Abstract
Human immunodeficiency virus type 1 particle assembly is directed by the Gag polyprotein Pr55gag, the precursor for the matrix (MA), capsid (CA), and nucleocapsid proteins of the mature virion. We now show that CA sequences N terminal to the major homology region (MHR), which form a distinct domain, are dispensable for particle formation. However, slightly larger deletions which extend into the MHR severely impair particle production. Remarkably, a deletion which removed essentially all MA and CA sequences between the N-terminal myristyl anchor and the MHR reduced the yield of extracellular particles only moderately. Particle formation even exceeded wild-type levels when additional MA sequences, either from the N or the C terminus of the domain, were retained. We conclude that no distinct region between the myristyl anchor and the MHR is required for efficient particle assembly or release.
Human immunodeficiency virus type 1 (HIV-1) morphogenesis is driven by the Gag precursor Pr55gag, which can assemble into virus-like particles even when expressed alone (11). Cleavage of Pr55gag by the viral protease (PR) initiates the maturation of the virus particle. PR and other viral enzymes, such as reverse transcriptase (RT) and integrase (IN), are brought into the virion as components of the Gag-Pol polyprotein, which results from ribosomal frameshifting near the end of the gag gene into the overlapping pol gene (16). The major proteolytic processing products derived from Pr55gag are matrix (MA), capsid (CA), and nucleocapsid (NC), which are common to all retroviruses (21). Additionally, Pr55gag yields a peptide designated p6, which has no homolog in oncoretroviruses. In the mature virion, MA forms a shell directly underneath the host cell-derived lipid envelope, CA forms the core structure, and NC is associated with the genomic RNA within the core (10).
Little is known about the interactions which govern the assembly of Gag precursor molecules into spherical protein shells. In Rous sarcoma virus (RSV), three distinct assembly domains have been defined which together comprise no more than 25% of the Gag precursor (4). Interestingly, despite its prominent structural role in the mature virion, RSV CA was not required for efficient particle assembly (3, 32, 33). In the case of HIV-1, the covalent attachment of myristic acid to the N terminus of the MA domain is required for Gag membrane binding and particle formation (2, 14). Furthermore, genetic analyses indicate that the major homology region (MHR), a uniquely conserved region within CA, forms the N terminus of an assembly domain which comprises the C-terminal third of CA and extends into the p2 peptide that separates CA from NC (1, 5, 17, 19). While deletions N terminal to the MHR had little or no effect on viral particle production (5, 22, 23, 30), numerous mutations within the C-terminal third of HIV-1 CA severely impaired virus assembly (5, 7, 19, 20, 23, 28).
Interestingly, recent structural studies have shown that the functionally distinct regions of CA form independently folding domains which are connected via a hinge region just proximal to the MHR (9, 12). In the present study, we demonstrate that the entire N-terminal domain of CA is dispensable for particle formation. However, particle release was dramatically reduced by deletions which extended into the MHR. In marked contrast, efficient production of particles of normal density was observed even when about 50% of Pr55gag between the myristylation signal and the MHR was deleted.
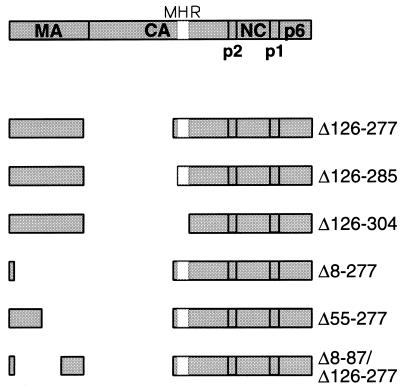
The N-terminal domain of HIV-1 CA is dispensable for particle production.
We reported previously that CA mutants with small deletions N terminal to the MHR retain the ability to form viral particles with wild-type efficiency (5). Furthermore, Wang and Barklis (30) showed that a mutant with a 56-amino-acid deletion in the N-terminal half of HIV-1 CA can assemble particles efficiently. Taken together, these results suggested that CA sequences proximal to the MHR may be dispensable for particle production. To explore this possibility, we used site-directed mutagenesis as described previously (14) to generate the Δ126-277 mutant proviral clone (Fig. 1), which encodes a Gag polyprotein that lacks the entire N-terminal CA domain. Of note, the Δ126-277 deletion, which also removed seven residues from the C terminus of MA, preserved the C-terminal domain of CA, including the MHR.
