems to point out the prospective impact on the interactions of the TCTE1 with other molecules (binding partners) known to be largely charge-based, including N-DRC elements or microtubule-associated proteins (Kubo and Oda, 2017; Drechsler et al., 2019). In the case of the TCTE1 protein, observed amino acid substitutions promote the abnormal aggregation of the TCTE1 protein, and decrease its structural stability, probably leading to the observed disturbances. The beating motion of flagella is maintained by an electrostatic cross-bridge between negatively charged tubulins and positively charged N-DRC (Kubo and Oda, 2017). Thus, changes in the charge potential of the TCTE1 surface, which is one of the N-DRC components, seem to provide modified (reduced) flagellar beating, similar to that with mutations in another N-DRC element, the DRC4 protein, where changes in charge were essential for proper flagellar motility (Kubo and Oda, 2017; Lewis et al., 2016). Also, the inflexibility of the midpiece observed in spermatozoa of Tcte1 knockout mice seems to be a consequence of the changed structure of LRRs in the Tcte1 protein.
It is also known that the lengths of the sperm midpiece and tail may be crucial in the determination of sperm swimming velocity, which is then followed by a decreased fertilization rate (Firman and Simmons, 2010; Gu et al., 2019). Available data show that sperm with a shorter midpiece may swim slowly and are observed in asthenozoospermic cases (Anderson and Dixson, 2002; Firman and Simmons, 2010; Gu et al., 2019). A meta-analysis of sperm shape features according to sperm competition theory showed that, in the majority of vertebrates, a longer sperm cell (including midpiece or flagellum) has been attributed to faster swimming and that the sperm size may be increased when sperm number is lowered (Lüpold et al., 2020). In our study, we have documented longer sperm tails and midpieces in Tcte1−/− animals that, in fact, were infertile. Thus, we can suggest that: (i) the observed longer sperm tails and midpieces may be co-responsible for the aberrant motility observed, and (ii) the documented longer sperm tails and midpieces may be an indirect result of the constrained sperm count observed, because of some sort of compensation for aberrant motility. However, this issue remains only a suggestion because of restricted literature data relative to similar observations. Bennison et al. (2016) revealed that, in a passerine bird, sperm tail velocity was correlated to total sperm length, but only up to a certain point of the length value, at which the velocity started to decrease. On the other hand, Gu et al. (2019) showed that in mammalian species, there were some changes in the axonemal slopes when the flagellar length increased. The results obtained in our study and supported by other research concerning sperm tail-beating pathways should be a good starting point for the creation of further mathematical computations for a better understanding of asthenozoospermia.
Constrained energy metabolism
Flagellar motility is determined by the coordinated activity of asymmetrically distributed dyneins in the axoneme. The driving factor for this activity is the hydrolysis of ATP released from the OXPHOS system (Mitochondrial Oxidative Phosphorylation System) (Ford 2006; Tourmente et al., 2015). ATP is being produced in mitochondria placed within the sperm midpiece. In our study, ATP level measurements revealed its decreased levels in sperm cells of Tcte1−/− mice, which can result in faulty dynein functioning, leading to asthenozoospermia via abnormal sperm tail beating. We have documented that Tcte1 KO resulted in significant changes in the expression level of testicular genes involved in mitochondrial energy transport. That leads to the conclusion that homozygous Tcte1 mutation results not only in an inflexible midpiece leading to asymmetric circular motility, as presented in this study and by others (Fung et al., 2018), and then to asthenozoospermia, but also to aberrant functioning of mitochondrial OXPHOS machinery leading to the disruption of ATP production. Thus, the sperm tail has also insufficient power for beating because of the decreased ATP content. Castaneda et al. (2017) suggested that the reason for the observed ATP decrease may result from disturbances in a glycolytic pathway. It is already known that glycolytic energy is enough for spermatozoa to survive (Nascimento et al., 2008; Zhang et al., 2022). However, for sperm motility and then fertilization, OXPHOS energy production is required (Ford 2006; Tourmente et al., 2015; Zhang et al., 2022). In our study, RNAseq results clearly revealed the influence of the Tcte1−/− mutation on genes involved directly in the electron transport chain, such as mt-Co2 and mt-Co3, crucial components of the respiratory chain complex IV, one of the OXPHOS elements. Observed reduced expression seems to clearly demonstrate decreased ATP production via disturbed electron chain transportation, and the reduction in the sperm motility observed in Tcte1−/− males, leading to the observed asthenozoospermia. Detailed STRING analysis revealed definite interactions between both listed proteins identified in our study, and a list of other mitochondrial ones (mt-Co1, mt-Cytb, Cox5a, Cox6a, Cox7c, mt-Atp6; Supplementary Data File S5 and Fig. S3) clearly related to proper processing of cytochrome energetic activities (Ganetzky et al., 2019; Brischigliaro and Zeviani, 2021). Overexpression or mutations in COX1, COX2, and COX3 were previously observed in infertile men with varicocele and OAT, or with diabetes (Perrotta et al., 2012; Heidari et al., 2016; Mostafa et al., 2016). Additionally, it was shown that the system of COX2 and prostaglandins plays a key role in the development of the male gonad, spermatogenesis, and steroidogenesis, especially in the functioning of Sertoli and Leydig cells, and was also found to be significant in the background of idiopathic infertility status (Frungieri et al., 2015). It seems that, in our study, the reason for the observed disturbed motility, represented as circular movement of spermatozoa, supported by increased ratio of immotile sperm cells in Tcte−/− mice, leading to asthenozoospermia, was a result of two factors: (i) inflexibility of the sperm midpiece resulting from a changed protein charge level in the Tcte1 protein, followed by elevated lengths of the sperm tail and midpiece, and (ii) the decreased amount of ATP produced, with an inability to get sufficient energy for proper sperm motility.
Protection and/or compensation?
