Abstract
Background and aims
We have identified a decreased abundance of microbial species known to have a potential anti-inflammatory, protective effect in subjects that developed Celiac Disease (CeD) compared to those who did not. We aim to confirm the potential protective role of one of these species, namely Bacteroides vulgatus, and to mechanistically establish the effect of bacterial bioproducts on gluten-dependent changes on human gut epithelial functions.
Methods
We identified, isolated, cultivated, and sequenced a unique novel strain (20220303-A2) of B. vulgatus found only in control subjects. Using a human gut organoid system developed from pre-celiac patients, we monitored epithelial phenotype and innate immune cytokines at baseline, after exposure to gliadin, or gliadin plus B. vulgatus cell free supernatant (CFS).
Results
Following gliadin exposure, we observed increases in epithelial cell death, epithelial monolayer permeability, and secretion of pro-inflammatory cytokines. These effects were mitigated upon exposure to B. vulgatus 20220303-A2 CFS, which had matched phenotype gene product mutations. These protective effects were mediated by epigenetic reprogramming of the organoids treated with B. vulgatus CFS.
Conclusions
We identified a unique strain of B. vulgatus that may exert a beneficial role by protecting CeD epithelium against a gluten-induced break of epithelial tolerance through miRNA reprogramming.
Impact
Gut dysbiosis precedes the onset of celiac disease in genetically at-risk infants.
This dysbiosis is characterized by the loss of protective bacterial strains in those children who will go on to develop celiac disease.
The paper reports the mechanism by which one of these protective strains, B. vulgatus, ameliorates the gluten-induced break of gut epithelial homeostasis by epigenetically re-programming the target intestinal epithelium involving pathways controlling permeability, immune response, and cell turnover.
Introduction
In the past two decades, we have acquired a deep understanding of the adaptive immune response mechanisms involved in celiac disease (CeD) pathogenesis.1–3 However, there is still limited information regarding the initial steps causing the loss of gut mucosal tolerance to gluten and the switch from genetic predisposition to onset of the disease. Environmental factors4,5 have been shown to affect both the composition and function of the gut microbiome, and several reports have suggested a pathogenic role of specific microorganisms in CeD pathogenesis.6–10 However, evidence to mechanistically link either a single pathogen or gut dysbiosis to CeD pathogenesis is lacking. Moreover, the emerging role of epigenetic modulations such as microRNAs (miRNAs), has been identified in the pathophysiology of CeD,11,12 but just how these modulations are triggered remains to be elucidated.
To overcome these limitations, we initiated a prospective, longitudinal birth cohort study called the Celiac Disease Genomic, Environmental, Microbiome and Metabolomic (CDGEMM) study, which enrolls infants at-risk of CeD.13 We utilized this study to examine the infants’ microbiome and metabolome during the first 6 months after birth in relation to environmental risk factors and genetic markers known to increase susceptibility to CeD development.14 In a subsequent study, we investigated changes in microbiome and metabolomic profiles preceding the onset of CeD by up to 18 months.15 Among the differences identified, we detected the decreased abundance of several microbial species/strains known to have a potential anti-inflammatory, protective effect in cases (defined as at-risk children who developed CeD) before CeD onset compared to controls (defined as age-matched, at-risk infants who did not develop CeD).15 With the present study, we aimed to further investigate the potential protective role of some of these bacterial strains isolated from children at risk of CeD who were protected from the onset of the disease. Among the identified bacterial strains, we used a prototype B. vulgatus strain 20220303 A2 (referred to as B. vulgatus-A2) to perform epigenetic profiling and functional studies using human, patient-derived gut organoids to mechanistically establish a protective effect of bacterial bioproducts against gluten-dependent changes on human gut epithelial functions.
Methods
Bacterial isolation from gut microbiota
We used stool samples from two CDGEMM cases and two matched controls, obtained at disease onset and 6 months prior in cases and the corresponding time points in controls. The Prospector®, GALT Inc culturomic platform (isolationbio, San Carlos, CA) was used to isolate bacterial strains previously shown to be potentially protective against CeD onset.15 Further details, including demographics of the four patients included in bacterial culture and isolation are presented in the Supplementary Material and Method Section.
Preparation of the bacterial cell free supernatant (CFS)
To optimize the growth conditions of strains of interest, we tested various bacterial culture media (see Supplemental data for more details). Given that B. vulgatus-A2 grew the most efficiently in comparison to the other four strains tested, it was selected as a prototype for a more in-depth genomic analysis and for additional functional studies. We collected CFS at four optical densities (ODs), namely 1.0, 0.8, 0.4, and 0.1. Based on the OD600 readings, the bacteria grew most efficiently in Brain Heart Infusion (BHI) Broth. The subcultures were then centrifuged at 4000 rpm; supernatants were collected and filtered through a 0.4 mm syringe filter and then filtered again using a 0.2 mm syringe filter. The CFS obtained was then aliquoted and stored at −80 °C for future experiments (see analysis of CFS below). The bacterial pellets were flash frozen and used for genomic DNA sequencing.
Bacterial sequencing and mutation analysis
Our isolated B. vulgatus-A2 strain underwent genomic DNA sequencing by the SeqCenter (Pittsburgh, PA). Basic variant analysis of paired-end reads was performed using breseq (version 0.36.1) to align and compare the genome to reference genomes as previously reported.16,17 Details on our mutation analysis are provided in the Supplementary Material and Methods Section.
Human gut organoids and establishment and characterization of a macrophage-epithelium organotypic model
Experiments using organoid-derived monolayer from human gut and human monocytes were conducted as previously described17 after obtaining approval from the Massachusetts General Hospital Institutional Review Board protocol #2014P000198 (for additional details and patients’ selection for organoid preparation, see Supplementary data).
CFS analysis design and procedures
After ~8 days in culture, organoid-derived monolayers reached confluency based on the reported Trans Epithelial Electrics Resistance (TEER) values as previously performed.17 The monolayers were then allowed to differentiate for 24 h. After differentiation, the media were changed to Dulbecco’s Modified Eagle Medium (MilliporeSigma, Darmstadt, Germany) (DMEM)/F12 (without FBS) both apically and basolaterally, and 10% CFS of the B. vulgatus-A2, 10% heat-inactivated CFS, or 10% BHI Broth (control) were added apically to the monolayers and then incubated at 37 °C for 24 h. Half of the CFS- and BHI-treated monolayers received apically 1 mg/ml of a peptic-tryptic digest of gliadin (PTG) 4 h into the incubation. Following the CFS and PTG exposure time, cellular permeability and cytotoxicity were measured, and the basolateral supernatants of the monolayers were collected for cytokine secretion analyses. The experiments were repeated six times.
Measurement of monolayer permeability
TEER was measured daily to monitor the organoid-derived monolayer development as previously reported [18]. Paracellular permeability was also measured based on fluorescein isothiocyanate-polyethylene glycol (FITC-PEG) 550 Da (#PG1-FC-550, Nanocs, Boston, MA) diffusion across the monolayers.17 As previously shown, fluorescence absorbance is directly proportional to the amount of FITC-PEG that crossed the monolayers and correlates directly with the paracellular permeability of the monolayers.18
Measurement of cellular viability
A lactase dehydrogenase (LDH) assay, Cytotoxcity Detection KitPLUS, #0474493001 (MilliporeSigma) was used according to the manufacturer’s protocol to evaluate the viability of the cell monolayers by testing the basolateral media from the transwell system. Cytotoxicity data are reported as a percentage of the absorbance of the untreated control. The background was subtracted from all the LDH release data. The percent cytotoxicity was calculated by dividing the experimental LDH release by the average of the untreated control with values greater than 100% interpreted as a reduction in cell viability.