FIG. 1.
Schematic representation of Gag deletion mutants. The domain structure of the Gag polyprotein and the position of the MHR within the CA domain are indicated. Numbers refer to the positions of the deleted amino acids relative to the initiating methionine.
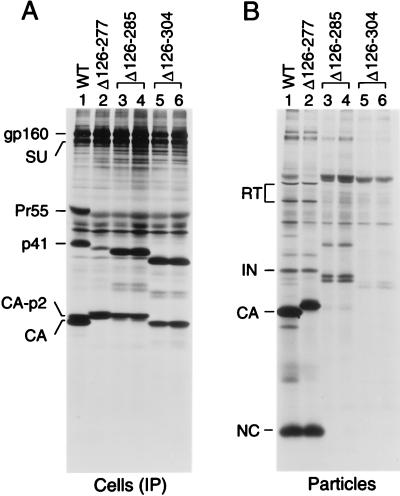
The parental and mutant proviral DNAs were transfected into HeLa cells, followed by metabolic labeling with [35S]methionine from 48 to 60 h posttransfection. Viral proteins in the transfected cells were then immunoprecipitated with serum from a patient infected with HIV-1 and examined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). As shown in Fig. 2A, the Δ126-277 mutant expressed a shortened Gag precursor with an electrophoretic mobility slightly faster than that of p41, a wild-type Gag cleavage intermediate. Immunorecognition of the Gag polyprotein by patient sera is frequently impaired even by minor alterations (6), which may explain why only relatively small amounts of the Δ126-277 Gag precursor were detected. The mutant also produced a prominent Gag species which migrated close to authentic CA, as expected for a fusion protein consisting of MA joined to the C-terminal domain of CA. A slightly more slowly migrating species presumably represented a form of the fusion protein which had p2 joined to its C terminus. An equivalent form of CA is found in cells transfected with the wild-type construct (Fig. 2A, lane 1).
FIG. 2.
Effects of large deletions in CA on particle production. HeLa cells were transfected with wild-type proviral DNA (WT) or with the indicated mutants. (A) The cells were metabolically labeled with [35S]methionine from 48 to 60 h posttransfection, and viral proteins were immunoprecipitated from the cell lysates. (B) Viral particles released during the labeling period were pelleted through 20% sucrose cushions, and the protein profile of the pelleted material was directly analyzed by SDS-PAGE. The positions of specific viral proteins are indicted on the left. The Δ126-285 and Δ126-304 mutants were analyzed in duplicate.
To examine the ability of the Δ126-277 mutant to produce extracellular particles, virions released during the labeling period were pelleted through 20% sucrose and their protein composition was directly analyzed by SDS-PAGE. As shown in Fig. 2B, cells transfected with the Δ126-277 mutant released wild-type amounts of pelletable gag- and pol-encoded products such as NC, RT, and IN. While authentic CA has 11 methionine residues, the MA-CA fusion protein in Δ126-277 virions was expected to contain only 3, explaining why the CA band in wild-type virions appeared more prominent. MA and p6 were not detected, because these products lack methionine residues. We conclude that deleting the entire N-terminal domain of CA was without effect on viral particle yield or Gag-Pol incorporation.
The MHR is required for efficient particle production.
Two additional mutants were generated by site directed-mutagenesis to determine whether deletions into the MHR would affect assembly. Both mutants lack the N-terminal CA domain and the interdomain connector region of CA; however, while the Δ126-285 mutant retains 19 of the 20 amino acids of the MHR, the Δ126-304 mutant lacks all of the MHR (Fig. 1). HeLa cells transfected with these constructs contained relatively high levels of the expected shortened Gag precursor molecules (Fig. 2A), suggesting that the mutant polyproteins were inefficiently exported and/or processed. Furthermore, cells transfected with each mutant contained a prominent Gag species that migrated at the position expected for a fusion protein consisting of MA and the remainder of the CA domain.