We have also documented an important role of the Fetub gene for the first time in the male testis. Besides its known role in female fertility (Bansho et al., 2017; Karmilin et al., 2019), a detailed later study suggested a role for it in the context of prostate cancer (Zhan et al., 2020). It was found that overexpression of FETUB increases the apoptosis of prostate cancer cells, followed by the inhibition of their proliferation, migration, and invasion, and also slows the growth of tumours when compared to controls (Zhan et al., 2020). Thus, in the overexpression mode, Fetub plays a protective role for the prostate. It seems to explain the results obtained in our study, where Fetub expression was increased almost 5-fold in Tcte1−/− mice, but no tumor signs were observed in the mouse testis at the tissue level. Zhan et al. (2020) found a connection between Fetub overexpression, and induction of Bax (Bcl-2-associated X protein) and caspases, resulting in the promotion of apoptosis. It is known that Bax/Bcl-2 induction/inhibition is directly related to the release of cytochrome c (Cyt c) in caspase-dependent signaling (Dadsena et al., 2021; Li et al., 2021; Samaiya et al., 2021). Cytochrome c is one of the two electron carriers in the electron transport chain responsible for transport directly to the respiratory chain complex IV (Alvarez-Paggi et al., 2017; Yin and O’Neil 2021). Due to Cyt c release, complex IV is essential for apoptosis and cell fate (Li et al., 2021; Samaiya et al., 2021). Following all these factors, our results seem to reveal a connection between Fetub and diminished mitochondrial chain IV activity in mice testis, which has not been reported so far. It can be also supported by the known relationship between FETUB and the inactivation of the PI3K/AKT signaling pathway (a key player in the regulation of cell proliferation and apoptosis in cancerogenesis), in which Bax/Bcl-2 is involved (Zhan et al., 2020).
The novel testis-protective role in our study can be also credited to three other genes with increased expression levels in testicular tissue: Hoxd10 (3-fold increase), Slc45a3 (2.2-fold), and Ube2l (2-fold). Overexpression/activation of HOXD10 has already been documented as an inhibitor of tumorigenesis (Mo et al., 2017; Zhang et al., 2019; Jonkers et al., 2020). Expression of Slc45a3, also known as ‘prostein’, has been documented as weaker in cancer tissues (Perner et al., 2013; Hernández-Llodrà et al. 2017; Pin et al., 2017). An elevated expression level of Ube2l is known to be connected with the degradation or suppression of tumor cells (Li et al., 2018). On the other hand, a 12-fold increased gene expression level was observed for Usp39 (ubiquitin specific peptidase 39), an oncogenic splicing factor whose expression is positively linked with cancerogenesis in prostate or ovaries, while its silencing has been shown to induce apoptosis and cell cycle arrest (Huang et al., 2016; Zhao et al., 2016; Yuan et al., 2021).
Taking together data obtained for the genes related to cancerogenesis and/or apoptosis described above, we can hypothesize that we observe an interesting novel network of interactions between two groups of genes that are reciprocally neutralizing the potential influence on the testis tissue. Mainly, in the first group, we have genes with a suggested novel anti-cancer role in the testis (Fetub, Hoxd10, Slc45a3, Ube2l6), with increased expression levels inhibiting cancer cell development, supported by the protective role of Mt3; while the Usp39 gene, with elevated expression levels (the most of all the observed changes), should lead to some tumor-like changes within the testis tissue. Following the fact that both histological evaluation, as well as apoptosis stainings (Fig. 11), showed no differences between heterozygous and homozygous mutant mice, our hypothesis seems to be justifiable. On the other hand, there is a prediction that many of the genes (especially so-called cancer-testis-associated antigens, CTA) related to spermatogenesis may function in tumorigenesis (CheNg et al., 2011; Bode et al., 2014; Wang et al., 2016; Babatunde et al., 2017; Nagirnaja et al., 2018; Yang et al., 2020; Aurrière et al., 2021). Due to known similarities between oncogenic and spermatogenic processes, especially in energy metabolism, we can carefully hypothesize that in our KO model, the genes with changed expression profiles, and known to be highly involved in tumorigenesis, may also be important in spermatogenesis. Obviously, there is a need for further studies in this direction to confirm these suggestions or to reveal the other (still unknown) mechanisms.
Conclusions
We have documented that the TCTE1 gene should be added to the ‘male infertility list’ of molecular background because of its crucial role in spermatogenesis and proper sperm functioning. The mutations in the mouse Tcte1 knockout model revealed two phenotypes, depending on zygosity with infertile oligoasthenoteratozoospermic homozygotes and fertile oligozoospermic heterozygotic males, suggesting the involvement of a haploinsufficiency mechanism. All collected data indicate that the Tcte1 protein is functioning in spermatogenesis and show how its mutations add a level of complexity to the clinical manifestation of decreased semen parameters. The disrupted energy machinery of the spermatozoa, followed by newly suggested roles of genes related to cancerogenesis and/or apoptosis (here in the context of their neutralizing or compensative effect on the testicular tissue), seems to underline the complex and devastating effect of the single-gene mutation on a variety of testicular molecular networks, leading to reproductive failure. Further advanced studies are required to reveal the details of the mechanisms responsible for the molecular pathways of the interactions between Tcte1 (the structural protein) and other non-structural networks whose expression levels were changed remarkably in this study.
Supplementary Material
Acknowledgements
We are grateful to Oliwia Kordyl, MSc, and Katarzyna Kurek, MSc (IHG PAS, Poznan) for their technical help in the preparation of the samples for sequencing, genotyping, and immunofluorescence staining.
Contributor Information
Marta Olszewska, Institute of Human Genetics, Polish Academy of Sciences, Poznan, Poland.
Agnieszka Malcher, Institute of Human Genetics, Polish Academy of Sciences, Poznan, Poland.
Tomasz Stokowy, Scientific Computing Group, IT Division, University of Bergen, Bergen, Norway.
Nijole Pollock, Department of OB/GYN and Reproductive Sciences, School of Medicine, University of Pittsburgh, Pittsburgh, PA, USA.
Andrea J Berman, Department of Biological Sciences, University of Pittsburgh, Pittsburgh, PA, USA.
Sylwia Budkiewicz, Institute of Human Genetics, Polish Academy of Sciences, Poznan, Poland.
Marzena Kamieniczna, Institute of Human Genetics, Polish Academy of Sciences, Poznan, Poland.
Hanna Jackowiak, Department of Histology and Embryology, Poznan University of Life Sciences, Poznan, Poland.
Joanna Suszynska-Zajczyk, Department of Biochemistry and Biotechnology, University of Life Sciences, Poznan, Poland.
Piotr Jedrzejczak, Division of Infertility and Reproductive Endocrinology, Department of Gynecology, Obstetrics and Gynecological Oncology, Poznan University of Medical Sciences, Poznan, Poland.
Alexander N Yatsenko, Department of OB/GYN and Reproductive Sciences, School of Medicine, University of Pittsburgh, Pittsburgh, PA, USA.