Baseline gene expression in organoids
Comparative gene expression analysis of healthy control (HC), acute CeD (ACD), celiac remission on a gluten-free diet (GFD), and potential CeD (PceD) were performed on organoids at baseline after 5 days in culture. RNA extraction was performed according to the manufacturer’s protocol. The RNA was purified with a Direct-zol RNA Miniprep Kit (#R2052, Zymo Research, Irvine, CA) per manufacturer’s instructions. cDNA was then synthesized from the purified RNA using the PrimeScript™ RT Master Mix (#RR036A, Takara Bio, San Jose, CA). The oligonucleotide primers employed for the RT-PCR analysis (Supplementary Table ST1) were designed by the MGH primer bank (Boston, MA) and synthesized by Integrative Device Technology (IDT, San Jose, CA). PerfeCTa SYBR Green SuperMix #95054-02 K (Quantabio, Beverly, MA), was used for the qPCR reactions. The quantitative RT-PCR was performed using a QuantStudio™ 3 Real-Time PCR System (Life Technologies, Carlsbad, CA). The 18S gene was used as an internal control for the experiments. The results were analyzed using the CT method (2-∆∆CT) and reported as a fold change value against the experimental control.
Cytokine assay
Innate immune response-related cytokines were measured in the basolateral supernatants of the human gut organoid-derived monolayers and were measured using the Human Innate Immune panel chips (Isoplexis, Inc product code CODEPLEX-2L03-2), which were loaded into the IsoSpark system as previously described.19 The panel of secreted cytokines was analyzed using the IsoSpeak software 2.9.0. The amount of secreted cytokines for each treatment is reported as picograms per milliliter (pg/ml).
Organoid monolayer microRNAs analysis
Total RNA from HC and PceD gut monolayers with or without B. vulgatus-A2 CFS treatment was extracted as previously reported17 and microRNA analysis performed as outlined in the supplementary data.
Statistical analysis
All statistical analyses were performed with GraphPad Prism 9.0. Data set outliers were identified with the Robust regression and Outlier removal (ROUT) Method with a Q value of 1%. Data were analyzed with the ordinary one-way ANOVA. The Dunnett’s multiple comparisons test with a single pool variance was used to identify significance levels within the data points. For the organoid monolayer microRNAs analysis, data were expressed as means ± standard error of the mean (SEM) and statistical significance were evaluated with two-tailed t-test. Significance levels were recognized by p < 0.001, p < 0.01, and p < 0.05.
Results
Bacterial isolation
Evidence suggests that the biological effect of commensal organisms may be associated with differences detected at the strain level.20 Thus, we decided to investigate the effect of specific strains isolated from stool samples of the CDGEMM subjects, rather than relying on publicly available strains. Using a high throughput, microbial cultivation system (Prospector®, GALT Inc.), we isolated 3500 bacterial colonies from stool samples from two CeD cases and two matched controls at the time of the onset of CeD and 6 months prior, resulting in approximately 150 unique strains. We identified five strains that we had previously found to be specifically enriched in control fecal samples compared to CeD cases,16 which were subcultured for further investigation. The selected strains included Bifidobacterium breve, shown to have significant immune regulatory activity;21 Bacteroides thetaiotaomicron, reported to be protective in a preclinical model of IBD;22 Bifidobacterium longum, recognized to have protective functions on cell barrier and cytokine modulation;23,24 Bacteroides vulgatus, reported to ameliorate intestinal inflammatory diseases in a DSS-induced model of colitis;25 and Bacteroides uniformis, reported to improve immune defense mechanisms in a preclinical model of obesity.26
Genotypic, molecular, and phenotypical characterization of pre-onset human gut organoid
Gene expression comparative analyses (Fig. 1) revealed that the prototype PceD organoids tested (PceD1) had altered gene expression levels for a subset of markers associated with goblet cells, tight junctions and chemokine signaling cascades consistent with our previous observations in acute celiac organoids (ACD).18 Similar alterations observed in PceD1 were confirmed in an organoid derived from a second pediatric PceD (PceD2) patient not enrolled through the CDGEMM study (Fig. 1). Other analyzed genes, including FUT-1, MUC2, and IL37 showed no differences (data not shown). These data suggest that, despite the absence of enteropathy, the duodenal epithelium from PceD patients had already shifted, at least partially, toward an altered gene expression profile typical of the acute phase of CeD months before its onset. Alternatively, subjects at risk of developing CeD may have a specific genetic predisposition (direct or indirect) that influences the basal expression of the aforementioned genes in PceD.
Fig. 1. Fold change (FC) of signature genes expressed in organoids of potential CeD (PCeD) compared to healthy controls (HC, N = 2), acute (ACD, N = 2) and remission (GFD, N = 2)).
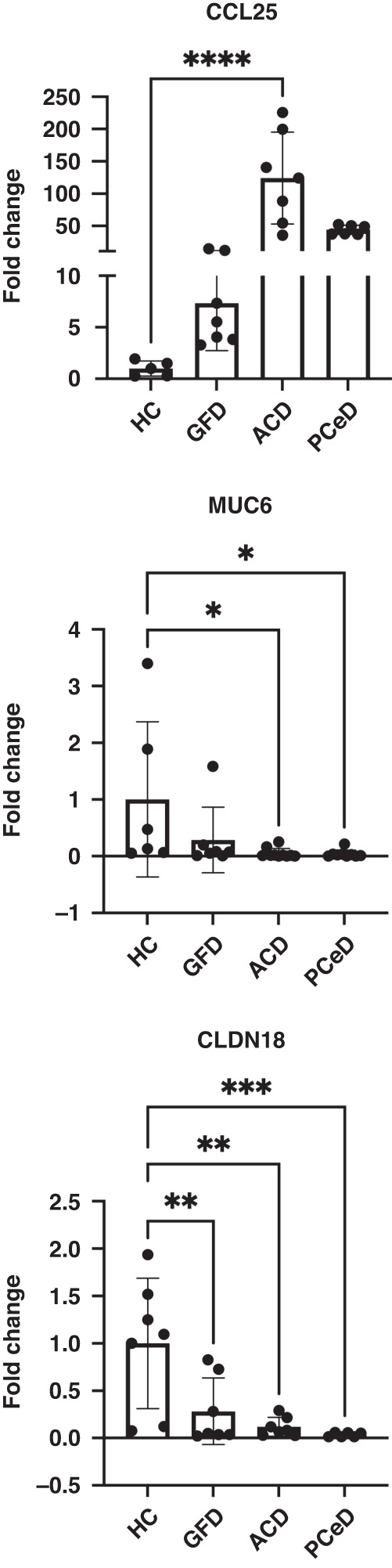
Markers of acute celiac activation related to goblet cells signature, chemokines and barrier [SDS1] were found already altered in both PCeDs. Triplicate for each organoid. One-way ANOVA. (P values: *<0.05, **<0.01, ***<0.001), ****<0.0001.