As shown in Fig. 2B, the Δ126-285 deletion reduced the yield of viral particles by at least 10-fold (lanes 3 and 4) and the Δ126-304 deletion practically prevented particle formation (lanes 5 and 6). Interestingly, the few particles that were released essentially lacked NC or the MA-CA fusion proteins that were prominent in the cell-associated fractions. Instead, the pelleted material contained unprocessed or incompletely processed Gag products. The latter migrated as two closely spaced bands which presumably represented forms of the mutant Gag precursors that lacked the C-terminal p6 domain or p6 plus the p1 interdomain peptide. Relative to the levels detected in the transfected cells, these Gag processing intermediates appeared selectively enriched in the particulate fractions. Moreover, these products were the only Gag proteins released by cells transfected with the Δ126-304 mutant (Fig. 2B, lanes 5 and 6). We also examined the effects of the deletions in CA after mutational inactivation of PR. Again, the Δ126-277 deletion was without effect on viral particle yields, while the Δ126-285 deletion significantly reduced and the Δ126-304 deletion essentially blocked particle production (data not shown). We infer that the defects in particle production caused by the Δ126-285 and Δ126-304 deletions were not secondary to a dysregulation of PR.
Efficient particle formation in the absence of both MA and the N-terminal CA domain.
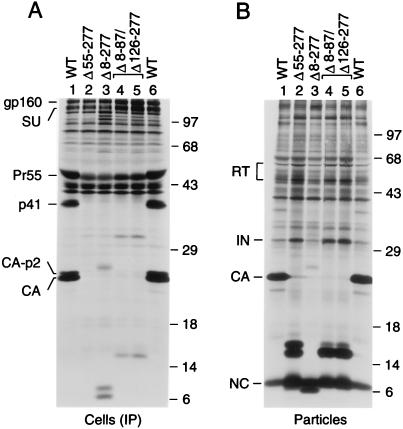
Efficient HIV-1 particle formation requires the N-terminal myristylation signal of MA (2, 14) but not its globular core domain (18, 24, 31). On the other hand, it has been reported that the MA protein of a simian immunodeficiency virus can form virus-like particles on its own (13), indicating that MA has the potential to contribute to the self-association of Gag precursor molecules. We therefore examined whether a role of MA in particle formation would become more apparent in the absence of the N-terminal domain of CA. To this end, we generated the Δ8-277 mutant, which retains a functional myristylation signal (24) but otherwise lacks MA sequences as well as the N-terminal domain of CA (Fig. 1). Additionally, we generated the Δ55-277 mutant (Fig. 1), which retains N-terminal MA sequences beyond the myristylation signal, including a conserved basic cluster that has been implicated in Gag membrane binding (34). A third MA-CA deletion mutant (Δ8-87/Δ126-277) retained sequences from the C rather than from the N terminus of MA (Fig. 1).
Immunoprecipitation from the lysate of cells transfected with the Δ8-277 mutant yielded a small amount of a 26-kDa product, the expected size of the mutant Gag polyprotein, as well as more prominent bands of about 6 and 7 kDa (Fig. 3A, lane 3). Because the patient serum used recognizes CA but not NC, the latter two bands presumably represented the mutant CA products expected from cleavage at the CA-p2 and p2-NC sites. Remarkably, significant amounts of Gag cleavage products of approximately 6 and 7 kDa were released into the supernatant and could be pelleted through 20% sucrose (Fig. 3B, lane 3). The pelleted material also contained a small quantity of the mutant Gag precursor, as well as IN and RT in amounts that were only modestly lower than those in wild-type particle preparations. Measurement of particle-associated RT activity released into the supernatant, as well as an analysis of particles released in the absence of active PR, confirmed that the Δ8-277 deletion did not affect Gag-Pol incorporation and only moderately reduced particle yields (data not shown).
FIG. 3.
Efficient particle production in the absence of up to 50% of Pr55gag. HeLa cells transfected with the indicated proviral constructs were labeled with [35S]methionine, and viral proteins were immunoprecipitated from the cell lysates with patient serum (A). Particles released into the supernatant were pelleted through sucrose and directly analyzed by SDS-PAGE (B). The positions of specific viral proteins are indicated on the left. The positions of migration of molecular mass markers (in kilodaltons) are indicated on the right.