Maciej Kurpisz, Institute of Human Genetics, Polish Academy of Sciences, Poznan, Poland.
Supplementary data
Supplementary data are available at Human Reproduction Open online.
Data availability
All data generated or analyzed during this study are included in this published article and its supplemental information files. RNAseq data are available in GEO database (https://www.ncbi.nlm.nih.gov/geo/) under the accession number: GSE207805. The results described in the publication are based on whole-genome or exome sequencing data which includes sensitive information in the form of patient-specific germline variants. Information regarding such variants must not be shared publicly following the European Union legislation described in the following document: https://publications.jrc.ec.europa.eu/repository/bitstream/JRC113479/policy_report_-_review_of_eu_national_legislation_on_genomics_-_with_identifiers_1.pdf, therefore the access to raw data that support findings of this study are available from the corresponding author upon reasonable request.
Authors’ roles
M.O.: design of the study, manuscript drafting and editing, coordination of the tasks, data interpretation, gene selection, preparation of human blood and semen samples, mice: mating, genotyping, gross sections and preparation of biological samples, RNAseq analysis, immunofluorescence, Sanger sequencing, and ATP measurements; A.M.: gene selection, data interpretation, genotyping, RNAseq analysis, collection and preparation of human testis samples, WGS and Sanger sequencing, and manuscript editing; T.S.: bioinformatic analysis of RNAseq and WGS results; N.P.: WES of azoospermia samples (including bioinformatics); A.J.B.: protein prediction modeling and its description; S.B.: preparation of DNA for sequencing and RNA for RNAseq, and screening of databases for RNAseq results; M.Ka.: collection of human blood and semen samples, semen analyses of human and mouse samples, and detailed morphology of mouse spermatozoa; J.S.-Z. and H.J.: histopathology of mouse tissue sections; P.J.: recruitment of patients and their medical history; A.N.Y.: gene selection, interpretation of azoospermia WES results, and data interpretation; M.Ku.: data interpretation, gene selection, recruitment of patients and their medical history, correction of the manuscript, funds collection and supervision. All authors critically reviewed and approved the final version of the article.
Funding
National Science Centre in Poland (2015/17/B/NZ2/01157, 2020/37/B/NZ5/00549 to M.K., 2017/26/D/NZ5/00789 to A.M., HD096723, GM127569-03, and NIH SAP #4100085736 PA DoH to A.N.Y.).
Conflict of interest
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
References
- Agarwal A, Baskaran S, Parekh N, Cho CL, Henkel R, Vij S, Arafa M, Panner Selvam MK, Shah R.. Male infertility. Lancet 2021;397:319–333. [DOI] [PubMed] [Google Scholar]
- Alvarez-Paggi D, Hannibal L, Castro MA, Oviedo-Rouco S, Demicheli V, Tórtora V, Tomasina F, Radi R, Murgida DH.. Multifunctional cytochrome C: learning new tricks from an old dog. Chem Rev 2017;117:13382–13460. [DOI] [PubMed] [Google Scholar]
- Anderson MJ, Dixson AF.. Sperm competition: motility and the midpiece in primates. Nature 2002;416:496. [DOI] [PubMed] [Google Scholar]
- Aprea I, Raidt J, Höben IM, Loges NT, Nöthe-Menchen T, Pennekamp P, Olbrich H, Kaiser T, Biebach L, Tüttelmann F. et al. Defects in the cytoplasmic assembly of axonemal dynein arms cause morphological abnormalities and dysmotility in sperm cells leading to male infertility. PLoS Genet 2021;17:e1009306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aurrière J, Goudenège D, Baris OR, Boguenet M, May-Panloup P, Lenaers G, Khiati S.. Cancer/testis antigens into mitochondria: a hub between spermatogenesis, tumorigenesis and mitochondrial physiology adaptation. Mitochondrion 2021;56:73–81. [DOI] [PubMed] [Google Scholar]
- Babatunde KA, Najafi A, Salehipour P, Modarressi MH, Mobasheri MB.. Cancer/testis genes in relation to sperm biology and function. Iran J Basic Med Sci 2017;20:967–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baklouti-Gargouri S, Ghorbel M, Mahmoud AB, Mkaouar-Rebai E, Cherif M, Chakroun N, Sellami A, Fakhfakh F, Ammar-Keskes L.. Mitochondrial DNA mutations and polymorphisms in asthenospermic infertile men. Mol Biol Rep 2013;40:4705–4712. [DOI] [PubMed] [Google Scholar]
- Bansho Y, Lee J, Nishida E, Nakajima-Koyama M.. Identification and characterization of secreted factors that are upregulated during somatic cell reprogramming. FEBS Lett 2017;591:1584–1600. [DOI] [PubMed] [Google Scholar]
- Barratt CLR, Björndahl L, De Jonge CJ, Lamb DJ, Martini FO, McLachlan R, Oates RD, van der Poel S, St John B, Sigman M. et al. The diagnosis of male infertility: an analysis of the evidence to support the development of global WHO guidance-challenges and future research opportunities. Hum Reprod Update 2017;23:660–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben Khelifa M, Coutton C, Zouari R, Karaouzène T, Rendu J, Bidart M, Yassine S, Pierre V, Delaroche J, Hennebicq S. et al. Mutations in DNAH1, which encodes an inner arm heavy chain dynein, lead to male infertility from multiple morphological abnormalities of the sperm flagella. Am J Hum Genet 2014;94:95–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennison C, Hemmings N, Brookes L, Slate J, Birkhead T.. Sperm morphology, adenosine triphosphate (ATP) concentration and swimming velocity: unexpected relationships in a passerine bird. Proc R Soc B 2016;283:20161558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binsila BK, Archana SS, Ramya L, Swathi D, Selvaraju S, Gowda NKS, Pal DT, Rafay A, Bhatta R.. Elucidating the processes and pathways enriched in buffalo sperm proteome in regulating semen quality. Cell Tissue Res 2021;383:881–903. [DOI] [PubMed] [Google Scholar]
- Bode PK, Thielken A, Brandt S, Barghorn A, Lohe B, Knuth A, Moch H.. Cancer testis antigen expression in testicular germ cell tumorigenesis. Mod Pathol 2014;27:899–905. [DOI] [PubMed] [Google Scholar]
- Bower R, Tritschler D, VanderWaal Mills K, Heuser T, Nicastro D, Porter ME.. DRC2/CCDC65 is a central hub for assembly of the nexin-dynein regulatory complex and other regulators of ciliary and flagellar motility. Mol Biol Cell 2018;29:137–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brischigliaro M, Zeviani M.. Cytochrome c oxidase deficiency. Biochim Biophys Acta Bioenerg 2021;1862:148335. [DOI] [PubMed] [Google Scholar]