Effect of microbial CFS on patients’ gut epithelium-MΦ co-culture in response to gliadin exposure
Gut mucosal response to environmental factors, including gliadin, is the result of a coordinated epithelial-immune action. Following the validation of our organoid-derived monolayer from human gut-MΦ co-culture (see supplemental data and Supplementary Fig. S1), we evaluated CFS derived from five candidate strains (see above) isolated from the CDGEMM control subjects to explore potential modulatory effects on the PceD GI epithelium-MΦ response when exposed to gliadin. When cell monolayers were treated with the five tested isolates CFS, B. longum, B thetaiotaomicron and B. vulgatus were the most consistent in mitigating the monolayer permeability, whether induced by gliadin (Supplementary Fig. S2) or at baseline (not shown) in PceD organoid-derived monolayer from human gut. Of note, B. vulgatus CFS was also able to improve the epithelial cell viability (Fig. 2a) in addition to ameliorating increases in gliadin-induced permeability (Fig. 2b). Protective activity of the B. vulgatus CFS correlated directly with the density (log phase) of the culture of origin (Fig. 2). Finally, when we compared the effect of microbial CFSs on organoids’ monolayers versus epithelium-macrophage organotypic cultures (data not shown), we did not observe differences in paracellular permeability and cytotoxicity readouts, suggesting that the observed protective effect was not mediated by macrophages. Combined, these observations prompted us to further investigate B. vulgatus-A2 CFS activity on the epithelium.
Fig. 2. The protective effect of B. Vulgatus-A2 CFS against PTG-induced cell death and increased epithelial paracellular permeability is dose-dependent.

Dose-dependent effect of B. vulgatus-A2 CFS on cell viability (a) and on epithelial paracellular permeability (b) of PCeD organoids (N = 2) exposed to PTG. N of experiments/organoid = 3. One-way ANOVA test (*) P < 0.05, (***)P < 0.001, (****) P < 0.0001. (**) P < 0.01, (***) P < 0.001.
B. vulgatus sequencing and genomic analysis
We assessed whether the B. vulgatus-A2 strain was the same strain available in reference databases or contained unique features. To this end, we sequenced the genome of our isolated B. vulgatus-A2 strain and compared its assembled genome with the genome of B. vulgatus ATCC 8482 reference strain. A list of over 20,000 mutations was identified in the newly isolated strain compared to B. vulgatus ATCC 8482. In particular, two large deletions were noted in this strain, a 78-gene segment starting at base pair 4,200,749 and an 85-gene segment starting at base pair 920,657. Notably, the latter included a gene for lipopolysaccharide (LPS) biosynthesis related polysaccharide transport and flippase (A6KY90).
The 20,000 mutations list was reviewed, and mutations in areas that were not of interest were removed from further investigation (see Supplementary Methods for details). A complete list of annotations for these removed genes is reported in Supplementary Fig. S3. This process yielded 1369 remaining mutations in 1342 distinct genes. These genes were involved in several functions in bacteria according to UniProt annotations (see Supplementary Methods and Supplementary Fig. S4). We further investigated these genes for either direct or indirect interactions of the associated gene products with host cells, such as modulating gut barrier function, or immune response and inflammation. Specifically, we found that several genes responsible for the secretion of bacterial proteins and metabolites contained transglutaminase domains (a key enzyme in CeD pathogenesis), or were involved in quorum sensing based on gene annotations or literature review. Furthermore, we conducted an automated search in PubMed (see Methods) to mine the literature for any evidence that these 1342 genes might be implicated in interactions with the human host. The identified articles from this automated search were then reviewed and the findings were combined with the results from the manual inspection of the mutations. The combined approaches identified a total of 39 genes involved in interactions with the host according to gene annotations and/or prior literature (Table 1). Out of these 39 genes, 13 had products that were secreted or exported, which could alter the microbe–host shared environment. Some examples include an AsmA-like protein involved in glycerophospholipid transport and bacterial outer membrane integrity (A6KWC8), the RND subunit of a putative metal resistance exported protein (A6L1Y6), and a sulfatase/hydrolase that can stimulate pro-inflammatory responses in the host (A6KZU2).27–29 Another notable example of a mutated gene encoding an exported protein in our isolated strain was a gene encoding a protein with transglutaminase activity that could be involved in regulating bacterial cell death and host cell apoptosis (A6KWH9).30 We additionally identified a second transglutaminase domain-containing protein involved in production of antibody and drug conjugates via transpeptidation (A6L738).31 Although mutations in these genes with transglutaminase activity were observed in B. vulgatus-A2, the proteins could impair the epithelial transglutaminase-driven deamidation reaction of gliadin that is presented by the MHC HLA DQ2/8 complexes to CD4+ T cells, a key step leading to the typical CeD autoimmune enteropathy. Examples of additional mutations are listed in the supplemental material. It should be noted that the protective change observed in our experiments for the isolated B. vulgatus-A2 strain might be due to the effect of mutations in a combination of these 39 genes and not necessarily due to mutations in individual genes.
Table 1.
The composite list of 39 genes from the automated PubMed search and manual review of the 1342 gene mutations.
| Uniprot ID | Base Pair | Identification Method | Annotation | Grouping | Citations |
|---|---|---|---|---|---|
| Gene Products with Direct Effects | |||||
| A6KWC8 | 5975 | Manual/Automated | Putative exported protein, AsmA-like - these are critical for glycerophospholipid transport and outer membrane integrity | Barrier Function | 35226662 |
| A6KWH9 | 91,425 | Manual | Conserved hypothetical exported protein with transglutaminase activity that could be involved in regulating cell death and apoptosis | Inflammation | 24464646 |
| A6KWJ8 | 117,182 | Manual | Putative secreted protein involved in glyocsidic bond hydrolysis | Unknown | 8535779 |
| A6KX36 | 412,558 | Manual | Putative exported protein | Unknown | |
| A6KZ75 | 1,395,966 | Manual | Putative secreted sulfatase | Inflammation, Metabolism and Nutrient Processing | 186484, 3982438 |
| A6KZD5 | 1,476,083 | Manual | Putative exported D-alanyl-D-alanine carboxypeptidase penicillin-binding protein (DacB) | Metabolism and Nutrient Processing | 32625007 |
| A6KZU2 | 1,700,527 | Manual | Putative exported sulfatase and hydrolase that can stimulate pro-inflammatory response in host | Barrier Function, Inflammation | 25974305 |
| A6L0C6 | 1,930,429 | Manual | Secreted protein involved in carbohydrate and protein metabolism, BACON domain containing | Metabolism and Nutrient Processing | 20416301 |