While cells transfected with the Δ55-277 and Δ8-87/Δ126-277 constructs expressed wild-type amounts of envelope glycoprotein, little Gag protein was detected in the cell-associated fractions by the patient serum used, particularly in the case of the Δ55-277 mutant (Fig. 3A). However, Gag clearly was efficiently expressed, because the transfected cells produced large amounts of extracellular particles (Fig. 3B). These particles contained the MA-CA fusion products expected from cleavage at the CA-p2 and p2-NC sites. Cleavage at the CA-p2 site appeared inefficient for the Δ55-277 mutant but was only moderately impaired in the case of the Δ8-87/Δ126-277 mutant. As judged from the relative amounts of pelletable RT, IN, and NC released into the medium, both mutants efficiently incorporated the Gag-Pol precursor and produced two- to threefold more particles than cells expressing the parental proviral construct (Fig. 3B).
Characterization of mutant particles.
Pelleted particles formed by the Δ8-277, Δ55-277, Δ126-277, or wild-type Gag precursor were pooled and layered on top of a preformed 20 to 60% sucrose gradient. Particles lacking active PR were used, because the wild-type and mutant Gag precursor molecules could be readily distinguished by SDS-PAGE. Following centrifugation at 40,000 rpm in an SW41 rotor for 16 h at 4°C, the gradient was fractionated into aliquots of equal size. Viral proteins were then immunoprecipitated from each fraction and separated by SDS-PAGE. Interestingly, particles formed by wild-type Pr55gag and by each of the mutant Gag precursors banded at a similar density (data not shown). We conclude that deleting up to 50% of the HIV-1 gag coding sequence had at most a minor effect on particle density.
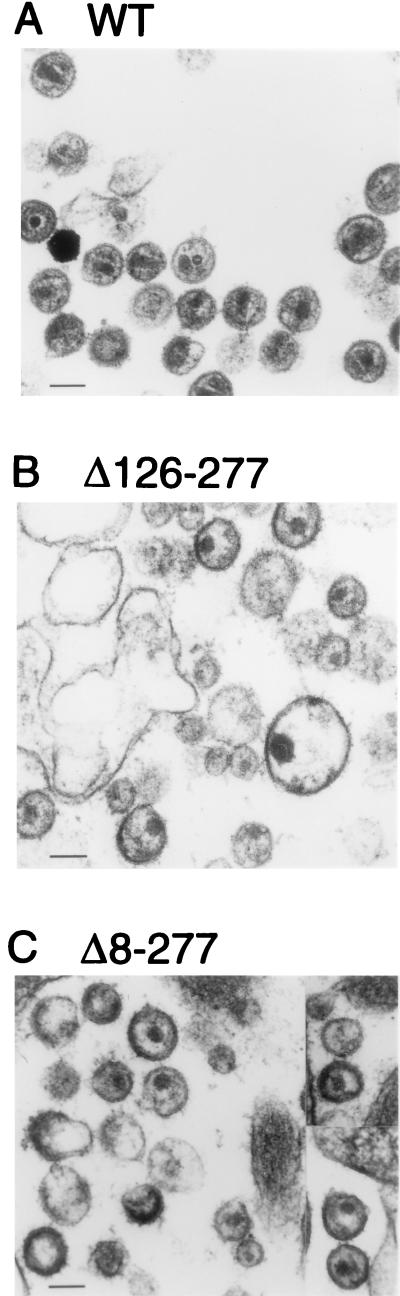
To compare the morphologies of wild-type and mutant particles, transfected HeLa cells were examined by electron microscopy. Cultures transfected with the Δ126-277 or the Δ8-277 mutant contained enveloped, roughly spherical extracellular particles which often resembled wild-type HIV-1 virions but always lacked a conical core (Fig. 4). Instead, a circular core structure was frequently visible. Furthermore, while wild-type particles ranged in diameter from 95 to 160 nm (n = 30), the mutant particles were more heterogeneous in size, with Δ126-277 particles varying between 80 and 310 nm and Δ8-277 particles between 90 and 250 nm.
FIG. 4.
Thin-section electron microscopy of HeLa cells transfected with the wild type provirus (A) or with the indicated Gag deletion mutants (B, C). Bars indicate a length of 100 nm.