- Cai J, Huang H, Hu X, Lang W, Fu W, Xu L, Qiu Z, Zhong H, Chen F.. Homoharringtonine synergized with gilteritinib results in the downregulation of myeloid cell leukemia-1 by upregulating UBE2L6 in FLT3-ITD-mutant acute myeloid (leukemia) cell lines. J Oncol 2021;2021:3766428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannarella R, Condorelli RA, La Vignera S, Bellucci C, Luca G, Calafiore R, Calogero AE.. IGF2 and IGF1R mRNAs are detectable in human spermatozoa. World J Mens Health 2020;38:545–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castaneda JM, Hua R, Miyata H, Oji A, Guo Y, Cheng Y, Zhou T, Guo X, Cui Y, Shen B.. TCTE1 is a conserved component of the dynein regulatory complex and is required for motility and metabolism in mouse spermatozoa. PNAS 2017;114:E5370–E5378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chemes HE, Olmedo SB, Carrere C, Oses R, Carizza C, Leisner M, Blaquier J.. Ultrastructural pathology of the sperm flagellum: association between flagellar pathology and fertility prognosis in severely asthenozoospermic men. Hum Reprod 1998;13:2521–2526. [DOI] [PubMed] [Google Scholar]
- Chen Y, Wang J, Pan C, Li D, Han X.. Microcystin-leucine-arginine causes blood-testis barrier disruption and degradation of occludin mediated by matrix metalloproteinase-8. Cell Mol Life Sci 2018;75:1117–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng YH, Wong EWP, Cheng CY.. Cancer/testis (CT) antigens, carcinogenesis and spermatogenesis. Spermatogenesis 2011;1:209–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang C, Layer RM, Faust GG, Lindberg MR, Rose DB, Garrison EP, Marth GT, Quinlan AR, Hall IM.. SpeedSeq: ultra-fast personal genome analysis and interpretation. Nat Methods 2015;12:966–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dadsena S, King LE, Garcia-Saez AJ.. Apoptosis regulation at the mitochondria membrane level. BBA Biomembranes 2021;1863:183718. [DOI] [PubMed] [Google Scholar]
- Damaschke NA, Yang B, Bhusari S, Avilla M, Zhong W, Blute ML, Huang W, Jarrard DF.. Loss of Igf2 gene imprinting in murine prostate promotes widespread neoplastic growth. Cancer Res 2017;77:5236–5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drechsler H, Xu Y, Geyer VF, Zhang Y, Diez S.. Multivalent electrostatic microtubule interactions of synthetic peptides are sufficient to mimic advanced MAP-like behavior. Mol Biol Cell 2019;30:2953–2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Plessis SS, Gokul S, Agarwal A.. Semen hyperviscosity: causes, consequences, and cures. Front Biosci (Elite Ed) 2013;5:224–231. [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K.. Features and development of coot. Acta Crystallogr D Biol Crystallogr 2010;66:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink E, Bhoola KD, Snyman C, Neth P, Figueroa CD.. Cellular expression of plasma prekallikrein in human tissues. Biol Chem 2007;388:957–963. [DOI] [PubMed] [Google Scholar]
- Firman RC, Simmons LW.. Sperm midpiece length predicts sperm swimming velocity in house mice. Biol Lett 2010;6:513–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford WC. Glycolysis and sperm motility: does a spoonful of sugar help the flagellum go round? Hum Reprod Update 2006;12:269–274. [DOI] [PubMed] [Google Scholar]
- Frungieri MB, Calandra RS, Mayerhofer A, Matzkin ME.. Correlation between testicular mast cell count and spermatogenic epithelium in non-obstructive azoospermia. Reproduction 2015;149:R169–R180.25504871 [Google Scholar]
- Fung E, Richter C, Yang H-B, Schäffer I, Fischer R, Kessler BM, Bassermann F, D’Angiolella V.. FBXL13 directs the proteolysis of CEP192 to regulate centrosome homeostasis and cell migration. EMBO Rep 2018;19:e44799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganetzky RD, Stendel C, McCormick EM, Zolkipli-Cunningham Z, Goldstein AC, Klopstock T, Falk MJ.. MT-ATP mitochondrial disease variants: phenotypic and biochemical features analysis in 218 published cases and cohort of 14 new cases. Hum Mutat 2019;40:499–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Hong J, Guo Y, Chen M, Chang AK, Xie L, Ying X.. Assessment spermatogenic cell apoptosis and the transcript levels of metallothionein and p53 in Meretrix meretrix induced by cadmium. Ecotoxicol Environ Saf 2021;217:112230. [DOI] [PubMed] [Google Scholar]
- Ghosh D, Najwa AR, Khan MA, Sengupta J.. IGF2, IGF binding protein 1, and matrix metalloproteinases 2 and 9 in implantation-stage endometrium following immunoneutralization of vascular endothelial growth factor in the rhesus monkey. Reproduction 2011;141:501–509. [DOI] [PubMed] [Google Scholar]
- Girardet L, Augière C, Asselin MP, Belleannée C.. Primary cilia: biosensors of the male reproductive tract. Andrology 2019;7:588–602. [DOI] [PubMed] [Google Scholar]
- Gu NH, Zhao WL, Wang GS, Sun F.. Comparative analysis of mammalian sperm ultrastructure reveals relationships between sperm morphology, mitochondrial functions and motility. Reprod Biol Endocrinol 2019;17:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui L, Song K, Tritschler D, Bower R, Yan S, Dai A, Augspurger K, Sakizadeh J, Grzemska M, Ni T. et al. Scaffold subunits support associated subunit assembly in the chlamydomonas ciliary nexin-dynein regulatory complex. Proc Natl Acad Sci USA 2019;116:23152–23162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha S, Lindsay AM, Timms AE, Beier DR.. Mutations in Dnaaf1 and Lrrc48 cause hydrocephalus, laterality defects, and sinusitis in mice. G3 (Bethesda) 2016;6:2479–2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy JJ, Wyrwoll MJ, Mcfadden W, Malcher A, Rotte N, Pollock NC, Munyoki S, Veroli MV, Houston BJ, Xavier MJ;. GEMINI Consortium Variants in GCNA, X-linked germ-cell genome integrity gene, identified in men with primary spermatogenic failure. Hum Genet 2021;140:1169–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidari MM, Khatami M, Danafar A, Dianat T, Farahmand G, Talebi AR.. 4Mitochondrial genetic variation in Iranian infertile men with varicocele. Int J Fertil Steril 2016;10:303–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández-Llodrà S, Juanpere N, de Muga S, Lorenzo M, Gil J, Font-Tello A, Agell L, Albero-González R, Segalés L, Merino J. et al. ERG overexpression plus SLC45A3 (prostein) and