| A6L0D9 | 1,945,454 | Manual | Putative secreted tripeptidyl aminopeptidase which cleaves terminal amino acids | Metabolism and Nutrient Processing | 8440407 |
| A6L198 | 2,298,035 | Manual | Secreted protein | Unknown | |
| A6L1Y6 | 2,636,133 | Manual | Putative metal resistance related exported protein, RND Subunit | Barrier Function | 27806930 |
| A6L240 | 2,706,777 | Manual | Putative exported cytochrome C biogenesis-related protein | Barrier Function, Metabolism and Nutrient Processing | 11967064, 12524212 |
| A6L2T7 | 3,018,133 | Automated | Peptidoglycan hydrolase (LysM) | Barrier Function, Cell Replication and Turnover, Inflammation, Metabolism and Nutrient Function | 27708039 |
| Gene Products with Indirect Effects | |||||
| A6KWC3 | 74,433 | Automated | NigD‚-like protein, a lipoprotein | Unknown | 19531060, 35871068 |
| A6KWI0 | 92,576 | Manual | Signal peptidase I, a protein that cleaves hydrophobic, N-terminal signal or leader sequences from secreted and periplasmic proteins allowing for release from the membrane | Inflammation | 22031009 |
| A6KWM3 | 153,245 | Automated | Putative outer membrane protein from the TonB receptor family probably involved in nutrient binding and shown to polarize host macrophages into the M2 state (DnaK) | Barrier Function | 25419575 |
| A6KXR1 | 732,950 | Manual | Neuraminidase, a BNR repeatcontaining protein and possible virulence factor | Inflammation, Metabolism and Nutrient Processing | 7463468 |
| A6KXX6 | 831,953 | Automated | 4-O-beta-D-mannosyl-D-glucose phosphorylase, converts 4-O-beta-D-mannopyranosyl-D-glucopyranose (Man-Glc) to mannose 1-phosphate (Man1P) and glucose | Barrier Function, Metabolism and Nutrient Processing | 23954514 |
| A6KYC8 | 1,014,004 | Automated | Alpha-galactosidase, Hydrolysis of terminal, non-reducing alpha-D-galactose residues in alpha-D-galactosides, including galactose oligosaccharides, galactomannans and galactolipids | Metabolism and Nutrient Processing | 36090029 |
| A6KYS6 | 1,202,941 | Manual | S-ribosylhomocysteine lyase, involved in the synthesis of secreted autoinducer 2 (AI-2) used in quorum sensing, biofilm formation, and virulence factor expression | Cell Replication and Turnover, Inflammation | 24026770 |
| A6KZ59 | 1,363,629 | Manual | Putative transporter, methyltransferase domain containing | Barrier Function | |
| A6KZ98 | 1,423,337 | Automated | Glycosyltransferase | Metabolism and Nutrient Processing | |
| A6KZV5 | 1,722,822 | Automated | Clostripain-related cysteine peptidase | Metabolism and Nutrient Processing | |
| A6L0P3 | 2,060,343 | Automated | Conserved protein (TraN) found in conjugation transposon involved in antagonistic secretion systems to compete with other Bacteroides species | Cell Replication and Turnover | 26768901 |
| A6L100 | 2,173,954 | Automated | GTPase Obg (ObgE), an essential GTPase which binds GTP, GDP and possibly (p)ppGpp. Plays a role in control of the cell cycle, stress response, ribosome biogenesis and in those bacteria that undergo differentiation, in morphogenesis control | Cell Replication and Turnover, Metabolism and Nutrient Processing | 34284824 |
| A6L2L7 | 2,939,217 | Automated | Amidohydrolase | Metabolism and Nutrient Processing | |
| A6L3C3 | 3,234,698 | Automated | Phosphotransferase, APH domain containing protein | Metabolism and Nutrient Processing | |
| A6L3P3 | 3,396,727 | Manual | Putative exopolymeric substances related membrane protein, involved in physiologic stress response | Barrier Function | |
| A6L3S7 | 3,416,804 | Automated | Transporter | Barrier Function | |
| A6L4A1 | 3,638,999 | Automated | Putative outer membrane protein, tryptophanrich sensory protein | Barrier Function | 32937127 |
| A6L4B2 | 3,650,452 | Automated | Ferrous iron transport protein B that participates in iron chelation, reducing inflammation in UC patients | Barrier Function | 27724868, 12379679 |
| A6L4G6 | 3,736,170 | Automated | Transcriptional regulator | Cell Replication and Turnover | |
| A6L4I5 | 3,756,653 | Automated | DNAbinding protein, HU-HIG domain containing protein | Cell Replication and Turnover | |
| A6L4R8 | 3,853,411 | Automated | LysM peptidoglycanbinding domaincontaining protein | Unknown | 27708039 |
| A6L4U7 | 3,889,712 | Manual | Putative exported periplasmic protein, ATP binding cassette transporter | Metabolism and Nutrient Processing | 26517916 |
| A6L4W1 | 3,908,142 | Manual | Putative exported periplasmic protein, ATP binding cassette transporter | Metabolism and Nutrient Processing | 26517916 |
| A6L738 | 4,807,737 | Manual | Transglutaminase domaincontaining protein involved in production of antibody and drug conjugates via transpeptidation | Metabolism and Nutrient Processing | 31643050 |
| A6L7K9 | 5,005,462 | Manual | Tricorn protease homolog | Metabolism and Nutrient Processing | 11347893 |
| A6L7U1 | 5,117,642 | Automated | ABC transporter ATP‚-binding protein/permease | Barrier Function | 19302324 |
The table is divided into gene products that could exert a protective phenotype either directly, e.g., an exported protein, or indirectly, e.g., a transcriptional activator affecting the level of a gene product being produced. Two large deletions of 78 and 85 genes described in the results section were not included in this table.
Effect of B. vulgatus-A2 CFS on gliadin-induced cytokine release in human gut epithelium
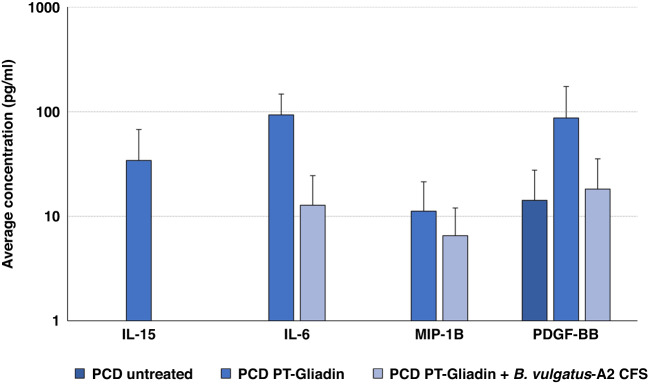
To establish the impact of B. vulgatus-A2 CFS on the response of the PceD gut epithelium, we measured innate immunity cytokines at baseline (before PT-gliadin exposure), after PT-gliadin exposure, and after PT-gliadin plus B. vulgatus-A2 CFS exposure. At baseline, several cytokines were constitutively secreted, including EGF, GM-CSF, IL4, PDGF-BB, sCD137, TNFa, and VEGF (Fig. 3 and Table 2). Following PT-gliadin exposure, there were increases in secretion of several cytokines, including IL-15, IL-6, MIP-1b, and PDGF-BB, which were completely prevented (IL-15) or reduced (IL-6, PDGF-BB, MIP-1b) by the exposure to B. vulgatus-A2 CFS (Fig. 3). Interestingly, B. vulgatus-A2 CFS was also able to completely ablate the constitutive TNFɑ secretion (Table 2). No effect was detected on the remaining cytokines analyzed (Table 2).
Fig. 3. Cytokine release analysis (N = 2 per treatment) with average bars in pg/ml from two bio replicates, each with technical duplicates.
Error bars are representative of the standard deviation of the averages.
Table 2.
Cytokine profiling at baseline, after PT-gliadin exposure and PT-gliadin plus B. vulgatus-A2 CFS in PCeD organoids.
| Experimental Condition | EGF | GM-CSF | IL-10 | IL-4 | IL-8 | IP-10 | MCP-1 | MIP-1a | sCD137 | TNF-a | VEGF |
|---|---|---|---|---|---|---|---|---|---|---|---|
| PCD Untreated | 47.91 | 35.09 | 2.27 | 15.60 | 2.40 | 7.73 | ND | ND | 53.43 | 23.35 | 171.11 |
| PCD PT-Gliadin | 135.9 | 15.37 | 13.39 | 13.96 | 71.13 | 15.95 | 5.74 | ND | 80.67 | 8.42 | 576.40 |
| PCD PT-Gliadin + B. vulgatus-A2 CFS | 92.11 | 7.62 | 15.15 | 13.06 | 76.18 | 29.94 | 10.71 | 18.24 | 63.49 | ND | 419.93 |
Analysis was performed by isoplex method and expressed in pg/ml of cytokine present in the basolateral organoid media.