Collectively, the results presented in this study show that the C-terminal half of Pr55gag contains all the protein-protein interaction domains required for efficient particle assembly. Interestingly, a 223-amino-acid deletion proximal to the MHR led to an increase in particle yield relative to the wild type. This deletion preserved a conserved N-proximal basic cluster in MA that has been shown to bind acidic phospholipids (34). However, the presence of this region was not specifically required to obtain an increase in particle yield, because the insertion of a 38-amino-acid fragment from the C terminus of MA between the myristylation signal and the MHR had a similar effect. Since the C-terminal third of MA appears to have an extended conformation (8, 15), we favor the interpretation that the retention of MA sequences between the myristyl anchor and the MHR primarily relieved steric constraints which resulted from tethering the C-terminal half of Pr55gag directly to the membrane.
A possible explanation for the increased particle yields relative to those of the wild type is offered by the recently proposed myristyl switch model of Gag membrane targeting (35). In this model, the myristyl group would be sequestered within MA until a conformational change makes it available for membrane insertion. In the absence of the globular core of MA, particle formation may be increased because the myristyl group is constitutively exposed. In support of this view, Spearman et al. (25) recently showed that deletions within α-helical regions in the globular core of MA can dramatically increase overall membrane binding, presumably because the myristyl group can no longer be sequestered.
It has been reported that even single amino acid substitutions in the N-terminal CA domain can reduce viral particle formation up to 20-fold (29). On the other hand, an earlier report showed that a 56-amino-acid deletion in the N-terminal half of CA had no effect on the efficiency of particle production (30). These results indicate that an incorrectly folded N-terminal CA domain can be more disruptive than the lack of a significant portion of the domain. This view is supported by the present study, which clearly shows that the entire N-terminal domain of CA is dispensable for particle assembly.
The role of the MHR in HIV-1 particle formation has been subject to controversy. We previously showed that small deletions (5) and even conservative single amino acid substitutions (19) in the MHR can severely impair HIV-1 particle production. Analogous results were obtained for Mason-Pfizer monkey virus (27). On the other hand, Srinivasakumar et al. (26) reported that an in-frame deletion which precisely removed the HIV-1 MHR had only a minor effect on particle formation. Furthermore, even deletions which essentially removed the entire C-terminal CA domain appeared to have only relatively small effects on particle budding and release (26). It seems possible that an assembly defect was mitigated by the overexpression system used, because several other groups have found that HIV-1 particle formation is highly sensitive to changes in the C-terminal domain of CA (5, 19, 20, 23, 28). In the present study, the lack of a requirement for the N-terminal domain of CA contrasted sharply with the effects of deletions that extended into the MHR, attesting to the importance of this region for HIV-1 particle assembly.
Acknowledgments
A.B. was supported by a fellowship from the Istituto Superiore di Sanitá (Rome, Italy). Å.Ö. was supported by a fellowship from the Swedish Medical Research Council. This work was supported by National Institutes of Health grants AI29873 and AI28691 (Center for AIDS Research) and by a gift from the G. Harold and Leila Y. Mathers Charitable Foundation.
REFERENCES
- 1.Accola M A, Höglund S, Göttlinger H G. A putative α-helical structure which overlaps the capsid-p2 boundary in the human immunodeficiency virus type 1 Gag precursor is crucial for viral particle assembly. J Virol. 1998;72:2072–2078. doi: 10.1128/jvi.72.3.2072-2078.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bryant M, Ratner L. Myristoylation-dependent replication and assembly of human immunodeficiency virus 1. Proc Natl Acad Sci USA. 1990;87:523–527. doi: 10.1073/pnas.87.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Craven R C, Leure-duPree A E, Weldon R A, Wills J W. Genetic analysis of the major homology region of the Rous sarcoma virus Gag protein. J Virol. 1995;69:4213–4227. doi: 10.1128/jvi.69.7.4213-4227.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Craven R C, Parent L J. Dynamic interactions of the Gag polyprotein. Curr Top Microbiol Immunol. 1996;214:65–94. doi: 10.1007/978-3-642-80145-7_3. [DOI] [PubMed] [Google Scholar]