PTEN expression loss: strong association of the triple hit phenotype with an aggressive pathway of prostate cancer progression. Oncotarget 2017;8:74106–74118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heshusius S, Grech L, Gillemans N, Brouwer RWW, den Dekker XT, van IJcken WFJ, Nota B, Felice AE, van Dijk TB, von Lindern M. et al. Epigenomic analysis of KLF1 haploinsufficiency in primary human erythroblasts. Sci Rep 2022;12:336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuser T, Raytchev M, Krell J, Porter ME, Nicastro D.. The dynein regulatory complex is the nexin link and a major regulatory node in cilia and flagella. J Cell Biol 2009;187:921–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda A, Komuro H, Shimada A, Hasegawa T, Seko Y, Nagase H, Hozumi I, Inuzuka T, Hara H, Fujiwara Y. et al. Attenuation of cadmium-induced testicular injury in metallothionein-III null mice. Life Sci 2010;87:545–550. [DOI] [PubMed] [Google Scholar]
- Houston BJ, Riera-Escamilla A, Wyrwoll MJ, Salas-Huetos A, Xavier MJ, Nagirnaja L, Friedrich C, Conrad DF, Aston AI, Krausz C. et al. A systematic review of the validated monogenic causes of human male infertility: 2020 update and a discussion of emerging gene–disease relationships. Hum Reprod Update 2021;28:15–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Huang Y, Pan XW, Li L, Lu Chen Liu X, Lu JL, Zhu XM, Huang H, Yang QW, Ye JQ. et al. Overexpression of USP39 predicts poor prognosis and promotes tumorigenesis of prostate cancer via promoting EGFR mRNA maturation and transcription elongation. Oncotarget 2016;7:22016–22030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull RL, Johnson PY, Braun KR, Day AJ, Wight TN.. Hyaluronan and hyaluronan binding proteins are normal components of mouse pancreatic islets and are differentially expressed by islet endocrine cell types. J Histochem Cytochem 2021;60:749–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang K, Smith JF, Coward RM, Penzias A, Bendikson K, Butts S, Coutifaris C, Falcone T, Fossum G, Gitlin S, et al. ; Practice Committee of the American Society for Reproductive Medicine in collaboration with the Society for Male Reproduction and Urology. Evaluation of the azoospermic male: a committee opinion. Fertil Steril 2018;109:777–782. [DOI] [PubMed] [Google Scholar]
- Ioannidis NM, Rothstein JH, Pejaver V, Middha S, McDonnell SK, Baheti S, Musolf A, Li Q, Holzinger E, Karyadi D. et al. REVEL: an ensemble method for predicting the pathogenicity of rare missense variants. Am J Hum Genet 2016;99:877–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa T. Axoneme structure from motile cilia. Cold Spring Harb Perspect Biol 2017;9:a028076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Y, Ohanyan A, Lue YH, Swerdloff RS, Liu PY, Cohen P, Wang C.. The effects of humanin and its analogues on male germ cell apoptosis induced by chemotherapeutic drugs. Apoptosis 2015;20:551–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jodar M, Oliva R.. Protamine alterations in human spermatozoa. Adv Exp Med Biol 2014;791:83–102. [DOI] [PubMed] [Google Scholar]
- Jonkers J, Pai P, Sukumar S.. Multiple roles of HOX proteins in metastasis: let me count the ways. Cancer Metastasis Rev 2020;39:661–679. [DOI] [PubMed] [Google Scholar]
- Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Žídek A, Potapenko A. et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021;596:583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karmilin K, Schmitz C, Kuske M, Körschgen H, Olf M, Meyer K, Hildebrand A, Felten M, Fridrich S, Yiallouros I. et al. Mammalian plasma fetuin-B is a selective inhibitor of ovastacin and meprin metalloproteinases. Sci Rep 2019;9:546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ.. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 2015;10:845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kherraf ZE, Christou-Kent M, Karaouzene T, Amiri-Yekta A, Martinez G, Vargas S, Lambert AS, Borel E, Dorphin C, Aknin B. et al. SPINK2 deficiency causes infertility by inducing sperm defects in heterozygotes and azoospermia in homozygotes. EMBO Mol Med 2017;9:1132–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein C, Troedsson MHT, Rutllant J.. Region-specific expression of aquaporin subtypes in equine testis, epididymis, and ductus deferens. Anat Rec (Hoboken) 2013;296:1115–1126. [DOI] [PubMed] [Google Scholar]
- Kobe B, Kajava AV.. The leucine-rich repeat as a protein recognition motif. Curr Opin Struct Biol 2001;11:725–732. [DOI] [PubMed] [Google Scholar]
- Krausz C, Riera-Escamilla A.. Genetics of male infertility. Nat Rev Urol 2018;15:369–384. [DOI] [PubMed] [Google Scholar]
- Krissinel E, Henrick K.. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D Biol Crystallogr 2004;60:2256–2268. [DOI] [PubMed] [Google Scholar]
- Kubo T, Oda T.. Electrostatic interaction between polyglutamylated tubulin and the nexin-dynein regulatory complex regulates flagellar motility. Mol Biol Cell 2017;28:2260–2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar D, Reiter J.. J. How the centriole builds its cilium: of mothers, daughters, and the acquisition of appendages. Curr Opin Struct Biol 2021;66:41–48. [DOI] [PubMed] [Google Scholar]
- Leal LF, Szarek E, Berthon A, Nesterova M, Faucz FR, London E, Mercier C, Abu-Asab M, Starost MF, Dye L. et al. Pde8b haploinsufficiency in mice is associated with modest adrenal defects, impaired steroidogenesis, and male infertility, unaltered by concurrent PKA or Wnt activation. Mol Cell Endocrinol 2021;522:111117. [DOI] [PubMed] [Google Scholar]
- Lei Q, Huang X, Zheng L, Zheng F, Dong J, Chen F, Zeng W.. Biosensors for caspase-3: from chemical methodologies to biomedical applications. Talanta 2022;240:123198. [DOI] [PubMed] [Google Scholar]
- Lewis WR, Malarkey EB, Tritschler D, Bower R, Pasek RC, Porath JD, Birket SE, Saunier S, Antignac C, Knowles MR. et al. Mutation of growth arrest specific 8 reveals a role in motile cilia function and human disease. PLoS Genet 2016;12:e1006220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Xing L, Zhao N, Wang J, Zheng N.. Furosine induced apoptosis by the regulation of STAT1/STAT2 and UBA7/UBE2L6 genes in HepG2 cells. Int J Mol Sci 2018;19:1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K, van Delft MF, Dewson G.. Too much death can kill you: inhibiting intrinsic apoptosis to treat disease. Embo J 2021;40:e107341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorès P, Kherraf Z-E, Amiri-Yekta A, Whitfield M, Daneshipour A, Stouvenel L, Cazin C, Cavarocchi E, Coutton C, Llabador M-A. et al. A missense mutation in IFT74, encoding for an essential component for intraflagellar transport of Tubulin, causes asthenozoospermia and male infertility without clinical signs of Bardet-Biedl syndrome. Hum Genet 2021;140:1031–1043. [DOI] [PubMed] [Google Scholar]