Effect of B. vulgatus-A2 CFS on human gut organoid monolayer miRNAs
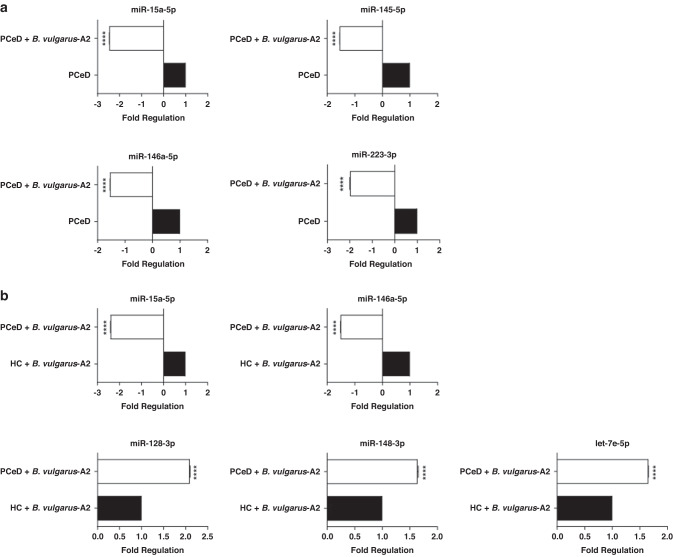
To complement the strain genome and functional analyses, we performed a high-throughput miRNAs analysis in organoid monolayers cultured from HC and PceD patients with or without B. vulgatus-A2 CFS treatment. Our results showed that miRNA modulations are mainly induced by this treatment; in fact, at basal condition the only significantly modulated miRNA is miR-152-3p whose expression is constitutively higher in PceD organoid monolayers when compared to HC organoid monolayers (Fig. 4a). Interestingly, the same miRNA seems to be specifically modulated by B. vulgatus-A2 CFS treatment. When the treatment with B. vulgatus-A2 CFS was performed, the expression of miR-152-3p showed less induction in PceD vs. HC organoid monolayers (Fig. 4b). Focusing on PceD organoid monolayers, an inverse trend for miR-152-3p modulation was reported after B. vulgatus-A2 CFS treatment (Fig. 4c). Furthermore, the B. vulgatus CFS treatment decreased the expression of miR-15a-5p, miR-145-5p, miR-146a-5p, and miR-223-3p in intestinal organoid monolayers from PceD (Fig. 5a). The same inhibitory effect on miRNAs expression was also reported for miR-146a-5p, and miR-15a-5p when organoid-derived monolayers from PceD were compared to those from HC, both exposed to B. vulgatus CFS treatment (Fig. 5b). The comparison between B. vulgatus-A2 CFS-treated organoids from PceD vs. B. vulgatus-A2 CFS-treated HC organoid monolayers also showed the induced expression of miR-128-3p, miR-148-3p, and let-7e-5p (Fig. 5b). On the contrary, another member of the let-7 family (let-7c-5p) was repressed in HC organoid monolayers after B. vulgatus-A2 CFS treatment (Supplementary Fig. S5). Of note, none of these miRNAs were modulated at baseline, highlighting the role of B. vulgatus-A2 CFS in regulating miRNAs expression in organoid monolayers. Based on target prediction and gene set enrichment analyses, all the miRNAs significantly modulated expression correlate with the involvement of signaling pathways regulating pluripotency of stem cells, MAPK, mTOR, and TGFβ signaling pathways (Supplementary Fig. S6). In addition, both these analyses showed the modulation of other inflammatory pathways for miR-146a-5p, such as the NF-κB and the T cell receptor signaling pathway (Supplementary Fig. S6).
Fig. 4. Effect of B. vulgatus-A2 CFS on miR-152-3p expression in both PCeD and HC organoids.
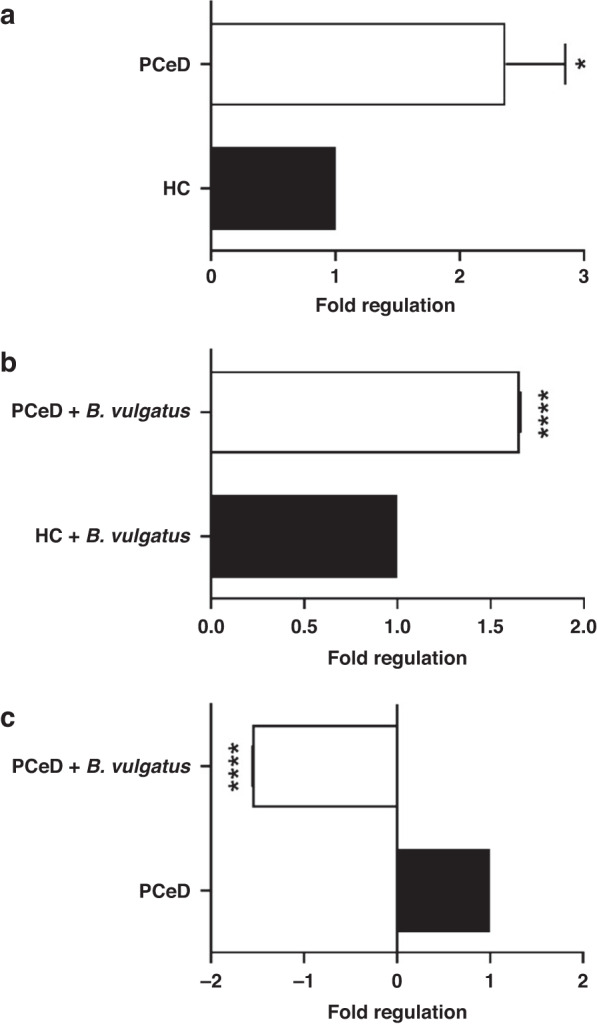
High-throughput miRNAs analysis was performed by Real-Time PCR using pre-custom plates for the inflammatory response and the autoimmunity focus in PCeD vs. HC organoids at basal condition (a) and after B. vulgatus-A2 CFS treatment (b), and in PCeD organoids treated with B. vulgatus-A2 CFS vs. untreated ones (c) (n = 3 organoids derived monolayers/group). Histograms represent the mean ± SEM. *P < 0.05, ****P < 0.0001.
Fig. 5. Effect of B. vulgatus-A2 CFS on miRNAs expression in both PCeD and HC organoids.
Modulation of miRNAs expression in B. vulgatus-A2 CFS-treated PCeD organoids (white bars) vs. untreated PCeD organoids (black bars) (a). Differential expression of miRNAs in HC (black bars) and PCeD organoids (white bars) after the treatment with B. vulgatus-A2 CFS (b). For both the analyses, a high-throughput miRNAs analysis was performed by Real-Time PCR using pre-custom plates for the inflammatory response and the autoimmunity focus (n = 3 organoids derived monolayers/group). Histograms represent the mean ± SEM. ****P < 0.0001.