- 5.Dorfman T, Bukovsky A, Öhagen A, Höglund S, Göttlinger H G. Functional domains of the capsid protein of human immunodeficiency virus type 1. J Virol. 1994;68:8180–8187. doi: 10.1128/jvi.68.12.8180-8187.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dorfman T, Mammano F, Haseltine W A, Göttlinger H G. Role of the matrix protein in the virion association of the human immunodeficiency virus type 1 envelope glycoprotein. J Virol. 1994;68:1689–1696. doi: 10.1128/jvi.68.3.1689-1696.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ebbets-Reed D, Scarlata S, Carter C A. The major homology region of the HIV-1 gag precursor influences membrane affinity. Biochemistry. 1996;35:14268–14275. doi: 10.1021/bi9606399. [DOI] [PubMed] [Google Scholar]
- 8.Fuller S D, Wilk T, Gowen B E, Kräusslich H-G, Vogt V M. Cryoelectron microscopy reveals ordered domains in the immature HIV-1 particle. Curr Biol. 1997;7:729–738. doi: 10.1016/s0960-9822(06)00331-9. [DOI] [PubMed] [Google Scholar]
- 9.Gamble T R, Yoo S, Vajdos F F, von Schwedler U K, Worthylake D K, Wang H, McCutcheon J P, Sundquist W I, Hill C P. Structure of the carboxy-terminal dimerization domain of the HIV-1 capsid protein. Science. 1997;278:849–853. doi: 10.1126/science.278.5339.849. [DOI] [PubMed] [Google Scholar]
- 10.Gelderblom H R. Assembly and morphology of HIV: potential effect of structure on viral function. AIDS. 1991;5:617–638. [PubMed] [Google Scholar]
- 11.Gheysen D, Jacobs E, de Foresta F, Thiriart C, Francotte M, Thines D, De Wilde M. Assembly and release of HIV-1 precursor Pr55gag virus-like particles from recombinant baculovirus-infected insect cells. Cell. 1989;59:103–112. doi: 10.1016/0092-8674(89)90873-8. [DOI] [PubMed] [Google Scholar]
- 12.Gitti R K, Lee B M, Walker J, Summers M F, Yoo S, Sundquist W I. Structure of the amino-terminal core domain of the HIV-1 capsid protein. Science. 1996;273:231–235. doi: 10.1126/science.273.5272.231. [DOI] [PubMed] [Google Scholar]
- 13.González S A, Affranchino J L, Gelderblom H R, Burny A. Assembly of the matrix protein of simian immunodeficiency virus into virus-like particles. Virology. 1993;194:548–556. doi: 10.1006/viro.1993.1293. [DOI] [PubMed] [Google Scholar]
- 14.Göttlinger H G, Sodroski J G, Haseltine W A. Role of capsid precursor processing and myristoylation in morphogenesis and infectivity of human immunodeficiency virus type 1. Proc Natl Acad Sci USA. 1989;86:5781–5785. doi: 10.1073/pnas.86.15.5781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hill C P, Worthylake D, Bancroft D P, Christensen A M, Sundquist W I. Crystal structures of the trimeric human immunodeficiency virus type 1 matrix protein: implications for membrane association and assembly. Proc Natl Acad Sci USA. 1996;93:3099–3104. doi: 10.1073/pnas.93.7.3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hunter E. Macromolecular interactions in the assembly of HIV and other retroviruses. Semin Virol. 1994;5:71–83. [Google Scholar]
- 17.Kräusslich H-G, Fäcke M, Heuser A-M, Konvalinka J, Zentgraf H. The spacer peptide between human immunodeficiency virus capsid and nucleocapsid proteins is essential for ordered assembly and viral infectivity. J Virol. 1995;69:3407–3419. doi: 10.1128/jvi.69.6.3407-3419.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee P P, Linial M L. Efficient particle formation can occur if the matrix domain of human immunodeficiency virus type 1 Gag is substituted by a myristylation signal. J Virol. 1994;68:6644–6654. doi: 10.1128/jvi.68.10.6644-6654.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mammano F, Öhagen Å, Höglund S, Göttlinger H G. Role of the major homology region of human immunodeficiency virus type 1 in virion morphogenesis. J Virol. 1994;68:4927–4936. doi: 10.1128/jvi.68.8.4927-4936.