- Lue Y, Swerdloff R, Jia Y, Wang C.. The emerging role of mitochondrial derived peptide humanin in the testis. Biochim Biophys Acta Gen Subj 2021;1865:130009. [DOI] [PubMed] [Google Scholar]
- Lüpold S, de Boer RA, Evans JP, Tomkins JL, Fitzpatrick JL.. How sperm competition shapes the evolution of testes and sperm: a meta-analysis. Philos Trans R Soc Lond B Biol Sci 2020;375:20200064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markou A, Unger L, Abir-Awan M, Saadallah A, Halsey A, Balklava Z, Conner M, Törnroth-Horsefield S, Greenhill SD, Conner A. et al. Molecular mechanisms governing aquaporin relocalisation. Biochim Biophys Acta Biomembr 2022;1864:183853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques PI, Fonseca F, Carvalho AS, Puente DA, Damião I, Almeida V, Barros N, Barros A, Carvalho F, Azkargorta M. et al. Sequence variation at KLK and WFDC clusters and its association to semen hyperviscosity and other male infertility phenotypes. Hum Reprod 2016;31:2881–2891. [DOI] [PubMed] [Google Scholar]
- Martinez G, Coutton C, Loeuillet C, Cazin C, Muroňová J, Boguenet M, Lambert E, Dhellemmes M, Chevalier G, Hograindleur JP. et al. Oligogenic heterozygous inheritance of sperm abnormalities in mouse. Elife 2022;11:e75373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushima N, Takatsuka S, Miyashita H, Kretsinger RH.. Leucine rich repeat proteins: sequences, mutations, structures and diseases. Protein Pept Lett 2019;26:108–131. [DOI] [PubMed] [Google Scholar]
- Matsuzaki H, Matsuzaki H, Kuramochi D, Okamura E, Hirakawa K, Ushiki A, Tanimoto K.. Recapitulation of gametic DNA methylation and its post-fertilization maintenance with reassembled DNA elements at the mouse Igf2/H19 locus. Epigenetics Chromatin 2020;13:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzuk MM, Lamb DJ.. The biology of infertility: research advances and clinical challenges. Nat Med 2008;14:1197–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GRS, Thormann A, Flicek P, Cunningham F.. The ensembl variant effect predictor. Genome Biol 2016;17:122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei S, Wu Y, Wang Y, Cui Y, Zhang M, Zhang T, Huang X, Yu S, Yu T, Zhao J.. Disruption of PIKFYVE causes congenital cataract in human and zebrafish. Elife 2022;11:e71256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelis M, Sobczak A, Koczan D, Langhammer M, Reinsch N, Schoen J, WeitzeL JM.. Selection for female traits of high fertility affects male reproductive performance and alters the testicular transcriptional profile. BMC Genomics 2017;18:889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo RJ, Lu JM, Wan YP, Hua W, Liang YX, Zhuo YJ, Kuang QW, Liu YL, He HC, Zhong WD.. Decreased HoxD10 expression promotes a proliferative and aggressive phenotype in prostate cancer. Curr Mol Med 2017;17:70–78. [DOI] [PubMed] [Google Scholar]
- Morohoshi A, Miyata H, Shimada K, Nozawa K, Matsumura T, Yanase R, Shiba K, Inaba K, Ikawa M.. Nexin-dynein regulatory complex component DRC7 but not FBXL13 is required for sperm flagellum formation and male fertility in mice. PLoS Genet 2020;16:e1008585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostafa T, Rashed L, Taymour M.. Seminal cyclooxygenase relationship with oxidative stress in infertile oligoasthenoteratozoospermic men with varicocele. Andrologia 2016;48:137–142. [DOI] [PubMed] [Google Scholar]
- Nagirnaja L, Aston KI, Conrad DF.. Genetic intersection of male infertility and cancer. Fertil Steril 2018;109:20–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nascimento JM, Shi LZ, Tam J, Chandsawangbhuwana C, Durrant B, Botvinick EL, Berns MW.. Comparison of glycolysis and oxidative phosphorylation as energy sources for mammalian sperm motility, using the combination of fluorescence imaging, laser tweezers, and real-time automated tracking and trapping. J Cellular Physiol 2008;217:745–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naz RK, Rajesh C.. Gene knockouts that cause female infertility: search for novel contraceptive targets. Front Biosci 2005;10:2447–2459. [DOI] [PubMed] [Google Scholar]
- Neirijnck Y, Kühne F, Mayère C, Pavlova E, Sararols P, Foti M, Atanassova N, Nef S.. Tumor suppressor PTEN regulates negatively Sertoli cell proliferation, testis size, and sperm production in vivo. Endocrinology 2019;160:387–398. [DOI] [PubMed] [Google Scholar]
- Ng ACY, Eisenberg JM, Heath RJ, Huett A, Robinson CM, Nau GJ, Xavier RJ.. Human leucine-rich repeat proteins: a genome-wide bioinformatic categorization and functional analysis in innate immunity. Proc Natl Acad Sci USA 2011;108(Suppl 1:4).631–4638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura Y, Uehara T, Ujibe K, Yoshihashi H, Yamada M, Suzuki H, Takenouchi T, Kosaki K, Hirata H.. The p.Thr395Met missense variant of NFIA found in a patient with intellectual disability is a defective variant. Am J Med Genet A 2022;188:1184–1192. [DOI] [PubMed] [Google Scholar]
- Oud MS, Houston BJ, Volozonoka L, Mastrorosa FK, Holt GS, Alobaidi BKS, deVries PF, Astuti G, Ramos L, Mclachlan RI. et al. Exome sequencing reveals variants in known and novel candidate genes for severe sperm motility disorders. Hum Reprod 2021;36:2597–2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan W, Wang K, Li J, Li H, Cai Y, Zhang M, Wang A, Wu Y, Gao W, Weng W.. Restoring HOXD10 exhibits therapeutic potential for ameliorating malignant progression and 5-fluorouracil resistance in colorectal cancer. Front Oncol 2021;11:771528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perner S, Rupp NJ, Braun M, Rubin MA, Moch H, Dietel M, Wernert N, Jung K, Stephan C, Kristiansen G.. Loss of SLC45A3 protein (prostein) expression in prostate cancer is associated with SLC45A3-ERG gene rearrangement and an unfavorable clinical course. Int J Cancer 2013;132:807–812. [DOI] [PubMed] [Google Scholar]