Discussion
While gluten serves as the main environmental trigger for CeD onset, we now appreciate that additional stimuli are necessary to lose tolerance to gluten and start the transition from genetic predisposition to clinical presentation. Among other environmental elements, special attention has been paid over the years to gastrointestinal infections. One of the first reports from almost 40 years ago hypothesized adenovirus infection as a possible trigger of CeD onset based on a possible antigen mimicry mechanism.32 Since then, several reports have been published claiming a pathogenic role of specific microorganisms in CeD onset.10 While this pathogenic mechanism remains a possibility, a likely alternative mechanism could involve a more complex disruption of the ecosystem of the gastrointestinal microbiota caused by infection rather than by the specific effect of a single pathogen on gut mucosal homeostasis. To explore this hypothesis, we developed a human pre-onset gut organoid model as a proof-of-concept approach to establish the epithelial response to gliadin exposure and to examine the potential protective role of bacterial strains found enriched in infants at risk of CeD who were protected against the onset of the disease compared to at-risk infants who lost tolerance to gluten and developed the disease. Among the five strains identified, we focused our studies mainly on B. vulgatus-A2, which showed sustainable growth and biological activity across all tested cultures in affecting the effect of gluten on gut epithelial homeostasis occurring during pre-clinical onset of CeD. Our data suggest that B. vulgatus-A2 strain may exert a protective role by acting on the gut epithelium with a coordinated action involving protection against loss of barrier function, cell death and pro-inflammatory cytokine production. These effects seem to correlate with an epigenetic reprogramming of the epithelial cells affecting these protective functions. Also interesting is the mutation of B. vulgatus-A2 transglutaminase activity, a feature that deserves further characterization, given the hypothesized role of bacterial-derived transglutaminases in CeD pathogenesis.33
At the molecular level, we have previously shown that gluten-derived peptides signal through MYD88-dependent activation of the CXCR3 receptor23 to promote secretion of IL-834 and upregulation of paracellular permeability in the epithelium via zonulin release.35 We also generated data suggesting the involvement of MΦ in the early steps of CeD pathogenesis.34,36 Thus, we have adopted an in vitro organotypic, co-culture model that incorporated both epithelial cells and MΦ to establish whether specific epithelium-MΦ interactions are necessary to maintain homeostasis in the context of a microbiome-established micromilieu. Our data suggest that mucosal homeostasis does not require this cooperation and is maintained by the action of specific microbiota-secreted components that exert the double action of epigenetically influencing key functions of the host epithelium involved in antigen trafficking, cell turnover, inflammatory immune response, and protecting against gliadin-induced cell death and increased permeability through secreted molecules present in conditioned media. Using B. vulgatus-A2 strain as a prototype, we have shown that this unique strain isolated from controls and not present in cases had several mutations compared to the ATCC 8482 reference strain that affects its capacity to regulate metabolic pathways of the host related to inflammatory response and barrier function. These data were complemented by the phenotypic observation in the gut epithelium in which we detected that B. vulgatus-A2 CFS was able to ameliorate the gliadin-induced cell damage and increased epithelial permeability.
Moreover, the epigenetic changes align with the phenotypic outcome. Specifically, we have shown that organoid-derived monolayers from PCeD downregulated the expression of miR-15a-5p, miR-145-5p, miR146a-5p, and miR-223-3p after treatment with B. vulgatus-A2 CFS. These data are in accordance with the previous literature,37–39 except for miR-145-5p for which a reduced expression was reported in CeD patients.40 Target prediction and gene set enrichment analyses linked the four miRNAs to signaling pathways, which have been previously shown to be involved in CeD pathogenesis.41–44 Specifically, in line with the literature, the reduction of miR-15a-5p and miR146a-5p after B. vulgatus-A2 CFS treatment in PCeD when compared to HC organoid monolayers impairs TGF-β-mediated signaling, thus promoting and sustaining intestinal inflammation in CeD.45 Furthermore, the downregulation of miR-15a-5p induced by B. vulgatus-A2 CFS treatment correlates with the bacterial ability to ameliorate the gliadin-induced increased mucosal permeability in gut organoid monolayers. In fact, miR-15a-5p negatively regulates intestinal epithelial tight junctions through cell division cycle (Cdc)42 in pediatric IBD.46 Additionally, the treatment with B. vulgatus-A2 CFS increased the expression of miR-128-3p, miR148-3p, and let-7e-5p in PCeD organoids relative to HC organoid monolayers thus supporting a possible role for these miRNAs in promoting CeD pathogenesis. In accordance with the let-7e-5p data, another member of the let-7 family (let-7c-5p), which is known to regulate proliferation, differentiation and apoptosis in development and cancer,47 was reduced in B. vulgatus-A2 CFS-treated HC organoids compared to untreated HC organoids. The effect on let-7 family members seems to be mediated by B. vulgatus-A2 CFS; in fact, a significant decrease of circulating let-7e-5p was reported between controls and samples taken <1 year before tissue transglutaminase antibodies positivity.48 In addition, our data demonstrated the ability of B. vulgatus-A2 CFS treatment to specifically affect miR-152-3p expression as indicated by less upregulation of this miRNA in PCeD organoids compared to HC organoids before and after B. vulgatus-A2 CFS treatment. This trend was confirmed by the reduced expression of miR-152-3p in B. vulgatus-A2 CFS-treated organoids from PCeD. Even if miR-152-3p has a positive effect on CeD signaling pathways in different pathological contexts,49,50 to the best of our knowledge, no studies have been reported in literature that specifically correlate this miRNA to CeD. Interestingly, in our dataset miR-152-3p is the only miRNA to be significantly modulated in PCeD versus HC organoids at basal condition.
Target prediction and gene set enrichment analyses linked the four miRNAs to signaling pathways regulating the pluripotency of stem cells, MAPK, mTOR, and TGFβ, which have been previously shown to be involved in CeD pathogenesis.41–48 These signaling pathways involving immune response and regulation of barrier function are complementary with our findings on the B. vulgatus-A2 strain genetic mutations and our functional studies that we have reported above, further supporting a possible probiotic profile of this strain. Thus, our data suggest that the B. vulgatus-A2 strain protective effect against gluten shown by the phenotypic experiments could act through miRNAs reprogramming in the celiac gut epithelium. However, more studies are needed to confirm and integrate the miRNAs data reported in this study and to validate the predicted targets.
In conclusion, our in vitro modeling of the pre-onset phase of CeD suggests a break of gut epithelial homeostasis secondary to gut dysbiosis characterized by loss of protective bacterial elements that both epigenetically and functionally influence the gut epithelium response aimed at maintaining a state of energy toward gluten. Perturbation of this microbiota balance, secondary to unknown stressors, may lead to a break of tolerance to gluten, starting the transition from genetic predisposition to active disease. Our data provide preliminary potential evidence of possible targets for disease interception in subjects at risk of CeD who present with elements of dysbiosis in their pre-clinical phase. However, to translate our proof-of-concept evidence into implementable therapeutic strategies, additional corroboration of our results also exploring the combined cooperation between protective vs. offensive elements that characterize the microbiome “signature” of children at-risk that went on to develop CeD is needed.
Disclaimer
This article was prepared while Stefania Senger was employed at Massachusetts General Hospital. The opinions expressed in this article are the author’s own and do not reflect the view of the National Institutes of Health, the Department of Health and Human Services, or the United States government.