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McDermott J, Farrel L, Ross R, Barklis E. Structural analysis of human immunodeficiency virus type 1 Gag protein interactions, using cysteine-specific reagents. J Virol. 1996;70:5106–5114. doi: 10.1128/jvi.70.8.5106-5114.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mervis R J, Ahmad N, Lillehoj E P, Raum M G, Salazar F H R, Chan H W, Venkatesan S. The gag gene products of human immunodeficiency virus type 1: alignment with the gag open reading frame, identification of posttranslational modifications, and evidence for alternative gag precursors. J Virol. 1988;62:3993–4002. doi: 10.1128/jvi.62.11.3993-4002.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reicin A S, Öhagen A, Yin L, Höglund S, Goff S P. The role of Gag in human immunodeficiency virus type 1 virion morphogenesis and early steps of the viral life cycle. J Virol. 1996;70:8645–8652. doi: 10.1128/jvi.70.12.8645-8652.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reicin A S, Paik S, Berkowitz R D, Luban J, Lowy I, Goff S P. Linker insertion mutations in the human immunodeficiency virus type 1 gag gene: effects on virion particle assembly, release, and infectivity. J Virol. 1995;69:642–650. doi: 10.1128/jvi.69.2.642-650.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reil H, Bukovsky A A, Gelderblom H R, Göttlinger H G. Efficient HIV-1 replication can occur in the absence of the viral matrix protein. EMBO J. 1998;17:2699–2708. doi: 10.1093/emboj/17.9.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spearman P, Horton R, Ratner L, Kuli-Zade I. Membrane binding of human immunodeficiency virus type 1 matrix protein in vivo supports a conformational myristyl switch mechanism. J Virol. 1997;71:6582–6592. doi: 10.1128/jvi.71.9.6582-6592.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Srinivasakumar N, Hammarskjöld M-L, Rekosh D. Characterization of deletion mutations in the capsid region of human immunodeficiency virus type 1 that affect particle formation and Gag-Pol precursor incorporation. J Virol. 1995;69:6106–6114. doi: 10.1128/jvi.69.10.6106-6114.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Strambio-de-Castillia K, Hunter E. Mutational analysis of the major homology region of Mason-Pfizer monkey virus by use of saturation mutagenesis. J Virol. 1992;66:7021–7032. doi: 10.1128/jvi.66.12.7021-7032.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.von Poblotzki A, Wagner R, Niedrig M, Wanner G, Wolf H, Modrow S. Identification of a region in the Pr55gag-polyprotein essential for HIV-1 particle formation. Virology. 1993;193:981–985. doi: 10.1006/viro.1993.1210. [DOI] [PubMed] [Google Scholar]
- 29.von Schwedler U K, Stemmler T L, Klishko V Y, Li S, Albertine K A, Davis D R, Sundquist W I. Proteolytic refolding of the HIV-1 capsid protein amino-terminus facilitates viral core assembly. EMBO J. 1998;17:1555–1568. doi: 10.1093/emboj/17.6.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang C-T, Barklis E. Assembly, processing, and infectivity of human immunodeficiency virus type 1 Gag mutants. J Virol. 1993;67:4264–4273. doi: 10.1128/jvi.67.7.4264-4273.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang C-T, Zhang Y, McDermott J, Barklis E. Conditional infectivity of a human immunodeficiency virus matrix domain deletion mutant. J Virol. 1993;67:7067–7076. doi: 10.1128/jvi.67.12.7067-7076.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weldon R A, Wills J W. Characterization of a small (25-kilodalton) derivative of the Rous sarcoma virus Gag protein competent for particle release. J Virol. 1993;67:5550–5561. doi: 10.1128/jvi.67.9.5550-5561.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wills J W, Craven R C. Form, function, and use of retroviral Gag proteins. AIDS. 1991;5:639–654. doi: 10.1097/00002030-199106000-00002. [DOI] [PubMed] [Google Scholar]
- 34.Zhou W, Parent L J, Wills J W, Resh M D. Identification of a membrane-binding domain within the amino-terminal region of human immunodeficiency virus type 1 Gag protein which interacts with acidic phospholipids. J Virol. 1994;68:2556–2569. doi: 10.1128/jvi.68.4.2556-2569.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou W, Resh M D. Differential membrane binding of the human immunodeficiency virus type 1 matrix protein. J Virol. 1996;70:8540–8548. doi: 10.1128/jvi.70.12.8540-8548.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]