- Perrotta I, Santoro M, Guido C, Avena P, Tripepi S, De Amicis F, Gervasi MC, Aquila S.. Expression of cyclooxygenase-1 (COX-1) and COX-2 in human male gametes from normal patients, and those with varicocele and diabetes: a potential molecular marker for diagnosing male infertility disorders. J Anat 2012;221:209–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pin E, Henjes F, Hong MG, Wiklund F, Magnusson P, Bjartell A, Uhlen M, Nilsson P, Schwenk JM.. Identification of a novel autoimmune peptide epitope of prostein in prostate cancer. J Proteome Res 2017;16:204–216. [DOI] [PubMed] [Google Scholar]
- Ramli NSK, Giribabu N, Karim K, Salleh N.. Hormonal control of vas deferens fluid volume and aquaporin expression in rats. J Mol Histol 2019;50:21–34. [DOI] [PubMed] [Google Scholar]
- Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, ACMG Laboratory Quality Assurance Committee. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 2015;17:405–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeis B, Mulisch M, Welsch U.. Romeis Mikroskopische Technik, Vol. xii. Dordrecht: Springer, 2010, 550. [Google Scholar]
- Roy A, Kucukural A, Zhang Y.. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc 2010;5:725–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salustri A, Campagnolo L, Klinger FG, Camaioni A.. Molecular organization and mechanical properties of the hyaluronan matrix surrounding the mammalian oocyte. Matrix Biol 2019;78-79:11–23. [DOI] [PubMed] [Google Scholar]
- Samaiya PK, Krishnamurthy S, Kumar A.. Mitochondrial dysfunction in perinatal asphyxia: role in pathogenesis and potential therapeutic interventions. Mol Cell Biochem 2021;476:4421–4434. [DOI] [PubMed] [Google Scholar]
- Sandhu N, Rana S, Meena K.. Nuclear receptor subfamily 5 group A member 2 (NR5A2): role in health and diseases. Mol Biol Rep 2021;48:8155–8170. [DOI] [PubMed] [Google Scholar]
- Sandy Z, da Costa IC, Schmidt CK.. More than meets the ISG15: emerging roles in the DNA damage response and beyond. Biomolecules 2020;10:1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satir P, Heuser T, Sale WS.. A structural basis for how motile cilia beat. Bioscience 2014;64:1073–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato JD, Kan M.. Media for culture of mammalian cells. Curr Protoc Cell Biol 2001;Chapter 1:Unit 1.2. [DOI] [PubMed] [Google Scholar]
- Schlegel PN, Sigman M, Collura B, De Jonge CJ, Eisenberg ML, Lamb DL, Sandlow JI, Sokol RZ, Spandorfer SD, Tanrikut C. et al. Diagnosis and treatment of infertility in men: AUA/ASRM guideline part I. Fertil Steril 2021;115:54–61. [DOI] [PubMed] [Google Scholar]
- Sironen A, Shoemark A, Patel M, Loebinger MR, Mitchison HM.. Sperm defects in primary ciliary dyskinesia and related causes of male infertility. Cell Mol Life Sci 2020;77:2029–2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CEL, Lake ARV, Johnson CA.. Primary cilia, ciliogenesis and the actin cytoskeleton: a little less resorption, a little more actin please. Front Cell Dev Biol 2020;8:622822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supernat A, Vidarsson OV, Steen VM, Stokowy T.. Comparison of three variant callers for human whole genome sequencing. Sci Rep 2018;8:17851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P. et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 2019;47:D607–D613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan JHL, Wollmann H, van Pelt AMM, Kaldis P, Messerschmidt DM.. Infertility-causing haploinsufficiency reveals TRIM28/KAP1 requirement in spermatogonia. Stem Cell Rep. 2020;14:818–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touré A, Martinez G, Kherraf ZE, Cazin C, Beurois J, Arnoult C, Ray PF, Coutton C.. The genetic architecture of morphological abnormalities of the sperm tail. Hum Genet 2021;140:21–42. [DOI] [PubMed] [Google Scholar]
- Tourmente M, Villar-Moya P, Rial E, Roldan ER.. 2015 Differences in ATP generation via glycolysis and oxidative phosphorylation and relationships with sperm motility in mouse species. J Biol Chem 2015;290:20613–20626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tunyasuvunakool K, Adler J, Wu Z, Green T, Zielinski M, Žídek A, Bridgland A, Cowie A, Meyer C, Laydon A. et al. Highly accurate protein structure prediction for the human proteome. Nature 2021;596:590–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UniProt Consortium. UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res 2019;47:D506–D15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valassina N, Brusco S, Salamone A, Serra L, Luoni M, Giannelli S, Bido S, Massimino L, Ungaro F, Mazzara PG. et al. Scn1a gene reactivation after symptom onset rescues pathological phenotypes in a mouse model of Dravet syndrome. Nat Commun 2022;13:161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Borght M, Wyns C.. Fertility and infertility: definition and epidemiology. Clin Biochem 2018;62:2–10. [DOI] [PubMed] [Google Scholar]
- Varadi M, Anyango S, Deshpande M, Nair S, Natassia C, Yordanova G, Yuan D, Stroe O, Wood G, Laydon A. et al. AlphaFold protein structure database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res 2022;50:D439–D444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Mering C, Jensen LJ, Snel B, Hooper SD, Krupp M, Foglierini M, Jouffre N, Huynen MA, Bork P.. STRING: known and predicted protein-protein associations, integrated and transferred across organisms. Nucleic Acids Res 2005;33:D433–D437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner K, Unger L, Salman MM, Kitchen P, Bill RM, Yool AJ.. Signaling mechanisms and pharmacological modulators governing diverse aquaporin functions in human health and disease. Int J Mol Sci 2022;23:1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Gu Y, Zhang K, Xie K, Zhu M, Dai N, Jiang Y, Guo X, Liu M, Dai J. et al. Systematic identification of genes with a cancer-testis expression pattern in 19 cancer types. Nat Commun 2016;7:10499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertheimer E, Krapf D, de la Vega-Beltran JL, Sánchez-Cárdenas C, Navarrete F, Haddad D, Escoffier J, Salicioni AM, Levin LR, Buck J. et al. Compartmentalization of distinct cAMP signaling pathways in mammalian sperm. J Biol Chem 2013;288:35307–35320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO. Infertility, 2020. https://www.who.int/news-room/fact-sheets/detail/infertility (15 March 2020, date last accessed).