Supplementary information
Author contributions
A.F. was responsible for the initial draft of the manuscript. All authors contributed to the reviewing and editing of this manuscript. The following contributed to the investigation and resources: T.T., S.S., M.B., F.C., M.C., S.DeS., L.E., C.S.F., R.F., M.M.L., F.M., M.M., A.M., L.N., T.P., P.P., J.C.R., N.S., F.V., A.R.Z., A.F. The following contributed to data curation, analysis, and interpretation: T.T., S.S., R.B., C.S.F., I.G.-M., R.S.L., J.C.R., A.R.Z., AF. The following contributed to obtaining funding: MM.L., A.F.
Funding
Research reported in this publication was supported by the National Institute of Allergy and Infectious Diseases (NIAID) Award Number R56AI169645 to A.F. and by National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) DK109620 and K23DK122127 to M.M.L. Additional support for C.S.F. and A.F. is from the National Institute of Diabetes and Digestive and Kidney Diseases grant Nutrition Obesity Research Center at Harvard (NORCH) 2P30DK040561-26.
Data availability
Materials described here and all relevant raw data will be freely available to any researcher wishing to use them for non-commercial purposes.
Competing interests
V.A.K. serves as a consultant for Takeda Pharmaceuticals. M.M.L. acted as a consultant for Anokion. A.F. is a stockholder at Alba Therapeutics, serves as a consultant for 9Meters, is an advisory board member for Axial Biotherapeutics, and has a speaker agreement with Mead Johnson Nutrition and Sanofi. Other authors have declared that no conflict of interest exists. None of the above financial disclosures are related to this work.
Informed consent
Informed Consent was obtained, and the study was approved by the Massachusetts General Brigham Institutional Review Board.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
A list of authors and their affiliations appears at the end of the paper.
Contributor Information
Alessio Fasano, Email: afasano@mgh.harvard.edu.
CDGEMM Team:
Maria Luisa Forchielli, Adelaide Serretiello, Corrado Vecchi, Gemma Castillejo de Villasante, Giorgia Venutolo, Basilio Malamisura, Angela Calvi, Maria Elena Lionetti, Mariella Baldassarre, Chiara Maria Trovato, Nicoletta Pietropaoli, Michela Perrone, Lidia Celeste Raguseo, and Carlo Catassi
Supplementary information
The online version contains supplementary material available at 10.1038/s41390-023-02960-0.
References
- 1.Fasano A, Catassi C. Clinical practice. Celiac disease. N. Engl. J. Med. 2012;367:2419–2426. doi: 10.1056/NEJMcp1113994. [DOI] [PubMed] [Google Scholar]
- 2.Kupfer SS, Jabri B. Pathophysiology of celiac disease. Gastrointest. Endosc. Clin. N. Am. 2012;22:639–660. doi: 10.1016/j.giec.2012.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jabri B, Sollid LM. Tissue-mediated control of immunopathology in coeliac disease. Nat. Rev. Immunol. 2009;9:858–870. doi: 10.1038/nri2670. [DOI] [PubMed] [Google Scholar]
- 4.Catassi C, et al. Natural history of celiac disease autoimmunity in a USA cohort followed since 1974. Ann. Med. 2010;42:530–538. doi: 10.3109/07853890.2010.514285. [DOI] [PubMed] [Google Scholar]
- 5.Choung RS, et al. Trends and racial/ethnic disparities in gluten-sensitive problems in the United States: findings from the National Health and Nutrition Examination Surveys from 1988 to 2012. Am. J. Gastroenterol. 2015;110:455–461. doi: 10.1038/ajg.2015.8. [DOI] [PubMed] [Google Scholar]
- 6.Cheng J, et al. Duodenal microbiota composition and mucosal homeostasis in pediatric celiac disease. BMC Gastroenterol. 2013;13:113. doi: 10.1186/1471-230X-13-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sanchez E, Donat E, Ribes-Koninckx C, Fernandez-Murga ML, Sanz Y. Duodenal-mucosal bacteria associated with celiac disease in children. Appl. Environ. Microbiol. 2013;79:5472–5479. doi: 10.1128/AEM.00869-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wacklin P, et al. The duodenal microbiota composition of adult celiac disease patients is associated with the clinical manifestation of the disease. Inflamm. Bowel Dis. 2013;19:934–941. doi: 10.1097/MIB.0b013e31828029a9. [DOI] [PubMed] [Google Scholar]
- 9.Pozo-Rubio T, et al. Immune development and intestinal microbiota in celiac disease. Clin. Dev. Immunol. 2012;2012:654143. doi: 10.1155/2012/654143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Verdu EF, Schuppan D. Co-factors, microbes, and immunogenetics in celiac disease to guide novel approaches for diagnosis and treatment. Gastroenterology. 2021;161:1395–1411.e1394. doi: 10.1053/j.gastro.2021.08.016. [DOI] [PubMed] [Google Scholar]
- 11.Felli C, Baldassarre A, Masotti A. Intestinal and circulating micrornas in coeliac disease. Int. J. Mol. Sci. 2017;18:1907. doi: 10.3390/ijms18091907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giuffrida P, Di Sabatino A. Micrornas in celiac disease diagnosis: a mir curiosity or game-changer? Dig. Dis. Sci. 2020;65:1877–1879. doi: 10.1007/s10620-020-06081-0. [DOI] [PubMed] [Google Scholar]
- 13.Leonard MM, Camhi S, Huedo-Medina TB, Fasano A. Celiac disease genomic, environmental, microbiome, and metabolomic (Cdgemm) study design: approach to the future of personalized prevention of celiac disease. Nutrients. 2015;7:9325–9336. doi: 10.3390/nu7115470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leonard MM, et al. Multi-omics analysis reveals the influence of genetic and environmental risk factors on developing gut microbiota in infants at risk of celiac disease. Microbiome. 2020;8:130. doi: 10.1186/s40168-020-00906-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leonard MM, et al. Microbiome signatures of progression toward celiac disease onset in at-risk children in a longitudinal prospective cohort study. Proc. Natl Acad. Sci. USA. 2021;118:e2020322118. doi: 10.1073/pnas.2020322118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Persyn A, Mueller A, Goormachtig S. Drops join to make a stream: high-throughput nanoscale cultivation to grasp the lettuce root microbiome. Environ. Microbiol. Rep. 2022;14:60–69. doi: 10.1111/1758-2229.13014. [DOI] [PubMed] [Google Scholar]
- 17.Freire R, et al. Human gut-derived-organoids provide model to study gluten response and effects of microbiota-derived molecules in celiac disease. Sci. Rep. 2019;9:7029. doi: 10.1038/s41598-019-43426-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fiorentino M, et al. Helicobacter pylori-induced disruption of monolayer permeability and proinflammatory cytokine secretion in polarized human gastric epithelial cells. Infect. Immun. 2013;81:876–883. doi: 10.1128/IAI.01406-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Szabo PA, et al. Longitudinal profiling of respiratory and systemic immune responses reveals myeloid cell-driven lung inflammation in severe Covid-19. Immunity. 2021;54:797–814.e796. doi: 10.1016/j.immuni.2021.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shanahan F, Ghosh TS, O’Toole PW. The healthy microbiome-what is the definition of a healthy gut microbiome? Gastroenterology. 2021;160:483–494. doi: 10.1053/j.gastro.2020.09.057. [DOI] [PubMed] [Google Scholar]