- WHO laboratory manual for the examination and processing of human semen. 6th edn. World Health Organization, Geneva, Switzerland. 2021. [Google Scholar]
- Wilson KS, Gonzalez O, Dutcher SK, Bayly PV.. Dynein-deficient flagella respond to increased viscosity with contrasting changes in power and recovery strokes. Cytoskeleton (Hoboken) 2015;72:477–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirschell M, Olbrich H, Werner C, Tritschler D, Bower R, Sale WS, Loges NT, Pennekamp P, Lindberg S, Stenram U. et al. The nexin-dynein regulatory complex subunit DRC1 is essential for motile cilia function in algae and humans. Nat Genet 2013;45:262–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyrwoll MJ, Temel SG, Nagirnaja L, Oud MS, Lopes AM, van der Heijden GW, Heald JS, Rotte N, Wistuba J, Wo ¨ste M, GEMINI Consortium et al. Bi-allelic mutations in M1AP are a frequent cause of meiotic arrest and severely impaired spermatogenesis leading to male infertility. Am J Hum Genet 2020;107:342–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xavier MJ, Salas-Huetos A, Oud MS, Aston KI, Veltman JA.. Disease gene discovery in male infertility: past, present and future. Hum Genet 2021;140:7–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Zhang Y, Qin D, Gui Y, Wang S, Du G, Yang F, Li L, Yuan S, Wang M. et al. Transcription factor-like 5 is a potential DNA- and RNA-binding protein essential for maintaining male fertility in mice. J Cell Sci 2022;135:jcs259036. [DOI] [PubMed] [Google Scholar]
- Xue M, Xue M, Fang Y, Sun G, Zhuo W, Zhong J, Qian C, Wang L, Wang L, Si J. et al. IGFBP3, a transcriptional target of homeobox D10, is correlated with the prognosis of gastric cancer. PLoS One 2013;8:e81423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y.. The I-TASSER suite: protein structure and function prediction. Nat Methods 2015;12:7–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Zhang Y.. I-TASSER server: new development for protein structure and function predictions. Nucleic Acids Res 2015;43:W174–W181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang LF, Yang F, Zhang FL, Xie YF, Hu ZX, Huang SL, Shao ZM, Li DQ.. Discrete functional and mechanistic roles of chromodomain Y-like 2 (CDYL2) transcript variants in breast cancer growth and metastasis. Theranostics 2020;10:5242–5258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin M, O’Neil LAJ.. The role of the electron transport chain in immunity. Faseb J 2021;35:e21974. [DOI] [PubMed] [Google Scholar]
- Yuan J, Li X, Zhang G, Cheng W, Wang W, Lei Y, Ma Q, Song G.. USP39 mediates p21-dependent proliferation and neoplasia of colon cancer cells by regulating the p53/p21/CDC2/cyclin B1 axis. Mol Carcinog 2021;60:265–278. [DOI] [PubMed] [Google Scholar]
- Zhan K, Liu R, Tong H, Gao S, Yang G, Hossain A, Li T, He W.. Fetuin B overexpression suppresses proliferation, migration, and invasion in prostate cancer by inhibiting the PI3K/AKT signaling pathway. Biomed Pharmacother 2020;131:110689. [DOI] [PubMed] [Google Scholar]
- Zhang J, Liu S, Zhang D, Ma Z, Sun L.. Homeobox D10, a tumor suppressor, inhibits the proliferation and migration of esophageal squamous cell carcinoma. J Cell Biochem 2019;120:13717–13725. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Miao J, Wang Y.. Mitochondrial regulation on spermatogenesis. Reproduction 2022;163:R55–R69. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Zhang B, Lei Y, Sun J, Zhang Y, Yang S, Zhang X.. Knockdown of USP39 induces cell cycle arrest and apoptosis in melanoma. Tumour Biol 2016;37:13167–13176. [DOI] [PubMed] [Google Scholar]
- Zhou S, Wu H, Zhang J, He X, Liu S, Zhou P, Hua R, Cao X, Liu M.. Bi-allelic variants in human TCTE1/DRC5 cause asthenozoospermia and male infertility. Eur J Hum Genet 2022;30:721–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorrilla M, Yatsenko AN.. The genetics of infertility: current status of the field. Curr Genet Med Rep 2013;1:247–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its supplemental information files. RNAseq data are available in GEO database (https://www.ncbi.nlm.nih.gov/geo/) under the accession number: GSE207805. The results described in the publication are based on whole-genome or exome sequencing data which includes sensitive information in the form of patient-specific germline variants. Information regarding such variants must not be shared publicly following the European Union legislation described in the following document: https://publications.jrc.ec.europa.eu/repository/bitstream/JRC113479/policy_report_-_review_of_eu_national_legislation_on_genomics_-_with_identifiers_1.pdf, therefore the access to raw data that support findings of this study are available from the corresponding author upon reasonable request.