- 21.Barrett E, Ross RP, Fitzgerald GF, Stanton C. Rapid screening method for analyzing the conjugated linoleic acid production capabilities of bacterial cultures. Appl. Environ. Microbiol. 2007;73:2333–2337. doi: 10.1128/AEM.01855-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Delday M, Mulder I, Logan ET, Grant G. Bacteroides the taiotaomicron ameliorates colon inflammation in preclinical models of Crohn’s disease. Inflamm. Bowel Dis. 2019;25:85–96. doi: 10.1093/ibd/izy281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thomas KE, Sapone A, Fasano A, Vogel SN. Gliadin stimulation of murine macrophage inflammatory gene expression and intestinal permeability are Myd88-dependent: role of the innate immune response in celiac disease. J. Immunol. 2006;176:2512–2521. doi: 10.4049/jimmunol.176.4.2512. [DOI] [PubMed] [Google Scholar]
- 24.Nakagaki BN, Vieira AT, Rezende RM, David BA, Menezes GB. Tissue macrophages as mediators of a healthy relationship with gut commensal microbiota. Cell Immunol. 2018;330:16–26. doi: 10.1016/j.cellimm.2018.01.017. [DOI] [PubMed] [Google Scholar]
- 25.Li S, et al. Evaluation of the effects of different bacteroides vulgatus strains against dss-induced colitis. J. Immunol. Res. 2021;2021:9117805. doi: 10.1155/2021/9117805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gauffin Cano P, Santacruz A, Moya A, Sanz Y. Bacteroides uniformis CECT 7771 ameliorates metabolic and immunological dysfunction in mice with high-fat-diet-induced obesity. PLoS One. 2012;7:e41079. doi: 10.1371/journal.pone.0041079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Douglass MV, McLean AB, Trent MS. Absence of Yhdp, Tamb, and Ydbh leads to defects in glycerophospholipid transport and cell morphology in gram-negative bacteria. PLoS Genet. 2022;18:e1010096. doi: 10.1371/journal.pgen.1010096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liang ZB, Chen YM, Chen Y, Cheng YY, Zhang LH. Rnd efflux pump and its interrelationship with quorum sensing system. Yi Chuan. 2016;38:894–901. doi: 10.16288/j.yczz.16-139. [DOI] [PubMed] [Google Scholar]
- 29.Hickey CA, et al. Colitogenic bacteroides thetaiotaomicron antigens access host immune cells in a sulfatase-dependent manner via outer membrane vesicles. Cell Host Microbe. 2015;17:672–680. doi: 10.1016/j.chom.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fraij BM. The 55 Kda tissue transglutaminase cross-linking active isoform tg induces cell death. Mol. Carcinog. 2015;54:720–729. doi: 10.1002/mc.22134. [DOI] [PubMed] [Google Scholar]
- 31.Anami Y, Tsuchikama K. Transglutaminase-mediated conjugations. Methods Mol. Biol. 2020;2078:71–82. doi: 10.1007/978-1-4939-9929-3_5. [DOI] [PubMed] [Google Scholar]
- 32.Kagnoff MF, Austin RK, Hubert JJ, Bernardin JE, Kasarda DD. Possible role for a human adenovirus in the pathogenesis of celiac disease. J. Exp. Med. 1984;160:1544–1557. doi: 10.1084/jem.160.5.1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lerner A, Matthias T. Possible association between celiac disease and bacterial transglutaminase in food processing: a hypothesis. Nutr. Rev. 2015;73:544–552. doi: 10.1093/nutrit/nuv011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lammers KM, et al. Identification of a novel immunomodulatory gliadin peptide that causes interleukin-8 release in a chemokine receptor cxcr3-dependent manner only in patients with coeliac disease. Immunology. 2011;132:432–440. doi: 10.1111/j.1365-2567.2010.03378.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lammers KM, et al. Gliadin induces an increase in intestinal permeability and zonulin release by binding to the chemokine receptor Cxcr3. Gastroenterology. 2008;135:194–204.e193. doi: 10.1053/j.gastro.2008.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mortha A, et al. Microbiota-dependent crosstalk between macrophages and Ilc3 promotes intestinal homeostasis. Science. 2014;343:1249288. doi: 10.1126/science.1249288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tan IL, et al. A combined mrna- and mirna-sequencing approach reveals mirnas as potential regulators of the small intestinal transcriptome in celiac disease. Int. J. Mol. Sci. 2021;22:11382. doi: 10.3390/ijms222111382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bascunan KA, et al. A Mirna-based blood and mucosal approach for detecting and monitoring celiac disease. Dig. Dis. Sci. 2020;65:1982–1991. doi: 10.1007/s10620-019-05966-z. [DOI] [PubMed] [Google Scholar]
- 39.Lemos DS, et al. Extracellular vesicle micrornas in celiac disease patients under a gluten-free diet, and in lactose intolerant individuals. BBA Adv. 2022;2:100053. doi: 10.1016/j.bbadva.2022.100053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Felli C, et al. Circulating Micrornas as novel non-invasive biomarkers of paediatric celiac disease and adherence to gluten-free diet. EBioMedicine. 2022;76:103851. doi: 10.1016/j.ebiom.2022.103851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Piscaglia AC, et al. Circulating hematopoietic stem cells and putative intestinal stem cells in coeliac disease. J. Transl. Med. 2015;13:220. doi: 10.1186/s12967-015-0591-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Broide E, et al. Evidence for aberrant regulation of map kinase signal transduction pathway in peripheral blood mononuclear cells in patients with active celiac disease. Dig. Dis. Sci. 2009;54:1270–1275. doi: 10.1007/s10620-008-0480-y. [DOI] [PubMed] [Google Scholar]
- 43.Sedda S, et al. Mtor sustains inflammatory response in celiac disease. Sci. Rep. 2020;10:10798. doi: 10.1038/s41598-020-67889-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hansson T, et al. Transforming growth factor-beta (Tgf-Beta) and tissue transglutaminase expression in the small intestine in children with coeliac disease. Scand. J. Immunol. 2002;56:530–537. doi: 10.1046/j.1365-3083.2002.01157.x. [DOI] [PubMed] [Google Scholar]
- 45.Benahmed M, et al. Inhibition of Tgf-beta signaling by Il-15: a new role for Il-15 in the loss of immune homeostasis in celiac disease. Gastroenterology. 2007;132:994–1008. doi: 10.1053/j.gastro.2006.12.025. [DOI] [PubMed] [Google Scholar]
- 46.Tang WJ, et al. Microrna-15a - cell division cycle 42 signaling pathway in pathogenesis of pediatric inflammatory bowel disease. World J. Gastroenterol. 2018;24:5234–5245. doi: 10.3748/wjg.v24.i46.5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roush S, Slack FJ. The Let-7 family of micrornas. Trends Cell Biol. 2008;18:505–516. doi: 10.1016/j.tcb.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 48.Tan IL, et al. Circulating Mirnas as potential biomarkers for celiac disease development. Front. Immunol. 2021;12:734763. doi: 10.3389/fimmu.2021.734763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ning YX, Wang XY, Wang JQ, Zeng R, Wang GQ. Mir‑152 regulates Tgf‑beta1‑induced epithelial‑mesenchymal transition by targeting Hpip in tubular epithelial cells. Mol. Med. Rep. 2018;17:7973–7979. doi: 10.3892/mmr.2018.8842. [DOI] [PubMed] [Google Scholar]
- 50.Wen YY, et al. Igf-1-mediated Pkm2/beta-catenin/Mir-152 regulatory circuit in breast cancer. Sci. Rep. 2017;7:15897. doi: 10.1038/s41598-017-15607-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Materials described here and all relevant raw data will be freely available to any researcher wishing to use them for non-commercial purposes.